This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-942-5p | ACADL | 1.91 | 0 | -4.56 | 0 | MirTarget | -0.64 | 0.0003 | NA | |
| 2 | hsa-miR-199a-3p | ACOX1 | 0.85 | 0.00036 | -0.89 | 0.00024 | PITA | -0.4 | 0 | NA | |
| 3 | hsa-miR-199b-3p | ACOX1 | 0.86 | 0.00032 | -0.89 | 0.00024 | PITA | -0.4 | 0 | NA | |
| 4 | hsa-miR-23a-3p | ACOX1 | 1.11 | 0 | -0.89 | 0.00024 | mirMAP | -0.46 | 0 | NA | |
| 5 | hsa-miR-409-3p | ACOX1 | 0.65 | 0.05151 | -0.89 | 0.00024 | mirMAP | -0.15 | 0.00302 | NA | |
| 6 | hsa-miR-424-5p | ACOX1 | 1.09 | 0.00042 | -0.89 | 0.00024 | miRNATAP | -0.16 | 0.00524 | NA | |
| 7 | hsa-miR-708-3p | ACOX1 | 0.62 | 0.27536 | -0.89 | 0.00024 | mirMAP | -0.2 | 0 | NA | |
| 8 | hsa-miR-429 | ACOX2 | 1.4 | 0.009 | -1.9 | 8.0E-5 | miRanda; miRNATAP | -0.21 | 0.00122 | NA | |
| 9 | hsa-miR-590-3p | ACOX2 | 1.12 | 0.00016 | -1.9 | 8.0E-5 | miRanda | -0.35 | 0.00303 | NA | |
| 10 | hsa-miR-455-5p | ACOX3 | 1.2 | 7.0E-5 | -1 | 4.0E-5 | miRanda | -0.2 | 0.00044 | NA | |
| 11 | hsa-miR-130b-3p | ACSL1 | 1.33 | 5.0E-5 | -0.82 | 0.0037 | MirTarget; miRNATAP | -0.17 | 0.00551 | NA | |
| 12 | hsa-miR-181b-5p | ACSL1 | 1.64 | 0 | -0.82 | 0.0037 | MirTarget | -0.22 | 0.00084 | NA | |
| 13 | hsa-miR-191-5p | ACSL1 | 0.96 | 0.00071 | -0.82 | 0.0037 | miRNAWalker2 validate | -0.36 | 0 | NA | |
| 14 | hsa-miR-2110 | ACSL1 | 0.76 | 0.0724 | -0.82 | 0.0037 | MirTarget | -0.15 | 0.00173 | NA | |
| 15 | hsa-miR-301a-3p | ACSL1 | 1.45 | 1.0E-5 | -0.82 | 0.0037 | MirTarget; miRNATAP | -0.17 | 0.00736 | NA | |
| 16 | hsa-miR-335-5p | ACSL1 | 1.6 | 6.0E-5 | -0.82 | 0.0037 | miRNAWalker2 validate | -0.17 | 0.00066 | NA | |
| 17 | hsa-miR-454-3p | ACSL1 | 0.63 | 0.01349 | -0.82 | 0.0037 | MirTarget | -0.23 | 0.00374 | NA | |
| 18 | hsa-miR-93-3p | ACSL1 | 1.89 | 0 | -0.82 | 0.0037 | miRNAWalker2 validate | -0.19 | 0.00106 | NA | |
| 19 | hsa-miR-421 | ACSL3 | 1.81 | 0 | -0.82 | 2.0E-5 | miRanda | -0.1 | 0.00915 | NA | |
| 20 | hsa-miR-224-3p | ACSL4 | 1.52 | 0.0065 | 0.01 | 0.96386 | MirTarget | -0.12 | 0.00051 | NA | |
| 21 | hsa-miR-3065-3p | ACSL4 | -1.04 | 0.02184 | 0.01 | 0.96386 | MirTarget; miRNATAP | -0.1 | 0.00804 | NA | |
| 22 | hsa-miR-378a-3p | ACSL4 | -0.19 | 0.5308 | 0.01 | 0.96386 | miRNAWalker2 validate | -0.21 | 0.0002 | NA | |
| 23 | hsa-miR-24-3p | ACSL6 | 1.09 | 0 | -1.14 | 0.09998 | MirTarget | -0.84 | 0.00014 | NA | |
| 24 | hsa-miR-455-3p | ACSL6 | 2.3 | 0 | -1.14 | 0.09998 | PITA | -0.4 | 0.0002 | NA | |
| 25 | hsa-miR-455-5p | ACSL6 | 1.2 | 7.0E-5 | -1.14 | 0.09998 | miRanda | -0.46 | 0.00437 | NA | |
| 26 | hsa-miR-21-3p | ADIPOQ | 1.75 | 0 | -5.93 | 0 | MirTarget | -0.77 | 4.0E-5 | NA | |
| 27 | hsa-miR-320b | ADIPOQ | 1.11 | 0.0005 | -5.93 | 0 | miRanda | -0.58 | 0.00065 | NA | |
| 28 | hsa-miR-3662 | ADIPOQ | 1.83 | 0.0054 | -5.93 | 0 | mirMAP; miRNATAP | -0.33 | 0.00215 | NA | |
| 29 | hsa-miR-429 | ADIPOQ | 1.4 | 0.009 | -5.93 | 0 | miRanda | -0.37 | 0.00026 | NA | |
| 30 | hsa-miR-940 | ADIPOQ | 1.64 | 0.00069 | -5.93 | 0 | MirTarget | -0.38 | 0.00425 | NA | |
| 31 | hsa-miR-214-3p | AQP7 | 1.01 | 0.00625 | -1.85 | 0.0207 | mirMAP | -0.61 | 7.0E-5 | NA | |
| 32 | hsa-miR-335-5p | CD36 | 1.6 | 6.0E-5 | -2.35 | 2.0E-5 | miRNAWalker2 validate | -0.53 | 0 | NA | |
| 33 | hsa-miR-361-5p | CD36 | 0.41 | 0.01813 | -2.35 | 2.0E-5 | miRanda | -0.98 | 3.0E-5 | NA | |
| 34 | hsa-miR-3662 | CD36 | 1.83 | 0.0054 | -2.35 | 2.0E-5 | mirMAP | -0.3 | 0.0001 | NA | |
| 35 | hsa-miR-421 | CD36 | 1.81 | 0 | -2.35 | 2.0E-5 | miRanda | -0.43 | 9.0E-5 | NA | |
| 36 | hsa-miR-629-3p | CD36 | 1.4 | 0.0001 | -2.35 | 2.0E-5 | MirTarget | -0.44 | 4.0E-5 | NA | |
| 37 | hsa-miR-96-5p | CD36 | 1.14 | 0.00943 | -2.35 | 2.0E-5 | MirTarget | -0.28 | 0.00282 | NA | |
| 38 | hsa-miR-543 | CPT1B | 0.03 | 0.92946 | -0.52 | 0.05906 | PITA | -0.17 | 0.00428 | NA | |
| 39 | hsa-miR-218-5p | FABP1 | -0.99 | 0.01083 | 2.57 | 0.08106 | mirMAP | -2.01 | 0 | NA | |
| 40 | hsa-miR-342-3p | FABP1 | 0.32 | 0.26915 | 2.57 | 0.08106 | miRanda | -1.04 | 0.00458 | NA | |
| 41 | hsa-miR-34c-5p | FABP1 | 2.21 | 0.00038 | 2.57 | 0.08106 | miRanda | -1.01 | 0 | NA | |
| 42 | hsa-miR-199a-5p | FABP2 | 0.37 | 0.23266 | 2.01 | 0.07561 | miRanda | -1.01 | 9.0E-5 | NA | |
| 43 | hsa-miR-944 | FABP2 | 3.33 | 0.01778 | 2.01 | 0.07561 | mirMAP | -0.48 | 0 | NA | |
| 44 | hsa-miR-335-5p | FABP4 | 1.6 | 6.0E-5 | -2.71 | 0.01064 | miRNAWalker2 validate | -1.04 | 0 | NA | |
| 45 | hsa-miR-338-5p | FABP4 | -1.2 | 0.01003 | -2.71 | 0.01064 | miRNATAP | -0.69 | 2.0E-5 | NA | |
| 46 | hsa-miR-589-3p | FABP4 | 1.02 | 0.00578 | -2.71 | 0.01064 | MirTarget | -0.8 | 7.0E-5 | NA | |
| 47 | hsa-miR-217 | FABP5 | -0.01 | 0.98082 | 1.56 | 0.04444 | miRanda | -0.46 | 3.0E-5 | NA | |
| 48 | hsa-miR-2110 | FADS2 | 0.76 | 0.0724 | 0.87 | 0.08837 | miRNATAP | -0.26 | 0.00326 | NA | |
| 49 | hsa-miR-944 | GK | 3.33 | 0.01778 | 1.1 | 0.00224 | MirTarget; PITA | -0.12 | 0 | NA | |
| 50 | hsa-miR-199a-5p | HMGCS2 | 0.37 | 0.23266 | -4.27 | 0.00932 | miRanda | -1.91 | 0 | NA | |
| 51 | hsa-miR-185-3p | ILK | 1.34 | 3.0E-5 | -0.7 | 0.00014 | MirTarget; miRNATAP | -0.12 | 0.00261 | NA | |
| 52 | hsa-miR-222-3p | ILK | 1.39 | 0 | -0.7 | 0.00014 | miRNAWalker2 validate | -0.19 | 3.0E-5 | NA | |
| 53 | hsa-miR-335-3p | ILK | 2.52 | 0 | -0.7 | 0.00014 | MirTarget | -0.13 | 2.0E-5 | NA | |
| 54 | hsa-miR-342-5p | ILK | 0.69 | 0.031 | -0.7 | 0.00014 | miRNATAP | -0.12 | 0.00265 | NA | |
| 55 | hsa-miR-142-3p | ME1 | 1.32 | 0.00015 | -0.01 | 0.9842 | PITA; miRanda | -0.24 | 0.00327 | NA | |
| 56 | hsa-miR-30a-5p | ME1 | -1.72 | 0 | -0.01 | 0.9842 | miRNATAP | -0.28 | 0.00074 | NA | |
| 57 | hsa-miR-30c-5p | ME1 | -0.98 | 5.0E-5 | -0.01 | 0.9842 | miRNATAP | -0.37 | 0.00135 | NA | |
| 58 | hsa-miR-145-5p | MMP1 | -1.75 | 2.0E-5 | 4.95 | 0 | miRNAWalker2 validate | -0.56 | 2.0E-5 | NA | |
| 59 | hsa-miR-326 | OLR1 | -0.43 | 0.26299 | 3.54 | 0 | miRanda | -0.36 | 0.00309 | NA | |
| 60 | hsa-miR-199a-5p | PCK1 | 0.37 | 0.23266 | 0.61 | 0.61617 | miRanda | -0.88 | 0.00165 | NA | |
| 61 | hsa-miR-22-5p | PDPK1 | 0.8 | 0.0008 | -0.59 | 4.0E-5 | miRNATAP | -0.13 | 0.00306 | NA | |
| 62 | hsa-miR-24-3p | PDPK1 | 1.09 | 0 | -0.59 | 4.0E-5 | miRNAWalker2 validate | -0.14 | 0.00295 | NA | |
| 63 | hsa-miR-27a-3p | PDPK1 | 1.3 | 0 | -0.59 | 4.0E-5 | miRNATAP | -0.13 | 0.00311 | NA | |
| 64 | hsa-miR-424-5p | PDPK1 | 1.09 | 0.00042 | -0.59 | 4.0E-5 | mirMAP | -0.13 | 0.00011 | NA | |
| 65 | hsa-miR-542-3p | PDPK1 | 0.76 | 0.00211 | -0.59 | 4.0E-5 | mirMAP | -0.16 | 0.00014 | NA | |
| 66 | hsa-let-7i-5p | PPARA | 0.8 | 3.0E-5 | -0.65 | 0.00315 | miRNATAP | -0.4 | 0 | NA | |
| 67 | hsa-miR-106b-5p | PPARA | 1.71 | 0 | -0.65 | 0.00315 | miRNATAP | -0.14 | 0.00437 | NA | |
| 68 | hsa-miR-127-5p | PPARA | 0.68 | 0.02163 | -0.65 | 0.00315 | mirMAP | -0.24 | 1.0E-5 | NA | |
| 69 | hsa-miR-136-3p | PPARA | -0.64 | 0.02189 | -0.65 | 0.00315 | mirMAP | -0.16 | 0.00585 | NA | |
| 70 | hsa-miR-136-5p | PPARA | -0.39 | 0.19926 | -0.65 | 0.00315 | mirMAP | -0.15 | 0.0038 | NA | |
| 71 | hsa-miR-146b-3p | PPARA | 1.1 | 4.0E-5 | -0.65 | 0.00315 | mirMAP | -0.19 | 0.00083 | NA | |
| 72 | hsa-miR-146b-5p | PPARA | 1.55 | 0 | -0.65 | 0.00315 | mirMAP | -0.19 | 0.0005 | NA | |
| 73 | hsa-miR-181a-2-3p | PPARA | 0.9 | 0.00083 | -0.65 | 0.00315 | mirMAP | -0.2 | 0.00051 | NA | |
| 74 | hsa-miR-181b-5p | PPARA | 1.64 | 0 | -0.65 | 0.00315 | miRNATAP | -0.14 | 0.00575 | NA | |
| 75 | hsa-miR-18a-5p | PPARA | 2.2 | 0 | -0.65 | 0.00315 | mirMAP | -0.11 | 0.00985 | NA | |
| 76 | hsa-miR-193b-5p | PPARA | 0.61 | 0.13924 | -0.65 | 0.00315 | mirMAP | -0.17 | 1.0E-5 | NA | |
| 77 | hsa-miR-21-5p | PPARA | 1.75 | 0 | -0.65 | 0.00315 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.25 | 0.00183 | 22244963 | In the final integrative analysis of lung cancer related miR-21-targets analysis 24 hub genes were identified by overlap calculation suggesting that miR-21 may play an important role in the development and progression of lung cancer through JAK/STAT signal pathway MAPK signaling pathway Wnt signaling pathway cell cycle PPAR signaling pathway apoptosis pathway and other pathways |
| 78 | hsa-miR-218-5p | PPARA | -0.99 | 0.01083 | -0.65 | 0.00315 | mirMAP | -0.11 | 0.00611 | NA | |
| 79 | hsa-miR-24-3p | PPARA | 1.09 | 0 | -0.65 | 0.00315 | MirTarget | -0.38 | 0 | NA | |
| 80 | hsa-miR-27a-3p | PPARA | 1.3 | 0 | -0.65 | 0.00315 | MirTarget; miRNATAP | -0.29 | 1.0E-5 | NA | |
| 81 | hsa-miR-369-3p | PPARA | 0.32 | 0.35899 | -0.65 | 0.00315 | mirMAP | -0.2 | 1.0E-5 | NA | |
| 82 | hsa-miR-382-5p | PPARA | 0.59 | 0.04514 | -0.65 | 0.00315 | mirMAP | -0.23 | 1.0E-5 | NA | |
| 83 | hsa-miR-409-3p | PPARA | 0.65 | 0.05151 | -0.65 | 0.00315 | mirMAP | -0.13 | 0.00519 | NA | |
| 84 | hsa-miR-409-5p | PPARA | 1.11 | 0.00089 | -0.65 | 0.00315 | MirTarget | -0.22 | 0 | NA | |
| 85 | hsa-miR-421 | PPARA | 1.81 | 0 | -0.65 | 0.00315 | miRanda; mirMAP | -0.12 | 0.00869 | NA | |
| 86 | hsa-miR-455-5p | PPARA | 1.2 | 7.0E-5 | -0.65 | 0.00315 | miRanda | -0.16 | 0.0018 | NA | |
| 87 | hsa-miR-543 | PPARA | 0.03 | 0.92946 | -0.65 | 0.00315 | miRanda | -0.18 | 0.00016 | NA | |
| 88 | hsa-miR-654-3p | PPARA | -0.18 | 0.60394 | -0.65 | 0.00315 | mirMAP | -0.15 | 0.00086 | NA | |
| 89 | hsa-miR-940 | PPARA | 1.64 | 0.00069 | -0.65 | 0.00315 | mirMAP | -0.17 | 0 | NA | |
| 90 | hsa-miR-129-5p | PPARD | -2.57 | 0 | -0.04 | 0.8611 | miRanda | -0.11 | 0.00037 | NA | |
| 91 | hsa-miR-148a-3p | PPARD | -1.54 | 4.0E-5 | -0.04 | 0.8611 | mirMAP | -0.15 | 0.00111 | NA | |
| 92 | hsa-miR-30b-3p | PPARD | -0.27 | 0.40085 | -0.04 | 0.8611 | mirMAP | -0.15 | 0.00807 | NA | |
| 93 | hsa-miR-326 | PPARD | -0.43 | 0.26299 | -0.04 | 0.8611 | miRanda; mirMAP | -0.12 | 0.00819 | NA | |
| 94 | hsa-miR-423-5p | PPARD | 0.83 | 0.00079 | -0.04 | 0.8611 | MirTarget; mirMAP | -0.28 | 7.0E-5 | NA | |
| 95 | hsa-miR-454-3p | PPARD | 0.63 | 0.01349 | -0.04 | 0.8611 | mirMAP | -0.19 | 0.00667 | NA | |
| 96 | hsa-miR-497-5p | PPARD | -0.8 | 0.0036 | -0.04 | 0.8611 | mirMAP | -0.17 | 0.00665 | NA | |
| 97 | hsa-miR-625-3p | PPARD | -0.19 | 0.62622 | -0.04 | 0.8611 | mirMAP | -0.14 | 0.00179 | NA | |
| 98 | hsa-miR-127-3p | PPARG | 0.21 | 0.46101 | -0.72 | 0.33883 | miRanda | -0.72 | 0.00011 | NA | |
| 99 | hsa-miR-130a-3p | PPARG | 0.15 | 0.63374 | -0.72 | 0.33883 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -1.01 | 0 | NA | |
| 100 | hsa-miR-199a-5p | PPARG | 0.37 | 0.23266 | -0.72 | 0.33883 | miRanda | -0.69 | 5.0E-5 | NA | |
| 101 | hsa-miR-27a-3p | PPARG | 1.3 | 0 | -0.72 | 0.33883 | MirTarget; miRNATAP | -1.49 | 0 | NA | |
| 102 | hsa-miR-27b-3p | PPARG | 0.08 | 0.72527 | -0.72 | 0.33883 | miRNAWalker2 validate; MirTarget; miRNATAP | -1.09 | 0 | NA | |
| 103 | hsa-miR-34c-5p | PPARG | 2.21 | 0.00038 | -0.72 | 0.33883 | miRanda | -0.5 | 0 | NA | |
| 104 | hsa-miR-455-5p | PPARG | 1.2 | 7.0E-5 | -0.72 | 0.33883 | miRanda | -0.79 | 0 | NA | |
| 105 | hsa-let-7g-3p | RXRG | 0.65 | 0.01031 | -3.75 | 0 | MirTarget; miRNATAP | -0.99 | 0 | NA | |
| 106 | hsa-miR-10a-5p | SCD | -0.47 | 0.1488 | 1.71 | 3.0E-5 | miRNAWalker2 validate | -0.34 | 0.00021 | NA | |
| 107 | hsa-miR-192-5p | SCD | 0.08 | 0.94106 | 1.71 | 3.0E-5 | miRNAWalker2 validate | -0.12 | 0 | NA | |
| 108 | hsa-miR-30a-5p | SCD | -1.72 | 0 | 1.71 | 3.0E-5 | miRNAWalker2 validate | -0.41 | 0 | NA | |
| 109 | hsa-miR-664a-5p | SCD | -0.13 | 0.62435 | 1.71 | 3.0E-5 | MirTarget | -0.4 | 0.00023 | NA | |
| 110 | hsa-miR-126-5p | SCD5 | 0.42 | 0.07532 | -1.16 | 0.00344 | mirMAP | -0.44 | 0.0003 | NA | |
| 111 | hsa-miR-140-3p | SCD5 | -0.3 | 0.12754 | -1.16 | 0.00344 | miRNAWalker2 validate | -0.41 | 0.0061 | NA | |
| 112 | hsa-miR-150-5p | SCD5 | 0.15 | 0.75372 | -1.16 | 0.00344 | MirTarget | -0.24 | 9.0E-5 | NA | |
| 113 | hsa-miR-17-5p | SCD5 | 1.66 | 0 | -1.16 | 0.00344 | TargetScan | -0.24 | 0.00677 | NA | |
| 114 | hsa-miR-320a | SCD5 | 0.59 | 0.0119 | -1.16 | 0.00344 | miRanda | -0.34 | 0.00522 | NA | |
| 115 | hsa-miR-330-3p | SCD5 | 0.8 | 0.00747 | -1.16 | 0.00344 | PITA | -0.33 | 0.00053 | NA | |
| 116 | hsa-miR-339-5p | SCD5 | 0.77 | 0.00817 | -1.16 | 0.00344 | miRanda | -0.25 | 0.00965 | NA | |
| 117 | hsa-miR-362-3p | SCD5 | -0.03 | 0.91378 | -1.16 | 0.00344 | miRanda | -0.36 | 6.0E-5 | NA | |
| 118 | hsa-miR-421 | SCD5 | 1.81 | 0 | -1.16 | 0.00344 | miRanda | -0.22 | 0.00702 | NA | |
| 119 | hsa-miR-590-5p | SCD5 | 1.04 | 0.00027 | -1.16 | 0.00344 | miRanda | -0.26 | 0.00994 | NA | |
| 120 | hsa-miR-592 | SCD5 | 0.78 | 0.2045 | -1.16 | 0.00344 | MirTarget | -0.22 | 1.0E-5 | NA | |
| 121 | hsa-miR-92a-3p | SCD5 | 1.22 | 1.0E-5 | -1.16 | 0.00344 | miRNAWalker2 validate | -0.27 | 0.00986 | NA | |
| 122 | hsa-miR-127-5p | SCP2 | 0.68 | 0.02163 | -0.54 | 0.0021 | PITA; miRNATAP | -0.2 | 0 | NA | |
| 123 | hsa-miR-146b-5p | SCP2 | 1.55 | 0 | -0.54 | 0.0021 | miRanda | -0.13 | 0.00335 | NA | |
| 124 | hsa-miR-193b-3p | SCP2 | 0.28 | 0.45126 | -0.54 | 0.0021 | MirTarget | -0.17 | 0 | NA | |
| 125 | hsa-miR-28-5p | SCP2 | -0.24 | 0.16732 | -0.54 | 0.0021 | miRanda | -0.2 | 0.00868 | NA | |
| 126 | hsa-miR-452-5p | SCP2 | 2.09 | 0.00026 | -0.54 | 0.0021 | MirTarget | -0.12 | 0 | NA | |
| 127 | hsa-miR-769-5p | SCP2 | 1.15 | 0 | -0.54 | 0.0021 | miRNAWalker2 validate | -0.15 | 0.0039 | NA | |
| 128 | hsa-miR-18a-5p | SLC27A1 | 2.2 | 0 | -0.62 | 0.01517 | mirMAP | -0.16 | 0.00056 | NA | |
| 129 | hsa-miR-324-5p | SLC27A1 | 1.09 | 3.0E-5 | -0.62 | 0.01517 | miRanda | -0.22 | 0.0015 | NA | |
| 130 | hsa-miR-421 | SLC27A1 | 1.81 | 0 | -0.62 | 0.01517 | miRanda; mirMAP | -0.21 | 4.0E-5 | NA | |
| 131 | hsa-miR-940 | SLC27A1 | 1.64 | 0.00069 | -0.62 | 0.01517 | MirTarget | -0.18 | 4.0E-5 | NA | |
| 132 | hsa-let-7b-5p | SLC27A2 | 0.06 | 0.7814 | 2.11 | 0.00358 | miRNAWalker2 validate | -1.14 | 0 | NA | |
| 133 | hsa-miR-125a-5p | SLC27A4 | -0.88 | 0.00021 | 0.39 | 0.15278 | mirMAP; miRNATAP | -0.44 | 0 | NA | |
| 134 | hsa-miR-125b-5p | SLC27A4 | -0.51 | 0.13327 | 0.39 | 0.15278 | mirMAP; miRNATAP | -0.25 | 1.0E-5 | NA | |
| 135 | hsa-miR-335-5p | SLC27A6 | 1.6 | 6.0E-5 | -1.91 | 0.0441 | miRNAWalker2 validate | -0.48 | 0.00446 | NA | |
| 136 | hsa-miR-577 | SLC27A6 | 0.91 | 0.22561 | -1.91 | 0.0441 | MirTarget | -0.55 | 0 | NA | |
| 137 | hsa-miR-590-3p | SLC27A6 | 1.12 | 0.00016 | -1.91 | 0.0441 | miRanda; mirMAP | -0.81 | 0.00034 | NA | |
| 138 | hsa-let-7a-3p | SORBS1 | 0.5 | 0.04111 | -3.44 | 0 | miRNATAP | -0.77 | 0 | NA | |
| 139 | hsa-let-7b-3p | SORBS1 | 0.22 | 0.29604 | -3.44 | 0 | miRNATAP | -0.85 | 1.0E-5 | NA | |
| 140 | hsa-let-7g-3p | SORBS1 | 0.65 | 0.01031 | -3.44 | 0 | miRNATAP | -1.02 | 0 | NA | |
| 141 | hsa-miR-142-5p | SORBS1 | 1.56 | 1.0E-5 | -3.44 | 0 | PITA; miRNATAP | -0.51 | 1.0E-5 | NA | |
| 142 | hsa-miR-146b-5p | SORBS1 | 1.55 | 0 | -3.44 | 0 | miRanda | -0.55 | 5.0E-5 | NA | |
| 143 | hsa-miR-186-5p | SORBS1 | 0.15 | 0.43471 | -3.44 | 0 | mirMAP | -0.76 | 0.0004 | NA | |
| 144 | hsa-miR-19b-1-5p | SORBS1 | 1.39 | 0 | -3.44 | 0 | mirMAP | -0.89 | 0 | NA | |
| 145 | hsa-miR-205-5p | SORBS1 | 3.14 | 0.02932 | -3.44 | 0 | miRNATAP | -0.12 | 3.0E-5 | NA | |
| 146 | hsa-miR-22-5p | SORBS1 | 0.8 | 0.0008 | -3.44 | 0 | mirMAP | -0.73 | 1.0E-5 | NA | |
| 147 | hsa-miR-3065-5p | SORBS1 | -0.24 | 0.63312 | -3.44 | 0 | mirMAP | -0.32 | 0.0001 | NA | |
| 148 | hsa-miR-320a | SORBS1 | 0.59 | 0.0119 | -3.44 | 0 | miRNAWalker2 validate; mirMAP | -0.54 | 0.00184 | NA | |
| 149 | hsa-miR-320b | SORBS1 | 1.11 | 0.0005 | -3.44 | 0 | mirMAP | -0.66 | 0 | NA | |
| 150 | hsa-miR-320c | SORBS1 | 0.46 | 0.24061 | -3.44 | 0 | mirMAP | -0.39 | 0.00138 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CELLULAR LIPID METABOLIC PROCESS | 28 | 913 | 5.965e-30 | 2.776e-26 |
| 2 | FATTY ACID METABOLIC PROCESS | 21 | 296 | 4.227e-29 | 9.834e-26 |
| 3 | LIPID METABOLIC PROCESS | 29 | 1158 | 9.237e-29 | 1.433e-25 |
| 4 | MONOCARBOXYLIC ACID METABOLIC PROCESS | 22 | 503 | 6.306e-26 | 7.335e-23 |
| 5 | FATTY ACID TRANSPORT | 13 | 56 | 7.428e-25 | 6.913e-22 |
| 6 | ORGANIC ACID METABOLIC PROCESS | 23 | 953 | 2.382e-21 | 1.848e-18 |
| 7 | CELLULAR LIPID CATABOLIC PROCESS | 14 | 151 | 8.746e-21 | 5.814e-18 |
| 8 | MONOCARBOXYLIC ACID TRANSPORT | 13 | 124 | 4.982e-20 | 2.897e-17 |
| 9 | LONG CHAIN FATTY ACID METABOLIC PROCESS | 12 | 88 | 5.954e-20 | 3.078e-17 |
| 10 | LONG CHAIN FATTY ACID TRANSPORT | 10 | 42 | 2.372e-19 | 1.104e-16 |
| 11 | SMALL MOLECULE METABOLIC PROCESS | 26 | 1767 | 3.192e-19 | 1.35e-16 |
| 12 | LIPID CATABOLIC PROCESS | 14 | 247 | 9.856e-18 | 3.822e-15 |
| 13 | LIPID LOCALIZATION | 14 | 264 | 2.516e-17 | 9.005e-15 |
| 14 | LIPID OXIDATION | 10 | 70 | 6.17e-17 | 2.051e-14 |
| 15 | FATTY ACID CATABOLIC PROCESS | 10 | 73 | 9.627e-17 | 2.986e-14 |
| 16 | FATTY ACID BETA OXIDATION | 9 | 51 | 3.333e-16 | 9.693e-14 |
| 17 | ORGANIC ACID TRANSPORT | 13 | 261 | 9.561e-16 | 2.617e-13 |
| 18 | MONOCARBOXYLIC ACID CATABOLIC PROCESS | 10 | 96 | 1.697e-15 | 4.387e-13 |
| 19 | ORGANIC ANION TRANSPORT | 14 | 387 | 5.182e-15 | 1.269e-12 |
| 20 | LIPID BIOSYNTHETIC PROCESS | 15 | 539 | 2.072e-14 | 4.82e-12 |
| 21 | ANION TRANSPORT | 14 | 507 | 2.098e-13 | 4.649e-11 |
| 22 | NEUTRAL LIPID METABOLIC PROCESS | 8 | 85 | 3.349e-12 | 7.083e-10 |
| 23 | CARBOXYLIC ACID CATABOLIC PROCESS | 10 | 205 | 3.782e-12 | 7.332e-10 |
| 24 | ORGANIC ACID CATABOLIC PROCESS | 10 | 205 | 3.782e-12 | 7.332e-10 |
| 25 | LIPID MODIFICATION | 10 | 210 | 4.808e-12 | 8.948e-10 |
| 26 | SINGLE ORGANISM CATABOLIC PROCESS | 16 | 957 | 5.502e-12 | 9.847e-10 |
| 27 | SMALL MOLECULE CATABOLIC PROCESS | 11 | 328 | 1.584e-11 | 2.729e-09 |
| 28 | OXIDATION REDUCTION PROCESS | 15 | 898 | 3.18e-11 | 5.285e-09 |
| 29 | CELLULAR CATABOLIC PROCESS | 17 | 1322 | 6.04e-11 | 9.69e-09 |
| 30 | SINGLE ORGANISM BIOSYNTHETIC PROCESS | 17 | 1340 | 7.468e-11 | 1.158e-08 |
| 31 | REGULATION OF LIPID METABOLIC PROCESS | 10 | 282 | 8.84e-11 | 1.327e-08 |
| 32 | ION TRANSPORT | 16 | 1262 | 3.405e-10 | 4.95e-08 |
| 33 | SMALL MOLECULE BIOSYNTHETIC PROCESS | 11 | 443 | 3.908e-10 | 5.511e-08 |
| 34 | FATTY ACYL COA METABOLIC PROCESS | 6 | 51 | 5.262e-10 | 7.201e-08 |
| 35 | CATABOLIC PROCESS | 18 | 1773 | 6.267e-10 | 8.332e-08 |
| 36 | ACYL COA BIOSYNTHETIC PROCESS | 6 | 54 | 7.515e-10 | 9.451e-08 |
| 37 | THIOESTER BIOSYNTHETIC PROCESS | 6 | 54 | 7.515e-10 | 9.451e-08 |
| 38 | RESPONSE TO NUTRIENT | 8 | 191 | 2.286e-09 | 2.799e-07 |
| 39 | COENZYME BIOSYNTHETIC PROCESS | 7 | 127 | 3.776e-09 | 4.505e-07 |
| 40 | RESPONSE TO INSULIN | 8 | 205 | 3.992e-09 | 4.53e-07 |
| 41 | POSITIVE REGULATION OF LIPID METABOLIC PROCESS | 7 | 128 | 3.989e-09 | 4.53e-07 |
| 42 | RESPONSE TO EXTRACELLULAR STIMULUS | 10 | 441 | 6.679e-09 | 7.4e-07 |
| 43 | FATTY ACID BETA OXIDATION USING ACYL COA OXIDASE | 4 | 13 | 7.823e-09 | 8.272e-07 |
| 44 | ALPHA LINOLENIC ACID METABOLIC PROCESS | 4 | 13 | 7.823e-09 | 8.272e-07 |
| 45 | THIOESTER METABOLIC PROCESS | 6 | 83 | 1.055e-08 | 1.068e-06 |
| 46 | ACYL COA METABOLIC PROCESS | 6 | 83 | 1.055e-08 | 1.068e-06 |
| 47 | RESPONSE TO OXYGEN CONTAINING COMPOUND | 15 | 1381 | 1.22e-08 | 1.208e-06 |
| 48 | REGULATION OF FATTY ACID METABOLIC PROCESS | 6 | 87 | 1.404e-08 | 1.361e-06 |
| 49 | ORGANIC HYDROXY COMPOUND METABOLIC PROCESS | 10 | 482 | 1.555e-08 | 1.476e-06 |
| 50 | GLYCEROLIPID METABOLIC PROCESS | 9 | 356 | 1.677e-08 | 1.561e-06 |
| 51 | HORMONE MEDIATED SIGNALING PATHWAY | 7 | 158 | 1.727e-08 | 1.575e-06 |
| 52 | COFACTOR BIOSYNTHETIC PROCESS | 7 | 166 | 2.429e-08 | 2.174e-06 |
| 53 | MONOCARBOXYLIC ACID BIOSYNTHETIC PROCESS | 7 | 172 | 3.103e-08 | 2.725e-06 |
| 54 | FATTY ACID BETA OXIDATION USING ACYL COA DEHYDROGENASE | 4 | 18 | 3.325e-08 | 2.865e-06 |
| 55 | ORGANIC HYDROXY COMPOUND BIOSYNTHETIC PROCESS | 7 | 175 | 3.496e-08 | 2.958e-06 |
| 56 | RESPONSE TO HORMONE | 12 | 893 | 5.394e-08 | 4.46e-06 |
| 57 | UNSATURATED FATTY ACID METABOLIC PROCESS | 6 | 109 | 5.463e-08 | 4.46e-06 |
| 58 | CELLULAR RESPONSE TO HORMONE STIMULUS | 10 | 552 | 5.576e-08 | 4.473e-06 |
| 59 | LIPID HOMEOSTASIS | 6 | 110 | 5.771e-08 | 4.551e-06 |
| 60 | TRIGLYCERIDE CATABOLIC PROCESS | 4 | 21 | 6.477e-08 | 5.023e-06 |
| 61 | NEUTRAL LIPID CATABOLIC PROCESS | 4 | 26 | 1.607e-07 | 1.206e-05 |
| 62 | ACYLGLYCEROL CATABOLIC PROCESS | 4 | 26 | 1.607e-07 | 1.206e-05 |
| 63 | RESPONSE TO ENDOGENOUS STIMULUS | 14 | 1450 | 1.922e-07 | 1.42e-05 |
| 64 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 12 | 1008 | 2.017e-07 | 1.466e-05 |
| 65 | REGULATION OF MACROPHAGE DERIVED FOAM CELL DIFFERENTIATION | 4 | 28 | 2.195e-07 | 1.547e-05 |
| 66 | REGULATION OF FATTY ACID OXIDATION | 4 | 28 | 2.195e-07 | 1.547e-05 |
| 67 | VERY LONG CHAIN FATTY ACID METABOLIC PROCESS | 4 | 29 | 2.543e-07 | 1.766e-05 |
| 68 | CHEMICAL HOMEOSTASIS | 11 | 874 | 4.242e-07 | 2.902e-05 |
| 69 | POSITIVE REGULATION OF FATTY ACID METABOLIC PROCESS | 4 | 33 | 4.357e-07 | 2.938e-05 |
| 70 | RESPONSE TO FATTY ACID | 5 | 83 | 4.911e-07 | 3.265e-05 |
| 71 | COENZYME METABOLIC PROCESS | 7 | 265 | 5.885e-07 | 3.857e-05 |
| 72 | NEGATIVE REGULATION OF SMOOTH MUSCLE CELL PROLIFERATION | 4 | 36 | 6.246e-07 | 4.037e-05 |
| 73 | CARBOXYLIC ACID BIOSYNTHETIC PROCESS | 7 | 270 | 6.672e-07 | 4.196e-05 |
| 74 | ORGANIC ACID BIOSYNTHETIC PROCESS | 7 | 270 | 6.672e-07 | 4.196e-05 |
| 75 | GLYCEROLIPID CATABOLIC PROCESS | 4 | 37 | 6.994e-07 | 4.339e-05 |
| 76 | RESPONSE TO PEPTIDE | 8 | 404 | 7.441e-07 | 4.556e-05 |
| 77 | REGULATION OF CELLULAR KETONE METABOLIC PROCESS | 6 | 173 | 8.427e-07 | 5.093e-05 |
| 78 | REGULATION OF LIPID TRANSPORT | 5 | 95 | 9.64e-07 | 5.751e-05 |
| 79 | NEGATIVE REGULATION OF MACROPHAGE DERIVED FOAM CELL DIFFERENTIATION | 3 | 13 | 1.786e-06 | 0.0001039 |
| 80 | REGULATION OF CHOLESTEROL STORAGE | 3 | 13 | 1.786e-06 | 0.0001039 |
| 81 | RESPONSE TO ABIOTIC STIMULUS | 11 | 1024 | 2.017e-06 | 0.0001159 |
| 82 | ALCOHOL BIOSYNTHETIC PROCESS | 5 | 111 | 2.086e-06 | 0.0001184 |
| 83 | SULFUR COMPOUND BIOSYNTHETIC PROCESS | 6 | 203 | 2.139e-06 | 0.0001199 |
| 84 | POSITIVE REGULATION OF FATTY ACID OXIDATION | 3 | 14 | 2.27e-06 | 0.0001258 |
| 85 | COFACTOR METABOLIC PROCESS | 7 | 334 | 2.751e-06 | 0.0001506 |
| 86 | NEGATIVE REGULATION OF SMOOTH MUSCLE CELL MIGRATION | 3 | 16 | 3.484e-06 | 0.0001885 |
| 87 | ALCOHOL METABOLIC PROCESS | 7 | 348 | 3.607e-06 | 0.0001929 |
| 88 | HOMEOSTATIC PROCESS | 12 | 1337 | 4.019e-06 | 0.0002125 |
| 89 | BILE ACID BIOSYNTHETIC PROCESS | 3 | 20 | 7.055e-06 | 0.0003688 |
| 90 | CELLULAR RESPONSE TO INSULIN STIMULUS | 5 | 146 | 8.001e-06 | 0.0004136 |
| 91 | REGULATION OF RECEPTOR BIOSYNTHETIC PROCESS | 3 | 21 | 8.22e-06 | 0.0004203 |
| 92 | REGULATION OF DEFENSE RESPONSE | 9 | 759 | 9.433e-06 | 0.0004771 |
| 93 | INTRACELLULAR RECEPTOR SIGNALING PATHWAY | 5 | 168 | 1.58e-05 | 0.0007906 |
| 94 | RESPONSE TO OXYGEN LEVELS | 6 | 311 | 2.446e-05 | 0.001211 |
| 95 | CARBOHYDRATE TRANSPORT | 4 | 95 | 3.115e-05 | 0.001526 |
| 96 | NEGATIVE REGULATION OF INFLAMMATORY RESPONSE | 4 | 100 | 3.811e-05 | 0.001828 |
| 97 | REGULATION OF SMOOTH MUSCLE CELL PROLIFERATION | 4 | 100 | 3.811e-05 | 0.001828 |
| 98 | BILE ACID METABOLIC PROCESS | 3 | 35 | 3.971e-05 | 0.001886 |
| 99 | RESPONSE TO ORGANIC CYCLIC COMPOUND | 9 | 917 | 4.221e-05 | 0.001984 |
| 100 | POSITIVE REGULATION OF PROTEIN LOCALIZATION TO CELL PERIPHERY | 3 | 37 | 4.702e-05 | 0.002166 |
| 101 | POSITIVE REGULATION OF PROTEIN LOCALIZATION TO PLASMA MEMBRANE | 3 | 37 | 4.702e-05 | 0.002166 |
| 102 | NEGATIVE REGULATION OF TRANSFERASE ACTIVITY | 6 | 351 | 4.81e-05 | 0.002194 |
| 103 | REGULATION OF STEROL TRANSPORT | 3 | 38 | 5.099e-05 | 0.002281 |
| 104 | REGULATION OF CHOLESTEROL TRANSPORT | 3 | 38 | 5.099e-05 | 0.002281 |
| 105 | SULFUR COMPOUND METABOLIC PROCESS | 6 | 359 | 5.452e-05 | 0.002416 |
| 106 | CELLULAR RESPONSE TO STEROID HORMONE STIMULUS | 5 | 218 | 5.503e-05 | 0.002416 |
| 107 | STEROID BIOSYNTHETIC PROCESS | 4 | 114 | 6.364e-05 | 0.002739 |
| 108 | REGULATION OF LIPID STORAGE | 3 | 41 | 6.417e-05 | 0.002739 |
| 109 | FATTY ACID BIOSYNTHETIC PROCESS | 4 | 114 | 6.364e-05 | 0.002739 |
| 110 | RESPONSE TO COLD | 3 | 43 | 7.41e-05 | 0.003134 |
| 111 | GLUCOSE METABOLIC PROCESS | 4 | 119 | 7.522e-05 | 0.003153 |
| 112 | STEROID METABOLIC PROCESS | 5 | 237 | 8.172e-05 | 0.003395 |
| 113 | REGULATION OF PLASMA LIPOPROTEIN PARTICLE LEVELS | 3 | 45 | 8.498e-05 | 0.003499 |
| 114 | RESPONSE TO EXTERNAL STIMULUS | 12 | 1821 | 8.945e-05 | 0.003651 |
| 115 | STEROID HORMONE MEDIATED SIGNALING PATHWAY | 4 | 125 | 9.105e-05 | 0.003684 |
| 116 | REGULATION OF LIPID BIOSYNTHETIC PROCESS | 4 | 128 | 9.982e-05 | 0.004004 |
| 117 | CELLULAR RESPONSE TO ORGANIC SUBSTANCE | 12 | 1848 | 0.0001031 | 0.004068 |
| 118 | REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION TO PLASMA MEMBRANE | 3 | 48 | 0.0001032 | 0.004068 |
| 119 | CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND | 8 | 799 | 0.0001055 | 0.004126 |
| 120 | REGULATION OF SMOOTH MUSCLE CELL MIGRATION | 3 | 49 | 0.0001098 | 0.004256 |
| 121 | REGULATION OF RESPONSE TO WOUNDING | 6 | 413 | 0.0001179 | 0.004536 |
| 122 | MONOSACCHARIDE TRANSPORT | 3 | 54 | 0.0001468 | 0.005553 |
| 123 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 11 | 1618 | 0.0001457 | 0.005553 |
| 124 | CELLULAR RESPONSE TO OXYGEN LEVELS | 4 | 143 | 0.0001531 | 0.005746 |
| 125 | NEGATIVE REGULATION OF DEFENSE RESPONSE | 4 | 144 | 0.0001573 | 0.005855 |
| 126 | CELLULAR RESPONSE TO PEPTIDE | 5 | 274 | 0.0001613 | 0.005958 |
| 127 | RESPONSE TO NITROGEN COMPOUND | 8 | 859 | 0.000174 | 0.006353 |
| 128 | RESPONSE TO TEMPERATURE STIMULUS | 4 | 148 | 0.0001748 | 0.006353 |
| 129 | UNSATURATED FATTY ACID BIOSYNTHETIC PROCESS | 3 | 58 | 0.0001817 | 0.006553 |
| 130 | POSITIVE REGULATION OF MEMBRANE INVAGINATION | 2 | 11 | 0.0001913 | 0.006691 |
| 131 | ORGANOPHOSPHATE BIOSYNTHETIC PROCESS | 6 | 450 | 0.0001881 | 0.006691 |
| 132 | LOW DENSITY LIPOPROTEIN PARTICLE CLEARANCE | 2 | 11 | 0.0001913 | 0.006691 |
| 133 | POSITIVE REGULATION OF PHAGOCYTOSIS ENGULFMENT | 2 | 11 | 0.0001913 | 0.006691 |
| 134 | TRANSCRIPTION INITIATION FROM RNA POLYMERASE II PROMOTER | 4 | 153 | 0.0001985 | 0.006893 |
| 135 | CELLULAR RESPONSE TO LIPID | 6 | 457 | 0.0002045 | 0.007048 |
| 136 | POSITIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 5 | 289 | 0.0002067 | 0.007071 |
| 137 | ORGANIC HYDROXY COMPOUND TRANSPORT | 4 | 155 | 0.0002086 | 0.007086 |
| 138 | RESPONSE TO LIPID | 8 | 888 | 0.0002183 | 0.00718 |
| 139 | CARBOHYDRATE METABOLIC PROCESS | 7 | 662 | 0.0002176 | 0.00718 |
| 140 | GENERATION OF PRECURSOR METABOLITES AND ENERGY | 5 | 292 | 0.0002168 | 0.00718 |
| 141 | HEXOSE METABOLIC PROCESS | 4 | 157 | 0.0002191 | 0.00718 |
| 142 | NEGATIVE REGULATION OF RESPONSE TO WOUNDING | 4 | 156 | 0.0002138 | 0.00718 |
| 143 | CELLULAR RESPONSE TO ORGANIC CYCLIC COMPOUND | 6 | 465 | 0.0002246 | 0.007256 |
| 144 | REGULATION OF INFLAMMATORY RESPONSE | 5 | 294 | 0.0002238 | 0.007256 |
| 145 | REGULATION OF SEQUESTERING OF TRIGLYCERIDE | 2 | 12 | 0.0002292 | 0.007306 |
| 146 | NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL METABOLIC PROCESS | 2 | 12 | 0.0002292 | 0.007306 |
| 147 | WHITE FAT CELL DIFFERENTIATION | 2 | 13 | 0.0002706 | 0.008176 |
| 148 | CELL SUBSTRATE ADHESION | 4 | 164 | 0.0002589 | 0.008176 |
| 149 | LIPOPROTEIN TRANSPORT | 2 | 13 | 0.0002706 | 0.008176 |
| 150 | REGULATION OF PHAGOCYTOSIS ENGULFMENT | 2 | 13 | 0.0002706 | 0.008176 |
| 151 | LIPOPROTEIN LOCALIZATION | 2 | 13 | 0.0002706 | 0.008176 |
| 152 | REGULATION OF PHOSPHOLIPID BIOSYNTHETIC PROCESS | 2 | 13 | 0.0002706 | 0.008176 |
| 153 | NEGATIVE REGULATION OF CARDIAC MUSCLE CELL APOPTOTIC PROCESS | 2 | 13 | 0.0002706 | 0.008176 |
| 154 | REGULATION OF MEMBRANE INVAGINATION | 2 | 13 | 0.0002706 | 0.008176 |
| 155 | POSITIVE REGULATION OF KINASE ACTIVITY | 6 | 482 | 0.0002724 | 0.008177 |
| 156 | RESPONSE TO CARBOHYDRATE | 4 | 168 | 0.0002838 | 0.008464 |
| 157 | REGULATION OF PHAGOCYTOSIS | 3 | 68 | 0.0002912 | 0.008631 |
| 158 | GLUCOSE HOMEOSTASIS | 4 | 170 | 0.0002968 | 0.008687 |
| 159 | CARBOHYDRATE HOMEOSTASIS | 4 | 170 | 0.0002968 | 0.008687 |
| 160 | RESPONSE TO ACTIVITY | 3 | 69 | 0.0003041 | 0.008842 |
| 161 | RESPONSE TO LOW DENSITY LIPOPROTEIN PARTICLE | 2 | 14 | 0.0003153 | 0.009001 |
| 162 | LINOLEIC ACID METABOLIC PROCESS | 2 | 14 | 0.0003153 | 0.009001 |
| 163 | POSITIVE REGULATION OF TRANSPORT | 8 | 936 | 0.000312 | 0.009001 |
| 164 | RESPONSE TO STEROID HORMONE | 6 | 497 | 0.000321 | 0.009106 |
| 165 | RESPONSE TO ACID CHEMICAL | 5 | 319 | 0.000326 | 0.009194 |
| 166 | REGULATION OF TRANSFERASE ACTIVITY | 8 | 946 | 0.0003352 | 0.009396 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | LONG CHAIN FATTY ACID COA LIGASE ACTIVITY | 8 | 13 | 9.861e-20 | 9.161e-17 |
| 2 | FATTY ACID LIGASE ACTIVITY | 8 | 16 | 9.821e-19 | 4.562e-16 |
| 3 | FATTY ACID BINDING | 8 | 31 | 5.901e-16 | 1.827e-13 |
| 4 | LIGASE ACTIVITY FORMING CARBON SULFUR BONDS | 8 | 40 | 5.684e-15 | 1.32e-12 |
| 5 | MONOCARBOXYLIC ACID BINDING | 8 | 65 | 3.608e-13 | 6.703e-11 |
| 6 | ORGANIC ACID BINDING | 10 | 209 | 4.585e-12 | 7.098e-10 |
| 7 | FATTY ACID TRANSPORTER ACTIVITY | 5 | 12 | 1.477e-11 | 1.96e-09 |
| 8 | FATTY ACYL COA BINDING | 5 | 31 | 3.087e-09 | 3.585e-07 |
| 9 | ACYL COA DEHYDROGENASE ACTIVITY | 4 | 17 | 2.59e-08 | 2.673e-06 |
| 10 | LIPID TRANSPORTER ACTIVITY | 6 | 108 | 5.17e-08 | 4.803e-06 |
| 11 | LIPID BINDING | 10 | 657 | 2.812e-07 | 2.375e-05 |
| 12 | LIGASE ACTIVITY | 8 | 406 | 7.723e-07 | 5.979e-05 |
| 13 | COENZYME BINDING | 6 | 179 | 1.029e-06 | 7.35e-05 |
| 14 | LONG CHAIN FATTY ACID BINDING | 3 | 14 | 2.27e-06 | 0.0001318 |
| 15 | TRANSCRIPTION FACTOR ACTIVITY DIRECT LIGAND REGULATED SEQUENCE SPECIFIC DNA BINDING | 4 | 48 | 2.03e-06 | 0.0001318 |
| 16 | ELECTRON CARRIER ACTIVITY | 5 | 112 | 2.18e-06 | 0.0001318 |
| 17 | OXIDOREDUCTASE ACTIVITY ACTING ON THE CH CH GROUP OF DONORS | 4 | 57 | 4.071e-06 | 0.0002224 |
| 18 | STEROID HORMONE RECEPTOR ACTIVITY | 4 | 59 | 4.677e-06 | 0.0002414 |
| 19 | COFACTOR BINDING | 6 | 263 | 9.472e-06 | 0.0004631 |
| 20 | FLAVIN ADENINE DINUCLEOTIDE BINDING | 4 | 74 | 1.159e-05 | 0.0005382 |
| 21 | OXIDOREDUCTASE ACTIVITY | 8 | 719 | 5.04e-05 | 0.00223 |
| 22 | DRUG BINDING | 4 | 109 | 5.342e-05 | 0.002256 |
| 23 | SULFUR COMPOUND BINDING | 5 | 234 | 7.695e-05 | 0.003108 |
| 24 | TRANSPORTER ACTIVITY | 10 | 1276 | 9.827e-05 | 0.003804 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | MICROBODY PART | 10 | 93 | 1.22e-15 | 7.126e-13 |
| 2 | MICROBODY | 10 | 134 | 5.226e-14 | 1.526e-11 |
| 3 | MICROBODY MEMBRANE | 6 | 58 | 1.171e-09 | 2.28e-07 |
| 4 | MICROBODY LUMEN | 5 | 45 | 2.178e-08 | 3.179e-06 |
| 5 | MITOCHONDRION | 15 | 1633 | 1.141e-07 | 1.333e-05 |
| 6 | OUTER MEMBRANE | 6 | 190 | 1.456e-06 | 0.0001417 |
| 7 | MITOCHONDRIAL ENVELOPE | 9 | 691 | 4.417e-06 | 0.0003685 |
| 8 | MITOCHONDRIAL PART | 9 | 953 | 5.695e-05 | 0.004158 |
| 9 | NUCLEAR OUTER MEMBRANE ENDOPLASMIC RETICULUM MEMBRANE NETWORK | 9 | 1005 | 8.583e-05 | 0.00557 |
| 10 | ENVELOPE | 9 | 1090 | 0.0001594 | 0.008462 |
| 11 | ENDOPLASMIC RETICULUM | 11 | 1631 | 0.0001564 | 0.008462 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
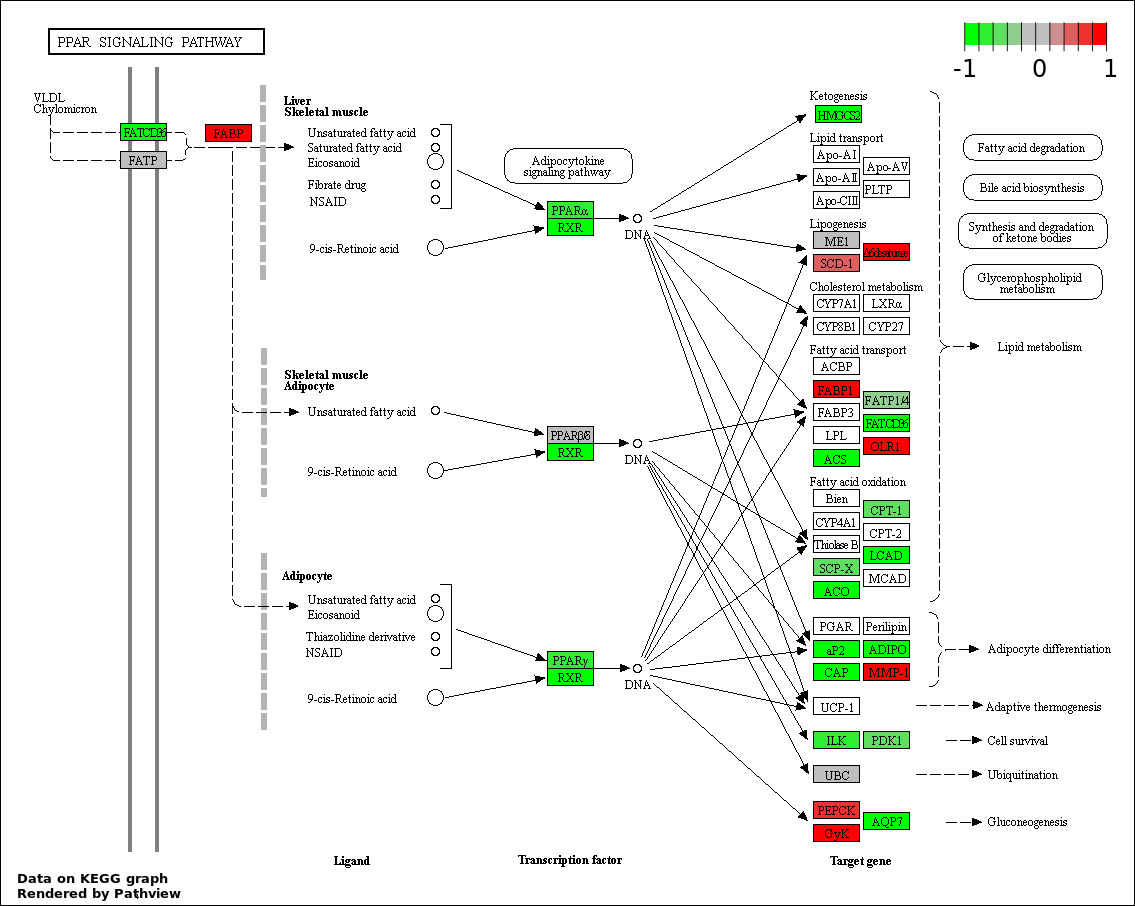
| 1 | hsa03320_PPAR_signaling_pathway | 38 | 70 | 1.715e-99 | 3.088e-97 | |
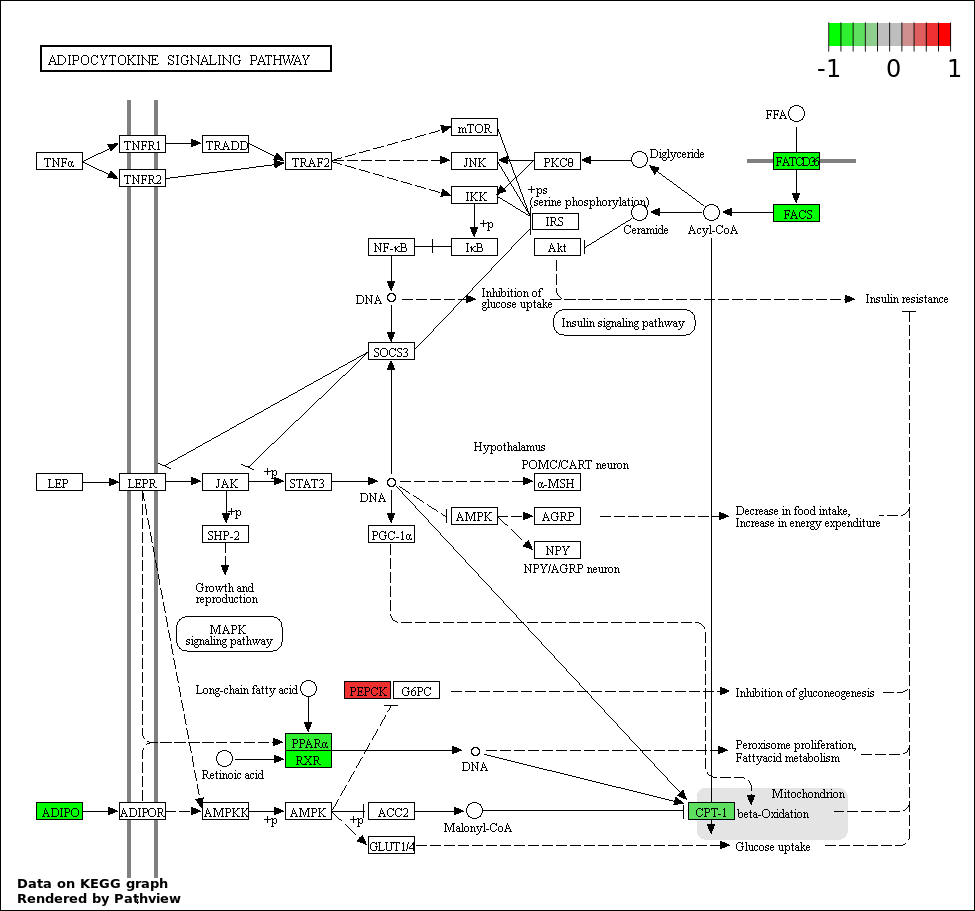
| 2 | hsa04920_Adipocytokine_signaling_pathway | 10 | 68 | 4.534e-17 | 4.08e-15 | |
| 3 | hsa00071_Fatty_acid_metabolism | 8 | 43 | 1.067e-14 | 6.404e-13 | |
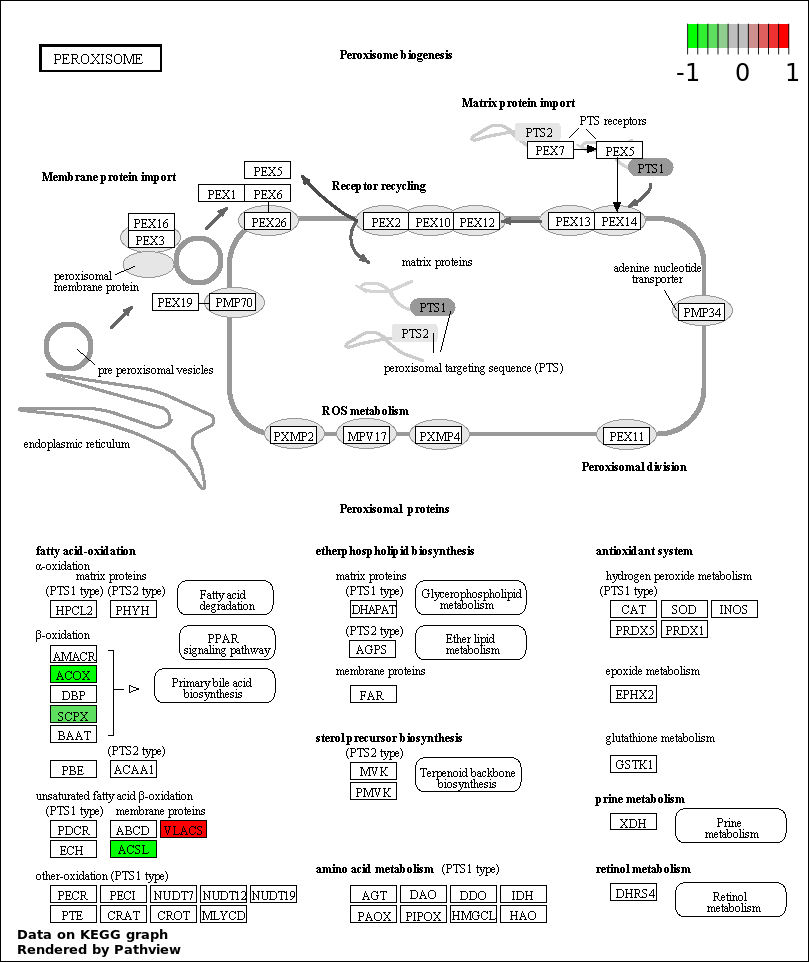
| 4 | hsa04146_Peroxisome | 9 | 79 | 2.174e-14 | 9.782e-13 | |
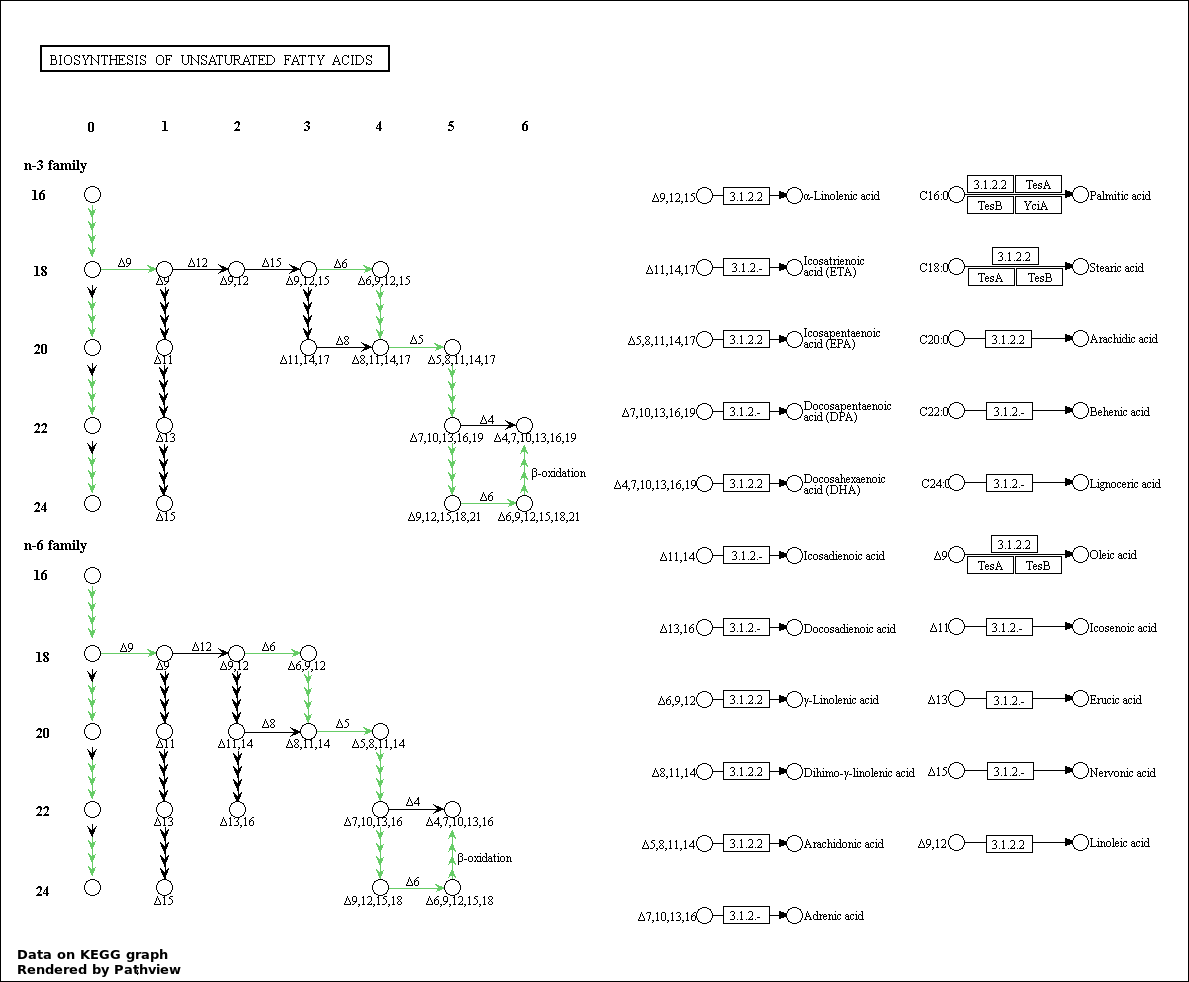
| 5 | hsa01040_Biosynthesis_of_unsaturated_fatty_acids | 5 | 21 | 3.749e-10 | 1.35e-08 | |
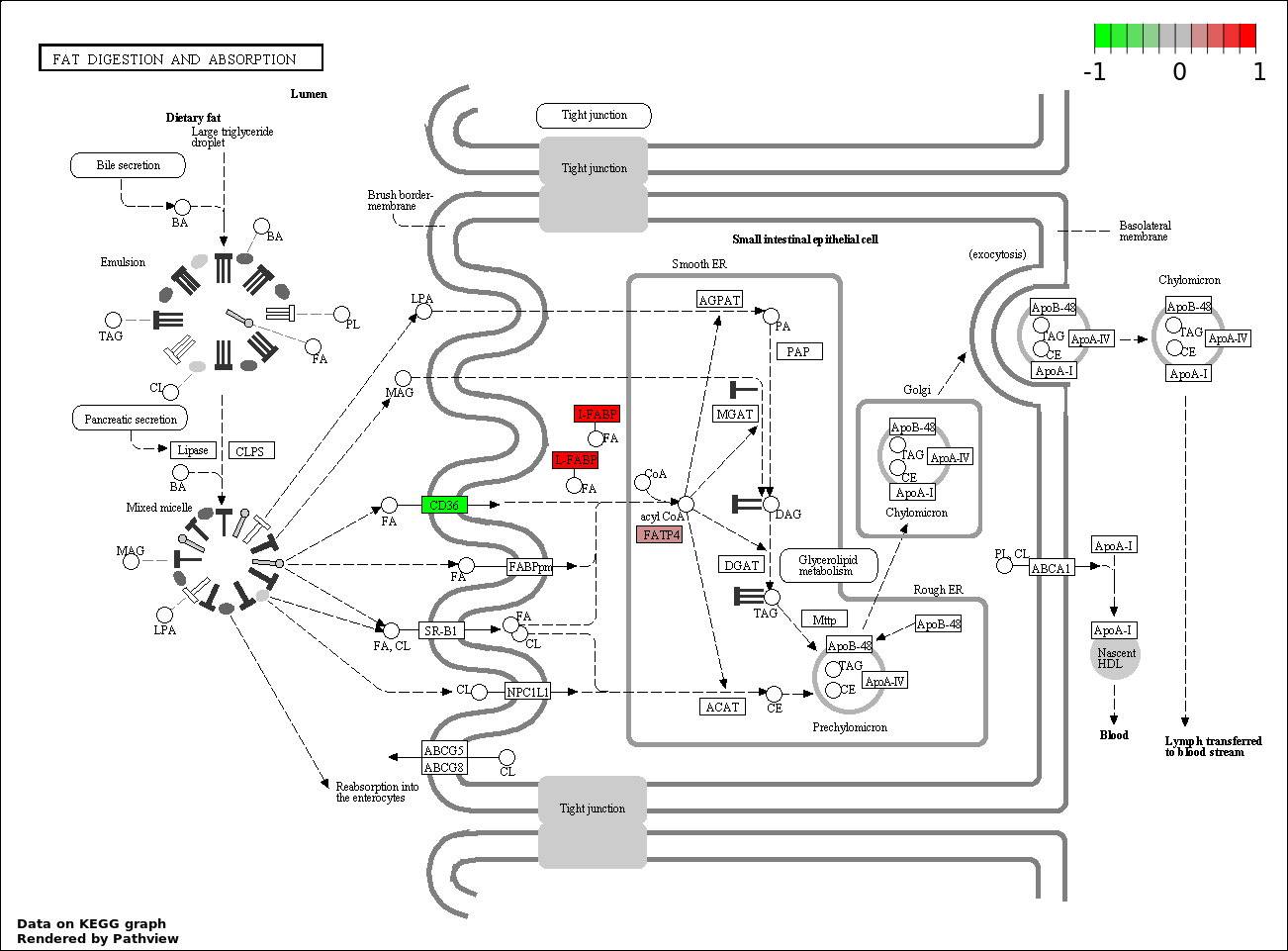
| 6 | hsa04975_Fat_digestion_and_absorption | 4 | 46 | 1.707e-06 | 5.121e-05 | |
| 7 | hsa00592_alpha.Linolenic_acid_metabolism | 3 | 20 | 7.055e-06 | 0.0001814 | |
| 8 | hsa00120_Primary_bile_acid_biosynthesis | 2 | 16 | 0.0004148 | 0.009333 | |
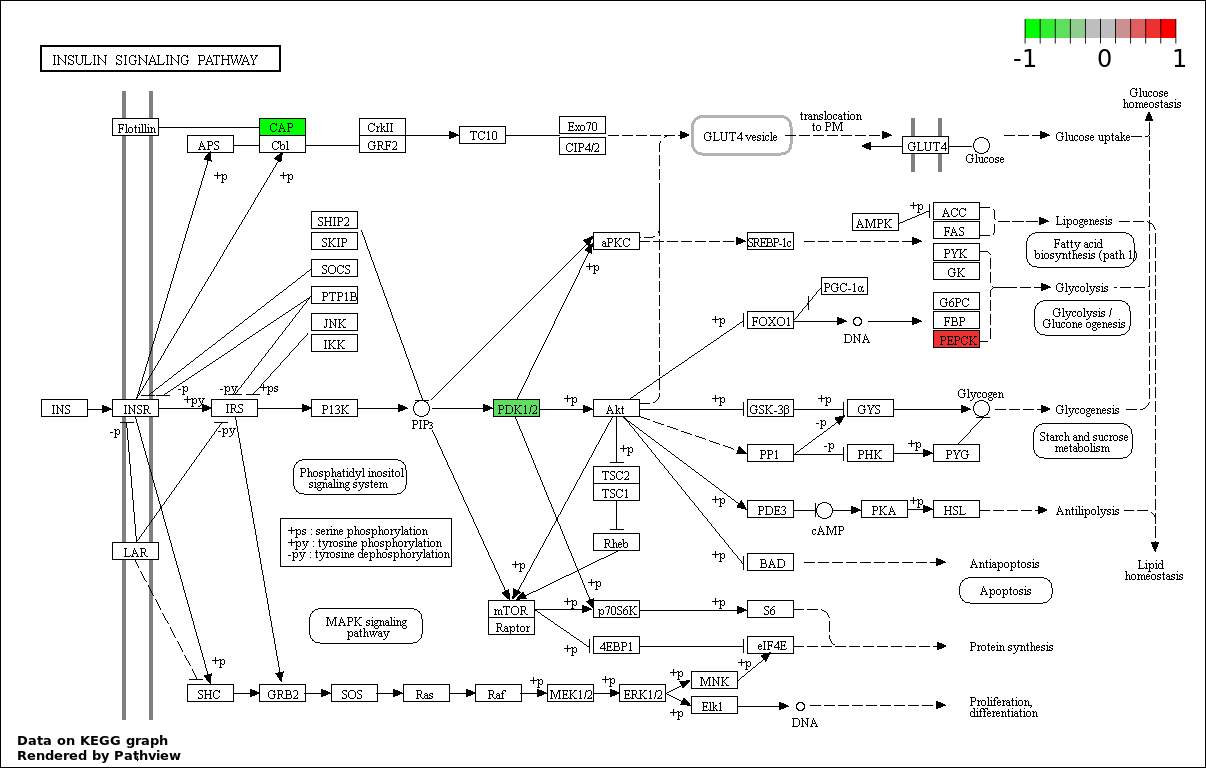
| 9 | hsa04910_Insulin_signaling_pathway | 3 | 138 | 0.002272 | 0.04545 | |
| 10 | hsa00620_Pyruvate_metabolism | 2 | 40 | 0.00262 | 0.04716 | |
| 11 | hsa04145_Phagosome | 2 | 156 | 0.03538 | 0.4899 | |
| 12 | hsa04510_Focal_adhesion | 2 | 200 | 0.0553 | 0.5876 | |
| 13 | hsa04151_PI3K_AKT_signaling_pathway | 2 | 351 | 0.1432 | 1 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-205-5p;hsa-miR-22-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-944 | 11 | SORBS1 | Sponge network | -2.778 | 8.0E-5 | -3.437 | 0 | 0.628 |
| 2 | CASC2 | hsa-miR-127-5p;hsa-miR-136-3p;hsa-miR-136-5p;hsa-miR-193b-5p;hsa-miR-218-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-654-3p;hsa-miR-940 | 11 | PPARA | Sponge network | -0.561 | 0.05962 | -0.648 | 0.00315 | 0.262 |