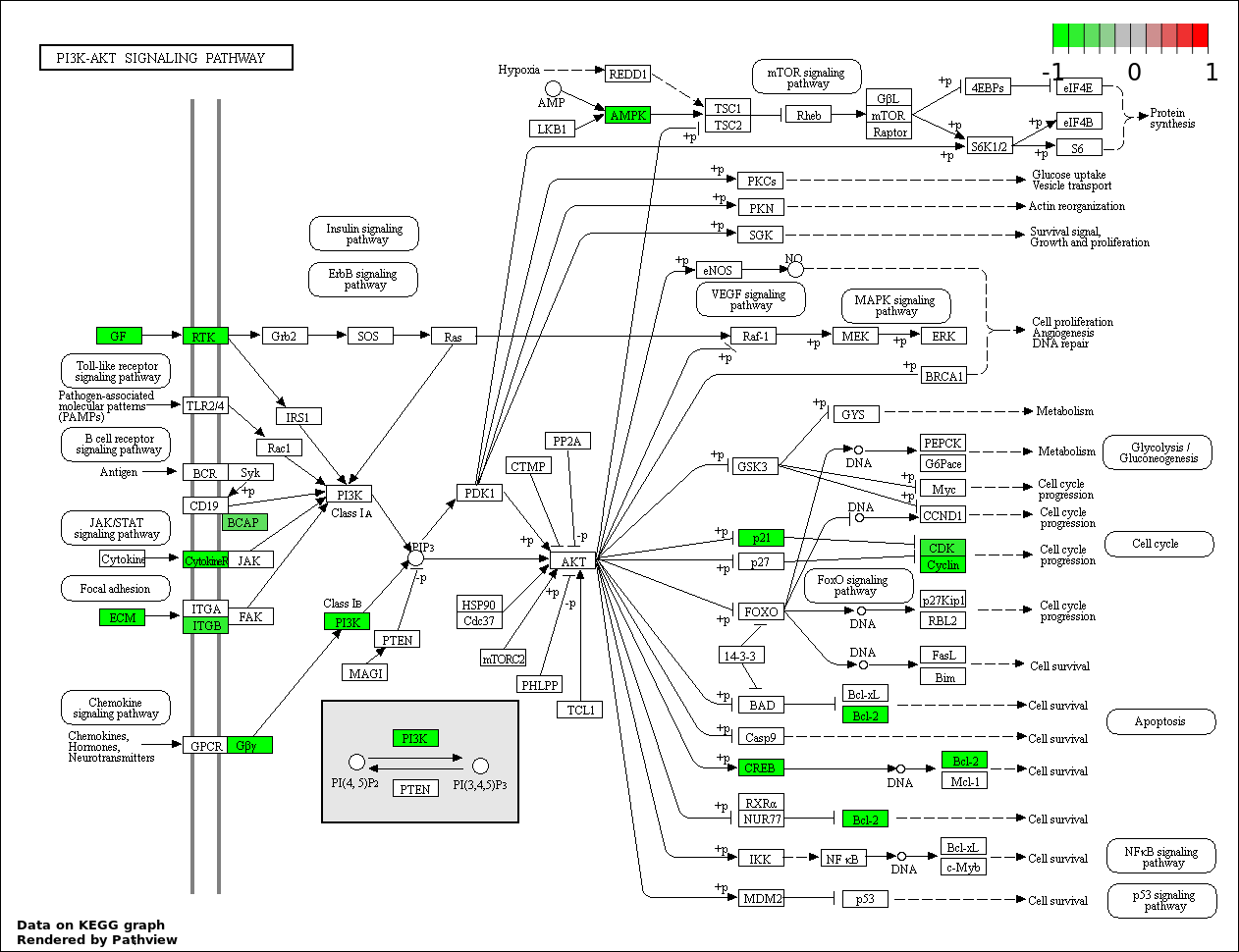
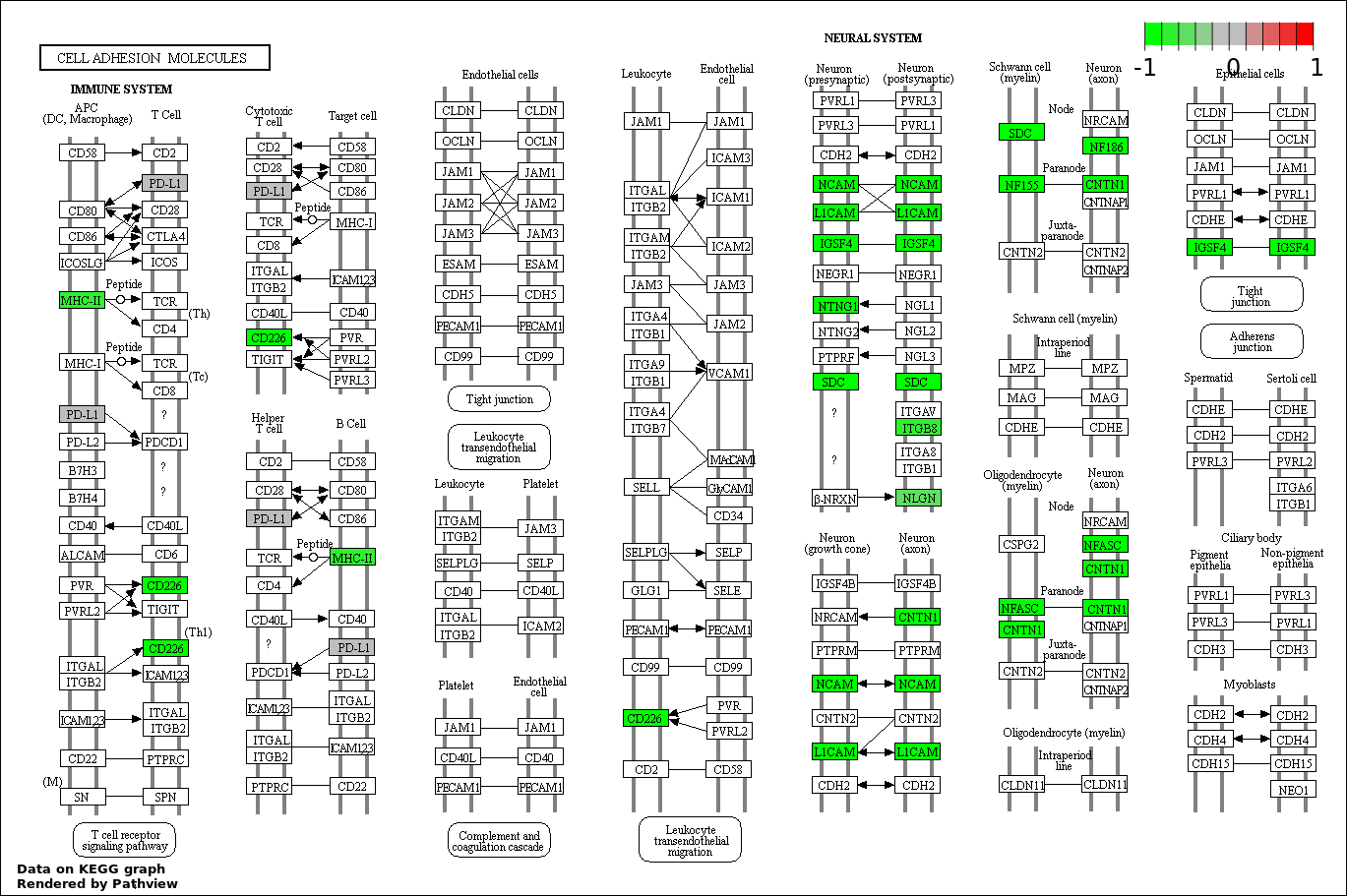
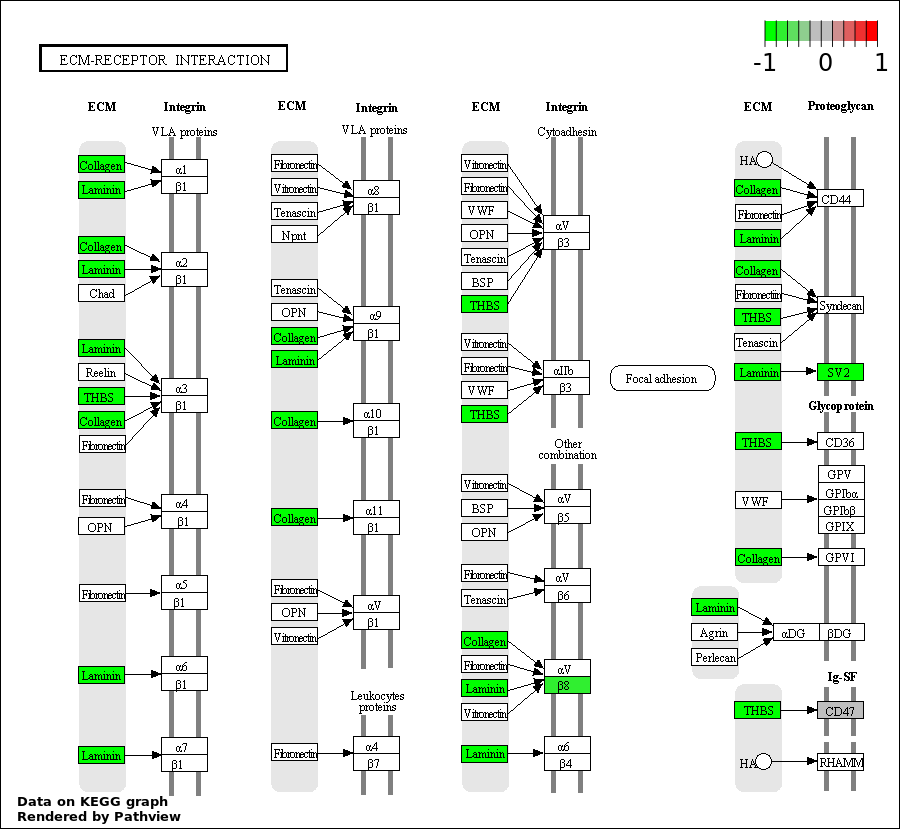
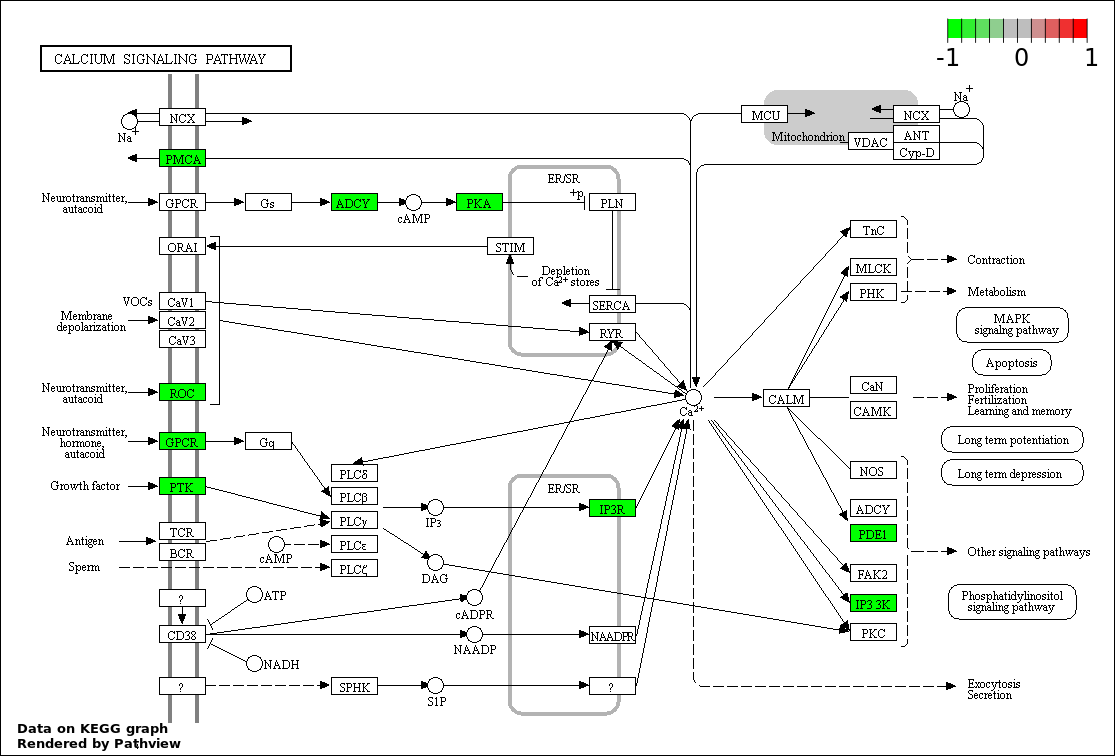
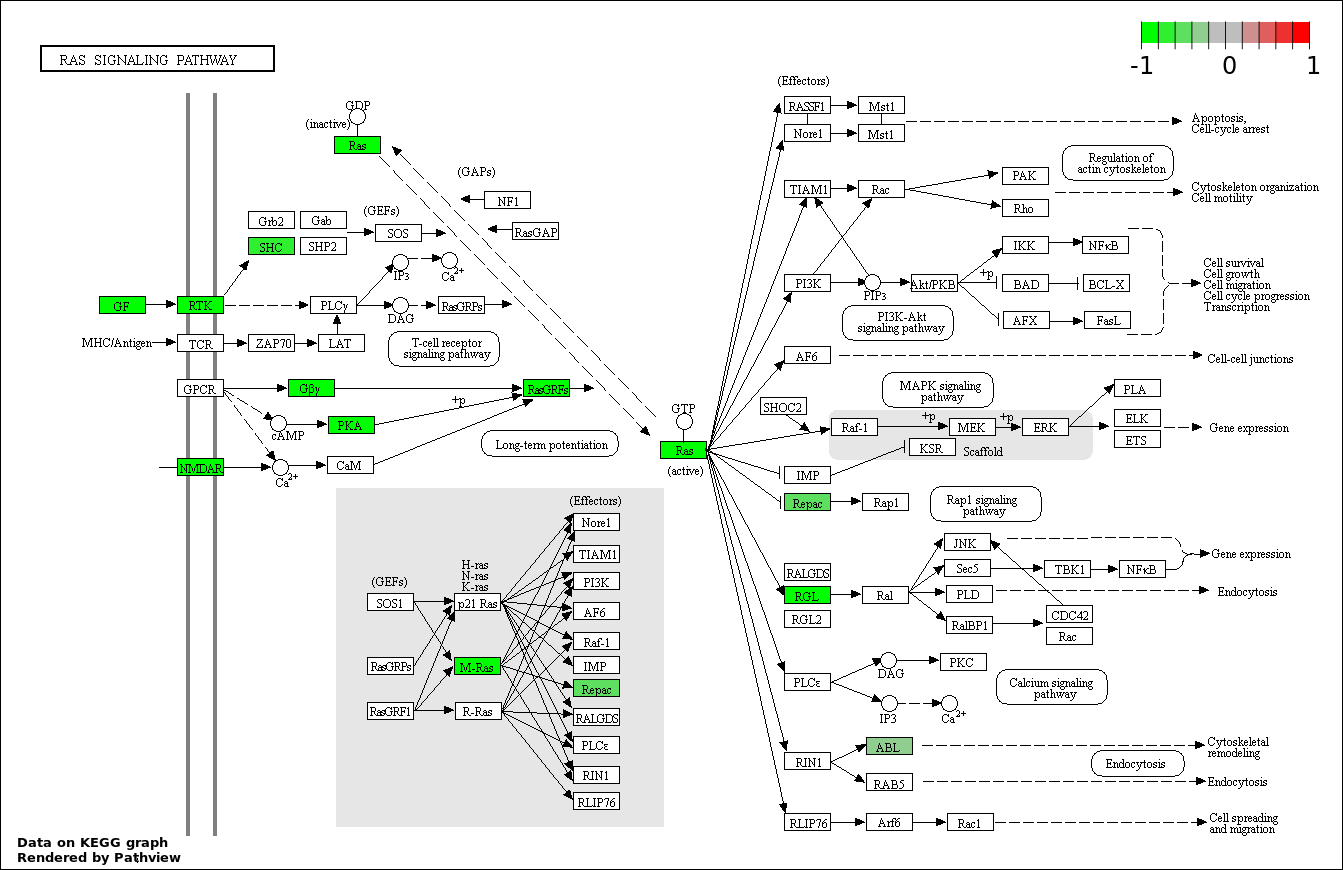
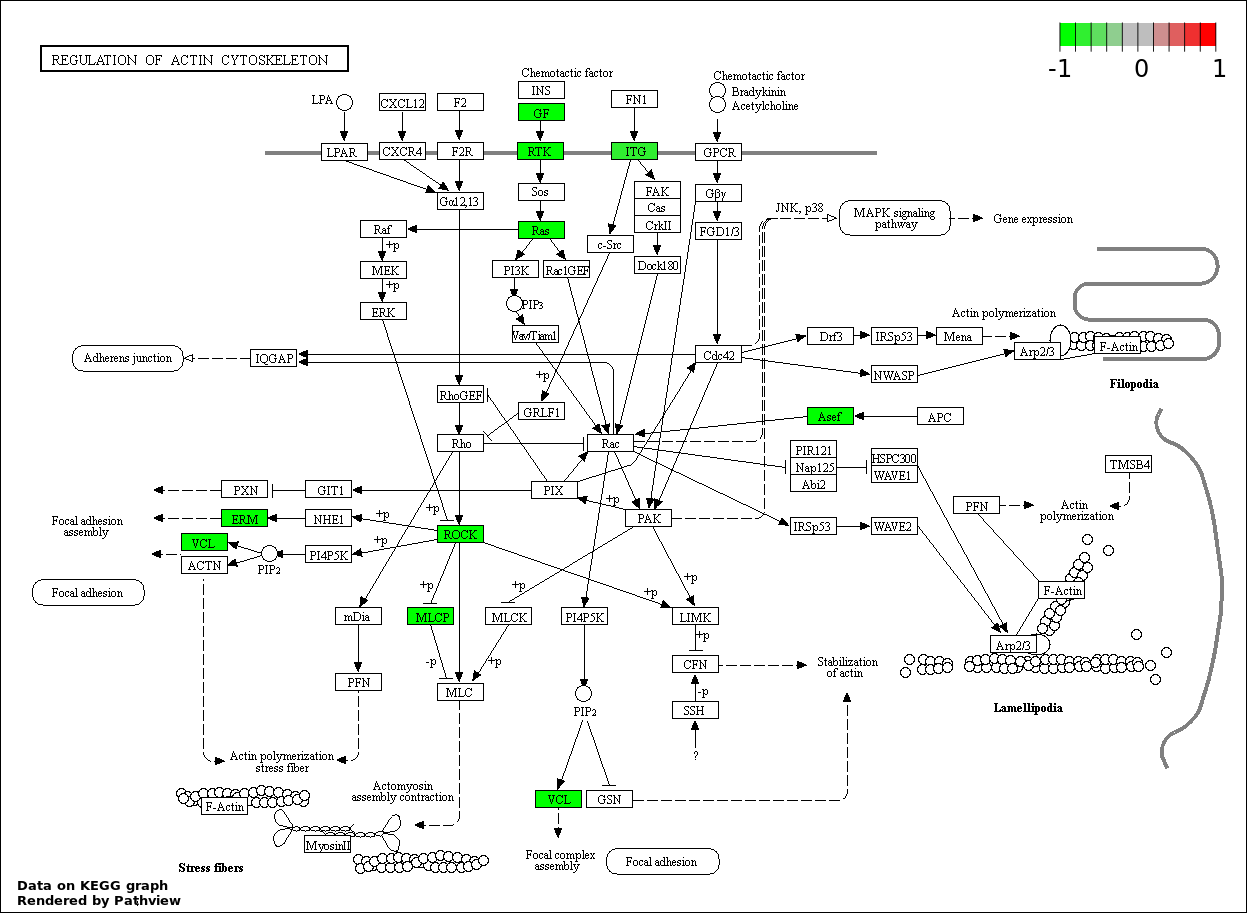
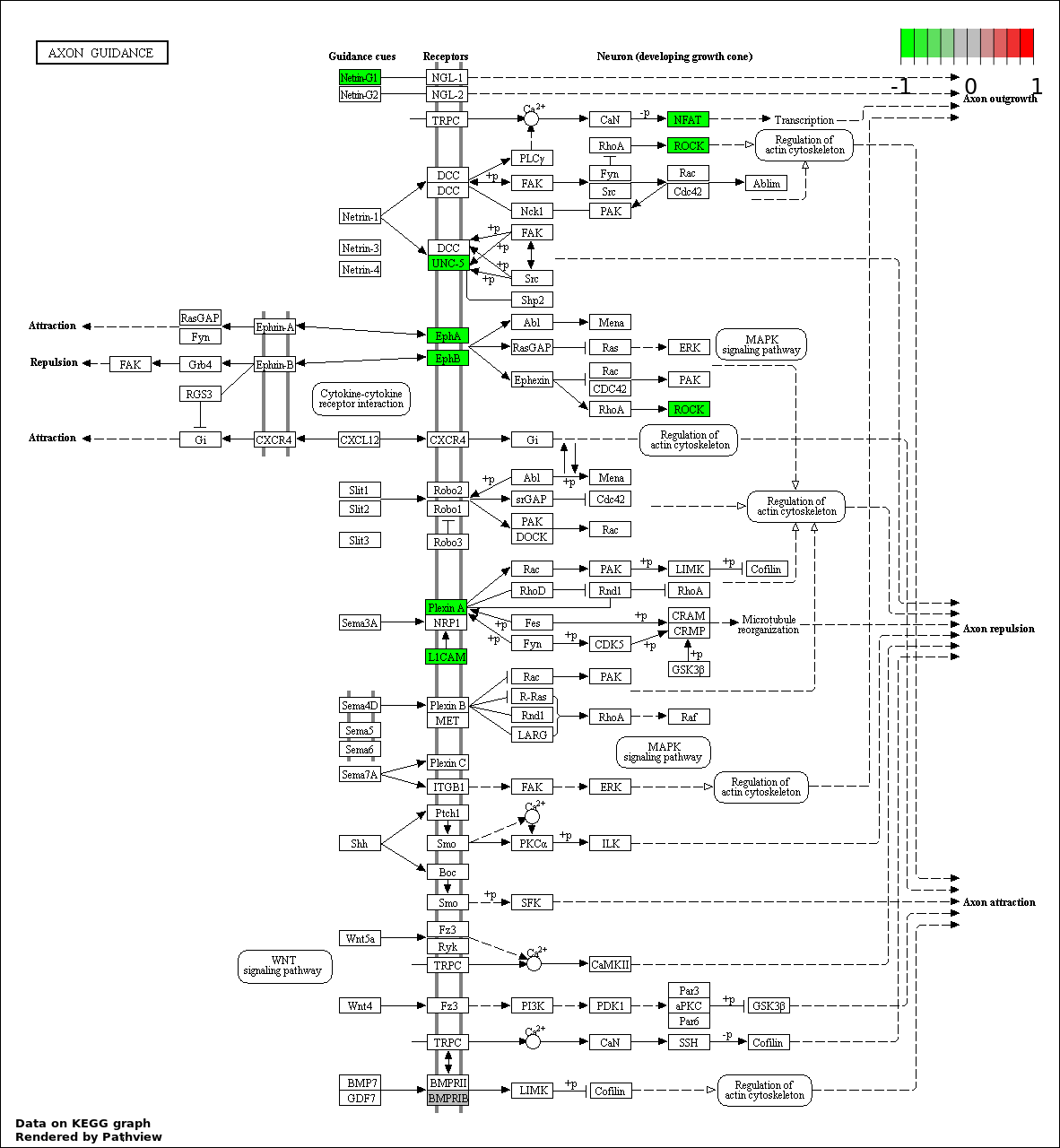
This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-182-5p | AASS | 3.54 | 0 | -1.35 | 5.0E-5 | MirTarget | -0.12 | 0.00483 | NA | |
| 2 | hsa-miR-182-5p | AATK | 3.54 | 0 | -0.62 | 0.02571 | MirTarget | -0.11 | 0.00069 | NA | |
| 3 | hsa-miR-182-5p | ABL2 | 3.54 | 0 | -0.32 | 0.04554 | mirMAP | -0.14 | 0 | NA | |
| 4 | hsa-miR-182-5p | ACVR1 | 3.54 | 0 | -0.44 | 0.01143 | MirTarget; miRNATAP | -0.15 | 0 | NA | |
| 5 | hsa-miR-182-5p | ACVRL1 | 3.54 | 0 | -1.25 | 0 | mirMAP | -0.25 | 0 | NA | |
| 6 | hsa-miR-182-5p | ADAM19 | 3.54 | 0 | -1.06 | 0.01895 | miRNATAP | -0.53 | 0 | NA | |
| 7 | hsa-miR-182-5p | ADAMTS18 | 3.54 | 0 | -0.74 | 0.14599 | miRNATAP | -0.22 | 0.00048 | NA | |
| 8 | hsa-miR-182-5p | ADAMTS4 | 3.54 | 0 | -2.17 | 0 | MirTarget | -0.42 | 0 | NA | |
| 9 | hsa-miR-182-5p | ADCY2 | 3.54 | 0 | -3.87 | 0 | miRNATAP | -0.4 | 0 | NA | |
| 10 | hsa-miR-182-5p | AFAP1 | 3.54 | 0 | -0.98 | 0.00012 | mirMAP | -0.26 | 0 | NA | |
| 11 | hsa-miR-182-5p | AFF2 | 3.54 | 0 | -1.73 | 0.00201 | mirMAP | -0.22 | 0.00162 | NA | |
| 12 | hsa-miR-182-5p | AK5 | 3.54 | 0 | -1.18 | 0.03248 | MirTarget | -0.16 | 0.02037 | NA | |
| 13 | hsa-miR-182-5p | AKAP7 | 3.54 | 0 | -1.26 | 1.0E-5 | miRNATAP | -0.14 | 6.0E-5 | NA | |
| 14 | hsa-miR-182-5p | AMOTL2 | 3.54 | 0 | -1.01 | 0.00015 | miRNATAP | -0.24 | 0 | NA | |
| 15 | hsa-miR-182-5p | ANK2 | 3.54 | 0 | -4.32 | 0 | miRNATAP | -0.75 | 0 | NA | |
| 16 | hsa-miR-182-5p | ANTXR2 | 3.54 | 0 | -2.31 | 0 | miRNATAP | -0.45 | 0 | NA | |
| 17 | hsa-miR-182-5p | ARHGAP6 | 3.54 | 0 | -3.25 | 0 | MirTarget | -0.21 | 0.00033 | NA | |
| 18 | hsa-miR-182-5p | ARHGEF3 | 3.54 | 0 | -0.76 | 0 | MirTarget | -0.11 | 0 | NA | |
| 19 | hsa-miR-182-5p | ARHGEF4 | 3.54 | 0 | -1.44 | 0.0131 | mirMAP | -0.35 | 0 | NA | |
| 20 | hsa-miR-182-5p | ARL4C | 3.54 | 0 | -0.28 | 0.33535 | miRNATAP | -0.19 | 0 | NA | |
| 21 | hsa-miR-182-5p | ASAP1 | 3.54 | 0 | 0.06 | 0.77944 | mirMAP | -0.11 | 2.0E-5 | NA | |
| 22 | hsa-miR-182-5p | ASB1 | 3.54 | 0 | -0.87 | 0 | mirMAP | -0.16 | 0 | NA | |
| 23 | hsa-miR-182-5p | ASXL3 | 3.54 | 0 | -3.34 | 0 | mirMAP | -0.37 | 0 | NA | |
| 24 | hsa-miR-182-5p | ATP10A | 3.54 | 0 | -0.81 | 0.03741 | mirMAP | -0.4 | 0 | NA | |
| 25 | hsa-miR-182-5p | ATP10D | 3.54 | 0 | -0.69 | 0.01004 | MirTarget | -0.22 | 0 | NA | |
| 26 | hsa-miR-182-5p | ATP2B4 | 3.54 | 0 | -1.72 | 0 | mirMAP | -0.29 | 0 | NA | |
| 27 | hsa-miR-182-5p | ATP8A1 | 3.54 | 0 | -1.48 | 8.0E-5 | MirTarget; miRNATAP | -0.16 | 0.00055 | NA | |
| 28 | hsa-miR-182-5p | ATXN1 | 3.54 | 0 | -1.51 | 0 | mirMAP; miRNATAP | -0.31 | 0 | NA | |
| 29 | hsa-miR-182-5p | B3GNT7 | 3.54 | 0 | -0.34 | 0.37613 | mirMAP | -0.13 | 0.00434 | NA | |
| 30 | hsa-miR-182-5p | BACE1 | 3.54 | 0 | -0.72 | 0.00096 | mirMAP | -0.23 | 0 | NA | |
| 31 | hsa-miR-182-5p | BACH2 | 3.54 | 0 | -1.52 | 0.00134 | MirTarget; mirMAP; miRNATAP | -0.44 | 0 | NA | |
| 32 | hsa-miR-182-5p | BCL2 | 3.54 | 0 | -2.02 | 0 | miRNAWalker2 validate; miRTarBase; mirMAP | -0.18 | 0 | 23936432; 26870290 | Inhibition of proliferation and induction of autophagy by atorvastatin in PC3 prostate cancer cells correlate with downregulation of Bcl2 and upregulation of miR 182 and p21; Bcl2 and p21 were identified to be potential target genes of miR-182 in PC3 cells;The expression levels of B-cell lymphoma-2 Bcl-2 and microRNA-182 miR-182 were detected using western blot analysis and quantitative reverse transcription-polymerase chain reaction respectively; Mangiferin treatment was also able to significantly reduce Bcl-2 expression levels and enhance miR-182 expression in PC3 cells; Finally it was observed that mangiferin inhibited proliferation and induced apoptosis in PC3 human prostate cancer cells and this effect was correlated with downregulation of Bcl-2 and upregulation of miR-182 |
| 33 | hsa-miR-182-5p | BDNF | 3.54 | 0 | -2.91 | 0 | MirTarget; miRNATAP | -0.3 | 0 | NA | |
| 34 | hsa-miR-182-5p | BHLHE22 | 3.54 | 0 | -1.78 | 1.0E-5 | MirTarget; miRNATAP | -0.13 | 0.01116 | NA | |
| 35 | hsa-miR-182-5p | BMPER | 3.54 | 0 | -2.38 | 0 | MirTarget | -0.31 | 0 | NA | |
| 36 | hsa-miR-182-5p | BMPR1B | 3.54 | 0 | 0.07 | 0.91404 | MirTarget | -0.35 | 1.0E-5 | NA | |
| 37 | hsa-miR-182-5p | BNC2 | 3.54 | 0 | -2.95 | 0 | MirTarget; miRNATAP | -0.72 | 0 | NA | |
| 38 | hsa-miR-182-5p | BRMS1L | 3.54 | 0 | -0.51 | 0.00391 | MirTarget | -0.12 | 0 | NA | |
| 39 | hsa-miR-182-5p | BTLA | 3.54 | 0 | -1.31 | 0.00559 | MirTarget | -0.38 | 0 | NA | |
| 40 | hsa-miR-182-5p | C15orf52 | 3.54 | 0 | -1.18 | 0.0026 | mirMAP | -0.16 | 0.00121 | NA | |
| 41 | hsa-miR-182-5p | C17orf51 | 3.54 | 0 | -0.68 | 0.02254 | mirMAP | -0.23 | 0 | NA | |
| 42 | hsa-miR-182-5p | C1orf21 | 3.54 | 0 | -1.51 | 0 | mirMAP | -0.14 | 0 | NA | |
| 43 | hsa-miR-182-5p | CACNA2D1 | 3.54 | 0 | -3.32 | 0 | miRNATAP | -0.8 | 0 | NA | |
| 44 | hsa-miR-182-5p | CACNB2 | 3.54 | 0 | -2.97 | 0 | miRNATAP | -0.37 | 0 | NA | |
| 45 | hsa-miR-182-5p | CACNB4 | 3.54 | 0 | -2.87 | 0 | miRNATAP | -0.32 | 0 | NA | |
| 46 | hsa-miR-182-5p | CADM1 | 3.54 | 0 | -1.19 | 0.01251 | miRNATAP | -0.14 | 0.01801 | 24445397 | TGF β upregulates miR 182 expression to promote gallbladder cancer metastasis by targeting CADM1; We further identified that the cell adhesion molecule1 CADM1 is a new target gene of miR-182 miR-182 negatively regulates CADM1 expression in vitro and in vivo |
| 47 | hsa-miR-182-5p | CADM2 | 3.54 | 0 | -3.84 | 0 | MirTarget; mirMAP; miRNATAP | -0.26 | 0.00033 | NA | |
| 48 | hsa-miR-182-5p | CALD1 | 3.54 | 0 | -2.47 | 0 | MirTarget | -0.57 | 0 | NA | |
| 49 | hsa-miR-182-5p | CBFA2T3 | 3.54 | 0 | -1.56 | 2.0E-5 | MirTarget; miRNATAP | -0.1 | 0.02527 | NA | |
| 50 | hsa-miR-182-5p | CBX6 | 3.54 | 0 | -1.54 | 0 | miRNAWalker2 validate | -0.19 | 0 | NA | |
| 51 | hsa-miR-182-5p | CCDC88A | 3.54 | 0 | -0.76 | 0.01199 | miRNATAP | -0.27 | 0 | NA | |
| 52 | hsa-miR-182-5p | CCND2 | 3.54 | 0 | -2.43 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.42 | 0 | NA | |
| 53 | hsa-miR-182-5p | CD209 | 3.54 | 0 | -1.78 | 0.00035 | mirMAP | -0.64 | 0 | NA | |
| 54 | hsa-miR-182-5p | CD226 | 3.54 | 0 | -1.61 | 0.00025 | MirTarget | -0.43 | 0 | NA | |
| 55 | hsa-miR-182-5p | CD274 | 3.54 | 0 | 0.06 | 0.90454 | mirMAP | -0.32 | 0 | NA | |
| 56 | hsa-miR-182-5p | CD47 | 3.54 | 0 | -0.16 | 0.4808 | MirTarget; miRNATAP | -0.13 | 0 | NA | |
| 57 | hsa-miR-182-5p | CD59 | 3.54 | 0 | -0.7 | 0.0013 | MirTarget | -0.14 | 0 | NA | |
| 58 | hsa-miR-182-5p | CDC42BPA | 3.54 | 0 | -1.41 | 0 | MirTarget; miRNATAP | -0.19 | 0 | NA | |
| 59 | hsa-miR-182-5p | CDC42EP3 | 3.54 | 0 | -1.35 | 0 | mirMAP | -0.25 | 0 | NA | |
| 60 | hsa-miR-182-5p | CDK6 | 3.54 | 0 | -0.77 | 0.06479 | mirMAP | -0.34 | 0 | NA | |
| 61 | hsa-miR-182-5p | CDKN1A | 3.54 | 0 | -1.29 | 0 | miRNAWalker2 validate | -0.11 | 0.00142 | NA | |
| 62 | hsa-miR-182-5p | CDKN2C | 3.54 | 0 | 0.23 | 0.43313 | MirTarget | -0.13 | 0.00017 | NA | |
| 63 | hsa-miR-182-5p | CDV3 | 3.54 | 0 | -0.42 | 0.0021 | MirTarget; miRNATAP | -0.13 | 0 | NA | |
| 64 | hsa-miR-182-5p | CECR1 | 3.54 | 0 | -0.68 | 0.05777 | mirMAP | -0.15 | 0.0004 | NA | |
| 65 | hsa-miR-182-5p | CELF2 | 3.54 | 0 | -3.05 | 0 | mirMAP; miRNATAP | -0.54 | 0 | NA | |
| 66 | hsa-miR-182-5p | CHL1 | 3.54 | 0 | -2.42 | 0.00043 | MirTarget; miRNATAP | -0.31 | 0.0003 | 24971532 | miR 182 targets CHL1 and controls tumor growth and invasion in papillary thyroid carcinoma; Bioinformatics analysis revealed close homolog of LI CHL1 as a potential target of miR-182; Upregulation of miR-182 was significantly correlated with CHL1 downregulation in human PTC tissues and cell lines; miR-182 suppressed the expression of CHL1 mRNA through direct targeting of the 3'-untranslated region 3'-UTR; Silencing of CHL1 counteracted the effects of miR-182 suppression while its overexpression mimicked these effects; Our data collectively indicate that miR-182 in PTC promotes cell proliferation and invasion through direct suppression of CHL1 supporting the potential utility of miR-182 inhibition as a novel therapeutic strategy against PTC |
| 67 | hsa-miR-182-5p | CHST10 | 3.54 | 0 | -0.67 | 0.06109 | miRNATAP | -0.16 | 0.00034 | NA | |
| 68 | hsa-miR-182-5p | CHST11 | 3.54 | 0 | 0.54 | 0.2311 | miRNATAP | -0.37 | 0 | NA | |
| 69 | hsa-miR-182-5p | CLIC5 | 3.54 | 0 | -1.44 | 0.00056 | miRNATAP | -0.31 | 0 | NA | |
| 70 | hsa-miR-182-5p | CLUAP1 | 3.54 | 0 | -0.82 | 3.0E-5 | miRNATAP | -0.12 | 0 | NA | |
| 71 | hsa-miR-182-5p | CNN3 | 3.54 | 0 | -0.66 | 0.00535 | MirTarget | -0.16 | 0 | NA | |
| 72 | hsa-miR-182-5p | CNTN1 | 3.54 | 0 | -4.98 | 0 | MirTarget; miRNATAP | -0.87 | 0 | NA | |
| 73 | hsa-miR-182-5p | COL4A4 | 3.54 | 0 | -2.67 | 0 | MirTarget | -0.36 | 0 | NA | |
| 74 | hsa-miR-182-5p | CORO1C | 3.54 | 0 | -1.12 | 0 | MirTarget; miRNATAP | -0.3 | 0 | NA | |
| 75 | hsa-miR-182-5p | CPEB1 | 3.54 | 0 | -4.68 | 0 | miRNATAP | -0.75 | 0 | NA | |
| 76 | hsa-miR-182-5p | CREB3L1 | 3.54 | 0 | -1.81 | 3.0E-5 | MirTarget | -0.39 | 0 | NA | |
| 77 | hsa-miR-182-5p | CREB5 | 3.54 | 0 | -2.28 | 0 | mirMAP | -0.46 | 0 | NA | |
| 78 | hsa-miR-182-5p | CRISPLD2 | 3.54 | 0 | -2.53 | 0 | mirMAP | -0.42 | 0 | NA | |
| 79 | hsa-miR-182-5p | CSMD2 | 3.54 | 0 | 0.91 | 0.0162 | mirMAP | -0.22 | 0 | NA | |
| 80 | hsa-miR-182-5p | CYLD | 3.54 | 0 | -0.89 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.17 | 0 | 27476169 | We then demonstrated that knockdown of miR-182 up-regulated the expression of cylindromatosis CYLD deubiquitinase which promoted the formation of death-inducing signaling complex DISC and subsequent caspase-8 activation in TNF-α-treated BT-549 cells |
| 81 | hsa-miR-182-5p | DAAM2 | 3.54 | 0 | -2.23 | 0 | mirMAP | -0.38 | 0 | NA | |
| 82 | hsa-miR-182-5p | DACT1 | 3.54 | 0 | -1.78 | 2.0E-5 | MirTarget | -0.43 | 0 | NA | |
| 83 | hsa-miR-182-5p | DENND5B | 3.54 | 0 | -1.35 | 0.0001 | MirTarget; miRNATAP | -0.33 | 0 | NA | |
| 84 | hsa-miR-182-5p | DLG2 | 3.54 | 0 | -3.8 | 0 | miRNATAP | -0.2 | 0.00137 | NA | |
| 85 | hsa-miR-182-5p | DPF3 | 3.54 | 0 | -1.67 | 1.0E-5 | mirMAP | -0.43 | 0 | NA | |
| 86 | hsa-miR-182-5p | DTNA | 3.54 | 0 | -4.06 | 0 | mirMAP | -0.71 | 0 | NA | |
| 87 | hsa-miR-182-5p | DUSP3 | 3.54 | 0 | -1.04 | 0 | mirMAP | -0.22 | 0 | NA | |
| 88 | hsa-miR-182-5p | DYNC1I1 | 3.54 | 0 | -2.05 | 0.0001 | MirTarget | -0.23 | 0.00047 | NA | |
| 89 | hsa-miR-182-5p | EBF1 | 3.54 | 0 | -2.89 | 0 | miRNATAP | -0.42 | 0 | NA | |
| 90 | hsa-miR-182-5p | EBF3 | 3.54 | 0 | -2.22 | 0 | MirTarget; miRNATAP | -0.27 | 0 | NA | |
| 91 | hsa-miR-182-5p | EDIL3 | 3.54 | 0 | -1.4 | 0.00541 | MirTarget | -0.29 | 0 | NA | |
| 92 | hsa-miR-182-5p | EDNRB | 3.54 | 0 | -2.52 | 0 | miRNATAP | -0.22 | 0 | NA | |
| 93 | hsa-miR-182-5p | EGR3 | 3.54 | 0 | -4.25 | 0 | MirTarget | -0.52 | 0 | NA | |
| 94 | hsa-miR-182-5p | EIF4E3 | 3.54 | 0 | -1.31 | 0 | mirMAP | -0.11 | 7.0E-5 | NA | |
| 95 | hsa-miR-182-5p | EIF5A2 | 3.54 | 0 | -1.12 | 0.00497 | mirMAP | -0.35 | 0 | NA | |
| 96 | hsa-miR-182-5p | ELL2 | 3.54 | 0 | -1.08 | 1.0E-5 | MirTarget; miRNATAP | -0.2 | 0 | NA | |
| 97 | hsa-miR-182-5p | ELMO1 | 3.54 | 0 | -1.21 | 6.0E-5 | MirTarget | -0.3 | 0 | NA | |
| 98 | hsa-miR-182-5p | ENTPD1 | 3.54 | 0 | -1.12 | 0 | mirMAP | -0.26 | 0 | NA | |
| 99 | hsa-miR-182-5p | EOMES | 3.54 | 0 | -0.65 | 0.19494 | MirTarget | -0.34 | 0 | NA | |
| 100 | hsa-miR-182-5p | EPDR1 | 3.54 | 0 | -1.64 | 0.00035 | MirTarget | -0.47 | 0 | NA | |
| 101 | hsa-miR-182-5p | EPHA3 | 3.54 | 0 | -3.18 | 0 | miRNATAP | -0.43 | 0 | NA | |
| 102 | hsa-miR-182-5p | EPHA7 | 3.54 | 0 | -4.2 | 0 | miRNATAP | -0.29 | 0.00012 | NA | |
| 103 | hsa-miR-182-5p | EPHB1 | 3.54 | 0 | -2.07 | 0 | MirTarget; miRNATAP | -0.41 | 0 | NA | |
| 104 | hsa-miR-182-5p | EPM2A | 3.54 | 0 | -1.9 | 0 | miRNATAP | -0.25 | 0 | NA | |
| 105 | hsa-miR-182-5p | EPYC | 3.54 | 0 | 2.07 | 0.00523 | MirTarget | -0.36 | 7.0E-5 | NA | |
| 106 | hsa-miR-182-5p | ERG | 3.54 | 0 | -1.35 | 0 | mirMAP | -0.27 | 0 | NA | |
| 107 | hsa-miR-182-5p | F13A1 | 3.54 | 0 | -2.67 | 0 | miRNATAP | -0.6 | 0 | NA | |
| 108 | hsa-miR-182-5p | FAM107A | 3.54 | 0 | -5.01 | 0 | MirTarget | -0.48 | 0 | NA | |
| 109 | hsa-miR-182-5p | FAM124A | 3.54 | 0 | -2.09 | 0 | mirMAP | -0.47 | 0 | NA | |
| 110 | hsa-miR-182-5p | FAM134B | 3.54 | 0 | -1.39 | 0.00067 | MirTarget; miRNATAP | -0.19 | 0.00013 | NA | |
| 111 | hsa-miR-182-5p | FAM167A | 3.54 | 0 | 0.27 | 0.48779 | miRNATAP | -0.17 | 0.00044 | NA | |
| 112 | hsa-miR-182-5p | FAM168A | 3.54 | 0 | -0.72 | 0.00481 | MirTarget | -0.16 | 0 | NA | |
| 113 | hsa-miR-182-5p | FAM171A1 | 3.54 | 0 | -1.6 | 4.0E-5 | miRNATAP | -0.2 | 2.0E-5 | NA | |
| 114 | hsa-miR-182-5p | FAM26E | 3.54 | 0 | -1.51 | 4.0E-5 | mirMAP | -0.43 | 0 | NA | |
| 115 | hsa-miR-182-5p | FAM78A | 3.54 | 0 | -0.18 | 0.57167 | MirTarget; miRNATAP | -0.27 | 0 | NA | |
| 116 | hsa-miR-182-5p | FAT3 | 3.54 | 0 | -2.79 | 0 | mirMAP | -0.31 | 3.0E-5 | NA | |
| 117 | hsa-miR-182-5p | FBN1 | 3.54 | 0 | -1.82 | 5.0E-5 | miRNATAP | -0.55 | 0 | NA | |
| 118 | hsa-miR-182-5p | FBXO32 | 3.54 | 0 | -0.93 | 0.00122 | mirMAP | -0.37 | 0 | NA | |
| 119 | hsa-miR-182-5p | FER | 3.54 | 0 | -0.96 | 0.00017 | mirMAP | -0.21 | 0 | NA | |
| 120 | hsa-miR-182-5p | FGF9 | 3.54 | 0 | -4.26 | 0 | miRTarBase; miRNATAP | -0.2 | 0.00291 | NA | |
| 121 | hsa-miR-182-5p | FMNL3 | 3.54 | 0 | 0.14 | 0.461 | miRNAWalker2 validate | -0.15 | 0 | NA | |
| 122 | hsa-miR-182-5p | FMO2 | 3.54 | 0 | -2.41 | 0.00014 | MirTarget | -0.65 | 0 | NA | |
| 123 | hsa-miR-182-5p | FOXN3 | 3.54 | 0 | -1.44 | 0 | MirTarget; miRNATAP | -0.15 | 0 | NA | |
| 124 | hsa-miR-182-5p | FOXO1 | 3.54 | 0 | -1.06 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.13 | 0 | 24865442; 26640590; 23313739 | Immunohistochemical analysis revealed that the levels of FOXO1 and FOXO3A two potential miR-182 targets are reduced in AA tumors;MiR 182 Is Associated with Growth Migration and Invasion in Prostate Cancer via Suppression of FOXO1; Our findings suggest that miR-182 may act to increase prostate cancer proliferation migration and invasion through suppression of FOXO1;Western blot flow cytometry and pathway analysis for the HeLa cells with miR-182 over/down-expression in vitro showed that miR-182 was involved in apoptosis and cell cycle pathways it also associated with the regulation of FOXO1 |
| 125 | hsa-miR-182-5p | FOXP2 | 3.54 | 0 | -3.99 | 0 | miRNATAP | -0.53 | 0 | NA | |
| 126 | hsa-miR-182-5p | FYCO1 | 3.54 | 0 | -1.62 | 0 | miRNATAP | -0.18 | 0 | NA | |
| 127 | hsa-miR-182-5p | GAS7 | 3.54 | 0 | -2.67 | 0 | mirMAP | -0.56 | 0 | NA | |
| 128 | hsa-miR-182-5p | GCNT1 | 3.54 | 0 | 0.04 | 0.92747 | miRNATAP | -0.27 | 0 | NA | |
| 129 | hsa-miR-182-5p | GFOD1 | 3.54 | 0 | -0.94 | 0.00661 | mirMAP | -0.33 | 0 | NA | |
| 130 | hsa-miR-182-5p | GIMAP1 | 3.54 | 0 | -1.24 | 6.0E-5 | MirTarget | -0.37 | 0 | NA | |
| 131 | hsa-miR-182-5p | GJC1 | 3.54 | 0 | -0.97 | 0.00222 | mirMAP | -0.24 | 0 | NA | |
| 132 | hsa-miR-182-5p | GLB1L | 3.54 | 0 | -0.51 | 0.00565 | MirTarget | -0.14 | 0 | NA | |
| 133 | hsa-miR-182-5p | GLI2 | 3.54 | 0 | -2.29 | 0 | MirTarget | -0.51 | 0 | NA | |
| 134 | hsa-miR-182-5p | GNG7 | 3.54 | 0 | -3.48 | 0 | mirMAP | -0.33 | 0 | NA | |
| 135 | hsa-miR-182-5p | GPR132 | 3.54 | 0 | -0.61 | 0.10569 | mirMAP | -0.34 | 0 | NA | |
| 136 | hsa-miR-182-5p | GPR183 | 3.54 | 0 | -2.34 | 0 | MirTarget | -0.51 | 0 | NA | |
| 137 | hsa-miR-182-5p | GPR3 | 3.54 | 0 | -0.96 | 0.00053 | mirMAP | -0.13 | 0.00022 | NA | |
| 138 | hsa-miR-182-5p | GPRASP2 | 3.54 | 0 | -1.46 | 0 | miRNATAP | -0.2 | 0 | NA | |
| 139 | hsa-miR-182-5p | GRIA3 | 3.54 | 0 | -2.07 | 7.0E-5 | miRNATAP | -0.45 | 0 | NA | |
| 140 | hsa-miR-182-5p | GRID1 | 3.54 | 0 | -0.62 | 0.16867 | miRNATAP | -0.19 | 0.00039 | NA | |
| 141 | hsa-miR-182-5p | GRIN2A | 3.54 | 0 | -3.32 | 0 | mirMAP | -0.43 | 0 | NA | |
| 142 | hsa-miR-182-5p | GRIN3A | 3.54 | 0 | -0.24 | 0.40319 | mirMAP | -0.2 | 0 | NA | |
| 143 | hsa-miR-182-5p | GXYLT2 | 3.54 | 0 | -1.72 | 0.00323 | MirTarget | -0.68 | 0 | NA | |
| 144 | hsa-miR-182-5p | HBEGF | 3.54 | 0 | -2.26 | 0 | MirTarget; miRNATAP | -0.25 | 0 | NA | |
| 145 | hsa-miR-182-5p | HDAC9 | 3.54 | 0 | -1.52 | 0.00072 | miRNATAP | -0.39 | 0 | 26858310 | HDAC Inhibition Induces MicroRNA 182 which Targets RAD51 and Impairs HR Repair to Sensitize Cells to Sapacitabine in Acute Myelogenous Leukemia; HR reporter assays apoptotic assays and colony-forming assays established that the miR-182 as well as the HDAC inhibition-mediated decreases in RAD51 inhibited HR repair and sensitized cells to sapacitabine; HDAC inhibition induced miR-182 in AML cell lines and primary AML blasts; Overexpression of miR-182 as well as HDAC inhibition-mediated induction of miR-182 were linked to time- and dose-dependent decreases in the levels of RAD51 an inhibition of HR increased levels of residual damage and decreased survival after exposure to double-strand damage-inducing agents; Our findings define the mechanism by which HDAC inhibition induces miR-182 to target RAD51 and highlights a novel pharmacologic strategy that compromises the ability of AML cells to conduct HR thereby sensitizing AML cells to DNA-damaging agents that activate HR as a repair and potential resistance mechanism |
| 146 | hsa-miR-182-5p | HIF3A | 3.54 | 0 | -4.98 | 0 | mirMAP | -0.6 | 0 | NA | |
| 147 | hsa-miR-182-5p | HLA-DOA | 3.54 | 0 | -0.79 | 0.07866 | mirMAP | -0.36 | 0 | NA | |
| 148 | hsa-miR-182-5p | HMCN1 | 3.54 | 0 | -0.31 | 0.47227 | MirTarget | -0.33 | 0 | NA | |
| 149 | hsa-miR-182-5p | HOXA9 | 3.54 | 0 | -1 | 0.05496 | miRNATAP | -0.15 | 0.02061 | NA | |
| 150 | hsa-miR-182-5p | HSPA13 | 3.54 | 0 | -0.29 | 0.10635 | miRNATAP | -0.1 | 0 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | MOVEMENT OF CELL OR SUBCELLULAR COMPONENT | 70 | 1275 | 6.771e-16 | 3.151e-12 |
| 2 | CELL DEVELOPMENT | 70 | 1426 | 1.755e-13 | 4.084e-10 |
| 3 | LOCOMOTION | 57 | 1114 | 8.73e-12 | 1.354e-08 |
| 4 | REGULATION OF CELL DIFFERENTIATION | 68 | 1492 | 1.245e-11 | 1.448e-08 |
| 5 | CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 36 | 513 | 1.796e-11 | 1.671e-08 |
| 6 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 71 | 1672 | 1.014e-10 | 7.717e-08 |
| 7 | CELLULAR COMPONENT MORPHOGENESIS | 48 | 900 | 1.161e-10 | 7.717e-08 |
| 8 | REGULATION OF CELLULAR COMPONENT MOVEMENT | 43 | 771 | 3.002e-10 | 1.746e-07 |
| 9 | CARDIOVASCULAR SYSTEM DEVELOPMENT | 43 | 788 | 5.88e-10 | 2.736e-07 |
| 10 | CIRCULATORY SYSTEM DEVELOPMENT | 43 | 788 | 5.88e-10 | 2.736e-07 |
| 11 | REGULATION OF ACTIN FILAMENT BASED PROCESS | 25 | 312 | 1.692e-09 | 7.156e-07 |
| 12 | CELL MOTILITY | 43 | 835 | 3.387e-09 | 1.145e-06 |
| 13 | LOCALIZATION OF CELL | 43 | 835 | 3.387e-09 | 1.145e-06 |
| 14 | TISSUE DEVELOPMENT | 63 | 1518 | 3.444e-09 | 1.145e-06 |
| 15 | REGULATION OF CELL ADHESION | 36 | 629 | 4.73e-09 | 1.467e-06 |
| 16 | NEUROGENESIS | 59 | 1402 | 7.123e-09 | 2.071e-06 |
| 17 | BIOLOGICAL ADHESION | 48 | 1032 | 1.023e-08 | 2.799e-06 |
| 18 | CELL MORPHOGENESIS INVOLVED IN NEURON DIFFERENTIATION | 26 | 368 | 1.112e-08 | 2.874e-06 |
| 19 | REGULATION OF ADHERENS JUNCTION ORGANIZATION | 10 | 50 | 2.917e-08 | 7.142e-06 |
| 20 | REGULATION OF CELL DEVELOPMENT | 41 | 836 | 3.169e-08 | 7.373e-06 |
| 21 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 49 | 1142 | 8.812e-08 | 1.952e-05 |
| 22 | REGULATION OF NEURON DIFFERENTIATION | 31 | 554 | 9.687e-08 | 2.049e-05 |
| 23 | VASCULATURE DEVELOPMENT | 28 | 469 | 1.1e-07 | 2.226e-05 |
| 24 | REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 37 | 750 | 1.346e-07 | 2.609e-05 |
| 25 | INTRACELLULAR SIGNAL TRANSDUCTION | 60 | 1572 | 1.708e-07 | 3.179e-05 |
| 26 | REGULATION OF CELLULAR COMPONENT BIOGENESIS | 37 | 767 | 2.345e-07 | 4.196e-05 |
| 27 | NEURON PROJECTION MORPHOGENESIS | 25 | 402 | 2.507e-07 | 4.32e-05 |
| 28 | NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 43 | 983 | 3.491e-07 | 5.801e-05 |
| 29 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 38 | 823 | 4.759e-07 | 7.636e-05 |
| 30 | CELL JUNCTION ORGANIZATION | 16 | 185 | 5.445e-07 | 8.446e-05 |
| 31 | MEMBRANE DEPOLARIZATION DURING ACTION POTENTIAL | 8 | 39 | 5.917e-07 | 8.851e-05 |
| 32 | REGULATION OF CELL JUNCTION ASSEMBLY | 10 | 68 | 6.087e-07 | 8.851e-05 |
| 33 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 43 | 1021 | 9.583e-07 | 0.0001351 |
| 34 | REGULATION OF CELL MATRIX ADHESION | 11 | 90 | 1.124e-06 | 0.0001538 |
| 35 | NEURON DEVELOPMENT | 33 | 687 | 1.194e-06 | 0.0001587 |
| 36 | ACTION POTENTIAL | 11 | 94 | 1.742e-06 | 0.0002191 |
| 37 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 42 | 1008 | 1.703e-06 | 0.0002191 |
| 38 | MEMBRANE DEPOLARIZATION | 9 | 61 | 2.175e-06 | 0.000231 |
| 39 | ACTIN FILAMENT BASED PROCESS | 25 | 450 | 1.984e-06 | 0.000231 |
| 40 | NEURON PROJECTION DEVELOPMENT | 28 | 545 | 2.213e-06 | 0.000231 |
| 41 | REGULATION OF CHONDROCYTE DIFFERENTIATION | 8 | 46 | 2.234e-06 | 0.000231 |
| 42 | PROTEIN PHOSPHORYLATION | 40 | 944 | 2.033e-06 | 0.000231 |
| 43 | MUSCLE SYSTEM PROCESS | 19 | 282 | 2.197e-06 | 0.000231 |
| 44 | NEURON DIFFERENTIATION | 38 | 874 | 2.034e-06 | 0.000231 |
| 45 | NEURON PROJECTION GUIDANCE | 16 | 205 | 2.13e-06 | 0.000231 |
| 46 | REGULATION OF CELL ACTIVATION | 26 | 484 | 2.293e-06 | 0.0002319 |
| 47 | POSITIVE REGULATION OF CELL PROLIFERATION | 36 | 814 | 2.611e-06 | 0.0002531 |
| 48 | MUSCLE CONTRACTION | 17 | 233 | 2.588e-06 | 0.0002531 |
| 49 | EPITHELIAL CELL DEVELOPMENT | 15 | 186 | 2.958e-06 | 0.0002809 |
| 50 | EPITHELIAL CELL DIFFERENTIATION | 26 | 495 | 3.45e-06 | 0.0003211 |
| 51 | RESPONSE TO ENDOGENOUS STIMULUS | 53 | 1450 | 3.576e-06 | 0.0003263 |
| 52 | POSITIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 24 | 437 | 3.881e-06 | 0.0003284 |
| 53 | CELLULAR RESPONSE TO ORGANIC SUBSTANCE | 63 | 1848 | 3.854e-06 | 0.0003284 |
| 54 | MESENCHYME DEVELOPMENT | 15 | 190 | 3.848e-06 | 0.0003284 |
| 55 | STEM CELL DIFFERENTIATION | 15 | 190 | 3.848e-06 | 0.0003284 |
| 56 | POSITIVE REGULATION OF GENE EXPRESSION | 60 | 1733 | 4.217e-06 | 0.0003407 |
| 57 | MEMBRANE DEPOLARIZATION DURING CARDIAC MUSCLE CELL ACTION POTENTIAL | 5 | 14 | 4.247e-06 | 0.0003407 |
| 58 | MULTICELLULAR ORGANISMAL SIGNALING | 12 | 123 | 4.135e-06 | 0.0003407 |
| 59 | CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND | 35 | 799 | 4.468e-06 | 0.0003507 |
| 60 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 57 | 1618 | 4.522e-06 | 0.0003507 |
| 61 | REGULATION OF CELL SUBSTRATE ADHESION | 14 | 173 | 6.071e-06 | 0.0004631 |
| 62 | REGULATION OF CELL MORPHOGENESIS | 27 | 552 | 8.29e-06 | 0.0006221 |
| 63 | REGULATION OF CELL PROLIFERATION | 53 | 1496 | 8.736e-06 | 0.0006452 |
| 64 | NEGATIVE REGULATION OF ADHERENS JUNCTION ORGANIZATION | 5 | 16 | 8.979e-06 | 0.0006528 |
| 65 | MESENCHYMAL CELL DIFFERENTIATION | 12 | 134 | 1.004e-05 | 0.0007189 |
| 66 | CELL PROJECTION ORGANIZATION | 37 | 902 | 1.04e-05 | 0.0007329 |
| 67 | TAXIS | 24 | 464 | 1.063e-05 | 0.0007385 |
| 68 | ACTIN FILAMENT BASED MOVEMENT | 10 | 93 | 1.109e-05 | 0.0007589 |
| 69 | HEART DEVELOPMENT | 24 | 466 | 1.142e-05 | 0.0007699 |
| 70 | EPITHELIUM DEVELOPMENT | 38 | 945 | 1.229e-05 | 0.0008166 |
| 71 | CARDIAC MUSCLE CELL CONTRACTION | 6 | 29 | 1.506e-05 | 0.0009871 |
| 72 | REGULATION OF SYNAPSE ASSEMBLY | 9 | 79 | 1.91e-05 | 0.001234 |
| 73 | REGULATION OF PROTEIN MODIFICATION PROCESS | 57 | 1710 | 2.295e-05 | 0.001463 |
| 74 | REGULATION OF HEART CONTRACTION | 15 | 221 | 2.364e-05 | 0.001467 |
| 75 | TUBE DEVELOPMENT | 26 | 552 | 2.337e-05 | 0.001467 |
| 76 | REGULATION OF CARTILAGE DEVELOPMENT | 8 | 63 | 2.501e-05 | 0.001511 |
| 77 | BLOOD VESSEL MORPHOGENESIS | 20 | 364 | 2.478e-05 | 0.001511 |
| 78 | REGULATION OF TRANSMEMBRANE TRANSPORT | 22 | 426 | 2.541e-05 | 0.001516 |
| 79 | POSITIVE REGULATION OF NEURON DIFFERENTIATION | 18 | 306 | 2.58e-05 | 0.00152 |
| 80 | REGULATION OF CELL PROJECTION ORGANIZATION | 26 | 558 | 2.808e-05 | 0.001633 |
| 81 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 33 | 801 | 2.924e-05 | 0.00168 |
| 82 | NEGATIVE REGULATION OF CELL JUNCTION ASSEMBLY | 5 | 20 | 2.993e-05 | 0.001698 |
| 83 | MUSCLE STRUCTURE DEVELOPMENT | 22 | 432 | 3.139e-05 | 0.00176 |
| 84 | CELL PART MORPHOGENESIS | 28 | 633 | 3.499e-05 | 0.001938 |
| 85 | CELL JUNCTION ASSEMBLY | 11 | 129 | 3.709e-05 | 0.00203 |
| 86 | REGULATION OF CYTOSKELETON ORGANIZATION | 24 | 502 | 3.808e-05 | 0.00206 |
| 87 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 58 | 1784 | 3.933e-05 | 0.002103 |
| 88 | REGULATION OF NEURON PROJECTION DEVELOPMENT | 21 | 408 | 4.067e-05 | 0.00215 |
| 89 | REGULATION OF SYSTEM PROCESS | 24 | 507 | 4.454e-05 | 0.002329 |
| 90 | REGULATION OF CELL DEATH | 50 | 1472 | 4.818e-05 | 0.002491 |
| 91 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 48 | 1395 | 5.045e-05 | 0.002551 |
| 92 | POST EMBRYONIC DEVELOPMENT | 9 | 89 | 4.991e-05 | 0.002551 |
| 93 | MUSCLE CELL DIFFERENTIATION | 15 | 237 | 5.299e-05 | 0.002651 |
| 94 | ENDOTHELIUM DEVELOPMENT | 9 | 90 | 5.453e-05 | 0.002671 |
| 95 | ESTABLISHMENT OF PROTEIN LOCALIZATION TO PLASMA MEMBRANE | 9 | 90 | 5.453e-05 | 0.002671 |
| 96 | REGULATION OF SYNAPSE ORGANIZATION | 10 | 113 | 6.087e-05 | 0.00295 |
| 97 | POSITIVE REGULATION OF LOCOMOTION | 21 | 420 | 6.166e-05 | 0.002958 |
| 98 | CELLULAR RESPONSE TO HORMONE STIMULUS | 25 | 552 | 6.328e-05 | 0.003005 |
| 99 | CARDIAC MUSCLE CELL ACTION POTENTIAL | 6 | 37 | 6.482e-05 | 0.003047 |
| 100 | ENDOTHELIAL CELL DIFFERENTIATION | 8 | 72 | 6.661e-05 | 0.003099 |
| 101 | NEGATIVE REGULATION OF GENE EXPRESSION | 50 | 1493 | 6.883e-05 | 0.003171 |
| 102 | CELLULAR RESPONSE TO LIPID | 22 | 457 | 7.23e-05 | 0.003298 |
| 103 | SENSORY ORGAN DEVELOPMENT | 23 | 493 | 7.983e-05 | 0.003606 |
| 104 | ACTIN MEDIATED CELL CONTRACTION | 8 | 74 | 8.115e-05 | 0.00363 |
| 105 | PHOSPHORYLATION | 43 | 1228 | 8.669e-05 | 0.003842 |
| 106 | SENSORY PERCEPTION OF PAIN | 8 | 75 | 8.935e-05 | 0.003903 |
| 107 | CARDIOCYTE DIFFERENTIATION | 9 | 96 | 9.058e-05 | 0.003903 |
| 108 | NEGATIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 30 | 740 | 9.046e-05 | 0.003903 |
| 109 | REGULATION OF MEMBRANE POTENTIAL | 18 | 343 | 0.0001124 | 0.004799 |
| 110 | REGULATION OF IMMUNE SYSTEM PROCESS | 47 | 1403 | 0.000115 | 0.004835 |
| 111 | POSITIVE REGULATION OF CELL DEVELOPMENT | 22 | 472 | 0.0001153 | 0.004835 |
| 112 | NEGATIVE REGULATION OF CELL DIFFERENTIATION | 26 | 609 | 0.0001183 | 0.004913 |
| 113 | REGULATION OF CELL CELL ADHESION | 19 | 380 | 0.0001372 | 0.00565 |
| 114 | POSITIVE REGULATION OF SYNAPSE ASSEMBLY | 7 | 61 | 0.0001552 | 0.006333 |
| 115 | CARDIAC CONDUCTION | 8 | 82 | 0.0001682 | 0.006804 |
| 116 | REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 13 | 207 | 0.0001786 | 0.007164 |
| 117 | REGULATION OF BLOOD CIRCULATION | 16 | 295 | 0.0001844 | 0.007333 |
| 118 | RESPONSE TO HORMONE | 33 | 893 | 0.0002288 | 0.009023 |
| 119 | SMOOTH MUSCLE CELL DIFFERENTIATION | 5 | 30 | 0.0002352 | 0.009045 |
| 120 | REGULATION OF HEART RATE BY CARDIAC CONDUCTION | 5 | 30 | 0.0002352 | 0.009045 |
| 121 | REGULATION OF MEMBRANE REPOLARIZATION | 5 | 30 | 0.0002352 | 0.009045 |
| 122 | CELL ACTIVATION | 24 | 568 | 0.0002508 | 0.009411 |
| 123 | ION TRANSMEMBRANE TRANSPORT | 31 | 822 | 0.0002501 | 0.009411 |
| 124 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 48 | 1492 | 0.0002495 | 0.009411 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | ENZYME BINDING | 64 | 1737 | 2.204e-07 | 0.0002047 |
| 2 | ACTIN BINDING | 24 | 393 | 6.066e-07 | 0.0002818 |
| 3 | PROTEIN KINASE ACTIVITY | 30 | 640 | 6.012e-06 | 0.001117 |
| 4 | KINASE ACTIVITY | 36 | 842 | 5.573e-06 | 0.001117 |
| 5 | TRANSCRIPTION FACTOR ACTIVITY RNA POLYMERASE II CORE PROMOTER PROXIMAL REGION SEQUENCE SPECIFIC BINDING | 20 | 328 | 5.426e-06 | 0.001117 |
| 6 | PASSIVE TRANSMEMBRANE TRANSPORTER ACTIVITY | 24 | 464 | 1.063e-05 | 0.001368 |
| 7 | RNA POLYMERASE II TRANSCRIPTION FACTOR ACTIVITY SEQUENCE SPECIFIC DNA BINDING | 29 | 629 | 1.178e-05 | 0.001368 |
| 8 | NUCLEIC ACID BINDING TRANSCRIPTION FACTOR ACTIVITY | 45 | 1199 | 1.088e-05 | 0.001368 |
| 9 | TRANSMEMBRANE RECEPTOR PROTEIN KINASE ACTIVITY | 9 | 81 | 2.341e-05 | 0.002417 |
| 10 | TRANSCRIPTIONAL ACTIVATOR ACTIVITY RNA POLYMERASE II CORE PROMOTER PROXIMAL REGION SEQUENCE SPECIFIC BINDING | 15 | 226 | 3.068e-05 | 0.00285 |
| 11 | CYTOSKELETAL PROTEIN BINDING | 33 | 819 | 4.519e-05 | 0.003498 |
| 12 | KINASE BINDING | 27 | 606 | 4.272e-05 | 0.003498 |
| 13 | GATED CHANNEL ACTIVITY | 18 | 325 | 5.665e-05 | 0.004049 |
| 14 | CATION CHANNEL ACTIVITY | 17 | 298 | 6.312e-05 | 0.004188 |
| 15 | MACROMOLECULAR COMPLEX BINDING | 47 | 1399 | 0.0001076 | 0.006661 |
| 16 | TRANSCRIPTIONAL ACTIVATOR ACTIVITY RNA POLYMERASE II TRANSCRIPTION REGULATORY REGION SEQUENCE SPECIFIC BINDING | 17 | 315 | 0.0001241 | 0.006794 |
| 17 | GROWTH FACTOR BINDING | 10 | 123 | 0.0001243 | 0.006794 |
| 18 | CALCIUM ION TRANSMEMBRANE TRANSPORTER ACTIVITY | 10 | 128 | 0.0001729 | 0.008453 |
| 19 | TRANSFERASE ACTIVITY TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 36 | 992 | 0.0001689 | 0.008453 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CELL JUNCTION | 61 | 1151 | 3.178e-13 | 1.856e-10 |
| 2 | INTRINSIC COMPONENT OF PLASMA MEMBRANE | 65 | 1649 | 1.396e-08 | 4.077e-06 |
| 3 | CELL PROJECTION | 68 | 1786 | 2.396e-08 | 4.664e-06 |
| 4 | ACTIN CYTOSKELETON | 27 | 444 | 1.291e-07 | 1.885e-05 |
| 5 | NEURON PART | 49 | 1265 | 1.769e-06 | 0.0001872 |
| 6 | PLASMA MEMBRANE PROTEIN COMPLEX | 27 | 510 | 1.924e-06 | 0.0001872 |
| 7 | NEURON PROJECTION | 39 | 942 | 4.766e-06 | 0.0003534 |
| 8 | CELL CELL JUNCTION | 22 | 383 | 4.841e-06 | 0.0003534 |
| 9 | SYNAPSE | 33 | 754 | 8.626e-06 | 0.0005037 |
| 10 | ANCHORING JUNCTION | 25 | 489 | 8.501e-06 | 0.0005037 |
| 11 | CELL CORTEX | 16 | 238 | 1.431e-05 | 0.0006963 |
| 12 | MEMBRANE REGION | 43 | 1134 | 1.355e-05 | 0.0006963 |
| 13 | T TUBULE | 7 | 45 | 2.096e-05 | 0.0009414 |
| 14 | CELL CORTEX PART | 10 | 119 | 9.43e-05 | 0.003671 |
| 15 | CYTOSKELETON | 61 | 1967 | 9.416e-05 | 0.003671 |
| 16 | VOLTAGE GATED SODIUM CHANNEL COMPLEX | 4 | 14 | 0.0001115 | 0.004071 |
| 17 | CYTOPLASMIC REGION | 16 | 287 | 0.0001347 | 0.004369 |
| 18 | POSTSYNAPSE | 19 | 378 | 0.0001282 | 0.004369 |
| 19 | SARCOLEMMA | 10 | 125 | 0.0001421 | 0.004369 |
| 20 | CORTICAL CYTOSKELETON | 8 | 81 | 0.0001543 | 0.004505 |
| 21 | POSTSYNAPTIC MEMBRANE | 13 | 205 | 0.0001623 | 0.004512 |
| 22 | RUFFLE | 11 | 156 | 0.0002055 | 0.005454 |
| 23 | SODIUM CHANNEL COMPLEX | 4 | 17 | 0.0002535 | 0.006168 |
| 24 | CELL SUBSTRATE JUNCTION | 19 | 398 | 0.0002474 | 0.006168 |
| 25 | CATION CHANNEL COMPLEX | 11 | 167 | 0.00037 | 0.008644 |
| 26 | INTERCALATED DISC | 6 | 51 | 0.0004013 | 0.009014 |
| 27 | CELL LEADING EDGE | 17 | 350 | 0.0004261 | 0.009215 |
| 28 | DENDRITE | 20 | 451 | 0.0004439 | 0.009258 |
| 29 | PLASMA MEMBRANE REGION | 33 | 929 | 0.0004614 | 0.009292 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
| 1 | hsa04510_Focal_adhesion | 15 | 200 | 7.199e-06 | 0.001296 | |
| 2 | hsa04151_PI3K_AKT_signaling_pathway | 20 | 351 | 1.47e-05 | 0.001323 | |
| 3 | hsa04514_Cell_adhesion_molecules_.CAMs. | 11 | 136 | 6.03e-05 | 0.003618 | |
| 4 | hsa04010_MAPK_signaling_pathway | 15 | 268 | 0.0002076 | 0.00778 | |
| 5 | hsa04512_ECM.receptor_interaction | 8 | 85 | 0.0002161 | 0.00778 | |
| 6 | hsa04020_Calcium_signaling_pathway | 11 | 177 | 0.0006045 | 0.01618 | |
| 7 | hsa04014_Ras_signaling_pathway | 13 | 236 | 0.0006294 | 0.01618 | |
| 8 | hsa04810_Regulation_of_actin_cytoskeleton | 11 | 214 | 0.002773 | 0.06239 | |
| 9 | hsa04360_Axon_guidance | 8 | 130 | 0.003488 | 0.06977 | |
| 10 | hsa04350_TGF.beta_signaling_pathway | 6 | 85 | 0.005681 | 0.1023 | |
| 11 | hsa04270_Vascular_smooth_muscle_contraction | 7 | 116 | 0.006819 | 0.1116 | |
| 12 | hsa04962_Vasopressin.regulated_water_reabsorption | 4 | 44 | 0.009657 | 0.1404 | |
| 13 | hsa04115_p53_signaling_pathway | 5 | 69 | 0.01014 | 0.1404 | |
| 14 | hsa04722_Neurotrophin_signaling_pathway | 7 | 127 | 0.01096 | 0.1409 | |
| 15 | hsa04012_ErbB_signaling_pathway | 5 | 87 | 0.02534 | 0.2876 | |
| 16 | hsa04310_Wnt_signaling_pathway | 7 | 151 | 0.02583 | 0.2876 | |
| 17 | hsa04390_Hippo_signaling_pathway | 7 | 154 | 0.02835 | 0.2876 | |
| 18 | hsa04062_Chemokine_signaling_pathway | 8 | 189 | 0.02876 | 0.2876 | |
| 19 | hsa00533_Glycosaminoglycan_biosynthesis_._keratan_sulfate | 2 | 15 | 0.03227 | 0.3057 | |
| 20 | hsa04912_GnRH_signaling_pathway | 5 | 101 | 0.04397 | 0.3644 | |
| 21 | hsa04916_Melanogenesis | 5 | 101 | 0.04397 | 0.3644 | |
| 22 | hsa04720_Long.term_potentiation | 4 | 70 | 0.04454 | 0.3644 | |
| 23 | hsa04910_Insulin_signaling_pathway | 6 | 138 | 0.04886 | 0.3798 | |
| 24 | hsa00310_Lysine_degradation | 3 | 44 | 0.05117 | 0.3798 | |
| 25 | hsa04971_Gastric_acid_secretion | 4 | 74 | 0.05274 | 0.3798 | |
| 26 | hsa04630_Jak.STAT_signaling_pathway | 6 | 155 | 0.07653 | 0.5102 | |
| 27 | hsa04914_Progesterone.mediated_oocyte_maturation | 4 | 87 | 0.08461 | 0.5403 | |
| 28 | hsa04210_Apoptosis | 4 | 89 | 0.09019 | 0.5403 | |
| 29 | hsa04970_Salivary_secretion | 4 | 89 | 0.09019 | 0.5403 | |
| 30 | hsa04540_Gap_junction | 4 | 90 | 0.09305 | 0.5403 | |
| 31 | hsa04380_Osteoclast_differentiation | 5 | 128 | 0.09816 | 0.5521 | |
| 32 | hsa04520_Adherens_junction | 3 | 73 | 0.1625 | 0.859 | |
| 33 | hsa04662_B_cell_receptor_signaling_pathway | 3 | 75 | 0.1719 | 0.859 | |
| 34 | hsa04114_Oocyte_meiosis | 4 | 114 | 0.1731 | 0.859 | |
| 35 | hsa04145_Phagosome | 5 | 156 | 0.1777 | 0.859 | |
| 36 | hsa04260_Cardiac_muscle_contraction | 3 | 77 | 0.1813 | 0.859 | |
| 37 | hsa04670_Leukocyte_transendothelial_migration | 4 | 117 | 0.1844 | 0.859 | |
| 38 | hsa04070_Phosphatidylinositol_signaling_system | 3 | 78 | 0.1861 | 0.859 | |
| 39 | hsa00230_Purine_metabolism | 5 | 162 | 0.1972 | 0.8875 | |
| 40 | hsa00514_Other_types_of_O.glycan_biosynthesis | 2 | 46 | 0.2183 | 0.9484 | |
| 41 | hsa04110_Cell_cycle | 4 | 128 | 0.2278 | 0.9535 | |
| 42 | hsa04150_mTOR_signaling_pathway | 2 | 52 | 0.2607 | 1 | |
| 43 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 4 | 136 | 0.2608 | 1 | |
| 44 | hsa04340_Hedgehog_signaling_pathway | 2 | 56 | 0.289 | 1 | |
| 45 | hsa04972_Pancreatic_secretion | 3 | 101 | 0.3025 | 1 | |
| 46 | hsa04620_Toll.like_receptor_signaling_pathway | 3 | 102 | 0.3077 | 1 | |
| 47 | hsa04144_Endocytosis | 5 | 203 | 0.3448 | 1 | |
| 48 | hsa04610_Complement_and_coagulation_cascades | 2 | 69 | 0.3795 | 1 | |
| 49 | hsa04730_Long.term_depression | 2 | 70 | 0.3863 | 1 | |
| 50 | hsa04622_RIG.I.like_receptor_signaling_pathway | 2 | 71 | 0.3931 | 1 | |
| 51 | hsa04976_Bile_secretion | 2 | 71 | 0.3931 | 1 | |
| 52 | hsa04370_VEGF_signaling_pathway | 2 | 76 | 0.4263 | 1 | |
| 53 | hsa04640_Hematopoietic_cell_lineage | 2 | 88 | 0.5019 | 1 | |
| 54 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 2 | 95 | 0.5429 | 1 | |
| 55 | hsa04660_T_cell_receptor_signaling_pathway | 2 | 108 | 0.6127 | 1 | |
| 56 | hsa04141_Protein_processing_in_endoplasmic_reticulum | 2 | 168 | 0.8328 | 1 | |
| 57 | hsa04740_Olfactory_transduction | 2 | 388 | 0.9954 | 1 |