This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-30a-5p | ABL2 | -0.92 | 0.00076 | 0.31 | 0.01567 | mirMAP; miRNATAP | -0.11 | 0 | NA | |
| 2 | hsa-miR-30a-5p | ADAM22 | -0.92 | 0.00076 | 0.04 | 0.92539 | MirTarget; miRNATAP | -0.16 | 0.01239 | NA | |
| 3 | hsa-miR-30a-5p | ADAMTS6 | -0.92 | 0.00076 | 0.37 | 0.18207 | MirTarget; miRNATAP | -0.15 | 0.00099 | NA | |
| 4 | hsa-miR-30a-5p | ALG10 | -0.92 | 0.00076 | 0.64 | 5.0E-5 | MirTarget | -0.13 | 0 | NA | |
| 5 | hsa-miR-30a-5p | ALG6 | -0.92 | 0.00076 | 0.8 | 0 | MirTarget | -0.11 | 0 | NA | |
| 6 | hsa-miR-30a-5p | AMMECR1 | -0.92 | 0.00076 | 0.59 | 1.0E-5 | miRNATAP | -0.11 | 0 | NA | |
| 7 | hsa-miR-30a-5p | ANTXR1 | -0.92 | 0.00076 | -0.82 | 0.00129 | mirMAP | -0.1 | 0.018 | NA | |
| 8 | hsa-miR-30a-5p | ARL4A | -0.92 | 0.00076 | -0.32 | 0.06979 | MirTarget | -0.1 | 0.00042 | NA | |
| 9 | hsa-miR-30a-5p | ARL4C | -0.92 | 0.00076 | 0.66 | 0.00161 | miRNATAP | -0.12 | 0.00063 | NA | |
| 10 | hsa-miR-30a-5p | ARL6IP6 | -0.92 | 0.00076 | 0.14 | 0.36045 | miRNATAP | -0.13 | 0 | NA | |
| 11 | hsa-miR-30a-5p | ATL2 | -0.92 | 0.00076 | 0.61 | 3.0E-5 | miRNAWalker2 validate; miRNATAP | -0.1 | 2.0E-5 | NA | |
| 12 | hsa-miR-30a-5p | ATL3 | -0.92 | 0.00076 | 0.11 | 0.46975 | mirMAP | -0.11 | 1.0E-5 | NA | |
| 13 | hsa-miR-30a-5p | ATP2A2 | -0.92 | 0.00076 | 0.63 | 1.0E-5 | miRNAWalker2 validate; mirMAP | -0.16 | 0 | NA | |
| 14 | hsa-miR-30a-5p | ATP6V1C1 | -0.92 | 0.00076 | 0.29 | 0.00966 | miRNAWalker2 validate | -0.12 | 0 | NA | |
| 15 | hsa-miR-30a-5p | ATP8B1 | -0.92 | 0.00076 | 0.31 | 0.17188 | miRNATAP | -0.11 | 0.00311 | NA | |
| 16 | hsa-miR-30a-5p | AVEN | -0.92 | 0.00076 | 0.61 | 4.0E-5 | miRNAWalker2 validate | -0.16 | 0 | 23445407 | At least one of these targets the anti-apoptotic protein AVEN was able to partially revert the effect of miR-30a overexpression |
| 17 | hsa-miR-30a-5p | AVL9 | -0.92 | 0.00076 | 0.94 | 0 | MirTarget; mirMAP; miRNATAP | -0.17 | 0 | NA | |
| 18 | hsa-miR-30a-5p | AZIN1 | -0.92 | 0.00076 | 0.34 | 0.00945 | miRNATAP | -0.14 | 0 | NA | |
| 19 | hsa-miR-30a-5p | B3GNT5 | -0.92 | 0.00076 | -0.33 | 0.14374 | MirTarget; miRNATAP | -0.13 | 0.00065 | NA | |
| 20 | hsa-miR-30a-5p | B4GALT1 | -0.92 | 0.00076 | 0.36 | 0.02384 | mirMAP | -0.18 | 0 | NA | |
| 21 | hsa-miR-30a-5p | BCL9 | -0.92 | 0.00076 | 1.24 | 0 | miRNATAP | -0.18 | 0 | NA | |
| 22 | hsa-miR-30a-5p | BEND3 | -0.92 | 0.00076 | 0.58 | 0.00022 | mirMAP; miRNATAP | -0.12 | 0 | NA | |
| 23 | hsa-miR-30a-5p | BMS1 | -0.92 | 0.00076 | 0.42 | 5.0E-5 | miRNAWalker2 validate | -0.11 | 0 | NA | |
| 24 | hsa-miR-30a-5p | BRAP | -0.92 | 0.00076 | 0.19 | 0.03293 | miRNATAP | -0.1 | 0 | NA | |
| 25 | hsa-miR-30a-5p | BRWD3 | -0.92 | 0.00076 | 0.68 | 0.00035 | MirTarget; miRNATAP | -0.12 | 0.00016 | NA | |
| 26 | hsa-miR-30a-5p | C16orf87 | -0.92 | 0.00076 | 0.57 | 1.0E-5 | miRNATAP | -0.16 | 0 | NA | |
| 27 | hsa-miR-30a-5p | C4orf46 | -0.92 | 0.00076 | 0.7 | 1.0E-5 | mirMAP | -0.25 | 0 | NA | |
| 28 | hsa-miR-30a-5p | CALU | -0.92 | 0.00076 | 0.7 | 3.0E-5 | MirTarget | -0.25 | 0 | NA | |
| 29 | hsa-miR-30a-5p | CAND1 | -0.92 | 0.00076 | 0.38 | 0.00711 | mirMAP | -0.12 | 0 | NA | |
| 30 | hsa-miR-30a-5p | CARS | -0.92 | 0.00076 | 0.48 | 1.0E-5 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.12 | 0 | NA | |
| 31 | hsa-miR-30a-5p | CBX2 | -0.92 | 0.00076 | 1.86 | 0 | miRNATAP | -0.4 | 0 | NA | |
| 32 | hsa-miR-30a-5p | CBX3 | -0.92 | 0.00076 | 1.01 | 0 | MirTarget | -0.22 | 0 | NA | |
| 33 | hsa-miR-30a-5p | CCDC43 | -0.92 | 0.00076 | 0.57 | 0 | MirTarget | -0.17 | 0 | NA | |
| 34 | hsa-miR-30a-5p | CCNE2 | -0.92 | 0.00076 | 2.15 | 0 | miRNATAP | -0.49 | 0 | NA | |
| 35 | hsa-miR-30a-5p | CDC123 | -0.92 | 0.00076 | 0.41 | 0.00055 | miRNAWalker2 validate | -0.1 | 0 | NA | |
| 36 | hsa-miR-30a-5p | CDC20 | -0.92 | 0.00076 | 3.63 | 0 | miRNAWalker2 validate | -0.66 | 0 | NA | |
| 37 | hsa-miR-30a-5p | CDCA7 | -0.92 | 0.00076 | 2.75 | 0 | MirTarget | -0.31 | 0 | NA | |
| 38 | hsa-miR-30a-5p | CDH2 | -0.92 | 0.00076 | 0.21 | 0.68235 | miRNAWalker2 validate | -0.38 | 1.0E-5 | 21633953 | Consistent with this microRNA-30a expression was found inversely proportional to the invasive potential of various NSCLC cell lines correlating positively with E-cadherin epithelial marker and negatively with N-cadherin mesenchymal marker expression |
| 39 | hsa-miR-30a-5p | CELF3 | -0.92 | 0.00076 | 1.15 | 0.11981 | MirTarget | -0.48 | 0.0001 | NA | |
| 40 | hsa-miR-30a-5p | CELSR3 | -0.92 | 0.00076 | 3.19 | 0 | MirTarget; miRNATAP | -0.41 | 0 | NA | |
| 41 | hsa-miR-30a-5p | CEP72 | -0.92 | 0.00076 | 1.46 | 0 | miRNAWalker2 validate | -0.23 | 0 | NA | |
| 42 | hsa-miR-30a-5p | CEP76 | -0.92 | 0.00076 | 0.44 | 0.0025 | MirTarget | -0.12 | 0 | NA | |
| 43 | hsa-miR-30a-5p | CHST1 | -0.92 | 0.00076 | -0.09 | 0.76687 | MirTarget | -0.2 | 0.0001 | NA | |
| 44 | hsa-miR-30a-5p | CIT | -0.92 | 0.00076 | 1.27 | 0.001 | miRNATAP | -0.18 | 0.00634 | NA | |
| 45 | hsa-miR-30a-5p | CLDN12 | -0.92 | 0.00076 | 1.02 | 0 | mirMAP | -0.12 | 2.0E-5 | NA | |
| 46 | hsa-miR-30a-5p | CMC1 | -0.92 | 0.00076 | 0.34 | 0.01376 | mirMAP | -0.12 | 0 | NA | |
| 47 | hsa-miR-30a-5p | CNP | -0.92 | 0.00076 | 0.68 | 0 | miRNAWalker2 validate | -0.14 | 0 | NA | |
| 48 | hsa-miR-30a-5p | COL12A1 | -0.92 | 0.00076 | -0.85 | 0.03497 | mirMAP | -0.37 | 0 | NA | |
| 49 | hsa-miR-30a-5p | COPS7B | -0.92 | 0.00076 | 0.71 | 0 | miRNAWalker2 validate; mirMAP; miRNATAP | -0.1 | 0 | NA | |
| 50 | hsa-miR-30a-5p | CPD | -0.92 | 0.00076 | 0.91 | 9.0E-5 | mirMAP | -0.25 | 0 | NA | |
| 51 | hsa-miR-30a-5p | CPLX1 | -0.92 | 0.00076 | 0.63 | 0.05262 | mirMAP | -0.33 | 0 | NA | |
| 52 | hsa-miR-30a-5p | CPOX | -0.92 | 0.00076 | 0.87 | 0 | miRNAWalker2 validate | -0.13 | 0 | NA | |
| 53 | hsa-miR-30a-5p | CPSF1 | -0.92 | 0.00076 | 0.84 | 0 | miRNAWalker2 validate | -0.11 | 0.00011 | NA | |
| 54 | hsa-miR-30a-5p | CPSF3 | -0.92 | 0.00076 | 0.74 | 0 | miRNAWalker2 validate | -0.16 | 0 | NA | |
| 55 | hsa-miR-30a-5p | CPSF4 | -0.92 | 0.00076 | 0.7 | 0 | miRNAWalker2 validate | -0.14 | 0 | NA | |
| 56 | hsa-miR-30a-5p | CPSF6 | -0.92 | 0.00076 | 0.62 | 0 | MirTarget; miRNATAP | -0.11 | 0 | NA | |
| 57 | hsa-miR-30a-5p | CTHRC1 | -0.92 | 0.00076 | 3.35 | 0 | MirTarget | -0.65 | 0 | NA | |
| 58 | hsa-miR-30a-5p | CUL2 | -0.92 | 0.00076 | 0.19 | 0.08607 | MirTarget; miRNATAP | -0.11 | 0 | NA | |
| 59 | hsa-miR-30a-5p | CYP24A1 | -0.92 | 0.00076 | 5.26 | 0 | miRNATAP | -0.69 | 0 | NA | |
| 60 | hsa-miR-30a-5p | DARS2 | -0.92 | 0.00076 | 1.03 | 0 | miRNAWalker2 validate | -0.24 | 0 | NA | |
| 61 | hsa-miR-30a-5p | DBF4 | -0.92 | 0.00076 | 1.26 | 0 | MirTarget | -0.32 | 0 | NA | |
| 62 | hsa-miR-30a-5p | DDX10 | -0.92 | 0.00076 | 0.39 | 0.00203 | miRNAWalker2 validate | -0.13 | 0 | NA | |
| 63 | hsa-miR-30a-5p | DDX52 | -0.92 | 0.00076 | 0.67 | 0 | mirMAP | -0.13 | 0 | NA | |
| 64 | hsa-miR-30a-5p | DERL1 | -0.92 | 0.00076 | 0.12 | 0.24598 | miRNAWalker2 validate | -0.12 | 0 | NA | |
| 65 | hsa-miR-30a-5p | DHX33 | -0.92 | 0.00076 | 0.21 | 0.19056 | miRNAWalker2 validate | -0.11 | 5.0E-5 | NA | |
| 66 | hsa-miR-30a-5p | DHX36 | -0.92 | 0.00076 | 0.08 | 0.48345 | miRNAWalker2 validate | -0.11 | 0 | NA | |
| 67 | hsa-miR-30a-5p | DIO2 | -0.92 | 0.00076 | 1.92 | 0 | mirMAP; miRNATAP | -0.47 | 0 | NA | |
| 68 | hsa-miR-30a-5p | DLG5 | -0.92 | 0.00076 | 1.13 | 0 | miRNATAP | -0.11 | 0.00011 | NA | |
| 69 | hsa-miR-30a-5p | DLGAP4 | -0.92 | 0.00076 | 0.27 | 0.02041 | MirTarget; miRNATAP | -0.11 | 0 | NA | |
| 70 | hsa-miR-30a-5p | DNAJA1 | -0.92 | 0.00076 | -0.13 | 0.31726 | miRNAWalker2 validate | -0.13 | 0 | NA | |
| 71 | hsa-miR-30a-5p | DNAJC2 | -0.92 | 0.00076 | 0.82 | 0 | miRNAWalker2 validate | -0.18 | 0 | NA | |
| 72 | hsa-miR-30a-5p | DNMT3A | -0.92 | 0.00076 | 1.2 | 0 | MirTarget; miRNATAP | -0.24 | 0 | NA | |
| 73 | hsa-miR-30a-5p | DPY19L1 | -0.92 | 0.00076 | 0.84 | 8.0E-5 | miRNAWalker2 validate; miRNATAP | -0.14 | 6.0E-5 | NA | |
| 74 | hsa-miR-30a-5p | DRP2 | -0.92 | 0.00076 | 2.45 | 0 | mirMAP | -0.65 | 0 | NA | |
| 75 | hsa-miR-30a-5p | DSG2 | -0.92 | 0.00076 | 0.96 | 0.00021 | miRNAWalker2 validate; MirTarget | -0.1 | 0.01959 | NA | |
| 76 | hsa-miR-30a-5p | DSP | -0.92 | 0.00076 | 2.27 | 0 | MirTarget | -0.37 | 0 | NA | |
| 77 | hsa-miR-30a-5p | DTL | -0.92 | 0.00076 | 1.88 | 0 | miRNAWalker2 validate; miRTarBase | -0.4 | 0 | 22287560 | MiR 30a 5p suppresses tumor growth in colon carcinoma by targeting DTL; Subsequent reporter gene assays confirmed the predicted miR-30a-5p binding site in the 3'untranslated region of DTL; In conclusion our data identified miR-30a-5p as a tumor-suppressing miRNA in colon cancer cells exerting its function via modulation of DTL expression which is frequently overexpressed in colorectal cancer |
| 78 | hsa-miR-30a-5p | E2F3 | -0.92 | 0.00076 | 1.31 | 0 | miRNATAP | -0.21 | 0 | NA | |
| 79 | hsa-miR-30a-5p | E2F7 | -0.92 | 0.00076 | 2.4 | 0 | miRNATAP | -0.33 | 0 | NA | |
| 80 | hsa-miR-30a-5p | EED | -0.92 | 0.00076 | 0.65 | 0 | MirTarget; miRNATAP | -0.15 | 0 | NA | |
| 81 | hsa-miR-30a-5p | EFNA3 | -0.92 | 0.00076 | 2.4 | 0 | MirTarget; mirMAP; miRNATAP | -0.32 | 0 | NA | |
| 82 | hsa-miR-30a-5p | EIF2AK2 | -0.92 | 0.00076 | 0.62 | 9.0E-5 | mirMAP | -0.15 | 0 | NA | |
| 83 | hsa-miR-30a-5p | EIF2B1 | -0.92 | 0.00076 | 0.35 | 0.0002 | miRNAWalker2 validate | -0.12 | 0 | NA | |
| 84 | hsa-miR-30a-5p | EIF5A2 | -0.92 | 0.00076 | -0.01 | 0.97137 | mirMAP; miRNATAP | -0.11 | 0.00111 | NA | |
| 85 | hsa-miR-30a-5p | ELMOD2 | -0.92 | 0.00076 | 0.46 | 6.0E-5 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.1 | 0 | NA | |
| 86 | hsa-miR-30a-5p | ENPP1 | -0.92 | 0.00076 | 0.22 | 0.5129 | mirMAP | -0.26 | 0 | NA | |
| 87 | hsa-miR-30a-5p | ERRFI1 | -0.92 | 0.00076 | 0.44 | 0.10678 | MirTarget | -0.17 | 0.00017 | NA | |
| 88 | hsa-miR-30a-5p | ESPN | -0.92 | 0.00076 | 2.31 | 0 | MirTarget; miRNATAP | -0.32 | 3.0E-5 | NA | |
| 89 | hsa-miR-30a-5p | FAM104A | -0.92 | 0.00076 | 0.38 | 3.0E-5 | MirTarget | -0.13 | 0 | NA | |
| 90 | hsa-miR-30a-5p | FAM126A | -0.92 | 0.00076 | 0.13 | 0.58359 | mirMAP | -0.14 | 0.0003 | NA | |
| 91 | hsa-miR-30a-5p | FAM196A | -0.92 | 0.00076 | 0.36 | 0.37 | MirTarget; miRNATAP | -0.18 | 0.00617 | NA | |
| 92 | hsa-miR-30a-5p | FAM199X | -0.92 | 0.00076 | 0.79 | 0 | mirMAP; miRNATAP | -0.15 | 0 | NA | |
| 93 | hsa-miR-30a-5p | FAM69A | -0.92 | 0.00076 | 0.75 | 1.0E-5 | MirTarget | -0.11 | 0.00018 | NA | |
| 94 | hsa-miR-30a-5p | FAM91A1 | -0.92 | 0.00076 | 0.6 | 0 | MirTarget | -0.15 | 0 | NA | |
| 95 | hsa-miR-30a-5p | FAP | -0.92 | 0.00076 | 1.77 | 0 | miRNATAP | -0.35 | 0 | NA | |
| 96 | hsa-miR-30a-5p | FBXL20 | -0.92 | 0.00076 | 0.58 | 7.0E-5 | miRNATAP | -0.1 | 2.0E-5 | NA | |
| 97 | hsa-miR-30a-5p | FBXO45 | -0.92 | 0.00076 | 0.67 | 2.0E-5 | miRNATAP | -0.21 | 0 | NA | |
| 98 | hsa-miR-30a-5p | FGF5 | -0.92 | 0.00076 | 1.42 | 0.00664 | mirMAP | -0.46 | 0 | NA | |
| 99 | hsa-miR-30a-5p | FKBP3 | -0.92 | 0.00076 | 0.93 | 0 | MirTarget | -0.22 | 0 | NA | |
| 100 | hsa-miR-30a-5p | FLVCR1 | -0.92 | 0.00076 | 1.37 | 0 | MirTarget | -0.23 | 0 | NA | |
| 101 | hsa-miR-30a-5p | FNDC3B | -0.92 | 0.00076 | 0.22 | 0.12432 | miRNAWalker2 validate; miRNATAP | -0.11 | 1.0E-5 | NA | |
| 102 | hsa-miR-30a-5p | FOXA1 | -0.92 | 0.00076 | 1.08 | 0.00032 | miRNATAP | -0.41 | 0 | NA | |
| 103 | hsa-miR-30a-5p | FOXP2 | -0.92 | 0.00076 | -0.82 | 0.04073 | mirMAP | -0.16 | 0.02004 | NA | |
| 104 | hsa-miR-30a-5p | FZD3 | -0.92 | 0.00076 | 0.79 | 0.00088 | MirTarget; miRNATAP | -0.21 | 0 | NA | |
| 105 | hsa-miR-30a-5p | GALNT7 | -0.92 | 0.00076 | 1.46 | 0 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.14 | 4.0E-5 | NA | |
| 106 | hsa-miR-30a-5p | GATC | -0.92 | 0.00076 | 0.23 | 0.19835 | miRNAWalker2 validate | -0.16 | 0 | NA | |
| 107 | hsa-miR-30a-5p | GCH1 | -0.92 | 0.00076 | 0.08 | 0.67135 | mirMAP | -0.17 | 0 | NA | |
| 108 | hsa-miR-30a-5p | GFPT2 | -0.92 | 0.00076 | -0.71 | 0.0142 | MirTarget; miRNATAP | -0.16 | 0.00123 | NA | |
| 109 | hsa-miR-30a-5p | GGCT | -0.92 | 0.00076 | 1.58 | 0 | miRNAWalker2 validate | -0.22 | 0 | NA | |
| 110 | hsa-miR-30a-5p | GLDC | -0.92 | 0.00076 | 1.59 | 0.0024 | MirTarget; miRNATAP | -0.48 | 0 | NA | |
| 111 | hsa-miR-30a-5p | GLRX3 | -0.92 | 0.00076 | 0.76 | 0 | miRNAWalker2 validate | -0.13 | 0 | NA | |
| 112 | hsa-miR-30a-5p | GMEB2 | -0.92 | 0.00076 | 0.23 | 0.05926 | MirTarget; miRNATAP | -0.1 | 0 | NA | |
| 113 | hsa-miR-30a-5p | GORASP2 | -0.92 | 0.00076 | 0.7 | 0 | miRNATAP | -0.14 | 0 | NA | |
| 114 | hsa-miR-30a-5p | GPR180 | -0.92 | 0.00076 | 0.52 | 0.00037 | mirMAP | -0.15 | 0 | NA | |
| 115 | hsa-miR-30a-5p | GRHL2 | -0.92 | 0.00076 | 0.7 | 0.00242 | miRNATAP | -0.1 | 0.00751 | NA | |
| 116 | hsa-miR-30a-5p | GTF2E2 | -0.92 | 0.00076 | 0.57 | 2.0E-5 | miRNATAP | -0.13 | 0 | NA | |
| 117 | hsa-miR-30a-5p | HDAC2 | -0.92 | 0.00076 | 0.48 | 0.00021 | mirMAP | -0.14 | 0 | NA | |
| 118 | hsa-miR-30a-5p | HDGF | -0.92 | 0.00076 | 0.95 | 0 | miRNAWalker2 validate | -0.22 | 0 | NA | |
| 119 | hsa-miR-30a-5p | HMGB3 | -0.92 | 0.00076 | 2.91 | 0 | miRNATAP | -0.65 | 0 | NA | |
| 120 | hsa-miR-30a-5p | HOXA11 | -0.92 | 0.00076 | 1.33 | 0.02898 | miRNATAP | -0.41 | 5.0E-5 | NA | |
| 121 | hsa-miR-30a-5p | HOXB8 | -0.92 | 0.00076 | 0.5 | 0.46283 | miRNATAP | -0.47 | 4.0E-5 | NA | |
| 122 | hsa-miR-30a-5p | HS2ST1 | -0.92 | 0.00076 | 0.16 | 0.27392 | mirMAP | -0.11 | 1.0E-5 | NA | |
| 123 | hsa-miR-30a-5p | HSPA5 | -0.92 | 0.00076 | 0.82 | 0 | miRNATAP | -0.15 | 0 | NA | |
| 124 | hsa-miR-30a-5p | HTRA3 | -0.92 | 0.00076 | 0.53 | 0.0926 | miRNATAP | -0.31 | 0 | NA | |
| 125 | hsa-miR-30a-5p | IFRD1 | -0.92 | 0.00076 | 0.35 | 0.07398 | miRNAWalker2 validate | -0.2 | 0 | NA | |
| 126 | hsa-miR-30a-5p | IKBIP | -0.92 | 0.00076 | 0.08 | 0.59742 | miRNAWalker2 validate; MirTarget | -0.14 | 0 | NA | |
| 127 | hsa-miR-30a-5p | IL1RAPL2 | -0.92 | 0.00076 | 2.45 | 0 | MirTarget | -0.26 | 0.0037 | NA | |
| 128 | hsa-miR-30a-5p | INHBA | -0.92 | 0.00076 | -0.96 | 0.00524 | mirMAP | -0.29 | 0 | NA | |
| 129 | hsa-miR-30a-5p | IPMK | -0.92 | 0.00076 | -0.02 | 0.93735 | mirMAP; miRNATAP | -0.11 | 0.0099 | NA | |
| 130 | hsa-miR-30a-5p | JAG2 | -0.92 | 0.00076 | 0.03 | 0.91715 | miRNATAP | -0.19 | 0 | NA | |
| 131 | hsa-miR-30a-5p | KCND2 | -0.92 | 0.00076 | 1.52 | 0.00037 | mirMAP | -0.7 | 0 | NA | |
| 132 | hsa-miR-30a-5p | KCTD5 | -0.92 | 0.00076 | 0.56 | 3.0E-5 | miRNATAP | -0.11 | 0 | NA | |
| 133 | hsa-miR-30a-5p | KIAA1549 | -0.92 | 0.00076 | 0.45 | 0.10267 | miRNATAP | -0.11 | 0.02172 | NA | |
| 134 | hsa-miR-30a-5p | KIF11 | -0.92 | 0.00076 | 2.52 | 0 | miRNAWalker2 validate; MirTarget | -0.48 | 0 | NA | |
| 135 | hsa-miR-128-3p | KLF4 | 1.04 | 0 | -2.55 | 0 | miRNAWalker2 validate; MirTarget | -0.24 | 0.01045 | NA | |
| 136 | hsa-miR-135b-5p | KLF4 | 3.25 | 0 | -2.55 | 0 | miRNAWalker2 validate; MirTarget | -0.11 | 0.00341 | NA | |
| 137 | hsa-miR-148a-3p | KLF4 | 2.31 | 0 | -2.55 | 0 | MirTarget; miRNATAP | -0.29 | 0 | NA | |
| 138 | hsa-miR-200b-3p | KLF4 | 1.55 | 0 | -2.55 | 0 | MirTarget; TargetScan | -0.19 | 0.00094 | NA | |
| 139 | hsa-miR-29b-3p | KLF4 | 3.11 | 0 | -2.55 | 0 | MirTarget; miRNATAP | -0.17 | 0.0016 | 22751119 | Progestin suppression of miR 29 potentiates dedifferentiation of breast cancer cells via KLF4; We demonstrate that miR-29 directly targets Krüppel-like factor 4 KLF4 a transcription factor required for the reprogramming of differentiated cells to pluripotent stem cells and for the maintenance of breast cancer stem cells; These results reveal a novel mechanism whereby progestins increase the stem cell-like population in hormone-responsive breast cancers by decreasing miR-29 to augment PR-mediated upregulation of KLF4 |
| 140 | hsa-miR-301a-3p | KLF4 | 2.7 | 0 | -2.55 | 0 | miRNATAP | -0.32 | 0 | NA | |
| 141 | hsa-miR-30b-5p | KLF4 | 0.36 | 0.13803 | -2.55 | 0 | miRNAWalker2 validate | -0.17 | 0.00836 | NA | |
| 142 | hsa-miR-429 | KLF4 | 2.38 | 0 | -2.55 | 0 | MirTarget; PITA; miRanda; miRNATAP | -0.22 | 1.0E-5 | NA | |
| 143 | hsa-miR-92b-3p | KLF4 | 0.05 | 0.83172 | -2.55 | 0 | MirTarget; miRNATAP | -0.23 | 0.00061 | NA | |
| 144 | hsa-miR-30a-5p | KPNA4 | -0.92 | 0.00076 | -0.01 | 0.93016 | miRNAWalker2 validate; mirMAP | -0.11 | 0 | NA | |
| 145 | hsa-miR-30a-5p | KRAS | -0.92 | 0.00076 | 0.54 | 0.00142 | mirMAP; miRNATAP | -0.16 | 0 | NA | |
| 146 | hsa-miR-30a-5p | KSR1 | -0.92 | 0.00076 | 0.25 | 0.33039 | MirTarget | -0.11 | 0.01192 | NA | |
| 147 | hsa-miR-30a-5p | LARP1 | -0.92 | 0.00076 | 0.42 | 0.0011 | miRNATAP | -0.1 | 0 | NA | |
| 148 | hsa-miR-30a-5p | LARP4 | -0.92 | 0.00076 | 0.16 | 0.18031 | miRNAWalker2 validate; mirMAP; miRNATAP | -0.12 | 0 | NA | |
| 149 | hsa-miR-30a-5p | LCLAT1 | -0.92 | 0.00076 | 0.68 | 0 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.16 | 0 | NA | |
| 150 | hsa-miR-30a-5p | LGSN | -0.92 | 0.00076 | 3.19 | 2.0E-5 | mirMAP | -0.59 | 0 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CELL CYCLE PHASE TRANSITION | 17 | 255 | 3.033e-06 | 0.003937 |
| 2 | EMBRYO DEVELOPMENT | 36 | 894 | 3.521e-06 | 0.003937 |
| 3 | CELL CYCLE G1 S PHASE TRANSITION | 11 | 111 | 4.23e-06 | 0.003937 |
| 4 | EMBRYONIC MORPHOGENESIS | 27 | 539 | 1.21e-06 | 0.003937 |
| 5 | G1 S TRANSITION OF MITOTIC CELL CYCLE | 11 | 111 | 4.23e-06 | 0.003937 |
| 6 | NEUROGENESIS | 48 | 1402 | 7.162e-06 | 0.005554 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | RNA BINDING | 62 | 1598 | 2.641e-09 | 2.453e-06 |
| 2 | POLY A RNA BINDING | 47 | 1170 | 1.058e-07 | 4.917e-05 |
| 3 | ENZYME BINDING | 60 | 1737 | 3.134e-07 | 9.706e-05 |
| 4 | TRANSCRIPTIONAL REPRESSOR ACTIVITY RNA POLYMERASE II ACTIVATING TRANSCRIPTION FACTOR BINDING | 8 | 53 | 3.779e-06 | 0.0008776 |
| 5 | RIBONUCLEOTIDE BINDING | 58 | 1860 | 1.243e-05 | 0.002309 |
| 6 | TRANSCRIPTIONAL REPRESSOR ACTIVITY RNA POLYMERASE II TRANSCRIPTION FACTOR BINDING | 9 | 86 | 2.043e-05 | 0.003163 |
| 7 | RNA POLYMERASE II TRANSCRIPTION COACTIVATOR ACTIVITY | 6 | 36 | 3.552e-05 | 0.004714 |
| 8 | IDENTICAL PROTEIN BINDING | 41 | 1209 | 4.399e-05 | 0.005109 |
| 9 | CORE PROMOTER BINDING | 11 | 152 | 8.16e-05 | 0.00758 |
| 10 | UBIQUITIN LIKE PROTEIN LIGASE BINDING | 15 | 264 | 7.377e-05 | 0.00758 |
| 11 | HISTONE DEACETYLASE BINDING | 9 | 105 | 9.967e-05 | 0.008418 |
| 12 | TRANSCRIPTION FACTOR ACTIVITY RNA POLYMERASE II TRANSCRIPTION FACTOR BINDING | 10 | 133 | 0.0001248 | 0.009658 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
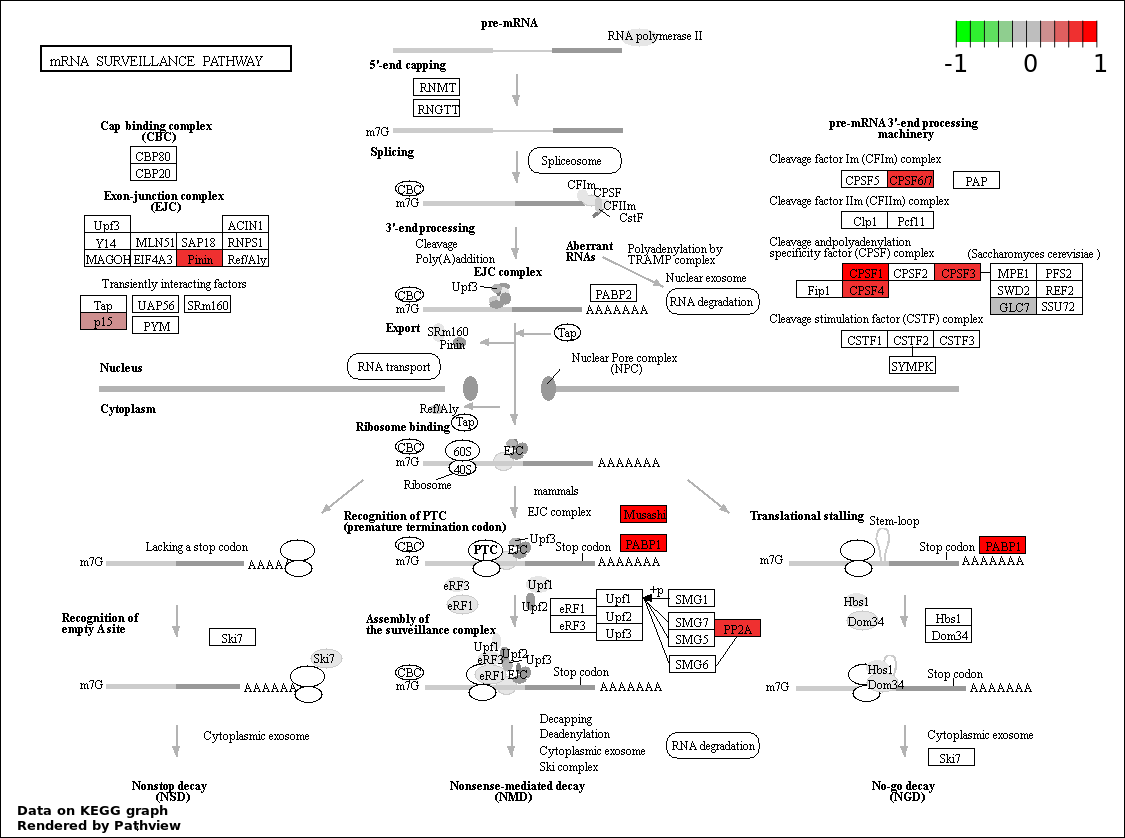
| 1 | hsa03015_mRNA_surveillance_pathway | 10 | 83 | 1.954e-06 | 0.0003516 | |
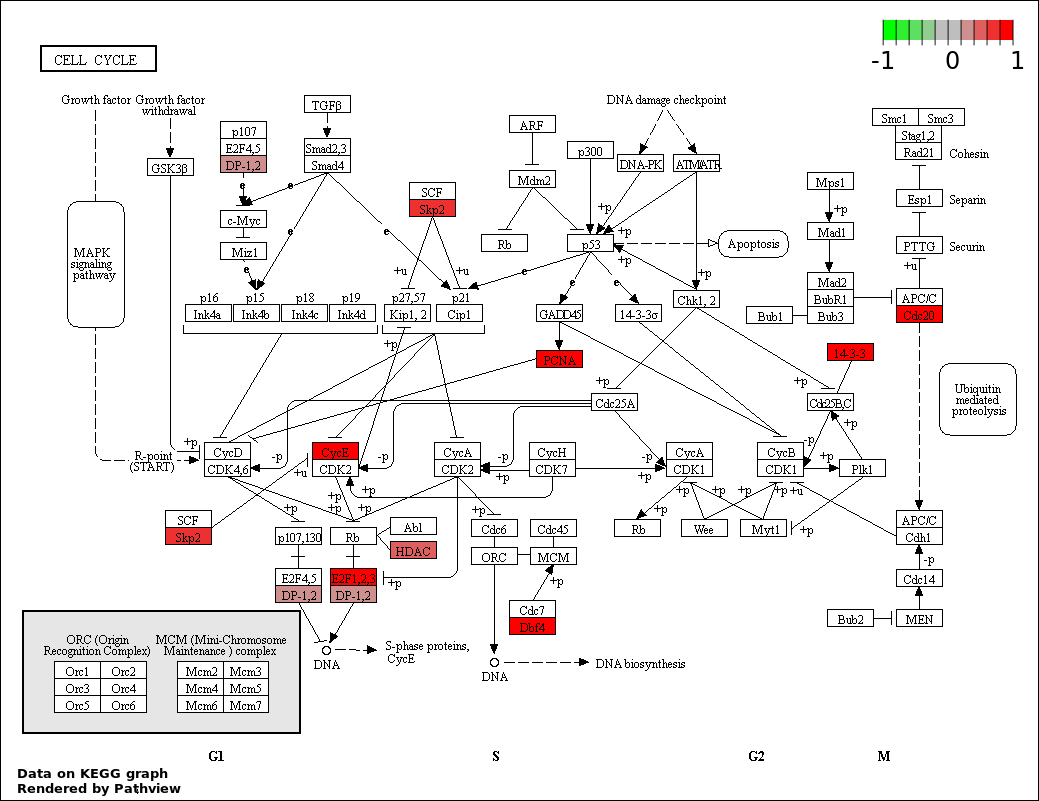
| 2 | hsa04110_Cell_cycle | 10 | 128 | 9.061e-05 | 0.008155 | |
| 3 | hsa03013_RNA_transport | 10 | 152 | 0.0003696 | 0.02218 | |
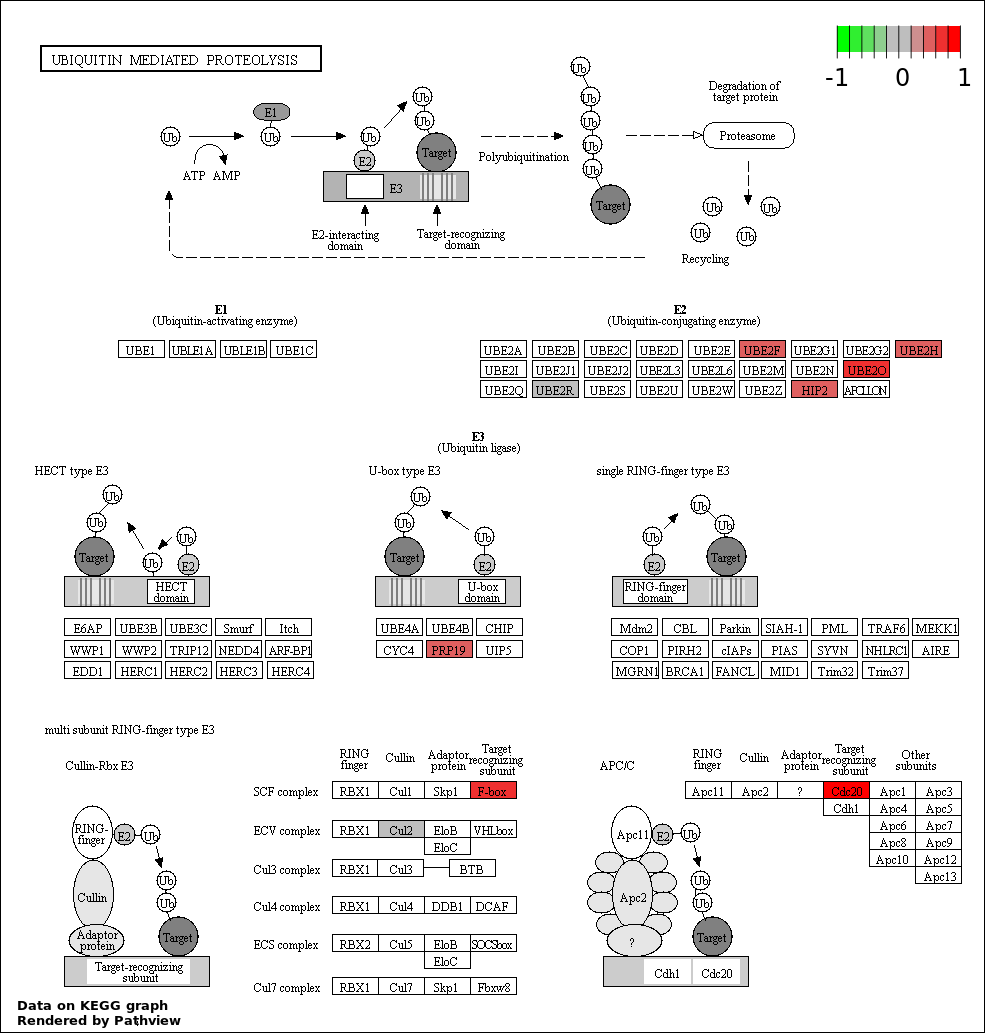
| 4 | hsa04120_Ubiquitin_mediated_proteolysis | 9 | 139 | 0.0008073 | 0.02913 | |
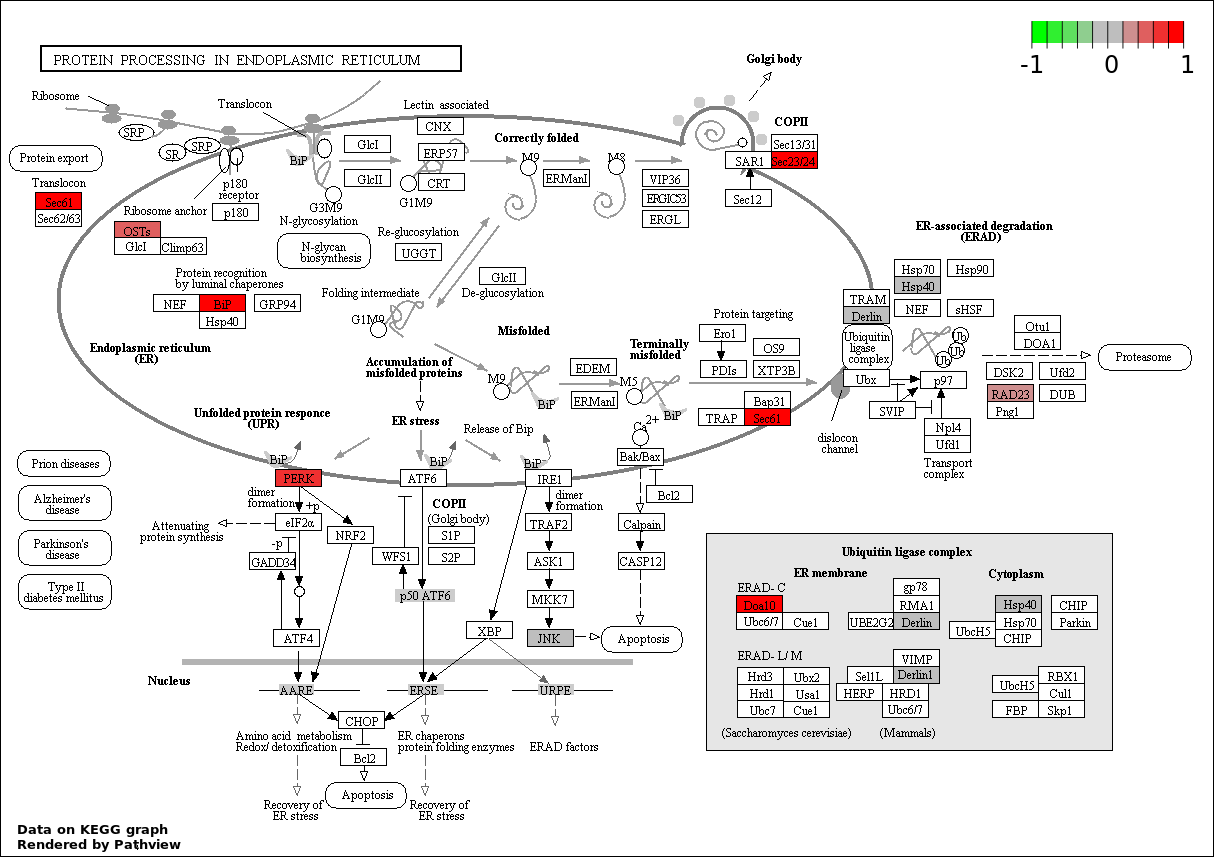
| 5 | hsa04141_Protein_processing_in_endoplasmic_reticulum | 10 | 168 | 0.0008092 | 0.02913 | |
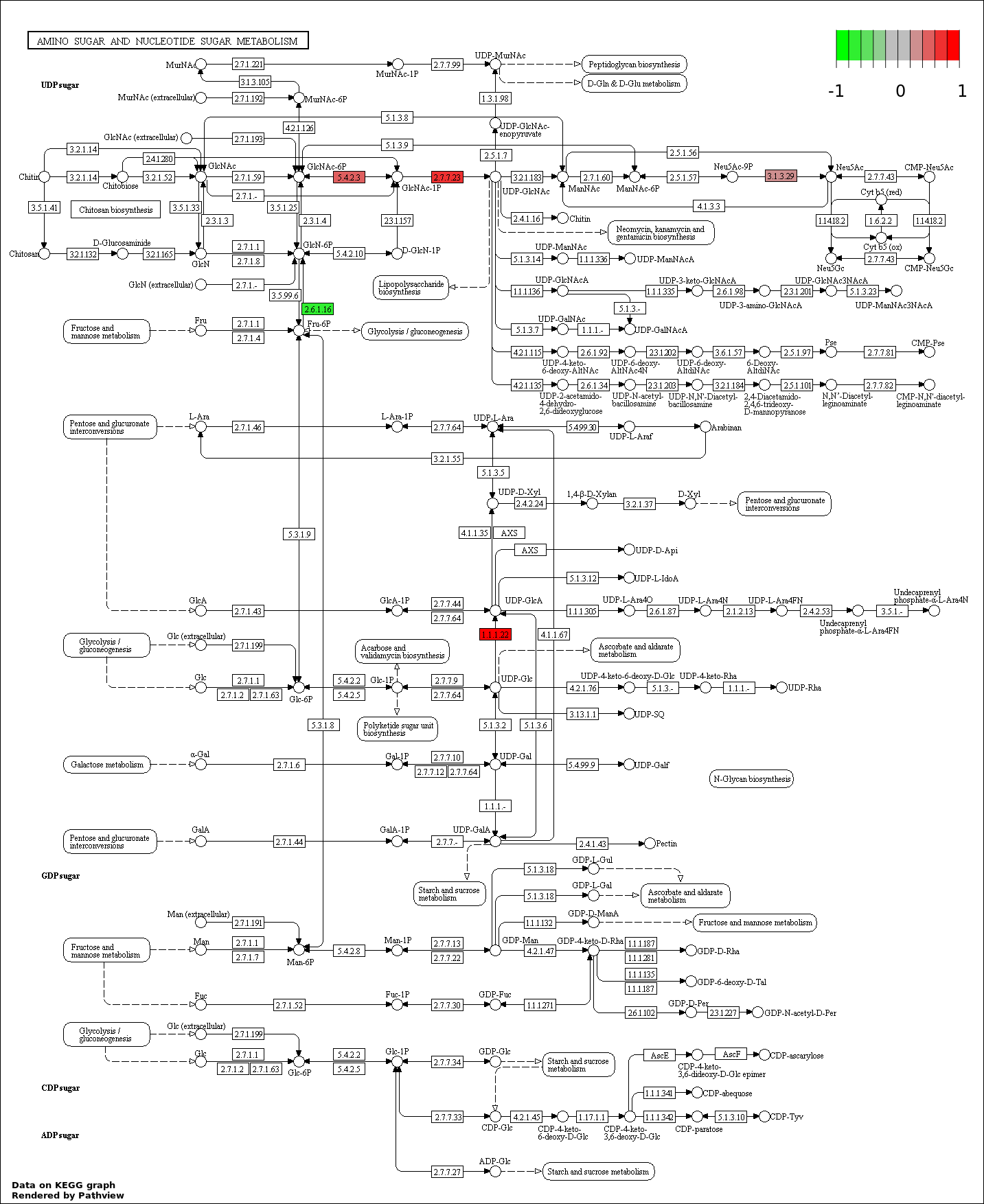
| 6 | hsa00520_Amino_sugar_and_nucleotide_sugar_metabolism | 5 | 48 | 0.001511 | 0.04532 | |
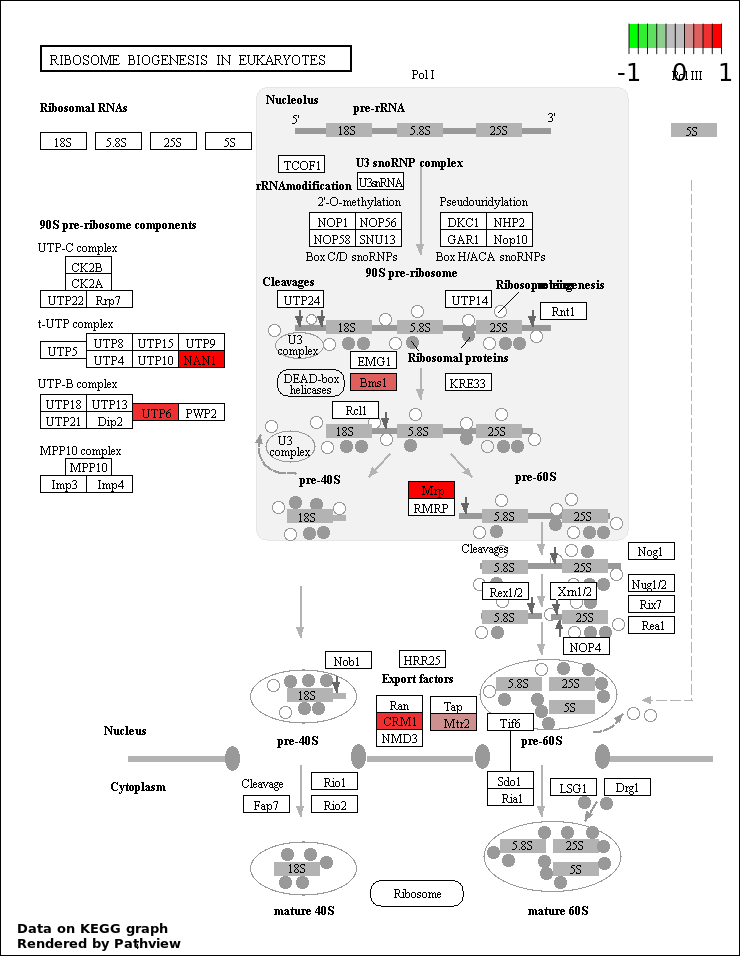
| 7 | hsa03008_Ribosome_biogenesis_in_eukaryotes | 6 | 81 | 0.003052 | 0.07849 | |
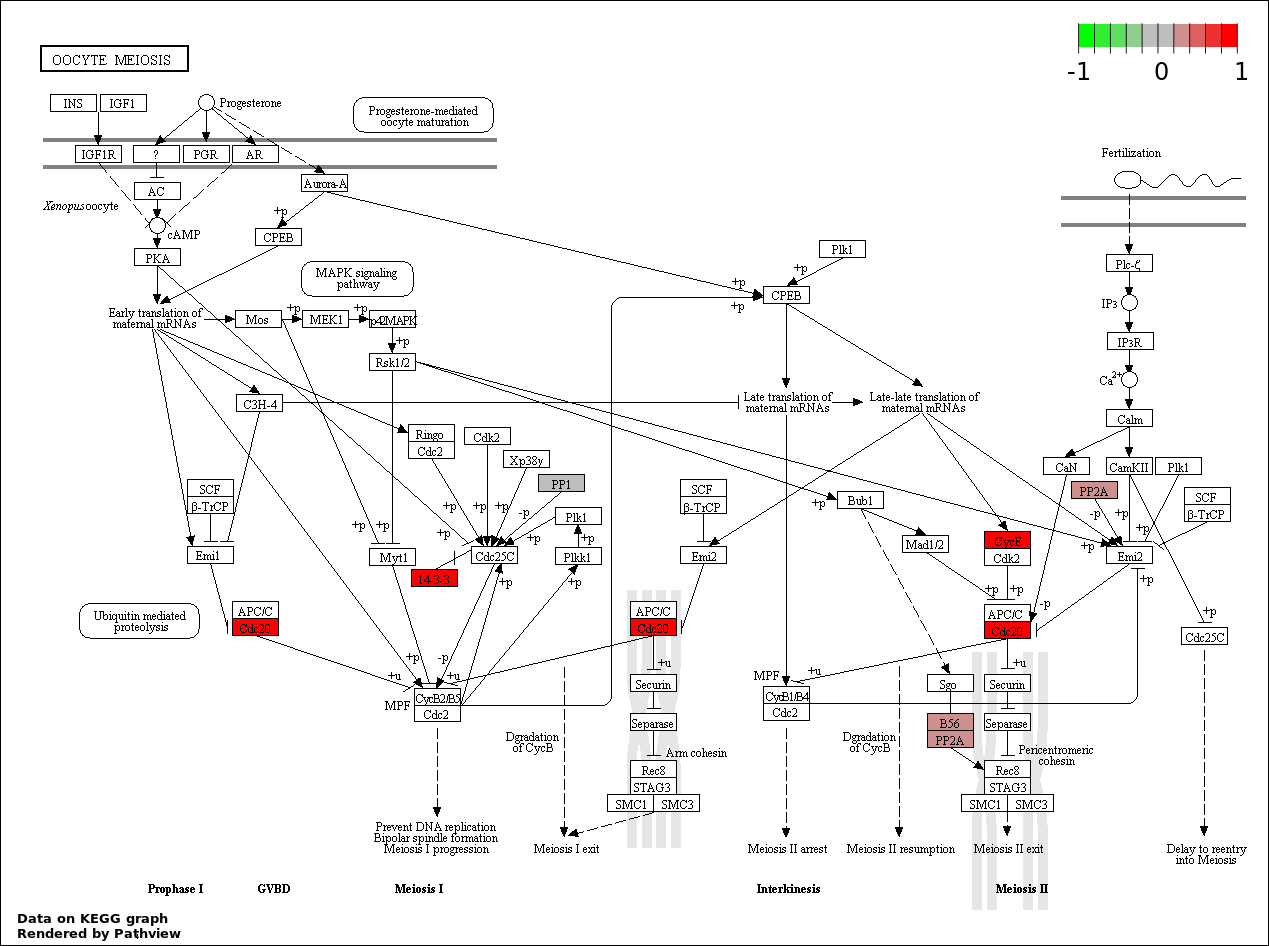
| 8 | hsa04114_Oocyte_meiosis | 7 | 114 | 0.004063 | 0.09141 | |
| 9 | hsa03060_Protein_export | 3 | 23 | 0.007372 | 0.1474 | |
| 10 | hsa00510_N.Glycan_biosynthesis | 4 | 49 | 0.01072 | 0.193 | |
| 11 | hsa04350_TGF.beta_signaling_pathway | 5 | 85 | 0.01712 | 0.2801 | |
| 12 | hsa03410_Base_excision_repair | 3 | 34 | 0.02162 | 0.3243 | |
| 13 | hsa00230_Purine_metabolism | 7 | 162 | 0.02482 | 0.3437 | |
| 14 | hsa00533_Glycosaminoglycan_biosynthesis_._keratan_sulfate | 2 | 15 | 0.02788 | 0.3585 | |
| 15 | hsa03050_Proteasome | 3 | 45 | 0.04456 | 0.5347 | |
| 16 | hsa04390_Hippo_signaling_pathway | 6 | 154 | 0.05521 | 0.6143 | |
| 17 | hsa04014_Ras_signaling_pathway | 8 | 236 | 0.05802 | 0.6143 | |
| 18 | hsa04012_ErbB_signaling_pathway | 4 | 87 | 0.06751 | 0.6751 | |
| 19 | hsa00534_Glycosaminoglycan_biosynthesis_._heparan_sulfate | 2 | 26 | 0.07611 | 0.677 | |
| 20 | hsa00601_Glycosphingolipid_biosynthesis_._lacto_and_neolacto_series | 2 | 26 | 0.07611 | 0.677 | |
| 21 | hsa00562_Inositol_phosphate_metabolism | 3 | 57 | 0.07898 | 0.677 | |
| 22 | hsa03020_RNA_polymerase | 2 | 29 | 0.09191 | 0.7014 | |
| 23 | hsa04151_PI3K_AKT_signaling_pathway | 10 | 351 | 0.09294 | 0.7014 | |
| 24 | hsa04910_Insulin_signaling_pathway | 5 | 138 | 0.09703 | 0.7014 | |
| 25 | hsa00240_Pyrimidine_metabolism | 4 | 99 | 0.09741 | 0.7014 | |
| 26 | hsa04730_Long.term_depression | 3 | 70 | 0.1257 | 0.8184 | |
| 27 | hsa03018_RNA_degradation | 3 | 71 | 0.1296 | 0.8184 | |
| 28 | hsa00051_Fructose_and_mannose_metabolism | 2 | 36 | 0.1319 | 0.8184 | |
| 29 | hsa03030_DNA_replication | 2 | 36 | 0.1319 | 0.8184 | |
| 30 | hsa04664_Fc_epsilon_RI_signaling_pathway | 3 | 79 | 0.1626 | 0.9646 | |
| 31 | hsa00564_Glycerophospholipid_metabolism | 3 | 80 | 0.1669 | 0.9646 | |
| 32 | hsa04722_Neurotrophin_signaling_pathway | 4 | 127 | 0.1857 | 0.9646 | |
| 33 | hsa03420_Nucleotide_excision_repair | 2 | 45 | 0.1876 | 0.9646 | |
| 34 | hsa00514_Other_types_of_O.glycan_biosynthesis | 2 | 46 | 0.194 | 0.9698 | |
| 35 | hsa04330_Notch_signaling_pathway | 2 | 47 | 0.2004 | 0.9748 | |
| 36 | hsa04540_Gap_junction | 3 | 90 | 0.2113 | 0.9849 | |
| 37 | hsa00480_Glutathione_metabolism | 2 | 50 | 0.2197 | 0.9849 | |
| 38 | hsa04150_mTOR_signaling_pathway | 2 | 52 | 0.2327 | 0.9849 | |
| 39 | hsa00330_Arginine_and_proline_metabolism | 2 | 54 | 0.2458 | 0.9849 | |
| 40 | hsa00500_Starch_and_sucrose_metabolism | 2 | 54 | 0.2458 | 0.9849 | |
| 41 | hsa04912_GnRH_signaling_pathway | 3 | 101 | 0.2627 | 0.9849 | |
| 42 | hsa04310_Wnt_signaling_pathway | 4 | 151 | 0.2756 | 0.9849 | |
| 43 | hsa04145_Phagosome | 4 | 156 | 0.2952 | 0.9849 | |
| 44 | hsa00970_Aminoacyl.tRNA_biosynthesis | 2 | 63 | 0.3046 | 0.9849 | |
| 45 | hsa04810_Regulation_of_actin_cytoskeleton | 5 | 214 | 0.325 | 0.9849 | |
| 46 | hsa04920_Adipocytokine_signaling_pathway | 2 | 68 | 0.337 | 0.9849 | |
| 47 | hsa04115_p53_signaling_pathway | 2 | 69 | 0.3434 | 0.9849 | |
| 48 | hsa04720_Long.term_potentiation | 2 | 70 | 0.3498 | 0.9849 | |
| 49 | hsa04520_Adherens_junction | 2 | 73 | 0.3689 | 0.9849 | |
| 50 | hsa04370_VEGF_signaling_pathway | 2 | 76 | 0.3877 | 0.9849 | |
| 51 | hsa03040_Spliceosome | 3 | 128 | 0.3922 | 0.9849 | |
| 52 | hsa04260_Cardiac_muscle_contraction | 2 | 77 | 0.3939 | 0.9849 | |
| 53 | hsa04612_Antigen_processing_and_presentation | 2 | 78 | 0.4001 | 0.9849 | |
| 54 | hsa04360_Axon_guidance | 3 | 130 | 0.4017 | 0.9849 | |
| 55 | hsa00190_Oxidative_phosphorylation | 3 | 132 | 0.4111 | 0.9849 | |
| 56 | hsa04530_Tight_junction | 3 | 133 | 0.4158 | 0.9849 | |
| 57 | hsa04914_Progesterone.mediated_oocyte_maturation | 2 | 87 | 0.4545 | 1 | |
| 58 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 2 | 95 | 0.5004 | 1 | |
| 59 | hsa04010_MAPK_signaling_pathway | 5 | 268 | 0.5105 | 1 | |
| 60 | hsa04916_Melanogenesis | 2 | 101 | 0.5331 | 1 | |
| 61 | hsa04972_Pancreatic_secretion | 2 | 101 | 0.5331 | 1 | |
| 62 | hsa04270_Vascular_smooth_muscle_contraction | 2 | 116 | 0.6084 | 1 | |
| 63 | hsa04020_Calcium_signaling_pathway | 2 | 177 | 0.8213 | 1 | |
| 64 | hsa04510_Focal_adhesion | 2 | 200 | 0.8698 | 1 |