This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-542-3p | ALG6 | 1.31 | 0 | -0.49 | 0 | miRanda | -0.18 | 0 | NA | |
| 2 | hsa-miR-30d-3p | ATM | 0.12 | 0.32955 | 0.24 | 0.01738 | mirMAP | -0.14 | 0.00043 | 24345332 | miR-30d has been observed to be significantly down-regulated in human anaplastic thyroid carcinoma ATC and is believed to be an important event in thyroid cell transformation; In this study we found that miR-30d has a critical role in modulating sensitivity of ATC cells to cisplatin a commonly used chemotherapeutic drug for treatment of this neoplasm; Using a mimic of miR-30d we demonstrated that miR-30d could negatively regulate the expression of beclin 1 a key autophagy gene leading to suppression of the cisplatin-activated autophagic response that protects ATC cells from apoptosis; We further showed that inhibition of the beclin 1-mediated autophagy by the miR-30d mimic sensitized ATC cells to cisplatin both in vitro cell culture and in vivo animal xenograft model; These results suggest that dysregulation of miR-30d in ATC cells is responsible for the insensitivity to cisplatin by promoting autophagic survival; Thus miR-30d may be exploited as a potential target for therapeutic intervention in the treatment of ATC |
| 3 | hsa-miR-339-5p | ATM | -0.28 | 0.03557 | 0.24 | 0.01738 | miRanda | -0.1 | 0.00399 | NA | |
| 4 | hsa-miR-455-5p | ATM | 0.27 | 0.05813 | 0.24 | 0.01738 | miRanda | -0.12 | 0.00045 | NA | |
| 5 | hsa-miR-361-5p | ATP5L | -0.23 | 0.00962 | -0.07 | 0.41425 | miRanda | -0.12 | 0.01395 | NA | |
| 6 | hsa-miR-130a-3p | CANX | 1.53 | 0 | -0.4 | 0 | miRNATAP | -0.15 | 0 | NA | |
| 7 | hsa-let-7b-5p | COX7B | 0.96 | 0 | -0.2 | 0.0881 | miRNAWalker2 validate | -0.16 | 3.0E-5 | NA | |
| 8 | hsa-miR-10a-5p | COX7B | 1.48 | 0 | -0.2 | 0.0881 | miRNAWalker2 validate | -0.13 | 1.0E-5 | NA | |
| 9 | hsa-miR-199b-5p | CRLS1 | 0.45 | 0.25461 | 0.22 | 0.0807 | miRanda; miRNATAP | -0.13 | 0 | NA | |
| 10 | hsa-miR-21-3p | CRLS1 | 0.48 | 0.003 | 0.22 | 0.0807 | MirTarget | -0.27 | 0 | NA | |
| 11 | hsa-miR-320a | CRLS1 | -0.33 | 0.02214 | 0.22 | 0.0807 | miRanda | -0.12 | 0.00555 | NA | |
| 12 | hsa-miR-320b | CRLS1 | -0.09 | 0.60798 | 0.22 | 0.0807 | miRanda | -0.14 | 2.0E-5 | NA | |
| 13 | hsa-miR-330-3p | CRLS1 | 0.33 | 0.03161 | 0.22 | 0.0807 | MirTarget | -0.22 | 0 | NA | |
| 14 | hsa-miR-335-3p | CRLS1 | 0.28 | 0.10663 | 0.22 | 0.0807 | mirMAP | -0.18 | 0 | NA | |
| 15 | hsa-miR-425-5p | CRLS1 | -0.59 | 2.0E-5 | 0.22 | 0.0807 | MirTarget | -0.13 | 0.00219 | NA | |
| 16 | hsa-miR-769-5p | CRLS1 | -0.22 | 0.03334 | 0.22 | 0.0807 | MirTarget | -0.18 | 0.00186 | NA | |
| 17 | hsa-miR-103a-3p | EIF5 | -0.77 | 0 | 0.72 | 0 | MirTarget; miRNATAP | -0.15 | 0.00015 | NA | |
| 18 | hsa-miR-140-3p | EIF5 | -0.55 | 0 | 0.72 | 0 | MirTarget | -0.2 | 0.00019 | NA | |
| 19 | hsa-miR-186-5p | EIF5 | 0.06 | 0.53529 | 0.72 | 0 | miRNAWalker2 validate; mirMAP | -0.11 | 0.02116 | NA | |
| 20 | hsa-miR-21-5p | EIF5 | -1.51 | 0 | 0.72 | 0 | miRNAWalker2 validate | -0.26 | 0 | NA | |
| 21 | hsa-miR-27a-3p | EIF5 | 0.37 | 0.00876 | 0.72 | 0 | MirTarget | -0.11 | 0.00044 | NA | |
| 22 | hsa-miR-500a-3p | EIF5 | -1.19 | 0 | 0.72 | 0 | MirTarget | -0.12 | 6.0E-5 | NA | |
| 23 | hsa-miR-125a-3p | HGD | 0.84 | 4.0E-5 | 1.1 | 6.0E-5 | miRanda | -0.43 | 0 | NA | |
| 24 | hsa-miR-320a | HGD | -0.33 | 0.02214 | 1.1 | 6.0E-5 | miRanda | -0.4 | 2.0E-5 | NA | |
| 25 | hsa-miR-320b | HGD | -0.09 | 0.60798 | 1.1 | 6.0E-5 | miRanda | -0.28 | 0.00015 | NA | |
| 26 | hsa-miR-330-3p | HGD | 0.33 | 0.03161 | 1.1 | 6.0E-5 | miRNATAP | -0.51 | 0 | NA | |
| 27 | hsa-miR-335-5p | HGD | 1.61 | 0 | 1.1 | 6.0E-5 | miRNAWalker2 validate | -0.15 | 0.03917 | NA | |
| 28 | hsa-miR-429 | HGD | 1.4 | 7.0E-5 | 1.1 | 6.0E-5 | miRNATAP | -0.12 | 0.00163 | NA | |
| 29 | hsa-miR-542-3p | HGD | 1.31 | 0 | 1.1 | 6.0E-5 | miRanda | -0.31 | 0.00078 | NA | |
| 30 | hsa-miR-326 | IL11RA | 1.88 | 0 | -0.13 | 0.31327 | miRanda | -0.11 | 0.00029 | NA | |
| 31 | hsa-miR-330-5p | IL11RA | -0.44 | 0.00533 | -0.13 | 0.31327 | miRanda | -0.19 | 0 | NA | |
| 32 | hsa-miR-140-3p | ITGB1 | -0.55 | 0 | -0.15 | 0.08751 | PITA | -0.18 | 0.00056 | NA | |
| 33 | hsa-miR-192-5p | ITGB1 | 0.5 | 0.00345 | -0.15 | 0.08751 | miRNAWalker2 validate | -0.16 | 0 | NA | |
| 34 | hsa-miR-590-5p | ITGB1 | 0.1 | 0.31003 | -0.15 | 0.08751 | miRanda | -0.14 | 0.00126 | NA | |
| 35 | hsa-miR-125a-3p | ITPR2 | 0.84 | 4.0E-5 | 0.68 | 5.0E-5 | miRanda | -0.27 | 0 | NA | |
| 36 | hsa-miR-1271-5p | ITPR2 | 1 | 0 | 0.68 | 5.0E-5 | MirTarget | -0.11 | 0.02788 | NA | |
| 37 | hsa-miR-148b-5p | ITPR2 | -0.3 | 0.02557 | 0.68 | 5.0E-5 | MirTarget | -0.24 | 0.00013 | NA | |
| 38 | hsa-miR-193a-3p | ITPR2 | 0.12 | 0.30939 | 0.68 | 5.0E-5 | miRanda | -0.25 | 0.00043 | NA | |
| 39 | hsa-miR-200b-3p | ITPR2 | 1.29 | 0.00027 | 0.68 | 5.0E-5 | MirTarget | -0.13 | 0 | NA | |
| 40 | hsa-miR-200c-3p | ITPR2 | 0.1 | 0.71696 | 0.68 | 5.0E-5 | MirTarget | -0.2 | 0 | NA | |
| 41 | hsa-miR-203a-3p | ITPR2 | 1.34 | 9.0E-5 | 0.68 | 5.0E-5 | MirTarget | -0.12 | 0 | NA | |
| 42 | hsa-miR-218-5p | ITPR2 | 0.5 | 0.03986 | 0.68 | 5.0E-5 | MirTarget | -0.22 | 0 | NA | |
| 43 | hsa-miR-324-5p | ITPR2 | -0.37 | 0.00592 | 0.68 | 5.0E-5 | miRNAWalker2 validate | -0.24 | 0.00011 | NA | |
| 44 | hsa-miR-330-5p | ITPR2 | -0.44 | 0.00533 | 0.68 | 5.0E-5 | miRanda | -0.44 | 0 | NA | |
| 45 | hsa-miR-335-3p | ITPR2 | 0.28 | 0.10663 | 0.68 | 5.0E-5 | mirMAP | -0.24 | 0 | NA | |
| 46 | hsa-miR-339-5p | ITPR2 | -0.28 | 0.03557 | 0.68 | 5.0E-5 | miRanda | -0.38 | 0 | NA | |
| 47 | hsa-miR-362-3p | ITPR2 | -0.81 | 0 | 0.68 | 5.0E-5 | MirTarget | -0.13 | 0.02545 | NA | |
| 48 | hsa-miR-429 | ITPR2 | 1.4 | 7.0E-5 | 0.68 | 5.0E-5 | MirTarget; miRanda | -0.12 | 0 | NA | |
| 49 | hsa-miR-590-5p | ITPR2 | 0.1 | 0.31003 | 0.68 | 5.0E-5 | miRanda | -0.25 | 0.00224 | NA | |
| 50 | hsa-miR-766-3p | ITPR2 | -0.2 | 0.25723 | 0.68 | 5.0E-5 | MirTarget | -0.26 | 0 | NA | |
| 51 | hsa-miR-940 | ITPR2 | -0.45 | 0.01771 | 0.68 | 5.0E-5 | MirTarget; PITA | -0.24 | 0 | NA | |
| 52 | hsa-miR-125b-5p | MAP3K1 | 1.36 | 0 | -0.02 | 0.87733 | miRNATAP | -0.11 | 0.00024 | NA | |
| 53 | hsa-miR-139-5p | MAP3K1 | 2.11 | 0 | -0.02 | 0.87733 | miRanda | -0.11 | 0 | NA | |
| 54 | hsa-miR-192-5p | MAP3K1 | 0.5 | 0.00345 | -0.02 | 0.87733 | miRNAWalker2 validate | -0.12 | 3.0E-5 | NA | |
| 55 | hsa-let-7c-5p | MDM4 | 1.71 | 0 | -0.73 | 0 | MirTarget | -0.16 | 0 | NA | |
| 56 | hsa-miR-125b-2-3p | MDM4 | 1.66 | 0 | -0.73 | 0 | mirMAP | -0.2 | 0 | NA | |
| 57 | hsa-miR-142-3p | MDM4 | 1.42 | 0 | -0.73 | 0 | miRanda | -0.11 | 9.0E-5 | NA | |
| 58 | hsa-miR-144-3p | MDM4 | 2.98 | 0 | -0.73 | 0 | miRNATAP | -0.11 | 0 | NA | |
| 59 | hsa-miR-152-3p | MDM4 | 0.96 | 0 | -0.73 | 0 | MirTarget | -0.12 | 0.00032 | NA | |
| 60 | hsa-miR-186-5p | MDM4 | 0.06 | 0.53529 | -0.73 | 0 | mirMAP | -0.13 | 0.02052 | NA | |
| 61 | hsa-miR-191-5p | MDM4 | 0.34 | 0.0015 | -0.73 | 0 | miRNAWalker2 validate; miRTarBase | -0.1 | 0.03541 | 23724042; 25670033; 26274820; 21084273 | Association of a genetic variation in a miR 191 binding site in MDM4 with risk of esophageal squamous cell carcinoma; It has been reported that an rs4245739 A>C polymorphism locating in the MDM4 3'-untranslated region creates a miR-191 target site and results in decreased MDM4 expression;Herein we show using reporter gene assays and endogenous MDM4 expression analyses that miR-191-5p and miR-887 have a specific affinity for the rs4245739 SNP C-allele in prostate cancer; By analysing gene expression datasets from patient cohorts we found that MDM4 is associated with metastasis and prostate cancer progression and that targeting this gene with miR-191-5p or miR-887 decreases in PC3 cell viability;After co-tranfection of miRNAs and different allelic-MDM4 reporter constructs into SCLC cells we found that the both miR-191-5p and miR-887-3p can lead to significantly decreased MDM4 expression activities in the construct with C-allelic 3'-UTR but not A-allelic 3'-UTR suggesting a consistent genotype-phenotype correlation;We identified an SNP SNP34091 in the 3'-UTR of MDM4 that creates a putative target site for hsa-miR-191 a microRNA that is highly expressed in normal and tumor tissues; We conclude that acquisition of an illegitimate miR-191 target site causes downregulation of MDM4 expression thereby significantly delaying ovarian carcinoma progression and tumor-related death |
| 62 | hsa-miR-29a-5p | MDM4 | 0.11 | 0.34962 | -0.73 | 0 | mirMAP | -0.15 | 0.00049 | NA | |
| 63 | hsa-miR-30a-3p | MDM4 | 1.53 | 0 | -0.73 | 0 | mirMAP | -0.16 | 0 | NA | |
| 64 | hsa-miR-30a-5p | MDM4 | 0.63 | 0.00011 | -0.73 | 0 | mirMAP; miRNATAP | -0.12 | 0.00024 | NA | |
| 65 | hsa-miR-30e-3p | MDM4 | 1.21 | 0 | -0.73 | 0 | mirMAP | -0.23 | 0 | NA | |
| 66 | hsa-miR-30e-5p | MDM4 | 0.63 | 0 | -0.73 | 0 | mirMAP | -0.1 | 0.04523 | NA | |
| 67 | hsa-miR-326 | MDM4 | 1.88 | 0 | -0.73 | 0 | miRanda | -0.16 | 0 | NA | |
| 68 | hsa-miR-33b-5p | MDM4 | 2.29 | 0 | -0.73 | 0 | MirTarget | -0.11 | 0 | NA | |
| 69 | hsa-miR-3607-3p | MDM4 | 2.16 | 0 | -0.73 | 0 | mirMAP | -0.13 | 0 | NA | |
| 70 | hsa-miR-374a-3p | MDM4 | 0.21 | 0.06235 | -0.73 | 0 | mirMAP | -0.17 | 0.0004 | NA | |
| 71 | hsa-miR-374b-5p | MDM4 | 0.31 | 0.00301 | -0.73 | 0 | mirMAP | -0.17 | 0.00052 | NA | |
| 72 | hsa-miR-424-5p | MDM4 | 2.63 | 0 | -0.73 | 0 | mirMAP | -0.15 | 0 | NA | |
| 73 | hsa-miR-542-3p | MDM4 | 1.31 | 0 | -0.73 | 0 | miRanda | -0.12 | 0.0013 | NA | |
| 74 | hsa-miR-590-3p | MRPL13 | 0.47 | 2.0E-5 | -0.59 | 0 | miRanda | -0.22 | 2.0E-5 | NA | |
| 75 | hsa-miR-15a-5p | NFATC3 | -0.35 | 0.00077 | 0.64 | 0 | MirTarget; miRNATAP | -0.14 | 0.00111 | NA | |
| 76 | hsa-miR-185-5p | NFATC3 | -0.48 | 0 | 0.64 | 0 | MirTarget; miRNATAP | -0.18 | 0.0001 | NA | |
| 77 | hsa-miR-222-3p | NFATC3 | -1.09 | 0 | 0.64 | 0 | MirTarget; miRNATAP | -0.13 | 0 | NA | |
| 78 | hsa-miR-30d-5p | NFATC3 | -0.72 | 0 | 0.64 | 0 | MirTarget; miRNATAP | -0.11 | 0.00162 | NA | |
| 79 | hsa-miR-324-3p | NFATC3 | -0.26 | 0.05061 | 0.64 | 0 | miRNAWalker2 validate | -0.11 | 0.00063 | NA | |
| 80 | hsa-miR-335-3p | NFATC3 | 0.28 | 0.10663 | 0.64 | 0 | mirMAP | -0.13 | 0 | NA | |
| 81 | hsa-miR-339-5p | NFATC3 | -0.28 | 0.03557 | 0.64 | 0 | miRanda | -0.17 | 0 | NA | |
| 82 | hsa-miR-361-3p | NFATC3 | -0.28 | 0.00549 | 0.64 | 0 | miRNAWalker2 validate | -0.12 | 0.01086 | NA | |
| 83 | hsa-miR-361-5p | NFATC3 | -0.23 | 0.00962 | 0.64 | 0 | miRanda | -0.11 | 0.03282 | NA | |
| 84 | hsa-miR-362-3p | NFATC3 | -0.81 | 0 | 0.64 | 0 | miRanda | -0.17 | 0 | NA | |
| 85 | hsa-miR-589-3p | NFATC3 | -1.17 | 0 | 0.64 | 0 | mirMAP | -0.15 | 0 | NA | |
| 86 | hsa-miR-181a-5p | PDCD6IP | -0.25 | 0.05519 | 0.1 | 0.09548 | miRNATAP | -0.11 | 0 | NA | |
| 87 | hsa-miR-181b-5p | PDCD6IP | -0.49 | 0.00105 | 0.1 | 0.09548 | miRNATAP | -0.1 | 0 | NA | |
| 88 | hsa-miR-103a-3p | PIK3R1 | -0.77 | 0 | 0.89 | 0 | MirTarget; miRNATAP | -0.19 | 0.00703 | NA | |
| 89 | hsa-miR-106b-5p | PIK3R1 | -0.65 | 0 | 0.89 | 0 | MirTarget; miRNATAP | -0.37 | 0 | NA | |
| 90 | hsa-miR-1301-3p | PIK3R1 | -1.12 | 0 | 0.89 | 0 | MirTarget | -0.26 | 0 | NA | |
| 91 | hsa-miR-132-3p | PIK3R1 | -0.32 | 0.00272 | 0.89 | 0 | MirTarget | -0.52 | 0 | NA | |
| 92 | hsa-miR-15b-5p | PIK3R1 | -0.23 | 0.08248 | 0.89 | 0 | MirTarget | -0.13 | 0.02495 | NA | |
| 93 | hsa-miR-17-5p | PIK3R1 | -0.7 | 2.0E-5 | 0.89 | 0 | MirTarget; TargetScan; miRNATAP | -0.21 | 0 | NA | |
| 94 | hsa-miR-185-5p | PIK3R1 | -0.48 | 0 | 0.89 | 0 | miRNATAP | -0.26 | 0.00154 | NA | |
| 95 | hsa-miR-188-5p | PIK3R1 | -1.12 | 0 | 0.89 | 0 | MirTarget | -0.18 | 5.0E-5 | NA | |
| 96 | hsa-miR-200c-3p | PIK3R1 | 0.1 | 0.71696 | 0.89 | 0 | mirMAP | -0.11 | 9.0E-5 | NA | |
| 97 | hsa-miR-20a-5p | PIK3R1 | -0.85 | 0 | 0.89 | 0 | MirTarget; miRNATAP | -0.14 | 0.00238 | NA | |
| 98 | hsa-miR-21-5p | PIK3R1 | -1.51 | 0 | 0.89 | 0 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.63 | 0 | 26676464 | PIK3R1 targeting by miR 21 suppresses tumor cell migration and invasion by reducing PI3K/AKT signaling and reversing EMT and predicts clinical outcome of breast cancer; Next we identified the PIK3R1 as a direct target of miR-21 and showed that it was negatively regulated by miR-21; Taken together we provide novel evidence that miR-21 knockdown suppresses cell growth migration and invasion partly by inhibiting PI3K/AKT activation via direct targeting PIK3R1 and reversing EMT in breast cancer |
| 99 | hsa-miR-212-3p | PIK3R1 | 0.29 | 0.10039 | 0.89 | 0 | MirTarget | -0.26 | 0 | NA | |
| 100 | hsa-miR-221-3p | PIK3R1 | -1.12 | 0 | 0.89 | 0 | MirTarget | -0.22 | 1.0E-5 | NA | |
| 101 | hsa-miR-222-3p | PIK3R1 | -1.09 | 0 | 0.89 | 0 | MirTarget | -0.26 | 0 | NA | |
| 102 | hsa-miR-324-3p | PIK3R1 | -0.26 | 0.05061 | 0.89 | 0 | MirTarget; PITA; miRNATAP | -0.28 | 0 | NA | |
| 103 | hsa-miR-330-3p | PIK3R1 | 0.33 | 0.03161 | 0.89 | 0 | MirTarget; PITA; miRNATAP | -0.21 | 3.0E-5 | NA | |
| 104 | hsa-miR-338-5p | PIK3R1 | 0.22 | 0.25239 | 0.89 | 0 | PITA | -0.14 | 0.00037 | NA | |
| 105 | hsa-miR-409-3p | PIK3R1 | 0.5 | 0.05268 | 0.89 | 0 | mirMAP | -0.11 | 0.00024 | NA | |
| 106 | hsa-miR-493-5p | PIK3R1 | 0.2 | 0.50287 | 0.89 | 0 | MirTarget; miRNATAP | -0.1 | 8.0E-5 | NA | |
| 107 | hsa-miR-582-5p | PIK3R1 | 0.68 | 0.00104 | 0.89 | 0 | mirMAP | -0.14 | 0.00021 | NA | |
| 108 | hsa-miR-589-3p | PIK3R1 | -1.17 | 0 | 0.89 | 0 | mirMAP | -0.21 | 0 | NA | |
| 109 | hsa-miR-590-5p | PIK3R1 | 0.1 | 0.31003 | 0.89 | 0 | MirTarget; PITA; miRanda; miRNATAP | -0.3 | 0.0001 | NA | |
| 110 | hsa-miR-629-3p | PIK3R1 | 0.32 | 0.11909 | 0.89 | 0 | MirTarget | -0.18 | 0 | NA | |
| 111 | hsa-miR-93-5p | PIK3R1 | -1.4 | 0 | 0.89 | 0 | MirTarget; miRNATAP | -0.32 | 0 | NA | |
| 112 | hsa-miR-106b-5p | PLA2G12A | -0.65 | 0 | 0.79 | 0 | mirMAP | -0.27 | 0 | NA | |
| 113 | hsa-miR-140-5p | PLA2G12A | 0.22 | 0.01407 | 0.79 | 0 | miRanda | -0.15 | 0.00839 | NA | |
| 114 | hsa-miR-17-5p | PLA2G12A | -0.7 | 2.0E-5 | 0.79 | 0 | mirMAP | -0.16 | 0 | NA | |
| 115 | hsa-miR-181a-5p | PLA2G12A | -0.25 | 0.05519 | 0.79 | 0 | mirMAP | -0.19 | 0 | NA | |
| 116 | hsa-miR-181b-5p | PLA2G12A | -0.49 | 0.00105 | 0.79 | 0 | mirMAP | -0.19 | 0 | NA | |
| 117 | hsa-miR-20a-5p | PLA2G12A | -0.85 | 0 | 0.79 | 0 | mirMAP | -0.13 | 1.0E-5 | NA | |
| 118 | hsa-miR-32-3p | PLA2G12A | -0.22 | 0.20722 | 0.79 | 0 | mirMAP | -0.1 | 0.00072 | NA | |
| 119 | hsa-miR-362-3p | PLA2G12A | -0.81 | 0 | 0.79 | 0 | miRanda | -0.14 | 0.00017 | NA | |
| 120 | hsa-miR-501-5p | PLA2G12A | -1.15 | 0 | 0.79 | 0 | miRNATAP | -0.16 | 0 | NA | |
| 121 | hsa-miR-589-3p | PLA2G12A | -1.17 | 0 | 0.79 | 0 | mirMAP | -0.19 | 0 | NA | |
| 122 | hsa-miR-625-3p | PLA2G12A | -0.24 | 0.18123 | 0.79 | 0 | mirMAP | -0.14 | 0 | NA | |
| 123 | hsa-miR-92b-3p | PLA2G12A | -0.22 | 0.29619 | 0.79 | 0 | miRNAWalker2 validate | -0.1 | 2.0E-5 | NA | |
| 124 | hsa-miR-93-5p | PLA2G12A | -1.4 | 0 | 0.79 | 0 | mirMAP | -0.32 | 0 | NA | |
| 125 | hsa-miR-33a-3p | PLA2G4A | 0.68 | 1.0E-5 | 1.52 | 0 | MirTarget | -0.19 | 0.0187 | NA | |
| 126 | hsa-miR-361-5p | PLA2G4A | -0.23 | 0.00962 | 1.52 | 0 | miRanda | -0.52 | 0.00029 | NA | |
| 127 | hsa-miR-362-3p | PLA2G4A | -0.81 | 0 | 1.52 | 0 | miRanda | -0.27 | 0.00355 | NA | |
| 128 | hsa-miR-374a-5p | PLA2G4A | -0.02 | 0.86978 | 1.52 | 0 | mirMAP | -0.28 | 0.03331 | NA | |
| 129 | hsa-miR-103a-2-5p | PLD1 | -1.17 | 0 | 1.16 | 0 | MirTarget | -0.1 | 0.03738 | NA | |
| 130 | hsa-miR-103a-3p | PLD1 | -0.77 | 0 | 1.16 | 0 | MirTarget | -0.26 | 0.0018 | NA | |
| 131 | hsa-miR-107 | PLD1 | -0.24 | 0.01708 | 1.16 | 0 | MirTarget; miRanda | -0.53 | 0 | NA | |
| 132 | hsa-miR-15a-5p | PLD1 | -0.35 | 0.00077 | 1.16 | 0 | MirTarget | -0.38 | 3.0E-5 | NA | |
| 133 | hsa-miR-32-3p | PLD1 | -0.22 | 0.20722 | 1.16 | 0 | mirMAP | -0.32 | 0 | NA | |
| 134 | hsa-miR-361-5p | PLD1 | -0.23 | 0.00962 | 1.16 | 0 | MirTarget; miRanda | -0.55 | 0 | NA | |
| 135 | hsa-miR-374a-3p | PLD1 | 0.21 | 0.06235 | 1.16 | 0 | MirTarget | -0.24 | 0.00437 | NA | |
| 136 | hsa-miR-374b-3p | PLD1 | 0.14 | 0.30532 | 1.16 | 0 | mirMAP | -0.24 | 0.00061 | NA | |
| 137 | hsa-miR-421 | PLD1 | -0.94 | 0 | 1.16 | 0 | miRanda | -0.16 | 0.00238 | NA | |
| 138 | hsa-miR-664a-3p | PLD1 | -0.49 | 0.00073 | 1.16 | 0 | mirMAP | -0.24 | 0.00022 | NA | |
| 139 | hsa-let-7a-3p | POLR2K | 0.57 | 0 | -0.85 | 0 | MirTarget | -0.25 | 0 | NA | |
| 140 | hsa-let-7b-3p | POLR2K | 1.22 | 0 | -0.85 | 0 | MirTarget | -0.22 | 0 | NA | |
| 141 | hsa-miR-590-3p | POLR2K | 0.47 | 2.0E-5 | -0.85 | 0 | miRanda | -0.16 | 0.00084 | NA | |
| 142 | hsa-miR-421 | PSAT1 | -0.94 | 0 | 1.76 | 0 | miRanda | -0.13 | 0.0494 | NA | |
| 143 | hsa-miR-148b-5p | RPS27L | -0.3 | 0.02557 | 0.3 | 0.00876 | MirTarget | -0.19 | 1.0E-5 | NA | |
| 144 | hsa-miR-320a | RPS27L | -0.33 | 0.02214 | 0.3 | 0.00876 | miRanda | -0.16 | 4.0E-5 | NA | |
| 145 | hsa-miR-361-5p | RPS27L | -0.23 | 0.00962 | 0.3 | 0.00876 | miRanda | -0.33 | 0 | NA | |
| 146 | hsa-miR-590-3p | RPS27L | 0.47 | 2.0E-5 | 0.3 | 0.00876 | MirTarget; miRanda; mirMAP | -0.17 | 0.00081 | NA | |
| 147 | hsa-let-7a-5p | RRM2 | 0.33 | 0.00046 | -3.28 | 0 | miRNAWalker2 validate; TargetScan; miRNATAP | -0.42 | 0.00212 | NA | |
| 148 | hsa-let-7b-5p | RRM2 | 0.96 | 0 | -3.28 | 0 | miRNAWalker2 validate; miRNATAP | -0.51 | 0 | NA | |
| 149 | hsa-let-7g-5p | RRM2 | 0.46 | 2.0E-5 | -3.28 | 0 | miRNAWalker2 validate; miRNATAP | -0.6 | 0 | NA | |
| 150 | hsa-miR-100-5p | RRM2 | 0.78 | 0.00022 | -3.28 | 0 | miRNAWalker2 validate | -0.47 | 0 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | ORGANOPHOSPHATE BIOSYNTHETIC PROCESS | 8 | 450 | 4.772e-08 | 0.000222 |
| 2 | SINGLE ORGANISM BIOSYNTHETIC PROCESS | 11 | 1340 | 2.178e-07 | 0.0003377 |
| 3 | GLYCEROLIPID BIOSYNTHETIC PROCESS | 6 | 211 | 1.924e-07 | 0.0003377 |
| 4 | PHOSPHOLIPID BIOSYNTHETIC PROCESS | 6 | 235 | 3.627e-07 | 0.0004219 |
| 5 | ORGANOPHOSPHATE METABOLIC PROCESS | 9 | 885 | 6.726e-07 | 0.0006259 |
| 6 | PHOSPHATIDYLGLYCEROL ACYL CHAIN REMODELING | 3 | 17 | 1.16e-06 | 0.0008993 |
| 7 | GLYCEROPHOSPHOLIPID METABOLIC PROCESS | 6 | 297 | 1.424e-06 | 0.0009464 |
| 8 | GLYCEROLIPID METABOLIC PROCESS | 6 | 356 | 4.057e-06 | 0.00236 |
| 9 | PHOSPHOLIPID METABOLIC PROCESS | 6 | 364 | 4.61e-06 | 0.002383 |
| 10 | PHOSPHATIDYLGLYCEROL METABOLIC PROCESS | 3 | 31 | 7.578e-06 | 0.003526 |
| 11 | PHOSPHATIDIC ACID METABOLIC PROCESS | 3 | 33 | 9.183e-06 | 0.003884 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
| 1 | hsa04912_GnRH_signaling_pathway | 5 | 101 | 1.457e-07 | 2.623e-05 | |
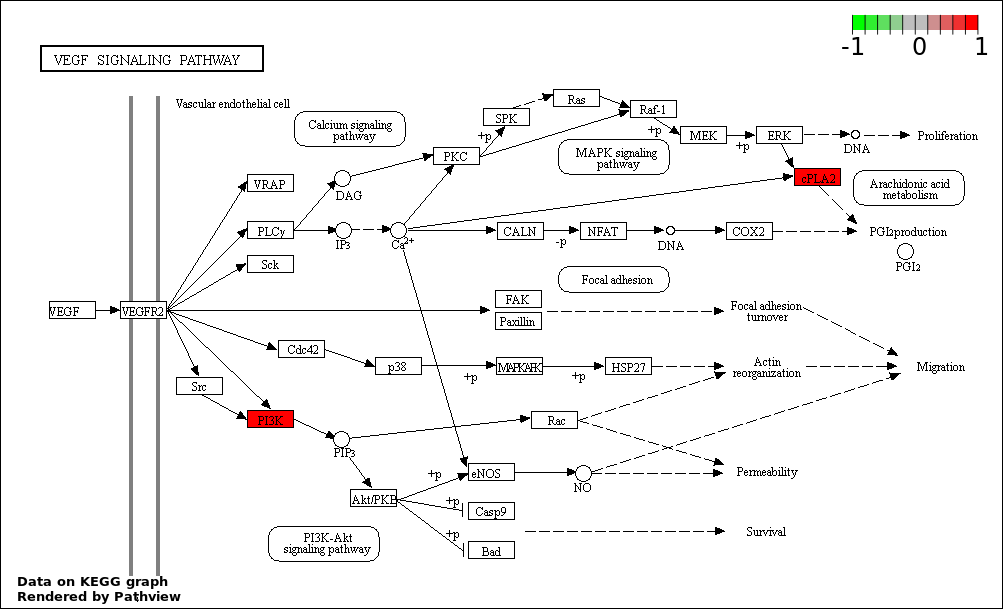
| 2 | hsa04370_VEGF_signaling_pathway | 4 | 76 | 2.292e-06 | 0.000169 | |
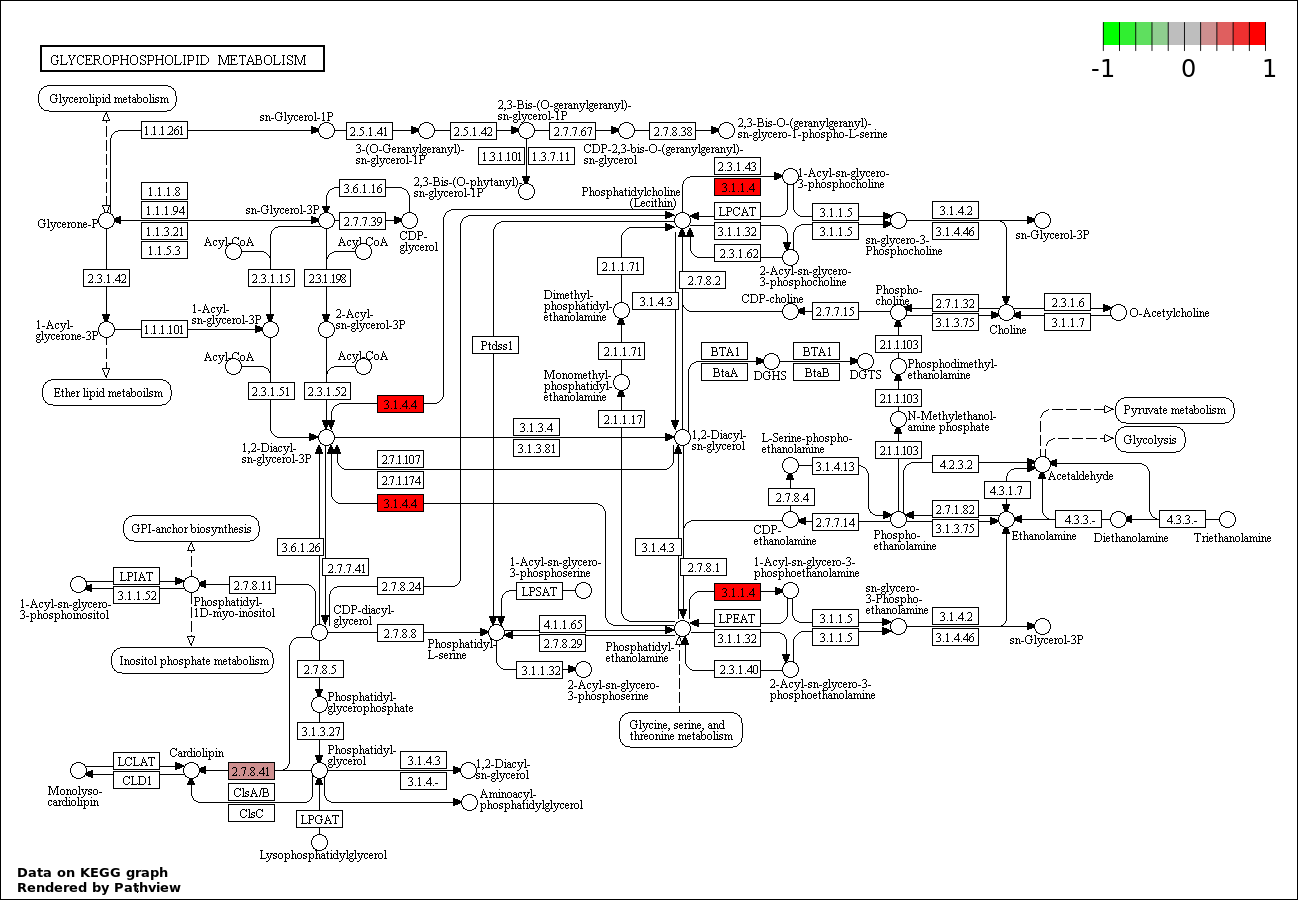
| 3 | hsa00564_Glycerophospholipid_metabolism | 4 | 80 | 2.816e-06 | 0.000169 | |
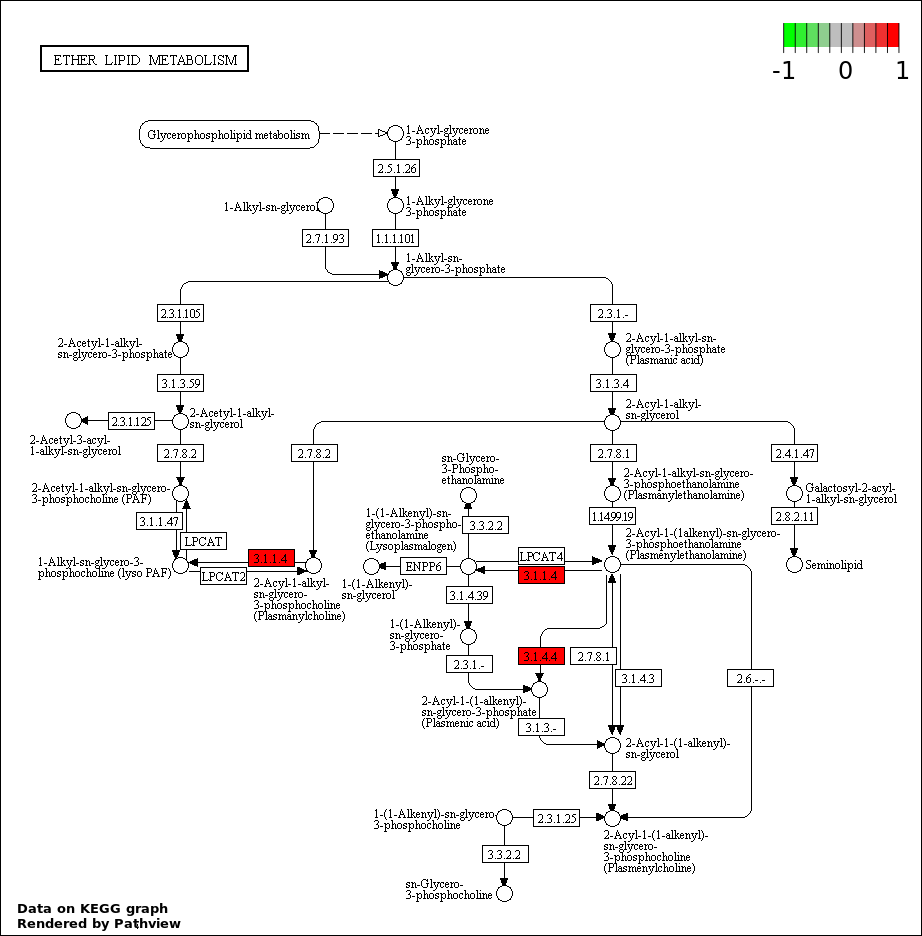
| 4 | hsa00565_Ether_lipid_metabolism | 3 | 36 | 1.199e-05 | 0.0005394 | |
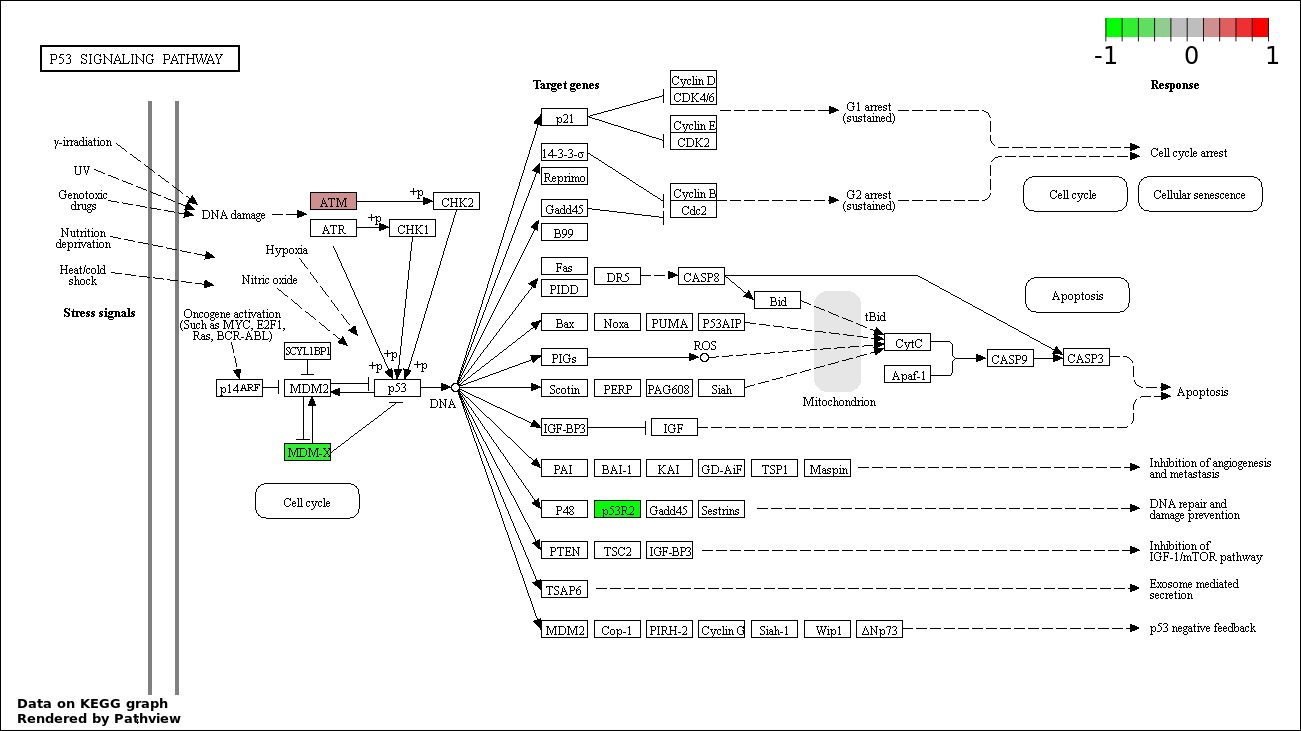
| 5 | hsa04115_p53_signaling_pathway | 3 | 69 | 8.56e-05 | 0.002681 | |
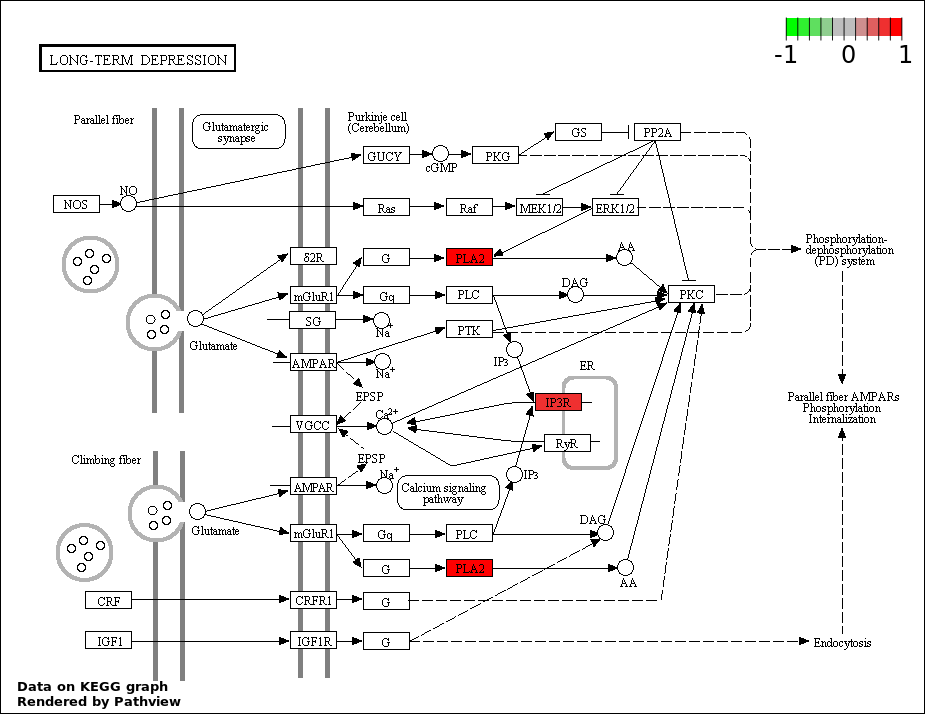
| 6 | hsa04730_Long.term_depression | 3 | 70 | 8.936e-05 | 0.002681 | |
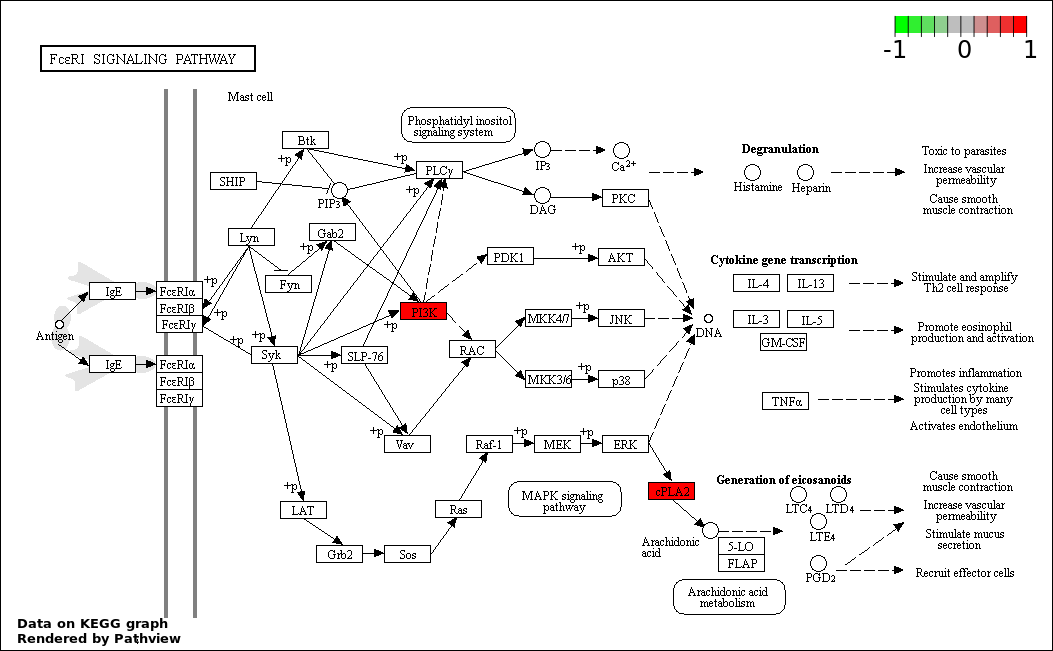
| 7 | hsa04664_Fc_epsilon_RI_signaling_pathway | 3 | 79 | 0.0001281 | 0.003295 | |
| 8 | hsa04014_Ras_signaling_pathway | 4 | 236 | 0.0001967 | 0.004427 | |
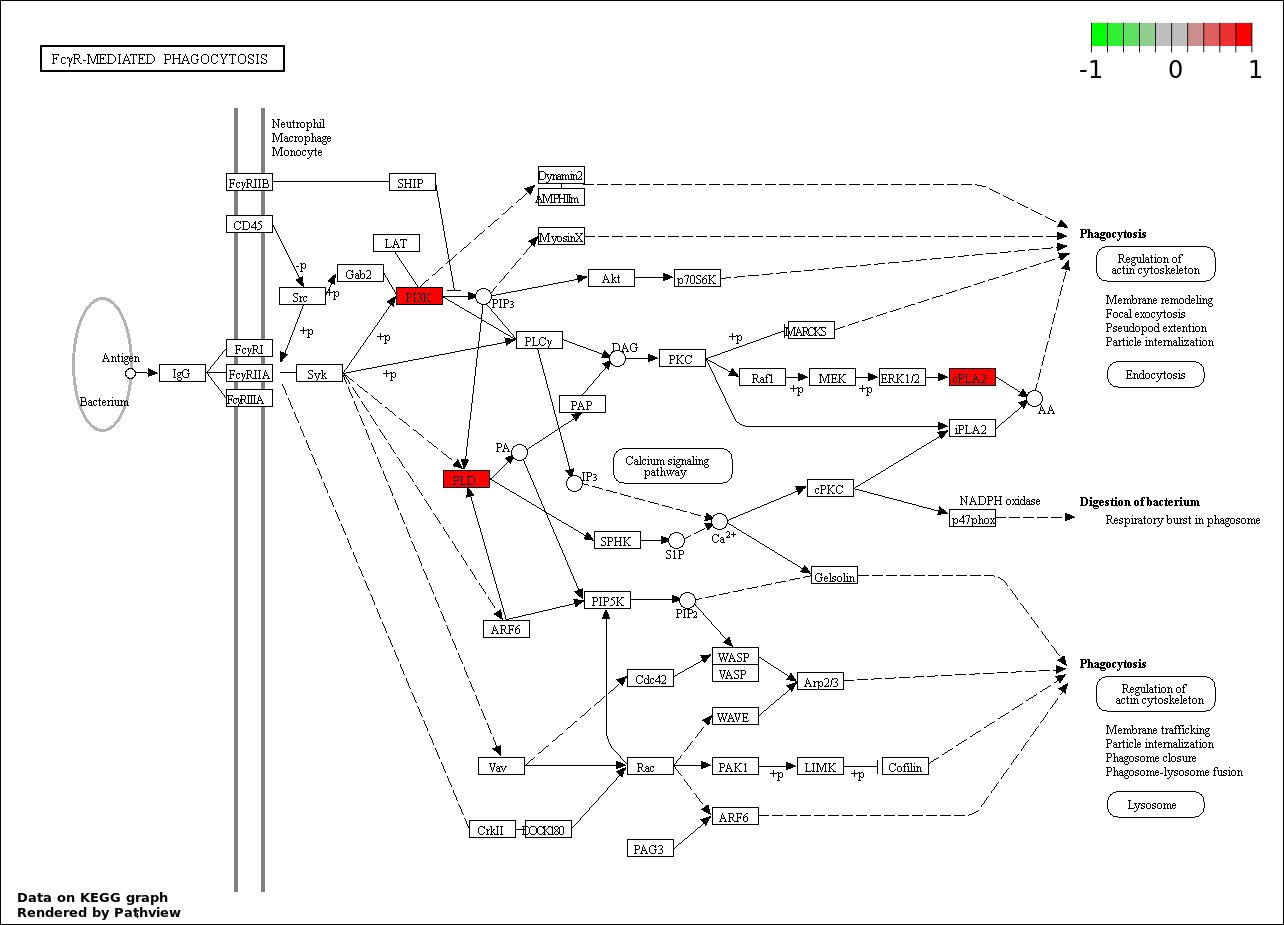
| 9 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 3 | 95 | 0.0002213 | 0.004427 | |
| 10 | hsa04972_Pancreatic_secretion | 3 | 101 | 0.0002652 | 0.0046 | |
| 11 | hsa00592_alpha.Linolenic_acid_metabolism | 2 | 20 | 0.0002811 | 0.0046 | |
| 12 | hsa04270_Vascular_smooth_muscle_contraction | 3 | 116 | 0.0003984 | 0.005976 | |
| 13 | hsa00591_Linoleic_acid_metabolism | 2 | 30 | 0.0006387 | 0.008843 | |
| 14 | hsa04975_Fat_digestion_and_absorption | 2 | 46 | 0.001501 | 0.0193 | |
| 15 | hsa04144_Endocytosis | 3 | 203 | 0.00201 | 0.02412 | |
| 16 | hsa00590_Arachidonic_acid_metabolism | 2 | 59 | 0.002457 | 0.02764 | |
| 17 | hsa04662_B_cell_receptor_signaling_pathway | 2 | 75 | 0.003936 | 0.04168 | |
| 18 | hsa04070_Phosphatidylinositol_signaling_system | 2 | 78 | 0.00425 | 0.04168 | |
| 19 | hsa04010_MAPK_signaling_pathway | 3 | 268 | 0.0044 | 0.04168 | |
| 20 | hsa04210_Apoptosis | 2 | 89 | 0.005496 | 0.04946 | |
| 21 | hsa03010_Ribosome | 2 | 92 | 0.005862 | 0.05024 | |
| 22 | hsa00240_Pyrimidine_metabolism | 2 | 99 | 0.006757 | 0.05528 | |
| 23 | hsa04660_T_cell_receptor_signaling_pathway | 2 | 108 | 0.007993 | 0.05994 | |
| 24 | hsa04670_Leukocyte_transendothelial_migration | 2 | 117 | 0.009323 | 0.06712 | |
| 25 | hsa04722_Neurotrophin_signaling_pathway | 2 | 127 | 0.01091 | 0.07549 | |
| 26 | hsa04360_Axon_guidance | 2 | 130 | 0.01141 | 0.07549 | |
| 27 | hsa00190_Oxidative_phosphorylation | 2 | 132 | 0.01174 | 0.07549 | |
| 28 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 2 | 136 | 0.01243 | 0.07716 | |
| 29 | hsa04120_Ubiquitin_mediated_proteolysis | 2 | 139 | 0.01296 | 0.07775 | |
| 30 | hsa04630_Jak.STAT_signaling_pathway | 2 | 155 | 0.01593 | 0.0907 | |
| 31 | hsa04145_Phagosome | 2 | 156 | 0.01612 | 0.0907 | |
| 32 | hsa00230_Purine_metabolism | 2 | 162 | 0.01731 | 0.09444 | |
| 33 | hsa04510_Focal_adhesion | 2 | 200 | 0.02567 | 0.1359 | |
| 34 | hsa04810_Regulation_of_actin_cytoskeleton | 2 | 214 | 0.02909 | 0.1496 | |
| 35 | hsa04151_PI3K_AKT_signaling_pathway | 2 | 351 | 0.0707 | 0.2892 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | MALAT1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p | 11 | MDM4 | Sponge network | -1.297 | 0 | -0.734 | 0 | 0.615 |
| 2 | KB-1572G7.2 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p | 11 | MDM4 | Sponge network | -2.124 | 0 | -0.734 | 0 | 0.611 |
| 3 | AC159540.1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-542-3p | 12 | MDM4 | Sponge network | -2.112 | 0 | -0.734 | 0 | 0.586 |
| 4 | GUSBP11 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p | 14 | MDM4 | Sponge network | -2.066 | 0 | -0.734 | 0 | 0.583 |
| 5 | DHRS4-AS1 | hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-582-5p;hsa-miR-589-3p | 10 | PIK3R1 | Sponge network | 0.646 | 0.01829 | 0.892 | 0 | 0.557 |
| 6 | RP11-434D9.1 | hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-17-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-409-3p;hsa-miR-629-3p | 13 | PIK3R1 | Sponge network | 2.913 | 0 | 0.892 | 0 | 0.525 |
| 7 | LINC00261 | hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-15b-5p;hsa-miR-188-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-589-3p;hsa-miR-629-3p | 13 | PIK3R1 | Sponge network | 1.194 | 0 | 0.892 | 0 | 0.513 |
| 8 | AC005154.6 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 12 | MDM4 | Sponge network | -1.75 | 0 | -0.734 | 0 | 0.509 |
| 9 | SNHG1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p | 10 | MDM4 | Sponge network | -2.013 | 0 | -0.734 | 0 | 0.493 |
| 10 | AC005562.1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | -1.127 | 0 | -0.734 | 0 | 0.493 |
| 11 | RP11-513G11.3 | hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-17-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-93-5p | 10 | PIK3R1 | Sponge network | 2.342 | 5.0E-5 | 0.892 | 0 | 0.492 |
| 12 | RP11-12A2.3 | hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-93-5p | 14 | PIK3R1 | Sponge network | 4.779 | 0 | 0.892 | 0 | 0.489 |
| 13 | LINC01018 | hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-409-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-93-5p | 18 | PIK3R1 | Sponge network | 3.231 | 0 | 0.892 | 0 | 0.472 |
| 14 | GAS5 | hsa-let-7c-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | -1.966 | 0 | -0.734 | 0 | 0.46 |
| 15 | KCNQ1OT1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-152-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p | 11 | MDM4 | Sponge network | -0.864 | 0 | -0.734 | 0 | 0.455 |
| 16 | CTD-2228K2.7 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-542-3p | 11 | MDM4 | Sponge network | -2.28 | 0 | -0.734 | 0 | 0.452 |
| 17 | RP11-600F24.7 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 12 | MDM4 | Sponge network | -2.603 | 0 | -0.734 | 0 | 0.451 |
| 18 | LDLRAD4-AS1 | hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-589-3p;hsa-miR-93-5p | 13 | PIK3R1 | Sponge network | 3.366 | 0 | 0.892 | 0 | 0.45 |
| 19 | MIR600HG | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 11 | MDM4 | Sponge network | -3.233 | 5.0E-5 | -0.734 | 0 | 0.447 |
| 20 | PSMD5-AS1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 12 | MDM4 | Sponge network | -1.538 | 0 | -0.734 | 0 | 0.436 |
| 21 | RP11-196G18.22 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-3607-3p | 10 | MDM4 | Sponge network | -2.705 | 0 | -0.734 | 0 | 0.431 |
| 22 | SNHG12 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-424-5p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | -1.791 | 0 | -0.734 | 0 | 0.419 |
| 23 | RP11-119D9.1 | hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-629-3p;hsa-miR-93-5p | 11 | PIK3R1 | Sponge network | 2.765 | 0 | 0.892 | 0 | 0.418 |
| 24 | RP11-196G18.24 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-424-5p | 10 | MDM4 | Sponge network | -2.972 | 0 | -0.734 | 0 | 0.406 |
| 25 | HCG18 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 12 | MDM4 | Sponge network | -1.42 | 0 | -0.734 | 0 | 0.405 |
| 26 | RP11-290F5.1 | hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-3p;hsa-miR-493-5p | 13 | PIK3R1 | Sponge network | 1.679 | 5.0E-5 | 0.892 | 0 | 0.402 |
| 27 | AP001469.9 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | -2.428 | 0 | -0.734 | 0 | 0.39 |
| 28 | RP11-89K21.1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p | 13 | MDM4 | Sponge network | -4.915 | 0 | -0.734 | 0 | 0.387 |
| 29 | DLEU2 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-152-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | -1.141 | 0 | -0.734 | 0 | 0.386 |
| 30 | MATN1-AS1 | hsa-let-7c-5p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | -1.031 | 0 | -0.734 | 0 | 0.386 |
| 31 | RP11-115J16.1 | hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-589-3p;hsa-miR-93-5p | 12 | PIK3R1 | Sponge network | 2.038 | 7.0E-5 | 0.892 | 0 | 0.382 |
| 32 | AC074117.10 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | -1.254 | 0 | -0.734 | 0 | 0.379 |
| 33 | LINC00238 | hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-629-3p | 12 | PIK3R1 | Sponge network | 4.997 | 0 | 0.892 | 0 | 0.369 |
| 34 | LINC00176 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | -3.423 | 0 | -0.734 | 0 | 0.368 |
| 35 | GS1-124K5.11 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | -1.5 | 0 | -0.734 | 0 | 0.365 |
| 36 | RP11-498C9.15 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-5p;hsa-miR-33b-5p;hsa-miR-374b-5p | 10 | MDM4 | Sponge network | -1.487 | 0 | -0.734 | 0 | 0.365 |
| 37 | ERVK3-1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-542-3p | 11 | MDM4 | Sponge network | -1.328 | 0 | -0.734 | 0 | 0.364 |
| 38 | RP1-228H13.5 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-542-3p | 11 | MDM4 | Sponge network | -1.554 | 0 | -0.734 | 0 | 0.363 |
| 39 | RP11-244O19.1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-191-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-424-5p | 11 | MDM4 | Sponge network | -1.429 | 0 | -0.734 | 0 | 0.363 |
| 40 | LINC00624 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | -3.71 | 0 | -0.734 | 0 | 0.359 |
| 41 | AC004862.6 | hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-93-5p | 13 | PIK3R1 | Sponge network | 2.202 | 0.00081 | 0.892 | 0 | 0.357 |
| 42 | SNHG11 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-3607-3p | 10 | MDM4 | Sponge network | -1.239 | 0 | -0.734 | 0 | 0.353 |
| 43 | RP11-250B2.6 | hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-93-5p | 12 | PIK3R1 | Sponge network | 0.98 | 2.0E-5 | 0.892 | 0 | 0.342 |
| 44 | AP000473.5 | hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-5p | 11 | PIK3R1 | Sponge network | 1.157 | 0.00884 | 0.892 | 0 | 0.341 |
| 45 | CTBP1-AS2 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 12 | MDM4 | Sponge network | -1.419 | 0 | -0.734 | 0 | 0.337 |
| 46 | RP11-963H4.3 | hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p | 10 | PIK3R1 | Sponge network | 1.857 | 0.00116 | 0.892 | 0 | 0.328 |
| 47 | RP11-727A23.5 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-5p;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-542-3p | 11 | MDM4 | Sponge network | -1.435 | 0 | -0.734 | 0 | 0.327 |
| 48 | RP11-407B7.1 | hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-188-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-93-5p | 13 | PIK3R1 | Sponge network | 0.818 | 0.00584 | 0.892 | 0 | 0.324 |
| 49 | LINC00864 | hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-15b-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-629-3p | 10 | PIK3R1 | Sponge network | 3.954 | 0 | 0.892 | 0 | 0.322 |
| 50 | SNHG7 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-424-5p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | -2.077 | 0 | -0.734 | 0 | 0.322 |
| 51 | LINC01011 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-424-5p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | -1.861 | 0 | -0.734 | 0 | 0.321 |
| 52 | RP11-166D19.1 | hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-324-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p | 14 | PIK3R1 | Sponge network | 0.244 | 0.28835 | 0.892 | 0 | 0.32 |
| 53 | STXBP5-AS1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p | 12 | MDM4 | Sponge network | -1.715 | 0 | -0.734 | 0 | 0.308 |
| 54 | SNHG17 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | -1.214 | 0 | -0.734 | 0 | 0.3 |
| 55 | LINC00885 | hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-3p | 10 | PIK3R1 | Sponge network | 4.686 | 0 | 0.892 | 0 | 0.3 |
| 56 | MAGI2-AS3 | hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-93-5p | 13 | PIK3R1 | Sponge network | 1.801 | 0 | 0.892 | 0 | 0.294 |
| 57 | CTD-3162L10.1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | -3.111 | 0 | -0.734 | 0 | 0.289 |
| 58 | RP11-540A21.2 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p | 10 | MDM4 | Sponge network | -1.758 | 0 | -0.734 | 0 | 0.285 |
| 59 | VAC14-AS1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-374b-5p | 10 | MDM4 | Sponge network | -2.836 | 5.0E-5 | -0.734 | 0 | 0.282 |
| 60 | AC005550.3 | hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-629-3p | 10 | PIK3R1 | Sponge network | 2.571 | 0.00132 | 0.892 | 0 | 0.281 |
| 61 | RP5-1165K10.2 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-424-5p | 10 | MDM4 | Sponge network | -1.401 | 0 | -0.734 | 0 | 0.272 |
| 62 | RP11-7F17.3 | hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-3p | 11 | PIK3R1 | Sponge network | 0.873 | 0.00204 | 0.892 | 0 | 0.271 |
| 63 | SMIM2-AS1 | hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-132-3p;hsa-miR-17-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-589-3p;hsa-miR-629-3p | 13 | PIK3R1 | Sponge network | 0.66 | 0.00587 | 0.892 | 0 | 0.27 |
| 64 | CTD-3220F14.1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 11 | MDM4 | Sponge network | -3.223 | 0 | -0.734 | 0 | 0.268 |
| 65 | ZNRD1-AS1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 11 | MDM4 | Sponge network | -1.284 | 0 | -0.734 | 0 | 0.266 |
| 66 | MIR4435-1HG | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-542-3p | 12 | MDM4 | Sponge network | -2.541 | 0 | -0.734 | 0 | 0.261 |
| 67 | LINC00665 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p | 11 | MDM4 | Sponge network | -2.394 | 0 | -0.734 | 0 | 0.256 |
| 68 | RP11-37B2.1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 12 | MDM4 | Sponge network | -1.504 | 0 | -0.734 | 0 | 0.253 |
| 69 | POLR2J4 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 12 | MDM4 | Sponge network | -1.386 | 0 | -0.734 | 0 | 0.251 |