This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-let-7a-3p | ABCA1 | 0.28 | 0.12701 | 0.61 | 0.0002 | mirMAP; miRNATAP | -0.3 | 0 | NA | |
| 2 | hsa-let-7a-3p | ABCA5 | 0.28 | 0.12701 | -0.62 | 0.00028 | mirMAP | -0.15 | 0.00733 | NA | |
| 3 | hsa-let-7a-3p | ABCC4 | 0.28 | 0.12701 | 0.37 | 0.06052 | mirMAP | -0.18 | 0.00536 | NA | |
| 4 | hsa-let-7a-3p | ABCC9 | 0.28 | 0.12701 | -0.47 | 0.14121 | mirMAP | -0.91 | 0 | NA | |
| 5 | hsa-let-7a-3p | ABCD2 | 0.28 | 0.12701 | -1.04 | 0.0005 | mirMAP | -0.45 | 0 | NA | |
| 6 | hsa-let-7a-3p | ABI3BP | 0.28 | 0.12701 | -2.06 | 0 | miRNATAP | -0.91 | 0 | NA | |
| 7 | hsa-let-7a-3p | ACAP2 | 0.28 | 0.12701 | 0.18 | 0.06624 | MirTarget; mirMAP; miRNATAP | -0.17 | 0 | NA | |
| 8 | hsa-let-7a-3p | ACVR1 | 0.28 | 0.12701 | 0.28 | 0.00234 | MirTarget; miRNATAP | -0.11 | 0.00039 | NA | |
| 9 | hsa-let-7a-3p | ACVR2B | 0.28 | 0.12701 | 0.48 | 0.00365 | MirTarget | -0.19 | 0.00039 | NA | |
| 10 | hsa-let-7a-3p | ADAM23 | 0.28 | 0.12701 | -0.71 | 0.00863 | mirMAP | -0.5 | 0 | NA | |
| 11 | hsa-let-7a-3p | ADAMTS5 | 0.28 | 0.12701 | 0.19 | 0.27716 | mirMAP | -0.33 | 0 | NA | |
| 12 | hsa-let-7a-3p | ADAMTSL3 | 0.28 | 0.12701 | -1.56 | 0 | MirTarget | -0.73 | 0 | NA | |
| 13 | hsa-let-7a-3p | ADCYAP1 | 0.28 | 0.12701 | -1.7 | 0.00038 | miRNATAP | -0.73 | 0 | NA | |
| 14 | hsa-let-7a-3p | ADD2 | 0.28 | 0.12701 | -0.33 | 0.34514 | mirMAP | -0.53 | 0 | NA | |
| 15 | hsa-let-7a-3p | ADH5 | 0.28 | 0.12701 | -0.37 | 0.00013 | MirTarget | -0.15 | 0 | NA | |
| 16 | hsa-let-7a-3p | AFF3 | 0.28 | 0.12701 | -2.37 | 0 | MirTarget | -0.64 | 0 | NA | |
| 17 | hsa-let-7a-3p | AFF4 | 0.28 | 0.12701 | -0.27 | 0.01565 | MirTarget; mirMAP | -0.24 | 0 | NA | |
| 18 | hsa-let-7a-3p | AHDC1 | 0.28 | 0.12701 | -0.37 | 0.00866 | miRNATAP | -0.2 | 1.0E-5 | NA | |
| 19 | hsa-let-7a-3p | AKAP11 | 0.28 | 0.12701 | 0.2 | 0.09825 | mirMAP | -0.29 | 0 | NA | |
| 20 | hsa-let-7a-3p | AKAP12 | 0.28 | 0.12701 | -0.93 | 0.0028 | miRNATAP | -0.7 | 0 | NA | |
| 21 | hsa-let-7a-3p | AKAP9 | 0.28 | 0.12701 | 0.25 | 0.08715 | MirTarget | -0.15 | 0.00208 | NA | |
| 22 | hsa-let-7a-3p | AKTIP | 0.28 | 0.12701 | -0.38 | 0.00082 | MirTarget | -0.22 | 0 | NA | |
| 23 | hsa-let-7a-3p | ALDH1L2 | 0.28 | 0.12701 | 0.23 | 0.32096 | MirTarget | -0.35 | 0 | NA | |
| 24 | hsa-let-7a-3p | ALG10B | 0.28 | 0.12701 | -0.05 | 0.81184 | MirTarget | -0.16 | 0.02105 | NA | |
| 25 | hsa-let-7a-3p | ALKBH8 | 0.28 | 0.12701 | 0.35 | 0.00043 | MirTarget | -0.14 | 4.0E-5 | NA | |
| 26 | hsa-let-7a-3p | AMIGO2 | 0.28 | 0.12701 | 0.61 | 0.01903 | MirTarget; mirMAP | -0.27 | 0.00207 | NA | |
| 27 | hsa-let-7a-3p | AMN1 | 0.28 | 0.12701 | -0.3 | 0.02124 | mirMAP | -0.1 | 0.01681 | NA | |
| 28 | hsa-let-7a-3p | AMOTL1 | 0.28 | 0.12701 | -0.49 | 0.0546 | mirMAP | -0.63 | 0 | NA | |
| 29 | hsa-let-7a-3p | AMOTL2 | 0.28 | 0.12701 | 0.15 | 0.30776 | miRNATAP | -0.11 | 0.02453 | NA | |
| 30 | hsa-let-7a-3p | ANK3 | 0.28 | 0.12701 | -0.55 | 0.00086 | MirTarget | -0.16 | 0.00316 | NA | |
| 31 | hsa-let-7a-3p | ANKRD50 | 0.28 | 0.12701 | 0.27 | 0.04034 | mirMAP | -0.25 | 0 | NA | |
| 32 | hsa-let-7a-3p | ANKS1B | 0.28 | 0.12701 | -0.22 | 0.12837 | miRNATAP | -0.28 | 0 | NA | |
| 33 | hsa-let-7a-3p | ANO4 | 0.28 | 0.12701 | 0.11 | 0.76908 | MirTarget; miRNATAP | -0.29 | 0.01822 | NA | |
| 34 | hsa-let-7a-3p | ANO5 | 0.28 | 0.12701 | -2.17 | 0 | mirMAP | -0.76 | 0 | NA | |
| 35 | hsa-let-7a-3p | ANTXR1 | 0.28 | 0.12701 | 0.82 | 0.00014 | mirMAP; miRNATAP | -0.42 | 0 | NA | |
| 36 | hsa-let-7a-3p | APC | 0.28 | 0.12701 | -0.26 | 0.04051 | mirMAP | -0.25 | 0 | NA | |
| 37 | hsa-let-7a-3p | APLF | 0.28 | 0.12701 | -0.24 | 0.12306 | MirTarget | -0.26 | 0 | NA | |
| 38 | hsa-let-7a-3p | APPBP2 | 0.28 | 0.12701 | -0.15 | 0.05098 | mirMAP; miRNATAP | -0.16 | 0 | NA | |
| 39 | hsa-let-7a-3p | APPL1 | 0.28 | 0.12701 | 0.04 | 0.68234 | MirTarget; mirMAP | -0.13 | 7.0E-5 | NA | |
| 40 | hsa-let-7a-3p | ARGLU1 | 0.28 | 0.12701 | 0.23 | 0.0598 | mirMAP | -0.13 | 0.00134 | NA | |
| 41 | hsa-let-7a-3p | ARHGAP20 | 0.28 | 0.12701 | -1.38 | 0 | MirTarget; miRNATAP | -0.69 | 0 | NA | |
| 42 | hsa-let-7a-3p | ARHGAP28 | 0.28 | 0.12701 | -0.91 | 0.00013 | mirMAP | -0.19 | 0.01861 | NA | |
| 43 | hsa-let-7a-3p | ARHGAP29 | 0.28 | 0.12701 | 0.13 | 0.42953 | mirMAP | -0.31 | 0 | NA | |
| 44 | hsa-let-7a-3p | ARHGAP5 | 0.28 | 0.12701 | -0.12 | 0.26304 | mirMAP | -0.19 | 0 | NA | |
| 45 | hsa-let-7a-3p | ARHGEF3 | 0.28 | 0.12701 | 0.02 | 0.82386 | MirTarget | -0.11 | 0.00239 | NA | |
| 46 | hsa-let-7a-3p | ARHGEF33 | 0.28 | 0.12701 | -0.33 | 0.12959 | miRNATAP | -0.46 | 0 | NA | |
| 47 | hsa-let-7a-3p | ARID4A | 0.28 | 0.12701 | -0.05 | 0.68424 | MirTarget; miRNATAP | -0.27 | 0 | NA | |
| 48 | hsa-let-7a-3p | ARID4B | 0.28 | 0.12701 | 0.37 | 0.00063 | MirTarget; miRNATAP | -0.18 | 0 | NA | |
| 49 | hsa-let-7a-3p | ARID5B | 0.28 | 0.12701 | 0.34 | 0.01676 | miRNATAP | -0.23 | 0 | NA | |
| 50 | hsa-let-7a-3p | ARIH1 | 0.28 | 0.12701 | 0.02 | 0.79347 | MirTarget | -0.17 | 0 | NA | |
| 51 | hsa-let-7a-3p | ARL10 | 0.28 | 0.12701 | -0.28 | 0.19832 | mirMAP | -0.46 | 0 | NA | |
| 52 | hsa-let-7a-3p | ARMC8 | 0.28 | 0.12701 | -0.11 | 0.17343 | miRNAWalker2 validate | -0.13 | 0 | NA | |
| 53 | hsa-let-7a-3p | ARPP19 | 0.28 | 0.12701 | 0.11 | 0.27444 | MirTarget | -0.2 | 0 | NA | |
| 54 | hsa-let-7a-3p | ARRDC3 | 0.28 | 0.12701 | -0.15 | 0.28741 | MirTarget | -0.29 | 0 | NA | |
| 55 | hsa-let-7a-3p | ARSB | 0.28 | 0.12701 | 0.33 | 0.02033 | mirMAP | -0.31 | 0 | NA | |
| 56 | hsa-let-7a-3p | ART4 | 0.28 | 0.12701 | -0.77 | 0.0772 | mirMAP | -0.46 | 0.00143 | NA | |
| 57 | hsa-let-7a-3p | ASAP1 | 0.28 | 0.12701 | 0.66 | 0 | mirMAP | -0.23 | 0 | NA | |
| 58 | hsa-let-7a-3p | ASTN2 | 0.28 | 0.12701 | -0.7 | 0.00014 | mirMAP | -0.19 | 0.00191 | NA | |
| 59 | hsa-let-7a-3p | ATE1 | 0.28 | 0.12701 | 0.13 | 0.35236 | mirMAP | -0.11 | 0.01096 | NA | |
| 60 | hsa-let-7a-3p | ATF2 | 0.28 | 0.12701 | 0.01 | 0.93568 | MirTarget; mirMAP; miRNATAP | -0.16 | 0 | NA | |
| 61 | hsa-let-7a-3p | ATG14 | 0.28 | 0.12701 | 0 | 0.96836 | mirMAP | -0.27 | 0 | NA | |
| 62 | hsa-let-7a-3p | ATG2B | 0.28 | 0.12701 | -0.11 | 0.28452 | MirTarget | -0.2 | 0 | NA | |
| 63 | hsa-let-7a-3p | ATL2 | 0.28 | 0.12701 | -0.38 | 0.00058 | MirTarget | -0.25 | 0 | NA | |
| 64 | hsa-let-7a-3p | ATRX | 0.28 | 0.12701 | 0.26 | 0.04408 | MirTarget; miRNATAP | -0.32 | 0 | NA | |
| 65 | hsa-let-7a-3p | AUTS2 | 0.28 | 0.12701 | -0.27 | 0.18068 | miRNATAP | -0.26 | 7.0E-5 | NA | |
| 66 | hsa-let-7a-3p | AXIN2 | 0.28 | 0.12701 | 0.01 | 0.97155 | miRNATAP | -0.21 | 0.02551 | NA | |
| 67 | hsa-let-7a-3p | B3GALNT1 | 0.28 | 0.12701 | 0.08 | 0.70688 | mirMAP | -0.15 | 0.02646 | NA | |
| 68 | hsa-let-7a-3p | BACH1 | 0.28 | 0.12701 | -0.04 | 0.68833 | MirTarget; mirMAP | -0.11 | 0.00162 | NA | |
| 69 | hsa-let-7a-3p | BCAP29 | 0.28 | 0.12701 | -0.09 | 0.29187 | MirTarget; mirMAP | -0.16 | 0 | NA | |
| 70 | hsa-let-7a-3p | BCL11A | 0.28 | 0.12701 | -0.93 | 0.0063 | mirMAP | -0.33 | 0.00333 | NA | |
| 71 | hsa-let-7a-3p | BCL6 | 0.28 | 0.12701 | -0.21 | 0.19489 | miRNATAP | -0.34 | 0 | NA | |
| 72 | hsa-let-7a-3p | BCLAF1 | 0.28 | 0.12701 | 0.21 | 0.00684 | mirMAP | -0.11 | 1.0E-5 | NA | |
| 73 | hsa-let-7a-3p | BDP1 | 0.28 | 0.12701 | 0.41 | 0.00065 | MirTarget; mirMAP | -0.27 | 0 | NA | |
| 74 | hsa-let-7a-3p | BEND6 | 0.28 | 0.12701 | 0.03 | 0.91694 | mirMAP | -0.47 | 0 | NA | |
| 75 | hsa-let-7a-3p | BICD2 | 0.28 | 0.12701 | -0.02 | 0.88611 | mirMAP | -0.27 | 0 | NA | |
| 76 | hsa-let-7a-3p | BMP2K | 0.28 | 0.12701 | 0.25 | 0.01328 | mirMAP | -0.1 | 0.00148 | NA | |
| 77 | hsa-let-7a-3p | BMP3 | 0.28 | 0.12701 | -2.36 | 0.00035 | MirTarget; mirMAP | -1.5 | 0 | NA | |
| 78 | hsa-let-7a-3p | BMPER | 0.28 | 0.12701 | -2.32 | 0 | MirTarget | -0.67 | 0 | NA | |
| 79 | hsa-let-7a-3p | BMPR1B | 0.28 | 0.12701 | -1.24 | 0.00562 | miRNATAP | -0.66 | 1.0E-5 | NA | |
| 80 | hsa-let-7a-3p | BMPR2 | 0.28 | 0.12701 | 0.21 | 0.0635 | MirTarget; miRNATAP | -0.28 | 0 | NA | |
| 81 | hsa-let-7a-3p | BNC2 | 0.28 | 0.12701 | -0.49 | 0.09988 | mirMAP | -0.77 | 0 | NA | |
| 82 | hsa-let-7a-3p | BNIP2 | 0.28 | 0.12701 | -0.05 | 0.63907 | mirMAP | -0.29 | 0 | NA | |
| 83 | hsa-let-7a-3p | BOD1L1 | 0.28 | 0.12701 | 0.23 | 0.03726 | MirTarget | -0.24 | 0 | NA | |
| 84 | hsa-let-7a-3p | BRWD1 | 0.28 | 0.12701 | 0.05 | 0.6598 | mirMAP | -0.2 | 0 | NA | |
| 85 | hsa-let-7a-3p | BTAF1 | 0.28 | 0.12701 | 0.28 | 0.01596 | MirTarget; miRNATAP | -0.19 | 0 | NA | |
| 86 | hsa-let-7a-3p | BTBD7 | 0.28 | 0.12701 | -0.17 | 0.03473 | mirMAP; miRNATAP | -0.12 | 1.0E-5 | NA | |
| 87 | hsa-let-7a-3p | BTC | 0.28 | 0.12701 | -1.69 | 0 | mirMAP | -0.43 | 1.0E-5 | NA | |
| 88 | hsa-let-7a-3p | BTF3L4 | 0.28 | 0.12701 | 0.18 | 0.05058 | MirTarget; miRNATAP | -0.15 | 0 | NA | |
| 89 | hsa-let-7a-3p | BTNL9 | 0.28 | 0.12701 | 0.41 | 0.12761 | MirTarget | -0.27 | 0.00215 | NA | |
| 90 | hsa-let-7a-3p | BVES | 0.28 | 0.12701 | -1.51 | 1.0E-5 | mirMAP | -0.95 | 0 | NA | |
| 91 | hsa-let-7a-3p | C16orf52 | 0.28 | 0.12701 | -0.07 | 0.3851 | mirMAP; miRNATAP | -0.18 | 0 | NA | |
| 92 | hsa-let-7a-3p | C4orf3 | 0.28 | 0.12701 | -0.9 | 0 | mirMAP | -0.16 | 5.0E-5 | NA | |
| 93 | hsa-let-7a-3p | C4orf32 | 0.28 | 0.12701 | -0.55 | 4.0E-5 | mirMAP | -0.11 | 0.01525 | NA | |
| 94 | hsa-let-7a-3p | C8orf46 | 0.28 | 0.12701 | -1.93 | 2.0E-5 | mirMAP | -0.98 | 0 | NA | |
| 95 | hsa-let-7a-3p | CACNA1C | 0.28 | 0.12701 | -0.18 | 0.557 | MirTarget | -0.79 | 0 | NA | |
| 96 | hsa-let-7a-3p | CACNA2D1 | 0.28 | 0.12701 | -0.85 | 0.00675 | mirMAP | -0.68 | 0 | NA | |
| 97 | hsa-let-7a-3p | CACNB2 | 0.28 | 0.12701 | -1.06 | 0.00045 | MirTarget; mirMAP | -0.9 | 0 | NA | |
| 98 | hsa-let-7a-3p | CACNG4 | 0.28 | 0.12701 | -0.7 | 0.19815 | MirTarget | -0.49 | 0.00654 | NA | |
| 99 | hsa-let-7a-3p | CAMSAP1 | 0.28 | 0.12701 | 0.24 | 0.03991 | mirMAP | -0.15 | 0.00012 | NA | |
| 100 | hsa-let-7a-3p | CAPZA2 | 0.28 | 0.12701 | -0.19 | 0.08777 | MirTarget | -0.15 | 6.0E-5 | NA | |
| 101 | hsa-let-7a-3p | CASD1 | 0.28 | 0.12701 | -0.06 | 0.67432 | MirTarget; miRNATAP | -0.26 | 0 | NA | |
| 102 | hsa-let-7a-3p | CASP8AP2 | 0.28 | 0.12701 | 0.47 | 0.00018 | MirTarget | -0.25 | 0 | NA | |
| 103 | hsa-let-7a-3p | CBL | 0.28 | 0.12701 | 0.29 | 0.01267 | MirTarget | -0.19 | 0 | NA | |
| 104 | hsa-let-7a-3p | CBLN4 | 0.28 | 0.12701 | 0.52 | 0.26155 | MirTarget | -0.78 | 0 | NA | |
| 105 | hsa-let-7a-3p | CBX5 | 0.28 | 0.12701 | 0.16 | 0.24446 | mirMAP | -0.16 | 0.00065 | NA | |
| 106 | hsa-let-7a-3p | CCDC102B | 0.28 | 0.12701 | 0.32 | 0.04327 | MirTarget | -0.17 | 0.00131 | NA | |
| 107 | hsa-let-7a-3p | CCDC141 | 0.28 | 0.12701 | -0 | 0.99219 | mirMAP | -0.34 | 0.00055 | NA | |
| 108 | hsa-let-7a-3p | CCDC50 | 0.28 | 0.12701 | -0.21 | 0.09064 | miRNATAP | -0.34 | 0 | NA | |
| 109 | hsa-let-7a-3p | CCND2 | 0.28 | 0.12701 | -0.27 | 0.3112 | mirMAP | -0.37 | 2.0E-5 | 20418948 | MicroRNA let 7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2 |
| 110 | hsa-let-7a-3p | CCNG2 | 0.28 | 0.12701 | -0.15 | 0.25163 | MirTarget; mirMAP | -0.12 | 0.00497 | NA | |
| 111 | hsa-let-7a-3p | CCPG1 | 0.28 | 0.12701 | -0.21 | 0.04535 | mirMAP | -0.13 | 0.00013 | NA | |
| 112 | hsa-let-7a-3p | CD109 | 0.28 | 0.12701 | 0.26 | 0.3195 | mirMAP | -0.51 | 0 | NA | |
| 113 | hsa-let-7a-3p | CD44 | 0.28 | 0.12701 | 0.22 | 0.30295 | miRNATAP | -0.19 | 0.00876 | NA | |
| 114 | hsa-let-7a-3p | CDC42EP3 | 0.28 | 0.12701 | -0.12 | 0.56698 | MirTarget; mirMAP | -0.47 | 0 | NA | |
| 115 | hsa-let-7a-3p | CDC73 | 0.28 | 0.12701 | 0.09 | 0.20463 | mirMAP; miRNATAP | -0.1 | 1.0E-5 | NA | |
| 116 | hsa-let-7a-3p | CDHR3 | 0.28 | 0.12701 | -0.98 | 4.0E-5 | mirMAP | -0.4 | 0 | NA | |
| 117 | hsa-let-7a-3p | CDK17 | 0.28 | 0.12701 | 0.07 | 0.48708 | MirTarget; miRNATAP | -0.26 | 0 | NA | |
| 118 | hsa-let-7a-3p | CDK19 | 0.28 | 0.12701 | 0.33 | 0.00181 | MirTarget; miRNATAP | -0.11 | 0.00141 | NA | |
| 119 | hsa-let-7a-3p | CDS2 | 0.28 | 0.12701 | -0.26 | 0.01248 | mirMAP | -0.26 | 0 | NA | |
| 120 | hsa-let-7a-3p | CELF2 | 0.28 | 0.12701 | -1.1 | 1.0E-5 | miRNATAP | -0.56 | 0 | NA | |
| 121 | hsa-let-7a-3p | CEP192 | 0.28 | 0.12701 | 0.5 | 2.0E-5 | MirTarget | -0.11 | 0.00483 | NA | |
| 122 | hsa-let-7a-3p | CFL2 | 0.28 | 0.12701 | -1.3 | 0 | mirMAP | -0.78 | 0 | NA | |
| 123 | hsa-let-7a-3p | CGGBP1 | 0.28 | 0.12701 | -0.06 | 0.38556 | mirMAP | -0.15 | 0 | NA | |
| 124 | hsa-let-7a-3p | CHD1 | 0.28 | 0.12701 | 0.32 | 0.00215 | MirTarget | -0.17 | 0 | NA | |
| 125 | hsa-let-7a-3p | CHD9 | 0.28 | 0.12701 | -0.26 | 0.02092 | mirMAP; miRNATAP | -0.22 | 0 | NA | |
| 126 | hsa-let-7a-3p | CHL1 | 0.28 | 0.12701 | -1.95 | 0 | MirTarget; miRNATAP | -0.47 | 4.0E-5 | NA | |
| 127 | hsa-let-7a-3p | CHM | 0.28 | 0.12701 | 0.11 | 0.24003 | MirTarget | -0.16 | 0 | NA | |
| 128 | hsa-let-7a-3p | CHMP1B | 0.28 | 0.12701 | -0.71 | 0 | mirMAP | -0.14 | 0.0001 | NA | |
| 129 | hsa-let-7a-3p | CLASP2 | 0.28 | 0.12701 | -0.04 | 0.71 | MirTarget; miRNATAP | -0.25 | 0 | NA | |
| 130 | hsa-let-7a-3p | CLIC4 | 0.28 | 0.12701 | -0.39 | 0.04124 | MirTarget | -0.57 | 0 | NA | |
| 131 | hsa-let-7a-3p | CLIP1 | 0.28 | 0.12701 | -0.22 | 0.08684 | miRNATAP | -0.37 | 0 | NA | |
| 132 | hsa-let-7a-3p | CLK4 | 0.28 | 0.12701 | -0.08 | 0.49185 | mirMAP | -0.22 | 0 | NA | |
| 133 | hsa-let-7a-3p | CLOCK | 0.28 | 0.12701 | -0.1 | 0.34735 | MirTarget | -0.12 | 0.001 | NA | |
| 134 | hsa-let-7a-3p | CNKSR2 | 0.28 | 0.12701 | -1.85 | 0.00014 | MirTarget; mirMAP; miRNATAP | -1.14 | 0 | NA | |
| 135 | hsa-let-7a-3p | CNKSR3 | 0.28 | 0.12701 | -0.14 | 0.34161 | MirTarget | -0.2 | 1.0E-5 | NA | |
| 136 | hsa-let-7a-3p | CNR1 | 0.28 | 0.12701 | -2.22 | 1.0E-5 | mirMAP | -0.86 | 0 | NA | |
| 137 | hsa-let-7a-3p | CNTN1 | 0.28 | 0.12701 | -2.59 | 0 | MirTarget; miRNATAP | -1.15 | 0 | NA | |
| 138 | hsa-let-7a-3p | CNTN5 | 0.28 | 0.12701 | -0.22 | 0.61957 | MirTarget; mirMAP | -0.36 | 0.01296 | NA | |
| 139 | hsa-let-7a-3p | CNTNAP3B | 0.28 | 0.12701 | -1.04 | 0.01401 | mirMAP | -0.82 | 0 | NA | |
| 140 | hsa-let-7a-3p | COL12A1 | 0.28 | 0.12701 | 1.85 | 0 | miRNATAP | -0.32 | 0.00013 | NA | |
| 141 | hsa-let-7a-3p | COL15A1 | 0.28 | 0.12701 | 0.68 | 0.00158 | MirTarget | -0.29 | 5.0E-5 | NA | |
| 142 | hsa-let-7a-3p | COL4A1 | 0.28 | 0.12701 | 1.79 | 0 | miRNATAP | -0.26 | 0.00016 | NA | |
| 143 | hsa-let-7a-3p | COL4A3BP | 0.28 | 0.12701 | -0.44 | 0 | MirTarget; mirMAP; miRNATAP | -0.16 | 0 | NA | |
| 144 | hsa-let-7a-3p | COL4A4 | 0.28 | 0.12701 | 0.2 | 0.56272 | mirMAP | -0.38 | 0.00099 | NA | |
| 145 | hsa-let-7a-3p | CORO1C | 0.28 | 0.12701 | -0.06 | 0.60556 | MirTarget; miRNATAP | -0.28 | 0 | NA | |
| 146 | hsa-let-7a-3p | CPEB2 | 0.28 | 0.12701 | -0.81 | 0 | mirMAP; miRNATAP | -0.24 | 0 | NA | |
| 147 | hsa-let-7a-3p | CPEB3 | 0.28 | 0.12701 | -0.73 | 0 | MirTarget; miRNATAP | -0.19 | 0 | NA | |
| 148 | hsa-let-7a-3p | CPEB4 | 0.28 | 0.12701 | -0.8 | 0 | mirMAP; miRNATAP | -0.19 | 0 | NA | |
| 149 | hsa-let-7a-3p | CREBBP | 0.28 | 0.12701 | 0.13 | 0.15186 | miRNATAP | -0.13 | 1.0E-5 | NA | |
| 150 | hsa-let-7a-3p | CREBL2 | 0.28 | 0.12701 | -0.29 | 0.00289 | mirMAP | -0.18 | 0 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 144 | 1672 | 2.516e-17 | 1.171e-13 |
| 2 | NEUROGENESIS | 122 | 1402 | 5.019e-15 | 1.168e-11 |
| 3 | REGULATION OF CELL DEVELOPMENT | 82 | 836 | 5.31e-13 | 8.236e-10 |
| 4 | REGULATION OF CELL DIFFERENTIATION | 121 | 1492 | 9.526e-13 | 9.244e-10 |
| 5 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 93 | 1021 | 9.933e-13 | 9.244e-10 |
| 6 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 136 | 1784 | 2.863e-12 | 2.22e-09 |
| 7 | CELL DEVELOPMENT | 114 | 1426 | 1.2e-11 | 7.978e-09 |
| 8 | REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 73 | 750 | 1.577e-11 | 8.071e-09 |
| 9 | ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY | 69 | 689 | 1.636e-11 | 8.071e-09 |
| 10 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 89 | 1008 | 1.735e-11 | 8.071e-09 |
| 11 | CARDIOVASCULAR SYSTEM DEVELOPMENT | 75 | 788 | 2.388e-11 | 9.258e-09 |
| 12 | CIRCULATORY SYSTEM DEVELOPMENT | 75 | 788 | 2.388e-11 | 9.258e-09 |
| 13 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 96 | 1142 | 4.09e-11 | 1.464e-08 |
| 14 | PROTEIN PHOSPHORYLATION | 84 | 944 | 4.566e-11 | 1.518e-08 |
| 15 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 125 | 1656 | 4.952e-11 | 1.536e-08 |
| 16 | REGULATION OF PROTEIN MODIFICATION PROCESS | 127 | 1710 | 9.814e-11 | 2.854e-08 |
| 17 | REGULATION OF CELLULAR COMPONENT BIOGENESIS | 72 | 767 | 1.15e-10 | 3.147e-08 |
| 18 | NEGATIVE REGULATION OF CELL COMMUNICATION | 96 | 1192 | 4.176e-10 | 1.079e-07 |
| 19 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 119 | 1618 | 7.665e-10 | 1.877e-07 |
| 20 | NEGATIVE REGULATION OF NITROGEN COMPOUND METABOLIC PROCESS | 113 | 1517 | 1.059e-09 | 2.371e-07 |
| 21 | INTRACELLULAR SIGNAL TRANSDUCTION | 116 | 1572 | 1.07e-09 | 2.371e-07 |
| 22 | GROWTH | 46 | 410 | 1.248e-09 | 2.639e-07 |
| 23 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE SIGNALING PATHWAY | 52 | 498 | 1.335e-09 | 2.7e-07 |
| 24 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 128 | 1805 | 1.579e-09 | 3.062e-07 |
| 25 | CELLULAR COMPONENT MORPHOGENESIS | 77 | 900 | 1.871e-09 | 3.482e-07 |
| 26 | CELL PROJECTION ORGANIZATION | 77 | 902 | 2.066e-09 | 3.697e-07 |
| 27 | NEGATIVE REGULATION OF GENE EXPRESSION | 110 | 1493 | 3.365e-09 | 5.399e-07 |
| 28 | POSITIVE REGULATION OF GENE EXPRESSION | 123 | 1733 | 3.321e-09 | 5.399e-07 |
| 29 | HEART DEVELOPMENT | 49 | 466 | 3.198e-09 | 5.399e-07 |
| 30 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 126 | 1791 | 3.527e-09 | 5.47e-07 |
| 31 | NEURON PROJECTION DEVELOPMENT | 54 | 545 | 4.153e-09 | 6.234e-07 |
| 32 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 82 | 1004 | 4.684e-09 | 6.811e-07 |
| 33 | NEGATIVE REGULATION OF RESPONSE TO STIMULUS | 102 | 1360 | 5.373e-09 | 7.576e-07 |
| 34 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 103 | 1395 | 1.018e-08 | 1.393e-06 |
| 35 | REGULATION OF HYDROLASE ACTIVITY | 99 | 1327 | 1.212e-08 | 1.525e-06 |
| 36 | REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 29 | 207 | 1.162e-08 | 1.525e-06 |
| 37 | REGULATION OF GTPASE ACTIVITY | 61 | 673 | 1.212e-08 | 1.525e-06 |
| 38 | CENTRAL NERVOUS SYSTEM DEVELOPMENT | 73 | 872 | 1.262e-08 | 1.545e-06 |
| 39 | MOVEMENT OF CELL OR SUBCELLULAR COMPONENT | 96 | 1275 | 1.316e-08 | 1.57e-06 |
| 40 | SINGLE ORGANISM BEHAVIOR | 42 | 384 | 1.391e-08 | 1.578e-06 |
| 41 | NEURON DIFFERENTIATION | 73 | 874 | 1.387e-08 | 1.578e-06 |
| 42 | HEAD DEVELOPMENT | 63 | 709 | 1.472e-08 | 1.631e-06 |
| 43 | TISSUE DEVELOPMENT | 109 | 1518 | 1.616e-08 | 1.748e-06 |
| 44 | PHOSPHORYLATION | 93 | 1228 | 1.721e-08 | 1.82e-06 |
| 45 | DEVELOPMENTAL GROWTH | 38 | 333 | 2.199e-08 | 2.274e-06 |
| 46 | REGULATION OF CELL DEATH | 106 | 1472 | 2.256e-08 | 2.282e-06 |
| 47 | REGULATION OF CELL PROJECTION ORGANIZATION | 53 | 558 | 2.432e-08 | 2.407e-06 |
| 48 | ANATOMICAL STRUCTURE FORMATION INVOLVED IN MORPHOGENESIS | 77 | 957 | 2.705e-08 | 2.517e-06 |
| 49 | POSITIVE REGULATION OF CELL COMMUNICATION | 109 | 1532 | 2.628e-08 | 2.517e-06 |
| 50 | CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 50 | 513 | 2.685e-08 | 2.517e-06 |
| 51 | REGULATION OF CELLULAR COMPONENT MOVEMENT | 66 | 771 | 2.805e-08 | 2.559e-06 |
| 52 | RESPONSE TO ENDOGENOUS STIMULUS | 104 | 1450 | 3.816e-08 | 3.415e-06 |
| 53 | REGULATION OF NEURON DIFFERENTIATION | 52 | 554 | 4.863e-08 | 4.269e-06 |
| 54 | CELLULAR RESPONSE TO ORGANIC SUBSTANCE | 124 | 1848 | 7.354e-08 | 6.222e-06 |
| 55 | NEGATIVE REGULATION OF PHOSPHORYLATION | 43 | 422 | 7.349e-08 | 6.222e-06 |
| 56 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 80 | 1036 | 8.316e-08 | 6.789e-06 |
| 57 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 80 | 1036 | 8.316e-08 | 6.789e-06 |
| 58 | RETINA VASCULATURE DEVELOPMENT IN CAMERA TYPE EYE | 8 | 16 | 8.688e-08 | 6.97e-06 |
| 59 | REGULATION OF CELL MORPHOGENESIS | 51 | 552 | 1.081e-07 | 8.524e-06 |
| 60 | POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 86 | 1152 | 1.175e-07 | 8.839e-06 |
| 61 | RESPONSE TO GROWTH FACTOR | 46 | 475 | 1.176e-07 | 8.839e-06 |
| 62 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 66 | 801 | 1.178e-07 | 8.839e-06 |
| 63 | UROGENITAL SYSTEM DEVELOPMENT | 34 | 299 | 1.317e-07 | 9.724e-06 |
| 64 | ORGAN MORPHOGENESIS | 68 | 841 | 1.531e-07 | 1.113e-05 |
| 65 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 104 | 1492 | 1.579e-07 | 1.13e-05 |
| 66 | REGULATION OF MAPK CASCADE | 57 | 660 | 1.909e-07 | 1.346e-05 |
| 67 | POSITIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 43 | 437 | 1.962e-07 | 1.362e-05 |
| 68 | BEHAVIOR | 48 | 516 | 2.099e-07 | 1.436e-05 |
| 69 | INOSITOL LIPID MEDIATED SIGNALING | 20 | 124 | 2.132e-07 | 1.438e-05 |
| 70 | BLOOD VESSEL MORPHOGENESIS | 38 | 364 | 2.281e-07 | 1.516e-05 |
| 71 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 84 | 1135 | 2.41e-07 | 1.579e-05 |
| 72 | POSITIVE REGULATION OF CELL DEVELOPMENT | 45 | 472 | 2.526e-07 | 1.632e-05 |
| 73 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 66 | 823 | 3.161e-07 | 2.015e-05 |
| 74 | NEGATIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 49 | 541 | 3.493e-07 | 2.167e-05 |
| 75 | NEGATIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 49 | 541 | 3.493e-07 | 2.167e-05 |
| 76 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 104 | 1518 | 3.631e-07 | 2.223e-05 |
| 77 | SIGNAL TRANSDUCTION BY PROTEIN PHOSPHORYLATION | 40 | 404 | 4.453e-07 | 2.691e-05 |
| 78 | VASCULATURE DEVELOPMENT | 44 | 469 | 5.354e-07 | 3.194e-05 |
| 79 | REGULATION OF NEURON PROJECTION DEVELOPMENT | 40 | 408 | 5.745e-07 | 3.384e-05 |
| 80 | CELL PART MORPHOGENESIS | 54 | 633 | 5.927e-07 | 3.447e-05 |
| 81 | NEGATIVE REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 19 | 121 | 6.541e-07 | 3.757e-05 |
| 82 | EMBRYO DEVELOPMENT | 69 | 894 | 6.915e-07 | 3.924e-05 |
| 83 | NEURON DEVELOPMENT | 57 | 687 | 7.065e-07 | 3.961e-05 |
| 84 | WNT SIGNALING PATHWAY | 36 | 351 | 7.246e-07 | 4.014e-05 |
| 85 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 124 | 1929 | 7.431e-07 | 4.039e-05 |
| 86 | REGULATION OF ADHERENS JUNCTION ORGANIZATION | 12 | 50 | 7.465e-07 | 4.039e-05 |
| 87 | NEGATIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 60 | 740 | 7.713e-07 | 4.125e-05 |
| 88 | PLATELET DERIVED GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 10 | 34 | 8.203e-07 | 4.338e-05 |
| 89 | REGULATION OF GLUCOSE IMPORT | 13 | 60 | 9.178e-07 | 4.799e-05 |
| 90 | NEGATIVE REGULATION OF CELL DIFFERENTIATION | 52 | 609 | 9.35e-07 | 4.834e-05 |
| 91 | NEURON PROJECTION MORPHOGENESIS | 39 | 402 | 1.033e-06 | 5.283e-05 |
| 92 | CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND | 63 | 799 | 1.052e-06 | 5.323e-05 |
| 93 | NEGATIVE REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 17 | 102 | 1.07e-06 | 5.355e-05 |
| 94 | PROTEIN UBIQUITINATION | 53 | 629 | 1.098e-06 | 5.437e-05 |
| 95 | REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 20 | 138 | 1.24e-06 | 6.073e-05 |
| 96 | REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 27 | 229 | 1.262e-06 | 6.116e-05 |
| 97 | NEGATIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 52 | 616 | 1.315e-06 | 6.308e-05 |
| 98 | NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 73 | 983 | 1.371e-06 | 6.512e-05 |
| 99 | CELLULAR RESPONSE TO HORMONE STIMULUS | 48 | 552 | 1.478e-06 | 6.875e-05 |
| 100 | TUBE DEVELOPMENT | 48 | 552 | 1.478e-06 | 6.875e-05 |
| 101 | POSITIVE REGULATION OF CELL PROJECTION ORGANIZATION | 32 | 303 | 1.584e-06 | 7.238e-05 |
| 102 | REGULATION OF OSSIFICATION | 23 | 178 | 1.587e-06 | 7.238e-05 |
| 103 | SENSORY ORGAN DEVELOPMENT | 44 | 493 | 2.05e-06 | 9.259e-05 |
| 104 | NEGATIVE REGULATION OF PROTEIN METABOLIC PROCESS | 78 | 1087 | 2.184e-06 | 9.772e-05 |
| 105 | NEGATIVE REGULATION OF CELL DEATH | 66 | 872 | 2.377e-06 | 0.0001053 |
| 106 | PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL | 66 | 873 | 2.47e-06 | 0.0001084 |
| 107 | CYTOSKELETON ORGANIZATION | 64 | 838 | 2.524e-06 | 0.0001098 |
| 108 | LOCOMOTION | 79 | 1114 | 2.925e-06 | 0.000126 |
| 109 | REGULATION OF CYTOSKELETON ORGANIZATION | 44 | 502 | 3.29e-06 | 0.0001392 |
| 110 | REGULATION OF BMP SIGNALING PATHWAY | 14 | 77 | 3.276e-06 | 0.0001392 |
| 111 | REGULATION OF TRANSFORMING GROWTH FACTOR BETA RECEPTOR SIGNALING PATHWAY | 16 | 99 | 3.353e-06 | 0.0001393 |
| 112 | REGULATION OF CELLULAR RESPONSE TO TRANSFORMING GROWTH FACTOR BETA STIMULUS | 16 | 99 | 3.353e-06 | 0.0001393 |
| 113 | REGULATION OF KINASE ACTIVITY | 60 | 776 | 3.563e-06 | 0.0001455 |
| 114 | REGULATION OF EPITHELIAL CELL PROLIFERATION | 30 | 285 | 3.565e-06 | 0.0001455 |
| 115 | REGULATION OF ORGAN MORPHOGENESIS | 27 | 242 | 3.657e-06 | 0.000148 |
| 116 | EMBRYONIC MORPHOGENESIS | 46 | 539 | 4.032e-06 | 0.0001617 |
| 117 | REGULATION OF CELL JUNCTION ASSEMBLY | 13 | 68 | 4.09e-06 | 0.0001627 |
| 118 | REGULATION OF CELL MATRIX ADHESION | 15 | 90 | 4.562e-06 | 0.0001794 |
| 119 | REGULATION OF STEM CELL DIFFERENTIATION | 17 | 113 | 4.587e-06 | 0.0001794 |
| 120 | POSITIVE REGULATION OF NEURON DIFFERENTIATION | 31 | 306 | 5.446e-06 | 0.0002112 |
| 121 | CELL MORPHOGENESIS INVOLVED IN NEURON DIFFERENTIATION | 35 | 368 | 5.708e-06 | 0.0002195 |
| 122 | TUBE MORPHOGENESIS | 32 | 323 | 6.16e-06 | 0.0002349 |
| 123 | ACTIVATION OF PROTEIN KINASE ACTIVITY | 29 | 279 | 6.606e-06 | 0.0002499 |
| 124 | TISSUE MORPHOGENESIS | 45 | 533 | 6.775e-06 | 0.0002528 |
| 125 | POSITIVE REGULATION OF GLUCOSE TRANSPORT | 10 | 42 | 6.792e-06 | 0.0002528 |
| 126 | PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS | 122 | 1977 | 6.985e-06 | 0.000258 |
| 127 | REGULATION OF WNT SIGNALING PATHWAY | 31 | 310 | 7.096e-06 | 0.00026 |
| 128 | NEGATIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 39 | 437 | 7.766e-06 | 0.0002823 |
| 129 | SKELETAL SYSTEM DEVELOPMENT | 40 | 455 | 8.519e-06 | 0.0003073 |
| 130 | EPITHELIUM DEVELOPMENT | 68 | 945 | 9.192e-06 | 0.000329 |
| 131 | REGULATION OF TRANSFERASE ACTIVITY | 68 | 946 | 9.517e-06 | 0.000338 |
| 132 | REGULATION OF CELL PROLIFERATION | 97 | 1496 | 9.785e-06 | 0.0003449 |
| 133 | HIPPO SIGNALING | 8 | 27 | 9.939e-06 | 0.0003477 |
| 134 | NEGATIVE REGULATION OF CELL JUNCTION ASSEMBLY | 7 | 20 | 1.054e-05 | 0.0003658 |
| 135 | REGULATION OF DEVELOPMENTAL GROWTH | 29 | 289 | 1.302e-05 | 0.0004486 |
| 136 | MORPHOGENESIS OF AN EPITHELIUM | 36 | 400 | 1.436e-05 | 0.0004876 |
| 137 | REGULATION OF POSTSYNAPTIC MEMBRANE POTENTIAL | 11 | 55 | 1.43e-05 | 0.0004876 |
| 138 | REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 32 | 337 | 1.469e-05 | 0.0004918 |
| 139 | MULTICELLULAR ORGANISMAL SIGNALING | 17 | 123 | 1.461e-05 | 0.0004918 |
| 140 | REGULATION OF SYNAPSE STRUCTURE OR ACTIVITY | 25 | 232 | 1.52e-05 | 0.000498 |
| 141 | TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 55 | 724 | 1.509e-05 | 0.000498 |
| 142 | LIPID PHOSPHORYLATION | 15 | 99 | 1.512e-05 | 0.000498 |
| 143 | POSITIVE REGULATION OF HYDROLASE ACTIVITY | 65 | 905 | 1.543e-05 | 0.000502 |
| 144 | OVULATION CYCLE PROCESS | 14 | 88 | 1.638e-05 | 0.0005292 |
| 145 | REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 40 | 470 | 1.817e-05 | 0.0005829 |
| 146 | REGULATION OF SYNAPSE ORGANIZATION | 16 | 113 | 1.905e-05 | 0.0006071 |
| 147 | EYE DEVELOPMENT | 31 | 326 | 1.934e-05 | 0.0006123 |
| 148 | CELLULAR RESPONSE TO NITROGEN COMPOUND | 42 | 505 | 1.956e-05 | 0.000615 |
| 149 | NEGATIVE REGULATION OF CELL PROLIFERATION | 50 | 643 | 2.005e-05 | 0.0006261 |
| 150 | REGULATION OF MEMBRANE POTENTIAL | 32 | 343 | 2.092e-05 | 0.000649 |
| 151 | REGULATION OF EPITHELIAL CELL MIGRATION | 20 | 166 | 2.138e-05 | 0.0006589 |
| 152 | RHYTHMIC PROCESS | 29 | 298 | 2.319e-05 | 0.0007099 |
| 153 | AMEBOIDAL TYPE CELL MIGRATION | 19 | 154 | 2.441e-05 | 0.0007414 |
| 154 | POSITIVE REGULATION OF EPITHELIAL CELL MIGRATION | 15 | 103 | 2.454e-05 | 0.0007414 |
| 155 | MEMBRANE DEPOLARIZATION DURING ACTION POTENTIAL | 9 | 39 | 2.578e-05 | 0.0007738 |
| 156 | APPENDAGE DEVELOPMENT | 20 | 169 | 2.78e-05 | 0.0008239 |
| 157 | LIMB DEVELOPMENT | 20 | 169 | 2.78e-05 | 0.0008239 |
| 158 | BONE DEVELOPMENT | 19 | 156 | 2.929e-05 | 0.0008624 |
| 159 | POSITIVE REGULATION OF KINASE ACTIVITY | 40 | 482 | 3.224e-05 | 0.0009436 |
| 160 | ACTION POTENTIAL | 14 | 94 | 3.519e-05 | 0.001023 |
| 161 | SYNAPSE ORGANIZATION | 18 | 145 | 3.648e-05 | 0.001054 |
| 162 | POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 62 | 876 | 3.753e-05 | 0.001078 |
| 163 | REGULATION OF CELL SUBSTRATE ADHESION | 20 | 173 | 3.902e-05 | 0.001114 |
| 164 | POSITIVE REGULATION OF LOCOMOTION | 36 | 420 | 4.061e-05 | 0.001145 |
| 165 | ENDOTHELIAL CELL DIFFERENTIATION | 12 | 72 | 4.057e-05 | 0.001145 |
| 166 | POSITIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY VIA DEATH DOMAIN RECEPTORS | 6 | 17 | 4.392e-05 | 0.001219 |
| 167 | POSITIVE REGULATION OF NEURON PROJECTION DEVELOPMENT | 24 | 232 | 4.356e-05 | 0.001219 |
| 168 | POSITIVE REGULATION OF OSSIFICATION | 13 | 84 | 4.403e-05 | 0.001219 |
| 169 | PROTEIN UBIQUITINATION INVOLVED IN UBIQUITIN DEPENDENT PROTEIN CATABOLIC PROCESS | 17 | 134 | 4.507e-05 | 0.001241 |
| 170 | POSITIVE REGULATION OF CELLULAR COMPONENT BIOGENESIS | 35 | 406 | 4.604e-05 | 0.001258 |
| 171 | REGULATION OF SYSTEM PROCESS | 41 | 507 | 4.646e-05 | 0.001258 |
| 172 | POSITIVE REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 11 | 62 | 4.651e-05 | 0.001258 |
| 173 | NEGATIVE REGULATION OF BMP SIGNALING PATHWAY | 9 | 42 | 4.846e-05 | 0.001303 |
| 174 | ADULT BEHAVIOR | 17 | 135 | 4.959e-05 | 0.001315 |
| 175 | PALATE DEVELOPMENT | 13 | 85 | 5.002e-05 | 0.001315 |
| 176 | MESENCHYME DEVELOPMENT | 21 | 190 | 4.93e-05 | 0.001315 |
| 177 | NEGATIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 51 | 684 | 4.996e-05 | 0.001315 |
| 178 | NEGATIVE REGULATION OF TRANSPORT | 38 | 458 | 5.071e-05 | 0.001325 |
| 179 | REGULATION OF TRANSPORT | 109 | 1804 | 5.526e-05 | 0.001437 |
| 180 | PROTEIN LOCALIZATION | 109 | 1805 | 5.652e-05 | 0.001461 |
| 181 | REGULATION OF OSTEOBLAST DIFFERENTIATION | 15 | 112 | 6.655e-05 | 0.001711 |
| 182 | TRANSMISSION OF NERVE IMPULSE | 10 | 54 | 7.012e-05 | 0.001793 |
| 183 | REGULATION OF GLUCOSE TRANSPORT | 14 | 100 | 7.076e-05 | 0.001793 |
| 184 | MODULATION OF SYNAPTIC TRANSMISSION | 28 | 301 | 7.092e-05 | 0.001793 |
| 185 | PEPTIDYL AMINO ACID MODIFICATION | 59 | 841 | 7.388e-05 | 0.001858 |
| 186 | REGULATION OF MAP KINASE ACTIVITY | 29 | 319 | 7.996e-05 | 0.002 |
| 187 | SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 31 | 352 | 8.317e-05 | 0.00207 |
| 188 | NEGATIVE REGULATION OF CELLULAR RESPONSE TO TRANSFORMING GROWTH FACTOR BETA STIMULUS | 11 | 66 | 8.441e-05 | 0.002078 |
| 189 | NEGATIVE REGULATION OF TRANSFORMING GROWTH FACTOR BETA RECEPTOR SIGNALING PATHWAY | 11 | 66 | 8.441e-05 | 0.002078 |
| 190 | HEART MORPHOGENESIS | 22 | 212 | 8.534e-05 | 0.00209 |
| 191 | POSITIVE REGULATION OF MAPK CASCADE | 38 | 470 | 8.757e-05 | 0.002133 |
| 192 | TELENCEPHALON DEVELOPMENT | 23 | 228 | 9.178e-05 | 0.002224 |
| 193 | POSITIVE REGULATION OF ENDOTHELIAL CELL MIGRATION | 11 | 67 | 9.722e-05 | 0.002308 |
| 194 | REGULATION OF DENDRITIC SPINE DEVELOPMENT | 10 | 56 | 9.674e-05 | 0.002308 |
| 195 | REGULATION OF EPITHELIAL TO MESENCHYMAL TRANSITION | 11 | 67 | 9.722e-05 | 0.002308 |
| 196 | HEAD MORPHOGENESIS | 8 | 36 | 9.698e-05 | 0.002308 |
| 197 | NEGATIVE REGULATION OF MOLECULAR FUNCTION | 71 | 1079 | 0.0001044 | 0.002465 |
| 198 | CELLULAR RESPONSE TO LIPID | 37 | 457 | 0.000105 | 0.002468 |
| 199 | FOREBRAIN DEVELOPMENT | 31 | 357 | 0.0001077 | 0.002519 |
| 200 | PEPTIDYL TYROSINE MODIFICATION | 20 | 186 | 0.0001084 | 0.002523 |
| 201 | RESPONSE TO TRANSFORMING GROWTH FACTOR BETA | 17 | 144 | 0.000112 | 0.002552 |
| 202 | POSITIVE REGULATION OF TRANSFERASE ACTIVITY | 46 | 616 | 0.0001118 | 0.002552 |
| 203 | ORGAN GROWTH | 11 | 68 | 0.0001117 | 0.002552 |
| 204 | ANGIOGENESIS | 27 | 293 | 0.0001111 | 0.002552 |
| 205 | DEVELOPMENT OF PRIMARY SEXUAL CHARACTERISTICS | 22 | 216 | 0.0001124 | 0.002552 |
| 206 | NEGATIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 25 | 262 | 0.0001138 | 0.002565 |
| 207 | REPRODUCTIVE SYSTEM DEVELOPMENT | 34 | 408 | 0.0001141 | 0.002565 |
| 208 | NEURONAL ACTION POTENTIAL | 7 | 28 | 0.00012 | 0.002685 |
| 209 | REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION | 15 | 118 | 0.0001216 | 0.002694 |
| 210 | CEREBRAL CORTEX DEVELOPMENT | 14 | 105 | 0.0001211 | 0.002694 |
| 211 | RESPONSE TO OXYGEN CONTAINING COMPOUND | 86 | 1381 | 0.0001325 | 0.002922 |
| 212 | POSITIVE REGULATION OF CATABOLIC PROCESS | 33 | 395 | 0.0001365 | 0.002996 |
| 213 | SEX DIFFERENTIATION | 25 | 266 | 0.0001446 | 0.003159 |
| 214 | REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 22 | 220 | 0.0001469 | 0.003185 |
| 215 | NEURON PROJECTION GUIDANCE | 21 | 205 | 0.0001472 | 0.003185 |
| 216 | REGULATION OF CANONICAL WNT SIGNALING PATHWAY | 23 | 236 | 0.0001542 | 0.003321 |
| 217 | MICROTUBULE CYTOSKELETON ORGANIZATION | 30 | 348 | 0.0001571 | 0.003369 |
| 218 | MUSCLE STRUCTURE DEVELOPMENT | 35 | 432 | 0.0001578 | 0.003369 |
| 219 | SYMPATHETIC NERVOUS SYSTEM DEVELOPMENT | 6 | 21 | 0.0001668 | 0.003544 |
| 220 | PHOSPHATIDYLINOSITOL 3 PHOSPHATE BIOSYNTHETIC PROCESS | 9 | 49 | 0.0001718 | 0.003633 |
| 221 | CELLULAR MACROMOLECULE LOCALIZATION | 78 | 1234 | 0.0001733 | 0.003648 |
| 222 | SECRETION BY CELL | 38 | 486 | 0.0001741 | 0.003649 |
| 223 | REGULATION OF PATHWAY RESTRICTED SMAD PROTEIN PHOSPHORYLATION | 10 | 60 | 0.0001761 | 0.003659 |
| 224 | POSITIVE REGULATION OF OSTEOBLAST DIFFERENTIATION | 10 | 60 | 0.0001761 | 0.003659 |
| 225 | PATTERN SPECIFICATION PROCESS | 34 | 418 | 0.0001809 | 0.003741 |
| 226 | NEGATIVE REGULATION OF CELL MATRIX ADHESION | 7 | 30 | 0.0001918 | 0.00395 |
| 227 | NEGATIVE REGULATION OF CELL DEVELOPMENT | 27 | 303 | 0.0001931 | 0.003959 |
| 228 | REGULATION OF SYNAPTIC TRANSMISSION GLUTAMATERGIC | 9 | 50 | 0.0002018 | 0.004101 |
| 229 | FACE DEVELOPMENT | 9 | 50 | 0.0002018 | 0.004101 |
| 230 | MEMBRANE DEPOLARIZATION | 10 | 61 | 0.0002029 | 0.004105 |
| 231 | REGULATION OF GROWTH | 46 | 633 | 0.0002082 | 0.004194 |
| 232 | MORPHOGENESIS OF A BRANCHING STRUCTURE | 18 | 167 | 0.0002284 | 0.004581 |
| 233 | REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 17 | 153 | 0.0002349 | 0.004672 |
| 234 | PALLIUM DEVELOPMENT | 17 | 153 | 0.0002349 | 0.004672 |
| 235 | REGULATION OF MICROTUBULE BASED PROCESS | 23 | 243 | 0.000237 | 0.004692 |
| 236 | REGULATION OF TRANSPORTER ACTIVITY | 20 | 198 | 0.0002529 | 0.004987 |
| 237 | OVULATION CYCLE | 14 | 113 | 0.0002661 | 0.005224 |
| 238 | SERTOLI CELL DEVELOPMENT | 5 | 15 | 0.0002682 | 0.005244 |
| 239 | POSITIVE REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 13 | 100 | 0.0002706 | 0.005269 |
| 240 | REGULATION OF BIOMINERAL TISSUE DEVELOPMENT | 11 | 75 | 0.0002735 | 0.005281 |
| 241 | NEURAL CREST CELL DIFFERENTIATION | 11 | 75 | 0.0002735 | 0.005281 |
| 242 | REGULATION OF ENDOTHELIAL CELL MIGRATION | 14 | 114 | 0.0002919 | 0.005612 |
| 243 | DENDRITE MORPHOGENESIS | 8 | 42 | 0.000303 | 0.005778 |
| 244 | POSITIVE REGULATION OF LIPID METABOLIC PROCESS | 15 | 128 | 0.0003018 | 0.005778 |
| 245 | REGULATION OF ORGANELLE ORGANIZATION | 74 | 1178 | 0.0003059 | 0.005809 |
| 246 | CELLULAR RESPONSE TO ORGANIC CYCLIC COMPOUND | 36 | 465 | 0.0003086 | 0.005813 |
| 247 | SKELETAL SYSTEM MORPHOGENESIS | 20 | 201 | 0.0003086 | 0.005813 |
| 248 | NEGATIVE REGULATION OF CELL SUBSTRATE ADHESION | 9 | 53 | 0.0003191 | 0.005988 |
| 249 | REGULATION OF PROTEIN COMPLEX DISASSEMBLY | 21 | 217 | 0.0003226 | 0.006028 |
| 250 | RAS PROTEIN SIGNAL TRANSDUCTION | 16 | 143 | 0.0003277 | 0.0061 |
| 251 | FEMALE SEX DIFFERENTIATION | 14 | 116 | 0.0003499 | 0.006487 |
| 252 | MICROTUBULE BASED PROCESS | 39 | 522 | 0.0003605 | 0.006639 |
| 253 | RESPONSE TO HORMONE | 59 | 893 | 0.000361 | 0.006639 |
| 254 | ENDOTHELIUM DEVELOPMENT | 12 | 90 | 0.0003636 | 0.006661 |
| 255 | COGNITION | 23 | 251 | 0.0003776 | 0.006863 |
| 256 | NEGATIVE REGULATION OF ADHERENS JUNCTION ORGANIZATION | 5 | 16 | 0.0003767 | 0.006863 |
| 257 | BODY MORPHOGENESIS | 8 | 44 | 0.0004226 | 0.007621 |
| 258 | MAINTENANCE OF CELL NUMBER | 15 | 132 | 0.0004215 | 0.007621 |
| 259 | REGULATION OF SYNAPSE ASSEMBLY | 11 | 79 | 0.0004337 | 0.007761 |
| 260 | BONE MORPHOGENESIS | 11 | 79 | 0.0004337 | 0.007761 |
| 261 | POSITIVE REGULATION OF VASCULATURE DEVELOPMENT | 15 | 133 | 0.0004571 | 0.008149 |
| 262 | DEVELOPMENTAL PROCESS INVOLVED IN REPRODUCTION | 43 | 602 | 0.000475 | 0.008437 |
| 263 | SMAD PROTEIN SIGNAL TRANSDUCTION | 9 | 56 | 0.0004878 | 0.008598 |
| 264 | OUTFLOW TRACT MORPHOGENESIS | 9 | 56 | 0.0004878 | 0.008598 |
| 265 | PHOSPHATIDYLINOSITOL METABOLIC PROCESS | 19 | 193 | 0.0004933 | 0.008648 |
| 266 | MESENCHYMAL CELL DIFFERENTIATION | 15 | 134 | 0.0004953 | 0.008648 |
| 267 | REGULATION OF DENDRITE DEVELOPMENT | 14 | 120 | 0.0004962 | 0.008648 |
| 268 | EPITHELIAL CELL DIFFERENTIATION | 37 | 495 | 0.0005009 | 0.008696 |
| 269 | REGULATION OF ENERGY HOMEOSTASIS | 5 | 17 | 0.0005154 | 0.008883 |
| 270 | REGULATION OF PROTEIN KINASE A SIGNALING | 5 | 17 | 0.0005154 | 0.008883 |
| 271 | POSITIVE REGULATION OF CELL DEATH | 43 | 605 | 0.0005261 | 0.009033 |
| 272 | POSITIVE REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 14 | 121 | 0.00054 | 0.009238 |
| 273 | LIPID MODIFICATION | 20 | 210 | 0.0005443 | 0.009278 |
| 274 | MUSCLE TISSUE DEVELOPMENT | 24 | 275 | 0.0005781 | 0.009818 |
| 275 | REGULATION OF CELL CYCLE | 61 | 949 | 0.0005825 | 0.009856 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | NUCLEIC ACID BINDING TRANSCRIPTION FACTOR ACTIVITY | 101 | 1199 | 1.057e-11 | 9.824e-09 |
| 2 | ENZYME BINDING | 124 | 1737 | 1.996e-09 | 9.273e-07 |
| 3 | RNA POLYMERASE II TRANSCRIPTION FACTOR ACTIVITY SEQUENCE SPECIFIC DNA BINDING | 59 | 629 | 6.1e-09 | 1.889e-06 |
| 4 | PROTEIN KINASE ACTIVITY | 59 | 640 | 1.15e-08 | 2.672e-06 |
| 5 | TRANSMEMBRANE RECEPTOR PROTEIN KINASE ACTIVITY | 17 | 81 | 3.266e-08 | 6.068e-06 |
| 6 | UBIQUITIN LIKE PROTEIN TRANSFERASE ACTIVITY | 43 | 420 | 6.419e-08 | 8.518e-06 |
| 7 | MOLECULAR FUNCTION REGULATOR | 98 | 1353 | 6.176e-08 | 8.518e-06 |
| 8 | TRANSCRIPTION FACTOR ACTIVITY RNA POLYMERASE II CORE PROMOTER PROXIMAL REGION SEQUENCE SPECIFIC BINDING | 36 | 328 | 1.381e-07 | 1.603e-05 |
| 9 | REGULATORY REGION NUCLEIC ACID BINDING | 66 | 818 | 2.537e-07 | 2.619e-05 |
| 10 | MACROMOLECULAR COMPLEX BINDING | 97 | 1399 | 5.508e-07 | 5.117e-05 |
| 11 | PROTEIN TYROSINE KINASE ACTIVITY | 23 | 176 | 1.301e-06 | 9.695e-05 |
| 12 | RAS GUANYL NUCLEOTIDE EXCHANGE FACTOR ACTIVITY | 27 | 228 | 1.158e-06 | 9.695e-05 |
| 13 | SEQUENCE SPECIFIC DNA BINDING | 76 | 1037 | 1.357e-06 | 9.695e-05 |
| 14 | KINASE ACTIVITY | 65 | 842 | 1.462e-06 | 9.701e-05 |
| 15 | ZINC ION BINDING | 82 | 1155 | 1.793e-06 | 0.0001111 |
| 16 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE ACTIVITY | 13 | 64 | 1.999e-06 | 0.0001161 |
| 17 | GROWTH FACTOR BINDING | 18 | 123 | 3.63e-06 | 0.0001984 |
| 18 | CYTOKINE BINDING | 15 | 92 | 6.038e-06 | 0.0002952 |
| 19 | PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 13 | 70 | 5.729e-06 | 0.0002952 |
| 20 | TRANSFERASE ACTIVITY TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 70 | 992 | 1.299e-05 | 0.0006032 |
| 21 | GTPASE BINDING | 29 | 295 | 1.919e-05 | 0.000849 |
| 22 | TRANSCRIPTIONAL ACTIVATOR ACTIVITY RNA POLYMERASE II CORE PROMOTER PROXIMAL REGION SEQUENCE SPECIFIC BINDING | 24 | 226 | 2.845e-05 | 0.001201 |
| 23 | GUANYL NUCLEOTIDE EXCHANGE FACTOR ACTIVITY | 29 | 303 | 3.156e-05 | 0.001275 |
| 24 | TRANSCRIPTION FACTOR ACTIVITY PROTEIN BINDING | 46 | 588 | 3.697e-05 | 0.001431 |
| 25 | SMAD BINDING | 12 | 72 | 4.057e-05 | 0.001462 |
| 26 | TRANSCRIPTION FACTOR ACTIVITY RNA POLYMERASE II TRANSCRIPTION FACTOR BINDING | 17 | 133 | 4.091e-05 | 0.001462 |
| 27 | TRANSCRIPTION COACTIVATOR BINDING | 5 | 11 | 4.747e-05 | 0.001633 |
| 28 | ENZYME REGULATOR ACTIVITY | 66 | 959 | 4.958e-05 | 0.001645 |
| 29 | PROTEIN SERINE THREONINE KINASE ACTIVITY | 37 | 445 | 6.067e-05 | 0.001944 |
| 30 | X1 PHOSPHATIDYLINOSITOL BINDING | 6 | 19 | 8.96e-05 | 0.002775 |
| 31 | UBIQUITIN LIKE PROTEIN LIGASE ACTIVITY | 21 | 199 | 9.655e-05 | 0.002893 |
| 32 | DOUBLE STRANDED DNA BINDING | 54 | 764 | 0.0001229 | 0.003568 |
| 33 | KINASE BINDING | 45 | 606 | 0.0001503 | 0.004162 |
| 34 | TRANSCRIPTIONAL ACTIVATOR ACTIVITY RNA POLYMERASE II TRANSCRIPTION REGULATORY REGION SEQUENCE SPECIFIC BINDING | 28 | 315 | 0.0001544 | 0.004162 |
| 35 | TRANSCRIPTION COREPRESSOR ACTIVITY | 22 | 221 | 0.0001568 | 0.004162 |
| 36 | PROTEIN COMPLEX BINDING | 62 | 935 | 0.0002332 | 0.005936 |
| 37 | TRANSCRIPTIONAL REPRESSOR ACTIVITY RNA POLYMERASE II TRANSCRIPTION FACTOR BINDING | 12 | 86 | 0.0002364 | 0.005936 |
| 38 | PROTEIN KINASE A BINDING | 8 | 42 | 0.000303 | 0.007166 |
| 39 | INSULIN RECEPTOR BINDING | 7 | 32 | 0.0002949 | 0.007166 |
| 40 | MICROTUBULE BINDING | 20 | 201 | 0.0003086 | 0.007166 |
| 41 | TRANSITION METAL ION BINDING | 85 | 1400 | 0.000331 | 0.007321 |
| 42 | GROWTH FACTOR RECEPTOR BINDING | 15 | 129 | 0.0003286 | 0.007321 |
| 43 | CYTOSKELETAL PROTEIN BINDING | 55 | 819 | 0.0003841 | 0.007503 |
| 44 | RHO GTPASE BINDING | 11 | 78 | 0.0003877 | 0.007503 |
| 45 | X1 PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 8 | 43 | 0.0003587 | 0.007503 |
| 46 | TRANSFORMING GROWTH FACTOR BETA BINDING | 5 | 16 | 0.0003767 | 0.007503 |
| 47 | TRANSCRIPTION COFACTOR BINDING | 6 | 24 | 0.0003715 | 0.007503 |
| 48 | CHROMATIN BINDING | 34 | 435 | 0.0003776 | 0.007503 |
| 49 | CORE PROMOTER PROXIMAL REGION DNA BINDING | 30 | 371 | 0.0004679 | 0.008871 |
| 50 | RAB GTPASE BINDING | 14 | 120 | 0.0004962 | 0.00922 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | SYNAPSE | 61 | 754 | 6.704e-07 | 0.0003302 |
| 2 | NEURON PROJECTION | 71 | 942 | 1.131e-06 | 0.0003302 |
| 3 | CELL PROJECTION | 115 | 1786 | 1.845e-06 | 0.0003592 |
| 4 | NEURON PART | 87 | 1265 | 3.085e-06 | 0.0004504 |
| 5 | SOMATODENDRITIC COMPARTMENT | 51 | 650 | 1.301e-05 | 0.00152 |
| 6 | POSTSYNAPSE | 34 | 378 | 2.497e-05 | 0.002083 |
| 7 | SYNAPSE PART | 48 | 610 | 2.167e-05 | 0.002083 |
| 8 | MICROTUBULE ORGANIZING CENTER | 48 | 623 | 3.696e-05 | 0.002698 |
| 9 | PML BODY | 14 | 97 | 5.028e-05 | 0.003263 |
| 10 | CELL PROJECTION PART | 65 | 946 | 5.933e-05 | 0.003465 |
| 11 | MICROTUBULE CYTOSKELETON | 71 | 1068 | 7.632e-05 | 0.003887 |
| 12 | EXCITATORY SYNAPSE | 21 | 197 | 8.351e-05 | 0.003887 |
| 13 | CYTOSKELETON | 116 | 1967 | 8.652e-05 | 0.003887 |
| 14 | TRANSFERASE COMPLEX | 51 | 703 | 9.977e-05 | 0.004162 |
| 15 | CENTROSOME | 38 | 487 | 0.0001815 | 0.006759 |
| 16 | VOLTAGE GATED SODIUM CHANNEL COMPLEX | 5 | 14 | 0.0001852 | 0.006759 |
| 17 | CELL JUNCTION | 73 | 1151 | 0.000254 | 0.008727 |
| 18 | APICAL JUNCTION COMPLEX | 15 | 128 | 0.0003018 | 0.009791 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
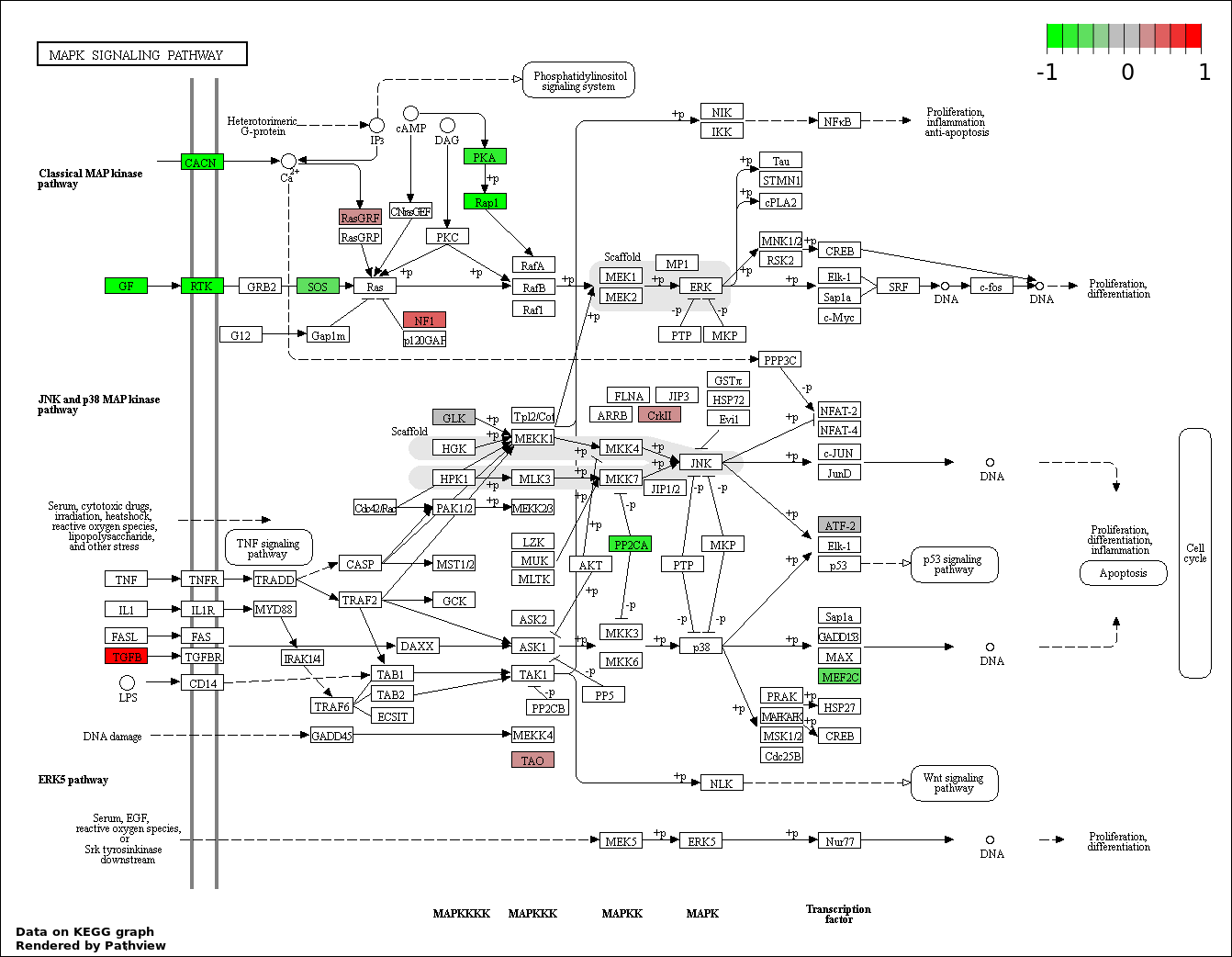
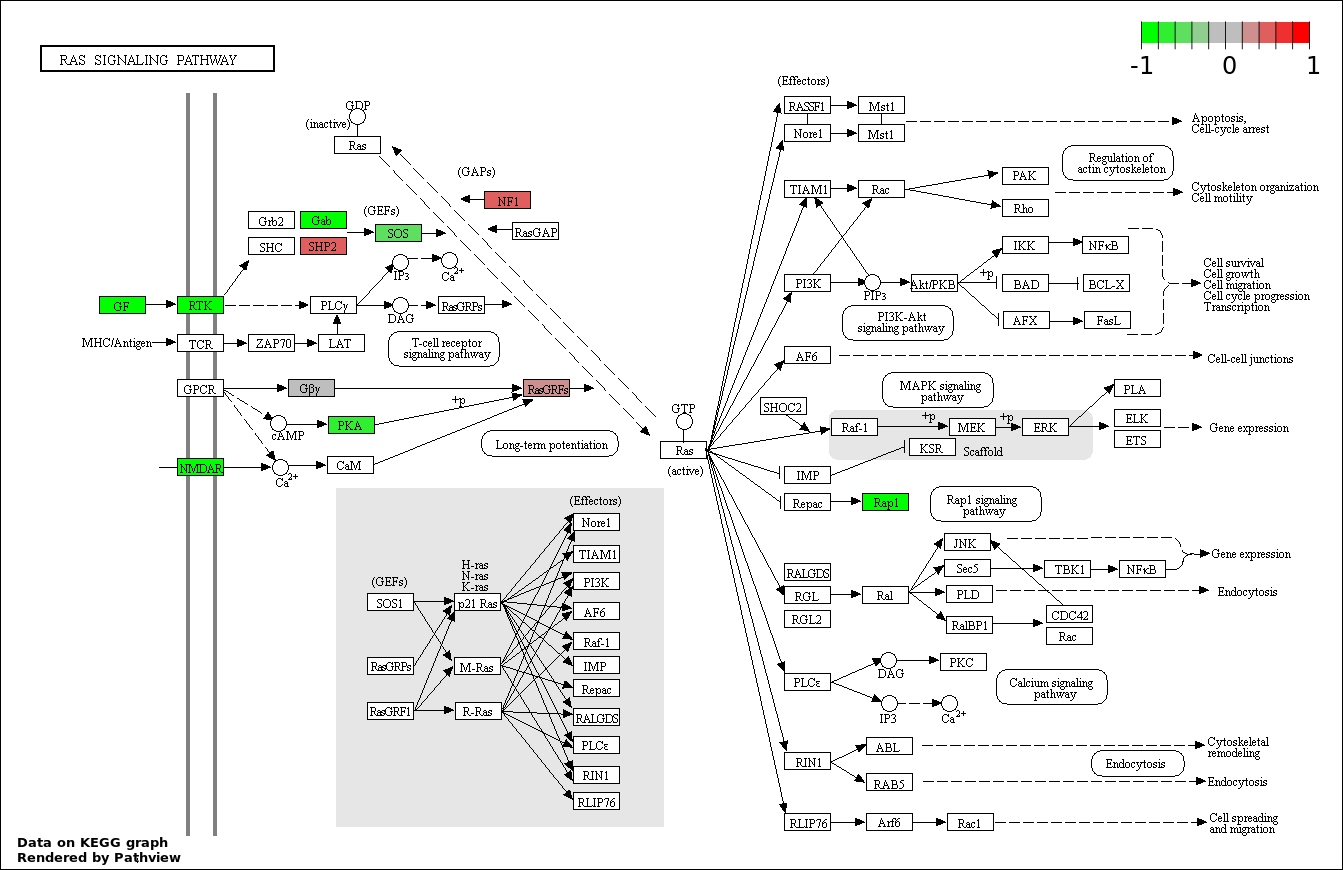
| 1 | hsa04350_TGF.beta_signaling_pathway | 15 | 85 | 2.176e-06 | 0.0003917 | |
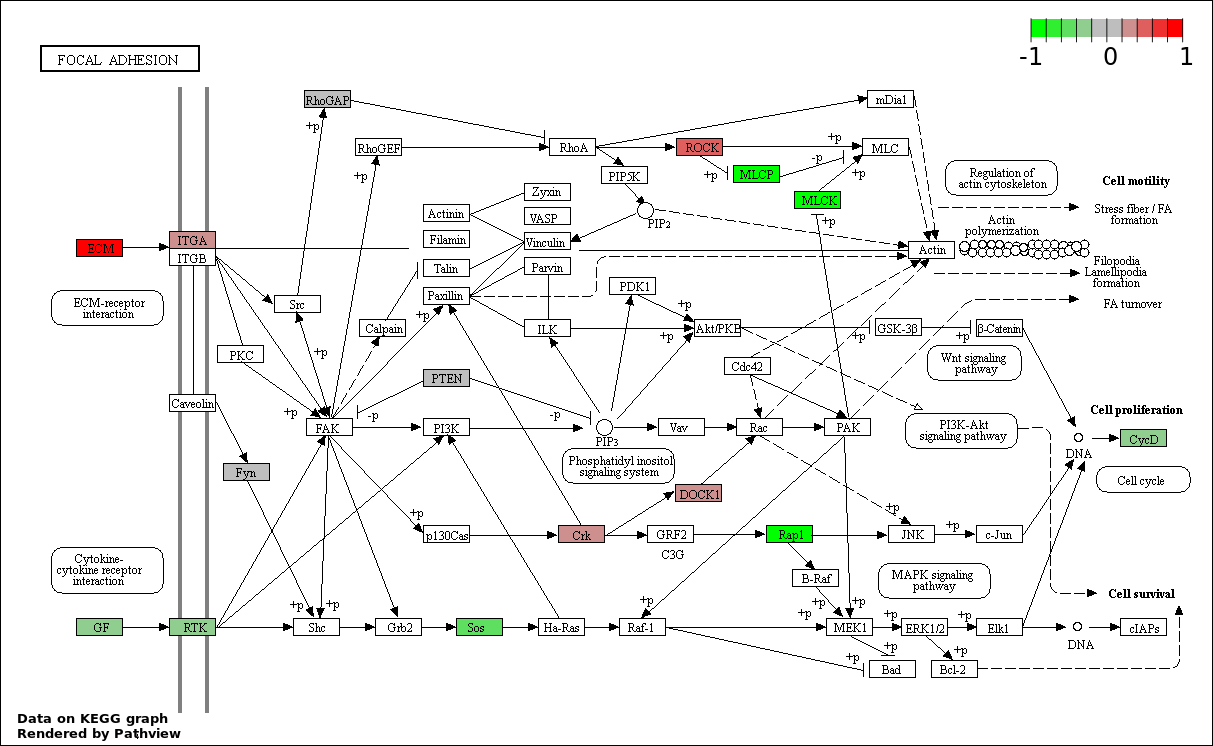
| 2 | hsa04510_Focal_adhesion | 23 | 200 | 1.15e-05 | 0.001035 | |
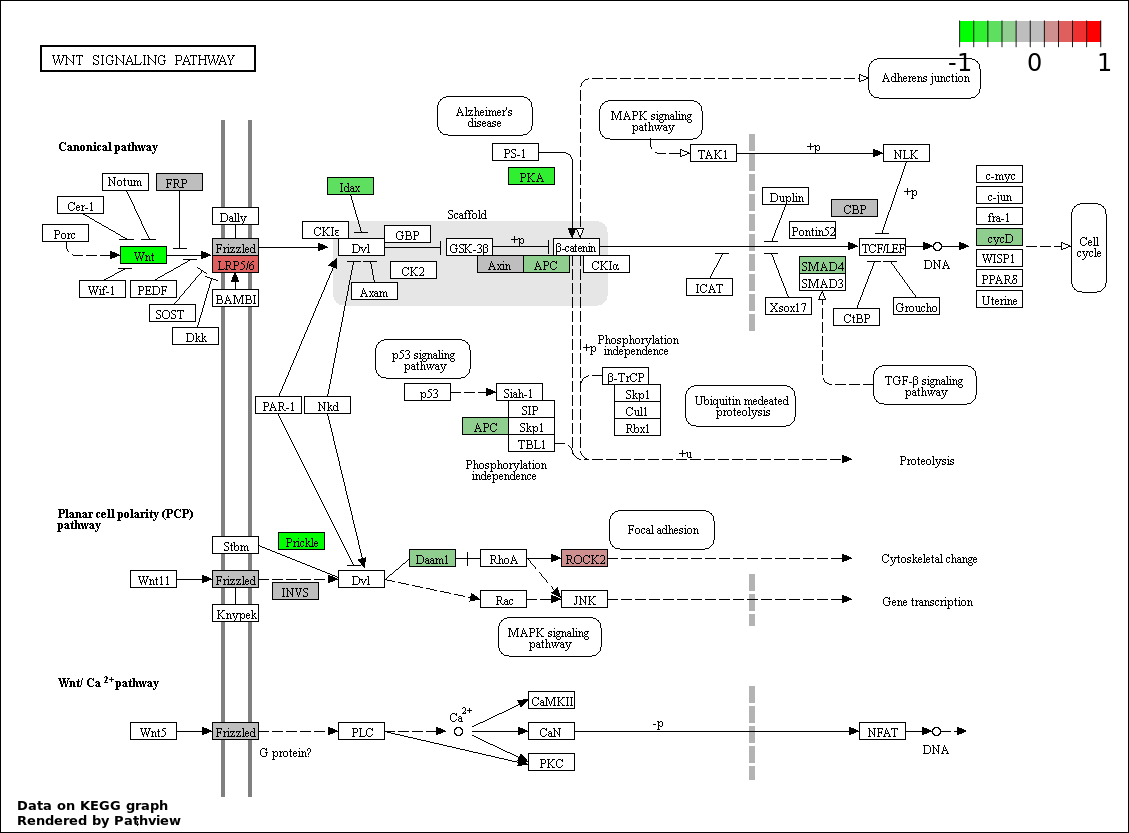
| 3 | hsa04310_Wnt_signaling_pathway | 19 | 151 | 1.844e-05 | 0.001098 | |
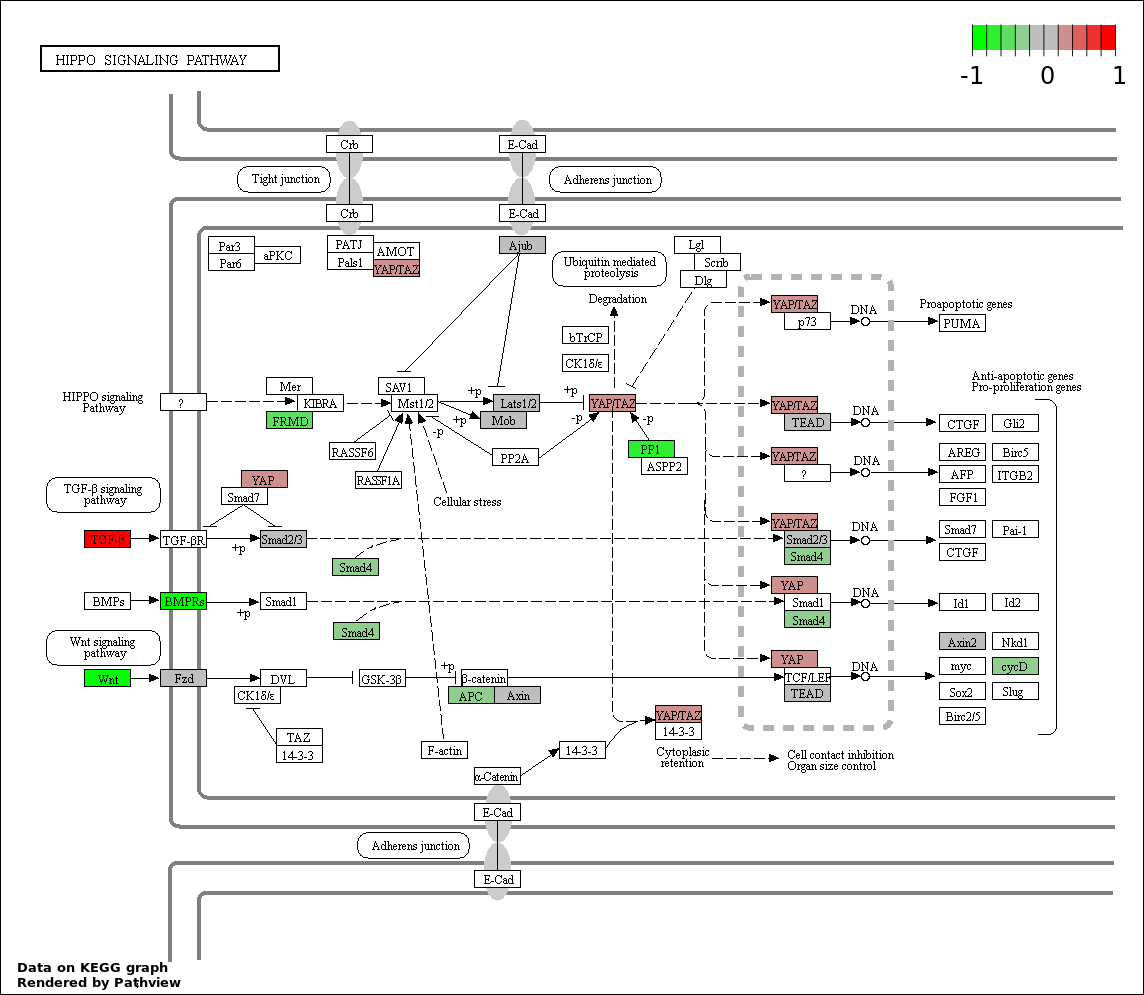
| 4 | hsa04390_Hippo_signaling_pathway | 19 | 154 | 2.441e-05 | 0.001098 | |
| 5 | hsa04810_Regulation_of_actin_cytoskeleton | 20 | 214 | 0.0006915 | 0.02186 | |
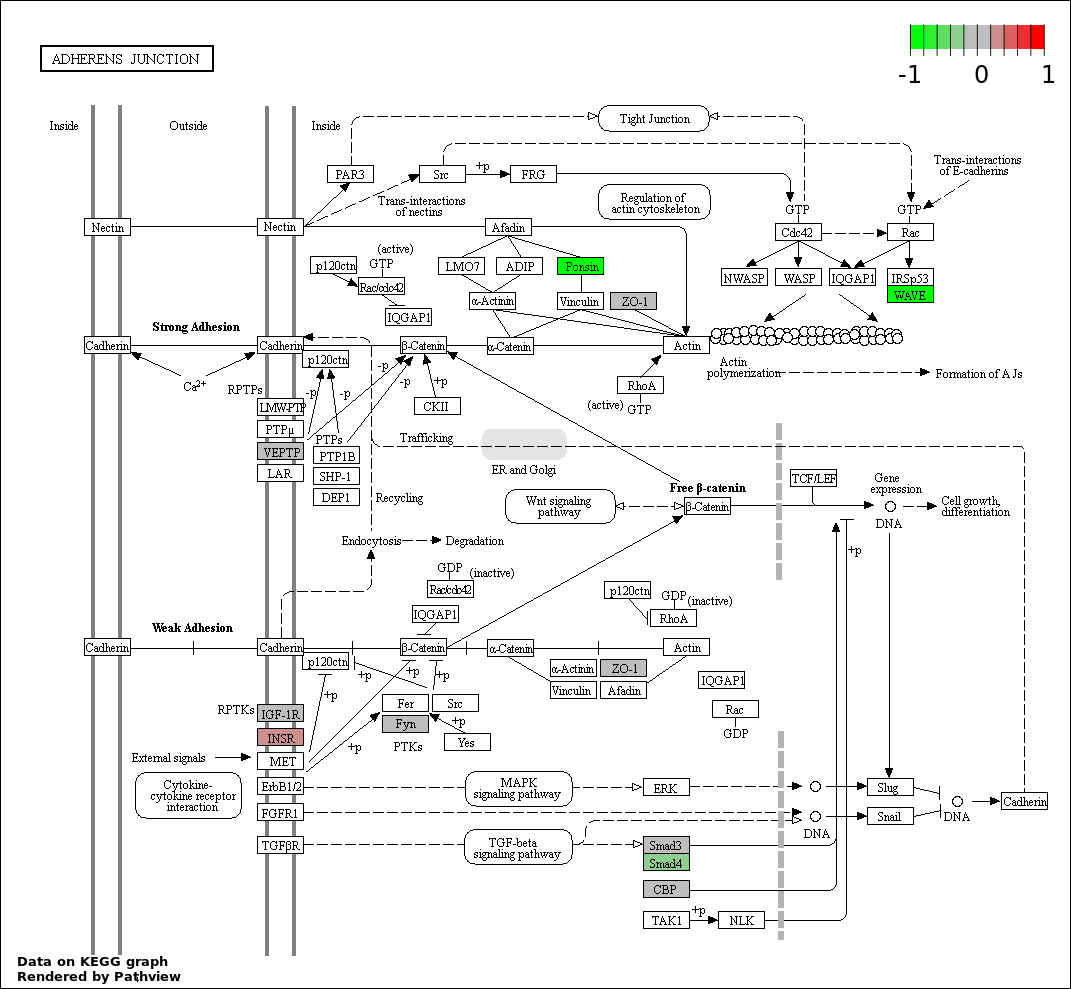
| 6 | hsa04520_Adherens_junction | 10 | 73 | 0.0008904 | 0.02186 | |
| 7 | hsa04010_MAPK_signaling_pathway | 23 | 268 | 0.0009364 | 0.02186 | |
| 8 | hsa04014_Ras_signaling_pathway | 21 | 236 | 0.0009714 | 0.02186 | |
| 9 | hsa04151_PI3K_AKT_signaling_pathway | 27 | 351 | 0.001824 | 0.03527 | |
| 10 | hsa04910_Insulin_signaling_pathway | 14 | 138 | 0.001959 | 0.03527 | |
| 11 | hsa04144_Endocytosis | 17 | 203 | 0.005254 | 0.08597 | |
| 12 | hsa04512_ECM.receptor_interaction | 9 | 85 | 0.009072 | 0.1361 | |
| 13 | hsa04630_Jak.STAT_signaling_pathway | 13 | 155 | 0.01344 | 0.1861 | |
| 14 | hsa04360_Axon_guidance | 11 | 130 | 0.02064 | 0.2654 | |
| 15 | hsa04270_Vascular_smooth_muscle_contraction | 10 | 116 | 0.02371 | 0.2845 | |
| 16 | hsa04720_Long.term_potentiation | 7 | 70 | 0.02681 | 0.3016 | |
| 17 | hsa04120_Ubiquitin_mediated_proteolysis | 11 | 139 | 0.03182 | 0.3369 | |
| 18 | hsa02010_ABC_transporters | 5 | 44 | 0.03582 | 0.3582 | |
| 19 | hsa04140_Regulation_of_autophagy | 4 | 34 | 0.05227 | 0.4952 | |
| 20 | hsa04514_Cell_adhesion_molecules_.CAMs. | 10 | 136 | 0.05958 | 0.5209 | |
| 21 | hsa04920_Adipocytokine_signaling_pathway | 6 | 68 | 0.06468 | 0.5209 | |
| 22 | hsa04150_mTOR_signaling_pathway | 5 | 52 | 0.06555 | 0.5209 | |
| 23 | hsa04115_p53_signaling_pathway | 6 | 69 | 0.06846 | 0.5209 | |
| 24 | hsa04710_Circadian_rhythm_._mammal | 3 | 23 | 0.06945 | 0.5209 | |
| 25 | hsa04340_Hedgehog_signaling_pathway | 5 | 56 | 0.08434 | 0.6073 | |
| 26 | hsa04916_Melanogenesis | 7 | 101 | 0.1307 | 0.9046 | |
| 27 | hsa04012_ErbB_signaling_pathway | 6 | 87 | 0.157 | 1 | |
| 28 | hsa04722_Neurotrophin_signaling_pathway | 8 | 127 | 0.1634 | 1 | |
| 29 | hsa04110_Cell_cycle | 8 | 128 | 0.1683 | 1 | |
| 30 | hsa00670_One_carbon_pool_by_folate | 2 | 18 | 0.1725 | 1 | |
| 31 | hsa00531_Glycosaminoglycan_degradation | 2 | 19 | 0.1878 | 1 | |
| 32 | hsa04260_Cardiac_muscle_contraction | 5 | 77 | 0.2201 | 1 | |
| 33 | hsa04974_Protein_digestion_and_absorption | 5 | 81 | 0.2512 | 1 | |
| 34 | hsa04977_Vitamin_digestion_and_absorption | 2 | 24 | 0.2658 | 1 | |
| 35 | hsa03015_mRNA_surveillance_pathway | 5 | 83 | 0.2672 | 1 | |
| 36 | hsa04540_Gap_junction | 5 | 90 | 0.3245 | 1 | |
| 37 | hsa04730_Long.term_depression | 4 | 70 | 0.3366 | 1 | |
| 38 | hsa00510_N.Glycan_biosynthesis | 3 | 49 | 0.3373 | 1 | |
| 39 | hsa04114_Oocyte_meiosis | 6 | 114 | 0.3429 | 1 | |
| 40 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 5 | 95 | 0.3663 | 1 | |
| 41 | hsa04062_Chemokine_signaling_pathway | 9 | 189 | 0.3945 | 1 | |
| 42 | hsa04070_Phosphatidylinositol_signaling_system | 4 | 78 | 0.4131 | 1 | |
| 43 | hsa00562_Inositol_phosphate_metabolism | 3 | 57 | 0.4288 | 1 | |
| 44 | hsa04660_T_cell_receptor_signaling_pathway | 5 | 108 | 0.474 | 1 | |
| 45 | hsa04960_Aldosterone.regulated_sodium_reabsorption | 2 | 42 | 0.5296 | 1 | |
| 46 | hsa04670_Leukocyte_transendothelial_migration | 5 | 117 | 0.545 | 1 | |
| 47 | hsa03018_RNA_degradation | 3 | 71 | 0.5754 | 1 | |
| 48 | hsa04330_Notch_signaling_pathway | 2 | 47 | 0.591 | 1 | |
| 49 | hsa04662_B_cell_receptor_signaling_pathway | 3 | 75 | 0.6128 | 1 | |
| 50 | hsa04020_Calcium_signaling_pathway | 7 | 177 | 0.6141 | 1 | |
| 51 | hsa04664_Fc_epsilon_RI_signaling_pathway | 3 | 79 | 0.6479 | 1 | |
| 52 | hsa00230_Purine_metabolism | 6 | 162 | 0.6764 | 1 | |
| 53 | hsa04914_Progesterone.mediated_oocyte_maturation | 3 | 87 | 0.7111 | 1 | |
| 54 | hsa04640_Hematopoietic_cell_lineage | 3 | 88 | 0.7184 | 1 | |
| 55 | hsa04210_Apoptosis | 3 | 89 | 0.7255 | 1 | |
| 56 | hsa00830_Retinol_metabolism | 2 | 64 | 0.7545 | 1 | |
| 57 | hsa04912_GnRH_signaling_pathway | 3 | 101 | 0.8002 | 1 | |
| 58 | hsa00980_Metabolism_of_xenobiotics_by_cytochrome_P450 | 2 | 71 | 0.8034 | 1 | |
| 59 | hsa04976_Bile_secretion | 2 | 71 | 0.8034 | 1 | |
| 60 | hsa04530_Tight_junction | 4 | 133 | 0.8126 | 1 | |
| 61 | hsa04971_Gastric_acid_secretion | 2 | 74 | 0.8216 | 1 | |
| 62 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 4 | 136 | 0.826 | 1 | |
| 63 | hsa04370_VEGF_signaling_pathway | 2 | 76 | 0.8329 | 1 | |
| 64 | hsa00564_Glycerophospholipid_metabolism | 2 | 80 | 0.8535 | 1 | |
| 65 | hsa04142_Lysosome | 3 | 121 | 0.8866 | 1 | |
| 66 | hsa04145_Phagosome | 4 | 156 | 0.8961 | 1 | |
| 67 | hsa04380_Osteoclast_differentiation | 3 | 128 | 0.9079 | 1 | |
| 68 | hsa04141_Protein_processing_in_endoplasmic_reticulum | 4 | 168 | 0.925 | 1 | |
| 69 | hsa03013_RNA_transport | 3 | 152 | 0.9561 | 1 | |
| 70 | hsa04740_Olfactory_transduction | 2 | 388 | 1 | 1 |