microRNA information: hsa-miR-155-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-155-5p | miRbase |
| Accession: | MIMAT0000646 | miRbase |
| Precursor name: | hsa-mir-155 | miRbase |
| Precursor accession: | MI0000681 | miRbase |
| Symbol: | MIR155 | HGNC |
| RefSeq ID: | NR_030784 | GenBank |
| Sequence: | UUAAUGCUAAUCGUGAUAGGGGU |
Reported expression in cancers: hsa-miR-155-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-155-5p | B cell lymphoma | upregulation | "BIC and miR 155 are highly expressed in Hodgkin pr ......" | 16041695 | qPCR; Northern blot |
| hsa-miR-155-5p | B cell lymphoma | deregulation | "Coordinated expression of microRNA 155 and predict ......" | 18262046 | Microarray |
| hsa-miR-155-5p | B cell lymphoma | deregulation | "Here we report a comprehensive miRNA-profiling stu ......" | 21062812 | |
| hsa-miR-155-5p | B cell lymphoma | downregulation | "We measured the levels of miRNAs miR-15a miR-16-1 ......" | 21987025 | Reverse transcription PCR; qPCR |
| hsa-miR-155-5p | B cell lymphoma | deregulation | "Quantitative proteomics reveals that miR 155 regul ......" | 22609116 | |
| hsa-miR-155-5p | B cell lymphoma | downregulation | "A set of 11 miRNAs associated with the differentia ......" | 22936066 | qPCR |
| hsa-miR-155-5p | B cell lymphoma | upregulation | "We also identified predictive miRNA biomarker sign ......" | 25498913 | |
| hsa-miR-155-5p | acute myeloid leukemia | upregulation | "FLT3-ITD+ samples were characterized by up-regulat ......" | 18308931 | |
| hsa-miR-155-5p | acute myeloid leukemia | upregulation | "Expression of miR-155 was not related to cytogenet ......" | 21818844 | |
| hsa-miR-155-5p | acute myeloid leukemia | upregulation | "Clinical role of microRNAs in cytogenetically norm ......" | 23650424 | Microarray |
| hsa-miR-155-5p | acute myeloid leukemia | upregulation | "MiRNA-155 MiR-155 was found to be upregulated in F ......" | 25092144 | |
| hsa-miR-155-5p | acute myeloid leukemia | upregulation | "By multivariate analysis we identified that high e ......" | 25428263 | |
| hsa-miR-155-5p | acute myeloid leukemia | upregulation | "Our previous data demonstrated specific up-regulat ......" | 26055960 | |
| hsa-miR-155-5p | bladder cancer | upregulation | "Correlation of Increased Expression of MicroRNA 15 ......" | 25918190 | qPCR |
| hsa-miR-155-5p | bladder cancer | deregulation | "In GBC miRNAs with tumor suppressor activity miR-1 ......" | 26855538 | |
| hsa-miR-155-5p | bladder cancer | downregulation | "Moreover miR-155 was downregulated whereas miR-200 ......" | 26927195 | |
| hsa-miR-155-5p | breast cancer | upregulation | "miR-155 microRNA-155 is frequently up-regulated in ......" | 20371610 | |
| hsa-miR-155-5p | breast cancer | upregulation | "Clinical significance of miR 155 expression in bre ......" | 22245916 | qPCR |
| hsa-miR-155-5p | breast cancer | upregulation | "Correlation of miR 155 on formalin fixed paraffin ......" | 23302487 | qPCR |
| hsa-miR-155-5p | breast cancer | upregulation | "MicroRNA-155 miR-155 is frequently upregulated in ......" | 23353819 | |
| hsa-miR-155-5p | breast cancer | upregulation | "MiR-155 has emerged as an "oncomiR" which is the m ......" | 23568502 | |
| hsa-miR-155-5p | breast cancer | upregulation | "MiR-155 has emerged as an "oncomiR" which is the m ......" | 24152184 | |
| hsa-miR-155-5p | breast cancer | upregulation | "In this study we identified that miR-155 is freque ......" | 26692956 | |
| hsa-miR-155-5p | cervical and endocervical cancer | upregulation | "In this study we aimed to investigate the biologic ......" | 25155037 | qPCR |
| hsa-miR-155-5p | cervical and endocervical cancer | upregulation | "Up regulated microRNA 155 expression is associated ......" | 27470551 | qPCR |
| hsa-miR-155-5p | chronic myeloid leukemia | deregulation | "Some onco-miRNAs were found to be downregulated mi ......" | 25833191 | |
| hsa-miR-155-5p | colon cancer | upregulation | "The expression of miR-155 in both tissues was test ......" | 24888652 | Reverse transcription PCR |
| hsa-miR-155-5p | colon cancer | upregulation | "MicroRNA 155 deletion promotes tumorigenesis in th ......" | 26744471 | |
| hsa-miR-155-5p | colorectal cancer | upregulation | "Human microRNA-155 miR-155 has been demonstrated t ......" | 23588589 | qPCR |
| hsa-miR-155-5p | colorectal cancer | upregulation | "In order to clarify the miRNA profile in CRC tissu ......" | 25421755 | Microarray |
| hsa-miR-155-5p | colorectal cancer | upregulation | "Investigation of microRNA 155 as a serum diagnosti ......" | 25528214 | Reverse transcription PCR; qPCR |
| hsa-miR-155-5p | gastric cancer | upregulation | "The expression levels of miR-20a miR-21 miR-25 miR ......" | 22996433 | qPCR |
| hsa-miR-155-5p | gastric cancer | upregulation | "Elevated expression of mature miR 21 and miR 155 i ......" | 23554630 | |
| hsa-miR-155-5p | gastric cancer | downregulation | "The clinical significance of downregulation of mir ......" | 24805774 | Microarray |
| hsa-miR-155-5p | gastric cancer | deregulation | "The level of miR-155 in 52 gastric carcinoma and c ......" | 24969405 | |
| hsa-miR-155-5p | gastric cancer | downregulation | "Inactivation of NF-κB by pyrrolidine dithiocarbam ......" | 26934326 | |
| hsa-miR-155-5p | head and neck cancer | downregulation | "Characterization of miR 146a and miR 155 in blood ......" | 26621153 | qPCR; Microarray |
| hsa-miR-155-5p | kidney renal cell cancer | upregulation | "We selected 28 clear-cell type human renal cell ca ......" | 20035975 | qPCR |
| hsa-miR-155-5p | kidney renal cell cancer | downregulation | "We analyzed 70 matched pairs of clear cell renal c ......" | 21784468 | qPCR; Microarray |
| hsa-miR-155-5p | liver cancer | upregulation | "Expression and survival prediction of microRNA 155 ......" | 23863669 | Reverse transcription PCR; qPCR |
| hsa-miR-155-5p | liver cancer | upregulation | "In liver cancer miR-155 up-regulation can regulate ......" | 25601564 | |
| hsa-miR-155-5p | liver cancer | upregulation | "The insulin-like growth factor IGF axis is importa ......" | 26722313 | qPCR |
| hsa-miR-155-5p | liver cancer | upregulation | "Aberrant expression of microRNA-155 miR-155 has be ......" | 27035278 | |
| hsa-miR-155-5p | liver cancer | upregulation | "MiR 155 and its functional variant rs767649 contri ......" | 27531892 | |
| hsa-miR-155-5p | lung cancer | upregulation | "The expression of 319 miRNAs was evaluated by Exiq ......" | 20818338 | Microarray |
| hsa-miR-155-5p | lung cancer | upregulation | "Potential diagnostic value of miR 155 in serum fro ......" | 24190459 | |
| hsa-miR-155-5p | lung cancer | upregulation | "Meta analysis of the association between the expre ......" | 25603615 | |
| hsa-miR-155-5p | lung cancer | upregulation | "A functional variant in miR 155 regulation region ......" | 26543233 | |
| hsa-miR-155-5p | lung cancer | upregulation | "Two miRNAs miR-21 and miR-155 were found to be sig ......" | 26867772 | |
| hsa-miR-155-5p | lung squamous cell cancer | upregulation | "Hsa-miR-155 miR-155 highly expressed in non-small- ......" | 22796123 | |
| hsa-miR-155-5p | lung squamous cell cancer | upregulation | "Association of MiR 155 expression with prognosis i ......" | 24854560 | qPCR |
| hsa-miR-155-5p | lymphoma | upregulation | "Using quantitative reverse transcription-polymeras ......" | 19177201 | Reverse transcription PCR; Microarray |
| hsa-miR-155-5p | lymphoma | upregulation | "The up-regulation of miR-155 miR-27b miR-30c and m ......" | 22776000 | Microarray |
| hsa-miR-155-5p | lymphoma | upregulation | "Overexpression of miR 142 5p and miR 155 in gastri ......" | 23209550 | Microarray |
| hsa-miR-155-5p | melanoma | upregulation | "Moreover as expected some discrepancies were also ......" | 20442294 | qPCR; Microarray |
| hsa-miR-155-5p | melanoma | upregulation | "Here we show that downregulation of MITF-M express ......" | 25853464 | |
| hsa-miR-155-5p | ovarian cancer | deregulation | "We found that in ovarian CAFs miR-31 and miR-214 w ......" | 23171795 | |
| hsa-miR-155-5p | pancreatic cancer | downregulation | "Eighty-eight samples of ductal pancreatic adenocar ......" | 22850622 | qPCR |
| hsa-miR-155-5p | pancreatic cancer | downregulation | "Four miRNAs miR-17-5p miR-21 miR-155 and miR-196a ......" | 24007214 | |
| hsa-miR-155-5p | prostate cancer | upregulation | "miR-155 has been implicated in the induction of br ......" | 25339368 | Reverse transcription PCR |
| hsa-miR-155-5p | prostate cancer | downregulation | "Among them 10 paired HGPIN and PCa were prepared t ......" | 27017949 | qPCR; Microarray |
| hsa-miR-155-5p | sarcoma | upregulation | "We found that KSHV-miR-K12-11 shares 100% seed seq ......" | 17881434 | |
| hsa-miR-155-5p | sarcoma | upregulation | "It has been demonstrated that mir-155 was the most ......" | 25888631 | |
| hsa-miR-155-5p | thyroid cancer | deregulation | "In this study we evaluated miRNA expression as a m ......" | 21537871 | Microarray |
Reported cancer pathway affected by hsa-miR-155-5p
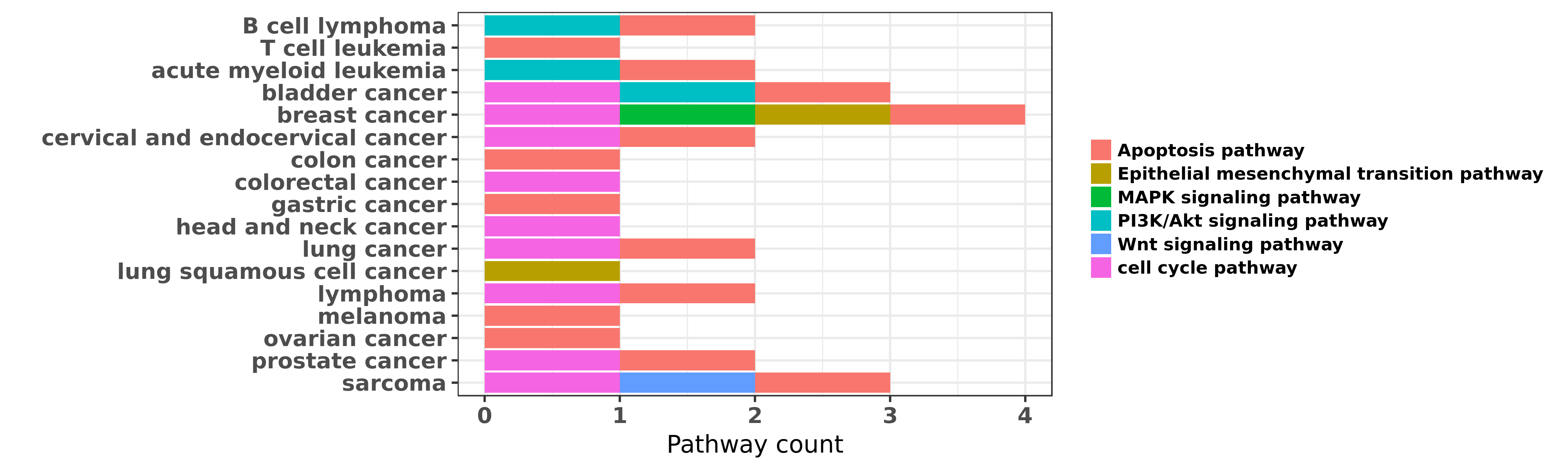
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-155-5p | B cell lymphoma | Apoptosis pathway | "Epstein Barr virus latent membrane protein 1 prote ......" | 22268450 | |
| hsa-miR-155-5p | B cell lymphoma | PI3K/Akt signaling pathway | "Quantitative proteomics reveals that miR 155 regul ......" | 22609116 | Luciferase |
| hsa-miR-155-5p | T cell leukemia | Apoptosis pathway | "MiR-155 is a kind of molecule with different funct ......" | 25598954 | MTT assay |
| hsa-miR-155-5p | acute myeloid leukemia | Apoptosis pathway | "Silvestrol exhibits significant in vivo and in vit ......" | 23497456 | Flow cytometry; RNA Immunoprecipitation; RNA immunoprecipitation; Colony formation |
| hsa-miR-155-5p | acute myeloid leukemia | Apoptosis pathway | "Clinical role of microRNAs in cytogenetically norm ......" | 23650424 | |
| hsa-miR-155-5p | acute myeloid leukemia | Apoptosis pathway | "MiRNA-155 MiR-155 was found to be upregulated in F ......" | 25092144 | |
| hsa-miR-155-5p | acute myeloid leukemia | Apoptosis pathway; PI3K/Akt signaling pathway | "SHIP1 is targeted by miR 155 in acute myeloid leuk ......" | 25175984 | |
| hsa-miR-155-5p | acute myeloid leukemia | Apoptosis pathway | "Pharmacological targeting of miR 155 via the NEDD8 ......" | 25971362 | |
| hsa-miR-155-5p | bladder cancer | cell cycle pathway | "MicroRNA 155 promotes bladder cancer growth by rep ......" | 25965824 | |
| hsa-miR-155-5p | bladder cancer | PI3K/Akt signaling pathway; Apoptosis pathway | "Hedyotis diffusa plus Scutellaria barbata Induce B ......" | 26989427 | |
| hsa-miR-155-5p | breast cancer | Apoptosis pathway | "MicroRNA 155 regulates cell survival growth and ch ......" | 20371610 | |
| hsa-miR-155-5p | breast cancer | Apoptosis pathway | "Clinical significance of microRNA 155 expression i ......" | 22105810 | |
| hsa-miR-155-5p | breast cancer | Apoptosis pathway | "Clinical significance of miR 155 expression in bre ......" | 22245916 | Flow cytometry |
| hsa-miR-155-5p | breast cancer | Apoptosis pathway | "17β estradiol up regulates miR 155 expression and ......" | 23568502 | |
| hsa-miR-155-5p | breast cancer | Apoptosis pathway | "Effects of miR 155 antisense oligonucleotide on br ......" | 23725141 | |
| hsa-miR-155-5p | breast cancer | Epithelial mesenchymal transition pathway | "MiR 155 mediated loss of C/EBPβ shifts the TGF β ......" | 23955085 | |
| hsa-miR-155-5p | breast cancer | cell cycle pathway; Apoptosis pathway | "MiR 155 promotes proliferation of human breast can ......" | 24152184 | Luciferase |
| hsa-miR-155-5p | breast cancer | Epithelial mesenchymal transition pathway | "Some miRNAs especially the miR-200 family miR-9 an ......" | 25086633 | |
| hsa-miR-155-5p | breast cancer | Apoptosis pathway; MAPK signaling pathway | "miR 155 induced transcriptome changes in the MCF 7 ......" | 25352952 | |
| hsa-miR-155-5p | breast cancer | MAPK signaling pathway | "Role of miR 155 in drug resistance of breast cance ......" | 25744731 | |
| hsa-miR-155-5p | breast cancer | Apoptosis pathway | "miR 155 5p antagonizes the apoptotic effect of buf ......" | 26398931 | |
| hsa-miR-155-5p | breast cancer | Apoptosis pathway | "In this study we identified that miR-155 is freque ......" | 26692956 | |
| hsa-miR-155-5p | breast cancer | Apoptosis pathway | "Deregulation of miR 21 and miR 155 and their putat ......" | 26877850 | |
| hsa-miR-155-5p | breast cancer | Apoptosis pathway | "Silibinin Induced Apoptosis and Downregulation of ......" | 27066095 | |
| hsa-miR-155-5p | cervical and endocervical cancer | cell cycle pathway; Apoptosis pathway | "Mir 155 promotes cervical cancer cell proliferatio ......" | 25155037 | MTT assay; Flow cytometry; Luciferase |
| hsa-miR-155-5p | colon cancer | Apoptosis pathway | "Adrenaline promotes cell proliferation and increas ......" | 23036199 | |
| hsa-miR-155-5p | colorectal cancer | cell cycle pathway | "miR 155 regulates the proliferation and cell cycle ......" | 24793496 | Western blot; Luciferase |
| hsa-miR-155-5p | gastric cancer | Apoptosis pathway | "MicroRNA 155 expression inversely correlates with ......" | 26955820 | |
| hsa-miR-155-5p | head and neck cancer | cell cycle pathway | "A global miRNA profiling was done on 51 formalin-f ......" | 20145181 | Flow cytometry |
| hsa-miR-155-5p | lung cancer | cell cycle pathway | "Effects of microRNA 155 on the growth of human lun ......" | 21762626 | MTT assay; Flow cytometry; Western blot |
| hsa-miR-155-5p | lung cancer | Apoptosis pathway | "MiR 155 inhibits the sensitivity of lung cancer ce ......" | 22996741 | |
| hsa-miR-155-5p | lung cancer | cell cycle pathway; Apoptosis pathway | "Effect of miR 155 knockdown on the reversal of dox ......" | 26893712 | Western blot |
| hsa-miR-155-5p | lung squamous cell cancer | Epithelial mesenchymal transition pathway | "Retrovirus-mediated RNA interference was employed ......" | 25373388 | RNAi; Western blot; Luciferase |
| hsa-miR-155-5p | lymphoma | cell cycle pathway | "Targeting of SMAD5 links microRNA 155 to the TGF b ......" | 20133617 | RNAi |
| hsa-miR-155-5p | lymphoma | Apoptosis pathway | "Nanoparticle based therapy in an in vivo microRNA ......" | 22685206 | |
| hsa-miR-155-5p | lymphoma | Apoptosis pathway | "Aberrant expression of oncogenic miRNAs including ......" | 24479800 | |
| hsa-miR-155-5p | lymphoma | Apoptosis pathway | "The expression of miRNA-155 was detected in lympho ......" | 25480585 | Flow cytometry; Western blot |
| hsa-miR-155-5p | melanoma | Apoptosis pathway | "MicroRNA 155 targets the SKI gene in human melanom ......" | 21466664 | Luciferase |
| hsa-miR-155-5p | ovarian cancer | Apoptosis pathway | "MicroRNA 155 promotes apoptosis in SKOV3 A2780 and ......" | 26779627 | Western blot; Luciferase |
| hsa-miR-155-5p | prostate cancer | cell cycle pathway; Apoptosis pathway | "microRNA 155 promotes the proliferation of prostat ......" | 25339368 | MTT assay; Western blot; Luciferase |
| hsa-miR-155-5p | sarcoma | cell cycle pathway | "MiR 155 is a liposarcoma oncogene that targets cas ......" | 22350414 | Colony formation |
| hsa-miR-155-5p | sarcoma | cell cycle pathway; Wnt signaling pathway | "miR 155 promotes the growth of osteosarcoma in a H ......" | 25666090 | |
| hsa-miR-155-5p | sarcoma | Apoptosis pathway | "Increased miR 155 5p and reduced miR 148a 3p contr ......" | 27041566 |
Reported cancer prognosis affected by hsa-miR-155-5p
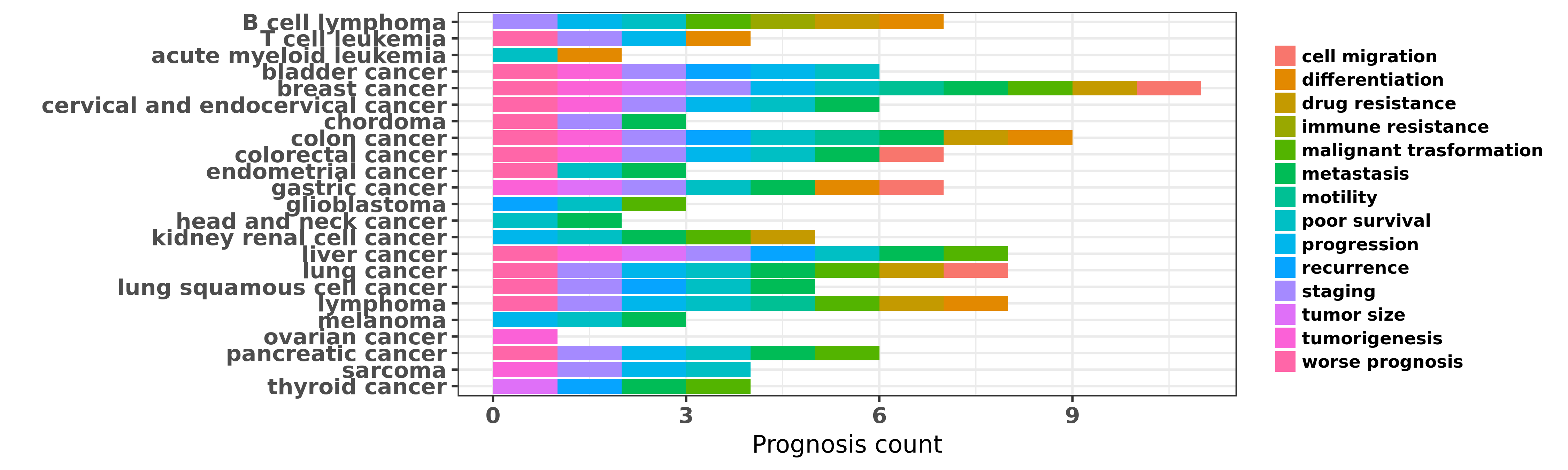
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-155-5p | B cell lymphoma | malignant trasformation | "Using microarray analysis on prototypic cell lines ......" | 17487835 | |
| hsa-miR-155-5p | B cell lymphoma | poor survival; drug resistance | "Epstein Barr virus latent membrane protein 1 prote ......" | 22268450 | |
| hsa-miR-155-5p | B cell lymphoma | staging; differentiation; progression; poor survival | "A set of 11 miRNAs associated with the differentia ......" | 22936066 | |
| hsa-miR-155-5p | B cell lymphoma | immune resistance; poor survival | "miR 155 targets histone deacetylase 4 HDAC4 and im ......" | 23169640 | |
| hsa-miR-155-5p | B cell lymphoma | poor survival | "Differential expression of miR 155 and miR 21 in t ......" | 25265435 | |
| hsa-miR-155-5p | B cell lymphoma | malignant trasformation | "In this review we discuss miRNAs with essential fu ......" | 25541152 | |
| hsa-miR-155-5p | T cell leukemia | progression; worse prognosis; staging | "Impact of miR 155 and miR 126 as novel biomarkers ......" | 22884882 | |
| hsa-miR-155-5p | T cell leukemia | differentiation | "MiR-155 is a kind of molecule with different funct ......" | 25598954 | MTT assay |
| hsa-miR-155-5p | acute myeloid leukemia | differentiation | "We studied miRNA expression of leukemic blasts of ......" | 20425795 | |
| hsa-miR-155-5p | acute myeloid leukemia | poor survival | "Clinical role of microRNAs in cytogenetically norm ......" | 23650424 | |
| hsa-miR-155-5p | acute myeloid leukemia | differentiation | "LIFRα CT3 induces differentiation of a human acut ......" | 25092123 | |
| hsa-miR-155-5p | acute myeloid leukemia | differentiation; poor survival | "Pharmacological targeting of miR 155 via the NEDD8 ......" | 25971362 | |
| hsa-miR-155-5p | acute myeloid leukemia | differentiation | "miR 155 regulative network in FLT3 mutated acute m ......" | 26055960 | |
| hsa-miR-155-5p | bladder cancer | worse prognosis; progression; poor survival | "Correlation of Increased Expression of MicroRNA 15 ......" | 25918190 | |
| hsa-miR-155-5p | bladder cancer | tumorigenesis; progression | "MicroRNA 155 promotes bladder cancer growth by rep ......" | 25965824 | |
| hsa-miR-155-5p | bladder cancer | worse prognosis; staging; progression; recurrence | "Direct quantitative detection for cell free miR 15 ......" | 26657502 | |
| hsa-miR-155-5p | bladder cancer | staging | "Deregulation of seven CpG island harboring miRNAs ......" | 27064870 | |
| hsa-miR-155-5p | breast cancer | poor survival | "To investigate the global expression profile of mi ......" | 18812439 | |
| hsa-miR-155-5p | breast cancer | drug resistance | "The drug resistance of MCF-7/ADR cells was evaluat ......" | 18971180 | Flow cytometry; MTT assay |
| hsa-miR-155-5p | breast cancer | tumorigenesis | "MicroRNA 155 functions as an OncomiR in breast can ......" | 20354188 | Colony formation; RNAi |
| hsa-miR-155-5p | breast cancer | poor survival; drug resistance | "MicroRNA 155 regulates cell survival growth and ch ......" | 20371610 | |
| hsa-miR-155-5p | breast cancer | staging; metastasis; worse prognosis | "Expression and its clinical significance of miR 15 ......" | 20388420 | |
| hsa-miR-155-5p | breast cancer | metastasis | "The relative concentrations of breast cancer-assoc ......" | 21047409 | |
| hsa-miR-155-5p | breast cancer | worse prognosis; staging; metastasis; poor survival | "Clinical significance of microRNA 155 expression i ......" | 22105810 | |
| hsa-miR-155-5p | breast cancer | staging | "Clinical significance of miR 155 expression in bre ......" | 22245916 | Flow cytometry |
| hsa-miR-155-5p | breast cancer | tumor size; metastasis | "Real Time RT-PCR was performed to identify the miR ......" | 22524830 | |
| hsa-miR-155-5p | breast cancer | progression | "The oncogenic role of miR 155 in breast cancer; mi ......" | 22736789 | |
| hsa-miR-155-5p | breast cancer | drug resistance | "Serum microRNA 155 as a potential biomarker to tra ......" | 23071695 | |
| hsa-miR-155-5p | breast cancer | staging; metastasis; tumor size | "miR 155 and miR 31 are differentially expressed in ......" | 23162645 | |
| hsa-miR-155-5p | breast cancer | worse prognosis; staging; tumor size; metastasis; poor survival | "Correlation of miR 155 on formalin fixed paraffin ......" | 23302487 | |
| hsa-miR-155-5p | breast cancer | staging; metastasis; worse prognosis; malignant trasformation | "MicroRNA-155 miR-155 is frequently upregulated in ......" | 23353819 | |
| hsa-miR-155-5p | breast cancer | staging | "Analysis of miR 205 and miR 155 expression in the ......" | 23372341 | |
| hsa-miR-155-5p | breast cancer | progression | "17β estradiol up regulates miR 155 expression and ......" | 23568502 | |
| hsa-miR-155-5p | breast cancer | tumorigenesis | "Effects of miR 155 antisense oligonucleotide on br ......" | 23725141 | |
| hsa-miR-155-5p | breast cancer | progression | "Deregulated serum concentrations of circulating ce ......" | 23748853 | |
| hsa-miR-155-5p | breast cancer | metastasis; drug resistance | "MiR 155 mediated loss of C/EBPβ shifts the TGF β ......" | 23955085 | |
| hsa-miR-155-5p | breast cancer | malignant trasformation | "Here we identified 33 miRNAs with similar deregula ......" | 24104550 | |
| hsa-miR-155-5p | breast cancer | drug resistance; poor survival | "Protective role of miR 155 in breast cancer throug ......" | 24616504 | |
| hsa-miR-155-5p | breast cancer | metastasis; poor survival | "miR 155 drives telomere fragility in human breast ......" | 24876105 | |
| hsa-miR-155-5p | breast cancer | worse prognosis | "Some miRNAs especially the miR-200 family miR-9 an ......" | 25086633 | |
| hsa-miR-155-5p | breast cancer | metastasis | "The clinicopathological significance of microRNA 1 ......" | 25157366 | |
| hsa-miR-155-5p | breast cancer | drug resistance; tumorigenesis | "miR 155 induced transcriptome changes in the MCF 7 ......" | 25352952 | |
| hsa-miR-155-5p | breast cancer | drug resistance | "Role of miR 155 in drug resistance of breast cance ......" | 25744731 | |
| hsa-miR-155-5p | breast cancer | metastasis | "Using real-time quantitative polymerase chain reac ......" | 26124926 | |
| hsa-miR-155-5p | breast cancer | malignant trasformation | "In order to obtain insight into miRNA de-regulatio ......" | 26408705 | |
| hsa-miR-155-5p | breast cancer | drug resistance; poor survival | "In this study we identified that miR-155 is freque ......" | 26692956 | |
| hsa-miR-155-5p | breast cancer | drug resistance; motility | "miR 155 Drives Metabolic Reprogramming of ER+ Brea ......" | 26795347 | |
| hsa-miR-155-5p | breast cancer | metastasis | "MiR 155 expression level changes might be associat ......" | 26889816 | |
| hsa-miR-155-5p | breast cancer | metastasis | "The connection of miR 21 and miR 155 with regulati ......" | 27420617 | |
| hsa-miR-155-5p | breast cancer | drug resistance; staging | "BRCA1 tumor suppressor is involved in DNA damage r ......" | 27596294 | |
| hsa-miR-155-5p | breast cancer | cell migration | "Here we investigate the anti-tumor effects of shor ......" | 27656894 | Western blot; Colony formation |
| hsa-miR-155-5p | cervical and endocervical cancer | tumorigenesis | "Mir 155 promotes cervical cancer cell proliferatio ......" | 25155037 | MTT assay; Flow cytometry; Luciferase |
| hsa-miR-155-5p | cervical and endocervical cancer | worse prognosis; staging; metastasis; poor survival; progression | "Up regulated microRNA 155 expression is associated ......" | 27470551 | |
| hsa-miR-155-5p | chordoma | staging; metastasis; worse prognosis | "MicroRNA 155 expression is independently predictiv ......" | 25823817 | |
| hsa-miR-155-5p | colon cancer | differentiation | "In the present study we isolated a population of c ......" | 21130073 | Flow cytometry |
| hsa-miR-155-5p | colon cancer | tumorigenesis; staging | "We performed microarray analysis of mRNA expressio ......" | 21806946 | Colony formation |
| hsa-miR-155-5p | colon cancer | drug resistance | "Adrenaline promotes cell proliferation and increas ......" | 23036199 | |
| hsa-miR-155-5p | colon cancer | motility | "S100P/RAGE signaling regulates microRNA 155 expres ......" | 23693020 | |
| hsa-miR-155-5p | colon cancer | metastasis; recurrence; worse prognosis | "Correlation between expression of miR 155 in colon ......" | 24888652 | |
| hsa-miR-155-5p | colon cancer | tumorigenesis; worse prognosis; drug resistance; poor survival | "MicroRNA 155 deletion promotes tumorigenesis in th ......" | 26744471 | |
| hsa-miR-155-5p | colorectal cancer | staging; metastasis; worse prognosis | "Clinicopathological and prognostic value of microR ......" | 21412018 | |
| hsa-miR-155-5p | colorectal cancer | progression | "The quantitative analysis by stem loop real time P ......" | 22562822 | |
| hsa-miR-155-5p | colorectal cancer | staging; metastasis; cell migration | "Upregulation of microRNA 155 promotes the migratio ......" | 23588589 | |
| hsa-miR-155-5p | colorectal cancer | tumorigenesis | "miR 155 regulates the proliferation and cell cycle ......" | 24793496 | Western blot; Luciferase |
| hsa-miR-155-5p | colorectal cancer | worse prognosis; progression; poor survival | "Investigation of microRNA 155 as a serum diagnosti ......" | 25528214 | |
| hsa-miR-155-5p | colorectal cancer | metastasis; tumorigenesis; staging | "Up regulated miR 155 5p promotes cell proliferatio ......" | 26261588 | |
| hsa-miR-155-5p | colorectal cancer | tumorigenesis | "Anti Warburg effect of rosmarinic acid via miR 155 ......" | 26340059 | |
| hsa-miR-155-5p | endometrial cancer | metastasis; worse prognosis | "Expression and its clinical significance of hsa mi ......" | 21176560 | |
| hsa-miR-155-5p | endometrial cancer | poor survival | "Angiotensin II type I receptor and miR 155 in endo ......" | 22525818 | MTT assay |
| hsa-miR-155-5p | gastric cancer | metastasis; cell migration | "microRNA 155 is downregulated in gastric cancer ce ......" | 22426647 | Luciferase |
| hsa-miR-155-5p | gastric cancer | metastasis; poor survival | "The expression levels of miR-20a miR-21 miR-25 miR ......" | 22996433 | |
| hsa-miR-155-5p | gastric cancer | tumorigenesis; staging; metastasis; differentiation | "The clinical significance of downregulation of mir ......" | 24805774 | |
| hsa-miR-155-5p | gastric cancer | tumorigenesis | "Anti Warburg effect of rosmarinic acid via miR 155 ......" | 26056431 | MTT assay; Western blot |
| hsa-miR-155-5p | gastric cancer | staging; tumor size | "MicroRNA 155 expression inversely correlates with ......" | 26955820 | |
| hsa-miR-155-5p | glioblastoma | malignant trasformation | "miR 155 is up regulated in primary and secondary g ......" | 22470130 | |
| hsa-miR-155-5p | glioblastoma | poor survival; recurrence | "Moreover miR-326 miR-130a miR-155 miR-210 and 4 mi ......" | 23302469 | |
| hsa-miR-155-5p | head and neck cancer | poor survival | "Our findings showed that significant elevated expr ......" | 25677760 | |
| hsa-miR-155-5p | head and neck cancer | metastasis | "Characterization of miR 146a and miR 155 in blood ......" | 26621153 | |
| hsa-miR-155-5p | kidney renal cell cancer | malignant trasformation | "We performed genome-wide expression profiling of m ......" | 19228262 | |
| hsa-miR-155-5p | kidney renal cell cancer | poor survival | "MicroRNA 155 is a predictive marker for survival i ......" | 23050614 | |
| hsa-miR-155-5p | kidney renal cell cancer | metastasis; malignant trasformation | "miR-122 miR-141 miR-155 miR-184 miR-200c miR-210 m ......" | 23178446 | |
| hsa-miR-155-5p | kidney renal cell cancer | progression; drug resistance | "miR 155 and miR 484 Are Associated with Time to Pr ......" | 26064968 | |
| hsa-miR-155-5p | liver cancer | metastasis; tumorigenesis; recurrence | "We examined expression patterns of 4 microRNAs mic ......" | 21458843 | |
| hsa-miR-155-5p | liver cancer | tumorigenesis | "Aberrant expression of microRNA 155 may accelerate ......" | 21989846 | Western blot; Luciferase |
| hsa-miR-155-5p | liver cancer | poor survival; worse prognosis; recurrence | "Up regulation of microRNA 155 promotes cancer cell ......" | 22071603 | |
| hsa-miR-155-5p | liver cancer | poor survival; worse prognosis; recurrence | "Expression and survival prediction of microRNA 155 ......" | 23863669 | |
| hsa-miR-155-5p | liver cancer | poor survival; recurrence | "We performed a pair-wise comparison of the miRNA t ......" | 25953724 | Colony formation |
| hsa-miR-155-5p | liver cancer | worse prognosis; staging; malignant trasformation; tumor size | "MicroRNA 155 promotes tumor growth of human hepato ......" | 27035278 | |
| hsa-miR-155-5p | liver cancer | poor survival; worse prognosis | "MiR 155 and its functional variant rs767649 contri ......" | 27531892 | |
| hsa-miR-155-5p | lung cancer | poor survival | "miRNA microarray analysis identified statistical u ......" | 16530703 | |
| hsa-miR-155-5p | lung cancer | staging; metastasis | "We searched the published literature for the miRNA ......" | 21904633 | |
| hsa-miR-155-5p | lung cancer | worse prognosis; poor survival | "Inhibition of hypoxia induced miR 155 radiosensiti ......" | 22027557 | |
| hsa-miR-155-5p | lung cancer | progression | "MiR 155 inhibits the sensitivity of lung cancer ce ......" | 22996741 | |
| hsa-miR-155-5p | lung cancer | malignant trasformation | "Using quantitative reverse transcriptase PCR analy ......" | 23462458 | |
| hsa-miR-155-5p | lung cancer | worse prognosis | "The latest deep sequencing of tumor genomes has re ......" | 25355421 | |
| hsa-miR-155-5p | lung cancer | poor survival; worse prognosis; metastasis | "A functional variant in miR 155 regulation region ......" | 26543233 | |
| hsa-miR-155-5p | lung cancer | drug resistance | "Effect of miR 155 knockdown on the reversal of dox ......" | 26893712 | Western blot |
| hsa-miR-155-5p | lung cancer | metastasis; cell migration | "Role of miR 155 in invasion and metastasis of lung ......" | 26899325 | Cell migration assay; Luciferase; Western blot |
| hsa-miR-155-5p | lung cancer | poor survival | "Tissues were microdissected to obtain >90% tumor o ......" | 27081085 | |
| hsa-miR-155-5p | lung cancer | worse prognosis; drug resistance | "Inhibition of microRNA 155 sensitizes lung cancer ......" | 27315591 | Western blot; Luciferase |
| hsa-miR-155-5p | lung squamous cell cancer | staging; recurrence | "In an exploratory study we determined whether expr ......" | 20975375 | |
| hsa-miR-155-5p | lung squamous cell cancer | staging; poor survival; metastasis | "Prognostic impact of MiR 155 in non small cell lun ......" | 21219656 | |
| hsa-miR-155-5p | lung squamous cell cancer | metastasis | "The miRs were quantified by microarray hybridizati ......" | 22295063 | |
| hsa-miR-155-5p | lung squamous cell cancer | poor survival; recurrence; worse prognosis | "High expression of miR 21 and miR 155 predicts rec ......" | 23099007 | |
| hsa-miR-155-5p | lung squamous cell cancer | worse prognosis; metastasis; poor survival | "The prognostic value of miR 21 and miR 155 in non ......" | 23817461 | |
| hsa-miR-155-5p | lung squamous cell cancer | worse prognosis; poor survival; recurrence | "MicroRNA 155 expression has prognostic value in pa ......" | 24460255 | |
| hsa-miR-155-5p | lung squamous cell cancer | staging; worse prognosis; poor survival | "Association of MiR 155 expression with prognosis i ......" | 24854560 | |
| hsa-miR-155-5p | lung squamous cell cancer | metastasis | "Retrovirus-mediated RNA interference was employed ......" | 25373388 | RNAi; Western blot; Luciferase |
| hsa-miR-155-5p | lung squamous cell cancer | staging | "In training set 25 early-stage NSCLC patients and ......" | 25421010 | |
| hsa-miR-155-5p | lung squamous cell cancer | poor survival | "The Value of MicroRNA 155 as a Prognostic Factor f ......" | 26322518 | |
| hsa-miR-155-5p | lung squamous cell cancer | metastasis | "Expression of miR-9 miR-10b miR-145 and miR-155 4 ......" | 26909466 | |
| hsa-miR-155-5p | lung squamous cell cancer | staging | "Expression of microRNA let 7a miR 155 and miR 205 ......" | 27296000 | |
| hsa-miR-155-5p | lymphoma | malignant trasformation | "We recently reported that overexpression of miR-21 ......" | 21502955 | |
| hsa-miR-155-5p | lymphoma | motility; differentiation | "miR 155 regulates HGAL expression and increases ly ......" | 22096245 | |
| hsa-miR-155-5p | lymphoma | worse prognosis | "Expressions of miR 21 miR 155 and miR 210 in plasm ......" | 22541087 | |
| hsa-miR-155-5p | lymphoma | malignant trasformation | "Nanoparticle based therapy in an in vivo microRNA ......" | 22685206 | |
| hsa-miR-155-5p | lymphoma | staging | "The miRNA expression profiles of skin biopsies fro ......" | 22776000 | |
| hsa-miR-155-5p | lymphoma | malignant trasformation | "STAT5 mediated expression of oncogenic miR 155 in ......" | 23676217 | |
| hsa-miR-155-5p | lymphoma | malignant trasformation | "Expression of miR 155 and miR 126 in situ in cutan ......" | 24033365 | |
| hsa-miR-155-5p | lymphoma | drug resistance | "Using quantitative real-time PCR qRT-PCR we tested ......" | 24222179 | |
| hsa-miR-155-5p | lymphoma | differentiation | "Aberrant expression of oncogenic miRNAs including ......" | 24479800 | |
| hsa-miR-155-5p | lymphoma | staging; progression | "Herein we used a global quantitative real-time pol ......" | 25503151 | |
| hsa-miR-155-5p | lymphoma | differentiation; poor survival; drug resistance | "Oncogenic microRNA 155 and its target PU.1: an int ......" | 26261072 | |
| hsa-miR-155-5p | melanoma | metastasis | "Six microRNAs miR-9 miR-145 miR-150 miR-155 miR-20 ......" | 23863473 | |
| hsa-miR-155-5p | melanoma | metastasis; poor survival | "SNPs at miR 155 binding sites of TYRP1 explain dis ......" | 26068396 | Western blot |
| hsa-miR-155-5p | melanoma | progression | "Small RNA deep sequencing discriminates subsets of ......" | 26176991 | |
| hsa-miR-155-5p | melanoma | progression | "These cells also expressed NK cell regulatory miRs ......" | 27565163 | |
| hsa-miR-155-5p | ovarian cancer | tumorigenesis | "MicroRNA 155 is a novel suppressor of ovarian canc ......" | 23523916 | Luciferase; Cell migration assay |
| hsa-miR-155-5p | pancreatic cancer | poor survival | "We measured the levels of miR-155 miR-203 miR-210 ......" | 19551852 | |
| hsa-miR-155-5p | pancreatic cancer | progression; staging | "Aberrant MicroRNA 155 expression is an early event ......" | 20332664 | |
| hsa-miR-155-5p | pancreatic cancer | malignant trasformation | "miR-148a/b and miR-375 expression were found decre ......" | 21738581 | |
| hsa-miR-155-5p | pancreatic cancer | staging; worse prognosis | "Eighty-eight samples of ductal pancreatic adenocar ......" | 22850622 | |
| hsa-miR-155-5p | pancreatic cancer | worse prognosis | "MLH1 as a direct target of MiR 155 and a potential ......" | 23715647 | Luciferase |
| hsa-miR-155-5p | pancreatic cancer | staging; metastasis | "Regulation of miR 155 affects pancreatic cancer ce ......" | 23817566 | Transwell assay |
| hsa-miR-155-5p | pancreatic cancer | staging | "Based on relevance to cancer a seven-miRNA signatu ......" | 25184537 | |
| hsa-miR-155-5p | sarcoma | staging | "Kaposi's sarcoma associated herpesvirus encodes an ......" | 17881434 | |
| hsa-miR-155-5p | sarcoma | tumorigenesis | "Kaposi's sarcoma-associated herpes virus KSHV the ......" | 18191785 | |
| hsa-miR-155-5p | sarcoma | progression | "MiR 155 is a liposarcoma oncogene that targets cas ......" | 22350414 | Colony formation |
| hsa-miR-155-5p | sarcoma | progression; poor survival | "miR 155 promotes the growth of osteosarcoma in a H ......" | 25666090 | |
| hsa-miR-155-5p | thyroid cancer | metastasis; recurrence | "In this study we evaluated miRNA expression as a m ......" | 21537871 | |
| hsa-miR-155-5p | thyroid cancer | tumor size; malignant trasformation | "Differential expression levels of plasma derived m ......" | 25456009 |
Reported gene related to hsa-miR-155-5p
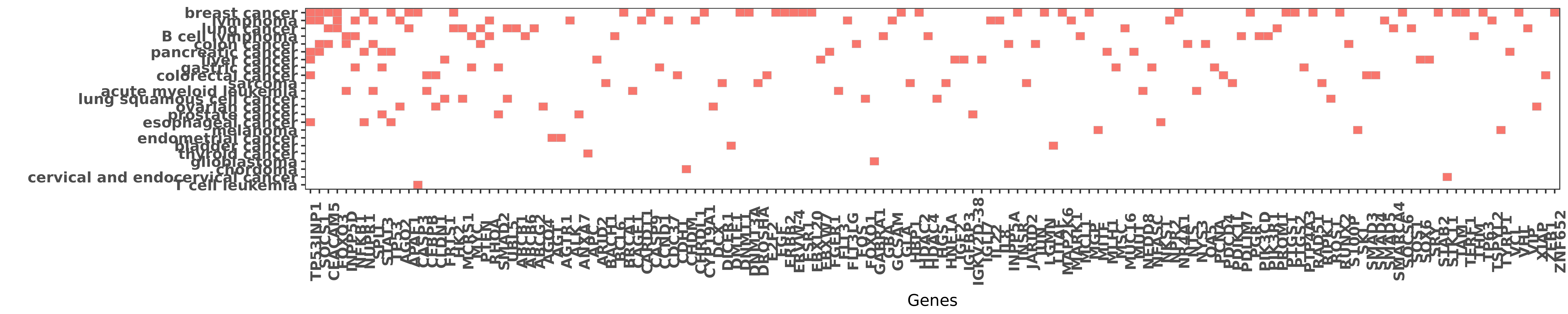
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-155-5p | breast cancer | TP53INP1 | "TP53INP1 is the direct target of miR-155; MiR-155 ......" | 24152184 |
| hsa-miR-155-5p | breast cancer | TP53INP1 | "17β estradiol up regulates miR 155 expression and ......" | 23568502 |
| hsa-miR-155-5p | colorectal cancer | TP53INP1 | "miR-21 and miR-155 expression was assessed in tumo ......" | 21412018 |
| hsa-miR-155-5p | esophageal cancer | TP53INP1 | "In addition miR-155 and TP53INP1 showed a negative ......" | 24551280 |
| hsa-miR-155-5p | liver cancer | TP53INP1 | "MiR 155 targets TP53INP1 to regulate liver cancer ......" | 25601564 |
| hsa-miR-155-5p | lymphoma | TP53INP1 | "In addition miR-142-5p and miR-155 suppress the pr ......" | 23209550 |
| hsa-miR-155-5p | pancreatic cancer | TP53INP1 | "TP53INP1 is a target of miR-155 in fibroblasts and ......" | 26195069 |
| hsa-miR-155-5p | pancreatic cancer | TP53INP1 | "Finally TP53INP1 expression is repressed by the on ......" | 17911264 |
| hsa-miR-155-5p | breast cancer | FOXO3 | "MiR-155 targets 3´UTR region of multiple componen ......" | 27596294 |
| hsa-miR-155-5p | breast cancer | FOXO3 | "MicroRNA 155 regulates cell survival growth and ch ......" | 20371610 |
| hsa-miR-155-5p | breast cancer | FOXO3 | "In contrast downregulation of miR-155-5p increased ......" | 26398931 |
| hsa-miR-155-5p | breast cancer | FOXO3 | "Our results further revealed that forkhead box O3a ......" | 27107135 |
| hsa-miR-155-5p | lung cancer | FOXO3 | "Here we show that hypoxic conditions induce miR-15 ......" | 22027557 |
| hsa-miR-155-5p | lymphoma | FOXO3 | "The effect of miRNA155 on FOXO3a expression was ex ......" | 25480585 |
| hsa-miR-155-5p | acute myeloid leukemia | SPI1 | "We also showed that PKC412-related FLT3 inhibition ......" | 26055960 |
| hsa-miR-155-5p | acute myeloid leukemia | SPI1 | "This results in the upregulation of the miR-155 ta ......" | 25971362 |
| hsa-miR-155-5p | acute myeloid leukemia | SPI1 | "Further we demonstrate that miR-155 targets the my ......" | 25092144 |
| hsa-miR-155-5p | colon cancer | SPI1 | "Treatment with TanshinoneIIA prevented increased P ......" | 22982040 |
| hsa-miR-155-5p | lymphoma | SPI1 | "Herein we show that increased expression of the NF ......" | 20947507 |
| hsa-miR-155-5p | lymphoma | SPI1 | "The transcription factor PU.1 and its inhibitory m ......" | 26261072 |
| hsa-miR-155-5p | breast cancer | TP53 | "Elevated miR-155 was found in Her-2 positive or ly ......" | 25157366 |
| hsa-miR-155-5p | breast cancer | TP53 | "Mutant p53 drives invasion in breast tumors throug ......" | 22797073 |
| hsa-miR-155-5p | breast cancer | TP53 | "MiR 155 promotes proliferation of human breast can ......" | 24152184 |
| hsa-miR-155-5p | breast cancer | TP53 | "High expression of miR155 was associated with clin ......" | 23372341 |
| hsa-miR-155-5p | esophageal cancer | TP53 | "microRNA 155 acts as an oncogene by targeting the ......" | 24551280 |
| hsa-miR-155-5p | pancreatic cancer | TP53 | "Tumor protein 53 induced nuclear protein 1 express ......" | 17911264 |
| hsa-miR-155-5p | acute myeloid leukemia | FLT3 | "miR 155 regulative network in FLT3 mutated acute m ......" | 26055960 |
| hsa-miR-155-5p | acute myeloid leukemia | FLT3 | "Further experiments demonstrated that the up-regul ......" | 18308931 |
| hsa-miR-155-5p | acute myeloid leukemia | FLT3 | "Pharmacological targeting of miR 155 via the NEDD8 ......" | 25971362 |
| hsa-miR-155-5p | acute myeloid leukemia | FLT3 | "Silvestrol exhibits significant in vivo and in vit ......" | 23497456 |
| hsa-miR-155-5p | B cell lymphoma | INPP5D | "Onco miR 155 targets SHIP1 to promote TNFalpha dep ......" | 19890474 |
| hsa-miR-155-5p | acute myeloid leukemia | INPP5D | "This results in the upregulation of the miR-155 ta ......" | 25971362 |
| hsa-miR-155-5p | acute myeloid leukemia | INPP5D | "SHIP1 is targeted by miR 155 in acute myeloid leuk ......" | 25175984 |
| hsa-miR-155-5p | colon cancer | INPP5D | "We identified Phosphatidylinositol-3 4 5-trisphosp ......" | 22982040 |
| hsa-miR-155-5p | breast cancer | SOCS1 | "Here we report that the tumor suppressor gene supp ......" | 20354188 |
| hsa-miR-155-5p | colon cancer | SOCS1 | "NTR1 activation stimulates expression of miR-21 an ......" | 21806946 |
| hsa-miR-155-5p | lymphoma | SOCS1 | "Ago2 immunoprecipitation revealed miR-155 as the m ......" | 25820993 |
| hsa-miR-155-5p | pancreatic cancer | SOCS1 | "Regulation of miR 155 affects pancreatic cancer ce ......" | 23817566 |
| hsa-miR-155-5p | breast cancer | BRCA1 | "BRCA1 tumor suppressor is involved in DNA damage r ......" | 27596294 |
| hsa-miR-155-5p | breast cancer | BRCA1 | "Tumor suppressor BRCA1 epigenetically controls onc ......" | 21946536 |
| hsa-miR-155-5p | breast cancer | BRCA1 | "The study was designed to investigate the molecula ......" | 27107135 |
| hsa-miR-155-5p | breast cancer | CEACAM5 | "Surprisingly a decreased level of serum miR-155 wa ......" | 23071695 |
| hsa-miR-155-5p | colon cancer | CEACAM5 | "Correlation between expression of miR 155 in colon ......" | 24888652 |
| hsa-miR-155-5p | lung cancer | CEACAM5 | "We assessed the differences in levels of miR-21 mi ......" | 24190459 |
| hsa-miR-155-5p | B cell lymphoma | GBA | "Interestingly expression levels of both miR-155 an ......" | 17487835 |
| hsa-miR-155-5p | B cell lymphoma | GBA | "We found no association of miR-155 and miR-21 with ......" | 25265435 |
| hsa-miR-155-5p | B cell lymphoma | GBA | "The expression level of miR-155 in non-GCB type wa ......" | 23450486 |
| hsa-miR-155-5p | B cell lymphoma | NFKB1 | "The expression of miR-155 is heterogeneous in thes ......" | 18262046 |
| hsa-miR-155-5p | gastric cancer | NFKB1 | "NF-kappa B p65 NF-κB p65 and inhibitor of NF-kapp ......" | 26934326 |
| hsa-miR-155-5p | lymphoma | NFKB1 | "Our data provide evidence for two levels of regula ......" | 17173072 |
| hsa-miR-155-5p | breast cancer | NUPR1 | "MiR 155 promotes proliferation of human breast can ......" | 24152184 |
| hsa-miR-155-5p | esophageal cancer | NUPR1 | "microRNA 155 acts as an oncogene by targeting the ......" | 24551280 |
| hsa-miR-155-5p | pancreatic cancer | NUPR1 | "Tumor protein 53 induced nuclear protein 1 express ......" | 17911264 |
| hsa-miR-155-5p | breast cancer | PGR | "It's indicated that the up-regulation of miR-155 e ......" | 20388420 |
| hsa-miR-155-5p | breast cancer | PGR | "While the expression of all three miRNAs was simil ......" | 19454029 |
| hsa-miR-155-5p | breast cancer | PGR | "miR 155 and miR 31 are differentially expressed in ......" | 23162645 |
| hsa-miR-155-5p | gastric cancer | STAT3 | "As expected RA inhibited proinflammatory cytokines ......" | 26056431 |
| hsa-miR-155-5p | pancreatic cancer | STAT3 | "Regulation of miR 155 affects pancreatic cancer ce ......" | 23817566 |
| hsa-miR-155-5p | prostate cancer | STAT3 | "Functional analysis was performed using a prostate ......" | 27017949 |
| hsa-miR-155-5p | lymphoma | TSPYL2 | "Quantitative qRT-PCR analysis of 103 patients with ......" | 21865341 |
| hsa-miR-155-5p | lymphoma | TSPYL2 | "Recent discoveries indicate that the oncogenic mic ......" | 23676217 |
| hsa-miR-155-5p | lymphoma | TSPYL2 | "Recently miR-155 has been implicated in cutaneous ......" | 24033365 |
| hsa-miR-155-5p | lymphoma | AGO2 | "Ago2 immunoprecipitation revealed miR-155 as the m ......" | 25820993 |
| hsa-miR-155-5p | ovarian cancer | AGO2 | "Endogenous processing of synthetic miR-155 favored ......" | 22307839 |
| hsa-miR-155-5p | breast cancer | APAF1 | "Also increase in the transcript level of APAF-1 an ......" | 26877850 |
| hsa-miR-155-5p | lung cancer | APAF1 | "MiR 155 inhibits the sensitivity of lung cancer ce ......" | 22996741 |
| hsa-miR-155-5p | T cell leukemia | CASP3 | "According to our results the use of miR-155 inhibi ......" | 25598954 |
| hsa-miR-155-5p | breast cancer | CASP3 | "Furthermore the effects of inhibitory effects of m ......" | 23725141 |
| hsa-miR-155-5p | acute myeloid leukemia | CEBPB | "We showed in experiments of miR-155 mimic in K562 ......" | 26055960 |
| hsa-miR-155-5p | colorectal cancer | CEBPB | "The upregulated miR-155 attenuated miR-143 by inhi ......" | 27626488 |
| hsa-miR-155-5p | colorectal cancer | CLDN1 | "Upregulation of microRNA 155 promotes the migratio ......" | 23588589 |
| hsa-miR-155-5p | ovarian cancer | CLDN1 | "MicroRNA 155 is a novel suppressor of ovarian canc ......" | 23523916 |
| hsa-miR-155-5p | breast cancer | EGF | "miR-155 was also significantly overexpressed in po ......" | 25232827 |
| hsa-miR-155-5p | breast cancer | EGF | "The tissue and plasma levels of miR-155 and miR-31 ......" | 23162645 |
| hsa-miR-155-5p | breast cancer | ESR1 | "It's indicated that the up-regulation of miR-155 e ......" | 20388420 |
| hsa-miR-155-5p | breast cancer | ESR1 | "miR 155 and miR 31 are differentially expressed in ......" | 23162645 |
| hsa-miR-155-5p | liver cancer | FRTS1 | "Patients with higher miR-155 expression had signif ......" | 22071603 |
| hsa-miR-155-5p | lung squamous cell cancer | FRTS1 | "Data were extracted from studies comparing overall ......" | 24460255 |
| hsa-miR-155-5p | lung squamous cell cancer | HGS | "The pooled hazard ratios HRs and 95% confidence in ......" | 24460255 |
| hsa-miR-155-5p | lung squamous cell cancer | HGS | "The hazard ratios HRs and 95% confidence intervals ......" | 26322518 |
| hsa-miR-155-5p | breast cancer | HK2 | "We further show that miR-155 acts to upregulate he ......" | 22354042 |
| hsa-miR-155-5p | lung cancer | HK2 | "Inhibition of microRNA 155 sensitizes lung cancer ......" | 27315591 |
| hsa-miR-155-5p | lung cancer | MCRS1 | "The candidate oncogene MCRS1 promotes the growth o ......" | 26467212 |
| hsa-miR-155-5p | lung squamous cell cancer | MCRS1 | "Furthermore we found that MCRS1 binds to the miR-1 ......" | 25373388 |
| hsa-miR-155-5p | pancreatic cancer | MLH1 | "MLH1 as a direct target of MiR 155 and a potential ......" | 23715647 |
| hsa-miR-155-5p | pancreatic cancer | MLH1 | "Predicted target mRNAs FGFR1 miR-10 and MLH1 miR-1 ......" | 21738581 |
| hsa-miR-155-5p | B cell lymphoma | MYC | "It was shown that the patients with MYC rearrangem ......" | 23450486 |
| hsa-miR-155-5p | gastric cancer | MYC | "Downregulation of microRNA 155 accelerates cell gr ......" | 24969405 |
| hsa-miR-155-5p | colon cancer | PTEN | "NTR1 activation stimulates expression of miR-21 an ......" | 21806946 |
| hsa-miR-155-5p | lung cancer | PTEN | "Western blot and real-time PCR were performed to c ......" | 26899325 |
| hsa-miR-155-5p | B cell lymphoma | RHOA | "MiR-155 can promote the occurrence of lymphoma thr ......" | 24989312 |
| hsa-miR-155-5p | lymphoma | RHOA | "We demonstrate that miR-155 directly down-regulate ......" | 22096245 |
| hsa-miR-155-5p | gastric cancer | SMAD2 | "Taken together these data suggest that miR-155 may ......" | 22426647 |
| hsa-miR-155-5p | prostate cancer | SMAD2 | "MiR-155 which targeted the SMAD2-3'UTR decreased t ......" | 25283513 |
| hsa-miR-155-5p | lung cancer | UBL5 | "Chitosan combined with molecular beacon for mir 15 ......" | 25230125 |
| hsa-miR-155-5p | lung squamous cell cancer | UBL5 | "A novel molecular beacon MB of miR-155 was designe ......" | 22796123 |
| hsa-miR-155-5p | lung cancer | ABCB1 | "Effect of miR 155 knockdown on the reversal of dox ......" | 26893712 |
| hsa-miR-155-5p | B cell lymphoma | ABCB6 | "Interestingly expression levels of both miR-155 an ......" | 17487835 |
| hsa-miR-155-5p | lung cancer | ABCG2 | "TICs expressing CD133 and CD338 were obtained from ......" | 24528019 |
| hsa-miR-155-5p | ovarian cancer | AGO4 | "Endogenous processing of synthetic miR-155 favored ......" | 22307839 |
| hsa-miR-155-5p | endometrial cancer | AGT | "Angiotensin II type I receptor and miR 155 in endo ......" | 22525818 |
| hsa-miR-155-5p | endometrial cancer | AGTR1 | "MicroRNA-155 miR-155 is one of the micro RNAs miRN ......" | 22525818 |
| hsa-miR-155-5p | lymphoma | ALK | "However no direct effect of the ALK kinase on miR- ......" | 25820993 |
| hsa-miR-155-5p | prostate cancer | ANXA7 | "microRNA 155 promotes the proliferation of prostat ......" | 25339368 |
| hsa-miR-155-5p | thyroid cancer | APC | "Upregulated miR 155 in papillary thyroid carcinoma ......" | 23796566 |
| hsa-miR-155-5p | liver cancer | ARID2 | "MicroRNA 155 promotes tumor growth of human hepato ......" | 27035278 |
| hsa-miR-155-5p | sarcoma | BACH1 | "Bioinformatic tools predicted the transcriptional ......" | 17881434 |
| hsa-miR-155-5p | B cell lymphoma | BCL6 | "We show that Bcl6 is indirectly regulated by miR-1 ......" | 23169640 |
| hsa-miR-155-5p | acute myeloid leukemia | CAGE1 | "LIFRα CT3 induces differentiation of a human acut ......" | 25092123 |
| hsa-miR-155-5p | lymphoma | CARD11 | "Additionally miR-155 is induced by LPS treatment o ......" | 20947507 |
| hsa-miR-155-5p | breast cancer | CASP9 | "Also increase in the transcript level of APAF-1 an ......" | 26877850 |
| hsa-miR-155-5p | gastric cancer | CCND1 | "MicroRNA 155 expression inversely correlates with ......" | 26955820 |
| hsa-miR-155-5p | lymphoma | CDC37 | "The definitive role of this miR-155 ortholog in on ......" | 21383974 |
| hsa-miR-155-5p | colorectal cancer | CDH1 | "E-cadherin expression levels decreased while ZEB1 ......" | 23588589 |
| hsa-miR-155-5p | chordoma | CHDM | "MicroRNA 155 expression is independently predictiv ......" | 25823817 |
| hsa-miR-155-5p | lymphoma | CHRDL1 | "Using quantitative real-time PCR qRT-PCR we tested ......" | 24222179 |
| hsa-miR-155-5p | breast cancer | CYP19A1 | "miR 155 Drives Metabolic Reprogramming of ER+ Brea ......" | 26795347 |
| hsa-miR-155-5p | ovarian cancer | DCX | "Here we took advantage of the spontaneous enhanced ......" | 22307839 |
| hsa-miR-155-5p | sarcoma | DICER1 | "Deregulation of dicer and mir 155 expression in li ......" | 25888631 |
| hsa-miR-155-5p | bladder cancer | DMTF1 | "MicroRNA 155 promotes bladder cancer growth by rep ......" | 25965824 |
| hsa-miR-155-5p | breast cancer | DNMT1 | "These results indicate that miR-155-5p antagonizes ......" | 26398931 |
| hsa-miR-155-5p | breast cancer | DNMT3A | "These results indicate that miR-155-5p antagonizes ......" | 26398931 |
| hsa-miR-155-5p | sarcoma | DROSHA | "The aim of this study is to evaluate the involveme ......" | 25888631 |
| hsa-miR-155-5p | colorectal cancer | E2F2 | "miR 155 regulates the proliferation and cell cycle ......" | 24793496 |
| hsa-miR-155-5p | breast cancer | ERBB2 | "miR-155 was also significantly overexpressed in po ......" | 25232827 |
| hsa-miR-155-5p | breast cancer | ERVW-4 | "Here we investigate the anti-tumor effects of shor ......" | 27656894 |
| hsa-miR-155-5p | breast cancer | FBXL20 | "Transfection efficiency detected by flow cytometry ......" | 22245916 |
| hsa-miR-155-5p | liver cancer | FBXW7 | "MicroRNA 155 3p promotes hepatocellular carcinoma ......" | 27306418 |
| hsa-miR-155-5p | pancreatic cancer | FGFR1 | "Predicted target mRNAs FGFR1 miR-10 and MLH1 miR-1 ......" | 21738581 |
| hsa-miR-155-5p | lymphoma | FLT3LG | "We studied expression of miR-155 and PU.1 in the s ......" | 26261072 |
| hsa-miR-155-5p | colon cancer | FOS | "Also S100P treatment stimulated the enrichment of ......" | 23693020 |
| hsa-miR-155-5p | lung squamous cell cancer | FOXO1 | "Moreover when N-acetylcysteine was present increas ......" | 26548866 |
| hsa-miR-155-5p | glioblastoma | GABRA1 | "We hypothesised that γ-aminobutyric acid A recept ......" | 22470130 |
| hsa-miR-155-5p | lymphoma | GCSAM | "miR 155 regulates HGAL expression and increases ly ......" | 22096245 |
| hsa-miR-155-5p | breast cancer | GLA | "Matrix Gla protein repression by miR 155 promotes ......" | 27009385 |
| hsa-miR-155-5p | sarcoma | HBP1 | "miR 155 promotes the growth of osteosarcoma in a H ......" | 25666090 |
| hsa-miR-155-5p | breast cancer | HDAC2 | "Mechanistically we found that BRCA1 epigenetically ......" | 21946536 |
| hsa-miR-155-5p | B cell lymphoma | HDAC4 | "Interestingly we found that miR-155 directly targe ......" | 23169640 |
| hsa-miR-155-5p | sarcoma | HNF1A | "HMG-box transcription factor 1 HBP1 a strong Wnt p ......" | 25666090 |
| hsa-miR-155-5p | liver cancer | IGF2 | "Induction of the expression of miR-155 in HCC cell ......" | 26722313 |
| hsa-miR-155-5p | liver cancer | IGFBP3 | "Transcriptional activation of the IGF II/IGF 1R ax ......" | 26722313 |
| hsa-miR-155-5p | prostate cancer | IGKV2D-38 | "In the present study As2 O3 attenuated angiogenic ......" | 25283513 |
| hsa-miR-155-5p | liver cancer | IGLJ7 | "Additionally expression knockdown of miR-155 in J7 ......" | 22629365 |
| hsa-miR-155-5p | lymphoma | IL22 | "Moreover miR-155 induces IL-22 expression and supp ......" | 25820993 |
| hsa-miR-155-5p | lymphoma | IL8 | "Moreover miR-155 induces IL-22 expression and supp ......" | 25820993 |
| hsa-miR-155-5p | colon cancer | INPP5A | "We identified Phosphatidylinositol-3 4 5-trisphosp ......" | 22982040 |
| hsa-miR-155-5p | breast cancer | INSR | "Overexpression of miR-155 decreased the efficiency ......" | 24616504 |
| hsa-miR-155-5p | sarcoma | JARID2 | "We furthermore identify Jarid2 a component of Poly ......" | 23185331 |
| hsa-miR-155-5p | colon cancer | JUN | "In the present study we demonstrated for the first ......" | 23693020 |
| hsa-miR-155-5p | breast cancer | LGMN | "Here we investigate the anti-tumor effects of shor ......" | 27656894 |
| hsa-miR-155-5p | bladder cancer | LITAF | "In conclusion detection of cell-free miR-155 in ur ......" | 26657502 |
| hsa-miR-155-5p | breast cancer | MAP2K6 | "miR 155 induced transcriptome changes in the MCF 7 ......" | 25352952 |
| hsa-miR-155-5p | lymphoma | MARK1 | "Upregulation of miR-155 and downregulation of PU.1 ......" | 26261072 |
| hsa-miR-155-5p | B cell lymphoma | MCL1 | "Epstein Barr virus latent membrane protein 1 prote ......" | 22268450 |
| hsa-miR-155-5p | breast cancer | MGP | "Matrix Gla protein repression by miR 155 promotes ......" | 27009385 |
| hsa-miR-155-5p | melanoma | MITF | "microRNA 155 induced by interleukin 1ß represses ......" | 25853464 |
| hsa-miR-155-5p | gastric cancer | MSC | "miR 155 5p inhibition promotes the transition of b ......" | 26934326 |
| hsa-miR-155-5p | lung cancer | MUC16 | "We assessed the differences in levels of miR-21 mi ......" | 24190459 |
| hsa-miR-155-5p | pancreatic cancer | MUT | "The regulation of Mut L homologue 1 MLH1 expressio ......" | 23715647 |
| hsa-miR-155-5p | acute myeloid leukemia | NEDD8 | "Pharmacological targeting of miR 155 via the NEDD8 ......" | 25971362 |
| hsa-miR-155-5p | gastric cancer | NFASC | "miR 155 5p inhibition promotes the transition of b ......" | 26934326 |
| hsa-miR-155-5p | esophageal cancer | NKS1 | "We also indicated that overexpression of miR-155 s ......" | 24551280 |
| hsa-miR-155-5p | lymphoma | NPR2 | "This inhibitory effect of miR-155 suggests that it ......" | 22096245 |
| hsa-miR-155-5p | breast cancer | NR4A1 | "A potential relationship between miR-155 levels an ......" | 23302487 |
| hsa-miR-155-5p | colon cancer | NTS | "Neurotensin stimulated differential expression of ......" | 21806946 |
| hsa-miR-155-5p | acute myeloid leukemia | NYS3 | "To evaluate the impact of miR-155 on the outcome o ......" | 23650424 |
| hsa-miR-155-5p | colon cancer | OA5 | "S100P/RAGE signaling regulates microRNA 155 expres ......" | 23693020 |
| hsa-miR-155-5p | gastric cancer | PCNA | "MicroRNA 155 expression inversely correlates with ......" | 26955820 |
| hsa-miR-155-5p | colorectal cancer | PDCD4 | "miR-21 and miR-155 expression was assessed in tumo ......" | 21412018 |
| hsa-miR-155-5p | sarcoma | PDIK1L | "MiR 155 is a liposarcoma oncogene that targets cas ......" | 22350414 |
| hsa-miR-155-5p | B cell lymphoma | PDLIM7 | "Here we show for the first time that LMP1 activate ......" | 22268450 |
| hsa-miR-155-5p | B cell lymphoma | PIK3CD | "PIK3R1 p85α a negative regulator of the phosphati ......" | 22609116 |
| hsa-miR-155-5p | B cell lymphoma | PIK3R1 | "PIK3R1 p85α a negative regulator of the phosphati ......" | 22609116 |
| hsa-miR-155-5p | lung cancer | PROM1 | "TICs expressing CD133 and CD338 were obtained from ......" | 24528019 |
| hsa-miR-155-5p | breast cancer | PTGS1 | "Correlations between the dysregulation of miR-21 m ......" | 27420617 |
| hsa-miR-155-5p | breast cancer | PTGS2 | "Correlations between the dysregulation of miR-21 m ......" | 27420617 |
| hsa-miR-155-5p | gastric cancer | PTP4A3 | "We applied the artificial miRNA pCMV-PRL3miRNA whi ......" | 16875667 |
| hsa-miR-155-5p | breast cancer | RAD51 | "Protective role of miR 155 in breast cancer throug ......" | 24616504 |
| hsa-miR-155-5p | sarcoma | RIPK1 | "In OS cells small interfering RNA against Ripk1 pr ......" | 27041566 |
| hsa-miR-155-5p | lung squamous cell cancer | ROS1 | "In conclusion we found that miR-155 promoted NSCLC ......" | 26548866 |
| hsa-miR-155-5p | breast cancer | RUNX2 | "MiR-155 targets 3´UTR region of multiple componen ......" | 27596294 |
| hsa-miR-155-5p | colon cancer | S100P | "In the present study we demonstrated for the first ......" | 23693020 |
| hsa-miR-155-5p | melanoma | SKI | "MicroRNA 155 targets the SKI gene in human melanom ......" | 21466664 |
| hsa-miR-155-5p | colorectal cancer | SMAD3 | "Here we demonstrated that TGF-β1 elevated the exp ......" | 27626488 |
| hsa-miR-155-5p | colorectal cancer | SMAD4 | "Here we demonstrated that TGF-β1 elevated the exp ......" | 27626488 |
| hsa-miR-155-5p | lymphoma | SMAD5 | "Targeting of SMAD5 links microRNA 155 to the TGF b ......" | 20133617 |
| hsa-miR-155-5p | lung cancer | SMARCA4 | "Importantly our experiments suggest that the oncog ......" | 25355421 |
| hsa-miR-155-5p | breast cancer | SOCS6 | "Further we identified SOCS6 as a new direct target ......" | 26692956 |
| hsa-miR-155-5p | lung cancer | SOS1 | "Furthermore the expression of SOS1 in miR-155-tran ......" | 21762626 |
| hsa-miR-155-5p | liver cancer | SOX6 | "Ectopic expression of sex-determining region Y box ......" | 21989846 |
| hsa-miR-155-5p | liver cancer | SRY | "Ectopic expression of sex-determining region Y box ......" | 21989846 |
| hsa-miR-155-5p | breast cancer | STAB2 | "We found that persistently elevated postoperative ......" | 25503185 |
| hsa-miR-155-5p | cervical and endocervical cancer | STK11 | "Mir 155 promotes cervical cancer cell proliferatio ......" | 25155037 |
| hsa-miR-155-5p | breast cancer | TAM | "Ectopic expression of miR-155 induces cell surviva ......" | 26692956 |
| hsa-miR-155-5p | breast cancer | TERF1 | "miR 155 drives telomere fragility in human breast ......" | 24876105 |
| hsa-miR-155-5p | B cell lymphoma | THM | "This finding was further supported by the observat ......" | 25498913 |
| hsa-miR-155-5p | breast cancer | TP63 | "The miR-155 host gene was directly repressed by p6 ......" | 22797073 |
| hsa-miR-155-5p | melanoma | TYRP1 | "SNPs at miR 155 binding sites of TYRP1 explain dis ......" | 26068396 |
| hsa-miR-155-5p | pancreatic cancer | VCL | "We identified that miR-155 was upregulated in PaC- ......" | 26195069 |
| hsa-miR-155-5p | breast cancer | VHL | "Here we demonstrate the role of miR-155 in angioge ......" | 23353819 |
| hsa-miR-155-5p | lung cancer | VIP | "7 case-control studies about the expression of mic ......" | 25603615 |
| hsa-miR-155-5p | ovarian cancer | XIAP | "Bioinformatic predictions the dual-luciferase repo ......" | 26779627 |
| hsa-miR-155-5p | colorectal cancer | ZEB1 | "E-cadherin expression levels decreased while ZEB1 ......" | 23588589 |
| hsa-miR-155-5p | breast cancer | ZNF652 | "A search for cancer-related target genes of miR-15 ......" | 22797073 |
Expression profile in cancer corhorts:
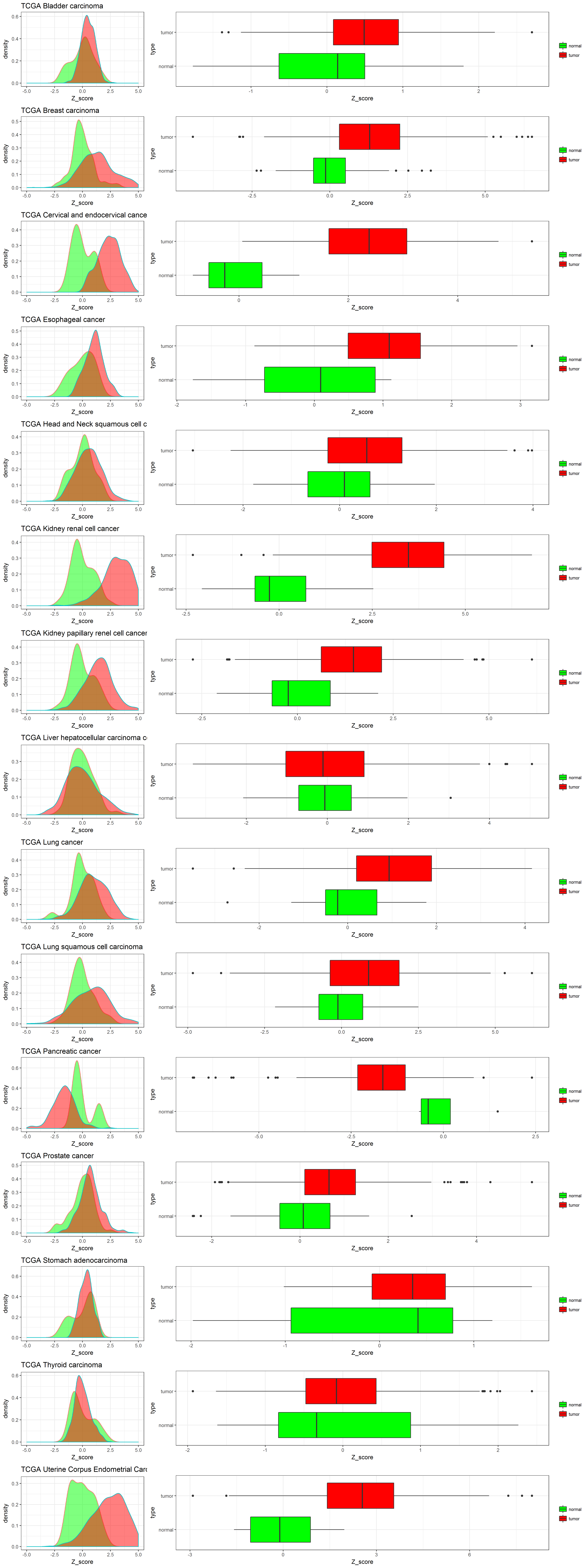
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-155-5p | ADAM10 | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LUAD; LUSC; PRAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.108; TCGA BRCA -0.079; TCGA CESC -0.127; TCGA HNSC -0.147; TCGA KIRP -0.139; TCGA LUAD -0.068; TCGA LUSC -0.113; TCGA PRAD -0.099; TCGA UCEC -0.09 |
hsa-miR-155-5p | ADH5 | 10 cancers: BLCA; BRCA; KIRC; KIRP; LIHC; LUAD; LUSC; SARC; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.066; TCGA BRCA -0.062; TCGA KIRC -0.13; TCGA KIRP -0.111; TCGA LIHC -0.138; TCGA LUAD -0.088; TCGA LUSC -0.112; TCGA SARC -0.068; TCGA THCA -0.067; TCGA STAD -0.102 |
hsa-miR-155-5p | AGL | 11 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; SARC; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.171; TCGA BRCA -0.135; TCGA HNSC -0.089; TCGA KIRC -0.21; TCGA KIRP -0.178; TCGA LIHC -0.155; TCGA LUAD -0.071; TCGA LUSC -0.107; TCGA SARC -0.095; TCGA THCA -0.106; TCGA STAD -0.128 |
hsa-miR-155-5p | AGTR1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; THCA; STAD; UCEC | miRNAWalker2 validate; miRTarBase | TCGA BLCA -0.184; TCGA BRCA -0.565; TCGA CESC -0.379; TCGA COAD -0.58; TCGA ESCA -0.374; TCGA HNSC -0.374; TCGA KIRC -0.26; TCGA KIRP -0.322; TCGA THCA -0.413; TCGA STAD -0.763; TCGA UCEC -0.371 |
hsa-miR-155-5p | AKR1C3 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; UCEC | miRNAWalker2 validate | TCGA BLCA -0.282; TCGA BRCA -0.17; TCGA CESC -0.42; TCGA ESCA -0.528; TCGA HNSC -0.66; TCGA LGG -0.366; TCGA LUAD -0.325; TCGA LUSC -0.547; TCGA UCEC -0.153 |
hsa-miR-155-5p | ALDH5A1 | 11 cancers: BLCA; BRCA; COAD; KIRC; KIRP; LGG; LIHC; LUAD; PAAD; SARC; THCA | miRNAWalker2 validate | TCGA BLCA -0.191; TCGA BRCA -0.086; TCGA COAD -0.134; TCGA KIRC -0.246; TCGA KIRP -0.225; TCGA LGG -0.29; TCGA LIHC -0.319; TCGA LUAD -0.14; TCGA PAAD -0.167; TCGA SARC -0.303; TCGA THCA -0.101 |
hsa-miR-155-5p | ANAPC16 | 10 cancers: BLCA; BRCA; ESCA; KIRC; LGG; LIHC; LUAD; PAAD; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.08; TCGA BRCA -0.051; TCGA ESCA -0.064; TCGA KIRC -0.07; TCGA LGG -0.07; TCGA LIHC -0.078; TCGA LUAD -0.101; TCGA PAAD -0.064; TCGA THCA -0.056; TCGA STAD -0.12 |
hsa-miR-155-5p | APC | 13 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; miRTarBase; miRNATAP | TCGA BLCA -0.062; TCGA BRCA -0.155; TCGA HNSC -0.085; TCGA KIRC -0.05; TCGA KIRP -0.135; TCGA LGG -0.071; TCGA LIHC -0.079; TCGA LUAD -0.08; TCGA LUSC -0.077; TCGA SARC -0.122; TCGA THCA -0.063; TCGA STAD -0.1; TCGA UCEC -0.087 |
hsa-miR-155-5p | ARID2 | 10 cancers: BLCA; BRCA; CESC; KIRP; LGG; LUAD; LUSC; PRAD; SARC; THCA | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | TCGA BLCA -0.159; TCGA BRCA -0.135; TCGA CESC -0.08; TCGA KIRP -0.056; TCGA LGG -0.068; TCGA LUAD -0.102; TCGA LUSC -0.073; TCGA PRAD -0.07; TCGA SARC -0.177; TCGA THCA -0.081 |
hsa-miR-155-5p | ARL15 | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LUAD; SARC; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.062; TCGA BRCA -0.166; TCGA CESC -0.099; TCGA COAD -0.148; TCGA HNSC -0.104; TCGA KIRC -0.144; TCGA KIRP -0.203; TCGA LUAD -0.073; TCGA SARC -0.085; TCGA UCEC -0.174 |
hsa-miR-155-5p | ATXN10 | 10 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD | miRNAWalker2 validate | TCGA BLCA -0.061; TCGA ESCA -0.101; TCGA HNSC -0.083; TCGA KIRC -0.114; TCGA KIRP -0.063; TCGA LGG -0.074; TCGA LIHC -0.078; TCGA LUAD -0.102; TCGA LUSC -0.079; TCGA PAAD -0.111 |
hsa-miR-155-5p | BCL6 | 9 cancers: BLCA; BRCA; ESCA; HNSC; LGG; LIHC; LUSC; STAD; UCEC | miRNAWalker2 validate; miRTarBase | TCGA BLCA -0.154; TCGA BRCA -0.102; TCGA ESCA -0.15; TCGA HNSC -0.146; TCGA LGG -0.122; TCGA LIHC -0.074; TCGA LUSC -0.092; TCGA STAD -0.188; TCGA UCEC -0.073 |
hsa-miR-155-5p | BRPF3 | 17 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; miRTarBase | TCGA BLCA -0.065; TCGA BRCA -0.104; TCGA CESC -0.152; TCGA COAD -0.17; TCGA ESCA -0.16; TCGA HNSC -0.087; TCGA KIRC -0.112; TCGA KIRP -0.156; TCGA LGG -0.146; TCGA LIHC -0.149; TCGA LUAD -0.12; TCGA LUSC -0.088; TCGA PAAD -0.088; TCGA PRAD -0.109; TCGA SARC -0.079; TCGA STAD -0.082; TCGA UCEC -0.099 |
hsa-miR-155-5p | CAB39L | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; OV; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.465; TCGA BRCA -0.288; TCGA CESC -0.242; TCGA COAD -0.569; TCGA ESCA -0.214; TCGA HNSC -0.085; TCGA KIRC -0.376; TCGA KIRP -0.22; TCGA LGG -0.108; TCGA LUAD -0.156; TCGA OV -0.119; TCGA SARC -0.147; TCGA THCA -0.098; TCGA STAD -0.548; TCGA UCEC -0.201 |
hsa-miR-155-5p | CAT | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LIHC; LUAD; LUSC; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.1; TCGA BRCA -0.184; TCGA CESC -0.23; TCGA COAD -0.131; TCGA ESCA -0.113; TCGA KIRC -0.312; TCGA KIRP -0.199; TCGA LIHC -0.253; TCGA LUAD -0.157; TCGA LUSC -0.143; TCGA THCA -0.128; TCGA UCEC -0.093 |
hsa-miR-155-5p | CBR4 | 11 cancers: BLCA; BRCA; KIRC; KIRP; LGG; LIHC; PAAD; PRAD; SARC; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.216; TCGA BRCA -0.17; TCGA KIRC -0.197; TCGA KIRP -0.134; TCGA LGG -0.157; TCGA LIHC -0.18; TCGA PAAD -0.141; TCGA PRAD -0.065; TCGA SARC -0.136; TCGA THCA -0.122; TCGA STAD -0.164 |
hsa-miR-155-5p | CDC42BPB | 16 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.095; TCGA BRCA -0.167; TCGA CESC -0.16; TCGA ESCA -0.106; TCGA HNSC -0.175; TCGA KIRC -0.143; TCGA KIRP -0.163; TCGA LGG -0.101; TCGA LIHC -0.086; TCGA LUAD -0.145; TCGA LUSC -0.221; TCGA PAAD -0.137; TCGA PRAD -0.07; TCGA SARC -0.06; TCGA THCA -0.059; TCGA STAD -0.09 |
hsa-miR-155-5p | CHD9 | 12 cancers: BLCA; BRCA; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUSC; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.114; TCGA BRCA -0.18; TCGA COAD -0.101; TCGA HNSC -0.143; TCGA KIRC -0.068; TCGA KIRP -0.17; TCGA LGG -0.132; TCGA LIHC -0.09; TCGA LUSC -0.096; TCGA SARC -0.187; TCGA STAD -0.13; TCGA UCEC -0.127 |
hsa-miR-155-5p | CNNM3 | 10 cancers: BLCA; BRCA; CESC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC | miRNAWalker2 validate | TCGA BLCA -0.082; TCGA BRCA -0.104; TCGA CESC -0.123; TCGA LGG -0.082; TCGA LIHC -0.195; TCGA LUAD -0.162; TCGA LUSC -0.057; TCGA PAAD -0.209; TCGA PRAD -0.066; TCGA SARC -0.069 |
hsa-miR-155-5p | COG2 | 10 cancers: BLCA; BRCA; CESC; COAD; KIRC; LGG; LIHC; LUAD; PAAD; SARC | miRNAWalker2 validate | TCGA BLCA -0.051; TCGA BRCA -0.098; TCGA CESC -0.055; TCGA COAD -0.057; TCGA KIRC -0.072; TCGA LGG -0.095; TCGA LIHC -0.061; TCGA LUAD -0.118; TCGA PAAD -0.119; TCGA SARC -0.08 |
hsa-miR-155-5p | DAG1 | 14 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA | miRNAWalker2 validate | TCGA BLCA -0.095; TCGA BRCA -0.099; TCGA CESC -0.112; TCGA ESCA -0.148; TCGA HNSC -0.053; TCGA KIRC -0.213; TCGA KIRP -0.107; TCGA LIHC -0.103; TCGA LUAD -0.162; TCGA LUSC -0.088; TCGA PAAD -0.103; TCGA PRAD -0.052; TCGA SARC -0.073; TCGA THCA -0.053 |
hsa-miR-155-5p | DCUN1D2 | 11 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; OV; PAAD; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.09; TCGA ESCA -0.099; TCGA HNSC -0.058; TCGA KIRC -0.095; TCGA KIRP -0.051; TCGA LGG -0.106; TCGA LIHC -0.091; TCGA OV -0.069; TCGA PAAD -0.174; TCGA THCA -0.057; TCGA STAD -0.181 |
hsa-miR-155-5p | DDX17 | 9 cancers: BLCA; BRCA; CESC; ESCA; KIRP; LGG; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.116; TCGA BRCA -0.091; TCGA CESC -0.081; TCGA ESCA -0.061; TCGA KIRP -0.096; TCGA LGG -0.119; TCGA SARC -0.088; TCGA STAD -0.094; TCGA UCEC -0.055 |
hsa-miR-155-5p | DET1 | 15 cancers: BLCA; BRCA; CESC; COAD; KIRC; KIRP; LGG; LIHC; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; miRTarBase | TCGA BLCA -0.137; TCGA BRCA -0.144; TCGA CESC -0.232; TCGA COAD -0.106; TCGA KIRC -0.071; TCGA KIRP -0.14; TCGA LGG -0.114; TCGA LIHC -0.09; TCGA LUAD -0.088; TCGA PAAD -0.099; TCGA PRAD -0.079; TCGA SARC -0.084; TCGA THCA -0.101; TCGA STAD -0.095; TCGA UCEC -0.115 |
hsa-miR-155-5p | DHX40 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; OV; PAAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; miRTarBase; MirTarget | TCGA BLCA -0.119; TCGA BRCA -0.127; TCGA CESC -0.165; TCGA COAD -0.091; TCGA ESCA -0.108; TCGA HNSC -0.09; TCGA LIHC -0.125; TCGA LUAD -0.11; TCGA LUSC -0.081; TCGA OV -0.098; TCGA PAAD -0.125; TCGA SARC -0.119; TCGA THCA -0.111; TCGA STAD -0.092; TCGA UCEC -0.074 |
hsa-miR-155-5p | DYNC2H1 | 15 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.154; TCGA BRCA -0.344; TCGA CESC -0.176; TCGA ESCA -0.212; TCGA HNSC -0.223; TCGA KIRC -0.148; TCGA KIRP -0.21; TCGA LGG -0.075; TCGA LUSC -0.188; TCGA OV -0.11; TCGA PRAD -0.075; TCGA SARC -0.156; TCGA THCA -0.151; TCGA STAD -0.395; TCGA UCEC -0.3 |
hsa-miR-155-5p | EDEM3 | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LIHC; LUAD; LUSC; UCEC | miRNAWalker2 validate | TCGA BLCA -0.057; TCGA BRCA -0.158; TCGA CESC -0.175; TCGA HNSC -0.137; TCGA KIRP -0.069; TCGA LIHC -0.058; TCGA LUAD -0.122; TCGA LUSC -0.084; TCGA UCEC -0.096 |
hsa-miR-155-5p | EEF1A2 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; OV; PAAD; SARC; STAD | miRNAWalker2 validate | TCGA BLCA -0.72; TCGA BRCA -0.632; TCGA CESC -0.29; TCGA COAD -0.345; TCGA ESCA -0.581; TCGA HNSC -0.724; TCGA LGG -0.105; TCGA OV -0.39; TCGA PAAD -0.823; TCGA SARC -0.586; TCGA STAD -0.475 |
hsa-miR-155-5p | FOXO3 | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; LGG; LUSC; SARC; STAD | miRNAWalker2 validate; miRTarBase | TCGA BLCA -0.07; TCGA BRCA -0.062; TCGA CESC -0.127; TCGA COAD -0.141; TCGA HNSC -0.118; TCGA LGG -0.102; TCGA LUSC -0.091; TCGA SARC -0.113; TCGA STAD -0.149 |
hsa-miR-155-5p | GCLC | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LIHC; LUSC; SARC; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.19; TCGA BRCA -0.057; TCGA COAD -0.081; TCGA ESCA -0.148; TCGA HNSC -0.191; TCGA LGG -0.222; TCGA LIHC -0.198; TCGA LUSC -0.25; TCGA SARC -0.068; TCGA THCA -0.065; TCGA STAD -0.079 |
hsa-miR-155-5p | GNAS | 11 cancers: BLCA; CESC; COAD; HNSC; LUAD; OV; PAAD; PRAD; SARC; THCA; STAD | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.117; TCGA CESC -0.118; TCGA COAD -0.142; TCGA HNSC -0.094; TCGA LUAD -0.088; TCGA OV -0.133; TCGA PAAD -0.366; TCGA PRAD -0.087; TCGA SARC -0.059; TCGA THCA -0.234; TCGA STAD -0.067 |
hsa-miR-155-5p | GPAM | 16 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.131; TCGA BRCA -0.365; TCGA CESC -0.099; TCGA HNSC -0.11; TCGA KIRC -0.177; TCGA KIRP -0.177; TCGA LGG -0.137; TCGA LIHC -0.33; TCGA LUAD -0.143; TCGA LUSC -0.084; TCGA OV -0.074; TCGA PRAD -0.067; TCGA SARC -0.155; TCGA THCA -0.23; TCGA STAD -0.157; TCGA UCEC -0.077 |
hsa-miR-155-5p | HBP1 | 10 cancers: BLCA; BRCA; COAD; HNSC; LIHC; LUAD; LUSC; SARC; STAD; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BLCA -0.055; TCGA BRCA -0.144; TCGA COAD -0.102; TCGA HNSC -0.084; TCGA LIHC -0.144; TCGA LUAD -0.093; TCGA LUSC -0.065; TCGA SARC -0.118; TCGA STAD -0.114; TCGA UCEC -0.122 |
hsa-miR-155-5p | HSDL1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PAAD; SARC; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.074; TCGA BRCA -0.106; TCGA CESC -0.082; TCGA COAD -0.155; TCGA ESCA -0.08; TCGA HNSC -0.055; TCGA KIRC -0.07; TCGA LGG -0.116; TCGA LUAD -0.117; TCGA LUSC -0.128; TCGA PAAD -0.068; TCGA SARC -0.098; TCGA THCA -0.089; TCGA STAD -0.138 |
hsa-miR-155-5p | KANK2 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRP; LIHC; LUAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.107; TCGA BRCA -0.219; TCGA CESC -0.278; TCGA COAD -0.161; TCGA ESCA -0.262; TCGA KIRP -0.158; TCGA LIHC -0.122; TCGA LUAD -0.111; TCGA SARC -0.244; TCGA STAD -0.431; TCGA UCEC -0.263 |
hsa-miR-155-5p | KDM3A | 11 cancers: BLCA; CESC; COAD; HNSC; LGG; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; miRTarBase; miRNATAP | TCGA BLCA -0.104; TCGA CESC -0.078; TCGA COAD -0.077; TCGA HNSC -0.096; TCGA LGG -0.094; TCGA LUAD -0.057; TCGA LUSC -0.101; TCGA SARC -0.086; TCGA THCA -0.059; TCGA STAD -0.133; TCGA UCEC -0.063 |
hsa-miR-155-5p | KIAA0430 | 10 cancers: BLCA; BRCA; CESC; COAD; KIRP; LGG; LUAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.126; TCGA BRCA -0.13; TCGA CESC -0.058; TCGA COAD -0.083; TCGA KIRP -0.082; TCGA LGG -0.138; TCGA LUAD -0.079; TCGA SARC -0.077; TCGA STAD -0.11; TCGA UCEC -0.102 |
hsa-miR-155-5p | LEMD3 | 12 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LGG; LUAD; LUSC; OV; SARC; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.051; TCGA BRCA -0.075; TCGA COAD -0.097; TCGA HNSC -0.057; TCGA KIRC -0.053; TCGA LGG -0.065; TCGA LUAD -0.075; TCGA LUSC -0.061; TCGA OV -0.055; TCGA SARC -0.132; TCGA THCA -0.064; TCGA UCEC -0.089 |
hsa-miR-155-5p | LNX2 | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; SARC; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.116; TCGA BRCA -0.108; TCGA COAD -0.223; TCGA ESCA -0.095; TCGA HNSC -0.115; TCGA KIRC -0.093; TCGA KIRP -0.103; TCGA LIHC -0.108; TCGA LUAD -0.251; TCGA LUSC -0.241; TCGA OV -0.1; TCGA SARC -0.119; TCGA THCA -0.079; TCGA STAD -0.074 |
hsa-miR-155-5p | LONP2 | 14 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; SARC; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.103; TCGA BRCA -0.118; TCGA CESC -0.11; TCGA ESCA -0.108; TCGA HNSC -0.172; TCGA KIRC -0.103; TCGA KIRP -0.136; TCGA LIHC -0.169; TCGA LUAD -0.089; TCGA LUSC -0.128; TCGA PRAD -0.083; TCGA SARC -0.062; TCGA THCA -0.074; TCGA STAD -0.074 |
hsa-miR-155-5p | LUC7L2 | 11 cancers: BLCA; COAD; ESCA; KIRP; LIHC; LUAD; LUSC; PRAD; SARC; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.062; TCGA COAD -0.065; TCGA ESCA -0.055; TCGA KIRP -0.076; TCGA LIHC -0.055; TCGA LUAD -0.062; TCGA LUSC -0.051; TCGA PRAD -0.055; TCGA SARC -0.078; TCGA THCA -0.106; TCGA UCEC -0.058 |
hsa-miR-155-5p | MAN1A2 | 13 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.061; TCGA BRCA -0.084; TCGA CESC -0.161; TCGA HNSC -0.164; TCGA KIRC -0.131; TCGA KIRP -0.199; TCGA LIHC -0.077; TCGA LUAD -0.135; TCGA LUSC -0.127; TCGA SARC -0.112; TCGA THCA -0.065; TCGA STAD -0.079; TCGA UCEC -0.071 |
hsa-miR-155-5p | MAVS | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.065; TCGA BRCA -0.111; TCGA CESC -0.103; TCGA COAD -0.102; TCGA ESCA -0.102; TCGA HNSC -0.121; TCGA KIRP -0.051; TCGA LIHC -0.05; TCGA LUAD -0.077; TCGA LUSC -0.153; TCGA PAAD -0.113; TCGA PRAD -0.092; TCGA SARC -0.058; TCGA THCA -0.07; TCGA STAD -0.139 |
hsa-miR-155-5p | MBLAC2 | 10 cancers: BLCA; BRCA; COAD; KIRP; LGG; LIHC; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.065; TCGA BRCA -0.185; TCGA COAD -0.12; TCGA KIRP -0.13; TCGA LGG -0.091; TCGA LIHC -0.208; TCGA SARC -0.177; TCGA THCA -0.112; TCGA STAD -0.105; TCGA UCEC -0.135 |
hsa-miR-155-5p | MECP2 | 13 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; miRTarBase; miRNATAP | TCGA BLCA -0.069; TCGA BRCA -0.103; TCGA COAD -0.073; TCGA ESCA -0.091; TCGA KIRC -0.088; TCGA KIRP -0.103; TCGA LUSC -0.07; TCGA PAAD -0.068; TCGA PRAD -0.065; TCGA SARC -0.088; TCGA THCA -0.072; TCGA STAD -0.178; TCGA UCEC -0.1 |
hsa-miR-155-5p | METTL7A | 9 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LIHC; SARC; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.392; TCGA BRCA -0.127; TCGA CESC -0.365; TCGA KIRC -0.242; TCGA KIRP -0.139; TCGA LIHC -0.292; TCGA SARC -0.157; TCGA THCA -0.196; TCGA UCEC -0.127 |
hsa-miR-155-5p | MSI2 | 14 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LUAD; OV; PAAD; PRAD; SARC; THCA | miRNAWalker2 validate | TCGA BLCA -0.129; TCGA BRCA -0.118; TCGA CESC -0.125; TCGA COAD -0.079; TCGA HNSC -0.113; TCGA KIRC -0.308; TCGA KIRP -0.243; TCGA LGG -0.06; TCGA LUAD -0.121; TCGA OV -0.108; TCGA PAAD -0.13; TCGA PRAD -0.072; TCGA SARC -0.168; TCGA THCA -0.069 |
hsa-miR-155-5p | MTFMT | 9 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD | miRNAWalker2 validate | TCGA BLCA -0.092; TCGA ESCA -0.07; TCGA HNSC -0.062; TCGA KIRC -0.054; TCGA KIRP -0.066; TCGA LIHC -0.084; TCGA LUAD -0.087; TCGA LUSC -0.092; TCGA PAAD -0.058 |
hsa-miR-155-5p | MYO6 | 10 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LUAD; LUSC; OV; PRAD; SARC | miRNAWalker2 validate | TCGA BLCA -0.175; TCGA BRCA -0.14; TCGA CESC -0.167; TCGA KIRC -0.255; TCGA KIRP -0.181; TCGA LUAD -0.116; TCGA LUSC -0.105; TCGA OV -0.074; TCGA PRAD -0.122; TCGA SARC -0.158 |
hsa-miR-155-5p | NCKAP1 | 15 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.091; TCGA CESC -0.111; TCGA COAD -0.071; TCGA ESCA -0.155; TCGA HNSC -0.242; TCGA KIRC -0.112; TCGA KIRP -0.131; TCGA LGG -0.082; TCGA LUAD -0.126; TCGA LUSC -0.212; TCGA PRAD -0.077; TCGA SARC -0.137; TCGA THCA -0.072; TCGA STAD -0.186; TCGA UCEC -0.087 |
hsa-miR-155-5p | NOVA1 | 10 cancers: BLCA; BRCA; KIRP; LGG; OV; PAAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.192; TCGA BRCA -0.683; TCGA KIRP -0.138; TCGA LGG -0.2; TCGA OV -0.435; TCGA PAAD -0.313; TCGA SARC -0.229; TCGA THCA -0.13; TCGA STAD -0.527; TCGA UCEC -0.41 |
hsa-miR-155-5p | OSBPL9 | 10 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.059; TCGA CESC -0.101; TCGA HNSC -0.053; TCGA KIRC -0.121; TCGA KIRP -0.122; TCGA LUAD -0.13; TCGA LUSC -0.088; TCGA PRAD -0.078; TCGA THCA -0.079; TCGA UCEC -0.07 |
hsa-miR-155-5p | PBRM1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LGG; LUAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.129; TCGA BRCA -0.101; TCGA CESC -0.102; TCGA COAD -0.088; TCGA ESCA -0.073; TCGA KIRC -0.072; TCGA KIRP -0.067; TCGA LGG -0.064; TCGA LUAD -0.118; TCGA STAD -0.15; TCGA UCEC -0.103 |
hsa-miR-155-5p | PCYOX1 | 17 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.059; TCGA BRCA -0.235; TCGA CESC -0.12; TCGA COAD -0.104; TCGA ESCA -0.137; TCGA HNSC -0.235; TCGA KIRC -0.18; TCGA KIRP -0.185; TCGA LIHC -0.228; TCGA LUAD -0.254; TCGA LUSC -0.24; TCGA OV -0.069; TCGA PAAD -0.147; TCGA SARC -0.126; TCGA THCA -0.113; TCGA STAD -0.231; TCGA UCEC -0.151 |
hsa-miR-155-5p | PEBP1 | 10 cancers: BLCA; BRCA; KIRC; KIRP; LIHC; LUAD; PAAD; PRAD; SARC; THCA | miRNAWalker2 validate | TCGA BLCA -0.148; TCGA BRCA -0.064; TCGA KIRC -0.258; TCGA KIRP -0.132; TCGA LIHC -0.233; TCGA LUAD -0.102; TCGA PAAD -0.198; TCGA PRAD -0.064; TCGA SARC -0.078; TCGA THCA -0.17 |
hsa-miR-155-5p | PGRMC2 | 9 cancers: BLCA; BRCA; ESCA; KIRP; LIHC; LUAD; LUSC; PAAD; THCA | miRNAWalker2 validate | TCGA BLCA -0.084; TCGA BRCA -0.144; TCGA ESCA -0.065; TCGA KIRP -0.077; TCGA LIHC -0.098; TCGA LUAD -0.119; TCGA LUSC -0.051; TCGA PAAD -0.148; TCGA THCA -0.07 |
hsa-miR-155-5p | PKN2 | 14 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | TCGA BLCA -0.111; TCGA BRCA -0.098; TCGA CESC -0.056; TCGA HNSC -0.063; TCGA KIRC -0.054; TCGA KIRP -0.108; TCGA LIHC -0.071; TCGA LUAD -0.095; TCGA LUSC -0.123; TCGA PRAD -0.056; TCGA SARC -0.062; TCGA THCA -0.088; TCGA STAD -0.054; TCGA UCEC -0.103 |
hsa-miR-155-5p | PLEKHA5 | 14 cancers: BLCA; BRCA; CESC; COAD; HNSC; LGG; LIHC; LUAD; LUSC; OV; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.17; TCGA BRCA -0.108; TCGA CESC -0.264; TCGA COAD -0.139; TCGA HNSC -0.134; TCGA LGG -0.122; TCGA LIHC -0.118; TCGA LUAD -0.11; TCGA LUSC -0.133; TCGA OV -0.102; TCGA PRAD -0.115; TCGA SARC -0.182; TCGA STAD -0.205; TCGA UCEC -0.065 |
hsa-miR-155-5p | PLS1 | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRC; LIHC; LUAD; LUSC; THCA | miRNAWalker2 validate | TCGA BLCA -0.134; TCGA BRCA -0.09; TCGA CESC -0.39; TCGA HNSC -0.201; TCGA KIRC -0.193; TCGA LIHC -0.173; TCGA LUAD -0.194; TCGA LUSC -0.211; TCGA THCA -0.07 |
hsa-miR-155-5p | PNPLA8 | 11 cancers: BLCA; BRCA; COAD; HNSC; KIRC; KIRP; LIHC; LUSC; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.094; TCGA BRCA -0.058; TCGA COAD -0.069; TCGA HNSC -0.14; TCGA KIRC -0.156; TCGA KIRP -0.138; TCGA LIHC -0.066; TCGA LUSC -0.08; TCGA SARC -0.052; TCGA STAD -0.161; TCGA UCEC -0.069 |
hsa-miR-155-5p | RAB11FIP2 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; OV; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BLCA -0.095; TCGA BRCA -0.135; TCGA CESC -0.112; TCGA COAD -0.214; TCGA ESCA -0.102; TCGA HNSC -0.06; TCGA KIRC -0.175; TCGA KIRP -0.201; TCGA LGG -0.14; TCGA LIHC -0.098; TCGA OV -0.157; TCGA PRAD -0.099; TCGA SARC -0.177; TCGA THCA -0.133; TCGA STAD -0.212; TCGA UCEC -0.243 |
hsa-miR-155-5p | RAPGEF2 | 13 cancers: BLCA; BRCA; CESC; COAD; KIRP; LGG; LIHC; LUAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.145; TCGA BRCA -0.113; TCGA CESC -0.149; TCGA COAD -0.102; TCGA KIRP -0.099; TCGA LGG -0.119; TCGA LIHC -0.127; TCGA LUAD -0.12; TCGA PRAD -0.068; TCGA SARC -0.065; TCGA THCA -0.132; TCGA STAD -0.074; TCGA UCEC -0.111 |
hsa-miR-155-5p | RBAK | 11 cancers: BLCA; COAD; HNSC; KIRP; LIHC; LUSC; OV; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.056; TCGA COAD -0.06; TCGA HNSC -0.086; TCGA KIRP -0.074; TCGA LIHC -0.086; TCGA LUSC -0.06; TCGA OV -0.086; TCGA SARC -0.117; TCGA THCA -0.076; TCGA STAD -0.07; TCGA UCEC -0.063 |
hsa-miR-155-5p | RCN2 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.058; TCGA CESC -0.084; TCGA COAD -0.104; TCGA ESCA -0.115; TCGA HNSC -0.126; TCGA KIRC -0.13; TCGA KIRP -0.129; TCGA LUAD -0.164; TCGA LUSC -0.116; TCGA PRAD -0.08; TCGA SARC -0.092; TCGA STAD -0.06; TCGA UCEC -0.071 |
hsa-miR-155-5p | RPRD1A | 13 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LGG; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.125; TCGA BRCA -0.081; TCGA ESCA -0.12; TCGA KIRC -0.107; TCGA KIRP -0.115; TCGA LGG -0.12; TCGA LUAD -0.127; TCGA PAAD -0.217; TCGA PRAD -0.102; TCGA SARC -0.157; TCGA THCA -0.061; TCGA STAD -0.194; TCGA UCEC -0.087 |
hsa-miR-155-5p | RREB1 | 10 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; THCA; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.055; TCGA BRCA -0.14; TCGA HNSC -0.116; TCGA KIRC -0.064; TCGA KIRP -0.135; TCGA LIHC -0.193; TCGA LUAD -0.14; TCGA LUSC -0.076; TCGA THCA -0.125; TCGA UCEC -0.061 |
hsa-miR-155-5p | RSF1 | 14 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.062; TCGA BRCA -0.078; TCGA CESC -0.129; TCGA HNSC -0.099; TCGA KIRC -0.079; TCGA KIRP -0.16; TCGA LGG -0.155; TCGA LUAD -0.098; TCGA LUSC -0.105; TCGA PRAD -0.056; TCGA SARC -0.106; TCGA THCA -0.051; TCGA STAD -0.148; TCGA UCEC -0.06 |
hsa-miR-155-5p | SACM1L | 9 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LIHC; LUAD; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.086; TCGA BRCA -0.098; TCGA CESC -0.059; TCGA KIRC -0.212; TCGA KIRP -0.136; TCGA LIHC -0.056; TCGA LUAD -0.072; TCGA THCA -0.104; TCGA UCEC -0.076 |
hsa-miR-155-5p | SCAMP1 | 18 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.089; TCGA BRCA -0.168; TCGA CESC -0.082; TCGA COAD -0.103; TCGA HNSC -0.103; TCGA KIRC -0.081; TCGA KIRP -0.076; TCGA LGG -0.079; TCGA LIHC -0.062; TCGA LUAD -0.094; TCGA LUSC -0.091; TCGA OV -0.072; TCGA PAAD -0.249; TCGA PRAD -0.067; TCGA SARC -0.155; TCGA THCA -0.102; TCGA STAD -0.149; TCGA UCEC -0.079 |
hsa-miR-155-5p | SEC24B | 10 cancers: BLCA; BRCA; HNSC; KIRP; LIHC; LUAD; LUSC; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.095; TCGA BRCA -0.107; TCGA HNSC -0.092; TCGA KIRP -0.099; TCGA LIHC -0.091; TCGA LUAD -0.074; TCGA LUSC -0.071; TCGA SARC -0.074; TCGA STAD -0.074; TCGA UCEC -0.062 |
hsa-miR-155-5p | SIN3A | 12 cancers: BLCA; BRCA; CESC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.069; TCGA BRCA -0.085; TCGA CESC -0.072; TCGA KIRP -0.074; TCGA LGG -0.06; TCGA LIHC -0.056; TCGA LUAD -0.098; TCGA LUSC -0.083; TCGA SARC -0.097; TCGA THCA -0.062; TCGA STAD -0.054; TCGA UCEC -0.069 |
hsa-miR-155-5p | SMAD3 | 9 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LIHC; LUSC; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.168; TCGA BRCA -0.161; TCGA HNSC -0.125; TCGA KIRC -0.059; TCGA KIRP -0.069; TCGA LIHC -0.144; TCGA LUSC -0.099; TCGA THCA -0.071; TCGA UCEC -0.118 |
hsa-miR-155-5p | SMAD4 | 13 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LGG; LIHC; LUAD; PAAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.09; TCGA BRCA -0.114; TCGA ESCA -0.099; TCGA KIRC -0.088; TCGA KIRP -0.088; TCGA LGG -0.118; TCGA LIHC -0.074; TCGA LUAD -0.118; TCGA PAAD -0.123; TCGA SARC -0.123; TCGA THCA -0.069; TCGA STAD -0.11; TCGA UCEC -0.125 |
hsa-miR-155-5p | SMAD5 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRP; LGG; LUAD; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; miRTarBase | TCGA BLCA -0.106; TCGA BRCA -0.166; TCGA CESC -0.114; TCGA COAD -0.091; TCGA ESCA -0.078; TCGA KIRP -0.089; TCGA LGG -0.058; TCGA LUAD -0.105; TCGA OV -0.078; TCGA PAAD -0.11; TCGA PRAD -0.06; TCGA SARC -0.169; TCGA THCA -0.096; TCGA STAD -0.16; TCGA UCEC -0.159 |
hsa-miR-155-5p | SYPL1 | 10 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.05; TCGA BRCA -0.062; TCGA HNSC -0.075; TCGA KIRC -0.109; TCGA KIRP -0.071; TCGA LIHC -0.056; TCGA LUAD -0.076; TCGA LUSC -0.09; TCGA THCA -0.08; TCGA UCEC -0.123 |
hsa-miR-155-5p | TBC1D14 | 15 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PAAD; PRAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.058; TCGA BRCA -0.129; TCGA CESC -0.071; TCGA ESCA -0.126; TCGA HNSC -0.116; TCGA KIRC -0.221; TCGA KIRP -0.076; TCGA LGG -0.138; TCGA LIHC -0.055; TCGA LUAD -0.095; TCGA LUSC -0.133; TCGA OV -0.062; TCGA PAAD -0.108; TCGA PRAD -0.075; TCGA UCEC -0.05 |
hsa-miR-155-5p | TBC1D8B | 11 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LIHC; LUAD; LUSC; PRAD; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.09; TCGA BRCA -0.167; TCGA CESC -0.244; TCGA HNSC -0.259; TCGA KIRP -0.152; TCGA LIHC -0.073; TCGA LUAD -0.108; TCGA LUSC -0.122; TCGA PRAD -0.072; TCGA THCA -0.083; TCGA UCEC -0.192 |
hsa-miR-155-5p | TCF12 | 11 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LGG; LUAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.081; TCGA BRCA -0.106; TCGA CESC -0.195; TCGA HNSC -0.101; TCGA KIRP -0.11; TCGA LGG -0.323; TCGA LUAD -0.087; TCGA PRAD -0.065; TCGA SARC -0.105; TCGA STAD -0.09; TCGA UCEC -0.107 |
hsa-miR-155-5p | TJP1 | 14 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.106; TCGA BRCA -0.139; TCGA CESC -0.133; TCGA HNSC -0.147; TCGA KIRC -0.114; TCGA KIRP -0.118; TCGA LIHC -0.116; TCGA LUAD -0.202; TCGA LUSC -0.174; TCGA PRAD -0.072; TCGA SARC -0.127; TCGA THCA -0.057; TCGA STAD -0.247; TCGA UCEC -0.127 |
hsa-miR-155-5p | TRAK1 | 11 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; THCA | miRNAWalker2 validate | TCGA BLCA -0.322; TCGA BRCA -0.185; TCGA CESC -0.196; TCGA HNSC -0.083; TCGA KIRC -0.15; TCGA KIRP -0.124; TCGA LGG -0.136; TCGA LIHC -0.053; TCGA LUAD -0.104; TCGA LUSC -0.092; TCGA THCA -0.093 |
hsa-miR-155-5p | TRIM24 | 11 cancers: BLCA; COAD; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; SARC; THCA | miRNAWalker2 validate | TCGA BLCA -0.152; TCGA COAD -0.129; TCGA KIRC -0.054; TCGA KIRP -0.133; TCGA LIHC -0.09; TCGA LUAD -0.137; TCGA LUSC -0.105; TCGA OV -0.056; TCGA PAAD -0.077; TCGA SARC -0.079; TCGA THCA -0.103 |
hsa-miR-155-5p | TTC37 | 12 cancers: BLCA; BRCA; CESC; COAD; KIRC; KIRP; LIHC; LUAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.071; TCGA BRCA -0.163; TCGA CESC -0.143; TCGA COAD -0.173; TCGA KIRC -0.071; TCGA KIRP -0.227; TCGA LIHC -0.126; TCGA LUAD -0.068; TCGA SARC -0.113; TCGA THCA -0.068; TCGA STAD -0.115; TCGA UCEC -0.136 |
hsa-miR-155-5p | VEZF1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.101; TCGA BRCA -0.169; TCGA CESC -0.203; TCGA COAD -0.105; TCGA ESCA -0.099; TCGA KIRC -0.105; TCGA KIRP -0.096; TCGA LGG -0.058; TCGA LUAD -0.086; TCGA LUSC -0.061; TCGA PAAD -0.088; TCGA STAD -0.162; TCGA UCEC -0.071 |
hsa-miR-155-5p | VPS36 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.115; TCGA BRCA -0.106; TCGA CESC -0.062; TCGA COAD -0.135; TCGA ESCA -0.083; TCGA HNSC -0.103; TCGA KIRC -0.107; TCGA KIRP -0.105; TCGA LGG -0.061; TCGA LIHC -0.102; TCGA LUAD -0.087; TCGA LUSC -0.08; TCGA SARC -0.052; TCGA THCA -0.065; TCGA STAD -0.101; TCGA UCEC -0.067 |
hsa-miR-155-5p | WDR11 | 9 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LIHC; PRAD; SARC; UCEC | miRNAWalker2 validate | TCGA BLCA -0.055; TCGA BRCA -0.082; TCGA CESC -0.051; TCGA KIRC -0.094; TCGA KIRP -0.114; TCGA LIHC -0.06; TCGA PRAD -0.069; TCGA SARC -0.07; TCGA UCEC -0.092 |
hsa-miR-155-5p | WEE1 | 11 cancers: BLCA; BRCA; CESC; COAD; KIRC; KIRP; LUAD; LUSC; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BLCA -0.119; TCGA BRCA -0.097; TCGA CESC -0.115; TCGA COAD -0.089; TCGA KIRC -0.135; TCGA KIRP -0.121; TCGA LUAD -0.089; TCGA LUSC -0.083; TCGA THCA -0.088; TCGA STAD -0.09; TCGA UCEC -0.087 |
hsa-miR-155-5p | WRB | 12 cancers: BLCA; BRCA; CESC; ESCA; LIHC; LUAD; OV; PAAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.114; TCGA BRCA -0.112; TCGA CESC -0.096; TCGA ESCA -0.169; TCGA LIHC -0.085; TCGA LUAD -0.093; TCGA OV -0.053; TCGA PAAD -0.2; TCGA SARC -0.119; TCGA THCA -0.075; TCGA STAD -0.076; TCGA UCEC -0.086 |
hsa-miR-155-5p | ZNF236 | 13 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LGG; LIHC; LUAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.093; TCGA BRCA -0.12; TCGA CESC -0.109; TCGA KIRC -0.056; TCGA KIRP -0.126; TCGA LGG -0.122; TCGA LIHC -0.064; TCGA LUAD -0.092; TCGA PRAD -0.082; TCGA SARC -0.161; TCGA THCA -0.067; TCGA STAD -0.069; TCGA UCEC -0.08 |
hsa-miR-155-5p | ZNF248 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; LGG; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.207; TCGA BRCA -0.09; TCGA CESC -0.164; TCGA COAD -0.093; TCGA ESCA -0.087; TCGA LGG -0.27; TCGA LUAD -0.084; TCGA PAAD -0.075; TCGA PRAD -0.059; TCGA SARC -0.156; TCGA THCA -0.128; TCGA STAD -0.239; TCGA UCEC -0.137 |
hsa-miR-155-5p | ZNF254 | 16 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; OV; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.256; TCGA BRCA -0.085; TCGA CESC -0.265; TCGA COAD -0.306; TCGA HNSC -0.117; TCGA KIRC -0.116; TCGA KIRP -0.108; TCGA LGG -0.108; TCGA LIHC -0.109; TCGA LUAD -0.151; TCGA OV -0.067; TCGA PRAD -0.067; TCGA SARC -0.149; TCGA THCA -0.101; TCGA STAD -0.23; TCGA UCEC -0.088 |
hsa-miR-155-5p | ZNF493 | 11 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LGG; OV; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.218; TCGA BRCA -0.194; TCGA CESC -0.207; TCGA KIRC -0.139; TCGA KIRP -0.217; TCGA LGG -0.15; TCGA OV -0.081; TCGA SARC -0.193; TCGA THCA -0.111; TCGA STAD -0.127; TCGA UCEC -0.131 |
hsa-miR-155-5p | ZNF561 | 10 cancers: BLCA; BRCA; KIRP; LIHC; OV; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.192; TCGA BRCA -0.109; TCGA KIRP -0.052; TCGA LIHC -0.087; TCGA OV -0.091; TCGA PAAD -0.107; TCGA PRAD -0.055; TCGA SARC -0.163; TCGA STAD -0.051; TCGA UCEC -0.062 |
hsa-miR-155-5p | ZNF652 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LGG; LIHC; LUAD; PAAD; SARC; THCA; STAD | miRNAWalker2 validate; miRTarBase; MirTarget | TCGA BLCA -0.181; TCGA BRCA -0.112; TCGA CESC -0.173; TCGA ESCA -0.085; TCGA HNSC -0.055; TCGA LGG -0.058; TCGA LIHC -0.091; TCGA LUAD -0.098; TCGA PAAD -0.062; TCGA SARC -0.115; TCGA THCA -0.06; TCGA STAD -0.097 |
hsa-miR-155-5p | RNF123 | 9 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LIHC; LUAD; PAAD; PRAD | miRTarBase; MirTarget | TCGA BLCA -0.104; TCGA BRCA -0.108; TCGA CESC -0.071; TCGA KIRC -0.146; TCGA KIRP -0.08; TCGA LIHC -0.092; TCGA LUAD -0.085; TCGA PAAD -0.108; TCGA PRAD -0.053 |
hsa-miR-155-5p | CEP68 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.087; TCGA BRCA -0.172; TCGA CESC -0.08; TCGA COAD -0.203; TCGA ESCA -0.135; TCGA HNSC -0.064; TCGA KIRC -0.068; TCGA KIRP -0.106; TCGA LGG -0.145; TCGA LUAD -0.09; TCGA LUSC -0.075; TCGA SARC -0.146; TCGA STAD -0.225; TCGA UCEC -0.172 |
hsa-miR-155-5p | LCA5 | 11 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRP; LUSC; OV; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.175; TCGA BRCA -0.294; TCGA CESC -0.205; TCGA COAD -0.165; TCGA HNSC -0.145; TCGA KIRP -0.088; TCGA LUSC -0.113; TCGA OV -0.137; TCGA SARC -0.192; TCGA STAD -0.266; TCGA UCEC -0.278 |
hsa-miR-155-5p | UNC80 | 10 cancers: BLCA; KIRC; LGG; LUSC; OV; PAAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.237; TCGA KIRC -0.198; TCGA LGG -0.336; TCGA LUSC -0.45; TCGA OV -0.529; TCGA PAAD -0.734; TCGA SARC -0.314; TCGA THCA -0.362; TCGA STAD -0.419; TCGA UCEC -0.424 |
hsa-miR-155-5p | STRN3 | 14 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LGG; LIHC; LUSC; PRAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.064; TCGA BRCA -0.156; TCGA CESC -0.1; TCGA HNSC -0.141; TCGA KIRC -0.052; TCGA KIRP -0.08; TCGA LGG -0.088; TCGA LIHC -0.086; TCGA LUSC -0.118; TCGA PRAD -0.073; TCGA SARC -0.109; TCGA THCA -0.066; TCGA STAD -0.201; TCGA UCEC -0.077 |
hsa-miR-155-5p | REPS2 | 12 cancers: BLCA; BRCA; CESC; COAD; LGG; LIHC; LUAD; LUSC; OV; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.133; TCGA BRCA -0.191; TCGA CESC -0.316; TCGA COAD -0.198; TCGA LGG -0.486; TCGA LIHC -0.198; TCGA LUAD -0.16; TCGA LUSC -0.239; TCGA OV -0.082; TCGA SARC -0.191; TCGA STAD -0.175; TCGA UCEC -0.11 |
hsa-miR-155-5p | XIAP | 11 cancers: BLCA; BRCA; COAD; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; STAD | MirTarget | TCGA BLCA -0.076; TCGA BRCA -0.099; TCGA COAD -0.071; TCGA HNSC -0.067; TCGA KIRC -0.072; TCGA KIRP -0.079; TCGA LIHC -0.052; TCGA LUAD -0.081; TCGA LUSC -0.073; TCGA PRAD -0.059; TCGA STAD -0.09 |
hsa-miR-155-5p | GDPD1 | 14 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LGG; LUAD; LUSC; OV; PAAD; PRAD; SARC; THCA; UCEC | MirTarget | TCGA BLCA -0.257; TCGA BRCA -0.161; TCGA CESC -0.3; TCGA KIRC -0.13; TCGA KIRP -0.162; TCGA LGG -0.305; TCGA LUAD -0.192; TCGA LUSC -0.115; TCGA OV -0.176; TCGA PAAD -0.313; TCGA PRAD -0.146; TCGA SARC -0.13; TCGA THCA -0.195; TCGA UCEC -0.079 |
hsa-miR-155-5p | SATB1 | 10 cancers: BLCA; CESC; COAD; KIRC; KIRP; LGG; OV; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.285; TCGA CESC -0.195; TCGA COAD -0.372; TCGA KIRC -0.177; TCGA KIRP -0.112; TCGA LGG -0.318; TCGA OV -0.216; TCGA THCA -0.108; TCGA STAD -0.3; TCGA UCEC -0.205 |
hsa-miR-155-5p | NDFIP1 | 13 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LIHC; LUAD; OV; PAAD; PRAD; SARC; THCA; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.06; TCGA BRCA -0.144; TCGA CESC -0.065; TCGA KIRC -0.12; TCGA KIRP -0.086; TCGA LIHC -0.11; TCGA LUAD -0.084; TCGA OV -0.062; TCGA PAAD -0.23; TCGA PRAD -0.075; TCGA SARC -0.108; TCGA THCA -0.144; TCGA UCEC -0.055 |
hsa-miR-155-5p | RUFY2 | 10 cancers: BLCA; BRCA; CESC; KIRP; LGG; LUAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.088; TCGA BRCA -0.153; TCGA CESC -0.055; TCGA KIRP -0.145; TCGA LGG -0.151; TCGA LUAD -0.08; TCGA SARC -0.11; TCGA THCA -0.059; TCGA STAD -0.16; TCGA UCEC -0.118 |
hsa-miR-155-5p | GPD1L | 13 cancers: BLCA; BRCA; CESC; ESCA; KIRC; KIRP; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.345; TCGA BRCA -0.323; TCGA CESC -0.172; TCGA ESCA -0.154; TCGA KIRC -0.399; TCGA KIRP -0.194; TCGA LUAD -0.221; TCGA PAAD -0.098; TCGA PRAD -0.076; TCGA SARC -0.173; TCGA THCA -0.168; TCGA STAD -0.094; TCGA UCEC -0.113 |
hsa-miR-155-5p | MAP4K3 | 17 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.228; TCGA BRCA -0.114; TCGA CESC -0.182; TCGA COAD -0.111; TCGA HNSC -0.139; TCGA KIRC -0.191; TCGA KIRP -0.174; TCGA LGG -0.067; TCGA LIHC -0.087; TCGA LUAD -0.136; TCGA LUSC -0.148; TCGA OV -0.106; TCGA PRAD -0.057; TCGA SARC -0.132; TCGA THCA -0.107; TCGA STAD -0.188; TCGA UCEC -0.135 |
hsa-miR-155-5p | KDM5B | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; OV; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.118; TCGA CESC -0.109; TCGA COAD -0.117; TCGA ESCA -0.094; TCGA HNSC -0.218; TCGA LGG -0.148; TCGA LUAD -0.17; TCGA LUSC -0.126; TCGA OV -0.142; TCGA SARC -0.254; TCGA STAD -0.137; TCGA UCEC -0.102 |
hsa-miR-155-5p | PLEKHA1 | 12 cancers: BLCA; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; OV; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.108; TCGA ESCA -0.084; TCGA HNSC -0.187; TCGA KIRP -0.074; TCGA LIHC -0.07; TCGA LUAD -0.197; TCGA LUSC -0.14; TCGA OV -0.069; TCGA SARC -0.121; TCGA THCA -0.108; TCGA STAD -0.163; TCGA UCEC -0.056 |
hsa-miR-155-5p | ANO5 | 13 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LGG; OV; PAAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.398; TCGA BRCA -0.273; TCGA CESC -0.434; TCGA HNSC -0.252; TCGA KIRC -0.667; TCGA KIRP -0.163; TCGA LGG -0.137; TCGA OV -0.313; TCGA PAAD -0.246; TCGA SARC -0.378; TCGA THCA -0.162; TCGA STAD -0.643; TCGA UCEC -0.329 |
hsa-miR-155-5p | TM9SF3 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; THCA; UCEC | MirTarget | TCGA BLCA -0.08; TCGA BRCA -0.057; TCGA CESC -0.142; TCGA ESCA -0.115; TCGA HNSC -0.098; TCGA KIRC -0.076; TCGA KIRP -0.071; TCGA LGG -0.101; TCGA LIHC -0.05; TCGA LUAD -0.132; TCGA THCA -0.063; TCGA UCEC -0.086 |
hsa-miR-155-5p | ZNF260 | 10 cancers: BLCA; CESC; COAD; KIRC; KIRP; LIHC; OV; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.068; TCGA CESC -0.107; TCGA COAD -0.154; TCGA KIRC -0.078; TCGA KIRP -0.084; TCGA LIHC -0.066; TCGA OV -0.082; TCGA SARC -0.128; TCGA THCA -0.066; TCGA STAD -0.079 |
hsa-miR-155-5p | ZMYM2 | 13 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRP; LGG; LUAD; OV; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.187; TCGA BRCA -0.11; TCGA CESC -0.161; TCGA COAD -0.202; TCGA HNSC -0.091; TCGA KIRP -0.06; TCGA LGG -0.092; TCGA LUAD -0.093; TCGA OV -0.096; TCGA SARC -0.154; TCGA THCA -0.094; TCGA STAD -0.161; TCGA UCEC -0.061 |
hsa-miR-155-5p | BRD1 | 12 cancers: BLCA; CESC; HNSC; LGG; LIHC; LUAD; LUSC; OV; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.123; TCGA CESC -0.079; TCGA HNSC -0.102; TCGA LGG -0.081; TCGA LIHC -0.055; TCGA LUAD -0.118; TCGA LUSC -0.11; TCGA OV -0.065; TCGA PRAD -0.061; TCGA SARC -0.131; TCGA STAD -0.088; TCGA UCEC -0.078 |
hsa-miR-155-5p | ATRNL1 | 12 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LGG; OV; PAAD; SARC; THCA; STAD; UCEC | MirTarget; mirMAP | TCGA BLCA -0.203; TCGA BRCA -0.342; TCGA COAD -0.472; TCGA HNSC -0.354; TCGA KIRC -0.641; TCGA LGG -0.318; TCGA OV -0.791; TCGA PAAD -0.538; TCGA SARC -0.384; TCGA THCA -0.226; TCGA STAD -0.565; TCGA UCEC -0.414 |
hsa-miR-155-5p | TTC21B | 16 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.119; TCGA BRCA -0.116; TCGA CESC -0.157; TCGA COAD -0.072; TCGA HNSC -0.08; TCGA KIRP -0.091; TCGA LGG -0.072; TCGA LIHC -0.09; TCGA LUAD -0.076; TCGA LUSC -0.132; TCGA OV -0.096; TCGA PAAD -0.06; TCGA PRAD -0.108; TCGA SARC -0.139; TCGA STAD -0.142; TCGA UCEC -0.148 |
hsa-miR-155-5p | ENTPD3 | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.436; TCGA BRCA -0.245; TCGA CESC -0.252; TCGA ESCA -0.545; TCGA HNSC -0.245; TCGA KIRC -0.743; TCGA KIRP -0.229; TCGA LUAD -0.226; TCGA LUSC -0.281; TCGA OV -0.152; TCGA SARC -0.305; TCGA STAD -0.475; TCGA UCEC -0.184 |
hsa-miR-155-5p | CCNT2 | 15 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; OV; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.196; TCGA BRCA -0.068; TCGA CESC -0.106; TCGA COAD -0.069; TCGA HNSC -0.08; TCGA KIRP -0.054; TCGA LGG -0.083; TCGA LIHC -0.062; TCGA LUAD -0.06; TCGA LUSC -0.096; TCGA OV -0.051; TCGA SARC -0.149; TCGA THCA -0.061; TCGA STAD -0.069; TCGA UCEC -0.081 |
hsa-miR-155-5p | DCLRE1A | 9 cancers: BLCA; KIRC; KIRP; LGG; LIHC; LUAD; PAAD; SARC; THCA | MirTarget; miRNATAP | TCGA BLCA -0.052; TCGA KIRC -0.056; TCGA KIRP -0.096; TCGA LGG -0.084; TCGA LIHC -0.073; TCGA LUAD -0.083; TCGA PAAD -0.136; TCGA SARC -0.087; TCGA THCA -0.063 |
hsa-miR-155-5p | SORT1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.134; TCGA BRCA -0.11; TCGA CESC -0.265; TCGA COAD -0.157; TCGA ESCA -0.189; TCGA HNSC -0.194; TCGA KIRC -0.282; TCGA KIRP -0.138; TCGA LUAD -0.217; TCGA LUSC -0.179; TCGA PAAD -0.108; TCGA PRAD -0.072; TCGA THCA -0.117; TCGA STAD -0.221 |
hsa-miR-155-5p | CNTN3 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LGG; OV; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.663; TCGA BRCA -0.221; TCGA CESC -0.374; TCGA COAD -0.382; TCGA ESCA -0.379; TCGA KIRC -0.903; TCGA KIRP -0.192; TCGA LGG -0.859; TCGA OV -0.325; TCGA PRAD -0.11; TCGA SARC -0.319; TCGA THCA -0.403; TCGA STAD -0.439; TCGA UCEC -0.289 |
hsa-miR-155-5p | SCUBE2 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LUAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.607; TCGA BRCA -0.723; TCGA CESC -0.404; TCGA COAD -0.442; TCGA ESCA -0.4; TCGA KIRC -0.191; TCGA KIRP -0.13; TCGA LUAD -0.236; TCGA SARC -0.428; TCGA STAD -0.411; TCGA UCEC -0.182 |
hsa-miR-155-5p | BSDC1 | 9 cancers: BLCA; BRCA; KIRP; LIHC; LUAD; PAAD; PRAD; THCA; UCEC | MirTarget | TCGA BLCA -0.058; TCGA BRCA -0.089; TCGA KIRP -0.051; TCGA LIHC -0.087; TCGA LUAD -0.094; TCGA PAAD -0.096; TCGA PRAD -0.06; TCGA THCA -0.053; TCGA UCEC -0.07 |
hsa-miR-155-5p | LRP1B | 10 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LGG; LUSC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.343; TCGA BRCA -0.351; TCGA HNSC -0.337; TCGA KIRC -0.711; TCGA KIRP -0.378; TCGA LGG -0.349; TCGA LUSC -0.539; TCGA THCA -0.348; TCGA STAD -0.621; TCGA UCEC -0.209 |
hsa-miR-155-5p | TAPT1 | 16 cancers: BLCA; BRCA; CESC; COAD; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.112; TCGA BRCA -0.224; TCGA CESC -0.095; TCGA COAD -0.081; TCGA KIRC -0.105; TCGA KIRP -0.109; TCGA LGG -0.157; TCGA LIHC -0.208; TCGA LUAD -0.178; TCGA LUSC -0.112; TCGA PAAD -0.087; TCGA PRAD -0.067; TCGA SARC -0.121; TCGA THCA -0.101; TCGA STAD -0.101; TCGA UCEC -0.063 |
hsa-miR-155-5p | MIER3 | 10 cancers: BLCA; BRCA; CESC; KIRP; LIHC; LUAD; OV; SARC; THCA; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.094; TCGA BRCA -0.142; TCGA CESC -0.135; TCGA KIRP -0.059; TCGA LIHC -0.098; TCGA LUAD -0.077; TCGA OV -0.08; TCGA SARC -0.171; TCGA THCA -0.075; TCGA UCEC -0.114 |
hsa-miR-155-5p | TRIM23 | 13 cancers: BLCA; BRCA; COAD; KIRP; LGG; LIHC; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.068; TCGA BRCA -0.207; TCGA COAD -0.092; TCGA KIRP -0.056; TCGA LGG -0.143; TCGA LIHC -0.117; TCGA OV -0.07; TCGA PAAD -0.084; TCGA PRAD -0.068; TCGA SARC -0.165; TCGA THCA -0.069; TCGA STAD -0.176; TCGA UCEC -0.141 |
hsa-miR-155-5p | TCF7L2 | 9 cancers: BLCA; CESC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; THCA | MirTarget; miRNATAP | TCGA BLCA -0.104; TCGA CESC -0.104; TCGA KIRP -0.109; TCGA LGG -0.147; TCGA LIHC -0.07; TCGA LUAD -0.174; TCGA LUSC -0.068; TCGA SARC -0.249; TCGA THCA -0.072 |
hsa-miR-155-5p | NEDD4L | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD | mirMAP | TCGA BLCA -0.173; TCGA BRCA -0.277; TCGA CESC -0.081; TCGA COAD -0.091; TCGA ESCA -0.255; TCGA HNSC -0.08; TCGA KIRC -0.359; TCGA KIRP -0.195; TCGA LIHC -0.07; TCGA LUAD -0.325; TCGA LUSC -0.216; TCGA PAAD -0.2; TCGA PRAD -0.16 |
hsa-miR-155-5p | ZNF275 | 13 cancers: BLCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LIHC; OV; PRAD; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.075; TCGA CESC -0.19; TCGA COAD -0.091; TCGA HNSC -0.098; TCGA KIRC -0.075; TCGA KIRP -0.154; TCGA LGG -0.198; TCGA LIHC -0.085; TCGA OV -0.083; TCGA PRAD -0.13; TCGA SARC -0.113; TCGA THCA -0.075; TCGA UCEC -0.089 |
hsa-miR-155-5p | DPP8 | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.065; TCGA BRCA -0.149; TCGA CESC -0.152; TCGA ESCA -0.075; TCGA HNSC -0.176; TCGA KIRC -0.146; TCGA KIRP -0.25; TCGA LGG -0.103; TCGA LUAD -0.06; TCGA LUSC -0.095; TCGA PRAD -0.095; TCGA STAD -0.114; TCGA UCEC -0.095 |
hsa-miR-155-5p | OPHN1 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.161; TCGA BRCA -0.128; TCGA CESC -0.18; TCGA COAD -0.104; TCGA ESCA -0.155; TCGA HNSC -0.1; TCGA KIRC -0.363; TCGA KIRP -0.258; TCGA LGG -0.459; TCGA LUAD -0.289; TCGA LUSC -0.16; TCGA PAAD -0.15; TCGA SARC -0.183; TCGA THCA -0.189; TCGA STAD -0.235; TCGA UCEC -0.118 |
hsa-miR-155-5p | COBLL1 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD | mirMAP | TCGA BLCA -0.219; TCGA BRCA -0.149; TCGA COAD -0.243; TCGA ESCA -0.218; TCGA HNSC -0.155; TCGA KIRC -0.527; TCGA KIRP -0.359; TCGA LIHC -0.286; TCGA LUAD -0.149; TCGA LUSC -0.289; TCGA PRAD -0.076 |
hsa-miR-155-5p | ZNF264 | 9 cancers: BLCA; BRCA; CESC; LIHC; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.114; TCGA BRCA -0.111; TCGA CESC -0.106; TCGA LIHC -0.059; TCGA PRAD -0.056; TCGA SARC -0.09; TCGA THCA -0.052; TCGA STAD -0.142; TCGA UCEC -0.056 |
hsa-miR-155-5p | ARHGAP5 | 14 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.11; TCGA BRCA -0.14; TCGA CESC -0.107; TCGA HNSC -0.133; TCGA KIRC -0.206; TCGA KIRP -0.195; TCGA LGG -0.089; TCGA LIHC -0.107; TCGA LUAD -0.103; TCGA LUSC -0.126; TCGA SARC -0.147; TCGA THCA -0.12; TCGA STAD -0.183; TCGA UCEC -0.152 |
hsa-miR-155-5p | ZC3H12B | 13 cancers: BLCA; BRCA; CESC; COAD; KIRC; KIRP; LGG; OV; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.275; TCGA BRCA -0.101; TCGA CESC -0.191; TCGA COAD -0.322; TCGA KIRC -0.196; TCGA KIRP -0.207; TCGA LGG -0.446; TCGA OV -0.124; TCGA PAAD -0.171; TCGA SARC -0.365; TCGA THCA -0.133; TCGA STAD -0.339; TCGA UCEC -0.283 |
hsa-miR-155-5p | ZDHHC15 | 12 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.179; TCGA BRCA -0.239; TCGA CESC -0.413; TCGA COAD -0.244; TCGA HNSC -0.171; TCGA KIRC -0.49; TCGA KIRP -0.277; TCGA PAAD -0.158; TCGA SARC -0.437; TCGA THCA -0.158; TCGA STAD -0.469; TCGA UCEC -0.372 |
hsa-miR-155-5p | NFAT5 | 12 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LUSC; SARC; THCA; STAD; UCEC | mirMAP; miRNATAP | TCGA BLCA -0.132; TCGA BRCA -0.101; TCGA CESC -0.083; TCGA COAD -0.152; TCGA HNSC -0.106; TCGA KIRC -0.153; TCGA KIRP -0.163; TCGA LUSC -0.107; TCGA SARC -0.118; TCGA THCA -0.075; TCGA STAD -0.177; TCGA UCEC -0.104 |
hsa-miR-155-5p | RPGRIP1L | 10 cancers: BLCA; BRCA; CESC; COAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.053; TCGA BRCA -0.05; TCGA CESC -0.149; TCGA COAD -0.12; TCGA PAAD -0.093; TCGA PRAD -0.097; TCGA SARC -0.08; TCGA THCA -0.076; TCGA STAD -0.097; TCGA UCEC -0.084 |
hsa-miR-155-5p | FZD3 | 11 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.223; TCGA CESC -0.36; TCGA HNSC -0.113; TCGA KIRC -0.191; TCGA KIRP -0.091; TCGA LUAD -0.253; TCGA LUSC -0.174; TCGA PAAD -0.236; TCGA SARC -0.202; TCGA THCA -0.121; TCGA STAD -0.3 |
hsa-miR-155-5p | ZFAND5 | 10 cancers: BLCA; ESCA; KIRC; LGG; LIHC; LUAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.066; TCGA ESCA -0.074; TCGA KIRC -0.076; TCGA LGG -0.063; TCGA LIHC -0.175; TCGA LUAD -0.075; TCGA PRAD -0.067; TCGA THCA -0.052; TCGA STAD -0.154; TCGA UCEC -0.074 |
hsa-miR-155-5p | SLAIN2 | 9 cancers: BLCA; BRCA; HNSC; KIRP; LIHC; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BLCA -0.06; TCGA BRCA -0.072; TCGA HNSC -0.084; TCGA KIRP -0.084; TCGA LIHC -0.061; TCGA LUAD -0.103; TCGA LUSC -0.081; TCGA STAD -0.133; TCGA UCEC -0.063 |
hsa-miR-155-5p | NNT | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; THCA; STAD | mirMAP | TCGA BLCA -0.081; TCGA BRCA -0.071; TCGA CESC -0.095; TCGA ESCA -0.152; TCGA HNSC -0.161; TCGA KIRC -0.335; TCGA KIRP -0.208; TCGA LIHC -0.237; TCGA LUAD -0.102; TCGA THCA -0.114; TCGA STAD -0.103 |
hsa-miR-155-5p | CPEB4 | 14 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LGG; LIHC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.093; TCGA BRCA -0.207; TCGA CESC -0.11; TCGA HNSC -0.145; TCGA KIRC -0.248; TCGA KIRP -0.187; TCGA LGG -0.078; TCGA LIHC -0.167; TCGA PAAD -0.15; TCGA PRAD -0.07; TCGA SARC -0.14; TCGA THCA -0.132; TCGA STAD -0.168; TCGA UCEC -0.13 |
hsa-miR-155-5p | SLC25A36 | 14 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LUSC; OV; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.124; TCGA BRCA -0.11; TCGA CESC -0.096; TCGA COAD -0.124; TCGA HNSC -0.095; TCGA KIRC -0.07; TCGA KIRP -0.117; TCGA LGG -0.129; TCGA LUSC -0.089; TCGA OV -0.05; TCGA SARC -0.098; TCGA THCA -0.143; TCGA STAD -0.126; TCGA UCEC -0.068 |
hsa-miR-155-5p | TRAK2 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.087; TCGA BRCA -0.143; TCGA CESC -0.201; TCGA COAD -0.069; TCGA ESCA -0.126; TCGA HNSC -0.076; TCGA KIRC -0.143; TCGA KIRP -0.129; TCGA LUAD -0.074; TCGA LUSC -0.138; TCGA SARC -0.106; TCGA THCA -0.066; TCGA STAD -0.182; TCGA UCEC -0.051 |
hsa-miR-155-5p | FAM20B | 16 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.115; TCGA BRCA -0.104; TCGA COAD -0.057; TCGA ESCA -0.193; TCGA HNSC -0.107; TCGA KIRC -0.135; TCGA KIRP -0.143; TCGA LIHC -0.081; TCGA LUAD -0.092; TCGA LUSC -0.144; TCGA PAAD -0.101; TCGA PRAD -0.066; TCGA SARC -0.083; TCGA THCA -0.072; TCGA STAD -0.221; TCGA UCEC -0.103 |
hsa-miR-155-5p | SMAD9 | 15 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.43; TCGA CESC -0.495; TCGA COAD -0.611; TCGA ESCA -0.464; TCGA HNSC -0.322; TCGA KIRP -0.336; TCGA LGG -0.39; TCGA LUAD -0.452; TCGA LUSC -0.336; TCGA OV -0.203; TCGA PRAD -0.164; TCGA SARC -0.489; TCGA THCA -0.266; TCGA STAD -0.606; TCGA UCEC -0.204 |
hsa-miR-155-5p | DACH2 | 9 cancers: BLCA; KIRC; KIRP; LGG; OV; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.748; TCGA KIRC -0.722; TCGA KIRP -0.465; TCGA LGG -0.707; TCGA OV -0.355; TCGA SARC -0.331; TCGA THCA -0.217; TCGA STAD -0.391; TCGA UCEC -0.279 |
hsa-miR-155-5p | MKLN1 | 16 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.056; TCGA BRCA -0.093; TCGA CESC -0.072; TCGA COAD -0.08; TCGA HNSC -0.076; TCGA KIRC -0.084; TCGA KIRP -0.088; TCGA LGG -0.088; TCGA LIHC -0.117; TCGA LUAD -0.07; TCGA LUSC -0.095; TCGA PRAD -0.073; TCGA SARC -0.081; TCGA THCA -0.061; TCGA STAD -0.104; TCGA UCEC -0.082 |
hsa-miR-155-5p | VPS13B | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRP; LGG; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.08; TCGA BRCA -0.09; TCGA CESC -0.103; TCGA COAD -0.113; TCGA HNSC -0.107; TCGA KIRP -0.142; TCGA LGG -0.094; TCGA SARC -0.122; TCGA STAD -0.13; TCGA UCEC -0.141 |
hsa-miR-155-5p | ARHGAP32 | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; UCEC | mirMAP | TCGA BLCA -0.209; TCGA BRCA -0.176; TCGA CESC -0.127; TCGA HNSC -0.178; TCGA KIRP -0.166; TCGA LGG -0.219; TCGA LIHC -0.194; TCGA LUAD -0.16; TCGA LUSC -0.175; TCGA UCEC -0.108 |
hsa-miR-155-5p | RC3H1 | 12 cancers: BLCA; BRCA; CESC; LGG; LIHC; LUAD; LUSC; OV; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.107; TCGA BRCA -0.067; TCGA CESC -0.093; TCGA LGG -0.057; TCGA LIHC -0.079; TCGA LUAD -0.095; TCGA LUSC -0.083; TCGA OV -0.059; TCGA SARC -0.083; TCGA THCA -0.07; TCGA STAD -0.132; TCGA UCEC -0.082 |
hsa-miR-155-5p | FAM8A1 | 16 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.084; TCGA BRCA -0.129; TCGA CESC -0.148; TCGA HNSC -0.09; TCGA KIRC -0.061; TCGA KIRP -0.094; TCGA LIHC -0.246; TCGA LUAD -0.172; TCGA LUSC -0.074; TCGA OV -0.057; TCGA PAAD -0.095; TCGA PRAD -0.067; TCGA SARC -0.183; TCGA THCA -0.132; TCGA STAD -0.156; TCGA UCEC -0.123 |
hsa-miR-155-5p | ABI2 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LGG; LUAD; LUSC; OV; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.076; TCGA BRCA -0.089; TCGA CESC -0.205; TCGA COAD -0.088; TCGA ESCA -0.122; TCGA KIRC -0.12; TCGA KIRP -0.144; TCGA LGG -0.093; TCGA LUAD -0.059; TCGA LUSC -0.07; TCGA OV -0.063; TCGA PAAD -0.109; TCGA SARC -0.225; TCGA THCA -0.105; TCGA STAD -0.23; TCGA UCEC -0.125 |
hsa-miR-155-5p | IGF1R | 15 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; OV; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.11; TCGA BRCA -0.39; TCGA CESC -0.119; TCGA COAD -0.157; TCGA HNSC -0.225; TCGA KIRC -0.137; TCGA KIRP -0.15; TCGA LGG -0.18; TCGA LUAD -0.269; TCGA LUSC -0.247; TCGA OV -0.132; TCGA PRAD -0.228; TCGA THCA -0.087; TCGA STAD -0.127; TCGA UCEC -0.208 |
hsa-miR-155-5p | HUNK | 11 cancers: BLCA; BRCA; CESC; COAD; KIRC; KIRP; LGG; LUAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.152; TCGA BRCA -0.142; TCGA CESC -0.371; TCGA COAD -0.402; TCGA KIRC -0.343; TCGA KIRP -0.17; TCGA LGG -0.09; TCGA LUAD -0.344; TCGA SARC -0.604; TCGA STAD -0.19; TCGA UCEC -0.183 |
hsa-miR-155-5p | POGK | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; OV | mirMAP | TCGA BLCA -0.1; TCGA COAD -0.089; TCGA ESCA -0.079; TCGA HNSC -0.062; TCGA KIRP -0.08; TCGA LGG -0.071; TCGA LUAD -0.144; TCGA LUSC -0.099; TCGA OV -0.06 |
hsa-miR-155-5p | PKP4 | 14 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; SARC; THCA | mirMAP | TCGA BLCA -0.053; TCGA BRCA -0.097; TCGA CESC -0.075; TCGA ESCA -0.119; TCGA HNSC -0.08; TCGA KIRC -0.216; TCGA KIRP -0.133; TCGA LGG -0.061; TCGA LIHC -0.062; TCGA LUAD -0.088; TCGA LUSC -0.145; TCGA PRAD -0.06; TCGA SARC -0.11; TCGA THCA -0.058 |
hsa-miR-155-5p | LPP | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; SARC; STAD; UCEC | mirMAP; miRNATAP | TCGA BLCA -0.165; TCGA BRCA -0.163; TCGA CESC -0.22; TCGA COAD -0.118; TCGA ESCA -0.238; TCGA HNSC -0.226; TCGA KIRP -0.163; TCGA LUAD -0.088; TCGA LUSC -0.195; TCGA SARC -0.148; TCGA STAD -0.475; TCGA UCEC -0.072 |
hsa-miR-155-5p | TMEM47 | 12 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.397; TCGA BRCA -0.192; TCGA CESC -0.541; TCGA COAD -0.19; TCGA HNSC -0.182; TCGA KIRC -0.071; TCGA KIRP -0.119; TCGA LGG -0.075; TCGA SARC -0.156; TCGA THCA -0.165; TCGA STAD -0.382; TCGA UCEC -0.241 |
hsa-miR-155-5p | UBN2 | 15 cancers: BLCA; BRCA; CESC; COAD; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.194; TCGA BRCA -0.114; TCGA CESC -0.13; TCGA COAD -0.101; TCGA HNSC -0.119; TCGA LGG -0.129; TCGA LIHC -0.091; TCGA LUAD -0.106; TCGA LUSC -0.097; TCGA PAAD -0.16; TCGA PRAD -0.065; TCGA SARC -0.154; TCGA THCA -0.121; TCGA STAD -0.104; TCGA UCEC -0.084 |
hsa-miR-155-5p | PPP1R9A | 13 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LGG; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.624; TCGA BRCA -0.195; TCGA CESC -1.035; TCGA KIRC -0.215; TCGA KIRP -0.14; TCGA LGG -0.235; TCGA LUAD -0.225; TCGA PAAD -0.391; TCGA PRAD -0.087; TCGA SARC -0.46; TCGA THCA -0.105; TCGA STAD -0.587; TCGA UCEC -0.089 |
hsa-miR-155-5p | UBR1 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.075; TCGA BRCA -0.143; TCGA CESC -0.179; TCGA ESCA -0.075; TCGA HNSC -0.087; TCGA KIRC -0.1; TCGA KIRP -0.136; TCGA LUAD -0.141; TCGA LUSC -0.056; TCGA SARC -0.119; TCGA STAD -0.119; TCGA UCEC -0.09 |
hsa-miR-155-5p | OTUD7B | 14 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.104; TCGA BRCA -0.18; TCGA CESC -0.127; TCGA ESCA -0.098; TCGA HNSC -0.122; TCGA KIRC -0.053; TCGA KIRP -0.147; TCGA LIHC -0.092; TCGA LUAD -0.192; TCGA LUSC -0.148; TCGA SARC -0.077; TCGA THCA -0.061; TCGA STAD -0.134; TCGA UCEC -0.091 |
hsa-miR-155-5p | PPM1L | 15 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.367; TCGA CESC -0.333; TCGA ESCA -0.381; TCGA HNSC -0.266; TCGA KIRC -0.251; TCGA KIRP -0.328; TCGA LGG -0.293; TCGA LIHC -0.121; TCGA LUAD -0.181; TCGA LUSC -0.27; TCGA OV -0.153; TCGA SARC -0.265; TCGA THCA -0.242; TCGA STAD -0.307; TCGA UCEC -0.217 |
hsa-miR-155-5p | SLMAP | 11 cancers: BLCA; BRCA; CESC; ESCA; KIRC; KIRP; LUAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.115; TCGA BRCA -0.11; TCGA CESC -0.072; TCGA ESCA -0.107; TCGA KIRC -0.171; TCGA KIRP -0.11; TCGA LUAD -0.066; TCGA SARC -0.188; TCGA THCA -0.081; TCGA STAD -0.36; TCGA UCEC -0.168 |
hsa-miR-155-5p | TSC1 | 12 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LGG; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.092; TCGA BRCA -0.137; TCGA CESC -0.078; TCGA KIRC -0.062; TCGA KIRP -0.121; TCGA LGG -0.164; TCGA PAAD -0.079; TCGA PRAD -0.062; TCGA SARC -0.098; TCGA THCA -0.1; TCGA STAD -0.103; TCGA UCEC -0.062 |
hsa-miR-155-5p | ZNF528 | 10 cancers: BLCA; BRCA; COAD; HNSC; OV; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.217; TCGA BRCA -0.227; TCGA COAD -0.306; TCGA HNSC -0.214; TCGA OV -0.086; TCGA PRAD -0.053; TCGA SARC -0.086; TCGA THCA -0.082; TCGA STAD -0.256; TCGA UCEC -0.078 |
hsa-miR-155-5p | PCMTD1 | 12 cancers: BLCA; BRCA; CESC; COAD; KIRC; KIRP; LGG; LUAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.205; TCGA BRCA -0.091; TCGA CESC -0.118; TCGA COAD -0.191; TCGA KIRC -0.133; TCGA KIRP -0.101; TCGA LGG -0.081; TCGA LUAD -0.078; TCGA SARC -0.153; TCGA THCA -0.093; TCGA STAD -0.218; TCGA UCEC -0.132 |
hsa-miR-155-5p | TET2 | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LGG; LUAD; LUSC; SARC; UCEC | mirMAP | TCGA BLCA -0.162; TCGA BRCA -0.07; TCGA CESC -0.109; TCGA HNSC -0.098; TCGA KIRP -0.084; TCGA LGG -0.14; TCGA LUAD -0.111; TCGA LUSC -0.119; TCGA SARC -0.107; TCGA UCEC -0.082 |
hsa-miR-155-5p | SPRY3 | 9 cancers: BLCA; CESC; COAD; HNSC; KIRP; LGG; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.114; TCGA CESC -0.277; TCGA COAD -0.256; TCGA HNSC -0.066; TCGA KIRP -0.103; TCGA LGG -0.064; TCGA SARC -0.261; TCGA STAD -0.142; TCGA UCEC -0.116 |
hsa-miR-155-5p | AR | 12 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LIHC; LUAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.596; TCGA BRCA -0.74; TCGA CESC -0.392; TCGA HNSC -0.188; TCGA KIRP -0.28; TCGA LIHC -0.469; TCGA LUAD -0.194; TCGA PRAD -0.144; TCGA SARC -0.4; TCGA THCA -0.234; TCGA STAD -0.562; TCGA UCEC -0.238 |
hsa-miR-155-5p | CA5B | 11 cancers: BLCA; BRCA; CESC; ESCA; KIRC; KIRP; LUSC; OV; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.096; TCGA BRCA -0.114; TCGA CESC -0.114; TCGA ESCA -0.101; TCGA KIRC -0.111; TCGA KIRP -0.141; TCGA LUSC -0.13; TCGA OV -0.082; TCGA THCA -0.053; TCGA STAD -0.214; TCGA UCEC -0.111 |
hsa-miR-155-5p | LONRF2 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.669; TCGA BRCA -0.644; TCGA CESC -0.961; TCGA COAD -0.412; TCGA ESCA -0.609; TCGA HNSC -0.331; TCGA KIRC -0.639; TCGA KIRP -0.519; TCGA LGG -0.117; TCGA LUAD -0.295; TCGA PAAD -0.301; TCGA PRAD -0.147; TCGA SARC -1.076; TCGA THCA -0.145; TCGA STAD -0.868; TCGA UCEC -0.316 |
hsa-miR-155-5p | ATF7 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.079; TCGA BRCA -0.167; TCGA CESC -0.083; TCGA COAD -0.096; TCGA ESCA -0.069; TCGA HNSC -0.101; TCGA LIHC -0.079; TCGA LUAD -0.135; TCGA LUSC -0.06; TCGA PRAD -0.069; TCGA SARC -0.155; TCGA STAD -0.15; TCGA UCEC -0.084 |
hsa-miR-155-5p | KCND3 | 9 cancers: BLCA; BRCA; KIRC; KIRP; LGG; LIHC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.158; TCGA BRCA -0.679; TCGA KIRC -0.335; TCGA KIRP -0.215; TCGA LGG -0.097; TCGA LIHC -0.236; TCGA SARC -0.53; TCGA STAD -0.224; TCGA UCEC -0.323 |
hsa-miR-155-5p | KSR2 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; PAAD; PRAD; THCA | mirMAP | TCGA BLCA -0.692; TCGA BRCA -0.388; TCGA CESC -0.547; TCGA ESCA -0.275; TCGA HNSC -0.311; TCGA KIRC -0.507; TCGA KIRP -0.191; TCGA LGG -0.724; TCGA LUAD -0.258; TCGA PAAD -0.587; TCGA PRAD -0.31; TCGA THCA -0.135 |
hsa-miR-155-5p | ZNF24 | 13 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LGG; LIHC; LUAD; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.074; TCGA BRCA -0.156; TCGA CESC -0.054; TCGA KIRC -0.072; TCGA KIRP -0.075; TCGA LGG -0.097; TCGA LIHC -0.054; TCGA LUAD -0.092; TCGA PAAD -0.124; TCGA SARC -0.084; TCGA THCA -0.072; TCGA STAD -0.099; TCGA UCEC -0.083 |
hsa-miR-155-5p | STOX2 | 13 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; OV; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.207; TCGA COAD -0.25; TCGA ESCA -0.274; TCGA HNSC -0.329; TCGA KIRP -0.189; TCGA LGG -0.122; TCGA LUAD -0.28; TCGA LUSC -0.29; TCGA OV -0.196; TCGA SARC -0.609; TCGA THCA -0.138; TCGA STAD -0.369; TCGA UCEC -0.265 |
hsa-miR-155-5p | ZNF354C | 10 cancers: BLCA; COAD; KIRC; KIRP; LGG; OV; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.128; TCGA COAD -0.249; TCGA KIRC -0.113; TCGA KIRP -0.41; TCGA LGG -0.12; TCGA OV -0.089; TCGA SARC -0.27; TCGA THCA -0.157; TCGA STAD -0.314; TCGA UCEC -0.356 |
hsa-miR-155-5p | ERBB4 | 12 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LGG; LUAD; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.473; TCGA BRCA -0.943; TCGA CESC -0.957; TCGA HNSC -0.222; TCGA KIRC -1.141; TCGA KIRP -0.733; TCGA LGG -0.231; TCGA LUAD -0.202; TCGA PRAD -0.167; TCGA SARC -0.238; TCGA THCA -0.208; TCGA STAD -0.44 |
hsa-miR-155-5p | TMTC2 | 11 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LUAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.216; TCGA BRCA -0.134; TCGA CESC -0.222; TCGA KIRC -0.082; TCGA KIRP -0.173; TCGA LUAD -0.103; TCGA PRAD -0.09; TCGA SARC -0.146; TCGA THCA -0.075; TCGA STAD -0.141; TCGA UCEC -0.097 |
hsa-miR-155-5p | ZADH2 | 11 cancers: BLCA; BRCA; KIRC; KIRP; LGG; LIHC; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.075; TCGA BRCA -0.148; TCGA KIRC -0.193; TCGA KIRP -0.144; TCGA LGG -0.13; TCGA LIHC -0.095; TCGA PAAD -0.18; TCGA SARC -0.122; TCGA THCA -0.131; TCGA STAD -0.064; TCGA UCEC -0.054 |
hsa-miR-155-5p | YOD1 | 9 cancers: BLCA; HNSC; KIRC; KIRP; LUAD; OV; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.058; TCGA HNSC -0.099; TCGA KIRC -0.06; TCGA KIRP -0.077; TCGA LUAD -0.087; TCGA OV -0.057; TCGA SARC -0.083; TCGA THCA -0.089; TCGA UCEC -0.053 |
hsa-miR-155-5p | CACNB4 | 11 cancers: BLCA; BRCA; CESC; COAD; KIRC; KIRP; LGG; SARC; THCA; STAD; UCEC | mirMAP; miRNATAP | TCGA BLCA -0.248; TCGA BRCA -0.169; TCGA CESC -0.317; TCGA COAD -0.323; TCGA KIRC -0.375; TCGA KIRP -0.209; TCGA LGG -0.103; TCGA SARC -0.264; TCGA THCA -0.275; TCGA STAD -0.285; TCGA UCEC -0.177 |
hsa-miR-155-5p | BRWD1 | 10 cancers: BLCA; BRCA; KIRP; LGG; LIHC; LUAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.163; TCGA BRCA -0.098; TCGA KIRP -0.083; TCGA LGG -0.11; TCGA LIHC -0.114; TCGA LUAD -0.065; TCGA SARC -0.117; TCGA THCA -0.099; TCGA STAD -0.146; TCGA UCEC -0.094 |
hsa-miR-155-5p | PPARA | 12 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.1; TCGA CESC -0.093; TCGA HNSC -0.161; TCGA KIRC -0.234; TCGA KIRP -0.234; TCGA LGG -0.116; TCGA LIHC -0.223; TCGA LUAD -0.071; TCGA LUSC -0.134; TCGA PRAD -0.06; TCGA STAD -0.15; TCGA UCEC -0.058 |
hsa-miR-155-5p | ZNF793 | 11 cancers: BLCA; BRCA; CESC; COAD; KIRC; KIRP; OV; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.246; TCGA BRCA -0.058; TCGA CESC -0.268; TCGA COAD -0.46; TCGA KIRC -0.135; TCGA KIRP -0.221; TCGA OV -0.09; TCGA SARC -0.389; TCGA THCA -0.137; TCGA STAD -0.306; TCGA UCEC -0.064 |
hsa-miR-155-5p | ACADSB | 13 cancers: BLCA; BRCA; CESC; ESCA; KIRC; KIRP; LGG; LIHC; LUAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.272; TCGA BRCA -0.442; TCGA CESC -0.148; TCGA ESCA -0.205; TCGA KIRC -0.407; TCGA KIRP -0.335; TCGA LGG -0.169; TCGA LIHC -0.274; TCGA LUAD -0.214; TCGA SARC -0.2; TCGA THCA -0.145; TCGA STAD -0.133; TCGA UCEC -0.111 |
hsa-miR-155-5p | PGAP1 | 15 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.289; TCGA BRCA -0.113; TCGA HNSC -0.122; TCGA KIRC -0.161; TCGA KIRP -0.235; TCGA LGG -0.069; TCGA LIHC -0.074; TCGA LUAD -0.103; TCGA LUSC -0.29; TCGA OV -0.136; TCGA PAAD -0.2; TCGA SARC -0.35; TCGA THCA -0.134; TCGA STAD -0.265; TCGA UCEC -0.233 |
hsa-miR-155-5p | DDI2 | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; UCEC | mirMAP | TCGA BLCA -0.103; TCGA BRCA -0.172; TCGA CESC -0.224; TCGA HNSC -0.15; TCGA KIRC -0.151; TCGA KIRP -0.335; TCGA LIHC -0.241; TCGA LUAD -0.148; TCGA LUSC -0.108; TCGA UCEC -0.134 |
hsa-miR-155-5p | ZNF677 | 9 cancers: BLCA; CESC; KIRC; KIRP; OV; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.167; TCGA CESC -0.205; TCGA KIRC -0.283; TCGA KIRP -0.283; TCGA OV -0.123; TCGA SARC -0.156; TCGA THCA -0.117; TCGA STAD -0.329; TCGA UCEC -0.187 |
hsa-miR-155-5p | MFAP3L | 9 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LIHC; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.523; TCGA CESC -0.412; TCGA HNSC -0.234; TCGA KIRC -0.336; TCGA KIRP -0.209; TCGA LIHC -0.206; TCGA SARC -0.187; TCGA THCA -0.249; TCGA STAD -0.377 |
hsa-miR-155-5p | ZBTB10 | 12 cancers: BLCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.196; TCGA CESC -0.532; TCGA COAD -0.604; TCGA HNSC -0.132; TCGA KIRC -0.185; TCGA KIRP -0.219; TCGA LGG -0.102; TCGA LIHC -0.112; TCGA LUAD -0.117; TCGA SARC -0.21; TCGA STAD -0.356; TCGA UCEC -0.171 |
hsa-miR-155-5p | ZNF709 | 13 cancers: BLCA; BRCA; CESC; COAD; KIRC; KIRP; LGG; OV; PAAD; PRAD; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.203; TCGA BRCA -0.11; TCGA CESC -0.257; TCGA COAD -0.162; TCGA KIRC -0.206; TCGA KIRP -0.14; TCGA LGG -0.059; TCGA OV -0.164; TCGA PAAD -0.158; TCGA PRAD -0.056; TCGA SARC -0.157; TCGA THCA -0.15; TCGA UCEC -0.133 |
hsa-miR-155-5p | NFIA | 10 cancers: BLCA; BRCA; COAD; LIHC; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.075; TCGA BRCA -0.203; TCGA COAD -0.117; TCGA LIHC -0.215; TCGA LUAD -0.116; TCGA LUSC -0.125; TCGA SARC -0.359; TCGA THCA -0.2; TCGA STAD -0.354; TCGA UCEC -0.088 |
hsa-miR-155-5p | ZNF664 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; THCA | miRNATAP | TCGA BLCA -0.143; TCGA BRCA -0.078; TCGA CESC -0.233; TCGA COAD -0.074; TCGA ESCA -0.105; TCGA HNSC -0.131; TCGA KIRC -0.148; TCGA KIRP -0.101; TCGA LUAD -0.158; TCGA LUSC -0.113; TCGA PAAD -0.232; TCGA PRAD -0.077; TCGA SARC -0.115; TCGA THCA -0.127 |
hsa-miR-155-5p | KLF3 | 12 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; THCA; UCEC | miRNATAP | TCGA BLCA -0.132; TCGA BRCA -0.068; TCGA CESC -0.076; TCGA HNSC -0.086; TCGA KIRC -0.053; TCGA KIRP -0.071; TCGA LIHC -0.07; TCGA LUAD -0.157; TCGA LUSC -0.13; TCGA PRAD -0.052; TCGA THCA -0.069; TCGA UCEC -0.073 |
hsa-miR-155-5p | FGF9 | 12 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; OV; STAD; UCEC | miRNATAP | TCGA BLCA -0.185; TCGA CESC -0.519; TCGA ESCA -0.327; TCGA HNSC -0.198; TCGA KIRC -1.456; TCGA KIRP -0.813; TCGA LGG -0.454; TCGA LUAD -0.256; TCGA LUSC -0.209; TCGA OV -0.608; TCGA STAD -0.324; TCGA UCEC -0.262 |
hsa-miR-155-5p | PROX1 | 12 cancers: BLCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LIHC; PAAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.163; TCGA CESC -0.525; TCGA COAD -0.412; TCGA HNSC -0.226; TCGA KIRC -0.892; TCGA KIRP -0.603; TCGA LGG -0.14; TCGA LIHC -0.276; TCGA PAAD -0.264; TCGA SARC -0.308; TCGA STAD -0.221; TCGA UCEC -0.181 |
hsa-miR-155-5p | LCOR | 12 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; UCEC | miRNATAP | TCGA BLCA -0.181; TCGA BRCA -0.213; TCGA CESC -0.193; TCGA HNSC -0.141; TCGA KIRC -0.182; TCGA KIRP -0.319; TCGA LGG -0.185; TCGA LIHC -0.118; TCGA LUAD -0.156; TCGA LUSC -0.094; TCGA SARC -0.136; TCGA UCEC -0.163 |
hsa-miR-155-5p | WNK3 | 10 cancers: BLCA; KIRC; KIRP; LGG; LIHC; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.231; TCGA KIRC -0.567; TCGA KIRP -0.427; TCGA LGG -0.159; TCGA LIHC -0.322; TCGA PRAD -0.147; TCGA SARC -0.297; TCGA THCA -0.157; TCGA STAD -0.434; TCGA UCEC -0.184 |
hsa-miR-155-5p | C3orf18 | 13 cancers: BLCA; BRCA; CESC; ESCA; KIRC; KIRP; LGG; LIHC; LUAD; PAAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.189; TCGA BRCA -0.294; TCGA CESC -0.209; TCGA ESCA -0.246; TCGA KIRC -0.286; TCGA KIRP -0.163; TCGA LGG -0.101; TCGA LIHC -0.118; TCGA LUAD -0.146; TCGA PAAD -0.187; TCGA THCA -0.17; TCGA STAD -0.281; TCGA UCEC -0.154 |
hsa-miR-155-5p | CEP350 | 9 cancers: BLCA; COAD; HNSC; KIRP; LGG; LIHC; LUAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.079; TCGA COAD -0.061; TCGA HNSC -0.078; TCGA KIRP -0.071; TCGA LGG -0.114; TCGA LIHC -0.081; TCGA LUAD -0.072; TCGA STAD -0.057; TCGA UCEC -0.078 |
hsa-miR-155-5p | ZNF703 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; STAD | miRNATAP | TCGA BLCA -0.217; TCGA BRCA -0.363; TCGA CESC -0.252; TCGA COAD -0.215; TCGA ESCA -0.221; TCGA HNSC -0.257; TCGA LUAD -0.163; TCGA LUSC -0.264; TCGA PRAD -0.164; TCGA STAD -0.301 |
hsa-miR-155-5p | STXBP5L | 12 cancers: BLCA; CESC; ESCA; LGG; LUSC; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.188; TCGA CESC -0.515; TCGA ESCA -0.371; TCGA LGG -0.193; TCGA LUSC -0.426; TCGA OV -0.178; TCGA PAAD -0.79; TCGA PRAD -0.232; TCGA SARC -0.363; TCGA THCA -0.588; TCGA STAD -0.627; TCGA UCEC -0.532 |
hsa-miR-155-5p | NR2F2 | 10 cancers: BLCA; BRCA; CESC; KIRC; KIRP; OV; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.203; TCGA BRCA -0.069; TCGA CESC -0.396; TCGA KIRC -0.135; TCGA KIRP -0.171; TCGA OV -0.1; TCGA PRAD -0.079; TCGA SARC -0.326; TCGA STAD -0.167; TCGA UCEC -0.119 |
hsa-miR-155-5p | SEC14L5 | 11 cancers: BLCA; BRCA; CESC; ESCA; KIRC; KIRP; LIHC; OV; PAAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.163; TCGA BRCA -0.21; TCGA CESC -0.208; TCGA ESCA -0.242; TCGA KIRC -0.238; TCGA KIRP -0.203; TCGA LIHC -0.265; TCGA OV -0.361; TCGA PAAD -0.305; TCGA STAD -0.389; TCGA UCEC -0.128 |
hsa-miR-155-5p | GOLPH3L | 10 cancers: BLCA; BRCA; CESC; HNSC; LIHC; LUAD; PAAD; PRAD; SARC; UCEC | miRNATAP | TCGA BLCA -0.1; TCGA BRCA -0.106; TCGA CESC -0.116; TCGA HNSC -0.089; TCGA LIHC -0.093; TCGA LUAD -0.135; TCGA PAAD -0.1; TCGA PRAD -0.051; TCGA SARC -0.051; TCGA UCEC -0.069 |
hsa-miR-155-5p | TRIM2 | 17 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PAAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.382; TCGA CESC -0.538; TCGA COAD -0.226; TCGA ESCA -0.226; TCGA HNSC -0.196; TCGA KIRC -0.498; TCGA KIRP -0.35; TCGA LGG -0.084; TCGA LIHC -0.111; TCGA LUAD -0.114; TCGA LUSC -0.193; TCGA OV -0.18; TCGA PAAD -0.258; TCGA SARC -0.215; TCGA THCA -0.141; TCGA STAD -0.136; TCGA UCEC -0.086 |
hsa-miR-155-5p | WDFY3 | 16 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.196; TCGA BRCA -0.194; TCGA CESC -0.17; TCGA ESCA -0.133; TCGA HNSC -0.17; TCGA KIRC -0.099; TCGA KIRP -0.118; TCGA LGG -0.162; TCGA LUAD -0.122; TCGA LUSC -0.165; TCGA PAAD -0.123; TCGA PRAD -0.069; TCGA SARC -0.079; TCGA THCA -0.102; TCGA STAD -0.187; TCGA UCEC -0.093 |
hsa-miR-155-5p | CHST9 | 9 cancers: BLCA; CESC; LGG; LUSC; OV; PAAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.338; TCGA CESC -0.507; TCGA LGG -0.329; TCGA LUSC -0.575; TCGA OV -0.415; TCGA PAAD -0.345; TCGA SARC -0.434; TCGA STAD -0.668; TCGA UCEC -0.341 |
hsa-miR-155-5p | SEMA5A | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LUAD; SARC; UCEC | miRNATAP | TCGA BLCA -0.457; TCGA BRCA -0.285; TCGA CESC -0.175; TCGA COAD -0.424; TCGA HNSC -0.152; TCGA KIRC -0.134; TCGA LUAD -0.162; TCGA SARC -0.249; TCGA UCEC -0.155 |
hsa-miR-155-5p | RNF146 | 10 cancers: BLCA; BRCA; CESC; COAD; KIRP; LGG; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.07; TCGA BRCA -0.053; TCGA CESC -0.073; TCGA COAD -0.062; TCGA KIRP -0.051; TCGA LGG -0.128; TCGA SARC -0.063; TCGA THCA -0.109; TCGA STAD -0.136; TCGA UCEC -0.089 |
hsa-miR-155-5p | RAPGEF4 | 15 cancers: BLCA; BRCA; CESC; COAD; HNSC; LGG; LIHC; LUAD; LUSC; OV; PAAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.159; TCGA BRCA -0.309; TCGA CESC -0.173; TCGA COAD -0.351; TCGA HNSC -0.183; TCGA LGG -0.373; TCGA LIHC -0.182; TCGA LUAD -0.176; TCGA LUSC -0.138; TCGA OV -0.124; TCGA PAAD -0.188; TCGA SARC -0.181; TCGA THCA -0.078; TCGA STAD -0.132; TCGA UCEC -0.278 |
hsa-miR-155-5p | CNTN4 | 10 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LGG; LUAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.2; TCGA BRCA -0.205; TCGA CESC -0.255; TCGA KIRC -0.16; TCGA KIRP -0.227; TCGA LGG -0.278; TCGA LUAD -0.28; TCGA SARC -0.576; TCGA STAD -0.267; TCGA UCEC -0.2 |
hsa-miR-155-5p | UBR3 | 18 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.136; TCGA BRCA -0.128; TCGA CESC -0.119; TCGA COAD -0.098; TCGA ESCA -0.123; TCGA HNSC -0.215; TCGA KIRC -0.127; TCGA KIRP -0.169; TCGA LGG -0.099; TCGA LIHC -0.136; TCGA LUAD -0.139; TCGA LUSC -0.182; TCGA OV -0.075; TCGA PRAD -0.059; TCGA SARC -0.146; TCGA THCA -0.125; TCGA STAD -0.182; TCGA UCEC -0.119 |
hsa-miR-155-5p | PEG3 | 14 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.188; TCGA BRCA -0.196; TCGA CESC -0.403; TCGA COAD -0.248; TCGA HNSC -0.24; TCGA KIRC -0.595; TCGA KIRP -0.475; TCGA OV -0.574; TCGA PAAD -0.391; TCGA PRAD -0.151; TCGA SARC -0.745; TCGA THCA -0.321; TCGA STAD -0.588; TCGA UCEC -0.528 |
hsa-miR-155-5p | ETNK2 | 12 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LIHC; LUSC; OV; PAAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.105; TCGA BRCA -0.113; TCGA COAD -0.557; TCGA HNSC -0.236; TCGA KIRC -0.28; TCGA LIHC -0.37; TCGA LUSC -0.151; TCGA OV -0.168; TCGA PAAD -0.274; TCGA SARC -0.446; TCGA STAD -0.489; TCGA UCEC -0.132 |
hsa-miR-155-5p | GRIP1 | 11 cancers: BLCA; COAD; KIRC; KIRP; LGG; OV; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.248; TCGA COAD -0.472; TCGA KIRC -0.322; TCGA KIRP -0.174; TCGA LGG -0.486; TCGA OV -0.168; TCGA PRAD -0.196; TCGA SARC -0.593; TCGA THCA -0.121; TCGA STAD -0.352; TCGA UCEC -0.084 |
hsa-miR-155-5p | ACVR2A | 14 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; OV; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.256; TCGA BRCA -0.072; TCGA CESC -0.095; TCGA HNSC -0.106; TCGA KIRC -0.117; TCGA KIRP -0.067; TCGA LGG -0.141; TCGA LUAD -0.098; TCGA LUSC -0.149; TCGA OV -0.082; TCGA SARC -0.1; TCGA THCA -0.066; TCGA STAD -0.149; TCGA UCEC -0.134 |
hsa-miR-155-5p | UST | 9 cancers: BLCA; HNSC; KIRC; LGG; LUSC; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.284; TCGA HNSC -0.193; TCGA KIRC -0.298; TCGA LGG -0.241; TCGA LUSC -0.236; TCGA SARC -0.137; TCGA THCA -0.23; TCGA STAD -0.283; TCGA UCEC -0.335 |
hsa-miR-155-5p | DCUN1D4 | 13 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.106; TCGA CESC -0.063; TCGA ESCA -0.101; TCGA HNSC -0.065; TCGA KIRP -0.063; TCGA LIHC -0.127; TCGA LUAD -0.051; TCGA LUSC -0.059; TCGA PAAD -0.052; TCGA SARC -0.085; TCGA THCA -0.114; TCGA STAD -0.191; TCGA UCEC -0.081 |
hsa-miR-155-5p | SOCS6 | 13 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; UCEC | miRNATAP | TCGA BLCA -0.068; TCGA BRCA -0.122; TCGA CESC -0.214; TCGA HNSC -0.084; TCGA KIRP -0.052; TCGA LIHC -0.104; TCGA LUAD -0.125; TCGA LUSC -0.137; TCGA PAAD -0.16; TCGA PRAD -0.059; TCGA SARC -0.094; TCGA THCA -0.057; TCGA UCEC -0.098 |
hsa-miR-155-5p | RNF111 | 9 cancers: BLCA; BRCA; HNSC; KIRP; LGG; LIHC; LUAD; PRAD; UCEC | miRNATAP | TCGA BLCA -0.08; TCGA BRCA -0.098; TCGA HNSC -0.099; TCGA KIRP -0.08; TCGA LGG -0.064; TCGA LIHC -0.073; TCGA LUAD -0.075; TCGA PRAD -0.061; TCGA UCEC -0.057 |
hsa-miR-155-5p | PRPF39 | 10 cancers: BLCA; BRCA; CESC; KIRP; LGG; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.143; TCGA BRCA -0.062; TCGA CESC -0.051; TCGA KIRP -0.067; TCGA LGG -0.112; TCGA PRAD -0.063; TCGA SARC -0.073; TCGA THCA -0.062; TCGA STAD -0.086; TCGA UCEC -0.065 |
hsa-miR-155-5p | SLC20A2 | 12 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LIHC; LUSC; OV; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.074; TCGA BRCA -0.071; TCGA HNSC -0.177; TCGA KIRC -0.274; TCGA KIRP -0.124; TCGA LIHC -0.21; TCGA LUSC -0.2; TCGA OV -0.104; TCGA SARC -0.189; TCGA THCA -0.109; TCGA STAD -0.175; TCGA UCEC -0.081 |
hsa-miR-155-5p | ZNRF3 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD | miRNATAP | TCGA BLCA -0.076; TCGA BRCA -0.162; TCGA CESC -0.161; TCGA COAD -0.317; TCGA ESCA -0.174; TCGA HNSC -0.173; TCGA KIRC -0.292; TCGA KIRP -0.228; TCGA LGG -0.064; TCGA LIHC -0.283; TCGA LUAD -0.208; TCGA LUSC -0.231; TCGA PAAD -0.092; TCGA PRAD -0.069; TCGA SARC -0.146; TCGA STAD -0.215 |
hsa-miR-155-5p | ALDH3A2 | 10 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; SARC; THCA | miRNAWalker2 validate; mirMAP | TCGA BRCA -0.208; TCGA ESCA -0.165; TCGA HNSC -0.093; TCGA KIRC -0.215; TCGA KIRP -0.154; TCGA LIHC -0.257; TCGA LUAD -0.325; TCGA LUSC -0.256; TCGA SARC -0.112; TCGA THCA -0.111 |
hsa-miR-155-5p | ALDH9A1 | 10 cancers: BRCA; KIRC; KIRP; LGG; LIHC; LUAD; OV; PAAD; THCA; UCEC | miRNAWalker2 validate | TCGA BRCA -0.1; TCGA KIRC -0.138; TCGA KIRP -0.124; TCGA LGG -0.066; TCGA LIHC -0.187; TCGA LUAD -0.105; TCGA OV -0.065; TCGA PAAD -0.074; TCGA THCA -0.158; TCGA UCEC -0.073 |
hsa-miR-155-5p | ATPAF1 | 10 cancers: BRCA; ESCA; KIRC; KIRP; LIHC; LUAD; PAAD; PRAD; THCA; STAD | miRNAWalker2 validate | TCGA BRCA -0.091; TCGA ESCA -0.11; TCGA KIRC -0.18; TCGA KIRP -0.12; TCGA LIHC -0.106; TCGA LUAD -0.08; TCGA PAAD -0.17; TCGA PRAD -0.068; TCGA THCA -0.08; TCGA STAD -0.052 |
hsa-miR-155-5p | CDH13 | 9 cancers: BRCA; COAD; HNSC; LGG; LUAD; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.27; TCGA COAD -0.325; TCGA HNSC -0.334; TCGA LGG -0.264; TCGA LUAD -0.117; TCGA PRAD -0.087; TCGA THCA -0.136; TCGA STAD -0.219; TCGA UCEC -0.268 |
hsa-miR-155-5p | CFL2 | 10 cancers: BRCA; CESC; COAD; ESCA; KIRC; KIRP; LIHC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.111; TCGA CESC -0.218; TCGA COAD -0.128; TCGA ESCA -0.15; TCGA KIRC -0.1; TCGA KIRP -0.122; TCGA LIHC -0.152; TCGA THCA -0.113; TCGA STAD -0.507; TCGA UCEC -0.167 |
hsa-miR-155-5p | CLUAP1 | 10 cancers: BRCA; KIRC; KIRP; LIHC; OV; PAAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.083; TCGA KIRC -0.067; TCGA KIRP -0.054; TCGA LIHC -0.08; TCGA OV -0.059; TCGA PAAD -0.146; TCGA SARC -0.103; TCGA THCA -0.081; TCGA STAD -0.086; TCGA UCEC -0.088 |
hsa-miR-155-5p | CPT1A | 12 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; SARC; THCA; UCEC | miRNAWalker2 validate | TCGA BRCA -0.082; TCGA CESC -0.226; TCGA HNSC -0.152; TCGA KIRC -0.194; TCGA KIRP -0.173; TCGA LIHC -0.17; TCGA LUAD -0.106; TCGA LUSC -0.159; TCGA PRAD -0.07; TCGA SARC -0.137; TCGA THCA -0.081; TCGA UCEC -0.075 |
hsa-miR-155-5p | CTAGE5 | 9 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUSC; PAAD; THCA | miRNAWalker2 validate | TCGA BRCA -0.054; TCGA CESC -0.104; TCGA HNSC -0.083; TCGA KIRC -0.112; TCGA KIRP -0.13; TCGA LIHC -0.188; TCGA LUSC -0.093; TCGA PAAD -0.188; TCGA THCA -0.138 |
hsa-miR-155-5p | CTNNB1 | 12 cancers: BRCA; CESC; COAD; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.074; TCGA CESC -0.166; TCGA COAD -0.071; TCGA HNSC -0.061; TCGA KIRC -0.071; TCGA KIRP -0.083; TCGA LIHC -0.077; TCGA LUAD -0.068; TCGA LUSC -0.079; TCGA SARC -0.088; TCGA STAD -0.054; TCGA UCEC -0.076 |
hsa-miR-155-5p | CUX1 | 12 cancers: BRCA; CESC; COAD; HNSC; LGG; LIHC; LUSC; PAAD; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.113; TCGA CESC -0.07; TCGA COAD -0.149; TCGA HNSC -0.148; TCGA LGG -0.059; TCGA LIHC -0.1; TCGA LUSC -0.094; TCGA PAAD -0.101; TCGA PRAD -0.06; TCGA THCA -0.091; TCGA STAD -0.11; TCGA UCEC -0.071 |
hsa-miR-155-5p | DNAJC19 | 11 cancers: BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; OV; PAAD; SARC; THCA | miRNAWalker2 validate | TCGA BRCA -0.061; TCGA COAD -0.07; TCGA ESCA -0.093; TCGA HNSC -0.056; TCGA KIRC -0.192; TCGA KIRP -0.111; TCGA LIHC -0.172; TCGA OV -0.057; TCGA PAAD -0.14; TCGA SARC -0.057; TCGA THCA -0.089 |
hsa-miR-155-5p | FAM177A1 | 12 cancers: BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; SARC | miRNAWalker2 validate | TCGA BRCA -0.085; TCGA COAD -0.084; TCGA ESCA -0.096; TCGA HNSC -0.064; TCGA KIRC -0.06; TCGA KIRP -0.077; TCGA LGG -0.073; TCGA LIHC -0.108; TCGA LUAD -0.16; TCGA LUSC -0.079; TCGA PAAD -0.075; TCGA SARC -0.051 |
hsa-miR-155-5p | FUBP3 | 16 cancers: BRCA; CESC; COAD; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.074; TCGA CESC -0.05; TCGA COAD -0.053; TCGA HNSC -0.063; TCGA KIRP -0.094; TCGA LGG -0.088; TCGA LIHC -0.082; TCGA LUAD -0.05; TCGA LUSC -0.079; TCGA OV -0.052; TCGA PAAD -0.108; TCGA PRAD -0.084; TCGA SARC -0.062; TCGA THCA -0.113; TCGA STAD -0.112; TCGA UCEC -0.056 |
hsa-miR-155-5p | GABARAPL1 | 14 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.084; TCGA ESCA -0.163; TCGA HNSC -0.166; TCGA KIRC -0.39; TCGA KIRP -0.146; TCGA LGG -0.126; TCGA LIHC -0.249; TCGA LUAD -0.107; TCGA LUSC -0.188; TCGA PAAD -0.108; TCGA PRAD -0.075; TCGA THCA -0.068; TCGA STAD -0.19; TCGA UCEC -0.116 |
hsa-miR-155-5p | ITGB5 | 10 cancers: BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.228; TCGA CESC -0.153; TCGA ESCA -0.18; TCGA HNSC -0.22; TCGA KIRC -0.051; TCGA LUAD -0.063; TCGA LUSC -0.123; TCGA PRAD -0.051; TCGA STAD -0.115; TCGA UCEC -0.086 |
hsa-miR-155-5p | L2HGDH | 11 cancers: BRCA; HNSC; KIRC; KIRP; LGG; LIHC; LUSC; PAAD; PRAD; SARC; THCA | miRNAWalker2 validate | TCGA BRCA -0.058; TCGA HNSC -0.071; TCGA KIRC -0.364; TCGA KIRP -0.272; TCGA LGG -0.17; TCGA LIHC -0.204; TCGA LUSC -0.107; TCGA PAAD -0.154; TCGA PRAD -0.065; TCGA SARC -0.104; TCGA THCA -0.134 |
hsa-miR-155-5p | MATR3 | 9 cancers: BRCA; CESC; KIRC; KIRP; LGG; LIHC; LUAD; SARC; UCEC | miRNAWalker2 validate; miRTarBase | TCGA BRCA -0.068; TCGA CESC -0.052; TCGA KIRC -0.06; TCGA KIRP -0.108; TCGA LGG -0.103; TCGA LIHC -0.051; TCGA LUAD -0.051; TCGA SARC -0.085; TCGA UCEC -0.096 |
hsa-miR-155-5p | MEST | 11 cancers: BRCA; CESC; COAD; HNSC; KIRC; KIRP; LUAD; LUSC; OV; SARC; THCA | miRNAWalker2 validate | TCGA BRCA -0.136; TCGA CESC -0.16; TCGA COAD -0.191; TCGA HNSC -0.122; TCGA KIRC -0.221; TCGA KIRP -0.184; TCGA LUAD -0.11; TCGA LUSC -0.17; TCGA OV -0.207; TCGA SARC -0.323; TCGA THCA -0.102 |
hsa-miR-155-5p | MPP2 | 13 cancers: BRCA; CESC; COAD; ESCA; KIRC; KIRP; LGG; OV; PAAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.345; TCGA CESC -0.139; TCGA COAD -0.157; TCGA ESCA -0.289; TCGA KIRC -0.474; TCGA KIRP -0.253; TCGA LGG -0.092; TCGA OV -0.203; TCGA PAAD -0.474; TCGA SARC -0.268; TCGA THCA -0.267; TCGA STAD -0.484; TCGA UCEC -0.221 |
hsa-miR-155-5p | MPP5 | 12 cancers: BRCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BRCA -0.135; TCGA HNSC -0.12; TCGA KIRC -0.314; TCGA KIRP -0.195; TCGA LIHC -0.11; TCGA LUAD -0.122; TCGA LUSC -0.129; TCGA PAAD -0.088; TCGA PRAD -0.069; TCGA THCA -0.147; TCGA STAD -0.081; TCGA UCEC -0.055 |
hsa-miR-155-5p | MUT | 15 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.096; TCGA CESC -0.069; TCGA COAD -0.073; TCGA ESCA -0.123; TCGA HNSC -0.118; TCGA KIRC -0.263; TCGA KIRP -0.214; TCGA LGG -0.074; TCGA LIHC -0.231; TCGA LUAD -0.153; TCGA LUSC -0.135; TCGA PAAD -0.077; TCGA THCA -0.075; TCGA STAD -0.091; TCGA UCEC -0.09 |
hsa-miR-155-5p | NES | 9 cancers: BRCA; COAD; HNSC; KIRC; KIRP; OV; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.073; TCGA COAD -0.241; TCGA HNSC -0.135; TCGA KIRC -0.082; TCGA KIRP -0.107; TCGA OV -0.162; TCGA SARC -0.131; TCGA STAD -0.171; TCGA UCEC -0.166 |
hsa-miR-155-5p | PALLD | 10 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUSC; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.059; TCGA CESC -0.142; TCGA ESCA -0.172; TCGA HNSC -0.214; TCGA KIRC -0.092; TCGA KIRP -0.092; TCGA LUSC -0.122; TCGA SARC -0.208; TCGA STAD -0.392; TCGA UCEC -0.175 |
hsa-miR-155-5p | PDE3A | 9 cancers: BRCA; CESC; COAD; HNSC; KIRP; LUAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.305; TCGA CESC -0.737; TCGA COAD -0.417; TCGA HNSC -0.435; TCGA KIRP -0.291; TCGA LUAD -0.305; TCGA SARC -0.373; TCGA STAD -0.554; TCGA UCEC -0.252 |
hsa-miR-155-5p | PDPR | 11 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.123; TCGA CESC -0.144; TCGA ESCA -0.146; TCGA HNSC -0.159; TCGA KIRC -0.083; TCGA KIRP -0.182; TCGA LGG -0.139; TCGA SARC -0.139; TCGA THCA -0.112; TCGA STAD -0.107; TCGA UCEC -0.153 |
hsa-miR-155-5p | PNPLA4 | 10 cancers: BRCA; COAD; HNSC; KIRC; KIRP; LIHC; PAAD; SARC; THCA; UCEC | miRNAWalker2 validate | TCGA BRCA -0.256; TCGA COAD -0.166; TCGA HNSC -0.056; TCGA KIRC -0.219; TCGA KIRP -0.125; TCGA LIHC -0.179; TCGA PAAD -0.21; TCGA SARC -0.119; TCGA THCA -0.139; TCGA UCEC -0.072 |
hsa-miR-155-5p | PPL | 10 cancers: BRCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.149; TCGA HNSC -0.163; TCGA KIRC -0.214; TCGA KIRP -0.127; TCGA LIHC -0.325; TCGA LUAD -0.181; TCGA LUSC -0.235; TCGA PAAD -0.284; TCGA STAD -0.404; TCGA UCEC -0.102 |
hsa-miR-155-5p | PRKAR1A | 13 cancers: BRCA; CESC; KIRC; KIRP; LIHC; LUAD; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.138; TCGA CESC -0.076; TCGA KIRC -0.11; TCGA KIRP -0.112; TCGA LIHC -0.114; TCGA LUAD -0.14; TCGA OV -0.054; TCGA PAAD -0.087; TCGA PRAD -0.056; TCGA SARC -0.074; TCGA THCA -0.068; TCGA STAD -0.11; TCGA UCEC -0.132 |
hsa-miR-155-5p | PRKAR2A | 12 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.172; TCGA CESC -0.217; TCGA ESCA -0.134; TCGA HNSC -0.189; TCGA KIRC -0.273; TCGA KIRP -0.336; TCGA LIHC -0.11; TCGA LUAD -0.146; TCGA LUSC -0.143; TCGA THCA -0.085; TCGA STAD -0.122; TCGA UCEC -0.102 |
hsa-miR-155-5p | RNF2 | 9 cancers: BRCA; CESC; COAD; HNSC; LUAD; LUSC; OV; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.055; TCGA CESC -0.055; TCGA COAD -0.118; TCGA HNSC -0.101; TCGA LUAD -0.127; TCGA LUSC -0.095; TCGA OV -0.089; TCGA STAD -0.067; TCGA UCEC -0.074 |
hsa-miR-155-5p | RPAP1 | 9 cancers: BRCA; CESC; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; SARC | miRNAWalker2 validate | TCGA BRCA -0.063; TCGA CESC -0.064; TCGA HNSC -0.078; TCGA KIRP -0.061; TCGA LGG -0.076; TCGA LIHC -0.058; TCGA LUAD -0.082; TCGA LUSC -0.06; TCGA SARC -0.07 |
hsa-miR-155-5p | RTN3 | 10 cancers: BRCA; CESC; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PAAD; THCA | miRNAWalker2 validate | TCGA BRCA -0.058; TCGA CESC -0.13; TCGA ESCA -0.068; TCGA HNSC -0.065; TCGA KIRC -0.105; TCGA LGG -0.149; TCGA LUAD -0.077; TCGA LUSC -0.097; TCGA PAAD -0.128; TCGA THCA -0.159 |
hsa-miR-155-5p | SAP30L | 9 cancers: BRCA; KIRC; KIRP; LGG; LIHC; PAAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.157; TCGA KIRC -0.068; TCGA KIRP -0.095; TCGA LGG -0.059; TCGA LIHC -0.132; TCGA PAAD -0.097; TCGA SARC -0.059; TCGA STAD -0.098; TCGA UCEC -0.089 |
hsa-miR-155-5p | SECISBP2 | 10 cancers: BRCA; COAD; KIRC; KIRP; LGG; LIHC; LUAD; PAAD; THCA; UCEC | miRNAWalker2 validate | TCGA BRCA -0.09; TCGA COAD -0.105; TCGA KIRC -0.116; TCGA KIRP -0.121; TCGA LGG -0.108; TCGA LIHC -0.094; TCGA LUAD -0.057; TCGA PAAD -0.129; TCGA THCA -0.068; TCGA UCEC -0.061 |
hsa-miR-155-5p | SH3BP4 | 14 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; THCA; STAD | miRNAWalker2 validate | TCGA BRCA -0.172; TCGA CESC -0.257; TCGA COAD -0.159; TCGA ESCA -0.223; TCGA HNSC -0.119; TCGA KIRC -0.273; TCGA KIRP -0.242; TCGA LGG -0.095; TCGA LIHC -0.271; TCGA LUAD -0.169; TCGA LUSC -0.185; TCGA OV -0.104; TCGA THCA -0.051; TCGA STAD -0.147 |
hsa-miR-155-5p | SLC30A1 | 10 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC | miRNAWalker2 validate | TCGA BRCA -0.108; TCGA CESC -0.157; TCGA COAD -0.101; TCGA ESCA -0.108; TCGA HNSC -0.082; TCGA KIRC -0.165; TCGA KIRP -0.199; TCGA LIHC -0.172; TCGA LUAD -0.07; TCGA LUSC -0.11 |
hsa-miR-155-5p | SLC39A10 | 10 cancers: BRCA; CESC; COAD; HNSC; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BRCA -0.065; TCGA CESC -0.168; TCGA COAD -0.225; TCGA HNSC -0.122; TCGA LUAD -0.061; TCGA LUSC -0.075; TCGA SARC -0.131; TCGA THCA -0.086; TCGA STAD -0.091; TCGA UCEC -0.088 |
hsa-miR-155-5p | SPIN1 | 13 cancers: BRCA; CESC; COAD; ESCA; KIRC; KIRP; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.076; TCGA CESC -0.087; TCGA COAD -0.106; TCGA ESCA -0.058; TCGA KIRC -0.097; TCGA KIRP -0.138; TCGA LUAD -0.074; TCGA LUSC -0.053; TCGA PRAD -0.062; TCGA SARC -0.126; TCGA THCA -0.122; TCGA STAD -0.173; TCGA UCEC -0.116 |
hsa-miR-155-5p | STAG2 | 14 cancers: BRCA; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.072; TCGA COAD -0.118; TCGA HNSC -0.064; TCGA KIRC -0.071; TCGA KIRP -0.132; TCGA LGG -0.07; TCGA LIHC -0.076; TCGA LUAD -0.059; TCGA LUSC -0.092; TCGA PRAD -0.074; TCGA SARC -0.122; TCGA THCA -0.081; TCGA STAD -0.068; TCGA UCEC -0.079 |
hsa-miR-155-5p | TFCP2 | 11 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; PAAD; SARC; UCEC | miRNAWalker2 validate | TCGA BRCA -0.076; TCGA CESC -0.058; TCGA HNSC -0.06; TCGA KIRC -0.056; TCGA KIRP -0.064; TCGA LGG -0.07; TCGA LIHC -0.077; TCGA LUAD -0.108; TCGA PAAD -0.078; TCGA SARC -0.089; TCGA UCEC -0.064 |
hsa-miR-155-5p | TOMM20 | 13 cancers: BRCA; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; PAAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BRCA -0.064; TCGA COAD -0.085; TCGA ESCA -0.077; TCGA HNSC -0.077; TCGA KIRP -0.079; TCGA LGG -0.122; TCGA LIHC -0.128; TCGA LUAD -0.126; TCGA PAAD -0.07; TCGA SARC -0.1; TCGA THCA -0.115; TCGA STAD -0.107; TCGA UCEC -0.061 |
hsa-miR-155-5p | TSPAN3 | 11 cancers: BRCA; CESC; HNSC; KIRC; LGG; LUAD; LUSC; OV; PRAD; SARC; THCA | miRNAWalker2 validate | TCGA BRCA -0.134; TCGA CESC -0.252; TCGA HNSC -0.056; TCGA KIRC -0.077; TCGA LGG -0.071; TCGA LUAD -0.117; TCGA LUSC -0.079; TCGA OV -0.068; TCGA PRAD -0.08; TCGA SARC -0.057; TCGA THCA -0.099 |
hsa-miR-155-5p | UBE2H | 13 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; SARC; STAD | miRNAWalker2 validate | TCGA BRCA -0.1; TCGA CESC -0.097; TCGA COAD -0.056; TCGA ESCA -0.091; TCGA HNSC -0.173; TCGA KIRC -0.115; TCGA KIRP -0.061; TCGA LIHC -0.114; TCGA LUAD -0.088; TCGA LUSC -0.156; TCGA PRAD -0.06; TCGA SARC -0.123; TCGA STAD -0.14 |
hsa-miR-155-5p | WNT5A | 9 cancers: BRCA; CESC; COAD; HNSC; KIRC; LUAD; OV; STAD; UCEC | miRNAWalker2 validate; mirMAP | TCGA BRCA -0.104; TCGA CESC -0.174; TCGA COAD -0.223; TCGA HNSC -0.292; TCGA KIRC -0.112; TCGA LUAD -0.144; TCGA OV -0.148; TCGA STAD -0.115; TCGA UCEC -0.171 |
hsa-miR-155-5p | TSPAN5 | 10 cancers: BRCA; COAD; HNSC; KIRC; KIRP; LGG; LUSC; OV; STAD; UCEC | MirTarget | TCGA BRCA -0.162; TCGA COAD -0.205; TCGA HNSC -0.149; TCGA KIRC -0.273; TCGA KIRP -0.274; TCGA LGG -0.072; TCGA LUSC -0.18; TCGA OV -0.122; TCGA STAD -0.179; TCGA UCEC -0.226 |
hsa-miR-155-5p | PSKH1 | 16 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.113; TCGA CESC -0.059; TCGA COAD -0.095; TCGA ESCA -0.126; TCGA HNSC -0.121; TCGA KIRC -0.17; TCGA KIRP -0.068; TCGA LIHC -0.069; TCGA LUAD -0.097; TCGA LUSC -0.133; TCGA PAAD -0.091; TCGA PRAD -0.098; TCGA SARC -0.115; TCGA THCA -0.101; TCGA STAD -0.083; TCGA UCEC -0.096 |
hsa-miR-155-5p | SLC35F5 | 12 cancers: BRCA; CESC; COAD; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; THCA; UCEC | MirTarget | TCGA BRCA -0.098; TCGA CESC -0.232; TCGA COAD -0.165; TCGA HNSC -0.215; TCGA KIRC -0.165; TCGA KIRP -0.069; TCGA LIHC -0.091; TCGA LUAD -0.075; TCGA LUSC -0.206; TCGA PRAD -0.075; TCGA THCA -0.119; TCGA UCEC -0.088 |
hsa-miR-155-5p | FAM168A | 10 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; SARC; STAD; UCEC | MirTarget | TCGA BRCA -0.102; TCGA CESC -0.254; TCGA ESCA -0.143; TCGA HNSC -0.178; TCGA KIRC -0.132; TCGA KIRP -0.259; TCGA LGG -0.191; TCGA SARC -0.112; TCGA STAD -0.129; TCGA UCEC -0.083 |
hsa-miR-155-5p | DIXDC1 | 11 cancers: BRCA; CESC; COAD; ESCA; HNSC; LUAD; OV; SARC; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.275; TCGA CESC -0.309; TCGA COAD -0.144; TCGA ESCA -0.163; TCGA HNSC -0.135; TCGA LUAD -0.128; TCGA OV -0.082; TCGA SARC -0.255; TCGA THCA -0.124; TCGA STAD -0.478; TCGA UCEC -0.273 |
hsa-miR-155-5p | BCORL1 | 12 cancers: BRCA; CESC; COAD; HNSC; KIRP; LUAD; LUSC; OV; SARC; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.11; TCGA CESC -0.181; TCGA COAD -0.104; TCGA HNSC -0.207; TCGA KIRP -0.052; TCGA LUAD -0.173; TCGA LUSC -0.175; TCGA OV -0.085; TCGA SARC -0.165; TCGA THCA -0.097; TCGA STAD -0.18; TCGA UCEC -0.126 |
hsa-miR-155-5p | FBXL2 | 11 cancers: BRCA; CESC; KIRC; KIRP; LGG; OV; PAAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.157; TCGA CESC -0.353; TCGA KIRC -0.22; TCGA KIRP -0.097; TCGA LGG -0.115; TCGA OV -0.095; TCGA PAAD -0.258; TCGA SARC -0.241; TCGA THCA -0.175; TCGA STAD -0.39; TCGA UCEC -0.171 |
hsa-miR-155-5p | KIAA2022 | 10 cancers: BRCA; CESC; KIRC; KIRP; LGG; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.278; TCGA CESC -0.426; TCGA KIRC -0.873; TCGA KIRP -0.568; TCGA LGG -0.293; TCGA PAAD -0.247; TCGA SARC -0.739; TCGA THCA -0.302; TCGA STAD -0.725; TCGA UCEC -0.371 |
hsa-miR-155-5p | SLC30A4 | 9 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LGG; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.172; TCGA CESC -0.14; TCGA HNSC -0.241; TCGA KIRC -0.093; TCGA KIRP -0.248; TCGA LGG -0.077; TCGA SARC -0.175; TCGA STAD -0.177; TCGA UCEC -0.213 |
hsa-miR-155-5p | RBM18 | 9 cancers: BRCA; ESCA; KIRC; KIRP; LIHC; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BRCA -0.078; TCGA ESCA -0.066; TCGA KIRC -0.11; TCGA KIRP -0.109; TCGA LIHC -0.068; TCGA LUSC -0.067; TCGA PAAD -0.089; TCGA STAD -0.082; TCGA UCEC -0.054 |
hsa-miR-155-5p | DCAF10 | 11 cancers: BRCA; COAD; KIRC; KIRP; LGG; LIHC; PAAD; PRAD; SARC; THCA; STAD | mirMAP | TCGA BRCA -0.081; TCGA COAD -0.067; TCGA KIRC -0.108; TCGA KIRP -0.126; TCGA LGG -0.051; TCGA LIHC -0.083; TCGA PAAD -0.106; TCGA PRAD -0.065; TCGA SARC -0.054; TCGA THCA -0.075; TCGA STAD -0.123 |
hsa-miR-155-5p | GTF3C4 | 10 cancers: BRCA; CESC; KIRC; KIRP; LGG; LUAD; LUSC; OV; THCA; UCEC | mirMAP | TCGA BRCA -0.052; TCGA CESC -0.082; TCGA KIRC -0.158; TCGA KIRP -0.18; TCGA LGG -0.122; TCGA LUAD -0.098; TCGA LUSC -0.105; TCGA OV -0.06; TCGA THCA -0.085; TCGA UCEC -0.057 |
hsa-miR-155-5p | TMEM132B | 10 cancers: BRCA; HNSC; KIRP; LGG; OV; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.196; TCGA HNSC -0.311; TCGA KIRP -0.132; TCGA LGG -0.266; TCGA OV -0.23; TCGA PAAD -0.276; TCGA SARC -0.225; TCGA THCA -0.192; TCGA STAD -0.494; TCGA UCEC -0.291 |
hsa-miR-155-5p | VPS53 | 9 cancers: BRCA; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PAAD; PRAD | mirMAP; miRNATAP | TCGA BRCA -0.07; TCGA ESCA -0.074; TCGA HNSC -0.058; TCGA KIRC -0.086; TCGA LGG -0.082; TCGA LUAD -0.083; TCGA LUSC -0.077; TCGA PAAD -0.156; TCGA PRAD -0.072 |
hsa-miR-155-5p | PTPRG | 12 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.26; TCGA CESC -0.365; TCGA HNSC -0.2; TCGA KIRC -0.12; TCGA KIRP -0.271; TCGA LGG -0.052; TCGA LIHC -0.116; TCGA LUAD -0.102; TCGA LUSC -0.2; TCGA THCA -0.069; TCGA STAD -0.131; TCGA UCEC -0.195 |
hsa-miR-155-5p | IL17RD | 13 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUSC; OV; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.144; TCGA CESC -0.268; TCGA COAD -0.397; TCGA ESCA -0.213; TCGA HNSC -0.102; TCGA KIRC -0.282; TCGA KIRP -0.21; TCGA LGG -0.144; TCGA LUSC -0.154; TCGA OV -0.137; TCGA SARC -0.482; TCGA STAD -0.635; TCGA UCEC -0.245 |
hsa-miR-155-5p | GABRB2 | 10 cancers: BRCA; CESC; COAD; ESCA; KIRC; KIRP; LGG; PAAD; STAD; UCEC | mirMAP | TCGA BRCA -0.262; TCGA CESC -0.636; TCGA COAD -0.579; TCGA ESCA -0.564; TCGA KIRC -0.612; TCGA KIRP -0.486; TCGA LGG -0.322; TCGA PAAD -0.218; TCGA STAD -0.52; TCGA UCEC -0.367 |
hsa-miR-155-5p | PURB | 10 cancers: BRCA; COAD; HNSC; KIRP; LUSC; OV; SARC; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.052; TCGA COAD -0.091; TCGA HNSC -0.077; TCGA KIRP -0.057; TCGA LUSC -0.1; TCGA OV -0.056; TCGA SARC -0.076; TCGA THCA -0.056; TCGA STAD -0.194; TCGA UCEC -0.071 |
hsa-miR-155-5p | MMP16 | 9 cancers: BRCA; CESC; COAD; HNSC; LGG; OV; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.322; TCGA CESC -0.38; TCGA COAD -0.274; TCGA HNSC -0.432; TCGA LGG -0.197; TCGA OV -0.149; TCGA SARC -0.209; TCGA STAD -0.476; TCGA UCEC -0.269 |
hsa-miR-155-5p | SNED1 | 12 cancers: BRCA; CESC; ESCA; KIRP; LIHC; LUAD; OV; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.241; TCGA CESC -0.317; TCGA ESCA -0.179; TCGA KIRP -0.133; TCGA LIHC -0.185; TCGA LUAD -0.141; TCGA OV -0.169; TCGA PRAD -0.064; TCGA SARC -0.16; TCGA THCA -0.169; TCGA STAD -0.169; TCGA UCEC -0.357 |
hsa-miR-155-5p | TBC1D16 | 11 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; PRAD; THCA; STAD | mirMAP | TCGA BRCA -0.158; TCGA CESC -0.309; TCGA COAD -0.126; TCGA ESCA -0.147; TCGA HNSC -0.212; TCGA KIRC -0.2; TCGA KIRP -0.178; TCGA LUSC -0.121; TCGA PRAD -0.093; TCGA THCA -0.067; TCGA STAD -0.066 |
hsa-miR-155-5p | AQP4 | 9 cancers: BRCA; CESC; KIRC; KIRP; LIHC; PAAD; SARC; THCA; UCEC | mirMAP | TCGA BRCA -0.146; TCGA CESC -0.472; TCGA KIRC -0.368; TCGA KIRP -0.326; TCGA LIHC -0.202; TCGA PAAD -0.394; TCGA SARC -0.392; TCGA THCA -0.196; TCGA UCEC -0.299 |
hsa-miR-155-5p | YIPF6 | 14 cancers: BRCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | mirMAP | TCGA BRCA -0.082; TCGA CESC -0.083; TCGA COAD -0.1; TCGA HNSC -0.172; TCGA KIRC -0.115; TCGA KIRP -0.156; TCGA LGG -0.06; TCGA LIHC -0.066; TCGA LUAD -0.148; TCGA LUSC -0.15; TCGA PAAD -0.099; TCGA PRAD -0.07; TCGA THCA -0.087; TCGA STAD -0.122 |
hsa-miR-155-5p | VKORC1L1 | 11 cancers: BRCA; CESC; COAD; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; UCEC | mirMAP | TCGA BRCA -0.108; TCGA CESC -0.061; TCGA COAD -0.096; TCGA HNSC -0.144; TCGA LIHC -0.118; TCGA LUAD -0.117; TCGA LUSC -0.137; TCGA PAAD -0.146; TCGA PRAD -0.065; TCGA THCA -0.052; TCGA UCEC -0.052 |
hsa-miR-155-5p | GFPT1 | 10 cancers: BRCA; CESC; COAD; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; THCA | mirMAP | TCGA BRCA -0.068; TCGA CESC -0.261; TCGA COAD -0.113; TCGA HNSC -0.173; TCGA LGG -0.063; TCGA LUAD -0.157; TCGA LUSC -0.08; TCGA PAAD -0.157; TCGA PRAD -0.08; TCGA THCA -0.083 |
hsa-miR-155-5p | LEPROT | 12 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BRCA -0.183; TCGA CESC -0.214; TCGA COAD -0.106; TCGA ESCA -0.082; TCGA HNSC -0.176; TCGA KIRC -0.17; TCGA KIRP -0.258; TCGA LIHC -0.15; TCGA LUAD -0.084; TCGA LUSC -0.144; TCGA STAD -0.125; TCGA UCEC -0.262 |
hsa-miR-155-5p | ATXN1L | 14 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BRCA -0.09; TCGA CESC -0.075; TCGA COAD -0.061; TCGA ESCA -0.089; TCGA HNSC -0.119; TCGA KIRC -0.123; TCGA KIRP -0.145; TCGA LIHC -0.092; TCGA LUAD -0.086; TCGA LUSC -0.108; TCGA SARC -0.113; TCGA THCA -0.085; TCGA STAD -0.091; TCGA UCEC -0.127 |
hsa-miR-155-5p | CYB561D1 | 9 cancers: BRCA; ESCA; HNSC; KIRC; LUSC; PAAD; PRAD; THCA; STAD | miRNATAP | TCGA BRCA -0.087; TCGA ESCA -0.106; TCGA HNSC -0.101; TCGA KIRC -0.071; TCGA LUSC -0.105; TCGA PAAD -0.095; TCGA PRAD -0.09; TCGA THCA -0.102; TCGA STAD -0.071 |
hsa-miR-155-5p | PRKG1 | 11 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BRCA -0.363; TCGA CESC -0.482; TCGA COAD -0.412; TCGA ESCA -0.243; TCGA HNSC -0.39; TCGA KIRC -0.1; TCGA KIRP -0.19; TCGA SARC -0.17; TCGA THCA -0.291; TCGA STAD -0.422; TCGA UCEC -0.423 |
hsa-miR-155-5p | SGCB | 9 cancers: BRCA; CESC; ESCA; HNSC; KIRP; LUSC; PAAD; STAD; UCEC | miRNATAP | TCGA BRCA -0.148; TCGA CESC -0.227; TCGA ESCA -0.132; TCGA HNSC -0.078; TCGA KIRP -0.061; TCGA LUSC -0.106; TCGA PAAD -0.118; TCGA STAD -0.217; TCGA UCEC -0.122 |
hsa-miR-155-5p | CDS1 | 9 cancers: BRCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; THCA | miRNATAP | TCGA BRCA -0.064; TCGA HNSC -0.133; TCGA KIRC -0.406; TCGA KIRP -0.191; TCGA LUAD -0.137; TCGA LUSC -0.257; TCGA OV -0.086; TCGA PAAD -0.18; TCGA THCA -0.104 |
hsa-miR-155-5p | AHCYL2 | 9 cancers: BRCA; HNSC; KIRC; KIRP; LIHC; LUAD; SARC; THCA; UCEC | miRNATAP | TCGA BRCA -0.126; TCGA HNSC -0.111; TCGA KIRC -0.177; TCGA KIRP -0.143; TCGA LIHC -0.092; TCGA LUAD -0.166; TCGA SARC -0.139; TCGA THCA -0.116; TCGA UCEC -0.122 |
hsa-miR-155-5p | ARMC2 | 9 cancers: CESC; COAD; HNSC; KIRP; LUSC; OV; PAAD; THCA; STAD | miRNAWalker2 validate | TCGA CESC -0.221; TCGA COAD -0.219; TCGA HNSC -0.071; TCGA KIRP -0.095; TCGA LUSC -0.089; TCGA OV -0.088; TCGA PAAD -0.169; TCGA THCA -0.073; TCGA STAD -0.143 |
hsa-miR-155-5p | ATL2 | 9 cancers: CESC; HNSC; KIRC; KIRP; LIHC; LUAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA CESC -0.075; TCGA HNSC -0.051; TCGA KIRC -0.077; TCGA KIRP -0.066; TCGA LIHC -0.133; TCGA LUAD -0.178; TCGA SARC -0.133; TCGA STAD -0.213; TCGA UCEC -0.115 |
hsa-miR-155-5p | CYP51A1 | 9 cancers: CESC; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; THCA | miRNAWalker2 validate | TCGA CESC -0.122; TCGA HNSC -0.171; TCGA KIRC -0.252; TCGA KIRP -0.188; TCGA LIHC -0.165; TCGA LUAD -0.145; TCGA LUSC -0.205; TCGA PAAD -0.198; TCGA THCA -0.07 |
hsa-miR-155-5p | GOLPH3 | 10 cancers: CESC; COAD; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA CESC -0.111; TCGA COAD -0.053; TCGA HNSC -0.066; TCGA KIRP -0.065; TCGA LUAD -0.12; TCGA LUSC -0.09; TCGA PAAD -0.059; TCGA PRAD -0.063; TCGA STAD -0.073; TCGA UCEC -0.057 |
hsa-miR-155-5p | GPM6B | 11 cancers: CESC; COAD; HNSC; KIRC; LGG; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA CESC -0.214; TCGA COAD -0.26; TCGA HNSC -0.128; TCGA KIRC -0.472; TCGA LGG -0.178; TCGA LUAD -0.163; TCGA LUSC -0.234; TCGA SARC -0.262; TCGA THCA -0.083; TCGA STAD -0.574; TCGA UCEC -0.194 |
hsa-miR-155-5p | HSD17B12 | 11 cancers: CESC; COAD; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; SARC; THCA; STAD | miRNAWalker2 validate | TCGA CESC -0.169; TCGA COAD -0.107; TCGA HNSC -0.129; TCGA KIRC -0.201; TCGA KIRP -0.102; TCGA LUAD -0.107; TCGA LUSC -0.112; TCGA PRAD -0.121; TCGA SARC -0.058; TCGA THCA -0.074; TCGA STAD -0.087 |
hsa-miR-155-5p | IMPAD1 | 11 cancers: CESC; HNSC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA CESC -0.055; TCGA HNSC -0.078; TCGA KIRP -0.077; TCGA LIHC -0.093; TCGA LUAD -0.087; TCGA LUSC -0.078; TCGA OV -0.064; TCGA PAAD -0.09; TCGA THCA -0.075; TCGA STAD -0.12; TCGA UCEC -0.051 |
hsa-miR-155-5p | KRAS | 9 cancers: CESC; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; OV; PRAD | miRNAWalker2 validate; miRNATAP | TCGA CESC -0.081; TCGA HNSC -0.107; TCGA KIRC -0.051; TCGA KIRP -0.051; TCGA LGG -0.063; TCGA LUAD -0.137; TCGA LUSC -0.079; TCGA OV -0.119; TCGA PRAD -0.055 |
hsa-miR-155-5p | LCLAT1 | 9 cancers: CESC; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | miRNAWalker2 validate | TCGA CESC -0.122; TCGA HNSC -0.139; TCGA LIHC -0.074; TCGA LUAD -0.106; TCGA LUSC -0.111; TCGA PAAD -0.191; TCGA PRAD -0.074; TCGA THCA -0.154; TCGA STAD -0.097 |
hsa-miR-155-5p | MYO10 | 11 cancers: CESC; COAD; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; SARC; THCA | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | TCGA CESC -0.143; TCGA COAD -0.225; TCGA HNSC -0.208; TCGA KIRC -0.305; TCGA KIRP -0.172; TCGA LGG -0.103; TCGA LUAD -0.237; TCGA LUSC -0.222; TCGA PAAD -0.26; TCGA SARC -0.112; TCGA THCA -0.133 |
hsa-miR-155-5p | PACSIN2 | 10 cancers: CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; STAD | miRNAWalker2 validate | TCGA CESC -0.201; TCGA ESCA -0.144; TCGA HNSC -0.087; TCGA KIRC -0.172; TCGA KIRP -0.194; TCGA LIHC -0.099; TCGA LUAD -0.189; TCGA LUSC -0.127; TCGA PRAD -0.059; TCGA STAD -0.072 |
hsa-miR-155-5p | PRKCI | 9 cancers: CESC; HNSC; KIRC; KIRP; LUAD; LUSC; SARC; THCA; UCEC | miRNAWalker2 validate | TCGA CESC -0.063; TCGA HNSC -0.105; TCGA KIRC -0.129; TCGA KIRP -0.069; TCGA LUAD -0.074; TCGA LUSC -0.208; TCGA SARC -0.091; TCGA THCA -0.06; TCGA UCEC -0.062 |
hsa-miR-155-5p | TROVE2 | 10 cancers: CESC; ESCA; HNSC; LIHC; LUAD; LUSC; OV; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA CESC -0.05; TCGA ESCA -0.057; TCGA HNSC -0.06; TCGA LIHC -0.062; TCGA LUAD -0.112; TCGA LUSC -0.068; TCGA OV -0.079; TCGA SARC -0.081; TCGA STAD -0.167; TCGA UCEC -0.117 |
hsa-miR-155-5p | TERF1 | 12 cancers: CESC; COAD; ESCA; KIRP; LGG; LUAD; OV; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA CESC -0.062; TCGA COAD -0.093; TCGA ESCA -0.067; TCGA KIRP -0.06; TCGA LGG -0.062; TCGA LUAD -0.089; TCGA OV -0.067; TCGA PRAD -0.051; TCGA SARC -0.119; TCGA THCA -0.054; TCGA STAD -0.072; TCGA UCEC -0.08 |
hsa-miR-155-5p | DYNC1I1 | 10 cancers: CESC; COAD; HNSC; LGG; LUAD; OV; PAAD; SARC; THCA; STAD | MirTarget; miRNATAP | TCGA CESC -0.435; TCGA COAD -0.418; TCGA HNSC -0.574; TCGA LGG -0.173; TCGA LUAD -0.411; TCGA OV -0.218; TCGA PAAD -0.394; TCGA SARC -0.399; TCGA THCA -0.208; TCGA STAD -0.639 |
hsa-miR-155-5p | G3BP2 | 9 cancers: CESC; ESCA; HNSC; KIRC; KIRP; LUAD; PAAD; SARC; STAD | MirTarget; miRNATAP | TCGA CESC -0.058; TCGA ESCA -0.073; TCGA HNSC -0.062; TCGA KIRC -0.104; TCGA KIRP -0.085; TCGA LUAD -0.05; TCGA PAAD -0.083; TCGA SARC -0.057; TCGA STAD -0.05 |
hsa-miR-155-5p | SP3 | 9 cancers: CESC; COAD; HNSC; LUAD; LUSC; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA CESC -0.085; TCGA COAD -0.114; TCGA HNSC -0.085; TCGA LUAD -0.065; TCGA LUSC -0.074; TCGA SARC -0.093; TCGA THCA -0.063; TCGA STAD -0.067; TCGA UCEC -0.124 |
hsa-miR-155-5p | WWTR1 | 10 cancers: CESC; ESCA; HNSC; KIRP; LUAD; LUSC; SARC; THCA; STAD; UCEC | mirMAP | TCGA CESC -0.271; TCGA ESCA -0.179; TCGA HNSC -0.16; TCGA KIRP -0.127; TCGA LUAD -0.097; TCGA LUSC -0.215; TCGA SARC -0.097; TCGA THCA -0.079; TCGA STAD -0.384; TCGA UCEC -0.091 |
hsa-miR-155-5p | SERTAD4 | 10 cancers: CESC; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; SARC; STAD; UCEC | mirMAP | TCGA CESC -0.583; TCGA ESCA -0.277; TCGA HNSC -0.265; TCGA KIRC -0.17; TCGA KIRP -0.273; TCGA LUAD -0.144; TCGA LUSC -0.34; TCGA SARC -0.611; TCGA STAD -0.516; TCGA UCEC -0.398 |
hsa-miR-155-5p | FSD1L | 9 cancers: CESC; COAD; KIRC; KIRP; LGG; OV; PAAD; THCA; UCEC | mirMAP | TCGA CESC -0.264; TCGA COAD -0.176; TCGA KIRC -0.182; TCGA KIRP -0.158; TCGA LGG -0.186; TCGA OV -0.083; TCGA PAAD -0.209; TCGA THCA -0.052; TCGA UCEC -0.084 |
hsa-miR-155-5p | SPIRE1 | 10 cancers: CESC; HNSC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD | mirMAP | TCGA CESC -0.139; TCGA HNSC -0.105; TCGA KIRP -0.178; TCGA LGG -0.229; TCGA LUAD -0.077; TCGA LUSC -0.167; TCGA PAAD -0.267; TCGA PRAD -0.065; TCGA SARC -0.064; TCGA STAD -0.309 |
hsa-miR-155-5p | FOXP4 | 12 cancers: CESC; COAD; ESCA; KIRC; LGG; LIHC; LUAD; OV; PAAD; SARC; THCA; UCEC | mirMAP | TCGA CESC -0.158; TCGA COAD -0.194; TCGA ESCA -0.146; TCGA KIRC -0.079; TCGA LGG -0.1; TCGA LIHC -0.082; TCGA LUAD -0.229; TCGA OV -0.101; TCGA PAAD -0.429; TCGA SARC -0.125; TCGA THCA -0.118; TCGA UCEC -0.075 |
hsa-miR-155-5p | SBNO1 | 9 cancers: CESC; HNSC; KIRC; KIRP; LUAD; LUSC; SARC; THCA; STAD | mirMAP | TCGA CESC -0.213; TCGA HNSC -0.194; TCGA KIRC -0.128; TCGA KIRP -0.237; TCGA LUAD -0.087; TCGA LUSC -0.118; TCGA SARC -0.136; TCGA THCA -0.111; TCGA STAD -0.082 |
hsa-miR-155-5p | FBXO11 | 10 cancers: CESC; HNSC; KIRP; LGG; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRNATAP | TCGA CESC -0.059; TCGA HNSC -0.079; TCGA KIRP -0.054; TCGA LGG -0.102; TCGA LUAD -0.062; TCGA LUSC -0.06; TCGA SARC -0.081; TCGA THCA -0.059; TCGA STAD -0.087; TCGA UCEC -0.095 |
hsa-miR-155-5p | SALL1 | 10 cancers: CESC; COAD; HNSC; KIRC; KIRP; LIHC; OV; SARC; THCA; UCEC | miRNATAP | TCGA CESC -0.587; TCGA COAD -0.377; TCGA HNSC -0.636; TCGA KIRC -0.179; TCGA KIRP -0.207; TCGA LIHC -0.329; TCGA OV -0.614; TCGA SARC -0.693; TCGA THCA -0.166; TCGA UCEC -0.23 |
hsa-miR-155-5p | PANK1 | 14 cancers: CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PAAD; SARC; THCA | miRNATAP | TCGA CESC -0.171; TCGA COAD -0.131; TCGA ESCA -0.139; TCGA HNSC -0.137; TCGA KIRC -0.403; TCGA KIRP -0.271; TCGA LGG -0.253; TCGA LIHC -0.28; TCGA LUAD -0.256; TCGA LUSC -0.094; TCGA OV -0.111; TCGA PAAD -0.209; TCGA SARC -0.132; TCGA THCA -0.149 |
hsa-miR-155-5p | MEX3B | 9 cancers: CESC; HNSC; LGG; LUAD; OV; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA CESC -0.326; TCGA HNSC -0.218; TCGA LGG -0.388; TCGA LUAD -0.109; TCGA OV -0.221; TCGA PRAD -0.117; TCGA SARC -0.228; TCGA STAD -0.091; TCGA UCEC -0.227 |
hsa-miR-155-5p | GSK3B | 13 cancers: COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PRAD; SARC; UCEC | miRNAWalker2 validate; miRNATAP | TCGA COAD -0.101; TCGA ESCA -0.091; TCGA HNSC -0.13; TCGA KIRC -0.065; TCGA KIRP -0.068; TCGA LGG -0.085; TCGA LIHC -0.07; TCGA LUAD -0.092; TCGA LUSC -0.146; TCGA OV -0.057; TCGA PRAD -0.073; TCGA SARC -0.057; TCGA UCEC -0.057 |
hsa-miR-155-5p | INTS6 | 10 cancers: COAD; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; SARC; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA COAD -0.074; TCGA ESCA -0.108; TCGA HNSC -0.13; TCGA KIRP -0.098; TCGA LGG -0.055; TCGA LUAD -0.089; TCGA LUSC -0.079; TCGA SARC -0.052; TCGA STAD -0.091; TCGA UCEC -0.056 |
hsa-miR-155-5p | PACSIN3 | 9 cancers: COAD; ESCA; KIRC; LIHC; LUAD; LUSC; OV; SARC; STAD | miRNAWalker2 validate | TCGA COAD -0.24; TCGA ESCA -0.232; TCGA KIRC -0.114; TCGA LIHC -0.234; TCGA LUAD -0.166; TCGA LUSC -0.177; TCGA OV -0.179; TCGA SARC -0.204; TCGA STAD -0.286 |
hsa-miR-155-5p | POLR2C | 9 cancers: COAD; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; THCA | miRNAWalker2 validate | TCGA COAD -0.058; TCGA HNSC -0.085; TCGA KIRP -0.055; TCGA LIHC -0.086; TCGA LUAD -0.074; TCGA LUSC -0.108; TCGA PAAD -0.099; TCGA PRAD -0.059; TCGA THCA -0.068 |
hsa-miR-155-5p | SOX6 | 11 cancers: COAD; KIRC; KIRP; LGG; LUAD; LUSC; OV; SARC; THCA; STAD; UCEC | miRTarBase; miRNATAP | TCGA COAD -0.28; TCGA KIRC -0.309; TCGA KIRP -0.108; TCGA LGG -0.372; TCGA LUAD -0.253; TCGA LUSC -0.249; TCGA OV -0.188; TCGA SARC -0.406; TCGA THCA -0.199; TCGA STAD -0.271; TCGA UCEC -0.151 |
hsa-miR-155-5p | IRF2BP2 | 10 cancers: COAD; ESCA; LIHC; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA COAD -0.172; TCGA ESCA -0.076; TCGA LIHC -0.085; TCGA LUAD -0.131; TCGA LUSC -0.065; TCGA PAAD -0.102; TCGA SARC -0.082; TCGA THCA -0.104; TCGA STAD -0.13; TCGA UCEC -0.089 |
hsa-miR-155-5p | PAPD5 | 11 cancers: COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; THCA; STAD; UCEC | mirMAP | TCGA COAD -0.076; TCGA HNSC -0.069; TCGA KIRC -0.057; TCGA KIRP -0.076; TCGA LGG -0.067; TCGA LIHC -0.072; TCGA LUAD -0.103; TCGA LUSC -0.095; TCGA THCA -0.063; TCGA STAD -0.053; TCGA UCEC -0.061 |
hsa-miR-155-5p | CDC73 | 10 cancers: ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA ESCA -0.086; TCGA HNSC -0.088; TCGA KIRP -0.088; TCGA LIHC -0.054; TCGA LUAD -0.106; TCGA LUSC -0.075; TCGA SARC -0.084; TCGA THCA -0.054; TCGA STAD -0.085; TCGA UCEC -0.073 |
hsa-miR-155-5p | PCCB | 9 cancers: ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; PAAD; THCA | miRNAWalker2 validate | TCGA ESCA -0.12; TCGA HNSC -0.051; TCGA KIRC -0.237; TCGA KIRP -0.117; TCGA LGG -0.053; TCGA LIHC -0.122; TCGA LUAD -0.056; TCGA PAAD -0.099; TCGA THCA -0.09 |
hsa-miR-155-5p | MED21 | 10 cancers: HNSC; KIRC; KIRP; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA HNSC -0.075; TCGA KIRC -0.188; TCGA KIRP -0.097; TCGA LUSC -0.087; TCGA OV -0.085; TCGA PAAD -0.105; TCGA PRAD -0.058; TCGA SARC -0.065; TCGA STAD -0.08; TCGA UCEC -0.052 |
hsa-miR-155-5p | NUCKS1 | 11 cancers: KIRC; KIRP; LGG; LIHC; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA KIRC -0.067; TCGA KIRP -0.107; TCGA LGG -0.059; TCGA LIHC -0.1; TCGA LUAD -0.091; TCGA PAAD -0.053; TCGA PRAD -0.05; TCGA SARC -0.094; TCGA THCA -0.087; TCGA STAD -0.144; TCGA UCEC -0.082 |
Enriched cancer pathways of putative targets