This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-let-7a-5p | ASPM | -0.03 | 0.25442 | 2.01 | 0 | miRNAWalker2 validate | -3.72 | 0 | ||
| 2 | hsa-let-7a-5p | CCNB2 | -0.03 | 0.25442 | 1.87 | 0 | miRNAWalker2 validate | -3.92 | 0 | ||
| 3 | hsa-let-7a-5p | RRM2 | -0.03 | 0.25442 | 1.6 | 0 | miRNAWalker2 validate; TargetScan; miRNATAP | -3.21 | 0 | ||
| 4 | hsa-let-7b-5p | BIRC5 | -0.12 | 0.00077 | 1.89 | 0 | miRNAWalker2 validate | -4.17 | 0 | ||
| 5 | hsa-let-7b-5p | CCNB1 | -0.12 | 0.00077 | 1.59 | 0 | miRNAWalker2 validate | -3.89 | 0 | ||
| 6 | hsa-let-7b-5p | CCNB2 | -0.12 | 0.00077 | 1.87 | 0 | miRNAWalker2 validate | -4.32 | 0 | ||
| 7 | hsa-let-7b-5p | CDCA8 | -0.12 | 0.00077 | 1.9 | 0 | miRNAWalker2 validate | -4.01 | 0 | ||
| 8 | hsa-let-7b-5p | RRM2 | -0.12 | 0.00077 | 1.6 | 0 | miRNAWalker2 validate; miRNATAP | -3.62 | 0 | ||
| 9 | hsa-let-7c-5p | CCNB2 | -0.13 | 0.00086 | 1.87 | 0 | miRNAWalker2 validate | -4.54 | 0 | ||
| 10 | hsa-let-7e-5p | MKI67 | -0.06 | 0.0129 | 1.67 | 0 | miRNAWalker2 validate | -4.4 | 0 | ||
| 11 | hsa-miR-100-5p | RRM2 | -0.14 | 0.00081 | 1.6 | 0 | miRNAWalker2 validate | -3.6 | 0 | ||
| 12 | hsa-miR-101-3p | BIRC5 | -0.13 | 4.0E-5 | 1.89 | 0 | miRNAWalker2 validate | -5.94 | 0 | ||
| 13 | hsa-miR-10a-5p | BIRC5 | -0.29 | 0 | 1.89 | 0 | miRNAWalker2 validate | -3.79 | 0 | ||
| 14 | hsa-miR-122-5p | CENPF | -0.14 | 0.00123 | 1.92 | 0 | miRNAWalker2 validate | -3.98 | 0 | ||
| 15 | hsa-miR-124-3p | BEX1 | 0.08 | 0.04068 | -1.68 | 0 | miRNAWalker2 validate | -1.46 | 0.02553 | ||
| 16 | hsa-miR-124-3p | ID1 | 0.08 | 0.04068 | -1.66 | 0 | miRNAWalker2 validate | -0.96 | 0.03411 | ||
| 17 | hsa-miR-124-3p | MFAP4 | 0.08 | 0.04068 | -1.79 | 0 | miRNAWalker2 validate | -1.66 | 0.00453 | ||
| 18 | hsa-miR-125a-5p | PRC1 | -0.13 | 0.00014 | 1.59 | 0 | miRNAWalker2 validate | -3.56 | 0 | ||
| 19 | hsa-miR-125b-5p | HOXA13 | -0.19 | 1.0E-5 | 1.81 | 0 | miRNAWalker2 validate | -2.15 | 0.00011 | ||
| 20 | hsa-miR-128-3p | LIFR | 0.08 | 0.0018 | -1.71 | 0 | miRNAWalker2 validate | -3.77 | 0 | ||
| 21 | hsa-miR-128-3p | RELN | 0.08 | 0.0018 | -1.76 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget | -4.11 | 1.0E-5 | ||
| 22 | hsa-miR-132-3p | CYP26A1 | 0.04 | 0.33877 | -1.78 | 0 | miRNAWalker2 validate | -1.32 | 0.03036 | ||
| 23 | hsa-miR-146a-5p | CDKN3 | -0.15 | 0.00019 | 1.99 | 0 | miRNAWalker2 validate | -2.83 | 0 | ||
| 24 | hsa-miR-148b-3p | CCL19 | 0.14 | 0 | -1.9 | 0 | miRNAWalker2 validate | -6.88 | 0 | ||
| 25 | hsa-miR-148b-3p | DPT | 0.14 | 0 | -2.5 | 0 | miRNAWalker2 validate | -8.78 | 0 | ||
| 26 | hsa-miR-148b-3p | IL13RA2 | 0.14 | 0 | -2.45 | 0 | miRNAWalker2 validate | -5.71 | 0 | ||
| 27 | hsa-miR-15b-5p | BEX1 | 0.17 | 0 | -1.68 | 0 | miRNAWalker2 validate | -5.39 | 0 | ||
| 28 | hsa-miR-16-5p | CRHBP | 0.05 | 0.02313 | -3.12 | 0 | miRNAWalker2 validate | -6.39 | 1.0E-5 | ||
| 29 | hsa-miR-192-5p | ANLN | 0.01 | 0.72622 | 2.11 | 0 | miRNAWalker2 validate | -1.76 | 0.01044 | ||
| 30 | hsa-miR-192-5p | ASPM | 0.01 | 0.72622 | 2.01 | 0 | miRNAWalker2 validate | -1.66 | 0.011 | ||
| 31 | hsa-miR-192-5p | CDKN3 | 0.01 | 0.72622 | 1.99 | 0 | miRNAWalker2 validate | -1.51 | 0.01409 | ||
| 32 | hsa-miR-192-5p | CENPA | 0.01 | 0.72622 | 1.91 | 0 | miRNAWalker2 validate | -1.82 | 0.00402 | ||
| 33 | hsa-miR-192-5p | CENPE | 0.01 | 0.72622 | 1.59 | 0 | miRNAWalker2 validate | -2.03 | 0.00015 | ||
| 34 | hsa-miR-192-5p | CENPF | 0.01 | 0.72622 | 1.92 | 0 | miRNAWalker2 validate | -1.9 | 0.00192 | ||
| 35 | hsa-miR-192-5p | DTL | 0.01 | 0.72622 | 1.64 | 0 | miRNAWalker2 validate | -1.34 | 0.01789 | ||
| 36 | hsa-miR-192-5p | E2F8 | 0.01 | 0.72622 | 1.83 | 0 | miRNAWalker2 validate | -1.16 | 0.04535 | ||
| 37 | hsa-miR-192-5p | KIF20A | 0.01 | 0.72622 | 1.86 | 0 | miRNAWalker2 validate | -1.83 | 0.00359 | ||
| 38 | hsa-miR-192-5p | MKI67 | 0.01 | 0.72622 | 1.67 | 0 | miRNAWalker2 validate; mirMAP | -2.04 | 0.00041 | ||
| 39 | hsa-miR-192-5p | NUF2 | 0.01 | 0.72622 | 1.95 | 0 | miRNAWalker2 validate | -2.03 | 0.00155 | ||
| 40 | hsa-miR-192-5p | PAK6 | 0.01 | 0.72622 | 1.6 | 0 | miRNAWalker2 validate | -1.72 | 0.00123 | ||
| 41 | hsa-miR-192-5p | RAB3B | 0.01 | 0.72622 | 1.83 | 0 | miRNAWalker2 validate; mirMAP | -2.28 | 0.00384 | ||
| 42 | hsa-miR-192-5p | TTK | 0.01 | 0.72622 | 1.73 | 0 | miRNAWalker2 validate | -2.05 | 0.00054 | ||
| 43 | hsa-miR-21-5p | LIFR | 0.23 | 0.00018 | -1.71 | 0 | miRNAWalker2 validate | -1.81 | 0 | ||
| 44 | hsa-miR-215-5p | CENPE | 0.04 | 0.21421 | 1.59 | 0 | miRNAWalker2 validate | -1.52 | 0.00355 | ||
| 45 | hsa-miR-215-5p | CENPF | 0.04 | 0.21421 | 1.92 | 0 | miRNAWalker2 validate | -1.44 | 0.01556 | ||
| 46 | hsa-miR-215-5p | ID1 | 0.04 | 0.21421 | -1.66 | 0 | miRNAWalker2 validate | -1.32 | 0.01002 | ||
| 47 | hsa-miR-215-5p | KIF20A | 0.04 | 0.21421 | 1.86 | 0 | miRNAWalker2 validate | -1.26 | 0.03958 | ||
| 48 | hsa-miR-215-5p | MKI67 | 0.04 | 0.21421 | 1.67 | 0 | miRNAWalker2 validate | -1.46 | 0.00933 | ||
| 49 | hsa-miR-215-5p | NUF2 | 0.04 | 0.21421 | 1.95 | 0 | miRNAWalker2 validate | -1.46 | 0.01959 | ||
| 50 | hsa-miR-215-5p | PAK6 | 0.04 | 0.21421 | 1.6 | 0 | miRNAWalker2 validate | -1.25 | 0.01669 | ||
| 51 | hsa-miR-215-5p | RAB3B | 0.04 | 0.21421 | 1.83 | 0 | miRNAWalker2 validate | -1.57 | 0.04108 | ||
| 52 | hsa-miR-215-5p | TTK | 0.04 | 0.21421 | 1.73 | 0 | miRNAWalker2 validate | -1.51 | 0.00889 | ||
| 53 | hsa-miR-23a-3p | KIF20A | -0.02 | 0.18092 | 1.86 | 0 | miRNAWalker2 validate | -4.7 | 0.00011 | ||
| 54 | hsa-miR-23b-3p | CCNB2 | 0.01 | 0.57764 | 1.87 | 0 | miRNAWalker2 validate | -3.74 | 0.00154 | ||
| 55 | hsa-miR-23b-3p | CDCA8 | 0.01 | 0.57764 | 1.9 | 0 | miRNAWalker2 validate | -2.69 | 0.01297 | ||
| 56 | hsa-miR-24-3p | BEX1 | -0.01 | 0.59303 | -1.68 | 0 | miRNAWalker2 validate | -3.13 | 0.02097 | ||
| 57 | hsa-miR-24-3p | CCNB1 | -0.01 | 0.59303 | 1.59 | 0 | miRNAWalker2 validate | -3.31 | 0.00049 | ||
| 58 | hsa-miR-24-3p | DTL | -0.01 | 0.59303 | 1.64 | 0 | miRNAWalker2 validate | -2.21 | 0.02677 | ||
| 59 | hsa-miR-24-3p | RRM2 | -0.01 | 0.59303 | 1.6 | 0 | miRNAWalker2 validate | -3.08 | 0.00198 | ||
| 60 | hsa-miR-24-3p | UBE2C | -0.01 | 0.59303 | 1.89 | 0 | miRNAWalker2 validate | -3.91 | 0.0003 | ||
| 61 | hsa-miR-26a-5p | RRM2 | -0.08 | 7.0E-5 | 1.6 | 0 | miRNAWalker2 validate | -5.75 | 0 | ||
| 62 | hsa-miR-26b-5p | ASPM | -0.05 | 0.04928 | 2.01 | 0 | miRNAWalker2 validate | -5.79 | 0 | ||
| 63 | hsa-miR-26b-5p | DTL | -0.05 | 0.04928 | 1.64 | 0 | miRNAWalker2 validate | -4.55 | 0 | ||
| 64 | hsa-miR-26b-5p | FOXM1 | -0.05 | 0.04928 | 1.79 | 0 | miRNAWalker2 validate | -5.76 | 0 | ||
| 65 | hsa-miR-26b-5p | PAK6 | -0.05 | 0.04928 | 1.6 | 0 | miRNAWalker2 validate | -5.13 | 0 | ||
| 66 | hsa-miR-26b-5p | SPC25 | -0.05 | 0.04928 | 1.93 | 0 | miRNAWalker2 validate | -5.75 | 0 | ||
| 67 | hsa-miR-30a-5p | DTL | -0.1 | 0.00105 | 1.64 | 0 | miRNAWalker2 validate; miRTarBase | -2.81 | 0 | 22287560 | Combining global gene expression analyses of miR-30a-5p transgenic line HCT116 with in silico miRNA target prediction we identified the denticleless protein homolog DTL as a potential miRNA-30a-5p target |
| 68 | hsa-miR-30a-5p | RRM2 | -0.1 | 0.00105 | 1.6 | 0 | miRNAWalker2 validate | -2.62 | 0 | ||
| 69 | hsa-miR-30c-5p | BIRC5 | -0.05 | 0.0598 | 1.89 | 0 | miRNAWalker2 validate | -5.67 | 0 | ||
| 70 | hsa-miR-30c-5p | CELSR3 | -0.05 | 0.0598 | 1.59 | 0 | miRNAWalker2 validate; MirTarget; miRNATAP | -3.83 | 0 | ||
| 71 | hsa-miR-30c-5p | RRM2 | -0.05 | 0.0598 | 1.6 | 0 | miRNAWalker2 validate | -5.2 | 0 | ||
| 72 | hsa-miR-30e-5p | EXO1 | -0.03 | 0.2867 | 1.64 | 0 | miRNAWalker2 validate | -3.08 | 0 | ||
| 73 | hsa-miR-324-5p | APOF | 0.23 | 0 | -1.83 | 0 | miRNAWalker2 validate | -5.34 | 0 | ||
| 74 | hsa-miR-335-5p | CCL19 | 0.02 | 0.79934 | -1.9 | 0 | miRNAWalker2 validate | -0.72 | 0.02147 | ||
| 75 | hsa-miR-335-5p | NGFR | 0.02 | 0.79934 | -1.59 | 0 | miRNAWalker2 validate | -0.65 | 0.00518 | ||
| 76 | hsa-miR-375 | CENPF | -0.55 | 0 | 1.92 | 0 | miRNAWalker2 validate | -1.22 | 0 | ||
| 77 | hsa-miR-375 | EXO1 | -0.55 | 0 | 1.64 | 0 | miRNAWalker2 validate | -1.09 | 0 | ||
| 78 | hsa-miR-375 | RAB3B | -0.55 | 0 | 1.83 | 0 | miRNAWalker2 validate | -0.56 | 0.02581 | ||
| 79 | hsa-miR-424-5p | ANLN | -0.16 | 0 | 2.11 | 0 | miRNAWalker2 validate; miRTarBase | -5.97 | 0 | ||
| 80 | hsa-miR-9-5p | CCL19 | 0.21 | 0.00475 | -1.9 | 0 | miRNAWalker2 validate | -1.05 | 0.00113 | ||
| 81 | hsa-miR-9-5p | CYP39A1 | 0.21 | 0.00475 | -1.72 | 0 | miRNAWalker2 validate | -1.02 | 0.00037 | ||
| 82 | hsa-miR-98-5p | LIFR | 0.11 | 0.00295 | -1.71 | 0 | miRNAWalker2 validate | -1.72 | 0.00018 | ||
| 83 | hsa-miR-221-3p | DIRAS3 | 0.19 | 0 | -1.7 | 0 | miRTarBase | -4.22 | 0 | 22117988; 22117988; 22117988; 22117988; 23801152; 23801152; 23801152; 23801152; 21071579; 21071579; 21071579 | The expression and clinical significance of GTP-binding RAS-like 3 ARHI and microRNA 221 and 222 in prostate cancer;MicroRNA 221 and 222 have been shown to regulate ARHI expression negatively;Expression of ARHI and microRNA 221 and 222 was measured by real-time reverse transcription-polymerase chain reaction;Whether ARHI and microRNA 221 and 222 could be considered as biomarkers for disease progression in prostate cancer requires further investigation;The downregulation of ARHI was regulated by miR-221 in prostate cancer cell lines;The inhibition of miR-221 induced a significant upregulation of ARHI in MCF-7 cells;To prove a direct interaction between miR-221 and ARHI mRNA ARHI 3'UTR which includes the potential target site for miR-221 was cloned downstream of the luciferase reporter gene of the pMIR-REPORT vector to generate the pMIR-ARHI-3'UTR vector;This study demonstrated for the first time that the downregulation of ARHI in breast cancer cells could be regulated by miR-221;Further studies on a new mechanism of ARHI downregulation showed a significant inverse relationship between ARHI and miR-221 and 222 which were upregulated in prostate cancer cell lines;Transfection of miR-221 and 222 inhibitors into PC-3 cells caused a significant induction of ARHI expression;Finally we also found that genistein upregulates ARHI by downregulating miR-221 and 222 in PC-3 cells |
| 84 | hsa-miR-222-3p | DIRAS3 | 0.15 | 3.0E-5 | -1.7 | 0 | miRTarBase | -4.33 | 0 | ||
| 85 | hsa-miR-218-5p | BIRC5 | -0.14 | 0.00268 | 1.89 | 0 | miRTarBase; MirTarget | -1.51 | 0.00025 | ||
| 86 | hsa-miR-203a-3p | BIRC5 | -0.27 | 3.0E-5 | 1.89 | 0 | miRTarBase | -1.62 | 0 | ||
| 87 | hsa-miR-21-5p | NTF3 | 0.23 | 0.00018 | -1.58 | 0 | miRTarBase; miRNATAP | -1.78 | 0 | ||
| 88 | hsa-miR-103a-3p | LRRN3 | 0.11 | 0 | -1.69 | 0 | MirTarget; miRNATAP | -4.41 | 0 | ||
| 89 | hsa-miR-105-5p | CLDN10 | 0.17 | 0.00159 | -2.25 | 0 | MirTarget | -1.51 | 0.00044 | ||
| 90 | hsa-miR-106a-5p | LRRC55 | 0.05 | 0.12421 | -1.67 | 0 | MirTarget | -1.81 | 0.00165 | ||
| 91 | hsa-miR-106a-5p | KCNJ10 | 0.05 | 0.12421 | -1.63 | 0 | MirTarget; miRNATAP | -1.31 | 0.01777 | ||
| 92 | hsa-miR-106b-5p | LRRC55 | 0.17 | 0 | -1.67 | 0 | MirTarget | -5.56 | 0 | ||
| 93 | hsa-miR-106b-5p | KCNJ10 | 0.17 | 0 | -1.63 | 0 | MirTarget; miRNATAP | -5 | 0 | ||
| 94 | hsa-miR-107 | LRRN3 | 0.13 | 0 | -1.69 | 0 | MirTarget; PITA; miRanda; miRNATAP | -2.98 | 0 | ||
| 95 | hsa-miR-126-5p | MELK | 0 | 0.93995 | 1.86 | 0 | MirTarget | -3.22 | 8.0E-5 | ||
| 96 | hsa-miR-128-3p | LRAT | 0.08 | 0.0018 | -2.15 | 0 | MirTarget | -4.55 | 0 | ||
| 97 | hsa-miR-128-3p | CLRN3 | 0.08 | 0.0018 | -1.93 | 0 | MirTarget | -3.78 | 0.00027 | ||
| 98 | hsa-miR-135a-5p | BMPER | 0.26 | 0.01554 | -2.99 | 0 | MirTarget; miRNATAP | -1.43 | 0 | ||
| 99 | hsa-miR-135a-5p | ST6GAL2 | 0.26 | 0.01554 | -2.01 | 0 | MirTarget; miRNATAP | -0.84 | 5.0E-5 | ||
| 100 | hsa-miR-140-5p | ST6GAL2 | 0.04 | 0.1479 | -2.01 | 0 | MirTarget; miRanda | -2.44 | 0.00178 | ||
| 101 | hsa-miR-142-5p | CCNB1 | -0.12 | 0.00783 | 1.59 | 0 | MirTarget | -1.38 | 0.0003 | ||
| 102 | hsa-miR-144-3p | E2F8 | -0.11 | 0.02244 | 1.83 | 0 | MirTarget; miRNATAP | -1.82 | 0 | ||
| 103 | hsa-miR-147a | SYT9 | 0.09 | 0.00179 | -1.93 | 0 | MirTarget | -2.3 | 0.00066 | ||
| 104 | hsa-miR-147a | CXCL14 | 0.09 | 0.00179 | -2.84 | 0 | MirTarget | -4.74 | 0 | ||
| 105 | hsa-miR-150-5p | NEK2 | -0.18 | 0.00075 | 2.04 | 0 | MirTarget | -2.19 | 0 | ||
| 106 | hsa-miR-15a-5p | LRRN3 | 0.01 | 0.57141 | -1.69 | 0 | MirTarget; miRNATAP | -1.87 | 0.00306 | ||
| 107 | hsa-miR-15a-5p | RELN | 0.01 | 0.57141 | -1.76 | 0 | MirTarget; miRNATAP | -2.6 | 0.00634 | ||
| 108 | hsa-miR-15a-5p | RSPO3 | 0.01 | 0.57141 | -2.56 | 0 | MirTarget | -4.07 | 6.0E-5 | ||
| 109 | hsa-miR-15b-5p | LRRN3 | 0.17 | 0 | -1.69 | 0 | MirTarget; miRNATAP | -3.92 | 0 | ||
| 110 | hsa-miR-15b-5p | RELN | 0.17 | 0 | -1.76 | 0 | MirTarget; miRNATAP | -6.6 | 0 | ||
| 111 | hsa-miR-15b-5p | RSPO3 | 0.17 | 0 | -2.56 | 0 | MirTarget | -7.32 | 0 | ||
| 112 | hsa-miR-16-5p | LRRN3 | 0.05 | 0.02313 | -1.69 | 0 | MirTarget; miRNATAP | -3.18 | 5.0E-5 | ||
| 113 | hsa-miR-16-5p | RELN | 0.05 | 0.02313 | -1.76 | 0 | MirTarget; miRNATAP | -6.34 | 0 | ||
| 114 | hsa-miR-16-5p | RSPO3 | 0.05 | 0.02313 | -2.56 | 0 | MirTarget | -6.2 | 0 | ||
| 115 | hsa-miR-17-3p | DPT | 0.01 | 0.63304 | -2.5 | 0 | MirTarget | -5.09 | 0.00054 | ||
| 116 | hsa-miR-17-5p | LRRC55 | 0.06 | 0.05778 | -1.67 | 0 | MirTarget; TargetScan | -2.35 | 0.00014 | ||
| 117 | hsa-miR-17-5p | KCNJ10 | 0.06 | 0.05778 | -1.63 | 0 | MirTarget; TargetScan; miRNATAP | -2.13 | 0.00031 | ||
| 118 | hsa-miR-181b-5p | TMEM27 | 0.04 | 0.18111 | -1.83 | 0 | MirTarget | -1.92 | 0.01189 | ||
| 119 | hsa-miR-181c-5p | ESM1 | -0.04 | 0.24874 | 2.3 | 0 | MirTarget; miRNATAP | -1.41 | 0.04857 | ||
| 120 | hsa-miR-182-5p | BMPER | 0.14 | 0.00166 | -2.99 | 0 | MirTarget | -3.49 | 0 | ||
| 121 | hsa-miR-182-5p | LRRC55 | 0.14 | 0.00166 | -1.67 | 0 | MirTarget | -2.09 | 0 | ||
| 122 | hsa-miR-185-5p | CLEC4M | 0.11 | 0 | -3.6 | 0 | MirTarget | -9.02 | 0 | ||
| 123 | hsa-miR-185-5p | ADH4 | 0.11 | 0 | -1.78 | 0 | MirTarget | -4.28 | 0.00025 | ||
| 124 | hsa-miR-185-5p | CFTR | 0.11 | 0 | -1.68 | 0 | MirTarget | -5.94 | 0 | ||
| 125 | hsa-miR-185-5p | KCNJ10 | 0.11 | 0 | -1.63 | 0 | MirTarget | -4.14 | 0 | ||
| 126 | hsa-miR-186-5p | COL6A6 | 0.12 | 0 | -1.8 | 0 | MirTarget | -3.66 | 0 | ||
| 127 | hsa-miR-188-5p | RSPO3 | 0.23 | 0 | -2.56 | 0 | MirTarget; PITA | -7.86 | 0 | ||
| 128 | hsa-miR-18a-5p | FCGR2B | 0.2 | 7.0E-5 | -1.93 | 0 | MirTarget | -2.31 | 0 | ||
| 129 | hsa-miR-194-5p | DUSP9 | 0.03 | 0.38268 | 1.59 | 0 | MirTarget; miRNATAP | -3.32 | 0 | ||
| 130 | hsa-miR-196b-5p | SYT9 | 0.35 | 0 | -1.93 | 0 | MirTarget | -1.6 | 0 | ||
| 131 | hsa-miR-199a-3p | PRC1 | -0.47 | 0 | 1.59 | 0 | MirTarget | -1.81 | 0 | ||
| 132 | hsa-miR-19a-3p | RSPO3 | 0.02 | 0.47729 | -2.56 | 0 | MirTarget | -2.79 | 0.00161 | ||
| 133 | hsa-miR-200a-3p | CDCA2 | -0.48 | 0 | 1.62 | 0 | MirTarget | -1.01 | 0 | ||
| 134 | hsa-miR-200a-3p | DTL | -0.48 | 0 | 1.64 | 0 | MirTarget | -1.08 | 0 | ||
| 135 | hsa-miR-200b-3p | ESM1 | -0.53 | 0 | 2.3 | 0 | MirTarget | -1.66 | 0 | ||
| 136 | hsa-miR-200b-3p | MAGEC2 | -0.53 | 0 | 1.62 | 0 | MirTarget | -1.57 | 0 | ||
| 137 | hsa-miR-200b-3p | ANLN | -0.53 | 0 | 2.11 | 0 | MirTarget; TargetScan | -1.4 | 0 | ||
| 138 | hsa-miR-202-3p | SULT1C2 | -0.19 | 0 | 1.58 | 1.0E-5 | MirTarget | -2.97 | 0 | ||
| 139 | hsa-miR-203a-3p | PRC1 | -0.27 | 3.0E-5 | 1.59 | 0 | MirTarget | -1.51 | 0 | ||
| 140 | hsa-miR-203a-3p | SPC25 | -0.27 | 3.0E-5 | 1.93 | 0 | MirTarget | -1.8 | 0 | ||
| 141 | hsa-miR-204-5p | GPC3 | 0.01 | 0.79023 | 1.83 | 0 | MirTarget | -0.95 | 0.02315 | ||
| 142 | hsa-miR-204-5p | CELSR3 | 0.01 | 0.79023 | 1.59 | 0 | MirTarget; miRNATAP | -0.74 | 0.01278 | ||
| 143 | hsa-miR-206 | RSPO3 | 0.03 | 0.13251 | -2.56 | 0 | MirTarget; PITA; miRanda | -4.73 | 0.00013 | ||
| 144 | hsa-miR-206 | INMT | 0.03 | 0.13251 | -1.68 | 0 | MirTarget; miRanda | -4.66 | 0 | ||
| 145 | hsa-miR-21-5p | LPA | 0.23 | 0.00018 | -1.67 | 0 | MirTarget | -2.1 | 0 | ||
| 146 | hsa-miR-211-5p | DBH | 0.09 | 0.01154 | -2.08 | 0 | MirTarget | -1.82 | 0.00218 | ||
| 147 | hsa-miR-212-3p | BMPER | 0.04 | 0.03172 | -2.99 | 0 | MirTarget | -4.89 | 0.00336 | ||
| 148 | hsa-miR-217 | COL6A6 | 0.09 | 0.11729 | -1.8 | 0 | MirTarget; miRanda | -1.23 | 0.00012 | ||
| 149 | hsa-miR-224-5p | NUDT10 | 0.35 | 6.0E-5 | -1.77 | 0 | MirTarget | -1.19 | 0 | ||
| 150 | hsa-miR-224-5p | RELN | 0.35 | 6.0E-5 | -1.76 | 0 | MirTarget | -1.65 | 0 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | MITOTIC CELL CYCLE | 29 | 766 | 2.272e-16 | 1.057e-12 |
| 2 | CELL CYCLE | 33 | 1316 | 1.892e-13 | 3.656e-10 |
| 3 | CELL CYCLE PROCESS | 30 | 1081 | 2.357e-13 | 3.656e-10 |
| 4 | SISTER CHROMATID SEGREGATION | 14 | 176 | 1.443e-12 | 1.678e-09 |
| 5 | CHROMOSOME SEGREGATION | 16 | 272 | 3.451e-12 | 3.212e-09 |
| 6 | CELL DIVISION | 19 | 460 | 1.397e-11 | 1.084e-08 |
| 7 | MITOTIC NUCLEAR DIVISION | 17 | 361 | 2.387e-11 | 1.587e-08 |
| 8 | ORGANELLE FISSION | 19 | 496 | 5.095e-11 | 2.634e-08 |
| 9 | NUCLEAR CHROMOSOME SEGREGATION | 14 | 228 | 4.793e-11 | 2.634e-08 |
| 10 | SISTER CHROMATID COHESION | 9 | 111 | 1.434e-08 | 6.674e-06 |
| 11 | CELL CYCLE PHASE TRANSITION | 12 | 255 | 2.491e-08 | 1.054e-05 |
| 12 | REGULATION OF CHROMOSOME SEGREGATION | 8 | 85 | 2.904e-08 | 1.126e-05 |
| 13 | REGULATION OF NUCLEAR DIVISION | 10 | 163 | 3.221e-08 | 1.153e-05 |
| 14 | REGULATION OF CELL DIVISION | 12 | 272 | 5.069e-08 | 1.685e-05 |
| 15 | REGULATION OF SISTER CHROMATID SEGREGATION | 7 | 67 | 1.065e-07 | 3.302e-05 |
| 16 | MITOTIC SISTER CHROMATID SEGREGATION | 7 | 91 | 8.855e-07 | 0.0002575 |
| 17 | CHROMOSOME LOCALIZATION | 6 | 61 | 1.292e-06 | 0.000334 |
| 18 | CELL CYCLE G2 M PHASE TRANSITION | 8 | 138 | 1.247e-06 | 0.000334 |
| 19 | REGULATION OF EXIT FROM MITOSIS | 4 | 16 | 1.728e-06 | 0.0004021 |
| 20 | REGULATION OF CELL CYCLE | 19 | 949 | 1.665e-06 | 0.0004021 |
| 21 | MITOTIC SPINDLE ORGANIZATION | 6 | 69 | 2.688e-06 | 0.0005956 |
| 22 | METAPHASE PLATE CONGRESSION | 5 | 42 | 3.96e-06 | 0.0008375 |
| 23 | SPINDLE CHECKPOINT | 4 | 25 | 1.155e-05 | 0.002336 |
| 24 | NEGATIVE REGULATION OF CELL DIVISION | 5 | 60 | 2.343e-05 | 0.004543 |
| 25 | CHROMOSOME CONDENSATION | 4 | 31 | 2.797e-05 | 0.005006 |
| 26 | HYALURONAN METABOLIC PROCESS | 4 | 31 | 2.797e-05 | 0.005006 |
| 27 | REGULATION OF CELL PROLIFERATION | 22 | 1496 | 3.287e-05 | 0.005664 |
| 28 | REGULATION OF ORGANELLE ORGANIZATION | 19 | 1178 | 3.586e-05 | 0.005959 |
| 29 | KINETOCHORE ORGANIZATION | 3 | 12 | 3.822e-05 | 0.006133 |
| 30 | AMINOGLYCAN CATABOLIC PROCESS | 5 | 68 | 4.313e-05 | 0.00669 |
| 31 | NEGATIVE REGULATION OF PROTEIN COMPLEX DISASSEMBLY | 7 | 170 | 5.433e-05 | 0.008155 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CONDENSED CHROMOSOME | 13 | 195 | 8.822e-11 | 5.152e-08 |
| 2 | CHROMOSOME CENTROMERIC REGION | 12 | 174 | 3.286e-10 | 9.596e-08 |
| 3 | CONDENSED CHROMOSOME CENTROMERIC REGION | 9 | 102 | 6.785e-09 | 1.321e-06 |
| 4 | KINETOCHORE | 9 | 120 | 2.844e-08 | 4.153e-06 |
| 5 | CHROMOSOMAL REGION | 12 | 330 | 4.091e-07 | 4.657e-05 |
| 6 | CONDENSED CHROMOSOME OUTER KINETOCHORE | 4 | 12 | 4.784e-07 | 4.657e-05 |
| 7 | MIDBODY | 8 | 132 | 8.903e-07 | 7.427e-05 |
| 8 | SPINDLE | 10 | 289 | 6.159e-06 | 0.0004496 |
| 9 | CHROMOSOME | 17 | 880 | 1e-05 | 0.000649 |
| 10 | MICROTUBULE CYTOSKELETON | 17 | 1068 | 0.0001137 | 0.006642 |
| 11 | CONDENSED NUCLEAR CHROMOSOME CENTROMERIC REGION | 3 | 18 | 0.0001383 | 0.007341 |
| 12 | EXTRACELLULAR MATRIX | 10 | 426 | 0.0001639 | 0.007979 |
| 13 | PROTEINACEOUS EXTRACELLULAR MATRIX | 9 | 356 | 0.0002036 | 0.009149 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
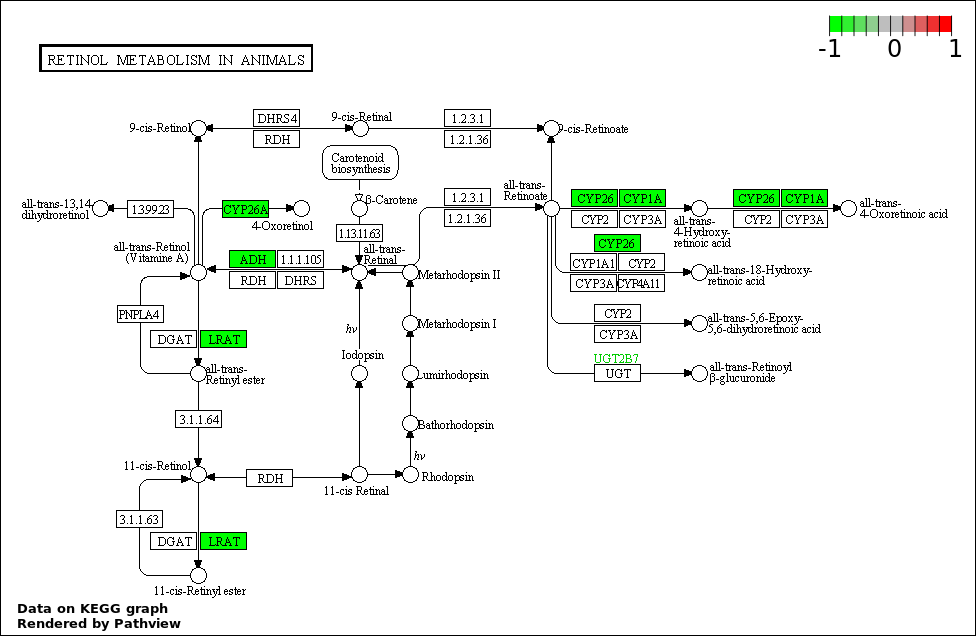
| 1 | hsa00830_Retinol_metabolism | 4 | 387 | 0.1798 | 1 | |
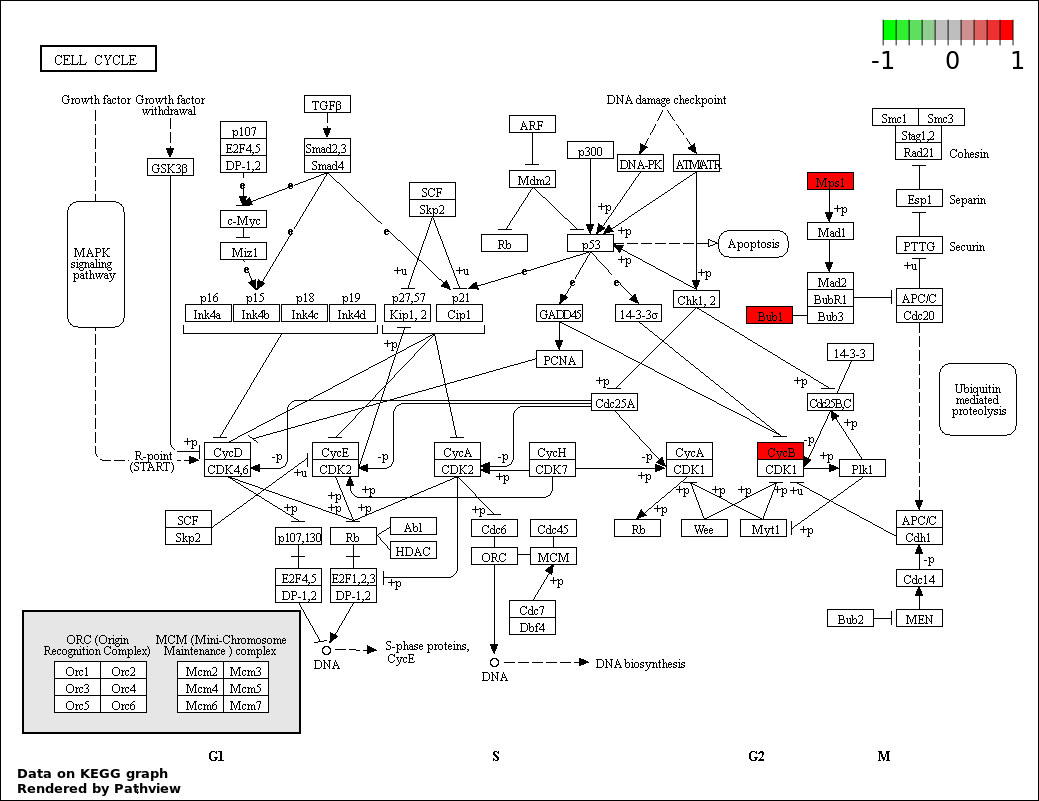
| 2 | hsa04110_Cell_cycle | 4 | 387 | 0.1798 | 1 | |
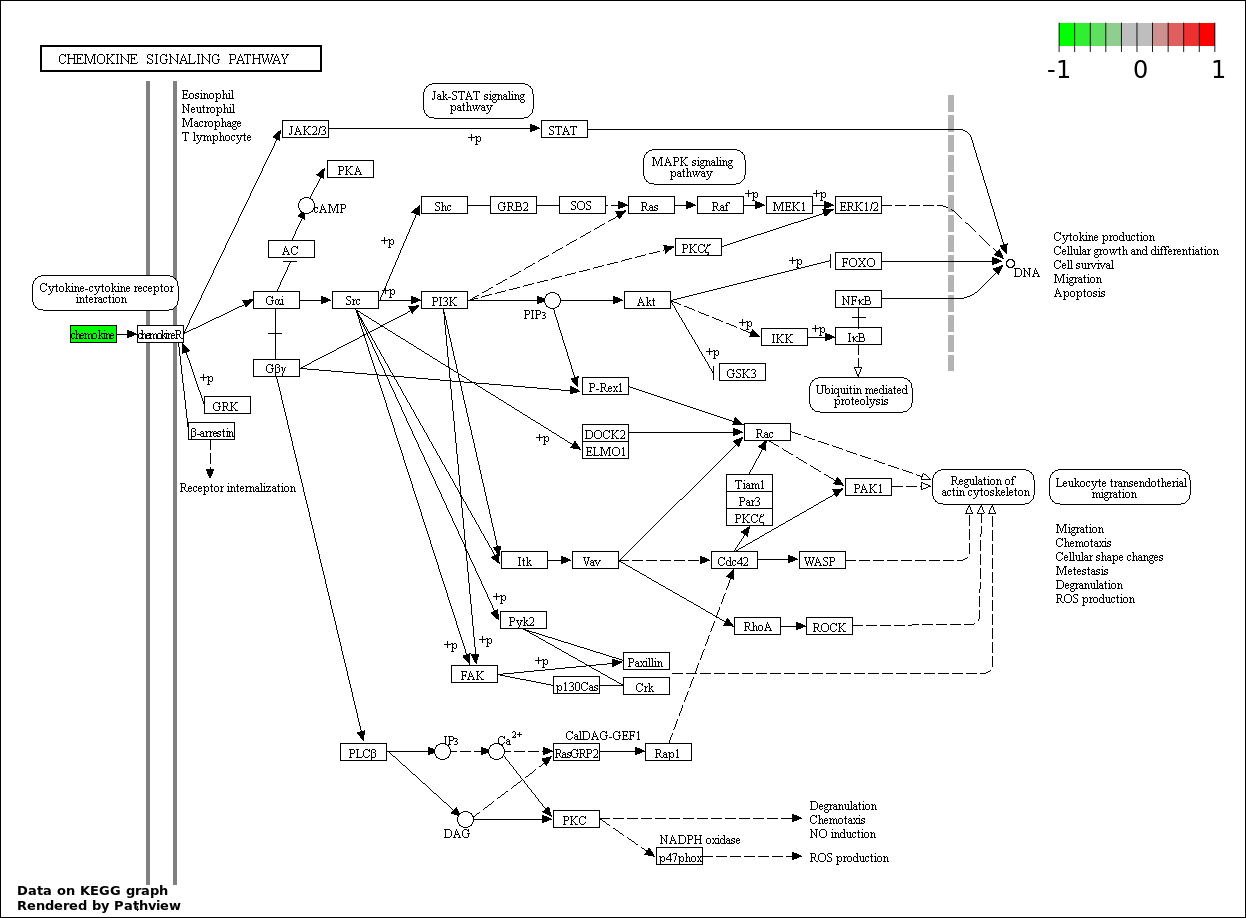
| 3 | hsa04062_Chemokine_signaling_pathway | 3 | 387 | 0.3795 | 1 | |
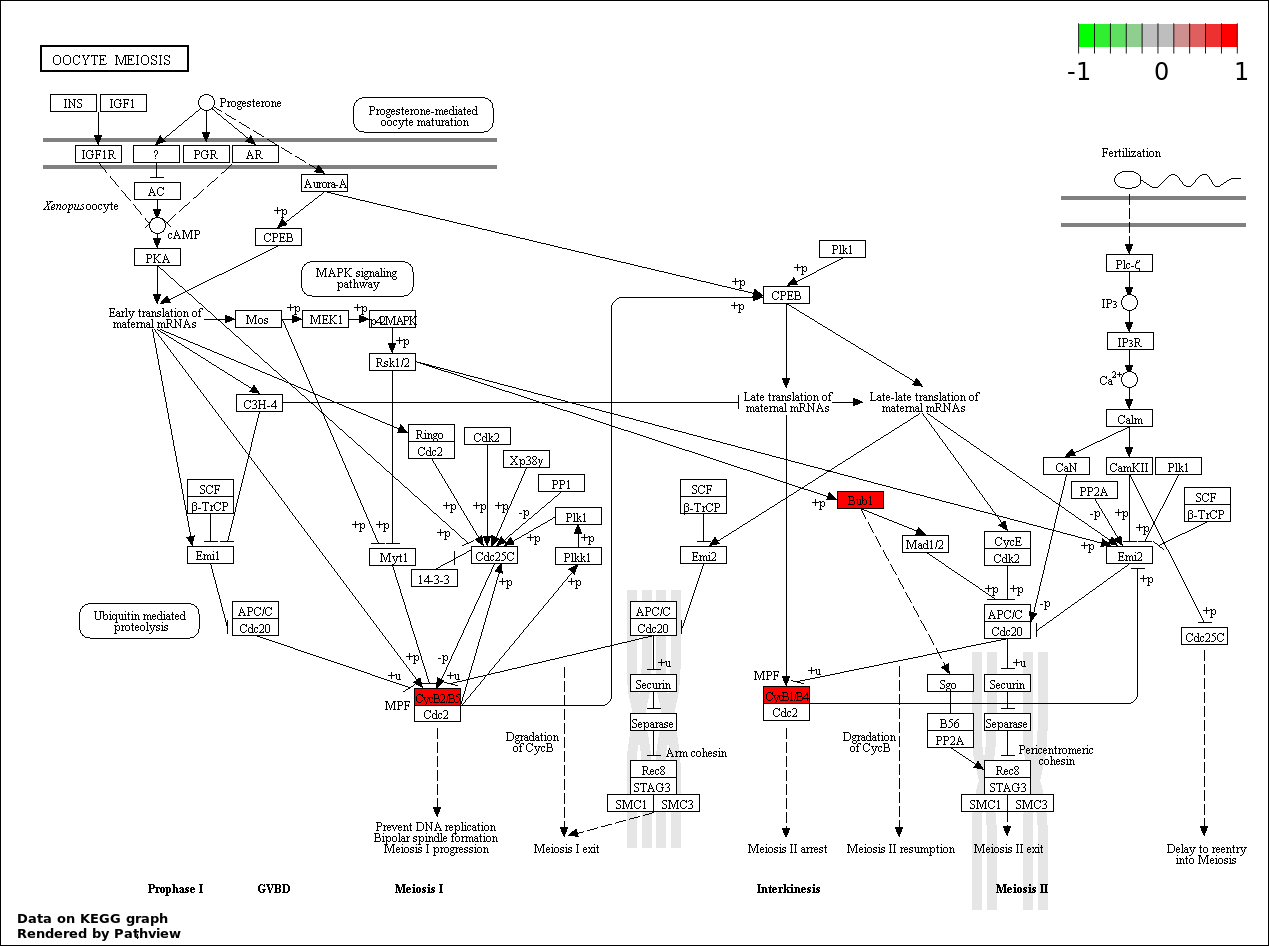
| 4 | hsa04114_Oocyte_meiosis | 3 | 387 | 0.3795 | 1 | |
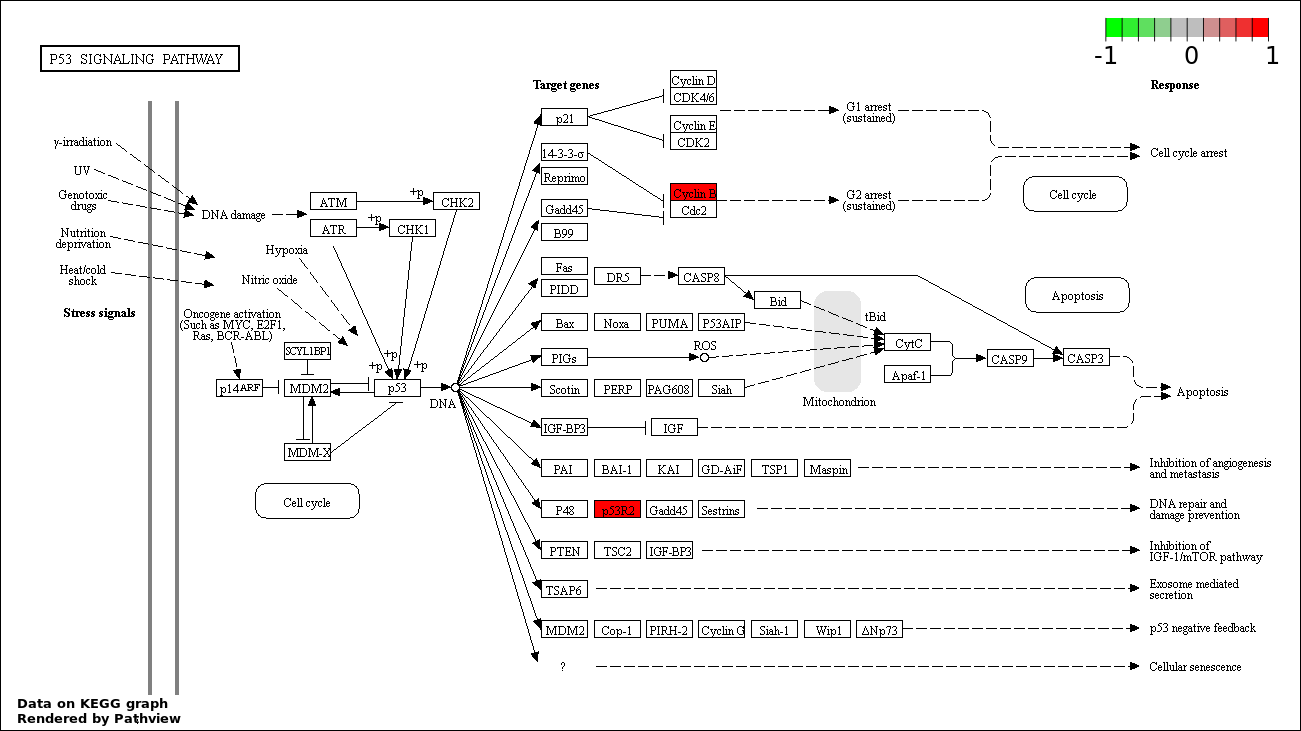
| 5 | hsa04115_p53_signaling_pathway | 3 | 387 | 0.3795 | 1 | |
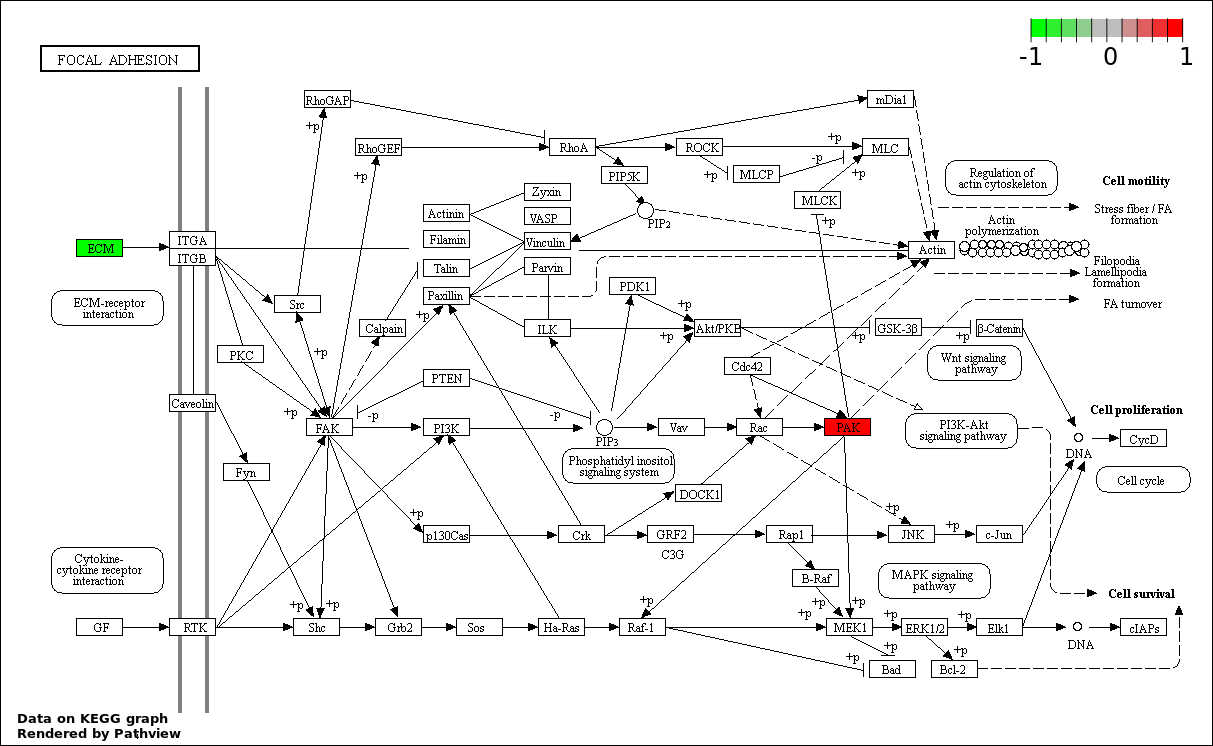
| 6 | hsa04510_Focal_adhesion | 3 | 387 | 0.3795 | 1 | |
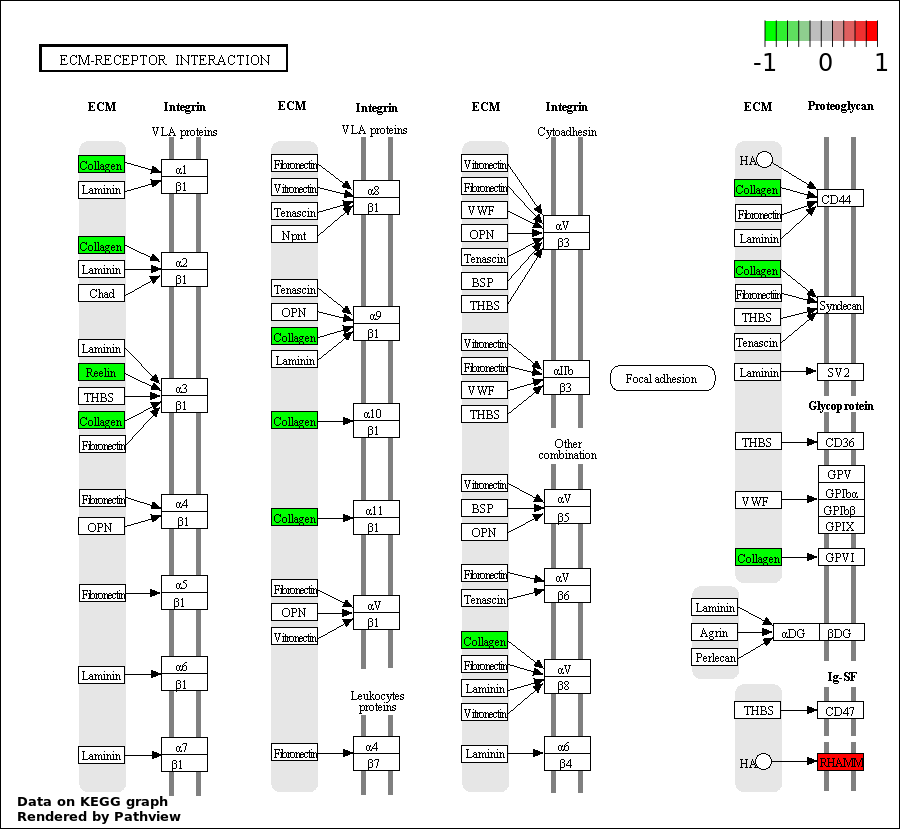
| 7 | hsa04512_ECM.receptor_interaction | 3 | 387 | 0.3795 | 1 | |
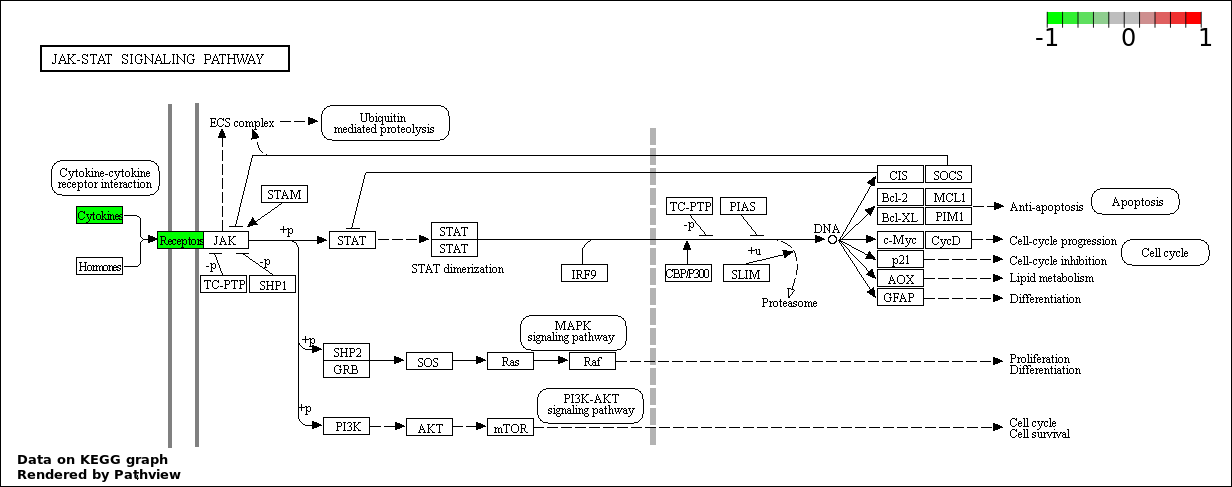
| 8 | hsa04630_Jak.STAT_signaling_pathway | 3 | 387 | 0.3795 | 1 | |
| 9 | hsa04914_Progesterone.mediated_oocyte_maturation | 3 | 387 | 0.3795 | 1 | |
| 10 | hsa04976_Bile_secretion | 3 | 387 | 0.3795 | 1 | |
| 11 | hsa00340_Histidine_metabolism | 2 | 387 | 0.6506 | 1 | |
| 12 | hsa00350_Tyrosine_metabolism | 2 | 387 | 0.6506 | 1 | |
| 13 | hsa00380_Tryptophan_metabolism | 2 | 387 | 0.6506 | 1 | |
| 14 | hsa00980_Metabolism_of_xenobiotics_by_cytochrome_P450 | 2 | 387 | 0.6506 | 1 | |
| 15 | hsa00982_Drug_metabolism_._cytochrome_P450 | 2 | 387 | 0.6506 | 1 | |
| 16 | hsa04010_MAPK_signaling_pathway | 2 | 387 | 0.6506 | 1 | |
| 17 | hsa04145_Phagosome | 2 | 387 | 0.6506 | 1 | |
| 18 | hsa04350_TGF.beta_signaling_pathway | 2 | 387 | 0.6506 | 1 | |
| 19 | hsa04530_Tight_junction | 2 | 387 | 0.6506 | 1 | |
| 20 | hsa04722_Neurotrophin_signaling_pathway | 2 | 387 | 0.6506 | 1 | |
| 21 | hsa04971_Gastric_acid_secretion | 2 | 387 | 0.6506 | 1 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | KCNQ1OT1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7e-5p;hsa-miR-100-5p;hsa-miR-133b;hsa-miR-194-5p;hsa-miR-202-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-99a-5p | 10 | DUSP9 | Sponge network | 0.459 | 0.00259 | 1.595 | 0 | 0.658 |
| 2 | FAM99A | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-21-5p | 10 | LRRC55 | Sponge network | -1.52 | 0 | -1.671 | 0 | 0.527 |
| 3 | GAS5 | hsa-miR-130a-3p;hsa-miR-146a-5p;hsa-miR-192-5p;hsa-miR-199a-3p;hsa-miR-204-5p;hsa-miR-214-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-373-5p | 13 | RAB3B | Sponge network | 0.239 | 0.00543 | 1.829 | 0 | 0.517 |
| 4 | SNHG6 | hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-133b;hsa-miR-192-5p;hsa-miR-199a-3p;hsa-miR-214-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-5p | 10 | RAB3B | Sponge network | 0.28 | 0.00038 | 1.829 | 0 | 0.474 |
| 5 | KCNQ1OT1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7e-5p;hsa-miR-100-5p;hsa-miR-125a-5p;hsa-miR-199a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-373-5p | 12 | RRM2 | Sponge network | 0.459 | 0.00259 | 1.604 | 0 | 0.467 |
| 6 | KTN1-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7e-5p;hsa-miR-100-5p;hsa-miR-133b;hsa-miR-194-5p;hsa-miR-202-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-99a-5p | 10 | DUSP9 | Sponge network | 0.382 | 0.00019 | 1.595 | 0 | 0.462 |
| 7 | KCNQ1OT1 |
hsa-miR-126-5p;hsa-miR-133b;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-192-5p;hsa-miR-199a-3p;hsa-miR-214-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-373-5p;hsa-miR-375 | 18 | RAB3B | Sponge network | 0.459 | 0.00259 | 1.829 | 0 | 0.416 |
| 8 | HCG18 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7e-5p;hsa-miR-100-5p;hsa-miR-125a-5p;hsa-miR-199a-5p;hsa-miR-26a-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-373-5p | 10 | RRM2 | Sponge network | 0.22 | 0.00721 | 1.604 | 0 | 0.41 |
| 9 | KTN1-AS1 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-133b;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-192-5p;hsa-miR-199a-3p;hsa-miR-214-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-373-5p | 14 | RAB3B | Sponge network | 0.382 | 0.00019 | 1.829 | 0 | 0.396 |
| 10 | MYCNOS | hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7e-5p;hsa-miR-125a-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-30a-5p;hsa-miR-30c-5p | 11 | RRM2 | Sponge network | 0.192 | 0.0041 | 1.604 | 0 | 0.384 |
| 11 | HCG18 |
hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-192-5p;hsa-miR-199a-3p;hsa-miR-204-5p;hsa-miR-214-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-373-5p;hsa-miR-375 | 17 | RAB3B | Sponge network | 0.22 | 0.00721 | 1.829 | 0 | 0.377 |
| 12 | SNHG5 | hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-34b-5p;hsa-miR-449a;hsa-miR-93-5p | 10 | LRRC55 | Sponge network | -0.338 | 1.0E-5 | -1.671 | 0 | 0.364 |
| 13 | SCARNA15 | hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-199a-3p;hsa-miR-200b-3p;hsa-miR-204-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-373-5p | 12 | RAB3B | Sponge network | 0.373 | 2.0E-5 | 1.829 | 0 | 0.32 |
| 14 | FAM87B | hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-375 | 10 | RAB3B | Sponge network | 0.318 | 0.00507 | 1.829 | 0 | 0.314 |
| 15 | PVT1 | hsa-miR-130a-3p;hsa-miR-192-5p;hsa-miR-199a-3p;hsa-miR-200b-3p;hsa-miR-214-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-373-5p | 12 | RAB3B | Sponge network | 0.192 | 0.00873 | 1.829 | 0 | 0.288 |