microRNA information: hsa-miR-125b-2-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-125b-2-3p | miRbase |
| Accession: | MIMAT0004603 | miRbase |
| Precursor name: | hsa-mir-125b-2 | miRbase |
| Precursor accession: | MI0000470 | miRbase |
| Symbol: | MIR125B2 | HGNC |
| RefSeq ID: | NR_029694 | GenBank |
| Sequence: | UCACAAGUCAGGCUCUUGGGAC |
Reported expression in cancers: hsa-miR-125b-2-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-125b-2-3p | glioblastoma | downregulation | "In this study we found that miR-125b-2 is overexpr ......" | 21879257 |
Reported cancer pathway affected by hsa-miR-125b-2-3p
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-125b-2-3p | glioblastoma | Apoptosis pathway | "MicroRNA 125b 2 confers human glioblastoma stem ce ......" | 21879257 |
Reported cancer prognosis affected by hsa-miR-125b-2-3p

| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-125b-2-3p | gastric cancer | cell migration | "miR 101 2 miR 125b 2 and miR 451a act as potential ......" | 26458815 | Western blot; Colony formation |
| hsa-miR-125b-2-3p | glioblastoma | drug resistance | "MicroRNA 125b 2 confers human glioblastoma stem ce ......" | 21879257 |
Reported gene related to hsa-miR-125b-2-3p
| miRNA | cancer | gene | reporting | PUBMED |
|---|
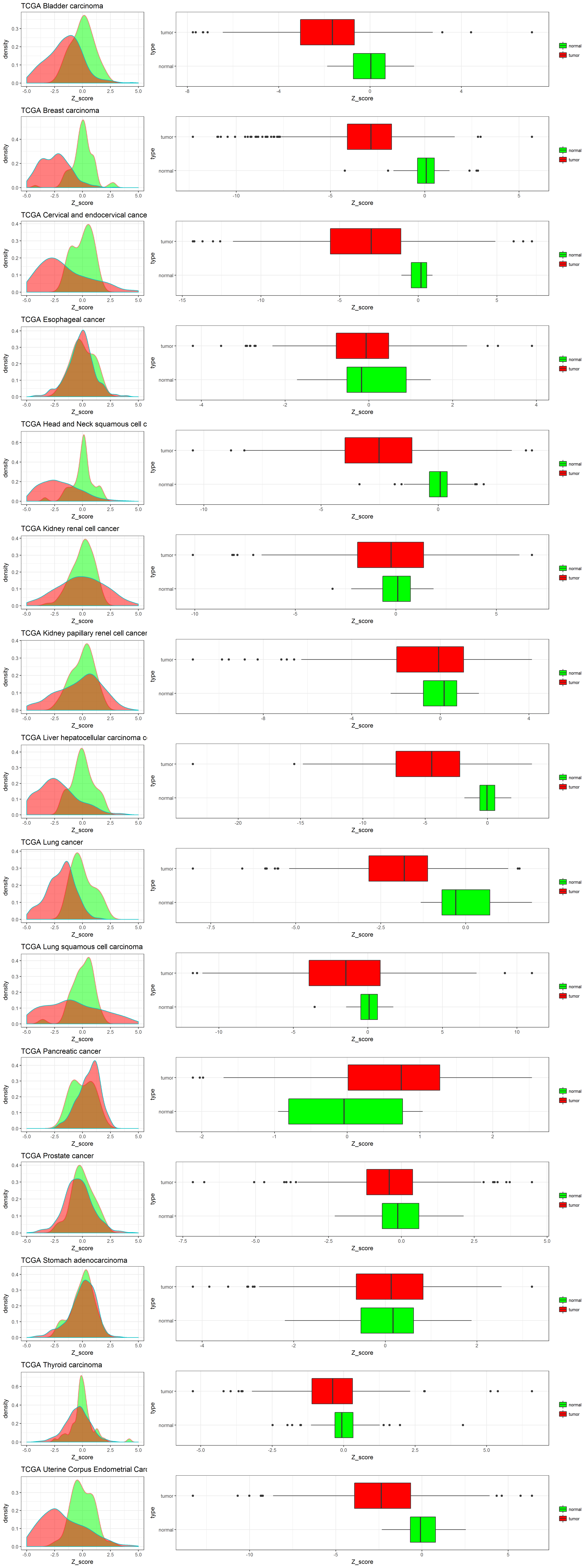
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-125b-2-3p | COX7A2 | 9 cancers: BLCA; BRCA; COAD; ESCA; LGG; LUAD; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.077; TCGA BRCA -0.173; TCGA COAD -0.138; TCGA ESCA -0.058; TCGA LGG -0.097; TCGA LUAD -0.055; TCGA PAAD -0.114; TCGA PRAD -0.259; TCGA STAD -0.171 |
hsa-miR-125b-2-3p | PRR13 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.054; TCGA BRCA -0.144; TCGA CESC -0.066; TCGA COAD -0.119; TCGA ESCA -0.173; TCGA HNSC -0.08; TCGA LIHC -0.065; TCGA LUAD -0.076; TCGA THCA -0.055; TCGA STAD -0.162; TCGA UCEC -0.073 |
hsa-miR-125b-2-3p | SNRPF | 11 cancers: BLCA; BRCA; COAD; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.07; TCGA BRCA -0.136; TCGA COAD -0.136; TCGA HNSC -0.102; TCGA LGG -0.129; TCGA LIHC -0.081; TCGA LUAD -0.15; TCGA LUSC -0.056; TCGA PAAD -0.167; TCGA PRAD -0.25; TCGA STAD -0.144 |
hsa-miR-125b-2-3p | OVCA2 | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; OV; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.07; TCGA BRCA -0.105; TCGA COAD -0.086; TCGA ESCA -0.075; TCGA HNSC -0.06; TCGA LGG -0.106; TCGA LUAD -0.055; TCGA LUSC -0.078; TCGA OV -0.075; TCGA PAAD -0.172; TCGA PRAD -0.104; TCGA THCA -0.083; TCGA STAD -0.084; TCGA UCEC -0.066 |
hsa-miR-125b-2-3p | DAZAP1 | 12 cancers: BLCA; BRCA; COAD; HNSC; KIRP; LGG; LIHC; LUAD; PAAD; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.061; TCGA BRCA -0.092; TCGA COAD -0.107; TCGA HNSC -0.065; TCGA KIRP -0.053; TCGA LGG -0.105; TCGA LIHC -0.102; TCGA LUAD -0.105; TCGA PAAD -0.227; TCGA PRAD -0.218; TCGA THCA -0.094; TCGA STAD -0.109 |
hsa-miR-125b-2-3p | RPUSD3 | 9 cancers: BLCA; BRCA; COAD; ESCA; LGG; LUAD; LUSC; PAAD; STAD | MirTarget | TCGA BLCA -0.052; TCGA BRCA -0.096; TCGA COAD -0.094; TCGA ESCA -0.075; TCGA LGG -0.06; TCGA LUAD -0.056; TCGA LUSC -0.105; TCGA PAAD -0.17; TCGA STAD -0.075 |
hsa-miR-125b-2-3p | GALNT7 | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LIHC; LUAD; OV; STAD; UCEC | MirTarget | TCGA BLCA -0.09; TCGA BRCA -0.263; TCGA COAD -0.113; TCGA ESCA -0.228; TCGA KIRC -0.134; TCGA LIHC -0.226; TCGA LUAD -0.142; TCGA OV -0.075; TCGA STAD -0.16; TCGA UCEC -0.074 |
hsa-miR-125b-2-3p | ELF4 | 9 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; LIHC; LUSC; THCA; STAD | MirTarget | TCGA BLCA -0.116; TCGA ESCA -0.075; TCGA HNSC -0.057; TCGA KIRC -0.12; TCGA KIRP -0.207; TCGA LIHC -0.211; TCGA LUSC -0.054; TCGA THCA -0.255; TCGA STAD -0.097 |
hsa-miR-125b-2-3p | SRD5A3 | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LIHC; LUAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.1; TCGA BRCA -0.126; TCGA COAD -0.102; TCGA ESCA -0.079; TCGA KIRP -0.092; TCGA LIHC -0.065; TCGA LUAD -0.119; TCGA PRAD -0.158; TCGA THCA -0.121; TCGA STAD -0.161; TCGA UCEC -0.169 |
hsa-miR-125b-2-3p | RPL37 | 10 cancers: BLCA; CESC; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD | mirMAP | TCGA BLCA -0.077; TCGA CESC -0.076; TCGA HNSC -0.083; TCGA KIRP -0.079; TCGA LGG -0.155; TCGA LIHC -0.122; TCGA LUAD -0.109; TCGA LUSC -0.062; TCGA PAAD -0.166; TCGA PRAD -0.273 |
hsa-miR-125b-2-3p | SLC19A1 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; PAAD; PRAD; STAD | mirMAP | TCGA BLCA -0.135; TCGA BRCA -0.184; TCGA CESC -0.185; TCGA COAD -0.116; TCGA ESCA -0.128; TCGA HNSC -0.075; TCGA LGG -0.114; TCGA PAAD -0.185; TCGA PRAD -0.244; TCGA STAD -0.245 |
hsa-miR-125b-2-3p | PCGF3 | 9 cancers: BLCA; BRCA; KIRC; LGG; LIHC; LUAD; PAAD; PRAD; STAD | mirMAP | TCGA BLCA -0.155; TCGA BRCA -0.057; TCGA KIRC -0.148; TCGA LGG -0.091; TCGA LIHC -0.118; TCGA LUAD -0.128; TCGA PAAD -0.172; TCGA PRAD -0.148; TCGA STAD -0.062 |
hsa-miR-125b-2-3p | MAFK | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; UCEC | mirMAP | TCGA BLCA -0.067; TCGA CESC -0.106; TCGA ESCA -0.095; TCGA HNSC -0.062; TCGA KIRP -0.068; TCGA LUAD -0.185; TCGA LUSC -0.137; TCGA PAAD -0.213; TCGA PRAD -0.132; TCGA SARC -0.134; TCGA UCEC -0.084 |
hsa-miR-125b-2-3p | HN1L | 10 cancers: BRCA; COAD; HNSC; LIHC; LUAD; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.23; TCGA COAD -0.09; TCGA HNSC -0.063; TCGA LIHC -0.082; TCGA LUAD -0.051; TCGA PAAD -0.141; TCGA PRAD -0.111; TCGA SARC -0.092; TCGA STAD -0.11; TCGA UCEC -0.066 |
hsa-miR-125b-2-3p | NUF2 | 14 cancers: BRCA; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.566; TCGA COAD -0.126; TCGA HNSC -0.119; TCGA KIRC -0.27; TCGA KIRP -0.315; TCGA LGG -0.19; TCGA LIHC -0.742; TCGA LUAD -0.533; TCGA LUSC -0.183; TCGA PAAD -0.174; TCGA PRAD -0.38; TCGA THCA -0.166; TCGA STAD -0.296; TCGA UCEC -0.123 |
hsa-miR-125b-2-3p | LLPH | 9 cancers: BRCA; CESC; COAD; HNSC; KIRP; LIHC; LUAD; PRAD; STAD | MirTarget | TCGA BRCA -0.103; TCGA CESC -0.065; TCGA COAD -0.098; TCGA HNSC -0.109; TCGA KIRP -0.066; TCGA LIHC -0.066; TCGA LUAD -0.095; TCGA PRAD -0.086; TCGA STAD -0.085 |
hsa-miR-125b-2-3p | FAM104A | 10 cancers: BRCA; COAD; KIRC; KIRP; LGG; LIHC; LUAD; OV; PAAD; STAD | MirTarget | TCGA BRCA -0.124; TCGA COAD -0.076; TCGA KIRC -0.053; TCGA KIRP -0.083; TCGA LGG -0.084; TCGA LIHC -0.068; TCGA LUAD -0.107; TCGA OV -0.052; TCGA PAAD -0.109; TCGA STAD -0.055 |
hsa-miR-125b-2-3p | CPD | 9 cancers: BRCA; CESC; ESCA; LIHC; LUAD; OV; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.067; TCGA CESC -0.085; TCGA ESCA -0.171; TCGA LIHC -0.225; TCGA LUAD -0.191; TCGA OV -0.078; TCGA THCA -0.087; TCGA STAD -0.122; TCGA UCEC -0.097 |
hsa-miR-125b-2-3p | ZNF706 | 10 cancers: BRCA; COAD; HNSC; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD | mirMAP | TCGA BRCA -0.1; TCGA COAD -0.105; TCGA HNSC -0.081; TCGA LIHC -0.153; TCGA LUAD -0.137; TCGA LUSC -0.081; TCGA OV -0.093; TCGA PAAD -0.078; TCGA PRAD -0.083; TCGA STAD -0.131 |
hsa-miR-125b-2-3p | PHKG2 | 9 cancers: BRCA; HNSC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.179; TCGA HNSC -0.057; TCGA LUAD -0.119; TCGA LUSC -0.07; TCGA PAAD -0.192; TCGA PRAD -0.154; TCGA THCA -0.075; TCGA STAD -0.096; TCGA UCEC -0.06 |
hsa-miR-125b-2-3p | NIPA1 | 9 cancers: BRCA; COAD; ESCA; KIRC; LIHC; LUAD; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.206; TCGA COAD -0.102; TCGA ESCA -0.122; TCGA KIRC -0.07; TCGA LIHC -0.165; TCGA LUAD -0.101; TCGA SARC -0.179; TCGA STAD -0.134; TCGA UCEC -0.142 |
hsa-miR-125b-2-3p | GMPPB | 9 cancers: BRCA; COAD; ESCA; LUAD; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.152; TCGA COAD -0.078; TCGA ESCA -0.132; TCGA LUAD -0.086; TCGA PAAD -0.138; TCGA SARC -0.064; TCGA THCA -0.198; TCGA STAD -0.177; TCGA UCEC -0.085 |
hsa-miR-125b-2-3p | GNB1L | 9 cancers: BRCA; HNSC; LGG; LIHC; LUAD; PAAD; PRAD; THCA; STAD | mirMAP | TCGA BRCA -0.067; TCGA HNSC -0.116; TCGA LGG -0.076; TCGA LIHC -0.148; TCGA LUAD -0.095; TCGA PAAD -0.248; TCGA PRAD -0.097; TCGA THCA -0.079; TCGA STAD -0.096 |
hsa-miR-125b-2-3p | SLC25A37 | 9 cancers: CESC; COAD; HNSC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | mirMAP | TCGA CESC -0.072; TCGA COAD -0.111; TCGA HNSC -0.104; TCGA LUAD -0.187; TCGA LUSC -0.077; TCGA PRAD -0.153; TCGA THCA -0.196; TCGA STAD -0.14; TCGA UCEC -0.095 |
hsa-miR-125b-2-3p | JMJD6 | 10 cancers: COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; OV; PAAD; SARC | mirMAP | TCGA COAD -0.059; TCGA ESCA -0.074; TCGA HNSC -0.09; TCGA LGG -0.055; TCGA LIHC -0.162; TCGA LUAD -0.135; TCGA LUSC -0.052; TCGA OV -0.053; TCGA PAAD -0.102; TCGA SARC -0.111 |
hsa-miR-125b-2-3p | TLL2 | 9 cancers: ESCA; HNSC; KIRP; LUAD; LUSC; OV; PRAD; THCA; STAD | mirMAP | TCGA ESCA -0.296; TCGA HNSC -0.122; TCGA KIRP -0.274; TCGA LUAD -0.196; TCGA LUSC -0.229; TCGA OV -0.167; TCGA PRAD -0.36; TCGA THCA -0.373; TCGA STAD -0.194 |
hsa-miR-125b-2-3p | ACTR3 | 10 cancers: KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA KIRC -0.083; TCGA KIRP -0.072; TCGA LIHC -0.057; TCGA LUAD -0.071; TCGA LUSC -0.092; TCGA PRAD -0.074; TCGA SARC -0.099; TCGA THCA -0.091; TCGA STAD -0.06; TCGA UCEC -0.051 |
Enriched cancer pathways of putative targets