This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-21-5p | AKT2 | 1.75 | 0 | -0.01 | 0.9227 | miRNAWalker2 validate | -0.15 | 0.00352 | NA | |
| 2 | hsa-miR-29b-3p | AKT2 | -0.23 | 0.36746 | -0.01 | 0.9227 | MirTarget | -0.14 | 0.00022 | 26512921; 26564501; 24076586 | MicroRNA 29B mir 29b regulates the Warburg effect in ovarian cancer by targeting AKT2 and AKT3;The expression of miR-29b was significantly upregualted by cisplatin treatmentwhile its target gene AKT2 was downregulated;Furthermore a feed-back loop comprising of c-Myc miR-29 family and Akt2 were found in myeloid leukemogenesis |
| 3 | hsa-miR-106b-5p | AKT3 | 1.71 | 0 | -0.75 | 0.06936 | miRNATAP | -0.37 | 7.0E-5 | NA | |
| 4 | hsa-miR-107 | AKT3 | 0.9 | 5.0E-5 | -0.75 | 0.06936 | PITA; miRanda | -0.6 | 0 | NA | |
| 5 | hsa-miR-15b-5p | AKT3 | 1.62 | 0 | -0.75 | 0.06936 | miRNATAP | -0.36 | 0.00041 | NA | |
| 6 | hsa-miR-16-5p | AKT3 | 1.01 | 1.0E-5 | -0.75 | 0.06936 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.47 | 0.00031 | NA | |
| 7 | hsa-miR-17-3p | AKT3 | 1.31 | 0 | -0.75 | 0.06936 | miRNATAP | -0.52 | 0 | NA | |
| 8 | hsa-miR-17-5p | AKT3 | 1.66 | 0 | -0.75 | 0.06936 | TargetScan; miRNATAP | -0.28 | 0.0027 | NA | |
| 9 | hsa-miR-29a-3p | AKT3 | -0.11 | 0.61501 | -0.75 | 0.06936 | miRNATAP | -0.41 | 0.00212 | NA | |
| 10 | hsa-miR-29b-3p | AKT3 | -0.23 | 0.36746 | -0.75 | 0.06936 | miRNATAP | -0.34 | 0.00361 | 26512921 | MicroRNA 29B mir 29b regulates the Warburg effect in ovarian cancer by targeting AKT2 and AKT3 |
| 11 | hsa-miR-32-3p | AKT3 | 0.58 | 0.11837 | -0.75 | 0.06936 | mirMAP | -0.32 | 0.00023 | NA | |
| 12 | hsa-miR-320b | AKT3 | 1.11 | 0.0005 | -0.75 | 0.06936 | PITA; miRanda; miRNATAP | -0.26 | 0.0039 | NA | |
| 13 | hsa-miR-335-3p | AKT3 | 2.52 | 0 | -0.75 | 0.06936 | mirMAP | -0.22 | 0.00074 | NA | |
| 14 | hsa-miR-33a-3p | AKT3 | 0.35 | 0.32171 | -0.75 | 0.06936 | mirMAP | -0.38 | 2.0E-5 | NA | |
| 15 | hsa-miR-362-3p | AKT3 | -0.03 | 0.91378 | -0.75 | 0.06936 | miRanda | -0.38 | 4.0E-5 | NA | |
| 16 | hsa-miR-362-5p | AKT3 | -0.35 | 0.35491 | -0.75 | 0.06936 | PITA; TargetScan; miRNATAP | -0.23 | 0.00253 | NA | |
| 17 | hsa-miR-501-3p | AKT3 | 0.73 | 0.02704 | -0.75 | 0.06936 | miRNATAP | -0.39 | 1.0E-5 | NA | |
| 18 | hsa-miR-502-3p | AKT3 | -0.16 | 0.55747 | -0.75 | 0.06936 | miRNATAP | -0.5 | 0 | NA | |
| 19 | hsa-miR-505-3p | AKT3 | 0.83 | 0.00112 | -0.75 | 0.06936 | mirMAP | -0.35 | 0.00235 | 22051041 | We also find that Akt3 correlate inversely with miR-505 modulates drug sensitivity in MCF7-ADR |
| 20 | hsa-miR-548v | AKT3 | -0.16 | 0.70867 | -0.75 | 0.06936 | miRNATAP | -0.27 | 0.00031 | NA | |
| 21 | hsa-miR-577 | AKT3 | 0.91 | 0.22561 | -0.75 | 0.06936 | mirMAP | -0.23 | 0 | NA | |
| 22 | hsa-miR-616-5p | AKT3 | 0.83 | 0.03478 | -0.75 | 0.06936 | mirMAP | -0.27 | 0.00041 | NA | |
| 23 | hsa-miR-103a-3p | CAB39 | 0.84 | 0 | -0.33 | 0.05024 | MirTarget; miRNATAP | -0.24 | 0.00078 | NA | |
| 24 | hsa-miR-107 | CAB39 | 0.9 | 5.0E-5 | -0.33 | 0.05024 | MirTarget; miRanda; miRNATAP | -0.14 | 0.00671 | NA | |
| 25 | hsa-miR-151a-3p | CAB39 | 1 | 1.0E-5 | -0.33 | 0.05024 | MirTarget | -0.17 | 0.00082 | NA | |
| 26 | hsa-miR-15b-5p | CAB39 | 1.62 | 0 | -0.33 | 0.05024 | miRNATAP | -0.16 | 5.0E-5 | NA | |
| 27 | hsa-miR-23b-3p | CAB39 | -0.29 | 0.18665 | -0.33 | 0.05024 | MirTarget; miRNATAP | -0.15 | 0.00624 | NA | |
| 28 | hsa-miR-27b-3p | CAB39 | 0.08 | 0.72527 | -0.33 | 0.05024 | MirTarget; miRNATAP | -0.13 | 0.00845 | NA | |
| 29 | hsa-miR-374b-3p | CAB39 | 1 | 0.00127 | -0.33 | 0.05024 | MirTarget | -0.13 | 0.00076 | NA | |
| 30 | hsa-miR-155-5p | CAB39L | 1.29 | 9.0E-5 | -2.75 | 0 | miRNAWalker2 validate | -0.21 | 0.00812 | NA | |
| 31 | hsa-miR-214-5p | CAB39L | 1.03 | 0.00141 | -2.75 | 0 | MirTarget | -0.34 | 3.0E-5 | NA | |
| 32 | hsa-miR-3662 | CAB39L | 1.83 | 0.0054 | -2.75 | 0 | MirTarget; miRNATAP | -0.16 | 0.00158 | NA | |
| 33 | hsa-miR-542-3p | CAB39L | 0.76 | 0.00211 | -2.75 | 0 | miRanda | -0.39 | 0.00033 | NA | |
| 34 | hsa-miR-590-3p | CAB39L | 1.12 | 0.00016 | -2.75 | 0 | miRanda | -0.35 | 0.0001 | NA | |
| 35 | hsa-miR-590-5p | CAB39L | 1.04 | 0.00027 | -2.75 | 0 | miRanda | -0.42 | 0 | NA | |
| 36 | hsa-miR-125a-5p | EIF4E2 | -0.88 | 0.00021 | -0.03 | 0.82669 | miRanda | -0.15 | 2.0E-5 | NA | |
| 37 | hsa-miR-125a-5p | EIF4EBP1 | -0.88 | 0.00021 | 1.1 | 0.00073 | miRNAWalker2 validate; MirTarget; miRanda; miRNATAP | -0.41 | 2.0E-5 | 26646586 | Increased expression of miR-125a and miR-125b inhibited invasion and migration of SKOV3 and OVCAR-429 ovarian cancer cells and was associated with a decrease in EIF4EBP1 expression |
| 38 | hsa-miR-106b-5p | HIF1A | 1.71 | 0 | 0.84 | 0.00084 | MirTarget | -0.17 | 0.00333 | NA | |
| 39 | hsa-miR-107 | HIF1A | 0.9 | 5.0E-5 | 0.84 | 0.00084 | miRNAWalker2 validate; miRTarBase; miRanda | -0.36 | 1.0E-5 | NA | |
| 40 | hsa-miR-139-5p | HIF1A | -1.83 | 0 | 0.84 | 0.00084 | miRanda | -0.16 | 0.00469 | NA | |
| 41 | hsa-miR-151a-3p | HIF1A | 1 | 1.0E-5 | 0.84 | 0.00084 | MirTarget | -0.23 | 0.00333 | NA | |
| 42 | hsa-miR-320c | HIF1A | 0.46 | 0.24061 | 0.84 | 0.00084 | miRanda | -0.15 | 0.00656 | NA | |
| 43 | hsa-miR-330-3p | HIF1A | 0.8 | 0.00747 | 0.84 | 0.00084 | MirTarget; PITA | -0.18 | 0.00313 | NA | |
| 44 | hsa-miR-338-3p | HIF1A | -0.96 | 0.01915 | 0.84 | 0.00084 | MirTarget; PITA; miRanda | -0.18 | 3.0E-5 | NA | |
| 45 | hsa-miR-338-5p | HIF1A | -1.2 | 0.01003 | 0.84 | 0.00084 | MirTarget; PITA; miRNATAP | -0.14 | 0.0002 | NA | |
| 46 | hsa-miR-361-5p | HIF1A | 0.41 | 0.01813 | 0.84 | 0.00084 | miRanda | -0.31 | 0.00432 | NA | |
| 47 | hsa-miR-660-5p | HIF1A | -0.02 | 0.93887 | 0.84 | 0.00084 | MirTarget | -0.28 | 2.0E-5 | NA | |
| 48 | hsa-let-7a-3p | IGF1 | 0.5 | 0.04111 | -2.08 | 0.00135 | mirMAP | -0.55 | 0.00422 | NA | |
| 49 | hsa-let-7b-3p | IGF1 | 0.22 | 0.29604 | -2.08 | 0.00135 | mirMAP | -0.64 | 0.00549 | NA | |
| 50 | hsa-let-7f-1-3p | IGF1 | 1.29 | 0 | -2.08 | 0.00135 | mirMAP | -0.85 | 0 | NA | |
| 51 | hsa-miR-103a-2-5p | IGF1 | 0.81 | 0.01999 | -2.08 | 0.00135 | mirMAP | -0.55 | 3.0E-5 | NA | |
| 52 | hsa-miR-130b-3p | IGF1 | 1.33 | 5.0E-5 | -2.08 | 0.00135 | MirTarget | -0.9 | 0 | NA | |
| 53 | hsa-miR-15b-3p | IGF1 | 1.76 | 0 | -2.08 | 0.00135 | mirMAP | -0.97 | 0 | NA | |
| 54 | hsa-miR-16-1-3p | IGF1 | 1.43 | 0 | -2.08 | 0.00135 | mirMAP | -0.89 | 0 | NA | |
| 55 | hsa-miR-181b-5p | IGF1 | 1.64 | 0 | -2.08 | 0.00135 | mirMAP | -0.48 | 0.00175 | NA | |
| 56 | hsa-miR-186-5p | IGF1 | 0.15 | 0.43471 | -2.08 | 0.00135 | mirMAP | -0.85 | 0.00061 | NA | |
| 57 | hsa-miR-19a-3p | IGF1 | 1.27 | 0.00011 | -2.08 | 0.00135 | MirTarget | -0.77 | 0 | NA | |
| 58 | hsa-miR-19b-1-5p | IGF1 | 1.39 | 0 | -2.08 | 0.00135 | mirMAP | -0.93 | 0 | NA | |
| 59 | hsa-miR-19b-3p | IGF1 | 0.76 | 0.00653 | -2.08 | 0.00135 | MirTarget | -0.82 | 0 | NA | |
| 60 | hsa-miR-20a-3p | IGF1 | 1.14 | 0.00045 | -2.08 | 0.00135 | mirMAP | -0.99 | 0 | NA | |
| 61 | hsa-miR-27a-3p | IGF1 | 1.3 | 0 | -2.08 | 0.00135 | miRNAWalker2 validate; miRTarBase | -0.62 | 0.00158 | NA | |
| 62 | hsa-miR-301a-3p | IGF1 | 1.45 | 1.0E-5 | -2.08 | 0.00135 | MirTarget | -0.44 | 0.00199 | NA | |
| 63 | hsa-miR-301b-3p | IGF1 | 1.1 | 0.07156 | -2.08 | 0.00135 | MirTarget | -0.49 | 0 | NA | |
| 64 | hsa-miR-3065-5p | IGF1 | -0.24 | 0.63312 | -2.08 | 0.00135 | mirMAP | -0.28 | 0.00304 | NA | |
| 65 | hsa-miR-32-3p | IGF1 | 0.58 | 0.11837 | -2.08 | 0.00135 | mirMAP | -0.53 | 0.00017 | NA | |
| 66 | hsa-miR-320a | IGF1 | 0.59 | 0.0119 | -2.08 | 0.00135 | miRNATAP | -0.57 | 0.00471 | NA | |
| 67 | hsa-miR-320b | IGF1 | 1.11 | 0.0005 | -2.08 | 0.00135 | miRNATAP | -0.64 | 1.0E-5 | NA | |
| 68 | hsa-miR-320c | IGF1 | 0.46 | 0.24061 | -2.08 | 0.00135 | miRNATAP | -0.42 | 0.00294 | NA | |
| 69 | hsa-miR-33a-3p | IGF1 | 0.35 | 0.32171 | -2.08 | 0.00135 | MirTarget | -0.68 | 0 | NA | |
| 70 | hsa-miR-3662 | IGF1 | 1.83 | 0.0054 | -2.08 | 0.00135 | MirTarget | -0.45 | 0 | NA | |
| 71 | hsa-miR-421 | IGF1 | 1.81 | 0 | -2.08 | 0.00135 | PITA | -0.45 | 0.00067 | NA | |
| 72 | hsa-miR-454-3p | IGF1 | 0.63 | 0.01349 | -2.08 | 0.00135 | MirTarget | -0.55 | 0.00337 | NA | |
| 73 | hsa-miR-486-5p | IGF1 | -0.63 | 0.15527 | -2.08 | 0.00135 | PITA; miRNATAP | -0.38 | 0.00034 | NA | |
| 74 | hsa-miR-576-5p | IGF1 | 0.63 | 0.04171 | -2.08 | 0.00135 | PITA; mirMAP; miRNATAP | -0.76 | 0 | NA | |
| 75 | hsa-miR-577 | IGF1 | 0.91 | 0.22561 | -2.08 | 0.00135 | PITA | -0.36 | 0 | NA | |
| 76 | hsa-miR-590-3p | IGF1 | 1.12 | 0.00016 | -2.08 | 0.00135 | MirTarget; miRanda; mirMAP; miRNATAP | -0.61 | 8.0E-5 | NA | |
| 77 | hsa-miR-592 | IGF1 | 0.78 | 0.2045 | -2.08 | 0.00135 | mirMAP | -0.41 | 0 | NA | |
| 78 | hsa-miR-629-5p | IGF1 | 1.06 | 0.00054 | -2.08 | 0.00135 | mirMAP | -0.88 | 0 | NA | |
| 79 | hsa-miR-940 | IGF1 | 1.64 | 0.00069 | -2.08 | 0.00135 | MirTarget; PITA; miRNATAP | -0.45 | 5.0E-5 | NA | |
| 80 | hsa-miR-126-5p | MAPK1 | 0.42 | 0.07532 | -0.5 | 0.00057 | mirMAP | -0.12 | 0.00476 | NA | |
| 81 | hsa-miR-140-3p | MAPK1 | -0.3 | 0.12754 | -0.5 | 0.00057 | PITA; miRNATAP | -0.15 | 0.00713 | NA | |
| 82 | hsa-miR-140-5p | MAPK1 | 0.21 | 0.303 | -0.5 | 0.00057 | miRanda | -0.17 | 0.00108 | NA | |
| 83 | hsa-miR-142-5p | MAPK1 | 1.56 | 1.0E-5 | -0.5 | 0.00057 | mirMAP | -0.12 | 2.0E-5 | NA | |
| 84 | hsa-miR-16-5p | MAPK1 | 1.01 | 1.0E-5 | -0.5 | 0.00057 | mirMAP | -0.13 | 0.00527 | NA | |
| 85 | hsa-miR-17-3p | MAPK1 | 1.31 | 0 | -0.5 | 0.00057 | mirMAP | -0.11 | 0.00661 | NA | |
| 86 | hsa-miR-32-5p | MAPK1 | 0.42 | 0.10646 | -0.5 | 0.00057 | mirMAP | -0.13 | 0.00101 | NA | |
| 87 | hsa-miR-320a | MAPK1 | 0.59 | 0.0119 | -0.5 | 0.00057 | miRNAWalker2 validate; PITA; miRanda; miRNATAP | -0.12 | 0.00441 | NA | |
| 88 | hsa-miR-320b | MAPK1 | 1.11 | 0.0005 | -0.5 | 0.00057 | PITA; miRanda; miRNATAP | -0.12 | 0.00017 | NA | |
| 89 | hsa-miR-3614-3p | MAPK1 | 0.82 | 0.01436 | -0.5 | 0.00057 | mirMAP | -0.14 | 0 | NA | |
| 90 | hsa-miR-22-5p | PDPK1 | 0.8 | 0.0008 | -0.59 | 4.0E-5 | miRNATAP | -0.13 | 0.00306 | NA | |
| 91 | hsa-miR-24-3p | PDPK1 | 1.09 | 0 | -0.59 | 4.0E-5 | miRNAWalker2 validate | -0.14 | 0.00295 | NA | |
| 92 | hsa-miR-27a-3p | PDPK1 | 1.3 | 0 | -0.59 | 4.0E-5 | miRNATAP | -0.13 | 0.00311 | NA | |
| 93 | hsa-miR-424-5p | PDPK1 | 1.09 | 0.00042 | -0.59 | 4.0E-5 | mirMAP | -0.13 | 0.00011 | NA | |
| 94 | hsa-miR-542-3p | PDPK1 | 0.76 | 0.00211 | -0.59 | 4.0E-5 | mirMAP | -0.16 | 0.00014 | NA | |
| 95 | hsa-miR-326 | PGF | -0.43 | 0.26299 | 2.29 | 0 | mirMAP | -0.25 | 0.00397 | NA | |
| 96 | hsa-miR-335-5p | PGF | 1.6 | 6.0E-5 | 2.29 | 0 | miRNATAP | -0.28 | 0.0004 | NA | |
| 97 | hsa-miR-429 | PGF | 1.4 | 0.009 | 2.29 | 0 | miRNATAP | -0.17 | 0.00414 | NA | |
| 98 | hsa-miR-126-5p | PIK3CA | 0.42 | 0.07532 | 0.18 | 0.37992 | mirMAP | -0.23 | 0.00023 | NA | |
| 99 | hsa-miR-146a-5p | PIK3CA | 1.82 | 2.0E-5 | 0.18 | 0.37992 | mirMAP | -0.13 | 8.0E-5 | NA | |
| 100 | hsa-miR-148a-5p | PIK3CA | -0.69 | 0.0793 | 0.18 | 0.37992 | mirMAP | -0.16 | 1.0E-5 | NA | |
| 101 | hsa-miR-148b-3p | PIK3CA | 0.3 | 0.17466 | 0.18 | 0.37992 | miRNAWalker2 validate | -0.27 | 6.0E-5 | NA | |
| 102 | hsa-miR-186-5p | PIK3CA | 0.15 | 0.43471 | 0.18 | 0.37992 | mirMAP | -0.28 | 0.00025 | NA | |
| 103 | hsa-miR-29b-1-5p | PIK3CA | 0.61 | 0.11636 | 0.18 | 0.37992 | mirMAP | -0.11 | 0.00515 | NA | |
| 104 | hsa-miR-320b | PIK3CA | 1.11 | 0.0005 | 0.18 | 0.37992 | miRanda | -0.13 | 0.00356 | NA | |
| 105 | hsa-miR-335-5p | PIK3CA | 1.6 | 6.0E-5 | 0.18 | 0.37992 | miRNAWalker2 validate | -0.14 | 7.0E-5 | NA | |
| 106 | hsa-miR-338-5p | PIK3CA | -1.2 | 0.01003 | 0.18 | 0.37992 | mirMAP | -0.13 | 3.0E-5 | NA | |
| 107 | hsa-miR-501-5p | PIK3CA | 0.27 | 0.45478 | 0.18 | 0.37992 | mirMAP | -0.18 | 0 | NA | |
| 108 | hsa-miR-7-1-3p | PIK3CA | 0.71 | 0.04123 | 0.18 | 0.37992 | mirMAP | -0.18 | 1.0E-5 | NA | |
| 109 | hsa-miR-130a-3p | PIK3CB | 0.15 | 0.63374 | 0.25 | 0.18038 | miRNATAP | -0.14 | 0.001 | NA | |
| 110 | hsa-miR-1468-5p | PIK3CD | -1.88 | 3.0E-5 | 0.39 | 0.25308 | MirTarget | -0.22 | 3.0E-5 | NA | |
| 111 | hsa-miR-30b-3p | PIK3CD | -0.27 | 0.40085 | 0.39 | 0.25308 | MirTarget | -0.43 | 0 | NA | |
| 112 | hsa-miR-30b-5p | PIK3CD | -0.43 | 0.05936 | 0.39 | 0.25308 | MirTarget | -0.48 | 1.0E-5 | NA | |
| 113 | hsa-miR-30d-5p | PIK3CD | -0.55 | 0.01401 | 0.39 | 0.25308 | MirTarget; miRNATAP | -0.67 | 0 | NA | |
| 114 | hsa-miR-616-5p | PIK3CD | 0.83 | 0.03478 | 0.39 | 0.25308 | MirTarget | -0.17 | 0.00628 | NA | |
| 115 | hsa-miR-7-5p | PIK3CD | 1.64 | 0.01244 | 0.39 | 0.25308 | miRTarBase; MirTarget; miRNATAP | -0.21 | 0 | 22234835 | We first screened and identified a novel miR-7 target phosphoinositide 3-kinase catalytic subunit delta PIK3CD; A correlation between miR-7 and PIK3CD expression was also confirmed in clinical samples of HCC; By targeting PIK3CD mTOR and p70S6K miR-7 efficiently regulates the PI3K/Akt pathway |
| 116 | hsa-miR-103a-3p | PIK3R1 | 0.84 | 0 | -1.09 | 2.0E-5 | MirTarget; miRNATAP | -0.32 | 0.00394 | NA | |
| 117 | hsa-miR-106b-5p | PIK3R1 | 1.71 | 0 | -1.09 | 2.0E-5 | MirTarget; miRNATAP | -0.25 | 2.0E-5 | NA | |
| 118 | hsa-miR-107 | PIK3R1 | 0.9 | 5.0E-5 | -1.09 | 2.0E-5 | MirTarget; PITA; miRanda; miRNATAP | -0.24 | 0.00469 | NA | |
| 119 | hsa-miR-128-3p | PIK3R1 | 0.98 | 6.0E-5 | -1.09 | 2.0E-5 | MirTarget | -0.25 | 0.00086 | 25962360 | miR 128 3p suppresses hepatocellular carcinoma proliferation by regulating PIK3R1 and is correlated with the prognosis of HCC patients; Mechanistically miR-128-3p was confirmed to regulate PIK3R1 p85α expression thereby suppressing phosphatidylinositol 3-kinase PI3K/AKT pathway activation using qRT-PCR and western blot analysis; Hence we conclude that miR-128-3p which is frequently downregulated in HCC inhibits HCC progression by regulating PIK3R1 and PI3K/AKT activation and is a prognostic marker for HCC patients |
| 120 | hsa-miR-144-3p | PIK3R1 | -0.13 | 0.79825 | -1.09 | 2.0E-5 | mirMAP | -0.14 | 0.00013 | NA | |
| 121 | hsa-miR-15b-5p | PIK3R1 | 1.62 | 0 | -1.09 | 2.0E-5 | MirTarget | -0.32 | 0 | NA | |
| 122 | hsa-miR-16-2-3p | PIK3R1 | 1.8 | 0 | -1.09 | 2.0E-5 | MirTarget | -0.27 | 0 | NA | |
| 123 | hsa-miR-16-5p | PIK3R1 | 1.01 | 1.0E-5 | -1.09 | 2.0E-5 | MirTarget | -0.41 | 0 | NA | |
| 124 | hsa-miR-17-5p | PIK3R1 | 1.66 | 0 | -1.09 | 2.0E-5 | MirTarget; TargetScan; miRNATAP | -0.19 | 0.00107 | NA | |
| 125 | hsa-miR-182-5p | PIK3R1 | 0.89 | 0.03106 | -1.09 | 2.0E-5 | miRNATAP | -0.16 | 0.0004 | NA | |
| 126 | hsa-miR-185-5p | PIK3R1 | 1.14 | 0 | -1.09 | 2.0E-5 | miRNATAP | -0.42 | 0 | NA | |
| 127 | hsa-miR-188-5p | PIK3R1 | 1.35 | 0.00038 | -1.09 | 2.0E-5 | MirTarget | -0.16 | 0.00078 | NA | |
| 128 | hsa-miR-200b-3p | PIK3R1 | 0.97 | 0.0595 | -1.09 | 2.0E-5 | mirMAP | -0.1 | 0.0041 | NA | |
| 129 | hsa-miR-200c-3p | PIK3R1 | 1.28 | 0.0037 | -1.09 | 2.0E-5 | mirMAP | -0.14 | 0.00064 | NA | |
| 130 | hsa-miR-20a-5p | PIK3R1 | 1.45 | 0 | -1.09 | 2.0E-5 | MirTarget; miRNATAP | -0.18 | 0.00561 | NA | |
| 131 | hsa-miR-21-5p | PIK3R1 | 1.75 | 0 | -1.09 | 2.0E-5 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.31 | 0.00155 | 26676464 | PIK3R1 targeting by miR 21 suppresses tumor cell migration and invasion by reducing PI3K/AKT signaling and reversing EMT and predicts clinical outcome of breast cancer; Next we identified the PIK3R1 as a direct target of miR-21 and showed that it was negatively regulated by miR-21; Taken together we provide novel evidence that miR-21 knockdown suppresses cell growth migration and invasion partly by inhibiting PI3K/AKT activation via direct targeting PIK3R1 and reversing EMT in breast cancer |
| 132 | hsa-miR-212-3p | PIK3R1 | 0.96 | 0.00133 | -1.09 | 2.0E-5 | MirTarget | -0.23 | 0.00022 | NA | |
| 133 | hsa-miR-22-5p | PIK3R1 | 0.8 | 0.0008 | -1.09 | 2.0E-5 | mirMAP | -0.27 | 0.0005 | NA | |
| 134 | hsa-miR-221-3p | PIK3R1 | 1.33 | 0 | -1.09 | 2.0E-5 | MirTarget | -0.31 | 1.0E-5 | NA | |
| 135 | hsa-miR-222-3p | PIK3R1 | 1.39 | 0 | -1.09 | 2.0E-5 | MirTarget | -0.28 | 1.0E-5 | NA | |
| 136 | hsa-miR-3065-5p | PIK3R1 | -0.24 | 0.63312 | -1.09 | 2.0E-5 | MirTarget; mirMAP; miRNATAP | -0.12 | 0.00106 | NA | |
| 137 | hsa-miR-32-3p | PIK3R1 | 0.58 | 0.11837 | -1.09 | 2.0E-5 | mirMAP | -0.17 | 0.00228 | NA | |
| 138 | hsa-miR-320b | PIK3R1 | 1.11 | 0.0005 | -1.09 | 2.0E-5 | miRNATAP | -0.16 | 0.00545 | NA | |
| 139 | hsa-miR-324-3p | PIK3R1 | 1.05 | 0.00039 | -1.09 | 2.0E-5 | MirTarget; PITA; miRNATAP | -0.18 | 0.00348 | NA | |
| 140 | hsa-miR-330-3p | PIK3R1 | 0.8 | 0.00747 | -1.09 | 2.0E-5 | MirTarget; PITA; miRNATAP | -0.26 | 2.0E-5 | NA | |
| 141 | hsa-miR-335-3p | PIK3R1 | 2.52 | 0 | -1.09 | 2.0E-5 | mirMAP | -0.15 | 0.0005 | NA | |
| 142 | hsa-miR-424-5p | PIK3R1 | 1.09 | 0.00042 | -1.09 | 2.0E-5 | MirTarget | -0.29 | 0 | NA | |
| 143 | hsa-miR-429 | PIK3R1 | 1.4 | 0.009 | -1.09 | 2.0E-5 | mirMAP; miRNATAP | -0.13 | 0.00023 | NA | |
| 144 | hsa-miR-455-3p | PIK3R1 | 2.3 | 0 | -1.09 | 2.0E-5 | MirTarget; PITA; miRNATAP | -0.18 | 1.0E-5 | NA | |
| 145 | hsa-miR-542-3p | PIK3R1 | 0.76 | 0.00211 | -1.09 | 2.0E-5 | MirTarget; PITA; miRanda | -0.24 | 0.00173 | NA | |
| 146 | hsa-miR-584-5p | PIK3R1 | 1.68 | 0.00017 | -1.09 | 2.0E-5 | mirMAP | -0.13 | 0.00171 | NA | |
| 147 | hsa-miR-590-3p | PIK3R1 | 1.12 | 0.00016 | -1.09 | 2.0E-5 | miRanda; mirMAP | -0.27 | 2.0E-5 | NA | |
| 148 | hsa-miR-590-5p | PIK3R1 | 1.04 | 0.00027 | -1.09 | 2.0E-5 | MirTarget; PITA; miRanda; miRNATAP | -0.3 | 0 | NA | |
| 149 | hsa-miR-629-3p | PIK3R1 | 1.4 | 0.0001 | -1.09 | 2.0E-5 | MirTarget | -0.17 | 0.00118 | NA | |
| 150 | hsa-miR-96-5p | PIK3R1 | 1.14 | 0.00943 | -1.09 | 2.0E-5 | TargetScan; miRNATAP | -0.19 | 1.0E-5 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | PHOSPHORYLATION | 25 | 1228 | 4.062e-23 | 1.89e-19 |
| 2 | PROTEIN PHOSPHORYLATION | 22 | 944 | 4.355e-21 | 1.013e-17 |
| 3 | PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS | 26 | 1977 | 1.852e-19 | 2.873e-16 |
| 4 | INTRACELLULAR SIGNAL TRANSDUCTION | 24 | 1572 | 4.685e-19 | 5.45e-16 |
| 5 | INOSITOL LIPID MEDIATED SIGNALING | 10 | 124 | 9.343e-15 | 8.695e-12 |
| 6 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 22 | 1929 | 1.724e-14 | 1.337e-11 |
| 7 | RESPONSE TO INSULIN | 11 | 205 | 3.372e-14 | 2.241e-11 |
| 8 | PEPTIDYL SERINE MODIFICATION | 10 | 148 | 5.673e-14 | 3.299e-11 |
| 9 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 20 | 1618 | 1.282e-13 | 6.628e-11 |
| 10 | CELLULAR RESPONSE TO HORMONE STIMULUS | 14 | 552 | 1.722e-13 | 8.014e-11 |
| 11 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE SIGNALING PATHWAY | 13 | 498 | 1.085e-12 | 4.588e-10 |
| 12 | RESPONSE TO PEPTIDE | 12 | 404 | 2.153e-12 | 8.349e-10 |
| 13 | CELLULAR RESPONSE TO INSULIN STIMULUS | 9 | 146 | 2.757e-12 | 9.869e-10 |
| 14 | PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 6 | 25 | 3.161e-12 | 1.051e-09 |
| 15 | RESPONSE TO HORMONE | 15 | 893 | 6.998e-12 | 2.171e-09 |
| 16 | REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 7 | 59 | 8.483e-12 | 2.467e-09 |
| 17 | REGULATION OF GLUCOSE IMPORT | 7 | 60 | 9.592e-12 | 2.625e-09 |
| 18 | REGULATION OF CELLULAR AMIDE METABOLIC PROCESS | 11 | 354 | 1.306e-11 | 3.376e-09 |
| 19 | REGULATION OF TOR SIGNALING | 7 | 68 | 2.384e-11 | 5.839e-09 |
| 20 | CELLULAR RESPONSE TO PEPTIDE | 10 | 274 | 2.684e-11 | 6.244e-09 |
| 21 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 18 | 1656 | 3.536e-11 | 7.835e-09 |
| 22 | VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 7 | 74 | 4.393e-11 | 9.292e-09 |
| 23 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 15 | 1036 | 5.741e-11 | 1.113e-08 |
| 24 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 15 | 1036 | 5.741e-11 | 1.113e-08 |
| 25 | ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY | 13 | 689 | 6.315e-11 | 1.175e-08 |
| 26 | POSITIVE REGULATION OF IMMUNE SYSTEM PROCESS | 14 | 867 | 7.401e-11 | 1.279e-08 |
| 27 | REGULATION OF GLUCOSE IMPORT IN RESPONSE TO INSULIN STIMULUS | 5 | 17 | 7.425e-11 | 1.279e-08 |
| 28 | T CELL RECEPTOR SIGNALING PATHWAY | 8 | 146 | 1.324e-10 | 2.201e-08 |
| 29 | PHOSPHATIDYLINOSITOL 3 PHOSPHATE BIOSYNTHETIC PROCESS | 6 | 49 | 2.422e-10 | 3.887e-08 |
| 30 | REGULATION OF KINASE ACTIVITY | 13 | 776 | 2.738e-10 | 4.247e-08 |
| 31 | RESPONSE TO OXYGEN CONTAINING COMPOUND | 16 | 1381 | 2.835e-10 | 4.256e-08 |
| 32 | REGULATION OF AUTOPHAGY | 9 | 249 | 3.312e-10 | 4.816e-08 |
| 33 | REGULATION OF GLUCOSE TRANSPORT | 7 | 100 | 3.785e-10 | 5.337e-08 |
| 34 | CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND | 13 | 799 | 3.919e-10 | 5.363e-08 |
| 35 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 14 | 1008 | 5.363e-10 | 7.13e-08 |
| 36 | CELLULAR RESPONSE TO NITROGEN COMPOUND | 11 | 505 | 5.758e-10 | 7.443e-08 |
| 37 | RESPONSE TO NITROGEN COMPOUND | 13 | 859 | 9.492e-10 | 1.194e-07 |
| 38 | ANTIGEN RECEPTOR MEDIATED SIGNALING PATHWAY | 8 | 195 | 1.329e-09 | 1.627e-07 |
| 39 | PHOSPHATIDYLINOSITOL BIOSYNTHETIC PROCESS | 7 | 120 | 1.372e-09 | 1.637e-07 |
| 40 | NEGATIVE REGULATION OF TOR SIGNALING | 5 | 30 | 1.682e-09 | 1.957e-07 |
| 41 | CELLULAR RESPONSE TO STRESS | 16 | 1565 | 1.777e-09 | 2.017e-07 |
| 42 | REGULATION OF TRANSFERASE ACTIVITY | 13 | 946 | 3.059e-09 | 3.389e-07 |
| 43 | POSTTRANSCRIPTIONAL REGULATION OF GENE EXPRESSION | 10 | 448 | 3.201e-09 | 3.456e-07 |
| 44 | PROTEIN KINASE B SIGNALING | 5 | 34 | 3.268e-09 | 3.456e-07 |
| 45 | POSITIVE REGULATION OF AUTOPHAGY | 6 | 75 | 3.377e-09 | 3.492e-07 |
| 46 | REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 7 | 138 | 3.656e-09 | 3.698e-07 |
| 47 | POSITIVE REGULATION OF GLUCOSE IMPORT IN RESPONSE TO INSULIN STIMULUS | 4 | 12 | 3.85e-09 | 3.812e-07 |
| 48 | INSULIN RECEPTOR SIGNALING PATHWAY | 6 | 80 | 5.008e-09 | 4.855e-07 |
| 49 | RESPONSE TO ENDOGENOUS STIMULUS | 15 | 1450 | 6.075e-09 | 5.769e-07 |
| 50 | CELL CYCLE ARREST | 7 | 154 | 7.853e-09 | 7.24e-07 |
| 51 | RESPONSE TO ABIOTIC STIMULUS | 13 | 1024 | 7.936e-09 | 7.24e-07 |
| 52 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 15 | 1492 | 8.958e-09 | 8.016e-07 |
| 53 | REGULATION OF GENERATION OF PRECURSOR METABOLITES AND ENERGY | 6 | 89 | 9.577e-09 | 8.408e-07 |
| 54 | POSITIVE REGULATION OF GLUCOSE TRANSPORT | 5 | 42 | 9.892e-09 | 8.524e-07 |
| 55 | POSITIVE REGULATION OF CELL COMMUNICATION | 15 | 1532 | 1.282e-08 | 1.085e-06 |
| 56 | NEGATIVE REGULATION OF CELL DEATH | 12 | 872 | 1.445e-08 | 1.2e-06 |
| 57 | GLUCOSE HOMEOSTASIS | 7 | 170 | 1.559e-08 | 1.25e-06 |
| 58 | CARBOHYDRATE HOMEOSTASIS | 7 | 170 | 1.559e-08 | 1.25e-06 |
| 59 | AUTOPHAGY | 9 | 394 | 1.831e-08 | 1.403e-06 |
| 60 | POSITIVE REGULATION OF NUCLEOTIDE CATABOLIC PROCESS | 4 | 17 | 1.84e-08 | 1.403e-06 |
| 61 | LIPID PHOSPHORYLATION | 6 | 99 | 1.824e-08 | 1.403e-06 |
| 62 | CELLULAR RESPONSE TO ORGANIC SUBSTANCE | 16 | 1848 | 1.955e-08 | 1.467e-06 |
| 63 | ACTIVATION OF PROTEIN KINASE ACTIVITY | 8 | 279 | 2.203e-08 | 1.627e-06 |
| 64 | REGULATION OF LIPID METABOLIC PROCESS | 8 | 282 | 2.394e-08 | 1.74e-06 |
| 65 | REGULATION OF CATABOLIC PROCESS | 11 | 731 | 2.7e-08 | 1.908e-06 |
| 66 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 13 | 1135 | 2.706e-08 | 1.908e-06 |
| 67 | CELL ACTIVATION | 10 | 568 | 3.054e-08 | 2.121e-06 |
| 68 | POSITIVE REGULATION OF LOCOMOTION | 9 | 420 | 3.174e-08 | 2.172e-06 |
| 69 | PHOSPHATIDYLINOSITOL METABOLIC PROCESS | 7 | 193 | 3.739e-08 | 2.422e-06 |
| 70 | REGULATION OF IMMUNE SYSTEM PROCESS | 14 | 1403 | 3.747e-08 | 2.422e-06 |
| 71 | REGULATION OF CELL CYCLE | 12 | 949 | 3.687e-08 | 2.422e-06 |
| 72 | ACTIVATION OF IMMUNE RESPONSE | 9 | 427 | 3.658e-08 | 2.422e-06 |
| 73 | REGULATION OF PROTEIN MODIFICATION PROCESS | 15 | 1710 | 5.613e-08 | 3.578e-06 |
| 74 | LIPID MODIFICATION | 7 | 210 | 6.671e-08 | 4.059e-06 |
| 75 | POSITIVE REGULATION OF GENE EXPRESSION | 15 | 1733 | 6.706e-08 | 4.059e-06 |
| 76 | POSITIVE REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 4 | 23 | 6.795e-08 | 4.059e-06 |
| 77 | GLYCEROLIPID BIOSYNTHETIC PROCESS | 7 | 211 | 6.891e-08 | 4.059e-06 |
| 78 | REGULATION OF CELL DEATH | 14 | 1472 | 6.85e-08 | 4.059e-06 |
| 79 | IMMUNE RESPONSE REGULATING CELL SURFACE RECEPTOR SIGNALING PATHWAY | 8 | 323 | 6.834e-08 | 4.059e-06 |
| 80 | POSITIVE REGULATION OF NUCLEOSIDE METABOLIC PROCESS | 4 | 24 | 8.144e-08 | 4.678e-06 |
| 81 | POSITIVE REGULATION OF ATP METABOLIC PROCESS | 4 | 24 | 8.144e-08 | 4.678e-06 |
| 82 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 14 | 1518 | 1.006e-07 | 5.71e-06 |
| 83 | POSITIVE REGULATION OF KINASE ACTIVITY | 9 | 482 | 1.03e-07 | 5.751e-06 |
| 84 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 15 | 1791 | 1.038e-07 | 5.751e-06 |
| 85 | RESPONSE TO ACTIVITY | 5 | 69 | 1.263e-07 | 6.916e-06 |
| 86 | RESPONSE TO EXTERNAL STIMULUS | 15 | 1821 | 1.293e-07 | 6.997e-06 |
| 87 | PHOSPHOLIPID BIOSYNTHETIC PROCESS | 7 | 235 | 1.437e-07 | 7.685e-06 |
| 88 | PLATELET ACTIVATION | 6 | 142 | 1.578e-07 | 8.252e-06 |
| 89 | FC EPSILON RECEPTOR SIGNALING PATHWAY | 6 | 142 | 1.578e-07 | 8.252e-06 |
| 90 | POSITIVE REGULATION OF CARBOHYDRATE METABOLIC PROCESS | 5 | 75 | 1.926e-07 | 9.957e-06 |
| 91 | POSITIVE REGULATION OF CELL ADHESION | 8 | 376 | 2.188e-07 | 1.119e-05 |
| 92 | POSITIVE REGULATION OF EPITHELIAL CELL PROLIFERATION | 6 | 154 | 2.551e-07 | 1.29e-05 |
| 93 | LIPID BIOSYNTHETIC PROCESS | 9 | 539 | 2.652e-07 | 1.327e-05 |
| 94 | POSITIVE REGULATION OF CATABOLIC PROCESS | 8 | 395 | 3.183e-07 | 1.576e-05 |
| 95 | RESPONSE TO WOUNDING | 9 | 563 | 3.825e-07 | 1.854e-05 |
| 96 | POSITIVE REGULATION OF IMMUNE RESPONSE | 9 | 563 | 3.825e-07 | 1.854e-05 |
| 97 | REGULATION OF NUCLEOTIDE CATABOLIC PROCESS | 4 | 36 | 4.448e-07 | 2.133e-05 |
| 98 | REGULATION OF CELL MATRIX ADHESION | 5 | 90 | 4.813e-07 | 2.285e-05 |
| 99 | REGULATION OF CELLULAR COMPONENT MOVEMENT | 10 | 771 | 5.229e-07 | 2.458e-05 |
| 100 | REGULATION OF EPITHELIAL CELL PROLIFERATION | 7 | 285 | 5.297e-07 | 2.465e-05 |
| 101 | POSITIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 7 | 289 | 5.817e-07 | 2.68e-05 |
| 102 | FC GAMMA RECEPTOR SIGNALING PATHWAY | 5 | 95 | 6.305e-07 | 2.829e-05 |
| 103 | CARDIOVASCULAR SYSTEM DEVELOPMENT | 10 | 788 | 6.383e-07 | 2.829e-05 |
| 104 | NEGATIVE REGULATION OF CELL CYCLE | 8 | 433 | 6.377e-07 | 2.829e-05 |
| 105 | CIRCULATORY SYSTEM DEVELOPMENT | 10 | 788 | 6.383e-07 | 2.829e-05 |
| 106 | REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 4 | 40 | 6.866e-07 | 3.014e-05 |
| 107 | GLYCEROPHOSPHOLIPID METABOLIC PROCESS | 7 | 297 | 6.987e-07 | 3.038e-05 |
| 108 | RESPONSE TO EXTRACELLULAR STIMULUS | 8 | 441 | 7.321e-07 | 3.154e-05 |
| 109 | POSITIVE REGULATION OF TRANSFERASE ACTIVITY | 9 | 616 | 8.114e-07 | 3.464e-05 |
| 110 | REGULATION OF CELLULAR CARBOHYDRATE CATABOLIC PROCESS | 4 | 42 | 8.389e-07 | 3.499e-05 |
| 111 | REGULATION OF TRANSPORT | 14 | 1804 | 8.423e-07 | 3.499e-05 |
| 112 | REGULATION OF CARBOHYDRATE CATABOLIC PROCESS | 4 | 42 | 8.389e-07 | 3.499e-05 |
| 113 | MOVEMENT OF CELL OR SUBCELLULAR COMPONENT | 12 | 1275 | 9.038e-07 | 3.722e-05 |
| 114 | RESPONSE TO OXYGEN LEVELS | 7 | 311 | 9.511e-07 | 3.882e-05 |
| 115 | REGULATION OF CELL ADHESION | 9 | 629 | 9.652e-07 | 3.905e-05 |
| 116 | REGULATION OF GROWTH | 9 | 633 | 1.017e-06 | 4.081e-05 |
| 117 | CELL MOTILITY | 10 | 835 | 1.082e-06 | 4.266e-05 |
| 118 | LOCALIZATION OF CELL | 10 | 835 | 1.082e-06 | 4.266e-05 |
| 119 | PEPTIDYL AMINO ACID MODIFICATION | 10 | 841 | 1.154e-06 | 4.506e-05 |
| 120 | VASCULATURE DEVELOPMENT | 8 | 469 | 1.162e-06 | 4.506e-05 |
| 121 | WOUND HEALING | 8 | 470 | 1.181e-06 | 4.541e-05 |
| 122 | CELL CYCLE | 12 | 1316 | 1.264e-06 | 4.822e-05 |
| 123 | FC RECEPTOR SIGNALING PATHWAY | 6 | 206 | 1.405e-06 | 5.314e-05 |
| 124 | REGULATION OF LIPID KINASE ACTIVITY | 4 | 48 | 1.447e-06 | 5.432e-05 |
| 125 | REGULATION OF NUCLEOSIDE METABOLIC PROCESS | 4 | 49 | 1.574e-06 | 5.813e-05 |
| 126 | REGULATION OF ATP METABOLIC PROCESS | 4 | 49 | 1.574e-06 | 5.813e-05 |
| 127 | REGULATION OF COENZYME METABOLIC PROCESS | 4 | 50 | 1.709e-06 | 6.212e-05 |
| 128 | REGULATION OF COFACTOR METABOLIC PROCESS | 4 | 50 | 1.709e-06 | 6.212e-05 |
| 129 | REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION | 5 | 118 | 1.85e-06 | 6.621e-05 |
| 130 | LOCOMOTION | 11 | 1114 | 1.844e-06 | 6.621e-05 |
| 131 | REGULATION OF BODY FLUID LEVELS | 8 | 506 | 2.049e-06 | 7.276e-05 |
| 132 | GLYCEROLIPID METABOLIC PROCESS | 7 | 356 | 2.337e-06 | 8.216e-05 |
| 133 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 11 | 1142 | 2.349e-06 | 8.216e-05 |
| 134 | POSITIVE REGULATION OF CELLULAR PROTEIN LOCALIZATION | 7 | 360 | 2.516e-06 | 8.737e-05 |
| 135 | PHOSPHOLIPID METABOLIC PROCESS | 7 | 364 | 2.707e-06 | 9.261e-05 |
| 136 | TOR SIGNALING | 3 | 16 | 2.707e-06 | 9.261e-05 |
| 137 | REGULATION OF PHOSPHOLIPID METABOLIC PROCESS | 4 | 61 | 3.82e-06 | 0.0001297 |
| 138 | RESPONSE TO OSMOTIC STRESS | 4 | 63 | 4.349e-06 | 0.0001467 |
| 139 | CELLULAR RESPONSE TO OXYGEN LEVELS | 5 | 143 | 4.758e-06 | 0.0001593 |
| 140 | REGULATION OF CELL PROLIFERATION | 12 | 1496 | 4.838e-06 | 0.0001608 |
| 141 | LEUKOCYTE MIGRATION | 6 | 259 | 5.272e-06 | 0.000174 |
| 142 | CELL DEATH | 10 | 1001 | 5.506e-06 | 0.0001792 |
| 143 | RESPONSE TO MUSCLE ACTIVITY | 3 | 20 | 5.484e-06 | 0.0001792 |
| 144 | REGULATION OF DNA TEMPLATED TRANSCRIPTION IN RESPONSE TO STRESS | 4 | 67 | 5.569e-06 | 0.0001799 |
| 145 | POSITIVE REGULATION OF ENDOTHELIAL CELL PROLIFERATION | 4 | 68 | 5.91e-06 | 0.0001896 |
| 146 | POSITIVE REGULATION OF NEUROBLAST PROLIFERATION | 3 | 21 | 6.39e-06 | 0.0002036 |
| 147 | POSITIVE REGULATION OF CELL PROLIFERATION | 9 | 814 | 7.975e-06 | 0.0002524 |
| 148 | CELLULAR GLUCOSE HOMEOSTASIS | 4 | 75 | 8.744e-06 | 0.0002731 |
| 149 | POSITIVE REGULATION OF PEPTIDYL TYROSINE PHOSPHORYLATION | 5 | 162 | 8.742e-06 | 0.0002731 |
| 150 | REGULATION OF POSITIVE CHEMOTAXIS | 3 | 24 | 9.689e-06 | 0.0003006 |
| 151 | LYMPHOCYTE COSTIMULATION | 4 | 78 | 1.022e-05 | 0.000315 |
| 152 | ANGIOGENESIS | 6 | 293 | 1.066e-05 | 0.0003264 |
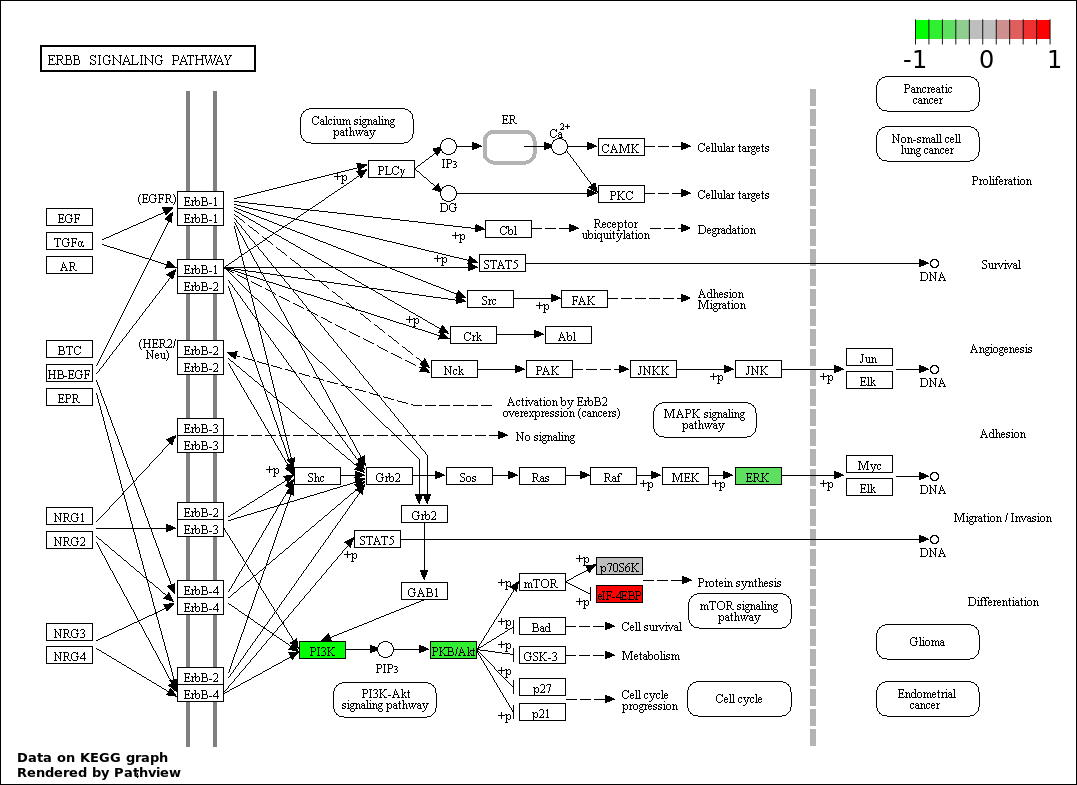
| 153 | ERBB SIGNALING PATHWAY | 4 | 79 | 1.076e-05 | 0.0003271 |
| 154 | ORGANOPHOSPHATE BIOSYNTHETIC PROCESS | 7 | 450 | 1.086e-05 | 0.0003281 |
| 155 | POSITIVE REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 6 | 296 | 1.13e-05 | 0.0003392 |
| 156 | REGULATION OF CARBOHYDRATE METABOLIC PROCESS | 5 | 172 | 1.169e-05 | 0.0003487 |
| 157 | REGULATION OF CELL SUBSTRATE ADHESION | 5 | 173 | 1.202e-05 | 0.0003563 |
| 158 | REGULATION OF IMMUNE RESPONSE | 9 | 858 | 1.218e-05 | 0.0003586 |
| 159 | REGULATION OF VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 3 | 27 | 1.395e-05 | 0.0004083 |
| 160 | CHEMICAL HOMEOSTASIS | 9 | 874 | 1.412e-05 | 0.0004105 |
| 161 | REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 7 | 470 | 1.439e-05 | 0.000416 |
| 162 | POSITIVE REGULATION OF CELL ACTIVATION | 6 | 311 | 1.496e-05 | 0.0004269 |
| 163 | HEMOSTASIS | 6 | 311 | 1.496e-05 | 0.0004269 |
| 164 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 12 | 1672 | 1.513e-05 | 0.0004293 |
| 165 | REGULATION OF NEUROBLAST PROLIFERATION | 3 | 28 | 1.561e-05 | 0.0004401 |
| 166 | IMMUNE SYSTEM PROCESS | 13 | 1984 | 1.57e-05 | 0.0004402 |
| 167 | NEGATIVE REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 4 | 88 | 1.651e-05 | 0.0004599 |
| 168 | IMMUNE EFFECTOR PROCESS | 7 | 486 | 1.786e-05 | 0.0004947 |
| 169 | POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 10 | 1152 | 1.888e-05 | 0.0005175 |
| 170 | PHAGOCYTOSIS | 5 | 190 | 1.891e-05 | 0.0005175 |
| 171 | CELL DEVELOPMENT | 11 | 1426 | 1.965e-05 | 0.0005347 |
| 172 | CELLULAR LIPID METABOLIC PROCESS | 9 | 913 | 1.998e-05 | 0.0005375 |
| 173 | PROTEIN AUTOPHOSPHORYLATION | 5 | 192 | 1.988e-05 | 0.0005375 |
| 174 | NEGATIVE REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 3 | 31 | 2.134e-05 | 0.0005706 |
| 175 | POSITIVE REGULATION OF TRANSPORT | 9 | 936 | 2.434e-05 | 0.0006471 |
| 176 | REGULATION OF ENDOTHELIAL CELL PROLIFERATION | 4 | 98 | 2.526e-05 | 0.0006669 |
| 177 | NEGATIVE REGULATION OF CELL COMMUNICATION | 10 | 1192 | 2.537e-05 | 0.0006669 |
| 178 | POSITIVE REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION | 7 | 514 | 2.56e-05 | 0.0006691 |
| 179 | REGULATION OF PROTEIN LOCALIZATION | 9 | 950 | 2.737e-05 | 0.0007114 |
| 180 | POSITIVE REGULATION OF MAP KINASE ACTIVITY | 5 | 207 | 2.853e-05 | 0.0007376 |
| 181 | REGULATION OF CELL DIFFERENTIATION | 11 | 1492 | 2.999e-05 | 0.0007709 |
| 182 | REGULATION OF NUCLEOTIDE METABOLIC PROCESS | 5 | 211 | 3.127e-05 | 0.0007995 |
| 183 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 12 | 1805 | 3.268e-05 | 0.0008273 |
| 184 | REGULATION OF PEPTIDYL TYROSINE PHOSPHORYLATION | 5 | 213 | 3.271e-05 | 0.0008273 |
| 185 | BLOOD VESSEL MORPHOGENESIS | 6 | 364 | 3.624e-05 | 0.0009114 |
| 186 | REGULATION OF CELLULAR PROTEIN LOCALIZATION | 7 | 552 | 4.034e-05 | 0.001009 |
| 187 | REGULATION OF CELLULAR LOCALIZATION | 10 | 1277 | 4.578e-05 | 0.001139 |
| 188 | REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 5 | 229 | 4.621e-05 | 0.001142 |
| 189 | POSITIVE REGULATION OF CELL MATRIX ADHESION | 3 | 40 | 4.64e-05 | 0.001142 |
| 190 | MAMMARY GLAND DEVELOPMENT | 4 | 117 | 5.063e-05 | 0.00124 |
| 191 | REGULATION OF CELL GROWTH | 6 | 391 | 5.397e-05 | 0.001308 |
| 192 | POSITIVE REGULATION OF NEURAL PRECURSOR CELL PROLIFERATION | 3 | 42 | 5.378e-05 | 0.001308 |
| 193 | POSITIVE REGULATION OF GROWTH | 5 | 238 | 5.55e-05 | 0.001338 |
| 194 | IMMUNE SYSTEM DEVELOPMENT | 7 | 582 | 5.641e-05 | 0.001353 |
| 195 | GLAND DEVELOPMENT | 6 | 395 | 5.711e-05 | 0.001363 |
| 196 | RESPONSE TO ELECTRICAL STIMULUS | 3 | 43 | 5.775e-05 | 0.001371 |
| 197 | POSITIVE REGULATION OF CELL CELL ADHESION | 5 | 243 | 6.125e-05 | 0.001447 |
| 198 | SINGLE ORGANISM BIOSYNTHETIC PROCESS | 10 | 1340 | 6.886e-05 | 0.001618 |
| 199 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 8 | 823 | 6.954e-05 | 0.001626 |
| 200 | POSITIVE REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 4 | 129 | 7.407e-05 | 0.001723 |
| 201 | CELL CYCLE PROCESS | 9 | 1081 | 7.505e-05 | 0.001737 |
| 202 | NEGATIVE REGULATION OF CELLULAR AMIDE METABOLIC PROCESS | 4 | 131 | 7.864e-05 | 0.001794 |
| 203 | NEGATIVE REGULATION OF RESPONSE TO STIMULUS | 10 | 1360 | 7.801e-05 | 0.001794 |
| 204 | NEGATIVE REGULATION OF PROTEIN METABOLIC PROCESS | 9 | 1087 | 7.832e-05 | 0.001794 |
| 205 | POSITIVE REGULATION OF RESPONSE TO EXTRACELLULAR STIMULUS | 3 | 48 | 8.045e-05 | 0.001817 |
| 206 | POSITIVE REGULATION OF RESPONSE TO NUTRIENT LEVELS | 3 | 48 | 8.045e-05 | 0.001817 |
| 207 | POSITIVE REGULATION OF CELL DIVISION | 4 | 132 | 8.099e-05 | 0.001821 |
| 208 | POSITIVE REGULATION OF PURINE NUCLEOTIDE METABOLIC PROCESS | 4 | 133 | 8.34e-05 | 0.001848 |
| 209 | POSITIVE REGULATION OF VASCULATURE DEVELOPMENT | 4 | 133 | 8.34e-05 | 0.001848 |
| 210 | POSITIVE REGULATION OF NUCLEOTIDE METABOLIC PROCESS | 4 | 133 | 8.34e-05 | 0.001848 |
| 211 | CELLULAR RESPONSE TO ABIOTIC STIMULUS | 5 | 263 | 8.899e-05 | 0.001962 |
| 212 | CELLULAR RESPONSE TO EXTERNAL STIMULUS | 5 | 264 | 9.059e-05 | 0.001988 |
| 213 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 10 | 1395 | 9.655e-05 | 0.002109 |
| 214 | NEGATIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 6 | 437 | 9.971e-05 | 0.002168 |
| 215 | REGULATION OF CELL DIVISION | 5 | 272 | 0.0001042 | 0.002256 |
| 216 | NEURON PROJECTION EXTENSION | 3 | 53 | 0.0001083 | 0.002312 |
| 217 | MAMMARY GLAND EPITHELIUM DEVELOPMENT | 3 | 53 | 0.0001083 | 0.002312 |
| 218 | POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 8 | 876 | 0.0001076 | 0.002312 |
| 219 | NEGATIVE REGULATION OF REPRODUCTIVE PROCESS | 3 | 54 | 0.0001145 | 0.002434 |
| 220 | EPIDERMAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 3 | 55 | 0.000121 | 0.002557 |
| 221 | MACROAUTOPHAGY | 5 | 281 | 0.0001214 | 0.002557 |
| 222 | REGULATION OF MAPK CASCADE | 7 | 660 | 0.000124 | 0.002587 |
| 223 | EMBRYO DEVELOPMENT | 8 | 894 | 0.0001239 | 0.002587 |
| 224 | POSITIVE REGULATION OF CELL GROWTH | 4 | 148 | 0.0001261 | 0.00262 |
| 225 | LIPID METABOLIC PROCESS | 9 | 1158 | 0.0001272 | 0.002631 |
| 226 | REGULATION OF VESICLE MEDIATED TRANSPORT | 6 | 462 | 0.0001352 | 0.002783 |
| 227 | REGULATION OF DEVELOPMENTAL GROWTH | 5 | 289 | 0.0001385 | 0.002839 |
| 228 | LEUKOCYTE DIFFERENTIATION | 5 | 292 | 0.0001453 | 0.002953 |
| 229 | REGULATION OF ORGANELLE ORGANIZATION | 9 | 1178 | 0.000145 | 0.002953 |
| 230 | REGULATION OF RESPONSE TO STRESS | 10 | 1468 | 0.0001476 | 0.002973 |
| 231 | RESPONSE TO STARVATION | 4 | 154 | 0.000147 | 0.002973 |
| 232 | POSITIVE REGULATION OF MAPK CASCADE | 6 | 470 | 0.0001484 | 0.002977 |
| 233 | POSITIVE REGULATION OF SMOOTH MUSCLE CELL PROLIFERATION | 3 | 60 | 0.0001569 | 0.003133 |
| 234 | REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 8 | 926 | 0.000158 | 0.003141 |
| 235 | REGULATION OF CELLULAR RESPONSE TO STRESS | 7 | 691 | 0.0001647 | 0.00325 |
| 236 | POSITIVE REGULATION OF STEM CELL PROLIFERATION | 3 | 61 | 0.0001648 | 0.00325 |
| 237 | REGULATION OF CELL ACTIVATION | 6 | 484 | 0.0001741 | 0.003418 |
| 238 | EPITHELIUM DEVELOPMENT | 8 | 945 | 0.0001816 | 0.003551 |
| 239 | REGULATION OF HOMOTYPIC CELL CELL ADHESION | 5 | 307 | 0.0001835 | 0.003573 |
| 240 | FATTY ACID HOMEOSTASIS | 2 | 12 | 0.0001942 | 0.003755 |
| 241 | TISSUE DEVELOPMENT | 10 | 1518 | 0.0001945 | 0.003755 |
| 242 | REGULATION OF EPITHELIAL CELL MIGRATION | 4 | 166 | 0.0001961 | 0.00377 |
| 243 | ANATOMICAL STRUCTURE FORMATION INVOLVED IN MORPHOGENESIS | 8 | 957 | 0.000198 | 0.003792 |
| 244 | RESPONSE TO CARBOHYDRATE | 4 | 168 | 0.0002053 | 0.003914 |
| 245 | VESICLE MEDIATED TRANSPORT | 9 | 1239 | 0.0002123 | 0.004032 |
| 246 | REGULATION OF RESPONSE TO EXTRACELLULAR STIMULUS | 4 | 171 | 0.0002196 | 0.004121 |
| 247 | REGULATION OF RESPONSE TO NUTRIENT LEVELS | 4 | 171 | 0.0002196 | 0.004121 |
| 248 | REGULATION OF MAP KINASE ACTIVITY | 5 | 319 | 0.0002193 | 0.004121 |
| 249 | REGULATION OF CELL SIZE | 4 | 172 | 0.0002246 | 0.004197 |
| 250 | RESPONSE TO MOLECULE OF BACTERIAL ORIGIN | 5 | 321 | 0.0002257 | 0.004201 |
| 251 | INDUCTION OF POSITIVE CHEMOTAXIS | 2 | 13 | 0.0002293 | 0.00425 |
| 252 | CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 6 | 513 | 0.0002385 | 0.004403 |
| 253 | CELLULAR RESPONSE TO ELECTRICAL STIMULUS | 2 | 14 | 0.0002672 | 0.004895 |
| 254 | INSULIN LIKE GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 2 | 14 | 0.0002672 | 0.004895 |
| 255 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 8 | 1004 | 0.0002746 | 0.005011 |
| 256 | REGULATION OF NEURAL PRECURSOR CELL PROLIFERATION | 3 | 73 | 0.0002808 | 0.005103 |
| 257 | REGULATION OF CELLULAR COMPONENT SIZE | 5 | 337 | 0.0002826 | 0.005116 |
| 258 | CELLULAR RESPONSE TO CARBOHYDRATE STIMULUS | 3 | 74 | 0.0002923 | 0.005271 |
| 259 | REGULATION OF CELLULAR COMPONENT BIOGENESIS | 7 | 767 | 0.000312 | 0.005606 |
| 260 | CELLULAR RESPONSE TO EXTRACELLULAR STIMULUS | 4 | 188 | 0.0003152 | 0.005641 |
| 261 | DEVELOPMENTAL CELL GROWTH | 3 | 77 | 0.0003286 | 0.005859 |
| 262 | DEVELOPMENTAL MATURATION | 4 | 193 | 0.0003483 | 0.006185 |
| 263 | MAMMARY GLAND EPITHELIAL CELL DIFFERENTIATION | 2 | 16 | 0.0003516 | 0.006196 |
| 264 | POSITIVE REGULATION OF VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 2 | 16 | 0.0003516 | 0.006196 |
| 265 | TUBE DEVELOPMENT | 6 | 552 | 0.0003532 | 0.006202 |
| 266 | HOMEOSTATIC PROCESS | 9 | 1337 | 0.0003746 | 0.006552 |
| 267 | REGULATION OF ENERGY HOMEOSTASIS | 2 | 17 | 0.000398 | 0.006808 |
| 268 | POSITIVE REGULATION OF GLYCOGEN METABOLIC PROCESS | 2 | 17 | 0.000398 | 0.006808 |
| 269 | MAMMARY GLAND ALVEOLUS DEVELOPMENT | 2 | 17 | 0.000398 | 0.006808 |
| 270 | MAMMARY GLAND LOBULE DEVELOPMENT | 2 | 17 | 0.000398 | 0.006808 |
| 271 | REGULATION OF TRANSLATIONAL INITIATION | 3 | 82 | 0.0003955 | 0.006808 |
| 272 | RESPONSE TO ALCOHOL | 5 | 362 | 0.0003925 | 0.006808 |
| 273 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 7 | 801 | 0.0004056 | 0.006914 |
| 274 | CIRCULATORY SYSTEM PROCESS | 5 | 366 | 0.0004127 | 0.007008 |
| 275 | CELL MORPHOGENESIS INVOLVED IN NEURON DIFFERENTIATION | 5 | 368 | 0.0004231 | 0.007158 |
| 276 | POSITIVE REGULATION OF ORGANELLE ORGANIZATION | 6 | 573 | 0.0004308 | 0.007263 |
| 277 | POSITIVE REGULATION OF INTRACELLULAR TRANSPORT | 5 | 370 | 0.0004337 | 0.007285 |
| 278 | CELLULAR RESPONSE TO PROSTAGLANDIN E STIMULUS | 2 | 18 | 0.0004473 | 0.007459 |
| 279 | RESPONSE TO CAFFEINE | 2 | 18 | 0.0004473 | 0.007459 |
| 280 | LYMPHOCYTE DIFFERENTIATION | 4 | 209 | 0.0004707 | 0.007821 |
| 281 | REGULATION OF STEM CELL PROLIFERATION | 3 | 88 | 0.0004866 | 0.008028 |
| 282 | POSITIVE REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION | 3 | 88 | 0.0004866 | 0.008028 |
| 283 | REGULATION OF CELL CELL ADHESION | 5 | 380 | 0.0004897 | 0.008052 |
| 284 | REGULATION OF TRANSLATION IN RESPONSE TO STRESS | 2 | 19 | 0.0004993 | 0.008181 |
| 285 | EPITHELIAL CELL PROLIFERATION | 3 | 89 | 0.0005029 | 0.008211 |
| 286 | REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 4 | 213 | 0.0005055 | 0.008224 |
| 287 | REGULATION OF CELL DEVELOPMENT | 7 | 836 | 0.0005244 | 0.008502 |
| 288 | NEUROGENESIS | 9 | 1402 | 0.000531 | 0.008579 |
| 289 | CELLULAR RESPONSE TO LIGHT STIMULUS | 3 | 91 | 0.0005367 | 0.008641 |
| 290 | EMBRYONIC HEMOPOIESIS | 2 | 20 | 0.0005542 | 0.008831 |
| 291 | CELLULAR RESPONSE TO GROWTH HORMONE STIMULUS | 2 | 20 | 0.0005542 | 0.008831 |
| 292 | REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 4 | 218 | 0.0005516 | 0.008831 |
| 293 | DEVELOPMENTAL PROCESS INVOLVED IN REPRODUCTION | 6 | 602 | 0.0005594 | 0.008883 |
| 294 | REGULATION OF PROTEIN STABILITY | 4 | 221 | 0.0005807 | 0.00919 |
| 295 | POSITIVE REGULATION OF ADHERENS JUNCTION ORGANIZATION | 2 | 21 | 0.0006119 | 0.009618 |
| 296 | REGULATED EXOCYTOSIS | 4 | 224 | 0.0006108 | 0.009618 |
| 297 | MORPHOGENESIS OF AN EPITHELIUM | 5 | 400 | 0.000618 | 0.009683 |
| 298 | CARDIOCYTE DIFFERENTIATION | 3 | 96 | 0.0006275 | 0.009797 |
| 299 | NEURON PROJECTION MORPHOGENESIS | 5 | 402 | 0.0006321 | 0.009837 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | KINASE ACTIVITY | 24 | 842 | 1.879e-25 | 1.745e-22 |
| 2 | TRANSFERASE ACTIVITY TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 24 | 992 | 9.302e-24 | 4.321e-21 |
| 3 | PROTEIN SERINE THREONINE KINASE ACTIVITY | 18 | 445 | 4.072e-21 | 1.261e-18 |
| 4 | PROTEIN KINASE ACTIVITY | 18 | 640 | 2.674e-18 | 6.21e-16 |
| 5 | ADENYL NUCLEOTIDE BINDING | 19 | 1514 | 5.676e-13 | 1.055e-10 |
| 6 | RIBONUCLEOTIDE BINDING | 20 | 1860 | 1.753e-12 | 2.714e-10 |
| 7 | PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 7 | 70 | 2.941e-11 | 3.903e-09 |
| 8 | X1 PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 6 | 43 | 1.064e-10 | 1.236e-08 |
| 9 | PHOSPHATIDYLINOSITOL KINASE ACTIVITY | 6 | 51 | 3.112e-10 | 3.212e-08 |
| 10 | KINASE BINDING | 11 | 606 | 3.89e-09 | 3.614e-07 |
| 11 | KINASE REGULATOR ACTIVITY | 7 | 186 | 2.9e-08 | 2.449e-06 |
| 12 | ENZYME REGULATOR ACTIVITY | 12 | 959 | 4.138e-08 | 3.204e-06 |
| 13 | ENZYME BINDING | 15 | 1737 | 6.915e-08 | 4.942e-06 |
| 14 | MOLECULAR FUNCTION REGULATOR | 13 | 1353 | 2.124e-07 | 1.41e-05 |
| 15 | PROTEIN SERINE THREONINE TYROSINE KINASE ACTIVITY | 4 | 39 | 6.187e-07 | 3.832e-05 |
| 16 | INSULIN RECEPTOR SUBSTRATE BINDING | 3 | 11 | 8.023e-07 | 4.658e-05 |
| 17 | PHOSPHATIDYLINOSITOL PHOSPHATE KINASE ACTIVITY | 3 | 16 | 2.707e-06 | 0.0001479 |
| 18 | PHOSPHATASE BINDING | 5 | 162 | 8.742e-06 | 0.0004512 |
| 19 | MAGNESIUM ION BINDING | 5 | 199 | 2.362e-05 | 0.001097 |
| 20 | INSULIN RECEPTOR BINDING | 3 | 32 | 2.352e-05 | 0.001097 |
| 21 | PROTEIN PHOSPHATASE BINDING | 4 | 120 | 5.589e-05 | 0.002472 |
| 22 | ENZYME ACTIVATOR ACTIVITY | 6 | 471 | 0.0001502 | 0.006341 |
| 23 | GROWTH FACTOR ACTIVITY | 4 | 160 | 0.0001702 | 0.006696 |
| 24 | KINASE ACTIVATOR ACTIVITY | 3 | 62 | 0.000173 | 0.006696 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | PHOSPHATIDYLINOSITOL 3 KINASE COMPLEX | 7 | 20 | 2.022e-15 | 1.181e-12 |
| 2 | TRANSFERASE COMPLEX TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 11 | 237 | 1.666e-13 | 4.864e-11 |
| 3 | EXTRINSIC COMPONENT OF MEMBRANE | 8 | 252 | 9.973e-09 | 1.941e-06 |
| 4 | TRANSFERASE COMPLEX | 11 | 703 | 1.808e-08 | 2.64e-06 |
| 5 | CATALYTIC COMPLEX | 11 | 1038 | 9.212e-07 | 0.0001076 |
| 6 | PROTEIN KINASE COMPLEX | 4 | 90 | 1.804e-05 | 0.001756 |
| 7 | PRE AUTOPHAGOSOMAL STRUCTURE | 3 | 31 | 2.134e-05 | 0.00178 |
| 8 | PLATELET ALPHA GRANULE LUMEN | 3 | 55 | 0.000121 | 0.008834 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
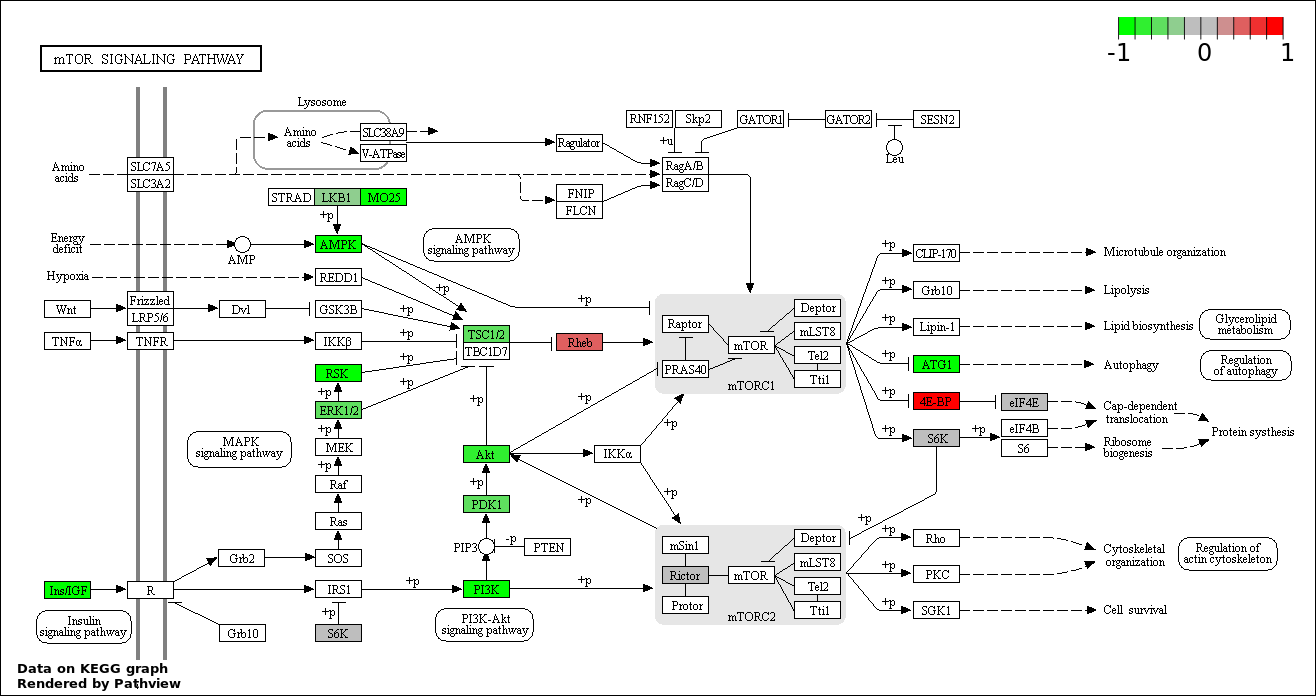
| 1 | hsa04150_mTOR_signaling_pathway | 35 | 52 | 6.799e-98 | 1.224e-95 | |
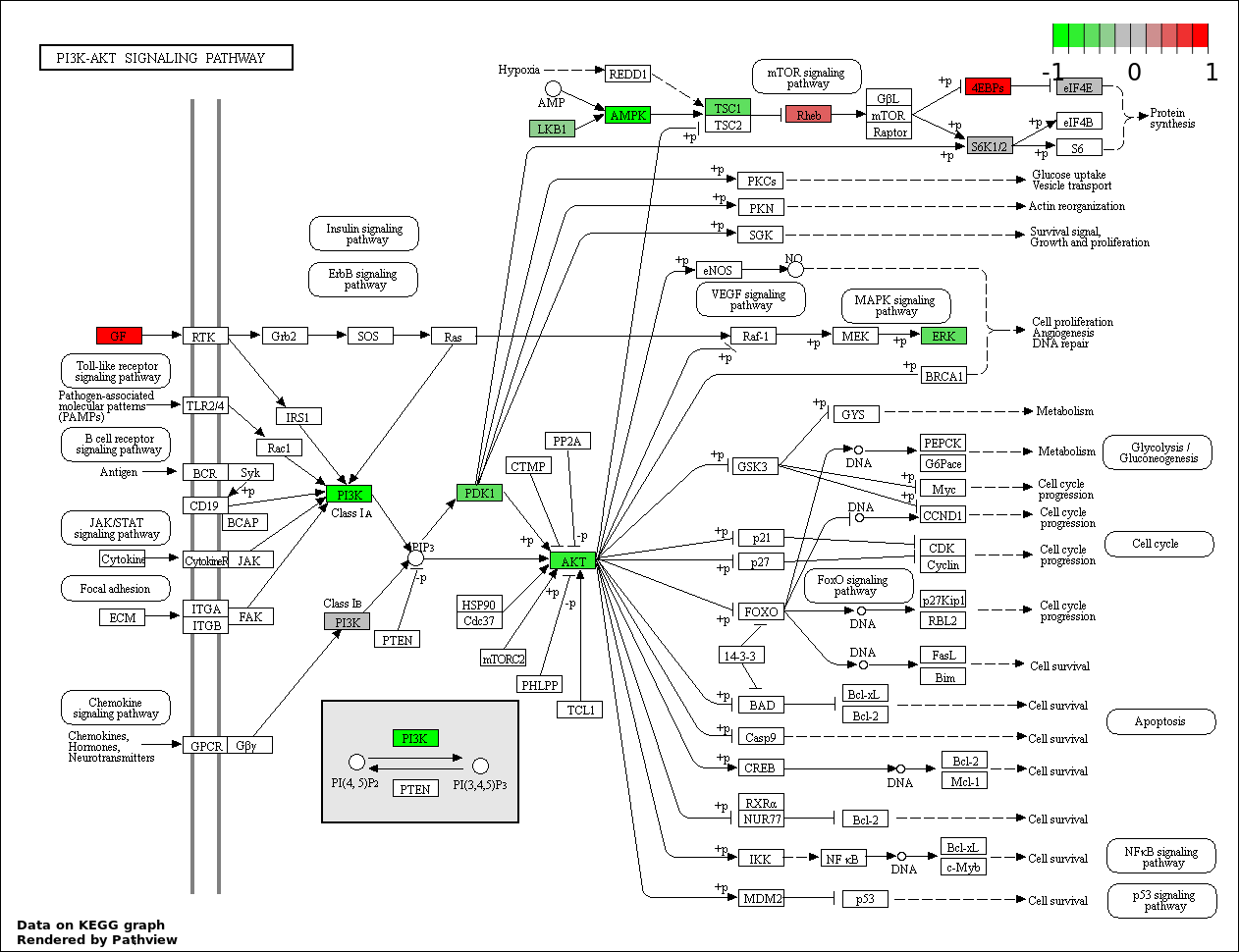
| 2 | hsa04151_PI3K_AKT_signaling_pathway | 24 | 351 | 1.159e-34 | 1.043e-32 | |
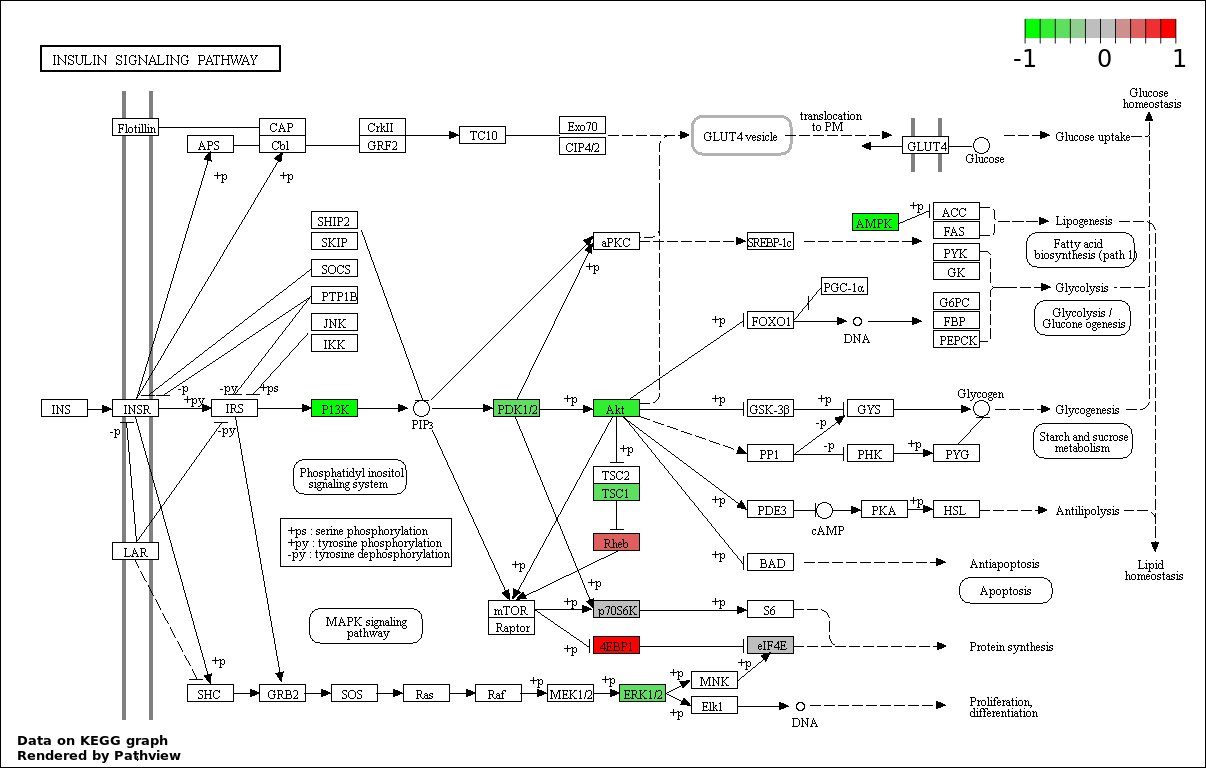
| 3 | hsa04910_Insulin_signaling_pathway | 19 | 138 | 8.851e-33 | 5.31e-31 | |
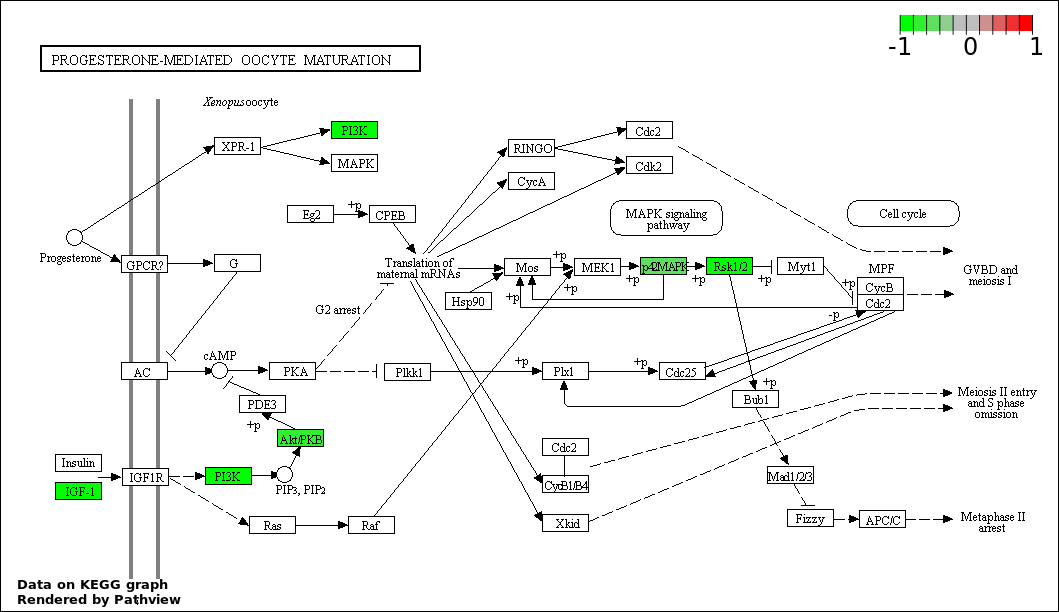
| 4 | hsa04914_Progesterone.mediated_oocyte_maturation | 15 | 87 | 3.205e-27 | 1.442e-25 | |
| 5 | hsa04012_ErbB_signaling_pathway | 13 | 87 | 1.069e-22 | 3.847e-21 | |
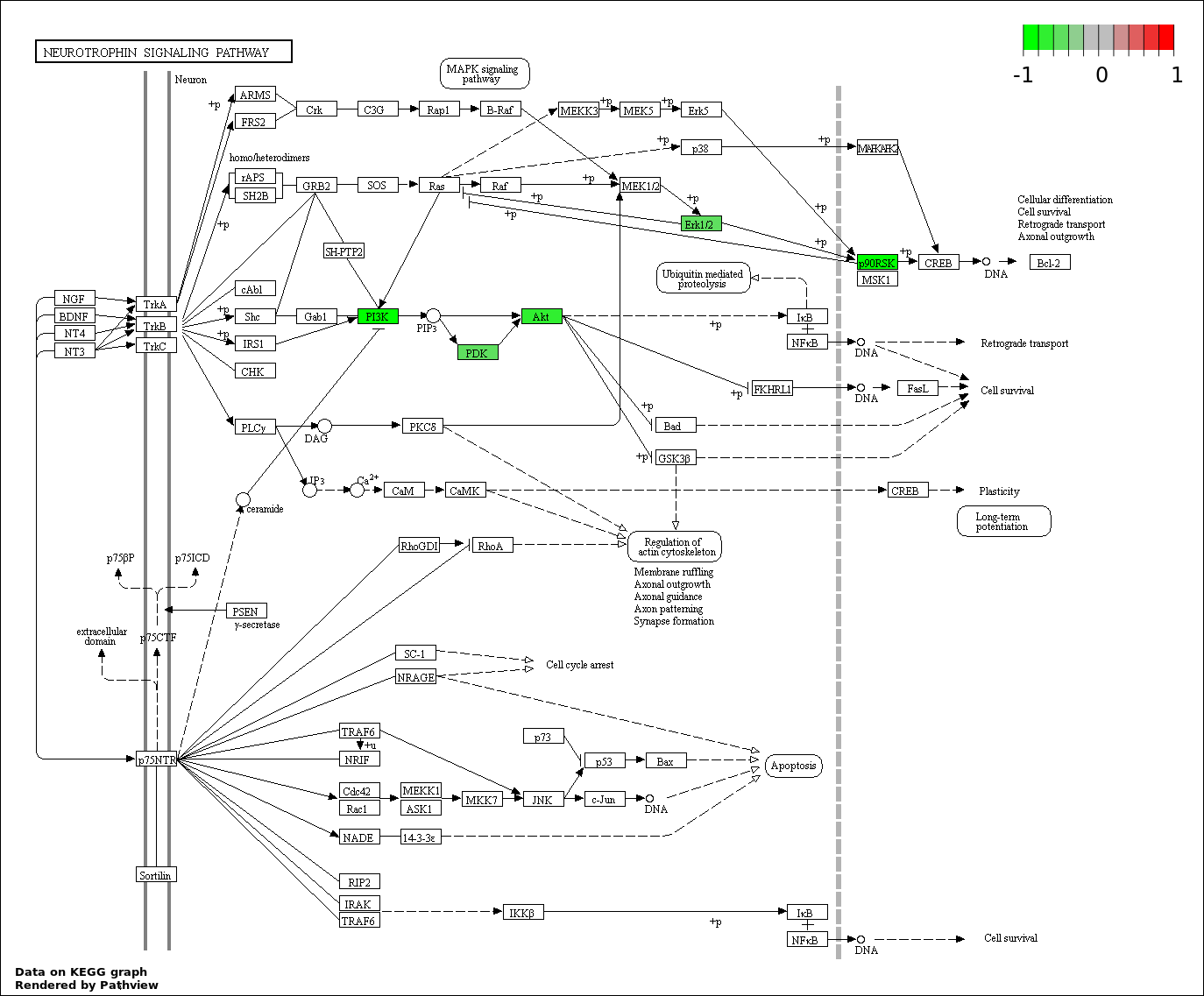
| 6 | hsa04722_Neurotrophin_signaling_pathway | 14 | 127 | 1.719e-22 | 5.156e-21 | |
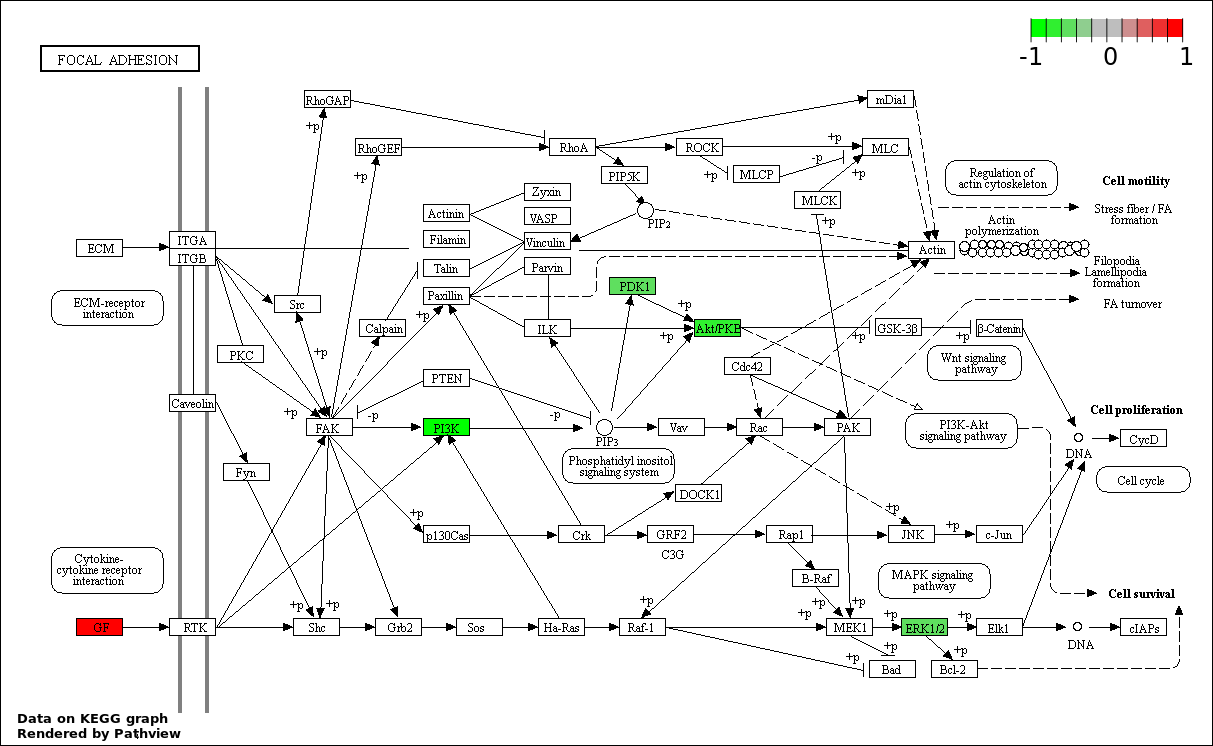
| 7 | hsa04510_Focal_adhesion | 15 | 200 | 1.601e-21 | 4.117e-20 | |
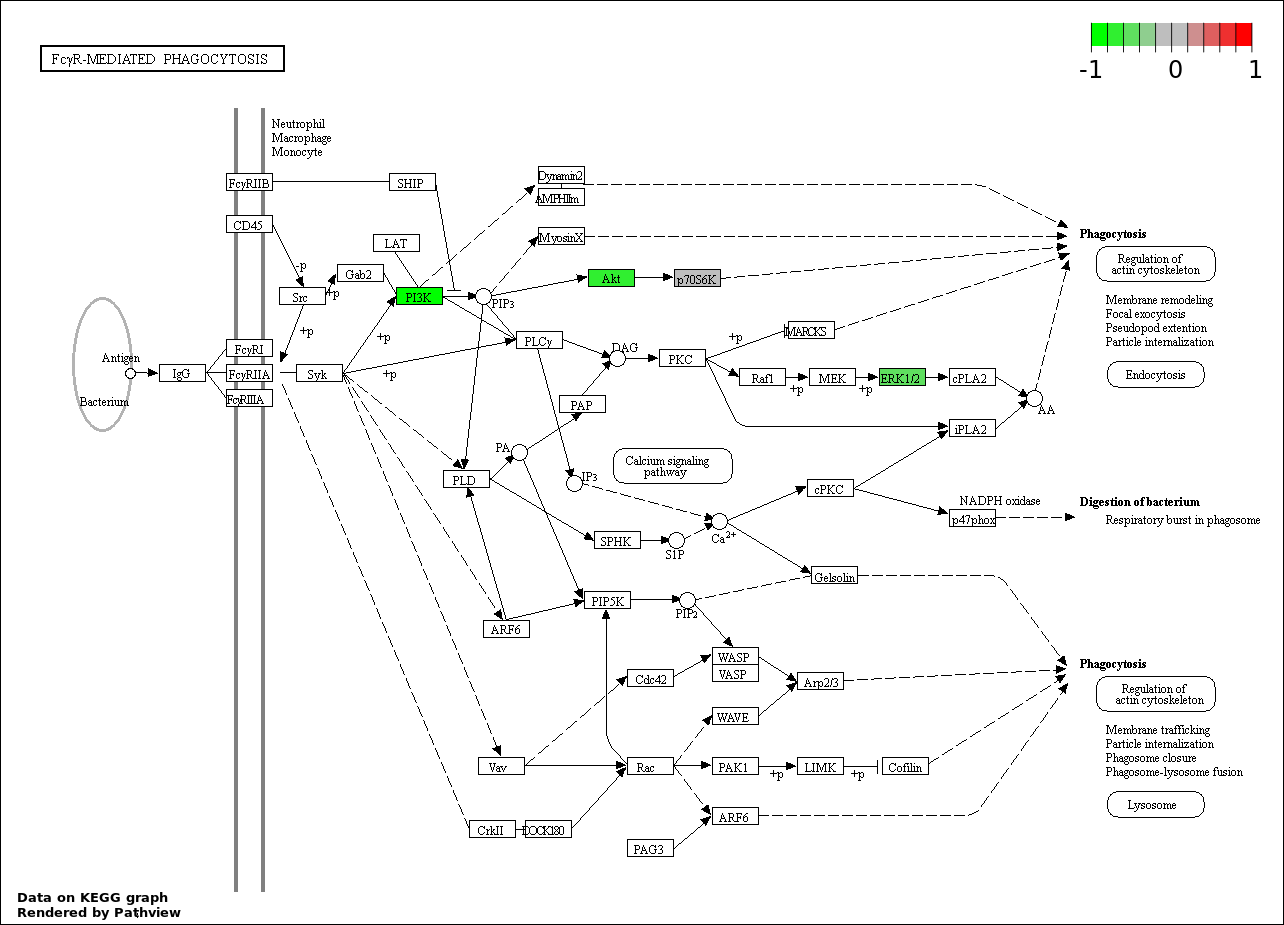
| 8 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 12 | 95 | 4.899e-20 | 1.102e-18 | |
| 9 | hsa04960_Aldosterone.regulated_sodium_reabsorption | 10 | 42 | 9.251e-20 | 1.85e-18 | |
| 10 | hsa04370_VEGF_signaling_pathway | 11 | 76 | 4.347e-19 | 7.825e-18 | |
| 11 | hsa04973_Carbohydrate_digestion_and_absorption | 9 | 44 | 3.411e-17 | 5.582e-16 | |
| 12 | hsa04662_B_cell_receptor_signaling_pathway | 10 | 75 | 5.019e-17 | 7.529e-16 | |
| 13 | hsa04014_Ras_signaling_pathway | 13 | 236 | 7.24e-17 | 1.002e-15 | |
| 14 | hsa04664_Fc_epsilon_RI_signaling_pathway | 10 | 79 | 8.684e-17 | 1.116e-15 | |
| 15 | hsa04620_Toll.like_receptor_signaling_pathway | 10 | 102 | 1.251e-15 | 1.501e-14 | |
| 16 | hsa04660_T_cell_receptor_signaling_pathway | 10 | 108 | 2.258e-15 | 2.54e-14 | |
| 17 | hsa04380_Osteoclast_differentiation | 10 | 128 | 1.293e-14 | 1.369e-13 | |
| 18 | hsa04210_Apoptosis | 9 | 89 | 2.901e-14 | 2.901e-13 | |
| 19 | hsa04062_Chemokine_signaling_pathway | 10 | 189 | 6.687e-13 | 6.335e-12 | |
| 20 | hsa04630_Jak.STAT_signaling_pathway | 9 | 155 | 4.744e-12 | 4.27e-11 | |
| 21 | hsa04070_Phosphatidylinositol_signaling_system | 7 | 78 | 6.418e-11 | 5.501e-10 | |
| 22 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 8 | 136 | 7.489e-11 | 6.127e-10 | |
| 23 | hsa04670_Leukocyte_transendothelial_migration | 7 | 117 | 1.148e-09 | 8.988e-09 | |
| 24 | hsa04810_Regulation_of_actin_cytoskeleton | 8 | 214 | 2.768e-09 | 2.076e-08 | |
| 25 | hsa04140_Regulation_of_autophagy | 5 | 34 | 3.268e-09 | 2.353e-08 | |
| 26 | hsa04114_Oocyte_meiosis | 6 | 114 | 4.26e-08 | 2.949e-07 | |
| 27 | hsa04920_Adipocytokine_signaling_pathway | 5 | 68 | 1.173e-07 | 7.823e-07 | |
| 28 | hsa04720_Long.term_potentiation | 5 | 70 | 1.359e-07 | 8.736e-07 | |
| 29 | hsa04010_MAPK_signaling_pathway | 7 | 268 | 3.5e-07 | 2.173e-06 | |
| 30 | hsa00562_Inositol_phosphate_metabolism | 3 | 57 | 0.0001346 | 0.0008078 | |
| 31 | hsa04350_TGF.beta_signaling_pathway | 3 | 85 | 0.0004395 | 0.002552 | |
| 32 | hsa04730_Long.term_depression | 2 | 70 | 0.006669 | 0.03751 | |
| 33 | hsa04530_Tight_junction | 2 | 133 | 0.02263 | 0.1234 | |
| 34 | hsa03013_RNA_transport | 2 | 152 | 0.02898 | 0.1534 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | RFPL1S |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p | 11 | AKT3 | Sponge network | -0.223 | 0.70704 | -0.749 | 0.06936 | 0.466 |
| 2 | MEG3 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3662;hsa-miR-577;hsa-miR-592;hsa-miR-629-5p | 13 | IGF1 | Sponge network | -1.645 | 0.00049 | -2.083 | 0.00135 | 0.434 |
| 3 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-32-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3662;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 24 | IGF1 | Sponge network | -2.778 | 8.0E-5 | -2.083 | 0.00135 | 0.404 |
| 4 | EMX2OS |
hsa-miR-107;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-32-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-577 | 12 | AKT3 | Sponge network | -1.088 | 0.10042 | -0.749 | 0.06936 | 0.354 |
| 5 | AGAP11 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-335-3p | 12 | PRKAA2 | Sponge network | -1.728 | 0.00016 | -3.711 | 4.0E-5 | 0.344 |
| 6 | AGAP11 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-3662;hsa-miR-421;hsa-miR-629-5p | 14 | IGF1 | Sponge network | -1.728 | 0.00016 | -2.083 | 0.00135 | 0.332 |
| 7 | EMX2OS |
hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-7-1-3p | 12 | PRKAA2 | Sponge network | -1.088 | 0.10042 | -3.711 | 4.0E-5 | 0.325 |
| 8 | MEG3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-616-5p | 11 | AKT3 | Sponge network | -1.645 | 0.00049 | -0.749 | 0.06936 | 0.312 |
| 9 | PCA3 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-32-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-96-5p | 28 | PIK3R1 | Sponge network | -2.778 | 8.0E-5 | -1.094 | 2.0E-5 | 0.309 |
| 10 | MEG3 |
hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-31-5p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-7-1-3p | 12 | PRKAA2 | Sponge network | -1.645 | 0.00049 | -3.711 | 4.0E-5 | 0.287 |