This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-34a-5p | ABHD2 | 0.13 | 0.13326 | 0.45 | 0.00053 | mirMAP | -0.12 | 0.01835 | NA | |
| 2 | hsa-miR-34a-5p | ACSL4 | 0.13 | 0.13326 | -1.48 | 0 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.11 | 0.02677 | NA | |
| 3 | hsa-miR-34a-5p | ADAM22 | 0.13 | 0.13326 | -1.22 | 0 | MirTarget; miRNATAP | -0.33 | 5.0E-5 | NA | |
| 4 | hsa-miR-34a-5p | ADCY1 | 0.13 | 0.13326 | -0.05 | 0.84149 | mirMAP | -0.24 | 0.01304 | NA | |
| 5 | hsa-miR-34a-5p | ADNP | 0.13 | 0.13326 | 0.29 | 6.0E-5 | miRNAWalker2 validate | -0.08 | 0.00869 | NA | |
| 6 | hsa-miR-34a-5p | AFAP1L2 | 0.13 | 0.13326 | -1.15 | 0 | miRNAWalker2 validate | -0.13 | 0.0145 | NA | |
| 7 | hsa-miR-34a-5p | AHNAK | 0.13 | 0.13326 | -1.77 | 0 | miRNAWalker2 validate | -0.15 | 0.00201 | NA | |
| 8 | hsa-miR-34a-5p | AK3 | 0.13 | 0.13326 | -0.93 | 0 | MirTarget | -0.07 | 0.01523 | NA | |
| 9 | hsa-miR-34a-5p | AKAP13 | 0.13 | 0.13326 | -0.61 | 0 | mirMAP | -0.11 | 0.0002 | NA | |
| 10 | hsa-miR-34a-5p | AKAP6 | 0.13 | 0.13326 | -2.28 | 0 | MirTarget | -0.13 | 0.0257 | NA | |
| 11 | hsa-miR-34a-5p | ALX4 | 0.13 | 0.13326 | -3.22 | 0 | mirMAP | -0.47 | 2.0E-5 | NA | |
| 12 | hsa-miR-34a-5p | AMOTL2 | 0.13 | 0.13326 | -1.57 | 0 | MirTarget | -0.09 | 0.03085 | NA | |
| 13 | hsa-miR-34a-5p | ANK2 | 0.13 | 0.13326 | -2.51 | 0 | MirTarget; miRNATAP | -0.32 | 0 | NA | |
| 14 | hsa-miR-34a-5p | ANK3 | 0.13 | 0.13326 | -0.03 | 0.84332 | miRNATAP | -0.14 | 0.00547 | NA | |
| 15 | hsa-miR-34a-5p | ANKRD52 | 0.13 | 0.13326 | 0.23 | 0.00024 | miRNATAP | -0.05 | 0.04535 | NA | |
| 16 | hsa-miR-34a-5p | ANKS1A | 0.13 | 0.13326 | -0.37 | 0 | MirTarget | -0.1 | 2.0E-5 | NA | |
| 17 | hsa-miR-34a-5p | ARHGAP19 | 0.13 | 0.13326 | -0.97 | 0 | mirMAP | -0.11 | 0.00019 | NA | |
| 18 | hsa-miR-34a-5p | ARHGEF33 | 0.13 | 0.13326 | -0.66 | 0 | MirTarget | -0.1 | 0.03642 | NA | |
| 19 | hsa-miR-34a-5p | ARID4A | 0.13 | 0.13326 | -0.86 | 0 | miRNATAP | -0.07 | 0.02073 | NA | |
| 20 | hsa-miR-34a-5p | ASB1 | 0.13 | 0.13326 | -0.93 | 0 | MirTarget; miRNATAP | -0.1 | 0.00018 | NA | |
| 21 | hsa-miR-34a-5p | ASTN1 | 0.13 | 0.13326 | -2.96 | 0 | MirTarget | -0.54 | 0 | NA | |
| 22 | hsa-miR-34a-5p | ASXL2 | 0.13 | 0.13326 | -0.86 | 0 | MirTarget | -0.17 | 0.00682 | NA | |
| 23 | hsa-miR-34a-5p | ATAD2B | 0.13 | 0.13326 | 0.12 | 0.0773 | miRNATAP | -0.06 | 0.02411 | NA | |
| 24 | hsa-miR-34a-5p | ATXN7 | 0.13 | 0.13326 | -0.52 | 0 | MirTarget; miRNATAP | -0.07 | 0.01757 | NA | |
| 25 | hsa-miR-34a-5p | BAZ2A | 0.13 | 0.13326 | -0.1 | 0.10091 | MirTarget; miRNATAP | -0.06 | 0.0086 | NA | |
| 26 | hsa-miR-34a-5p | BCL2 | 0.13 | 0.13326 | -0.9 | 0 | miRNAWalker2 validate; miRTarBase | -0.22 | 0.00143 | 24565525; 22623155; 21399894; 23155233; 24444609; 20687223; 24988056; 18803879; 19714243; 25053345; 20433755; 22964582; 23862748 | In vitro and in vivo experiments showed that miR-34a and DOX can be efficiently encapsulated into HA-CS NPs and delivered into tumor cells or tumor tissues and enhance anti-tumor effects of DOX by suppressing the expression of non-pump resistance and anti-apoptosis proto-oncogene Bcl-2;MiR 34a inhibits proliferation and migration of breast cancer through down regulation of Bcl 2 and SIRT1; In this study we aimed to determine the effect of miR-34a on the growth of breast cancer and to investigate whether its effect is achieved by targeting Bcl-2 and SIRT1; Bcl-2 and SIRT1 as the targets of miR-34a were found to be in reverse correlation with ectopic expression of miR-34a;Quantitative PCR and western analysis confirmed decreased expression of two genes BCL-2 and CCND1 in docetaxel-resistant cells which are both targeted by miR-34a;The miR-34a expression levels in cells after irradiation at 30 and 60 Gy were 0.17- and 18.7-times the BCL2 and caspase-9 expression levels respectively;Functional analyses further indicate that restoration of miR-34a inhibits B cell lymphoma-2 Bcl-2 protein expression to withdraw the survival advantage of these resistant NSCLC cells;Thus in PC3PR cells reduced expression of miR-34a confers paclitaxel resistance via up-regulating SIRT1 and Bcl2 expression; MiR-34a and its downstream targets SIRT1 and Bcl2 play important roles in the development of paclitaxel resistance all of which can be useful biomarkers and promising therapeutic targets for the drug resistance in hormone-refractory prostate cancer;Target analysis indicated that micro RNA miR-34a directly regulates Bcl-2 and miR-34a overexpression decreased Bcl-2 protein level in gastric cancer cells; We also found that luteolin upregulates miR-34a expression and downregulates Bcl-2 expression; Based on these results we can draw the conclusion that luteolin partly decreases Bcl-2 expression through upregulating miR-34a expression;miR-34 targets Notch HMGA2 and Bcl-2 genes involved in the self-renewal and survival of cancer stem cells; Human gastric cancer cells were transfected with miR-34 mimics or infected with the lentiviral miR-34-MIF expression system and validated by miR-34 reporter assay using Bcl-2 3'UTR reporter; Human gastric cancer Kato III cells with miR-34 restoration reduced the expression of target genes Bcl-2 Notch and HMGA2; Bcl-2 3'UTR reporter assay showed that the transfected miR-34s were functional and confirmed that Bcl-2 is a direct target of miR-34; The mechanism of miR-34-mediated suppression of self-renewal appears to be related to the direct modulation of downstream targets Bcl-2 Notch and HMGA2 indicating that miR-34 may be involved in gastric cancer stem cell self-renewal/differentiation decision-making;Among the target proteins regulated by miR-34 are Notch pathway proteins and Bcl-2 suggesting the possibility of a role for miR-34 in the maintenance and survival of cancer stem cells; Our data support the view that miR-34 may be involved in pancreatic cancer stem cell self-renewal potentially via the direct modulation of downstream targets Bcl-2 and Notch implying that miR-34 may play an important role in pancreatic cancer stem cell self-renewal and/or cell fate determination;Manipulating miR-34a in prostate cancer cells confirms that this miRNA regulates BCL-2 and may in part regulate response to docetaxel;For instance miR-34a up-regulation corresponded with a down-regulation of BCL2 protein; Treating Par-4-overexpressing HT29 cells with a miR-34a antagomir functionally reversed both BCL2 down-regulation and apoptosis by 5-FU;Tumors harvested from these lungs have elevated levels of oncogenic miRNAs miR-21 and miR-155; are deficient for p53-regulated miRNAs; and have heightened expression of miR-34 target genes such as Met and Bcl-2;MicroRNA 34a targets Bcl 2 and sensitizes human hepatocellular carcinoma cells to sorafenib treatment; HCC tissues with lower miR-34a expression displayed higher expression of Bcl-2 protein than those with high expression of miR-34a; therefore an inverse correlation is evident between the miR-34a level and Bcl-2 expression; Bioinformatics and luciferase reporter assays revealed that miR-34a binds the 3'-UTR of the Bcl-2 mRNA and represses its translation; Western blotting analysis and qRT-PCR confirmed that Bcl-2 is inhibited by miR-34a overexpression; Functional analyses indicated that the restoration of miR-34a reduced cell viability promoted cell apoptosis and potentiated sorafenib-induced apoptosis and toxicity in HCC cell lines by inhibiting Bcl-2 expression |
| 27 | hsa-miR-34a-5p | BCLAF1 | 0.13 | 0.13326 | -0.27 | 1.0E-5 | miRNAWalker2 validate | -0.06 | 0.01233 | NA | |
| 28 | hsa-miR-34a-5p | BIRC6 | 0.13 | 0.13326 | -0.31 | 0.0001 | miRNAWalker2 validate | -0.07 | 0.02909 | NA | |
| 29 | hsa-miR-34a-5p | BNC2 | 0.13 | 0.13326 | -0.78 | 0 | miRNATAP | -0.13 | 0.03676 | NA | |
| 30 | hsa-miR-34a-5p | BRPF3 | 0.13 | 0.13326 | 0.09 | 0.20587 | MirTarget; miRNATAP | -0.12 | 3.0E-5 | NA | |
| 31 | hsa-miR-34a-5p | C17orf51 | 0.13 | 0.13326 | -1.38 | 0 | mirMAP | -0.18 | 0.00028 | NA | |
| 32 | hsa-miR-34a-5p | C1orf21 | 0.13 | 0.13326 | -0.47 | 0.00106 | mirMAP | -0.15 | 0.01067 | NA | |
| 33 | hsa-miR-34a-5p | CACHD1 | 0.13 | 0.13326 | -2.57 | 0 | MirTarget | -0.38 | 0 | NA | |
| 34 | hsa-miR-34a-5p | CAMSAP1 | 0.13 | 0.13326 | -0.1 | 0.08639 | miRNATAP | -0.07 | 0.00353 | NA | |
| 35 | hsa-miR-34a-5p | CAPN6 | 0.13 | 0.13326 | -3.62 | 0 | miRNATAP | -0.8 | 0 | NA | |
| 36 | hsa-miR-34a-5p | CAPRIN1 | 0.13 | 0.13326 | 0.34 | 0 | miRNAWalker2 validate | -0.09 | 0.00079 | NA | |
| 37 | hsa-miR-34a-5p | CCDC50 | 0.13 | 0.13326 | -1.33 | 0 | miRNATAP | -0.11 | 0.00025 | NA | |
| 38 | hsa-miR-34a-5p | CDKN1C | 0.13 | 0.13326 | -2.11 | 0 | miRNATAP | -0.28 | 0 | NA | |
| 39 | hsa-miR-34a-5p | CDKN2AIP | 0.13 | 0.13326 | -0.39 | 0 | miRNAWalker2 validate | -0.06 | 0.01838 | NA | |
| 40 | hsa-miR-34a-5p | CDKN2C | 0.13 | 0.13326 | -1.08 | 0 | miRNAWalker2 validate | -0.09 | 0.03462 | NA | |
| 41 | hsa-miR-34a-5p | CDON | 0.13 | 0.13326 | -0.99 | 0 | miRNAWalker2 validate | -0.13 | 0.01996 | NA | |
| 42 | hsa-miR-34a-5p | CELF2 | 0.13 | 0.13326 | -2.12 | 0 | miRNATAP | -0.14 | 0.01317 | NA | |
| 43 | hsa-miR-34a-5p | CEP170 | 0.13 | 0.13326 | -0.11 | 0.22962 | miRNAWalker2 validate | -0.07 | 0.03809 | NA | |
| 44 | hsa-miR-34a-5p | CHM | 0.13 | 0.13326 | -0.44 | 0 | MirTarget | -0.09 | 0.00582 | NA | |
| 45 | hsa-miR-34a-5p | CLOCK | 0.13 | 0.13326 | -0.44 | 0.00109 | MirTarget | -0.11 | 0.04508 | NA | |
| 46 | hsa-miR-34a-5p | CNTN2 | 0.13 | 0.13326 | -1.15 | 0 | MirTarget; miRNATAP | -0.27 | 0.00433 | NA | |
| 47 | hsa-miR-34a-5p | CPEB2 | 0.13 | 0.13326 | -0.3 | 0.01548 | miRNATAP | -0.1 | 0.04497 | NA | |
| 48 | hsa-miR-34a-5p | CR2 | 0.13 | 0.13326 | -0.58 | 0.1206 | MirTarget | -0.33 | 0.0271 | NA | |
| 49 | hsa-miR-34a-5p | CRTC1 | 0.13 | 0.13326 | 0.03 | 0.71583 | miRNATAP | -0.07 | 0.02068 | NA | |
| 50 | hsa-miR-34a-5p | CRY2 | 0.13 | 0.13326 | -1.2 | 0 | MirTarget | -0.13 | 7.0E-5 | NA | |
| 51 | hsa-miR-34a-5p | CTDSP2 | 0.13 | 0.13326 | -0.73 | 0 | miRNATAP | -0.14 | 0 | NA | |
| 52 | hsa-miR-34a-5p | CTNNB1 | 0.13 | 0.13326 | -0.34 | 0 | miRNAWalker2 validate | -0.06 | 0.02022 | NA | |
| 53 | hsa-miR-34a-5p | DAPK2 | 0.13 | 0.13326 | -1.03 | 0 | MirTarget | -0.1 | 0.02207 | 27704360 | E2F 1 promotes DAPK2 induced anti tumor immunity of gastric cancer cells by targeting miR 34a |
| 54 | hsa-miR-34a-5p | DCX | 0.13 | 0.13326 | -2.75 | 0 | MirTarget; miRNATAP | -0.68 | 0 | NA | |
| 55 | hsa-miR-34a-5p | DDAH2 | 0.13 | 0.13326 | 0.19 | 0.03999 | miRNAWalker2 validate | -0.08 | 0.044 | NA | |
| 56 | hsa-miR-34a-5p | DIXDC1 | 0.13 | 0.13326 | -1.64 | 0 | MirTarget | -0.09 | 0.04466 | NA | |
| 57 | hsa-miR-34a-5p | DLL1 | 0.13 | 0.13326 | -0.95 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.18 | 0.0003 | 22438124 | Delta tocotrienol suppresses Notch 1 pathway by upregulating miR 34a in nonsmall cell lung cancer cells |
| 58 | hsa-miR-34a-5p | DNMBP | 0.13 | 0.13326 | -0.67 | 0 | MirTarget | -0.1 | 0.04209 | NA | |
| 59 | hsa-miR-34a-5p | EDAR | 0.13 | 0.13326 | -2.79 | 0 | miRNATAP | -0.46 | 4.0E-5 | NA | |
| 60 | hsa-miR-34a-5p | EEA1 | 0.13 | 0.13326 | -0.92 | 0 | miRNATAP | -0.08 | 0.03427 | NA | |
| 61 | hsa-miR-34a-5p | EFNB3 | 0.13 | 0.13326 | -1.46 | 0 | miRNATAP | -0.22 | 0.00215 | NA | |
| 62 | hsa-miR-34a-5p | EPB41L2 | 0.13 | 0.13326 | -1.76 | 0 | miRNAWalker2 validate | -0.18 | 0.00016 | NA | |
| 63 | hsa-miR-34a-5p | EPHA4 | 0.13 | 0.13326 | -1.55 | 0 | miRNATAP | -0.15 | 0.00985 | NA | |
| 64 | hsa-miR-34a-5p | ERC1 | 0.13 | 0.13326 | -0.36 | 0 | mirMAP; miRNATAP | -0.15 | 0 | NA | |
| 65 | hsa-miR-34a-5p | EVC | 0.13 | 0.13326 | -0.7 | 0 | mirMAP | -0.11 | 0.01474 | NA | |
| 66 | hsa-miR-34a-5p | EXT2 | 0.13 | 0.13326 | -0.08 | 0.15651 | miRNAWalker2 validate | -0.05 | 0.01949 | NA | |
| 67 | hsa-miR-34a-5p | F2RL2 | 0.13 | 0.13326 | 1.83 | 0 | MirTarget; miRNATAP | -0.27 | 0.00474 | NA | |
| 68 | hsa-miR-34a-5p | FAM117B | 0.13 | 0.13326 | 0.06 | 0.3918 | MirTarget | -0.13 | 0 | NA | |
| 69 | hsa-miR-34a-5p | FAM126B | 0.13 | 0.13326 | -0.48 | 0 | miRNATAP | -0.06 | 0.03998 | NA | |
| 70 | hsa-miR-34a-5p | FAM168A | 0.13 | 0.13326 | -0.6 | 0 | mirMAP | -0.19 | 0.00014 | NA | |
| 71 | hsa-miR-34a-5p | FAM53B | 0.13 | 0.13326 | 0.27 | 0.00126 | mirMAP | -0.16 | 0 | NA | |
| 72 | hsa-miR-34a-5p | FAT3 | 0.13 | 0.13326 | -1.4 | 0 | miRNATAP | -0.18 | 0.03489 | NA | |
| 73 | hsa-miR-34a-5p | FAT4 | 0.13 | 0.13326 | -2.33 | 0 | MirTarget | -0.35 | 0 | NA | |
| 74 | hsa-miR-34a-5p | FIGN | 0.13 | 0.13326 | -2.44 | 0 | miRNAWalker2 validate | -0.2 | 0.01621 | NA | |
| 75 | hsa-miR-34a-5p | FOXJ2 | 0.13 | 0.13326 | -0.39 | 0 | MirTarget; miRNATAP | -0.09 | 3.0E-5 | NA | |
| 76 | hsa-miR-34a-5p | FOXK1 | 0.13 | 0.13326 | -0.35 | 1.0E-5 | mirMAP | -0.11 | 0.00056 | NA | |
| 77 | hsa-miR-34a-5p | FOXN3 | 0.13 | 0.13326 | -1.7 | 0 | miRNATAP | -0.19 | 0 | NA | |
| 78 | hsa-miR-34a-5p | FRMD4A | 0.13 | 0.13326 | -1.02 | 0 | MirTarget | -0.1 | 0.00464 | NA | |
| 79 | hsa-miR-34a-5p | GAB1 | 0.13 | 0.13326 | -0.69 | 0 | MirTarget | -0.08 | 0.02198 | NA | |
| 80 | hsa-miR-34a-5p | GDPD5 | 0.13 | 0.13326 | -1.42 | 0 | mirMAP | -0.13 | 0.01726 | NA | |
| 81 | hsa-miR-34a-5p | GFRA1 | 0.13 | 0.13326 | -0.69 | 0.0647 | miRNATAP | -0.66 | 1.0E-5 | NA | |
| 82 | hsa-miR-34a-5p | GIGYF2 | 0.13 | 0.13326 | -0.19 | 0.00053 | miRNAWalker2 validate | -0.06 | 0.00713 | NA | |
| 83 | hsa-miR-34a-5p | GLCE | 0.13 | 0.13326 | -0.01 | 0.95505 | MirTarget; miRNATAP | -0.09 | 0.026 | NA | |
| 84 | hsa-miR-34a-5p | GPR156 | 0.13 | 0.13326 | -0.93 | 3.0E-5 | MirTarget | -0.19 | 0.03103 | NA | |
| 85 | hsa-miR-34a-5p | GREM2 | 0.13 | 0.13326 | -2.65 | 0 | MirTarget; miRNATAP | -0.2 | 0.0442 | NA | |
| 86 | hsa-miR-34a-5p | GRID1 | 0.13 | 0.13326 | -1.27 | 0 | miRNATAP | -0.23 | 2.0E-5 | NA | |
| 87 | hsa-miR-34a-5p | HIC2 | 0.13 | 0.13326 | -0.3 | 3.0E-5 | mirMAP | -0.12 | 2.0E-5 | NA | |
| 88 | hsa-miR-34a-5p | HIPK2 | 0.13 | 0.13326 | -0.95 | 0 | mirMAP | -0.16 | 0.00145 | NA | |
| 89 | hsa-miR-34a-5p | HNRNPUL1 | 0.13 | 0.13326 | 0.26 | 0 | miRNAWalker2 validate | -0.06 | 0.00052 | NA | |
| 90 | hsa-miR-34a-5p | IGF2 | 0.13 | 0.13326 | -1.81 | 0 | mirMAP | -0.24 | 0.0023 | NA | |
| 91 | hsa-miR-34a-5p | IGFBP5 | 0.13 | 0.13326 | -0.62 | 0.001 | mirMAP | -0.17 | 0.02346 | NA | |
| 92 | hsa-miR-34a-5p | IGSF3 | 0.13 | 0.13326 | 0.89 | 0 | MirTarget | -0.1 | 0.02882 | NA | |
| 93 | hsa-miR-34a-5p | ING5 | 0.13 | 0.13326 | -0.24 | 0.00024 | mirMAP | -0.1 | 0.0001 | NA | |
| 94 | hsa-miR-34a-5p | ITPKB | 0.13 | 0.13326 | -0.46 | 0 | mirMAP | -0.09 | 0.00433 | NA | |
| 95 | hsa-miR-34a-5p | ITPR2 | 0.13 | 0.13326 | -0.98 | 0 | MirTarget | -0.18 | 0.00066 | NA | |
| 96 | hsa-miR-34a-5p | ITSN1 | 0.13 | 0.13326 | -1.55 | 0 | mirMAP | -0.12 | 0.00146 | NA | |
| 97 | hsa-miR-34a-5p | JAG1 | 0.13 | 0.13326 | -0.64 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.11 | 0.00421 | NA | |
| 98 | hsa-miR-34a-5p | JMJD1C | 0.13 | 0.13326 | -0.95 | 0 | miRNATAP | -0.1 | 0.00704 | NA | |
| 99 | hsa-miR-34a-5p | JRK | 0.13 | 0.13326 | -0.4 | 1.0E-5 | MirTarget | -0.1 | 0.00725 | NA | |
| 100 | hsa-miR-34a-5p | KBTBD6 | 0.13 | 0.13326 | -0.65 | 0 | miRNAWalker2 validate | -0.08 | 0.00679 | NA | |
| 101 | hsa-miR-34a-5p | KCNA1 | 0.13 | 0.13326 | -2.93 | 0 | miRNATAP | -0.59 | 0 | NA | |
| 102 | hsa-miR-34a-5p | KCNQ4 | 0.13 | 0.13326 | -2.01 | 0 | mirMAP | -0.31 | 0.00177 | NA | |
| 103 | hsa-miR-34a-5p | KIAA1462 | 0.13 | 0.13326 | -0.89 | 0 | MirTarget | -0.13 | 0.00232 | NA | |
| 104 | hsa-miR-34a-5p | KIT | 0.13 | 0.13326 | -3.56 | 0 | MirTarget | -0.44 | 0 | NA | |
| 105 | hsa-miR-34a-5p | KITLG | 0.13 | 0.13326 | -0.14 | 0.34864 | MirTarget | -0.19 | 0.0018 | NA | |
| 106 | hsa-miR-34a-5p | KLB | 0.13 | 0.13326 | -4.4 | 0 | miRNAWalker2 validate; miRTarBase | -0.23 | 0.01748 | NA | |
| 107 | hsa-miR-34a-5p | KLF13 | 0.13 | 0.13326 | -0.9 | 0 | mirMAP | -0.13 | 0.00022 | NA | |
| 108 | hsa-miR-34a-5p | KLF4 | 0.13 | 0.13326 | -2.57 | 0 | MirTarget; miRNATAP | -0.16 | 0.01412 | NA | |
| 109 | hsa-miR-34a-5p | KLF7 | 0.13 | 0.13326 | -0.96 | 0 | miRNATAP | -0.16 | 0.00405 | NA | |
| 110 | hsa-miR-34a-5p | KLHDC3 | 0.13 | 0.13326 | 0.17 | 0.02381 | MirTarget | -0.07 | 0.02782 | NA | |
| 111 | hsa-miR-34a-5p | LGR4 | 0.13 | 0.13326 | -1.8 | 0 | MirTarget; miRNATAP | -0.17 | 0.00747 | NA | |
| 112 | hsa-miR-34a-5p | LRCH1 | 0.13 | 0.13326 | -0.85 | 0 | MirTarget | -0.09 | 0.00309 | NA | |
| 113 | hsa-miR-34a-5p | LRP4 | 0.13 | 0.13326 | -1.81 | 0 | MirTarget | -0.25 | 7.0E-5 | NA | |
| 114 | hsa-miR-34a-5p | LRRC40 | 0.13 | 0.13326 | -0.18 | 0.00762 | miRNAWalker2 validate; MirTarget | -0.06 | 0.02253 | NA | |
| 115 | hsa-miR-34a-5p | LRRC55 | 0.13 | 0.13326 | 0.6 | 7.0E-5 | MirTarget; miRNATAP | -0.25 | 3.0E-5 | NA | |
| 116 | hsa-miR-34a-5p | MAGI1 | 0.13 | 0.13326 | -1.26 | 0 | miRNAWalker2 validate | -0.14 | 0.00144 | NA | |
| 117 | hsa-miR-34a-5p | MBP | 0.13 | 0.13326 | -1.06 | 0 | mirMAP | -0.13 | 0.00583 | NA | |
| 118 | hsa-miR-34a-5p | MCF2L | 0.13 | 0.13326 | -0.02 | 0.81188 | mirMAP | -0.09 | 0.01889 | NA | |
| 119 | hsa-miR-34a-5p | MCTP1 | 0.13 | 0.13326 | -1.6 | 0 | miRNATAP | -0.23 | 0.0002 | NA | |
| 120 | hsa-miR-34a-5p | MDM4 | 0.13 | 0.13326 | 0.11 | 0.0961 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.08 | 0.00397 | NA | |
| 121 | hsa-miR-34a-5p | MECP2 | 0.13 | 0.13326 | -0.46 | 0 | mirMAP | -0.07 | 0.00047 | NA | |
| 122 | hsa-miR-34a-5p | MET | 0.13 | 0.13326 | -2.37 | 0 | miRNAWalker2 validate; miRTarBase | -0.26 | 0.00075 | 23243217; 26104764; 22457788; 23593387; 19006648; 19773441; 19029026 | Detection of miR 34a promoter methylation in combination with elevated expression of c Met and β catenin predicts distant metastasis of colon cancer; Using a case-control study design of 94 primary colon cancer samples with and without liver metastases we determined CpG methylation frequencies of miR-34a and miR-34b/c promoters expression of miR-34a and its targets c-Met Snail and β-catenin and their prognostic value; Decreased miR-34a expression was associated with upregulation of c-Met Snail and β-catenin protein levels P = 0.031 0.132 and 0.004 which were associated with distant metastases P = 0.001 0.017 and 0.005; In a confounder-adjusted multivariate regression model miR-34a methylation high c-Met and β-catenin levels provided the most significant prognostic information about metastases to the liver P = 0.014 0.031 and 0.058 and matched pairs showed a higher prevalence of these risk factors in the samples with distant spread P = 0.029; Silencing of miR-34a and upregulation of c-Met Snail and β-catenin expression is associated with liver metastases of colon cancer; Detection of miR-34a silencing in resected primary colon cancer may be of prognostic value especially in combination with detection of c-Met and β-catenin expression;The expression of microRNA 34a is inversely correlated with c MET and CDK6 and has a prognostic significance in lung adenocarcinoma patients; We found significant inverse correlations between miR-34a and c-MET R = -0.316 P = 0.028 and CDK6 expression R = -0.4582 P = 0.004;c-Met is a target of miR-34a and regulates the migration and invasion of osteosarcoma cells; Osteosarcoma cells over-expressing miR-34a exhibited a significant decrease in the expression levels of c-Met mRNA and protein simultaneously; The results presented in this study demonstrated that over-expression of miR-34a could inhibit the tumor growth and metastasis of osteosarcoma probably through down regulating c-Met; And there are other putative miR-34a target genes beside c-Met which could potentially be key players in the development of osteosarcoma;Underexpression of miR 34a in hepatocellular carcinoma and its contribution towards enhancement of proliferating inhibitory effects of agents targeting c MET; In addition miR-34a mimic enhanced the effect of cell proliferation inhibition and caspase activity induction of agents targeting c-MET siRNAs and small molecular inhibitor su11274;miR 34a inhibits migration and invasion by down regulation of c Met expression in human hepatocellular carcinoma cells; miR-34a directly targeted c-Met and reduced both mRNA and protein levels of c-Met; thus decreased c-Met-induced phosphorylation of extracellular signal-regulated kinases 1 and 2 ERK1/2; Taken together these results provide evidence to show the suppression role of miR-34a in tumor migration and invasion through modulation of the c-Met signaling pathway;miR-34a levels in human gliomas inversely correlated to c-Met levels measured in the same tumors; Forced expression of c-Met or Notch-1/Notch-2 transcripts lacking the 3'-untranslated region sequences partially reversed the effects of miR-34a on cell cycle arrest and cell death in glioma cells and stem cells respectively; They show that miR-34a suppresses brain tumor growth by targeting c-Met and Notch;MicroRNA 34a inhibits uveal melanoma cell proliferation and migration through downregulation of c Met; In addition expression of c-Met and cell cycle-related proteins was determined by Western blotting and immunofluorescence after the introduction of miR-34a miR-34a is actively expressed in melanocytes but not in uveal melanoma cells based on Northern blot analysis; After identification of two putative miR-34a binding sites within the 3' UTR of the human c-Met mRNA miR-34a was shown to suppress luciferase activity using HEK293 cells with a luciferase reporter construct containing the binding sites; miR-34a was confirmed to downregulate the expression of c-Met protein by Western blotting and immunofluorescence; These results demonstrate that miR-34a acts as a tumor suppressor in uveal melanoma cell proliferation and migration through the downregulation of c-Met |
| 123 | hsa-miR-34a-5p | MEX3C | 0.13 | 0.13326 | -0.51 | 0 | MirTarget | -0.07 | 0.02381 | NA | |
| 124 | hsa-miR-34a-5p | MLC1 | 0.13 | 0.13326 | -0.01 | 0.98183 | mirMAP | -0.42 | 6.0E-5 | NA | |
| 125 | hsa-miR-34a-5p | MLLT3 | 0.13 | 0.13326 | -0.38 | 0.00144 | MirTarget; miRNATAP | -0.1 | 0.04219 | NA | |
| 126 | hsa-miR-34a-5p | MON2 | 0.13 | 0.13326 | -0.53 | 0 | MirTarget | -0.06 | 0.02136 | NA | |
| 127 | hsa-miR-34a-5p | MSL2 | 0.13 | 0.13326 | -0.19 | 0.00012 | MirTarget; miRNATAP | -0.07 | 0.00035 | NA | |
| 128 | hsa-miR-34a-5p | MTMR9 | 0.13 | 0.13326 | -0.63 | 0 | miRNAWalker2 validate | -0.06 | 0.04385 | NA | |
| 129 | hsa-miR-34a-5p | MYADM | 0.13 | 0.13326 | -0.98 | 0 | MirTarget | -0.07 | 0.04033 | NA | |
| 130 | hsa-miR-34a-5p | MYC | 0.13 | 0.13326 | -1.13 | 0 | miRNAWalker2 validate; miRTarBase | -0.22 | 0.00013 | 25572695; 25686834; 21460242; 22159222; 23640973; 22830357; 22235332 | The c-Myc and CD44 were confirmed as direct targets of miR-34a in EJ cell apoptosis induced by PRE;miR 34a induces cellular senescence via modulation of telomerase activity in human hepatocellular carcinoma by targeting FoxM1/c Myc pathway;Myc mediated repression of microRNA 34a promotes high grade transformation of B cell lymphoma by dysregulation of FoxP1;MicroRNA 34a suppresses malignant transformation by targeting c Myc transcriptional complexes in human renal cell carcinoma; We investigated the functional effects of microRNA-34a miR-34a on c-Myc transcriptional complexes in renal cell carcinoma; miR-34a down-regulated expression of multiple oncogenes including c-Myc by targeting its 3' untranslated region which was revealed by luciferase reporter assays; Our results demonstrate that miR-34a suppresses assembly and function of the c-Myc complex that activates or elongates transcription indicating a novel role of miR-34a in the regulation of transcription by c-Myc;Among them miR-34a was also associated with poor prognosis in 2 independent series of leukemic and nodal MCL and in cooperation with high expression of the MYC oncogene;We report that miR-34a did not inhibit cell proliferation notwithstanding a marked down-regulation of c-MYC;MicroRNA 34a modulates c Myc transcriptional complexes to suppress malignancy in human prostate cancer cells; We studied the functional effects of miR-34a on c-Myc transcriptional complexes in PC-3 prostate cancer cells; miR-34a downregulated expression of c-Myc oncogene by targeting its 3' UTR as shown by luciferase reporter assays; This is the first report to document that miR-34a suppresses assembly and function of the c-Myc-Skp2-Miz1 complex that activates RhoA and the c-Myc-pTEFB complex that elongates transcription of various genes suggesting a novel role of miR-34a in the regulation of transcription by c-Myc complex |
| 131 | hsa-miR-34a-5p | MYNN | 0.13 | 0.13326 | -0.15 | 0.00944 | MirTarget | -0.05 | 0.01698 | NA | |
| 132 | hsa-miR-34a-5p | MYRIP | 0.13 | 0.13326 | -2.17 | 0 | miRNATAP | -0.53 | 0 | NA | |
| 133 | hsa-miR-34a-5p | NAP1L5 | 0.13 | 0.13326 | -1.1 | 0 | miRNATAP | -0.12 | 0.00826 | NA | |
| 134 | hsa-miR-34a-5p | NAT8L | 0.13 | 0.13326 | -2.94 | 0 | MirTarget | -0.3 | 0.00125 | NA | |
| 135 | hsa-miR-34a-5p | NAV3 | 0.13 | 0.13326 | -1.78 | 0 | MirTarget | -0.18 | 0.02613 | NA | |
| 136 | hsa-miR-34a-5p | NCDN | 0.13 | 0.13326 | 1.04 | 0 | MirTarget | -0.09 | 0.01045 | NA | |
| 137 | hsa-miR-34a-5p | NCOR2 | 0.13 | 0.13326 | 0.25 | 7.0E-5 | MirTarget | -0.08 | 0.00171 | NA | |
| 138 | hsa-miR-34a-5p | NDST1 | 0.13 | 0.13326 | -0.62 | 0 | MirTarget; miRNATAP | -0.07 | 0.0117 | NA | |
| 139 | hsa-miR-34a-5p | NFIC | 0.13 | 0.13326 | -0.32 | 0.00149 | mirMAP | -0.12 | 0.00361 | NA | |
| 140 | hsa-miR-34a-5p | NGFR | 0.13 | 0.13326 | -2.5 | 0 | mirMAP | -0.49 | 0 | NA | |
| 141 | hsa-miR-34a-5p | NNT | 0.13 | 0.13326 | -0.33 | 0.00102 | miRNAWalker2 validate | -0.1 | 0.00912 | NA | |
| 142 | hsa-miR-34a-5p | NOS1AP | 0.13 | 0.13326 | -0.68 | 0.00068 | miRNATAP | -0.34 | 2.0E-5 | NA | |
| 143 | hsa-miR-34a-5p | NOTCH1 | 0.13 | 0.13326 | -0.55 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.08 | 0.02917 | 24565525; 25783790; 27082152; 25623761; 23085450; 22438124; 23140286; 24349627; 20351093; 23642368; 22347519; 23430952; 23902763; 21743299; 22992310; 23145211 | In addition intracellular restoration of miR-34a inhibited breast cancer cell migration via targeting Notch-1 signaling;MicroRNA 34a suppresses the breast cancer stem cell like characteristics by downregulating Notch1 pathway; In this study we verified that miR-34a directly and functionally targeted Notch1 in MCF-7 cells; We reported that miR-34a negatively regulated cell proliferation migration and invasion and breast cancer stem cell propagation by downregulating Notch1; The expression of miR-34a was negatively correlated with tumor stages metastasis and Notch1 expression in breast cancer tissues; Furthermore overexpression of miR-34a increased chemosensitivity of breast cancer cells to paclitaxel PTX by downregulating the Notch1 pathway; Taken together our results indicate that miR-34a inhibited breast cancer stemness and increased the chemosensitivity to PTX partially by downregulating the Notch1 pathway suggesting that miR-34a/Notch1 play an important role in regulating breast cancer stem cells;We showed that miR-34a as a tumor suppressor could separately reduce the stemness of BCSCs and activate the cytotoxic susceptibility of BCSCs to natural killer NK cells in vitro via down regulating the expression of Notch1 signaling molecules;miR 34a may regulate sensitivity of breast cancer cells to adriamycin via targeting Notch1; To explore the influence of miR-34a on Notch1 expression in breast cancer cells and to explore the role of miR-34a in the sensitivity of breast cancer cells to Adriamycin ADR; The expression levels of Notch1 mRNA and protein in MCF-7/ADR cells transfected with miR-34a mimics were significantly up-regulated; On the contrary the expressions of Notch1 mRNA and protein in MCF-7 cells transfected with miR-34a inhibitor were down-regulated; miR-34a negatively regulates the expression of Notch1 at both mRNA and protein levels;MicroRNA 34a modulates chemosensitivity of breast cancer cells to adriamycin by targeting Notch1; The association of miR-34a and Notch1 was analyzed by dual-luciferase reporter assay and Notch1-siRNA technology; Real-time PCR assay was performed to test the expression of miR-34a and Notch1 in 38 selective breast cancer tissue samples; MiR-34a mimic could inhibit the luciferase activity of the construct containing wild-type 3' UTR of Notch1 in MCF-7/ADR cells; Further there was an inverse association between Notch1 and miR-34a expression in breast cancer; Dysregulation of miR-34a plays critical roles in the acquired ADR resistance of breast cancer at least in part via targeting Notch1;Delta tocotrienol suppresses Notch 1 pathway by upregulating miR 34a in nonsmall cell lung cancer cells; In our study using miRNA microarray we observed that downregulation of the Notch-1 pathway by delta-tocotrienol correlated with upregulation of miR-34a in nonsmall cell lung cancer cells NSCLC;We found that Re-expression forced expression of miR-34a inhibits cell growth and induces apoptosis with concomitant down-regulation of Notch-1 signaling pathway one of the target of miR-34a; Moreover treatment of PC cells with a natural compound genistein led to the up-regulation of miR-34a resulting in the down-regulation of Notch-1 which was correlated with inhibition of cell growth and induction of apoptosis;Most importantly BR-DIM intervention in PCa patients prior to radical prostatectomy showed reexpression of miR-34a which was consistent with decreased expression of AR PSA and Notch-1 in PCa tissue specimens;MicroRNA 34a suppresses invasion through downregulation of Notch1 and Jagged1 in cervical carcinoma and choriocarcinoma cells; Computational miRNA target prediction suggested that Notch1 and Jagged1 were targets of miR-34a; By using functional assays miR-34a was demonstrated to bind to the 3' untranslated regions of Notch1 and Jagged1; Forced expression of miR-34a altered the expression of Notch1 and Jagged1 protein as well as Notch signaling as shown by the response of Hairy Enhancer of Split-1 protein to these treatments using western blot analysis;Mechanistically miR-34a sequesters Notch1 mRNA to generate a sharp threshold response where a bimodal Notch signal specifies the choice between self-renewal and differentiation;Most importantly BR-DIM intervention in PCa patients prior to radical prostatectomy showed re-expression of miR-34a which was consistent with decreased expression of AR PSA and Notch-1 in PCa tissue specimens;We also found that reexpression of miR-34a and miR-200b by transfection led to reduced expression of Notch-1 resulting in the inhibition of osteosarcoma cell proliferation invasion and angiogenesis;Rhamnetin and cirsiliol induce radiosensitization and inhibition of epithelial mesenchymal transition EMT by miR 34a mediated suppression of Notch 1 expression in non small cell lung cancer cell lines; Indeed rhamnetin and cirsiliol increased the expression of tumor-suppressive microRNA miR-34a in a p53-dependent manner leading to inhibition of Notch-1 expression;MicroRNA 34a targets notch1 and inhibits cell proliferation in glioblastoma multiforme; Also we identified notch1 as a direct target gene of miR-34a; Knockdown of notch1 showed similar cellular functions as overexpression of miR-34a both in vitro and in vivo; Collectively our findings show that miR-34a is downregulated in GBM cells and inhibits GBM growth by targeting notch1;The re-expression of miR-34 led to a marked reduction in the expression of its target gene Notch-1;Inactivation of AR and Notch 1 signaling by miR 34a attenuates prostate cancer aggressiveness; We found that over-expression of miR-34a led to reduced expression of AR PSA and Notch-1; These findings suggest that the loss of miR-34a is directly linked with up-regulation of AR and Notch-1 both of which are highly expressed in PCa and thus finding innovative approaches by which miR-34a expression could be up-regulated will have a huge impact on the treatment of PCa especially for the treatment of mCRPC |
| 144 | hsa-miR-34a-5p | NOTCH2 | 0.13 | 0.13326 | -0.57 | 0 | miRNAWalker2 validate; miRNATAP | -0.11 | 0.00302 | NA | |
| 145 | hsa-miR-34a-5p | NSD1 | 0.13 | 0.13326 | -0.07 | 0.30526 | miRNATAP | -0.08 | 0.00253 | NA | |
| 146 | hsa-miR-34a-5p | NTN1 | 0.13 | 0.13326 | -0.34 | 0.04597 | mirMAP | -0.2 | 0.00412 | NA | |
| 147 | hsa-miR-34a-5p | NUCKS1 | 0.13 | 0.13326 | 0.12 | 0.06394 | miRNAWalker2 validate | -0.07 | 0.00814 | NA | |
| 148 | hsa-miR-34a-5p | NUP98 | 0.13 | 0.13326 | -0.11 | 0.03227 | miRNAWalker2 validate | -0.07 | 0.0005 | NA | |
| 149 | hsa-miR-34a-5p | P2RY14 | 0.13 | 0.13326 | -1.72 | 0 | MirTarget | -0.17 | 0.00367 | NA | |
| 150 | hsa-miR-34a-5p | PAX5 | 0.13 | 0.13326 | 0.19 | 0.53615 | mirMAP | -0.37 | 0.00242 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | POSITIVE REGULATION OF GENE EXPRESSION | 62 | 1733 | 8.148e-14 | 1.896e-10 |
| 2 | NEUROGENESIS | 55 | 1402 | 7.153e-14 | 1.896e-10 |
| 3 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 63 | 1805 | 1.485e-13 | 2.303e-10 |
| 4 | POSITIVE REGULATION OF CELL COMMUNICATION | 53 | 1532 | 2.823e-11 | 3.213e-08 |
| 5 | REGULATION OF CELL DIFFERENTIATION | 52 | 1492 | 3.453e-11 | 3.213e-08 |
| 6 | NEURON DIFFERENTIATION | 38 | 874 | 4.891e-11 | 3.793e-08 |
| 7 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 57 | 1784 | 9.646e-11 | 6.412e-08 |
| 8 | NEGATIVE REGULATION OF GENE EXPRESSION | 51 | 1493 | 1.14e-10 | 6.633e-08 |
| 9 | CELL DEVELOPMENT | 49 | 1426 | 2.331e-10 | 1.205e-07 |
| 10 | NEGATIVE REGULATION OF NITROGEN COMPOUND METABOLIC PROCESS | 49 | 1517 | 1.869e-09 | 8.435e-07 |
| 11 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 52 | 1672 | 1.994e-09 | 8.435e-07 |
| 12 | REGULATION OF CELL DEVELOPMENT | 34 | 836 | 3.146e-09 | 1.22e-06 |
| 13 | ORGAN MORPHOGENESIS | 34 | 841 | 3.656e-09 | 1.309e-06 |
| 14 | TISSUE DEVELOPMENT | 48 | 1518 | 5.611e-09 | 1.865e-06 |
| 15 | MUSCLE STRUCTURE DEVELOPMENT | 23 | 432 | 1.073e-08 | 3.329e-06 |
| 16 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 55 | 1929 | 1.344e-08 | 3.678e-06 |
| 17 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 37 | 1021 | 1.298e-08 | 3.678e-06 |
| 18 | INTRACELLULAR SIGNAL TRANSDUCTION | 48 | 1572 | 1.696e-08 | 4.385e-06 |
| 19 | NEURON DEVELOPMENT | 29 | 687 | 2.238e-08 | 5.482e-06 |
| 20 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 44 | 1395 | 2.934e-08 | 6.336e-06 |
| 21 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 36 | 1004 | 2.735e-08 | 6.336e-06 |
| 22 | POSITIVE REGULATION OF SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 8 | 39 | 2.996e-08 | 6.336e-06 |
| 23 | NEURON PROJECTION DEVELOPMENT | 25 | 545 | 4.569e-08 | 9.243e-06 |
| 24 | CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 24 | 513 | 5.971e-08 | 1.158e-05 |
| 25 | CELL MORPHOGENESIS INVOLVED IN NEURON DIFFERENTIATION | 20 | 368 | 7.252e-08 | 1.35e-05 |
| 26 | REGULATION OF CELLULAR COMPONENT MOVEMENT | 30 | 771 | 7.647e-08 | 1.368e-05 |
| 27 | REGULATION OF PROTEIN MODIFICATION PROCESS | 49 | 1710 | 8.552e-08 | 1.474e-05 |
| 28 | TISSUE MORPHOGENESIS | 24 | 533 | 1.214e-07 | 2.018e-05 |
| 29 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 47 | 1656 | 2.195e-07 | 3.461e-05 |
| 30 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 37 | 1142 | 2.242e-07 | 3.461e-05 |
| 31 | REGULATION OF CELL MORPHOGENESIS | 24 | 552 | 2.306e-07 | 3.461e-05 |
| 32 | REGULATION OF NEURON DIFFERENTIATION | 24 | 554 | 2.462e-07 | 3.58e-05 |
| 33 | NEPHRON DEVELOPMENT | 11 | 115 | 2.901e-07 | 4.09e-05 |
| 34 | NEURON PROJECTION MORPHOGENESIS | 20 | 402 | 3.008e-07 | 4.116e-05 |
| 35 | SKIN EPIDERMIS DEVELOPMENT | 9 | 71 | 3.191e-07 | 4.242e-05 |
| 36 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 49 | 1791 | 3.464e-07 | 4.477e-05 |
| 37 | ENDOTHELIAL CELL DIFFERENTIATION | 9 | 72 | 3.606e-07 | 4.522e-05 |
| 38 | POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 31 | 876 | 3.693e-07 | 4.522e-05 |
| 39 | UROGENITAL SYSTEM DEVELOPMENT | 17 | 299 | 3.79e-07 | 4.522e-05 |
| 40 | REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 28 | 750 | 4.939e-07 | 5.745e-05 |
| 41 | REGULATION OF MEMBRANE POTENTIAL | 18 | 343 | 5.547e-07 | 6.296e-05 |
| 42 | MUSCLE ORGAN DEVELOPMENT | 16 | 277 | 6.815e-07 | 7.375e-05 |
| 43 | KIDNEY EPITHELIUM DEVELOPMENT | 11 | 125 | 6.749e-07 | 7.375e-05 |
| 44 | RENAL TUBULE DEVELOPMENT | 9 | 78 | 7.224e-07 | 7.57e-05 |
| 45 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 45 | 1618 | 7.321e-07 | 7.57e-05 |
| 46 | HEART DEVELOPMENT | 21 | 466 | 7.624e-07 | 7.712e-05 |
| 47 | FOREBRAIN DEVELOPMENT | 18 | 357 | 9.884e-07 | 9.785e-05 |
| 48 | CENTRAL NERVOUS SYSTEM DEVELOPMENT | 30 | 872 | 1.038e-06 | 0.0001007 |
| 49 | ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY | 26 | 689 | 1.063e-06 | 0.000101 |
| 50 | MAINTENANCE OF CELL NUMBER | 11 | 132 | 1.164e-06 | 0.0001083 |
| 51 | NEGATIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 27 | 740 | 1.242e-06 | 0.0001133 |
| 52 | NEGATIVE REGULATION OF CELL DIFFERENTIATION | 24 | 609 | 1.331e-06 | 0.0001191 |
| 53 | REGULATION OF CELL PROLIFERATION | 42 | 1496 | 1.436e-06 | 0.0001261 |
| 54 | CELL FATE COMMITMENT | 14 | 227 | 1.591e-06 | 0.0001371 |
| 55 | RETINA MORPHOGENESIS IN CAMERA TYPE EYE | 7 | 45 | 1.637e-06 | 0.0001385 |
| 56 | DEVELOPMENTAL GROWTH | 17 | 333 | 1.68e-06 | 0.0001396 |
| 57 | SENSORY ORGAN DEVELOPMENT | 21 | 493 | 1.872e-06 | 0.0001528 |
| 58 | NEGATIVE REGULATION OF CELL DEVELOPMENT | 16 | 303 | 2.209e-06 | 0.0001772 |
| 59 | ENDOTHELIUM DEVELOPMENT | 9 | 90 | 2.446e-06 | 0.0001929 |
| 60 | SINGLE ORGANISM BEHAVIOR | 18 | 384 | 2.779e-06 | 0.0002119 |
| 61 | NEURON PROJECTION GUIDANCE | 13 | 205 | 2.772e-06 | 0.0002119 |
| 62 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 28 | 823 | 3.023e-06 | 0.0002269 |
| 63 | NEPHRON EPITHELIUM DEVELOPMENT | 9 | 93 | 3.221e-06 | 0.0002379 |
| 64 | NEURONAL STEM CELL POPULATION MAINTENANCE | 5 | 19 | 3.447e-06 | 0.0002506 |
| 65 | POSITIVE REGULATION OF CELL DEVELOPMENT | 20 | 472 | 3.589e-06 | 0.0002569 |
| 66 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 41 | 1518 | 5.089e-06 | 0.0003588 |
| 67 | EPIDERMIS DEVELOPMENT | 14 | 253 | 5.631e-06 | 0.0003911 |
| 68 | HEAD DEVELOPMENT | 25 | 709 | 5.715e-06 | 0.0003911 |
| 69 | REGULATION OF CELL DEATH | 40 | 1472 | 5.842e-06 | 0.0003939 |
| 70 | CAMERA TYPE EYE MORPHOGENESIS | 9 | 101 | 6.394e-06 | 0.000425 |
| 71 | GROWTH | 18 | 410 | 6.882e-06 | 0.000451 |
| 72 | NEGATIVE REGULATION OF NEURON DIFFERENTIATION | 12 | 191 | 7.391e-06 | 0.0004725 |
| 73 | REGULATION OF HISTONE METHYLATION | 7 | 56 | 7.413e-06 | 0.0004725 |
| 74 | RHYTHMIC PROCESS | 15 | 298 | 8.204e-06 | 0.0005159 |
| 75 | NEGATIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 14 | 262 | 8.396e-06 | 0.0005209 |
| 76 | REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 16 | 337 | 8.552e-06 | 0.0005236 |
| 77 | ADULT LOCOMOTORY BEHAVIOR | 8 | 80 | 8.947e-06 | 0.000527 |
| 78 | NEGATIVE REGULATION OF CELL DEATH | 28 | 872 | 8.942e-06 | 0.000527 |
| 79 | TELENCEPHALON DEVELOPMENT | 13 | 228 | 8.846e-06 | 0.000527 |
| 80 | ADULT BEHAVIOR | 10 | 135 | 1.028e-05 | 0.0005978 |
| 81 | HAIR CYCLE | 8 | 83 | 1.178e-05 | 0.0006602 |
| 82 | MOLTING CYCLE | 8 | 83 | 1.178e-05 | 0.0006602 |
| 83 | NEGATIVE REGULATION OF PROTEIN METABOLIC PROCESS | 32 | 1087 | 1.163e-05 | 0.0006602 |
| 84 | REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 10 | 138 | 1.247e-05 | 0.0006909 |
| 85 | BEHAVIOR | 20 | 516 | 1.334e-05 | 0.0007301 |
| 86 | RESPONSE TO OXYGEN LEVELS | 15 | 311 | 1.362e-05 | 0.000737 |
| 87 | CARDIAC SEPTUM DEVELOPMENT | 8 | 85 | 1.405e-05 | 0.0007431 |
| 88 | POSITIVE REGULATION OF GROWTH | 13 | 238 | 1.4e-05 | 0.0007431 |
| 89 | MUSCLE TISSUE DEVELOPMENT | 14 | 275 | 1.45e-05 | 0.0007582 |
| 90 | REGULATION OF CELL CYCLE | 29 | 949 | 1.556e-05 | 0.0008042 |
| 91 | CELLULAR COMPONENT MORPHOGENESIS | 28 | 900 | 1.594e-05 | 0.000806 |
| 92 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 26 | 801 | 1.582e-05 | 0.000806 |
| 93 | POSITIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 18 | 437 | 1.63e-05 | 0.0008153 |
| 94 | CELL PROJECTION ORGANIZATION | 28 | 902 | 1.659e-05 | 0.0008212 |
| 95 | GLIOGENESIS | 11 | 175 | 1.771e-05 | 0.0008611 |
| 96 | CELL FATE DETERMINATION | 6 | 43 | 1.777e-05 | 0.0008611 |
| 97 | CARDIAC CHAMBER DEVELOPMENT | 10 | 144 | 1.81e-05 | 0.0008626 |
| 98 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 30 | 1008 | 1.817e-05 | 0.0008626 |
| 99 | SKIN DEVELOPMENT | 12 | 211 | 2.013e-05 | 0.0009462 |
| 100 | REGULATION OF HOMEOSTATIC PROCESS | 18 | 447 | 2.201e-05 | 0.001024 |
| 101 | EYE DEVELOPMENT | 15 | 326 | 2.364e-05 | 0.001089 |
| 102 | CHROMATIN MODIFICATION | 20 | 539 | 2.49e-05 | 0.001125 |
| 103 | EMBRYONIC MORPHOGENESIS | 20 | 539 | 2.49e-05 | 0.001125 |
| 104 | REGULATION OF MYELOID CELL DIFFERENTIATION | 11 | 183 | 2.683e-05 | 0.001201 |
| 105 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE SIGNALING PATHWAY | 19 | 498 | 2.733e-05 | 0.001211 |
| 106 | REGULATION OF MUSCLE CELL DIFFERENTIATION | 10 | 152 | 2.89e-05 | 0.001269 |
| 107 | PALLIUM DEVELOPMENT | 10 | 153 | 3.057e-05 | 0.00133 |
| 108 | EPIDERMIS MORPHOGENESIS | 5 | 29 | 3.168e-05 | 0.001365 |
| 109 | NEGATIVE REGULATION OF CELL PROLIFERATION | 22 | 643 | 3.352e-05 | 0.001431 |
| 110 | CARDIOCYTE DIFFERENTIATION | 8 | 96 | 3.428e-05 | 0.00145 |
| 111 | TAXIS | 18 | 464 | 3.592e-05 | 0.001506 |
| 112 | EMBRYO DEVELOPMENT | 27 | 894 | 3.809e-05 | 0.001573 |
| 113 | GLOMERULUS DEVELOPMENT | 6 | 49 | 3.82e-05 | 0.001573 |
| 114 | MODULATION OF SYNAPTIC TRANSMISSION | 14 | 301 | 3.933e-05 | 0.001605 |
| 115 | MEMORY | 8 | 98 | 3.979e-05 | 0.00161 |
| 116 | REGULATION OF CELL PROJECTION ORGANIZATION | 20 | 558 | 4.052e-05 | 0.001625 |
| 117 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 38 | 1492 | 4.307e-05 | 0.001713 |
| 118 | CELL SURFACE RECEPTOR SIGNALING PATHWAY INVOLVED IN HEART DEVELOPMENT | 4 | 16 | 4.358e-05 | 0.001719 |
| 119 | ADULT WALKING BEHAVIOR | 5 | 31 | 4.438e-05 | 0.001721 |
| 120 | WALKING BEHAVIOR | 5 | 31 | 4.438e-05 | 0.001721 |
| 121 | LOCOMOTION | 31 | 1114 | 4.706e-05 | 0.00181 |
| 122 | ARTERY MORPHOGENESIS | 6 | 51 | 4.815e-05 | 0.001836 |
| 123 | POSITIVE REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 10 | 162 | 4.978e-05 | 0.001883 |
| 124 | ARTERY DEVELOPMENT | 7 | 75 | 5.152e-05 | 0.001927 |
| 125 | RETINA DEVELOPMENT IN CAMERA TYPE EYE | 9 | 131 | 5.176e-05 | 0.001927 |
| 126 | CHROMATIN ORGANIZATION | 22 | 663 | 5.285e-05 | 0.001952 |
| 127 | REGULATION OF GENE SILENCING | 6 | 52 | 5.384e-05 | 0.001973 |
| 128 | REGULATION OF ACTIN FILAMENT BASED PROCESS | 14 | 312 | 5.798e-05 | 0.002108 |
| 129 | REGULATION OF TRANSPORT | 43 | 1804 | 6.043e-05 | 0.00218 |
| 130 | CENTRAL NERVOUS SYSTEM NEURON DIFFERENTIATION | 10 | 166 | 6.117e-05 | 0.002189 |
| 131 | MUSCLE CELL DIFFERENTIATION | 12 | 237 | 6.264e-05 | 0.002225 |
| 132 | CELL PROLIFERATION | 22 | 672 | 6.443e-05 | 0.002271 |
| 133 | CEREBRAL CORTEX DEVELOPMENT | 8 | 105 | 6.525e-05 | 0.002283 |
| 134 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 31 | 1135 | 6.652e-05 | 0.00231 |
| 135 | GLIAL CELL DIFFERENTIATION | 9 | 136 | 6.931e-05 | 0.002371 |
| 136 | EYE MORPHOGENESIS | 9 | 136 | 6.931e-05 | 0.002371 |
| 137 | DENDRITE DEVELOPMENT | 7 | 79 | 7.203e-05 | 0.002446 |
| 138 | REGULATION OF GROWTH | 21 | 633 | 7.867e-05 | 0.002653 |
| 139 | EMBRYONIC ORGAN DEVELOPMENT | 16 | 406 | 8.137e-05 | 0.002724 |
| 140 | POSITIVE REGULATION OF MUSCLE TISSUE DEVELOPMENT | 6 | 56 | 8.225e-05 | 0.002734 |
| 141 | CARDIAC MUSCLE TISSUE DEVELOPMENT | 9 | 140 | 8.672e-05 | 0.002862 |
| 142 | CIRCADIAN REGULATION OF GENE EXPRESSION | 6 | 57 | 9.095e-05 | 0.00293 |
| 143 | REGULATION OF CARDIAC MUSCLE CELL ACTION POTENTIAL | 4 | 19 | 9.003e-05 | 0.00293 |
| 144 | KIDNEY MORPHOGENESIS | 7 | 82 | 9.142e-05 | 0.00293 |
| 145 | CARDIOVASCULAR SYSTEM DEVELOPMENT | 24 | 788 | 9.193e-05 | 0.00293 |
| 146 | CIRCULATORY SYSTEM DEVELOPMENT | 24 | 788 | 9.193e-05 | 0.00293 |
| 147 | PROTEIN PHOSPHORYLATION | 27 | 944 | 9.524e-05 | 0.002994 |
| 148 | HOMEOSTASIS OF NUMBER OF CELLS | 10 | 175 | 9.511e-05 | 0.002994 |
| 149 | REGULATION OF HYDROLASE ACTIVITY | 34 | 1327 | 0.0001013 | 0.003158 |
| 150 | HEART MORPHOGENESIS | 11 | 212 | 0.0001018 | 0.003158 |
| 151 | POSITIVE REGULATION OF MUSCLE CELL DIFFERENTIATION | 7 | 84 | 0.0001066 | 0.003284 |
| 152 | TUBE DEVELOPMENT | 19 | 552 | 0.0001076 | 0.003295 |
| 153 | GENETIC IMPRINTING | 4 | 20 | 0.0001114 | 0.003344 |
| 154 | REGULATION OF SYSTEM PROCESS | 18 | 507 | 0.0001113 | 0.003344 |
| 155 | REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 6 | 59 | 0.0001105 | 0.003344 |
| 156 | SYNAPSE ORGANIZATION | 9 | 145 | 0.0001135 | 0.003386 |
| 157 | RESPONSE TO ENDOGENOUS STIMULUS | 36 | 1450 | 0.000117 | 0.003466 |
| 158 | POSITIVE REGULATION OF HYDROLASE ACTIVITY | 26 | 905 | 0.0001201 | 0.003538 |
| 159 | REGULATION OF MITOTIC CELL CYCLE | 17 | 468 | 0.0001315 | 0.003847 |
| 160 | DIGESTIVE SYSTEM DEVELOPMENT | 9 | 148 | 0.0001327 | 0.003859 |
| 161 | REGULATION OF CHROMATIN SILENCING | 4 | 21 | 0.0001362 | 0.003937 |
| 162 | MYELOID CELL HOMEOSTASIS | 7 | 88 | 0.000143 | 0.004106 |
| 163 | CARDIAC VENTRICLE MORPHOGENESIS | 6 | 62 | 0.000146 | 0.004168 |
| 164 | RESPONSE TO GROWTH FACTOR | 17 | 475 | 0.0001569 | 0.004451 |
| 165 | REGULATION OF CHROMATIN ORGANIZATION | 9 | 152 | 0.0001624 | 0.004581 |
| 166 | RETINA LAYER FORMATION | 4 | 22 | 0.0001648 | 0.00462 |
| 167 | NEGATIVE REGULATION OF CELL CYCLE | 16 | 433 | 0.0001706 | 0.004753 |
| 168 | REGULATION OF MEMBRANE DEPOLARIZATION | 5 | 41 | 0.0001762 | 0.00488 |
| 169 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 28 | 1036 | 0.0001821 | 0.004928 |
| 170 | POSITIVE REGULATION OF NEURON DIFFERENTIATION | 13 | 306 | 0.0001827 | 0.004928 |
| 171 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 28 | 1036 | 0.0001821 | 0.004928 |
| 172 | REGULATION OF CELL GROWTH | 15 | 391 | 0.0001818 | 0.004928 |
| 173 | NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 27 | 983 | 0.0001843 | 0.004928 |
| 174 | REGULATION OF GTPASE ACTIVITY | 21 | 673 | 0.0001836 | 0.004928 |
| 175 | POSITIVE REGULATION OF DEVELOPMENTAL GROWTH | 9 | 156 | 0.0001975 | 0.005163 |
| 176 | PROTEIN TARGETING TO PLASMA MEMBRANE | 4 | 23 | 0.0001975 | 0.005163 |
| 177 | REGULATION OF METANEPHROS DEVELOPMENT | 4 | 23 | 0.0001975 | 0.005163 |
| 178 | HEPARAN SULFATE PROTEOGLYCAN BIOSYNTHETIC PROCESS | 4 | 23 | 0.0001975 | 0.005163 |
| 179 | REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 11 | 229 | 0.0002003 | 0.005207 |
| 180 | POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 30 | 1152 | 0.0002022 | 0.005228 |
| 181 | IMMUNE SYSTEM DEVELOPMENT | 19 | 582 | 0.000212 | 0.00545 |
| 182 | ACTION POTENTIAL | 7 | 94 | 0.0002159 | 0.00552 |
| 183 | CELL PART MORPHOGENESIS | 20 | 633 | 0.0002224 | 0.005593 |
| 184 | PEPTIDYL LYSINE MODIFICATION | 13 | 312 | 0.0002208 | 0.005593 |
| 185 | REGULATION OF INSULIN RECEPTOR SIGNALING PATHWAY | 5 | 43 | 0.0002216 | 0.005593 |
| 186 | REGULATION OF MYELOID CELL APOPTOTIC PROCESS | 4 | 24 | 0.0002346 | 0.005715 |
| 187 | MORPHOGENESIS OF AN EPITHELIUM | 15 | 400 | 0.0002322 | 0.005715 |
| 188 | POSITIVE REGULATION OF CELLULAR RESPONSE TO TRANSFORMING GROWTH FACTOR BETA STIMULUS | 4 | 24 | 0.0002346 | 0.005715 |
| 189 | EAR DEVELOPMENT | 10 | 195 | 0.0002305 | 0.005715 |
| 190 | POSITIVE REGULATION OF TRANSFORMING GROWTH FACTOR BETA RECEPTOR SIGNALING PATHWAY | 4 | 24 | 0.0002346 | 0.005715 |
| 191 | REGULATION OF MUSCLE SYSTEM PROCESS | 10 | 195 | 0.0002305 | 0.005715 |
| 192 | EPITHELIUM DEVELOPMENT | 26 | 945 | 0.0002372 | 0.005747 |
| 193 | MOVEMENT OF CELL OR SUBCELLULAR COMPONENT | 32 | 1275 | 0.0002384 | 0.005747 |
| 194 | PEPTIDYL AMINO ACID MODIFICATION | 24 | 841 | 0.0002439 | 0.00582 |
| 195 | ORGAN GROWTH | 6 | 68 | 0.0002437 | 0.00582 |
| 196 | BRANCHING INVOLVED IN URETERIC BUD MORPHOGENESIS | 5 | 44 | 0.0002474 | 0.005874 |
| 197 | HOMEOSTATIC PROCESS | 33 | 1337 | 0.0002565 | 0.006059 |
| 198 | REGULATION OF TRANSPORTER ACTIVITY | 10 | 198 | 0.0002606 | 0.006124 |
| 199 | VENTRICULAR CARDIAC MUSCLE TISSUE DEVELOPMENT | 5 | 45 | 0.0002755 | 0.006408 |
| 200 | POSITIVE REGULATION OF RECEPTOR ACTIVITY | 5 | 45 | 0.0002755 | 0.006408 |
| 201 | REGULATION OF SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 12 | 278 | 0.0002788 | 0.006453 |
| 202 | ANATOMICAL STRUCTURE FORMATION INVOLVED IN MORPHOGENESIS | 26 | 957 | 0.0002882 | 0.0066 |
| 203 | REPRODUCTIVE SYSTEM DEVELOPMENT | 15 | 408 | 0.0002868 | 0.0066 |
| 204 | SENSORY ORGAN MORPHOGENESIS | 11 | 239 | 0.0002894 | 0.0066 |
| 205 | REGULATION OF TRANSFORMING GROWTH FACTOR BETA RECEPTOR SIGNALING PATHWAY | 7 | 99 | 0.0002974 | 0.006695 |
| 206 | REGULATION OF CELLULAR RESPONSE TO TRANSFORMING GROWTH FACTOR BETA STIMULUS | 7 | 99 | 0.0002974 | 0.006695 |
| 207 | REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 14 | 365 | 0.0002978 | 0.006695 |
| 208 | TUBE MORPHOGENESIS | 13 | 323 | 0.0003085 | 0.006876 |
| 209 | CELL FATE SPECIFICATION | 6 | 71 | 0.0003088 | 0.006876 |
| 210 | POSITIVE REGULATION OF SKELETAL MUSCLE TISSUE DEVELOPMENT | 4 | 26 | 0.0003234 | 0.007151 |
| 211 | ENDOCARDIUM DEVELOPMENT | 3 | 11 | 0.0003243 | 0.007151 |
| 212 | REGULATION OF BINDING | 12 | 283 | 0.0003277 | 0.007193 |
| 213 | RESPONSE TO ABIOTIC STIMULUS | 27 | 1024 | 0.0003518 | 0.007686 |
| 214 | REGULATION OF CELL CYCLE PROCESS | 18 | 558 | 0.0003583 | 0.007754 |
| 215 | NEGATIVE REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 7 | 102 | 0.000357 | 0.007754 |
| 216 | REGULATION OF CARDIAC MUSCLE CELL CONTRACTION | 4 | 27 | 0.0003759 | 0.008051 |
| 217 | REGULATION OF MUSCLE TISSUE DEVELOPMENT | 7 | 103 | 0.0003789 | 0.008051 |
| 218 | REGULATION OF ASTROCYTE DIFFERENTIATION | 4 | 27 | 0.0003759 | 0.008051 |
| 219 | POSITIVE REGULATION OF CELL PROLIFERATION | 23 | 814 | 0.0003782 | 0.008051 |
| 220 | POSITIVE REGULATION OF LOCOMOTION | 15 | 420 | 0.0003893 | 0.008233 |
| 221 | CARDIAC CHAMBER MORPHOGENESIS | 7 | 104 | 0.0004019 | 0.008461 |
| 222 | CIRCADIAN RHYTHM | 8 | 137 | 0.0004109 | 0.008564 |
| 223 | CARDIAC SEPTUM MORPHOGENESIS | 5 | 49 | 0.0004123 | 0.008564 |
| 224 | CARDIAC CELL DEVELOPMENT | 5 | 49 | 0.0004123 | 0.008564 |
| 225 | REGULATION OF TIMING OF CELL DIFFERENTIATION | 3 | 12 | 0.0004283 | 0.008779 |
| 226 | POSITIVE REGULATION OF INSULIN RECEPTOR SIGNALING PATHWAY | 3 | 12 | 0.0004283 | 0.008779 |
| 227 | REGULATION OF DEVELOPMENT HETEROCHRONIC | 3 | 12 | 0.0004283 | 0.008779 |
| 228 | HEPARAN SULFATE PROTEOGLYCAN METABOLIC PROCESS | 4 | 28 | 0.0004341 | 0.00882 |
| 229 | REGULATION OF SPROUTING ANGIOGENESIS | 4 | 28 | 0.0004341 | 0.00882 |
| 230 | POSITIVE REGULATION OF TRANSPORTER ACTIVITY | 6 | 76 | 0.0004468 | 0.00904 |
| 231 | REGULATION OF TRANSMEMBRANE TRANSPORT | 15 | 426 | 0.0004514 | 0.009092 |
| 232 | CEREBRAL CORTEX RADIALLY ORIENTED CELL MIGRATION | 4 | 29 | 0.0004985 | 0.009998 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | REGULATORY REGION NUCLEIC ACID BINDING | 34 | 818 | 1.811e-09 | 1.683e-06 |
| 2 | DOUBLE STRANDED DNA BINDING | 30 | 764 | 6.265e-08 | 2.91e-05 |
| 3 | SEQUENCE SPECIFIC DNA BINDING | 35 | 1037 | 1.889e-07 | 5.849e-05 |
| 4 | TRANSCRIPTION FACTOR ACTIVITY PROTEIN BINDING | 23 | 588 | 2.531e-06 | 0.0005878 |
| 5 | NUCLEIC ACID BINDING TRANSCRIPTION FACTOR ACTIVITY | 35 | 1199 | 5.222e-06 | 0.0008085 |
| 6 | TRANSCRIPTION FACTOR BINDING | 21 | 524 | 4.844e-06 | 0.0008085 |
| 7 | RAS GUANYL NUCLEOTIDE EXCHANGE FACTOR ACTIVITY | 13 | 228 | 8.846e-06 | 0.001174 |
| 8 | ENZYME BINDING | 44 | 1737 | 1.118e-05 | 0.001298 |
| 9 | CHROMATIN BINDING | 18 | 435 | 1.533e-05 | 0.001582 |
| 10 | RAB GTPASE BINDING | 9 | 120 | 2.589e-05 | 0.002396 |
| 11 | MOLECULAR FUNCTION REGULATOR | 36 | 1353 | 2.837e-05 | 0.002396 |
| 12 | NOTCH BINDING | 4 | 18 | 7.18e-05 | 0.005558 |
| 13 | TRANSMEMBRANE RECEPTOR PROTEIN KINASE ACTIVITY | 7 | 81 | 8.453e-05 | 0.006041 |
| 14 | ION CHANNEL BINDING | 8 | 111 | 9.669e-05 | 0.006416 |
| 15 | CALMODULIN BINDING | 10 | 179 | 0.0001147 | 0.007102 |
| 16 | CORE PROMOTER BINDING | 9 | 152 | 0.0001624 | 0.008995 |
| 17 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE ACTIVITY | 6 | 64 | 0.0001743 | 0.008995 |
| 18 | GUANYL NUCLEOTIDE EXCHANGE FACTOR ACTIVITY | 13 | 303 | 0.0001659 | 0.008995 |
| 19 | RNA POLYMERASE II TRANSCRIPTION FACTOR ACTIVITY SEQUENCE SPECIFIC DNA BINDING | 20 | 629 | 0.0002047 | 0.009822 |
| 20 | ZINC ION BINDING | 30 | 1155 | 0.0002114 | 0.009822 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CELL JUNCTION | 36 | 1151 | 7.702e-07 | 0.0004498 |
| 2 | CELL CELL CONTACT ZONE | 7 | 64 | 1.819e-05 | 0.002124 |
| 3 | NEURON PART | 35 | 1265 | 1.649e-05 | 0.002124 |
| 4 | SYNAPSE | 25 | 754 | 1.63e-05 | 0.002124 |
| 5 | MEMBRANE REGION | 33 | 1134 | 1.064e-05 | 0.002124 |
| 6 | T TUBULE | 6 | 45 | 2.323e-05 | 0.002261 |
| 7 | PLASMA MEMBRANE REGION | 28 | 929 | 2.815e-05 | 0.002348 |
| 8 | CELL CELL JUNCTION | 16 | 383 | 4.089e-05 | 0.002985 |
| 9 | INTERCALATED DISC | 6 | 51 | 4.815e-05 | 0.003124 |
| 10 | NEURON PROJECTION | 27 | 942 | 9.195e-05 | 0.005328 |
| 11 | MAIN AXON | 6 | 58 | 0.0001004 | 0.005328 |
| 12 | NUCLEAR CHROMATIN | 13 | 291 | 0.0001112 | 0.005413 |
| 13 | CHROMATIN | 16 | 441 | 0.0002098 | 0.009423 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
| 1 | hsa00534_Glycosaminoglycan_biosynthesis_._heparan_sulfate | 4 | 26 | 0.0003234 | 0.02997 | |
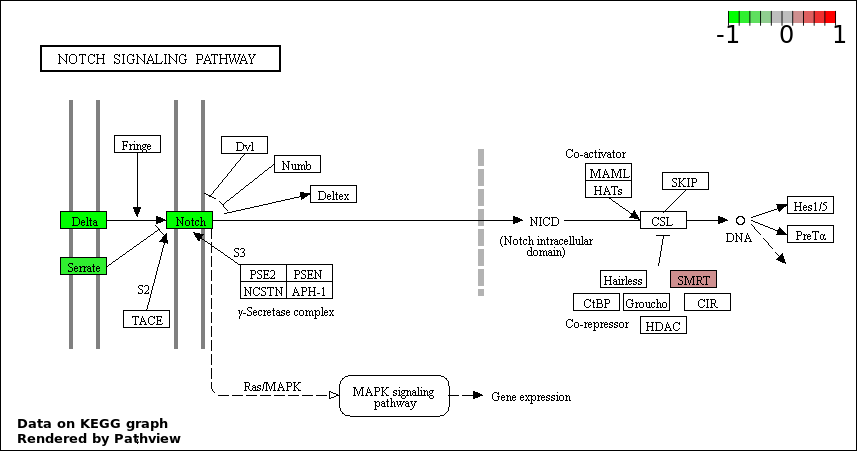
| 2 | hsa04330_Notch_signaling_pathway | 5 | 47 | 0.0003387 | 0.02997 | |
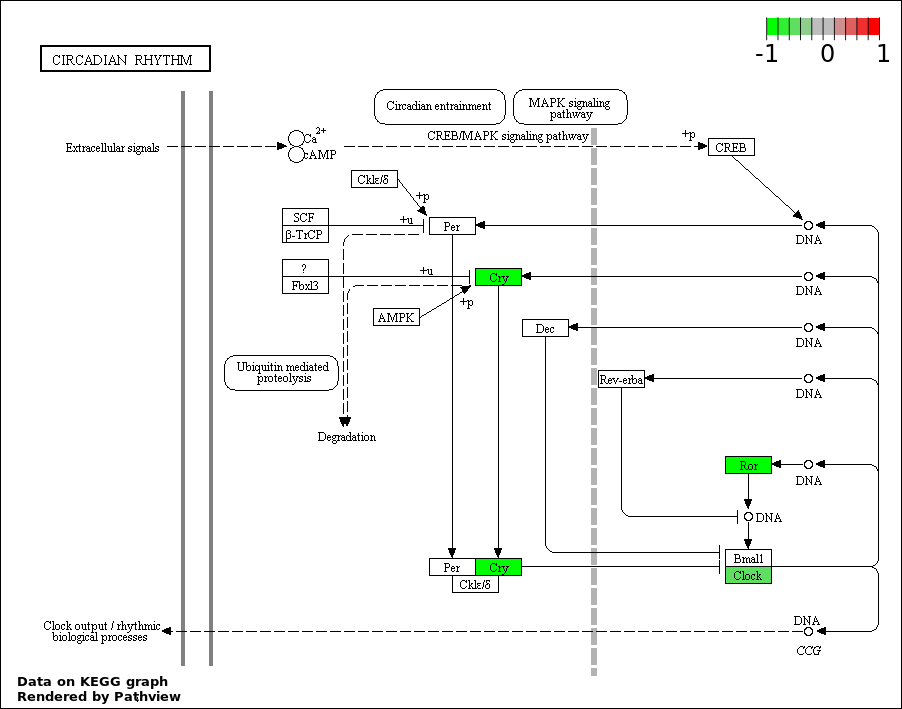
| 3 | hsa04710_Circadian_rhythm_._mammal | 3 | 23 | 0.003105 | 0.1832 | |
| 4 | hsa04540_Gap_junction | 5 | 90 | 0.006202 | 0.2256 | |
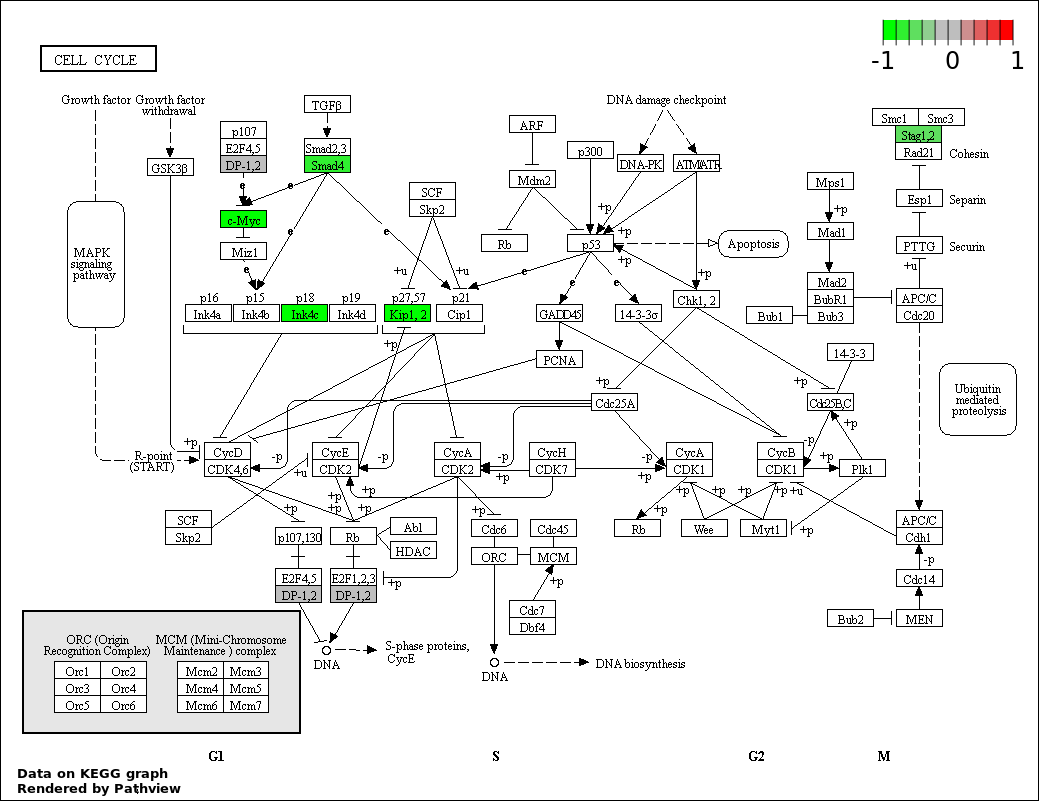
| 5 | hsa04110_Cell_cycle | 6 | 128 | 0.006374 | 0.2256 | |
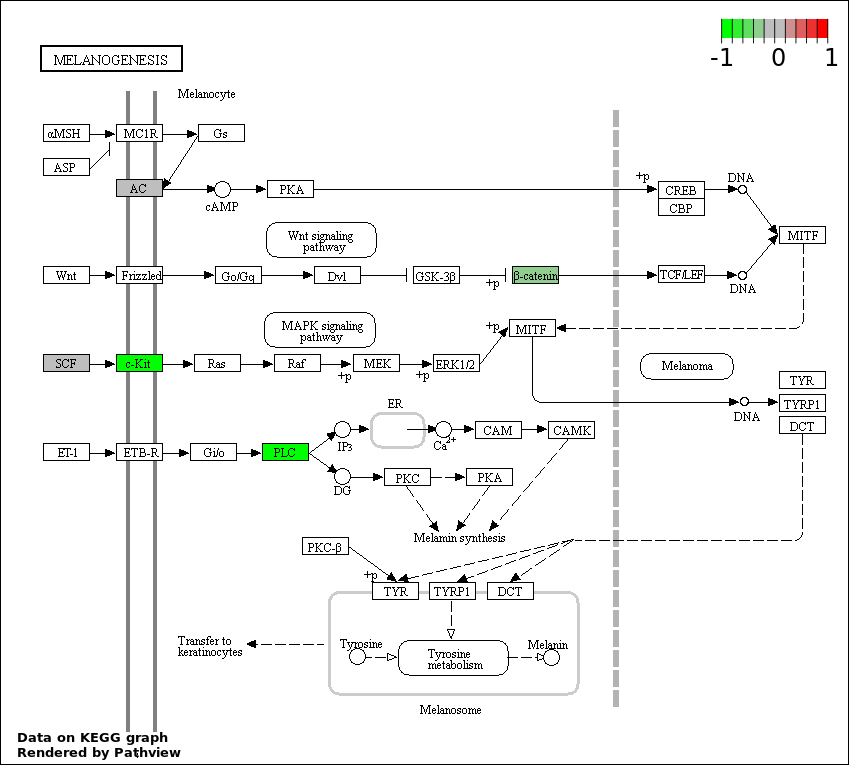
| 6 | hsa04916_Melanogenesis | 5 | 101 | 0.009979 | 0.2944 | |
| 7 | hsa04520_Adherens_junction | 4 | 73 | 0.0147 | 0.3655 | |
| 8 | hsa04144_Endocytosis | 7 | 203 | 0.01652 | 0.3655 | |
| 9 | hsa04020_Calcium_signaling_pathway | 6 | 177 | 0.02754 | 0.5416 | |
| 10 | hsa04320_Dorso.ventral_axis_formation | 2 | 25 | 0.04093 | 0.7244 | |
| 11 | hsa04510_Focal_adhesion | 6 | 200 | 0.04558 | 0.7334 | |
| 12 | hsa04720_Long.term_potentiation | 3 | 70 | 0.06185 | 0.8635 | |
| 13 | hsa04270_Vascular_smooth_muscle_contraction | 4 | 116 | 0.06342 | 0.8635 | |
| 14 | hsa04971_Gastric_acid_secretion | 3 | 74 | 0.0706 | 0.8926 | |
| 15 | hsa04070_Phosphatidylinositol_signaling_system | 3 | 78 | 0.07988 | 0.9426 | |
| 16 | hsa04360_Axon_guidance | 4 | 130 | 0.08788 | 0.9548 | |
| 17 | hsa04530_Tight_junction | 4 | 133 | 0.09363 | 0.9548 | |
| 18 | hsa04350_TGF.beta_signaling_pathway | 3 | 85 | 0.0973 | 0.9548 | |
| 19 | hsa04640_Hematopoietic_cell_lineage | 3 | 88 | 0.1052 | 0.9548 | |
| 20 | hsa04970_Salivary_secretion | 3 | 89 | 0.1079 | 0.9548 | |
| 21 | hsa04310_Wnt_signaling_pathway | 4 | 151 | 0.1317 | 1 | |
| 22 | hsa04912_GnRH_signaling_pathway | 3 | 101 | 0.1421 | 1 | |
| 23 | hsa04972_Pancreatic_secretion | 3 | 101 | 0.1421 | 1 | |
| 24 | hsa00562_Inositol_phosphate_metabolism | 2 | 57 | 0.1674 | 1 | |
| 25 | hsa04920_Adipocytokine_signaling_pathway | 2 | 68 | 0.2188 | 1 | |
| 26 | hsa04722_Neurotrophin_signaling_pathway | 3 | 127 | 0.2257 | 1 | |
| 27 | hsa04730_Long.term_depression | 2 | 70 | 0.2283 | 1 | |
| 28 | hsa04012_ErbB_signaling_pathway | 2 | 87 | 0.3095 | 1 | |
| 29 | hsa00230_Purine_metabolism | 3 | 162 | 0.3479 | 1 | |
| 30 | hsa00240_Pyrimidine_metabolism | 2 | 99 | 0.3659 | 1 | |
| 31 | hsa04114_Oocyte_meiosis | 2 | 114 | 0.4338 | 1 | |
| 32 | hsa04514_Cell_adhesion_molecules_.CAMs. | 2 | 136 | 0.5257 | 1 | |
| 33 | hsa04120_Ubiquitin_mediated_proteolysis | 2 | 139 | 0.5375 | 1 | |
| 34 | hsa04010_MAPK_signaling_pathway | 3 | 268 | 0.6748 | 1 | |
| 35 | hsa04062_Chemokine_signaling_pathway | 2 | 189 | 0.7032 | 1 | |
| 36 | hsa04810_Regulation_of_actin_cytoskeleton | 2 | 214 | 0.7658 | 1 |