Help
1. Ten categories of knowledge
1.1 Function
1.2 Literature
1.3 High throughput screening
1.4 Immunotherapy
1.5 Tumor-infiltrating lymphocytes (TILs)
1.6 Immunomodulators
1.7 Chemokines
1.8 Subtype
1.9 Clinical
1.10 Drug
2. Browse by
2.1 Literature
2.2 High throughput screening
2.3 Cohorts with immunotherapy
2.4 Function
3. Search database
3.1 Simple search
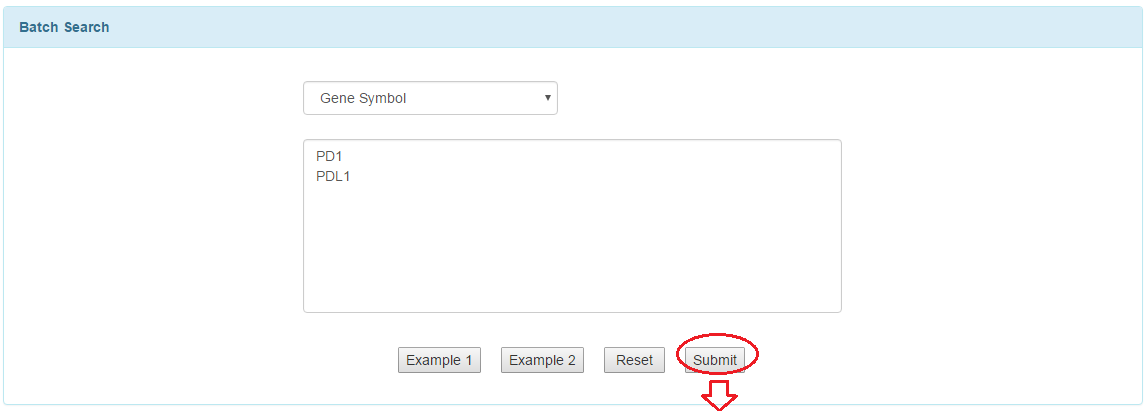
3.2 Batch search
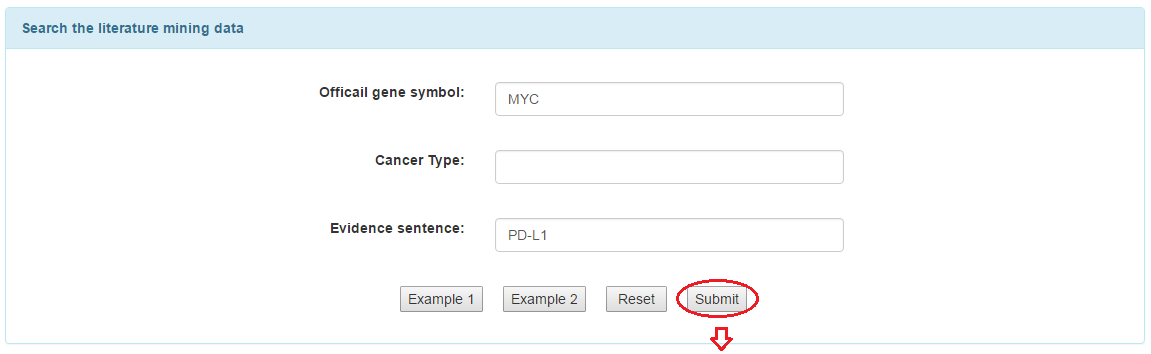
3.3 Text mining search
Users can obtain basic infomation for each gene, including gene symbol, name, location and several links to public database.
Additionally, subcellular location, protein domain and function as well as GO and pathways are also included in this part.
 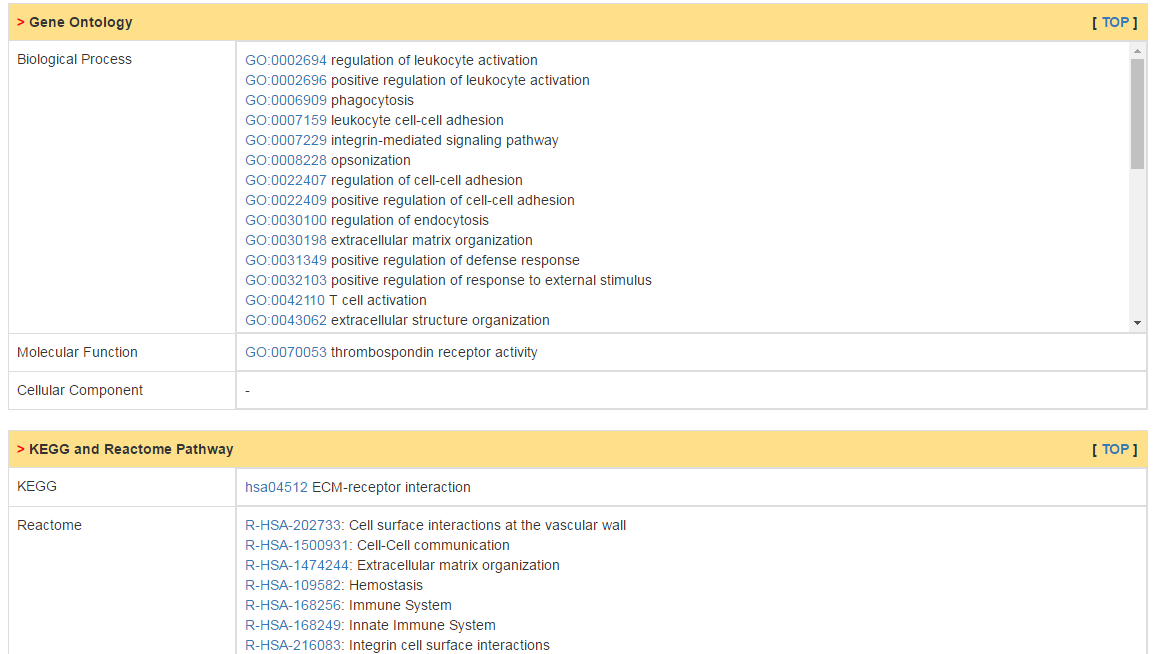
|
In this part, users can review the literatures that report relations between current gene and anti-tumor immunity (e.g. infiltration and T/NK cell function).
 |
In this tab, users can check whether their selected genes cause resistance or increase sensitivity to T cell-mediated killing in various high-throughput screening data sets. Nine groups of screening data sets were collected in the current database.
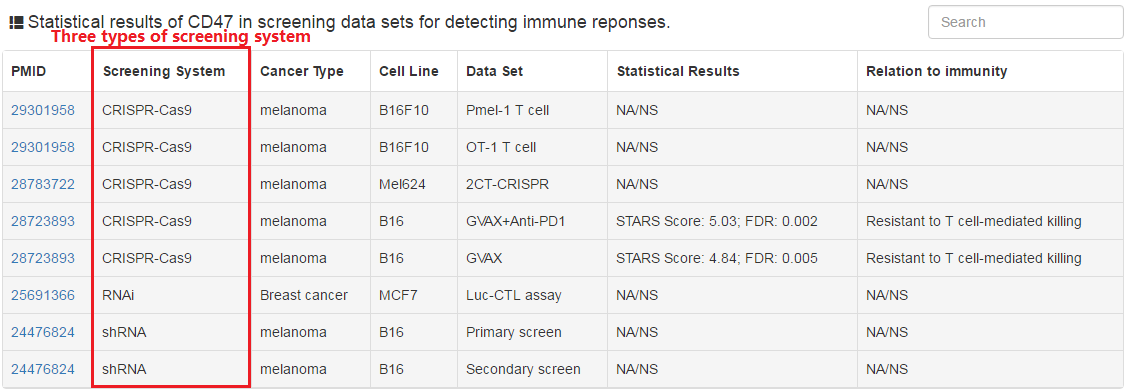 |
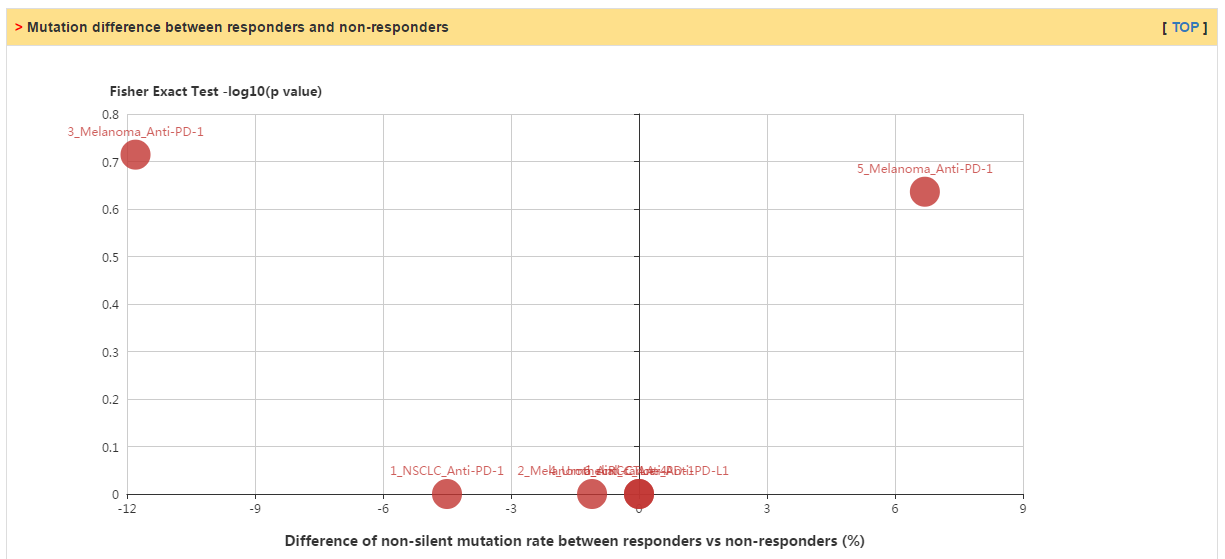
In this tab, users can check whether their selected genes have significant difference of expression or mutation between responders and non-responders in various patient cohorts with immunotherapy. Currently, we collected 5 and 6 of transcriptomic and genomic data sets, respectively.
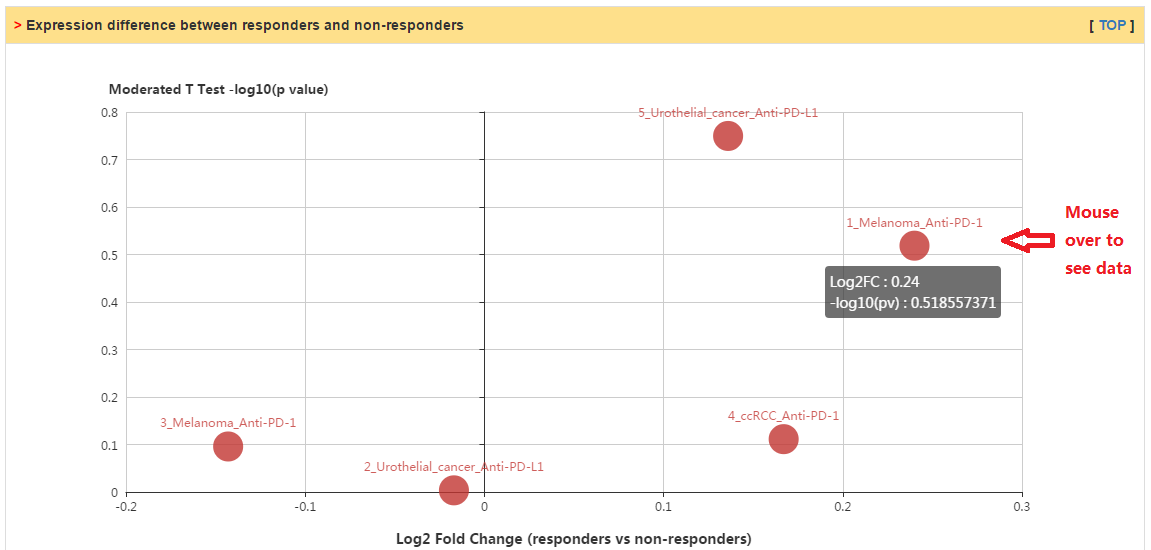
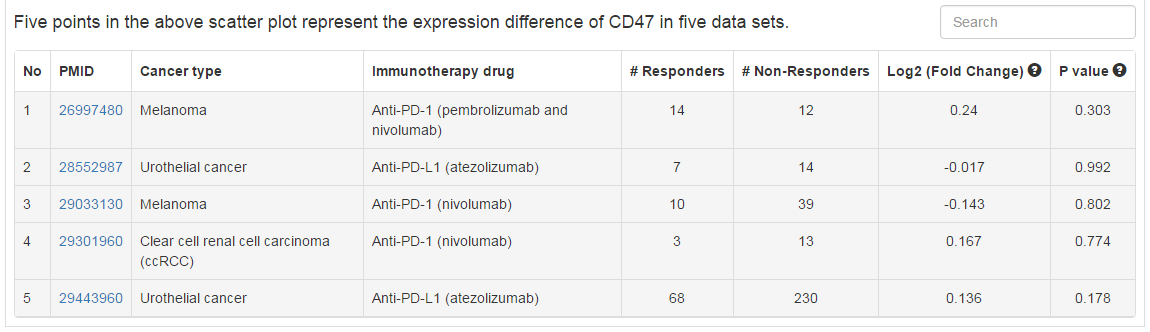 
 |
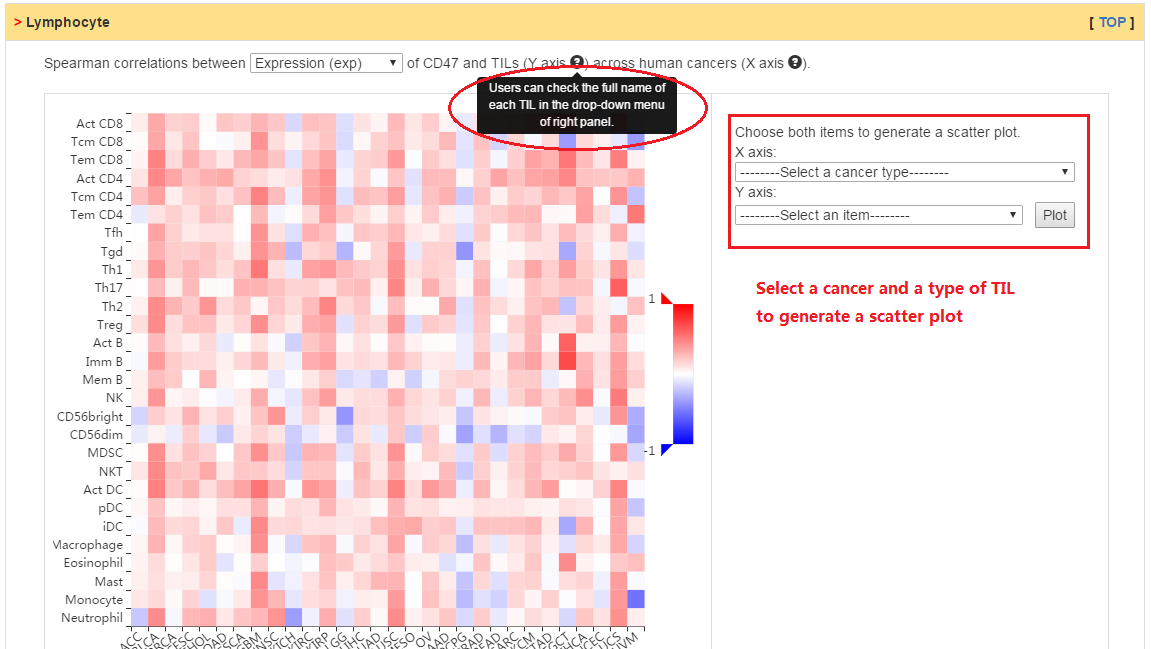
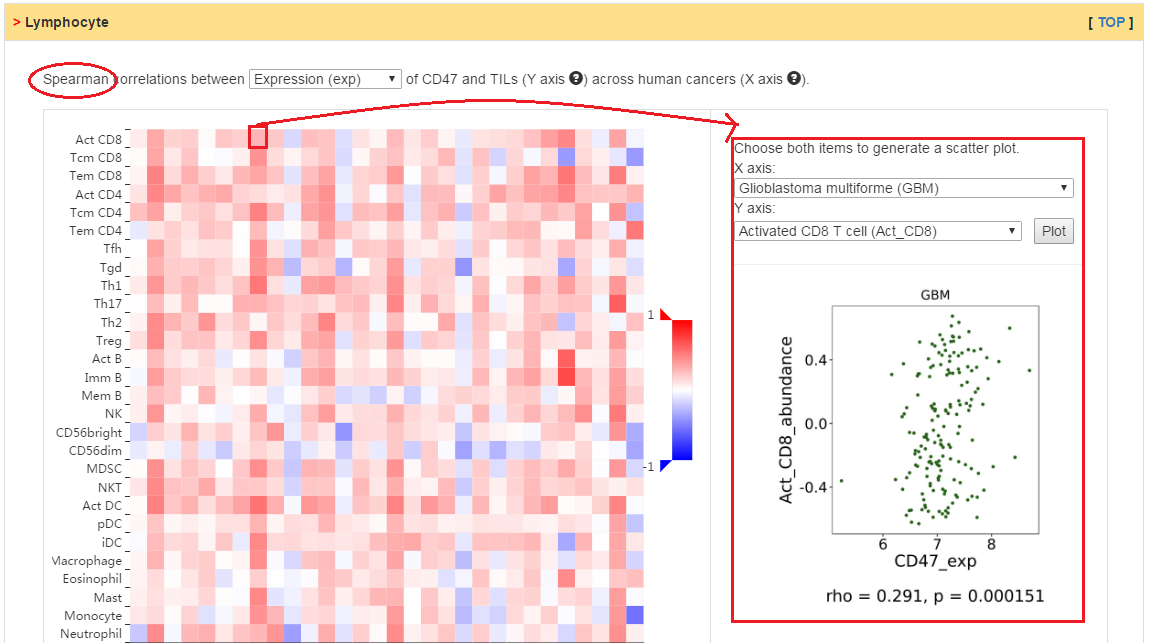
Users can explore the correlation between abundance of tumor-infiltrating lymphocytes (TILs) and expression, copy number, or methylation of current gene.
 |
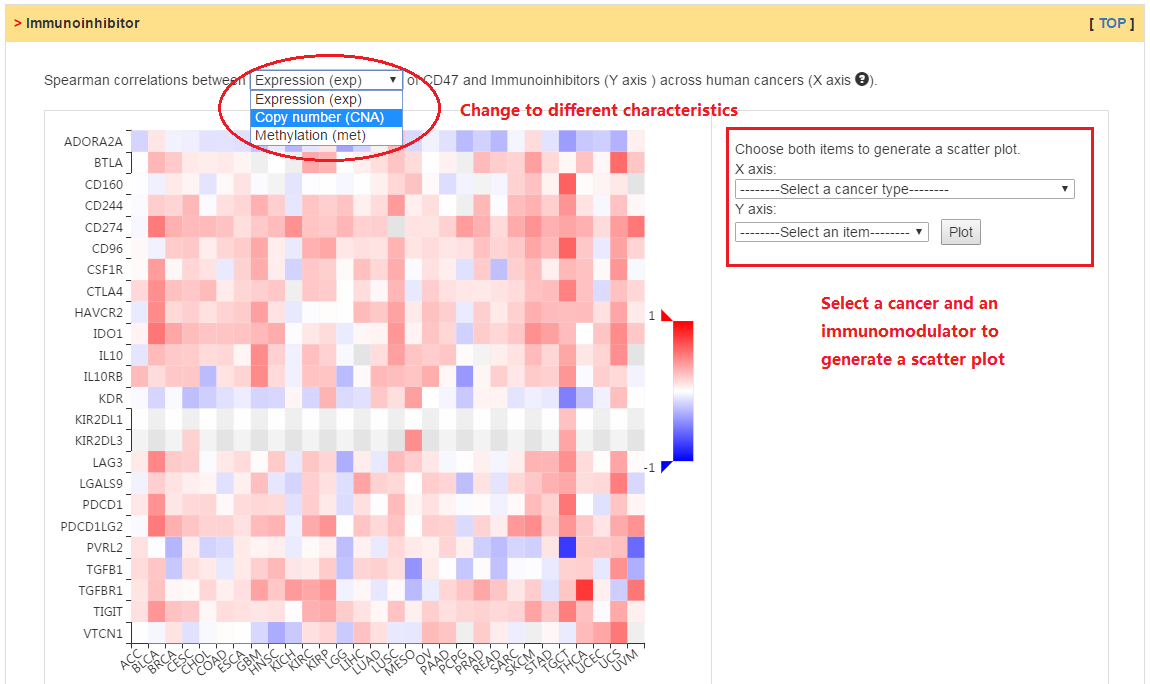
Users can explore the correlation between abundance of immunomodulators (i.e., immunoinhibitor, immunostimulator, and MHC molecules) and expression, copy number, or methylation of current gene.
  |
Users can explore the correlation between abundance of chemokines (or receptors) and expression, copy number, or methylation of current gene.
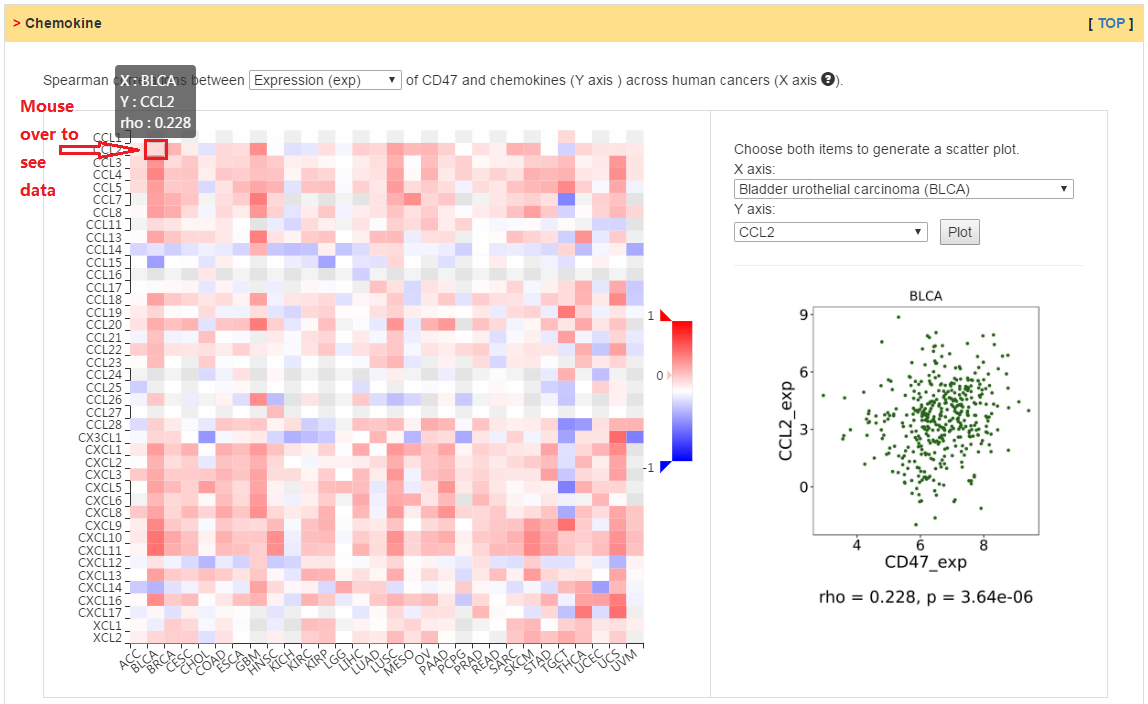 |
Distribution of expression, copy number, and methylation for current gene across various cancer immune and molecular subtypes derived from TCGA datasets.
 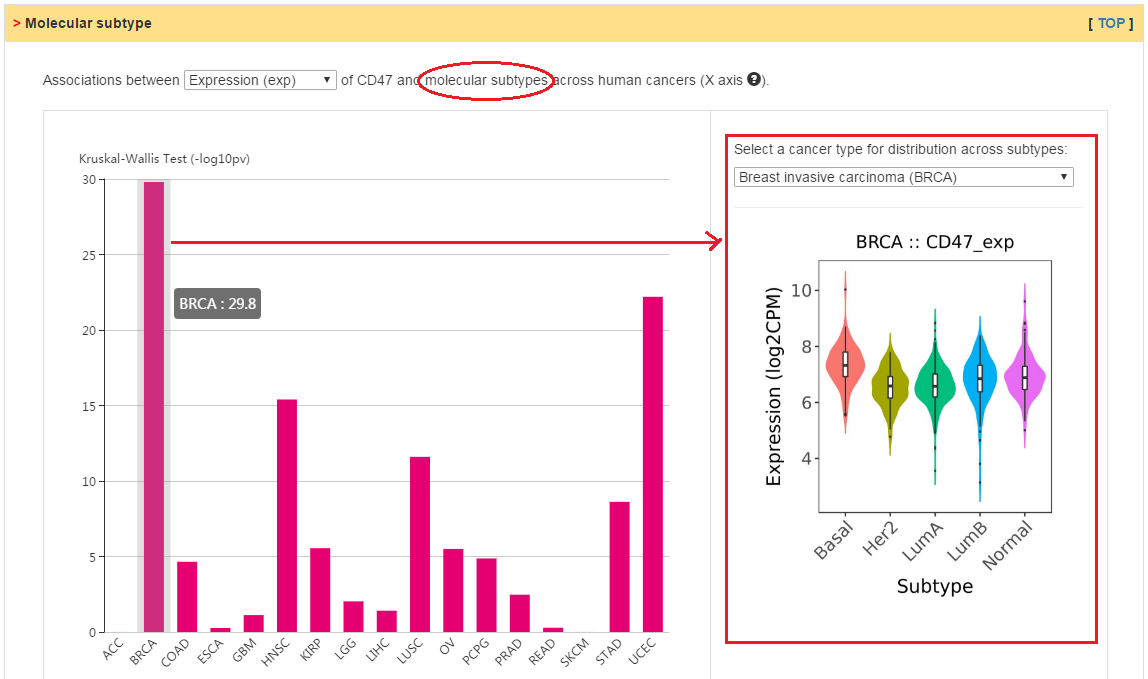 |
Relation between clinical features and the expression, copy number or methylation of current gene.
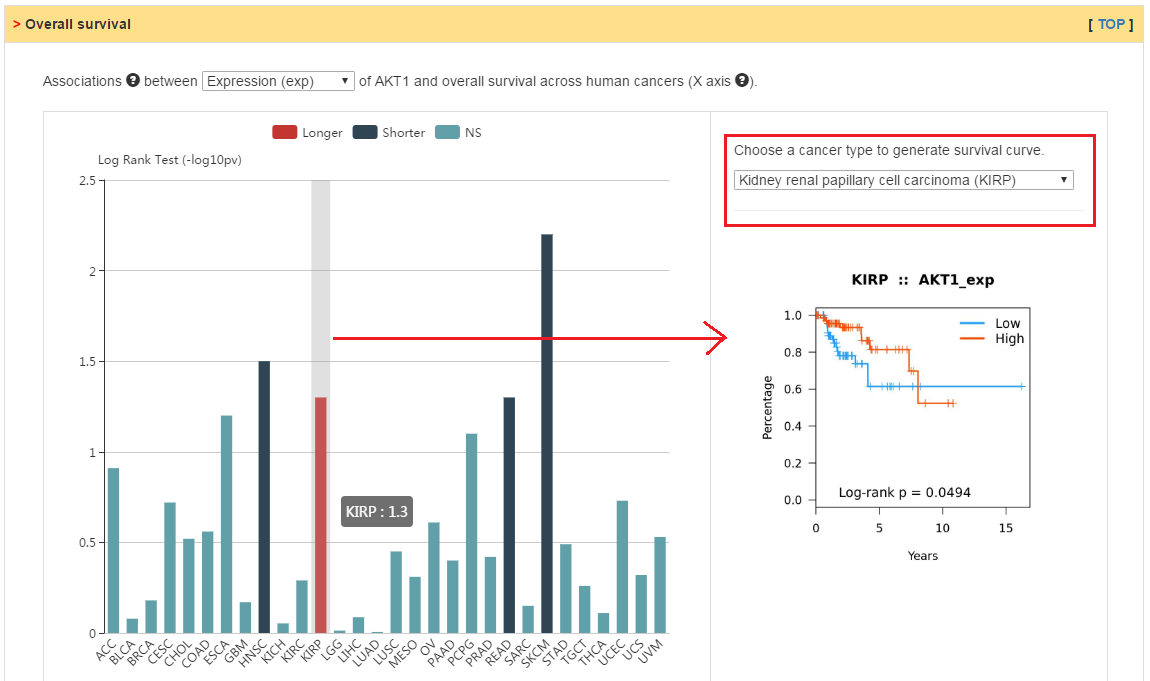 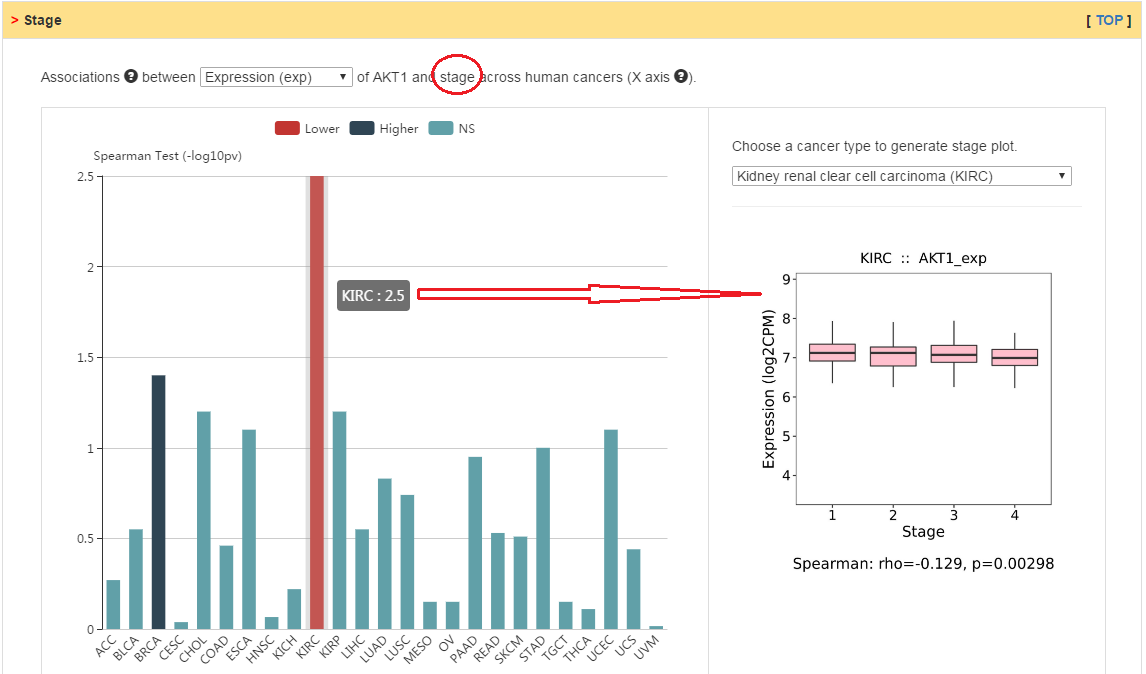 |
Drugs targeting current gene collected from DrugBank database.
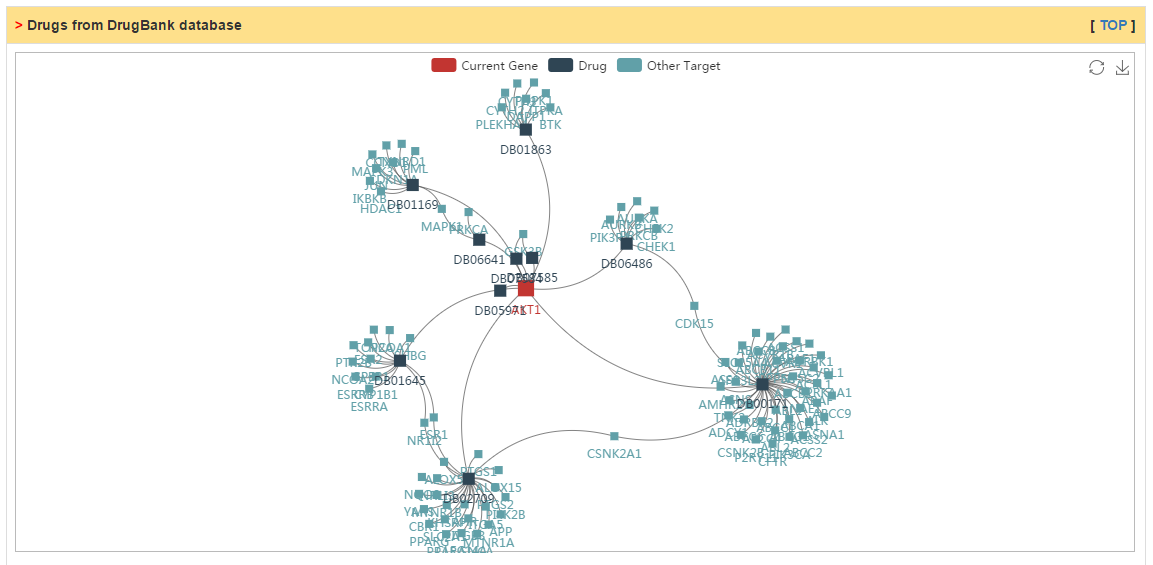
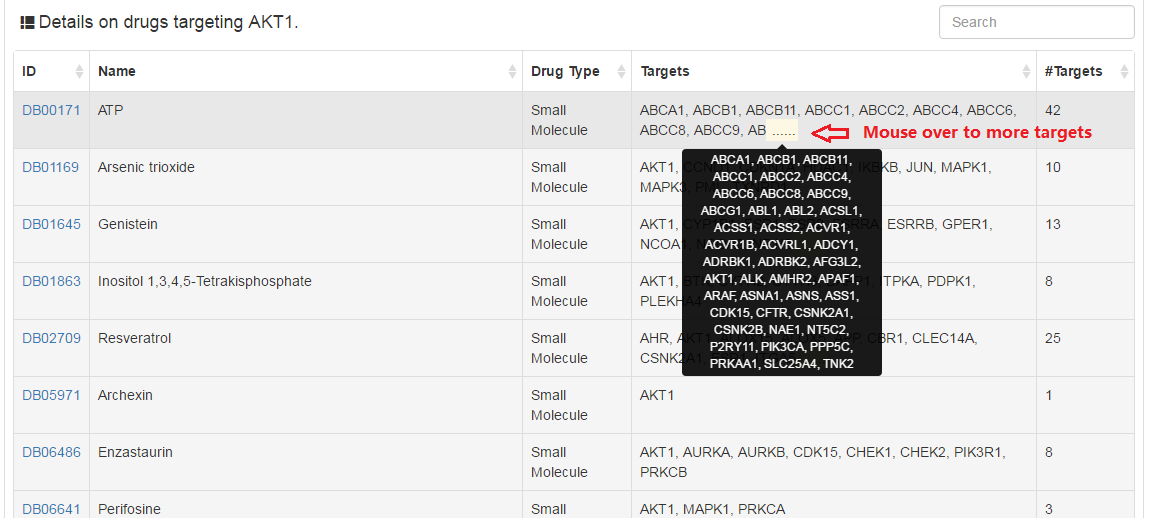 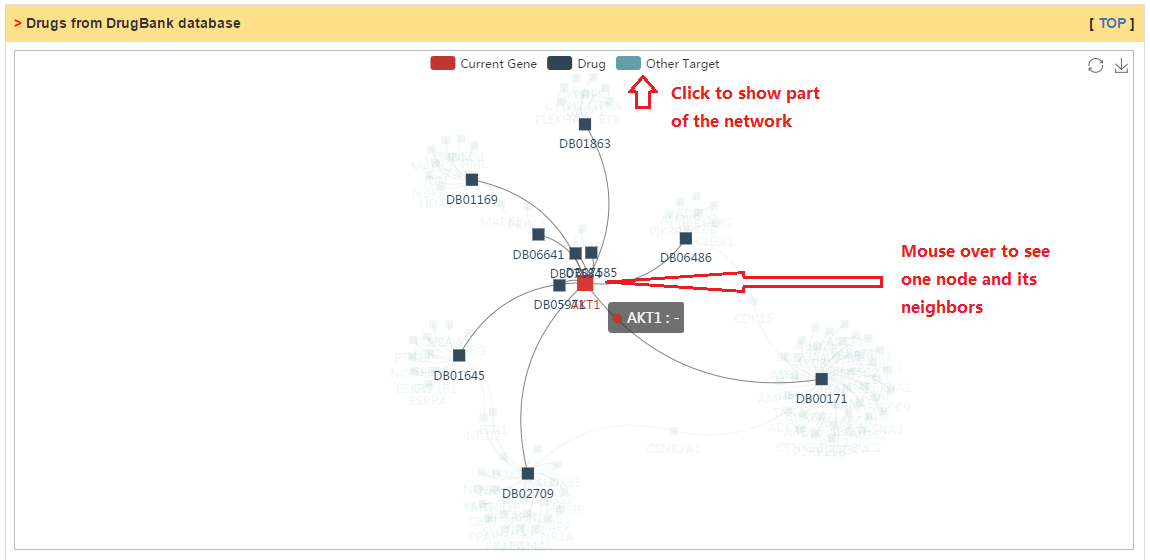 |
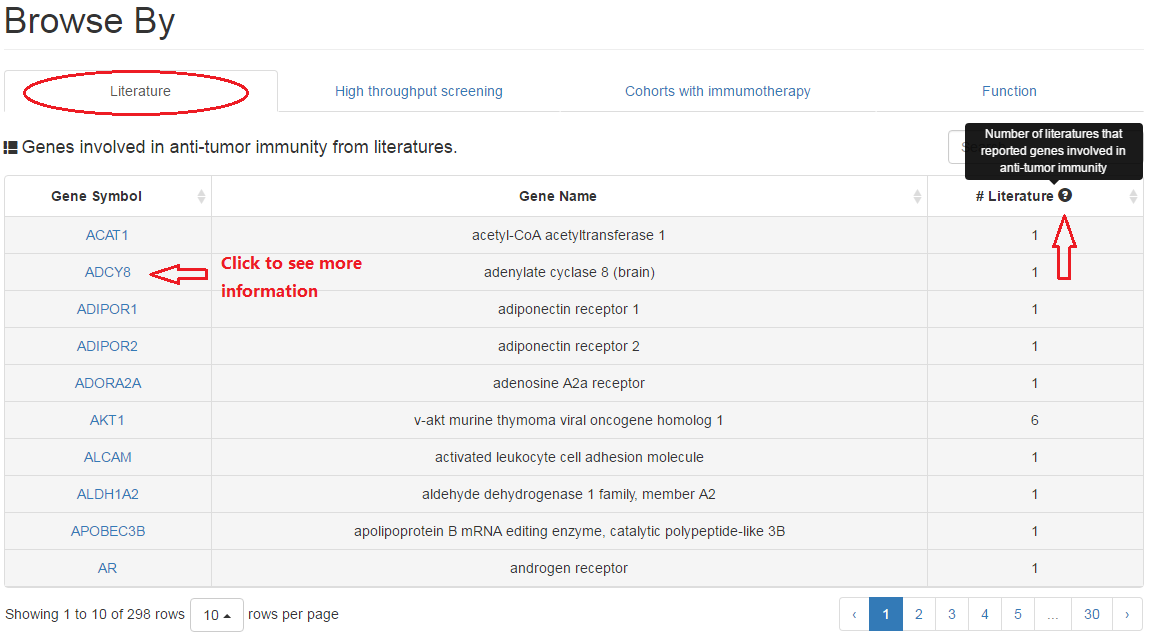 |
 |
 |
 |
|
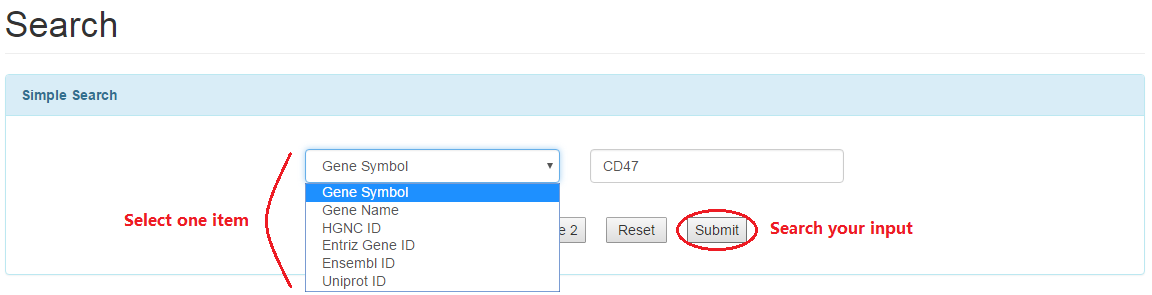
3.1 Simple search: users can search one gene based on basic terms such as gene symbol, ensembl ID and so on.
[ TOP ]
|
|---|
 |
  |
  |