This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-let-7a-3p | PTEN | 0.89 | 0 | -0.59 | 0 | mirMAP | -0.3 | 0 | NA | |
| 2 | hsa-let-7b-3p | PTEN | 0.51 | 0 | -0.59 | 0 | mirMAP | -0.26 | 0 | NA | |
| 3 | hsa-miR-103a-3p | PTEN | 1.21 | 0 | -0.59 | 0 | miRNAWalker2 validate; miRTarBase | -0.27 | 0 | 26511107; 24828205 | LncRNA GAS5 induces PTEN expression through inhibiting miR 103 in endometrial cancer cells; To investigate the expression of GAS5 PTEN and miR-103 RT-PCR was performed; Finally we found that miR-103 mimic could decrease the mRNA and protein levels of PTEN through luciferase reporter assay and western blotting and GAS5 plasmid may reverse this regulation effect in endometrial cancer cells; Through inhibiting the expression of miR-103 GAS5 significantly enhanced the expression of PTEN to promote cancer cell apoptosis and thus could be an important mediator in the pathogenesis of endometrial cancer;Our data collectively demonstrate that miR-103 is an oncogene miRNA that promotes colorectal cancer proliferation and migration through down-regulation of the tumor suppressor genes DICER and PTEN |
| 4 | hsa-miR-106b-5p | PTEN | 1.03 | 0 | -0.59 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.17 | 0.00044 | 24842611; 26238857; 26722252 | MicroRNA 106b in cancer associated fibroblasts from gastric cancer promotes cell migration and invasion by targeting PTEN;We further identified PTEN and p21 as novel direct targets of miR-106b by using target prediction algorithms and a luciferase assay; Overexpression of miR-106b reduced the expression of PTEN and p21 and increased the expression of p-AKT which is a downstream of PTEN; Restoring the expression of PTEN or p21 in stably miR-106b-overexpressed cells could rescue the effect of miR-106b on cell radioresistance; These observations illustrated that miR-106b could induce cell radioresistance by directly targeting PTEN and p21 this process was accompanied by tumour-initiating cell capacity enhancement which is universally confirmed to be associated with radioresistance;Cantharidin modulates the E2F1/MCM7 miR 106b 93/p21 PTEN signaling axis in MCF 7 breast cancer cells |
| 5 | hsa-miR-130b-3p | PTEN | 1.01 | 0 | -0.59 | 0 | MirTarget; miRNATAP | -0.12 | 0.00364 | 26837847; 25637514 | The miR 130 family promotes cell migration and invasion in bladder cancer through FAK and Akt phosphorylation by regulating PTEN; In clinical bladder cancer specimens downregulation of PTEN was found to be closely correlated with miR-130 family expression levels;MiR 130b plays an oncogenic role by repressing PTEN expression in esophageal squamous cell carcinoma cells; We confirmed that miR-130b interacted with the 3'-untranslated region of PTEN and that an increase in the expression level of miR-130b negatively affected the protein level of PTEN; However the dysregulation of miR-130b had no obvious impact on PTEN mRNA; As Akt is a downstream effector of PTEN we explored if miR-130b affected Akt expression and found that miR-130b indirectly regulated the level of phosphorylated Akt while total Akt protein remained unchanged; The results indicate that miR-130b plays an oncogenic role in ESCC cells by repressing PTEN expression and Akt phosphorylation which would be helpful in developing miRNA-based treatments for ESCC |
| 6 | hsa-miR-141-3p | PTEN | 1.8 | 0 | -0.59 | 0 | miRNAWalker2 validate; miRTarBase; TargetScan; miRNATAP | -0.15 | 0 | 27644195; 24742567 | Involvement of microRNA 141 3p in 5 fluorouracil and oxaliplatin chemo resistance in esophageal cancer cells via regulation of PTEN; Western blot exhibited altered protein levels of PTEN Akt and PI3k with miR-141-3p inhibitor; An inverse correlation between PTEN expression and miR-141-3p expression was also observed in tissue samples; Our study demonstrated that miR-141-3p contributed to an acquired chemo-resistance through PTEN modulation both in vitro and in vivo;PTEN might be a potential target of miR-141 and miR-200a in endometrial carcinogenesis |
| 7 | hsa-miR-146b-5p | PTEN | 1.7 | 0 | -0.59 | 0 | miRanda | -0.14 | 4.0E-5 | NA | |
| 8 | hsa-miR-148a-3p | PTEN | 1.85 | 0 | -0.59 | 0 | MirTarget; miRNATAP | -0.12 | 0.00262 | 22496917 | Introduction of anti-miR-148a increased PTEN protein and mRNA expression suggesting that PTEN was targeted by miR-148a |
| 9 | hsa-miR-15b-3p | PTEN | 0.7 | 0 | -0.59 | 0 | mirMAP | -0.22 | 0 | NA | |
| 10 | hsa-miR-16-1-3p | PTEN | 0.67 | 0 | -0.59 | 0 | MirTarget | -0.13 | 0.00036 | NA | |
| 11 | hsa-miR-182-5p | PTEN | 2.58 | 0 | -0.59 | 0 | mirMAP | -0.17 | 0 | NA | |
| 12 | hsa-miR-195-3p | PTEN | 0.6 | 0 | -0.59 | 0 | mirMAP | -0.19 | 8.0E-5 | NA | |
| 13 | hsa-miR-200a-3p | PTEN | 0.48 | 0.03921 | -0.59 | 0 | miRNATAP | -0.12 | 0 | 22637745; 24742567; 21408027 | Re expression of miR 200 by novel approaches regulates the expression of PTEN and MT1 MMP in pancreatic cancer; We initially compared the expression profile of miR-200 family PTEN and MT1-MMP expression in six pancreatic cancer PC cell lines by qRT-PCR and western blot analysis; We found loss of expression of miR-200a b and c in chemo-resistant PC cell lines which was correlated with loss of PTEN and over-expression of MT1-MMP; The expression of miR-200 family and PTEN was significantly re-expressed whereas the expression of MT1-MMP was down-regulated by CDF and BR-DIM treatment; These results provide strong experimental evidence showing that the loss of miR-200 family and PTEN expression and increased level of MT1-MMP leads to aggressive behavior of PC cells which could be attenuated through re-expression of miR-200c by CDF and/or BR-DIM treatment suggesting that these agents could be useful for PC treatment;PTEN might be a potential target of miR-141 and miR-200a in endometrial carcinogenesis;Anti tumor activity of a novel compound CDF is mediated by regulating miR 21 miR 200 and PTEN in pancreatic cancer; In a xenograft mouse model of human PC CDF treatment significantly inhibited tumor growth which was associated with decreased NF-κB DNA binding activity COX-2 and miR-21 expression and increased PTEN and miR-200 expression in tumor remnants |
| 14 | hsa-miR-200b-3p | PTEN | 0.52 | 0.02018 | -0.59 | 0 | TargetScan; mirMAP | -0.12 | 0 | 22637745; 21408027 | Re expression of miR 200 by novel approaches regulates the expression of PTEN and MT1 MMP in pancreatic cancer; We initially compared the expression profile of miR-200 family PTEN and MT1-MMP expression in six pancreatic cancer PC cell lines by qRT-PCR and western blot analysis; The expression of miR-200 family and PTEN was significantly re-expressed whereas the expression of MT1-MMP was down-regulated by CDF and BR-DIM treatment; These results provide strong experimental evidence showing that the loss of miR-200 family and PTEN expression and increased level of MT1-MMP leads to aggressive behavior of PC cells which could be attenuated through re-expression of miR-200c by CDF and/or BR-DIM treatment suggesting that these agents could be useful for PC treatment;Anti tumor activity of a novel compound CDF is mediated by regulating miR 21 miR 200 and PTEN in pancreatic cancer; In a xenograft mouse model of human PC CDF treatment significantly inhibited tumor growth which was associated with decreased NF-κB DNA binding activity COX-2 and miR-21 expression and increased PTEN and miR-200 expression in tumor remnants |
| 15 | hsa-miR-200c-3p | PTEN | 2.04 | 0 | -0.59 | 0 | mirMAP; miRNATAP | -0.18 | 0 | 24682933; 22637745 | In conclusion miR-200c functions as an oncogene in colon cancer cells through regulating tumor cell apoptosis survival invasion and metastasis as well as xenograft tumor growth through inhibition of PTEN expression and p53 phosphorylation;Forced over-expression or silencing of miR-200c followed by either CDF or BR-DIM treatment of MIAPaCa-2 cells altered the morphology of cells wound-healing capacity colony formation and the expression of MT1-MMP and PTEN; These results provide strong experimental evidence showing that the loss of miR-200 family and PTEN expression and increased level of MT1-MMP leads to aggressive behavior of PC cells which could be attenuated through re-expression of miR-200c by CDF and/or BR-DIM treatment suggesting that these agents could be useful for PC treatment |
| 16 | hsa-miR-21-5p | PTEN | 1.03 | 0 | -0.59 | 0 | miRNAWalker2 validate; miRTarBase; mirMAP | -0.38 | 0 | 20092645; 27725205; 26387181; 22267008; 26384051; 22322462; 27644439; 21471222; 21468550; 20113523; 26905520; 21806946; 24154840; 27611950; 24780321; 25027758; 23036707; 23684551; 26559642; 26731559; 26847601; 27350731; 24331411; 21820606; 25909227; 24930006; 24293118; 26289851; 22547075; 26666820; 24324076; 23201752; 17681183; 25963606; 27188433; 22958183; 22956424; 19730150; 21842656; 21104017; 23894315; 23548551; 26236156; 23951172; 24460329; 25973032; 25563770; 24659669; 25647415; 26230405; 25543482; 20048743; 23466500; 22922228; 25058005; 20223231; 27220494; 22678116; 24763002; 24221338; 22120473; 21408027; 23174819; 22832383; 19212625; 22978663; 25799148; 26741162; 23226804; 26787105; 26864640; 26311740; 25603978; 26975392 | In the present study we investigated the role of miR-21 and its potential as a therapeutic target in two prostate cancer cell lines characterized by different miR-21 expression levels and PTEN gene status;The results showed that upon exposure to mUA miR-21 expression was decreased and the expression of PTEN and Pdcd4 protein was elevated; In conclusion our data suggest that mUA can suppress cell viability in DU145 cells through modulating miR-21 and its downstream series-wound targets including PTEN Akt and Wnt/β-catenin signaling;In addition treated with 5-AZA resulted in significant increases of miR-21 expression in both MCF-7 and MDA-MB-231 cells P < 0.01 with the protein level of PTEN increased in MCF-7 cell which was further involved in the downregulation of AKT;microRNA 21 promotes tumor proliferation and invasion in gastric cancer by targeting PTEN; Thus in this study we focused on the expression and significance of miR-21 in gastric cancer tissues and the role of miR-21 in the biological behaviour and the expression of PTEN in gastric cancer cells; Western blotting and the Luciferase Reporter Assay were used to evaluate the change of PTEN expression after lowered expression of miR-21 in gastric cancer cell lines; The western blot results and Luciferase Reporter Assay demonstrated that PTEN expression was remarkably increased after miR-21 inhibition P<0.05 microRNA-21 expression was upregulated in gastric carcinoma tissues and was significantly associated with the degree of differentiation of tumour tissues local invasion and lymph node metastasis;Furthermore we identified that miR-21 overexpression could promote Hela and U2OS cells proliferation by targeting phosphatase-tensin homolog PTEN the result of which can be rescued by miR-21 inhibitor;Inhibition of microRNA-21 mir‑21 induced upregulation of Spry2 and PTEN which underscores the importance of mir-21 in Spry2-associated tumorigenesis of the colon;Bmi-1 also regulates p53 and PTEN via miR-21;Here we demonstrated that miR-21 expression was up-regulated and its function was elevated in HER2+ BT474 SKBR3 and MDA-MB-453 breast cancer cells that are induced to acquire trastuzumab resistance by long-term exposure to the antibody whereas protein expression of the PTEN gene a miR-21 target was reduced; Rescuing PTEN expression with a p3XFLAG-PTEN-mut construct with deleted miR-21 targeting sequence at its 3' UTR restored the growth inhibition of trastuzumab in the resistant cells by inducing PTEN activation and AKT inhibition; In vivo administering miR-21 antisense oligonucleotides restored trastuzumab sensitivity in the resistant breast cancer xenografts by inducing PTEN expression whereas injection of miR-21 mimics conferred trastuzumab resistant in the sensitive breast tumors via PTEN silence;The expression of miR-21 and its target PTEN was determined by real-time qRT-PCR and western blotting respectively in tumor tissues as well as adjacent non-tumor mucosa; miR-21 was significantly up-regulated in tumor tissues while PTEN was expressed in lower levels compared to non-tumor tissues; A negative correlation between expression of miR-21 and PTEN was established in vivo;MicroRNA 21 inhibitor sensitizes human glioblastoma cells U251 PTEN mutant and LN229 PTEN wild type to taxol; Human glioblastoma U251 PTEN-mutant and LN229 PTEN wild-type cells were treated with taxol and the miR-21 inhibitor in a poly amidoamine PAMAM dendrimer alone or in combination; Interestingly the above data suggested that in both the PTEN mutant and the wild-type GBM cells miR-21 blockage increased the chemosensitivity to taxol; Thus the miR-21 inhibitor might interrupt the activity of EGFR pathways independently of PTEN status;The effect of ZA on miR-21 expression was quantified by qRT-PCR and the amount of PTEN protein and its targets were analyzed by Western blot;NTR1 activation stimulates expression of miR-21 and miR-155 in colonocytes via Akt and NF-κB to down-regulate PTEN and SOCS1 and promote growth of tumors in mice;Correlations between the expression levels of miR-21 PTEN and p-AKT were analyzed by real-time PCR and Western blot test in HER2-positive GC cell lines; Overexpression of miR-21 down-regulated PTEN expression increased AKT phosphorylation and did not affect HER2 expression; Inversely suppression of miR-21 increased PTEN expression and down-regulated AKT phosphorylation but still did not affect HER2 expression;MicroRNA 21 promotes TGF β1 induced epithelial mesenchymal transition in gastric cancer through up regulating PTEN expression; In GC tissues the expressions of miR-21 Akt and p-Akt were up-regulated while PTEN expression was down-regulated; These results suggest that miR-21 could promote TGF-β1-induced EMT in GC cells through up-regulating PTEN expression;Expressions of PTEN were significantly down-regulated in CRC tissues and negatively correlated with expressions of Notch-1 r2=0.5207 p<0.01 and miR-21 r2=0.6996 p<0.01; These data indicate that the crosstalk between Notch-1 and miR-21 is involved in CRC development through degradation of PTEN;In the same patients we also compared miR-21 expression with the expression of its presumed target PTEN; miR-21 expression levels were found to significantly correlate with tumour size r = 0.403 p = 0.009; Spearman's rank whereas no relation was found between miR-21 and PTEN expression levels Kruskal-Wallis test;Over-expression of miR-21 suppressed its target PTEN and disrupted acinar morphogenesis;MicroRNA 21 suppresses PTEN and hSulf 1 expression and promotes hepatocellular carcinoma progression through AKT/ERK pathways;There was overexpression of the miR-21 target genes PTEN by 67% and caspase-3 by 15% upon cotreatment;We previously reported that microRNA-21 miR-21 was strongly expressed in melanoma relative to naevi and now sought to further assess the significance of this by assessing its relationship with its putative target PTEN; Clinical melanoma samples were analysed by immunohistochemical analysis for PTEN stem-loop qRT-PCR for miR-21 and PCR for BRAF/NRAS mutation status; miR-21 expression was inversely associated with nuclear PTEN expression but not with cytoplasmic PTEN expression; These data suggest miR-21 may exert an oncogenic effect in melanoma by favouring redistribution of PTEN to the nucleus;Triptolide reduces proliferation and enhances apoptosis of human non small cell lung cancer cells through PTEN by targeting miR 21; To the best of our knowledge the present study is the first to demonstrate that triptolide reduced the proliferation and enhanced the apoptosis of human NSCLC cells through PTEN by targeting miR-21;Quantitative real-time PCR qRT-PCR and Western blot were used to detect the expression levels of miR-21 and PTEN in HCT116 HT29 Colo32 and SW480 CRC cell lines; Also the expression levels of PTEN mRNA and its downstream proteins AKT and PI3K in HCT116 cells after downregulating miR-21 were investigated; In comparing the levels of PTEN protein and downstream AKT and PI3K in HCT116 cells after downregulation of miR-21 expression the levels of AKT and PI3K protein expression significantly decreased P < 0.05; PTEN is one of the direct target genes of miR-21;The level of miR-21 was reversely correlated with the expression of PTEN and PDCD4 and positive correlated with PI3K/Akt pathway; miR-21 is involved in acquired resistance of EGFR-TKI in NSCLC which is mediated by down-regulating PTEN and PDCD4 and activating PI3K/Akt pathway;MicroRNA 21 modulates chemosensitivity of breast cancer cells to doxorubicin by targeting PTEN; TaqMan RT-PCR or Western blot assay was performed to detect the expression of mature miR-21 and tumor suppressor gene PTEN protein; We showed that upregulation of miR-21 in MCF-7/ADR cells was concurrent with downregulation of PTEN protein; Overexpression of PTEN could mimic the same effects of miR-21 inhibitor in MCF-7/ADR cells and PTEN-siRNA could increase the resistance of MCF-7 cells to ADR; MiR-21 inhibitor could increase PTEN protein expression and the luciferase activity of a PTEN 3' untranslated region-based reporter construct in MCF-7/ADR cells; Dysregulation of miR-21 plays critical roles in the ADR resistance of breast cancer at least in part via targeting PTEN;MiR-21 level was inversely correlated with the levels of FOXO1 and PTEN in DLBCL cell lines; MiR-21 also down-regulated PTEN expression and consequently activated the PI3K/AKT/mTOR pathway which further decreased FOXO1 expression;In present study we determined the miR-21 levels in TNBC specimens and TNBC cell levels in vitro and then identified the role of miR-21 on tumor cell proliferation apoptosis and then identified PTEN as the possible target of the microRNA;MiR 21 suppresses the anticancer activities of curcumin by targeting PTEN gene in human non small cell lung cancer A549 cells; Transfection of A549 cells with microRNA-21 mimic or PTEN small interfering RNA was performed to modulate the expression of microRNA-21 and PTEN under the treatment of curcumin; Moreover the protein level of PTEN a putative target of microRNA-21 was significantly elevated in curcumin-treated A549 cells as determined by Western blot analysis; Transfection of A549 cells with microRNA-21 mimic or PTEN small interfering RNA significantly P < 0.05 reversed the growth suppression and apoptosis induction by curcumin compared to corresponding controls; Our data suggest a novel molecular mechanism in which inhibition of microRNA-21 and upregulation of PTEN mediate the anticancer activities of curcumin in NSCLC cells;PTEN gene expression was performed as a known target of miR-21;Induction of miR-21 may enable cancer cells to elude DNA damage-induced apoptosis and enhance the metastatic potential of breast cancer cells through repressing expression of PTEN and PDCD4;PTEN a direct target gene of miR-21 was significantly downregulated in gemcitabine-resistant breast cancer cells and restoration of PTEN expression blocked miR-21-induced EMT and gemcitabine resistance;We also found that Rawq01 up-regulated the expression of PTEN through mir-21 inhibition and therefore inhibited the PI3K-AKT pathway;Moreover we demonstrate that oligonucleotide-mediated miR-21 silencing in U87 human GBM cells resulted in increased levels of the tumor suppressors PTEN and PDCD4 caspase 3/7 activation and decreased tumor cell proliferation;MicroRNA 21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer; PTEN was shown to be a direct target of miR-21 and to contribute to miR-21 effects on cell invasion; Modulation of miR-21 altered focal adhesion kinase phosphorylation and expression of matrix metalloproteases 2 and 9 both downstream mediators of PTEN involved in cell migration and invasion; Aberrant expression of miR-21 can contribute to HCC growth and spread by modulating PTEN expression and PTEN-dependent pathways involved in mediating phenotypic characteristics of cancer cells such as cell growth migration and invasion;Overexpression of miR 21 promotes the proliferation and migration of cervical cancer cells via the inhibition of PTEN; The aim of this study was to examine the expression of miR-21 and PTEN in cervical cancer specimens using quantitative PCR; miR-21 was upregulated in the cervical cancer specimens negatively correlating with the PTEN mRNA level; Transfection of the miR-21 mimics was markedly promoted whereas the miR-21 inhibitor suppressed the proliferation migration and invasion of cervical cancer cells with a significant inhibition of PTEN expression; The present study showed the upregulation of miR-21 in invasive cervical cancers and confirmed the promotion of miR-21 with regard to the proliferation migration and invasion in cervical cancer cells via inhibiting the PTEN expression;The expressions of miR-21 PTEN PI3K and AKT were detected in 89 esophageal cancer samples and 58 adjacent normal tissues respectively; MiR-21 PI3K and AKT have higher expressions but PTEN has lower expression in esophageal cancer tissues compared with adjacent normal tissues; Further PTEN was a target gene of miR-21;The expression level of miR-21 and PTEN messenger RNA were measured by quantitative real-time reverse transcription polymerase chain reaction or reverse transcription polymerase chain reaction; miR-21 was overexpressed and PTEN was suppressed in established radioresistant TE-R60 cells compared with the parent cells 1.3-fold and 70.83%; The inhibition of miR-21 significantly increased the cells' radiosensitivity P < 0.05 and the PTEN protein expression 2.3-fold in TE-1 cells; Knockdown of PTEN in anti-miR-21 TE-1 cells could abrogate the miR-21 inhibition-induced radiosensitization P < 0.05; Inhibition of miR-21 increased radiosensitivity of esophageal cancer TE-1 cells and this effect was possibly through the activation of PTEN;MicroRNA 21 miR 21 expression promotes growth metastasis and chemo or radioresistance in non small cell lung cancer cells by targeting PTEN; Taken together these results provide evidence to show the promotion role of miR-21 in NSCLC development through modulation of the PTEN signaling pathway;Protein levels of tumor suppressor targets of the miRNAs were increased by antisense to miR-21 PTEN and RECK and miR-221 p27;The expression of miR-21 PTEN and PDCD4 were determined by Real-time PCR; Glossy ganoderma spore oil down-regulated the expression of miR-21 and up-regulated the expression of PTEN and PDCD4 significantly;Role of microRNA 21 and effect on PTEN in Kazakh's esophageal squamous cell carcinoma; To evaluate the role of miR-21 and PTEN cell proliferations were analyzed with miR-21 mimics or their inhibitor-transfected cells; In Eca109 when transfected with miR-21 mimics accumulation of miR-21 was obviously increased and expression of PTEN protein was decreased to be approximately 40% which resulted in the promotion of cell proliferation; However when transfected with miR-21 inhibitor expression of miR-21 was declined and PTEN protein was overexpressed to be approximately 79% which resulted in the suppression of cell proliferation; Furthermore there was a significantly inverse correlation between miR-21 expression and PTEN protein levels p < 0.05; The author concluded that MiR-21 was overexpressed in vitro and ESCC and promoted the cell proliferation might target PTEN at post-transcriptional level and regulated the cancer invasion in Kazakh's ESCC;Difluorinated curcumin CDF restores PTEN expression in colon cancer cells by down regulating miR 21; Indeed our current data demonstrate a marked downregulation of PTEN in SCID mice xenografts of miR-21 over-expressing colon cancer HCT116 cells; Colonospheres that are highly enriched in cancer stem/stem like cells reveal increased miR-21 expression and decreased PTEN; Difluorinated curcumin CDF a novel analog of the dietary ingredient curcumin which has been shown to inhibit the growth of 5-Flurouracil + Oxaliplatin resistant colon cancer cells downregulated miR-21 in chemo-resistant colon cancer HCT116 and HT-29 cells and restored PTEN levels with subsequent reduction in Akt phosphorylation;Moreover knockdown of miR-21 increased PDCD4 and PTEN expression at the protein level but not at the mRNA level;Mechanistic evidence showed that down-regulation of miR-21 increased the expression of its target molecule PTEN in HCT116 cells;The results showed that knockdown of miR-21 by antagomir-21 decreased cell proliferation and induced apoptosis via targeting PTEN both in 4T1 cells and HUVECs;Finally invasion and metastasis assays were performed and alteration in mir-21 PTEN AKT and pAKT level was evaluated in these cells; Enhanced invasion and metastasis increased miR-21 expression decreased PTEN elevated pAKT level were demonstrated in gemcitabine-resistant HPAC and PANC-1 cells;PDCD4 and PTEN expression was decreased gradually after tumor induction and negatively correlated with miR-21 expression; Our in vivo experiments further confirmed that miR-21 plays an important role in promoting the occurrence and development of HCC by regulating PDCD4 and PTEN;Expressions of microRNA-21 miR-21 PTEN MMP9 and p47 were detected by qPCR;The protein levels of miR-21 targets PTEN and PDCD4 were estimated; In the control experiment miR-21 mimic significantly inhibited the expression of PTEN and PDCD4 proteins in the two gastric cell lines leading to an increase in cell invasion and migration; miR-21 is overexpressed in gastric cancer and its aberrant expression may have important role in gastric cancer growth and dissemination by modulating the expression of the tumor suppressors PTEN and PDCD4 as well as by modulating the pathways involved in mediating cell growth migration invasion and apoptosis;MiR-21 upregulation contributes to PTEN downregulation which is beneficial for the activation of PI3K/AKT signaling;microRNA 21 Regulates Cell Proliferation and Migration and Cross Talk with PTEN and p53 in Bladder Cancer; MicroRNA-21 regulates proliferation and migration of bladder cancer cells and cross talk with PTEN and p53 in bladder cancer;Moreover knockdown of miR-21 increased the expressions of PDCD4 and PTEN at the protein level but not at the mRNA level;Downregulation of miR 21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status; To explore whether miR-21 can serve as a therapeutic target for glioblastoma we downregulated miR-21 with a specific antisense oligonucleotide and found that apoptosis was induced and cell-cycle progression was inhibited in vitro in U251 PTEN mutant and LN229 PTEN wild-type GBM cells; xenograft tumors from antisense-treated U251 cells were suppressed in vivo; Taken together our studies provide evidence that miR-21 may serve as a novel therapeutic target for malignant gliomas independent of PTEN status;miR 21 confers cisplatin resistance in gastric cancer cells by regulating PTEN; In addition miR-21 induced cell survival and cisplatin resistance through downregulating the expression of phosphatase and tension homolog deleted on chromosome 10 PTEN and activation of Akt pathway;Finally we demonstrate that modulation of tumor suppressors PTEN and p53 in U87 cells does not affect the decrease in miR-21 levels associated with PDGF-B overexpression;This study aimed to investigate whether NSCLC miR-21 mediated resistance to TKIs also results from Pten targeting; Here we show miR-21 promotes cancer by negatively regulating Pten expression in human NSCLC tissues: high miR-21 expression levels were associated with shorter DFS in 47 NSCLC patients; high miR-21/low Pten expression levels indicated a poor TKI clinical response and shorter overall survival in another 46 NSCLC patients undergoing TKI treatment; In vitro assays showed that miR-21 was up-regulated concomitantly to down-regulation of Pten in pc-9/GR cells in comparison with pc-9 cells; Moreover over-expression of miR-21 significantly decreased gefitinib sensitivity by down-regulating Pten expression and activating Akt and ERK pathways in pc-9 cells while miR-21 knockdown dramatically restored gefitinib sensitivity of pc-9/GR cells by up-regulation of Pten expression and inactivation of AKT and ERK pathways in vivo and in vitro;MicroRNA 21 miR 21 represses tumor suppressor PTEN and promotes growth and invasion in non small cell lung cancer NSCLC; We identified the role of miR-21 in non-small cell lung cancer NSCLC and to clarify the regulation of PTEN by miR-21 and determine mechanisms of this regulation; Expression of miR-21 and PTEN in 20 paired NSCLC and adjacent non-tumor lung tissues was investigated by qRT-PCR and western blot respectively; Tumor tissues showed an inverse correlation between miR-21 and PTEN protein; miR-21 inhibitor transfection increased a luciferase-reporter activity containing the PTEN-3'-UTR construct and increased PTEN protein but not PTEN-mRNA levels in NSCLC cell lines; miR-21 post-transcriptionally down-regulates the expression of tumor suppressor PTEN and stimulates growth and invasion in NSCLC;MiR-21 overexpression decreases PTEN increases p-Akt and subsequently increases HIF-1α expression while miR-21 inhibition results in increased PTEN decreased p-Akt and then decreased HIF-1α;Furthermore miR-21-induced upregulation of CSF-1 mRNA and its transcription were prevented by expression of PTEN mRNA lacking 3'-untranslated region UTR and miR-21 recognition sequence; Our results reveal a novel mechanism for the therapeutic function of fish oil diet that blocks miR-21 thereby increasing PTEN levels to prevent expression of CSF-1 in breast cancer;This study was aimed to investigate the expression of microRNA-21 and its correlation with PTEN in diffuse large B cell lymphoma DLBCL paraffin-embedded tissues and evaluate its potential relevance with clinical characteristics; In patients with DLBCL the expression level of miR-21 was negatively correlated with the level of PTEN protein; These findings suggest that PTEN is possibly one of the targets of miR-21 in DLBCL;Down regulation of PTEN expression modulated by dysregulated miR 21 contributes to the progression of esophageal cancer; We demonstrated that knockdown of miR-21 significantly increased expression of PTEN protein; Our findings suggest that miR-21 could be a potential oncomiR probably by regulation of PTEN and a novel prognostic factor for ESCC patients;The levels of mir-21 did not associate with the expression of PTEN an important tumour suppressor in CRC and one of many putative targets of miR-21 but interestingly was associated with stage of disease in the PTEN expressing tumours;Anti tumor activity of a novel compound CDF is mediated by regulating miR 21 miR 200 and PTEN in pancreatic cancer; In a xenograft mouse model of human PC CDF treatment significantly inhibited tumor growth which was associated with decreased NF-κB DNA binding activity COX-2 and miR-21 expression and increased PTEN and miR-200 expression in tumor remnants;The PTEN protein levels in CRC tissues and cells had an inverse correlation with miR-21 expression; MiR-21 targets PTEN at the post-transcriptional level and regulates cell proliferation and invasion in CRC;After miR-21 was transfected in MCF-7 cells PTEN protein level was measured by Western blot; Matrine up-regulated PTEN by downregulating miR-21 which in turn dephosphorylated Akt resulting in accumulation of Bad p21/WAF1/CIP1 and p27/KIP1;To further determine the potential involvement of miR-21 in breast cancer we have evaluated the expression level of miR-21 by stem-loop real-time RT-PCR based on SYBR-Green I in human invasive ductal carcinoma of the breast and we have correlated the results with clinicopathologic features and PTEN protein expression; The expression levels of miR-21 were correlated with PTEN and commonly used clinicopathologic features of breast cancer; Expression of miR-21 was negatively correlated with expression of PTEN P=0.013; These findings suggest that PTEN is possibly one of the targets of miR-21 in breast cancer and high expression of mir-21 indicates a more aggressive phenotype;These findings demonstrate a novel role of AR in the regulation of miR-21 and its target PTEN in growth factor-induced colon cancer cell growth;Whether miR-21 regulated PTEN expression was assessed by luciferase assay; The RNA and protein levels of PTEN were significantly decreased by exogenous miR-21 and the 3'-untranslated region of PTEN was shown to be a target of miR-21;In vitro study showed QTsome/AM-21 induced upregulation of miR-21 targets including PTEN and DDAH1 in A549 cells while increasing their sensitivity toward paclitaxel PTX;microRNA 21 overexpression contributes to cell proliferation by targeting PTEN in endometrioid endometrial cancer; We performed a qRT-PCR assay with miR-21 and PTEN in 16 paired EEC tumor tissues and adjacent non-tumor endometrium; To validate the putative binding site of miR-21 in the 3' untranslated region 3'-UTR of PTEN messenger RNA mRNA a dual-luciferase reporter assay was carried out; The upregulation of miR-21 led to a significant decrease in the PTEN protein expression level P=0.007; The downregulation of miR-21 led to a significant increase in PTEN protein P=0.002; In conclusion we demonstrated that the expression of PTEN protein but not mRNA was negatively directly regulated by miR-21 in the KLE cell line; The overexpression of miR-21 modulated EEC cell proliferation through the downregulation of PTEN;The prognostic effect of PTEN expression status in colorectal cancer development and evaluation of factors affecting it: miR 21 and promoter methylation; In this study we investigated the effect of miR-21 and promoter methylation on the PTEN expression status in CRC tissues and analyzed association of the PTEN expression status with clinicopathological features in patients with CRC; PTEN mRNA level was negatively correlated with miR-21 level r = -0.595 P < 0.001; PTEN expression was also correlated directly with the PTEN mRNA level r = 0.583 P < 0.001 and conversely with miR-21 level r = -0.632 P < 0.001; This study suggests a high frequency of miR-21 overexpression and aberrant promoter methylation in down-regulation of PTEN expression in colorectal carcinoma;MicroRNA 21 induces 5 fluorouracil resistance in human pancreatic cancer cells by regulating PTEN and PDCD4; The proresistance effects of miR-21 were attributed to the attenuated expression of tumor suppressor genes including PTEN and PDCD4;Exposure of HCC cells to sorafenib led to an increase in miR-21 expression a decrease in PTEN expression and sequential Akt activation;MicroRNA 21 controls hTERT via PTEN in human colorectal cancer cell proliferation; The aim of this study was to determine a role of microRNA-21 miRNA-21 in colorectal cancer CRC and to elucidate miRNA-21 regulation of hTERT by phosphatase and tensin homologue PTEN;Relevance of miR 21 in regulation of tumor suppressor gene PTEN in human cervical cancer cells; We identified the tumor suppressor gene PTEN as a target of miR-21 and determined the mechanism of its regulation throughout reporter construct plasmids; Using this model we analyzed the expression of miR-21 and PTEN as well as functional effects such as autophagy and apoptosis induction; In SiHa cells there was an inverse correlation between miR-21 expression and PTEN mRNA level as well as PTEN protein expression in cervical cancer cells; We conclude that miR-21 post-transcriptionally down-regulates the expression of PTEN to promote cell proliferation and cervical cancer cell survival |
| 17 | hsa-miR-217 | PTEN | 0.65 | 0.00062 | -0.59 | 0 | mir2Disease; miRNAWalker2 validate; miRTarBase | -0.11 | 4.0E-5 | 26109338 | MicroRNA 217 overexpression induces drug resistance and invasion of breast cancer cells by targeting PTEN signaling; MiR-217 activates AKT by downregulation of PTEN in breast cancer cells |
| 18 | hsa-miR-25-3p | PTEN | 1.49 | 0 | -0.59 | 0 | miRTarBase; MirTarget; miRNATAP | -0.19 | 0.0001 | NA | |
| 19 | hsa-miR-32-5p | PTEN | 0.92 | 0 | -0.59 | 0 | MirTarget; miRNATAP | -0.13 | 0.00035 | 24123284; 25647261; 23617834 | In this study we determined the levels of the correlation between and the clinical significance of the expression of miR-32 and phosphatase and tensin homologue PTEN a tumor suppressor targeted by miR-32 in CRC; The levels of miR-32 and PTEN gene expression in 35 colorectal carcinoma samples 35 corresponding cancer-adjacent tissue samples 27 colorectal adenoma samples and 16 normal tissue samples were quantified using real-time quantitative reverse transcriptase-polymerase chain reaction; The relationship between the miR-32 and PTEN protein expression and clinicopathological factors was analyzed; Significant upregulation of miR-32 expression and reduction of PTEN were identified in CRC tissues; An inverse relationship between miR-32 and PTEN protein expression was identified; MiR-32 and PTEN expression were inversely correlated and miR-32 may be associated with the development of CRC;MiR 32 induces cell proliferation migration and invasion in hepatocellular carcinoma by targeting PTEN; Besides miRNA-32 down-regulates PTEN through binding to 3'-UTR of PTEN mRNA from luciferase reporter assay and the expression level of miR-32 could affect the proliferation migration and invasion of liver cancer cell lines via PTEN/Akt signaling pathway; Down-expression of PTEN could significantly attenuate the inhibitory effects of knockdown miR-32 on the proliferation migration and invasion of liver cancer cells suggesting that miR-32 could be a potential target for HCC treatment;MicroRNA 32 miR 32 regulates phosphatase and tensin homologue PTEN expression and promotes growth migration and invasion in colorectal carcinoma cells; In this study we identified the potential effects of miR-32 on some important biological properties of CRC cells and clarified the regulation of PTEN by miR-32; The 3'-untranslated region 3'-UTR of PTEN combined with miR-32 was verified by dual-luciferase reporter assay; Gain-of-function and loss-of-function studies showed that overexpression of miR-32 promoted SW480 cell proliferation migration and invasion reduced apoptosis and resulted in downregulation of PTEN at a posttranscriptional level; However miR-32 knock-down inhibited these processes in HCT-116 cells and enhanced the expression of PTEN protein; In addition we further identified PTEN as the functional downstream target of miR-32 by directly targeting the 3'-UTR of PTEN; Our results demonstrated that miR-32 was involved in tumorigenesis of CRC at least in part by suppression of PTEN |
| 20 | hsa-miR-320a | PTEN | 1 | 0 | -0.59 | 0 | MirTarget; PITA; miRanda; miRNATAP | -0.17 | 0.00027 | NA | |
| 21 | hsa-miR-335-3p | PTEN | 0.63 | 0 | -0.59 | 0 | MirTarget | -0.12 | 0.00309 | NA | |
| 22 | hsa-miR-340-5p | PTEN | 0.42 | 1.0E-5 | -0.59 | 0 | miRNATAP | -0.17 | 0.00165 | NA | |
| 23 | hsa-miR-342-3p | PTEN | 1.26 | 0 | -0.59 | 0 | miRanda | -0.13 | 0.00462 | NA | |
| 24 | hsa-miR-3613-5p | PTEN | 0.72 | 0 | -0.59 | 0 | miRNATAP | -0.12 | 0.00155 | NA | |
| 25 | hsa-miR-425-5p | PTEN | 1.75 | 0 | -0.59 | 0 | miRNATAP | -0.19 | 0 | 25154996 | An increase in miR-425 depended upon IL-1β-induced NF-kappaB activation.Repression of PTEN by miR-425 promoted gastric cancer cell proliferation |
| 26 | hsa-miR-429 | PTEN | 0.68 | 0.00462 | -0.59 | 0 | PITA; miRanda; mirMAP; miRNATAP | -0.12 | 0 | NA | |
| 27 | hsa-miR-454-3p | PTEN | 1.05 | 0 | -0.59 | 0 | MirTarget; miRNATAP | -0.12 | 0.0035 | 26296312; 27261580 | MicroRNA 454 functions as an oncogene by regulating PTEN in uveal melanoma; Furthermore we identified PTEN as a direct target of miR-454; Our data revealed that ectopic expression of PTEN restored the effects of miR-454 on cell proliferation and invasion in uveal melanoma cells;MiR 454 promotes the progression of human non small cell lung cancer and directly targets PTEN; At last the potential regulatory function of miR-454 on PTEN expression was confirmed; Further PTEN was confirmed as a direct target of miR-454 by using Luciferase Reporter Assay |
| 28 | hsa-miR-484 | PTEN | 0.97 | 0 | -0.59 | 0 | miRNATAP | -0.25 | 0 | NA | |
| 29 | hsa-miR-7-1-3p | PTEN | 1.46 | 0 | -0.59 | 0 | mirMAP | -0.12 | 0.00167 | NA | |
| 30 | hsa-miR-708-3p | PTEN | 1.69 | 0 | -0.59 | 0 | mirMAP | -0.21 | 0 | NA | |
| 31 | hsa-miR-92a-3p | PTEN | 1.46 | 0 | -0.59 | 0 | MirTarget; miRNATAP | -0.14 | 0.00134 | 26432332; 25515201; 24137349; 23546593; 23133552; 24026406 | Downregulation of PTEN could mimic the same effects of miR-92a mimic in NSCLC cells and rescue the effects on NSCLC cells induced by miR-92a inhibitor; Taken together these findings suggested that miR-92a could promote growth metastasis and chemoresistance in NSCLC cells at least partially by targeting PTEN;MiR 92a Promotes Cell Metastasis of Colorectal Cancer Through PTEN Mediated PI3K/AKT Pathway; The expression of miR-92a PTEN and E-cadherin was analyzed by real-time PCR; In addition there was a negative correlation between levels of miR-92a and the PTEN gene p < 0.0001; The association of levels of miR-92a and PTEN with tumor cell migration in CRC was also confirmed in CRC cell models;MicroRNA miR-92 is overexpressed in a number of tumors and has been proven to negatively regulate a number of tumor suppressor genes including phosphatase and tensin homologue PTEN; PTEN protein expression was decreased in the SiHa cells that were transfected with the miR-92 mimic; The data indicated that miR-92 may increase the migration and invasion of SiHa cells partially through the downregulation of PTEN protein expression;Expression and significance of PTEN and miR 92 in hepatocellular carcinoma; Immunohistochemistry streptavidin-peroxidase SP and quantitative reverse transcriptase-polymerase chain reaction qRT‑PCR were used to detect the expression of PTEN and miR-92 in 15 cases of HCC and the corresponding paracancerous tissues; The correlation between PTEN and miR-92 was analyzed; Additionally the mRNA levels of PTEN and miR-92 showed a significantly negative correlation with each other r=-0.858 P<0.05; In conclusion PTEN and miR-92 have different roles in the development of HCC; The combined detection of PTEN and miR-92 may provide critical clinical evidence for the early diagnosis and prognosis of HCC;PTEN mRNA correlated inversely with miR-92a and members of the miR-17 and miR-130/301 families;The expression levels of miR-92a and phosphatase and tensin homologue PTEN were detected by qRT-PCR and western blot; In addition the regulation of PTEN by miR-92a was evaluated by qRT-PCR western blot and luciferase reporter assays; There was an inverse correlation between the levels of miR-92a and PTEN in CRC tissues; The overexpression of miR-92a in CRC cells decreased PTEN expression at the translational level and decreased PTEN-driven luciferase-reporter activity; Our results demonstrated that miR-92a induced EMT and regulated cell growth migration and invasion in the SW480 cells at least partially via suppression of PTEN expression |
| 32 | hsa-miR-92b-3p | PTEN | 0.05 | 0.65846 | -0.59 | 0 | MirTarget; miRNATAP | -0.13 | 0.00447 | 24099768; 26878388; 24137349; 23546593 | MiR 92b regulates the cell growth cisplatin chemosensitivity of A549 non small cell lung cancer cell line and target PTEN; Furthermore we found miR-92b could directly target PTEN a unique tumor suppressor gene which was downregulated in lung cancer tissues compared to the matched adjacent normal tissues;We revealed that patients exhibiting an upregulation of hsa-miR-92b and patients with deletions of PTEN did not tend to overlap and hsa-miR-92b and PTEN coordinately regulated the pathway of 'cell cycle' and so on;MicroRNA miR-92 is overexpressed in a number of tumors and has been proven to negatively regulate a number of tumor suppressor genes including phosphatase and tensin homologue PTEN; PTEN protein expression was decreased in the SiHa cells that were transfected with the miR-92 mimic; The data indicated that miR-92 may increase the migration and invasion of SiHa cells partially through the downregulation of PTEN protein expression;Expression and significance of PTEN and miR 92 in hepatocellular carcinoma; Immunohistochemistry streptavidin-peroxidase SP and quantitative reverse transcriptase-polymerase chain reaction qRT‑PCR were used to detect the expression of PTEN and miR-92 in 15 cases of HCC and the corresponding paracancerous tissues; The correlation between PTEN and miR-92 was analyzed; Additionally the mRNA levels of PTEN and miR-92 showed a significantly negative correlation with each other r=-0.858 P<0.05; In conclusion PTEN and miR-92 have different roles in the development of HCC; The combined detection of PTEN and miR-92 may provide critical clinical evidence for the early diagnosis and prognosis of HCC |
| 33 | hsa-miR-93-5p | PTEN | 2.03 | 0 | -0.59 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.19 | 0 | 25633810; 26243299; 22465665; 26087719 | MicroRNA 93 activates c Met/PI3K/Akt pathway activity in hepatocellular carcinoma by directly inhibiting PTEN and CDKN1A; We confirmed that miR-93 directly bound with the 3' untranslated regions of the tumor-suppressor genes PTEN and CDKN1A respectivelyand inhibited their expression; We concluded that miR-93 stimulated cell proliferation migration and invasion through the oncogenic c-Met/PI3K/Akt pathway and also inhibited apoptosis by directly inhibiting PTEN and CDKN1A expression in human HCC;microRNA 93 promotes cell proliferation via targeting of PTEN in Osteosarcoma cells; An miRNA miR-93 was significantly up-regulated whereas phosphatase and tensin homologue PTEN expression was significantly down-regulated in all tested OS cells when compared with hMSCs; Ectopic expression of miR-93 decreased PTEN protein levels; Taking these observations together miR-93 can be seen to play a critical role in carcinogenesis through suppression of PTEN and may serve as a therapeutic target for the treatment of OS;Furthermore we found that miR-93 can directly target PTEN and participates in the regulation of the AKT signaling pathway; MiR-93 inversely correlates with PTEN expression in CDDP-resistant and sensitive human ovarian cancer tissues;Furthermore our study found berberine could inhibit miR-93 expression and function in ovarian cancer as shown by an increase of its target PTEN an important tumor suppressor in ovarian cancer; More importantly A2780 cells that were treated with PTEN siRNA had a survival pattern that is similar to cells with miR-93 overexpression |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | NEURON DIFFERENTIATION | 29 | 874 | 2.405e-11 | 1.119e-07 |
| 2 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 39 | 1656 | 1.706e-10 | 1.985e-07 |
| 3 | NEUROGENESIS | 36 | 1402 | 9.628e-11 | 1.985e-07 |
| 4 | CELL DEVELOPMENT | 36 | 1426 | 1.543e-10 | 1.985e-07 |
| 5 | NEURON DEVELOPMENT | 24 | 687 | 5.642e-10 | 5.249e-07 |
| 6 | PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS | 42 | 1977 | 6.768e-10 | 5.249e-07 |
| 7 | INTRACELLULAR SIGNAL TRANSDUCTION | 36 | 1572 | 2.189e-09 | 1.455e-06 |
| 8 | CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 20 | 513 | 2.783e-09 | 1.619e-06 |
| 9 | REGULATION OF CELL DIFFERENTIATION | 34 | 1492 | 7.705e-09 | 3.983e-06 |
| 10 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 37 | 1791 | 1.972e-08 | 8.459e-06 |
| 11 | REGULATION OF CELL DEATH | 33 | 1472 | 2e-08 | 8.459e-06 |
| 12 | CELL PROJECTION ORGANIZATION | 25 | 902 | 2.544e-08 | 9.516e-06 |
| 13 | CELL MORPHOGENESIS INVOLVED IN NEURON DIFFERENTIATION | 16 | 368 | 2.659e-08 | 9.516e-06 |
| 14 | ORGAN MORPHOGENESIS | 24 | 841 | 2.895e-08 | 9.623e-06 |
| 15 | POSITIVE REGULATION OF CELL COMMUNICATION | 33 | 1532 | 5.201e-08 | 1.537e-05 |
| 16 | EPITHELIAL CELL DIFFERENTIATION | 18 | 495 | 5.287e-08 | 1.537e-05 |
| 17 | REGULATION OF TRANSFERASE ACTIVITY | 25 | 946 | 6.404e-08 | 1.732e-05 |
| 18 | REGULATION OF CELL CYCLE | 25 | 949 | 6.806e-08 | 1.732e-05 |
| 19 | GLAND DEVELOPMENT | 16 | 395 | 7.072e-08 | 1.732e-05 |
| 20 | CELLULAR COMPONENT MORPHOGENESIS | 24 | 900 | 1.029e-07 | 2.28e-05 |
| 21 | REGULATION OF CELL PROLIFERATION | 32 | 1496 | 1.017e-07 | 2.28e-05 |
| 22 | REGULATION OF CELLULAR COMPONENT MOVEMENT | 22 | 771 | 1.169e-07 | 2.473e-05 |
| 23 | PARASYMPATHETIC NERVOUS SYSTEM DEVELOPMENT | 5 | 17 | 1.456e-07 | 2.947e-05 |
| 24 | CATABOLIC PROCESS | 35 | 1773 | 1.605e-07 | 3.112e-05 |
| 25 | CELL FATE COMMITMENT | 12 | 227 | 1.983e-07 | 3.69e-05 |
| 26 | REGULATION OF PROTEIN MODIFICATION PROCESS | 34 | 1710 | 2.13e-07 | 3.812e-05 |
| 27 | NEURON PROJECTION DEVELOPMENT | 18 | 545 | 2.217e-07 | 3.82e-05 |
| 28 | CELL CYCLE | 29 | 1316 | 2.446e-07 | 4.065e-05 |
| 29 | POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 23 | 876 | 2.579e-07 | 4.138e-05 |
| 30 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 25 | 1021 | 2.723e-07 | 4.223e-05 |
| 31 | MITOTIC CELL CYCLE | 21 | 766 | 4.499e-07 | 6.752e-05 |
| 32 | REGULATION OF KINASE ACTIVITY | 21 | 776 | 5.554e-07 | 8.076e-05 |
| 33 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 32 | 1618 | 5.92e-07 | 8.347e-05 |
| 34 | RESPONSE TO OXYGEN CONTAINING COMPOUND | 29 | 1381 | 6.63e-07 | 9.073e-05 |
| 35 | CELLULAR RESPONSE TO STRESS | 31 | 1565 | 8.924e-07 | 0.0001186 |
| 36 | RESPONSE TO INORGANIC SUBSTANCE | 16 | 479 | 9.4e-07 | 0.0001215 |
| 37 | NEGATIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 20 | 740 | 1.081e-06 | 0.000136 |
| 38 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 35 | 1929 | 1.183e-06 | 0.0001449 |
| 39 | REGULATION OF NEURON DIFFERENTIATION | 17 | 554 | 1.353e-06 | 0.0001614 |
| 40 | MOVEMENT OF CELL OR SUBCELLULAR COMPONENT | 27 | 1275 | 1.466e-06 | 0.0001642 |
| 41 | ESTABLISHMENT OR MAINTENANCE OF CELL POLARITY | 9 | 141 | 1.482e-06 | 0.0001642 |
| 42 | TISSUE DEVELOPMENT | 30 | 1518 | 1.455e-06 | 0.0001642 |
| 43 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 25 | 1142 | 2.112e-06 | 0.0002285 |
| 44 | CELL DIVISION | 15 | 460 | 2.813e-06 | 0.0002974 |
| 45 | NEURON PROJECTION MORPHOGENESIS | 14 | 402 | 2.88e-06 | 0.0002978 |
| 46 | HEART DEVELOPMENT | 15 | 466 | 3.294e-06 | 0.0003332 |
| 47 | REGULATION OF NEURON PROJECTION DEVELOPMENT | 14 | 408 | 3.418e-06 | 0.0003363 |
| 48 | REGULATION OF MITOTIC CELL CYCLE | 15 | 468 | 3.47e-06 | 0.0003363 |
| 49 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 31 | 1672 | 3.568e-06 | 0.0003366 |
| 50 | GROWTH | 14 | 410 | 3.617e-06 | 0.0003366 |
| 51 | ECTODERMAL PLACODE DEVELOPMENT | 4 | 15 | 4.32e-06 | 0.0003722 |
| 52 | ECTODERMAL PLACODE MORPHOGENESIS | 4 | 15 | 4.32e-06 | 0.0003722 |
| 53 | ECTODERMAL PLACODE FORMATION | 4 | 15 | 4.32e-06 | 0.0003722 |
| 54 | ACTIVATION OF ANAPHASE PROMOTING COMPLEX ACTIVITY | 4 | 15 | 4.32e-06 | 0.0003722 |
| 55 | MITOTIC NUCLEAR DIVISION | 13 | 361 | 4.607e-06 | 0.0003828 |
| 56 | LOCOMOTION | 24 | 1114 | 4.56e-06 | 0.0003828 |
| 57 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 32 | 1784 | 4.818e-06 | 0.0003933 |
| 58 | EMBRYO DEVELOPMENT | 21 | 894 | 5.205e-06 | 0.0004175 |
| 59 | NEGATIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 17 | 616 | 5.601e-06 | 0.0004418 |
| 60 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 32 | 1805 | 6.147e-06 | 0.0004767 |
| 61 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 24 | 1135 | 6.255e-06 | 0.0004772 |
| 62 | REGULATION OF CELL PROJECTION ORGANIZATION | 16 | 558 | 6.664e-06 | 0.0004956 |
| 63 | REGULATION OF RESPONSE TO STRESS | 28 | 1468 | 6.816e-06 | 0.0004956 |
| 64 | REGULATION OF CELL DEVELOPMENT | 20 | 836 | 6.813e-06 | 0.0004956 |
| 65 | PHOSPHORYLATION | 25 | 1228 | 7.56e-06 | 0.0005412 |
| 66 | HEAD DEVELOPMENT | 18 | 709 | 9.187e-06 | 0.000638 |
| 67 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 22 | 1004 | 9.157e-06 | 0.000638 |
| 68 | PHARYNGEAL SYSTEM DEVELOPMENT | 4 | 18 | 9.511e-06 | 0.0006508 |
| 69 | CELLULAR RESPONSE TO RETINOIC ACID | 6 | 65 | 1.073e-05 | 0.0006967 |
| 70 | CARDIOVASCULAR SYSTEM DEVELOPMENT | 19 | 788 | 1.057e-05 | 0.0006967 |
| 71 | CIRCULATORY SYSTEM DEVELOPMENT | 19 | 788 | 1.057e-05 | 0.0006967 |
| 72 | DEVELOPMENTAL GROWTH | 12 | 333 | 1.078e-05 | 0.0006967 |
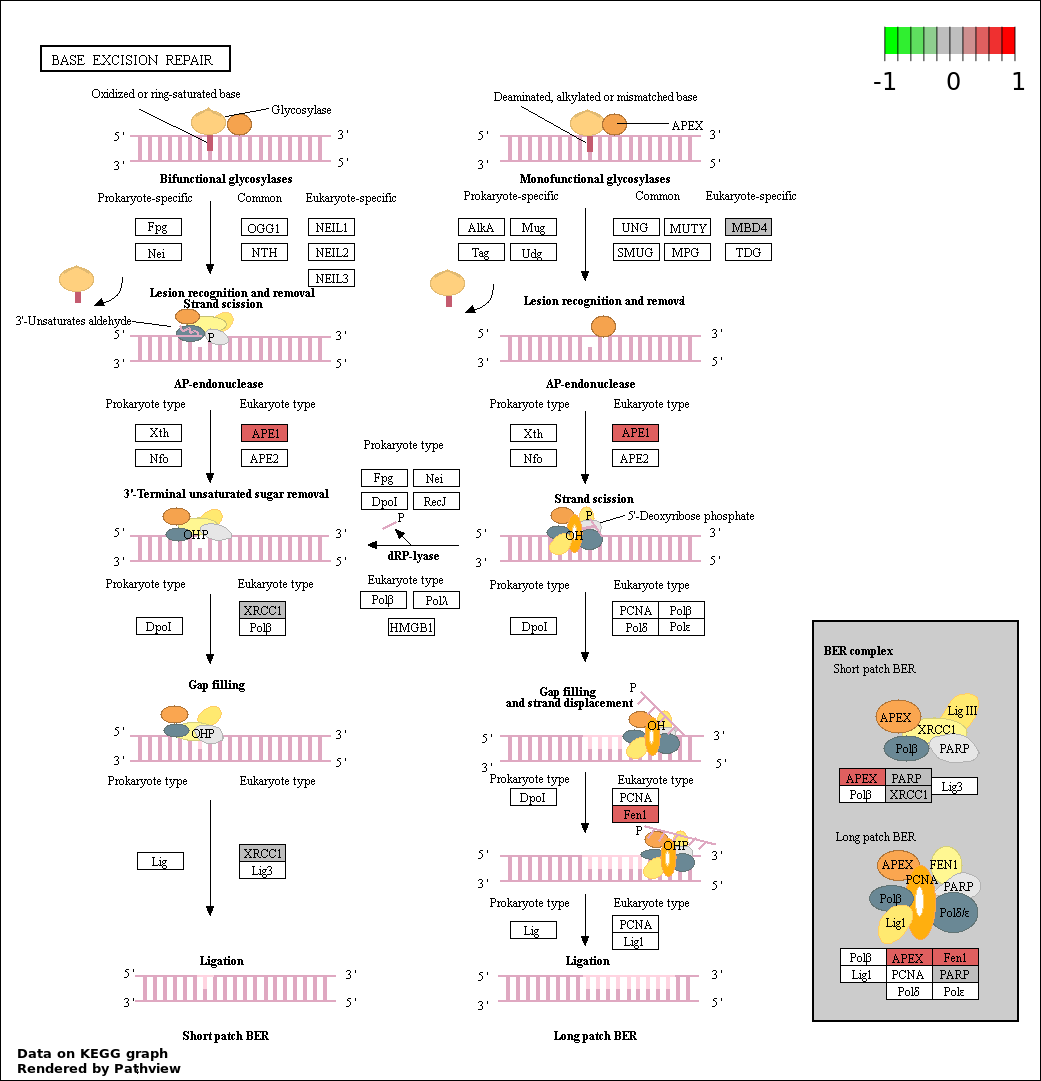
| 73 | BASE EXCISION REPAIR | 5 | 39 | 1.182e-05 | 0.0007535 |
| 74 | EPITHELIUM DEVELOPMENT | 21 | 945 | 1.21e-05 | 0.000761 |
| 75 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 28 | 1518 | 1.271e-05 | 0.0007639 |
| 76 | NEGATIVE REGULATION OF CELL DEATH | 20 | 872 | 1.262e-05 | 0.0007639 |
| 77 | NEURON FATE COMMITMENT | 6 | 67 | 1.281e-05 | 0.0007639 |
| 78 | CENTRAL NERVOUS SYSTEM DEVELOPMENT | 20 | 872 | 1.262e-05 | 0.0007639 |
| 79 | SINGLE ORGANISM CATABOLIC PROCESS | 21 | 957 | 1.462e-05 | 0.0008613 |
| 80 | TONGUE DEVELOPMENT | 4 | 20 | 1.488e-05 | 0.0008654 |
| 81 | EMBRYONIC ORGAN DEVELOPMENT | 13 | 406 | 1.616e-05 | 0.0009286 |
| 82 | POSITIVE REGULATION OF CELL PROLIFERATION | 19 | 814 | 1.662e-05 | 0.0009433 |
| 83 | AUTONOMIC NERVOUS SYSTEM DEVELOPMENT | 5 | 42 | 1.714e-05 | 0.0009611 |
| 84 | RESPONSE TO HORMONE | 20 | 893 | 1.779e-05 | 0.0009848 |
| 85 | NEGATIVE REGULATION OF TRANSFERASE ACTIVITY | 12 | 351 | 1.82e-05 | 0.0009848 |
| 86 | POSITIVE REGULATION OF CELL DEATH | 16 | 605 | 1.814e-05 | 0.0009848 |
| 87 | REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 18 | 750 | 1.953e-05 | 0.0009986 |
| 88 | CRANIAL NERVE DEVELOPMENT | 5 | 43 | 1.928e-05 | 0.0009986 |
| 89 | NEGATIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 15 | 541 | 1.945e-05 | 0.0009986 |
| 90 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 19 | 823 | 1.935e-05 | 0.0009986 |
| 91 | NEGATIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 15 | 541 | 1.945e-05 | 0.0009986 |
| 92 | POSITIVE REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL | 9 | 196 | 2.161e-05 | 0.001093 |
| 93 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 26 | 1395 | 2.283e-05 | 0.00113 |
| 94 | POSITIVE REGULATION OF TRANSFERASE ACTIVITY | 16 | 616 | 2.259e-05 | 0.00113 |
| 95 | CELL MOTILITY | 19 | 835 | 2.362e-05 | 0.001145 |
| 96 | LOCALIZATION OF CELL | 19 | 835 | 2.362e-05 | 0.001145 |
| 97 | NEGATIVE REGULATION OF PHOSPHORYLATION | 13 | 422 | 2.422e-05 | 0.001162 |
| 98 | POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 23 | 1152 | 2.493e-05 | 0.001184 |
| 99 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 27 | 1492 | 2.611e-05 | 0.001227 |
| 100 | ESTABLISHMENT OF EPITHELIAL CELL POLARITY | 4 | 23 | 2.671e-05 | 0.00123 |
| 101 | CRANIAL NERVE MORPHOGENESIS | 4 | 23 | 2.671e-05 | 0.00123 |
| 102 | NEGATIVE REGULATION OF MOLECULAR FUNCTION | 22 | 1079 | 2.779e-05 | 0.001268 |
| 103 | SENSORY ORGAN DEVELOPMENT | 14 | 493 | 2.872e-05 | 0.001297 |
| 104 | CELL PART MORPHOGENESIS | 16 | 633 | 3.138e-05 | 0.001391 |
| 105 | REGULATION OF GROWTH | 16 | 633 | 3.138e-05 | 0.001391 |
| 106 | NEGATIVE REGULATION OF LOCOMOTION | 10 | 263 | 3.804e-05 | 0.001654 |
| 107 | NEGATIVE REGULATION OF CELL PROLIFERATION | 16 | 643 | 3.787e-05 | 0.001654 |
| 108 | IMMUNE SYSTEM PROCESS | 32 | 1984 | 4.119e-05 | 0.001774 |
| 109 | RESPONSE TO ENDOGENOUS STIMULUS | 26 | 1450 | 4.4e-05 | 0.001878 |
| 110 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 18 | 801 | 4.622e-05 | 0.001955 |
| 111 | REGULATION OF CATABOLIC PROCESS | 17 | 731 | 4.993e-05 | 0.002093 |
| 112 | REGULATION OF MAPK CASCADE | 16 | 660 | 5.164e-05 | 0.002142 |
| 113 | PROTEOLYSIS | 23 | 1208 | 5.201e-05 | 0.002142 |
| 114 | POSITIVE REGULATION OF GENE EXPRESSION | 29 | 1733 | 5.274e-05 | 0.002153 |
| 115 | RESPONSE TO LIPID | 19 | 888 | 5.432e-05 | 0.002185 |
| 116 | POSITIVE REGULATION OF CELL CYCLE | 11 | 332 | 5.446e-05 | 0.002185 |
| 117 | ESTABLISHMENT OF CELL POLARITY | 6 | 88 | 6.09e-05 | 0.002422 |
| 118 | CELLULAR RESPONSE TO ORGANIC SUBSTANCE | 30 | 1848 | 6.693e-05 | 0.002639 |
| 119 | LOW DENSITY LIPOPROTEIN PARTICLE REMODELING | 3 | 11 | 7.06e-05 | 0.002761 |
| 120 | NEGATIVE REGULATION OF GENE EXPRESSION | 26 | 1493 | 7.153e-05 | 0.002761 |
| 121 | NEGATIVE REGULATION OF CATALYTIC ACTIVITY | 18 | 829 | 7.181e-05 | 0.002761 |
| 122 | EMBRYONIC MORPHOGENESIS | 14 | 539 | 7.513e-05 | 0.002842 |
| 123 | NEGATIVE REGULATION OF CELL DIFFERENTIATION | 15 | 609 | 7.501e-05 | 0.002842 |
| 124 | INNER EAR MORPHOGENESIS | 6 | 92 | 7.813e-05 | 0.002932 |
| 125 | REGULATION OF CELLULAR RESPONSE TO STRESS | 16 | 691 | 8.846e-05 | 0.003293 |
| 126 | NEGATIVE REGULATION OF GROWTH | 9 | 236 | 9.151e-05 | 0.00336 |
| 127 | CELLULAR RESPONSE TO OXIDATIVE STRESS | 8 | 184 | 9.17e-05 | 0.00336 |
| 128 | NEGATIVE REGULATION OF PROTEIN METABOLIC PROCESS | 21 | 1087 | 9.281e-05 | 0.003374 |
| 129 | NEGATIVE REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 4 | 32 | 0.0001028 | 0.003707 |
| 130 | REGULATION OF ORGAN MORPHOGENESIS | 9 | 242 | 0.0001107 | 0.003964 |
| 131 | NEGATIVE REGULATION OF NEURON DIFFERENTIATION | 8 | 191 | 0.0001188 | 0.004187 |
| 132 | PROTEIN PHOSPHORYLATION | 19 | 944 | 0.0001215 | 0.004187 |
| 133 | SEMAPHORIN PLEXIN SIGNALING PATHWAY INVOLVED IN NEURON PROJECTION GUIDANCE | 3 | 13 | 0.000121 | 0.004187 |
| 134 | NEGATIVE REGULATION OF CELL COMMUNICATION | 22 | 1192 | 0.0001209 | 0.004187 |
| 135 | RESPONSE TO REACTIVE OXYGEN SPECIES | 8 | 191 | 0.0001188 | 0.004187 |
| 136 | ORGANELLE FISSION | 13 | 496 | 0.000125 | 0.004216 |
| 137 | BLOOD VESSEL MORPHOGENESIS | 11 | 364 | 0.0001233 | 0.004216 |
| 138 | LIMBIC SYSTEM DEVELOPMENT | 6 | 100 | 0.0001242 | 0.004216 |
| 139 | EAR DEVELOPMENT | 8 | 195 | 0.0001371 | 0.00459 |
| 140 | T CELL RECEPTOR SIGNALING PATHWAY | 7 | 146 | 0.0001389 | 0.004615 |
| 141 | NEGATIVE REGULATION OF KINASE ACTIVITY | 9 | 250 | 0.0001416 | 0.004672 |
| 142 | DEVELOPMENTAL GROWTH INVOLVED IN MORPHOGENESIS | 6 | 104 | 0.0001541 | 0.005015 |
| 143 | CELLULAR RESPONSE TO REACTIVE OXYGEN SPECIES | 6 | 104 | 0.0001541 | 0.005015 |
| 144 | FOREBRAIN GENERATION OF NEURONS | 5 | 66 | 0.0001553 | 0.005017 |
| 145 | ORGANOPHOSPHATE METABOLIC PROCESS | 18 | 885 | 0.0001629 | 0.005228 |
| 146 | NERVE DEVELOPMENT | 5 | 68 | 0.0001789 | 0.005701 |
| 147 | RESPONSE TO RETINOIC ACID | 6 | 107 | 0.0001801 | 0.005702 |
| 148 | ESTABLISHMENT OR MAINTENANCE OF APICAL BASAL CELL POLARITY | 4 | 37 | 0.0001832 | 0.005719 |
| 149 | ESTABLISHMENT OR MAINTENANCE OF BIPOLAR CELL POLARITY | 4 | 37 | 0.0001832 | 0.005719 |
| 150 | BEHAVIOR | 13 | 516 | 0.0001844 | 0.005719 |
| 151 | RESPONSE TO HYDROGEN PEROXIDE | 6 | 109 | 0.0001993 | 0.006142 |
| 152 | POSITIVE REGULATION OF LIGASE ACTIVITY | 6 | 110 | 0.0002095 | 0.006413 |
| 153 | COCHLEA DEVELOPMENT | 4 | 39 | 0.0002254 | 0.006856 |
| 154 | EAR MORPHOGENESIS | 6 | 112 | 0.0002311 | 0.006982 |
| 155 | CELL CYCLE PROCESS | 20 | 1081 | 0.0002448 | 0.007348 |
| 156 | HIPPOCAMPUS DEVELOPMENT | 5 | 73 | 0.0002499 | 0.007454 |
| 157 | CELL DEATH | 19 | 1001 | 0.0002568 | 0.007611 |
| 158 | REGULATION OF SKELETAL MUSCLE CELL DIFFERENTIATION | 3 | 17 | 0.0002812 | 0.008282 |
| 159 | VASCULATURE DEVELOPMENT | 12 | 469 | 0.0002848 | 0.008334 |
| 160 | REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 10 | 337 | 0.0002909 | 0.008406 |
| 161 | REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 12 | 470 | 0.0002903 | 0.008406 |
| 162 | NEGATIVE REGULATION OF RESPONSE TO STIMULUS | 23 | 1360 | 0.0002999 | 0.008615 |
| 163 | REGULATION OF EPITHELIAL CELL MIGRATION | 7 | 166 | 0.0003056 | 0.008723 |
| 164 | EMBRYONIC ORGAN MORPHOGENESIS | 9 | 279 | 0.0003197 | 0.008936 |
| 165 | REGULATION OF CELLULAR LOCALIZATION | 22 | 1277 | 0.0003188 | 0.008936 |
| 166 | RESPONSE TO GROWTH FACTOR | 12 | 475 | 0.0003195 | 0.008936 |
| 167 | DEVELOPMENTAL CELL GROWTH | 5 | 77 | 0.0003207 | 0.008936 |
| 168 | REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL | 9 | 280 | 0.0003282 | 0.00909 |
| 169 | CELL DEATH IN RESPONSE TO OXIDATIVE STRESS | 3 | 18 | 0.0003356 | 0.009185 |
| 170 | REGULATION OF DENDRITE DEVELOPMENT | 6 | 120 | 0.0003355 | 0.009185 |
| 171 | REGULATION OF BLOOD PRESSURE | 7 | 169 | 0.0003406 | 0.009227 |
| 172 | RESPONSE TO ABIOTIC STIMULUS | 19 | 1024 | 0.0003411 | 0.009227 |
| 173 | REGULATION OF BINDING | 9 | 283 | 0.0003548 | 0.009542 |
| 174 | EMBRYO DEVELOPMENT ENDING IN BIRTH OR EGG HATCHING | 13 | 554 | 0.0003654 | 0.009772 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | ENZYME BINDING | 44 | 1737 | 6.55e-13 | 6.085e-10 |
| 2 | RIBONUCLEOTIDE BINDING | 37 | 1860 | 5.28e-08 | 2.453e-05 |
| 3 | HISTONE DEACETYLASE BINDING | 9 | 105 | 1.205e-07 | 2.969e-05 |
| 4 | KINASE ACTIVITY | 23 | 842 | 1.278e-07 | 2.969e-05 |
| 5 | TRANSFERASE ACTIVITY TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 24 | 992 | 6.036e-07 | 0.0001121 |
| 6 | KINASE BINDING | 18 | 606 | 1.032e-06 | 0.0001598 |
| 7 | PROTEIN KINASE ACTIVITY | 18 | 640 | 2.236e-06 | 0.0002968 |
| 8 | PROTEIN COMPLEX BINDING | 22 | 935 | 2.95e-06 | 0.0003426 |
| 9 | MACROMOLECULAR COMPLEX BINDING | 26 | 1399 | 2.397e-05 | 0.002475 |
| 10 | ADENYL NUCLEOTIDE BINDING | 27 | 1514 | 3.372e-05 | 0.003132 |
| 11 | MOLECULAR FUNCTION REGULATOR | 25 | 1353 | 3.867e-05 | 0.003266 |
| 12 | TRANSCRIPTIONAL REPRESSOR ACTIVITY RNA POLYMERASE II TRANSCRIPTION REGULATORY REGION SEQUENCE SPECIFIC BINDING | 8 | 168 | 4.838e-05 | 0.003746 |
| 13 | IDENTICAL PROTEIN BINDING | 23 | 1209 | 5.267e-05 | 0.003764 |
| 14 | PROTEIN TYROSINE KINASE ACTIVITY | 8 | 176 | 6.718e-05 | 0.004099 |
| 15 | CHANNEL REGULATOR ACTIVITY | 7 | 131 | 7.035e-05 | 0.004099 |
| 16 | SEMAPHORIN RECEPTOR ACTIVITY | 3 | 11 | 7.06e-05 | 0.004099 |
| 17 | MAGNESIUM ION BINDING | 8 | 199 | 0.0001576 | 0.007943 |
| 18 | TRANSCRIPTIONAL REPRESSOR ACTIVITY RNA POLYMERASE II CORE PROMOTER PROXIMAL REGION SEQUENCE SPECIFIC BINDING | 6 | 105 | 0.0001625 | 0.007943 |
| 19 | THIOESTERASE BINDING | 3 | 14 | 0.0001531 | 0.007943 |
| 20 | DNA N GLYCOSYLASE ACTIVITY | 3 | 15 | 0.0001903 | 0.008437 |
| 21 | REGULATORY REGION NUCLEIC ACID BINDING | 17 | 818 | 0.0001941 | 0.008437 |
| 22 | HYDROLASE ACTIVITY ACTING ON ACID ANHYDRIDES | 17 | 820 | 0.0001998 | 0.008437 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CELL PROJECTION | 33 | 1786 | 1.733e-06 | 0.001012 |
| 2 | CELL PROJECTION PART | 21 | 946 | 1.23e-05 | 0.003591 |
| 3 | RUFFLE | 8 | 156 | 2.852e-05 | 0.005552 |
| 4 | SOMATODENDRITIC COMPARTMENT | 16 | 650 | 4.308e-05 | 0.00629 |
| 5 | SEMAPHORIN RECEPTOR COMPLEX | 3 | 11 | 7.06e-05 | 0.008246 |
| 6 | CELL LEADING EDGE | 11 | 350 | 8.726e-05 | 0.008493 |
| 7 | NEURON PROJECTION | 19 | 942 | 0.0001182 | 0.009859 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
| 1 | hsa03410_Base_excision_repair | 5 | 34 | 5.893e-06 | 0.001061 | |
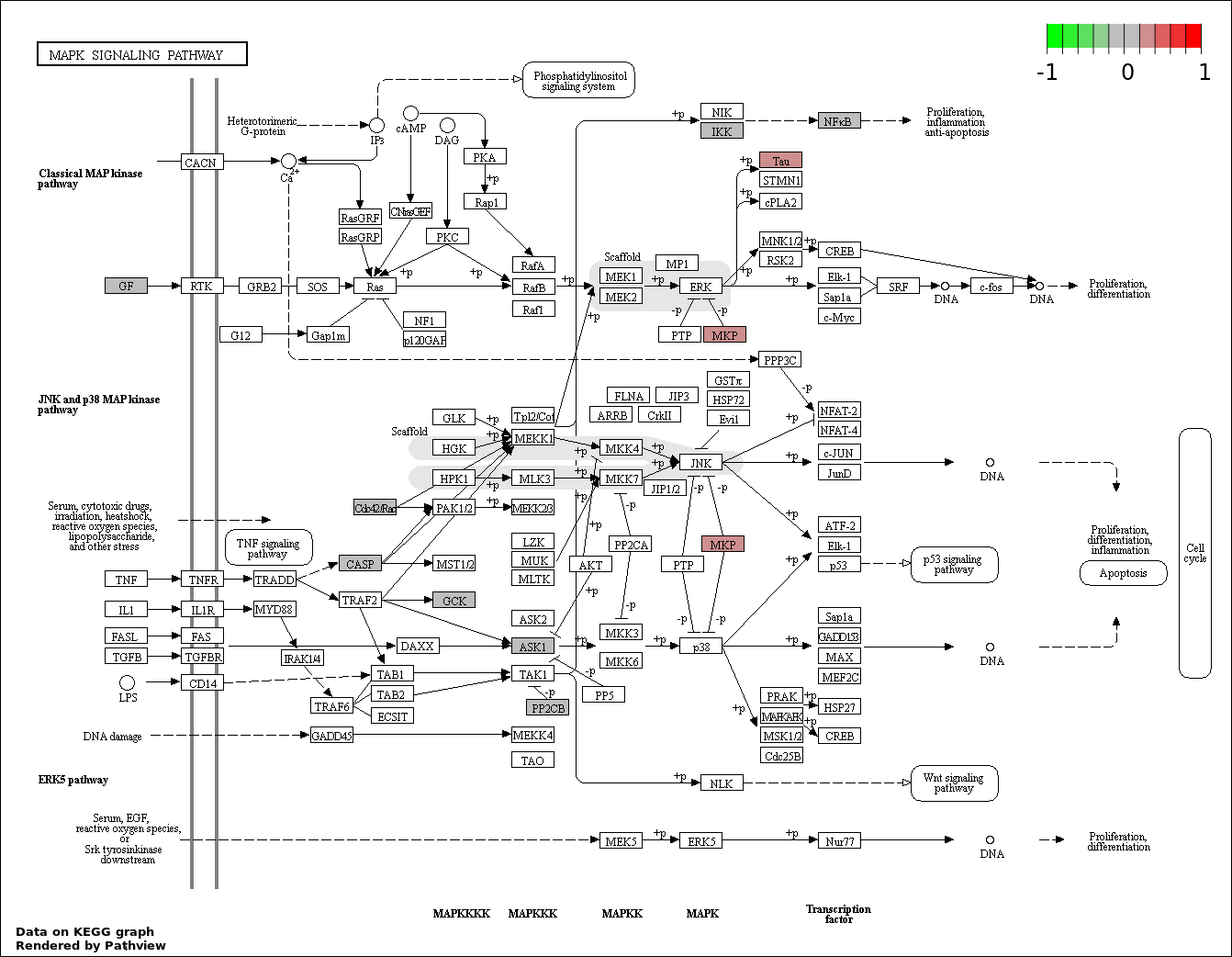
| 2 | hsa04010_MAPK_signaling_pathway | 10 | 268 | 4.459e-05 | 0.003679 | |
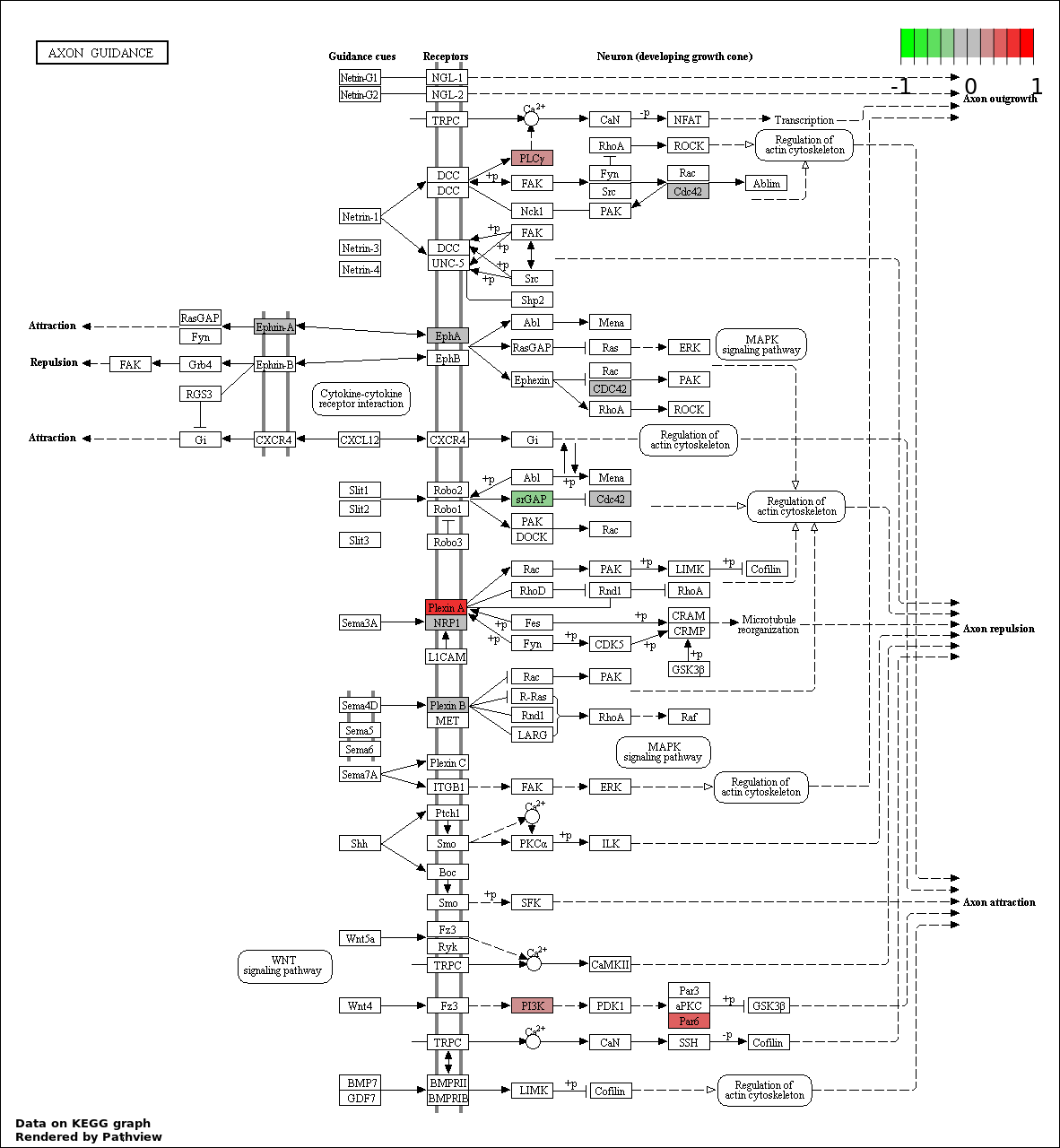
| 3 | hsa04360_Axon_guidance | 7 | 130 | 6.702e-05 | 0.003679 | |
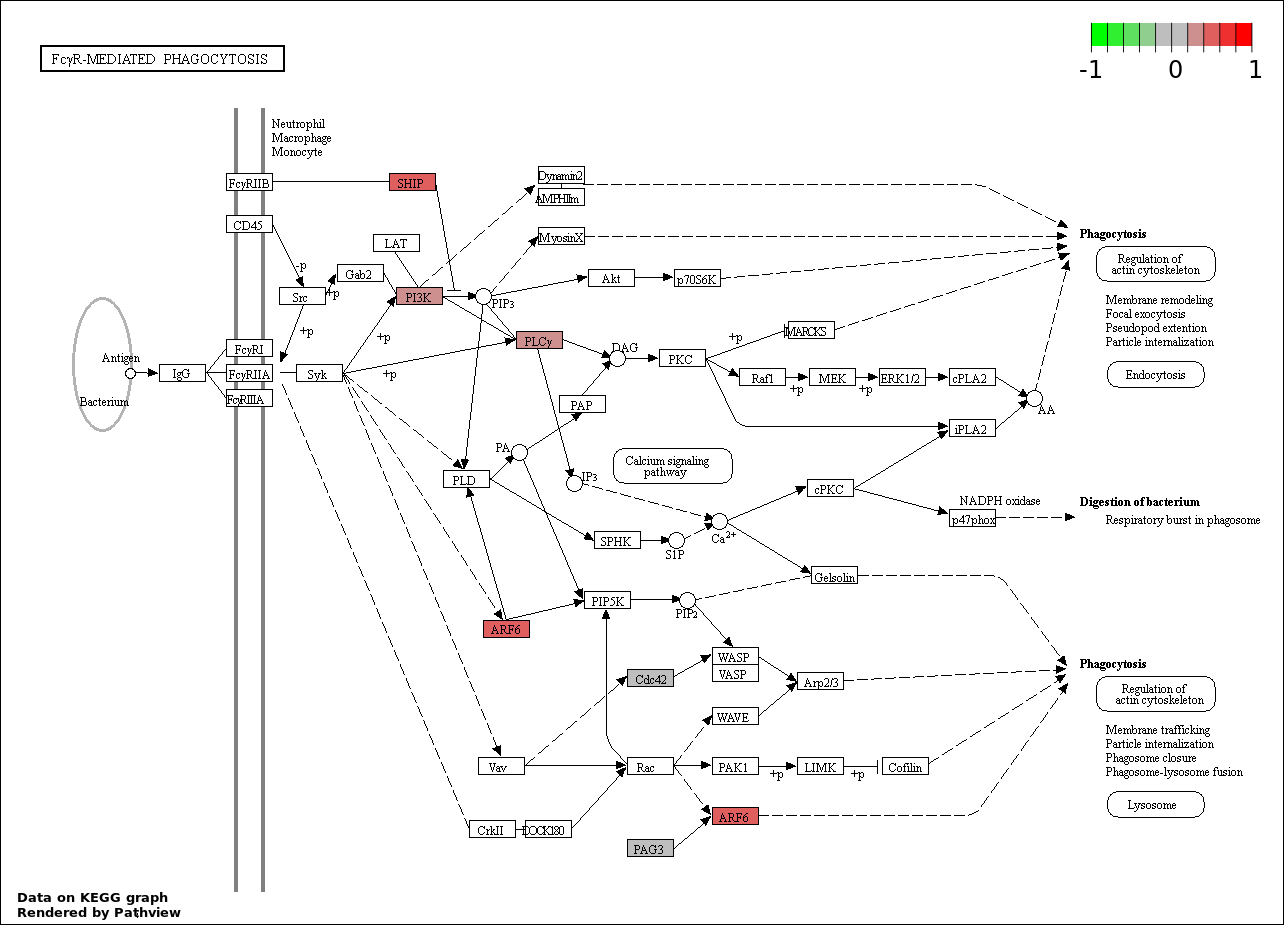
| 4 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 6 | 95 | 9.344e-05 | 0.003679 | |
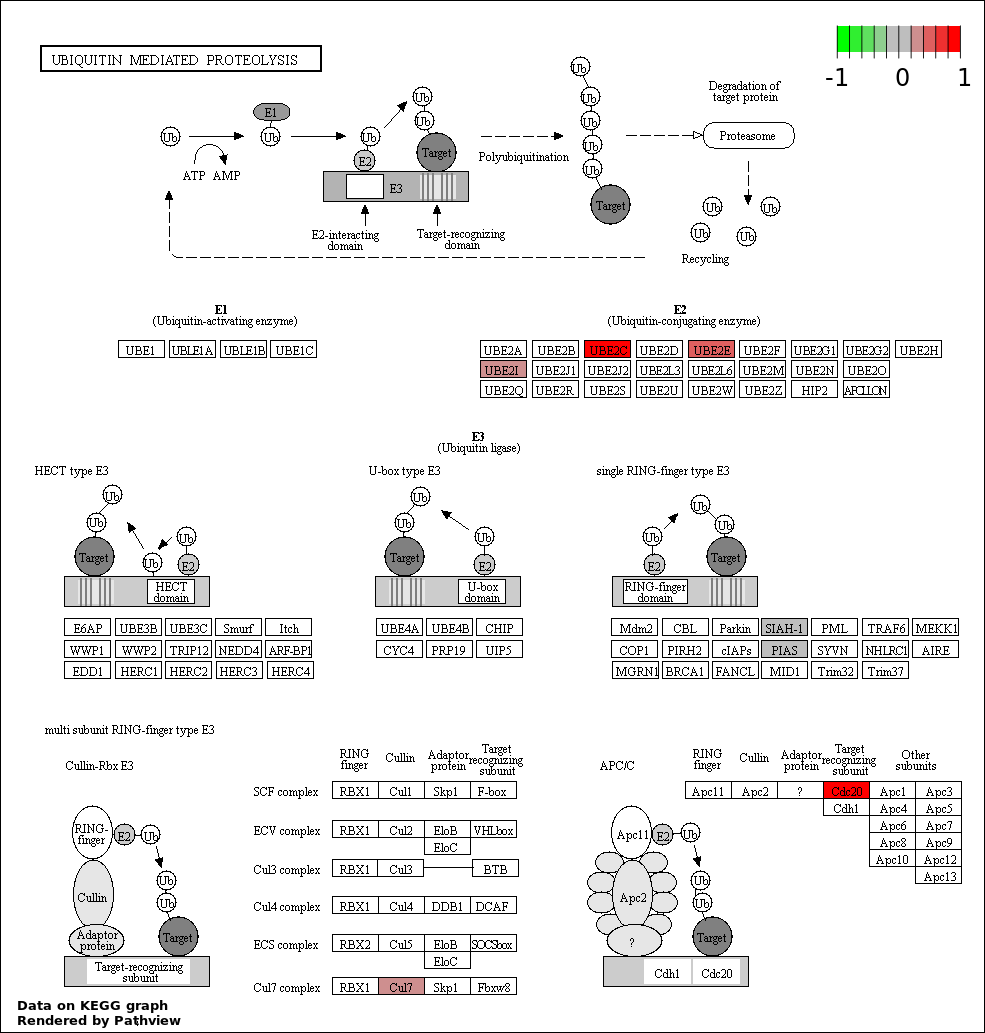
| 5 | hsa04120_Ubiquitin_mediated_proteolysis | 7 | 139 | 0.0001022 | 0.003679 | |
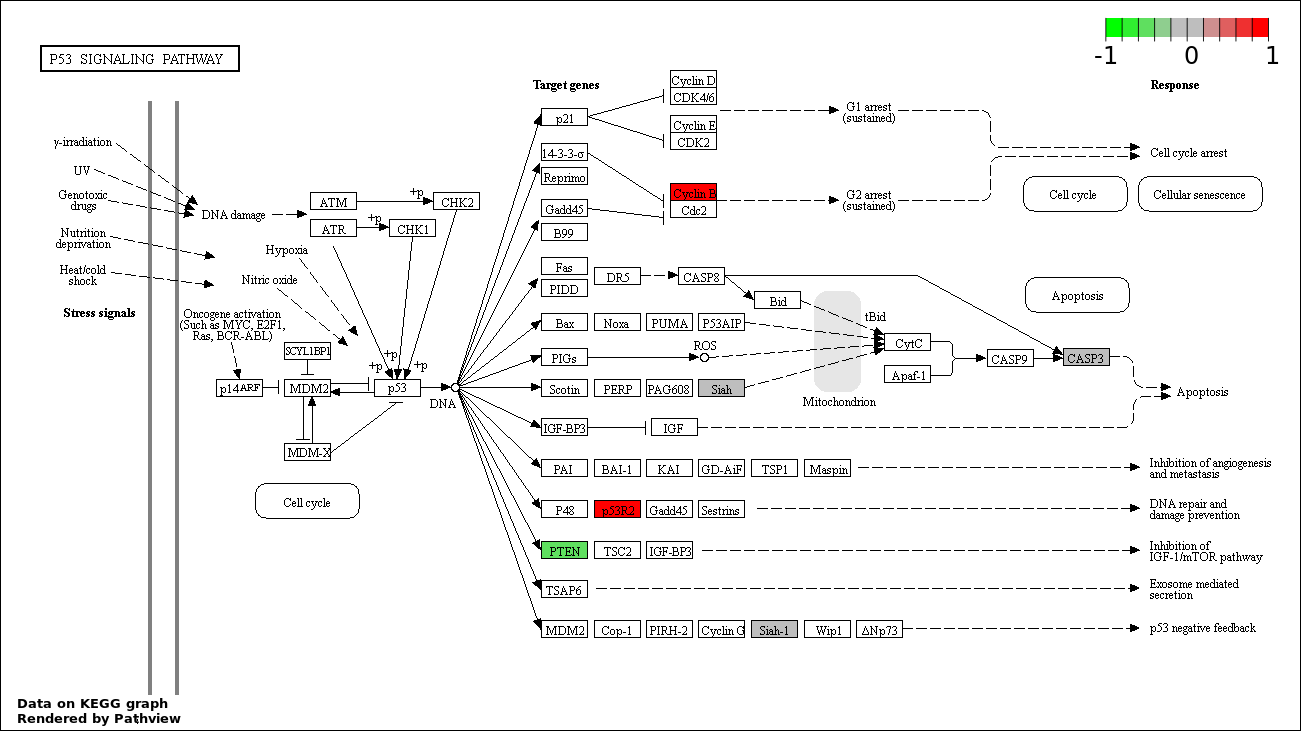
| 6 | hsa04115_p53_signaling_pathway | 5 | 69 | 0.0001917 | 0.00575 | |
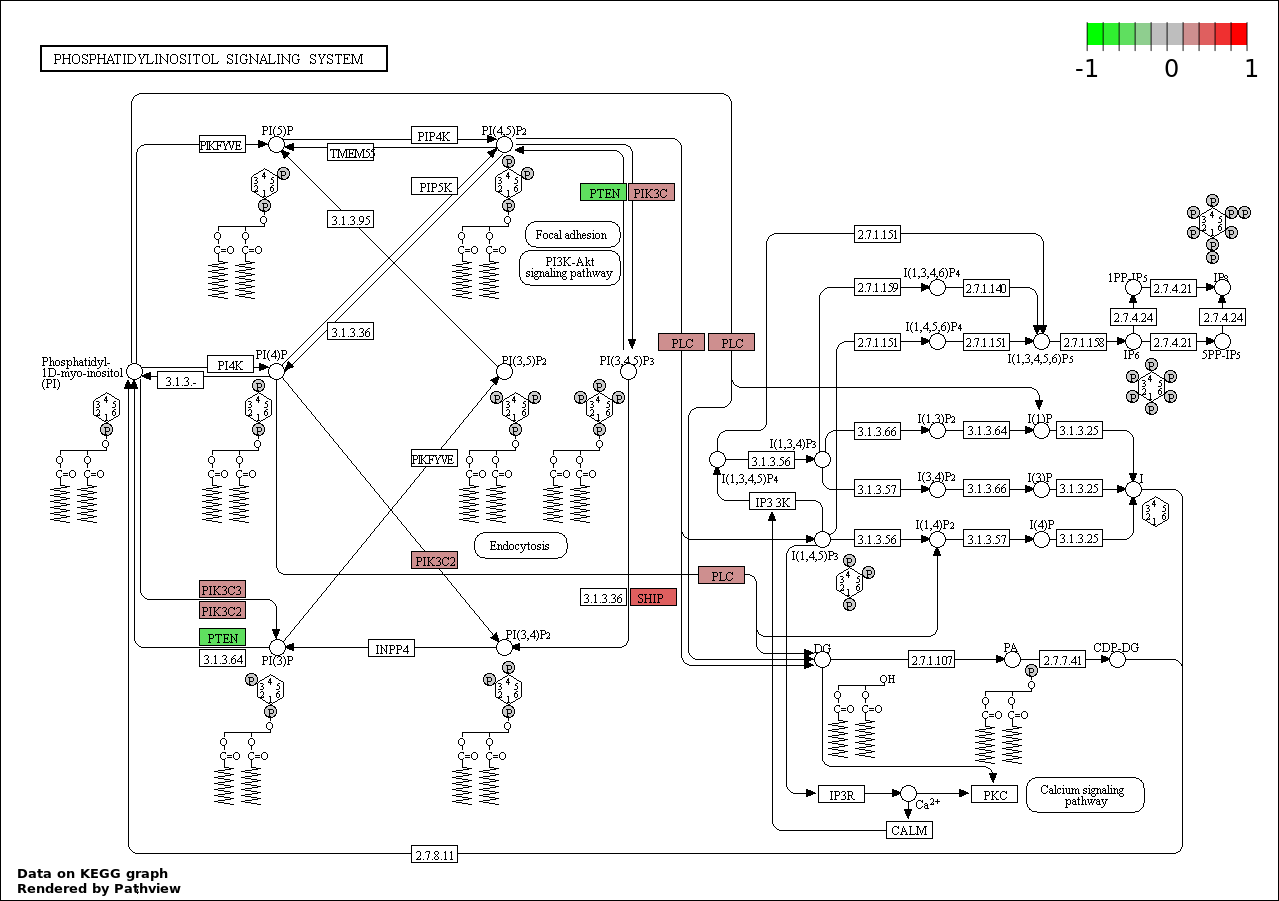
| 7 | hsa04070_Phosphatidylinositol_signaling_system | 5 | 78 | 0.0003406 | 0.008758 | |
| 8 | hsa04722_Neurotrophin_signaling_pathway | 6 | 127 | 0.0004543 | 0.00994 | |
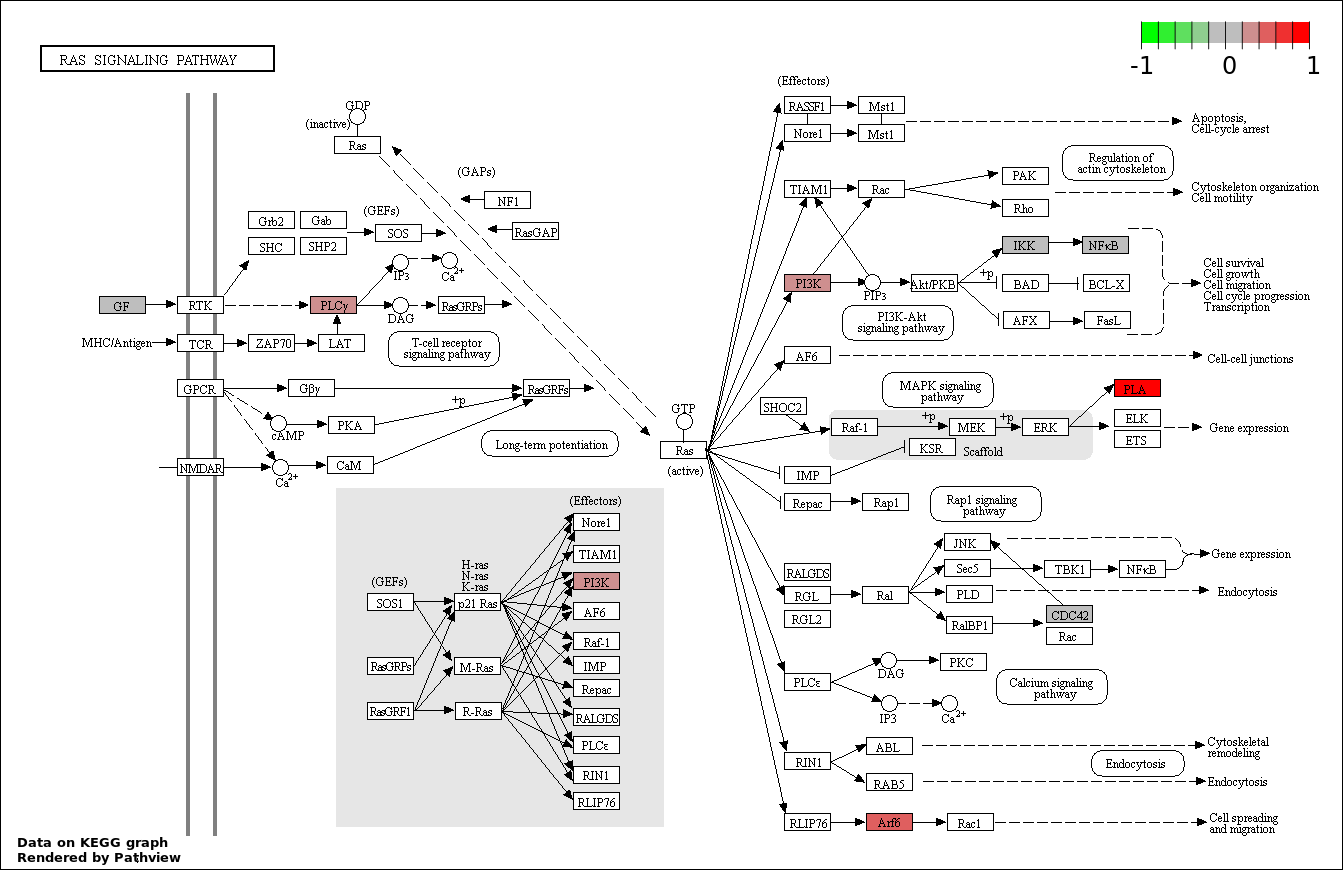
| 9 | hsa04014_Ras_signaling_pathway | 8 | 236 | 0.000497 | 0.00994 | |
| 10 | hsa00562_Inositol_phosphate_metabolism | 4 | 57 | 0.0009725 | 0.0175 | |
| 11 | hsa04660_T_cell_receptor_signaling_pathway | 5 | 108 | 0.001495 | 0.02447 | |
| 12 | hsa04670_Leukocyte_transendothelial_migration | 5 | 117 | 0.002127 | 0.03191 | |
| 13 | hsa04370_VEGF_signaling_pathway | 4 | 76 | 0.002822 | 0.03768 | |
| 14 | hsa04110_Cell_cycle | 5 | 128 | 0.00314 | 0.03768 | |
| 15 | hsa04380_Osteoclast_differentiation | 5 | 128 | 0.00314 | 0.03768 | |
| 16 | hsa04210_Apoptosis | 4 | 89 | 0.004973 | 0.05594 | |
| 17 | hsa04916_Melanogenesis | 4 | 101 | 0.007747 | 0.08203 | |
| 18 | hsa04710_Circadian_rhythm_._mammal | 2 | 23 | 0.0134 | 0.1308 | |
| 19 | hsa00010_Glycolysis_._Gluconeogenesis | 3 | 65 | 0.0138 | 0.1308 | |
| 20 | hsa04062_Chemokine_signaling_pathway | 5 | 189 | 0.01561 | 0.1405 | |
| 21 | hsa04530_Tight_junction | 4 | 133 | 0.0196 | 0.1611 | |
| 22 | hsa04662_B_cell_receptor_signaling_pathway | 3 | 75 | 0.02019 | 0.1611 | |
| 23 | hsa04144_Endocytosis | 5 | 203 | 0.02058 | 0.1611 | |
| 24 | hsa04664_Fc_epsilon_RI_signaling_pathway | 3 | 79 | 0.02313 | 0.1735 | |
| 25 | hsa04310_Wnt_signaling_pathway | 4 | 151 | 0.02948 | 0.2058 | |
| 26 | hsa00565_Ether_lipid_metabolism | 2 | 36 | 0.03128 | 0.2058 | |
| 27 | hsa04390_Hippo_signaling_pathway | 4 | 154 | 0.03137 | 0.2058 | |
| 28 | hsa04630_Jak.STAT_signaling_pathway | 4 | 155 | 0.03201 | 0.2058 | |
| 29 | hsa00620_Pyruvate_metabolism | 2 | 40 | 0.03796 | 0.2356 | |
| 30 | hsa00350_Tyrosine_metabolism | 2 | 41 | 0.03971 | 0.2383 | |
| 31 | hsa00240_Pyrimidine_metabolism | 3 | 99 | 0.04109 | 0.2386 | |
| 32 | hsa04912_GnRH_signaling_pathway | 3 | 101 | 0.04318 | 0.2414 | |
| 33 | hsa04620_Toll.like_receptor_signaling_pathway | 3 | 102 | 0.04425 | 0.2414 | |
| 34 | hsa04330_Notch_signaling_pathway | 2 | 47 | 0.05082 | 0.2691 | |
| 35 | hsa04151_PI3K_AKT_signaling_pathway | 6 | 351 | 0.0546 | 0.2808 | |
| 36 | hsa04510_Focal_adhesion | 4 | 200 | 0.06907 | 0.3281 | |
| 37 | hsa04340_Hedgehog_signaling_pathway | 2 | 56 | 0.06926 | 0.3281 | |
| 38 | hsa04623_Cytosolic_DNA.sensing_pathway | 2 | 56 | 0.06926 | 0.3281 | |
| 39 | hsa04621_NOD.like_receptor_signaling_pathway | 2 | 59 | 0.07583 | 0.35 | |
| 40 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 3 | 136 | 0.08784 | 0.3953 | |
| 41 | hsa04920_Adipocytokine_signaling_pathway | 2 | 68 | 0.09661 | 0.4242 | |
| 42 | hsa04730_Long.term_depression | 2 | 70 | 0.1014 | 0.4249 | |
| 43 | hsa04622_RIG.I.like_receptor_signaling_pathway | 2 | 71 | 0.1039 | 0.4249 | |
| 44 | hsa00564_Glycerophospholipid_metabolism | 2 | 80 | 0.1264 | 0.4948 | |
| 45 | hsa04350_TGF.beta_signaling_pathway | 2 | 85 | 0.1394 | 0.5108 | |
| 46 | hsa04012_ErbB_signaling_pathway | 2 | 87 | 0.1447 | 0.5108 | |
| 47 | hsa04914_Progesterone.mediated_oocyte_maturation | 2 | 87 | 0.1447 | 0.5108 | |
| 48 | hsa04540_Gap_junction | 2 | 90 | 0.1527 | 0.5286 | |
| 49 | hsa04972_Pancreatic_secretion | 2 | 101 | 0.1827 | 0.6205 | |
| 50 | hsa04114_Oocyte_meiosis | 2 | 114 | 0.2191 | 0.7048 | |
| 51 | hsa04270_Vascular_smooth_muscle_contraction | 2 | 116 | 0.2248 | 0.7097 | |
| 52 | hsa03040_Spliceosome | 2 | 128 | 0.2589 | 0.7516 | |
| 53 | hsa04910_Insulin_signaling_pathway | 2 | 138 | 0.2874 | 0.7991 | |
| 54 | hsa03013_RNA_transport | 2 | 152 | 0.3271 | 0.8283 | |
| 55 | hsa04145_Phagosome | 2 | 156 | 0.3384 | 0.831 | |
| 56 | hsa00230_Purine_metabolism | 2 | 162 | 0.3551 | 0.8523 | |
| 57 | hsa04141_Protein_processing_in_endoplasmic_reticulum | 2 | 168 | 0.3717 | 0.869 | |
| 58 | hsa04020_Calcium_signaling_pathway | 2 | 177 | 0.3963 | 0.9146 | |
| 59 | hsa04810_Regulation_of_actin_cytoskeleton | 2 | 214 | 0.4923 | 1 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | RP11-887P2.5 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-340-5p;hsa-miR-3613-5p;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | PTEN | Sponge network | -2.139 | 0 | -0.594 | 0 | 0.452 |
| 2 | MAGI2-AS3 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-708-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 26 | PTEN | Sponge network | -1.613 | 0 | -0.594 | 0 | 0.44 |
| 3 | AF131217.1 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-217;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-425-5p;hsa-miR-7-1-3p;hsa-miR-708-3p;hsa-miR-93-5p | 25 | PTEN | Sponge network | -1.829 | 0 | -0.594 | 0 | 0.435 |
| 4 | DNM3OS | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | PTEN | Sponge network | -1.145 | 0 | -0.594 | 0 | 0.432 |
| 5 | ADAMTS9-AS1 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-217;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 29 | PTEN | Sponge network | -2.702 | 0 | -0.594 | 0 | 0.43 |
| 6 | RP11-166D19.1 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-708-3p;hsa-miR-92a-3p | 26 | PTEN | Sponge network | -1.306 | 0 | -0.594 | 0 | 0.411 |
| 7 | ADAMTS9-AS2 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-708-3p;hsa-miR-92a-3p | 23 | PTEN | Sponge network | -1.864 | 0 | -0.594 | 0 | 0.405 |
| 8 | CTC-296K1.3 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-217;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-454-3p | 17 | PTEN | Sponge network | -2.102 | 0 | -0.594 | 0 | 0.404 |
| 9 | RP11-356J5.12 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-195-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-217;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-7-1-3p;hsa-miR-708-3p;hsa-miR-92a-3p | 24 | PTEN | Sponge network | -1.453 | 0 | -0.594 | 0 | 0.389 |
| 10 | RP11-60A24.3 | hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-21-5p;hsa-miR-217;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-3613-5p;hsa-miR-484 | 11 | PTEN | Sponge network | -1.965 | 0 | -0.594 | 0 | 0.376 |
| 11 | RP13-514E23.1 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | PTEN | Sponge network | -0.744 | 0 | -0.594 | 0 | 0.374 |
| 12 | MIR143HG | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-708-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 24 | PTEN | Sponge network | -1.391 | 0 | -0.594 | 0 | 0.371 |
| 13 | HCG11 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-195-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-217;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-340-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | PTEN | Sponge network | -1.393 | 0 | -0.594 | 0 | 0.363 |
| 14 | LINC00672 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-32-5p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-93-5p | 13 | PTEN | Sponge network | -1.467 | 0 | -0.594 | 0 | 0.362 |
| 15 | GAS6-AS2 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-708-3p;hsa-miR-92a-3p | 21 | PTEN | Sponge network | -1.442 | 0 | -0.594 | 0 | 0.356 |
| 16 | MIR133A1 | hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-217;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-7-1-3p | 11 | PTEN | Sponge network | -1.721 | 0 | -0.594 | 0 | 0.355 |
| 17 | PGM5-AS1 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-217;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-484;hsa-miR-7-1-3p | 16 | PTEN | Sponge network | -2.475 | 0 | -0.594 | 0 | 0.35 |
| 18 | BOLA3-AS1 | hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-454-3p;hsa-miR-7-1-3p | 14 | PTEN | Sponge network | -1.462 | 0 | -0.594 | 0 | 0.348 |
| 19 | RP11-571L19.8 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-3613-5p;hsa-miR-7-1-3p | 10 | PTEN | Sponge network | -0.496 | 0 | -0.594 | 0 | 0.345 |
| 20 | LINC00595 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-217;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-484;hsa-miR-7-1-3p | 17 | PTEN | Sponge network | -2.01 | 0 | -0.594 | 0 | 0.344 |
| 21 | BZRAP1-AS1 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-93-5p | 14 | PTEN | Sponge network | -0.876 | 0 | -0.594 | 0 | 0.341 |
| 22 | FENDRR | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-217;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | PTEN | Sponge network | -1.142 | 0 | -0.594 | 0 | 0.328 |
| 23 | LINC01018 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-195-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-217;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-425-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | PTEN | Sponge network | -1.956 | 0 | -0.594 | 0 | 0.327 |
| 24 | C20orf166-AS1 | hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-93-5p | 16 | PTEN | Sponge network | -1.771 | 0 | -0.594 | 0 | 0.327 |
| 25 | RP11-532F6.3 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p | 12 | PTEN | Sponge network | -1.109 | 0 | -0.594 | 0 | 0.323 |
| 26 | RP11-401P9.4 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-708-3p;hsa-miR-93-5p | 21 | PTEN | Sponge network | -1.073 | 0 | -0.594 | 0 | 0.323 |
| 27 | RP11-1006G14.4 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 10 | PTEN | Sponge network | -0.455 | 0 | -0.594 | 0 | 0.317 |
| 28 | RP11-705C15.3 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-425-5p;hsa-miR-484;hsa-miR-7-1-3p | 15 | PTEN | Sponge network | -0.62 | 0 | -0.594 | 0 | 0.317 |
| 29 | RP11-399O19.9 | hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-3613-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 11 | PTEN | Sponge network | -0.764 | 3.0E-5 | -0.594 | 0 | 0.316 |
| 30 | ACTA2-AS1 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-7-1-3p | 18 | PTEN | Sponge network | -1.544 | 0 | -0.594 | 0 | 0.315 |
| 31 | EMX2OS | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-195-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-217;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 24 | PTEN | Sponge network | -3.747 | 0 | -0.594 | 0 | 0.309 |
| 32 | RP11-389C8.2 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | PTEN | Sponge network | -0.511 | 0.00014 | -0.594 | 0 | 0.309 |
| 33 | RP11-822E23.8 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-93-5p | 18 | PTEN | Sponge network | -1.845 | 0 | -0.594 | 0 | 0.304 |
| 34 | RP11-999E24.3 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-340-5p;hsa-miR-425-5p | 13 | PTEN | Sponge network | -1.094 | 0 | -0.594 | 0 | 0.3 |
| 35 | TBX5-AS1 | hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-217;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 18 | PTEN | Sponge network | -0.969 | 0.00412 | -0.594 | 0 | 0.3 |
| 36 | RP11-13K12.5 | hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | PTEN | Sponge network | -1.385 | 0 | -0.594 | 0 | 0.294 |
| 37 | FRMD6-AS2 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-217 | 11 | PTEN | Sponge network | -2.797 | 0 | -0.594 | 0 | 0.293 |
| 38 | CTD-2013N24.2 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-708-3p;hsa-miR-93-5p | 18 | PTEN | Sponge network | -0.665 | 0 | -0.594 | 0 | 0.291 |
| 39 | U91328.19 | hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-425-5p;hsa-miR-484 | 12 | PTEN | Sponge network | -0.418 | 2.0E-5 | -0.594 | 0 | 0.286 |
| 40 | CTB-92J24.3 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | PTEN | Sponge network | -1.626 | 0.00109 | -0.594 | 0 | 0.281 |
| 41 | AC003090.1 | hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | PTEN | Sponge network | -1.216 | 0 | -0.594 | 0 | 0.277 |
| 42 | RP11-13K12.1 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-93-5p | 19 | PTEN | Sponge network | -1.408 | 0 | -0.594 | 0 | 0.275 |
| 43 | AP001046.5 | hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-425-5p;hsa-miR-484 | 13 | PTEN | Sponge network | -1.665 | 0 | -0.594 | 0 | 0.273 |
| 44 | RP11-686D22.8 | hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-335-3p | 10 | PTEN | Sponge network | -1.003 | 0 | -0.594 | 0 | 0.268 |
| 45 | RP11-284N8.3 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-3613-5p;hsa-miR-425-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | PTEN | Sponge network | -0.322 | 0.14616 | -0.594 | 0 | 0.268 |
| 46 | RP11-696N14.1 | hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-340-5p;hsa-miR-93-5p | 10 | PTEN | Sponge network | -0.577 | 0.00046 | -0.594 | 0 | 0.266 |
| 47 | RP11-121A8.1 | hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-340-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-7-1-3p | 15 | PTEN | Sponge network | -0.598 | 0.06897 | -0.594 | 0 | 0.265 |
| 48 | H19 | hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-484;hsa-miR-93-5p | 12 | PTEN | Sponge network | -2.173 | 0 | -0.594 | 0 | 0.263 |
| 49 | SNHG18 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-195-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-340-5p;hsa-miR-454-3p;hsa-miR-93-5p | 18 | PTEN | Sponge network | -1.812 | 0 | -0.594 | 0 | 0.261 |
| 50 | CTD-3064M3.3 | hsa-miR-103a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-429 | 10 | PTEN | Sponge network | -0.671 | 2.0E-5 | -0.594 | 0 | 0.261 |
| 51 | RP11-456K23.1 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-484;hsa-miR-92a-3p;hsa-miR-93-5p | 17 | PTEN | Sponge network | -0.554 | 0 | -0.594 | 0 | 0.258 |
| 52 | CTD-2314G24.2 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | PTEN | Sponge network | -0.781 | 2.0E-5 | -0.594 | 0 | 0.257 |
| 53 | LINC00702 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 26 | PTEN | Sponge network | -1.189 | 0 | -0.594 | 0 | 0.256 |
| 54 | LINC00641 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-708-3p | 20 | PTEN | Sponge network | -0.825 | 0 | -0.594 | 0 | 0.254 |
| 55 | LINC00883 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-425-5p;hsa-miR-484 | 18 | PTEN | Sponge network | -1.026 | 0 | -0.594 | 0 | 0.254 |
| 56 | CTD-2521M24.9 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | PTEN | Sponge network | -0.843 | 0 | -0.594 | 0 | 0.252 |
| 57 | ZNF883 | hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-340-5p | 10 | PTEN | Sponge network | -1.034 | 1.0E-5 | -0.594 | 0 | 0.25 |