| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|
| 1 |
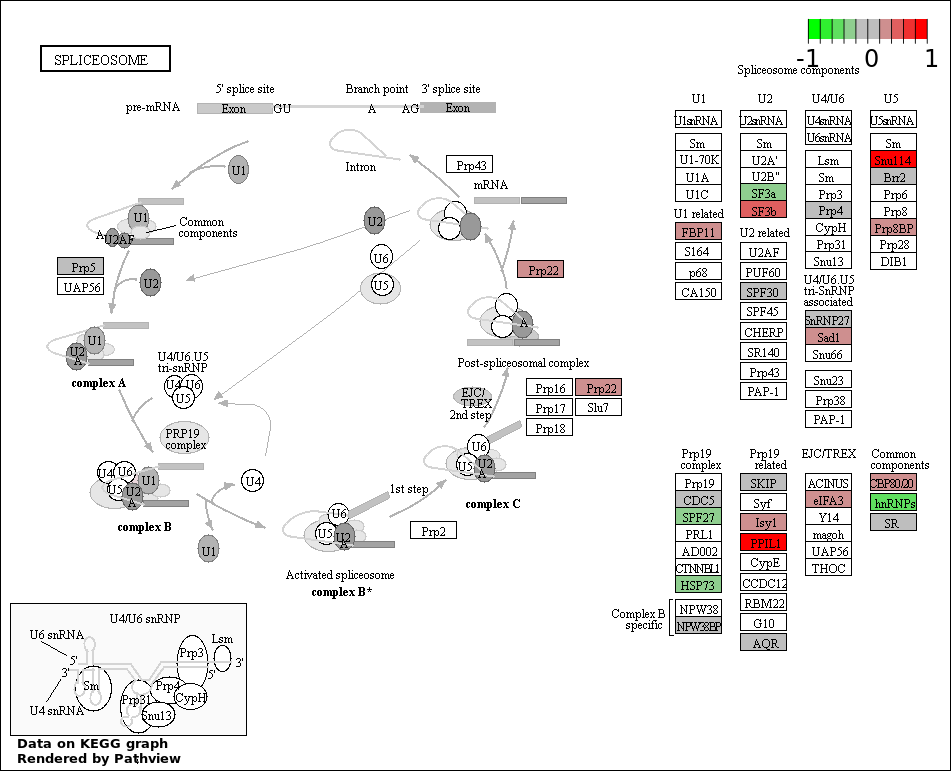
RNA PROCESSING |
61 |
835 |
2.575e-33 |
1.198e-29 |
| 2 |
MRNA METABOLIC PROCESS |
53 |
611 |
1.785e-32 |
4.152e-29 |
| 3 |
MRNA PROCESSING |
46 |
432 |
4.887e-32 |
7.579e-29 |
| 4 |
RNA SPLICING VIA TRANSESTERIFICATION REACTIONS |
38 |
267 |
3.236e-31 |
3.765e-28 |
| 5 |
RNA SPLICING |
42 |
367 |
1.6e-30 |
1.489e-27 |
| 6 |
RIBONUCLEOPROTEIN COMPLEX BIOGENESIS |
29 |
440 |
1.009e-14 |
7.826e-12 |
| 7 |
NLS BEARING PROTEIN IMPORT INTO NUCLEUS |
9 |
22 |
8.794e-13 |
5.845e-10 |
| 8 |
MITOTIC CELL CYCLE |
34 |
766 |
3.879e-12 |
2.256e-09 |
| 9 |
CELL CYCLE PHASE TRANSITION |
20 |
255 |
7.54e-12 |
3.509e-09 |
| 10 |
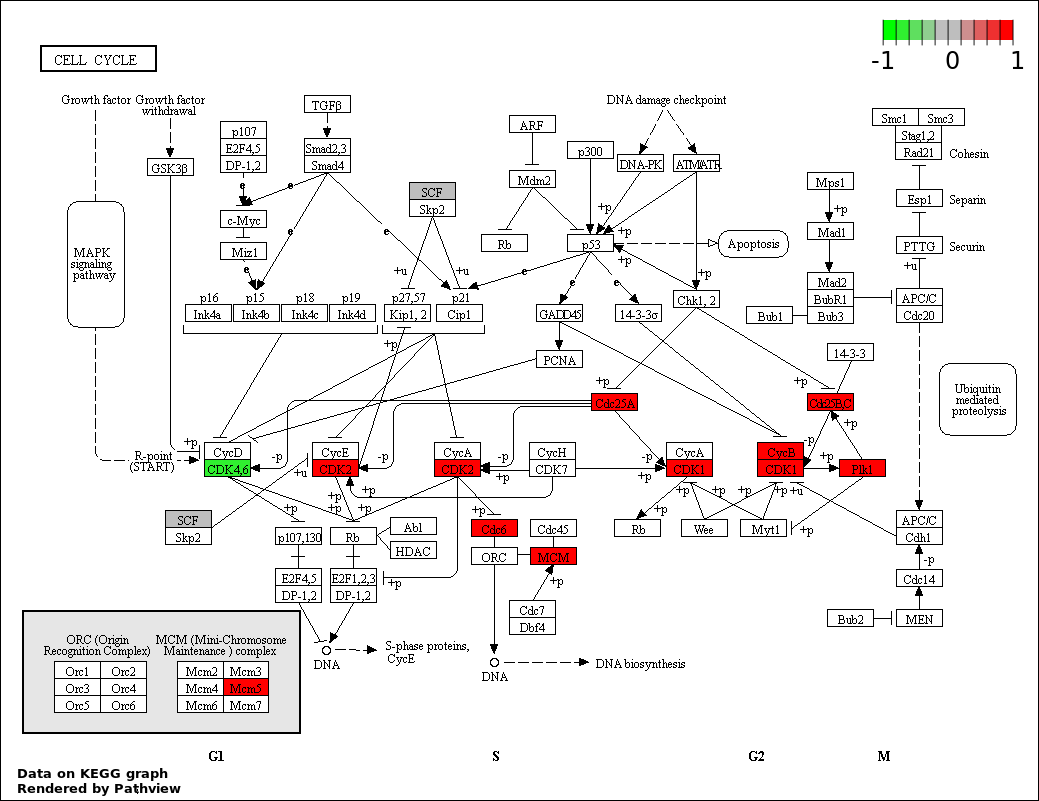
CELL CYCLE |
45 |
1316 |
7.026e-12 |
3.509e-09 |
| 11 |
NUCLEAR IMPORT |
15 |
129 |
1.28e-11 |
5.416e-09 |
| 12 |
REGULATION OF CELL CYCLE |
37 |
949 |
1.751e-11 |
6.788e-09 |
| 13 |
CELL CYCLE G1 S PHASE TRANSITION |
14 |
111 |
2.058e-11 |
6.84e-09 |
| 14 |
G1 S TRANSITION OF MITOTIC CELL CYCLE |
14 |
111 |
2.058e-11 |
6.84e-09 |
| 15 |
CELL CYCLE PROCESS |
39 |
1081 |
4.706e-11 |
1.46e-08 |
| 16 |
POSITIVE REGULATION OF GENE EXPRESSION |
51 |
1733 |
5.365e-11 |
1.56e-08 |
| 17 |
NUCLEAR TRANSPORT |
22 |
355 |
6.696e-11 |
1.833e-08 |
| 18 |
REGULATION OF TELOMERASE RNA LOCALIZATION TO CAJAL BODY |
7 |
15 |
1.058e-10 |
2.59e-08 |
| 19 |
POSITIVE REGULATION OF TELOMERASE RNA LOCALIZATION TO CAJAL BODY |
7 |
15 |
1.058e-10 |
2.59e-08 |
| 20 |
PROTEIN IMPORT |
15 |
155 |
1.811e-10 |
4.214e-08 |
| 21 |
PROTEIN LOCALIZATION TO NUCLEUS |
15 |
156 |
1.985e-10 |
4.398e-08 |
| 22 |
PROTEIN FOLDING |
17 |
224 |
4.965e-10 |
1.05e-07 |
| 23 |
POSTTRANSCRIPTIONAL REGULATION OF GENE EXPRESSION |
23 |
448 |
9.861e-10 |
1.995e-07 |
| 24 |
REGULATION OF MRNA METABOLIC PROCESS |
12 |
118 |
7.121e-09 |
1.381e-06 |
| 25 |
CELL DIVISION |
22 |
460 |
8.502e-09 |
1.582e-06 |
| 26 |
REGULATION OF DNA REPLICATION |
13 |
161 |
2.757e-08 |
4.933e-06 |
| 27 |
RIBONUCLEOPROTEIN COMPLEX SUBUNIT ORGANIZATION |
14 |
199 |
4.588e-08 |
7.907e-06 |
| 28 |
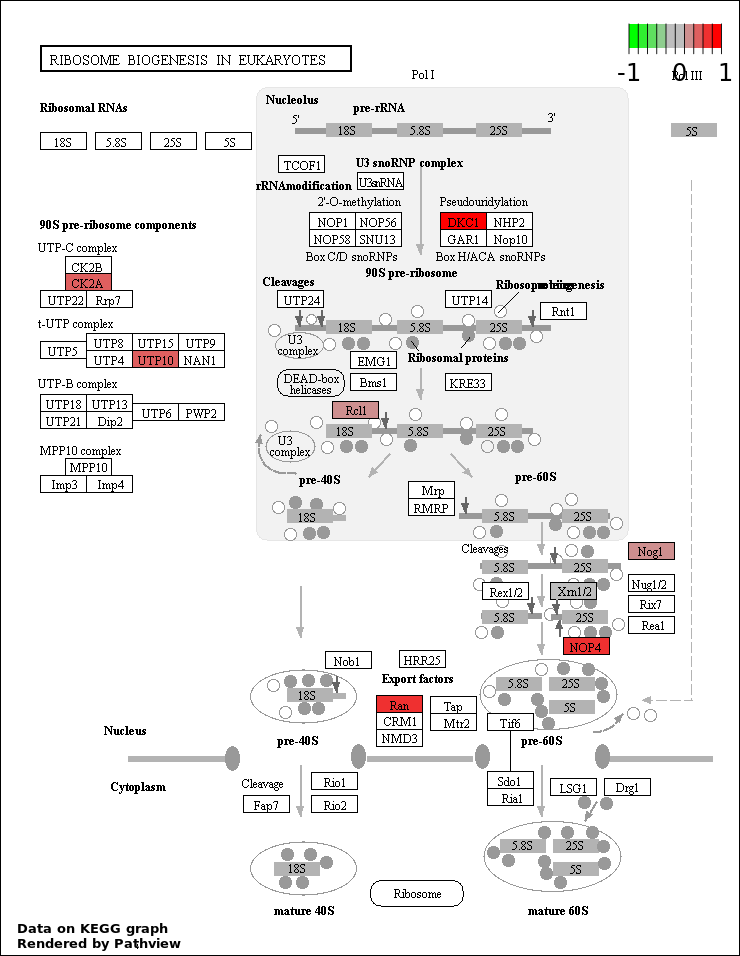
RIBOSOME BIOGENESIS |
17 |
308 |
5.879e-08 |
8.629e-06 |
| 29 |
MULTI ORGANISM TRANSPORT |
9 |
68 |
5.621e-08 |
8.629e-06 |
| 30 |
MULTI ORGANISM LOCALIZATION |
9 |
68 |
5.621e-08 |
8.629e-06 |
| 31 |
POSITIVE REGULATION OF TELOMERE MAINTENANCE VIA TELOMERE LENGTHENING |
7 |
33 |
5.934e-08 |
8.629e-06 |
| 32 |
PROTEIN LOCALIZATION TO ORGANELLE |
23 |
556 |
5.611e-08 |
8.629e-06 |
| 33 |
REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION TO CHROMOSOME |
5 |
11 |
6.738e-08 |
9.322e-06 |
| 34 |
REGULATION OF TELOMERE MAINTENANCE VIA TELOMERE LENGTHENING |
8 |
50 |
6.811e-08 |
9.322e-06 |
| 35 |
INTERSPECIES INTERACTION BETWEEN ORGANISMS |
25 |
662 |
8.351e-08 |
1.05e-05 |
| 36 |
SYMBIOSIS ENCOMPASSING MUTUALISM THROUGH PARASITISM |
25 |
662 |
8.351e-08 |
1.05e-05 |
| 37 |
DNA METABOLIC PROCESS |
27 |
758 |
8.015e-08 |
1.05e-05 |
| 38 |
MITOTIC NUCLEAR DIVISION |
18 |
361 |
1.101e-07 |
1.314e-05 |
| 39 |
SPLICEOSOMAL COMPLEX ASSEMBLY |
8 |
53 |
1.093e-07 |
1.314e-05 |
| 40 |
RNA LOCALIZATION |
13 |
185 |
1.422e-07 |
1.654e-05 |
| 41 |
RRNA METABOLIC PROCESS |
15 |
255 |
1.595e-07 |
1.81e-05 |
| 42 |
MACROMOLECULAR COMPLEX ASSEMBLY |
38 |
1398 |
1.909e-07 |
2.115e-05 |
| 43 |
REGULATION OF DNA METABOLIC PROCESS |
17 |
340 |
2.417e-07 |
2.616e-05 |
| 44 |
DNA GEOMETRIC CHANGE |
9 |
81 |
2.63e-07 |
2.781e-05 |
| 45 |
REGULATION OF PROTEIN LOCALIZATION TO CHROMOSOME TELOMERIC REGION |
5 |
14 |
2.842e-07 |
2.939e-05 |
| 46 |
CELL CYCLE G2 M PHASE TRANSITION |
11 |
138 |
3.765e-07 |
3.808e-05 |
| 47 |
MACROMOLECULE CATABOLIC PROCESS |
29 |
926 |
3.857e-07 |
3.819e-05 |
| 48 |
REGULATION OF RNA STABILITY |
11 |
139 |
4.049e-07 |
3.846e-05 |
| 49 |
CELLULAR RESPONSE TO DNA DAMAGE STIMULUS |
25 |
720 |
4.05e-07 |
3.846e-05 |
| 50 |
POSITIVE REGULATION OF DNA REPLICATION |
9 |
86 |
4.42e-07 |
4.113e-05 |
| 51 |
RNA SECONDARY STRUCTURE UNWINDING |
7 |
44 |
4.804e-07 |
4.383e-05 |
| 52 |
MODULATION BY SYMBIONT OF HOST CELLULAR PROCESS |
6 |
28 |
5.093e-07 |
4.557e-05 |
| 53 |
ESTABLISHMENT OF PROTEIN LOCALIZATION TO ORGANELLE |
17 |
361 |
5.597e-07 |
4.914e-05 |
| 54 |
REGULATION OF TELOMERE MAINTENANCE |
8 |
67 |
7.049e-07 |
6.074e-05 |
| 55 |
POSITIVE REGULATION OF TELOMERE MAINTENANCE |
7 |
47 |
7.666e-07 |
6.486e-05 |
| 56 |
POSITIVE REGULATION OF BIOSYNTHETIC PROCESS |
43 |
1805 |
9.815e-07 |
8.155e-05 |
| 57 |
NEGATIVE REGULATION OF PROTEIN METABOLIC PROCESS |
31 |
1087 |
1.1e-06 |
8.976e-05 |
| 58 |
REGULATION OF RNA SPLICING |
9 |
97 |
1.237e-06 |
9.927e-05 |
| 59 |
RNA 3 END PROCESSING |
9 |
98 |
1.35e-06 |
0.0001064 |
| 60 |
NCRNA PROCESSING |
17 |
386 |
1.407e-06 |
0.0001091 |
| 61 |
SEXUAL REPRODUCTION |
24 |
730 |
1.831e-06 |
0.0001397 |
| 62 |
REGULATION OF TRANSFERASE ACTIVITY |
28 |
946 |
1.887e-06 |
0.0001416 |
| 63 |
REGULATION OF CELLULAR AMIDE METABOLIC PROCESS |
16 |
354 |
2.075e-06 |
0.0001532 |
| 64 |
CHROMOSOME ORGANIZATION |
29 |
1009 |
2.176e-06 |
0.0001582 |
| 65 |
ORGANELLE FISSION |
19 |
496 |
2.641e-06 |
0.0001862 |
| 66 |
REGULATION OF CHROMOSOME ORGANIZATION |
14 |
278 |
2.64e-06 |
0.0001862 |
| 67 |
PROTEIN TARGETING |
17 |
406 |
2.785e-06 |
0.0001934 |
| 68 |
MODULATION BY VIRUS OF HOST MORPHOLOGY OR PHYSIOLOGY |
6 |
37 |
2.893e-06 |
0.000198 |
| 69 |
REGULATION OF CELLULAR PROTEIN LOCALIZATION |
20 |
552 |
3.316e-06 |
0.0002236 |
| 70 |
POSITIVE REGULATION OF DNA BIOSYNTHETIC PROCESS |
7 |
59 |
3.718e-06 |
0.0002472 |
| 71 |
MULTICELLULAR ORGANISM REPRODUCTION |
24 |
768 |
4.372e-06 |
0.0002865 |
| 72 |
REGULATION OF MRNA SPLICING VIA SPLICEOSOME |
7 |
62 |
5.213e-06 |
0.0003369 |
| 73 |
REPRODUCTION |
33 |
1297 |
5.654e-06 |
0.0003604 |
| 74 |
MULTI ORGANISM REPRODUCTIVE PROCESS |
26 |
891 |
5.848e-06 |
0.0003677 |
| 75 |
REGULATION OF PROTEIN LOCALIZATION |
27 |
950 |
6.185e-06 |
0.0003837 |
| 76 |
POSITIVE REGULATION OF CHROMOSOME ORGANIZATION |
10 |
150 |
6.471e-06 |
0.0003956 |
| 77 |
NEGATIVE REGULATION OF CELL CYCLE |
17 |
433 |
6.547e-06 |
0.0003956 |
| 78 |
POSITIVE REGULATION OF RNA SPLICING |
5 |
25 |
6.833e-06 |
0.0004076 |
| 79 |
NCRNA METABOLIC PROCESS |
19 |
533 |
7.422e-06 |
0.0004372 |
| 80 |
POSITIVE REGULATION OF MRNA METABOLIC PROCESS |
6 |
45 |
9.42e-06 |
0.0005345 |
| 81 |
MODIFICATION BY SYMBIONT OF HOST MORPHOLOGY OR PHYSIOLOGY |
6 |
45 |
9.42e-06 |
0.0005345 |
| 82 |
REGULATION OF DNA BIOSYNTHETIC PROCESS |
8 |
94 |
9.33e-06 |
0.0005345 |
| 83 |
RESPONSE TO ORGANIC CYCLIC COMPOUND |
26 |
917 |
9.716e-06 |
0.0005447 |
| 84 |
CELL CYCLE CHECKPOINT |
11 |
194 |
1.052e-05 |
0.0005827 |
| 85 |
POSITIVE REGULATION OF CELLULAR PROTEIN LOCALIZATION |
15 |
360 |
1.154e-05 |
0.0006317 |
| 86 |
POSITIVE REGULATION OF TELOMERASE ACTIVITY |
5 |
28 |
1.23e-05 |
0.0006658 |
| 87 |
POSITIVE REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS |
9 |
129 |
1.31e-05 |
0.0007005 |
| 88 |
NUCLEOBASE CONTAINING COMPOUND TRANSPORT |
11 |
199 |
1.336e-05 |
0.0007067 |
| 89 |
MRNA 3 END PROCESSING |
7 |
72 |
1.423e-05 |
0.0007438 |
| 90 |
NEGATIVE REGULATION OF CELLULAR AMIDE METABOLIC PROCESS |
9 |
131 |
1.483e-05 |
0.0007581 |
| 91 |
RNA POLYADENYLATION |
5 |
29 |
1.474e-05 |
0.0007581 |
| 92 |
DNA TEMPLATED TRANSCRIPTION TERMINATION |
8 |
101 |
1.586e-05 |
0.0008021 |
| 93 |
PROTEIN PHOSPHORYLATION |
26 |
944 |
1.609e-05 |
0.0008048 |
| 94 |
CELLULAR MACROMOLECULAR COMPLEX ASSEMBLY |
22 |
727 |
1.84e-05 |
0.0009109 |
| 95 |
CELLULAR RESPONSE TO STRESS |
36 |
1565 |
1.875e-05 |
0.0009182 |
| 96 |
RNA STABILIZATION |
5 |
31 |
2.071e-05 |
0.001004 |
| 97 |
POSITIVE REGULATION OF MRNA PROCESSING |
5 |
32 |
2.433e-05 |
0.001167 |
| 98 |
CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND |
23 |
799 |
2.59e-05 |
0.001226 |
| 99 |
NUCLEIC ACID PHOSPHODIESTER BOND HYDROLYSIS |
12 |
254 |
2.609e-05 |
0.001226 |
| 100 |
DNA INTEGRITY CHECKPOINT |
9 |
146 |
3.515e-05 |
0.001635 |
| 101 |
PEPTIDYL SERINE MODIFICATION |
9 |
148 |
3.912e-05 |
0.001802 |
| 102 |
CELLULAR RESPONSE TO HORMONE STIMULUS |
18 |
552 |
4.21e-05 |
0.00192 |
| 103 |
CELLULAR RESPONSE TO LIPID |
16 |
457 |
4.932e-05 |
0.002228 |
| 104 |
NEGATIVE REGULATION OF PROTEIN MODIFICATION PROCESS |
19 |
616 |
5.431e-05 |
0.00243 |
| 105 |
CELLULAR COMPONENT DISASSEMBLY |
17 |
515 |
5.944e-05 |
0.002634 |
| 106 |
REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL |
12 |
280 |
6.71e-05 |
0.002946 |
| 107 |
REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY |
16 |
470 |
6.864e-05 |
0.002985 |
| 108 |
ANDROGEN RECEPTOR SIGNALING PATHWAY |
5 |
41 |
8.353e-05 |
0.003599 |
| 109 |
DNA REPAIR |
16 |
480 |
8.776e-05 |
0.003746 |
| 110 |
CELLULAR MACROMOLECULE LOCALIZATION |
29 |
1234 |
9.216e-05 |
0.003898 |
| 111 |
REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY |
7 |
97 |
9.838e-05 |
0.004124 |
| 112 |
INTRACELLULAR RECEPTOR SIGNALING PATHWAY |
9 |
168 |
0.0001044 |
0.004338 |
| 113 |
REGULATION OF TELOMERASE ACTIVITY |
5 |
43 |
0.0001054 |
0.00434 |
| 114 |
GAMETE GENERATION |
18 |
595 |
0.0001093 |
0.004463 |
| 115 |
MODIFICATION OF MORPHOLOGY OR PHYSIOLOGY OF OTHER ORGANISM |
7 |
100 |
0.0001192 |
0.004824 |
| 116 |
CELLULAR RESPONSE TO ORGANIC SUBSTANCE |
38 |
1848 |
0.0001207 |
0.00484 |
| 117 |
CELLULAR CATABOLIC PROCESS |
30 |
1322 |
0.0001286 |
0.005071 |
| 118 |
CELLULAR RESPONSE TO ENDOGENOUS STIMULUS |
25 |
1008 |
0.0001275 |
0.005071 |
| 119 |
NUCLEUS ORGANIZATION |
8 |
136 |
0.000133 |
0.0052 |
| 120 |
NEGATIVE REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL |
8 |
139 |
0.0001547 |
0.00595 |
| 121 |
TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER |
20 |
724 |
0.0001547 |
0.00595 |
| 122 |
REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS |
10 |
218 |
0.0001569 |
0.005985 |
| 123 |
MEMBRANE DISASSEMBLY |
5 |
47 |
0.0001621 |
0.006081 |
| 124 |
NUCLEAR ENVELOPE DISASSEMBLY |
5 |
47 |
0.0001621 |
0.006081 |
| 125 |
MRNA SPLICE SITE SELECTION |
4 |
26 |
0.0001761 |
0.006452 |
| 126 |
REGULATION OF CELLULAR SENESCENCE |
4 |
26 |
0.0001761 |
0.006452 |
| 127 |
REGULATION OF PROTEIN STABILITY |
10 |
221 |
0.0001754 |
0.006452 |
| 128 |
REGULATION OF CELLULAR RESPONSE TO HEAT |
6 |
76 |
0.0001905 |
0.006924 |
| 129 |
DNA CATABOLIC PROCESS |
4 |
27 |
0.000205 |
0.007393 |
| 130 |
POSITIVE REGULATION OF DNA METABOLIC PROCESS |
9 |
185 |
0.0002161 |
0.007675 |
| 131 |
PROTEASOMAL PROTEIN CATABOLIC PROCESS |
11 |
271 |
0.0002164 |
0.007675 |
| 132 |
RNA CATABOLIC PROCESS |
10 |
227 |
0.0002177 |
0.007675 |
| 133 |
MICROTUBULE BASED PROCESS |
16 |
522 |
0.0002285 |
0.007933 |
| 134 |
NUCLEAR EXPORT |
8 |
147 |
0.0002271 |
0.007933 |
| 135 |
DNA CONFORMATION CHANGE |
11 |
273 |
0.0002306 |
0.007947 |
| 136 |
REGULATION OF NERVOUS SYSTEM DEVELOPMENT |
20 |
750 |
0.0002458 |
0.00841 |
| 137 |
NEGATIVE REGULATION OF PHOSPHORYLATION |
14 |
422 |
0.0002548 |
0.008655 |
| 138 |
PROTEIN IMPORT INTO NUCLEUS TRANSLOCATION |
4 |
29 |
0.0002727 |
0.009193 |
| 139 |
NEGATIVE REGULATION OF NITROGEN COMPOUND METABOLIC PROCESS |
32 |
1517 |
0.000284 |
0.009439 |
| 140 |
RESPONSE TO ENDOGENOUS STIMULUS |
31 |
1450 |
0.0002831 |
0.009439 |
| 141 |
POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER |
24 |
1004 |
0.0003015 |
0.00995 |