This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-30d-3p | ATM | -0.12 | 0.32955 | -0.24 | 0.01738 | mirMAP | -0.14 | 0.00043 | 24345332 | miR-30d has been observed to be significantly down-regulated in human anaplastic thyroid carcinoma ATC and is believed to be an important event in thyroid cell transformation; In this study we found that miR-30d has a critical role in modulating sensitivity of ATC cells to cisplatin a commonly used chemotherapeutic drug for treatment of this neoplasm; Using a mimic of miR-30d we demonstrated that miR-30d could negatively regulate the expression of beclin 1 a key autophagy gene leading to suppression of the cisplatin-activated autophagic response that protects ATC cells from apoptosis; We further showed that inhibition of the beclin 1-mediated autophagy by the miR-30d mimic sensitized ATC cells to cisplatin both in vitro cell culture and in vivo animal xenograft model; These results suggest that dysregulation of miR-30d in ATC cells is responsible for the insensitivity to cisplatin by promoting autophagic survival; Thus miR-30d may be exploited as a potential target for therapeutic intervention in the treatment of ATC |
| 2 | hsa-miR-339-5p | ATM | 0.28 | 0.03557 | -0.24 | 0.01738 | miRanda | -0.1 | 0.00399 | NA | |
| 3 | hsa-miR-455-5p | ATM | -0.27 | 0.05813 | -0.24 | 0.01738 | miRanda | -0.12 | 0.00045 | NA | |
| 4 | hsa-miR-30a-5p | BAX | -0.63 | 0.00011 | 0.8 | 0 | miRNAWalker2 validate | -0.12 | 0.00058 | NA | |
| 5 | hsa-miR-365a-3p | BAX | 0.16 | 0.15325 | 0.8 | 0 | miRNAWalker2 validate | -0.23 | 0 | 24216611 | MiR 365 induces gemcitabine resistance in pancreatic cancer cells by targeting the adaptor protein SHC1 and pro apoptotic regulator BAX |
| 6 | hsa-let-7a-2-3p | BBC3 | -1.19 | 0 | 0.8 | 0 | MirTarget | -0.19 | 0 | NA | |
| 7 | hsa-let-7g-3p | BBC3 | -1.14 | 0 | 0.8 | 0 | MirTarget; miRNATAP | -0.18 | 0.0001 | NA | |
| 8 | hsa-miR-101-3p | BBC3 | -1.48 | 0 | 0.8 | 0 | miRNATAP | -0.32 | 0 | NA | |
| 9 | hsa-miR-125b-5p | BBC3 | -1.36 | 0 | 0.8 | 0 | miRNAWalker2 validate; miRTarBase | -0.25 | 0 | 25184537 | Thus far two of these target genes BBC3 and NEU1 that are tumor suppressor genes but not yet studied in PDAC appear to be functional targets of miR-125b since knockdown of miR125b caused their up regulation |
| 10 | hsa-miR-139-5p | BBC3 | -2.11 | 0 | 0.8 | 0 | miRNATAP | -0.23 | 0 | NA | |
| 11 | hsa-miR-140-5p | BBC3 | -0.22 | 0.01407 | 0.8 | 0 | miRNATAP | -0.25 | 0.0003 | NA | |
| 12 | hsa-miR-144-3p | BBC3 | -2.98 | 0 | 0.8 | 0 | miRNATAP | -0.11 | 0 | NA | |
| 13 | hsa-miR-27b-3p | BBC3 | -0.82 | 0 | 0.8 | 0 | miRNATAP | -0.2 | 0.00022 | NA | |
| 14 | hsa-miR-345-5p | BBC3 | -0.71 | 0 | 0.8 | 0 | miRNATAP | -0.17 | 4.0E-5 | NA | |
| 15 | hsa-miR-590-3p | BBC3 | -0.47 | 2.0E-5 | 0.8 | 0 | miRanda | -0.25 | 1.0E-5 | NA | |
| 16 | hsa-miR-140-5p | BID | -0.22 | 0.01407 | 0.21 | 0.07516 | miRanda | -0.21 | 0.00101 | NA | |
| 17 | hsa-let-7g-5p | CASP3 | -0.46 | 2.0E-5 | 0.31 | 0.00047 | MirTarget; miRNATAP | -0.11 | 0.00694 | NA | |
| 18 | hsa-miR-374b-5p | CASP3 | -0.31 | 0.00301 | 0.31 | 0.00047 | mirMAP | -0.14 | 0.00058 | NA | |
| 19 | hsa-miR-455-5p | CASP8 | -0.27 | 0.05813 | 0.33 | 0.00029 | miRanda | -0.15 | 0 | NA | |
| 20 | hsa-miR-542-3p | CASP8 | -1.31 | 0 | 0.33 | 0.00029 | miRanda | -0.13 | 4.0E-5 | NA | |
| 21 | hsa-let-7b-5p | CCNB1 | -0.96 | 0 | 3.16 | 0 | miRNAWalker2 validate | -0.54 | 0 | NA | |
| 22 | hsa-miR-139-5p | CCNB1 | -2.11 | 0 | 3.16 | 0 | miRanda | -0.8 | 0 | NA | |
| 23 | hsa-let-7a-5p | CCNB2 | -0.33 | 0.00046 | 4.24 | 0 | miRNAWalker2 validate | -0.45 | 0.00714 | NA | |
| 24 | hsa-let-7b-5p | CCNB2 | -0.96 | 0 | 4.24 | 0 | miRNAWalker2 validate | -0.59 | 0 | NA | |
| 25 | hsa-let-7c-5p | CCNB2 | -1.71 | 0 | 4.24 | 0 | miRNAWalker2 validate | -0.85 | 0 | NA | |
| 26 | hsa-miR-23b-3p | CCNB2 | -0.53 | 0 | 4.24 | 0 | miRNAWalker2 validate | -0.64 | 1.0E-5 | NA | |
| 27 | hsa-miR-339-5p | CCNB3 | 0.28 | 0.03557 | 0.19 | 0.27484 | miRanda | -0.21 | 0.00149 | NA | |
| 28 | hsa-miR-106a-5p | CCND1 | -0.46 | 0.00972 | -0.9 | 1.0E-5 | MirTarget; miRNATAP | -0.26 | 0 | NA | |
| 29 | hsa-miR-106b-5p | CCND1 | 0.65 | 0 | -0.9 | 1.0E-5 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.43 | 0 | NA | |
| 30 | hsa-miR-1266-5p | CCND1 | 1.63 | 0 | -0.9 | 1.0E-5 | MirTarget | -0.23 | 0 | NA | |
| 31 | hsa-miR-15b-5p | CCND1 | 0.23 | 0.08248 | -0.9 | 1.0E-5 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.54 | 0 | NA | |
| 32 | hsa-miR-16-5p | CCND1 | -0.4 | 0.0001 | -0.9 | 1.0E-5 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.31 | 0.00178 | 23991964; 22922827; 18483394 | At the molecular level our results further revealed that cyclin D1 expression was negatively regulated by miR-16;CCND1 has been found to be a target of miR-15a and miR-16-1 through analysis of complementary sequences between microRNAs and CCND1 mRNA; Moreover the transcription of CCND1 is suppressed by miR-15a and miR-16-1 via direct binding to the CCND1 3'-untranslated region 3'-UTR;Truncation in CCND1 mRNA alters miR 16 1 regulation in mantle cell lymphoma; Furthermore we demonstrated that this truncation alters miR-16-1 binding sites and through the use of reporter constructs we were able to show that miR-16-1 regulates CCND1 mRNA expression; This study introduces the role of miR-16-1 in the regulation of CCND1 in MCL |
| 33 | hsa-miR-17-5p | CCND1 | 0.7 | 2.0E-5 | -0.9 | 1.0E-5 | miRNAWalker2 validate; MirTarget; TargetScan; miRNATAP | -0.34 | 0 | 26431674 | Bioinformatics Prediction and In Vitro Analysis Revealed That miR 17 Targets Cyclin D1 mRNA in Triple Negative Breast Cancer Cells; In this study using bioinformatic analyses miR-17 was selected as it targets the 3'UTR of CCND1 gene with the highest score; After lentiviral transduction of miR-17 to the target cells gene expression analysis showed decreased expression of CCND1 gene |
| 34 | hsa-miR-186-5p | CCND1 | -0.06 | 0.53529 | -0.9 | 1.0E-5 | mirMAP | -0.32 | 0.00286 | NA | |
| 35 | hsa-miR-19a-3p | CCND1 | 1.02 | 0 | -0.9 | 1.0E-5 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.28 | 0 | 25985117 | Moreover miR-19a might play inhibitory roles in HCC malignancy via regulating Cyclin D1 expression |
| 36 | hsa-miR-19b-1-5p | CCND1 | -0.28 | 0.07831 | -0.9 | 1.0E-5 | miRNAWalker2 validate; miRTarBase | -0.31 | 0 | NA | |
| 37 | hsa-miR-19b-3p | CCND1 | 0.6 | 0.00017 | -0.9 | 1.0E-5 | miRNATAP | -0.34 | 0 | NA | |
| 38 | hsa-miR-20a-5p | CCND1 | 0.85 | 0 | -0.9 | 1.0E-5 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.33 | 0 | NA | |
| 39 | hsa-miR-20b-5p | CCND1 | 0.46 | 0.02859 | -0.9 | 1.0E-5 | MirTarget; miRNATAP | -0.23 | 0 | NA | |
| 40 | hsa-miR-340-5p | CCND1 | -0 | 0.9685 | -0.9 | 1.0E-5 | mirMAP | -0.32 | 0.00013 | NA | |
| 41 | hsa-miR-425-5p | CCND1 | 0.59 | 2.0E-5 | -0.9 | 1.0E-5 | miRNAWalker2 validate | -0.39 | 0 | NA | |
| 42 | hsa-miR-503-5p | CCND1 | 0.19 | 0.26842 | -0.9 | 1.0E-5 | miRNAWalker2 validate; miRTarBase; MirTarget | -0.16 | 0.00815 | 26047605; 23731275 | MiR 503 inhibited cell proliferation of human breast cancer cells by suppressing CCND1 expression; Overexpression of miR-503 in breast cancer cell lines reduced cell proliferation through inducing G0/G1 cell cycle arrest by targeting CCND1;MicroRNA 503 suppresses proliferation and cell cycle progression of endometrioid endometrial cancer by negatively regulating cyclin D1; CCND1 has a binding sequence of miR-503 within its 3' untranslated region and was confirmed to be a direct target of miR-503 by the fluorescent reporter assays; Increasing the miR-503 level in EEC cells suppressed cell viability colon formation activity and cell-cycle progression and the inhibited oncogenic phenotypes induced by miR-503 were alleviated by ectopic expression of CCND1 without the untranslated region sequence; Collectively this study suggested that miR-503 plays a tumor-suppressor role by targeting CCND1; Abnormal suppression of miR-503 leads to an increase in the CCND1 level which may promote carcinogenesis and progression of EEC |
| 43 | hsa-miR-589-3p | CCND1 | 1.17 | 0 | -0.9 | 1.0E-5 | MirTarget | -0.18 | 0.00124 | NA | |
| 44 | hsa-miR-616-5p | CCND1 | 0.15 | 0.40284 | -0.9 | 1.0E-5 | mirMAP | -0.26 | 1.0E-5 | NA | |
| 45 | hsa-miR-7-1-3p | CCND1 | -0.57 | 2.0E-5 | -0.9 | 1.0E-5 | mirMAP | -0.26 | 0.00057 | NA | |
| 46 | hsa-miR-9-5p | CCND1 | 1.26 | 9.0E-5 | -0.9 | 1.0E-5 | miRNAWalker2 validate | -0.14 | 1.0E-5 | NA | |
| 47 | hsa-miR-92a-3p | CCND1 | 0.21 | 0.13429 | -0.9 | 1.0E-5 | miRNAWalker2 validate | -0.41 | 0 | NA | |
| 48 | hsa-miR-93-5p | CCND1 | 1.4 | 0 | -0.9 | 1.0E-5 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.34 | 0 | NA | |
| 49 | hsa-miR-942-5p | CCND1 | 0.35 | 0.02833 | -0.9 | 1.0E-5 | MirTarget | -0.25 | 0.00012 | NA | |
| 50 | hsa-miR-130b-5p | CCND2 | 0.17 | 0.33761 | 0.36 | 0.03656 | mirMAP | -0.17 | 0.00018 | NA | |
| 51 | hsa-miR-20a-5p | CCND2 | 0.85 | 0 | 0.36 | 0.03656 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.16 | 0.00121 | NA | |
| 52 | hsa-miR-28-5p | CCND2 | -0.43 | 0 | 0.36 | 0.03656 | miRanda | -0.48 | 0 | NA | |
| 53 | hsa-miR-33a-3p | CCND2 | -0.68 | 1.0E-5 | 0.36 | 0.03656 | MirTarget | -0.26 | 0 | NA | |
| 54 | hsa-miR-3607-3p | CCND2 | -2.16 | 0 | 0.36 | 0.03656 | mirMAP | -0.12 | 0.0007 | NA | |
| 55 | hsa-miR-378a-3p | CCND2 | -1.19 | 0 | 0.36 | 0.03656 | miRNAWalker2 validate | -0.18 | 2.0E-5 | NA | |
| 56 | hsa-miR-548v | CCND2 | -0.27 | 0.17626 | 0.36 | 0.03656 | MirTarget | -0.15 | 0.00034 | NA | |
| 57 | hsa-miR-616-5p | CCND2 | 0.15 | 0.40284 | 0.36 | 0.03656 | mirMAP | -0.32 | 0 | NA | |
| 58 | hsa-miR-618 | CCND2 | 0.14 | 0.51715 | 0.36 | 0.03656 | mirMAP | -0.23 | 0 | NA | |
| 59 | hsa-miR-27b-3p | CCND3 | -0.82 | 0 | 0.08 | 0.47843 | miRNAWalker2 validate | -0.24 | 0 | NA | |
| 60 | hsa-miR-320a | CCND3 | 0.33 | 0.02214 | 0.08 | 0.47843 | miRanda | -0.12 | 0.00135 | NA | |
| 61 | hsa-miR-125b-5p | CCNE1 | -1.36 | 0 | 3.05 | 0 | miRNAWalker2 validate | -0.8 | 0 | NA | |
| 62 | hsa-miR-192-5p | CCNE1 | -0.5 | 0.00345 | 3.05 | 0 | miRNAWalker2 validate | -0.35 | 2.0E-5 | NA | |
| 63 | hsa-miR-195-5p | CCNE1 | -1.86 | 0 | 3.05 | 0 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.3 | 4.0E-5 | 24402230 | Furthermore through qPCR and western blot assays we showed that overexpression of miR-195-5p reduced CCNE1 mRNA and protein levels respectively |
| 64 | hsa-miR-26a-5p | CCNE1 | -0.96 | 0 | 3.05 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.6 | 2.0E-5 | 22094936 | Cell cycle regulation and CCNE1 and CDC2 were the only significant overlapping pathway and genes differentially expressed between tumors with high and low levels of miR-26a and EZH2 respectively; Low mRNA levels of EZH2 CCNE1 and CDC2 and high levels of miR-26a are associated with favorable outcome on tamoxifen |
| 65 | hsa-miR-26b-5p | CCNE1 | -1.11 | 0 | 3.05 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.89 | 0 | NA | |
| 66 | hsa-miR-424-5p | CCNE1 | -2.63 | 0 | 3.05 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.37 | 0 | NA | |
| 67 | hsa-miR-497-5p | CCNE1 | -1.41 | 0 | 3.05 | 0 | MirTarget; miRNATAP | -0.27 | 0.00125 | 24112607; 25909221; 24909281 | Western blot assays confirmed that overexpression of miR-497 reduced cyclin E1 protein levels; Inhibited cellular growth suppressed cellular migration and invasion and G1 cell cycle arrest were observed upon overexpression of miR-497 in cells possibly by targeting cyclin E1;The effect of simultaneous overexpression of miR-497 and miR-34a on the inhibition of cell proliferation colony formation and tumor growth and the downregulation of cyclin E1 was stronger than the effect of each miRNA alone; The synergistic actions of miR-497 and miR-34a partly correlated with cyclin E1 levels; These results indicate cyclin E1 is downregulated by both miR-497 and miR-34a which synergistically retard the growth of human lung cancer cells;miR 497 suppresses proliferation of human cervical carcinoma HeLa cells by targeting cyclin E1; Furthermore the target effect of miR-497 on the CCNE1 was identified by dual-luciferase reporter assay system qRT-PCR and Western blotting; Over-expressed miR-497 in HeLa cells could suppress cell proliferation by targeting CCNE1 |
| 68 | hsa-let-7b-3p | CCNE2 | -1.22 | 0 | 2.02 | 0 | mirMAP | -0.27 | 0.00021 | NA | |
| 69 | hsa-miR-126-3p | CCNE2 | -0.65 | 0 | 2.02 | 0 | miRNAWalker2 validate | -0.39 | 5.0E-5 | NA | |
| 70 | hsa-miR-26a-5p | CCNE2 | -0.96 | 0 | 2.02 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.49 | 0 | 24116110; 21901171 | The loss of miR 26a mediated post transcriptional regulation of cyclin E2 in pancreatic cancer cell proliferation and decreased patient survival; The in vitro and in vivo assays showed that overexpression of miR-26a resulted in cell cycle arrest inhibited cell proliferation and decreased tumor growth which was associated with cyclin E2 downregulation;We also show that enforced expression of miR-26a in AML cells is able to inhibit cell cycle progression by downregulating cyclin E2 expression |
| 71 | hsa-miR-26b-5p | CCNE2 | -1.11 | 0 | 2.02 | 0 | miRNATAP | -0.58 | 0 | NA | |
| 72 | hsa-miR-30a-5p | CCNE2 | -0.63 | 0.00011 | 2.02 | 0 | miRNATAP | -0.36 | 0 | NA | |
| 73 | hsa-let-7e-5p | CCNG1 | 0.04 | 0.81107 | -0.05 | 0.64033 | miRNAWalker2 validate | -0.2 | 0 | NA | |
| 74 | hsa-miR-132-3p | CCNG1 | 0.32 | 0.00272 | -0.05 | 0.64033 | MirTarget | -0.19 | 1.0E-5 | NA | |
| 75 | hsa-miR-142-3p | CCNG1 | -1.42 | 0 | -0.05 | 0.64033 | miRanda | -0.12 | 0 | NA | |
| 76 | hsa-miR-142-5p | CCNG1 | -1.45 | 0 | -0.05 | 0.64033 | mirMAP | -0.12 | 0 | NA | |
| 77 | hsa-miR-181a-5p | CCNG1 | 0.25 | 0.05519 | -0.05 | 0.64033 | miRNAWalker2 validate | -0.22 | 0 | NA | |
| 78 | hsa-miR-21-5p | CCNG1 | 1.51 | 0 | -0.05 | 0.64033 | miRNAWalker2 validate | -0.13 | 0.00024 | NA | |
| 79 | hsa-miR-212-3p | CCNG1 | -0.29 | 0.10039 | -0.05 | 0.64033 | MirTarget | -0.13 | 0 | NA | |
| 80 | hsa-miR-23a-3p | CCNG1 | -0.18 | 0.13598 | -0.05 | 0.64033 | MirTarget; miRNATAP | -0.32 | 0 | NA | |
| 81 | hsa-miR-24-3p | CCNG1 | -0.26 | 0.0069 | -0.05 | 0.64033 | miRNAWalker2 validate | -0.34 | 0 | NA | |
| 82 | hsa-miR-27a-3p | CCNG1 | -0.37 | 0.00876 | -0.05 | 0.64033 | MirTarget; miRNATAP | -0.28 | 0 | NA | |
| 83 | hsa-miR-339-5p | CCNG1 | 0.28 | 0.03557 | -0.05 | 0.64033 | miRanda | -0.18 | 0 | NA | |
| 84 | hsa-miR-590-5p | CCNG1 | -0.1 | 0.31003 | -0.05 | 0.64033 | miRanda | -0.18 | 0.00014 | NA | |
| 85 | hsa-miR-101-3p | CCNG2 | -1.48 | 0 | 0.2 | 0.09986 | miRNATAP | -0.15 | 0.00064 | NA | |
| 86 | hsa-miR-139-5p | CCNG2 | -2.11 | 0 | 0.2 | 0.09986 | miRanda | -0.12 | 4.0E-5 | NA | |
| 87 | hsa-miR-192-3p | CCNG2 | -0.64 | 0.00027 | 0.2 | 0.09986 | MirTarget | -0.17 | 0 | NA | |
| 88 | hsa-miR-365a-3p | CCNG2 | 0.16 | 0.15325 | 0.2 | 0.09986 | MirTarget | -0.2 | 8.0E-5 | NA | |
| 89 | hsa-miR-374b-5p | CCNG2 | -0.31 | 0.00301 | 0.2 | 0.09986 | mirMAP | -0.16 | 0.00526 | NA | |
| 90 | hsa-miR-590-5p | CCNG2 | -0.1 | 0.31003 | 0.2 | 0.09986 | mirMAP | -0.17 | 0.00377 | NA | |
| 91 | hsa-miR-362-3p | CD82 | 0.81 | 0 | -1.1 | 0 | miRanda | -0.15 | 0.00572 | 25652145 | Anti miR 362 3p Inhibits Migration and Invasion of Human Gastric Cancer Cells by Its Target CD82; Next we analyzed the level of miR-362-3p expression and CD82 in different differentiated GC cells compared with a normal gastric mucosa cell by RT-PCR and Western blot; Dual-luciferase reporter assay and Western blot confirmed a direct interaction between miR-362-3p and CD82 3'UTR; After miR-362-3p and CD82 were silenced in GC cells we compared the transfected GC cells migration and invasion capacity by transwell assay; Western blot was used to detect the impact of CD82 and miR-362-3p on epithelial-to-mesenchymal transition markers in treated GC cells; Level of miR-362-3p expression was much higher in GC cells than in normal gastric mucosa cell and miR-362-3p expression negatively correlated with CD82 mRNA expression in these cell lines; Furthermore miR-362-3p expression induced corrected GC cell metastasis capacity by suppression of CD82 expression; This study illuminated that downregulation of miR-362-3p along with the upregulation of CD82 in GC cells resulted in the inhibition of GC migration and invasion; Thus our results suggested that miR-362-3p or CD82 can be exploited as a new potential target for control of GC in the future |
| 92 | hsa-miR-122-5p | CDK4 | -1.24 | 0 | 0.67 | 0 | miRNAWalker2 validate | -0.12 | 0 | NA | |
| 93 | hsa-miR-145-5p | CDK4 | -1.48 | 0 | 0.67 | 0 | miRNAWalker2 validate; miRTarBase | -0.15 | 0 | 21092188 | Furthermore we found that CDK4 was regulated by miR-145 in cell cycle control |
| 94 | hsa-miR-193b-3p | CDK4 | -0.17 | 0.27202 | 0.67 | 0 | miRNAWalker2 validate | -0.15 | 0 | NA | |
| 95 | hsa-miR-195-5p | CDK4 | -1.86 | 0 | 0.67 | 0 | miRNAWalker2 validate; miRTarBase | -0.18 | 0 | NA | |
| 96 | hsa-let-7a-3p | CDK6 | -0.57 | 0 | -0.31 | 0.22057 | miRNATAP | -0.34 | 0.00261 | NA | |
| 97 | hsa-let-7b-5p | CDK6 | -0.96 | 0 | -0.31 | 0.22057 | miRNAWalker2 validate; miRTarBase | -0.22 | 0.00756 | NA | |
| 98 | hsa-miR-106a-5p | CDK6 | -0.46 | 0.00972 | -0.31 | 0.22057 | mirMAP | -0.28 | 4.0E-5 | NA | |
| 99 | hsa-miR-106b-5p | CDK6 | 0.65 | 0 | -0.31 | 0.22057 | mirMAP | -0.39 | 0.00029 | NA | |
| 100 | hsa-miR-141-3p | CDK6 | -0.35 | 0.257 | -0.31 | 0.22057 | TargetScan; miRNATAP | -0.14 | 0.00031 | NA | |
| 101 | hsa-miR-148b-3p | CDK6 | 0.27 | 0.00185 | -0.31 | 0.22057 | mirMAP | -0.76 | 0 | NA | |
| 102 | hsa-miR-16-5p | CDK6 | -0.4 | 0.0001 | -0.31 | 0.22057 | miRNAWalker2 validate; miRTarBase | -0.5 | 3.0E-5 | NA | |
| 103 | hsa-miR-17-5p | CDK6 | 0.7 | 2.0E-5 | -0.31 | 0.22057 | TargetScan; mirMAP | -0.33 | 0 | NA | |
| 104 | hsa-miR-182-5p | CDK6 | 1.97 | 0 | -0.31 | 0.22057 | mirMAP | -0.11 | 0.00279 | NA | |
| 105 | hsa-miR-195-5p | CDK6 | -1.86 | 0 | -0.31 | 0.22057 | miRNAWalker2 validate; miRTarBase | -0.26 | 2.0E-5 | 23333942 | Expression of cyclin-dependent kinase 6 and vascular endothelial growth factor was down-regulated by exogenous miR-195 and miR-378 respectively |
| 106 | hsa-miR-200c-3p | CDK6 | -0.1 | 0.71696 | -0.31 | 0.22057 | mirMAP | -0.12 | 0.00446 | NA | |
| 107 | hsa-miR-20a-5p | CDK6 | 0.85 | 0 | -0.31 | 0.22057 | mirMAP | -0.34 | 0 | NA | |
| 108 | hsa-miR-20b-5p | CDK6 | 0.46 | 0.02859 | -0.31 | 0.22057 | mirMAP | -0.21 | 0.0003 | 26166554 | The transfection of miR-20b into EJ cells induced G1 phase cell cycle arrest via the decreased expression of cyclin D1 CDK2 and CDK6 without affecting another G1 phase cell cycle regulator cyclin E |
| 109 | hsa-miR-217 | CDK6 | 1.06 | 0.0401 | -0.31 | 0.22057 | mirMAP | -0.18 | 0 | NA | |
| 110 | hsa-miR-224-3p | CDK6 | 1.41 | 0 | -0.31 | 0.22057 | mirMAP | -0.14 | 0.00324 | NA | |
| 111 | hsa-miR-26a-5p | CDK6 | -0.96 | 0 | -0.31 | 0.22057 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.45 | 0.00013 | 26314438 | Maxvision immunohistochemistry technique was used to detect the expression level of CDK6 and miR-26a in tissue of 20 ENKTCL cases 10 cases of proliferative lymphadenitis and 10 samples of normal lymph node respectively; The possible role of miR-26a and its target gene CDK6 in genesis and development of ENKTCL were analyzed according to the clinical features of ENKTCL patients; Correlation analysis showed that there was significant negative correlation between miR-26a expression and CDK6 expression r = -0.54 P = 0.04 |
| 112 | hsa-miR-338-3p | CDK6 | 0.54 | 0.00461 | -0.31 | 0.22057 | mirMAP | -0.19 | 0.00245 | NA | |
| 113 | hsa-miR-340-5p | CDK6 | -0 | 0.9685 | -0.31 | 0.22057 | mirMAP | -0.59 | 0 | NA | |
| 114 | hsa-miR-425-5p | CDK6 | 0.59 | 2.0E-5 | -0.31 | 0.22057 | mirMAP | -0.37 | 2.0E-5 | NA | |
| 115 | hsa-miR-452-5p | CDK6 | 1.92 | 0 | -0.31 | 0.22057 | mirMAP | -0.15 | 0.00162 | NA | |
| 116 | hsa-miR-497-5p | CDK6 | -1.41 | 0 | -0.31 | 0.22057 | miRNATAP | -0.34 | 0 | NA | |
| 117 | hsa-miR-502-3p | CDK6 | 0.66 | 0 | -0.31 | 0.22057 | PITA; miRNATAP | -0.49 | 0 | NA | |
| 118 | hsa-miR-616-5p | CDK6 | 0.15 | 0.40284 | -0.31 | 0.22057 | mirMAP | -0.22 | 0.00134 | NA | |
| 119 | hsa-miR-885-5p | CDK6 | -0.94 | 0.00119 | -0.31 | 0.22057 | mirMAP | -0.31 | 0 | NA | |
| 120 | hsa-miR-92a-3p | CDK6 | 0.21 | 0.13429 | -0.31 | 0.22057 | miRNATAP | -0.48 | 0 | NA | |
| 121 | hsa-miR-106b-5p | CDKN1A | 0.65 | 0 | -0.77 | 6.0E-5 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.44 | 0 | NA | |
| 122 | hsa-miR-146b-5p | CDKN1A | 0.42 | 0.04574 | -0.77 | 6.0E-5 | miRNAWalker2 validate | -0.15 | 0.00059 | 27602131 | During the search for potential targets of miR-146b in ATC p21 also known as p21Waf1/Cip1 or CDKN1A was noted for its role in cell cycle progression and tumor pathogenesis |
| 123 | hsa-miR-17-5p | CDKN1A | 0.7 | 2.0E-5 | -0.77 | 6.0E-5 | miRNAWalker2 validate; miRTarBase; MirTarget; TargetScan; miRNATAP | -0.3 | 0 | 26482648; 24989082 | The low expressions of miR-17 and miR-92 families can maintain cisplatin resistance through the regulation of CDKN1A and RAD21;According to PicTar and Miranda algorithms which predicted CDKN1A p21 as a putative target of miR-17 a luciferase assay was performed and revealed that miR-17 directly targets the 3'-UTR of p21 mRNA |
| 124 | hsa-miR-20a-5p | CDKN1A | 0.85 | 0 | -0.77 | 6.0E-5 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.28 | 0 | 26012475 | Using the poorly tumorigenic and TGF-β-sensitive FET cell line that expresses low miR-20a levels we first confirmed that miR-20a downmodulated CDKN1A expression both at mRNA and protein level through direct binding to its 3'-UTR; Moreover besides modulating CDKN1A miR-20a blocked TGF-β-induced transactivation of its promoter without affecting the post-receptor activation of Smad3/4 effectors directly; Finally miR-20a abrogated the TGF-β-mediated c-Myc repression a direct inhibitor of the CDKN1A promoter activation most likely by reducing the expression of specific MYC-regulating genes from the Smad/E2F-based core repressor complex |
| 125 | hsa-miR-423-3p | CDKN1A | 0.3 | 0.00067 | -0.77 | 6.0E-5 | miRNAWalker2 validate; miRTarBase | -0.42 | 6.0E-5 | NA | |
| 126 | hsa-miR-423-5p | CDKN1A | 0.7 | 0 | -0.77 | 6.0E-5 | MirTarget | -0.37 | 3.0E-5 | NA | |
| 127 | hsa-miR-503-5p | CDKN1A | 0.19 | 0.26842 | -0.77 | 6.0E-5 | miRNAWalker2 validate; miRTarBase | -0.14 | 0.00973 | NA | |
| 128 | hsa-miR-93-5p | CDKN1A | 1.4 | 0 | -0.77 | 6.0E-5 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.45 | 0 | 25633810 | MicroRNA 93 activates c Met/PI3K/Akt pathway activity in hepatocellular carcinoma by directly inhibiting PTEN and CDKN1A; We confirmed that miR-93 directly bound with the 3' untranslated regions of the tumor-suppressor genes PTEN and CDKN1A respectivelyand inhibited their expression; We concluded that miR-93 stimulated cell proliferation migration and invasion through the oncogenic c-Met/PI3K/Akt pathway and also inhibited apoptosis by directly inhibiting PTEN and CDKN1A expression in human HCC |
| 129 | hsa-miR-942-5p | CDKN1A | 0.35 | 0.02833 | -0.77 | 6.0E-5 | miRNAWalker2 validate | -0.26 | 1.0E-5 | NA | |
| 130 | hsa-let-7g-5p | CDKN2A | -0.46 | 2.0E-5 | 4 | 0 | miRNAWalker2 validate; miRTarBase | -0.48 | 0.002 | NA | |
| 131 | hsa-miR-125a-5p | CDKN2A | -0.91 | 0 | 4 | 0 | miRanda | -0.54 | 0 | NA | |
| 132 | hsa-miR-125b-5p | CDKN2A | -1.36 | 0 | 4 | 0 | miRNAWalker2 validate | -0.65 | 0 | 23585871 | In this study we further extend our studies by showing that miR-125b represses the protein product of the ink4a/ARF locus p14ARF in two prostate cancer cell lines LNCaP wild type-p53 and 22Rv1 both wild type and mutant p53 as well as in the PC-346C prostate cancer xenograft model that lentivirally overexpressed miR-125b; Conversely treatment of prostate cancer cells with an inhibitor of miR-125b anti-miR-125b resulted in increased expression of p14ARF decreased level of Mdm2 and induction of apoptosis; In addition overexpression of miR-125b in p53-deficient PC3 cells induced down-regulation of p14ARF which leads to increased cell proliferation through a p53-independent manner |
| 133 | hsa-miR-455-3p | CDKN2A | -1.4 | 0 | 4 | 0 | miRNAWalker2 validate | -0.42 | 1.0E-5 | NA | |
| 134 | hsa-miR-139-5p | CHEK1 | -2.11 | 0 | 1.38 | 0 | miRanda | -0.41 | 0 | NA | |
| 135 | hsa-miR-195-5p | CHEK1 | -1.86 | 0 | 1.38 | 0 | MirTarget; miRNATAP | -0.27 | 0 | 25840419 | MiR 195 suppresses non small cell lung cancer by targeting CHEK1; We discovered that CHEK1 was a direct target of miR-195 which decreased CHEK1 expression in lung cancer cells |
| 136 | hsa-miR-326 | CHEK1 | -1.88 | 0 | 1.38 | 0 | miRanda | -0.11 | 0.00505 | NA | |
| 137 | hsa-miR-424-5p | CHEK1 | -2.63 | 0 | 1.38 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.28 | 0 | 22469983 | Suppressed miR 424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer; Moreover miR-424 expression levels were inversely correlated with Chk1 and p-Chk1 protein levels in both cervical cancer and normal tissues; Furthermore RNAi-mediated knockdown of Chk1 decreased matrix metalloproteinase 9 expression and phenocopied the tumor suppressive effects of miR-424 in cell models; Taken together our results identify a crucial tumor suppressive role of miR-424 in the progression of cervical cancer at least partly via upreglating the expression of Chk1 and p-Chk1 and suggest that miR-424 might be a candidate of prognostic predictor or an anticancer therapeutic target for cervical cancer patients |
| 138 | hsa-miR-497-5p | CHEK1 | -1.41 | 0 | 1.38 | 0 | MirTarget; miRNATAP | -0.29 | 0 | 24464213 | Checkpoint kinase 1 is negatively regulated by miR 497 in hepatocellular carcinoma; In silico analysis showed that CHEK1 was a candidate target of miR-497 which was previously found to be downregulated in HCC by us; To test whether miR-497 could bind to 3'untranslated region 3'UTR of CHEK1 luciferase reporter assay was conducted; The result revealed that miR-497 could bind to the 3'untranslated region 3'UTR of CHEK1 mRNA; Western blot showed that ectopic expression of miR-497 suppressed the CHEK1 expression and inhibition of miR-497 led to significant upregulation of CHEK1; Finally miR-497 expression was measured in the same 30 HCC samples and the correlation between miR-497 and CHEK1 was analyzed; The results indicated that miR-497 was downregulated in HCC and had a significant negative correlation with CHEK1; Taken together these results demonstrated that CHEK1 was negatively regulated by miR-497 and the overexpressed CHEK1 was resulted from the downregulated miR-497 in HCC which provided a potential molecular target for HCC therapy |
| 139 | hsa-miR-511-5p | CHEK1 | -1.75 | 0 | 1.38 | 0 | MirTarget | -0.26 | 0 | NA | |
| 140 | hsa-miR-542-3p | CHEK2 | -1.31 | 0 | 0.99 | 0 | miRanda | -0.18 | 0.00279 | NA | |
| 141 | hsa-miR-361-5p | CYCS | 0.23 | 0.00962 | 0.26 | 0.01519 | miRNAWalker2 validate | -0.17 | 0.00331 | NA | |
| 142 | hsa-miR-330-5p | DDB2 | 0.44 | 0.00533 | -0.47 | 0.00025 | miRanda | -0.15 | 0.00021 | NA | |
| 143 | hsa-miR-27a-3p | EI24 | -0.37 | 0.00876 | -0.75 | 0 | miRNATAP | -0.12 | 0.00041 | NA | |
| 144 | hsa-miR-330-5p | EI24 | 0.44 | 0.00533 | -0.75 | 0 | miRanda | -0.13 | 2.0E-5 | NA | |
| 145 | hsa-miR-106a-5p | FAS | -0.46 | 0.00972 | -1.02 | 0 | miRNAWalker2 validate; miRTarBase | -0.3 | 0 | 22431000; 27142596 | miR 106a is frequently upregulated in gastric cancer and inhibits the extrinsic apoptotic pathway by targeting FAS; Bioinformatic analysis combining with validation experiments identified FAS as a direct target of miR-106a; Moreover a significant inverse correlation was found between miR-106a and FAS expression not only in gastric cancer cell lines but also in gastric cancer specimens; Taken together these findings suggest that ectopicly overexpressed miR-106a may play an oncogenic role in gastric carcinogenesis and impair extrinsic apoptotic pathway through targeting FAS;Functional experiment ascertained that miR-106a interacted with FAS and mediated caspase3 pathway |
| 146 | hsa-miR-21-5p | FAS | 1.51 | 0 | -1.02 | 0 | miRNAWalker2 validate | -0.35 | 0 | 24710931 | miR 21 targets Fas ligand mediated apoptosis in breast cancer cell line MCF 7 |
| 147 | hsa-miR-338-3p | FAS | 0.54 | 0.00461 | -1.02 | 0 | miRanda | -0.23 | 0 | NA | |
| 148 | hsa-miR-361-5p | FAS | 0.23 | 0.00962 | -1.02 | 0 | miRanda | -0.4 | 0.00016 | NA | |
| 149 | hsa-miR-590-5p | FAS | -0.1 | 0.31003 | -1.02 | 0 | miRanda | -0.56 | 0 | NA | |
| 150 | hsa-miR-98-5p | FAS | -0.05 | 0.71591 | -1.02 | 0 | miRNAWalker2 validate | -0.18 | 0.00981 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | REGULATION OF CELL CYCLE | 33 | 949 | 1.344e-29 | 6.255e-26 |
| 2 | CELL CYCLE | 32 | 1316 | 9.932e-24 | 2.311e-20 |
| 3 | POSITIVE REGULATION OF CELL DEATH | 24 | 605 | 3.873e-22 | 4.506e-19 |
| 4 | REGULATION OF CELL DEATH | 32 | 1472 | 3.069e-22 | 4.506e-19 |
| 5 | NEGATIVE REGULATION OF CELL CYCLE | 21 | 433 | 4.594e-21 | 4.275e-18 |
| 6 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 31 | 1492 | 7.534e-21 | 5.843e-18 |
| 7 | CELL DEATH | 27 | 1001 | 9.994e-21 | 6.643e-18 |
| 8 | REGULATION OF CELL CYCLE ARREST | 14 | 108 | 3.604e-20 | 2.096e-17 |
| 9 | CELL CYCLE PROCESS | 27 | 1081 | 7.273e-20 | 3.76e-17 |
| 10 | MITOTIC CELL CYCLE | 24 | 766 | 9.531e-20 | 4.435e-17 |
| 11 | SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 14 | 127 | 3.848e-19 | 1.628e-16 |
| 12 | NEGATIVE REGULATION OF CELL CYCLE G1 S PHASE TRANSITION | 13 | 98 | 6.537e-19 | 2.482e-16 |
| 13 | POSITIVE REGULATION OF CELL CYCLE | 18 | 332 | 6.934e-19 | 2.482e-16 |
| 14 | G1 DNA DAMAGE CHECKPOINT | 12 | 73 | 1.064e-18 | 3.536e-16 |
| 15 | REGULATION OF CELL CYCLE G1 S PHASE TRANSITION | 14 | 147 | 3.181e-18 | 9.869e-16 |
| 16 | CELL CYCLE CHECKPOINT | 15 | 194 | 4.202e-18 | 1.222e-15 |
| 17 | POSITIVE REGULATION OF CELL CYCLE PROCESS | 16 | 247 | 4.822e-18 | 1.275e-15 |
| 18 | RESPONSE TO ABIOTIC STIMULUS | 25 | 1024 | 4.932e-18 | 1.275e-15 |
| 19 | REGULATION OF PROTEIN MODIFICATION PROCESS | 30 | 1710 | 5.268e-18 | 1.29e-15 |
| 20 | POSITIVE REGULATION OF CELL CYCLE ARREST | 12 | 85 | 7.429e-18 | 1.728e-15 |
| 21 | CELLULAR RESPONSE TO DNA DAMAGE STIMULUS | 22 | 720 | 8.553e-18 | 1.895e-15 |
| 22 | REGULATION OF CELL CYCLE PHASE TRANSITION | 17 | 321 | 1.135e-17 | 2.4e-15 |
| 23 | REGULATION OF TRANSFERASE ACTIVITY | 24 | 946 | 1.214e-17 | 2.457e-15 |
| 24 | REGULATION OF MITOTIC CELL CYCLE | 19 | 468 | 1.378e-17 | 2.672e-15 |
| 25 | NEGATIVE REGULATION OF CELL CYCLE PROCESS | 15 | 214 | 1.857e-17 | 3.457e-15 |
| 26 | SIGNAL TRANSDUCTION IN RESPONSE TO DNA DAMAGE | 12 | 96 | 3.449e-17 | 6.172e-15 |
| 27 | APOPTOTIC SIGNALING PATHWAY | 16 | 289 | 5.911e-17 | 9.822e-15 |
| 28 | MITOTIC DNA INTEGRITY CHECKPOINT | 12 | 100 | 5.752e-17 | 9.822e-15 |
| 29 | CELLULAR RESPONSE TO STRESS | 28 | 1565 | 7.339e-17 | 1.178e-14 |
| 30 | NEGATIVE REGULATION OF CELL CYCLE PHASE TRANSITION | 13 | 146 | 1.402e-16 | 2.104e-14 |
| 31 | DNA INTEGRITY CHECKPOINT | 13 | 146 | 1.402e-16 | 2.104e-14 |
| 32 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 28 | 1618 | 1.74e-16 | 2.53e-14 |
| 33 | NEGATIVE REGULATION OF MITOTIC CELL CYCLE | 14 | 199 | 2.365e-16 | 3.334e-14 |
| 34 | REGULATION OF CELL CYCLE PROCESS | 19 | 558 | 3.521e-16 | 4.818e-14 |
| 35 | REGULATION OF KINASE ACTIVITY | 21 | 776 | 6.647e-16 | 8.837e-14 |
| 36 | ACTIVATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 11 | 95 | 1.91e-15 | 2.469e-13 |
| 37 | POSITIVE REGULATION OF PROTEOLYSIS | 16 | 363 | 2.15e-15 | 2.704e-13 |
| 38 | REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 11 | 97 | 2.423e-15 | 2.967e-13 |
| 39 | MITOTIC CELL CYCLE CHECKPOINT | 12 | 139 | 3.37e-15 | 4.02e-13 |
| 40 | REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 17 | 470 | 6.438e-15 | 7.489e-13 |
| 41 | CELL CYCLE PHASE TRANSITION | 14 | 255 | 7.563e-15 | 8.583e-13 |
| 42 | INTRINSIC APOPTOTIC SIGNALING PATHWAY | 12 | 152 | 1.001e-14 | 1.109e-12 |
| 43 | ZYMOGEN ACTIVATION | 11 | 112 | 1.239e-14 | 1.341e-12 |
| 44 | REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 13 | 213 | 1.982e-14 | 2.096e-12 |
| 45 | REGULATION OF PROTEOLYSIS | 19 | 711 | 2.852e-14 | 2.949e-12 |
| 46 | REGULATION OF CELL PROLIFERATION | 25 | 1496 | 3.35e-14 | 3.388e-12 |
| 47 | POSITIVE REGULATION OF APOPTOTIC SIGNALING PATHWAY | 12 | 171 | 4.165e-14 | 4.123e-12 |
| 48 | REGULATION OF APOPTOTIC SIGNALING PATHWAY | 15 | 363 | 4.743e-14 | 4.598e-12 |
| 49 | INTRACELLULAR SIGNAL TRANSDUCTION | 25 | 1572 | 1.034e-13 | 9.815e-12 |
| 50 | RESPONSE TO OXYGEN LEVELS | 14 | 311 | 1.165e-13 | 1.084e-11 |
| 51 | RESPONSE TO IONIZING RADIATION | 11 | 145 | 2.231e-13 | 2.035e-11 |
| 52 | CELLULAR RESPONSE TO ABIOTIC STIMULUS | 13 | 263 | 2.987e-13 | 2.672e-11 |
| 53 | RESPONSE TO RADIATION | 15 | 413 | 3.09e-13 | 2.712e-11 |
| 54 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 25 | 1656 | 3.35e-13 | 2.834e-11 |
| 55 | REPLICATIVE SENESCENCE | 6 | 12 | 3.334e-13 | 2.834e-11 |
| 56 | POSITIVE REGULATION OF PEPTIDASE ACTIVITY | 11 | 154 | 4.347e-13 | 3.612e-11 |
| 57 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 24 | 1518 | 4.543e-13 | 3.709e-11 |
| 58 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 21 | 1135 | 1.138e-12 | 9.127e-11 |
| 59 | POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 19 | 876 | 1.166e-12 | 9.196e-11 |
| 60 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 25 | 1791 | 1.937e-12 | 1.477e-10 |
| 61 | RESPONSE TO UV | 10 | 126 | 1.91e-12 | 1.477e-10 |
| 62 | REGULATION OF PEPTIDASE ACTIVITY | 14 | 392 | 2.696e-12 | 2.023e-10 |
| 63 | AGING | 12 | 264 | 7.269e-12 | 5.285e-10 |
| 64 | POSITIVE REGULATION OF MITOCHONDRIAL OUTER MEMBRANE PERMEABILIZATION INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 7 | 36 | 7.17e-12 | 5.285e-10 |
| 65 | RESPONSE TO DRUG | 14 | 431 | 9.616e-12 | 6.883e-10 |
| 66 | REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 10 | 153 | 1.344e-11 | 9.474e-10 |
| 67 | NEGATIVE REGULATION OF CELL PROLIFERATION | 16 | 643 | 1.366e-11 | 9.484e-10 |
| 68 | CELL CYCLE ARREST | 10 | 154 | 1.434e-11 | 9.813e-10 |
| 69 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 19 | 1036 | 2.172e-11 | 1.444e-09 |
| 70 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 19 | 1036 | 2.172e-11 | 1.444e-09 |
| 71 | CELL DIVISION | 14 | 460 | 2.289e-11 | 1.5e-09 |
| 72 | REGULATION OF SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 10 | 162 | 2.376e-11 | 1.535e-09 |
| 73 | POSITIVE REGULATION OF PROTEIN OLIGOMERIZATION | 6 | 22 | 2.635e-11 | 1.68e-09 |
| 74 | REGULATION OF MITOCHONDRIAL OUTER MEMBRANE PERMEABILIZATION INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 7 | 43 | 2.727e-11 | 1.714e-09 |
| 75 | REGULATION OF FIBROBLAST PROLIFERATION | 8 | 81 | 6.165e-11 | 3.825e-09 |
| 76 | INTRINSIC APOPTOTIC SIGNALING PATHWAY BY P53 CLASS MEDIATOR | 7 | 53 | 1.277e-10 | 7.815e-09 |
| 77 | REGULATION OF RESPONSE TO STRESS | 21 | 1468 | 1.46e-10 | 8.762e-09 |
| 78 | CELLULAR RESPONSE TO RADIATION | 9 | 137 | 1.469e-10 | 8.762e-09 |
| 79 | CELLULAR RESPONSE TO EXTERNAL STIMULUS | 11 | 264 | 1.516e-10 | 8.932e-09 |
| 80 | PROTEIN MATURATION | 11 | 265 | 1.579e-10 | 9.183e-09 |
| 81 | REGULATION OF PROTEIN INSERTION INTO MITOCHONDRIAL MEMBRANE INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 6 | 29 | 1.653e-10 | 9.379e-09 |
| 82 | POSITIVE REGULATION OF PROTEIN INSERTION INTO MITOCHONDRIAL MEMBRANE INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 6 | 29 | 1.653e-10 | 9.379e-09 |
| 83 | REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY VIA DEATH DOMAIN RECEPTORS | 7 | 55 | 1.673e-10 | 9.381e-09 |
| 84 | RESPONSE TO X RAY | 6 | 30 | 2.062e-10 | 1.142e-08 |
| 85 | REGULATION OF CELL CYCLE G2 M PHASE TRANSITION | 7 | 59 | 2.789e-10 | 1.527e-08 |
| 86 | REGULATION OF CELLULAR RESPONSE TO STRESS | 15 | 691 | 4.474e-10 | 2.421e-08 |
| 87 | REGULATION OF PROTEIN OLIGOMERIZATION | 6 | 35 | 5.575e-10 | 2.948e-08 |
| 88 | NEURON APOPTOTIC PROCESS | 6 | 35 | 5.575e-10 | 2.948e-08 |
| 89 | CELLULAR RESPONSE TO UV | 7 | 66 | 6.272e-10 | 3.279e-08 |
| 90 | CELL AGING | 7 | 67 | 6.989e-10 | 3.613e-08 |
| 91 | CELL CYCLE G1 S PHASE TRANSITION | 8 | 111 | 7.937e-10 | 4.014e-08 |
| 92 | G1 S TRANSITION OF MITOTIC CELL CYCLE | 8 | 111 | 7.937e-10 | 4.014e-08 |
| 93 | RESPONSE TO STEROID HORMONE | 13 | 497 | 8.407e-10 | 4.206e-08 |
| 94 | REGULATION OF MEMBRANE PERMEABILITY | 7 | 70 | 9.572e-10 | 4.738e-08 |
| 95 | INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO DNA DAMAGE | 7 | 71 | 1.06e-09 | 5.19e-08 |
| 96 | RESPONSE TO LIPID | 16 | 888 | 1.565e-09 | 7.584e-08 |
| 97 | DNA METABOLIC PROCESS | 15 | 758 | 1.592e-09 | 7.636e-08 |
| 98 | NEGATIVE REGULATION OF CELL CYCLE ARREST | 5 | 20 | 2.152e-09 | 1.022e-07 |
| 99 | REGULATION OF RELEASE OF CYTOCHROME C FROM MITOCHONDRIA | 6 | 44 | 2.378e-09 | 1.118e-07 |
| 100 | RESPONSE TO ORGANIC CYCLIC COMPOUND | 16 | 917 | 2.483e-09 | 1.155e-07 |
| 101 | RESPONSE TO OXYGEN CONTAINING COMPOUND | 19 | 1381 | 2.811e-09 | 1.295e-07 |
| 102 | NEURON DEATH | 6 | 47 | 3.595e-09 | 1.624e-07 |
| 103 | POSITIVE REGULATION OF NEURON APOPTOTIC PROCESS | 6 | 47 | 3.595e-09 | 1.624e-07 |
| 104 | NEGATIVE REGULATION OF PROTEIN METABOLIC PROCESS | 17 | 1087 | 3.641e-09 | 1.629e-07 |
| 105 | POSITIVE REGULATION OF CELLULAR PROTEIN LOCALIZATION | 11 | 360 | 3.984e-09 | 1.765e-07 |
| 106 | RESPONSE TO ALCOHOL | 11 | 362 | 4.22e-09 | 1.852e-07 |
| 107 | RESPONSE TO LIGHT STIMULUS | 10 | 280 | 4.974e-09 | 2.163e-07 |
| 108 | CELLULAR RESPONSE TO OXYGEN LEVELS | 8 | 143 | 5.964e-09 | 2.569e-07 |
| 109 | CELLULAR RESPONSE TO LIGHT STIMULUS | 7 | 91 | 6.178e-09 | 2.637e-07 |
| 110 | MITOCHONDRIAL MEMBRANE ORGANIZATION | 7 | 92 | 6.672e-09 | 2.822e-07 |
| 111 | POSITIVE REGULATION OF INTRINSIC APOPTOTIC SIGNALING PATHWAY | 6 | 52 | 6.742e-09 | 2.826e-07 |
| 112 | POSITIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 6 | 53 | 7.587e-09 | 3.152e-07 |
| 113 | REGULATION OF MITOCHONDRION ORGANIZATION | 9 | 218 | 8.947e-09 | 3.652e-07 |
| 114 | RESPONSE TO ESTROGEN | 9 | 218 | 8.947e-09 | 3.652e-07 |
| 115 | REGULATION OF PROTEIN STABILITY | 9 | 221 | 1.008e-08 | 4.077e-07 |
| 116 | NEGATIVE REGULATION OF CELL DEATH | 15 | 872 | 1.057e-08 | 4.241e-07 |
| 117 | POSITIVE REGULATION OF TRANSFERASE ACTIVITY | 13 | 616 | 1.108e-08 | 4.406e-07 |
| 118 | POSITIVE REGULATION OF LEUKOCYTE APOPTOTIC PROCESS | 5 | 28 | 1.341e-08 | 5.244e-07 |
| 119 | POSITIVE REGULATION OF RELEASE OF CYTOCHROME C FROM MITOCHONDRIA | 5 | 28 | 1.341e-08 | 5.244e-07 |
| 120 | POSITIVE REGULATION OF CELL COMMUNICATION | 19 | 1532 | 1.54e-08 | 5.971e-07 |
| 121 | PROTEOLYSIS | 17 | 1208 | 1.755e-08 | 6.749e-07 |
| 122 | NEGATIVE REGULATION OF PHOSPHORYLATION | 11 | 422 | 2.056e-08 | 7.842e-07 |
| 123 | RESPONSE TO TOXIC SUBSTANCE | 9 | 241 | 2.135e-08 | 8.078e-07 |
| 124 | NEGATIVE REGULATION OF MOLECULAR FUNCTION | 16 | 1079 | 2.489e-08 | 9.339e-07 |
| 125 | RESPONSE TO METAL ION | 10 | 333 | 2.584e-08 | 9.619e-07 |
| 126 | POSITIVE REGULATION OF NEURON DEATH | 6 | 67 | 3.2e-08 | 1.174e-06 |
| 127 | REGULATION OF CELLULAR PROTEIN LOCALIZATION | 12 | 552 | 3.203e-08 | 1.174e-06 |
| 128 | T CELL HOMEOSTASIS | 5 | 34 | 3.749e-08 | 1.363e-06 |
| 129 | ACTIVATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 4 | 13 | 3.867e-08 | 1.395e-06 |
| 130 | RESPONSE TO KETONE | 8 | 182 | 3.944e-08 | 1.412e-06 |
| 131 | NEGATIVE REGULATION OF TRANSFERASE ACTIVITY | 10 | 351 | 4.241e-08 | 1.506e-06 |
| 132 | RESPONSE TO EXTERNAL STIMULUS | 20 | 1821 | 4.375e-08 | 1.542e-06 |
| 133 | NEGATIVE REGULATION OF CATALYTIC ACTIVITY | 14 | 829 | 4.544e-08 | 1.59e-06 |
| 134 | REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION TO MITOCHONDRION | 7 | 128 | 6.663e-08 | 2.314e-06 |
| 135 | POSITIVE REGULATION OF INTRACELLULAR TRANSPORT | 10 | 370 | 6.949e-08 | 2.395e-06 |
| 136 | EXTRINSIC APOPTOTIC SIGNALING PATHWAY VIA DEATH DOMAIN RECEPTORS | 5 | 39 | 7.676e-08 | 2.626e-06 |
| 137 | PROTEIN STABILIZATION | 7 | 131 | 7.816e-08 | 2.655e-06 |
| 138 | POSITIVE REGULATION OF KINASE ACTIVITY | 11 | 482 | 7.956e-08 | 2.683e-06 |
| 139 | NEGATIVE REGULATION OF APOPTOTIC SIGNALING PATHWAY | 8 | 200 | 8.185e-08 | 2.74e-06 |
| 140 | REGULATION OF LEUKOCYTE APOPTOTIC PROCESS | 6 | 79 | 8.688e-08 | 2.887e-06 |
| 141 | NEGATIVE REGULATION OF RESPONSE TO STIMULUS | 17 | 1360 | 9.919e-08 | 3.273e-06 |
| 142 | POSITIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 9 | 289 | 1.014e-07 | 3.322e-06 |
| 143 | CELL CYCLE G2 M PHASE TRANSITION | 7 | 138 | 1.118e-07 | 3.637e-06 |
| 144 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 20 | 1929 | 1.139e-07 | 3.657e-06 |
| 145 | RESPONSE TO HORMONE | 14 | 893 | 1.14e-07 | 3.657e-06 |
| 146 | REGULATION OF INTRACELLULAR TRANSPORT | 12 | 621 | 1.156e-07 | 3.686e-06 |
| 147 | POSITIVE REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 4 | 17 | 1.277e-07 | 4.041e-06 |
| 148 | REGULATION OF INTRINSIC APOPTOTIC SIGNALING PATHWAY | 7 | 145 | 1.569e-07 | 4.932e-06 |
| 149 | RESPONSE TO ESTRADIOL | 7 | 146 | 1.644e-07 | 5.1e-06 |
| 150 | POSITIVE REGULATION OF MITOCHONDRIAL MEMBRANE PERMEABILITY | 4 | 18 | 1.638e-07 | 5.1e-06 |
| 151 | NEGATIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 11 | 541 | 2.536e-07 | 7.762e-06 |
| 152 | NEGATIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 11 | 541 | 2.536e-07 | 7.762e-06 |
| 153 | POSITIVE REGULATION OF LYMPHOCYTE APOPTOTIC PROCESS | 4 | 20 | 2.583e-07 | 7.854e-06 |
| 154 | LYMPHOCYTE HOMEOSTASIS | 5 | 50 | 2.759e-07 | 8.283e-06 |
| 155 | RESPONSE TO GAMMA RADIATION | 5 | 50 | 2.759e-07 | 8.283e-06 |
| 156 | NEGATIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 6 | 98 | 3.159e-07 | 9.421e-06 |
| 157 | REGENERATION | 7 | 161 | 3.201e-07 | 9.486e-06 |
| 158 | EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 6 | 99 | 3.355e-07 | 9.88e-06 |
| 159 | POSITIVE REGULATION OF FIBROBLAST PROLIFERATION | 5 | 53 | 3.714e-07 | 1.087e-05 |
| 160 | REGULATION OF MITOCHONDRIAL MEMBRANE PERMEABILITY INVOLVED IN APOPTOTIC PROCESS | 4 | 22 | 3.883e-07 | 1.115e-05 |
| 161 | RELEASE OF CYTOCHROME C FROM MITOCHONDRIA | 4 | 22 | 3.883e-07 | 1.115e-05 |
| 162 | REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 4 | 22 | 3.883e-07 | 1.115e-05 |
| 163 | REGULATION OF LYMPHOCYTE APOPTOTIC PROCESS | 5 | 54 | 4.084e-07 | 1.164e-05 |
| 164 | POSITIVE REGULATION OF MITOCHONDRION ORGANIZATION | 7 | 167 | 4.102e-07 | 1.164e-05 |
| 165 | REGULATION OF HYDROLASE ACTIVITY | 16 | 1327 | 4.262e-07 | 1.202e-05 |
| 166 | NEGATIVE REGULATION OF KINASE ACTIVITY | 8 | 250 | 4.51e-07 | 1.257e-05 |
| 167 | POSITIVE REGULATION OF ORGANELLE ORGANIZATION | 11 | 573 | 4.487e-07 | 1.257e-05 |
| 168 | REGULATION OF NEURON DEATH | 8 | 252 | 4.791e-07 | 1.327e-05 |
| 169 | RESPONSE TO NITROGEN COMPOUND | 13 | 859 | 5.261e-07 | 1.449e-05 |
| 170 | APOPTOTIC MITOCHONDRIAL CHANGES | 5 | 57 | 5.372e-07 | 1.47e-05 |
| 171 | RESPONSE TO CORTICOSTEROID | 7 | 176 | 5.848e-07 | 1.591e-05 |
| 172 | MITOCHONDRIAL TRANSPORT | 7 | 177 | 6.076e-07 | 1.644e-05 |
| 173 | NEGATIVE REGULATION OF CELL COMMUNICATION | 15 | 1192 | 6.258e-07 | 1.683e-05 |
| 174 | MITOCHONDRION ORGANIZATION | 11 | 594 | 6.4e-07 | 1.712e-05 |
| 175 | REGULATION OF CATABOLIC PROCESS | 12 | 731 | 6.618e-07 | 1.76e-05 |
| 176 | LEUKOCYTE HOMEOSTASIS | 5 | 60 | 6.963e-07 | 1.841e-05 |
| 177 | RESPONSE TO INORGANIC SUBSTANCE | 10 | 479 | 7.503e-07 | 1.972e-05 |
| 178 | RESPONSE TO CORTICOSTERONE | 4 | 26 | 7.87e-07 | 2.057e-05 |
| 179 | NEGATIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 11 | 616 | 9.148e-07 | 2.378e-05 |
| 180 | REGULATION OF NEURON APOPTOTIC PROCESS | 7 | 192 | 1.05e-06 | 2.713e-05 |
| 181 | REGULATION OF DNA DAMAGE RESPONSE SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 4 | 28 | 1.073e-06 | 2.759e-05 |
| 182 | REGULATION OF GROWTH | 11 | 633 | 1.194e-06 | 3.052e-05 |
| 183 | POSITIVE REGULATION OF MITOTIC CELL CYCLE | 6 | 123 | 1.208e-06 | 3.073e-05 |
| 184 | POSITIVE REGULATION OF PROTEIN COMPLEX ASSEMBLY | 7 | 197 | 1.247e-06 | 3.152e-05 |
| 185 | REGULATION OF PROTEIN CATABOLIC PROCESS | 9 | 393 | 1.335e-06 | 3.359e-05 |
| 186 | RESPONSE TO ENDOGENOUS STIMULUS | 16 | 1450 | 1.388e-06 | 3.473e-05 |
| 187 | INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO DNA DAMAGE BY P53 CLASS MEDIATOR | 4 | 30 | 1.431e-06 | 3.522e-05 |
| 188 | NEGATIVE REGULATION OF B CELL ACTIVATION | 4 | 30 | 1.431e-06 | 3.522e-05 |
| 189 | NEGATIVE REGULATION OF CELL MATRIX ADHESION | 4 | 30 | 1.431e-06 | 3.522e-05 |
| 190 | DNA REPLICATION | 7 | 208 | 1.791e-06 | 4.386e-05 |
| 191 | NEGATIVE REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 4 | 32 | 1.87e-06 | 4.531e-05 |
| 192 | INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO ENDOPLASMIC RETICULUM STRESS | 4 | 32 | 1.87e-06 | 4.531e-05 |
| 193 | RESPONSE TO MECHANICAL STIMULUS | 7 | 210 | 1.909e-06 | 4.602e-05 |
| 194 | REGULATION OF PROTEIN EXPORT FROM NUCLEUS | 4 | 33 | 2.123e-06 | 5.092e-05 |
| 195 | PROTEIN KINASE B SIGNALING | 4 | 34 | 2.401e-06 | 5.7e-05 |
| 196 | NEGATIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY VIA DEATH DOMAIN RECEPTORS | 4 | 34 | 2.401e-06 | 5.7e-05 |
| 197 | POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 14 | 1152 | 2.432e-06 | 5.744e-05 |
| 198 | RESPONSE TO MINERALOCORTICOID | 4 | 35 | 2.705e-06 | 6.357e-05 |
| 199 | CELLULAR RESPONSE TO MECHANICAL STIMULUS | 5 | 80 | 2.937e-06 | 6.868e-05 |
| 200 | POSITIVE REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 4 | 36 | 3.037e-06 | 7.066e-05 |
| 201 | REGULATION OF ORGANELLE ORGANIZATION | 14 | 1178 | 3.157e-06 | 7.272e-05 |
| 202 | REGULATION OF RESPONSE TO DNA DAMAGE STIMULUS | 6 | 145 | 3.154e-06 | 7.272e-05 |
| 203 | ACTIVATION OF MAPKKK ACTIVITY | 3 | 11 | 3.377e-06 | 7.74e-05 |
| 204 | RESPONSE TO EXTRACELLULAR STIMULUS | 9 | 441 | 3.434e-06 | 7.834e-05 |
| 205 | REGULATION OF PROTEASOMAL UBIQUITIN DEPENDENT PROTEIN CATABOLIC PROCESS | 6 | 148 | 3.551e-06 | 7.981e-05 |
| 206 | ORGAN REGENERATION | 5 | 83 | 3.525e-06 | 7.981e-05 |
| 207 | RESPONSE TO TRANSITION METAL NANOPARTICLE | 6 | 148 | 3.551e-06 | 7.981e-05 |
| 208 | RESPONSE TO ENDOPLASMIC RETICULUM STRESS | 7 | 233 | 3.799e-06 | 8.498e-05 |
| 209 | PROTEIN CATABOLIC PROCESS | 10 | 579 | 4.107e-06 | 9.144e-05 |
| 210 | NEGATIVE REGULATION OF GROWTH | 7 | 236 | 4.133e-06 | 9.157e-05 |
| 211 | REGULATION OF DNA METABOLIC PROCESS | 8 | 340 | 4.484e-06 | 9.889e-05 |
| 212 | REGULATION OF CELL MATRIX ADHESION | 5 | 90 | 5.257e-06 | 0.0001154 |
| 213 | POSITIVE REGULATION OF MAPK CASCADE | 9 | 470 | 5.755e-06 | 0.0001257 |
| 214 | POSITIVE REGULATION OF DNA DAMAGE RESPONSE SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 3 | 13 | 5.83e-06 | 0.0001262 |
| 215 | RESPONSE TO COBALT ION | 3 | 13 | 5.83e-06 | 0.0001262 |
| 216 | POSITIVE REGULATION OF HYDROLASE ACTIVITY | 12 | 905 | 6.071e-06 | 0.0001308 |
| 217 | DNA REPAIR | 9 | 480 | 6.82e-06 | 0.0001462 |
| 218 | POSITIVE REGULATION OF P38MAPK CASCADE | 3 | 14 | 7.405e-06 | 0.0001566 |
| 219 | RESPONSE TO CARBOHYDRATE | 6 | 168 | 7.367e-06 | 0.0001566 |
| 220 | DETERMINATION OF ADULT LIFESPAN | 3 | 14 | 7.405e-06 | 0.0001566 |
| 221 | REGULATION OF CELLULAR LOCALIZATION | 14 | 1277 | 8.021e-06 | 0.0001689 |
| 222 | RESPONSE TO ANTIBIOTIC | 4 | 47 | 8.988e-06 | 0.0001884 |
| 223 | NEGATIVE REGULATION OF B CELL PROLIFERATION | 3 | 15 | 9.238e-06 | 0.0001928 |
| 224 | REGULATION OF PROTEIN LOCALIZATION | 12 | 950 | 9.925e-06 | 0.0002062 |
| 225 | REGULATION OF CELLULAR PROTEIN CATABOLIC PROCESS | 7 | 274 | 1.097e-05 | 0.0002268 |
| 226 | REGULATION OF PROTEASOMAL PROTEIN CATABOLIC PROCESS | 6 | 181 | 1.128e-05 | 0.0002322 |
| 227 | REGULATION OF PROTEIN HOMOOLIGOMERIZATION | 3 | 16 | 1.135e-05 | 0.0002326 |
| 228 | POSITIVE REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION | 9 | 514 | 1.18e-05 | 0.0002408 |
| 229 | CELLULAR RESPONSE TO IONIZING RADIATION | 4 | 52 | 1.35e-05 | 0.0002743 |
| 230 | POSITIVE REGULATION OF SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 3 | 17 | 1.375e-05 | 0.000277 |
| 231 | POSITIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY VIA DEATH DOMAIN RECEPTORS | 3 | 17 | 1.375e-05 | 0.000277 |
| 232 | CELLULAR RESPONSE TO EXTRACELLULAR STIMULUS | 6 | 188 | 1.4e-05 | 0.0002807 |
| 233 | NEGATIVE REGULATION OF CELL SUBSTRATE ADHESION | 4 | 53 | 1.457e-05 | 0.000291 |
| 234 | POSITIVE REGULATION OF CELLULAR COMPONENT BIOGENESIS | 8 | 406 | 1.631e-05 | 0.0003243 |
| 235 | REGULATION OF B CELL PROLIFERATION | 4 | 55 | 1.69e-05 | 0.0003347 |
| 236 | RHYTHMIC PROCESS | 7 | 298 | 1.886e-05 | 0.0003719 |
| 237 | POSITIVE REGULATION OF PROTEIN EXPORT FROM NUCLEUS | 3 | 19 | 1.952e-05 | 0.0003832 |
| 238 | REGULATION OF B CELL ACTIVATION | 5 | 121 | 2.226e-05 | 0.0004353 |
| 239 | REGULATION OF TRANSPORT | 16 | 1804 | 2.281e-05 | 0.0004442 |
| 240 | NEGATIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 5 | 126 | 2.706e-05 | 0.0005247 |
| 241 | NEGATIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 8 | 437 | 2.762e-05 | 0.0005333 |
| 242 | MEMBRANE ORGANIZATION | 11 | 899 | 3.311e-05 | 0.0006367 |
| 243 | REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 6 | 220 | 3.396e-05 | 0.0006502 |
| 244 | RESPONSE TO MAGNESIUM ION | 3 | 23 | 3.539e-05 | 0.0006667 |
| 245 | RESPONSE TO INCREASED OXYGEN LEVELS | 3 | 23 | 3.539e-05 | 0.0006667 |
| 246 | RESPONSE TO HYPEROXIA | 3 | 23 | 3.539e-05 | 0.0006667 |
| 247 | PROTEIN INSERTION INTO MEMBRANE | 3 | 23 | 3.539e-05 | 0.0006667 |
| 248 | POSITIVE REGULATION OF CELL CYCLE PHASE TRANSITION | 4 | 68 | 3.929e-05 | 0.0007371 |
| 249 | CIRCADIAN RHYTHM | 5 | 137 | 4.044e-05 | 0.0007527 |
| 250 | REGULATION OF EXECUTION PHASE OF APOPTOSIS | 3 | 24 | 4.036e-05 | 0.0007527 |
| 251 | CELLULAR RESPONSE TO ORGANIC CYCLIC COMPOUND | 8 | 465 | 4.291e-05 | 0.0007954 |
| 252 | MACROMOLECULE CATABOLIC PROCESS | 11 | 926 | 4.338e-05 | 0.000801 |
| 253 | PROTEIN LOCALIZATION TO MITOCHONDRION | 4 | 70 | 4.405e-05 | 0.0008101 |
| 254 | HISTONE PHOSPHORYLATION | 3 | 25 | 4.578e-05 | 0.0008386 |
| 255 | REGULATION OF CELLULAR COMPONENT BIOGENESIS | 10 | 767 | 4.668e-05 | 0.0008518 |
| 256 | POSITIVE REGULATION OF TRANSPORT | 11 | 936 | 4.782e-05 | 0.0008692 |
| 257 | REGULATION OF P38MAPK CASCADE | 3 | 26 | 5.165e-05 | 0.0009351 |
| 258 | NEGATIVE REGULATION OF FIBROBLAST PROLIFERATION | 3 | 27 | 5.799e-05 | 0.001046 |
| 259 | PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS | 16 | 1977 | 6.988e-05 | 0.001255 |
| 260 | REGULATION OF PROTEIN COMPLEX ASSEMBLY | 7 | 375 | 8.104e-05 | 0.00145 |
| 261 | REGULATION OF MAPK CASCADE | 9 | 660 | 8.315e-05 | 0.001482 |
| 262 | MULTICELLULAR ORGANISM AGING | 3 | 31 | 8.841e-05 | 0.00157 |
| 263 | REGULATION OF NUCLEAR DIVISION | 5 | 163 | 9.234e-05 | 0.001634 |
| 264 | POSITIVE REGULATION OF REACTIVE OXYGEN SPECIES METABOLIC PROCESS | 4 | 86 | 9.869e-05 | 0.001739 |
| 265 | MACROMOLECULAR COMPLEX ASSEMBLY | 13 | 1398 | 9.987e-05 | 0.001754 |
| 266 | CELLULAR SENESCENCE | 3 | 33 | 0.0001069 | 0.001849 |
| 267 | SIGNAL TRANSDUCTION IN ABSENCE OF LIGAND | 3 | 33 | 0.0001069 | 0.001849 |
| 268 | EXTRINSIC APOPTOTIC SIGNALING PATHWAY IN ABSENCE OF LIGAND | 3 | 33 | 0.0001069 | 0.001849 |
| 269 | REGULATION OF CELL AGING | 3 | 33 | 0.0001069 | 0.001849 |
| 270 | POSITIVE REGULATION OF CATABOLIC PROCESS | 7 | 395 | 0.000112 | 0.00193 |
| 271 | PROTEIN DESTABILIZATION | 3 | 34 | 0.000117 | 0.002009 |
| 272 | REGULATION OF CELL SUBSTRATE ADHESION | 5 | 173 | 0.0001222 | 0.00209 |
| 273 | NEGATIVE REGULATION OF PROTEIN PROCESSING | 3 | 35 | 0.0001277 | 0.002145 |
| 274 | ACTIVATION OF PROTEIN KINASE ACTIVITY | 6 | 279 | 0.0001263 | 0.002145 |
| 275 | NEGATIVE REGULATION OF PROTEIN MATURATION | 3 | 35 | 0.0001277 | 0.002145 |
| 276 | RESPONSE TO IRON ION | 3 | 35 | 0.0001277 | 0.002145 |
| 277 | REGULATION OF RESPONSE TO REACTIVE OXYGEN SPECIES | 3 | 35 | 0.0001277 | 0.002145 |
| 278 | HOMEOSTASIS OF NUMBER OF CELLS | 5 | 175 | 0.000129 | 0.002158 |
| 279 | REGULATION OF EPITHELIAL CELL PROLIFERATION | 6 | 285 | 0.0001418 | 0.002365 |
| 280 | RESPONSE TO BIOTIC STIMULUS | 10 | 886 | 0.0001543 | 0.002565 |
| 281 | POSITIVE REGULATION OF PROTEASOMAL PROTEIN CATABOLIC PROCESS | 4 | 98 | 0.0001637 | 0.002711 |
| 282 | LIMBIC SYSTEM DEVELOPMENT | 4 | 100 | 0.000177 | 0.002921 |
| 283 | RESPONSE TO NUTRIENT | 5 | 191 | 0.000194 | 0.00319 |
| 284 | POSITIVE REGULATION OF CELLULAR PROTEIN CATABOLIC PROCESS | 5 | 192 | 0.0001988 | 0.003257 |
| 285 | PROTEIN OLIGOMERIZATION | 7 | 434 | 2e-04 | 0.003266 |
| 286 | CELLULAR RESPONSE TO ESTROGEN STIMULUS | 3 | 41 | 0.0002055 | 0.003344 |
| 287 | REGULATION OF GLUCOSE METABOLIC PROCESS | 4 | 106 | 0.0002215 | 0.003591 |
| 288 | POSITIVE REGULATION OF GENE EXPRESSION | 14 | 1733 | 0.0002282 | 0.003687 |
| 289 | CELLULAR CATABOLIC PROCESS | 12 | 1322 | 0.0002436 | 0.003923 |
| 290 | PROTEIN COMPLEX BIOGENESIS | 11 | 1132 | 0.0002578 | 0.004094 |
| 291 | PROTEIN PHOSPHORYLATION | 10 | 944 | 0.0002578 | 0.004094 |
| 292 | DEVELOPMENTAL PROCESS INVOLVED IN REPRODUCTION | 8 | 602 | 0.0002556 | 0.004094 |
| 293 | PROTEIN COMPLEX ASSEMBLY | 11 | 1132 | 0.0002578 | 0.004094 |
| 294 | RESPONSE TO MOLECULE OF BACTERIAL ORIGIN | 6 | 321 | 0.0002697 | 0.004268 |
| 295 | REGULATION OF LEUKOCYTE PROLIFERATION | 5 | 206 | 0.0002753 | 0.004343 |
| 296 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 11 | 1142 | 0.0002781 | 0.004372 |
| 297 | CATABOLIC PROCESS | 14 | 1773 | 0.0002892 | 0.004531 |
| 298 | NEGATIVE REGULATION OF PROTEOLYSIS | 6 | 329 | 0.0003076 | 0.004804 |
| 299 | CARDIOVASCULAR SYSTEM DEVELOPMENT | 9 | 788 | 0.0003141 | 0.004872 |
| 300 | CIRCULATORY SYSTEM DEVELOPMENT | 9 | 788 | 0.0003141 | 0.004872 |
| 301 | CELLULAR RESPONSE TO STARVATION | 4 | 117 | 0.0003231 | 0.004995 |
| 302 | PROTEIN UBIQUITINATION | 8 | 629 | 0.0003433 | 0.005289 |
| 303 | REGULATION OF SMOOTH MUSCLE CELL MIGRATION | 3 | 49 | 0.0003497 | 0.005369 |
| 304 | POSITIVE REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 4 | 121 | 0.0003672 | 0.00562 |
| 305 | REGULATION OF CYTOPLASMIC TRANSPORT | 7 | 481 | 0.0003731 | 0.005692 |
| 306 | LYMPHOCYTE ACTIVATION | 6 | 342 | 0.0003782 | 0.005751 |
| 307 | REGULATION OF CELL ACTIVATION | 7 | 484 | 0.0003873 | 0.00587 |
| 308 | NEGATIVE REGULATION OF CELL ADHESION | 5 | 223 | 0.0003963 | 0.005987 |
| 309 | REGULATION OF IMMUNE SYSTEM PROCESS | 12 | 1403 | 0.00042 | 0.006324 |
| 310 | TELENCEPHALON DEVELOPMENT | 5 | 228 | 0.0004385 | 0.006582 |
| 311 | RESPONSE TO OXIDATIVE STRESS | 6 | 352 | 0.0004406 | 0.006592 |
| 312 | ORGANELLE FISSION | 7 | 496 | 0.0004485 | 0.006688 |
| 313 | REGULATION OF MITOCHONDRIAL MEMBRANE POTENTIAL | 3 | 54 | 0.0004661 | 0.006929 |
| 314 | EXECUTION PHASE OF APOPTOSIS | 3 | 55 | 0.000492 | 0.007291 |
| 315 | REGULATION OF THYMOCYTE APOPTOTIC PROCESS | 2 | 12 | 0.0004992 | 0.007304 |
| 316 | DEOXYRIBONUCLEOTIDE BIOSYNTHETIC PROCESS | 2 | 12 | 0.0004992 | 0.007304 |
| 317 | POSITIVE REGULATION OF EXECUTION PHASE OF APOPTOSIS | 2 | 12 | 0.0004992 | 0.007304 |
| 318 | POSITIVE REGULATION OF INSULIN LIKE GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 2 | 12 | 0.0004992 | 0.007304 |
| 319 | MITOTIC NUCLEAR DIVISION | 6 | 361 | 0.0005034 | 0.007343 |
| 320 | PHOSPHORYLATION | 11 | 1228 | 0.0005161 | 0.007505 |
| 321 | CELL PROLIFERATION | 8 | 672 | 0.0005325 | 0.007719 |
| 322 | PROTEIN UBIQUITINATION INVOLVED IN UBIQUITIN DEPENDENT PROTEIN CATABOLIC PROCESS | 4 | 134 | 0.0005403 | 0.007807 |
| 323 | RESPONSE TO ETHANOL | 4 | 136 | 0.0005713 | 0.008229 |
| 324 | MITOTIC CELL CYCLE ARREST | 2 | 13 | 0.0005889 | 0.008311 |
| 325 | REGULATION OF IRE1 MEDIATED UNFOLDED PROTEIN RESPONSE | 2 | 13 | 0.0005889 | 0.008311 |
| 326 | RESPONSE TO ALKALOID | 4 | 137 | 0.0005872 | 0.008311 |
| 327 | HEPATOCYTE APOPTOTIC PROCESS | 2 | 13 | 0.0005889 | 0.008311 |
| 328 | POSITIVE REGULATION OF ENDOPLASMIC RETICULUM UNFOLDED PROTEIN RESPONSE | 2 | 13 | 0.0005889 | 0.008311 |
| 329 | REGULATION OF HISTONE PHOSPHORYLATION | 2 | 13 | 0.0005889 | 0.008311 |
| 330 | NEGATIVE REGULATION OF IMMUNE SYSTEM PROCESS | 6 | 372 | 0.0005894 | 0.008311 |
| 331 | REGULATION OF EPITHELIAL CELL APOPTOTIC PROCESS | 3 | 59 | 0.0006049 | 0.008503 |
| 332 | RESPONSE TO BACTERIUM | 7 | 528 | 0.0006502 | 0.009112 |
| 333 | PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL | 9 | 873 | 0.0006607 | 0.009231 |
| 334 | REGULATION OF INTRACELLULAR PROTEIN TRANSPORT | 6 | 381 | 0.000668 | 0.009306 |
| 335 | MITOCHONDRIAL DNA METABOLIC PROCESS | 2 | 14 | 0.0006858 | 0.009413 |
| 336 | INSULIN LIKE GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 2 | 14 | 0.0006858 | 0.009413 |
| 337 | REGULATION OF FIBRINOLYSIS | 2 | 14 | 0.0006858 | 0.009413 |
| 338 | POSITIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY IN ABSENCE OF LIGAND | 2 | 14 | 0.0006858 | 0.009413 |
| 339 | REGULATION OF SMOOTH MUSCLE CELL APOPTOTIC PROCESS | 2 | 14 | 0.0006858 | 0.009413 |
| 340 | RAS PROTEIN SIGNAL TRANSDUCTION | 4 | 143 | 0.0006897 | 0.009439 |
| 341 | LEUKOCYTE CELL CELL ADHESION | 5 | 255 | 0.0007282 | 0.009937 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CYCLIN DEPENDENT PROTEIN SERINE THREONINE KINASE REGULATOR ACTIVITY | 7 | 28 | 1.035e-12 | 9.612e-10 |
| 2 | ENZYME BINDING | 23 | 1737 | 6.414e-11 | 2.979e-08 |
| 3 | KINASE BINDING | 14 | 606 | 8.507e-10 | 2.634e-07 |
| 4 | CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 5 | 34 | 3.749e-08 | 6.966e-06 |
| 5 | P53 BINDING | 6 | 67 | 3.2e-08 | 6.966e-06 |
| 6 | KINASE REGULATOR ACTIVITY | 7 | 186 | 8.483e-07 | 0.0001313 |
| 7 | CYCLIN DEPENDENT PROTEIN SERINE THREONINE KINASE INHIBITOR ACTIVITY | 3 | 12 | 4.493e-06 | 0.0005963 |
| 8 | PROTEIN COMPLEX BINDING | 12 | 935 | 8.452e-06 | 0.0009815 |
| 9 | PROTEIN KINASE ACTIVITY | 10 | 640 | 9.896e-06 | 0.001021 |
| 10 | CYCLIN BINDING | 3 | 19 | 1.952e-05 | 0.001511 |
| 11 | KINASE ACTIVITY | 11 | 842 | 1.81e-05 | 0.001511 |
| 12 | DEATH RECEPTOR BINDING | 3 | 18 | 1.647e-05 | 0.001511 |
| 13 | MACROMOLECULAR COMPLEX BINDING | 14 | 1399 | 2.252e-05 | 0.001609 |
| 14 | PROTEIN SERINE THREONINE KINASE ACTIVITY | 8 | 445 | 3.143e-05 | 0.002085 |
| 15 | PROTEIN SERINE THREONINE KINASE INHIBITOR ACTIVITY | 3 | 30 | 8.001e-05 | 0.004687 |
| 16 | TRANSFERASE ACTIVITY TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 11 | 992 | 8.073e-05 | 0.004687 |
| 17 | UBIQUITIN LIKE PROTEIN LIGASE BINDING | 6 | 264 | 9.334e-05 | 0.005101 |
| 18 | IDENTICAL PROTEIN BINDING | 12 | 1209 | 0.0001054 | 0.005441 |
| 19 | UBIQUITIN LIKE PROTEIN TRANSFERASE ACTIVITY | 7 | 420 | 0.0001636 | 0.008 |
| 20 | PROTEASE BINDING | 4 | 104 | 0.0002059 | 0.009562 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CYCLIN DEPENDENT PROTEIN KINASE HOLOENZYME COMPLEX | 7 | 31 | 2.283e-12 | 1.333e-09 |
| 2 | PROTEIN KINASE COMPLEX | 7 | 90 | 5.715e-09 | 1.669e-06 |
| 3 | TRANSFERASE COMPLEX TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 8 | 237 | 3.006e-07 | 5.852e-05 |
| 4 | CATALYTIC COMPLEX | 14 | 1038 | 7.075e-07 | 0.0001033 |
| 5 | TRANSFERASE COMPLEX | 10 | 703 | 2.227e-05 | 0.002601 |
| 6 | NUCLEAR BODY | 7 | 349 | 5.161e-05 | 0.005024 |
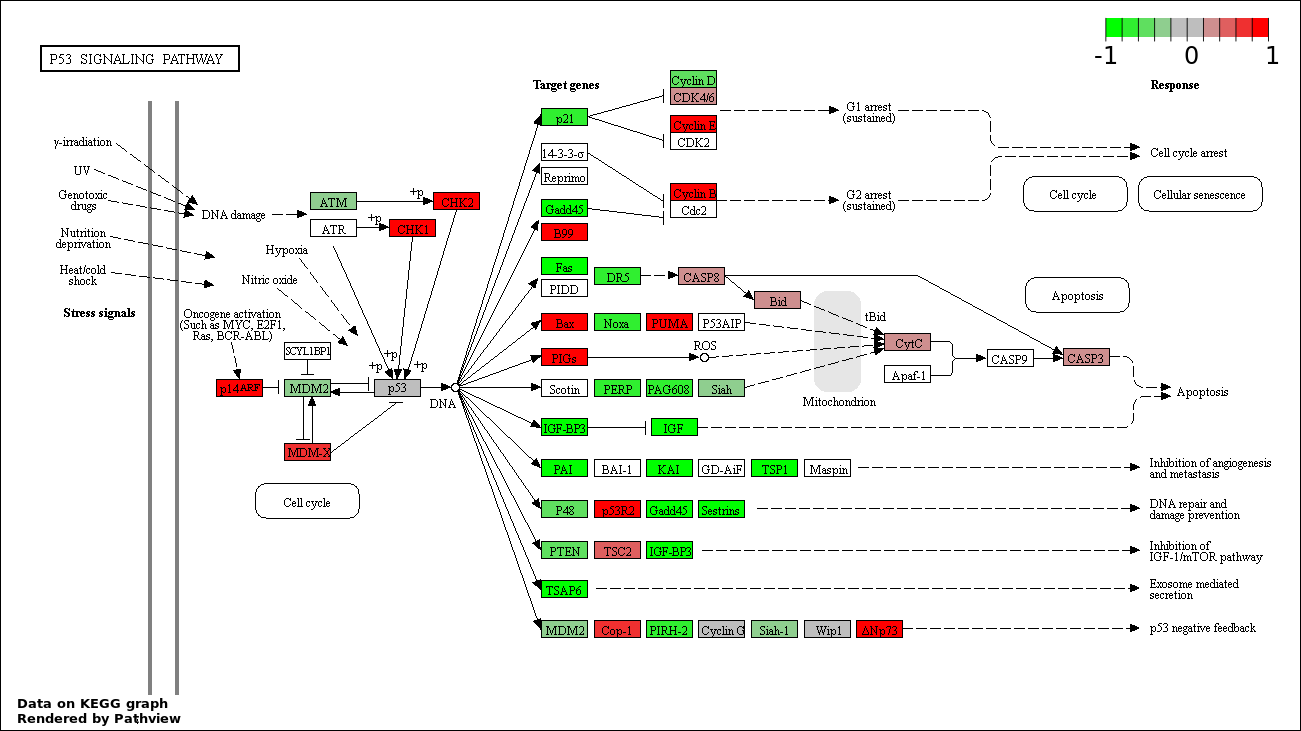
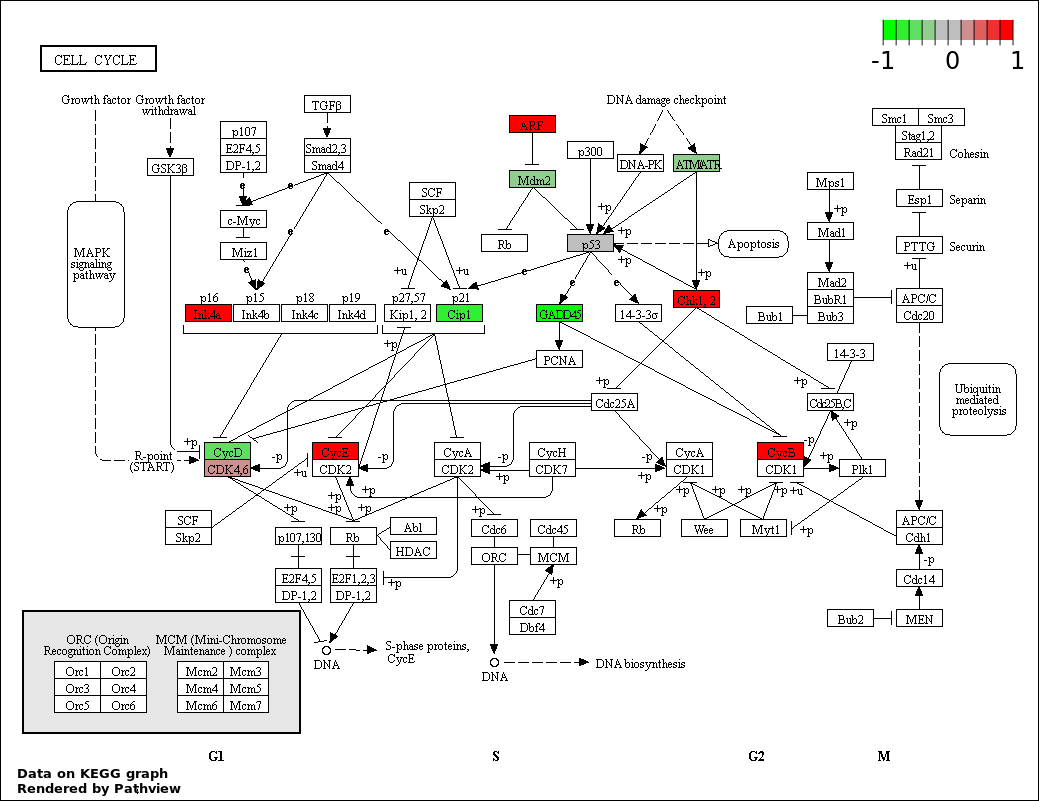
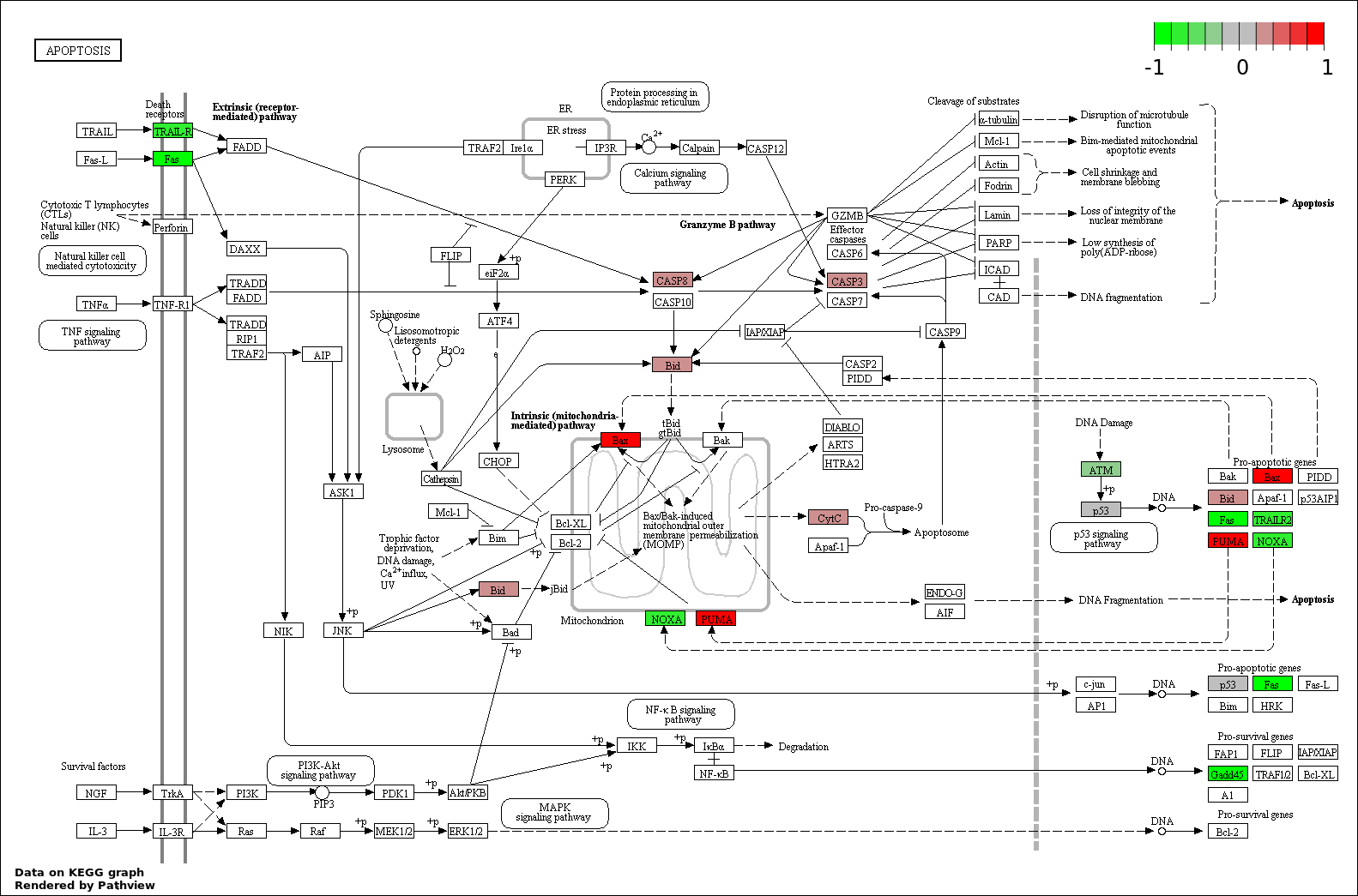
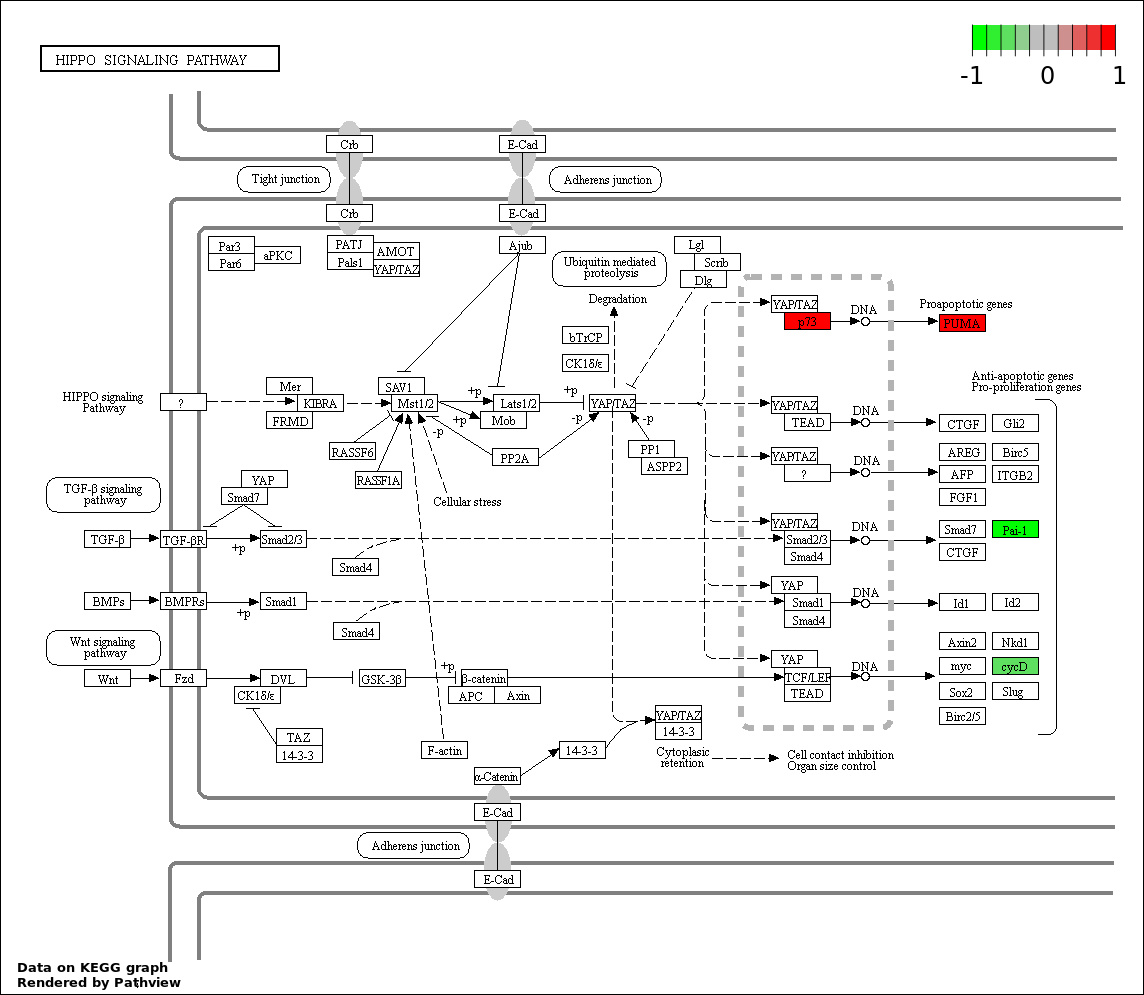
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
| 1 | hsa04115_p53_signaling_pathway | 56 | 69 | 4.119e-153 | 7.415e-151 | |
| 2 | hsa04110_Cell_cycle | 20 | 128 | 1.828e-30 | 1.645e-28 | |
| 3 | hsa04151_PI3K_AKT_signaling_pathway | 14 | 351 | 6.059e-13 | 3.635e-11 | |
| 4 | hsa04210_Apoptosis | 9 | 89 | 2.886e-12 | 1.299e-10 | |
| 5 | hsa04390_Hippo_signaling_pathway | 6 | 154 | 4.467e-06 | 0.0001608 | |
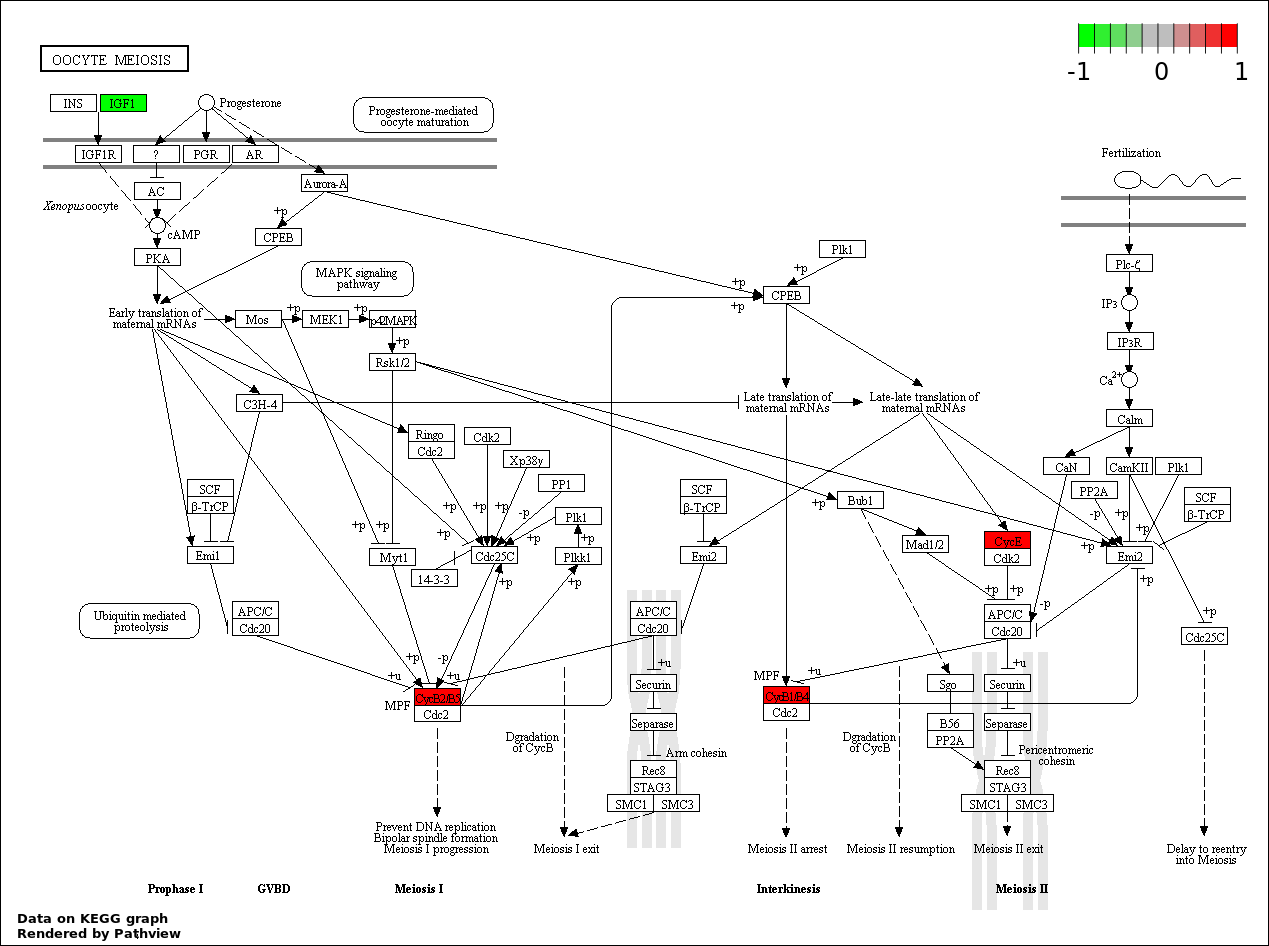
| 6 | hsa04114_Oocyte_meiosis | 5 | 114 | 1.669e-05 | 0.0005006 | |
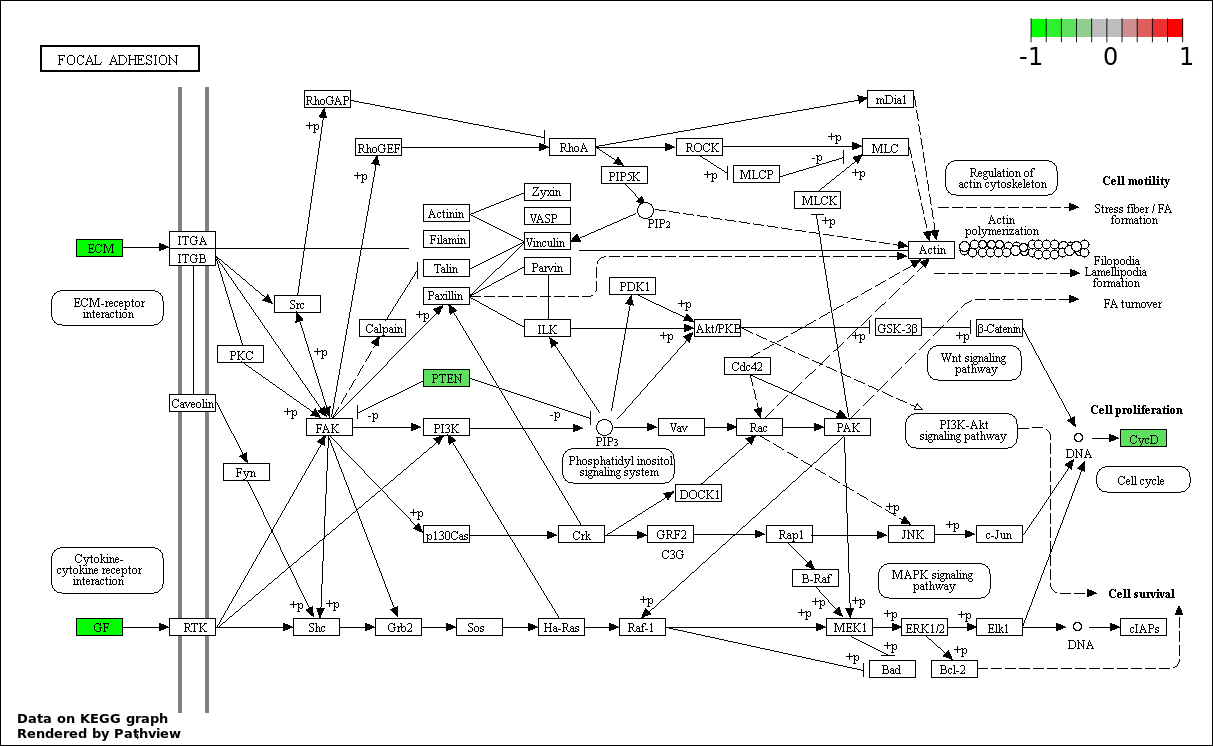
| 7 | hsa04510_Focal_adhesion | 6 | 200 | 1.987e-05 | 0.0005109 | |
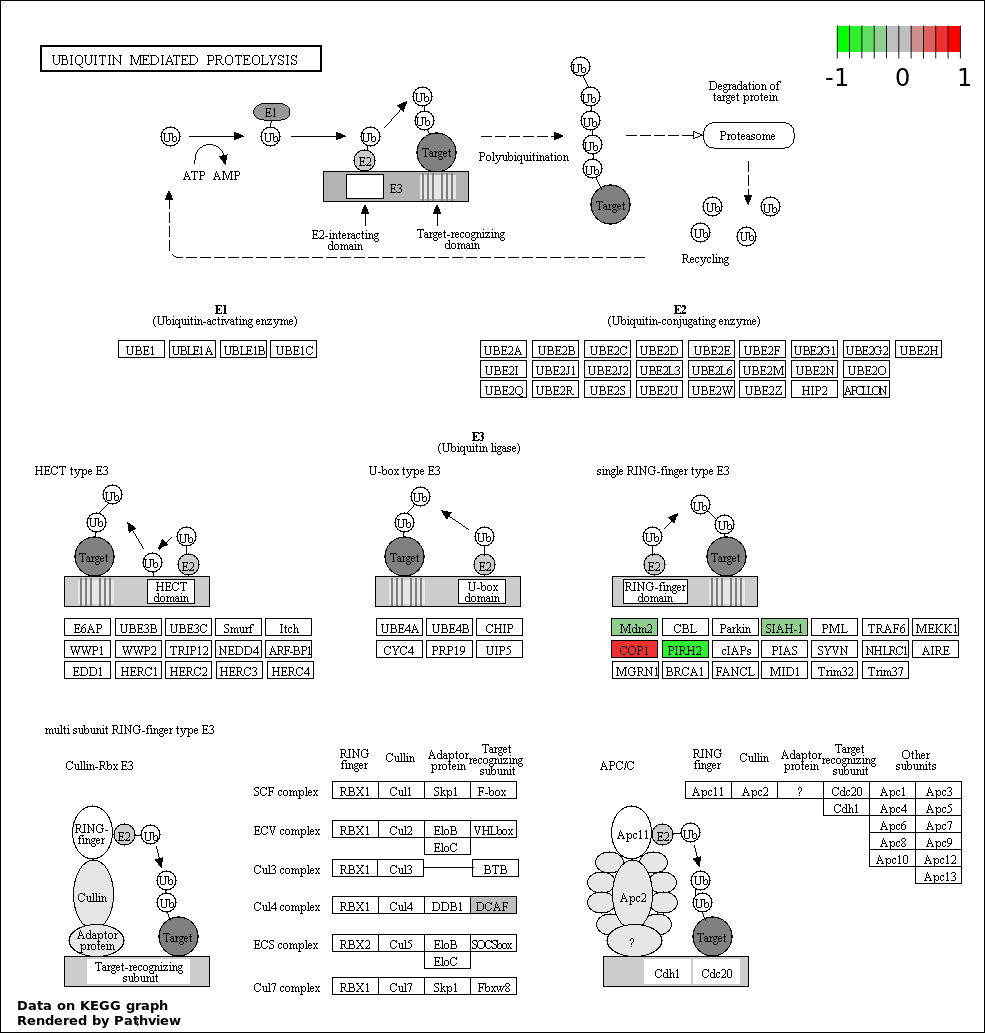
| 8 | hsa04120_Ubiquitin_mediated_proteolysis | 5 | 139 | 4.334e-05 | 0.0009752 | |
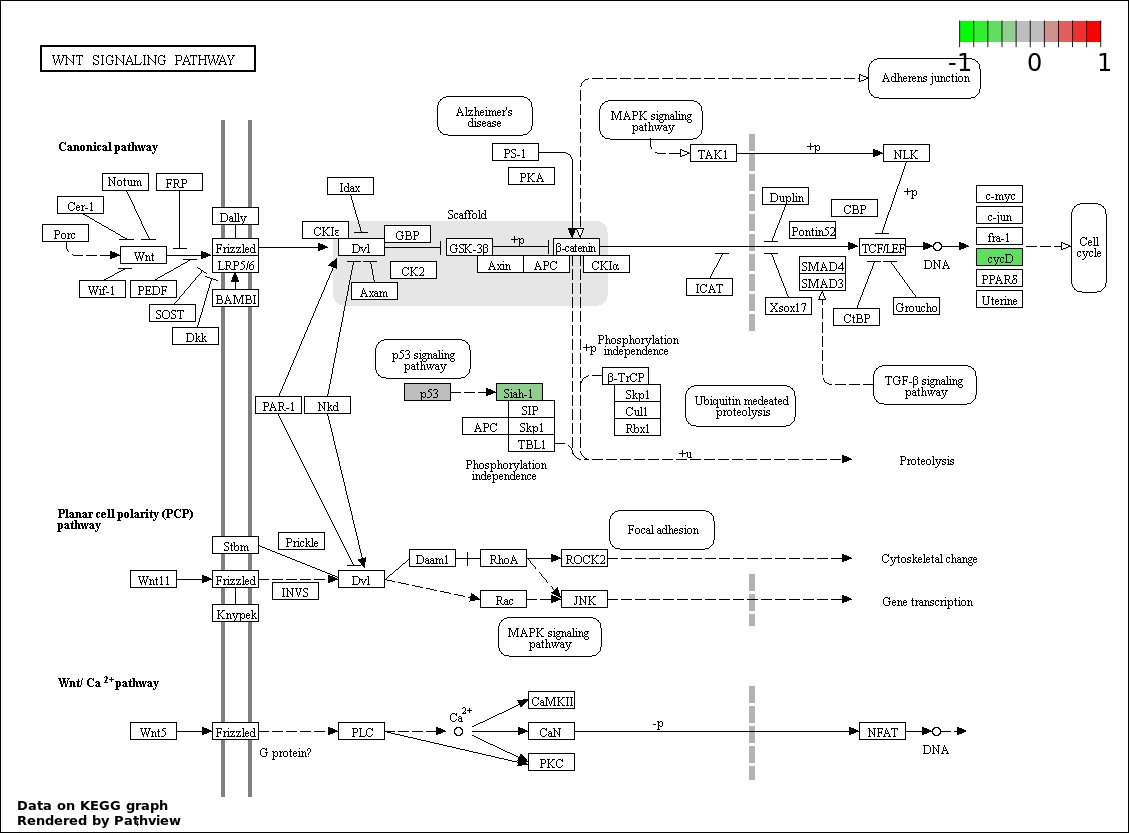
| 9 | hsa04310_Wnt_signaling_pathway | 5 | 151 | 6.43e-05 | 0.001286 | |
| 10 | hsa04010_MAPK_signaling_pathway | 6 | 268 | 0.0001014 | 0.001689 | |
| 11 | hsa04914_Progesterone.mediated_oocyte_maturation | 4 | 87 | 0.0001032 | 0.001689 | |
| 12 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 4 | 136 | 0.0005713 | 0.008569 | |
| 13 | hsa04722_Neurotrophin_signaling_pathway | 3 | 127 | 0.005424 | 0.0751 | |
| 14 | hsa00480_Glutathione_metabolism | 2 | 50 | 0.008655 | 0.1055 | |
| 15 | hsa04150_mTOR_signaling_pathway | 2 | 52 | 0.009335 | 0.1055 | |
| 16 | hsa04630_Jak.STAT_signaling_pathway | 3 | 155 | 0.009374 | 0.1055 | |
| 17 | hsa00240_Pyrimidine_metabolism | 2 | 99 | 0.03142 | 0.3326 | |
| 18 | hsa04530_Tight_junction | 2 | 133 | 0.05354 | 0.5354 | |
| 19 | hsa00230_Purine_metabolism | 2 | 162 | 0.07561 | 0.7163 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | MALAT1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p | 10 | MDM4 | Sponge network | 1.297 | 0 | 0.734 | 0 | 0.615 |
| 2 | KB-1572G7.2 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p | 10 | MDM4 | Sponge network | 2.124 | 0 | 0.734 | 0 | 0.611 |
| 3 | AC159540.1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p | 11 | MDM4 | Sponge network | 2.112 | 0 | 0.734 | 0 | 0.586 |
| 4 | GUSBP11 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p | 10 | MDM4 | Sponge network | 2.066 | 0 | 0.734 | 0 | 0.583 |
| 5 | AC005154.6 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 11 | MDM4 | Sponge network | 1.75 | 0 | 0.734 | 0 | 0.509 |
| 6 | SNHG1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p | 10 | MDM4 | Sponge network | 2.013 | 0 | 0.734 | 0 | 0.493 |
| 7 | GAS5 |
hsa-let-7c-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | 1.966 | 0 | 0.734 | 0 | 0.46 |
| 8 | GAS5 |
hsa-let-7a-2-3p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-125b-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-27b-3p;hsa-miR-345-5p;hsa-miR-590-3p | 10 | BBC3 | Sponge network | 1.966 | 0 | 0.805 | 0 | 0.454 |
| 9 | CTD-2228K2.7 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-542-3p | 11 | MDM4 | Sponge network | 2.28 | 0 | 0.734 | 0 | 0.452 |
| 10 | RP11-600F24.7 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 11 | MDM4 | Sponge network | 2.603 | 0 | 0.734 | 0 | 0.451 |
| 11 | PSMD5-AS1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 11 | MDM4 | Sponge network | 1.538 | 0 | 0.734 | 0 | 0.436 |
| 12 | SNHG12 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-424-5p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | 1.791 | 0 | 0.734 | 0 | 0.419 |
| 13 | HCG18 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p | 10 | MDM4 | Sponge network | 1.42 | 0 | 0.734 | 0 | 0.405 |
| 14 | RP11-12A2.3 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-589-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 13 | SESN3 | Sponge network | -4.779 | 0 | -0.822 | 0.00384 | 0.389 |
| 15 | RP11-89K21.1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-374b-5p;hsa-miR-424-5p | 10 | MDM4 | Sponge network | 4.915 | 0 | 0.734 | 0 | 0.387 |
| 16 | AC074117.10 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | 1.254 | 0 | 0.734 | 0 | 0.379 |
| 17 | LINC00176 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | 3.423 | 0 | 0.734 | 0 | 0.368 |
| 18 | GS1-124K5.11 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | 1.5 | 0 | 0.734 | 0 | 0.365 |
| 19 | RP1-228H13.5 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | 1.554 | 0 | 0.734 | 0 | 0.363 |
| 20 | MAGI2-AS3 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-589-3p;hsa-miR-616-5p;hsa-miR-9-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 13 | CCND1 | Sponge network | -1.801 | 0 | -0.902 | 1.0E-5 | 0.359 |
| 21 | CTBP1-AS2 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 11 | MDM4 | Sponge network | 1.419 | 0 | 0.734 | 0 | 0.337 |
| 22 | RP11-727A23.5 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | 1.435 | 0 | 0.734 | 0 | 0.327 |
| 23 | SNHG7 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-424-5p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | 2.077 | 0 | 0.734 | 0 | 0.322 |
| 24 | ALDH1L1-AS2 | hsa-let-7e-5p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-181a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p | 10 | CCNG1 | Sponge network | 0.116 | 0.79006 | -0.046 | 0.64033 | 0.317 |
| 25 | MAGI2-AS3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 13 | SESN3 | Sponge network | -1.801 | 0 | -0.822 | 0.00384 | 0.314 |
| 26 | RP11-166D19.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-877-5p;hsa-miR-92a-3p | 13 | SESN3 | Sponge network | -0.244 | 0.28835 | -0.822 | 0.00384 | 0.285 |
| 27 | RP11-166D19.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-5p | 12 | PTEN | Sponge network | -0.244 | 0.28835 | -0.546 | 0 | 0.28 |
| 28 | RP5-1165K10.2 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-424-5p | 10 | MDM4 | Sponge network | 1.401 | 0 | 0.734 | 0 | 0.272 |
| 29 | CTD-3220F14.1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 10 | MDM4 | Sponge network | 3.223 | 0 | 0.734 | 0 | 0.268 |
| 30 | ZNRD1-AS1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 11 | MDM4 | Sponge network | 1.284 | 0 | 0.734 | 0 | 0.266 |
| 31 | MIR4435-1HG | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 11 | MDM4 | Sponge network | 2.541 | 0 | 0.734 | 0 | 0.261 |
| 32 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-425-5p;hsa-miR-589-3p;hsa-miR-616-5p;hsa-miR-92a-3p;hsa-miR-942-5p | 16 | CCND1 | Sponge network | -0.244 | 0.28835 | -0.902 | 1.0E-5 | 0.253 |
| 33 | RP11-37B2.1 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 11 | MDM4 | Sponge network | 1.504 | 0 | 0.734 | 0 | 0.253 |
| 34 | POLR2J4 | hsa-let-7c-5p;hsa-miR-125b-2-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p | 12 | MDM4 | Sponge network | 1.386 | 0 | 0.734 | 0 | 0.251 |