This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-let-7b-5p | DENND4B | 0.78 | 0.88055 | -0.12 | 0.95409 | miRNAWalker2 validate | -0.12 | 0.03676 | ||
| 2 | hsa-let-7b-5p | PVR | 0.78 | 0.88055 | -0.37 | 0.86991 | miRNAWalker2 validate | -0.31 | 0.00012 | ||
| 3 | hsa-let-7b-5p | SCML2 | 0.78 | 0.88055 | -1.56 | 0.00352 | miRNAWalker2 validate | -0.64 | 2.0E-5 | ||
| 4 | hsa-let-7b-5p | SLC35F1 | 0.78 | 0.88055 | -1.39 | 0.00581 | miRNAWalker2 validate | -0.48 | 0.00235 | ||
| 5 | hsa-let-7e-5p | PYCR1 | -0.26 | 0.9437 | 0.91 | 0.67259 | miRNAWalker2 validate | -0.19 | 0.01919 | ||
| 6 | hsa-let-7e-5p | SLCO4A1 | -0.26 | 0.9437 | 0.42 | 0.80102 | miRNAWalker2 validate | -0.26 | 0.03962 | ||
| 7 | hsa-miR-101-3p | CAPN2 | 0.13 | 0.97919 | 0.39 | 0.90197 | miRNAWalker2 validate; MirTarget | -0.31 | 2.0E-5 | ||
| 8 | hsa-miR-10a-5p | ORAI2 | 0.68 | 0.90388 | -0.47 | 0.81719 | miRNAWalker2 validate | -0.17 | 0.00201 | ||
| 9 | hsa-miR-10b-5p | SDC1 | 0.49 | 0.92285 | 1.28 | 0.68069 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.28 | 0.0237 | 22573479; 22573479; 22573479 | Targeting of syndecan-1 by microRNA miR-10b promotes breast cancer cell motility and invasiveness via a Rho-GTPase- and E-cadherin-dependent mechanism;In breast cancer overexpression of the transmembrane heparan sulfate proteoglycan syndecan-1 a predicted target of the oncomiR miR-10b correlates with poor clinical outcome;miR-10b overexpression induced post-transcriptional downregulation of syndecan-1 as demonstrated by quantitative real-time PCR qPCR flow cytometry and 3'UTR luciferase assays resulting in increased cancer cell migration and matrigel invasiveness |
| 10 | hsa-miR-125a-5p | CLDN12 | -0.16 | 0.96341 | 0.18 | 0.9345 | miRNAWalker2 validate | -0.19 | 0.02444 | ||
| 11 | hsa-miR-125a-5p | ERBB3 | -0.16 | 0.96341 | 0.89 | 0.74708 | miRNAWalker2 validate; miRanda | -0.38 | 0.01284 | 23519125 | Indeed entinostat significantly upregulated miR-125a miR-125b and miR-205 that have been reported to target erbB2 and/or erbB3 |
| 12 | hsa-miR-125a-5p | FERMT1 | -0.16 | 0.96341 | 1.51 | 0.48743 | miRNAWalker2 validate | -0.99 | 0 | ||
| 13 | hsa-miR-125a-5p | PLA2G4F | -0.16 | 0.96341 | 1.78 | 0.03327 | miRNAWalker2 validate | -1.08 | 0 | ||
| 14 | hsa-miR-125a-5p | ZDHHC9 | -0.16 | 0.96341 | 0.66 | 0.76379 | miRNAWalker2 validate | -0.45 | 0 | ||
| 15 | hsa-miR-128-3p | AZGP1 | -0.47 | 0.84408 | 1.21 | 0.58016 | miRNAWalker2 validate | -0.96 | 3.0E-5 | ||
| 16 | hsa-miR-128-3p | CD300A | -0.47 | 0.84408 | 0.33 | 0.82001 | miRNAWalker2 validate | -0.26 | 0.01716 | ||
| 17 | hsa-miR-128-3p | EPB41L1 | -0.47 | 0.84408 | 0.54 | 0.83266 | miRNAWalker2 validate | -0.16 | 0.04487 | ||
| 18 | hsa-miR-128-3p | HMOX1 | -0.47 | 0.84408 | 0.69 | 0.71708 | miRNAWalker2 validate | -0.26 | 0.01304 | ||
| 19 | hsa-miR-128-3p | HTRA4 | -0.47 | 0.84408 | 1.17 | 0.1189 | miRNAWalker2 validate | -0.7 | 0.0001 | ||
| 20 | hsa-miR-128-3p | KLF5 | -0.47 | 0.84408 | 1.42 | 0.6032 | miRNAWalker2 validate | -1.03 | 0 | ||
| 21 | hsa-miR-128-3p | RASSF6 | -0.47 | 0.84408 | 0.79 | 0.62567 | miRNAWalker2 validate | -0.31 | 0.01773 | ||
| 22 | hsa-miR-128-3p | SLC7A11 | -0.47 | 0.84408 | 0.3 | 0.83928 | miRNAWalker2 validate | -0.75 | 0 | ||
| 23 | hsa-miR-129-5p | HIST1H2BJ | -1.35 | 0.49148 | 1.2 | 0.0475 | miRNAWalker2 validate | -0.16 | 0.00077 | ||
| 24 | hsa-miR-130b-3p | HOXC8 | -0.13 | 0.92172 | 2.34 | 0.00215 | miRNAWalker2 validate | -0.44 | 0.00878 | ||
| 25 | hsa-miR-132-3p | ARHGAP32 | -0.64 | 0.81464 | 0.43 | 0.84674 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.34 | 0 | ||
| 26 | hsa-miR-132-3p | CHRNA5 | -0.64 | 0.81464 | -0.8 | 0.42367 | miRNAWalker2 validate | -0.26 | 0.03118 | ||
| 27 | hsa-miR-132-3p | ECT2 | -0.64 | 0.81464 | 0.68 | 0.71315 | miRNAWalker2 validate | -0.45 | 0 | ||
| 28 | hsa-miR-132-3p | MUC13 | -0.64 | 0.81464 | 0.62 | 0.81719 | miRNAWalker2 validate; MirTarget | -0.69 | 0.00081 | ||
| 29 | hsa-miR-132-3p | RNF128 | -0.64 | 0.81464 | 0.68 | 0.75648 | miRNAWalker2 validate | -0.46 | 0.00041 | ||
| 30 | hsa-miR-142-3p | CNIH4 | 0.7 | 0.85325 | 0.1 | 0.9591 | miRNAWalker2 validate; miRanda | -0.07 | 0.01027 | ||
| 31 | hsa-miR-142-3p | FAM177A1 | 0.7 | 0.85325 | 0.23 | 0.91217 | miRNAWalker2 validate; miRanda | -0.13 | 0 | ||
| 32 | hsa-miR-142-3p | NCKAP1 | 0.7 | 0.85325 | 0.3 | 0.91282 | miRNAWalker2 validate; miRanda | -0.11 | 0.00032 | ||
| 33 | hsa-miR-142-3p | PPP2R2C | 0.7 | 0.85325 | -1.49 | 0.22611 | miRNAWalker2 validate | -0.36 | 0.00407 | ||
| 34 | hsa-miR-148a-3p | HOXC8 | 0.06 | 0.99183 | 2.34 | 0.00215 | miRNAWalker2 validate; miRNATAP | -0.38 | 0.00341 | ||
| 35 | hsa-miR-148b-3p | ABCA13 | -0.73 | 0.76331 | 1.84 | 0.02908 | miRNAWalker2 validate | -1.24 | 0 | ||
| 36 | hsa-miR-148b-3p | CD300A | -0.73 | 0.76331 | 0.33 | 0.82001 | miRNAWalker2 validate | -0.48 | 1.0E-5 | ||
| 37 | hsa-miR-148b-3p | DNAH3 | -0.73 | 0.76331 | 1.54 | 0.04542 | miRNAWalker2 validate | -0.52 | 0.01295 | ||
| 38 | hsa-miR-148b-3p | HMOX1 | -0.73 | 0.76331 | 0.69 | 0.71708 | miRNAWalker2 validate | -0.51 | 0 | ||
| 39 | hsa-miR-148b-3p | TUBAL3 | -0.73 | 0.76331 | 0.39 | 0.72672 | miRNAWalker2 validate | -0.4 | 0.04986 | ||
| 40 | hsa-miR-149-5p | TOX3 | 0.01 | 0.99674 | 0.24 | 0.8787 | miRNAWalker2 validate | -0.48 | 2.0E-5 | ||
| 41 | hsa-miR-155-5p | CPD | 0.52 | 0.85485 | 0 | 0.99977 | miRNAWalker2 validate | -0.14 | 0.00258 | ||
| 42 | hsa-miR-155-5p | FAM177A1 | 0.52 | 0.85485 | 0.23 | 0.91217 | miRNAWalker2 validate | -0.07 | 0.02305 | ||
| 43 | hsa-miR-155-5p | PPL | 0.52 | 0.85485 | -0.12 | 0.96435 | miRNAWalker2 validate | -0.28 | 0.00121 | ||
| 44 | hsa-miR-155-5p | SLC27A2 | 0.52 | 0.85485 | 0.28 | 0.771 | miRNAWalker2 validate | -0.25 | 0.01653 | ||
| 45 | hsa-miR-15a-5p | GOLPH3L | -0.62 | 0.8034 | 0.24 | 0.9064 | miRNAWalker2 validate | -0.22 | 0.00099 | ||
| 46 | hsa-miR-15b-3p | RND2 | 0.46 | 0.73254 | -1.4 | 0.00676 | miRNAWalker2 validate | -0.39 | 0.00043 | ||
| 47 | hsa-miR-16-5p | EPHA2 | -0.14 | 0.96015 | 1.8 | 0.49987 | miRNAWalker2 validate | -0.67 | 0.00029 | ||
| 48 | hsa-miR-16-5p | GFPT1 | -0.14 | 0.96015 | 0.22 | 0.93408 | miRNAWalker2 validate | -0.21 | 0.00864 | ||
| 49 | hsa-miR-16-5p | GOLPH3L | -0.14 | 0.96015 | 0.24 | 0.9064 | miRNAWalker2 validate | -0.14 | 0.04228 | ||
| 50 | hsa-miR-16-5p | LMO7 | -0.14 | 0.96015 | 0.73 | 0.79354 | miRNAWalker2 validate | -0.52 | 2.0E-5 | ||
| 51 | hsa-miR-16-5p | PHLDA2 | -0.14 | 0.96015 | 1.6 | 0.39646 | miRNAWalker2 validate | -0.49 | 0.03165 | ||
| 52 | hsa-miR-16-5p | PTPN3 | -0.14 | 0.96015 | -0.45 | 0.81625 | miRNAWalker2 validate | -0.22 | 0.04315 | ||
| 53 | hsa-miR-16-5p | SERPINB5 | -0.14 | 0.96015 | 2.81 | 0.19467 | miRNAWalker2 validate | -1.59 | 2.0E-5 | ||
| 54 | hsa-miR-16-5p | SLC9A2 | -0.14 | 0.96015 | 2.58 | 0.03655 | miRNAWalker2 validate | -0.85 | 0.00744 | ||
| 55 | hsa-miR-17-5p | BCL2 | 0.34 | 0.90424 | -0.05 | 0.9708 | miRNAWalker2 validate; miRTarBase | -0.29 | 0.02016 | 25435430; 25435430; 25435430 | MiR-16 targets Bcl-2 in paclitaxel-resistant lung cancer cells and overexpression of miR-16 along with miR-17 causes unprecedented sensitivity by simultaneously modulating autophagy and apoptosis;Combined overexpression of miR-16 and miR-17 greatly reduced Beclin-1 and Bcl-2 expressions respectively;miR-17 overexpression reduced cytoprotective autophagy by targeting Beclin-1 whereas overexpression of miR-16 potentiated paclitaxel induced apoptotic cell death by inhibiting anti-apoptotic protein Bcl-2 |
| 56 | hsa-miR-181a-5p | PLA2G4C | 0.71 | 0.86075 | -0.36 | 0.74959 | miRNAWalker2 validate | -0.33 | 0.00061 | ||
| 57 | hsa-miR-181b-5p | DDHD1 | 0.88 | 0.7882 | -0.32 | 0.83008 | miRNAWalker2 validate | -0.12 | 0.01157 | ||
| 58 | hsa-miR-182-5p | BCL2 | -0.2 | 0.96662 | -0.05 | 0.9708 | miRNAWalker2 validate; miRTarBase; mirMAP | -0.4 | 0 | 23936432 | Inhibition of proliferation and induction of autophagy by atorvastatin in PC3 prostate cancer cells correlate with downregulation of Bcl2 and upregulation of miR-182 and p21 |
| 59 | hsa-miR-182-5p | FMNL3 | -0.2 | 0.96662 | 0.28 | 0.89206 | miRNAWalker2 validate | -0.24 | 0 | ||
| 60 | hsa-miR-185-5p | CTTN | -0.28 | 0.89678 | 0.34 | 0.91143 | miRNAWalker2 validate | -0.19 | 0.00299 | ||
| 61 | hsa-miR-186-5p | CGN | -0.18 | 0.94756 | 1.04 | 0.68969 | miRNAWalker2 validate; miRNATAP | -0.77 | 8.0E-5 | ||
| 62 | hsa-miR-186-5p | FAM177A1 | -0.18 | 0.94756 | 0.23 | 0.91217 | miRNAWalker2 validate; MirTarget | -0.22 | 0.00221 | ||
| 63 | hsa-miR-186-5p | HIST1H1C | -0.18 | 0.94756 | 1.21 | 0.53653 | miRNAWalker2 validate | -0.44 | 0.02144 | ||
| 64 | hsa-miR-186-5p | PLS1 | -0.18 | 0.94756 | 1.02 | 0.66167 | miRNAWalker2 validate; miRNATAP | -0.74 | 0.00014 | ||
| 65 | hsa-miR-186-5p | PPL | -0.18 | 0.94756 | -0.12 | 0.96435 | miRNAWalker2 validate | -0.71 | 0.00026 | ||
| 66 | hsa-miR-192-5p | ANKRD13A | 0.42 | 0.92883 | -0.18 | 0.93034 | miRNAWalker2 validate | -0.09 | 0.00013 | ||
| 67 | hsa-miR-192-5p | BCL2 | 0.42 | 0.92883 | -0.05 | 0.9708 | miRNAWalker2 validate | -0.31 | 0 | 24213572; 24213572; 25566965 | Further studies indicate that miR-192 downregulates expression of Bcl-2 Zeb2 and VEGFA in vitro and in vivo which is responsible for enhanced apoptosis increased expression of E-cadherin and decreased angiogenesis in vivo respectively;Therefore a major implication of our studies is that restoration of miR-192 expression or antagonism of its target genes Bcl-2 Zeb2 or VEGFA may have considerable therapeutic potential for anti-metastatic therapy in patients with colon cancer;Moreover overexpression of miR-192 significantly induced apoptotic death in bladder cancer cells increased the levels of p21 p27 and Bax and decreased the levels of cyclin D1 Bcl-2 and Mcl-1 |
| 68 | hsa-miR-192-5p | BMP2K | 0.42 | 0.92883 | 0.38 | 0.76933 | miRNAWalker2 validate | -0.11 | 0.00682 | ||
| 69 | hsa-miR-192-5p | CD83 | 0.42 | 0.92883 | 0.64 | 0.65493 | miRNAWalker2 validate | -0.28 | 0 | ||
| 70 | hsa-miR-192-5p | FNBP1 | 0.42 | 0.92883 | -0.01 | 0.99641 | miRNAWalker2 validate | -0.26 | 0 | ||
| 71 | hsa-miR-192-5p | HSPBAP1 | 0.42 | 0.92883 | 0.15 | 0.89776 | miRNAWalker2 validate | -0.1 | 1.0E-5 | ||
| 72 | hsa-miR-192-5p | LIPA | 0.42 | 0.92883 | 0.07 | 0.97853 | miRNAWalker2 validate | -0.08 | 0.02358 | ||
| 73 | hsa-miR-192-5p | NT5M | 0.42 | 0.92883 | -1.19 | 0.00449 | miRNAWalker2 validate | -0.17 | 0.00062 | ||
| 74 | hsa-miR-192-5p | SH2B3 | 0.42 | 0.92883 | 0.03 | 0.9889 | miRNAWalker2 validate | -0.23 | 0 | ||
| 75 | hsa-miR-192-5p | SLC25A30 | 0.42 | 0.92883 | -0.05 | 0.97228 | miRNAWalker2 validate | -0.1 | 0.00075 | ||
| 76 | hsa-miR-192-5p | SNX10 | 0.42 | 0.92883 | 0.08 | 0.96005 | miRNAWalker2 validate | -0.21 | 4.0E-5 | ||
| 77 | hsa-miR-192-5p | TRAF3IP3 | 0.42 | 0.92883 | 0.55 | 0.60507 | miRNAWalker2 validate | -0.42 | 0 | ||
| 78 | hsa-miR-192-5p | VASH2 | 0.42 | 0.92883 | 0.55 | 0.41186 | miRNAWalker2 validate | -0.25 | 1.0E-5 | ||
| 79 | hsa-miR-193b-3p | RAPGEF1 | 0.9 | 0.65512 | -0.11 | 0.96508 | miRNAWalker2 validate | -0.07 | 0.02653 | ||
| 80 | hsa-miR-196a-5p | ARHGAP28 | 2.52 | 0.15332 | 0.7 | 0.38581 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.16 | 4.0E-5 | ||
| 81 | hsa-miR-196a-5p | ATP6V1B2 | 2.52 | 0.15332 | -0.02 | 0.99471 | miRNAWalker2 validate | -0.08 | 0 | ||
| 82 | hsa-miR-196a-5p | HMOX1 | 2.52 | 0.15332 | 0.69 | 0.71708 | miRNAWalker2 validate | -0.07 | 0.02366 | ||
| 83 | hsa-miR-196a-5p | SCN11A | 2.52 | 0.15332 | -0.46 | 0.62422 | miRNAWalker2 validate | -0.2 | 0 | ||
| 84 | hsa-miR-196b-5p | BCL2 | 2.33 | 0.29399 | -0.05 | 0.9708 | miRNAWalker2 validate | -0.13 | 0.00097 | ||
| 85 | hsa-miR-197-3p | AGR2 | 0.2 | 0.94188 | 3.02 | 0.34586 | miRNAWalker2 validate | -0.99 | 0.00474 | ||
| 86 | hsa-miR-197-3p | HNF4A | 0.2 | 0.94188 | 0.48 | 0.79622 | miRNAWalker2 validate | -0.71 | 0.00133 | ||
| 87 | hsa-miR-19a-3p | BCL2L11 | -0.51 | 0.73956 | -0.05 | 0.979 | miRNAWalker2 validate; miRTarBase | -0.08 | 0.02031 | ||
| 88 | hsa-miR-19b-3p | BCL2L11 | -0.24 | 0.91761 | -0.05 | 0.979 | miRNAWalker2 validate; miRTarBase | -0.11 | 0.00587 | ||
| 89 | hsa-miR-19b-3p | CPD | -0.24 | 0.91761 | 0 | 0.99977 | miRNAWalker2 validate; miRNATAP | -0.22 | 0.00019 | ||
| 90 | hsa-miR-19b-3p | PSAP | -0.24 | 0.91761 | 0.05 | 0.98976 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.13 | 0.00443 | ||
| 91 | hsa-miR-19b-3p | STEAP2 | -0.24 | 0.91761 | 0.19 | 0.92446 | miRNAWalker2 validate; MirTarget | -0.17 | 0.01283 | ||
| 92 | hsa-miR-200b-3p | BCL2 | 0.03 | 0.99376 | -0.05 | 0.9708 | miRNAWalker2 validate; miRTarBase; TargetScan; mirMAP | -0.46 | 0 | ||
| 93 | hsa-miR-200c-3p | BCL2 | -0.17 | 0.96964 | -0.05 | 0.9708 | miRNAWalker2 validate; miRTarBase; mirMAP | -0.4 | 0 | ||
| 94 | hsa-miR-200c-3p | FHOD1 | -0.17 | 0.96964 | 0.22 | 0.90156 | miRNAWalker2 validate | -0.15 | 0 | 22144583; 22144583; 22144583 | MicroRNA-200c represses migration and invasion of breast cancer cells by targeting actin-regulatory proteins FHOD1 and PPM1F;Remarkably expression levels of FHOD1 and PPM1F were inversely correlated with the level of miR-200c in breast cancer cell lines breast cancer patient samples and 58 cancer cell lines of various origins;Mechanistically targeting of FHOD1 by miR-200c resulted in decreased expression and transcriptional activity of serum response factor SRF mediated by interference with the translocation of the SRF coactivator mycocardin-related transcription factor A MRTF-A |
| 95 | hsa-miR-200c-3p | PPM1F | -0.17 | 0.96964 | -0.06 | 0.97696 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.22 | 0 | 22144583; 22144583 | MicroRNA-200c represses migration and invasion of breast cancer cells by targeting actin-regulatory proteins FHOD1 and PPM1F;Remarkably expression levels of FHOD1 and PPM1F were inversely correlated with the level of miR-200c in breast cancer cell lines breast cancer patient samples and 58 cancer cell lines of various origins |
| 96 | hsa-miR-204-5p | AKAP1 | -1.1 | 0.45392 | -0.05 | 0.98191 | miRNAWalker2 validate; miRNATAP | -0.09 | 0.00148 | ||
| 97 | hsa-miR-204-5p | ELOVL6 | -1.1 | 0.45392 | 0.45 | 0.76863 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.11 | 0.00645 | ||
| 98 | hsa-miR-204-5p | SPDEF | -1.1 | 0.45392 | 1.98 | 0.2557 | miRNAWalker2 validate | -0.48 | 0 | ||
| 99 | hsa-miR-20a-5p | BCL2 | 0.29 | 0.91626 | -0.05 | 0.9708 | miRNAWalker2 validate; miRTarBase | -0.29 | 0.01076 | ||
| 100 | hsa-miR-21-5p | DOCK10 | 1.04 | 0.87764 | -0.33 | 0.82898 | miRNAWalker2 validate | -0.23 | 0.02734 | ||
| 101 | hsa-miR-21-5p | PTPN3 | 1.04 | 0.87764 | -0.45 | 0.81625 | miRNAWalker2 validate | -0.19 | 0.00465 | ||
| 102 | hsa-miR-215-5p | BCL2 | 1.07 | 0.65842 | -0.05 | 0.9708 | miRNAWalker2 validate | -0.11 | 0.00607 | ||
| 103 | hsa-miR-215-5p | CD83 | 1.07 | 0.65842 | 0.64 | 0.65493 | miRNAWalker2 validate | -0.12 | 0.00447 | ||
| 104 | hsa-miR-215-5p | HSPBAP1 | 1.07 | 0.65842 | 0.15 | 0.89776 | miRNAWalker2 validate | -0.06 | 0.00022 | ||
| 105 | hsa-miR-215-5p | NT5M | 1.07 | 0.65842 | -1.19 | 0.00449 | miRNAWalker2 validate | -0.17 | 0 | ||
| 106 | hsa-miR-215-5p | SH2B3 | 1.07 | 0.65842 | 0.03 | 0.9889 | miRNAWalker2 validate | -0.11 | 1.0E-5 | ||
| 107 | hsa-miR-218-5p | EFNA1 | -0.19 | 0.92936 | 0.24 | 0.92512 | miRNAWalker2 validate | -0.29 | 0.00125 | ||
| 108 | hsa-miR-218-5p | TOB1 | -0.19 | 0.92936 | 0.49 | 0.84261 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.17 | 0.01066 | ||
| 109 | hsa-miR-221-3p | EVL | 0.22 | 0.94091 | -0.45 | 0.83273 | miRNAWalker2 validate | -0.24 | 0.00023 | ||
| 110 | hsa-miR-222-3p | AP1B1 | 0.23 | 0.91635 | -0.22 | 0.93279 | miRNAWalker2 validate | -0.08 | 0.00624 | ||
| 111 | hsa-miR-24-3p | CORO1A | 0.07 | 0.98634 | 0.18 | 0.93433 | miRNAWalker2 validate | -0.47 | 0.00374 | ||
| 112 | hsa-miR-24-3p | GGA2 | 0.07 | 0.98634 | -0.28 | 0.90519 | miRNAWalker2 validate | -0.25 | 0.00012 | ||
| 113 | hsa-miR-24-3p | PPM1F | 0.07 | 0.98634 | -0.06 | 0.97696 | miRNAWalker2 validate | -0.29 | 1.0E-5 | ||
| 114 | hsa-miR-24-3p | SCML2 | 0.07 | 0.98634 | -1.56 | 0.00352 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.68 | 1.0E-5 | ||
| 115 | hsa-miR-25-3p | PLEKHA5 | 0.02 | 0.99673 | 0.37 | 0.84945 | miRNAWalker2 validate | -0.39 | 0.00011 | ||
| 116 | hsa-miR-26a-5p | CDC6 | -0.16 | 0.96904 | 0.46 | 0.71504 | miRNAWalker2 validate | -0.43 | 0.00486 | ||
| 117 | hsa-miR-26b-5p | ECT2 | -0.2 | 0.95056 | 0.68 | 0.71315 | miRNAWalker2 validate | -0.28 | 0.00626 | ||
| 118 | hsa-miR-27a-3p | TMEM170B | 0.31 | 0.93601 | -0.51 | 0.59649 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.57 | 0 | ||
| 119 | hsa-miR-296-3p | RAPGEFL1 | -0.26 | 0.63073 | 0.65 | 0.75783 | miRNAWalker2 validate | -0.33 | 0.00312 | ||
| 120 | hsa-miR-29b-3p | BCL2 | -0.59 | 0.86279 | -0.05 | 0.9708 | miRNAWalker2 validate; miRTarBase | -0.21 | 0.02912 | ||
| 121 | hsa-miR-301a-3p | KDELR2 | -0.99 | 0.24262 | 0.32 | 0.91672 | miRNAWalker2 validate | -0.18 | 1.0E-5 | ||
| 122 | hsa-miR-30a-5p | ATP6V1B2 | 0.38 | 0.94113 | -0.02 | 0.99471 | miRNAWalker2 validate; MirTarget | -0.1 | 0.02939 | ||
| 123 | hsa-miR-30a-5p | DNAJA4 | 0.38 | 0.94113 | -0.09 | 0.96442 | miRNAWalker2 validate | -0.19 | 0.00501 | ||
| 124 | hsa-miR-30a-5p | UAP1 | 0.38 | 0.94113 | 0.69 | 0.75682 | miRNAWalker2 validate | -0.11 | 0.03577 | ||
| 125 | hsa-miR-30b-5p | PIP4K2A | -0.72 | 0.81119 | 0.4 | 0.83929 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.25 | 8.0E-5 | ||
| 126 | hsa-miR-30b-5p | SH2B3 | -0.72 | 0.81119 | 0.03 | 0.9889 | miRNAWalker2 validate; MirTarget | -0.15 | 0.01662 | ||
| 127 | hsa-miR-30c-5p | ECT2 | -0.39 | 0.90259 | 0.68 | 0.71315 | miRNAWalker2 validate | -0.51 | 0 | ||
| 128 | hsa-miR-30d-5p | ADAP2 | -0.44 | 0.92148 | 0.21 | 0.86301 | miRNAWalker2 validate | -0.37 | 0.00039 | ||
| 129 | hsa-miR-324-3p | HIST1H1C | -0.4 | 0.75296 | 1.21 | 0.53653 | miRNAWalker2 validate | -0.47 | 0.00018 | ||
| 130 | hsa-miR-324-5p | FAM83H | -0.46 | 0.76026 | 0.27 | 0.91528 | miRNAWalker2 validate | -0.23 | 0.02314 | ||
| 131 | hsa-miR-330-5p | ZDHHC9 | -0.3 | 0.85073 | 0.66 | 0.76379 | miRNAWalker2 validate; PITA; miRanda; miRNATAP | -0.16 | 0.0069 | ||
| 132 | hsa-miR-331-3p | PITX1 | 0.15 | 0.91479 | 1.66 | 0.34783 | miRNAWalker2 validate | -0.59 | 0.04171 | ||
| 133 | hsa-miR-335-5p | ABCA12 | -0.51 | 0.71837 | 1.35 | 0.18118 | miRNAWalker2 validate | -0.66 | 0.0005 | ||
| 134 | hsa-miR-335-5p | AGR2 | -0.51 | 0.71837 | 3.02 | 0.34586 | miRNAWalker2 validate | -0.56 | 0.00578 | ||
| 135 | hsa-miR-335-5p | AGR3 | -0.51 | 0.71837 | 3.18 | 0.09122 | miRNAWalker2 validate | -0.4 | 0.03706 | ||
| 136 | hsa-miR-335-5p | ANO1 | -0.51 | 0.71837 | 0.74 | 0.77937 | miRNAWalker2 validate | -0.26 | 0.00472 | ||
| 137 | hsa-miR-335-5p | AQP5 | -0.51 | 0.71837 | 2.63 | 0.21089 | miRNAWalker2 validate | -0.57 | 0.01636 | ||
| 138 | hsa-miR-335-5p | AREG | -0.51 | 0.71837 | 1.31 | 0.41326 | miRNAWalker2 validate | -0.44 | 0.00143 | ||
| 139 | hsa-miR-335-5p | ATP6V0D2 | -0.51 | 0.71837 | 1.49 | 0.0092 | miRNAWalker2 validate | -0.38 | 0.00052 | ||
| 140 | hsa-miR-335-5p | B3GNT3 | -0.51 | 0.71837 | 1.15 | 0.63224 | miRNAWalker2 validate | -0.3 | 0.02061 | ||
| 141 | hsa-miR-335-5p | C1QA | -0.51 | 0.71837 | 0.32 | 0.90418 | miRNAWalker2 validate | -0.18 | 0.0435 | ||
| 142 | hsa-miR-335-5p | CD33 | -0.51 | 0.71837 | 0.62 | 0.42975 | miRNAWalker2 validate | -0.2 | 0.03391 | ||
| 143 | hsa-miR-335-5p | CDCP1 | -0.51 | 0.71837 | 0.14 | 0.954 | miRNAWalker2 validate | -0.25 | 0.0019 | ||
| 144 | hsa-miR-335-5p | CDHR2 | -0.51 | 0.71837 | 0.95 | 0.6251 | miRNAWalker2 validate | -0.48 | 0.02006 | ||
| 145 | hsa-miR-335-5p | CEACAM3 | -0.51 | 0.71837 | 0.2 | 0.82999 | miRNAWalker2 validate | -0.25 | 0.03895 | ||
| 146 | hsa-miR-335-5p | DUOXA1 | -0.51 | 0.71837 | 1.72 | 0.05661 | miRNAWalker2 validate | -0.37 | 0.01675 | ||
| 147 | hsa-miR-335-5p | EREG | -0.51 | 0.71837 | 1.95 | 0.1318 | miRNAWalker2 validate | -0.89 | 3.0E-5 | ||
| 148 | hsa-miR-335-5p | FAM49A | -0.51 | 0.71837 | 0.91 | 0.34061 | miRNAWalker2 validate | -0.33 | 0.0004 | ||
| 149 | hsa-miR-335-5p | GALNT5 | -0.51 | 0.71837 | 3.05 | 0.10125 | miRNAWalker2 validate | -0.7 | 0.00014 | ||
| 150 | hsa-miR-335-5p | GCNT3 | -0.51 | 0.71837 | 1.71 | 0.50856 | miRNAWalker2 validate | -0.36 | 0.02683 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | IMMUNE SYSTEM PROCESS | 125 | 1984 | 6.283e-12 | 1.462e-08 |
| 2 | BIOLOGICAL ADHESION | 80 | 1032 | 4.353e-12 | 1.462e-08 |
| 3 | REGULATION OF CELL ADHESION | 57 | 629 | 1.793e-11 | 2.782e-08 |
| 4 | REGULATION OF IMMUNE RESPONSE | 68 | 858 | 7.829e-11 | 9.107e-08 |
| 5 | REGULATION OF IMMUNE SYSTEM PROCESS | 93 | 1403 | 3.837e-10 | 3.57e-07 |
| 6 | IMMUNE RESPONSE | 76 | 1100 | 3.138e-09 | 2.434e-06 |
| 7 | REGULATION OF CELL ACTIVATION | 43 | 484 | 1.066e-08 | 7.021e-06 |
| 8 | POSITIVE REGULATION OF IMMUNE SYSTEM PROCESS | 63 | 867 | 1.207e-08 | 7.021e-06 |
| 9 | TISSUE DEVELOPMENT | 93 | 1518 | 1.974e-08 | 1.021e-05 |
| 10 | ACTIN FILAMENT BASED PROCESS | 40 | 450 | 3.456e-08 | 1.608e-05 |
| 11 | REGULATION OF CELL CELL ADHESION | 35 | 380 | 1.057e-07 | 4.368e-05 |
| 12 | POSITIVE REGULATION OF IMMUNE RESPONSE | 45 | 563 | 1.126e-07 | 4.368e-05 |
| 13 | POSITIVE REGULATION OF CELL ADHESION | 34 | 376 | 2.486e-07 | 8.896e-05 |
| 14 | LEUKOCYTE ACTIVATION | 36 | 414 | 2.885e-07 | 8.95e-05 |
| 15 | EPITHELIUM DEVELOPMENT | 63 | 945 | 2.758e-07 | 8.95e-05 |
| 16 | DEFENSE RESPONSE | 76 | 1231 | 3.35e-07 | 9.741e-05 |
| 17 | CELL CELL ADHESION | 46 | 608 | 3.959e-07 | 0.0001083 |
| 18 | SINGLE ORGANISM CELL ADHESION | 38 | 459 | 4.664e-07 | 0.0001206 |
| 19 | REGULATION OF LEUKOCYTE PROLIFERATION | 23 | 206 | 6.151e-07 | 0.0001506 |
| 20 | POSITIVE REGULATION OF CELL CELL ADHESION | 25 | 243 | 9.457e-07 | 0.00022 |
| 21 | CELL MOTILITY | 56 | 835 | 1.088e-06 | 0.0002201 |
| 22 | LOCALIZATION OF CELL | 56 | 835 | 1.088e-06 | 0.0002201 |
| 23 | LOCOMOTION | 69 | 1114 | 1.086e-06 | 0.0002201 |
| 24 | EPITHELIAL CELL DIFFERENTIATION | 39 | 495 | 1.142e-06 | 0.0002214 |
| 25 | REGULATION OF IMMUNE EFFECTOR PROCESS | 35 | 424 | 1.416e-06 | 0.0002636 |
| 26 | REGULATION OF LEUKOCYTE MEDIATED IMMUNITY | 19 | 156 | 1.568e-06 | 0.0002703 |
| 27 | POSITIVE REGULATION OF IMMUNE EFFECTOR PROCESS | 19 | 156 | 1.568e-06 | 0.0002703 |
| 28 | ACTIN FILAMENT ORGANIZATION | 20 | 174 | 2.095e-06 | 0.0003482 |
| 29 | CELL ACTIVATION | 42 | 568 | 2.293e-06 | 0.0003608 |
| 30 | LYMPHOCYTE ACTIVATION | 30 | 342 | 2.358e-06 | 0.0003608 |
| 31 | REGULATION OF HOMOTYPIC CELL CELL ADHESION | 28 | 307 | 2.404e-06 | 0.0003608 |
| 32 | REGULATION OF LYMPHOCYTE DIFFERENTIATION | 17 | 132 | 2.55e-06 | 0.0003708 |
| 33 | LEUKOCYTE MIGRATION | 25 | 259 | 3.03e-06 | 0.0004229 |
| 34 | POSITIVE REGULATION OF CELL ACTIVATION | 28 | 311 | 3.09e-06 | 0.0004229 |
| 35 | REGULATION OF T CELL DIFFERENTIATION | 15 | 107 | 3.399e-06 | 0.0004519 |
| 36 | REGULATION OF MICROVILLUS ORGANIZATION | 6 | 14 | 3.624e-06 | 0.0004669 |
| 37 | REGULATION OF CELL PROLIFERATION | 84 | 1496 | 3.713e-06 | 0.0004669 |
| 38 | REGULATION OF LEUKOCYTE DIFFERENTIATION | 23 | 232 | 4.789e-06 | 0.0005864 |
| 39 | CELLULAR DEFENSE RESPONSE | 11 | 60 | 4.933e-06 | 0.0005886 |
| 40 | IMMUNE SYSTEM DEVELOPMENT | 41 | 582 | 9.813e-06 | 0.001141 |
| 41 | REGULATION OF HEMOPOIESIS | 27 | 314 | 1.077e-05 | 0.001222 |
| 42 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 100 | 1929 | 1.281e-05 | 0.001368 |
| 43 | RESPONSE TO XENOBIOTIC STIMULUS | 14 | 105 | 1.293e-05 | 0.001368 |
| 44 | ORGAN MORPHOGENESIS | 53 | 841 | 1.263e-05 | 0.001368 |
| 45 | MOVEMENT OF CELL OR SUBCELLULAR COMPONENT | 72 | 1275 | 1.614e-05 | 0.001669 |
| 46 | NEGATIVE REGULATION OF LEUKOCYTE PROLIFERATION | 11 | 69 | 1.989e-05 | 0.001932 |
| 47 | REGULATION OF CYTOKINE PRODUCTION INVOLVED IN IMMUNE RESPONSE | 10 | 57 | 1.993e-05 | 0.001932 |
| 48 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE SIGNALING PATHWAY | 36 | 498 | 1.973e-05 | 0.001932 |
| 49 | LEUKOCYTE CELL CELL ADHESION | 23 | 255 | 2.248e-05 | 0.002097 |
| 50 | T CELL SELECTION | 8 | 36 | 2.254e-05 | 0.002097 |
| 51 | LEUKOCYTE DIFFERENTIATION | 25 | 292 | 2.435e-05 | 0.002222 |
| 52 | NEGATIVE REGULATION OF CELL ACTIVATION | 17 | 158 | 2.867e-05 | 0.002565 |
| 53 | REGULATION OF CELLULAR COMPONENT SIZE | 27 | 337 | 3.775e-05 | 0.003302 |
| 54 | REGULATION OF PRODUCTION OF MOLECULAR MEDIATOR OF IMMUNE RESPONSE | 13 | 101 | 3.832e-05 | 0.003302 |
| 55 | LYMPHOCYTE HOMEOSTASIS | 9 | 50 | 4.187e-05 | 0.003406 |
| 56 | POSITIVE REGULATION OF LEUKOCYTE DIFFERENTIATION | 15 | 131 | 4.052e-05 | 0.003406 |
| 57 | REGULATION OF T CELL PROLIFERATION | 16 | 147 | 4.245e-05 | 0.003406 |
| 58 | PEPTIDYL TYROSINE AUTOPHOSPHORYLATION | 8 | 39 | 4.187e-05 | 0.003406 |
| 59 | CELL SUBSTRATE ADHESION | 17 | 164 | 4.625e-05 | 0.003648 |
| 60 | B CELL HOMEOSTASIS | 6 | 21 | 5.339e-05 | 0.00414 |
| 61 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 75 | 1395 | 5.502e-05 | 0.004197 |
| 62 | REGULATION OF CELL DIFFERENTIATION | 79 | 1492 | 5.743e-05 | 0.00431 |
| 63 | EPIDERMIS DEVELOPMENT | 22 | 253 | 5.849e-05 | 0.00432 |
| 64 | POSITIVE REGULATION OF LEUKOCYTE PROLIFERATION | 15 | 136 | 6.273e-05 | 0.004561 |
| 65 | REGULATION OF PROTEIN HOMODIMERIZATION ACTIVITY | 6 | 22 | 7.131e-05 | 0.005104 |
| 66 | TUBE DEVELOPMENT | 37 | 552 | 7.552e-05 | 0.005324 |
| 67 | REGULATION OF ADAPTIVE IMMUNE RESPONSE | 14 | 123 | 7.753e-05 | 0.005384 |
| 68 | REGULATION OF ANATOMICAL STRUCTURE SIZE | 33 | 472 | 8.285e-05 | 0.005507 |
| 69 | POSITIVE REGULATION OF LYMPHOCYTE DIFFERENTIATION | 11 | 80 | 8.142e-05 | 0.005507 |
| 70 | NEGATIVE REGULATION OF CELL ADHESION | 20 | 223 | 8.186e-05 | 0.005507 |
| 71 | CYTOSKELETON ORGANIZATION | 50 | 838 | 8.939e-05 | 0.005777 |
| 72 | POSITIVE REGULATION OF HYDROLASE ACTIVITY | 53 | 905 | 8.895e-05 | 0.005777 |
| 73 | POSITIVE REGULATION OF T CELL PROLIFERATION | 12 | 95 | 9.095e-05 | 0.005797 |
| 74 | REGULATION OF CELL PROJECTION ORGANIZATION | 37 | 558 | 9.42e-05 | 0.005923 |
| 75 | POSITIVE REGULATION OF INTERLEUKIN 6 PRODUCTION | 10 | 68 | 9.586e-05 | 0.005947 |
| 76 | ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY | 43 | 689 | 0.0001041 | 0.006375 |
| 77 | CELL DEVELOPMENT | 75 | 1426 | 0.0001107 | 0.006688 |
| 78 | MYELOID LEUKOCYTE ACTIVATION | 12 | 98 | 0.0001232 | 0.007349 |
| 79 | POSITIVE REGULATION OF HEMOPOIESIS | 16 | 163 | 0.0001462 | 0.008505 |
| 80 | REGULATION OF TRANSPORT | 90 | 1804 | 0.0001449 | 0.008505 |
| 81 | POSITIVE REGULATION OF CELLULAR COMPONENT BIOGENESIS | 29 | 406 | 0.0001529 | 0.008781 |
| 82 | ACTIVATION OF IMMUNE RESPONSE | 30 | 427 | 0.0001581 | 0.008973 |
| 83 | REGULATION OF PROTEIN LOCALIZATION | 54 | 950 | 0.0001634 | 0.009158 |
| 84 | NEGATIVE REGULATION OF HOMOTYPIC CELL CELL ADHESION | 12 | 102 | 0.0001811 | 0.00999 |
| 85 | LEUKOCYTE HOMEOSTASIS | 9 | 60 | 0.0001825 | 0.00999 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | ACTIN BINDING | 32 | 393 | 5.307e-06 | 0.001643 |
| 2 | MHC PROTEIN BINDING | 8 | 30 | 5.222e-06 | 0.001643 |
| 3 | LIPID BINDING | 46 | 657 | 3.315e-06 | 0.001643 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | INTRINSIC COMPONENT OF PLASMA MEMBRANE | 106 | 1649 | 1.109e-10 | 6.476e-08 |
| 2 | MEMBRANE REGION | 78 | 1134 | 2.327e-09 | 6.795e-07 |
| 3 | CELL JUNCTION | 77 | 1151 | 1.002e-08 | 1.951e-06 |
| 4 | PLASMA MEMBRANE REGION | 65 | 929 | 2.965e-08 | 4.328e-06 |
| 5 | CELL CELL JUNCTION | 36 | 383 | 4.156e-08 | 4.855e-06 |
| 6 | CELL LEADING EDGE | 31 | 350 | 1.308e-06 | 0.0001273 |
| 7 | APICAL PART OF CELL | 31 | 361 | 2.504e-06 | 0.0002044 |
| 8 | APICAL PLASMA MEMBRANE | 27 | 292 | 2.8e-06 | 0.0002044 |
| 9 | ACTIN CYTOSKELETON | 35 | 444 | 3.998e-06 | 0.0002595 |
| 10 | INTRACELLULAR VESICLE | 73 | 1259 | 5.672e-06 | 0.0003312 |
| 11 | VACUOLE | 69 | 1180 | 7.877e-06 | 0.0004182 |
| 12 | GOLGI APPARATUS | 79 | 1445 | 1.925e-05 | 0.0009368 |
| 13 | RUFFLE | 17 | 156 | 2.43e-05 | 0.001091 |
| 14 | CELL PROJECTION MEMBRANE | 25 | 298 | 3.418e-05 | 0.001426 |
| 15 | SIDE OF MEMBRANE | 31 | 428 | 7.096e-05 | 0.002763 |
| 16 | BASOLATERAL PLASMA MEMBRANE | 19 | 211 | 0.000116 | 0.004234 |
| 17 | MHC CLASS II PROTEIN COMPLEX | 5 | 16 | 0.0001441 | 0.004951 |
| 18 | BRUSH BORDER | 12 | 102 | 0.0001811 | 0.005782 |
| 19 | CYTOSKELETON | 96 | 1967 | 0.0001881 | 0.005782 |
| 20 | CELL CORTEX | 20 | 238 | 0.0001982 | 0.005788 |
| 21 | MICROVILLUS | 10 | 75 | 0.0002208 | 0.006139 |
| 22 | MHC PROTEIN COMPLEX | 6 | 27 | 0.0002447 | 0.006495 |
| 23 | ANCHORING JUNCTION | 32 | 489 | 0.0003469 | 0.008441 |
| 24 | PLASMA MEMBRANE PROTEIN COMPLEX | 33 | 510 | 0.0003449 | 0.008441 |
| 25 | RUFFLE MEMBRANE | 10 | 80 | 0.0003766 | 0.008459 |
| 26 | MEMBRANE MICRODOMAIN | 22 | 288 | 0.0003749 | 0.008459 |
| 27 | MYOSIN COMPLEX | 9 | 67 | 0.0004273 | 0.008604 |
| 28 | CELL SURFACE | 44 | 757 | 0.0004111 | 0.008604 |
| 29 | EXTRINSIC COMPONENT OF MEMBRANE | 20 | 252 | 0.0004188 | 0.008604 |
| 30 | ACTIN BASED CELL PROJECTION | 16 | 181 | 0.0004809 | 0.009362 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
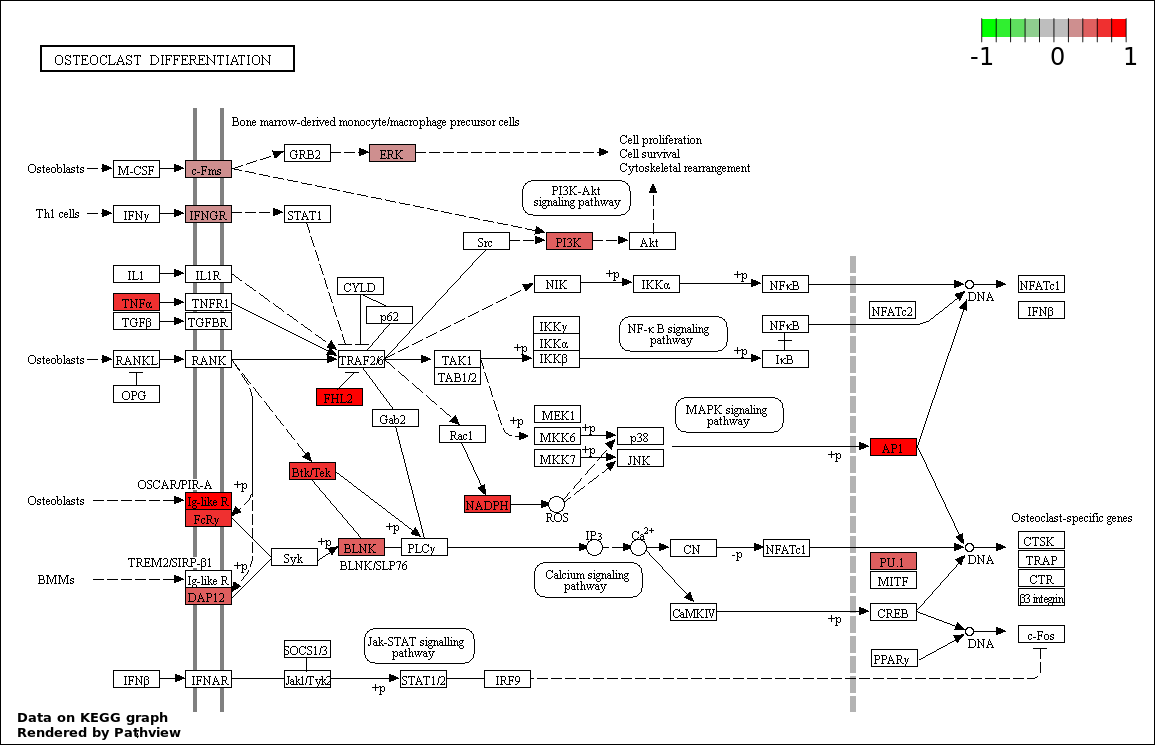
| 1 | hsa04380_Osteoclast_differentiation | 19 | 387 | 0.07162 | 1 | |
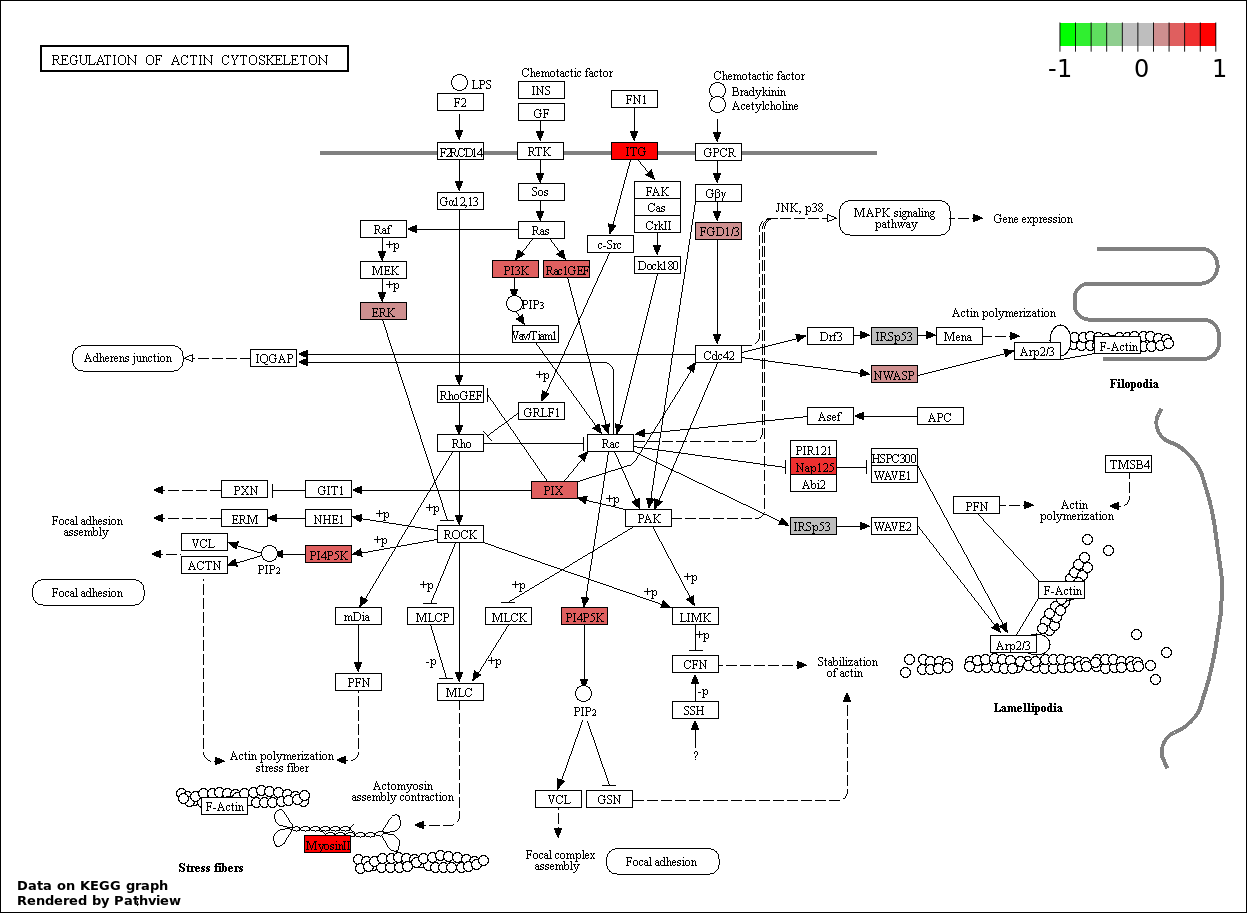
| 2 | hsa04810_Regulation_of_actin_cytoskeleton | 16 | 387 | 0.2477 | 1 | |
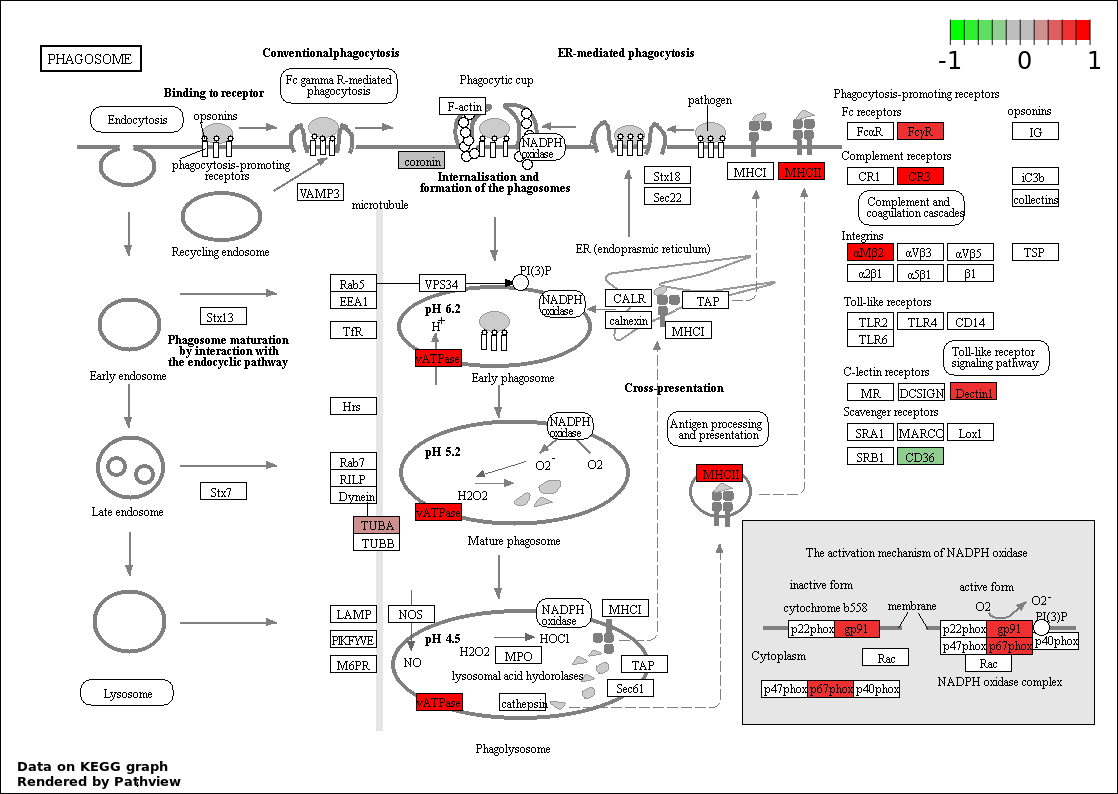
| 3 | hsa04145_Phagosome | 14 | 387 | 0.4469 | 1 | |
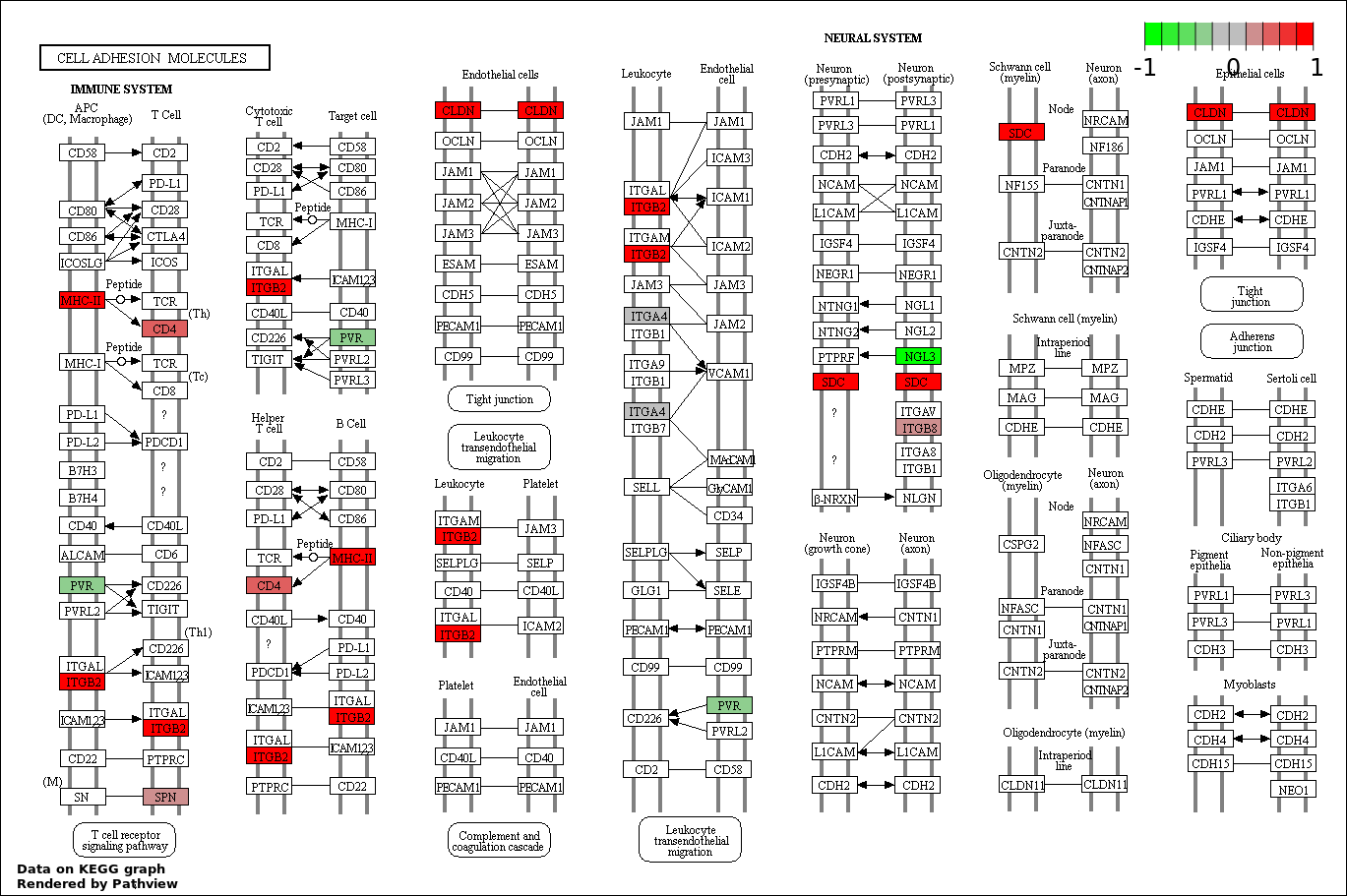
| 4 | hsa04514_Cell_adhesion_molecules_.CAMs. | 13 | 387 | 0.5597 | 1 | |
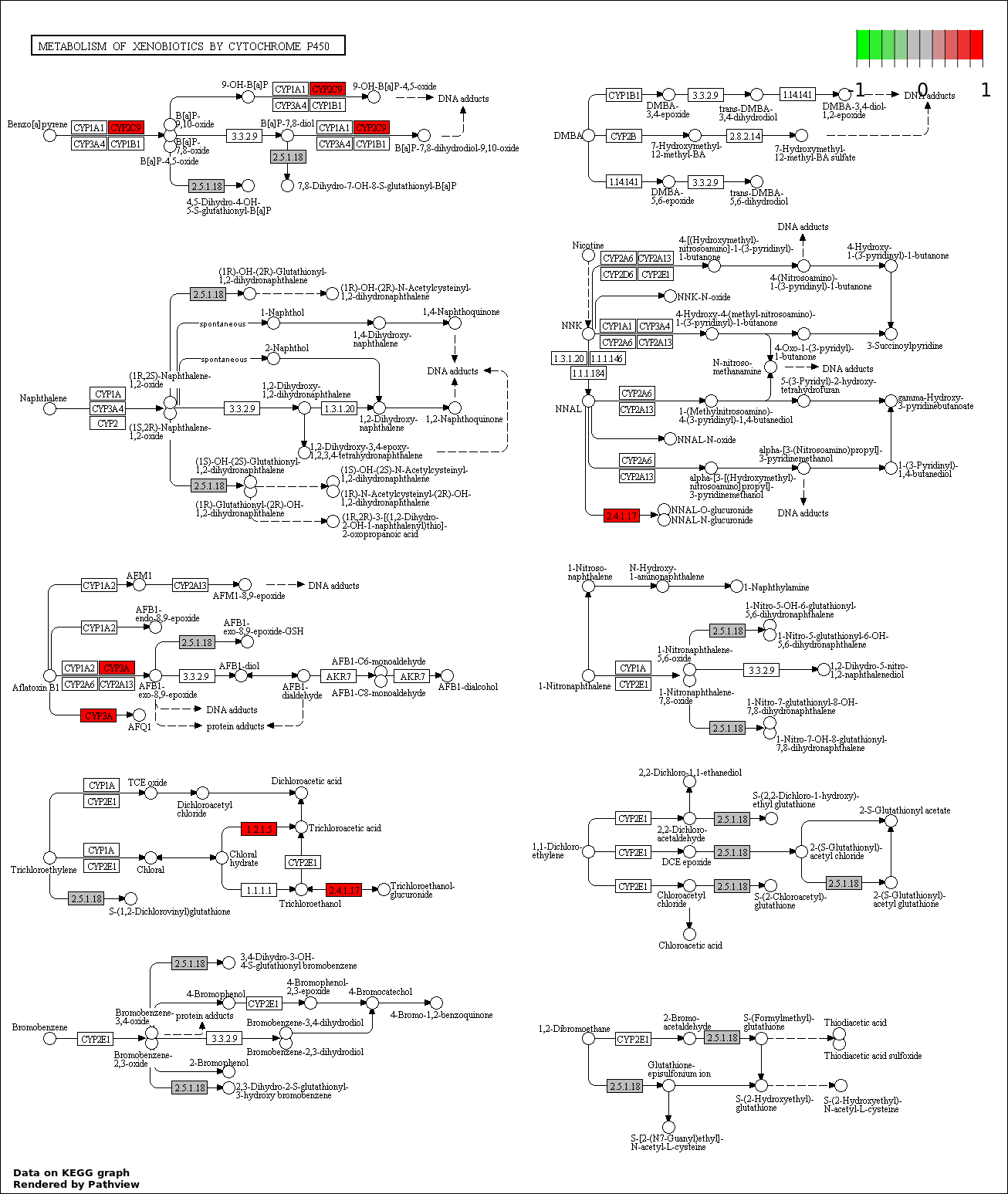
| 5 | hsa00980_Metabolism_of_xenobiotics_by_cytochrome_P450 | 11 | 387 | 0.7703 | 1 | |
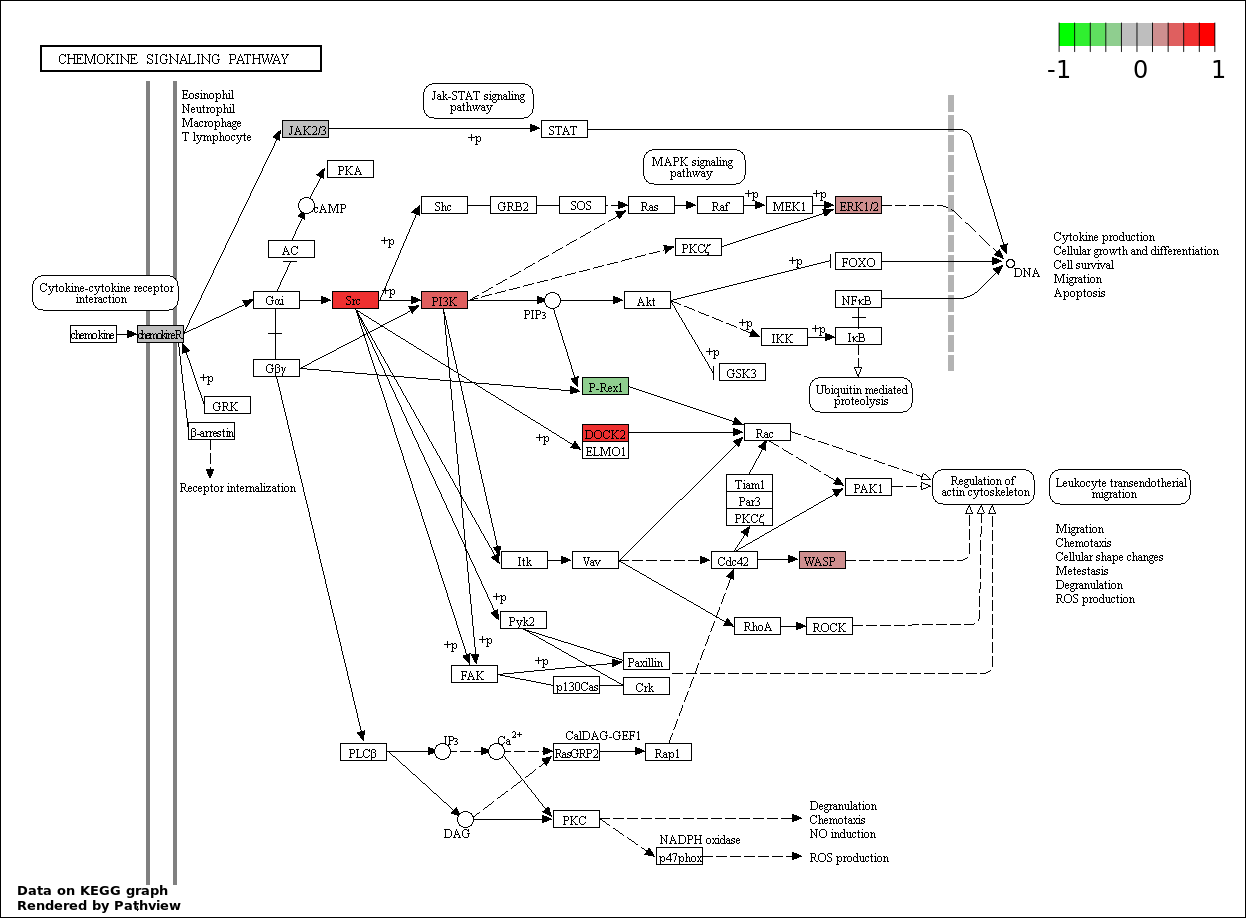
| 6 | hsa04062_Chemokine_signaling_pathway | 11 | 387 | 0.7703 | 1 | |
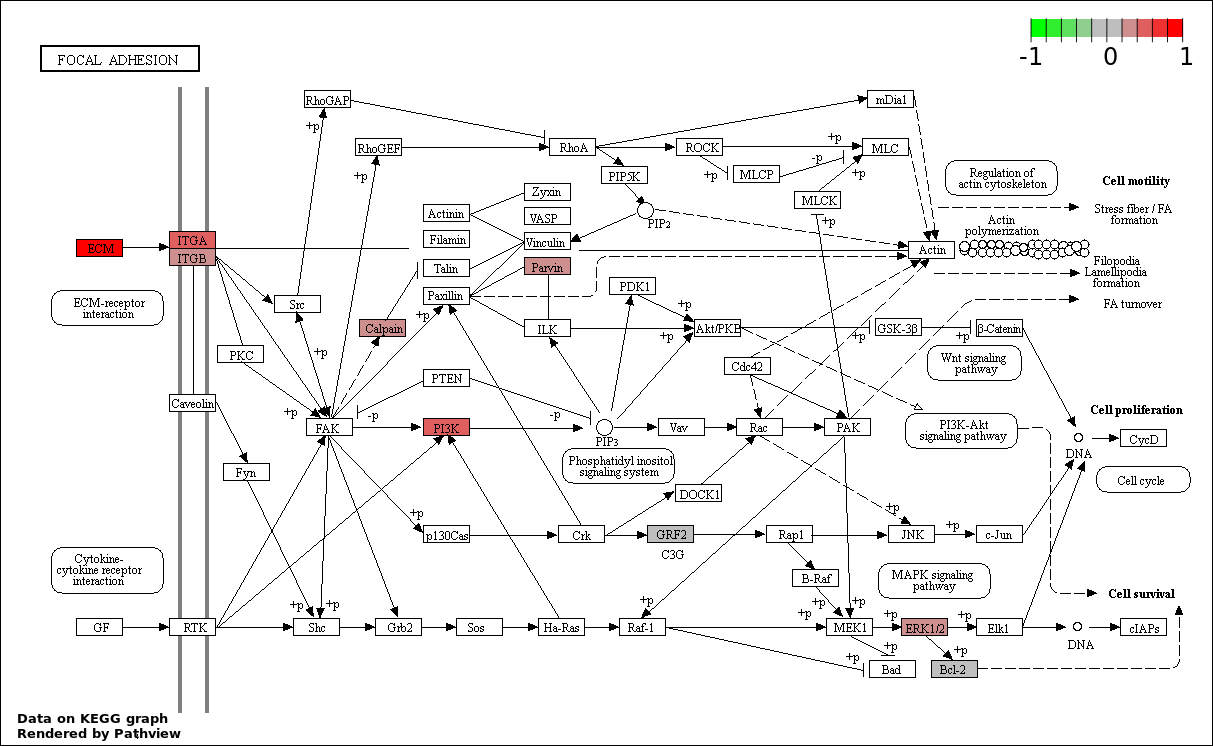
| 7 | hsa04510_Focal_adhesion | 11 | 387 | 0.7703 | 1 | |
| 8 | hsa00982_Drug_metabolism_._cytochrome_P450 | 10 | 387 | 0.8526 | 1 | |
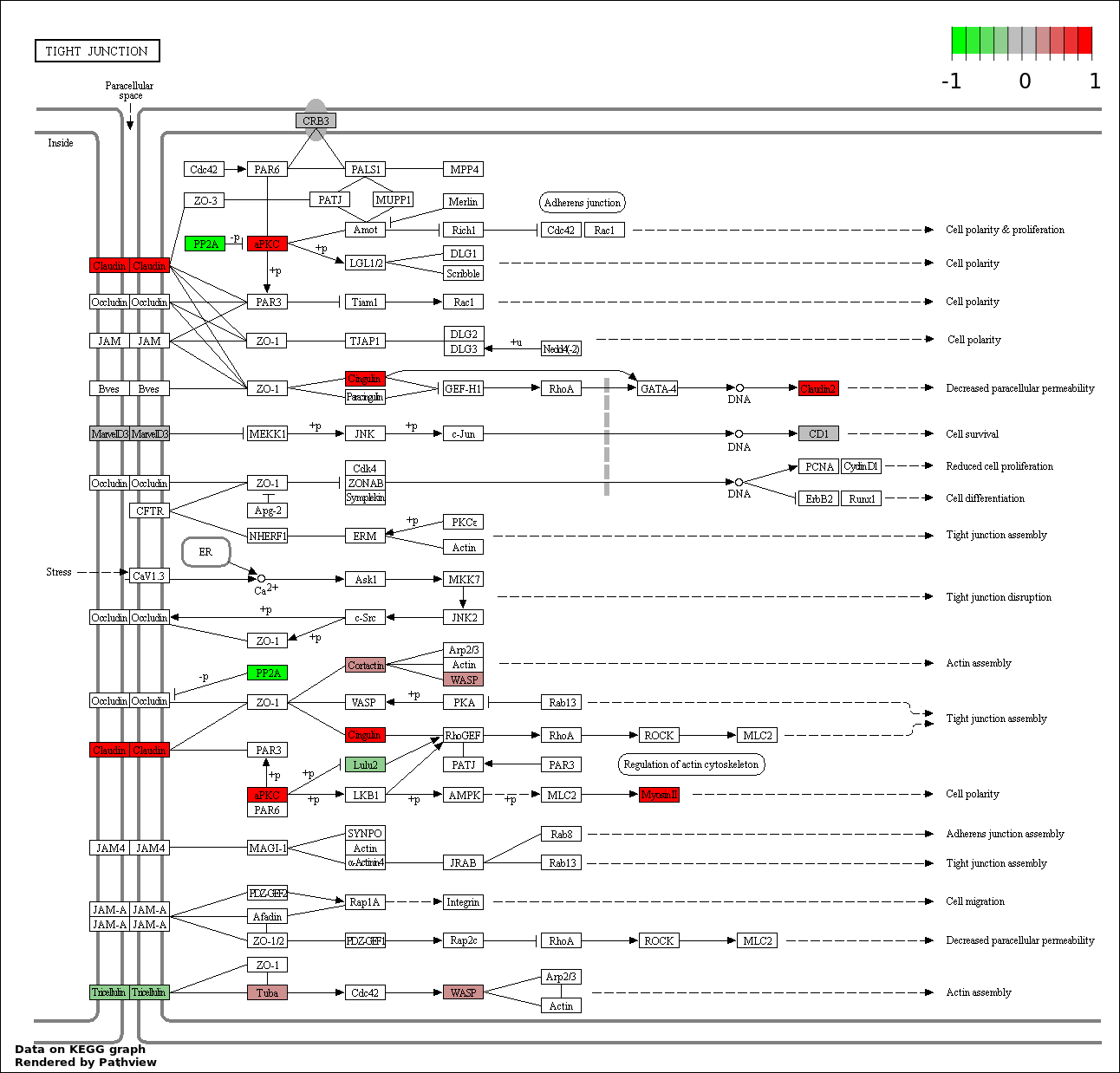
| 9 | hsa04530_Tight_junction | 10 | 387 | 0.8526 | 1 | |
| 10 | hsa04640_Hematopoietic_cell_lineage | 10 | 387 | 0.8526 | 1 | |
| 11 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 10 | 387 | 0.8526 | 1 | |
| 12 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 10 | 387 | 0.8526 | 1 | |
| 13 | hsa00140_Steroid_hormone_biosynthesis | 9 | 387 | 0.914 | 1 | |
| 14 | hsa00830_Retinol_metabolism | 9 | 387 | 0.914 | 1 | |
| 15 | hsa04142_Lysosome | 9 | 387 | 0.914 | 1 | |
| 16 | hsa04612_Antigen_processing_and_presentation | 9 | 387 | 0.914 | 1 | |
| 17 | hsa04630_Jak.STAT_signaling_pathway | 9 | 387 | 0.914 | 1 | |
| 18 | hsa04660_T_cell_receptor_signaling_pathway | 9 | 387 | 0.914 | 1 | |
| 19 | hsa00230_Purine_metabolism | 8 | 387 | 0.955 | 1 | |
| 20 | hsa00983_Drug_metabolism_._other_enzymes | 8 | 387 | 0.955 | 1 | |
| 21 | hsa04360_Axon_guidance | 8 | 387 | 0.955 | 1 | |
| 22 | hsa04670_Leukocyte_transendothelial_migration | 8 | 387 | 0.955 | 1 | |
| 23 | hsa04722_Neurotrophin_signaling_pathway | 8 | 387 | 0.955 | 1 | |
| 24 | hsa04010_MAPK_signaling_pathway | 7 | 387 | 0.9792 | 1 | |
| 25 | hsa04012_ErbB_signaling_pathway | 7 | 387 | 0.9792 | 1 | |
| 26 | hsa04620_Toll.like_receptor_signaling_pathway | 7 | 387 | 0.9792 | 1 | |
| 27 | hsa04662_B_cell_receptor_signaling_pathway | 7 | 387 | 0.9792 | 1 | |
| 28 | hsa04910_Insulin_signaling_pathway | 7 | 387 | 0.9792 | 1 | |
| 29 | hsa04020_Calcium_signaling_pathway | 6 | 387 | 0.9918 | 1 | |
| 30 | hsa04070_Phosphatidylinositol_signaling_system | 6 | 387 | 0.9918 | 1 | |
| 31 | hsa04210_Apoptosis | 6 | 387 | 0.9918 | 1 | |
| 32 | hsa04512_ECM.receptor_interaction | 6 | 387 | 0.9918 | 1 | |
| 33 | hsa04664_Fc_epsilon_RI_signaling_pathway | 6 | 387 | 0.9918 | 1 | |
| 34 | hsa04672_Intestinal_immune_network_for_IgA_production | 6 | 387 | 0.9918 | 1 | |
| 35 | hsa00500_Starch_and_sucrose_metabolism | 5 | 387 | 0.9973 | 1 | |
| 36 | hsa00514_Other_types_of_O.glycan_biosynthesis | 5 | 387 | 0.9973 | 1 | |
| 37 | hsa00591_Linoleic_acid_metabolism | 5 | 387 | 0.9973 | 1 | |
| 38 | hsa00860_Porphyrin_and_chlorophyll_metabolism | 5 | 387 | 0.9973 | 1 | |
| 39 | hsa04144_Endocytosis | 5 | 387 | 0.9973 | 1 | |
| 40 | hsa04970_Salivary_secretion | 5 | 387 | 0.9973 | 1 | |
| 41 | hsa00040_Pentose_and_glucuronate_interconversions | 4 | 387 | 0.9993 | 1 | |
| 42 | hsa00053_Ascorbate_and_aldarate_metabolism | 4 | 387 | 0.9993 | 1 | |
| 43 | hsa00512_Mucin_type_O.Glycan_biosynthesis | 4 | 387 | 0.9993 | 1 | |
| 44 | hsa00520_Amino_sugar_and_nucleotide_sugar_metabolism | 4 | 387 | 0.9993 | 1 | |
| 45 | hsa00562_Inositol_phosphate_metabolism | 4 | 387 | 0.9993 | 1 | |
| 46 | hsa00590_Arachidonic_acid_metabolism | 4 | 387 | 0.9993 | 1 | |
| 47 | hsa04115_p53_signaling_pathway | 4 | 387 | 0.9993 | 1 | |
| 48 | hsa04340_Hedgehog_signaling_pathway | 4 | 387 | 0.9993 | 1 | |
| 49 | hsa04350_TGF.beta_signaling_pathway | 4 | 387 | 0.9993 | 1 | |
| 50 | hsa04520_Adherens_junction | 4 | 387 | 0.9993 | 1 | |
| 51 | hsa04621_NOD.like_receptor_signaling_pathway | 4 | 387 | 0.9993 | 1 | |
| 52 | hsa04960_Aldosterone.regulated_sodium_reabsorption | 4 | 387 | 0.9993 | 1 | |
| 53 | hsa00240_Pyrimidine_metabolism | 3 | 387 | 0.9999 | 1 | |
| 54 | hsa00330_Arginine_and_proline_metabolism | 3 | 387 | 0.9999 | 1 | |
| 55 | hsa00564_Glycerophospholipid_metabolism | 3 | 387 | 0.9999 | 1 | |
| 56 | hsa00601_Glycosphingolipid_biosynthesis_._lacto_and_neolacto_series | 3 | 387 | 0.9999 | 1 | |
| 57 | hsa04130_SNARE_interactions_in_vesicular_transport | 3 | 387 | 0.9999 | 1 | |
| 58 | hsa04141_Protein_processing_in_endoplasmic_reticulum | 3 | 387 | 0.9999 | 1 | |
| 59 | hsa04146_Peroxisome | 3 | 387 | 0.9999 | 1 | |
| 60 | hsa04150_mTOR_signaling_pathway | 3 | 387 | 0.9999 | 1 | |
| 61 | hsa04270_Vascular_smooth_muscle_contraction | 3 | 387 | 0.9999 | 1 | |
| 62 | hsa04370_VEGF_signaling_pathway | 3 | 387 | 0.9999 | 1 | |
| 63 | hsa04610_Complement_and_coagulation_cascades | 3 | 387 | 0.9999 | 1 | |
| 64 | hsa04914_Progesterone.mediated_oocyte_maturation | 3 | 387 | 0.9999 | 1 | |
| 65 | hsa04920_Adipocytokine_signaling_pathway | 3 | 387 | 0.9999 | 1 | |
| 66 | hsa04962_Vasopressin.regulated_water_reabsorption | 3 | 387 | 0.9999 | 1 | |
| 67 | hsa04973_Carbohydrate_digestion_and_absorption | 3 | 387 | 0.9999 | 1 | |
| 68 | hsa00051_Fructose_and_mannose_metabolism | 2 | 387 | 1 | 1 | |
| 69 | hsa00190_Oxidative_phosphorylation | 2 | 387 | 1 | 1 | |
| 70 | hsa00260_Glycine._serine_and_threonine_metabolism | 2 | 387 | 1 | 1 | |
| 71 | hsa00480_Glutathione_metabolism | 2 | 387 | 1 | 1 | |
| 72 | hsa00603_Glycosphingolipid_biosynthesis_._globo_series | 2 | 387 | 1 | 1 | |
| 73 | hsa00650_Butanoate_metabolism | 2 | 387 | 1 | 1 | |
| 74 | hsa00910_Nitrogen_metabolism | 2 | 387 | 1 | 1 | |
| 75 | hsa02010_ABC_transporters | 2 | 387 | 1 | 1 | |
| 76 | hsa03008_Ribosome_biogenesis_in_eukaryotes | 2 | 387 | 1 | 1 | |
| 77 | hsa03015_mRNA_surveillance_pathway | 2 | 387 | 1 | 1 | |
| 78 | hsa03018_RNA_degradation | 2 | 387 | 1 | 1 | |
| 79 | hsa03320_PPAR_signaling_pathway | 2 | 387 | 1 | 1 | |
| 80 | hsa04110_Cell_cycle | 2 | 387 | 1 | 1 | |
| 81 | hsa04310_Wnt_signaling_pathway | 2 | 387 | 1 | 1 | |
| 82 | hsa04320_Dorso.ventral_axis_formation | 2 | 387 | 1 | 1 | |
| 83 | hsa04540_Gap_junction | 2 | 387 | 1 | 1 | |
| 84 | hsa04916_Melanogenesis | 2 | 387 | 1 | 1 | |
| 85 | hsa04964_Proximal_tubule_bicarbonate_reclamation | 2 | 387 | 1 | 1 | |
| 86 | hsa04966_Collecting_duct_acid_secretion | 2 | 387 | 1 | 1 | |
| 87 | hsa04971_Gastric_acid_secretion | 2 | 387 | 1 | 1 | |
| 88 | hsa04974_Protein_digestion_and_absorption | 2 | 387 | 1 | 1 | |
| 89 | hsa04975_Fat_digestion_and_absorption | 2 | 387 | 1 | 1 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HCG11 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7f-2-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-3065-5p;hsa-miR-501-5p | 17 | MEF2C | Sponge network | 0.443 | 0.74476 | 0.324 | 0.85064 | 0.653 |
| 2 | UCA1 |
hsa-miR-129-5p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20b-5p;hsa-miR-26a-5p;hsa-miR-29b-2-5p;hsa-miR-3065-5p;hsa-miR-362-5p;hsa-miR-432-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | GPRC5A | Sponge network | 1.304 | 0.38107 | 3.241 | 0.27992 | 0.622 |
| 3 | UCA1 |
hsa-miR-101-5p;hsa-miR-103a-2-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | TMPRSS4 | Sponge network | 1.304 | 0.38107 | 3.334 | 0.24362 | 0.619 |
| 4 | RFPL1S |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-31-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-96-5p | 20 | CELF2 | Sponge network | -0.397 | 0.54233 | -0.1 | 0.9529 | 0.595 |
| 5 | DIO3OS |
hsa-miR-103a-2-5p;hsa-miR-125a-5p;hsa-miR-129-5p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-330-5p;hsa-miR-3613-3p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-769-5p | 10 | TMPRSS4 | Sponge network | 1.09 | 0.12548 | 3.334 | 0.24362 | 0.584 |
| 6 | RFPL1S |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7f-2-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 19 | MEF2C | Sponge network | -0.397 | 0.54233 | 0.324 | 0.85064 | 0.582 |
| 7 | DIO3OS |
hsa-let-7g-3p;hsa-miR-129-5p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20b-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30c-1-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-93-3p | 19 | GPRC5A | Sponge network | 1.09 | 0.12548 | 3.241 | 0.27992 | 0.573 |
| 8 | UCA1 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-135a-5p;hsa-miR-299-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-3p | 11 | EREG | Sponge network | 1.304 | 0.38107 | 1.952 | 0.1318 | 0.56 |
| 9 | RFPL1S |
hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-877-5p;hsa-miR-92a-3p | 10 | FLI1 | Sponge network | -0.397 | 0.54233 | 0.253 | 0.87092 | 0.552 |
| 10 | PART1 |
hsa-miR-10a-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-214-3p;hsa-miR-2355-5p;hsa-miR-27a-3p;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-93-5p | 11 | CHD5 | Sponge network | -2.652 | 0.02777 | -2.067 | 0.00463 | 0.552 |
| 11 | HCG11 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-590-3p | 12 | ARHGAP28 | Sponge network | 0.443 | 0.74476 | 0.702 | 0.38581 | 0.546 |
| 12 | HCG11 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-455-5p | 15 | CELF2 | Sponge network | 0.443 | 0.74476 | -0.1 | 0.9529 | 0.545 |
| 13 | HCG11 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-135a-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-369-3p;hsa-miR-664a-3p | 12 | VASH2 | Sponge network | 0.443 | 0.74476 | 0.553 | 0.41186 | 0.54 |
| 14 | RFPL1S |
hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-96-5p | 17 | BCL2 | Sponge network | -0.397 | 0.54233 | -0.051 | 0.9708 | 0.539 |
| 15 | PART1 |
hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-28-5p;hsa-miR-34b-5p;hsa-miR-93-5p | 13 | RND2 | Sponge network | -2.652 | 0.02777 | -1.402 | 0.00676 | 0.53 |
| 16 | HCG11 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-29b-3p | 10 | MARCH1 | Sponge network | 0.443 | 0.74476 | 0.152 | 0.84595 | 0.528 |
| 17 | HCP5 |
hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-154-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3065-3p;hsa-miR-330-3p;hsa-miR-33b-5p;hsa-miR-377-3p;hsa-miR-539-5p;hsa-miR-654-5p;hsa-miR-7-5p;hsa-miR-744-3p | 15 | CIITA | Sponge network | 0.397 | 0.83984 | 0.291 | 0.86137 | 0.525 |
| 18 | RFPL1S |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-93-5p | 13 | SH2B3 | Sponge network | -0.397 | 0.54233 | 0.028 | 0.9889 | 0.524 |
| 19 | PVT1 |
hsa-let-7g-3p;hsa-miR-129-5p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-432-5p;hsa-miR-532-3p;hsa-miR-592 | 12 | GPRC5A | Sponge network | 0.549 | 0.51449 | 3.241 | 0.27992 | 0.522 |
| 20 | HCG11 |
hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3065-5p;hsa-miR-455-5p | 14 | BCL2 | Sponge network | 0.443 | 0.74476 | -0.051 | 0.9708 | 0.503 |
| 21 | PART1 |
hsa-miR-10a-5p;hsa-miR-138-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-193a-5p;hsa-miR-224-3p;hsa-miR-2355-3p;hsa-miR-27a-5p;hsa-miR-31-5p;hsa-miR-3614-5p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-629-3p | 13 | CELF5 | Sponge network | -2.652 | 0.02777 | -1.99 | 0.00411 | 0.5 |
| 22 | CECR7 |
hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-429 | 11 | BCL2 | Sponge network | -0.265 | 0.77134 | -0.051 | 0.9708 | 0.495 |
| 23 | HCG11 |
hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-183-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-29b-3p;hsa-miR-3065-5p;hsa-miR-455-5p;hsa-miR-550a-5p | 13 | CIITA | Sponge network | 0.443 | 0.74476 | 0.291 | 0.86137 | 0.493 |
| 24 | RFPL1S |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7f-2-3p;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-224-3p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-96-5p | 19 | TMEM170B | Sponge network | -0.397 | 0.54233 | -0.514 | 0.59649 | 0.493 |
| 25 | RFPL1S |
hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-31-5p;hsa-miR-429;hsa-miR-96-5p | 10 | BACH2 | Sponge network | -0.397 | 0.54233 | -0.19 | 0.85383 | 0.492 |
| 26 | RFPL1S |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3934-5p;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-96-5p | 13 | GIT2 | Sponge network | -0.397 | 0.54233 | -0.143 | 0.94486 | 0.489 |
| 27 | HCG11 |
hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-3065-5p;hsa-miR-320b;hsa-miR-320c;hsa-miR-655-3p | 13 | FRMD4A | Sponge network | 0.443 | 0.74476 | 0.146 | 0.93001 | 0.485 |
| 28 | CASC2 |
hsa-miR-10a-5p;hsa-miR-10b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-193a-5p;hsa-miR-224-3p;hsa-miR-2355-3p;hsa-miR-27a-5p;hsa-miR-31-5p;hsa-miR-3614-5p;hsa-miR-505-5p;hsa-miR-629-3p | 12 | CELF5 | Sponge network | -0.659 | 0.19467 | -1.99 | 0.00411 | 0.478 |
| 29 | CECR7 |
hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-425-5p;hsa-miR-429 | 15 | CELF2 | Sponge network | -0.265 | 0.77134 | -0.1 | 0.9529 | 0.475 |
| 30 | CECR7 |
hsa-let-7f-1-3p;hsa-let-7f-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200c-5p;hsa-miR-27b-3p;hsa-miR-33b-5p;hsa-miR-425-5p;hsa-miR-429 | 10 | MEF2C | Sponge network | -0.265 | 0.77134 | 0.324 | 0.85064 | 0.474 |
| 31 | PVT1 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-29b-2-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-432-5p;hsa-miR-541-3p;hsa-miR-654-5p;hsa-miR-7-5p;hsa-miR-769-3p | 11 | ZDHHC9 | Sponge network | 0.549 | 0.51449 | 0.658 | 0.76379 | 0.455 |
| 32 | EGOT |
hsa-let-7g-3p;hsa-miR-137;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-495-3p;hsa-miR-592;hsa-miR-7-1-3p | 11 | GPRC5A | Sponge network | 1.405 | 0.04702 | 3.241 | 0.27992 | 0.451 |
| 33 | RFPL1S |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-32-5p;hsa-miR-429 | 11 | MARCH1 | Sponge network | -0.397 | 0.54233 | 0.152 | 0.84595 | 0.448 |
| 34 | RFPL1S |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-16-2-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-345-5p;hsa-miR-429 | 11 | ARHGAP28 | Sponge network | -0.397 | 0.54233 | 0.702 | 0.38581 | 0.438 |
| 35 | DIO3OS |
hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-204-5p;hsa-miR-30b-3p;hsa-miR-3613-3p;hsa-miR-491-5p;hsa-miR-642a-5p | 11 | CORO2A | Sponge network | 1.09 | 0.12548 | 0.908 | 0.67669 | 0.437 |
| 36 | CASC2 |
hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-28-5p;hsa-miR-493-3p | 12 | RND2 | Sponge network | -0.659 | 0.19467 | -1.402 | 0.00676 | 0.426 |
| 37 | RFPL1S |
hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-96-5p | 11 | PPM1F | Sponge network | -0.397 | 0.54233 | -0.056 | 0.97696 | 0.422 |
| 38 | PLAC4 |
hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-139-5p;hsa-miR-15a-5p;hsa-miR-212-3p;hsa-miR-218-5p;hsa-miR-324-5p;hsa-miR-421;hsa-miR-543;hsa-miR-769-3p | 11 | ARHGAP32 | Sponge network | 1.229 | 0.11833 | 0.429 | 0.84674 | 0.414 |
| 39 | UCA1 |
hsa-miR-101-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-296-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | LANCL3 | Sponge network | 1.304 | 0.38107 | 0.575 | 0.31897 | 0.412 |
| 40 | UCA1 |
hsa-miR-132-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-299-3p;hsa-miR-339-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | PPP1R1C | Sponge network | 1.304 | 0.38107 | 0.447 | 0.43628 | 0.407 |
| 41 | UCA1 |
hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-3065-5p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-487a-3p;hsa-miR-500a-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-3p;hsa-miR-7-1-3p | 12 | ABCA13 | Sponge network | 1.304 | 0.38107 | 1.838 | 0.02908 | 0.403 |
| 42 | UCA1 |
hsa-miR-101-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-204-5p;hsa-miR-20b-5p;hsa-miR-296-3p;hsa-miR-324-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | EPB41L1 | Sponge network | 1.304 | 0.38107 | 0.536 | 0.83266 | 0.401 |
| 43 | DIO3OS |
hsa-miR-128-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-204-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-376a-3p;hsa-miR-376b-3p;hsa-miR-582-5p;hsa-miR-628-5p | 11 | EMP2 | Sponge network | 1.09 | 0.12548 | 0.298 | 0.90368 | 0.399 |
| 44 | EMX2OS |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p | 11 | VASH2 | Sponge network | 1.611 | 0.09138 | 0.553 | 0.41186 | 0.398 |
| 45 | MIAT | hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-96-5p | 11 | CELF2 | Sponge network | 0.718 | 0.59601 | -0.1 | 0.9529 | 0.394 |
| 46 | UCA1 |
hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-212-3p;hsa-miR-29b-2-5p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-377-5p;hsa-miR-423-5p;hsa-miR-432-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | ARHGAP32 | Sponge network | 1.304 | 0.38107 | 0.429 | 0.84674 | 0.39 |
| 47 | PVT1 |
hsa-miR-1224-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-204-5p;hsa-miR-216a-5p;hsa-miR-324-3p;hsa-miR-501-5p;hsa-miR-628-5p;hsa-miR-758-3p | 11 | DNAH3 | Sponge network | 0.549 | 0.51449 | 1.542 | 0.04542 | 0.387 |
| 48 | CASC2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-150-3p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-497-5p | 13 | NEBL | Sponge network | -0.659 | 0.19467 | -0.34 | 0.8605 | 0.386 |
| 49 | CECR7 |
hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-429;hsa-miR-550a-5p | 11 | CIITA | Sponge network | -0.265 | 0.77134 | 0.291 | 0.86137 | 0.384 |
| 50 | EGOT |
hsa-miR-139-5p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-320c;hsa-miR-324-5p;hsa-miR-421;hsa-miR-487a-3p;hsa-miR-491-5p;hsa-miR-7-1-3p | 10 | ABCA13 | Sponge network | 1.405 | 0.04702 | 1.838 | 0.02908 | 0.378 |
| 51 | PART1 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-24-3p;hsa-miR-30a-5p;hsa-miR-34a-5p;hsa-miR-34c-5p;hsa-miR-450b-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 14 | SCML2 | Sponge network | -2.652 | 0.02777 | -1.557 | 0.00352 | 0.377 |
| 52 | CASC2 |
hsa-miR-146b-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-221-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-450b-5p | 10 | TTC39A | Sponge network | -0.659 | 0.19467 | -0.448 | 0.80896 | 0.373 |
| 53 | H19 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-423-3p | 10 | FRMD4A | Sponge network | 0.885 | 0.7435 | 0.146 | 0.93001 | 0.372 |
| 54 | H19 |
hsa-let-7g-3p;hsa-miR-129-5p;hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-592 | 14 | GPRC5A | Sponge network | 0.885 | 0.7435 | 3.241 | 0.27992 | 0.366 |
| 55 | HCP5 |
hsa-miR-129-5p;hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-539-5p | 10 | GPRC5A | Sponge network | 0.397 | 0.83984 | 3.241 | 0.27992 | 0.361 |
| 56 | DIO3OS |
hsa-miR-1224-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-204-5p;hsa-miR-324-3p;hsa-miR-377-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-628-5p;hsa-miR-98-5p | 13 | DNAH3 | Sponge network | 1.09 | 0.12548 | 1.542 | 0.04542 | 0.361 |
| 57 | NEAT1 |
hsa-miR-1270;hsa-miR-132-5p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-204-5p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-382-5p;hsa-miR-421;hsa-miR-431-5p;hsa-miR-532-3p;hsa-miR-543;hsa-miR-628-5p;hsa-miR-744-3p | 14 | ONECUT3 | Sponge network | 0.61 | 0.85461 | 2.227 | 0.06101 | 0.36 |
| 58 | CASC2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-374a-3p;hsa-miR-424-5p | 12 | CPD | Sponge network | -0.659 | 0.19467 | 0.001 | 0.99977 | 0.356 |
| 59 | DIO3OS |
hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-154-5p;hsa-miR-183-5p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-377-3p;hsa-miR-382-5p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-7-5p | 15 | CIITA | Sponge network | 1.09 | 0.12548 | 0.291 | 0.86137 | 0.345 |
| 60 | AGAP11 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-147b;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-30b-3p;hsa-miR-455-3p;hsa-miR-455-5p | 10 | WDFY4 | Sponge network | 0.61 | 0.2086 | 0.645 | 0.65009 | 0.342 |
| 61 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-1270;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-185-3p;hsa-miR-204-5p;hsa-miR-20b-5p;hsa-miR-296-5p;hsa-miR-324-3p;hsa-miR-382-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-628-5p;hsa-miR-769-3p;hsa-miR-93-3p;hsa-miR-98-5p | 19 | ONECUT3 | Sponge network | 1.09 | 0.12548 | 2.227 | 0.06101 | 0.342 |
| 62 | NEAT1 |
hsa-miR-101-3p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-139-5p;hsa-miR-212-3p;hsa-miR-377-5p;hsa-miR-421;hsa-miR-432-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-543;hsa-miR-7-1-3p | 13 | ARHGAP32 | Sponge network | 0.61 | 0.85461 | 0.429 | 0.84674 | 0.339 |
| 63 | KCNQ1OT1 |
hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-374a-5p;hsa-miR-376a-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-543;hsa-miR-590-3p;hsa-miR-628-5p | 12 | PLEKHA5 | Sponge network | 0.517 | 0.63991 | 0.368 | 0.84945 | 0.332 |
| 64 | CASC2 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-224-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p | 12 | RAB3B | Sponge network | -0.659 | 0.19467 | -0.385 | 0.52757 | 0.331 |
| 65 | DIO3OS |
hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-204-5p;hsa-miR-20b-5p;hsa-miR-296-3p;hsa-miR-324-3p;hsa-miR-539-5p | 13 | EPB41L1 | Sponge network | 1.09 | 0.12548 | 0.536 | 0.83266 | 0.331 |
| 66 | FAM66C |
hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-10a-5p;hsa-miR-15b-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-224-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-452-5p;hsa-miR-92a-3p | 13 | TMEM170B | Sponge network | -0.683 | 0.08339 | -0.514 | 0.59649 | 0.329 |
| 67 | EMX2OS |
hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200c-5p;hsa-miR-3065-5p;hsa-miR-33b-5p;hsa-miR-425-5p;hsa-miR-940 | 11 | MEF2C | Sponge network | 1.611 | 0.09138 | 0.324 | 0.85064 | 0.326 |
| 68 | UCA1 |
hsa-miR-101-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-33a-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | SIX4 | Sponge network | 1.304 | 0.38107 | 0.184 | 0.86111 | 0.325 |
| 69 | KCNQ1OT1 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-139-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-212-3p;hsa-miR-29b-1-5p;hsa-miR-30e-5p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-377-3p;hsa-miR-377-5p;hsa-miR-421;hsa-miR-432-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-543;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 23 | ARHGAP32 | Sponge network | 0.517 | 0.63991 | 0.429 | 0.84674 | 0.324 |
| 70 | HCG11 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7f-2-3p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-29a-5p | 13 | TMEM170B | Sponge network | 0.443 | 0.74476 | -0.514 | 0.59649 | 0.321 |
| 71 | UCA1 |
hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-185-5p;hsa-miR-20b-5p;hsa-miR-29b-2-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-423-5p;hsa-miR-432-5p;hsa-miR-539-5p;hsa-miR-590-3p | 12 | ZDHHC9 | Sponge network | 1.304 | 0.38107 | 0.658 | 0.76379 | 0.32 |
| 72 | AGAP11 |
hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-30d-5p;hsa-miR-455-5p;hsa-miR-550a-3p | 12 | CELF2 | Sponge network | 0.61 | 0.2086 | -0.1 | 0.9529 | 0.319 |
| 73 | KCNQ1OT1 |
hsa-miR-126-5p;hsa-miR-154-3p;hsa-miR-186-5p;hsa-miR-20b-5p;hsa-miR-3200-3p;hsa-miR-323a-3p;hsa-miR-374a-5p;hsa-miR-377-5p;hsa-miR-411-3p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-487a-3p;hsa-miR-496;hsa-miR-539-5p;hsa-miR-543;hsa-miR-590-3p;hsa-miR-7-5p | 17 | REPS2 | Sponge network | 0.517 | 0.63991 | 0.566 | 0.76424 | 0.307 |
| 74 | CASC2 |
hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-193a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-452-3p;hsa-miR-452-5p | 16 | PLXNA2 | Sponge network | -0.659 | 0.19467 | 0.364 | 0.87338 | 0.307 |
| 75 | PART1 |
hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-10a-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-205-5p;hsa-miR-21-5p;hsa-miR-224-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-34b-3p;hsa-miR-625-3p;hsa-miR-92a-3p | 15 | TMEM170B | Sponge network | -2.652 | 0.02777 | -0.514 | 0.59649 | 0.307 |
| 76 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-185-5p;hsa-miR-20b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-3p;hsa-miR-330-5p;hsa-miR-539-5p;hsa-miR-7-5p;hsa-miR-769-3p | 11 | ZDHHC9 | Sponge network | 1.09 | 0.12548 | 0.658 | 0.76379 | 0.306 |
| 77 | H19 |
hsa-miR-129-2-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-654-5p | 13 | CIITA | Sponge network | 0.885 | 0.7435 | 0.291 | 0.86137 | 0.303 |
| 78 | PVT1 |
hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-204-5p;hsa-miR-324-3p;hsa-miR-382-5p;hsa-miR-431-5p;hsa-miR-532-3p;hsa-miR-543;hsa-miR-628-5p;hsa-miR-769-3p | 13 | ONECUT3 | Sponge network | 0.549 | 0.51449 | 2.227 | 0.06101 | 0.302 |
| 79 | HCP5 |
hsa-let-7f-5p;hsa-miR-125a-5p;hsa-miR-129-5p;hsa-miR-148b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-539-5p | 10 | VASH2 | Sponge network | 0.397 | 0.83984 | 0.553 | 0.41186 | 0.299 |
| 80 | KCNQ1OT1 |
hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-376a-5p;hsa-miR-485-3p;hsa-miR-496;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-769-5p | 12 | TMPRSS4 | Sponge network | 0.517 | 0.63991 | 3.334 | 0.24362 | 0.299 |
| 81 | KCNQ1OT1 |
hsa-let-7e-5p;hsa-miR-132-3p;hsa-miR-185-3p;hsa-miR-20b-5p;hsa-miR-212-3p;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-543;hsa-miR-7-5p;hsa-miR-98-5p | 10 | SEMA4G | Sponge network | 0.517 | 0.63991 | 0.784 | 0.68976 | 0.293 |
| 82 | UCA1 |
hsa-miR-1224-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148a-5p;hsa-miR-204-5p;hsa-miR-324-3p;hsa-miR-377-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 11 | DNAH3 | Sponge network | 1.304 | 0.38107 | 1.542 | 0.04542 | 0.29 |
| 83 | H19 |
hsa-let-7g-3p;hsa-miR-125a-5p;hsa-miR-129-5p;hsa-miR-148b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-539-5p | 11 | VASH2 | Sponge network | 0.885 | 0.7435 | 0.553 | 0.41186 | 0.287 |
| 84 | EMX2OS |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-425-5p | 12 | CELF2 | Sponge network | 1.611 | 0.09138 | -0.1 | 0.9529 | 0.28 |
| 85 | DGCR5 |
hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-28-5p;hsa-miR-34b-5p;hsa-miR-3614-3p;hsa-miR-625-5p | 11 | RND2 | Sponge network | -0.899 | 0.13441 | -1.402 | 0.00676 | 0.279 |
| 86 | KCNQ1OT1 |
hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-543;hsa-miR-590-3p;hsa-miR-616-5p;hsa-miR-98-5p | 10 | LMO7 | Sponge network | 0.517 | 0.63991 | 0.732 | 0.79354 | 0.275 |
| 87 | DGCR5 |
hsa-miR-10a-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-214-3p;hsa-miR-2355-5p;hsa-miR-27a-3p;hsa-miR-345-5p;hsa-miR-625-5p | 10 | CHD5 | Sponge network | -0.899 | 0.13441 | -2.067 | 0.00463 | 0.275 |
| 88 | UCA1 |
hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-148b-5p;hsa-miR-204-5p;hsa-miR-20b-5p;hsa-miR-296-5p;hsa-miR-324-3p;hsa-miR-382-5p;hsa-miR-590-3p;hsa-miR-744-3p | 10 | ONECUT3 | Sponge network | 1.304 | 0.38107 | 2.227 | 0.06101 | 0.274 |
| 89 | FAM66C |
hsa-let-7b-3p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-34a-5p;hsa-miR-425-5p;hsa-miR-615-3p | 10 | CELF2 | Sponge network | -0.683 | 0.08339 | -0.1 | 0.9529 | 0.27 |
| 90 | UCA1 |
hsa-miR-129-2-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-154-5p;hsa-miR-3065-5p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-382-5p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-744-3p | 11 | CIITA | Sponge network | 1.304 | 0.38107 | 0.291 | 0.86137 | 0.269 |
| 91 | PLAC4 |
hsa-miR-125a-5p;hsa-miR-128-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-296-3p;hsa-miR-3200-3p;hsa-miR-323b-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-543 | 10 | LANCL3 | Sponge network | 1.229 | 0.11833 | 0.575 | 0.31897 | 0.268 |
| 92 | PVT1 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-139-5p;hsa-miR-212-3p;hsa-miR-29b-2-5p;hsa-miR-432-5p;hsa-miR-543;hsa-miR-769-3p | 10 | ARHGAP32 | Sponge network | 0.549 | 0.51449 | 0.429 | 0.84674 | 0.265 |
| 93 | UCA1 |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-204-5p;hsa-miR-3065-5p;hsa-miR-323a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-7-1-3p | 12 | SLC7A11 | Sponge network | 1.304 | 0.38107 | 0.302 | 0.83928 | 0.265 |
| 94 | PART1 |
hsa-miR-10a-5p;hsa-miR-181a-2-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30c-2-3p;hsa-miR-31-5p;hsa-miR-452-3p;hsa-miR-625-3p | 12 | ORAI2 | Sponge network | -2.652 | 0.02777 | -0.472 | 0.81719 | 0.262 |
| 95 | HCG11 |
hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-5p;hsa-miR-3613-3p;hsa-miR-590-3p | 10 | CREB5 | Sponge network | 0.443 | 0.74476 | 0.493 | 0.69026 | 0.256 |
| 96 | DIO3OS |
hsa-miR-154-3p;hsa-miR-186-5p;hsa-miR-20b-5p;hsa-miR-324-5p;hsa-miR-377-5p;hsa-miR-487a-3p;hsa-miR-494-3p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-5p | 11 | REPS2 | Sponge network | 1.09 | 0.12548 | 0.566 | 0.76424 | 0.252 |
| 97 | FAM66C |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-34a-5p | 10 | BCL2 | Sponge network | -0.683 | 0.08339 | -0.051 | 0.9708 | 0.251 |