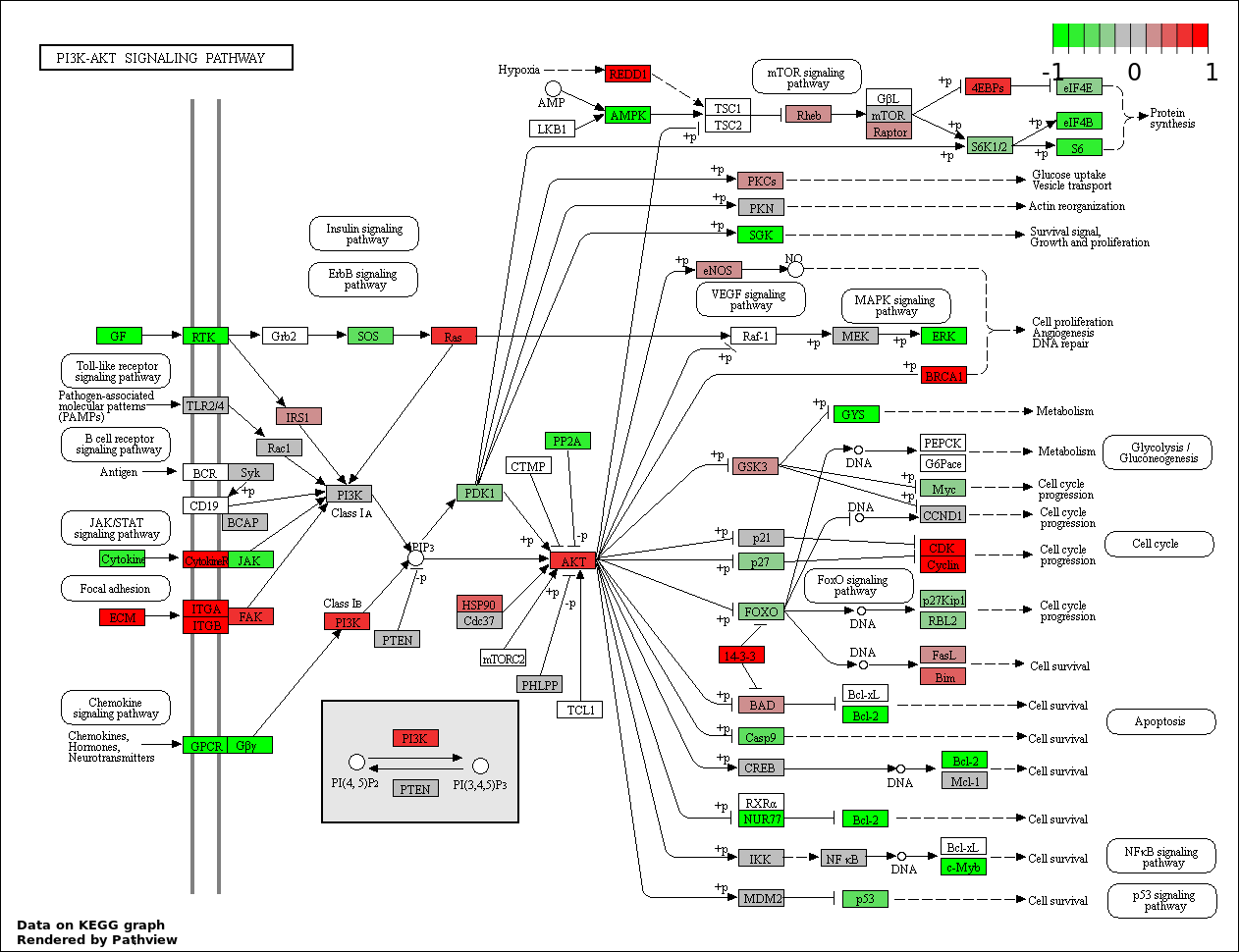
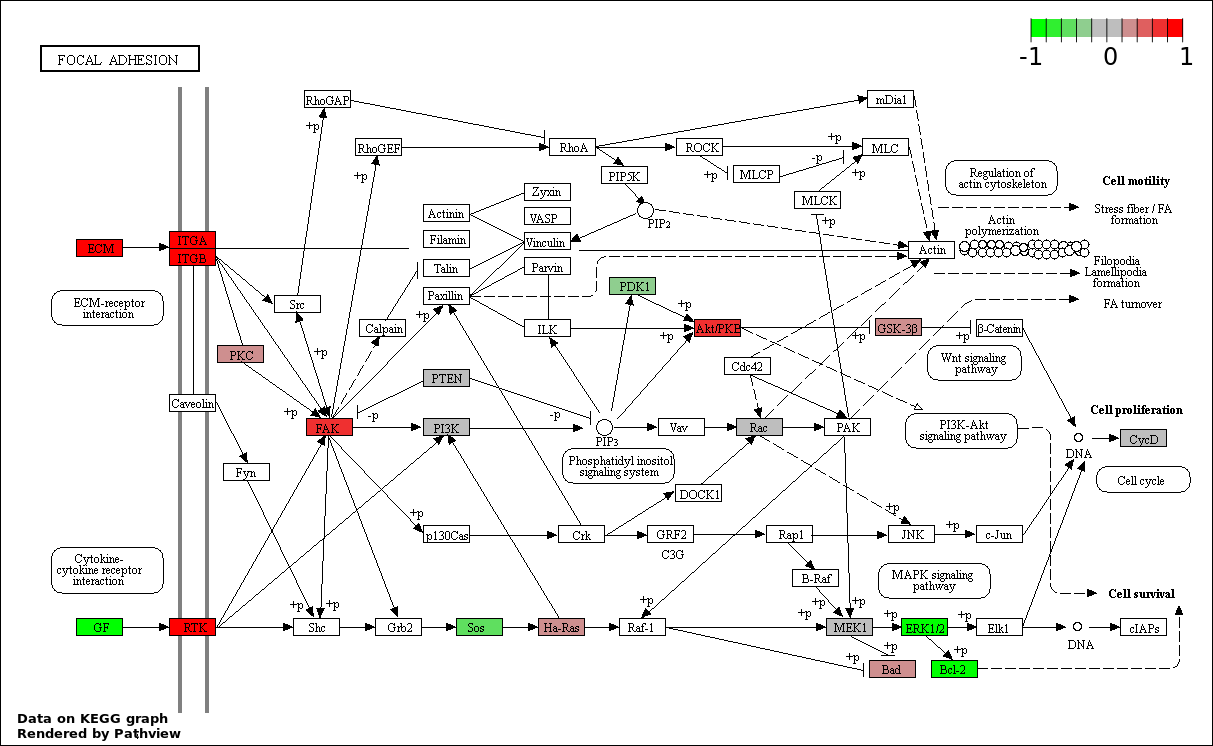
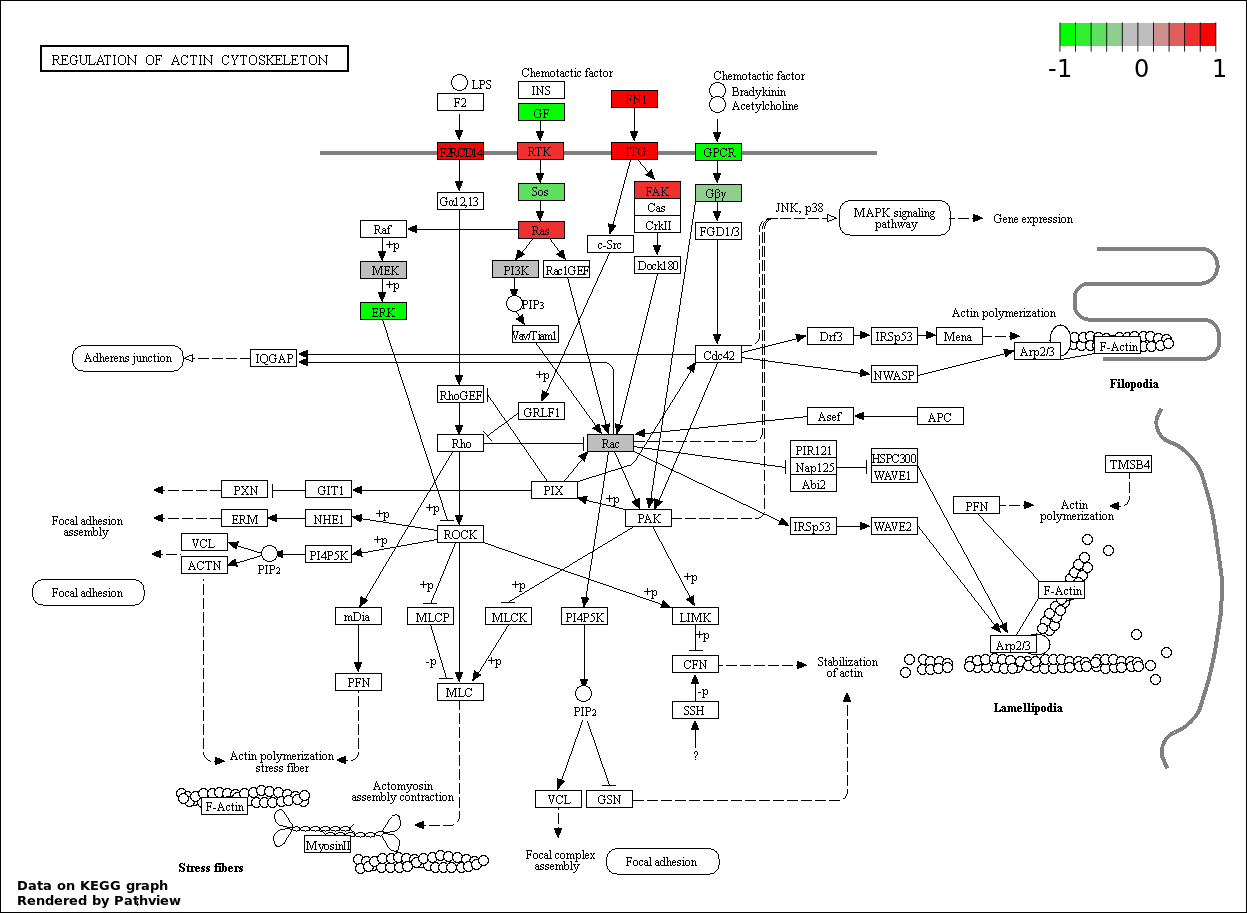
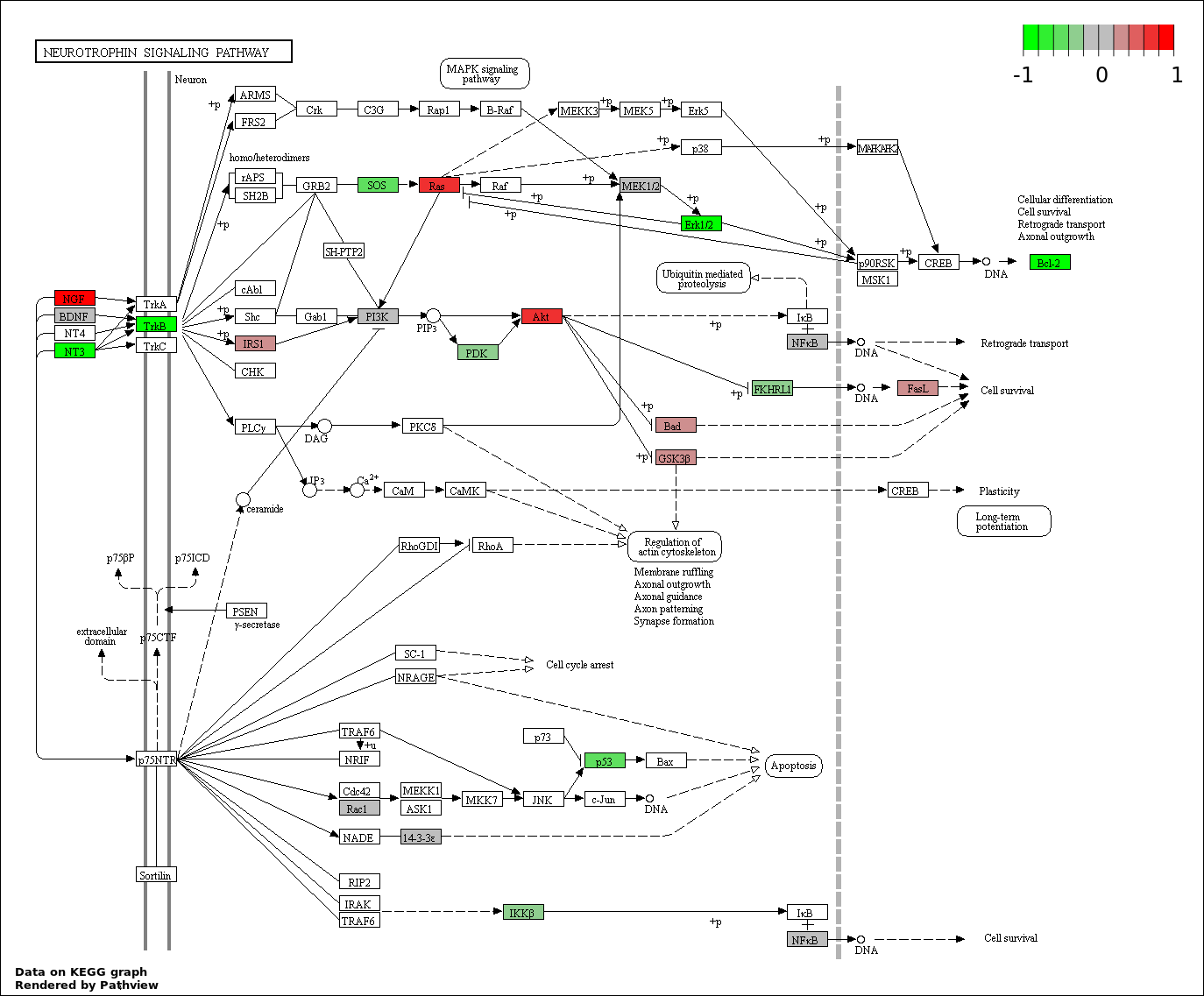
This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-139-5p | AKT3 | -1.46 | 0 | 0.76 | 5.0E-5 | PITA; miRanda | -0.2 | 7.0E-5 | NA | |
| 2 | hsa-miR-15a-5p | AKT3 | 0.78 | 0 | 0.76 | 5.0E-5 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.27 | 0.00011 | NA | |
| 3 | hsa-miR-17-5p | AKT3 | 1.42 | 0 | 0.76 | 5.0E-5 | TargetScan; miRNATAP | -0.21 | 1.0E-5 | NA | |
| 4 | hsa-miR-205-3p | AKT3 | 1.3 | 0 | 0.76 | 5.0E-5 | mirMAP | -0.25 | 0 | NA | |
| 5 | hsa-miR-20a-5p | AKT3 | 1.15 | 0 | 0.76 | 5.0E-5 | miRNATAP | -0.24 | 0 | NA | |
| 6 | hsa-miR-29b-3p | AKT3 | -0.11 | 0.51126 | 0.76 | 5.0E-5 | miRNATAP | -0.26 | 0 | 26512921 | MicroRNA 29B mir 29b regulates the Warburg effect in ovarian cancer by targeting AKT2 and AKT3 |
| 7 | hsa-miR-362-3p | AKT3 | -0.7 | 0 | 0.76 | 5.0E-5 | miRanda | -0.2 | 0.00031 | NA | |
| 8 | hsa-miR-362-5p | AKT3 | 0.41 | 0.04043 | 0.76 | 5.0E-5 | PITA; TargetScan; miRNATAP | -0.16 | 0.00012 | NA | |
| 9 | hsa-miR-429 | ANGPT1 | 0.56 | 0.00564 | -0.99 | 0.0001 | miRanda | -0.28 | 0 | NA | |
| 10 | hsa-miR-590-3p | ANGPT1 | 0.22 | 0.09659 | -0.99 | 0.0001 | PITA; miRanda; mirMAP | -0.26 | 0.00204 | NA | |
| 11 | hsa-miR-944 | ANGPT1 | 1.47 | 0 | -0.99 | 0.0001 | PITA; mirMAP | -0.32 | 0 | NA | |
| 12 | hsa-miR-148a-5p | ANGPT2 | -1 | 0 | 1.61 | 0 | mirMAP | -0.11 | 0.00532 | NA | |
| 13 | hsa-miR-19b-3p | ANGPT2 | 0.54 | 0.00062 | 1.61 | 0 | MirTarget | -0.14 | 0.00344 | NA | |
| 14 | hsa-miR-374a-5p | ANGPT2 | -0.62 | 0 | 1.61 | 0 | mirMAP | -0.41 | 0 | NA | |
| 15 | hsa-miR-374b-5p | ANGPT2 | -0.29 | 0.00415 | 1.61 | 0 | mirMAP | -0.37 | 0 | NA | |
| 16 | hsa-miR-429 | ANGPT2 | 0.56 | 0.00564 | 1.61 | 0 | miRanda | -0.19 | 0 | NA | |
| 17 | hsa-miR-582-5p | ANGPT2 | -0.04 | 0.82183 | 1.61 | 0 | mirMAP | -0.17 | 0.00036 | NA | |
| 18 | hsa-miR-664a-3p | ANGPT2 | -0.26 | 0.05132 | 1.61 | 0 | mirMAP | -0.31 | 0 | NA | |
| 19 | hsa-let-7a-3p | ATF2 | 0.1 | 0.42376 | 0.1 | 0.47373 | MirTarget; mirMAP; miRNATAP | -0.2 | 0.00011 | NA | |
| 20 | hsa-miR-10b-5p | ATF2 | -0.58 | 0 | 0.1 | 0.47373 | MirTarget; miRNATAP | -0.27 | 0 | NA | |
| 21 | hsa-miR-15a-5p | ATF2 | 0.78 | 0 | 0.1 | 0.47373 | miRNAWalker2 validate | -0.45 | 0 | NA | |
| 22 | hsa-miR-181a-5p | ATF2 | 0.82 | 0 | 0.1 | 0.47373 | mirMAP; miRNATAP | -0.2 | 0.00034 | NA | |
| 23 | hsa-miR-181b-5p | ATF2 | 0.97 | 0 | 0.1 | 0.47373 | mirMAP; miRNATAP | -0.14 | 0.00584 | NA | |
| 24 | hsa-miR-19a-3p | ATF2 | 1.36 | 0 | 0.1 | 0.47373 | MirTarget; miRNATAP | -0.27 | 0 | NA | |
| 25 | hsa-miR-19b-3p | ATF2 | 0.54 | 0.00062 | 0.1 | 0.47373 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.36 | 0 | NA | |
| 26 | hsa-miR-222-3p | ATF2 | 1.08 | 0 | 0.1 | 0.47373 | MirTarget | -0.18 | 5.0E-5 | NA | |
| 27 | hsa-miR-26b-5p | ATF2 | -0.25 | 0.02852 | 0.1 | 0.47373 | MirTarget; miRNATAP | -0.38 | 0 | 21901137 | Coordinated regulation of ATF2 by miR 26b in γ irradiated lung cancer cells; Concurrent analysis of time-series mRNA and microRNA profiles uncovered that expression of miR-26b was down regulated and its target activating transcription factor 2 ATF2 mRNA was up regulated in γ-irradiated H1299 cells; IR in miR-26b overexpressed H1299 cells could not induce expression of ATF2; From these results we concluded that IR-induced up-regulation of ATF2 was coordinately enhanced by suppression of miR-26b in lung cancer cells which may enhance the effect of IR in the MAPK signaling pathway |
| 28 | hsa-miR-29a-5p | ATF2 | 0.32 | 0.03704 | 0.1 | 0.47373 | MirTarget; miRNATAP | -0.22 | 0 | NA | |
| 29 | hsa-miR-30a-5p | ATF2 | -1.8 | 0 | 0.1 | 0.47373 | mirMAP | -0.12 | 0.0008 | NA | |
| 30 | hsa-miR-30b-5p | ATF2 | -0.4 | 0.00347 | 0.1 | 0.47373 | mirMAP | -0.32 | 0 | NA | |
| 31 | hsa-miR-30c-5p | ATF2 | 0.02 | 0.88637 | 0.1 | 0.47373 | mirMAP | -0.36 | 0 | NA | |
| 32 | hsa-miR-320a | ATF2 | 0.8 | 0 | 0.1 | 0.47373 | miRanda | -0.15 | 0.00077 | NA | |
| 33 | hsa-miR-338-3p | ATF2 | -0.95 | 0 | 0.1 | 0.47373 | miRanda | -0.31 | 0 | NA | |
| 34 | hsa-miR-3607-3p | ATF2 | 1.01 | 9.0E-5 | 0.1 | 0.47373 | mirMAP | -0.13 | 0 | NA | |
| 35 | hsa-miR-374b-5p | ATF2 | -0.29 | 0.00415 | 0.1 | 0.47373 | miRNAWalker2 validate; mirMAP; miRNATAP | -0.24 | 0.00016 | NA | |
| 36 | hsa-miR-582-5p | ATF2 | -0.04 | 0.82183 | 0.1 | 0.47373 | MirTarget; miRNATAP | -0.17 | 1.0E-5 | NA | |
| 37 | hsa-miR-590-3p | ATF2 | 0.22 | 0.09659 | 0.1 | 0.47373 | MirTarget; miRanda; mirMAP; miRNATAP | -0.12 | 0.00962 | NA | |
| 38 | hsa-miR-127-3p | BAD | -1.47 | 0 | 0.23 | 0.0411 | miRanda; miRNATAP | -0.11 | 1.0E-5 | NA | |
| 39 | hsa-miR-326 | BAD | -0.43 | 0.0415 | 0.23 | 0.0411 | miRanda | -0.16 | 0 | NA | |
| 40 | hsa-miR-17-5p | BCL2 | 1.42 | 0 | -0.91 | 1.0E-5 | miRNAWalker2 validate; miRTarBase | -0.29 | 0 | 25435430 | Combined overexpression of miR-16 and miR-17 greatly reduced Beclin-1 and Bcl-2 expressions respectively; miR-17 overexpression reduced cytoprotective autophagy by targeting Beclin-1 whereas overexpression of miR-16 potentiated paclitaxel induced apoptotic cell death by inhibiting anti-apoptotic protein Bcl-2 |
| 41 | hsa-miR-181b-5p | BCL2 | 0.97 | 0 | -0.91 | 1.0E-5 | miRNAWalker2 validate; miRTarBase | -0.37 | 0 | 25524579 | Moreover we also found miR-181 reduction was associated with increased Bcl-2 levels and miR-181 was further suggested to exert its pro-apoptotic function mainly through targeting Bcl-2 expression |
| 42 | hsa-miR-205-3p | BCL2 | 1.3 | 0 | -0.91 | 1.0E-5 | mirMAP | -0.23 | 0 | 23612742 | MicroRNA 205 a novel regulator of the anti apoptotic protein Bcl2 is downregulated in prostate cancer; In addition we describe the anti-apoptotic protein BCL2 as a target of miR-205 relevant for prostate cancer due to its role in prognosis of primary tumours and in the appearance of androgen independence; The repression of BCL2 by miR-205 was confirmed using reporter assays and western blotting; Consistent with its anti-apoptotic target BCL2 miR-205 promoted apoptosis in prostate cancer cells in response to DNA damage by cisplatin and doxorubicin in the prostate cancer cell lines PC3 and LnCap |
| 43 | hsa-miR-20a-3p | BCL2 | 1.35 | 0 | -0.91 | 1.0E-5 | mirMAP | -0.21 | 1.0E-5 | NA | |
| 44 | hsa-miR-20a-5p | BCL2 | 1.15 | 0 | -0.91 | 1.0E-5 | miRNAWalker2 validate; miRTarBase | -0.21 | 8.0E-5 | NA | |
| 45 | hsa-miR-21-5p | BCL2 | 1.62 | 0 | -0.91 | 1.0E-5 | miRNAWalker2 validate; miRTarBase | -0.44 | 0 | 21468550; 25994220; 25381586; 26555418; 23359184; 22964582; 21376256 | BCL-2 up-regulation could be achieved by miR-21 overexpression which prevented T24 cells from apoptosis induced by doxorubicin; Furthermore the miR-21 induced BCL-2 up-regulation could be cancelled by the PI3K inhibitor LY294002;Meanwhile miR-21 loss reduced STAT3 and Bcl-2 activation causing an increase in the apoptosis of tumour cells in CAC mice;Changes in the sensitivity of osteosarcoma cells to CDDP were examined after transfection with miR-21 mimics or anti-miR-21 or bcl-2 siRNA in combination with CDDP;The expression of Bax Bcl-2 and miR-21 in parental and paclitaxel-resistant cells was detected by RT-PCR and Western blotting;Resveratrol induces apoptosis of pancreatic cancers cells by inhibiting miR 21 regulation of BCL 2 expression; We also used Western blot to measure BCL-2 protein levels after down-regulation of miR-21 expression; Besides down-regulation of miR-21 expression can inhibit BCL-2 expression in PANC-1 CFPAC-1 and MIA Paca-2 cells; Over-expression of miR-21 expression can reverse down-regulation of BCL-2 expression and apoptosis induced by resveratrol; In this study we demonstrated that the effect of resveratrol on apoptosis is due to inhibiting miR-21 regulation of BCL-2 expression;Tumors harvested from these lungs have elevated levels of oncogenic miRNAs miR-21 and miR-155; are deficient for p53-regulated miRNAs; and have heightened expression of miR-34 target genes such as Met and Bcl-2;Bcl 2 upregulation induced by miR 21 via a direct interaction is associated with apoptosis and chemoresistance in MIA PaCa 2 pancreatic cancer cells; However the roles and mechanisms of miRNA miR-21 in regulation of Bcl-2 in pancreatic cancer remain to be elucidated; Then luciferase activity was observed after miR-21 mimics and pRL-TK plasmids containing wild-type and mutant 3'UTRs of Bcl-2 mRNA were co-transfected; Cells transfected with miR-21 inhibitor revealed an opposite trend. There was a significant increase in luciferase activity in the cells transfected with the wild-type pRL-TK plasmid in contrast to those transfected with the mutant one indicating that miR-21 promotes Bcl-2 expression by binding directly to the 3'UTR of Bcl-2 mRNA; Upregulation of Bcl-2 directly induced by miR-21 is associated with apoptosis chemoresistance and proliferation of MIA PaCa-2 pancreatic cancer cells |
| 46 | hsa-miR-224-3p | BCL2 | 0.5 | 0.00811 | -0.91 | 1.0E-5 | mirMAP | -0.15 | 0.00211 | 24796455 | In addition the expressions of Bcl2 mRNA and protein were 1.05 ± 0.04 and 0.21 ± 0.03 in the miR-224 ASO group significantly lower than that in the control group 4.87 ± 0.96 and 0.88 ± 0.09 P < 0.01 |
| 47 | hsa-miR-224-5p | BCL2 | 1.95 | 0 | -0.91 | 1.0E-5 | mirMAP | -0.35 | 0 | 24796455 | In addition the expressions of Bcl2 mRNA and protein were 1.05 ± 0.04 and 0.21 ± 0.03 in the miR-224 ASO group significantly lower than that in the control group 4.87 ± 0.96 and 0.88 ± 0.09 P < 0.01 |
| 48 | hsa-miR-24-2-5p | BCL2 | 1.27 | 0 | -0.91 | 1.0E-5 | miRNAWalker2 validate; miRTarBase | -0.49 | 0 | NA | |
| 49 | hsa-miR-29a-5p | BCL2 | 0.32 | 0.03704 | -0.91 | 1.0E-5 | mirMAP | -0.16 | 0.00917 | 20041405 | Subsequent investigation characterized two antiapoptotic molecules Bcl-2 and Mcl-1 as direct targets of miR-29; Furthermore silencing of Bcl-2 and Mcl-1 phenocopied the proapoptotic effect of miR-29 whereas overexpression of these proteins attenuated the effect of miR-29 |
| 50 | hsa-miR-365a-3p | BCL2 | 0.45 | 0.00234 | -0.91 | 1.0E-5 | miRNAWalker2 validate; miRTarBase | -0.62 | 0 | NA | |
| 51 | hsa-miR-450b-5p | BCL2 | 1.26 | 0 | -0.91 | 1.0E-5 | mirMAP | -0.3 | 0 | NA | |
| 52 | hsa-miR-452-5p | BCL2 | 0.37 | 0.03485 | -0.91 | 1.0E-5 | mirMAP | -0.29 | 0 | NA | |
| 53 | hsa-miR-455-5p | BCL2 | 1.41 | 0 | -0.91 | 1.0E-5 | mirMAP | -0.67 | 0 | NA | |
| 54 | hsa-miR-542-3p | BCL2 | 0.64 | 0 | -0.91 | 1.0E-5 | mirMAP | -0.3 | 2.0E-5 | NA | |
| 55 | hsa-miR-590-5p | BCL2 | 0.76 | 0 | -0.91 | 1.0E-5 | miRanda | -0.21 | 0.00251 | NA | |
| 56 | hsa-miR-7-5p | BCL2 | 1.56 | 0 | -0.91 | 1.0E-5 | miRNAWalker2 validate; miRTarBase; mirMAP | -0.24 | 0 | 26464649; 25862909; 21750649 | Western blotting was used to evaluate the effect of miR-7 on Bcl2 in A549 and H460 cells; Moreover subsequent experiments showed that BCL-2 was downregulated by miR-7 at both transcriptional and translational levels; This study further extends the biological role of miR-7 in NSCLC A549 and H460 cells and identifies BCL-2 as a novel target possibly involved in miR-7-mediated growth suppression and apoptosis induction of NSCLC cells;miR-7 overexpression correlated with diminished BCL2 expression but there was no relationship between miR-7 and EGFR expression neither in tumour samples nor in the cell lines; Of the two postulated miR-7 target genes we examined BCL2 but not EGFR seems to be a possible miR-7 target in OC;Bioinformatics predictions revealed a potential binding site of miR-7 on 3'UTR of BCL-2 and it was further confirmed by luciferase assay; Moreover subsequent experiments showed that BCL-2 was downregulated by miR-7 at both transcriptional and translational levels; These results suggest that miR-7 regulates the expression of BCL-2 through direct 3'UTR interactions |
| 57 | hsa-miR-369-3p | BCL2L11 | -1.23 | 0 | 0.43 | 2.0E-5 | mirMAP | -0.13 | 0 | NA | |
| 58 | hsa-miR-495-3p | BCL2L11 | -1.48 | 0 | 0.43 | 2.0E-5 | mirMAP | -0.13 | 0 | 26375442 | Combination of miR-130a and miR-495 significantly decreased apoptosis determined by Annexin V-FITC/propidium iodide staining and flow cytometric analysis and the expression of Bim in SNU484 gastric cancer cells; In addition p21 and Bim RUNX3 target genes were completely downregulated by the combination of miR-130a and miR-495 |
| 59 | hsa-let-7b-3p | BDNF | 0.12 | 0.29922 | 0.04 | 0.88345 | MirTarget; miRNATAP | -0.49 | 0 | NA | |
| 60 | hsa-miR-142-5p | BDNF | 0.29 | 0.09 | 0.04 | 0.88345 | mirMAP | -0.31 | 1.0E-5 | NA | |
| 61 | hsa-miR-146b-5p | BDNF | 0.92 | 0 | 0.04 | 0.88345 | miRanda | -0.3 | 0.00012 | NA | |
| 62 | hsa-miR-155-5p | BDNF | 0.53 | 0.00249 | 0.04 | 0.88345 | miRNATAP | -0.23 | 0.00094 | NA | |
| 63 | hsa-miR-22-3p | BDNF | 0.01 | 0.92636 | 0.04 | 0.88345 | miRNAWalker2 validate; miRTarBase | -0.43 | 0.00361 | NA | |
| 64 | hsa-miR-29b-1-5p | BDNF | 1.4 | 0 | 0.04 | 0.88345 | mirMAP | -0.23 | 5.0E-5 | NA | |
| 65 | hsa-miR-338-5p | BDNF | -1.72 | 0 | 0.04 | 0.88345 | miRNATAP | -0.23 | 0.00053 | NA | |
| 66 | hsa-miR-365a-3p | BDNF | 0.45 | 0.00234 | 0.04 | 0.88345 | miRNATAP | -0.32 | 0.00013 | NA | |
| 67 | hsa-miR-424-5p | BDNF | 1.48 | 0 | 0.04 | 0.88345 | MirTarget; miRNATAP | -0.28 | 5.0E-5 | NA | |
| 68 | hsa-miR-199a-5p | BRCA1 | -0.36 | 0.04351 | 1.08 | 0 | miRanda | -0.21 | 0 | NA | |
| 69 | hsa-miR-199b-5p | BRCA1 | -1.08 | 0 | 1.08 | 0 | miRanda | -0.24 | 0 | NA | |
| 70 | hsa-miR-493-5p | BRCA1 | -0.47 | 0.02213 | 1.08 | 0 | MirTarget | -0.13 | 0 | NA | |
| 71 | hsa-miR-193b-3p | CASP9 | 1.73 | 0 | -0.46 | 0 | miRNAWalker2 validate | -0.15 | 0 | NA | |
| 72 | hsa-miR-424-5p | CASP9 | 1.48 | 0 | -0.46 | 0 | mirMAP | -0.14 | 0 | NA | |
| 73 | hsa-miR-7-5p | CASP9 | 1.56 | 0 | -0.46 | 0 | miRNAWalker2 validate | -0.1 | 0 | NA | |
| 74 | hsa-let-7g-5p | CCND1 | -0.25 | 0.0094 | -0.01 | 0.95947 | miRNATAP | -0.42 | 1.0E-5 | NA | |
| 75 | hsa-miR-106a-5p | CCND1 | 0.68 | 0.00222 | -0.01 | 0.95947 | MirTarget; miRNATAP | -0.29 | 0 | NA | |
| 76 | hsa-miR-1266-5p | CCND1 | -0.6 | 0.02013 | -0.01 | 0.95947 | MirTarget | -0.14 | 0.00019 | NA | |
| 77 | hsa-miR-142-3p | CCND1 | 1.13 | 0 | -0.01 | 0.95947 | miRanda | -0.21 | 0 | 23619912 | Transfection of miR-142-3p mimics in colon cancer cells downregulated cyclin D1 expression induced G1 phase cell cycle arrest and elevated the sensitivity of the cells to 5-fluorouracil |
| 78 | hsa-miR-142-5p | CCND1 | 0.29 | 0.09 | -0.01 | 0.95947 | PITA | -0.19 | 0.00053 | NA | |
| 79 | hsa-miR-150-5p | CCND1 | -0.24 | 0.33155 | -0.01 | 0.95947 | mirMAP | -0.18 | 0 | NA | |
| 80 | hsa-miR-155-5p | CCND1 | 0.53 | 0.00249 | -0.01 | 0.95947 | miRNAWalker2 validate | -0.21 | 0.0001 | 26955820 | MicroRNA 155 expression inversely correlates with pathologic stage of gastric cancer and it inhibits gastric cancer cell growth by targeting cyclin D1 |
| 81 | hsa-miR-15a-5p | CCND1 | 0.78 | 0 | -0.01 | 0.95947 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.32 | 5.0E-5 | 22922827 | CCND1 has been found to be a target of miR-15a and miR-16-1 through analysis of complementary sequences between microRNAs and CCND1 mRNA; Moreover the transcription of CCND1 is suppressed by miR-15a and miR-16-1 via direct binding to the CCND1 3'-untranslated region 3'-UTR |
| 82 | hsa-miR-15b-5p | CCND1 | 0.85 | 0 | -0.01 | 0.95947 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.2 | 0.00913 | NA | |
| 83 | hsa-miR-16-1-3p | CCND1 | 0.96 | 0 | -0.01 | 0.95947 | miRNAWalker2 validate; miRTarBase | -0.3 | 0 | 22922827; 18483394 | CCND1 has been found to be a target of miR-15a and miR-16-1 through analysis of complementary sequences between microRNAs and CCND1 mRNA; Moreover the transcription of CCND1 is suppressed by miR-15a and miR-16-1 via direct binding to the CCND1 3'-untranslated region 3'-UTR;Truncation in CCND1 mRNA alters miR 16 1 regulation in mantle cell lymphoma; Furthermore we demonstrated that this truncation alters miR-16-1 binding sites and through the use of reporter constructs we were able to show that miR-16-1 regulates CCND1 mRNA expression; This study introduces the role of miR-16-1 in the regulation of CCND1 in MCL |
| 84 | hsa-miR-16-5p | CCND1 | 0.52 | 0 | -0.01 | 0.95947 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.36 | 3.0E-5 | 23991964; 22922827; 18483394 | At the molecular level our results further revealed that cyclin D1 expression was negatively regulated by miR-16;CCND1 has been found to be a target of miR-15a and miR-16-1 through analysis of complementary sequences between microRNAs and CCND1 mRNA; Moreover the transcription of CCND1 is suppressed by miR-15a and miR-16-1 via direct binding to the CCND1 3'-untranslated region 3'-UTR;Truncation in CCND1 mRNA alters miR 16 1 regulation in mantle cell lymphoma; Furthermore we demonstrated that this truncation alters miR-16-1 binding sites and through the use of reporter constructs we were able to show that miR-16-1 regulates CCND1 mRNA expression; This study introduces the role of miR-16-1 in the regulation of CCND1 in MCL |
| 85 | hsa-miR-19a-3p | CCND1 | 1.36 | 0 | -0.01 | 0.95947 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.13 | 0.00806 | 25985117 | Moreover miR-19a might play inhibitory roles in HCC malignancy via regulating Cyclin D1 expression |
| 86 | hsa-miR-19b-1-5p | CCND1 | 0.76 | 0 | -0.01 | 0.95947 | miRNAWalker2 validate; miRTarBase | -0.2 | 0.00133 | NA | |
| 87 | hsa-miR-19b-3p | CCND1 | 0.54 | 0.00062 | -0.01 | 0.95947 | miRNATAP | -0.23 | 0.0001 | NA | |
| 88 | hsa-miR-20a-5p | CCND1 | 1.15 | 0 | -0.01 | 0.95947 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.15 | 0.0057 | NA | |
| 89 | hsa-miR-20b-5p | CCND1 | 0.35 | 0.23201 | -0.01 | 0.95947 | MirTarget; miRNATAP | -0.25 | 0 | NA | |
| 90 | hsa-miR-29b-3p | CCND1 | -0.11 | 0.51126 | -0.01 | 0.95947 | mirMAP | -0.18 | 0.00109 | NA | |
| 91 | hsa-miR-29c-3p | CCND1 | -2.08 | 0 | -0.01 | 0.95947 | mirMAP | -0.16 | 0.00015 | NA | |
| 92 | hsa-miR-33a-3p | CCND1 | 0.2 | 0.21234 | -0.01 | 0.95947 | MirTarget | -0.2 | 0.00047 | NA | |
| 93 | hsa-miR-34a-5p | CCND1 | -0.12 | 0.364 | -0.01 | 0.95947 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.31 | 2.0E-5 | 25792709; 21399894 | This inhibition of proliferation was associated with a decrease in cyclin D1 levels orchestrated principally by HNF-4α a target of miR-34a considered to act as a tumour suppressor in the liver;Quantitative PCR and western analysis confirmed decreased expression of two genes BCL-2 and CCND1 in docetaxel-resistant cells which are both targeted by miR-34a |
| 94 | hsa-miR-425-5p | CCND1 | 0.93 | 0 | -0.01 | 0.95947 | miRNAWalker2 validate | -0.15 | 0.00573 | NA | |
| 95 | hsa-miR-497-5p | CCND1 | -0.97 | 0 | -0.01 | 0.95947 | MirTarget; miRNATAP | -0.27 | 5.0E-5 | 21350001 | Raf-1 and Ccnd1 were identified as novel direct targets of miR-195 and miR-497 miR-195/497 expression levels in clinical specimens were found to be correlated inversely with malignancy of breast cancer |
| 96 | hsa-miR-590-3p | CCND1 | 0.22 | 0.09659 | -0.01 | 0.95947 | mirMAP | -0.24 | 0.00077 | NA | |
| 97 | hsa-miR-7-1-3p | CCND1 | 1.14 | 0 | -0.01 | 0.95947 | mirMAP | -0.22 | 0.00036 | NA | |
| 98 | hsa-miR-9-5p | CCND1 | 1.66 | 2.0E-5 | -0.01 | 0.95947 | miRNAWalker2 validate | -0.14 | 0 | NA | |
| 99 | hsa-miR-92a-3p | CCND1 | 0.99 | 0 | -0.01 | 0.95947 | miRNAWalker2 validate | -0.24 | 0.002 | NA | |
| 100 | hsa-miR-942-5p | CCND1 | 0.68 | 0 | -0.01 | 0.95947 | MirTarget | -0.23 | 0.00111 | NA | |
| 101 | hsa-miR-224-3p | CCND2 | 0.5 | 0.00811 | 0.15 | 0.53242 | mirMAP | -0.31 | 0 | NA | |
| 102 | hsa-miR-27b-3p | CCND3 | -0.73 | 0 | -0.05 | 0.63156 | miRNAWalker2 validate | -0.13 | 0.00016 | NA | |
| 103 | hsa-miR-125b-5p | CCNE1 | -0.99 | 0 | 1.47 | 0 | miRNAWalker2 validate | -0.28 | 0 | NA | |
| 104 | hsa-miR-151a-3p | CCNE1 | 0.13 | 0.29527 | 1.47 | 0 | miRNAWalker2 validate | -0.15 | 0.00299 | NA | |
| 105 | hsa-miR-195-5p | CCNE1 | -1.84 | 0 | 1.47 | 0 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.31 | 0 | 24402230 | Furthermore through qPCR and western blot assays we showed that overexpression of miR-195-5p reduced CCNE1 mRNA and protein levels respectively |
| 106 | hsa-miR-215-5p | CCNE1 | -0.76 | 6.0E-5 | 1.47 | 0 | miRNAWalker2 validate | -0.13 | 0.0001 | NA | |
| 107 | hsa-miR-26a-5p | CCNE1 | -1.09 | 0 | 1.47 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.49 | 0 | 22094936 | Cell cycle regulation and CCNE1 and CDC2 were the only significant overlapping pathway and genes differentially expressed between tumors with high and low levels of miR-26a and EZH2 respectively; Low mRNA levels of EZH2 CCNE1 and CDC2 and high levels of miR-26a are associated with favorable outcome on tamoxifen |
| 108 | hsa-miR-497-5p | CCNE1 | -0.97 | 0 | 1.47 | 0 | MirTarget; miRNATAP | -0.16 | 0.00018 | 24112607; 25909221; 24909281 | Western blot assays confirmed that overexpression of miR-497 reduced cyclin E1 protein levels; Inhibited cellular growth suppressed cellular migration and invasion and G1 cell cycle arrest were observed upon overexpression of miR-497 in cells possibly by targeting cyclin E1;The effect of simultaneous overexpression of miR-497 and miR-34a on the inhibition of cell proliferation colony formation and tumor growth and the downregulation of cyclin E1 was stronger than the effect of each miRNA alone; The synergistic actions of miR-497 and miR-34a partly correlated with cyclin E1 levels; These results indicate cyclin E1 is downregulated by both miR-497 and miR-34a which synergistically retard the growth of human lung cancer cells;miR 497 suppresses proliferation of human cervical carcinoma HeLa cells by targeting cyclin E1; Furthermore the target effect of miR-497 on the CCNE1 was identified by dual-luciferase reporter assay system qRT-PCR and Western blotting; Over-expressed miR-497 in HeLa cells could suppress cell proliferation by targeting CCNE1 |
| 109 | hsa-miR-30a-5p | CCNE2 | -1.8 | 0 | 1.24 | 0 | miRNATAP | -0.16 | 5.0E-5 | NA | |
| 110 | hsa-miR-369-3p | CCNE2 | -1.23 | 0 | 1.24 | 0 | PITA | -0.24 | 0 | NA | |
| 111 | hsa-miR-330-3p | CDC37 | -0.05 | 0.71986 | 0.15 | 0.05107 | PITA; miRNATAP | -0.14 | 0 | NA | |
| 112 | hsa-miR-10b-5p | CDK2 | -0.58 | 0 | 0.83 | 0 | miRNAWalker2 validate | -0.16 | 0 | NA | |
| 113 | hsa-miR-495-3p | CDK2 | -1.48 | 0 | 0.83 | 0 | mirMAP | -0.12 | 0 | NA | |
| 114 | hsa-miR-101-3p | CDK6 | -1.91 | 0 | 0.94 | 0 | mirMAP | -0.33 | 0 | NA | |
| 115 | hsa-miR-106a-5p | CDK6 | 0.68 | 0.00222 | 0.94 | 0 | mirMAP | -0.27 | 0 | NA | |
| 116 | hsa-miR-140-3p | CDK6 | -0.97 | 0 | 0.94 | 0 | miRNATAP | -0.27 | 0.00057 | NA | |
| 117 | hsa-miR-141-3p | CDK6 | 0.73 | 0.00012 | 0.94 | 0 | TargetScan; miRNATAP | -0.14 | 0.00069 | NA | |
| 118 | hsa-miR-142-5p | CDK6 | 0.29 | 0.09 | 0.94 | 0 | PITA | -0.14 | 0.00345 | NA | |
| 119 | hsa-miR-146a-5p | CDK6 | -0.02 | 0.90588 | 0.94 | 0 | mirMAP | -0.22 | 1.0E-5 | NA | |
| 120 | hsa-miR-148a-3p | CDK6 | -0.43 | 0.00954 | 0.94 | 0 | mirMAP | -0.35 | 0 | NA | |
| 121 | hsa-miR-148a-5p | CDK6 | -1 | 0 | 0.94 | 0 | mirMAP | -0.17 | 6.0E-5 | NA | |
| 122 | hsa-miR-15a-5p | CDK6 | 0.78 | 0 | 0.94 | 0 | miRNATAP | -0.29 | 3.0E-5 | NA | |
| 123 | hsa-miR-16-1-3p | CDK6 | 0.96 | 0 | 0.94 | 0 | mirMAP | -0.19 | 0.00047 | NA | |
| 124 | hsa-miR-16-5p | CDK6 | 0.52 | 0 | 0.94 | 0 | miRNAWalker2 validate; miRTarBase | -0.39 | 0 | NA | |
| 125 | hsa-miR-191-5p | CDK6 | 0.65 | 0 | 0.94 | 0 | miRNAWalker2 validate; miRTarBase | -0.3 | 0 | NA | |
| 126 | hsa-miR-195-5p | CDK6 | -1.84 | 0 | 0.94 | 0 | miRNAWalker2 validate; miRTarBase | -0.15 | 0.00222 | 23333942 | Expression of cyclin-dependent kinase 6 and vascular endothelial growth factor was down-regulated by exogenous miR-195 and miR-378 respectively |
| 127 | hsa-miR-200a-3p | CDK6 | 0.61 | 0.0029 | 0.94 | 0 | miRNATAP | -0.14 | 0.00037 | 24009066 | microRNA 200a is an independent prognostic factor of hepatocellular carcinoma and induces cell cycle arrest by targeting CDK6 |
| 128 | hsa-miR-200b-3p | CDK6 | 0.07 | 0.68286 | 0.94 | 0 | mirMAP | -0.17 | 0.0002 | NA | |
| 129 | hsa-miR-20b-5p | CDK6 | 0.35 | 0.23201 | 0.94 | 0 | mirMAP | -0.32 | 0 | 26166554 | The transfection of miR-20b into EJ cells induced G1 phase cell cycle arrest via the decreased expression of cyclin D1 CDK2 and CDK6 without affecting another G1 phase cell cycle regulator cyclin E |
| 130 | hsa-miR-217 | CDK6 | -1.35 | 0 | 0.94 | 0 | mirMAP | -0.1 | 0.00385 | NA | |
| 131 | hsa-miR-218-5p | CDK6 | -1.07 | 0 | 0.94 | 0 | miRNAWalker2 validate | -0.16 | 0.00036 | 23996750 | Ectopic expression of miR-218 in HepG2 cells resulted in suppressed cell proliferation and enhanced cell apoptosis as well as the down-regulation of Bmi-1 and CDK6 mRNA and protein expressions P<0.05; The low-expression of miR-218 is correlated with malignant clinicopathological characteristics of HCC and miR-218 may inhibit cell proliferation and promote cell apoptosis by down-regulating Bmi-1 and CDK6 in HCC |
| 132 | hsa-miR-23b-3p | CDK6 | -0.5 | 3.0E-5 | 0.94 | 0 | mirMAP | -0.19 | 0.00569 | NA | |
| 133 | hsa-miR-26a-5p | CDK6 | -1.09 | 0 | 0.94 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.3 | 2.0E-5 | 26314438 | Maxvision immunohistochemistry technique was used to detect the expression level of CDK6 and miR-26a in tissue of 20 ENKTCL cases 10 cases of proliferative lymphadenitis and 10 samples of normal lymph node respectively; The possible role of miR-26a and its target gene CDK6 in genesis and development of ENKTCL were analyzed according to the clinical features of ENKTCL patients; Correlation analysis showed that there was significant negative correlation between miR-26a expression and CDK6 expression r = -0.54 P = 0.04 |
| 134 | hsa-miR-26b-5p | CDK6 | -0.25 | 0.02852 | 0.94 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.37 | 0 | NA | |
| 135 | hsa-miR-27b-3p | CDK6 | -0.73 | 0 | 0.94 | 0 | mirMAP | -0.25 | 0.0001 | NA | |
| 136 | hsa-miR-27b-5p | CDK6 | 0.58 | 0.00018 | 0.94 | 0 | mirMAP | -0.14 | 0.00915 | NA | |
| 137 | hsa-miR-29b-3p | CDK6 | -0.11 | 0.51126 | 0.94 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.23 | 0 | 23245396; 25472644; 26180082; 23591808; 27230400; 20086245 | The IFN-γ-induced G1-arrest of melanoma cells involves down-regulation of CDK6 which we proved to be a direct target of miR-29 in these cells;Moreover miR-29b inhibited the expression of MCL1 and CDK6;Knockdown of NTSR1 increased the expression of miR-29b-1 and miR-129-3p which were responsible for the decreased CDK6 expression;Here we have identified the oncogene cyclin-dependent protein kinase 6 CDK6 as a direct target of miR-29b in lung cancer;MiR 29b suppresses the proliferation and migration of osteosarcoma cells by targeting CDK6; In this study we investigated the role of miR-29b as a novel regulator of CDK6 using bioinformatics methods; We demonstrated that CDK6 can be downregulated by miR-29b via binding to the 3'-UTR region in osteosarcoma cells; Furthermore we identified an inverse correlation between miR-29b and CDK6 protein levels in osteosarcoma tissues; The results revealed that miR-29b acts as a tumor suppressor of osteosarcoma by targeting CDK6 in the proliferation and migration processes;microRNA expression profile and identification of miR 29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma; Furthermore we demonstrate miR-29 inhibition of CDK6 protein and mRNA levels by direct binding to 3'-untranslated region; Inverse correlation between miR-29 and CDK6 was observed in MCL |
| 138 | hsa-miR-29c-3p | CDK6 | -2.08 | 0 | 0.94 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.26 | 0 | 26396669 | Furthermore through qPCR and Western blot assays confirmed that overexpression of miR-29c reduced CDK6 mRNA and protein levels; miR-29c could inhibit the proliferation migration and invasion of bladder cancer cells via regulating CDK6 |
| 139 | hsa-miR-30a-3p | CDK6 | -2.26 | 0 | 0.94 | 0 | miRNAWalker2 validate; miRTarBase; mirMAP | -0.14 | 0.00032 | NA | |
| 140 | hsa-miR-30a-5p | CDK6 | -1.8 | 0 | 0.94 | 0 | mirMAP | -0.24 | 0 | NA | |
| 141 | hsa-miR-30b-5p | CDK6 | -0.4 | 0.00347 | 0.94 | 0 | mirMAP | -0.43 | 0 | NA | |
| 142 | hsa-miR-30c-5p | CDK6 | 0.02 | 0.88637 | 0.94 | 0 | mirMAP | -0.34 | 0 | NA | |
| 143 | hsa-miR-30d-3p | CDK6 | -0.58 | 0 | 0.94 | 0 | mirMAP | -0.5 | 0 | NA | |
| 144 | hsa-miR-30d-5p | CDK6 | -0.38 | 0.00017 | 0.94 | 0 | mirMAP | -0.48 | 0 | NA | |
| 145 | hsa-miR-30e-3p | CDK6 | -0.89 | 0 | 0.94 | 0 | mirMAP | -0.49 | 0 | NA | |
| 146 | hsa-miR-30e-5p | CDK6 | -1.02 | 0 | 0.94 | 0 | mirMAP | -0.53 | 0 | NA | |
| 147 | hsa-miR-32-5p | CDK6 | -0.32 | 0.01025 | 0.94 | 0 | miRNATAP | -0.34 | 0 | NA | |
| 148 | hsa-miR-320a | CDK6 | 0.8 | 0 | 0.94 | 0 | PITA; miRNATAP | -0.22 | 0.00023 | NA | |
| 149 | hsa-miR-335-3p | CDK6 | 0.34 | 0.05424 | 0.94 | 0 | mirMAP | -0.13 | 0.00396 | NA | |
| 150 | hsa-miR-335-5p | CDK6 | -1.17 | 0 | 0.94 | 0 | mirMAP | -0.18 | 5.0E-5 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 116 | 1618 | 2.033e-63 | 9.46e-60 |
| 2 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE SIGNALING PATHWAY | 71 | 498 | 6.402e-57 | 1.489e-53 |
| 3 | REGULATION OF PROTEIN MODIFICATION PROCESS | 112 | 1710 | 1.112e-56 | 1.724e-53 |
| 4 | REGULATION OF KINASE ACTIVITY | 82 | 776 | 1.025e-55 | 1.192e-52 |
| 5 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 91 | 1036 | 2.233e-55 | 1.732e-52 |
| 6 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 91 | 1036 | 2.233e-55 | 1.732e-52 |
| 7 | POSITIVE REGULATION OF CELL PROLIFERATION | 82 | 814 | 4.735e-54 | 3.147e-51 |
| 8 | INTRACELLULAR SIGNAL TRANSDUCTION | 105 | 1572 | 2.542e-53 | 1.351e-50 |
| 9 | PROTEIN PHOSPHORYLATION | 86 | 944 | 2.614e-53 | 1.351e-50 |
| 10 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 114 | 1929 | 3.408e-53 | 1.464e-50 |
| 11 | CELLULAR RESPONSE TO ORGANIC SUBSTANCE | 112 | 1848 | 3.461e-53 | 1.464e-50 |
| 12 | POSITIVE REGULATION OF KINASE ACTIVITY | 67 | 482 | 8.563e-53 | 3.32e-50 |
| 13 | ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY | 75 | 689 | 1.553e-51 | 5.559e-49 |
| 14 | REGULATION OF TRANSFERASE ACTIVITY | 84 | 946 | 4.792e-51 | 1.593e-48 |
| 15 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 100 | 1492 | 1.667e-50 | 5.172e-48 |
| 16 | POSITIVE REGULATION OF CELL COMMUNICATION | 101 | 1532 | 2.017e-50 | 5.847e-48 |
| 17 | REGULATION OF CELL PROLIFERATION | 100 | 1496 | 2.136e-50 | 5.847e-48 |
| 18 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 107 | 1791 | 9.64e-50 | 2.492e-47 |
| 19 | POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 80 | 876 | 2.537e-49 | 6.213e-47 |
| 20 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 103 | 1656 | 3.431e-49 | 7.982e-47 |
| 21 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 88 | 1135 | 8.112e-49 | 1.797e-46 |
| 22 | LOCOMOTION | 86 | 1114 | 1.974e-47 | 4.175e-45 |
| 23 | PHOSPHORYLATION | 89 | 1228 | 5.343e-47 | 1.081e-44 |
| 24 | POSITIVE REGULATION OF TRANSFERASE ACTIVITY | 68 | 616 | 7.831e-47 | 1.518e-44 |
| 25 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 95 | 1518 | 5.2e-45 | 9.678e-43 |
| 26 | REGULATION OF MAPK CASCADE | 68 | 660 | 7.634e-45 | 1.312e-42 |
| 27 | REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 40 | 138 | 8.179e-45 | 1.312e-42 |
| 28 | POSITIVE REGULATION OF MAPK CASCADE | 60 | 470 | 8.097e-45 | 1.312e-42 |
| 29 | RESPONSE TO ENDOGENOUS STIMULUS | 93 | 1450 | 7.867e-45 | 1.312e-42 |
| 30 | REGULATION OF CELLULAR COMPONENT MOVEMENT | 72 | 771 | 1.101e-44 | 1.708e-42 |
| 31 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 98 | 1672 | 4.031e-44 | 6.05e-42 |
| 32 | EXTRACELLULAR STRUCTURE ORGANIZATION | 51 | 304 | 4.508e-44 | 6.554e-42 |
| 33 | POSITIVE REGULATION OF LOCOMOTION | 56 | 420 | 8.605e-43 | 1.213e-40 |
| 34 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 78 | 1008 | 1.136e-42 | 1.555e-40 |
| 35 | CELL MOTILITY | 72 | 835 | 2.619e-42 | 3.385e-40 |
| 36 | LOCALIZATION OF CELL | 72 | 835 | 2.619e-42 | 3.385e-40 |
| 37 | REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 57 | 470 | 3.071e-41 | 3.862e-39 |
| 38 | MOVEMENT OF CELL OR SUBCELLULAR COMPONENT | 84 | 1275 | 7.335e-41 | 8.982e-39 |
| 39 | REGULATION OF CELL DEATH | 88 | 1472 | 1.12e-39 | 1.336e-37 |
| 40 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 85 | 1395 | 8.632e-39 | 1.004e-36 |
| 41 | POSITIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 46 | 289 | 1.294e-38 | 1.468e-36 |
| 42 | PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS | 98 | 1977 | 8.666e-38 | 9.6e-36 |
| 43 | RESPONSE TO EXTERNAL STIMULUS | 94 | 1821 | 1.942e-37 | 2.102e-35 |
| 44 | NEGATIVE REGULATION OF CELL DEATH | 68 | 872 | 5.039e-37 | 5.329e-35 |
| 45 | RESPONSE TO OXYGEN CONTAINING COMPOUND | 82 | 1381 | 1.917e-36 | 1.982e-34 |
| 46 | RESPONSE TO HORMONE | 68 | 893 | 2.26e-36 | 2.286e-34 |
| 47 | PEPTIDYL TYROSINE MODIFICATION | 38 | 186 | 3.243e-36 | 3.21e-34 |
| 48 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 75 | 1142 | 5.559e-36 | 5.389e-34 |
| 49 | REGULATION OF MAP KINASE ACTIVITY | 45 | 319 | 2.285e-35 | 2.17e-33 |
| 50 | RESPONSE TO GROWTH FACTOR | 52 | 475 | 2.903e-35 | 2.702e-33 |
| 51 | BIOLOGICAL ADHESION | 71 | 1032 | 3.27e-35 | 2.983e-33 |
| 52 | POSITIVE REGULATION OF MAP KINASE ACTIVITY | 38 | 207 | 2.391e-34 | 2.139e-32 |
| 53 | TISSUE DEVELOPMENT | 83 | 1518 | 2.639e-34 | 2.317e-32 |
| 54 | INOSITOL LIPID MEDIATED SIGNALING | 32 | 124 | 4.16e-34 | 3.585e-32 |
| 55 | CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND | 61 | 799 | 1.681e-32 | 1.422e-30 |
| 56 | SIGNAL TRANSDUCTION BY PROTEIN PHOSPHORYLATION | 46 | 404 | 6.565e-32 | 5.455e-30 |
| 57 | RESPONSE TO NITROGEN COMPOUND | 61 | 859 | 9.601e-31 | 7.838e-29 |
| 58 | REGULATION OF CELL ADHESION | 53 | 629 | 3.288e-30 | 2.638e-28 |
| 59 | VASCULATURE DEVELOPMENT | 47 | 469 | 4.364e-30 | 3.442e-28 |
| 60 | ANATOMICAL STRUCTURE FORMATION INVOLVED IN MORPHOGENESIS | 63 | 957 | 6.32e-30 | 4.901e-28 |
| 61 | CELLULAR RESPONSE TO NITROGEN COMPOUND | 48 | 505 | 1.112e-29 | 8.485e-28 |
| 62 | PEPTIDYL AMINO ACID MODIFICATION | 59 | 841 | 2.063e-29 | 1.548e-27 |
| 63 | ACTIVATION OF PROTEIN KINASE ACTIVITY | 38 | 279 | 2.657e-29 | 1.963e-27 |
| 64 | REGULATION OF CELL DIFFERENTIATION | 76 | 1492 | 4.249e-29 | 3.089e-27 |
| 65 | CARDIOVASCULAR SYSTEM DEVELOPMENT | 57 | 788 | 4.559e-29 | 3.214e-27 |
| 66 | CIRCULATORY SYSTEM DEVELOPMENT | 57 | 788 | 4.559e-29 | 3.214e-27 |
| 67 | REGULATION OF PEPTIDYL TYROSINE PHOSPHORYLATION | 34 | 213 | 1.22e-28 | 8.471e-27 |
| 68 | ANGIOGENESIS | 38 | 293 | 1.699e-28 | 1.162e-26 |
| 69 | TAXIS | 45 | 464 | 3.447e-28 | 2.324e-26 |
| 70 | CELLULAR RESPONSE TO HORMONE STIMULUS | 48 | 552 | 6.249e-28 | 4.154e-26 |
| 71 | REGULATION OF HYDROLASE ACTIVITY | 70 | 1327 | 1.59e-27 | 1.042e-25 |
| 72 | POSITIVE REGULATION OF PEPTIDYL TYROSINE PHOSPHORYLATION | 30 | 162 | 2.349e-27 | 1.518e-25 |
| 73 | CELL SUBSTRATE ADHESION | 30 | 164 | 3.452e-27 | 2.201e-25 |
| 74 | BLOOD VESSEL MORPHOGENESIS | 40 | 364 | 3.728e-27 | 2.344e-25 |
| 75 | LIPID PHOSPHORYLATION | 25 | 99 | 1.454e-26 | 9.023e-25 |
| 76 | RESPONSE TO PEPTIDE | 41 | 404 | 1.834e-26 | 1.123e-24 |
| 77 | ORGAN MORPHOGENESIS | 55 | 841 | 7.543e-26 | 4.558e-24 |
| 78 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 60 | 1021 | 8.118e-26 | 4.843e-24 |
| 79 | CELL DEVELOPMENT | 70 | 1426 | 1.168e-25 | 6.88e-24 |
| 80 | REGULATION OF EPITHELIAL CELL PROLIFERATION | 35 | 285 | 1.83e-25 | 1.064e-23 |
| 81 | CELL DEATH | 59 | 1001 | 1.95e-25 | 1.12e-23 |
| 82 | POSITIVE REGULATION OF EPITHELIAL CELL PROLIFERATION | 28 | 154 | 2.36e-25 | 1.339e-23 |
| 83 | POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 62 | 1152 | 1.157e-24 | 6.488e-23 |
| 84 | LEUKOCYTE MIGRATION | 33 | 259 | 1.451e-24 | 8.036e-23 |
| 85 | RESPONSE TO LIPID | 54 | 888 | 7.365e-24 | 4.032e-22 |
| 86 | REGULATION OF IMMUNE SYSTEM PROCESS | 67 | 1403 | 8.225e-24 | 4.45e-22 |
| 87 | CELLULAR RESPONSE TO PEPTIDE | 33 | 274 | 9.038e-24 | 4.834e-22 |
| 88 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 51 | 823 | 7.059e-23 | 3.732e-21 |
| 89 | PHOSPHATIDYLINOSITOL METABOLIC PROCESS | 28 | 193 | 1.54e-22 | 8.052e-21 |
| 90 | POSITIVE REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 19 | 62 | 3.049e-22 | 1.576e-20 |
| 91 | RESPONSE TO WOUNDING | 42 | 563 | 7.254e-22 | 3.709e-20 |
| 92 | CELL MATRIX ADHESION | 23 | 119 | 1.22e-21 | 6.102e-20 |
| 93 | NEUROGENESIS | 64 | 1402 | 1.207e-21 | 6.102e-20 |
| 94 | CELL PROLIFERATION | 45 | 672 | 1.619e-21 | 8.013e-20 |
| 95 | POSITIVE REGULATION OF ERK1 AND ERK2 CASCADE | 26 | 172 | 1.828e-21 | 8.953e-20 |
| 96 | POSITIVE REGULATION OF CELL ADHESION | 35 | 376 | 2.077e-21 | 1.007e-19 |
| 97 | REGULATION OF VASCULATURE DEVELOPMENT | 29 | 233 | 2.167e-21 | 1.039e-19 |
| 98 | IMMUNE SYSTEM PROCESS | 76 | 1984 | 2.508e-21 | 1.191e-19 |
| 99 | REGULATION OF RESPONSE TO STRESS | 65 | 1468 | 2.702e-21 | 1.27e-19 |
| 100 | REGULATION OF ERK1 AND ERK2 CASCADE | 29 | 238 | 3.968e-21 | 1.828e-19 |
| 101 | INTEGRIN MEDIATED SIGNALING PATHWAY | 20 | 82 | 3.946e-21 | 1.828e-19 |
| 102 | PHOSPHATIDYLINOSITOL 3 PHOSPHATE BIOSYNTHETIC PROCESS | 17 | 49 | 4.334e-21 | 1.977e-19 |
| 103 | POSITIVE REGULATION OF IMMUNE SYSTEM PROCESS | 50 | 867 | 4.594e-21 | 2.075e-19 |
| 104 | POSITIVE REGULATION OF HYDROLASE ACTIVITY | 51 | 905 | 4.765e-21 | 2.132e-19 |
| 105 | RESPONSE TO ABIOTIC STIMULUS | 54 | 1024 | 5.64e-21 | 2.499e-19 |
| 106 | REGULATION OF TRANSPORT | 71 | 1804 | 1.832e-20 | 8.043e-19 |
| 107 | REGULATION OF NEURON DEATH | 29 | 252 | 2.002e-20 | 8.704e-19 |
| 108 | LIPID MODIFICATION | 27 | 210 | 2.359e-20 | 1.016e-18 |
| 109 | POSITIVE REGULATION OF GENE EXPRESSION | 69 | 1733 | 4.04e-20 | 1.725e-18 |
| 110 | WOUND HEALING | 37 | 470 | 4.227e-20 | 1.788e-18 |
| 111 | EMBRYO DEVELOPMENT | 49 | 894 | 1.035e-19 | 4.339e-18 |
| 112 | REGULATION OF LIPID KINASE ACTIVITY | 16 | 48 | 1.365e-19 | 5.672e-18 |
| 113 | REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 15 | 40 | 2.369e-19 | 9.754e-18 |
| 114 | RESPONSE TO STEROID HORMONE | 37 | 497 | 2.769e-19 | 1.13e-17 |
| 115 | RESPONSE TO ORGANIC CYCLIC COMPOUND | 49 | 917 | 2.982e-19 | 1.207e-17 |
| 116 | ACTIVATION OF MAPK ACTIVITY | 22 | 137 | 6.543e-19 | 2.625e-17 |
| 117 | POSITIVE REGULATION OF TRANSPORT | 49 | 936 | 6.973e-19 | 2.773e-17 |
| 118 | CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 37 | 513 | 7.969e-19 | 3.142e-17 |
| 119 | PLATELET ACTIVATION | 22 | 142 | 1.461e-18 | 5.712e-17 |
| 120 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 68 | 1805 | 1.588e-18 | 6.156e-17 |
| 121 | REGULATION OF EPITHELIAL CELL MIGRATION | 23 | 166 | 3.122e-18 | 1.201e-16 |
| 122 | CELL ACTIVATION | 38 | 568 | 3.274e-18 | 1.249e-16 |
| 123 | POSITIVE REGULATION OF CELL DEATH | 39 | 605 | 4.024e-18 | 1.522e-16 |
| 124 | REGULATION OF ENDOTHELIAL CELL MIGRATION | 20 | 114 | 4.386e-18 | 1.646e-16 |
| 125 | POSITIVE REGULATION OF CELL DIVISION | 21 | 132 | 5.146e-18 | 1.915e-16 |
| 126 | PROTEIN AUTOPHOSPHORYLATION | 24 | 192 | 6.479e-18 | 2.392e-16 |
| 127 | REGULATION OF CELL CYCLE | 48 | 949 | 6.759e-18 | 2.476e-16 |
| 128 | TUBE DEVELOPMENT | 37 | 552 | 8.953e-18 | 3.254e-16 |
| 129 | PHOSPHATIDYLINOSITOL BIOSYNTHETIC PROCESS | 20 | 120 | 1.263e-17 | 4.52e-16 |
| 130 | POSITIVE REGULATION OF CHEMOTAXIS | 20 | 120 | 1.263e-17 | 4.52e-16 |
| 131 | REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 47 | 926 | 1.384e-17 | 4.915e-16 |
| 132 | POSITIVE REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 28 | 296 | 1.795e-17 | 6.328e-16 |
| 133 | GLYCEROPHOSPHOLIPID METABOLIC PROCESS | 28 | 297 | 1.962e-17 | 6.863e-16 |
| 134 | REGULATION OF CHEMOTAXIS | 23 | 180 | 1.981e-17 | 6.88e-16 |
| 135 | CELLULAR COMPONENT MORPHOGENESIS | 46 | 900 | 2.484e-17 | 8.562e-16 |
| 136 | REGULATION OF BODY FLUID LEVELS | 35 | 506 | 2.839e-17 | 9.713e-16 |
| 137 | FORMATION OF PRIMARY GERM LAYER | 19 | 110 | 4.177e-17 | 1.419e-15 |
| 138 | PEPTIDYL SERINE MODIFICATION | 21 | 148 | 5.832e-17 | 1.966e-15 |
| 139 | NEGATIVE REGULATION OF RESPONSE TO STIMULUS | 56 | 1360 | 7.909e-17 | 2.647e-15 |
| 140 | REGULATION OF DEVELOPMENTAL GROWTH | 27 | 289 | 9.563e-17 | 3.178e-15 |
| 141 | POSITIVE REGULATION OF VASCULATURE DEVELOPMENT | 20 | 133 | 1.026e-16 | 3.387e-15 |
| 142 | REGULATION OF CELL SUBSTRATE ADHESION | 22 | 173 | 1.126e-16 | 3.691e-15 |
| 143 | RESPONSE TO ESTROGEN | 24 | 218 | 1.258e-16 | 4.093e-15 |
| 144 | RESPONSE TO ACID CHEMICAL | 28 | 319 | 1.272e-16 | 4.112e-15 |
| 145 | GLYCEROLIPID METABOLIC PROCESS | 29 | 356 | 2.582e-16 | 8.287e-15 |
| 146 | NEURON PROJECTION DEVELOPMENT | 35 | 545 | 2.819e-16 | 8.984e-15 |
| 147 | REGULATION OF PHOSPHOLIPID METABOLIC PROCESS | 15 | 61 | 3.338e-16 | 1.056e-14 |
| 148 | POSITIVE REGULATION OF CELL CYCLE | 28 | 332 | 3.582e-16 | 1.126e-14 |
| 149 | REGULATION OF APOPTOTIC SIGNALING PATHWAY | 29 | 363 | 4.329e-16 | 1.352e-14 |
| 150 | EPITHELIUM DEVELOPMENT | 45 | 945 | 8.132e-16 | 2.523e-14 |
| 151 | REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 40 | 750 | 9.692e-16 | 2.987e-14 |
| 152 | REGULATION OF CELL DEVELOPMENT | 42 | 836 | 1.362e-15 | 4.17e-14 |
| 153 | GASTRULATION | 20 | 155 | 2.181e-15 | 6.633e-14 |
| 154 | REGULATION OF INTRACELLULAR TRANSPORT | 36 | 621 | 2.466e-15 | 7.451e-14 |
| 155 | POSITIVE REGULATION OF CELLULAR PROTEIN LOCALIZATION | 28 | 360 | 2.851e-15 | 8.559e-14 |
| 156 | REGULATION OF IMMUNE RESPONSE | 42 | 858 | 3.354e-15 | 1e-13 |
| 157 | PHOSPHOLIPID METABOLIC PROCESS | 28 | 364 | 3.775e-15 | 1.119e-13 |
| 158 | SINGLE ORGANISM CELL ADHESION | 31 | 459 | 4.182e-15 | 1.232e-13 |
| 159 | POSITIVE REGULATION OF EPITHELIAL CELL MIGRATION | 17 | 103 | 4.367e-15 | 1.278e-13 |
| 160 | REGULATION OF GROWTH | 36 | 633 | 4.442e-15 | 1.292e-13 |
| 161 | REGULATION OF LIPID METABOLIC PROCESS | 25 | 282 | 4.751e-15 | 1.373e-13 |
| 162 | REGULATION OF GTPASE ACTIVITY | 37 | 673 | 5.129e-15 | 1.473e-13 |
| 163 | RESPONSE TO OXYGEN LEVELS | 26 | 311 | 5.358e-15 | 1.52e-13 |
| 164 | HEMOSTASIS | 26 | 311 | 5.358e-15 | 1.52e-13 |
| 165 | RESPONSE TO CYTOKINE | 38 | 714 | 5.933e-15 | 1.673e-13 |
| 166 | NEURON PROJECTION MORPHOGENESIS | 29 | 402 | 6.288e-15 | 1.763e-13 |
| 167 | POSITIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 30 | 437 | 7.786e-15 | 2.169e-13 |
| 168 | APOPTOTIC SIGNALING PATHWAY | 25 | 289 | 8.386e-15 | 2.323e-13 |
| 169 | EMBRYONIC MORPHOGENESIS | 33 | 539 | 8.559e-15 | 2.356e-13 |
| 170 | POSITIVE REGULATION OF CELL DEVELOPMENT | 31 | 472 | 8.969e-15 | 2.455e-13 |
| 171 | POSITIVE REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION | 32 | 514 | 1.425e-14 | 3.877e-13 |
| 172 | POSITIVE REGULATION OF SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 12 | 39 | 1.625e-14 | 4.385e-13 |
| 173 | NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 44 | 983 | 1.631e-14 | 4.385e-13 |
| 174 | UROGENITAL SYSTEM DEVELOPMENT | 25 | 299 | 1.837e-14 | 4.913e-13 |
| 175 | RESPONSE TO AMINO ACID | 17 | 112 | 1.859e-14 | 4.93e-13 |
| 176 | CELL PROJECTION ORGANIZATION | 42 | 902 | 1.865e-14 | 4.93e-13 |
| 177 | POSITIVE REGULATION OF CELL PROJECTION ORGANIZATION | 25 | 303 | 2.493e-14 | 6.552e-13 |
| 178 | CELLULAR RESPONSE TO LIPID | 30 | 457 | 2.545e-14 | 6.653e-13 |
| 179 | RESPONSE TO ALCOHOL | 27 | 362 | 2.566e-14 | 6.67e-13 |
| 180 | HEAD DEVELOPMENT | 37 | 709 | 2.59e-14 | 6.696e-13 |
| 181 | CENTRAL NERVOUS SYSTEM DEVELOPMENT | 41 | 872 | 2.897e-14 | 7.447e-13 |
| 182 | GLAND DEVELOPMENT | 28 | 395 | 2.953e-14 | 7.55e-13 |
| 183 | CELL SUBSTRATE JUNCTION ASSEMBLY | 12 | 41 | 3.215e-14 | 8.174e-13 |
| 184 | POSITIVE REGULATION OF DEVELOPMENTAL GROWTH | 19 | 156 | 3.285e-14 | 8.307e-13 |
| 185 | RESPONSE TO FIBROBLAST GROWTH FACTOR | 17 | 116 | 3.392e-14 | 8.527e-13 |
| 186 | REGULATION OF ENDOTHELIAL CELL PROLIFERATION | 16 | 98 | 3.408e-14 | 8.527e-13 |
| 187 | POSITIVE REGULATION OF ENDOTHELIAL CELL MIGRATION | 14 | 67 | 3.875e-14 | 9.643e-13 |
| 188 | EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 16 | 99 | 4.023e-14 | 9.958e-13 |
| 189 | POSITIVE REGULATION OF INTRACELLULAR TRANSPORT | 27 | 370 | 4.36e-14 | 1.073e-12 |
| 190 | POSITIVE REGULATION OF LIPID KINASE ACTIVITY | 11 | 32 | 5.073e-14 | 1.242e-12 |
| 191 | NEURON DEVELOPMENT | 36 | 687 | 5.335e-14 | 1.3e-12 |
| 192 | CELL CHEMOTAXIS | 19 | 162 | 6.607e-14 | 1.601e-12 |
| 193 | POSITIVE REGULATION OF DNA METABOLIC PROCESS | 20 | 185 | 6.82e-14 | 1.644e-12 |
| 194 | POSITIVE REGULATION OF DNA REPLICATION | 15 | 86 | 7.942e-14 | 1.905e-12 |
| 195 | REGULATION OF NEURON DIFFERENTIATION | 32 | 554 | 1.123e-13 | 2.68e-12 |
| 196 | RESPONSE TO ESTRADIOL | 18 | 146 | 1.282e-13 | 3.029e-12 |
| 197 | CELLULAR RESPONSE TO STRESS | 55 | 1565 | 1.282e-13 | 3.029e-12 |
| 198 | REGULATION OF CELL PROJECTION ORGANIZATION | 32 | 558 | 1.367e-13 | 3.212e-12 |
| 199 | REGULATION OF NEURON APOPTOTIC PROCESS | 20 | 192 | 1.388e-13 | 3.245e-12 |
| 200 | REGULATION OF CELLULAR LOCALIZATION | 49 | 1277 | 1.481e-13 | 3.445e-12 |
| 201 | NEURON DIFFERENTIATION | 40 | 874 | 1.496e-13 | 3.464e-12 |
| 202 | REGULATION OF CELL MATRIX ADHESION | 15 | 90 | 1.601e-13 | 3.689e-12 |
| 203 | VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 14 | 74 | 1.679e-13 | 3.848e-12 |
| 204 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 38 | 801 | 2.192e-13 | 4.999e-12 |
| 205 | POSITIVE REGULATION OF MITOCHONDRIAL OUTER MEMBRANE PERMEABILIZATION INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 11 | 36 | 2.266e-13 | 5.144e-12 |
| 206 | MULTICELLULAR ORGANISM METABOLIC PROCESS | 15 | 93 | 2.648e-13 | 5.98e-12 |
| 207 | POSITIVE REGULATION OF ORGANELLE ORGANIZATION | 32 | 573 | 2.81e-13 | 6.316e-12 |
| 208 | REGULATION OF DNA METABOLIC PROCESS | 25 | 340 | 3.399e-13 | 7.603e-12 |
| 209 | MULTICELLULAR ORGANISMAL MACROMOLECULE METABOLIC PROCESS | 14 | 79 | 4.348e-13 | 9.68e-12 |
| 210 | REGULATION OF CELLULAR PROTEIN LOCALIZATION | 31 | 552 | 5.911e-13 | 1.31e-11 |
| 211 | REGULATION OF PROTEIN INSERTION INTO MITOCHONDRIAL MEMBRANE INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 10 | 29 | 7.117e-13 | 1.562e-11 |
| 212 | POSITIVE REGULATION OF PROTEIN INSERTION INTO MITOCHONDRIAL MEMBRANE INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 10 | 29 | 7.117e-13 | 1.562e-11 |
| 213 | GLYCEROLIPID BIOSYNTHETIC PROCESS | 20 | 211 | 8.271e-13 | 1.807e-11 |
| 214 | ENDODERMAL CELL DIFFERENTIATION | 11 | 40 | 8.366e-13 | 1.819e-11 |
| 215 | POSITIVE REGULATION OF GROWTH | 21 | 238 | 8.817e-13 | 1.908e-11 |
| 216 | POSITIVE REGULATION OF NEURON DEATH | 13 | 67 | 8.916e-13 | 1.921e-11 |
| 217 | POSITIVE REGULATION OF PHOSPHOLIPASE ACTIVITY | 12 | 53 | 9.585e-13 | 2.055e-11 |
| 218 | FIBROBLAST GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 14 | 84 | 1.053e-12 | 2.247e-11 |
| 219 | POSITIVE REGULATION OF ENDOTHELIAL CELL PROLIFERATION | 13 | 68 | 1.091e-12 | 2.318e-11 |
| 220 | REGULATION OF CELL DIVISION | 22 | 272 | 1.434e-12 | 3.034e-11 |
| 221 | POSITIVE REGULATION OF PHOSPHOLIPID METABOLIC PROCESS | 11 | 42 | 1.517e-12 | 3.195e-11 |
| 222 | REGULATION OF MEMBRANE PERMEABILITY | 13 | 70 | 1.617e-12 | 3.389e-11 |
| 223 | RESPONSE TO DRUG | 27 | 431 | 1.652e-12 | 3.446e-11 |
| 224 | REGULATION OF CELLULAR RESPONSE TO STRESS | 34 | 691 | 1.697e-12 | 3.524e-11 |
| 225 | REGULATION OF MITOCHONDRIAL OUTER MEMBRANE PERMEABILIZATION INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 11 | 43 | 2.018e-12 | 4.173e-11 |
| 226 | REGULATION OF PROTEIN LOCALIZATION | 40 | 950 | 2.117e-12 | 4.359e-11 |
| 227 | REPRODUCTIVE SYSTEM DEVELOPMENT | 26 | 408 | 2.982e-12 | 6.085e-11 |
| 228 | REGULATION OF NEURON PROJECTION DEVELOPMENT | 26 | 408 | 2.982e-12 | 6.085e-11 |
| 229 | REGULATION OF REACTIVE OXYGEN SPECIES METABOLIC PROCESS | 17 | 152 | 3.125e-12 | 6.35e-11 |
| 230 | GROWTH | 26 | 410 | 3.33e-12 | 6.738e-11 |
| 231 | REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 17 | 153 | 3.48e-12 | 7.009e-11 |
| 232 | REGULATION OF RESPONSE TO WOUNDING | 26 | 413 | 3.927e-12 | 7.875e-11 |
| 233 | POSITIVE REGULATION OF SMOOTH MUSCLE CELL PROLIFERATION | 12 | 60 | 4.675e-12 | 9.333e-11 |
| 234 | RESPONSE TO INSULIN | 19 | 205 | 4.693e-12 | 9.333e-11 |
| 235 | POSITIVE REGULATION OF NEURON PROJECTION DEVELOPMENT | 20 | 232 | 4.836e-12 | 9.575e-11 |
| 236 | FC RECEPTOR SIGNALING PATHWAY | 19 | 206 | 5.117e-12 | 1.009e-10 |
| 237 | PHOSPHOLIPID BIOSYNTHETIC PROCESS | 20 | 235 | 6.129e-12 | 1.203e-10 |
| 238 | CELLULAR RESPONSE TO EXTERNAL STIMULUS | 21 | 264 | 6.51e-12 | 1.273e-10 |
| 239 | CELLULAR RESPONSE TO CYTOKINE STIMULUS | 31 | 606 | 6.65e-12 | 1.295e-10 |
| 240 | REGULATION OF ENDOTHELIAL CELL CHEMOTAXIS | 8 | 17 | 7.406e-12 | 1.436e-10 |
| 241 | RESPONSE TO GLUCAGON | 11 | 48 | 7.526e-12 | 1.453e-10 |
| 242 | TISSUE MORPHOGENESIS | 29 | 533 | 7.615e-12 | 1.464e-10 |
| 243 | REGULATION OF DNA REPLICATION | 17 | 161 | 7.984e-12 | 1.529e-10 |
| 244 | ERBB SIGNALING PATHWAY | 13 | 79 | 8.186e-12 | 1.561e-10 |
| 245 | NEGATIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 14 | 98 | 9.354e-12 | 1.777e-10 |
| 246 | REGULATION OF PHOSPHOLIPASE ACTIVITY | 12 | 64 | 1.053e-11 | 1.991e-10 |
| 247 | ENDODERM FORMATION | 11 | 50 | 1.219e-11 | 2.296e-10 |
| 248 | FC EPSILON RECEPTOR SIGNALING PATHWAY | 16 | 142 | 1.234e-11 | 2.316e-10 |
| 249 | IMMUNE SYSTEM DEVELOPMENT | 30 | 582 | 1.241e-11 | 2.319e-10 |
| 250 | CELL MORPHOGENESIS INVOLVED IN NEURON DIFFERENTIATION | 24 | 368 | 1.326e-11 | 2.467e-10 |
| 251 | REGULATION OF PROTEIN KINASE B SIGNALING | 15 | 121 | 1.367e-11 | 2.534e-10 |
| 252 | MORPHOGENESIS OF A BRANCHING STRUCTURE | 17 | 167 | 1.443e-11 | 2.665e-10 |
| 253 | POSITIVE REGULATION OF NEURON DIFFERENTIATION | 22 | 306 | 1.486e-11 | 2.733e-10 |
| 254 | SUBSTRATE ADHESION DEPENDENT CELL SPREADING | 10 | 38 | 1.531e-11 | 2.804e-10 |
| 255 | POSITIVE REGULATION OF LIPASE ACTIVITY | 12 | 66 | 1.545e-11 | 2.82e-10 |
| 256 | POSITIVE REGULATION OF MITOTIC CELL CYCLE | 15 | 123 | 1.738e-11 | 3.16e-10 |
| 257 | ERBB2 SIGNALING PATHWAY | 10 | 39 | 2.038e-11 | 3.681e-10 |
| 258 | POSITIVE REGULATION OF CELL ACTIVATION | 22 | 311 | 2.041e-11 | 3.681e-10 |
| 259 | POSITIVE REGULATION OF APOPTOTIC SIGNALING PATHWAY | 17 | 171 | 2.113e-11 | 3.796e-10 |
| 260 | RESPIRATORY SYSTEM DEVELOPMENT | 18 | 197 | 2.217e-11 | 3.968e-10 |
| 261 | REGULATION OF CELL ACTIVATION | 27 | 484 | 2.406e-11 | 4.29e-10 |
| 262 | REGULATION OF HEMOPOIESIS | 22 | 314 | 2.462e-11 | 4.373e-10 |
| 263 | REGULATION OF CELL CELL ADHESION | 24 | 380 | 2.593e-11 | 4.587e-10 |
| 264 | DEVELOPMENTAL PROCESS INVOLVED IN REPRODUCTION | 30 | 602 | 2.859e-11 | 5.04e-10 |
| 265 | REGULATION OF ORGANELLE ORGANIZATION | 43 | 1178 | 3.028e-11 | 5.317e-10 |
| 266 | CELLULAR RESPONSE TO ACID CHEMICAL | 17 | 175 | 3.061e-11 | 5.355e-10 |
| 267 | POSITIVE REGULATION OF LIPID METABOLIC PROCESS | 15 | 128 | 3.11e-11 | 5.42e-10 |
| 268 | CELL JUNCTION ASSEMBLY | 15 | 129 | 3.483e-11 | 6.047e-10 |
| 269 | INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO DNA DAMAGE | 12 | 71 | 3.815e-11 | 6.575e-10 |
| 270 | ENDODERM DEVELOPMENT | 12 | 71 | 3.815e-11 | 6.575e-10 |
| 271 | REGULATION OF LEUKOCYTE DIFFERENTIATION | 19 | 232 | 4.122e-11 | 7.077e-10 |
| 272 | IMMUNE RESPONSE REGULATING CELL SURFACE RECEPTOR SIGNALING PATHWAY | 22 | 323 | 4.267e-11 | 7.299e-10 |
| 273 | NEURON PROJECTION GUIDANCE | 18 | 205 | 4.313e-11 | 7.352e-10 |
| 274 | NEGATIVE REGULATION OF CELL COMMUNICATION | 43 | 1192 | 4.418e-11 | 7.502e-10 |
| 275 | CELLULAR LIPID METABOLIC PROCESS | 37 | 913 | 4.776e-11 | 8.08e-10 |
| 276 | POSITIVE REGULATION OF STAT CASCADE | 12 | 73 | 5.366e-11 | 9.014e-10 |
| 277 | POSITIVE REGULATION OF JAK STAT CASCADE | 12 | 73 | 5.366e-11 | 9.014e-10 |
| 278 | RESPONSE TO KETONE | 17 | 182 | 5.723e-11 | 9.579e-10 |
| 279 | MITOCHONDRIAL MEMBRANE ORGANIZATION | 13 | 92 | 6.049e-11 | 1.009e-09 |
| 280 | REGULATION OF MITOTIC CELL CYCLE | 26 | 468 | 6.327e-11 | 1.051e-09 |
| 281 | RESPONSE TO MECHANICAL STIMULUS | 18 | 210 | 6.438e-11 | 1.066e-09 |
| 282 | CELL JUNCTION ORGANIZATION | 17 | 185 | 7.419e-11 | 1.224e-09 |
| 283 | NEPHRON DEVELOPMENT | 14 | 115 | 8.549e-11 | 1.406e-09 |
| 284 | POSITIVE REGULATION OF CELL CELL ADHESION | 19 | 243 | 9.181e-11 | 1.504e-09 |
| 285 | SPROUTING ANGIOGENESIS | 10 | 45 | 9.611e-11 | 1.569e-09 |
| 286 | RESPONSE TO EXTRACELLULAR STIMULUS | 25 | 441 | 9.866e-11 | 1.605e-09 |
| 287 | REGULATION OF PHOSPHOPROTEIN PHOSPHATASE ACTIVITY | 11 | 60 | 1.008e-10 | 1.634e-09 |
| 288 | SIGNAL TRANSDUCTION IN ABSENCE OF LIGAND | 9 | 33 | 1.148e-10 | 1.848e-09 |
| 289 | EXTRINSIC APOPTOTIC SIGNALING PATHWAY IN ABSENCE OF LIGAND | 9 | 33 | 1.148e-10 | 1.848e-09 |
| 290 | MEMORY | 13 | 98 | 1.368e-10 | 2.195e-09 |
| 291 | RESPONSE TO LIGHT STIMULUS | 20 | 280 | 1.463e-10 | 2.34e-09 |
| 292 | MYELOID LEUKOCYTE MIGRATION | 13 | 99 | 1.558e-10 | 2.475e-09 |
| 293 | POSITIVE REGULATION OF CELL SUBSTRATE ADHESION | 13 | 99 | 1.558e-10 | 2.475e-09 |
| 294 | REGULATION OF JAK STAT CASCADE | 15 | 144 | 1.699e-10 | 2.68e-09 |
| 295 | REGULATION OF STAT CASCADE | 15 | 144 | 1.699e-10 | 2.68e-09 |
| 296 | REGULATION OF SMOOTH MUSCLE CELL PROLIFERATION | 13 | 100 | 1.773e-10 | 2.786e-09 |
| 297 | REGULATION OF FIBROBLAST PROLIFERATION | 12 | 81 | 1.9e-10 | 2.976e-09 |
| 298 | NEGATIVE REGULATION OF NEURON DEATH | 16 | 171 | 2.081e-10 | 3.249e-09 |
| 299 | REGULATION OF LIPASE ACTIVITY | 12 | 83 | 2.548e-10 | 3.965e-09 |
| 300 | POSITIVE CHEMOTAXIS | 9 | 36 | 2.717e-10 | 4.214e-09 |
| 301 | GLIOGENESIS | 16 | 175 | 2.939e-10 | 4.518e-09 |
| 302 | TISSUE MIGRATION | 12 | 84 | 2.942e-10 | 4.518e-09 |
| 303 | HOMEOSTASIS OF NUMBER OF CELLS | 16 | 175 | 2.939e-10 | 4.518e-09 |
| 304 | CELLULAR RESPONSE TO ORGANIC CYCLIC COMPOUND | 25 | 465 | 3.003e-10 | 4.596e-09 |
| 305 | PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 8 | 25 | 3.037e-10 | 4.633e-09 |
| 306 | CELLULAR RESPONSE TO ABIOTIC STIMULUS | 19 | 263 | 3.544e-10 | 5.389e-09 |
| 307 | REGULATION OF BLOOD VESSEL ENDOTHELIAL CELL MIGRATION | 10 | 51 | 3.618e-10 | 5.484e-09 |
| 308 | INTRINSIC APOPTOTIC SIGNALING PATHWAY | 15 | 152 | 3.665e-10 | 5.538e-09 |
| 309 | NEGATIVE REGULATION OF CELL CYCLE | 24 | 433 | 3.771e-10 | 5.679e-09 |
| 310 | POSITIVE REGULATION OF REACTIVE OXYGEN SPECIES METABOLIC PROCESS | 12 | 86 | 3.899e-10 | 5.853e-09 |
| 311 | REGULATION OF TYROSINE PHOSPHORYLATION OF STAT PROTEIN | 11 | 68 | 4.151e-10 | 6.211e-09 |
| 312 | AMEBOIDAL TYPE CELL MIGRATION | 15 | 154 | 4.41e-10 | 6.576e-09 |
| 313 | CELL PART MORPHOGENESIS | 29 | 633 | 4.523e-10 | 6.724e-09 |
| 314 | REGULATION OF CELL MORPHOGENESIS | 27 | 552 | 4.558e-10 | 6.754e-09 |
| 315 | CELLULAR RESPONSE TO GLUCAGON STIMULUS | 9 | 38 | 4.609e-10 | 6.808e-09 |
| 316 | HOMEOSTATIC PROCESS | 44 | 1337 | 4.758e-10 | 7.006e-09 |
| 317 | POSITIVE REGULATION OF LEUKOCYTE DIFFERENTIATION | 14 | 131 | 4.976e-10 | 7.281e-09 |
| 318 | BRANCHING MORPHOGENESIS OF AN EPITHELIAL TUBE | 14 | 131 | 4.976e-10 | 7.281e-09 |
| 319 | NEGATIVE REGULATION OF ANOIKIS | 7 | 17 | 5.126e-10 | 7.477e-09 |
| 320 | POSITIVE REGULATION OF FIBROBLAST PROLIFERATION | 10 | 53 | 5.409e-10 | 7.865e-09 |
| 321 | CELL CELL SIGNALING | 32 | 767 | 5.498e-10 | 7.97e-09 |
| 322 | REGULATION OF PHOSPHOLIPASE C ACTIVITY | 9 | 39 | 5.931e-10 | 8.57e-09 |
| 323 | REGULATION OF CYTOPLASMIC TRANSPORT | 25 | 481 | 6.062e-10 | 8.732e-09 |
| 324 | RESPONSE TO TOXIC SUBSTANCE | 18 | 241 | 6.134e-10 | 8.809e-09 |
| 325 | REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 21 | 337 | 6.153e-10 | 8.81e-09 |
| 326 | EPIDERMAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 10 | 55 | 7.948e-10 | 1.134e-08 |
| 327 | RESPONSE TO RADIATION | 23 | 413 | 8.164e-10 | 1.162e-08 |
| 328 | GLIAL CELL DIFFERENTIATION | 14 | 136 | 8.202e-10 | 1.164e-08 |
| 329 | POSITIVE REGULATION OF CELL CYCLE PROCESS | 18 | 247 | 9.117e-10 | 1.289e-08 |
| 330 | CELL CYCLE | 43 | 1316 | 9.687e-10 | 1.366e-08 |
| 331 | INFLAMMATORY RESPONSE | 24 | 454 | 9.747e-10 | 1.37e-08 |
| 332 | POSITIVE REGULATION OF HEMOPOIESIS | 15 | 163 | 9.797e-10 | 1.373e-08 |
| 333 | SENSORY ORGAN DEVELOPMENT | 25 | 493 | 1.007e-09 | 1.407e-08 |
| 334 | RESPONSE TO NUTRIENT | 16 | 191 | 1.072e-09 | 1.493e-08 |
| 335 | POSITIVE REGULATION OF ENDOTHELIAL CELL CHEMOTAXIS | 6 | 11 | 1.114e-09 | 1.547e-08 |
| 336 | SINGLE ORGANISM BEHAVIOR | 22 | 384 | 1.146e-09 | 1.587e-08 |
| 337 | COGNITION | 18 | 251 | 1.18e-09 | 1.624e-08 |
| 338 | OSSIFICATION | 18 | 251 | 1.18e-09 | 1.624e-08 |
| 339 | LEUKOCYTE CHEMOTAXIS | 13 | 117 | 1.293e-09 | 1.769e-08 |
| 340 | MAMMARY GLAND DEVELOPMENT | 13 | 117 | 1.293e-09 | 1.769e-08 |
| 341 | CELL CYCLE PROCESS | 38 | 1081 | 1.459e-09 | 1.99e-08 |
| 342 | REGULATION OF AXONOGENESIS | 15 | 168 | 1.493e-09 | 2.032e-08 |
| 343 | REGULATION OF POLYSACCHARIDE METABOLIC PROCESS | 9 | 43 | 1.515e-09 | 2.055e-08 |
| 344 | ACTIVATION OF IMMUNE RESPONSE | 23 | 427 | 1.552e-09 | 2.099e-08 |
| 345 | POSITIVE REGULATION OF SMOOTH MUSCLE CELL MIGRATION | 8 | 30 | 1.562e-09 | 2.101e-08 |
| 346 | CELLULAR RESPONSE TO VASCULAR ENDOTHELIAL GROWTH FACTOR STIMULUS | 8 | 30 | 1.562e-09 | 2.101e-08 |
| 347 | HEART DEVELOPMENT | 24 | 466 | 1.636e-09 | 2.194e-08 |
| 348 | REGULATION OF TYROSINE PHOSPHORYLATION OF STAT3 PROTEIN | 9 | 44 | 1.885e-09 | 2.52e-08 |
| 349 | POSITIVE REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 13 | 121 | 1.966e-09 | 2.621e-08 |
| 350 | CELLULAR RESPONSE TO GROWTH HORMONE STIMULUS | 7 | 20 | 1.982e-09 | 2.636e-08 |
| 351 | REPRODUCTION | 42 | 1297 | 2.054e-09 | 2.722e-08 |
| 352 | REGULATION OF CELL SIZE | 15 | 172 | 2.07e-09 | 2.736e-08 |
| 353 | HEMIDESMOSOME ASSEMBLY | 6 | 12 | 2.206e-09 | 2.908e-08 |
| 354 | MORPHOGENESIS OF AN EPITHELIUM | 22 | 400 | 2.443e-09 | 3.211e-08 |
| 355 | LIPID METABOLIC PROCESS | 39 | 1158 | 2.871e-09 | 3.752e-08 |
| 356 | POSITIVE REGULATION OF PROTEIN KINASE B SIGNALING | 11 | 81 | 2.87e-09 | 3.752e-08 |
| 357 | POSITIVE REGULATION OF IMMUNE RESPONSE | 26 | 563 | 3.269e-09 | 4.26e-08 |
| 358 | RESPONSE TO ANTIBIOTIC | 9 | 47 | 3.513e-09 | 4.554e-08 |
| 359 | POSITIVE REGULATION OF NEURON APOPTOTIC PROCESS | 9 | 47 | 3.513e-09 | 4.554e-08 |
| 360 | NEGATIVE REGULATION OF CELL DIFFERENTIATION | 27 | 609 | 3.821e-09 | 4.938e-08 |
| 361 | PALLIUM DEVELOPMENT | 14 | 153 | 3.869e-09 | 4.987e-08 |
| 362 | ERK1 AND ERK2 CASCADE | 7 | 22 | 4.275e-09 | 5.494e-08 |
| 363 | CYTOKINE MEDIATED SIGNALING PATHWAY | 23 | 452 | 4.583e-09 | 5.875e-08 |
| 364 | PLATELET DERIVED GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 8 | 34 | 4.652e-09 | 5.947e-08 |
| 365 | REGULATION OF HOMOTYPIC CELL CELL ADHESION | 19 | 307 | 4.669e-09 | 5.952e-08 |
| 366 | RESPONSE TO BIOTIC STIMULUS | 33 | 886 | 4.94e-09 | 6.28e-08 |
| 367 | PLATELET DEGRANULATION | 12 | 107 | 5.083e-09 | 6.445e-08 |
| 368 | GLOMERULUS DEVELOPMENT | 9 | 49 | 5.19e-09 | 6.562e-08 |
| 369 | REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 16 | 213 | 5.221e-09 | 6.583e-08 |
| 370 | NEGATIVE REGULATION OF PROTEIN METABOLIC PROCESS | 37 | 1087 | 5.859e-09 | 7.368e-08 |
| 371 | REGULATION OF DEPHOSPHORYLATION | 14 | 158 | 5.878e-09 | 7.371e-08 |
| 372 | LIPID BIOSYNTHETIC PROCESS | 25 | 539 | 6.137e-09 | 7.676e-08 |
| 373 | STAT CASCADE | 9 | 50 | 6.264e-09 | 7.793e-08 |
| 374 | JAK STAT CASCADE | 9 | 50 | 6.264e-09 | 7.793e-08 |
| 375 | POSITIVE REGULATION OF LEUKOCYTE MIGRATION | 12 | 109 | 6.294e-09 | 7.81e-08 |
| 376 | VASCULAR ENDOTHELIAL GROWTH FACTOR SIGNALING PATHWAY | 6 | 14 | 7.03e-09 | 8.699e-08 |
| 377 | CELL GROWTH | 13 | 135 | 7.578e-09 | 9.353e-08 |
| 378 | CELL CYCLE G1 S PHASE TRANSITION | 12 | 111 | 7.758e-09 | 9.514e-08 |
| 379 | G1 S TRANSITION OF MITOTIC CELL CYCLE | 12 | 111 | 7.758e-09 | 9.514e-08 |
| 380 | RESPONSE TO OXIDATIVE STRESS | 20 | 352 | 7.77e-09 | 9.514e-08 |
| 381 | EPITHELIAL CELL PROLIFERATION | 11 | 89 | 7.957e-09 | 9.717e-08 |
| 382 | POSITIVE REGULATION OF AXONOGENESIS | 10 | 69 | 7.998e-09 | 9.742e-08 |
| 383 | POSITIVE REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 14 | 162 | 8.122e-09 | 9.867e-08 |
| 384 | REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 16 | 220 | 8.297e-09 | 1.005e-07 |
| 385 | REGULATION OF ANOIKIS | 7 | 24 | 8.502e-09 | 1.025e-07 |
| 386 | REGULATION OF PROTEIN PHOSPHATASE TYPE 2A ACTIVITY | 7 | 24 | 8.502e-09 | 1.025e-07 |
| 387 | RESPONSE TO REACTIVE OXYGEN SPECIES | 15 | 191 | 8.718e-09 | 1.048e-07 |
| 388 | REGULATION OF CELLULAR COMPONENT BIOGENESIS | 30 | 767 | 8.83e-09 | 1.059e-07 |
| 389 | POSITIVE REGULATION OF TYROSINE PHOSPHORYLATION OF STAT3 PROTEIN | 8 | 37 | 9.596e-09 | 1.148e-07 |
| 390 | CELL CYCLE PHASE TRANSITION | 17 | 255 | 1.035e-08 | 1.235e-07 |
| 391 | CELLULAR RESPONSE TO AMINO ACID STIMULUS | 9 | 53 | 1.074e-08 | 1.279e-07 |
| 392 | CONNECTIVE TISSUE DEVELOPMENT | 15 | 194 | 1.077e-08 | 1.279e-07 |
| 393 | POSITIVE REGULATION OF BLOOD VESSEL ENDOTHELIAL CELL MIGRATION | 7 | 25 | 1.169e-08 | 1.384e-07 |
| 394 | REGULATION OF METAL ION TRANSPORT | 19 | 325 | 1.18e-08 | 1.393e-07 |
| 395 | BEHAVIOR | 24 | 516 | 1.193e-08 | 1.405e-07 |
| 396 | REGULATION OF STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE | 15 | 197 | 1.326e-08 | 1.558e-07 |
| 397 | RESPONSE TO INORGANIC SUBSTANCE | 23 | 479 | 1.358e-08 | 1.592e-07 |
| 398 | TELENCEPHALON DEVELOPMENT | 16 | 228 | 1.379e-08 | 1.612e-07 |
| 399 | REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 20 | 365 | 1.43e-08 | 1.667e-07 |
| 400 | ASTROCYTE DIFFERENTIATION | 8 | 39 | 1.498e-08 | 1.739e-07 |
| 401 | PEPTIDYL TYROSINE AUTOPHOSPHORYLATION | 8 | 39 | 1.498e-08 | 1.739e-07 |
| 402 | MESODERM DEVELOPMENT | 12 | 118 | 1.561e-08 | 1.807e-07 |
| 403 | CELL CELL ADHESION | 26 | 608 | 1.583e-08 | 1.824e-07 |
| 404 | MESODERMAL CELL DIFFERENTIATION | 7 | 26 | 1.584e-08 | 1.824e-07 |
| 405 | NEGATIVE REGULATION OF APOPTOTIC SIGNALING PATHWAY | 15 | 200 | 1.625e-08 | 1.867e-07 |
| 406 | REGULATION OF CARBOHYDRATE METABOLIC PROCESS | 14 | 172 | 1.752e-08 | 2.008e-07 |
| 407 | REGULATION OF SYNAPSE STRUCTURE OR ACTIVITY | 16 | 232 | 1.764e-08 | 2.016e-07 |
| 408 | POSITIVE REGULATION OF CARBOHYDRATE METABOLIC PROCESS | 10 | 75 | 1.833e-08 | 2.086e-07 |
| 409 | REGULATION OF HOMEOSTATIC PROCESS | 22 | 447 | 1.838e-08 | 2.086e-07 |
| 410 | POSITIVE REGULATION OF VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 6 | 16 | 1.838e-08 | 2.086e-07 |
| 411 | POSITIVE REGULATION OF CELL MATRIX ADHESION | 8 | 40 | 1.854e-08 | 2.099e-07 |
| 412 | CELLULAR RESPONSE TO INSULIN STIMULUS | 13 | 146 | 1.96e-08 | 2.214e-07 |
| 413 | ENDOTHELIAL CELL MIGRATION | 9 | 57 | 2.094e-08 | 2.359e-07 |
| 414 | REGULATION OF VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 7 | 27 | 2.117e-08 | 2.379e-07 |
| 415 | INTERSPECIES INTERACTION BETWEEN ORGANISMS | 27 | 662 | 2.204e-08 | 2.465e-07 |
| 416 | SYMBIOSIS ENCOMPASSING MUTUALISM THROUGH PARASITISM | 27 | 662 | 2.204e-08 | 2.465e-07 |
| 417 | RESPONSE TO VITAMIN | 11 | 98 | 2.224e-08 | 2.482e-07 |
| 418 | RESPONSE TO CORTICOSTEROID | 14 | 176 | 2.349e-08 | 2.614e-07 |
| 419 | MITOCHONDRIAL TRANSPORT | 14 | 177 | 2.524e-08 | 2.803e-07 |
| 420 | REGULATION OF DEFENSE RESPONSE | 29 | 759 | 2.664e-08 | 2.951e-07 |
| 421 | NEGATIVE REGULATION OF TRANSPORT | 22 | 458 | 2.835e-08 | 3.133e-07 |
| 422 | NEGATIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 24 | 541 | 2.938e-08 | 3.232e-07 |
| 423 | NEGATIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 24 | 541 | 2.938e-08 | 3.232e-07 |
| 424 | CELLULAR RESPONSE TO MECHANICAL STIMULUS | 10 | 80 | 3.458e-08 | 3.795e-07 |
| 425 | CELL CYCLE ARREST | 13 | 154 | 3.716e-08 | 4.068e-07 |
| 426 | REGULATION OF PROTEIN IMPORT | 14 | 183 | 3.849e-08 | 4.204e-07 |
| 427 | REGULATION OF ION TRANSPORT | 25 | 592 | 3.889e-08 | 4.238e-07 |
| 428 | REGULATION OF PHOSPHATASE ACTIVITY | 12 | 128 | 3.913e-08 | 4.254e-07 |
| 429 | REGULATION OF CELL GROWTH | 20 | 391 | 4.475e-08 | 4.854e-07 |
| 430 | CEREBRAL CORTEX DEVELOPMENT | 11 | 105 | 4.6e-08 | 4.966e-07 |
| 431 | ODONTOGENESIS | 11 | 105 | 4.6e-08 | 4.966e-07 |
| 432 | RESPONSE TO GROWTH HORMONE | 7 | 30 | 4.708e-08 | 5.071e-07 |
| 433 | FOREBRAIN DEVELOPMENT | 19 | 357 | 5.285e-08 | 5.68e-07 |
| 434 | RESPONSE TO MOLECULE OF BACTERIAL ORIGIN | 18 | 321 | 5.47e-08 | 5.865e-07 |
| 435 | TUBE MORPHOGENESIS | 18 | 323 | 6.01e-08 | 6.428e-07 |
| 436 | REGENERATION | 13 | 161 | 6.303e-08 | 6.727e-07 |
| 437 | RESPONSE TO HYDROGEN PEROXIDE | 11 | 109 | 6.798e-08 | 7.238e-07 |
| 438 | REGULATED EXOCYTOSIS | 15 | 224 | 7.348e-08 | 7.806e-07 |
| 439 | POSITIVE REGULATION OF LEUKOCYTE PROLIFERATION | 12 | 136 | 7.698e-08 | 8.16e-07 |
| 440 | REGULATION OF CARBOHYDRATE BIOSYNTHETIC PROCESS | 10 | 87 | 7.818e-08 | 8.268e-07 |
| 441 | POSITIVE REGULATION OF CELLULAR COMPONENT BIOGENESIS | 20 | 406 | 8.279e-08 | 8.735e-07 |
| 442 | RESPONSE TO ALKALOID | 12 | 137 | 8.35e-08 | 8.791e-07 |
| 443 | ANTIGEN RECEPTOR MEDIATED SIGNALING PATHWAY | 14 | 195 | 8.535e-08 | 8.965e-07 |
| 444 | POSITIVE REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 15 | 228 | 9.271e-08 | 9.694e-07 |
| 445 | POSITIVE REGULATION OF CYTOKINE PRODUCTION | 19 | 370 | 9.266e-08 | 9.694e-07 |
| 446 | DEVELOPMENTAL GROWTH | 18 | 333 | 9.513e-08 | 9.924e-07 |
| 447 | REGULATION OF GENERATION OF PRECURSOR METABOLITES AND ENERGY | 10 | 89 | 9.734e-08 | 1.013e-06 |
| 448 | REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 15 | 229 | 9.818e-08 | 1.02e-06 |
| 449 | REGULATION OF SMOOTH MUSCLE CELL MIGRATION | 8 | 49 | 9.923e-08 | 1.028e-06 |
| 450 | REGULATION OF CELL JUNCTION ASSEMBLY | 9 | 68 | 1.025e-07 | 1.06e-06 |
| 451 | RESPONSE TO CARBOHYDRATE | 13 | 168 | 1.041e-07 | 1.074e-06 |
| 452 | REGULATION OF SYNAPTIC PLASTICITY | 12 | 140 | 1.061e-07 | 1.093e-06 |
| 453 | AGING | 16 | 264 | 1.066e-07 | 1.095e-06 |
| 454 | RESPONSE TO ACTIVITY | 9 | 69 | 1.166e-07 | 1.195e-06 |
| 455 | REGULATION OF ADHERENS JUNCTION ORGANIZATION | 8 | 50 | 1.169e-07 | 1.196e-06 |
| 456 | REGULATION OF PROTEIN IMPORT INTO NUCLEUS TRANSLOCATION | 6 | 21 | 1.185e-07 | 1.209e-06 |
| 457 | CELLULAR RESPONSE TO OXYGEN LEVELS | 12 | 143 | 1.341e-07 | 1.365e-06 |
| 458 | POSITIVE REGULATION OF MITOTIC NUCLEAR DIVISION | 8 | 51 | 1.373e-07 | 1.395e-06 |
| 459 | SECRETION | 24 | 588 | 1.388e-07 | 1.407e-06 |
| 460 | DEFENSE RESPONSE | 37 | 1231 | 1.448e-07 | 1.465e-06 |
| 461 | REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION | 11 | 118 | 1.545e-07 | 1.559e-06 |
| 462 | CHEMICAL HOMEOSTASIS | 30 | 874 | 1.582e-07 | 1.593e-06 |
| 463 | CELL ADHESION MEDIATED BY INTEGRIN | 5 | 12 | 1.623e-07 | 1.628e-06 |
| 464 | ACTIVATION OF TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE ACTIVITY | 5 | 12 | 1.623e-07 | 1.628e-06 |
| 465 | NEGATIVE REGULATION OF MOLECULAR FUNCTION | 34 | 1079 | 1.662e-07 | 1.663e-06 |
| 466 | T CELL RECEPTOR SIGNALING PATHWAY | 12 | 146 | 1.684e-07 | 1.682e-06 |
| 467 | POSITIVE REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 7 | 36 | 1.818e-07 | 1.811e-06 |
| 468 | MAMMARY GLAND EPITHELIUM DEVELOPMENT | 8 | 53 | 1.873e-07 | 1.862e-06 |
| 469 | REGULATION OF ORGAN GROWTH | 9 | 73 | 1.918e-07 | 1.903e-06 |
| 470 | RESPONSE TO TRANSITION METAL NANOPARTICLE | 12 | 148 | 1.955e-07 | 1.935e-06 |
| 471 | SIGNAL TRANSDUCTION IN RESPONSE TO DNA DAMAGE | 10 | 96 | 2.009e-07 | 1.985e-06 |
| 472 | REGULATION OF LEUKOCYTE MIGRATION | 12 | 149 | 2.104e-07 | 2.074e-06 |
| 473 | GLAND MORPHOGENESIS | 10 | 97 | 2.217e-07 | 2.177e-06 |
| 474 | REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 10 | 97 | 2.217e-07 | 2.177e-06 |
| 475 | SKIN DEVELOPMENT | 14 | 211 | 2.262e-07 | 2.216e-06 |
| 476 | NEGATIVE REGULATION OF CELL CYCLE G1 S PHASE TRANSITION | 10 | 98 | 2.444e-07 | 2.389e-06 |
| 477 | POSITIVE REGULATION OF PROTEIN IMPORT INTO NUCLEUS TRANSLOCATION | 5 | 13 | 2.613e-07 | 2.538e-06 |
| 478 | CELLULAR RESPONSE TO HEPATOCYTE GROWTH FACTOR STIMULUS | 5 | 13 | 2.613e-07 | 2.538e-06 |
| 479 | RESPONSE TO HEPATOCYTE GROWTH FACTOR | 5 | 13 | 2.613e-07 | 2.538e-06 |
| 480 | REGULATION OF INNATE IMMUNE RESPONSE | 18 | 357 | 2.676e-07 | 2.594e-06 |
| 481 | NEGATIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 20 | 437 | 2.701e-07 | 2.613e-06 |
| 482 | CELLULAR RESPONSE TO PROSTAGLANDIN STIMULUS | 6 | 24 | 2.854e-07 | 2.75e-06 |
| 483 | REGULATION OF POSITIVE CHEMOTAXIS | 6 | 24 | 2.854e-07 | 2.75e-06 |
| 484 | POSITIVE REGULATION OF MUSCLE TISSUE DEVELOPMENT | 8 | 56 | 2.911e-07 | 2.799e-06 |
| 485 | REGULATION OF RAS PROTEIN SIGNAL TRANSDUCTION | 13 | 184 | 3.001e-07 | 2.873e-06 |
| 486 | CELLULAR RESPONSE TO OXIDATIVE STRESS | 13 | 184 | 3.001e-07 | 2.873e-06 |
| 487 | DEVELOPMENT OF PRIMARY SEXUAL CHARACTERISTICS | 14 | 216 | 3.011e-07 | 2.877e-06 |
| 488 | RESPONSE TO UV | 11 | 126 | 3.019e-07 | 2.879e-06 |
| 489 | SYSTEM PROCESS | 46 | 1785 | 3.296e-07 | 3.137e-06 |
| 490 | REGULATION OF MITOCHONDRION ORGANIZATION | 14 | 218 | 3.369e-07 | 3.193e-06 |
| 491 | REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 14 | 218 | 3.369e-07 | 3.193e-06 |
| 492 | POSITIVE REGULATION OF DEFENSE RESPONSE | 18 | 364 | 3.559e-07 | 3.365e-06 |
| 493 | CELLULAR RESPONSE TO EXTRACELLULAR STIMULUS | 13 | 188 | 3.844e-07 | 3.628e-06 |
| 494 | LEUKOCYTE CELL CELL ADHESION | 15 | 255 | 3.945e-07 | 3.716e-06 |
| 495 | INSULIN LIKE GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 5 | 14 | 4.025e-07 | 3.784e-06 |
| 496 | MITOTIC CELL CYCLE | 27 | 766 | 4.153e-07 | 3.896e-06 |
| 497 | LEUKOCYTE DIFFERENTIATION | 16 | 292 | 4.18e-07 | 3.913e-06 |
| 498 | NEGATIVE REGULATION OF VASCULATURE DEVELOPMENT | 9 | 80 | 4.265e-07 | 3.969e-06 |
| 499 | POSITIVE REGULATION OF LYMPHOCYTE DIFFERENTIATION | 9 | 80 | 4.265e-07 | 3.969e-06 |
| 500 | INSULIN RECEPTOR SIGNALING PATHWAY | 9 | 80 | 4.265e-07 | 3.969e-06 |
| 501 | POSITIVE REGULATION OF PROTEIN IMPORT | 10 | 104 | 4.282e-07 | 3.977e-06 |
| 502 | ORGANOPHOSPHATE BIOSYNTHETIC PROCESS | 20 | 450 | 4.295e-07 | 3.981e-06 |
| 503 | MESENCHYME DEVELOPMENT | 13 | 190 | 4.34e-07 | 4.015e-06 |
| 504 | REGULATION OF EPITHELIAL CELL APOPTOTIC PROCESS | 8 | 59 | 4.408e-07 | 4.07e-06 |
| 505 | REGULATION OF GLUCOSE IMPORT | 8 | 60 | 5.035e-07 | 4.63e-06 |
| 506 | PROTEIN COMPLEX BIOGENESIS | 34 | 1132 | 5.045e-07 | 4.63e-06 |
| 507 | PROTEIN COMPLEX ASSEMBLY | 34 | 1132 | 5.045e-07 | 4.63e-06 |
| 508 | SKELETAL SYSTEM DEVELOPMENT | 20 | 455 | 5.109e-07 | 4.68e-06 |
| 509 | RHYTHMIC PROCESS | 16 | 298 | 5.483e-07 | 5.012e-06 |
| 510 | CELL CYCLE CHECKPOINT | 13 | 194 | 5.507e-07 | 5.024e-06 |
| 511 | CELLULAR RESPONSE TO BIOTIC STIMULUS | 12 | 163 | 5.565e-07 | 5.067e-06 |
| 512 | REGULATION OF T CELL DIFFERENTIATION | 10 | 107 | 5.589e-07 | 5.08e-06 |
| 513 | PROTEIN COMPLEX SUBUNIT ORGANIZATION | 41 | 1527 | 5.689e-07 | 5.16e-06 |
| 514 | RESPONSE TO FATTY ACID | 9 | 83 | 5.863e-07 | 5.307e-06 |
| 515 | CELL MIGRATION INVOLVED IN SPROUTING ANGIOGENESIS | 5 | 15 | 5.98e-07 | 5.392e-06 |
| 516 | JAK STAT CASCADE INVOLVED IN GROWTH HORMONE SIGNALING PATHWAY | 5 | 15 | 5.98e-07 | 5.392e-06 |
| 517 | NEGATIVE REGULATION OF NEURON APOPTOTIC PROCESS | 11 | 135 | 6.06e-07 | 5.454e-06 |
| 518 | HETEROTYPIC CELL CELL ADHESION | 6 | 27 | 6.094e-07 | 5.474e-06 |
| 519 | RESPONSE TO ETHANOL | 11 | 136 | 6.526e-07 | 5.84e-06 |
| 520 | POSITIVE REGULATION OF NUCLEAR DIVISION | 8 | 62 | 6.52e-07 | 5.84e-06 |
| 521 | ORGANOPHOSPHATE METABOLIC PROCESS | 29 | 885 | 6.728e-07 | 6.009e-06 |
| 522 | REGULATION OF SYSTEM PROCESS | 21 | 507 | 6.903e-07 | 6.154e-06 |
| 523 | LYMPHOCYTE ACTIVATION | 17 | 342 | 7.07e-07 | 6.29e-06 |
| 524 | POSITIVE REGULATION OF MITOCHONDRION ORGANIZATION | 12 | 167 | 7.215e-07 | 6.407e-06 |
| 525 | NEGATIVE REGULATION OF MITOTIC CELL CYCLE | 13 | 199 | 7.355e-07 | 6.519e-06 |
| 526 | MITOTIC CELL CYCLE CHECKPOINT | 11 | 139 | 8.117e-07 | 7.181e-06 |
| 527 | TOR SIGNALING | 5 | 16 | 8.614e-07 | 7.592e-06 |
| 528 | POSITIVE REGULATION OF TYROSINE PHOSPHORYLATION OF STAT5 PROTEIN | 5 | 16 | 8.614e-07 | 7.592e-06 |
| 529 | LUNG MORPHOGENESIS | 7 | 45 | 9.029e-07 | 7.941e-06 |
| 530 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 31 | 1004 | 9.539e-07 | 8.374e-06 |
| 531 | OVULATION CYCLE PROCESS | 9 | 88 | 9.683e-07 | 8.485e-06 |
| 532 | REGULATION OF CYTOKINE PRODUCTION | 22 | 563 | 9.812e-07 | 8.582e-06 |
| 533 | REGULATION OF PEPTIDASE ACTIVITY | 18 | 392 | 1.04e-06 | 9.082e-06 |
| 534 | MESODERM MORPHOGENESIS | 8 | 66 | 1.064e-06 | 9.267e-06 |
| 535 | RAS PROTEIN SIGNAL TRANSDUCTION | 11 | 143 | 1.077e-06 | 9.364e-06 |
| 536 | REGULATION OF LEUKOCYTE PROLIFERATION | 13 | 206 | 1.087e-06 | 9.437e-06 |
| 537 | FEMALE SEX DIFFERENTIATION | 10 | 116 | 1.183e-06 | 1.025e-05 |
| 538 | NEGATIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 23 | 616 | 1.2e-06 | 1.037e-05 |
| 539 | REGULATION OF CALCIUM ION TRANSPORT | 13 | 209 | 1.279e-06 | 1.104e-05 |
| 540 | POSITIVE REGULATION OF CYTOPLASMIC TRANSPORT | 15 | 282 | 1.404e-06 | 1.21e-05 |
| 541 | SECRETION BY CELL | 20 | 486 | 1.42e-06 | 1.216e-05 |
| 542 | POSITIVE REGULATION OF REACTIVE OXYGEN SPECIES BIOSYNTHETIC PROCESS | 7 | 48 | 1.422e-06 | 1.216e-05 |
| 543 | REGULATION OF CELL CYCLE G1 S PHASE TRANSITION | 11 | 147 | 1.415e-06 | 1.216e-05 |
| 544 | IMMUNE EFFECTOR PROCESS | 20 | 486 | 1.42e-06 | 1.216e-05 |
| 545 | HOMEOSTASIS OF NUMBER OF CELLS WITHIN A TISSUE | 6 | 31 | 1.457e-06 | 1.242e-05 |
| 546 | REGULATION OF PLATELET ACTIVATION | 6 | 31 | 1.457e-06 | 1.242e-05 |
| 547 | CELLULAR RESPONSE TO DNA DAMAGE STIMULUS | 25 | 720 | 1.505e-06 | 1.28e-05 |
| 548 | DIGESTIVE SYSTEM DEVELOPMENT | 11 | 148 | 1.513e-06 | 1.282e-05 |
| 549 | POSITIVE REGULATION OF CELL GROWTH | 11 | 148 | 1.513e-06 | 1.282e-05 |
| 550 | REGULATION OF COLLATERAL SPROUTING | 5 | 18 | 1.657e-06 | 1.4e-05 |
| 551 | CELLULAR RESPONSE TO PROSTAGLANDIN E STIMULUS | 5 | 18 | 1.657e-06 | 1.4e-05 |
| 552 | REGULATION OF DNA BIOSYNTHETIC PROCESS | 9 | 94 | 1.696e-06 | 1.429e-05 |
| 553 | EMBRYONIC ORGAN DEVELOPMENT | 18 | 406 | 1.714e-06 | 1.442e-05 |
| 554 | FC GAMMA RECEPTOR SIGNALING PATHWAY | 9 | 95 | 1.854e-06 | 1.554e-05 |
| 555 | ACTIVATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 9 | 95 | 1.854e-06 | 1.554e-05 |
| 556 | REGULATION OF MYELOID CELL DIFFERENTIATION | 12 | 183 | 1.9e-06 | 1.59e-05 |
| 557 | REGULATION OF CATABOLIC PROCESS | 25 | 731 | 1.974e-06 | 1.649e-05 |
| 558 | EPIDERMIS DEVELOPMENT | 14 | 253 | 1.999e-06 | 1.667e-05 |
| 559 | ENDOCRINE SYSTEM DEVELOPMENT | 10 | 123 | 2.025e-06 | 1.685e-05 |
| 560 | RESPONSE TO NICOTINE | 7 | 51 | 2.169e-06 | 1.799e-05 |
| 561 | CELLULAR RESPONSE TO FATTY ACID | 7 | 51 | 2.169e-06 | 1.799e-05 |
| 562 | KIDNEY VASCULATURE DEVELOPMENT | 5 | 19 | 2.228e-06 | 1.838e-05 |
| 563 | RENAL SYSTEM VASCULATURE DEVELOPMENT | 5 | 19 | 2.228e-06 | 1.838e-05 |
| 564 | ASTROCYTE DEVELOPMENT | 5 | 19 | 2.228e-06 | 1.838e-05 |
| 565 | POSITIVE REGULATION OF PEPTIDASE ACTIVITY | 11 | 154 | 2.235e-06 | 1.841e-05 |
| 566 | LEUKOCYTE ACTIVATION | 18 | 414 | 2.257e-06 | 1.856e-05 |
| 567 | G1 DNA DAMAGE CHECKPOINT | 8 | 73 | 2.319e-06 | 1.903e-05 |
| 568 | NEGATIVE REGULATION OF CELL PROLIFERATION | 23 | 643 | 2.472e-06 | 2.025e-05 |
| 569 | POSITIVE REGULATION OF INTRACELLULAR PROTEIN TRANSPORT | 14 | 258 | 2.514e-06 | 2.056e-05 |
| 570 | REGULATION OF WOUND HEALING | 10 | 126 | 2.521e-06 | 2.058e-05 |
| 571 | RESPONSE TO PROSTAGLANDIN | 6 | 34 | 2.584e-06 | 2.102e-05 |
| 572 | PROTEIN KINASE B SIGNALING | 6 | 34 | 2.584e-06 | 2.102e-05 |
| 573 | SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 10 | 127 | 2.709e-06 | 2.199e-05 |
| 574 | STEM CELL DIFFERENTIATION | 12 | 190 | 2.812e-06 | 2.279e-05 |
| 575 | REGULATION OF NITRIC OXIDE BIOSYNTHETIC PROCESS | 7 | 53 | 2.831e-06 | 2.291e-05 |
| 576 | MITOTIC DNA INTEGRITY CHECKPOINT | 9 | 100 | 2.852e-06 | 2.3e-05 |
| 577 | LIMBIC SYSTEM DEVELOPMENT | 9 | 100 | 2.852e-06 | 2.3e-05 |
| 578 | REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION TO MITOCHONDRION | 10 | 128 | 2.908e-06 | 2.341e-05 |
| 579 | REGULATION OF NEURON PROJECTION REGENERATION | 5 | 20 | 2.942e-06 | 2.36e-05 |
| 580 | REGULATION OF TYROSINE PHOSPHORYLATION OF STAT5 PROTEIN | 5 | 20 | 2.942e-06 | 2.36e-05 |
| 581 | NEGATIVE REGULATION OF PHOSPHORYLATION | 18 | 422 | 2.952e-06 | 2.364e-05 |
| 582 | REGULATION OF INTRACELLULAR PROTEIN TRANSPORT | 17 | 381 | 3.078e-06 | 2.461e-05 |
| 583 | REGULATION OF GLYCOGEN METABOLIC PROCESS | 6 | 35 | 3.088e-06 | 2.464e-05 |
| 584 | POSITIVE REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 10 | 129 | 3.12e-06 | 2.486e-05 |
| 585 | GLIAL CELL DEVELOPMENT | 8 | 76 | 3.155e-06 | 2.51e-05 |
| 586 | CELLULAR MACROMOLECULE LOCALIZATION | 34 | 1234 | 3.455e-06 | 2.743e-05 |
| 587 | SEX DIFFERENTIATION | 14 | 266 | 3.586e-06 | 2.843e-05 |
| 588 | REGULATION OF ANATOMICAL STRUCTURE SIZE | 19 | 472 | 3.618e-06 | 2.863e-05 |
| 589 | REGULATION OF MUSCLE ORGAN DEVELOPMENT | 9 | 103 | 3.649e-06 | 2.883e-05 |
| 590 | NEUROMUSCULAR JUNCTION DEVELOPMENT | 6 | 36 | 3.669e-06 | 2.893e-05 |
| 591 | EAR DEVELOPMENT | 12 | 195 | 3.68e-06 | 2.893e-05 |
| 592 | REGULATION OF MUSCLE SYSTEM PROCESS | 12 | 195 | 3.68e-06 | 2.893e-05 |
| 593 | REGULATION OF LYMPHOCYTE DIFFERENTIATION | 10 | 132 | 3.839e-06 | 3.013e-05 |
| 594 | CELLULAR RESPONSE TO REACTIVE OXYGEN SPECIES | 9 | 104 | 3.954e-06 | 3.098e-05 |
| 595 | REGULATION OF TRANSPORTER ACTIVITY | 12 | 198 | 4.308e-06 | 3.369e-05 |
| 596 | MESENCHYMAL CELL DIFFERENTIATION | 10 | 134 | 4.395e-06 | 3.426e-05 |
| 597 | INTERACTION WITH HOST | 10 | 134 | 4.395e-06 | 3.426e-05 |
| 598 | EXOCYTOSIS | 15 | 310 | 4.492e-06 | 3.496e-05 |
| 599 | INNATE IMMUNE RESPONSE ACTIVATING CELL SURFACE RECEPTOR SIGNALING PATHWAY | 9 | 106 | 4.631e-06 | 3.597e-05 |
| 600 | POSITIVE REGULATION OF PROTEIN AUTOPHOSPHORYLATION | 5 | 22 | 4.902e-06 | 3.801e-05 |
| 601 | RESPONSE TO BACTERIUM | 20 | 528 | 4.963e-06 | 3.843e-05 |
| 602 | POSITIVE REGULATION OF ORGAN GROWTH | 6 | 38 | 5.099e-06 | 3.934e-05 |
| 603 | REGULATION OF MULTICELLULAR ORGANISMAL METABOLIC PROCESS | 6 | 38 | 5.099e-06 | 3.934e-05 |
| 604 | POSITIVE REGULATION OF LEUKOCYTE CHEMOTAXIS | 8 | 81 | 5.116e-06 | 3.941e-05 |
| 605 | REGULATION OF CELL CYCLE ARREST | 9 | 108 | 5.404e-06 | 4.15e-05 |
| 606 | REGULATION OF MYELOID LEUKOCYTE DIFFERENTIATION | 9 | 108 | 5.404e-06 | 4.15e-05 |
| 607 | SENSORY ORGAN MORPHOGENESIS | 13 | 239 | 5.623e-06 | 4.311e-05 |
| 608 | CELLULAR HOMEOSTASIS | 23 | 676 | 5.65e-06 | 4.324e-05 |
| 609 | GLUCOSE HOMEOSTASIS | 11 | 170 | 5.823e-06 | 4.441e-05 |
| 610 | CARBOHYDRATE HOMEOSTASIS | 11 | 170 | 5.823e-06 | 4.441e-05 |
| 611 | NEGATIVE REGULATION OF CATALYTIC ACTIVITY | 26 | 829 | 5.848e-06 | 4.449e-05 |
| 612 | ACTIVATION OF INNATE IMMUNE RESPONSE | 12 | 204 | 5.852e-06 | 4.449e-05 |
| 613 | POSITIVE REGULATION OF DNA BIOSYNTHETIC PROCESS | 7 | 59 | 5.899e-06 | 4.471e-05 |
| 614 | REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 7 | 59 | 5.899e-06 | 4.471e-05 |
| 615 | REGULATION OF SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 14 | 278 | 5.962e-06 | 4.51e-05 |
| 616 | ORGAN REGENERATION | 8 | 83 | 6.149e-06 | 4.644e-05 |
| 617 | TISSUE HOMEOSTASIS | 11 | 171 | 6.159e-06 | 4.645e-05 |
| 618 | LEUKOCYTE APOPTOTIC PROCESS | 5 | 23 | 6.203e-06 | 4.67e-05 |
| 619 | NEURON MIGRATION | 9 | 110 | 6.286e-06 | 4.725e-05 |
| 620 | POSTTRANSCRIPTIONAL REGULATION OF GENE EXPRESSION | 18 | 448 | 6.746e-06 | 5.063e-05 |
| 621 | POSITIVE REGULATION OF PROTEOLYSIS | 16 | 363 | 7.049e-06 | 5.282e-05 |
| 622 | ZYMOGEN ACTIVATION | 9 | 112 | 7.288e-06 | 5.452e-05 |
| 623 | POSITIVE REGULATION OF CELL CYCLE ARREST | 8 | 85 | 7.352e-06 | 5.491e-05 |
| 624 | OVARIAN FOLLICLE DEVELOPMENT | 7 | 61 | 7.395e-06 | 5.505e-05 |
| 625 | POSITIVE REGULATION OF STEM CELL PROLIFERATION | 7 | 61 | 7.395e-06 | 5.505e-05 |
| 626 | LYMPHOCYTE DIFFERENTIATION | 12 | 209 | 7.491e-06 | 5.568e-05 |
| 627 | BLOOD VESSEL ENDOTHELIAL CELL MIGRATION | 5 | 24 | 7.76e-06 | 5.759e-05 |
| 628 | CIRCULATORY SYSTEM PROCESS | 16 | 366 | 7.816e-06 | 5.791e-05 |
| 629 | OVULATION CYCLE | 9 | 113 | 7.839e-06 | 5.799e-05 |
| 630 | EYE DEVELOPMENT | 15 | 326 | 8.231e-06 | 6.079e-05 |
| 631 | RESPONSE TO TRANSFORMING GROWTH FACTOR BETA | 10 | 144 | 8.348e-06 | 6.146e-05 |
| 632 | NEGATIVE REGULATION OF CELL PROJECTION ORGANIZATION | 10 | 144 | 8.348e-06 | 6.146e-05 |
| 633 | MEMBRANE ORGANIZATION | 27 | 899 | 8.424e-06 | 6.193e-05 |
| 634 | REGULATION OF GOLGI ORGANIZATION | 4 | 12 | 8.686e-06 | 6.365e-05 |
| 635 | TRACHEA MORPHOGENESIS | 4 | 12 | 8.686e-06 | 6.365e-05 |
| 636 | TISSUE REMODELING | 8 | 87 | 8.748e-06 | 6.4e-05 |
| 637 | CELLULAR RESPONSE TO ALCOHOL | 9 | 115 | 9.047e-06 | 6.608e-05 |
| 638 | MULTI MULTICELLULAR ORGANISM PROCESS | 12 | 213 | 9.079e-06 | 6.621e-05 |
| 639 | POSITIVE REGULATION OF GLUCOSE TRANSPORT | 6 | 42 | 9.314e-06 | 6.782e-05 |
| 640 | NEGATIVE REGULATION OF CELL CYCLE PHASE TRANSITION | 10 | 146 | 9.43e-06 | 6.846e-05 |
| 641 | DNA INTEGRITY CHECKPOINT | 10 | 146 | 9.43e-06 | 6.846e-05 |
| 642 | REGULATION OF COAGULATION | 8 | 88 | 9.526e-06 | 6.882e-05 |
| 643 | LEUKOCYTE PROLIFERATION | 8 | 88 | 9.526e-06 | 6.882e-05 |
| 644 | POSITIVE REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION | 8 | 88 | 9.526e-06 | 6.882e-05 |
| 645 | RESPONSE TO PROSTAGLANDIN E | 5 | 25 | 9.607e-06 | 6.909e-05 |
| 646 | CELLULAR EXTRAVASATION | 5 | 25 | 9.607e-06 | 6.909e-05 |
| 647 | HISTONE PHOSPHORYLATION | 5 | 25 | 9.607e-06 | 6.909e-05 |
| 648 | CARTILAGE DEVELOPMENT | 10 | 147 | 1.002e-05 | 7.192e-05 |
| 649 | REGULATION OF VESICLE MEDIATED TRANSPORT | 18 | 462 | 1.025e-05 | 7.35e-05 |
| 650 | RESPONSE TO HEAT | 8 | 89 | 1.036e-05 | 7.417e-05 |
| 651 | NEGATIVE REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 9 | 117 | 1.041e-05 | 7.441e-05 |
| 652 | RESPONSE TO METAL ION | 15 | 333 | 1.06e-05 | 7.563e-05 |
| 653 | CEREBRAL CORTEX CELL MIGRATION | 6 | 43 | 1.072e-05 | 7.638e-05 |
| 654 | REGULATION OF CELL CYCLE PROCESS | 20 | 558 | 1.118e-05 | 7.953e-05 |
| 655 | MIDBRAIN DEVELOPMENT | 8 | 90 | 1.126e-05 | 7.997e-05 |
| 656 | REGULATION OF REACTIVE OXYGEN SPECIES BIOSYNTHETIC PROCESS | 7 | 65 | 1.134e-05 | 8.042e-05 |
| 657 | CELLULAR RESPONSE TO STEROID HORMONE STIMULUS | 12 | 218 | 1.147e-05 | 8.126e-05 |
| 658 | DEVELOPMENTAL PROGRAMMED CELL DEATH | 5 | 26 | 1.178e-05 | 8.293e-05 |
| 659 | REPLACEMENT OSSIFICATION | 5 | 26 | 1.178e-05 | 8.293e-05 |
| 660 | NEGATIVE REGULATION OF LIPID TRANSPORT | 5 | 26 | 1.178e-05 | 8.293e-05 |
| 661 | ENDOCHONDRAL OSSIFICATION | 5 | 26 | 1.178e-05 | 8.293e-05 |
| 662 | REGULATION OF CELLULAR COMPONENT SIZE | 15 | 337 | 1.221e-05 | 8.58e-05 |
| 663 | REGULATION OF MULTICELLULAR ORGANISM GROWTH | 7 | 66 | 1.256e-05 | 8.812e-05 |
| 664 | REGULATION OF DENDRITE DEVELOPMENT | 9 | 120 | 1.278e-05 | 8.958e-05 |
| 665 | REGULATION OF TRANSMEMBRANE TRANSPORT | 17 | 426 | 1.335e-05 | 9.341e-05 |
| 666 | EXOCRINE SYSTEM DEVELOPMENT | 6 | 45 | 1.404e-05 | 9.81e-05 |
| 667 | POSITIVE REGULATION OF HEART GROWTH | 5 | 27 | 1.432e-05 | 9.975e-05 |
| 668 | POSITIVE REGULATION OF MESENCHYMAL CELL PROLIFERATION | 5 | 27 | 1.432e-05 | 9.975e-05 |
| 669 | NEGATIVE REGULATION OF CELL ADHESION | 12 | 223 | 1.44e-05 | 0.0001002 |
| 670 | MODULATION OF SYNAPTIC TRANSMISSION | 14 | 301 | 1.466e-05 | 0.0001018 |
| 671 | CELLULAR CHEMICAL HOMEOSTASIS | 20 | 570 | 1.521e-05 | 0.0001054 |
| 672 | MULTICELLULAR ORGANISMAL RESPONSE TO STRESS | 7 | 68 | 1.532e-05 | 0.0001059 |
| 673 | REGULATION OF NEUROLOGICAL SYSTEM PROCESS | 7 | 68 | 1.532e-05 | 0.0001059 |
| 674 | T CELL DIFFERENTIATION | 9 | 123 | 1.56e-05 | 0.0001077 |
| 675 | NEGATIVE REGULATION OF CELL DEVELOPMENT | 14 | 303 | 1.578e-05 | 0.0001088 |
| 676 | PEPTIDYL THREONINE MODIFICATION | 6 | 46 | 1.599e-05 | 0.0001099 |
| 677 | REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY IN ABSENCE OF LIGAND | 6 | 46 | 1.599e-05 | 0.0001099 |
| 678 | REGULATION OF PROTEIN SECRETION | 16 | 389 | 1.664e-05 | 0.0001142 |
| 679 | PHAGOCYTOSIS | 11 | 190 | 1.669e-05 | 0.0001143 |
| 680 | BONE DEVELOPMENT | 10 | 156 | 1.685e-05 | 0.0001151 |
| 681 | PROTEIN LOCALIZATION TO NUCLEUS | 10 | 156 | 1.685e-05 | 0.0001151 |
| 682 | POSITIVE REGULATION OF NITRIC OXIDE SYNTHASE BIOSYNTHETIC PROCESS | 4 | 14 | 1.724e-05 | 0.0001171 |
| 683 | PROTEIN HETEROTRIMERIZATION | 4 | 14 | 1.724e-05 | 0.0001171 |
| 684 | LONG TERM MEMORY | 5 | 28 | 1.727e-05 | 0.0001171 |
| 685 | POSITIVE REGULATION OF SPROUTING ANGIOGENESIS | 4 | 14 | 1.724e-05 | 0.0001171 |
| 686 | REGULATION OF SPROUTING ANGIOGENESIS | 5 | 28 | 1.727e-05 | 0.0001171 |
| 687 | THYMUS DEVELOPMENT | 6 | 47 | 1.815e-05 | 0.0001227 |
| 688 | REGULATION OF LEUKOCYTE CHEMOTAXIS | 8 | 96 | 1.812e-05 | 0.0001227 |
| 689 | REGULATION OF PROTEIN TARGETING | 14 | 307 | 1.827e-05 | 0.0001234 |
| 690 | RESPONSE TO PURINE CONTAINING COMPOUND | 10 | 158 | 1.883e-05 | 0.0001269 |
| 691 | POSITIVE REGULATION OF CATABOLIC PROCESS | 16 | 395 | 2.006e-05 | 0.0001351 |
| 692 | SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 15 | 352 | 2.034e-05 | 0.0001368 |
| 693 | POSITIVE REGULATION OF CARDIAC MUSCLE TISSUE DEVELOPMENT | 5 | 29 | 2.066e-05 | 0.0001387 |
| 694 | REGULATION OF LIPID BIOSYNTHETIC PROCESS | 9 | 128 | 2.148e-05 | 0.000144 |
| 695 | REGULATION OF CELLULAR AMIDE METABOLIC PROCESS | 15 | 354 | 2.173e-05 | 0.0001455 |
| 696 | POSITIVE REGULATION OF CHEMOKINE PRODUCTION | 6 | 49 | 2.318e-05 | 0.0001549 |
| 697 | NEGATIVE REGULATION OF DENDRITE MORPHOGENESIS | 4 | 15 | 2.329e-05 | 0.0001553 |
| 698 | T CELL APOPTOTIC PROCESS | 4 | 15 | 2.329e-05 | 0.0001553 |
| 699 | POSITIVE REGULATION OF RESPONSE TO WOUNDING | 10 | 162 | 2.337e-05 | 0.0001556 |
| 700 | NEGATIVE REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 13 | 274 | 2.414e-05 | 0.0001604 |
| 701 | ASSOCIATIVE LEARNING | 7 | 73 | 2.45e-05 | 0.0001619 |
| 702 | HIPPOCAMPUS DEVELOPMENT | 7 | 73 | 2.45e-05 | 0.0001619 |
| 703 | CELLULAR RESPONSE TO KETONE | 7 | 73 | 2.45e-05 | 0.0001619 |
| 704 | REGULATION OF GLUCOSE TRANSPORT | 8 | 100 | 2.443e-05 | 0.0001619 |
| 705 | RESPONSE TO EPIDERMAL GROWTH FACTOR | 5 | 30 | 2.456e-05 | 0.0001621 |
| 706 | POSITIVE REGULATION OF ION TRANSPORT | 12 | 236 | 2.528e-05 | 0.0001666 |
| 707 | EPITHELIAL CELL DIFFERENTIATION | 18 | 495 | 2.573e-05 | 0.0001693 |
| 708 | LEARNING | 9 | 131 | 2.584e-05 | 0.0001698 |
| 709 | VISUAL BEHAVIOR | 6 | 50 | 2.608e-05 | 0.0001709 |
| 710 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 41 | 1784 | 2.607e-05 | 0.0001709 |
| 711 | REGULATION OF EXTENT OF CELL GROWTH | 8 | 101 | 2.626e-05 | 0.0001719 |
| 712 | MITOCHONDRION ORGANIZATION | 20 | 594 | 2.738e-05 | 0.0001789 |
| 713 | POSITIVE REGULATION OF NF KAPPAB TRANSCRIPTION FACTOR ACTIVITY | 9 | 132 | 2.745e-05 | 0.0001791 |
| 714 | PROTEIN TARGETING | 16 | 406 | 2.8e-05 | 0.0001824 |
| 715 | ESTABLISHMENT OF PROTEIN LOCALIZATION | 35 | 1423 | 2.814e-05 | 0.0001831 |
| 716 | LIPOPOLYSACCHARIDE MEDIATED SIGNALING PATHWAY | 5 | 31 | 2.9e-05 | 0.0001885 |
| 717 | GRANULOCYTE MIGRATION | 7 | 75 | 2.925e-05 | 0.0001898 |
| 718 | REGULATION OF MUSCLE TISSUE DEVELOPMENT | 8 | 103 | 3.028e-05 | 0.0001962 |
| 719 | REGULATION OF EARLY ENDOSOME TO LATE ENDOSOME TRANSPORT | 4 | 16 | 3.077e-05 | 0.0001986 |
| 720 | BRANCHING INVOLVED IN SALIVARY GLAND MORPHOGENESIS | 4 | 16 | 3.077e-05 | 0.0001986 |
| 721 | REGULATION OF DEVELOPMENTAL PIGMENTATION | 4 | 16 | 3.077e-05 | 0.0001986 |
| 722 | REGULATION OF CYTOSKELETON ORGANIZATION | 18 | 502 | 3.093e-05 | 0.0001993 |
| 723 | REGULATION OF CELLULAR RESPONSE TO HEAT | 7 | 76 | 3.19e-05 | 0.0002053 |
| 724 | REGULATION OF ORGAN MORPHOGENESIS | 12 | 242 | 3.234e-05 | 0.0002078 |
| 725 | DEVELOPMENTAL GROWTH INVOLVED IN MORPHOGENESIS | 8 | 104 | 3.247e-05 | 0.0002084 |
| 726 | ACTIVATION OF MAPKK ACTIVITY | 6 | 52 | 3.276e-05 | 0.00021 |
| 727 | EMBRYO DEVELOPMENT ENDING IN BIRTH OR EGG HATCHING | 19 | 554 | 3.408e-05 | 0.0002175 |
| 728 | SALIVARY GLAND DEVELOPMENT | 5 | 32 | 3.404e-05 | 0.0002175 |
| 729 | PROTEIN LOCALIZATION | 41 | 1805 | 3.412e-05 | 0.0002175 |
| 730 | REGULATION OF ACTIN CYTOSKELETON REORGANIZATION | 5 | 32 | 3.404e-05 | 0.0002175 |
| 731 | REGULATION OF MORPHOGENESIS OF A BRANCHING STRUCTURE | 6 | 53 | 3.658e-05 | 0.0002325 |
| 732 | NEGATIVE REGULATION OF CELL SUBSTRATE ADHESION | 6 | 53 | 3.658e-05 | 0.0002325 |
| 733 | REGULATION OF GLUCOSE METABOLIC PROCESS | 8 | 106 | 3.726e-05 | 0.0002365 |
| 734 | POSITIVE REGULATION OF INNATE IMMUNE RESPONSE | 12 | 246 | 3.795e-05 | 0.0002406 |
| 735 | REGULATION OF PROTEOLYSIS | 22 | 711 | 3.882e-05 | 0.0002457 |
| 736 | PLACENTA DEVELOPMENT | 9 | 138 | 3.9e-05 | 0.0002466 |
| 737 | POSITIVE REGULATION OF GLYCOGEN METABOLIC PROCESS | 4 | 17 | 3.987e-05 | 0.0002503 |
| 738 | REGULATION OF GLUCOSE IMPORT IN RESPONSE TO INSULIN STIMULUS | 4 | 17 | 3.987e-05 | 0.0002503 |
| 739 | POSITIVE REGULATION OF NUCLEOTIDE CATABOLIC PROCESS | 4 | 17 | 3.987e-05 | 0.0002503 |
| 740 | MAMMARY GLAND ALVEOLUS DEVELOPMENT | 4 | 17 | 3.987e-05 | 0.0002503 |
| 741 | MAMMARY GLAND LOBULE DEVELOPMENT | 4 | 17 | 3.987e-05 | 0.0002503 |
| 742 | REGULATION OF CELL SHAPE | 9 | 139 | 4.128e-05 | 0.0002589 |
| 743 | REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY VIA DEATH DOMAIN RECEPTORS | 6 | 55 | 4.529e-05 | 0.0002837 |
| 744 | HEART MORPHOGENESIS | 11 | 212 | 4.581e-05 | 0.0002865 |
| 745 | ORGAN FORMATION | 5 | 34 | 4.614e-05 | 0.0002878 |
| 746 | REGULATION OF MESENCHYMAL CELL PROLIFERATION | 5 | 34 | 4.614e-05 | 0.0002878 |
| 747 | MACROMOLECULAR COMPLEX ASSEMBLY | 34 | 1398 | 4.65e-05 | 0.0002897 |
| 748 | NEGATIVE REGULATION OF CELL CYCLE PROCESS | 11 | 214 | 4.988e-05 | 0.0003103 |
| 749 | REGULATION OF INFLAMMATORY RESPONSE | 13 | 294 | 5.002e-05 | 0.0003107 |
| 750 | REGULATION OF CELL MATURATION | 4 | 18 | 5.078e-05 | 0.0003142 |
| 751 | RESPONSE TO PLATELET DERIVED GROWTH FACTOR | 4 | 18 | 5.078e-05 | 0.0003142 |
| 752 | LYMPHOCYTE APOPTOTIC PROCESS | 4 | 18 | 5.078e-05 | 0.0003142 |
| 753 | POSITIVE REGULATION OF CELLULAR AMIDE METABOLIC PROCESS | 8 | 111 | 5.187e-05 | 0.0003201 |
| 754 | COLUMNAR CUBOIDAL EPITHELIAL CELL DIFFERENTIATION | 8 | 111 | 5.187e-05 | 0.0003201 |
| 755 | REGULATION OF OSSIFICATION | 10 | 178 | 5.222e-05 | 0.0003218 |
| 756 | NEGATIVE REGULATION OF EPITHELIAL CELL APOPTOTIC PROCESS | 5 | 35 | 5.332e-05 | 0.0003282 |
| 757 | REGULATION OF RESPONSE TO CYTOKINE STIMULUS | 9 | 144 | 5.443e-05 | 0.0003346 |
| 758 | EAR MORPHOGENESIS | 8 | 112 | 5.53e-05 | 0.000339 |
| 759 | REGULATION OF OSTEOBLAST DIFFERENTIATION | 8 | 112 | 5.53e-05 | 0.000339 |
| 760 | APOPTOTIC MITOCHONDRIAL CHANGES | 6 | 57 | 5.559e-05 | 0.0003404 |
| 761 | SYNAPSE ORGANIZATION | 9 | 145 | 5.745e-05 | 0.0003508 |
| 762 | ION HOMEOSTASIS | 19 | 576 | 5.745e-05 | 0.0003508 |
| 763 | MUSCLE STRUCTURE DEVELOPMENT | 16 | 432 | 5.871e-05 | 0.0003581 |
| 764 | PROTEIN HETEROOLIGOMERIZATION | 8 | 113 | 5.891e-05 | 0.0003588 |
| 765 | REGULATION OF NUCLEOTIDE CATABOLIC PROCESS | 5 | 36 | 6.133e-05 | 0.000373 |
| 766 | REGULATION OF NITRIC OXIDE SYNTHASE BIOSYNTHETIC PROCESS | 4 | 19 | 6.373e-05 | 0.0003866 |
| 767 | POSITIVE REGULATION OF CARDIAC MUSCLE CELL PROLIFERATION | 4 | 19 | 6.373e-05 | 0.0003866 |
| 768 | EPHRIN RECEPTOR SIGNALING PATHWAY | 7 | 85 | 6.583e-05 | 0.0003988 |
| 769 | RESPONSE TO INTERLEUKIN 1 | 8 | 115 | 6.674e-05 | 0.0004033 |
| 770 | REGULATION OF PROTEIN STABILITY | 11 | 221 | 6.669e-05 | 0.0004033 |
| 771 | RESPONSE TO TEMPERATURE STIMULUS | 9 | 148 | 6.735e-05 | 0.0004065 |
| 772 | VASCULOGENESIS | 6 | 59 | 6.769e-05 | 0.000408 |
| 773 | POSITIVE REGULATION OF PROTEIN LOCALIZATION TO CELL PERIPHERY | 5 | 37 | 7.023e-05 | 0.0004211 |
| 774 | POSITIVE REGULATION OF PROTEIN TYROSINE KINASE ACTIVITY | 5 | 37 | 7.023e-05 | 0.0004211 |
| 775 | POSITIVE REGULATION OF PROTEIN LOCALIZATION TO PLASMA MEMBRANE | 5 | 37 | 7.023e-05 | 0.0004211 |
| 776 | REGULATION OF PROTEIN AUTOPHOSPHORYLATION | 5 | 37 | 7.023e-05 | 0.0004211 |
| 777 | REGULATION OF CYTOKINE SECRETION | 9 | 149 | 7.096e-05 | 0.0004244 |
| 778 | POSITIVE REGULATION OF B CELL ACTIVATION | 7 | 86 | 7.095e-05 | 0.0004244 |
| 779 | REGULATION OF MONOOXYGENASE ACTIVITY | 6 | 60 | 7.447e-05 | 0.0004437 |
| 780 | CHONDROCYTE DIFFERENTIATION | 6 | 60 | 7.447e-05 | 0.0004437 |
| 781 | LEUKOCYTE HOMEOSTASIS | 6 | 60 | 7.447e-05 | 0.0004437 |
| 782 | MOVEMENT IN ENVIRONMENT OF OTHER ORGANISM INVOLVED IN SYMBIOTIC INTERACTION | 7 | 87 | 7.639e-05 | 0.0004511 |
| 783 | ENTRY INTO CELL OF OTHER ORGANISM INVOLVED IN SYMBIOTIC INTERACTION | 7 | 87 | 7.639e-05 | 0.0004511 |
| 784 | VIRAL ENTRY INTO HOST CELL | 7 | 87 | 7.639e-05 | 0.0004511 |
| 785 | ENTRY INTO OTHER ORGANISM INVOLVED IN SYMBIOTIC INTERACTION | 7 | 87 | 7.639e-05 | 0.0004511 |
| 786 | MOVEMENT IN HOST ENVIRONMENT | 7 | 87 | 7.639e-05 | 0.0004511 |
| 787 | ENTRY INTO HOST | 7 | 87 | 7.639e-05 | 0.0004511 |
| 788 | ENTRY INTO HOST CELL | 7 | 87 | 7.639e-05 | 0.0004511 |
| 789 | EMBRYONIC HEMOPOIESIS | 4 | 20 | 7.893e-05 | 0.0004643 |
| 790 | TRACHEA DEVELOPMENT | 4 | 20 | 7.893e-05 | 0.0004643 |
| 791 | AMELOGENESIS | 4 | 20 | 7.893e-05 | 0.0004643 |
| 792 | CELLULAR RESPONSE TO HYDROGEN PEROXIDE | 6 | 61 | 8.179e-05 | 0.0004805 |
| 793 | MYELOID CELL HOMEOSTASIS | 7 | 88 | 8.217e-05 | 0.0004809 |
| 794 | NEGATIVE REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 7 | 88 | 8.217e-05 | 0.0004809 |
| 795 | REGULATION OF STEM CELL PROLIFERATION | 7 | 88 | 8.217e-05 | 0.0004809 |
| 796 | MYELOID CELL DIFFERENTIATION | 10 | 189 | 8.63e-05 | 0.0005044 |
| 797 | REGULATION OF SECRETION | 21 | 699 | 8.876e-05 | 0.0005182 |
| 798 | FOREBRAIN CELL MIGRATION | 6 | 62 | 8.967e-05 | 0.0005222 |
| 799 | REGULATION OF TISSUE REMODELING | 6 | 62 | 8.967e-05 | 0.0005222 |
| 800 | REGULATION OF CELL ADHESION MEDIATED BY INTEGRIN | 5 | 39 | 9.101e-05 | 0.000528 |
| 801 | EXTRINSIC APOPTOTIC SIGNALING PATHWAY VIA DEATH DOMAIN RECEPTORS | 5 | 39 | 9.101e-05 | 0.000528 |
| 802 | CELLULAR RESPONSE TO NUTRIENT | 5 | 39 | 9.101e-05 | 0.000528 |
| 803 | NEGATIVE REGULATION OF NEURON DIFFERENTIATION | 10 | 191 | 9.418e-05 | 0.0005457 |
| 804 | MESONEPHROS DEVELOPMENT | 7 | 90 | 9.48e-05 | 0.0005486 |
| 805 | PROTEIN IMPORT | 9 | 155 | 9.617e-05 | 0.0005559 |
| 806 | POSITIVE REGULATION OF ADHERENS JUNCTION ORGANIZATION | 4 | 21 | 9.66e-05 | 0.0005577 |
| 807 | REGULATION OF OSTEOCLAST DIFFERENTIATION | 6 | 63 | 9.814e-05 | 0.0005645 |
| 808 | REGULATION OF MUSCLE ADAPTATION | 6 | 63 | 9.814e-05 | 0.0005645 |
| 809 | RESPONSE TO OSMOTIC STRESS | 6 | 63 | 9.814e-05 | 0.0005645 |
| 810 | MULTICELLULAR ORGANISMAL HOMEOSTASIS | 12 | 272 | 9.948e-05 | 0.0005715 |
| 811 | NEGATIVE REGULATION OF RESPONSE TO WOUNDING | 9 | 156 | 0.000101 | 0.0005796 |
| 812 | REGULATION OF EPITHELIAL CELL DIFFERENTIATION | 8 | 122 | 0.0001012 | 0.0005801 |
| 813 | CELLULAR RESPONSE TO LIGHT STIMULUS | 7 | 91 | 0.0001017 | 0.000582 |
| 814 | RESPONSE TO TUMOR NECROSIS FACTOR | 11 | 233 | 0.0001069 | 0.0006109 |
| 815 | POSITIVE REGULATION OF KIDNEY DEVELOPMENT | 5 | 41 | 0.0001162 | 0.0006634 |
| 816 | REGULATION OF CHEMOKINE PRODUCTION | 6 | 65 | 0.000117 | 0.0006646 |
| 817 | SOMATIC STEM CELL DIVISION | 4 | 22 | 0.000117 | 0.0006646 |
| 818 | ACTIVATION OF PROTEIN KINASE B ACTIVITY | 4 | 22 | 0.000117 | 0.0006646 |
| 819 | CELLULAR RESPONSE TO INTERLEUKIN 6 | 4 | 22 | 0.000117 | 0.0006646 |
| 820 | NEGATIVE REGULATION OF GROWTH | 11 | 236 | 0.0001196 | 0.0006789 |
| 821 | KIDNEY EPITHELIUM DEVELOPMENT | 8 | 125 | 0.00012 | 0.00068 |
| 822 | OSTEOBLAST DIFFERENTIATION | 8 | 126 | 0.0001268 | 0.0007179 |
| 823 | CELLULAR RESPONSE TO UV | 6 | 66 | 0.0001274 | 0.0007196 |
| 824 | NEURAL NUCLEUS DEVELOPMENT | 6 | 66 | 0.0001274 | 0.0007196 |
| 825 | REGULATION OF ENDOTHELIAL CELL APOPTOTIC PROCESS | 5 | 42 | 0.0001306 | 0.0007342 |
| 826 | REGULATION OF CELLULAR CARBOHYDRATE CATABOLIC PROCESS | 5 | 42 | 0.0001306 | 0.0007342 |
| 827 | REGULATION OF HEART GROWTH | 5 | 42 | 0.0001306 | 0.0007342 |
| 828 | REGULATION OF CARBOHYDRATE CATABOLIC PROCESS | 5 | 42 | 0.0001306 | 0.0007342 |
| 829 | REGULATION OF ENDOCYTOSIS | 10 | 199 | 0.0001321 | 0.0007414 |
| 830 | NEGATIVE REGULATION OF ION TRANSPORT | 8 | 127 | 0.000134 | 0.0007512 |
| 831 | CELL AGING | 6 | 67 | 0.0001386 | 0.0007761 |
| 832 | RESPONSE TO INCREASED OXYGEN LEVELS | 4 | 23 | 0.0001403 | 0.0007809 |
| 833 | POSITIVE REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 4 | 23 | 0.0001403 | 0.0007809 |
| 834 | POSITIVE REGULATION OF NEUROLOGICAL SYSTEM PROCESS | 4 | 23 | 0.0001403 | 0.0007809 |
| 835 | RESPONSE TO HYPEROXIA | 4 | 23 | 0.0001403 | 0.0007809 |
| 836 | NEUROTROPHIN SIGNALING PATHWAY | 4 | 23 | 0.0001403 | 0.0007809 |
| 837 | CARDIOCYTE DIFFERENTIATION | 7 | 96 | 0.0001425 | 0.000792 |
| 838 | RESPONSE TO ELECTRICAL STIMULUS | 5 | 43 | 0.0001464 | 0.0008115 |
| 839 | VESICLE MEDIATED TRANSPORT | 30 | 1239 | 0.0001465 | 0.0008115 |
| 840 | MYELOID LEUKOCYTE MEDIATED IMMUNITY | 5 | 43 | 0.0001464 | 0.0008115 |
| 841 | NUCLEAR IMPORT | 8 | 129 | 0.0001493 | 0.0008263 |
| 842 | ORGAN GROWTH | 6 | 68 | 0.0001505 | 0.0008309 |
| 843 | REGULATION OF TOR SIGNALING | 6 | 68 | 0.0001505 | 0.0008309 |
| 844 | NEUROLOGICAL SYSTEM PROCESS | 30 | 1242 | 0.0001528 | 0.0008424 |
| 845 | ANATOMICAL STRUCTURE HOMEOSTASIS | 12 | 285 | 0.0001541 | 0.0008485 |
| 846 | NEGATIVE REGULATION OF LIPID BIOSYNTHETIC PROCESS | 5 | 44 | 0.0001636 | 0.0008978 |
| 847 | BRANCHING INVOLVED IN URETERIC BUD MORPHOGENESIS | 5 | 44 | 0.0001636 | 0.0008978 |
| 848 | REGULATION OF RELEASE OF CYTOCHROME C FROM MITOCHONDRIA | 5 | 44 | 0.0001636 | 0.0008978 |
| 849 | REGULATION OF MYELOID CELL APOPTOTIC PROCESS | 4 | 24 | 0.0001668 | 0.0009088 |
| 850 | AXON REGENERATION | 4 | 24 | 0.0001668 | 0.0009088 |
| 851 | POSITIVE REGULATION OF NUCLEOSIDE METABOLIC PROCESS | 4 | 24 | 0.0001668 | 0.0009088 |
| 852 | POSITIVE REGULATION OF CELL JUNCTION ASSEMBLY | 4 | 24 | 0.0001668 | 0.0009088 |
| 853 | POSITIVE REGULATION OF RECEPTOR INTERNALIZATION | 4 | 24 | 0.0001668 | 0.0009088 |
| 854 | POSITIVE REGULATION OF ATP METABOLIC PROCESS | 4 | 24 | 0.0001668 | 0.0009088 |
| 855 | B CELL ACTIVATION | 8 | 132 | 0.0001751 | 0.0009527 |
| 856 | CENTRAL NERVOUS SYSTEM NEURON DEVELOPMENT | 6 | 70 | 0.0001768 | 0.0009612 |
| 857 | INTRACELLULAR RECEPTOR SIGNALING PATHWAY | 9 | 168 | 0.0001772 | 0.0009623 |
| 858 | SUBSTANTIA NIGRA DEVELOPMENT | 5 | 45 | 0.0001823 | 0.0009876 |
| 859 | ENDOCHONDRAL BONE MORPHOGENESIS | 5 | 45 | 0.0001823 | 0.0009876 |
| 860 | NEGATIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 20 | 684 | 0.0001876 | 0.001015 |
| 861 | SKIN EPIDERMIS DEVELOPMENT | 6 | 71 | 0.0001913 | 0.001034 |
| 862 | THYROID GLAND DEVELOPMENT | 4 | 25 | 0.0001967 | 0.001062 |
| 863 | REGULATION OF NUCLEOTIDE METABOLIC PROCESS | 10 | 211 | 0.0002127 | 0.001147 |
| 864 | POSITIVE REGULATION OF GLIOGENESIS | 5 | 47 | 0.0002246 | 0.001209 |
| 865 | CELLULAR RESPONSE TO RADIATION | 8 | 137 | 0.0002259 | 0.001215 |
| 866 | RESPONSE TO INTERLEUKIN 6 | 4 | 26 | 0.0002304 | 0.001236 |
| 867 | CELLULAR RESPONSE TO VITAMIN | 4 | 26 | 0.0002304 | 0.001236 |
| 868 | REGULATION OF DENDRITE MORPHOGENESIS | 6 | 74 | 0.0002402 | 0.001288 |
| 869 | REGULATION OF CARDIAC MUSCLE TISSUE DEVELOPMENT | 5 | 48 | 0.0002483 | 0.001325 |
| 870 | RESPONSE TO AXON INJURY | 5 | 48 | 0.0002483 | 0.001325 |
| 871 | COLUMNAR CUBOIDAL EPITHELIAL CELL DEVELOPMENT | 5 | 48 | 0.0002483 | 0.001325 |
| 872 | REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION TO PLASMA MEMBRANE | 5 | 48 | 0.0002483 | 0.001325 |
| 873 | PROSTATE GLAND GROWTH | 3 | 11 | 0.0002496 | 0.00133 |
| 874 | SINGLE ORGANISM BIOSYNTHETIC PROCESS | 31 | 1340 | 0.0002524 | 0.001344 |
| 875 | BIOMINERAL TISSUE DEVELOPMENT | 6 | 75 | 0.0002586 | 0.001372 |
| 876 | ODONTOGENESIS OF DENTIN CONTAINING TOOTH | 6 | 75 | 0.0002586 | 0.001372 |
| 877 | POSITIVE REGULATION OF AUTOPHAGY | 6 | 75 | 0.0002586 | 0.001372 |
| 878 | IMMUNE RESPONSE | 27 | 1100 | 0.0002589 | 0.001372 |
| 879 | FAT CELL DIFFERENTIATION | 7 | 106 | 0.0002637 | 0.001396 |
| 880 | REGULATION OF LAMELLIPODIUM ASSEMBLY | 4 | 27 | 0.000268 | 0.001415 |
| 881 | SUBSTRATE DEPENDENT CELL MIGRATION | 4 | 27 | 0.000268 | 0.001415 |
| 882 | REGULATION OF NITRIC OXIDE SYNTHASE ACTIVITY | 5 | 49 | 0.0002739 | 0.001442 |
| 883 | REGULATION OF NUCLEOSIDE METABOLIC PROCESS | 5 | 49 | 0.0002739 | 0.001442 |
| 884 | REGULATION OF ATP METABOLIC PROCESS | 5 | 49 | 0.0002739 | 0.001442 |
| 885 | EXTRACELLULAR MATRIX DISASSEMBLY | 6 | 76 | 0.000278 | 0.001462 |
| 886 | REGULATION OF SODIUM ION TRANSPORT | 6 | 77 | 0.0002986 | 0.001568 |
| 887 | LYMPHOCYTE HOMEOSTASIS | 5 | 50 | 0.0003014 | 0.001574 |
| 888 | REGULATION OF COENZYME METABOLIC PROCESS | 5 | 50 | 0.0003014 | 0.001574 |
| 889 | FACE DEVELOPMENT | 5 | 50 | 0.0003014 | 0.001574 |
| 890 | REGULATION OF COFACTOR METABOLIC PROCESS | 5 | 50 | 0.0003014 | 0.001574 |
| 891 | POSITIVE REGULATION OF MYELOID LEUKOCYTE DIFFERENTIATION | 5 | 50 | 0.0003014 | 0.001574 |
| 892 | REGULATION OF MACROPHAGE DERIVED FOAM CELL DIFFERENTIATION | 4 | 28 | 0.0003097 | 0.001608 |
| 893 | NEGATIVE REGULATION OF DENDRITE DEVELOPMENT | 4 | 28 | 0.0003097 | 0.001608 |
| 894 | POSITIVE REGULATION OF ACTIVATED T CELL PROLIFERATION | 4 | 28 | 0.0003097 | 0.001608 |
| 895 | MAMMARY GLAND DUCT MORPHOGENESIS | 4 | 28 | 0.0003097 | 0.001608 |
| 896 | POSITIVE REGULATION OF PHOSPHATASE ACTIVITY | 4 | 28 | 0.0003097 | 0.001608 |
| 897 | REGULATION OF RHO PROTEIN SIGNAL TRANSDUCTION | 7 | 109 | 0.0003129 | 0.001623 |
| 898 | REGULATION OF RECEPTOR MEDIATED ENDOCYTOSIS | 6 | 78 | 0.0003203 | 0.001657 |
| 899 | RENAL TUBULE DEVELOPMENT | 6 | 78 | 0.0003203 | 0.001657 |
| 900 | MULTICELLULAR ORGANISM REPRODUCTION | 21 | 768 | 0.0003204 | 0.001657 |
| 901 | ESTABLISHMENT OF LOCALIZATION IN CELL | 36 | 1676 | 0.0003269 | 0.001688 |
| 902 | PRODUCTION OF MOLECULAR MEDIATOR INVOLVED IN INFLAMMATORY RESPONSE | 3 | 12 | 0.0003299 | 0.001696 |
| 903 | I KAPPAB PHOSPHORYLATION | 3 | 12 | 0.0003299 | 0.001696 |
| 904 | MAMMARY GLAND EPITHELIAL CELL PROLIFERATION | 3 | 12 | 0.0003299 | 0.001696 |
| 905 | POSITIVE REGULATION OF GLUCOSE IMPORT IN RESPONSE TO INSULIN STIMULUS | 3 | 12 | 0.0003299 | 0.001696 |
| 906 | REGULATION OF INTERLEUKIN 12 PRODUCTION | 5 | 51 | 0.000331 | 0.0017 |
| 907 | RESPONSE TO IONIZING RADIATION | 8 | 145 | 0.0003322 | 0.001704 |
| 908 | IN UTERO EMBRYONIC DEVELOPMENT | 12 | 311 | 0.0003428 | 0.001757 |
| 909 | BONE MORPHOGENESIS | 6 | 79 | 0.0003433 | 0.001757 |
| 910 | REGULATION OF CARDIAC MUSCLE CELL PROLIFERATION | 4 | 29 | 0.000356 | 0.001816 |
| 911 | REGULATION OF VACUOLAR TRANSPORT | 4 | 29 | 0.000356 | 0.001816 |
| 912 | STEM CELL DIVISION | 4 | 29 | 0.000356 | 0.001816 |
| 913 | NEGATIVE REGULATION OF GENE EXPRESSION | 33 | 1493 | 0.0003591 | 0.00183 |
| 914 | POSITIVE REGULATION OF INTRINSIC APOPTOTIC SIGNALING PATHWAY | 5 | 52 | 0.0003627 | 0.001846 |
| 915 | NEGATIVE REGULATION OF LIPID METABOLIC PROCESS | 6 | 80 | 0.0003675 | 0.001869 |
| 916 | EPITHELIAL CELL DEVELOPMENT | 9 | 186 | 0.0003771 | 0.001915 |
| 917 | ESTABLISHMENT OF PROTEIN LOCALIZATION TO ORGANELLE | 13 | 361 | 0.0003794 | 0.001923 |
| 918 | ENDOCYTOSIS | 16 | 509 | 0.0003795 | 0.001923 |
| 919 | MALE SEX DIFFERENTIATION | 8 | 148 | 0.0003812 | 0.00193 |
| 920 | REGULATION OF SYNAPSE ORGANIZATION | 7 | 113 | 0.0003898 | 0.001969 |
| 921 | POSITIVE REGULATION OF INFLAMMATORY RESPONSE | 7 | 113 | 0.0003898 | 0.001969 |
| 922 | METANEPHROS DEVELOPMENT | 6 | 81 | 0.000393 | 0.001981 |
| 923 | POSITIVE REGULATION OF MYELOID CELL DIFFERENTIATION | 6 | 81 | 0.000393 | 0.001981 |
| 924 | MESONEPHRIC TUBULE MORPHOGENESIS | 5 | 53 | 0.0003967 | 0.001998 |
| 925 | INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO DNA DAMAGE BY P53 CLASS MEDIATOR | 4 | 30 | 0.000407 | 0.002041 |
| 926 | RESPONSE TO MORPHINE | 4 | 30 | 0.000407 | 0.002041 |
| 927 | NEGATIVE REGULATION OF MUSCLE CELL APOPTOTIC PROCESS | 4 | 30 | 0.000407 | 0.002041 |
| 928 | RESPONSE TO ISOQUINOLINE ALKALOID | 4 | 30 | 0.000407 | 0.002041 |
| 929 | POSITIVE REGULATION OF ENDOCYTOSIS | 7 | 114 | 0.0004112 | 0.002057 |
| 930 | REGULATION OF EMBRYONIC DEVELOPMENT | 7 | 114 | 0.0004112 | 0.002057 |
| 931 | KIDNEY MORPHOGENESIS | 6 | 82 | 0.0004199 | 0.002096 |
| 932 | SINGLE ORGANISM CELLULAR LOCALIZATION | 23 | 898 | 0.0004197 | 0.002096 |
| 933 | NEURONAL STEM CELL DIVISION | 3 | 13 | 0.0004251 | 0.002113 |
| 934 | INDUCTION OF POSITIVE CHEMOTAXIS | 3 | 13 | 0.0004251 | 0.002113 |
| 935 | REGULATION OF CELL PROLIFERATION INVOLVED IN KIDNEY DEVELOPMENT | 3 | 13 | 0.0004251 | 0.002113 |
| 936 | NEUROBLAST DIVISION | 3 | 13 | 0.0004251 | 0.002113 |
| 937 | REGULATION OF ERBB SIGNALING PATHWAY | 6 | 83 | 0.0004482 | 0.002221 |
| 938 | HAIR CYCLE | 6 | 83 | 0.0004482 | 0.002221 |
| 939 | MOLTING CYCLE | 6 | 83 | 0.0004482 | 0.002221 |
| 940 | NEGATIVE REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 4 | 31 | 0.000463 | 0.002289 |
| 941 | CELLULAR RESPONSE TO GLUCOSE STARVATION | 4 | 31 | 0.000463 | 0.002289 |
| 942 | RESPONSE TO ENDOPLASMIC RETICULUM STRESS | 10 | 233 | 0.0004677 | 0.002308 |
| 943 | REGULATION OF I KAPPAB KINASE NF KAPPAB SIGNALING | 10 | 233 | 0.0004677 | 0.002308 |
| 944 | REGULATION OF KIDNEY DEVELOPMENT | 5 | 55 | 0.0004718 | 0.002325 |
| 945 | EMBRYONIC ORGAN MORPHOGENESIS | 11 | 279 | 0.0005034 | 0.002477 |
| 946 | NEGATIVE REGULATION OF IMMUNE SYSTEM PROCESS | 13 | 372 | 0.0005036 | 0.002477 |
| 947 | REGULATION OF DENDRITIC SPINE DEVELOPMENT | 5 | 56 | 0.0005131 | 0.002521 |
| 948 | SENSORY PERCEPTION OF MECHANICAL STIMULUS | 8 | 155 | 0.0005188 | 0.002547 |
| 949 | POSITIVE REGULATION OF GLIAL CELL DIFFERENTIATION | 4 | 32 | 0.0005242 | 0.00256 |
| 950 | NEGATIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY IN ABSENCE OF LIGAND | 4 | 32 | 0.0005242 | 0.00256 |
| 951 | METANEPHRIC NEPHRON DEVELOPMENT | 4 | 32 | 0.0005242 | 0.00256 |
| 952 | POSITIVE REGULATION OF MULTICELLULAR ORGANISM GROWTH | 4 | 32 | 0.0005242 | 0.00256 |
| 953 | NEGATIVE REGULATION OF SIGNAL TRANSDUCTION IN ABSENCE OF LIGAND | 4 | 32 | 0.0005242 | 0.00256 |
| 954 | POSITIVE REGULATION OF METANEPHROS DEVELOPMENT | 3 | 14 | 0.0005364 | 0.002602 |
| 955 | POSITIVE REGULATION OF RHO PROTEIN SIGNAL TRANSDUCTION | 3 | 14 | 0.0005364 | 0.002602 |
| 956 | EPITHELIAL CELL CELL ADHESION | 3 | 14 | 0.0005364 | 0.002602 |
| 957 | REGULATION OF FIBROBLAST APOPTOTIC PROCESS | 3 | 14 | 0.0005364 | 0.002602 |
| 958 | NEGATIVE REGULATION OF MYELOID CELL APOPTOTIC PROCESS | 3 | 14 | 0.0005364 | 0.002602 |
| 959 | POSITIVE REGULATION OF CYTOSOLIC CALCIUM ION CONCENTRATION INVOLVED IN PHOSPHOLIPASE C ACTIVATING G PROTEIN COUPLED SIGNALING PATHWAY | 3 | 14 | 0.0005364 | 0.002602 |
| 960 | REGULATION OF IMMUNE EFFECTOR PROCESS | 14 | 424 | 0.0005423 | 0.002629 |
| 961 | POSITIVE REGULATION OF TUMOR NECROSIS FACTOR SUPERFAMILY CYTOKINE PRODUCTION | 5 | 57 | 0.0005571 | 0.002697 |
| 962 | POSITIVE REGULATION OF PROTEIN COMPLEX ASSEMBLY | 9 | 197 | 0.0005721 | 0.002767 |
| 963 | NEGATIVE REGULATION OF TRANSMEMBRANE TRANSPORT | 6 | 87 | 0.0005767 | 0.002787 |
| 964 | CELLULAR SENESCENCE | 4 | 33 | 0.0005911 | 0.002841 |
| 965 | REGULATION OF B CELL ACTIVATION | 7 | 121 | 0.0005887 | 0.002841 |
| 966 | RESPONSE TO VITAMIN D | 4 | 33 | 0.0005911 | 0.002841 |
| 967 | NEURON PROJECTION REGENERATION | 4 | 33 | 0.0005911 | 0.002841 |
| 968 | REGULATION OF BONE RESORPTION | 4 | 33 | 0.0005911 | 0.002841 |
| 969 | CELLULAR RESPONSE TO INTERLEUKIN 1 | 6 | 88 | 0.0006129 | 0.002943 |
| 970 | REGULATION OF JNK CASCADE | 8 | 159 | 0.000614 | 0.002945 |
| 971 | T CELL HOMEOSTASIS | 4 | 34 | 0.0006637 | 0.003165 |
| 972 | POSITIVE REGULATION OF SODIUM ION TRANSPORT | 4 | 34 | 0.0006637 | 0.003165 |
| 973 | RESPONSE TO VITAMIN E | 3 | 15 | 0.0006646 | 0.003165 |
| 974 | NEGATIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY VIA DEATH DOMAIN RECEPTORS | 4 | 34 | 0.0006637 | 0.003165 |
| 975 | T CELL LINEAGE COMMITMENT | 3 | 15 | 0.0006646 | 0.003165 |
| 976 | NEUROTROPHIN TRK RECEPTOR SIGNALING PATHWAY | 3 | 15 | 0.0006646 | 0.003165 |
| 977 | CELLULAR RESPONSE TO ALKALOID | 4 | 34 | 0.0006637 | 0.003165 |
| 978 | REGULATION OF OXIDOREDUCTASE ACTIVITY | 6 | 90 | 0.0006905 | 0.003282 |
| 979 | REGULATION OF GLIOGENESIS | 6 | 90 | 0.0006905 | 0.003282 |
| 980 | MATERNAL PROCESS INVOLVED IN FEMALE PREGNANCY | 5 | 60 | 0.0007061 | 0.003353 |
| 981 | REGULATION OF CYTOSOLIC CALCIUM ION CONCENTRATION | 9 | 203 | 0.0007093 | 0.003361 |
| 982 | NEGATIVE REGULATION OF CATABOLIC PROCESS | 9 | 203 | 0.0007093 | 0.003361 |
| 983 | REGULATION OF NUCLEAR DIVISION | 8 | 163 | 0.0007228 | 0.003421 |
| 984 | RESPONSE TO VIRUS | 10 | 247 | 0.000735 | 0.003475 |
| 985 | BONE REMODELING | 4 | 35 | 0.0007425 | 0.0035 |
| 986 | RESPONSE TO MINERALOCORTICOID | 4 | 35 | 0.0007425 | 0.0035 |
| 987 | RESPONSE TO IRON ION | 4 | 35 | 0.0007425 | 0.0035 |
| 988 | REGULATION OF PROTEIN TYROSINE KINASE ACTIVITY | 5 | 61 | 0.0007619 | 0.003588 |
| 989 | INNER EAR MORPHOGENESIS | 6 | 92 | 0.0007756 | 0.003649 |
| 990 | REGULATION OF AUTOPHAGY | 10 | 249 | 0.0007818 | 0.003674 |
| 991 | NEGATIVE REGULATION OF KINASE ACTIVITY | 10 | 250 | 0.0008061 | 0.003781 |
| 992 | POSITIVE REGULATION OF ACTIN CYTOSKELETON REORGANIZATION | 3 | 16 | 0.0008109 | 0.003781 |
| 993 | REGULATION OF GENE EXPRESSION BY GENETIC IMPRINTING | 3 | 16 | 0.0008109 | 0.003781 |
| 994 | POSITIVE REGULATION OF PHOSPHOPROTEIN PHOSPHATASE ACTIVITY | 3 | 16 | 0.0008109 | 0.003781 |
| 995 | POSITIVE REGULATION OF LAMELLIPODIUM ASSEMBLY | 3 | 16 | 0.0008109 | 0.003781 |
| 996 | POSITIVE REGULATION OF TRANSFORMING GROWTH FACTOR BETA PRODUCTION | 3 | 16 | 0.0008109 | 0.003781 |
| 997 | RETINA VASCULATURE DEVELOPMENT IN CAMERA TYPE EYE | 3 | 16 | 0.0008109 | 0.003781 |
| 998 | RESPONSE TO UV B | 3 | 16 | 0.0008109 | 0.003781 |
| 999 | DIVALENT INORGANIC CATION HOMEOSTASIS | 12 | 343 | 0.0008152 | 0.003797 |
| 1000 | NEPHRON EPITHELIUM DEVELOPMENT | 6 | 93 | 0.000821 | 0.00382 |
| 1001 | POSITIVE REGULATION OF ERBB SIGNALING PATHWAY | 4 | 36 | 0.0008276 | 0.003843 |
| 1002 | POSITIVE REGULATION OF GLUCOSE METABOLIC PROCESS | 4 | 36 | 0.0008276 | 0.003843 |
| 1003 | REGULATION OF CATION TRANSMEMBRANE TRANSPORT | 9 | 208 | 0.000843 | 0.003911 |
| 1004 | AUTOPHAGY | 13 | 394 | 0.0008572 | 0.003969 |
| 1005 | CELLULAR PROCESS INVOLVED IN REPRODUCTION IN MULTICELLULAR ORGANISM | 10 | 252 | 0.0008567 | 0.003969 |
| 1006 | REGULATION OF CYTOKINE BIOSYNTHETIC PROCESS | 6 | 94 | 0.0008685 | 0.004017 |
| 1007 | REGULATION OF CELL KILLING | 5 | 63 | 0.0008831 | 0.004081 |
| 1008 | MULTI ORGANISM REPRODUCTIVE PROCESS | 22 | 891 | 0.0009071 | 0.004187 |
| 1009 | POSITIVE REGULATION OF T CELL PROLIFERATION | 6 | 95 | 0.000918 | 0.004203 |
| 1010 | POSITIVE REGULATION OF B CELL PROLIFERATION | 4 | 37 | 0.0009194 | 0.004203 |
| 1011 | REGULATION OF RECEPTOR INTERNALIZATION | 4 | 37 | 0.0009194 | 0.004203 |
| 1012 | REGULATION OF MUSCLE HYPERTROPHY | 4 | 37 | 0.0009194 | 0.004203 |
| 1013 | REGULATION OF LIPID TRANSPORT | 6 | 95 | 0.000918 | 0.004203 |
| 1014 | TRANSFORMING GROWTH FACTOR BETA RECEPTOR SIGNALING PATHWAY | 6 | 95 | 0.000918 | 0.004203 |
| 1015 | NEGATIVE REGULATION OF HYDROLASE ACTIVITY | 13 | 397 | 0.0009186 | 0.004203 |
| 1016 | REGULATION OF LAMELLIPODIUM ORGANIZATION | 4 | 37 | 0.0009194 | 0.004203 |
| 1017 | REGULATION OF PEPTIDYL THREONINE PHOSPHORYLATION | 4 | 37 | 0.0009194 | 0.004203 |
| 1018 | POSITIVE REGULATION OF ALPHA BETA T CELL DIFFERENTIATION | 4 | 37 | 0.0009194 | 0.004203 |
| 1019 | POSITIVE REGULATION OF PROTEIN SECRETION | 9 | 211 | 0.0009327 | 0.004255 |
| 1020 | NEGATIVE REGULATION OF CYTOKINE PRODUCTION | 9 | 211 | 0.0009327 | 0.004255 |
| 1021 | PROTEIN STABILIZATION | 7 | 131 | 0.0009423 | 0.00429 |
| 1022 | RETINA DEVELOPMENT IN CAMERA TYPE EYE | 7 | 131 | 0.0009423 | 0.00429 |
| 1023 | NEGATIVE REGULATION OF CELL GROWTH | 8 | 170 | 0.0009502 | 0.004322 |
| 1024 | ACTIN FILAMENT BASED PROCESS | 14 | 450 | 0.0009656 | 0.004387 |
| 1025 | MYELOID LEUKOCYTE DIFFERENTIATION | 6 | 96 | 0.0009697 | 0.004398 |
| 1026 | FEMALE GAMETE GENERATION | 6 | 96 | 0.0009697 | 0.004398 |
| 1027 | BRANCH ELONGATION OF AN EPITHELIUM | 3 | 17 | 0.0009761 | 0.004406 |
| 1028 | NEGATIVE REGULATION OF PLATELET ACTIVATION | 3 | 17 | 0.0009761 | 0.004406 |
| 1029 | REGULATION OF DNA METHYLATION | 3 | 17 | 0.0009761 | 0.004406 |
| 1030 | NEGATIVE REGULATION OF LIPID STORAGE | 3 | 17 | 0.0009761 | 0.004406 |
| 1031 | PROTEIN LOCALIZATION TO ORGANELLE | 16 | 556 | 0.0009763 | 0.004406 |
| 1032 | WNT SIGNALING PATHWAY | 12 | 351 | 0.0009946 | 0.00448 |
| 1033 | NEGATIVE REGULATION OF TRANSFERASE ACTIVITY | 12 | 351 | 0.0009946 | 0.00448 |
| 1034 | INTRACELLULAR PROTEIN TRANSPORT | 20 | 781 | 0.001002 | 0.004508 |
| 1035 | COLLAGEN FIBRIL ORGANIZATION | 4 | 38 | 0.001018 | 0.004569 |
| 1036 | RESPONSE TO TESTOSTERONE | 4 | 38 | 0.001018 | 0.004569 |
| 1037 | BONE MINERALIZATION | 4 | 38 | 0.001018 | 0.004569 |
| 1038 | STRIATED MUSCLE CELL DIFFERENTIATION | 8 | 173 | 0.001064 | 0.004768 |
| 1039 | MYELOID LEUKOCYTE ACTIVATION | 6 | 98 | 0.00108 | 0.004836 |
| 1040 | REGULATION OF GRANULOCYTE CHEMOTAXIS | 4 | 39 | 0.001124 | 0.00502 |
| 1041 | LONG TERM SYNAPTIC POTENTIATION | 4 | 39 | 0.001124 | 0.00502 |
| 1042 | PLATELET AGGREGATION | 4 | 39 | 0.001124 | 0.00502 |
| 1043 | STRIATED MUSCLE CONTRACTION | 6 | 99 | 0.001138 | 0.005078 |
| 1044 | NEGATIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 10 | 262 | 0.00115 | 0.005125 |
| 1045 | POSITIVE REGULATION OF MITOCHONDRIAL MEMBRANE PERMEABILITY | 3 | 18 | 0.001161 | 0.005141 |
| 1046 | OVULATION | 3 | 18 | 0.001161 | 0.005141 |
| 1047 | RESPONSE TO CAFFEINE | 3 | 18 | 0.001161 | 0.005141 |
| 1048 | POSITIVE REGULATION OF T HELPER CELL DIFFERENTIATION | 3 | 18 | 0.001161 | 0.005141 |
| 1049 | MAST CELL MEDIATED IMMUNITY | 3 | 18 | 0.001161 | 0.005141 |
| 1050 | NOTOCHORD DEVELOPMENT | 3 | 18 | 0.001161 | 0.005141 |
| 1051 | MUSCLE CELL MIGRATION | 3 | 18 | 0.001161 | 0.005141 |
| 1052 | CELL DIVISION | 14 | 460 | 0.00119 | 0.005262 |
| 1053 | REGULATION OF WNT SIGNALING PATHWAY | 11 | 310 | 0.001192 | 0.005267 |
| 1054 | MAMMARY GLAND MORPHOGENESIS | 4 | 40 | 0.001238 | 0.005448 |
| 1055 | REGULATION OF ACTIVATED T CELL PROLIFERATION | 4 | 40 | 0.001238 | 0.005448 |
| 1056 | PROTEIN TRIMERIZATION | 4 | 40 | 0.001238 | 0.005448 |
| 1057 | ENDOCRINE PANCREAS DEVELOPMENT | 4 | 40 | 0.001238 | 0.005448 |
| 1058 | POSITIVE REGULATION OF INTERLEUKIN 6 PRODUCTION | 5 | 68 | 0.001249 | 0.005492 |
| 1059 | REGULATION OF ACTIN FILAMENT BASED PROCESS | 11 | 312 | 0.001255 | 0.005513 |
| 1060 | REGULATION OF TUMOR NECROSIS FACTOR SUPERFAMILY CYTOKINE PRODUCTION | 6 | 101 | 0.001263 | 0.005543 |
| 1061 | POSITIVE REGULATION OF I KAPPAB KINASE NF KAPPAB SIGNALING | 8 | 179 | 0.001323 | 0.005803 |
| 1062 | RESPONSE TO ORGANOPHOSPHORUS | 7 | 139 | 0.001331 | 0.005826 |
| 1063 | CELL ACTIVATION INVOLVED IN IMMUNE RESPONSE | 7 | 139 | 0.001331 | 0.005826 |
| 1064 | REGULATION OF LIPID STORAGE | 4 | 41 | 0.001359 | 0.005904 |
| 1065 | MULTICELLULAR ORGANISMAL MOVEMENT | 4 | 41 | 0.001359 | 0.005904 |
| 1066 | CELLULAR RESPONSE TO ESTROGEN STIMULUS | 4 | 41 | 0.001359 | 0.005904 |
| 1067 | MYELOID CELL ACTIVATION INVOLVED IN IMMUNE RESPONSE | 4 | 41 | 0.001359 | 0.005904 |
| 1068 | MUSCULOSKELETAL MOVEMENT | 4 | 41 | 0.001359 | 0.005904 |
| 1069 | MONOCYTE CHEMOTAXIS | 4 | 41 | 0.001359 | 0.005904 |
| 1070 | PROSTATE GLAND DEVELOPMENT | 4 | 41 | 0.001359 | 0.005904 |
| 1071 | LACTATION | 4 | 41 | 0.001359 | 0.005904 |
| 1072 | CELLULAR SODIUM ION HOMEOSTASIS | 3 | 19 | 0.001367 | 0.005922 |
| 1073 | MACROPHAGE DIFFERENTIATION | 3 | 19 | 0.001367 | 0.005922 |
| 1074 | POSITIVE REGULATION OF PROTEIN EXPORT FROM NUCLEUS | 3 | 19 | 0.001367 | 0.005922 |
| 1075 | CARDIAC MUSCLE TISSUE DEVELOPMENT | 7 | 140 | 0.001387 | 0.006004 |
| 1076 | REGULATION OF CIRCADIAN RHYTHM | 6 | 103 | 0.001397 | 0.006036 |
| 1077 | STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE | 6 | 103 | 0.001397 | 0.006036 |
| 1078 | POSITIVE REGULATION OF TYPE I INTERFERON PRODUCTION | 5 | 70 | 0.001423 | 0.006135 |
| 1079 | I KAPPAB KINASE NF KAPPAB SIGNALING | 5 | 70 | 0.001423 | 0.006135 |
| 1080 | REGULATION OF INTERLEUKIN 6 PRODUCTION | 6 | 104 | 0.001468 | 0.006326 |
| 1081 | POSITIVE REGULATION OF DNA BINDING | 4 | 42 | 0.001488 | 0.006394 |
| 1082 | POSITIVE REGULATION OF NEURAL PRECURSOR CELL PROLIFERATION | 4 | 42 | 0.001488 | 0.006394 |
| 1083 | REGULATION OF BONE REMODELING | 4 | 42 | 0.001488 | 0.006394 |
| 1084 | BODY FLUID SECRETION | 5 | 71 | 0.001516 | 0.006507 |
| 1085 | REGULATION OF CELL CYCLE PHASE TRANSITION | 11 | 321 | 0.001573 | 0.006745 |
| 1086 | REGULATION OF SMOOTH MUSCLE CELL DIFFERENTIATION | 3 | 20 | 0.001594 | 0.006799 |
| 1087 | HYPEROSMOTIC RESPONSE | 3 | 20 | 0.001594 | 0.006799 |
| 1088 | NEGATIVE REGULATION OF CELL JUNCTION ASSEMBLY | 3 | 20 | 0.001594 | 0.006799 |
| 1089 | RESPONSE TO PROTOZOAN | 3 | 20 | 0.001594 | 0.006799 |
| 1090 | GENETIC IMPRINTING | 3 | 20 | 0.001594 | 0.006799 |
| 1091 | LYMPH VESSEL DEVELOPMENT | 3 | 20 | 0.001594 | 0.006799 |
| 1092 | ENERGY RESERVE METABOLIC PROCESS | 5 | 72 | 0.001614 | 0.006876 |
| 1093 | POSITIVE REGULATION OF CALCIUM ION TRANSPORT | 6 | 106 | 0.001619 | 0.006892 |
| 1094 | REGULATION OF MUSCLE CELL APOPTOTIC PROCESS | 4 | 43 | 0.001626 | 0.006902 |
| 1095 | REGULATION OF INSULIN RECEPTOR SIGNALING PATHWAY | 4 | 43 | 0.001626 | 0.006902 |
| 1096 | ACUTE PHASE RESPONSE | 4 | 43 | 0.001626 | 0.006902 |
| 1097 | ERYTHROCYTE HOMEOSTASIS | 5 | 73 | 0.001716 | 0.007258 |
| 1098 | PANCREAS DEVELOPMENT | 5 | 73 | 0.001716 | 0.007258 |
| 1099 | REGULATION OF PLASMA MEMBRANE ORGANIZATION | 5 | 73 | 0.001716 | 0.007258 |
| 1100 | REGULATION OF NEURAL PRECURSOR CELL PROLIFERATION | 5 | 73 | 0.001716 | 0.007258 |
| 1101 | REGULATION OF PROTEIN COMPLEX ASSEMBLY | 12 | 375 | 0.00174 | 0.007355 |
| 1102 | CARDIAC MUSCLE CELL DIFFERENTIATION | 5 | 74 | 0.001823 | 0.007696 |
| 1103 | REGULATION OF T CELL PROLIFERATION | 7 | 147 | 0.001835 | 0.007741 |
| 1104 | B CELL HOMEOSTASIS | 3 | 21 | 0.001844 | 0.00775 |
| 1105 | MAST CELL ACTIVATION | 3 | 21 | 0.001844 | 0.00775 |
| 1106 | POSITIVE REGULATION OF EXCITATORY POSTSYNAPTIC POTENTIAL | 3 | 21 | 0.001844 | 0.00775 |
| 1107 | RUFFLE ORGANIZATION | 3 | 21 | 0.001844 | 0.00775 |
| 1108 | PATTERN RECOGNITION RECEPTOR SIGNALING PATHWAY | 6 | 109 | 0.001867 | 0.007838 |
| 1109 | G PROTEIN COUPLED RECEPTOR SIGNALING PATHWAY | 26 | 1193 | 0.001905 | 0.007993 |
| 1110 | MODIFICATION BY SYMBIONT OF HOST MORPHOLOGY OR PHYSIOLOGY | 4 | 45 | 0.001927 | 0.008067 |
| 1111 | POSITIVE REGULATION OF RECEPTOR ACTIVITY | 4 | 45 | 0.001927 | 0.008067 |
| 1112 | PROTEIN DEPHOSPHORYLATION | 8 | 190 | 0.001928 | 0.008067 |
| 1113 | MACROAUTOPHAGY | 10 | 281 | 0.001932 | 0.008077 |
| 1114 | POSITIVE REGULATION OF SYNAPTIC TRANSMISSION | 6 | 110 | 0.001955 | 0.008166 |
| 1115 | MUSCLE SYSTEM PROCESS | 10 | 282 | 0.001983 | 0.008275 |
| 1116 | PROTEIN OLIGOMERIZATION | 13 | 434 | 0.002033 | 0.008476 |
| 1117 | REGULATION OF BINDING | 10 | 283 | 0.002035 | 0.008476 |
| 1118 | POSITIVE REGULATION OF TRANSPORTER ACTIVITY | 5 | 76 | 0.002051 | 0.008537 |
| 1119 | POSITIVE REGULATION OF CHROMOSOME ORGANIZATION | 7 | 150 | 0.002058 | 0.008558 |
| 1120 | MUSCLE CELL DIFFERENTIATION | 9 | 237 | 0.002081 | 0.008647 |
| 1121 | INTRASPECIES INTERACTION BETWEEN ORGANISMS | 4 | 46 | 0.002091 | 0.008665 |
| 1122 | REGULATION OF ALPHA BETA T CELL DIFFERENTIATION | 4 | 46 | 0.002091 | 0.008665 |
| 1123 | SOCIAL BEHAVIOR | 4 | 46 | 0.002091 | 0.008665 |
| 1124 | REGULATION OF MITOCHONDRIAL MEMBRANE PERMEABILITY INVOLVED IN APOPTOTIC PROCESS | 3 | 22 | 0.002117 | 0.008739 |
| 1125 | POSITIVE REGULATION OF MUSCLE HYPERTROPHY | 3 | 22 | 0.002117 | 0.008739 |
| 1126 | POSITIVE REGULATION OF CARDIAC MUSCLE HYPERTROPHY | 3 | 22 | 0.002117 | 0.008739 |
| 1127 | REGULATION OF RESPONSE TO INTERFERON GAMMA | 3 | 22 | 0.002117 | 0.008739 |
| 1128 | DEVELOPMENTAL MATURATION | 8 | 193 | 0.002126 | 0.008769 |
| 1129 | POSITIVE REGULATION OF RECEPTOR MEDIATED ENDOCYTOSIS | 4 | 47 | 0.002265 | 0.009326 |
| 1130 | POSITIVE REGULATION OF DEPHOSPHORYLATION | 4 | 47 | 0.002265 | 0.009326 |
| 1131 | LYMPHOCYTE COSTIMULATION | 5 | 78 | 0.0023 | 0.009461 |
| 1132 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL METABOLIC PROCESS | 3 | 23 | 0.002413 | 0.009857 |
| 1133 | RESPONSE TO MAGNESIUM ION | 3 | 23 | 0.002413 | 0.009857 |
| 1134 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE III PROMOTER | 3 | 23 | 0.002413 | 0.009857 |
| 1135 | LYSOSOME LOCALIZATION | 3 | 23 | 0.002413 | 0.009857 |
| 1136 | REGULATION OF OSTEOBLAST PROLIFERATION | 3 | 23 | 0.002413 | 0.009857 |
| 1137 | REGULATION OF METANEPHROS DEVELOPMENT | 3 | 23 | 0.002413 | 0.009857 |
| 1138 | PROTEIN INSERTION INTO MEMBRANE | 3 | 23 | 0.002413 | 0.009857 |
| 1139 | POSITIVE REGULATION OF COLLAGEN METABOLIC PROCESS | 3 | 23 | 0.002413 | 0.009857 |
| 1140 | REGULATION OF SYNAPSE ASSEMBLY | 5 | 79 | 0.002432 | 0.009926 |
| 1141 | NEGATIVE REGULATION OF EPITHELIAL CELL PROLIFERATION | 6 | 115 | 0.002447 | 0.009966 |
| 1142 | DIGESTIVE TRACT MORPHOGENESIS | 4 | 48 | 0.002448 | 0.009966 |
| 1143 | POSITIVE REGULATION OF WOUND HEALING | 4 | 48 | 0.002448 | 0.009966 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | RECEPTOR BINDING | 88 | 1476 | 1.388e-39 | 1.29e-36 |
| 2 | KINASE ACTIVITY | 69 | 842 | 5.424e-39 | 2.519e-36 |
| 3 | TRANSFERASE ACTIVITY TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 71 | 992 | 2.515e-36 | 7.788e-34 |
| 4 | PROTEIN KINASE ACTIVITY | 58 | 640 | 7.123e-35 | 1.654e-32 |
| 5 | PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 26 | 70 | 1.284e-32 | 2.387e-30 |
| 6 | PROTEIN TYROSINE KINASE ACTIVITY | 33 | 176 | 3.297e-30 | 5.105e-28 |
| 7 | MOLECULAR FUNCTION REGULATOR | 72 | 1353 | 1.306e-28 | 1.734e-26 |
| 8 | GROWTH FACTOR ACTIVITY | 30 | 160 | 1.589e-27 | 1.845e-25 |
| 9 | GROWTH FACTOR BINDING | 26 | 123 | 2.096e-25 | 2.164e-23 |
| 10 | ENZYME BINDING | 75 | 1737 | 3.684e-24 | 3.423e-22 |
| 11 | GROWTH FACTOR RECEPTOR BINDING | 25 | 129 | 1.831e-23 | 1.547e-21 |
| 12 | X1 PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 17 | 43 | 2.989e-22 | 2.314e-20 |
| 13 | PROTEIN COMPLEX BINDING | 53 | 935 | 5.305e-22 | 3.791e-20 |
| 14 | KINASE BINDING | 43 | 606 | 1.551e-21 | 1.029e-19 |
| 15 | PHOSPHATIDYLINOSITOL KINASE ACTIVITY | 17 | 51 | 9.647e-21 | 5.974e-19 |
| 16 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE ACTIVITY | 18 | 64 | 2.277e-20 | 1.322e-18 |
| 17 | MACROMOLECULAR COMPLEX BINDING | 62 | 1399 | 2.775e-20 | 1.517e-18 |
| 18 | SIGNAL TRANSDUCER ACTIVITY | 69 | 1731 | 3.793e-20 | 1.957e-18 |
| 19 | RAS GUANYL NUCLEOTIDE EXCHANGE FACTOR ACTIVITY | 27 | 228 | 2.088e-19 | 1.021e-17 |
| 20 | TRANSMEMBRANE RECEPTOR PROTEIN KINASE ACTIVITY | 18 | 81 | 2.419e-18 | 1.124e-16 |
| 21 | GUANYL NUCLEOTIDE EXCHANGE FACTOR ACTIVITY | 27 | 303 | 3.155e-16 | 1.396e-14 |
| 22 | INTEGRIN BINDING | 18 | 105 | 3.307e-16 | 1.396e-14 |
| 23 | CELL ADHESION MOLECULE BINDING | 21 | 186 | 6.554e-15 | 2.647e-13 |
| 24 | PLATELET DERIVED GROWTH FACTOR RECEPTOR BINDING | 9 | 15 | 1.792e-14 | 6.936e-13 |
| 25 | PLATELET DERIVED GROWTH FACTOR BINDING | 8 | 11 | 5.343e-14 | 1.985e-12 |
| 26 | KINASE REGULATOR ACTIVITY | 20 | 186 | 7.563e-14 | 2.702e-12 |
| 27 | COLLAGEN BINDING | 13 | 65 | 5.892e-13 | 2.027e-11 |
| 28 | PHOSPHATASE BINDING | 18 | 162 | 7.889e-13 | 2.617e-11 |
| 29 | PROTEIN PHOSPHATASE BINDING | 16 | 120 | 8.876e-13 | 2.843e-11 |
| 30 | ENZYME REGULATOR ACTIVITY | 40 | 959 | 2.844e-12 | 8.806e-11 |
| 31 | EXTRACELLULAR MATRIX STRUCTURAL CONSTITUENT | 13 | 76 | 4.887e-12 | 1.465e-10 |
| 32 | PROTEIN SERINE THREONINE KINASE ACTIVITY | 26 | 445 | 2.085e-11 | 6.052e-10 |
| 33 | FIBROBLAST GROWTH FACTOR RECEPTOR BINDING | 9 | 28 | 2.164e-11 | 6.092e-10 |
| 34 | HEPARIN BINDING | 16 | 157 | 5.742e-11 | 1.569e-09 |
| 35 | INSULIN LIKE GROWTH FACTOR RECEPTOR BINDING | 7 | 15 | 1.731e-10 | 4.593e-09 |
| 36 | ADENYL NUCLEOTIDE BINDING | 48 | 1514 | 2.317e-10 | 5.978e-09 |
| 37 | RIBONUCLEOTIDE BINDING | 54 | 1860 | 3.49e-10 | 8.762e-09 |
| 38 | EXTRACELLULAR MATRIX BINDING | 10 | 51 | 3.618e-10 | 8.846e-09 |
| 39 | GLYCOSAMINOGLYCAN BINDING | 17 | 205 | 3.721e-10 | 8.865e-09 |
| 40 | RECEPTOR ACTIVITY | 50 | 1649 | 4.229e-10 | 9.822e-09 |
| 41 | SIGNALING RECEPTOR ACTIVITY | 45 | 1393 | 5.253e-10 | 1.19e-08 |
| 42 | FIBRONECTIN BINDING | 8 | 28 | 8.465e-10 | 1.854e-08 |
| 43 | IDENTICAL PROTEIN BINDING | 41 | 1209 | 8.583e-10 | 1.854e-08 |
| 44 | INSULIN RECEPTOR SUBSTRATE BINDING | 6 | 11 | 1.114e-09 | 2.352e-08 |
| 45 | PROTEIN HETERODIMERIZATION ACTIVITY | 24 | 468 | 1.781e-09 | 3.676e-08 |
| 46 | PROTEIN DIMERIZATION ACTIVITY | 39 | 1149 | 2.308e-09 | 4.661e-08 |
| 47 | SULFUR COMPOUND BINDING | 17 | 234 | 2.843e-09 | 5.62e-08 |
| 48 | PROTEIN PHOSPHATASE TYPE 2A REGULATOR ACTIVITY | 7 | 21 | 2.944e-09 | 5.698e-08 |
| 49 | CYTOKINE RECEPTOR BINDING | 18 | 271 | 3.991e-09 | 7.566e-08 |
| 50 | PROTEIN DOMAIN SPECIFIC BINDING | 27 | 624 | 6.402e-09 | 1.189e-07 |
| 51 | CYTOKINE BINDING | 11 | 92 | 1.135e-08 | 2.068e-07 |
| 52 | CHEMOATTRACTANT ACTIVITY | 7 | 27 | 2.117e-08 | 3.782e-07 |
| 53 | KINASE ACTIVATOR ACTIVITY | 9 | 62 | 4.486e-08 | 7.862e-07 |
| 54 | PHOSPHATIDYLINOSITOL 3 KINASE BINDING | 7 | 30 | 4.708e-08 | 8.1e-07 |
| 55 | CYTOKINE RECEPTOR ACTIVITY | 10 | 89 | 9.734e-08 | 1.644e-06 |
| 56 | NEUROTROPHIN RECEPTOR BINDING | 5 | 14 | 4.025e-07 | 6.678e-06 |
| 57 | RECEPTOR SIGNALING PROTEIN ACTIVITY | 12 | 172 | 9.879e-07 | 1.61e-05 |
| 58 | LAMININ BINDING | 6 | 30 | 1.187e-06 | 1.901e-05 |
| 59 | PROTEIN BINDING INVOLVED IN CELL ADHESION | 5 | 17 | 1.209e-06 | 1.903e-05 |
| 60 | INSULIN RECEPTOR BINDING | 6 | 32 | 1.776e-06 | 2.704e-05 |
| 61 | SODIUM CHANNEL REGULATOR ACTIVITY | 6 | 32 | 1.776e-06 | 2.704e-05 |
| 62 | HISTONE KINASE ACTIVITY | 5 | 19 | 2.228e-06 | 3.338e-05 |
| 63 | PROTEIN TYROSINE KINASE BINDING | 7 | 54 | 3.22e-06 | 4.748e-05 |
| 64 | PROTEASE BINDING | 9 | 104 | 3.954e-06 | 5.74e-05 |
| 65 | PROTEIN SERINE THREONINE TYROSINE KINASE ACTIVITY | 6 | 39 | 5.967e-06 | 8.528e-05 |
| 66 | ENZYME ACTIVATOR ACTIVITY | 18 | 471 | 1.329e-05 | 0.0001871 |
| 67 | PROTEIN HOMODIMERIZATION ACTIVITY | 23 | 722 | 1.628e-05 | 0.0002231 |
| 68 | UBIQUITIN LIKE PROTEIN LIGASE BINDING | 13 | 264 | 1.633e-05 | 0.0002231 |
| 69 | CYCLIN DEPENDENT PROTEIN SERINE THREONINE KINASE REGULATOR ACTIVITY | 5 | 28 | 1.727e-05 | 0.0002291 |
| 70 | PROTEIN PHOSPHATASE 2A BINDING | 5 | 28 | 1.727e-05 | 0.0002291 |
| 71 | VIRUS RECEPTOR ACTIVITY | 7 | 70 | 1.857e-05 | 0.000243 |
| 72 | CHANNEL REGULATOR ACTIVITY | 9 | 131 | 2.584e-05 | 0.0003334 |
| 73 | PHOSPHATIDYLINOSITOL PHOSPHATE KINASE ACTIVITY | 4 | 16 | 3.077e-05 | 0.0003916 |
| 74 | PEPTIDE HORMONE BINDING | 5 | 36 | 6.133e-05 | 0.0007699 |
| 75 | X14 3 3 PROTEIN BINDING | 4 | 19 | 6.373e-05 | 0.0007894 |
| 76 | PHOSPHATASE REGULATOR ACTIVITY | 7 | 87 | 7.639e-05 | 0.0009338 |
| 77 | CHEMOKINE BINDING | 4 | 21 | 9.66e-05 | 0.001165 |
| 78 | FIBROBLAST GROWTH FACTOR BINDING | 4 | 23 | 0.0001403 | 0.001671 |
| 79 | PROTEIN PHOSPHORYLATED AMINO ACID BINDING | 4 | 24 | 0.0001668 | 0.001937 |
| 80 | EPHRIN RECEPTOR BINDING | 4 | 24 | 0.0001668 | 0.001937 |
| 81 | SCAFFOLD PROTEIN BINDING | 5 | 45 | 0.0001823 | 0.002091 |
| 82 | HISTONE DEACETYLASE BINDING | 7 | 105 | 0.0002487 | 0.002818 |
| 83 | TAU PROTEIN BINDING | 3 | 12 | 0.0003299 | 0.003692 |
| 84 | CAMP RESPONSE ELEMENT BINDING | 3 | 13 | 0.0004251 | 0.004702 |
| 85 | STRUCTURAL MOLECULE ACTIVITY | 20 | 732 | 0.0004496 | 0.004914 |
| 86 | RECEPTOR ACTIVATOR ACTIVITY | 4 | 32 | 0.0005242 | 0.005598 |
| 87 | TRANSCRIPTION FACTOR BINDING | 16 | 524 | 0.0005203 | 0.005598 |
| 88 | BIOACTIVE LIPID RECEPTOR ACTIVITY | 3 | 14 | 0.0005364 | 0.005662 |
| 89 | HEAT SHOCK PROTEIN BINDING | 6 | 89 | 0.0006508 | 0.006793 |
| 90 | PDZ DOMAIN BINDING | 6 | 90 | 0.0006905 | 0.007128 |
| 91 | PHOSPHOPROTEIN BINDING | 5 | 60 | 0.0007061 | 0.007194 |
| 92 | GTPASE ACTIVITY | 10 | 246 | 0.0007124 | 0.007194 |
| 93 | PROTEIN KINASE C ACTIVITY | 3 | 16 | 0.0008109 | 0.008014 |
| 94 | CHLORIDE CHANNEL REGULATOR ACTIVITY | 3 | 16 | 0.0008109 | 0.008014 |
| 95 | HORMONE BINDING | 5 | 65 | 0.001018 | 0.009752 |
| 96 | CORECEPTOR ACTIVITY | 4 | 38 | 0.001018 | 0.009752 |
| 97 | CALCIUM CHANNEL REGULATOR ACTIVITY | 4 | 38 | 0.001018 | 0.009752 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | EXTRACELLULAR MATRIX COMPONENT | 29 | 125 | 1.681e-29 | 9.816e-27 |
| 2 | PROTEIN COMPLEX INVOLVED IN CELL ADHESION | 18 | 30 | 7.785e-28 | 2.273e-25 |
| 3 | EXTRACELLULAR MATRIX | 43 | 426 | 1.193e-27 | 2.323e-25 |
| 4 | PROTEINACEOUS EXTRACELLULAR MATRIX | 39 | 356 | 1.965e-26 | 2.869e-24 |
| 5 | BASEMENT MEMBRANE | 24 | 93 | 8.621e-26 | 1.007e-23 |
| 6 | RECEPTOR COMPLEX | 36 | 327 | 1.653e-24 | 1.609e-22 |
| 7 | CELL SURFACE | 48 | 757 | 6.076e-22 | 5.069e-20 |
| 8 | CELL SUBSTRATE JUNCTION | 36 | 398 | 1.393e-21 | 1.017e-19 |
| 9 | PLASMA MEMBRANE RECEPTOR COMPLEX | 26 | 175 | 2.881e-21 | 1.869e-19 |
| 10 | COMPLEX OF COLLAGEN TRIMERS | 12 | 23 | 6.635e-18 | 3.875e-16 |
| 11 | SIDE OF MEMBRANE | 33 | 428 | 9.967e-18 | 4.85e-16 |
| 12 | ANCHORING JUNCTION | 35 | 489 | 9.752e-18 | 4.85e-16 |
| 13 | PLASMA MEMBRANE PROTEIN COMPLEX | 35 | 510 | 3.628e-17 | 1.63e-15 |
| 14 | BASAL LAMINA | 11 | 21 | 1.554e-16 | 6.484e-15 |
| 15 | MEMBRANE PROTEIN COMPLEX | 46 | 1020 | 2.806e-15 | 1.092e-13 |
| 16 | EXTRINSIC COMPONENT OF MEMBRANE | 24 | 252 | 3.449e-15 | 1.259e-13 |
| 17 | TRANSFERASE COMPLEX TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 23 | 237 | 8.869e-15 | 3.047e-13 |
| 18 | COLLAGEN TRIMER | 15 | 88 | 1.133e-13 | 3.676e-12 |
| 19 | EXTRACELLULAR SPACE | 51 | 1376 | 1.62e-13 | 4.979e-12 |
| 20 | PHOSPHATIDYLINOSITOL 3 KINASE COMPLEX | 9 | 20 | 5.712e-13 | 1.668e-11 |
| 21 | CELL JUNCTION | 45 | 1151 | 9.293e-13 | 2.584e-11 |
| 22 | INTRINSIC COMPONENT OF PLASMA MEMBRANE | 55 | 1649 | 1.064e-12 | 2.825e-11 |
| 23 | CATALYTIC COMPLEX | 42 | 1038 | 2.003e-12 | 5.086e-11 |
| 24 | MEMBRANE REGION | 44 | 1134 | 2.25e-12 | 5.474e-11 |
| 25 | INTRACELLULAR VESICLE | 46 | 1259 | 5.238e-12 | 1.224e-10 |
| 26 | CYTOPLASMIC SIDE OF MEMBRANE | 17 | 170 | 1.923e-11 | 4.319e-10 |
| 27 | PROTEIN PHOSPHATASE TYPE 2A COMPLEX | 8 | 20 | 3.722e-11 | 8.051e-10 |
| 28 | PROTEIN KINASE COMPLEX | 13 | 90 | 4.546e-11 | 9.481e-10 |
| 29 | PLATELET ALPHA GRANULE | 12 | 75 | 7.468e-11 | 1.504e-09 |
| 30 | EXTRINSIC COMPONENT OF CYTOPLASMIC SIDE OF PLASMA MEMBRANE | 13 | 98 | 1.368e-10 | 2.663e-09 |
| 31 | MEMBRANE MICRODOMAIN | 20 | 288 | 2.413e-10 | 4.545e-09 |
| 32 | ENDOPLASMIC RETICULUM LUMEN | 17 | 201 | 2.737e-10 | 4.995e-09 |
| 33 | PLASMA MEMBRANE REGION | 36 | 929 | 3.093e-10 | 5.473e-09 |
| 34 | VESICLE LUMEN | 13 | 106 | 3.727e-10 | 6.401e-09 |
| 35 | PLATELET ALPHA GRANULE LUMEN | 10 | 55 | 7.948e-10 | 1.326e-08 |
| 36 | EXTRINSIC COMPONENT OF PLASMA MEMBRANE | 14 | 136 | 8.202e-10 | 1.331e-08 |
| 37 | HETEROTRIMERIC G PROTEIN COMPLEX | 8 | 32 | 2.751e-09 | 4.342e-08 |
| 38 | EXTERNAL SIDE OF PLASMA MEMBRANE | 17 | 238 | 3.676e-09 | 5.649e-08 |
| 39 | PERINUCLEAR REGION OF CYTOPLASM | 27 | 642 | 1.164e-08 | 1.743e-07 |
| 40 | CYCLIN DEPENDENT PROTEIN KINASE HOLOENZYME COMPLEX | 7 | 31 | 6.021e-08 | 8.791e-07 |
| 41 | SECRETORY GRANULE LUMEN | 10 | 85 | 6.242e-08 | 8.891e-07 |
| 42 | PHOSPHATASE COMPLEX | 8 | 48 | 8.387e-08 | 1.166e-06 |
| 43 | CYTOPLASMIC VESICLE PART | 24 | 601 | 2.071e-07 | 2.813e-06 |
| 44 | PLASMA MEMBRANE RAFT | 9 | 86 | 7.954e-07 | 1.056e-05 |
| 45 | TRANSFERASE COMPLEX | 25 | 703 | 9.783e-07 | 1.27e-05 |
| 46 | VACUOLE | 34 | 1180 | 1.289e-06 | 1.637e-05 |
| 47 | SECRETORY GRANULE | 16 | 352 | 4.781e-06 | 5.94e-05 |
| 48 | BANDED COLLAGEN FIBRIL | 4 | 12 | 8.686e-06 | 0.0001057 |
| 49 | CELL PROJECTION | 42 | 1786 | 1.177e-05 | 0.0001403 |
| 50 | BASAL PART OF CELL | 6 | 51 | 2.927e-05 | 0.0003418 |
| 51 | RUFFLE MEMBRANE | 7 | 80 | 4.453e-05 | 0.00051 |
| 52 | ENDOPLASMIC RETICULUM | 37 | 1631 | 8.963e-05 | 0.001007 |
| 53 | SECRETORY VESICLE | 16 | 461 | 0.000125 | 0.001377 |
| 54 | NEURON PROJECTION | 25 | 942 | 0.0001361 | 0.001472 |
| 55 | MYELIN SHEATH | 9 | 168 | 0.0001772 | 0.001882 |
| 56 | LEADING EDGE MEMBRANE | 8 | 134 | 0.0001941 | 0.002025 |
| 57 | BASOLATERAL PLASMA MEMBRANE | 10 | 211 | 0.0002127 | 0.002141 |
| 58 | NEURON PART | 30 | 1265 | 0.0002098 | 0.002141 |
| 59 | PIGMENT GRANULE | 7 | 103 | 0.0002209 | 0.002186 |
| 60 | SOMATODENDRITIC COMPARTMENT | 19 | 650 | 0.000273 | 0.002658 |
| 61 | CELL LEADING EDGE | 13 | 350 | 0.0002823 | 0.002703 |
| 62 | RUFFLE | 8 | 156 | 0.0005414 | 0.0051 |
| 63 | BASAL PLASMA MEMBRANE | 4 | 33 | 0.0005911 | 0.005479 |
| 64 | SYNAPSE | 20 | 754 | 0.0006512 | 0.005942 |
| 65 | CELL PROJECTION MEMBRANE | 11 | 298 | 0.0008665 | 0.007785 |
| 66 | ENDOCYTIC VESICLE | 10 | 256 | 0.0009655 | 0.008543 |
| 67 | DENDRITE | 14 | 451 | 0.0009862 | 0.008596 |
| 68 | LAMELLIPODIUM | 8 | 172 | 0.001025 | 0.0088 |
| 69 | FILOPODIUM MEMBRANE | 3 | 18 | 0.001161 | 0.009828 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
| 1 | hsa04151_PI3K_AKT_signaling_pathway | 236 | 351 | 0 | 0 | |
| 2 | hsa04510_Focal_adhesion | 94 | 200 | 8.737e-133 | 7.863e-131 | |
| 3 | hsa04014_Ras_signaling_pathway | 83 | 236 | 3.787e-103 | 2.272e-101 | |
| 4 | hsa04512_ECM.receptor_interaction | 52 | 85 | 3.988e-80 | 1.795e-78 | |
| 5 | hsa04810_Regulation_of_actin_cytoskeleton | 61 | 214 | 2.973e-68 | 1.07e-66 | |
| 6 | hsa04722_Neurotrophin_signaling_pathway | 38 | 127 | 3.509e-43 | 1.053e-41 | |
| 7 | hsa04010_MAPK_signaling_pathway | 45 | 268 | 7.568e-39 | 1.946e-37 | |
| 8 | hsa04150_mTOR_signaling_pathway | 26 | 52 | 6.848e-37 | 1.541e-35 | |
| 9 | hsa04012_ErbB_signaling_pathway | 30 | 87 | 2.364e-36 | 4.727e-35 | |
| 10 | hsa04910_Insulin_signaling_pathway | 34 | 138 | 1.91e-35 | 3.439e-34 | |
| 11 | hsa04062_Chemokine_signaling_pathway | 36 | 189 | 3.634e-33 | 5.947e-32 | |
| 12 | hsa04662_B_cell_receptor_signaling_pathway | 25 | 75 | 5.375e-30 | 8.062e-29 | |
| 13 | hsa04630_Jak.STAT_signaling_pathway | 29 | 155 | 1.32e-26 | 1.827e-25 | |
| 14 | hsa04370_VEGF_signaling_pathway | 23 | 76 | 1.49e-26 | 1.915e-25 | |
| 15 | hsa04620_Toll.like_receptor_signaling_pathway | 22 | 102 | 7.362e-22 | 8.835e-21 | |
| 16 | hsa04664_Fc_epsilon_RI_signaling_pathway | 20 | 79 | 1.739e-21 | 1.956e-20 | |
| 17 | hsa04660_T_cell_receptor_signaling_pathway | 22 | 108 | 2.817e-21 | 2.982e-20 | |
| 18 | hsa04380_Osteoclast_differentiation | 23 | 128 | 7.051e-21 | 7.051e-20 | |
| 19 | hsa04960_Aldosterone.regulated_sodium_reabsorption | 16 | 42 | 1.073e-20 | 1.017e-19 | |
| 20 | hsa04640_Hematopoietic_cell_lineage | 19 | 88 | 4.843e-19 | 4.359e-18 | |
| 21 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 21 | 136 | 9.736e-18 | 8.345e-17 | |
| 22 | hsa04210_Apoptosis | 18 | 89 | 1.478e-17 | 1.209e-16 | |
| 23 | hsa04540_Gap_junction | 18 | 90 | 1.828e-17 | 1.43e-16 | |
| 24 | hsa04110_Cell_cycle | 20 | 128 | 4.72e-17 | 3.54e-16 | |
| 25 | hsa04114_Oocyte_meiosis | 19 | 114 | 8.398e-17 | 6.047e-16 | |
| 26 | hsa04914_Progesterone.mediated_oocyte_maturation | 16 | 87 | 4.777e-15 | 3.307e-14 | |
| 27 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 16 | 95 | 2.048e-14 | 1.365e-13 | |
| 28 | hsa04115_p53_signaling_pathway | 14 | 69 | 5.996e-14 | 3.855e-13 | |
| 29 | hsa04390_Hippo_signaling_pathway | 18 | 154 | 3.268e-13 | 2.028e-12 | |
| 30 | hsa04730_Long.term_depression | 13 | 70 | 1.617e-12 | 9.701e-12 | |
| 31 | hsa04974_Protein_digestion_and_absorption | 13 | 81 | 1.14e-11 | 6.621e-11 | |
| 32 | hsa04916_Melanogenesis | 13 | 101 | 2.013e-10 | 1.132e-09 | |
| 33 | hsa04350_TGF.beta_signaling_pathway | 12 | 85 | 3.39e-10 | 1.849e-09 | |
| 34 | hsa04360_Axon_guidance | 14 | 130 | 4.491e-10 | 2.377e-09 | |
| 35 | hsa04144_Endocytosis | 16 | 203 | 2.607e-09 | 1.341e-08 | |
| 36 | hsa04320_Dorso.ventral_axis_formation | 7 | 25 | 1.169e-08 | 5.845e-08 | |
| 37 | hsa04670_Leukocyte_transendothelial_migration | 12 | 117 | 1.417e-08 | 6.892e-08 | |
| 38 | hsa04310_Wnt_signaling_pathway | 13 | 151 | 2.937e-08 | 1.391e-07 | |
| 39 | hsa04912_GnRH_signaling_pathway | 11 | 101 | 3.059e-08 | 1.412e-07 | |
| 40 | hsa04973_Carbohydrate_digestion_and_absorption | 8 | 44 | 4.103e-08 | 1.846e-07 | |
| 41 | hsa04530_Tight_junction | 12 | 133 | 6.006e-08 | 2.637e-07 | |
| 42 | hsa04920_Adipocytokine_signaling_pathway | 9 | 68 | 1.025e-07 | 4.392e-07 | |
| 43 | hsa04070_Phosphatidylinositol_signaling_system | 9 | 78 | 3.423e-07 | 1.418e-06 | |
| 44 | hsa04145_Phagosome | 12 | 156 | 3.467e-07 | 1.418e-06 | |
| 45 | hsa04621_NOD.like_receptor_signaling_pathway | 8 | 59 | 4.408e-07 | 1.763e-06 | |
| 46 | hsa03015_mRNA_surveillance_pathway | 9 | 83 | 5.863e-07 | 2.294e-06 | |
| 47 | hsa04720_Long.term_potentiation | 8 | 70 | 1.679e-06 | 6.429e-06 | |
| 48 | hsa04520_Adherens_junction | 8 | 73 | 2.319e-06 | 8.695e-06 | |
| 49 | hsa04623_Cytosolic_DNA.sensing_pathway | 5 | 56 | 0.0005131 | 0.001885 | |
| 50 | hsa00562_Inositol_phosphate_metabolism | 5 | 57 | 0.0005571 | 0.002006 | |
| 51 | hsa04514_Cell_adhesion_molecules_.CAMs. | 7 | 136 | 0.001173 | 0.004139 | |
| 52 | hsa04020_Calcium_signaling_pathway | 7 | 177 | 0.005137 | 0.01778 | |
| 53 | hsa04622_RIG.I.like_receptor_signaling_pathway | 4 | 71 | 0.009909 | 0.03365 | |
| 54 | hsa04270_Vascular_smooth_muscle_contraction | 5 | 116 | 0.01221 | 0.04069 | |
| 55 | hsa04962_Vasopressin.regulated_water_reabsorption | 3 | 44 | 0.01505 | 0.04927 | |
| 56 | hsa04140_Regulation_of_autophagy | 2 | 34 | 0.06077 | 0.1953 | |
| 57 | hsa03013_RNA_transport | 4 | 152 | 0.1058 | 0.3341 | |
| 58 | hsa04672_Intestinal_immune_network_for_IgA_production | 2 | 49 | 0.1138 | 0.3531 | |
| 59 | hsa00500_Starch_and_sucrose_metabolism | 2 | 54 | 0.1334 | 0.407 | |
| 60 | hsa04610_Complement_and_coagulation_cascades | 2 | 69 | 0.1958 | 0.5874 | |
| 61 | hsa04612_Antigen_processing_and_presentation | 2 | 78 | 0.2347 | 0.6927 | |
| 62 | hsa04972_Pancreatic_secretion | 2 | 101 | 0.335 | 0.9571 | |
| 63 | hsa04120_Ubiquitin_mediated_proteolysis | 2 | 139 | 0.4899 | 1 | |
| 64 | hsa04141_Protein_processing_in_endoplasmic_reticulum | 2 | 168 | 0.5919 | 1 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CTD-2171N6.1 |
hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-20b-5p;hsa-miR-23b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p | 10 | COL1A1 | Sponge network | 5.89 | 0 | 3.608 | 0 | 0.772 |
| 2 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-31-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-708-3p;hsa-miR-944 | 23 | FGF7 | Sponge network | -0.939 | 4.0E-5 | -2.015 | 0 | 0.755 |
| 3 | RP11-815I9.4 |
hsa-miR-103a-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-2355-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-589-3p;hsa-miR-590-5p | 12 | PDPK1 | Sponge network | 0.004 | 0.98116 | -0.375 | 0.00067 | 0.744 |
| 4 | RP11-815I9.4 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-15a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-320a | 11 | ATF2 | Sponge network | 0.004 | 0.98116 | 0.104 | 0.47373 | 0.74 |
| 5 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-149-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-944 | 38 | PDGFRA | Sponge network | -0.939 | 4.0E-5 | -0.463 | 0.04818 | 0.727 |
| 6 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-301a-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | FGFR1 | Sponge network | -0.939 | 4.0E-5 | -0.285 | 0.17112 | 0.723 |
| 7 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-224-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 26 | IGF1 | Sponge network | -0.939 | 4.0E-5 | -1.836 | 0 | 0.709 |
| 8 | RP11-815I9.4 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-664a-3p | 12 | FOXO3 | Sponge network | 0.004 | 0.98116 | -0.326 | 0.00103 | 0.708 |
| 9 | SOX2-OT | hsa-miR-125a-3p;hsa-miR-146b-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-22-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-455-3p;hsa-miR-576-5p | 10 | NTRK2 | Sponge network | 0.38 | 0.29773 | -1.115 | 0.01397 | 0.706 |
| 10 | RP11-815I9.4 |
hsa-miR-103a-2-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-30c-5p;hsa-miR-365a-3p;hsa-miR-576-5p;hsa-miR-590-5p | 10 | SOS1 | Sponge network | 0.004 | 0.98116 | -0.144 | 0.07887 | 0.69 |
| 11 | MAGI2-AS3 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-200b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-429;hsa-miR-452-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p | 18 | ITGA9 | Sponge network | -0.939 | 4.0E-5 | -1.022 | 1.0E-5 | 0.689 |
| 12 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-200a-3p;hsa-miR-224-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-944 | 12 | HGF | Sponge network | -0.939 | 4.0E-5 | -0.629 | 0.02489 | 0.681 |
| 13 | CASC15 |
hsa-let-7g-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-363-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | COL1A2 | Sponge network | 1.485 | 0 | 2.674 | 0 | 0.679 |
| 14 | CASC15 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-23b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-590-3p | 11 | COL1A1 | Sponge network | 1.485 | 0 | 3.608 | 0 | 0.666 |
| 15 | FAM83A-AS1 |
hsa-miR-127-3p;hsa-miR-148a-3p;hsa-miR-181c-5p;hsa-miR-199a-5p;hsa-miR-23b-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-335-3p;hsa-miR-491-5p;hsa-miR-495-3p | 10 | TGFA | Sponge network | 1.064 | 0.00307 | 0.472 | 0.01138 | 0.666 |
| 16 | MAGI2-AS3 |
hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p | 10 | KDR | Sponge network | -0.939 | 4.0E-5 | -0.064 | 0.72302 | 0.661 |
| 17 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-505-3p | 11 | FGF18 | Sponge network | -0.939 | 4.0E-5 | -1.888 | 0 | 0.659 |
| 18 | LINC00941 |
hsa-miR-106a-5p;hsa-miR-140-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-497-5p | 22 | CDK6 | Sponge network | 4.292 | 0 | 0.944 | 0 | 0.658 |
| 19 | MAGI2-AS3 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-205-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-944 | 13 | ITGA8 | Sponge network | -0.939 | 4.0E-5 | -0.747 | 0.01533 | 0.654 |
| 20 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-671-5p | 18 | KIT | Sponge network | -0.939 | 4.0E-5 | -1.942 | 0 | 0.652 |
| 21 | MAGI2-AS3 |
hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p | 11 | ITGB3 | Sponge network | -0.939 | 4.0E-5 | -0.16 | 0.54839 | 0.639 |
| 22 | SOX21-AS1 |
hsa-miR-125a-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-22-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p | 14 | NTRK2 | Sponge network | -1.061 | 0.01562 | -1.115 | 0.01397 | 0.635 |
| 23 | RAMP2-AS1 |
hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-365a-3p;hsa-miR-455-5p;hsa-miR-7-5p | 12 | BCL2 | Sponge network | -1.258 | 0.00026 | -0.91 | 1.0E-5 | 0.623 |
| 24 | RP1-193H18.2 |
hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-542-3p;hsa-miR-7-5p | 11 | BCL2 | Sponge network | -1.539 | 0 | -0.91 | 1.0E-5 | 0.62 |
| 25 | RP11-166D19.1 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-301a-3p | 10 | FGFR1 | Sponge network | -0.882 | 5.0E-5 | -0.285 | 0.17112 | 0.62 |
| 26 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-31-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-944 | 19 | FGF7 | Sponge network | -0.882 | 5.0E-5 | -2.015 | 0 | 0.611 |
| 27 | DNM3OS |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-944;hsa-miR-96-5p | 18 | PDGFRA | Sponge network | 0.932 | 0.00442 | -0.463 | 0.04818 | 0.601 |
| 28 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-149-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-944;hsa-miR-96-5p | 27 | PDGFRA | Sponge network | -0.882 | 5.0E-5 | -0.463 | 0.04818 | 0.6 |
| 29 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-149-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-93-5p | 11 | PDGFRA | Sponge network | -0.317 | 0.39052 | -0.463 | 0.04818 | 0.6 |
| 30 | ZNF667-AS1 |
hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-452-5p;hsa-miR-590-5p;hsa-miR-7-5p | 12 | BCL2 | Sponge network | -1.395 | 0 | -0.91 | 1.0E-5 | 0.598 |
| 31 | RP11-400K9.4 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-93-5p | 13 | CSF1 | Sponge network | -0.304 | 0.38627 | -0.395 | 0.03662 | 0.598 |
| 32 | CASC15 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-7-1-3p | 10 | ITGA1 | Sponge network | 1.485 | 0 | 1.673 | 0 | 0.597 |
| 33 | AC007879.5 | hsa-miR-101-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-34a-5p;hsa-miR-502-3p | 12 | CDK6 | Sponge network | 2.671 | 0 | 0.944 | 0 | 0.595 |
| 34 | RP11-193H5.1 |
hsa-miR-103a-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-2355-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-374a-3p;hsa-miR-93-3p | 14 | PDPK1 | Sponge network | -0.426 | 0.01062 | -0.375 | 0.00067 | 0.59 |
| 35 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 25 | PIK3R1 | Sponge network | -0.939 | 4.0E-5 | -1.219 | 0 | 0.589 |
| 36 | RP11-389C8.2 |
hsa-miR-130a-3p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-93-5p | 10 | CSF1 | Sponge network | 0.187 | 0.31051 | -0.395 | 0.03662 | 0.587 |
| 37 | AC007879.7 |
hsa-miR-101-3p;hsa-miR-140-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-340-5p | 18 | CDK6 | Sponge network | 2.7 | 0 | 0.944 | 0 | 0.587 |
| 38 | RP11-426C22.4 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-200b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-942-5p | 10 | LAMC1 | Sponge network | 3.117 | 0 | 1.059 | 0 | 0.578 |
| 39 | MAGI2-AS3 |
hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-31-5p;hsa-miR-330-5p;hsa-miR-3615;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-939-5p;hsa-miR-940 | 16 | GNG7 | Sponge network | -0.939 | 4.0E-5 | -1.969 | 0 | 0.571 |
| 40 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-452-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 23 | PRKAA2 | Sponge network | -4.323 | 0 | -3.347 | 0 | 0.567 |
| 41 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-205-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | INSR | Sponge network | -0.939 | 4.0E-5 | -0.194 | 0.13669 | 0.566 |
| 42 | FAM225B |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-92a-3p | 11 | LAMC1 | Sponge network | 2.186 | 0 | 1.059 | 0 | 0.565 |
| 43 | RP11-221N13.3 | hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-20b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-9-5p | 15 | CDK6 | Sponge network | 5.876 | 0 | 0.944 | 0 | 0.56 |
| 44 | SH3RF3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-200b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-877-5p | 11 | ITGA9 | Sponge network | -0.175 | 0.58985 | -1.022 | 1.0E-5 | 0.552 |
| 45 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-671-5p | 12 | KIT | Sponge network | -1.258 | 0.00026 | -1.942 | 0 | 0.55 |
| 46 | ZNF667-AS1 |
hsa-miR-130a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-671-5p | 11 | KIT | Sponge network | -1.395 | 0 | -1.942 | 0 | 0.545 |
| 47 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 10 | COL1A2 | Sponge network | -0.939 | 4.0E-5 | 2.674 | 0 | 0.543 |
| 48 | RP11-594N15.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-452-3p;hsa-miR-93-5p | 20 | PRKAA2 | Sponge network | -2.86 | 0 | -3.347 | 0 | 0.543 |
| 49 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-224-3p;hsa-miR-27a-3p;hsa-miR-301a-3p | 11 | IGF1 | Sponge network | 0.187 | 0.31051 | -1.836 | 0 | 0.541 |
| 50 | MAGI2-AS3 |
hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-205-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 15 | LPAR1 | Sponge network | -0.939 | 4.0E-5 | -0.679 | 0.00029 | 0.539 |
| 51 | RP5-1185I7.1 | hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-576-5p | 10 | NTRK2 | Sponge network | -1.805 | 0.01092 | -1.115 | 0.01397 | 0.538 |
| 52 | RP11-815I9.4 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-590-5p | 14 | IL6R | Sponge network | 0.004 | 0.98116 | -0.049 | 0.80939 | 0.537 |
| 53 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-149-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | PDGFRA | Sponge network | -0.175 | 0.58985 | -0.463 | 0.04818 | 0.535 |
| 54 | RP11-150O12.1 |
hsa-miR-106a-5p;hsa-miR-140-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p | 17 | CDK6 | Sponge network | 1.523 | 0 | 0.944 | 0 | 0.534 |
| 55 | RP11-284N8.3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-338-3p | 10 | KIT | Sponge network | 0.003 | 0.99478 | -1.942 | 0 | 0.533 |
| 56 | MAGI2-AS3 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-205-5p;hsa-miR-224-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | RELN | Sponge network | -0.939 | 4.0E-5 | -2.339 | 0 | 0.532 |
| 57 | AC007743.1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-501-5p;hsa-miR-671-5p | 15 | KIT | Sponge network | -1.053 | 0.00923 | -1.942 | 0 | 0.531 |
| 58 | RP11-30P6.6 |
hsa-miR-148a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-429;hsa-miR-628-5p;hsa-miR-660-5p | 12 | IRS1 | Sponge network | 2.847 | 0 | 0.369 | 0.06704 | 0.531 |
| 59 | RP11-30P6.6 |
hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-140-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-374a-5p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-660-5p | 28 | CDK6 | Sponge network | 2.847 | 0 | 0.944 | 0 | 0.53 |
| 60 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-576-5p | 14 | IGF1 | Sponge network | -0.175 | 0.58985 | -1.836 | 0 | 0.528 |
| 61 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-503-5p;hsa-miR-589-3p;hsa-miR-92a-3p;hsa-miR-944 | 16 | FGF2 | Sponge network | -0.939 | 4.0E-5 | -0.874 | 0.00075 | 0.527 |
| 62 | CTA-221G9.11 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-93-5p | 12 | PRKAA2 | Sponge network | -2.198 | 0 | -3.347 | 0 | 0.527 |
| 63 | DNM3OS |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-200a-3p;hsa-miR-224-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-944 | 11 | HGF | Sponge network | 0.932 | 0.00442 | -0.629 | 0.02489 | 0.521 |
| 64 | RP11-760H22.2 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-331-3p;hsa-miR-454-3p;hsa-miR-589-5p;hsa-miR-944 | 12 | PRKAA2 | Sponge network | -2.79 | 0 | -3.347 | 0 | 0.52 |
| 65 | LINC00707 |
hsa-miR-148a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-628-5p | 12 | IRS1 | Sponge network | 2.018 | 1.0E-5 | 0.369 | 0.06704 | 0.517 |
| 66 | MAGI2-AS3 |
hsa-miR-141-5p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p | 10 | ITGA1 | Sponge network | -0.939 | 4.0E-5 | 1.673 | 0 | 0.517 |
| 67 | SNHG14 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-342-3p;hsa-miR-452-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-944 | 17 | PRKAA2 | Sponge network | -1.125 | 0.0001 | -3.347 | 0 | 0.515 |
| 68 | RP11-815I9.4 |
hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-29b-1-5p;hsa-miR-671-5p;hsa-miR-92a-1-5p | 12 | CREB3L2 | Sponge network | 0.004 | 0.98116 | -0.271 | 0.03509 | 0.513 |
| 69 | PWAR6 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-944 | 16 | PRKAA2 | Sponge network | -1.629 | 0 | -3.347 | 0 | 0.512 |
| 70 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-200a-3p;hsa-miR-224-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-944 | 10 | HGF | Sponge network | -0.882 | 5.0E-5 | -0.629 | 0.02489 | 0.51 |
| 71 | PWAR6 |
hsa-miR-17-5p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-7-5p | 10 | BCL2 | Sponge network | -1.629 | 0 | -0.91 | 1.0E-5 | 0.509 |
| 72 | PCBP1-AS1 | hsa-miR-181b-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-365a-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-7-5p | 10 | BCL2 | Sponge network | -0.746 | 0 | -0.91 | 1.0E-5 | 0.509 |
| 73 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-944 | 13 | FGF2 | Sponge network | -0.882 | 5.0E-5 | -0.874 | 0.00075 | 0.509 |
| 74 | RP11-774O3.3 |
hsa-miR-17-5p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-365a-3p;hsa-miR-542-3p;hsa-miR-7-5p | 10 | BCL2 | Sponge network | -1.712 | 0 | -0.91 | 1.0E-5 | 0.509 |
| 75 | CTD-2547L24.4 | hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-378a-3p | 13 | CDK6 | Sponge network | 1.587 | 0 | 0.944 | 0 | 0.508 |
| 76 | RP11-166D19.1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-200b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-429;hsa-miR-452-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-877-5p | 16 | ITGA9 | Sponge network | -0.882 | 5.0E-5 | -1.022 | 1.0E-5 | 0.506 |
| 77 | SNHG14 |
hsa-miR-17-5p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-365a-3p;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-7-5p | 10 | BCL2 | Sponge network | -1.125 | 0.0001 | -0.91 | 1.0E-5 | 0.505 |
| 78 | LINC00707 |
hsa-miR-106a-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-664a-3p | 31 | CDK6 | Sponge network | 2.018 | 1.0E-5 | 0.944 | 0 | 0.503 |
| 79 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-671-5p | 11 | KIT | Sponge network | 0.187 | 0.31051 | -1.942 | 0 | 0.502 |
| 80 | RP11-193H5.1 |
hsa-miR-103a-2-5p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-501-5p;hsa-miR-576-5p | 13 | IL6R | Sponge network | -0.426 | 0.01062 | -0.049 | 0.80939 | 0.502 |
| 81 | PWAR6 |
hsa-miR-130a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-671-5p | 11 | KIT | Sponge network | -1.629 | 0 | -1.942 | 0 | 0.501 |
| 82 | SNHG14 |
hsa-miR-130a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-671-5p | 12 | KIT | Sponge network | -1.125 | 0.0001 | -1.942 | 0 | 0.498 |
| 83 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-205-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | FGF7 | Sponge network | -0.175 | 0.58985 | -2.015 | 0 | 0.496 |
| 84 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-224-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-629-5p;hsa-miR-940 | 19 | IGF1 | Sponge network | -0.882 | 5.0E-5 | -1.836 | 0 | 0.496 |
| 85 | SNHG14 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-629-5p | 12 | IGF1 | Sponge network | -1.125 | 0.0001 | -1.836 | 0 | 0.495 |
| 86 | MAGI2-AS3 |
hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-224-3p;hsa-miR-23a-5p;hsa-miR-23b-5p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-339-5p;hsa-miR-454-3p;hsa-miR-93-5p | 15 | IGF2 | Sponge network | -0.939 | 4.0E-5 | 0.131 | 0.60661 | 0.494 |
| 87 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-452-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-944 | 31 | PRKAA2 | Sponge network | -0.939 | 4.0E-5 | -3.347 | 0 | 0.494 |
| 88 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-93-5p;hsa-miR-944 | 16 | PDGFRA | Sponge network | 0.187 | 0.31051 | -0.463 | 0.04818 | 0.493 |
| 89 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-452-3p;hsa-miR-500a-5p | 15 | PRKAA2 | Sponge network | -1.258 | 0.00026 | -3.347 | 0 | 0.491 |
| 90 | AC007743.1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-342-3p;hsa-miR-500a-5p;hsa-miR-7-1-3p | 15 | PRKAA2 | Sponge network | -1.053 | 0.00923 | -3.347 | 0 | 0.491 |
| 91 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-454-3p;hsa-miR-671-5p | 13 | KIT | Sponge network | -0.233 | 0.50729 | -1.942 | 0 | 0.49 |
| 92 | SNHG14 |
hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-31-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-944 | 12 | FGF7 | Sponge network | -1.125 | 0.0001 | -2.015 | 0 | 0.49 |
| 93 | AC011738.4 |
hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-140-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-20b-5p;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-497-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-9-5p | 25 | CDK6 | Sponge network | 3.153 | 0 | 0.944 | 0 | 0.489 |
| 94 | MAGI2-AS3 |
hsa-miR-17-5p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-365a-3p;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-7-5p | 13 | BCL2 | Sponge network | -0.939 | 4.0E-5 | -0.91 | 1.0E-5 | 0.488 |
| 95 | RP1-193H18.2 |
hsa-miR-181a-2-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-31-5p;hsa-miR-3614-5p;hsa-miR-96-5p | 10 | GNG7 | Sponge network | -1.539 | 0 | -1.969 | 0 | 0.488 |
| 96 | FGF14-AS2 | hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-452-5p;hsa-miR-7-5p | 10 | BCL2 | Sponge network | -1.914 | 0 | -0.91 | 1.0E-5 | 0.488 |
| 97 | MIR133A1 |
hsa-miR-103a-2-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-452-3p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-944 | 11 | PRKAA2 | Sponge network | -3.563 | 0 | -3.347 | 0 | 0.488 |
| 98 | MAGI2-AS3 |
hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-708-5p;hsa-miR-93-5p;hsa-miR-940 | 20 | CSF1 | Sponge network | -0.939 | 4.0E-5 | -0.395 | 0.03662 | 0.488 |
| 99 | MAGI2-AS3 |
hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | FLT1 | Sponge network | -0.939 | 4.0E-5 | 0.739 | 0 | 0.487 |
| 100 | RP11-193H5.1 |
hsa-miR-130a-3p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-1-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-501-5p | 12 | MAPK1 | Sponge network | -0.426 | 0.01062 | -0.025 | 0.75566 | 0.487 |
| 101 | ZNF667-AS1 |
hsa-miR-141-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-31-5p;hsa-miR-330-5p;hsa-miR-3615;hsa-miR-671-5p;hsa-miR-939-5p;hsa-miR-96-5p | 12 | GNG7 | Sponge network | -1.395 | 0 | -1.969 | 0 | 0.487 |
| 102 | RP1-122P22.2 | hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-29b-1-5p;hsa-miR-92a-1-5p | 10 | CREB3L2 | Sponge network | -1.073 | 6.0E-5 | -0.271 | 0.03509 | 0.485 |
| 103 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | THBS2 | Sponge network | -0.939 | 4.0E-5 | 1.626 | 0 | 0.485 |
| 104 | RP11-594N15.3 |
hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-93-5p | 15 | PIK3R1 | Sponge network | -2.86 | 0 | -1.219 | 0 | 0.483 |
| 105 | RP11-284N8.3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-944;hsa-miR-96-5p | 16 | PDGFRA | Sponge network | 0.003 | 0.99478 | -0.463 | 0.04818 | 0.483 |
| 106 | PWAR6 |
hsa-miR-141-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-31-3p;hsa-miR-31-5p;hsa-miR-330-5p;hsa-miR-671-5p;hsa-miR-939-5p | 11 | GNG7 | Sponge network | -1.629 | 0 | -1.969 | 0 | 0.483 |
| 107 | AC003090.1 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-31-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-452-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | FGF7 | Sponge network | -4.323 | 0 | -2.015 | 0 | 0.483 |
| 108 | AC007743.1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p | 12 | IGF1 | Sponge network | -1.053 | 0.00923 | -1.836 | 0 | 0.482 |
| 109 | SNHG14 |
hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-455-3p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-629-3p | 11 | PIK3R1 | Sponge network | -1.125 | 0.0001 | -1.219 | 0 | 0.482 |
| 110 | MAGI2-AS3 |
hsa-miR-135b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-429;hsa-miR-93-5p;hsa-miR-944 | 10 | COL4A3 | Sponge network | -0.939 | 4.0E-5 | -0.782 | 0.02643 | 0.482 |
| 111 | RP5-1198O20.4 |
hsa-miR-125a-5p;hsa-miR-140-3p;hsa-miR-217;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-328-3p;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-582-3p;hsa-miR-664a-3p | 10 | YWHAZ | Sponge network | 0.114 | 0.54508 | 0.345 | 0.0016 | 0.481 |
| 112 | AC007743.1 |
hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p | 13 | PIK3R1 | Sponge network | -1.053 | 0.00923 | -1.219 | 0 | 0.478 |
| 113 | RP11-145A3.1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-20b-5p;hsa-miR-23b-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p | 11 | COL1A1 | Sponge network | 2.565 | 0 | 3.608 | 0 | 0.478 |
| 114 | PWAR6 |
hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-31-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-944 | 11 | FGF7 | Sponge network | -1.629 | 0 | -2.015 | 0 | 0.477 |
| 115 | CTA-221G9.11 |
hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-450b-5p;hsa-miR-629-3p;hsa-miR-93-5p | 10 | PIK3R1 | Sponge network | -2.198 | 0 | -1.219 | 0 | 0.477 |
| 116 | RP1-193H18.2 |
hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-96-5p | 11 | PIK3R1 | Sponge network | -1.539 | 0 | -1.219 | 0 | 0.476 |
| 117 | RP11-815I9.4 |
hsa-miR-103a-3p;hsa-miR-130a-3p;hsa-miR-15a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-324-5p;hsa-miR-590-5p;hsa-miR-664a-3p | 13 | ITGA2 | Sponge network | 0.004 | 0.98116 | 0.346 | 0.05579 | 0.475 |
| 118 | CTD-2015G9.2 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-2355-5p;hsa-miR-342-3p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-708-3p;hsa-miR-93-5p | 15 | PDPK1 | Sponge network | -3.112 | 5.0E-5 | -0.375 | 0.00067 | 0.474 |
| 119 | RP11-389C8.2 |
hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-3065-3p;hsa-miR-429;hsa-miR-93-5p;hsa-miR-944 | 10 | COL4A3 | Sponge network | 0.187 | 0.31051 | -0.782 | 0.02643 | 0.474 |
| 120 | AC108142.1 |
hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27b-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-532-3p | 10 | PRKCA | Sponge network | 2.31 | 0 | 0.318 | 0.04622 | 0.474 |
| 121 | RP11-890B15.2 |
hsa-miR-106a-5p;hsa-miR-140-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-497-5p;hsa-miR-9-5p | 21 | CDK6 | Sponge network | 4.029 | 0 | 0.944 | 0 | 0.474 |
| 122 | CTA-221G9.11 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-224-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-576-5p | 12 | IGF1 | Sponge network | -2.198 | 0 | -1.836 | 0 | 0.473 |
| 123 | EMX2OS |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-224-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 21 | IGF1 | Sponge network | -1.459 | 1.0E-5 | -1.836 | 0 | 0.472 |
| 124 | LINC00702 |
hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 17 | THBS1 | Sponge network | -0.573 | 0.0699 | -0.06 | 0.8171 | 0.472 |
| 125 | PWAR6 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-205-3p;hsa-miR-224-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-629-5p | 13 | IGF1 | Sponge network | -1.629 | 0 | -1.836 | 0 | 0.471 |
| 126 | RP5-884M6.1 | hsa-miR-101-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-378a-3p | 18 | CDK6 | Sponge network | 6.548 | 0 | 0.944 | 0 | 0.471 |
| 127 | BHLHE40-AS1 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-940;hsa-miR-96-5p | 11 | CSF1 | Sponge network | -0.932 | 0 | -0.395 | 0.03662 | 0.47 |
| 128 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-324-3p;hsa-miR-93-5p | 10 | PIK3R1 | Sponge network | -0.304 | 0.38627 | -1.219 | 0 | 0.469 |
| 129 | RP11-166D19.1 |
hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-205-5p;hsa-miR-20a-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 11 | LPAR1 | Sponge network | -0.882 | 5.0E-5 | -0.679 | 0.00029 | 0.468 |
| 130 | AC007743.1 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-31-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | FGF7 | Sponge network | -1.053 | 0.00923 | -2.015 | 0 | 0.466 |
| 131 | PWAR6 |
hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-629-3p | 11 | PIK3R1 | Sponge network | -1.629 | 0 | -1.219 | 0 | 0.466 |
| 132 | AC003090.1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-629-5p;hsa-miR-940 | 18 | IGF1 | Sponge network | -4.323 | 0 | -1.836 | 0 | 0.465 |
| 133 | RP11-815I9.4 |
hsa-miR-103a-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-589-3p;hsa-miR-590-5p | 11 | PIK3R1 | Sponge network | 0.004 | 0.98116 | -1.219 | 0 | 0.464 |
| 134 | RP13-463N16.6 | hsa-miR-106a-5p;hsa-miR-140-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-217;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30a-5p | 11 | CDK6 | Sponge network | 3.312 | 0 | 0.944 | 0 | 0.463 |
| 135 | C10orf71-AS1 |
hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 10 | PRKAA2 | Sponge network | -3.382 | 0.00013 | -3.347 | 0 | 0.463 |
| 136 | RP11-193H5.1 |
hsa-miR-125a-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-320a;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-501-5p;hsa-miR-7-1-3p | 14 | EGFR | Sponge network | -0.426 | 0.01062 | 0.422 | 0.07321 | 0.463 |
| 137 | MIR31HG | hsa-miR-106a-5p;hsa-miR-140-3p;hsa-miR-148a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-23b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-34a-5p | 11 | CDK6 | Sponge network | 0.768 | 0.15574 | 0.944 | 0 | 0.463 |
| 138 | RP11-594N15.3 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-31-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-484 | 11 | FGF7 | Sponge network | -2.86 | 0 | -2.015 | 0 | 0.463 |
| 139 | ZNF667-AS1 |
hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-31-5p;hsa-miR-424-5p;hsa-miR-452-5p;hsa-miR-484;hsa-miR-590-5p | 11 | FGF7 | Sponge network | -1.395 | 0 | -2.015 | 0 | 0.463 |
| 140 | RP1-193H18.2 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-205-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-450b-5p | 10 | KIT | Sponge network | -1.539 | 0 | -1.942 | 0 | 0.461 |
| 141 | CTA-221G9.11 |
hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-450b-5p | 11 | BCL2 | Sponge network | -2.198 | 0 | -0.91 | 1.0E-5 | 0.461 |
| 142 | RP11-284N8.3 |
hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-96-5p | 10 | GNG7 | Sponge network | 0.003 | 0.99478 | -1.969 | 0 | 0.46 |
| 143 | RP11-594N15.3 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-940 | 16 | IGF1 | Sponge network | -2.86 | 0 | -1.836 | 0 | 0.46 |
| 144 | EMX2OS |
hsa-miR-138-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-205-5p;hsa-miR-224-5p;hsa-miR-331-5p;hsa-miR-34a-5p;hsa-miR-34c-5p;hsa-miR-589-3p;hsa-miR-590-3p | 13 | RELN | Sponge network | -1.459 | 1.0E-5 | -2.339 | 0 | 0.459 |
| 145 | RP11-760H22.2 |
hsa-miR-15b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-224-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-629-5p | 10 | IGF1 | Sponge network | -2.79 | 0 | -1.836 | 0 | 0.459 |
| 146 | RAMP2-AS1 |
hsa-miR-141-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-31-3p;hsa-miR-3614-5p;hsa-miR-671-5p | 10 | GNG7 | Sponge network | -1.258 | 0.00026 | -1.969 | 0 | 0.458 |
| 147 | SH3RF3-AS1 |
hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-205-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 11 | LPAR1 | Sponge network | -0.175 | 0.58985 | -0.679 | 0.00029 | 0.457 |
| 148 | RP11-284N8.3 |
hsa-miR-103a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-455-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-96-5p | 12 | PIK3R1 | Sponge network | 0.003 | 0.99478 | -1.219 | 0 | 0.457 |
| 149 | AC006116.24 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 10 | IGF1 | Sponge network | -1.832 | 1.0E-5 | -1.836 | 0 | 0.456 |
| 150 | RP11-166D19.1 |
hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-205-3p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-944 | 11 | ITGA8 | Sponge network | -0.882 | 5.0E-5 | -0.747 | 0.01533 | 0.456 |
| 151 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p | 12 | KIT | Sponge network | -1.712 | 0 | -1.942 | 0 | 0.455 |
| 152 | RP11-284N8.3 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-224-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-452-5p;hsa-miR-576-5p | 12 | IGF1 | Sponge network | 0.003 | 0.99478 | -1.836 | 0 | 0.454 |
| 153 | MIR497HG |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-224-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p | 14 | IGF1 | Sponge network | -1.263 | 0.00248 | -1.836 | 0 | 0.454 |
| 154 | TPTEP1 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-31-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-944 | 15 | FGF7 | Sponge network | -2.193 | 0 | -2.015 | 0 | 0.454 |
| 155 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-944 | 17 | PDGFRA | Sponge network | -1.293 | 0.01561 | -0.463 | 0.04818 | 0.453 |
| 156 | RP11-462L8.1 | hsa-miR-101-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-34a-5p;hsa-miR-9-5p | 13 | CDK6 | Sponge network | 4.262 | 0 | 0.944 | 0 | 0.453 |
| 157 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 25 | CREB3L2 | Sponge network | -0.939 | 4.0E-5 | -0.271 | 0.03509 | 0.452 |
| 158 | AC003090.1 |
hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-31-5p;hsa-miR-3614-5p;hsa-miR-940;hsa-miR-96-5p | 12 | GNG7 | Sponge network | -4.323 | 0 | -1.969 | 0 | 0.452 |
| 159 | RP11-815I9.4 |
hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-340-3p;hsa-miR-576-5p | 10 | ITGB8 | Sponge network | 0.004 | 0.98116 | 0.348 | 0.04508 | 0.451 |
| 160 | RP11-384L8.1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-671-5p | 11 | KIT | Sponge network | -1.152 | 0.00016 | -1.942 | 0 | 0.45 |
| 161 | RP11-774O3.3 |
hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-31-5p;hsa-miR-330-5p;hsa-miR-3614-5p | 11 | GNG7 | Sponge network | -1.712 | 0 | -1.969 | 0 | 0.45 |
| 162 | RP11-417E7.1 | hsa-miR-106a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-30a-5p | 11 | CDK6 | Sponge network | 3.987 | 0 | 0.944 | 0 | 0.449 |
| 163 | CASC15 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-200b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 13 | LAMC1 | Sponge network | 1.485 | 0 | 1.059 | 0 | 0.448 |
| 164 | ZNF667-AS1 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-331-3p;hsa-miR-452-3p;hsa-miR-589-5p;hsa-miR-96-5p | 17 | PRKAA2 | Sponge network | -1.395 | 0 | -3.347 | 0 | 0.448 |
| 165 | AL035610.1 |
hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-342-3p | 11 | NTRK2 | Sponge network | -5.893 | 0 | -1.115 | 0.01397 | 0.447 |
| 166 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-940 | 11 | IGF1 | Sponge network | -0.793 | 0 | -1.836 | 0 | 0.447 |
| 167 | CTD-2066L21.3 |
hsa-miR-101-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-200b-3p;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-9-5p | 19 | CDK6 | Sponge network | 5.449 | 0 | 0.944 | 0 | 0.447 |
| 168 | SNHG14 |
hsa-miR-141-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-224-5p;hsa-miR-31-3p;hsa-miR-31-5p;hsa-miR-3614-5p;hsa-miR-3615;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-939-5p | 12 | GNG7 | Sponge network | -1.125 | 0.0001 | -1.969 | 0 | 0.446 |
| 169 | RP11-58E21.3 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-217;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30d-3p;hsa-miR-500a-3p | 10 | PPP2R5E | Sponge network | 0.445 | 0.04375 | 0.181 | 0.07679 | 0.444 |
| 170 | RP11-693J15.4 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-940 | 11 | IGF1 | Sponge network | -0.732 | 0.18487 | -1.836 | 0 | 0.444 |
| 171 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-671-5p | 14 | KIT | Sponge network | -0.882 | 5.0E-5 | -1.942 | 0 | 0.444 |
| 172 | RP1-27K12.2 | hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-576-5p | 11 | NTRK2 | Sponge network | -0.134 | 0.89838 | -1.115 | 0.01397 | 0.443 |
| 173 | AC108142.1 |
hsa-miR-148a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27b-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-429;hsa-miR-628-5p;hsa-miR-96-5p | 10 | IRS1 | Sponge network | 2.31 | 0 | 0.369 | 0.06704 | 0.443 |
| 174 | PSMG3-AS1 |
hsa-miR-141-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-31-5p;hsa-miR-92a-3p;hsa-miR-940 | 10 | GNG7 | Sponge network | -0.793 | 0 | -1.969 | 0 | 0.442 |
| 175 | MIR497HG |
hsa-let-7a-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-31-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | FGF7 | Sponge network | -1.263 | 0.00248 | -2.015 | 0 | 0.442 |
| 176 | TPTEP1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-671-5p | 13 | KIT | Sponge network | -2.193 | 0 | -1.942 | 0 | 0.442 |
| 177 | LINC00284 |
hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-31-3p;hsa-miR-31-5p;hsa-miR-3614-5p;hsa-miR-3615;hsa-miR-671-5p | 10 | GNG7 | Sponge network | -4.159 | 0 | -1.969 | 0 | 0.441 |
| 178 | FAM83A-AS1 |
hsa-let-7a-5p;hsa-let-7c-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-101-3p;hsa-miR-106b-5p;hsa-miR-20b-5p;hsa-miR-28-5p;hsa-miR-491-5p | 11 | CDKN1A | Sponge network | 1.064 | 0.00307 | 0.128 | 0.35863 | 0.44 |
| 179 | RP11-693J15.4 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-671-5p | 10 | KIT | Sponge network | -0.732 | 0.18487 | -1.942 | 0 | 0.44 |
| 180 | BHLHE40-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-501-5p | 13 | KIT | Sponge network | -0.932 | 0 | -1.942 | 0 | 0.44 |
| 181 | FAM66C |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-452-3p;hsa-miR-576-5p;hsa-miR-944 | 10 | PRKAA2 | Sponge network | -0.798 | 0.00038 | -3.347 | 0 | 0.44 |
| 182 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-7-1-3p;hsa-miR-944 | 16 | PRKAA2 | Sponge network | -1.293 | 0.01561 | -3.347 | 0 | 0.439 |
| 183 | LINC00702 |
hsa-miR-138-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-205-5p;hsa-miR-224-5p;hsa-miR-34a-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p | 12 | RELN | Sponge network | -0.573 | 0.0699 | -2.339 | 0 | 0.439 |
| 184 | BHLHE40-AS1 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-96-5p | 10 | PIK3R1 | Sponge network | -0.932 | 0 | -1.219 | 0 | 0.438 |
| 185 | AC007743.1 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 12 | FGF2 | Sponge network | -1.053 | 0.00923 | -0.874 | 0.00075 | 0.438 |
| 186 | LINC00941 |
hsa-let-7g-5p;hsa-miR-125b-5p;hsa-miR-139-5p;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-29b-2-5p;hsa-miR-29c-3p;hsa-miR-497-5p;hsa-miR-532-5p | 10 | IGF1R | Sponge network | 4.292 | 0 | 0.659 | 0 | 0.438 |
| 187 | EMX2OS |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-31-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-944 | 16 | FGF7 | Sponge network | -1.459 | 1.0E-5 | -2.015 | 0 | 0.437 |
| 188 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-224-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 11 | IGF1 | Sponge network | -0.33 | 0.08136 | -1.836 | 0 | 0.436 |
| 189 | LINC00702 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | FGF7 | Sponge network | -0.573 | 0.0699 | -2.015 | 0 | 0.436 |
| 190 | LINC00518 | hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-140-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-20b-5p;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p | 13 | CDK6 | Sponge network | 2.747 | 4.0E-5 | 0.944 | 0 | 0.436 |
| 191 | LINC00707 |
hsa-miR-195-3p;hsa-miR-217;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-328-3p;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-664a-3p | 13 | YWHAZ | Sponge network | 2.018 | 1.0E-5 | 0.345 | 0.0016 | 0.436 |
| 192 | RP1-193H18.2 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-576-5p;hsa-miR-944;hsa-miR-96-5p | 14 | PRKAA2 | Sponge network | -1.539 | 0 | -3.347 | 0 | 0.435 |
| 193 | RP11-445F12.1 | hsa-miR-140-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-340-5p;hsa-miR-497-5p | 15 | CDK6 | Sponge network | 7.107 | 0 | 0.944 | 0 | 0.435 |
| 194 | CASC15 |
hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PDGFRA | Sponge network | 1.485 | 0 | -0.463 | 0.04818 | 0.435 |
| 195 | SH3RF3-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-5p | 13 | PIK3R1 | Sponge network | -0.175 | 0.58985 | -1.219 | 0 | 0.435 |
| 196 | DNM3OS |
hsa-let-7a-3p;hsa-miR-15b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-224-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-629-5p | 11 | IGF1 | Sponge network | 0.932 | 0.00442 | -1.836 | 0 | 0.434 |
| 197 | ZNF667-AS1 |
hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-96-5p | 14 | PIK3R1 | Sponge network | -1.395 | 0 | -1.219 | 0 | 0.434 |
| 198 | SSTR5-AS1 |
hsa-miR-103a-2-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-454-3p | 13 | PRKAA2 | Sponge network | -1.443 | 0.04871 | -3.347 | 0 | 0.434 |
| 199 | RP11-218E20.3 |
hsa-miR-101-3p;hsa-miR-140-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-497-5p | 24 | CDK6 | Sponge network | 4.613 | 0 | 0.944 | 0 | 0.434 |
| 200 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | THBS1 | Sponge network | -0.175 | 0.58985 | -0.06 | 0.8171 | 0.433 |
| 201 | SNHG14 |
hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-452-5p;hsa-miR-503-5p;hsa-miR-589-3p;hsa-miR-92a-3p;hsa-miR-944 | 11 | FGF2 | Sponge network | -1.125 | 0.0001 | -0.874 | 0.00075 | 0.433 |
| 202 | RP11-42I10.1 | hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-140-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30a-5p | 15 | CDK6 | Sponge network | 2.1 | 0 | 0.944 | 0 | 0.432 |
| 203 | LINC00704 | hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-140-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-20b-5p;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p | 12 | CDK6 | Sponge network | 2.465 | 0 | 0.944 | 0 | 0.43 |
| 204 | TPTEP1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-342-3p;hsa-miR-452-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-93-5p;hsa-miR-944 | 20 | PRKAA2 | Sponge network | -2.193 | 0 | -3.347 | 0 | 0.43 |
| 205 | AC093627.10 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-454-3p | 13 | KIT | Sponge network | -0.893 | 0.00123 | -1.942 | 0 | 0.43 |
| 206 | AC007743.1 |
hsa-miR-17-5p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-365a-3p;hsa-miR-455-5p;hsa-miR-590-5p;hsa-miR-7-5p | 12 | BCL2 | Sponge network | -1.053 | 0.00923 | -0.91 | 1.0E-5 | 0.429 |
| 207 | LINC00702 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-224-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 18 | IGF1 | Sponge network | -0.573 | 0.0699 | -1.836 | 0 | 0.429 |
| 208 | TPTEP1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-15a-5p;hsa-miR-181b-5p;hsa-miR-205-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-590-5p | 10 | INSR | Sponge network | -2.193 | 0 | -0.194 | 0.13669 | 0.429 |
| 209 | TPTEP1 |
hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-31-3p;hsa-miR-31-5p;hsa-miR-3614-5p;hsa-miR-3615;hsa-miR-671-5p;hsa-miR-92a-3p | 12 | GNG7 | Sponge network | -2.193 | 0 | -1.969 | 0 | 0.428 |
| 210 | EMX2OS |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-205-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-944 | 12 | ITGA8 | Sponge network | -1.459 | 1.0E-5 | -0.747 | 0.01533 | 0.427 |
| 211 | EMX2OS |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-149-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-944 | 33 | PDGFRA | Sponge network | -1.459 | 1.0E-5 | -0.463 | 0.04818 | 0.427 |
| 212 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-452-5p;hsa-miR-454-3p | 10 | IGF1 | Sponge network | -0.233 | 0.50729 | -1.836 | 0 | 0.427 |
| 213 | AC007743.1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | PDGFRA | Sponge network | -1.053 | 0.00923 | -0.463 | 0.04818 | 0.426 |
| 214 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 17 | THBS1 | Sponge network | -0.882 | 5.0E-5 | -0.06 | 0.8171 | 0.426 |
| 215 | RP11-166D19.1 |
hsa-miR-130a-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-3p;hsa-miR-940;hsa-miR-96-5p | 15 | CSF1 | Sponge network | -0.882 | 5.0E-5 | -0.395 | 0.03662 | 0.426 |
| 216 | RP4-798P15.3 |
hsa-miR-103a-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-2355-5p;hsa-miR-589-3p;hsa-miR-590-5p | 11 | PDPK1 | Sponge network | -1.213 | 0.00098 | -0.375 | 0.00067 | 0.426 |
| 217 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-331-3p;hsa-miR-452-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-944 | 22 | PRKAA2 | Sponge network | -1.712 | 0 | -3.347 | 0 | 0.425 |
| 218 | LINC00460 | hsa-miR-140-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-497-5p | 16 | CDK6 | Sponge network | 7.222 | 0 | 0.944 | 0 | 0.425 |
| 219 | PCED1B-AS1 |
hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 12 | GNG7 | Sponge network | 0.634 | 0.00501 | -1.969 | 0 | 0.424 |
| 220 | TPTEP1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-576-5p | 13 | IGF1 | Sponge network | -2.193 | 0 | -1.836 | 0 | 0.423 |
| 221 | LINC00958 | hsa-miR-140-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-340-5p;hsa-miR-374a-3p;hsa-miR-497-5p | 11 | CDK6 | Sponge network | 1.836 | 0 | 0.944 | 0 | 0.422 |
| 222 | RP11-282O18.3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-301a-3p | 11 | KIT | Sponge network | -0.266 | 0.05794 | -1.942 | 0 | 0.42 |
| 223 | RP4-639F20.1 | hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-331-3p;hsa-miR-96-5p | 10 | PRKAA2 | Sponge network | -0.199 | 0.36619 | -3.347 | 0 | 0.42 |
| 224 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-455-5p;hsa-miR-589-3p | 13 | PDGFRA | Sponge network | -1.258 | 0.00026 | -0.463 | 0.04818 | 0.42 |
| 225 | RP11-166D19.1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-205-5p;hsa-miR-224-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-589-3p | 10 | RELN | Sponge network | -0.882 | 5.0E-5 | -2.339 | 0 | 0.419 |
| 226 | RASSF8-AS1 |
hsa-miR-141-3p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-429;hsa-miR-944 | 12 | PDGFRA | Sponge network | 0.121 | 0.52445 | -0.463 | 0.04818 | 0.419 |
| 227 | LINC00900 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-576-5p | 17 | IGF1 | Sponge network | -1.803 | 0 | -1.836 | 0 | 0.419 |
| 228 | AC108142.1 |
hsa-miR-141-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-7-1-3p | 12 | ITGA1 | Sponge network | 2.31 | 0 | 1.673 | 0 | 0.418 |
| 229 | XXyac-YX65C7_A.3 |
hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-140-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p | 17 | CDK6 | Sponge network | 6.112 | 0 | 0.944 | 0 | 0.418 |
| 230 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-501-5p | 10 | KIT | Sponge network | -0.72 | 0.01036 | -1.942 | 0 | 0.418 |
| 231 | KB-1732A1.1 |
hsa-let-7a-5p;hsa-let-7c-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-20b-5p;hsa-miR-28-5p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-491-5p | 12 | CDKN1A | Sponge network | 1.697 | 0 | 0.128 | 0.35863 | 0.417 |
| 232 | CTD-2587H24.5 | hsa-miR-101-3p;hsa-miR-140-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-217;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-497-5p;hsa-miR-582-5p | 20 | CDK6 | Sponge network | 7.138 | 0 | 0.944 | 0 | 0.417 |
| 233 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p | 11 | PRKAA2 | Sponge network | -1.447 | 0.0004 | -3.347 | 0 | 0.417 |
| 234 | RP11-193H5.1 |
hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29b-2-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 14 | CREB3L2 | Sponge network | -0.426 | 0.01062 | -0.271 | 0.03509 | 0.417 |
| 235 | C10orf71-AS1 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-224-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-576-5p | 12 | IGF1 | Sponge network | -3.382 | 0.00013 | -1.836 | 0 | 0.416 |
| 236 | TP73-AS1 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-452-3p | 13 | PRKAA2 | Sponge network | -0.533 | 0.00643 | -3.347 | 0 | 0.416 |
| 237 | RP11-218E20.3 |
hsa-miR-127-3p;hsa-miR-140-3p;hsa-miR-148a-3p;hsa-miR-181c-5p;hsa-miR-199b-5p;hsa-miR-23b-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-491-5p;hsa-miR-495-3p | 10 | TGFA | Sponge network | 4.613 | 0 | 0.472 | 0.01138 | 0.415 |
| 238 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-589-3p | 15 | PIK3R1 | Sponge network | -1.712 | 0 | -1.219 | 0 | 0.415 |
| 239 | RP11-693J15.4 |
hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-31-5p;hsa-miR-671-5p;hsa-miR-940 | 11 | GNG7 | Sponge network | -0.732 | 0.18487 | -1.969 | 0 | 0.413 |
| 240 | AF127936.7 |
hsa-miR-17-3p;hsa-miR-181c-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27b-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-532-3p | 12 | PRKCA | Sponge network | 0.453 | 0.00811 | 0.318 | 0.04622 | 0.412 |
| 241 | KTN1-AS1 | hsa-miR-101-3p;hsa-miR-140-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-34a-5p;hsa-miR-378a-3p | 16 | CDK6 | Sponge network | 1.107 | 0 | 0.944 | 0 | 0.412 |
| 242 | AF131215.2 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-671-5p | 10 | KIT | Sponge network | -1.136 | 0.00092 | -1.942 | 0 | 0.411 |
| 243 | RP11-594N15.3 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-200b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-452-3p;hsa-miR-484;hsa-miR-877-5p | 11 | ITGA9 | Sponge network | -2.86 | 0 | -1.022 | 1.0E-5 | 0.411 |
| 244 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 17 | THBS1 | Sponge network | -0.939 | 4.0E-5 | -0.06 | 0.8171 | 0.41 |
| 245 | LINC01018 |
hsa-miR-125a-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-452-3p;hsa-miR-455-3p;hsa-miR-590-3p | 11 | NTRK2 | Sponge network | -1.613 | 0.00396 | -1.115 | 0.01397 | 0.409 |
| 246 | TPTEP1 |
hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-365a-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-5p | 14 | BCL2 | Sponge network | -2.193 | 0 | -0.91 | 1.0E-5 | 0.409 |
| 247 | HHIP-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 12 | IGF1 | Sponge network | -1.293 | 0.01561 | -1.836 | 0 | 0.408 |
| 248 | AC006116.24 |
hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-93-5p | 13 | PDGFRA | Sponge network | -1.832 | 1.0E-5 | -0.463 | 0.04818 | 0.408 |
| 249 | ZNF667-AS1 |
hsa-miR-15a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-452-5p;hsa-miR-503-5p;hsa-miR-589-3p;hsa-miR-590-5p | 12 | FGF2 | Sponge network | -1.395 | 0 | -0.874 | 0.00075 | 0.408 |
| 250 | OIP5-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-454-3p;hsa-miR-671-5p | 10 | KIT | Sponge network | -0.06 | 0.55122 | -1.942 | 0 | 0.407 |
| 251 | RP1-193H18.2 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-19a-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-576-5p | 11 | IGF1 | Sponge network | -1.539 | 0 | -1.836 | 0 | 0.407 |
| 252 | LINC00639 |
hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-18a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-31-5p;hsa-miR-424-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-484 | 11 | FGF7 | Sponge network | -0.91 | 0.00848 | -2.015 | 0 | 0.407 |
| 253 | APCDD1L-AS1 |
hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27b-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-628-5p | 10 | IRS1 | Sponge network | 2.077 | 0 | 0.369 | 0.06704 | 0.407 |
| 254 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-33b-5p | 14 | PDGFRA | Sponge network | -1.447 | 0.0004 | -0.463 | 0.04818 | 0.407 |
| 255 | RP11-218E20.3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-20b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p | 10 | COL1A1 | Sponge network | 4.613 | 0 | 3.608 | 0 | 0.406 |
| 256 | EMX2OS |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-452-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-944 | 27 | PRKAA2 | Sponge network | -1.459 | 1.0E-5 | -3.347 | 0 | 0.406 |
| 257 | RP11-594N15.3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-671-5p | 11 | KIT | Sponge network | -2.86 | 0 | -1.942 | 0 | 0.405 |
| 258 | RP11-1020M18.10 | hsa-miR-106a-5p;hsa-miR-146a-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-497-5p;hsa-miR-582-5p | 10 | CDK6 | Sponge network | 6.911 | 0 | 0.944 | 0 | 0.404 |
| 259 | SH3RF3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-2-5p;hsa-miR-339-5p;hsa-miR-505-5p | 10 | PRKCA | Sponge network | -0.175 | 0.58985 | 0.318 | 0.04622 | 0.404 |
| 260 | EMX2OS |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-200b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-452-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p | 17 | ITGA9 | Sponge network | -1.459 | 1.0E-5 | -1.022 | 1.0E-5 | 0.404 |
| 261 | EMX2OS |
hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-224-3p;hsa-miR-23a-5p;hsa-miR-23b-5p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-93-5p | 17 | IGF2 | Sponge network | -1.459 | 1.0E-5 | 0.131 | 0.60661 | 0.402 |
| 262 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p | 11 | KIT | Sponge network | -0.793 | 0 | -1.942 | 0 | 0.402 |
| 263 | SNHG14 |
hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-944 | 18 | PDGFRA | Sponge network | -1.125 | 0.0001 | -0.463 | 0.04818 | 0.402 |
| 264 | TP73-AS1 |
hsa-miR-130a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p | 10 | KIT | Sponge network | -0.533 | 0.00643 | -1.942 | 0 | 0.402 |
| 265 | RP11-166D19.1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-96-5p | 19 | PIK3R1 | Sponge network | -0.882 | 5.0E-5 | -1.219 | 0 | 0.401 |
| 266 | RP1-193H18.2 |
hsa-let-7a-3p;hsa-miR-18a-3p;hsa-miR-205-3p;hsa-miR-27a-3p;hsa-miR-31-5p;hsa-miR-424-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-708-3p;hsa-miR-944 | 10 | FGF7 | Sponge network | -1.539 | 0 | -2.015 | 0 | 0.4 |
| 267 | RP11-757G1.6 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-576-5p | 13 | IGF1 | Sponge network | -1.346 | 0.00088 | -1.836 | 0 | 0.4 |
| 268 | RP11-166D19.1 |
hsa-miR-141-5p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | ITGA1 | Sponge network | -0.882 | 5.0E-5 | 1.673 | 0 | 0.399 |
| 269 | MEG3 |
hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-205-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-944 | 12 | FGF7 | Sponge network | -0.286 | 0.30726 | -2.015 | 0 | 0.398 |
| 270 | DNM3OS |
hsa-let-7a-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-944 | 12 | FGF7 | Sponge network | 0.932 | 0.00442 | -2.015 | 0 | 0.398 |
| 271 | MIR497HG |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-149-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | PDGFRA | Sponge network | -1.263 | 0.00248 | -0.463 | 0.04818 | 0.397 |
| 272 | ZNF667-AS1 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-224-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-452-5p;hsa-miR-629-5p | 12 | IGF1 | Sponge network | -1.395 | 0 | -1.836 | 0 | 0.397 |
| 273 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-598-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 16 | THBS2 | Sponge network | -0.573 | 0.0699 | 1.626 | 0 | 0.397 |
| 274 | CTD-2015G9.2 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-93-5p | 16 | PIK3R1 | Sponge network | -3.112 | 5.0E-5 | -1.219 | 0 | 0.397 |
| 275 | TTLL11-IT1 | hsa-miR-140-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-200b-3p;hsa-miR-217;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30a-5p | 11 | CDK6 | Sponge network | 5.5 | 0 | 0.944 | 0 | 0.397 |
| 276 | AC108142.1 |
hsa-miR-101-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-7-1-3p | 24 | CDK6 | Sponge network | 2.31 | 0 | 0.944 | 0 | 0.396 |
| 277 | TPTEP1 |
hsa-miR-181b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-452-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p | 11 | NTRK2 | Sponge network | -2.193 | 0 | -1.115 | 0.01397 | 0.395 |
| 278 | LINC00900 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-671-5p | 12 | KIT | Sponge network | -1.803 | 0 | -1.942 | 0 | 0.395 |
| 279 | AC074286.1 | hsa-miR-125a-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-576-5p | 10 | NTRK2 | Sponge network | 0.146 | 0.43972 | -1.115 | 0.01397 | 0.395 |
| 280 | KB-1732A1.1 |
hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-140-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-497-5p | 23 | CDK6 | Sponge network | 1.697 | 0 | 0.944 | 0 | 0.394 |
| 281 | RP11-166D19.1 |
hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-31-5p;hsa-miR-3615;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-939-5p;hsa-miR-940;hsa-miR-96-5p | 15 | GNG7 | Sponge network | -0.882 | 5.0E-5 | -1.969 | 0 | 0.394 |
| 282 | RP4-798P15.3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-671-5p | 12 | KIT | Sponge network | -1.213 | 0.00098 | -1.942 | 0 | 0.394 |
| 283 | RP11-30P6.6 |
hsa-miR-101-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-217;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-374a-5p | 14 | PPP2R5E | Sponge network | 2.847 | 0 | 0.181 | 0.07679 | 0.394 |
| 284 | RP11-557H15.3 |
hsa-miR-106a-5p;hsa-miR-140-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-374b-5p;hsa-miR-378a-3p | 15 | CDK6 | Sponge network | 1.673 | 8.0E-5 | 0.944 | 0 | 0.393 |
| 285 | DNM3OS |
hsa-let-7a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | THBS1 | Sponge network | 0.932 | 0.00442 | -0.06 | 0.8171 | 0.393 |
| 286 | RP4-798P15.3 |
hsa-miR-103a-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-96-5p | 14 | PIK3R1 | Sponge network | -1.213 | 0.00098 | -1.219 | 0 | 0.393 |
| 287 | LINC00639 |
hsa-miR-130a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p | 10 | KIT | Sponge network | -0.91 | 0.00848 | -1.942 | 0 | 0.392 |
| 288 | EMX2OS |
hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-31-5p;hsa-miR-330-5p;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-939-5p;hsa-miR-940 | 13 | GNG7 | Sponge network | -1.459 | 1.0E-5 | -1.969 | 0 | 0.392 |
| 289 | RP11-13K12.5 |
hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-589-3p | 11 | PDGFRA | Sponge network | -0.552 | 0.43847 | -0.463 | 0.04818 | 0.392 |
| 290 | RP11-166D19.1 |
hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-224-3p;hsa-miR-23a-5p;hsa-miR-23b-5p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-339-5p;hsa-miR-454-3p | 14 | IGF2 | Sponge network | -0.882 | 5.0E-5 | 0.131 | 0.60661 | 0.391 |
| 291 | RP11-366H4.1 | hsa-miR-101-3p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-30a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-497-5p | 12 | CDK6 | Sponge network | 4.872 | 0 | 0.944 | 0 | 0.39 |
| 292 | KB-1732A1.1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-20b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p | 11 | COL1A1 | Sponge network | 1.697 | 0 | 3.608 | 0 | 0.39 |
| 293 | RP11-554I8.2 |
hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-20b-5p;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-29c-3p;hsa-miR-378a-3p | 14 | CDK6 | Sponge network | 0.342 | 0.50007 | 0.944 | 0 | 0.389 |
| 294 | CTD-2555C10.3 |
hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-664a-3p | 14 | CDK6 | Sponge network | 1.037 | 0.0004 | 0.944 | 0 | 0.389 |
| 295 | SH3RF3-AS1 |
hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-2355-5p;hsa-miR-29b-1-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 11 | CREB3L2 | Sponge network | -0.175 | 0.58985 | -0.271 | 0.03509 | 0.389 |
| 296 | AC093627.10 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-452-3p;hsa-miR-454-3p | 12 | PRKAA2 | Sponge network | -0.893 | 0.00123 | -3.347 | 0 | 0.387 |
| 297 | AC007743.1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-15a-5p;hsa-miR-205-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | INSR | Sponge network | -1.053 | 0.00923 | -0.194 | 0.13669 | 0.386 |
| 298 | AC011738.4 |
hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-29b-2-5p;hsa-miR-320a;hsa-miR-374a-3p;hsa-miR-7-1-3p;hsa-miR-9-5p | 10 | EGFR | Sponge network | 3.153 | 0 | 0.422 | 0.07321 | 0.386 |
| 299 | LINC00284 |
hsa-miR-103a-2-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-629-5p | 10 | IGF1 | Sponge network | -4.159 | 0 | -1.836 | 0 | 0.385 |
| 300 | RP11-757G1.6 |
hsa-miR-130a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 10 | KIT | Sponge network | -1.346 | 0.00088 | -1.942 | 0 | 0.385 |
| 301 | CTD-2008P7.9 |
hsa-miR-103a-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-2355-5p | 11 | PDPK1 | Sponge network | -1.202 | 0.00093 | -0.375 | 0.00067 | 0.385 |
| 302 | MIR497HG |
hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-31-5p;hsa-miR-3615;hsa-miR-671-5p;hsa-miR-939-5p;hsa-miR-96-5p | 13 | GNG7 | Sponge network | -1.263 | 0.00248 | -1.969 | 0 | 0.384 |
| 303 | TP73-AS1 |
hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-589-3p | 10 | PDGFRA | Sponge network | -0.533 | 0.00643 | -0.463 | 0.04818 | 0.384 |
| 304 | CYP4F35P | hsa-miR-181a-2-3p;hsa-miR-18a-3p;hsa-miR-193b-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-31-5p;hsa-miR-3614-5p;hsa-miR-671-5p;hsa-miR-939-5p | 10 | GNG7 | Sponge network | -5.012 | 0 | -1.969 | 0 | 0.384 |
| 305 | RP11-13K12.5 |
hsa-miR-130a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-671-5p | 10 | KIT | Sponge network | -0.552 | 0.43847 | -1.942 | 0 | 0.384 |
| 306 | LINC00702 |
hsa-miR-141-5p;hsa-miR-200b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-452-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-877-5p | 14 | ITGA9 | Sponge network | -0.573 | 0.0699 | -1.022 | 1.0E-5 | 0.383 |
| 307 | MIR497HG |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-671-5p | 11 | KIT | Sponge network | -1.263 | 0.00248 | -1.942 | 0 | 0.382 |
| 308 | RP11-890B15.2 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-150-5p;hsa-miR-20b-5p;hsa-miR-23b-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p | 10 | COL1A1 | Sponge network | 4.029 | 0 | 3.608 | 0 | 0.382 |
| 309 | LINC00639 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-452-5p | 10 | IGF1 | Sponge network | -0.91 | 0.00848 | -1.836 | 0 | 0.381 |
| 310 | ZNF667-AS1 |
hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-96-5p | 15 | PDGFRA | Sponge network | -1.395 | 0 | -0.463 | 0.04818 | 0.381 |
| 311 | RP11-346D19.1 | hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-340-5p | 11 | CDK6 | Sponge network | 6.549 | 0 | 0.944 | 0 | 0.38 |
| 312 | CTD-2357A8.3 | hsa-miR-101-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-335-5p;hsa-miR-34a-5p | 11 | CDK6 | Sponge network | 7.162 | 0 | 0.944 | 0 | 0.38 |
| 313 | APCDD1L-AS1 |
hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-140-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-335-3p;hsa-miR-378a-3p;hsa-miR-497-5p;hsa-miR-7-1-3p;hsa-miR-9-5p | 24 | CDK6 | Sponge network | 2.077 | 0 | 0.944 | 0 | 0.379 |
| 314 | CTD-2135D7.5 |
hsa-miR-103a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p | 11 | PIK3R1 | Sponge network | -2.579 | 0.0001 | -1.219 | 0 | 0.379 |
| 315 | RP11-815I9.4 |
hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-34a-5p | 10 | IGF1R | Sponge network | 0.004 | 0.98116 | 0.659 | 0 | 0.379 |
| 316 | LINC01018 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-22-5p;hsa-miR-31-5p;hsa-miR-452-3p;hsa-miR-944 | 10 | PRKAA2 | Sponge network | -1.613 | 0.00396 | -3.347 | 0 | 0.379 |
| 317 | CTD-2555C10.3 |
hsa-let-7a-5p;hsa-let-7c-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-20b-5p;hsa-miR-28-5p;hsa-miR-30b-3p;hsa-miR-491-5p | 10 | CDKN1A | Sponge network | 1.037 | 0.0004 | 0.128 | 0.35863 | 0.379 |
| 318 | TPTEP1 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-944 | 14 | FGF2 | Sponge network | -2.193 | 0 | -0.874 | 0.00075 | 0.378 |
| 319 | TPTEP1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-944 | 19 | PDGFRA | Sponge network | -2.193 | 0 | -0.463 | 0.04818 | 0.377 |
| 320 | RP11-815I9.4 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-671-5p | 12 | KIT | Sponge network | 0.004 | 0.98116 | -1.942 | 0 | 0.377 |
| 321 | LINC00284 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-31-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-5p;hsa-miR-944 | 13 | FGF7 | Sponge network | -4.159 | 0 | -2.015 | 0 | 0.377 |
| 322 | AC011738.4 |
hsa-miR-106a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20b-5p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-9-5p | 10 | CREB5 | Sponge network | 3.153 | 0 | -0.062 | 0.77912 | 0.375 |
| 323 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-96-5p | 16 | PDGFRA | Sponge network | -0.33 | 0.08136 | -0.463 | 0.04818 | 0.374 |
| 324 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | PDGFRA | Sponge network | -0.218 | 0.34607 | -0.463 | 0.04818 | 0.374 |
| 325 | NOP14-AS1 |
hsa-miR-101-3p;hsa-miR-140-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-217;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-497-5p;hsa-miR-502-3p | 25 | CDK6 | Sponge network | 0.939 | 0 | 0.944 | 0 | 0.374 |
| 326 | CTD-2171N6.1 |
hsa-miR-139-5p;hsa-miR-140-3p;hsa-miR-150-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-497-5p | 14 | IGF1R | Sponge network | 5.89 | 0 | 0.659 | 0 | 0.374 |
| 327 | RP4-798P15.3 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-576-5p | 12 | IGF1 | Sponge network | -1.213 | 0.00098 | -1.836 | 0 | 0.374 |
| 328 | PSMG3-AS1 |
hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-708-5p;hsa-miR-940 | 10 | CSF1 | Sponge network | -0.793 | 0 | -0.395 | 0.03662 | 0.374 |
| 329 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-7-1-3p | 19 | PRKAA2 | Sponge network | -0.218 | 0.34607 | -3.347 | 0 | 0.374 |
| 330 | AC006262.4 |
hsa-miR-106a-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-497-5p;hsa-miR-582-5p | 15 | CDK6 | Sponge network | 1.721 | 0.01526 | 0.944 | 0 | 0.373 |
| 331 | LINC00900 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-93-5p | 19 | PIK3R1 | Sponge network | -1.803 | 0 | -1.219 | 0 | 0.373 |
| 332 | RP11-497E19.1 |
hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-29c-3p;hsa-miR-335-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 19 | CDK6 | Sponge network | 2.366 | 0.00063 | 0.944 | 0 | 0.373 |
| 333 | DHRS4-AS1 |
hsa-miR-130a-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-454-3p;hsa-miR-501-5p | 10 | KIT | Sponge network | -0.068 | 0.72321 | -1.942 | 0 | 0.373 |
| 334 | RP11-815I9.4 |
hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30c-5p;hsa-miR-589-3p;hsa-miR-664a-3p | 10 | KITLG | Sponge network | 0.004 | 0.98116 | -0.019 | 0.91044 | 0.372 |
| 335 | PWAR6 |
hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-944 | 18 | PDGFRA | Sponge network | -1.629 | 0 | -0.463 | 0.04818 | 0.372 |
| 336 | BZRAP1-AS1 |
hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-31-5p;hsa-miR-671-5p | 10 | GNG7 | Sponge network | -0.233 | 0.50729 | -1.969 | 0 | 0.37 |
| 337 | RP11-150O12.1 |
hsa-let-7c-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-20b-5p;hsa-miR-28-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-423-5p | 11 | CDKN1A | Sponge network | 1.523 | 0 | 0.128 | 0.35863 | 0.369 |
| 338 | FZD10-AS1 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | NTRK2 | Sponge network | -0.218 | 0.34607 | -1.115 | 0.01397 | 0.369 |
| 339 | AC003090.1 |
hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 18 | PIK3R1 | Sponge network | -4.323 | 0 | -1.219 | 0 | 0.369 |
| 340 | BHLHE40-AS1 |
hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-92a-3p;hsa-miR-939-5p;hsa-miR-940;hsa-miR-96-5p | 12 | GNG7 | Sponge network | -0.932 | 0 | -1.969 | 0 | 0.369 |
| 341 | LINC00957 |
hsa-miR-130a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-671-5p | 11 | KIT | Sponge network | -0.677 | 2.0E-5 | -1.942 | 0 | 0.369 |
| 342 | AC003090.1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p | 11 | KIT | Sponge network | -4.323 | 0 | -1.942 | 0 | 0.368 |
| 343 | CTD-2135D7.5 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-224-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-629-5p | 14 | IGF1 | Sponge network | -2.579 | 0.0001 | -1.836 | 0 | 0.368 |
| 344 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p | 10 | KIT | Sponge network | -1.447 | 0.0004 | -1.942 | 0 | 0.368 |
| 345 | DHRS4-AS1 |
hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-29b-1-5p;hsa-miR-362-5p;hsa-miR-501-5p | 11 | CREB3L2 | Sponge network | -0.068 | 0.72321 | -0.271 | 0.03509 | 0.368 |
| 346 | RP11-757G1.6 |
hsa-miR-103a-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-93-5p | 11 | PIK3R1 | Sponge network | -1.346 | 0.00088 | -1.219 | 0 | 0.367 |
| 347 | MAGI2-AS3 |
hsa-miR-15a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 10 | LAMC1 | Sponge network | -0.939 | 4.0E-5 | 1.059 | 0 | 0.367 |
| 348 | AL035610.1 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-342-3p | 14 | PRKAA2 | Sponge network | -5.893 | 0 | -3.347 | 0 | 0.365 |
| 349 | AC003090.1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-205-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | INSR | Sponge network | -4.323 | 0 | -0.194 | 0.13669 | 0.365 |
| 350 | BDNF-AS |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-452-3p;hsa-miR-944 | 10 | PRKAA2 | Sponge network | -1.18 | 0 | -3.347 | 0 | 0.365 |
| 351 | RP11-58E21.3 |
hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-148a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-218-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-5p | 14 | CDK6 | Sponge network | 0.445 | 0.04375 | 0.944 | 0 | 0.365 |
| 352 | LINC00957 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-452-3p;hsa-miR-576-5p | 14 | PRKAA2 | Sponge network | -0.677 | 2.0E-5 | -3.347 | 0 | 0.365 |
| 353 | APCDD1L-AS1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-20b-5p;hsa-miR-23b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-92a-1-5p | 12 | COL1A1 | Sponge network | 2.077 | 0 | 3.608 | 0 | 0.365 |
| 354 | AC093627.10 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-454-3p | 11 | IGF1 | Sponge network | -0.893 | 0.00123 | -1.836 | 0 | 0.364 |
| 355 | RP11-757G1.6 |
hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-18a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-31-5p;hsa-miR-424-5p;hsa-miR-452-5p;hsa-miR-944 | 10 | FGF7 | Sponge network | -1.346 | 0.00088 | -2.015 | 0 | 0.364 |
| 356 | AC006116.24 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-452-3p;hsa-miR-454-3p;hsa-miR-93-5p | 14 | PRKAA2 | Sponge network | -1.832 | 1.0E-5 | -3.347 | 0 | 0.364 |
| 357 | TPTEP1 |
hsa-miR-103a-2-5p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-452-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p | 11 | ITGA9 | Sponge network | -2.193 | 0 | -1.022 | 1.0E-5 | 0.364 |
| 358 | LINC00605 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-7-1-3p;hsa-miR-944 | 11 | PRKAA2 | Sponge network | -2.602 | 0.00047 | -3.347 | 0 | 0.361 |
| 359 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-944 | 17 | PDGFRA | Sponge network | -0.233 | 0.50729 | -0.463 | 0.04818 | 0.361 |
| 360 | LINC00900 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-200b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-452-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p | 13 | ITGA9 | Sponge network | -1.803 | 0 | -1.022 | 1.0E-5 | 0.361 |
| 361 | AF127936.7 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-362-5p;hsa-miR-7-1-3p | 11 | EGFR | Sponge network | 0.453 | 0.00811 | 0.422 | 0.07321 | 0.361 |
| 362 | RP11-166D19.1 |
hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-205-5p;hsa-miR-2355-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-944 | 10 | FGF1 | Sponge network | -0.882 | 5.0E-5 | -0.258 | 0.2676 | 0.361 |
| 363 | CTD-2015G9.2 |
hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 16 | CREB3L2 | Sponge network | -3.112 | 5.0E-5 | -0.271 | 0.03509 | 0.361 |
| 364 | AC007743.1 |
hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-29b-1-5p;hsa-miR-501-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 16 | CREB3L2 | Sponge network | -1.053 | 0.00923 | -0.271 | 0.03509 | 0.361 |
| 365 | RP11-58E21.3 |
hsa-let-7c-5p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-20b-5p;hsa-miR-28-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-345-5p;hsa-miR-491-5p;hsa-miR-93-5p | 11 | CDKN1A | Sponge network | 0.445 | 0.04375 | 0.128 | 0.35863 | 0.36 |
| 366 | LINC00702 |
hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27b-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 14 | PRKCA | Sponge network | -0.573 | 0.0699 | 0.318 | 0.04622 | 0.359 |
| 367 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-576-5p | 14 | IGF1 | Sponge network | -1.712 | 0 | -1.836 | 0 | 0.359 |
| 368 | RP11-517P14.2 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-93-5p | 10 | IL6R | Sponge network | -0.402 | 0.1503 | -0.049 | 0.80939 | 0.359 |
| 369 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p | 12 | IGF1 | Sponge network | -1.447 | 0.0004 | -1.836 | 0 | 0.359 |
| 370 | RP1-193H18.2 |
hsa-let-7a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-503-5p;hsa-miR-589-3p;hsa-miR-944 | 12 | FGF2 | Sponge network | -1.539 | 0 | -0.874 | 0.00075 | 0.359 |
| 371 | AC005682.5 | hsa-miR-101-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-340-5p | 12 | CDK6 | Sponge network | 1.074 | 0 | 0.944 | 0 | 0.358 |
| 372 | LINC00900 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-31-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-708-3p | 18 | FGF7 | Sponge network | -1.803 | 0 | -2.015 | 0 | 0.358 |
| 373 | LINC00973 | hsa-miR-101-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-200b-3p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-335-5p | 10 | CDK6 | Sponge network | 5.015 | 0 | 0.944 | 0 | 0.357 |
| 374 | LINC00284 |
hsa-miR-103a-2-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-452-3p;hsa-miR-576-5p;hsa-miR-93-5p;hsa-miR-944 | 14 | PRKAA2 | Sponge network | -4.159 | 0 | -3.347 | 0 | 0.357 |
| 375 | LINC00702 |
hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-224-3p;hsa-miR-23b-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-93-5p | 16 | IGF2 | Sponge network | -0.573 | 0.0699 | 0.131 | 0.60661 | 0.356 |
| 376 | RP11-693J15.4 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-590-5p | 15 | PDGFRA | Sponge network | -0.732 | 0.18487 | -0.463 | 0.04818 | 0.356 |
| 377 | RP4-798P15.3 |
hsa-miR-103a-2-5p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p | 10 | ITGA9 | Sponge network | -1.213 | 0.00098 | -1.022 | 1.0E-5 | 0.355 |
| 378 | RP11-594N15.3 |
hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-29b-1-5p;hsa-miR-671-5p | 12 | CREB3L2 | Sponge network | -2.86 | 0 | -0.271 | 0.03509 | 0.354 |
| 379 | VPS9D1-AS1 | hsa-miR-101-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-378a-3p;hsa-miR-497-5p | 10 | CDK6 | Sponge network | 1.07 | 1.0E-5 | 0.944 | 0 | 0.353 |
| 380 | LINC00702 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-27b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 16 | CSF1 | Sponge network | -0.573 | 0.0699 | -0.395 | 0.03662 | 0.353 |
| 381 | RP11-145A3.1 |
hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-140-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-20b-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p | 14 | CDK6 | Sponge network | 2.565 | 0 | 0.944 | 0 | 0.353 |
| 382 | RP11-30P6.6 |
hsa-miR-140-3p;hsa-miR-148a-3p;hsa-miR-181c-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p | 10 | YWHAG | Sponge network | 2.847 | 0 | 0.33 | 0.00014 | 0.353 |
| 383 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-331-3p;hsa-miR-452-3p | 16 | PRKAA2 | Sponge network | -0.793 | 0 | -3.347 | 0 | 0.353 |
| 384 | RP11-693J15.4 |
hsa-miR-130a-3p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-940 | 10 | CSF1 | Sponge network | -0.732 | 0.18487 | -0.395 | 0.03662 | 0.353 |
| 385 | RP11-227H15.4 |
hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-140-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-497-5p | 22 | CDK6 | Sponge network | 4.405 | 0 | 0.944 | 0 | 0.352 |
| 386 | RP11-594N15.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-149-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-93-5p | 20 | PDGFRA | Sponge network | -2.86 | 0 | -0.463 | 0.04818 | 0.352 |
| 387 | RP11-384L8.1 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p | 10 | FGF2 | Sponge network | -1.152 | 0.00016 | -0.874 | 0.00075 | 0.352 |
| 388 | MIR497HG |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-200b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-452-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-877-5p | 11 | ITGA9 | Sponge network | -1.263 | 0.00248 | -1.022 | 1.0E-5 | 0.351 |
| 389 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p | 10 | CREB3L2 | Sponge network | 0.517 | 0.04893 | -0.271 | 0.03509 | 0.351 |
| 390 | MAGI2-AS3 |
hsa-miR-103a-2-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-2-5p;hsa-miR-27b-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 20 | PRKCA | Sponge network | -0.939 | 4.0E-5 | 0.318 | 0.04622 | 0.351 |
| 391 | AC003090.1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-452-3p;hsa-miR-576-5p;hsa-miR-590-5p | 10 | ITGA9 | Sponge network | -4.323 | 0 | -1.022 | 1.0E-5 | 0.351 |
| 392 | LINC00702 |
hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27b-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | IRS1 | Sponge network | -0.573 | 0.0699 | 0.369 | 0.06704 | 0.351 |
| 393 | LUCAT1 | hsa-miR-101-3p;hsa-miR-148a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-660-5p | 16 | CDK6 | Sponge network | 1.475 | 0 | 0.944 | 0 | 0.351 |
| 394 | LINC00284 |
hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-503-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-944 | 12 | FGF2 | Sponge network | -4.159 | 0 | -0.874 | 0.00075 | 0.351 |
| 395 | BHLHE40-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-940 | 13 | IGF1 | Sponge network | -0.932 | 0 | -1.836 | 0 | 0.35 |
| 396 | RP11-693J15.4 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-31-5p | 15 | PRKAA2 | Sponge network | -0.732 | 0.18487 | -3.347 | 0 | 0.35 |
| 397 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 22 | PIK3R1 | Sponge network | -1.459 | 1.0E-5 | -1.219 | 0 | 0.349 |
| 398 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-454-3p | 10 | IGF1 | Sponge network | -0.72 | 0.01036 | -1.836 | 0 | 0.349 |
| 399 | LINC00900 |
hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-708-5p;hsa-miR-93-5p | 15 | CSF1 | Sponge network | -1.803 | 0 | -0.395 | 0.03662 | 0.349 |
| 400 | MIR22HG |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-28-5p;hsa-miR-345-5p;hsa-miR-582-5p;hsa-miR-93-5p | 10 | CDKN1A | Sponge network | -0.601 | 4.0E-5 | 0.128 | 0.35863 | 0.348 |
| 401 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-944 | 22 | PRLR | Sponge network | -0.939 | 4.0E-5 | -1.308 | 0.00047 | 0.348 |
| 402 | FAM66C |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-18a-3p;hsa-miR-205-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-944 | 11 | FGF7 | Sponge network | -0.798 | 0.00038 | -2.015 | 0 | 0.348 |
| 403 | AC007879.7 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-20b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-361-3p | 10 | COL1A1 | Sponge network | 2.7 | 0 | 3.608 | 0 | 0.347 |
| 404 | FGD5-AS1 | hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p | 12 | PRKAA2 | Sponge network | -0.292 | 0.01482 | -3.347 | 0 | 0.347 |
| 405 | AC003090.1 |
hsa-miR-17-5p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-365a-3p;hsa-miR-452-5p;hsa-miR-590-5p | 10 | BCL2 | Sponge network | -4.323 | 0 | -0.91 | 1.0E-5 | 0.347 |
| 406 | LINC00900 |
hsa-let-7a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-149-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | PDGFRA | Sponge network | -1.803 | 0 | -0.463 | 0.04818 | 0.347 |
| 407 | LINC00702 |
hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-452-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 20 | PRKAA2 | Sponge network | -0.573 | 0.0699 | -3.347 | 0 | 0.346 |
| 408 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-301a-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | FGFR1 | Sponge network | -1.459 | 1.0E-5 | -0.285 | 0.17112 | 0.346 |
| 409 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-452-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-944;hsa-miR-96-5p | 25 | PRKAA2 | Sponge network | -0.882 | 5.0E-5 | -3.347 | 0 | 0.345 |
| 410 | PCED1B-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-944;hsa-miR-96-5p | 14 | PDGFRA | Sponge network | 0.634 | 0.00501 | -0.463 | 0.04818 | 0.344 |
| 411 | WDR86-AS1 |
hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 10 | CSF1 | Sponge network | -0.396 | 0.35778 | -0.395 | 0.03662 | 0.344 |
| 412 | MIR497HG |
hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-455-3p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-96-5p | 13 | PIK3R1 | Sponge network | -1.263 | 0.00248 | -1.219 | 0 | 0.343 |
| 413 | FAM83A-AS1 |
hsa-miR-101-3p;hsa-miR-148a-3p;hsa-miR-20b-5p;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-378a-3p | 14 | CDK6 | Sponge network | 1.064 | 0.00307 | 0.944 | 0 | 0.343 |
| 414 | RASSF8-AS1 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27b-5p;hsa-miR-429;hsa-miR-944 | 11 | PRKAA2 | Sponge network | 0.121 | 0.52445 | -3.347 | 0 | 0.342 |
| 415 | LINC00639 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-31-5p | 11 | PRKAA2 | Sponge network | -0.91 | 0.00848 | -3.347 | 0 | 0.342 |
| 416 | RP11-594N15.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-374b-3p | 13 | PRLR | Sponge network | -2.86 | 0 | -1.308 | 0.00047 | 0.342 |
| 417 | RP11-479G22.8 |
hsa-miR-101-3p;hsa-miR-140-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-429 | 17 | CDK6 | Sponge network | 0.955 | 0 | 0.944 | 0 | 0.342 |
| 418 | LINC00865 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p | 10 | IGF1 | Sponge network | -0.336 | 0.22355 | -1.836 | 0 | 0.341 |
| 419 | CTC-297N7.7 |
hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-589-5p;hsa-miR-96-5p | 11 | PRKAA2 | Sponge network | -1.666 | 0.01558 | -3.347 | 0 | 0.341 |
| 420 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-590-3p | 12 | ATF2 | Sponge network | -0.939 | 4.0E-5 | 0.104 | 0.47373 | 0.341 |
| 421 | RP4-798P15.3 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-944;hsa-miR-96-5p | 19 | PRKAA2 | Sponge network | -1.213 | 0.00098 | -3.347 | 0 | 0.34 |
| 422 | EMX2OS |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-671-5p | 15 | KIT | Sponge network | -1.459 | 1.0E-5 | -1.942 | 0 | 0.339 |
| 423 | LINC00707 |
hsa-miR-101-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-195-3p;hsa-miR-217;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-374a-3p;hsa-miR-500a-3p | 16 | PPP2R5E | Sponge network | 2.018 | 1.0E-5 | 0.181 | 0.07679 | 0.339 |
| 424 | MAGI2-AS3 |
hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | GHR | Sponge network | -0.939 | 4.0E-5 | -1.225 | 0 | 0.339 |
| 425 | FZD10-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 17 | IGF1 | Sponge network | -0.218 | 0.34607 | -1.836 | 0 | 0.339 |
| 426 | LINC00865 |
hsa-let-7a-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-301a-3p | 10 | KIT | Sponge network | -0.336 | 0.22355 | -1.942 | 0 | 0.338 |
| 427 | LINC00707 |
hsa-let-7c-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-20b-5p;hsa-miR-28-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-345-5p;hsa-miR-363-3p;hsa-miR-429;hsa-miR-491-5p | 12 | CDKN1A | Sponge network | 2.018 | 1.0E-5 | 0.128 | 0.35863 | 0.338 |
| 428 | RP4-798P15.3 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-576-5p | 10 | NTRK2 | Sponge network | -1.213 | 0.00098 | -1.115 | 0.01397 | 0.338 |
| 429 | CTD-2015G9.2 |
hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-29b-1-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 12 | EGFR | Sponge network | -3.112 | 5.0E-5 | 0.422 | 0.07321 | 0.337 |
| 430 | LINC01006 | hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-31-5p;hsa-miR-331-3p;hsa-miR-452-3p;hsa-miR-576-5p | 13 | PRKAA2 | Sponge network | -0.754 | 0.09126 | -3.347 | 0 | 0.336 |
| 431 | RP11-285E9.6 | hsa-miR-101-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-217;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-497-5p | 10 | CDK6 | Sponge network | 1.44 | 1.0E-5 | 0.944 | 0 | 0.336 |
| 432 | RP11-321G12.1 |
hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-576-5p | 10 | NTRK2 | Sponge network | -3.552 | 0 | -1.115 | 0.01397 | 0.334 |
| 433 | RP11-384L8.1 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-484 | 10 | FGF7 | Sponge network | -1.152 | 0.00016 | -2.015 | 0 | 0.333 |
| 434 | AC011738.4 |
hsa-let-7g-5p;hsa-miR-125b-5p;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-34a-5p;hsa-miR-497-5p;hsa-miR-532-5p | 13 | IGF1R | Sponge network | 3.153 | 0 | 0.659 | 0 | 0.333 |
| 435 | AC006116.24 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-93-5p | 10 | IL6R | Sponge network | -1.832 | 1.0E-5 | -0.049 | 0.80939 | 0.333 |
| 436 | LINC00284 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p | 17 | PIK3R1 | Sponge network | -4.159 | 0 | -1.219 | 0 | 0.333 |
| 437 | RP4-798P15.3 |
hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-29b-1-5p;hsa-miR-362-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 15 | CREB3L2 | Sponge network | -1.213 | 0.00098 | -0.271 | 0.03509 | 0.332 |
| 438 | RP11-10H3.1 |
hsa-miR-130a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-454-3p | 13 | PRKAA2 | Sponge network | -5.781 | 0 | -3.347 | 0 | 0.332 |
| 439 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-130a-3p;hsa-miR-15a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p | 11 | ITGA2 | Sponge network | -0.304 | 0.38627 | 0.346 | 0.05579 | 0.331 |
| 440 | AC003090.1 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-93-5p | 10 | FGFR1 | Sponge network | -4.323 | 0 | -0.285 | 0.17112 | 0.329 |
| 441 | RP11-594N15.3 |
hsa-miR-130a-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-93-5p;hsa-miR-940 | 11 | CSF1 | Sponge network | -2.86 | 0 | -0.395 | 0.03662 | 0.329 |
| 442 | RP11-11N5.1 | hsa-miR-140-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p | 10 | CDK6 | Sponge network | 5.005 | 0 | 0.944 | 0 | 0.328 |
| 443 | RP11-557H15.3 |
hsa-let-7a-5p;hsa-let-7c-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-20b-5p;hsa-miR-28-5p;hsa-miR-30b-3p | 10 | CDKN1A | Sponge network | 1.673 | 8.0E-5 | 0.128 | 0.35863 | 0.328 |
| 444 | AC144831.1 |
hsa-miR-17-3p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-31-5p;hsa-miR-330-5p;hsa-miR-671-5p;hsa-miR-939-5p;hsa-miR-940 | 11 | GNG7 | Sponge network | -1.546 | 0 | -1.969 | 0 | 0.328 |
| 445 | RP11-384L8.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-452-3p;hsa-miR-576-5p | 14 | PRKAA2 | Sponge network | -1.152 | 0.00016 | -3.347 | 0 | 0.328 |
| 446 | CTD-2015G9.2 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-93-5p | 15 | IL6R | Sponge network | -3.112 | 5.0E-5 | -0.049 | 0.80939 | 0.328 |
| 447 | TBX5-AS1 |
hsa-miR-16-1-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | PDGFRA | Sponge network | 0.014 | 0.98501 | -0.463 | 0.04818 | 0.328 |
| 448 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-582-5p | 11 | ATF2 | Sponge network | -0.175 | 0.58985 | 0.104 | 0.47373 | 0.328 |
| 449 | DNM3OS |
hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-224-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-96-5p | 11 | PIK3R1 | Sponge network | 0.932 | 0.00442 | -1.219 | 0 | 0.328 |
| 450 | RP11-362F19.1 |
hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-944 | 10 | PDGFRA | Sponge network | -0.803 | 0.19311 | -0.463 | 0.04818 | 0.327 |
| 451 | SNHG14 |
hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-29b-1-5p;hsa-miR-671-5p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 14 | CREB3L2 | Sponge network | -1.125 | 0.0001 | -0.271 | 0.03509 | 0.326 |
| 452 | FZD10-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-505-3p;hsa-miR-671-5p | 15 | KIT | Sponge network | -0.218 | 0.34607 | -1.942 | 0 | 0.326 |
| 453 | RP11-524H19.2 | hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-191-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-340-5p;hsa-miR-374a-3p | 10 | CDK6 | Sponge network | 0.759 | 0.01839 | 0.944 | 0 | 0.326 |
| 454 | RP4-798P15.3 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-944 | 12 | FGF7 | Sponge network | -1.213 | 0.00098 | -2.015 | 0 | 0.326 |
| 455 | BHLHE40-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-96-5p | 15 | PRKAA2 | Sponge network | -0.932 | 0 | -3.347 | 0 | 0.325 |
| 456 | LINC00284 |
hsa-miR-103a-2-5p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-452-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-877-5p | 10 | ITGA9 | Sponge network | -4.159 | 0 | -1.022 | 1.0E-5 | 0.325 |
| 457 | RP11-78C3.1 | hsa-miR-101-3p;hsa-miR-140-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-217;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p | 10 | CDK6 | Sponge network | 5.978 | 0 | 0.944 | 0 | 0.324 |
| 458 | CTD-2620I22.1 |
hsa-miR-106a-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-339-5p | 10 | PDGFRA | Sponge network | -1.967 | 0.00106 | -0.463 | 0.04818 | 0.324 |
| 459 | HHIP-AS1 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-31-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-944 | 13 | FGF7 | Sponge network | -1.293 | 0.01561 | -2.015 | 0 | 0.324 |
| 460 | RP11-398K22.12 | hsa-miR-101-3p;hsa-miR-140-3p;hsa-miR-195-5p;hsa-miR-30a-5p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-378a-3p;hsa-miR-664a-3p | 11 | CDK6 | Sponge network | 1.036 | 0 | 0.944 | 0 | 0.324 |
| 461 | RP11-62F24.2 |
hsa-miR-103a-2-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-452-3p;hsa-miR-7-1-3p | 10 | PRKAA2 | Sponge network | -3.195 | 1.0E-5 | -3.347 | 0 | 0.324 |
| 462 | RP11-25I15.3 | hsa-miR-106a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30a-5p | 11 | CDK6 | Sponge network | 5.655 | 0 | 0.944 | 0 | 0.323 |
| 463 | LINC00911 | hsa-let-7a-5p;hsa-let-7c-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-20b-5p;hsa-miR-28-5p;hsa-miR-491-5p | 10 | CDKN1A | Sponge network | 4.956 | 0 | 0.128 | 0.35863 | 0.323 |
| 464 | CTD-2620I22.1 |
hsa-miR-106a-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29b-1-5p;hsa-miR-362-5p;hsa-miR-501-5p | 12 | CREB3L2 | Sponge network | -1.967 | 0.00106 | -0.271 | 0.03509 | 0.322 |
| 465 | APCDD1L-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-20b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-335-3p;hsa-miR-598-3p;hsa-miR-7-1-3p;hsa-miR-9-5p | 10 | THBS2 | Sponge network | 2.077 | 0 | 1.626 | 0 | 0.322 |
| 466 | RP11-757G1.6 |
hsa-miR-130a-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-93-5p | 11 | CSF1 | Sponge network | -1.346 | 0.00088 | -0.395 | 0.03662 | 0.322 |
| 467 | WDR86-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p | 10 | IGF1 | Sponge network | -0.396 | 0.35778 | -1.836 | 0 | 0.321 |
| 468 | NOP14-AS1 |
hsa-miR-140-3p;hsa-miR-195-3p;hsa-miR-217;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-374a-3p;hsa-miR-378a-3p | 10 | YWHAZ | Sponge network | 0.939 | 0 | 0.345 | 0.0016 | 0.321 |
| 469 | RP11-62F24.2 |
hsa-let-7g-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 10 | PDGFRA | Sponge network | -3.195 | 1.0E-5 | -0.463 | 0.04818 | 0.32 |
| 470 | RP11-384L8.1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-629-5p | 12 | IGF1 | Sponge network | -1.152 | 0.00016 | -1.836 | 0 | 0.32 |
| 471 | RP11-757G1.6 |
hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-93-5p;hsa-miR-944 | 15 | PDGFRA | Sponge network | -1.346 | 0.00088 | -0.463 | 0.04818 | 0.32 |
| 472 | LINC00702 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-149-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 27 | PDGFRA | Sponge network | -0.573 | 0.0699 | -0.463 | 0.04818 | 0.319 |
| 473 | XXyac-YX65C7_A.2 | hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p | 10 | PDGFRA | Sponge network | 0.549 | 0.00949 | -0.463 | 0.04818 | 0.318 |
| 474 | RP11-456K23.1 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 10 | PPP2R3A | Sponge network | -0.223 | 0.27461 | -0.147 | 0.34058 | 0.317 |
| 475 | WDR86-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p | 11 | KIT | Sponge network | -0.396 | 0.35778 | -1.942 | 0 | 0.317 |
| 476 | CTD-2015G9.2 |
hsa-miR-130a-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-671-5p | 13 | KIT | Sponge network | -3.112 | 5.0E-5 | -1.942 | 0 | 0.317 |
| 477 | LINC00284 |
hsa-miR-17-5p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-365a-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-5p | 12 | BCL2 | Sponge network | -4.159 | 0 | -0.91 | 1.0E-5 | 0.316 |
| 478 | AF127936.7 |
hsa-miR-106a-5p;hsa-miR-140-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-23b-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-378a-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 22 | CDK6 | Sponge network | 0.453 | 0.00811 | 0.944 | 0 | 0.315 |
| 479 | DLGAP1-AS5 |
hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-589-3p | 10 | PIK3R1 | Sponge network | -6.207 | 0 | -1.219 | 0 | 0.315 |
| 480 | LINC00900 |
hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-31-5p;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-939-5p | 12 | GNG7 | Sponge network | -1.803 | 0 | -1.969 | 0 | 0.314 |
| 481 | RP11-284N8.3 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-944;hsa-miR-96-5p | 10 | PRLR | Sponge network | 0.003 | 0.99478 | -1.308 | 0.00047 | 0.314 |
| 482 | RP11-150O12.1 |
hsa-let-7g-5p;hsa-miR-125b-5p;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-532-5p | 11 | IGF1R | Sponge network | 1.523 | 0 | 0.659 | 0 | 0.313 |
| 483 | AC093627.10 |
hsa-let-7a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-503-5p;hsa-miR-589-3p | 10 | FGF2 | Sponge network | -0.893 | 0.00123 | -0.874 | 0.00075 | 0.313 |
| 484 | RP11-815I9.4 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-34a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p | 16 | PDGFRA | Sponge network | 0.004 | 0.98116 | -0.463 | 0.04818 | 0.313 |
| 485 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-9-5p;hsa-miR-93-5p | 13 | THBS2 | Sponge network | -1.459 | 1.0E-5 | 1.626 | 0 | 0.313 |
| 486 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 13 | PRKAA2 | Sponge network | -0.175 | 0.58985 | -3.347 | 0 | 0.312 |
| 487 | FAM225B |
hsa-miR-139-5p;hsa-miR-19a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-362-3p;hsa-miR-500a-3p | 11 | IGF1R | Sponge network | 2.186 | 0 | 0.659 | 0 | 0.312 |
| 488 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-576-5p | 13 | IGF1 | Sponge network | -0.304 | 0.38627 | -1.836 | 0 | 0.312 |
| 489 | LINC00639 |
hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-452-5p;hsa-miR-503-5p;hsa-miR-589-3p | 10 | FGF2 | Sponge network | -0.91 | 0.00848 | -0.874 | 0.00075 | 0.311 |
| 490 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-31-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-944 | 12 | FGF7 | Sponge network | -1.712 | 0 | -2.015 | 0 | 0.31 |
| 491 | RP11-517P14.2 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-500a-5p;hsa-miR-93-5p | 11 | PRKAA2 | Sponge network | -0.402 | 0.1503 | -3.347 | 0 | 0.31 |
| 492 | AC003090.1 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-452-5p;hsa-miR-590-5p | 11 | FGF2 | Sponge network | -4.323 | 0 | -0.874 | 0.00075 | 0.31 |
| 493 | CTD-2171N6.1 |
hsa-miR-101-3p;hsa-miR-140-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-34a-5p;hsa-miR-497-5p;hsa-miR-502-3p;hsa-miR-582-5p | 28 | CDK6 | Sponge network | 5.89 | 0 | 0.944 | 0 | 0.31 |
| 494 | AF127936.7 |
hsa-miR-125b-5p;hsa-miR-139-5p;hsa-miR-140-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-29b-2-5p;hsa-miR-320a;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-532-5p | 10 | IGF1R | Sponge network | 0.453 | 0.00811 | 0.659 | 0 | 0.309 |
| 495 | MIR497HG |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-331-3p;hsa-miR-452-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | PRKAA2 | Sponge network | -1.263 | 0.00248 | -3.347 | 0 | 0.309 |
| 496 | RP11-757G1.6 |
hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-944 | 11 | FGF2 | Sponge network | -1.346 | 0.00088 | -0.874 | 0.00075 | 0.309 |
| 497 | RP11-282O18.3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-590-5p | 12 | PDGFRA | Sponge network | -0.266 | 0.05794 | -0.463 | 0.04818 | 0.308 |
| 498 | RP11-218E20.3 |
hsa-miR-101-5p;hsa-miR-181c-5p;hsa-miR-195-3p;hsa-miR-217;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-374a-5p;hsa-miR-374b-5p | 14 | PPP2R5E | Sponge network | 4.613 | 0 | 0.181 | 0.07679 | 0.308 |
| 499 | CLDN10-AS1 |
hsa-miR-103a-2-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-452-3p;hsa-miR-944 | 12 | PRKAA2 | Sponge network | -5.599 | 0 | -3.347 | 0 | 0.308 |
| 500 | CTD-2620I22.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-501-5p | 10 | IL6R | Sponge network | -1.967 | 0.00106 | -0.049 | 0.80939 | 0.308 |
| 501 | TPTEP1 |
hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p | 13 | PIK3R1 | Sponge network | -2.193 | 0 | -1.219 | 0 | 0.308 |
| 502 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181c-5p;hsa-miR-181d-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-93-5p | 10 | PPP2R3A | Sponge network | -0.601 | 4.0E-5 | -0.147 | 0.34058 | 0.308 |
| 503 | FZD10-AS1 |
hsa-miR-130a-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-23b-5p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-339-5p | 10 | IGF2 | Sponge network | -0.218 | 0.34607 | 0.131 | 0.60661 | 0.307 |
| 504 | RP11-166D19.1 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | CREB5 | Sponge network | -0.882 | 5.0E-5 | -0.062 | 0.77912 | 0.306 |
| 505 | LINC00957 |
hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p | 10 | PIK3R1 | Sponge network | -0.677 | 2.0E-5 | -1.219 | 0 | 0.306 |
| 506 | MAGI2-AS3 |
hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 11 | ITGAV | Sponge network | -0.939 | 4.0E-5 | 0.843 | 0 | 0.305 |
| 507 | AC108142.1 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20b-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-7-1-3p | 12 | CREB5 | Sponge network | 2.31 | 0 | -0.062 | 0.77912 | 0.304 |
| 508 | CTA-221G9.11 |
hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-93-5p | 10 | PDGFRA | Sponge network | -2.198 | 0 | -0.463 | 0.04818 | 0.304 |
| 509 | AF127936.7 |
hsa-miR-141-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-7-1-3p | 10 | ITGA1 | Sponge network | 0.453 | 0.00811 | 1.673 | 0 | 0.304 |
| 510 | RP4-798P15.3 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-503-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-944 | 11 | FGF2 | Sponge network | -1.213 | 0.00098 | -0.874 | 0.00075 | 0.303 |
| 511 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-34a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | PPP2R3A | Sponge network | -1.459 | 1.0E-5 | -0.147 | 0.34058 | 0.303 |
| 512 | LINC00520 | hsa-miR-101-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-217;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-335-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-497-5p | 15 | CDK6 | Sponge network | 3.383 | 0 | 0.944 | 0 | 0.303 |
| 513 | SH3RF3-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29b-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | CREB5 | Sponge network | -0.175 | 0.58985 | -0.062 | 0.77912 | 0.302 |
| 514 | RP11-58E21.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-25-3p;hsa-miR-28-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-582-3p;hsa-miR-93-5p | 14 | PTEN | Sponge network | 0.445 | 0.04375 | -0.106 | 0.25948 | 0.301 |
| 515 | RP11-863P13.3 | hsa-miR-140-3p;hsa-miR-148a-5p;hsa-miR-20b-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-502-3p | 12 | CDK6 | Sponge network | 4.189 | 0 | 0.944 | 0 | 0.3 |
| 516 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-3p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | CREB5 | Sponge network | -0.939 | 4.0E-5 | -0.062 | 0.77912 | 0.3 |
| 517 | BZRAP1-AS1 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-29b-1-5p;hsa-miR-671-5p | 10 | CREB3L2 | Sponge network | -0.233 | 0.50729 | -0.271 | 0.03509 | 0.3 |
| 518 | DNM3OS |
hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-429;hsa-miR-7-1-3p | 10 | CREB3L2 | Sponge network | 0.932 | 0.00442 | -0.271 | 0.03509 | 0.3 |
| 519 | RP11-218E20.3 |
hsa-let-7g-5p;hsa-miR-125b-5p;hsa-miR-139-5p;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-29b-2-5p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-532-5p | 16 | IGF1R | Sponge network | 4.613 | 0 | 0.659 | 0 | 0.299 |
| 520 | CTD-2135D7.5 |
hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-590-5p | 10 | FGF2 | Sponge network | -2.579 | 0.0001 | -0.874 | 0.00075 | 0.299 |
| 521 | RP11-815I9.4 |
hsa-let-7a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-664a-3p | 11 | THBS1 | Sponge network | 0.004 | 0.98116 | -0.06 | 0.8171 | 0.299 |
| 522 | LINC00605 |
hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p;hsa-miR-944 | 10 | PDGFRA | Sponge network | -2.602 | 0.00047 | -0.463 | 0.04818 | 0.298 |
| 523 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p;hsa-miR-944 | 11 | PRLR | Sponge network | -1.293 | 0.01561 | -1.308 | 0.00047 | 0.298 |
| 524 | RP4-794H19.1 |
hsa-let-7c-5p;hsa-let-7g-5p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-20b-5p;hsa-miR-28-5p;hsa-miR-30b-3p;hsa-miR-345-5p;hsa-miR-363-3p;hsa-miR-491-5p | 11 | CDKN1A | Sponge network | 1.019 | 8.0E-5 | 0.128 | 0.35863 | 0.298 |
| 525 | RP11-477H21.2 | hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-944 | 10 | PRKAA2 | Sponge network | -2.192 | 0.00014 | -3.347 | 0 | 0.298 |
| 526 | RP11-166D19.1 |
hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-29b-1-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 16 | CREB3L2 | Sponge network | -0.882 | 5.0E-5 | -0.271 | 0.03509 | 0.297 |
| 527 | RP11-193H5.1 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-29b-3p;hsa-miR-320a | 11 | PIK3R1 | Sponge network | -0.426 | 0.01062 | -1.219 | 0 | 0.297 |
| 528 | FAM66C |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-944 | 11 | PDGFRA | Sponge network | -0.798 | 0.00038 | -0.463 | 0.04818 | 0.296 |
| 529 | RP11-384L8.1 |
hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-365a-3p;hsa-miR-7-5p | 11 | BCL2 | Sponge network | -1.152 | 0.00016 | -0.91 | 1.0E-5 | 0.296 |
| 530 | RP11-282O18.3 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-452-3p | 11 | PRKAA2 | Sponge network | -0.266 | 0.05794 | -3.347 | 0 | 0.295 |
| 531 | CTD-2015G9.2 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p | 13 | IGF1 | Sponge network | -3.112 | 5.0E-5 | -1.836 | 0 | 0.295 |
| 532 | AC007743.1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 11 | PRLR | Sponge network | -1.053 | 0.00923 | -1.308 | 0.00047 | 0.295 |
| 533 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-224-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 21 | PIK3R1 | Sponge network | -0.573 | 0.0699 | -1.219 | 0 | 0.294 |
| 534 | DLGAP1-AS5 |
hsa-miR-15a-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-503-5p;hsa-miR-589-3p;hsa-miR-944 | 10 | FGF2 | Sponge network | -6.207 | 0 | -0.874 | 0.00075 | 0.294 |
| 535 | CASC15 |
hsa-let-7g-5p;hsa-miR-139-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p | 10 | IGF1R | Sponge network | 1.485 | 0 | 0.659 | 0 | 0.292 |
| 536 | WDR86-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 11 | PRKAA2 | Sponge network | -0.396 | 0.35778 | -3.347 | 0 | 0.291 |
| 537 | LINC00900 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-205-5p;hsa-miR-224-5p;hsa-miR-331-5p;hsa-miR-455-3p;hsa-miR-589-3p | 10 | RELN | Sponge network | -1.803 | 0 | -2.339 | 0 | 0.291 |
| 538 | LINC00702 |
hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 10 | ITGAV | Sponge network | -0.573 | 0.0699 | 0.843 | 0 | 0.29 |
| 539 | FZD10-AS1 |
hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-205-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 10 | LPAR1 | Sponge network | -0.218 | 0.34607 | -0.679 | 0.00029 | 0.29 |
| 540 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-944;hsa-miR-96-5p | 16 | PRLR | Sponge network | -0.882 | 5.0E-5 | -1.308 | 0.00047 | 0.289 |
| 541 | OIP5-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-146b-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-454-3p;hsa-miR-576-5p | 10 | PRKAA2 | Sponge network | -0.06 | 0.55122 | -3.347 | 0 | 0.289 |
| 542 | RP11-30P6.6 |
hsa-let-7g-5p;hsa-miR-125b-5p;hsa-miR-140-3p;hsa-miR-29b-2-5p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-374a-5p | 11 | IGF1R | Sponge network | 2.847 | 0 | 0.659 | 0 | 0.288 |
| 543 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p | 10 | PDGFRA | Sponge network | -0.793 | 0 | -0.463 | 0.04818 | 0.288 |
| 544 | RP11-297P16.3 | hsa-miR-101-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-20b-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-340-5p | 14 | CDK6 | Sponge network | 6.381 | 0 | 0.944 | 0 | 0.288 |
| 545 | EMX2OS |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-205-5p;hsa-miR-21-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | INSR | Sponge network | -1.459 | 1.0E-5 | -0.194 | 0.13669 | 0.288 |
| 546 | RP4-798P15.3 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-590-5p | 14 | IL6R | Sponge network | -1.213 | 0.00098 | -0.049 | 0.80939 | 0.286 |
| 547 | CTA-392E5.1 | hsa-miR-106a-5p;hsa-miR-140-3p;hsa-miR-195-5p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-378a-3p | 16 | CDK6 | Sponge network | 5.137 | 0 | 0.944 | 0 | 0.285 |
| 548 | CTD-2015G9.2 |
hsa-miR-130a-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-708-5p;hsa-miR-93-5p | 10 | CSF1 | Sponge network | -3.112 | 5.0E-5 | -0.395 | 0.03662 | 0.285 |
| 549 | RP11-166D19.1 |
hsa-miR-103a-2-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-2-5p;hsa-miR-27b-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 13 | PRKCA | Sponge network | -0.882 | 5.0E-5 | 0.318 | 0.04622 | 0.285 |
| 550 | CASC15 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-7-1-3p | 12 | CREB5 | Sponge network | 1.485 | 0 | -0.062 | 0.77912 | 0.284 |
| 551 | AC003090.1 |
hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-205-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | ITGA8 | Sponge network | -4.323 | 0 | -0.747 | 0.01533 | 0.284 |
| 552 | CTD-2008P7.9 |
hsa-miR-103a-2-5p;hsa-miR-125a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-576-5p | 11 | IL6R | Sponge network | -1.202 | 0.00093 | -0.049 | 0.80939 | 0.283 |
| 553 | LINC00900 |
hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-205-5p;hsa-miR-20a-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 10 | LPAR1 | Sponge network | -1.803 | 0 | -0.679 | 0.00029 | 0.283 |
| 554 | DHRS4-AS1 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-454-3p;hsa-miR-500a-5p | 13 | PRKAA2 | Sponge network | -0.068 | 0.72321 | -3.347 | 0 | 0.283 |
| 555 | RP11-362F19.1 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-224-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-452-5p;hsa-miR-454-3p | 11 | IGF1 | Sponge network | -0.803 | 0.19311 | -1.836 | 0 | 0.283 |
| 556 | CTD-2171N6.1 |
hsa-miR-148a-3p;hsa-miR-17-3p;hsa-miR-200b-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-628-5p | 10 | IRS1 | Sponge network | 5.89 | 0 | 0.369 | 0.06704 | 0.282 |
| 557 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-944 | 11 | FGF2 | Sponge network | -1.712 | 0 | -0.874 | 0.00075 | 0.282 |
| 558 | LINC00639 |
hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-589-3p | 13 | PDGFRA | Sponge network | -0.91 | 0.00848 | -0.463 | 0.04818 | 0.282 |
| 559 | HHIP-AS1 |
hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 14 | CREB3L2 | Sponge network | -1.293 | 0.01561 | -0.271 | 0.03509 | 0.281 |
| 560 | LINC00900 |
hsa-miR-17-5p;hsa-miR-205-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-365a-3p;hsa-miR-455-5p;hsa-miR-590-5p;hsa-miR-7-5p | 11 | BCL2 | Sponge network | -1.803 | 0 | -0.91 | 1.0E-5 | 0.281 |
| 561 | LINC00707 |
hsa-let-7g-5p;hsa-miR-125b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-34a-5p;hsa-miR-497-5p;hsa-miR-500a-3p | 11 | IGF1R | Sponge network | 2.018 | 1.0E-5 | 0.659 | 0 | 0.281 |
| 562 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-33b-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 24 | PDGFRA | Sponge network | -4.323 | 0 | -0.463 | 0.04818 | 0.281 |
| 563 | RP11-193H5.1 |
hsa-miR-103a-2-5p;hsa-miR-186-5p;hsa-miR-24-2-5p;hsa-miR-29b-2-5p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-93-3p | 10 | PRKCA | Sponge network | -0.426 | 0.01062 | 0.318 | 0.04622 | 0.281 |
| 564 | LINC00702 |
hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-205-3p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | ITGA8 | Sponge network | -0.573 | 0.0699 | -0.747 | 0.01533 | 0.28 |
| 565 | RP4-794H19.1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-20b-5p;hsa-miR-23b-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p | 10 | COL1A1 | Sponge network | 1.019 | 8.0E-5 | 3.608 | 0 | 0.279 |
| 566 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-576-5p | 10 | PRLR | Sponge network | -0.304 | 0.38627 | -1.308 | 0.00047 | 0.279 |
| 567 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-452-3p;hsa-miR-454-3p;hsa-miR-96-5p | 16 | PRKAA2 | Sponge network | -0.33 | 0.08136 | -3.347 | 0 | 0.279 |
| 568 | RP11-815I9.4 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-576-5p | 12 | IGF1 | Sponge network | 0.004 | 0.98116 | -1.836 | 0 | 0.279 |
| 569 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | PRLR | Sponge network | -4.323 | 0 | -1.308 | 0.00047 | 0.278 |
| 570 | RP11-774O3.3 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-708-5p | 10 | CSF1 | Sponge network | -1.712 | 0 | -0.395 | 0.03662 | 0.275 |
| 571 | RP11-879F14.2 | hsa-miR-101-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-378a-3p | 14 | CDK6 | Sponge network | 1.149 | 9.0E-5 | 0.944 | 0 | 0.274 |
| 572 | RP11-244O19.1 |
hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-34a-5p;hsa-miR-378a-3p | 12 | CDK6 | Sponge network | 0.52 | 0.05156 | 0.944 | 0 | 0.274 |
| 573 | RP11-362F19.1 |
hsa-miR-130a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 11 | KIT | Sponge network | -0.803 | 0.19311 | -1.942 | 0 | 0.274 |
| 574 | XXyac-YX65C7_A.3 |
hsa-let-7g-5p;hsa-miR-125b-5p;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-29b-2-5p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p | 10 | IGF1R | Sponge network | 6.112 | 0 | 0.659 | 0 | 0.274 |
| 575 | RP4-798P15.3 |
hsa-miR-130a-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-23a-5p;hsa-miR-301a-3p;hsa-miR-708-5p;hsa-miR-96-5p | 10 | CSF1 | Sponge network | -1.213 | 0.00098 | -0.395 | 0.03662 | 0.273 |
| 576 | RP11-384L8.1 |
hsa-let-7d-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-29b-1-5p;hsa-miR-671-5p | 12 | CREB3L2 | Sponge network | -1.152 | 0.00016 | -0.271 | 0.03509 | 0.273 |
| 577 | CTD-2354A18.1 | hsa-miR-101-3p;hsa-miR-140-3p;hsa-miR-195-5p;hsa-miR-217;hsa-miR-26a-5p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-34a-5p;hsa-miR-374a-5p | 11 | CDK6 | Sponge network | 4.214 | 0 | 0.944 | 0 | 0.273 |
| 578 | RP11-389C8.2 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-92a-1-5p | 12 | CREB3L2 | Sponge network | 0.187 | 0.31051 | -0.271 | 0.03509 | 0.272 |
| 579 | RP11-497E19.1 |
hsa-miR-106a-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20b-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-582-3p;hsa-miR-7-1-3p | 13 | CREB5 | Sponge network | 2.366 | 0.00063 | -0.062 | 0.77912 | 0.272 |
| 580 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | FGFR1 | Sponge network | -0.573 | 0.0699 | -0.285 | 0.17112 | 0.271 |
| 581 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-944 | 15 | PRLR | Sponge network | -1.712 | 0 | -1.308 | 0.00047 | 0.271 |
| 582 | RP11-91K9.1 | hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-141-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-29c-3p;hsa-miR-9-5p | 13 | CDK6 | Sponge network | 3.763 | 0 | 0.944 | 0 | 0.271 |
| 583 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | PPP2R3A | Sponge network | -0.573 | 0.0699 | -0.147 | 0.34058 | 0.269 |
| 584 | MEG3 |
hsa-let-7f-1-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-452-3p;hsa-miR-7-1-3p;hsa-miR-944 | 12 | PRKAA2 | Sponge network | -0.286 | 0.30726 | -3.347 | 0 | 0.269 |
| 585 | CASC15 |
hsa-miR-17-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-27b-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-590-3p | 10 | PRKCA | Sponge network | 1.485 | 0 | 0.318 | 0.04622 | 0.269 |
| 586 | RP4-798P15.3 |
hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | EGFR | Sponge network | -1.213 | 0.00098 | 0.422 | 0.07321 | 0.269 |
| 587 | RP11-244O19.1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-28-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-532-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 16 | PTEN | Sponge network | 0.52 | 0.05156 | -0.106 | 0.25948 | 0.269 |
| 588 | RBPMS-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-454-3p | 12 | PRKAA2 | Sponge network | -2.717 | 0 | -3.347 | 0 | 0.269 |
| 589 | CTC-297N7.7 |
hsa-miR-103a-3p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-224-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-96-5p | 11 | PIK3R1 | Sponge network | -1.666 | 0.01558 | -1.219 | 0 | 0.267 |
| 590 | AC007743.1 |
hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-2355-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-589-3p;hsa-miR-590-5p | 12 | PDPK1 | Sponge network | -1.053 | 0.00923 | -0.375 | 0.00067 | 0.267 |
| 591 | CTD-2554C21.3 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-455-3p;hsa-miR-629-3p | 10 | PIK3R1 | Sponge network | -2.118 | 6.0E-5 | -1.219 | 0 | 0.267 |
| 592 | RP11-64C12.8 | hsa-let-7a-5p;hsa-let-7c-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-20b-5p;hsa-miR-28-5p;hsa-miR-30b-3p | 11 | CDKN1A | Sponge network | -1.035 | 0.07653 | 0.128 | 0.35863 | 0.267 |
| 593 | CTD-2015G9.2 |
hsa-miR-103a-3p;hsa-miR-130a-3p;hsa-miR-15a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-454-3p | 10 | ITGA2 | Sponge network | -3.112 | 5.0E-5 | 0.346 | 0.05579 | 0.266 |
| 594 | CTD-2015G9.2 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | PRKAA2 | Sponge network | -3.112 | 5.0E-5 | -3.347 | 0 | 0.265 |
| 595 | EMX2OS |
hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-23a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-708-5p;hsa-miR-93-5p;hsa-miR-940 | 17 | CSF1 | Sponge network | -1.459 | 1.0E-5 | -0.395 | 0.03662 | 0.264 |
| 596 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-93-3p;hsa-miR-93-5p | 10 | PDPK1 | Sponge network | -0.304 | 0.38627 | -0.375 | 0.00067 | 0.264 |
| 597 | SH3RF3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-576-5p;hsa-miR-590-5p | 11 | IL6R | Sponge network | -0.175 | 0.58985 | -0.049 | 0.80939 | 0.264 |
| 598 | EMX2OS |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 23 | CREB3L2 | Sponge network | -1.459 | 1.0E-5 | -0.271 | 0.03509 | 0.263 |
| 599 | LINC01018 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-193a-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-944 | 10 | PDGFRA | Sponge network | -1.613 | 0.00396 | -0.463 | 0.04818 | 0.263 |
| 600 | AF127936.7 |
hsa-miR-106a-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-29b-2-5p;hsa-miR-362-5p;hsa-miR-7-1-3p | 10 | CREB3L2 | Sponge network | 0.453 | 0.00811 | -0.271 | 0.03509 | 0.263 |
| 601 | CTD-2554C21.3 |
hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-455-5p | 11 | PDGFRA | Sponge network | -2.118 | 6.0E-5 | -0.463 | 0.04818 | 0.262 |
| 602 | CTA-221G9.11 |
hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29b-1-5p;hsa-miR-671-5p | 10 | CREB3L2 | Sponge network | -2.198 | 0 | -0.271 | 0.03509 | 0.262 |
| 603 | LINC00900 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-205-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | ITGA8 | Sponge network | -1.803 | 0 | -0.747 | 0.01533 | 0.261 |
| 604 | AC006262.5 | hsa-miR-106a-5p;hsa-miR-140-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-497-5p | 15 | CDK6 | Sponge network | 2.376 | 0 | 0.944 | 0 | 0.261 |
| 605 | RP11-456K23.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-205-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-708-5p;hsa-miR-93-5p | 14 | CSF1 | Sponge network | -0.223 | 0.27461 | -0.395 | 0.03662 | 0.261 |
| 606 | LINC00900 |
hsa-miR-106a-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-301a-3p;hsa-miR-455-5p;hsa-miR-93-5p | 13 | FGFR1 | Sponge network | -1.803 | 0 | -0.285 | 0.17112 | 0.26 |
| 607 | LINC00865 |
hsa-let-7a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-339-5p | 11 | PDGFRA | Sponge network | -0.336 | 0.22355 | -0.463 | 0.04818 | 0.26 |
| 608 | AC016995.3 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-454-3p;hsa-miR-7-1-3p | 10 | PRKAA2 | Sponge network | -0.803 | 0.01494 | -3.347 | 0 | 0.259 |
| 609 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-454-3p | 11 | PRKAA2 | Sponge network | -0.72 | 0.01036 | -3.347 | 0 | 0.259 |
| 610 | RP4-798P15.3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-944;hsa-miR-96-5p | 19 | PDGFRA | Sponge network | -1.213 | 0.00098 | -0.463 | 0.04818 | 0.259 |
| 611 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 14 | PRLR | Sponge network | -0.175 | 0.58985 | -1.308 | 0.00047 | 0.258 |
| 612 | RP11-384L8.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-576-5p | 14 | PDGFRA | Sponge network | -1.152 | 0.00016 | -0.463 | 0.04818 | 0.258 |
| 613 | RP11-321G12.1 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-576-5p | 11 | PRKAA2 | Sponge network | -3.552 | 0 | -3.347 | 0 | 0.257 |
| 614 | RP4-798P15.3 |
hsa-miR-103a-3p;hsa-miR-130a-3p;hsa-miR-15a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-590-5p | 11 | ITGA2 | Sponge network | -1.213 | 0.00098 | 0.346 | 0.05579 | 0.256 |
| 615 | TPTEP1 |
hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-29b-1-5p;hsa-miR-671-5p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 14 | CREB3L2 | Sponge network | -2.193 | 0 | -0.271 | 0.03509 | 0.256 |
| 616 | MAGI2-AS3 |
hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-452-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | NTRK2 | Sponge network | -0.939 | 4.0E-5 | -1.115 | 0.01397 | 0.256 |
| 617 | CASC15 |
hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-205-3p;hsa-miR-29b-2-5p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CREB3L2 | Sponge network | 1.485 | 0 | -0.271 | 0.03509 | 0.255 |
| 618 | RP1-193H18.2 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-944;hsa-miR-96-5p | 12 | PDGFRA | Sponge network | -1.539 | 0 | -0.463 | 0.04818 | 0.254 |
| 619 | RP11-321G12.1 |
hsa-miR-103a-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-2355-5p;hsa-miR-542-3p;hsa-miR-589-3p | 10 | PDPK1 | Sponge network | -3.552 | 0 | -0.375 | 0.00067 | 0.254 |
| 620 | LINC00668 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-361-3p | 10 | COL1A1 | Sponge network | 3.431 | 0 | 3.608 | 0 | 0.254 |
| 621 | SSTR5-AS1 |
hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-455-3p | 10 | NTRK2 | Sponge network | -1.443 | 0.04871 | -1.115 | 0.01397 | 0.254 |
| 622 | PART1 |
hsa-miR-125a-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p | 12 | NTRK2 | Sponge network | -3.265 | 0 | -1.115 | 0.01397 | 0.254 |
| 623 | CTD-2015G9.2 |
hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 10 | THBS1 | Sponge network | -3.112 | 5.0E-5 | -0.06 | 0.8171 | 0.254 |
| 624 | RP11-554I8.2 |
hsa-let-7c-5p;hsa-let-7g-5p;hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-20b-5p;hsa-miR-28-5p;hsa-miR-30b-3p;hsa-miR-345-5p;hsa-miR-363-3p | 10 | CDKN1A | Sponge network | 0.342 | 0.50007 | 0.128 | 0.35863 | 0.253 |
| 625 | RP11-356J5.12 | hsa-miR-101-3p;hsa-miR-106a-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-200b-3p;hsa-miR-20b-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-29c-3p;hsa-miR-30d-3p | 13 | CDK6 | Sponge network | 0.472 | 0.02484 | 0.944 | 0 | 0.253 |
| 626 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-454-3p;hsa-miR-944 | 15 | PRKAA2 | Sponge network | -0.233 | 0.50729 | -3.347 | 0 | 0.252 |
| 627 | RP11-362F19.1 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-452-3p;hsa-miR-454-3p;hsa-miR-944 | 10 | PRKAA2 | Sponge network | -0.803 | 0.19311 | -3.347 | 0 | 0.252 |
| 628 | RAMP2-AS1 |
hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-29b-1-5p;hsa-miR-671-5p | 13 | CREB3L2 | Sponge network | -1.258 | 0.00026 | -0.271 | 0.03509 | 0.252 |
| 629 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-26b-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-532-3p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | CREB5 | Sponge network | -0.573 | 0.0699 | -0.062 | 0.77912 | 0.251 |
| 630 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 14 | PRLR | Sponge network | -0.218 | 0.34607 | -1.308 | 0.00047 | 0.251 |
| 631 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-205-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29b-2-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 17 | CREB3L2 | Sponge network | -0.573 | 0.0699 | -0.271 | 0.03509 | 0.251 |
| 632 | LINC01018 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-205-3p;hsa-miR-27a-3p;hsa-miR-31-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-944 | 10 | FGF7 | Sponge network | -1.613 | 0.00396 | -2.015 | 0 | 0.251 |
| 633 | RP11-757G1.6 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-31-5p;hsa-miR-452-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-93-5p;hsa-miR-944 | 16 | PRKAA2 | Sponge network | -1.346 | 0.00088 | -3.347 | 0 | 0.25 |