microRNA information: hsa-miR-195-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-195-3p | miRbase |
| Accession: | MIMAT0004615 | miRbase |
| Precursor name: | hsa-mir-195 | miRbase |
| Precursor accession: | MI0000489 | miRbase |
| Symbol: | MIR195 | HGNC |
| RefSeq ID: | NR_029712 | GenBank |
| Sequence: | CCAAUAUUGGCUGUGCUGCUCC |
Reported expression in cancers: hsa-miR-195-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-195-3p | bladder cancer | downregulation | "We identified a subset of 7 miRNAs miR-145 miR-30a ......" | 19378336 | qPCR |
| hsa-miR-195-3p | bladder cancer | downregulation | "microRNA 195 inhibits cell proliferation in bladde ......" | 26622447 | qPCR |
| hsa-miR-195-3p | breast cancer | downregulation | "Analysis of MiR 195 and MiR 497 expression regulat ......" | 21350001 | Northern blot; qPCR; RNA-Seq |
| hsa-miR-195-3p | breast cancer | downregulation | "Serum microRNA 195 is down regulated in breast can ......" | 25103018 | qPCR |
| hsa-miR-195-3p | cervical and endocervical cancer | downregulation | "The present study investigated the role of miR-195 ......" | 26622903 | |
| hsa-miR-195-3p | cervical and endocervical cancer | downregulation | "Here we found that miR-195 expression was down-reg ......" | 26631043 | |
| hsa-miR-195-3p | cervical and endocervical cancer | downregulation | "Quantitative real-time polymerase chain reaction w ......" | 27206216 | qPCR |
| hsa-miR-195-3p | colorectal cancer | downregulation | "In the present study we analyzed the roles of miR- ......" | 20727858 | |
| hsa-miR-195-3p | colorectal cancer | downregulation | "Downregulation of miR 195 correlates with lymph no ......" | 21390519 | qPCR |
| hsa-miR-195-3p | colorectal cancer | downregulation | "MiR-497 and miR-195 of the miR-15/16/195/424/497 f ......" | 22710713 | |
| hsa-miR-195-3p | endometrial cancer | deregulation | "A set of EEC-associated miRNAs in tissue and plasm ......" | 24491411 | RNA-Seq |
| hsa-miR-195-3p | esophageal cancer | downregulation | "In this study Agilent miRNA microarray was used to ......" | 24025765 | Microarray |
| hsa-miR-195-3p | gastric cancer | downregulation | "The low expression of miR-195 and miR-378 was clos ......" | 23333942 | |
| hsa-miR-195-3p | gastric cancer | downregulation | "Expression level of microRNA 195 in the serum of p ......" | 27097947 | qPCR |
| hsa-miR-195-3p | gastric cancer | downregulation | "In our previous study we demonstrated that four mi ......" | 27529338 | Reverse transcription PCR; qPCR |
| hsa-miR-195-3p | glioblastoma | downregulation | "Previous studies have reported that microRNA-195 m ......" | 22217655 | |
| hsa-miR-195-3p | liver cancer | downregulation | "Down-regulation of miR-195 has been observed in va ......" | 19441017 | |
| hsa-miR-195-3p | liver cancer | downregulation | "There is frequent down-regulation of miR-195 expre ......" | 23468064 | |
| hsa-miR-195-3p | liver cancer | downregulation | "In this study we experimentally demonstrated throu ......" | 23487264 | |
| hsa-miR-195-3p | liver cancer | downregulation | "Among them two clustered miRNAs miR-195 and miR-49 ......" | 23544130 | RNA-Seq |
| hsa-miR-195-3p | liver cancer | downregulation | "The result of Real-time PCR reveals the expression ......" | 26823724 | qPCR |
| hsa-miR-195-3p | liver cancer | downregulation | "MiR-195 expression is frequently reduced in variou ......" | 27179445 | |
| hsa-miR-195-3p | lung squamous cell cancer | upregulation | "MiR-195 suppresses tumor growth and is associated ......" | 25840419 | |
| hsa-miR-195-3p | prostate cancer | downregulation | "miR-195 is downregulated in many human cancers. Ho ......" | 26650737 | |
| hsa-miR-195-3p | sarcoma | upregulation | "miR-195 one of the miR-16/15/195/424/497 family me ......" | 23162665 | |
| hsa-miR-195-3p | thyroid cancer | downregulation | "We also find that the expression of miR-195 is dow ......" | 26527888 |
Reported cancer pathway affected by hsa-miR-195-3p
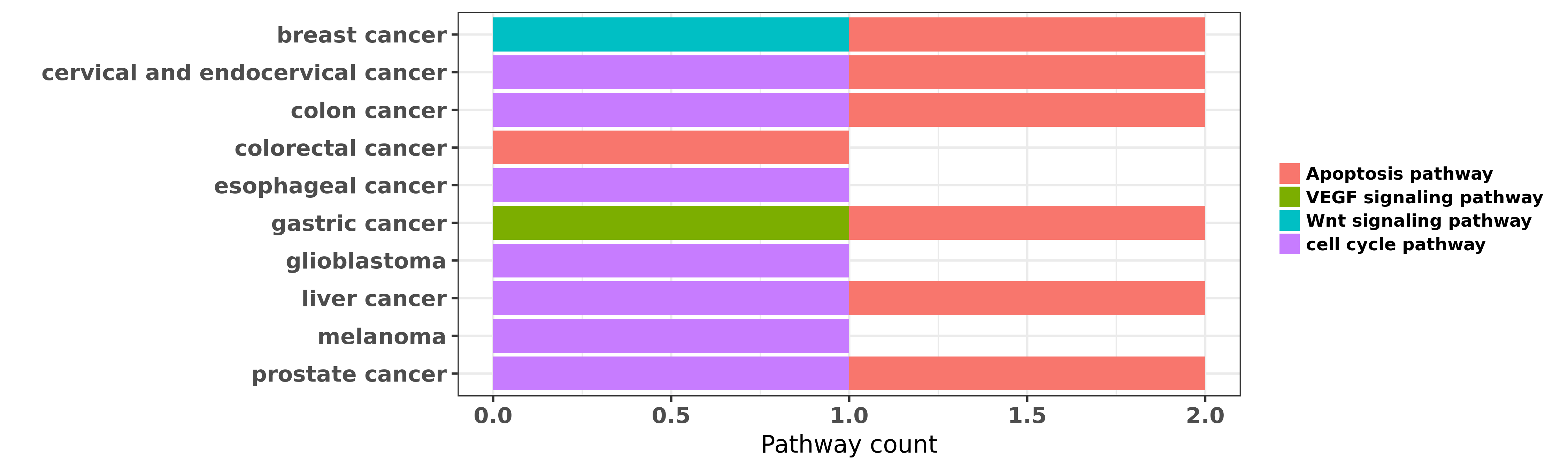
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-195-3p | breast cancer | Apoptosis pathway | "Upregulation of miR 195 increases the sensitivity ......" | 23760062 | |
| hsa-miR-195-3p | breast cancer | Apoptosis pathway | "Upregulation of miR 195 enhances the radiosensitiv ......" | 26309570 | Colony formation; Western blot |
| hsa-miR-195-3p | breast cancer | Wnt signaling pathway | "Eribulin upregulates miR 195 expression and downre ......" | 26573286 | |
| hsa-miR-195-3p | breast cancer | Apoptosis pathway | "MicroRNA 195 inhibits proliferation invasion and m ......" | 26632252 | |
| hsa-miR-195-3p | cervical and endocervical cancer | cell cycle pathway | "MicroRNA 195 inhibits proliferation of cervical ca ......" | 26511972 | Western blot; Luciferase |
| hsa-miR-195-3p | cervical and endocervical cancer | Apoptosis pathway | "MiR 195 inhibits the proliferation of human cervic ......" | 26631043 | Luciferase |
| hsa-miR-195-3p | colon cancer | Apoptosis pathway | "MicroRNA 195 chemosensitizes colon cancer cells to ......" | 23526568 | |
| hsa-miR-195-3p | colon cancer | cell cycle pathway | "Tumor suppressive microRNA 195 5p regulates cell g ......" | 27347317 | |
| hsa-miR-195-3p | colorectal cancer | Apoptosis pathway | "microRNA 195 promotes apoptosis and suppresses tum ......" | 20727858 | |
| hsa-miR-195-3p | esophageal cancer | cell cycle pathway | "Differential expression of miR 195 in esophageal s ......" | 24025765 | |
| hsa-miR-195-3p | gastric cancer | VEGF signaling pathway | "MicroRNA 195 and microRNA 378 mediate tumor growth ......" | 23333942 | |
| hsa-miR-195-3p | gastric cancer | Apoptosis pathway | "In our previous study we demonstrated that four mi ......" | 27529338 | Colony formation |
| hsa-miR-195-3p | glioblastoma | cell cycle pathway | "MicroRNA 195 plays a tumor suppressor role in huma ......" | 22217655 | |
| hsa-miR-195-3p | liver cancer | cell cycle pathway | "MicroRNA 195 suppresses tumorigenicity and regulat ......" | 19441017 | |
| hsa-miR-195-3p | liver cancer | Apoptosis pathway | "The role of microRNA-195 in developing acquired dr ......" | 21947305 | Luciferase |
| hsa-miR-195-3p | liver cancer | Apoptosis pathway | "MiR 195 regulates cell apoptosis of human hepatoce ......" | 22888524 | Western blot; Flow cytometry; Luciferase |
| hsa-miR-195-3p | liver cancer | cell cycle pathway | "Among them two clustered miRNAs miR-195 and miR-49 ......" | 23544130 | |
| hsa-miR-195-3p | liver cancer | cell cycle pathway; Apoptosis pathway | "MicroRNA 195 acts as a tumor suppressor by directl ......" | 25174704 | Luciferase; Western blot |
| hsa-miR-195-3p | melanoma | cell cycle pathway | "Regulation of cell cycle checkpoint kinase WEE1 by ......" | 22847610 | |
| hsa-miR-195-3p | prostate cancer | Apoptosis pathway | "miR 195 Inhibits Tumor Progression by Targeting RP ......" | 26080838 | Luciferase |
| hsa-miR-195-3p | prostate cancer | cell cycle pathway | "Downregulation of miR 195 promotes prostate cancer ......" | 27175617 |
Reported cancer prognosis affected by hsa-miR-195-3p
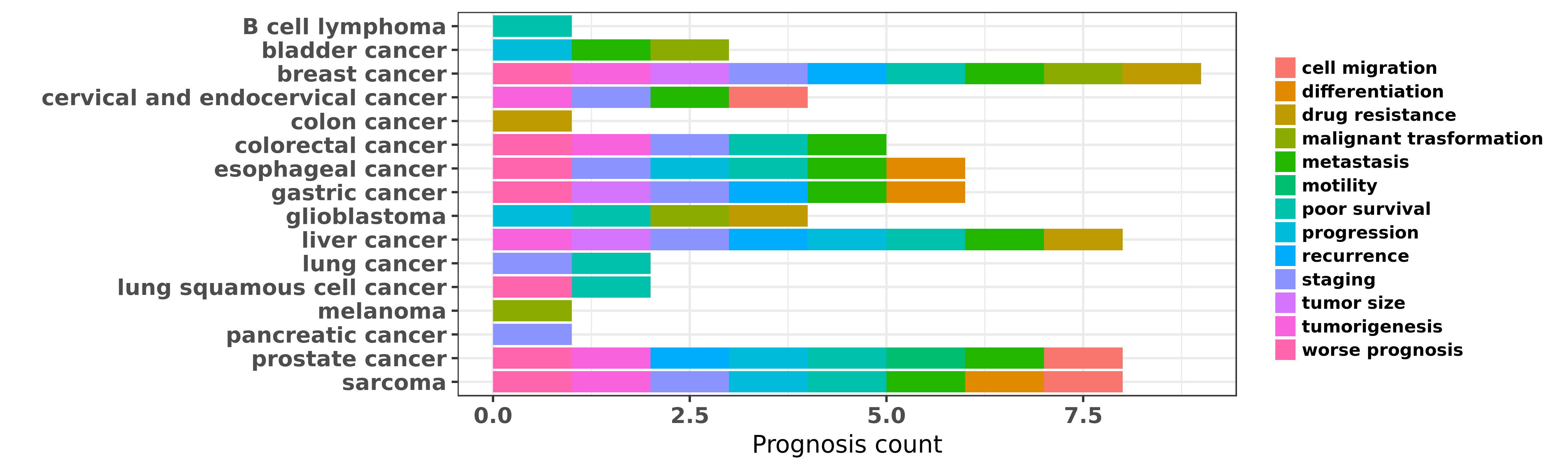
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-195-3p | B cell lymphoma | poor survival | "Finally eight miRNAs were found to correlate with ......" | 18537969 | |
| hsa-miR-195-3p | bladder cancer | metastasis; progression | "The microRNA expression signature of bladder cance ......" | 24520312 | Western blot; Luciferase |
| hsa-miR-195-3p | bladder cancer | malignant trasformation | "microRNA 195 inhibits cell proliferation in bladde ......" | 26622447 | Western blot; Luciferase; MTT assay |
| hsa-miR-195-3p | breast cancer | worse prognosis; metastasis; tumor size; staging; recurrence; poor survival | "Correlation of miR 195 with invasiveness and progn ......" | 22800791 | |
| hsa-miR-195-3p | breast cancer | poor survival | "Upregulation of miR 195 increases the sensitivity ......" | 23760062 | |
| hsa-miR-195-3p | breast cancer | malignant trasformation | "In support of these findings in vitro functional s ......" | 24104550 | |
| hsa-miR-195-3p | breast cancer | tumor size | "The aim of the present study was to assess the eff ......" | 25009660 | |
| hsa-miR-195-3p | breast cancer | tumorigenesis; staging; metastasis; drug resistance | "Serum microRNA 195 is down regulated in breast can ......" | 25103018 | |
| hsa-miR-195-3p | breast cancer | metastasis | "Using real-time quantitative polymerase chain reac ......" | 26124926 | |
| hsa-miR-195-3p | breast cancer | drug resistance | "New miRNA-based drugs are also promising therapy f ......" | 26199650 | |
| hsa-miR-195-3p | breast cancer | poor survival; drug resistance | "Upregulation of miR 195 enhances the radiosensitiv ......" | 26309570 | Colony formation; Western blot |
| hsa-miR-195-3p | breast cancer | metastasis | "MicroRNA 195 inhibits proliferation invasion and m ......" | 26632252 | |
| hsa-miR-195-3p | breast cancer | staging | "We report for the first time an extremely high pre ......" | 27404381 | |
| hsa-miR-195-3p | cervical and endocervical cancer | tumorigenesis | "Serum miRNAs panel miR 16 2* miR 195 miR 2861 miR ......" | 26656154 | |
| hsa-miR-195-3p | cervical and endocervical cancer | staging; metastasis; cell migration | "MiR 195 Suppresses Cervical Cancer Migration and I ......" | 27206216 | Luciferase |
| hsa-miR-195-3p | colon cancer | drug resistance | "MicroRNA 195 chemosensitizes colon cancer cells to ......" | 23526568 | |
| hsa-miR-195-3p | colorectal cancer | metastasis; worse prognosis; tumorigenesis; poor survival; staging | "Downregulation of miR 195 correlates with lymph no ......" | 21390519 | |
| hsa-miR-195-3p | colorectal cancer | staging | "A meta-analysis was performed to review three miRN ......" | 25027346 | |
| hsa-miR-195-3p | colorectal cancer | metastasis | "Study on the molecular regulatory mechanism of Mic ......" | 26064276 | Western blot |
| hsa-miR-195-3p | colorectal cancer | poor survival | "In this study ten candidates were identified using ......" | 27126129 | |
| hsa-miR-195-3p | colorectal cancer | poor survival | "IGF2BP2 promotes colorectal cancer cell proliferat ......" | 27153315 | |
| hsa-miR-195-3p | esophageal cancer | worse prognosis; staging; metastasis; differentiation; progression; poor survival | "microRNA 195 Cdc42 axis acts as a prognostic facto ......" | 25400770 | |
| hsa-miR-195-3p | gastric cancer | metastasis; differentiation; tumor size | "The expressions of 60 candidate miRNAs in 30 gastr ......" | 21987613 | |
| hsa-miR-195-3p | gastric cancer | recurrence; worse prognosis | "Initial profiling using miR microarrays was perfor ......" | 22046085 | |
| hsa-miR-195-3p | gastric cancer | staging; metastasis; differentiation; worse prognosis | "Expression level of microRNA 195 in the serum of p ......" | 27097947 | |
| hsa-miR-195-3p | glioblastoma | drug resistance | "miR 195 miR 455 3p and miR 10a * are implicated in ......" | 20444541 | |
| hsa-miR-195-3p | glioblastoma | poor survival | "MiR 195 miR 196b miR 181c miR 21 expression levels ......" | 21895872 | |
| hsa-miR-195-3p | glioblastoma | progression; malignant trasformation | "MicroRNA 195 plays a tumor suppressor role in huma ......" | 22217655 | |
| hsa-miR-195-3p | liver cancer | progression | "MicroRNA 195 suppresses tumorigenicity and regulat ......" | 19441017 | |
| hsa-miR-195-3p | liver cancer | drug resistance | "The role of microRNA-195 in developing acquired dr ......" | 21947305 | Luciferase |
| hsa-miR-195-3p | liver cancer | staging | "MiR 195 regulates cell apoptosis of human hepatoce ......" | 22888524 | Western blot; Flow cytometry; Luciferase |
| hsa-miR-195-3p | liver cancer | metastasis; poor survival; recurrence | "MicroRNA 195 suppresses angiogenesis and metastasi ......" | 23468064 | Transwell assay |
| hsa-miR-195-3p | liver cancer | tumor size; metastasis; progression | "Genome wide screening reveals that miR 195 targets ......" | 23487264 | Luciferase |
| hsa-miR-195-3p | liver cancer | progression; tumorigenesis | "Among them two clustered miRNAs miR-195 and miR-49 ......" | 23544130 | |
| hsa-miR-195-3p | liver cancer | metastasis | "MicroRNA 195 functions as a tumor suppressor by in ......" | 25607636 | Western blot; Luciferase |
| hsa-miR-195-3p | liver cancer | metastasis; staging; poor survival; tumor size | "MiR 195 is a key negative regulator of hepatocellu ......" | 26823724 | Luciferase |
| hsa-miR-195-3p | lung cancer | staging; poor survival | "Using real-time RT-PCR we analyzed the expression ......" | 24282590 | |
| hsa-miR-195-3p | lung squamous cell cancer | poor survival; worse prognosis | "MiR 195 suppresses non small cell lung cancer by t ......" | 25840419 | |
| hsa-miR-195-3p | melanoma | malignant trasformation | "Regulation of cell cycle checkpoint kinase WEE1 by ......" | 22847610 | |
| hsa-miR-195-3p | pancreatic cancer | staging | "In addition from 10 to 50 weeks of age stage-speci ......" | 26516699 | |
| hsa-miR-195-3p | prostate cancer | progression; worse prognosis; metastasis; recurrence; poor survival | "miR 195 Inhibits Tumor Progression by Targeting RP ......" | 26080838 | Luciferase |
| hsa-miR-195-3p | prostate cancer | progression; motility | "MicroRNA 195 5p a new regulator of Fra 1 suppresse ......" | 26337460 | Transwell assay; Wound Healing Assay; Western blot; Luciferase |
| hsa-miR-195-3p | prostate cancer | metastasis; poor survival; recurrence | "MicroRNA 195 suppresses tumor cell proliferation a ......" | 26338045 | |
| hsa-miR-195-3p | prostate cancer | metastasis; poor survival; cell migration | "miR 195 Inhibits EMT by Targeting FGF2 in Prostate ......" | 26650737 | Western blot; Luciferase |
| hsa-miR-195-3p | prostate cancer | progression; tumorigenesis; worse prognosis | "Downregulation of miR 195 promotes prostate cancer ......" | 27175617 | |
| hsa-miR-195-3p | sarcoma | differentiation | "In order to investigate the involvement of miRNAs ......" | 23133552 | |
| hsa-miR-195-3p | sarcoma | tumorigenesis; cell migration | "microRNA 195 suppresses osteosarcoma cell invasion ......" | 23162665 | Western blot; Luciferase |
| hsa-miR-195-3p | sarcoma | progression; worse prognosis; poor survival; staging; metastasis | "Serum miR 195 is a diagnostic and prognostic marke ......" | 25498513 | |
| hsa-miR-195-3p | sarcoma | metastasis; poor survival | "MicroRNA profiling identifies MiR 195 suppresses o ......" | 25823925 |
Reported gene related to hsa-miR-195-3p

| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-195-3p | breast cancer | BCL2 | "Previously we demonstrated that hsa-miR-195 target ......" | 26632252 |
| hsa-miR-195-3p | breast cancer | BCL2 | "Moreover knockdown of Raf-1 expression had similar ......" | 23760062 |
| hsa-miR-195-3p | breast cancer | BCL2 | "Upregulation of miR 195 enhances the radiosensitiv ......" | 26309570 |
| hsa-miR-195-3p | colorectal cancer | BCL2 | "Moreover important antiapoptotic Bcl-2 was identif ......" | 20727858 |
| hsa-miR-195-3p | bladder cancer | CDC42 | "A luciferase reporter assay was then conducted whi ......" | 26622447 |
| hsa-miR-195-3p | esophageal cancer | CDC42 | "microRNA 195 Cdc42 axis acts as a prognostic facto ......" | 25400770 |
| hsa-miR-195-3p | esophageal cancer | CDC42 | "Differential expression of miR 195 in esophageal s ......" | 24025765 |
| hsa-miR-195-3p | liver cancer | CDC42 | "Additionally miR-195 down-regulation led to increa ......" | 23468064 |
| hsa-miR-195-3p | breast cancer | RAF1 | "Raf-1 and Ccnd1 were identified as novel direct ta ......" | 21350001 |
| hsa-miR-195-3p | breast cancer | RAF1 | "Upregulation of miR 195 increases the sensitivity ......" | 23760062 |
| hsa-miR-195-3p | colorectal cancer | RAF1 | "IGF2BP2 promotes colorectal cancer cell proliferat ......" | 27153315 |
| hsa-miR-195-3p | thyroid cancer | RAF1 | "miR 195 is a key regulator of Raf1 in thyroid canc ......" | 26527888 |
| hsa-miR-195-3p | breast cancer | CCND1 | "Raf-1 and Ccnd1 were identified as novel direct ta ......" | 21350001 |
| hsa-miR-195-3p | cervical and endocervical cancer | CCND1 | "MiR 195 inhibits the proliferation of human cervic ......" | 26631043 |
| hsa-miR-195-3p | sarcoma | CCND1 | "MicroRNA profiling identifies MiR 195 suppresses o ......" | 25823925 |
| hsa-miR-195-3p | cervical and endocervical cancer | PCNA | "MicroRNA 195 inhibits proliferation of cervical ca ......" | 26511972 |
| hsa-miR-195-3p | cervical and endocervical cancer | PCNA | "MiR 195 inhibits the proliferation of human cervic ......" | 26631043 |
| hsa-miR-195-3p | colon cancer | PCNA | "Tumor suppressive microRNA 195 5p regulates cell g ......" | 27347317 |
| hsa-miR-195-3p | liver cancer | VEGFA | "In this study we used a new cationic peptide disul ......" | 27574422 |
| hsa-miR-195-3p | liver cancer | VEGFA | "Furthermore we revealed that miR-195 down-regulati ......" | 23468064 |
| hsa-miR-195-3p | liver cancer | VEGFA | "MiR 195 is a key negative regulator of hepatocellu ......" | 26823724 |
| hsa-miR-195-3p | colon cancer | BCL2L2 | "MicroRNA 195 chemosensitizes colon cancer cells to ......" | 23526568 |
| hsa-miR-195-3p | liver cancer | BCL2L2 | "These results indicate that miR-195 could improve ......" | 21947305 |
| hsa-miR-195-3p | liver cancer | FGF2 | "MiR 195 is a key negative regulator of hepatocellu ......" | 26823724 |
| hsa-miR-195-3p | prostate cancer | FGF2 | "miR 195 Inhibits EMT by Targeting FGF2 in Prostate ......" | 26650737 |
| hsa-miR-195-3p | cervical and endocervical cancer | MYB | "microRNA 195 inhibits the proliferation migration ......" | 26622903 |
| hsa-miR-195-3p | lung squamous cell cancer | MYB | "MicroRNA 195 inhibits non small cell lung cancer c ......" | 24486218 |
| hsa-miR-195-3p | breast cancer | WNT3A | "Eribulin upregulates miR 195 expression and downre ......" | 26573286 |
| hsa-miR-195-3p | liver cancer | WNT3A | "MicroRNA 195 acts as a tumor suppressor by directl ......" | 25174704 |
| hsa-miR-195-3p | colorectal cancer | CARD10 | "MicroRNA 195 inhibits colorectal cancer cell proli ......" | 24787958 |
| hsa-miR-195-3p | liver cancer | CBX4 | "MicroRNA 195 functions as a tumor suppressor by in ......" | 25607636 |
| hsa-miR-195-3p | cervical and endocervical cancer | CCND2 | "microRNA 195 inhibits the proliferation migration ......" | 26622903 |
| hsa-miR-195-3p | glioblastoma | CCND3 | "We identified E2F3 and CCND3 as functional downstr ......" | 22217655 |
| hsa-miR-195-3p | bladder cancer | CDC37 | "The aim of the present study was to investigate th ......" | 26622447 |
| hsa-miR-195-3p | gastric cancer | CDK6 | "Expression of cyclin-dependent kinase 6 and vascul ......" | 23333942 |
| hsa-miR-195-3p | breast cancer | CEACAM5 | "The sensitivity and specificity of miR-195 in the ......" | 25103018 |
| hsa-miR-195-3p | lung squamous cell cancer | CHEK1 | "MiR 195 suppresses non small cell lung cancer by t ......" | 25840419 |
| hsa-miR-195-3p | glioblastoma | E2F3 | "We identified E2F3 and CCND3 as functional downstr ......" | 22217655 |
| hsa-miR-195-3p | lung cancer | EGFR | "Plasma levels of miR-195 and miR-122 expression we ......" | 24282590 |
| hsa-miR-195-3p | sarcoma | FASN | "microRNA 195 suppresses osteosarcoma cell invasion ......" | 23162665 |
| hsa-miR-195-3p | prostate cancer | FOSL1 | "MicroRNA 195 5p a new regulator of Fra 1 suppresse ......" | 26337460 |
| hsa-miR-195-3p | prostate cancer | FRTS1 | "Low miR-195 expression was characterised with a sh ......" | 26650737 |
| hsa-miR-195-3p | liver cancer | HDAC3 | "Expression of microRNA 195 is transactivated by Sp ......" | 27179445 |
| hsa-miR-195-3p | prostate cancer | HMGA1 | "Downregulation of miR 195 promotes prostate cancer ......" | 27175617 |
| hsa-miR-195-3p | colorectal cancer | IGF2BP2 | "IGF2BP2 promotes colorectal cancer cell proliferat ......" | 27153315 |
| hsa-miR-195-3p | breast cancer | INSR | "We show that miR-195 is inversely related with Ins ......" | 27133044 |
| hsa-miR-195-3p | breast cancer | IRS1 | "miR 195 inhibits tumor growth and angiogenesis thr ......" | 27133044 |
| hsa-miR-195-3p | prostate cancer | KIAA0100 | "MicroRNA 195 suppresses tumor cell proliferation a ......" | 26338045 |
| hsa-miR-195-3p | liver cancer | LATS2 | "MiR 195 regulates cell apoptosis of human hepatoce ......" | 22888524 |
| hsa-miR-195-3p | liver cancer | NCOA3 | "In the current study by way of informatics softwar ......" | 24740565 |
| hsa-miR-195-3p | liver cancer | PAEP | "ST21-H3R5-polyethylene glycol PEG exhibited excell ......" | 27574422 |
| hsa-miR-195-3p | liver cancer | PCMT1 | "Hsa miR 195 targets PCMT1 in hepatocellular carcin ......" | 25119594 |
| hsa-miR-195-3p | liver cancer | PHF19 | "MicroRNA 195 5p acts as an anti oncogene by target ......" | 25955388 |
| hsa-miR-195-3p | liver cancer | ROCK2 | "In this study we used a new cationic peptide disul ......" | 27574422 |
| hsa-miR-195-3p | prostate cancer | RPS6 | "The roles of miR-195 and its candidate target gene ......" | 26080838 |
| hsa-miR-195-3p | prostate cancer | RPS6KB1 | "miR 195 Inhibits Tumor Progression by Targeting RP ......" | 26080838 |
| hsa-miR-195-3p | cervical and endocervical cancer | SMAD3 | "MiR 195 Suppresses Cervical Cancer Migration and I ......" | 27206216 |
| hsa-miR-195-3p | liver cancer | SP1 | "Expression of microRNA 195 is transactivated by Sp ......" | 27179445 |
| hsa-miR-195-3p | liver cancer | SRA1 | "MicroRNA 195 regulates steroid receptor coactivato ......" | 24740565 |
| hsa-miR-195-3p | breast cancer | STAB2 | "We found that persistently elevated postoperative ......" | 25503185 |
| hsa-miR-195-3p | bladder cancer | STAT3 | "The aim of the present study was to investigate th ......" | 26622447 |
| hsa-miR-195-3p | liver cancer | TAB3 | "Genome wide screening reveals that miR 195 targets ......" | 23487264 |
| hsa-miR-195-3p | liver cancer | TNF | "Genome wide screening reveals that miR 195 targets ......" | 23487264 |
| hsa-miR-195-3p | liver cancer | VAV2 | "Additionally miR-195 down-regulation led to increa ......" | 23468064 |
| hsa-miR-195-3p | melanoma | WEE1 | "Regulation of cell cycle checkpoint kinase WEE1 by ......" | 22847610 |
Expression profile in cancer corhorts:

Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-195-3p | UBE2I | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRP; LIHC; LUAD; LUSC; OV; PAAD; THCA; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.068; TCGA BRCA -0.141; TCGA CESC -0.055; TCGA COAD -0.087; TCGA ESCA -0.098; TCGA KIRP -0.135; TCGA LIHC -0.084; TCGA LUAD -0.075; TCGA LUSC -0.157; TCGA OV -0.09; TCGA PAAD -0.114; TCGA THCA -0.145; TCGA UCEC -0.052 |
hsa-miR-195-3p | CDK1 | 9 cancers: BLCA; BRCA; COAD; ESCA; LIHC; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.207; TCGA BRCA -0.583; TCGA COAD -0.205; TCGA ESCA -0.362; TCGA LIHC -0.373; TCGA LUAD -0.506; TCGA LUSC -0.626; TCGA STAD -0.691; TCGA UCEC -0.254 |
hsa-miR-195-3p | FAM135A | 9 cancers: BLCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; STAD; UCEC | MirTarget | TCGA BLCA -0.169; TCGA HNSC -0.098; TCGA KIRC -0.118; TCGA KIRP -0.127; TCGA LUAD -0.138; TCGA LUSC -0.111; TCGA OV -0.12; TCGA STAD -0.166; TCGA UCEC -0.083 |
hsa-miR-195-3p | GNAI3 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.083; TCGA BRCA -0.105; TCGA COAD -0.061; TCGA ESCA -0.121; TCGA HNSC -0.084; TCGA LUAD -0.103; TCGA LUSC -0.087; TCGA PRAD -0.052; TCGA STAD -0.112; TCGA UCEC -0.061 |
hsa-miR-195-3p | RPE | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.085; TCGA BRCA -0.083; TCGA COAD -0.1; TCGA ESCA -0.119; TCGA HNSC -0.075; TCGA KIRC -0.077; TCGA LUAD -0.18; TCGA LUSC -0.2; TCGA STAD -0.178; TCGA UCEC -0.131 |
hsa-miR-195-3p | FBXO28 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.108; TCGA BRCA -0.081; TCGA COAD -0.078; TCGA ESCA -0.096; TCGA HNSC -0.051; TCGA LUAD -0.089; TCGA LUSC -0.087; TCGA THCA -0.074; TCGA STAD -0.109; TCGA UCEC -0.061 |
hsa-miR-195-3p | CASP3 | 9 cancers: BLCA; BRCA; COAD; KIRP; LUAD; LUSC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.054; TCGA BRCA -0.161; TCGA COAD -0.064; TCGA KIRP -0.103; TCGA LUAD -0.122; TCGA LUSC -0.091; TCGA THCA -0.088; TCGA STAD -0.162; TCGA UCEC -0.113 |
hsa-miR-195-3p | ABCE1 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.051; TCGA BRCA -0.055; TCGA COAD -0.096; TCGA ESCA -0.108; TCGA HNSC -0.055; TCGA LUAD -0.161; TCGA LUSC -0.107; TCGA STAD -0.266; TCGA UCEC -0.071 |
hsa-miR-195-3p | SASS6 | 9 cancers: BLCA; BRCA; COAD; ESCA; LIHC; LUAD; LUSC; STAD; UCEC | MirTarget; mirMAP | TCGA BLCA -0.164; TCGA BRCA -0.153; TCGA COAD -0.146; TCGA ESCA -0.102; TCGA LIHC -0.125; TCGA LUAD -0.299; TCGA LUSC -0.375; TCGA STAD -0.304; TCGA UCEC -0.107 |
hsa-miR-195-3p | KIF18A | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; LIHC; LUAD; LUSC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.194; TCGA BRCA -0.52; TCGA CESC -0.15; TCGA COAD -0.179; TCGA ESCA -0.356; TCGA LIHC -0.358; TCGA LUAD -0.431; TCGA LUSC -0.459; TCGA THCA -0.082; TCGA STAD -0.594; TCGA UCEC -0.273 |
hsa-miR-195-3p | TMX1 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; STAD | MirTarget | TCGA BLCA -0.086; TCGA CESC -0.057; TCGA COAD -0.072; TCGA ESCA -0.145; TCGA HNSC -0.091; TCGA LUAD -0.09; TCGA LUSC -0.096; TCGA PRAD -0.066; TCGA STAD -0.189 |
hsa-miR-195-3p | GRHL2 | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LUAD; LUSC; OV; STAD | MirTarget | TCGA BLCA -0.323; TCGA BRCA -0.26; TCGA ESCA -0.198; TCGA HNSC -0.124; TCGA KIRC -1.173; TCGA LUAD -0.095; TCGA LUSC -0.317; TCGA OV -0.125; TCGA STAD -0.374 |
hsa-miR-195-3p | AHCTF1 | 9 cancers: BLCA; BRCA; COAD; ESCA; LIHC; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.068; TCGA BRCA -0.107; TCGA COAD -0.087; TCGA ESCA -0.075; TCGA LIHC -0.088; TCGA LUAD -0.131; TCGA LUSC -0.175; TCGA STAD -0.183; TCGA UCEC -0.063 |
hsa-miR-195-3p | LMNB1 | 9 cancers: BLCA; BRCA; COAD; ESCA; LUAD; LUSC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.124; TCGA BRCA -0.469; TCGA COAD -0.116; TCGA ESCA -0.18; TCGA LUAD -0.397; TCGA LUSC -0.403; TCGA THCA -0.128; TCGA STAD -0.441; TCGA UCEC -0.207 |
hsa-miR-195-3p | FAM91A1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.081; TCGA BRCA -0.149; TCGA CESC -0.108; TCGA COAD -0.073; TCGA ESCA -0.175; TCGA HNSC -0.096; TCGA LIHC -0.057; TCGA LUAD -0.177; TCGA LUSC -0.178; TCGA STAD -0.253; TCGA UCEC -0.101 |
hsa-miR-195-3p | GALNT1 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LUAD; LUSC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.088; TCGA BRCA -0.066; TCGA CESC -0.112; TCGA ESCA -0.105; TCGA HNSC -0.113; TCGA KIRP -0.1; TCGA LUAD -0.068; TCGA LUSC -0.135; TCGA STAD -0.146 |
hsa-miR-195-3p | BLOC1S2 | 11 cancers: BLCA; BRCA; ESCA; KIRC; LGG; LIHC; LUAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.085; TCGA BRCA -0.091; TCGA ESCA -0.162; TCGA KIRC -0.057; TCGA LGG -0.067; TCGA LIHC -0.075; TCGA LUAD -0.05; TCGA SARC -0.105; TCGA THCA -0.093; TCGA STAD -0.214; TCGA UCEC -0.102 |
hsa-miR-195-3p | YWHAZ | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.079; TCGA BRCA -0.217; TCGA CESC -0.103; TCGA COAD -0.085; TCGA ESCA -0.258; TCGA HNSC -0.179; TCGA LIHC -0.123; TCGA LUAD -0.222; TCGA LUSC -0.251; TCGA THCA -0.07; TCGA STAD -0.221; TCGA UCEC -0.112 |
hsa-miR-195-3p | DUS4L | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; STAD | MirTarget | TCGA BLCA -0.078; TCGA BRCA -0.125; TCGA ESCA -0.129; TCGA HNSC -0.072; TCGA KIRC -0.093; TCGA KIRP -0.101; TCGA LIHC -0.086; TCGA LUAD -0.225; TCGA LUSC -0.294; TCGA PAAD -0.212; TCGA STAD -0.198 |
hsa-miR-195-3p | CHAC2 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.126; TCGA BRCA -0.195; TCGA CESC -0.131; TCGA COAD -0.129; TCGA ESCA -0.229; TCGA HNSC -0.104; TCGA KIRC -0.231; TCGA LUAD -0.25; TCGA LUSC -0.332; TCGA STAD -0.288; TCGA UCEC -0.226 |
hsa-miR-195-3p | COMMD8 | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; OV; STAD; UCEC | MirTarget | TCGA BLCA -0.081; TCGA COAD -0.103; TCGA ESCA -0.096; TCGA HNSC -0.084; TCGA KIRC -0.175; TCGA LIHC -0.089; TCGA LUAD -0.132; TCGA OV -0.07; TCGA STAD -0.151; TCGA UCEC -0.054 |
hsa-miR-195-3p | NAB1 | 9 cancers: BLCA; COAD; ESCA; HNSC; LUAD; LUSC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.101; TCGA COAD -0.108; TCGA ESCA -0.135; TCGA HNSC -0.129; TCGA LUAD -0.098; TCGA LUSC -0.142; TCGA THCA -0.094; TCGA STAD -0.118; TCGA UCEC -0.113 |
hsa-miR-195-3p | OLA1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; OV; PAAD; THCA; STAD; UCEC | MirTarget; mirMAP | TCGA BLCA -0.068; TCGA BRCA -0.118; TCGA CESC -0.151; TCGA COAD -0.132; TCGA ESCA -0.167; TCGA HNSC -0.071; TCGA LIHC -0.131; TCGA LUAD -0.231; TCGA LUSC -0.262; TCGA OV -0.099; TCGA PAAD -0.098; TCGA THCA -0.082; TCGA STAD -0.202; TCGA UCEC -0.137 |
hsa-miR-195-3p | DBF4 | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LIHC; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BLCA -0.079; TCGA BRCA -0.271; TCGA COAD -0.098; TCGA ESCA -0.232; TCGA KIRP -0.104; TCGA LIHC -0.123; TCGA LUAD -0.325; TCGA LUSC -0.355; TCGA STAD -0.38; TCGA UCEC -0.086 |
hsa-miR-195-3p | ZNF207 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; LIHC; LUAD; LUSC; OV; THCA; STAD | mirMAP | TCGA BLCA -0.066; TCGA BRCA -0.069; TCGA CESC -0.055; TCGA COAD -0.077; TCGA ESCA -0.086; TCGA LIHC -0.096; TCGA LUAD -0.109; TCGA LUSC -0.106; TCGA OV -0.053; TCGA THCA -0.062; TCGA STAD -0.129 |
hsa-miR-195-3p | SPAST | 11 cancers: BLCA; BRCA; COAD; LIHC; LUAD; LUSC; OV; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.053; TCGA BRCA -0.079; TCGA COAD -0.058; TCGA LIHC -0.075; TCGA LUAD -0.141; TCGA LUSC -0.232; TCGA OV -0.087; TCGA PRAD -0.064; TCGA THCA -0.065; TCGA STAD -0.171; TCGA UCEC -0.09 |
hsa-miR-195-3p | EIF2AK2 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BLCA -0.108; TCGA BRCA -0.224; TCGA CESC -0.189; TCGA COAD -0.083; TCGA ESCA -0.176; TCGA HNSC -0.122; TCGA LUAD -0.213; TCGA LUSC -0.208; TCGA STAD -0.121; TCGA UCEC -0.238 |
hsa-miR-195-3p | MRPL22 | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LIHC; LUAD; LUSC; PAAD; STAD | mirMAP | TCGA BLCA -0.05; TCGA BRCA -0.129; TCGA COAD -0.134; TCGA ESCA -0.163; TCGA KIRP -0.08; TCGA LIHC -0.064; TCGA LUAD -0.129; TCGA LUSC -0.122; TCGA PAAD -0.172; TCGA STAD -0.119 |
hsa-miR-195-3p | G2E3 | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BLCA -0.078; TCGA BRCA -0.066; TCGA CESC -0.076; TCGA COAD -0.094; TCGA HNSC -0.08; TCGA LUAD -0.223; TCGA LUSC -0.186; TCGA STAD -0.197; TCGA UCEC -0.055 |
hsa-miR-195-3p | TLL2 | 10 cancers: BLCA; BRCA; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; SARC; UCEC | mirMAP | TCGA BLCA -0.28; TCGA BRCA -0.336; TCGA KIRC -0.36; TCGA KIRP -0.217; TCGA LGG -0.352; TCGA LUAD -0.243; TCGA LUSC -0.451; TCGA PAAD -0.278; TCGA SARC -0.203; TCGA UCEC -0.224 |
hsa-miR-195-3p | ERMP1 | 10 cancers: BLCA; BRCA; ESCA; KIRC; LIHC; LUAD; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.186; TCGA BRCA -0.159; TCGA ESCA -0.136; TCGA KIRC -0.364; TCGA LIHC -0.144; TCGA LUAD -0.107; TCGA PAAD -0.141; TCGA PRAD -0.094; TCGA STAD -0.255; TCGA UCEC -0.123 |
hsa-miR-195-3p | AMMECR1 | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.128; TCGA BRCA -0.154; TCGA ESCA -0.208; TCGA HNSC -0.169; TCGA KIRC -0.053; TCGA KIRP -0.071; TCGA LUAD -0.142; TCGA LUSC -0.251; TCGA THCA -0.097; TCGA STAD -0.239; TCGA UCEC -0.063 |
hsa-miR-195-3p | PGK1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.134; TCGA BRCA -0.246; TCGA CESC -0.12; TCGA COAD -0.098; TCGA ESCA -0.167; TCGA HNSC -0.12; TCGA LIHC -0.102; TCGA LUAD -0.181; TCGA LUSC -0.242; TCGA PAAD -0.193; TCGA STAD -0.224; TCGA UCEC -0.14 |
hsa-miR-195-3p | CYB5B | 10 cancers: BLCA; CESC; COAD; ESCA; KIRC; KIRP; LIHC; LUSC; PAAD; STAD | mirMAP | TCGA BLCA -0.092; TCGA CESC -0.054; TCGA COAD -0.141; TCGA ESCA -0.194; TCGA KIRC -0.106; TCGA KIRP -0.075; TCGA LIHC -0.087; TCGA LUSC -0.056; TCGA PAAD -0.123; TCGA STAD -0.26 |
hsa-miR-195-3p | GSPT1 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; OV; STAD; UCEC | mirMAP | TCGA BLCA -0.066; TCGA BRCA -0.088; TCGA CESC -0.099; TCGA ESCA -0.092; TCGA HNSC -0.069; TCGA KIRC -0.096; TCGA OV -0.055; TCGA STAD -0.109; TCGA UCEC -0.059 |
hsa-miR-195-3p | ESRP1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.26; TCGA BRCA -0.414; TCGA CESC -0.146; TCGA COAD -0.109; TCGA ESCA -0.31; TCGA HNSC -0.181; TCGA KIRC -1.151; TCGA LUAD -0.189; TCGA LUSC -0.36; TCGA PAAD -0.418; TCGA THCA -0.101; TCGA STAD -0.464; TCGA UCEC -0.259 |
hsa-miR-195-3p | C1GALT1 | 11 cancers: BLCA; CESC; ESCA; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.072; TCGA CESC -0.186; TCGA ESCA -0.196; TCGA KIRC -0.068; TCGA KIRP -0.078; TCGA LIHC -0.104; TCGA LUAD -0.159; TCGA LUSC -0.125; TCGA PAAD -0.198; TCGA STAD -0.255; TCGA UCEC -0.201 |
hsa-miR-195-3p | PLEKHA1 | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LGG; LUAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.142; TCGA COAD -0.078; TCGA ESCA -0.083; TCGA HNSC -0.087; TCGA KIRP -0.084; TCGA LGG -0.223; TCGA LUAD -0.061; TCGA PRAD -0.102; TCGA STAD -0.099; TCGA UCEC -0.094 |
hsa-miR-195-3p | TMEM33 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.108; TCGA BRCA -0.126; TCGA CESC -0.089; TCGA COAD -0.11; TCGA ESCA -0.093; TCGA HNSC -0.085; TCGA KIRC -0.184; TCGA LUAD -0.183; TCGA LUSC -0.117; TCGA PAAD -0.096; TCGA PRAD -0.067; TCGA SARC -0.06; TCGA STAD -0.298; TCGA UCEC -0.067 |
hsa-miR-195-3p | FANCE | 12 cancers: BLCA; COAD; HNSC; LIHC; LUAD; LUSC; OV; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.097; TCGA COAD -0.123; TCGA HNSC -0.132; TCGA LIHC -0.152; TCGA LUAD -0.118; TCGA LUSC -0.318; TCGA OV -0.102; TCGA PAAD -0.113; TCGA PRAD -0.056; TCGA THCA -0.177; TCGA STAD -0.207; TCGA UCEC -0.105 |
hsa-miR-195-3p | ASCC3 | 9 cancers: BLCA; BRCA; ESCA; HNSC; LUAD; LUSC; OV; THCA; STAD | mirMAP | TCGA BLCA -0.089; TCGA BRCA -0.077; TCGA ESCA -0.134; TCGA HNSC -0.084; TCGA LUAD -0.108; TCGA LUSC -0.084; TCGA OV -0.083; TCGA THCA -0.153; TCGA STAD -0.185 |
hsa-miR-195-3p | POLR3G | 9 cancers: BLCA; CESC; ESCA; HNSC; LIHC; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BLCA -0.174; TCGA CESC -0.235; TCGA ESCA -0.365; TCGA HNSC -0.119; TCGA LIHC -0.092; TCGA LUAD -0.161; TCGA LUSC -0.267; TCGA STAD -0.381; TCGA UCEC -0.072 |
hsa-miR-195-3p | SLC35F5 | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRC; LUAD; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.064; TCGA BRCA -0.064; TCGA CESC -0.085; TCGA HNSC -0.123; TCGA KIRC -0.145; TCGA LUAD -0.142; TCGA LUSC -0.177; TCGA SARC -0.071; TCGA STAD -0.076; TCGA UCEC -0.212 |
hsa-miR-195-3p | ACTR3 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BLCA -0.05; TCGA BRCA -0.157; TCGA CESC -0.098; TCGA ESCA -0.133; TCGA HNSC -0.086; TCGA LUAD -0.131; TCGA LUSC -0.146; TCGA STAD -0.115; TCGA UCEC -0.164 |
hsa-miR-195-3p | CACYBP | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.062; TCGA BRCA -0.233; TCGA CESC -0.058; TCGA COAD -0.152; TCGA ESCA -0.203; TCGA KIRC -0.051; TCGA LGG -0.063; TCGA LIHC -0.153; TCGA LUAD -0.221; TCGA LUSC -0.274; TCGA PAAD -0.184; TCGA PRAD -0.067; TCGA STAD -0.263; TCGA UCEC -0.083 |
hsa-miR-195-3p | SSX2IP | 9 cancers: BLCA; BRCA; KIRC; LUAD; LUSC; OV; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.076; TCGA BRCA -0.228; TCGA KIRC -0.084; TCGA LUAD -0.228; TCGA LUSC -0.301; TCGA OV -0.107; TCGA PRAD -0.081; TCGA STAD -0.195; TCGA UCEC -0.107 |
hsa-miR-195-3p | CD46 | 11 cancers: BLCA; BRCA; CESC; COAD; KIRC; LIHC; LUAD; PAAD; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.107; TCGA BRCA -0.059; TCGA CESC -0.078; TCGA COAD -0.168; TCGA KIRC -0.077; TCGA LIHC -0.086; TCGA LUAD -0.098; TCGA PAAD -0.208; TCGA PRAD -0.07; TCGA THCA -0.083; TCGA STAD -0.287 |
hsa-miR-195-3p | DR1 | 9 cancers: BLCA; COAD; HNSC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.062; TCGA COAD -0.064; TCGA HNSC -0.058; TCGA LUAD -0.104; TCGA LUSC -0.085; TCGA PRAD -0.053; TCGA THCA -0.079; TCGA STAD -0.064; TCGA UCEC -0.054 |
hsa-miR-195-3p | RAD23B | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUAD; LUSC; THCA; STAD | mirMAP | TCGA BLCA -0.058; TCGA BRCA -0.101; TCGA CESC -0.065; TCGA COAD -0.091; TCGA ESCA -0.181; TCGA LUAD -0.169; TCGA LUSC -0.134; TCGA THCA -0.227; TCGA STAD -0.13 |
hsa-miR-195-3p | HELLS | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LIHC; LUAD; LUSC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.137; TCGA BRCA -0.37; TCGA COAD -0.126; TCGA ESCA -0.217; TCGA KIRC -0.14; TCGA LIHC -0.252; TCGA LUAD -0.47; TCGA LUSC -0.621; TCGA THCA -0.273; TCGA STAD -0.51; TCGA UCEC -0.205 |
hsa-miR-195-3p | LYPLA1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.073; TCGA BRCA -0.188; TCGA CESC -0.129; TCGA COAD -0.107; TCGA ESCA -0.2; TCGA HNSC -0.129; TCGA KIRC -0.144; TCGA LUAD -0.243; TCGA LUSC -0.13; TCGA OV -0.102; TCGA SARC -0.056; TCGA STAD -0.256; TCGA UCEC -0.134 |
hsa-miR-195-3p | RNF2 | 11 cancers: BLCA; BRCA; HNSC; LGG; LIHC; LUAD; LUSC; OV; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.073; TCGA BRCA -0.059; TCGA HNSC -0.109; TCGA LGG -0.078; TCGA LIHC -0.067; TCGA LUAD -0.16; TCGA LUSC -0.154; TCGA OV -0.059; TCGA PRAD -0.069; TCGA STAD -0.149; TCGA UCEC -0.062 |
hsa-miR-195-3p | ACSL3 | 12 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; STAD | mirMAP | TCGA BLCA -0.097; TCGA BRCA -0.076; TCGA CESC -0.085; TCGA COAD -0.084; TCGA HNSC -0.063; TCGA KIRC -0.138; TCGA LGG -0.08; TCGA LIHC -0.126; TCGA LUAD -0.153; TCGA LUSC -0.117; TCGA PAAD -0.091; TCGA STAD -0.171 |
hsa-miR-195-3p | RAB22A | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.082; TCGA BRCA -0.085; TCGA ESCA -0.074; TCGA HNSC -0.117; TCGA KIRP -0.058; TCGA LUAD -0.089; TCGA LUSC -0.121; TCGA PAAD -0.084; TCGA SARC -0.054; TCGA STAD -0.077; TCGA UCEC -0.063 |
hsa-miR-195-3p | STAMBP | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; OV; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.066; TCGA BRCA -0.119; TCGA COAD -0.103; TCGA ESCA -0.073; TCGA HNSC -0.07; TCGA LIHC -0.051; TCGA LUAD -0.134; TCGA LUSC -0.219; TCGA OV -0.052; TCGA PRAD -0.074; TCGA STAD -0.103; TCGA UCEC -0.06 |
hsa-miR-195-3p | SSR1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.071; TCGA BRCA -0.11; TCGA CESC -0.068; TCGA COAD -0.107; TCGA ESCA -0.083; TCGA HNSC -0.088; TCGA LIHC -0.121; TCGA LUAD -0.147; TCGA LUSC -0.12; TCGA PRAD -0.113; TCGA SARC -0.06; TCGA STAD -0.126 |
hsa-miR-195-3p | KRAS | 9 cancers: BLCA; BRCA; HNSC; LGG; LUAD; LUSC; OV; STAD; UCEC | mirMAP | TCGA BLCA -0.066; TCGA BRCA -0.144; TCGA HNSC -0.094; TCGA LGG -0.083; TCGA LUAD -0.261; TCGA LUSC -0.147; TCGA OV -0.156; TCGA STAD -0.125; TCGA UCEC -0.13 |
hsa-miR-195-3p | ARHGAP32 | 10 cancers: BLCA; BRCA; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; THCA; STAD | mirMAP | TCGA BLCA -0.197; TCGA BRCA -0.075; TCGA ESCA -0.2; TCGA HNSC -0.125; TCGA LGG -0.143; TCGA LUAD -0.097; TCGA LUSC -0.131; TCGA PAAD -0.16; TCGA THCA -0.083; TCGA STAD -0.208 |
hsa-miR-195-3p | MRPL50 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LIHC; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BLCA -0.067; TCGA BRCA -0.053; TCGA CESC -0.059; TCGA COAD -0.136; TCGA ESCA -0.233; TCGA KIRC -0.082; TCGA LIHC -0.058; TCGA LUAD -0.113; TCGA LUSC -0.116; TCGA STAD -0.205; TCGA UCEC -0.055 |
hsa-miR-195-3p | G3BP1 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BLCA -0.085; TCGA BRCA -0.091; TCGA CESC -0.109; TCGA COAD -0.087; TCGA ESCA -0.153; TCGA LUAD -0.11; TCGA LUSC -0.149; TCGA STAD -0.217; TCGA UCEC -0.105 |
hsa-miR-195-3p | TPBG | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; SARC; THCA | mirMAP | TCGA BLCA -0.137; TCGA BRCA -0.091; TCGA ESCA -0.207; TCGA HNSC -0.259; TCGA KIRC -0.132; TCGA KIRP -0.102; TCGA LGG -0.381; TCGA LUAD -0.188; TCGA LUSC -0.407; TCGA SARC -0.197; TCGA THCA -0.073 |
hsa-miR-195-3p | HMGA2 | 15 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.35; TCGA BRCA -0.358; TCGA CESC -0.458; TCGA ESCA -0.481; TCGA HNSC -0.726; TCGA KIRC -0.37; TCGA KIRP -0.725; TCGA LIHC -0.515; TCGA LUAD -0.433; TCGA LUSC -0.953; TCGA OV -0.648; TCGA SARC -1.447; TCGA THCA -1.239; TCGA STAD -0.419; TCGA UCEC -0.63 |
hsa-miR-195-3p | ASAP2 | 11 cancers: BLCA; BRCA; HNSC; KIRC; LGG; LIHC; LUSC; OV; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.072; TCGA BRCA -0.059; TCGA HNSC -0.125; TCGA KIRC -0.094; TCGA LGG -0.1; TCGA LIHC -0.086; TCGA LUSC -0.105; TCGA OV -0.077; TCGA THCA -0.152; TCGA STAD -0.103; TCGA UCEC -0.171 |
hsa-miR-195-3p | SCOC | 9 cancers: BLCA; BRCA; KIRC; LGG; LUAD; LUSC; PAAD; PRAD; UCEC | mirMAP | TCGA BLCA -0.057; TCGA BRCA -0.069; TCGA KIRC -0.2; TCGA LGG -0.077; TCGA LUAD -0.153; TCGA LUSC -0.096; TCGA PAAD -0.109; TCGA PRAD -0.061; TCGA UCEC -0.067 |
hsa-miR-195-3p | TMEM87B | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; LUAD; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.077; TCGA BRCA -0.092; TCGA CESC -0.068; TCGA COAD -0.058; TCGA HNSC -0.106; TCGA LUAD -0.107; TCGA PRAD -0.066; TCGA SARC -0.115; TCGA THCA -0.088; TCGA STAD -0.158 |
hsa-miR-195-3p | SFXN1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.09; TCGA BRCA -0.224; TCGA CESC -0.082; TCGA COAD -0.164; TCGA ESCA -0.098; TCGA HNSC -0.101; TCGA LUAD -0.256; TCGA LUSC -0.366; TCGA PAAD -0.14; TCGA STAD -0.264; TCGA UCEC -0.093 |
hsa-miR-195-3p | REEP3 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUAD; PRAD; THCA; UCEC | mirMAP | TCGA BLCA -0.092; TCGA BRCA -0.063; TCGA CESC -0.142; TCGA ESCA -0.098; TCGA HNSC -0.128; TCGA LUAD -0.1; TCGA PRAD -0.139; TCGA THCA -0.082; TCGA UCEC -0.118 |
hsa-miR-195-3p | TMEM167A | 9 cancers: BLCA; BRCA; CESC; LIHC; LUAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.085; TCGA BRCA -0.081; TCGA CESC -0.054; TCGA LIHC -0.121; TCGA LUAD -0.068; TCGA PRAD -0.063; TCGA SARC -0.081; TCGA STAD -0.112; TCGA UCEC -0.079 |
hsa-miR-195-3p | PAWR | 9 cancers: BLCA; CESC; HNSC; KIRC; LUAD; LUSC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.102; TCGA CESC -0.102; TCGA HNSC -0.161; TCGA KIRC -0.144; TCGA LUAD -0.206; TCGA LUSC -0.231; TCGA THCA -0.072; TCGA STAD -0.206; TCGA UCEC -0.065 |
hsa-miR-195-3p | PLAG1 | 13 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; OV; PRAD; SARC; THCA; UCEC | mirMAP; miRNATAP | TCGA BLCA -0.287; TCGA CESC -0.235; TCGA HNSC -0.203; TCGA KIRC -0.511; TCGA KIRP -0.323; TCGA LGG -0.204; TCGA LIHC -0.351; TCGA LUAD -0.125; TCGA OV -0.386; TCGA PRAD -0.282; TCGA SARC -0.773; TCGA THCA -0.604; TCGA UCEC -0.305 |
hsa-miR-195-3p | PRPF40A | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BLCA -0.073; TCGA BRCA -0.075; TCGA CESC -0.055; TCGA COAD -0.112; TCGA ESCA -0.062; TCGA HNSC -0.053; TCGA LIHC -0.051; TCGA LUAD -0.164; TCGA LUSC -0.142; TCGA STAD -0.208; TCGA UCEC -0.107 |
hsa-miR-195-3p | HDAC2 | 10 cancers: BLCA; BRCA; COAD; ESCA; LGG; LUAD; LUSC; OV; STAD; UCEC | mirMAP | TCGA BLCA -0.058; TCGA BRCA -0.174; TCGA COAD -0.088; TCGA ESCA -0.141; TCGA LGG -0.062; TCGA LUAD -0.195; TCGA LUSC -0.26; TCGA OV -0.161; TCGA STAD -0.154; TCGA UCEC -0.104 |
hsa-miR-195-3p | CD2AP | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.087; TCGA BRCA -0.075; TCGA CESC -0.084; TCGA COAD -0.073; TCGA ESCA -0.159; TCGA HNSC -0.051; TCGA KIRC -0.103; TCGA LIHC -0.124; TCGA LUAD -0.203; TCGA THCA -0.076; TCGA STAD -0.273; TCGA UCEC -0.16 |
hsa-miR-195-3p | WDHD1 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.13; TCGA BRCA -0.23; TCGA COAD -0.107; TCGA ESCA -0.22; TCGA HNSC -0.11; TCGA KIRP -0.072; TCGA LIHC -0.124; TCGA LUAD -0.355; TCGA LUSC -0.471; TCGA THCA -0.187; TCGA STAD -0.429; TCGA UCEC -0.171 |
hsa-miR-195-3p | TSN | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; OV; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.062; TCGA BRCA -0.053; TCGA COAD -0.134; TCGA ESCA -0.102; TCGA HNSC -0.061; TCGA LIHC -0.097; TCGA LUAD -0.144; TCGA LUSC -0.185; TCGA OV -0.056; TCGA THCA -0.089; TCGA STAD -0.2; TCGA UCEC -0.11 |
hsa-miR-195-3p | TRIM59 | 10 cancers: BLCA; BRCA; COAD; LGG; LIHC; LUAD; LUSC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.1; TCGA BRCA -0.355; TCGA COAD -0.146; TCGA LGG -0.373; TCGA LIHC -0.173; TCGA LUAD -0.232; TCGA LUSC -0.344; TCGA THCA -0.177; TCGA STAD -0.322; TCGA UCEC -0.08 |
hsa-miR-195-3p | NRAS | 9 cancers: BLCA; BRCA; CESC; HNSC; LIHC; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BLCA -0.084; TCGA BRCA -0.081; TCGA CESC -0.051; TCGA HNSC -0.106; TCGA LIHC -0.082; TCGA LUAD -0.161; TCGA LUSC -0.087; TCGA STAD -0.178; TCGA UCEC -0.12 |
hsa-miR-195-3p | TMEM189 | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; UCEC | mirMAP | TCGA BLCA -0.119; TCGA BRCA -0.189; TCGA ESCA -0.192; TCGA HNSC -0.093; TCGA KIRP -0.087; TCGA LIHC -0.074; TCGA LUAD -0.125; TCGA LUSC -0.316; TCGA PAAD -0.189; TCGA UCEC -0.151 |
hsa-miR-195-3p | UBE2V1 | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; OV; PAAD; SARC; UCEC | mirMAP | TCGA BLCA -0.05; TCGA BRCA -0.125; TCGA CESC -0.098; TCGA ESCA -0.129; TCGA HNSC -0.077; TCGA LGG -0.057; TCGA LIHC -0.054; TCGA LUAD -0.095; TCGA LUSC -0.101; TCGA OV -0.104; TCGA PAAD -0.089; TCGA SARC -0.079; TCGA UCEC -0.102 |
hsa-miR-195-3p | PHF6 | 9 cancers: BLCA; BRCA; COAD; KIRC; LIHC; LUAD; LUSC; STAD; UCEC | miRNATAP | TCGA BLCA -0.082; TCGA BRCA -0.1; TCGA COAD -0.092; TCGA KIRC -0.109; TCGA LIHC -0.07; TCGA LUAD -0.161; TCGA LUSC -0.2; TCGA STAD -0.271; TCGA UCEC -0.087 |
hsa-miR-195-3p | HSPA5 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.052; TCGA BRCA -0.162; TCGA CESC -0.122; TCGA COAD -0.086; TCGA ESCA -0.103; TCGA HNSC -0.072; TCGA LUAD -0.119; TCGA LUSC -0.146; TCGA PAAD -0.226; TCGA SARC -0.099; TCGA STAD -0.183; TCGA UCEC -0.072 |
hsa-miR-195-3p | C5orf30 | 10 cancers: BLCA; BRCA; CESC; KIRC; LGG; LIHC; LUAD; LUSC; STAD; UCEC | miRNATAP | TCGA BLCA -0.122; TCGA BRCA -0.083; TCGA CESC -0.144; TCGA KIRC -0.134; TCGA LGG -0.223; TCGA LIHC -0.157; TCGA LUAD -0.128; TCGA LUSC -0.213; TCGA STAD -0.256; TCGA UCEC -0.151 |
hsa-miR-195-3p | YES1 | 10 cancers: BLCA; BRCA; HNSC; LGG; LUAD; LUSC; OV; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.085; TCGA BRCA -0.054; TCGA HNSC -0.129; TCGA LGG -0.078; TCGA LUAD -0.174; TCGA LUSC -0.136; TCGA OV -0.055; TCGA SARC -0.091; TCGA STAD -0.177; TCGA UCEC -0.086 |
hsa-miR-195-3p | INTS2 | 9 cancers: BLCA; BRCA; COAD; HNSC; KIRP; LIHC; LUAD; LUSC; STAD | miRNATAP | TCGA BLCA -0.093; TCGA BRCA -0.093; TCGA COAD -0.068; TCGA HNSC -0.096; TCGA KIRP -0.06; TCGA LIHC -0.062; TCGA LUAD -0.119; TCGA LUSC -0.189; TCGA STAD -0.179 |
hsa-miR-195-3p | PFDN4 | 9 cancers: BRCA; CESC; ESCA; KIRP; LIHC; LUAD; LUSC; PAAD; UCEC | MirTarget | TCGA BRCA -0.125; TCGA CESC -0.108; TCGA ESCA -0.145; TCGA KIRP -0.105; TCGA LIHC -0.126; TCGA LUAD -0.252; TCGA LUSC -0.28; TCGA PAAD -0.125; TCGA UCEC -0.163 |
hsa-miR-195-3p | FAM49B | 10 cancers: BRCA; CESC; ESCA; HNSC; LIHC; LUAD; LUSC; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.19; TCGA CESC -0.142; TCGA ESCA -0.265; TCGA HNSC -0.1; TCGA LIHC -0.144; TCGA LUAD -0.149; TCGA LUSC -0.135; TCGA SARC -0.096; TCGA STAD -0.237; TCGA UCEC -0.211 |
hsa-miR-195-3p | KIAA0907 | 10 cancers: BRCA; COAD; KIRP; LIHC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.075; TCGA COAD -0.096; TCGA KIRP -0.158; TCGA LIHC -0.155; TCGA LUAD -0.112; TCGA LUSC -0.313; TCGA PAAD -0.173; TCGA THCA -0.106; TCGA STAD -0.208; TCGA UCEC -0.082 |
hsa-miR-195-3p | CLTC | 12 cancers: BRCA; CESC; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.236; TCGA CESC -0.069; TCGA ESCA -0.081; TCGA HNSC -0.1; TCGA KIRC -0.074; TCGA LIHC -0.108; TCGA LUAD -0.12; TCGA LUSC -0.082; TCGA PRAD -0.074; TCGA SARC -0.058; TCGA STAD -0.121; TCGA UCEC -0.053 |
hsa-miR-195-3p | RAB1A | 11 cancers: BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.122; TCGA CESC -0.078; TCGA ESCA -0.087; TCGA HNSC -0.096; TCGA KIRC -0.061; TCGA LUAD -0.117; TCGA LUSC -0.113; TCGA PAAD -0.051; TCGA PRAD -0.054; TCGA SARC -0.074; TCGA UCEC -0.067 |
hsa-miR-195-3p | RAD21 | 10 cancers: BRCA; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.183; TCGA COAD -0.105; TCGA ESCA -0.117; TCGA HNSC -0.068; TCGA KIRC -0.065; TCGA LIHC -0.077; TCGA LUAD -0.197; TCGA LUSC -0.185; TCGA STAD -0.204; TCGA UCEC -0.084 |
hsa-miR-195-3p | DCUN1D5 | 10 cancers: BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA BRCA -0.147; TCGA CESC -0.262; TCGA COAD -0.08; TCGA ESCA -0.222; TCGA HNSC -0.136; TCGA LIHC -0.091; TCGA LUAD -0.19; TCGA LUSC -0.279; TCGA STAD -0.086; TCGA UCEC -0.123 |
hsa-miR-195-3p | NOL10 | 10 cancers: BRCA; CESC; COAD; ESCA; LIHC; LUAD; LUSC; OV; STAD; UCEC | MirTarget | TCGA BRCA -0.128; TCGA CESC -0.064; TCGA COAD -0.068; TCGA ESCA -0.092; TCGA LIHC -0.069; TCGA LUAD -0.16; TCGA LUSC -0.239; TCGA OV -0.055; TCGA STAD -0.178; TCGA UCEC -0.126 |
hsa-miR-195-3p | RCN2 | 10 cancers: BRCA; COAD; KIRC; LIHC; LUAD; LUSC; OV; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.109; TCGA COAD -0.09; TCGA KIRC -0.065; TCGA LIHC -0.113; TCGA LUAD -0.134; TCGA LUSC -0.242; TCGA OV -0.084; TCGA THCA -0.111; TCGA STAD -0.146; TCGA UCEC -0.091 |
hsa-miR-195-3p | DSN1 | 9 cancers: BRCA; COAD; ESCA; KIRP; LUAD; LUSC; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.233; TCGA COAD -0.088; TCGA ESCA -0.154; TCGA KIRP -0.128; TCGA LUAD -0.239; TCGA LUSC -0.332; TCGA THCA -0.074; TCGA STAD -0.27; TCGA UCEC -0.142 |
hsa-miR-195-3p | UBE2V2 | 10 cancers: BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; UCEC | MirTarget | TCGA BRCA -0.189; TCGA CESC -0.092; TCGA COAD -0.069; TCGA ESCA -0.135; TCGA HNSC -0.09; TCGA LGG -0.06; TCGA LIHC -0.054; TCGA LUAD -0.225; TCGA LUSC -0.219; TCGA UCEC -0.099 |
hsa-miR-195-3p | ARFGEF1 | 9 cancers: BRCA; HNSC; KIRC; LIHC; LUAD; LUSC; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.116; TCGA HNSC -0.077; TCGA KIRC -0.105; TCGA LIHC -0.064; TCGA LUAD -0.075; TCGA LUSC -0.082; TCGA THCA -0.085; TCGA STAD -0.093; TCGA UCEC -0.075 |
hsa-miR-195-3p | HIF1A | 10 cancers: BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; THCA; STAD | MirTarget | TCGA BRCA -0.066; TCGA CESC -0.119; TCGA ESCA -0.137; TCGA HNSC -0.175; TCGA KIRC -0.198; TCGA LUAD -0.138; TCGA LUSC -0.112; TCGA PRAD -0.099; TCGA THCA -0.118; TCGA STAD -0.075 |
hsa-miR-195-3p | NLK | 10 cancers: BRCA; CESC; KIRC; LGG; LIHC; LUAD; LUSC; OV; STAD; UCEC | mirMAP | TCGA BRCA -0.145; TCGA CESC -0.057; TCGA KIRC -0.091; TCGA LGG -0.146; TCGA LIHC -0.051; TCGA LUAD -0.08; TCGA LUSC -0.091; TCGA OV -0.102; TCGA STAD -0.081; TCGA UCEC -0.08 |
hsa-miR-195-3p | TPD52L2 | 12 cancers: BRCA; CESC; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.071; TCGA CESC -0.092; TCGA ESCA -0.13; TCGA HNSC -0.098; TCGA KIRP -0.137; TCGA LIHC -0.139; TCGA LUAD -0.102; TCGA LUSC -0.109; TCGA PAAD -0.158; TCGA THCA -0.061; TCGA STAD -0.098; TCGA UCEC -0.079 |
hsa-miR-195-3p | CAND1 | 10 cancers: BRCA; CESC; COAD; HNSC; LUAD; LUSC; OV; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.102; TCGA CESC -0.083; TCGA COAD -0.054; TCGA HNSC -0.069; TCGA LUAD -0.155; TCGA LUSC -0.146; TCGA OV -0.116; TCGA THCA -0.1; TCGA STAD -0.17; TCGA UCEC -0.061 |
hsa-miR-195-3p | SSR3 | 9 cancers: BRCA; CESC; COAD; HNSC; LIHC; LUAD; PRAD; SARC; STAD | mirMAP | TCGA BRCA -0.054; TCGA CESC -0.09; TCGA COAD -0.087; TCGA HNSC -0.078; TCGA LIHC -0.082; TCGA LUAD -0.101; TCGA PRAD -0.1; TCGA SARC -0.093; TCGA STAD -0.114 |
hsa-miR-195-3p | MRPL19 | 10 cancers: BRCA; CESC; COAD; KIRC; LUAD; LUSC; OV; PRAD; STAD; UCEC | mirMAP | TCGA BRCA -0.104; TCGA CESC -0.07; TCGA COAD -0.078; TCGA KIRC -0.158; TCGA LUAD -0.164; TCGA LUSC -0.174; TCGA OV -0.065; TCGA PRAD -0.078; TCGA STAD -0.112; TCGA UCEC -0.107 |
hsa-miR-195-3p | YIPF4 | 12 cancers: BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; OV; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.091; TCGA CESC -0.099; TCGA COAD -0.086; TCGA ESCA -0.064; TCGA HNSC -0.077; TCGA LUAD -0.156; TCGA LUSC -0.206; TCGA OV -0.057; TCGA PRAD -0.062; TCGA SARC -0.071; TCGA STAD -0.06; TCGA UCEC -0.152 |
hsa-miR-195-3p | TCP1 | 10 cancers: BRCA; CESC; COAD; ESCA; LGG; LUAD; LUSC; OV; STAD; UCEC | mirMAP | TCGA BRCA -0.128; TCGA CESC -0.104; TCGA COAD -0.137; TCGA ESCA -0.201; TCGA LGG -0.078; TCGA LUAD -0.142; TCGA LUSC -0.196; TCGA OV -0.155; TCGA STAD -0.194; TCGA UCEC -0.06 |
hsa-miR-195-3p | NUDCD1 | 9 cancers: BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; STAD | mirMAP | TCGA BRCA -0.14; TCGA CESC -0.141; TCGA COAD -0.096; TCGA ESCA -0.223; TCGA HNSC -0.066; TCGA LIHC -0.134; TCGA LUAD -0.228; TCGA LUSC -0.207; TCGA STAD -0.3 |
hsa-miR-195-3p | VAPB | 9 cancers: BRCA; CESC; HNSC; KIRC; LUAD; LUSC; OV; PAAD; UCEC | mirMAP | TCGA BRCA -0.084; TCGA CESC -0.05; TCGA HNSC -0.065; TCGA KIRC -0.089; TCGA LUAD -0.12; TCGA LUSC -0.121; TCGA OV -0.068; TCGA PAAD -0.089; TCGA UCEC -0.092 |
hsa-miR-195-3p | MRS2 | 10 cancers: BRCA; CESC; COAD; KIRC; KIRP; LIHC; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BRCA -0.124; TCGA CESC -0.115; TCGA COAD -0.138; TCGA KIRC -0.107; TCGA KIRP -0.059; TCGA LIHC -0.132; TCGA LUAD -0.098; TCGA LUSC -0.092; TCGA STAD -0.103; TCGA UCEC -0.074 |
hsa-miR-195-3p | NAA38 | 9 cancers: BRCA; ESCA; KIRP; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BRCA -0.119; TCGA ESCA -0.088; TCGA KIRP -0.097; TCGA LIHC -0.093; TCGA LUAD -0.159; TCGA LUSC -0.201; TCGA PAAD -0.065; TCGA STAD -0.119; TCGA UCEC -0.083 |
hsa-miR-195-3p | UBQLN1 | 9 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; STAD | mirMAP | TCGA BRCA -0.077; TCGA CESC -0.056; TCGA COAD -0.072; TCGA ESCA -0.144; TCGA HNSC -0.052; TCGA KIRC -0.086; TCGA LGG -0.059; TCGA LUAD -0.101; TCGA STAD -0.128 |
hsa-miR-195-3p | GSTCD | 9 cancers: BRCA; COAD; HNSC; KIRC; LUAD; LUSC; OV; STAD; UCEC | mirMAP | TCGA BRCA -0.059; TCGA COAD -0.09; TCGA HNSC -0.07; TCGA KIRC -0.073; TCGA LUAD -0.182; TCGA LUSC -0.135; TCGA OV -0.107; TCGA STAD -0.185; TCGA UCEC -0.102 |
hsa-miR-195-3p | TPM3 | 11 cancers: BRCA; CESC; COAD; ESCA; LIHC; LUAD; LUSC; SARC; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.246; TCGA CESC -0.122; TCGA COAD -0.081; TCGA ESCA -0.224; TCGA LIHC -0.066; TCGA LUAD -0.15; TCGA LUSC -0.055; TCGA SARC -0.1; TCGA THCA -0.102; TCGA STAD -0.24; TCGA UCEC -0.091 |
hsa-miR-195-3p | HAPLN1 | 11 cancers: BRCA; CESC; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.765; TCGA CESC -0.705; TCGA HNSC -0.315; TCGA LIHC -0.403; TCGA LUAD -0.223; TCGA LUSC -0.296; TCGA PAAD -0.938; TCGA PRAD -0.566; TCGA THCA -0.876; TCGA STAD -0.309; TCGA UCEC -0.44 |
hsa-miR-195-3p | SURF4 | 9 cancers: BRCA; CESC; COAD; ESCA; HNSC; LUAD; PAAD; SARC; STAD | mirMAP | TCGA BRCA -0.105; TCGA CESC -0.105; TCGA COAD -0.069; TCGA ESCA -0.141; TCGA HNSC -0.052; TCGA LUAD -0.087; TCGA PAAD -0.117; TCGA SARC -0.1; TCGA STAD -0.128 |
hsa-miR-195-3p | SLC7A11 | 11 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BRCA -0.293; TCGA CESC -0.329; TCGA COAD -0.177; TCGA ESCA -0.341; TCGA HNSC -0.432; TCGA KIRP -0.232; TCGA LIHC -0.694; TCGA LUAD -0.478; TCGA LUSC -0.702; TCGA STAD -0.553; TCGA UCEC -0.191 |
hsa-miR-195-3p | ATP6V1C1 | 9 cancers: BRCA; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BRCA -0.09; TCGA ESCA -0.081; TCGA HNSC -0.077; TCGA LGG -0.064; TCGA LIHC -0.191; TCGA LUAD -0.107; TCGA LUSC -0.109; TCGA STAD -0.132; TCGA UCEC -0.058 |
hsa-miR-195-3p | OTUD6B | 10 cancers: BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BRCA -0.056; TCGA CESC -0.088; TCGA COAD -0.144; TCGA ESCA -0.135; TCGA HNSC -0.051; TCGA LIHC -0.082; TCGA LUAD -0.154; TCGA LUSC -0.207; TCGA STAD -0.17; TCGA UCEC -0.059 |
hsa-miR-195-3p | C16orf87 | 9 cancers: BRCA; CESC; HNSC; KIRC; LUAD; LUSC; OV; STAD; UCEC | mirMAP; miRNATAP | TCGA BRCA -0.15; TCGA CESC -0.091; TCGA HNSC -0.091; TCGA KIRC -0.073; TCGA LUAD -0.141; TCGA LUSC -0.167; TCGA OV -0.091; TCGA STAD -0.102; TCGA UCEC -0.061 |
hsa-miR-195-3p | WDYHV1 | 10 cancers: BRCA; CESC; COAD; ESCA; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BRCA -0.154; TCGA CESC -0.087; TCGA COAD -0.104; TCGA ESCA -0.118; TCGA LIHC -0.133; TCGA LUAD -0.197; TCGA LUSC -0.248; TCGA PAAD -0.152; TCGA STAD -0.122; TCGA UCEC -0.052 |
hsa-miR-195-3p | IGF2BP1 | 9 cancers: BRCA; CESC; HNSC; LIHC; LUAD; LUSC; OV; SARC; UCEC | mirMAP | TCGA BRCA -0.553; TCGA CESC -0.876; TCGA HNSC -0.453; TCGA LIHC -0.832; TCGA LUAD -0.942; TCGA LUSC -0.824; TCGA OV -0.323; TCGA SARC -0.588; TCGA UCEC -0.618 |
hsa-miR-195-3p | KIAA1841 | 9 cancers: BRCA; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; THCA; STAD | mirMAP | TCGA BRCA -0.161; TCGA KIRP -0.161; TCGA LGG -0.125; TCGA LIHC -0.179; TCGA LUAD -0.127; TCGA LUSC -0.117; TCGA PAAD -0.134; TCGA THCA -0.1; TCGA STAD -0.204 |
hsa-miR-195-3p | GMPS | 9 cancers: BRCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BRCA -0.21; TCGA COAD -0.128; TCGA ESCA -0.14; TCGA HNSC -0.069; TCGA LIHC -0.081; TCGA LUAD -0.246; TCGA LUSC -0.335; TCGA STAD -0.239; TCGA UCEC -0.128 |
hsa-miR-195-3p | NAA15 | 9 cancers: BRCA; COAD; ESCA; HNSC; LUAD; LUSC; OV; STAD; UCEC | mirMAP | TCGA BRCA -0.107; TCGA COAD -0.059; TCGA ESCA -0.07; TCGA HNSC -0.057; TCGA LUAD -0.166; TCGA LUSC -0.124; TCGA OV -0.07; TCGA STAD -0.172; TCGA UCEC -0.068 |
hsa-miR-195-3p | TMEM68 | 12 cancers: BRCA; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; OV; PAAD; THCA; STAD | mirMAP | TCGA BRCA -0.116; TCGA COAD -0.11; TCGA ESCA -0.113; TCGA HNSC -0.073; TCGA KIRC -0.113; TCGA LIHC -0.096; TCGA LUAD -0.155; TCGA LUSC -0.151; TCGA OV -0.065; TCGA PAAD -0.21; TCGA THCA -0.091; TCGA STAD -0.228 |
hsa-miR-195-3p | TMEM99 | 9 cancers: BRCA; COAD; HNSC; KIRC; KIRP; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BRCA -0.078; TCGA COAD -0.13; TCGA HNSC -0.076; TCGA KIRC -0.055; TCGA KIRP -0.077; TCGA LUSC -0.319; TCGA PRAD -0.093; TCGA STAD -0.239; TCGA UCEC -0.079 |
hsa-miR-195-3p | E2F6 | 10 cancers: BRCA; COAD; KIRP; LIHC; LUAD; LUSC; OV; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.065; TCGA COAD -0.093; TCGA KIRP -0.06; TCGA LIHC -0.11; TCGA LUAD -0.165; TCGA LUSC -0.208; TCGA OV -0.118; TCGA THCA -0.057; TCGA STAD -0.134; TCGA UCEC -0.06 |
hsa-miR-195-3p | ESCO2 | 9 cancers: BRCA; COAD; ESCA; KIRP; LUAD; LUSC; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.496; TCGA COAD -0.186; TCGA ESCA -0.257; TCGA KIRP -0.146; TCGA LUAD -0.514; TCGA LUSC -0.656; TCGA THCA -0.155; TCGA STAD -0.549; TCGA UCEC -0.287 |
hsa-miR-195-3p | ELMOD2 | 9 cancers: BRCA; COAD; HNSC; KIRC; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BRCA -0.06; TCGA COAD -0.05; TCGA HNSC -0.053; TCGA KIRC -0.061; TCGA LUAD -0.124; TCGA LUSC -0.051; TCGA PRAD -0.068; TCGA STAD -0.103; TCGA UCEC -0.059 |
hsa-miR-195-3p | C14orf119 | 11 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; OV; THCA | mirMAP | TCGA BRCA -0.086; TCGA CESC -0.072; TCGA COAD -0.061; TCGA ESCA -0.147; TCGA HNSC -0.095; TCGA KIRC -0.06; TCGA LIHC -0.067; TCGA LUAD -0.094; TCGA LUSC -0.075; TCGA OV -0.055; TCGA THCA -0.127 |
hsa-miR-195-3p | GNB1L | 10 cancers: BRCA; ESCA; HNSC; LGG; LIHC; LUSC; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.148; TCGA ESCA -0.129; TCGA HNSC -0.066; TCGA LGG -0.101; TCGA LIHC -0.176; TCGA LUSC -0.265; TCGA PAAD -0.205; TCGA THCA -0.101; TCGA STAD -0.149; TCGA UCEC -0.057 |
hsa-miR-195-3p | CMC1 | 11 cancers: BRCA; CESC; COAD; ESCA; KIRC; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BRCA -0.156; TCGA CESC -0.077; TCGA COAD -0.086; TCGA ESCA -0.176; TCGA KIRC -0.187; TCGA LIHC -0.086; TCGA LUAD -0.135; TCGA LUSC -0.082; TCGA PAAD -0.178; TCGA STAD -0.126; TCGA UCEC -0.064 |
hsa-miR-195-3p | GFPT1 | 12 cancers: BRCA; COAD; HNSC; KIRC; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.129; TCGA COAD -0.08; TCGA HNSC -0.064; TCGA KIRC -0.066; TCGA LUAD -0.197; TCGA LUSC -0.112; TCGA OV -0.069; TCGA PAAD -0.145; TCGA PRAD -0.108; TCGA SARC -0.063; TCGA STAD -0.269; TCGA UCEC -0.064 |
hsa-miR-195-3p | EIF5AL1 | 9 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; STAD | mirMAP | TCGA BRCA -0.112; TCGA CESC -0.122; TCGA COAD -0.108; TCGA ESCA -0.224; TCGA HNSC -0.087; TCGA KIRP -0.097; TCGA LUAD -0.162; TCGA LUSC -0.197; TCGA STAD -0.239 |
hsa-miR-195-3p | PDCD10 | 9 cancers: BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; STAD; UCEC | miRNATAP | TCGA BRCA -0.142; TCGA COAD -0.09; TCGA ESCA -0.176; TCGA HNSC -0.111; TCGA KIRC -0.068; TCGA LUAD -0.143; TCGA LUSC -0.248; TCGA STAD -0.133; TCGA UCEC -0.145 |
hsa-miR-195-3p | CCL20 | 9 cancers: BRCA; COAD; ESCA; HNSC; LIHC; LUAD; SARC; THCA; STAD | miRNATAP | TCGA BRCA -0.426; TCGA COAD -0.243; TCGA ESCA -0.457; TCGA HNSC -0.248; TCGA LIHC -0.339; TCGA LUAD -0.321; TCGA SARC -0.368; TCGA THCA -0.737; TCGA STAD -0.651 |
hsa-miR-195-3p | CSNK1E | 10 cancers: CESC; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; THCA | miRNATAP | TCGA CESC -0.078; TCGA ESCA -0.097; TCGA HNSC -0.054; TCGA KIRP -0.06; TCGA LIHC -0.104; TCGA LUAD -0.09; TCGA LUSC -0.165; TCGA OV -0.053; TCGA PAAD -0.169; TCGA THCA -0.128 |
hsa-miR-195-3p | IGF2BP2 | 15 cancers: CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA CESC -0.542; TCGA COAD -0.101; TCGA ESCA -0.319; TCGA HNSC -0.461; TCGA KIRC -0.583; TCGA KIRP -0.392; TCGA LIHC -0.294; TCGA LUAD -0.285; TCGA LUSC -0.442; TCGA OV -0.296; TCGA PRAD -0.184; TCGA SARC -1.171; TCGA THCA -0.829; TCGA STAD -0.281; TCGA UCEC -0.763 |
Enriched cancer pathways of putative targets