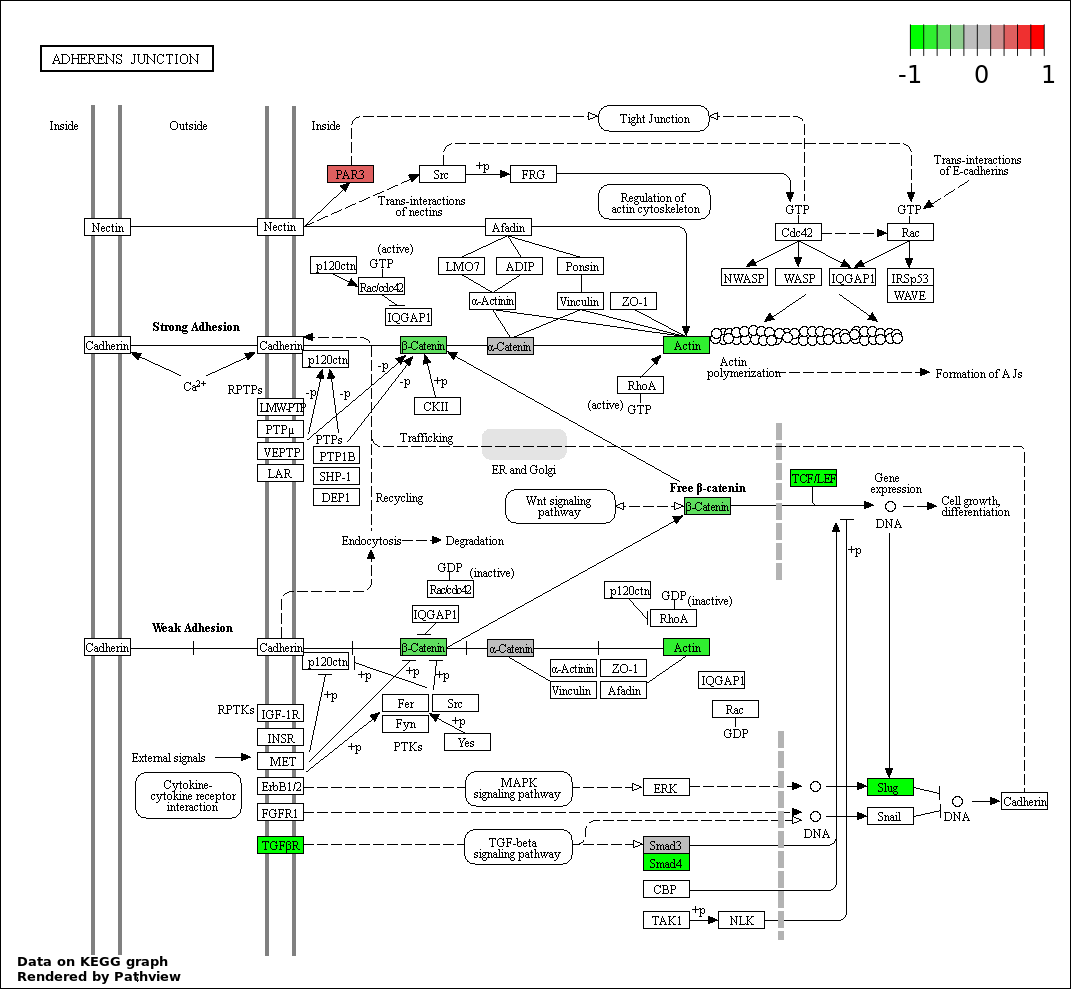
This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-378a-3p | ACTB | 1.47 | 0.04667 | -0.79 | 0.01089 | miRNAWalker2 validate | -0.11 | 0 | NA | |
| 2 | hsa-let-7a-3p | AMOT | 0.83 | 0.04681 | -1.94 | 0.20701 | mirMAP | -0.59 | 0.00568 | NA | |
| 3 | hsa-let-7f-1-3p | AMOT | 1.62 | 0.00069 | -1.94 | 0.20701 | mirMAP | -1.03 | 0 | NA | |
| 4 | hsa-miR-203a-3p | AMOT | 6.35 | 0 | -1.94 | 0.20701 | MirTarget | -0.4 | 0 | NA | |
| 5 | hsa-miR-205-5p | AMOT | 8.08 | 0 | -1.94 | 0.20701 | MirTarget; miRNATAP | -0.39 | 0 | 26239614 | MicroRNA 205 inhibits the proliferation and invasion of breast cancer by regulating AMOT expression; In the present study miR-205 was significantly downregulated in breast cancer samples and it was identified to directly target the 3'-untranslated region 3'-UTR of AMOT in breast cancer MCF-7 cells by luciferase assay; miR-205 and small interfering RNA siRNA-mediated AMOT-knockdown experiments revealed that miR-205 significantly inhibited the proliferation and the invasion of MCF-7 cells through a decrease in the expression of AMOT yet had no effect on apoptosis; Furthermore we observed that the overexpression of AMOT partially reversed the inhibitory effect of miR-205 on the growth and the invasion of MCF-7 cells; The data indicated that miR-205 regulated the proliferation and the invasion of breast cancer cells through suppression of AMOT expression at least partly; Therefore the disordered decreased expression of miR-205 and the resulting AMOT upregulation contributes to breast carcinogenesis and miR-205-AMOT represents a new potential therapeutic target for the treatment of breast carcinoma |
| 6 | hsa-miR-21-3p | AMOT | 3.5 | 0 | -1.94 | 0.20701 | mirMAP | -0.74 | 0 | NA | |
| 7 | hsa-miR-212-3p | AMOT | 0.74 | 0.2589 | -1.94 | 0.20701 | MirTarget | -0.43 | 0.00122 | NA | |
| 8 | hsa-miR-22-3p | AMOT | 0.85 | 0.01489 | -1.94 | 0.20701 | MirTarget; miRNATAP | -1.19 | 0 | NA | |
| 9 | hsa-miR-221-3p | AMOT | 0.94 | 0.17475 | -1.94 | 0.20701 | miRNAWalker2 validate | -0.85 | 0 | NA | |
| 10 | hsa-miR-2355-3p | AMOT | 0.49 | 0.48372 | -1.94 | 0.20701 | MirTarget; miRNATAP | -0.72 | 0 | NA | |
| 11 | hsa-miR-29a-3p | AMOT | -0.64 | 0.25192 | -1.94 | 0.20701 | MirTarget; miRNATAP | -0.56 | 0.00034 | NA | |
| 12 | hsa-miR-29b-3p | AMOT | 0.67 | 0.23406 | -1.94 | 0.20701 | MirTarget; miRNATAP | -0.59 | 0.00013 | NA | |
| 13 | hsa-miR-330-3p | AMOT | 2.49 | 0.00013 | -1.94 | 0.20701 | MirTarget; PITA; miRNATAP | -0.63 | 0 | NA | |
| 14 | hsa-miR-330-5p | AMOT | 2.25 | 0.00028 | -1.94 | 0.20701 | miRanda | -0.64 | 0 | NA | |
| 15 | hsa-miR-342-3p | AMOT | 1.31 | 0.02072 | -1.94 | 0.20701 | miRanda | -0.68 | 1.0E-5 | NA | |
| 16 | hsa-miR-34c-3p | AMOT | 4.04 | 0.0002 | -1.94 | 0.20701 | MirTarget; mirMAP | -0.25 | 0.00175 | NA | |
| 17 | hsa-miR-34c-5p | AMOT | 2.65 | 0.01574 | -1.94 | 0.20701 | miRanda | -0.23 | 0.00315 | NA | |
| 18 | hsa-miR-944 | AMOT | 7.21 | 0.00082 | -1.94 | 0.20701 | mirMAP; miRNATAP | -0.38 | 0 | NA | |
| 19 | hsa-miR-186-5p | APC | 0.45 | 0.18545 | -0.08 | 0.84234 | miRNAWalker2 validate | -0.19 | 0.00487 | NA | |
| 20 | hsa-miR-30a-3p | APC | -1.22 | 0.16757 | -0.08 | 0.84234 | MirTarget; miRNATAP | -0.11 | 1.0E-5 | NA | |
| 21 | hsa-miR-320b | APC | 0.2 | 0.72722 | -0.08 | 0.84234 | miRanda | -0.12 | 0.00135 | NA | |
| 22 | hsa-miR-374b-5p | APC | -0.11 | 0.76489 | -0.08 | 0.84234 | mirMAP | -0.19 | 0.00128 | NA | |
| 23 | hsa-miR-590-3p | APC | 2.35 | 0 | -0.08 | 0.84234 | PITA; miRanda; mirMAP; miRNATAP | -0.17 | 0.0001 | NA | |
| 24 | hsa-miR-7-1-3p | APC | 1.43 | 0.00471 | -0.08 | 0.84234 | MirTarget | -0.15 | 0.00061 | NA | |
| 25 | hsa-miR-125a-5p | APC2 | -1.32 | 0.00714 | 3.13 | 0.00737 | mirMAP | -0.85 | 0 | NA | |
| 26 | hsa-miR-194-3p | APC2 | 1.92 | 0.10538 | 3.13 | 0.00737 | mirMAP | -0.16 | 0.00507 | NA | |
| 27 | hsa-miR-28-5p | APC2 | -0.82 | 0.02212 | 3.13 | 0.00737 | mirMAP | -0.67 | 0.00043 | NA | |
| 28 | hsa-let-7a-3p | AXIN2 | 0.83 | 0.04681 | -3.82 | 0.00033 | miRNATAP | -0.49 | 0.00085 | NA | |
| 29 | hsa-let-7b-3p | AXIN2 | 0.59 | 0.20051 | -3.82 | 0.00033 | miRNATAP | -0.5 | 0.00019 | NA | |
| 30 | hsa-let-7f-2-3p | AXIN2 | 1.03 | 0.07873 | -3.82 | 0.00033 | MirTarget | -0.36 | 0.00084 | NA | |
| 31 | hsa-miR-15a-5p | AXIN2 | 2.05 | 0 | -3.82 | 0.00033 | MirTarget; miRNATAP | -0.55 | 8.0E-5 | 26252081 | The combination of miR-15a and AXIN2 significantly improved the diagnostic accuracy |
| 32 | hsa-miR-15b-5p | AXIN2 | 3.32 | 0 | -3.82 | 0.00033 | miRTarBase; MirTarget; miRNATAP | -0.47 | 0.00022 | NA | |
| 33 | hsa-miR-16-2-3p | AXIN2 | 3.8 | 0 | -3.82 | 0.00033 | mirMAP | -0.68 | 0 | NA | |
| 34 | hsa-miR-16-5p | AXIN2 | 2.94 | 0 | -3.82 | 0.00033 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.7 | 0 | NA | |
| 35 | hsa-miR-205-5p | AXIN2 | 8.08 | 0 | -3.82 | 0.00033 | miRNATAP | -0.3 | 0 | NA | |
| 36 | hsa-miR-221-3p | AXIN2 | 0.94 | 0.17475 | -3.82 | 0.00033 | miRNATAP | -0.52 | 0 | NA | |
| 37 | hsa-miR-222-3p | AXIN2 | 1.55 | 0.0223 | -3.82 | 0.00033 | miRNATAP | -0.53 | 0 | NA | |
| 38 | hsa-miR-3614-5p | AXIN2 | 4.5 | 0 | -3.82 | 0.00033 | MirTarget; miRNATAP | -0.26 | 0.00012 | NA | |
| 39 | hsa-miR-590-3p | AXIN2 | 2.35 | 0 | -3.82 | 0.00033 | MirTarget; PITA; miRanda; miRNATAP | -0.5 | 3.0E-5 | NA | |
| 40 | hsa-miR-944 | AXIN2 | 7.21 | 0.00082 | -3.82 | 0.00033 | MirTarget; PITA; miRNATAP | -0.28 | 0 | NA | |
| 41 | hsa-miR-139-5p | BBC3 | -2.09 | 0.00038 | 0.48 | 0.3579 | miRNATAP | -0.16 | 0.00143 | NA | |
| 42 | hsa-miR-101-3p | BIRC5 | -1.12 | 0.02009 | 6.15 | 0 | miRNAWalker2 validate | -0.2 | 0.0054 | NA | |
| 43 | hsa-miR-106a-5p | BMP2 | 3.99 | 0 | 0.6 | 0.59614 | miRNATAP | -0.28 | 0.0002 | NA | |
| 44 | hsa-miR-362-3p | BMP2 | 0.68 | 0.22615 | 0.6 | 0.59614 | miRanda | -0.39 | 0.0006 | NA | |
| 45 | hsa-miR-378c | BMP2 | 0.45 | 0.49938 | 0.6 | 0.59614 | miRNATAP | -0.28 | 0.0034 | NA | |
| 46 | hsa-miR-429 | BMP2 | 6.4 | 0 | 0.6 | 0.59614 | miRNATAP | -0.32 | 0.00011 | NA | |
| 47 | hsa-miR-142-3p | BMP4 | 4.35 | 0 | -1.95 | 0.0822 | PITA; miRanda | -0.38 | 2.0E-5 | NA | |
| 48 | hsa-miR-590-3p | BMP4 | 2.35 | 0 | -1.95 | 0.0822 | miRanda | -0.37 | 0.00319 | NA | |
| 49 | hsa-miR-125a-3p | BMP7 | -0.07 | 0.92074 | 2.47 | 0.14183 | miRanda | -0.6 | 0 | NA | |
| 50 | hsa-miR-30a-5p | BMP7 | -0.77 | 0.32049 | 2.47 | 0.14183 | mirMAP; miRNATAP | -0.84 | 0 | NA | |
| 51 | hsa-miR-338-3p | BMP7 | 0.45 | 0.55849 | 2.47 | 0.14183 | miRanda | -0.68 | 0 | NA | |
| 52 | hsa-miR-142-3p | BMP8A | 4.35 | 0 | -0.91 | 0.14941 | miRNAWalker2 validate | -0.15 | 0.00247 | NA | |
| 53 | hsa-miR-30e-3p | BMP8A | -0.04 | 0.93258 | -0.91 | 0.14941 | MirTarget | -0.27 | 0.00121 | NA | |
| 54 | hsa-miR-326 | BMP8A | 1.77 | 0.03673 | -0.91 | 0.14941 | miRanda | -0.12 | 0.00459 | NA | |
| 55 | hsa-miR-125a-5p | BMP8B | -1.32 | 0.00714 | 2.55 | 0.00041 | miRanda | -0.22 | 0.00973 | NA | |
| 56 | hsa-miR-26b-5p | BMP8B | 0.31 | 0.46163 | 2.55 | 0.00041 | miRNAWalker2 validate | -0.34 | 0.00051 | NA | |
| 57 | hsa-miR-455-5p | BMP8B | -0.32 | 0.6163 | 2.55 | 0.00041 | miRanda | -0.19 | 0.00365 | NA | |
| 58 | hsa-miR-142-5p | BMPR1B | 3.96 | 0 | -1.52 | 0.34339 | mirMAP | -0.73 | 0 | NA | |
| 59 | hsa-miR-146b-5p | BMPR1B | 1.88 | 0.00074 | -1.52 | 0.34339 | miRanda | -0.67 | 4.0E-5 | NA | |
| 60 | hsa-miR-150-5p | BMPR1B | 1.77 | 0.05938 | -1.52 | 0.34339 | MirTarget | -0.47 | 0 | NA | |
| 61 | hsa-miR-205-5p | BMPR1B | 8.08 | 0 | -1.52 | 0.34339 | miRNATAP | -0.31 | 0 | NA | |
| 62 | hsa-miR-23b-3p | BMPR1B | -0.58 | 0.19048 | -1.52 | 0.34339 | mirMAP | -0.74 | 0.00035 | NA | |
| 63 | hsa-miR-24-3p | BMPR1B | 1.56 | 0.00052 | -1.52 | 0.34339 | miRNATAP | -0.55 | 0.00635 | NA | |
| 64 | hsa-miR-374a-5p | BMPR1B | 0.28 | 0.45888 | -1.52 | 0.34339 | MirTarget | -0.88 | 0.00036 | NA | |
| 65 | hsa-miR-590-3p | BMPR1B | 2.35 | 0 | -1.52 | 0.34339 | miRanda; mirMAP | -0.55 | 0.00205 | NA | |
| 66 | hsa-miR-629-3p | BMPR1B | 3.48 | 0 | -1.52 | 0.34339 | MirTarget | -0.42 | 0.00285 | NA | |
| 67 | hsa-let-7f-1-3p | BMPR2 | 1.62 | 0.00069 | -1.13 | 0.00109 | MirTarget | -0.12 | 0.00286 | NA | |
| 68 | hsa-miR-106b-5p | BMPR2 | 2.81 | 0 | -1.13 | 0.00109 | MirTarget; miRNATAP | -0.12 | 0.00443 | NA | |
| 69 | hsa-miR-146b-3p | BMPR2 | 1.35 | 0.00936 | -1.13 | 0.00109 | MirTarget; PITA | -0.13 | 0.00059 | NA | |
| 70 | hsa-miR-148b-3p | BMPR2 | 1.76 | 0 | -1.13 | 0.00109 | mirMAP | -0.2 | 0.00099 | NA | |
| 71 | hsa-miR-16-2-3p | BMPR2 | 3.8 | 0 | -1.13 | 0.00109 | mirMAP | -0.15 | 6.0E-5 | NA | |
| 72 | hsa-miR-17-5p | BMPR2 | 2.33 | 2.0E-5 | -1.13 | 0.00109 | miRNAWalker2 validate; MirTarget; TargetScan; miRNATAP | -0.12 | 0.00049 | NA | |
| 73 | hsa-miR-19b-3p | BMPR2 | 1.68 | 0.00086 | -1.13 | 0.00109 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.15 | 0.0001 | NA | |
| 74 | hsa-miR-20a-5p | BMPR2 | 2.14 | 0.00018 | -1.13 | 0.00109 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.11 | 0.00126 | NA | |
| 75 | hsa-miR-25-3p | BMPR2 | 1.13 | 0.00311 | -1.13 | 0.00109 | miRNATAP | -0.16 | 0.00181 | NA | |
| 76 | hsa-miR-30b-5p | BMPR2 | 0.02 | 0.95322 | -1.13 | 0.00109 | mirMAP | -0.17 | 0.00068 | NA | |
| 77 | hsa-miR-30e-5p | BMPR2 | 0.78 | 0.03467 | -1.13 | 0.00109 | mirMAP | -0.15 | 0.00461 | NA | |
| 78 | hsa-miR-32-5p | BMPR2 | 2.93 | 0 | -1.13 | 0.00109 | miRNATAP | -0.15 | 0.00016 | NA | |
| 79 | hsa-miR-590-3p | BMPR2 | 2.35 | 0 | -1.13 | 0.00109 | MirTarget; miRanda; mirMAP; miRNATAP | -0.18 | 0 | NA | |
| 80 | hsa-miR-590-5p | BMPR2 | 1.51 | 0.00239 | -1.13 | 0.00109 | MirTarget; PITA; miRNATAP | -0.15 | 0.00015 | NA | |
| 81 | hsa-miR-92a-3p | BMPR2 | 1.88 | 1.0E-5 | -1.13 | 0.00109 | miRNAWalker2 validate; miRNATAP | -0.2 | 1.0E-5 | NA | |
| 82 | hsa-miR-93-3p | BMPR2 | 2.63 | 0 | -1.13 | 0.00109 | miRNAWalker2 validate | -0.11 | 0.00334 | NA | |
| 83 | hsa-miR-93-5p | BMPR2 | 2.66 | 0 | -1.13 | 0.00109 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.13 | 0.00184 | NA | |
| 84 | hsa-let-7e-5p | BTRC | -0.11 | 0.81474 | -0.39 | 0.22952 | miRNAWalker2 validate | -0.16 | 4.0E-5 | NA | |
| 85 | hsa-miR-125a-3p | BTRC | -0.07 | 0.92074 | -0.39 | 0.22952 | miRanda | -0.11 | 0 | NA | |
| 86 | hsa-miR-15b-5p | BTRC | 3.32 | 0 | -0.39 | 0.22952 | MirTarget; miRNATAP | -0.14 | 0.00015 | NA | |
| 87 | hsa-miR-16-5p | BTRC | 2.94 | 0 | -0.39 | 0.22952 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.13 | 0.00072 | NA | |
| 88 | hsa-miR-28-5p | BTRC | -0.82 | 0.02212 | -0.39 | 0.22952 | miRanda | -0.24 | 0 | NA | |
| 89 | hsa-miR-339-5p | BTRC | 1.23 | 0.03075 | -0.39 | 0.22952 | MirTarget; miRanda | -0.11 | 0.00101 | NA | |
| 90 | hsa-miR-3913-5p | BTRC | 0.15 | 0.73484 | -0.39 | 0.22952 | mirMAP | -0.16 | 0.00018 | NA | |
| 91 | hsa-miR-505-3p | BTRC | 0.59 | 0.22694 | -0.39 | 0.22952 | MirTarget | -0.1 | 0.00592 | NA | |
| 92 | hsa-let-7a-5p | CCND1 | 0.15 | 0.64531 | 0.15 | 0.87753 | TargetScan; miRNATAP | -0.54 | 0.00245 | NA | |
| 93 | hsa-miR-106a-5p | CCND1 | 3.99 | 0 | 0.15 | 0.87753 | MirTarget; miRNATAP | -0.43 | 0 | NA | |
| 94 | hsa-miR-15a-5p | CCND1 | 2.05 | 0 | 0.15 | 0.87753 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.35 | 0.00776 | 22922827 | CCND1 has been found to be a target of miR-15a and miR-16-1 through analysis of complementary sequences between microRNAs and CCND1 mRNA; Moreover the transcription of CCND1 is suppressed by miR-15a and miR-16-1 via direct binding to the CCND1 3'-untranslated region 3'-UTR |
| 95 | hsa-miR-15b-5p | CCND1 | 3.32 | 0 | 0.15 | 0.87753 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.42 | 0.00029 | NA | |
| 96 | hsa-miR-20b-5p | CCND1 | 4.57 | 5.0E-5 | 0.15 | 0.87753 | MirTarget; miRNATAP | -0.31 | 0 | NA | |
| 97 | hsa-miR-34a-5p | CCND1 | 0.83 | 0.04775 | 0.15 | 0.87753 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.42 | 0.00174 | 25792709; 21399894 | This inhibition of proliferation was associated with a decrease in cyclin D1 levels orchestrated principally by HNF-4α a target of miR-34a considered to act as a tumour suppressor in the liver;Quantitative PCR and western analysis confirmed decreased expression of two genes BCL-2 and CCND1 in docetaxel-resistant cells which are both targeted by miR-34a |
| 98 | hsa-miR-497-5p | CCND1 | -1.44 | 0.02251 | 0.15 | 0.87753 | MirTarget; miRNATAP | -0.25 | 0.00498 | 21350001 | Raf-1 and Ccnd1 were identified as novel direct targets of miR-195 and miR-497 miR-195/497 expression levels in clinical specimens were found to be correlated inversely with malignancy of breast cancer |
| 99 | hsa-miR-106a-5p | CCND2 | 3.99 | 0 | -2.81 | 0.0014 | miRNATAP | -0.44 | 0 | NA | |
| 100 | hsa-miR-106b-5p | CCND2 | 2.81 | 0 | -2.81 | 0.0014 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.31 | 0.00374 | NA | |
| 101 | hsa-miR-10a-3p | CCND2 | 0.97 | 0.31667 | -2.81 | 0.0014 | mirMAP | -0.2 | 0.00011 | NA | |
| 102 | hsa-miR-130b-5p | CCND2 | 3.74 | 0 | -2.81 | 0.0014 | mirMAP | -0.46 | 0 | NA | |
| 103 | hsa-miR-141-3p | CCND2 | 7.3 | 0 | -2.81 | 0.0014 | MirTarget; TargetScan | -0.24 | 0.00021 | NA | |
| 104 | hsa-miR-15b-5p | CCND2 | 3.32 | 0 | -2.81 | 0.0014 | miRNATAP | -0.53 | 0 | NA | |
| 105 | hsa-miR-16-2-3p | CCND2 | 3.8 | 0 | -2.81 | 0.0014 | mirMAP | -0.29 | 0.00207 | NA | |
| 106 | hsa-miR-182-5p | CCND2 | 5.87 | 0 | -2.81 | 0.0014 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.28 | 2.0E-5 | NA | |
| 107 | hsa-miR-183-5p | CCND2 | 6.62 | 0 | -2.81 | 0.0014 | miRNATAP | -0.29 | 0 | NA | |
| 108 | hsa-miR-191-5p | CCND2 | 1.59 | 0.00074 | -2.81 | 0.0014 | MirTarget | -0.32 | 0.00314 | NA | |
| 109 | hsa-miR-200a-3p | CCND2 | 6.34 | 0 | -2.81 | 0.0014 | MirTarget | -0.41 | 0 | NA | |
| 110 | hsa-miR-20b-5p | CCND2 | 4.57 | 5.0E-5 | -2.81 | 0.0014 | miRNATAP | -0.3 | 0 | NA | |
| 111 | hsa-miR-224-3p | CCND2 | 2.85 | 0.00018 | -2.81 | 0.0014 | mirMAP | -0.22 | 0.00101 | NA | |
| 112 | hsa-miR-28-5p | CCND2 | -0.82 | 0.02212 | -2.81 | 0.0014 | miRanda | -0.43 | 0.00273 | NA | |
| 113 | hsa-miR-3065-3p | CCND2 | 1.89 | 0.03082 | -2.81 | 0.0014 | MirTarget; miRNATAP | -0.21 | 0.00027 | NA | |
| 114 | hsa-miR-3065-5p | CCND2 | 2.14 | 0.06094 | -2.81 | 0.0014 | mirMAP | -0.2 | 0.00026 | NA | |
| 115 | hsa-miR-30d-3p | CCND2 | -0.07 | 0.85742 | -2.81 | 0.0014 | mirMAP | -0.55 | 1.0E-5 | NA | |
| 116 | hsa-miR-324-3p | CCND2 | 1.51 | 0.00384 | -2.81 | 0.0014 | miRNAWalker2 validate | -0.44 | 0 | NA | |
| 117 | hsa-miR-33a-3p | CCND2 | 2.06 | 0.00156 | -2.81 | 0.0014 | MirTarget | -0.27 | 0.00041 | NA | |
| 118 | hsa-miR-378a-3p | CCND2 | 1.47 | 0.04667 | -2.81 | 0.0014 | miRNAWalker2 validate | -0.19 | 0.00601 | NA | |
| 119 | hsa-miR-429 | CCND2 | 6.4 | 0 | -2.81 | 0.0014 | miRNATAP | -0.46 | 0 | NA | |
| 120 | hsa-miR-497-5p | CCND2 | -1.44 | 0.02251 | -2.81 | 0.0014 | MirTarget; miRNATAP | -0.27 | 0.00058 | NA | |
| 121 | hsa-miR-550a-5p | CCND2 | 1.22 | 0.06138 | -2.81 | 0.0014 | MirTarget | -0.22 | 0.00363 | NA | |
| 122 | hsa-miR-660-5p | CCND2 | -0.07 | 0.88525 | -2.81 | 0.0014 | mirMAP | -0.29 | 0.00793 | NA | |
| 123 | hsa-miR-9-3p | CCND2 | 1.69 | 0.12517 | -2.81 | 0.0014 | MirTarget; mirMAP; miRNATAP | -0.14 | 0.00185 | NA | |
| 124 | hsa-miR-93-5p | CCND2 | 2.66 | 0 | -2.81 | 0.0014 | miRNATAP | -0.48 | 0 | NA | |
| 125 | hsa-miR-96-5p | CCND2 | 5.63 | 0 | -2.81 | 0.0014 | TargetScan; miRNATAP | -0.24 | 0.0003 | NA | |
| 126 | hsa-miR-27b-3p | CCND3 | -0.09 | 0.85847 | -0.54 | 0.12437 | miRNAWalker2 validate | -0.14 | 0.00019 | NA | |
| 127 | hsa-miR-429 | CCND3 | 6.4 | 0 | -0.54 | 0.12437 | miRNATAP | -0.11 | 1.0E-5 | NA | |
| 128 | hsa-miR-96-5p | CCND3 | 5.63 | 0 | -0.54 | 0.12437 | TargetScan | -0.12 | 1.0E-5 | NA | |
| 129 | hsa-let-7b-5p | CRB2 | -0.19 | 0.65188 | -2.32 | 0.05811 | miRNATAP | -0.46 | 0.00608 | NA | |
| 130 | hsa-miR-16-2-3p | CTGF | 3.8 | 0 | -4.34 | 2.0E-5 | MirTarget | -0.74 | 0 | NA | |
| 131 | hsa-miR-18a-5p | CTGF | 3.91 | 0 | -4.34 | 2.0E-5 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.47 | 0 | 23249750 | Targeting of TGFβ signature and its essential component CTGF by miR 18 correlates with improved survival in glioblastoma; Indeed microRNA-18a but not other miR-17∼92 members has a functional site in the CTGF 3' UTR and its forced reexpression sharply reduces CTGF protein and mRNA levels; The unexpected effects of miR-18a on CTGF transcription are mediated in part by direct targeting of Smad3 and ensuing weakening of TGFβ signaling |
| 132 | hsa-miR-19a-3p | CTGF | 2.17 | 0.00122 | -4.34 | 2.0E-5 | MirTarget; miRNATAP | -0.48 | 0 | NA | |
| 133 | hsa-miR-19b-3p | CTGF | 1.68 | 0.00086 | -4.34 | 2.0E-5 | MirTarget; miRNATAP | -0.63 | 0 | NA | |
| 134 | hsa-miR-205-5p | CTGF | 8.08 | 0 | -4.34 | 2.0E-5 | miRNAWalker2 validate; miRTarBase | -0.25 | 0 | NA | |
| 135 | hsa-miR-330-3p | CTGF | 2.49 | 0.00013 | -4.34 | 2.0E-5 | MirTarget; PITA; miRNATAP | -0.27 | 0.00261 | NA | |
| 136 | hsa-miR-486-5p | CTGF | 1.11 | 0.30929 | -4.34 | 2.0E-5 | miRanda | -0.14 | 0.00686 | NA | |
| 137 | hsa-miR-590-3p | CTGF | 2.35 | 0 | -4.34 | 2.0E-5 | PITA; miRanda | -0.39 | 0.00059 | NA | |
| 138 | hsa-miR-140-5p | CTNNA1 | -0.63 | 0.12667 | 0.14 | 0.59553 | miRanda | -0.13 | 0.00029 | NA | |
| 139 | hsa-miR-150-5p | CTNNB1 | 1.77 | 0.05938 | -0.6 | 0.15222 | MirTarget | -0.12 | 0 | NA | |
| 140 | hsa-miR-155-5p | CTNNB1 | 2.81 | 7.0E-5 | -0.6 | 0.15222 | miRNAWalker2 validate | -0.17 | 0 | NA | |
| 141 | hsa-miR-221-3p | CTNNB1 | 0.94 | 0.17475 | -0.6 | 0.15222 | miRNAWalker2 validate | -0.15 | 1.0E-5 | NA | |
| 142 | hsa-miR-330-3p | CTNNB1 | 2.49 | 0.00013 | -0.6 | 0.15222 | MirTarget; PITA; miRNATAP | -0.11 | 0.00159 | NA | |
| 143 | hsa-miR-590-3p | CTNNB1 | 2.35 | 0 | -0.6 | 0.15222 | miRanda | -0.16 | 0.00059 | NA | |
| 144 | hsa-miR-146b-5p | DLG1 | 1.88 | 0.00074 | 0.98 | 0.01314 | MirTarget; miRanda | -0.14 | 0.00052 | NA | |
| 145 | hsa-miR-186-5p | DLG1 | 0.45 | 0.18545 | 0.98 | 0.01314 | MirTarget; mirMAP; miRNATAP | -0.25 | 0.00032 | NA | |
| 146 | hsa-miR-320b | DLG1 | 0.2 | 0.72722 | 0.98 | 0.01314 | miRanda | -0.1 | 0.00732 | NA | |
| 147 | hsa-let-7g-3p | DLG2 | 2.1 | 5.0E-5 | -4.94 | 0.00023 | miRNATAP | -0.46 | 0.00167 | NA | |
| 148 | hsa-miR-130a-3p | DLG2 | 0.18 | 0.75775 | -4.94 | 0.00023 | miRNATAP | -0.46 | 0.0006 | NA | |
| 149 | hsa-miR-130a-5p | DLG2 | 1.58 | 0.02435 | -4.94 | 0.00023 | MirTarget; miRNATAP | -0.49 | 1.0E-5 | NA | |
| 150 | hsa-miR-135b-5p | DLG2 | 4.81 | 0 | -4.94 | 0.00023 | MirTarget | -0.43 | 0 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CANONICAL WNT SIGNALING PATHWAY | 28 | 95 | 5.866e-43 | 2.73e-39 |
| 2 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 61 | 1672 | 5.379e-40 | 1.251e-36 |
| 3 | ORGAN MORPHOGENESIS | 48 | 841 | 5.083e-39 | 7.884e-36 |
| 4 | WNT SIGNALING PATHWAY | 36 | 351 | 1.124e-37 | 1.307e-34 |
| 5 | TISSUE DEVELOPMENT | 57 | 1518 | 1.817e-37 | 1.691e-34 |
| 6 | EPITHELIUM DEVELOPMENT | 48 | 945 | 1.204e-36 | 9.334e-34 |
| 7 | REGULATION OF PROTEIN MODIFICATION PROCESS | 58 | 1710 | 7.948e-36 | 5.283e-33 |
| 8 | TISSUE MORPHOGENESIS | 38 | 533 | 7.83e-34 | 4.554e-31 |
| 9 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 55 | 1618 | 1.246e-33 | 6.443e-31 |
| 10 | REGULATION OF WNT SIGNALING PATHWAY | 32 | 310 | 1.971e-33 | 9.172e-31 |
| 11 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 46 | 1021 | 1.265e-32 | 5.351e-30 |
| 12 | REGULATION OF CANONICAL WNT SIGNALING PATHWAY | 29 | 236 | 1.737e-32 | 6.734e-30 |
| 13 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 47 | 1142 | 1.177e-31 | 4.213e-29 |
| 14 | EMBRYONIC MORPHOGENESIS | 36 | 539 | 6.037e-31 | 2.006e-28 |
| 15 | REGULATION OF CELL DIFFERENTIATION | 51 | 1492 | 7.349e-31 | 2.28e-28 |
| 16 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 56 | 1929 | 9.045e-31 | 2.63e-28 |
| 17 | POSITIVE REGULATION OF GENE EXPRESSION | 53 | 1733 | 6.807e-30 | 1.863e-27 |
| 18 | CELL FATE COMMITMENT | 27 | 227 | 7.921e-30 | 2.048e-27 |
| 19 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 50 | 1492 | 9.438e-30 | 2.311e-27 |
| 20 | TUBE DEVELOPMENT | 35 | 552 | 2.879e-29 | 6.698e-27 |
| 21 | NEUROGENESIS | 48 | 1402 | 8.529e-29 | 1.804e-26 |
| 22 | RESPONSE TO GROWTH FACTOR | 33 | 475 | 8.191e-29 | 1.804e-26 |
| 23 | MORPHOGENESIS OF AN EPITHELIUM | 31 | 400 | 1.799e-28 | 3.488e-26 |
| 24 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 39 | 801 | 1.795e-28 | 3.488e-26 |
| 25 | POSITIVE REGULATION OF CELL COMMUNICATION | 49 | 1532 | 3.896e-28 | 7.251e-26 |
| 26 | EMBRYO DEVELOPMENT | 40 | 894 | 7.438e-28 | 1.331e-25 |
| 27 | CARDIOVASCULAR SYSTEM DEVELOPMENT | 38 | 788 | 1.541e-27 | 2.561e-25 |
| 28 | CIRCULATORY SYSTEM DEVELOPMENT | 38 | 788 | 1.541e-27 | 2.561e-25 |
| 29 | NEGATIVE REGULATION OF CELL COMMUNICATION | 44 | 1192 | 1.952e-27 | 3.133e-25 |
| 30 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 43 | 1135 | 3.367e-27 | 5.222e-25 |
| 31 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 38 | 823 | 7.472e-27 | 1.122e-24 |
| 32 | REGULATION OF CELL DEATH | 47 | 1472 | 8.748e-27 | 1.272e-24 |
| 33 | CELLULAR RESPONSE TO ORGANIC SUBSTANCE | 51 | 1848 | 1.845e-26 | 2.602e-24 |
| 34 | REGULATION OF ORGAN MORPHOGENESIS | 25 | 242 | 4.785e-26 | 6.548e-24 |
| 35 | SENSORY ORGAN DEVELOPMENT | 31 | 493 | 1.07e-25 | 1.422e-23 |
| 36 | NEGATIVE REGULATION OF CANONICAL WNT SIGNALING PATHWAY | 22 | 162 | 1.213e-25 | 1.568e-23 |
| 37 | ANATOMICAL STRUCTURE FORMATION INVOLVED IN MORPHOGENESIS | 39 | 957 | 1.323e-25 | 1.664e-23 |
| 38 | REGULATION OF CARTILAGE DEVELOPMENT | 17 | 63 | 1.843e-25 | 2.256e-23 |
| 39 | NEGATIVE REGULATION OF WNT SIGNALING PATHWAY | 23 | 197 | 3.146e-25 | 3.753e-23 |
| 40 | NEGATIVE REGULATION OF RESPONSE TO STIMULUS | 44 | 1360 | 4.335e-25 | 5.043e-23 |
| 41 | NEURON DIFFERENTIATION | 37 | 874 | 8.905e-25 | 1.011e-22 |
| 42 | REGULATION OF CELL PROLIFERATION | 45 | 1496 | 2.024e-24 | 2.243e-22 |
| 43 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 39 | 1036 | 2.412e-24 | 2.55e-22 |
| 44 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 39 | 1036 | 2.412e-24 | 2.55e-22 |
| 45 | REGULATION OF CELL DEVELOPMENT | 36 | 836 | 2.56e-24 | 2.647e-22 |
| 46 | CELL DEVELOPMENT | 44 | 1426 | 2.959e-24 | 2.993e-22 |
| 47 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 48 | 1805 | 6.092e-24 | 6.031e-22 |
| 48 | CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 30 | 513 | 6.261e-24 | 6.069e-22 |
| 49 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 38 | 1008 | 1.076e-23 | 1.022e-21 |
| 50 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 43 | 1395 | 1.271e-23 | 1.183e-21 |
| 51 | REGULATION OF CELLULAR PROTEIN LOCALIZATION | 30 | 552 | 5.231e-23 | 4.712e-21 |
| 52 | NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 37 | 983 | 5.266e-23 | 4.712e-21 |
| 53 | EMBRYONIC ORGAN DEVELOPMENT | 27 | 406 | 5.622e-23 | 4.936e-21 |
| 54 | CELL PROLIFERATION | 32 | 672 | 7.981e-23 | 6.877e-21 |
| 55 | POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 39 | 1152 | 1.125e-22 | 9.52e-21 |
| 56 | REGULATION OF STEM CELL DIFFERENTIATION | 18 | 113 | 2.129e-22 | 1.769e-20 |
| 57 | EMBRYO DEVELOPMENT ENDING IN BIRTH OR EGG HATCHING | 29 | 554 | 8.959e-22 | 7.313e-20 |
| 58 | TUBE MORPHOGENESIS | 24 | 323 | 1.435e-21 | 1.151e-19 |
| 59 | POSITIVE REGULATION OF STEM CELL DIFFERENTIATION | 14 | 50 | 2.221e-21 | 1.751e-19 |
| 60 | POSITIVE REGULATION OF CELL DEVELOPMENT | 27 | 472 | 2.927e-21 | 2.27e-19 |
| 61 | STEM CELL DIFFERENTIATION | 20 | 190 | 4.437e-21 | 3.384e-19 |
| 62 | RESPONSE TO ENDOGENOUS STIMULUS | 41 | 1450 | 5.16e-21 | 3.872e-19 |
| 63 | POSITIVE REGULATION OF CELL DEATH | 29 | 605 | 1.023e-20 | 7.555e-19 |
| 64 | NON CANONICAL WNT SIGNALING PATHWAY | 18 | 140 | 1.207e-20 | 8.778e-19 |
| 65 | NEGATIVE REGULATION OF CELL DIFFERENTIATION | 29 | 609 | 1.226e-20 | 8.779e-19 |
| 66 | EPITHELIAL TO MESENCHYMAL TRANSITION | 14 | 56 | 1.341e-20 | 9.456e-19 |
| 67 | CARTILAGE DEVELOPMENT | 18 | 147 | 2.991e-20 | 2.077e-18 |
| 68 | REGULATION OF OSSIFICATION | 19 | 178 | 3.559e-20 | 2.435e-18 |
| 69 | CELLULAR COMPONENT MORPHOGENESIS | 33 | 900 | 4.773e-20 | 3.219e-18 |
| 70 | NEGATIVE REGULATION OF CELL PROLIFERATION | 29 | 643 | 5.441e-20 | 3.617e-18 |
| 71 | POSITIVE REGULATION OF EPITHELIAL TO MESENCHYMAL TRANSITION | 12 | 34 | 7.005e-20 | 4.591e-18 |
| 72 | REGULATION OF MAPK CASCADE | 29 | 660 | 1.109e-19 | 7.17e-18 |
| 73 | MESENCHYME DEVELOPMENT | 19 | 190 | 1.251e-19 | 7.869e-18 |
| 74 | TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 19 | 190 | 1.251e-19 | 7.869e-18 |
| 75 | CONNECTIVE TISSUE DEVELOPMENT | 19 | 194 | 1.867e-19 | 1.158e-17 |
| 76 | MESENCHYMAL CELL DIFFERENTIATION | 17 | 134 | 1.934e-19 | 1.184e-17 |
| 77 | REGULATION OF EPITHELIAL TO MESENCHYMAL TRANSITION | 14 | 67 | 2.162e-19 | 1.307e-17 |
| 78 | SKELETAL SYSTEM DEVELOPMENT | 25 | 455 | 2.864e-19 | 1.708e-17 |
| 79 | REGULATION OF OSTEOBLAST DIFFERENTIATION | 16 | 112 | 3.324e-19 | 1.958e-17 |
| 80 | HIPPO SIGNALING | 11 | 27 | 3.791e-19 | 2.2e-17 |
| 81 | GROWTH | 24 | 410 | 3.829e-19 | 2.2e-17 |
| 82 | ESTABLISHMENT OR MAINTENANCE OF CELL POLARITY | 17 | 141 | 4.722e-19 | 2.679e-17 |
| 83 | EMBRYONIC ORGAN MORPHOGENESIS | 21 | 279 | 4.846e-19 | 2.717e-17 |
| 84 | HEART DEVELOPMENT | 25 | 466 | 5.075e-19 | 2.811e-17 |
| 85 | EYE DEVELOPMENT | 22 | 326 | 6.713e-19 | 3.675e-17 |
| 86 | UROGENITAL SYSTEM DEVELOPMENT | 21 | 299 | 2.024e-18 | 1.095e-16 |
| 87 | GASTRULATION | 17 | 155 | 2.453e-18 | 1.297e-16 |
| 88 | REGULATION OF PROTEIN LOCALIZATION | 32 | 950 | 2.453e-18 | 1.297e-16 |
| 89 | GLAND DEVELOPMENT | 23 | 395 | 2.615e-18 | 1.367e-16 |
| 90 | PATTERN SPECIFICATION PROCESS | 23 | 418 | 9.133e-18 | 4.722e-16 |
| 91 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 41 | 1784 | 9.776e-18 | 4.998e-16 |
| 92 | REGULATION OF EPITHELIAL CELL PROLIFERATION | 20 | 285 | 1.451e-17 | 7.336e-16 |
| 93 | REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 18 | 207 | 1.545e-17 | 7.73e-16 |
| 94 | REGULATION OF EMBRYONIC DEVELOPMENT | 15 | 114 | 1.666e-17 | 8.248e-16 |
| 95 | REGULATION OF TRANSFERASE ACTIVITY | 31 | 946 | 2.068e-17 | 1.013e-15 |
| 96 | REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 21 | 337 | 2.34e-17 | 1.134e-15 |
| 97 | HEART MORPHOGENESIS | 18 | 212 | 2.371e-17 | 1.137e-15 |
| 98 | OSSIFICATION | 19 | 251 | 2.463e-17 | 1.169e-15 |
| 99 | REGULATION OF CELL MORPHOGENESIS | 25 | 552 | 2.815e-17 | 1.323e-15 |
| 100 | EPITHELIAL CELL DIFFERENTIATION | 24 | 495 | 2.881e-17 | 1.34e-15 |
| 101 | CELL JUNCTION ORGANIZATION | 17 | 185 | 5.128e-17 | 2.362e-15 |
| 102 | REGULATION OF KINASE ACTIVITY | 28 | 776 | 8.986e-17 | 4.099e-15 |
| 103 | HEAD DEVELOPMENT | 27 | 709 | 9.359e-17 | 4.228e-15 |
| 104 | BRANCHING MORPHOGENESIS OF AN EPITHELIAL TUBE | 15 | 131 | 1.425e-16 | 6.374e-15 |
| 105 | REGULATION OF STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE | 17 | 197 | 1.492e-16 | 6.549e-15 |
| 106 | RESPIRATORY SYSTEM DEVELOPMENT | 17 | 197 | 1.492e-16 | 6.549e-15 |
| 107 | REGULATION OF PATHWAY RESTRICTED SMAD PROTEIN PHOSPHORYLATION | 12 | 60 | 1.606e-16 | 6.984e-15 |
| 108 | NEGATIVE REGULATION OF CELL DEATH | 29 | 872 | 1.94e-16 | 8.356e-15 |
| 109 | MORPHOGENESIS OF A BRANCHING STRUCTURE | 16 | 167 | 2.331e-16 | 9.952e-15 |
| 110 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 38 | 1656 | 2.65e-16 | 1.121e-14 |
| 111 | POSITIVE REGULATION OF OSSIFICATION | 13 | 84 | 2.913e-16 | 1.221e-14 |
| 112 | REGULATION OF CHONDROCYTE DIFFERENTIATION | 11 | 46 | 3.586e-16 | 1.49e-14 |
| 113 | REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 27 | 750 | 3.774e-16 | 1.554e-14 |
| 114 | NEGATIVE REGULATION OF GENE EXPRESSION | 36 | 1493 | 4.646e-16 | 1.896e-14 |
| 115 | RESPONSE TO TRANSFORMING GROWTH FACTOR BETA | 15 | 144 | 6.039e-16 | 2.423e-14 |
| 116 | POSITIVE REGULATION OF PATHWAY RESTRICTED SMAD PROTEIN PHOSPHORYLATION | 11 | 48 | 6.023e-16 | 2.423e-14 |
| 117 | MESONEPHROS DEVELOPMENT | 13 | 90 | 7.463e-16 | 2.968e-14 |
| 118 | NEGATIVE REGULATION OF NITROGEN COMPOUND METABOLIC PROCESS | 36 | 1517 | 7.654e-16 | 3.018e-14 |
| 119 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 30 | 1004 | 9.355e-16 | 3.658e-14 |
| 120 | RESPONSE TO BMP | 13 | 94 | 1.346e-15 | 5.175e-14 |
| 121 | CELLULAR RESPONSE TO BMP STIMULUS | 13 | 94 | 1.346e-15 | 5.175e-14 |
| 122 | VASCULATURE DEVELOPMENT | 22 | 469 | 1.447e-15 | 5.518e-14 |
| 123 | POSITIVE REGULATION OF MAPK CASCADE | 22 | 470 | 1.512e-15 | 5.674e-14 |
| 124 | REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 22 | 470 | 1.512e-15 | 5.674e-14 |
| 125 | REGULATION OF APOPTOTIC SIGNALING PATHWAY | 20 | 363 | 1.567e-15 | 5.833e-14 |
| 126 | REGULATION OF CELLULAR LOCALIZATION | 33 | 1277 | 1.587e-15 | 5.86e-14 |
| 127 | REGULATION OF CELL CYCLE | 29 | 949 | 1.773e-15 | 6.496e-14 |
| 128 | KIDNEY EPITHELIUM DEVELOPMENT | 14 | 125 | 2.136e-15 | 7.765e-14 |
| 129 | POSITIVE REGULATION OF CELL PROLIFERATION | 27 | 814 | 2.822e-15 | 1.018e-13 |
| 130 | POSITIVE REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 15 | 162 | 3.582e-15 | 1.282e-13 |
| 131 | SMAD PROTEIN SIGNAL TRANSDUCTION | 11 | 56 | 3.84e-15 | 1.364e-13 |
| 132 | POSITIVE REGULATION OF STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE | 14 | 135 | 6.383e-15 | 2.25e-13 |
| 133 | DEVELOPMENTAL INDUCTION | 9 | 27 | 6.449e-15 | 2.256e-13 |
| 134 | FORMATION OF PRIMARY GERM LAYER | 13 | 110 | 1.109e-14 | 3.852e-13 |
| 135 | REPRODUCTIVE SYSTEM DEVELOPMENT | 20 | 408 | 1.443e-14 | 4.975e-13 |
| 136 | REGIONALIZATION | 18 | 311 | 2.015e-14 | 6.892e-13 |
| 137 | BLOOD VESSEL MORPHOGENESIS | 19 | 364 | 2.294e-14 | 7.792e-13 |
| 138 | MAMMARY GLAND DEVELOPMENT | 13 | 117 | 2.516e-14 | 8.484e-13 |
| 139 | LENS DEVELOPMENT IN CAMERA TYPE EYE | 11 | 66 | 2.657e-14 | 8.896e-13 |
| 140 | REGULATION OF INTRACELLULAR TRANSPORT | 23 | 621 | 4.633e-14 | 1.54e-12 |
| 141 | NEGATIVE REGULATION OF PROTEIN METABOLIC PROCESS | 29 | 1087 | 5.807e-14 | 1.916e-12 |
| 142 | DEVELOPMENTAL GROWTH | 18 | 333 | 6.532e-14 | 2.14e-12 |
| 143 | OSTEOBLAST DIFFERENTIATION | 13 | 126 | 6.688e-14 | 2.176e-12 |
| 144 | SENSORY ORGAN MORPHOGENESIS | 16 | 239 | 6.785e-14 | 2.192e-12 |
| 145 | REGULATION OF GROWTH | 23 | 633 | 6.926e-14 | 2.222e-12 |
| 146 | SKELETAL SYSTEM MORPHOGENESIS | 15 | 201 | 8.877e-14 | 2.829e-12 |
| 147 | CELL JUNCTION ASSEMBLY | 13 | 129 | 9.11e-14 | 2.884e-12 |
| 148 | POSITIVE REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 12 | 100 | 1.022e-13 | 3.214e-12 |
| 149 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 36 | 1791 | 1.259e-13 | 3.931e-12 |
| 150 | MORPHOGENESIS OF EMBRYONIC EPITHELIUM | 13 | 134 | 1.499e-13 | 4.649e-12 |
| 151 | POSITIVE REGULATION OF APOPTOTIC SIGNALING PATHWAY | 14 | 171 | 1.754e-13 | 5.404e-12 |
| 152 | RESPONSE TO LIPID | 26 | 888 | 1.866e-13 | 5.712e-12 |
| 153 | CELLULAR MACROMOLECULE LOCALIZATION | 30 | 1234 | 2.15e-13 | 6.497e-12 |
| 154 | MESENCHYME MORPHOGENESIS | 9 | 38 | 2.143e-13 | 6.497e-12 |
| 155 | WNT SIGNALING PATHWAY CALCIUM MODULATING PATHWAY | 9 | 39 | 2.774e-13 | 8.328e-12 |
| 156 | REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 15 | 218 | 2.922e-13 | 8.716e-12 |
| 157 | POSITIVE REGULATION OF KINASE ACTIVITY | 20 | 482 | 3.257e-13 | 9.59e-12 |
| 158 | KIDNEY MORPHOGENESIS | 11 | 82 | 3.242e-13 | 9.59e-12 |
| 159 | REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 15 | 220 | 3.339e-13 | 9.773e-12 |
| 160 | APICAL JUNCTION ASSEMBLY | 9 | 40 | 3.565e-13 | 1.037e-11 |
| 161 | POSITIVE REGULATION OF TRANSFERASE ACTIVITY | 22 | 616 | 3.722e-13 | 1.076e-11 |
| 162 | STEM CELL PROLIFERATION | 10 | 60 | 4.181e-13 | 1.201e-11 |
| 163 | REGULATION OF ORGANELLE ORGANIZATION | 29 | 1178 | 4.409e-13 | 1.258e-11 |
| 164 | REGULATION OF PROTEIN IMPORT | 14 | 183 | 4.477e-13 | 1.27e-11 |
| 165 | NEGATIVE REGULATION OF CARTILAGE DEVELOPMENT | 8 | 26 | 4.622e-13 | 1.303e-11 |
| 166 | PALATE DEVELOPMENT | 11 | 85 | 4.879e-13 | 1.368e-11 |
| 167 | NEGATIVE REGULATION OF CELL CYCLE | 19 | 433 | 5.103e-13 | 1.422e-11 |
| 168 | NEPHRON DEVELOPMENT | 12 | 115 | 5.64e-13 | 1.562e-11 |
| 169 | REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 15 | 229 | 5.991e-13 | 1.649e-11 |
| 170 | REGULATION OF STEM CELL PROLIFERATION | 11 | 88 | 7.229e-13 | 1.979e-11 |
| 171 | LOCOMOTION | 28 | 1114 | 7.502e-13 | 2.041e-11 |
| 172 | MESODERM DEVELOPMENT | 12 | 118 | 7.708e-13 | 2.085e-11 |
| 173 | POSITIVE REGULATION OF EPITHELIAL CELL PROLIFERATION | 13 | 154 | 9.127e-13 | 2.455e-11 |
| 174 | NEGATIVE REGULATION OF GROWTH | 15 | 236 | 9.277e-13 | 2.481e-11 |
| 175 | CENTRAL NERVOUS SYSTEM DEVELOPMENT | 25 | 872 | 9.511e-13 | 2.529e-11 |
| 176 | CELLULAR RESPONSE TO RETINOIC ACID | 10 | 65 | 9.724e-13 | 2.571e-11 |
| 177 | ANTERIOR POSTERIOR PATTERN SPECIFICATION | 14 | 194 | 9.982e-13 | 2.624e-11 |
| 178 | POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 25 | 876 | 1.052e-12 | 2.75e-11 |
| 179 | MESODERM MORPHOGENESIS | 10 | 66 | 1.141e-12 | 2.967e-11 |
| 180 | POSITIVE REGULATION OF CARTILAGE DEVELOPMENT | 8 | 29 | 1.254e-12 | 3.242e-11 |
| 181 | POSITIVE REGULATION OF CELLULAR PROTEIN LOCALIZATION | 17 | 360 | 3.039e-12 | 7.644e-11 |
| 182 | REGULATION OF ORGAN FORMATION | 8 | 32 | 3.035e-12 | 7.644e-11 |
| 183 | APPENDAGE DEVELOPMENT | 13 | 169 | 3.012e-12 | 7.644e-11 |
| 184 | BICELLULAR TIGHT JUNCTION ASSEMBLY | 8 | 32 | 3.035e-12 | 7.644e-11 |
| 185 | LIMB DEVELOPMENT | 13 | 169 | 3.012e-12 | 7.644e-11 |
| 186 | ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY | 22 | 689 | 3.43e-12 | 8.58e-11 |
| 187 | REGULATION OF CELLULAR RESPONSE TO STRESS | 22 | 691 | 3.631e-12 | 9.034e-11 |
| 188 | CELL CELL JUNCTION ASSEMBLY | 10 | 74 | 3.759e-12 | 9.304e-11 |
| 189 | RESPONSE TO OXYGEN CONTAINING COMPOUND | 30 | 1381 | 3.821e-12 | 9.407e-11 |
| 190 | IN UTERO EMBRYONIC DEVELOPMENT | 16 | 311 | 3.867e-12 | 9.47e-11 |
| 191 | REGULATION OF CELLULAR COMPONENT MOVEMENT | 23 | 771 | 4.085e-12 | 9.951e-11 |
| 192 | ODONTOGENESIS | 11 | 105 | 5.245e-12 | 1.271e-10 |
| 193 | MAMMARY GLAND EPITHELIUM DEVELOPMENT | 9 | 53 | 5.474e-12 | 1.313e-10 |
| 194 | MESONEPHRIC TUBULE MORPHOGENESIS | 9 | 53 | 5.474e-12 | 1.313e-10 |
| 195 | POSITIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 18 | 437 | 6.431e-12 | 1.53e-10 |
| 196 | RENAL TUBULE DEVELOPMENT | 10 | 78 | 6.476e-12 | 1.53e-10 |
| 197 | RESPONSE TO RETINOIC ACID | 11 | 107 | 6.469e-12 | 1.53e-10 |
| 198 | POSITIVE REGULATION OF ORGANELLE ORGANIZATION | 20 | 573 | 7.707e-12 | 1.811e-10 |
| 199 | REGULATION OF ESTABLISHMENT OF PLANAR POLARITY | 11 | 110 | 8.791e-12 | 2.056e-10 |
| 200 | BETA CATENIN DESTRUCTION COMPLEX DISASSEMBLY | 7 | 22 | 1.087e-11 | 2.529e-10 |
| 201 | REGULATION OF BINDING | 15 | 283 | 1.261e-11 | 2.919e-10 |
| 202 | POSITIVE REGULATION OF BIOMINERAL TISSUE DEVELOPMENT | 8 | 38 | 1.377e-11 | 3.171e-10 |
| 203 | NEGATIVE REGULATION OF MOLECULAR FUNCTION | 26 | 1079 | 1.535e-11 | 3.519e-10 |
| 204 | POSITIVE REGULATION OF WNT SIGNALING PATHWAY | 12 | 152 | 1.597e-11 | 3.642e-10 |
| 205 | REGULATION OF DEVELOPMENTAL GROWTH | 15 | 289 | 1.699e-11 | 3.857e-10 |
| 206 | CHONDROCYTE DIFFERENTIATION | 9 | 60 | 1.774e-11 | 3.987e-10 |
| 207 | POSITIVE REGULATION OF OSTEOBLAST DIFFERENTIATION | 9 | 60 | 1.774e-11 | 3.987e-10 |
| 208 | DEVELOPMENTAL PROCESS INVOLVED IN REPRODUCTION | 20 | 602 | 1.875e-11 | 4.195e-10 |
| 209 | RESPONSE TO ORGANIC CYCLIC COMPOUND | 24 | 917 | 1.999e-11 | 4.451e-10 |
| 210 | CARDIAC EPITHELIAL TO MESENCHYMAL TRANSITION | 7 | 24 | 2.188e-11 | 4.847e-10 |
| 211 | PATHWAY RESTRICTED SMAD PROTEIN PHOSPHORYLATION | 6 | 13 | 2.379e-11 | 5.247e-10 |
| 212 | NEGATIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 19 | 541 | 2.477e-11 | 5.412e-10 |
| 213 | NEGATIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 19 | 541 | 2.477e-11 | 5.412e-10 |
| 214 | RHYTHMIC PROCESS | 15 | 298 | 2.625e-11 | 5.707e-10 |
| 215 | REGULATION OF JNK CASCADE | 12 | 159 | 2.716e-11 | 5.879e-10 |
| 216 | EMBRYONIC SKELETAL SYSTEM DEVELOPMENT | 11 | 122 | 2.753e-11 | 5.931e-10 |
| 217 | NEGATIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 20 | 616 | 2.83e-11 | 6.069e-10 |
| 218 | NEGATIVE REGULATION OF CELL DEVELOPMENT | 15 | 303 | 3.32e-11 | 7.087e-10 |
| 219 | PROTEIN PHOSPHORYLATION | 24 | 944 | 3.648e-11 | 7.75e-10 |
| 220 | NEPHRON EPITHELIUM DEVELOPMENT | 10 | 93 | 3.907e-11 | 8.264e-10 |
| 221 | CELL CYCLE | 28 | 1316 | 3.959e-11 | 8.336e-10 |
| 222 | REGULATION OF PROTEIN TARGETING | 15 | 307 | 3.996e-11 | 8.375e-10 |
| 223 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 30 | 1518 | 4.058e-11 | 8.467e-10 |
| 224 | REGULATION OF CELL ADHESION | 20 | 629 | 4.109e-11 | 8.535e-10 |
| 225 | REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 16 | 365 | 4.27e-11 | 8.83e-10 |
| 226 | BRANCHING INVOLVED IN URETERIC BUD MORPHOGENESIS | 8 | 44 | 4.866e-11 | 9.975e-10 |
| 227 | TRANSFORMING GROWTH FACTOR BETA RECEPTOR SIGNALING PATHWAY | 10 | 95 | 4.846e-11 | 9.975e-10 |
| 228 | SKIN DEVELOPMENT | 13 | 211 | 4.998e-11 | 1.02e-09 |
| 229 | REGULATION OF CATENIN IMPORT INTO NUCLEUS | 7 | 27 | 5.544e-11 | 1.126e-09 |
| 230 | CELL ACTIVATION | 19 | 568 | 5.699e-11 | 1.153e-09 |
| 231 | REGULATION OF INTRACELLULAR PROTEIN TRANSPORT | 16 | 381 | 8.063e-11 | 1.624e-09 |
| 232 | EYE MORPHOGENESIS | 11 | 136 | 9.014e-11 | 1.808e-09 |
| 233 | POSITIVE REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION | 18 | 514 | 9.253e-11 | 1.848e-09 |
| 234 | REGULATION OF RESPONSE TO STRESS | 29 | 1468 | 9.552e-11 | 1.899e-09 |
| 235 | MOVEMENT OF CELL OR SUBCELLULAR COMPONENT | 27 | 1275 | 1.077e-10 | 2.132e-09 |
| 236 | ORGAN INDUCTION | 6 | 16 | 1.097e-10 | 2.163e-09 |
| 237 | REGULATION OF ORGAN GROWTH | 9 | 73 | 1.104e-10 | 2.167e-09 |
| 238 | PROTEIN LOCALIZATION | 32 | 1805 | 1.238e-10 | 2.42e-09 |
| 239 | ESTABLISHMENT OR MAINTENANCE OF EPITHELIAL CELL APICAL BASAL POLARITY | 7 | 30 | 1.255e-10 | 2.444e-09 |
| 240 | CELLULAR RESPONSE TO LIPID | 17 | 457 | 1.289e-10 | 2.499e-09 |
| 241 | CELL DIVISION | 17 | 460 | 1.426e-10 | 2.753e-09 |
| 242 | RESPONSE TO ABIOTIC STIMULUS | 24 | 1024 | 1.931e-10 | 3.714e-09 |
| 243 | POSITIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 14 | 289 | 2.105e-10 | 4.03e-09 |
| 244 | DIGESTIVE SYSTEM DEVELOPMENT | 11 | 148 | 2.25e-10 | 4.292e-09 |
| 245 | REGULATION OF HYDROLASE ACTIVITY | 27 | 1327 | 2.631e-10 | 4.996e-09 |
| 246 | REGULATION OF NEURON DIFFERENTIATION | 18 | 554 | 3.104e-10 | 5.871e-09 |
| 247 | REGULATION OF KIDNEY DEVELOPMENT | 8 | 55 | 3.194e-10 | 6.017e-09 |
| 248 | REGULATION OF MESENCHYMAL CELL PROLIFERATION | 7 | 34 | 3.263e-10 | 6.122e-09 |
| 249 | POSITIVE REGULATION OF LOCOMOTION | 16 | 420 | 3.373e-10 | 6.302e-09 |
| 250 | NEGATIVE REGULATION OF DEVELOPMENTAL GROWTH | 9 | 84 | 3.998e-10 | 7.442e-09 |
| 251 | CARDIAC SEPTUM DEVELOPMENT | 9 | 85 | 4.453e-10 | 8.255e-09 |
| 252 | EPIDERMIS DEVELOPMENT | 13 | 253 | 4.744e-10 | 8.76e-09 |
| 253 | EMBRYONIC PATTERN SPECIFICATION | 8 | 58 | 4.967e-10 | 9.134e-09 |
| 254 | POSITIVE REGULATION OF MITOCHONDRIAL OUTER MEMBRANE PERMEABILIZATION INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 7 | 36 | 5.022e-10 | 9.199e-09 |
| 255 | NEGATIVE REGULATION OF CHONDROCYTE DIFFERENTIATION | 6 | 20 | 5.223e-10 | 9.531e-09 |
| 256 | ESTABLISHMENT OR MAINTENANCE OF APICAL BASAL CELL POLARITY | 7 | 37 | 6.168e-10 | 1.117e-08 |
| 257 | ESTABLISHMENT OR MAINTENANCE OF BIPOLAR CELL POLARITY | 7 | 37 | 6.168e-10 | 1.117e-08 |
| 258 | NEGATIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 20 | 740 | 7.148e-10 | 1.289e-08 |
| 259 | NEGATIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 13 | 262 | 7.281e-10 | 1.308e-08 |
| 260 | CELL DEATH | 23 | 1001 | 7.315e-10 | 1.309e-08 |
| 261 | MIDBRAIN DEVELOPMENT | 9 | 90 | 7.482e-10 | 1.334e-08 |
| 262 | REGULATION OF MAP KINASE ACTIVITY | 14 | 319 | 7.647e-10 | 1.358e-08 |
| 263 | NEGATIVE REGULATION OF CATALYTIC ACTIVITY | 21 | 829 | 8.081e-10 | 1.43e-08 |
| 264 | REPRODUCTION | 26 | 1297 | 8.429e-10 | 1.486e-08 |
| 265 | EMBRYONIC SKELETAL SYSTEM MORPHOGENESIS | 9 | 93 | 1.006e-09 | 1.767e-08 |
| 266 | TUBE FORMATION | 10 | 129 | 1.024e-09 | 1.791e-08 |
| 267 | PHOSPHORYLATION | 25 | 1228 | 1.404e-09 | 2.447e-08 |
| 268 | GLAND MORPHOGENESIS | 9 | 97 | 1.471e-09 | 2.553e-08 |
| 269 | REGULATION OF MITOCHONDRIAL OUTER MEMBRANE PERMEABILIZATION INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 7 | 43 | 1.883e-09 | 3.258e-08 |
| 270 | POSITIVE REGULATION OF CELLULAR COMPONENT BIOGENESIS | 15 | 406 | 1.923e-09 | 3.315e-08 |
| 271 | CAMERA TYPE EYE MORPHOGENESIS | 9 | 101 | 2.114e-09 | 3.629e-08 |
| 272 | INTRACELLULAR SIGNAL TRANSDUCTION | 28 | 1572 | 2.297e-09 | 3.929e-08 |
| 273 | REGULATION OF CYTOPLASMIC TRANSPORT | 16 | 481 | 2.391e-09 | 4.076e-08 |
| 274 | REGULATION OF MUSCLE TISSUE DEVELOPMENT | 9 | 103 | 2.519e-09 | 4.262e-08 |
| 275 | REGULATION OF MUSCLE ORGAN DEVELOPMENT | 9 | 103 | 2.519e-09 | 4.262e-08 |
| 276 | NEGATIVE REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 8 | 71 | 2.612e-09 | 4.388e-08 |
| 277 | REGULATION OF TRANSPORT | 30 | 1804 | 2.607e-09 | 4.388e-08 |
| 278 | ANGIOGENESIS | 13 | 293 | 2.832e-09 | 4.741e-08 |
| 279 | CARDIAC CHAMBER DEVELOPMENT | 10 | 144 | 3.003e-09 | 5.009e-08 |
| 280 | NEGATIVE REGULATION OF EMBRYONIC DEVELOPMENT | 6 | 26 | 3.028e-09 | 5.031e-08 |
| 281 | CELL CYCLE PROCESS | 23 | 1081 | 3.197e-09 | 5.293e-08 |
| 282 | NEGATIVE REGULATION OF PHOSPHORYLATION | 15 | 422 | 3.249e-09 | 5.361e-08 |
| 283 | REGULATION OF BIOMINERAL TISSUE DEVELOPMENT | 8 | 75 | 4.074e-09 | 6.699e-08 |
| 284 | DIGESTIVE TRACT MORPHOGENESIS | 7 | 48 | 4.216e-09 | 6.907e-08 |
| 285 | CELL MORPHOGENESIS INVOLVED IN NEURON DIFFERENTIATION | 14 | 368 | 4.807e-09 | 7.847e-08 |
| 286 | VENTRICULAR SEPTUM MORPHOGENESIS | 6 | 28 | 4.914e-09 | 7.967e-08 |
| 287 | CARDIAC SEPTUM MORPHOGENESIS | 7 | 49 | 4.898e-09 | 7.967e-08 |
| 288 | POSITIVE REGULATION OF INTRACELLULAR TRANSPORT | 14 | 370 | 5.15e-09 | 8.321e-08 |
| 289 | PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS | 31 | 1977 | 5.316e-09 | 8.56e-08 |
| 290 | REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 10 | 153 | 5.407e-09 | 8.676e-08 |
| 291 | REGULATION OF PROTEIN INSERTION INTO MITOCHONDRIAL MEMBRANE INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 6 | 29 | 6.171e-09 | 9.766e-08 |
| 292 | POSITIVE REGULATION OF PROTEIN INSERTION INTO MITOCHONDRIAL MEMBRANE INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 6 | 29 | 6.171e-09 | 9.766e-08 |
| 293 | STEM CELL DIVISION | 6 | 29 | 6.171e-09 | 9.766e-08 |
| 294 | REGULATION OF HEART MORPHOGENESIS | 6 | 29 | 6.171e-09 | 9.766e-08 |
| 295 | REGULATION OF DEPHOSPHORYLATION | 10 | 158 | 7.376e-09 | 1.163e-07 |
| 296 | RESPONSE TO ACID CHEMICAL | 13 | 319 | 7.857e-09 | 1.235e-07 |
| 297 | CELL CELL SIGNALING | 19 | 767 | 8.378e-09 | 1.308e-07 |
| 298 | REGULATION OF CELLULAR COMPONENT BIOGENESIS | 19 | 767 | 8.378e-09 | 1.308e-07 |
| 299 | REGULATION OF MORPHOGENESIS OF A BRANCHING STRUCTURE | 7 | 53 | 8.646e-09 | 1.345e-07 |
| 300 | SEX DIFFERENTIATION | 12 | 266 | 9.979e-09 | 1.548e-07 |
| 301 | SINGLE ORGANISM CELL ADHESION | 15 | 459 | 1.006e-08 | 1.555e-07 |
| 302 | CELLULAR RESPONSE TO ORGANIC CYCLIC COMPOUND | 15 | 465 | 1.196e-08 | 1.842e-07 |
| 303 | ENDOCRINE SYSTEM DEVELOPMENT | 9 | 123 | 1.216e-08 | 1.867e-07 |
| 304 | REGULATION OF SMAD PROTEIN IMPORT INTO NUCLEUS | 5 | 16 | 1.242e-08 | 1.895e-07 |
| 305 | PARAXIAL MESODERM DEVELOPMENT | 5 | 16 | 1.242e-08 | 1.895e-07 |
| 306 | REGULATION OF PROTEIN BINDING | 10 | 168 | 1.33e-08 | 2.023e-07 |
| 307 | RESPONSE TO ESTROGEN | 11 | 218 | 1.35e-08 | 2.046e-07 |
| 308 | ESTABLISHMENT OF CELL POLARITY | 8 | 88 | 1.471e-08 | 2.222e-07 |
| 309 | REGULATION OF PROTEIN STABILITY | 11 | 221 | 1.555e-08 | 2.342e-07 |
| 310 | SEGMENTATION | 8 | 89 | 1.609e-08 | 2.408e-07 |
| 311 | EPITHELIAL CELL PROLIFERATION | 8 | 89 | 1.609e-08 | 2.408e-07 |
| 312 | REGULATION OF PHOSPHATASE ACTIVITY | 9 | 128 | 1.725e-08 | 2.572e-07 |
| 313 | AXIS SPECIFICATION | 8 | 90 | 1.759e-08 | 2.599e-07 |
| 314 | NEGATIVE REGULATION OF STEM CELL PROLIFERATION | 5 | 17 | 1.753e-08 | 2.599e-07 |
| 315 | ENDOTHELIUM DEVELOPMENT | 8 | 90 | 1.759e-08 | 2.599e-07 |
| 316 | VASCULOGENESIS | 7 | 59 | 1.867e-08 | 2.749e-07 |
| 317 | DORSAL VENTRAL PATTERN FORMATION | 8 | 91 | 1.921e-08 | 2.819e-07 |
| 318 | CELLULAR RESPONSE TO ACID CHEMICAL | 10 | 175 | 1.966e-08 | 2.876e-07 |
| 319 | EMBRYONIC DIGIT MORPHOGENESIS | 7 | 61 | 2.368e-08 | 3.443e-07 |
| 320 | POSITIVE REGULATION OF STEM CELL PROLIFERATION | 7 | 61 | 2.368e-08 | 3.443e-07 |
| 321 | NOTOCHORD DEVELOPMENT | 5 | 18 | 2.417e-08 | 3.504e-07 |
| 322 | NEGATIVE REGULATION OF MUSCLE ORGAN DEVELOPMENT | 6 | 36 | 2.459e-08 | 3.542e-07 |
| 323 | NEGATIVE REGULATION OF MUSCLE TISSUE DEVELOPMENT | 6 | 36 | 2.459e-08 | 3.542e-07 |
| 324 | NEURAL TUBE FORMATION | 8 | 94 | 2.485e-08 | 3.568e-07 |
| 325 | PROTEIN COMPLEX SUBUNIT ORGANIZATION | 26 | 1527 | 2.521e-08 | 3.61e-07 |
| 326 | REGULATION OF VASCULATURE DEVELOPMENT | 11 | 233 | 2.682e-08 | 3.828e-07 |
| 327 | NEGATIVE REGULATION OF INTRACELLULAR PROTEIN TRANSPORT | 8 | 95 | 2.702e-08 | 3.845e-07 |
| 328 | FOREBRAIN DEVELOPMENT | 13 | 357 | 2.979e-08 | 4.227e-07 |
| 329 | CELL MOTILITY | 19 | 835 | 3.261e-08 | 4.598e-07 |
| 330 | LOCALIZATION OF CELL | 19 | 835 | 3.261e-08 | 4.598e-07 |
| 331 | IMMUNE SYSTEM DEVELOPMENT | 16 | 582 | 3.495e-08 | 4.913e-07 |
| 332 | RESPONSE TO ALCOHOL | 13 | 362 | 3.508e-08 | 4.916e-07 |
| 333 | TRACHEA DEVELOPMENT | 5 | 20 | 4.339e-08 | 6.062e-07 |
| 334 | MAMMARY GLAND MORPHOGENESIS | 6 | 40 | 4.768e-08 | 6.642e-07 |
| 335 | CARDIAC CHAMBER MORPHOGENESIS | 8 | 104 | 5.516e-08 | 7.617e-07 |
| 336 | EAR DEVELOPMENT | 10 | 195 | 5.486e-08 | 7.617e-07 |
| 337 | DEVELOPMENTAL GROWTH INVOLVED IN MORPHOGENESIS | 8 | 104 | 5.516e-08 | 7.617e-07 |
| 338 | RESPONSE TO EXTERNAL STIMULUS | 28 | 1821 | 5.648e-08 | 7.775e-07 |
| 339 | NEGATIVE REGULATION OF ORGAN GROWTH | 5 | 21 | 5.672e-08 | 7.785e-07 |
| 340 | NEURON DEVELOPMENT | 17 | 687 | 5.744e-08 | 7.861e-07 |
| 341 | REGULATION OF CELL CELL ADHESION | 13 | 380 | 6.184e-08 | 8.438e-07 |
| 342 | NEURAL PRECURSOR CELL PROLIFERATION | 7 | 70 | 6.269e-08 | 8.505e-07 |
| 343 | REGULATION OF MEMBRANE PERMEABILITY | 7 | 70 | 6.269e-08 | 8.505e-07 |
| 344 | REGULATION OF FAT CELL DIFFERENTIATION | 8 | 106 | 6.406e-08 | 8.664e-07 |
| 345 | PITUITARY GLAND DEVELOPMENT | 6 | 42 | 6.463e-08 | 8.695e-07 |
| 346 | NEURAL TUBE DEVELOPMENT | 9 | 149 | 6.466e-08 | 8.695e-07 |
| 347 | NEGATIVE REGULATION OF MITOTIC CELL CYCLE | 10 | 199 | 6.642e-08 | 8.906e-07 |
| 348 | LEUKOCYTE CELL CELL ADHESION | 11 | 255 | 6.745e-08 | 9.018e-07 |
| 349 | SOMATIC STEM CELL DIVISION | 5 | 22 | 7.311e-08 | 9.747e-07 |
| 350 | BETA CATENIN TCF COMPLEX ASSEMBLY | 6 | 43 | 7.48e-08 | 9.945e-07 |
| 351 | CELL CYCLE ARREST | 9 | 154 | 8.593e-08 | 1.139e-06 |
| 352 | TAXIS | 14 | 464 | 8.758e-08 | 1.158e-06 |
| 353 | CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND | 18 | 799 | 9.177e-08 | 1.21e-06 |
| 354 | REGULATION OF METANEPHROS DEVELOPMENT | 5 | 23 | 9.304e-08 | 1.223e-06 |
| 355 | RESPONSE TO HORMONE | 19 | 893 | 9.374e-08 | 1.229e-06 |
| 356 | NEURON PROJECTION DEVELOPMENT | 15 | 545 | 9.639e-08 | 1.256e-06 |
| 357 | POSITIVE REGULATION OF MAP KINASE ACTIVITY | 10 | 207 | 9.615e-08 | 1.256e-06 |
| 358 | LUNG MORPHOGENESIS | 6 | 45 | 9.913e-08 | 1.288e-06 |
| 359 | ODONTOGENESIS OF DENTIN CONTAINING TOOTH | 7 | 75 | 1.017e-07 | 1.318e-06 |
| 360 | OVULATION CYCLE | 8 | 113 | 1.056e-07 | 1.365e-06 |
| 361 | EMBRYONIC CRANIAL SKELETON MORPHOGENESIS | 6 | 46 | 1.135e-07 | 1.463e-06 |
| 362 | POSITIVE REGULATION OF CELL CYCLE | 12 | 332 | 1.141e-07 | 1.467e-06 |
| 363 | NEURON PROJECTION MORPHOGENESIS | 13 | 402 | 1.187e-07 | 1.522e-06 |
| 364 | NEGATIVE REGULATION OF EPITHELIAL CELL PROLIFERATION | 8 | 115 | 1.21e-07 | 1.547e-06 |
| 365 | DIENCEPHALON DEVELOPMENT | 7 | 77 | 1.222e-07 | 1.553e-06 |
| 366 | REGULATION OF BMP SIGNALING PATHWAY | 7 | 77 | 1.222e-07 | 1.553e-06 |
| 367 | SIGNAL TRANSDUCTION BY PROTEIN PHOSPHORYLATION | 13 | 404 | 1.257e-07 | 1.594e-06 |
| 368 | REGULATION OF CELL CYCLE PROCESS | 15 | 558 | 1.307e-07 | 1.653e-06 |
| 369 | SOMITE DEVELOPMENT | 7 | 78 | 1.337e-07 | 1.686e-06 |
| 370 | NEGATIVE REGULATION OF CYTOPLASMIC TRANSPORT | 8 | 117 | 1.384e-07 | 1.736e-06 |
| 371 | NEGATIVE REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 8 | 117 | 1.384e-07 | 1.736e-06 |
| 372 | DEVELOPMENT OF PRIMARY SEXUAL CHARACTERISTICS | 10 | 216 | 1.431e-07 | 1.79e-06 |
| 373 | REGULATION OF CARDIAC MUSCLE TISSUE DEVELOPMENT | 6 | 48 | 1.475e-07 | 1.826e-06 |
| 374 | RESPONSE TO WOUNDING | 15 | 563 | 1.467e-07 | 1.826e-06 |
| 375 | ANTERIOR POSTERIOR AXIS SPECIFICATION | 6 | 48 | 1.475e-07 | 1.826e-06 |
| 376 | POSITIVE REGULATION OF ACTIN FILAMENT BUNDLE ASSEMBLY | 6 | 48 | 1.475e-07 | 1.826e-06 |
| 377 | LYMPHOCYTE ACTIVATION | 12 | 342 | 1.573e-07 | 1.941e-06 |
| 378 | POSITIVE REGULATION OF CANONICAL WNT SIGNALING PATHWAY | 8 | 119 | 1.578e-07 | 1.943e-06 |
| 379 | LEUKOCYTE ACTIVATION | 13 | 414 | 1.665e-07 | 2.045e-06 |
| 380 | METANEPHROS DEVELOPMENT | 7 | 81 | 1.738e-07 | 2.123e-06 |
| 381 | REGULATION OF JUN KINASE ACTIVITY | 7 | 81 | 1.738e-07 | 2.123e-06 |
| 382 | NEGATIVE REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 8 | 121 | 1.796e-07 | 2.188e-06 |
| 383 | POSITIVE REGULATION OF TRANSPORT | 19 | 936 | 1.943e-07 | 2.361e-06 |
| 384 | RESPONSE TO STEROID HORMONE | 14 | 497 | 2.027e-07 | 2.456e-06 |
| 385 | POSITIVE REGULATION OF MESENCHYMAL CELL PROLIFERATION | 5 | 27 | 2.197e-07 | 2.648e-06 |
| 386 | AXIS ELONGATION | 5 | 27 | 2.197e-07 | 2.648e-06 |
| 387 | REGULATION OF CYTOSKELETON ORGANIZATION | 14 | 502 | 2.288e-07 | 2.751e-06 |
| 388 | NEGATIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 8 | 126 | 2.456e-07 | 2.945e-06 |
| 389 | POSITIVE REGULATION OF SMAD PROTEIN IMPORT INTO NUCLEUS | 4 | 12 | 2.94e-07 | 3.498e-06 |
| 390 | TRACHEA MORPHOGENESIS | 4 | 12 | 2.94e-07 | 3.498e-06 |
| 391 | MAMMARY GLAND EPITHELIAL CELL PROLIFERATION | 4 | 12 | 2.94e-07 | 3.498e-06 |
| 392 | VENTRICULAR SEPTUM DEVELOPMENT | 6 | 54 | 3.03e-07 | 3.597e-06 |
| 393 | OVULATION CYCLE PROCESS | 7 | 88 | 3.083e-07 | 3.65e-06 |
| 394 | CRANIAL SKELETAL SYSTEM DEVELOPMENT | 6 | 55 | 3.387e-07 | 4e-06 |
| 395 | POSITIVE REGULATION OF CELL PROJECTION ORGANIZATION | 11 | 303 | 3.816e-07 | 4.495e-06 |
| 396 | NEGATIVE REGULATION OF CELLULAR PROTEIN LOCALIZATION | 8 | 135 | 4.172e-07 | 4.903e-06 |
| 397 | MITOCHONDRIAL MEMBRANE ORGANIZATION | 7 | 92 | 4.185e-07 | 4.905e-06 |
| 398 | POSITIVE REGULATION OF NEURON DIFFERENTIATION | 11 | 306 | 4.207e-07 | 4.919e-06 |
| 399 | MESENCHYMAL CELL PROLIFERATION | 4 | 13 | 4.23e-07 | 4.933e-06 |
| 400 | EPITHELIAL CELL DEVELOPMENT | 9 | 186 | 4.29e-07 | 4.99e-06 |
| 401 | RESPONSE TO OXYGEN LEVELS | 11 | 311 | 4.94e-07 | 5.732e-06 |
| 402 | REGULATION OF ACTIN FILAMENT BASED PROCESS | 11 | 312 | 5.099e-07 | 5.902e-06 |
| 403 | REGULATION OF PROTEOLYSIS | 16 | 711 | 5.272e-07 | 6.087e-06 |
| 404 | NEGATIVE REGULATION OF NEURON DIFFERENTIATION | 9 | 191 | 5.363e-07 | 6.171e-06 |
| 405 | ENDOCARDIAL CUSHION DEVELOPMENT | 5 | 32 | 5.371e-07 | 6.171e-06 |
| 406 | NEGATIVE REGULATION OF KINASE ACTIVITY | 10 | 250 | 5.518e-07 | 6.324e-06 |
| 407 | REGULATION OF ESTABLISHMENT OF PLANAR POLARITY INVOLVED IN NEURAL TUBE CLOSURE | 4 | 14 | 5.899e-07 | 6.727e-06 |
| 408 | EMBRYONIC SKELETAL JOINT DEVELOPMENT | 4 | 14 | 5.899e-07 | 6.727e-06 |
| 409 | EPIDERMAL CELL DIFFERENTIATION | 8 | 142 | 6.138e-07 | 6.983e-06 |
| 410 | EMBRYONIC AXIS SPECIFICATION | 5 | 33 | 6.305e-07 | 7.138e-06 |
| 411 | POSITIVE REGULATION OF EMBRYONIC DEVELOPMENT | 5 | 33 | 6.305e-07 | 7.138e-06 |
| 412 | NEGATIVE REGULATION OF INTRACELLULAR TRANSPORT | 8 | 143 | 6.475e-07 | 7.313e-06 |
| 413 | CELL CYCLE PHASE TRANSITION | 10 | 255 | 6.612e-07 | 7.449e-06 |
| 414 | REGULATION OF MITOTIC CELL CYCLE | 13 | 468 | 6.703e-07 | 7.534e-06 |
| 415 | SOMITOGENESIS | 6 | 62 | 6.981e-07 | 7.827e-06 |
| 416 | WOUND HEALING | 13 | 470 | 7.032e-07 | 7.866e-06 |
| 417 | ORGAN FORMATION | 5 | 34 | 7.363e-07 | 8.215e-06 |
| 418 | RESPONSE TO ESTRADIOL | 8 | 146 | 7.582e-07 | 8.441e-06 |
| 419 | POSITIVE REGULATION OF JUN KINASE ACTIVITY | 6 | 63 | 7.685e-07 | 8.534e-06 |
| 420 | REGULATION OF CELL PROJECTION ORGANIZATION | 14 | 558 | 8.133e-07 | 9.01e-06 |
| 421 | MALE SEX DIFFERENTIATION | 8 | 148 | 8.407e-07 | 9.292e-06 |
| 422 | NEGATIVE REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 7 | 102 | 8.466e-07 | 9.335e-06 |
| 423 | AGING | 10 | 264 | 9.065e-07 | 9.948e-06 |
| 424 | POSITIVE REGULATION OF EPITHELIAL CELL MIGRATION | 7 | 103 | 9.046e-07 | 9.948e-06 |
| 425 | NEURON PROJECTION GUIDANCE | 9 | 205 | 9.692e-07 | 1.061e-05 |
| 426 | NEURON FATE COMMITMENT | 6 | 67 | 1.11e-06 | 1.213e-05 |
| 427 | NEGATIVE REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION | 9 | 209 | 1.138e-06 | 1.236e-05 |
| 428 | REGULATION OF PEPTIDYL THREONINE PHOSPHORYLATION | 5 | 37 | 1.14e-06 | 1.236e-05 |
| 429 | NEGATIVE REGULATION OF EPITHELIAL CELL DIFFERENTIATION | 5 | 37 | 1.14e-06 | 1.236e-05 |
| 430 | REGULATION OF CELL DIVISION | 10 | 272 | 1.188e-06 | 1.286e-05 |
| 431 | POSITIVE REGULATION OF ENDOTHELIAL CELL PROLIFERATION | 6 | 68 | 1.213e-06 | 1.31e-05 |
| 432 | BONE DEVELOPMENT | 8 | 156 | 1.252e-06 | 1.348e-05 |
| 433 | BRANCH ELONGATION OF AN EPITHELIUM | 4 | 17 | 1.386e-06 | 1.483e-05 |
| 434 | ESTABLISHMENT OF TISSUE POLARITY | 4 | 17 | 1.386e-06 | 1.483e-05 |
| 435 | NEGATIVE REGULATION OF CARDIAC MUSCLE TISSUE DEVELOPMENT | 4 | 17 | 1.386e-06 | 1.483e-05 |
| 436 | ACTIVATION OF PROTEIN KINASE ACTIVITY | 10 | 279 | 1.494e-06 | 1.595e-05 |
| 437 | COLUMNAR CUBOIDAL EPITHELIAL CELL DIFFERENTIATION | 7 | 111 | 1.501e-06 | 1.598e-05 |
| 438 | SKIN EPIDERMIS DEVELOPMENT | 6 | 71 | 1.568e-06 | 1.666e-05 |
| 439 | NEGATIVE REGULATION OF TRANSFERASE ACTIVITY | 11 | 351 | 1.616e-06 | 1.713e-05 |
| 440 | ENDOTHELIAL CELL DIFFERENTIATION | 6 | 72 | 1.704e-06 | 1.797e-05 |
| 441 | NEGATIVE REGULATION OF OSTEOBLAST DIFFERENTIATION | 5 | 40 | 1.7e-06 | 1.797e-05 |
| 442 | KIDNEY MESENCHYME DEVELOPMENT | 4 | 18 | 1.775e-06 | 1.869e-05 |
| 443 | POSITIVE REGULATION OF PROTEIN BINDING | 6 | 73 | 1.849e-06 | 1.937e-05 |
| 444 | REGULATION OF NEURAL PRECURSOR CELL PROLIFERATION | 6 | 73 | 1.849e-06 | 1.937e-05 |
| 445 | MUSCLE STRUCTURE DEVELOPMENT | 12 | 432 | 1.862e-06 | 1.946e-05 |
| 446 | LUNG ALVEOLUS DEVELOPMENT | 5 | 41 | 1.928e-06 | 2.007e-05 |
| 447 | RECEPTOR CLUSTERING | 5 | 41 | 1.928e-06 | 2.007e-05 |
| 448 | REGULATION OF EPITHELIAL CELL MIGRATION | 8 | 166 | 1.997e-06 | 2.074e-05 |
| 449 | FEMALE SEX DIFFERENTIATION | 7 | 116 | 2.018e-06 | 2.091e-05 |
| 450 | POSITIVE REGULATION OF MITOCHONDRION ORGANIZATION | 8 | 167 | 2.088e-06 | 2.159e-05 |
| 451 | NEGATIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 12 | 437 | 2.097e-06 | 2.164e-05 |
| 452 | EPITHELIAL CELL MORPHOGENESIS | 5 | 42 | 2.18e-06 | 2.244e-05 |
| 453 | CELL CELL ADHESION | 14 | 608 | 2.23e-06 | 2.29e-05 |
| 454 | LEUKOCYTE DIFFERENTIATION | 10 | 292 | 2.249e-06 | 2.305e-05 |
| 455 | POSITIVE REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 9 | 228 | 2.336e-06 | 2.389e-05 |
| 456 | MEMBRANE ORGANIZATION | 17 | 899 | 2.453e-06 | 2.503e-05 |
| 457 | REGULATION OF ACTIN FILAMENT BUNDLE ASSEMBLY | 6 | 77 | 2.533e-06 | 2.579e-05 |
| 458 | POSITIVE REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 7 | 121 | 2.677e-06 | 2.72e-05 |
| 459 | TONGUE DEVELOPMENT | 4 | 20 | 2.789e-06 | 2.821e-05 |
| 460 | DORSAL VENTRAL AXIS SPECIFICATION | 4 | 20 | 2.789e-06 | 2.821e-05 |
| 461 | REGULATION OF EPITHELIAL CELL DIFFERENTIATION | 7 | 122 | 2.828e-06 | 2.855e-05 |
| 462 | BONE MORPHOGENESIS | 6 | 79 | 2.946e-06 | 2.967e-05 |
| 463 | THYMOCYTE AGGREGATION | 5 | 45 | 3.094e-06 | 3.096e-05 |
| 464 | T CELL DIFFERENTIATION IN THYMUS | 5 | 45 | 3.094e-06 | 3.096e-05 |
| 465 | ENDOCHONDRAL BONE MORPHOGENESIS | 5 | 45 | 3.094e-06 | 3.096e-05 |
| 466 | POSITIVE REGULATION OF CELL ADHESION | 11 | 376 | 3.138e-06 | 3.134e-05 |
| 467 | POSITIVE REGULATION OF GROWTH | 9 | 238 | 3.32e-06 | 3.308e-05 |
| 468 | CELL AGGREGATION | 4 | 21 | 3.432e-06 | 3.398e-05 |
| 469 | COCHLEA MORPHOGENESIS | 4 | 21 | 3.432e-06 | 3.398e-05 |
| 470 | CARTILAGE CONDENSATION | 4 | 21 | 3.432e-06 | 3.398e-05 |
| 471 | CELL PART MORPHOGENESIS | 14 | 633 | 3.556e-06 | 3.513e-05 |
| 472 | POSITIVE REGULATION OF BINDING | 7 | 127 | 3.696e-06 | 3.644e-05 |
| 473 | BIOLOGICAL ADHESION | 18 | 1032 | 3.721e-06 | 3.66e-05 |
| 474 | REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION TO MITOCHONDRION | 7 | 128 | 3.894e-06 | 3.823e-05 |
| 475 | HAIR CYCLE | 6 | 83 | 3.935e-06 | 3.847e-05 |
| 476 | MOLTING CYCLE | 6 | 83 | 3.935e-06 | 3.847e-05 |
| 477 | ENDOCARDIAL CUSHION MORPHOGENESIS | 4 | 22 | 4.179e-06 | 4.068e-05 |
| 478 | ACTIVIN RECEPTOR SIGNALING PATHWAY | 4 | 22 | 4.179e-06 | 4.068e-05 |
| 479 | PROTEIN STABILIZATION | 7 | 131 | 4.541e-06 | 4.411e-05 |
| 480 | REGULATION OF CELL GROWTH | 11 | 391 | 4.56e-06 | 4.42e-05 |
| 481 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL METABOLIC PROCESS | 4 | 23 | 5.039e-06 | 4.864e-05 |
| 482 | POSITIVE REGULATION OF COLLAGEN METABOLIC PROCESS | 4 | 23 | 5.039e-06 | 4.864e-05 |
| 483 | POSITIVE REGULATION OF FAT CELL DIFFERENTIATION | 5 | 51 | 5.807e-06 | 5.593e-05 |
| 484 | NEGATIVE REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 7 | 136 | 5.818e-06 | 5.593e-05 |
| 485 | EMBRYONIC CAMERA TYPE EYE MORPHOGENESIS | 4 | 24 | 6.023e-06 | 5.767e-05 |
| 486 | REGULATION OF PROTEIN PHOSPHATASE TYPE 2A ACTIVITY | 4 | 24 | 6.023e-06 | 5.767e-05 |
| 487 | REGULATION OF GLIOGENESIS | 6 | 90 | 6.309e-06 | 6.028e-05 |
| 488 | CELL CYCLE G2 M PHASE TRANSITION | 7 | 138 | 6.406e-06 | 6.108e-05 |
| 489 | REGULATION OF NEURON PROJECTION DEVELOPMENT | 11 | 408 | 6.826e-06 | 6.495e-05 |
| 490 | CARDIAC MUSCLE TISSUE DEVELOPMENT | 7 | 140 | 7.043e-06 | 6.674e-05 |
| 491 | POSITIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 5 | 53 | 7.038e-06 | 6.674e-05 |
| 492 | FOREBRAIN REGIONALIZATION | 4 | 25 | 7.143e-06 | 6.741e-05 |
| 493 | REGULATION OF TRANSFORMING GROWTH FACTOR BETA PRODUCTION | 4 | 25 | 7.143e-06 | 6.741e-05 |
| 494 | INNER EAR MORPHOGENESIS | 6 | 92 | 7.167e-06 | 6.751e-05 |
| 495 | NEGATIVE REGULATION OF APOPTOTIC SIGNALING PATHWAY | 8 | 200 | 7.932e-06 | 7.456e-05 |
| 496 | NEGATIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 14 | 684 | 8.614e-06 | 8.081e-05 |
| 497 | SYNAPSE ORGANIZATION | 7 | 145 | 8.868e-06 | 8.302e-05 |
| 498 | REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 6 | 97 | 9.733e-06 | 9.094e-05 |
| 499 | RESPONSE TO LITHIUM ION | 4 | 27 | 9.833e-06 | 9.169e-05 |
| 500 | NEGATIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 6 | 98 | 1.033e-05 | 9.59e-05 |
| 501 | REGULATION OF ENDOTHELIAL CELL PROLIFERATION | 6 | 98 | 1.033e-05 | 9.59e-05 |
| 502 | MUSCLE TISSUE DEVELOPMENT | 9 | 275 | 1.067e-05 | 9.894e-05 |
| 503 | REGULATION OF TRANSFORMING GROWTH FACTOR BETA RECEPTOR SIGNALING PATHWAY | 6 | 99 | 1.095e-05 | 0.0001009 |
| 504 | REGULATION OF CELLULAR RESPONSE TO TRANSFORMING GROWTH FACTOR BETA STIMULUS | 6 | 99 | 1.095e-05 | 0.0001009 |
| 505 | LYMPHOCYTE DIFFERENTIATION | 8 | 209 | 1.094e-05 | 0.0001009 |
| 506 | CELL PROJECTION ORGANIZATION | 16 | 902 | 1.123e-05 | 0.0001032 |
| 507 | GASTRULATION WITH MOUTH FORMING SECOND | 4 | 28 | 1.143e-05 | 0.0001039 |
| 508 | CELL FATE COMMITMENT INVOLVED IN FORMATION OF PRIMARY GERM LAYER | 4 | 28 | 1.143e-05 | 0.0001039 |
| 509 | MORPHOGENESIS OF A POLARIZED EPITHELIUM | 4 | 28 | 1.143e-05 | 0.0001039 |
| 510 | DOPAMINERGIC NEURON DIFFERENTIATION | 4 | 28 | 1.143e-05 | 0.0001039 |
| 511 | MAMMARY GLAND DUCT MORPHOGENESIS | 4 | 28 | 1.143e-05 | 0.0001039 |
| 512 | METANEPHROS MORPHOGENESIS | 4 | 28 | 1.143e-05 | 0.0001039 |
| 513 | REGULATION OF SMOOTH MUSCLE CELL PROLIFERATION | 6 | 100 | 1.16e-05 | 0.0001052 |
| 514 | REGULATION OF GLIAL CELL DIFFERENTIATION | 5 | 59 | 1.199e-05 | 0.0001085 |
| 515 | KERATINOCYTE DIFFERENTIATION | 6 | 101 | 1.228e-05 | 0.000111 |
| 516 | REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 8 | 213 | 1.255e-05 | 0.0001132 |
| 517 | REGULATION OF CARDIAC MUSCLE CELL PROLIFERATION | 4 | 29 | 1.32e-05 | 0.000118 |
| 518 | AMEBOIDAL TYPE CELL MIGRATION | 7 | 154 | 1.314e-05 | 0.000118 |
| 519 | PROTEIN COMPLEX BIOGENESIS | 18 | 1132 | 1.324e-05 | 0.000118 |
| 520 | EMBRYONIC HINDLIMB MORPHOGENESIS | 4 | 29 | 1.32e-05 | 0.000118 |
| 521 | REGULATION OF OLIGODENDROCYTE DIFFERENTIATION | 4 | 29 | 1.32e-05 | 0.000118 |
| 522 | PROTEIN COMPLEX ASSEMBLY | 18 | 1132 | 1.324e-05 | 0.000118 |
| 523 | REGULATION OF MITOCHONDRION ORGANIZATION | 8 | 218 | 1.485e-05 | 0.0001321 |
| 524 | HORMONE MEDIATED SIGNALING PATHWAY | 7 | 158 | 1.552e-05 | 0.0001378 |
| 525 | CARDIAC VENTRICLE DEVELOPMENT | 6 | 106 | 1.62e-05 | 0.0001436 |
| 526 | TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 14 | 724 | 1.629e-05 | 0.0001441 |
| 527 | NEGATIVE REGULATION OF CELL ADHESION | 8 | 223 | 1.749e-05 | 0.0001544 |
| 528 | REGENERATION | 7 | 161 | 1.754e-05 | 0.0001545 |
| 529 | POSITIVE REGULATION OF BMP SIGNALING PATHWAY | 4 | 32 | 1.976e-05 | 0.0001735 |
| 530 | EMBRYONIC FORELIMB MORPHOGENESIS | 4 | 32 | 1.976e-05 | 0.0001735 |
| 531 | REGULATION OF ODONTOGENESIS OF DENTIN CONTAINING TOOTH | 3 | 11 | 2.003e-05 | 0.0001755 |
| 532 | PROTEIN LOCALIZATION TO MEMBRANE | 10 | 376 | 2.059e-05 | 0.0001801 |
| 533 | NEGATIVE REGULATION OF CELLULAR RESPONSE TO TRANSFORMING GROWTH FACTOR BETA STIMULUS | 5 | 66 | 2.081e-05 | 0.0001813 |
| 534 | NEGATIVE REGULATION OF TRANSFORMING GROWTH FACTOR BETA RECEPTOR SIGNALING PATHWAY | 5 | 66 | 2.081e-05 | 0.0001813 |
| 535 | EAR MORPHOGENESIS | 6 | 112 | 2.217e-05 | 0.0001928 |
| 536 | POSITIVE REGULATION OF ENDOTHELIAL CELL MIGRATION | 5 | 67 | 2.24e-05 | 0.0001937 |
| 537 | REGULATION OF T CELL APOPTOTIC PROCESS | 4 | 33 | 2.24e-05 | 0.0001937 |
| 538 | EMBRYONIC EYE MORPHOGENESIS | 4 | 33 | 2.24e-05 | 0.0001937 |
| 539 | POSITIVE REGULATION OF NEURON PROJECTION DEVELOPMENT | 8 | 232 | 2.324e-05 | 0.0002006 |
| 540 | REGULATION OF ENDOTHELIAL CELL MIGRATION | 6 | 114 | 2.451e-05 | 0.0002112 |
| 541 | NEGATIVE REGULATION OF CELL GROWTH | 7 | 170 | 2.491e-05 | 0.0002142 |
| 542 | LUNG EPITHELIUM DEVELOPMENT | 4 | 34 | 2.529e-05 | 0.0002167 |
| 543 | PROTEIN DESTABILIZATION | 4 | 34 | 2.529e-05 | 0.0002167 |
| 544 | NEGATIVE REGULATION OF OSSIFICATION | 5 | 69 | 2.586e-05 | 0.0002207 |
| 545 | SYNAPSE ASSEMBLY | 5 | 69 | 2.586e-05 | 0.0002207 |
| 546 | RESPONSE TO NITROGEN COMPOUND | 15 | 859 | 2.613e-05 | 0.0002226 |
| 547 | NEGATIVE REGULATION OF CARDIAC MUSCLE TISSUE GROWTH | 3 | 12 | 2.661e-05 | 0.0002247 |
| 548 | ANATOMICAL STRUCTURE REGRESSION | 3 | 12 | 2.661e-05 | 0.0002247 |
| 549 | REGULATION OF THYMOCYTE APOPTOTIC PROCESS | 3 | 12 | 2.661e-05 | 0.0002247 |
| 550 | REGULATION OF MACROPHAGE CYTOKINE PRODUCTION | 3 | 12 | 2.661e-05 | 0.0002247 |
| 551 | NEGATIVE REGULATION OF HEART GROWTH | 3 | 12 | 2.661e-05 | 0.0002247 |
| 552 | MUSCLE ORGAN MORPHOGENESIS | 5 | 70 | 2.773e-05 | 0.0002338 |
| 553 | EMBRYONIC CAMERA TYPE EYE DEVELOPMENT | 4 | 35 | 2.844e-05 | 0.0002393 |
| 554 | REGULATION OF PEPTIDASE ACTIVITY | 10 | 392 | 2.937e-05 | 0.0002466 |
| 555 | ENDODERM DEVELOPMENT | 5 | 71 | 2.972e-05 | 0.0002487 |
| 556 | CELL FATE SPECIFICATION | 5 | 71 | 2.972e-05 | 0.0002487 |
| 557 | CELLULAR RESPONSE TO STRESS | 21 | 1565 | 2.982e-05 | 0.0002491 |
| 558 | GLIOGENESIS | 7 | 175 | 3e-05 | 0.0002493 |
| 559 | POSITIVE REGULATION OF CYTOSKELETON ORGANIZATION | 7 | 175 | 3e-05 | 0.0002493 |
| 560 | REGULATION OF PROTEIN CATABOLIC PROCESS | 10 | 393 | 3.001e-05 | 0.0002493 |
| 561 | MITOTIC CELL CYCLE | 14 | 766 | 3.036e-05 | 0.0002518 |
| 562 | MULTICELLULAR ORGANISM REPRODUCTION | 14 | 768 | 3.124e-05 | 0.0002587 |
| 563 | HEAD MORPHOGENESIS | 4 | 36 | 3.187e-05 | 0.0002634 |
| 564 | MITOCHONDRIAL TRANSPORT | 7 | 177 | 3.226e-05 | 0.0002662 |
| 565 | NEGATIVE REGULATION OF HYDROLASE ACTIVITY | 10 | 397 | 3.27e-05 | 0.0002693 |
| 566 | PANCREAS DEVELOPMENT | 5 | 73 | 3.401e-05 | 0.0002791 |
| 567 | EMBRYONIC HEART TUBE DEVELOPMENT | 5 | 73 | 3.401e-05 | 0.0002791 |
| 568 | NEGATIVE REGULATION OF OLIGODENDROCYTE DIFFERENTIATION | 3 | 13 | 3.446e-05 | 0.0002813 |
| 569 | REGULATION OF CELL PROLIFERATION INVOLVED IN KIDNEY DEVELOPMENT | 3 | 13 | 3.446e-05 | 0.0002813 |
| 570 | GDP METABOLIC PROCESS | 3 | 13 | 3.446e-05 | 0.0002813 |
| 571 | HINDLIMB MORPHOGENESIS | 4 | 37 | 3.56e-05 | 0.0002901 |
| 572 | POSITIVE REGULATION OF CELL CYCLE PROCESS | 8 | 247 | 3.633e-05 | 0.0002955 |
| 573 | T CELL DIFFERENTIATION | 6 | 123 | 3.766e-05 | 0.0003058 |
| 574 | MACROMOLECULAR COMPLEX DISASSEMBLY | 7 | 182 | 3.854e-05 | 0.0003124 |
| 575 | ARTERY DEVELOPMENT | 5 | 75 | 3.877e-05 | 0.0003137 |
| 576 | REGULATION OF MULTICELLULAR ORGANISMAL METABOLIC PROCESS | 4 | 38 | 3.963e-05 | 0.0003201 |
| 577 | RENAL VESICLE DEVELOPMENT | 3 | 14 | 4.37e-05 | 0.0003494 |
| 578 | POSITIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY IN ABSENCE OF LIGAND | 3 | 14 | 4.37e-05 | 0.0003494 |
| 579 | CELL MIGRATION INVOLVED IN GASTRULATION | 3 | 14 | 4.37e-05 | 0.0003494 |
| 580 | METANEPHRIC MESENCHYME DEVELOPMENT | 3 | 14 | 4.37e-05 | 0.0003494 |
| 581 | CONVERGENT EXTENSION | 3 | 14 | 4.37e-05 | 0.0003494 |
| 582 | ADENOHYPOPHYSIS DEVELOPMENT | 3 | 14 | 4.37e-05 | 0.0003494 |
| 583 | COCHLEA DEVELOPMENT | 4 | 39 | 4.399e-05 | 0.0003505 |
| 584 | GLANDULAR EPITHELIAL CELL DIFFERENTIATION | 4 | 39 | 4.399e-05 | 0.0003505 |
| 585 | FORELIMB MORPHOGENESIS | 4 | 40 | 4.869e-05 | 0.0003873 |
| 586 | POSITIVE REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 6 | 129 | 4.92e-05 | 0.00039 |
| 587 | REGULATION OF REPRODUCTIVE PROCESS | 6 | 129 | 4.92e-05 | 0.00039 |
| 588 | POSITIVE REGULATION OF INTRACELLULAR PROTEIN TRANSPORT | 8 | 258 | 4.943e-05 | 0.0003912 |
| 589 | NEGATIVE REGULATION OF VASCULATURE DEVELOPMENT | 5 | 80 | 5.293e-05 | 0.0004182 |
| 590 | POSITIVE REGULATION OF KIDNEY DEVELOPMENT | 4 | 41 | 5.374e-05 | 0.0004231 |
| 591 | PROSTATE GLAND DEVELOPMENT | 4 | 41 | 5.374e-05 | 0.0004231 |
| 592 | MESENCHYMAL TO EPITHELIAL TRANSITION | 3 | 15 | 5.442e-05 | 0.0004242 |
| 593 | NEGATIVE REGULATION OF DENDRITE MORPHOGENESIS | 3 | 15 | 5.442e-05 | 0.0004242 |
| 594 | REGULATION OF EPITHELIAL CELL DIFFERENTIATION INVOLVED IN KIDNEY DEVELOPMENT | 3 | 15 | 5.442e-05 | 0.0004242 |
| 595 | REGULATION OF NEURON APOPTOTIC PROCESS | 7 | 192 | 5.412e-05 | 0.0004242 |
| 596 | REGULATION OF CELL PROLIFERATION INVOLVED IN HEART MORPHOGENESIS | 3 | 15 | 5.442e-05 | 0.0004242 |
| 597 | POSITIVE REGULATION OF T CELL APOPTOTIC PROCESS | 3 | 15 | 5.442e-05 | 0.0004242 |
| 598 | POSITIVE REGULATION OF CELL DIVISION | 6 | 132 | 5.594e-05 | 0.0004346 |
| 599 | MAINTENANCE OF CELL NUMBER | 6 | 132 | 5.594e-05 | 0.0004346 |
| 600 | POSITIVE REGULATION OF PROTEIN CATABOLIC PROCESS | 8 | 263 | 5.658e-05 | 0.000438 |
| 601 | NEGATIVE REGULATION OF LOCOMOTION | 8 | 263 | 5.658e-05 | 0.000438 |
| 602 | POSTTRANSCRIPTIONAL GENE SILENCING | 4 | 42 | 5.917e-05 | 0.0004566 |
| 603 | REGULATION OF HEART GROWTH | 4 | 42 | 5.917e-05 | 0.0004566 |
| 604 | EMBRYONIC PLACENTA DEVELOPMENT | 5 | 83 | 6.317e-05 | 0.0004867 |
| 605 | MACROMOLECULAR COMPLEX ASSEMBLY | 19 | 1398 | 6.445e-05 | 0.0004956 |
| 606 | NEGATIVE REGULATION OF STEM CELL DIFFERENTIATION | 4 | 43 | 6.499e-05 | 0.0004982 |
| 607 | MORPHOGENESIS OF AN EPITHELIAL SHEET | 4 | 43 | 6.499e-05 | 0.0004982 |
| 608 | RESPONSE TO DRUG | 10 | 431 | 6.515e-05 | 0.0004986 |
| 609 | GLIAL CELL DIFFERENTIATION | 6 | 136 | 6.607e-05 | 0.0005048 |
| 610 | PROTEIN LOCALIZATION TO SYNAPSE | 3 | 16 | 6.674e-05 | 0.0005082 |
| 611 | APOPTOTIC PROCESS INVOLVED IN MORPHOGENESIS | 3 | 16 | 6.674e-05 | 0.0005082 |
| 612 | HINDBRAIN DEVELOPMENT | 6 | 137 | 6.881e-05 | 0.0005232 |
| 613 | LABYRINTHINE LAYER DEVELOPMENT | 4 | 44 | 7.121e-05 | 0.0005397 |
| 614 | BODY MORPHOGENESIS | 4 | 44 | 7.121e-05 | 0.0005397 |
| 615 | PLACENTA DEVELOPMENT | 6 | 138 | 7.165e-05 | 0.0005421 |
| 616 | ENDOTHELIAL CELL DEVELOPMENT | 4 | 45 | 7.786e-05 | 0.0005871 |
| 617 | REGULATION OF ALCOHOL BIOSYNTHETIC PROCESS | 4 | 45 | 7.786e-05 | 0.0005871 |
| 618 | TISSUE REMODELING | 5 | 87 | 7.912e-05 | 0.0005957 |
| 619 | NEGATIVE REGULATION OF KIDNEY DEVELOPMENT | 3 | 17 | 8.074e-05 | 0.000604 |
| 620 | CELLULAR RESPONSE TO LITHIUM ION | 3 | 17 | 8.074e-05 | 0.000604 |
| 621 | NEGATIVE REGULATION OF ALCOHOL BIOSYNTHETIC PROCESS | 3 | 17 | 8.074e-05 | 0.000604 |
| 622 | REGULATION OF STEM CELL POPULATION MAINTENANCE | 3 | 17 | 8.074e-05 | 0.000604 |
| 623 | REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY IN ABSENCE OF LIGAND | 4 | 46 | 8.494e-05 | 0.0006344 |
| 624 | POSITIVE REGULATION OF CYTOPLASMIC TRANSPORT | 8 | 282 | 9.206e-05 | 0.0006865 |
| 625 | POSITIVE REGULATION OF NEURON APOPTOTIC PROCESS | 4 | 47 | 9.249e-05 | 0.0006874 |
| 626 | POSITIVE REGULATION OF GLIOGENESIS | 4 | 47 | 9.249e-05 | 0.0006874 |
| 627 | POSITIVE REGULATION OF PROTEOLYSIS | 9 | 363 | 9.312e-05 | 0.000691 |
| 628 | NEGATIVE REGULATION OF MAPK CASCADE | 6 | 145 | 9.422e-05 | 0.0006981 |
| 629 | PERICARDIUM DEVELOPMENT | 3 | 18 | 9.653e-05 | 0.0007118 |
| 630 | OSTEOBLAST DEVELOPMENT | 3 | 18 | 9.653e-05 | 0.0007118 |
| 631 | UTERUS DEVELOPMENT | 3 | 18 | 9.653e-05 | 0.0007118 |
| 632 | POSITIVE REGULATION OF CELL GROWTH | 6 | 148 | 0.0001055 | 0.0007766 |
| 633 | REGULATION OF STEROID BIOSYNTHETIC PROCESS | 4 | 49 | 0.000109 | 0.0008013 |
| 634 | POSITIVE REGULATION OF CHONDROCYTE DIFFERENTIATION | 3 | 19 | 0.0001142 | 0.0008382 |
| 635 | FACE DEVELOPMENT | 4 | 50 | 0.000118 | 0.0008649 |
| 636 | ARTERY MORPHOGENESIS | 4 | 51 | 0.0001276 | 0.0009335 |
| 637 | REGULATION OF CYTOKINE PRODUCTION | 11 | 563 | 0.0001285 | 0.0009386 |
| 638 | POSITIVE REGULATION OF LYMPHOCYTE APOPTOTIC PROCESS | 3 | 20 | 0.0001339 | 0.0009733 |
| 639 | GMP METABOLIC PROCESS | 3 | 20 | 0.0001339 | 0.0009733 |
| 640 | BRANCHING INVOLVED IN MAMMARY GLAND DUCT MORPHOGENESIS | 3 | 20 | 0.0001339 | 0.0009733 |
| 641 | POSITIVE REGULATION OF DEVELOPMENTAL GROWTH | 6 | 156 | 0.0001408 | 0.00102 |
| 642 | PROTEIN LOCALIZATION TO NUCLEUS | 6 | 156 | 0.0001408 | 0.00102 |
| 643 | CHONDROCYTE DEVELOPMENT | 3 | 21 | 0.0001556 | 0.001123 |
| 644 | APOPTOTIC PROCESS INVOLVED IN DEVELOPMENT | 3 | 21 | 0.0001556 | 0.001123 |
| 645 | NEGATIVE REGULATION OF NEURAL PRECURSOR CELL PROLIFERATION | 3 | 21 | 0.0001556 | 0.001123 |
| 646 | TELENCEPHALON DEVELOPMENT | 7 | 228 | 0.0001583 | 0.00114 |
| 647 | NEGATIVE REGULATION OF REPRODUCTIVE PROCESS | 4 | 54 | 0.0001596 | 0.001144 |
| 648 | REGULATION OF LYMPHOCYTE APOPTOTIC PROCESS | 4 | 54 | 0.0001596 | 0.001144 |
| 649 | CARDIAC MUSCLE TISSUE MORPHOGENESIS | 4 | 54 | 0.0001596 | 0.001144 |
| 650 | REGULATION OF GENE EXPRESSION EPIGENETIC | 7 | 229 | 0.0001626 | 0.001164 |
| 651 | SINGLE ORGANISM CELLULAR LOCALIZATION | 14 | 898 | 0.0001655 | 0.001183 |
| 652 | REGULATION OF LEUKOCYTE DIFFERENTIATION | 7 | 232 | 0.0001761 | 0.001257 |
| 653 | REGULATION OF NUCLEAR DIVISION | 6 | 163 | 0.0001788 | 0.001274 |
| 654 | EMBRYONIC PLACENTA MORPHOGENESIS | 3 | 22 | 0.0001795 | 0.001277 |
| 655 | POSITIVE REGULATION OF PROTEIN IMPORT | 5 | 104 | 0.000184 | 0.001305 |
| 656 | OUTFLOW TRACT MORPHOGENESIS | 4 | 56 | 0.0001839 | 0.001305 |
| 657 | CELL SUBSTRATE ADHESION | 6 | 164 | 0.0001849 | 0.001309 |
| 658 | REGULATION OF CYTOKINE PRODUCTION INVOLVED IN IMMUNE RESPONSE | 4 | 57 | 0.0001971 | 0.001391 |
| 659 | ENDOTHELIAL CELL MIGRATION | 4 | 57 | 0.0001971 | 0.001391 |
| 660 | CENTRAL NERVOUS SYSTEM NEURON DIFFERENTIATION | 6 | 166 | 0.0001975 | 0.001392 |
| 661 | MUSCLE CELL DIFFERENTIATION | 7 | 237 | 0.0002007 | 0.001413 |
| 662 | FAT CELL DIFFERENTIATION | 5 | 106 | 0.0002012 | 0.001414 |
| 663 | ESTABLISHMENT OF EPITHELIAL CELL POLARITY | 3 | 23 | 0.0002057 | 0.001441 |
| 664 | SCF DEPENDENT PROTEASOMAL UBIQUITIN DEPENDENT PROTEIN CATABOLIC PROCESS | 3 | 23 | 0.0002057 | 0.001441 |
| 665 | REGULATION OF IMMUNE SYSTEM PROCESS | 18 | 1403 | 0.0002126 | 0.001488 |
| 666 | REGULATION OF EPITHELIAL CELL APOPTOTIC PROCESS | 4 | 59 | 0.0002253 | 0.001574 |
| 667 | NEGATIVE REGULATION OF STEROID METABOLIC PROCESS | 3 | 24 | 0.0002342 | 0.001627 |
| 668 | REGULATION OF ODONTOGENESIS | 3 | 24 | 0.0002342 | 0.001627 |
| 669 | ENDOTHELIAL CELL PROLIFERATION | 3 | 24 | 0.0002342 | 0.001627 |
| 670 | POSITIVE REGULATION OF CELL CELL ADHESION | 7 | 243 | 0.0002338 | 0.001627 |
| 671 | NEGATIVE REGULATION OF CELL DIVISION | 4 | 60 | 0.0002405 | 0.001663 |
| 672 | POSITIVE REGULATION OF SMOOTH MUSCLE CELL PROLIFERATION | 4 | 60 | 0.0002405 | 0.001663 |
| 673 | REGULATION OF PHOSPHOPROTEIN PHOSPHATASE ACTIVITY | 4 | 60 | 0.0002405 | 0.001663 |
| 674 | REGULATION OF CELL SUBSTRATE ADHESION | 6 | 173 | 0.0002468 | 0.001704 |
| 675 | OVARIAN FOLLICLE DEVELOPMENT | 4 | 61 | 0.0002563 | 0.001767 |
| 676 | LENS FIBER CELL DIFFERENTIATION | 3 | 25 | 0.0002652 | 0.001812 |
| 677 | EPITHELIAL TUBE BRANCHING INVOLVED IN LUNG MORPHOGENESIS | 3 | 25 | 0.0002652 | 0.001812 |
| 678 | LUNG CELL DIFFERENTIATION | 3 | 25 | 0.0002652 | 0.001812 |
| 679 | REGULATION OF FIBROBLAST GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 3 | 25 | 0.0002652 | 0.001812 |
| 680 | POSITIVE REGULATION OF PEPTIDYL THREONINE PHOSPHORYLATION | 3 | 25 | 0.0002652 | 0.001812 |
| 681 | EPITHELIAL CELL APOPTOTIC PROCESS | 3 | 25 | 0.0002652 | 0.001812 |
| 682 | REGULATION OF SMOOTHENED SIGNALING PATHWAY | 4 | 62 | 0.000273 | 0.001862 |
| 683 | NEUROEPITHELIAL CELL DIFFERENTIATION | 4 | 63 | 0.0002903 | 0.001978 |
| 684 | REGULATION OF NEURON DEATH | 7 | 252 | 0.0002916 | 0.001984 |
| 685 | DEVELOPMENTAL PROGRAMMED CELL DEATH | 3 | 26 | 0.0002987 | 0.00202 |
| 686 | NEGATIVE REGULATION OF GLIAL CELL DIFFERENTIATION | 3 | 26 | 0.0002987 | 0.00202 |
| 687 | REGULATION OF HORMONE METABOLIC PROCESS | 3 | 26 | 0.0002987 | 0.00202 |
| 688 | REGULATION OF P38MAPK CASCADE | 3 | 26 | 0.0002987 | 0.00202 |
| 689 | REGULATION OF CHEMOTAXIS | 6 | 180 | 0.0003055 | 0.002063 |
| 690 | REGULATION OF CELLULAR COMPONENT SIZE | 8 | 337 | 0.0003103 | 0.002092 |
| 691 | IMMUNE SYSTEM PROCESS | 22 | 1984 | 0.0003109 | 0.002094 |
| 692 | REGULATION OF CATABOLIC PROCESS | 12 | 731 | 0.0003153 | 0.00212 |
| 693 | REGULATION OF DNA METABOLIC PROCESS | 8 | 340 | 0.0003292 | 0.00221 |
| 694 | REGULATION OF ENDOTHELIAL CELL DIFFERENTIATION | 3 | 27 | 0.0003348 | 0.002241 |
| 695 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER INVOLVED IN CELLULAR RESPONSE TO CHEMICAL STIMULUS | 3 | 27 | 0.0003348 | 0.002241 |
| 696 | SOMATIC STEM CELL POPULATION MAINTENANCE | 4 | 66 | 0.0003473 | 0.002322 |
| 697 | POSITIVE REGULATION OF DNA METABOLIC PROCESS | 6 | 185 | 0.0003536 | 0.002361 |
| 698 | POSITIVE REGULATION OF NEURON DEATH | 4 | 67 | 0.0003679 | 0.002452 |
| 699 | POSITIVE REGULATION OF LEUKOCYTE APOPTOTIC PROCESS | 3 | 28 | 0.0003736 | 0.00248 |
| 700 | NEGATIVE REGULATION OF DENDRITE DEVELOPMENT | 3 | 28 | 0.0003736 | 0.00248 |
| 701 | REGULATION OF NEUROBLAST PROLIFERATION | 3 | 28 | 0.0003736 | 0.00248 |
| 702 | CELLULAR RESPONSE TO ABIOTIC STIMULUS | 7 | 263 | 0.0003773 | 0.002501 |
| 703 | CELLULAR RESPONSE TO EXTERNAL STIMULUS | 7 | 264 | 0.000386 | 0.002552 |
| 704 | LOCALIZATION WITHIN MEMBRANE | 5 | 122 | 0.0003862 | 0.002552 |
| 705 | REGULATION OF CELL JUNCTION ASSEMBLY | 4 | 68 | 0.0003894 | 0.00257 |
| 706 | RESPONSE TO EXTRACELLULAR STIMULUS | 9 | 441 | 0.0003955 | 0.002607 |
| 707 | PROTEIN DEPHOSPHORYLATION | 6 | 190 | 0.0004076 | 0.002682 |
| 708 | EPIDERMIS MORPHOGENESIS | 3 | 29 | 0.0004152 | 0.002725 |
| 709 | NEUROBLAST PROLIFERATION | 3 | 29 | 0.0004152 | 0.002725 |
| 710 | STEROID HORMONE MEDIATED SIGNALING PATHWAY | 5 | 125 | 0.0004318 | 0.00283 |
| 711 | PROTEASOMAL PROTEIN CATABOLIC PROCESS | 7 | 271 | 0.0004515 | 0.002955 |
| 712 | CELL CYCLE CHECKPOINT | 6 | 194 | 0.0004552 | 0.002975 |
| 713 | SMOOTH MUSCLE CELL DIFFERENTIATION | 3 | 30 | 0.0004596 | 0.002991 |
| 714 | CELL SURFACE RECEPTOR SIGNALING PATHWAY INVOLVED IN CELL CELL SIGNALING | 4 | 71 | 0.0004592 | 0.002991 |
| 715 | ESTABLISHMENT OF ENDOTHELIAL BARRIER | 3 | 30 | 0.0004596 | 0.002991 |
| 716 | CELLULAR RESPONSE TO HORMONE STIMULUS | 10 | 552 | 0.0004789 | 0.003112 |
| 717 | NEGATIVE REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 7 | 274 | 0.0004821 | 0.003129 |
| 718 | NEGATIVE REGULATION OF MAP KINASE ACTIVITY | 4 | 73 | 0.0005105 | 0.003308 |
| 719 | NEGATIVE REGULATION OF TRANSPORT | 9 | 458 | 0.0005197 | 0.003363 |
| 720 | NEGATIVE REGULATION OF BINDING | 5 | 131 | 0.000535 | 0.003457 |
| 721 | REGULATION OF STEROID METABOLIC PROCESS | 4 | 74 | 0.0005376 | 0.003469 |
| 722 | NEGATIVE REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 3 | 32 | 0.0005574 | 0.003577 |
| 723 | SALIVARY GLAND DEVELOPMENT | 3 | 32 | 0.0005574 | 0.003577 |
| 724 | POSITIVE REGULATION OF GLIAL CELL DIFFERENTIATION | 3 | 32 | 0.0005574 | 0.003577 |
| 725 | PATTERNING OF BLOOD VESSELS | 3 | 32 | 0.0005574 | 0.003577 |
| 726 | NEURAL CREST CELL DIFFERENTIATION | 4 | 75 | 0.0005657 | 0.003625 |
| 727 | POSITIVE REGULATION OF VASCULATURE DEVELOPMENT | 5 | 133 | 0.0005732 | 0.003669 |
| 728 | REGULATION OF MYELINATION | 3 | 33 | 0.0006109 | 0.003899 |
| 729 | EMBRYONIC DIGESTIVE TRACT DEVELOPMENT | 3 | 33 | 0.0006109 | 0.003899 |
| 730 | NEGATIVE REGULATION OF DEPHOSPHORYLATION | 4 | 77 | 0.000625 | 0.003984 |
| 731 | POSITIVE REGULATION OF HYDROLASE ACTIVITY | 13 | 905 | 0.0006261 | 0.003985 |
| 732 | CIRCADIAN RHYTHM | 5 | 137 | 0.0006557 | 0.004168 |
| 733 | HEART VALVE DEVELOPMENT | 3 | 34 | 0.0006676 | 0.004226 |
| 734 | NEGATIVE REGULATION OF MITOTIC NUCLEAR DIVISION | 3 | 34 | 0.0006676 | 0.004226 |
| 735 | BRAIN MORPHOGENESIS | 3 | 34 | 0.0006676 | 0.004226 |
| 736 | NEGATIVE REGULATION OF CELL CELL ADHESION | 5 | 138 | 0.0006777 | 0.004284 |
| 737 | REGULATION OF LEUKOCYTE APOPTOTIC PROCESS | 4 | 79 | 0.0006886 | 0.004341 |
| 738 | NEGATIVE REGULATION OF PROTEIN BINDING | 4 | 79 | 0.0006886 | 0.004341 |
| 739 | MITOTIC CELL CYCLE CHECKPOINT | 5 | 139 | 0.0007002 | 0.004409 |
| 740 | BONE REMODELING | 3 | 35 | 0.0007275 | 0.004574 |
| 741 | REGULATION OF FIBROBLAST PROLIFERATION | 4 | 81 | 0.0007566 | 0.004751 |
| 742 | NEGATIVE REGULATION OF CELL CYCLE PROCESS | 6 | 214 | 0.000762 | 0.004779 |
| 743 | POSITIVE REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 3 | 36 | 0.0007907 | 0.004952 |
| 744 | NEGATIVE REGULATION OF LEUKOCYTE DIFFERENTIATION | 4 | 82 | 0.0007923 | 0.004955 |
| 745 | NEGATIVE REGULATION OF CELL PROJECTION ORGANIZATION | 5 | 144 | 0.0008212 | 0.005129 |
| 746 | RESPONSE TO IONIZING RADIATION | 5 | 145 | 0.0008472 | 0.005284 |
| 747 | NEGATIVE REGULATION OF GLIOGENESIS | 3 | 37 | 0.0008574 | 0.00534 |
| 748 | TISSUE MIGRATION | 4 | 84 | 0.0008674 | 0.005396 |
| 749 | POSITIVE REGULATION OF CATABOLIC PROCESS | 8 | 395 | 0.0008812 | 0.005474 |
| 750 | BONE MINERALIZATION | 3 | 38 | 0.0009275 | 0.005754 |
| 751 | PEPTIDYL SERINE MODIFICATION | 5 | 148 | 0.0009287 | 0.005754 |
| 752 | POSITIVE REGULATION OF DNA REPLICATION | 4 | 86 | 0.0009473 | 0.005861 |
| 753 | NEGATIVE REGULATION OF TRANSCRIPTION FACTOR IMPORT INTO NUCLEUS | 3 | 39 | 0.001001 | 0.00617 |
| 754 | REGULATION OF AXON GUIDANCE | 3 | 39 | 0.001001 | 0.00617 |
| 755 | REGULATION OF CELL ADHESION MEDIATED BY INTEGRIN | 3 | 39 | 0.001001 | 0.00617 |
| 756 | RESPONSE TO PEPTIDE | 8 | 404 | 0.001018 | 0.006268 |
| 757 | CELLULAR RESPONSE TO NITROGEN COMPOUND | 9 | 505 | 0.001038 | 0.006382 |
| 758 | PROTEIN TARGETING | 8 | 406 | 0.001051 | 0.006452 |
| 759 | REGULATION OF HEMOPOIESIS | 7 | 314 | 0.001073 | 0.00658 |
| 760 | POSITIVE REGULATION OF CELL MATRIX ADHESION | 3 | 40 | 0.001078 | 0.006584 |
| 761 | ENDODERMAL CELL DIFFERENTIATION | 3 | 40 | 0.001078 | 0.006584 |
| 762 | TRANSCRIPTION INITIATION FROM RNA POLYMERASE II PROMOTER | 5 | 153 | 0.001078 | 0.006584 |
| 763 | REGULATION OF CELL MATRIX ADHESION | 4 | 90 | 0.001122 | 0.006845 |
| 764 | NEGATIVE REGULATION OF FAT CELL DIFFERENTIATION | 3 | 41 | 0.001159 | 0.007041 |
| 765 | NEGATIVE REGULATION OF STRESS ACTIVATED MAPK CASCADE | 3 | 41 | 0.001159 | 0.007041 |
| 766 | NEGATIVE REGULATION OF STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE | 3 | 41 | 0.001159 | 0.007041 |
| 767 | REGULATION OF ENDOTHELIAL CELL APOPTOTIC PROCESS | 3 | 42 | 0.001244 | 0.007526 |
| 768 | NEGATIVE REGULATION OF BMP SIGNALING PATHWAY | 3 | 42 | 0.001244 | 0.007526 |
| 769 | POSITIVE REGULATION OF NEURAL PRECURSOR CELL PROLIFERATION | 3 | 42 | 0.001244 | 0.007526 |
| 770 | REGULATION OF DNA BINDING | 4 | 93 | 0.001268 | 0.007662 |
| 771 | REGULATION OF DNA BIOSYNTHETIC PROCESS | 4 | 94 | 0.001319 | 0.007961 |
| 772 | ENDOCARDIUM DEVELOPMENT | 2 | 11 | 0.001348 | 0.008084 |
| 773 | NEGATIVE REGULATION OF FIBROBLAST GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 2 | 11 | 0.001348 | 0.008084 |
| 774 | POSITIVE REGULATION OF NON CANONICAL WNT SIGNALING PATHWAY | 2 | 11 | 0.001348 | 0.008084 |
| 775 | EPITHELIAL TO MESENCHYMAL TRANSITION INVOLVED IN ENDOCARDIAL CUSHION FORMATION | 2 | 11 | 0.001348 | 0.008084 |
| 776 | ENDOTHELIAL TUBE MORPHOGENESIS | 2 | 11 | 0.001348 | 0.008084 |
| 777 | REGULATION OF TRANSCRIPTION FACTOR IMPORT INTO NUCLEUS | 4 | 95 | 0.001372 | 0.008215 |
| 778 | POSITIVE REGULATION OF IMMUNE SYSTEM PROCESS | 12 | 867 | 0.001406 | 0.008412 |
| 779 | MYELOID LEUKOCYTE DIFFERENTIATION | 4 | 96 | 0.001426 | 0.008507 |
| 780 | NEGATIVE REGULATION OF LIPID BIOSYNTHETIC PROCESS | 3 | 44 | 0.001424 | 0.008507 |
| 781 | REGULATION OF MICROTUBULE BASED PROCESS | 6 | 243 | 0.001466 | 0.008732 |
| 782 | EXOCRINE SYSTEM DEVELOPMENT | 3 | 45 | 0.001521 | 0.009048 |
| 783 | ANTERIOR POSTERIOR AXIS SPECIFICATION EMBRYO | 2 | 12 | 0.001612 | 0.009485 |
| 784 | CARTILAGE MORPHOGENESIS | 2 | 12 | 0.001612 | 0.009485 |
| 785 | HEART FORMATION | 2 | 12 | 0.001612 | 0.009485 |
| 786 | MAINTENANCE OF CELL POLARITY | 2 | 12 | 0.001612 | 0.009485 |
| 787 | TRIPARTITE REGIONAL SUBDIVISION | 2 | 12 | 0.001612 | 0.009485 |
| 788 | COLLECTING DUCT DEVELOPMENT | 2 | 12 | 0.001612 | 0.009485 |
| 789 | POSITIVE REGULATION OF ASTROCYTE DIFFERENTIATION | 2 | 12 | 0.001612 | 0.009485 |
| 790 | EMBRYONIC CAMERA TYPE EYE FORMATION | 2 | 12 | 0.001612 | 0.009485 |
| 791 | LENS FIBER CELL DEVELOPMENT | 2 | 12 | 0.001612 | 0.009485 |
| 792 | NEGATIVE REGULATION OF DNA BINDING | 3 | 46 | 0.001621 | 0.009487 |
| 793 | GUANOSINE CONTAINING COMPOUND METABOLIC PROCESS | 3 | 46 | 0.001621 | 0.009487 |
| 794 | PEPTIDYL THREONINE MODIFICATION | 3 | 46 | 0.001621 | 0.009487 |
| 795 | NEGATIVE REGULATION OF NUCLEAR DIVISION | 3 | 46 | 0.001621 | 0.009487 |
| 796 | REGULATION OF AXONOGENESIS | 5 | 168 | 0.00163 | 0.009531 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | FRIZZLED BINDING | 15 | 36 | 6.105e-26 | 5.671e-23 |
| 2 | RECEPTOR BINDING | 40 | 1476 | 8.647e-20 | 4.016e-17 |
| 3 | TRANSFORMING GROWTH FACTOR BETA RECEPTOR BINDING | 12 | 50 | 1.452e-17 | 4.496e-15 |
| 4 | BETA CATENIN BINDING | 13 | 84 | 2.913e-16 | 6.765e-14 |
| 5 | WNT ACTIVATED RECEPTOR ACTIVITY | 9 | 22 | 6.988e-16 | 1.298e-13 |
| 6 | ENZYME BINDING | 38 | 1737 | 1.267e-15 | 1.961e-13 |
| 7 | KINASE BINDING | 23 | 606 | 2.768e-14 | 3.214e-12 |
| 8 | WNT PROTEIN BINDING | 9 | 31 | 2.728e-14 | 3.214e-12 |
| 9 | SMAD BINDING | 10 | 72 | 2.829e-12 | 2.92e-10 |
| 10 | G PROTEIN COUPLED RECEPTOR BINDING | 15 | 259 | 3.552e-12 | 3.3e-10 |
| 11 | I SMAD BINDING | 6 | 11 | 6.459e-12 | 5.455e-10 |
| 12 | PROTEIN DOMAIN SPECIFIC BINDING | 19 | 624 | 2.797e-10 | 2.165e-08 |
| 13 | GAMMA CATENIN BINDING | 5 | 12 | 2.289e-09 | 1.635e-07 |
| 14 | GROWTH FACTOR ACTIVITY | 10 | 160 | 8.327e-09 | 5.31e-07 |
| 15 | RECEPTOR SERINE THREONINE KINASE BINDING | 5 | 15 | 8.574e-09 | 5.31e-07 |
| 16 | RECEPTOR AGONIST ACTIVITY | 5 | 16 | 1.242e-08 | 6.788e-07 |
| 17 | CYTOKINE RECEPTOR BINDING | 12 | 271 | 1.228e-08 | 6.788e-07 |
| 18 | CYTOKINE ACTIVITY | 11 | 219 | 1.416e-08 | 7.306e-07 |
| 19 | RECEPTOR SIGNALING PROTEIN ACTIVITY | 10 | 172 | 1.666e-08 | 8.148e-07 |
| 20 | SIGNAL TRANSDUCER ACTIVITY | 27 | 1731 | 7.818e-08 | 3.631e-06 |
| 21 | IONOTROPIC GLUTAMATE RECEPTOR BINDING | 5 | 23 | 9.304e-08 | 4.116e-06 |
| 22 | MOLECULAR FUNCTION REGULATOR | 23 | 1353 | 2.006e-07 | 8.471e-06 |
| 23 | PROTEIN DIMERIZATION ACTIVITY | 21 | 1149 | 2.307e-07 | 9.319e-06 |
| 24 | PDZ DOMAIN BINDING | 7 | 90 | 3.599e-07 | 1.393e-05 |
| 25 | RECEPTOR ACTIVATOR ACTIVITY | 5 | 32 | 5.371e-07 | 1.996e-05 |
| 26 | REGULATORY REGION NUCLEIC ACID BINDING | 17 | 818 | 6.759e-07 | 2.415e-05 |
| 27 | UBIQUITIN LIKE PROTEIN LIGASE BINDING | 10 | 264 | 9.065e-07 | 3.119e-05 |
| 28 | GLUTAMATE RECEPTOR BINDING | 5 | 37 | 1.14e-06 | 3.781e-05 |
| 29 | TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE ACTIVITY | 4 | 17 | 1.386e-06 | 4.441e-05 |
| 30 | MACROMOLECULAR COMPLEX BINDING | 22 | 1399 | 1.447e-06 | 4.48e-05 |
| 31 | TRANSCRIPTION FACTOR BINDING | 13 | 524 | 2.353e-06 | 7.05e-05 |
| 32 | RECEPTOR REGULATOR ACTIVITY | 5 | 45 | 3.094e-06 | 8.981e-05 |
| 33 | PROTEIN PHOSPHATASE TYPE 2A REGULATOR ACTIVITY | 4 | 21 | 3.432e-06 | 9.662e-05 |
| 34 | PROTEIN HETERODIMERIZATION ACTIVITY | 12 | 468 | 4.239e-06 | 0.0001125 |
| 35 | PROTEIN COMPLEX BINDING | 17 | 935 | 4.151e-06 | 0.0001125 |
| 36 | KINASE ACTIVITY | 16 | 842 | 4.732e-06 | 0.0001221 |
| 37 | RECEPTOR SIGNALING PROTEIN SERINE THREONINE KINASE ACTIVITY | 6 | 92 | 7.167e-06 | 0.00018 |
| 38 | RNA POLYMERASE II TRANSCRIPTION FACTOR BINDING | 6 | 104 | 1.453e-05 | 0.0003551 |
| 39 | PROTEIN SERINE THREONINE KINASE ACTIVITY | 11 | 445 | 1.538e-05 | 0.0003664 |
| 40 | CORE PROMOTER PROXIMAL REGION DNA BINDING | 10 | 371 | 1.836e-05 | 0.0004263 |
| 41 | PROTEIN KINASE ACTIVITY | 13 | 640 | 2.012e-05 | 0.0004558 |
| 42 | GUANYLATE KINASE ACTIVITY | 3 | 12 | 2.661e-05 | 0.0005885 |
| 43 | ARMADILLO REPEAT DOMAIN BINDING | 3 | 13 | 3.446e-05 | 0.0007445 |
| 44 | TRANSFERASE ACTIVITY TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 16 | 992 | 3.592e-05 | 0.0007583 |
| 45 | TRANSCRIPTION FACTOR ACTIVITY RNA POLYMERASE II CORE PROMOTER PROXIMAL REGION SEQUENCE SPECIFIC BINDING | 9 | 328 | 4.277e-05 | 0.0008829 |
| 46 | PROTEIN C TERMINUS BINDING | 7 | 186 | 4.426e-05 | 0.0008938 |
| 47 | TRANSFORMING GROWTH FACTOR BETA BINDING | 3 | 16 | 6.674e-05 | 0.001319 |
| 48 | PHOSPHATASE REGULATOR ACTIVITY | 5 | 87 | 7.912e-05 | 0.001531 |
| 49 | ENZYME REGULATOR ACTIVITY | 15 | 959 | 9.125e-05 | 0.00173 |
| 50 | NUCLEIC ACID BINDING TRANSCRIPTION FACTOR ACTIVITY | 17 | 1199 | 9.929e-05 | 0.001845 |
| 51 | CYTOKINE BINDING | 5 | 92 | 0.0001032 | 0.00188 |
| 52 | IDENTICAL PROTEIN BINDING | 17 | 1209 | 0.0001098 | 0.001962 |
| 53 | TRANSCRIPTION COREPRESSOR ACTIVITY | 7 | 221 | 0.0001305 | 0.002288 |
| 54 | TRANSCRIPTIONAL ACTIVATOR ACTIVITY RNA POLYMERASE II CORE PROMOTER PROXIMAL REGION SEQUENCE SPECIFIC BINDING | 7 | 226 | 0.0001499 | 0.002579 |
| 55 | PHOSPHATASE BINDING | 6 | 162 | 0.000173 | 0.002921 |
| 56 | TRANSCRIPTION FACTOR ACTIVITY PROTEIN BINDING | 11 | 588 | 0.0001875 | 0.003111 |
| 57 | R SMAD BINDING | 3 | 23 | 0.0002057 | 0.003353 |
| 58 | NUCLEOTIDE KINASE ACTIVITY | 3 | 24 | 0.0002342 | 0.003752 |
| 59 | CHROMATIN BINDING | 9 | 435 | 0.000358 | 0.005635 |
| 60 | CELL ADHESION MOLECULE BINDING | 6 | 186 | 0.000364 | 0.005635 |
| 61 | CADHERIN BINDING | 3 | 28 | 0.0003736 | 0.00569 |
| 62 | GROWTH FACTOR BINDING | 5 | 123 | 0.000401 | 0.006008 |
| 63 | DOUBLE STRANDED DNA BINDING | 12 | 764 | 0.0004686 | 0.006911 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | APICAL JUNCTION COMPLEX | 15 | 128 | 9.983e-17 | 2.915e-14 |
| 2 | WNT SIGNALOSOME | 8 | 11 | 5.194e-17 | 2.915e-14 |
| 3 | CELL JUNCTION | 29 | 1151 | 2.465e-13 | 4.798e-11 |
| 4 | EXTRACELLULAR MATRIX | 19 | 426 | 3.824e-13 | 5.582e-11 |
| 5 | CELL CELL JUNCTION | 18 | 383 | 7.062e-13 | 8.248e-11 |
| 6 | PROTEINACEOUS EXTRACELLULAR MATRIX | 15 | 356 | 3.166e-10 | 3.081e-08 |
| 7 | EXTRACELLULAR SPACE | 26 | 1376 | 2.939e-09 | 2.452e-07 |
| 8 | BETA CATENIN DESTRUCTION COMPLEX | 5 | 14 | 5.739e-09 | 4.189e-07 |
| 9 | TRANSCRIPTION FACTOR COMPLEX | 12 | 298 | 3.511e-08 | 2.278e-06 |
| 10 | PROTEIN PHOSPHATASE TYPE 2A COMPLEX | 5 | 20 | 4.339e-08 | 2.534e-06 |
| 11 | PHOSPHATASE COMPLEX | 6 | 48 | 1.475e-07 | 7.833e-06 |
| 12 | ANCHORING JUNCTION | 14 | 489 | 1.664e-07 | 8.098e-06 |
| 13 | CELL SURFACE | 17 | 757 | 2.29e-07 | 1.029e-05 |
| 14 | PLASMA MEMBRANE PROTEIN COMPLEX | 14 | 510 | 2.771e-07 | 1.156e-05 |
| 15 | CYTOPLASMIC VESICLE PART | 15 | 601 | 3.382e-07 | 1.317e-05 |
| 16 | PLASMA MEMBRANE RECEPTOR COMPLEX | 8 | 175 | 2.961e-06 | 0.0001081 |
| 17 | INTRACELLULAR VESICLE | 20 | 1259 | 4.094e-06 | 0.0001407 |
| 18 | LATERAL PLASMA MEMBRANE | 5 | 50 | 5.259e-06 | 0.0001706 |
| 19 | VESICLE MEMBRANE | 12 | 512 | 1.051e-05 | 0.000323 |
| 20 | ENDOCYTIC VESICLE MEMBRANE | 7 | 152 | 1.207e-05 | 0.0003524 |
| 21 | MEMBRANE REGION | 18 | 1134 | 1.356e-05 | 0.0003771 |
| 22 | MEMBRANE MICRODOMAIN | 9 | 288 | 1.542e-05 | 0.0004093 |
| 23 | MICROTUBULE CYTOSKELETON | 17 | 1068 | 2.349e-05 | 0.0005965 |
| 24 | CELL SUBSTRATE JUNCTION | 10 | 398 | 3.34e-05 | 0.0008127 |
| 25 | RECEPTOR COMPLEX | 9 | 327 | 4.177e-05 | 0.0009757 |
| 26 | ENDOCYTIC VESICLE | 8 | 256 | 4.679e-05 | 0.001051 |
| 27 | MICROTUBULE ORGANIZING CENTER | 12 | 623 | 7.143e-05 | 0.001545 |
| 28 | PROTEIN KINASE COMPLEX | 5 | 90 | 9.299e-05 | 0.00194 |
| 29 | CYTOSKELETON | 23 | 1967 | 9.96e-05 | 0.002006 |
| 30 | GOLGI LUMEN | 5 | 94 | 0.0001143 | 0.002225 |
| 31 | INTERCALATED DISC | 4 | 51 | 0.0001276 | 0.002404 |
| 32 | CELL CELL ADHERENS JUNCTION | 4 | 54 | 0.0001596 | 0.002913 |
| 33 | CELL BODY | 10 | 494 | 0.0001989 | 0.003416 |
| 34 | CYTOPLASMIC MICROTUBULE | 4 | 57 | 0.0001971 | 0.003416 |
| 35 | CELL CORTEX | 7 | 238 | 0.000206 | 0.003437 |
| 36 | PLASMA MEMBRANE REGION | 14 | 929 | 0.0002347 | 0.003808 |
| 37 | CELL PROJECTION PART | 14 | 946 | 0.0002824 | 0.004458 |
| 38 | CELL CELL CONTACT ZONE | 4 | 64 | 0.0003085 | 0.004741 |
| 39 | CHROMATIN | 9 | 441 | 0.0003955 | 0.005923 |
| 40 | SPINDLE POLE | 5 | 126 | 0.0004478 | 0.006379 |
| 41 | SOMATODENDRITIC COMPARTMENT | 11 | 650 | 0.0004404 | 0.006379 |
| 42 | CHROMOSOME | 13 | 880 | 0.0004816 | 0.006696 |
| 43 | EXCITATORY SYNAPSE | 6 | 197 | 0.0004937 | 0.006705 |
| 44 | CYCLIN DEPENDENT PROTEIN KINASE HOLOENZYME COMPLEX | 3 | 31 | 0.000507 | 0.006729 |
| 45 | MEMBRANE PROTEIN COMPLEX | 14 | 1020 | 6e-04 | 0.007787 |
| 46 | CYTOPLASMIC REGION | 7 | 287 | 0.0006346 | 0.007885 |
| 47 | EXTRINSIC COMPONENT OF PLASMA MEMBRANE | 5 | 136 | 0.0006343 | 0.007885 |
| 48 | NUCLEAR CHROMATIN | 7 | 291 | 0.0006885 | 0.008377 |
| 49 | CATALYTIC COMPLEX | 14 | 1038 | 0.0007123 | 0.00849 |
| 50 | CENTROSOME | 9 | 487 | 0.0008047 | 0.009326 |
| 51 | CYTOSKELETAL PART | 17 | 1436 | 0.0008145 | 0.009326 |
| 52 | DENDRITIC SHAFT | 3 | 37 | 0.0008574 | 0.009629 |
| 53 | NEURON PROJECTION | 13 | 942 | 0.0009066 | 0.00999 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
| 1 | hsa04390_Hippo_signaling_pathway | 101 | 154 | 3.671e-233 | 6.608e-231 | |
| 2 | hsa04310_Wnt_signaling_pathway | 41 | 151 | 6.302e-62 | 5.672e-60 | |
| 3 | hsa04916_Melanogenesis | 27 | 101 | 4.337e-40 | 2.602e-38 | |
| 4 | hsa04340_Hedgehog_signaling_pathway | 20 | 56 | 1.07e-32 | 4.814e-31 | |
| 5 | hsa04350_TGF.beta_signaling_pathway | 21 | 85 | 2.158e-30 | 7.768e-29 | |
| 6 | hsa04520_Adherens_junction | 12 | 73 | 2e-15 | 6e-14 | |
| 7 | hsa04110_Cell_cycle | 13 | 128 | 8.226e-14 | 2.115e-12 | |
| 8 | hsa04530_Tight_junction | 13 | 133 | 1.359e-13 | 3.058e-12 | |
| 9 | hsa04151_PI3K_AKT_signaling_pathway | 15 | 351 | 2.602e-10 | 5.204e-09 | |
| 10 | hsa04114_Oocyte_meiosis | 9 | 114 | 6.216e-09 | 1.119e-07 | |
| 11 | hsa04144_Endocytosis | 10 | 203 | 8.007e-08 | 1.31e-06 | |
| 12 | hsa04115_p53_signaling_pathway | 6 | 69 | 1.323e-06 | 1.985e-05 | |
| 13 | hsa04722_Neurotrophin_signaling_pathway | 6 | 127 | 4.507e-05 | 0.0006241 | |
| 14 | hsa03015_mRNA_surveillance_pathway | 5 | 83 | 6.317e-05 | 0.0008122 | |
| 15 | hsa04510_Focal_adhesion | 7 | 200 | 7.001e-05 | 0.0008401 | |
| 16 | hsa04010_MAPK_signaling_pathway | 7 | 268 | 0.0004224 | 0.004752 | |
| 17 | hsa04810_Regulation_of_actin_cytoskeleton | 6 | 214 | 0.000762 | 0.008069 | |
| 18 | hsa04670_Leukocyte_transendothelial_migration | 4 | 117 | 0.002936 | 0.02936 | |
| 19 | hsa04710_Circadian_rhythm_._mammal | 2 | 23 | 0.005962 | 0.05648 | |
| 20 | hsa04630_Jak.STAT_signaling_pathway | 4 | 155 | 0.007922 | 0.0713 | |
| 21 | hsa04330_Notch_signaling_pathway | 2 | 47 | 0.02355 | 0.2019 | |
| 22 | hsa04380_Osteoclast_differentiation | 3 | 128 | 0.0271 | 0.2217 | |
| 23 | hsa04910_Insulin_signaling_pathway | 3 | 138 | 0.03282 | 0.2569 | |
| 24 | hsa04730_Long.term_depression | 2 | 70 | 0.04885 | 0.3663 | |
| 25 | hsa04012_ErbB_signaling_pathway | 2 | 87 | 0.07165 | 0.5159 | |
| 26 | hsa04660_T_cell_receptor_signaling_pathway | 2 | 108 | 0.1035 | 0.7166 | |
| 27 | hsa04120_Ubiquitin_mediated_proteolysis | 2 | 139 | 0.1558 | 1 | |
| 28 | hsa04145_Phagosome | 2 | 156 | 0.1863 | 1 | |
| 29 | hsa04062_Chemokine_signaling_pathway | 2 | 189 | 0.2474 | 1 | |
| 30 | hsa04014_Ras_signaling_pathway | 2 | 236 | 0.335 | 1 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | EMX2OS |
hsa-let-7a-3p;hsa-let-7f-2-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-205-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-944 | 12 | AXIN2 | Sponge network | -6.205 | 0.00015 | -3.824 | 0.00033 | 0.595 |
| 2 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7f-2-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-205-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 11 | AXIN2 | Sponge network | -4.563 | 0 | -3.824 | 0.00033 | 0.525 |
| 3 | AC073283.4 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-205-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-34c-3p;hsa-miR-944 | 11 | AMOT | Sponge network | -2.801 | 0.08856 | -1.936 | 0.20701 | 0.524 |
| 4 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7f-2-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-205-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-944 | 12 | AXIN2 | Sponge network | -7.871 | 0 | -3.824 | 0.00033 | 0.523 |
| 5 | RP11-384L8.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-205-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-330-5p | 10 | AMOT | Sponge network | -1.784 | 0.21615 | -1.936 | 0.20701 | 0.501 |
| 6 | MEG3 |
hsa-let-7b-3p;hsa-let-7f-2-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-205-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-3614-5p;hsa-miR-944 | 11 | AXIN2 | Sponge network | -3.613 | 0.00075 | -3.824 | 0.00033 | 0.496 |
| 7 | FRMD6-AS2 | hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20b-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-375 | 10 | FRMD6 | Sponge network | -12.644 | 0 | -1.608 | 0.08207 | 0.494 |
| 8 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-9-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 13 | TGFBR2 | Sponge network | -4.563 | 0 | -2.733 | 2.0E-5 | 0.479 |
| 9 | TRHDE-AS1 |
hsa-let-7g-3p;hsa-miR-130a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-32-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-3613-5p;hsa-miR-3928-3p;hsa-miR-484;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-769-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 25 | DLG2 | Sponge network | -6.205 | 0.01165 | -4.94 | 0.00023 | 0.476 |
| 10 | ADAMTS9-AS1 |
hsa-let-7g-3p;hsa-miR-130a-5p;hsa-miR-135b-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-769-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 25 | DLG2 | Sponge network | -8.573 | 0.00012 | -4.94 | 0.00023 | 0.474 |
| 11 | RP11-166D19.1 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 15 | FZD4 | Sponge network | -4.209 | 2.0E-5 | -2.839 | 4.0E-5 | 0.456 |
| 12 | EMX2OS |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-205-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-2355-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-944 | 10 | AMOT | Sponge network | -6.205 | 0.00015 | -1.936 | 0.20701 | 0.452 |
| 13 | MAGI2-AS3 |
hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 11 | LATS2 | Sponge network | -4.563 | 0 | -1.641 | 0.00068 | 0.434 |
| 14 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-205-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-590-3p;hsa-miR-944 | 10 | AXIN2 | Sponge network | -2.562 | 0.00163 | -3.824 | 0.00033 | 0.431 |
| 15 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-9-5p;hsa-miR-92a-3p | 13 | TGFBR2 | Sponge network | -4.209 | 2.0E-5 | -2.733 | 2.0E-5 | 0.422 |
| 16 | RP5-1042I8.7 | hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-32-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 10 | DLG2 | Sponge network | -0.504 | 0.36806 | -4.94 | 0.00023 | 0.414 |
| 17 | RP11-166D19.1 |
hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-3613-5p;hsa-miR-3928-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-944 | 27 | DLG2 | Sponge network | -4.209 | 2.0E-5 | -4.94 | 0.00023 | 0.406 |
| 18 | HAND2-AS1 |
hsa-let-7g-3p;hsa-miR-130a-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-32-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-3613-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-944 | 28 | DLG2 | Sponge network | -7.871 | 0 | -4.94 | 0.00023 | 0.395 |
| 19 | AC005682.5 | hsa-miR-106a-5p;hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-20b-5p;hsa-miR-28-5p;hsa-miR-3065-3p;hsa-miR-3065-5p | 11 | CCND2 | Sponge network | -2.193 | 0.07184 | -2.811 | 0.0014 | 0.394 |
| 20 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 12 | TGFBR2 | Sponge network | -7.871 | 0 | -2.733 | 2.0E-5 | 0.394 |
| 21 | RP11-774O3.3 | hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-3928-3p;hsa-miR-93-3p | 16 | DLG2 | Sponge network | -1.989 | 0.00136 | -4.94 | 0.00023 | 0.394 |
| 22 | RP11-999E24.3 | hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-93-3p | 15 | DLG2 | Sponge network | -4.893 | 2.0E-5 | -4.94 | 0.00023 | 0.393 |
| 23 | AC003090.1 | hsa-miR-130a-5p;hsa-miR-135b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-582-5p;hsa-miR-590-5p | 18 | DLG2 | Sponge network | -7.817 | 0.00161 | -4.94 | 0.00023 | 0.391 |
| 24 | ACTA2-AS1 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-590-3p;hsa-miR-93-3p | 10 | BMPR2 | Sponge network | -6.142 | 0.00223 | -1.126 | 0.00109 | 0.39 |
| 25 | RP4-607I7.1 | hsa-miR-106a-5p;hsa-miR-141-3p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20b-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-671-5p;hsa-miR-93-5p | 12 | FRMD6 | Sponge network | -0.625 | 0.69827 | -1.608 | 0.08207 | 0.384 |
| 26 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-3065-3p;hsa-miR-3065-5p;hsa-miR-30d-3p;hsa-miR-93-5p | 12 | CCND2 | Sponge network | -1.318 | 0.0924 | -2.811 | 0.0014 | 0.383 |
| 27 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-miR-130a-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-32-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-3613-5p;hsa-miR-3928-3p;hsa-miR-590-3p;hsa-miR-642a-5p;hsa-miR-769-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 32 | DLG2 | Sponge network | -4.563 | 0 | -4.94 | 0.00023 | 0.382 |
| 28 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 15 | FZD4 | Sponge network | -4.563 | 0 | -2.839 | 4.0E-5 | 0.382 |
| 29 | DNM3OS |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-93-5p | 12 | BMPR2 | Sponge network | -3.933 | 0.00059 | -1.126 | 0.00109 | 0.382 |
| 30 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7f-2-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-205-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-944 | 12 | AXIN2 | Sponge network | -6.142 | 0.00223 | -3.824 | 0.00033 | 0.379 |
| 31 | RP11-166D19.1 |
hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-96-5p | 10 | LATS2 | Sponge network | -4.209 | 2.0E-5 | -1.641 | 0.00068 | 0.378 |
| 32 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-miR-130a-3p;hsa-miR-130a-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-3613-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-628-3p;hsa-miR-769-3p;hsa-miR-93-3p;hsa-miR-944 | 28 | DLG2 | Sponge network | -6.142 | 0.00223 | -4.94 | 0.00023 | 0.377 |
| 33 | MAGI2-AS3 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-93-5p | 15 | BMPR2 | Sponge network | -4.563 | 0 | -1.126 | 0.00109 | 0.376 |
| 34 | DIO3OS | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-20b-5p;hsa-miR-324-3p;hsa-miR-378a-3p | 13 | CCND2 | Sponge network | -4.295 | 0.00689 | -2.811 | 0.0014 | 0.374 |
| 35 | MEG3 |
hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-629-3p | 13 | FZD4 | Sponge network | -3.613 | 0.00075 | -2.839 | 4.0E-5 | 0.369 |
| 36 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7f-2-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 10 | AXIN2 | Sponge network | -4.398 | 5.0E-5 | -3.824 | 0.00033 | 0.368 |
| 37 | ZNF667-AS1 | hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3613-5p;hsa-miR-484 | 15 | DLG2 | Sponge network | -4.019 | 0.00137 | -4.94 | 0.00023 | 0.367 |
| 38 | RP11-554A11.4 |
hsa-miR-135b-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-32-3p;hsa-miR-33a-3p;hsa-miR-3928-3p | 13 | DLG2 | Sponge network | -5.361 | 2.0E-5 | -4.94 | 0.00023 | 0.366 |
| 39 | LINC00899 | hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-484;hsa-miR-769-3p;hsa-miR-93-3p | 12 | DLG2 | Sponge network | -1.597 | 0.00258 | -4.94 | 0.00023 | 0.366 |
| 40 | MIR143HG |
hsa-let-7g-3p;hsa-miR-130a-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-642a-5p;hsa-miR-769-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 29 | DLG2 | Sponge network | -6.51 | 0 | -4.94 | 0.00023 | 0.365 |
| 41 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20b-5p;hsa-miR-429 | 10 | FRMD6 | Sponge network | -1.941 | 0.0681 | -1.608 | 0.08207 | 0.363 |
| 42 | RP11-119F7.5 | hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-22-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-92b-3p | 13 | DLG2 | Sponge network | -2.406 | 0.03687 | -4.94 | 0.00023 | 0.362 |
| 43 | MIR143HG |
hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-2110;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-5p;hsa-miR-616-5p;hsa-miR-92a-3p | 12 | WNT5A | Sponge network | -6.51 | 0 | -0.335 | 0.71496 | 0.356 |
| 44 | RP11-389C8.2 |
hsa-let-7g-3p;hsa-miR-130a-5p;hsa-miR-135b-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-33a-3p | 16 | DLG2 | Sponge network | -3.089 | 2.0E-5 | -4.94 | 0.00023 | 0.355 |
| 45 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 12 | TGFBR2 | Sponge network | -6.51 | 0 | -2.733 | 2.0E-5 | 0.353 |
| 46 | DNM3OS |
hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 11 | LATS2 | Sponge network | -3.933 | 0.00059 | -1.641 | 0.00068 | 0.348 |
| 47 | MIR143HG |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-3p;hsa-miR-25-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 13 | FZD4 | Sponge network | -6.51 | 0 | -2.839 | 4.0E-5 | 0.346 |
| 48 | CTB-92J24.3 | hsa-let-7g-3p;hsa-miR-130a-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-33a-5p;hsa-miR-944 | 12 | DLG2 | Sponge network | -7.226 | 0.0046 | -4.94 | 0.00023 | 0.345 |
| 49 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-224-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-708-5p | 13 | FZD4 | Sponge network | -6.142 | 0.00223 | -2.839 | 4.0E-5 | 0.343 |
| 50 | DNM3OS |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 13 | FZD4 | Sponge network | -3.933 | 0.00059 | -2.839 | 4.0E-5 | 0.341 |
| 51 | EMX2OS |
hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-32-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-944 | 23 | DLG2 | Sponge network | -6.205 | 0.00015 | -4.94 | 0.00023 | 0.341 |
| 52 | RP11-344E13.3 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-93-5p | 11 | BMPR2 | Sponge network | -4.307 | 3.0E-5 | -1.126 | 0.00109 | 0.34 |
| 53 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-20b-5p;hsa-miR-224-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-96-5p | 18 | CCND2 | Sponge network | -4.209 | 2.0E-5 | -2.811 | 0.0014 | 0.339 |
| 54 | RP11-819C21.1 | hsa-miR-130a-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-590-3p | 16 | DLG2 | Sponge network | -1.571 | 0.00379 | -4.94 | 0.00023 | 0.337 |
| 55 | RP11-439M11.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-20b-5p;hsa-miR-324-3p | 10 | CCND2 | Sponge network | -2.662 | 0.21003 | -2.811 | 0.0014 | 0.337 |
| 56 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 11 | TGFBR2 | Sponge network | -3.933 | 0.00059 | -2.733 | 2.0E-5 | 0.336 |
| 57 | BDNF-AS | hsa-miR-130a-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-484;hsa-miR-590-3p | 12 | DLG2 | Sponge network | -1.712 | 0.02515 | -4.94 | 0.00023 | 0.336 |
| 58 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-671-5p | 12 | FRMD6 | Sponge network | -1.585 | 0.19508 | -1.608 | 0.08207 | 0.334 |
| 59 | RP11-344E13.3 |
hsa-let-7g-3p;hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-3613-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-642a-5p;hsa-miR-877-5p;hsa-miR-93-3p | 23 | DLG2 | Sponge network | -4.307 | 3.0E-5 | -4.94 | 0.00023 | 0.331 |
| 60 | CTD-2554C21.3 | hsa-miR-130a-3p;hsa-miR-130a-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-3613-5p;hsa-miR-484;hsa-miR-93-3p | 16 | DLG2 | Sponge network | -6.258 | 0.00703 | -4.94 | 0.00023 | 0.33 |
| 61 | NDUFA6-AS1 | hsa-miR-130a-3p;hsa-miR-130a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-484;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-877-5p | 13 | DLG2 | Sponge network | -1.026 | 0.04234 | -4.94 | 0.00023 | 0.326 |
| 62 | NR2F1-AS1 | hsa-let-7g-3p;hsa-miR-130a-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-484;hsa-miR-590-3p | 18 | DLG2 | Sponge network | -2.961 | 0.00154 | -4.94 | 0.00023 | 0.323 |
| 63 | RP11-166D19.1 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 13 | BMPR2 | Sponge network | -4.209 | 2.0E-5 | -1.126 | 0.00109 | 0.321 |
| 64 | RP11-116O18.1 | hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | DLG2 | Sponge network | -5.007 | 0.06008 | -4.94 | 0.00023 | 0.321 |
| 65 | MIR497HG |
hsa-miR-130a-5p;hsa-miR-135b-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-590-5p;hsa-miR-769-3p;hsa-miR-877-5p;hsa-miR-93-3p | 21 | DLG2 | Sponge network | -6.146 | 0.00024 | -4.94 | 0.00023 | 0.32 |
| 66 | RP11-359B12.2 | hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-3613-5p;hsa-miR-484;hsa-miR-590-3p | 13 | DLG2 | Sponge network | -2.094 | 0.00033 | -4.94 | 0.00023 | 0.32 |
| 67 | HAND2-AS1 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-93-5p | 13 | BMPR2 | Sponge network | -7.871 | 0 | -1.126 | 0.00109 | 0.317 |
| 68 | RP11-887P2.5 | hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-3613-5p;hsa-miR-590-5p | 14 | DLG2 | Sponge network | -9.865 | 1.0E-5 | -4.94 | 0.00023 | 0.316 |
| 69 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-20b-5p;hsa-miR-224-3p;hsa-miR-3065-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-9-3p;hsa-miR-93-5p | 19 | CCND2 | Sponge network | -4.563 | 0 | -2.811 | 0.0014 | 0.315 |
| 70 | WT1-AS |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-32-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3928-3p;hsa-miR-590-3p;hsa-miR-769-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-944 | 27 | DLG2 | Sponge network | -6.875 | 2.0E-5 | -4.94 | 0.00023 | 0.314 |
| 71 | RP11-284N8.3 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-3065-5p;hsa-miR-324-3p;hsa-miR-93-5p;hsa-miR-96-5p | 10 | CCND2 | Sponge network | -0.845 | 0.52848 | -2.811 | 0.0014 | 0.313 |
| 72 | TPTEP1 |
hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-33b-5p;hsa-miR-3613-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 12 | DLG2 | Sponge network | -4.398 | 5.0E-5 | -4.94 | 0.00023 | 0.312 |
| 73 | RP11-130L8.1 | hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-944 | 13 | DLG2 | Sponge network | -4.329 | 1.0E-5 | -4.94 | 0.00023 | 0.311 |
| 74 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-629-3p | 12 | FZD4 | Sponge network | -3.089 | 2.0E-5 | -2.839 | 4.0E-5 | 0.311 |
| 75 | FAM66C |
hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-3613-5p;hsa-miR-3928-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 26 | DLG2 | Sponge network | -2.927 | 0.00012 | -4.94 | 0.00023 | 0.311 |
| 76 | MAGI2-AS3 |
hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-2110;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-5p;hsa-miR-616-5p;hsa-miR-92a-3p | 11 | WNT5A | Sponge network | -4.563 | 0 | -0.335 | 0.71496 | 0.31 |
| 77 | USP3-AS1 | hsa-miR-130a-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-29b-3p | 16 | DLG2 | Sponge network | -4.151 | 0 | -4.94 | 0.00023 | 0.31 |
| 78 | NR2F2-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-16-5p;hsa-miR-224-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-452-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-708-5p | 11 | FZD4 | Sponge network | -3.785 | 0.00281 | -2.839 | 4.0E-5 | 0.309 |
| 79 | CTA-204B4.2 | hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-3613-5p | 10 | DLG2 | Sponge network | -1.612 | 0.00122 | -4.94 | 0.00023 | 0.303 |
| 80 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-5p;hsa-miR-93-5p | 10 | FRMD6 | Sponge network | -1.318 | 0.0924 | -1.608 | 0.08207 | 0.303 |
| 81 | RP11-401P9.4 | hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-484 | 15 | DLG2 | Sponge network | -3.793 | 0.00144 | -4.94 | 0.00023 | 0.302 |
| 82 | MEG3 |
hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-93-3p;hsa-miR-944 | 13 | DLG2 | Sponge network | -3.613 | 0.00075 | -4.94 | 0.00023 | 0.302 |
| 83 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-20b-5p;hsa-miR-28-5p;hsa-miR-3065-5p;hsa-miR-324-3p;hsa-miR-429 | 14 | CCND2 | Sponge network | -1.941 | 0.0681 | -2.811 | 0.0014 | 0.301 |
| 84 | PGM5-AS1 | hsa-miR-130a-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-484 | 13 | DLG2 | Sponge network | -14.107 | 0 | -4.94 | 0.00023 | 0.301 |
| 85 | DNM3OS |
hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-3p;hsa-miR-769-3p;hsa-miR-93-3p | 25 | DLG2 | Sponge network | -3.933 | 0.00059 | -4.94 | 0.00023 | 0.298 |
| 86 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20b-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-93-5p | 13 | FRMD6 | Sponge network | -2.494 | 0.07597 | -1.608 | 0.08207 | 0.297 |
| 87 | RP11-13K12.1 | hsa-miR-130a-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-944 | 14 | DLG2 | Sponge network | -5.093 | 0.01151 | -4.94 | 0.00023 | 0.295 |
| 88 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-224-3p;hsa-miR-3065-3p;hsa-miR-3065-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-93-5p | 17 | CCND2 | Sponge network | -3.089 | 2.0E-5 | -2.811 | 0.0014 | 0.293 |
| 89 | SOCS2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20b-5p;hsa-miR-3613-5p;hsa-miR-93-5p | 11 | FRMD6 | Sponge network | -4.167 | 1.0E-5 | -1.608 | 0.08207 | 0.292 |
| 90 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-20b-5p;hsa-miR-324-3p;hsa-miR-33a-3p | 10 | CCND2 | Sponge network | -5.361 | 2.0E-5 | -2.811 | 0.0014 | 0.29 |
| 91 | MEG3 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-93-3p;hsa-miR-93-5p | 10 | BMPR2 | Sponge network | -3.613 | 0.00075 | -1.126 | 0.00109 | 0.29 |
| 92 | PSMD5-AS1 | hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-3613-5p;hsa-miR-484;hsa-miR-590-5p | 11 | DLG2 | Sponge network | -0.973 | 0.02357 | -4.94 | 0.00023 | 0.29 |
| 93 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-224-3p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-629-3p | 10 | FZD4 | Sponge network | -4.398 | 5.0E-5 | -2.839 | 4.0E-5 | 0.287 |
| 94 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-20b-5p;hsa-miR-224-3p;hsa-miR-3065-3p;hsa-miR-3065-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-93-5p | 21 | CCND2 | Sponge network | -6.51 | 0 | -2.811 | 0.0014 | 0.285 |
| 95 | MIR143HG |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-93-5p | 16 | BMPR2 | Sponge network | -6.51 | 0 | -1.126 | 0.00109 | 0.282 |
| 96 | CTD-2554C21.2 | hsa-let-7g-3p;hsa-miR-130a-3p;hsa-miR-130a-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-29b-3p;hsa-miR-944 | 17 | DLG2 | Sponge network | -6.968 | 0.00817 | -4.94 | 0.00023 | 0.279 |
| 97 | RP11-344E13.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-200a-3p;hsa-miR-20b-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-3613-5p;hsa-miR-454-3p;hsa-miR-671-5p;hsa-miR-93-5p | 12 | FRMD6 | Sponge network | -4.307 | 3.0E-5 | -1.608 | 0.08207 | 0.276 |
| 98 | RP11-981G7.6 | hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-3928-3p;hsa-miR-484;hsa-miR-590-3p | 11 | DLG2 | Sponge network | -0.726 | 0.44765 | -4.94 | 0.00023 | 0.274 |
| 99 | CTA-109P11.4 | hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-330-3p;hsa-miR-484 | 10 | DLG2 | Sponge network | -7.746 | 0.00804 | -4.94 | 0.00023 | 0.272 |
| 100 | NR2F2-AS1 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-93-5p | 11 | BMPR2 | Sponge network | -3.785 | 0.00281 | -1.126 | 0.00109 | 0.265 |
| 101 | CKMT2-AS1 | hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 10 | DLG2 | Sponge network | -1.384 | 0.001 | -4.94 | 0.00023 | 0.265 |
| 102 | SOCS2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20b-5p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-93-5p | 12 | CCND2 | Sponge network | -4.167 | 1.0E-5 | -2.811 | 0.0014 | 0.258 |
| 103 | TRHDE-AS1 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-93-5p | 12 | BMPR2 | Sponge network | -6.205 | 0.01165 | -1.126 | 0.00109 | 0.257 |
| 104 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-93-5p | 10 | TGFBR2 | Sponge network | -3.089 | 2.0E-5 | -2.733 | 2.0E-5 | 0.253 |
| 105 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20b-5p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-93-5p;hsa-miR-96-5p | 14 | CCND2 | Sponge network | -2.494 | 0.07597 | -2.811 | 0.0014 | 0.252 |
| 106 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-3065-3p;hsa-miR-3065-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-93-5p;hsa-miR-96-5p | 19 | CCND2 | Sponge network | -3.933 | 0.00059 | -2.811 | 0.0014 | 0.25 |