This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-let-7a-3p | AXIN2 | 1.42 | 0 | -1.7 | 6.0E-5 | miRNATAP | -0.58 | 0 | NA | |
| 2 | hsa-miR-15b-5p | AXIN2 | 1.57 | 0 | -1.7 | 6.0E-5 | miRTarBase; MirTarget; miRNATAP | -0.56 | 0 | NA | |
| 3 | hsa-miR-16-2-3p | AXIN2 | 2.32 | 0 | -1.7 | 6.0E-5 | mirMAP | -0.59 | 0 | NA | |
| 4 | hsa-miR-16-5p | AXIN2 | 1.76 | 0 | -1.7 | 6.0E-5 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.75 | 0 | NA | |
| 5 | hsa-miR-10a-5p | CAMK2A | 1.15 | 0.00372 | -4.14 | 0 | mirMAP | -0.69 | 0 | NA | |
| 6 | hsa-miR-10b-5p | CAMK2A | -0.16 | 0.55501 | -4.14 | 0 | mirMAP | -0.81 | 0 | NA | |
| 7 | hsa-miR-1226-3p | CAMK2A | 1.38 | 0.00021 | -4.14 | 0 | mirMAP | -0.68 | 0 | NA | |
| 8 | hsa-miR-125a-5p | CAMK2A | 0.6 | 0.01023 | -4.14 | 0 | mirMAP | -0.75 | 0 | NA | |
| 9 | hsa-miR-130b-5p | CAMK2A | 3 | 0 | -4.14 | 0 | mirMAP | -0.79 | 0 | NA | |
| 10 | hsa-miR-148b-5p | CAMK2A | 2.18 | 0 | -4.14 | 0 | mirMAP | -1.01 | 0 | NA | |
| 11 | hsa-miR-15b-3p | CAMK2A | 2.34 | 0 | -4.14 | 0 | mirMAP | -1.07 | 0 | NA | |
| 12 | hsa-miR-185-5p | CAMK2A | 2.34 | 0 | -4.14 | 0 | mirMAP | -1.21 | 0 | NA | |
| 13 | hsa-miR-19b-1-5p | CAMK2A | 2.58 | 0 | -4.14 | 0 | mirMAP | -0.86 | 0 | NA | |
| 14 | hsa-miR-25-3p | CAMK2A | 1.36 | 0 | -4.14 | 0 | MirTarget | -1.13 | 0 | NA | |
| 15 | hsa-miR-26b-5p | CAMK2A | 0.89 | 0 | -4.14 | 0 | miRNATAP | -1.08 | 0 | NA | |
| 16 | hsa-miR-30b-5p | CAMK2A | 0.8 | 0.00013 | -4.14 | 0 | mirMAP | -0.91 | 0 | NA | |
| 17 | hsa-miR-30c-5p | CAMK2A | 0.78 | 0.00029 | -4.14 | 0 | mirMAP | -1.03 | 0 | NA | |
| 18 | hsa-miR-30d-5p | CAMK2A | 0.68 | 0.00271 | -4.14 | 0 | mirMAP | -1.36 | 0 | NA | |
| 19 | hsa-miR-30e-5p | CAMK2A | 1.24 | 0 | -4.14 | 0 | mirMAP | -1.66 | 0 | NA | |
| 20 | hsa-miR-32-5p | CAMK2A | 2.34 | 0 | -4.14 | 0 | MirTarget | -0.86 | 0 | NA | |
| 21 | hsa-miR-331-3p | CAMK2A | 2.28 | 0 | -4.14 | 0 | MirTarget | -1.16 | 0 | NA | |
| 22 | hsa-miR-34a-3p | CAMK2A | 1.76 | 0 | -4.14 | 0 | mirMAP | -0.71 | 0 | NA | |
| 23 | hsa-miR-361-3p | CAMK2A | 1.07 | 0 | -4.14 | 0 | mirMAP | -0.9 | 0 | NA | |
| 24 | hsa-miR-3913-5p | CAMK2A | 2.44 | 0 | -4.14 | 0 | mirMAP | -0.79 | 0 | NA | |
| 25 | hsa-miR-429 | CAMK2A | 4.49 | 0 | -4.14 | 0 | miRNATAP | -0.62 | 0 | NA | |
| 26 | hsa-miR-582-3p | CAMK2A | 0.01 | 0.97713 | -4.14 | 0 | mirMAP | -0.52 | 0 | NA | |
| 27 | hsa-miR-7-1-3p | CAMK2A | 1.85 | 0 | -4.14 | 0 | MirTarget | -0.81 | 0 | NA | |
| 28 | hsa-miR-92a-3p | CAMK2A | 2.06 | 0 | -4.14 | 0 | MirTarget | -1.24 | 0 | NA | |
| 29 | hsa-miR-340-5p | CCND1 | 0.3 | 0.15774 | 0.08 | 0.84033 | mirMAP | -0.53 | 0 | NA | |
| 30 | hsa-let-7a-5p | CCND2 | 0.62 | 3.0E-5 | -2.43 | 0 | miRNAWalker2 validate; miRTarBase; TargetScan; miRNATAP | -0.5 | 0.0001 | 20418948 | MicroRNA let 7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2 |
| 31 | hsa-let-7g-5p | CCND2 | 1.2 | 0 | -2.43 | 0 | miRNATAP | -0.78 | 0 | NA | |
| 32 | hsa-miR-106b-5p | CCND2 | 2.47 | 0 | -2.43 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.72 | 0 | NA | |
| 33 | hsa-miR-130b-5p | CCND2 | 3 | 0 | -2.43 | 0 | mirMAP | -0.52 | 0 | NA | |
| 34 | hsa-miR-151a-3p | CCND2 | 1.24 | 0 | -2.43 | 0 | mirMAP | -0.62 | 0 | NA | |
| 35 | hsa-miR-15a-5p | CCND2 | 2.35 | 0 | -2.43 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.56 | 0 | NA | |
| 36 | hsa-miR-15b-5p | CCND2 | 1.57 | 0 | -2.43 | 0 | miRNATAP | -0.81 | 0 | NA | |
| 37 | hsa-miR-16-2-3p | CCND2 | 2.32 | 0 | -2.43 | 0 | mirMAP | -0.52 | 0 | NA | |
| 38 | hsa-miR-16-5p | CCND2 | 1.76 | 0 | -2.43 | 0 | miRNAWalker2 validate; miRNATAP | -0.62 | 0 | NA | |
| 39 | hsa-miR-17-5p | CCND2 | 3.27 | 0 | -2.43 | 0 | miRNAWalker2 validate; miRTarBase; TargetScan; miRNATAP | -0.58 | 0 | NA | |
| 40 | hsa-miR-185-5p | CCND2 | 2.34 | 0 | -2.43 | 0 | MirTarget; miRNATAP | -0.6 | 0 | NA | |
| 41 | hsa-miR-186-5p | CCND2 | 1.47 | 0 | -2.43 | 0 | mirMAP; miRNATAP | -0.75 | 0 | NA | |
| 42 | hsa-miR-191-5p | CCND2 | 2.3 | 0 | -2.43 | 0 | MirTarget | -0.63 | 0 | NA | |
| 43 | hsa-miR-19b-3p | CCND2 | 2.5 | 0 | -2.43 | 0 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.64 | 0 | NA | |
| 44 | hsa-miR-20a-5p | CCND2 | 3.16 | 0 | -2.43 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.55 | 0 | NA | |
| 45 | hsa-miR-30d-3p | CCND2 | 0.98 | 4.0E-5 | -2.43 | 0 | mirMAP | -0.51 | 0 | NA | |
| 46 | hsa-miR-324-3p | CCND2 | 2.09 | 0 | -2.43 | 0 | miRNAWalker2 validate | -0.57 | 0 | NA | |
| 47 | hsa-miR-331-5p | CCND2 | 2.17 | 0 | -2.43 | 0 | miRNATAP | -0.55 | 0 | NA | |
| 48 | hsa-miR-423-5p | CCND2 | 0.96 | 0 | -2.43 | 0 | miRNAWalker2 validate | -0.68 | 0 | NA | |
| 49 | hsa-miR-590-5p | CCND2 | 3.18 | 0 | -2.43 | 0 | mirMAP | -0.51 | 0 | NA | |
| 50 | hsa-miR-7-1-3p | CCND2 | 1.85 | 0 | -2.43 | 0 | mirMAP | -0.52 | 0 | NA | |
| 51 | hsa-miR-93-5p | CCND2 | 3.04 | 0 | -2.43 | 0 | miRNATAP | -0.64 | 0 | NA | |
| 52 | hsa-miR-1976 | DAAM2 | 2.07 | 0 | -2.23 | 0 | mirMAP | -0.64 | 0 | NA | |
| 53 | hsa-miR-19b-3p | DAAM2 | 2.5 | 0 | -2.23 | 0 | miRNAWalker2 validate | -0.6 | 0 | NA | |
| 54 | hsa-miR-361-3p | DAAM2 | 1.07 | 0 | -2.23 | 0 | mirMAP | -0.61 | 0 | NA | |
| 55 | hsa-miR-590-3p | DAAM2 | 2.59 | 0 | -2.23 | 0 | miRanda; mirMAP | -0.54 | 0 | NA | |
| 56 | hsa-miR-361-5p | DKK2 | 0.97 | 0 | 1.45 | 0.00748 | miRanda | -0.53 | 0.00074 | NA | |
| 57 | hsa-miR-146b-5p | MAPK10 | 1.76 | 0 | -1.4 | 0.00097 | miRanda | -0.59 | 0 | NA | |
| 58 | hsa-let-7g-5p | MYC | 1.2 | 0 | -1.77 | 0 | miRNAWalker2 validate; miRTarBase | -0.65 | 0 | NA | |
| 59 | hsa-miR-423-5p | MYC | 0.96 | 0 | -1.77 | 0 | miRNAWalker2 validate | -0.59 | 0 | NA | |
| 60 | hsa-miR-98-5p | MYC | 1.11 | 0 | -1.77 | 0 | miRNAWalker2 validate; miRTarBase | -0.6 | 0 | NA | |
| 61 | hsa-miR-3613-5p | NFATC2 | 2.64 | 0 | -2.1 | 3.0E-5 | mirMAP | -0.58 | 0 | NA | |
| 62 | hsa-miR-361-5p | NKD1 | 0.97 | 0 | -1.7 | 4.0E-5 | miRanda | -0.66 | 0 | NA | |
| 63 | hsa-let-7a-3p | PLCB4 | 1.42 | 0 | -3.77 | 0 | mirMAP; miRNATAP | -0.83 | 0 | NA | |
| 64 | hsa-let-7b-3p | PLCB4 | 0.87 | 1.0E-5 | -3.77 | 0 | mirMAP; miRNATAP | -0.71 | 0 | NA | |
| 65 | hsa-let-7f-1-3p | PLCB4 | 1.55 | 0 | -3.77 | 0 | mirMAP | -0.71 | 0 | NA | |
| 66 | hsa-let-7g-5p | PLCB4 | 1.2 | 0 | -3.77 | 0 | miRNATAP | -0.61 | 2.0E-5 | NA | |
| 67 | hsa-let-7i-5p | PLCB4 | 0.41 | 0.00623 | -3.77 | 0 | miRNATAP | -0.76 | 1.0E-5 | NA | |
| 68 | hsa-miR-16-2-3p | PLCB4 | 2.32 | 0 | -3.77 | 0 | mirMAP | -0.82 | 0 | NA | |
| 69 | hsa-miR-23a-3p | PLCB4 | 1 | 0 | -3.77 | 0 | miRNATAP | -1.16 | 0 | NA | |
| 70 | hsa-miR-26b-5p | PLCB4 | 0.89 | 0 | -3.77 | 0 | miRNAWalker2 validate | -0.77 | 0 | NA | |
| 71 | hsa-miR-454-3p | PLCB4 | 2.47 | 0 | -3.77 | 0 | miRNATAP | -0.58 | 0 | NA | |
| 72 | hsa-miR-455-5p | PLCB4 | 2.26 | 0 | -3.77 | 0 | PITA; miRanda | -0.61 | 0 | NA | |
| 73 | hsa-miR-590-3p | PLCB4 | 2.59 | 0 | -3.77 | 0 | miRanda | -0.61 | 0 | NA | |
| 74 | hsa-miR-30b-5p | PRICKLE1 | 0.8 | 0.00013 | -1.64 | 0.00013 | mirMAP | -0.56 | 0 | NA | |
| 75 | hsa-miR-30c-5p | PRICKLE1 | 0.78 | 0.00029 | -1.64 | 0.00013 | miRNATAP | -0.69 | 0 | NA | |
| 76 | hsa-miR-30d-5p | PRICKLE1 | 0.68 | 0.00271 | -1.64 | 0.00013 | miRNATAP | -0.8 | 0 | NA | |
| 77 | hsa-miR-330-3p | PRICKLE1 | 0.88 | 0.00074 | -1.64 | 0.00013 | mirMAP; miRNATAP | -0.62 | 0 | NA | |
| 78 | hsa-miR-361-5p | PRICKLE1 | 0.97 | 0 | -1.64 | 0.00013 | miRanda | -0.96 | 0 | NA | |
| 79 | hsa-miR-532-5p | PRICKLE1 | 1.56 | 0 | -1.64 | 0.00013 | miRNATAP | -0.57 | 0 | NA | |
| 80 | hsa-let-7a-3p | PRICKLE2 | 1.42 | 0 | -2.5 | 0 | mirMAP | -0.54 | 0 | NA | |
| 81 | hsa-miR-148b-3p | PRICKLE2 | 1.98 | 0 | -2.5 | 0 | MirTarget; miRNATAP | -0.79 | 0 | NA | |
| 82 | hsa-miR-15b-3p | PRICKLE2 | 2.34 | 0 | -2.5 | 0 | mirMAP | -0.63 | 0 | NA | |
| 83 | hsa-miR-16-2-3p | PRICKLE2 | 2.32 | 0 | -2.5 | 0 | mirMAP | -0.63 | 0 | NA | |
| 84 | hsa-miR-186-5p | PRICKLE2 | 1.47 | 0 | -2.5 | 0 | MirTarget | -0.65 | 0 | NA | |
| 85 | hsa-miR-197-3p | PRICKLE2 | 1.76 | 0 | -2.5 | 0 | mirMAP | -0.52 | 0 | NA | |
| 86 | hsa-miR-19b-3p | PRICKLE2 | 2.5 | 0 | -2.5 | 0 | MirTarget; miRNATAP | -0.58 | 0 | NA | |
| 87 | hsa-miR-361-5p | PRICKLE2 | 0.97 | 0 | -2.5 | 0 | MirTarget; PITA; miRanda; miRNATAP | -0.72 | 0 | NA | |
| 88 | hsa-miR-484 | PRICKLE2 | 1.82 | 0 | -2.5 | 0 | miRNATAP | -0.61 | 0 | NA | |
| 89 | hsa-miR-590-3p | PRICKLE2 | 2.59 | 0 | -2.5 | 0 | MirTarget; PITA; miRanda; mirMAP; miRNATAP | -0.51 | 0 | NA | |
| 90 | hsa-miR-7-1-3p | PRICKLE2 | 1.85 | 0 | -2.5 | 0 | MirTarget | -0.55 | 0 | NA | |
| 91 | hsa-miR-16-2-3p | PRKCB | 2.32 | 0 | -3.26 | 0 | mirMAP | -0.51 | 0 | NA | |
| 92 | hsa-miR-186-5p | PRKCB | 1.47 | 0 | -3.26 | 0 | mirMAP | -0.79 | 0 | NA | |
| 93 | hsa-miR-30d-3p | PRKCB | 0.98 | 4.0E-5 | -3.26 | 0 | mirMAP | -0.53 | 0 | NA | |
| 94 | hsa-miR-30d-5p | PRKCB | 0.68 | 0.00271 | -3.26 | 0 | mirMAP | -0.6 | 0 | NA | |
| 95 | hsa-miR-361-5p | PRKCB | 0.97 | 0 | -3.26 | 0 | MirTarget; miRanda; miRNATAP | -1.09 | 0 | NA | |
| 96 | hsa-miR-484 | PRKCB | 1.82 | 0 | -3.26 | 0 | MirTarget; miRNATAP | -0.7 | 0 | NA | |
| 97 | hsa-miR-589-5p | PRKCB | 1.72 | 0 | -3.26 | 0 | mirMAP | -0.54 | 0 | NA | |
| 98 | hsa-miR-590-5p | PRKCB | 3.18 | 0 | -3.26 | 0 | miRanda | -0.55 | 0 | NA | |
| 99 | hsa-miR-1301-3p | SFRP1 | 2.55 | 0 | -4.72 | 0 | MirTarget | -0.93 | 0 | NA | |
| 100 | hsa-miR-16-1-3p | SFRP1 | 2.57 | 0 | -4.72 | 0 | MirTarget | -0.99 | 0 | NA | |
| 101 | hsa-miR-182-5p | SFRP1 | 3.54 | 0 | -4.72 | 0 | mirMAP | -0.69 | 0 | NA | |
| 102 | hsa-miR-19b-1-5p | SFRP1 | 2.58 | 0 | -4.72 | 0 | MirTarget | -0.81 | 0 | NA | |
| 103 | hsa-miR-26b-3p | SFRP1 | 2.18 | 0 | -4.72 | 0 | MirTarget | -1.18 | 0 | NA | |
| 104 | hsa-miR-27a-3p | SFRP1 | 1.67 | 0 | -4.72 | 0 | MirTarget; miRNATAP | -0.52 | 0.00046 | NA | |
| 105 | hsa-miR-30d-3p | SFRP1 | 0.98 | 4.0E-5 | -4.72 | 0 | miRNATAP | -0.81 | 0 | NA | |
| 106 | hsa-miR-328-3p | SFRP1 | 0.6 | 0.01386 | -4.72 | 0 | MirTarget | -0.81 | 0 | NA | |
| 107 | hsa-miR-33a-3p | SFRP1 | 1.73 | 0 | -4.72 | 0 | MirTarget | -0.83 | 0 | NA | |
| 108 | hsa-miR-429 | SFRP1 | 4.49 | 0 | -4.72 | 0 | miRNATAP | -0.53 | 0 | NA | |
| 109 | hsa-miR-484 | SFRP1 | 1.82 | 0 | -4.72 | 0 | MirTarget | -1 | 0 | NA | |
| 110 | hsa-miR-576-5p | SFRP1 | 2.2 | 0 | -4.72 | 0 | MirTarget; PITA | -1 | 0 | NA | |
| 111 | hsa-miR-7-1-3p | SFRP1 | 1.85 | 0 | -4.72 | 0 | MirTarget; mirMAP | -0.76 | 0 | NA | |
| 112 | hsa-miR-940 | SFRP1 | 3.21 | 0 | -4.72 | 0 | MirTarget | -0.69 | 0 | NA | |
| 113 | hsa-miR-96-5p | SFRP1 | 4.89 | 0 | -4.72 | 0 | mirMAP | -0.67 | 0 | NA | |
| 114 | hsa-let-7a-3p | SFRP2 | 1.42 | 0 | -4.16 | 3.0E-5 | miRNATAP | -0.89 | 0.00023 | NA | |
| 115 | hsa-miR-129-5p | SFRP2 | -0.52 | 0.22258 | -4.16 | 3.0E-5 | miRanda | -0.63 | 0 | NA | |
| 116 | hsa-miR-33a-3p | SFRP2 | 1.73 | 0 | -4.16 | 3.0E-5 | miRNATAP | -1.63 | 0 | NA | |
| 117 | hsa-miR-421 | SFRP2 | 1.18 | 1.0E-5 | -4.16 | 3.0E-5 | miRanda | -1.02 | 0 | NA | |
| 118 | hsa-miR-582-5p | SFRP2 | 0.61 | 0.03299 | -4.16 | 3.0E-5 | miRNATAP | -1.18 | 0 | NA | |
| 119 | hsa-miR-590-3p | SFRP2 | 2.59 | 0 | -4.16 | 3.0E-5 | miRanda | -0.97 | 0 | NA | |
| 120 | hsa-miR-129-5p | SFRP4 | -0.52 | 0.22258 | -1.42 | 0.14414 | miRanda | -0.64 | 0 | NA | |
| 121 | hsa-miR-144-5p | SFRP4 | 0.2 | 0.61856 | -1.42 | 0.14414 | MirTarget | -0.94 | 0 | NA | |
| 122 | hsa-miR-26b-5p | SFRP4 | 0.89 | 0 | -1.42 | 0.14414 | miRNAWalker2 validate | -1.21 | 0 | NA | |
| 123 | hsa-miR-3607-3p | SFRP4 | 2.69 | 0 | -1.42 | 0.14414 | MirTarget | -0.85 | 0 | NA | |
| 124 | hsa-miR-429 | SFRP4 | 4.49 | 0 | -1.42 | 0.14414 | miRanda | -0.66 | 0 | NA | |
| 125 | hsa-miR-532-5p | SFRP4 | 1.56 | 0 | -1.42 | 0.14414 | MirTarget | -0.93 | 0 | NA | |
| 126 | hsa-miR-664a-3p | SFRP4 | 0.63 | 0.0052 | -1.42 | 0.14414 | mirMAP | -1.04 | 0 | NA | |
| 127 | hsa-miR-944 | SFRP4 | 2.91 | 0 | -1.42 | 0.14414 | mirMAP | -0.54 | 0 | NA | |
| 128 | hsa-miR-330-3p | WNT11 | 0.88 | 0.00074 | -0.82 | 0.17434 | miRNATAP | -0.63 | 0 | NA | |
| 129 | hsa-miR-186-5p | WNT2B | 1.47 | 0 | -2.77 | 0 | mirMAP | -0.53 | 0 | NA | |
| 130 | hsa-miR-185-5p | WNT5B | 2.34 | 0 | -2.11 | 3.0E-5 | miRNAWalker2 validate | -0.61 | 0 | NA | |
| 131 | hsa-miR-28-5p | WNT5B | 0.23 | 0.07429 | -2.11 | 3.0E-5 | miRanda | -0.53 | 0.0071 | NA | |
| 132 | hsa-miR-15b-5p | WNT7A | 1.57 | 0 | 1.48 | 0.07649 | MirTarget | -0.55 | 0.00235 | 27195958 | WNT7A Regulation by miR 15b in Ovarian Cancer; In the present study we report that WNT7A is a direct target of miR-15b in ovarian cancer; We showed that a luciferase reporter containing the putative binding site of miR-15b in the WNT7A 3'-UTR was significantly repressed by miR-15b; Mutation of the putative binding site of miR-15b in the WNT7A 3'-UTR restored luciferase activity; Furthermore miR-15b was able to repress increased levels of TOPFLASH activity by WNT7A but not those induced by S33Y; Additionally miR-15b dose-dependently decreased WNT7A expression; When we evaluated the prognostic impact of WNT7A and miR-15b expression using TCGA datasets a significant inverse correlation in which high-expression of WNT7A and low-expression of miR-15b was associated with reduced survival rates of ovarian cancer patients; These results suggest that WNT7A is post-transcriptionally regulated by miR-15b which could be down-regulated by promoter hypermethylation potentially via DNMT1 in ovarian cancer |
| 133 | hsa-miR-361-5p | WNT7A | 0.97 | 0 | 1.48 | 0.07649 | MirTarget; PITA; miRanda | -0.78 | 0.00129 | NA | |
| 134 | hsa-miR-140-3p | WNT9A | 0.06 | 0.72772 | -2.07 | 0 | mirMAP | -0.58 | 0 | NA | |
| 135 | hsa-miR-148b-3p | WNT9A | 1.98 | 0 | -2.07 | 0 | mirMAP | -0.8 | 0 | NA | |
| 136 | hsa-miR-15a-5p | WNT9A | 2.35 | 0 | -2.07 | 0 | mirMAP | -0.52 | 0 | NA | |
| 137 | hsa-miR-15b-5p | WNT9A | 1.57 | 0 | -2.07 | 0 | mirMAP | -0.54 | 0 | NA | |
| 138 | hsa-miR-16-5p | WNT9A | 1.76 | 0 | -2.07 | 0 | mirMAP | -0.53 | 0 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | WNT SIGNALING PATHWAY | 16 | 351 | 3.906e-24 | 1.817e-20 |
| 2 | REGULATION OF WNT SIGNALING PATHWAY | 12 | 310 | 8.755e-17 | 2.037e-13 |
| 3 | REGULATION OF CANONICAL WNT SIGNALING PATHWAY | 11 | 236 | 3.077e-16 | 4.772e-13 |
| 4 | NEGATIVE REGULATION OF WNT SIGNALING PATHWAY | 10 | 197 | 3.987e-15 | 4.638e-12 |
| 5 | NEGATIVE REGULATION OF CANONICAL WNT SIGNALING PATHWAY | 9 | 162 | 5.453e-14 | 5.074e-11 |
| 6 | NON CANONICAL WNT SIGNALING PATHWAY | 8 | 140 | 1.387e-12 | 1.075e-09 |
| 7 | POSITIVE REGULATION OF WNT SIGNALING PATHWAY | 8 | 152 | 2.701e-12 | 1.796e-09 |
| 8 | CANONICAL WNT SIGNALING PATHWAY | 7 | 95 | 7.01e-12 | 4.077e-09 |
| 9 | POSITIVE REGULATION OF CANONICAL WNT SIGNALING PATHWAY | 7 | 119 | 3.498e-11 | 1.809e-08 |
| 10 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 12 | 1021 | 1.183e-10 | 5.003e-08 |
| 11 | REGULATION OF ORGAN MORPHOGENESIS | 8 | 242 | 1.131e-10 | 5.003e-08 |
| 12 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 14 | 1672 | 1.301e-10 | 5.045e-08 |
| 13 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 12 | 1142 | 4.306e-10 | 1.541e-07 |
| 14 | NEGATIVE REGULATION OF CELL COMMUNICATION | 12 | 1192 | 7.046e-10 | 2.342e-07 |
| 15 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 14 | 1929 | 8.74e-10 | 2.711e-07 |
| 16 | REGULATION OF ESTABLISHMENT OF PLANAR POLARITY | 6 | 110 | 1.675e-09 | 4.871e-07 |
| 17 | NEGATIVE REGULATION OF RESPONSE TO STIMULUS | 12 | 1360 | 3.18e-09 | 8.703e-07 |
| 18 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 10 | 801 | 4.193e-09 | 1.084e-06 |
| 19 | CHONDROCYTE DIFFERENTIATION | 5 | 60 | 5.19e-09 | 1.271e-06 |
| 20 | POSITIVE REGULATION OF CELL COMMUNICATION | 12 | 1532 | 1.226e-08 | 2.853e-06 |
| 21 | SKELETAL SYSTEM DEVELOPMENT | 8 | 455 | 1.631e-08 | 3.615e-06 |
| 22 | ORGAN MORPHOGENESIS | 9 | 841 | 1.196e-07 | 2.487e-05 |
| 23 | REGULATION OF CELL DIFFERENTIATION | 11 | 1492 | 1.255e-07 | 2.487e-05 |
| 24 | CELL FATE COMMITMENT | 6 | 227 | 1.283e-07 | 2.487e-05 |
| 25 | NEGATIVE REGULATION OF CELL DIFFERENTIATION | 8 | 609 | 1.548e-07 | 2.881e-05 |
| 26 | REGULATION OF WNT SIGNALING PATHWAY PLANAR CELL POLARITY PATHWAY | 3 | 12 | 2.525e-07 | 4.519e-05 |
| 27 | EPITHELIUM DEVELOPMENT | 9 | 945 | 3.222e-07 | 5.552e-05 |
| 28 | CONVERGENT EXTENSION | 3 | 14 | 4.172e-07 | 6.933e-05 |
| 29 | CARTILAGE DEVELOPMENT | 5 | 147 | 4.77e-07 | 7.654e-05 |
| 30 | REGULATION OF NON CANONICAL WNT SIGNALING PATHWAY | 3 | 19 | 1.107e-06 | 0.0001716 |
| 31 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 10 | 1492 | 1.457e-06 | 0.0002105 |
| 32 | SOMITE DEVELOPMENT | 4 | 78 | 1.484e-06 | 0.0002105 |
| 33 | REGULATION OF CELL PROLIFERATION | 10 | 1496 | 1.493e-06 | 0.0002105 |
| 34 | TISSUE DEVELOPMENT | 10 | 1518 | 1.707e-06 | 0.0002336 |
| 35 | CONNECTIVE TISSUE DEVELOPMENT | 5 | 194 | 1.878e-06 | 0.0002497 |
| 36 | SKELETAL SYSTEM MORPHOGENESIS | 5 | 201 | 2.235e-06 | 0.0002889 |
| 37 | POSITIVE REGULATION OF CELL DEATH | 7 | 605 | 2.57e-06 | 0.0003232 |
| 38 | AXIS ELONGATION | 3 | 27 | 3.322e-06 | 0.0004067 |
| 39 | MORPHOGENESIS OF AN EPITHELIUM | 6 | 400 | 3.507e-06 | 0.0004184 |
| 40 | NEGATIVE REGULATION OF CELL PROLIFERATION | 7 | 643 | 3.846e-06 | 0.0004474 |
| 41 | RESPONSE TO X RAY | 3 | 30 | 4.601e-06 | 0.0005097 |
| 42 | PATTERN SPECIFICATION PROCESS | 6 | 418 | 4.517e-06 | 0.0005097 |
| 43 | REGULATION OF PROTEIN MODIFICATION PROCESS | 10 | 1710 | 5.037e-06 | 0.000545 |
| 44 | NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 8 | 983 | 5.711e-06 | 0.000604 |
| 45 | REGULATION OF STEM CELL DIFFERENTIATION | 4 | 113 | 6.533e-06 | 0.0006755 |
| 46 | REGULATION OF EMBRYONIC DEVELOPMENT | 4 | 114 | 6.766e-06 | 0.0006844 |
| 47 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 10 | 1791 | 7.638e-06 | 0.0007562 |
| 48 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 9 | 1395 | 8.239e-06 | 0.0007987 |
| 49 | SEX DIFFERENTIATION | 5 | 266 | 8.77e-06 | 0.0008328 |
| 50 | NEGATIVE REGULATION OF STRESS ACTIVATED MAPK CASCADE | 3 | 41 | 1.199e-05 | 0.001093 |
| 51 | NEGATIVE REGULATION OF STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE | 3 | 41 | 1.199e-05 | 0.001093 |
| 52 | REGULATION OF EPITHELIAL CELL PROLIFERATION | 5 | 285 | 1.225e-05 | 0.001096 |
| 53 | REGULATION OF CELL DEATH | 9 | 1472 | 1.276e-05 | 0.00112 |
| 54 | NEGATIVE REGULATION OF STEM CELL DIFFERENTIATION | 3 | 43 | 1.386e-05 | 0.001194 |
| 55 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 9 | 1518 | 1.637e-05 | 0.001364 |
| 56 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 8 | 1135 | 1.641e-05 | 0.001364 |
| 57 | RESPONSE TO IONIZING RADIATION | 4 | 145 | 1.752e-05 | 0.001429 |
| 58 | TISSUE MORPHOGENESIS | 6 | 533 | 1.806e-05 | 0.001429 |
| 59 | DIGESTIVE TRACT MORPHOGENESIS | 3 | 48 | 1.935e-05 | 0.001429 |
| 60 | EMBRYONIC MORPHOGENESIS | 6 | 539 | 1.924e-05 | 0.001429 |
| 61 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 7 | 823 | 1.931e-05 | 0.001429 |
| 62 | REGIONALIZATION | 5 | 311 | 1.866e-05 | 0.001429 |
| 63 | MALE SEX DIFFERENTIATION | 4 | 148 | 1.899e-05 | 0.001429 |
| 64 | EMBRYO DEVELOPMENT ENDING IN BIRTH OR EGG HATCHING | 6 | 554 | 2.247e-05 | 0.001561 |
| 65 | POSITIVE REGULATION OF EPITHELIAL CELL PROLIFERATION | 4 | 154 | 2.22e-05 | 0.001561 |
| 66 | TUBE DEVELOPMENT | 6 | 552 | 2.202e-05 | 0.001561 |
| 67 | TUBE MORPHOGENESIS | 5 | 323 | 2.238e-05 | 0.001561 |
| 68 | POSITIVE REGULATION OF FAT CELL DIFFERENTIATION | 3 | 51 | 2.325e-05 | 0.001578 |
| 69 | EYE DEVELOPMENT | 5 | 326 | 2.34e-05 | 0.001578 |
| 70 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 9 | 1618 | 2.735e-05 | 0.001818 |
| 71 | NEURON DIFFERENTIATION | 7 | 874 | 2.846e-05 | 0.001865 |
| 72 | EMBRYO DEVELOPMENT | 7 | 894 | 3.292e-05 | 0.002127 |
| 73 | CELLULAR RESPONSE TO ACID CHEMICAL | 4 | 175 | 3.663e-05 | 0.002335 |
| 74 | SOMITOGENESIS | 3 | 62 | 4.189e-05 | 0.002634 |
| 75 | REGULATION OF CARTILAGE DEVELOPMENT | 3 | 63 | 4.395e-05 | 0.002727 |
| 76 | POSITIVE REGULATION OF GENE EXPRESSION | 9 | 1733 | 4.728e-05 | 0.002895 |
| 77 | CELLULAR RESPONSE TO RETINOIC ACID | 3 | 65 | 4.828e-05 | 0.002917 |
| 78 | LENS DEVELOPMENT IN CAMERA TYPE EYE | 3 | 66 | 5.054e-05 | 0.003015 |
| 79 | REGULATION OF EPITHELIAL TO MESENCHYMAL TRANSITION | 3 | 67 | 5.287e-05 | 0.003114 |
| 80 | REGULATION OF STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE | 4 | 197 | 5.812e-05 | 0.00338 |
| 81 | POSITIVE REGULATION OF NON CANONICAL WNT SIGNALING PATHWAY | 2 | 11 | 6.315e-05 | 0.003628 |
| 82 | REPRODUCTIVE SYSTEM DEVELOPMENT | 5 | 408 | 6.817e-05 | 0.003822 |
| 83 | RESPONSE TO OXYGEN CONTAINING COMPOUND | 8 | 1381 | 6.745e-05 | 0.003822 |
| 84 | RESPONSE TO RADIATION | 5 | 413 | 7.222e-05 | 0.004 |
| 85 | NEUROGENESIS | 8 | 1402 | 7.509e-05 | 0.004097 |
| 86 | LENS FIBER CELL DEVELOPMENT | 2 | 12 | 7.573e-05 | 0.004097 |
| 87 | REGULATION OF CELLULAR RESPONSE TO STRESS | 6 | 691 | 7.731e-05 | 0.004135 |
| 88 | CELLULAR RESPONSE TO ORGANIC SUBSTANCE | 9 | 1848 | 7.851e-05 | 0.004151 |
| 89 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 7 | 1036 | 8.417e-05 | 0.004304 |
| 90 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 7 | 1036 | 8.417e-05 | 0.004304 |
| 91 | DEVELOPMENT OF PRIMARY SEXUAL CHARACTERISTICS | 4 | 216 | 8.308e-05 | 0.004304 |
| 92 | RESPONSE TO DRUG | 5 | 431 | 8.833e-05 | 0.004467 |
| 93 | HEMATOPOIETIC STEM CELL PROLIFERATION | 2 | 13 | 8.944e-05 | 0.004475 |
| 94 | RESPONSE TO EXTRACELLULAR STIMULUS | 5 | 441 | 9.841e-05 | 0.004871 |
| 95 | REGULATION OF ESTABLISHMENT OF PLANAR POLARITY INVOLVED IN NEURAL TUBE CLOSURE | 2 | 14 | 0.0001043 | 0.005002 |
| 96 | NEGATIVE REGULATION OF JUN KINASE ACTIVITY | 2 | 14 | 0.0001043 | 0.005002 |
| 97 | NEGATIVE REGULATION OF DEVELOPMENTAL GROWTH | 3 | 84 | 0.0001039 | 0.005002 |
| 98 | NEGATIVE REGULATION OF GENE EXPRESSION | 8 | 1493 | 0.0001171 | 0.00547 |
| 99 | NEGATIVE REGULATION OF TRANSPORT | 5 | 458 | 0.0001176 | 0.00547 |
| 100 | NEGATIVE REGULATION OF GROWTH | 4 | 236 | 0.000117 | 0.00547 |
| 101 | SEGMENTATION | 3 | 89 | 0.0001234 | 0.005684 |
| 102 | AXIS SPECIFICATION | 3 | 90 | 0.0001276 | 0.005762 |
| 103 | MESONEPHROS DEVELOPMENT | 3 | 90 | 0.0001276 | 0.005762 |
| 104 | NEGATIVE REGULATION OF NITROGEN COMPOUND METABOLIC PROCESS | 8 | 1517 | 0.000131 | 0.005826 |
| 105 | DORSAL VENTRAL PATTERN FORMATION | 3 | 91 | 0.0001318 | 0.005826 |
| 106 | REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 5 | 470 | 0.0001327 | 0.005826 |
| 107 | REGULATION OF CELLULAR COMPONENT MOVEMENT | 6 | 771 | 0.0001414 | 0.006147 |
| 108 | REGULATION OF KINASE ACTIVITY | 6 | 776 | 0.0001465 | 0.006252 |
| 109 | NEURAL TUBE FORMATION | 3 | 94 | 0.0001451 | 0.006252 |
| 110 | SENSORY ORGAN DEVELOPMENT | 5 | 493 | 0.0001659 | 0.00702 |
| 111 | CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND | 6 | 799 | 0.0001718 | 0.007202 |
| 112 | POSITIVE REGULATION OF CELL PROLIFERATION | 6 | 814 | 0.0001901 | 0.007899 |
| 113 | DEVELOPMENTAL GROWTH INVOLVED IN MORPHOGENESIS | 3 | 104 | 0.0001958 | 0.008061 |
| 114 | REGULATION OF FAT CELL DIFFERENTIATION | 3 | 106 | 0.0002071 | 0.008452 |
| 115 | RESPONSE TO RETINOIC ACID | 3 | 107 | 0.0002129 | 0.008614 |
| 116 | DORSAL VENTRAL AXIS SPECIFICATION | 2 | 20 | 0.0002168 | 0.008698 |
| 117 | REGULATION OF OSTEOBLAST DIFFERENTIATION | 3 | 112 | 0.0002436 | 0.009688 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | FRIZZLED BINDING | 6 | 36 | 1.603e-12 | 1.489e-09 |
| 2 | G PROTEIN COUPLED RECEPTOR BINDING | 6 | 259 | 2.791e-07 | 0.0001296 |
| 3 | WNT ACTIVATED RECEPTOR ACTIVITY | 3 | 22 | 1.755e-06 | 0.0005435 |
| 4 | WNT PROTEIN BINDING | 3 | 31 | 5.09e-06 | 0.001182 |
| 5 | RECEPTOR BINDING | 9 | 1476 | 1.304e-05 | 0.002423 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | EXTRACELLULAR MATRIX | 7 | 426 | 2.45e-07 | 0.0001431 |
| 2 | PROTEINACEOUS EXTRACELLULAR MATRIX | 6 | 356 | 1.788e-06 | 0.0005222 |
| 3 | EXTRACELLULAR SPACE | 9 | 1376 | 7.366e-06 | 0.001434 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
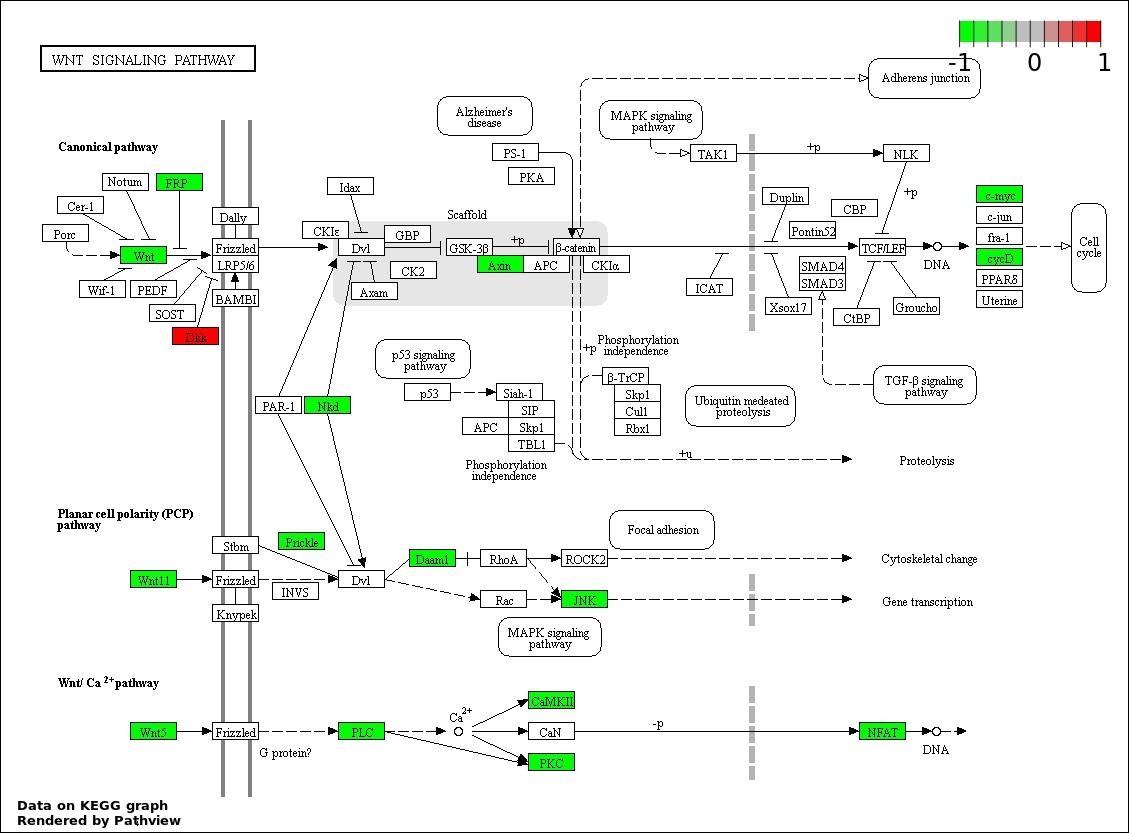
| 1 | hsa04310_Wnt_signaling_pathway | 22 | 151 | 4.183e-48 | 7.404e-46 | |
| 2 | hsa04916_Melanogenesis | 8 | 101 | 9.618e-14 | 8.512e-12 | |
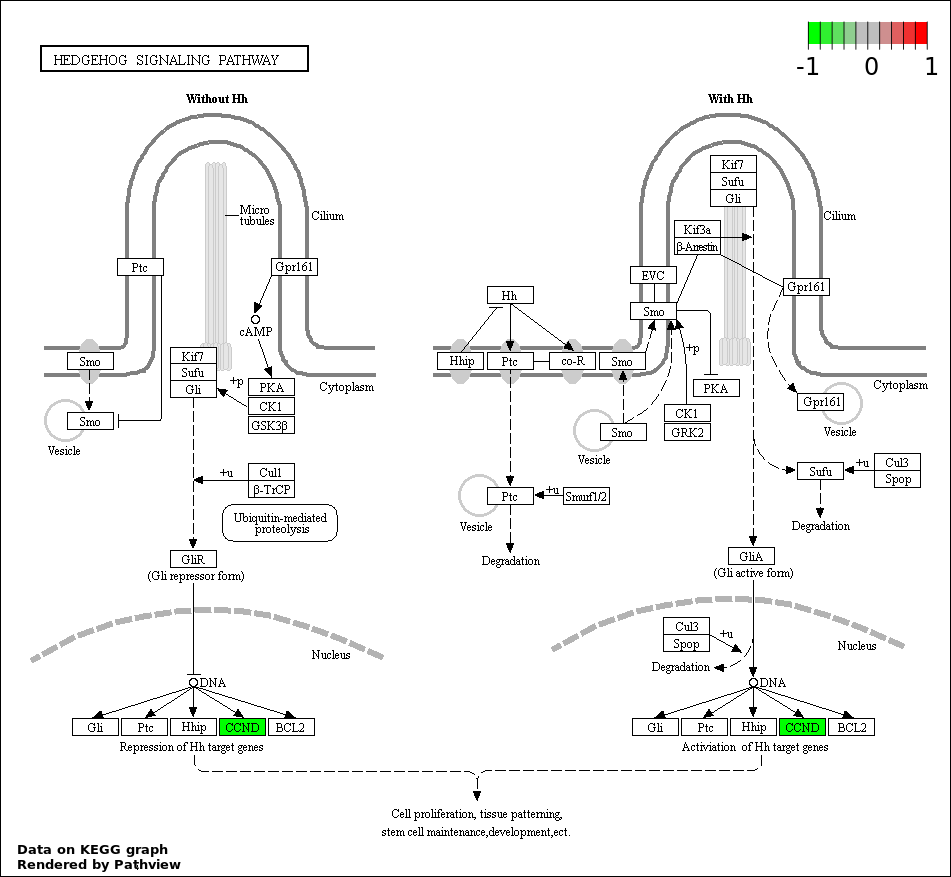
| 3 | hsa04340_Hedgehog_signaling_pathway | 5 | 56 | 3.64e-09 | 2.148e-07 | |
| 4 | hsa04012_ErbB_signaling_pathway | 4 | 87 | 2.301e-06 | 0.0001018 | |
| 5 | hsa04912_GnRH_signaling_pathway | 4 | 101 | 4.179e-06 | 0.0001479 | |
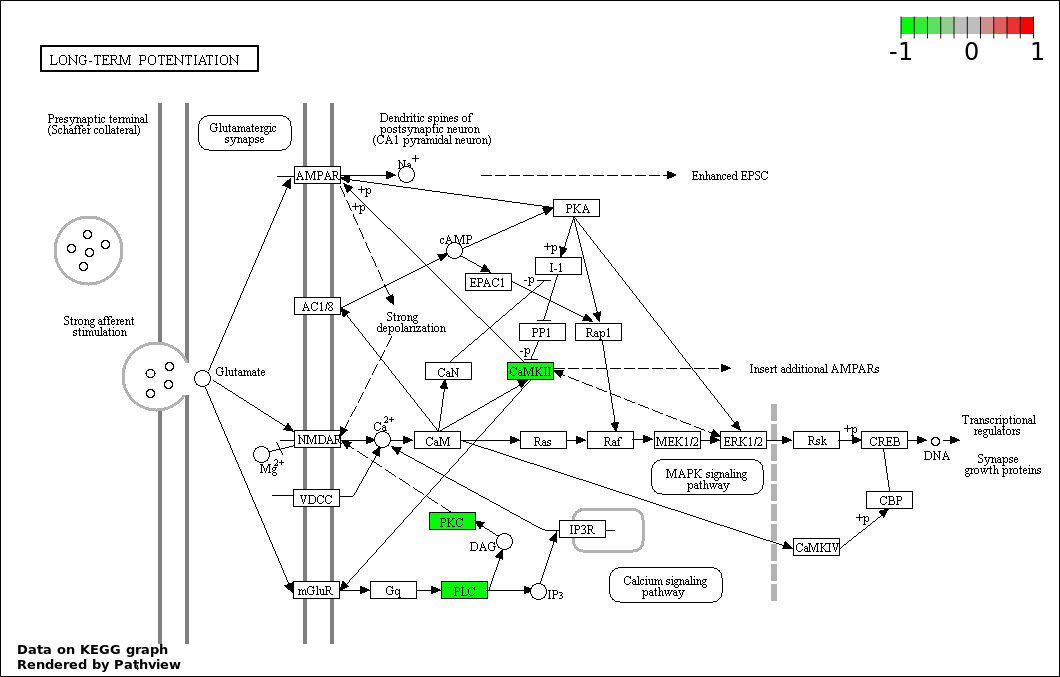
| 6 | hsa04720_Long.term_potentiation | 3 | 70 | 6.029e-05 | 0.001559 | |
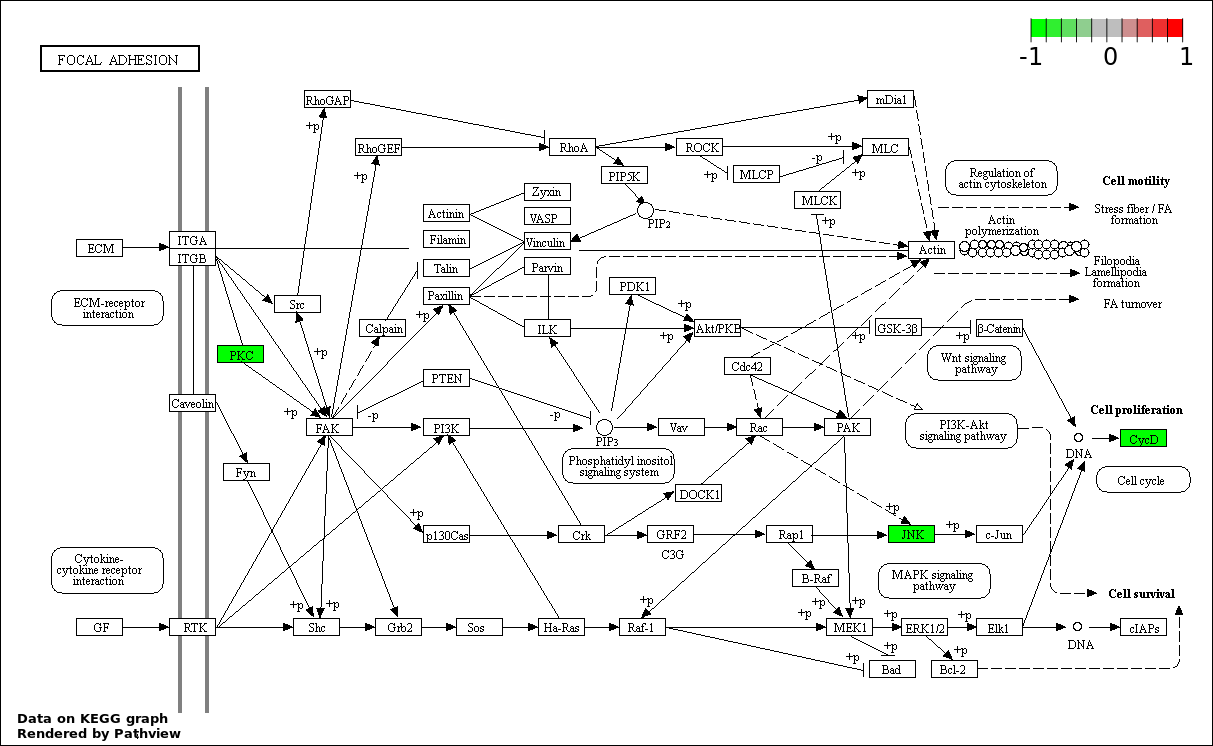
| 7 | hsa04510_Focal_adhesion | 4 | 200 | 6.164e-05 | 0.001559 | |
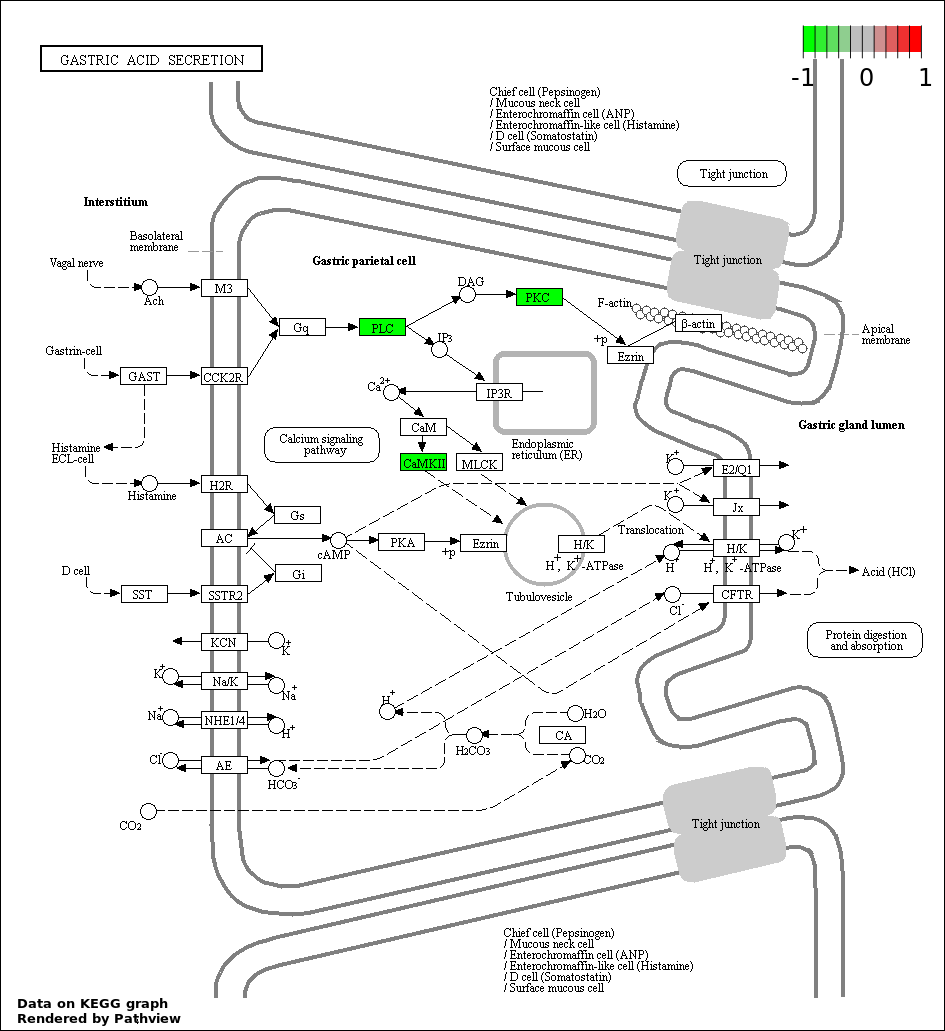
| 8 | hsa04971_Gastric_acid_secretion | 3 | 74 | 7.119e-05 | 0.001575 | |
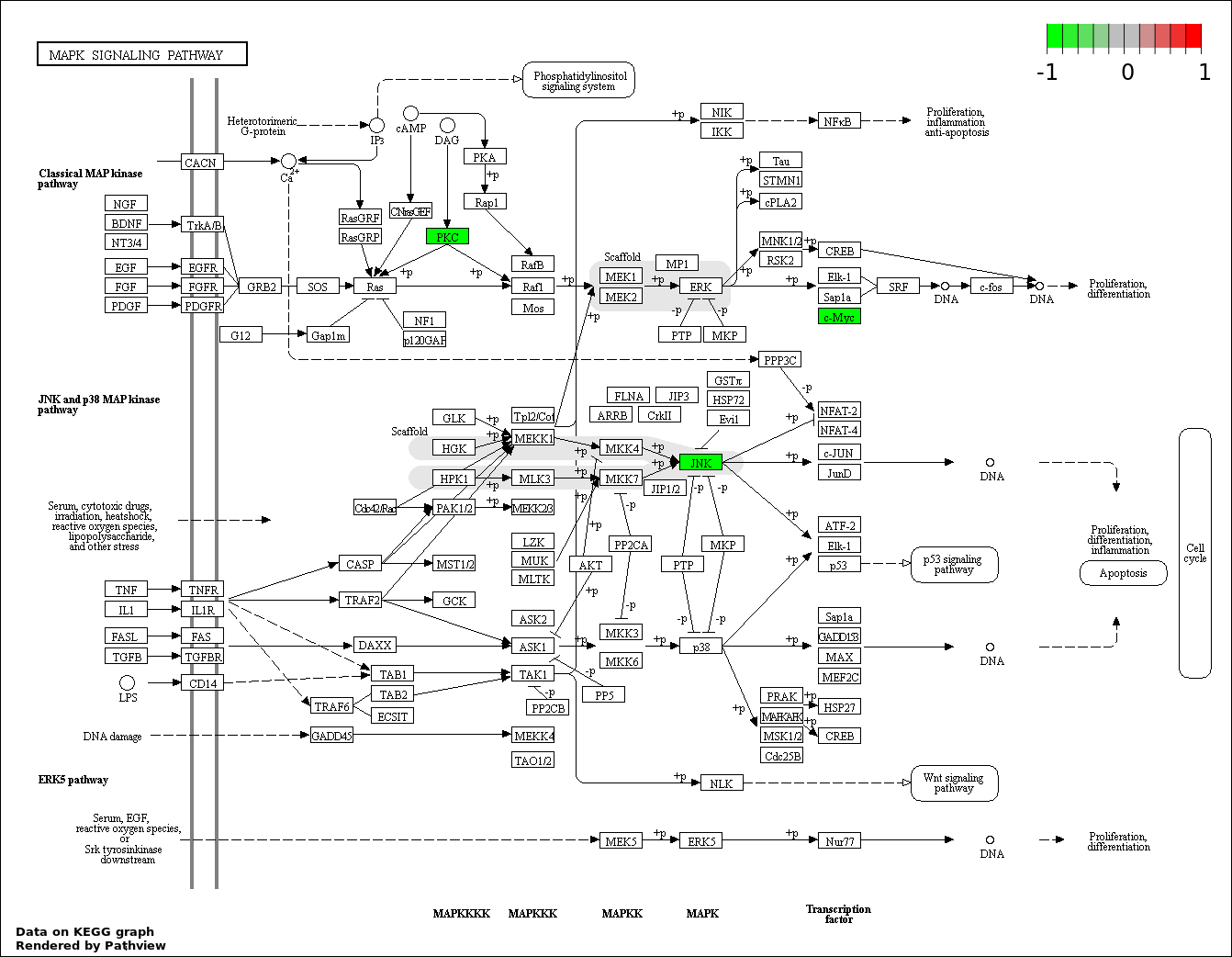
| 9 | hsa04010_MAPK_signaling_pathway | 4 | 268 | 0.0001907 | 0.00375 | |
| 10 | hsa04110_Cell_cycle | 3 | 128 | 0.0003607 | 0.006385 | |
| 11 | hsa04630_Jak.STAT_signaling_pathway | 3 | 155 | 0.000631 | 0.01015 | |
| 12 | hsa04020_Calcium_signaling_pathway | 3 | 177 | 0.0009272 | 0.01368 | |
| 13 | hsa04115_p53_signaling_pathway | 2 | 69 | 0.002592 | 0.03325 | |
| 14 | hsa04730_Long.term_depression | 2 | 70 | 0.002666 | 0.03325 | |
| 15 | hsa04662_B_cell_receptor_signaling_pathway | 2 | 75 | 0.003053 | 0.03325 | |
| 16 | hsa04370_VEGF_signaling_pathway | 2 | 76 | 0.003134 | 0.03325 | |
| 17 | hsa04070_Phosphatidylinositol_signaling_system | 2 | 78 | 0.003297 | 0.03325 | |
| 18 | hsa04664_Fc_epsilon_RI_signaling_pathway | 2 | 79 | 0.003381 | 0.03325 | |
| 19 | hsa04970_Salivary_secretion | 2 | 89 | 0.004269 | 0.03861 | |
| 20 | hsa04540_Gap_junction | 2 | 90 | 0.004363 | 0.03861 | |
| 21 | hsa04972_Pancreatic_secretion | 2 | 101 | 0.005461 | 0.04603 | |
| 22 | hsa04270_Vascular_smooth_muscle_contraction | 2 | 116 | 0.007142 | 0.05746 | |
| 23 | hsa04722_Neurotrophin_signaling_pathway | 2 | 127 | 0.008504 | 0.06367 | |
| 24 | hsa04380_Osteoclast_differentiation | 2 | 128 | 0.008634 | 0.06367 | |
| 25 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 2 | 136 | 0.0097 | 0.06867 | |
| 26 | hsa04062_Chemokine_signaling_pathway | 2 | 189 | 0.01812 | 0.1234 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | RP11-166D19.1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-331-3p;hsa-miR-3913-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 11 | CAMK2A | Sponge network | -3.855 | 0 | -4.139 | 0 | 0.733 |
| 2 | MIR143HG |
hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 14 | CAMK2A | Sponge network | -4.237 | 0 | -4.139 | 0 | 0.721 |
| 3 | LINC00702 |
hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-331-3p;hsa-miR-3913-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 16 | CAMK2A | Sponge network | -2.704 | 0 | -4.139 | 0 | 0.72 |
| 4 | HAND2-AS1 |
hsa-miR-1226-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-331-3p;hsa-miR-34a-3p;hsa-miR-361-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 18 | CAMK2A | Sponge network | -5.605 | 0 | -4.139 | 0 | 0.647 |
| 5 | MIR143HG |
hsa-let-7a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PRICKLE2 | Sponge network | -4.237 | 0 | -2.505 | 0 | 0.639 |
| 6 | RP11-166D19.1 |
hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-423-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | CCND2 | Sponge network | -3.855 | 0 | -2.427 | 0 | 0.629 |
| 7 | ADAMTS9-AS1 |
hsa-miR-1301-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 12 | SFRP1 | Sponge network | -7.614 | 0 | -4.722 | 0 | 0.619 |
| 8 | RP11-166D19.1 |
hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-361-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PRICKLE2 | Sponge network | -3.855 | 0 | -2.505 | 0 | 0.598 |
| 9 | LINC00702 |
hsa-miR-1301-3p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-328-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-7-1-3p | 10 | SFRP1 | Sponge network | -2.704 | 0 | -4.722 | 0 | 0.59 |
| 10 | ADAMTS9-AS1 |
hsa-miR-1226-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-3913-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 14 | CAMK2A | Sponge network | -7.614 | 0 | -4.139 | 0 | 0.577 |
| 11 | HAND2-AS1 |
hsa-miR-1301-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-328-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940 | 12 | SFRP1 | Sponge network | -5.605 | 0 | -4.722 | 0 | 0.571 |
| 12 | LINC00163 |
hsa-miR-10a-5p;hsa-miR-10b-5p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-331-3p;hsa-miR-3913-5p;hsa-miR-582-3p | 12 | CAMK2A | Sponge network | -4.312 | 0.00014 | -4.139 | 0 | 0.568 |
| 13 | HAND2-AS1 |
hsa-let-7a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PRICKLE2 | Sponge network | -5.605 | 0 | -2.505 | 0 | 0.555 |
| 14 | RP5-1172A22.1 | hsa-miR-10a-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-3913-5p;hsa-miR-582-3p | 10 | CAMK2A | Sponge network | -3.373 | 0.00572 | -4.139 | 0 | 0.552 |
| 15 | LINC00702 |
hsa-let-7a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-361-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PRICKLE2 | Sponge network | -2.704 | 0 | -2.505 | 0 | 0.549 |
| 16 | ACTA2-AS1 |
hsa-miR-1301-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-7-1-3p | 10 | SFRP1 | Sponge network | -3.838 | 0 | -4.722 | 0 | 0.547 |
| 17 | C20orf166-AS1 |
hsa-miR-1226-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-3913-5p | 11 | CAMK2A | Sponge network | -6.333 | 0 | -4.139 | 0 | 0.545 |
| 18 | MIR143HG |
hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-423-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | CCND2 | Sponge network | -4.237 | 0 | -2.427 | 0 | 0.544 |
| 19 | PGM5-AS1 |
hsa-miR-1301-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-940 | 10 | SFRP1 | Sponge network | -11.072 | 0 | -4.722 | 0 | 0.527 |
| 20 | C20orf166-AS1 |
hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-324-3p;hsa-miR-93-5p | 16 | CCND2 | Sponge network | -6.333 | 0 | -2.427 | 0 | 0.513 |
| 21 | PDZRN3-AS1 |
hsa-miR-10b-5p;hsa-miR-1226-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-7-1-3p | 10 | CAMK2A | Sponge network | -5.049 | 1.0E-5 | -4.139 | 0 | 0.512 |
| 22 | FENDRR |
hsa-let-7a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-361-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PRICKLE2 | Sponge network | -4.793 | 0 | -2.505 | 0 | 0.509 |
| 23 | LINC00702 |
hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | CCND2 | Sponge network | -2.704 | 0 | -2.427 | 0 | 0.509 |
| 24 | HAND2-AS1 |
hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-423-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | CCND2 | Sponge network | -5.605 | 0 | -2.427 | 0 | 0.497 |
| 25 | RP11-720L2.4 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-93-5p | 11 | CCND2 | Sponge network | -3.686 | 2.0E-5 | -2.427 | 0 | 0.476 |
| 26 | RP11-389G6.3 |
hsa-miR-10a-5p;hsa-miR-1226-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26b-5p | 10 | CAMK2A | Sponge network | -7.573 | 0 | -4.139 | 0 | 0.473 |
| 27 | PDZRN3-AS1 |
hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-361-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PRICKLE2 | Sponge network | -5.049 | 1.0E-5 | -2.505 | 0 | 0.469 |
| 28 | ADAMTS9-AS1 |
hsa-let-7a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PRICKLE2 | Sponge network | -7.614 | 0 | -2.505 | 0 | 0.467 |
| 29 | RP11-243M5.1 | hsa-let-7a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-331-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | CCND2 | Sponge network | -8.824 | 0 | -2.427 | 0 | 0.464 |
| 30 | ADAMTS9-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-16-2-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p | 10 | PLCB4 | Sponge network | -7.614 | 0 | -3.769 | 0 | 0.46 |
| 31 | GAS6-AS2 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-324-3p | 11 | CCND2 | Sponge network | -2.655 | 0 | -2.427 | 0 | 0.458 |
| 32 | AC104654.2 |
hsa-miR-1226-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-34a-3p;hsa-miR-7-1-3p | 11 | CAMK2A | Sponge network | -3.012 | 0.00731 | -4.139 | 0 | 0.454 |
| 33 | BVES-AS1 |
hsa-miR-10a-5p;hsa-miR-10b-5p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-331-3p | 10 | CAMK2A | Sponge network | -4.161 | 1.0E-5 | -4.139 | 0 | 0.45 |
| 34 | AC093627.8 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-324-3p | 13 | CCND2 | Sponge network | -5.744 | 0 | -2.427 | 0 | 0.445 |
| 35 | CTC-296K1.3 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-5p | 10 | CCND2 | Sponge network | -5.677 | 0 | -2.427 | 0 | 0.443 |
| 36 | ADAMTS9-AS1 |
hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | CCND2 | Sponge network | -7.614 | 0 | -2.427 | 0 | 0.443 |
| 37 | AC104654.2 |
hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-7-1-3p | 13 | CCND2 | Sponge network | -3.012 | 0.00731 | -2.427 | 0 | 0.443 |
| 38 | RP11-867G23.10 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-423-5p;hsa-miR-93-5p | 14 | CCND2 | Sponge network | -5.684 | 0 | -2.427 | 0 | 0.437 |
| 39 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-361-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PRICKLE2 | Sponge network | -3.838 | 0 | -2.505 | 0 | 0.436 |
| 40 | ZFHX4-AS1 |
hsa-miR-125a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-3p;hsa-miR-34a-3p;hsa-miR-7-1-3p | 14 | CAMK2A | Sponge network | -2.966 | 0.00743 | -4.139 | 0 | 0.426 |
| 41 | RP11-384P7.7 |
hsa-miR-125a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-331-3p;hsa-miR-34a-3p;hsa-miR-361-3p;hsa-miR-7-1-3p | 10 | CAMK2A | Sponge network | -3.649 | 3.0E-5 | -4.139 | 0 | 0.425 |
| 42 | C4A-AS1 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-423-5p | 11 | CCND2 | Sponge network | -1.76 | 0.00265 | -2.427 | 0 | 0.416 |
| 43 | RP11-753H16.3 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-93-5p | 12 | CCND2 | Sponge network | -5.702 | 0 | -2.427 | 0 | 0.402 |
| 44 | PGM5-AS1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-5p;hsa-miR-324-3p;hsa-miR-423-5p;hsa-miR-7-1-3p | 11 | CCND2 | Sponge network | -11.072 | 0 | -2.427 | 0 | 0.4 |
| 45 | LINC00694 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-30d-3p | 13 | CCND2 | Sponge network | -4.485 | 1.0E-5 | -2.427 | 0 | 0.395 |
| 46 | RP11-20J15.3 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-20a-5p | 14 | CCND2 | Sponge network | -5.104 | 8.0E-5 | -2.427 | 0 | 0.394 |
| 47 | RP11-672A2.4 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-5p;hsa-miR-30d-3p | 12 | CCND2 | Sponge network | -2.885 | 0.00052 | -2.427 | 0 | 0.39 |
| 48 | AP000473.5 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-20a-5p | 10 | CCND2 | Sponge network | -5.021 | 3.0E-5 | -2.427 | 0 | 0.389 |
| 49 | TBX5-AS1 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-3p | 10 | CCND2 | Sponge network | -2.557 | 2.0E-5 | -2.427 | 0 | 0.387 |
| 50 | RP11-887P2.5 |
hsa-miR-1301-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940 | 10 | SFRP1 | Sponge network | -6.751 | 0 | -4.722 | 0 | 0.382 |
| 51 | RP11-554A11.4 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-20a-5p;hsa-miR-324-3p;hsa-miR-423-5p | 15 | CCND2 | Sponge network | -3.989 | 0 | -2.427 | 0 | 0.379 |
| 52 | RP11-401O9.4 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-590-5p | 11 | CCND2 | Sponge network | -0.359 | 0.78702 | -2.427 | 0 | 0.375 |
| 53 | RP11-693J15.4 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | CCND2 | Sponge network | -3.319 | 0.00281 | -2.427 | 0 | 0.375 |
| 54 | RP11-180N14.1 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-7-1-3p | 16 | CCND2 | Sponge network | -4.46 | 0 | -2.427 | 0 | 0.371 |
| 55 | AF131217.1 | hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-331-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | CCND2 | Sponge network | -5.31 | 0 | -2.427 | 0 | 0.363 |
| 56 | PDZRN3-AS1 |
hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | CCND2 | Sponge network | -5.049 | 1.0E-5 | -2.427 | 0 | 0.353 |
| 57 | RP11-887P2.5 |
hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | CCND2 | Sponge network | -6.751 | 0 | -2.427 | 0 | 0.352 |
| 58 | FENDRR |
hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-423-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | CCND2 | Sponge network | -4.793 | 0 | -2.427 | 0 | 0.35 |
| 59 | ACTA2-AS1 |
hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-324-3p;hsa-miR-7-1-3p | 17 | CCND2 | Sponge network | -3.838 | 0 | -2.427 | 0 | 0.348 |
| 60 | RP4-800J21.3 | hsa-miR-1226-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-331-3p;hsa-miR-3913-5p | 10 | CAMK2A | Sponge network | -2.473 | 0.00577 | -4.139 | 0 | 0.345 |
| 61 | MIR497HG | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-324-3p;hsa-miR-423-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | CCND2 | Sponge network | -3.802 | 0 | -2.427 | 0 | 0.341 |
| 62 | CTC-296K1.4 | hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-30d-3p;hsa-miR-590-5p | 12 | CCND2 | Sponge network | -6.559 | 0 | -2.427 | 0 | 0.339 |
| 63 | LINC00473 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-423-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | CCND2 | Sponge network | -5.53 | 0 | -2.427 | 0 | 0.337 |
| 64 | LINC00163 |
hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-20a-5p;hsa-miR-324-3p | 14 | CCND2 | Sponge network | -4.312 | 0.00014 | -2.427 | 0 | 0.333 |
| 65 | RP11-62F24.2 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 11 | CCND2 | Sponge network | -4.034 | 0.00034 | -2.427 | 0 | 0.323 |
| 66 | RP1-65J11.1 |
hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30e-5p;hsa-miR-331-3p;hsa-miR-34a-3p;hsa-miR-7-1-3p | 11 | CAMK2A | Sponge network | -5.989 | 0 | -4.139 | 0 | 0.321 |
| 67 | RP11-88I18.3 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-5p | 13 | CCND2 | Sponge network | -7.88 | 0 | -2.427 | 0 | 0.319 |
| 68 | RP11-805I24.3 |
hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-324-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | CCND2 | Sponge network | -5.815 | 0 | -2.427 | 0 | 0.312 |
| 69 | RP11-6O2.3 |
hsa-miR-10b-5p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-361-3p | 10 | CAMK2A | Sponge network | -4.533 | 0 | -4.139 | 0 | 0.308 |
| 70 | RP11-805I24.3 |
hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30e-5p;hsa-miR-361-3p;hsa-miR-7-1-3p | 10 | CAMK2A | Sponge network | -5.815 | 0 | -4.139 | 0 | 0.306 |
| 71 | LINC00327 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-30d-3p | 12 | CCND2 | Sponge network | -1.951 | 0.01135 | -2.427 | 0 | 0.306 |
| 72 | BVES-AS1 |
hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-20a-5p | 11 | CCND2 | Sponge network | -4.161 | 1.0E-5 | -2.427 | 0 | 0.303 |
| 73 | RP1-65J11.1 |
hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-324-3p;hsa-miR-7-1-3p | 12 | CCND2 | Sponge network | -5.989 | 0 | -2.427 | 0 | 0.302 |
| 74 | RP11-403B2.7 | hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-423-5p | 12 | CCND2 | Sponge network | -2.507 | 0.00687 | -2.427 | 0 | 0.299 |
| 75 | RP11-514D23.3 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-93-5p | 11 | CCND2 | Sponge network | -3.847 | 0 | -2.427 | 0 | 0.284 |
| 76 | GATA6-AS1 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-324-3p;hsa-miR-93-5p | 16 | CCND2 | Sponge network | -3.855 | 0 | -2.427 | 0 | 0.282 |
| 77 | DIO3OS | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-423-5p | 15 | CCND2 | Sponge network | -3.619 | 0 | -2.427 | 0 | 0.278 |
| 78 | RP11-389G6.3 |
hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-423-5p;hsa-miR-93-5p | 18 | CCND2 | Sponge network | -7.573 | 0 | -2.427 | 0 | 0.272 |
| 79 | RP11-120J1.1 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p | 10 | CCND2 | Sponge network | -6.067 | 0 | -2.427 | 0 | 0.272 |
| 80 | LINC00982 | hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-423-5p;hsa-miR-7-1-3p | 11 | CCND2 | Sponge network | -3.353 | 3.0E-5 | -2.427 | 0 | 0.254 |