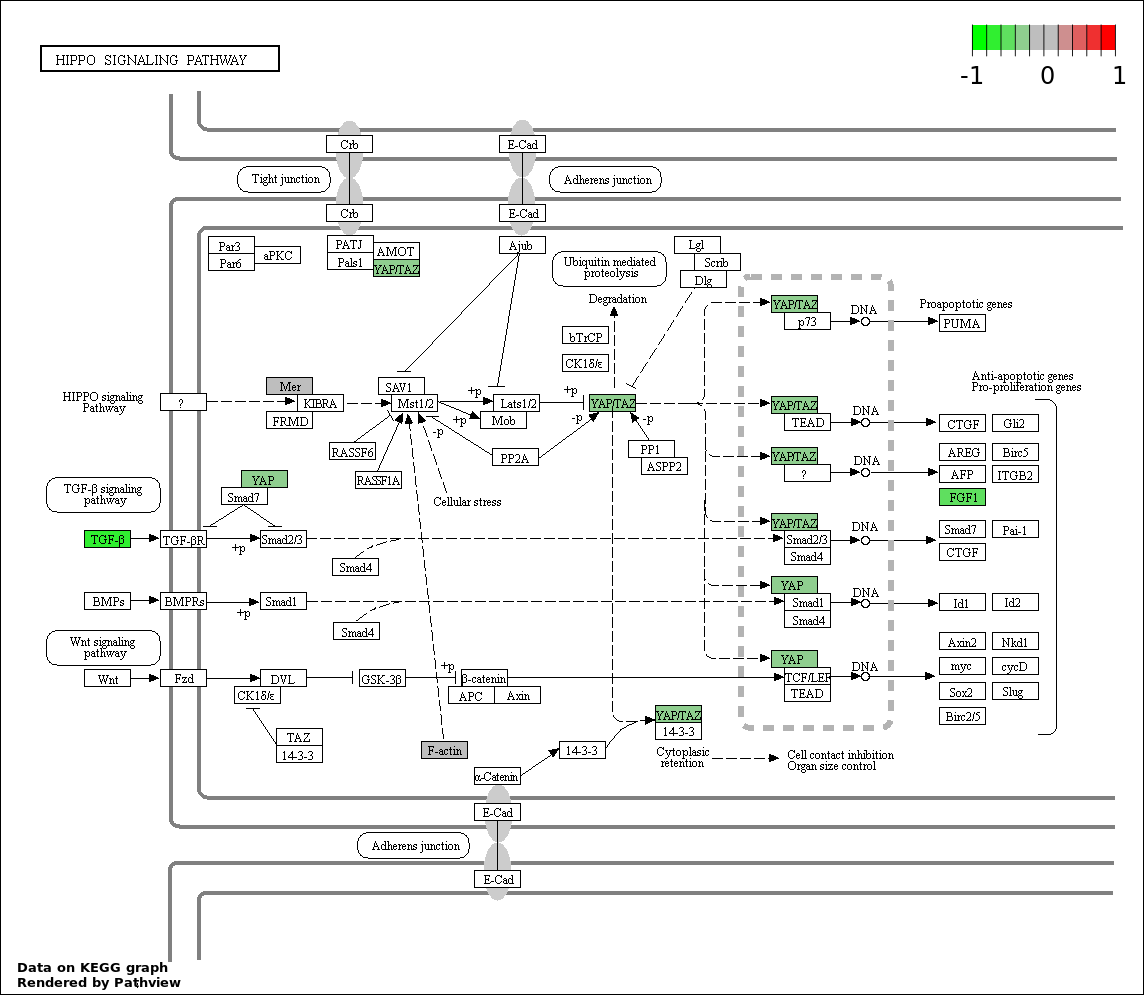
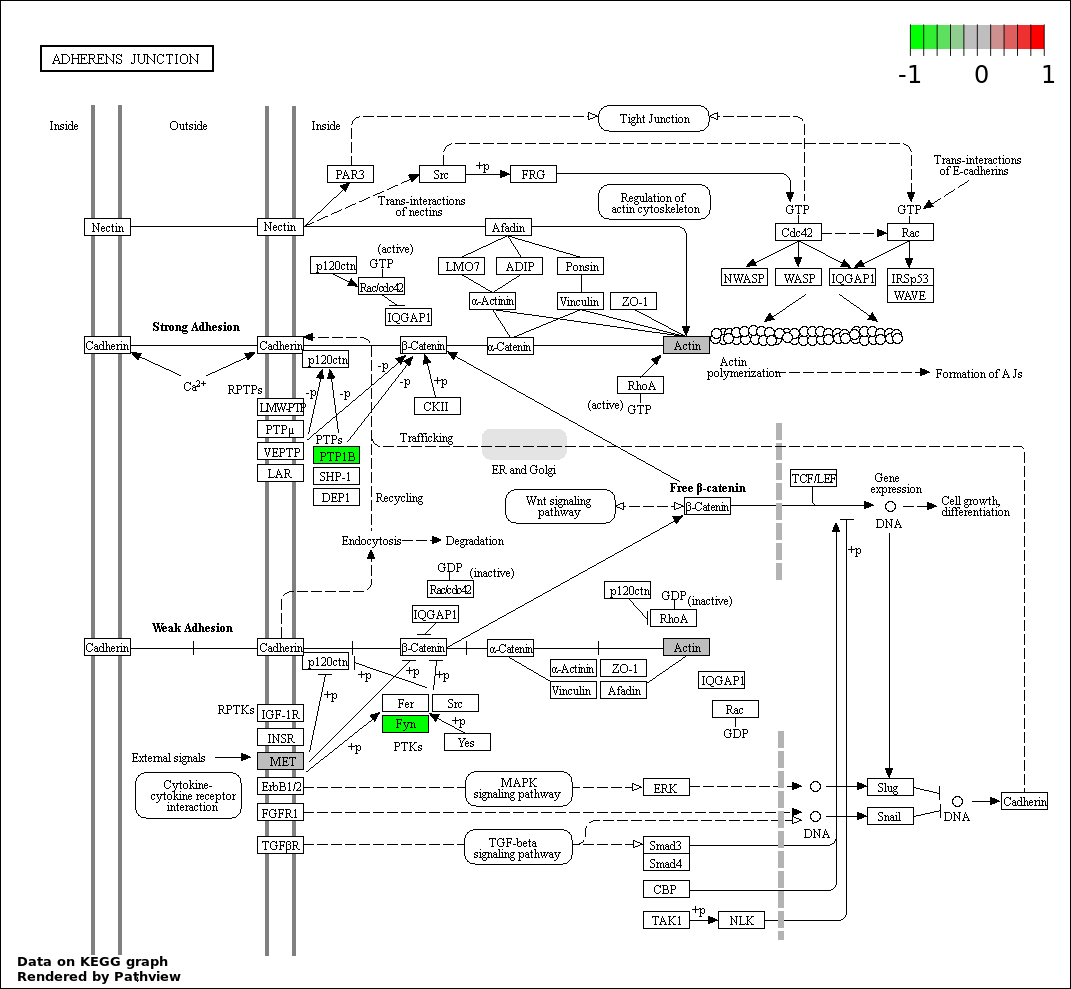
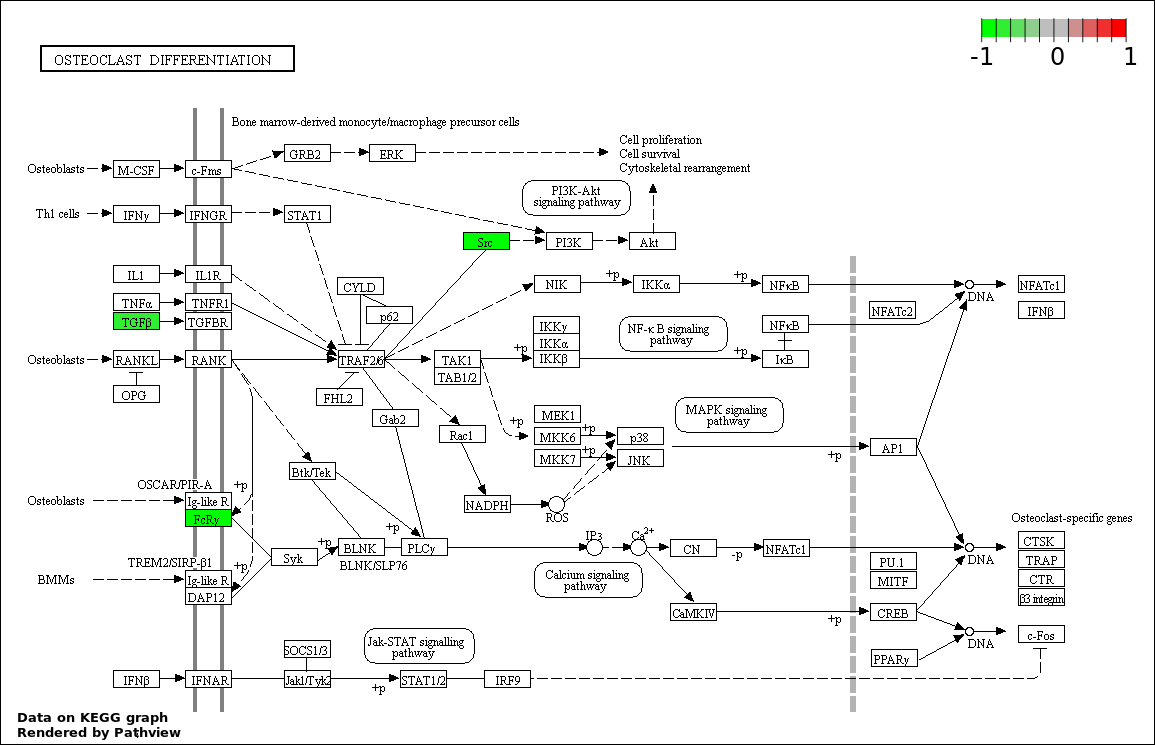
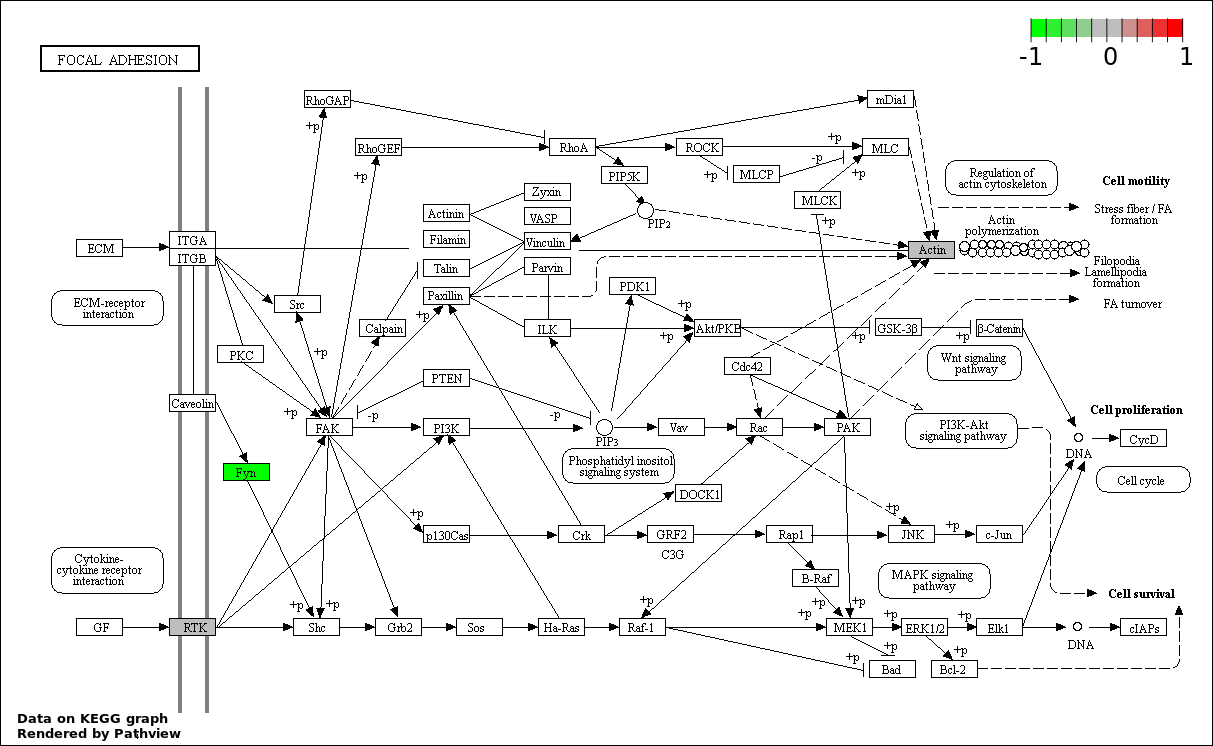
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|
| 1 |
POSITIVE REGULATION OF GENE EXPRESSION |
17 |
1733 |
8.088e-10 |
3.764e-06 |
| 2 |
BRANCHING MORPHOGENESIS OF AN EPITHELIAL TUBE |
7 |
131 |
2.54e-09 |
5.91e-06 |
| 3 |
ORGAN MORPHOGENESIS |
12 |
841 |
9.655e-09 |
1.497e-05 |
| 4 |
MORPHOGENESIS OF A BRANCHING STRUCTURE |
7 |
167 |
1.378e-08 |
1.603e-05 |
| 5 |
LEUKOCYTE ACTIVATION |
9 |
414 |
2.805e-08 |
2.369e-05 |
| 6 |
CELL ACTIVATION |
10 |
568 |
3.054e-08 |
2.369e-05 |
| 7 |
RESPONSE TO ENDOGENOUS STIMULUS |
14 |
1450 |
5.671e-08 |
3.77e-05 |
| 8 |
POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER |
12 |
1004 |
6.847e-08 |
3.983e-05 |
| 9 |
LYMPHOCYTE ACTIVATION |
8 |
342 |
1.06e-07 |
4.933e-05 |
| 10 |
REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER |
15 |
1784 |
9.857e-08 |
4.933e-05 |
| 11 |
TISSUE MORPHOGENESIS |
9 |
533 |
2.413e-07 |
0.0001021 |
| 12 |
POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS |
13 |
1395 |
3.026e-07 |
0.0001173 |
| 13 |
EPITHELIUM DEVELOPMENT |
11 |
945 |
3.629e-07 |
0.0001206 |
| 14 |
MORPHOGENESIS OF AN EPITHELIUM |
8 |
400 |
3.502e-07 |
0.0001206 |
| 15 |
REGULATION OF EPITHELIAL CELL MIGRATION |
6 |
166 |
3.971e-07 |
0.0001232 |
| 16 |
REGULATION OF CELLULAR COMPONENT MOVEMENT |
10 |
771 |
5.229e-07 |
0.0001521 |
| 17 |
CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND |
10 |
799 |
7.245e-07 |
0.0001774 |
| 18 |
CELLULAR RESPONSE TO ENDOGENOUS STIMULUS |
11 |
1008 |
6.894e-07 |
0.0001774 |
| 19 |
REGULATION OF CELL PROLIFERATION |
13 |
1496 |
6.753e-07 |
0.0001774 |
| 20 |
POSITIVE REGULATION OF BIOSYNTHETIC PROCESS |
14 |
1805 |
8.48e-07 |
0.0001973 |
| 21 |
REGULATION OF CELL ADHESION |
9 |
629 |
9.652e-07 |
0.0002139 |
| 22 |
WOUND HEALING |
8 |
470 |
1.181e-06 |
0.0002477 |
| 23 |
TUBE MORPHOGENESIS |
7 |
323 |
1.225e-06 |
0.0002477 |
| 24 |
LYMPHOCYTE DIFFERENTIATION |
6 |
209 |
1.528e-06 |
0.0002962 |
| 25 |
REGULATION OF PEPTIDYL TYROSINE PHOSPHORYLATION |
6 |
213 |
1.706e-06 |
0.0003175 |
| 26 |
POSITIVE REGULATION OF RESPONSE TO STIMULUS |
14 |
1929 |
1.89e-06 |
0.0003383 |
| 27 |
RESPONSE TO OXYGEN CONTAINING COMPOUND |
12 |
1381 |
2.101e-06 |
0.0003621 |
| 28 |
NEGATIVE REGULATION OF HOMEOSTATIC PROCESS |
5 |
124 |
2.363e-06 |
0.0003926 |
| 29 |
REGULATION OF IMMUNE SYSTEM PROCESS |
12 |
1403 |
2.48e-06 |
0.0003979 |
| 30 |
IMMUNE SYSTEM PROCESS |
14 |
1984 |
2.646e-06 |
0.0004104 |
| 31 |
TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER |
9 |
724 |
3.078e-06 |
0.000462 |
| 32 |
BRANCH ELONGATION OF AN EPITHELIUM |
3 |
17 |
3.283e-06 |
0.0004773 |
| 33 |
NEGATIVE REGULATION OF CELL CELL ADHESION |
5 |
138 |
3.997e-06 |
0.000547 |
| 34 |
TUBE DEVELOPMENT |
8 |
552 |
3.903e-06 |
0.000547 |
| 35 |
RESPONSE TO WOUNDING |
8 |
563 |
4.514e-06 |
0.0006001 |
| 36 |
LEUKOCYTE CELL CELL ADHESION |
6 |
255 |
4.822e-06 |
0.0006232 |
| 37 |
TISSUE DEVELOPMENT |
12 |
1518 |
5.627e-06 |
0.0007076 |
| 38 |
CARDIOVASCULAR SYSTEM DEVELOPMENT |
9 |
788 |
6.135e-06 |
0.0007319 |
| 39 |
CIRCULATORY SYSTEM DEVELOPMENT |
9 |
788 |
6.135e-06 |
0.0007319 |
| 40 |
POSITIVE REGULATION OF LOCOMOTION |
7 |
420 |
6.931e-06 |
0.0008062 |
| 41 |
NEGATIVE REGULATION OF CELL ACTIVATION |
5 |
158 |
7.741e-06 |
0.0008786 |
| 42 |
LEUKOCYTE DIFFERENTIATION |
6 |
292 |
1.046e-05 |
0.001159 |
| 43 |
LENS FIBER CELL DIFFERENTIATION |
3 |
25 |
1.1e-05 |
0.00119 |
| 44 |
CELLULAR RESPONSE TO LIPID |
7 |
457 |
1.2e-05 |
0.001219 |
| 45 |
SINGLE ORGANISM CELL ADHESION |
7 |
459 |
1.235e-05 |
0.001219 |
| 46 |
NEGATIVE REGULATION OF RESPONSE TO STIMULUS |
11 |
1360 |
1.257e-05 |
0.001219 |
| 47 |
KIDNEY MORPHOGENESIS |
4 |
82 |
1.247e-05 |
0.001219 |
| 48 |
NEGATIVE REGULATION OF CELL PROLIFERATION |
8 |
643 |
1.192e-05 |
0.001219 |
| 49 |
POSITIVE REGULATION OF IMMUNE SYSTEM PROCESS |
9 |
867 |
1.324e-05 |
0.001257 |
| 50 |
AXIS ELONGATION |
3 |
27 |
1.395e-05 |
0.001298 |
| 51 |
POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION |
9 |
876 |
1.438e-05 |
0.001312 |
| 52 |
RESPONSE TO LIPID |
9 |
888 |
1.602e-05 |
0.001434 |
| 53 |
RESPONSE TO HORMONE |
9 |
893 |
1.676e-05 |
0.001444 |
| 54 |
REGULATION OF STEM CELL PROLIFERATION |
4 |
88 |
1.651e-05 |
0.001444 |
| 55 |
REGULATION OF CELL ACTIVATION |
7 |
484 |
1.739e-05 |
0.001455 |
| 56 |
POSITIVE REGULATION OF DEVELOPMENTAL PROCESS |
10 |
1142 |
1.751e-05 |
0.001455 |
| 57 |
MESONEPHROS DEVELOPMENT |
4 |
90 |
1.804e-05 |
0.001473 |
| 58 |
ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY |
8 |
689 |
1.965e-05 |
0.001576 |
| 59 |
EPITHELIAL CELL DIFFERENTIATION |
7 |
495 |
2.01e-05 |
0.001585 |
| 60 |
TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE SIGNALING PATHWAY |
7 |
498 |
2.09e-05 |
0.001621 |
| 61 |
LIPOPOLYSACCHARIDE MEDIATED SIGNALING PATHWAY |
3 |
31 |
2.134e-05 |
0.001628 |
| 62 |
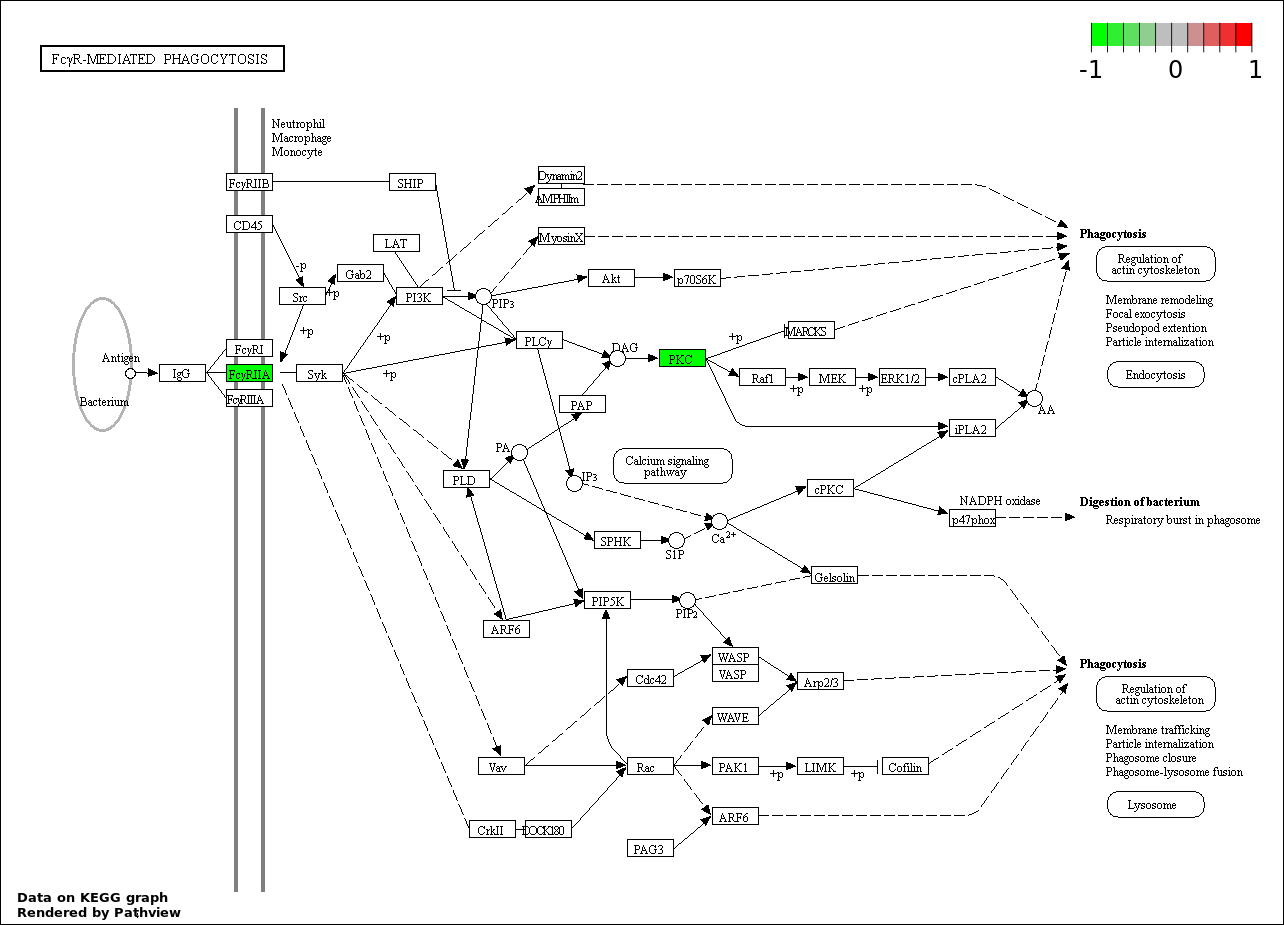
FC GAMMA RECEPTOR SIGNALING PATHWAY |
4 |
95 |
2.234e-05 |
0.001677 |
| 63 |
REGULATION OF CELL DEATH |
11 |
1472 |
2.645e-05 |
0.001953 |
| 64 |
REGULATION OF PROTEIN LOCALIZATION |
9 |
950 |
2.737e-05 |
0.00199 |
| 65 |
REGULATION OF CELL DIFFERENTIATION |
11 |
1492 |
2.999e-05 |
0.002114 |
| 66 |
NEGATIVE REGULATION OF HOMOTYPIC CELL CELL ADHESION |
4 |
102 |
2.956e-05 |
0.002114 |
| 67 |
POSITIVE REGULATION OF EPITHELIAL CELL MIGRATION |
4 |
103 |
3.072e-05 |
0.002134 |
| 68 |
RESPONSE TO ALCOHOL |
6 |
362 |
3.514e-05 |
0.002404 |
| 69 |
NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS |
9 |
983 |
3.581e-05 |
0.002415 |
| 70 |
REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS |
5 |
218 |
3.655e-05 |
0.002429 |
| 71 |
CELLULAR RESPONSE TO HORMONE STIMULUS |
7 |
552 |
4.034e-05 |
0.002632 |
| 72 |
NEGATIVE REGULATION OF CELL ADHESION |
5 |
223 |
4.072e-05 |
0.002632 |
| 73 |
CELLULAR RESPONSE TO ORGANIC SUBSTANCE |
12 |
1848 |
4.13e-05 |
0.002633 |
| 74 |
MAMMARY GLAND MORPHOGENESIS |
3 |
40 |
4.64e-05 |
0.002804 |
| 75 |
REGULATION OF ENDOTHELIAL CELL MIGRATION |
4 |
114 |
4.574e-05 |
0.002804 |
| 76 |
REGULATION OF CELLULAR LOCALIZATION |
10 |
1277 |
4.578e-05 |
0.002804 |
| 77 |
REGULATION OF CELL CELL ADHESION |
6 |
380 |
4.606e-05 |
0.002804 |
| 78 |
POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS |
9 |
1036 |
5.397e-05 |
0.003179 |
| 79 |
POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS |
9 |
1036 |
5.397e-05 |
0.003179 |
| 80 |
IMMUNE SYSTEM DEVELOPMENT |
7 |
582 |
5.641e-05 |
0.00328 |
| 81 |
GLAND DEVELOPMENT |
6 |
395 |
5.711e-05 |
0.00328 |
| 82 |
BRANCHING INVOLVED IN URETERIC BUD MORPHOGENESIS |
3 |
44 |
6.19e-05 |
0.00347 |
| 83 |
T CELL DIFFERENTIATION |
4 |
123 |
6.154e-05 |
0.00347 |
| 84 |
KIDNEY EPITHELIUM DEVELOPMENT |
4 |
125 |
6.553e-05 |
0.00363 |
| 85 |
NEGATIVE REGULATION OF ION TRANSPORT |
4 |
127 |
6.971e-05 |
0.003771 |
| 86 |
POSITIVE REGULATION OF CELL DIFFERENTIATION |
8 |
823 |
6.954e-05 |
0.003771 |
| 87 |
CELL CELL ADHESION |
7 |
608 |
7.427e-05 |
0.003972 |
| 88 |
REGULATION OF CELL DEVELOPMENT |
8 |
836 |
7.762e-05 |
0.004104 |
| 89 |
REGULATION OF IMMUNE RESPONSE |
8 |
858 |
9.308e-05 |
0.004866 |
| 90 |
LOCOMOTION |
9 |
1114 |
9.46e-05 |
0.004891 |
| 91 |
NEGATIVE REGULATION OF CALCIUM ION TRANSPORT |
3 |
51 |
9.652e-05 |
0.004935 |
| 92 |
CENTRAL NERVOUS SYSTEM DEVELOPMENT |
8 |
872 |
0.0001042 |
0.00527 |
| 93 |
MESONEPHRIC TUBULE MORPHOGENESIS |
3 |
53 |
0.0001083 |
0.005404 |
| 94 |
POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS |
9 |
1135 |
0.0001092 |
0.005404 |
| 95 |
EMBRYONIC ORGAN MORPHOGENESIS |
5 |
279 |
0.0001174 |
0.005752 |
| 96 |
REGULATION OF MAPK CASCADE |
7 |
660 |
0.000124 |
0.00601 |
| 97 |
NEGATIVE REGULATION OF TRANSPORT |
6 |
458 |
0.0001289 |
0.006184 |
| 98 |
CELL PROLIFERATION |
7 |
672 |
0.0001387 |
0.006583 |
| 99 |
HEART DEVELOPMENT |
6 |
466 |
0.0001417 |
0.006659 |
| 100 |
VASCULATURE DEVELOPMENT |
6 |
469 |
0.0001467 |
0.006827 |
| 101 |
POSITIVE REGULATION OF MAPK CASCADE |
6 |
470 |
0.0001484 |
0.006838 |
| 102 |
NEGATIVE REGULATION OF CELL COMMUNICATION |
9 |
1192 |
0.0001586 |
0.007233 |
| 103 |
POSITIVE REGULATION OF FIBROBLAST MIGRATION |
2 |
11 |
0.000162 |
0.007262 |
| 104 |
UROGENITAL SYSTEM DEVELOPMENT |
5 |
299 |
0.0001623 |
0.007262 |
| 105 |
NEGATIVE REGULATION OF CATION TRANSMEMBRANE TRANSPORT |
3 |
61 |
0.0001648 |
0.007304 |
| 106 |
NEGATIVE REGULATION OF GENE EXPRESSION |
10 |
1493 |
0.0001697 |
0.007448 |
| 107 |
POSITIVE REGULATION OF PEPTIDYL TYROSINE PHOSPHORYLATION |
4 |
162 |
0.0001786 |
0.007693 |
| 108 |
IMMUNE EFFECTOR PROCESS |
6 |
486 |
0.000178 |
0.007693 |
| 109 |
CELLULAR RESPONSE TO BIOTIC STIMULUS |
4 |
163 |
0.0001828 |
0.007763 |
| 110 |
REGULATION OF HOMOTYPIC CELL CELL ADHESION |
5 |
307 |
0.0001835 |
0.007763 |
| 111 |
NEGATIVE REGULATION OF NITROGEN COMPOUND METABOLIC PROCESS |
10 |
1517 |
0.0001934 |
0.008097 |
| 112 |
HEMOSTASIS |
5 |
311 |
0.0001949 |
0.008097 |
| 113 |
POSITIVE REGULATION OF CELL COMMUNICATION |
10 |
1532 |
0.0002097 |
0.008559 |
| 114 |
LENS DEVELOPMENT IN CAMERA TYPE EYE |
3 |
66 |
0.0002083 |
0.008559 |
| 115 |
POSITIVE REGULATION OF ENDOTHELIAL CELL MIGRATION |
3 |
67 |
0.0002178 |
0.008813 |
| 116 |
REGULATION OF BODY FLUID LEVELS |
6 |
506 |
0.0002214 |
0.008882 |
| 117 |
RESPONSE TO MOLECULE OF BACTERIAL ORIGIN |
5 |
321 |
0.0002257 |
0.008976 |
| 118 |
ENDOCYTOSIS |
6 |
509 |
0.0002286 |
0.009015 |
| 119 |
GLIOGENESIS |
4 |
175 |
0.0002399 |
0.009382 |
| 120 |
NEGATIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER |
7 |
740 |
0.0002509 |
0.009727 |