This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-23a-3p | APAF1 | 0.11 | 0.39309 | 0.05 | 0.6283 | miRNATAP | -0.13 | 0.00069 | 24992592; 24249161 | Luciferase assay was performed to verify a putative target site of miR-23a in the 3'-UTR of apoptosis protease activating factor 1 APAF1 mRNA; The expression levels of miR-23a and APAF1 in CRC cell lines SW480 and SW620 and clinical samples were assessed using reverse transcription-quantitative real-time PCR RT-qPCR and Western blot; Moreover miR-23a up-regulation was coupled with APAF1 down-regulation in CRC tissue samples;We found that the expression of miR-23a was increased and the level of apoptosis-activating factor-1 APAF-1 was decreased in 5-FU-treated colon cancer cells compared to untreated cells; APAF-1 as a target gene of miR-23a was identified and miR-23a antisense-induced increase in the activation of caspase-9 was observed |
| 2 | hsa-miR-23b-3p | APAF1 | -0.07 | 0.62059 | 0.05 | 0.6283 | miRNATAP | -0.11 | 0.00216 | NA | |
| 3 | hsa-miR-27a-3p | APAF1 | 0.43 | 0.00737 | 0.05 | 0.6283 | miRNATAP | -0.16 | 0 | NA | |
| 4 | hsa-miR-27b-3p | APAF1 | 0.24 | 0.12264 | 0.05 | 0.6283 | miRNATAP | -0.17 | 0 | NA | |
| 5 | hsa-miR-664a-3p | APAF1 | 0.44 | 0.02142 | 0.05 | 0.6283 | mirMAP | -0.11 | 2.0E-5 | NA | |
| 6 | hsa-miR-664a-5p | ATR | -0.09 | 0.66227 | 0.55 | 0 | mirMAP | -0.1 | 9.0E-5 | NA | |
| 7 | hsa-miR-143-3p | BBC3 | -1.21 | 1.0E-5 | 0.73 | 0.00024 | miRNATAP | -0.26 | 0 | NA | |
| 8 | hsa-miR-423-5p | BBC3 | -1.8 | 0 | 0.73 | 0.00024 | PITA | -0.19 | 2.0E-5 | NA | |
| 9 | hsa-miR-139-5p | CASP3 | -2.27 | 0 | 0.75 | 0 | miRanda | -0.1 | 0 | NA | |
| 10 | hsa-miR-195-3p | CASP3 | -1.33 | 0 | 0.75 | 0 | MirTarget | -0.12 | 0 | NA | |
| 11 | hsa-miR-30c-5p | CASP3 | -0.33 | 0.1236 | 0.75 | 0 | miRNATAP | -0.15 | 0 | NA | |
| 12 | hsa-miR-30d-5p | CASP3 | -0.92 | 4.0E-5 | 0.75 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.17 | 0 | NA | |
| 13 | hsa-let-7b-5p | CCNB1 | -1.62 | 0 | 2.59 | 0 | miRNAWalker2 validate | -0.6 | 0 | NA | |
| 14 | hsa-miR-139-5p | CCNB1 | -2.27 | 0 | 2.59 | 0 | miRanda | -0.4 | 0 | NA | |
| 15 | hsa-let-7a-5p | CCNB2 | -1.37 | 0 | 3.05 | 0 | miRNAWalker2 validate | -0.57 | 0 | NA | |
| 16 | hsa-let-7b-5p | CCNB2 | -1.62 | 0 | 3.05 | 0 | miRNAWalker2 validate | -0.61 | 0 | NA | |
| 17 | hsa-let-7c-5p | CCNB2 | -2.14 | 0 | 3.05 | 0 | miRNAWalker2 validate | -0.62 | 0 | NA | |
| 18 | hsa-miR-23b-3p | CCNB2 | -0.07 | 0.62059 | 3.05 | 0 | miRNAWalker2 validate | -0.6 | 0 | NA | |
| 19 | hsa-miR-106a-5p | CCND1 | 1.39 | 6.0E-5 | -0.3 | 0.2554 | MirTarget; miRNATAP | -0.25 | 0 | NA | |
| 20 | hsa-miR-106b-5p | CCND1 | 1.47 | 0 | -0.3 | 0.2554 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.26 | 1.0E-5 | NA | |
| 21 | hsa-miR-142-3p | CCND1 | 3.98 | 0 | -0.3 | 0.2554 | miRanda | -0.12 | 0.00053 | 23619912 | Transfection of miR-142-3p mimics in colon cancer cells downregulated cyclin D1 expression induced G1 phase cell cycle arrest and elevated the sensitivity of the cells to 5-fluorouracil |
| 22 | hsa-miR-15a-5p | CCND1 | 1.63 | 0 | -0.3 | 0.2554 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.2 | 0.00193 | 22922827 | CCND1 has been found to be a target of miR-15a and miR-16-1 through analysis of complementary sequences between microRNAs and CCND1 mRNA; Moreover the transcription of CCND1 is suppressed by miR-15a and miR-16-1 via direct binding to the CCND1 3'-untranslated region 3'-UTR |
| 23 | hsa-miR-15b-5p | CCND1 | -1.26 | 0 | -0.3 | 0.2554 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.3 | 0 | NA | |
| 24 | hsa-miR-16-1-3p | CCND1 | 1.5 | 0 | -0.3 | 0.2554 | miRNAWalker2 validate; miRTarBase | -0.25 | 3.0E-5 | 22922827; 18483394 | CCND1 has been found to be a target of miR-15a and miR-16-1 through analysis of complementary sequences between microRNAs and CCND1 mRNA; Moreover the transcription of CCND1 is suppressed by miR-15a and miR-16-1 via direct binding to the CCND1 3'-untranslated region 3'-UTR;Truncation in CCND1 mRNA alters miR 16 1 regulation in mantle cell lymphoma; Furthermore we demonstrated that this truncation alters miR-16-1 binding sites and through the use of reporter constructs we were able to show that miR-16-1 regulates CCND1 mRNA expression; This study introduces the role of miR-16-1 in the regulation of CCND1 in MCL |
| 25 | hsa-miR-16-5p | CCND1 | 0.75 | 0 | -0.3 | 0.2554 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.37 | 0 | 23991964; 22922827; 18483394 | At the molecular level our results further revealed that cyclin D1 expression was negatively regulated by miR-16;CCND1 has been found to be a target of miR-15a and miR-16-1 through analysis of complementary sequences between microRNAs and CCND1 mRNA; Moreover the transcription of CCND1 is suppressed by miR-15a and miR-16-1 via direct binding to the CCND1 3'-untranslated region 3'-UTR;Truncation in CCND1 mRNA alters miR 16 1 regulation in mantle cell lymphoma; Furthermore we demonstrated that this truncation alters miR-16-1 binding sites and through the use of reporter constructs we were able to show that miR-16-1 regulates CCND1 mRNA expression; This study introduces the role of miR-16-1 in the regulation of CCND1 in MCL |
| 26 | hsa-miR-17-5p | CCND1 | 2.07 | 0 | -0.3 | 0.2554 | miRNAWalker2 validate; MirTarget; TargetScan; miRNATAP | -0.31 | 0 | 26431674 | Bioinformatics Prediction and In Vitro Analysis Revealed That miR 17 Targets Cyclin D1 mRNA in Triple Negative Breast Cancer Cells; In this study using bioinformatic analyses miR-17 was selected as it targets the 3'UTR of CCND1 gene with the highest score; After lentiviral transduction of miR-17 to the target cells gene expression analysis showed decreased expression of CCND1 gene |
| 27 | hsa-miR-186-5p | CCND1 | 0.85 | 0 | -0.3 | 0.2554 | mirMAP | -0.38 | 1.0E-5 | NA | |
| 28 | hsa-miR-193a-3p | CCND1 | 0.55 | 0.0319 | -0.3 | 0.2554 | MirTarget; PITA; miRanda | -0.19 | 0.00016 | NA | |
| 29 | hsa-miR-193b-3p | CCND1 | 1.1 | 0.00082 | -0.3 | 0.2554 | miRNAWalker2 validate; miRTarBase; MirTarget | -0.15 | 4.0E-5 | 27071318; 20655737; 20304954; 21893020; 26129688 | MicroRNA 193b inhibits the proliferation migration and invasion of gastric cancer cells via targeting cyclin D1; Further mechanism study indicated that CCND1 was a direct target of miR-193b in GC;CCND1 and ETS1 were revealed to be regulated by miR-193b directly;MicroRNA 193b represses cell proliferation and regulates cyclin D1 in melanoma; Overexpression of miR-193b in Malme-3M cells down-regulated CCND1 mRNA and protein by > or = 50%; A luciferase reporter assay confirmed that miR-193b directly regulates CCND1 by binding to the 3'untranslated region of CCND1 mRNA; These studies indicate that miR-193b represses cell proliferation and regulates CCND1 expression and suggest that dysregulation of miR-193b may play an important role in melanoma development;In a previous study we reported that miR-193b represses cell proliferation and regulates cyclin D1 in melanoma cells suggesting that miR-193b could act as a tumor suppressor;Epigenetically altered miR 193b targets cyclin D1 in prostate cancer; It has been suggested that miR-193b targets cyclin D1 in several malignancies; Here our aim was to determine if miR-193b targets cyclin D1 in prostate cancer; Furthermore the PC cell lines 22Rv1 and VCaP which express low levels of miR-193b and high levels of CCND1 showed significant growth retardation when treated with a CDK4/6 inhibitor; In contrast the inhibitor had no effect on the growth of PC-3 and DU145 cells with high miR-193b and low CCND1 expression; Taken together our data demonstrate that miR-193b targets cyclin D1 in prostate cancer |
| 30 | hsa-miR-19a-3p | CCND1 | 2.12 | 0 | -0.3 | 0.2554 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.22 | 0 | 25985117 | Moreover miR-19a might play inhibitory roles in HCC malignancy via regulating Cyclin D1 expression |
| 31 | hsa-miR-19b-1-5p | CCND1 | 1.71 | 0 | -0.3 | 0.2554 | miRNAWalker2 validate; miRTarBase | -0.3 | 0 | NA | |
| 32 | hsa-miR-19b-3p | CCND1 | 2.11 | 0 | -0.3 | 0.2554 | miRNATAP | -0.18 | 0.0002 | NA | |
| 33 | hsa-miR-20a-5p | CCND1 | 2.65 | 0 | -0.3 | 0.2554 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.24 | 0 | NA | |
| 34 | hsa-miR-20b-5p | CCND1 | 1.36 | 0.00261 | -0.3 | 0.2554 | MirTarget; miRNATAP | -0.18 | 0 | NA | |
| 35 | hsa-miR-365a-3p | CCND1 | 0.01 | 0.9536 | -0.3 | 0.2554 | miRNAWalker2 validate; miRTarBase | -0.19 | 0.00023 | NA | |
| 36 | hsa-miR-374a-5p | CCND1 | -0.2 | 0.29808 | -0.3 | 0.2554 | MirTarget | -0.31 | 1.0E-5 | 27191497 | microRNA 374a suppresses colon cancer progression by directly reducing CCND1 to inactivate the PI3K/AKT pathway; Furthermore luciferase reporter assays confirmed that miR-374a could directly reduce CCND1; We examined miR-374a levels by in situ hybridization and its correlation with CCND1 expression in CRC tumor tissues; High miR-374a expression with low level of CCND1 was protective factor in CRC; Together these findings indicate that miR-374a inactivates the PI3K/AKT axis by inhibiting CCND1 suppressing of colon cancer progression |
| 37 | hsa-miR-374b-5p | CCND1 | 0.47 | 0.01092 | -0.3 | 0.2554 | miRNAWalker2 validate; MirTarget | -0.26 | 0.00012 | NA | |
| 38 | hsa-miR-425-5p | CCND1 | 1.22 | 0 | -0.3 | 0.2554 | miRNAWalker2 validate | -0.25 | 1.0E-5 | NA | |
| 39 | hsa-miR-589-3p | CCND1 | 1.34 | 2.0E-5 | -0.3 | 0.2554 | MirTarget | -0.13 | 0.00079 | NA | |
| 40 | hsa-miR-590-3p | CCND1 | 0.84 | 0.00129 | -0.3 | 0.2554 | mirMAP | -0.18 | 0.00086 | NA | |
| 41 | hsa-miR-769-3p | CCND1 | 0.45 | 0.07482 | -0.3 | 0.2554 | mirMAP | -0.14 | 0.00277 | NA | |
| 42 | hsa-miR-92a-3p | CCND1 | -0.14 | 0.49341 | -0.3 | 0.2554 | miRNAWalker2 validate | -0.34 | 0 | NA | |
| 43 | hsa-miR-93-5p | CCND1 | 1.51 | 0 | -0.3 | 0.2554 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.32 | 0 | NA | |
| 44 | hsa-miR-942-5p | CCND1 | -0.04 | 0.87063 | -0.3 | 0.2554 | MirTarget | -0.18 | 0.00098 | NA | |
| 45 | hsa-miR-106a-5p | CCND2 | 1.39 | 6.0E-5 | -1.64 | 0 | miRNATAP | -0.15 | 2.0E-5 | NA | |
| 46 | hsa-miR-106b-5p | CCND2 | 1.47 | 0 | -1.64 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.38 | 0 | NA | |
| 47 | hsa-miR-130b-5p | CCND2 | 1.54 | 0 | -1.64 | 0 | mirMAP | -0.27 | 0 | NA | |
| 48 | hsa-miR-141-3p | CCND2 | 3.37 | 0 | -1.64 | 0 | MirTarget; TargetScan | -0.24 | 0 | NA | |
| 49 | hsa-miR-151a-3p | CCND2 | 0.67 | 0.00028 | -1.64 | 0 | mirMAP | -0.28 | 0 | NA | |
| 50 | hsa-miR-15a-5p | CCND2 | 1.63 | 0 | -1.64 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.16 | 0.00712 | NA | |
| 51 | hsa-miR-17-5p | CCND2 | 2.07 | 0 | -1.64 | 0 | miRNAWalker2 validate; miRTarBase; TargetScan; miRNATAP | -0.37 | 0 | NA | |
| 52 | hsa-miR-182-5p | CCND2 | 3.22 | 0 | -1.64 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.37 | 0 | NA | |
| 53 | hsa-miR-183-5p | CCND2 | 2.39 | 0 | -1.64 | 0 | miRNATAP | -0.41 | 0 | NA | |
| 54 | hsa-miR-185-5p | CCND2 | 1.14 | 0 | -1.64 | 0 | MirTarget; miRNATAP | -0.24 | 0.00048 | NA | |
| 55 | hsa-miR-186-5p | CCND2 | 0.85 | 0 | -1.64 | 0 | mirMAP; miRNATAP | -0.36 | 1.0E-5 | NA | |
| 56 | hsa-miR-191-5p | CCND2 | 0.34 | 0.06681 | -1.64 | 0 | MirTarget | -0.2 | 0.00075 | NA | |
| 57 | hsa-miR-19a-3p | CCND2 | 2.12 | 0 | -1.64 | 0 | MirTarget; miRNATAP | -0.17 | 1.0E-5 | NA | |
| 58 | hsa-miR-19b-3p | CCND2 | 2.11 | 0 | -1.64 | 0 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.19 | 1.0E-5 | NA | |
| 59 | hsa-miR-200a-3p | CCND2 | 3.15 | 0 | -1.64 | 0 | MirTarget | -0.17 | 0 | NA | |
| 60 | hsa-miR-20a-5p | CCND2 | 2.65 | 0 | -1.64 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.28 | 0 | NA | |
| 61 | hsa-miR-21-3p | CCND2 | 2.54 | 0 | -1.64 | 0 | mirMAP | -0.21 | 0 | NA | |
| 62 | hsa-miR-2355-3p | CCND2 | 1.11 | 1.0E-5 | -1.64 | 0 | miRNATAP | -0.22 | 0 | NA | |
| 63 | hsa-miR-28-5p | CCND2 | 1.2 | 0 | -1.64 | 0 | miRanda | -0.29 | 0.00018 | NA | |
| 64 | hsa-miR-301a-3p | CCND2 | 2.7 | 0 | -1.64 | 0 | miRNAWalker2 validate | -0.27 | 0 | NA | |
| 65 | hsa-miR-320b | CCND2 | 0.23 | 0.37882 | -1.64 | 0 | mirMAP; miRNATAP | -0.12 | 0.00373 | NA | |
| 66 | hsa-miR-324-3p | CCND2 | -0.08 | 0.68923 | -1.64 | 0 | miRNAWalker2 validate | -0.28 | 0 | NA | |
| 67 | hsa-miR-331-5p | CCND2 | 0.58 | 0.00131 | -1.64 | 0 | miRNATAP | -0.28 | 1.0E-5 | NA | |
| 68 | hsa-miR-429 | CCND2 | 2.38 | 0 | -1.64 | 0 | miRNATAP | -0.21 | 0 | NA | |
| 69 | hsa-miR-450b-5p | CCND2 | 1.69 | 0 | -1.64 | 0 | MirTarget; PITA; miRNATAP | -0.15 | 9.0E-5 | NA | |
| 70 | hsa-miR-501-5p | CCND2 | 0.41 | 0.10435 | -1.64 | 0 | PITA; mirMAP; miRNATAP | -0.14 | 0.00086 | NA | |
| 71 | hsa-miR-503-5p | CCND2 | 1.97 | 0 | -1.64 | 0 | MirTarget | -0.13 | 5.0E-5 | 25860935 | We then identified two targets of miR-503 CCND2 and CCND3 |
| 72 | hsa-miR-550a-5p | CCND2 | 0.6 | 0.03148 | -1.64 | 0 | MirTarget | -0.22 | 0 | NA | |
| 73 | hsa-miR-589-3p | CCND2 | 1.34 | 2.0E-5 | -1.64 | 0 | mirMAP | -0.19 | 0 | NA | |
| 74 | hsa-miR-590-3p | CCND2 | 0.84 | 0.00129 | -1.64 | 0 | miRanda; mirMAP | -0.13 | 0.00821 | NA | |
| 75 | hsa-miR-590-5p | CCND2 | 2.07 | 0 | -1.64 | 0 | mirMAP | -0.26 | 0 | NA | |
| 76 | hsa-miR-660-5p | CCND2 | 2.05 | 0 | -1.64 | 0 | mirMAP | -0.15 | 0.00131 | NA | |
| 77 | hsa-miR-7-1-3p | CCND2 | 2.61 | 0 | -1.64 | 0 | mirMAP | -0.26 | 0 | NA | |
| 78 | hsa-miR-877-5p | CCND2 | -0.37 | 0.20671 | -1.64 | 0 | miRNAWalker2 validate | -0.15 | 6.0E-5 | NA | |
| 79 | hsa-miR-93-5p | CCND2 | 1.51 | 0 | -1.64 | 0 | miRNATAP | -0.41 | 0 | NA | |
| 80 | hsa-miR-96-5p | CCND2 | 3.04 | 0 | -1.64 | 0 | TargetScan; miRNATAP | -0.36 | 0 | NA | |
| 81 | hsa-miR-320b | CCND3 | 0.23 | 0.37882 | -0.86 | 1.0E-5 | miRanda | -0.11 | 0.00235 | NA | |
| 82 | hsa-miR-421 | CCND3 | 0.17 | 0.53528 | -0.86 | 1.0E-5 | PITA; miRanda | -0.12 | 0.00073 | NA | |
| 83 | hsa-miR-96-5p | CCND3 | 3.04 | 0 | -0.86 | 1.0E-5 | TargetScan | -0.12 | 0.00015 | NA | |
| 84 | hsa-miR-125b-5p | CCNE1 | -0.55 | 0.01072 | 3 | 0 | miRNAWalker2 validate | -0.21 | 0.00408 | NA | |
| 85 | hsa-miR-195-5p | CCNE1 | -1.02 | 5.0E-5 | 3 | 0 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.52 | 0 | 24402230 | Furthermore through qPCR and western blot assays we showed that overexpression of miR-195-5p reduced CCNE1 mRNA and protein levels respectively |
| 86 | hsa-miR-26a-5p | CCNE1 | -0.13 | 0.44003 | 3 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.7 | 0 | 22094936 | Cell cycle regulation and CCNE1 and CDC2 were the only significant overlapping pathway and genes differentially expressed between tumors with high and low levels of miR-26a and EZH2 respectively; Low mRNA levels of EZH2 CCNE1 and CDC2 and high levels of miR-26a are associated with favorable outcome on tamoxifen |
| 87 | hsa-miR-497-5p | CCNE1 | -0.05 | 0.78621 | 3 | 0 | MirTarget; miRNATAP | -0.5 | 0 | 25909221; 24112607; 24909281 | The effect of simultaneous overexpression of miR-497 and miR-34a on the inhibition of cell proliferation colony formation and tumor growth and the downregulation of cyclin E1 was stronger than the effect of each miRNA alone; The synergistic actions of miR-497 and miR-34a partly correlated with cyclin E1 levels; These results indicate cyclin E1 is downregulated by both miR-497 and miR-34a which synergistically retard the growth of human lung cancer cells;Western blot assays confirmed that overexpression of miR-497 reduced cyclin E1 protein levels; Inhibited cellular growth suppressed cellular migration and invasion and G1 cell cycle arrest were observed upon overexpression of miR-497 in cells possibly by targeting cyclin E1;miR 497 suppresses proliferation of human cervical carcinoma HeLa cells by targeting cyclin E1; Furthermore the target effect of miR-497 on the CCNE1 was identified by dual-luciferase reporter assay system qRT-PCR and Western blotting; Over-expressed miR-497 in HeLa cells could suppress cell proliferation by targeting CCNE1 |
| 88 | hsa-let-7a-3p | CCNE2 | 0.17 | 0.43183 | 2.15 | 0 | mirMAP | -0.22 | 0.00307 | NA | |
| 89 | hsa-let-7b-3p | CCNE2 | -1.82 | 0 | 2.15 | 0 | mirMAP | -0.49 | 0 | NA | |
| 90 | hsa-miR-126-3p | CCNE2 | 0.4 | 0.11564 | 2.15 | 0 | miRNAWalker2 validate | -0.15 | 0.00656 | NA | |
| 91 | hsa-miR-26a-5p | CCNE2 | -0.13 | 0.44003 | 2.15 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.51 | 0 | 24116110; 21901171 | The loss of miR 26a mediated post transcriptional regulation of cyclin E2 in pancreatic cancer cell proliferation and decreased patient survival; The in vitro and in vivo assays showed that overexpression of miR-26a resulted in cell cycle arrest inhibited cell proliferation and decreased tumor growth which was associated with cyclin E2 downregulation;We also show that enforced expression of miR-26a in AML cells is able to inhibit cell cycle progression by downregulating cyclin E2 expression |
| 92 | hsa-miR-26b-5p | CCNE2 | 0.72 | 5.0E-5 | 2.15 | 0 | miRNATAP | -0.22 | 0.00477 | NA | |
| 93 | hsa-miR-3065-5p | CCNE2 | 0.65 | 0.09995 | 2.15 | 0 | mirMAP | -0.2 | 0 | NA | |
| 94 | hsa-miR-30a-5p | CCNE2 | -0.92 | 0.00076 | 2.15 | 0 | miRNATAP | -0.49 | 0 | NA | |
| 95 | hsa-miR-30b-5p | CCNE2 | 0.36 | 0.13803 | 2.15 | 0 | miRNAWalker2 validate; miRTarBase | -0.2 | 0.0005 | 22384020 | A luciferase-based reporter assay demonstrated that miR-30b post-transcriptionally reduced 27% p = 0.005 of the gene expression by interacting with two binding sites in the 3'-UTR of CCNE2; The upregulation of miR-30b by trastuzumab may play a biological role in trastuzumab-induced cell growth inhibition by targeting CCNE2 |
| 96 | hsa-miR-30c-5p | CCNE2 | -0.33 | 0.1236 | 2.15 | 0 | miRNATAP | -0.44 | 0 | NA | |
| 97 | hsa-miR-30d-5p | CCNE2 | -0.92 | 4.0E-5 | 2.15 | 0 | miRNATAP | -0.35 | 0 | 25843294 | MicroRNA 30d 5p inhibits tumour cell proliferation and motility by directly targeting CCNE2 in non small cell lung cancer; In addition the re-introduction of CCNE2 expression antagonised the inhibitory effects of miR-30d-5p on the capacity of NSCLC cells for proliferation and motility |
| 98 | hsa-miR-34a-5p | CCNE2 | 1.41 | 0 | 2.15 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.2 | 0.00337 | NA | |
| 99 | hsa-miR-34c-5p | CCNE2 | -1 | 0.07244 | 2.15 | 0 | miRNAWalker2 validate; miRTarBase; PITA; miRanda; miRNATAP | -0.11 | 4.0E-5 | NA | |
| 100 | hsa-miR-514a-3p | CCNE2 | -0.62 | 0.31374 | 2.15 | 0 | miRNATAP | -0.17 | 0 | NA | |
| 101 | hsa-miR-132-3p | CCNG1 | 0.01 | 0.93403 | -0.38 | 0.02147 | MirTarget | -0.16 | 0.00185 | NA | |
| 102 | hsa-miR-197-3p | CCNG1 | -1.3 | 0 | -0.38 | 0.02147 | miRNAWalker2 validate | -0.12 | 6.0E-5 | NA | |
| 103 | hsa-miR-21-5p | CCNG1 | 4.38 | 0 | -0.38 | 0.02147 | miRNAWalker2 validate | -0.13 | 1.0E-5 | NA | |
| 104 | hsa-miR-23a-3p | CCNG1 | 0.11 | 0.39309 | -0.38 | 0.02147 | MirTarget; miRNATAP | -0.18 | 0.00203 | NA | |
| 105 | hsa-miR-27a-3p | CCNG1 | 0.43 | 0.00737 | -0.38 | 0.02147 | MirTarget; miRNATAP | -0.25 | 0 | NA | |
| 106 | hsa-miR-27b-3p | CCNG1 | 0.24 | 0.12264 | -0.38 | 0.02147 | MirTarget; miRNATAP | -0.17 | 0.00064 | 26623719 | Moreover miR-27b directly targets the 3' untranslated regions 3'-UTRs of CCNG1 a well-known negative regulator of P53 stability; Interestingly miR-27b up-regulation leads to increased miR-508-5p expression and this phenomenon is mediated by CCNG1 and P53 |
| 107 | hsa-miR-339-5p | CCNG1 | 0.54 | 0.04881 | -0.38 | 0.02147 | miRanda | -0.15 | 0 | NA | |
| 108 | hsa-miR-590-5p | CCNG1 | 2.07 | 0 | -0.38 | 0.02147 | miRanda | -0.19 | 0 | NA | |
| 109 | hsa-miR-96-5p | CCNG1 | 3.04 | 0 | -0.38 | 0.02147 | MirTarget; TargetScan | -0.13 | 1.0E-5 | NA | |
| 110 | hsa-miR-195-3p | CCNG2 | -1.33 | 0 | 0.22 | 0.10548 | mirMAP | -0.1 | 0.00014 | NA | |
| 111 | hsa-miR-365a-3p | CCNG2 | 0.01 | 0.9536 | 0.22 | 0.10548 | MirTarget | -0.11 | 3.0E-5 | NA | |
| 112 | hsa-miR-374a-5p | CCNG2 | -0.2 | 0.29808 | 0.22 | 0.10548 | mirMAP | -0.14 | 0.00021 | NA | |
| 113 | hsa-miR-664a-3p | CCNG2 | 0.44 | 0.02142 | 0.22 | 0.10548 | MirTarget; mirMAP | -0.16 | 0 | NA | |
| 114 | hsa-miR-23b-3p | CDK2 | -0.07 | 0.62059 | 0.34 | 0.01969 | miRNAWalker2 validate | -0.32 | 0 | NA | |
| 115 | hsa-miR-26b-5p | CDK2 | 0.72 | 5.0E-5 | 0.34 | 0.01969 | miRNAWalker2 validate | -0.13 | 0.00058 | NA | |
| 116 | hsa-miR-145-5p | CDK4 | -1.35 | 0 | 0.74 | 1.0E-5 | miRNAWalker2 validate; miRTarBase | -0.15 | 0 | 21092188 | Furthermore we found that CDK4 was regulated by miR-145 in cell cycle control |
| 117 | hsa-miR-195-5p | CDK4 | -1.02 | 5.0E-5 | 0.74 | 1.0E-5 | miRNAWalker2 validate; miRTarBase | -0.11 | 0.00037 | NA | |
| 118 | hsa-miR-320a | CDK4 | -0.96 | 0 | 0.74 | 1.0E-5 | miRNAWalker2 validate | -0.2 | 1.0E-5 | NA | |
| 119 | hsa-miR-141-3p | CDK6 | 3.37 | 0 | -0.63 | 0.01747 | TargetScan; miRNATAP | -0.16 | 6.0E-5 | NA | |
| 120 | hsa-miR-148a-3p | CDK6 | 2.31 | 0 | -0.63 | 0.01747 | mirMAP | -0.14 | 0.00378 | NA | |
| 121 | hsa-miR-15a-5p | CDK6 | 1.63 | 0 | -0.63 | 0.01747 | miRNATAP | -0.21 | 0.00126 | NA | |
| 122 | hsa-miR-16-1-3p | CDK6 | 1.5 | 0 | -0.63 | 0.01747 | mirMAP | -0.19 | 0.0025 | NA | |
| 123 | hsa-miR-182-5p | CDK6 | 3.22 | 0 | -0.63 | 0.01747 | mirMAP | -0.2 | 2.0E-5 | NA | |
| 124 | hsa-miR-200a-3p | CDK6 | 3.15 | 0 | -0.63 | 0.01747 | miRNATAP | -0.17 | 0 | 24009066 | microRNA 200a is an independent prognostic factor of hepatocellular carcinoma and induces cell cycle arrest by targeting CDK6 |
| 125 | hsa-miR-200b-3p | CDK6 | 1.55 | 0 | -0.63 | 0.01747 | mirMAP | -0.17 | 0.00022 | NA | |
| 126 | hsa-miR-21-5p | CDK6 | 4.38 | 0 | -0.63 | 0.01747 | miRNAWalker2 validate; mirMAP | -0.15 | 0.00158 | NA | |
| 127 | hsa-miR-23b-3p | CDK6 | -0.07 | 0.62059 | -0.63 | 0.01747 | mirMAP | -0.24 | 0.00674 | NA | |
| 128 | hsa-miR-27b-3p | CDK6 | 0.24 | 0.12264 | -0.63 | 0.01747 | mirMAP | -0.29 | 0.00035 | NA | |
| 129 | hsa-miR-27b-5p | CDK6 | 1.04 | 0 | -0.63 | 0.01747 | mirMAP | -0.22 | 0.00423 | NA | |
| 130 | hsa-miR-29b-3p | CDK6 | 3.11 | 0 | -0.63 | 0.01747 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.18 | 4.0E-5 | 23591808; 23245396; 25472644; 26180082; 27230400; 20086245 | Here we have identified the oncogene cyclin-dependent protein kinase 6 CDK6 as a direct target of miR-29b in lung cancer;The IFN-γ-induced G1-arrest of melanoma cells involves down-regulation of CDK6 which we proved to be a direct target of miR-29 in these cells;Moreover miR-29b inhibited the expression of MCL1 and CDK6;Knockdown of NTSR1 increased the expression of miR-29b-1 and miR-129-3p which were responsible for the decreased CDK6 expression;MiR 29b suppresses the proliferation and migration of osteosarcoma cells by targeting CDK6; In this study we investigated the role of miR-29b as a novel regulator of CDK6 using bioinformatics methods; We demonstrated that CDK6 can be downregulated by miR-29b via binding to the 3'-UTR region in osteosarcoma cells; Furthermore we identified an inverse correlation between miR-29b and CDK6 protein levels in osteosarcoma tissues; The results revealed that miR-29b acts as a tumor suppressor of osteosarcoma by targeting CDK6 in the proliferation and migration processes;microRNA expression profile and identification of miR 29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma; Furthermore we demonstrate miR-29 inhibition of CDK6 protein and mRNA levels by direct binding to 3'-untranslated region; Inverse correlation between miR-29 and CDK6 was observed in MCL |
| 131 | hsa-miR-3065-5p | CDK6 | 0.65 | 0.09995 | -0.63 | 0.01747 | mirMAP | -0.23 | 0 | NA | |
| 132 | hsa-miR-30b-5p | CDK6 | 0.36 | 0.13803 | -0.63 | 0.01747 | mirMAP | -0.3 | 0 | NA | |
| 133 | hsa-miR-30c-5p | CDK6 | -0.33 | 0.1236 | -0.63 | 0.01747 | mirMAP | -0.16 | 0.00457 | NA | |
| 134 | hsa-miR-30d-3p | CDK6 | 0 | 0.98646 | -0.63 | 0.01747 | mirMAP | -0.37 | 0 | NA | |
| 135 | hsa-miR-30d-5p | CDK6 | -0.92 | 4.0E-5 | -0.63 | 0.01747 | mirMAP | -0.32 | 0 | NA | |
| 136 | hsa-miR-30e-5p | CDK6 | 1.6 | 0 | -0.63 | 0.01747 | mirMAP | -0.29 | 1.0E-5 | NA | |
| 137 | hsa-miR-338-3p | CDK6 | 0.73 | 0.05063 | -0.63 | 0.01747 | mirMAP | -0.15 | 0 | NA | |
| 138 | hsa-miR-34a-5p | CDK6 | 1.41 | 0 | -0.63 | 0.01747 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.24 | 3.0E-5 | 26104764; 21702042 | The expression of microRNA 34a is inversely correlated with c MET and CDK6 and has a prognostic significance in lung adenocarcinoma patients; We found significant inverse correlations between miR-34a and c-MET R = -0.316 P = 0.028 and CDK6 expression R = -0.4582 P = 0.004;Molecular analyses identified Cdk6 and sirtuin SIRT-1 as being targeted by miR-34a in MI-TCC cells however inhibition of Cdk6 and SIRT-1 was not as effective as pre-miR-34a in mediating chemosensitization |
| 139 | hsa-miR-362-5p | CDK6 | 0.66 | 0.02433 | -0.63 | 0.01747 | mirMAP | -0.18 | 2.0E-5 | NA | |
| 140 | hsa-miR-429 | CDK6 | 2.38 | 0 | -0.63 | 0.01747 | mirMAP; miRNATAP | -0.15 | 0.00018 | NA | |
| 141 | hsa-miR-660-5p | CDK6 | 2.05 | 0 | -0.63 | 0.01747 | mirMAP | -0.16 | 0.00164 | NA | |
| 142 | hsa-miR-664a-3p | CDK6 | 0.44 | 0.02142 | -0.63 | 0.01747 | mirMAP | -0.3 | 0 | NA | |
| 143 | hsa-miR-106b-5p | CDKN1A | 1.47 | 0 | -0.99 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.23 | 0 | NA | |
| 144 | hsa-miR-17-5p | CDKN1A | 2.07 | 0 | -0.99 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget; TargetScan; miRNATAP | -0.16 | 3.0E-5 | 26482648; 24989082 | The low expressions of miR-17 and miR-92 families can maintain cisplatin resistance through the regulation of CDKN1A and RAD21;According to PicTar and Miranda algorithms which predicted CDKN1A p21 as a putative target of miR-17 a luciferase assay was performed and revealed that miR-17 directly targets the 3'-UTR of p21 mRNA |
| 145 | hsa-miR-182-5p | CDKN1A | 3.22 | 0 | -0.99 | 0 | miRNAWalker2 validate | -0.19 | 0 | NA | |
| 146 | hsa-miR-20a-5p | CDKN1A | 2.65 | 0 | -0.99 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.14 | 7.0E-5 | 26012475 | Using the poorly tumorigenic and TGF-β-sensitive FET cell line that expresses low miR-20a levels we first confirmed that miR-20a downmodulated CDKN1A expression both at mRNA and protein level through direct binding to its 3'-UTR; Moreover besides modulating CDKN1A miR-20a blocked TGF-β-induced transactivation of its promoter without affecting the post-receptor activation of Smad3/4 effectors directly; Finally miR-20a abrogated the TGF-β-mediated c-Myc repression a direct inhibitor of the CDKN1A promoter activation most likely by reducing the expression of specific MYC-regulating genes from the Smad/E2F-based core repressor complex |
| 147 | hsa-miR-28-5p | CDKN1A | 1.2 | 0 | -0.99 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget; miRanda; miRNATAP | -0.2 | 0.00151 | NA | |
| 148 | hsa-miR-93-5p | CDKN1A | 1.51 | 0 | -0.99 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.17 | 6.0E-5 | 25633810 | MicroRNA 93 activates c Met/PI3K/Akt pathway activity in hepatocellular carcinoma by directly inhibiting PTEN and CDKN1A; We confirmed that miR-93 directly bound with the 3' untranslated regions of the tumor-suppressor genes PTEN and CDKN1A respectivelyand inhibited their expression; We concluded that miR-93 stimulated cell proliferation migration and invasion through the oncogenic c-Met/PI3K/Akt pathway and also inhibited apoptosis by directly inhibiting PTEN and CDKN1A expression in human HCC |
| 149 | hsa-miR-96-5p | CDKN1A | 3.04 | 0 | -0.99 | 0 | miRNAWalker2 validate; miRTarBase | -0.15 | 1.0E-5 | 26582573 | Upregulation of microRNA 96 and its oncogenic functions by targeting CDKN1A in bladder cancer; Bioinformatics prediction combined with luciferase reporter assay were used to verify whether the cyclin-dependent kinase inhibitor CDKN1A was a potential target gene of miR-96; According to the data of miRTarBase CDKN1A might be a candidate target gene of miR-96; In addition luciferase reporter and Western blot assays respectively demonstrated that miR-96 could bind to the putative seed region in CDKN1A mRNA 3'UTR and significantly reduce the expression level of CDKN1A protein; Moreover we found that the inhibition of miR-96 expression remarkably decreased cell proliferation and promoted cell apoptosis of BC cell lines which was consistent with the findings observed following the introduction of CDKN1A cDNA without 3'UTR restored miR-96; Upregulation of miR-96 may contribute to aggressive malignancy partly through suppressing CDKN1A protein expression in BC cells |
| 150 | hsa-miR-215-5p | CDKN2A | 0.12 | 0.83847 | 1.92 | 0.00062 | miRNAWalker2 validate | -0.18 | 0.00071 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | REGULATION OF CELL CYCLE | 27 | 949 | 1.626e-25 | 7.564e-22 |
| 2 | CELL CYCLE | 27 | 1316 | 9.127e-22 | 2.123e-18 |
| 3 | CELL CYCLE PROCESS | 24 | 1081 | 9.01e-20 | 1.397e-16 |
| 4 | MITOTIC CELL CYCLE | 21 | 766 | 6.419e-19 | 7.467e-16 |
| 5 | REGULATION OF CELL DEATH | 25 | 1472 | 6.45e-18 | 6.002e-15 |
| 6 | NEGATIVE REGULATION OF CELL CYCLE | 17 | 433 | 9.286e-18 | 7.202e-15 |
| 7 | CELL CYCLE PHASE TRANSITION | 14 | 255 | 1.183e-16 | 6.272e-14 |
| 8 | POSITIVE REGULATION OF CELL DEATH | 18 | 605 | 1.055e-16 | 6.272e-14 |
| 9 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 24 | 1492 | 1.483e-16 | 6.272e-14 |
| 10 | CELL CYCLE CHECKPOINT | 13 | 194 | 1.272e-16 | 6.272e-14 |
| 11 | POSITIVE REGULATION OF CELL CYCLE | 15 | 332 | 1.455e-16 | 6.272e-14 |
| 12 | RESPONSE TO ABIOTIC STIMULUS | 21 | 1024 | 2.305e-16 | 8.938e-14 |
| 13 | REGULATION OF CELL CYCLE ARREST | 11 | 108 | 3.363e-16 | 1.204e-13 |
| 14 | CELLULAR RESPONSE TO STRESS | 24 | 1565 | 4.381e-16 | 1.456e-13 |
| 15 | SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 11 | 127 | 2.108e-15 | 6.539e-13 |
| 16 | CELL DEATH | 20 | 1001 | 2.593e-15 | 7.54e-13 |
| 17 | POSITIVE REGULATION OF CELL CYCLE PROCESS | 13 | 247 | 2.983e-15 | 8.165e-13 |
| 18 | REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 10 | 97 | 7.514e-15 | 1.942e-12 |
| 19 | NEGATIVE REGULATION OF CELL CYCLE G1 S PHASE TRANSITION | 10 | 98 | 8.355e-15 | 2.046e-12 |
| 20 | DNA INTEGRITY CHECKPOINT | 11 | 146 | 1.008e-14 | 2.346e-12 |
| 21 | REGULATION OF TRANSFERASE ACTIVITY | 19 | 946 | 1.517e-14 | 3.361e-12 |
| 22 | NEGATIVE REGULATION OF CELL CYCLE PROCESS | 12 | 214 | 1.893e-14 | 4.005e-12 |
| 23 | REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 15 | 470 | 2.447e-14 | 4.949e-12 |
| 24 | G1 DNA DAMAGE CHECKPOINT | 9 | 73 | 3.524e-14 | 6.832e-12 |
| 25 | REGULATION OF PROTEIN MODIFICATION PROCESS | 23 | 1710 | 4.232e-14 | 7.573e-12 |
| 26 | CELLULAR RESPONSE TO DNA DAMAGE STIMULUS | 17 | 720 | 4.175e-14 | 7.573e-12 |
| 27 | RESPONSE TO OXYGEN LEVELS | 13 | 311 | 5.827e-14 | 1.004e-11 |
| 28 | REGULATION OF KINASE ACTIVITY | 17 | 776 | 1.409e-13 | 2.341e-11 |
| 29 | POSITIVE REGULATION OF CELL CYCLE ARREST | 9 | 85 | 1.467e-13 | 2.354e-11 |
| 30 | NEGATIVE REGULATION OF MITOTIC CELL CYCLE | 11 | 199 | 3.123e-13 | 4.844e-11 |
| 31 | ACTIVATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 9 | 95 | 4.123e-13 | 6.061e-11 |
| 32 | REGULATION OF CELL PROLIFERATION | 21 | 1496 | 4.168e-13 | 6.061e-11 |
| 33 | NEGATIVE REGULATION OF CELL CYCLE PHASE TRANSITION | 10 | 146 | 4.911e-13 | 6.924e-11 |
| 34 | REGULATION OF CELL CYCLE G1 S PHASE TRANSITION | 10 | 147 | 5.261e-13 | 6.994e-11 |
| 35 | REGULATION OF MITOTIC CELL CYCLE | 14 | 468 | 5.131e-13 | 6.994e-11 |
| 36 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 21 | 1518 | 5.536e-13 | 7.155e-11 |
| 37 | MITOTIC DNA INTEGRITY CHECKPOINT | 9 | 100 | 6.624e-13 | 8.111e-11 |
| 38 | REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 11 | 213 | 6.585e-13 | 8.111e-11 |
| 39 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 22 | 1791 | 1.289e-12 | 1.538e-10 |
| 40 | CELL CYCLE G1 S PHASE TRANSITION | 9 | 111 | 1.73e-12 | 1.964e-10 |
| 41 | G1 S TRANSITION OF MITOTIC CELL CYCLE | 9 | 111 | 1.73e-12 | 1.964e-10 |
| 42 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 21 | 1618 | 1.904e-12 | 2.06e-10 |
| 43 | ZYMOGEN ACTIVATION | 9 | 112 | 1.878e-12 | 2.06e-10 |
| 44 | REGULATION OF CELL CYCLE PHASE TRANSITION | 12 | 321 | 2.34e-12 | 2.474e-10 |
| 45 | RESPONSE TO DRUG | 13 | 431 | 3.673e-12 | 3.798e-10 |
| 46 | REGULATION OF CELL CYCLE PROCESS | 14 | 558 | 5.489e-12 | 5.552e-10 |
| 47 | CELL DIVISION | 13 | 460 | 8.315e-12 | 8.232e-10 |
| 48 | POSITIVE REGULATION OF PROTEOLYSIS | 12 | 363 | 9.865e-12 | 9.563e-10 |
| 49 | MITOTIC CELL CYCLE CHECKPOINT | 9 | 139 | 1.344e-11 | 1.276e-09 |
| 50 | APOPTOTIC SIGNALING PATHWAY | 11 | 289 | 1.81e-11 | 1.684e-09 |
| 51 | REGULATION OF PEPTIDASE ACTIVITY | 12 | 392 | 2.412e-11 | 2.2e-09 |
| 52 | SIGNAL TRANSDUCTION IN RESPONSE TO DNA DAMAGE | 8 | 96 | 2.644e-11 | 2.366e-09 |
| 53 | INTRINSIC APOPTOTIC SIGNALING PATHWAY | 9 | 152 | 3.014e-11 | 2.646e-09 |
| 54 | POSITIVE REGULATION OF PEPTIDASE ACTIVITY | 9 | 154 | 3.391e-11 | 2.922e-09 |
| 55 | REGULATION OF SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 9 | 162 | 5.347e-11 | 4.523e-09 |
| 56 | INTRACELLULAR SIGNAL TRANSDUCTION | 19 | 1572 | 1.188e-10 | 9.869e-09 |
| 57 | REGULATION OF PROTEOLYSIS | 14 | 711 | 1.367e-10 | 1.116e-08 |
| 58 | AGING | 10 | 264 | 1.771e-10 | 1.42e-08 |
| 59 | REGULATION OF APOPTOTIC SIGNALING PATHWAY | 11 | 363 | 2.072e-10 | 1.634e-08 |
| 60 | RESPONSE TO UV | 8 | 126 | 2.39e-10 | 1.853e-08 |
| 61 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 19 | 1656 | 2.896e-10 | 2.209e-08 |
| 62 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 16 | 1135 | 6.456e-10 | 4.845e-08 |
| 63 | REGULATION OF CELLULAR RESPONSE TO STRESS | 13 | 691 | 1.257e-09 | 9.281e-08 |
| 64 | REGENERATION | 8 | 161 | 1.69e-09 | 1.229e-07 |
| 65 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 15 | 1036 | 1.811e-09 | 1.277e-07 |
| 66 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 15 | 1036 | 1.811e-09 | 1.277e-07 |
| 67 | REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY VIA DEATH DOMAIN RECEPTORS | 6 | 55 | 1.841e-09 | 1.278e-07 |
| 68 | POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 14 | 876 | 2.066e-09 | 1.414e-07 |
| 69 | RESPONSE TO LIPID | 14 | 888 | 2.461e-09 | 1.66e-07 |
| 70 | POSITIVE REGULATION OF APOPTOTIC SIGNALING PATHWAY | 8 | 171 | 2.723e-09 | 1.81e-07 |
| 71 | REGULATION OF RESPONSE TO STRESS | 17 | 1468 | 3.189e-09 | 2.09e-07 |
| 72 | CELLULAR RESPONSE TO ABIOTIC STIMULUS | 9 | 263 | 3.914e-09 | 2.529e-07 |
| 73 | CELLULAR RESPONSE TO EXTERNAL STIMULUS | 9 | 264 | 4.046e-09 | 2.579e-07 |
| 74 | PROTEIN MATURATION | 9 | 265 | 4.182e-09 | 2.604e-07 |
| 75 | POSITIVE REGULATION OF TRANSFERASE ACTIVITY | 12 | 616 | 4.197e-09 | 2.604e-07 |
| 76 | RESPONSE TO X RAY | 5 | 30 | 4.947e-09 | 3.029e-07 |
| 77 | RESPONSE TO STEROID HORMONE | 11 | 497 | 5.605e-09 | 3.387e-07 |
| 78 | NEGATIVE REGULATION OF CELL PROLIFERATION | 12 | 643 | 6.78e-09 | 4.045e-07 |
| 79 | POSITIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 9 | 289 | 8.895e-09 | 5.239e-07 |
| 80 | REPLICATIVE SENESCENCE | 4 | 12 | 9.052e-09 | 5.265e-07 |
| 81 | DNA REPLICATION | 8 | 208 | 1.269e-08 | 7.29e-07 |
| 82 | RESPONSE TO RADIATION | 10 | 413 | 1.32e-08 | 7.49e-07 |
| 83 | CELL CYCLE G2 M PHASE TRANSITION | 7 | 138 | 1.673e-08 | 9.381e-07 |
| 84 | RESPONSE TO ESTROGEN | 8 | 218 | 1.831e-08 | 1.014e-06 |
| 85 | REGULATION OF FIBROBLAST PROLIFERATION | 6 | 81 | 1.977e-08 | 1.082e-06 |
| 86 | CELLULAR RESPONSE TO OXYGEN LEVELS | 7 | 143 | 2.142e-08 | 1.159e-06 |
| 87 | ORGAN REGENERATION | 6 | 83 | 2.292e-08 | 1.226e-06 |
| 88 | RESPONSE TO IONIZING RADIATION | 7 | 145 | 2.358e-08 | 1.247e-06 |
| 89 | RESPONSE TO METAL ION | 9 | 333 | 3.025e-08 | 1.582e-06 |
| 90 | NEGATIVE REGULATION OF PROTEIN METABOLIC PROCESS | 14 | 1087 | 3.213e-08 | 1.661e-06 |
| 91 | REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 7 | 153 | 3.417e-08 | 1.747e-06 |
| 92 | CELL CYCLE ARREST | 7 | 154 | 3.574e-08 | 1.808e-06 |
| 93 | RESPONSE TO ORGANIC CYCLIC COMPOUND | 13 | 917 | 3.696e-08 | 1.849e-06 |
| 94 | NEGATIVE REGULATION OF TRANSFERASE ACTIVITY | 9 | 351 | 4.752e-08 | 2.352e-06 |
| 95 | POSITIVE REGULATION OF KINASE ACTIVITY | 10 | 482 | 5.659e-08 | 2.772e-06 |
| 96 | RESPONSE TO ALCOHOL | 9 | 362 | 6.187e-08 | 2.999e-06 |
| 97 | RESPONSE TO EXTERNAL STIMULUS | 17 | 1821 | 7.94e-08 | 3.809e-06 |
| 98 | RESPONSE TO OXYGEN CONTAINING COMPOUND | 15 | 1381 | 8.6e-08 | 4.083e-06 |
| 99 | NEGATIVE REGULATION OF CELL CYCLE ARREST | 4 | 20 | 8.75e-08 | 4.113e-06 |
| 100 | POSITIVE REGULATION OF FIBROBLAST PROLIFERATION | 5 | 53 | 9.605e-08 | 4.425e-06 |
| 101 | INTRINSIC APOPTOTIC SIGNALING PATHWAY BY P53 CLASS MEDIATOR | 5 | 53 | 9.605e-08 | 4.425e-06 |
| 102 | NEGATIVE REGULATION OF CATALYTIC ACTIVITY | 12 | 829 | 1.113e-07 | 5.078e-06 |
| 103 | RESPONSE TO LIGHT STIMULUS | 8 | 280 | 1.267e-07 | 5.721e-06 |
| 104 | RESPONSE TO NITROGEN COMPOUND | 12 | 859 | 1.636e-07 | 7.319e-06 |
| 105 | NEGATIVE REGULATION OF CELL DEATH | 12 | 872 | 1.924e-07 | 8.526e-06 |
| 106 | NEGATIVE REGULATION OF PHOSPHORYLATION | 9 | 422 | 2.274e-07 | 9.982e-06 |
| 107 | POSITIVE REGULATION OF MITOTIC CELL CYCLE | 6 | 123 | 2.422e-07 | 1.053e-05 |
| 108 | NEGATIVE REGULATION OF MOLECULAR FUNCTION | 13 | 1079 | 2.454e-07 | 1.057e-05 |
| 109 | CELLULAR RESPONSE TO UV | 5 | 66 | 2.93e-07 | 1.251e-05 |
| 110 | CELL AGING | 5 | 67 | 3.161e-07 | 1.337e-05 |
| 111 | RESPONSE TO EXTRACELLULAR STIMULUS | 9 | 441 | 3.294e-07 | 1.381e-05 |
| 112 | POSITIVE REGULATION OF CELL COMMUNICATION | 15 | 1532 | 3.328e-07 | 1.382e-05 |
| 113 | PROTEIN STABILIZATION | 6 | 131 | 3.517e-07 | 1.448e-05 |
| 114 | REGULATION OF DNA DAMAGE RESPONSE SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 4 | 28 | 3.652e-07 | 1.491e-05 |
| 115 | DNA METABOLIC PROCESS | 11 | 758 | 4.089e-07 | 1.654e-05 |
| 116 | REGULATION OF PROTEIN STABILITY | 7 | 221 | 4.204e-07 | 1.686e-05 |
| 117 | CELLULAR RESPONSE TO RADIATION | 6 | 137 | 4.581e-07 | 1.822e-05 |
| 118 | NEGATIVE REGULATION OF CELL MATRIX ADHESION | 4 | 30 | 4.873e-07 | 1.921e-05 |
| 119 | NEGATIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 10 | 616 | 5.471e-07 | 2.139e-05 |
| 120 | POSITIVE REGULATION OF MAPK CASCADE | 9 | 470 | 5.615e-07 | 2.177e-05 |
| 121 | NEGATIVE REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 4 | 32 | 6.374e-07 | 2.431e-05 |
| 122 | INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO ENDOPLASMIC RETICULUM STRESS | 4 | 32 | 6.374e-07 | 2.431e-05 |
| 123 | RESPONSE TO INORGANIC SUBSTANCE | 9 | 479 | 6.578e-07 | 2.488e-05 |
| 124 | RESPONSE TO ESTRADIOL | 6 | 146 | 6.661e-07 | 2.5e-05 |
| 125 | RESPONSE TO TOXIC SUBSTANCE | 7 | 241 | 7.53e-07 | 2.803e-05 |
| 126 | T CELL HOMEOSTASIS | 4 | 34 | 8.195e-07 | 3.026e-05 |
| 127 | POSITIVE REGULATION OF CELLULAR PROTEIN LOCALIZATION | 8 | 360 | 8.532e-07 | 3.126e-05 |
| 128 | NEGATIVE REGULATION OF KINASE ACTIVITY | 7 | 250 | 9.626e-07 | 3.499e-05 |
| 129 | POSITIVE REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 4 | 36 | 1.038e-06 | 3.743e-05 |
| 130 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 16 | 1929 | 1.111e-06 | 3.946e-05 |
| 131 | RESPONSE TO ENDOGENOUS STIMULUS | 14 | 1450 | 1.104e-06 | 3.946e-05 |
| 132 | REGULATION OF CELL MATRIX ADHESION | 5 | 90 | 1.387e-06 | 4.89e-05 |
| 133 | CELLULAR RESPONSE TO LIGHT STIMULUS | 5 | 91 | 1.466e-06 | 5.127e-05 |
| 134 | ACTIVATION OF MAPKKK ACTIVITY | 3 | 11 | 1.509e-06 | 5.241e-05 |
| 135 | NEGATIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 9 | 541 | 1.802e-06 | 6.165e-05 |
| 136 | NEGATIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 9 | 541 | 1.802e-06 | 6.165e-05 |
| 137 | RESPONSE TO CORTICOSTEROID | 6 | 176 | 1.984e-06 | 6.739e-05 |
| 138 | RESPONSE TO HORMONE | 11 | 893 | 2.046e-06 | 6.897e-05 |
| 139 | NEGATIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 5 | 98 | 2.117e-06 | 7.066e-05 |
| 140 | REGULATION OF CELLULAR PROTEIN LOCALIZATION | 9 | 552 | 2.126e-06 | 7.066e-05 |
| 141 | RESPONSE TO KETONE | 6 | 182 | 2.41e-06 | 7.954e-05 |
| 142 | POSITIVE REGULATION OF DNA DAMAGE RESPONSE SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 3 | 13 | 2.608e-06 | 8.546e-05 |
| 143 | RESPONSE TO NUTRIENT | 6 | 191 | 3.187e-06 | 0.0001037 |
| 144 | POSITIVE REGULATION OF P38MAPK CASCADE | 3 | 14 | 3.314e-06 | 0.0001071 |
| 145 | LYMPHOCYTE HOMEOSTASIS | 4 | 50 | 3.969e-06 | 0.0001274 |
| 146 | NEGATIVE REGULATION OF APOPTOTIC SIGNALING PATHWAY | 6 | 200 | 4.156e-06 | 0.0001324 |
| 147 | NEGATIVE REGULATION OF CELL SUBSTRATE ADHESION | 4 | 53 | 5.023e-06 | 0.0001579 |
| 148 | POSITIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 4 | 53 | 5.023e-06 | 0.0001579 |
| 149 | RESPONSE TO MECHANICAL STIMULUS | 6 | 210 | 5.501e-06 | 0.0001718 |
| 150 | PROTEOLYSIS | 12 | 1208 | 5.954e-06 | 0.0001847 |
| 151 | POSITIVE REGULATION OF SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 3 | 17 | 6.164e-06 | 0.0001887 |
| 152 | POSITIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY VIA DEATH DOMAIN RECEPTORS | 3 | 17 | 6.164e-06 | 0.0001887 |
| 153 | REGULATION OF GROWTH | 9 | 633 | 6.481e-06 | 0.0001971 |
| 154 | NEGATIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 5 | 126 | 7.281e-06 | 0.00022 |
| 155 | REGULATION OF DNA METABOLIC PROCESS | 7 | 340 | 7.346e-06 | 0.0002205 |
| 156 | REGULATION OF CELL CYCLE G2 M PHASE TRANSITION | 4 | 59 | 7.734e-06 | 0.0002307 |
| 157 | PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS | 15 | 1977 | 8.255e-06 | 0.0002437 |
| 158 | LEUKOCYTE HOMEOSTASIS | 4 | 60 | 8.274e-06 | 0.0002437 |
| 159 | REGULATION OF MAPK CASCADE | 9 | 660 | 9.07e-06 | 0.0002654 |
| 160 | RESPONSE TO ENDOPLASMIC RETICULUM STRESS | 6 | 233 | 9.966e-06 | 0.0002898 |
| 161 | POSITIVE REGULATION OF CELL CYCLE PHASE TRANSITION | 4 | 68 | 1.365e-05 | 0.0003944 |
| 162 | POSITIVE REGULATION OF PROTEIN OLIGOMERIZATION | 3 | 22 | 1.386e-05 | 0.000398 |
| 163 | REGULATION OF RESPONSE TO DNA DAMAGE STIMULUS | 5 | 145 | 1.441e-05 | 0.0004114 |
| 164 | REGULATION OF HYDROLASE ACTIVITY | 12 | 1327 | 1.545e-05 | 0.0004384 |
| 165 | RESPONSE TO MAGNESIUM ION | 3 | 23 | 1.591e-05 | 0.0004407 |
| 166 | RESPONSE TO INCREASED OXYGEN LEVELS | 3 | 23 | 1.591e-05 | 0.0004407 |
| 167 | RESPONSE TO TRANSITION METAL NANOPARTICLE | 5 | 148 | 1.591e-05 | 0.0004407 |
| 168 | RESPONSE TO HYPEROXIA | 3 | 23 | 1.591e-05 | 0.0004407 |
| 169 | NEGATIVE REGULATION OF RESPONSE TO STIMULUS | 12 | 1360 | 1.977e-05 | 0.0005443 |
| 170 | HISTONE PHOSPHORYLATION | 3 | 25 | 2.06e-05 | 0.0005638 |
| 171 | RESPONSE TO CORTICOSTERONE | 3 | 26 | 2.325e-05 | 0.000629 |
| 172 | REGULATION OF P38MAPK CASCADE | 3 | 26 | 2.325e-05 | 0.000629 |
| 173 | PROTEIN PHOSPHORYLATION | 10 | 944 | 2.42e-05 | 0.0006509 |
| 174 | REGULATION OF LEUKOCYTE APOPTOTIC PROCESS | 4 | 79 | 2.475e-05 | 0.0006618 |
| 175 | CELLULAR RESPONSE TO MECHANICAL STIMULUS | 4 | 80 | 2.601e-05 | 0.0006916 |
| 176 | ACTIVATION OF PROTEIN KINASE ACTIVITY | 6 | 279 | 2.76e-05 | 0.0007296 |
| 177 | POSITIVE REGULATION OF LEUKOCYTE APOPTOTIC PROCESS | 3 | 28 | 2.921e-05 | 0.0007658 |
| 178 | RESPONSE TO CARBOHYDRATE | 5 | 168 | 2.93e-05 | 0.0007658 |
| 179 | NEGATIVE REGULATION OF CELL COMMUNICATION | 11 | 1192 | 3.159e-05 | 0.0008212 |
| 180 | REGULATION OF PROTEIN INSERTION INTO MITOCHONDRIAL MEMBRANE INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 3 | 29 | 3.253e-05 | 0.0008363 |
| 181 | POSITIVE REGULATION OF PROTEIN INSERTION INTO MITOCHONDRIAL MEMBRANE INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 3 | 29 | 3.253e-05 | 0.0008363 |
| 182 | REGULATION OF CELL SUBSTRATE ADHESION | 5 | 173 | 3.371e-05 | 0.0008619 |
| 183 | INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO DNA DAMAGE BY P53 CLASS MEDIATOR | 3 | 30 | 3.609e-05 | 0.0009127 |
| 184 | NEGATIVE REGULATION OF B CELL ACTIVATION | 3 | 30 | 3.609e-05 | 0.0009127 |
| 185 | PHOSPHORYLATION | 11 | 1228 | 4.153e-05 | 0.001045 |
| 186 | POSITIVE REGULATION OF GENE EXPRESSION | 13 | 1733 | 4.555e-05 | 0.00114 |
| 187 | REGULATION OF INTRACELLULAR TRANSPORT | 8 | 621 | 4.583e-05 | 0.00114 |
| 188 | POSITIVE REGULATION OF CELL PROLIFERATION | 9 | 814 | 4.757e-05 | 0.001177 |
| 189 | REGULATION OF PROTEIN EXPORT FROM NUCLEUS | 3 | 33 | 4.828e-05 | 0.001182 |
| 190 | REGULATION OF CELL AGING | 3 | 33 | 4.828e-05 | 0.001182 |
| 191 | CELLULAR RESPONSE TO EXTRACELLULAR STIMULUS | 5 | 188 | 5.013e-05 | 0.001221 |
| 192 | NEGATIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY VIA DEATH DOMAIN RECEPTORS | 3 | 34 | 5.288e-05 | 0.001281 |
| 193 | RESPONSE TO REACTIVE OXYGEN SPECIES | 5 | 191 | 5.405e-05 | 0.001303 |
| 194 | NEGATIVE REGULATION OF PROTEIN PROCESSING | 3 | 35 | 5.775e-05 | 0.00135 |
| 195 | REGULATION OF PROTEIN OLIGOMERIZATION | 3 | 35 | 5.775e-05 | 0.00135 |
| 196 | NEURON APOPTOTIC PROCESS | 3 | 35 | 5.775e-05 | 0.00135 |
| 197 | RESPONSE TO MINERALOCORTICOID | 3 | 35 | 5.775e-05 | 0.00135 |
| 198 | NEGATIVE REGULATION OF PROTEIN MATURATION | 3 | 35 | 5.775e-05 | 0.00135 |
| 199 | RESPONSE TO IRON ION | 3 | 35 | 5.775e-05 | 0.00135 |
| 200 | POSITIVE REGULATION OF MITOCHONDRIAL OUTER MEMBRANE PERMEABILIZATION INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 3 | 36 | 6.29e-05 | 0.001463 |
| 201 | RESPONSE TO BIOTIC STIMULUS | 9 | 886 | 9.143e-05 | 0.002117 |
| 202 | CELLULAR RESPONSE TO ESTROGEN STIMULUS | 3 | 41 | 9.321e-05 | 0.002147 |
| 203 | RESPONSE TO OXIDATIVE STRESS | 6 | 352 | 0.0001002 | 0.002297 |
| 204 | REGULATION OF MITOCHONDRION ORGANIZATION | 5 | 218 | 0.000101 | 0.002303 |
| 205 | REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 5 | 220 | 0.0001054 | 0.002392 |
| 206 | POSITIVE REGULATION OF HYDROLASE ACTIVITY | 9 | 905 | 0.0001075 | 0.002418 |
| 207 | REGULATION OF MITOCHONDRIAL OUTER MEMBRANE PERMEABILIZATION INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 3 | 43 | 0.0001076 | 0.002418 |
| 208 | NEGATIVE REGULATION OF CELL ADHESION | 5 | 223 | 0.0001123 | 0.002512 |
| 209 | MITOTIC NUCLEAR DIVISION | 6 | 361 | 0.0001151 | 0.002555 |
| 210 | REGULATION OF RELEASE OF CYTOCHROME C FROM MITOCHONDRIA | 3 | 44 | 0.0001153 | 0.002555 |
| 211 | POSITIVE REGULATION OF INTRACELLULAR TRANSPORT | 6 | 370 | 0.0001316 | 0.002889 |
| 212 | REGULATION OF B CELL ACTIVATION | 4 | 121 | 0.0001311 | 0.002889 |
| 213 | NEURON DEATH | 3 | 47 | 0.0001405 | 0.003055 |
| 214 | POSITIVE REGULATION OF NEURON APOPTOTIC PROCESS | 3 | 47 | 0.0001405 | 0.003055 |
| 215 | REGULATION OF CATABOLIC PROCESS | 8 | 731 | 0.0001429 | 0.003093 |
| 216 | NEGATIVE REGULATION OF GROWTH | 5 | 236 | 0.0001464 | 0.003154 |
| 217 | REGULATION OF PROTEIN LOCALIZATION | 9 | 950 | 0.0001553 | 0.003331 |
| 218 | REGULATION OF SMOOTH MUSCLE CELL MIGRATION | 3 | 49 | 0.0001592 | 0.003398 |
| 219 | REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION TO MITOCHONDRION | 4 | 128 | 0.0001628 | 0.003459 |
| 220 | REGULATION OF LIGASE ACTIVITY | 4 | 130 | 0.0001728 | 0.003655 |
| 221 | CELLULAR RESPONSE TO IONIZING RADIATION | 3 | 52 | 0.0001901 | 0.004002 |
| 222 | REGULATION OF NEURON DEATH | 5 | 252 | 0.0001987 | 0.004165 |
| 223 | POSITIVE REGULATION OF ORGANELLE ORGANIZATION | 7 | 573 | 0.0002009 | 0.004192 |
| 224 | RESPONSE TO ETHANOL | 4 | 136 | 0.0002055 | 0.004269 |
| 225 | CIRCADIAN RHYTHM | 4 | 137 | 0.0002114 | 0.004371 |
| 226 | REGULATION OF LYMPHOCYTE APOPTOTIC PROCESS | 3 | 54 | 0.0002127 | 0.004379 |
| 227 | REGULATION OF B CELL PROLIFERATION | 3 | 55 | 0.0002246 | 0.004605 |
| 228 | RAS PROTEIN SIGNAL TRANSDUCTION | 4 | 143 | 0.000249 | 0.005076 |
| 229 | APOPTOTIC MITOCHONDRIAL CHANGES | 3 | 57 | 0.0002498 | 0.005076 |
| 230 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 8 | 801 | 0.0002668 | 0.005397 |
| 231 | REGULATION OF EPITHELIAL CELL APOPTOTIC PROCESS | 3 | 59 | 0.0002767 | 0.005574 |
| 232 | DEOXYRIBONUCLEOTIDE BIOSYNTHETIC PROCESS | 2 | 12 | 0.000294 | 0.00587 |
| 233 | POSITIVE REGULATION OF INSULIN LIKE GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 2 | 12 | 0.000294 | 0.00587 |
| 234 | REGULATION OF CELLULAR LOCALIZATION | 10 | 1277 | 0.0002996 | 0.005958 |
| 235 | MITOTIC CELL CYCLE ARREST | 2 | 13 | 0.0003469 | 0.006811 |
| 236 | ACTIVATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 2 | 13 | 0.0003469 | 0.006811 |
| 237 | RESPONSE TO COBALT ION | 2 | 13 | 0.0003469 | 0.006811 |
| 238 | REGULATION OF EPITHELIAL CELL PROLIFERATION | 5 | 285 | 0.0003506 | 0.006853 |
| 239 | POSITIVE REGULATION OF RESPONSE TO DNA DAMAGE STIMULUS | 3 | 64 | 0.000352 | 0.006853 |
| 240 | REGULATION OF CELL ADHESION | 7 | 629 | 0.0003542 | 0.006866 |
| 241 | RESPONSE TO PURINE CONTAINING COMPOUND | 4 | 158 | 0.000364 | 0.007028 |
| 242 | REGULATION OF JNK CASCADE | 4 | 159 | 0.0003728 | 0.007169 |
| 243 | POSITIVE REGULATION OF NEURON DEATH | 3 | 67 | 0.0004029 | 0.007677 |
| 244 | REGULATION OF FIBRINOLYSIS | 2 | 14 | 0.0004042 | 0.007677 |
| 245 | REGULATION OF SMOOTH MUSCLE CELL APOPTOTIC PROCESS | 2 | 14 | 0.0004042 | 0.007677 |
| 246 | REGULATION OF NUCLEAR DIVISION | 4 | 163 | 0.0004096 | 0.007725 |
| 247 | CELLULAR RESPONSE TO LIPID | 6 | 457 | 0.0004101 | 0.007725 |
| 248 | RHYTHMIC PROCESS | 5 | 298 | 0.0004299 | 0.008066 |
| 249 | CELLULAR RESPONSE TO ORGANIC CYCLIC COMPOUND | 6 | 465 | 0.0004496 | 0.008401 |
| 250 | REGULATION OF MEMBRANE PERMEABILITY | 3 | 70 | 0.0004583 | 0.008531 |
| 251 | DENTATE GYRUS DEVELOPMENT | 2 | 15 | 0.0004658 | 0.008566 |
| 252 | NEGATIVE REGULATION OF B CELL PROLIFERATION | 2 | 15 | 0.0004658 | 0.008566 |
| 253 | RESPONSE TO VITAMIN E | 2 | 15 | 0.0004658 | 0.008566 |
| 254 | INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO DNA DAMAGE | 3 | 71 | 0.0004778 | 0.008754 |
| 255 | HIPPOCAMPUS DEVELOPMENT | 3 | 73 | 0.0005184 | 0.009423 |
| 256 | REGULATION OF ORGAN GROWTH | 3 | 73 | 0.0005184 | 0.009423 |
| 257 | NEGATIVE REGULATION OF ADHERENS JUNCTION ORGANIZATION | 2 | 16 | 0.0005316 | 0.009514 |
| 258 | NEGATIVE REGULATION OF SMOOTH MUSCLE CELL MIGRATION | 2 | 16 | 0.0005316 | 0.009514 |
| 259 | DNA REPAIR | 6 | 480 | 0.0005316 | 0.009514 |
| 260 | REGULATION OF PROTEIN HOMOOLIGOMERIZATION | 2 | 16 | 0.0005316 | 0.009514 |
| 261 | HOMEOSTASIS OF NUMBER OF CELLS | 4 | 175 | 0.0005356 | 0.009548 |
| 262 | REGULATION OF CELL ACTIVATION | 6 | 484 | 0.0005554 | 0.009863 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CYCLIN DEPENDENT PROTEIN SERINE THREONINE KINASE REGULATOR ACTIVITY | 7 | 28 | 1.455e-13 | 1.352e-10 |
| 2 | CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 6 | 34 | 8.83e-11 | 4.101e-08 |
| 3 | KINASE BINDING | 11 | 606 | 4.302e-08 | 9.993e-06 |
| 4 | ENZYME BINDING | 17 | 1737 | 3.964e-08 | 9.993e-06 |
| 5 | CYCLIN BINDING | 4 | 19 | 7.011e-08 | 1.303e-05 |
| 6 | KINASE REGULATOR ACTIVITY | 7 | 186 | 1.305e-07 | 2.021e-05 |
| 7 | PROTEIN KINASE ACTIVITY | 10 | 640 | 7.75e-07 | 0.0001029 |
| 8 | KINASE ACTIVITY | 11 | 842 | 1.153e-06 | 0.0001339 |
| 9 | CYCLIN DEPENDENT PROTEIN SERINE THREONINE KINASE INHIBITOR ACTIVITY | 3 | 12 | 2.009e-06 | 0.0002074 |
| 10 | PROTEIN COMPLEX BINDING | 11 | 935 | 3.191e-06 | 0.0002965 |
| 11 | PROTEIN SERINE THREONINE KINASE ACTIVITY | 8 | 445 | 4.13e-06 | 0.0003488 |
| 12 | TRANSFERASE ACTIVITY TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 11 | 992 | 5.63e-06 | 0.0004359 |
| 13 | HISTONE KINASE ACTIVITY | 3 | 19 | 8.757e-06 | 0.0006258 |
| 14 | P53 BINDING | 4 | 67 | 1.286e-05 | 0.0008536 |
| 15 | ENZYME REGULATOR ACTIVITY | 10 | 959 | 2.77e-05 | 0.001609 |
| 16 | MACROMOLECULAR COMPLEX BINDING | 12 | 1399 | 2.621e-05 | 0.001609 |
| 17 | PROTEIN SERINE THREONINE KINASE INHIBITOR ACTIVITY | 3 | 30 | 3.609e-05 | 0.001972 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CYCLIN DEPENDENT PROTEIN KINASE HOLOENZYME COMPLEX | 8 | 31 | 1.741e-15 | 1.017e-12 |
| 2 | PROTEIN KINASE COMPLEX | 8 | 90 | 1.56e-11 | 4.555e-09 |
| 3 | TRANSFERASE COMPLEX TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 8 | 237 | 3.504e-08 | 6.821e-06 |
| 4 | CATALYTIC COMPLEX | 11 | 1038 | 8.665e-06 | 0.001265 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
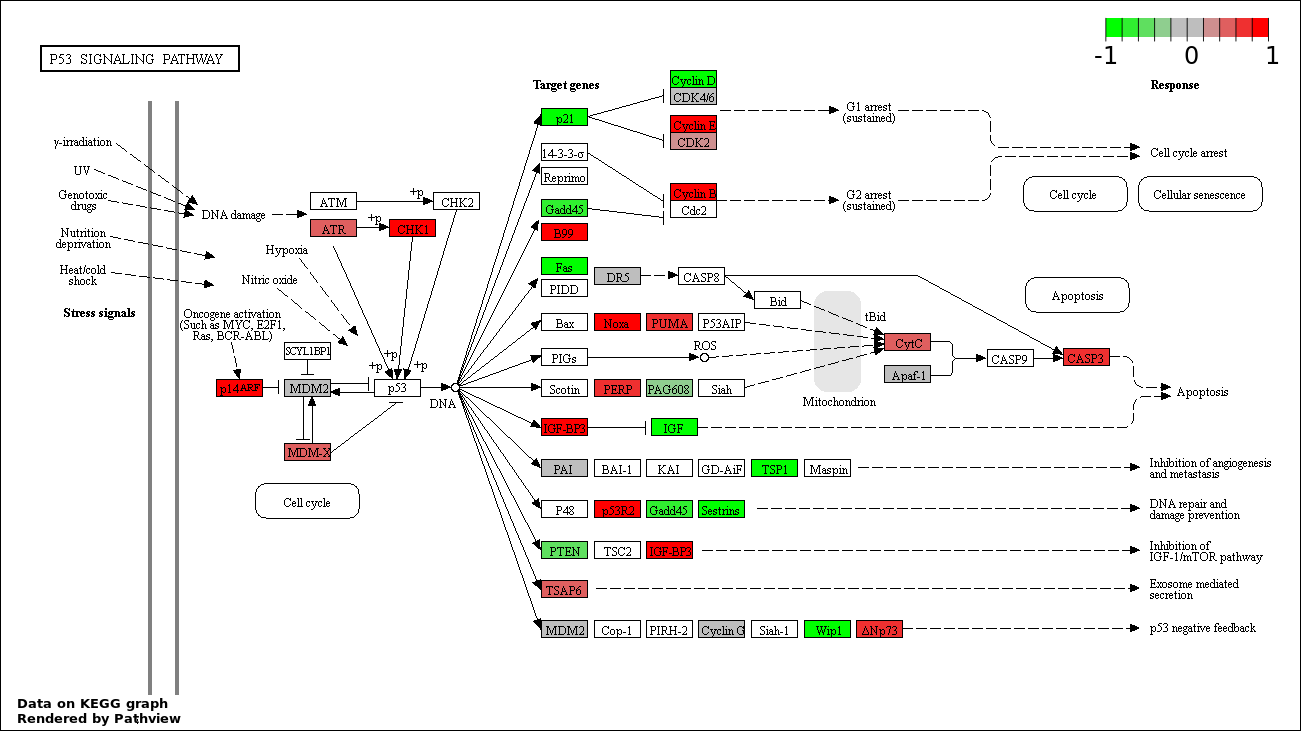
| 1 | hsa04115_p53_signaling_pathway | 43 | 69 | 5.047e-114 | 9.084e-112 | |
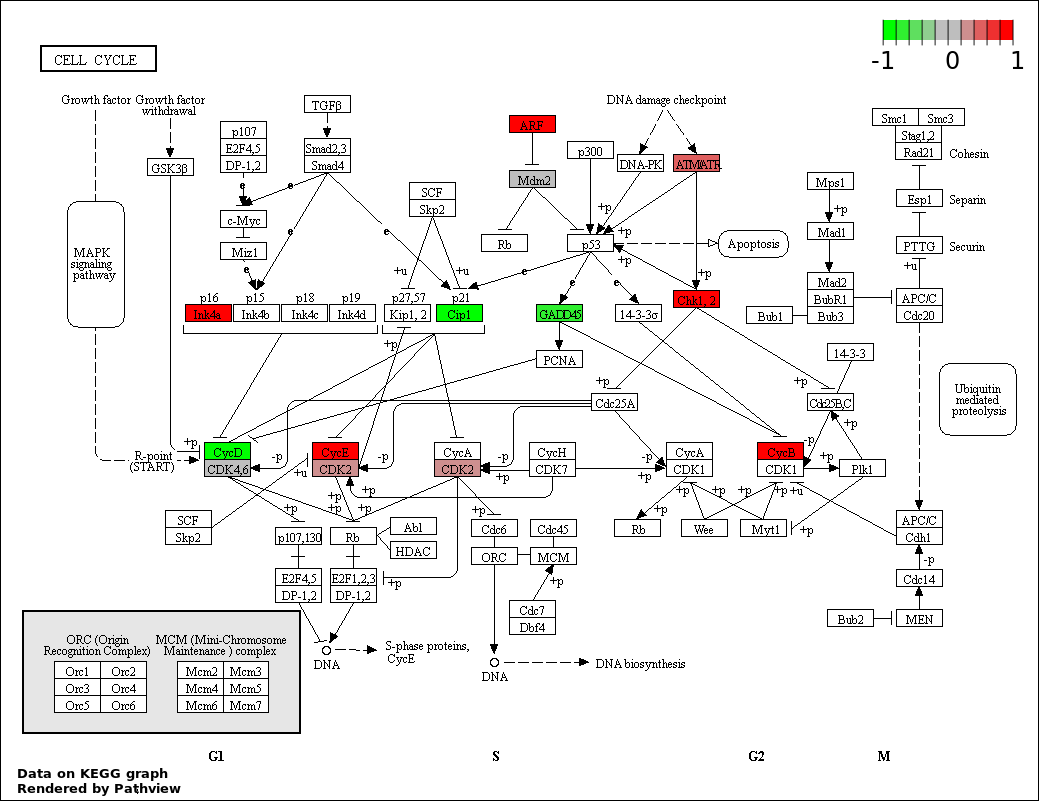
| 2 | hsa04110_Cell_cycle | 18 | 128 | 4.982e-29 | 4.484e-27 | |
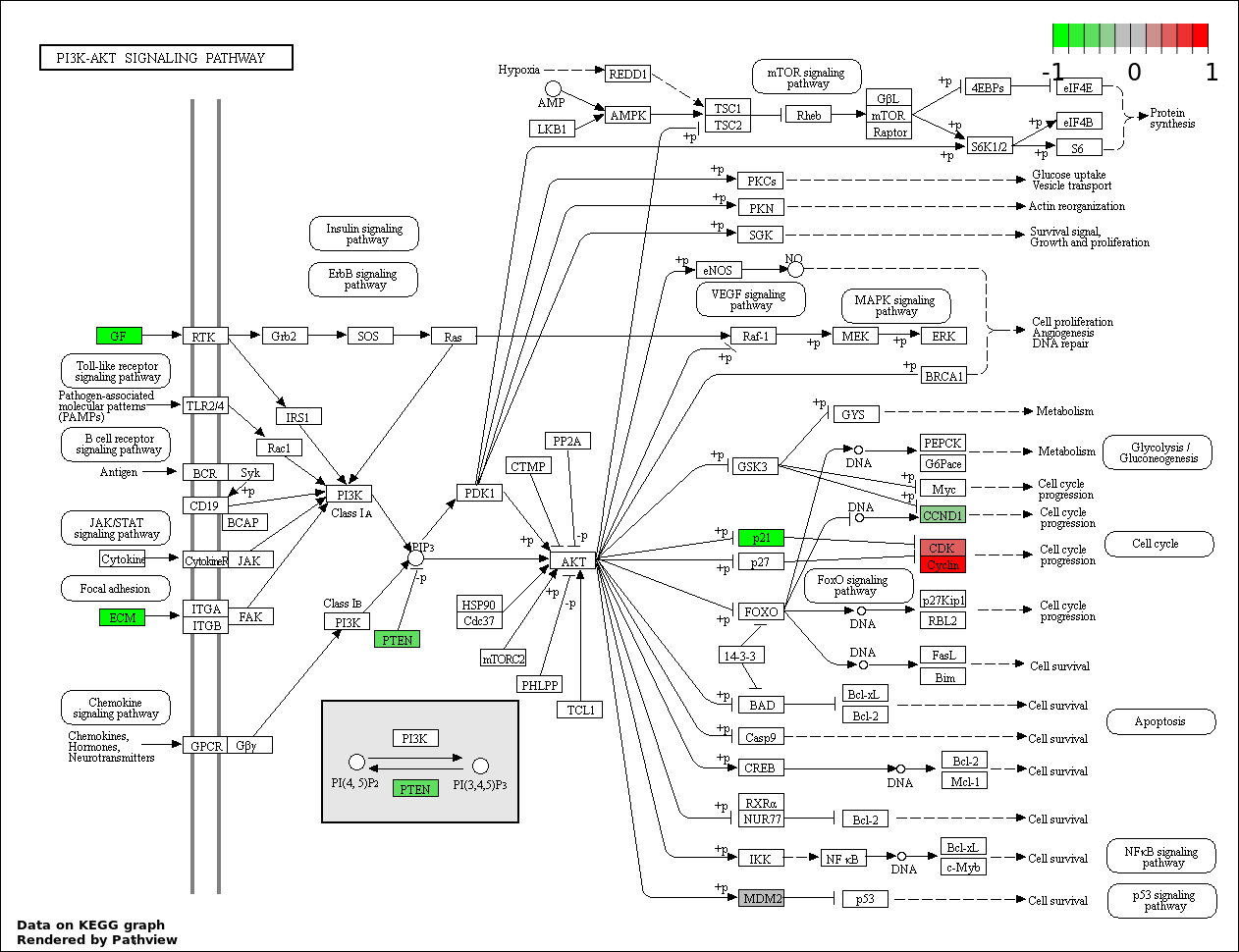
| 3 | hsa04151_PI3K_AKT_signaling_pathway | 13 | 351 | 2.734e-13 | 1.64e-11 | |
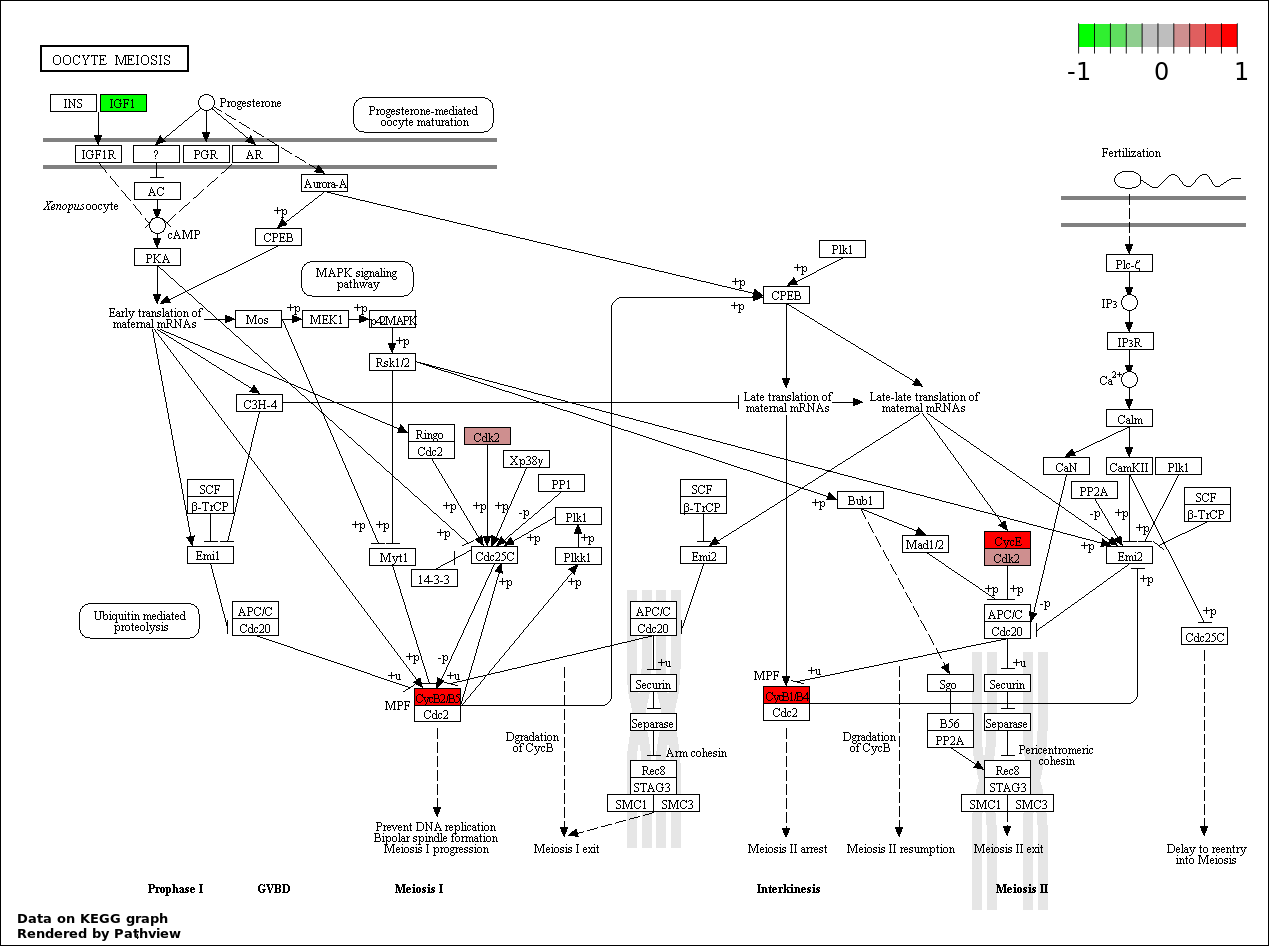
| 4 | hsa04114_Oocyte_meiosis | 6 | 114 | 1.542e-07 | 6.939e-06 | |
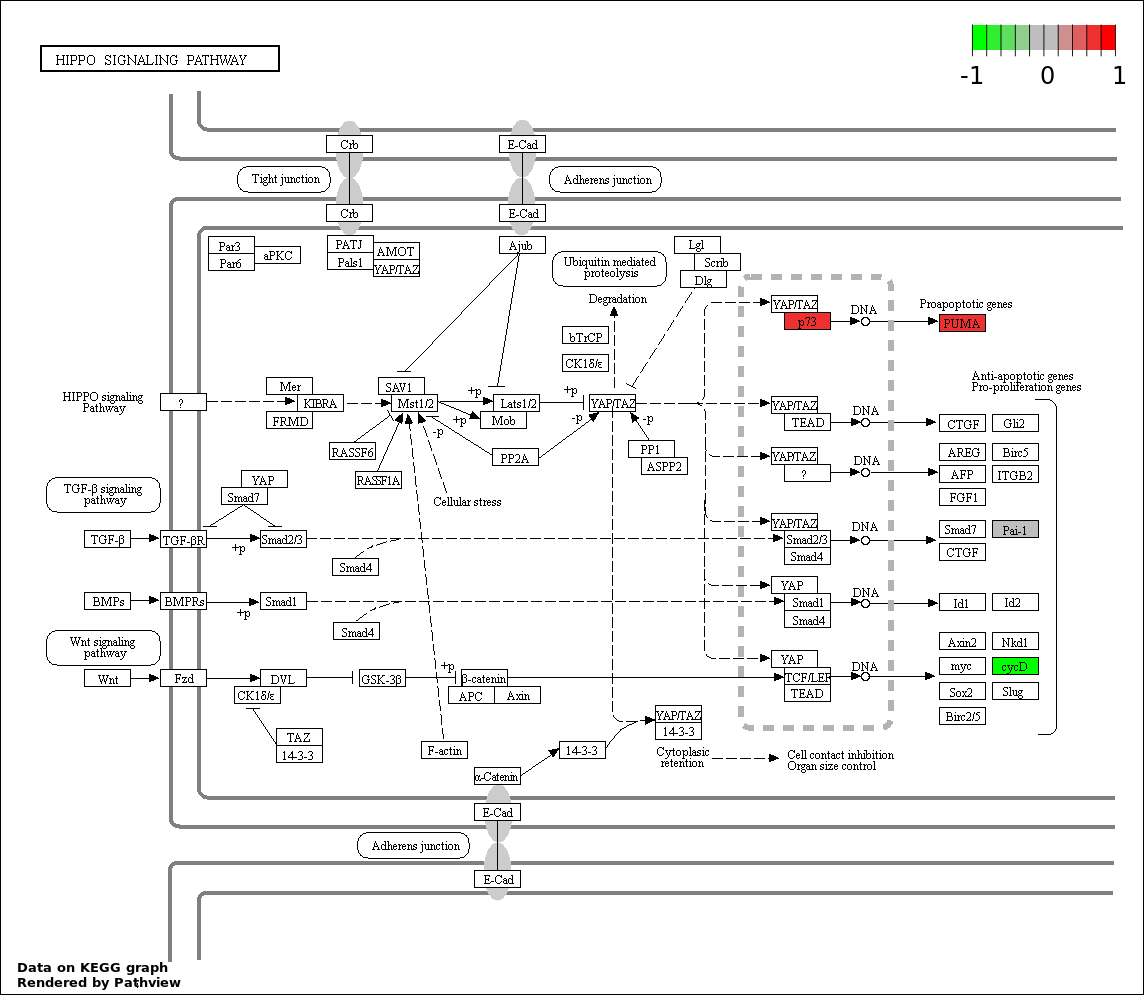
| 5 | hsa04390_Hippo_signaling_pathway | 6 | 154 | 9.108e-07 | 3.279e-05 | |
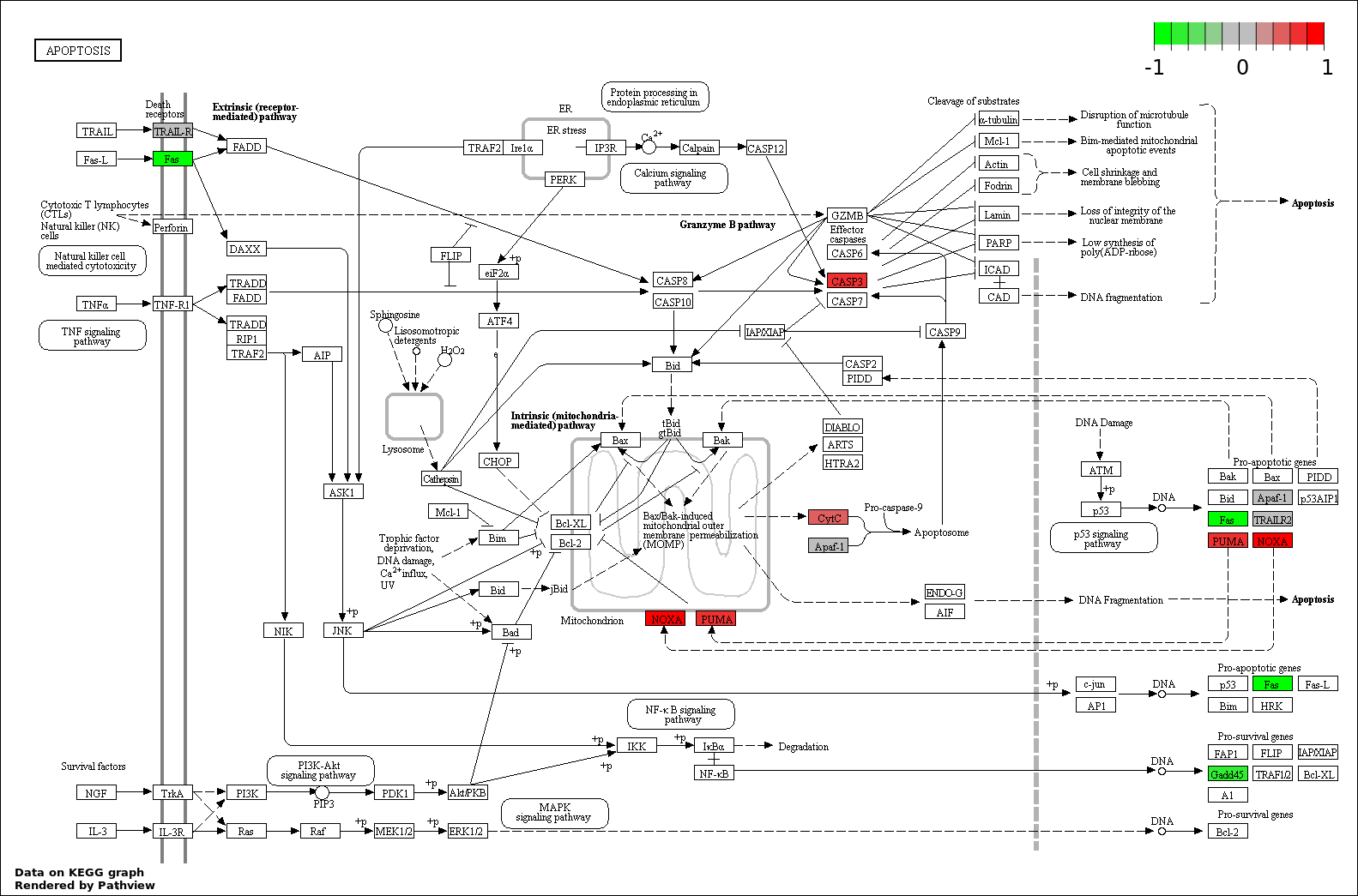
| 6 | hsa04210_Apoptosis | 5 | 89 | 1.312e-06 | 3.937e-05 | |
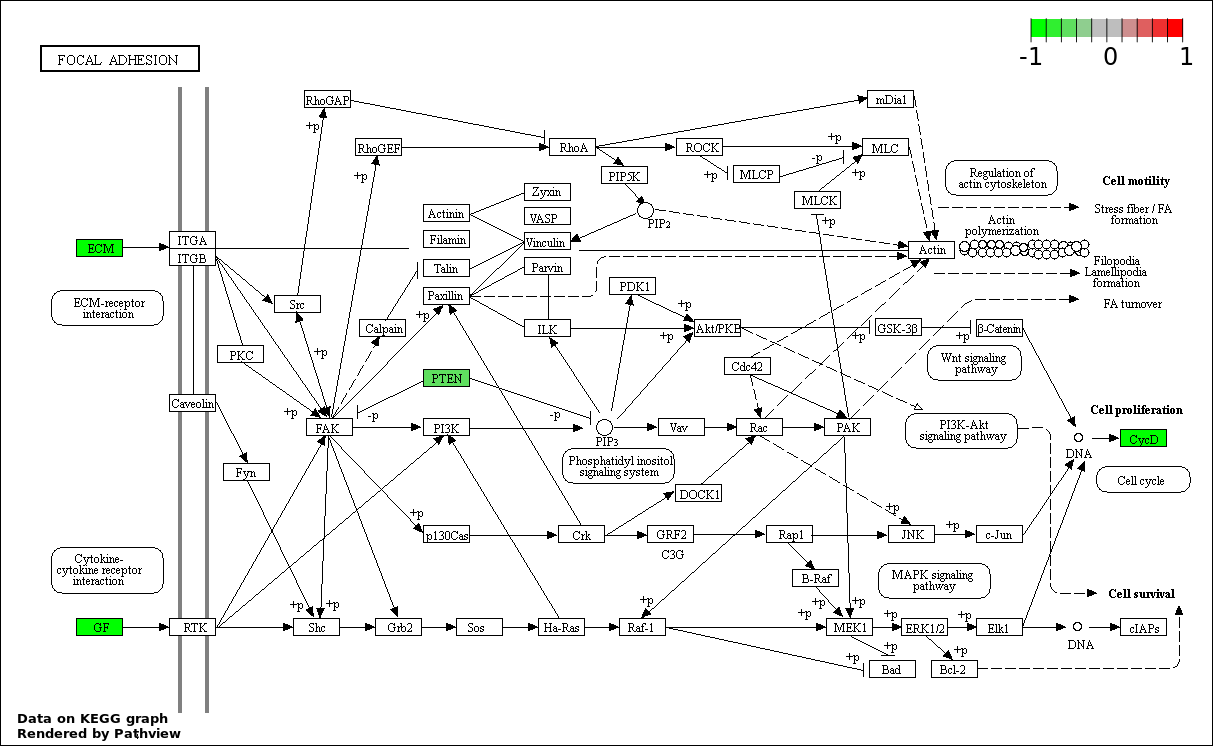
| 7 | hsa04510_Focal_adhesion | 6 | 200 | 4.156e-06 | 0.0001069 | |
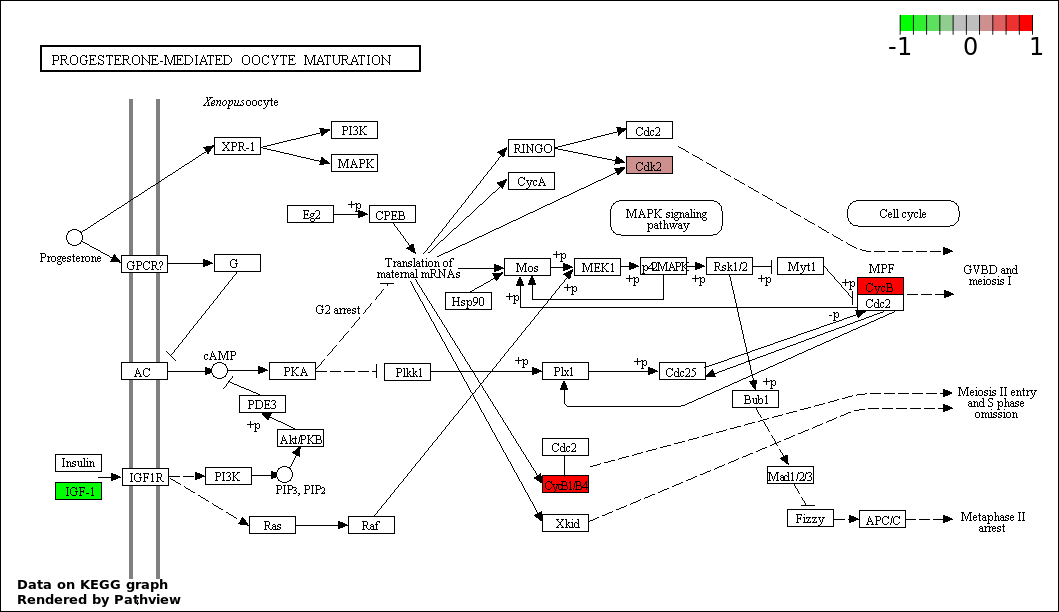
| 8 | hsa04914_Progesterone.mediated_oocyte_maturation | 4 | 87 | 3.621e-05 | 0.0008148 | |
| 9 | hsa04010_MAPK_signaling_pathway | 5 | 268 | 0.0002642 | 0.005284 | |
| 10 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 3 | 136 | 0.003111 | 0.056 | |
| 11 | hsa04310_Wnt_signaling_pathway | 3 | 151 | 0.004174 | 0.06735 | |
| 12 | hsa04630_Jak.STAT_signaling_pathway | 3 | 155 | 0.00449 | 0.06735 | |
| 13 | hsa00480_Glutathione_metabolism | 2 | 50 | 0.005181 | 0.07173 | |
| 14 | hsa00240_Pyrimidine_metabolism | 2 | 99 | 0.0192 | 0.2468 | |
| 15 | hsa04530_Tight_junction | 2 | 133 | 0.03319 | 0.3982 | |
| 16 | hsa00230_Purine_metabolism | 2 | 162 | 0.04742 | 0.5335 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | CCND2 | Sponge network | -2.039 | 0 | -1.641 | 0 | 0.52 |
| 2 | AF131215.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-550a-5p;hsa-miR-589-3p | 14 | CCND2 | Sponge network | -2.09 | 0 | -1.641 | 0 | 0.505 |
| 3 | LINC00702 |
hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 19 | THBS1 | Sponge network | -2.856 | 0 | -0.931 | 0.0014 | 0.501 |
| 4 | RP11-750H9.5 | hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-550a-5p;hsa-miR-877-5p | 10 | CCND2 | Sponge network | -1.959 | 0 | -1.641 | 0 | 0.493 |
| 5 | PCED1B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 18 | CCND2 | Sponge network | -0.672 | 0.02084 | -1.641 | 0 | 0.488 |
| 6 | SH3RF3-AS1 |
hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | THBS1 | Sponge network | -1.583 | 0 | -0.931 | 0.0014 | 0.469 |
| 7 | RP11-399O19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | CCND2 | Sponge network | -0.873 | 0.00072 | -1.641 | 0 | 0.469 |
| 8 | MAGI2-AS3 |
hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 18 | THBS1 | Sponge network | -1.892 | 0 | -0.931 | 0.0014 | 0.461 |
| 9 | AF131215.9 | hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-503-5p;hsa-miR-590-3p | 13 | CCND2 | Sponge network | -1.808 | 0 | -1.641 | 0 | 0.46 |
| 10 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-550a-5p;hsa-miR-877-5p | 14 | CCND2 | Sponge network | -0.785 | 0.00723 | -1.641 | 0 | 0.456 |
| 11 | LINC00996 | hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-320b;hsa-miR-550a-5p | 10 | CCND2 | Sponge network | -1.372 | 0.00025 | -1.641 | 0 | 0.454 |
| 12 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-301a-3p | 13 | CCND2 | Sponge network | -2.028 | 0 | -1.641 | 0 | 0.445 |
| 13 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 34 | CCND2 | Sponge network | -2.856 | 0 | -1.641 | 0 | 0.433 |
| 14 | TBX5-AS1 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-629-5p | 14 | IGF1 | Sponge network | -2.108 | 0 | -0.879 | 0.00545 | 0.429 |
| 15 | RP11-456K23.1 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | IGF1 | Sponge network | -1.488 | 0 | -0.879 | 0.00545 | 0.429 |
| 16 | RP11-166D19.1 |
hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 15 | THBS1 | Sponge network | -0.582 | 0.05253 | -0.931 | 0.0014 | 0.426 |
| 17 | LINC00702 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p | 16 | IGF1 | Sponge network | -2.856 | 0 | -0.879 | 0.00545 | 0.42 |
| 18 | AC109642.1 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-629-5p | 12 | IGF1 | Sponge network | -2.791 | 0 | -0.879 | 0.00545 | 0.41 |
| 19 | TBX5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 26 | CCND2 | Sponge network | -2.108 | 0 | -1.641 | 0 | 0.404 |
| 20 | CTD-2269F5.1 |
hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-32-5p;hsa-miR-7-1-3p | 11 | THBS1 | Sponge network | -1.576 | 0.00334 | -0.931 | 0.0014 | 0.404 |
| 21 | AC011899.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | CCND2 | Sponge network | -2.611 | 0 | -1.641 | 0 | 0.404 |
| 22 | RP11-253E3.3 | hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-27b-5p;hsa-miR-29b-3p;hsa-miR-30e-5p;hsa-miR-660-5p | 11 | CDK6 | Sponge network | -2.404 | 0 | -0.635 | 0.01747 | 0.403 |
| 23 | TBX5-AS1 |
hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 15 | THBS1 | Sponge network | -2.108 | 0 | -0.931 | 0.0014 | 0.398 |
| 24 | LINC00968 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-450b-5p;hsa-miR-503-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-96-5p | 27 | CCND2 | Sponge network | -4.19 | 0 | -1.641 | 0 | 0.396 |
| 25 | RP11-720L2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-93-5p | 12 | CCND2 | Sponge network | -2.305 | 0 | -1.641 | 0 | 0.394 |
| 26 | CTD-2013N24.2 |
hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | THBS1 | Sponge network | -1.745 | 0 | -0.931 | 0.0014 | 0.393 |
| 27 | MAGI2-AS3 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p | 15 | IGF1 | Sponge network | -1.892 | 0 | -0.879 | 0.00545 | 0.384 |
| 28 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-501-5p;hsa-miR-503-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 27 | CCND2 | Sponge network | -1.892 | 0 | -1.641 | 0 | 0.378 |
| 29 | SH3RF3-AS1 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-576-5p | 11 | IGF1 | Sponge network | -1.583 | 0 | -0.879 | 0.00545 | 0.374 |
| 30 | RP11-1024P17.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p | 21 | CCND2 | Sponge network | -2.062 | 0 | -1.641 | 0 | 0.374 |
| 31 | AC007743.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 10 | SESN3 | Sponge network | -2.595 | 0 | -0.097 | 0.73838 | 0.366 |
| 32 | RP11-1008C21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-550a-5p;hsa-miR-590-3p | 17 | CCND2 | Sponge network | -1.826 | 3.0E-5 | -1.641 | 0 | 0.36 |
| 33 | RP11-284N8.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 20 | CCND2 | Sponge network | -0.761 | 0.05061 | -1.641 | 0 | 0.36 |
| 34 | GAS6-AS2 |
hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 13 | THBS1 | Sponge network | -1.761 | 0 | -0.931 | 0.0014 | 0.355 |
| 35 | CTD-2013N24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | CCND2 | Sponge network | -1.745 | 0 | -1.641 | 0 | 0.354 |
| 36 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-450b-5p;hsa-miR-503-5p;hsa-miR-93-5p;hsa-miR-96-5p | 10 | CCND2 | Sponge network | -2.783 | 0 | -1.641 | 0 | 0.352 |
| 37 | RP11-389C8.2 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-629-5p | 11 | IGF1 | Sponge network | -2.039 | 0 | -0.879 | 0.00545 | 0.345 |
| 38 | CTD-2003C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-550a-5p | 11 | CCND2 | Sponge network | -3.403 | 0 | -1.641 | 0 | 0.345 |
| 39 | RP11-1223D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | SESN3 | Sponge network | -0.862 | 0.05389 | -0.097 | 0.73838 | 0.34 |
| 40 | RP11-23P13.6 | hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-590-3p;hsa-miR-877-5p | 11 | CCND2 | Sponge network | -0.705 | 0.00072 | -1.641 | 0 | 0.34 |
| 41 | AC109642.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 24 | CCND2 | Sponge network | -2.791 | 0 | -1.641 | 0 | 0.339 |
| 42 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-550a-5p;hsa-miR-590-5p;hsa-miR-93-5p | 19 | CCND2 | Sponge network | -3.04 | 0 | -1.641 | 0 | 0.337 |
| 43 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | CCND2 | Sponge network | -1.761 | 0 | -1.641 | 0 | 0.337 |
| 44 | RP11-354E11.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-96-5p | 17 | CCND2 | Sponge network | -2.138 | 0 | -1.641 | 0 | 0.335 |
| 45 | MIR497HG |
hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 13 | THBS1 | Sponge network | -2.142 | 0 | -0.931 | 0.0014 | 0.33 |
| 46 | AC109642.1 |
hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 13 | THBS1 | Sponge network | -2.791 | 0 | -0.931 | 0.0014 | 0.328 |
| 47 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | CCND2 | Sponge network | -2.952 | 0 | -1.641 | 0 | 0.327 |
| 48 | AC144831.1 | hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-28-5p;hsa-miR-324-3p;hsa-miR-550a-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 14 | CCND2 | Sponge network | -2.063 | 0 | -1.641 | 0 | 0.327 |
| 49 | RP11-1024P17.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-301a-3p | 15 | PTEN | Sponge network | -2.062 | 0 | -0.419 | 0.00014 | 0.325 |
| 50 | CTD-2013N24.2 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | IGF1 | Sponge network | -1.745 | 0 | -0.879 | 0.00545 | 0.323 |
| 51 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-96-5p | 22 | CCND2 | Sponge network | -0.582 | 0.05253 | -1.641 | 0 | 0.322 |
| 52 | LINC00092 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-503-5p;hsa-miR-589-3p;hsa-miR-590-3p | 14 | CCND2 | Sponge network | -2.383 | 0 | -1.641 | 0 | 0.321 |
| 53 | RP11-166D19.1 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-629-5p | 10 | IGF1 | Sponge network | -0.582 | 0.05253 | -0.879 | 0.00545 | 0.319 |
| 54 | NR2F1-AS1 |
hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 12 | THBS1 | Sponge network | -0.427 | 0.1559 | -0.931 | 0.0014 | 0.317 |
| 55 | BAIAP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | SESN3 | Sponge network | -0.182 | 0.51705 | -0.097 | 0.73838 | 0.317 |
| 56 | RP5-1042I8.7 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 13 | CCND2 | Sponge network | -0.733 | 0.00018 | -1.641 | 0 | 0.316 |
| 57 | AC079630.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 19 | CCND2 | Sponge network | -3.758 | 0 | -1.641 | 0 | 0.314 |
| 58 | RP11-462G12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-25-3p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 13 | SESN3 | Sponge network | -1.071 | 0.01175 | -0.097 | 0.73838 | 0.313 |
| 59 | RP11-354E11.2 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p | 11 | IGF1 | Sponge network | -2.138 | 0 | -0.879 | 0.00545 | 0.313 |
| 60 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 21 | PTEN | Sponge network | -2.856 | 0 | -0.419 | 0.00014 | 0.312 |
| 61 | DIO3OS | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-550a-5p;hsa-miR-877-5p | 14 | CCND2 | Sponge network | -1.936 | 0.00085 | -1.641 | 0 | 0.307 |
| 62 | RP11-283G6.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-450b-5p;hsa-miR-93-5p;hsa-miR-96-5p | 12 | CCND2 | Sponge network | -3.669 | 1.0E-5 | -1.641 | 0 | 0.305 |
| 63 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-671-5p;hsa-miR-93-5p | 22 | ZMAT3 | Sponge network | -1.892 | 0 | -0.319 | 0.07814 | 0.305 |
| 64 | CTC-366B18.4 | hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-93-5p | 10 | CCND2 | Sponge network | -0.652 | 0.01265 | -1.641 | 0 | 0.303 |
| 65 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | PTEN | Sponge network | -2.039 | 0 | -0.419 | 0.00014 | 0.303 |
| 66 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-331-5p;hsa-miR-450b-5p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 22 | CCND2 | Sponge network | -4.222 | 0 | -1.641 | 0 | 0.301 |
| 67 | RP11-378A13.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 20 | CCND2 | Sponge network | -1.713 | 0 | -1.641 | 0 | 0.301 |
| 68 | BAIAP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-365a-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-942-5p | 12 | CCND1 | Sponge network | -0.182 | 0.51705 | -0.296 | 0.2554 | 0.3 |
| 69 | RP4-647J21.1 | hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-550a-5p;hsa-miR-93-5p | 10 | CCND2 | Sponge network | -0.153 | 0.73575 | -1.641 | 0 | 0.299 |
| 70 | RP11-815I9.4 | hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-29b-1-5p;hsa-miR-590-5p;hsa-miR-671-5p | 10 | ZMAT3 | Sponge network | -0.881 | 0.0002 | -0.319 | 0.07814 | 0.299 |
| 71 | LINC00922 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-324-3p;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 14 | CCND2 | Sponge network | -0.842 | 0.11239 | -1.641 | 0 | 0.298 |
| 72 | RP11-476D10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | CCND2 | Sponge network | -4.519 | 0 | -1.641 | 0 | 0.296 |
| 73 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 22 | SESN3 | Sponge network | -1.892 | 0 | -0.097 | 0.73838 | 0.295 |
| 74 | RP11-166D19.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 19 | PTEN | Sponge network | -0.582 | 0.05253 | -0.419 | 0.00014 | 0.295 |
| 75 | RP11-416I2.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-769-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-942-5p | 13 | CCND1 | Sponge network | 3.177 | 1.0E-5 | -0.296 | 0.2554 | 0.294 |
| 76 | CTC-523E23.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-671-5p | 13 | ZMAT3 | Sponge network | -1.636 | 0.00051 | -0.319 | 0.07814 | 0.294 |
| 77 | RP11-456K23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 23 | CCND2 | Sponge network | -1.488 | 0 | -1.641 | 0 | 0.292 |
| 78 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-425-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | PTEN | Sponge network | -1.297 | 0 | -0.419 | 0.00014 | 0.292 |
| 79 | LINC00961 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-96-5p | 16 | CCND2 | Sponge network | -2.724 | 0 | -1.641 | 0 | 0.292 |
| 80 | RP11-284N8.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 15 | SESN3 | Sponge network | -0.761 | 0.05061 | -0.097 | 0.73838 | 0.291 |
| 81 | RP11-1008C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-92a-3p;hsa-miR-93-5p | 13 | SESN3 | Sponge network | -1.249 | 0 | -0.097 | 0.73838 | 0.291 |
| 82 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-93-5p | 12 | CCND2 | Sponge network | -1.193 | 0.00359 | -1.641 | 0 | 0.29 |
| 83 | AC004947.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 17 | CCND2 | Sponge network | -3.94 | 0 | -1.641 | 0 | 0.289 |
| 84 | AC006129.1 | hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-877-5p | 11 | CCND2 | Sponge network | -1.587 | 0.00086 | -1.641 | 0 | 0.287 |
| 85 | LINC00968 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | IGF1 | Sponge network | -4.19 | 0 | -0.879 | 0.00545 | 0.287 |
| 86 | SH3RF3-AS1 |
hsa-miR-15a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-503-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | CCND2 | Sponge network | -1.583 | 0 | -1.641 | 0 | 0.286 |
| 87 | TBX5-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 19 | PTEN | Sponge network | -2.108 | 0 | -0.419 | 0.00014 | 0.286 |
| 88 | TBX5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-363-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 20 | SESN3 | Sponge network | -2.108 | 0 | -0.097 | 0.73838 | 0.283 |
| 89 | LINC00968 |
hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p | 14 | THBS1 | Sponge network | -4.19 | 0 | -0.931 | 0.0014 | 0.283 |
| 90 | AC010226.4 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-93-5p | 10 | CCND2 | Sponge network | -1.081 | 2.0E-5 | -1.641 | 0 | 0.272 |
| 91 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-503-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 27 | CCND2 | Sponge network | -2.142 | 0 | -1.641 | 0 | 0.271 |
| 92 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-501-5p;hsa-miR-550a-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | CCND2 | Sponge network | 0.053 | 0.85755 | -1.641 | 0 | 0.27 |
| 93 | C1orf132 | hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-28-5p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 15 | CCND2 | Sponge network | -0.86 | 0.02429 | -1.641 | 0 | 0.269 |
| 94 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 20 | PTEN | Sponge network | -1.892 | 0 | -0.419 | 0.00014 | 0.268 |
| 95 | RP11-462G12.2 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 11 | SESN3 | Sponge network | -0.602 | 0.22954 | -0.097 | 0.73838 | 0.268 |
| 96 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | SESN3 | Sponge network | -2.039 | 0 | -0.097 | 0.73838 | 0.265 |
| 97 | RP11-399O19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 13 | PTEN | Sponge network | -0.873 | 0.00072 | -0.419 | 0.00014 | 0.264 |
| 98 | RP11-20J15.3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-550a-5p;hsa-miR-877-5p | 11 | CCND2 | Sponge network | -1.709 | 0.0268 | -1.641 | 0 | 0.264 |
| 99 | AC022182.3 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-877-5p;hsa-miR-93-5p | 10 | CCND2 | Sponge network | -0.559 | 0.20451 | -1.641 | 0 | 0.263 |
| 100 | RP11-536K7.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-877-5p;hsa-miR-93-5p | 13 | CCND2 | Sponge network | -1.239 | 5.0E-5 | -1.641 | 0 | 0.263 |
| 101 | RP11-517P14.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | ZMAT3 | Sponge network | -0.795 | 6.0E-5 | -0.319 | 0.07814 | 0.261 |
| 102 | AC109642.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 19 | SESN3 | Sponge network | -2.791 | 0 | -0.097 | 0.73838 | 0.261 |
| 103 | RP11-456K23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 20 | SESN3 | Sponge network | -1.488 | 0 | -0.097 | 0.73838 | 0.256 |
| 104 | AC003991.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-365a-3p;hsa-miR-425-5p;hsa-miR-769-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 14 | CCND1 | Sponge network | -0.787 | 0.08132 | -0.296 | 0.2554 | 0.255 |
| 105 | LIPE-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-425-5p;hsa-miR-590-5p;hsa-miR-93-5p | 13 | PTEN | Sponge network | -0.734 | 0.00039 | -0.419 | 0.00014 | 0.254 |
| 106 | RP11-53M11.3 | hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 10 | CCND2 | Sponge network | -2.058 | 0.01204 | -1.641 | 0 | 0.253 |