This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-22-3p | A4GNT | -0.63 | 0 | -1.24 | 0.00042 | MirTarget | -0.76 | 0 | NA | |
| 2 | hsa-miR-22-3p | ACLY | -0.63 | 0 | 1.02 | 0 | MirTarget | -0.34 | 0 | 27317765; 26477310 | Herein four different types of tumor cells including osteosarcoma prostate cervical and lung cancers were adopted in our study and we have demonstrated that miR-22 directly downregulated ACLY; Moreover miR-22 was proved to attenuate cancer cell proliferation and invasion as well as promote cell apoptosis via inhibiting ACLY; Additionally we confirmed the higher ACLY protein levels and the lower miR-22 expressions in hundreds of clinical samples of the four primary tumors and a negative correlation relationship between ACLY and miR-22 was clarified; These findings provide unequivocal proofs that miR-22 is responsible for the posttranscriptional regulation of ACLY which yields promising therapeutic effects in osteosarcoma prostate cervical and lung cancers;Within this cluster the cancer-associated and cardioprotective miR-22 was shown to repress fatty acid synthesis and elongation in tumour cells by targeting ATP citrate lyase and fatty acid elongase 6 as well as impairing mitochondrial one-carbon metabolism by suppression of methylene tetrahydrofolate dehydrogenase/cyclohydrolase |
| 3 | hsa-miR-22-3p | ADAP1 | -0.63 | 0 | -0.91 | 0.00084 | mirMAP | -0.52 | 5.0E-5 | NA | |
| 4 | hsa-miR-22-3p | ADCK2 | -0.63 | 0 | 0.53 | 0 | MirTarget | -0.15 | 0.00087 | NA | |
| 5 | hsa-miR-22-3p | ADM2 | -0.63 | 0 | 1.72 | 0 | mirMAP | -0.86 | 0 | NA | |
| 6 | hsa-miR-22-3p | AGBL5 | -0.63 | 0 | 0.72 | 0 | MirTarget; miRNATAP | -0.33 | 0 | NA | |
| 7 | hsa-miR-22-3p | ALMS1 | -0.63 | 0 | 0.54 | 0 | miRNAWalker2 validate | -0.23 | 0 | NA | |
| 8 | hsa-miR-22-3p | ANXA13 | -0.63 | 0 | -0.63 | 0.168 | MirTarget | -0.72 | 0.00066 | NA | |
| 9 | hsa-miR-22-3p | ARFIP2 | -0.63 | 0 | 0.37 | 0 | MirTarget; miRNATAP | -0.17 | 0 | NA | |
| 10 | hsa-miR-22-3p | ARID3B | -0.63 | 0 | 0.46 | 0.00021 | MirTarget; miRNATAP | -0.39 | 0 | NA | |
| 11 | hsa-miR-22-3p | ARID5B | -0.63 | 0 | -0.52 | 0.00012 | miRNAWalker2 validate | -0.16 | 0.01242 | NA | |
| 12 | hsa-miR-22-3p | ASB6 | -0.63 | 0 | 0.35 | 0 | MirTarget | -0.21 | 0 | NA | |
| 13 | hsa-miR-22-3p | ATP9A | -0.63 | 0 | 0.35 | 0.03032 | MirTarget | -0.32 | 3.0E-5 | NA | |
| 14 | hsa-miR-22-3p | B4GALT5 | -0.63 | 0 | -0.52 | 0 | MirTarget | -0.21 | 5.0E-5 | NA | |
| 15 | hsa-miR-22-3p | BCL9 | -0.63 | 0 | 1.18 | 0 | MirTarget; miRNATAP | -0.61 | 0 | NA | |
| 16 | hsa-miR-22-3p | BIN1 | -0.63 | 0 | 0.29 | 0.02481 | MirTarget; miRNATAP | -0.14 | 0.01955 | NA | |
| 17 | hsa-miR-22-3p | BMP7 | -0.63 | 0 | 0.83 | 0.04887 | miRNAWalker2 validate; miRTarBase | -0.87 | 1.0E-5 | NA | |
| 18 | hsa-miR-22-3p | BRSK2 | -0.63 | 0 | 1.39 | 0.0004 | MirTarget; miRNATAP | -1.07 | 0 | NA | |
| 19 | hsa-miR-22-3p | BRWD3 | -0.63 | 0 | 0.1 | 0.5367 | MirTarget; miRNATAP | -0.22 | 0.00248 | NA | |
| 20 | hsa-miR-22-3p | BSPRY | -0.63 | 0 | -0.09 | 0.79035 | MirTarget | -0.86 | 0 | NA | |
| 21 | hsa-miR-22-3p | BTBD10 | -0.63 | 0 | 0.28 | 2.0E-5 | MirTarget; miRNATAP | -0.15 | 0 | NA | |
| 22 | hsa-miR-22-3p | BTF3 | -0.63 | 0 | 0.14 | 0.09744 | miRNAWalker2 validate | -0.11 | 0.00766 | NA | |
| 23 | hsa-miR-22-3p | BUB1B | -0.63 | 0 | 3.86 | 0 | miRNAWalker2 validate | -1.6 | 0 | NA | |
| 24 | hsa-miR-22-3p | C17orf58 | -0.63 | 0 | 0.66 | 0 | miRNATAP | -0.17 | 0.00135 | NA | |
| 25 | hsa-miR-22-3p | C20orf194 | -0.63 | 0 | 0.24 | 0.07938 | mirMAP | -0.28 | 1.0E-5 | NA | |
| 26 | hsa-miR-22-3p | CBFA2T2 | -0.63 | 0 | 1.21 | 0 | mirMAP | -0.53 | 0 | NA | |
| 27 | hsa-miR-22-3p | CBL | -0.63 | 0 | 0.05 | 0.65471 | MirTarget | -0.29 | 0 | NA | |
| 28 | hsa-miR-22-3p | CBX6 | -0.63 | 0 | -0.14 | 0.56236 | MirTarget | -0.47 | 1.0E-5 | NA | |
| 29 | hsa-miR-22-3p | CCNA2 | -0.63 | 0 | 3.37 | 0 | MirTarget | -1.22 | 0 | 25596928 | The sequence of miR-22 which is conserved in mice rats humans and other mammalians aligns with the sequence of 3'-UTR of CCNA2; Chenodeoxycholic acid treatment and miR-22 mimics reduced CCNA2 protein and increased the number of G0/G1 Huh7 and HCT116 cells; In humans the expression levels of miR-22 and CCNA2 are inversely correlated in liver and colon cancers |
| 30 | hsa-miR-22-3p | CCNT2 | -0.63 | 0 | -0.13 | 0.25328 | miRNAWalker2 validate | -0.13 | 0.01181 | NA | |
| 31 | hsa-miR-22-3p | CDCA7L | -0.63 | 0 | 0.48 | 0.01422 | MirTarget | -0.66 | 0 | NA | |
| 32 | hsa-miR-22-3p | CLIP2 | -0.63 | 0 | 0.62 | 0.00364 | MirTarget | -0.36 | 0.00029 | NA | |
| 33 | hsa-miR-22-3p | COPS7B | -0.63 | 0 | 0.64 | 0 | MirTarget; miRNATAP | -0.34 | 0 | NA | |
| 34 | hsa-miR-22-3p | CREB1 | -0.63 | 0 | -0.01 | 0.8838 | MirTarget | -0.21 | 0 | NA | |
| 35 | hsa-miR-22-3p | CSMD2 | -0.63 | 0 | 2.32 | 0 | MirTarget | -0.45 | 0.00633 | NA | |
| 36 | hsa-miR-22-3p | CSNK2A1 | -0.63 | 0 | 0.31 | 6.0E-5 | miRNAWalker2 validate | -0.31 | 0 | NA | |
| 37 | hsa-miR-22-3p | CTSC | -0.63 | 0 | 0.24 | 0.17137 | miRNATAP | -0.3 | 0.00023 | NA | |
| 38 | hsa-miR-22-3p | DAGLA | -0.63 | 0 | 1.16 | 1.0E-5 | MirTarget | -0.6 | 0 | NA | |
| 39 | hsa-miR-22-3p | DIDO1 | -0.63 | 0 | 0.25 | 0.00079 | mirMAP | -0.2 | 0 | NA | |
| 40 | hsa-miR-22-3p | DNAJC5 | -0.63 | 0 | 0.73 | 0 | MirTarget | -0.28 | 0 | NA | |
| 41 | hsa-miR-22-3p | DPF2 | -0.63 | 0 | 0.62 | 0 | MirTarget; miRNATAP | -0.29 | 0 | NA | |
| 42 | hsa-miR-22-3p | DPM2 | -0.63 | 0 | 0.58 | 0 | MirTarget | -0.24 | 1.0E-5 | NA | |
| 43 | hsa-miR-22-3p | DSTYK | -0.63 | 0 | 1.01 | 0 | MirTarget | -0.27 | 0 | NA | |
| 44 | hsa-miR-22-3p | E2F2 | -0.63 | 0 | 2.82 | 0 | miRNAWalker2 validate | -1.18 | 0 | NA | |
| 45 | hsa-miR-22-3p | EDARADD | -0.63 | 0 | 0.94 | 4.0E-5 | MirTarget | -0.45 | 2.0E-5 | NA | |
| 46 | hsa-miR-22-3p | EDC3 | -0.63 | 0 | 0.73 | 0 | miRNAWalker2 validate; MirTarget | -0.32 | 0 | NA | |
| 47 | hsa-miR-22-3p | EFR3B | -0.63 | 0 | 0.42 | 0.00614 | MirTarget | -0.22 | 0.00208 | NA | |
| 48 | hsa-miR-22-3p | ENO1 | -0.63 | 0 | 0.29 | 0.02864 | MirTarget | -0.17 | 0.0055 | NA | |
| 49 | hsa-miR-22-3p | EP300 | -0.63 | 0 | -0.2 | 0.07049 | MirTarget; miRNATAP | -0.1 | 0.04632 | NA | |
| 50 | hsa-miR-22-3p | ERBB3 | -0.63 | 0 | 0.58 | 0.00109 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.46 | 0 | 22484852; 22510010; 21057539; 27569217 | Tumor suppressor miR 22 suppresses lung cancer cell progression through post transcriptional regulation of ErbB3; Furthermore miR-22 was demonstrated to inhibit the expression of ErbB3 through post-transcriptional regulation via binding to ErbB3 3'-UTR; miR-22 exhibited excellent anti-lung cancer activity in vitro and in vivo and post-transcriptional regulation of ErbB3 might be a potential mechanism;Transfected miR-22 Y79 cells inhibited the cell proliferation and reduced the migration and erythoblastic leukemia viral oncogene homolog 3 Erbb3 was confirmed to be the target gene of miR-22;These metastatic cells expressed higher levels of putative/proven miR-22 target oncogenes ERBB3 CDC25C and EVI-1; Introduction of miR-22 into cancer cells reduced the levels of ERBB3 and EVI-1 as well as phospho-AKT an EVI-1 downstream target;Finally we demonstrated the ability of miR-22 to intercept and overcome the intrinsic resistance to MEK inhibition based on ERBB3 upregulation |
| 51 | hsa-miR-22-3p | FAM49B | -0.63 | 0 | 0.71 | 0 | MirTarget; miRNATAP | -0.51 | 0 | NA | |
| 52 | hsa-miR-22-3p | FAM53C | -0.63 | 0 | 0.38 | 0 | MirTarget; miRNATAP | -0.14 | 2.0E-5 | NA | |
| 53 | hsa-miR-22-3p | FBRS | -0.63 | 0 | 0.35 | 0 | miRNATAP | -0.16 | 0 | NA | |
| 54 | hsa-miR-22-3p | FBXL19 | -0.63 | 0 | 1.09 | 0 | miRNATAP | -0.49 | 0 | NA | |
| 55 | hsa-miR-22-3p | FGD6 | -0.63 | 0 | 0.31 | 0.04392 | MirTarget | -0.36 | 0 | NA | |
| 56 | hsa-miR-22-3p | FIBCD1 | -0.63 | 0 | 1.89 | 0 | MirTarget | -0.62 | 0.00031 | NA | |
| 57 | hsa-miR-22-3p | FMNL2 | -0.63 | 0 | 0.05 | 0.80772 | MirTarget | -0.34 | 0.00061 | NA | |
| 58 | hsa-miR-22-3p | FOXP1 | -0.63 | 0 | 0.13 | 0.18315 | miRNAWalker2 validate; miRNATAP | -0.14 | 0.00251 | NA | |
| 59 | hsa-miR-22-3p | FRAT2 | -0.63 | 0 | 0.23 | 0.05124 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.23 | 1.0E-5 | NA | |
| 60 | hsa-miR-22-3p | FUBP1 | -0.63 | 0 | 0.25 | 0.02191 | miRNAWalker2 validate | -0.36 | 0 | NA | |
| 61 | hsa-miR-22-3p | GALNT4 | -0.63 | 0 | -0.65 | 0 | MirTarget | -0.18 | 0.0015 | NA | |
| 62 | hsa-miR-22-3p | GIGYF2 | -0.63 | 0 | -0.02 | 0.78691 | MirTarget | -0.13 | 1.0E-5 | NA | |
| 63 | hsa-miR-22-3p | GLIS2 | -0.63 | 0 | 0.56 | 0.01316 | miRNAWalker2 validate | -0.6 | 0 | NA | |
| 64 | hsa-miR-22-3p | GMEB2 | -0.63 | 0 | 0.65 | 0 | mirMAP | -0.26 | 0 | NA | |
| 65 | hsa-miR-22-3p | GNA12 | -0.63 | 0 | 0.6 | 0 | mirMAP | -0.11 | 0.01718 | NA | |
| 66 | hsa-miR-22-3p | GNB1L | -0.63 | 0 | 0.97 | 0 | mirMAP | -0.25 | 4.0E-5 | NA | |
| 67 | hsa-miR-22-3p | GPBP1 | -0.63 | 0 | 0.37 | 0 | MirTarget; miRNATAP | -0.14 | 0 | NA | |
| 68 | hsa-miR-22-3p | GPSM2 | -0.63 | 0 | 1.45 | 0 | MirTarget | -0.82 | 0 | NA | |
| 69 | hsa-miR-22-3p | H3F3B | -0.63 | 0 | 0.01 | 0.92585 | miRNAWalker2 validate; MirTarget | -0.17 | 0 | NA | |
| 70 | hsa-miR-22-3p | H3F3C | -0.63 | 0 | -0.33 | 0.00085 | MirTarget | -0.11 | 0.0142 | NA | |
| 71 | hsa-miR-22-3p | HDAC4 | -0.63 | 0 | 1.01 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.39 | 0 | 20842113 | Furthermore histone deacetylase 4 HDAC4 known to have critical roles in cancer development was proved to be directly targeted and regulated by miR-22; Furthermore HDAC4 was upregulated in miR-22-downregulated HCC tissues suggesting that downregulation of miR-22 might participate in HCC carcinogenesis and progression through potentiation of HDAC4 expression |
| 72 | hsa-miR-22-3p | HES2 | -0.63 | 0 | 1.81 | 0 | mirMAP | -0.59 | 0.0001 | NA | |
| 73 | hsa-miR-22-3p | HIF1AN | -0.63 | 0 | 0.59 | 0 | MirTarget | -0.22 | 0 | NA | |
| 74 | hsa-miR-22-3p | HNF4G | -0.63 | 0 | -0.51 | 0.00622 | MirTarget | -0.32 | 0.00019 | NA | |
| 75 | hsa-miR-22-3p | HNRNPH1 | -0.63 | 0 | 0.42 | 0 | miRNATAP | -0.33 | 0 | NA | |
| 76 | hsa-miR-22-3p | HNRNPUL2 | -0.63 | 0 | 0.28 | 0 | miRNATAP | -0.11 | 5.0E-5 | NA | |
| 77 | hsa-miR-22-3p | HOMER1 | -0.63 | 0 | -0.08 | 0.79615 | MirTarget | -0.52 | 0.00013 | NA | |
| 78 | hsa-miR-22-3p | HSF2BP | -0.63 | 0 | 2.11 | 0 | MirTarget | -0.97 | 0 | NA | |
| 79 | hsa-miR-22-3p | HSPA1B | -0.63 | 0 | 0.5 | 0.01779 | miRNAWalker2 validate | -0.3 | 0.00219 | NA | |
| 80 | hsa-miR-22-3p | HTR3A | -0.63 | 0 | 0.75 | 0.03159 | MirTarget | -0.6 | 0.00019 | NA | |
| 81 | hsa-miR-22-3p | HUNK | -0.63 | 0 | -0.72 | 0.05314 | MirTarget | -0.97 | 0 | NA | |
| 82 | hsa-miR-22-3p | IKZF4 | -0.63 | 0 | 0.32 | 0.00089 | MirTarget; miRNATAP | -0.23 | 0 | NA | |
| 83 | hsa-miR-22-3p | IL17RD | -0.63 | 0 | -0.13 | 0.6098 | MirTarget; miRNATAP | -0.3 | 0.00842 | NA | |
| 84 | hsa-miR-22-3p | IL27RA | -0.63 | 0 | 0.52 | 0.00116 | MirTarget | -0.35 | 0 | NA | |
| 85 | hsa-miR-22-3p | IPO7 | -0.63 | 0 | -0.13 | 0.12689 | MirTarget; miRNATAP | -0.18 | 0 | NA | |
| 86 | hsa-miR-22-3p | IRF5 | -0.63 | 0 | 0.55 | 0 | miRNAWalker2 validate | -0.17 | 0.00212 | NA | |
| 87 | hsa-miR-22-3p | ISY1 | -0.63 | 0 | 0.2 | 0.00017 | mirMAP | -0.18 | 0 | NA | |
| 88 | hsa-miR-22-3p | JARID2 | -0.63 | 0 | 0.39 | 3.0E-5 | miRNATAP | -0.36 | 0 | NA | |
| 89 | hsa-miR-22-3p | KCTD10 | -0.63 | 0 | 0.32 | 0 | MirTarget | -0.15 | 0 | NA | |
| 90 | hsa-miR-22-3p | KDM3A | -0.63 | 0 | 0.26 | 0.00489 | MirTarget; miRNATAP | -0.3 | 0 | NA | |
| 91 | hsa-miR-22-3p | KIAA1257 | -0.63 | 0 | 0.43 | 0.10986 | mirMAP | -0.4 | 0.00137 | NA | |
| 92 | hsa-miR-22-3p | KIF21B | -0.63 | 0 | 1.18 | 0 | mirMAP | -0.22 | 0.04318 | NA | |
| 93 | hsa-miR-22-3p | LAMC1 | -0.63 | 0 | 1.18 | 0 | MirTarget; miRNATAP | -0.44 | 0 | 26052614 | The identification of laminin gamma 1 LAMC1 and myeloid cell leukemia 1 MCL1 as direct targets of miR-22 and miR-29a respectively suggested a tumor-suppressive role of these miRNAs |
| 94 | hsa-miR-22-3p | LEMD3 | -0.63 | 0 | -0.24 | 0.00152 | miRNAWalker2 validate | -0.11 | 0.00227 | NA | |
| 95 | hsa-miR-22-3p | LIG3 | -0.63 | 0 | 0.62 | 0 | MirTarget | -0.23 | 0 | NA | |
| 96 | hsa-miR-22-3p | LRRC1 | -0.63 | 0 | 2.14 | 0 | miRNAWalker2 validate; miRNATAP | -1.34 | 0 | NA | |
| 97 | hsa-miR-22-3p | LRRC14 | -0.63 | 0 | 1.37 | 0 | mirMAP | -0.34 | 0 | NA | |
| 98 | hsa-miR-22-3p | LRRC59 | -0.63 | 0 | 0.44 | 0 | MirTarget | -0.16 | 1.0E-5 | NA | |
| 99 | hsa-miR-22-3p | MAP3K3 | -0.63 | 0 | 0.01 | 0.88843 | MirTarget; miRNATAP | -0.1 | 0.00322 | NA | |
| 100 | hsa-miR-22-3p | MAPK8IP3 | -0.63 | 0 | 1.01 | 0 | MirTarget | -0.31 | 0 | NA | |
| 101 | hsa-miR-22-3p | MAT2A | -0.63 | 0 | -0.01 | 0.92186 | MirTarget; miRNATAP | -0.15 | 0.00115 | NA | |
| 102 | hsa-miR-22-3p | MDC1 | -0.63 | 0 | 0.59 | 0 | MirTarget | -0.42 | 0 | NA | |
| 103 | hsa-miR-22-3p | MECOM | -0.63 | 0 | 0.96 | 0 | miRNATAP | -0.18 | 0.03306 | NA | |
| 104 | hsa-miR-22-3p | MECP2 | -0.63 | 0 | 0.26 | 0.00043 | miRNATAP | -0.14 | 4.0E-5 | NA | |
| 105 | hsa-miR-22-3p | MED22 | -0.63 | 0 | 0.9 | 0 | mirMAP | -0.42 | 0 | NA | |
| 106 | hsa-miR-22-3p | MEIS2 | -0.63 | 0 | 0.16 | 0.32841 | MirTarget | -0.23 | 0.00193 | NA | |
| 107 | hsa-miR-22-3p | MFGE8 | -0.63 | 0 | 0.98 | 0 | MirTarget | -0.24 | 0.00194 | NA | |
| 108 | hsa-miR-22-3p | MTF2 | -0.63 | 0 | 0.17 | 0.07103 | MirTarget; miRNATAP | -0.42 | 0 | NA | |
| 109 | hsa-miR-22-3p | MTMR2 | -0.63 | 0 | 0.4 | 0.001 | MirTarget | -0.49 | 0 | NA | |
| 110 | hsa-miR-22-3p | MURC | -0.63 | 0 | 1.58 | 0 | MirTarget | -0.37 | 6.0E-5 | NA | |
| 111 | hsa-miR-22-3p | MXD1 | -0.63 | 0 | -0.23 | 0.05533 | miRNATAP | -0.18 | 0.00079 | NA | |
| 112 | hsa-miR-22-3p | MYCBP | -0.63 | 0 | 0.27 | 0.00288 | miRNAWalker2 validate; miRTarBase | -0.17 | 8.0E-5 | NA | |
| 113 | hsa-miR-22-3p | MYO6 | -0.63 | 0 | -0.01 | 0.93833 | miRNAWalker2 validate | -0.17 | 0.00684 | NA | |
| 114 | hsa-miR-22-3p | N4BP3 | -0.63 | 0 | 1.82 | 0 | MirTarget | -0.61 | 0 | NA | |
| 115 | hsa-miR-22-3p | NET1 | -0.63 | 0 | -0.26 | 0.0069 | MirTarget; miRNATAP | -0.15 | 0.00068 | NA | |
| 116 | hsa-miR-22-3p | NHP2 | -0.63 | 0 | 0.77 | 0 | MirTarget | -0.12 | 0.0246 | NA | |
| 117 | hsa-miR-22-3p | NPNT | -0.63 | 0 | 0.53 | 0.0583 | miRNATAP | -0.59 | 1.0E-5 | NA | |
| 118 | hsa-miR-22-3p | NRAS | -0.63 | 0 | 0.3 | 0.00029 | MirTarget; miRNATAP | -0.29 | 0 | NA | |
| 119 | hsa-miR-22-3p | NUP210 | -0.63 | 0 | 0.78 | 0 | MirTarget | -0.54 | 0 | NA | |
| 120 | hsa-miR-22-3p | NUP214 | -0.63 | 0 | 0.17 | 0.00488 | miRNAWalker2 validate | -0.12 | 4.0E-5 | 20081105 | In addition to these specific effects reversing the expression of mir-22 and the putative oncogene mir-182 had widespread effects on target and nontarget gene populations that ultimately caused a global shift in the cancer gene signature toward a more normal state |
| 121 | hsa-miR-22-3p | NUSAP1 | -0.63 | 0 | 2.13 | 0 | MirTarget | -1.02 | 0 | NA | |
| 122 | hsa-miR-22-3p | ORAI2 | -0.63 | 0 | 0.15 | 0.33665 | mirMAP | -0.28 | 9.0E-5 | NA | |
| 123 | hsa-miR-22-3p | OTUD6B | -0.63 | 0 | 0.72 | 0 | MirTarget | -0.2 | 9.0E-5 | NA | |
| 124 | hsa-miR-22-3p | PACS2 | -0.63 | 0 | 0.78 | 0 | mirMAP | -0.21 | 0 | NA | |
| 125 | hsa-miR-22-3p | PHC1 | -0.63 | 0 | -0.35 | 0.02268 | MirTarget | -0.19 | 0.00881 | NA | |
| 126 | hsa-miR-22-3p | PIK3R6 | -0.63 | 0 | 0.97 | 0 | MirTarget | -0.38 | 1.0E-5 | NA | |
| 127 | hsa-miR-22-3p | PIP4K2B | -0.63 | 0 | 0.58 | 0 | MirTarget | -0.28 | 0 | NA | |
| 128 | hsa-miR-22-3p | PLAGL2 | -0.63 | 0 | 0.34 | 0.003 | miRNATAP | -0.49 | 0 | NA | |
| 129 | hsa-miR-22-3p | PLEKHA6 | -0.63 | 0 | -0.15 | 0.29725 | MirTarget | -0.14 | 0.03513 | NA | |
| 130 | hsa-miR-22-3p | PLEKHG4B | -0.63 | 0 | 0.79 | 0.03377 | mirMAP | -0.39 | 0.02379 | NA | |
| 131 | hsa-miR-22-3p | PMAIP1 | -0.63 | 0 | -0.75 | 0.00471 | MirTarget | -0.38 | 0.00196 | NA | |
| 132 | hsa-miR-22-3p | POGK | -0.63 | 0 | 0.63 | 0 | MirTarget | -0.45 | 0 | NA | |
| 133 | hsa-miR-22-3p | PRDM4 | -0.63 | 0 | 0.27 | 0.00011 | MirTarget | -0.24 | 0 | NA | |
| 134 | hsa-miR-22-3p | PRPF38A | -0.63 | 0 | 0.13 | 0.06066 | MirTarget | -0.32 | 0 | NA | |
| 135 | hsa-miR-22-3p | PRRT2 | -0.63 | 0 | 1.35 | 0 | miRNATAP | -0.59 | 0 | NA | |
| 136 | hsa-miR-22-3p | PRX | -0.63 | 0 | 0.97 | 0 | mirMAP | -0.36 | 0 | NA | |
| 137 | hsa-miR-22-3p | PTP4A3 | -0.63 | 0 | 1.98 | 0 | MirTarget | -0.73 | 0 | NA | |
| 138 | hsa-miR-22-3p | PTPN1 | -0.63 | 0 | 0.12 | 0.12184 | miRNATAP | -0.11 | 0.00191 | NA | |
| 139 | hsa-miR-22-3p | PURB | -0.63 | 0 | 0.17 | 0.00583 | miRNATAP | -0.2 | 0 | NA | |
| 140 | hsa-miR-22-3p | RAB11FIP4 | -0.63 | 0 | 1.54 | 0 | miRNATAP | -0.61 | 0 | NA | |
| 141 | hsa-miR-22-3p | RAPGEFL1 | -0.63 | 0 | 0.51 | 0.0061 | miRNATAP | -0.52 | 0 | NA | |
| 142 | hsa-miR-22-3p | RBM33 | -0.63 | 0 | 0.43 | 0 | mirMAP | -0.17 | 0 | NA | |
| 143 | hsa-miR-22-3p | RBM39 | -0.63 | 0 | 0.35 | 0 | miRNAWalker2 validate | -0.17 | 0 | NA | |
| 144 | hsa-miR-22-3p | RCC2 | -0.63 | 0 | 0.69 | 0 | MirTarget | -0.6 | 0 | NA | |
| 145 | hsa-miR-22-3p | RFXANK | -0.63 | 0 | 1.01 | 0 | MirTarget | -0.35 | 0 | NA | |
| 146 | hsa-miR-22-3p | RNF38 | -0.63 | 0 | -0.08 | 0.42663 | miRNATAP | -0.2 | 0 | NA | |
| 147 | hsa-miR-22-3p | RPL24 | -0.63 | 0 | 0.12 | 0.31552 | miRNAWalker2 validate | -0.15 | 0.00627 | NA | |
| 148 | hsa-miR-22-3p | RPL35A | -0.63 | 0 | 0.4 | 0.00057 | miRNAWalker2 validate | -0.29 | 0 | NA | |
| 149 | hsa-miR-22-3p | RPS2 | -0.63 | 0 | 0.39 | 0.00217 | miRNAWalker2 validate | -0.32 | 0 | NA | |
| 150 | hsa-miR-22-3p | RPS4X | -0.63 | 0 | 0.17 | 0.13378 | miRNAWalker2 validate | -0.25 | 0 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | NUCLEIC ACID BINDING TRANSCRIPTION FACTOR ACTIVITY | 29 | 1199 | 9.068e-06 | 0.004212 |
| 2 | POLY A RNA BINDING | 29 | 1170 | 5.681e-06 | 0.004212 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
| 1 | hsa04520_Adherens_junction | 5 | 73 | 0.0008235 | 0.1482 | |
| 2 | hsa03010_Ribosome | 5 | 92 | 0.002315 | 0.2083 | |
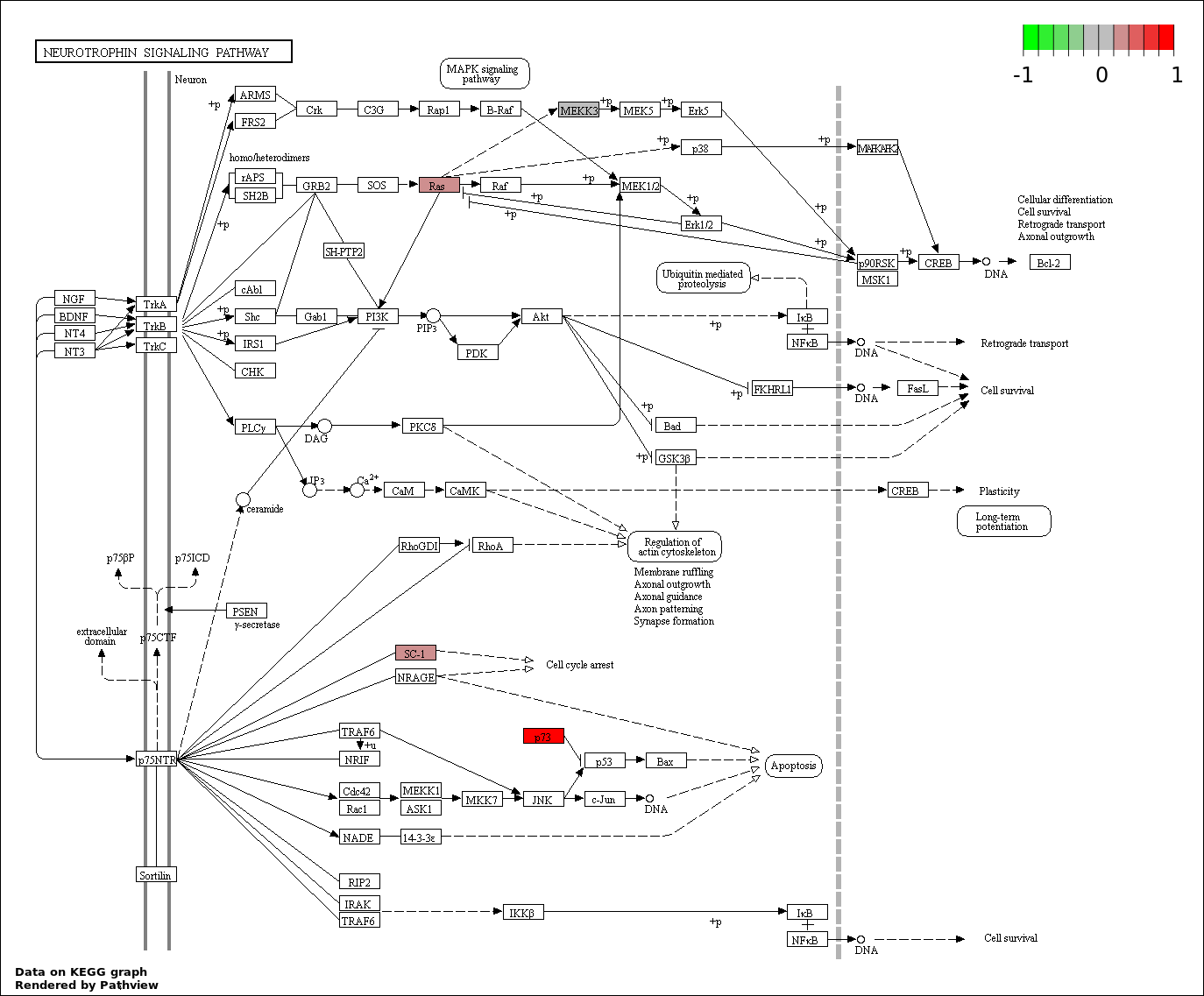
| 3 | hsa04722_Neurotrophin_signaling_pathway | 5 | 127 | 0.009051 | 0.3768 | |
| 4 | hsa04110_Cell_cycle | 5 | 128 | 0.009345 | 0.3768 | |
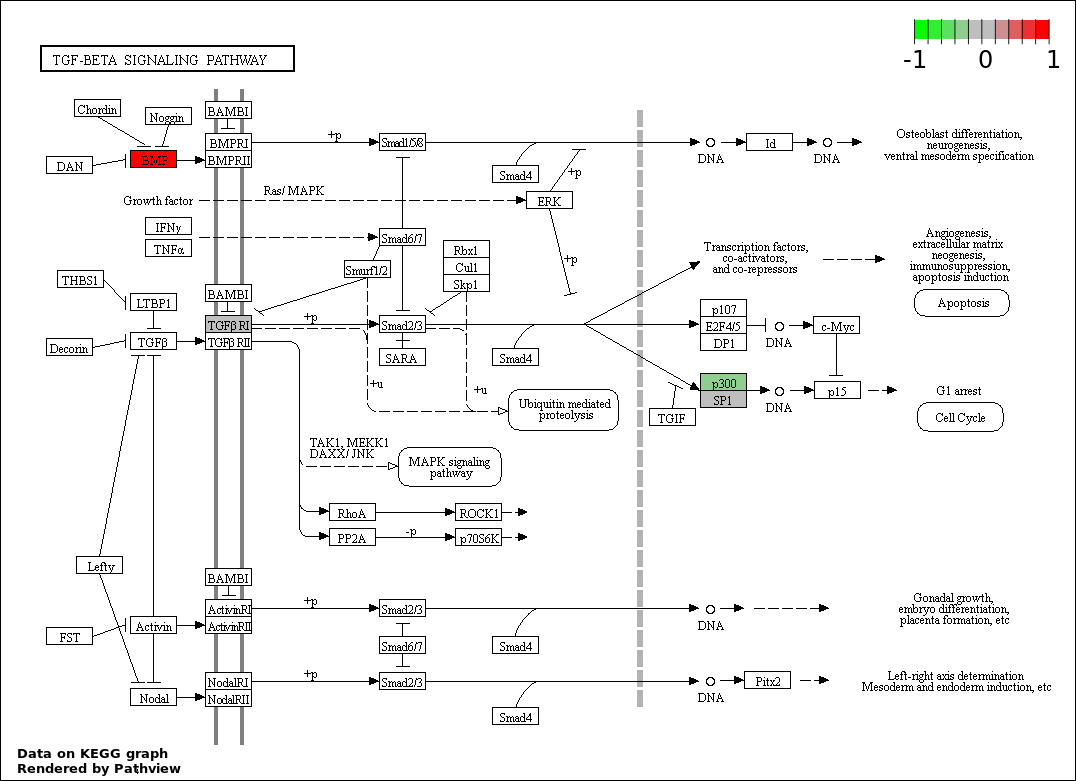
| 5 | hsa04350_TGF.beta_signaling_pathway | 4 | 85 | 0.01047 | 0.3768 | |
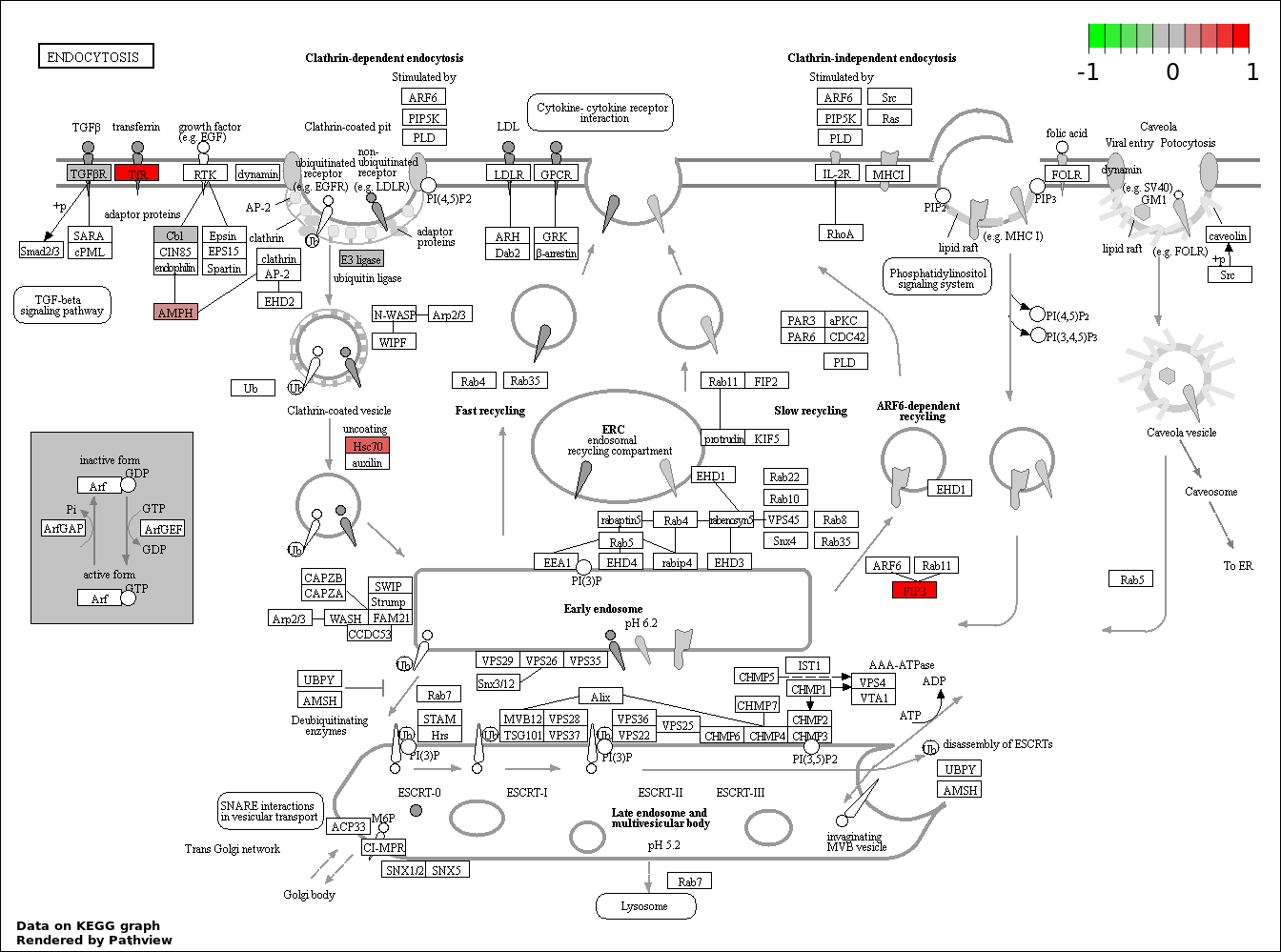
| 6 | hsa04144_Endocytosis | 6 | 203 | 0.01655 | 0.4966 | |
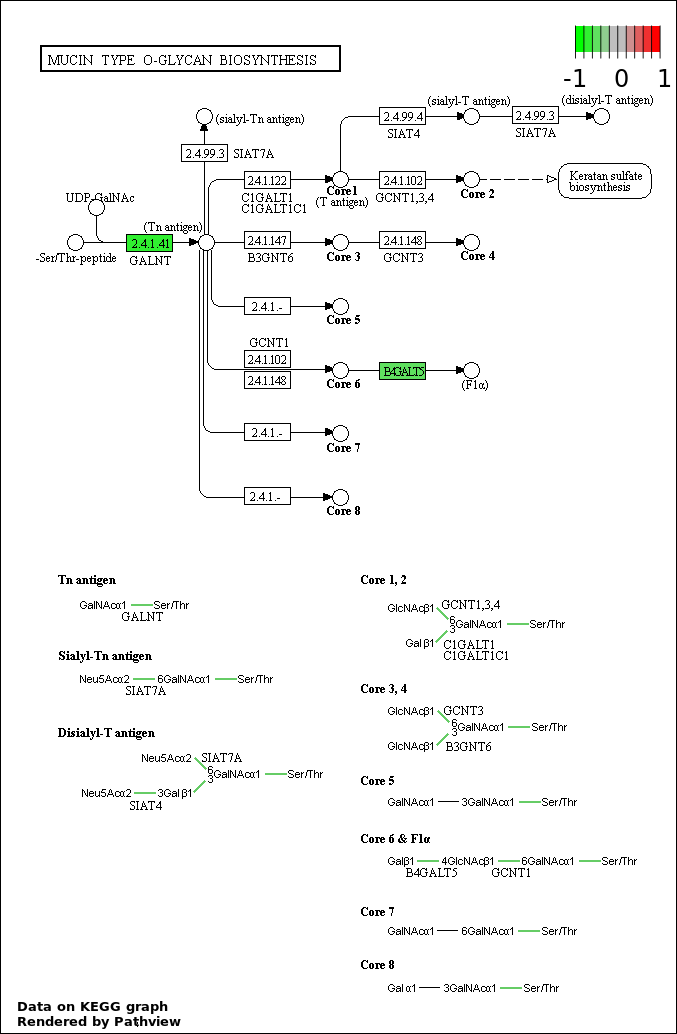
| 7 | hsa00512_Mucin_type_O.Glycan_biosynthesis | 2 | 30 | 0.03603 | 0.888 | |
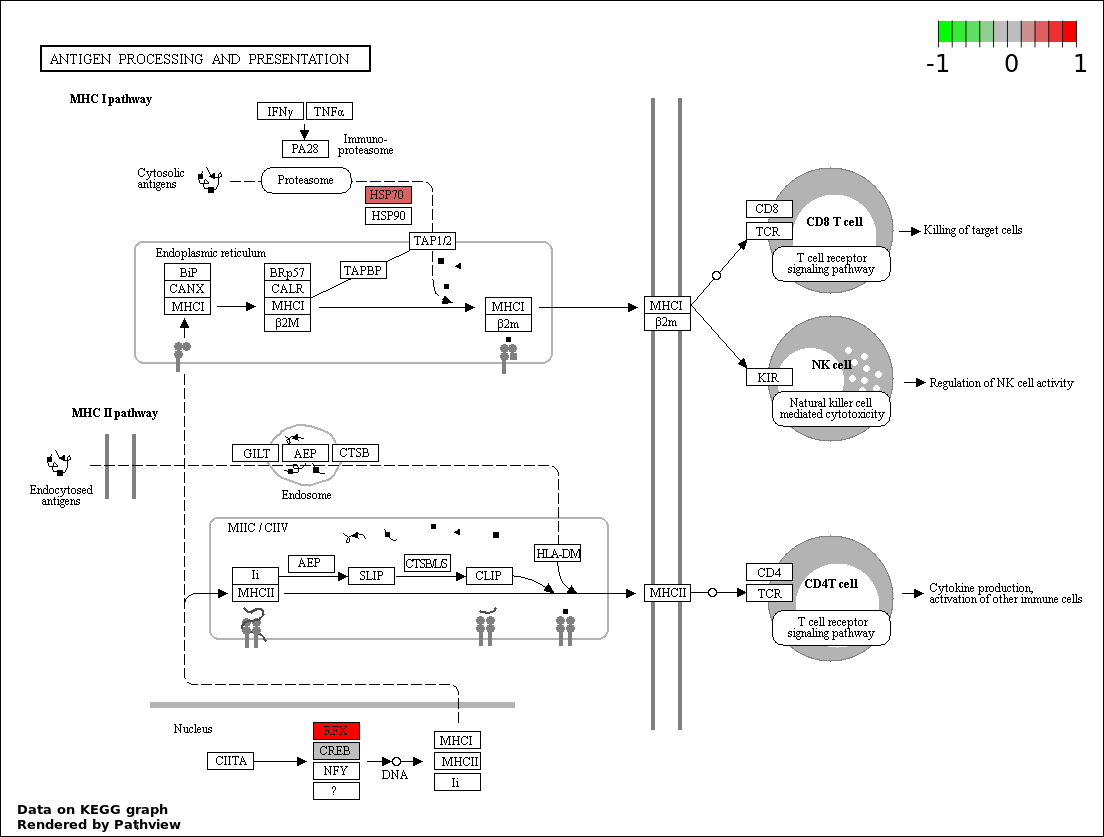
| 8 | hsa04612_Antigen_processing_and_presentation | 3 | 78 | 0.0434 | 0.888 | |
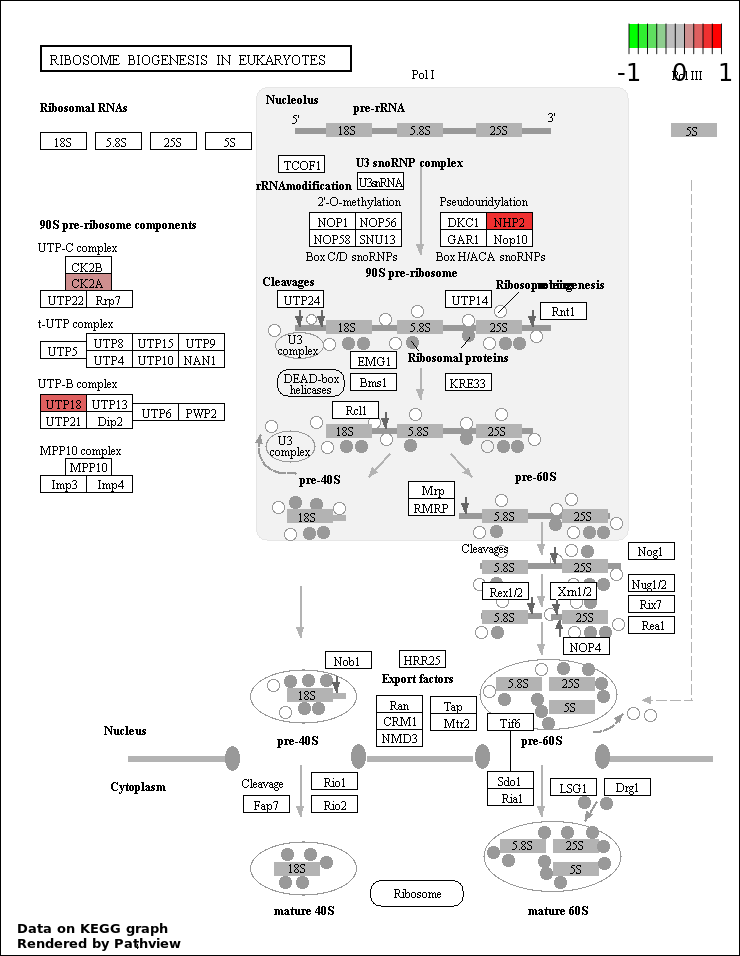
| 9 | hsa03008_Ribosome_biogenesis_in_eukaryotes | 3 | 81 | 0.04764 | 0.888 | |
| 10 | hsa04010_MAPK_signaling_pathway | 6 | 268 | 0.05299 | 0.888 | |
| 11 | hsa04012_ErbB_signaling_pathway | 3 | 87 | 0.05671 | 0.888 | |
| 12 | hsa04810_Regulation_of_actin_cytoskeleton | 5 | 214 | 0.06418 | 0.888 | |
| 13 | hsa04310_Wnt_signaling_pathway | 4 | 151 | 0.06524 | 0.888 | |
| 14 | hsa04390_Hippo_signaling_pathway | 4 | 154 | 0.06907 | 0.888 | |
| 15 | hsa04916_Melanogenesis | 3 | 101 | 0.08075 | 0.969 | |
| 16 | hsa03040_Spliceosome | 3 | 128 | 0.1369 | 1 | |
| 17 | hsa04530_Tight_junction | 3 | 133 | 0.1485 | 1 | |
| 18 | hsa04115_p53_signaling_pathway | 2 | 69 | 0.1516 | 1 | |
| 19 | hsa04720_Long.term_potentiation | 2 | 70 | 0.1551 | 1 | |
| 20 | hsa04730_Long.term_depression | 2 | 70 | 0.1551 | 1 | |
| 21 | hsa03018_RNA_degradation | 2 | 71 | 0.1586 | 1 | |
| 22 | hsa04976_Bile_secretion | 2 | 71 | 0.1586 | 1 | |
| 23 | hsa04910_Insulin_signaling_pathway | 3 | 138 | 0.1604 | 1 | |
| 24 | hsa04260_Cardiac_muscle_contraction | 2 | 77 | 0.1799 | 1 | |
| 25 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 2 | 95 | 0.2457 | 1 | |
| 26 | hsa04912_GnRH_signaling_pathway | 2 | 101 | 0.2679 | 1 | |
| 27 | hsa04151_PI3K_AKT_signaling_pathway | 5 | 351 | 0.2751 | 1 | |
| 28 | hsa04660_T_cell_receptor_signaling_pathway | 2 | 108 | 0.2939 | 1 | |
| 29 | hsa04380_Osteoclast_differentiation | 2 | 128 | 0.3669 | 1 | |
| 30 | hsa04120_Ubiquitin_mediated_proteolysis | 2 | 139 | 0.4058 | 1 | |
| 31 | hsa03013_RNA_transport | 2 | 152 | 0.4504 | 1 | |
| 32 | hsa04630_Jak.STAT_signaling_pathway | 2 | 155 | 0.4604 | 1 | |
| 33 | hsa04141_Protein_processing_in_endoplasmic_reticulum | 2 | 168 | 0.5025 | 1 | |
| 34 | hsa04510_Focal_adhesion | 2 | 200 | 0.5967 | 1 |