This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-125a-3p | ACPP | -0.07 | 0.92074 | 3.93 | 3.0E-5 | MirTarget; miRanda | -0.32 | 1.0E-5 | NA | |
| 2 | hsa-miR-125a-5p | ACPP | -1.32 | 0.00714 | 3.93 | 3.0E-5 | MirTarget; miRanda | -0.27 | 0.0146 | NA | |
| 3 | hsa-miR-192-5p | ACPP | 1.78 | 0.11349 | 3.93 | 3.0E-5 | MirTarget | -0.1 | 0.03284 | NA | |
| 4 | hsa-miR-326 | ACPP | 1.77 | 0.03673 | 3.93 | 3.0E-5 | miRanda | -0.13 | 0.04646 | NA | |
| 5 | hsa-miR-491-5p | ACPP | 0.57 | 0.31331 | 3.93 | 3.0E-5 | miRanda | -0.31 | 0.00154 | NA | |
| 6 | hsa-miR-338-3p | AIM2 | 0.45 | 0.55849 | 5.33 | 0.00329 | miRanda | -0.5 | 0.0002 | NA | |
| 7 | hsa-miR-125a-3p | ALOX12 | -0.07 | 0.92074 | 3.5 | 0.02136 | miRanda | -0.37 | 0.00117 | NA | |
| 8 | hsa-miR-335-5p | APOC1 | 0.17 | 0.8039 | 3.77 | 0.00044 | miRNAWalker2 validate | -0.19 | 0.03662 | NA | |
| 9 | hsa-miR-140-5p | CCNB1 | -0.63 | 0.12667 | 4.14 | 0 | miRanda | -0.13 | 0.02333 | NA | |
| 10 | hsa-miR-125b-5p | CCNE1 | -2.01 | 0.00516 | 3.91 | 0 | miRNAWalker2 validate | -0.13 | 0.00065 | NA | |
| 11 | hsa-miR-195-5p | CCNE1 | -1.59 | 0.01691 | 3.91 | 0 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.12 | 0.0047 | 24402230 | Furthermore through qPCR and western blot assays we showed that overexpression of miR-195-5p reduced CCNE1 mRNA and protein levels respectively |
| 12 | hsa-miR-26a-5p | CCNE1 | -0.35 | 0.36204 | 3.91 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.18 | 0.00988 | 22094936 | Cell cycle regulation and CCNE1 and CDC2 were the only significant overlapping pathway and genes differentially expressed between tumors with high and low levels of miR-26a and EZH2 respectively; Low mRNA levels of EZH2 CCNE1 and CDC2 and high levels of miR-26a are associated with favorable outcome on tamoxifen |
| 13 | hsa-miR-497-5p | CCNE1 | -1.44 | 0.02251 | 3.91 | 0 | MirTarget; miRNATAP | -0.13 | 0.00315 | 24909281; 24112607; 25909221 | miR 497 suppresses proliferation of human cervical carcinoma HeLa cells by targeting cyclin E1; Furthermore the target effect of miR-497 on the CCNE1 was identified by dual-luciferase reporter assay system qRT-PCR and Western blotting; Over-expressed miR-497 in HeLa cells could suppress cell proliferation by targeting CCNE1;Western blot assays confirmed that overexpression of miR-497 reduced cyclin E1 protein levels; Inhibited cellular growth suppressed cellular migration and invasion and G1 cell cycle arrest were observed upon overexpression of miR-497 in cells possibly by targeting cyclin E1;The effect of simultaneous overexpression of miR-497 and miR-34a on the inhibition of cell proliferation colony formation and tumor growth and the downregulation of cyclin E1 was stronger than the effect of each miRNA alone; The synergistic actions of miR-497 and miR-34a partly correlated with cyclin E1 levels; These results indicate cyclin E1 is downregulated by both miR-497 and miR-34a which synergistically retard the growth of human lung cancer cells |
| 14 | hsa-miR-10b-5p | CDKN2A | -3.08 | 0 | 7.38 | 0 | miRNAWalker2 validate; miRTarBase | -0.36 | 0.00054 | NA | |
| 15 | hsa-miR-125a-5p | CDKN2A | -1.32 | 0.00714 | 7.38 | 0 | miRanda | -0.39 | 0.00048 | NA | |
| 16 | hsa-miR-192-5p | CDKN2A | 1.78 | 0.11349 | 7.38 | 0 | miRNAWalker2 validate | -0.11 | 0.02051 | NA | |
| 17 | hsa-miR-497-5p | CEP55 | -1.44 | 0.02251 | 6.54 | 0 | MirTarget | -0.11 | 0.0371 | NA | |
| 18 | hsa-miR-338-3p | CHAF1A | 0.45 | 0.55849 | 2.75 | 0 | MirTarget; PITA; miRanda | -0.11 | 7.0E-5 | NA | |
| 19 | hsa-miR-125a-5p | CKS1B | -1.32 | 0.00714 | 2.11 | 0 | miRanda | -0.17 | 0.00056 | NA | |
| 20 | hsa-miR-542-3p | CKS1B | -0.38 | 0.438 | 2.11 | 0 | miRNATAP | -0.15 | 0.00451 | NA | |
| 21 | hsa-miR-335-5p | CRNN | 0.17 | 0.8039 | 3.51 | 0.2192 | miRNAWalker2 validate | -1.44 | 0 | NA | |
| 22 | hsa-miR-125a-3p | CYP2C9 | -0.07 | 0.92074 | 2.39 | 0.17341 | miRanda | -0.27 | 0.04044 | NA | |
| 23 | hsa-miR-128-3p | CYP2C9 | 1.36 | 0.00408 | 2.39 | 0.17341 | MirTarget | -0.44 | 0.03887 | 25704921 | To investigate the molecular mechanisms that control CYP2C9 expression we applied integrative approaches including in silico in vitro and in vivo analyses to elucidate the role of microRNA hsa-miR-128-3p in the regulation of CYP2C9 expression and translation; RNA electrophoresis mobility shift assays demonstrated a direct interaction between hsa-miR-128-3p and its cognate target the CYP2C9 transcript; Furthermore the expression of a luciferase reporter gene containing the 3'-UTR of CYP2C9 and the endogenous expression of CYP2C9 were suppressed by transfection of hsa-miR-128-3p; Importantly chemically-induced up- or down-regulation of hsa-miR-128-3p correlated inversely with the expression of CYP2C9; Finally an association analysis revealed that the expression of hsa-miR-128-3p is inversely correlated with the expression of CYP2C9 in HCC tumor tissues; Altogether the study helped to elucidate the mechanism of CYP2C9 regulation by hsa-miR-128-3p and the inverse association in HCC |
| 24 | hsa-miR-30a-5p | DTL | -0.77 | 0.32049 | 4.64 | 0 | miRNAWalker2 validate; miRTarBase | -0.11 | 0.00023 | 22287560 | MiR 30a 5p suppresses tumor growth in colon carcinoma by targeting DTL; Subsequent reporter gene assays confirmed the predicted miR-30a-5p binding site in the 3'untranslated region of DTL; In conclusion our data identified miR-30a-5p as a tumor-suppressing miRNA in colon cancer cells exerting its function via modulation of DTL expression which is frequently overexpressed in colorectal cancer |
| 25 | hsa-miR-10b-3p | ECT2 | -2.52 | 0 | 3.71 | 0 | MirTarget | -0.13 | 0.01178 | NA | |
| 26 | hsa-miR-140-5p | ECT2 | -0.63 | 0.12667 | 3.71 | 0 | miRanda | -0.16 | 0.01722 | NA | |
| 27 | hsa-miR-664a-3p | ECT2 | 0.25 | 0.56171 | 3.71 | 0 | MirTarget; mirMAP | -0.14 | 0.02816 | NA | |
| 28 | hsa-miR-330-5p | EDN3 | 2.25 | 0.00028 | -3.46 | 0.03889 | miRanda | -0.61 | 5.0E-5 | NA | |
| 29 | hsa-miR-146b-5p | ENDOU | 1.88 | 0.00074 | -1.71 | 0.28835 | miRanda | -0.34 | 0.04045 | NA | |
| 30 | hsa-miR-26b-3p | ENDOU | 0.99 | 0.03514 | -1.71 | 0.28835 | miRNATAP | -0.51 | 0.00943 | NA | |
| 31 | hsa-miR-335-5p | ENDOU | 0.17 | 0.8039 | -1.71 | 0.28835 | miRNAWalker2 validate | -0.63 | 0 | NA | |
| 32 | hsa-miR-491-5p | ENDOU | 0.57 | 0.31331 | -1.71 | 0.28835 | miRanda | -0.44 | 0.00719 | NA | |
| 33 | hsa-let-7f-1-3p | ESR1 | 1.62 | 0.00069 | -4.87 | 9.0E-5 | mirMAP | -0.51 | 0.00059 | NA | |
| 34 | hsa-miR-107 | ESR1 | 1.49 | 0.00013 | -4.87 | 9.0E-5 | PITA; miRanda; miRNATAP | -0.49 | 0.00802 | 26033453 | Estrogen-receptor negative tumors displayed higher concentrations of circulating miR-107 than their counterparts p = 0.035; However overexpression of miR-107 in MCF-7 cells did not downregulate estrogen receptor protein |
| 35 | hsa-miR-130b-3p | ESR1 | 3.92 | 0 | -4.87 | 9.0E-5 | miRNATAP | -0.66 | 0 | NA | |
| 36 | hsa-miR-130b-5p | ESR1 | 3.74 | 0 | -4.87 | 9.0E-5 | mirMAP | -0.46 | 8.0E-5 | NA | |
| 37 | hsa-miR-17-5p | ESR1 | 2.33 | 2.0E-5 | -4.87 | 9.0E-5 | TargetScan | -0.71 | 0 | NA | |
| 38 | hsa-miR-181b-5p | ESR1 | 1.11 | 0.02734 | -4.87 | 9.0E-5 | miRNATAP | -0.31 | 0.02925 | NA | |
| 39 | hsa-miR-188-5p | ESR1 | 1.39 | 0.02516 | -4.87 | 9.0E-5 | PITA; miRNATAP | -0.29 | 0.01114 | NA | |
| 40 | hsa-miR-18a-3p | ESR1 | 3.65 | 0 | -4.87 | 9.0E-5 | miRNATAP | -0.58 | 0 | 19027010; 24975878 | MicroRNA 18a prevents estrogen receptor alpha expression promoting proliferation of hepatocellular carcinoma cells; The gene ESR1 which encodes the estrogen receptor-alpha ERalpha was identified as a target of miR-18a miR-18a can repress ERalpha translation by binding to its mRNA at the 3' untranslated region;Elevated p53 promotes the processing of miR 18a to decrease estrogen receptor α in female hepatocellular carcinoma; Our previous study identified that estrogen receptor alpha ERα protein is downregulated in 60% of female HCC cases via a miR-18a elevation mediated suppression of ERα translation |
| 41 | hsa-miR-18a-5p | ESR1 | 3.91 | 0 | -4.87 | 9.0E-5 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.72 | 0 | 19027010; 24975878 | MicroRNA 18a prevents estrogen receptor alpha expression promoting proliferation of hepatocellular carcinoma cells; The gene ESR1 which encodes the estrogen receptor-alpha ERalpha was identified as a target of miR-18a miR-18a can repress ERalpha translation by binding to its mRNA at the 3' untranslated region;Elevated p53 promotes the processing of miR 18a to decrease estrogen receptor α in female hepatocellular carcinoma; Our previous study identified that estrogen receptor alpha ERα protein is downregulated in 60% of female HCC cases via a miR-18a elevation mediated suppression of ERα translation |
| 42 | hsa-miR-193a-3p | ESR1 | 0.65 | 0.20713 | -4.87 | 9.0E-5 | miRanda | -0.51 | 0.00024 | NA | |
| 43 | hsa-miR-193a-5p | ESR1 | 0.51 | 0.23928 | -4.87 | 9.0E-5 | miRNATAP | -0.33 | 0.04839 | NA | |
| 44 | hsa-miR-193b-3p | ESR1 | 1.35 | 0.08749 | -4.87 | 9.0E-5 | miRNAWalker2 validate; miRTarBase | -0.28 | 0.00155 | NA | |
| 45 | hsa-miR-19a-3p | ESR1 | 2.17 | 0.00122 | -4.87 | 9.0E-5 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.41 | 0.00011 | 26416600 | We speculate that miR-19a might be co-expressed with lncRNA-DLEU1 to co-regulate the expression of ESR1 which influences the occurrence and development of breast cancer cells with different levels of ER expression |
| 46 | hsa-miR-19b-3p | ESR1 | 1.68 | 0.00086 | -4.87 | 9.0E-5 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.6 | 2.0E-5 | NA | |
| 47 | hsa-miR-21-3p | ESR1 | 3.5 | 0 | -4.87 | 9.0E-5 | mirMAP | -0.48 | 8.0E-5 | 19264808; 25337203; 25969534; 21131358 | miR-21 is higher in ER alpha positive than negative tumors but no one has examined how estradiol E2 regulates miR-21 in breast cancer cells; The E2-induced reduction in miR-21 was inhibited by 4-hydroxytamoxifen 4-OHT ICI 182 780 Faslodex and siRNA ER alpha indicating that the suppression is ER alpha-mediated; ER alpha and ER beta agonists PPT and DPN inhibited and 4-OHT increased miR-21 expression; These results are the first to demonstrate that E2 represses the expression of an oncogenic miRNA miR-21 by activating estrogen receptor in MCF-7 cells;Odds ratios ORs showed that miR-21 expression was closely associated with estrogen receptor ER progesterone receptor PR lymph node metastasis histological grade Her2/neu;Dehydroepiandrosterone Activation of G protein coupled Estrogen Receptor Rapidly Stimulates MicroRNA 21 Transcription in Human Hepatocellular Carcinoma Cells;Induction of miR 21 by retinoic acid in estrogen receptor positive breast carcinoma cells: biological correlates and molecular targets |
| 48 | hsa-miR-221-3p | ESR1 | 0.94 | 0.17475 | -4.87 | 9.0E-5 | miRNAWalker2 validate; miRNATAP | -0.3 | 0.00321 | 25483016 | HBx protein induced upregulation of microRNA 221 promotes aberrant proliferation in HBV‑related hepatocellular carcinoma by targeting estrogen receptor α |
| 49 | hsa-miR-222-3p | ESR1 | 1.55 | 0.0223 | -4.87 | 9.0E-5 | miRNAWalker2 validate; miRNATAP | -0.36 | 0.00063 | NA | |
| 50 | hsa-miR-301a-3p | ESR1 | 1.99 | 0.00081 | -4.87 | 9.0E-5 | miRNATAP | -0.52 | 1.0E-5 | NA | |
| 51 | hsa-miR-31-5p | ESR1 | 7.16 | 0 | -4.87 | 9.0E-5 | mirMAP | -0.19 | 0.00142 | 23162645 | miR 155 and miR 31 are differentially expressed in breast cancer patients and are correlated with the estrogen receptor and progesterone receptor status; The expression levels of miR-155 but not miR-31 were inversely correlated with estrogen receptor ER and progesterone receptor PR expression ER r=-0.353 P=0.003; PR r=-0.357 P=0.003 |
| 52 | hsa-miR-32-3p | ESR1 | 2.2 | 0.03928 | -4.87 | 9.0E-5 | mirMAP | -0.6 | 0 | NA | |
| 53 | hsa-miR-324-5p | ESR1 | 1.31 | 0.01168 | -4.87 | 9.0E-5 | PITA | -0.48 | 0.00049 | NA | |
| 54 | hsa-miR-330-3p | ESR1 | 2.49 | 0.00013 | -4.87 | 9.0E-5 | PITA; mirMAP; miRNATAP | -0.4 | 0.00024 | NA | |
| 55 | hsa-miR-33a-3p | ESR1 | 2.06 | 0.00156 | -4.87 | 9.0E-5 | mirMAP | -0.3 | 0.00664 | NA | |
| 56 | hsa-miR-454-3p | ESR1 | 1.4 | 0.00366 | -4.87 | 9.0E-5 | miRNATAP | -0.52 | 0.00045 | NA | |
| 57 | hsa-miR-589-3p | ESR1 | 1.33 | 0.05263 | -4.87 | 9.0E-5 | mirMAP | -0.34 | 0.00105 | NA | |
| 58 | hsa-miR-590-3p | ESR1 | 2.35 | 0 | -4.87 | 9.0E-5 | miRanda; mirMAP; miRNATAP | -0.46 | 0.00091 | NA | |
| 59 | hsa-miR-629-3p | ESR1 | 3.48 | 0 | -4.87 | 9.0E-5 | mirMAP | -0.52 | 0 | NA | |
| 60 | hsa-miR-7-5p | ESR1 | 3.6 | 0.00068 | -4.87 | 9.0E-5 | mirMAP | -0.45 | 0 | NA | |
| 61 | hsa-miR-9-3p | ESR1 | 1.69 | 0.12517 | -4.87 | 9.0E-5 | mirMAP | -0.24 | 0.0002 | 22723919 | Higher expression of miR-9 was significantly associated with breast cancer LR in all cases as well as the subset of estrogen receptor ER positive cases p = 0.02 |
| 62 | hsa-miR-9-5p | ESR1 | 1.8 | 0.14527 | -4.87 | 9.0E-5 | mirMAP; miRNATAP | -0.19 | 0.00106 | 22723919 | Higher expression of miR-9 was significantly associated with breast cancer LR in all cases as well as the subset of estrogen receptor ER positive cases p = 0.02 |
| 63 | hsa-let-7e-5p | EZH2 | -0.11 | 0.81474 | 4.69 | 0 | miRNATAP | -0.21 | 0.00023 | NA | |
| 64 | hsa-miR-139-5p | EZH2 | -2.09 | 0.00038 | 4.69 | 0 | miRanda | -0.15 | 0.00087 | NA | |
| 65 | hsa-miR-140-3p | FANCA | -1.98 | 0 | 4.78 | 0 | miRNATAP | -0.29 | 1.0E-5 | NA | |
| 66 | hsa-miR-26a-5p | FANCA | -0.35 | 0.36204 | 4.78 | 0 | miRNATAP | -0.13 | 0.04673 | NA | |
| 67 | hsa-miR-505-5p | FANCA | -0.55 | 0.33141 | 4.78 | 0 | miRNATAP | -0.11 | 0.01457 | NA | |
| 68 | hsa-miR-542-3p | FANCA | -0.38 | 0.438 | 4.78 | 0 | miRanda | -0.13 | 0.01254 | NA | |
| 69 | hsa-miR-140-5p | FEN1 | -0.63 | 0.12667 | 3 | 0 | PITA; miRanda; miRNATAP | -0.14 | 0.00531 | NA | |
| 70 | hsa-let-7f-1-3p | FGFR2 | 1.62 | 0.00069 | -1.55 | 0.07059 | mirMAP | -0.23 | 0.02313 | NA | |
| 71 | hsa-miR-146b-5p | FGFR2 | 1.88 | 0.00074 | -1.55 | 0.07059 | miRanda | -0.3 | 0.00059 | NA | |
| 72 | hsa-miR-186-5p | FGFR2 | 0.45 | 0.18545 | -1.55 | 0.07059 | miRNAWalker2 validate | -0.37 | 0.01225 | NA | |
| 73 | hsa-miR-324-5p | FGFR2 | 1.31 | 0.01168 | -1.55 | 0.07059 | miRanda | -0.22 | 0.01943 | NA | |
| 74 | hsa-miR-330-5p | FGFR2 | 2.25 | 0.00028 | -1.55 | 0.07059 | miRanda | -0.39 | 0 | NA | |
| 75 | hsa-miR-338-3p | FGFR2 | 0.45 | 0.55849 | -1.55 | 0.07059 | miRanda; miRNATAP | -0.2 | 0.00168 | NA | |
| 76 | hsa-miR-125a-3p | FUT6 | -0.07 | 0.92074 | 3.63 | 0.02084 | MirTarget; miRanda; miRNATAP | -0.35 | 0.00305 | NA | |
| 77 | hsa-miR-2355-5p | FUT6 | 0.42 | 0.50215 | 3.63 | 0.02084 | MirTarget | -0.52 | 0.00028 | NA | |
| 78 | hsa-miR-324-5p | IFI44 | 1.31 | 0.01168 | 2.01 | 0.03461 | miRanda | -0.41 | 9.0E-5 | NA | |
| 79 | hsa-miR-589-3p | IFI44 | 1.33 | 0.05263 | 2.01 | 0.03461 | MirTarget | -0.28 | 0.00043 | NA | |
| 80 | hsa-miR-616-5p | IFI6 | 2.48 | 0.00318 | 1.65 | 0.10931 | MirTarget | -0.22 | 0.01287 | NA | |
| 81 | hsa-miR-10a-3p | INHBA | 0.97 | 0.31667 | 0.6 | 0.64331 | mirMAP | -0.23 | 0.00289 | NA | |
| 82 | hsa-miR-10b-5p | INHBA | -3.08 | 0 | 0.6 | 0.64331 | miRNAWalker2 validate | -0.38 | 0.00596 | NA | |
| 83 | hsa-miR-125b-2-3p | INHBA | -1.76 | 0.12262 | 0.6 | 0.64331 | mirMAP | -0.16 | 0.01384 | NA | |
| 84 | hsa-miR-148b-3p | INHBA | 1.76 | 0 | 0.6 | 0.64331 | miRNAWalker2 validate | -0.67 | 0.00272 | NA | |
| 85 | hsa-miR-16-2-3p | INHBA | 3.8 | 0 | 0.6 | 0.64331 | mirMAP | -0.46 | 0.00075 | NA | |
| 86 | hsa-miR-181c-5p | INHBA | -0.3 | 0.53753 | 0.6 | 0.64331 | mirMAP | -0.33 | 0.02957 | NA | |
| 87 | hsa-miR-195-3p | INHBA | -2.39 | 0.00019 | 0.6 | 0.64331 | mirMAP | -0.36 | 0.00157 | NA | |
| 88 | hsa-miR-200b-3p | INHBA | 5.56 | 0 | 0.6 | 0.64331 | TargetScan | -0.56 | 0 | NA | |
| 89 | hsa-miR-224-3p | INHBA | 2.85 | 0.00018 | 0.6 | 0.64331 | mirMAP | -0.24 | 0.01071 | NA | |
| 90 | hsa-miR-26a-5p | INHBA | -0.35 | 0.36204 | 0.6 | 0.64331 | mirMAP | -0.48 | 0.01215 | NA | |
| 91 | hsa-miR-3065-5p | INHBA | 2.14 | 0.06094 | 0.6 | 0.64331 | mirMAP | -0.36 | 0 | NA | |
| 92 | hsa-miR-30b-5p | INHBA | 0.02 | 0.95322 | 0.6 | 0.64331 | mirMAP | -0.64 | 0.00041 | NA | |
| 93 | hsa-miR-30c-5p | INHBA | 0.39 | 0.34861 | 0.6 | 0.64331 | mirMAP | -0.83 | 0 | NA | |
| 94 | hsa-miR-30d-5p | INHBA | 0.3 | 0.38019 | 0.6 | 0.64331 | mirMAP | -0.81 | 0.00017 | NA | |
| 95 | hsa-miR-30e-5p | INHBA | 0.78 | 0.03467 | 0.6 | 0.64331 | mirMAP | -1.27 | 0 | NA | |
| 96 | hsa-miR-3607-3p | INHBA | 1.38 | 0.02401 | 0.6 | 0.64331 | mirMAP | -0.39 | 0.00118 | NA | |
| 97 | hsa-miR-374a-5p | INHBA | 0.28 | 0.45888 | 0.6 | 0.64331 | mirMAP | -0.53 | 0.00725 | NA | |
| 98 | hsa-miR-374b-5p | INHBA | -0.11 | 0.76489 | 0.6 | 0.64331 | mirMAP | -0.4 | 0.04085 | NA | |
| 99 | hsa-miR-664a-3p | INHBA | 0.25 | 0.56171 | 0.6 | 0.64331 | mirMAP | -0.47 | 0.00659 | NA | |
| 100 | hsa-miR-30a-5p | KIF11 | -0.77 | 0.32049 | 4.88 | 0 | miRNAWalker2 validate; MirTarget | -0.12 | 0.0005 | NA | |
| 101 | hsa-miR-381-3p | KIF11 | -1.9 | 0.00523 | 4.88 | 0 | MirTarget | -0.11 | 0.00567 | NA | |
| 102 | hsa-miR-129-5p | KIF20A | -2.67 | 0.00696 | 6.75 | 0 | miRanda | -0.11 | 0.00129 | NA | |
| 103 | hsa-miR-140-5p | KIF20A | -0.63 | 0.12667 | 6.75 | 0 | miRanda | -0.17 | 0.04331 | NA | |
| 104 | hsa-miR-125a-5p | KIF23 | -1.32 | 0.00714 | 3.87 | 0 | miRanda | -0.13 | 0.00816 | NA | |
| 105 | hsa-miR-101-3p | KIF2C | -1.12 | 0.02009 | 5.28 | 0 | miRNAWalker2 validate | -0.14 | 0.0101 | NA | |
| 106 | hsa-let-7e-5p | LOR | -0.11 | 0.81474 | 1.54 | 0.24951 | MirTarget | -0.64 | 7.0E-5 | NA | |
| 107 | hsa-miR-30b-3p | LOR | 0.17 | 0.76608 | 1.54 | 0.24951 | MirTarget; miRNATAP | -0.43 | 0.0012 | NA | |
| 108 | hsa-miR-139-5p | MAD2L1 | -2.09 | 0.00038 | 3.34 | 0 | miRanda | -0.1 | 0.01194 | NA | |
| 109 | hsa-miR-199a-5p | MCM10 | -1.25 | 0.07478 | 6.99 | 0 | miRanda | -0.14 | 0.0018 | NA | |
| 110 | hsa-miR-199b-5p | MCM10 | -0.54 | 0.47689 | 6.99 | 0 | miRanda | -0.14 | 0.00136 | NA | |
| 111 | hsa-miR-140-5p | MCM4 | -0.63 | 0.12667 | 4.07 | 0 | MirTarget | -0.18 | 0.00533 | NA | |
| 112 | hsa-miR-181c-5p | MELK | -0.3 | 0.53753 | 7.07 | 0 | MirTarget | -0.19 | 0.00355 | NA | |
| 113 | hsa-miR-181d-5p | MELK | -0.53 | 0.32526 | 7.07 | 0 | MirTarget | -0.18 | 0.00345 | NA | |
| 114 | hsa-miR-361-5p | MMP1 | 0.01 | 0.9852 | 5.12 | 0.00282 | miRanda | -0.71 | 0.01253 | NA | |
| 115 | hsa-miR-145-5p | MMP12 | -3.56 | 0 | 4.87 | 0.00038 | miRNAWalker2 validate | -0.42 | 0.0001 | NA | |
| 116 | hsa-let-7d-5p | MXD1 | 0.83 | 0.0127 | 1.61 | 0.03992 | MirTarget; miRNATAP | -0.34 | 0.01369 | NA | |
| 117 | hsa-let-7e-5p | MXD1 | -0.11 | 0.81474 | 1.61 | 0.03992 | MirTarget; miRNATAP | -0.59 | 0 | NA | |
| 118 | hsa-miR-106a-5p | MXD1 | 3.99 | 0 | 1.61 | 0.03992 | mirMAP | -0.23 | 2.0E-5 | NA | |
| 119 | hsa-miR-125a-3p | MXD1 | -0.07 | 0.92074 | 1.61 | 0.03992 | miRanda | -0.29 | 0 | NA | |
| 120 | hsa-miR-148b-3p | MXD1 | 1.76 | 0 | 1.61 | 0.03992 | MirTarget | -0.29 | 0.03551 | NA | |
| 121 | hsa-miR-195-3p | MXD1 | -2.39 | 0.00019 | 1.61 | 0.03992 | mirMAP | -0.23 | 0.00071 | NA | |
| 122 | hsa-miR-200b-3p | MXD1 | 5.56 | 0 | 1.61 | 0.03992 | mirMAP | -0.23 | 0.00024 | NA | |
| 123 | hsa-miR-20b-5p | MXD1 | 4.57 | 5.0E-5 | 1.61 | 0.03992 | mirMAP | -0.16 | 6.0E-5 | NA | |
| 124 | hsa-miR-217 | MXD1 | -0.38 | 0.71741 | 1.61 | 0.03992 | miRanda | -0.12 | 0.00407 | NA | |
| 125 | hsa-miR-30a-3p | MXD1 | -1.22 | 0.16757 | 1.61 | 0.03992 | mirMAP | -0.35 | 0 | NA | |
| 126 | hsa-miR-30d-3p | MXD1 | -0.07 | 0.85742 | 1.61 | 0.03992 | mirMAP | -0.35 | 0.00126 | NA | |
| 127 | hsa-miR-324-3p | MXD1 | 1.51 | 0.00384 | 1.61 | 0.03992 | MirTarget | -0.32 | 0.00015 | NA | |
| 128 | hsa-miR-335-5p | MXD1 | 0.17 | 0.8039 | 1.61 | 0.03992 | miRNAWalker2 validate | -0.4 | 0 | NA | |
| 129 | hsa-miR-3607-3p | MXD1 | 1.38 | 0.02401 | 1.61 | 0.03992 | mirMAP | -0.21 | 0.00394 | NA | |
| 130 | hsa-miR-3913-5p | MXD1 | 0.15 | 0.73484 | 1.61 | 0.03992 | MirTarget | -0.51 | 0 | NA | |
| 131 | hsa-miR-429 | MXD1 | 6.4 | 0 | 1.61 | 0.03992 | mirMAP; miRNATAP | -0.19 | 0.00069 | NA | |
| 132 | hsa-miR-501-5p | MXD1 | 1.04 | 0.07772 | 1.61 | 0.03992 | mirMAP | -0.19 | 0.01075 | NA | |
| 133 | hsa-miR-508-3p | MXD1 | 0.98 | 0.43953 | 1.61 | 0.03992 | miRNATAP | -0.16 | 1.0E-5 | NA | |
| 134 | hsa-miR-589-3p | MXD1 | 1.33 | 0.05263 | 1.61 | 0.03992 | MirTarget | -0.2 | 0.00222 | NA | |
| 135 | hsa-miR-592 | MXD1 | 2.8 | 0.02935 | 1.61 | 0.03992 | mirMAP | -0.23 | 0 | NA | |
| 136 | hsa-miR-625-5p | MXD1 | 2.03 | 0.00094 | 1.61 | 0.03992 | MirTarget | -0.25 | 0.00051 | NA | |
| 137 | hsa-miR-148b-3p | NDN | 1.76 | 0 | -5.07 | 0 | miRNAWalker2 validate | -0.65 | 0 | NA | |
| 138 | hsa-miR-200b-3p | NDN | 5.56 | 0 | -5.07 | 0 | MirTarget; TargetScan | -0.35 | 0 | NA | |
| 139 | hsa-miR-200c-3p | NDN | 6.47 | 0 | -5.07 | 0 | MirTarget; miRNATAP | -0.46 | 0 | NA | |
| 140 | hsa-miR-330-5p | NDN | 2.25 | 0.00028 | -5.07 | 0 | miRanda | -0.22 | 0.00378 | NA | |
| 141 | hsa-miR-33a-3p | NDN | 2.06 | 0.00156 | -5.07 | 0 | MirTarget | -0.29 | 5.0E-5 | NA | |
| 142 | hsa-miR-3607-3p | NDN | 1.38 | 0.02401 | -5.07 | 0 | MirTarget | -0.16 | 0.03573 | NA | |
| 143 | hsa-miR-429 | NDN | 6.4 | 0 | -5.07 | 0 | MirTarget; PITA; miRanda; miRNATAP | -0.33 | 0 | NA | |
| 144 | hsa-miR-486-5p | NDN | 1.11 | 0.30929 | -5.07 | 0 | miRanda | -0.14 | 0.00079 | NA | |
| 145 | hsa-miR-30a-5p | PCNA | -0.77 | 0.32049 | 2.73 | 0 | miRNAWalker2 validate | -0.11 | 0.00013 | NA | |
| 146 | hsa-miR-125a-5p | PRC1 | -1.32 | 0.00714 | 3.91 | 0 | miRNAWalker2 validate | -0.15 | 0.00355 | NA | |
| 147 | hsa-miR-125a-5p | RACGAP1 | -1.32 | 0.00714 | 3.59 | 0 | miRanda | -0.2 | 0 | NA | |
| 148 | hsa-miR-342-3p | SLC15A1 | 1.31 | 0.02072 | 0.71 | 0.66939 | miRanda | -0.69 | 2.0E-5 | NA | |
| 149 | hsa-miR-590-3p | SLC15A1 | 2.35 | 0 | 0.71 | 0.66939 | miRanda | -0.36 | 0.04905 | NA | |
| 150 | hsa-miR-192-5p | SNX10 | 1.78 | 0.11349 | 0.37 | 0.6156 | miRNAWalker2 validate | -0.12 | 0.00181 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CELL CYCLE | 24 | 1316 | 2.47e-16 | 1.149e-12 |
| 2 | CELL CYCLE PROCESS | 22 | 1081 | 7.647e-16 | 1.779e-12 |
| 3 | CELL DIVISION | 15 | 460 | 1.16e-13 | 1.8e-10 |
| 4 | MITOTIC CELL CYCLE | 17 | 766 | 9.611e-13 | 1.118e-09 |
| 5 | REGULATION OF CELL CYCLE | 16 | 949 | 3.14e-10 | 2.922e-07 |
| 6 | CHROMOSOME SEGREGATION | 10 | 272 | 7.615e-10 | 5.906e-07 |
| 7 | MITOTIC SISTER CHROMATID SEGREGATION | 7 | 91 | 2.02e-09 | 1.343e-06 |
| 8 | CHROMOSOME ORGANIZATION | 15 | 1009 | 7.176e-09 | 4.174e-06 |
| 9 | REGULATION OF CELL DIVISION | 9 | 272 | 1.471e-08 | 6.879e-06 |
| 10 | MITOTIC SPINDLE ORGANIZATION | 6 | 69 | 1.478e-08 | 6.879e-06 |
| 11 | REGULATION OF CHROMOSOME SEGREGATION | 6 | 85 | 5.24e-08 | 2.032e-05 |
| 12 | CYTOKINESIS | 6 | 84 | 4.879e-08 | 2.032e-05 |
| 13 | REGULATION OF CELL CYCLE PROCESS | 11 | 558 | 6.406e-08 | 2.137e-05 |
| 14 | NUCLEAR CHROMOSOME SEGREGATION | 8 | 228 | 6.429e-08 | 2.137e-05 |
| 15 | POSITIVE REGULATION OF CELL CYCLE | 9 | 332 | 8.151e-08 | 2.529e-05 |
| 16 | POSITIVE REGULATION OF CELL CYCLE PROCESS | 8 | 247 | 1.19e-07 | 3.462e-05 |
| 17 | CELL CYCLE PHASE TRANSITION | 8 | 255 | 1.52e-07 | 4.16e-05 |
| 18 | SISTER CHROMATID SEGREGATION | 7 | 176 | 1.97e-07 | 5.092e-05 |
| 19 | ORGANELLE FISSION | 10 | 496 | 2.258e-07 | 5.529e-05 |
| 20 | CELL CYCLE G1 S PHASE TRANSITION | 6 | 111 | 2.589e-07 | 5.736e-05 |
| 21 | G1 S TRANSITION OF MITOTIC CELL CYCLE | 6 | 111 | 2.589e-07 | 5.736e-05 |
| 22 | DNA REPLICATION | 7 | 208 | 6.101e-07 | 0.000129 |
| 23 | MITOTIC CYTOKINESIS | 4 | 31 | 8.76e-07 | 0.0001772 |
| 24 | MICROTUBULE CYTOSKELETON ORGANIZATION | 8 | 348 | 1.595e-06 | 0.0003092 |
| 25 | REGULATION OF MICROTUBULE BASED PROCESS | 7 | 243 | 1.727e-06 | 0.0003214 |
| 26 | MITOTIC NUCLEAR DIVISION | 8 | 361 | 2.096e-06 | 0.000364 |
| 27 | REGULATION OF ATTACHMENT OF SPINDLE MICROTUBULES TO KINETOCHORE | 3 | 11 | 2.112e-06 | 0.000364 |
| 28 | CYTOSKELETON DEPENDENT CYTOKINESIS | 4 | 39 | 2.258e-06 | 0.0003752 |
| 29 | NEGATIVE REGULATION OF PROTEIN COMPLEX DISASSEMBLY | 6 | 170 | 3.152e-06 | 0.0005057 |
| 30 | CELL PROLIFERATION | 10 | 672 | 3.536e-06 | 0.0005484 |
| 31 | DNA DEPENDENT DNA REPLICATION | 5 | 99 | 3.882e-06 | 0.0005827 |
| 32 | REGULATION OF MICROTUBULE POLYMERIZATION OR DEPOLYMERIZATION | 6 | 178 | 4.111e-06 | 0.0005977 |
| 33 | GROWTH | 8 | 410 | 5.363e-06 | 0.0007562 |
| 34 | REGULATION OF CELL PROLIFERATION | 14 | 1496 | 6.921e-06 | 0.0009472 |
| 35 | REGULATION OF PROTEIN COMPLEX DISASSEMBLY | 6 | 217 | 1.277e-05 | 0.001698 |
| 36 | NEGATIVE REGULATION OF CYTOSKELETON ORGANIZATION | 6 | 221 | 1.417e-05 | 0.001735 |
| 37 | REGULATION OF MITOTIC CELL CYCLE | 8 | 468 | 1.405e-05 | 0.001735 |
| 38 | DEVELOPMENTAL GROWTH | 7 | 333 | 1.364e-05 | 0.001735 |
| 39 | POSITIVE REGULATION OF CELL DIVISION | 5 | 132 | 1.583e-05 | 0.001888 |
| 40 | NEGATIVE REGULATION OF CELL CYCLE PHASE TRANSITION | 5 | 146 | 2.574e-05 | 0.002994 |
| 41 | MICROTUBULE BASED PROCESS | 8 | 522 | 3.073e-05 | 0.003487 |
| 42 | RETROGRADE VESICLE MEDIATED TRANSPORT GOLGI TO ER | 4 | 77 | 3.475e-05 | 0.003849 |
| 43 | NEGATIVE REGULATION OF ORGANELLE ORGANIZATION | 7 | 387 | 3.576e-05 | 0.003869 |
| 44 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 13 | 1518 | 4.106e-05 | 0.004342 |
| 45 | REGULATION OF NUCLEAR DIVISION | 5 | 163 | 4.362e-05 | 0.004511 |
| 46 | DNA REPLICATION INITIATION | 3 | 29 | 4.537e-05 | 0.004589 |
| 47 | DNA METABOLIC PROCESS | 9 | 758 | 6.841e-05 | 0.006773 |
| 48 | NEGATIVE REGULATION OF CELL CYCLE | 7 | 433 | 7.266e-05 | 0.007043 |
| 49 | ANTIGEN PROCESSING AND PRESENTATION OF PEPTIDE OR POLYSACCHARIDE ANTIGEN VIA MHC CLASS II | 4 | 94 | 7.599e-05 | 0.007072 |
| 50 | ANTIGEN PROCESSING AND PRESENTATION OF PEPTIDE ANTIGEN VIA MHC CLASS II | 4 | 94 | 7.599e-05 | 0.007072 |
| 51 | POSITIVE REGULATION OF CYTOKINESIS | 3 | 36 | 8.762e-05 | 0.007994 |
| 52 | CELL CYCLE CHECKPOINT | 5 | 194 | 9.954e-05 | 0.008907 |
| 53 | NEGATIVE REGULATION OF MITOTIC CELL CYCLE | 5 | 199 | 0.0001122 | 0.009779 |
| 54 | REGULATION OF CELL CYCLE PHASE TRANSITION | 6 | 321 | 0.0001135 | 0.009779 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | TUBULIN BINDING | 7 | 273 | 3.731e-06 | 0.003466 |
| 2 | MICROTUBULE BINDING | 6 | 201 | 8.257e-06 | 0.003835 |
| 3 | MICROTUBULE MOTOR ACTIVITY | 4 | 77 | 3.475e-05 | 0.009463 |
| 4 | CYCLIN DEPENDENT PROTEIN SERINE THREONINE KINASE REGULATOR ACTIVITY | 3 | 28 | 4.075e-05 | 0.009463 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | MICROTUBULE CYTOSKELETON | 16 | 1068 | 1.749e-09 | 1.021e-06 |
| 2 | CHROMOSOME | 14 | 880 | 1.098e-08 | 3.205e-06 |
| 3 | SPINDLE | 9 | 289 | 2.483e-08 | 4.834e-06 |
| 4 | CYTOSKELETAL PART | 16 | 1436 | 1.145e-07 | 1.671e-05 |
| 5 | NUCLEAR CHROMOSOME | 10 | 523 | 3.676e-07 | 4.293e-05 |
| 6 | MIDBODY | 6 | 132 | 7.208e-07 | 7.015e-05 |
| 7 | KINESIN COMPLEX | 4 | 55 | 9.102e-06 | 0.0005906 |
| 8 | MITOTIC SPINDLE | 4 | 55 | 9.102e-06 | 0.0005906 |
| 9 | CYTOSKELETON | 16 | 1967 | 7.613e-06 | 0.0005906 |
| 10 | CENTROSOME | 8 | 487 | 1.871e-05 | 0.001093 |
| 11 | MICROTUBULE ORGANIZING CENTER | 8 | 623 | 0.0001064 | 0.005648 |
| 12 | CHROMOSOMAL REGION | 6 | 330 | 0.000132 | 0.006423 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
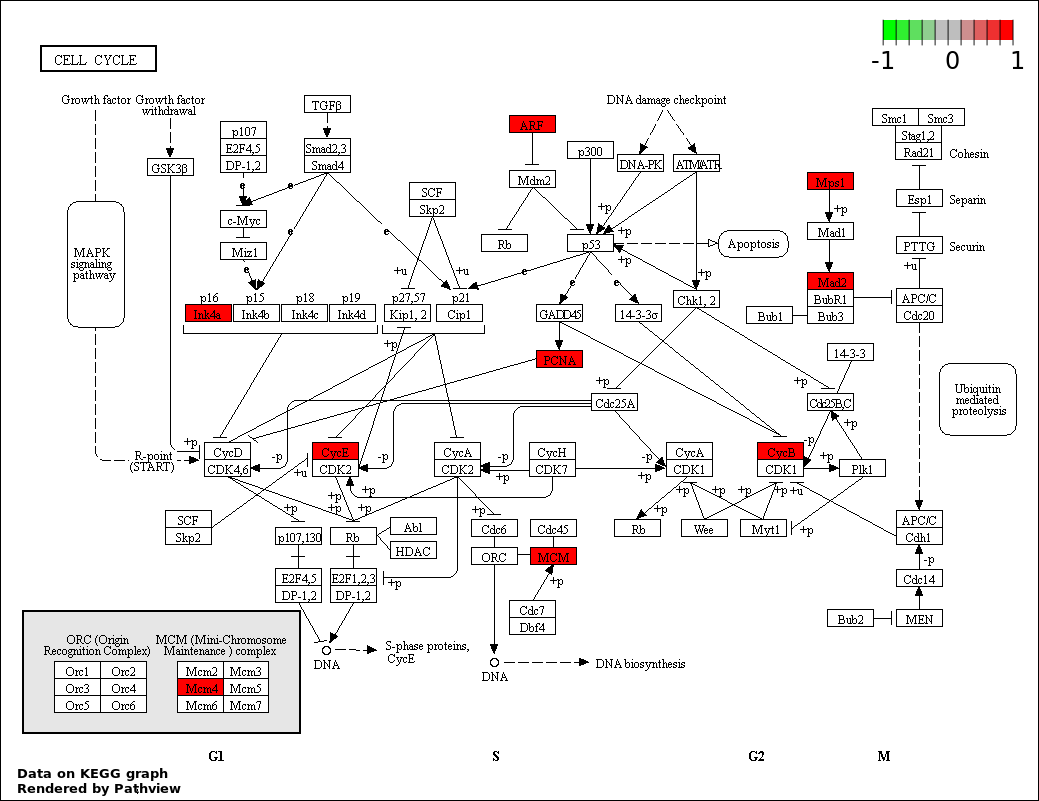
| 1 | hsa04110_Cell_cycle | 7 | 128 | 2.207e-08 | 3.973e-06 | |
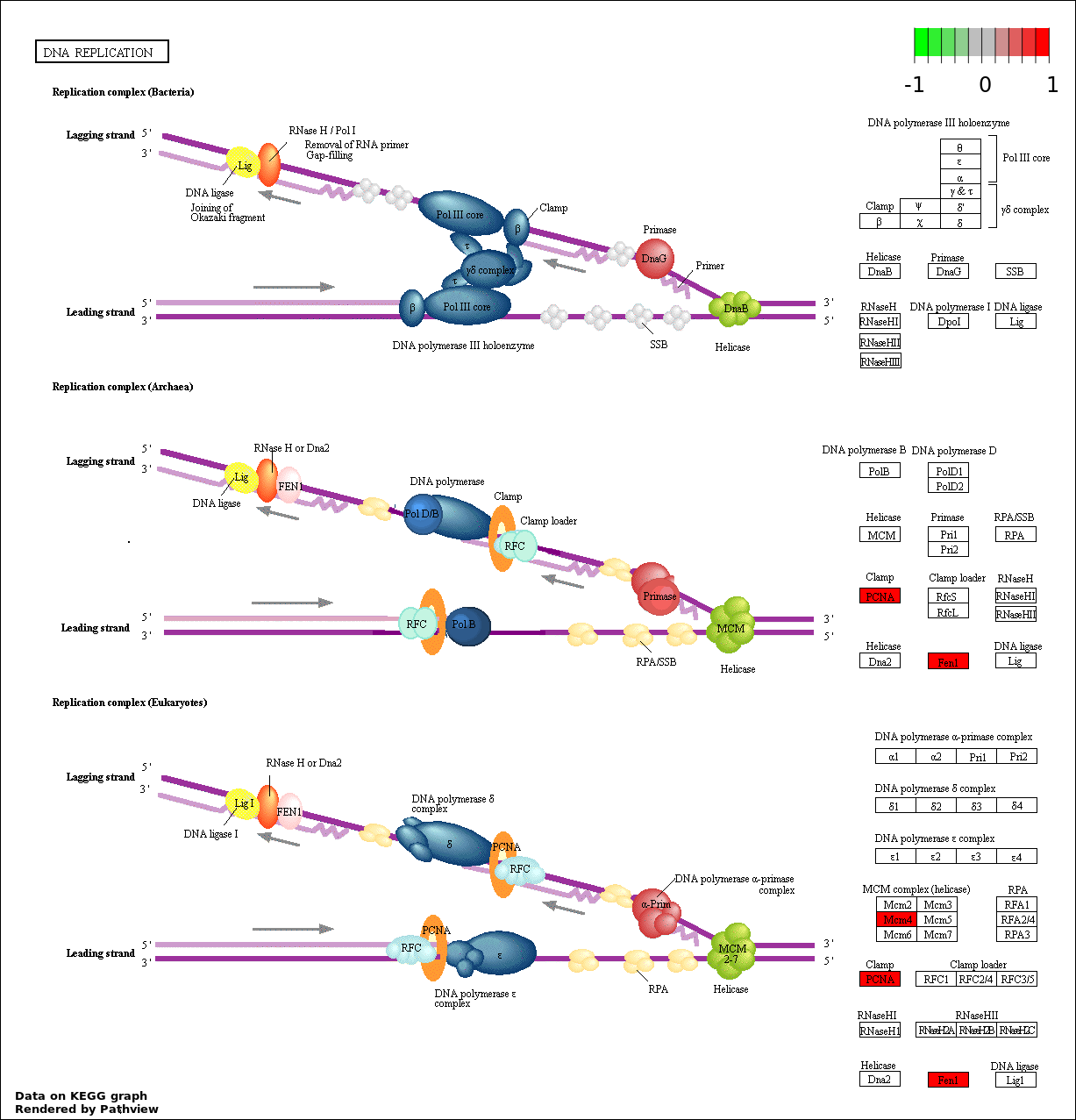
| 2 | hsa03030_DNA_replication | 3 | 36 | 8.762e-05 | 0.007886 | |
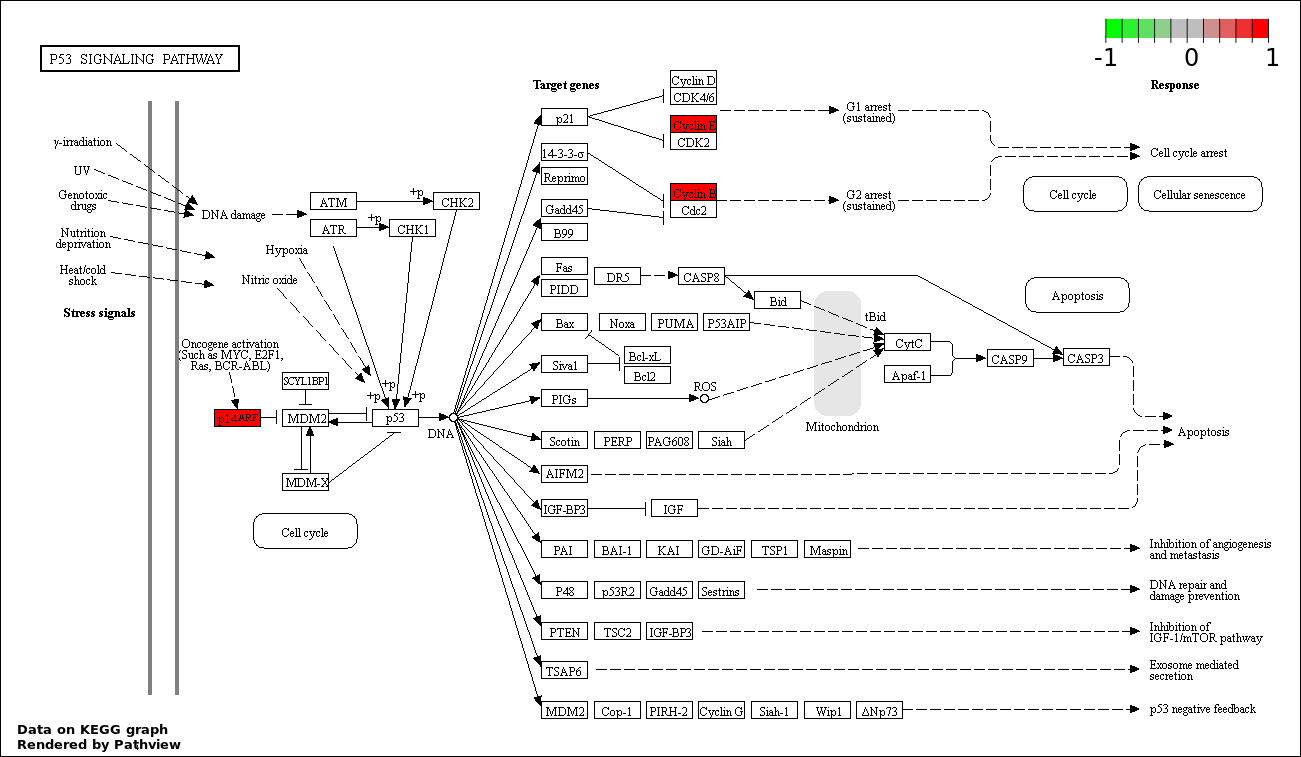
| 3 | hsa04115_p53_signaling_pathway | 3 | 69 | 0.0006082 | 0.03649 | |
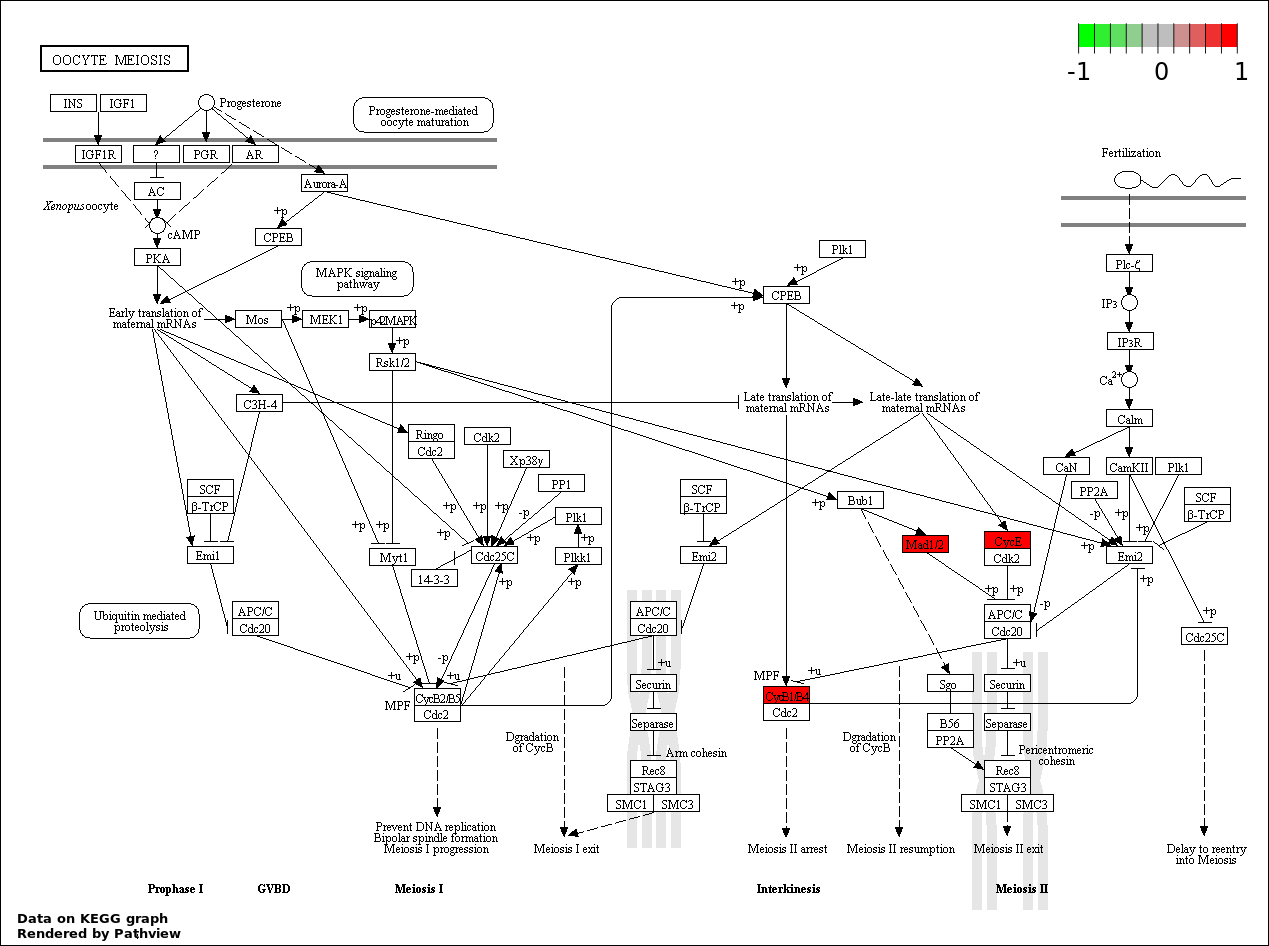
| 4 | hsa04114_Oocyte_meiosis | 3 | 114 | 0.002588 | 0.1085 | |
| 5 | hsa03410_Base_excision_repair | 2 | 34 | 0.003013 | 0.1085 | |
| 6 | hsa00590_Arachidonic_acid_metabolism | 2 | 59 | 0.008846 | 0.2654 | |
| 7 | hsa04914_Progesterone.mediated_oocyte_maturation | 2 | 87 | 0.01854 | 0.4766 | |
| 8 | hsa04151_PI3K_AKT_signaling_pathway | 2 | 351 | 0.2059 | 1 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | TINCR | hsa-let-7e-5p;hsa-miR-106a-5p;hsa-miR-125a-3p;hsa-miR-148b-3p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-30a-3p;hsa-miR-335-5p;hsa-miR-592 | 10 | MXD1 | Sponge network | 3.51 | 0.03668 | 1.612 | 0.03992 | 0.575 |
| 2 | CALML3-AS1 | hsa-let-7e-5p;hsa-miR-106a-5p;hsa-miR-125a-3p;hsa-miR-148b-3p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-30a-3p;hsa-miR-335-5p;hsa-miR-589-3p;hsa-miR-592 | 10 | MXD1 | Sponge network | 4.924 | 0.00575 | 1.612 | 0.03992 | 0.526 |
| 3 | RP11-760H22.2 | hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-31-5p;hsa-miR-32-3p;hsa-miR-7-5p;hsa-miR-9-3p | 11 | ESR1 | Sponge network | -3.418 | 0.00912 | -4.867 | 9.0E-5 | 0.515 |
| 4 | HAND2-AS1 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-32-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 25 | ESR1 | Sponge network | -7.871 | 0 | -4.867 | 9.0E-5 | 0.509 |
| 5 | RP11-130L8.1 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-590-3p | 12 | ESR1 | Sponge network | -4.329 | 1.0E-5 | -4.867 | 9.0E-5 | 0.491 |
| 6 | EMX2OS |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-32-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 25 | ESR1 | Sponge network | -6.205 | 0.00015 | -4.867 | 9.0E-5 | 0.471 |
| 7 | LINC00284 |
hsa-let-7f-1-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-31-5p;hsa-miR-32-3p;hsa-miR-330-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 19 | ESR1 | Sponge network | -5.478 | 0.02716 | -4.867 | 9.0E-5 | 0.454 |
| 8 | BDNF-AS | hsa-let-7f-1-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-31-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 15 | ESR1 | Sponge network | -1.712 | 0.02515 | -4.867 | 9.0E-5 | 0.441 |
| 9 | ZNF667-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-627-5p | 14 | TGFBR3 | Sponge network | -4.019 | 0.00137 | -4.817 | 0 | 0.44 |
| 10 | RP11-757G1.6 |
hsa-let-7f-1-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-31-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-9-3p | 13 | ESR1 | Sponge network | -2.705 | 0.04664 | -4.867 | 9.0E-5 | 0.412 |
| 11 | CTB-92J24.3 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-33a-3p;hsa-miR-627-5p;hsa-miR-944 | 11 | TGFBR3 | Sponge network | -7.226 | 0.0046 | -4.817 | 0 | 0.405 |
| 12 | RP11-355F16.1 | hsa-let-7f-1-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-32-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-629-3p | 14 | ESR1 | Sponge network | -2.178 | 0.07502 | -4.867 | 9.0E-5 | 0.402 |
| 13 | RP11-1036E20.9 | hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-32-3p;hsa-miR-7-5p | 13 | ESR1 | Sponge network | -0.331 | 0.90522 | -4.867 | 9.0E-5 | 0.4 |
| 14 | CTD-2554C21.2 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-944 | 11 | TGFBR3 | Sponge network | -6.968 | 0.00817 | -4.817 | 0 | 0.396 |
| 15 | RASSF8-AS1 | hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-944 | 13 | TGFBR3 | Sponge network | -2.562 | 0.00163 | -4.817 | 0 | 0.393 |
| 16 | RP1-193H18.2 | hsa-let-7f-1-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-31-5p;hsa-miR-629-3p;hsa-miR-7-5p | 12 | ESR1 | Sponge network | -0.388 | 0.6877 | -4.867 | 9.0E-5 | 0.39 |
| 17 | RP11-359B12.2 |
hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 13 | TGFBR3 | Sponge network | -2.094 | 0.00033 | -4.817 | 0 | 0.38 |
| 18 | NR2F2-AS1 |
hsa-let-7f-1-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-31-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-9-3p | 17 | ESR1 | Sponge network | -3.785 | 0.00281 | -4.867 | 9.0E-5 | 0.376 |
| 19 | RP11-166D19.1 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-32-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-9-5p | 23 | ESR1 | Sponge network | -4.209 | 2.0E-5 | -4.867 | 9.0E-5 | 0.374 |
| 20 | RP11-747H7.3 | hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-32-3p;hsa-miR-7-5p;hsa-miR-9-3p;hsa-miR-9-5p | 11 | ESR1 | Sponge network | -0.36 | 0.74172 | -4.867 | 9.0E-5 | 0.371 |
| 21 | RP11-680F20.6 |
hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-9-5p | 11 | ESR1 | Sponge network | -3.912 | 0.17114 | -4.867 | 9.0E-5 | 0.37 |
| 22 | MAGI2-AS3 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | TGFBR3 | Sponge network | -4.563 | 0 | -4.817 | 0 | 0.366 |
| 23 | RP11-627G23.1 | hsa-let-7f-1-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-31-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-629-3p;hsa-miR-9-3p;hsa-miR-9-5p | 15 | ESR1 | Sponge network | -4.055 | 0.12629 | -4.867 | 9.0E-5 | 0.365 |
| 24 | RP11-554A11.4 | hsa-miR-107;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-222-3p;hsa-miR-32-3p;hsa-miR-33a-3p;hsa-miR-629-3p | 11 | ESR1 | Sponge network | -5.361 | 2.0E-5 | -4.867 | 9.0E-5 | 0.363 |
| 25 | MIR143HG |
hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-224-3p;hsa-miR-3065-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p | 10 | INHBA | Sponge network | -6.51 | 0 | 0.597 | 0.64331 | 0.362 |
| 26 | NR2F2-AS1 |
hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-590-3p;hsa-miR-627-5p;hsa-miR-944 | 13 | TGFBR3 | Sponge network | -3.785 | 0.00281 | -4.817 | 0 | 0.36 |
| 27 | RP11-166D19.1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-627-5p;hsa-miR-944 | 18 | TGFBR3 | Sponge network | -4.209 | 2.0E-5 | -4.817 | 0 | 0.359 |
| 28 | ACVR2B-AS1 | hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-484;hsa-miR-590-3p | 10 | TGFBR3 | Sponge network | -2.253 | 0.00072 | -4.817 | 0 | 0.355 |
| 29 | CTB-92J24.3 |
hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-589-3p | 14 | ESR1 | Sponge network | -7.226 | 0.0046 | -4.867 | 9.0E-5 | 0.355 |
| 30 | RP11-774O3.3 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-31-5p;hsa-miR-32-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 18 | ESR1 | Sponge network | -1.989 | 0.00136 | -4.867 | 9.0E-5 | 0.351 |
| 31 | HAND2-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-944 | 14 | TGFBR3 | Sponge network | -7.871 | 0 | -4.817 | 0 | 0.35 |
| 32 | MIR497HG |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-5p | 23 | ESR1 | Sponge network | -6.146 | 0.00024 | -4.867 | 9.0E-5 | 0.349 |
| 33 | RP11-150O12.3 |
hsa-let-7f-1-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-33a-3p;hsa-miR-629-3p;hsa-miR-9-3p | 13 | ESR1 | Sponge network | -4.03 | 0.14448 | -4.867 | 9.0E-5 | 0.348 |
| 34 | RP11-887P2.5 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-7-5p | 15 | ESR1 | Sponge network | -9.865 | 1.0E-5 | -4.867 | 9.0E-5 | 0.342 |
| 35 | AC016582.2 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-590-3p | 11 | TGFBR3 | Sponge network | -5.41 | 0.03371 | -4.817 | 0 | 0.34 |
| 36 | NR2F1-AS1 |
hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-484;hsa-miR-590-3p | 12 | TGFBR3 | Sponge network | -2.961 | 0.00154 | -4.817 | 0 | 0.336 |
| 37 | MAGI2-AS3 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-32-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-9-3p | 27 | ESR1 | Sponge network | -4.563 | 0 | -4.867 | 9.0E-5 | 0.335 |
| 38 | ADAMTS9-AS1 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-324-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 25 | ESR1 | Sponge network | -8.573 | 0.00012 | -4.867 | 9.0E-5 | 0.335 |
| 39 | WDR86-AS1 | hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-32-3p;hsa-miR-629-3p;hsa-miR-9-3p | 14 | ESR1 | Sponge network | -2.587 | 0.08454 | -4.867 | 9.0E-5 | 0.333 |
| 40 | RP11-887P2.5 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-590-5p;hsa-miR-944 | 13 | TGFBR3 | Sponge network | -9.865 | 1.0E-5 | -4.817 | 0 | 0.332 |
| 41 | HOTTIP |
hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-32-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p | 11 | ESR1 | Sponge network | 0.232 | 0.87041 | -4.867 | 9.0E-5 | 0.33 |
| 42 | CTC-297N7.9 | hsa-miR-130b-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-7-5p;hsa-miR-9-5p | 10 | ESR1 | Sponge network | -3.463 | 0.00542 | -4.867 | 9.0E-5 | 0.33 |
| 43 | RP11-532F6.3 | hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-629-3p;hsa-miR-9-5p | 12 | ESR1 | Sponge network | -2.663 | 0.00676 | -4.867 | 9.0E-5 | 0.329 |
| 44 | TRHDE-AS1 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-31-5p;hsa-miR-32-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 21 | ESR1 | Sponge network | -6.205 | 0.01165 | -4.867 | 9.0E-5 | 0.329 |
| 45 | CTD-2554C21.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-629-3p;hsa-miR-7-5p | 13 | ESR1 | Sponge network | -6.968 | 0.00817 | -4.867 | 9.0E-5 | 0.326 |
| 46 | RP11-757G1.6 |
hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-627-5p;hsa-miR-944 | 13 | TGFBR3 | Sponge network | -2.705 | 0.04664 | -4.817 | 0 | 0.325 |
| 47 | MIR143HG |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-627-5p | 16 | TGFBR3 | Sponge network | -6.51 | 0 | -4.817 | 0 | 0.323 |
| 48 | EMX2OS |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-627-5p;hsa-miR-944 | 17 | TGFBR3 | Sponge network | -6.205 | 0.00015 | -4.817 | 0 | 0.319 |
| 49 | AC073283.4 |
hsa-let-7f-1-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-31-5p;hsa-miR-7-5p;hsa-miR-9-3p | 11 | ESR1 | Sponge network | -2.801 | 0.08856 | -4.867 | 9.0E-5 | 0.316 |
| 50 | TPTEP1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-627-5p | 13 | TGFBR3 | Sponge network | -4.398 | 5.0E-5 | -4.817 | 0 | 0.316 |
| 51 | RP11-359B12.2 |
hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-629-3p | 13 | ESR1 | Sponge network | -2.094 | 0.00033 | -4.867 | 9.0E-5 | 0.314 |
| 52 | PGM5-AS1 | hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-301a-3p;hsa-miR-33a-3p | 13 | ESR1 | Sponge network | -14.107 | 0 | -4.867 | 9.0E-5 | 0.311 |
| 53 | RP11-389C8.2 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-629-3p | 19 | ESR1 | Sponge network | -3.089 | 2.0E-5 | -4.867 | 9.0E-5 | 0.309 |
| 54 | CTD-2314G24.2 | hsa-let-7f-1-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-32-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-9-3p | 12 | ESR1 | Sponge network | 2.041 | 0.35788 | -4.867 | 9.0E-5 | 0.306 |
| 55 | RP11-597D13.9 | hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-32-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-7-5p;hsa-miR-9-5p | 15 | ESR1 | Sponge network | -2.494 | 0.07597 | -4.867 | 9.0E-5 | 0.305 |
| 56 | RP11-981G7.6 | hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 14 | ESR1 | Sponge network | -0.726 | 0.44765 | -4.867 | 9.0E-5 | 0.303 |
| 57 | RP11-161M6.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-330-3p | 12 | ESR1 | Sponge network | -2.608 | 0.00296 | -4.867 | 9.0E-5 | 0.303 |
| 58 | MIR497HG |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-590-5p | 13 | TGFBR3 | Sponge network | -6.146 | 0.00024 | -4.817 | 0 | 0.303 |
| 59 | RP11-13K12.1 |
hsa-let-7f-1-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-31-5p;hsa-miR-330-3p;hsa-miR-629-3p | 15 | ESR1 | Sponge network | -5.093 | 0.01151 | -4.867 | 9.0E-5 | 0.303 |
| 60 | FAM66C |
hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 16 | ESR1 | Sponge network | -2.927 | 0.00012 | -4.867 | 9.0E-5 | 0.302 |
| 61 | RP11-326C3.11 | hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p | 10 | ESR1 | Sponge network | -3.026 | 0 | -4.867 | 9.0E-5 | 0.3 |
| 62 | USP3-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p | 11 | TGFBR3 | Sponge network | -4.151 | 0 | -4.817 | 0 | 0.297 |
| 63 | RP11-116O18.1 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-590-3p;hsa-miR-7-5p | 14 | ESR1 | Sponge network | -5.007 | 0.06008 | -4.867 | 9.0E-5 | 0.293 |
| 64 | LINC00899 | hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-21-3p;hsa-miR-324-5p;hsa-miR-590-3p;hsa-miR-7-5p | 11 | ESR1 | Sponge network | -1.597 | 0.00258 | -4.867 | 9.0E-5 | 0.292 |
| 65 | RP11-161M6.2 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-484 | 11 | TGFBR3 | Sponge network | -2.608 | 0.00296 | -4.817 | 0 | 0.292 |
| 66 | FAM66C |
hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 14 | TGFBR3 | Sponge network | -2.927 | 0.00012 | -4.817 | 0 | 0.292 |
| 67 | LINC00323 | hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-629-3p;hsa-miR-7-5p | 13 | ESR1 | Sponge network | -3.25 | 0.07329 | -4.867 | 9.0E-5 | 0.291 |
| 68 | RP11-774O3.3 |
hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-590-3p | 11 | TGFBR3 | Sponge network | -1.989 | 0.00136 | -4.817 | 0 | 0.288 |
| 69 | AC141928.1 | hsa-let-7f-1-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-629-3p | 14 | ESR1 | Sponge network | -4.805 | 0.00102 | -4.867 | 9.0E-5 | 0.286 |
| 70 | RP11-389C8.2 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-33a-3p | 11 | TGFBR3 | Sponge network | -3.089 | 2.0E-5 | -4.817 | 0 | 0.284 |
| 71 | LINC00861 | hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-33a-3p;hsa-miR-589-3p | 13 | ESR1 | Sponge network | 0.999 | 0.45301 | -4.867 | 9.0E-5 | 0.284 |
| 72 | RP11-983P16.4 |
hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-484 | 11 | TGFBR3 | Sponge network | -2.087 | 0.00021 | -4.817 | 0 | 0.281 |
| 73 | AC003090.1 |
hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-629-3p | 19 | ESR1 | Sponge network | -7.817 | 0.00161 | -4.867 | 9.0E-5 | 0.28 |
| 74 | ADAMTS9-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-627-5p | 14 | TGFBR3 | Sponge network | -8.573 | 0.00012 | -4.817 | 0 | 0.279 |
| 75 | RP11-130L8.1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-627-5p;hsa-miR-944 | 12 | TGFBR3 | Sponge network | -4.329 | 1.0E-5 | -4.817 | 0 | 0.278 |
| 76 | BZRAP1-AS1 | hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-222-3p;hsa-miR-31-5p | 13 | ESR1 | Sponge network | -1.931 | 0.08861 | -4.867 | 9.0E-5 | 0.278 |
| 77 | ACTA2-AS1 |
hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p | 22 | ESR1 | Sponge network | -6.142 | 0.00223 | -4.867 | 9.0E-5 | 0.277 |
| 78 | RP11-116O18.1 |
hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-944 | 12 | TGFBR3 | Sponge network | -5.007 | 0.06008 | -4.817 | 0 | 0.277 |
| 79 | PWAR6 |
hsa-let-7f-1-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-31-5p;hsa-miR-330-3p;hsa-miR-629-3p | 14 | ESR1 | Sponge network | -3.15 | 0.0082 | -4.867 | 9.0E-5 | 0.275 |
| 80 | CTD-2554C21.3 |
hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-629-3p;hsa-miR-7-5p | 15 | ESR1 | Sponge network | -6.258 | 0.00703 | -4.867 | 9.0E-5 | 0.275 |
| 81 | RP11-822E23.8 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-484 | 12 | TGFBR3 | Sponge network | -8.351 | 0.00374 | -4.817 | 0 | 0.275 |
| 82 | A2M-AS1 | hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-627-5p | 12 | TGFBR3 | Sponge network | -5.049 | 0.03435 | -4.817 | 0 | 0.274 |
| 83 | RP11-13K12.5 | hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-629-3p | 10 | ESR1 | Sponge network | -5.713 | 0.01127 | -4.867 | 9.0E-5 | 0.274 |
| 84 | USP3-AS1 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p | 13 | ESR1 | Sponge network | -4.151 | 0 | -4.867 | 9.0E-5 | 0.272 |
| 85 | SOCS2-AS1 | hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-21-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-629-3p | 12 | ESR1 | Sponge network | -4.167 | 1.0E-5 | -4.867 | 9.0E-5 | 0.271 |
| 86 | ZNF667-AS1 |
hsa-let-7f-1-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-629-3p;hsa-miR-7-5p | 16 | ESR1 | Sponge network | -4.019 | 0.00137 | -4.867 | 9.0E-5 | 0.27 |
| 87 | CD27-AS1 | hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-330-3p;hsa-miR-589-3p;hsa-miR-629-3p | 10 | ESR1 | Sponge network | -1.311 | 0.00574 | -4.867 | 9.0E-5 | 0.269 |
| 88 | DNM3OS |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 14 | TGFBR3 | Sponge network | -3.933 | 0.00059 | -4.817 | 0 | 0.267 |
| 89 | RP11-983P16.4 |
hsa-let-7f-1-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-31-5p;hsa-miR-629-3p | 11 | ESR1 | Sponge network | -2.087 | 0.00021 | -4.867 | 9.0E-5 | 0.264 |
| 90 | LINC00284 |
hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-627-5p;hsa-miR-944 | 13 | TGFBR3 | Sponge network | -5.478 | 0.02716 | -4.817 | 0 | 0.263 |
| 91 | LINC00982 | hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-629-3p | 13 | ESR1 | Sponge network | -3.897 | 0.03858 | -4.867 | 9.0E-5 | 0.263 |
| 92 | CTD-2554C21.3 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-484 | 12 | TGFBR3 | Sponge network | -6.258 | 0.00703 | -4.817 | 0 | 0.258 |
| 93 | WT1-AS |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-32-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-9-3p;hsa-miR-9-5p | 24 | ESR1 | Sponge network | -6.875 | 2.0E-5 | -4.867 | 9.0E-5 | 0.258 |
| 94 | RP11-54O7.3 | hsa-let-7f-1-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-31-5p;hsa-miR-330-3p;hsa-miR-629-3p | 10 | ESR1 | Sponge network | -2.864 | 0.01902 | -4.867 | 9.0E-5 | 0.257 |
| 95 | RP11-354E11.2 | hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-629-3p;hsa-miR-7-5p | 11 | ESR1 | Sponge network | -3.495 | 0.03784 | -4.867 | 9.0E-5 | 0.256 |
| 96 | DNM3OS |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-32-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 21 | ESR1 | Sponge network | -3.933 | 0.00059 | -4.867 | 9.0E-5 | 0.256 |
| 97 | LINC00461 | hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-944 | 11 | TGFBR3 | Sponge network | -2.019 | 0.3186 | -4.817 | 0 | 0.253 |
| 98 | ZNF582-AS1 |
hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-9-3p;hsa-miR-9-5p | 16 | ESR1 | Sponge network | -4.925 | 0.00112 | -4.867 | 9.0E-5 | 0.25 |