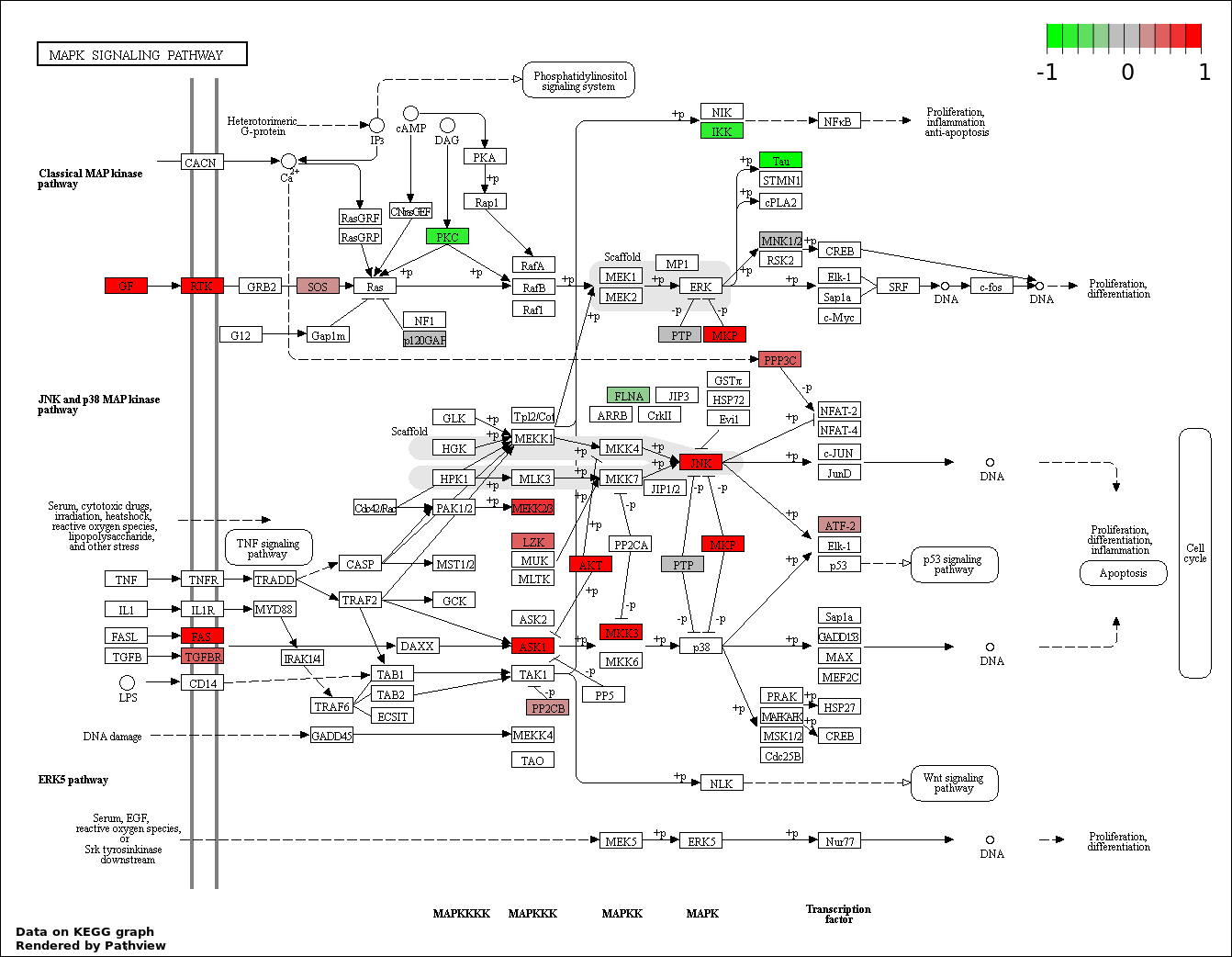
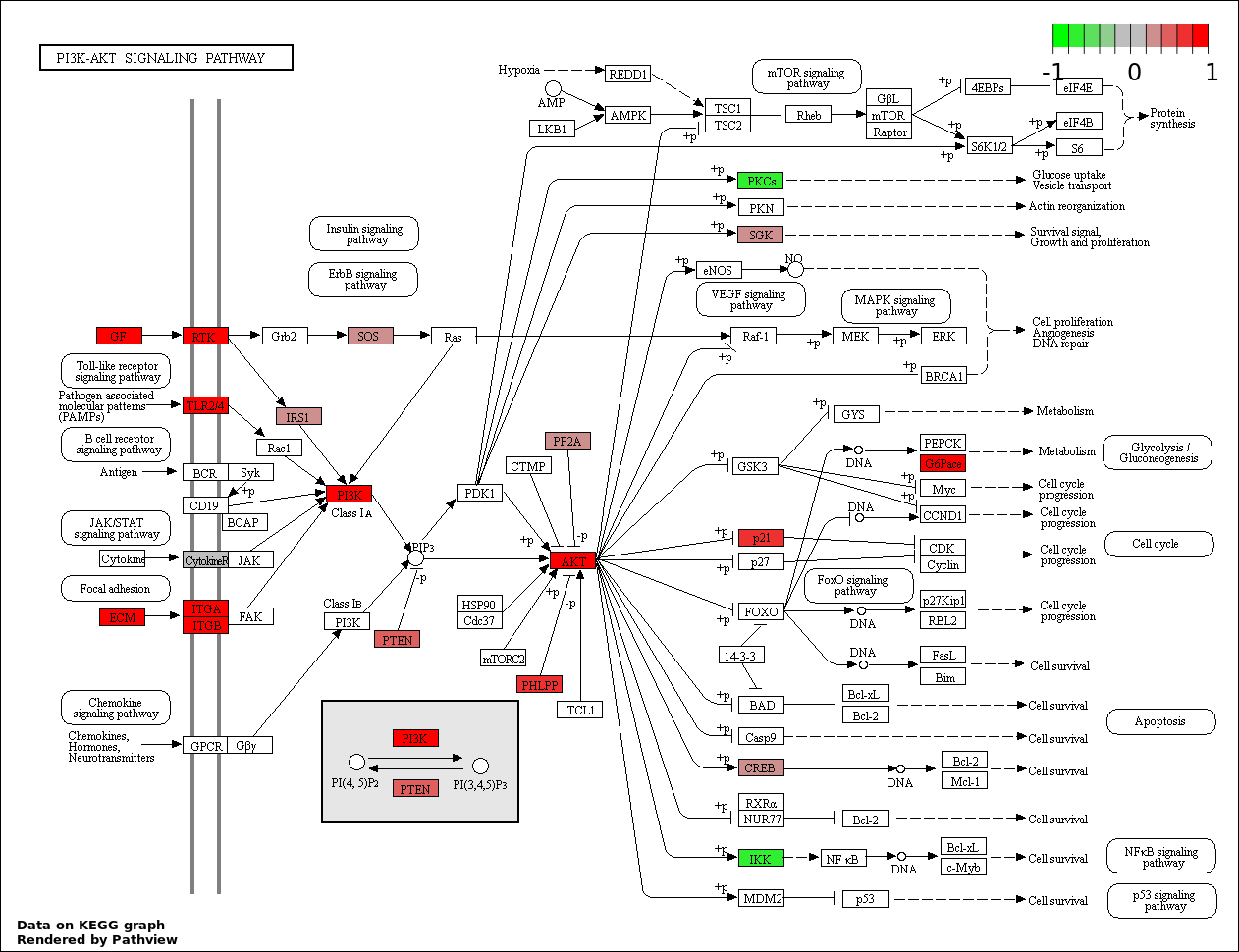
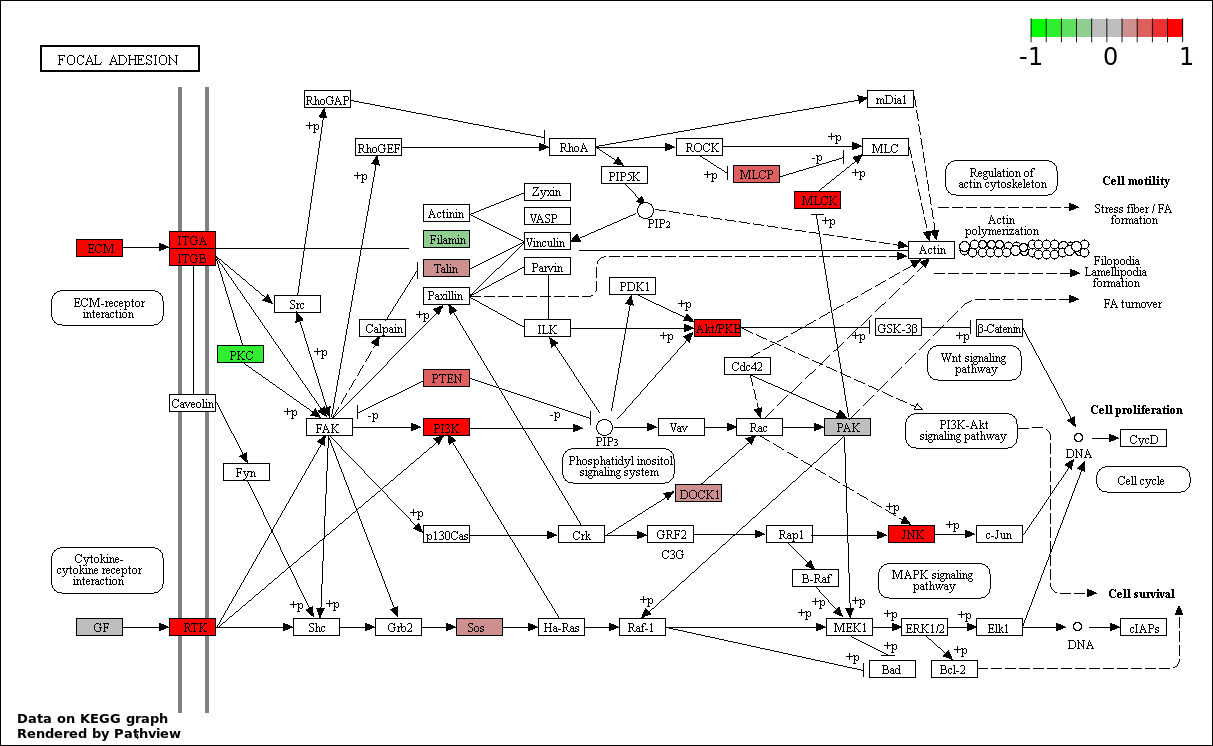
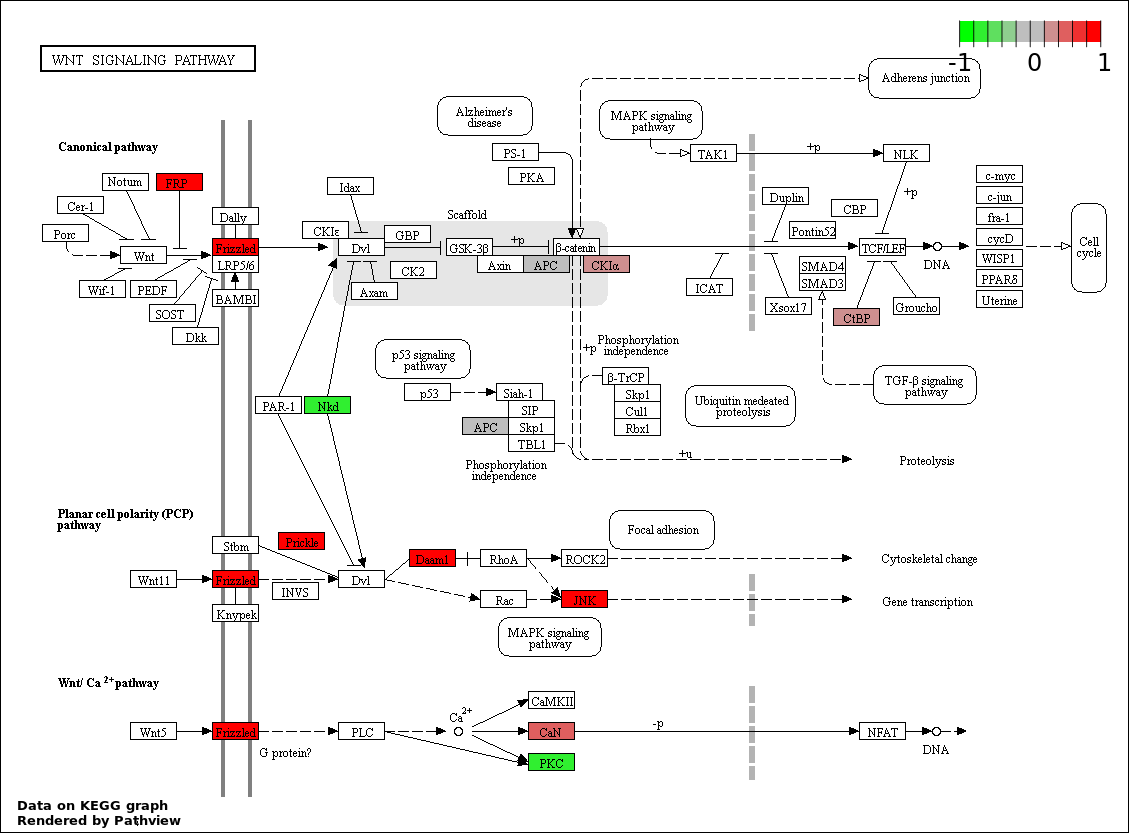
This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-146b-5p | AASS | -0.42 | 0.04574 | 1.86 | 0 | miRanda | -0.17 | 0.01702 | NA | |
| 2 | hsa-miR-200b-3p | ABAT | 1.29 | 0.00027 | 2.16 | 0 | MirTarget; TargetScan | -0.2 | 0 | NA | |
| 3 | hsa-miR-21-5p | ABAT | -1.51 | 0 | 2.16 | 0 | MirTarget | -0.93 | 0 | NA | |
| 4 | hsa-miR-21-5p | ABCA1 | -1.51 | 0 | 1.1 | 0 | mirMAP | -0.29 | 0 | NA | |
| 5 | hsa-miR-21-5p | ABCA10 | -1.51 | 0 | 1.06 | 4.0E-5 | mirMAP | -0.45 | 0 | NA | |
| 6 | hsa-miR-21-5p | ABCD3 | -1.51 | 0 | 0.9 | 0 | miRNAWalker2 validate | -0.38 | 0 | NA | |
| 7 | hsa-miR-877-5p | ABHD2 | -1.36 | 0 | 1.2 | 0 | mirMAP | -0.31 | 0 | NA | |
| 8 | hsa-miR-21-5p | ACAT1 | -1.51 | 0 | 1.62 | 0 | miRNAWalker2 validate | -0.62 | 0 | NA | |
| 9 | hsa-miR-21-5p | ACBD5 | -1.51 | 0 | 0.75 | 0 | miRNAWalker2 validate; miRNATAP | -0.33 | 0 | NA | |
| 10 | hsa-miR-146b-5p | ACO1 | -0.42 | 0.04574 | 0.9 | 0 | miRanda | -0.12 | 9.0E-5 | NA | |
| 11 | hsa-miR-200b-3p | ACVR1C | 1.29 | 0.00027 | 2.2 | 0 | TargetScan | -0.18 | 0 | NA | |
| 12 | hsa-miR-21-5p | ACVR2A | -1.51 | 0 | 0.55 | 0 | miRNATAP | -0.17 | 0 | NA | |
| 13 | hsa-miR-146b-5p | ADCY1 | -0.42 | 0.04574 | 2.7 | 0 | mirMAP | -0.41 | 0 | NA | |
| 14 | hsa-miR-146b-5p | AFAP1L2 | -0.42 | 0.04574 | 0.55 | 0.00077 | miRanda; miRNATAP | -0.13 | 0.00063 | NA | |
| 15 | hsa-miR-877-5p | AFF1 | -1.36 | 0 | 0.71 | 0 | miRNAWalker2 validate | -0.16 | 0 | NA | |
| 16 | hsa-miR-877-5p | AGL | -1.36 | 0 | 1.74 | 0 | MirTarget | -0.34 | 0 | NA | |
| 17 | hsa-miR-21-5p | AIM1 | -1.51 | 0 | 1.46 | 0 | miRNAWalker2 validate | -0.35 | 0 | NA | |
| 18 | hsa-miR-146b-5p | AKAP13 | -0.42 | 0.04574 | 0.32 | 0.00142 | miRanda | -0.12 | 0 | NA | |
| 19 | hsa-miR-21-5p | AKAP6 | -1.51 | 0 | 0.97 | 0 | MirTarget | -0.54 | 0 | NA | |
| 20 | hsa-miR-146b-5p | AKAP9 | -0.42 | 0.04574 | 0.79 | 0 | miRanda | -0.18 | 0 | NA | |
| 21 | hsa-miR-21-5p | AKAP9 | -1.51 | 0 | 0.79 | 0 | miRNAWalker2 validate | -0.29 | 0 | NA | |
| 22 | hsa-miR-21-5p | AKIRIN1 | -1.51 | 0 | 0.73 | 0 | miRNATAP | -0.13 | 0.00038 | NA | |
| 23 | hsa-miR-150-5p | AKR1C2 | 1.15 | 0 | -0.25 | 0.39842 | mirMAP | -0.19 | 0.00177 | NA | |
| 24 | hsa-miR-21-5p | AKT2 | -1.51 | 0 | 0.34 | 1.0E-5 | miRNAWalker2 validate | -0.25 | 0 | NA | |
| 25 | hsa-miR-146b-5p | AKT3 | -0.42 | 0.04574 | 0.66 | 0.00047 | miRNAWalker2 validate | -0.16 | 0.00026 | NA | |
| 26 | hsa-miR-877-5p | AMOTL1 | -1.36 | 0 | 0.53 | 0.02603 | mirMAP | -0.14 | 0.01575 | NA | |
| 27 | hsa-miR-146b-5p | AMOTL2 | -0.42 | 0.04574 | 1.29 | 0 | miRanda | -0.14 | 0.0002 | NA | |
| 28 | hsa-miR-146b-5p | ANGPTL1 | -0.42 | 0.04574 | 2.91 | 0 | miRanda | -0.4 | 0 | NA | |
| 29 | hsa-miR-200b-3p | ANGPTL3 | 1.29 | 0.00027 | 1.81 | 0 | TargetScan | -0.13 | 0.00277 | NA | |
| 30 | hsa-miR-877-5p | ANKFY1 | -1.36 | 0 | 0.58 | 0 | MirTarget | -0.13 | 0 | NA | |
| 31 | hsa-miR-21-5p | ANKRD28 | -1.51 | 0 | 0.36 | 1.0E-5 | miRNAWalker2 validate | -0.17 | 0 | NA | |
| 32 | hsa-miR-21-5p | ANKRD46 | -1.51 | 0 | 0.78 | 0 | miRNAWalker2 validate; miRTarBase | -0.41 | 0 | 21219636 | Knockdown of miR-21 significantly increased the expression of ANKRD46 at both mRNA and protein levels; ANKRD46 is newly identified as a direct target of miR-21 in BC |
| 33 | hsa-miR-21-5p | ANO3 | -1.51 | 0 | 2.3 | 0 | mirMAP | -0.98 | 0 | NA | |
| 34 | hsa-miR-200a-3p | ANO5 | 1.5 | 3.0E-5 | 1.28 | 0.00055 | mirMAP | -0.12 | 0.01856 | NA | |
| 35 | hsa-miR-200b-3p | ANO5 | 1.29 | 0.00027 | 1.28 | 0.00055 | TargetScan | -0.13 | 0.00908 | NA | |
| 36 | hsa-miR-146b-5p | ANXA3 | -0.42 | 0.04574 | 2.62 | 0 | miRanda | -0.29 | 0.00067 | NA | |
| 37 | hsa-miR-146b-5p | AOC3 | -0.42 | 0.04574 | 0.85 | 0 | miRanda | -0.1 | 0.00112 | NA | |
| 38 | hsa-miR-21-5p | AP1AR | -1.51 | 0 | 0.61 | 0 | MirTarget; miRNATAP | -0.25 | 0 | NA | |
| 39 | hsa-miR-652-3p | AP1G1 | -0.14 | 0.3917 | 0.48 | 0 | miRNAWalker2 validate | -0.1 | 3.0E-5 | NA | |
| 40 | hsa-miR-877-5p | AP3S2 | -1.36 | 0 | 0.33 | 4.0E-5 | mirMAP | -0.14 | 0 | NA | |
| 41 | hsa-miR-21-5p | APC | -1.51 | 0 | 0.18 | 0.06792 | miRNAWalker2 validate | -0.17 | 0 | 23773491; 24832083 | The prognostic significance of APC gene mutation and miR 21 expression in advanced stage colorectal cancer; The aim of this study was to analyse the association of APC gene mutation and miR-21 expression with clinical outcome in CRC patients; APC gene mutation and expression of APC and miR-21 were analysed by direct DNA sequencing and real-time reverse transcription polymerase chain reaction; APC gene expression was low in CRC and negatively correlated with miR-21 expression and gene mutation; In Taiwan downregulation of the APC gene in CRC correlated with gene mutation and miR-21 upregulation; APC mutation and miR-21 expression could be used to predict the clinical outcome of CRC especially in patients with advanced disease;MicroRNA 21 promotes tumour malignancy via increased nuclear translocation of β catenin and predicts poor outcome in APC mutated but not in APC wild type colorectal cancer; However in our preliminary data the prognostic value of miR-21 levels was observed only in adenomatous polyposis coli APC-mutated tumours not in APC-wild-type tumours; We enrolled 165 colorectal tumour to determine APC mutation miR-21 levels and nuclear β-catenin expression by direct sequencing real-time PCR and immunohistochemistry |
| 42 | hsa-miR-21-5p | APOLD1 | -1.51 | 0 | -0.45 | 0.00929 | miRNAWalker2 validate | -0.18 | 0.00545 | NA | |
| 43 | hsa-miR-146b-5p | AQP1 | -0.42 | 0.04574 | 0.67 | 0.0031 | miRanda | -0.18 | 0.00079 | NA | |
| 44 | hsa-miR-200a-3p | AR | 1.5 | 3.0E-5 | 2.66 | 0 | mirMAP | -0.32 | 0 | 24391862 | We identified miR-200 b as a downstream target of androgen receptor and linked its expression to decreased tumorigenicity and metastatic capacity of the prostate cancer cells |
| 45 | hsa-miR-877-5p | AR | -1.36 | 0 | 2.66 | 0 | MirTarget | -0.92 | 0 | NA | |
| 46 | hsa-miR-21-5p | ARHGAP21 | -1.51 | 0 | 0.09 | 0.37274 | miRNAWalker2 validate | -0.11 | 0.00286 | NA | |
| 47 | hsa-miR-21-5p | ARHGAP24 | -1.51 | 0 | 0.99 | 0 | MirTarget; miRNATAP | -0.36 | 0 | NA | |
| 48 | hsa-miR-21-5p | ARHGAP32 | -1.51 | 0 | 0.2 | 0.09921 | MirTarget | -0.29 | 0 | NA | |
| 49 | hsa-miR-146b-5p | ARHGAP6 | -0.42 | 0.04574 | -0.22 | 0.19399 | PITA; miRanda | -0.11 | 0.00647 | NA | |
| 50 | hsa-miR-21-5p | ARHGEF12 | -1.51 | 0 | 0.86 | 0 | miRNAWalker2 validate; MirTarget | -0.26 | 0 | NA | |
| 51 | hsa-miR-146b-5p | ARHGEF4 | -0.42 | 0.04574 | 1.69 | 3.0E-5 | miRanda | -0.27 | 0.0043 | NA | |
| 52 | hsa-miR-877-5p | ARHGEF6 | -1.36 | 0 | 0.58 | 0.00012 | miRNAWalker2 validate | -0.1 | 0.0076 | NA | |
| 53 | hsa-miR-21-5p | ARID4A | -1.51 | 0 | 0.9 | 0 | miRNAWalker2 validate | -0.43 | 0 | NA | |
| 54 | hsa-miR-200b-3p | ARPP21 | 1.29 | 0.00027 | 2.21 | 0 | MirTarget | -0.14 | 0.01222 | NA | |
| 55 | hsa-miR-21-5p | ART4 | -1.51 | 0 | 1.74 | 0 | mirMAP | -0.48 | 1.0E-5 | NA | |
| 56 | hsa-miR-21-5p | ASPN | -1.51 | 0 | 1.64 | 0 | MirTarget; miRNATAP | -0.2 | 0.04902 | NA | |
| 57 | hsa-miR-877-5p | ASTN1 | -1.36 | 0 | 1.44 | 0 | MirTarget | -0.26 | 0.00014 | NA | |
| 58 | hsa-miR-146b-5p | ASXL2 | -0.42 | 0.04574 | 0.29 | 0.18051 | miRanda | -0.12 | 0.01657 | NA | |
| 59 | hsa-miR-200b-3p | ASXL3 | 1.29 | 0.00027 | 2.69 | 0 | TargetScan | -0.17 | 0.00117 | NA | |
| 60 | hsa-miR-21-5p | ATAD2B | -1.51 | 0 | 0.06 | 0.4791 | miRNAWalker2 validate | -0.14 | 3.0E-5 | NA | |
| 61 | hsa-miR-877-5p | ATE1 | -1.36 | 0 | 0.6 | 0.0009 | MirTarget | -0.14 | 0.00256 | NA | |
| 62 | hsa-miR-21-5p | ATF2 | -1.51 | 0 | 0.24 | 0.15619 | miRNAWalker2 validate | -0.18 | 0.00336 | NA | |
| 63 | hsa-miR-21-5p | ATMIN | -1.51 | 0 | 0.37 | 0 | miRNAWalker2 validate | -0.12 | 0 | NA | |
| 64 | hsa-miR-146b-5p | ATP10B | -0.42 | 0.04574 | 0.24 | 0.56639 | miRanda; miRNATAP | -0.27 | 0.00487 | NA | |
| 65 | hsa-miR-21-5p | ATP11B | -1.51 | 0 | 0.35 | 0.00133 | miRNAWalker2 validate | -0.16 | 4.0E-5 | NA | |
| 66 | hsa-miR-200b-3p | ATP11C | 1.29 | 0.00027 | 1.6 | 0 | MirTarget; TargetScan | -0.15 | 0 | NA | |
| 67 | hsa-miR-21-5p | ATP2B2 | -1.51 | 0 | 0.4 | 0.15518 | mirMAP | -0.24 | 0.01837 | NA | |
| 68 | hsa-miR-21-5p | ATRN | -1.51 | 0 | -0.12 | 0.34173 | miRNATAP | -0.17 | 0.00015 | NA | |
| 69 | hsa-miR-21-5p | ATRNL1 | -1.51 | 0 | 2.28 | 0 | MirTarget | -0.65 | 8.0E-5 | NA | |
| 70 | hsa-miR-21-5p | ATRX | -1.51 | 0 | 0.07 | 0.48759 | miRNAWalker2 validate | -0.1 | 0.00335 | NA | |
| 71 | hsa-miR-21-5p | ATXN3 | -1.51 | 0 | 0.29 | 0.00037 | mirMAP | -0.22 | 0 | NA | |
| 72 | hsa-miR-21-5p | AUTS2 | -1.51 | 0 | 1.2 | 0 | miRNAWalker2 validate | -0.85 | 0 | NA | |
| 73 | hsa-miR-146b-5p | B3GNT5 | -0.42 | 0.04574 | -0.79 | 0.0009 | miRanda | -0.15 | 0.00816 | NA | |
| 74 | hsa-miR-21-5p | BACE1 | -1.51 | 0 | 0.45 | 0 | mirMAP | -0.29 | 0 | NA | |
| 75 | hsa-miR-200b-3p | BAGE | 1.29 | 0.00027 | -1.25 | 0.00088 | MirTarget | -0.23 | 1.0E-5 | NA | |
| 76 | hsa-miR-146b-5p | BAZ2B | -0.42 | 0.04574 | 0.61 | 0 | miRanda | -0.1 | 0.00011 | NA | |
| 77 | hsa-miR-146b-5p | BCAT2 | -0.42 | 0.04574 | -1.58 | 0 | miRNATAP | -0.11 | 0.00603 | NA | |
| 78 | hsa-miR-21-5p | BCL6 | -1.51 | 0 | 0.38 | 0.00305 | miRNAWalker2 validate | -0.19 | 6.0E-5 | NA | |
| 79 | hsa-miR-21-5p | BDH2 | -1.51 | 0 | 1.59 | 0 | miRNAWalker2 validate | -0.57 | 0 | NA | |
| 80 | hsa-miR-146b-5p | BHLHB9 | -0.42 | 0.04574 | -0.02 | 0.89414 | miRanda | -0.14 | 8.0E-5 | NA | |
| 81 | hsa-miR-877-5p | BIVM | -1.36 | 0 | 0.29 | 0.00612 | miRNATAP | -0.11 | 1.0E-5 | NA | |
| 82 | hsa-miR-21-5p | BMP2K | -1.51 | 0 | 0.82 | 0 | mirMAP | -0.12 | 0.00963 | NA | |
| 83 | hsa-miR-146b-5p | BMP8A | -0.42 | 0.04574 | -1.39 | 0 | miRanda | -0.12 | 0.02024 | NA | |
| 84 | hsa-miR-21-5p | BMPR2 | -1.51 | 0 | 0.74 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget | -0.28 | 0 | NA | |
| 85 | hsa-miR-877-5p | BMPR2 | -1.36 | 0 | 0.74 | 0 | MirTarget | -0.15 | 0 | NA | |
| 86 | hsa-miR-21-5p | BRD1 | -1.51 | 0 | 0.22 | 0.01173 | MirTarget | -0.1 | 0.0018 | NA | |
| 87 | hsa-miR-877-5p | BTD | -1.36 | 0 | 0.71 | 0 | MirTarget | -0.24 | 0 | NA | |
| 88 | hsa-miR-21-5p | C16orf52 | -1.51 | 0 | 0.5 | 0 | miRNATAP | -0.29 | 0 | NA | |
| 89 | hsa-miR-146b-5p | C1orf21 | -0.42 | 0.04574 | 0.75 | 0.00045 | mirMAP | -0.1 | 0.03512 | NA | |
| 90 | hsa-miR-200a-3p | C1orf21 | 1.5 | 3.0E-5 | 0.75 | 0.00045 | miRNATAP | -0.12 | 4.0E-5 | NA | |
| 91 | hsa-miR-877-5p | C1orf21 | -1.36 | 0 | 0.75 | 0.00045 | mirMAP | -0.32 | 0 | NA | |
| 92 | hsa-miR-21-5p | C1orf226 | -1.51 | 0 | -0.29 | 0.07217 | mirMAP | -0.13 | 0.03584 | NA | |
| 93 | hsa-miR-150-5p | C3orf18 | 1.15 | 0 | -0.57 | 0.00014 | miRNATAP | -0.13 | 1.0E-5 | NA | |
| 94 | hsa-miR-150-5p | C6orf223 | 1.15 | 0 | -0.67 | 0.18925 | mirMAP | -0.26 | 0.01153 | NA | |
| 95 | hsa-miR-200a-3p | CADM1 | 1.5 | 3.0E-5 | 0.03 | 0.88629 | MirTarget | -0.12 | 0 | NA | |
| 96 | hsa-miR-200b-3p | CADM1 | 1.29 | 0.00027 | 0.03 | 0.88629 | TargetScan | -0.13 | 0 | NA | |
| 97 | hsa-miR-21-5p | CADM2 | -1.51 | 0 | 2.32 | 0 | mirMAP; miRNATAP | -0.38 | 0.00648 | NA | |
| 98 | hsa-miR-146b-5p | CALCRL | -0.42 | 0.04574 | -0.11 | 0.54853 | miRanda | -0.11 | 0.01138 | NA | |
| 99 | hsa-miR-21-5p | CALD1 | -1.51 | 0 | 0.53 | 0 | miRNAWalker2 validate; MirTarget | -0.27 | 0 | NA | |
| 100 | hsa-miR-146b-5p | CAPS | -0.42 | 0.04574 | -0.58 | 0.00027 | miRanda | -0.11 | 0.00402 | NA | |
| 101 | hsa-miR-146b-5p | CAPS2 | -0.42 | 0.04574 | 0.81 | 8.0E-5 | miRanda | -0.18 | 0.00014 | NA | |
| 102 | hsa-miR-21-5p | CASC4 | -1.51 | 0 | 0.46 | 0 | miRNATAP | -0.14 | 8.0E-5 | NA | |
| 103 | hsa-miR-146b-5p | CASR | -0.42 | 0.04574 | 3.09 | 0 | miRanda | -0.19 | 0.04519 | 26178670 | miR 135b and miR 146b dependent silencing of calcium sensing receptor expression in colorectal tumors; Inhibition of miR-135b and miR-146b expression led to high CaSR levels and significantly reduced proliferation; In samples of colorectal tumors we observed overexpression of miR-135b and miR-146b and this correlated inversely with CaSR expression miR-135b: r = -0.684 p < 0.001 and miR-146b: r = -0.448 p < 0.001 supporting our in vitro findings; We demonstrate that miR-135b and miR-146b target the CaSR and reduce its expression in colorectal tumors reducing the antiproliferative and prodifferentiating actions of calcium |
| 104 | hsa-miR-877-5p | CBFA2T3 | -1.36 | 0 | 2.47 | 0 | miRNATAP | -0.29 | 0 | NA | |
| 105 | hsa-miR-21-5p | CCDC121 | -1.51 | 0 | -0.09 | 0.43018 | MirTarget | -0.21 | 0 | NA | |
| 106 | hsa-miR-21-5p | CCDC152 | -1.51 | 0 | 1.09 | 0 | mirMAP | -0.61 | 0 | NA | |
| 107 | hsa-miR-146b-5p | CCDC85A | -0.42 | 0.04574 | -0.74 | 9.0E-5 | miRanda | -0.13 | 0.00257 | NA | |
| 108 | hsa-miR-21-5p | CCNG1 | -1.51 | 0 | 0.05 | 0.64033 | miRNAWalker2 validate | -0.13 | 0.00024 | NA | |
| 109 | hsa-miR-146b-5p | CCNJ | -0.42 | 0.04574 | 0.4 | 0.0818 | PITA; miRanda; miRNATAP | -0.13 | 0.01784 | NA | |
| 110 | hsa-miR-877-5p | CD302 | -1.36 | 0 | 1.76 | 0 | MirTarget | -0.3 | 0 | NA | |
| 111 | hsa-miR-652-3p | CD46 | -0.14 | 0.3917 | -0.36 | 0.00059 | miRNAWalker2 validate | -0.12 | 0.00015 | NA | |
| 112 | hsa-miR-200a-3p | CDC14B | 1.5 | 3.0E-5 | 1.19 | 0 | mirMAP | -0.15 | 0 | NA | |
| 113 | hsa-miR-150-5p | CDCA5 | 1.15 | 0 | -3.36 | 0 | mirMAP | -0.14 | 0.00793 | NA | |
| 114 | hsa-miR-146b-5p | CDH1 | -0.42 | 0.04574 | 0.93 | 4.0E-5 | miRanda | -0.21 | 6.0E-5 | NA | |
| 115 | hsa-miR-877-5p | CDH19 | -1.36 | 0 | 4.11 | 0 | MirTarget | -0.28 | 0.00793 | NA | |
| 116 | hsa-miR-150-5p | CDHR3 | 1.15 | 0 | -0.83 | 1.0E-5 | mirMAP | -0.2 | 0 | NA | |
| 117 | hsa-miR-146b-5p | CDKN1A | -0.42 | 0.04574 | 0.77 | 6.0E-5 | miRNAWalker2 validate | -0.15 | 0.00059 | 27602131 | During the search for potential targets of miR-146b in ATC p21 also known as p21Waf1/Cip1 or CDKN1A was noted for its role in cell cycle progression and tumor pathogenesis |
| 118 | hsa-miR-877-5p | CDO1 | -1.36 | 0 | 1.67 | 0 | MirTarget | -0.43 | 0 | NA | |
| 119 | hsa-miR-146b-5p | CDS1 | -0.42 | 0.04574 | 0.32 | 0.40167 | miRanda; miRNATAP | -0.22 | 0.01361 | NA | |
| 120 | hsa-miR-200b-3p | CECR2 | 1.29 | 0.00027 | 0.78 | 0.01282 | TargetScan | -0.3 | 0 | NA | |
| 121 | hsa-miR-877-5p | CFHR5 | -1.36 | 0 | 1.75 | 0.00027 | MirTarget | -0.64 | 0 | NA | |
| 122 | hsa-miR-200b-3p | CFL2 | 1.29 | 0.00027 | 1.18 | 0 | MirTarget; TargetScan | -0.15 | 0 | NA | |
| 123 | hsa-miR-200b-3p | CHN2 | 1.29 | 0.00027 | 0.87 | 0 | MirTarget; TargetScan | -0.15 | 0 | NA | |
| 124 | hsa-miR-21-5p | CLCN5 | -1.51 | 0 | 0.78 | 0 | miRNAWalker2 validate; mirMAP | -0.33 | 0 | NA | |
| 125 | hsa-miR-146b-5p | CLDN10 | -0.42 | 0.04574 | 5.18 | 0 | miRanda | -0.26 | 0.04926 | NA | |
| 126 | hsa-miR-877-5p | CLEC14A | -1.36 | 0 | -0.26 | 0.07318 | MirTarget | -0.17 | 0 | NA | |
| 127 | hsa-miR-146b-5p | CLIC5 | -0.42 | 0.04574 | -0.65 | 0.00496 | miRanda | -0.15 | 0.00602 | NA | |
| 128 | hsa-miR-146b-5p | CLOCK | -0.42 | 0.04574 | 0.53 | 0.00745 | mirMAP | -0.19 | 5.0E-5 | NA | |
| 129 | hsa-miR-21-5p | CLOCK | -1.51 | 0 | 0.53 | 0.00745 | miRNAWalker2 validate | -0.26 | 0.00029 | NA | |
| 130 | hsa-miR-652-3p | CNN3 | -0.14 | 0.3917 | -0 | 0.99816 | miRNAWalker2 validate | -0.1 | 0.00282 | NA | |
| 131 | hsa-miR-146b-5p | CNTFR | -0.42 | 0.04574 | 3.51 | 0 | miRNATAP | -0.26 | 0.03741 | NA | |
| 132 | hsa-miR-21-5p | CNTFR | -1.51 | 0 | 3.51 | 0 | miRNATAP | -1.1 | 0 | NA | |
| 133 | hsa-miR-146b-5p | CNTN1 | -0.42 | 0.04574 | 2.32 | 0 | miRanda | -0.23 | 0.02389 | NA | |
| 134 | hsa-miR-877-5p | CNTN3 | -1.36 | 0 | 3.79 | 0 | MirTarget | -0.75 | 0 | NA | |
| 135 | hsa-miR-21-5p | COBLL1 | -1.51 | 0 | 1.47 | 0 | miRNAWalker2 validate | -0.8 | 0 | NA | |
| 136 | hsa-miR-146b-5p | COL6A3 | -0.42 | 0.04574 | -0.17 | 0.43229 | miRanda | -0.13 | 0.00943 | NA | |
| 137 | hsa-miR-146b-5p | COQ9 | -0.42 | 0.04574 | 0.62 | 0 | miRanda | -0.1 | 0.00021 | NA | |
| 138 | hsa-miR-21-5p | CPEB3 | -1.51 | 0 | 2.74 | 0 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.98 | 0 | NA | |
| 139 | hsa-miR-200a-3p | CPLX2 | 1.5 | 3.0E-5 | -5.16 | 0 | mirMAP | -0.38 | 0.00019 | NA | |
| 140 | hsa-miR-146b-5p | CRB3 | -0.42 | 0.04574 | 0.07 | 0.72072 | PITA; miRanda | -0.14 | 0.00125 | NA | |
| 141 | hsa-miR-21-5p | CREBL2 | -1.51 | 0 | 0.86 | 0 | miRNATAP | -0.28 | 0 | NA | |
| 142 | hsa-miR-21-5p | CRIM1 | -1.51 | 0 | 0.43 | 0.03963 | miRNATAP | -0.15 | 0.04655 | NA | |
| 143 | hsa-miR-21-5p | CSNK1A1 | -1.51 | 0 | 0.34 | 0 | miRNAWalker2 validate | -0.1 | 0 | NA | |
| 144 | hsa-miR-146b-5p | CTBP2 | -0.42 | 0.04574 | 0.39 | 0.02251 | miRanda | -0.18 | 1.0E-5 | NA | |
| 145 | hsa-miR-877-5p | CTBS | -1.36 | 0 | 1.15 | 0 | MirTarget | -0.25 | 0 | NA | |
| 146 | hsa-miR-150-5p | CTH | 1.15 | 0 | 2.13 | 0 | MirTarget | -0.16 | 0.01441 | NA | |
| 147 | hsa-miR-146b-5p | CTNNA3 | -0.42 | 0.04574 | 2.94 | 0 | miRanda | -0.23 | 0.00499 | NA | |
| 148 | hsa-miR-877-5p | CTTNBP2 | -1.36 | 0 | 2.02 | 0 | MirTarget | -0.42 | 1.0E-5 | NA | |
| 149 | hsa-miR-146b-5p | CYB5D1 | -0.42 | 0.04574 | 0.74 | 0 | mirMAP | -0.1 | 1.0E-5 | NA | |
| 150 | hsa-miR-21-5p | CYBRD1 | -1.51 | 0 | 1.29 | 0 | miRNAWalker2 validate | -0.31 | 6.0E-5 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 125 | 1784 | 7.966e-10 | 3.706e-06 |
| 2 | CARDIOVASCULAR SYSTEM DEVELOPMENT | 67 | 788 | 8.708e-09 | 1.351e-05 |
| 3 | CIRCULATORY SYSTEM DEVELOPMENT | 67 | 788 | 8.708e-09 | 1.351e-05 |
| 4 | INTRACELLULAR SIGNAL TRANSDUCTION | 109 | 1572 | 2.018e-08 | 2.102e-05 |
| 5 | TISSUE DEVELOPMENT | 106 | 1518 | 2.259e-08 | 2.102e-05 |
| 6 | PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS | 129 | 1977 | 3.203e-08 | 2.285e-05 |
| 7 | NEGATIVE REGULATION OF CELL COMMUNICATION | 88 | 1192 | 3.438e-08 | 2.285e-05 |
| 8 | PROTEIN PHOSPHORYLATION | 74 | 944 | 4.305e-08 | 2.504e-05 |
| 9 | REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 29 | 229 | 5.841e-08 | 3.02e-05 |
| 10 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 111 | 1656 | 8.789e-08 | 4.09e-05 |
| 11 | REGULATION OF CELL DEATH | 101 | 1472 | 1.194e-07 | 4.848e-05 |
| 12 | NEGATIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 61 | 740 | 1.25e-07 | 4.848e-05 |
| 13 | MUSCLE STRUCTURE DEVELOPMENT | 42 | 432 | 1.671e-07 | 5.981e-05 |
| 14 | NEGATIVE REGULATION OF GENE EXPRESSION | 101 | 1493 | 2.35e-07 | 7.812e-05 |
| 15 | MUSCLE TISSUE DEVELOPMENT | 31 | 275 | 2.954e-07 | 9.164e-05 |
| 16 | NEUROGENESIS | 95 | 1402 | 5.196e-07 | 0.0001511 |
| 17 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 115 | 1805 | 6.85e-07 | 0.0001875 |
| 18 | GROWTH | 39 | 410 | 8.01e-07 | 0.0002071 |
| 19 | NEGATIVE REGULATION OF NITROGEN COMPOUND METABOLIC PROCESS | 100 | 1517 | 8.954e-07 | 0.0002193 |
| 20 | REGULATION OF HORMONE LEVELS | 43 | 478 | 9.929e-07 | 0.000231 |
| 21 | MUSCLE ORGAN DEVELOPMENT | 30 | 277 | 1.072e-06 | 0.0002376 |
| 22 | REGULATION OF CELL DIFFERENTIATION | 98 | 1492 | 1.366e-06 | 0.000289 |
| 23 | RESPONSE TO ENDOGENOUS STIMULUS | 95 | 1450 | 2.248e-06 | 0.0004265 |
| 24 | BLOOD VESSEL MORPHOGENESIS | 35 | 364 | 2.291e-06 | 0.0004265 |
| 25 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 72 | 1008 | 2.205e-06 | 0.0004265 |
| 26 | POSITIVE REGULATION OF GENE EXPRESSION | 109 | 1733 | 2.547e-06 | 0.0004558 |
| 27 | ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY | 54 | 689 | 3.105e-06 | 0.0005312 |
| 28 | PHOSPHORYLATION | 83 | 1228 | 3.196e-06 | 0.0005312 |
| 29 | REGULATION OF TRANSFERASE ACTIVITY | 68 | 946 | 3.45e-06 | 0.0005448 |
| 30 | CELL DEVELOPMENT | 93 | 1426 | 3.513e-06 | 0.0005448 |
| 31 | NEGATIVE REGULATION OF PHOSPHORYLATION | 38 | 422 | 4.119e-06 | 0.000599 |
| 32 | LUNG ALVEOLUS DEVELOPMENT | 10 | 41 | 4.109e-06 | 0.000599 |
| 33 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 105 | 1672 | 4.272e-06 | 0.0006024 |
| 34 | CELLULAR COMPONENT MORPHOGENESIS | 65 | 900 | 4.896e-06 | 0.00067 |
| 35 | RESPIRATORY SYSTEM DEVELOPMENT | 23 | 197 | 5.474e-06 | 0.0007277 |
| 36 | REGULATION OF DEVELOPMENTAL GROWTH | 29 | 289 | 7.353e-06 | 0.0009503 |
| 37 | TUBE DEVELOPMENT | 45 | 552 | 7.688e-06 | 0.0009668 |
| 38 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 101 | 1618 | 8.574e-06 | 0.00105 |
| 39 | ANATOMICAL STRUCTURE FORMATION INVOLVED IN MORPHOGENESIS | 67 | 957 | 9.735e-06 | 0.001161 |
| 40 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 109 | 1791 | 1.116e-05 | 0.001298 |
| 41 | REGULATION OF KINASE ACTIVITY | 57 | 776 | 1.165e-05 | 0.001322 |
| 42 | NEGATIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 17 | 126 | 1.366e-05 | 0.001514 |
| 43 | REGULATION OF CELL PROLIFERATION | 94 | 1496 | 1.403e-05 | 0.001518 |
| 44 | REGULATION OF GTPASE ACTIVITY | 51 | 673 | 1.472e-05 | 0.001557 |
| 45 | ORGAN MORPHOGENESIS | 60 | 841 | 1.648e-05 | 0.001667 |
| 46 | DEVELOPMENTAL GROWTH | 31 | 333 | 1.635e-05 | 0.001667 |
| 47 | HEART DEVELOPMENT | 39 | 466 | 1.709e-05 | 0.001692 |
| 48 | REGULATION OF MAP KINASE ACTIVITY | 30 | 319 | 1.835e-05 | 0.001776 |
| 49 | NEGATIVE REGULATION OF CELL PROLIFERATION | 49 | 643 | 1.871e-05 | 0.001776 |
| 50 | REGULATION OF CELLULAR COMPONENT MOVEMENT | 56 | 771 | 1.915e-05 | 0.001782 |
| 51 | VASCULATURE DEVELOPMENT | 39 | 469 | 1.976e-05 | 0.001803 |
| 52 | REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 39 | 470 | 2.074e-05 | 0.001855 |
| 53 | REGULATION OF SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 27 | 278 | 2.698e-05 | 0.002368 |
| 54 | REGULATION OF PROTEIN MODIFICATION PROCESS | 103 | 1710 | 3.055e-05 | 0.002632 |
| 55 | NEGATIVE REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 16 | 121 | 3.132e-05 | 0.002649 |
| 56 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 68 | 1021 | 4.097e-05 | 0.003404 |
| 57 | EPITHELIUM DEVELOPMENT | 64 | 945 | 4.236e-05 | 0.003458 |
| 58 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 67 | 1004 | 4.399e-05 | 0.003529 |
| 59 | NEGATIVE REGULATION OF RESPONSE TO STIMULUS | 85 | 1360 | 4.505e-05 | 0.003553 |
| 60 | NEGATIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 42 | 541 | 4.812e-05 | 0.003671 |
| 61 | NEGATIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 42 | 541 | 4.812e-05 | 0.003671 |
| 62 | NEGATIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 36 | 437 | 4.965e-05 | 0.003726 |
| 63 | CELL JUNCTION ORGANIZATION | 20 | 185 | 6.662e-05 | 0.004884 |
| 64 | ANGIOGENESIS | 27 | 293 | 6.718e-05 | 0.004884 |
| 65 | NEGATIVE REGULATION OF MOLECULAR FUNCTION | 70 | 1079 | 7.144e-05 | 0.005114 |
| 66 | POSITIVE REGULATION OF ENDOTHELIAL CELL MIGRATION | 11 | 67 | 7.422e-05 | 0.005232 |
| 67 | GOLGI TO VACUOLE TRANSPORT | 7 | 27 | 7.717e-05 | 0.005359 |
| 68 | ORGAN GROWTH | 11 | 68 | 8.533e-05 | 0.005839 |
| 69 | NEGATIVE REGULATION OF KINASE ACTIVITY | 24 | 250 | 8.903e-05 | 0.005939 |
| 70 | REGULATION OF HYDROLASE ACTIVITY | 82 | 1327 | 9.062e-05 | 0.005939 |
| 71 | REGULATION OF LIPID METABOLIC PROCESS | 26 | 282 | 9.025e-05 | 0.005939 |
| 72 | NEURON DIFFERENTIATION | 59 | 874 | 9.234e-05 | 0.005968 |
| 73 | UROGENITAL SYSTEM DEVELOPMENT | 27 | 299 | 9.467e-05 | 0.006034 |
| 74 | POSITIVE REGULATION OF VASCULATURE DEVELOPMENT | 16 | 133 | 9.973e-05 | 0.006271 |
| 75 | PHOSPHATIDYLINOSITOL BIOSYNTHETIC PROCESS | 15 | 120 | 0.0001055 | 0.006542 |
| 76 | REGULATION OF EPITHELIAL CELL PROLIFERATION | 26 | 285 | 0.0001073 | 0.006571 |
| 77 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 91 | 1518 | 0.0001091 | 0.006595 |
| 78 | REGULATION OF PEPTIDE SECRETION | 21 | 209 | 0.0001272 | 0.00759 |
| 79 | NEGATIVE REGULATION OF PEPTIDE SECRETION | 9 | 49 | 0.0001367 | 0.007952 |
| 80 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 72 | 1142 | 0.0001356 | 0.007952 |
| 81 | REGULATION OF MAPK CASCADE | 47 | 660 | 0.0001416 | 0.008074 |
| 82 | NEGATIVE REGULATION OF DEVELOPMENTAL GROWTH | 12 | 84 | 0.0001423 | 0.008074 |
| 83 | NEGATIVE REGULATION OF CELL DEATH | 58 | 872 | 0.0001567 | 0.008534 |
| 84 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 89 | 1492 | 0.0001545 | 0.008534 |
| 85 | REGULATION OF PROTEIN LOCALIZATION | 62 | 950 | 0.0001577 | 0.008534 |
| 86 | BEHAVIOR | 39 | 516 | 0.0001555 | 0.008534 |
| 87 | NEGATIVE REGULATION OF JUN KINASE ACTIVITY | 5 | 14 | 0.0001606 | 0.00859 |
| 88 | KIDNEY EPITHELIUM DEVELOPMENT | 15 | 125 | 0.0001675 | 0.008858 |
| 89 | CARDIAC MUSCLE TISSUE DEVELOPMENT | 16 | 140 | 0.0001829 | 0.009522 |
| 90 | REGULATION OF RAS PROTEIN SIGNAL TRANSDUCTION | 19 | 184 | 0.0001842 | 0.009522 |
| 91 | NEURON DEVELOPMENT | 48 | 687 | 0.0001885 | 0.009637 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | ENZYME BINDING | 118 | 1737 | 1.524e-08 | 1.416e-05 |
| 2 | PROTEIN KINASE ACTIVITY | 55 | 640 | 1.389e-07 | 6.453e-05 |
| 3 | KINASE ACTIVITY | 66 | 842 | 2.411e-07 | 7.467e-05 |
| 4 | PROTEIN SERINE THREONINE KINASE ACTIVITY | 42 | 445 | 3.737e-07 | 8.678e-05 |
| 5 | GROWTH FACTOR BINDING | 19 | 123 | 5.371e-07 | 9.632e-05 |
| 6 | TRANSFERASE ACTIVITY TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 73 | 992 | 6.221e-07 | 9.632e-05 |
| 7 | NUCLEIC ACID BINDING TRANSCRIPTION FACTOR ACTIVITY | 82 | 1199 | 2.341e-06 | 0.0003107 |
| 8 | ACTIN BINDING | 36 | 393 | 5.007e-06 | 0.0005815 |
| 9 | CYTOSKELETAL PROTEIN BINDING | 60 | 819 | 7.414e-06 | 0.0007296 |
| 10 | KINASE BINDING | 48 | 606 | 8.316e-06 | 0.0007296 |
| 11 | TRANSLATION REPRESSOR ACTIVITY | 7 | 20 | 8.639e-06 | 0.0007296 |
| 12 | LIPID BINDING | 50 | 657 | 1.594e-05 | 0.001139 |
| 13 | MACROMOLECULAR COMPLEX BINDING | 89 | 1399 | 1.519e-05 | 0.001139 |
| 14 | RECEPTOR SIGNALING PROTEIN SERINE THREONINE KINASE ACTIVITY | 14 | 92 | 1.967e-05 | 0.001305 |
| 15 | TRANSMEMBRANE RECEPTOR PROTEIN KINASE ACTIVITY | 13 | 81 | 2.161e-05 | 0.001338 |
| 16 | MOLECULAR FUNCTION REGULATOR | 84 | 1353 | 6.308e-05 | 0.003447 |
| 17 | PHOSPHATASE ACTIVITY | 26 | 275 | 5.95e-05 | 0.003447 |
| 18 | RECEPTOR SIGNALING PROTEIN ACTIVITY | 19 | 172 | 7.515e-05 | 0.003878 |
| 19 | GUANYL NUCLEOTIDE EXCHANGE FACTOR ACTIVITY | 27 | 303 | 0.0001182 | 0.00553 |
| 20 | PHOSPHOPROTEIN PHOSPHATASE ACTIVITY | 19 | 178 | 0.0001191 | 0.00553 |
| 21 | RNA POLYMERASE II TRANSCRIPTION FACTOR ACTIVITY SEQUENCE SPECIFIC DNA BINDING | 45 | 629 | 0.0001761 | 0.007791 |
| 22 | RETINOIC ACID BINDING | 6 | 23 | 0.0002452 | 0.00949 |
| 23 | INSULIN RECEPTOR BINDING | 7 | 32 | 0.000245 | 0.00949 |
| 24 | PROTEIN SERINE THREONINE PHOSPHATASE ACTIVITY | 10 | 64 | 0.0002389 | 0.00949 |
| 25 | IDENTICAL PROTEIN BINDING | 74 | 1209 | 0.0002679 | 0.009872 |
| 26 | PDZ DOMAIN BINDING | 12 | 90 | 0.0002763 | 0.009872 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | PLASMA MEMBRANE REGION | 67 | 929 | 3.652e-06 | 0.001066 |
| 2 | MEMBRANE REGION | 78 | 1134 | 3.397e-06 | 0.001066 |
| 3 | PML BODY | 15 | 97 | 8.19e-06 | 0.001594 |
| 4 | EXCITATORY SYNAPSE | 22 | 197 | 1.768e-05 | 0.002581 |
| 5 | CELL PROJECTION | 107 | 1786 | 2.635e-05 | 0.003078 |
| 6 | CELL JUNCTION | 73 | 1151 | 0.0001014 | 0.009873 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
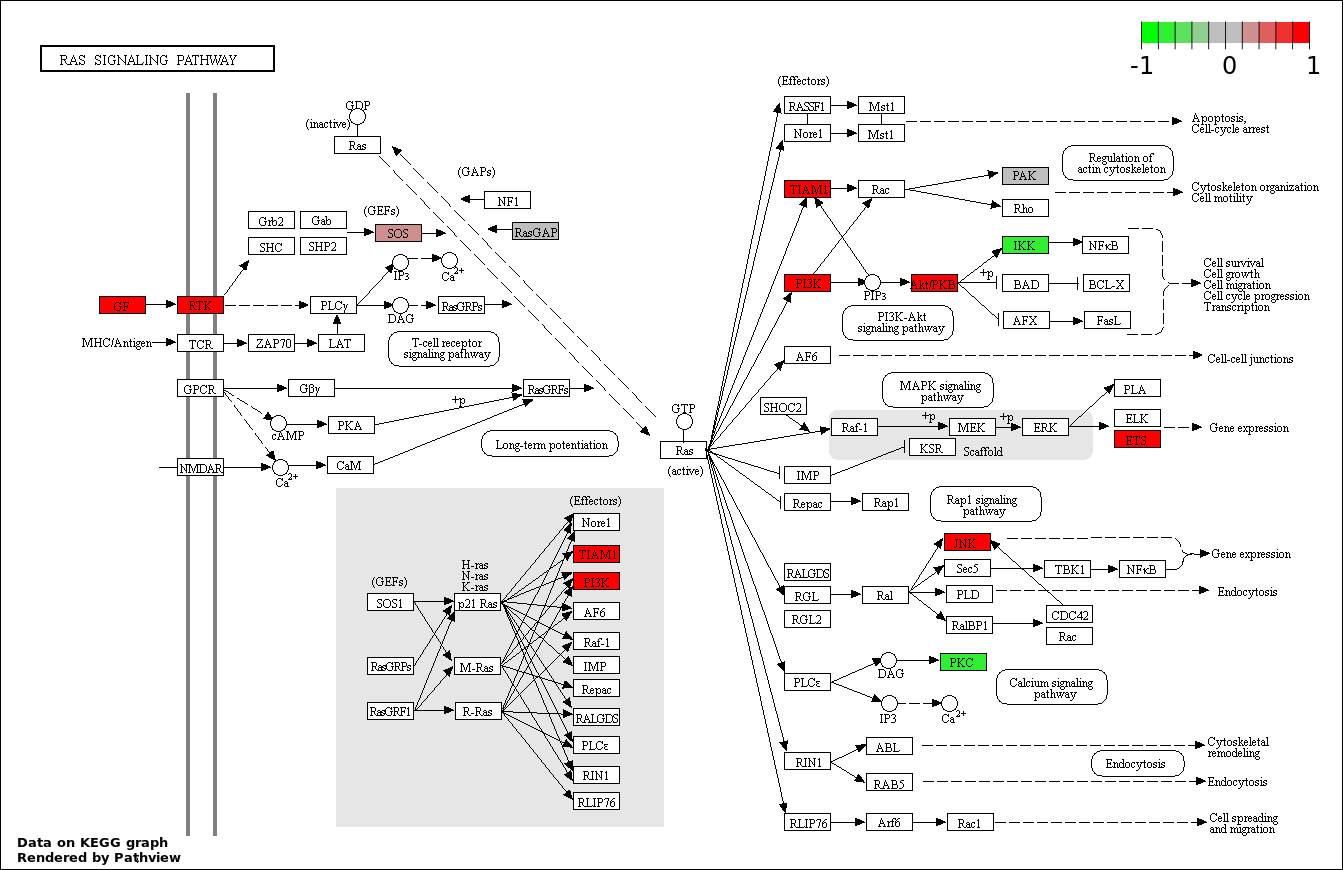
| 1 | hsa04010_MAPK_signaling_pathway | 27 | 268 | 1.4e-05 | 0.002521 | |
| 2 | hsa04151_PI3K_AKT_signaling_pathway | 31 | 351 | 4.512e-05 | 0.003306 | |
| 3 | hsa04390_Hippo_signaling_pathway | 18 | 154 | 5.51e-05 | 0.003306 | |
| 4 | hsa04510_Focal_adhesion | 20 | 200 | 0.0001947 | 0.008761 | |
| 5 | hsa04920_Adipocytokine_signaling_pathway | 10 | 68 | 0.0003963 | 0.01216 | |
| 6 | hsa04310_Wnt_signaling_pathway | 16 | 151 | 0.0004341 | 0.01216 | |
| 7 | hsa00514_Other_types_of_O.glycan_biosynthesis | 8 | 46 | 0.0004728 | 0.01216 | |
| 8 | hsa04014_Ras_signaling_pathway | 21 | 236 | 0.0006619 | 0.01489 | |
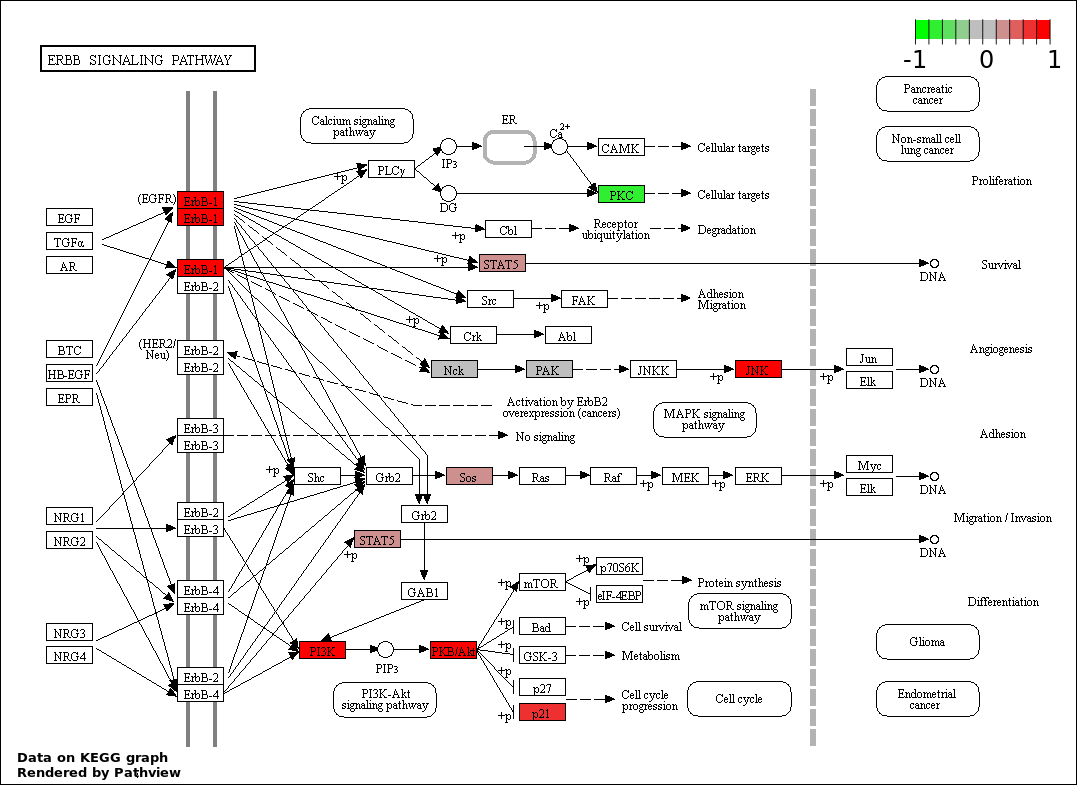
| 9 | hsa04012_ErbB_signaling_pathway | 11 | 87 | 0.0007756 | 0.01551 | |
| 10 | hsa04270_Vascular_smooth_muscle_contraction | 13 | 116 | 0.0008621 | 0.01552 | |
| 11 | hsa04810_Regulation_of_actin_cytoskeleton | 18 | 214 | 0.00289 | 0.04412 | |
| 12 | hsa04320_Dorso.ventral_axis_formation | 5 | 25 | 0.002941 | 0.04412 | |
| 13 | hsa00053_Ascorbate_and_aldarate_metabolism | 5 | 26 | 0.003522 | 0.04876 | |
| 14 | hsa04910_Insulin_signaling_pathway | 13 | 138 | 0.004107 | 0.05281 | |
| 15 | hsa00500_Starch_and_sucrose_metabolism | 7 | 54 | 0.005959 | 0.07151 | |
| 16 | hsa00980_Metabolism_of_xenobiotics_by_cytochrome_P450 | 8 | 71 | 0.007941 | 0.08016 | |
| 17 | hsa04530_Tight_junction | 12 | 133 | 0.007988 | 0.08016 | |
| 18 | hsa00140_Steroid_hormone_biosynthesis | 7 | 57 | 0.008016 | 0.08016 | |
| 19 | hsa00040_Pentose_and_glucuronate_interconversions | 5 | 32 | 0.008825 | 0.0836 | |
| 20 | hsa04520_Adherens_junction | 8 | 73 | 0.009351 | 0.08416 | |
| 21 | hsa04971_Gastric_acid_secretion | 8 | 74 | 0.01012 | 0.08677 | |
| 22 | hsa04660_T_cell_receptor_signaling_pathway | 10 | 108 | 0.01244 | 0.1018 | |
| 23 | hsa04070_Phosphatidylinositol_signaling_system | 8 | 78 | 0.01369 | 0.1023 | |
| 24 | hsa04722_Neurotrophin_signaling_pathway | 11 | 127 | 0.01441 | 0.1023 | |
| 25 | hsa04664_Fc_epsilon_RI_signaling_pathway | 8 | 79 | 0.0147 | 0.1023 | |
| 26 | hsa00830_Retinol_metabolism | 7 | 64 | 0.01478 | 0.1023 | |
| 27 | hsa04115_p53_signaling_pathway | 7 | 69 | 0.02161 | 0.1389 | |
| 28 | hsa04912_GnRH_signaling_pathway | 9 | 101 | 0.02161 | 0.1389 | |
| 29 | hsa04630_Jak.STAT_signaling_pathway | 12 | 155 | 0.02434 | 0.1511 | |
| 30 | hsa04960_Aldosterone.regulated_sodium_reabsorption | 5 | 42 | 0.02679 | 0.1607 | |
| 31 | hsa00982_Drug_metabolism_._cytochrome_P450 | 7 | 73 | 0.02845 | 0.1624 | |
| 32 | hsa00860_Porphyrin_and_chlorophyll_metabolism | 5 | 43 | 0.02933 | 0.1624 | |
| 33 | hsa04540_Gap_junction | 8 | 90 | 0.02977 | 0.1624 | |
| 34 | hsa00650_Butanoate_metabolism | 4 | 30 | 0.03196 | 0.1664 | |
| 35 | hsa04662_B_cell_receptor_signaling_pathway | 7 | 75 | 0.03236 | 0.1664 | |
| 36 | hsa04370_VEGF_signaling_pathway | 7 | 76 | 0.03445 | 0.1722 | |
| 37 | hsa04380_Osteoclast_differentiation | 10 | 128 | 0.03577 | 0.174 | |
| 38 | hsa04141_Protein_processing_in_endoplasmic_reticulum | 12 | 168 | 0.0415 | 0.1966 | |
| 39 | hsa00510_N.Glycan_biosynthesis | 5 | 49 | 0.04777 | 0.2205 | |
| 40 | hsa00072_Synthesis_and_degradation_of_ketone_bodies | 2 | 9 | 0.04903 | 0.2207 | |
| 41 | hsa04916_Melanogenesis | 8 | 101 | 0.05306 | 0.233 | |
| 42 | hsa04350_TGF.beta_signaling_pathway | 7 | 85 | 0.05726 | 0.2454 | |
| 43 | hsa00983_Drug_metabolism_._other_enzymes | 5 | 52 | 0.05907 | 0.2473 | |
| 44 | hsa04914_Progesterone.mediated_oocyte_maturation | 7 | 87 | 0.06335 | 0.2527 | |
| 45 | hsa04730_Long.term_depression | 6 | 70 | 0.06446 | 0.2527 | |
| 46 | hsa04710_Circadian_rhythm_._mammal | 3 | 23 | 0.06459 | 0.2527 | |
| 47 | hsa04210_Apoptosis | 7 | 89 | 0.06982 | 0.2674 | |
| 48 | hsa04977_Vitamin_digestion_and_absorption | 3 | 24 | 0.07167 | 0.2688 | |
| 49 | hsa04360_Axon_guidance | 9 | 130 | 0.08278 | 0.298 | |
| 50 | hsa00534_Glycosaminoglycan_biosynthesis_._heparan_sulfate | 3 | 26 | 0.08682 | 0.3064 | |
| 51 | hsa00380_Tryptophan_metabolism | 4 | 42 | 0.08968 | 0.3104 | |
| 52 | hsa04114_Oocyte_meiosis | 8 | 114 | 0.09259 | 0.3144 | |
| 53 | hsa00280_Valine._leucine_and_isoleucine_degradation | 4 | 44 | 0.1023 | 0.3287 | |
| 54 | hsa00310_Lysine_degradation | 4 | 44 | 0.1023 | 0.3287 | |
| 55 | hsa04973_Carbohydrate_digestion_and_absorption | 4 | 44 | 0.1023 | 0.3287 | |
| 56 | hsa04020_Calcium_signaling_pathway | 11 | 177 | 0.1068 | 0.3373 | |
| 57 | hsa04620_Toll.like_receptor_signaling_pathway | 7 | 102 | 0.121 | 0.3693 | |
| 58 | hsa04512_ECM.receptor_interaction | 6 | 85 | 0.1313 | 0.394 | |
| 59 | hsa00120_Primary_bile_acid_biosynthesis | 2 | 16 | 0.1359 | 0.3945 | |
| 60 | hsa00770_Pantothenate_and_CoA_biosynthesis | 2 | 16 | 0.1359 | 0.3945 | |
| 61 | hsa00260_Glycine._serine_and_threonine_metabolism | 3 | 32 | 0.1391 | 0.3976 | |
| 62 | hsa00360_Phenylalanine_metabolism | 2 | 17 | 0.15 | 0.4155 | |
| 63 | hsa00450_Selenocompound_metabolism | 2 | 17 | 0.15 | 0.4155 | |
| 64 | hsa04970_Salivary_secretion | 6 | 89 | 0.1533 | 0.4157 | |
| 65 | hsa04720_Long.term_potentiation | 5 | 70 | 0.1547 | 0.4157 | |
| 66 | hsa04150_mTOR_signaling_pathway | 4 | 52 | 0.1597 | 0.4227 | |
| 67 | hsa00630_Glyoxylate_and_dicarboxylate_metabolism | 2 | 18 | 0.1645 | 0.4291 | |
| 68 | hsa03022_Basal_transcription_factors | 3 | 37 | 0.1889 | 0.4789 | |
| 69 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 6 | 95 | 0.1889 | 0.4789 | |
| 70 | hsa04340_Hedgehog_signaling_pathway | 4 | 56 | 0.192 | 0.48 | |
| 71 | hsa00562_Inositol_phosphate_metabolism | 4 | 57 | 0.2004 | 0.4941 | |
| 72 | hsa04144_Endocytosis | 11 | 203 | 0.204 | 0.4962 | |
| 73 | hsa00410_beta.Alanine_metabolism | 2 | 22 | 0.2241 | 0.5308 | |
| 74 | hsa00910_Nitrogen_metabolism | 2 | 23 | 0.2393 | 0.5523 | |
| 75 | hsa03060_Protein_export | 2 | 23 | 0.2393 | 0.5523 | |
| 76 | hsa02010_ABC_transporters | 3 | 44 | 0.2644 | 0.6025 | |
| 77 | hsa00601_Glycosphingolipid_biosynthesis_._lacto_and_neolacto_series | 2 | 26 | 0.2851 | 0.6414 | |
| 78 | hsa04330_Notch_signaling_pathway | 3 | 47 | 0.2979 | 0.662 | |
| 79 | hsa04610_Complement_and_coagulation_cascades | 4 | 69 | 0.3073 | 0.6746 | |
| 80 | hsa00250_Alanine._aspartate_and_glutamate_metabolism | 2 | 32 | 0.3751 | 0.7698 | |
| 81 | hsa00640_Propanoate_metabolism | 2 | 32 | 0.3751 | 0.7698 | |
| 82 | hsa00330_Arginine_and_proline_metabolism | 3 | 54 | 0.3763 | 0.7698 | |
| 83 | hsa04972_Pancreatic_secretion | 5 | 101 | 0.3912 | 0.7912 | |
| 84 | hsa04146_Peroxisome | 4 | 79 | 0.4001 | 0.8003 | |
| 85 | hsa04974_Protein_digestion_and_absorption | 4 | 81 | 0.4186 | 0.8279 | |
| 86 | hsa00270_Cysteine_and_methionine_metabolism | 2 | 36 | 0.4326 | 0.8464 | |
| 87 | hsa04514_Cell_adhesion_molecules_.CAMs. | 6 | 136 | 0.4769 | 0.894 | |
| 88 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 6 | 136 | 0.4769 | 0.894 | |
| 89 | hsa04640_Hematopoietic_cell_lineage | 4 | 88 | 0.4818 | 0.894 | |
| 90 | hsa00350_Tyrosine_metabolism | 2 | 41 | 0.5002 | 0.9105 | |
| 91 | hsa04062_Chemokine_signaling_pathway | 8 | 189 | 0.5031 | 0.9105 | |
| 92 | hsa00071_Fatty_acid_metabolism | 2 | 43 | 0.5258 | 0.9281 | |
| 93 | hsa04142_Lysosome | 5 | 121 | 0.5479 | 0.9483 | |
| 94 | hsa04622_RIG.I.like_receptor_signaling_pathway | 3 | 71 | 0.5546 | 0.9508 | |
| 95 | hsa04110_Cell_cycle | 5 | 128 | 0.5982 | 1 | |
| 96 | hsa00564_Glycerophospholipid_metabolism | 3 | 80 | 0.6358 | 1 | |
| 97 | hsa04623_Cytosolic_DNA.sensing_pathway | 2 | 56 | 0.6696 | 1 | |
| 98 | hsa04621_NOD.like_receptor_signaling_pathway | 2 | 59 | 0.6973 | 1 | |
| 99 | hsa04670_Leukocyte_transendothelial_migration | 4 | 117 | 0.7045 | 1 | |
| 100 | hsa03320_PPAR_signaling_pathway | 2 | 70 | 0.7828 | 1 | |
| 101 | hsa04976_Bile_secretion | 2 | 71 | 0.7894 | 1 | |
| 102 | hsa00230_Purine_metabolism | 5 | 162 | 0.7917 | 1 | |
| 103 | hsa04120_Ubiquitin_mediated_proteolysis | 4 | 139 | 0.8208 | 1 | |
| 104 | hsa04145_Phagosome | 4 | 156 | 0.8823 | 1 | |
| 105 | hsa00240_Pyrimidine_metabolism | 2 | 99 | 0.9148 | 1 | |
| 106 | hsa03013_RNA_transport | 2 | 152 | 0.9866 | 1 | |
| 107 | hsa04740_Olfactory_transduction | 2 | 388 | 1 | 1 |