This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-139-5p | AKT3 | -1.46 | 0 | 0.76 | 5.0E-5 | PITA; miRanda | -0.2 | 7.0E-5 | NA | |
| 2 | hsa-miR-15a-5p | AKT3 | 0.78 | 0 | 0.76 | 5.0E-5 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.27 | 0.00011 | NA | |
| 3 | hsa-miR-17-5p | AKT3 | 1.42 | 0 | 0.76 | 5.0E-5 | TargetScan; miRNATAP | -0.21 | 1.0E-5 | NA | |
| 4 | hsa-miR-205-3p | AKT3 | 1.3 | 0 | 0.76 | 5.0E-5 | mirMAP | -0.25 | 0 | NA | |
| 5 | hsa-miR-20a-5p | AKT3 | 1.15 | 0 | 0.76 | 5.0E-5 | miRNATAP | -0.24 | 0 | NA | |
| 6 | hsa-miR-29b-3p | AKT3 | -0.11 | 0.51126 | 0.76 | 5.0E-5 | miRNATAP | -0.26 | 0 | 26512921 | MicroRNA 29B mir 29b regulates the Warburg effect in ovarian cancer by targeting AKT2 and AKT3 |
| 7 | hsa-miR-362-3p | AKT3 | -0.7 | 0 | 0.76 | 5.0E-5 | miRanda | -0.2 | 0.00031 | NA | |
| 8 | hsa-miR-362-5p | AKT3 | 0.41 | 0.04043 | 0.76 | 5.0E-5 | PITA; TargetScan; miRNATAP | -0.16 | 0.00012 | NA | |
| 9 | hsa-miR-141-3p | CBL | 0.73 | 0.00012 | 0.36 | 0.00252 | MirTarget; TargetScan; mirMAP | -0.12 | 2.0E-5 | NA | |
| 10 | hsa-miR-199a-5p | CBL | -0.36 | 0.04351 | 0.36 | 0.00252 | MirTarget; PITA; miRNATAP | -0.12 | 4.0E-5 | NA | |
| 11 | hsa-miR-199b-5p | CBL | -1.08 | 0 | 0.36 | 0.00252 | MirTarget; PITA | -0.13 | 1.0E-5 | NA | |
| 12 | hsa-miR-19a-3p | CBL | 1.36 | 0 | 0.36 | 0.00252 | mirMAP | -0.13 | 0 | NA | |
| 13 | hsa-miR-19b-3p | CBL | 0.54 | 0.00062 | 0.36 | 0.00252 | mirMAP | -0.19 | 0 | NA | |
| 14 | hsa-miR-200a-3p | CBL | 0.61 | 0.0029 | 0.36 | 0.00252 | MirTarget; mirMAP | -0.13 | 0 | NA | |
| 15 | hsa-miR-20a-5p | CBL | 1.15 | 0 | 0.36 | 0.00252 | mirMAP | -0.12 | 6.0E-5 | NA | |
| 16 | hsa-miR-29b-3p | CBL | -0.11 | 0.51126 | 0.36 | 0.00252 | mirMAP | -0.16 | 0 | NA | |
| 17 | hsa-miR-30d-3p | CBL | -0.58 | 0 | 0.36 | 0.00252 | MirTarget | -0.21 | 0 | NA | |
| 18 | hsa-miR-320a | CBL | 0.8 | 0 | 0.36 | 0.00252 | miRNATAP | -0.11 | 0.00396 | NA | |
| 19 | hsa-miR-338-3p | CBL | -0.95 | 0 | 0.36 | 0.00252 | miRNATAP | -0.17 | 0 | NA | |
| 20 | hsa-miR-342-3p | CBL | 1 | 0 | 0.36 | 0.00252 | mirMAP | -0.15 | 0 | NA | |
| 21 | hsa-miR-374a-3p | CBL | -0.3 | 0.00683 | 0.36 | 0.00252 | mirMAP | -0.16 | 0.00085 | NA | |
| 22 | hsa-miR-374b-5p | CBL | -0.29 | 0.00415 | 0.36 | 0.00252 | mirMAP | -0.21 | 4.0E-5 | NA | |
| 23 | hsa-miR-429 | CBL | 0.56 | 0.00564 | 0.36 | 0.00252 | MirTarget; PITA; mirMAP; miRNATAP | -0.1 | 5.0E-5 | NA | |
| 24 | hsa-miR-497-5p | CBL | -0.97 | 0 | 0.36 | 0.00252 | mirMAP | -0.12 | 0.00174 | NA | |
| 25 | hsa-miR-501-3p | CBL | 0.82 | 0 | 0.36 | 0.00252 | TargetScan | -0.1 | 0.00347 | NA | |
| 26 | hsa-miR-576-5p | CBL | 0.94 | 0 | 0.36 | 0.00252 | mirMAP | -0.1 | 0.00548 | NA | |
| 27 | hsa-miR-590-5p | CBL | 0.76 | 0 | 0.36 | 0.00252 | mirMAP | -0.11 | 0.00884 | NA | |
| 28 | hsa-miR-664a-3p | CBL | -0.26 | 0.05132 | 0.36 | 0.00252 | mirMAP | -0.23 | 0 | NA | |
| 29 | hsa-miR-126-5p | CBLB | -0.95 | 0 | 1 | 0 | mirMAP | -0.13 | 0.00505 | NA | |
| 30 | hsa-miR-139-5p | CBLB | -1.46 | 0 | 1 | 0 | miRanda | -0.14 | 5.0E-5 | NA | |
| 31 | hsa-miR-30a-5p | CBLB | -1.8 | 0 | 1 | 0 | MirTarget | -0.15 | 0 | NA | |
| 32 | hsa-miR-30b-5p | CBLB | -0.4 | 0.00347 | 1 | 0 | MirTarget; mirMAP | -0.16 | 8.0E-5 | NA | |
| 33 | hsa-miR-30c-5p | CBLB | 0.02 | 0.88637 | 1 | 0 | MirTarget; mirMAP | -0.15 | 3.0E-5 | NA | |
| 34 | hsa-miR-30e-5p | CBLB | -1.02 | 0 | 1 | 0 | MirTarget | -0.2 | 0 | NA | |
| 35 | hsa-miR-576-5p | CBLB | 0.94 | 0 | 1 | 0 | mirMAP | -0.11 | 0.00606 | NA | |
| 36 | hsa-let-7g-5p | CCND1 | -0.25 | 0.0094 | -0.01 | 0.95947 | miRNATAP | -0.42 | 1.0E-5 | NA | |
| 37 | hsa-miR-106a-5p | CCND1 | 0.68 | 0.00222 | -0.01 | 0.95947 | MirTarget; miRNATAP | -0.29 | 0 | NA | |
| 38 | hsa-miR-1266-5p | CCND1 | -0.6 | 0.02013 | -0.01 | 0.95947 | MirTarget | -0.14 | 0.00019 | NA | |
| 39 | hsa-miR-142-3p | CCND1 | 1.13 | 0 | -0.01 | 0.95947 | miRanda | -0.21 | 0 | 23619912 | Transfection of miR-142-3p mimics in colon cancer cells downregulated cyclin D1 expression induced G1 phase cell cycle arrest and elevated the sensitivity of the cells to 5-fluorouracil |
| 40 | hsa-miR-142-5p | CCND1 | 0.29 | 0.09 | -0.01 | 0.95947 | PITA | -0.19 | 0.00053 | NA | |
| 41 | hsa-miR-150-5p | CCND1 | -0.24 | 0.33155 | -0.01 | 0.95947 | mirMAP | -0.18 | 0 | NA | |
| 42 | hsa-miR-155-5p | CCND1 | 0.53 | 0.00249 | -0.01 | 0.95947 | miRNAWalker2 validate | -0.21 | 0.0001 | 26955820 | MicroRNA 155 expression inversely correlates with pathologic stage of gastric cancer and it inhibits gastric cancer cell growth by targeting cyclin D1 |
| 43 | hsa-miR-15a-5p | CCND1 | 0.78 | 0 | -0.01 | 0.95947 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.32 | 5.0E-5 | 22922827 | CCND1 has been found to be a target of miR-15a and miR-16-1 through analysis of complementary sequences between microRNAs and CCND1 mRNA; Moreover the transcription of CCND1 is suppressed by miR-15a and miR-16-1 via direct binding to the CCND1 3'-untranslated region 3'-UTR |
| 44 | hsa-miR-15b-5p | CCND1 | 0.85 | 0 | -0.01 | 0.95947 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.2 | 0.00913 | NA | |
| 45 | hsa-miR-16-1-3p | CCND1 | 0.96 | 0 | -0.01 | 0.95947 | miRNAWalker2 validate; miRTarBase | -0.3 | 0 | 22922827; 18483394 | CCND1 has been found to be a target of miR-15a and miR-16-1 through analysis of complementary sequences between microRNAs and CCND1 mRNA; Moreover the transcription of CCND1 is suppressed by miR-15a and miR-16-1 via direct binding to the CCND1 3'-untranslated region 3'-UTR;Truncation in CCND1 mRNA alters miR 16 1 regulation in mantle cell lymphoma; Furthermore we demonstrated that this truncation alters miR-16-1 binding sites and through the use of reporter constructs we were able to show that miR-16-1 regulates CCND1 mRNA expression; This study introduces the role of miR-16-1 in the regulation of CCND1 in MCL |
| 46 | hsa-miR-16-5p | CCND1 | 0.52 | 0 | -0.01 | 0.95947 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.36 | 3.0E-5 | 23991964; 22922827; 18483394 | At the molecular level our results further revealed that cyclin D1 expression was negatively regulated by miR-16;CCND1 has been found to be a target of miR-15a and miR-16-1 through analysis of complementary sequences between microRNAs and CCND1 mRNA; Moreover the transcription of CCND1 is suppressed by miR-15a and miR-16-1 via direct binding to the CCND1 3'-untranslated region 3'-UTR;Truncation in CCND1 mRNA alters miR 16 1 regulation in mantle cell lymphoma; Furthermore we demonstrated that this truncation alters miR-16-1 binding sites and through the use of reporter constructs we were able to show that miR-16-1 regulates CCND1 mRNA expression; This study introduces the role of miR-16-1 in the regulation of CCND1 in MCL |
| 47 | hsa-miR-19a-3p | CCND1 | 1.36 | 0 | -0.01 | 0.95947 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.13 | 0.00806 | 25985117 | Moreover miR-19a might play inhibitory roles in HCC malignancy via regulating Cyclin D1 expression |
| 48 | hsa-miR-19b-1-5p | CCND1 | 0.76 | 0 | -0.01 | 0.95947 | miRNAWalker2 validate; miRTarBase | -0.2 | 0.00133 | NA | |
| 49 | hsa-miR-19b-3p | CCND1 | 0.54 | 0.00062 | -0.01 | 0.95947 | miRNATAP | -0.23 | 0.0001 | NA | |
| 50 | hsa-miR-20a-5p | CCND1 | 1.15 | 0 | -0.01 | 0.95947 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.15 | 0.0057 | NA | |
| 51 | hsa-miR-20b-5p | CCND1 | 0.35 | 0.23201 | -0.01 | 0.95947 | MirTarget; miRNATAP | -0.25 | 0 | NA | |
| 52 | hsa-miR-29b-3p | CCND1 | -0.11 | 0.51126 | -0.01 | 0.95947 | mirMAP | -0.18 | 0.00109 | NA | |
| 53 | hsa-miR-29c-3p | CCND1 | -2.08 | 0 | -0.01 | 0.95947 | mirMAP | -0.16 | 0.00015 | NA | |
| 54 | hsa-miR-33a-3p | CCND1 | 0.2 | 0.21234 | -0.01 | 0.95947 | MirTarget | -0.2 | 0.00047 | NA | |
| 55 | hsa-miR-34a-5p | CCND1 | -0.12 | 0.364 | -0.01 | 0.95947 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.31 | 2.0E-5 | 25792709; 21399894 | This inhibition of proliferation was associated with a decrease in cyclin D1 levels orchestrated principally by HNF-4α a target of miR-34a considered to act as a tumour suppressor in the liver;Quantitative PCR and western analysis confirmed decreased expression of two genes BCL-2 and CCND1 in docetaxel-resistant cells which are both targeted by miR-34a |
| 56 | hsa-miR-425-5p | CCND1 | 0.93 | 0 | -0.01 | 0.95947 | miRNAWalker2 validate | -0.15 | 0.00573 | NA | |
| 57 | hsa-miR-497-5p | CCND1 | -0.97 | 0 | -0.01 | 0.95947 | MirTarget; miRNATAP | -0.27 | 5.0E-5 | 21350001 | Raf-1 and Ccnd1 were identified as novel direct targets of miR-195 and miR-497 miR-195/497 expression levels in clinical specimens were found to be correlated inversely with malignancy of breast cancer |
| 58 | hsa-miR-590-3p | CCND1 | 0.22 | 0.09659 | -0.01 | 0.95947 | mirMAP | -0.24 | 0.00077 | NA | |
| 59 | hsa-miR-7-1-3p | CCND1 | 1.14 | 0 | -0.01 | 0.95947 | mirMAP | -0.22 | 0.00036 | NA | |
| 60 | hsa-miR-9-5p | CCND1 | 1.66 | 2.0E-5 | -0.01 | 0.95947 | miRNAWalker2 validate | -0.14 | 0 | NA | |
| 61 | hsa-miR-92a-3p | CCND1 | 0.99 | 0 | -0.01 | 0.95947 | miRNAWalker2 validate | -0.24 | 0.002 | NA | |
| 62 | hsa-miR-942-5p | CCND1 | 0.68 | 0 | -0.01 | 0.95947 | MirTarget | -0.23 | 0.00111 | NA | |
| 63 | hsa-miR-224-3p | CCND2 | 0.5 | 0.00811 | 0.15 | 0.53242 | mirMAP | -0.31 | 0 | NA | |
| 64 | hsa-miR-27b-3p | CCND3 | -0.73 | 0 | -0.05 | 0.63156 | miRNAWalker2 validate | -0.13 | 0.00016 | NA | |
| 65 | hsa-miR-24-3p | CISH | 0.13 | 0.20389 | -0.86 | 0 | MirTarget | -0.15 | 0.00933 | NA | |
| 66 | hsa-miR-324-5p | CISH | 1.67 | 0 | -0.86 | 0 | miRanda | -0.2 | 0 | NA | |
| 67 | hsa-miR-361-5p | CISH | 0.08 | 0.31404 | -0.86 | 0 | miRanda | -0.27 | 0.00018 | NA | |
| 68 | hsa-miR-30a-5p | CLCF1 | -1.8 | 0 | 0.65 | 0.00137 | miRNATAP | -0.15 | 0.00281 | NA | |
| 69 | hsa-miR-30d-5p | CLCF1 | -0.38 | 0.00017 | 0.65 | 0.00137 | miRNATAP | -0.37 | 1.0E-5 | NA | |
| 70 | hsa-miR-146b-5p | CNTF | 0.92 | 0 | -0.57 | 0.00215 | MirTarget; miRanda | -0.19 | 0.0003 | NA | |
| 71 | hsa-miR-146b-5p | CNTFR | 0.92 | 0 | -3.32 | 0 | miRNATAP | -0.3 | 0.00716 | NA | |
| 72 | hsa-miR-1976 | CNTFR | 0.64 | 0 | -3.32 | 0 | miRNATAP | -0.58 | 3.0E-5 | NA | |
| 73 | hsa-miR-19a-3p | CNTFR | 1.36 | 0 | -3.32 | 0 | miRNATAP | -0.36 | 3.0E-5 | NA | |
| 74 | hsa-miR-200c-3p | CNTFR | 0.63 | 0.00013 | -3.32 | 0 | miRNATAP | -0.6 | 0 | NA | |
| 75 | hsa-miR-21-5p | CNTFR | 1.62 | 0 | -3.32 | 0 | miRNATAP | -0.97 | 0 | NA | |
| 76 | hsa-miR-24-3p | CNTFR | 0.13 | 0.20389 | -3.32 | 0 | miRNATAP | -0.45 | 0.00747 | NA | |
| 77 | hsa-miR-429 | CNTFR | 0.56 | 0.00564 | -3.32 | 0 | PITA; miRanda; miRNATAP | -0.25 | 0.00333 | NA | |
| 78 | hsa-miR-589-5p | CNTFR | 0.81 | 0 | -3.32 | 0 | miRNATAP | -0.59 | 0.00023 | NA | |
| 79 | hsa-miR-708-5p | CNTFR | 1.54 | 0 | -3.32 | 0 | miRNATAP | -0.32 | 0.00097 | NA | |
| 80 | hsa-let-7a-3p | CREBBP | 0.1 | 0.42376 | 0.02 | 0.83362 | miRNATAP | -0.14 | 9.0E-5 | NA | |
| 81 | hsa-miR-186-5p | CREBBP | 0.35 | 0.00015 | 0.02 | 0.83362 | mirMAP; miRNATAP | -0.27 | 0 | NA | |
| 82 | hsa-miR-26b-5p | CREBBP | -0.25 | 0.02852 | 0.02 | 0.83362 | miRNATAP | -0.21 | 0 | NA | |
| 83 | hsa-miR-30d-3p | CREBBP | -0.58 | 0 | 0.02 | 0.83362 | MirTarget; miRNATAP | -0.13 | 0.00047 | NA | |
| 84 | hsa-miR-590-3p | CREBBP | 0.22 | 0.09659 | 0.02 | 0.83362 | PITA; miRanda; mirMAP; miRNATAP | -0.1 | 0.00208 | NA | |
| 85 | hsa-miR-34c-5p | CSF2RA | 0.57 | 0.02241 | -0.24 | 0.17016 | miRanda | -0.11 | 0.00039 | NA | |
| 86 | hsa-miR-361-5p | CSF2RA | 0.08 | 0.31404 | -0.24 | 0.17016 | miRanda | -0.38 | 0.0001 | NA | |
| 87 | hsa-miR-429 | CSF2RA | 0.56 | 0.00564 | -0.24 | 0.17016 | miRanda | -0.18 | 0 | NA | |
| 88 | hsa-miR-19a-3p | CSF2RB | 1.36 | 0 | -0.37 | 0.08597 | MirTarget | -0.32 | 0 | NA | |
| 89 | hsa-miR-19b-3p | CSF2RB | 0.54 | 0.00062 | -0.37 | 0.08597 | MirTarget | -0.36 | 0 | NA | |
| 90 | hsa-miR-452-3p | CSF2RB | 1.1 | 0 | -0.37 | 0.08597 | mirMAP | -0.2 | 1.0E-5 | NA | |
| 91 | hsa-miR-455-5p | CSF2RB | 1.41 | 0 | -0.37 | 0.08597 | miRanda | -0.35 | 0 | NA | |
| 92 | hsa-miR-485-3p | CSF2RB | 0.36 | 0.09294 | -0.37 | 0.08597 | MirTarget | -0.12 | 0.0086 | NA | |
| 93 | hsa-miR-493-3p | CSF2RB | -0.59 | 0.00436 | -0.37 | 0.08597 | MirTarget | -0.12 | 0.00918 | NA | |
| 94 | hsa-miR-576-5p | CSF2RB | 0.94 | 0 | -0.37 | 0.08597 | miRNATAP | -0.19 | 0.00484 | NA | |
| 95 | hsa-miR-361-5p | CSF3R | 0.08 | 0.31404 | 0.72 | 0.00194 | miRanda | -0.5 | 9.0E-5 | NA | |
| 96 | hsa-miR-429 | CSF3R | 0.56 | 0.00564 | 0.72 | 0.00194 | miRanda | -0.2 | 9.0E-5 | NA | |
| 97 | hsa-miR-185-5p | CTF1 | 0.68 | 0 | -0.59 | 0.0065 | MirTarget | -0.33 | 0.0013 | NA | |
| 98 | hsa-miR-342-3p | CTF1 | 1 | 0 | -0.59 | 0.0065 | miRanda | -0.17 | 0.00265 | NA | |
| 99 | hsa-miR-140-5p | EP300 | -0.41 | 0.00014 | -0.04 | 0.77828 | miRNAWalker2 validate | -0.14 | 0.00917 | NA | |
| 100 | hsa-miR-181b-5p | EP300 | 0.97 | 0 | -0.04 | 0.77828 | miRNAWalker2 validate | -0.25 | 0 | NA | |
| 101 | hsa-miR-182-5p | EP300 | 1.15 | 0 | -0.04 | 0.77828 | miRNAWalker2 validate; miRNATAP | -0.11 | 0.00038 | NA | |
| 102 | hsa-miR-186-5p | EP300 | 0.35 | 0.00015 | -0.04 | 0.77828 | miRNATAP | -0.39 | 0 | NA | |
| 103 | hsa-miR-193b-3p | EP300 | 1.73 | 0 | -0.04 | 0.77828 | miRNAWalker2 validate | -0.14 | 0 | NA | |
| 104 | hsa-miR-20a-5p | EP300 | 1.15 | 0 | -0.04 | 0.77828 | miRNATAP | -0.26 | 0 | NA | |
| 105 | hsa-miR-2355-3p | EP300 | 1.58 | 0 | -0.04 | 0.77828 | MirTarget; miRNATAP | -0.13 | 2.0E-5 | NA | |
| 106 | hsa-miR-25-3p | EP300 | 0.77 | 0 | -0.04 | 0.77828 | miRNAWalker2 validate | -0.17 | 0.00053 | NA | |
| 107 | hsa-miR-26b-5p | EP300 | -0.25 | 0.02852 | -0.04 | 0.77828 | miRNAWalker2 validate; miRNATAP | -0.32 | 0 | NA | |
| 108 | hsa-miR-29b-1-5p | EP300 | 1.4 | 0 | -0.04 | 0.77828 | MirTarget | -0.18 | 0 | NA | |
| 109 | hsa-miR-30c-5p | EP300 | 0.02 | 0.88637 | -0.04 | 0.77828 | miRNAWalker2 validate | -0.35 | 0 | NA | |
| 110 | hsa-miR-30d-3p | EP300 | -0.58 | 0 | -0.04 | 0.77828 | MirTarget; miRNATAP | -0.18 | 0.00016 | NA | |
| 111 | hsa-miR-339-5p | EP300 | 1.08 | 0 | -0.04 | 0.77828 | miRanda | -0.25 | 0 | NA | |
| 112 | hsa-miR-342-3p | EP300 | 1 | 0 | -0.04 | 0.77828 | MirTarget; PITA; miRanda; miRNATAP | -0.28 | 0 | NA | |
| 113 | hsa-miR-361-3p | EP300 | 0.27 | 0.01479 | -0.04 | 0.77828 | PITA | -0.22 | 3.0E-5 | NA | |
| 114 | hsa-miR-374b-5p | EP300 | -0.29 | 0.00415 | -0.04 | 0.77828 | mirMAP; miRNATAP | -0.29 | 0 | NA | |
| 115 | hsa-miR-582-3p | EP300 | -0.25 | 0.1438 | -0.04 | 0.77828 | MirTarget; PITA | -0.11 | 0.00136 | NA | |
| 116 | hsa-miR-590-3p | EP300 | 0.22 | 0.09659 | -0.04 | 0.77828 | MirTarget; PITA; miRanda; mirMAP; miRNATAP | -0.12 | 0.00472 | NA | |
| 117 | hsa-miR-92a-3p | EP300 | 0.99 | 0 | -0.04 | 0.77828 | miRNAWalker2 validate | -0.19 | 5.0E-5 | NA | |
| 118 | hsa-miR-96-5p | EP300 | 1.42 | 0 | -0.04 | 0.77828 | TargetScan; miRNATAP | -0.12 | 8.0E-5 | NA | |
| 119 | hsa-miR-326 | EPO | -0.43 | 0.0415 | 0.95 | 0.00515 | miRanda | -0.38 | 0 | NA | |
| 120 | hsa-miR-142-3p | GHR | 1.13 | 0 | -1.23 | 0 | MirTarget; PITA; miRanda; miRNATAP | -0.3 | 0 | NA | |
| 121 | hsa-miR-15a-5p | GHR | 0.78 | 0 | -1.23 | 0 | MirTarget; miRNATAP | -0.63 | 0 | NA | |
| 122 | hsa-miR-15b-5p | GHR | 0.85 | 0 | -1.23 | 0 | MirTarget; miRNATAP | -0.38 | 3.0E-5 | NA | |
| 123 | hsa-miR-16-2-3p | GHR | 1.16 | 0 | -1.23 | 0 | mirMAP | -0.49 | 0 | NA | |
| 124 | hsa-miR-16-5p | GHR | 0.52 | 0 | -1.23 | 0 | MirTarget; miRNATAP | -0.71 | 0 | NA | |
| 125 | hsa-miR-29a-5p | GHR | 0.32 | 0.03704 | -1.23 | 0 | mirMAP; miRNATAP | -0.33 | 1.0E-5 | NA | |
| 126 | hsa-miR-3607-3p | GHR | 1.01 | 9.0E-5 | -1.23 | 0 | mirMAP | -0.19 | 2.0E-5 | NA | |
| 127 | hsa-miR-424-5p | GHR | 1.48 | 0 | -1.23 | 0 | MirTarget; miRNATAP | -0.28 | 3.0E-5 | NA | |
| 128 | hsa-miR-429 | GHR | 0.56 | 0.00564 | -1.23 | 0 | miRanda; miRNATAP | -0.22 | 0.00013 | NA | |
| 129 | hsa-miR-455-5p | GHR | 1.41 | 0 | -1.23 | 0 | miRanda | -0.26 | 0.00055 | NA | |
| 130 | hsa-miR-590-3p | GHR | 0.22 | 0.09659 | -1.23 | 0 | mirMAP | -0.44 | 0 | NA | |
| 131 | hsa-miR-7-1-3p | GHR | 1.14 | 0 | -1.23 | 0 | MirTarget; mirMAP | -0.41 | 0 | NA | |
| 132 | hsa-miR-141-3p | IFNAR2 | 0.73 | 0.00012 | 0.42 | 1.0E-5 | mirMAP | -0.1 | 0 | NA | |
| 133 | hsa-miR-330-3p | IFNE | -0.05 | 0.71986 | 1.72 | 0 | MirTarget | -0.49 | 2.0E-5 | NA | |
| 134 | hsa-miR-369-3p | IFNG | -1.23 | 0 | 1.84 | 0 | PITA; miRNATAP | -0.29 | 0.00026 | NA | |
| 135 | hsa-miR-409-3p | IFNG | -0.34 | 0.05388 | 1.84 | 0 | miRNAWalker2 validate | -0.53 | 0 | NA | |
| 136 | hsa-miR-429 | IFNG | 0.56 | 0.00564 | 1.84 | 0 | miRanda | -0.22 | 0.00439 | NA | |
| 137 | hsa-miR-129-5p | IFNGR1 | -0.25 | 0.43645 | 0.24 | 0.02434 | miRanda | -0.1 | 0 | NA | |
| 138 | hsa-miR-98-5p | IFNGR1 | 0.02 | 0.85827 | 0.24 | 0.02434 | miRNAWalker2 validate | -0.23 | 0 | NA | |
| 139 | hsa-miR-375 | IFNK | -4.24 | 0 | 1.36 | 0.00046 | miRanda | -0.12 | 0.00375 | NA | |
| 140 | hsa-miR-27b-3p | IL10 | -0.73 | 0 | 0.69 | 0.00084 | miRNATAP | -0.32 | 1.0E-5 | NA | |
| 141 | hsa-miR-361-5p | IL10 | 0.08 | 0.31404 | 0.69 | 0.00084 | MirTarget; PITA; miRanda; miRNATAP | -0.48 | 3.0E-5 | NA | |
| 142 | hsa-miR-590-5p | IL10 | 0.76 | 0 | 0.69 | 0.00084 | miRanda | -0.19 | 0.00473 | NA | |
| 143 | hsa-miR-769-5p | IL10 | 0.09 | 0.4454 | 0.69 | 0.00084 | miRNATAP | -0.32 | 7.0E-5 | NA | |
| 144 | hsa-miR-455-5p | IL10RA | 1.41 | 0 | 0.21 | 0.30121 | miRanda | -0.26 | 1.0E-5 | NA | |
| 145 | hsa-miR-140-5p | IL11 | -0.41 | 0.00014 | 4.18 | 0 | miRanda | -0.43 | 0.00154 | NA | |
| 146 | hsa-miR-23b-3p | IL11 | -0.5 | 3.0E-5 | 4.18 | 0 | miRNATAP | -0.82 | 0 | NA | |
| 147 | hsa-miR-125a-5p | IL12RB2 | 0.22 | 0.11955 | 1.31 | 7.0E-5 | miRanda | -0.36 | 0.00057 | NA | |
| 148 | hsa-miR-129-5p | IL12RB2 | -0.25 | 0.43645 | 1.31 | 7.0E-5 | miRanda | -0.2 | 2.0E-5 | NA | |
| 149 | hsa-miR-199a-5p | IL12RB2 | -0.36 | 0.04351 | 1.31 | 7.0E-5 | miRanda | -0.42 | 0 | NA | |
| 150 | hsa-miR-199b-5p | IL12RB2 | -1.08 | 0 | 1.31 | 7.0E-5 | miRanda | -0.45 | 0 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CYTOKINE MEDIATED SIGNALING PATHWAY | 48 | 452 | 6.49e-56 | 3.02e-52 |
| 2 | RESPONSE TO CYTOKINE | 51 | 714 | 6.689e-51 | 1.556e-47 |
| 3 | CELLULAR RESPONSE TO CYTOKINE STIMULUS | 48 | 606 | 1.211e-49 | 1.879e-46 |
| 4 | CELLULAR RESPONSE TO ORGANIC SUBSTANCE | 64 | 1848 | 5.998e-47 | 6.978e-44 |
| 5 | REGULATION OF JAK STAT CASCADE | 25 | 144 | 1.264e-33 | 9.806e-31 |
| 6 | REGULATION OF STAT CASCADE | 25 | 144 | 1.264e-33 | 9.806e-31 |
| 7 | STAT CASCADE | 17 | 50 | 1.476e-28 | 8.587e-26 |
| 8 | JAK STAT CASCADE | 17 | 50 | 1.476e-28 | 8.587e-26 |
| 9 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 46 | 1656 | 7.199e-27 | 3.722e-24 |
| 10 | REGULATION OF TYROSINE PHOSPHORYLATION OF STAT PROTEIN | 17 | 68 | 6.343e-26 | 2.952e-23 |
| 11 | POSITIVE REGULATION OF STAT CASCADE | 17 | 73 | 2.453e-25 | 9.483e-23 |
| 12 | REGULATION OF TYROSINE PHOSPHORYLATION OF STAT3 PROTEIN | 15 | 44 | 2.649e-25 | 9.483e-23 |
| 13 | POSITIVE REGULATION OF JAK STAT CASCADE | 17 | 73 | 2.453e-25 | 9.483e-23 |
| 14 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 43 | 1618 | 4.545e-24 | 1.51e-21 |
| 15 | POSITIVE REGULATION OF TYROSINE PHOSPHORYLATION OF STAT3 PROTEIN | 13 | 37 | 3.002e-22 | 9.311e-20 |
| 16 | REGULATION OF PROTEIN MODIFICATION PROCESS | 42 | 1710 | 4.166e-22 | 1.212e-19 |
| 17 | REGULATION OF PEPTIDYL TYROSINE PHOSPHORYLATION | 20 | 213 | 2.322e-21 | 6.355e-19 |
| 18 | REGULATION OF IMMUNE SYSTEM PROCESS | 38 | 1403 | 3.268e-21 | 8.448e-19 |
| 19 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 43 | 1929 | 4.526e-21 | 1.108e-18 |
| 20 | REGULATION OF IMMUNE RESPONSE | 31 | 858 | 1.288e-20 | 2.996e-18 |
| 21 | CELL ACTIVATION | 26 | 568 | 9.832e-20 | 2.179e-17 |
| 22 | IMMUNE SYSTEM PROCESS | 42 | 1984 | 1.169e-19 | 2.473e-17 |
| 23 | INTRACELLULAR SIGNAL TRANSDUCTION | 38 | 1572 | 1.67e-19 | 3.378e-17 |
| 24 | POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 30 | 876 | 2.848e-19 | 5.521e-17 |
| 25 | LYMPHOCYTE ACTIVATION | 21 | 342 | 1.491e-18 | 2.774e-16 |
| 26 | POSITIVE REGULATION OF PEPTIDYL TYROSINE PHOSPHORYLATION | 16 | 162 | 1.415e-17 | 2.532e-15 |
| 27 | REGULATION OF KINASE ACTIVITY | 27 | 776 | 1.809e-17 | 3.118e-15 |
| 28 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 35 | 1492 | 2.061e-17 | 3.425e-15 |
| 29 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 30 | 1036 | 2.952e-17 | 4.578e-15 |
| 30 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 30 | 1036 | 2.952e-17 | 4.578e-15 |
| 31 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 31 | 1135 | 3.862e-17 | 5.797e-15 |
| 32 | LEUKOCYTE ACTIVATION | 21 | 414 | 7.282e-17 | 1.059e-14 |
| 33 | REGULATION OF DEFENSE RESPONSE | 26 | 759 | 1.188e-16 | 1.675e-14 |
| 34 | REGULATION OF CELL PROLIFERATION | 34 | 1496 | 1.845e-16 | 2.525e-14 |
| 35 | LYMPHOCYTE DIFFERENTIATION | 16 | 209 | 8.467e-16 | 1.126e-13 |
| 36 | REGULATION OF TRANSFERASE ACTIVITY | 27 | 946 | 2.456e-15 | 3.175e-13 |
| 37 | REGULATION OF RESPONSE TO STRESS | 32 | 1468 | 6.379e-15 | 8.023e-13 |
| 38 | REGULATION OF CELL ACTIVATION | 20 | 484 | 2.163e-14 | 2.516e-12 |
| 39 | POSITIVE REGULATION OF CELL COMMUNICATION | 32 | 1532 | 2.103e-14 | 2.516e-12 |
| 40 | REGULATION OF HOMOTYPIC CELL CELL ADHESION | 17 | 307 | 2.152e-14 | 2.516e-12 |
| 41 | POSITIVE REGULATION OF CELL ACTIVATION | 17 | 311 | 2.661e-14 | 3.02e-12 |
| 42 | REGULATION OF RESPONSE TO INTERFERON GAMMA | 8 | 22 | 3.084e-14 | 3.417e-12 |
| 43 | REGULATION OF CELL CELL ADHESION | 18 | 380 | 5.019e-14 | 5.431e-12 |
| 44 | RESPONSE TO HORMONE | 25 | 893 | 5.227e-14 | 5.527e-12 |
| 45 | POSITIVE REGULATION OF CELL PROLIFERATION | 24 | 814 | 6.09e-14 | 6.297e-12 |
| 46 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 30 | 1395 | 8.524e-14 | 8.622e-12 |
| 47 | LEUKOCYTE DIFFERENTIATION | 16 | 292 | 1.593e-13 | 1.577e-11 |
| 48 | ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY | 22 | 689 | 1.69e-13 | 1.639e-11 |
| 49 | PHOSPHORYLATION | 28 | 1228 | 1.78e-13 | 1.69e-11 |
| 50 | POSITIVE REGULATION OF IMMUNE SYSTEM PROCESS | 24 | 867 | 2.387e-13 | 2.221e-11 |
| 51 | NEGATIVE REGULATION OF CELL DEATH | 24 | 872 | 2.702e-13 | 2.465e-11 |
| 52 | REGULATION OF CELL ADHESION | 21 | 629 | 2.81e-13 | 2.515e-11 |
| 53 | NEGATIVE REGULATION OF RESPONSE TO STIMULUS | 29 | 1360 | 3.135e-13 | 2.752e-11 |
| 54 | REGULATION OF CELL DEATH | 30 | 1472 | 3.446e-13 | 2.97e-11 |
| 55 | IMMUNE SYSTEM DEVELOPMENT | 20 | 582 | 6.682e-13 | 5.653e-11 |
| 56 | PROTEIN PHOSPHORYLATION | 24 | 944 | 1.476e-12 | 1.227e-10 |
| 57 | RESPONSE TO ENDOGENOUS STIMULUS | 29 | 1450 | 1.557e-12 | 1.258e-10 |
| 58 | REGULATION OF RESPONSE TO CYTOKINE STIMULUS | 12 | 144 | 1.569e-12 | 1.258e-10 |
| 59 | REGULATION OF LEUKOCYTE DIFFERENTIATION | 14 | 232 | 1.622e-12 | 1.279e-10 |
| 60 | RESPONSE TO NITROGEN COMPOUND | 23 | 859 | 1.659e-12 | 1.286e-10 |
| 61 | CELLULAR RESPONSE TO GROWTH HORMONE STIMULUS | 7 | 20 | 1.852e-12 | 1.413e-10 |
| 62 | POSITIVE REGULATION OF CELL CELL ADHESION | 14 | 243 | 3.042e-12 | 2.283e-10 |
| 63 | REGULATION OF CELL DIFFERENTIATION | 29 | 1492 | 3.16e-12 | 2.334e-10 |
| 64 | NEGATIVE REGULATION OF CELL COMMUNICATION | 26 | 1192 | 4.499e-12 | 3.271e-10 |
| 65 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE SIGNALING PATHWAY | 18 | 498 | 4.832e-12 | 3.459e-10 |
| 66 | LEUKOCYTE CELL CELL ADHESION | 14 | 255 | 5.834e-12 | 4.113e-10 |
| 67 | REGULATION OF HEMOPOIESIS | 15 | 314 | 7.032e-12 | 4.884e-10 |
| 68 | CELL PROLIFERATION | 20 | 672 | 9.188e-12 | 6.287e-10 |
| 69 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 25 | 1142 | 1.196e-11 | 8.068e-10 |
| 70 | POSITIVE REGULATION OF LEUKOCYTE DIFFERENTIATION | 11 | 131 | 1.297e-11 | 8.624e-10 |
| 71 | RESPONSE TO OXYGEN CONTAINING COMPOUND | 27 | 1381 | 1.957e-11 | 1.282e-09 |
| 72 | RESPONSE TO PEPTIDE | 16 | 404 | 2.198e-11 | 1.42e-09 |
| 73 | CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND | 21 | 799 | 2.621e-11 | 1.665e-09 |
| 74 | CELLULAR RESPONSE TO HORMONE STIMULUS | 18 | 552 | 2.647e-11 | 1.665e-09 |
| 75 | JAK STAT CASCADE INVOLVED IN GROWTH HORMONE SIGNALING PATHWAY | 6 | 15 | 2.958e-11 | 1.835e-09 |
| 76 | IMMUNE RESPONSE | 24 | 1100 | 3.661e-11 | 2.242e-09 |
| 77 | REGULATION OF INFLAMMATORY RESPONSE | 14 | 294 | 3.922e-11 | 2.37e-09 |
| 78 | REGULATION OF T CELL DIFFERENTIATION | 10 | 107 | 3.99e-11 | 2.38e-09 |
| 79 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 23 | 1008 | 4.233e-11 | 2.493e-09 |
| 80 | REGULATION OF INNATE IMMUNE RESPONSE | 15 | 357 | 4.33e-11 | 2.518e-09 |
| 81 | RESPONSE TO GROWTH HORMONE | 7 | 30 | 4.695e-11 | 2.697e-09 |
| 82 | NEGATIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 16 | 437 | 7.05e-11 | 4.001e-09 |
| 83 | POSITIVE REGULATION OF CYTOKINE PRODUCTION | 15 | 370 | 7.153e-11 | 4.01e-09 |
| 84 | POSITIVE REGULATION OF LYMPHOCYTE DIFFERENTIATION | 9 | 80 | 7.304e-11 | 4.046e-09 |
| 85 | POSITIVE REGULATION OF CELL ADHESION | 15 | 376 | 8.959e-11 | 4.904e-09 |
| 86 | REGULATION OF LEUKOCYTE PROLIFERATION | 12 | 206 | 1.07e-10 | 5.777e-09 |
| 87 | LEUKOCYTE MIGRATION | 13 | 259 | 1.08e-10 | 5.777e-09 |
| 88 | POSITIVE REGULATION OF HEMOPOIESIS | 11 | 163 | 1.398e-10 | 7.392e-09 |
| 89 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 28 | 1672 | 2.766e-10 | 1.446e-08 |
| 90 | HOMEOSTASIS OF NUMBER OF CELLS | 11 | 175 | 2.998e-10 | 1.55e-08 |
| 91 | REGULATION OF CYTOKINE PRODUCTION | 17 | 563 | 3.317e-10 | 1.642e-08 |
| 92 | REGULATION OF RESPONSE TO WOUNDING | 15 | 413 | 3.304e-10 | 1.642e-08 |
| 93 | RESPONSE TO ORGANIC CYCLIC COMPOUND | 21 | 917 | 3.309e-10 | 1.642e-08 |
| 94 | REGULATION OF LYMPHOCYTE DIFFERENTIATION | 10 | 132 | 3.242e-10 | 1.642e-08 |
| 95 | DEFENSE RESPONSE | 24 | 1231 | 3.665e-10 | 1.795e-08 |
| 96 | NEGATIVE REGULATION OF PHOSPHORYLATION | 15 | 422 | 4.449e-10 | 2.156e-08 |
| 97 | PEPTIDYL TYROSINE MODIFICATION | 11 | 186 | 5.748e-10 | 2.757e-08 |
| 98 | CELLULAR RESPONSE TO NITROGEN COMPOUND | 16 | 505 | 5.857e-10 | 2.781e-08 |
| 99 | NEGATIVE REGULATION OF ERBB SIGNALING PATHWAY | 7 | 44 | 8.408e-10 | 3.952e-08 |
| 100 | REGULATION OF T CELL PROLIFERATION | 10 | 147 | 9.35e-10 | 4.351e-08 |
| 101 | RESPONSE TO BIOTIC STIMULUS | 20 | 886 | 1.212e-09 | 5.586e-08 |
| 102 | RESPONSE TO LIPID | 20 | 888 | 1.261e-09 | 5.75e-08 |
| 103 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 28 | 1791 | 1.325e-09 | 5.985e-08 |
| 104 | SINGLE ORGANISM CELL ADHESION | 15 | 459 | 1.409e-09 | 6.302e-08 |
| 105 | PHOSPHATIDYLINOSITOL 3 PHOSPHATE BIOSYNTHETIC PROCESS | 7 | 49 | 1.852e-09 | 8.206e-08 |
| 106 | RESPONSE TO EXTERNAL STIMULUS | 28 | 1821 | 1.926e-09 | 8.454e-08 |
| 107 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 19 | 823 | 2.345e-09 | 1.02e-07 |
| 108 | PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS | 29 | 1977 | 2.643e-09 | 1.139e-07 |
| 109 | POSITIVE REGULATION OF IMMUNE RESPONSE | 16 | 563 | 2.798e-09 | 1.195e-07 |
| 110 | REGULATION OF ERBB SIGNALING PATHWAY | 8 | 83 | 3.061e-09 | 1.283e-07 |
| 111 | IMMUNE EFFECTOR PROCESS | 15 | 486 | 3.062e-09 | 1.283e-07 |
| 112 | REGULATION OF ADAPTIVE IMMUNE RESPONSE | 9 | 123 | 3.556e-09 | 1.464e-07 |
| 113 | T CELL DIFFERENTIATION | 9 | 123 | 3.556e-09 | 1.464e-07 |
| 114 | REGULATION OF MAPK CASCADE | 17 | 660 | 3.676e-09 | 1.5e-07 |
| 115 | NATURAL KILLER CELL ACTIVATION | 7 | 55 | 4.282e-09 | 1.733e-07 |
| 116 | POSITIVE REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION | 8 | 88 | 4.9e-09 | 1.966e-07 |
| 117 | B CELL ACTIVATION | 9 | 132 | 6.639e-09 | 2.64e-07 |
| 118 | CELL CELL ADHESION | 16 | 608 | 8.328e-09 | 3.284e-07 |
| 119 | POSITIVE REGULATION OF LEUKOCYTE PROLIFERATION | 9 | 136 | 8.634e-09 | 3.376e-07 |
| 120 | NEGATIVE REGULATION OF KINASE ACTIVITY | 11 | 250 | 1.281e-08 | 4.923e-07 |
| 121 | NEGATIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 15 | 541 | 1.291e-08 | 4.923e-07 |
| 122 | NEGATIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 15 | 541 | 1.291e-08 | 4.923e-07 |
| 123 | REGULATION OF GROWTH | 16 | 633 | 1.467e-08 | 5.548e-07 |
| 124 | REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 14 | 470 | 1.698e-08 | 6.373e-07 |
| 125 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 24 | 1518 | 2.291e-08 | 8.527e-07 |
| 126 | POSITIVE REGULATION OF KINASE ACTIVITY | 14 | 482 | 2.329e-08 | 8.601e-07 |
| 127 | CELLULAR RESPONSE TO PEPTIDE | 11 | 274 | 3.288e-08 | 1.205e-06 |
| 128 | RESPONSE TO STEROID HORMONE | 14 | 497 | 3.413e-08 | 1.241e-06 |
| 129 | NEGATIVE REGULATION OF JAK STAT CASCADE | 6 | 44 | 3.768e-08 | 1.349e-06 |
| 130 | NEGATIVE REGULATION OF STAT CASCADE | 6 | 44 | 3.768e-08 | 1.349e-06 |
| 131 | RESPONSE TO ESTROGEN | 10 | 218 | 4.143e-08 | 1.46e-06 |
| 132 | REGULATION OF IMMUNE EFFECTOR PROCESS | 13 | 424 | 4.133e-08 | 1.46e-06 |
| 133 | REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION | 8 | 118 | 5.011e-08 | 1.753e-06 |
| 134 | REGULATION OF IMMUNOGLOBULIN PRODUCTION | 6 | 47 | 5.671e-08 | 1.969e-06 |
| 135 | PHOSPHATIDYLINOSITOL BIOSYNTHETIC PROCESS | 8 | 120 | 5.715e-08 | 1.97e-06 |
| 136 | RESPONSE TO ALCOHOL | 12 | 362 | 6.157e-08 | 2.107e-06 |
| 137 | PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 5 | 25 | 7.287e-08 | 2.475e-06 |
| 138 | CELL MOTILITY | 17 | 835 | 1.145e-07 | 3.831e-06 |
| 139 | LOCALIZATION OF CELL | 17 | 835 | 1.145e-07 | 3.831e-06 |
| 140 | LEUKOCYTE PROLIFERATION | 7 | 88 | 1.192e-07 | 3.96e-06 |
| 141 | PEPTIDYL AMINO ACID MODIFICATION | 17 | 841 | 1.268e-07 | 4.183e-06 |
| 142 | B CELL DIFFERENTIATION | 7 | 89 | 1.289e-07 | 4.223e-06 |
| 143 | EPIDERMAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 6 | 55 | 1.489e-07 | 4.844e-06 |
| 144 | NEGATIVE REGULATION OF MOLECULAR FUNCTION | 19 | 1079 | 1.77e-07 | 5.721e-06 |
| 145 | POSITIVE REGULATION OF T CELL PROLIFERATION | 7 | 95 | 2.023e-07 | 6.493e-06 |
| 146 | POSITIVE REGULATION OF IMMUNOGLOBULIN PRODUCTION | 5 | 31 | 2.282e-07 | 7.224e-06 |
| 147 | REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 6 | 59 | 2.281e-07 | 7.224e-06 |
| 148 | HOMEOSTATIC PROCESS | 21 | 1337 | 2.391e-07 | 7.519e-06 |
| 149 | MYELOID LEUKOCYTE MIGRATION | 7 | 99 | 2.687e-07 | 8.336e-06 |
| 150 | LIPID PHOSPHORYLATION | 7 | 99 | 2.687e-07 | 8.336e-06 |
| 151 | NEGATIVE REGULATION OF TRANSFERASE ACTIVITY | 11 | 351 | 3.987e-07 | 1.229e-05 |
| 152 | REGULATION OF CHEMOKINE PRODUCTION | 6 | 65 | 4.095e-07 | 1.254e-05 |
| 153 | REGULATION OF LEUKOCYTE MEDIATED IMMUNITY | 8 | 156 | 4.337e-07 | 1.319e-05 |
| 154 | REGULATION OF MULTICELLULAR ORGANISM GROWTH | 6 | 66 | 4.489e-07 | 1.356e-05 |
| 155 | POSITIVE REGULATION OF TRANSFERASE ACTIVITY | 14 | 616 | 4.701e-07 | 1.402e-05 |
| 156 | NEGATIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 14 | 616 | 4.701e-07 | 1.402e-05 |
| 157 | REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 17 | 926 | 4.925e-07 | 1.46e-05 |
| 158 | RESPONSE TO BACTERIUM | 13 | 528 | 5.142e-07 | 1.514e-05 |
| 159 | POSITIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 10 | 289 | 5.68e-07 | 1.662e-05 |
| 160 | POSITIVE REGULATION OF RESPONSE TO WOUNDING | 8 | 162 | 5.781e-07 | 1.681e-05 |
| 161 | REGULATION OF TYROSINE PHOSPHORYLATION OF STAT1 PROTEIN | 4 | 16 | 6.116e-07 | 1.768e-05 |
| 162 | INTERFERON GAMMA MEDIATED SIGNALING PATHWAY | 6 | 70 | 6.387e-07 | 1.834e-05 |
| 163 | POSITIVE REGULATION OF INFLAMMATORY RESPONSE | 7 | 113 | 6.634e-07 | 1.894e-05 |
| 164 | INFLAMMATORY RESPONSE | 12 | 454 | 6.985e-07 | 1.982e-05 |
| 165 | REGULATION OF LYMPHOCYTE MEDIATED IMMUNITY | 7 | 114 | 7.043e-07 | 1.986e-05 |
| 166 | REGULATION OF TYPE I INTERFERON MEDIATED SIGNALING PATHWAY | 5 | 39 | 7.523e-07 | 2.109e-05 |
| 167 | NEGATIVE REGULATION OF CELL PROLIFERATION | 14 | 643 | 7.841e-07 | 2.185e-05 |
| 168 | LEUKOCYTE CHEMOTAXIS | 7 | 117 | 8.399e-07 | 2.326e-05 |
| 169 | NEGATIVE REGULATION OF NEURON DEATH | 8 | 171 | 8.71e-07 | 2.398e-05 |
| 170 | POSITIVE REGULATION OF MAPK CASCADE | 12 | 470 | 1.005e-06 | 2.751e-05 |
| 171 | REGULATION OF B CELL ACTIVATION | 7 | 121 | 1.054e-06 | 2.852e-05 |
| 172 | NEGATIVE REGULATION OF IMMUNE RESPONSE | 7 | 121 | 1.054e-06 | 2.852e-05 |
| 173 | POSITIVE REGULATION OF GENE EXPRESSION | 23 | 1733 | 1.108e-06 | 2.98e-05 |
| 174 | REGULATION OF EPITHELIAL CELL DIFFERENTIATION | 7 | 122 | 1.114e-06 | 2.98e-05 |
| 175 | NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 17 | 983 | 1.125e-06 | 2.992e-05 |
| 176 | ACUTE PHASE RESPONSE | 5 | 43 | 1.24e-06 | 3.269e-05 |
| 177 | INOSITOL LIPID MEDIATED SIGNALING | 7 | 124 | 1.244e-06 | 3.269e-05 |
| 178 | ERBB SIGNALING PATHWAY | 6 | 79 | 1.312e-06 | 3.429e-05 |
| 179 | REGULATION OF MAP KINASE ACTIVITY | 10 | 319 | 1.391e-06 | 3.616e-05 |
| 180 | RESPONSE TO VIRUS | 9 | 247 | 1.413e-06 | 3.652e-05 |
| 181 | RESPONSE TO MOLECULE OF BACTERIAL ORIGIN | 10 | 321 | 1.471e-06 | 3.783e-05 |
| 182 | RESPONSE TO PROTOZOAN | 4 | 20 | 1.606e-06 | 4.107e-05 |
| 183 | REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION OF STAT PROTEIN | 4 | 21 | 1.978e-06 | 5.001e-05 |
| 184 | POSITIVE REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION OF STAT PROTEIN | 4 | 21 | 1.978e-06 | 5.001e-05 |
| 185 | PHOSPHATIDYLINOSITOL METABOLIC PROCESS | 8 | 193 | 2.162e-06 | 5.389e-05 |
| 186 | REGULATION OF BODY FLUID LEVELS | 12 | 506 | 2.166e-06 | 5.389e-05 |
| 187 | POSITIVE REGULATION OF B CELL ACTIVATION | 6 | 86 | 2.165e-06 | 5.389e-05 |
| 188 | BIOLOGICAL ADHESION | 17 | 1032 | 2.184e-06 | 5.405e-05 |
| 189 | CELLULAR RESPONSE TO INTERLEUKIN 6 | 4 | 22 | 2.409e-06 | 5.931e-05 |
| 190 | REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 7 | 138 | 2.547e-06 | 6.238e-05 |
| 191 | CELL ACTIVATION INVOLVED IN IMMUNE RESPONSE | 7 | 139 | 2.673e-06 | 6.511e-05 |
| 192 | PLATELET ACTIVATION | 7 | 142 | 3.081e-06 | 7.466e-05 |
| 193 | RESPONSE TO INSULIN | 8 | 205 | 3.385e-06 | 8.161e-05 |
| 194 | REGULATION OF ISOTYPE SWITCHING | 4 | 24 | 3.476e-06 | 8.337e-05 |
| 195 | NEGATIVE REGULATION OF MAPK CASCADE | 7 | 145 | 3.54e-06 | 8.447e-05 |
| 196 | RESPONSE TO ESTRADIOL | 7 | 146 | 3.705e-06 | 8.796e-05 |
| 197 | GLYCEROLIPID BIOSYNTHETIC PROCESS | 8 | 211 | 4.191e-06 | 9.898e-05 |
| 198 | NEGATIVE REGULATION OF PROTEIN METABOLIC PROCESS | 17 | 1087 | 4.387e-06 | 0.0001031 |
| 199 | POSITIVE REGULATION OF DEFENSE RESPONSE | 10 | 364 | 4.512e-06 | 0.0001055 |
| 200 | REGULATION OF TYPE 2 IMMUNE RESPONSE | 4 | 26 | 4.858e-06 | 0.0001125 |
| 201 | RESPONSE TO INTERLEUKIN 6 | 4 | 26 | 4.858e-06 | 0.0001125 |
| 202 | ADAPTIVE IMMUNE RESPONSE | 9 | 288 | 4.966e-06 | 0.0001144 |
| 203 | NEGATIVE REGULATION OF INFLAMMATORY RESPONSE | 6 | 100 | 5.226e-06 | 0.0001192 |
| 204 | REGULATION OF SMOOTH MUSCLE CELL PROLIFERATION | 6 | 100 | 5.226e-06 | 0.0001192 |
| 205 | NEGATIVE REGULATION OF IMMUNE SYSTEM PROCESS | 10 | 372 | 5.465e-06 | 0.000124 |
| 206 | REGULATION OF PRODUCTION OF MOLECULAR MEDIATOR OF IMMUNE RESPONSE | 6 | 101 | 5.537e-06 | 0.0001251 |
| 207 | REGULATION OF DNA RECOMBINATION | 5 | 58 | 5.606e-06 | 0.000126 |
| 208 | POSITIVE REGULATION OF CD4 POSITIVE ALPHA BETA T CELL ACTIVATION | 4 | 27 | 5.684e-06 | 0.0001271 |
| 209 | POSITIVE REGULATION OF IMMUNE EFFECTOR PROCESS | 7 | 156 | 5.74e-06 | 0.0001278 |
| 210 | NEGATIVE REGULATION OF IMMUNE EFFECTOR PROCESS | 6 | 102 | 5.862e-06 | 0.0001299 |
| 211 | CELLULAR RESPONSE TO ORGANIC CYCLIC COMPOUND | 11 | 465 | 6.093e-06 | 0.0001337 |
| 212 | LOCOMOTION | 17 | 1114 | 6.078e-06 | 0.0001337 |
| 213 | POSITIVE REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 9 | 296 | 6.197e-06 | 0.0001354 |
| 214 | HORMONE MEDIATED SIGNALING PATHWAY | 7 | 158 | 6.242e-06 | 0.0001357 |
| 215 | NEGATIVE REGULATION OF CELL ADHESION | 8 | 223 | 6.296e-06 | 0.0001363 |
| 216 | T CELL ACTIVATION INVOLVED IN IMMUNE RESPONSE | 5 | 60 | 6.636e-06 | 0.0001416 |
| 217 | POSITIVE REGULATION OF ACUTE INFLAMMATORY RESPONSE | 4 | 28 | 6.609e-06 | 0.0001416 |
| 218 | LEUKOCYTE HOMEOSTASIS | 5 | 60 | 6.636e-06 | 0.0001416 |
| 219 | CELL CHEMOTAXIS | 7 | 162 | 7.356e-06 | 0.0001556 |
| 220 | FAT CELL DIFFERENTIATION | 6 | 106 | 7.323e-06 | 0.0001556 |
| 221 | DEFENSE RESPONSE TO VIRUS | 7 | 164 | 7.972e-06 | 0.0001679 |
| 222 | REGULATION OF CELL KILLING | 5 | 63 | 8.453e-06 | 0.0001772 |
| 223 | MOVEMENT OF CELL OR SUBCELLULAR COMPONENT | 18 | 1275 | 9.069e-06 | 0.0001892 |
| 224 | POSITIVE REGULATION OF PRODUCTION OF MOLECULAR MEDIATOR OF IMMUNE RESPONSE | 5 | 64 | 9.138e-06 | 0.0001898 |
| 225 | PHOSPHOLIPID BIOSYNTHETIC PROCESS | 8 | 235 | 9.235e-06 | 0.000191 |
| 226 | TYROSINE PHOSPHORYLATION OF STAT PROTEIN | 3 | 10 | 9.66e-06 | 0.0001989 |
| 227 | REGULATION OF REACTIVE OXYGEN SPECIES BIOSYNTHETIC PROCESS | 5 | 65 | 9.865e-06 | 0.0002022 |
| 228 | GLUCOSE HOMEOSTASIS | 7 | 170 | 1.008e-05 | 0.0002049 |
| 229 | CARBOHYDRATE HOMEOSTASIS | 7 | 170 | 1.008e-05 | 0.0002049 |
| 230 | RESPONSE TO TYPE I INTERFERON | 5 | 68 | 1.232e-05 | 0.0002493 |
| 231 | GROWTH | 10 | 410 | 1.277e-05 | 0.0002571 |
| 232 | MAMMARY GLAND DEVELOPMENT | 6 | 117 | 1.292e-05 | 0.0002591 |
| 233 | POSITIVE REGULATION OF TYROSINE PHOSPHORYLATION OF STAT1 PROTEIN | 3 | 11 | 1.324e-05 | 0.0002599 |
| 234 | OXALOACETATE METABOLIC PROCESS | 3 | 11 | 1.324e-05 | 0.0002599 |
| 235 | CREATINE METABOLIC PROCESS | 3 | 11 | 1.324e-05 | 0.0002599 |
| 236 | REGULATION OF T HELPER 2 CELL DIFFERENTIATION | 3 | 11 | 1.324e-05 | 0.0002599 |
| 237 | DEFENSE RESPONSE TO OTHER ORGANISM | 11 | 505 | 1.322e-05 | 0.0002599 |
| 238 | NEGATIVE REGULATION OF CATALYTIC ACTIVITY | 14 | 829 | 1.471e-05 | 0.0002876 |
| 239 | REGULATION OF NEURON DEATH | 8 | 252 | 1.532e-05 | 0.0002984 |
| 240 | RESPONSE TO KETONE | 7 | 182 | 1.571e-05 | 0.0003045 |
| 241 | REGULATION OF MYELOID CELL DIFFERENTIATION | 7 | 183 | 1.628e-05 | 0.0003142 |
| 242 | CELLULAR RESPONSE TO INTERFERON GAMMA | 6 | 122 | 1.64e-05 | 0.0003154 |
| 243 | NATURAL KILLER CELL DIFFERENTIATION | 3 | 12 | 1.76e-05 | 0.0003315 |
| 244 | POSITIVE REGULATION OF ADAPTIVE IMMUNE RESPONSE | 5 | 73 | 1.745e-05 | 0.0003315 |
| 245 | ACUTE INFLAMMATORY RESPONSE | 5 | 73 | 1.745e-05 | 0.0003315 |
| 246 | POSITIVE REGULATION OF ASTROCYTE DIFFERENTIATION | 3 | 12 | 1.76e-05 | 0.0003315 |
| 247 | POSITIVE REGULATION OF GLUCOSE IMPORT IN RESPONSE TO INSULIN STIMULUS | 3 | 12 | 1.76e-05 | 0.0003315 |
| 248 | POSITIVE REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 4 | 36 | 1.851e-05 | 0.0003473 |
| 249 | REGULATION OF ACUTE INFLAMMATORY RESPONSE | 5 | 74 | 1.865e-05 | 0.0003485 |
| 250 | REGULATION OF DNA METABOLIC PROCESS | 9 | 340 | 1.873e-05 | 0.0003487 |
| 251 | POSITIVE REGULATION OF ALPHA BETA T CELL DIFFERENTIATION | 4 | 37 | 2.068e-05 | 0.0003834 |
| 252 | POSITIVE REGULATION OF PROTEIN IMPORT INTO NUCLEUS TRANSLOCATION | 3 | 13 | 2.28e-05 | 0.0004194 |
| 253 | ENERGY HOMEOSTASIS | 3 | 13 | 2.28e-05 | 0.0004194 |
| 254 | B CELL PROLIFERATION | 4 | 38 | 2.304e-05 | 0.0004204 |
| 255 | REGULATION OF CD4 POSITIVE ALPHA BETA T CELL ACTIVATION | 4 | 38 | 2.304e-05 | 0.0004204 |
| 256 | ASTROCYTE DIFFERENTIATION | 4 | 39 | 2.559e-05 | 0.000465 |
| 257 | INSULIN RECEPTOR SIGNALING PATHWAY | 5 | 80 | 2.726e-05 | 0.0004936 |
| 258 | REGULATION OF B CELL MEDIATED IMMUNITY | 4 | 41 | 3.129e-05 | 0.0005621 |
| 259 | LACTATION | 4 | 41 | 3.129e-05 | 0.0005621 |
| 260 | ORGAN REGENERATION | 5 | 83 | 3.259e-05 | 0.0005832 |
| 261 | NEGATIVE REGULATION OF CELL CELL ADHESION | 6 | 138 | 3.297e-05 | 0.0005877 |
| 262 | NEGATIVE REGULATION OF PEPTIDYL THREONINE PHOSPHORYLATION | 3 | 15 | 3.605e-05 | 0.0006377 |
| 263 | T CELL LINEAGE COMMITMENT | 3 | 15 | 3.605e-05 | 0.0006377 |
| 264 | POSITIVE REGULATION OF SECRETION | 9 | 370 | 3.634e-05 | 0.0006405 |
| 265 | POSITIVE REGULATION OF LEUKOCYTE MEDIATED IMMUNITY | 5 | 85 | 3.656e-05 | 0.000642 |
| 266 | MYELOID LEUKOCYTE MEDIATED IMMUNITY | 4 | 43 | 3.788e-05 | 0.0006626 |
| 267 | LIPID MODIFICATION | 7 | 210 | 3.935e-05 | 0.0006858 |
| 268 | REGULATION OF DEVELOPMENTAL GROWTH | 8 | 289 | 4.078e-05 | 0.0007081 |
| 269 | NEGATIVE REGULATION OF DEFENSE RESPONSE | 6 | 144 | 4.187e-05 | 0.0007215 |
| 270 | RESPONSE TO INTERFERON GAMMA | 6 | 144 | 4.187e-05 | 0.0007215 |
| 271 | NEGATIVE REGULATION OF CHEMOKINE PRODUCTION | 3 | 16 | 4.422e-05 | 0.0007565 |
| 272 | POSITIVE REGULATION OF TYROSINE PHOSPHORYLATION OF STAT5 PROTEIN | 3 | 16 | 4.422e-05 | 0.0007565 |
| 273 | RESPONSE TO GROWTH FACTOR | 10 | 475 | 4.476e-05 | 0.0007629 |
| 274 | REGULATION OF ALPHA BETA T CELL DIFFERENTIATION | 4 | 46 | 4.958e-05 | 0.0008389 |
| 275 | GLYCEROPHOSPHOLIPID METABOLIC PROCESS | 8 | 297 | 4.945e-05 | 0.0008389 |
| 276 | REGULATION OF GLUCOSE IMPORT IN RESPONSE TO INSULIN STIMULUS | 3 | 17 | 5.353e-05 | 0.0009024 |
| 277 | RESPONSE TO ANTIBIOTIC | 4 | 47 | 5.401e-05 | 0.0009073 |
| 278 | REGULATION OF HYDROLASE ACTIVITY | 17 | 1327 | 5.755e-05 | 0.0009633 |
| 279 | GLAND DEVELOPMENT | 9 | 395 | 6.025e-05 | 0.001005 |
| 280 | POSITIVE REGULATION OF CHEMOKINE PRODUCTION | 4 | 49 | 6.373e-05 | 0.001057 |
| 281 | POSITIVE REGULATION OF T HELPER CELL DIFFERENTIATION | 3 | 18 | 6.403e-05 | 0.001057 |
| 282 | POSITIVE REGULATION OF DNA RECOMBINATION | 3 | 18 | 6.403e-05 | 0.001057 |
| 283 | NEGATIVE REGULATION OF RESPONSE TO WOUNDING | 6 | 156 | 6.542e-05 | 0.001076 |
| 284 | HEMOSTASIS | 8 | 311 | 6.83e-05 | 0.001119 |
| 285 | SIGNAL TRANSDUCTION BY PROTEIN PHOSPHORYLATION | 9 | 404 | 7.161e-05 | 0.001169 |
| 286 | LYMPHOCYTE ACTIVATION INVOLVED IN IMMUNE RESPONSE | 5 | 98 | 7.238e-05 | 0.001178 |
| 287 | NEGATIVE REGULATION OF CELL DIFFERENTIATION | 11 | 609 | 7.29e-05 | 0.001182 |
| 288 | POSITIVE REGULATION OF ALPHA BETA T CELL ACTIVATION | 4 | 51 | 7.467e-05 | 0.001206 |
| 289 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 20 | 1784 | 7.541e-05 | 0.001214 |
| 290 | REGENERATION | 6 | 161 | 7.792e-05 | 0.00125 |
| 291 | NEGATIVE REGULATION OF ERK1 AND ERK2 CASCADE | 4 | 52 | 8.062e-05 | 0.001289 |
| 292 | INNATE IMMUNE RESPONSE | 11 | 619 | 8.427e-05 | 0.001343 |
| 293 | MAMMARY GLAND EPITHELIUM DEVELOPMENT | 4 | 53 | 8.691e-05 | 0.001371 |
| 294 | REGULATION OF NITRIC OXIDE BIOSYNTHETIC PROCESS | 4 | 53 | 8.691e-05 | 0.001371 |
| 295 | REGULATION OF ERK1 AND ERK2 CASCADE | 7 | 238 | 8.662e-05 | 0.001371 |
| 296 | MEGAKARYOCYTE DIFFERENTIATION | 3 | 20 | 8.889e-05 | 0.001374 |
| 297 | REGULATION OF TYROSINE PHOSPHORYLATION OF STAT5 PROTEIN | 3 | 20 | 8.889e-05 | 0.001374 |
| 298 | IMMUNE RESPONSE REGULATING CELL SURFACE RECEPTOR SIGNALING PATHWAY | 8 | 323 | 8.889e-05 | 0.001374 |
| 299 | X2 OXOGLUTARATE METABOLIC PROCESS | 3 | 20 | 8.889e-05 | 0.001374 |
| 300 | NEGATIVE REGULATION OF HOMOTYPIC CELL CELL ADHESION | 5 | 102 | 8.756e-05 | 0.001374 |
| 301 | RESPONSE TO LEPTIN | 3 | 20 | 8.889e-05 | 0.001374 |
| 302 | ALPHA BETA T CELL ACTIVATION | 4 | 54 | 9.355e-05 | 0.001441 |
| 303 | MORPHOGENESIS OF A BRANCHING STRUCTURE | 6 | 167 | 9.536e-05 | 0.001464 |
| 304 | REGULATION OF INTERLEUKIN 6 PRODUCTION | 5 | 104 | 9.601e-05 | 0.00147 |
| 305 | REGULATION OF PROTEIN IMPORT INTO NUCLEUS TRANSLOCATION | 3 | 21 | 0.0001034 | 0.001577 |
| 306 | POSITIVE REGULATION OF CELL CYCLE | 8 | 332 | 0.0001075 | 0.001635 |
| 307 | REGULATION OF MYELOID LEUKOCYTE DIFFERENTIATION | 5 | 108 | 0.0001148 | 0.00174 |
| 308 | RESPONSE TO DRUG | 9 | 431 | 0.0001171 | 0.001768 |
| 309 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 14 | 1004 | 0.0001177 | 0.001773 |
| 310 | POSITIVE REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 3 | 23 | 0.0001368 | 0.002033 |
| 311 | LEUKOCYTE APOPTOTIC PROCESS | 3 | 23 | 0.0001368 | 0.002033 |
| 312 | REGULATION OF METANEPHROS DEVELOPMENT | 3 | 23 | 0.0001368 | 0.002033 |
| 313 | BRANCHED CHAIN AMINO ACID METABOLIC PROCESS | 3 | 23 | 0.0001368 | 0.002033 |
| 314 | POSITIVE REGULATION OF SMOOTH MUSCLE CELL PROLIFERATION | 4 | 60 | 0.0001414 | 0.002095 |
| 315 | CELLULAR RESPONSE TO ALCOHOL | 5 | 115 | 0.0001543 | 0.002279 |
| 316 | POSITIVE REGULATION OF DNA METABOLIC PROCESS | 6 | 185 | 0.000167 | 0.002458 |
| 317 | GLYCEROLIPID METABOLIC PROCESS | 8 | 356 | 0.0001735 | 0.002547 |
| 318 | CELLULAR RESPONSE TO LIPID | 9 | 457 | 0.0001816 | 0.002658 |
| 319 | MYELOID CELL DIFFERENTIATION | 6 | 189 | 0.0001875 | 0.002735 |
| 320 | REGULATION OF T HELPER CELL DIFFERENTIATION | 3 | 26 | 0.0001989 | 0.002856 |
| 321 | POSITIVE REGULATION OF B CELL MEDIATED IMMUNITY | 3 | 26 | 0.0001989 | 0.002856 |
| 322 | POSITIVE REGULATION OF IMMUNOGLOBULIN MEDIATED IMMUNE RESPONSE | 3 | 26 | 0.0001989 | 0.002856 |
| 323 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 12 | 801 | 0.0001967 | 0.002856 |
| 324 | POSITIVE REGULATION OF PROTEOLYSIS | 8 | 363 | 0.0001981 | 0.002856 |
| 325 | PHOSPHOLIPID METABOLIC PROCESS | 8 | 364 | 0.0002019 | 0.00289 |
| 326 | TAXIS | 9 | 464 | 0.0002034 | 0.002903 |
| 327 | CELLULAR RESPONSE TO UV | 4 | 66 | 0.0002048 | 0.002914 |
| 328 | REGULATION OF ASTROCYTE DIFFERENTIATION | 3 | 27 | 0.000223 | 0.003164 |
| 329 | REGULATION OF ALPHA BETA T CELL ACTIVATION | 4 | 68 | 0.0002299 | 0.003241 |
| 330 | ORGAN GROWTH | 4 | 68 | 0.0002299 | 0.003241 |
| 331 | ACTIVATION OF PROTEIN KINASE ACTIVITY | 7 | 279 | 0.000231 | 0.003247 |
| 332 | RESPONSE TO UV | 5 | 126 | 0.0002364 | 0.003313 |
| 333 | EPITHELIUM DEVELOPMENT | 13 | 945 | 0.0002413 | 0.003372 |
| 334 | REGULATION OF SECRETION | 11 | 699 | 0.0002436 | 0.003383 |
| 335 | POSITIVE REGULATION OF LYMPHOCYTE MEDIATED IMMUNITY | 4 | 69 | 0.0002432 | 0.003383 |
| 336 | POSITIVE REGULATION OF ACTIVATED T CELL PROLIFERATION | 3 | 28 | 0.000249 | 0.003448 |
| 337 | NEGATIVE REGULATION OF HEMOPOIESIS | 5 | 128 | 0.0002543 | 0.003511 |
| 338 | POSITIVE REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 5 | 129 | 0.0002637 | 0.003619 |
| 339 | NUCLEAR IMPORT | 5 | 129 | 0.0002637 | 0.003619 |
| 340 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 19 | 1805 | 0.0002686 | 0.003675 |
| 341 | BODY FLUID SECRETION | 4 | 71 | 0.0002715 | 0.003705 |
| 342 | T CELL DIFFERENTIATION INVOLVED IN IMMUNE RESPONSE | 3 | 29 | 0.0002769 | 0.003767 |
| 343 | NEGATIVE REGULATION OF MAP KINASE ACTIVITY | 4 | 73 | 0.0003021 | 0.004099 |
| 344 | ORGAN MORPHOGENESIS | 12 | 841 | 0.0003073 | 0.004156 |
| 345 | REGULATION OF PROTEIN SECRETION | 8 | 389 | 0.0003158 | 0.004259 |
| 346 | NEGATIVE REGULATION OF HORMONE SECRETION | 4 | 74 | 0.0003183 | 0.004281 |
| 347 | NEGATIVE REGULATION OF NEURON APOPTOTIC PROCESS | 5 | 135 | 0.0003253 | 0.004363 |
| 348 | RESPONSE TO INTERLEUKIN 4 | 3 | 31 | 0.0003384 | 0.004469 |
| 349 | NEGATIVE REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 3 | 31 | 0.0003384 | 0.004469 |
| 350 | POSITIVE REGULATION OF PROTEIN SECRETION | 6 | 211 | 0.000339 | 0.004469 |
| 351 | GRANULOCYTE MIGRATION | 4 | 75 | 0.0003351 | 0.004469 |
| 352 | HOMEOSTASIS OF NUMBER OF CELLS WITHIN A TISSUE | 3 | 31 | 0.0003384 | 0.004469 |
| 353 | NEGATIVE REGULATION OF CYTOKINE PRODUCTION | 6 | 211 | 0.000339 | 0.004469 |
| 354 | NEUROGENESIS | 16 | 1402 | 0.0003677 | 0.004833 |
| 355 | POSITIVE REGULATION OF GLIAL CELL DIFFERENTIATION | 3 | 32 | 0.0003723 | 0.004865 |
| 356 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER IN RESPONSE TO HYPOXIA | 3 | 32 | 0.0003723 | 0.004865 |
| 357 | REGULATION OF SYSTEM PROCESS | 9 | 507 | 0.0003899 | 0.005082 |
| 358 | CELLULAR RESPONSE TO STEROID HORMONE STIMULUS | 6 | 218 | 0.0004033 | 0.005227 |
| 359 | REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 6 | 218 | 0.0004033 | 0.005227 |
| 360 | REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 6 | 220 | 0.0004233 | 0.005471 |
| 361 | CHEMICAL HOMEOSTASIS | 12 | 874 | 0.0004347 | 0.005603 |
| 362 | REGULATION OF PROTEIN DEACETYLATION | 3 | 34 | 0.0004463 | 0.005736 |
| 363 | POSITIVE REGULATION OF PROTEIN KINASE B SIGNALING | 4 | 81 | 0.0004497 | 0.005764 |
| 364 | CELLULAR RESPONSE TO INSULIN STIMULUS | 5 | 146 | 0.0004661 | 0.005942 |
| 365 | T CELL RECEPTOR SIGNALING PATHWAY | 5 | 146 | 0.0004661 | 0.005942 |
| 366 | NEGATIVE REGULATION OF LEUKOCYTE DIFFERENTIATION | 4 | 82 | 0.0004711 | 0.00599 |
| 367 | LIPID METABOLIC PROCESS | 14 | 1158 | 0.0005077 | 0.006437 |
| 368 | REGULATION OF CYTOKINE SECRETION | 5 | 149 | 0.0005115 | 0.006467 |
| 369 | FIBROBLAST GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 4 | 84 | 0.0005163 | 0.00651 |
| 370 | T CELL PROLIFERATION | 3 | 36 | 0.0005291 | 0.006636 |
| 371 | T CELL SELECTION | 3 | 36 | 0.0005291 | 0.006636 |
| 372 | REGULATION OF REACTIVE OXYGEN SPECIES METABOLIC PROCESS | 5 | 152 | 0.00056 | 0.007005 |
| 373 | NEGATIVE REGULATION OF ADAPTIVE IMMUNE RESPONSE | 3 | 37 | 0.000574 | 0.007103 |
| 374 | TRICARBOXYLIC ACID METABOLIC PROCESS | 3 | 37 | 0.000574 | 0.007103 |
| 375 | REGULATION OF PEPTIDYL THREONINE PHOSPHORYLATION | 3 | 37 | 0.000574 | 0.007103 |
| 376 | NEGATIVE REGULATION OF EPITHELIAL CELL DIFFERENTIATION | 3 | 37 | 0.000574 | 0.007103 |
| 377 | LIPID BIOSYNTHETIC PROCESS | 9 | 539 | 0.000606 | 0.007479 |
| 378 | PROTEIN IMPORT | 5 | 155 | 0.000612 | 0.007533 |
| 379 | MYELOID CELL HOMEOSTASIS | 4 | 88 | 0.0006157 | 0.007559 |
| 380 | REGULATION OF MULTICELLULAR ORGANISMAL METABOLIC PROCESS | 3 | 38 | 0.0006212 | 0.007606 |
| 381 | DEFENSE RESPONSE TO BACTERIUM | 6 | 237 | 0.0006266 | 0.007653 |
| 382 | PROTEIN LOCALIZATION TO NUCLEUS | 5 | 156 | 0.0006301 | 0.007675 |
| 383 | CELLULAR LIPID METABOLIC PROCESS | 12 | 913 | 0.0006405 | 0.007781 |
| 384 | EPITHELIAL CELL PROLIFERATION | 4 | 89 | 0.0006425 | 0.007785 |
| 385 | NEGATIVE REGULATION OF PEPTIDYL TYROSINE PHOSPHORYLATION | 3 | 39 | 0.0006708 | 0.008024 |
| 386 | ERBB2 SIGNALING PATHWAY | 3 | 39 | 0.0006708 | 0.008024 |
| 387 | NEGATIVE REGULATION OF CELL ACTIVATION | 5 | 158 | 0.0006675 | 0.008024 |
| 388 | POSITIVE REGULATION OF SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 3 | 39 | 0.0006708 | 0.008024 |
| 389 | EMBRYO IMPLANTATION | 3 | 39 | 0.0006708 | 0.008024 |
| 390 | POSITIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 8 | 437 | 0.0006809 | 0.008123 |
| 391 | CELLULAR RESPONSE TO LIGHT STIMULUS | 4 | 91 | 0.0006986 | 0.008296 |
| 392 | REGULATION OF ORGAN MORPHOGENESIS | 6 | 242 | 0.0006989 | 0.008296 |
| 393 | REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 3 | 40 | 0.0007229 | 0.008537 |
| 394 | REGULATION OF ACTIVATED T CELL PROLIFERATION | 3 | 40 | 0.0007229 | 0.008537 |
| 395 | REGULATION OF GTPASE ACTIVITY | 10 | 673 | 0.0007397 | 0.008714 |
| 396 | REGULATION OF CYTOKINE BIOSYNTHETIC PROCESS | 4 | 94 | 0.0007892 | 0.009273 |
| 397 | POSITIVE REGULATION OF TRANSPORT | 12 | 936 | 0.0007966 | 0.009336 |
| 398 | ORGANOPHOSPHATE BIOSYNTHETIC PROCESS | 8 | 450 | 0.0008235 | 0.009628 |
| 399 | RESPONSE TO WOUNDING | 9 | 563 | 0.0008257 | 0.009628 |
| 400 | POSITIVE REGULATION OF GLUCOSE TRANSPORT | 3 | 42 | 0.0008347 | 0.009709 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CYTOKINE RECEPTOR ACTIVITY | 27 | 89 | 1.111e-43 | 1.033e-40 |
| 2 | CYTOKINE RECEPTOR BINDING | 24 | 271 | 5.713e-25 | 2.654e-22 |
| 3 | CYTOKINE BINDING | 14 | 92 | 3.306e-18 | 1.024e-15 |
| 4 | RECEPTOR BINDING | 35 | 1476 | 1.471e-17 | 3.416e-15 |
| 5 | SIGNAL TRANSDUCER ACTIVITY | 37 | 1731 | 3.588e-17 | 6.666e-15 |
| 6 | KINASE REGULATOR ACTIVITY | 15 | 186 | 3.34e-15 | 5.171e-13 |
| 7 | CYTOKINE ACTIVITY | 13 | 219 | 1.326e-11 | 1.76e-09 |
| 8 | SIGNALING RECEPTOR ACTIVITY | 27 | 1393 | 2.386e-11 | 2.771e-09 |
| 9 | KINASE BINDING | 18 | 606 | 1.213e-10 | 1.252e-08 |
| 10 | KINASE INHIBITOR ACTIVITY | 9 | 89 | 1.939e-10 | 1.801e-08 |
| 11 | X1 PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 7 | 43 | 7.096e-10 | 5.484e-08 |
| 12 | PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 8 | 70 | 7.673e-10 | 5.484e-08 |
| 13 | INSULIN RECEPTOR SUBSTRATE BINDING | 5 | 11 | 6.651e-10 | 5.484e-08 |
| 14 | RECEPTOR ACTIVITY | 27 | 1649 | 1.053e-09 | 6.986e-08 |
| 15 | PHOSPHATIDYLINOSITOL KINASE ACTIVITY | 7 | 51 | 2.478e-09 | 1.439e-07 |
| 16 | MOLECULAR FUNCTION REGULATOR | 24 | 1353 | 2.429e-09 | 1.439e-07 |
| 17 | GROWTH FACTOR RECEPTOR BINDING | 9 | 129 | 5.42e-09 | 2.962e-07 |
| 18 | KINASE ACTIVITY | 18 | 842 | 2.169e-08 | 1.119e-06 |
| 19 | ENZYME BINDING | 25 | 1737 | 6.849e-08 | 3.219e-06 |
| 20 | GROWTH FACTOR BINDING | 8 | 123 | 6.931e-08 | 3.219e-06 |
| 21 | TRANSFERASE ACTIVITY TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 18 | 992 | 2.547e-07 | 1.127e-05 |
| 22 | PHOSPHATIDYLINOSITOL PHOSPHATE KINASE ACTIVITY | 4 | 16 | 6.116e-07 | 2.583e-05 |
| 23 | HORMONE RECEPTOR BINDING | 8 | 168 | 7.618e-07 | 3.077e-05 |
| 24 | ENZYME REGULATOR ACTIVITY | 17 | 959 | 8.006e-07 | 3.099e-05 |
| 25 | GROWTH FACTOR ACTIVITY | 7 | 160 | 6.78e-06 | 0.000252 |
| 26 | RAS GUANYL NUCLEOTIDE EXCHANGE FACTOR ACTIVITY | 8 | 228 | 7.406e-06 | 0.0002646 |
| 27 | PROTEIN KINASE ACTIVITY | 12 | 640 | 2.317e-05 | 0.0007971 |
| 28 | GLUCOCORTICOID RECEPTOR BINDING | 3 | 14 | 2.893e-05 | 0.0009276 |
| 29 | STEROID HORMONE RECEPTOR BINDING | 5 | 81 | 2.896e-05 | 0.0009276 |
| 30 | ENZYME INHIBITOR ACTIVITY | 9 | 378 | 4.291e-05 | 0.001329 |
| 31 | GUANYL NUCLEOTIDE EXCHANGE FACTOR ACTIVITY | 8 | 303 | 5.691e-05 | 0.001705 |
| 32 | PHOSPHATASE BINDING | 6 | 162 | 8.064e-05 | 0.002341 |
| 33 | RNA POLYMERASE II CORE PROMOTER SEQUENCE SPECIFIC DNA BINDING | 4 | 53 | 8.691e-05 | 0.002447 |
| 34 | PROTEIN TYROSINE KINASE ACTIVITY | 6 | 176 | 0.0001272 | 0.003476 |
| 35 | PROTEIN PHOSPHATASE BINDING | 5 | 120 | 0.0001883 | 0.004998 |
| 36 | PROTEIN SERINE THREONINE KINASE INHIBITOR ACTIVITY | 3 | 30 | 0.0003067 | 0.007913 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | PHOSPHATIDYLINOSITOL 3 KINASE COMPLEX | 7 | 20 | 1.852e-12 | 1.082e-09 |
| 2 | EXTRACELLULAR SPACE | 23 | 1376 | 1.739e-08 | 5.078e-06 |
| 3 | TRANSFERASE COMPLEX TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 10 | 237 | 9.082e-08 | 1.768e-05 |
| 4 | RECEPTOR COMPLEX | 11 | 327 | 1.969e-07 | 2.3e-05 |
| 5 | EXTRINSIC COMPONENT OF MEMBRANE | 10 | 252 | 1.609e-07 | 2.3e-05 |
| 6 | CELL SURFACE | 14 | 757 | 5.264e-06 | 0.0005124 |
| 7 | PLASMA MEMBRANE RECEPTOR COMPLEX | 7 | 175 | 1.218e-05 | 0.001016 |
| 8 | MEMBRANE PROTEIN COMPLEX | 15 | 1020 | 3.558e-05 | 0.002598 |
| 9 | TRANSFERASE COMPLEX | 12 | 703 | 5.763e-05 | 0.00374 |
| 10 | INTRINSIC COMPONENT OF PLASMA MEMBRANE | 19 | 1649 | 8.247e-05 | 0.004599 |
| 11 | EXTERNAL SIDE OF PLASMA MEMBRANE | 7 | 238 | 8.662e-05 | 0.004599 |
| 12 | SIDE OF MEMBRANE | 9 | 428 | 0.000111 | 0.005404 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
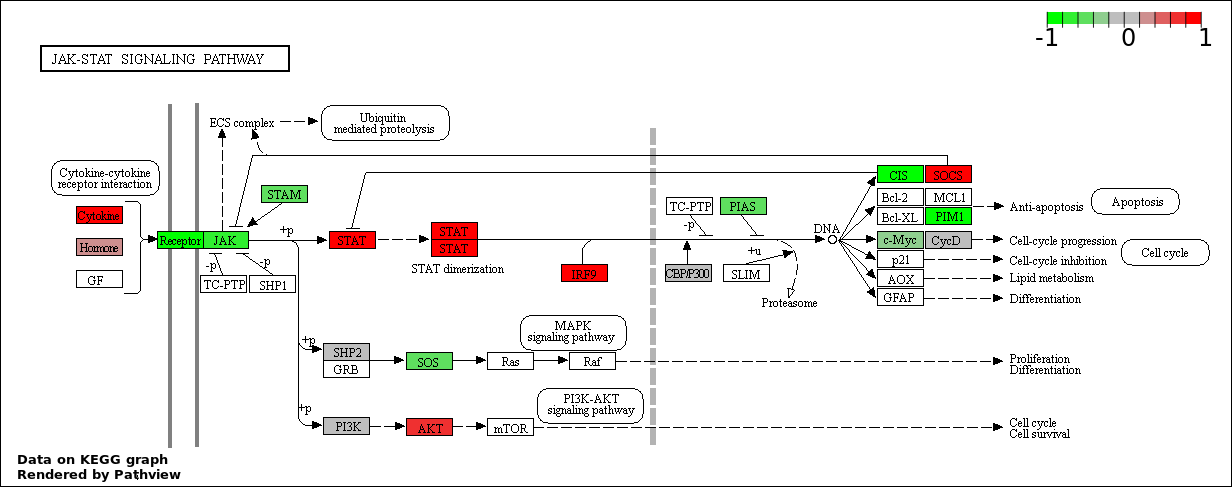
| 1 | hsa04630_Jak.STAT_signaling_pathway | 88 | 155 | 5.139e-200 | 9.251e-198 | |
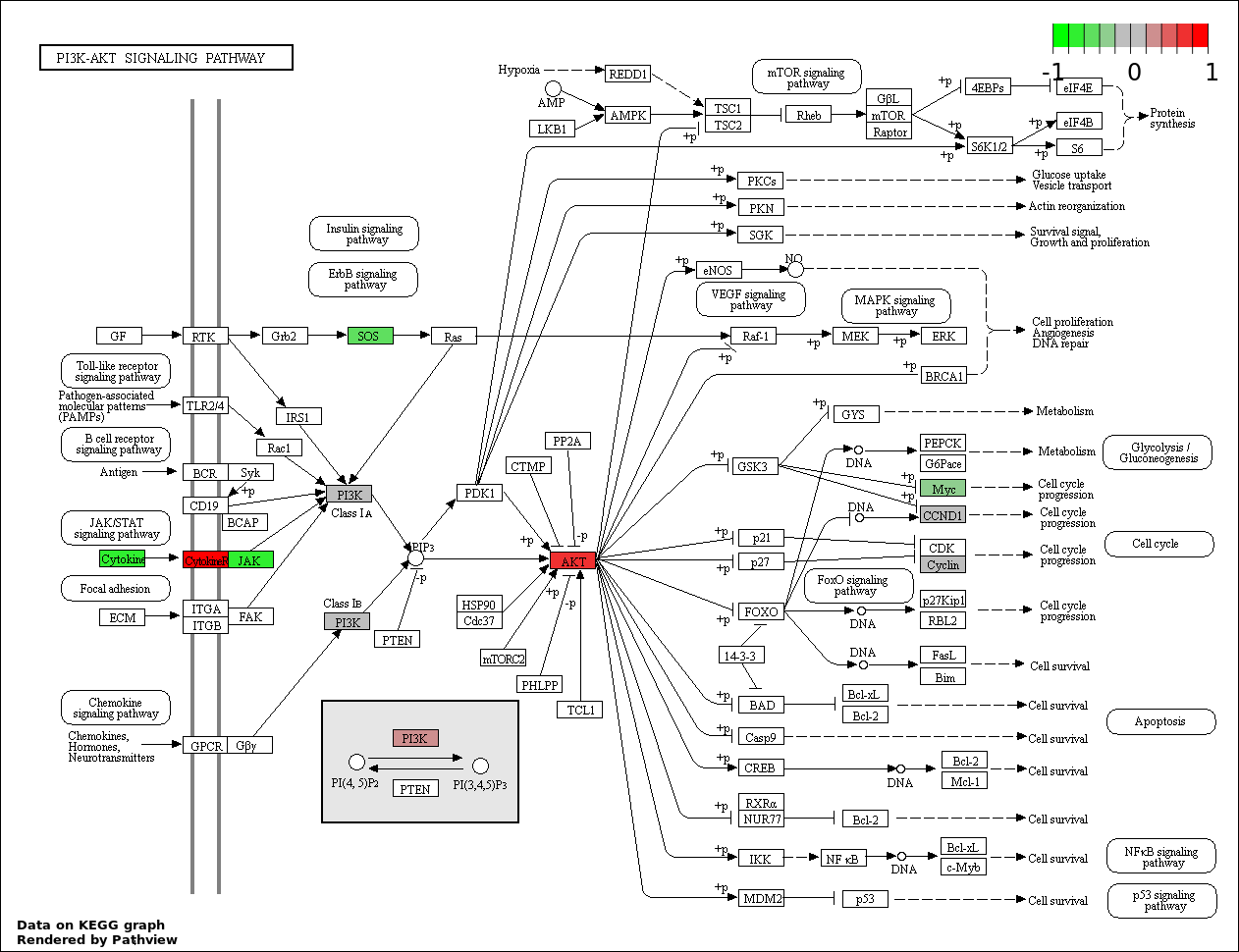
| 2 | hsa04151_PI3K_AKT_signaling_pathway | 29 | 351 | 2.255e-29 | 2.03e-27 | |
| 3 | hsa04380_Osteoclast_differentiation | 17 | 128 | 7.089e-21 | 4.253e-19 | |
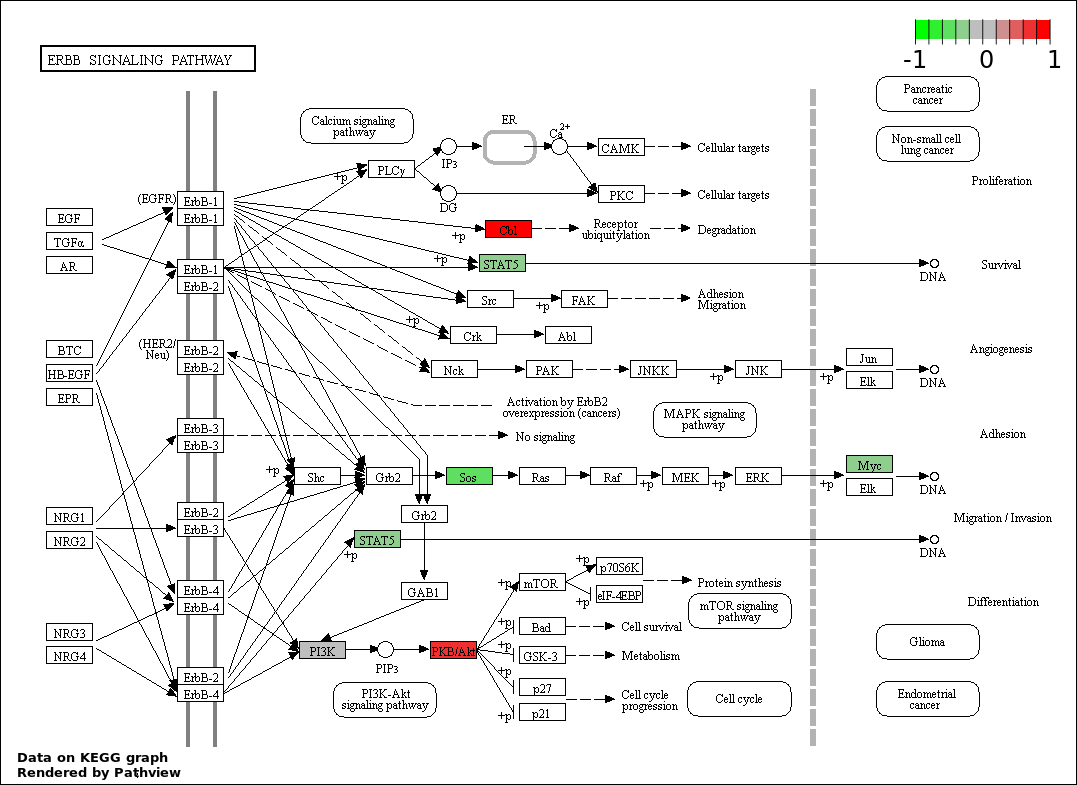
| 4 | hsa04012_ErbB_signaling_pathway | 15 | 87 | 2.617e-20 | 1.178e-18 | |
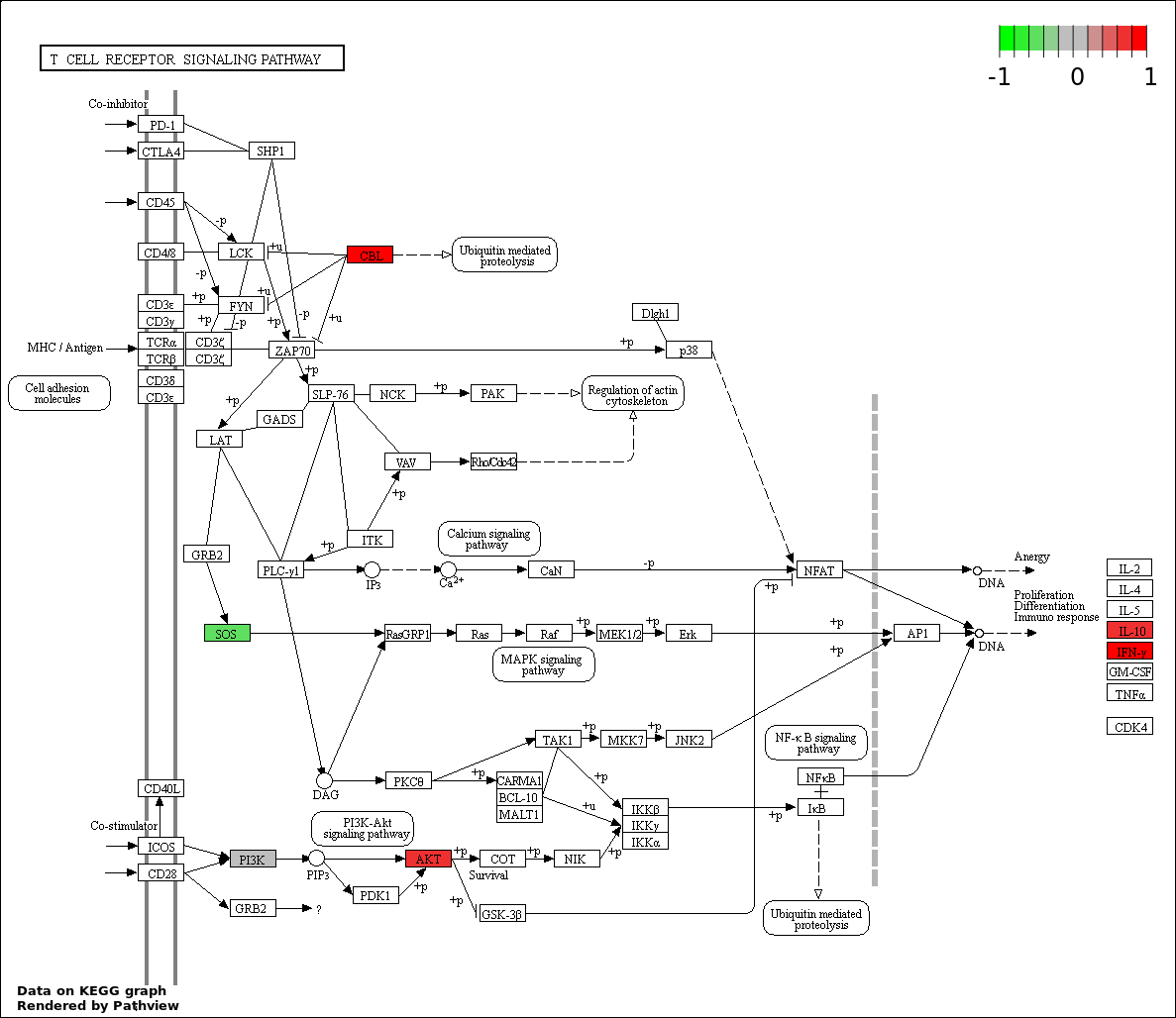
| 5 | hsa04660_T_cell_receptor_signaling_pathway | 14 | 108 | 3.468e-17 | 1.09e-15 | |
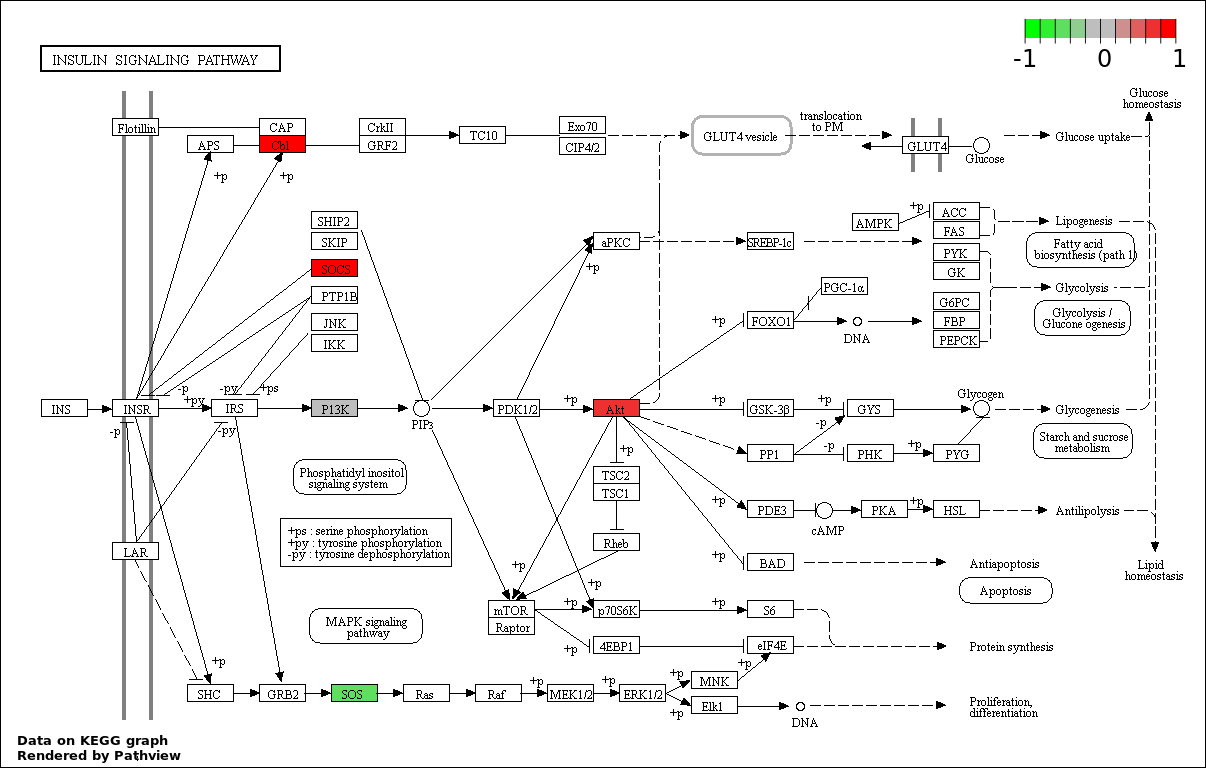
| 6 | hsa04910_Insulin_signaling_pathway | 15 | 138 | 3.633e-17 | 1.09e-15 | |
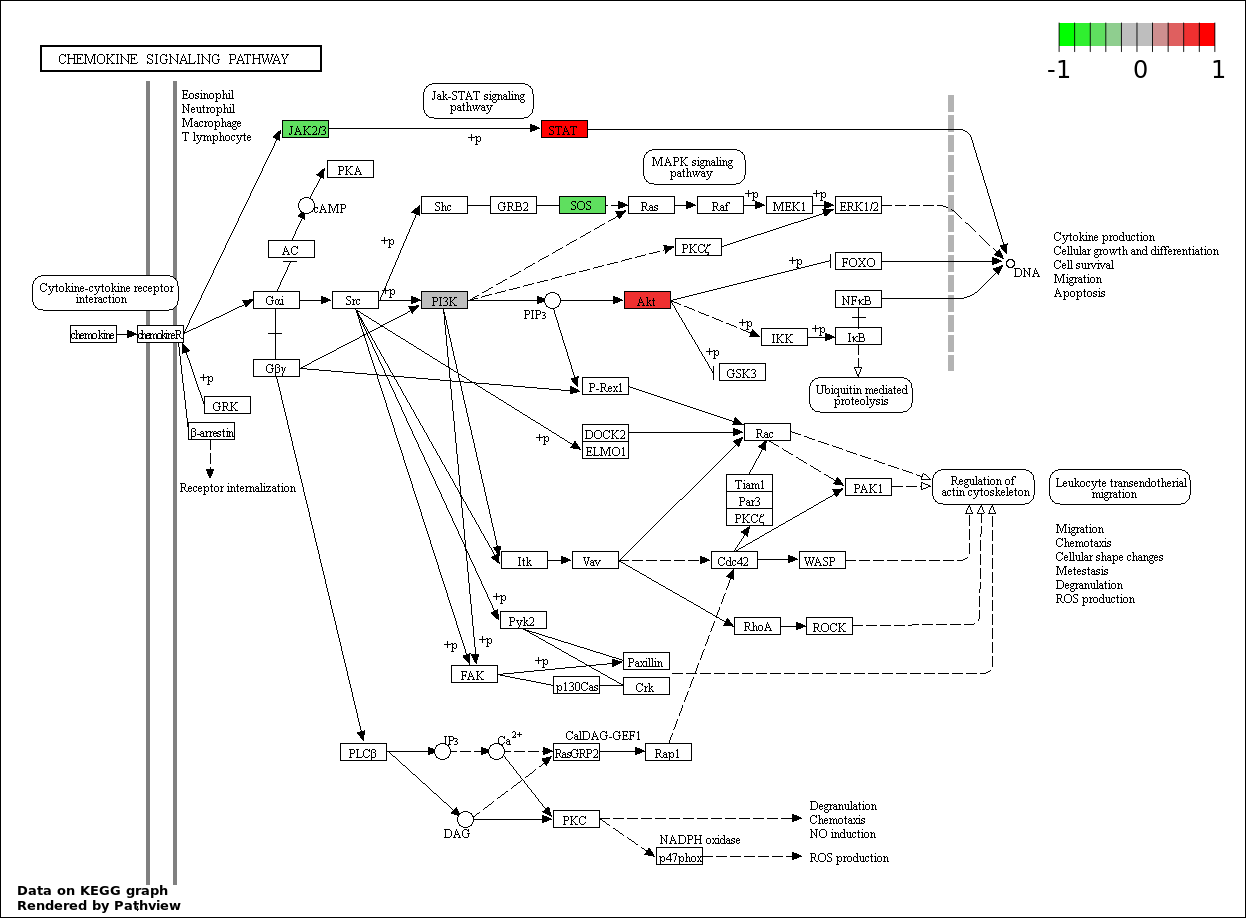
| 7 | hsa04062_Chemokine_signaling_pathway | 15 | 189 | 4.242e-15 | 1.091e-13 | |
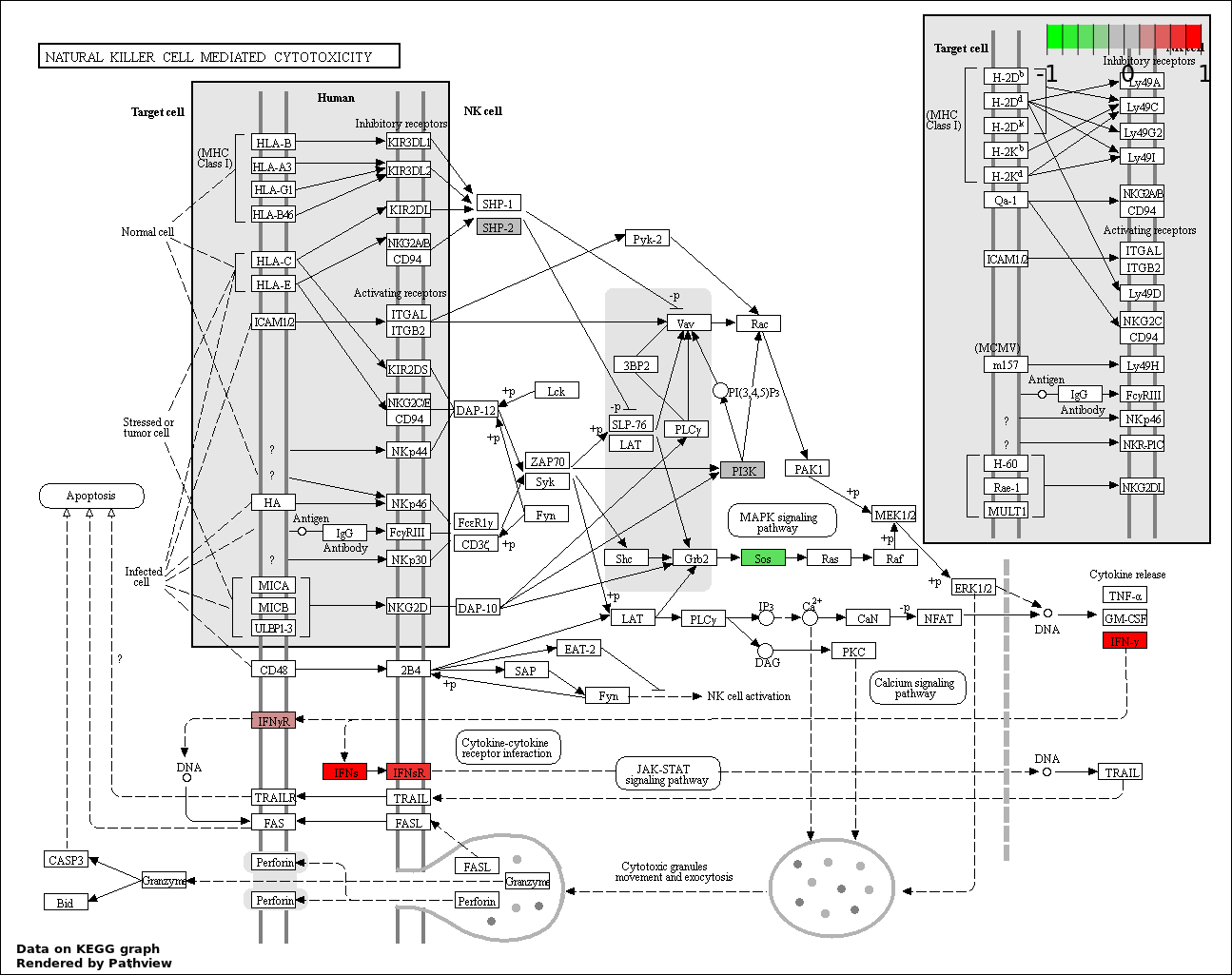
| 8 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 13 | 136 | 2.883e-14 | 6.487e-13 | |
| 9 | hsa04640_Hematopoietic_cell_lineage | 11 | 88 | 1.524e-13 | 3.049e-12 | |
| 10 | hsa04620_Toll.like_receptor_signaling_pathway | 11 | 102 | 8.074e-13 | 1.453e-11 | |
| 11 | hsa04662_B_cell_receptor_signaling_pathway | 10 | 75 | 1.055e-12 | 1.726e-11 | |
| 12 | hsa04664_Fc_epsilon_RI_signaling_pathway | 10 | 79 | 1.807e-12 | 2.711e-11 | |
| 13 | hsa04510_Focal_adhesion | 13 | 200 | 4.206e-12 | 5.824e-11 | |
| 14 | hsa04722_Neurotrophin_signaling_pathway | 11 | 127 | 9.226e-12 | 1.186e-10 | |
| 15 | hsa04973_Carbohydrate_digestion_and_absorption | 8 | 44 | 1.58e-11 | 1.897e-10 | |
| 16 | hsa04150_mTOR_signaling_pathway | 8 | 52 | 6.522e-11 | 7.337e-10 | |
| 17 | hsa04210_Apoptosis | 9 | 89 | 1.939e-10 | 2.053e-09 | |
| 18 | hsa04960_Aldosterone.regulated_sodium_reabsorption | 7 | 42 | 5.962e-10 | 5.962e-09 | |
| 19 | hsa04370_VEGF_signaling_pathway | 8 | 76 | 1.5e-09 | 1.421e-08 | |
| 20 | hsa04914_Progesterone.mediated_oocyte_maturation | 8 | 87 | 4.47e-09 | 4.023e-08 | |
| 21 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 8 | 95 | 9.038e-09 | 7.747e-08 | |
| 22 | hsa04670_Leukocyte_transendothelial_migration | 8 | 117 | 4.688e-08 | 3.836e-07 | |
| 23 | hsa04070_Phosphatidylinositol_signaling_system | 7 | 78 | 5.138e-08 | 4.021e-07 | |
| 24 | hsa04810_Regulation_of_actin_cytoskeleton | 9 | 214 | 4.272e-07 | 3.204e-06 | |
| 25 | hsa04920_Adipocytokine_signaling_pathway | 6 | 68 | 5.369e-07 | 3.866e-06 | |
| 26 | hsa04014_Ras_signaling_pathway | 9 | 236 | 9.683e-07 | 6.704e-06 | |
| 27 | hsa04110_Cell_cycle | 6 | 128 | 2.155e-05 | 0.0001437 | |
| 28 | hsa04120_Ubiquitin_mediated_proteolysis | 6 | 139 | 3.433e-05 | 0.0002207 | |
| 29 | hsa04310_Wnt_signaling_pathway | 6 | 151 | 5.458e-05 | 0.0003388 | |
| 30 | hsa04672_Intestinal_immune_network_for_IgA_production | 4 | 49 | 6.373e-05 | 0.0003824 | |
| 31 | hsa00562_Inositol_phosphate_metabolism | 4 | 57 | 0.0001157 | 0.0006717 | |
| 32 | hsa04144_Endocytosis | 6 | 203 | 0.0002757 | 0.001551 | |
| 33 | hsa04350_TGF.beta_signaling_pathway | 4 | 85 | 0.00054 | 0.002945 | |
| 34 | hsa04115_p53_signaling_pathway | 3 | 69 | 0.003498 | 0.01852 | |
| 35 | hsa04390_Hippo_signaling_pathway | 4 | 154 | 0.004777 | 0.02457 | |
| 36 | hsa04320_Dorso.ventral_axis_formation | 2 | 25 | 0.005377 | 0.02688 | |
| 37 | hsa04330_Notch_signaling_pathway | 2 | 47 | 0.0182 | 0.08854 | |
| 38 | hsa04010_MAPK_signaling_pathway | 4 | 268 | 0.03063 | 0.1451 | |
| 39 | hsa04720_Long.term_potentiation | 2 | 70 | 0.03811 | 0.1759 | |
| 40 | hsa04622_RIG.I.like_receptor_signaling_pathway | 2 | 71 | 0.0391 | 0.176 | |
| 41 | hsa04520_Adherens_junction | 2 | 73 | 0.04112 | 0.1805 | |
| 42 | hsa04540_Gap_junction | 2 | 90 | 0.05976 | 0.2561 | |
| 43 | hsa04912_GnRH_signaling_pathway | 2 | 101 | 0.07308 | 0.299 | |
| 44 | hsa04916_Melanogenesis | 2 | 101 | 0.07308 | 0.299 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | RP11-815I9.4 |
hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-130a-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-664a-3p | 26 | IL6ST | Sponge network | 0.004 | 0.98116 | -1.519 | 0 | 0.787 |
| 2 | AC090587.5 |
hsa-miR-103a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-93-3p | 16 | IL6ST | Sponge network | -0.612 | 0.00023 | -1.519 | 0 | 0.75 |
| 3 | RP11-815I9.4 |
hsa-miR-103a-2-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-30c-5p;hsa-miR-365a-3p;hsa-miR-576-5p;hsa-miR-590-5p | 10 | SOS1 | Sponge network | 0.004 | 0.98116 | -0.144 | 0.07887 | 0.69 |
| 4 | RP11-193H5.1 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130a-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 25 | IL6ST | Sponge network | -0.426 | 0.01062 | -1.519 | 0 | 0.657 |
| 5 | RP11-750H9.5 | hsa-miR-103a-3p;hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p | 12 | IL6ST | Sponge network | -0.173 | 0.60204 | -1.519 | 0 | 0.646 |
| 6 | CTC-429P9.5 | hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-501-5p | 14 | IL6ST | Sponge network | -0.72 | 0.01036 | -1.519 | 0 | 0.637 |
| 7 | RP1-122P22.2 | hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-135b-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 16 | IL6ST | Sponge network | -1.073 | 6.0E-5 | -1.519 | 0 | 0.633 |
| 8 | LINC00941 | hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-181c-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-20b-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-497-5p | 15 | SPRY4 | Sponge network | 4.292 | 0 | 1.651 | 0 | 0.621 |
| 9 | AC006116.24 |
hsa-miR-103a-3p;hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-455-5p | 15 | IL6ST | Sponge network | -1.832 | 1.0E-5 | -1.519 | 0 | 0.62 |
| 10 | RP11-182J1.1 | hsa-miR-103a-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p | 10 | IL6ST | Sponge network | -0.401 | 0.08289 | -1.519 | 0 | 0.589 |
| 11 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 25 | PIK3R1 | Sponge network | -0.939 | 4.0E-5 | -1.219 | 0 | 0.589 |
| 12 | RP11-876N24.4 | hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-24-2-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-589-3p | 10 | IL6ST | Sponge network | 0.037 | 0.92383 | -1.519 | 0 | 0.586 |
| 13 | RP11-517P14.2 |
hsa-miR-103a-3p;hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p | 12 | IL6ST | Sponge network | -0.402 | 0.1503 | -1.519 | 0 | 0.585 |
| 14 | RP11-282O18.3 | hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-590-5p | 12 | IL6ST | Sponge network | -0.266 | 0.05794 | -1.519 | 0 | 0.577 |
| 15 | FGD5-AS1 | hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-29b-1-5p;hsa-miR-454-3p;hsa-miR-576-5p | 12 | IL6ST | Sponge network | -0.292 | 0.01482 | -1.519 | 0 | 0.571 |
| 16 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | SOCS5 | Sponge network | -0.939 | 4.0E-5 | -0.086 | 0.31771 | 0.561 |
| 17 | CTD-2171N6.1 |
hsa-miR-140-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-20b-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-497-5p | 11 | SPRY4 | Sponge network | 5.89 | 0 | 1.651 | 0 | 0.558 |
| 18 | CTD-2620I22.1 |
hsa-miR-103a-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-2355-3p;hsa-miR-29b-1-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-505-3p | 12 | IL6ST | Sponge network | -1.967 | 0.00106 | -1.519 | 0 | 0.551 |
| 19 | RP11-1049A21.2 | hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-362-5p;hsa-miR-501-5p | 15 | IL6ST | Sponge network | -0.334 | 0.61847 | -1.519 | 0 | 0.55 |
| 20 | DHRS4-AS1 | hsa-miR-130a-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-29b-1-5p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p | 15 | IL6ST | Sponge network | -0.068 | 0.72321 | -1.519 | 0 | 0.543 |
| 21 | RP11-815I9.4 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-590-5p | 14 | IL6R | Sponge network | 0.004 | 0.98116 | -0.049 | 0.80939 | 0.537 |
| 22 | CTD-3099C6.9 | hsa-miR-130a-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-576-5p | 11 | IL6ST | Sponge network | -0.515 | 0.15915 | -1.519 | 0 | 0.52 |
| 23 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-93-5p | 23 | SPRED1 | Sponge network | -0.939 | 4.0E-5 | -0.094 | 0.38958 | 0.513 |
| 24 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-205-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-708-3p;hsa-miR-944 | 16 | LIFR | Sponge network | -0.939 | 4.0E-5 | -1.519 | 0 | 0.506 |
| 25 | OIP5-AS1 | hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-146b-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-29b-1-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p | 13 | IL6ST | Sponge network | -0.06 | 0.55122 | -1.519 | 0 | 0.506 |
| 26 | NNT-AS1 | hsa-let-7a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-205-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-944 | 10 | LIFR | Sponge network | -0.2 | 0.23102 | -1.519 | 0 | 0.504 |
| 27 | RP11-193H5.1 |
hsa-miR-103a-2-5p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-501-5p;hsa-miR-576-5p | 13 | IL6R | Sponge network | -0.426 | 0.01062 | -0.049 | 0.80939 | 0.502 |
| 28 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-205-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-500a-5p | 11 | LIFR | Sponge network | -1.258 | 0.00026 | -1.519 | 0 | 0.495 |
| 29 | TP73-AS1 |
hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-205-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-708-3p | 10 | LIFR | Sponge network | -0.533 | 0.00643 | -1.519 | 0 | 0.487 |
| 30 | RP11-594N15.3 |
hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-93-5p | 15 | PIK3R1 | Sponge network | -2.86 | 0 | -1.219 | 0 | 0.483 |
| 31 | CTD-2015G9.2 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130a-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-708-3p | 25 | IL6ST | Sponge network | -3.112 | 5.0E-5 | -1.519 | 0 | 0.482 |
| 32 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 19 | SPRED1 | Sponge network | -0.882 | 5.0E-5 | -0.094 | 0.38958 | 0.482 |
| 33 | SNHG14 |
hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-455-3p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-629-3p | 11 | PIK3R1 | Sponge network | -1.125 | 0.0001 | -1.219 | 0 | 0.482 |
| 34 | AC007743.1 |
hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p | 13 | PIK3R1 | Sponge network | -1.053 | 0.00923 | -1.219 | 0 | 0.478 |
| 35 | CTA-221G9.11 |
hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-450b-5p;hsa-miR-629-3p;hsa-miR-93-5p | 10 | PIK3R1 | Sponge network | -2.198 | 0 | -1.219 | 0 | 0.477 |
| 36 | RP1-193H18.2 |
hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-96-5p | 11 | PIK3R1 | Sponge network | -1.539 | 0 | -1.219 | 0 | 0.476 |
| 37 | DNM3OS |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-429 | 12 | SPRED1 | Sponge network | 0.932 | 0.00442 | -0.094 | 0.38958 | 0.472 |
| 38 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-455-5p | 13 | IL6ST | Sponge network | -0.233 | 0.50729 | -1.519 | 0 | 0.471 |
| 39 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-324-3p;hsa-miR-93-5p | 10 | PIK3R1 | Sponge network | -0.304 | 0.38627 | -1.219 | 0 | 0.469 |
| 40 | PWAR6 |
hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-629-3p | 11 | PIK3R1 | Sponge network | -1.629 | 0 | -1.219 | 0 | 0.466 |
| 41 | RP11-815I9.4 |
hsa-miR-103a-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-589-3p;hsa-miR-590-5p | 11 | PIK3R1 | Sponge network | 0.004 | 0.98116 | -1.219 | 0 | 0.464 |
| 42 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-205-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-944;hsa-miR-96-5p | 15 | LIFR | Sponge network | -0.882 | 5.0E-5 | -1.519 | 0 | 0.46 |
| 43 | ZNF667-AS1 |
hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-205-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-590-5p;hsa-miR-96-5p | 12 | LIFR | Sponge network | -1.395 | 0 | -1.519 | 0 | 0.459 |
| 44 | RP4-798P15.3 |
hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | IL6ST | Sponge network | -1.213 | 0.00098 | -1.519 | 0 | 0.458 |
| 45 | RP11-284N8.3 |
hsa-miR-103a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-455-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-96-5p | 12 | PIK3R1 | Sponge network | 0.003 | 0.99478 | -1.219 | 0 | 0.457 |
| 46 | AF131215.2 | hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-589-3p | 14 | IL6ST | Sponge network | -1.136 | 0.00092 | -1.519 | 0 | 0.452 |
| 47 | RP11-284N8.3 |
hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-590-5p | 13 | IL6ST | Sponge network | 0.003 | 0.99478 | -1.519 | 0 | 0.45 |
| 48 | RASSF8-AS1 | hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-205-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-944 | 11 | LIFR | Sponge network | 0.121 | 0.52445 | -1.519 | 0 | 0.45 |
| 49 | BHLHE40-AS1 |
hsa-let-7a-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-500a-5p;hsa-miR-590-5p;hsa-miR-96-5p | 11 | LIFR | Sponge network | -0.932 | 0 | -1.519 | 0 | 0.449 |
| 50 | SNHG14 |
hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-205-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-944 | 10 | LIFR | Sponge network | -1.125 | 0.0001 | -1.519 | 0 | 0.442 |
| 51 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-93-5p | 12 | SPRED1 | Sponge network | 0.187 | 0.31051 | -0.094 | 0.38958 | 0.442 |
| 52 | BHLHE40-AS1 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-96-5p | 10 | PIK3R1 | Sponge network | -0.932 | 0 | -1.219 | 0 | 0.438 |
| 53 | CTC-429P9.1 | hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-20a-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p | 12 | IL6ST | Sponge network | -0.57 | 0.00453 | -1.519 | 0 | 0.436 |
| 54 | SH3RF3-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-5p | 13 | PIK3R1 | Sponge network | -0.175 | 0.58985 | -1.219 | 0 | 0.435 |
| 55 | ZNF667-AS1 |
hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-96-5p | 14 | PIK3R1 | Sponge network | -1.395 | 0 | -1.219 | 0 | 0.434 |
| 56 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-708-3p;hsa-miR-93-3p | 42 | IL6ST | Sponge network | -0.939 | 4.0E-5 | -1.519 | 0 | 0.433 |
| 57 | RP1-193H18.2 |
hsa-let-7a-3p;hsa-miR-19a-3p;hsa-miR-205-3p;hsa-miR-205-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-708-3p;hsa-miR-944;hsa-miR-96-5p | 12 | LIFR | Sponge network | -1.539 | 0 | -1.519 | 0 | 0.433 |
| 58 | AC007879.7 | hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-181c-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-340-5p;hsa-miR-361-3p | 12 | SPRY4 | Sponge network | 2.7 | 0 | 1.651 | 0 | 0.428 |
| 59 | KB-1732A1.1 | hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-181c-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-20b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-497-5p | 13 | SPRY4 | Sponge network | 1.697 | 0 | 1.651 | 0 | 0.427 |
| 60 | RP11-594N15.3 |
hsa-let-7a-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-205-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p | 11 | LIFR | Sponge network | -2.86 | 0 | -1.519 | 0 | 0.422 |
| 61 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-96-5p | 10 | LIFR | Sponge network | -0.33 | 0.08136 | -1.519 | 0 | 0.421 |
| 62 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-205-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-944 | 12 | LIFR | Sponge network | -1.712 | 0 | -1.519 | 0 | 0.419 |
| 63 | CTA-221G9.11 |
hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-576-5p | 17 | IL6ST | Sponge network | -2.198 | 0 | -1.519 | 0 | 0.416 |
| 64 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-589-3p | 15 | PIK3R1 | Sponge network | -1.712 | 0 | -1.219 | 0 | 0.415 |
| 65 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-26b-5p | 11 | SPRED1 | Sponge network | -0.175 | 0.58985 | -0.094 | 0.38958 | 0.415 |
| 66 | RP11-218E20.3 | hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-181c-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-497-5p | 11 | SPRY4 | Sponge network | 4.613 | 0 | 1.651 | 0 | 0.412 |
| 67 | RP11-284N8.3 |
hsa-let-7a-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-27a-3p;hsa-miR-590-5p;hsa-miR-944;hsa-miR-96-5p | 11 | LIFR | Sponge network | 0.003 | 0.99478 | -1.519 | 0 | 0.406 |
| 68 | AC003090.1 |
hsa-let-7a-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-205-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-590-5p;hsa-miR-96-5p | 13 | LIFR | Sponge network | -4.323 | 0 | -1.519 | 0 | 0.405 |
| 69 | PWAR6 |
hsa-miR-19a-3p;hsa-miR-205-3p;hsa-miR-205-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-944 | 10 | LIFR | Sponge network | -1.629 | 0 | -1.519 | 0 | 0.404 |
| 70 | RP11-166D19.1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-96-5p | 19 | PIK3R1 | Sponge network | -0.882 | 5.0E-5 | -1.219 | 0 | 0.401 |
| 71 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-93-3p | 23 | IL6ST | Sponge network | -0.304 | 0.38627 | -1.519 | 0 | 0.399 |
| 72 | AC007743.1 |
hsa-let-7a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-205-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-500a-5p;hsa-miR-590-5p | 12 | LIFR | Sponge network | -1.053 | 0.00923 | -1.519 | 0 | 0.397 |
| 73 | CTD-2015G9.2 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-93-5p | 16 | PIK3R1 | Sponge network | -3.112 | 5.0E-5 | -1.219 | 0 | 0.397 |
| 74 | RP4-798P15.3 |
hsa-miR-103a-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-96-5p | 14 | PIK3R1 | Sponge network | -1.213 | 0.00098 | -1.219 | 0 | 0.393 |
| 75 | WDR86-AS1 |
hsa-let-7a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-205-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-96-5p | 10 | LIFR | Sponge network | -0.396 | 0.35778 | -1.519 | 0 | 0.389 |
| 76 | APCDD1L-AS1 |
hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181c-5p;hsa-miR-200a-3p;hsa-miR-20b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-497-5p | 11 | SPRY4 | Sponge network | 2.077 | 0 | 1.651 | 0 | 0.383 |
| 77 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | IL6ST | Sponge network | -0.175 | 0.58985 | -1.519 | 0 | 0.382 |
| 78 | CTD-2135D7.5 |
hsa-miR-103a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p | 11 | PIK3R1 | Sponge network | -2.579 | 0.0001 | -1.219 | 0 | 0.379 |
| 79 | FAM66C |
hsa-let-7a-3p;hsa-miR-19a-3p;hsa-miR-205-3p;hsa-miR-205-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-944 | 10 | LIFR | Sponge network | -0.798 | 0.00038 | -1.519 | 0 | 0.378 |
| 80 | RP11-284N8.3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-93-5p | 10 | SPRED1 | Sponge network | 0.003 | 0.99478 | -0.094 | 0.38958 | 0.374 |
| 81 | LINC00900 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-93-5p | 19 | PIK3R1 | Sponge network | -1.803 | 0 | -1.219 | 0 | 0.373 |
| 82 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-455-5p;hsa-miR-589-3p | 18 | IL6ST | Sponge network | -1.258 | 0.00026 | -1.519 | 0 | 0.373 |
| 83 | AC007743.1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | IL6ST | Sponge network | -1.053 | 0.00923 | -1.519 | 0 | 0.372 |
| 84 | RP4-798P15.3 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-182-5p;hsa-miR-205-3p;hsa-miR-21-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-708-5p;hsa-miR-96-5p | 13 | SPRY3 | Sponge network | -1.213 | 0.00098 | -0.516 | 0.00013 | 0.371 |
| 85 | AC003090.1 |
hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 18 | PIK3R1 | Sponge network | -4.323 | 0 | -1.219 | 0 | 0.369 |
| 86 | RP11-757G1.6 |
hsa-miR-103a-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-93-5p | 11 | PIK3R1 | Sponge network | -1.346 | 0.00088 | -1.219 | 0 | 0.367 |
| 87 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-205-5p;hsa-miR-224-3p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-944 | 10 | LIFR | Sponge network | 0.187 | 0.31051 | -1.519 | 0 | 0.359 |
| 88 | RP11-815I9.4 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-664a-3p | 10 | SPRED1 | Sponge network | 0.004 | 0.98116 | -0.094 | 0.38958 | 0.359 |
| 89 | RP11-517P14.2 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-93-5p | 10 | IL6R | Sponge network | -0.402 | 0.1503 | -0.049 | 0.80939 | 0.359 |
| 90 | TPTEP1 |
hsa-let-7a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-205-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-590-5p;hsa-miR-944 | 12 | LIFR | Sponge network | -2.193 | 0 | -1.519 | 0 | 0.353 |
| 91 | BHLHE40-AS1 |
hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-93-3p | 21 | IL6ST | Sponge network | -0.932 | 0 | -1.519 | 0 | 0.35 |
| 92 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 22 | PIK3R1 | Sponge network | -1.459 | 1.0E-5 | -1.219 | 0 | 0.349 |
| 93 | CTD-2135D7.5 |
hsa-miR-103a-3p;hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-590-5p | 17 | IL6ST | Sponge network | -2.579 | 0.0001 | -1.519 | 0 | 0.349 |
| 94 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-454-3p | 11 | SPRED1 | Sponge network | -0.33 | 0.08136 | -0.094 | 0.38958 | 0.349 |
| 95 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-944 | 22 | PRLR | Sponge network | -0.939 | 4.0E-5 | -1.308 | 0.00047 | 0.348 |
| 96 | MIR497HG |
hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-455-3p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-96-5p | 13 | PIK3R1 | Sponge network | -1.263 | 0.00248 | -1.219 | 0 | 0.343 |
| 97 | MAGI2-AS3 |
hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 13 | LEPR | Sponge network | -0.939 | 4.0E-5 | -1.236 | 0 | 0.343 |
| 98 | RP11-594N15.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-374b-3p | 13 | PRLR | Sponge network | -2.86 | 0 | -1.308 | 0.00047 | 0.342 |
| 99 | MAGI2-AS3 |
hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | GHR | Sponge network | -0.939 | 4.0E-5 | -1.225 | 0 | 0.339 |
| 100 | AC006116.24 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-93-5p | 10 | IL6R | Sponge network | -1.832 | 1.0E-5 | -0.049 | 0.80939 | 0.333 |
| 101 | CTD-2008P7.9 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-28-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-454-3p;hsa-miR-576-5p | 24 | IL6ST | Sponge network | -1.202 | 0.00093 | -1.519 | 0 | 0.333 |
| 102 | LINC00284 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p | 17 | PIK3R1 | Sponge network | -4.159 | 0 | -1.219 | 0 | 0.333 |
| 103 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-589-3p | 17 | IL6ST | Sponge network | 0.187 | 0.31051 | -1.519 | 0 | 0.331 |
| 104 | CTD-2015G9.2 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-93-5p | 15 | IL6R | Sponge network | -3.112 | 5.0E-5 | -0.049 | 0.80939 | 0.328 |
| 105 | DNM3OS |
hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-224-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-96-5p | 11 | PIK3R1 | Sponge network | 0.932 | 0.00442 | -1.219 | 0 | 0.328 |
| 106 | NOP14-AS1 | hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-181c-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-497-5p | 12 | SPRY4 | Sponge network | 0.939 | 0 | 1.651 | 0 | 0.327 |
| 107 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-205-3p;hsa-miR-21-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-708-3p;hsa-miR-708-5p | 18 | SPRY3 | Sponge network | -0.939 | 4.0E-5 | -0.516 | 0.00013 | 0.325 |
| 108 | SNHG14 |
hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-92a-3p | 10 | SPRED1 | Sponge network | -1.125 | 0.0001 | -0.094 | 0.38958 | 0.318 |
| 109 | DLGAP1-AS5 |
hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-589-3p | 10 | PIK3R1 | Sponge network | -6.207 | 0 | -1.219 | 0 | 0.315 |
| 110 | RP1-193H18.2 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-183-5p;hsa-miR-205-3p;hsa-miR-21-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-708-3p;hsa-miR-96-5p | 10 | SPRY3 | Sponge network | -1.539 | 0 | -0.516 | 0.00013 | 0.315 |
| 111 | MIR497HG |
hsa-let-7a-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-205-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-590-5p;hsa-miR-96-5p | 13 | LIFR | Sponge network | -1.263 | 0.00248 | -1.519 | 0 | 0.314 |
| 112 | RP11-284N8.3 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-944;hsa-miR-96-5p | 10 | PRLR | Sponge network | 0.003 | 0.99478 | -1.308 | 0.00047 | 0.314 |
| 113 | RP11-594N15.3 |
hsa-let-7a-3p;hsa-miR-1301-3p;hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-374b-3p | 23 | IL6ST | Sponge network | -2.86 | 0 | -1.519 | 0 | 0.313 |
| 114 | LINC00707 |
hsa-miR-148a-5p;hsa-miR-181c-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-20b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-497-5p | 11 | SPRY4 | Sponge network | 2.018 | 1.0E-5 | 1.651 | 0 | 0.311 |
| 115 | CTD-2620I22.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-501-5p | 10 | IL6R | Sponge network | -1.967 | 0.00106 | -0.049 | 0.80939 | 0.308 |
| 116 | TPTEP1 |
hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p | 13 | PIK3R1 | Sponge network | -2.193 | 0 | -1.219 | 0 | 0.308 |
| 117 | LINC00702 |
hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 12 | LEPR | Sponge network | -0.573 | 0.0699 | -1.236 | 0 | 0.307 |
| 118 | LINC00957 |
hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p | 10 | PIK3R1 | Sponge network | -0.677 | 2.0E-5 | -1.219 | 0 | 0.306 |
| 119 | EMX2OS |
hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 11 | LEPR | Sponge network | -1.459 | 1.0E-5 | -1.236 | 0 | 0.304 |
| 120 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p;hsa-miR-944 | 11 | PRLR | Sponge network | -1.293 | 0.01561 | -1.308 | 0.00047 | 0.298 |
| 121 | MAGI2-AS3 |
hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-30c-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 14 | EP300 | Sponge network | -0.939 | 4.0E-5 | -0.038 | 0.77828 | 0.297 |
| 122 | RP11-193H5.1 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-320a | 11 | PIK3R1 | Sponge network | -0.426 | 0.01062 | -1.219 | 0 | 0.297 |
| 123 | AC007743.1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 11 | PRLR | Sponge network | -1.053 | 0.00923 | -1.308 | 0.00047 | 0.295 |
| 124 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 21 | PIK3R1 | Sponge network | -0.573 | 0.0699 | -1.219 | 0 | 0.294 |
| 125 | RP4-798P15.3 |
hsa-let-7a-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-205-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-29b-1-5p;hsa-miR-500a-5p;hsa-miR-590-5p;hsa-miR-944;hsa-miR-96-5p | 13 | LIFR | Sponge network | -1.213 | 0.00098 | -1.519 | 0 | 0.293 |
| 126 | RP11-693J15.4 |
hsa-let-7a-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-205-5p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-590-5p | 10 | LIFR | Sponge network | -0.732 | 0.18487 | -1.519 | 0 | 0.29 |
| 127 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-944;hsa-miR-96-5p | 16 | PRLR | Sponge network | -0.882 | 5.0E-5 | -1.308 | 0.00047 | 0.289 |
| 128 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 26 | IL6ST | Sponge network | -0.882 | 5.0E-5 | -1.519 | 0 | 0.288 |
| 129 | FAM225B | hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-1-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p | 14 | IL6ST | Sponge network | 2.186 | 0 | -1.519 | 0 | 0.287 |
| 130 | RP4-798P15.3 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-590-5p | 14 | IL6R | Sponge network | -1.213 | 0.00098 | -0.049 | 0.80939 | 0.286 |
| 131 | CTD-2008P7.9 |
hsa-miR-103a-2-5p;hsa-miR-125a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-576-5p | 11 | IL6R | Sponge network | -1.202 | 0.00093 | -0.049 | 0.80939 | 0.283 |
| 132 | LINC00900 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 18 | SPRED1 | Sponge network | -1.803 | 0 | -0.094 | 0.38958 | 0.283 |
| 133 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-183-5p;hsa-miR-205-3p;hsa-miR-21-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-708-5p | 10 | SPRY3 | Sponge network | -0.793 | 0 | -0.516 | 0.00013 | 0.282 |
| 134 | RP11-193H5.1 |
hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-500a-5p;hsa-miR-93-3p | 10 | SPRED1 | Sponge network | -0.426 | 0.01062 | -0.094 | 0.38958 | 0.281 |
| 135 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-576-5p | 10 | PRLR | Sponge network | -0.304 | 0.38627 | -1.308 | 0.00047 | 0.279 |
| 136 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-205-3p;hsa-miR-21-3p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-708-5p | 12 | SPRY3 | Sponge network | -1.712 | 0 | -0.516 | 0.00013 | 0.279 |
| 137 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | PRLR | Sponge network | -4.323 | 0 | -1.308 | 0.00047 | 0.278 |
| 138 | AC007743.1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-500a-5p;hsa-miR-92a-3p | 10 | SPRED1 | Sponge network | -1.053 | 0.00923 | -0.094 | 0.38958 | 0.277 |
| 139 | AF127936.7 |
hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-532-5p;hsa-miR-7-1-3p | 13 | IL6ST | Sponge network | 0.453 | 0.00811 | -1.519 | 0 | 0.276 |
| 140 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-339-5p;hsa-miR-576-5p | 10 | SPRY3 | Sponge network | -0.304 | 0.38627 | -0.516 | 0.00013 | 0.276 |
| 141 | CTD-2540L5.5 | hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-589-3p | 10 | IL6ST | Sponge network | -3.856 | 0 | -1.519 | 0 | 0.275 |
| 142 | CTC-205M6.5 | hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-1-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-7-1-3p | 10 | IL6ST | Sponge network | 0.381 | 0.01756 | -1.519 | 0 | 0.273 |
| 143 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-944 | 15 | PRLR | Sponge network | -1.712 | 0 | -1.308 | 0.00047 | 0.271 |
| 144 | RP11-321G12.1 | hsa-miR-103a-3p;hsa-miR-130a-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-589-3p | 16 | IL6ST | Sponge network | -3.552 | 0 | -1.519 | 0 | 0.27 |
| 145 | CTC-297N7.7 |
hsa-miR-103a-3p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-224-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-96-5p | 11 | PIK3R1 | Sponge network | -1.666 | 0.01558 | -1.219 | 0 | 0.267 |
| 146 | CTD-2554C21.3 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-455-3p;hsa-miR-629-3p | 10 | PIK3R1 | Sponge network | -2.118 | 6.0E-5 | -1.219 | 0 | 0.267 |
| 147 | RP11-227H15.4 |
hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-497-5p | 10 | SPRY4 | Sponge network | 4.405 | 0 | 1.651 | 0 | 0.267 |
| 148 | SNHG14 |
hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-589-3p | 18 | IL6ST | Sponge network | -1.125 | 0.0001 | -1.519 | 0 | 0.264 |
| 149 | SH3RF3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-576-5p;hsa-miR-590-5p | 11 | IL6R | Sponge network | -0.175 | 0.58985 | -0.049 | 0.80939 | 0.264 |
| 150 | RP11-757G1.6 |
hsa-miR-103a-3p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-93-3p | 16 | IL6ST | Sponge network | -1.346 | 0.00088 | -1.519 | 0 | 0.263 |
| 151 | LINC00900 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-205-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-708-3p;hsa-miR-708-5p | 17 | SPRY3 | Sponge network | -1.803 | 0 | -0.516 | 0.00013 | 0.261 |
| 152 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 14 | PRLR | Sponge network | -0.175 | 0.58985 | -1.308 | 0.00047 | 0.258 |
| 153 | AC108142.1 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-429 | 10 | SPRED1 | Sponge network | 2.31 | 0 | -0.094 | 0.38958 | 0.256 |
| 154 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-589-3p | 18 | IL6ST | Sponge network | -0.33 | 0.08136 | -1.519 | 0 | 0.254 |
| 155 | RP11-456K23.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-205-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-708-5p | 11 | SPRY3 | Sponge network | -0.223 | 0.27461 | -0.516 | 0.00013 | 0.252 |
| 156 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 14 | PRLR | Sponge network | -0.218 | 0.34607 | -1.308 | 0.00047 | 0.251 |
| 157 | RP11-594N15.3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-93-5p | 14 | SPRED1 | Sponge network | -2.86 | 0 | -0.094 | 0.38958 | 0.251 |