This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-let-7b-5p | AARSD1 | -0.96 | 0 | 0.39 | 0.0002 | miRNAWalker2 validate | -0.22 | 0 | NA | |
| 2 | hsa-let-7b-5p | AATF | -0.96 | 0 | 0.62 | 0 | miRNAWalker2 validate | -0.25 | 0 | NA | |
| 3 | hsa-let-7a-5p | ABCB9 | -0.33 | 0.00046 | 1.01 | 0 | MirTarget; TargetScan | -0.2 | 0.02597 | NA | |
| 4 | hsa-let-7c-5p | ABCB9 | -1.71 | 0 | 1.01 | 0 | MirTarget | -0.27 | 0 | NA | |
| 5 | hsa-let-7c-5p | ABCC10 | -1.71 | 0 | 1.42 | 0 | MirTarget | -0.3 | 0 | NA | |
| 6 | hsa-let-7a-5p | ABCC5 | -0.33 | 0.00046 | 1.23 | 0 | TargetScan; miRNATAP | -0.27 | 0 | NA | |
| 7 | hsa-let-7b-5p | ABCC5 | -0.96 | 0 | 1.23 | 0 | miRNATAP | -0.2 | 0 | NA | |
| 8 | hsa-let-7b-5p | ABCF1 | -0.96 | 0 | 0.32 | 1.0E-5 | miRNAWalker2 validate | -0.12 | 0 | NA | |
| 9 | hsa-let-7d-5p | ABHD14B | 0.05 | 0.70258 | -0.88 | 0 | MirTarget | -0.16 | 0.00188 | NA | |
| 10 | hsa-let-7b-5p | ACACA | -0.96 | 0 | 0.78 | 0 | miRNAWalker2 validate | -0.25 | 0 | NA | |
| 11 | hsa-let-7a-5p | ACAP3 | -0.33 | 0.00046 | 0.47 | 1.0E-5 | TargetScan | -0.12 | 0.02739 | NA | |
| 12 | hsa-let-7d-5p | ACVR1C | 0.05 | 0.70258 | -2.2 | 0 | MirTarget; miRNATAP | -0.47 | 0 | NA | |
| 13 | hsa-let-7d-5p | ADRB2 | 0.05 | 0.70258 | -1.63 | 0 | MirTarget | -0.53 | 0 | NA | |
| 14 | hsa-let-7b-5p | ADRM1 | -0.96 | 0 | 0.19 | 0.01952 | miRNAWalker2 validate | -0.16 | 0 | NA | |
| 15 | hsa-let-7a-5p | ADSL | -0.33 | 0.00046 | 0.46 | 0 | miRNAWalker2 validate | -0.17 | 0.00014 | NA | |
| 16 | hsa-let-7a-5p | AEN | -0.33 | 0.00046 | 0.28 | 0.01239 | MirTarget; TargetScan | -0.17 | 0.00498 | NA | |
| 17 | hsa-let-7b-5p | AEN | -0.96 | 0 | 0.28 | 0.01239 | MirTarget | -0.16 | 2.0E-5 | NA | |
| 18 | hsa-let-7d-5p | AEN | 0.05 | 0.70258 | 0.28 | 0.01239 | MirTarget | -0.13 | 0.00174 | NA | |
| 19 | hsa-let-7b-5p | AFF2 | -0.96 | 0 | 1.4 | 0.00046 | MirTarget | -0.39 | 0.00293 | NA | |
| 20 | hsa-let-7d-5p | AFF2 | 0.05 | 0.70258 | 1.4 | 0.00046 | MirTarget | -0.33 | 0.02581 | NA | |
| 21 | hsa-let-7b-5p | AGFG2 | -0.96 | 0 | -0.02 | 0.87061 | miRNAWalker2 validate | -0.17 | 1.0E-5 | NA | |
| 22 | hsa-let-7a-5p | AIDA | -0.33 | 0.00046 | 0.39 | 0 | miRNAWalker2 validate | -0.11 | 0.01007 | NA | |
| 23 | hsa-let-7a-5p | AIFM1 | -0.33 | 0.00046 | -0.39 | 0.00049 | TargetScan; miRNATAP | -0.16 | 0.00689 | NA | |
| 24 | hsa-let-7d-5p | AIFM1 | 0.05 | 0.70258 | -0.39 | 0.00049 | miRNATAP | -0.23 | 0 | NA | |
| 25 | hsa-let-7d-5p | AKAP6 | 0.05 | 0.70258 | -0.97 | 0 | MirTarget | -0.3 | 1.0E-5 | NA | |
| 26 | hsa-let-7b-5p | ALG3 | -0.96 | 0 | 0.6 | 0 | miRNAWalker2 validate | -0.23 | 0 | NA | |
| 27 | hsa-let-7b-5p | AMT | -0.96 | 0 | -0.85 | 0 | MirTarget | -0.16 | 0.00457 | NA | |
| 28 | hsa-let-7d-5p | AMT | 0.05 | 0.70258 | -0.85 | 0 | MirTarget | -0.3 | 0 | NA | |
| 29 | hsa-let-7c-5p | ANKRD49 | -1.71 | 0 | 0.4 | 0 | MirTarget | -0.1 | 0 | NA | |
| 30 | hsa-let-7a-5p | ANKRD52 | -0.33 | 0.00046 | 1.46 | 0 | TargetScan; mirMAP; miRNATAP | -0.18 | 0.00575 | NA | |
| 31 | hsa-let-7b-5p | ANKRD52 | -0.96 | 0 | 1.46 | 0 | miRNAWalker2 validate; mirMAP; miRNATAP | -0.21 | 0 | NA | |
| 32 | hsa-let-7b-5p | ANKZF1 | -0.96 | 0 | 0.85 | 0 | miRNAWalker2 validate | -0.25 | 0 | NA | |
| 33 | hsa-let-7a-5p | AP1S1 | -0.33 | 0.00046 | 0.55 | 0 | MirTarget; TargetScan; miRNATAP | -0.23 | 1.0E-5 | NA | |
| 34 | hsa-let-7b-5p | AP1S1 | -0.96 | 0 | 0.55 | 0 | MirTarget; miRNATAP | -0.17 | 0 | NA | |
| 35 | hsa-let-7d-5p | AP1S1 | 0.05 | 0.70258 | 0.55 | 0 | MirTarget; miRNATAP | -0.17 | 0 | NA | |
| 36 | hsa-let-7a-5p | APC2 | -0.33 | 0.00046 | 1.72 | 0 | TargetScan | -0.3 | 0.00538 | NA | |
| 37 | hsa-let-7b-5p | APRT | -0.96 | 0 | 0.19 | 0.12663 | miRNAWalker2 validate | -0.27 | 0 | NA | |
| 38 | hsa-let-7b-5p | ARFIP2 | -0.96 | 0 | 0.37 | 0 | miRNAWalker2 validate | -0.16 | 0 | NA | |
| 39 | hsa-let-7b-5p | ARG2 | -0.96 | 0 | 0.15 | 0.43591 | MirTarget | -0.18 | 0.00562 | NA | |
| 40 | hsa-let-7d-5p | ARG2 | 0.05 | 0.70258 | 0.15 | 0.43591 | MirTarget | -0.24 | 0.00086 | NA | |
| 41 | hsa-let-7d-5p | ARHGAP28 | 0.05 | 0.70258 | 0.59 | 0.03308 | MirTarget; miRNATAP | -0.21 | 0.03867 | NA | |
| 42 | hsa-let-7a-5p | ARID3A | -0.33 | 0.00046 | 1.53 | 0 | TargetScan | -0.63 | 0 | NA | |
| 43 | hsa-let-7b-5p | ARID3A | -0.96 | 0 | 1.53 | 0 | miRNAWalker2 validate | -0.38 | 1.0E-5 | NA | |
| 44 | hsa-let-7a-5p | ARID3B | -0.33 | 0.00046 | 0.46 | 0.00021 | miRNAWalker2 validate; TargetScan; miRNATAP | -0.17 | 0.01059 | NA | |
| 45 | hsa-let-7b-5p | ARID3B | -0.96 | 0 | 0.46 | 0.00021 | miRNAWalker2 validate; miRNATAP | -0.14 | 0.00075 | NA | |
| 46 | hsa-let-7b-5p | ASNA1 | -0.96 | 0 | 0.58 | 0 | miRNAWalker2 validate | -0.19 | 0 | NA | |
| 47 | hsa-let-7b-5p | ASPSCR1 | -0.96 | 0 | 0.72 | 1.0E-5 | miRNAWalker2 validate | -0.43 | 0 | NA | |
| 48 | hsa-let-7b-5p | ATAD3B | -0.96 | 0 | 0.76 | 0 | miRNAWalker2 validate | -0.29 | 0 | NA | |
| 49 | hsa-let-7a-5p | ATG10 | -0.33 | 0.00046 | 0.35 | 0.00052 | MirTarget | -0.21 | 6.0E-5 | NA | |
| 50 | hsa-let-7b-5p | ATG10 | -0.96 | 0 | 0.35 | 0.00052 | MirTarget | -0.2 | 0 | NA | |
| 51 | hsa-let-7d-5p | ATG10 | 0.05 | 0.70258 | 0.35 | 0.00052 | MirTarget | -0.16 | 1.0E-5 | NA | |
| 52 | hsa-let-7a-5p | ATG4B | -0.33 | 0.00046 | 0.37 | 0 | TargetScan | -0.17 | 3.0E-5 | NA | |
| 53 | hsa-let-7b-5p | ATG4B | -0.96 | 0 | 0.37 | 0 | miRNAWalker2 validate; miRNATAP | -0.12 | 0 | NA | |
| 54 | hsa-let-7b-5p | ATOX1 | -0.96 | 0 | 0.43 | 0.00496 | miRNAWalker2 validate | -0.25 | 0 | NA | |
| 55 | hsa-let-7b-5p | ATP6V0A1 | -0.96 | 0 | 0.28 | 0.00395 | miRNAWalker2 validate | -0.12 | 0.00032 | NA | |
| 56 | hsa-let-7a-5p | ATP6V1C1 | -0.33 | 0.00046 | 0.88 | 0 | TargetScan | -0.28 | 1.0E-5 | NA | |
| 57 | hsa-let-7b-5p | ATP6V1F | -0.96 | 0 | 0.67 | 0 | miRNAWalker2 validate | -0.2 | 0 | NA | |
| 58 | hsa-let-7b-5p | ATXN2L | -0.96 | 0 | 0.61 | 0 | miRNAWalker2 validate | -0.1 | 3.0E-5 | NA | |
| 59 | hsa-let-7a-5p | ATXN7L2 | -0.33 | 0.00046 | 1.07 | 0 | miRNATAP | -0.18 | 0.0043 | NA | |
| 60 | hsa-let-7b-5p | ATXN7L2 | -0.96 | 0 | 1.07 | 0 | miRNATAP | -0.19 | 0 | NA | |
| 61 | hsa-let-7b-5p | ATXN7L3 | -0.96 | 0 | 0.7 | 0 | miRNATAP | -0.1 | 0.00048 | NA | |
| 62 | hsa-let-7b-5p | AUP1 | -0.96 | 0 | 0.15 | 0.05579 | miRNAWalker2 validate | -0.17 | 0 | NA | |
| 63 | hsa-let-7b-5p | AURKA | -0.96 | 0 | 2.71 | 0 | miRNAWalker2 validate | -0.44 | 0 | NA | |
| 64 | hsa-let-7b-5p | AURKB | -0.96 | 0 | 3.49 | 0 | miRNAWalker2 validate | -0.6 | 0 | NA | |
| 65 | hsa-let-7a-5p | B3GAT3 | -0.33 | 0.00046 | 0.5 | 1.0E-5 | TargetScan | -0.38 | 0 | NA | |
| 66 | hsa-let-7b-5p | BBS7 | -0.96 | 0 | 0.27 | 0.09013 | miRNAWalker2 validate | -0.1 | 0.04332 | NA | |
| 67 | hsa-let-7a-5p | BCAT1 | -0.33 | 0.00046 | 1.33 | 0 | miRNAWalker2 validate; TargetScan | -0.28 | 0.00571 | NA | |
| 68 | hsa-let-7b-5p | BFSP1 | -0.96 | 0 | 1.36 | 0 | miRNAWalker2 validate | -0.2 | 0.00047 | NA | |
| 69 | hsa-let-7b-5p | BGLAP | -0.96 | 0 | 0.57 | 0.00109 | miRNAWalker2 validate | -0.28 | 0 | NA | |
| 70 | hsa-let-7a-5p | BIN3 | -0.33 | 0.00046 | -0.4 | 6.0E-5 | TargetScan | -0.12 | 0.0229 | NA | |
| 71 | hsa-let-7b-5p | BIRC5 | -0.96 | 0 | 4.5 | 0 | miRNAWalker2 validate | -0.71 | 0 | NA | |
| 72 | hsa-let-7a-5p | BRI3BP | -0.33 | 0.00046 | 0.53 | 0 | mirMAP | -0.19 | 0.00226 | NA | |
| 73 | hsa-let-7b-5p | BRI3BP | -0.96 | 0 | 0.53 | 0 | mirMAP | -0.16 | 3.0E-5 | NA | |
| 74 | hsa-let-7a-5p | BZW2 | -0.33 | 0.00046 | 0.58 | 0 | MirTarget; TargetScan; miRNATAP | -0.21 | 0.00015 | NA | |
| 75 | hsa-let-7b-5p | BZW2 | -0.96 | 0 | 0.58 | 0 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.2 | 0 | NA | |
| 76 | hsa-let-7c-5p | BZW2 | -1.71 | 0 | 0.58 | 0 | MirTarget | -0.2 | 0 | NA | |
| 77 | hsa-let-7b-5p | C15orf41 | -0.96 | 0 | 0.9 | 0 | miRNATAP | -0.14 | 0.00043 | NA | |
| 78 | hsa-let-7b-5p | C22orf29 | -0.96 | 0 | 1.09 | 0 | mirMAP | -0.1 | 0.00283 | NA | |
| 79 | hsa-let-7a-5p | CACNA1E | -0.33 | 0.00046 | 1.61 | 4.0E-5 | TargetScan; miRNATAP | -1.16 | 0 | NA | |
| 80 | hsa-let-7b-5p | CACNA1E | -0.96 | 0 | 1.61 | 4.0E-5 | miRNATAP | -0.58 | 1.0E-5 | NA | |
| 81 | hsa-let-7a-5p | CACNB4 | -0.33 | 0.00046 | 1.93 | 0 | TargetScan; miRNATAP | -0.32 | 0.04341 | NA | |
| 82 | hsa-let-7b-5p | CACNB4 | -0.96 | 0 | 1.93 | 0 | miRNATAP | -0.43 | 1.0E-5 | NA | |
| 83 | hsa-let-7d-5p | CADM2 | 0.05 | 0.70258 | -2.32 | 0 | MirTarget; miRNATAP | -0.51 | 0.00035 | NA | |
| 84 | hsa-let-7d-5p | CALCOCO2 | 0.05 | 0.70258 | -0.31 | 1.0E-5 | miRNAWalker2 validate | -0.12 | 1.0E-5 | NA | |
| 85 | hsa-let-7a-5p | CANT1 | -0.33 | 0.00046 | 0.65 | 0 | TargetScan | -0.15 | 0.00046 | NA | |
| 86 | hsa-let-7a-5p | CAP1 | -0.33 | 0.00046 | 0.04 | 0.49169 | miRNATAP | -0.1 | 0.00115 | NA | |
| 87 | hsa-let-7b-5p | CASKIN1 | -0.96 | 0 | 1.73 | 0 | miRNATAP | -0.37 | 0.00036 | NA | |
| 88 | hsa-let-7d-5p | CBR4 | 0.05 | 0.70258 | -1.38 | 0 | miRNAWalker2 validate | -0.22 | 6.0E-5 | NA | |
| 89 | hsa-let-7a-5p | CBX2 | -0.33 | 0.00046 | 2.25 | 0 | TargetScan; miRNATAP | -0.31 | 0.01532 | NA | |
| 90 | hsa-let-7b-5p | CBX2 | -0.96 | 0 | 2.25 | 0 | miRNATAP | -0.29 | 0.0003 | NA | |
| 91 | hsa-let-7c-5p | CBX5 | -1.71 | 0 | 0.35 | 0.00158 | MirTarget | -0.12 | 3.0E-5 | NA | |
| 92 | hsa-let-7c-5p | CBX6 | -1.71 | 0 | -0.14 | 0.56236 | miRNAWalker2 validate | -0.15 | 0.01207 | NA | |
| 93 | hsa-let-7b-5p | CCDC134 | -0.96 | 0 | 0.72 | 0 | miRNAWalker2 validate | -0.14 | 0.00081 | NA | |
| 94 | hsa-let-7b-5p | CCL16 | -0.96 | 0 | -1.6 | 0.00036 | MirTarget | -0.39 | 0.00879 | NA | |
| 95 | hsa-let-7a-5p | CCL3 | -0.33 | 0.00046 | -1.83 | 0 | TargetScan | -0.29 | 0.01759 | NA | |
| 96 | hsa-let-7b-5p | CCNA2 | -0.96 | 0 | 3.37 | 0 | miRNAWalker2 validate; miRTarBase | -0.52 | 0 | NA | |
| 97 | hsa-let-7b-5p | CCNB1 | -0.96 | 0 | 3.16 | 0 | miRNAWalker2 validate | -0.54 | 0 | NA | |
| 98 | hsa-let-7a-5p | CCNB2 | -0.33 | 0.00046 | 4.24 | 0 | miRNAWalker2 validate | -0.45 | 0.00714 | NA | |
| 99 | hsa-let-7b-5p | CCNB2 | -0.96 | 0 | 4.24 | 0 | miRNAWalker2 validate | -0.59 | 0 | NA | |
| 100 | hsa-let-7c-5p | CCNB2 | -1.71 | 0 | 4.24 | 0 | miRNAWalker2 validate | -0.85 | 0 | NA | |
| 101 | hsa-let-7a-5p | CCNF | -0.33 | 0.00046 | 2.17 | 0 | MirTarget; TargetScan | -0.33 | 0.00114 | NA | |
| 102 | hsa-let-7b-5p | CCNF | -0.96 | 0 | 2.17 | 0 | miRNAWalker2 validate; MirTarget | -0.34 | 0 | NA | |
| 103 | hsa-let-7c-5p | CCNF | -1.71 | 0 | 2.17 | 0 | miRNAWalker2 validate; MirTarget | -0.5 | 0 | NA | |
| 104 | hsa-let-7b-5p | CD2BP2 | -0.96 | 0 | 0.56 | 0 | miRNAWalker2 validate | -0.15 | 0 | NA | |
| 105 | hsa-let-7b-5p | CD99 | -0.96 | 0 | 0.06 | 0.61503 | miRNAWalker2 validate | -0.14 | 0.00045 | NA | |
| 106 | hsa-let-7a-5p | CDC25A | -0.33 | 0.00046 | 1.92 | 0 | MirTarget; TargetScan; miRNATAP | -0.26 | 0.02869 | NA | |
| 107 | hsa-let-7b-5p | CDC25A | -0.96 | 0 | 1.92 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.37 | 0 | NA | |
| 108 | hsa-let-7c-5p | CDC25A | -1.71 | 0 | 1.92 | 0 | MirTarget | -0.52 | 0 | 25909324 | MicroRNA let 7c Inhibits Cell Proliferation and Induces Cell Cycle Arrest by Targeting CDC25A in Human Hepatocellular Carcinoma; The aim of the present study was to determine whether the cell cycle regulator CDC25A is involved in the antitumor effect of let-7c in HCC; The luciferase reporter assay showed that CDC25A was a direct target of let-7c and that let-7c inhibited the expression of CDC25A protein by directly targeting its 3' UTR; In conclusion this study indicates that let-7c suppresses HCC progression possibly by directly targeting the cell cycle regulator CDC25A and indirectly affecting its downstream target molecules |
| 109 | hsa-let-7a-5p | CDC25B | -0.33 | 0.00046 | 0.8 | 0 | miRNAWalker2 validate | -0.21 | 0.00571 | NA | |
| 110 | hsa-let-7a-5p | CDC34 | -0.33 | 0.00046 | -0.1 | 0.33032 | miRTarBase; MirTarget; TargetScan; miRNATAP | -0.23 | 2.0E-5 | NA | |
| 111 | hsa-let-7b-5p | CDC34 | -0.96 | 0 | -0.1 | 0.33032 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.13 | 0.00017 | NA | |
| 112 | hsa-let-7d-5p | CDC34 | 0.05 | 0.70258 | -0.1 | 0.33032 | MirTarget; miRNATAP | -0.1 | 0.00857 | NA | |
| 113 | hsa-let-7a-5p | CDCA8 | -0.33 | 0.00046 | 3.15 | 0 | TargetScan | -0.29 | 0.02597 | NA | |
| 114 | hsa-let-7b-5p | CDCA8 | -0.96 | 0 | 3.15 | 0 | miRNAWalker2 validate | -0.42 | 0 | NA | |
| 115 | hsa-let-7b-5p | CDIPT | -0.96 | 0 | -0.02 | 0.81886 | miRNAWalker2 validate | -0.11 | 0.00035 | NA | |
| 116 | hsa-let-7b-5p | CDK6 | -0.96 | 0 | -0.31 | 0.22057 | miRNAWalker2 validate; miRTarBase | -0.22 | 0.00756 | NA | |
| 117 | hsa-let-7c-5p | CECR6 | -1.71 | 0 | 0.39 | 0.03999 | MirTarget | -0.15 | 0.0024 | NA | |
| 118 | hsa-let-7d-5p | CEP120 | 0.05 | 0.70258 | -0.39 | 2.0E-5 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.11 | 0.00124 | NA | |
| 119 | hsa-let-7c-5p | CEP135 | -1.71 | 0 | 0.28 | 0.03781 | MirTarget | -0.13 | 0.00011 | NA | |
| 120 | hsa-let-7c-5p | CERCAM | -1.71 | 0 | -0.12 | 0.64816 | MirTarget | -0.14 | 0.03365 | NA | |
| 121 | hsa-let-7d-5p | CGNL1 | 0.05 | 0.70258 | -0.69 | 0.00039 | MirTarget; miRNATAP | -0.16 | 0.0289 | NA | |
| 122 | hsa-let-7b-5p | CHAF1A | -0.96 | 0 | 1.47 | 0 | miRNAWalker2 validate | -0.21 | 0 | NA | |
| 123 | hsa-let-7b-5p | CHD7 | -0.96 | 0 | 0.37 | 0.00251 | miRNAWalker2 validate | -0.13 | 0.00098 | NA | |
| 124 | hsa-let-7b-5p | CHMP2A | -0.96 | 0 | 0.29 | 0.01082 | miRNAWalker2 validate | -0.17 | 0 | NA | |
| 125 | hsa-let-7b-5p | CHRAC1 | -0.96 | 0 | 0.33 | 0.00039 | miRNAWalker2 validate | -0.12 | 0.0001 | NA | |
| 126 | hsa-let-7a-5p | CHRD | -0.33 | 0.00046 | -0.03 | 0.86098 | MirTarget; TargetScan; miRNATAP | -0.31 | 0.0027 | NA | |
| 127 | hsa-let-7b-5p | CHRD | -0.96 | 0 | -0.03 | 0.86098 | MirTarget; miRNATAP | -0.19 | 0.00276 | NA | |
| 128 | hsa-let-7d-5p | CHRD | 0.05 | 0.70258 | -0.03 | 0.86098 | MirTarget; miRNATAP | -0.27 | 0.00016 | NA | |
| 129 | hsa-let-7b-5p | CIZ1 | -0.96 | 0 | 0.66 | 0 | miRNAWalker2 validate | -0.15 | 0 | NA | |
| 130 | hsa-let-7b-5p | CKAP2 | -0.96 | 0 | 1.29 | 0 | miRNAWalker2 validate | -0.18 | 0.00023 | NA | |
| 131 | hsa-let-7b-5p | CKS2 | -0.96 | 0 | 1.22 | 0 | miRNAWalker2 validate | -0.3 | 0 | NA | |
| 132 | hsa-let-7b-5p | CLDN12 | -0.96 | 0 | -0.37 | 0.00035 | MirTarget; miRNATAP | -0.1 | 0.00328 | NA | |
| 133 | hsa-let-7d-5p | CLDN12 | 0.05 | 0.70258 | -0.37 | 0.00035 | MirTarget; miRNATAP | -0.17 | 2.0E-5 | NA | |
| 134 | hsa-let-7b-5p | CMC1 | -0.96 | 0 | 0.29 | 0.00197 | miRNAWalker2 validate | -0.27 | 0 | NA | |
| 135 | hsa-let-7c-5p | CNOT3 | -1.71 | 0 | 0.58 | 0 | miRNAWalker2 validate | -0.14 | 0 | NA | |
| 136 | hsa-let-7b-5p | COIL | -0.96 | 0 | 0.38 | 0 | miRNAWalker2 validate; miRNATAP | -0.14 | 0 | NA | |
| 137 | hsa-let-7a-5p | COL24A1 | -0.33 | 0.00046 | 2.07 | 0 | MirTarget; TargetScan | -0.59 | 0.00301 | NA | |
| 138 | hsa-let-7b-5p | COL24A1 | -0.96 | 0 | 2.07 | 0 | MirTarget | -0.58 | 0 | NA | |
| 139 | hsa-let-7c-5p | COL24A1 | -1.71 | 0 | 2.07 | 0 | MirTarget | -0.32 | 0.00072 | NA | |
| 140 | hsa-let-7d-5p | COL24A1 | 0.05 | 0.70258 | 2.07 | 0 | MirTarget | -0.61 | 2.0E-5 | NA | |
| 141 | hsa-let-7b-5p | COL4A5 | -0.96 | 0 | 0.06 | 0.88138 | miRNATAP | -0.31 | 0.02448 | NA | |
| 142 | hsa-let-7a-5p | COL9A1 | -0.33 | 0.00046 | 2.79 | 0 | TargetScan | -0.37 | 0.01712 | NA | |
| 143 | hsa-let-7b-5p | COMMD9 | -0.96 | 0 | 0.25 | 0.00305 | miRNAWalker2 validate | -0.17 | 0 | NA | |
| 144 | hsa-let-7a-5p | COPZ1 | -0.33 | 0.00046 | 0.35 | 0 | miRNAWalker2 validate | -0.23 | 0 | NA | |
| 145 | hsa-let-7b-5p | COX7B | -0.96 | 0 | 0.2 | 0.0881 | miRNAWalker2 validate | -0.16 | 3.0E-5 | NA | |
| 146 | hsa-let-7b-5p | CPSF1 | -0.96 | 0 | 0.75 | 0 | miRNAWalker2 validate | -0.19 | 0 | NA | |
| 147 | hsa-let-7b-5p | CPSF3L | -0.96 | 0 | 0.17 | 0.03017 | miRNAWalker2 validate | -0.16 | 0 | NA | |
| 148 | hsa-let-7a-5p | CPSF4 | -0.33 | 0.00046 | 0.78 | 0 | TargetScan | -0.27 | 0 | NA | |
| 149 | hsa-let-7a-5p | CS | -0.33 | 0.00046 | 0.63 | 0 | miRNAWalker2 validate | -0.1 | 0.02829 | NA | |
| 150 | hsa-let-7b-5p | CS | -0.96 | 0 | 0.63 | 0 | miRNAWalker2 validate | -0.12 | 3.0E-5 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | NCRNA METABOLIC PROCESS | 60 | 533 | 2.055e-19 | 9.564e-16 |
| 2 | AMIDE BIOSYNTHETIC PROCESS | 52 | 507 | 2.977e-15 | 6.927e-12 |
| 3 | RNA PROCESSING | 69 | 835 | 4.875e-15 | 7.561e-12 |
| 4 | NCRNA PROCESSING | 44 | 386 | 1.052e-14 | 1.223e-11 |
| 5 | RIBONUCLEOPROTEIN COMPLEX BIOGENESIS | 47 | 440 | 1.497e-14 | 1.393e-11 |
| 6 | PEPTIDE METABOLIC PROCESS | 53 | 571 | 9.585e-14 | 7.433e-11 |
| 7 | CELLULAR AMIDE METABOLIC PROCESS | 60 | 727 | 3.625e-13 | 2.41e-10 |
| 8 | RIBOSOME BIOGENESIS | 35 | 308 | 6.581e-12 | 3.828e-09 |
| 9 | ORGANONITROGEN COMPOUND BIOSYNTHETIC PROCESS | 71 | 1024 | 9.775e-12 | 5.054e-09 |
| 10 | MITOTIC CELL CYCLE | 58 | 766 | 3.058e-11 | 1.423e-08 |
| 11 | TRANSLATIONAL INITIATION | 23 | 146 | 4.17e-11 | 1.764e-08 |
| 12 | CELL CYCLE PROCESS | 71 | 1081 | 1.138e-10 | 4.414e-08 |
| 13 | RRNA METABOLIC PROCESS | 29 | 255 | 4.25e-10 | 1.521e-07 |
| 14 | CELL CYCLE | 79 | 1316 | 6.624e-10 | 2.201e-07 |
| 15 | MRNA METABOLIC PROCESS | 46 | 611 | 4.51e-09 | 1.399e-06 |
| 16 | CELL DIVISION | 38 | 460 | 8.927e-09 | 2.596e-06 |
| 17 | TRNA METABOLIC PROCESS | 22 | 176 | 9.754e-09 | 2.67e-06 |
| 18 | NEGATIVE REGULATION OF CELL CYCLE PROCESS | 24 | 214 | 1.811e-08 | 4.681e-06 |
| 19 | ESTABLISHMENT OF LOCALIZATION IN CELL | 88 | 1676 | 4.104e-08 | 1.005e-05 |
| 20 | NEGATIVE REGULATION OF CELL CYCLE | 34 | 433 | 1.843e-07 | 4.289e-05 |
| 21 | ESTABLISHMENT OF PROTEIN LOCALIZATION TO ENDOPLASMIC RETICULUM | 15 | 104 | 3.396e-07 | 7.183e-05 |
| 22 | NUCLEAR TRANSCRIBED MRNA CATABOLIC PROCESS NONSENSE MEDIATED DECAY | 16 | 118 | 3.299e-07 | 7.183e-05 |
| 23 | NEGATIVE REGULATION OF MITOTIC CELL CYCLE | 21 | 199 | 3.943e-07 | 7.976e-05 |
| 24 | CELL CYCLE PHASE TRANSITION | 24 | 255 | 4.984e-07 | 9.662e-05 |
| 25 | PROTEIN LOCALIZATION TO ENDOPLASMIC RETICULUM | 16 | 123 | 5.861e-07 | 0.000101 |
| 26 | MULTI ORGANISM METABOLIC PROCESS | 17 | 138 | 5.749e-07 | 0.000101 |
| 27 | CELLULAR RESPONSE TO DNA DAMAGE STIMULUS | 46 | 720 | 5.791e-07 | 0.000101 |
| 28 | ORGANONITROGEN COMPOUND METABOLIC PROCESS | 88 | 1796 | 8.465e-07 | 0.0001407 |
| 29 | MACROMOLECULE CATABOLIC PROCESS | 54 | 926 | 1.035e-06 | 0.000166 |
| 30 | CELL CYCLE CHECKPOINT | 20 | 194 | 1.077e-06 | 0.000167 |
| 31 | NEGATIVE REGULATION OF CELL CYCLE PHASE TRANSITION | 17 | 146 | 1.281e-06 | 0.0001923 |
| 32 | TRNA AMINOACYLATION | 10 | 52 | 2.153e-06 | 0.0002963 |
| 33 | AMINO ACID ACTIVATION | 10 | 52 | 2.153e-06 | 0.0002963 |
| 34 | REGULATION OF CELL CYCLE | 54 | 949 | 2.165e-06 | 0.0002963 |
| 35 | NITROGEN COMPOUND TRANSPORT | 35 | 507 | 2.457e-06 | 0.0003266 |
| 36 | MITOTIC CELL CYCLE CHECKPOINT | 16 | 139 | 3.049e-06 | 0.0003941 |
| 37 | MACROMOLECULAR COMPLEX ASSEMBLY | 71 | 1398 | 3.253e-06 | 0.0003983 |
| 38 | ESTABLISHMENT OF PROTEIN LOCALIZATION TO MEMBRANE | 23 | 264 | 3.239e-06 | 0.0003983 |
| 39 | RNA CATABOLIC PROCESS | 21 | 227 | 3.369e-06 | 0.000402 |
| 40 | POSITIVE REGULATION OF CELL CYCLE PROCESS | 22 | 247 | 3.689e-06 | 0.0004292 |
| 41 | CELLULAR RESPONSE TO STRESS | 77 | 1565 | 3.888e-06 | 0.0004413 |
| 42 | PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL | 50 | 873 | 4.38e-06 | 0.0004853 |
| 43 | RIBONUCLEOPROTEIN COMPLEX SUBUNIT ORGANIZATION | 19 | 199 | 6.202e-06 | 0.0006712 |
| 44 | CELLULAR CATABOLIC PROCESS | 67 | 1322 | 6.769e-06 | 0.0006999 |
| 45 | MITOTIC DNA INTEGRITY CHECKPOINT | 13 | 100 | 6.7e-06 | 0.0006999 |
| 46 | REGULATION OF MITOTIC CELL CYCLE | 32 | 468 | 8.067e-06 | 0.000816 |
| 47 | MITOTIC NUCLEAR DIVISION | 27 | 361 | 8.334e-06 | 0.0008251 |
| 48 | POSITIVE REGULATION OF PROTEIN CATABOLIC PROCESS | 22 | 263 | 1.011e-05 | 0.0009798 |
| 49 | PROTEIN TARGETING TO MEMBRANE | 16 | 157 | 1.469e-05 | 0.001395 |
| 50 | REGULATION OF CELL CYCLE PROCESS | 35 | 558 | 1.96e-05 | 0.001824 |
| 51 | CELL CYCLE G1 S PHASE TRANSITION | 13 | 111 | 2.113e-05 | 0.001891 |
| 52 | G1 S TRANSITION OF MITOTIC CELL CYCLE | 13 | 111 | 2.113e-05 | 0.001891 |
| 53 | DNA INTEGRITY CHECKPOINT | 15 | 146 | 2.454e-05 | 0.002154 |
| 54 | INTERSPECIES INTERACTION BETWEEN ORGANISMS | 39 | 662 | 2.734e-05 | 0.002313 |
| 55 | SYMBIOSIS ENCOMPASSING MUTUALISM THROUGH PARASITISM | 39 | 662 | 2.734e-05 | 0.002313 |
| 56 | CATABOLIC PROCESS | 81 | 1773 | 3.215e-05 | 0.002672 |
| 57 | POSITIVE REGULATION OF CELL CYCLE ARREST | 11 | 85 | 3.556e-05 | 0.002902 |
| 58 | REGULATION OF PROTEIN CATABOLIC PROCESS | 27 | 393 | 3.76e-05 | 0.003017 |
| 59 | POSITIVE REGULATION OF CATABOLIC PROCESS | 27 | 395 | 4.104e-05 | 0.003237 |
| 60 | POSITIVE REGULATION OF CELL CYCLE | 24 | 332 | 4.528e-05 | 0.003512 |
| 61 | G1 DNA DAMAGE CHECKPOINT | 10 | 73 | 4.887e-05 | 0.003726 |
| 62 | PROTEASOMAL PROTEIN CATABOLIC PROCESS | 21 | 271 | 4.964e-05 | 0.003726 |
| 63 | DNA METABOLIC PROCESS | 42 | 758 | 5.554e-05 | 0.004102 |
| 64 | POSTTRANSCRIPTIONAL REGULATION OF GENE EXPRESSION | 29 | 448 | 5.759e-05 | 0.004187 |
| 65 | ESTABLISHMENT OF PROTEIN LOCALIZATION TO ORGANELLE | 25 | 361 | 6.364e-05 | 0.004556 |
| 66 | ESTABLISHMENT OF PROTEIN LOCALIZATION | 67 | 1423 | 6.825e-05 | 0.004811 |
| 67 | MRNA PROCESSING | 28 | 432 | 7.532e-05 | 0.005154 |
| 68 | REGULATION OF CELL CYCLE PHASE TRANSITION | 23 | 321 | 7.424e-05 | 0.005154 |
| 69 | CELLULAR RESPONSE TO OXYGEN LEVELS | 14 | 143 | 7.723e-05 | 0.005208 |
| 70 | SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 13 | 127 | 8.76e-05 | 0.005823 |
| 71 | CELLULAR MACROMOLECULAR COMPLEX ASSEMBLY | 40 | 727 | 9.696e-05 | 0.006355 |
| 72 | PROTEIN UBIQUITINATION | 36 | 629 | 9.955e-05 | 0.006433 |
| 73 | SIGNAL TRANSDUCTION IN RESPONSE TO DNA DAMAGE | 11 | 96 | 0.0001102 | 0.006926 |
| 74 | REGULATION OF CATABOLIC PROCESS | 40 | 731 | 0.0001089 | 0.006926 |
| 75 | VIRAL LIFE CYCLE | 21 | 290 | 0.0001301 | 0.008074 |
| 76 | NEGATIVE REGULATION OF CELL CYCLE G1 S PHASE TRANSITION | 11 | 98 | 0.0001328 | 0.008131 |
| 77 | ORGANELLE FISSION | 30 | 496 | 0.0001453 | 0.008778 |
| 78 | ORGANIC CYCLIC COMPOUND CATABOLIC PROCESS | 27 | 427 | 0.0001512 | 0.009022 |
| 79 | MITOCHONDRION ORGANIZATION | 34 | 594 | 0.0001543 | 0.009088 |
| 80 | PROTEIN TARGETING | 26 | 406 | 0.0001632 | 0.009494 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | RNA BINDING | 114 | 1598 | 1.438e-19 | 1.336e-16 |
| 2 | POLY A RNA BINDING | 86 | 1170 | 1.582e-15 | 7.351e-13 |
| 3 | STRUCTURAL CONSTITUENT OF RIBOSOME | 27 | 212 | 1.326e-10 | 4.106e-08 |
| 4 | LIGASE ACTIVITY | 31 | 406 | 1.168e-06 | 0.0002713 |
| 5 | RIBONUCLEOTIDE BINDING | 88 | 1860 | 3.557e-06 | 0.0006608 |
| 6 | NUCLEOTIDYLTRANSFERASE ACTIVITY | 15 | 131 | 6.595e-06 | 0.001021 |
| 7 | HYDROLASE ACTIVITY ACTING ON ACID ANHYDRIDES | 47 | 820 | 8.401e-06 | 0.001115 |
| 8 | STRUCTURAL MOLECULE ACTIVITY | 43 | 732 | 1.135e-05 | 0.001318 |
| 9 | LIGASE ACTIVITY FORMING CARBON OXYGEN BONDS | 8 | 44 | 3.488e-05 | 0.00324 |
| 10 | RNA POLYMERASE ACTIVITY | 8 | 44 | 3.488e-05 | 0.00324 |
| 11 | ADENYL NUCLEOTIDE BINDING | 71 | 1514 | 4.62e-05 | 0.003902 |
| 12 | TRANSLATION REGULATOR ACTIVITY | 7 | 35 | 5.702e-05 | 0.004414 |
| 13 | ATPASE ACTIVITY COUPLED | 22 | 313 | 0.0001395 | 0.009971 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | RIBONUCLEOPROTEIN COMPLEX | 61 | 721 | 7.597e-14 | 4.437e-11 |
| 2 | NUCLEOLUS | 59 | 848 | 5.662e-10 | 1.653e-07 |
| 3 | RIBOSOMAL SUBUNIT | 22 | 163 | 2.289e-09 | 4.456e-07 |
| 4 | RIBOSOME | 25 | 226 | 1.209e-08 | 1.443e-06 |
| 5 | MITOCHONDRION | 88 | 1633 | 1.235e-08 | 1.443e-06 |
| 6 | MITOCHONDRIAL MATRIX | 33 | 412 | 1.772e-07 | 1.499e-05 |
| 7 | CYTOSOLIC RIBOSOME | 16 | 113 | 1.796e-07 | 1.499e-05 |
| 8 | MITOCHONDRIAL PART | 55 | 953 | 1.126e-06 | 8.22e-05 |
| 9 | SMALL RIBOSOMAL SUBUNIT | 11 | 68 | 3.994e-06 | 0.0002592 |
| 10 | CHROMOSOME | 47 | 880 | 4.964e-05 | 0.002899 |
| 11 | LARGE RIBOSOMAL SUBUNIT | 11 | 95 | 0.0001001 | 0.004534 |
| 12 | TRANSFERASE COMPLEX | 39 | 703 | 9.966e-05 | 0.004534 |
| 13 | ENVELOPE | 54 | 1090 | 0.0001009 | 0.004534 |
| 14 | NUCLEOPLASM PART | 39 | 708 | 0.0001155 | 0.00482 |
| 15 | HETEROCHROMATIN | 9 | 67 | 0.0001354 | 0.005273 |
| 16 | PCG PROTEIN COMPLEX | 7 | 43 | 0.0002233 | 0.008152 |
| 17 | CYTOSOLIC SMALL RIBOSOMAL SUBUNIT | 7 | 44 | 0.000259 | 0.008897 |
| 18 | CYTOSOLIC PART | 17 | 223 | 0.0003067 | 0.00995 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
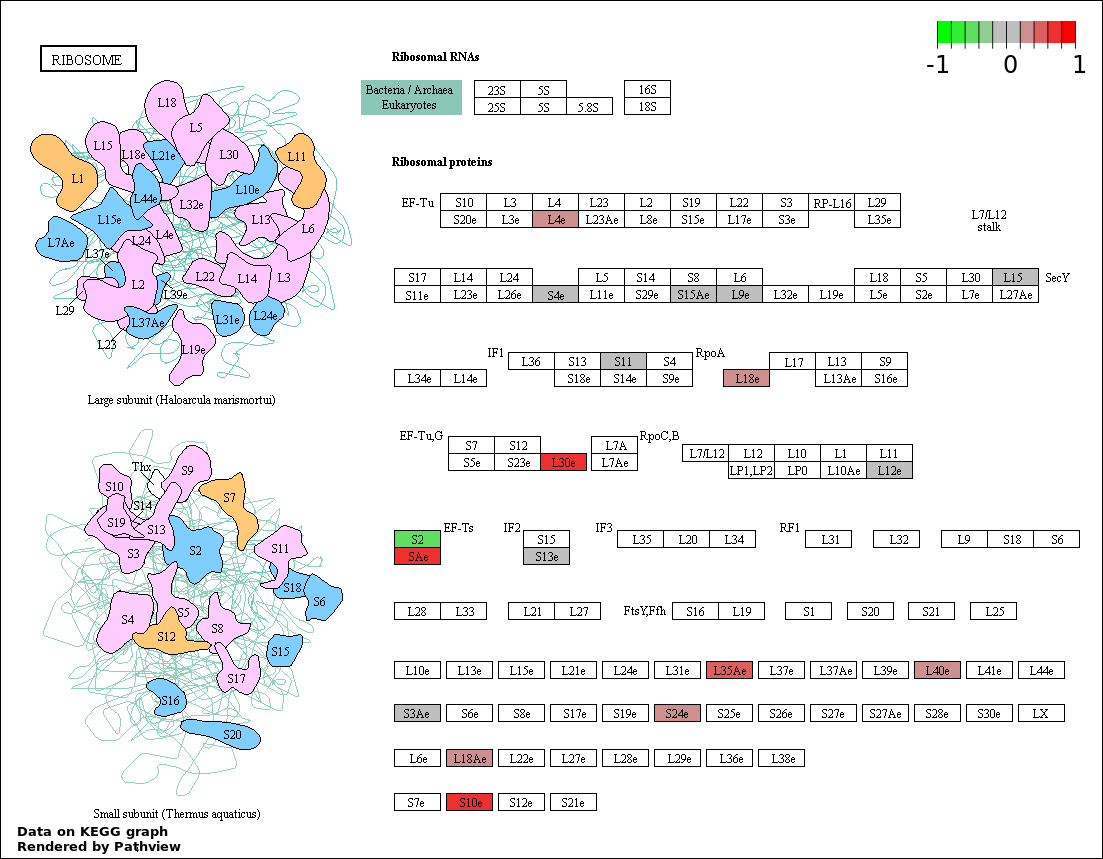
| 1 | hsa03010_Ribosome | 15 | 92 | 6.436e-08 | 1.159e-05 | |
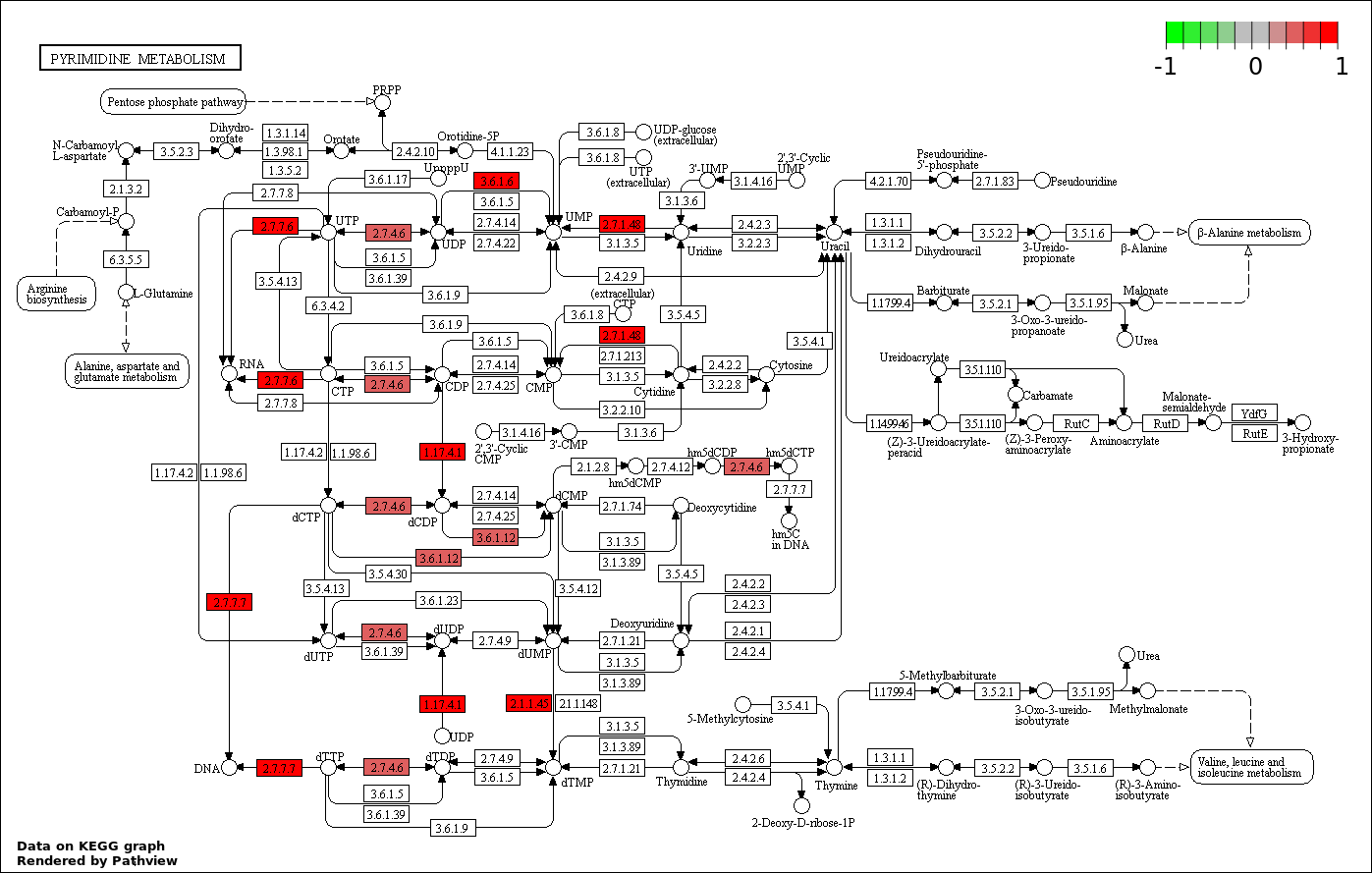
| 2 | hsa00240_Pyrimidine_metabolism | 15 | 99 | 1.753e-07 | 1.578e-05 | |
| 3 | hsa00230_Purine_metabolism | 17 | 162 | 5.388e-06 | 0.0003233 | |
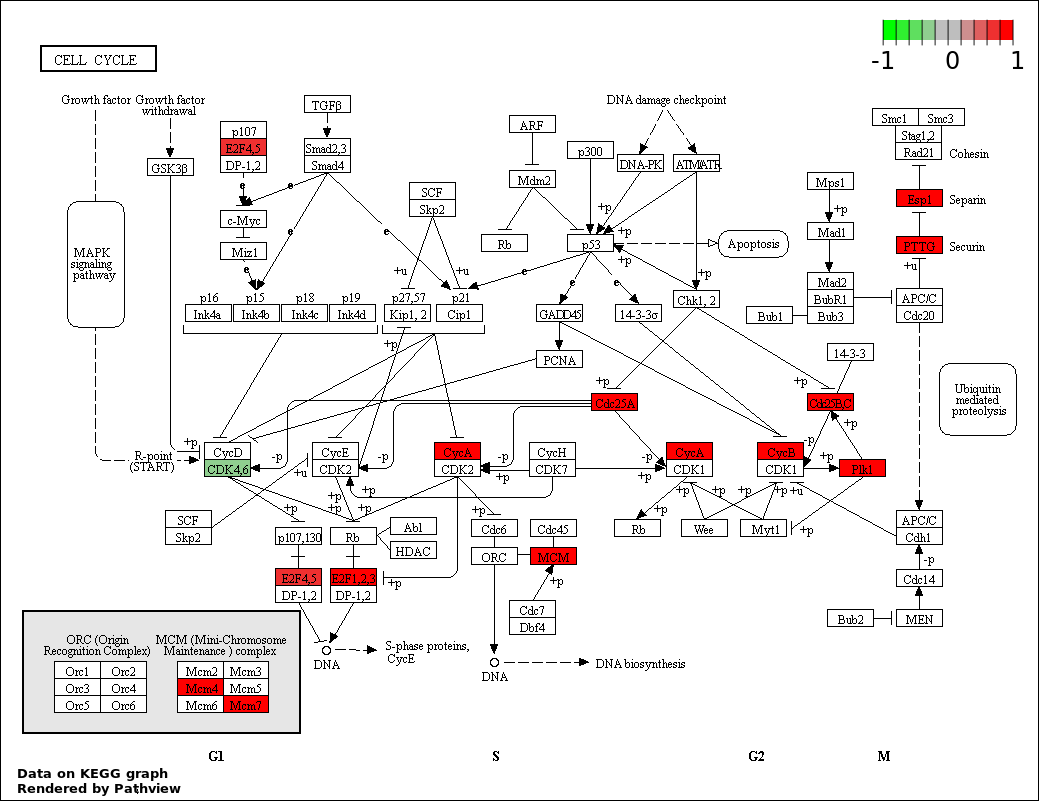
| 4 | hsa04110_Cell_cycle | 14 | 128 | 2.252e-05 | 0.001013 | |
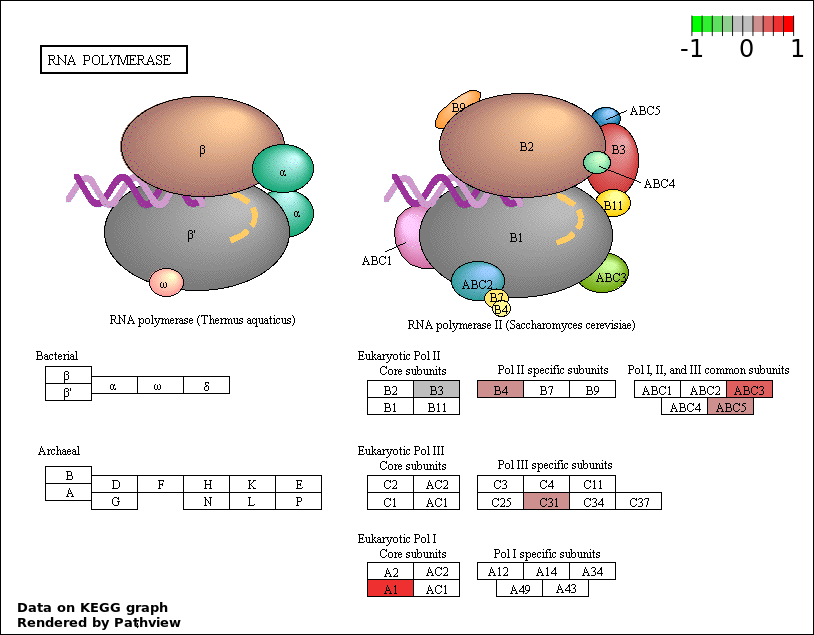
| 5 | hsa03020_RNA_polymerase | 6 | 29 | 0.0001603 | 0.00577 | |
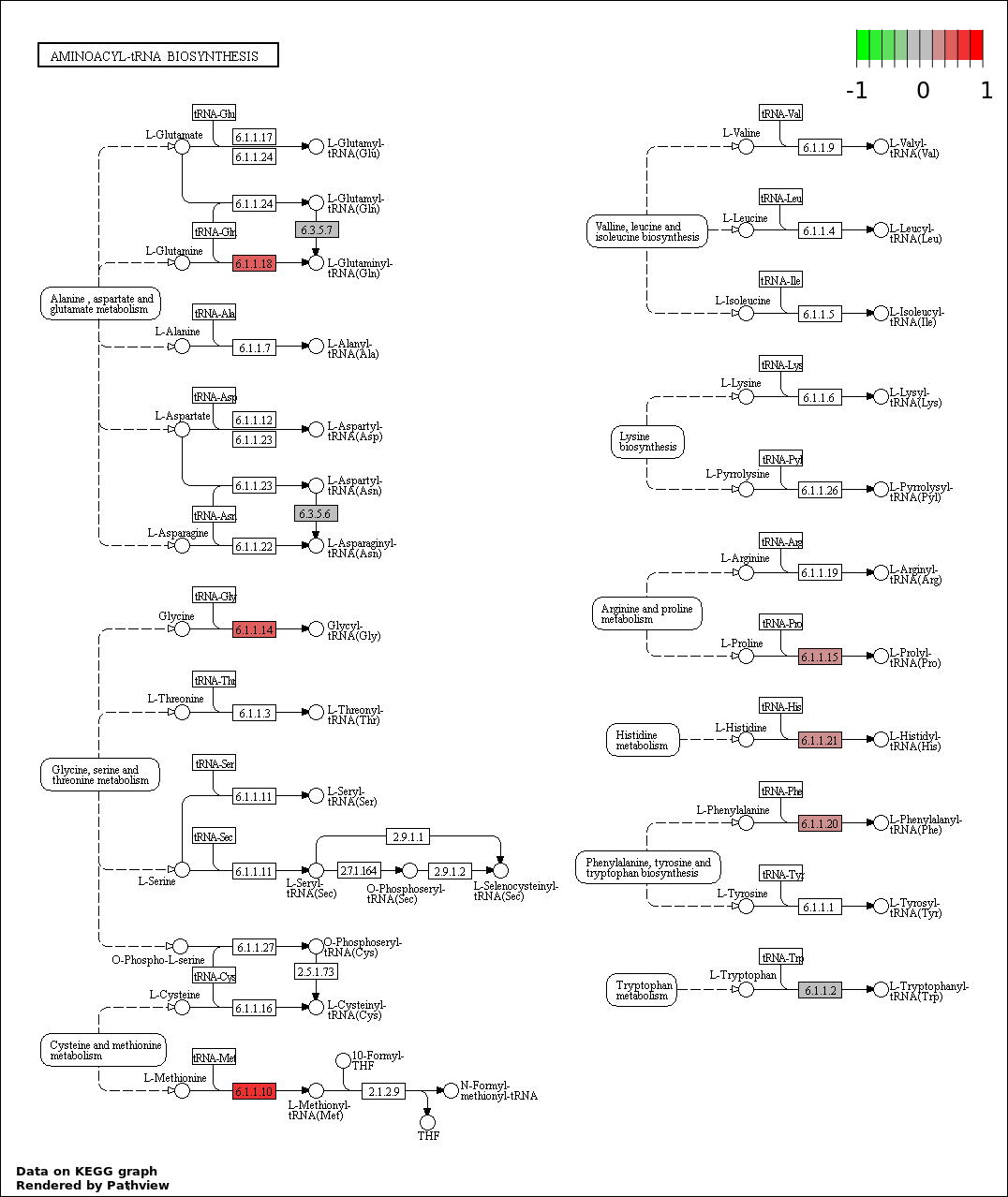
| 6 | hsa00970_Aminoacyl.tRNA_biosynthesis | 7 | 63 | 0.002324 | 0.06971 | |
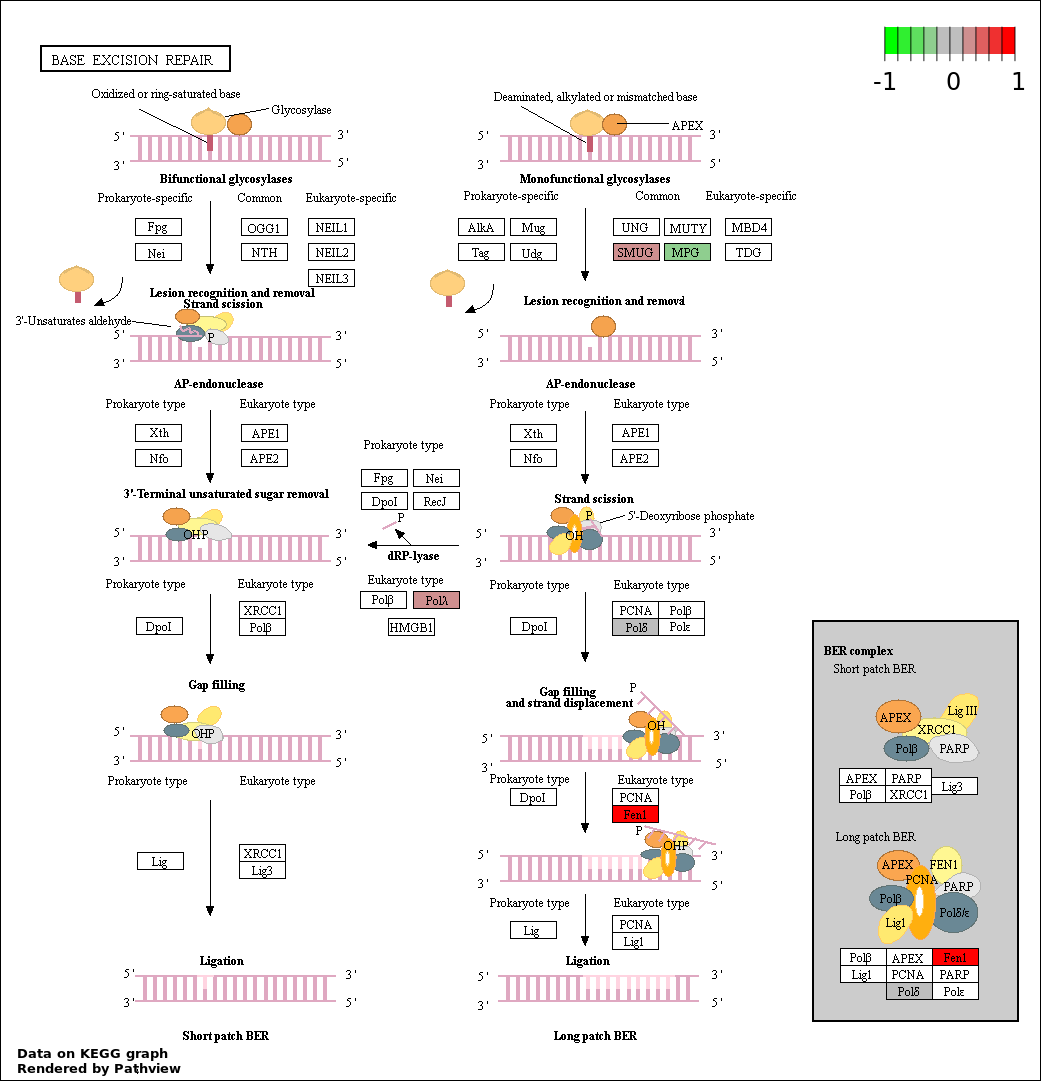
| 7 | hsa03410_Base_excision_repair | 5 | 34 | 0.00286 | 0.07353 | |
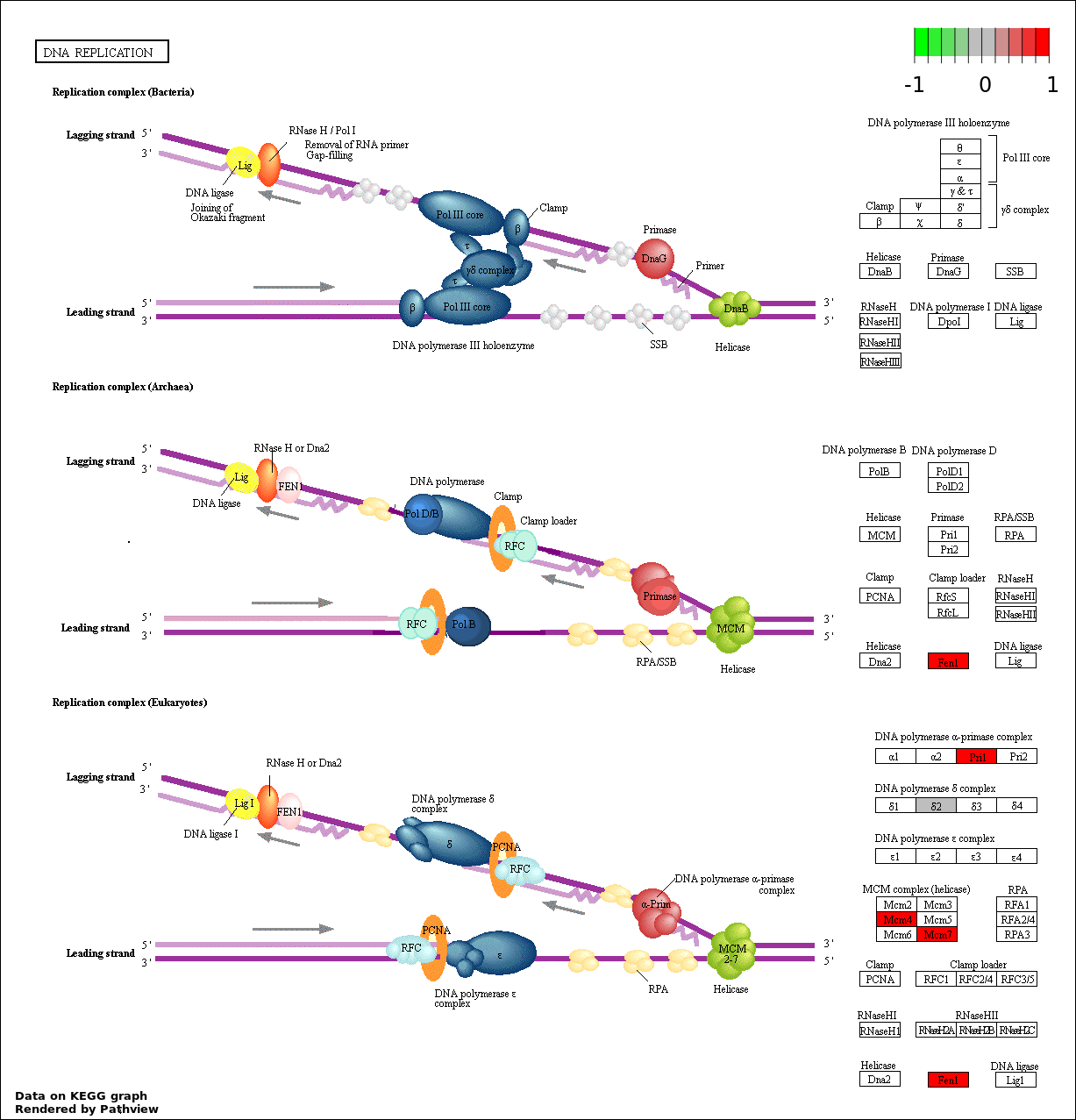
| 8 | hsa03030_DNA_replication | 5 | 36 | 0.003694 | 0.08311 | |
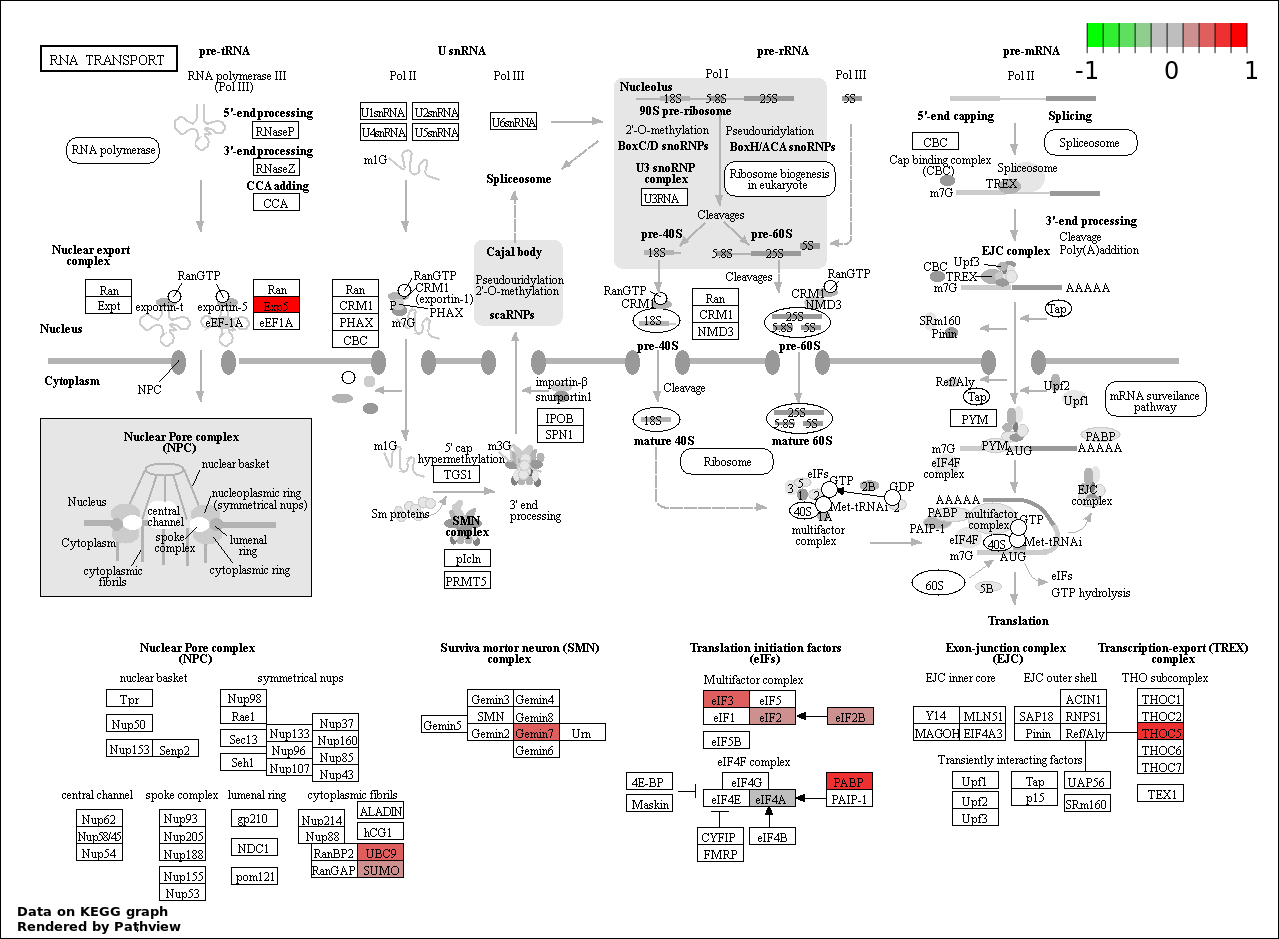
| 9 | hsa03013_RNA_transport | 11 | 152 | 0.005039 | 0.1008 | |
| 10 | hsa03040_Spliceosome | 9 | 128 | 0.01273 | 0.2291 | |
| 11 | hsa00670_One_carbon_pool_by_folate | 3 | 18 | 0.01451 | 0.2374 | |
| 12 | hsa04540_Gap_junction | 7 | 90 | 0.01609 | 0.2413 | |
| 13 | hsa03008_Ribosome_biogenesis_in_eukaryotes | 6 | 81 | 0.03085 | 0.4272 | |
| 14 | hsa04144_Endocytosis | 11 | 203 | 0.03618 | 0.4652 | |
| 15 | hsa04914_Progesterone.mediated_oocyte_maturation | 6 | 87 | 0.0416 | 0.4828 | |
| 16 | hsa04966_Collecting_duct_acid_secretion | 3 | 27 | 0.04292 | 0.4828 | |
| 17 | hsa04115_p53_signaling_pathway | 5 | 69 | 0.05069 | 0.5097 | |
| 18 | hsa04120_Ubiquitin_mediated_proteolysis | 8 | 139 | 0.05097 | 0.5097 | |
| 19 | hsa03450_Non.homologous_end.joining | 2 | 14 | 0.06124 | 0.5802 | |
| 20 | hsa00900_Terpenoid_backbone_biosynthesis | 2 | 15 | 0.06934 | 0.624 | |
| 21 | hsa00564_Glycerophospholipid_metabolism | 5 | 80 | 0.08417 | 0.7052 | |
| 22 | hsa04360_Axon_guidance | 7 | 130 | 0.08619 | 0.7052 | |
| 23 | hsa04660_T_cell_receptor_signaling_pathway | 6 | 108 | 0.09599 | 0.7512 | |
| 24 | hsa04012_ErbB_signaling_pathway | 5 | 87 | 0.1103 | 0.7948 | |
| 25 | hsa00620_Pyruvate_metabolism | 3 | 40 | 0.1104 | 0.7948 | |
| 26 | hsa04114_Oocyte_meiosis | 6 | 114 | 0.1163 | 0.8051 | |
| 27 | hsa00532_Glycosaminoglycan_biosynthesis_._chondroitin_sulfate | 2 | 22 | 0.1338 | 0.8759 | |
| 28 | hsa02010_ABC_transporters | 3 | 44 | 0.1363 | 0.8759 | |
| 29 | hsa04010_MAPK_signaling_pathway | 11 | 268 | 0.1614 | 0.9534 | |
| 30 | hsa04145_Phagosome | 7 | 156 | 0.1715 | 0.9534 | |
| 31 | hsa04810_Regulation_of_actin_cytoskeleton | 9 | 214 | 0.1743 | 0.9534 | |
| 32 | hsa00534_Glycosaminoglycan_biosynthesis_._heparan_sulfate | 2 | 26 | 0.1748 | 0.9534 | |
| 33 | hsa04370_VEGF_signaling_pathway | 4 | 76 | 0.181 | 0.9585 | |
| 34 | hsa04530_Tight_junction | 6 | 133 | 0.1928 | 0.9641 | |
| 35 | hsa04664_Fc_epsilon_RI_signaling_pathway | 4 | 79 | 0.1988 | 0.9672 | |
| 36 | hsa00330_Arginine_and_proline_metabolism | 3 | 54 | 0.2082 | 0.986 | |
| 37 | hsa00020_Citrate_cycle_.TCA_cycle. | 2 | 30 | 0.2174 | 0.9895 | |
| 38 | hsa04141_Protein_processing_in_endoplasmic_reticulum | 7 | 168 | 0.2199 | 0.9895 | |
| 39 | hsa00260_Glycine._serine_and_threonine_metabolism | 2 | 32 | 0.2391 | 1 | |
| 40 | hsa00590_Arachidonic_acid_metabolism | 3 | 59 | 0.2468 | 1 | |
| 41 | hsa04140_Regulation_of_autophagy | 2 | 34 | 0.2609 | 1 | |
| 42 | hsa04142_Lysosome | 5 | 121 | 0.278 | 1 | |
| 43 | hsa04062_Chemokine_signaling_pathway | 7 | 189 | 0.3139 | 1 | |
| 44 | hsa04920_Adipocytokine_signaling_pathway | 3 | 68 | 0.3184 | 1 | |
| 45 | hsa03320_PPAR_signaling_pathway | 3 | 70 | 0.3344 | 1 | |
| 46 | hsa04720_Long.term_potentiation | 3 | 70 | 0.3344 | 1 | |
| 47 | hsa04730_Long.term_depression | 3 | 70 | 0.3344 | 1 | |
| 48 | hsa04916_Melanogenesis | 4 | 101 | 0.34 | 1 | |
| 49 | hsa00280_Valine._leucine_and_isoleucine_degradation | 2 | 44 | 0.3686 | 1 | |
| 50 | hsa00310_Lysine_degradation | 2 | 44 | 0.3686 | 1 | |
| 51 | hsa04662_B_cell_receptor_signaling_pathway | 3 | 75 | 0.3744 | 1 | |
| 52 | hsa04910_Insulin_signaling_pathway | 5 | 138 | 0.3756 | 1 | |
| 53 | hsa03420_Nucleotide_excision_repair | 2 | 45 | 0.3791 | 1 | |
| 54 | hsa04146_Peroxisome | 3 | 79 | 0.406 | 1 | |
| 55 | hsa00510_N.Glycan_biosynthesis | 2 | 49 | 0.4202 | 1 | |
| 56 | hsa00480_Glutathione_metabolism | 2 | 50 | 0.4303 | 1 | |
| 57 | hsa03015_mRNA_surveillance_pathway | 3 | 83 | 0.4372 | 1 | |
| 58 | hsa00983_Drug_metabolism_._other_enzymes | 2 | 52 | 0.4502 | 1 | |
| 59 | hsa04350_TGF.beta_signaling_pathway | 3 | 85 | 0.4525 | 1 | |
| 60 | hsa04390_Hippo_signaling_pathway | 5 | 154 | 0.4675 | 1 | |
| 61 | hsa04623_Cytosolic_DNA.sensing_pathway | 2 | 56 | 0.4887 | 1 | |
| 62 | hsa04722_Neurotrophin_signaling_pathway | 4 | 127 | 0.5089 | 1 | |
| 63 | hsa00190_Oxidative_phosphorylation | 4 | 132 | 0.5393 | 1 | |
| 64 | hsa04912_GnRH_signaling_pathway | 3 | 101 | 0.5679 | 1 | |
| 65 | hsa00010_Glycolysis_._Gluconeogenesis | 2 | 65 | 0.5689 | 1 | |
| 66 | hsa04151_PI3K_AKT_signaling_pathway | 10 | 351 | 0.5742 | 1 | |
| 67 | hsa04620_Toll.like_receptor_signaling_pathway | 3 | 102 | 0.5746 | 1 | |
| 68 | hsa03018_RNA_degradation | 2 | 71 | 0.6171 | 1 | |
| 69 | hsa04622_RIG.I.like_receptor_signaling_pathway | 2 | 71 | 0.6171 | 1 | |
| 70 | hsa04260_Cardiac_muscle_contraction | 2 | 77 | 0.6611 | 1 | |
| 71 | hsa04612_Antigen_processing_and_presentation | 2 | 78 | 0.668 | 1 | |
| 72 | hsa04974_Protein_digestion_and_absorption | 2 | 81 | 0.6881 | 1 | |
| 73 | hsa04014_Ras_signaling_pathway | 6 | 236 | 0.6888 | 1 | |
| 74 | hsa04210_Apoptosis | 2 | 89 | 0.7368 | 1 | |
| 75 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 3 | 136 | 0.7619 | 1 | |
| 76 | hsa04020_Calcium_signaling_pathway | 4 | 177 | 0.7621 | 1 | |
| 77 | hsa04310_Wnt_signaling_pathway | 3 | 151 | 0.8203 | 1 | |
| 78 | hsa04510_Focal_adhesion | 4 | 200 | 0.8384 | 1 | |
| 79 | hsa04670_Leukocyte_transendothelial_migration | 2 | 117 | 0.8591 | 1 | |
| 80 | hsa04514_Cell_adhesion_molecules_.CAMs. | 2 | 136 | 0.9098 | 1 |