This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-let-7d-5p | APP | 1.33 | 0 | -0.4 | 0.04288 | miRNAWalker2 validate | -0.31 | 0 | NA | |
| 2 | hsa-let-7g-3p | APP | 3.26 | 0 | -0.4 | 0.04288 | miRNATAP | -0.19 | 0 | NA | |
| 3 | hsa-miR-101-3p | APP | 0.52 | 0.00376 | -0.4 | 0.04288 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.37 | 0 | NA | |
| 4 | hsa-miR-106a-5p | APP | 2.49 | 0 | -0.4 | 0.04288 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.14 | 0 | NA | |
| 5 | hsa-miR-106b-5p | APP | 2.47 | 0 | -0.4 | 0.04288 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.26 | 0 | NA | |
| 6 | hsa-miR-15a-5p | APP | 2.35 | 0 | -0.4 | 0.04288 | miRNAWalker2 validate; miRNATAP | -0.22 | 0 | NA | |
| 7 | hsa-miR-15b-5p | APP | 1.57 | 0 | -0.4 | 0.04288 | miRNATAP | -0.19 | 1.0E-5 | NA | |
| 8 | hsa-miR-16-5p | APP | 1.76 | 0 | -0.4 | 0.04288 | miRNAWalker2 validate; miRNATAP | -0.18 | 4.0E-5 | NA | |
| 9 | hsa-miR-17-5p | APP | 3.27 | 0 | -0.4 | 0.04288 | miRNAWalker2 validate; miRTarBase; TargetScan; miRNATAP | -0.16 | 0 | NA | |
| 10 | hsa-miR-185-5p | APP | 2.34 | 0 | -0.4 | 0.04288 | miRNATAP | -0.26 | 0 | NA | |
| 11 | hsa-miR-20a-5p | APP | 3.16 | 0 | -0.4 | 0.04288 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.18 | 0 | 20458444 | miR 20a promotes proliferation and invasion by targeting APP in human ovarian cancer cells; Furthermore suppression of APP expression could also promote ovarian cancer cell proliferation and invasion which is consistent with the results of miR-20a overexpression; Therefore we concluded that the regulation of APP is an important mechanism for miR-20a to promote proliferation and invasion in ovarian cancer cells |
| 12 | hsa-miR-324-5p | APP | 2.96 | 0 | -0.4 | 0.04288 | PITA; miRanda; miRNATAP | -0.14 | 0 | NA | |
| 13 | hsa-miR-328-3p | APP | 0.6 | 0.01386 | -0.4 | 0.04288 | miRNAWalker2 validate | -0.14 | 0.00029 | NA | |
| 14 | hsa-miR-361-3p | APP | 1.07 | 0 | -0.4 | 0.04288 | miRNATAP | -0.21 | 8.0E-5 | NA | |
| 15 | hsa-miR-423-3p | APP | 2.58 | 0 | -0.4 | 0.04288 | miRNAWalker2 validate | -0.22 | 0 | NA | |
| 16 | hsa-miR-429 | APP | 4.49 | 0 | -0.4 | 0.04288 | miRanda | -0.12 | 0 | NA | |
| 17 | hsa-miR-484 | APP | 1.82 | 0 | -0.4 | 0.04288 | miRNAWalker2 validate | -0.19 | 0 | NA | |
| 18 | hsa-miR-532-3p | APP | 1.97 | 0 | -0.4 | 0.04288 | miRNAWalker2 validate; PITA | -0.15 | 1.0E-5 | NA | |
| 19 | hsa-miR-582-5p | APP | 0.61 | 0.03299 | -0.4 | 0.04288 | miRNATAP | -0.13 | 6.0E-5 | NA | |
| 20 | hsa-miR-590-3p | APP | 2.59 | 0 | -0.4 | 0.04288 | miRanda | -0.17 | 0 | NA | |
| 21 | hsa-miR-660-5p | APP | 1.83 | 0 | -0.4 | 0.04288 | MirTarget | -0.17 | 0 | NA | |
| 22 | hsa-miR-93-5p | APP | 3.04 | 0 | -0.4 | 0.04288 | miRNATAP | -0.22 | 0 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | REGULATION OF CELL DEATH | 33 | 1472 | 3.804e-10 | 5.9e-07 |
| 2 | NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 27 | 983 | 2.664e-10 | 5.9e-07 |
| 3 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 35 | 1618 | 2.538e-10 | 5.9e-07 |
| 4 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 32 | 1492 | 2.243e-09 | 2.609e-06 |
| 5 | REGULATION OF PROTEIN MODIFICATION PROCESS | 34 | 1710 | 4.341e-09 | 4.04e-06 |
| 6 | RESPONSE TO ORGANIC CYCLIC COMPOUND | 24 | 917 | 7.832e-09 | 4.556e-06 |
| 7 | RESPONSE TO OXYGEN CONTAINING COMPOUND | 30 | 1381 | 5.979e-09 | 4.556e-06 |
| 8 | CELLULAR RESPONSE TO STRESS | 32 | 1565 | 7.162e-09 | 4.556e-06 |
| 9 | NEGATIVE REGULATION OF GENE EXPRESSION | 31 | 1493 | 9.124e-09 | 4.717e-06 |
| 10 | NEGATIVE REGULATION OF NITROGEN COMPOUND METABOLIC PROCESS | 30 | 1517 | 4.998e-08 | 2.114e-05 |
| 11 | RESPONSE TO LIGHT STIMULUS | 13 | 280 | 4.804e-08 | 2.114e-05 |
| 12 | RESPONSE TO ABIOTIC STIMULUS | 24 | 1024 | 6.384e-08 | 2.476e-05 |
| 13 | POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 22 | 876 | 7.411e-08 | 2.652e-05 |
| 14 | CELLULAR RESPONSE TO ORGANIC SUBSTANCE | 33 | 1848 | 1.041e-07 | 3.459e-05 |
| 15 | REGULATION OF CELL DIFFERENTIATION | 29 | 1492 | 1.272e-07 | 3.945e-05 |
| 16 | PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS | 34 | 1977 | 1.557e-07 | 4.528e-05 |
| 17 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 32 | 1791 | 1.693e-07 | 4.608e-05 |
| 18 | RESPONSE TO DRUG | 15 | 431 | 1.882e-07 | 4.608e-05 |
| 19 | CELL DEVELOPMENT | 28 | 1426 | 1.785e-07 | 4.608e-05 |
| 20 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 23 | 1036 | 3.272e-07 | 6.362e-05 |
| 21 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 30 | 1656 | 3.38e-07 | 6.362e-05 |
| 22 | CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND | 20 | 799 | 3.286e-07 | 6.362e-05 |
| 23 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 23 | 1036 | 3.272e-07 | 6.362e-05 |
| 24 | REGULATION OF RESPONSE TO STRESS | 28 | 1468 | 3.232e-07 | 6.362e-05 |
| 25 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 20 | 801 | 3.418e-07 | 6.362e-05 |
| 26 | REPRODUCTION | 26 | 1297 | 3.599e-07 | 6.442e-05 |
| 27 | RESPONSE TO KETONE | 10 | 182 | 3.903e-07 | 6.726e-05 |
| 28 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 30 | 1672 | 4.15e-07 | 6.781e-05 |
| 29 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 24 | 1135 | 4.226e-07 | 6.781e-05 |
| 30 | REGULATION OF CELL PROLIFERATION | 28 | 1496 | 4.739e-07 | 7.349e-05 |
| 31 | CELLULAR RESPONSE TO ORGANIC CYCLIC COMPOUND | 15 | 465 | 4.947e-07 | 7.426e-05 |
| 32 | POSITIVE REGULATION OF CELL DEATH | 17 | 605 | 5.649e-07 | 8.214e-05 |
| 33 | RESPONSE TO ENDOGENOUS STIMULUS | 27 | 1450 | 8.754e-07 | 0.0001234 |
| 34 | REGULATION OF KINASE ACTIVITY | 19 | 776 | 9.297e-07 | 0.0001272 |
| 35 | RESPONSE TO NITROGEN COMPOUND | 20 | 859 | 1.02e-06 | 0.0001356 |
| 36 | REGULATION OF TRANSFERASE ACTIVITY | 21 | 946 | 1.142e-06 | 0.0001475 |
| 37 | CHEMICAL HOMEOSTASIS | 20 | 874 | 1.331e-06 | 0.0001639 |
| 38 | RESPONSE TO FATTY ACID | 7 | 83 | 1.338e-06 | 0.0001639 |
| 39 | REGULATION OF PROTEIN BINDING | 9 | 168 | 1.876e-06 | 0.0002238 |
| 40 | GLUCOSE HOMEOSTASIS | 9 | 170 | 2.069e-06 | 0.0002348 |
| 41 | CARBOHYDRATE HOMEOSTASIS | 9 | 170 | 2.069e-06 | 0.0002348 |
| 42 | CYTOPLASMIC PATTERN RECOGNITION RECEPTOR SIGNALING PATHWAY | 5 | 33 | 2.464e-06 | 0.000273 |
| 43 | POSITIVE REGULATION OF GENE EXPRESSION | 29 | 1733 | 2.771e-06 | 0.0002846 |
| 44 | NEGATIVE REGULATION OF PROTEIN METABOLIC PROCESS | 22 | 1087 | 2.814e-06 | 0.0002846 |
| 45 | ACTIVATION OF PROTEIN KINASE ACTIVITY | 11 | 279 | 2.68e-06 | 0.0002846 |
| 46 | REGULATION OF CELL DEVELOPMENT | 19 | 836 | 2.785e-06 | 0.0002846 |
| 47 | POSITIVE REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 10 | 228 | 3.023e-06 | 0.0002931 |
| 48 | NEGATIVE REGULATION OF CELL DIFFERENTIATION | 16 | 609 | 2.971e-06 | 0.0002931 |
| 49 | REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 14 | 470 | 3.086e-06 | 0.0002931 |
| 50 | POSITIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 11 | 289 | 3.759e-06 | 0.0003498 |
| 51 | RESPONSE TO RADIATION | 13 | 413 | 3.904e-06 | 0.0003561 |
| 52 | POSITIVE REGULATION OF KINASE ACTIVITY | 14 | 482 | 4.126e-06 | 0.0003623 |
| 53 | SMALL MOLECULE METABOLIC PROCESS | 29 | 1767 | 4.068e-06 | 0.0003623 |
| 54 | SYNAPTIC SIGNALING | 13 | 424 | 5.19e-06 | 0.0004472 |
| 55 | ORGANONITROGEN COMPOUND METABOLIC PROCESS | 29 | 1796 | 5.593e-06 | 0.0004732 |
| 56 | PHOSPHORYLATION | 23 | 1228 | 5.897e-06 | 0.00049 |
| 57 | REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 12 | 365 | 6.034e-06 | 0.0004926 |
| 58 | PROTEIN LOCALIZATION | 29 | 1805 | 6.164e-06 | 0.0004945 |
| 59 | CELLULAR RESPONSE TO NITROGEN COMPOUND | 14 | 505 | 7.026e-06 | 0.0005541 |
| 60 | REGULATION OF CATABOLIC PROCESS | 17 | 731 | 7.241e-06 | 0.0005615 |
| 61 | HOMEOSTATIC PROCESS | 24 | 1337 | 7.368e-06 | 0.000562 |
| 62 | POSITIVE REGULATION OF CELL COMMUNICATION | 26 | 1532 | 7.872e-06 | 0.0005908 |
| 63 | REGULATION OF MAPK CASCADE | 16 | 660 | 8.21e-06 | 0.0006063 |
| 64 | NEGATIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 17 | 740 | 8.497e-06 | 0.0006177 |
| 65 | REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 17 | 750 | 1.012e-05 | 0.0007243 |
| 66 | DEVELOPMENTAL PROCESS INVOLVED IN REPRODUCTION | 15 | 602 | 1.163e-05 | 0.0008196 |
| 67 | MULTI MULTICELLULAR ORGANISM PROCESS | 9 | 213 | 1.294e-05 | 0.000899 |
| 68 | GUANOSINE CONTAINING COMPOUND METABOLIC PROCESS | 5 | 46 | 1.328e-05 | 0.0009088 |
| 69 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 24 | 1395 | 1.495e-05 | 0.001008 |
| 70 | POSITIVE REGULATION OF TRANSFERASE ACTIVITY | 15 | 616 | 1.523e-05 | 0.001013 |
| 71 | POSITIVE REGULATION OF MAPK CASCADE | 13 | 470 | 1.557e-05 | 0.001015 |
| 72 | PROTEIN PHOSPHORYLATION | 19 | 944 | 1.57e-05 | 0.001015 |
| 73 | INTRACELLULAR RECEPTOR SIGNALING PATHWAY | 8 | 168 | 1.687e-05 | 0.001075 |
| 74 | REGULATION OF NEURON PROJECTION DEVELOPMENT | 12 | 408 | 1.839e-05 | 0.001156 |
| 75 | NEGATIVE REGULATION OF CELL DEATH | 18 | 872 | 1.923e-05 | 0.001193 |
| 76 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 25 | 1518 | 2.023e-05 | 0.001223 |
| 77 | REGULATION OF BINDING | 10 | 283 | 2.006e-05 | 0.001223 |
| 78 | RESPONSE TO EXTERNAL STIMULUS | 28 | 1821 | 2.082e-05 | 0.001242 |
| 79 | REGULATION OF EPITHELIAL CELL PROLIFERATION | 10 | 285 | 2.131e-05 | 0.001255 |
| 80 | CELLULAR RESPONSE TO FATTY ACID | 5 | 51 | 2.216e-05 | 0.001289 |
| 81 | RESPONSE TO LIPID | 18 | 888 | 2.446e-05 | 0.001405 |
| 82 | NEGATIVE REGULATION OF CELL PROLIFERATION | 15 | 643 | 2.509e-05 | 0.001424 |
| 83 | MULTI ORGANISM REPRODUCTIVE PROCESS | 18 | 891 | 2.558e-05 | 0.001434 |
| 84 | MOTOR NEURON AXON GUIDANCE | 4 | 27 | 2.913e-05 | 0.001614 |
| 85 | NEGATIVE REGULATION OF MOLECULAR FUNCTION | 20 | 1079 | 3e-05 | 0.001641 |
| 86 | NEGATIVE REGULATION OF RESPONSE TO STIMULUS | 23 | 1360 | 3.033e-05 | 0.001641 |
| 87 | NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR SIGNALING PATHWAY | 4 | 28 | 3.381e-05 | 0.001808 |
| 88 | INTERSPECIES INTERACTION BETWEEN ORGANISMS | 15 | 662 | 3.507e-05 | 0.001833 |
| 89 | SYMBIOSIS ENCOMPASSING MUTUALISM THROUGH PARASITISM | 15 | 662 | 3.507e-05 | 0.001833 |
| 90 | NEGATIVE REGULATION OF CELL DEVELOPMENT | 10 | 303 | 3.589e-05 | 0.001835 |
| 91 | INTRACELLULAR SIGNAL TRANSDUCTION | 25 | 1572 | 3.624e-05 | 0.001835 |
| 92 | NEGATIVE REGULATION OF CATALYTIC ACTIVITY | 17 | 829 | 3.629e-05 | 0.001835 |
| 93 | MULTI ORGANISM METABOLIC PROCESS | 7 | 138 | 3.859e-05 | 0.001912 |
| 94 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 19 | 1008 | 3.863e-05 | 0.001912 |
| 95 | TRANSFORMING GROWTH FACTOR BETA RECEPTOR SIGNALING PATHWAY | 6 | 95 | 4.134e-05 | 0.002025 |
| 96 | PEPTIDYL AMINO ACID MODIFICATION | 17 | 841 | 4.342e-05 | 0.002104 |
| 97 | REGULATION OF NEURON APOPTOTIC PROCESS | 8 | 192 | 4.391e-05 | 0.002106 |
| 98 | MYD88 INDEPENDENT TOLL LIKE RECEPTOR SIGNALING PATHWAY | 4 | 30 | 4.479e-05 | 0.002127 |
| 99 | PROTEIN HOMOTETRAMERIZATION | 5 | 59 | 4.526e-05 | 0.002127 |
| 100 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 27 | 1805 | 4.876e-05 | 0.002246 |
| 101 | REGULATION OF NEURON DEATH | 9 | 252 | 4.858e-05 | 0.002246 |
| 102 | RESPONSE TO TRANSFORMING GROWTH FACTOR BETA | 7 | 144 | 5.062e-05 | 0.002287 |
| 103 | REGULATION OF RESPONSE TO CYTOKINE STIMULUS | 7 | 144 | 5.062e-05 | 0.002287 |
| 104 | CELL CELL SIGNALING | 16 | 767 | 5.113e-05 | 0.002288 |
| 105 | SINGLE ORGANISM BEHAVIOR | 11 | 384 | 5.275e-05 | 0.002316 |
| 106 | RESPIRATORY SYSTEM DEVELOPMENT | 8 | 197 | 5.266e-05 | 0.002316 |
| 107 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 28 | 1929 | 5.849e-05 | 0.00252 |
| 108 | RESPONSE TO MOLECULE OF BACTERIAL ORIGIN | 10 | 321 | 5.828e-05 | 0.00252 |
| 109 | NUCLEOBASE CONTAINING SMALL MOLECULE METABOLIC PROCESS | 13 | 535 | 5.942e-05 | 0.002536 |
| 110 | ESTABLISHMENT OF PROTEIN LOCALIZATION | 23 | 1423 | 6.108e-05 | 0.002561 |
| 111 | NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL METABOLIC PROCESS | 3 | 12 | 6.054e-05 | 0.002561 |
| 112 | NEGATIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 9 | 262 | 6.559e-05 | 0.002677 |
| 113 | REGULATION OF PROTEIN CATABOLIC PROCESS | 11 | 393 | 6.493e-05 | 0.002677 |
| 114 | STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE | 6 | 103 | 6.512e-05 | 0.002677 |
| 115 | POSITIVE REGULATION OF MAP KINASE ACTIVITY | 8 | 207 | 7.456e-05 | 0.003017 |
| 116 | INNATE IMMUNE RESPONSE ACTIVATING CELL SURFACE RECEPTOR SIGNALING PATHWAY | 6 | 106 | 7.644e-05 | 0.003066 |
| 117 | REGULATION OF PROTEOLYSIS | 15 | 711 | 7.858e-05 | 0.003125 |
| 118 | ORGANOPHOSPHATE METABOLIC PROCESS | 17 | 885 | 8.137e-05 | 0.003186 |
| 119 | TUBE DEVELOPMENT | 13 | 552 | 8.147e-05 | 0.003186 |
| 120 | POSITIVE REGULATION OF NEURON DEATH | 5 | 67 | 8.368e-05 | 0.003218 |
| 121 | RESPONSE TO MONOAMINE | 4 | 35 | 8.34e-05 | 0.003218 |
| 122 | REGULATION OF NEURON DIFFERENTIATION | 13 | 554 | 8.448e-05 | 0.003222 |
| 123 | REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 10 | 337 | 8.726e-05 | 0.003274 |
| 124 | RESPONSE TO INORGANIC SUBSTANCE | 12 | 479 | 8.67e-05 | 0.003274 |
| 125 | REGULATION OF CELL PROJECTION ORGANIZATION | 13 | 558 | 9.079e-05 | 0.003353 |
| 126 | RESPONSE TO HORMONE | 17 | 893 | 9.078e-05 | 0.003353 |
| 127 | REGULATION OF OSTEOBLAST DIFFERENTIATION | 6 | 112 | 0.0001038 | 0.003802 |
| 128 | NOTCH SIGNALING PATHWAY | 6 | 114 | 0.0001144 | 0.00416 |
| 129 | RESPONSE TO TESTOSTERONE | 4 | 38 | 0.0001158 | 0.004176 |
| 130 | NEGATIVE REGULATION OF CELL COMMUNICATION | 20 | 1192 | 0.0001189 | 0.004256 |
| 131 | NEGATIVE REGULATION OF TELOMERASE ACTIVITY | 3 | 15 | 0.0001234 | 0.004284 |
| 132 | ECTODERMAL PLACODE DEVELOPMENT | 3 | 15 | 0.0001234 | 0.004284 |
| 133 | ECTODERMAL PLACODE MORPHOGENESIS | 3 | 15 | 0.0001234 | 0.004284 |
| 134 | ECTODERMAL PLACODE FORMATION | 3 | 15 | 0.0001234 | 0.004284 |
| 135 | CELLULAR RESPONSE TO KETONE | 5 | 73 | 0.0001261 | 0.004345 |
| 136 | ERBB2 SIGNALING PATHWAY | 4 | 39 | 0.0001283 | 0.004391 |
| 137 | REGULATION OF AXONOGENESIS | 7 | 168 | 0.0001334 | 0.00453 |
| 138 | NEUROGENESIS | 22 | 1402 | 0.0001386 | 0.004672 |
| 139 | SECRETION | 13 | 588 | 0.0001526 | 0.005036 |
| 140 | GUANOSINE CONTAINING COMPOUND BIOSYNTHETIC PROCESS | 3 | 16 | 0.0001511 | 0.005036 |
| 141 | ORGANONITROGEN COMPOUND BIOSYNTHETIC PROCESS | 18 | 1024 | 0.0001518 | 0.005036 |
| 142 | REGULATION OF DEFENSE RESPONSE | 15 | 759 | 0.0001613 | 0.005287 |
| 143 | PROTEIN LOCALIZATION TO ENDOPLASMIC RETICULUM | 6 | 123 | 0.0001737 | 0.005651 |
| 144 | SYNAPTIC TRANSMISSION DOPAMINERGIC | 3 | 17 | 0.0001826 | 0.005883 |
| 145 | MULTICELLULAR ORGANISM REPRODUCTION | 15 | 768 | 0.0001833 | 0.005883 |
| 146 | NEGATIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 14 | 684 | 0.0001884 | 0.006004 |
| 147 | RESPONSE TO UV | 6 | 126 | 0.0001981 | 0.006269 |
| 148 | REGULATION OF CELLULAR RESPONSE TO STRESS | 14 | 691 | 0.0002092 | 0.006576 |
| 149 | PROTEIN LOCALIZATION TO MEMBRANE | 10 | 376 | 0.0002127 | 0.006643 |
| 150 | RESPONSE TO BACTERIUM | 12 | 528 | 0.0002146 | 0.006658 |
| 151 | JNK CASCADE | 5 | 82 | 0.0002183 | 0.006728 |
| 152 | REGULATION OF ERBB SIGNALING PATHWAY | 5 | 83 | 0.0002311 | 0.007075 |
| 153 | NEGATIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 13 | 616 | 0.0002402 | 0.007304 |
| 154 | CELLULAR RESPONSE TO LIPID | 11 | 457 | 0.0002432 | 0.007349 |
| 155 | POSITIVE REGULATION OF NF KAPPAB TRANSCRIPTION FACTOR ACTIVITY | 6 | 132 | 0.0002549 | 0.007651 |
| 156 | TOLL LIKE RECEPTOR SIGNALING PATHWAY | 5 | 85 | 0.0002583 | 0.007706 |
| 157 | ESTABLISHMENT OF LOCALIZATION IN CELL | 24 | 1676 | 0.0002661 | 0.007866 |
| 158 | POSITIVE REGULATION OF NEURON APOPTOTIC PROCESS | 4 | 47 | 0.0002671 | 0.007866 |
| 159 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 25 | 1784 | 0.0002706 | 0.00792 |
| 160 | NEURON PROJECTION DEVELOPMENT | 12 | 545 | 0.0002865 | 0.00822 |
| 161 | HEART DEVELOPMENT | 11 | 466 | 0.0002873 | 0.00822 |
| 162 | REGULATION OF MAP KINASE ACTIVITY | 9 | 319 | 0.000288 | 0.00822 |
| 163 | PROTEIN TETRAMERIZATION | 6 | 135 | 0.0002877 | 0.00822 |
| 164 | REGULATION OF CELLULAR LOCALIZATION | 20 | 1277 | 0.000296 | 0.008364 |
| 165 | REGULATION OF PEPTIDASE ACTIVITY | 10 | 392 | 0.0002966 | 0.008364 |
| 166 | PROTEIN AUTOPHOSPHORYLATION | 7 | 192 | 0.0003026 | 0.008482 |
| 167 | REGULATION OF GROWTH | 13 | 633 | 0.000312 | 0.008524 |
| 168 | RESPONSE TO ALKALOID | 6 | 137 | 0.0003114 | 0.008524 |
| 169 | PURINE NUCLEOSIDE BIOSYNTHETIC PROCESS | 5 | 89 | 0.0003201 | 0.008524 |
| 170 | REGULATION OF TRANSPORT | 25 | 1804 | 0.0003206 | 0.008524 |
| 171 | PURINE CONTAINING COMPOUND METABOLIC PROCESS | 10 | 394 | 0.0003088 | 0.008524 |
| 172 | GLAND DEVELOPMENT | 10 | 395 | 0.000315 | 0.008524 |
| 173 | ACTIVATION OF MAPK ACTIVITY | 6 | 137 | 0.0003114 | 0.008524 |
| 174 | PURINE RIBONUCLEOSIDE BIOSYNTHETIC PROCESS | 5 | 89 | 0.0003201 | 0.008524 |
| 175 | CELLULAR RESPONSE TO DNA DAMAGE STIMULUS | 14 | 720 | 0.0003176 | 0.008524 |
| 176 | PURINE CONTAINING COMPOUND BIOSYNTHETIC PROCESS | 6 | 139 | 0.0003366 | 0.008899 |
| 177 | CELL DEATH | 17 | 1001 | 0.0003503 | 0.009209 |
| 178 | NEGATIVE REGULATION OF PROTEOLYSIS | 9 | 329 | 0.0003609 | 0.009433 |
| 179 | RESPONSE TO AMMONIUM ION | 4 | 51 | 0.0003666 | 0.00953 |
| 180 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 15 | 823 | 0.0003832 | 0.009906 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | RECEPTOR BINDING | 28 | 1476 | 3.609e-07 | 0.0003353 |
| 2 | ENZYME BINDING | 30 | 1737 | 9.281e-07 | 0.0004311 |
| 3 | TRANSCRIPTION FACTOR BINDING | 15 | 524 | 2.188e-06 | 0.0006777 |
| 4 | KINASE ACTIVITY | 18 | 842 | 1.203e-05 | 0.002795 |
| 5 | IDENTICAL PROTEIN BINDING | 22 | 1209 | 1.519e-05 | 0.002822 |
| 6 | TRANSFERASE ACTIVITY TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 19 | 992 | 3.109e-05 | 0.004814 |
| 7 | KINASE BINDING | 14 | 606 | 5.271e-05 | 0.006512 |
| 8 | ADENYL NUCLEOTIDE BINDING | 24 | 1514 | 5.608e-05 | 0.006512 |
| 9 | BETA AMYLOID BINDING | 4 | 35 | 8.34e-05 | 0.008609 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | NEURON PROJECTION | 23 | 942 | 5.994e-08 | 3.501e-05 |
| 2 | NEURON PART | 25 | 1265 | 8.175e-07 | 0.0002387 |
| 3 | AXON | 13 | 418 | 4.448e-06 | 0.000866 |
| 4 | SOMATODENDRITIC COMPARTMENT | 16 | 650 | 6.782e-06 | 0.0009902 |
| 5 | CELL PROJECTION | 28 | 1786 | 1.458e-05 | 0.001703 |
| 6 | PERINUCLEAR REGION OF CYTOPLASM | 15 | 642 | 2.464e-05 | 0.001914 |
| 7 | CELL BODY | 13 | 494 | 2.621e-05 | 0.001914 |
| 8 | CELL LEADING EDGE | 11 | 350 | 2.27e-05 | 0.001914 |
| 9 | CELL PROJECTION PART | 18 | 946 | 5.58e-05 | 0.003193 |
| 10 | MEMBRANE REGION | 20 | 1134 | 6.015e-05 | 0.003193 |
| 11 | SYNAPSE PART | 14 | 610 | 5.657e-05 | 0.003193 |
| 12 | MITOCHONDRION | 25 | 1633 | 6.748e-05 | 0.003284 |
| 13 | PERIKARYON | 6 | 108 | 8.482e-05 | 0.00381 |
| 14 | CYTOPLASMIC SIDE OF MEMBRANE | 7 | 170 | 0.0001435 | 0.005988 |
| 15 | POSTSYNAPSE | 10 | 378 | 0.000222 | 0.008102 |
| 16 | DENDRITE | 11 | 451 | 0.0002172 | 0.008102 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
| 1 | hsa03010_Ribosome | 5 | 92 | 0.0003732 | 0.06718 | |
| 2 | hsa00230_Purine_metabolism | 6 | 162 | 0.0007569 | 0.06812 | |
| 3 | hsa04912_GnRH_signaling_pathway | 4 | 101 | 0.004641 | 0.2784 | |
| 4 | hsa04151_PI3K_AKT_signaling_pathway | 7 | 351 | 0.00914 | 0.3652 | |
| 5 | hsa04730_Long.term_depression | 3 | 70 | 0.01138 | 0.3652 | |
| 6 | hsa04662_B_cell_receptor_signaling_pathway | 3 | 75 | 0.01371 | 0.3652 | |
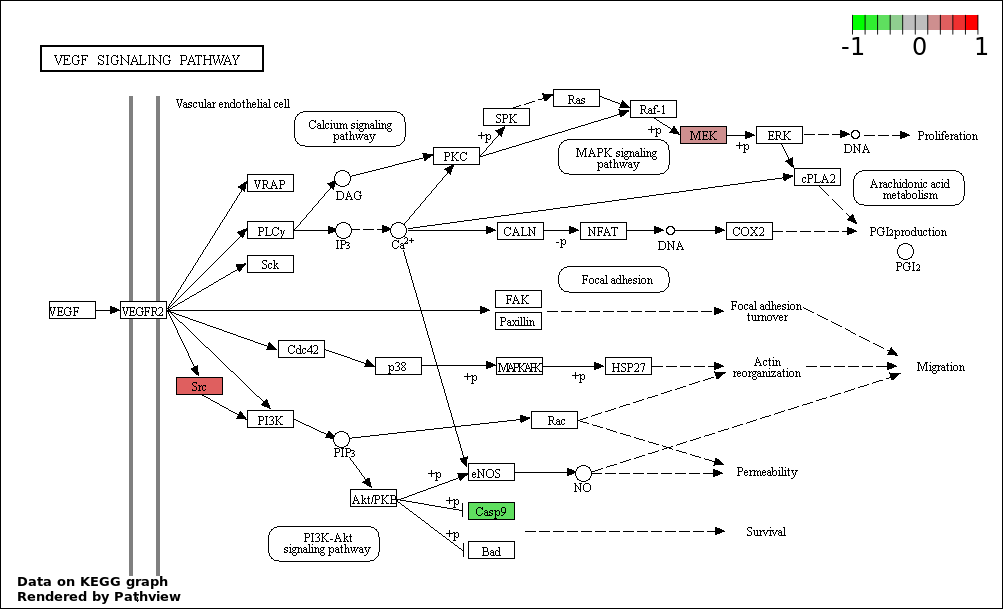
| 7 | hsa04370_VEGF_signaling_pathway | 3 | 76 | 0.0142 | 0.3652 | |
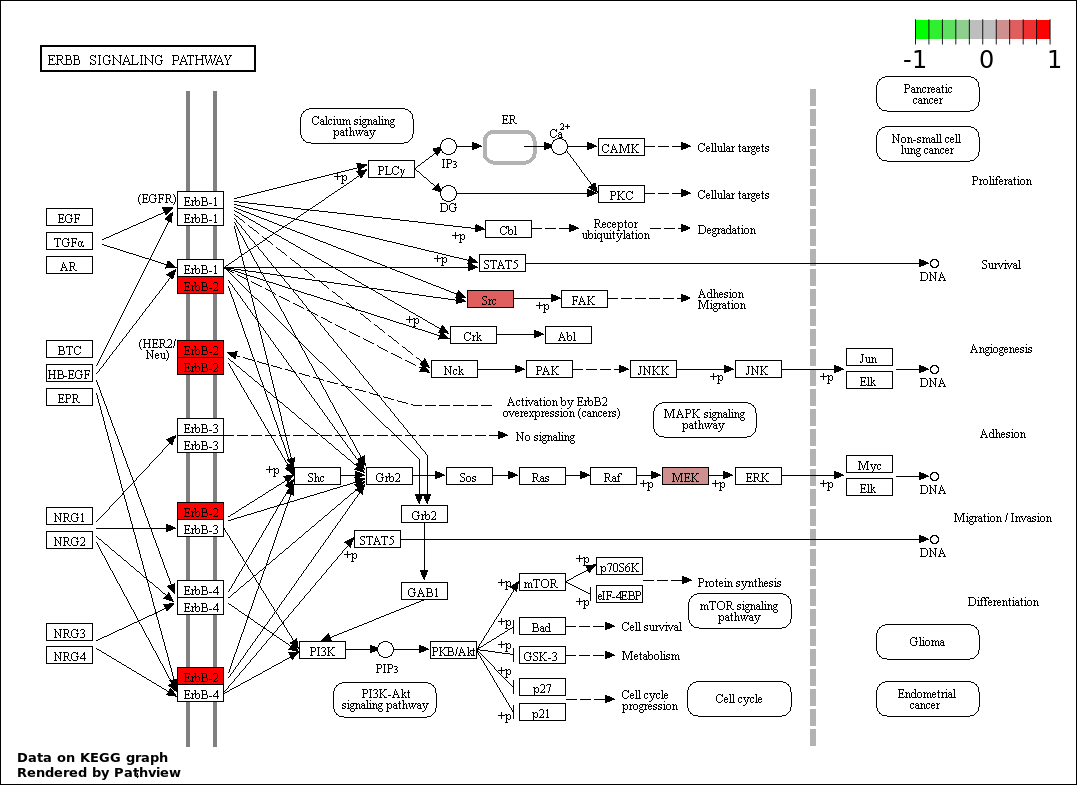
| 8 | hsa04012_ErbB_signaling_pathway | 3 | 87 | 0.02032 | 0.3951 | |
| 9 | hsa03410_Base_excision_repair | 2 | 34 | 0.02143 | 0.3951 | |
| 10 | hsa04540_Gap_junction | 3 | 90 | 0.0222 | 0.3951 | |
| 11 | hsa04620_Toll.like_receptor_signaling_pathway | 3 | 102 | 0.03065 | 0.4244 | |
| 12 | hsa04010_MAPK_signaling_pathway | 5 | 268 | 0.03356 | 0.425 | |
| 13 | hsa04660_T_cell_receptor_signaling_pathway | 3 | 108 | 0.03542 | 0.425 | |
| 14 | hsa00480_Glutathione_metabolism | 2 | 50 | 0.04369 | 0.4689 | |
| 15 | hsa04144_Endocytosis | 4 | 203 | 0.04677 | 0.4689 | |
| 16 | hsa04150_mTOR_signaling_pathway | 2 | 52 | 0.04689 | 0.4689 | |
| 17 | hsa04623_Cytosolic_DNA.sensing_pathway | 2 | 56 | 0.05354 | 0.4796 | |
| 18 | hsa04380_Osteoclast_differentiation | 3 | 128 | 0.0539 | 0.4796 | |
| 19 | hsa04360_Axon_guidance | 3 | 130 | 0.05596 | 0.4796 | |
| 20 | hsa04530_Tight_junction | 3 | 133 | 0.05912 | 0.4837 | |
| 21 | hsa04910_Insulin_signaling_pathway | 3 | 138 | 0.06456 | 0.5053 | |
| 22 | hsa04920_Adipocytokine_signaling_pathway | 2 | 68 | 0.07527 | 0.5558 | |
| 23 | hsa04115_p53_signaling_pathway | 2 | 69 | 0.07719 | 0.5558 | |
| 24 | hsa04622_RIG.I.like_receptor_signaling_pathway | 2 | 71 | 0.08108 | 0.5613 | |
| 25 | hsa04520_Adherens_junction | 2 | 73 | 0.08502 | 0.5668 | |
| 26 | hsa03015_mRNA_surveillance_pathway | 2 | 83 | 0.1056 | 0.6229 | |
| 27 | hsa04210_Apoptosis | 2 | 89 | 0.1185 | 0.6462 | |
| 28 | hsa04970_Salivary_secretion | 2 | 89 | 0.1185 | 0.6462 | |
| 29 | hsa00240_Pyrimidine_metabolism | 2 | 99 | 0.1408 | 0.6885 | |
| 30 | hsa04916_Melanogenesis | 2 | 101 | 0.1453 | 0.6885 | |
| 31 | hsa04972_Pancreatic_secretion | 2 | 101 | 0.1453 | 0.6885 | |
| 32 | hsa04270_Vascular_smooth_muscle_contraction | 2 | 116 | 0.1804 | 0.778 | |
| 33 | hsa04722_Neurotrophin_signaling_pathway | 2 | 127 | 0.2069 | 0.8192 | |
| 34 | hsa03040_Spliceosome | 2 | 128 | 0.2094 | 0.8192 | |
| 35 | hsa04310_Wnt_signaling_pathway | 2 | 151 | 0.2658 | 0.9105 | |
| 36 | hsa04390_Hippo_signaling_pathway | 2 | 154 | 0.2732 | 0.9105 | |
| 37 | hsa04141_Protein_processing_in_endoplasmic_reticulum | 2 | 168 | 0.3076 | 0.9384 | |
| 38 | hsa04020_Calcium_signaling_pathway | 2 | 177 | 0.3296 | 0.9416 | |
| 39 | hsa04510_Focal_adhesion | 2 | 200 | 0.3847 | 1 | |
| 40 | hsa04014_Ras_signaling_pathway | 2 | 236 | 0.4668 | 1 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | RP3-439F8.1 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-328-3p;hsa-miR-423-3p;hsa-miR-484;hsa-miR-93-5p | 14 | APP | Sponge network | -0.149 | 0.78031 | -0.401 | 0.04288 | 0.44 |
| 2 | LINC00883 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-423-3p;hsa-miR-484;hsa-miR-582-5p | 12 | APP | Sponge network | -0.614 | 0.0511 | -0.401 | 0.04288 | 0.424 |
| 3 | RP11-426C22.4 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-361-3p | 12 | APP | Sponge network | -0.045 | 0.93351 | -0.401 | 0.04288 | 0.404 |
| 4 | RP11-967K21.1 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-429;hsa-miR-93-5p | 10 | APP | Sponge network | -1.206 | 0.01654 | -0.401 | 0.04288 | 0.4 |
| 5 | LINC00702 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-328-3p;hsa-miR-423-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-93-5p | 19 | APP | Sponge network | -2.704 | 0 | -0.401 | 0.04288 | 0.399 |
| 6 | RP11-517P14.2 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | APP | Sponge network | -0.825 | 0.01106 | -0.401 | 0.04288 | 0.383 |
| 7 | LINC00163 | hsa-let-7d-5p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-328-3p;hsa-miR-423-3p;hsa-miR-484 | 14 | APP | Sponge network | -4.312 | 0.00014 | -0.401 | 0.04288 | 0.378 |
| 8 | GAS6-AS2 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-423-3p;hsa-miR-429 | 13 | APP | Sponge network | -2.655 | 0 | -0.401 | 0.04288 | 0.372 |
| 9 | MAGI2-AS3 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-423-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-93-5p | 19 | APP | Sponge network | -2.414 | 0 | -0.401 | 0.04288 | 0.36 |
| 10 | HCG11 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | APP | Sponge network | -1.194 | 0.00331 | -0.401 | 0.04288 | 0.356 |
| 11 | RP11-399O19.9 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-93-5p | 14 | APP | Sponge network | -0.911 | 0.02612 | -0.401 | 0.04288 | 0.353 |
| 12 | RP11-835E18.5 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-324-5p | 10 | APP | Sponge network | -0.943 | 0.00219 | -0.401 | 0.04288 | 0.353 |
| 13 | PDZRN3-AS1 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p | 15 | APP | Sponge network | -5.049 | 1.0E-5 | -0.401 | 0.04288 | 0.344 |
| 14 | RP11-166D19.1 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-328-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p | 17 | APP | Sponge network | -3.855 | 0 | -0.401 | 0.04288 | 0.34 |
| 15 | AC020571.3 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-423-3p;hsa-miR-582-5p | 14 | APP | Sponge network | -0.862 | 0.18422 | -0.401 | 0.04288 | 0.334 |
| 16 | DNM3OS | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-328-3p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | APP | Sponge network | -2.298 | 1.0E-5 | -0.401 | 0.04288 | 0.327 |
| 17 | RP11-6O2.3 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-328-3p;hsa-miR-361-3p;hsa-miR-660-5p | 14 | APP | Sponge network | -4.533 | 0 | -0.401 | 0.04288 | 0.318 |
| 18 | NR2F1-AS1 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-328-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | APP | Sponge network | -1.881 | 0 | -0.401 | 0.04288 | 0.317 |
| 19 | RP11-532F6.3 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-423-3p;hsa-miR-590-3p | 13 | APP | Sponge network | -1.772 | 1.0E-5 | -0.401 | 0.04288 | 0.315 |
| 20 | CTD-2013N24.2 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-93-5p | 16 | APP | Sponge network | -1.002 | 1.0E-5 | -0.401 | 0.04288 | 0.307 |
| 21 | RP11-356J5.12 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-532-3p;hsa-miR-590-3p | 15 | APP | Sponge network | -2.015 | 0 | -0.401 | 0.04288 | 0.305 |
| 22 | TBX5-AS1 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-328-3p;hsa-miR-532-3p | 14 | APP | Sponge network | -2.557 | 2.0E-5 | -0.401 | 0.04288 | 0.303 |
| 23 | RP11-1223D19.1 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-423-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-93-5p | 14 | APP | Sponge network | -1.23 | 0.05444 | -0.401 | 0.04288 | 0.3 |
| 24 | RP11-359E10.1 | hsa-let-7d-5p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-484;hsa-miR-582-5p;hsa-miR-93-5p | 13 | APP | Sponge network | -1.216 | 0.0438 | -0.401 | 0.04288 | 0.298 |
| 25 | RP4-798P15.3 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-423-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-660-5p | 16 | APP | Sponge network | -0.958 | 0.06952 | -0.401 | 0.04288 | 0.296 |
| 26 | VIM-AS1 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-93-5p | 11 | APP | Sponge network | -1.424 | 0.00627 | -0.401 | 0.04288 | 0.272 |
| 27 | RP11-456K23.1 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p;hsa-miR-93-5p | 16 | APP | Sponge network | -1.962 | 1.0E-5 | -0.401 | 0.04288 | 0.271 |
| 28 | MIR143HG | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-328-3p;hsa-miR-361-3p;hsa-miR-423-3p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-93-5p | 22 | APP | Sponge network | -4.237 | 0 | -0.401 | 0.04288 | 0.268 |
| 29 | RASSF8-AS1 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | APP | Sponge network | -0.877 | 0.00508 | -0.401 | 0.04288 | 0.267 |
| 30 | RP11-426C22.5 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-532-3p;hsa-miR-582-5p;hsa-miR-660-5p | 13 | APP | Sponge network | -0.559 | 0.08048 | -0.401 | 0.04288 | 0.264 |
| 31 | AC016747.3 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-328-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-93-5p | 15 | APP | Sponge network | 0.026 | 0.94846 | -0.401 | 0.04288 | 0.264 |
| 32 | ZNF667-AS1 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-361-3p;hsa-miR-423-3p;hsa-miR-484;hsa-miR-93-5p | 14 | APP | Sponge network | -1.846 | 0.00013 | -0.401 | 0.04288 | 0.258 |
| 33 | RP11-554A11.4 | hsa-let-7d-5p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p | 10 | APP | Sponge network | -3.989 | 0 | -0.401 | 0.04288 | 0.253 |
| 34 | LINC00565 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-484 | 11 | APP | Sponge network | -1.493 | 0.05998 | -0.401 | 0.04288 | 0.253 |
| 35 | RP11-389G6.3 | hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | APP | Sponge network | -7.573 | 0 | -0.401 | 0.04288 | 0.251 |