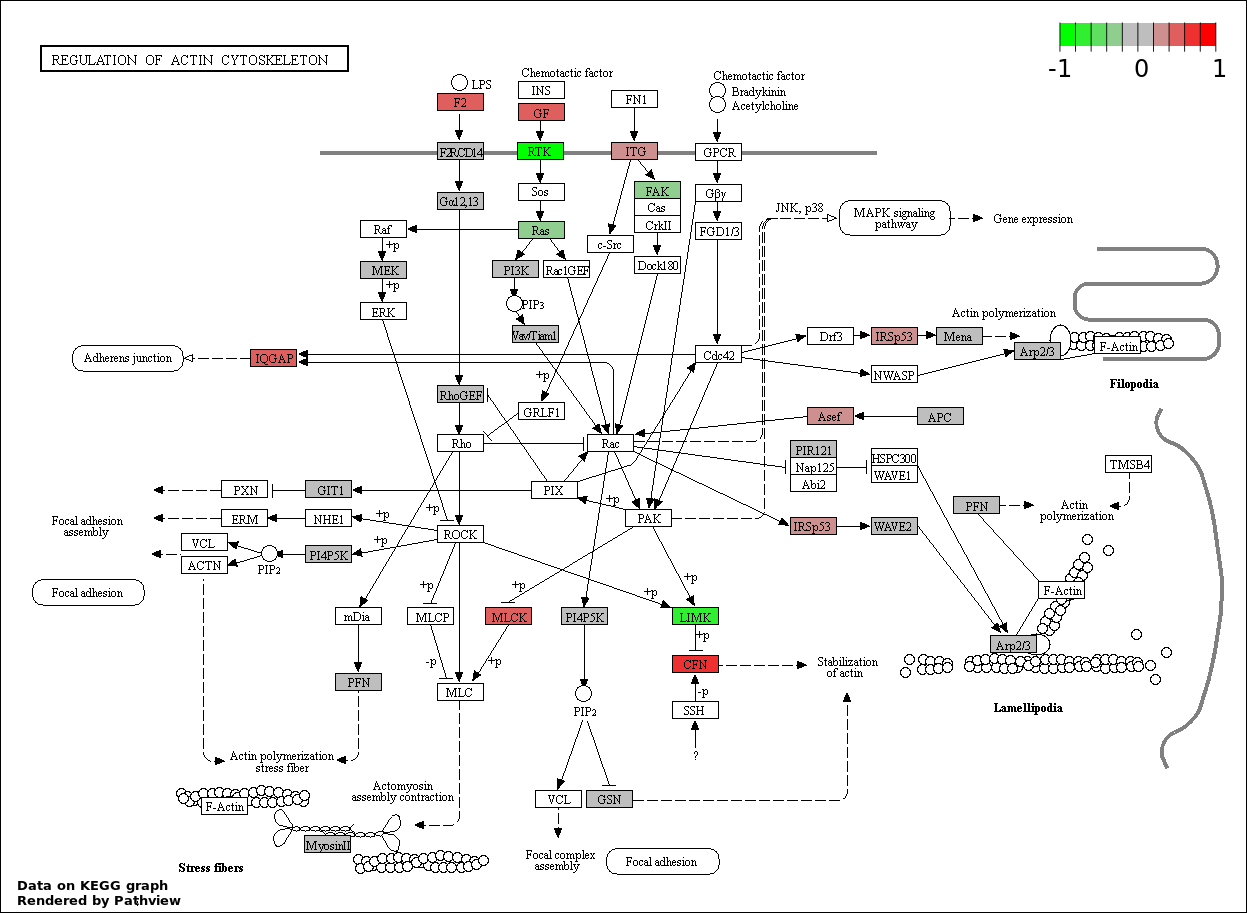
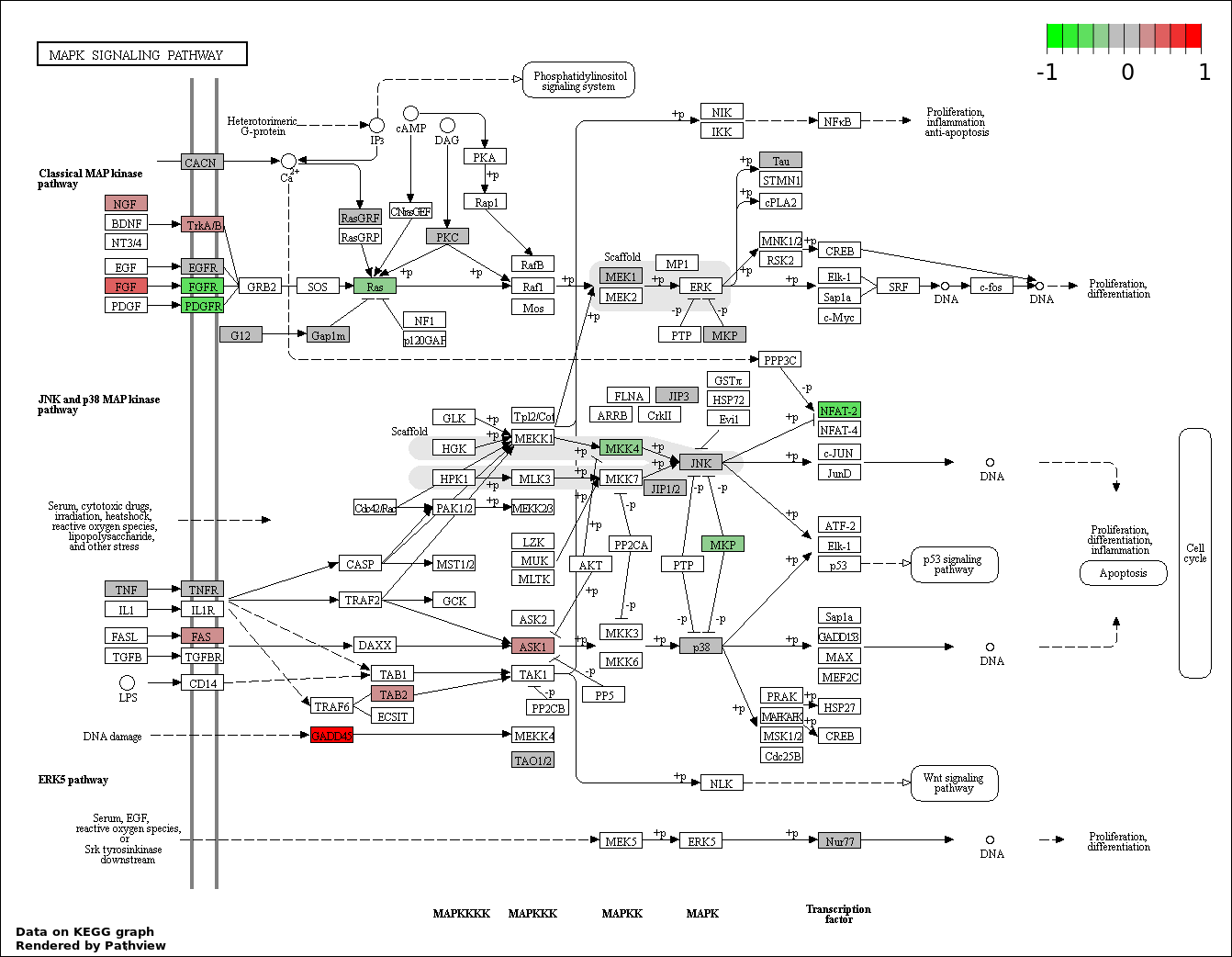
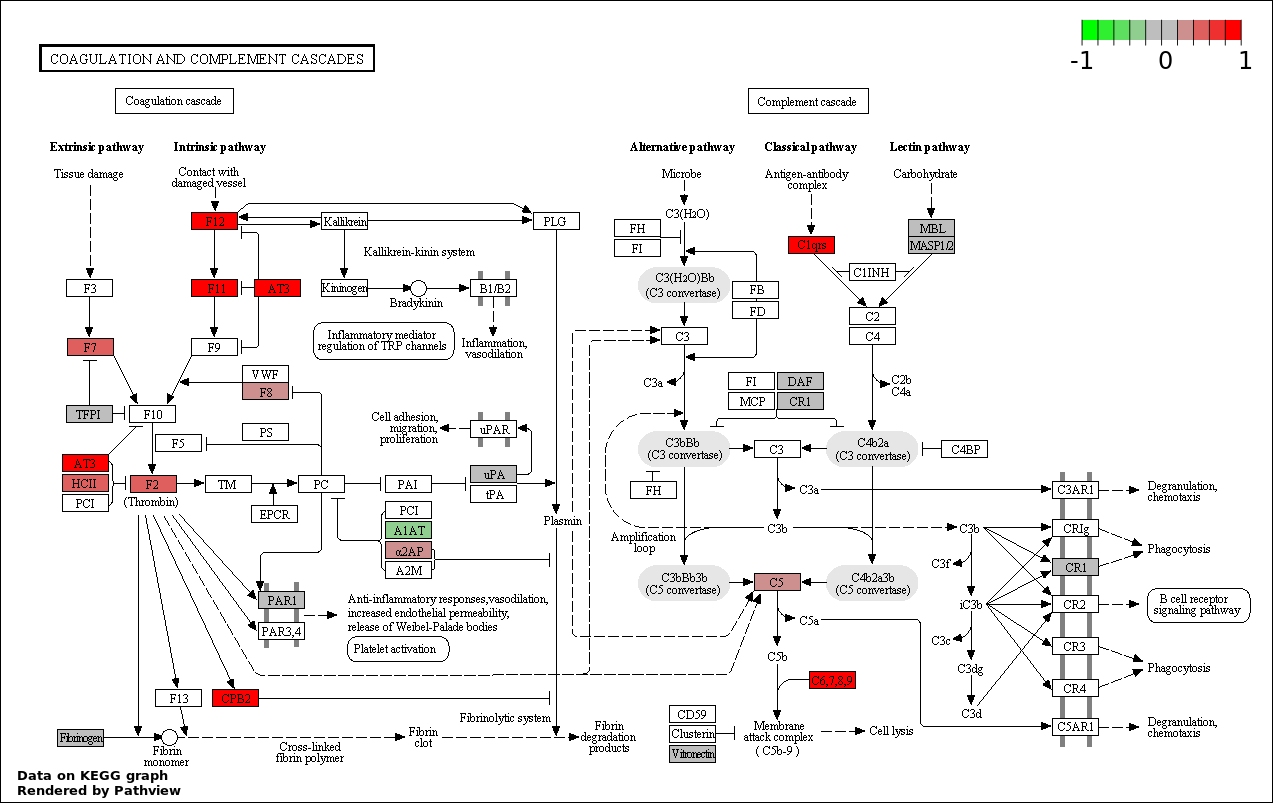
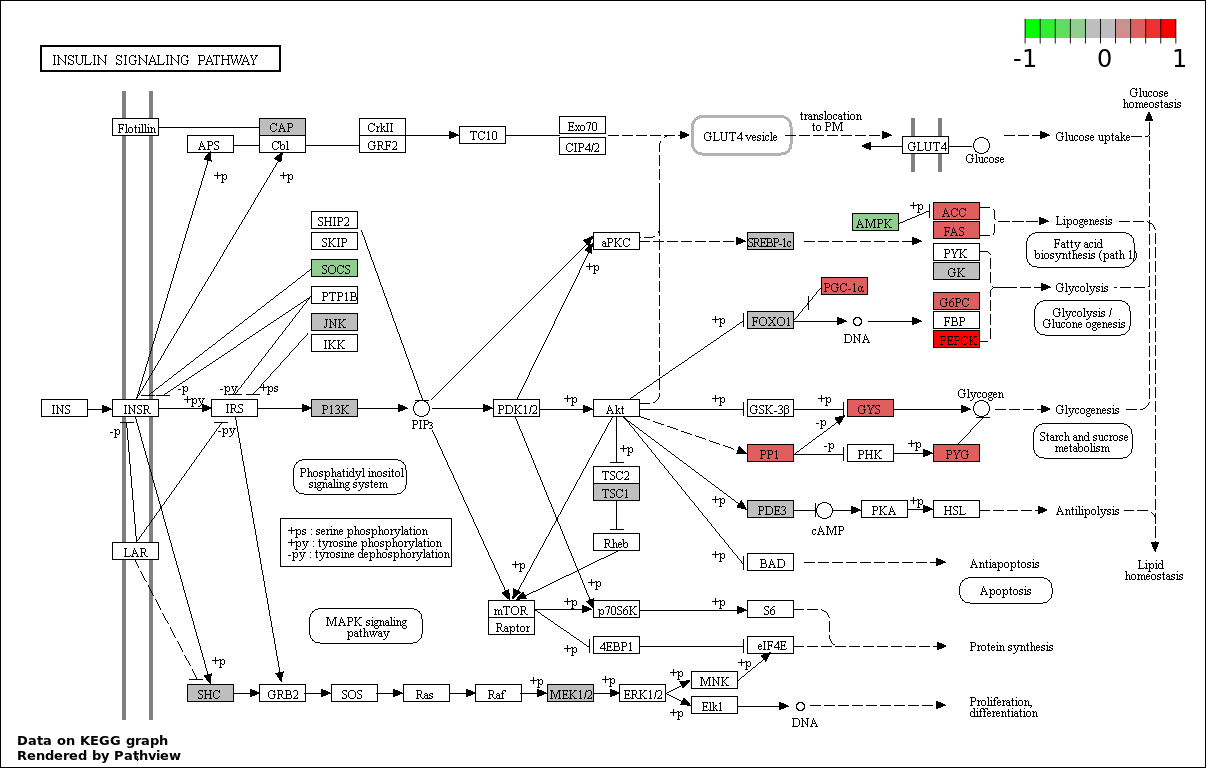
This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-100-5p | SMAD7 | 0.23 | 0.34446 | -0 | 0.98392 | miRNAWalker2 validate | -0.11 | 0.04468 | ||
| 2 | hsa-miR-106b-5p | PRRG1 | 0.14 | 0.51414 | 0.14 | 0.03454 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.07 | 0.02884 | ||
| 3 | hsa-miR-107 | DAPK1 | 0.33 | 0.15378 | 0.09 | 0.61779 | miRNAWalker2 validate; miRTarBase | -0.16 | 0.04008 | ||
| 4 | hsa-miR-10a-5p | CYP8B1 | -0.27 | 0.17524 | 1.39 | 0.01301 | miRNAWalker2 validate | -0.7 | 0.01373 | ||
| 5 | hsa-miR-10a-5p | MAPK8 | -0.27 | 0.17524 | 0.06 | 0.17218 | miRNAWalker2 validate | -0.05 | 0.0181 | ||
| 6 | hsa-miR-10a-5p | PYGL | -0.27 | 0.17524 | 0.55 | 0.04912 | miRNAWalker2 validate | -0.28 | 0.04613 | ||
| 7 | hsa-miR-10a-5p | ZNF618 | -0.27 | 0.17524 | 0.08 | 0.11003 | miRNAWalker2 validate; mirMAP | -0.07 | 0.00856 | ||
| 8 | hsa-miR-10b-5p | NRP2 | 0.02 | 0.84842 | -0.1 | 0.43013 | miRNAWalker2 validate; miRNATAP | -0.34 | 0.00069 | ||
| 9 | hsa-miR-122-5p | AACS | 0.56 | 0.1942 | -0.15 | 0.35856 | miRNAWalker2 validate; miRTarBase | -0.11 | 0.00342 | ||
| 10 | hsa-miR-122-5p | ALDOA | 0.56 | 0.1942 | -0.22 | 0.25911 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.11 | 0.02076 | ||
| 11 | hsa-miR-122-5p | CD83 | 0.56 | 0.1942 | -0.04 | 0.89012 | miRNAWalker2 validate | -0.13 | 0.02836 | ||
| 12 | hsa-miR-122-5p | CPA3 | 0.56 | 0.1942 | -0.56 | 0.019 | miRNAWalker2 validate | -0.13 | 0.01612 | ||
| 13 | hsa-miR-122-5p | DSTYK | 0.56 | 0.1942 | -0.23 | 0.02966 | miRNAWalker2 validate; miRTarBase | -0.06 | 0.01802 | ||
| 14 | hsa-miR-122-5p | DZIP1L | 0.56 | 0.1942 | -0.19 | 0.16432 | miRNAWalker2 validate | -0.09 | 0.00265 | ||
| 15 | hsa-miR-122-5p | EGLN3 | 0.56 | 0.1942 | -0.2 | 0.51644 | miRNAWalker2 validate; miRTarBase | -0.2 | 0.0047 | ||
| 16 | hsa-miR-122-5p | LMNB2 | 0.56 | 0.1942 | -0.2 | 0.25708 | miRNAWalker2 validate | -0.11 | 0.00881 | ||
| 17 | hsa-miR-122-5p | MPV17 | 0.56 | 0.1942 | -0.18 | 0.16125 | miRNAWalker2 validate | -0.06 | 0.04828 | ||
| 18 | hsa-miR-122-5p | PHPT1 | 0.56 | 0.1942 | -0.04 | 0.80549 | miRNAWalker2 validate | -0.07 | 0.0396 | ||
| 19 | hsa-miR-122-5p | PSMD10 | 0.56 | 0.1942 | -0.21 | 0.25615 | miRNAWalker2 validate | -0.1 | 0.02354 | ||
| 20 | hsa-miR-122-5p | RAB6B | 0.56 | 0.1942 | -0.13 | 0.28733 | miRNAWalker2 validate; miRTarBase | -0.1 | 0.00041 | ||
| 21 | hsa-miR-122-5p | TPD52L2 | 0.56 | 0.1942 | -0.01 | 0.92778 | miRNAWalker2 validate; miRTarBase | -0.06 | 0.01096 | ||
| 22 | hsa-miR-1226-3p | DDT | -0.06 | 0.23641 | 0.44 | 0.07272 | miRNAWalker2 validate | -1.14 | 0.03118 | ||
| 23 | hsa-miR-1236-3p | LDOC1L | 0.03 | 0.54197 | -0.55 | 0.037 | miRNAWalker2 validate | -1.07 | 0.03226 | ||
| 24 | hsa-miR-124-3p | AURKB | 0.11 | 0.13453 | -0.04 | 0.63356 | miRNAWalker2 validate | -0.29 | 0.02536 | ||
| 25 | hsa-miR-124-3p | CKS2 | 0.11 | 0.13453 | -0.29 | 0.20016 | miRNAWalker2 validate | -0.96 | 0.00263 | ||
| 26 | hsa-miR-124-3p | EFNA1 | 0.11 | 0.13453 | -0.12 | 0.63145 | miRNAWalker2 validate | -0.81 | 0.02575 | ||
| 27 | hsa-miR-124-3p | FAM83H | 0.11 | 0.13453 | -0.08 | 0.66576 | miRNAWalker2 validate | -0.62 | 0.02117 | ||
| 28 | hsa-miR-124-3p | IL18R1 | 0.11 | 0.13453 | -0.04 | 0.64416 | miRNAWalker2 validate | -0.26 | 0.04561 | ||
| 29 | hsa-miR-124-3p | SDC4 | 0.11 | 0.13453 | -0.12 | 0.52279 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.8 | 0.00274 | ||
| 30 | hsa-miR-125a-3p | IP6K2 | 0.22 | 0.22546 | 0.18 | 0.25596 | miRNAWalker2 validate | -0.2 | 0.01892 | ||
| 31 | hsa-miR-125a-5p | CDKN1A | -0.2 | 0.44397 | 0.06 | 0.775 | miRNAWalker2 validate; miRTarBase | -0.18 | 0.02762 | ||
| 32 | hsa-miR-125a-5p | CGNL1 | -0.2 | 0.44397 | 0.89 | 0.00929 | miRNAWalker2 validate; miRanda | -0.28 | 0.0393 | ||
| 33 | hsa-miR-125a-5p | NDEL1 | -0.2 | 0.44397 | 0.01 | 0.91143 | miRNAWalker2 validate | -0.07 | 0.04288 | ||
| 34 | hsa-miR-125b-5p | FUS | 0.25 | 0.38797 | -0.04 | 0.77376 | miRNAWalker2 validate | -0.09 | 0.04787 | ||
| 35 | hsa-miR-125b-5p | HMGA2 | 0.25 | 0.38797 | -0.6 | 0.02419 | miRNAWalker2 validate; miRTarBase | -0.2 | 0.03693 | ||
| 36 | hsa-miR-125b-5p | LIN28B | 0.25 | 0.38797 | -0.01 | 0.92264 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.07 | 0.01184 | ||
| 37 | hsa-miR-125b-5p | PLXND1 | 0.25 | 0.38797 | -0.16 | 0.08894 | miRNAWalker2 validate | -0.1 | 0.00284 | ||
| 38 | hsa-miR-125b-5p | PSAT1 | 0.25 | 0.38797 | -0.05 | 0.89936 | miRNAWalker2 validate | -0.31 | 0.01538 | ||
| 39 | hsa-miR-1301-3p | HIGD1A | -0.01 | 0.71722 | 0.24 | 0.34854 | miRNAWalker2 validate | -2.23 | 0.00692 | ||
| 40 | hsa-miR-130a-3p | BMPR2 | -0.02 | 0.92297 | 0.05 | 0.64147 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.11 | 0.0293 | ||
| 41 | hsa-miR-130b-3p | AKAP11 | 0.05 | 0.68208 | -0.13 | 0.58853 | miRNAWalker2 validate; miRNATAP | -0.35 | 0.04984 | ||
| 42 | hsa-miR-130b-3p | OTUD4 | 0.05 | 0.68208 | -0.08 | 0.45645 | miRNAWalker2 validate; miRNATAP | -0.18 | 0.0309 | ||
| 43 | hsa-miR-130b-3p | PTP4A1 | 0.05 | 0.68208 | 0.07 | 0.78319 | miRNAWalker2 validate; miRNATAP | -0.41 | 0.03394 | ||
| 44 | hsa-miR-130b-5p | FASN | 0.05 | 0.19689 | 0.53 | 0.057 | miRNAWalker2 validate | -1.67 | 0.0352 | ||
| 45 | hsa-miR-132-3p | ETNK2 | 0.03 | 0.63073 | 0.34 | 0.09005 | miRNAWalker2 validate | -0.94 | 0.01377 | ||
| 46 | hsa-miR-133a-3p | BCAN | 0.04 | 0.51945 | 0.09 | 0.05682 | miRNAWalker2 validate | -0.2 | 0.01057 | ||
| 47 | hsa-miR-133a-3p | EGFR | 0.04 | 0.51945 | -0.07 | 0.75424 | miRNAWalker2 validate; miRTarBase; MirTarget | -1.29 | 0.0009 | ||
| 48 | hsa-miR-133a-3p | HCN2 | 0.04 | 0.51945 | 0.14 | 0.51559 | miRNAWalker2 validate; miRTarBase | -0.71 | 0.04658 | ||
| 49 | hsa-miR-141-3p | PPARA | -0.44 | 8.0E-5 | 0.18 | 0.1046 | miRNAWalker2 validate; miRTarBase | -0.21 | 0.03014 | ||
| 50 | hsa-miR-142-3p | ABCC9 | 0.04 | 0.72618 | 0.41 | 0.12279 | miRNAWalker2 validate | -0.65 | 0.01135 | ||
| 51 | hsa-miR-142-3p | ETNK2 | 0.04 | 0.72618 | 0.34 | 0.09005 | miRNAWalker2 validate | -0.47 | 0.015 | ||
| 52 | hsa-miR-146a-5p | EGFR | 0.14 | 0.40939 | -0.07 | 0.75424 | miRNAWalker2 validate | -0.48 | 0.00056 | ||
| 53 | hsa-miR-148a-3p | CCNI | 0.17 | 0.27929 | -0.07 | 0.63194 | miRNAWalker2 validate | -0.26 | 0.00264 | ||
| 54 | hsa-miR-149-5p | AATF | -0.09 | 0.30273 | -0.02 | 0.83277 | miRNAWalker2 validate | -0.24 | 0.04933 | ||
| 55 | hsa-miR-149-5p | DDT | -0.09 | 0.30273 | 0.44 | 0.07272 | miRNAWalker2 validate | -0.81 | 0.00587 | ||
| 56 | hsa-miR-149-5p | ETS2 | -0.09 | 0.30273 | -0.02 | 0.92787 | miRNAWalker2 validate | -0.74 | 0.01714 | ||
| 57 | hsa-miR-151a-3p | CCNE1 | 0.04 | 0.16538 | -0.1 | 0.60975 | miRNAWalker2 validate | -1.43 | 0.03246 | ||
| 58 | hsa-miR-155-5p | AGL | 0.06 | 0.18374 | 0.51 | 0.11552 | miRNAWalker2 validate | -1.42 | 0.04025 | ||
| 59 | hsa-miR-155-5p | ANPEP | 0.06 | 0.18374 | 0.19 | 0.49064 | miRNAWalker2 validate | -1.36 | 0.02062 | ||
| 60 | hsa-miR-155-5p | B4GALT1 | 0.06 | 0.18374 | -0.12 | 0.46339 | miRNAWalker2 validate; mirMAP | -0.71 | 0.04268 | ||
| 61 | hsa-miR-155-5p | CPT1A | 0.06 | 0.18374 | 0.14 | 0.44457 | miRNAWalker2 validate | -0.83 | 0.02801 | ||
| 62 | hsa-miR-155-5p | CRAT | 0.06 | 0.18374 | 0.35 | 0.00789 | miRNAWalker2 validate | -0.67 | 0.01965 | ||
| 63 | hsa-miR-155-5p | CTAGE5 | 0.06 | 0.18374 | 0.18 | 0.39722 | miRNAWalker2 validate | -1.2 | 0.00793 | ||
| 64 | hsa-miR-155-5p | DMD | 0.06 | 0.18374 | 0 | 0.87312 | miRNAWalker2 validate | -0.14 | 0.02014 | ||
| 65 | hsa-miR-155-5p | EGFR | 0.06 | 0.18374 | -0.07 | 0.75424 | miRNAWalker2 validate; MirTarget | -1.74 | 0.00045 | 22213426 | Furthermore miR-155 knock-down in RInk-1 cells resulted in the inhibition of cell growth and colony formation consistent with down-regulation of EGFR MT1-MMP and K-Ras expression |
| 66 | hsa-miR-155-5p | GABARAPL1 | 0.06 | 0.18374 | 0.12 | 0.67753 | miRNAWalker2 validate | -1.92 | 0.00119 | ||
| 67 | hsa-miR-155-5p | GNPNAT1 | 0.06 | 0.18374 | -0.12 | 0.56006 | miRNAWalker2 validate | -1.29 | 0.00251 | ||
| 68 | hsa-miR-155-5p | GPT2 | 0.06 | 0.18374 | 0.59 | 0.14834 | miRNAWalker2 validate | -2.33 | 0.00781 | ||
| 69 | hsa-miR-155-5p | IL13RA1 | 0.06 | 0.18374 | 0.24 | 0.33165 | miRNAWalker2 validate; miRTarBase | -1.15 | 0.02879 | ||
| 70 | hsa-miR-155-5p | LNX2 | 0.06 | 0.18374 | -0.1 | 0.59732 | miRNAWalker2 validate | -1.2 | 0.00344 | ||
| 71 | hsa-miR-155-5p | LONP2 | 0.06 | 0.18374 | 0.3 | 0.1493 | miRNAWalker2 validate | -0.89 | 0.04417 | ||
| 72 | hsa-miR-155-5p | MYD88 | 0.06 | 0.18374 | 0.35 | 0.00943 | miRNAWalker2 validate | -0.63 | 0.03325 | ||
| 73 | hsa-miR-155-5p | MYO1E | 0.06 | 0.18374 | -0.11 | 0.56076 | miRNAWalker2 validate | -0.99 | 0.01525 | ||
| 74 | hsa-miR-155-5p | NAMPT | 0.06 | 0.18374 | -0.24 | 0.45229 | miRNAWalker2 validate; miRNATAP | -1.45 | 0.03271 | ||
| 75 | hsa-miR-155-5p | PHGDH | 0.06 | 0.18374 | 0.33 | 0.29662 | miRNAWalker2 validate | -1.84 | 0.00648 | 24938880 | Aim of this project was to determine expression of miR-155 miR-19a miR-181b miR-24 relative to let-7a in sera of 63 patients with EBC and 21 healthy controls |
| 76 | hsa-miR-155-5p | PPP2R2A | 0.06 | 0.18374 | -0.13 | 0.43859 | miRNAWalker2 validate | -0.74 | 0.03366 | ||
| 77 | hsa-miR-155-5p | PSAT1 | 0.06 | 0.18374 | -0.05 | 0.89936 | miRNAWalker2 validate | -1.83 | 0.02019 | ||
| 78 | hsa-miR-155-5p | RETSAT | 0.06 | 0.18374 | 0.2 | 0.27423 | miRNAWalker2 validate | -1.02 | 0.00927 | ||
| 79 | hsa-miR-155-5p | SLC27A2 | 0.06 | 0.18374 | 0.57 | 0.14759 | miRNAWalker2 validate | -2.29 | 0.00646 | ||
| 80 | hsa-miR-155-5p | TMBIM6 | 0.06 | 0.18374 | 0.18 | 0.28991 | miRNAWalker2 validate | -1.05 | 0.00395 | ||
| 81 | hsa-miR-15a-5p | HERC6 | 0.18 | 0.3674 | 0.04 | 0.83964 | miRNAWalker2 validate | -0.19 | 0.04047 | ||
| 82 | hsa-miR-15b-5p | CCND1 | 0.28 | 0.23748 | -0.28 | 0.35282 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.27 | 0.03772 | ||
| 83 | hsa-miR-15b-5p | PLSCR4 | 0.28 | 0.23748 | 0.02 | 0.93743 | miRNAWalker2 validate; MirTarget | -0.28 | 0.01393 | ||
| 84 | hsa-miR-15b-5p | SMAD7 | 0.28 | 0.23748 | -0 | 0.98392 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.12 | 0.04409 | ||
| 85 | hsa-miR-16-5p | CD274 | 0.28 | 0.3893 | -0.02 | 0.85592 | miRNAWalker2 validate; MirTarget | -0.06 | 0.04602 | ||
| 86 | hsa-miR-16-5p | HERC6 | 0.28 | 0.3893 | 0.04 | 0.83964 | miRNAWalker2 validate | -0.12 | 0.04806 | ||
| 87 | hsa-miR-16-5p | PLSCR4 | 0.28 | 0.3893 | 0.02 | 0.93743 | miRNAWalker2 validate; MirTarget | -0.18 | 0.02943 | ||
| 88 | hsa-miR-16-5p | PTGS1 | 0.28 | 0.3893 | -0.09 | 0.70066 | miRNAWalker2 validate | -0.15 | 0.04 | ||
| 89 | hsa-miR-16-5p | SAV1 | 0.28 | 0.3893 | 0.05 | 0.50417 | miRNAWalker2 validate | -0.05 | 0.02719 | ||
| 90 | hsa-miR-16-5p | SMAD7 | 0.28 | 0.3893 | -0 | 0.98392 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.11 | 0.00962 | ||
| 91 | hsa-miR-16-5p | USP53 | 0.28 | 0.3893 | 0.02 | 0.89833 | miRNAWalker2 validate; miRNATAP | -0.11 | 0.04178 | ||
| 92 | hsa-miR-17-5p | CCND1 | -0.07 | 0.79091 | -0.28 | 0.35282 | miRNAWalker2 validate; MirTarget; TargetScan; miRNATAP | -0.22 | 0.04711 | 18695042 | Mammary epithelial cell-targeted cyclin D1 expression induced miR-17-5p and miR-20a expression in vivo and cyclin D1 bound the miR-17/20 cluster promoter regulatory region |
| 93 | hsa-miR-17-5p | GPR137B | -0.07 | 0.79091 | -0.37 | 0.14479 | miRNAWalker2 validate; miRTarBase; TargetScan; miRNATAP | -0.21 | 0.02912 | ||
| 94 | hsa-miR-17-5p | NAT8L | -0.07 | 0.79091 | -0 | 0.99449 | miRNAWalker2 validate; mirMAP | -0.14 | 0.00713 | ||
| 95 | hsa-miR-181a-5p | AKAP12 | -0.11 | 0.64975 | 0.16 | 0.49994 | miRNAWalker2 validate | -0.23 | 0.0228 | ||
| 96 | hsa-miR-181a-5p | GADD45G | -0.11 | 0.64975 | 0.54 | 0.17956 | miRNAWalker2 validate | -0.42 | 0.01643 | ||
| 97 | hsa-miR-181a-5p | NKX3-2 | -0.11 | 0.64975 | 0.03 | 0.77376 | miRNAWalker2 validate; miRNATAP | -0.11 | 0.02381 | ||
| 98 | hsa-miR-181a-5p | TGFBR3 | -0.11 | 0.64975 | -0.01 | 0.97548 | miRNAWalker2 validate; mirMAP; miRNATAP | -0.21 | 0.00644 | ||
| 99 | hsa-miR-181d-5p | DHRS4 | -0.01 | 0.83654 | 0.53 | 0.00982 | miRNAWalker2 validate | -1.12 | 0.00529 | ||
| 100 | hsa-miR-183-5p | FAM188A | -0.05 | 0.42998 | -0.12 | 0.39256 | miRNAWalker2 validate | -0.45 | 0.02503 | ||
| 101 | hsa-miR-183-5p | INSIG1 | -0.05 | 0.42998 | 0.68 | 0.12159 | miRNAWalker2 validate | -1.33 | 0.04205 | ||
| 102 | hsa-miR-183-5p | ZFHX4 | -0.05 | 0.42998 | 0.4 | 0.0213 | miRNAWalker2 validate | -0.55 | 0.03655 | ||
| 103 | hsa-miR-185-5p | EPAS1 | 0.1 | 0.46192 | 0.02 | 0.92425 | miRNAWalker2 validate; miRTarBase; MirTarget | -0.43 | 0.00371 | ||
| 104 | hsa-miR-186-5p | TMBIM6 | 0.12 | 0.14218 | 0.18 | 0.28991 | miRNAWalker2 validate | -0.45 | 0.03688 | ||
| 105 | hsa-miR-186-5p | WLS | 0.12 | 0.14218 | 0.23 | 0.41764 | miRNAWalker2 validate | -0.69 | 0.04766 | ||
| 106 | hsa-miR-187-3p | TUBG1 | 0.02 | 0.70394 | 0.1 | 0.41207 | miRNAWalker2 validate | -0.57 | 0.03603 | ||
| 107 | hsa-miR-1914-3p | PTCD1 | 0.09 | 0.35632 | 0.19 | 0.21488 | miRNAWalker2 validate | -0.36 | 0.02266 | ||
| 108 | hsa-miR-192-5p | ARL4C | 0.45 | 0.16782 | -0.45 | 0.10109 | miRNAWalker2 validate | -0.19 | 0.01876 | ||
| 109 | hsa-miR-192-5p | B3GALNT1 | 0.45 | 0.16782 | -0.32 | 0.00364 | miRNAWalker2 validate | -0.11 | 0.0013 | ||
| 110 | hsa-miR-192-5p | CA8 | 0.45 | 0.16782 | -0.46 | 8.0E-5 | miRNAWalker2 validate | -0.08 | 0.02364 | ||
| 111 | hsa-miR-192-5p | CD83 | 0.45 | 0.16782 | -0.04 | 0.89012 | miRNAWalker2 validate | -0.16 | 0.04747 | ||
| 112 | hsa-miR-192-5p | CKLF | 0.45 | 0.16782 | -0.27 | 0.09823 | miRNAWalker2 validate | -0.1 | 0.0403 | ||
| 113 | hsa-miR-192-5p | CLIC1 | 0.45 | 0.16782 | -0.39 | 0.0253 | miRNAWalker2 validate | -0.16 | 0.00175 | ||
| 114 | hsa-miR-192-5p | ELOVL1 | 0.45 | 0.16782 | -0.14 | 0.20144 | miRNAWalker2 validate | -0.1 | 0.00187 | ||
| 115 | hsa-miR-192-5p | ENC1 | 0.45 | 0.16782 | 0.03 | 0.89957 | miRNAWalker2 validate | -0.17 | 0.02518 | ||
| 116 | hsa-miR-192-5p | FAIM | 0.45 | 0.16782 | -0.51 | 0.01338 | miRNAWalker2 validate | -0.13 | 0.04925 | ||
| 117 | hsa-miR-192-5p | FAM171B | 0.45 | 0.16782 | -0.38 | 0.02071 | miRNAWalker2 validate | -0.14 | 0.00707 | ||
| 118 | hsa-miR-192-5p | FHDC1 | 0.45 | 0.16782 | -0.22 | 0.03374 | miRNAWalker2 validate; MirTarget | -0.11 | 0.00028 | ||
| 119 | hsa-miR-192-5p | FZD7 | 0.45 | 0.16782 | -0.23 | 0.30795 | miRNAWalker2 validate | -0.23 | 0.00077 | ||
| 120 | hsa-miR-192-5p | GPC4 | 0.45 | 0.16782 | -0.29 | 0.16621 | miRNAWalker2 validate | -0.2 | 0.00121 | ||
| 121 | hsa-miR-192-5p | GSG2 | 0.45 | 0.16782 | -0.13 | 0.03853 | miRNAWalker2 validate | -0.05 | 0.00932 | ||
| 122 | hsa-miR-192-5p | ID4 | 0.45 | 0.16782 | -0.41 | 0.26313 | miRNAWalker2 validate | -0.38 | 0.00042 | ||
| 123 | hsa-miR-192-5p | KCTD12 | 0.45 | 0.16782 | -0.23 | 0.26808 | miRNAWalker2 validate | -0.15 | 0.02149 | ||
| 124 | hsa-miR-192-5p | LIMCH1 | 0.45 | 0.16782 | -0.11 | 0.422 | miRNAWalker2 validate | -0.09 | 0.02158 | ||
| 125 | hsa-miR-192-5p | LMNB2 | 0.45 | 0.16782 | -0.2 | 0.25708 | miRNAWalker2 validate | -0.14 | 0.00895 | ||
| 126 | hsa-miR-192-5p | LRP8 | 0.45 | 0.16782 | -0.33 | 0.13394 | miRNAWalker2 validate | -0.24 | 0.00019 | ||
| 127 | hsa-miR-192-5p | MCM3 | 0.45 | 0.16782 | -0.24 | 0.2025 | miRNAWalker2 validate | -0.12 | 0.0373 | ||
| 128 | hsa-miR-192-5p | MED22 | 0.45 | 0.16782 | -0.15 | 0.33307 | miRNAWalker2 validate; mirMAP | -0.15 | 0.00117 | ||
| 129 | hsa-miR-192-5p | MYB | 0.45 | 0.16782 | -0.31 | 0.10306 | miRNAWalker2 validate | -0.18 | 0.00142 | ||
| 130 | hsa-miR-192-5p | PLAU | 0.45 | 0.16782 | -0.07 | 0.82417 | miRNAWalker2 validate | -0.26 | 0.0047 | ||
| 131 | hsa-miR-192-5p | PODXL | 0.45 | 0.16782 | -0.05 | 0.87612 | miRNAWalker2 validate | -0.2 | 0.0299 | ||
| 132 | hsa-miR-192-5p | PRICKLE1 | 0.45 | 0.16782 | -0.05 | 0.80973 | miRNAWalker2 validate | -0.17 | 0.00325 | ||
| 133 | hsa-miR-192-5p | RASSF8 | 0.45 | 0.16782 | -0.06 | 0.59041 | miRNAWalker2 validate | -0.07 | 0.02772 | ||
| 134 | hsa-miR-192-5p | RCC2 | 0.45 | 0.16782 | -0.37 | 0.05119 | miRNAWalker2 validate | -0.21 | 0.00032 | ||
| 135 | hsa-miR-192-5p | SNRPD1 | 0.45 | 0.16782 | -0.4 | 0.00671 | miRNAWalker2 validate | -0.1 | 0.03412 | ||
| 136 | hsa-miR-192-5p | SPARC | 0.45 | 0.16782 | -0.24 | 0.42268 | miRNAWalker2 validate | -0.18 | 0.04775 | ||
| 137 | hsa-miR-192-5p | STIL | 0.45 | 0.16782 | -0.24 | 0.01568 | miRNAWalker2 validate | -0.07 | 0.02535 | ||
| 138 | hsa-miR-192-5p | TBC1D22B | 0.45 | 0.16782 | -0.03 | 0.69509 | miRNAWalker2 validate | -0.06 | 0.03101 | ||
| 139 | hsa-miR-192-5p | TNFSF13 | 0.45 | 0.16782 | -0.11 | 0.48782 | miRNAWalker2 validate | -0.16 | 0.00123 | ||
| 140 | hsa-miR-192-5p | TTF2 | 0.45 | 0.16782 | -0.12 | 0.20487 | miRNAWalker2 validate | -0.08 | 0.00614 | ||
| 141 | hsa-miR-192-5p | ZNF519 | 0.45 | 0.16782 | -0.24 | 0.0018 | miRNAWalker2 validate | -0.08 | 0.00047 | ||
| 142 | hsa-miR-193b-3p | APOBEC3B | 0.02 | 0.92751 | 0.09 | 0.52251 | miRNAWalker2 validate | -0.14 | 0.03945 | ||
| 143 | hsa-miR-193b-3p | ATP13A2 | 0.02 | 0.92751 | -0.03 | 0.64481 | miRNAWalker2 validate | -0.06 | 0.02788 | ||
| 144 | hsa-miR-193b-3p | CDK4 | 0.02 | 0.92751 | -0.12 | 0.45614 | miRNAWalker2 validate | -0.19 | 0.0174 | ||
| 145 | hsa-miR-193b-3p | DOK2 | 0.02 | 0.92751 | 0.05 | 0.65109 | miRNAWalker2 validate | -0.12 | 0.03329 | ||
| 146 | hsa-miR-193b-3p | HM13 | 0.02 | 0.92751 | -0 | 0.98157 | miRNAWalker2 validate | -0.16 | 0.0119 | ||
| 147 | hsa-miR-193b-3p | IRAK1 | 0.02 | 0.92751 | -0.15 | 0.29128 | miRNAWalker2 validate | -0.15 | 0.04031 | ||
| 148 | hsa-miR-193b-3p | NLRP3 | 0.02 | 0.92751 | 0.03 | 0.89168 | miRNAWalker2 validate | -0.26 | 0.01848 | ||
| 149 | hsa-miR-193b-3p | SNRPB | 0.02 | 0.92751 | -0.08 | 0.62898 | miRNAWalker2 validate | -0.17 | 0.04746 | ||
| 150 | hsa-miR-194-5p | PTPN13 | 0.47 | 0.15528 | -0.51 | 0.07673 | miRNAWalker2 validate; miRTarBase | -0.27 | 0.00152 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | ORGANIC ACID METABOLIC PROCESS | 224 | 953 | 2.859e-31 | 1.33e-27 |
| 2 | SMALL MOLECULE METABOLIC PROCESS | 345 | 1767 | 7.482e-31 | 1.741e-27 |
| 3 | MONOCARBOXYLIC ACID METABOLIC PROCESS | 138 | 503 | 2.996e-26 | 4.647e-23 |
| 4 | CARBOXYLIC ACID CATABOLIC PROCESS | 74 | 205 | 3.656e-22 | 3.402e-19 |
| 5 | ORGANIC ACID CATABOLIC PROCESS | 74 | 205 | 3.656e-22 | 3.402e-19 |
| 6 | RESPONSE TO ENDOGENOUS STIMULUS | 272 | 1450 | 1.676e-21 | 1.299e-18 |
| 7 | RESPONSE TO OXYGEN CONTAINING COMPOUND | 260 | 1381 | 8.902e-21 | 5.917e-18 |
| 8 | SMALL MOLECULE CATABOLIC PROCESS | 94 | 328 | 1.231e-19 | 7.158e-17 |
| 9 | RESPONSE TO ORGANIC CYCLIC COMPOUND | 188 | 917 | 4.097e-19 | 2.118e-16 |
| 10 | SINGLE ORGANISM CATABOLIC PROCESS | 193 | 957 | 8.655e-19 | 4.027e-16 |
| 11 | RESPONSE TO EXTERNAL STIMULUS | 313 | 1821 | 1.186e-18 | 5.018e-16 |
| 12 | SMALL MOLECULE BIOSYNTHETIC PROCESS | 112 | 443 | 1.601e-18 | 6.21e-16 |
| 13 | CELLULAR RESPONSE TO ORGANIC SUBSTANCE | 313 | 1848 | 1.075e-17 | 3.846e-15 |
| 14 | RESPONSE TO LIPID | 179 | 888 | 1.849e-17 | 6.146e-15 |
| 15 | LIPID METABOLIC PROCESS | 217 | 1158 | 3.674e-17 | 1.14e-14 |
| 16 | RESPONSE TO HORMONE | 178 | 893 | 7.289e-17 | 2.12e-14 |
| 17 | CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND | 163 | 799 | 1.781e-16 | 4.876e-14 |
| 18 | CARDIOVASCULAR SYSTEM DEVELOPMENT | 160 | 788 | 5.358e-16 | 1.312e-13 |
| 19 | CIRCULATORY SYSTEM DEVELOPMENT | 160 | 788 | 5.358e-16 | 1.312e-13 |
| 20 | CARBOXYLIC ACID BIOSYNTHETIC PROCESS | 76 | 270 | 1.167e-15 | 2.586e-13 |
| 21 | ORGANIC ACID BIOSYNTHETIC PROCESS | 76 | 270 | 1.167e-15 | 2.586e-13 |
| 22 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 190 | 1008 | 2.246e-15 | 4.75e-13 |
| 23 | VASCULATURE DEVELOPMENT | 109 | 469 | 2.796e-15 | 5.655e-13 |
| 24 | ORGANIC HYDROXY COMPOUND METABOLIC PROCESS | 111 | 482 | 3.046e-15 | 5.905e-13 |
| 25 | RESPONSE TO NITROGEN COMPOUND | 167 | 859 | 7.492e-15 | 1.394e-12 |
| 26 | OXIDATION REDUCTION PROCESS | 171 | 898 | 2.451e-14 | 4.36e-12 |
| 27 | FATTY ACID METABOLIC PROCESS | 78 | 296 | 2.53e-14 | 4.36e-12 |
| 28 | ORGANONITROGEN COMPOUND CATABOLIC PROCESS | 86 | 343 | 2.701e-14 | 4.488e-12 |
| 29 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 275 | 1672 | 5.845e-14 | 9.378e-12 |
| 30 | CELLULAR AMINO ACID CATABOLIC PROCESS | 42 | 112 | 7.641e-14 | 1.185e-11 |
| 31 | CELLULAR LIPID METABOLIC PROCESS | 171 | 913 | 1.122e-13 | 1.685e-11 |
| 32 | REGULATION OF CELL DIFFERENTIATION | 250 | 1492 | 1.205e-13 | 1.752e-11 |
| 33 | CHEMICAL HOMEOSTASIS | 165 | 874 | 1.552e-13 | 2.189e-11 |
| 34 | RESPONSE TO EXTRACELLULAR STIMULUS | 100 | 441 | 2.047e-13 | 2.802e-11 |
| 35 | CELLULAR RESPONSE TO HORMONE STIMULUS | 117 | 552 | 2.616e-13 | 3.478e-11 |
| 36 | EMBRYO DEVELOPMENT | 166 | 894 | 5.616e-13 | 7.259e-11 |
| 37 | BLOOD VESSEL MORPHOGENESIS | 86 | 364 | 9.562e-13 | 1.202e-10 |
| 38 | CELL DEVELOPMENT | 237 | 1426 | 1.369e-12 | 1.676e-10 |
| 39 | CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 109 | 513 | 1.482e-12 | 1.768e-10 |
| 40 | REGULATION OF TRANSPORT | 286 | 1804 | 1.55e-12 | 1.803e-10 |
| 41 | CELLULAR RESPONSE TO LIPID | 100 | 457 | 2.023e-12 | 2.296e-10 |
| 42 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 232 | 1395 | 2.274e-12 | 2.519e-10 |
| 43 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 261 | 1618 | 2.573e-12 | 2.784e-10 |
| 44 | MONOCARBOXYLIC ACID CATABOLIC PROCESS | 36 | 96 | 4.542e-12 | 4.804e-10 |
| 45 | STEROID METABOLIC PROCESS | 63 | 237 | 5.14e-12 | 5.314e-10 |
| 46 | CELLULAR RESPONSE TO NITROGEN COMPOUND | 106 | 505 | 6.643e-12 | 6.719e-10 |
| 47 | TISSUE DEVELOPMENT | 246 | 1518 | 7.399e-12 | 7.325e-10 |
| 48 | SINGLE ORGANISM BIOSYNTHETIC PROCESS | 222 | 1340 | 1.049e-11 | 1.017e-09 |
| 49 | RESPONSE TO STEROID HORMONE | 104 | 497 | 1.277e-11 | 1.213e-09 |
| 50 | CELLULAR RESPONSE TO ORGANIC CYCLIC COMPOUND | 99 | 465 | 1.413e-11 | 1.315e-09 |
| 51 | RESPONSE TO PEPTIDE | 89 | 404 | 2.295e-11 | 2.094e-09 |
| 52 | ORGAN MORPHOGENESIS | 153 | 841 | 2.408e-11 | 2.155e-09 |
| 53 | RESPONSE TO DRUG | 93 | 431 | 2.733e-11 | 2.4e-09 |
| 54 | PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS | 302 | 1977 | 2.945e-11 | 2.537e-09 |
| 55 | LOCOMOTION | 190 | 1114 | 3.067e-11 | 2.595e-09 |
| 56 | REGULATION OF CELLULAR COMPONENT MOVEMENT | 142 | 771 | 5.311e-11 | 4.413e-09 |
| 57 | RESPONSE TO ALCOHOL | 81 | 362 | 8.11e-11 | 6.62e-09 |
| 58 | BIOLOGICAL ADHESION | 177 | 1032 | 9.672e-11 | 7.759e-09 |
| 59 | NEURON PROJECTION MORPHOGENESIS | 87 | 402 | 1.019e-10 | 8.035e-09 |
| 60 | REGULATION OF CELL PROLIFERATION | 238 | 1496 | 1.081e-10 | 8.381e-09 |
| 61 | INTRACELLULAR SIGNAL TRANSDUCTION | 247 | 1572 | 1.604e-10 | 1.223e-08 |
| 62 | REGULATION OF KINASE ACTIVITY | 140 | 776 | 3.179e-10 | 2.386e-08 |
| 63 | TUBE DEVELOPMENT | 108 | 552 | 3.444e-10 | 2.543e-08 |
| 64 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 235 | 1492 | 3.765e-10 | 2.737e-08 |
| 65 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 175 | 1036 | 4.248e-10 | 2.995e-08 |
| 66 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 175 | 1036 | 4.248e-10 | 2.995e-08 |
| 67 | NEURON DEVELOPMENT | 127 | 687 | 4.489e-10 | 3.118e-08 |
| 68 | CELL MORPHOGENESIS INVOLVED IN NEURON DIFFERENTIATION | 80 | 368 | 4.6e-10 | 3.148e-08 |
| 69 | CENTRAL NERVOUS SYSTEM DEVELOPMENT | 152 | 872 | 6.905e-10 | 4.656e-08 |
| 70 | TAXIS | 94 | 464 | 7.539e-10 | 5.011e-08 |
| 71 | COENZYME METABOLIC PROCESS | 63 | 265 | 7.976e-10 | 5.227e-08 |
| 72 | LONG CHAIN FATTY ACID METABOLIC PROCESS | 31 | 88 | 8.434e-10 | 5.451e-08 |
| 73 | HEART DEVELOPMENT | 94 | 466 | 9.526e-10 | 5.99e-08 |
| 74 | RESPONSE TO ABIOTIC STIMULUS | 172 | 1024 | 9.399e-10 | 5.99e-08 |
| 75 | CELLULAR COMPONENT MORPHOGENESIS | 155 | 900 | 1.124e-09 | 6.974e-08 |
| 76 | GLAND DEVELOPMENT | 83 | 395 | 1.232e-09 | 7.543e-08 |
| 77 | CELLULAR RESPONSE TO PEPTIDE | 64 | 274 | 1.26e-09 | 7.616e-08 |
| 78 | CATABOLIC PROCESS | 268 | 1773 | 1.349e-09 | 8.046e-08 |
| 79 | ALPHA AMINO ACID CATABOLIC PROCESS | 32 | 95 | 1.656e-09 | 9.752e-08 |
| 80 | ANATOMICAL STRUCTURE FORMATION INVOLVED IN MORPHOGENESIS | 162 | 957 | 1.684e-09 | 9.797e-08 |
| 81 | COFACTOR METABOLIC PROCESS | 73 | 334 | 2.065e-09 | 1.186e-07 |
| 82 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 170 | 1021 | 2.304e-09 | 1.307e-07 |
| 83 | ALCOHOL METABOLIC PROCESS | 75 | 348 | 2.403e-09 | 1.347e-07 |
| 84 | HOMEOSTATIC PROCESS | 211 | 1337 | 2.78e-09 | 1.54e-07 |
| 85 | NEURON PROJECTION DEVELOPMENT | 104 | 545 | 3.047e-09 | 1.668e-07 |
| 86 | REGULATION OF CELLULAR LOCALIZATION | 203 | 1277 | 3.14e-09 | 1.699e-07 |
| 87 | REGULATION OF CELL DEATH | 228 | 1472 | 3.239e-09 | 1.732e-07 |
| 88 | HEAD DEVELOPMENT | 127 | 709 | 3.414e-09 | 1.805e-07 |
| 89 | CELLULAR AMINO ACID METABOLIC PROCESS | 72 | 332 | 3.778e-09 | 1.975e-07 |
| 90 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 185 | 1142 | 3.888e-09 | 2.01e-07 |
| 91 | ION HOMEOSTASIS | 108 | 576 | 4.16e-09 | 2.127e-07 |
| 92 | REGULATION OF LIPID METABOLIC PROCESS | 64 | 282 | 4.272e-09 | 2.16e-07 |
| 93 | NEURON DIFFERENTIATION | 149 | 874 | 4.835e-09 | 2.419e-07 |
| 94 | ION TRANSPORT | 200 | 1262 | 5.332e-09 | 2.64e-07 |
| 95 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 267 | 1791 | 5.657e-09 | 2.771e-07 |
| 96 | REGULATION OF CELL ADHESION | 115 | 629 | 5.844e-09 | 2.833e-07 |
| 97 | ARTERY DEVELOPMENT | 27 | 75 | 5.945e-09 | 2.852e-07 |
| 98 | ALPHA AMINO ACID METABOLIC PROCESS | 55 | 229 | 6.342e-09 | 3.011e-07 |
| 99 | EMBRYONIC MORPHOGENESIS | 102 | 539 | 6.83e-09 | 3.21e-07 |
| 100 | ORGANONITROGEN COMPOUND METABOLIC PROCESS | 267 | 1796 | 7.396e-09 | 3.442e-07 |
| 101 | ANGIOGENESIS | 65 | 293 | 8.43e-09 | 3.884e-07 |
| 102 | RESPONSE TO ACID CHEMICAL | 69 | 319 | 8.894e-09 | 4.057e-07 |
| 103 | MONOCARBOXYLIC ACID BIOSYNTHETIC PROCESS | 45 | 172 | 9.319e-09 | 4.21e-07 |
| 104 | MUSCLE STRUCTURE DEVELOPMENT | 86 | 432 | 9.432e-09 | 4.22e-07 |
| 105 | DEVELOPMENTAL GROWTH | 71 | 333 | 1.022e-08 | 4.528e-07 |
| 106 | LIPID LOCALIZATION | 60 | 264 | 1.221e-08 | 5.359e-07 |
| 107 | RESPIRATORY SYSTEM DEVELOPMENT | 49 | 197 | 1.256e-08 | 5.463e-07 |
| 108 | ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY | 122 | 689 | 1.397e-08 | 6.02e-07 |
| 109 | RESPONSE TO FATTY ACID | 28 | 83 | 1.642e-08 | 7.008e-07 |
| 110 | GROWTH | 82 | 410 | 1.673e-08 | 7.079e-07 |
| 111 | ORGANIC ACID TRANSPORT | 59 | 261 | 1.982e-08 | 8.276e-07 |
| 112 | NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 161 | 983 | 1.992e-08 | 8.276e-07 |
| 113 | BILE ACID METABOLIC PROCESS | 17 | 35 | 2.088e-08 | 8.596e-07 |
| 114 | REGULATION OF TRANSFERASE ACTIVITY | 156 | 946 | 2.11e-08 | 8.61e-07 |
| 115 | REGULATION OF PROTEIN MODIFICATION PROCESS | 253 | 1710 | 2.987e-08 | 1.209e-06 |
| 116 | POSITIVE REGULATION OF TRANSPORT | 154 | 936 | 3.037e-08 | 1.218e-06 |
| 117 | MOVEMENT OF CELL OR SUBCELLULAR COMPONENT | 198 | 1275 | 3.173e-08 | 1.262e-06 |
| 118 | RESPONSE TO NUTRIENT | 47 | 191 | 3.535e-08 | 1.394e-06 |
| 119 | REGULATION OF ANATOMICAL STRUCTURE SIZE | 90 | 472 | 3.656e-08 | 1.429e-06 |
| 120 | ANION TRANSPORT | 95 | 507 | 3.703e-08 | 1.436e-06 |
| 121 | TELENCEPHALON DEVELOPMENT | 53 | 228 | 3.854e-08 | 1.482e-06 |
| 122 | NEGATIVE REGULATION OF CELL DEATH | 145 | 872 | 3.99e-08 | 1.522e-06 |
| 123 | REGULATION OF RESPONSE TO WOUNDING | 81 | 413 | 4.999e-08 | 1.891e-06 |
| 124 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 245 | 1656 | 5.107e-08 | 1.904e-06 |
| 125 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 179 | 1135 | 5.114e-08 | 1.904e-06 |
| 126 | EMBRYO DEVELOPMENT ENDING IN BIRTH OR EGG HATCHING | 101 | 554 | 5.75e-08 | 2.107e-06 |
| 127 | CARBOHYDRATE METABOLIC PROCESS | 116 | 662 | 5.722e-08 | 2.107e-06 |
| 128 | RESPONSE TO CORTICOSTEROID | 44 | 176 | 5.823e-08 | 2.117e-06 |
| 129 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE SIGNALING PATHWAY | 93 | 498 | 6.069e-08 | 2.189e-06 |
| 130 | CELLULAR CHEMICAL HOMEOSTASIS | 103 | 570 | 6.733e-08 | 2.41e-06 |
| 131 | REGULATION OF MEMBRANE POTENTIAL | 70 | 343 | 8.123e-08 | 2.863e-06 |
| 132 | CELLULAR RESPONSE TO ALCOHOL | 33 | 115 | 8.069e-08 | 2.863e-06 |
| 133 | BEHAVIOR | 95 | 516 | 8.833e-08 | 3.09e-06 |
| 134 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 137 | 823 | 8.951e-08 | 3.108e-06 |
| 135 | CELLULAR RESPONSE TO FATTY ACID | 20 | 51 | 1.064e-07 | 3.669e-06 |
| 136 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 226 | 1518 | 1.081e-07 | 3.699e-06 |
| 137 | SULFUR COMPOUND METABOLIC PROCESS | 72 | 359 | 1.1e-07 | 3.738e-06 |
| 138 | TISSUE MORPHOGENESIS | 97 | 533 | 1.147e-07 | 3.846e-06 |
| 139 | NEGATIVE REGULATION OF RESPONSE TO STIMULUS | 206 | 1360 | 1.149e-07 | 3.846e-06 |
| 140 | CELL MOTILITY | 138 | 835 | 1.24e-07 | 4.008e-06 |
| 141 | BILE ACID BIOSYNTHETIC PROCESS | 12 | 20 | 1.234e-07 | 4.008e-06 |
| 142 | LOCALIZATION OF CELL | 138 | 835 | 1.24e-07 | 4.008e-06 |
| 143 | ORGANIC ANION TRANSPORT | 76 | 387 | 1.216e-07 | 4.008e-06 |
| 144 | NEGATIVE REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 59 | 274 | 1.228e-07 | 4.008e-06 |
| 145 | REGULATION OF NEURON PROJECTION DEVELOPMENT | 79 | 408 | 1.278e-07 | 4.099e-06 |
| 146 | NEUROGENESIS | 211 | 1402 | 1.309e-07 | 4.172e-06 |
| 147 | CELLULAR RESPONSE TO ACID CHEMICAL | 43 | 175 | 1.39e-07 | 4.399e-06 |
| 148 | RESPONSE TO KETONE | 44 | 182 | 1.646e-07 | 5.175e-06 |
| 149 | SINGLE ORGANISM BEHAVIOR | 75 | 384 | 1.846e-07 | 5.764e-06 |
| 150 | REGULATION OF MUSCLE SYSTEM PROCESS | 46 | 195 | 1.888e-07 | 5.857e-06 |
| 151 | REGULATION OF SECRETION | 119 | 699 | 1.919e-07 | 5.914e-06 |
| 152 | PHOSPHORYLATION | 188 | 1228 | 2.029e-07 | 6.21e-06 |
| 153 | EMBRYONIC ORGAN DEVELOPMENT | 78 | 406 | 2.139e-07 | 6.504e-06 |
| 154 | CIRCADIAN RHYTHM | 36 | 137 | 2.435e-07 | 7.31e-06 |
| 155 | VITAMIN METABOLIC PROCESS | 33 | 120 | 2.432e-07 | 7.31e-06 |
| 156 | CELLULAR AMINO ACID BIOSYNTHETIC PROCESS | 28 | 93 | 2.54e-07 | 7.575e-06 |
| 157 | REGULATION OF HORMONE LEVELS | 88 | 478 | 2.63e-07 | 7.793e-06 |
| 158 | EPITHELIUM DEVELOPMENT | 151 | 945 | 2.675e-07 | 7.877e-06 |
| 159 | CARDIAC SEPTUM MORPHOGENESIS | 19 | 49 | 2.728e-07 | 7.984e-06 |
| 160 | RESPONSE TO MECHANICAL STIMULUS | 48 | 210 | 2.842e-07 | 8.266e-06 |
| 161 | FATTY ACID CATABOLIC PROCESS | 24 | 73 | 2.956e-07 | 8.543e-06 |
| 162 | NEGATIVE REGULATION OF RESPONSE TO WOUNDING | 39 | 156 | 3.229e-07 | 9.273e-06 |
| 163 | MUSCLE TISSUE DEVELOPMENT | 58 | 275 | 3.275e-07 | 9.35e-06 |
| 164 | RESPONSE TO XENOBIOTIC STIMULUS | 30 | 105 | 3.429e-07 | 9.611e-06 |
| 165 | RESPONSE TO INSULIN | 47 | 205 | 3.426e-07 | 9.611e-06 |
| 166 | NEURON PROJECTION GUIDANCE | 47 | 205 | 3.426e-07 | 9.611e-06 |
| 167 | REGULATION OF GROWTH | 109 | 633 | 3.537e-07 | 9.855e-06 |
| 168 | AORTA DEVELOPMENT | 17 | 41 | 3.71e-07 | 1.01e-05 |
| 169 | REGULATION OF PROTEIN LOCALIZATION | 151 | 950 | 3.701e-07 | 1.01e-05 |
| 170 | CELLULAR RESPONSE TO STEROID HORMONE STIMULUS | 49 | 218 | 3.672e-07 | 1.01e-05 |
| 171 | MUSCLE CELL DIFFERENTIATION | 52 | 237 | 3.649e-07 | 1.01e-05 |
| 172 | MUSCLE CELL DEVELOPMENT | 34 | 128 | 3.951e-07 | 1.069e-05 |
| 173 | REGULATION OF GLUCOSE METABOLIC PROCESS | 30 | 106 | 4.296e-07 | 1.156e-05 |
| 174 | POSITIVE REGULATION OF LOCOMOTION | 79 | 420 | 4.39e-07 | 1.174e-05 |
| 175 | REGULATION OF REACTIVE OXYGEN SPECIES METABOLIC PROCESS | 38 | 152 | 4.549e-07 | 1.21e-05 |
| 176 | RESPONSE TO PURINE CONTAINING COMPOUND | 39 | 158 | 4.598e-07 | 1.216e-05 |
| 177 | CARDIAC SEPTUM DEVELOPMENT | 26 | 85 | 4.784e-07 | 1.258e-05 |
| 178 | RHYTHMIC PROCESS | 61 | 298 | 4.873e-07 | 1.274e-05 |
| 179 | PALLIUM DEVELOPMENT | 38 | 153 | 5.434e-07 | 1.413e-05 |
| 180 | RESPONSE TO INORGANIC SUBSTANCE | 87 | 479 | 5.589e-07 | 1.445e-05 |
| 181 | LIPID CATABOLIC PROCESS | 53 | 247 | 5.922e-07 | 1.523e-05 |
| 182 | REGULATION OF NEUROTRANSMITTER LEVELS | 44 | 190 | 5.975e-07 | 1.528e-05 |
| 183 | RESPONSE TO TRANSITION METAL NANOPARTICLE | 37 | 148 | 6.411e-07 | 1.63e-05 |
| 184 | SECOND MESSENGER MEDIATED SIGNALING | 39 | 160 | 6.494e-07 | 1.633e-05 |
| 185 | REGULATION OF CELL DEVELOPMENT | 135 | 836 | 6.475e-07 | 1.633e-05 |
| 186 | CELLULAR CATABOLIC PROCESS | 197 | 1322 | 7.13e-07 | 1.784e-05 |
| 187 | POSITIVE REGULATION OF KINASE ACTIVITY | 87 | 482 | 7.342e-07 | 1.827e-05 |
| 188 | FOREBRAIN DEVELOPMENT | 69 | 357 | 8.412e-07 | 2.082e-05 |
| 189 | CARDIAC CHAMBER DEVELOPMENT | 36 | 144 | 9.035e-07 | 2.213e-05 |
| 190 | REGULATION OF AXONOGENESIS | 40 | 168 | 8.992e-07 | 2.213e-05 |
| 191 | REGULATION OF HYDROLASE ACTIVITY | 197 | 1327 | 9.276e-07 | 2.26e-05 |
| 192 | TRANSMEMBRANE TRANSPORT | 168 | 1098 | 9.699e-07 | 2.35e-05 |
| 193 | REGULATION OF SYSTEM PROCESS | 90 | 507 | 9.953e-07 | 2.4e-05 |
| 194 | REGULATION OF CELL PROJECTION ORGANIZATION | 97 | 558 | 1.009e-06 | 2.408e-05 |
| 195 | WATER SOLUBLE VITAMIN METABOLIC PROCESS | 26 | 88 | 1.008e-06 | 2.408e-05 |
| 196 | NEGATIVE REGULATION OF WOUND HEALING | 20 | 58 | 1.21e-06 | 2.871e-05 |
| 197 | EAR DEVELOPMENT | 44 | 195 | 1.27e-06 | 3e-05 |
| 198 | CELLULAR RESPONSE TO INSULIN STIMULUS | 36 | 146 | 1.287e-06 | 3.024e-05 |
| 199 | MONOSACCHARIDE METABOLIC PROCESS | 45 | 202 | 1.404e-06 | 3.282e-05 |
| 200 | CATION TRANSPORT | 128 | 796 | 1.586e-06 | 3.69e-05 |
| 201 | CELLULAR HOMEOSTASIS | 112 | 676 | 1.714e-06 | 3.967e-05 |
| 202 | RESPONSE TO STARVATION | 37 | 154 | 1.805e-06 | 4.157e-05 |
| 203 | ADIPOSE TISSUE DEVELOPMENT | 14 | 32 | 1.837e-06 | 4.211e-05 |
| 204 | GLUCOSE METABOLIC PROCESS | 31 | 119 | 1.987e-06 | 4.532e-05 |
| 205 | LIPID OXIDATION | 22 | 70 | 2.156e-06 | 4.895e-05 |
| 206 | RESPONSE TO ESTROGEN | 47 | 218 | 2.19e-06 | 4.947e-05 |
| 207 | HEART MORPHOGENESIS | 46 | 212 | 2.326e-06 | 5.207e-05 |
| 208 | LIPID BIOSYNTHETIC PROCESS | 93 | 539 | 2.328e-06 | 5.207e-05 |
| 209 | REGULATION OF EPITHELIAL CELL PROLIFERATION | 57 | 285 | 2.459e-06 | 5.474e-05 |
| 210 | REGULATION OF WOUND HEALING | 32 | 126 | 2.483e-06 | 5.502e-05 |
| 211 | RESPONSE TO METAL ION | 64 | 333 | 2.526e-06 | 5.569e-05 |
| 212 | ORGANIC HYDROXY COMPOUND BIOSYNTHETIC PROCESS | 40 | 175 | 2.7e-06 | 5.925e-05 |
| 213 | CELLULAR LIPID CATABOLIC PROCESS | 36 | 151 | 2.997e-06 | 6.548e-05 |
| 214 | CARDIAC CHAMBER MORPHOGENESIS | 28 | 104 | 3.053e-06 | 6.638e-05 |
| 215 | ION TRANSMEMBRANE TRANSPORT | 130 | 822 | 3.171e-06 | 6.862e-05 |
| 216 | CELL SUBSTRATE ADHESION | 38 | 164 | 3.361e-06 | 7.241e-05 |
| 217 | CELL PART MORPHOGENESIS | 105 | 633 | 3.41e-06 | 7.313e-05 |
| 218 | HEPATICOBILIARY SYSTEM DEVELOPMENT | 32 | 128 | 3.573e-06 | 7.59e-05 |
| 219 | REGULATION OF COAGULATION | 25 | 88 | 3.563e-06 | 7.59e-05 |
| 220 | METAL ION TRANSPORT | 98 | 582 | 3.742e-06 | 7.878e-05 |
| 221 | MORPHOGENESIS OF AN EPITHELIUM | 73 | 400 | 3.733e-06 | 7.878e-05 |
| 222 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 267 | 1929 | 3.811e-06 | 7.949e-05 |
| 223 | REGULATION OF DEVELOPMENTAL GROWTH | 57 | 289 | 3.86e-06 | 7.949e-05 |
| 224 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 252 | 1805 | 3.861e-06 | 7.949e-05 |
| 225 | REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 143 | 926 | 3.859e-06 | 7.949e-05 |
| 226 | AMINO ACID BETAINE METABOLIC PROCESS | 10 | 18 | 3.813e-06 | 7.949e-05 |
| 227 | REGULATION OF PEPTIDE TRANSPORT | 52 | 256 | 4.184e-06 | 8.576e-05 |
| 228 | REGULATION OF CARBOHYDRATE METABOLIC PROCESS | 39 | 172 | 4.397e-06 | 8.974e-05 |
| 229 | PROTEIN PHOSPHORYLATION | 145 | 944 | 4.433e-06 | 9.007e-05 |
| 230 | EXTRACELLULAR STRUCTURE ORGANIZATION | 59 | 304 | 4.47e-06 | 9.043e-05 |
| 231 | TRANSITION METAL ION HOMEOSTASIS | 28 | 106 | 4.57e-06 | 9.205e-05 |
| 232 | REGULATION OF MAPK CASCADE | 108 | 660 | 4.674e-06 | 9.375e-05 |
| 233 | DICARBOXYLIC ACID METABOLIC PROCESS | 27 | 101 | 5.267e-06 | 0.0001052 |
| 234 | UROGENITAL SYSTEM DEVELOPMENT | 58 | 299 | 5.455e-06 | 0.0001085 |
| 235 | SKELETAL SYSTEM DEVELOPMENT | 80 | 455 | 5.682e-06 | 0.0001125 |
| 236 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 126 | 801 | 5.886e-06 | 0.0001161 |
| 237 | NEUTRAL LIPID METABOLIC PROCESS | 24 | 85 | 6.233e-06 | 0.0001224 |
| 238 | CARDIAC CELL DEVELOPMENT | 17 | 49 | 6.876e-06 | 0.0001344 |
| 239 | POSITIVE REGULATION OF HYDROLASE ACTIVITY | 139 | 905 | 7.029e-06 | 0.0001368 |
| 240 | CARBOHYDRATE DERIVATIVE METABOLIC PROCESS | 157 | 1047 | 7.106e-06 | 0.0001372 |
| 241 | PLACENTA DEVELOPMENT | 33 | 138 | 7.108e-06 | 0.0001372 |
| 242 | REGULATION OF BLOOD CIRCULATION | 57 | 295 | 7.405e-06 | 0.0001424 |
| 243 | REGULATION OF NEURON DIFFERENTIATION | 93 | 554 | 7.471e-06 | 0.0001425 |
| 244 | NEGATIVE REGULATION OF PHOSPHORYLATION | 75 | 422 | 7.446e-06 | 0.0001425 |
| 245 | REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 63 | 337 | 7.674e-06 | 0.0001451 |
| 246 | REPRODUCTIVE SYSTEM DEVELOPMENT | 73 | 408 | 7.704e-06 | 0.0001451 |
| 247 | TUBE MORPHOGENESIS | 61 | 323 | 7.651e-06 | 0.0001451 |
| 248 | HEXOSE METABOLIC PROCESS | 36 | 157 | 7.733e-06 | 0.0001451 |
| 249 | REGULATION OF CELL CELL ADHESION | 69 | 380 | 8.135e-06 | 0.000152 |
| 250 | UNSATURATED FATTY ACID METABOLIC PROCESS | 28 | 109 | 8.165e-06 | 0.000152 |
| 251 | GLUCOSE HOMEOSTASIS | 38 | 170 | 8.26e-06 | 0.0001525 |
| 252 | CARBOHYDRATE HOMEOSTASIS | 38 | 170 | 8.26e-06 | 0.0001525 |
| 253 | CARBOHYDRATE BIOSYNTHETIC PROCESS | 30 | 121 | 8.501e-06 | 0.0001563 |
| 254 | COENZYME BIOSYNTHETIC PROCESS | 31 | 127 | 8.533e-06 | 0.0001563 |
| 255 | REGULATION OF GLUCOSE IMPORT | 19 | 60 | 9.286e-06 | 0.0001688 |
| 256 | CELL PROJECTION ORGANIZATION | 138 | 902 | 9.286e-06 | 0.0001688 |
| 257 | ARACHIDONIC ACID METABOLIC PROCESS | 17 | 50 | 9.383e-06 | 0.0001699 |
| 258 | DEVELOPMENTAL GROWTH INVOLVED IN MORPHOGENESIS | 27 | 104 | 9.525e-06 | 0.0001718 |
| 259 | REGULATION OF LIPID BIOSYNTHETIC PROCESS | 31 | 128 | 1.012e-05 | 0.0001811 |
| 260 | POSITIVE REGULATION OF LIPID METABOLIC PROCESS | 31 | 128 | 1.012e-05 | 0.0001811 |
| 261 | CELL PROLIFERATION | 108 | 672 | 1.059e-05 | 0.0001887 |
| 262 | REGULATION OF CELL SIZE | 38 | 172 | 1.1e-05 | 0.0001953 |
| 263 | REGULATION OF CELL MORPHOGENESIS | 92 | 552 | 1.12e-05 | 0.0001982 |
| 264 | REGULATION OF CARDIAC MUSCLE CONTRACTION | 20 | 66 | 1.145e-05 | 0.0002018 |
| 265 | REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 118 | 750 | 1.15e-05 | 0.000202 |
| 266 | POSITIVE REGULATION OF CELL DEATH | 99 | 605 | 1.155e-05 | 0.0002021 |
| 267 | ADULT BEHAVIOR | 32 | 135 | 1.18e-05 | 0.0002056 |
| 268 | STEROL METABOLIC PROCESS | 30 | 123 | 1.204e-05 | 0.0002091 |
| 269 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 246 | 1784 | 1.257e-05 | 0.0002175 |
| 270 | STRIATED MUSCLE CELL DIFFERENTIATION | 38 | 173 | 1.266e-05 | 0.0002176 |
| 271 | ARTERY MORPHOGENESIS | 17 | 51 | 1.268e-05 | 0.0002176 |
| 272 | POSITIVE REGULATION OF CELL PROLIFERATION | 126 | 814 | 1.299e-05 | 0.0002221 |
| 273 | VENTRICULAR SEPTUM MORPHOGENESIS | 12 | 28 | 1.309e-05 | 0.0002224 |
| 274 | DIGESTIVE SYSTEM DEVELOPMENT | 34 | 148 | 1.31e-05 | 0.0002224 |
| 275 | AROMATIC AMINO ACID FAMILY CATABOLIC PROCESS | 10 | 20 | 1.315e-05 | 0.0002225 |
| 276 | RESPONSE TO CYTOKINE | 113 | 714 | 1.347e-05 | 0.0002271 |
| 277 | REGULATION OF FAT CELL DIFFERENTIATION | 27 | 106 | 1.391e-05 | 0.0002327 |
| 278 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 150 | 1004 | 1.389e-05 | 0.0002327 |
| 279 | POSITIVE REGULATION OF CELL COMMUNICATION | 215 | 1532 | 1.509e-05 | 0.0002515 |
| 280 | CELLULAR RESPONSE TO STRESS | 219 | 1565 | 1.513e-05 | 0.0002515 |
| 281 | NEGATIVE REGULATION OF MOLECULAR FUNCTION | 159 | 1079 | 1.644e-05 | 0.0002723 |
| 282 | POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 168 | 1152 | 1.708e-05 | 0.0002819 |
| 283 | REGULATION OF BODY FLUID LEVELS | 85 | 506 | 1.773e-05 | 0.0002915 |
| 284 | CELLULAR CARBOHYDRATE METABOLIC PROCESS | 33 | 144 | 1.84e-05 | 0.0003015 |
| 285 | IN UTERO EMBRYONIC DEVELOPMENT | 58 | 311 | 1.864e-05 | 0.0003024 |
| 286 | RESPONSE TO OXYGEN LEVELS | 58 | 311 | 1.864e-05 | 0.0003024 |
| 287 | VASCULAR PROCESS IN CIRCULATORY SYSTEM | 36 | 163 | 1.865e-05 | 0.0003024 |
| 288 | SYNAPSE ORGANIZATION | 33 | 145 | 2.141e-05 | 0.0003433 |
| 289 | CARDIAC MUSCLE CELL DIFFERENTIATION | 21 | 74 | 2.138e-05 | 0.0003433 |
| 290 | REGULATION OF STEROID METABOLIC PROCESS | 21 | 74 | 2.138e-05 | 0.0003433 |
| 291 | HORMONE METABOLIC PROCESS | 36 | 164 | 2.147e-05 | 0.0003433 |
| 292 | CELLULAR RESPONSE TO CORTICOSTEROID STIMULUS | 18 | 58 | 2.163e-05 | 0.0003447 |
| 293 | LIPID MODIFICATION | 43 | 210 | 2.235e-05 | 0.0003549 |
| 294 | NEGATIVE REGULATION OF COAGULATION | 16 | 48 | 2.27e-05 | 0.0003593 |
| 295 | POSITIVE REGULATION OF GENE EXPRESSION | 238 | 1733 | 2.33e-05 | 0.0003676 |
| 296 | REGULATION OF MUSCLE TISSUE DEVELOPMENT | 26 | 103 | 2.363e-05 | 0.0003715 |
| 297 | EMBRYONIC ORGAN MORPHOGENESIS | 53 | 279 | 2.488e-05 | 0.0003898 |
| 298 | POSITIVE REGULATION OF REACTIVE OXYGEN SPECIES METABOLIC PROCESS | 23 | 86 | 2.544e-05 | 0.0003972 |
| 299 | CARDIAC MUSCLE TISSUE DEVELOPMENT | 32 | 140 | 2.586e-05 | 0.0004024 |
| 300 | INNER EAR MORPHOGENESIS | 24 | 92 | 2.681e-05 | 0.0004159 |
| 301 | RESPONSE TO CAMP | 26 | 104 | 2.833e-05 | 0.0004365 |
| 302 | COFACTOR BIOSYNTHETIC PROCESS | 36 | 166 | 2.83e-05 | 0.0004365 |
| 303 | BIOTIN METABOLIC PROCESS | 8 | 14 | 2.85e-05 | 0.0004377 |
| 304 | REGULATION OF MUSCLE CONTRACTION | 33 | 147 | 2.88e-05 | 0.0004408 |
| 305 | VENTRICULAR SEPTUM DEVELOPMENT | 17 | 54 | 2.958e-05 | 0.0004512 |
| 306 | CIRCULATORY SYSTEM PROCESS | 65 | 366 | 3.013e-05 | 0.0004581 |
| 307 | REGULATION OF FATTY ACID METABOLIC PROCESS | 23 | 87 | 3.111e-05 | 0.0004715 |
| 308 | CEREBRAL CORTEX DEVELOPMENT | 26 | 105 | 3.386e-05 | 0.0005116 |
| 309 | CELLULAR RESPONSE TO CYTOKINE STIMULUS | 97 | 606 | 3.46e-05 | 0.000521 |
| 310 | SMOOTH MUSCLE TISSUE DEVELOPMENT | 9 | 18 | 3.621e-05 | 0.0005383 |
| 311 | REGULATION OF CERAMIDE BIOSYNTHETIC PROCESS | 7 | 11 | 3.597e-05 | 0.0005383 |
| 312 | CONNECTIVE TISSUE DEVELOPMENT | 40 | 194 | 3.602e-05 | 0.0005383 |
| 313 | NEGATIVE REGULATION OF CELL COMMUNICATION | 171 | 1192 | 3.611e-05 | 0.0005383 |
| 314 | POSITIVE REGULATION OF MUSCLE HYPERTROPHY | 10 | 22 | 3.76e-05 | 0.0005519 |
| 315 | POSITIVE REGULATION OF CARDIAC MUSCLE HYPERTROPHY | 10 | 22 | 3.76e-05 | 0.0005519 |
| 316 | REGULATION OF ION TRANSPORT | 95 | 592 | 3.76e-05 | 0.0005519 |
| 317 | PYRIDINE CONTAINING COMPOUND BIOSYNTHETIC PROCESS | 10 | 22 | 3.76e-05 | 0.0005519 |
| 318 | POSITIVE REGULATION OF CELL ADHESION | 66 | 376 | 3.82e-05 | 0.0005589 |
| 319 | AMINO ACID TRANSPORT | 29 | 124 | 3.877e-05 | 0.0005638 |
| 320 | MONOCARBOXYLIC ACID TRANSPORT | 29 | 124 | 3.877e-05 | 0.0005638 |
| 321 | EAR MORPHOGENESIS | 27 | 112 | 4.018e-05 | 0.0005825 |
| 322 | CELLULAR RESPONSE TO OXYGEN LEVELS | 32 | 143 | 4.038e-05 | 0.0005834 |
| 323 | MUSCLE ORGAN DEVELOPMENT | 52 | 277 | 4.106e-05 | 0.0005905 |
| 324 | CELLULAR TRANSITION METAL ION HOMEOSTASIS | 21 | 77 | 4.112e-05 | 0.0005905 |
| 325 | RESPONSE TO WOUNDING | 91 | 563 | 4.14e-05 | 0.0005927 |
| 326 | REGULATION OF PEPTIDE SECRETION | 42 | 209 | 4.387e-05 | 0.0006262 |
| 327 | CELLULAR RESPONSE TO EXTERNAL STIMULUS | 50 | 264 | 4.513e-05 | 0.0006422 |
| 328 | CELL MATRIX ADHESION | 28 | 119 | 4.66e-05 | 0.0006598 |
| 329 | NEGATIVE REGULATION OF CELL PROJECTION ORGANIZATION | 32 | 144 | 4.665e-05 | 0.0006598 |
| 330 | REGULATION OF EXTENT OF CELL GROWTH | 25 | 101 | 4.789e-05 | 0.0006753 |
| 331 | REGULATED EXOCYTOSIS | 44 | 224 | 5.18e-05 | 0.0007259 |
| 332 | NEGATIVE REGULATION OF CELL PROLIFERATION | 101 | 643 | 5.173e-05 | 0.0007259 |
| 333 | ISOPRENOID METABOLIC PROCESS | 29 | 126 | 5.3e-05 | 0.0007405 |
| 334 | REGULATION OF CELLULAR COMPONENT SIZE | 60 | 337 | 5.556e-05 | 0.0007671 |
| 335 | CELLULAR RESPONSE TO STEROL | 8 | 15 | 5.532e-05 | 0.0007671 |
| 336 | REGULATION OF INTRACELLULAR TRANSPORT | 98 | 621 | 5.547e-05 | 0.0007671 |
| 337 | AMINO ACID IMPORT | 8 | 15 | 5.532e-05 | 0.0007671 |
| 338 | STEROID BIOSYNTHETIC PROCESS | 27 | 114 | 5.594e-05 | 0.0007701 |
| 339 | REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 43 | 218 | 5.663e-05 | 0.0007773 |
| 340 | HIPPOCAMPUS DEVELOPMENT | 20 | 73 | 5.795e-05 | 0.0007907 |
| 341 | CELLULAR RESPONSE TO KETONE | 20 | 73 | 5.795e-05 | 0.0007907 |
| 342 | BRANCHED CHAIN AMINO ACID METABOLIC PROCESS | 10 | 23 | 6.014e-05 | 0.0008182 |
| 343 | SENSORY ORGAN DEVELOPMENT | 81 | 493 | 6.098e-05 | 0.0008273 |
| 344 | REGULATION OF PROTEIN SECRETION | 67 | 389 | 6.117e-05 | 0.0008273 |
| 345 | RESPONSE TO ESTRADIOL | 32 | 146 | 6.193e-05 | 0.0008347 |
| 346 | DRUG TRANSMEMBRANE TRANSPORT | 9 | 19 | 6.225e-05 | 0.0008347 |
| 347 | REGULATION OF STRIATED MUSCLE CONTRACTION | 21 | 79 | 6.209e-05 | 0.0008347 |
| 348 | POSITIVE REGULATION OF TRANSFERASE ACTIVITY | 97 | 616 | 6.571e-05 | 0.0008786 |
| 349 | POSITIVE REGULATION OF CELL CYCLE | 59 | 332 | 6.734e-05 | 0.0008978 |
| 350 | PRIMARY ALCOHOL METABOLIC PROCESS | 15 | 47 | 7.215e-05 | 0.0009592 |
| 351 | NEGATIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 87 | 541 | 7.282e-05 | 0.0009626 |
| 352 | NEGATIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 87 | 541 | 7.282e-05 | 0.0009626 |
| 353 | REGULATION OF MUSCLE ADAPTATION | 18 | 63 | 7.375e-05 | 0.0009721 |
| 354 | ORGANOPHOSPHATE METABOLIC PROCESS | 131 | 885 | 7.453e-05 | 0.0009796 |
| 355 | ADULT LOCOMOTORY BEHAVIOR | 21 | 80 | 7.578e-05 | 0.0009933 |
| 356 | L AMINO ACID IMPORT | 7 | 12 | 7.83e-05 | 0.001023 |
| 357 | CELL GROWTH | 30 | 135 | 7.935e-05 | 0.001034 |
| 358 | CELLULAR GLUCAN METABOLIC PROCESS | 17 | 58 | 8.164e-05 | 0.001058 |
| 359 | GLUCAN METABOLIC PROCESS | 17 | 58 | 8.164e-05 | 0.001058 |
| 360 | MONOVALENT INORGANIC CATION HOMEOSTASIS | 28 | 123 | 8.685e-05 | 0.001123 |
| 361 | NEGATIVE REGULATION OF GROWTH | 45 | 236 | 8.97e-05 | 0.001156 |
| 362 | STEROID CATABOLIC PROCESS | 10 | 24 | 9.321e-05 | 0.001198 |
| 363 | REGULATION OF HOMEOSTATIC PROCESS | 74 | 447 | 9.753e-05 | 0.00125 |
| 364 | ACTIVATION OF PROTEIN KINASE ACTIVITY | 51 | 279 | 9.835e-05 | 0.001257 |
| 365 | POSITIVE REGULATION OF MAPK CASCADE | 77 | 470 | 0.0001008 | 0.001281 |
| 366 | WOUND HEALING | 77 | 470 | 0.0001008 | 0.001281 |
| 367 | INTRACELLULAR LIPID TRANSPORT | 9 | 20 | 0.0001024 | 0.001299 |
| 368 | PROTEIN HOMOTETRAMERIZATION | 17 | 59 | 0.0001033 | 0.0013 |
| 369 | SULFUR COMPOUND BIOSYNTHETIC PROCESS | 40 | 203 | 0.0001034 | 0.0013 |
| 370 | REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 17 | 59 | 0.0001033 | 0.0013 |
| 371 | REGULATION OF INFLAMMATORY RESPONSE | 53 | 294 | 0.0001055 | 0.001323 |
| 372 | NEGATIVE REGULATION OF CELL DIFFERENTIATION | 95 | 609 | 0.0001107 | 0.001384 |
| 373 | TERPENOID METABOLIC PROCESS | 25 | 106 | 0.0001116 | 0.001392 |
| 374 | MONOSACCHARIDE BIOSYNTHETIC PROCESS | 16 | 54 | 0.0001138 | 0.001405 |
| 375 | REGULATION OF SMOOTH MUSCLE CELL PROLIFERATION | 24 | 100 | 0.0001135 | 0.001405 |
| 376 | LIMBIC SYSTEM DEVELOPMENT | 24 | 100 | 0.0001135 | 0.001405 |
| 377 | SODIUM ION TRANSPORT | 31 | 144 | 0.0001138 | 0.001405 |
| 378 | NEGATIVE REGULATION OF AXONOGENESIS | 18 | 65 | 0.0001151 | 0.001417 |
| 379 | POSITIVE REGULATION OF NEURON PROJECTION DEVELOPMENT | 44 | 232 | 0.0001213 | 0.00149 |
| 380 | REGULATION OF STEROID BIOSYNTHETIC PROCESS | 15 | 49 | 0.000123 | 0.001506 |
| 381 | NEGATIVE REGULATION OF CELL DEVELOPMENT | 54 | 303 | 0.0001243 | 0.001518 |
| 382 | RESPONSE TO FLUID SHEAR STRESS | 12 | 34 | 0.0001278 | 0.001552 |
| 383 | REGULATION OF CELL GROWTH | 66 | 391 | 0.0001276 | 0.001552 |
| 384 | POSITIVE REGULATION OF SMOOTH MUSCLE CELL PROLIFERATION | 17 | 60 | 0.0001299 | 0.001575 |
| 385 | DEVELOPMENTAL CELL GROWTH | 20 | 77 | 0.0001305 | 0.001577 |
| 386 | THIOESTER METABOLIC PROCESS | 21 | 83 | 0.0001343 | 0.001615 |
| 387 | ACYL COA METABOLIC PROCESS | 21 | 83 | 0.0001343 | 0.001615 |
| 388 | EPITHELIAL CELL PROLIFERATION | 22 | 89 | 0.000136 | 0.00163 |
| 389 | RESPONSE TO ORGANOPHOSPHORUS | 30 | 139 | 0.0001388 | 0.00166 |
| 390 | DRUG TRANSPORT | 10 | 25 | 0.0001405 | 0.001676 |
| 391 | REGULATION OF ACTIN FILAMENT BASED PROCESS | 55 | 312 | 0.0001451 | 0.001727 |
| 392 | REGULATION OF HORMONE SECRETION | 48 | 262 | 0.0001468 | 0.001742 |
| 393 | GLYCEROLIPID METABOLIC PROCESS | 61 | 356 | 0.000148 | 0.001753 |
| 394 | ENERGY RESERVE METABOLIC PROCESS | 19 | 72 | 0.0001519 | 0.001782 |
| 395 | REGULATION OF SPHINGOLIPID BIOSYNTHETIC PROCESS | 7 | 13 | 0.0001539 | 0.001782 |
| 396 | REGULATION OF MEMBRANE LIPID METABOLIC PROCESS | 7 | 13 | 0.0001539 | 0.001782 |
| 397 | ORGANIC HYDROXY COMPOUND CATABOLIC PROCESS | 19 | 72 | 0.0001519 | 0.001782 |
| 398 | REGULATION OF CHOLESTEROL BIOSYNTHETIC PROCESS | 7 | 13 | 0.0001539 | 0.001782 |
| 399 | DICARBOXYLIC ACID BIOSYNTHETIC PROCESS | 7 | 13 | 0.0001539 | 0.001782 |
| 400 | CARNITINE METABOLIC PROCESS | 7 | 13 | 0.0001539 | 0.001782 |
| 401 | SEMAPHORIN PLEXIN SIGNALING PATHWAY INVOLVED IN NEURON PROJECTION GUIDANCE | 7 | 13 | 0.0001539 | 0.001782 |
| 402 | VENTRICULAR CARDIAC MUSCLE CELL DEVELOPMENT | 7 | 13 | 0.0001539 | 0.001782 |
| 403 | REGULATION OF COENZYME METABOLIC PROCESS | 15 | 50 | 0.0001584 | 0.001816 |
| 404 | CELLULAR RESPONSE TO CAMP | 15 | 50 | 0.0001584 | 0.001816 |
| 405 | POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 128 | 876 | 0.0001584 | 0.001816 |
| 406 | REGULATION OF COFACTOR METABOLIC PROCESS | 15 | 50 | 0.0001584 | 0.001816 |
| 407 | REGULATION OF CELLULAR KETONE METABOLIC PROCESS | 35 | 173 | 0.0001591 | 0.001819 |
| 408 | NEGATIVE REGULATION OF LOCOMOTION | 48 | 263 | 0.0001612 | 0.001821 |
| 409 | REGULATION OF SYSTEMIC ARTERIAL BLOOD PRESSURE | 21 | 84 | 0.0001612 | 0.001821 |
| 410 | CARDIOCYTE DIFFERENTIATION | 23 | 96 | 0.0001606 | 0.001821 |
| 411 | ICOSANOID METABOLIC PROCESS | 23 | 96 | 0.0001606 | 0.001821 |
| 412 | FATTY ACID DERIVATIVE METABOLIC PROCESS | 23 | 96 | 0.0001606 | 0.001821 |
| 413 | REGULATION OF HEART CONTRACTION | 42 | 221 | 0.0001632 | 0.001838 |
| 414 | REGULATION OF ORGAN MORPHOGENESIS | 45 | 242 | 0.0001641 | 0.001844 |
| 415 | MORPHOGENESIS OF A BRANCHING STRUCTURE | 34 | 167 | 0.0001727 | 0.001937 |
| 416 | RESPONSE TO IRON ION | 12 | 35 | 0.0001756 | 0.001964 |
| 417 | REGULATION OF ORGAN GROWTH | 19 | 73 | 0.000185 | 0.002059 |
| 418 | REGULATION OF MUSCLE ORGAN DEVELOPMENT | 24 | 103 | 0.0001849 | 0.002059 |
| 419 | CELL DEATH | 143 | 1001 | 0.0001904 | 0.002114 |
| 420 | FORMATION OF PRIMARY GERM LAYER | 25 | 110 | 0.0002084 | 0.002308 |
| 421 | ORGAN GROWTH | 18 | 68 | 0.0002153 | 0.00238 |
| 422 | CELLULAR HORMONE METABOLIC PROCESS | 24 | 104 | 0.0002163 | 0.002385 |
| 423 | CYCLIC NUCLEOTIDE MEDIATED SIGNALING | 14 | 46 | 0.00022 | 0.00242 |
| 424 | CIRCADIAN REGULATION OF GENE EXPRESSION | 16 | 57 | 0.0002289 | 0.002512 |
| 425 | NEGATIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 110 | 740 | 0.0002342 | 0.002564 |
| 426 | REGULATION OF NUCLEOTIDE METABOLIC PROCESS | 40 | 211 | 0.0002433 | 0.002657 |
| 427 | POSITIVE REGULATION OF SECRETION | 62 | 370 | 0.0002468 | 0.002689 |
| 428 | REGULATION OF CHOLESTEROL METABOLIC PROCESS | 9 | 22 | 0.0002485 | 0.002702 |
| 429 | HINDBRAIN DEVELOPMENT | 29 | 137 | 0.0002525 | 0.002739 |
| 430 | IRON ION HOMEOSTASIS | 18 | 69 | 0.0002625 | 0.00284 |
| 431 | NEGATIVE REGULATION OF DEFENSE RESPONSE | 30 | 144 | 0.0002672 | 0.002884 |
| 432 | EPOXYGENASE P450 PATHWAY | 8 | 18 | 0.0002798 | 0.003014 |
| 433 | REGULATION OF OSSIFICATION | 35 | 178 | 0.0002827 | 0.003038 |
| 434 | CARDIAC VENTRICLE DEVELOPMENT | 24 | 106 | 0.0002936 | 0.003148 |
| 435 | REGULATION OF FATTY ACID TRANSPORT | 10 | 27 | 0.0002965 | 0.003172 |
| 436 | REGULATION OF PEPTIDYL TYROSINE PHOSPHORYLATION | 40 | 213 | 0.000298 | 0.003173 |
| 437 | CELLULAR MODIFIED AMINO ACID METABOLIC PROCESS | 41 | 220 | 0.000298 | 0.003173 |
| 438 | REGULATION OF HEART GROWTH | 13 | 42 | 0.0003045 | 0.003235 |
| 439 | REGULATION OF MUSCLE HYPERTROPHY | 12 | 37 | 0.000318 | 0.003363 |
| 440 | CAMP MEDIATED SIGNALING | 12 | 37 | 0.000318 | 0.003363 |
| 441 | AMINOGLYCAN BIOSYNTHETIC PROCESS | 24 | 107 | 0.0003406 | 0.003594 |
| 442 | POSITIVE REGULATION OF CELL CELL ADHESION | 44 | 243 | 0.0003531 | 0.003717 |
| 443 | REGULATION OF CARDIAC MUSCLE TISSUE DEVELOPMENT | 14 | 48 | 0.0003601 | 0.003783 |
| 444 | REGULATION OF LIPID TRANSPORT | 22 | 95 | 0.0003694 | 0.003865 |
| 445 | TRABECULA FORMATION | 9 | 23 | 0.0003696 | 0.003865 |
| 446 | GLUTAMINE FAMILY AMINO ACID METABOLIC PROCESS | 17 | 65 | 0.0003728 | 0.003889 |
| 447 | ANION TRANSMEMBRANE TRANSPORT | 45 | 251 | 0.0003816 | 0.003972 |
| 448 | CARBOHYDRATE DERIVATIVE CATABOLIC PROCESS | 34 | 174 | 0.0003866 | 0.004007 |
| 449 | REGULATION OF LIPASE ACTIVITY | 20 | 83 | 0.0003866 | 0.004007 |
| 450 | ALPHA AMINO ACID BIOSYNTHETIC PROCESS | 19 | 77 | 0.0003885 | 0.004017 |
| 451 | LOCOMOTORY BEHAVIOR | 35 | 181 | 0.0003928 | 0.004052 |
| 452 | ACUTE PHASE RESPONSE | 13 | 43 | 0.0003941 | 0.004057 |
| 453 | NEGATIVE REGULATION OF CELL ADHESION | 41 | 223 | 0.0003988 | 0.004096 |
| 454 | POSITIVE REGULATION OF EPITHELIAL CELL PROLIFERATION | 31 | 154 | 0.0004017 | 0.004117 |
| 455 | CELL CELL SIGNALING | 112 | 767 | 0.0004097 | 0.00418 |
| 456 | REGULATION OF PROTEOLYSIS | 105 | 711 | 0.0004089 | 0.00418 |
| 457 | GLUTAMATE METABOLIC PROCESS | 10 | 28 | 0.0004173 | 0.00423 |
| 458 | AROMATIC AMINO ACID FAMILY METABOLIC PROCESS | 10 | 28 | 0.0004173 | 0.00423 |
| 459 | REGULATION OF FATTY ACID OXIDATION | 10 | 28 | 0.0004173 | 0.00423 |
| 460 | REGULATION OF GLUCONEOGENESIS | 12 | 38 | 0.0004198 | 0.004247 |
| 461 | RESPONSE TO CARBOHYDRATE | 33 | 168 | 0.0004235 | 0.004275 |
| 462 | REGULATION OF RESPONSE TO STRESS | 197 | 1468 | 0.0004268 | 0.004298 |
| 463 | NEURON RECOGNITION | 11 | 33 | 0.0004305 | 0.004327 |
| 464 | CALCIUM MEDIATED SIGNALING | 21 | 90 | 0.0004462 | 0.004475 |
| 465 | REGULATION OF GTPASE ACTIVITY | 100 | 673 | 0.0004483 | 0.004486 |
| 466 | NEUROTRANSMITTER TRANSPORT | 31 | 155 | 0.0004515 | 0.0045 |
| 467 | DEVELOPMENTAL PROCESS INVOLVED IN REPRODUCTION | 91 | 602 | 0.000452 | 0.0045 |
| 468 | POSITIVE REGULATION OF LIPID BIOSYNTHETIC PROCESS | 17 | 66 | 0.0004526 | 0.0045 |
| 469 | NEGATIVE REGULATION OF PROTEIN METABOLIC PROCESS | 151 | 1087 | 0.0004606 | 0.00457 |
| 470 | CARBOHYDRATE DERIVATIVE BIOSYNTHETIC PROCESS | 90 | 595 | 0.0004742 | 0.004695 |
| 471 | POSITIVE REGULATION OF MEMBRANE PROTEIN ECTODOMAIN PROTEOLYSIS | 7 | 15 | 0.0004754 | 0.004696 |
| 472 | ORGANIC ACID TRANSMEMBRANE TRANSPORT | 23 | 103 | 0.0004795 | 0.004727 |
| 473 | REGULATION OF PROTEIN IMPORT | 35 | 183 | 0.0004858 | 0.004779 |
| 474 | POSITIVE REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 27 | 129 | 0.0004892 | 0.004802 |
| 475 | RESPONSE TO ETHANOL | 28 | 136 | 0.0005139 | 0.005034 |
| 476 | LIPID HOMEOSTASIS | 24 | 110 | 0.0005237 | 0.005119 |
| 477 | IMMUNE SYSTEM PROCESS | 257 | 1984 | 0.0005279 | 0.005149 |
| 478 | CELLULAR RESPONSE TO OXIDATIVE STRESS | 35 | 184 | 0.0005392 | 0.005249 |
| 479 | RESPONSE TO MOLECULE OF BACTERIAL ORIGIN | 54 | 321 | 0.000544 | 0.005285 |
| 480 | REGULATION OF AXON GUIDANCE | 12 | 39 | 0.0005478 | 0.00531 |
| 481 | ACUTE INFLAMMATORY RESPONSE | 18 | 73 | 0.0005524 | 0.005344 |
| 482 | POSITIVE REGULATION OF PROTEIN IMPORT | 23 | 104 | 0.0005543 | 0.005351 |
| 483 | NEGATIVE REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 25 | 117 | 0.0005634 | 0.005406 |
| 484 | MAMMARY GLAND DEVELOPMENT | 25 | 117 | 0.0005634 | 0.005406 |
| 485 | RESPONSE TO TOXIC SUBSTANCE | 43 | 241 | 0.0005631 | 0.005406 |
| 486 | GENERATION OF PRECURSOR METABOLITES AND ENERGY | 50 | 292 | 0.0005678 | 0.005436 |
| 487 | REGULATION OF ORGANIC ACID TRANSPORT | 14 | 50 | 0.0005704 | 0.005449 |
| 488 | NEGATIVE REGULATION OF TRANSPORT | 72 | 458 | 0.0005716 | 0.00545 |
| 489 | MONOVALENT INORGANIC CATION TRANSPORT | 69 | 435 | 0.0005733 | 0.005455 |
| 490 | REGULATION OF CARDIAC MUSCLE CELL PROLIFERATION | 10 | 29 | 0.0005761 | 0.005471 |
| 491 | INORGANIC ION TRANSMEMBRANE TRANSPORT | 88 | 583 | 0.0005831 | 0.005526 |
| 492 | RESPONSE TO OXIDATIVE STRESS | 58 | 352 | 0.000592 | 0.005598 |
| 493 | OUTFLOW TRACT MORPHOGENESIS | 15 | 56 | 0.0006161 | 0.005815 |
| 494 | POSITIVE REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION | 79 | 514 | 0.0006356 | 0.005987 |
| 495 | REGULATION OF SYSTEMIC ARTERIAL BLOOD PRESSURE MEDIATED BY A CHEMICAL SIGNAL | 13 | 45 | 0.0006413 | 0.006028 |
| 496 | POLYSACCHARIDE METABOLIC PROCESS | 19 | 80 | 0.0006487 | 0.006086 |
| 497 | REGULATION OF ICOSANOID SECRETION | 8 | 20 | 0.0006618 | 0.006136 |
| 498 | REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 28 | 138 | 0.0006557 | 0.006136 |
| 499 | REGULATION OF EXOCYTOSIS | 35 | 186 | 0.000662 | 0.006136 |
| 500 | REFLEX | 8 | 20 | 0.0006618 | 0.006136 |
| 501 | REGULATION OF ACUTE INFLAMMATORY RESPONSE | 18 | 74 | 0.0006579 | 0.006136 |
| 502 | RESPONSE TO LEPTIN | 8 | 20 | 0.0006618 | 0.006136 |
| 503 | REGULATION OF CELL ACTIVATION | 75 | 484 | 0.000679 | 0.006281 |
| 504 | POSITIVE REGULATION OF ION TRANSPORT | 42 | 236 | 0.000684 | 0.006315 |
| 505 | REGULATION OF MUSCLE CELL DIFFERENTIATION | 30 | 152 | 0.0006955 | 0.006408 |
| 506 | FATTY ACID BETA OXIDATION | 14 | 51 | 0.0007096 | 0.006499 |
| 507 | POSITIVE REGULATION OF LIPID TRANSPORT | 14 | 51 | 0.0007096 | 0.006499 |
| 508 | FATTY ACYL COA METABOLIC PROCESS | 14 | 51 | 0.0007096 | 0.006499 |
| 509 | COGNITION | 44 | 251 | 0.0007172 | 0.006556 |
| 510 | REGULATION OF EPITHELIAL CELL MIGRATION | 32 | 166 | 0.0007198 | 0.006568 |
| 511 | MOVEMENT IN ENVIRONMENT OF OTHER ORGANISM INVOLVED IN SYMBIOTIC INTERACTION | 20 | 87 | 0.0007393 | 0.006641 |
| 512 | ENTRY INTO CELL OF OTHER ORGANISM INVOLVED IN SYMBIOTIC INTERACTION | 20 | 87 | 0.0007393 | 0.006641 |
| 513 | VIRAL ENTRY INTO HOST CELL | 20 | 87 | 0.0007393 | 0.006641 |
| 514 | ENTRY INTO OTHER ORGANISM INVOLVED IN SYMBIOTIC INTERACTION | 20 | 87 | 0.0007393 | 0.006641 |
| 515 | FAT CELL DIFFERENTIATION | 23 | 106 | 0.0007346 | 0.006641 |
| 516 | MOVEMENT IN HOST ENVIRONMENT | 20 | 87 | 0.0007393 | 0.006641 |
| 517 | ENTRY INTO HOST | 20 | 87 | 0.0007393 | 0.006641 |
| 518 | ENTRY INTO HOST CELL | 20 | 87 | 0.0007393 | 0.006641 |
| 519 | REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 73 | 470 | 0.0007437 | 0.006668 |
| 520 | GLYCOGEN BIOSYNTHETIC PROCESS | 9 | 25 | 0.0007576 | 0.006749 |
| 521 | POSITIVE REGULATION OF PROTEOLYSIS | 59 | 363 | 0.0007563 | 0.006749 |
| 522 | GLUCAN BIOSYNTHETIC PROCESS | 9 | 25 | 0.0007576 | 0.006749 |
| 523 | RESPONSE TO MONOAMINE | 11 | 35 | 0.0007586 | 0.006749 |
| 524 | SECRETION BY CELL | 75 | 486 | 0.000765 | 0.00677 |
| 525 | SECRETION | 88 | 588 | 0.0007653 | 0.00677 |
| 526 | CALCIUM ION REGULATED EXOCYTOSIS | 19 | 81 | 0.0007637 | 0.00677 |
| 527 | DICARBOXYLIC ACID CATABOLIC PROCESS | 7 | 16 | 0.0007671 | 0.006773 |
| 528 | SMOOTH MUSCLE CELL DIFFERENTIATION | 10 | 30 | 0.0007819 | 0.006852 |
| 529 | CELLULAR GLUCOSE HOMEOSTASIS | 18 | 75 | 0.0007803 | 0.006852 |
| 530 | REGULATION OF GLUCOSE TRANSPORT | 22 | 100 | 0.0007796 | 0.006852 |
| 531 | REGULATION OF DENDRITIC SPINE MORPHOGENESIS | 10 | 30 | 0.0007819 | 0.006852 |
| 532 | CELLULAR RESPONSE TO CHOLESTEROL | 6 | 12 | 0.0007853 | 0.006852 |
| 533 | POSITIVE REGULATION OF AXONOGENESIS | 17 | 69 | 0.0007863 | 0.006852 |
| 534 | POSITIVE REGULATION OF INSULIN LIKE GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 6 | 12 | 0.0007853 | 0.006852 |
| 535 | POSITIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 49 | 289 | 0.0008021 | 0.006976 |
| 536 | POSITIVE REGULATION OF PEPTIDASE ACTIVITY | 30 | 154 | 0.0008692 | 0.007545 |
| 537 | REGULATION OF LIPID CATABOLIC PROCESS | 14 | 52 | 0.0008765 | 0.007595 |
| 538 | REGULATION OF CYTOPLASMIC TRANSPORT | 74 | 481 | 0.0009017 | 0.007798 |
| 539 | LUNG ALVEOLUS DEVELOPMENT | 12 | 41 | 0.0009033 | 0.007798 |
| 540 | ACTIN FILAMENT BASED PROCESS | 70 | 450 | 0.0009054 | 0.007799 |
| 541 | REGULATION OF SYNAPSE STRUCTURE OR ACTIVITY | 41 | 232 | 0.0009068 | 0.007799 |
| 542 | MULTICELLULAR ORGANISM GROWTH | 18 | 76 | 0.0009216 | 0.007912 |
| 543 | REGULATION OF PHOSPHOLIPASE ACTIVITY | 16 | 64 | 0.0009367 | 0.008011 |
| 544 | CENTRAL NERVOUS SYSTEM NEURON DEVELOPMENT | 17 | 70 | 0.0009364 | 0.008011 |
| 545 | POSITIVE REGULATION OF NEURON DIFFERENTIATION | 51 | 306 | 0.000945 | 0.008068 |
| 546 | CELL CELL ADHESION | 90 | 608 | 0.0009516 | 0.00811 |
| 547 | POSITIVE REGULATION OF PROTEIN SECRETION | 38 | 211 | 0.0009538 | 0.008114 |
| 548 | TRANSITION METAL ION TRANSPORT | 23 | 108 | 0.0009639 | 0.00817 |
| 549 | REGULATION OF MEMBRANE PROTEIN ECTODOMAIN PROTEOLYSIS | 8 | 21 | 0.0009693 | 0.00817 |
| 550 | CELLULAR BIOGENIC AMINE CATABOLIC PROCESS | 8 | 21 | 0.0009693 | 0.00817 |
| 551 | AMINE CATABOLIC PROCESS | 8 | 21 | 0.0009693 | 0.00817 |
| 552 | FIBRINOLYSIS | 8 | 21 | 0.0009693 | 0.00817 |
| 553 | SINGLE ORGANISM CELL ADHESION | 71 | 459 | 0.0009761 | 0.008198 |
| 554 | NEGATIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 45 | 262 | 0.0009752 | 0.008198 |
| 555 | AMMONIUM ION METABOLIC PROCESS | 32 | 169 | 0.0009868 | 0.008236 |
| 556 | REGULATION OF BLOOD PRESSURE | 32 | 169 | 0.0009868 | 0.008236 |
| 557 | REGULATION OF TRANSCRIPTION REGULATORY REGION DNA BINDING | 11 | 36 | 0.0009877 | 0.008236 |
| 558 | POSITIVE REGULATION OF GLUCOSE METABOLIC PROCESS | 11 | 36 | 0.0009877 | 0.008236 |
| 559 | NEGATIVE REGULATION OF GENE EXPRESSION | 197 | 1493 | 0.000999 | 0.008308 |
| 560 | REGULATION OF PH | 20 | 89 | 0.001002 | 0.008308 |
| 561 | RESPONSE TO GROWTH FACTOR | 73 | 475 | 0.001002 | 0.008308 |
| 562 | REGULATION OF PROTEIN TARGETING | 51 | 307 | 0.001018 | 0.008427 |
| 563 | ORGAN REGENERATION | 19 | 83 | 0.001047 | 0.008627 |
| 564 | EMBRYONIC PLACENTA DEVELOPMENT | 19 | 83 | 0.001047 | 0.008627 |
| 565 | MESODERMAL CELL DIFFERENTIATION | 9 | 26 | 0.001049 | 0.008627 |
| 566 | INACTIVATION OF MAPK ACTIVITY | 9 | 26 | 0.001049 | 0.008627 |
| 567 | POSITIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 68 | 437 | 0.001054 | 0.008646 |
| 568 | EMBRYONIC SKELETAL SYSTEM DEVELOPMENT | 25 | 122 | 0.00107 | 0.008768 |
| 569 | REGULATION OF TRANSPORTER ACTIVITY | 36 | 198 | 0.001072 | 0.008769 |
| 570 | NEURON PROJECTION EXTENSION | 14 | 53 | 0.001075 | 0.008778 |
| 571 | BONE DEVELOPMENT | 30 | 156 | 0.00108 | 0.008782 |
| 572 | CELLULAR RESPONSE TO INORGANIC SUBSTANCE | 30 | 156 | 0.00108 | 0.008782 |
| 573 | NEGATIVE REGULATION OF NEURON DIFFERENTIATION | 35 | 191 | 0.001082 | 0.00879 |
| 574 | REGULATION OF CATABOLIC PROCESS | 105 | 731 | 0.001085 | 0.008791 |
| 575 | REACTIVE OXYGEN SPECIES METABOLIC PROCESS | 21 | 96 | 0.001097 | 0.008874 |
| 576 | NEGATIVE REGULATION OF CATALYTIC ACTIVITY | 117 | 829 | 0.001109 | 0.008963 |
| 577 | MODULATION OF SYNAPTIC TRANSMISSION | 50 | 301 | 0.001135 | 0.009153 |
| 578 | AUTONOMIC NERVOUS SYSTEM DEVELOPMENT | 12 | 42 | 0.001143 | 0.009184 |
| 579 | SERINE FAMILY AMINO ACID METABOLIC PROCESS | 12 | 42 | 0.001143 | 0.009184 |
| 580 | INFLAMMATORY RESPONSE | 70 | 454 | 0.001151 | 0.009237 |
| 581 | SODIUM ION TRANSMEMBRANE TRANSPORT | 20 | 90 | 0.00116 | 0.009293 |
| 582 | REGULATION OF CIRCADIAN RHYTHM | 22 | 103 | 0.00118 | 0.009418 |
| 583 | REGULATION OF PLATELET AGGREGATION | 7 | 17 | 0.001184 | 0.009418 |
| 584 | POSITIVE REGULATION OF EPITHELIAL CELL MIGRATION | 22 | 103 | 0.00118 | 0.009418 |
| 585 | PARASYMPATHETIC NERVOUS SYSTEM DEVELOPMENT | 7 | 17 | 0.001184 | 0.009418 |
| 586 | NEGATIVE REGULATION OF KINASE ACTIVITY | 43 | 250 | 0.001211 | 0.009619 |
| 587 | REGULATION OF CYTOSKELETON ORGANIZATION | 76 | 502 | 0.001225 | 0.009714 |
| 588 | NEURON MIGRATION | 23 | 110 | 0.001252 | 0.00991 |
| 589 | GLAND MORPHOGENESIS | 21 | 97 | 0.001261 | 0.009961 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | COFACTOR BINDING | 69 | 263 | 9.868e-13 | 9.167e-10 |
| 2 | CYTOSKELETAL PROTEIN BINDING | 146 | 819 | 3.111e-10 | 1.293e-07 |
| 3 | IDENTICAL PROTEIN BINDING | 198 | 1209 | 4.175e-10 | 1.293e-07 |
| 4 | OXIDOREDUCTASE ACTIVITY | 128 | 719 | 4.362e-09 | 1.013e-06 |
| 5 | TRANSITION METAL ION BINDING | 216 | 1400 | 1.22e-08 | 1.889e-06 |
| 6 | LIPID BINDING | 118 | 657 | 1.061e-08 | 1.889e-06 |
| 7 | KINASE ACTIVITY | 142 | 842 | 2.268e-08 | 3.01e-06 |
| 8 | MONOOXYGENASE ACTIVITY | 30 | 95 | 2.879e-08 | 3.343e-06 |
| 9 | PROTEIN DIMERIZATION ACTIVITY | 179 | 1149 | 1.22e-07 | 1.259e-05 |
| 10 | PYRIDOXAL PHOSPHATE BINDING | 20 | 52 | 1.556e-07 | 1.314e-05 |
| 11 | RECEPTOR BINDING | 220 | 1476 | 1.483e-07 | 1.314e-05 |
| 12 | PROTEIN HOMODIMERIZATION ACTIVITY | 122 | 722 | 2.034e-07 | 1.575e-05 |
| 13 | RIBONUCLEOTIDE BINDING | 265 | 1860 | 3.929e-07 | 2.808e-05 |
| 14 | METAL ION TRANSMEMBRANE TRANSPORTER ACTIVITY | 78 | 417 | 6.518e-07 | 4.325e-05 |
| 15 | OXIDOREDUCTASE ACTIVITY ACTING ON PAIRED DONORS WITH INCORPORATION OR REDUCTION OF MOLECULAR OXYGEN | 38 | 155 | 7.706e-07 | 4.773e-05 |
| 16 | PROTEIN COMPLEX BINDING | 147 | 935 | 1.011e-06 | 5.87e-05 |
| 17 | TETRAPYRROLE BINDING | 34 | 134 | 1.249e-06 | 6.825e-05 |
| 18 | TRANSMEMBRANE TRANSPORTER ACTIVITY | 154 | 997 | 1.604e-06 | 7.844e-05 |
| 19 | ADENYL NUCLEOTIDE BINDING | 219 | 1514 | 1.521e-06 | 7.844e-05 |
| 20 | COENZYME BINDING | 41 | 179 | 1.924e-06 | 8.939e-05 |
| 21 | IRON ION BINDING | 38 | 163 | 2.876e-06 | 0.0001206 |
| 22 | TRANSPORTER ACTIVITY | 188 | 1276 | 2.744e-06 | 0.0001206 |
| 23 | TRANSFERASE ACTIVITY TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 152 | 992 | 2.986e-06 | 0.0001206 |
| 24 | MACROMOLECULAR COMPLEX BINDING | 202 | 1399 | 4.513e-06 | 0.0001747 |
| 25 | SODIUM ION TRANSMEMBRANE TRANSPORTER ACTIVITY | 33 | 136 | 5.087e-06 | 0.000189 |
| 26 | STEROID HYDROXYLASE ACTIVITY | 13 | 31 | 7.619e-06 | 0.0002722 |
| 27 | MONOSACCHARIDE BINDING | 21 | 70 | 8.297e-06 | 0.0002855 |
| 28 | GROWTH FACTOR BINDING | 30 | 123 | 1.204e-05 | 0.0003995 |
| 29 | CATION TRANSMEMBRANE TRANSPORTER ACTIVITY | 101 | 622 | 1.315e-05 | 0.0004213 |
| 30 | OXIDOREDUCTASE ACTIVITY ACTING ON PAIRED DONORS WITH INCORPORATION OR REDUCTION OF MOLECULAR OXYGEN NAD P H AS ONE DONOR AND INCORPORATION OF ONE ATOM OF OXYGEN | 14 | 37 | 1.417e-05 | 0.0004389 |
| 31 | SULFUR COMPOUND BINDING | 47 | 234 | 1.613e-05 | 0.0004834 |
| 32 | ORGANIC ACID BINDING | 43 | 209 | 1.979e-05 | 0.0005746 |
| 33 | TRANSCRIPTION FACTOR ACTIVITY DIRECT LIGAND REGULATED SEQUENCE SPECIFIC DNA BINDING | 16 | 48 | 2.27e-05 | 0.0006392 |
| 34 | ACTIVE TRANSMEMBRANE TRANSPORTER ACTIVITY | 64 | 356 | 2.349e-05 | 0.0006419 |
| 35 | INORGANIC CATION TRANSMEMBRANE TRANSPORTER ACTIVITY | 87 | 527 | 2.766e-05 | 0.0007341 |
| 36 | KINASE BINDING | 97 | 606 | 3.46e-05 | 0.0008773 |
| 37 | OXIDOREDUCTASE ACTIVITY ACTING ON PAIRED DONORS WITH INCORPORATION OR REDUCTION OF MOLECULAR OXYGEN REDUCED FLAVIN OR FLAVOPROTEIN AS ONE DONOR AND INCORPORATION OF ONE ATOM OF OXYGEN | 11 | 26 | 3.494e-05 | 0.0008773 |
| 38 | ENZYME BINDING | 237 | 1737 | 3.759e-05 | 0.0009189 |
| 39 | PROTEIN KINASE ACTIVITY | 101 | 640 | 4.288e-05 | 0.0009959 |
| 40 | ZINC ION BINDING | 166 | 1155 | 4.258e-05 | 0.0009959 |
| 41 | MOLECULAR FUNCTION REGULATOR | 190 | 1353 | 4.678e-05 | 0.001059 |
| 42 | GLYCOPROTEIN BINDING | 25 | 101 | 4.789e-05 | 0.001059 |
| 43 | APOLIPOPROTEIN BINDING | 8 | 15 | 5.532e-05 | 0.001195 |
| 44 | ORGANIC ANION TRANSMEMBRANE TRANSPORTER ACTIVITY | 37 | 178 | 5.793e-05 | 0.001223 |
| 45 | PHOSPHATIDYLINOSITOL BINDING | 40 | 200 | 7.356e-05 | 0.001519 |
| 46 | GLYCOSAMINOGLYCAN BINDING | 40 | 205 | 0.0001289 | 0.002603 |
| 47 | TRANSITION METAL ION TRANSMEMBRANE TRANSPORTER ACTIVITY | 13 | 39 | 0.0001318 | 0.002605 |
| 48 | ACTIN BINDING | 66 | 393 | 0.0001486 | 0.002858 |
| 49 | KINASE REGULATOR ACTIVITY | 37 | 186 | 0.0001508 | 0.002858 |
| 50 | OXIDIZED NAD BINDING | 7 | 13 | 0.0001539 | 0.00286 |
| 51 | TRANSCRIPTION FACTOR ACTIVITY RNA POLYMERASE II CORE PROMOTER PROXIMAL REGION SEQUENCE SPECIFIC BINDING | 57 | 328 | 0.0001643 | 0.002993 |
| 52 | PROTEIN BINDING INVOLVED IN CELL ADHESION | 8 | 17 | 0.0001716 | 0.003065 |
| 53 | TRANSCRIPTION FACTOR BINDING | 83 | 524 | 0.0001766 | 0.003095 |
| 54 | ANION TRANSMEMBRANE TRANSPORTER ACTIVITY | 52 | 293 | 0.0001827 | 0.003144 |
| 55 | TRANSCRIPTIONAL REPRESSOR ACTIVITY RNA POLYMERASE II TRANSCRIPTION REGULATORY REGION SEQUENCE SPECIFIC BINDING | 34 | 168 | 0.0001946 | 0.003288 |
| 56 | PHOSPHATIDYLINOSITOL PHOSPHATE BINDING | 26 | 116 | 0.0001993 | 0.003299 |
| 57 | CARBON CARBON LYASE ACTIVITY | 15 | 51 | 0.0002024 | 0.003299 |
| 58 | ORGANIC ACID TRANSMEMBRANE TRANSPORTER ACTIVITY | 30 | 142 | 0.0002067 | 0.003311 |
| 59 | OXIDOREDUCTASE ACTIVITY ACTING ON THE CH CH GROUP OF DONORS | 16 | 57 | 0.0002289 | 0.00357 |
| 60 | FATTY ACYL COA BINDING | 11 | 31 | 0.0002306 | 0.00357 |
| 61 | ION CHANNEL BINDING | 25 | 111 | 0.000242 | 0.003685 |
| 62 | TRANSCRIPTIONAL REPRESSOR ACTIVITY RNA POLYMERASE II CORE PROMOTER PROXIMAL REGION SEQUENCE SPECIFIC BINDING | 24 | 105 | 0.0002524 | 0.003781 |
| 63 | FILAMIN BINDING | 7 | 14 | 0.0002793 | 0.004104 |
| 64 | OXYGEN BINDING | 14 | 47 | 0.0002827 | 0.004104 |
| 65 | ANION CATION SYMPORTER ACTIVITY | 15 | 53 | 0.0003228 | 0.004613 |
| 66 | STEROID HORMONE RECEPTOR ACTIVITY | 16 | 59 | 0.0003526 | 0.00486 |
| 67 | RNA POLYMERASE II TRANSCRIPTION FACTOR ACTIVITY SEQUENCE SPECIFIC DNA BINDING | 95 | 629 | 0.0003529 | 0.00486 |
| 68 | PHOSPHOLIPID BINDING | 60 | 360 | 0.0003558 | 0.00486 |
| 69 | TRANSFERASE ACTIVITY TRANSFERRING NITROGENOUS GROUPS | 9 | 23 | 0.0003696 | 0.004933 |
| 70 | ENZYME REGULATOR ACTIVITY | 136 | 959 | 0.0003717 | 0.004933 |
| 71 | PROTEIN BINDING INVOLVED IN CELL CELL ADHESION | 6 | 11 | 0.0004318 | 0.005571 |
| 72 | SEMAPHORIN RECEPTOR ACTIVITY | 6 | 11 | 0.0004318 | 0.005571 |
| 73 | SYMPORTER ACTIVITY | 29 | 142 | 0.0004743 | 0.006036 |
| 74 | SECONDARY ACTIVE TRANSMEMBRANE TRANSPORTER ACTIVITY | 42 | 233 | 0.0005224 | 0.006558 |
| 75 | MONOVALENT INORGANIC CATION TRANSMEMBRANE TRANSPORTER ACTIVITY | 60 | 366 | 0.0005498 | 0.00681 |
| 76 | PROTEIN SERINE THREONINE KINASE ACTIVITY | 70 | 445 | 0.0006645 | 0.008123 |
| 77 | SOLUTE CATION SYMPORTER ACTIVITY | 22 | 99 | 0.0006753 | 0.008147 |
| 78 | EXTRACELLULAR MATRIX BINDING | 14 | 51 | 0.0007096 | 0.008451 |
| 79 | SODIUM AMINO ACID SYMPORTER ACTIVITY | 6 | 12 | 0.0007853 | 0.009006 |
| 80 | ORGANIC ACID SODIUM SYMPORTER ACTIVITY | 10 | 30 | 0.0007819 | 0.009006 |
| 81 | CATION AMINO ACID SYMPORTER ACTIVITY | 7 | 16 | 0.0007671 | 0.009006 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | NEURON PART | 217 | 1265 | 5.812e-13 | 3.394e-10 |
| 2 | MEMBRANE REGION | 198 | 1134 | 1.258e-12 | 3.673e-10 |
| 3 | INTRINSIC COMPONENT OF PLASMA MEMBRANE | 264 | 1649 | 4.396e-12 | 8.558e-10 |
| 4 | CELL JUNCTION | 193 | 1151 | 9.483e-11 | 1.127e-08 |
| 5 | PLASMA MEMBRANE REGION | 163 | 929 | 9.645e-11 | 1.127e-08 |
| 6 | NEURON PROJECTION | 164 | 942 | 1.553e-10 | 1.512e-08 |
| 7 | SYNAPSE | 137 | 754 | 3.038e-10 | 2.534e-08 |
| 8 | CELL SURFACE | 137 | 757 | 3.998e-10 | 2.918e-08 |
| 9 | ENDOPLASMIC RETICULUM | 248 | 1631 | 3.511e-09 | 2.278e-07 |
| 10 | APICAL PART OF CELL | 74 | 361 | 2.88e-08 | 1.682e-06 |
| 11 | SYNAPSE PART | 109 | 610 | 5.118e-08 | 2.717e-06 |
| 12 | MEMBRANE MICRODOMAIN | 62 | 288 | 5.975e-08 | 2.908e-06 |
| 13 | APICAL PLASMA MEMBRANE | 61 | 292 | 2.332e-07 | 9.844e-06 |
| 14 | SOMATODENDRITIC COMPARTMENT | 112 | 650 | 2.36e-07 | 9.844e-06 |
| 15 | CELL PROJECTION | 257 | 1786 | 2.627e-07 | 1.023e-05 |
| 16 | ENDOPLASMIC RETICULUM PART | 178 | 1163 | 4.446e-07 | 1.623e-05 |
| 17 | CELL CELL JUNCTION | 73 | 383 | 7.103e-07 | 2.44e-05 |
| 18 | MICROBODY | 34 | 134 | 1.249e-06 | 4.052e-05 |
| 19 | MICROBODY PART | 26 | 93 | 3.183e-06 | 9.785e-05 |
| 20 | NUCLEAR OUTER MEMBRANE ENDOPLASMIC RETICULUM MEMBRANE NETWORK | 153 | 1005 | 4.005e-06 | 0.000117 |
| 21 | AXON PART | 46 | 219 | 5.844e-06 | 0.0001625 |
| 22 | PROTEINACEOUS EXTRACELLULAR MATRIX | 66 | 356 | 6.276e-06 | 0.0001666 |
| 23 | DENDRITE | 79 | 451 | 7.477e-06 | 0.0001899 |
| 24 | AXON | 74 | 418 | 9.835e-06 | 0.0002393 |
| 25 | SIDE OF MEMBRANE | 75 | 428 | 1.243e-05 | 0.0002605 |
| 26 | RECEPTOR COMPLEX | 61 | 327 | 1.138e-05 | 0.0002605 |
| 27 | CELL BODY | 84 | 494 | 1.249e-05 | 0.0002605 |
| 28 | CELL PROJECTION PART | 143 | 946 | 1.208e-05 | 0.0002605 |
| 29 | EXTRACELLULAR MATRIX | 74 | 426 | 1.928e-05 | 0.0003882 |
| 30 | VACUOLE | 171 | 1180 | 2.055e-05 | 4e-04 |
| 31 | EXTRACELLULAR SPACE | 195 | 1376 | 2.143e-05 | 0.0004037 |
| 32 | PRESYNAPSE | 52 | 283 | 7.357e-05 | 0.001343 |
| 33 | MICROBODY MEMBRANE | 17 | 58 | 8.164e-05 | 0.001368 |
| 34 | PERINUCLEAR REGION OF CYTOPLASM | 100 | 642 | 7.819e-05 | 0.001368 |
| 35 | POSTSYNAPSE | 65 | 378 | 8.213e-05 | 0.001368 |
| 36 | MITOCHONDRION | 222 | 1633 | 8.43e-05 | 0.001368 |
| 37 | EXTERNAL SIDE OF PLASMA MEMBRANE | 45 | 238 | 0.0001101 | 0.001738 |
| 38 | BRUSH BORDER | 24 | 102 | 0.0001576 | 0.002423 |
| 39 | NEURON PROJECTION TERMINUS | 28 | 129 | 0.0002062 | 0.003087 |
| 40 | ACTIN CYTOSKELETON | 72 | 444 | 0.0002282 | 0.003332 |
| 41 | BASOLATERAL PLASMA MEMBRANE | 40 | 211 | 0.0002433 | 0.003466 |
| 42 | PLATELET ALPHA GRANULE | 19 | 75 | 0.0002705 | 0.003761 |
| 43 | CYTOSKELETON | 257 | 1967 | 0.000313 | 0.004106 |
| 44 | PLASMA MEMBRANE PROTEIN COMPLEX | 80 | 510 | 0.0003129 | 0.004106 |
| 45 | ANCHORED COMPONENT OF MEMBRANE | 31 | 152 | 0.0003164 | 0.004106 |
| 46 | CLUSTER OF ACTIN BASED CELL PROJECTIONS | 29 | 139 | 0.0003268 | 0.004148 |
| 47 | ANCHORING JUNCTION | 77 | 489 | 0.000357 | 0.004436 |
| 48 | ACTIN BASED CELL PROJECTION | 35 | 181 | 0.0003928 | 0.004778 |
| 49 | PLASMA MEMBRANE RECEPTOR COMPLEX | 34 | 175 | 0.0004313 | 0.005043 |
| 50 | SEMAPHORIN RECEPTOR COMPLEX | 6 | 11 | 0.0004318 | 0.005043 |
| 51 | ENDOSOME | 115 | 793 | 0.0004492 | 0.005144 |
| 52 | PLATELET ALPHA GRANULE LUMEN | 15 | 55 | 5e-04 | 0.005615 |
| 53 | TRANSPORT VESICLE | 56 | 338 | 0.0006361 | 0.007009 |
| 54 | AXONAL GROWTH CONE | 8 | 20 | 0.0006618 | 0.007157 |
| 55 | EXTRINSIC COMPONENT OF MEMBRANE | 44 | 252 | 0.0007806 | 0.008289 |
| 56 | CELL LEADING EDGE | 57 | 350 | 0.0008762 | 0.009138 |
| 57 | NEURON SPINE | 25 | 121 | 0.0009455 | 0.009358 |
| 58 | MAIN AXON | 15 | 58 | 0.0009182 | 0.009358 |
| 59 | TERMINAL BOUTON | 16 | 64 | 0.0009367 | 0.009358 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
| 1 | hsa04810_Regulation_of_actin_cytoskeleton | 41 | 387 | 0.5569 | 1 | |
| 2 | hsa04010_MAPK_signaling_pathway | 40 | 387 | 0.622 | 1 | |
| 3 | hsa04510_Focal_adhesion | 38 | 387 | 0.742 | 1 | |
| 4 | hsa04360_Axon_guidance | 29 | 387 | 0.9876 | 1 | |
| 5 | hsa04610_Complement_and_coagulation_cascades | 26 | 387 | 0.9975 | 1 | |
| 6 | hsa04910_Insulin_signaling_pathway | 25 | 387 | 0.9986 | 1 | |
| 7 | hsa04020_Calcium_signaling_pathway | 24 | 387 | 0.9993 | 1 | |
| 8 | hsa03320_PPAR_signaling_pathway | 23 | 387 | 0.9997 | 1 | |
| 9 | hsa04062_Chemokine_signaling_pathway | 23 | 387 | 0.9997 | 1 | |
| 10 | hsa04380_Osteoclast_differentiation | 22 | 387 | 0.9998 | 1 | |
| 11 | hsa04514_Cell_adhesion_molecules_.CAMs. | 22 | 387 | 0.9998 | 1 | |
| 12 | hsa04630_Jak.STAT_signaling_pathway | 22 | 387 | 0.9998 | 1 | |
| 13 | hsa04146_Peroxisome | 21 | 387 | 0.9999 | 1 | |
| 14 | hsa04920_Adipocytokine_signaling_pathway | 21 | 387 | 0.9999 | 1 | |
| 15 | hsa04974_Protein_digestion_and_absorption | 21 | 387 | 0.9999 | 1 | |
| 16 | hsa00010_Glycolysis_._Gluconeogenesis | 15 | 387 | 1 | 1 | |
| 17 | hsa00020_Citrate_cycle_.TCA_cycle. | 6 | 387 | 1 | 1 | |
| 18 | hsa00030_Pentose_phosphate_pathway | 6 | 387 | 1 | 1 | |
| 19 | hsa00040_Pentose_and_glucuronate_interconversions | 6 | 387 | 1 | 1 | |
| 20 | hsa00051_Fructose_and_mannose_metabolism | 8 | 387 | 1 | 1 | |
| 21 | hsa00052_Galactose_metabolism | 7 | 387 | 1 | 1 | |
| 22 | hsa00053_Ascorbate_and_aldarate_metabolism | 5 | 387 | 1 | 1 | |
| 23 | hsa00061_Fatty_acid_biosynthesis | 3 | 387 | 1 | 1 | |
| 24 | hsa00071_Fatty_acid_metabolism | 18 | 387 | 1 | 1 | |
| 25 | hsa00072_Synthesis_and_degradation_of_ketone_bodies | 3 | 387 | 1 | 1 | |
| 26 | hsa00120_Primary_bile_acid_biosynthesis | 10 | 387 | 1 | 1 | |
| 27 | hsa00140_Steroid_hormone_biosynthesis | 14 | 387 | 1 | 1 | |
| 28 | hsa00190_Oxidative_phosphorylation | 8 | 387 | 1 | 1 | |
| 29 | hsa00230_Purine_metabolism | 17 | 387 | 1 | 1 | |
| 30 | hsa00240_Pyrimidine_metabolism | 13 | 387 | 1 | 1 | |
| 31 | hsa00250_Alanine._aspartate_and_glutamate_metabolism | 12 | 387 | 1 | 1 | |
| 32 | hsa00260_Glycine._serine_and_threonine_metabolism | 14 | 387 | 1 | 1 | |
| 33 | hsa00270_Cysteine_and_methionine_metabolism | 9 | 387 | 1 | 1 | |
| 34 | hsa00280_Valine._leucine_and_isoleucine_degradation | 19 | 387 | 1 | 1 | |
| 35 | hsa00290_Valine._leucine_and_isoleucine_biosynthesis | 2 | 387 | 1 | 1 | |
| 36 | hsa00300_Lysine_biosynthesis | 2 | 387 | 1 | 1 | |
| 37 | hsa00310_Lysine_degradation | 12 | 387 | 1 | 1 | |
| 38 | hsa00330_Arginine_and_proline_metabolism | 14 | 387 | 1 | 1 | |
| 39 | hsa00340_Histidine_metabolism | 8 | 387 | 1 | 1 | |
| 40 | hsa00350_Tyrosine_metabolism | 9 | 387 | 1 | 1 | |
| 41 | hsa00360_Phenylalanine_metabolism | 7 | 387 | 1 | 1 | |
| 42 | hsa00380_Tryptophan_metabolism | 13 | 387 | 1 | 1 | |
| 43 | hsa00400_Phenylalanine._tyrosine_and_tryptophan_biosynthesis | 4 | 387 | 1 | 1 | |
| 44 | hsa00410_beta.Alanine_metabolism | 9 | 387 | 1 | 1 | |
| 45 | hsa00430_Taurine_and_hypotaurine_metabolism | 4 | 387 | 1 | 1 | |
| 46 | hsa00450_Selenocompound_metabolism | 3 | 387 | 1 | 1 | |
| 47 | hsa00471_D.Glutamine_and_D.glutamate_metabolism | 2 | 387 | 1 | 1 | |
| 48 | hsa00480_Glutathione_metabolism | 6 | 387 | 1 | 1 | |
| 49 | hsa00500_Starch_and_sucrose_metabolism | 14 | 387 | 1 | 1 | |
| 50 | hsa00510_N.Glycan_biosynthesis | 5 | 387 | 1 | 1 | |
| 51 | hsa00511_Other_glycan_degradation | 2 | 387 | 1 | 1 | |
| 52 | hsa00512_Mucin_type_O.Glycan_biosynthesis | 5 | 387 | 1 | 1 | |
| 53 | hsa00514_Other_types_of_O.glycan_biosynthesis | 6 | 387 | 1 | 1 | |
| 54 | hsa00520_Amino_sugar_and_nucleotide_sugar_metabolism | 7 | 387 | 1 | 1 | |
| 55 | hsa00531_Glycosaminoglycan_degradation | 2 | 387 | 1 | 1 | |
| 56 | hsa00533_Glycosaminoglycan_biosynthesis_._keratan_sulfate | 4 | 387 | 1 | 1 | |
| 57 | hsa00534_Glycosaminoglycan_biosynthesis_._heparan_sulfate | 4 | 387 | 1 | 1 | |
| 58 | hsa00561_Glycerolipid_metabolism | 15 | 387 | 1 | 1 | |
| 59 | hsa00562_Inositol_phosphate_metabolism | 9 | 387 | 1 | 1 | |
| 60 | hsa00563_Glycosylphosphatidylinositol.GPI..anchor_biosynthesis | 3 | 387 | 1 | 1 | |
| 61 | hsa00564_Glycerophospholipid_metabolism | 13 | 387 | 1 | 1 | |
| 62 | hsa00565_Ether_lipid_metabolism | 3 | 387 | 1 | 1 | |
| 63 | hsa00590_Arachidonic_acid_metabolism | 14 | 387 | 1 | 1 | |
| 64 | hsa00591_Linoleic_acid_metabolism | 9 | 387 | 1 | 1 | |
| 65 | hsa00592_alpha.Linolenic_acid_metabolism | 3 | 387 | 1 | 1 | |
| 66 | hsa00600_Sphingolipid_metabolism | 4 | 387 | 1 | 1 | |
| 67 | hsa00601_Glycosphingolipid_biosynthesis_._lacto_and_neolacto_series | 5 | 387 | 1 | 1 | |
| 68 | hsa00603_Glycosphingolipid_biosynthesis_._globo_series | 3 | 387 | 1 | 1 | |
| 69 | hsa00604_Glycosphingolipid_biosynthesis_._ganglio_series | 3 | 387 | 1 | 1 | |
| 70 | hsa00620_Pyruvate_metabolism | 10 | 387 | 1 | 1 | |
| 71 | hsa00630_Glyoxylate_and_dicarboxylate_metabolism | 5 | 387 | 1 | 1 | |
| 72 | hsa00640_Propanoate_metabolism | 11 | 387 | 1 | 1 | |
| 73 | hsa00650_Butanoate_metabolism | 10 | 387 | 1 | 1 | |
| 74 | hsa00670_One_carbon_pool_by_folate | 3 | 387 | 1 | 1 | |
| 75 | hsa00740_Riboflavin_metabolism | 2 | 387 | 1 | 1 | |
| 76 | hsa00750_Vitamin_B6_metabolism | 3 | 387 | 1 | 1 | |
| 77 | hsa00760_Nicotinate_and_nicotinamide_metabolism | 6 | 387 | 1 | 1 | |
| 78 | hsa00770_Pantothenate_and_CoA_biosynthesis | 7 | 387 | 1 | 1 | |
| 79 | hsa00790_Folate_biosynthesis | 2 | 387 | 1 | 1 | |
| 80 | hsa00830_Retinol_metabolism | 19 | 387 | 1 | 1 | |
| 81 | hsa00860_Porphyrin_and_chlorophyll_metabolism | 6 | 387 | 1 | 1 | |
| 82 | hsa00900_Terpenoid_backbone_biosynthesis | 4 | 387 | 1 | 1 | |
| 83 | hsa00910_Nitrogen_metabolism | 6 | 387 | 1 | 1 | |
| 84 | hsa00920_Sulfur_metabolism | 2 | 387 | 1 | 1 | |
| 85 | hsa00970_Aminoacyl.tRNA_biosynthesis | 2 | 387 | 1 | 1 | |
| 86 | hsa00980_Metabolism_of_xenobiotics_by_cytochrome_P450 | 18 | 387 | 1 | 1 | |
| 87 | hsa00982_Drug_metabolism_._cytochrome_P450 | 19 | 387 | 1 | 1 | |
| 88 | hsa00983_Drug_metabolism_._other_enzymes | 13 | 387 | 1 | 1 | |
| 89 | hsa01040_Biosynthesis_of_unsaturated_fatty_acids | 8 | 387 | 1 | 1 | |
| 90 | hsa02010_ABC_transporters | 12 | 387 | 1 | 1 | |
| 91 | hsa03008_Ribosome_biogenesis_in_eukaryotes | 6 | 387 | 1 | 1 | |
| 92 | hsa03013_RNA_transport | 8 | 387 | 1 | 1 | |
| 93 | hsa03015_mRNA_surveillance_pathway | 8 | 387 | 1 | 1 | |
| 94 | hsa03018_RNA_degradation | 6 | 387 | 1 | 1 | |
| 95 | hsa03020_RNA_polymerase | 2 | 387 | 1 | 1 | |
| 96 | hsa03030_DNA_replication | 2 | 387 | 1 | 1 | |
| 97 | hsa03040_Spliceosome | 8 | 387 | 1 | 1 | |
| 98 | hsa03410_Base_excision_repair | 2 | 387 | 1 | 1 | |
| 99 | hsa03440_Homologous_recombination | 4 | 387 | 1 | 1 | |
| 100 | hsa04012_ErbB_signaling_pathway | 17 | 387 | 1 | 1 | |
| 101 | hsa04070_Phosphatidylinositol_signaling_system | 15 | 387 | 1 | 1 | |
| 102 | hsa04110_Cell_cycle | 19 | 387 | 1 | 1 | |
| 103 | hsa04114_Oocyte_meiosis | 15 | 387 | 1 | 1 | |
| 104 | hsa04115_p53_signaling_pathway | 20 | 387 | 1 | 1 | |
| 105 | hsa04120_Ubiquitin_mediated_proteolysis | 9 | 387 | 1 | 1 | |
| 106 | hsa04130_SNARE_interactions_in_vesicular_transport | 3 | 387 | 1 | 1 | |
| 107 | hsa04140_Regulation_of_autophagy | 3 | 387 | 1 | 1 | |
| 108 | hsa04141_Protein_processing_in_endoplasmic_reticulum | 13 | 387 | 1 | 1 | |
| 109 | hsa04142_Lysosome | 10 | 387 | 1 | 1 | |
| 110 | hsa04144_Endocytosis | 19 | 387 | 1 | 1 | |
| 111 | hsa04145_Phagosome | 20 | 387 | 1 | 1 | |
| 112 | hsa04150_mTOR_signaling_pathway | 7 | 387 | 1 | 1 | |
| 113 | hsa04210_Apoptosis | 16 | 387 | 1 | 1 | |
| 114 | hsa04260_Cardiac_muscle_contraction | 4 | 387 | 1 | 1 | |
| 115 | hsa04270_Vascular_smooth_muscle_contraction | 18 | 387 | 1 | 1 | |
| 116 | hsa04310_Wnt_signaling_pathway | 20 | 387 | 1 | 1 | |
| 117 | hsa04320_Dorso.ventral_axis_formation | 5 | 387 | 1 | 1 | |
| 118 | hsa04330_Notch_signaling_pathway | 2 | 387 | 1 | 1 | |
| 119 | hsa04340_Hedgehog_signaling_pathway | 5 | 387 | 1 | 1 | |
| 120 | hsa04350_TGF.beta_signaling_pathway | 16 | 387 | 1 | 1 | |
| 121 | hsa04370_VEGF_signaling_pathway | 13 | 387 | 1 | 1 | |
| 122 | hsa04512_ECM.receptor_interaction | 19 | 387 | 1 | 1 | |
| 123 | hsa04520_Adherens_junction | 10 | 387 | 1 | 1 | |
| 124 | hsa04530_Tight_junction | 19 | 387 | 1 | 1 | |
| 125 | hsa04540_Gap_junction | 13 | 387 | 1 | 1 | |
| 126 | hsa04612_Antigen_processing_and_presentation | 6 | 387 | 1 | 1 | |
| 127 | hsa04614_Renin.angiotensin_system | 8 | 387 | 1 | 1 | |
| 128 | hsa04620_Toll.like_receptor_signaling_pathway | 16 | 387 | 1 | 1 | |
| 129 | hsa04621_NOD.like_receptor_signaling_pathway | 6 | 387 | 1 | 1 | |
| 130 | hsa04622_RIG.I.like_receptor_signaling_pathway | 5 | 387 | 1 | 1 | |
| 131 | hsa04623_Cytosolic_DNA.sensing_pathway | 2 | 387 | 1 | 1 | |
| 132 | hsa04640_Hematopoietic_cell_lineage | 14 | 387 | 1 | 1 | |
| 133 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 18 | 387 | 1 | 1 | |
| 134 | hsa04660_T_cell_receptor_signaling_pathway | 15 | 387 | 1 | 1 | |
| 135 | hsa04662_B_cell_receptor_signaling_pathway | 11 | 387 | 1 | 1 | |
| 136 | hsa04664_Fc_epsilon_RI_signaling_pathway | 16 | 387 | 1 | 1 | |
| 137 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 18 | 387 | 1 | 1 | |
| 138 | hsa04670_Leukocyte_transendothelial_migration | 14 | 387 | 1 | 1 | |
| 139 | hsa04672_Intestinal_immune_network_for_IgA_production | 4 | 387 | 1 | 1 | |
| 140 | hsa04710_Circadian_rhythm_._mammal | 6 | 387 | 1 | 1 | |
| 141 | hsa04720_Long.term_potentiation | 9 | 387 | 1 | 1 | |
| 142 | hsa04722_Neurotrophin_signaling_pathway | 19 | 387 | 1 | 1 | |
| 143 | hsa04730_Long.term_depression | 12 | 387 | 1 | 1 | |
| 144 | hsa04740_Olfactory_transduction | 3 | 388 | 1 | 1 | |
| 145 | hsa04742_Taste_transduction | 3 | 387 | 1 | 1 | |
| 146 | hsa04912_GnRH_signaling_pathway | 16 | 387 | 1 | 1 | |
| 147 | hsa04914_Progesterone.mediated_oocyte_maturation | 14 | 387 | 1 | 1 | |
| 148 | hsa04916_Melanogenesis | 15 | 387 | 1 | 1 | |
| 149 | hsa04960_Aldosterone.regulated_sodium_reabsorption | 9 | 387 | 1 | 1 | |
| 150 | hsa04962_Vasopressin.regulated_water_reabsorption | 4 | 387 | 1 | 1 | |
| 151 | hsa04964_Proximal_tubule_bicarbonate_reclamation | 7 | 387 | 1 | 1 | |
| 152 | hsa04966_Collecting_duct_acid_secretion | 5 | 387 | 1 | 1 | |
| 153 | hsa04970_Salivary_secretion | 12 | 387 | 1 | 1 | |
| 154 | hsa04971_Gastric_acid_secretion | 11 | 387 | 1 | 1 | |
| 155 | hsa04972_Pancreatic_secretion | 14 | 387 | 1 | 1 | |
| 156 | hsa04973_Carbohydrate_digestion_and_absorption | 6 | 387 | 1 | 1 | |
| 157 | hsa04975_Fat_digestion_and_absorption | 9 | 387 | 1 | 1 | |
| 158 | hsa04976_Bile_secretion | 19 | 387 | 1 | 1 | |
| 159 | hsa04977_Vitamin_digestion_and_absorption | 6 | 387 | 1 | 1 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | FAM201A |
hsa-miR-100-3p;hsa-miR-105-5p;hsa-miR-1178-3p;hsa-miR-1271-5p;hsa-miR-1299;hsa-miR-135b-5p;hsa-miR-147a;hsa-miR-149-5p;hsa-miR-558;hsa-miR-561-3p;hsa-miR-708-5p | 11 | CACNA1E | Sponge network | 0.028 | 0.75302 | 0.17 | 0.40772 | 0.755 |
| 2 | SNX29P2 | hsa-miR-1237-3p;hsa-miR-125a-5p;hsa-miR-133a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-145-5p;hsa-miR-146a-5p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-544a | 10 | EGFR | Sponge network | 0.195 | 0.26721 | -0.075 | 0.75424 | 0.581 |
| 3 | FAM201A |
hsa-miR-147a;hsa-miR-217;hsa-miR-326;hsa-miR-506-3p;hsa-miR-509-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-607;hsa-miR-641;hsa-miR-7-1-3p | 10 | AKAP6 | Sponge network | 0.028 | 0.75302 | 0.08 | 0.08966 | 0.54 |
| 4 | CCDC26 |
hsa-miR-1255a;hsa-miR-1270;hsa-miR-1291;hsa-miR-1304-5p;hsa-miR-141-3p;hsa-miR-1538;hsa-miR-1911-3p;hsa-miR-448;hsa-miR-507;hsa-miR-590-3p;hsa-miR-641;hsa-miR-661 | 12 | CLYBL | Sponge network | 0.054 | 0.50959 | 0.345 | 0.14058 | 0.5 |
| 5 | CCDC26 |
hsa-miR-1200;hsa-miR-1256;hsa-miR-1257;hsa-miR-1262;hsa-miR-1271-5p;hsa-miR-1294;hsa-miR-1305;hsa-miR-141-3p;hsa-miR-199b-5p;hsa-miR-548n;hsa-miR-548o-3p | 11 | ZNF618 | Sponge network | 0.054 | 0.50959 | 0.083 | 0.11003 | 0.483 |
| 6 | CCDC26 |
hsa-miR-105-5p;hsa-miR-1299;hsa-miR-133a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-182-3p;hsa-miR-29b-2-5p;hsa-miR-302e;hsa-miR-620 | 10 | EGFR | Sponge network | 0.054 | 0.50959 | -0.075 | 0.75424 | 0.45 |
| 7 | CCDC26 |
hsa-miR-1224-5p;hsa-miR-1226-3p;hsa-miR-1271-5p;hsa-miR-1294;hsa-miR-150-5p;hsa-miR-200c-3p;hsa-miR-206;hsa-miR-31-5p;hsa-miR-541-5p;hsa-miR-661 | 10 | NDST1 | Sponge network | 0.054 | 0.50959 | 0.015 | 0.85538 | 0.416 |
| 8 | CCDC26 |
hsa-miR-1253;hsa-miR-126-5p;hsa-miR-1290;hsa-miR-1297;hsa-miR-1299;hsa-miR-1305;hsa-miR-181d-5p;hsa-miR-200a-5p;hsa-miR-302e;hsa-miR-548a-5p;hsa-miR-548e-3p;hsa-miR-548h-5p | 12 | MCC | Sponge network | 0.054 | 0.50959 | 0.041 | 0.8141 | 0.415 |