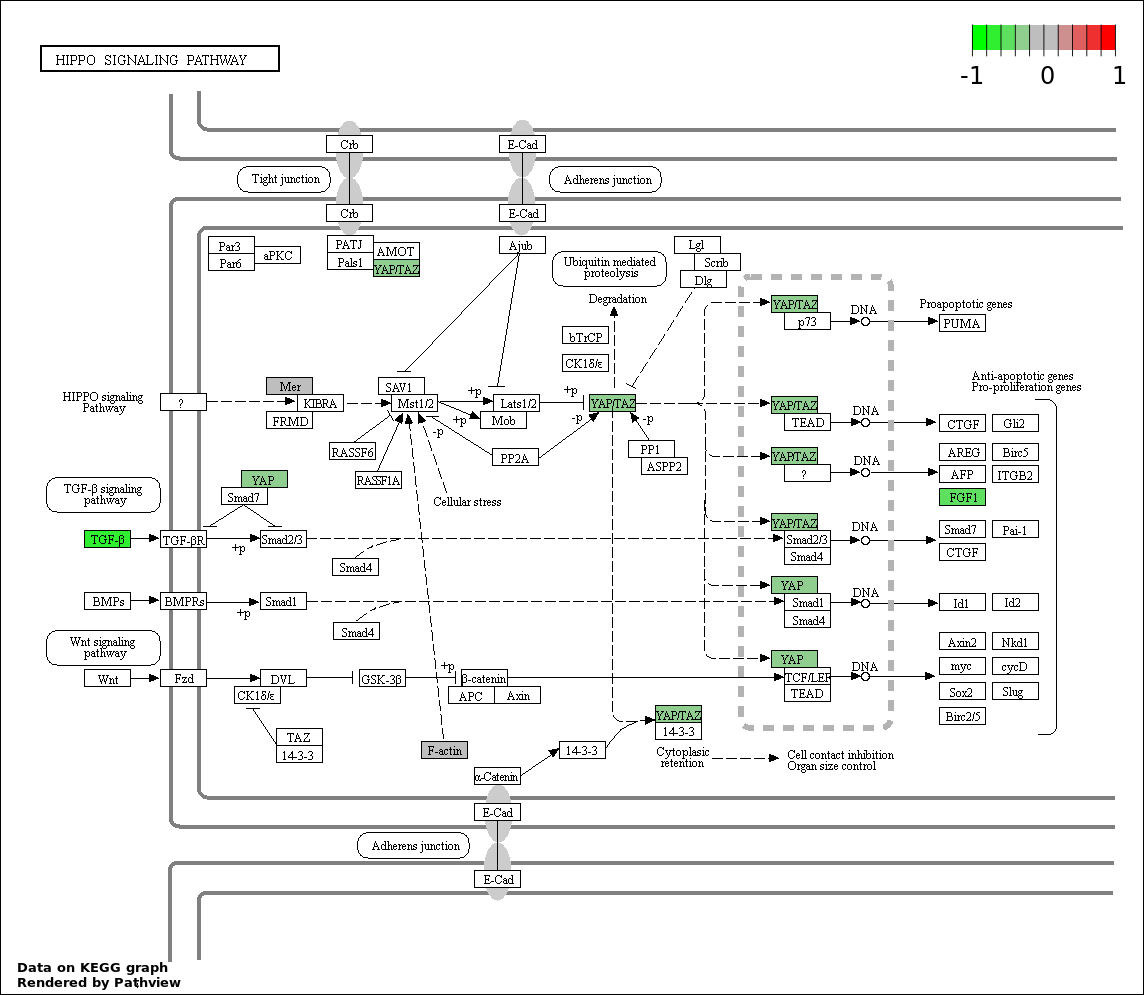
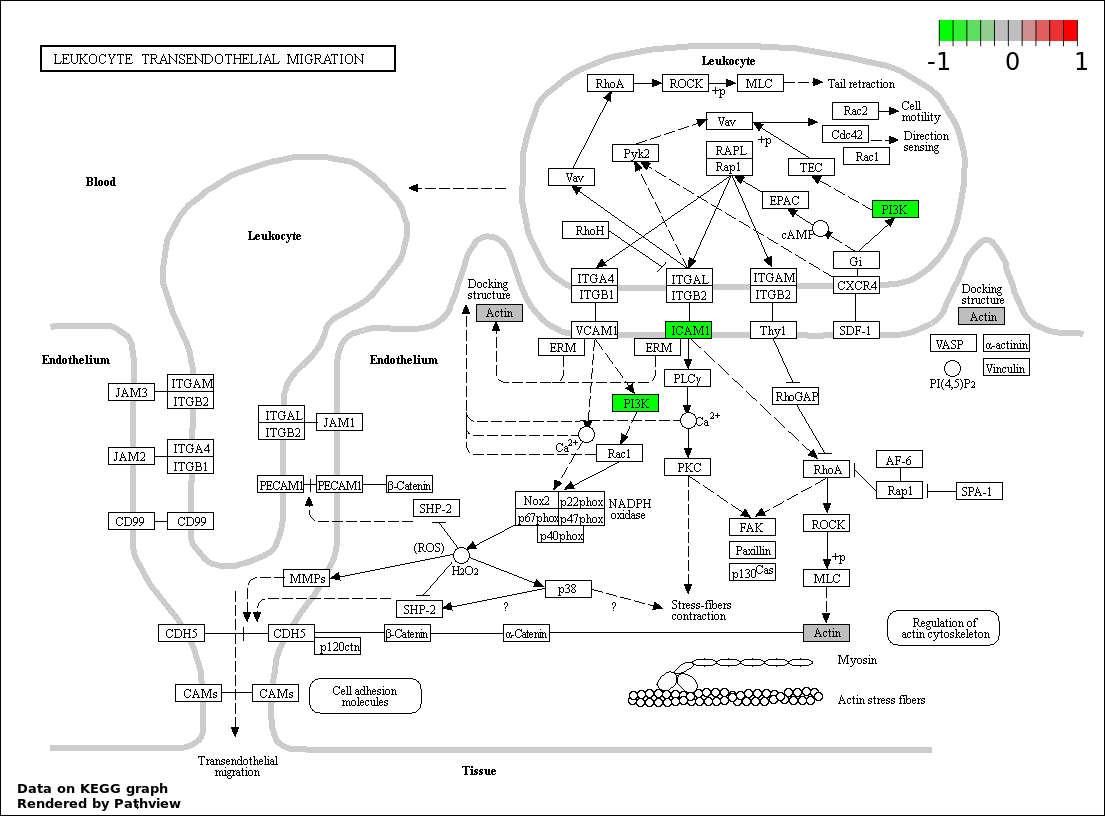
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|
| 1 |
REGULATION OF CELLULAR COMPONENT MOVEMENT |
20 |
771 |
1.65e-10 |
7.679e-07 |
| 2 |
CELL ACTIVATION |
16 |
568 |
4.466e-09 |
7.72e-06 |
| 3 |
ORGAN MORPHOGENESIS |
19 |
841 |
4.978e-09 |
7.72e-06 |
| 4 |
LEUKOCYTE ACTIVATION |
13 |
414 |
4.12e-08 |
3.316e-05 |
| 5 |
LYMPHOCYTE ACTIVATION |
12 |
342 |
4.276e-08 |
3.316e-05 |
| 6 |
LYMPHOCYTE DIFFERENTIATION |
10 |
209 |
3.461e-08 |
3.316e-05 |
| 7 |
REGULATION OF EPITHELIAL CELL MIGRATION |
9 |
166 |
5.975e-08 |
3.66e-05 |
| 8 |
MORPHOGENESIS OF A BRANCHING STRUCTURE |
9 |
167 |
6.293e-08 |
3.66e-05 |
| 9 |
SINGLE ORGANISM CELL ADHESION |
13 |
459 |
1.364e-07 |
6.347e-05 |
| 10 |
BRANCHING MORPHOGENESIS OF AN EPITHELIAL TUBE |
8 |
131 |
1.35e-07 |
6.347e-05 |
| 11 |
WOUND HEALING |
13 |
470 |
1.79e-07 |
6.942e-05 |
| 12 |
TUBE DEVELOPMENT |
14 |
552 |
1.664e-07 |
6.942e-05 |
| 13 |
RESPONSE TO WOUNDING |
14 |
563 |
2.115e-07 |
7.571e-05 |
| 14 |
ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY |
15 |
689 |
4.088e-07 |
0.0001119 |
| 15 |
TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE SIGNALING PATHWAY |
13 |
498 |
3.465e-07 |
0.0001119 |
| 16 |
CARDIOVASCULAR SYSTEM DEVELOPMENT |
16 |
788 |
4.072e-07 |
0.0001119 |
| 17 |
CIRCULATORY SYSTEM DEVELOPMENT |
16 |
788 |
4.072e-07 |
0.0001119 |
| 18 |
POSITIVE REGULATION OF GENE EXPRESSION |
24 |
1733 |
4.363e-07 |
0.0001128 |
| 19 |
LEUKOCYTE DIFFERENTIATION |
10 |
292 |
7.71e-07 |
0.0001888 |
| 20 |
EPITHELIUM DEVELOPMENT |
17 |
945 |
9.108e-07 |
0.0002119 |
| 21 |
T CELL DIFFERENTIATION |
7 |
123 |
1.372e-06 |
0.000304 |
| 22 |
CELL PROLIFERATION |
14 |
672 |
1.745e-06 |
0.000369 |
| 23 |
IMMUNE SYSTEM DEVELOPMENT |
13 |
582 |
1.982e-06 |
0.000401 |
| 24 |
LEUKOCYTE CELL CELL ADHESION |
9 |
255 |
2.222e-06 |
0.0004308 |
| 25 |
POSITIVE REGULATION OF LOCOMOTION |
11 |
420 |
2.886e-06 |
0.0005112 |
| 26 |
POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS |
20 |
1395 |
2.967e-06 |
0.0005112 |
| 27 |
TISSUE DEVELOPMENT |
21 |
1518 |
2.797e-06 |
0.0005112 |
| 28 |
CELL CELL ADHESION |
13 |
608 |
3.202e-06 |
0.0005174 |
| 29 |
MESONEPHROS DEVELOPMENT |
6 |
90 |
3.225e-06 |
0.0005174 |
| 30 |
REGULATION OF CELL ADHESION |
13 |
629 |
4.632e-06 |
0.0007184 |
| 31 |
POSITIVE REGULATION OF EPITHELIAL CELL MIGRATION |
6 |
103 |
7.067e-06 |
0.001061 |
| 32 |
DEVELOPMENTAL GROWTH INVOLVED IN MORPHOGENESIS |
6 |
104 |
7.473e-06 |
0.001087 |
| 33 |
LOCOMOTION |
17 |
1114 |
8.337e-06 |
0.001128 |
| 34 |
VASCULATURE DEVELOPMENT |
11 |
469 |
8.235e-06 |
0.001128 |
| 35 |
REGULATION OF CELL PROLIFERATION |
20 |
1496 |
8.483e-06 |
0.001128 |
| 36 |
POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER |
16 |
1004 |
9.342e-06 |
0.001208 |
| 37 |
MORPHOGENESIS OF AN EPITHELIUM |
10 |
400 |
1.261e-05 |
0.001416 |
| 38 |
POSITIVE REGULATION OF ENDOTHELIAL CELL MIGRATION |
5 |
67 |
1.278e-05 |
0.001416 |
| 39 |
REGULATION OF ENDOTHELIAL CELL MIGRATION |
6 |
114 |
1.267e-05 |
0.001416 |
| 40 |
REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT |
21 |
1672 |
1.254e-05 |
0.001416 |
| 41 |
MOVEMENT OF CELL OR SUBCELLULAR COMPONENT |
18 |
1275 |
1.259e-05 |
0.001416 |
| 42 |
GLAND DEVELOPMENT |
10 |
395 |
1.131e-05 |
0.001416 |
| 43 |
BIOLOGICAL ADHESION |
16 |
1032 |
1.316e-05 |
0.001424 |
| 44 |
TUBE MORPHOGENESIS |
9 |
323 |
1.499e-05 |
0.001585 |
| 45 |
DEVELOPMENTAL GROWTH |
9 |
333 |
1.908e-05 |
0.001973 |
| 46 |
RESPONSE TO ENDOGENOUS STIMULUS |
19 |
1450 |
1.979e-05 |
0.002001 |
| 47 |
VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY |
5 |
74 |
2.08e-05 |
0.00206 |
| 48 |
KIDNEY EPITHELIUM DEVELOPMENT |
6 |
125 |
2.142e-05 |
0.002077 |
| 49 |
TISSUE MORPHOGENESIS |
11 |
533 |
2.699e-05 |
0.002563 |
| 50 |
DENDRITE DEVELOPMENT |
5 |
79 |
2.86e-05 |
0.002661 |
| 51 |
REGULATION OF CELL DIFFERENTIATION |
19 |
1492 |
2.94e-05 |
0.002682 |
| 52 |
POSITIVE REGULATION OF PROTEIN COMPLEX ASSEMBLY |
7 |
197 |
3.029e-05 |
0.002683 |
| 53 |
NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS |
15 |
983 |
3.056e-05 |
0.002683 |
| 54 |
POSITIVE REGULATION OF RESPONSE TO STIMULUS |
22 |
1929 |
3.348e-05 |
0.002832 |
| 55 |
REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER |
21 |
1784 |
3.322e-05 |
0.002832 |
| 56 |
KIDNEY MORPHOGENESIS |
5 |
82 |
3.426e-05 |
0.002846 |
| 57 |
REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION |
20 |
1656 |
3.715e-05 |
0.003033 |
| 58 |
NEUROGENESIS |
18 |
1402 |
4.459e-05 |
0.003578 |
| 59 |
POSITIVE REGULATION OF MAPK CASCADE |
10 |
470 |
4.978e-05 |
0.003639 |
| 60 |
POSITIVE REGULATION OF HYDROLASE ACTIVITY |
14 |
905 |
5.005e-05 |
0.003639 |
| 61 |
THYMOCYTE AGGREGATION |
4 |
45 |
4.961e-05 |
0.003639 |
| 62 |
REGULATION OF PEPTIDYL TYROSINE PHOSPHORYLATION |
7 |
213 |
4.978e-05 |
0.003639 |
| 63 |
HEART DEVELOPMENT |
10 |
466 |
4.634e-05 |
0.003639 |
| 64 |
T CELL DIFFERENTIATION IN THYMUS |
4 |
45 |
4.961e-05 |
0.003639 |
| 65 |
REGULATION OF CELL CELL ADHESION |
9 |
380 |
5.344e-05 |
0.003826 |
| 66 |
BRANCH ELONGATION OF AN EPITHELIUM |
3 |
17 |
5.725e-05 |
0.003986 |
| 67 |
CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND |
13 |
799 |
5.739e-05 |
0.003986 |
| 68 |
UROGENITAL SYSTEM DEVELOPMENT |
8 |
299 |
6.092e-05 |
0.004169 |
| 69 |
NEGATIVE REGULATION OF CELL ADHESION |
7 |
223 |
6.648e-05 |
0.004483 |
| 70 |
FC GAMMA RECEPTOR SIGNALING PATHWAY |
5 |
95 |
6.948e-05 |
0.004618 |
| 71 |
EPITHELIAL CELL DIFFERENTIATION |
10 |
495 |
7.665e-05 |
0.005023 |
| 72 |
NEGATIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY |
5 |
98 |
8.058e-05 |
0.005067 |
| 73 |
PROTEIN PHOSPHORYLATION |
14 |
944 |
7.872e-05 |
0.005067 |
| 74 |
NEGATIVE REGULATION OF CELL ACTIVATION |
6 |
158 |
7.963e-05 |
0.005067 |
| 75 |
REGULATION OF CELL DEATH |
18 |
1472 |
8.378e-05 |
0.005198 |
| 76 |
CELL MOTILITY |
13 |
835 |
8.962e-05 |
0.00521 |
| 77 |
POSITIVE REGULATION OF CELLULAR COMPONENT BIOGENESIS |
9 |
406 |
8.87e-05 |
0.00521 |
| 78 |
LOCALIZATION OF CELL |
13 |
835 |
8.962e-05 |
0.00521 |
| 79 |
EMBRYONIC ORGAN DEVELOPMENT |
9 |
406 |
8.87e-05 |
0.00521 |
| 80 |
REGULATION OF CELL DEVELOPMENT |
13 |
836 |
9.07e-05 |
0.00521 |
| 81 |
NEGATIVE REGULATION OF CELL DIFFERENTIATION |
11 |
609 |
8.971e-05 |
0.00521 |
| 82 |
MESONEPHRIC TUBULE MORPHOGENESIS |
4 |
53 |
9.486e-05 |
0.005357 |
| 83 |
GROWTH |
9 |
410 |
9.556e-05 |
0.005357 |
| 84 |
EPIDERMAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY |
4 |
55 |
0.0001097 |
0.006078 |
| 85 |
POSITIVE REGULATION OF MOLECULAR FUNCTION |
20 |
1791 |
0.000111 |
0.006078 |
| 86 |
POSITIVE REGULATION OF CELL CELL ADHESION |
7 |
243 |
0.0001137 |
0.006152 |
| 87 |
POSITIVE REGULATION OF CATALYTIC ACTIVITY |
18 |
1518 |
0.0001239 |
0.006628 |
| 88 |
POSITIVE REGULATION OF IMMUNE SYSTEM PROCESS |
13 |
867 |
0.0001305 |
0.006898 |
| 89 |
POSITIVE REGULATION OF CELL COMMUNICATION |
18 |
1532 |
0.0001391 |
0.007114 |
| 90 |
NEGATIVE REGULATION OF CELL DEATH |
13 |
872 |
0.0001381 |
0.007114 |
| 91 |
CENTRAL NERVOUS SYSTEM DEVELOPMENT |
13 |
872 |
0.0001381 |
0.007114 |
| 92 |
POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION |
13 |
876 |
0.0001445 |
0.007308 |
| 93 |
IMMUNE SYSTEM PROCESS |
21 |
1984 |
0.0001539 |
0.007544 |
| 94 |
REGULATION OF IMMUNE SYSTEM PROCESS |
17 |
1403 |
0.0001523 |
0.007544 |
| 95 |
EMBRYONIC MORPHOGENESIS |
10 |
539 |
0.000154 |
0.007544 |
| 96 |
CELLULAR RESPONSE TO ENDOGENOUS STIMULUS |
14 |
1008 |
0.0001571 |
0.007615 |
| 97 |
NEGATIVE REGULATION OF CATION TRANSMEMBRANE TRANSPORT |
4 |
61 |
0.0001645 |
0.007833 |
| 98 |
POSITIVE REGULATION OF DEVELOPMENTAL PROCESS |
15 |
1142 |
0.000165 |
0.007833 |
| 99 |
RESPONSE TO HORMONE |
13 |
893 |
0.0001746 |
0.008208 |
| 100 |
REGULATION OF MAPK CASCADE |
11 |
660 |
0.0001815 |
0.008281 |
| 101 |
POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION |
15 |
1152 |
0.0001814 |
0.008281 |
| 102 |
REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS |
14 |
1021 |
0.0001795 |
0.008281 |
| 103 |
LENS FIBER CELL DIFFERENTIATION |
3 |
25 |
0.0001886 |
0.00836 |
| 104 |
NEGATIVE REGULATION OF LOCOMOTION |
7 |
263 |
0.0001852 |
0.00836 |
| 105 |
CELLULAR COMPONENT MORPHOGENESIS |
13 |
900 |
0.0001885 |
0.00836 |
| 106 |
POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS |
14 |
1036 |
0.0002087 |
0.009075 |
| 107 |
POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS |
14 |
1036 |
0.0002087 |
0.009075 |
| 108 |
MESENCHYME DEVELOPMENT |
6 |
190 |
0.0002182 |
0.009402 |
| 109 |
LENS DEVELOPMENT IN CAMERA TYPE EYE |
4 |
66 |
0.0002233 |
0.009533 |
| 110 |
BLOOD VESSEL MORPHOGENESIS |
8 |
364 |
0.0002359 |
0.009978 |
| 111 |
AXIS ELONGATION |
3 |
27 |
0.0002384 |
0.009992 |