This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-15a-5p | ACSL4 | 0.01 | 0.57141 | 0.66 | 0.00114 | MirTarget; miRNATAP | -1.42 | 0.01374 | NA | |
| 2 | hsa-miR-15a-5p | ACVR2A | 0.01 | 0.57141 | -0.17 | 0.00276 | MirTarget; miRNATAP | -0.49 | 0.00283 | NA | |
| 3 | hsa-miR-15a-5p | ADAMTS5 | 0.01 | 0.57141 | -0.31 | 0.00979 | miRNATAP | -1.09 | 0.00103 | NA | |
| 4 | hsa-miR-15a-5p | ADCY1 | 0.01 | 0.57141 | 0.12 | 0.37601 | mirMAP | -1.27 | 0.00049 | NA | |
| 5 | hsa-miR-15a-5p | ADCY5 | 0.01 | 0.57141 | -0.52 | 2.0E-5 | MirTarget; miRNATAP | -1.66 | 0 | NA | |
| 6 | hsa-miR-15a-5p | ADGRD1 | 0.01 | 0.57141 | -0.17 | 0.42537 | MirTarget | -3.61 | 0 | NA | |
| 7 | hsa-miR-15a-5p | ADGRL2 | 0.01 | 0.57141 | -0.73 | 0 | MirTarget; miRNATAP | -1.01 | 0.00949 | NA | |
| 8 | hsa-miR-15a-5p | AHRR | 0.01 | 0.57141 | 0.04 | 0.54652 | mirMAP | -0.42 | 0.01078 | NA | |
| 9 | hsa-miR-15a-5p | AKT3 | 0.01 | 0.57141 | -0.32 | 0.00594 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.89 | 0.00617 | NA | |
| 10 | hsa-miR-126-5p | ALCAM | 0 | 0.93995 | 0.26 | 0.00012 | mirMAP; miRNATAP | -0.54 | 0.01139 | NA | |
| 11 | hsa-miR-142-5p | ALCAM | -0.12 | 0.00783 | 0.26 | 0.00012 | MirTarget; PITA; mirMAP; miRNATAP | -0.25 | 0.02509 | NA | |
| 12 | hsa-miR-148a-3p | ALCAM | -0.01 | 0.73806 | 0.26 | 0.00012 | MirTarget | -0.39 | 0.00086 | NA | |
| 13 | hsa-miR-150-5p | ALCAM | -0.18 | 0.00075 | 0.26 | 0.00012 | miRNATAP | -0.38 | 3.0E-5 | NA | |
| 14 | hsa-miR-152-3p | ALCAM | -0 | 0.97674 | 0.26 | 0.00012 | MirTarget | -0.34 | 0.02225 | NA | |
| 15 | hsa-miR-199a-5p | ALCAM | -0.44 | 0 | 0.26 | 0.00012 | miRanda | -0.38 | 0 | NA | |
| 16 | hsa-miR-199b-5p | ALCAM | -0.22 | 0.00068 | 0.26 | 0.00012 | miRanda | -0.21 | 0.00438 | NA | |
| 17 | hsa-miR-223-3p | ALCAM | -0.15 | 0.00045 | 0.26 | 0.00012 | MirTarget | -0.31 | 0.00612 | NA | |
| 18 | hsa-miR-15a-5p | ANK2 | 0.01 | 0.57141 | -0.79 | 2.0E-5 | MirTarget; miRNATAP | -2.09 | 6.0E-5 | NA | |
| 19 | hsa-miR-15a-5p | ANKS1A | 0.01 | 0.57141 | -0.37 | 0 | MirTarget | -0.59 | 0.0032 | NA | |
| 20 | hsa-miR-15a-5p | ARHGAP20 | 0.01 | 0.57141 | -0.69 | 6.0E-5 | MirTarget; miRNATAP | -2.07 | 2.0E-5 | NA | |
| 21 | hsa-miR-15a-5p | ARL2 | 0.01 | 0.57141 | 0.2 | 0.0061 | MirTarget; miRNATAP | -0.79 | 8.0E-5 | NA | |
| 22 | hsa-miR-15a-5p | ARMCX2 | 0.01 | 0.57141 | -0.59 | 0 | MirTarget; miRNATAP | -1.3 | 0.00015 | NA | |
| 23 | hsa-miR-15a-5p | ASB1 | 0.01 | 0.57141 | 0.05 | 0.5086 | MirTarget; miRNATAP | -0.6 | 0.00566 | NA | |
| 24 | hsa-miR-15a-5p | ASTN1 | 0.01 | 0.57141 | -0.43 | 5.0E-5 | MirTarget | -0.68 | 0.0248 | NA | |
| 25 | hsa-miR-15a-5p | BAG5 | 0.01 | 0.57141 | 0.01 | 0.7689 | miRNATAP | -0.23 | 0.03793 | NA | |
| 26 | hsa-miR-15a-5p | BCL11B | 0.01 | 0.57141 | -0.7 | 0.0001 | MirTarget | -1.28 | 0.01211 | NA | |
| 27 | hsa-miR-15a-5p | BRSK2 | 0.01 | 0.57141 | 0.27 | 0.0036 | mirMAP | -0.86 | 0.00086 | NA | |
| 28 | hsa-miR-15a-5p | BTG2 | 0.01 | 0.57141 | -0.27 | 0.03814 | MirTarget; miRNATAP | -0.77 | 0.03312 | NA | |
| 29 | hsa-miR-15a-5p | BVES | 0.01 | 0.57141 | -0.09 | 0.34596 | MirTarget | -1.26 | 0 | NA | |
| 30 | hsa-miR-15a-5p | C8orf58 | 0.01 | 0.57141 | -0.19 | 0.02594 | miRNATAP | -0.63 | 0.01023 | NA | |
| 31 | hsa-miR-141-3p | CADM1 | -0.14 | 0.00512 | 0.01 | 0.91974 | MirTarget; TargetScan | -0.63 | 0.0007 | NA | |
| 32 | hsa-miR-142-5p | CADM1 | -0.12 | 0.00783 | 0.01 | 0.91974 | PITA | -0.65 | 0.00307 | NA | |
| 33 | hsa-miR-182-3p | CADM1 | 0.08 | 0.03587 | 0.01 | 0.91974 | MirTarget; miRNATAP | -0.58 | 0.03482 | 24445397 | TGF β upregulates miR 182 expression to promote gallbladder cancer metastasis by targeting CADM1; We further identified that the cell adhesion molecule1 CADM1 is a new target gene of miR-182 miR-182 negatively regulates CADM1 expression in vitro and in vivo |
| 34 | hsa-miR-199b-5p | CADM1 | -0.22 | 0.00068 | 0.01 | 0.91974 | PITA; miRanda | -0.47 | 0.00135 | NA | |
| 35 | hsa-miR-34c-5p | CADM1 | 0.2 | 5.0E-5 | 0.01 | 0.91974 | miRanda | -0.59 | 0.00234 | NA | |
| 36 | hsa-miR-92a-3p | CADM1 | -0.03 | 0.37888 | 0.01 | 0.91974 | miRNATAP | -1.87 | 0 | NA | |
| 37 | hsa-miR-107 | CADM3 | 0.13 | 0 | -0.1 | 0.01289 | miRanda | -0.28 | 0.00674 | NA | |
| 38 | hsa-miR-324-3p | CADM3 | 0.02 | 0.24561 | -0.1 | 0.01289 | MirTarget | -0.46 | 0.00346 | NA | |
| 39 | hsa-miR-330-3p | CADM3 | 0.05 | 0.00086 | -0.1 | 0.01289 | mirMAP; miRNATAP | -0.53 | 0.01298 | NA | |
| 40 | hsa-miR-331-3p | CADM3 | 0.14 | 0 | -0.1 | 0.01289 | MirTarget; mirMAP | -0.36 | 0.00148 | NA | |
| 41 | hsa-miR-346 | CADM3 | 0.09 | 1.0E-5 | -0.1 | 0.01289 | miRanda; miRNATAP | -0.32 | 0.0304 | NA | |
| 42 | hsa-miR-15a-5p | CALM1 | 0.01 | 0.57141 | -0.13 | 0.00385 | miRNATAP | -0.61 | 0 | NA | |
| 43 | hsa-miR-15a-5p | CCNJL | 0.01 | 0.57141 | -0.47 | 0.00012 | miRNATAP | -1.04 | 0.00256 | NA | |
| 44 | hsa-miR-15a-5p | CCNYL1 | 0.01 | 0.57141 | 0.04 | 0.59551 | miRNAWalker2 validate | -0.52 | 0.02602 | NA | |
| 45 | hsa-miR-15a-5p | CD164 | 0.01 | 0.57141 | -0.41 | 0 | MirTarget; miRNATAP | -0.52 | 0.03639 | NA | |
| 46 | hsa-miR-140-5p | CD226 | 0.04 | 0.1479 | -0.77 | 0 | miRanda | -0.82 | 0.00924 | NA | |
| 47 | hsa-miR-17-5p | CD226 | 0.06 | 0.05778 | -0.77 | 0 | TargetScan | -1.31 | 0 | NA | |
| 48 | hsa-miR-182-5p | CD226 | 0.14 | 0.00166 | -0.77 | 0 | MirTarget | -1.35 | 0 | NA | |
| 49 | hsa-miR-211-5p | CD226 | 0.09 | 0.01154 | -0.77 | 0 | MirTarget | -0.65 | 0.01589 | NA | |
| 50 | hsa-miR-217 | CD226 | 0.09 | 0.11729 | -0.77 | 0 | miRanda | -0.38 | 0.02038 | NA | |
| 51 | hsa-miR-331-3p | CD226 | 0.14 | 0 | -0.77 | 0 | miRNATAP | -2.43 | 0 | NA | |
| 52 | hsa-miR-422a | CD226 | 0.04 | 0.07853 | -0.77 | 0 | MirTarget; miRanda | -1.36 | 0.00017 | NA | |
| 53 | hsa-miR-448 | CD226 | 0.03 | 0.16644 | -0.77 | 0 | miRanda | -1.39 | 0.00113 | NA | |
| 54 | hsa-miR-103a-3p | CD274 | 0.11 | 0 | -0.74 | 0 | miRNAWalker2 validate | -2.21 | 0 | NA | |
| 55 | hsa-miR-106a-5p | CD274 | 0.05 | 0.12421 | -0.74 | 0 | MirTarget; miRNATAP | -1.21 | 0.00039 | NA | |
| 56 | hsa-miR-106b-5p | CD274 | 0.17 | 0 | -0.74 | 0 | MirTarget; miRNATAP | -2.78 | 0 | NA | |
| 57 | hsa-miR-107 | CD274 | 0.13 | 0 | -0.74 | 0 | miRanda | -1.95 | 0 | NA | |
| 58 | hsa-miR-137 | CD274 | 0.03 | 0.28838 | -0.74 | 0 | miRanda | -0.87 | 0.02966 | NA | |
| 59 | hsa-miR-140-5p | CD274 | 0.04 | 0.1479 | -0.74 | 0 | miRanda | -0.84 | 0.03715 | NA | |
| 60 | hsa-miR-15b-5p | CD274 | 0.17 | 0 | -0.74 | 0 | MirTarget | -1.48 | 2.0E-5 | NA | |
| 61 | hsa-miR-17-5p | CD274 | 0.06 | 0.05778 | -0.74 | 0 | MirTarget; TargetScan; miRNATAP | -1.43 | 0.0001 | NA | |
| 62 | hsa-miR-182-5p | CD274 | 0.14 | 0.00166 | -0.74 | 0 | mirMAP | -0.84 | 0.00144 | NA | |
| 63 | hsa-miR-20a-5p | CD274 | -0.02 | 0.56346 | -0.74 | 0 | MirTarget; miRNATAP | -0.99 | 0.00635 | NA | |
| 64 | hsa-miR-302c-5p | CD274 | 0.09 | 0.0137 | -0.74 | 0 | MirTarget | -1.84 | 0 | NA | |
| 65 | hsa-miR-324-5p | CD274 | 0.23 | 0 | -0.74 | 0 | miRanda | -1.66 | 0 | NA | |
| 66 | hsa-miR-339-5p | CD274 | 0.14 | 0 | -0.74 | 0 | miRanda | -2.1 | 0 | NA | |
| 67 | hsa-miR-93-5p | CD274 | 0.19 | 0 | -0.74 | 0 | MirTarget; miRNATAP | -2.38 | 0 | NA | |
| 68 | hsa-let-7a-5p | CD276 | -0.03 | 0.25442 | -0.01 | 0.89577 | miRNAWalker2 validate | -0.5 | 0.01745 | NA | |
| 69 | hsa-miR-28-5p | CD276 | 0.01 | 0.57396 | -0.01 | 0.89577 | miRNAWalker2 validate; miRanda | -0.81 | 0.02525 | NA | |
| 70 | hsa-miR-29a-3p | CD276 | -0.02 | 0.42365 | -0.01 | 0.89577 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -1.05 | 0 | NA | |
| 71 | hsa-miR-29b-3p | CD276 | 0.03 | 0.47094 | -0.01 | 0.89577 | MirTarget; miRNATAP | -0.82 | 0 | NA | |
| 72 | hsa-miR-29c-3p | CD276 | -0.12 | 0.01035 | -0.01 | 0.89577 | MirTarget; miRNATAP | -0.67 | 0 | 24577056 | Identifying microRNAs regulating B7 H3 in breast cancer: the clinical impact of microRNA 29c |
| 73 | hsa-miR-335-5p | CD276 | 0.02 | 0.79934 | -0.01 | 0.89577 | miRNAWalker2 validate | -0.15 | 0.03377 | NA | |
| 74 | hsa-miR-448 | CD276 | 0.03 | 0.16644 | -0.01 | 0.89577 | miRNATAP | -0.69 | 0.00847 | NA | |
| 75 | hsa-miR-449a | CD276 | 0.03 | 0.11286 | -0.01 | 0.89577 | mirMAP | -0.62 | 0.02052 | NA | |
| 76 | hsa-miR-15a-5p | CD3E | 0.01 | 0.57141 | -0.63 | 0.00056 | MirTarget | -1.03 | 0.04802 | NA | |
| 77 | hsa-miR-15a-5p | CD80 | 0.01 | 0.57141 | -0.47 | 0.00188 | MirTarget | -1 | 0.02025 | NA | |
| 78 | hsa-miR-15a-5p | CDC42EP2 | 0.01 | 0.57141 | -0.21 | 0.00054 | miRNATAP | -0.54 | 0.00136 | NA | |
| 79 | hsa-miR-129-5p | CDH2 | -0.01 | 0.79472 | -0.09 | 0.08311 | miRanda; miRNATAP | -0.26 | 0.02505 | NA | |
| 80 | hsa-miR-155-5p | CDH2 | 0.04 | 0.14794 | -0.09 | 0.08311 | miRNAWalker2 validate | -0.3 | 0.01004 | NA | |
| 81 | hsa-miR-206 | CDH2 | 0.03 | 0.13251 | -0.09 | 0.08311 | miRanda | -0.65 | 8.0E-5 | NA | |
| 82 | hsa-miR-185-5p | CDH4 | 0.11 | 0 | -0.51 | 1.0E-5 | MirTarget | -1.37 | 0.00017 | NA | |
| 83 | hsa-miR-34c-5p | CDH4 | 0.2 | 5.0E-5 | -0.51 | 1.0E-5 | miRanda | -1.02 | 0 | NA | |
| 84 | hsa-miR-107 | CDH5 | 0.13 | 0 | -0.41 | 1.0E-5 | miRanda | -0.78 | 0.00097 | NA | |
| 85 | hsa-miR-128-3p | CDH5 | 0.08 | 0.0018 | -0.41 | 1.0E-5 | miRNAWalker2 validate | -1.21 | 0 | NA | |
| 86 | hsa-miR-335-5p | CDH5 | 0.02 | 0.79934 | -0.41 | 1.0E-5 | miRNAWalker2 validate | -0.4 | 1.0E-5 | NA | |
| 87 | hsa-miR-15a-5p | CECR6 | 0.01 | 0.57141 | 0.12 | 0.15753 | MirTarget; miRNATAP | -0.58 | 0.01123 | NA | |
| 88 | hsa-miR-186-5p | CLDN11 | 0.12 | 0 | -1.02 | 7.0E-5 | mirMAP | -2.97 | 0.00012 | NA | |
| 89 | hsa-miR-217 | CLDN11 | 0.09 | 0.11729 | -1.02 | 7.0E-5 | miRanda | -0.97 | 0.00363 | NA | |
| 90 | hsa-miR-221-3p | CLDN11 | 0.19 | 0 | -1.02 | 7.0E-5 | MirTarget | -1.34 | 0.00217 | NA | |
| 91 | hsa-miR-222-3p | CLDN11 | 0.15 | 3.0E-5 | -1.02 | 7.0E-5 | MirTarget | -1.3 | 0.00867 | NA | |
| 92 | hsa-miR-25-3p | CLDN11 | 0.11 | 0 | -1.02 | 7.0E-5 | MirTarget | -5.17 | 0 | NA | |
| 93 | hsa-miR-32-5p | CLDN11 | 0.16 | 0 | -1.02 | 7.0E-5 | MirTarget | -2.95 | 0 | NA | |
| 94 | hsa-miR-331-3p | CLDN11 | 0.14 | 0 | -1.02 | 7.0E-5 | PITA | -3.63 | 0 | NA | |
| 95 | hsa-miR-346 | CLDN11 | 0.09 | 1.0E-5 | -1.02 | 7.0E-5 | MirTarget; miRanda | -6.81 | 0 | NA | |
| 96 | hsa-miR-365a-3p | CLDN11 | 0.1 | 0.00092 | -1.02 | 7.0E-5 | MirTarget | -2.01 | 0.00076 | NA | |
| 97 | hsa-miR-367-3p | CLDN11 | 0.08 | 0.00094 | -1.02 | 7.0E-5 | MirTarget | -3.26 | 1.0E-5 | NA | |
| 98 | hsa-miR-448 | CLDN11 | 0.03 | 0.16644 | -1.02 | 7.0E-5 | miRanda | -3.43 | 0.0001 | NA | |
| 99 | hsa-miR-92a-3p | CLDN11 | -0.03 | 0.37888 | -1.02 | 7.0E-5 | MirTarget | -1.59 | 0.01732 | NA | |
| 100 | hsa-miR-125a-5p | CLDN16 | -0.13 | 0.00014 | 0.06 | 0.37863 | miRanda | -0.33 | 0.01625 | NA | |
| 101 | hsa-miR-186-5p | CLDN18 | 0.12 | 0 | 0.05 | 0.35128 | MirTarget | -0.43 | 0.01307 | NA | |
| 102 | hsa-miR-30e-3p | CLDN18 | -0.08 | 0.0159 | 0.05 | 0.35128 | mirMAP | -0.29 | 0.02966 | NA | |
| 103 | hsa-miR-125a-5p | CLDN19 | -0.13 | 0.00014 | 0.11 | 0.07529 | mirMAP | -0.31 | 0.01478 | NA | |
| 104 | hsa-miR-125b-5p | CLDN19 | -0.19 | 1.0E-5 | 0.11 | 0.07529 | mirMAP | -0.29 | 0.00409 | NA | |
| 105 | hsa-miR-195-5p | CLDN19 | -0.18 | 1.0E-5 | 0.11 | 0.07529 | mirMAP | -0.43 | 3.0E-5 | NA | |
| 106 | hsa-miR-214-3p | CLDN19 | -0.32 | 0 | 0.11 | 0.07529 | mirMAP | -0.33 | 5.0E-5 | NA | |
| 107 | hsa-miR-424-5p | CLDN19 | -0.16 | 0 | 0.11 | 0.07529 | mirMAP | -0.41 | 0.00229 | NA | |
| 108 | hsa-miR-142-3p | CLDN20 | -0.06 | 0.14584 | 0.02 | 0.55738 | miRanda | -0.18 | 0.00237 | NA | |
| 109 | hsa-miR-199b-5p | CLDN22 | -0.22 | 0.00068 | -0 | 0.99119 | miRanda | -0.1 | 0.02081 | NA | |
| 110 | hsa-miR-218-5p | CLDN23 | -0.14 | 0.00268 | 0.05 | 0.55692 | MirTarget | -0.31 | 0.01512 | NA | |
| 111 | hsa-miR-222-3p | CLDN23 | 0.15 | 3.0E-5 | 0.05 | 0.55692 | miRNAWalker2 validate | -0.41 | 0.00774 | NA | |
| 112 | hsa-miR-129-5p | CLDN3 | -0.01 | 0.79472 | -0.12 | 0.0706 | MirTarget | -0.39 | 0.01539 | NA | |
| 113 | hsa-miR-139-5p | CLDN4 | -0.08 | 0.00239 | -0.38 | 0.0378 | miRanda | -1.31 | 0.01495 | NA | |
| 114 | hsa-miR-128-3p | CLDN5 | 0.08 | 0.0018 | -0.2 | 0.08862 | miRNAWalker2 validate | -0.9 | 0.0071 | NA | |
| 115 | hsa-miR-15a-5p | CNN1 | 0.01 | 0.57141 | -0.07 | 0.52402 | miRNATAP | -0.86 | 0.00868 | NA | |
| 116 | hsa-miR-32-5p | CNTN1 | 0.16 | 0 | -0.09 | 0.27962 | miRNATAP | -0.37 | 0.029 | NA | |
| 117 | hsa-miR-324-5p | CNTN1 | 0.23 | 0 | -0.09 | 0.27962 | miRanda | -0.29 | 0.03174 | NA | |
| 118 | hsa-miR-33a-5p | CNTN1 | 0.16 | 0 | -0.09 | 0.27962 | MirTarget; miRNATAP | -0.43 | 0.01667 | NA | |
| 119 | hsa-miR-369-3p | CNTN1 | 0.06 | 0.0174 | -0.09 | 0.27962 | mirMAP | -0.47 | 0.03585 | NA | |
| 120 | hsa-miR-138-5p | CNTN2 | -0.21 | 0.00013 | -0.02 | 0.53811 | mirMAP | -0.14 | 0.00193 | NA | |
| 121 | hsa-miR-150-5p | CNTN2 | -0.18 | 0.00075 | -0.02 | 0.53811 | mirMAP | -0.12 | 0.01264 | NA | |
| 122 | hsa-miR-205-5p | CNTN2 | 0.01 | 0.71055 | -0.02 | 0.53811 | mirMAP | -0.19 | 0.02556 | NA | |
| 123 | hsa-miR-195-5p | CNTNAP1 | -0.18 | 1.0E-5 | 0.57 | 2.0E-5 | MirTarget | -1.08 | 0 | NA | |
| 124 | hsa-miR-214-3p | CNTNAP1 | -0.32 | 0 | 0.57 | 2.0E-5 | MirTarget | -0.38 | 0.04452 | NA | |
| 125 | hsa-miR-424-5p | CNTNAP1 | -0.16 | 0 | 0.57 | 2.0E-5 | MirTarget | -1.07 | 0.00038 | NA | |
| 126 | hsa-miR-448 | CNTNAP1 | 0.03 | 0.16644 | 0.57 | 2.0E-5 | MirTarget; miRanda; miRNATAP | -1.7 | 0.00026 | NA | |
| 127 | hsa-miR-449a | CNTNAP1 | 0.03 | 0.11286 | 0.57 | 2.0E-5 | MirTarget; PITA; miRanda; miRNATAP | -1.34 | 0.00454 | NA | |
| 128 | hsa-miR-211-5p | CNTNAP2 | 0.09 | 0.01154 | -0.13 | 0.52501 | miRNATAP | -0.83 | 0.04927 | NA | |
| 129 | hsa-miR-330-3p | CNTNAP2 | 0.05 | 0.00086 | -0.13 | 0.52501 | mirMAP | -2.65 | 0.00988 | NA | |
| 130 | hsa-miR-34c-5p | CNTNAP2 | 0.2 | 5.0E-5 | -0.13 | 0.52501 | PITA; miRanda; miRNATAP | -0.7 | 0.01303 | NA | |
| 131 | hsa-miR-15a-5p | COL12A1 | 0.01 | 0.57141 | -0.32 | 0.02386 | MirTarget; miRNATAP | -1.36 | 0.00058 | NA | |
| 132 | hsa-miR-15a-5p | COMT | 0.01 | 0.57141 | -0.61 | 0 | MirTarget | -0.82 | 0.01531 | NA | |
| 133 | hsa-miR-15a-5p | CREB5 | 0.01 | 0.57141 | -0.08 | 0.5143 | miRNATAP | -0.67 | 0.04607 | NA | |
| 134 | hsa-miR-449a | CTLA4 | 0.03 | 0.11286 | -0.18 | 0.37956 | miRanda | -3.65 | 0 | NA | |
| 135 | hsa-miR-15a-5p | CYP26B1 | 0.01 | 0.57141 | -0.29 | 0.08677 | MirTarget; miRNATAP | -1.48 | 0.00175 | NA | |
| 136 | hsa-miR-15a-5p | CYS1 | 0.01 | 0.57141 | -1.3 | 0 | mirMAP | -2.35 | 3.0E-5 | NA | |
| 137 | hsa-miR-15a-5p | DBH | 0.01 | 0.57141 | -2.08 | 0 | mirMAP | -1.94 | 0.01347 | NA | |
| 138 | hsa-miR-15a-5p | DENND2C | 0.01 | 0.57141 | -0.78 | 0 | MirTarget | -0.82 | 0.03035 | NA | |
| 139 | hsa-miR-15a-5p | DLK1 | 0.01 | 0.57141 | -0.13 | 0.75273 | miRNAWalker2 validate | -5.61 | 0 | NA | |
| 140 | hsa-miR-15a-5p | EDA | 0.01 | 0.57141 | -0.07 | 0.50152 | MirTarget; miRNATAP | -0.74 | 0.01184 | NA | |
| 141 | hsa-miR-15a-5p | EIF4B | 0.01 | 0.57141 | 0.04 | 0.51431 | miRNATAP | -0.61 | 0.00078 | NA | |
| 142 | hsa-miR-15a-5p | ELL | 0.01 | 0.57141 | -0.01 | 0.86744 | MirTarget; miRNATAP | -0.33 | 0.02568 | NA | |
| 143 | hsa-miR-15a-5p | EPB41L1 | 0.01 | 0.57141 | -0.04 | 0.81245 | MirTarget | -1.11 | 0.01005 | NA | |
| 144 | hsa-miR-15a-5p | EPHA1 | 0.01 | 0.57141 | 0.16 | 0.32601 | miRNATAP | -1.06 | 0.0209 | NA | |
| 145 | hsa-miR-15a-5p | EYA4 | 0.01 | 0.57141 | -0.05 | 0.19647 | MirTarget | -0.23 | 0.03713 | NA | |
| 146 | hsa-miR-181a-5p | F11R | -0.02 | 0.5042 | 0.08 | 0.44822 | mirMAP | -0.65 | 0.02062 | NA | |
| 147 | hsa-miR-335-5p | F11R | 0.02 | 0.79934 | 0.08 | 0.44822 | miRNAWalker2 validate | -0.24 | 0.01205 | NA | |
| 148 | hsa-miR-15a-5p | FAT4 | 0.01 | 0.57141 | -0.98 | 0 | MirTarget | -0.97 | 0.03663 | NA | |
| 149 | hsa-miR-15a-5p | FCHSD1 | 0.01 | 0.57141 | -0.1 | 0.18807 | MirTarget; miRNATAP | -0.75 | 0.00037 | NA | |
| 150 | hsa-miR-15a-5p | FCHSD2 | 0.01 | 0.57141 | -0.32 | 0 | miRNATAP | -0.78 | 4.0E-5 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | BIOLOGICAL ADHESION | 78 | 1032 | 1.155e-43 | 5.373e-40 |
| 2 | CELL CELL ADHESION | 56 | 608 | 3.364e-35 | 7.827e-32 |
| 3 | CELL CELL ADHESION VIA PLASMA MEMBRANE ADHESION MOLECULES | 28 | 204 | 1.938e-22 | 3.006e-19 |
| 4 | SINGLE ORGANISM CELL ADHESION | 36 | 459 | 3.142e-20 | 3.655e-17 |
| 5 | LOCOMOTION | 49 | 1114 | 1.026e-16 | 9.545e-14 |
| 6 | POSITIVE REGULATION OF CELL CELL ADHESION | 24 | 243 | 5.091e-16 | 3.948e-13 |
| 7 | HETEROPHILIC CELL CELL ADHESION VIA PLASMA MEMBRANE CELL ADHESION MOLECULES | 13 | 45 | 1.734e-15 | 1.153e-12 |
| 8 | POSITIVE REGULATION OF CELL ADHESION | 28 | 376 | 2.496e-15 | 1.452e-12 |
| 9 | REGULATION OF CELL ADHESION | 35 | 629 | 4.828e-15 | 2.496e-12 |
| 10 | CALCIUM INDEPENDENT CELL CELL ADHESION VIA PLASMA MEMBRANE CELL ADHESION MOLECULES | 10 | 21 | 8.412e-15 | 3.914e-12 |
| 11 | REGULATION OF CELL CELL ADHESION | 27 | 380 | 2.556e-14 | 1.081e-11 |
| 12 | MOVEMENT OF CELL OR SUBCELLULAR COMPONENT | 48 | 1275 | 8.482e-14 | 3.289e-11 |
| 13 | REGULATION OF HOMOTYPIC CELL CELL ADHESION | 24 | 307 | 9.569e-14 | 3.425e-11 |
| 14 | LYMPHOCYTE COSTIMULATION | 14 | 78 | 1.878e-13 | 6.243e-11 |
| 15 | CELL MOTILITY | 37 | 835 | 8.156e-13 | 2.372e-10 |
| 16 | LOCALIZATION OF CELL | 37 | 835 | 8.156e-13 | 2.372e-10 |
| 17 | CELL DEVELOPMENT | 49 | 1426 | 1.331e-12 | 3.643e-10 |
| 18 | CELL JUNCTION ORGANIZATION | 18 | 185 | 3.436e-12 | 8.882e-10 |
| 19 | NEURON PROJECTION DEVELOPMENT | 28 | 545 | 2.184e-11 | 5.348e-09 |
| 20 | CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 27 | 513 | 2.966e-11 | 6.901e-09 |
| 21 | NEURON DEVELOPMENT | 31 | 687 | 4.56e-11 | 1.01e-08 |
| 22 | POSITIVE REGULATION OF T CELL PROLIFERATION | 13 | 95 | 5.054e-11 | 1.069e-08 |
| 23 | POSITIVE REGULATION OF CELL ACTIVATION | 21 | 311 | 5.782e-11 | 1.17e-08 |
| 24 | REGULATION OF T CELL PROLIFERATION | 15 | 147 | 1.167e-10 | 2.262e-08 |
| 25 | REGULATION OF CELL ACTIVATION | 25 | 484 | 2.5e-10 | 4.653e-08 |
| 26 | INTERFERON GAMMA MEDIATED SIGNALING PATHWAY | 11 | 70 | 3.457e-10 | 6.187e-08 |
| 27 | NEUROGENESIS | 44 | 1402 | 4.322e-10 | 7.448e-08 |
| 28 | CELL SUBSTRATE ADHESION | 15 | 164 | 5.508e-10 | 9.154e-08 |
| 29 | LEUKOCYTE MIGRATION | 18 | 259 | 9.054e-10 | 1.453e-07 |
| 30 | NEURON DIFFERENTIATION | 33 | 874 | 1.025e-09 | 1.59e-07 |
| 31 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 48 | 1672 | 1.167e-09 | 1.751e-07 |
| 32 | REGULATION OF IMMUNE SYSTEM PROCESS | 43 | 1403 | 1.464e-09 | 2.129e-07 |
| 33 | REGULATION OF LEUKOCYTE PROLIFERATION | 16 | 206 | 1.616e-09 | 2.279e-07 |
| 34 | CELL PROJECTION ORGANIZATION | 33 | 902 | 2.256e-09 | 3.087e-07 |
| 35 | REGULATION OF CELL DIFFERENTIATION | 44 | 1492 | 2.967e-09 | 3.945e-07 |
| 36 | IMMUNE SYSTEM PROCESS | 52 | 1984 | 4.77e-09 | 5.998e-07 |
| 37 | POSITIVE REGULATION OF LEUKOCYTE PROLIFERATION | 13 | 136 | 4.671e-09 | 5.998e-07 |
| 38 | CELLULAR COMPONENT MORPHOGENESIS | 32 | 900 | 8.051e-09 | 9.858e-07 |
| 39 | CELL MATRIX ADHESION | 12 | 119 | 1.009e-08 | 1.185e-06 |
| 40 | CELL CELL JUNCTION ASSEMBLY | 10 | 74 | 1.019e-08 | 1.185e-06 |
| 41 | CELL JUNCTION ASSEMBLY | 12 | 129 | 2.52e-08 | 2.86e-06 |
| 42 | LEUKOCYTE CELL CELL ADHESION | 16 | 255 | 3.399e-08 | 3.766e-06 |
| 43 | POSITIVE REGULATION OF IMMUNE SYSTEM PROCESS | 30 | 867 | 4.536e-08 | 4.908e-06 |
| 44 | EXTRACELLULAR STRUCTURE ORGANIZATION | 17 | 304 | 6.754e-08 | 7.142e-06 |
| 45 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 39 | 1395 | 1.137e-07 | 1.176e-05 |
| 46 | CELLULAR RESPONSE TO INTERFERON GAMMA | 11 | 122 | 1.344e-07 | 1.36e-05 |
| 47 | TRANSMISSION OF NERVE IMPULSE | 8 | 54 | 1.51e-07 | 1.495e-05 |
| 48 | NEURON PROJECTION MORPHOGENESIS | 19 | 402 | 1.586e-07 | 1.538e-05 |
| 49 | HOMOPHILIC CELL ADHESION VIA PLASMA MEMBRANE ADHESION MOLECULES | 12 | 153 | 1.678e-07 | 1.593e-05 |
| 50 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 34 | 1142 | 1.963e-07 | 1.827e-05 |
| 51 | HETEROTYPIC CELL CELL ADHESION | 6 | 27 | 4.607e-07 | 4.204e-05 |
| 52 | RESPONSE TO INTERFERON GAMMA | 11 | 144 | 7.214e-07 | 6.455e-05 |
| 53 | ANTIGEN PROCESSING AND PRESENTATION OF PEPTIDE ANTIGEN | 12 | 177 | 8.092e-07 | 7.104e-05 |
| 54 | ANTIGEN PROCESSING AND PRESENTATION | 13 | 213 | 9.262e-07 | 7.981e-05 |
| 55 | CELL MORPHOGENESIS INVOLVED IN NEURON DIFFERENTIATION | 17 | 368 | 9.99e-07 | 8.451e-05 |
| 56 | MULTICELLULAR ORGANISMAL SIGNALING | 10 | 123 | 1.317e-06 | 0.0001094 |
| 57 | TAXIS | 19 | 464 | 1.388e-06 | 0.0001133 |
| 58 | NEGATIVE REGULATION OF CELL ADHESION | 13 | 223 | 1.55e-06 | 0.0001243 |
| 59 | DEVELOPMENTAL MATURATION | 12 | 193 | 2.017e-06 | 0.000159 |
| 60 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 26 | 823 | 2.12e-06 | 0.0001644 |
| 61 | NEGATIVE REGULATION OF HOMOTYPIC CELL CELL ADHESION | 9 | 102 | 2.276e-06 | 0.0001736 |
| 62 | REGULATION OF CELL DEVELOPMENT | 26 | 836 | 2.821e-06 | 0.0002117 |
| 63 | NEURON PROJECTION GUIDANCE | 12 | 205 | 3.773e-06 | 0.0002787 |
| 64 | SUBSTRATE ADHESION DEPENDENT CELL SPREADING | 6 | 38 | 3.875e-06 | 0.0002817 |
| 65 | INTEGRIN MEDIATED SIGNALING PATHWAY | 8 | 82 | 3.949e-06 | 0.0002827 |
| 66 | REGULATION OF IMMUNE RESPONSE | 26 | 858 | 4.505e-06 | 0.0003176 |
| 67 | LEUKOCYTE ACTIVATION | 17 | 414 | 4.892e-06 | 0.0003397 |
| 68 | MOVEMENT IN ENVIRONMENT OF OTHER ORGANISM INVOLVED IN SYMBIOTIC INTERACTION | 8 | 87 | 6.17e-06 | 0.0003828 |
| 69 | ENTRY INTO CELL OF OTHER ORGANISM INVOLVED IN SYMBIOTIC INTERACTION | 8 | 87 | 6.17e-06 | 0.0003828 |
| 70 | VIRAL ENTRY INTO HOST CELL | 8 | 87 | 6.17e-06 | 0.0003828 |
| 71 | ENTRY INTO OTHER ORGANISM INVOLVED IN SYMBIOTIC INTERACTION | 8 | 87 | 6.17e-06 | 0.0003828 |
| 72 | MOVEMENT IN HOST ENVIRONMENT | 8 | 87 | 6.17e-06 | 0.0003828 |
| 73 | ENTRY INTO HOST | 8 | 87 | 6.17e-06 | 0.0003828 |
| 74 | CENTRAL NERVOUS SYSTEM DEVELOPMENT | 26 | 872 | 6.013e-06 | 0.0003828 |
| 75 | ENTRY INTO HOST CELL | 8 | 87 | 6.17e-06 | 0.0003828 |
| 76 | NEURONAL ION CHANNEL CLUSTERING | 4 | 12 | 7.193e-06 | 0.0004404 |
| 77 | CELLULAR EXTRAVASATION | 5 | 25 | 7.622e-06 | 0.0004606 |
| 78 | LYMPHOCYTE ACTIVATION | 15 | 342 | 8.214e-06 | 0.00049 |
| 79 | ANTIGEN PROCESSING AND PRESENTATION OF PEPTIDE OR POLYSACCHARIDE ANTIGEN VIA MHC CLASS II | 8 | 94 | 1.099e-05 | 0.0006391 |
| 80 | ANTIGEN PROCESSING AND PRESENTATION OF PEPTIDE ANTIGEN VIA MHC CLASS II | 8 | 94 | 1.099e-05 | 0.0006391 |
| 81 | REGULATION OF CELL PROLIFERATION | 36 | 1496 | 1.172e-05 | 0.0006735 |
| 82 | NEGATIVE REGULATION OF LEUKOCYTE PROLIFERATION | 7 | 69 | 1.24e-05 | 0.0007035 |
| 83 | ADHERENS JUNCTION ORGANIZATION | 7 | 71 | 1.499e-05 | 0.0008406 |
| 84 | REGULATION OF CELL MORPHOGENESIS | 19 | 552 | 1.67e-05 | 0.000925 |
| 85 | REGULATION OF NEURON DIFFERENTIATION | 19 | 554 | 1.755e-05 | 0.000948 |
| 86 | LEARNING | 9 | 131 | 1.773e-05 | 0.000948 |
| 87 | IMMUNE RESPONSE | 29 | 1100 | 1.77e-05 | 0.000948 |
| 88 | NEURON MATURATION | 5 | 30 | 1.953e-05 | 0.001033 |
| 89 | INTERACTION WITH HOST | 9 | 134 | 2.124e-05 | 0.001111 |
| 90 | NEGATIVE REGULATION OF IMMUNE SYSTEM PROCESS | 15 | 372 | 2.21e-05 | 0.001143 |
| 91 | REGULATION OF TRANSPORT | 40 | 1804 | 2.428e-05 | 0.001241 |
| 92 | NEGATIVE REGULATION OF T CELL PROLIFERATION | 6 | 52 | 2.506e-05 | 0.001268 |
| 93 | NEURON CELL CELL ADHESION | 4 | 16 | 2.553e-05 | 0.001277 |
| 94 | NEGATIVE REGULATION OF CELL CELL ADHESION | 9 | 138 | 2.685e-05 | 0.001329 |
| 95 | REGULATION OF LEUKOCYTE MEDIATED CYTOTOXICITY | 6 | 53 | 2.8e-05 | 0.001371 |
| 96 | COGNITION | 12 | 251 | 2.896e-05 | 0.001404 |
| 97 | ANATOMICAL STRUCTURE FORMATION INVOLVED IN MORPHOGENESIS | 26 | 957 | 3.012e-05 | 0.001416 |
| 98 | CARDIOVASCULAR SYSTEM DEVELOPMENT | 23 | 788 | 2.986e-05 | 0.001416 |
| 99 | CIRCULATORY SYSTEM DEVELOPMENT | 23 | 788 | 2.986e-05 | 0.001416 |
| 100 | CELL PART MORPHOGENESIS | 20 | 633 | 3.382e-05 | 0.001574 |
| 101 | REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 22 | 750 | 4.163e-05 | 0.001918 |
| 102 | HEAD DEVELOPMENT | 21 | 709 | 5.455e-05 | 0.002488 |
| 103 | FOREBRAIN DEVELOPMENT | 14 | 357 | 5.621e-05 | 0.002539 |
| 104 | POSITIVE REGULATION OF TRANSPORT | 25 | 936 | 5.693e-05 | 0.002547 |
| 105 | TELENCEPHALON DEVELOPMENT | 11 | 228 | 5.747e-05 | 0.002547 |
| 106 | RESPONSE TO CYTOKINE | 21 | 714 | 6.029e-05 | 0.002647 |
| 107 | SKELETAL SYSTEM DEVELOPMENT | 16 | 455 | 6.126e-05 | 0.002664 |
| 108 | REGULATION OF SYSTEM PROCESS | 17 | 507 | 6.488e-05 | 0.002795 |
| 109 | ENSHEATHMENT OF NEURONS | 7 | 91 | 7.546e-05 | 0.003164 |
| 110 | AXON ENSHEATHMENT | 7 | 91 | 7.546e-05 | 0.003164 |
| 111 | REGULATION OF CELL KILLING | 6 | 63 | 7.547e-05 | 0.003164 |
| 112 | NEGATIVE REGULATION OF CELL ACTIVATION | 9 | 158 | 7.74e-05 | 0.003216 |
| 113 | APICAL JUNCTION ASSEMBLY | 5 | 40 | 8.228e-05 | 0.003272 |
| 114 | CELL ACTIVATION | 18 | 568 | 8.125e-05 | 0.003272 |
| 115 | ENDODERMAL CELL DIFFERENTIATION | 5 | 40 | 8.228e-05 | 0.003272 |
| 116 | POSITIVE REGULATION OF NATURAL KILLER CELL MEDIATED IMMUNITY | 4 | 21 | 8.031e-05 | 0.003272 |
| 117 | BEHAVIOR | 17 | 516 | 8.042e-05 | 0.003272 |
| 118 | VASCULATURE DEVELOPMENT | 16 | 469 | 8.732e-05 | 0.003436 |
| 119 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 26 | 1021 | 8.788e-05 | 0.003436 |
| 120 | TISSUE DEVELOPMENT | 34 | 1518 | 8.895e-05 | 0.003449 |
| 121 | REGULATION OF CYTOKINE BIOSYNTHETIC PROCESS | 7 | 94 | 9.275e-05 | 0.003542 |
| 122 | RECEPTOR CLUSTERING | 5 | 41 | 9.286e-05 | 0.003542 |
| 123 | REGULATION OF INTERFERON GAMMA PRODUCTION | 7 | 97 | 0.0001131 | 0.00428 |
| 124 | REGULATION OF SECRETION | 20 | 699 | 0.0001315 | 0.004933 |
| 125 | POSTSYNAPTIC MEMBRANE ORGANIZATION | 4 | 25 | 0.0001639 | 0.006099 |
| 126 | ODONTOGENESIS | 7 | 105 | 0.0001858 | 0.006862 |
| 127 | POSITIVE REGULATION OF NEURON DIFFERENTIATION | 12 | 306 | 0.000191 | 0.006997 |
| 128 | ORGAN MORPHOGENESIS | 22 | 841 | 0.0002168 | 0.007822 |
| 129 | LEUKOCYTE MIGRATION INVOLVED IN INFLAMMATORY RESPONSE | 3 | 11 | 0.0002169 | 0.007822 |
| 130 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 39 | 1929 | 0.000224 | 0.008017 |
| 131 | REGULATION OF CYTOKINE PRODUCTION | 17 | 563 | 0.0002272 | 0.008027 |
| 132 | REGULATION OF NEURON PROJECTION DEVELOPMENT | 14 | 408 | 0.0002277 | 0.008027 |
| 133 | SYNAPSE ORGANIZATION | 8 | 145 | 0.0002409 | 0.008335 |
| 134 | REGULATION OF SYNAPTIC TRANSMISSION GLUTAMATERGIC | 5 | 50 | 0.0002418 | 0.008335 |
| 135 | ENDODERM FORMATION | 5 | 50 | 0.0002418 | 0.008335 |
| 136 | BLOOD VESSEL MORPHOGENESIS | 13 | 364 | 0.0002594 | 0.008874 |
| 137 | CELL RECOGNITION | 8 | 148 | 0.0002769 | 0.009403 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CELL ADHESION MOLECULE BINDING | 18 | 186 | 3.767e-12 | 3.499e-09 |
| 2 | MHC CLASS II RECEPTOR ACTIVITY | 6 | 12 | 1.656e-09 | 7.694e-07 |
| 3 | PEPTIDE ANTIGEN BINDING | 7 | 31 | 4.342e-08 | 1.345e-05 |
| 4 | RECEPTOR ACTIVITY | 42 | 1649 | 4.331e-07 | 0.0001006 |
| 5 | ANTIGEN BINDING | 10 | 114 | 6.523e-07 | 0.0001212 |
| 6 | VIRUS RECEPTOR ACTIVITY | 8 | 70 | 1.174e-06 | 0.0001818 |
| 7 | RECEPTOR BINDING | 37 | 1476 | 3.435e-06 | 0.0004558 |
| 8 | PROTEIN COMPLEX BINDING | 26 | 935 | 2.029e-05 | 0.002356 |
| 9 | CORECEPTOR ACTIVITY | 5 | 38 | 6.392e-05 | 0.006598 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CELL SURFACE | 45 | 757 | 2.451e-20 | 1.432e-17 |
| 2 | INTRINSIC COMPONENT OF PLASMA MEMBRANE | 63 | 1649 | 1.605e-18 | 4.41e-16 |
| 3 | SIDE OF MEMBRANE | 33 | 428 | 2.265e-18 | 4.41e-16 |
| 4 | PLASMA MEMBRANE PROTEIN COMPLEX | 34 | 510 | 5.758e-17 | 8.406e-15 |
| 5 | CELL CELL JUNCTION | 28 | 383 | 3.986e-15 | 3.879e-13 |
| 6 | EXTERNAL SIDE OF PLASMA MEMBRANE | 23 | 238 | 3.437e-15 | 3.879e-13 |
| 7 | CELL JUNCTION | 46 | 1151 | 3.643e-14 | 3.039e-12 |
| 8 | MHC PROTEIN COMPLEX | 10 | 27 | 1.897e-13 | 1.385e-11 |
| 9 | APICAL JUNCTION COMPLEX | 16 | 128 | 1.182e-12 | 7.669e-11 |
| 10 | MHC CLASS II PROTEIN COMPLEX | 7 | 16 | 2.181e-10 | 1.273e-08 |
| 11 | PROTEIN COMPLEX INVOLVED IN CELL ADHESION | 8 | 30 | 1.071e-09 | 5.215e-08 |
| 12 | MEMBRANE PROTEIN COMPLEX | 36 | 1020 | 1.004e-09 | 5.215e-08 |
| 13 | ANCHORING JUNCTION | 24 | 489 | 1.617e-09 | 7.265e-08 |
| 14 | LUMENAL SIDE OF MEMBRANE | 8 | 33 | 2.469e-09 | 1.03e-07 |
| 15 | ER TO GOLGI TRANSPORT VESICLE MEMBRANE | 9 | 55 | 9.98e-09 | 3.886e-07 |
| 16 | ER TO GOLGI TRANSPORT VESICLE | 9 | 71 | 1e-07 | 3.651e-06 |
| 17 | NEURON PROJECTION | 30 | 942 | 2.731e-07 | 9.383e-06 |
| 18 | RECEPTOR COMPLEX | 16 | 327 | 9.916e-07 | 3.217e-05 |
| 19 | ENDOCYTIC VESICLE | 14 | 256 | 1.307e-06 | 4.017e-05 |
| 20 | NEURON PART | 34 | 1265 | 2.01e-06 | 5.833e-05 |
| 21 | GOLGI APPARATUS | 37 | 1445 | 2.098e-06 | 5.833e-05 |
| 22 | CELL CELL ADHERENS JUNCTION | 7 | 54 | 2.348e-06 | 6.232e-05 |
| 23 | CELL PROJECTION | 42 | 1786 | 3.431e-06 | 8.711e-05 |
| 24 | COATED VESICLE MEMBRANE | 10 | 139 | 3.998e-06 | 9.729e-05 |
| 25 | MEMBRANE REGION | 31 | 1134 | 4.265e-06 | 9.962e-05 |
| 26 | CLATHRIN COATED ENDOCYTIC VESICLE | 7 | 65 | 8.311e-06 | 0.0001809 |
| 27 | TRANSPORT VESICLE MEMBRANE | 10 | 151 | 8.362e-06 | 0.0001809 |
| 28 | ENDOCYTIC VESICLE MEMBRANE | 10 | 152 | 8.864e-06 | 0.0001849 |
| 29 | CELL SUBSTRATE JUNCTION | 16 | 398 | 1.218e-05 | 0.0002452 |
| 30 | CLATHRIN COATED ENDOCYTIC VESICLE MEMBRANE | 6 | 48 | 1.568e-05 | 0.0003053 |
| 31 | MYELIN SHEATH | 10 | 168 | 2.124e-05 | 0.0004002 |
| 32 | APICOLATERAL PLASMA MEMBRANE | 4 | 16 | 2.553e-05 | 0.0004658 |
| 33 | PLASMA MEMBRANE RECEPTOR COMPLEX | 10 | 175 | 3.018e-05 | 0.0005341 |
| 34 | TRANS GOLGI NETWORK MEMBRANE | 7 | 81 | 3.565e-05 | 0.0006123 |
| 35 | COATED VESICLE | 11 | 234 | 7.258e-05 | 0.001211 |
| 36 | VACUOLAR PART | 20 | 694 | 0.0001194 | 0.001938 |
| 37 | PLASMA MEMBRANE REGION | 24 | 929 | 0.0001342 | 0.002118 |
| 38 | INTRINSIC COMPONENT OF ENDOPLASMIC RETICULUM MEMBRANE | 8 | 136 | 0.0001552 | 0.002385 |
| 39 | GOLGI APPARATUS PART | 23 | 893 | 0.0001945 | 0.002912 |
| 40 | MHC CLASS I PROTEIN COMPLEX | 3 | 11 | 0.0002169 | 0.003166 |
| 41 | BASAL PART OF CELL | 5 | 51 | 0.0002657 | 0.003784 |
| 42 | AXON | 14 | 418 | 0.0002912 | 0.004045 |
| 43 | CORTICAL CYTOSKELETON | 6 | 81 | 0.0003047 | 0.004045 |
| 44 | CLATHRIN COATED VESICLE MEMBRANE | 6 | 81 | 0.0003047 | 0.004045 |
| 45 | INTRINSIC COMPONENT OF ORGANELLE MEMBRANE | 11 | 281 | 0.0003576 | 0.00464 |
| 46 | ENDOSOMAL PART | 14 | 430 | 0.0003869 | 0.004912 |
| 47 | CLATHRIN COATED VESICLE | 8 | 157 | 0.000412 | 0.005119 |
| 48 | SOMATODENDRITIC COMPARTMENT | 18 | 650 | 0.0004227 | 0.005143 |
| 49 | MEMBRANE MICRODOMAIN | 11 | 288 | 0.0004399 | 0.005243 |
| 50 | BASAL PLASMA MEMBRANE | 4 | 33 | 0.000494 | 0.005657 |
| 51 | MAIN AXON | 5 | 58 | 0.0004863 | 0.005657 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
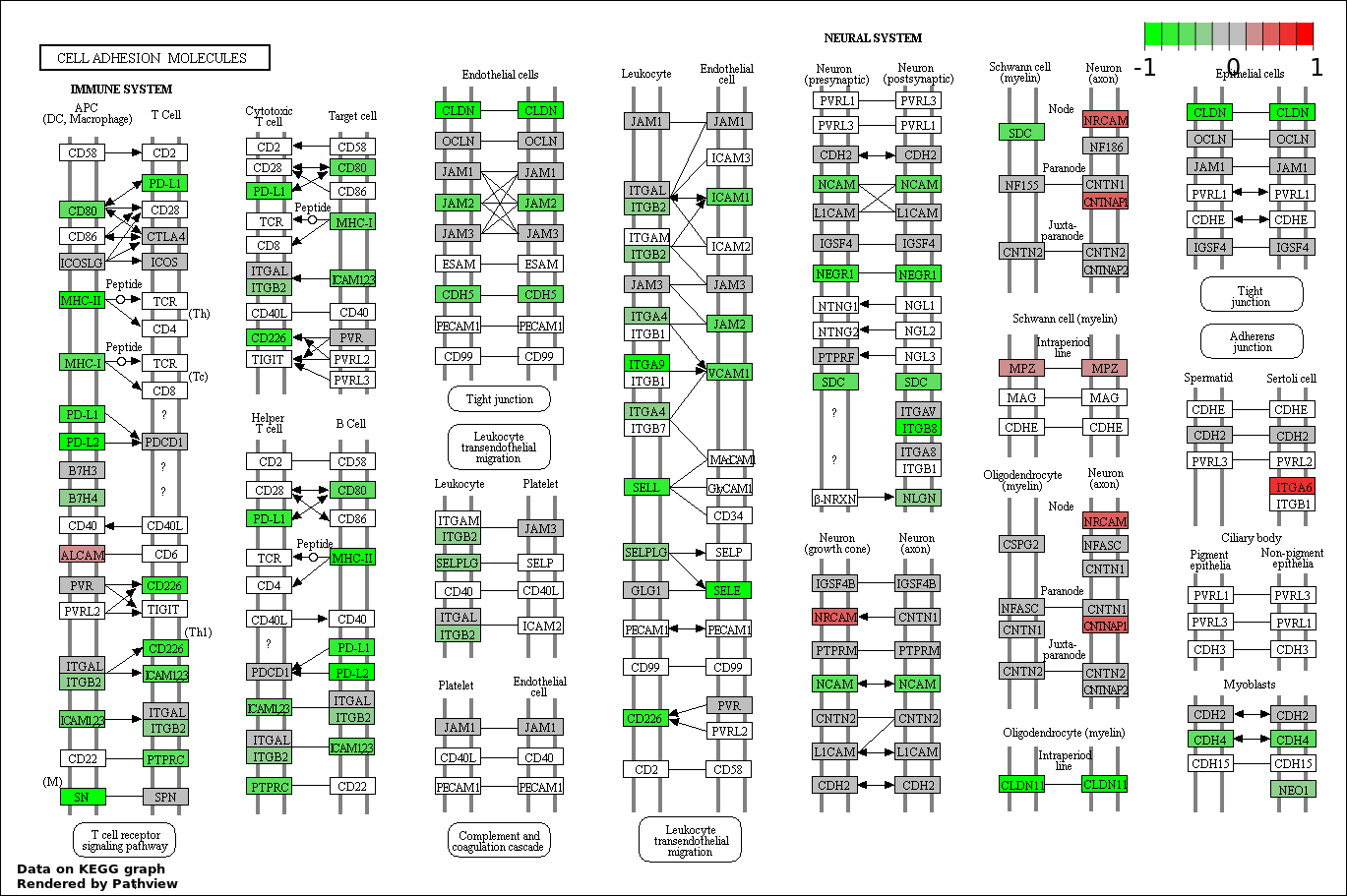
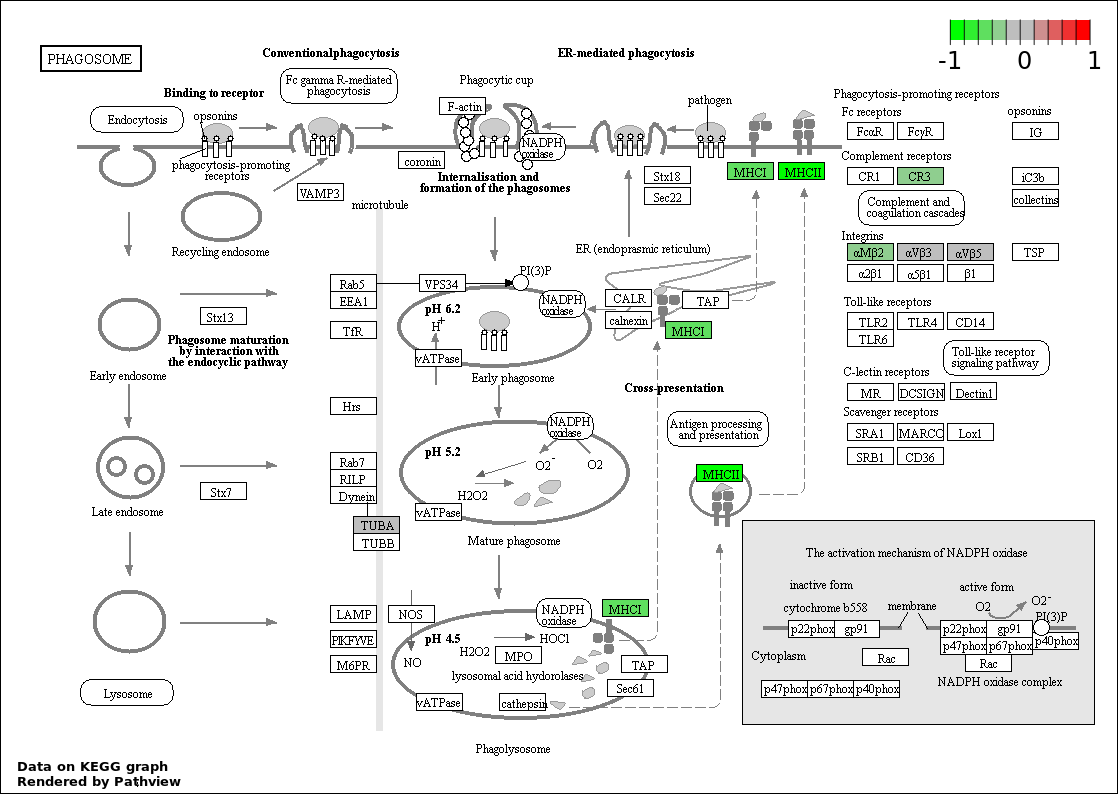
| 1 | hsa04514_Cell_adhesion_molecules_.CAMs. | 78 | 136 | 2.521e-120 | 4.537e-118 | |
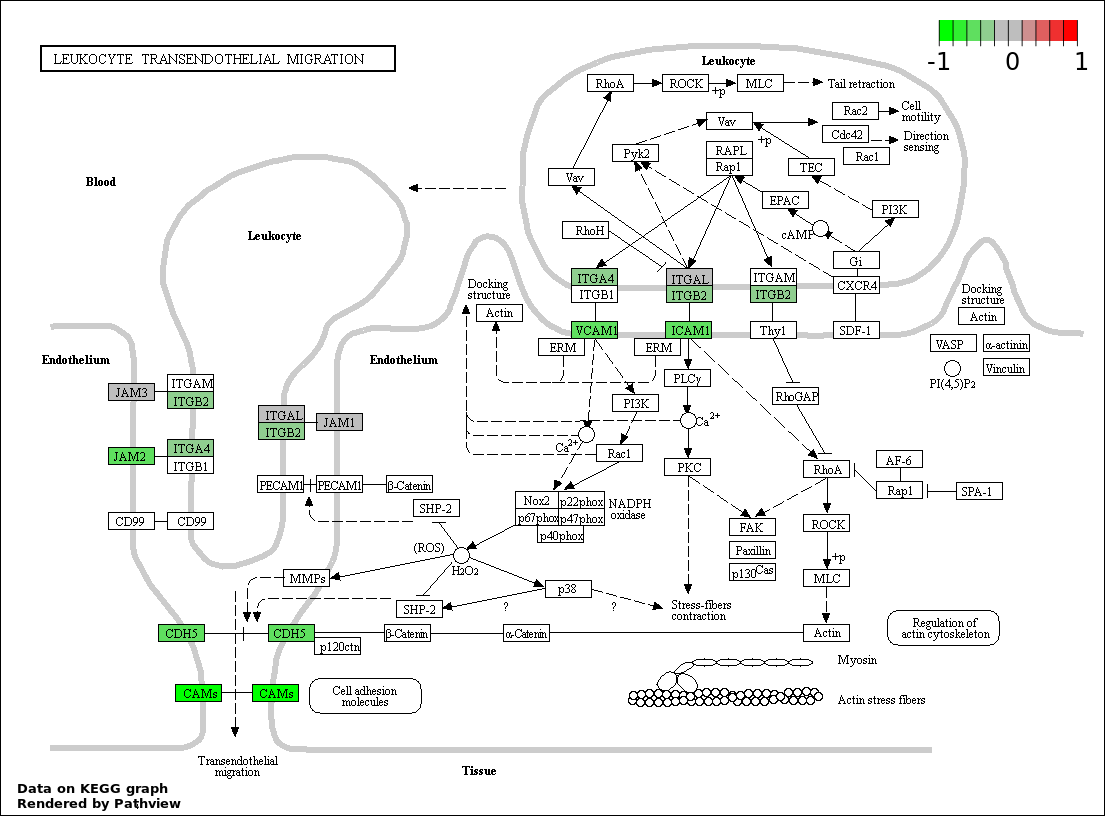
| 2 | hsa04670_Leukocyte_transendothelial_migration | 20 | 117 | 2.916e-18 | 2.624e-16 | |
| 3 | hsa04530_Tight_junction | 16 | 133 | 2.157e-12 | 1.294e-10 | |
| 4 | hsa04672_Intestinal_immune_network_for_IgA_production | 11 | 49 | 5.725e-12 | 2.576e-10 | |
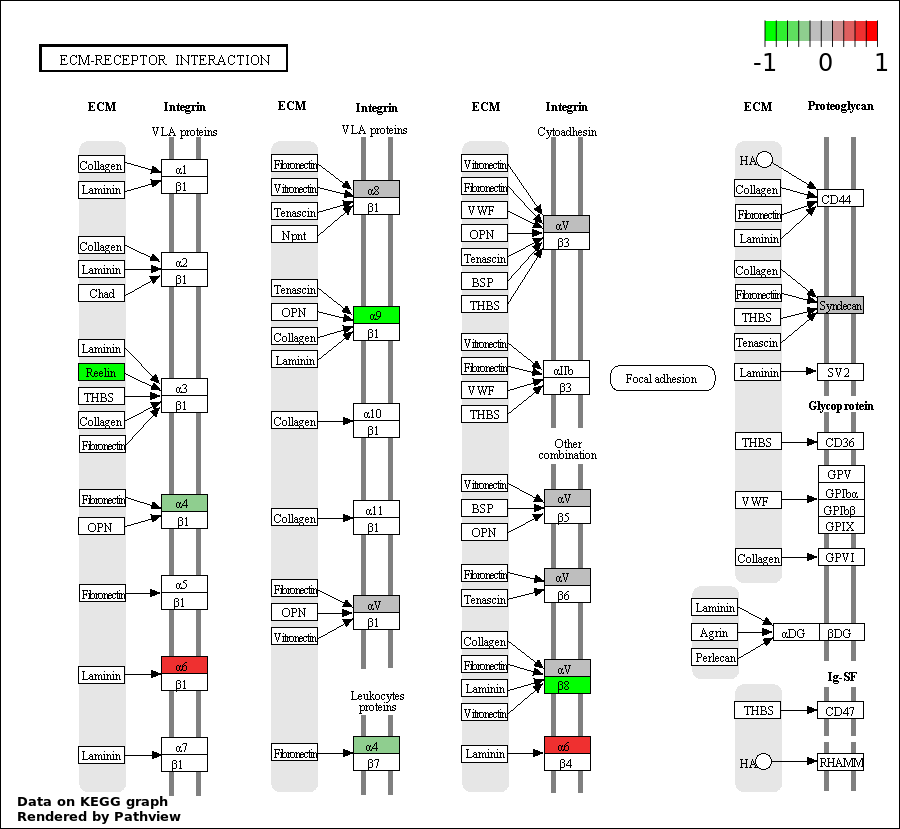
| 5 | hsa04512_ECM.receptor_interaction | 11 | 85 | 2.941e-09 | 1.059e-07 | |
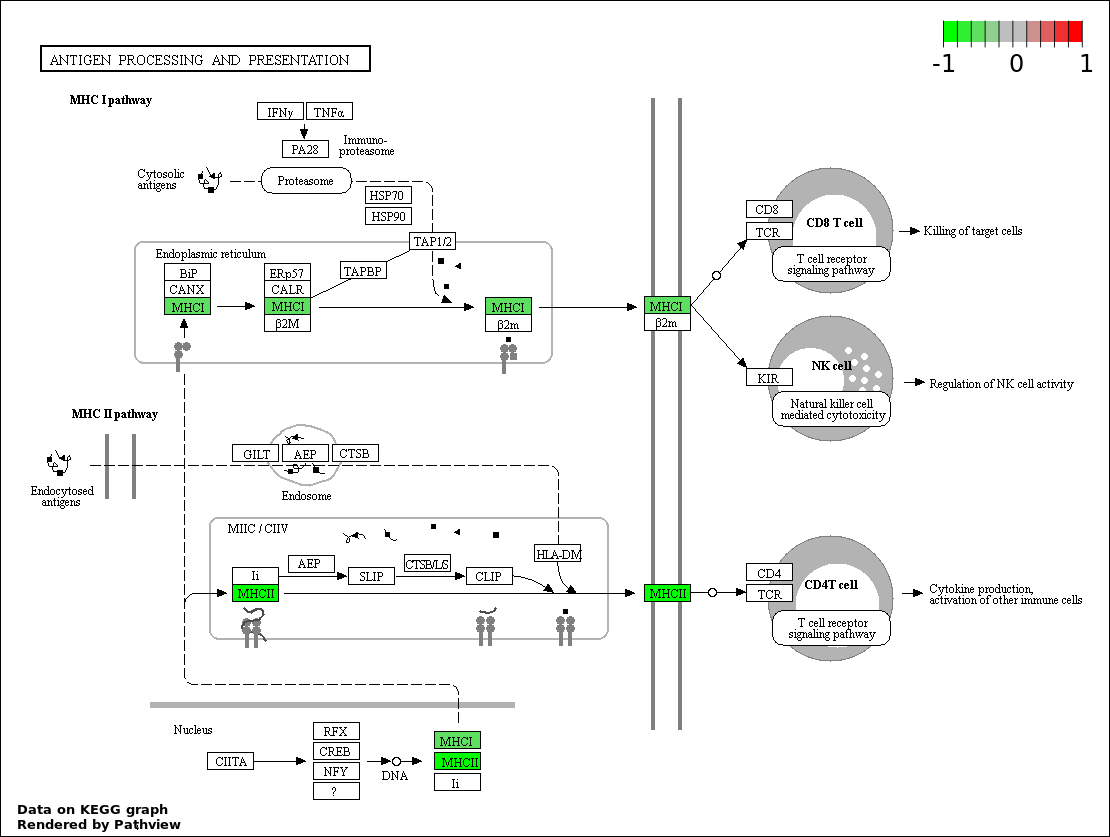
| 6 | hsa04612_Antigen_processing_and_presentation | 10 | 78 | 1.716e-08 | 5.147e-07 | |
| 7 | hsa04145_Phagosome | 13 | 156 | 2.466e-08 | 6.342e-07 | |
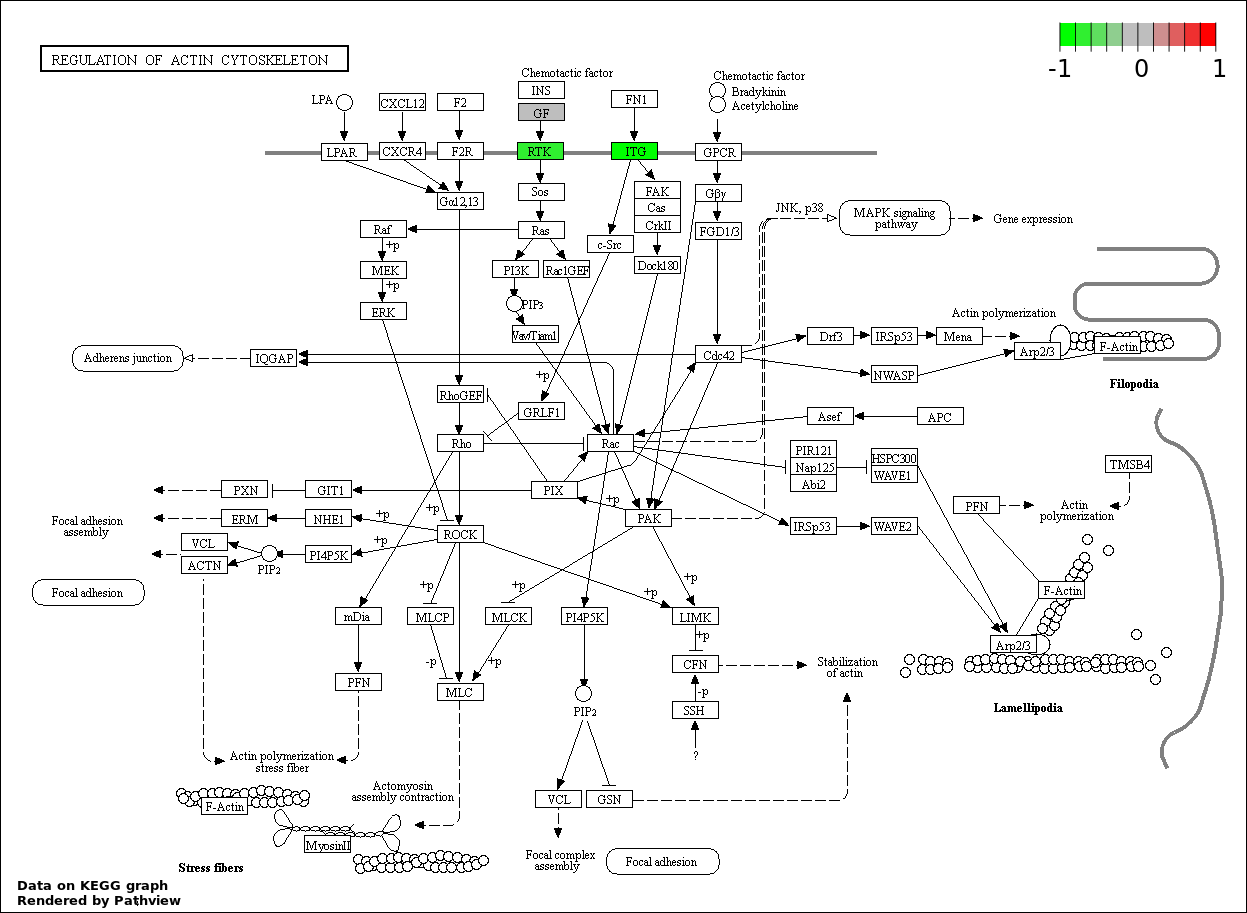
| 8 | hsa04810_Regulation_of_actin_cytoskeleton | 10 | 214 | 0.000162 | 0.003646 | |
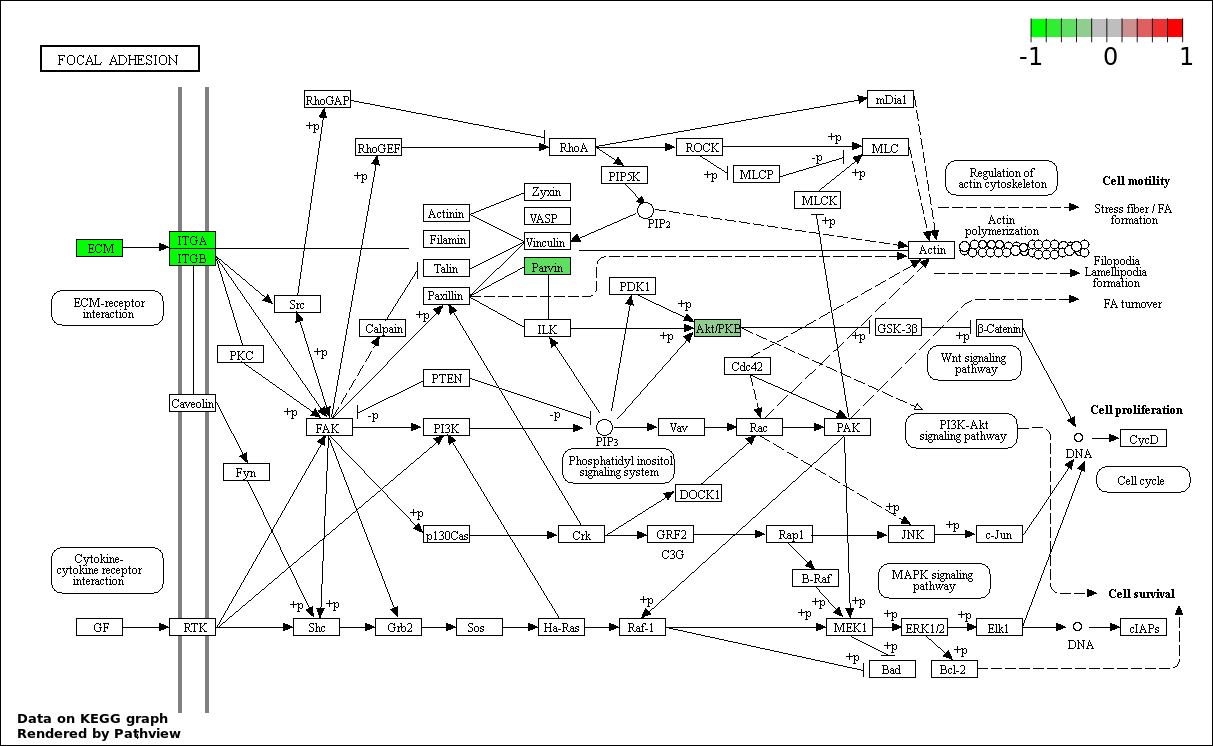
| 9 | hsa04510_Focal_adhesion | 9 | 200 | 0.0004523 | 0.009046 | |
| 10 | hsa04151_PI3K_AKT_signaling_pathway | 12 | 351 | 0.000656 | 0.01181 | |
| 11 | hsa04660_T_cell_receptor_signaling_pathway | 6 | 108 | 0.001398 | 0.02288 | |
| 12 | hsa04970_Salivary_secretion | 5 | 89 | 0.003326 | 0.04989 | |
| 13 | hsa04916_Melanogenesis | 5 | 101 | 0.005697 | 0.07889 | |
| 14 | hsa04914_Progesterone.mediated_oocyte_maturation | 4 | 87 | 0.01679 | 0.2003 | |
| 15 | hsa04640_Hematopoietic_cell_lineage | 4 | 88 | 0.01744 | 0.2003 | |
| 16 | hsa04540_Gap_junction | 4 | 90 | 0.01879 | 0.2003 | |
| 17 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 5 | 136 | 0.01893 | 0.2003 | |
| 18 | hsa04910_Insulin_signaling_pathway | 5 | 138 | 0.02003 | 0.2003 | |
| 19 | hsa04912_GnRH_signaling_pathway | 4 | 101 | 0.02731 | 0.2364 | |
| 20 | hsa04972_Pancreatic_secretion | 4 | 101 | 0.02731 | 0.2364 | |
| 21 | hsa04144_Endocytosis | 6 | 203 | 0.02759 | 0.2364 | |
| 22 | hsa04270_Vascular_smooth_muscle_contraction | 4 | 116 | 0.04214 | 0.3344 | |
| 23 | hsa00512_Mucin_type_O.Glycan_biosynthesis | 2 | 30 | 0.04459 | 0.3344 | |
| 24 | hsa04520_Adherens_junction | 3 | 73 | 0.04922 | 0.3523 | |
| 25 | hsa04971_Gastric_acid_secretion | 3 | 74 | 0.05088 | 0.3523 | |
| 26 | hsa00350_Tyrosine_metabolism | 2 | 41 | 0.07763 | 0.4991 | |
| 27 | hsa04390_Hippo_signaling_pathway | 4 | 154 | 0.09625 | 0.5974 | |
| 28 | hsa04620_Toll.like_receptor_signaling_pathway | 3 | 102 | 0.1079 | 0.6471 | |
| 29 | hsa04150_mTOR_signaling_pathway | 2 | 52 | 0.116 | 0.6737 | |
| 30 | hsa04114_Oocyte_meiosis | 3 | 114 | 0.1375 | 0.7733 | |
| 31 | hsa04920_Adipocytokine_signaling_pathway | 2 | 68 | 0.1781 | 0.951 | |
| 32 | hsa04720_Long.term_potentiation | 2 | 70 | 0.1861 | 0.951 | |
| 33 | hsa04730_Long.term_depression | 2 | 70 | 0.1861 | 0.951 | |
| 34 | hsa04976_Bile_secretion | 2 | 71 | 0.1902 | 0.951 | |
| 35 | hsa04370_VEGF_signaling_pathway | 2 | 76 | 0.2107 | 1 | |
| 36 | hsa04664_Fc_epsilon_RI_signaling_pathway | 2 | 79 | 0.2231 | 1 | |
| 37 | hsa04350_TGF.beta_signaling_pathway | 2 | 85 | 0.248 | 1 | |
| 38 | hsa00230_Purine_metabolism | 3 | 162 | 0.2749 | 1 | |
| 39 | hsa04014_Ras_signaling_pathway | 4 | 236 | 0.2751 | 1 | |
| 40 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 2 | 95 | 0.2897 | 1 | |
| 41 | hsa04020_Calcium_signaling_pathway | 3 | 177 | 0.3209 | 1 | |
| 42 | hsa04010_MAPK_signaling_pathway | 4 | 268 | 0.3561 | 1 | |
| 43 | hsa04062_Chemokine_signaling_pathway | 3 | 189 | 0.3578 | 1 | |
| 44 | hsa04722_Neurotrophin_signaling_pathway | 2 | 127 | 0.4193 | 1 | |
| 45 | hsa04380_Osteoclast_differentiation | 2 | 128 | 0.4232 | 1 | |
| 46 | hsa04360_Axon_guidance | 2 | 130 | 0.4309 | 1 | |
| 47 | hsa04310_Wnt_signaling_pathway | 2 | 151 | 0.5084 | 1 | |
| 48 | hsa04740_Olfactory_transduction | 2 | 388 | 0.9346 | 1 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | SNHG5 | hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-137;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-301a-3p;hsa-miR-302d-3p;hsa-miR-373-3p;hsa-miR-449a;hsa-miR-93-5p | 10 | ITGB8 | Sponge network | -0.338 | 1.0E-5 | -1.454 | 0 | 0.3 |