Help
- Mutation status by literature mining, and mutation data by sequencing from TCGA, CCLE, COSMIC
- Expression status by literature mining from various methods including qPCR, northern blot, in situ hybridization; Quantitative expression levels of tumor samples and cell-lines by RNA-Seq, proteomics and microArray from TCGA and 318 GEO studies
- Copy number variations from sequencing
- Methylation changes from literature mining and microArray
- miRNA and lncRNA-mediated sponge regulations from literature mining, as well as inferred from expression and multiple miRNA-target database including miRNAWalker2, TarBase, TargetScan, PITA, mirMAP, miRanda, and miRNATAP
- The gene regulatory network inferred from expression correlation and interaction data of Pathway commons, HumanNet and STRING
- Effected cancer pathways from literature mining, MigSig, KEGG and gene ontology
- Drugs that target on the gene or target on the enzyme or excellular proteins that interacted with the gene in protein level
The following sections introduce the usage of GastricCancerMAP step by step
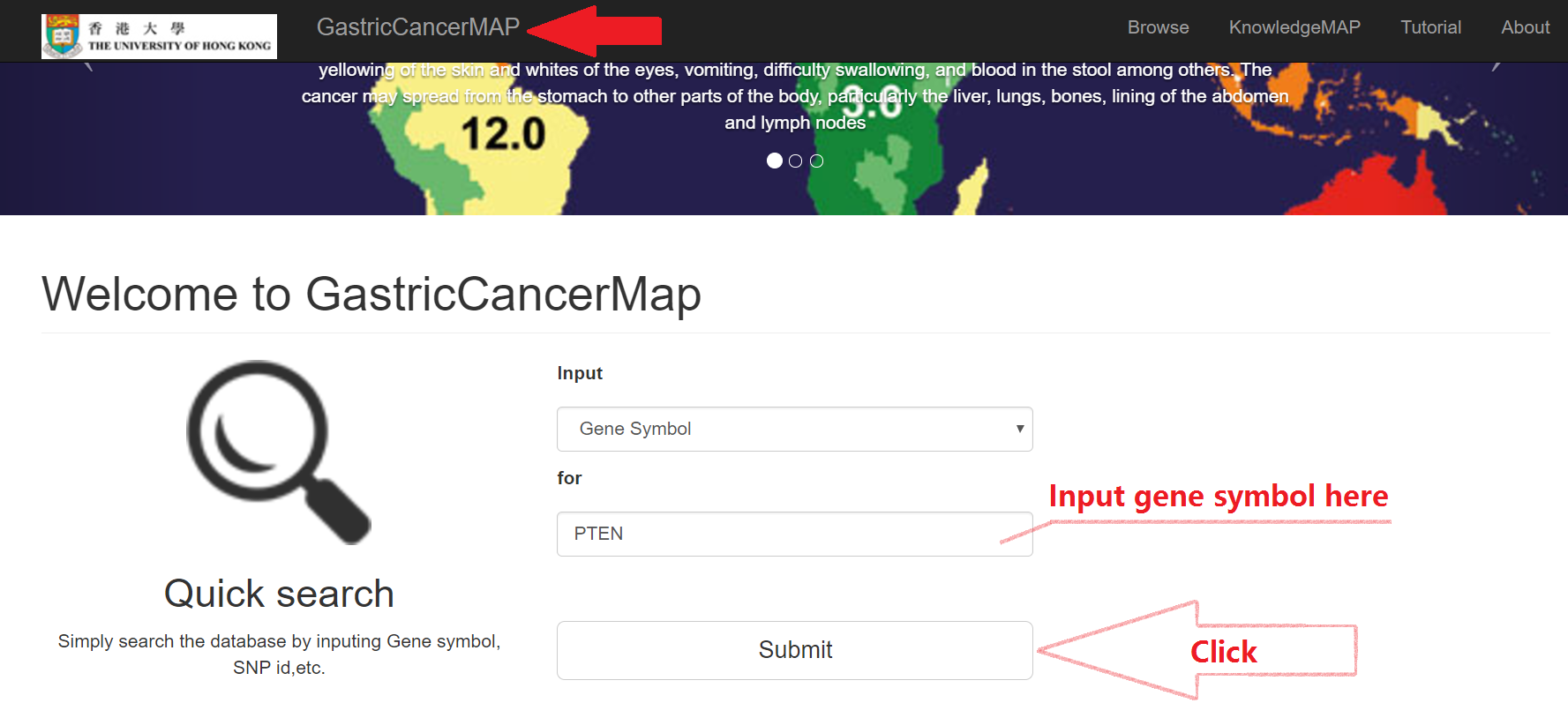
Click "GastricCancerMAP" button
- Input the Gene Symbol to search the database
- Click "Submit"
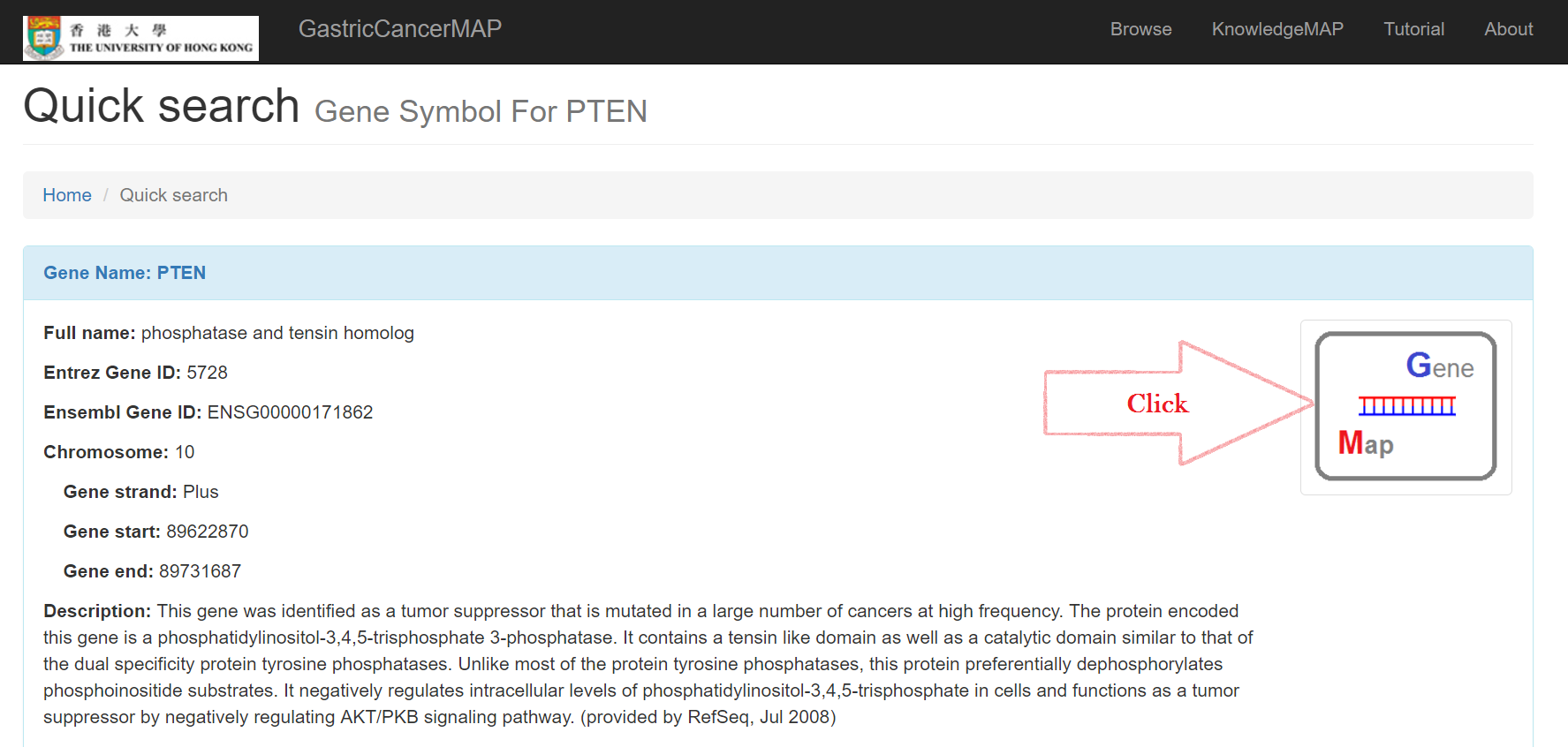
Click on the symbol on the right for detailed information about the gene
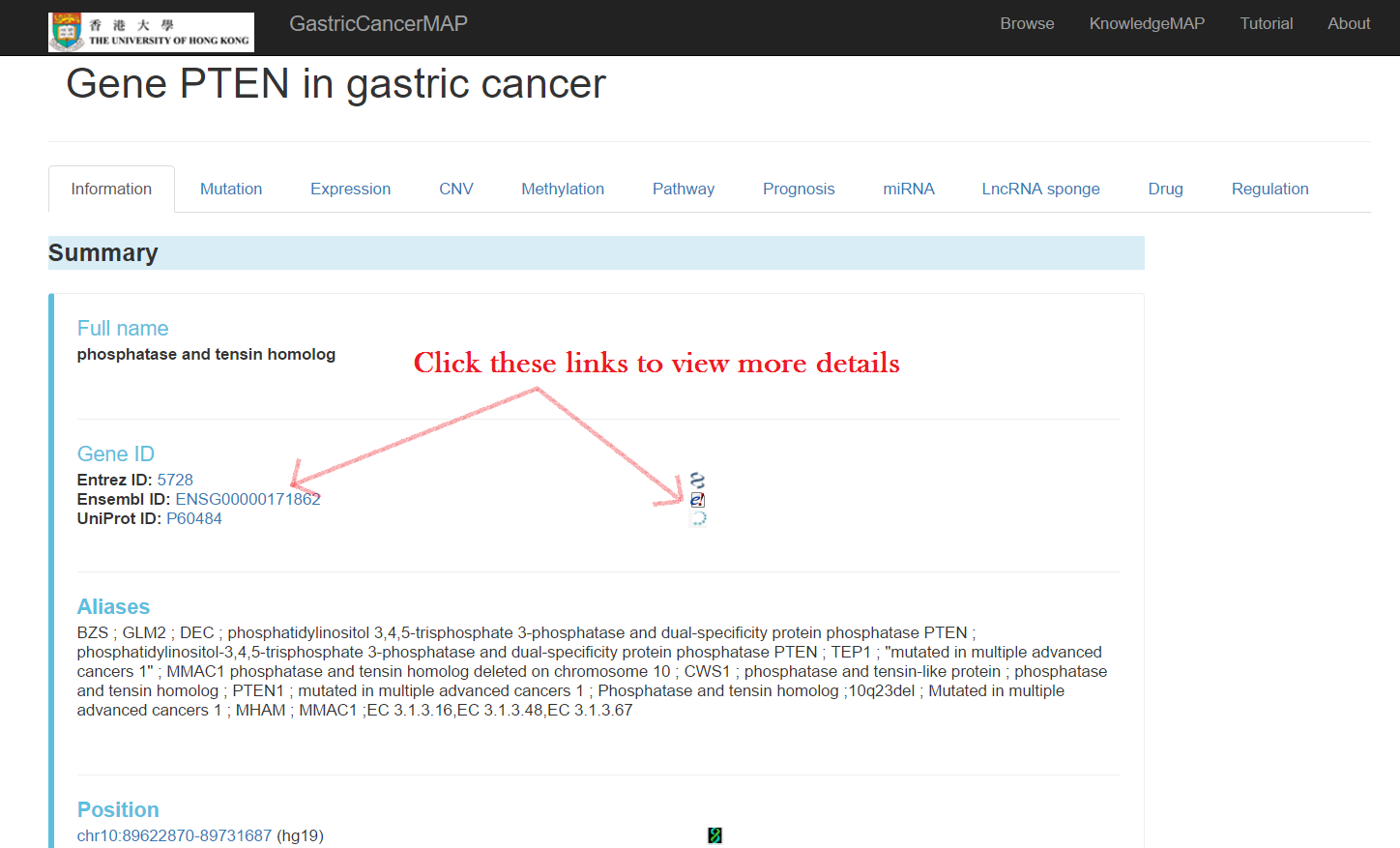
The "information" tab includes:
Summary of the gene including full name, gene ID, aliases, and position with links to follow
Function of the gene
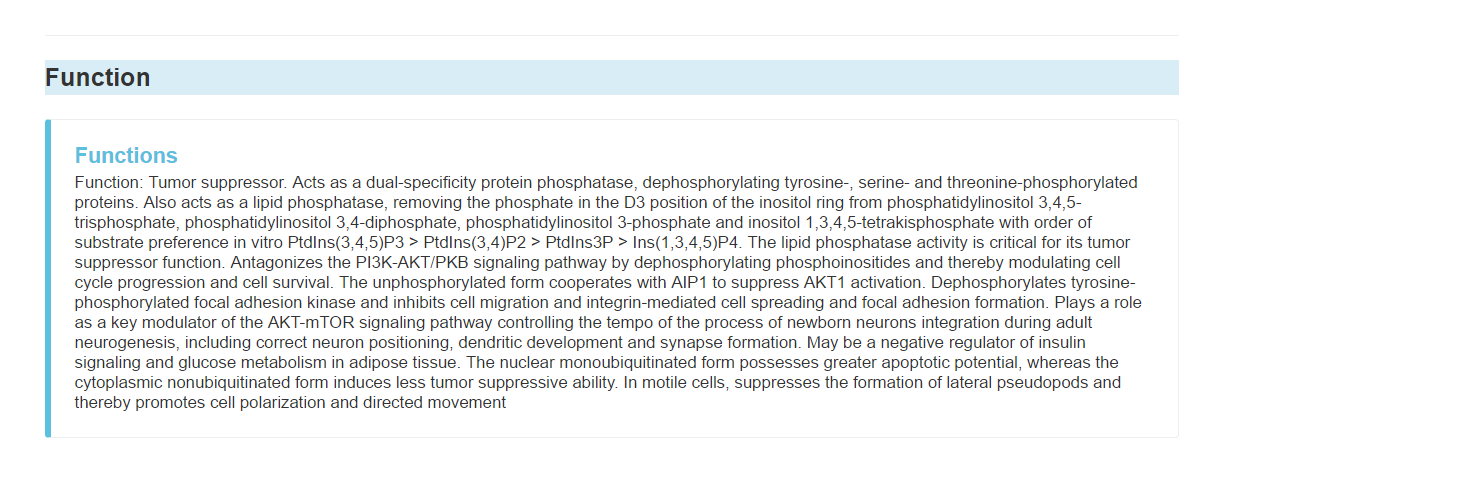
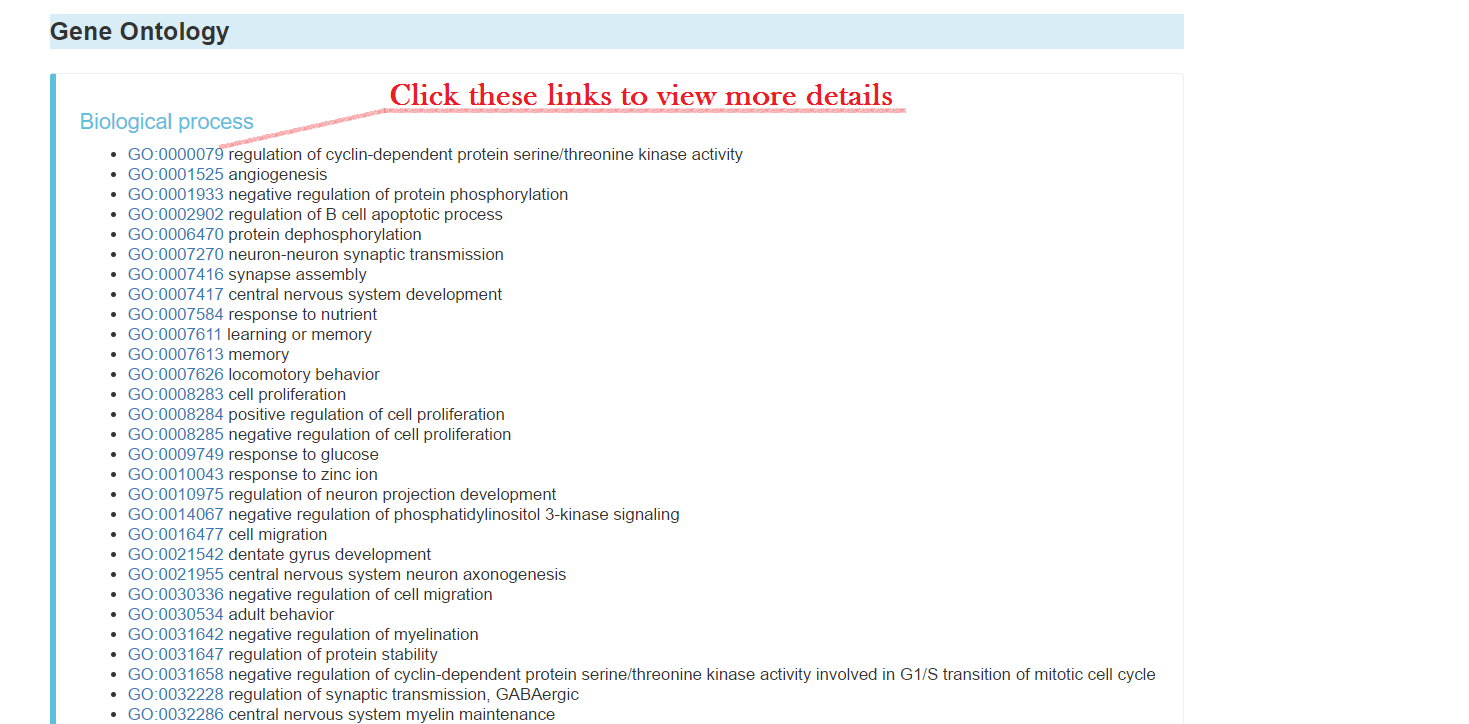
Gene ontology, including:
- Biological process with links to follow
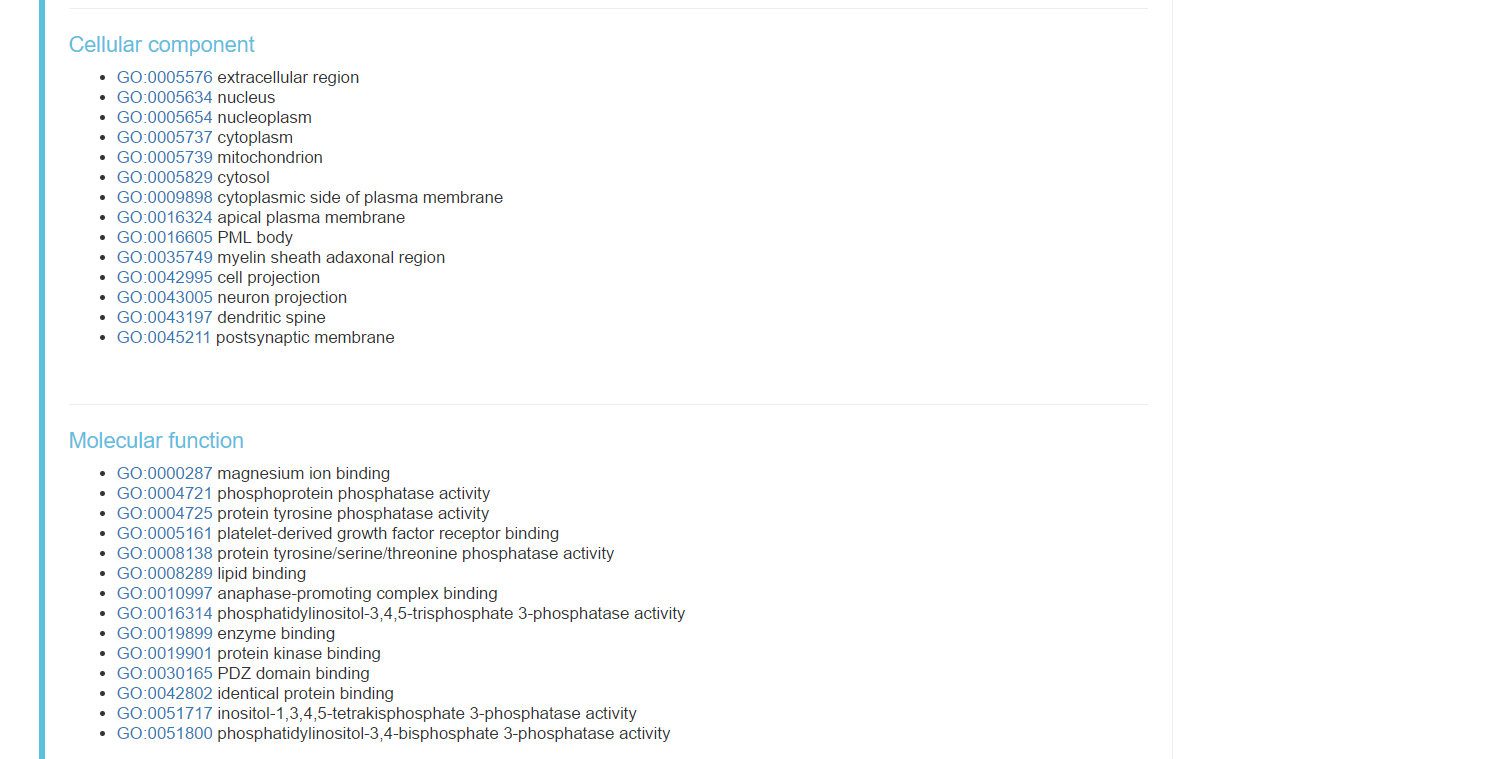
- Cellular component with link to follow
- Molecular function with links to follow
KEGG pathway of the gene
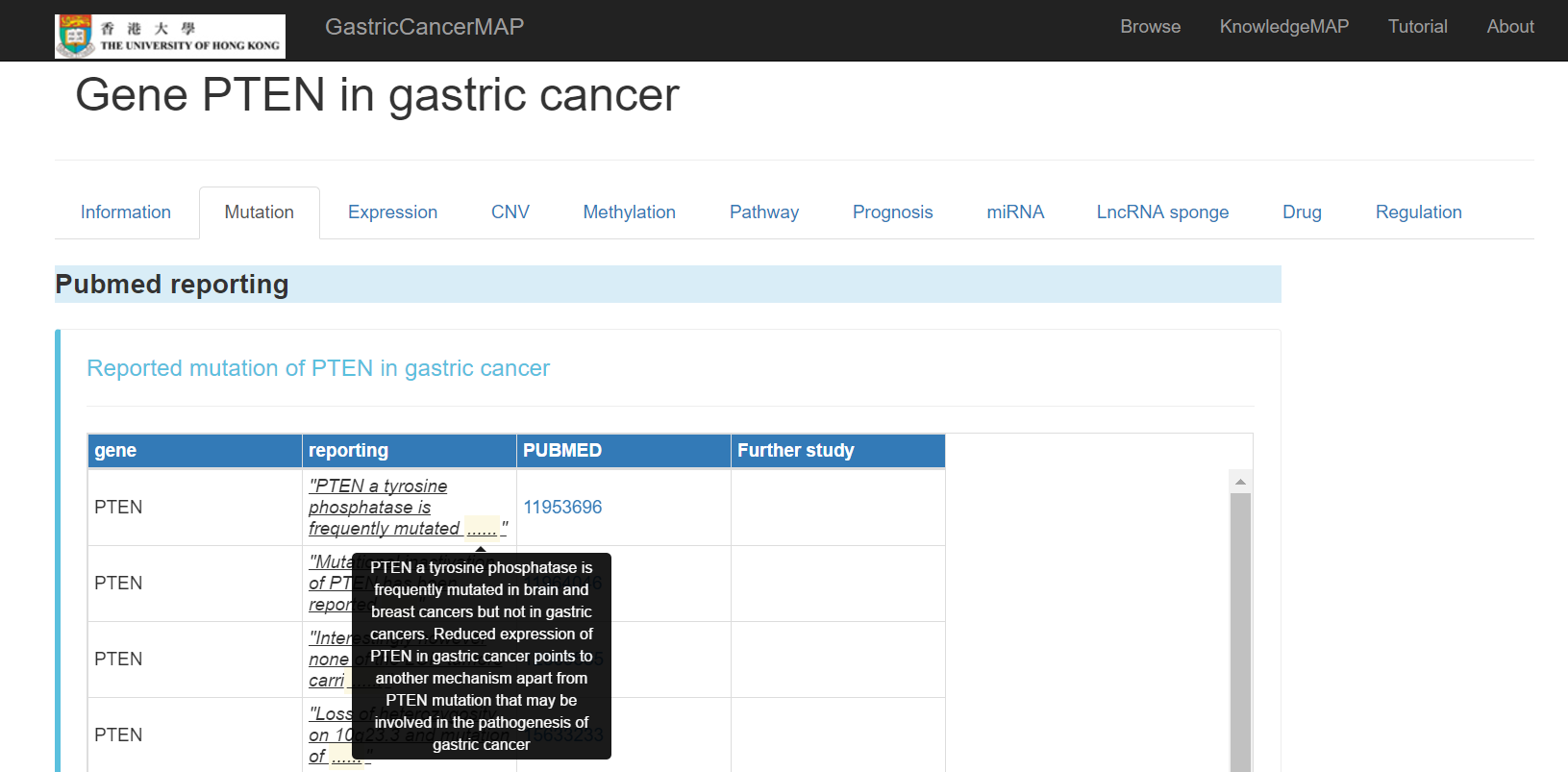
You can click the "mutation" tab to see the mutation information of the gene in gastric cancer, including:
Pubmed reported mutation of the gene with Pubmed links to follow
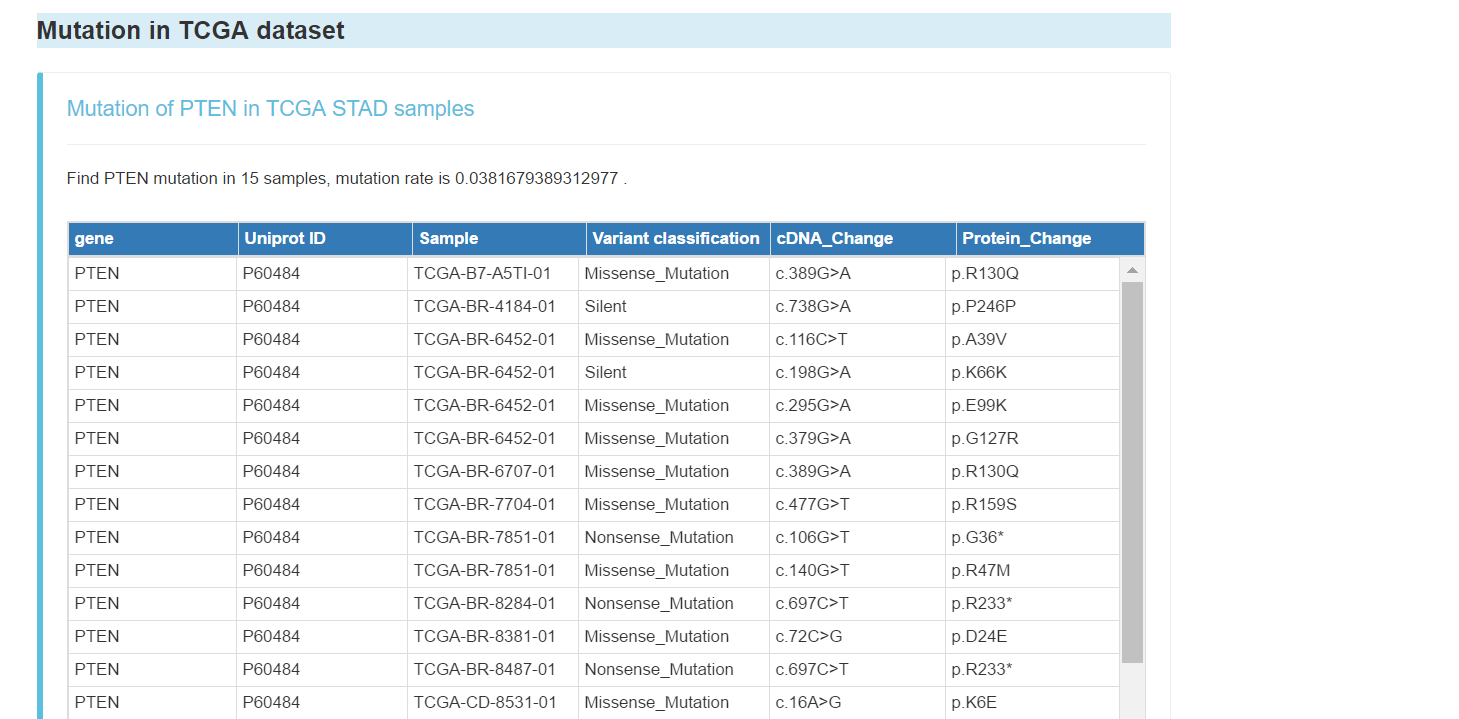
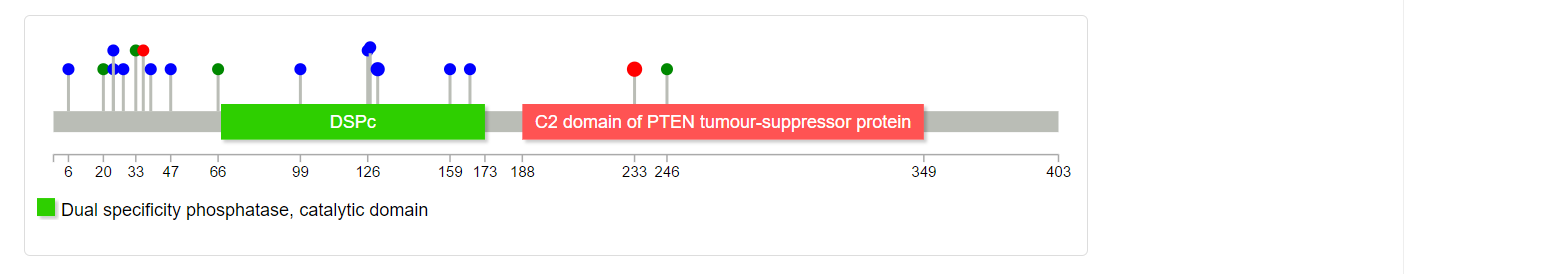
Mutation of the gene in TCGA dataset
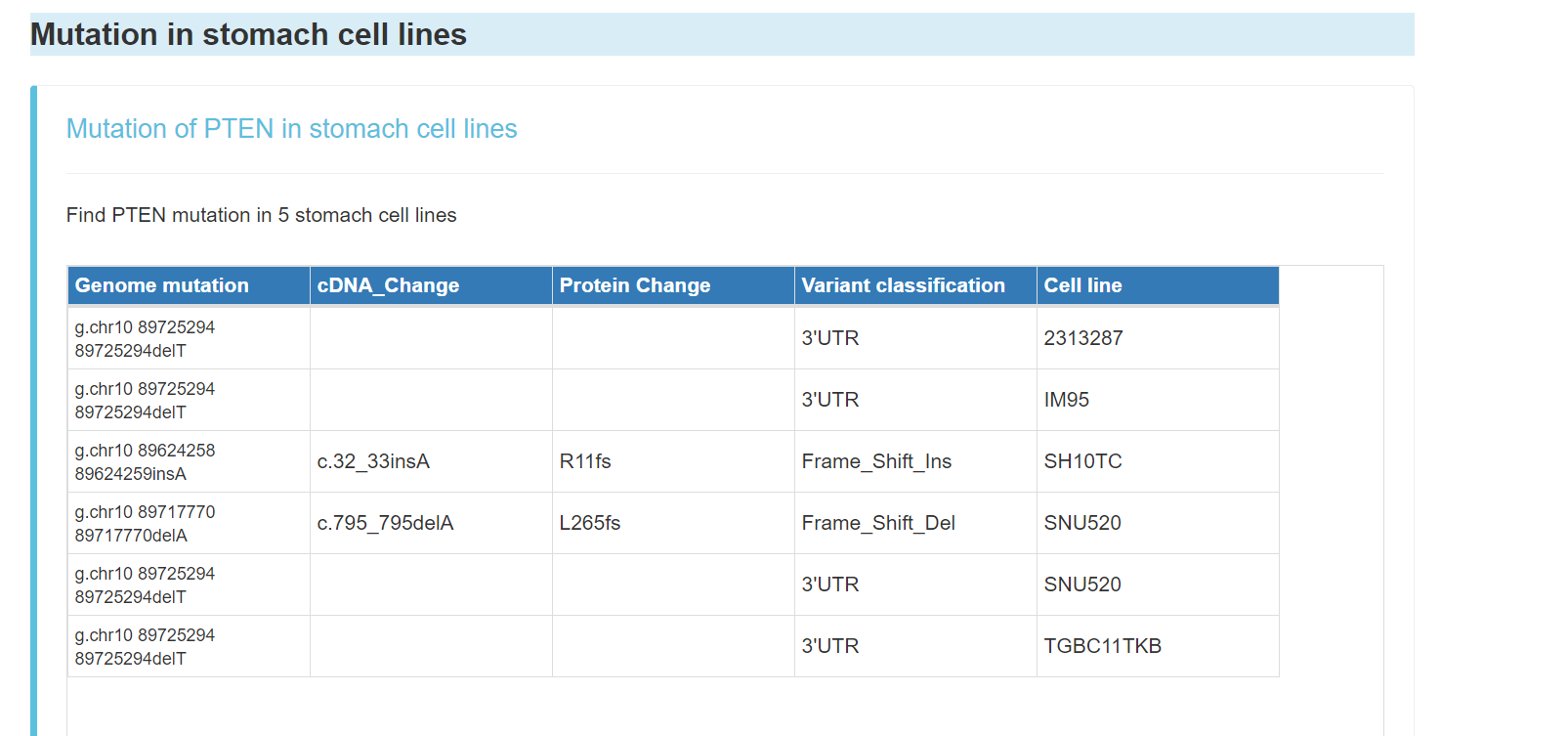
Mutation of the gene in stomach cell lines
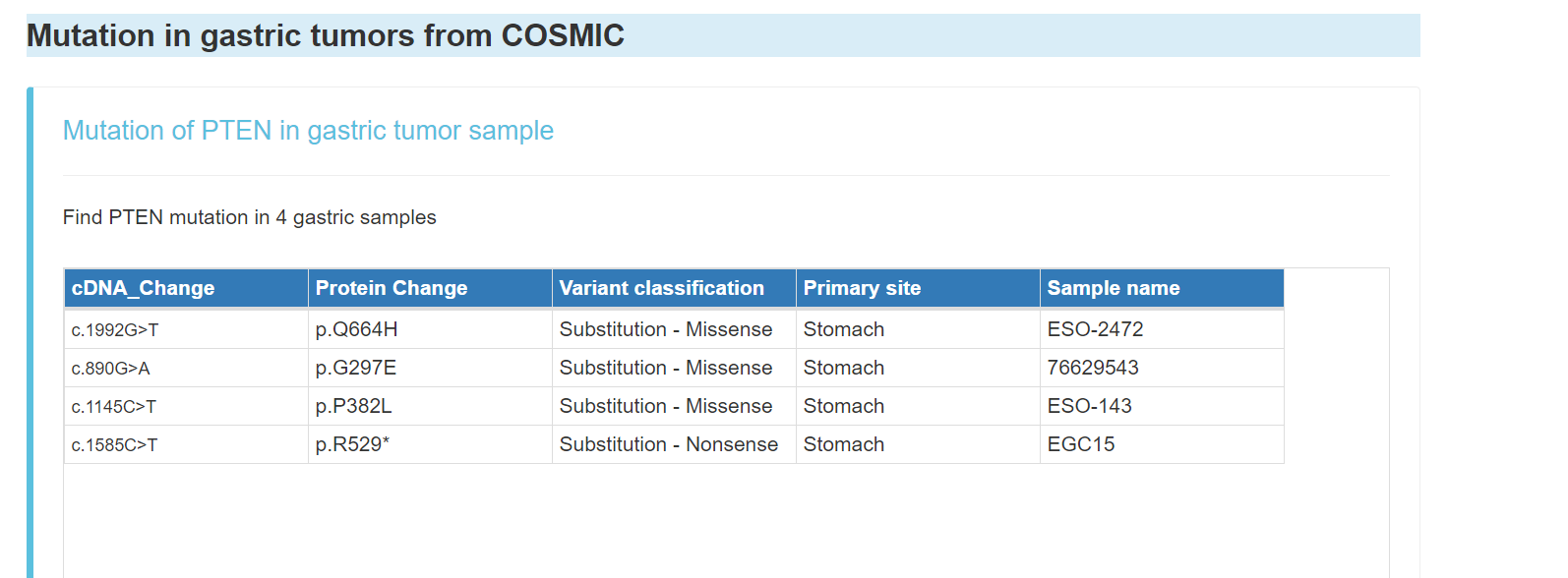
Mutation of gene in gastric tumors from COSMIC
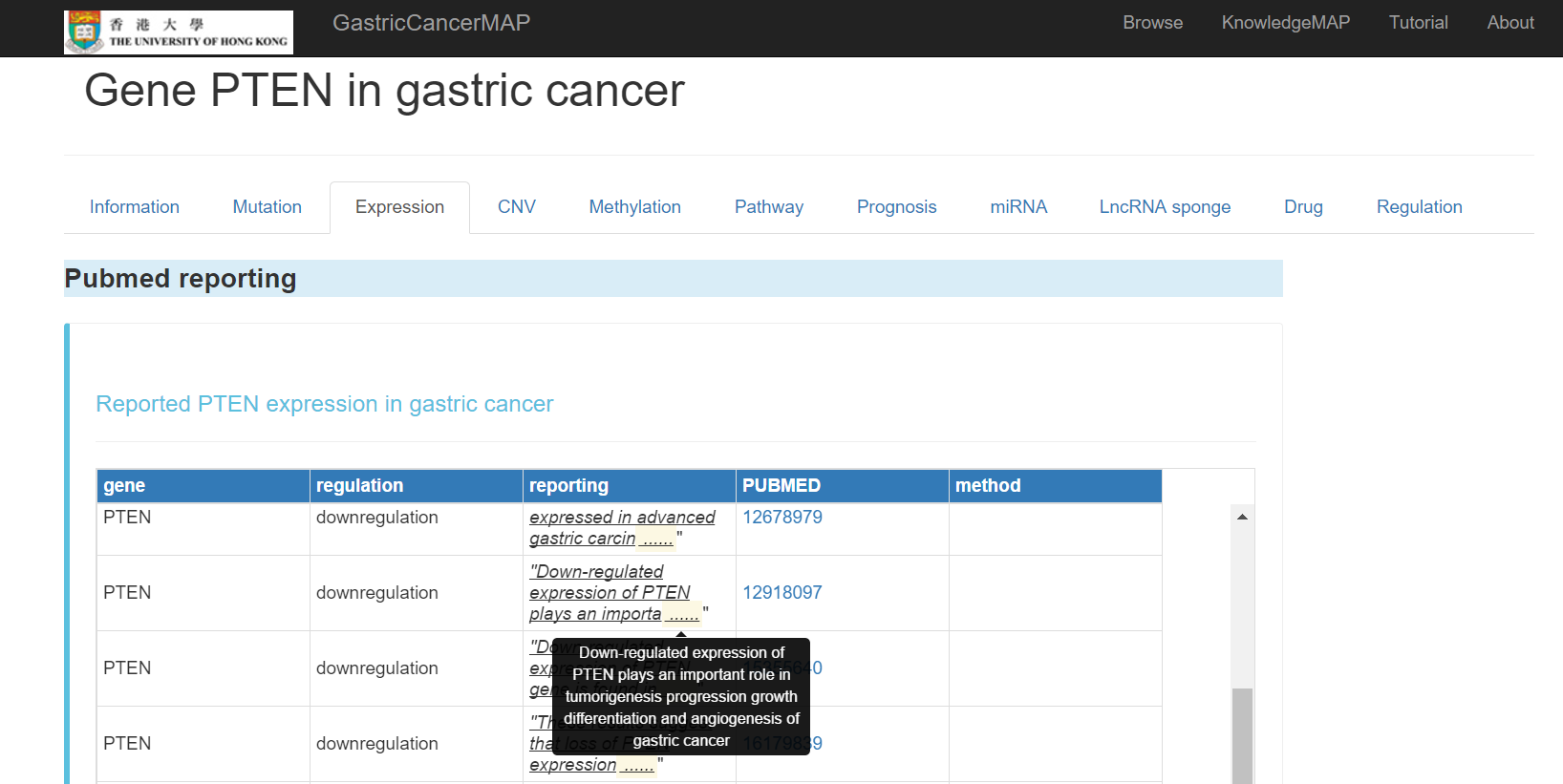
You can click the "Expression" tab to see the expression information about the gene, including:
Pubmed reported expression of the gene in gastric cancer
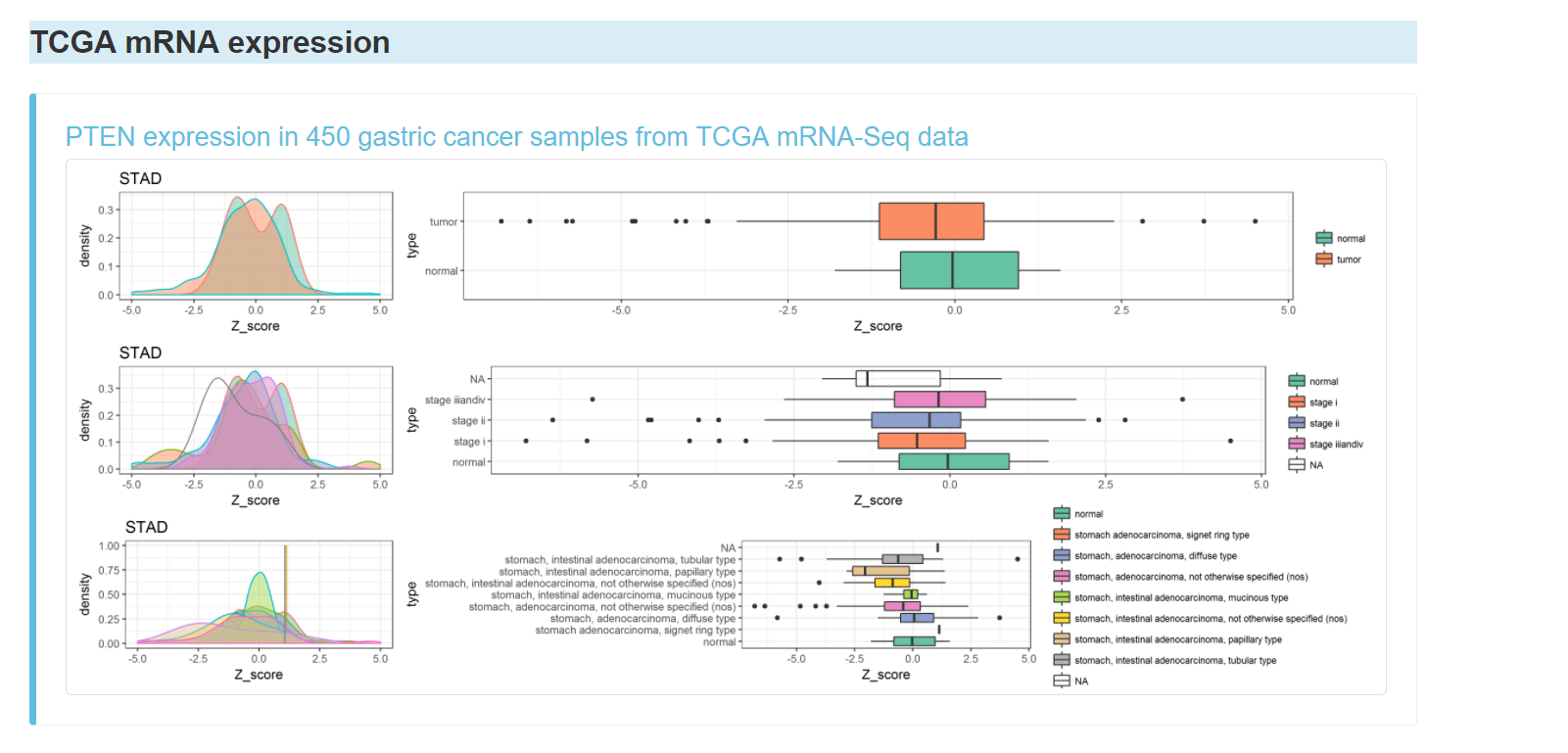
TCGA mRNA expression of the gene
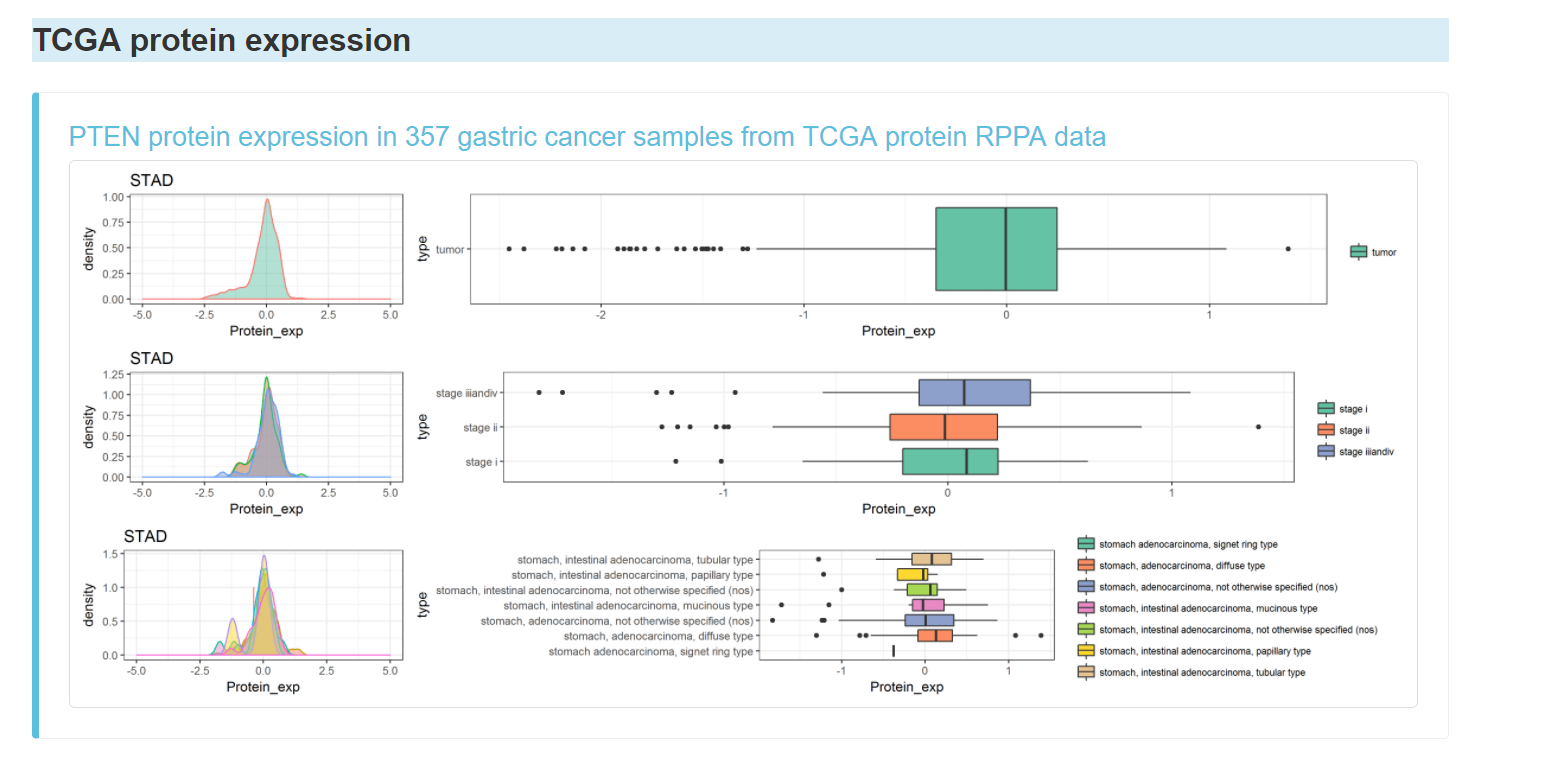
TCGA protein expression of the gene
Expression of the gene in Gene Expression Omnibus datasets
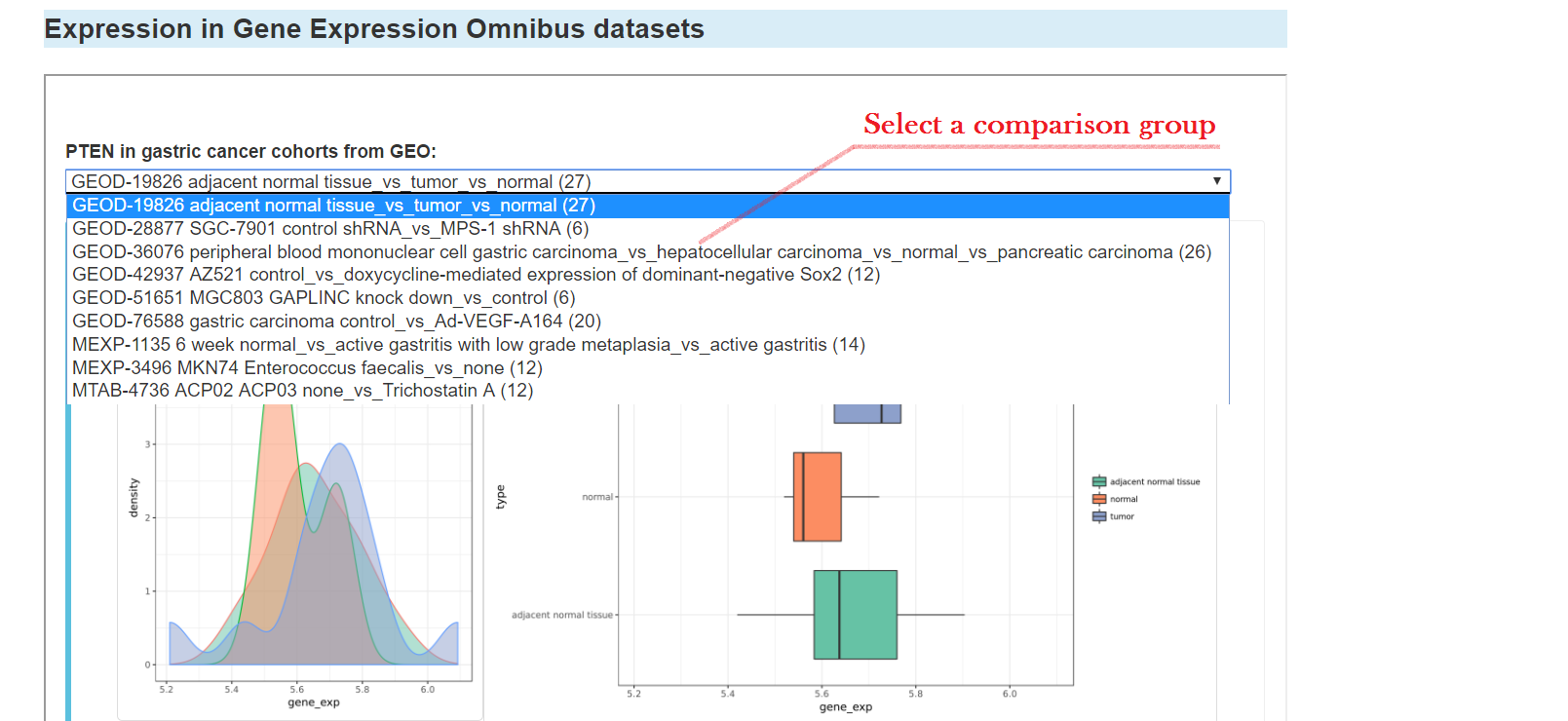
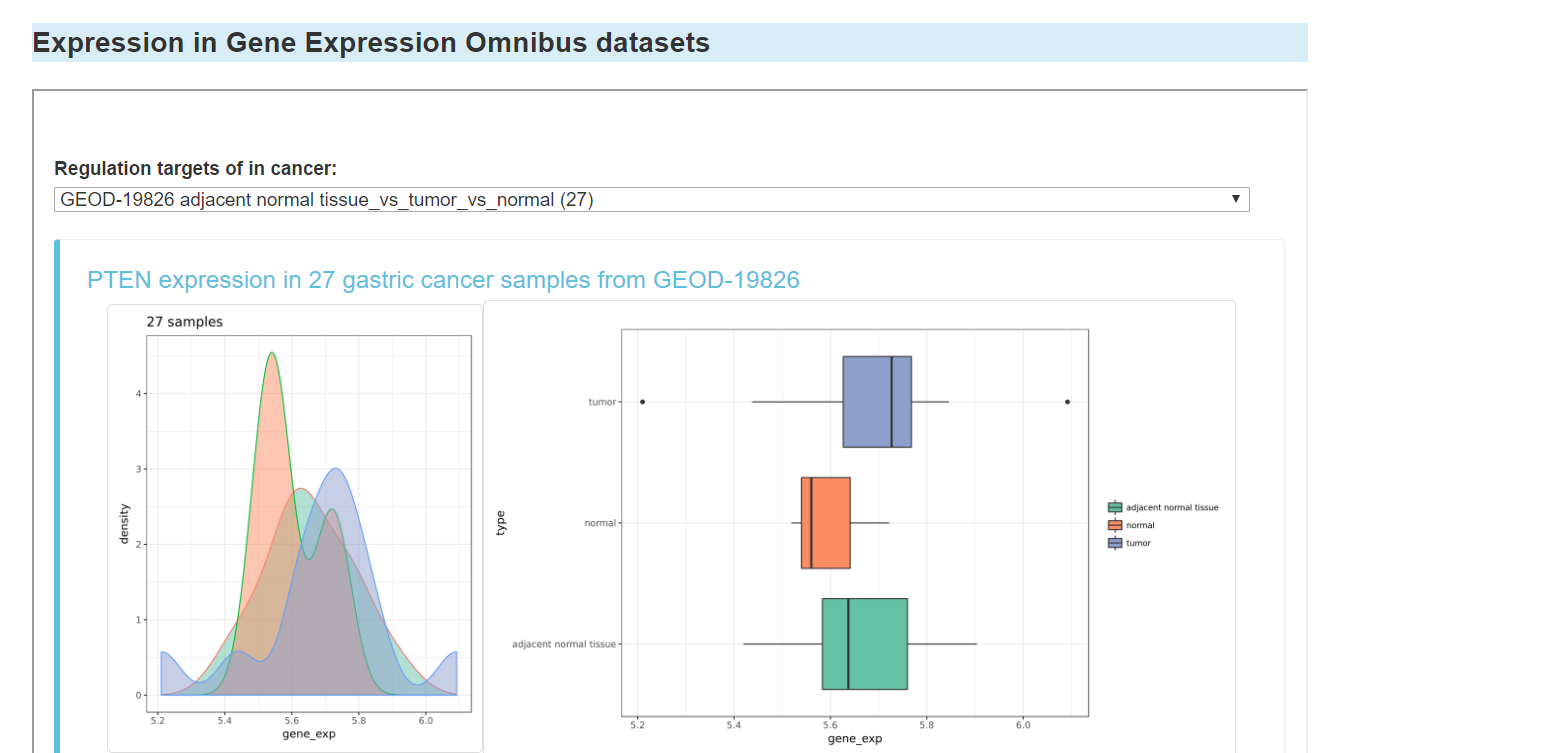
You can click the "CNV" tab to see CNV information of the gene, including:
Copy number alteration of the gene in TCGA STAD
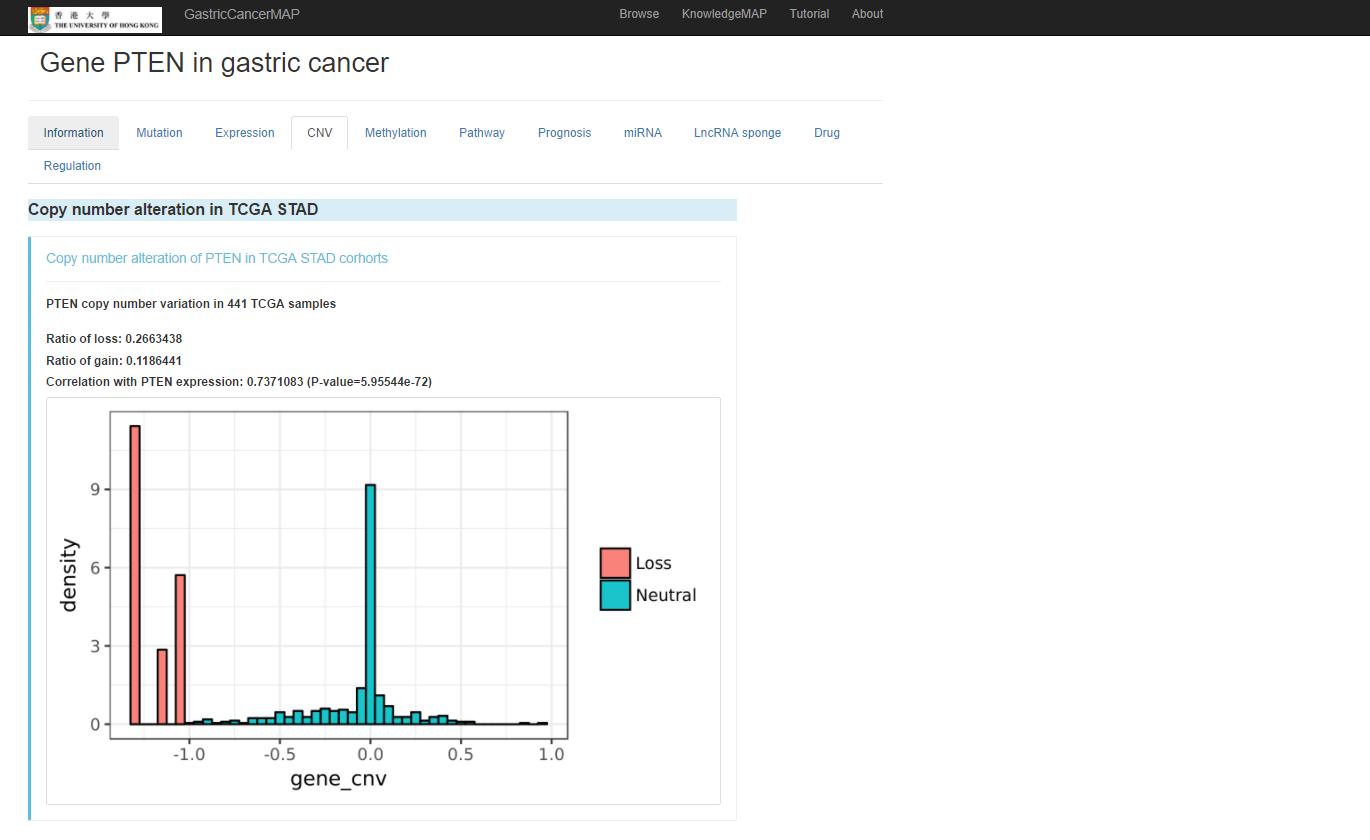
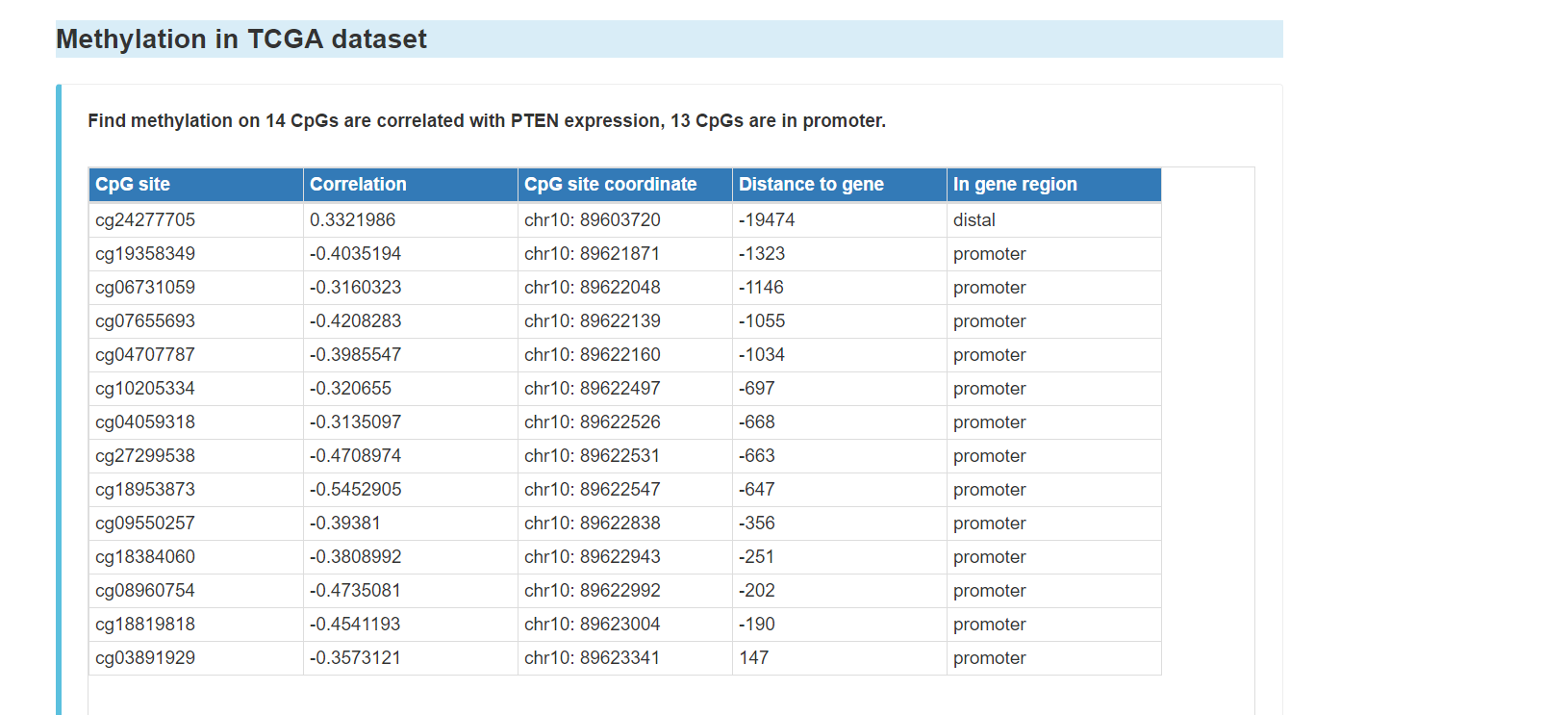
You can click the "Methylation" tab to see methylation information of the gene, including:
Pubmed reported methylation of the gene in gastric cancer
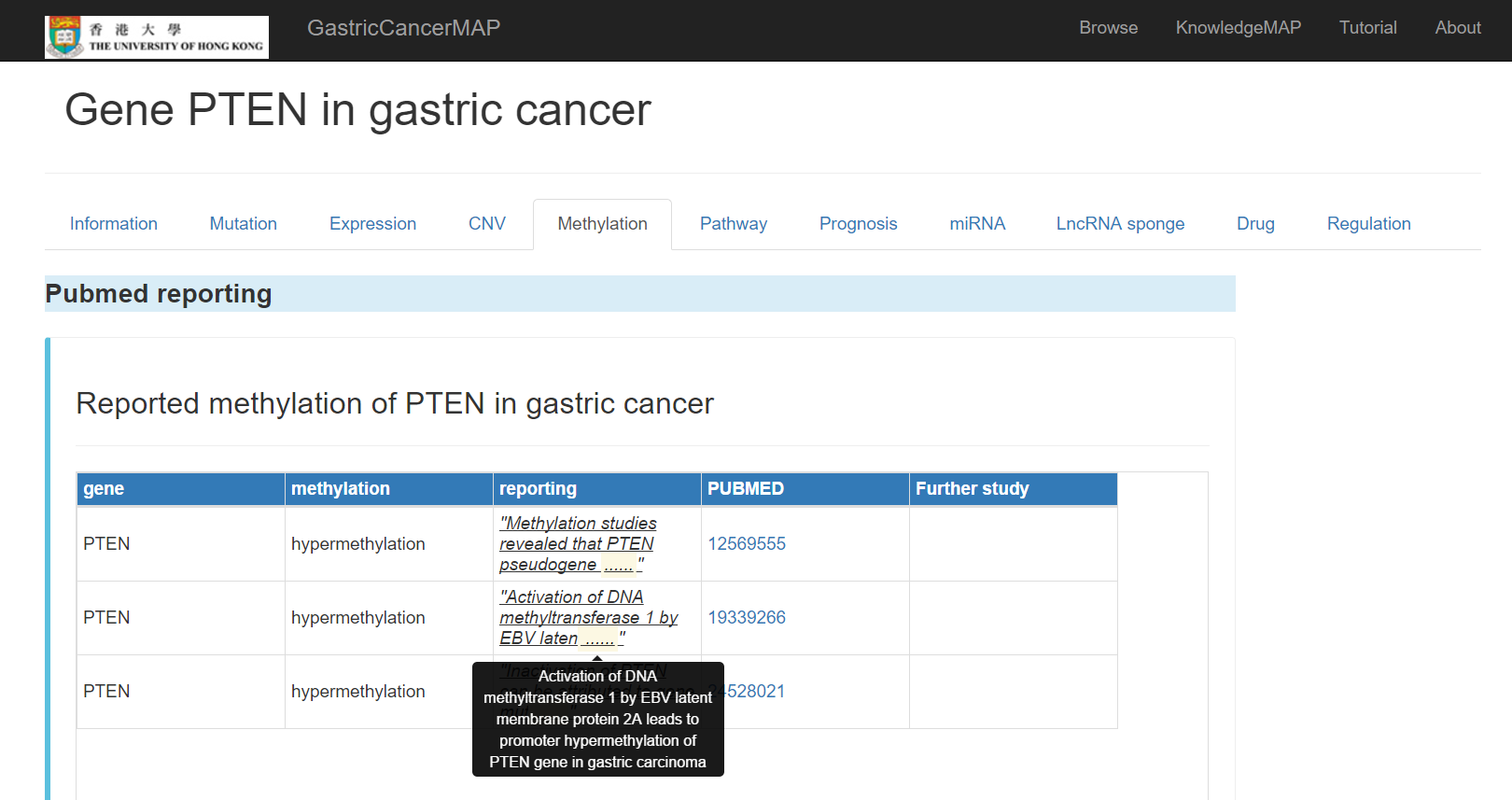
Methylation in TCGA dataset
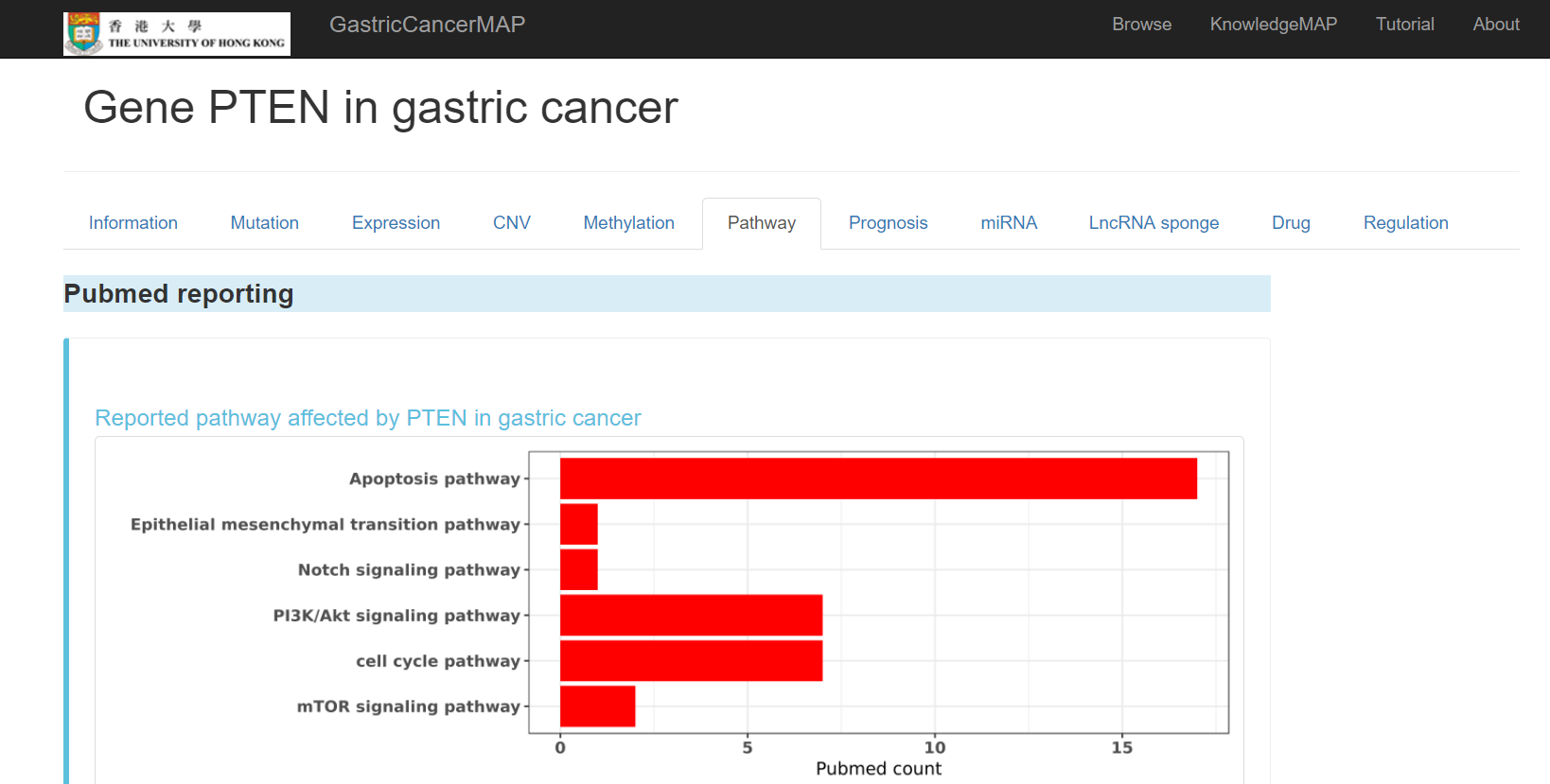
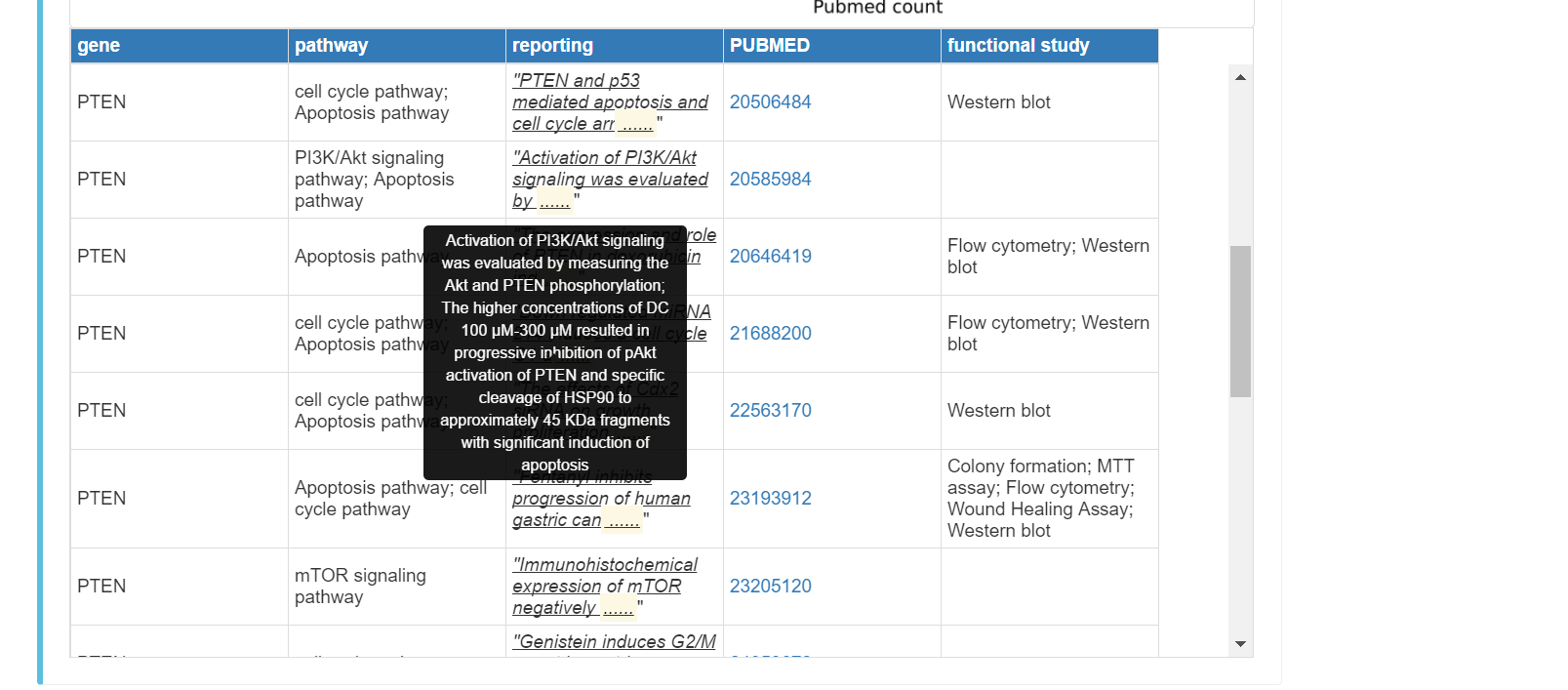
You can click the "Pathway" tab to see pathway information of the gene, including:
Pubmed reported pathways affected by the gene in gastric cancer
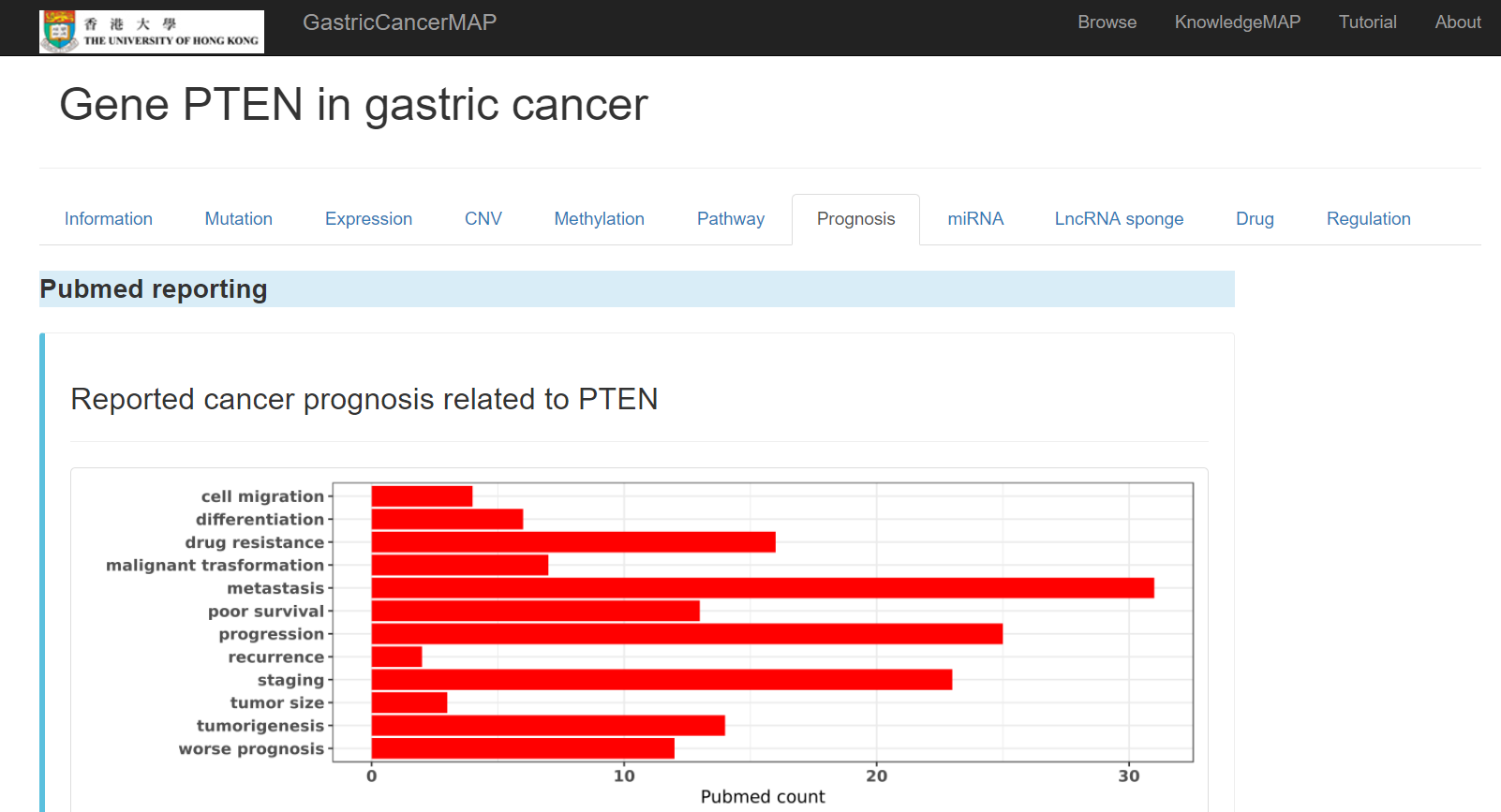
You can click the "Prognosis" tab to see prognosis information of the gene, including:
Pubmed reported cancer prognosis related to the gene

You can click the "miRNA" tab to see miRNA information of the gene, including:
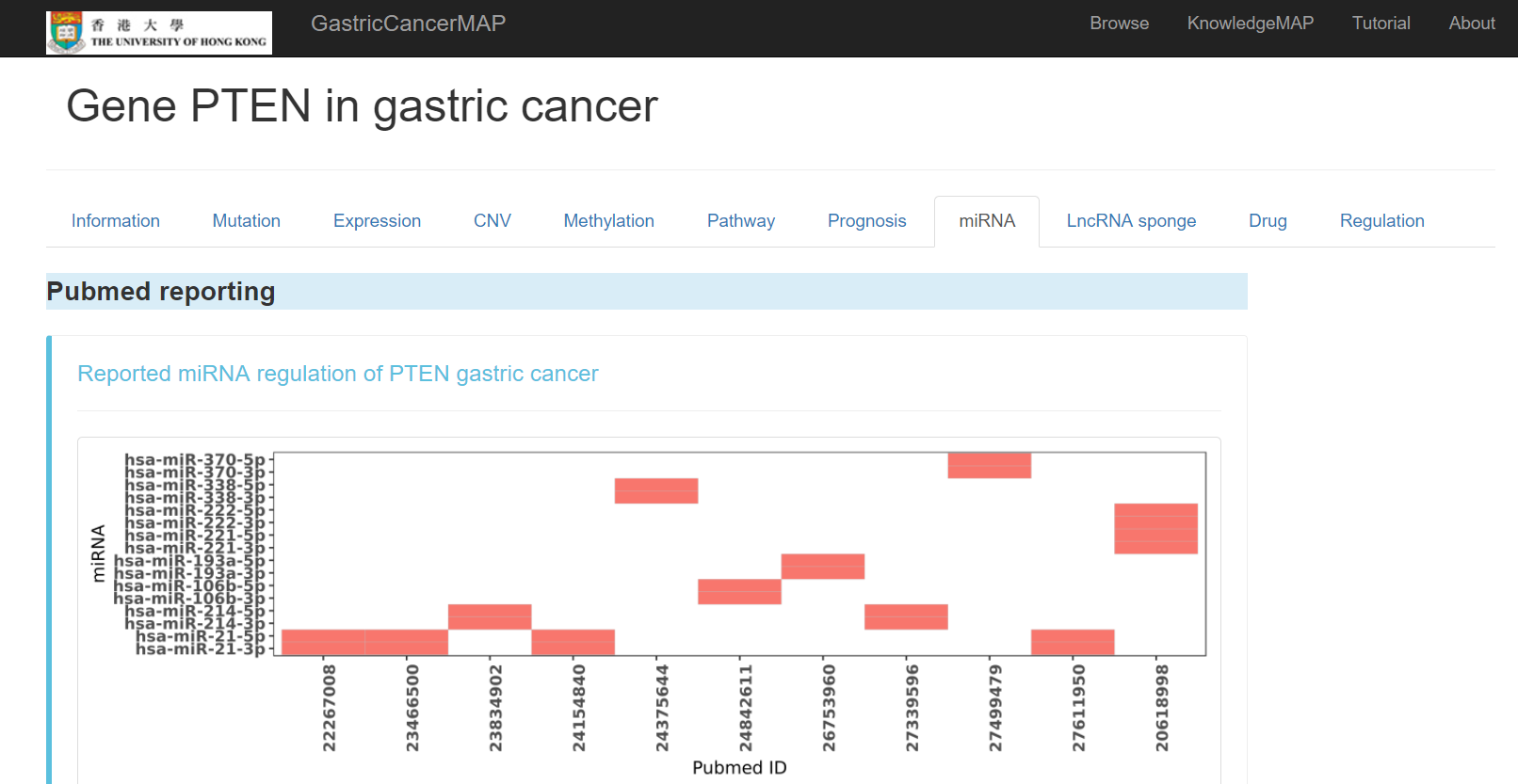
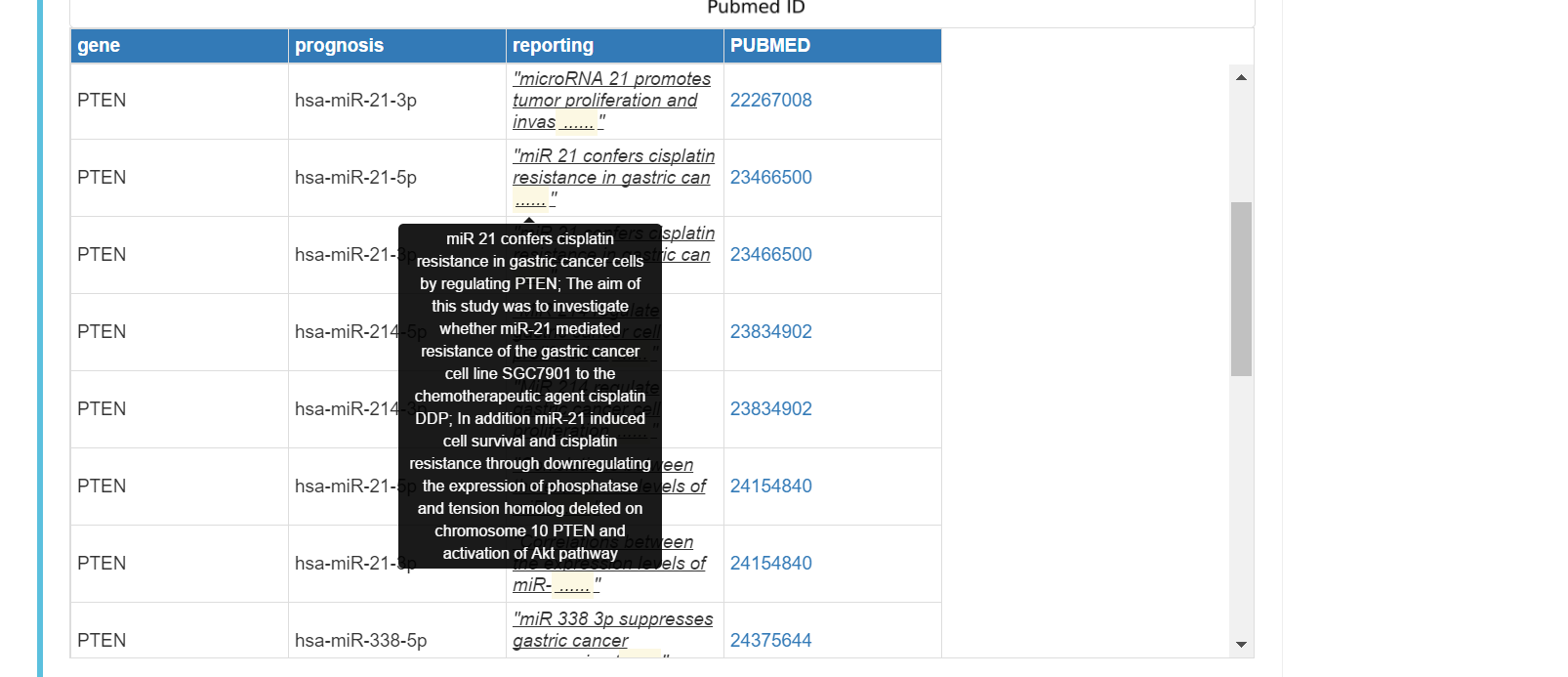
Pubmed reported miRNA regulation of the gene in gastric cancer
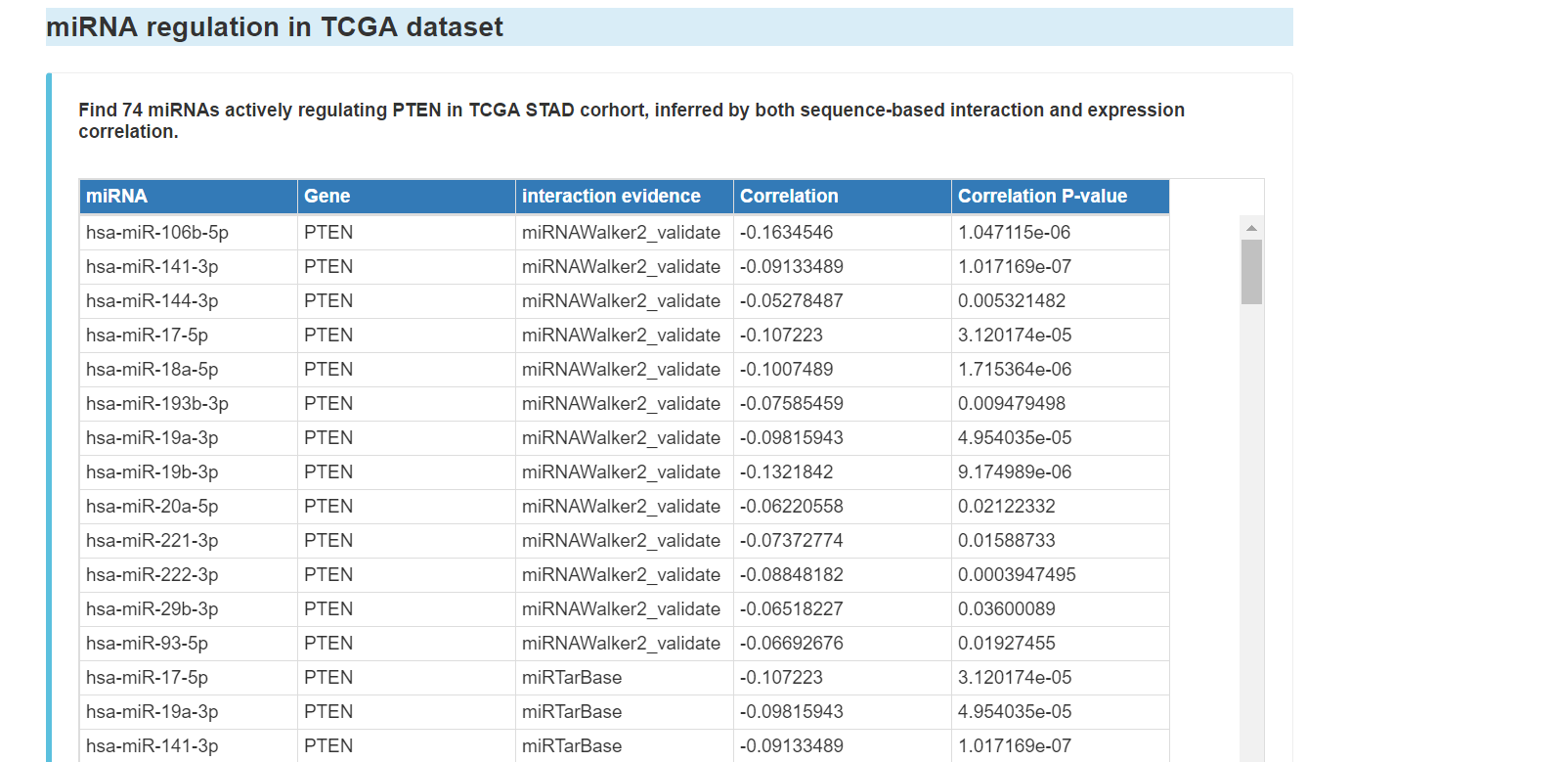
miRNA regulation of the gene in TCGA dataset
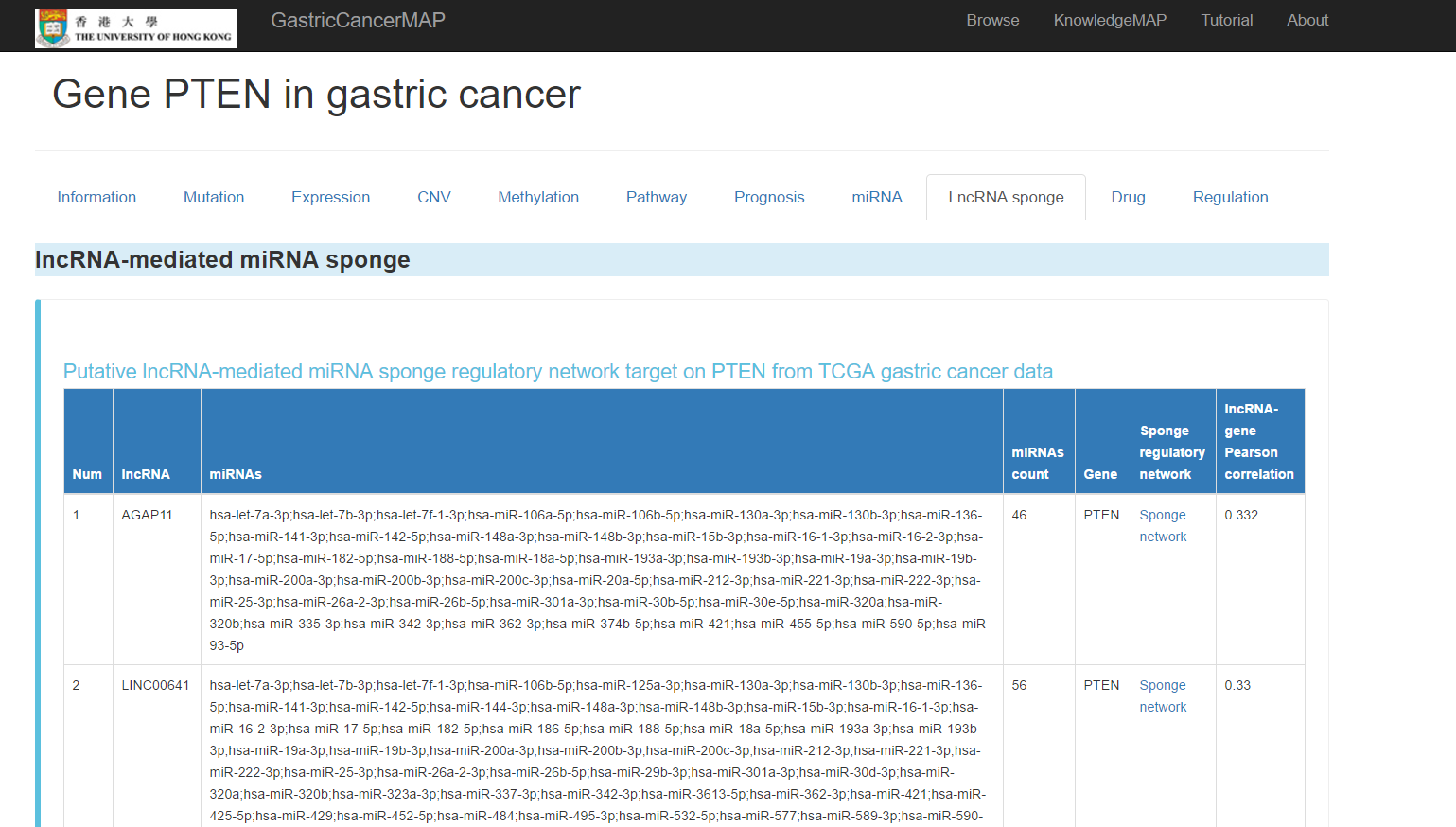
You can click the "LncRNA sponge" tab to see lncRNA sponge information of the gene, including:
Putative lncRNA-mediated miRNA sponge
You can click the "Drug" tab to see drug information of the gene, including:
Drug target on the gene and interacted proteins
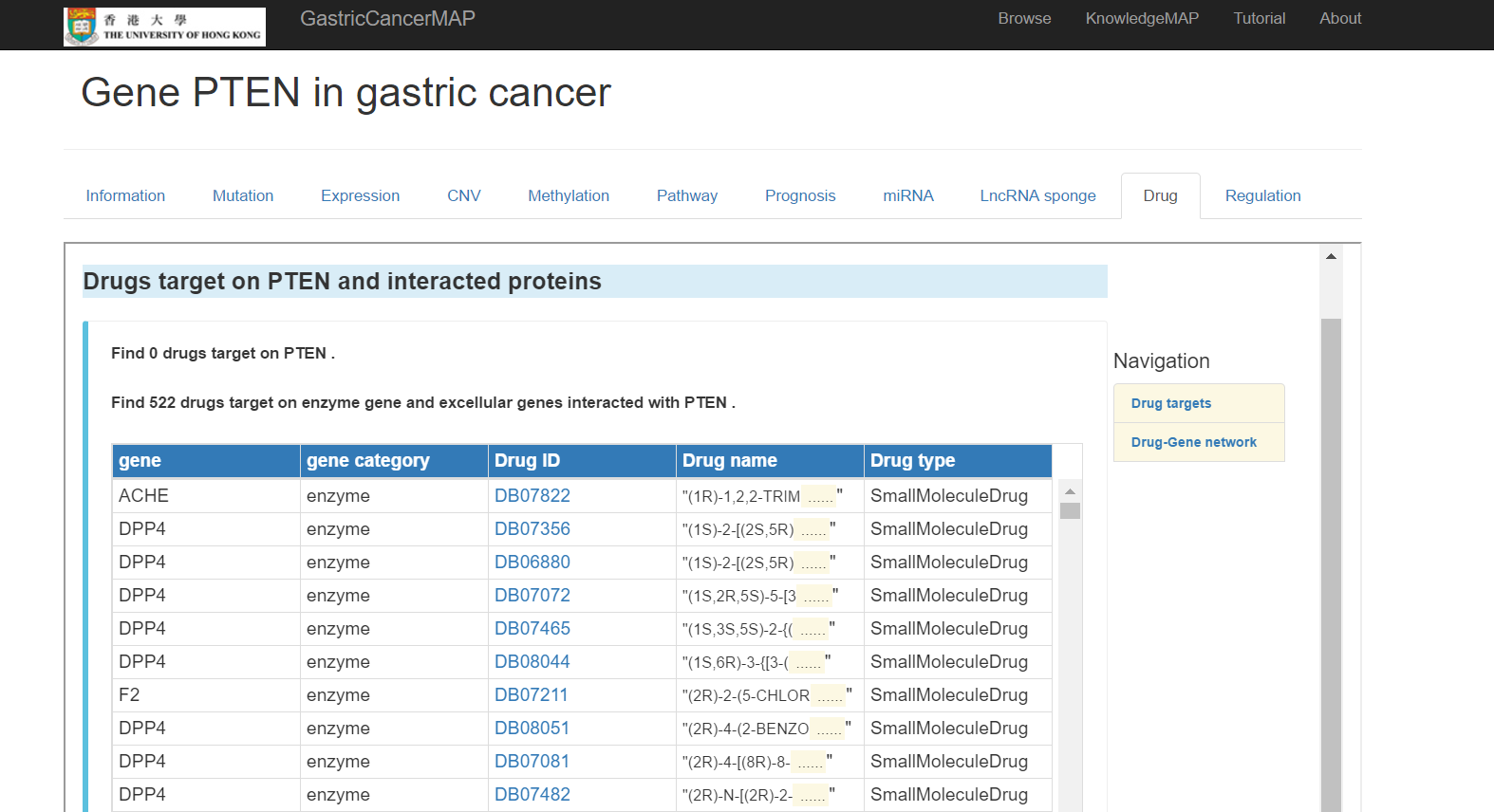
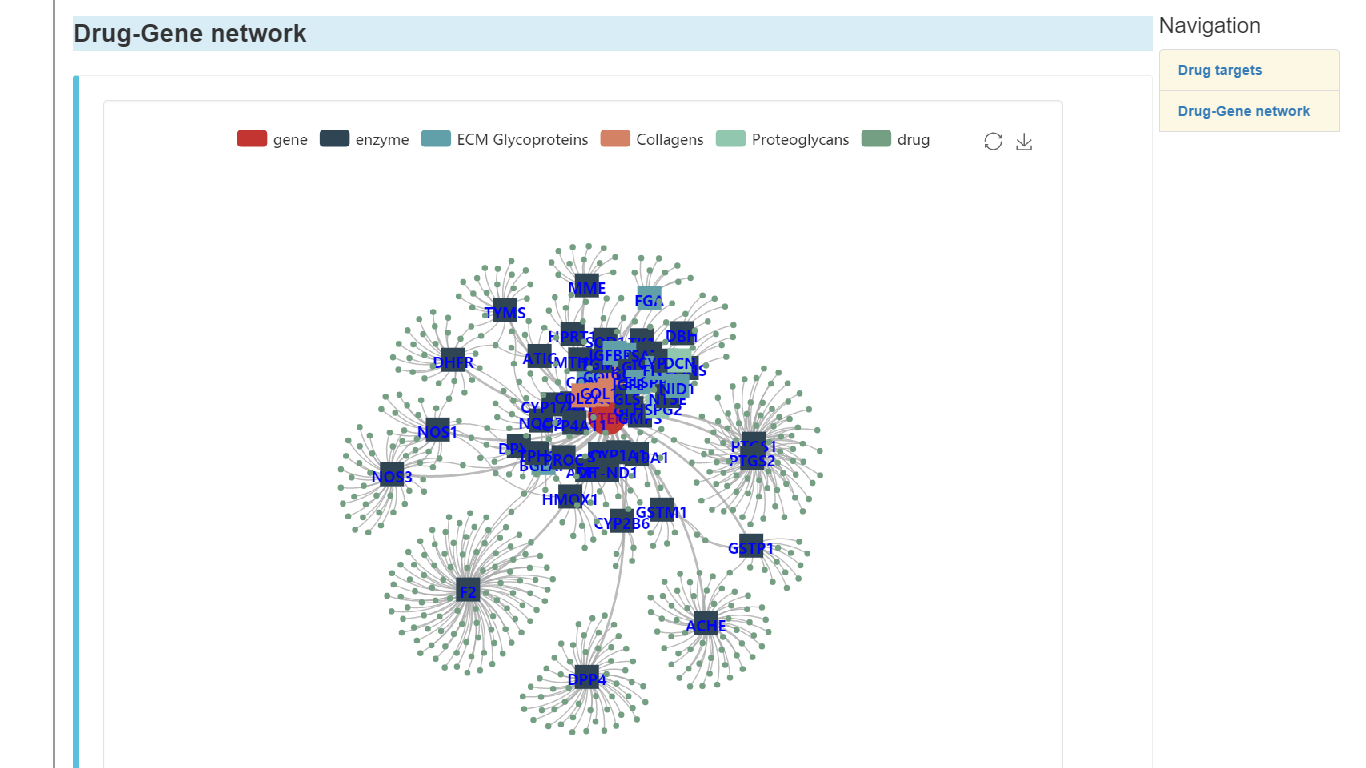
Drug-gene network
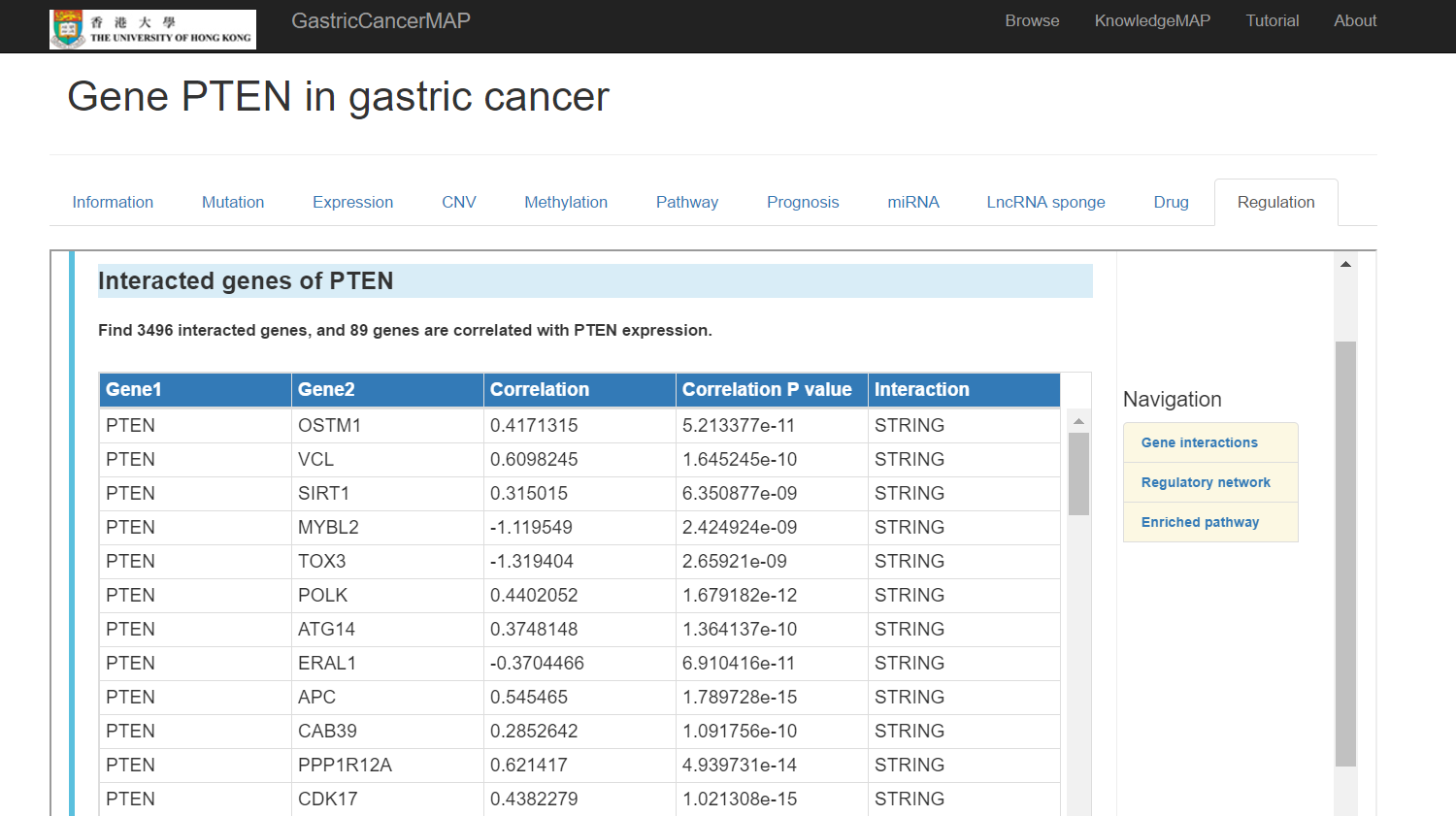
You can click the "Regulation" tab to see regulation information of the gene, including:
Gene interactions
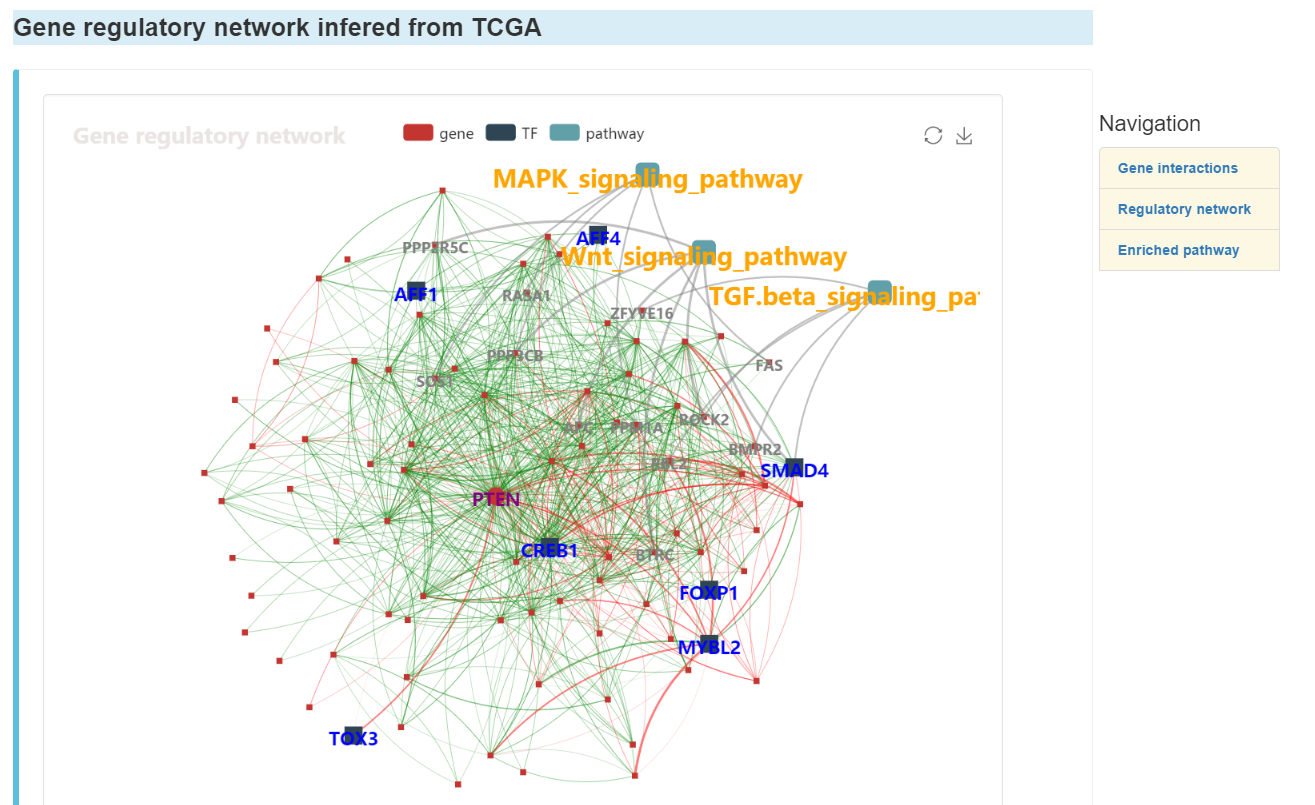
Gene regulatory network
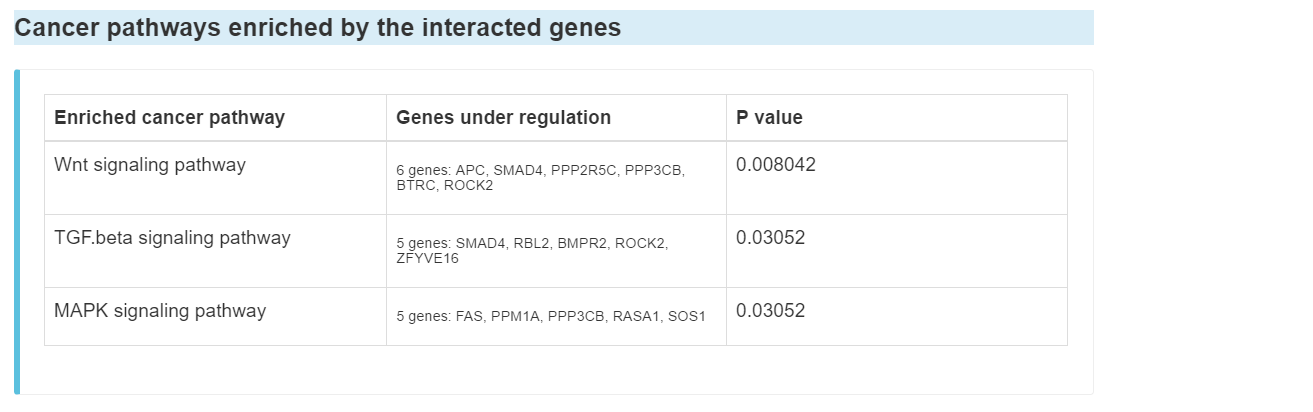
Cancer pathways enriched by the interacted genes