microRNA information: hsa-miR-197-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-197-3p | miRbase |
| Accession: | MIMAT0000227 | miRbase |
| Precursor name: | hsa-mir-197 | miRbase |
| Precursor accession: | MI0000239 | miRbase |
| Symbol: | MIR197 | HGNC |
| RefSeq ID: | NR_029583 | GenBank |
| Sequence: | UUCACCACCUUCUCCACCCAGC |
Reported expression in cancers: hsa-miR-197-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-197-3p | breast cancer | deregulation | "For identification of differentially expressed mic ......" | 20331864 | qPCR |
| hsa-miR-197-3p | esophageal cancer | downregulation | "Implications of microRNA 197 downregulated express ......" | 25117314 | Microarray |
| hsa-miR-197-3p | gastric cancer | deregulation | "miRNA expression signature was first analyzed by r ......" | 21628394 | qPCR |
| hsa-miR-197-3p | glioblastoma | downregulation | "Northern blotting and reverse transcription‑quan ......" | 27035789 | Northern blot; qPCR |
| hsa-miR-197-3p | lung squamous cell cancer | downregulation | "miR-197 is downregulated in platinum-resistant NSC ......" | 25597412 | |
| hsa-miR-197-3p | lymphoma | deregulation | "In addition we derived an 11-miRNA signature 4 upr ......" | 23801630 |
Reported cancer pathway affected by hsa-miR-197-3p
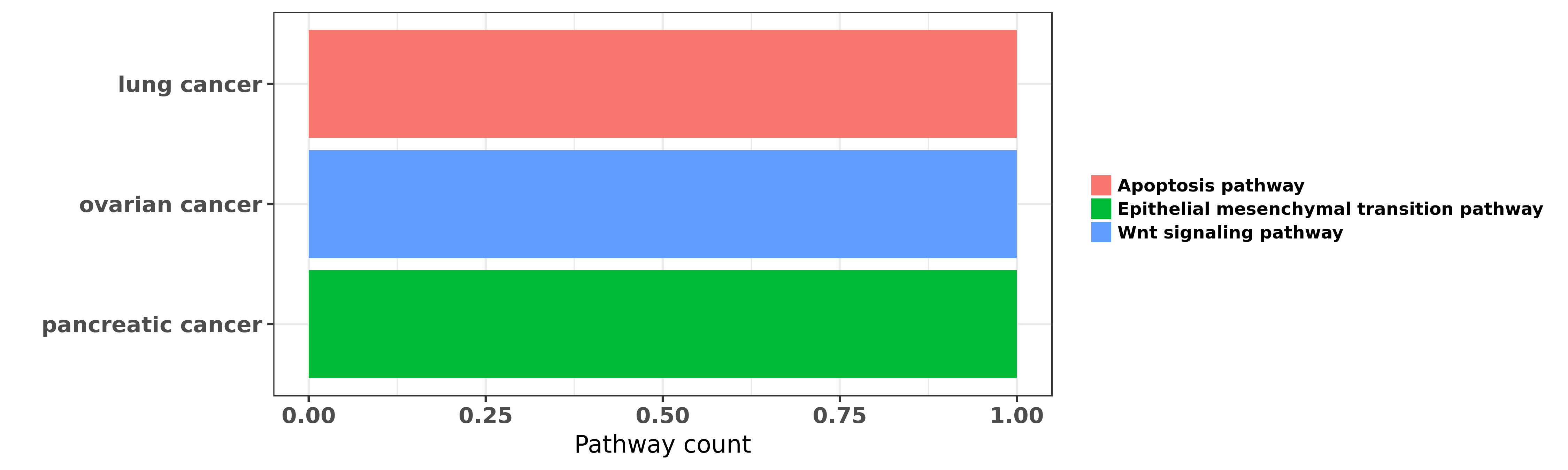
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-197-3p | lung cancer | Apoptosis pathway | "Antitumor effect of miR 197 targeting in p53 wild ......" | 24488097 | |
| hsa-miR-197-3p | ovarian cancer | Wnt signaling pathway | "MiR 197 induces Taxol resistance in human ovarian ......" | 25833695 | |
| hsa-miR-197-3p | pancreatic cancer | Epithelial mesenchymal transition pathway | "miR 197 induces epithelial mesenchymal transition ......" | 23139153 |
Reported cancer prognosis affected by hsa-miR-197-3p
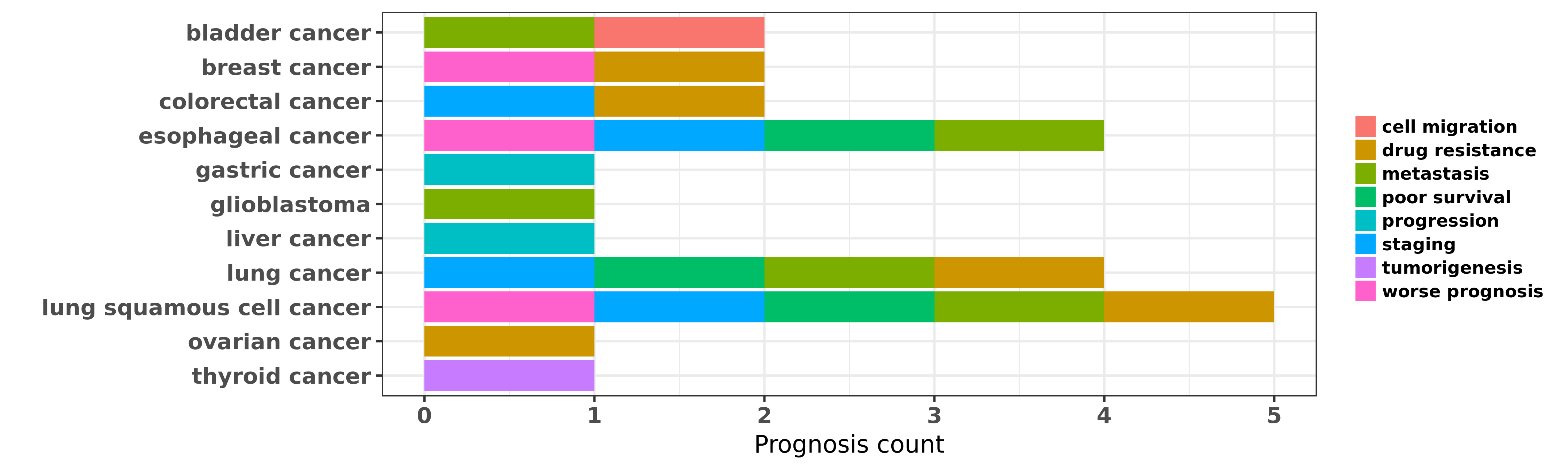
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-197-3p | bladder cancer | metastasis; cell migration | "LINC00312 inhibits the migration and invasion of b ......" | 27631965 | Transwell assay; Western blot |
| hsa-miR-197-3p | breast cancer | worse prognosis; drug resistance | "The aim of this study was to investigate the role ......" | 27746365 | |
| hsa-miR-197-3p | colorectal cancer | drug resistance; staging | "MicroRNA 197 influences 5 fluorouracil resistance ......" | 26055341 | Luciferase; Cell Proliferation Assay; Western blot |
| hsa-miR-197-3p | esophageal cancer | worse prognosis; staging; metastasis; poor survival | "Implications of microRNA 197 downregulated express ......" | 25117314 | |
| hsa-miR-197-3p | gastric cancer | progression | "Associations of Il 1 Family Related Polymorphisms ......" | 26632693 | Western blot |
| hsa-miR-197-3p | glioblastoma | metastasis | "FUS1 acts as a tumor suppressor gene by upregulati ......" | 26081814 | |
| hsa-miR-197-3p | liver cancer | progression | "Reciprocal control of miR 197 and IL 6/STAT3 pathw ......" | 26451302 | |
| hsa-miR-197-3p | lung cancer | staging; metastasis | "We searched the published literature for the miRNA ......" | 21904633 | |
| hsa-miR-197-3p | lung cancer | poor survival | "Antitumor effect of miR 197 targeting in p53 wild ......" | 24488097 | |
| hsa-miR-197-3p | lung cancer | drug resistance | "For better understanding of the cellular response ......" | 25451482 | |
| hsa-miR-197-3p | lung squamous cell cancer | metastasis; drug resistance; poor survival | "miR-197 is downregulated in platinum-resistant NSC ......" | 25597412 | |
| hsa-miR-197-3p | lung squamous cell cancer | worse prognosis; staging | "The oncomiR miR 197 is a novel prognostic indicato ......" | 25867273 | |
| hsa-miR-197-3p | ovarian cancer | drug resistance | "MiR 197 induces Taxol resistance in human ovarian ......" | 25833695 | |
| hsa-miR-197-3p | thyroid cancer | tumorigenesis | "Validation was done by quantitative RT-PCR; Two sp ......" | 16822819 |
Reported gene related to hsa-miR-197-3p
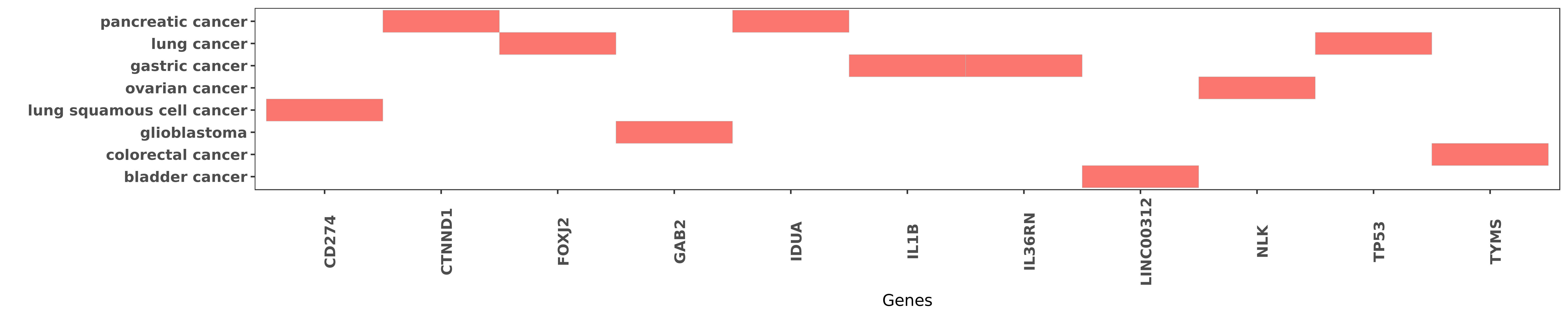
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-197-3p | lung squamous cell cancer | CD274 | "More importantly expression levels of miR-197 are ......" | 25597412 |
| hsa-miR-197-3p | pancreatic cancer | CTNND1 | "miR 197 induces epithelial mesenchymal transition ......" | 23139153 |
| hsa-miR-197-3p | lung cancer | FOXJ2 | "We here show that miR-197 not only negatively regu ......" | 25451482 |
| hsa-miR-197-3p | glioblastoma | GAB2 | "MicroRNA 197 inhibits cell proliferation by target ......" | 27035789 |
| hsa-miR-197-3p | pancreatic cancer | IDUA | "Assessing the difference of miRNA expression profi ......" | 23139153 |
| hsa-miR-197-3p | gastric cancer | IL1B | "To explore whether the roles of IL-1 family single ......" | 26632693 |
| hsa-miR-197-3p | gastric cancer | IL36RN | "The recombinant plasmid-pGenesil-1-miR-197 could u ......" | 26632693 |
| hsa-miR-197-3p | bladder cancer | LINC00312 | "LINC00312 inhibits the migration and invasion of b ......" | 27631965 |
| hsa-miR-197-3p | ovarian cancer | NLK | "MiR 197 induces Taxol resistance in human ovarian ......" | 25833695 |
| hsa-miR-197-3p | lung cancer | TP53 | "Antitumor effect of miR 197 targeting in p53 wild ......" | 24488097 |
| hsa-miR-197-3p | colorectal cancer | TYMS | "MicroRNA 197 influences 5 fluorouracil resistance ......" | 26055341 |
Expression profile in cancer corhorts:
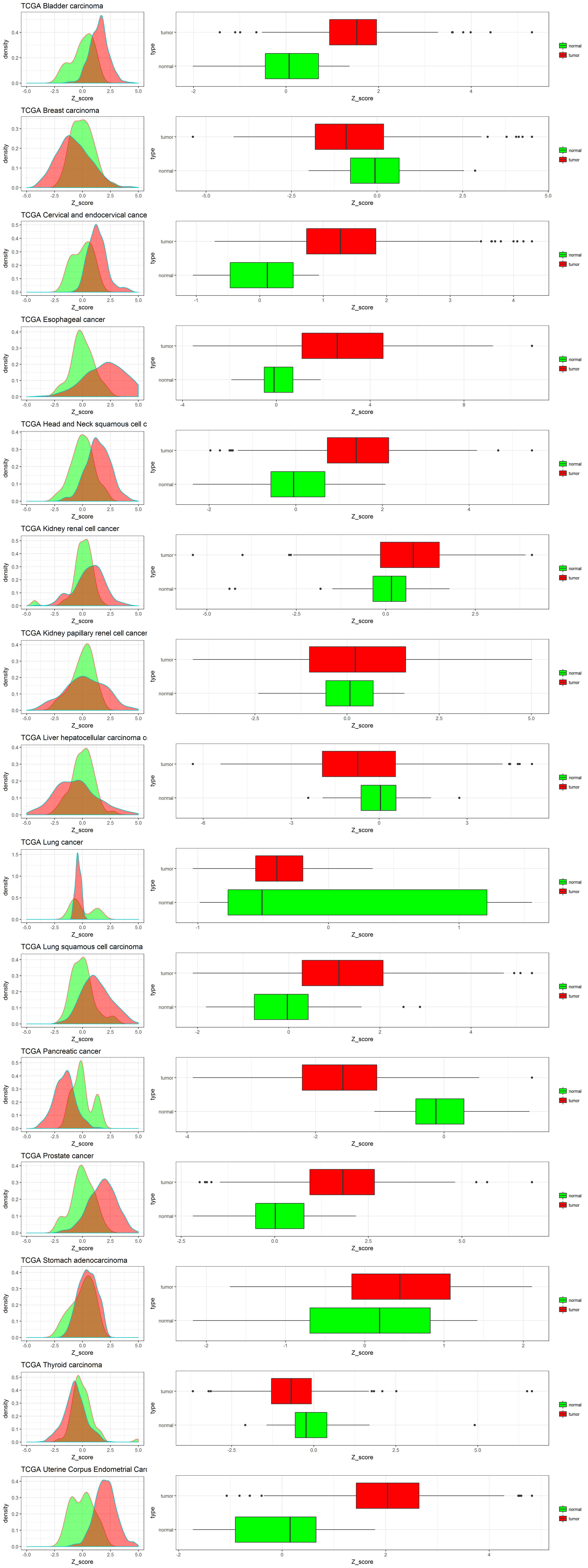
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-197-3p | ACVR1 | 13 cancers: BLCA; CESC; COAD; ESCA; LGG; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.262; TCGA CESC -0.106; TCGA COAD -0.123; TCGA ESCA -0.115; TCGA LGG -0.054; TCGA LUAD -0.094; TCGA LUSC -0.1; TCGA PAAD -0.18; TCGA PRAD -0.183; TCGA SARC -0.201; TCGA THCA -0.28; TCGA STAD -0.128; TCGA UCEC -0.073 |
hsa-miR-197-3p | CYLD | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.234; TCGA CESC -0.183; TCGA COAD -0.114; TCGA ESCA -0.293; TCGA HNSC -0.107; TCGA LUSC -0.274; TCGA PRAD -0.181; TCGA SARC -0.125; TCGA STAD -0.224; TCGA UCEC -0.195 |
hsa-miR-197-3p | DPYSL3 | 12 cancers: BLCA; COAD; ESCA; HNSC; LGG; LIHC; LUSC; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.702; TCGA COAD -0.682; TCGA ESCA -0.575; TCGA HNSC -0.268; TCGA LGG -0.173; TCGA LIHC -0.325; TCGA LUSC -0.305; TCGA PRAD -0.443; TCGA SARC -0.386; TCGA THCA -0.448; TCGA STAD -0.786; TCGA UCEC -0.551 |
hsa-miR-197-3p | EHD2 | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LIHC; LUSC; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.34; TCGA BRCA -0.142; TCGA COAD -0.443; TCGA ESCA -0.34; TCGA KIRP -0.191; TCGA LIHC -0.186; TCGA LUSC -0.267; TCGA PRAD -0.425; TCGA THCA -0.26; TCGA STAD -0.487; TCGA UCEC -0.522 |
hsa-miR-197-3p | IL1R1 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUSC; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.245; TCGA CESC -0.202; TCGA COAD -0.45; TCGA ESCA -0.496; TCGA HNSC -0.206; TCGA LUSC -0.375; TCGA PRAD -0.099; TCGA THCA -0.107; TCGA STAD -0.393; TCGA UCEC -0.22 |
hsa-miR-197-3p | KLF10 | 10 cancers: BLCA; ESCA; HNSC; KIRP; LGG; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.191; TCGA ESCA -0.291; TCGA HNSC -0.115; TCGA KIRP -0.214; TCGA LGG -0.187; TCGA LUSC -0.162; TCGA PRAD -0.315; TCGA SARC -0.113; TCGA STAD -0.296; TCGA UCEC -0.163 |
hsa-miR-197-3p | PDZRN3 | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUSC; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.809; TCGA CESC -0.318; TCGA COAD -0.326; TCGA ESCA -0.547; TCGA HNSC -0.32; TCGA LUSC -0.632; TCGA PRAD -0.097; TCGA SARC -0.529; TCGA THCA -0.355; TCGA STAD -0.817; TCGA UCEC -0.257 |
hsa-miR-197-3p | SNX1 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LGG; OV; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.137; TCGA BRCA -0.093; TCGA CESC -0.074; TCGA ESCA -0.118; TCGA HNSC -0.064; TCGA LGG -0.081; TCGA OV -0.093; TCGA PRAD -0.106; TCGA SARC -0.084; TCGA THCA -0.166; TCGA STAD -0.175; TCGA UCEC -0.151 |
hsa-miR-197-3p | TMX4 | 11 cancers: BLCA; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.218; TCGA KIRC -0.175; TCGA KIRP -0.313; TCGA LIHC -0.127; TCGA LUAD -0.114; TCGA LUSC -0.123; TCGA PRAD -0.106; TCGA SARC -0.169; TCGA THCA -0.077; TCGA STAD -0.19; TCGA UCEC -0.117 |
hsa-miR-197-3p | ZNF208 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.304; TCGA CESC -0.379; TCGA COAD -0.505; TCGA ESCA -0.988; TCGA HNSC -0.306; TCGA KIRC -0.518; TCGA LGG -0.247; TCGA LUSC -0.253; TCGA PRAD -0.408; TCGA SARC -0.425; TCGA STAD -0.585; TCGA UCEC -0.57 |
hsa-miR-197-3p | PPP2R5A | 11 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; SARC; UCEC | MirTarget | TCGA BLCA -0.168; TCGA ESCA -0.128; TCGA HNSC -0.073; TCGA KIRC -0.229; TCGA KIRP -0.176; TCGA LIHC -0.16; TCGA LUAD -0.114; TCGA LUSC -0.141; TCGA PRAD -0.133; TCGA SARC -0.31; TCGA UCEC -0.13 |
hsa-miR-197-3p | STXBP4 | 10 cancers: BLCA; COAD; KIRP; LGG; LUAD; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.107; TCGA COAD -0.092; TCGA KIRP -0.233; TCGA LGG -0.11; TCGA LUAD -0.103; TCGA PAAD -0.374; TCGA PRAD -0.257; TCGA SARC -0.202; TCGA STAD -0.158; TCGA UCEC -0.113 |
hsa-miR-197-3p | PITPNM3 | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.189; TCGA CESC -0.19; TCGA ESCA -0.393; TCGA HNSC -0.24; TCGA KIRC -0.473; TCGA KIRP -0.476; TCGA PRAD -0.382; TCGA SARC -0.643; TCGA THCA -0.184; TCGA STAD -0.627; TCGA UCEC -0.123 |
hsa-miR-197-3p | PPP1R13B | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; SARC | MirTarget | TCGA BLCA -0.095; TCGA BRCA -0.121; TCGA CESC -0.112; TCGA ESCA -0.149; TCGA HNSC -0.134; TCGA KIRC -0.21; TCGA KIRP -0.119; TCGA LIHC -0.094; TCGA LUAD -0.071; TCGA SARC -0.114 |
hsa-miR-197-3p | ARHGEF12 | 12 cancers: BLCA; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.122; TCGA ESCA -0.194; TCGA HNSC -0.189; TCGA KIRP -0.21; TCGA LIHC -0.169; TCGA LUAD -0.091; TCGA LUSC -0.071; TCGA PAAD -0.142; TCGA PRAD -0.167; TCGA SARC -0.182; TCGA STAD -0.133; TCGA UCEC -0.104 |
hsa-miR-197-3p | CALD1 | 9 cancers: BLCA; COAD; ESCA; LIHC; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.635; TCGA COAD -0.627; TCGA ESCA -0.645; TCGA LIHC -0.081; TCGA LUSC -0.394; TCGA PRAD -0.546; TCGA SARC -0.617; TCGA STAD -0.8; TCGA UCEC -0.613 |
hsa-miR-197-3p | DIP2C | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.189; TCGA CESC -0.228; TCGA COAD -0.287; TCGA ESCA -0.325; TCGA HNSC -0.172; TCGA LIHC -0.276; TCGA LUSC -0.132; TCGA PAAD -0.228; TCGA PRAD -0.16; TCGA SARC -0.292; TCGA STAD -0.425; TCGA UCEC -0.159 |
hsa-miR-197-3p | TAL1 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; LIHC; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.204; TCGA BRCA -0.199; TCGA CESC -0.318; TCGA COAD -0.312; TCGA ESCA -0.516; TCGA LIHC -0.326; TCGA LUSC -0.387; TCGA PRAD -0.131; TCGA STAD -0.313; TCGA UCEC -0.691 |
hsa-miR-197-3p | DOCK9 | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LIHC; LUSC; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.115; TCGA CESC -0.179; TCGA ESCA -0.253; TCGA HNSC -0.259; TCGA KIRP -0.238; TCGA LIHC -0.129; TCGA LUSC -0.237; TCGA PRAD -0.26; TCGA THCA -0.305; TCGA STAD -0.144; TCGA UCEC -0.189 |
hsa-miR-197-3p | FAM110B | 9 cancers: BLCA; BRCA; COAD; HNSC; KIRP; LGG; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.529; TCGA BRCA -0.35; TCGA COAD -0.499; TCGA HNSC -0.186; TCGA KIRP -0.283; TCGA LGG -0.226; TCGA LUSC -0.262; TCGA STAD -0.435; TCGA UCEC -0.236 |
hsa-miR-197-3p | MMRN1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.958; TCGA BRCA -0.534; TCGA CESC -0.636; TCGA COAD -1.096; TCGA ESCA -1.393; TCGA HNSC -0.782; TCGA LUSC -0.771; TCGA PAAD -0.759; TCGA PRAD -0.641; TCGA SARC -0.655; TCGA STAD -0.917; TCGA UCEC -0.756 |
hsa-miR-197-3p | ZMYND11 | 11 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.079; TCGA ESCA -0.116; TCGA HNSC -0.115; TCGA KIRC -0.107; TCGA KIRP -0.188; TCGA LUAD -0.065; TCGA LUSC -0.095; TCGA PRAD -0.18; TCGA SARC -0.165; TCGA STAD -0.204; TCGA UCEC -0.136 |
hsa-miR-197-3p | PLCL1 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.46; TCGA BRCA -0.171; TCGA COAD -0.439; TCGA ESCA -0.632; TCGA HNSC -0.326; TCGA KIRC -0.459; TCGA LUSC -0.416; TCGA PRAD -0.546; TCGA SARC -0.312; TCGA STAD -0.607; TCGA UCEC -0.533 |
hsa-miR-197-3p | FOXP2 | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.785; TCGA CESC -0.475; TCGA COAD -0.998; TCGA ESCA -0.865; TCGA HNSC -0.676; TCGA KIRP -0.409; TCGA PAAD -0.496; TCGA PRAD -0.972; TCGA SARC -0.339; TCGA STAD -1.267; TCGA UCEC -0.941 |
hsa-miR-197-3p | USP4 | 9 cancers: BLCA; ESCA; HNSC; KIRC; LUSC; OV; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.074; TCGA ESCA -0.093; TCGA HNSC -0.062; TCGA KIRC -0.109; TCGA LUSC -0.089; TCGA OV -0.086; TCGA PRAD -0.09; TCGA THCA -0.105; TCGA STAD -0.101 |
hsa-miR-197-3p | XPC | 13 cancers: BLCA; BRCA; CESC; ESCA; KIRC; LGG; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.065; TCGA BRCA -0.154; TCGA CESC -0.092; TCGA ESCA -0.137; TCGA KIRC -0.134; TCGA LGG -0.078; TCGA LUSC -0.134; TCGA OV -0.079; TCGA PRAD -0.068; TCGA SARC -0.128; TCGA THCA -0.126; TCGA STAD -0.071; TCGA UCEC -0.137 |
hsa-miR-197-3p | AFAP1L2 | 9 cancers: BLCA; BRCA; CESC; LGG; LUSC; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.157; TCGA BRCA -0.214; TCGA CESC -0.287; TCGA LGG -0.261; TCGA LUSC -0.358; TCGA PRAD -0.544; TCGA THCA -0.169; TCGA STAD -0.326; TCGA UCEC -0.422 |
hsa-miR-197-3p | SEPT10 | 9 cancers: BLCA; COAD; HNSC; KIRP; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.106; TCGA COAD -0.126; TCGA HNSC -0.15; TCGA KIRP -0.232; TCGA LUSC -0.12; TCGA PRAD -0.298; TCGA SARC -0.236; TCGA STAD -0.196; TCGA UCEC -0.281 |
hsa-miR-197-3p | SERTAD4 | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.304; TCGA CESC -0.514; TCGA COAD -0.427; TCGA ESCA -0.372; TCGA HNSC -0.402; TCGA KIRP -0.277; TCGA PAAD -0.443; TCGA PRAD -0.865; TCGA SARC -0.543; TCGA STAD -0.84; TCGA UCEC -0.37 |
hsa-miR-197-3p | CILP | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -1.182; TCGA BRCA -0.493; TCGA CESC -0.484; TCGA COAD -0.952; TCGA ESCA -1.686; TCGA HNSC -0.767; TCGA LUSC -0.626; TCGA STAD -0.85; TCGA UCEC -0.384 |
hsa-miR-197-3p | DACT1 | 9 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.622; TCGA BRCA -0.095; TCGA COAD -0.369; TCGA ESCA -0.481; TCGA KIRC -0.223; TCGA LUSC -0.499; TCGA PRAD -0.423; TCGA STAD -0.326; TCGA UCEC -0.416 |
hsa-miR-197-3p | CELF2 | 10 cancers: BLCA; CESC; ESCA; HNSC; LUSC; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.775; TCGA CESC -0.258; TCGA ESCA -0.761; TCGA HNSC -0.328; TCGA LUSC -0.559; TCGA PRAD -0.5; TCGA SARC -0.163; TCGA THCA -0.166; TCGA STAD -0.598; TCGA UCEC -0.439 |
hsa-miR-197-3p | H6PD | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.111; TCGA BRCA -0.053; TCGA CESC -0.139; TCGA ESCA -0.16; TCGA HNSC -0.148; TCGA PRAD -0.142; TCGA THCA -0.061; TCGA STAD -0.15; TCGA UCEC -0.178 |
hsa-miR-197-3p | TNS1 | 11 cancers: BLCA; COAD; ESCA; HNSC; LGG; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.984; TCGA COAD -0.691; TCGA ESCA -0.965; TCGA HNSC -0.518; TCGA LGG -0.061; TCGA LUSC -0.514; TCGA PAAD -0.325; TCGA PRAD -0.755; TCGA SARC -0.753; TCGA STAD -0.927; TCGA UCEC -0.515 |
hsa-miR-197-3p | PRX | 9 cancers: BLCA; BRCA; ESCA; LIHC; LUSC; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.194; TCGA BRCA -0.362; TCGA ESCA -0.18; TCGA LIHC -0.198; TCGA LUSC -0.223; TCGA SARC -0.148; TCGA THCA -0.089; TCGA STAD -0.254; TCGA UCEC -0.354 |
hsa-miR-197-3p | SPOCK2 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUSC; THCA; STAD | mirMAP | TCGA BLCA -0.296; TCGA CESC -0.224; TCGA COAD -0.328; TCGA ESCA -0.324; TCGA HNSC -0.221; TCGA KIRP -0.253; TCGA LIHC -0.219; TCGA LUSC -0.7; TCGA THCA -0.752; TCGA STAD -0.177 |
hsa-miR-197-3p | DUSP3 | 11 cancers: BLCA; COAD; ESCA; HNSC; LIHC; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.195; TCGA COAD -0.111; TCGA ESCA -0.237; TCGA HNSC -0.107; TCGA LIHC -0.127; TCGA LUSC -0.162; TCGA PAAD -0.215; TCGA PRAD -0.22; TCGA SARC -0.33; TCGA STAD -0.313; TCGA UCEC -0.21 |
hsa-miR-197-3p | KIAA1614 | 9 cancers: BLCA; COAD; KIRP; LGG; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.403; TCGA COAD -0.274; TCGA KIRP -0.226; TCGA LGG -0.114; TCGA LUSC -0.197; TCGA PRAD -0.528; TCGA SARC -0.228; TCGA STAD -0.459; TCGA UCEC -0.23 |
hsa-miR-197-3p | IGF1R | 10 cancers: BLCA; BRCA; KIRC; KIRP; LGG; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.184; TCGA BRCA -0.347; TCGA KIRC -0.141; TCGA KIRP -0.158; TCGA LGG -0.092; TCGA PRAD -0.116; TCGA SARC -0.287; TCGA THCA -0.192; TCGA STAD -0.146; TCGA UCEC -0.135 |
hsa-miR-197-3p | ASXL3 | 10 cancers: BLCA; ESCA; KIRC; KIRP; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.519; TCGA ESCA -0.909; TCGA KIRC -0.673; TCGA KIRP -0.659; TCGA LUSC -0.463; TCGA PAAD -0.383; TCGA PRAD -0.842; TCGA SARC -0.377; TCGA STAD -0.723; TCGA UCEC -0.368 |
hsa-miR-197-3p | MR1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.23; TCGA BRCA -0.143; TCGA CESC -0.274; TCGA COAD -0.144; TCGA ESCA -0.228; TCGA HNSC -0.155; TCGA LIHC -0.158; TCGA LUAD -0.137; TCGA LUSC -0.279; TCGA PRAD -0.588; TCGA SARC -0.126; TCGA STAD -0.242; TCGA UCEC -0.189 |
hsa-miR-197-3p | PRICKLE2 | 9 cancers: BLCA; BRCA; COAD; ESCA; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.524; TCGA BRCA -0.26; TCGA COAD -0.494; TCGA ESCA -0.714; TCGA LUSC -0.522; TCGA PRAD -0.674; TCGA SARC -0.239; TCGA STAD -0.844; TCGA UCEC -0.458 |
hsa-miR-197-3p | COL8A2 | 9 cancers: BLCA; BRCA; CESC; COAD; LUSC; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.551; TCGA BRCA -0.204; TCGA CESC -0.262; TCGA COAD -0.532; TCGA LUSC -0.415; TCGA PRAD -0.375; TCGA SARC -0.298; TCGA THCA -0.651; TCGA STAD -0.549 |
hsa-miR-197-3p | CREB3L2 | 9 cancers: BLCA; ESCA; HNSC; KIRP; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.129; TCGA ESCA -0.228; TCGA HNSC -0.117; TCGA KIRP -0.152; TCGA LUSC -0.158; TCGA PRAD -0.07; TCGA SARC -0.085; TCGA STAD -0.196; TCGA UCEC -0.232 |
hsa-miR-197-3p | P2RY8 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUSC; STAD; UCEC | mirMAP | TCGA BLCA -0.289; TCGA BRCA -0.101; TCGA CESC -0.405; TCGA COAD -0.305; TCGA ESCA -0.448; TCGA HNSC -0.241; TCGA LIHC -0.443; TCGA LUSC -0.627; TCGA STAD -0.252; TCGA UCEC -0.266 |
hsa-miR-197-3p | SFMBT2 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.489; TCGA CESC -0.41; TCGA COAD -0.383; TCGA ESCA -0.695; TCGA HNSC -0.384; TCGA LIHC -0.21; TCGA LUSC -0.471; TCGA PRAD -0.503; TCGA STAD -0.249; TCGA UCEC -0.428 |
hsa-miR-197-3p | NEURL1B | 9 cancers: BLCA; CESC; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.289; TCGA CESC -0.252; TCGA LUSC -0.255; TCGA PAAD -0.311; TCGA PRAD -0.314; TCGA SARC -0.5; TCGA THCA -0.125; TCGA STAD -0.353; TCGA UCEC -0.195 |
hsa-miR-197-3p | NHLRC4 | 9 cancers: BLCA; BRCA; COAD; KIRC; LUSC; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.274; TCGA BRCA -0.708; TCGA COAD -0.282; TCGA KIRC -0.634; TCGA LUSC -0.621; TCGA PRAD -0.143; TCGA THCA -0.243; TCGA STAD -0.294; TCGA UCEC -0.309 |
hsa-miR-197-3p | PAFAH1B1 | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LIHC; LUSC; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.085; TCGA COAD -0.063; TCGA ESCA -0.108; TCGA HNSC -0.081; TCGA KIRP -0.172; TCGA LIHC -0.147; TCGA LUSC -0.102; TCGA PRAD -0.149; TCGA STAD -0.145; TCGA UCEC -0.093 |
hsa-miR-197-3p | XYLT1 | 10 cancers: BLCA; CESC; ESCA; HNSC; LGG; LIHC; LUSC; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.32; TCGA CESC -0.467; TCGA ESCA -0.386; TCGA HNSC -0.227; TCGA LGG -0.3; TCGA LIHC -0.268; TCGA LUSC -0.385; TCGA PRAD -0.268; TCGA STAD -0.167; TCGA UCEC -0.552 |
hsa-miR-197-3p | ANK2 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUSC; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.654; TCGA CESC -0.541; TCGA COAD -0.798; TCGA ESCA -1.273; TCGA HNSC -0.641; TCGA LUSC -0.491; TCGA PRAD -0.69; TCGA SARC -0.295; TCGA STAD -1.031; TCGA UCEC -0.396 |
hsa-miR-197-3p | IGFBP5 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.715; TCGA BRCA -0.131; TCGA CESC -0.308; TCGA COAD -0.559; TCGA ESCA -0.754; TCGA HNSC -0.506; TCGA KIRC -0.334; TCGA LIHC -0.325; TCGA PRAD -0.299; TCGA SARC -0.286; TCGA THCA -0.581; TCGA STAD -0.493; TCGA UCEC -0.391 |
hsa-miR-197-3p | NLGN1 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.471; TCGA CESC -0.391; TCGA COAD -0.882; TCGA ESCA -0.735; TCGA HNSC -0.467; TCGA KIRP -0.346; TCGA LGG -0.272; TCGA PAAD -0.787; TCGA PRAD -0.439; TCGA SARC -0.531; TCGA STAD -1.307; TCGA UCEC -0.359 |
hsa-miR-197-3p | KCNMA1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.817; TCGA BRCA -0.272; TCGA CESC -0.339; TCGA COAD -0.91; TCGA ESCA -1.226; TCGA HNSC -0.3; TCGA LUSC -0.557; TCGA PAAD -0.439; TCGA PRAD -0.591; TCGA SARC -0.829; TCGA STAD -1.37; TCGA UCEC -0.747 |
hsa-miR-197-3p | RCAN2 | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUSC; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.687; TCGA CESC -0.253; TCGA COAD -0.497; TCGA ESCA -0.927; TCGA HNSC -0.393; TCGA KIRC -0.252; TCGA LIHC -0.401; TCGA LUSC -0.587; TCGA PRAD -0.472; TCGA STAD -0.588; TCGA UCEC -0.408 |
hsa-miR-197-3p | NBEA | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.741; TCGA BRCA -0.436; TCGA CESC -0.503; TCGA COAD -0.863; TCGA ESCA -0.981; TCGA HNSC -0.632; TCGA KIRC -0.197; TCGA KIRP -0.29; TCGA LUAD -0.13; TCGA LUSC -0.188; TCGA PAAD -0.4; TCGA PRAD -0.296; TCGA SARC -0.445; TCGA STAD -1.056; TCGA UCEC -0.529 |
hsa-miR-197-3p | TCF4 | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUSC; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.403; TCGA CESC -0.191; TCGA COAD -0.461; TCGA ESCA -0.208; TCGA HNSC -0.168; TCGA LGG -0.15; TCGA LIHC -0.186; TCGA LUSC -0.277; TCGA PRAD -0.332; TCGA STAD -0.349; TCGA UCEC -0.307 |
hsa-miR-197-3p | CPEB2 | 10 cancers: BLCA; BRCA; ESCA; KIRP; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.292; TCGA BRCA -0.351; TCGA ESCA -0.382; TCGA KIRP -0.126; TCGA LUAD -0.138; TCGA LUSC -0.21; TCGA PRAD -0.188; TCGA SARC -0.3; TCGA STAD -0.238; TCGA UCEC -0.296 |
hsa-miR-197-3p | NCAM1 | 9 cancers: BLCA; COAD; ESCA; HNSC; LGG; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -1.176; TCGA COAD -0.591; TCGA ESCA -1.151; TCGA HNSC -0.365; TCGA LGG -0.126; TCGA PRAD -0.809; TCGA SARC -0.442; TCGA STAD -0.932; TCGA UCEC -0.642 |
hsa-miR-197-3p | GRIK2 | 10 cancers: BLCA; COAD; ESCA; KIRC; KIRP; LGG; LUAD; PAAD; PRAD; STAD | miRNATAP | TCGA BLCA -0.29; TCGA COAD -0.281; TCGA ESCA -0.464; TCGA KIRC -0.362; TCGA KIRP -0.85; TCGA LGG -0.443; TCGA LUAD -0.213; TCGA PAAD -0.427; TCGA PRAD -0.426; TCGA STAD -0.677 |
hsa-miR-197-3p | EBF1 | 9 cancers: BLCA; CESC; COAD; ESCA; LIHC; LUSC; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.544; TCGA CESC -0.316; TCGA COAD -0.366; TCGA ESCA -0.477; TCGA LIHC -0.293; TCGA LUSC -0.458; TCGA PRAD -0.253; TCGA STAD -0.492; TCGA UCEC -0.373 |
hsa-miR-197-3p | PLXNC1 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUSC; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.607; TCGA CESC -0.471; TCGA COAD -0.565; TCGA ESCA -0.455; TCGA HNSC -0.49; TCGA LUSC -0.718; TCGA PRAD -0.628; TCGA THCA -0.519; TCGA STAD -0.148; TCGA UCEC -0.322 |
hsa-miR-197-3p | ADCYAP1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUSC; PAAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.762; TCGA BRCA -0.216; TCGA CESC -0.547; TCGA COAD -1.099; TCGA ESCA -1.145; TCGA HNSC -0.358; TCGA KIRP -0.448; TCGA LIHC -0.304; TCGA LUSC -0.413; TCGA PAAD -0.912; TCGA SARC -0.572; TCGA STAD -1.1; TCGA UCEC -0.728 |
hsa-miR-197-3p | ADAMTS1 | 10 cancers: BLCA; COAD; ESCA; KIRP; LIHC; LUSC; OV; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.581; TCGA COAD -0.371; TCGA ESCA -0.654; TCGA KIRP -0.343; TCGA LIHC -0.217; TCGA LUSC -0.181; TCGA OV -0.204; TCGA SARC -0.359; TCGA STAD -0.493; TCGA UCEC -0.372 |
hsa-miR-197-3p | DCTN4 | 9 cancers: BLCA; BRCA; HNSC; LGG; LUAD; LUSC; OV; SARC; STAD | miRNATAP | TCGA BLCA -0.058; TCGA BRCA -0.101; TCGA HNSC -0.125; TCGA LGG -0.174; TCGA LUAD -0.092; TCGA LUSC -0.083; TCGA OV -0.085; TCGA SARC -0.094; TCGA STAD -0.111 |
hsa-miR-197-3p | KLHL14 | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LUSC; PRAD; STAD | miRNATAP | TCGA BLCA -0.473; TCGA BRCA -0.388; TCGA CESC -0.512; TCGA COAD -0.867; TCGA HNSC -0.347; TCGA KIRC -0.628; TCGA KIRP -0.74; TCGA LUSC -0.417; TCGA PRAD -0.796; TCGA STAD -0.764 |
hsa-miR-197-3p | SETBP1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUSC; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.519; TCGA BRCA -0.218; TCGA CESC -0.313; TCGA COAD -0.344; TCGA ESCA -0.571; TCGA HNSC -0.288; TCGA KIRC -0.159; TCGA KIRP -0.255; TCGA LGG -0.107; TCGA LUSC -0.193; TCGA PRAD -0.33; TCGA SARC -0.293; TCGA STAD -0.825; TCGA UCEC -0.38 |
hsa-miR-197-3p | TAOK3 | 9 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUSC; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.069; TCGA CESC -0.113; TCGA ESCA -0.13; TCGA HNSC -0.13; TCGA LGG -0.1; TCGA LUSC -0.186; TCGA PRAD -0.136; TCGA STAD -0.069; TCGA UCEC -0.232 |
hsa-miR-197-3p | NAV2 | 9 cancers: BLCA; HNSC; KIRC; KIRP; LGG; PRAD; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.176; TCGA HNSC -0.141; TCGA KIRC -0.254; TCGA KIRP -0.254; TCGA LGG -0.083; TCGA PRAD -0.74; TCGA SARC -0.449; TCGA THCA -0.322; TCGA STAD -0.305 |
hsa-miR-197-3p | PALM2-AKAP2 | 9 cancers: BLCA; COAD; ESCA; LIHC; LUSC; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.322; TCGA COAD -0.325; TCGA ESCA -0.372; TCGA LIHC -0.134; TCGA LUSC -0.496; TCGA PRAD -0.186; TCGA SARC -0.195; TCGA STAD -0.349; TCGA UCEC -0.501 |
hsa-miR-197-3p | NRP1 | 9 cancers: BLCA; COAD; ESCA; KIRP; LGG; LUAD; LUSC; STAD; UCEC | miRNATAP | TCGA BLCA -0.159; TCGA COAD -0.328; TCGA ESCA -0.264; TCGA KIRP -0.156; TCGA LGG -0.092; TCGA LUAD -0.125; TCGA LUSC -0.375; TCGA STAD -0.156; TCGA UCEC -0.346 |
hsa-miR-197-3p | AGR2 | 9 cancers: BRCA; HNSC; KIRC; KIRP; LUAD; PAAD; PRAD; THCA; UCEC | miRNAWalker2 validate | TCGA BRCA -1.686; TCGA HNSC -0.403; TCGA KIRC -0.915; TCGA KIRP -0.595; TCGA LUAD -0.32; TCGA PAAD -0.995; TCGA PRAD -0.347; TCGA THCA -1.045; TCGA UCEC -0.365 |
hsa-miR-197-3p | MEGF9 | 10 cancers: BRCA; HNSC; KIRC; KIRP; LUAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.237; TCGA HNSC -0.19; TCGA KIRC -0.193; TCGA KIRP -0.164; TCGA LUAD -0.118; TCGA PRAD -0.185; TCGA SARC -0.109; TCGA THCA -0.25; TCGA STAD -0.227; TCGA UCEC -0.115 |
hsa-miR-197-3p | ZNF181 | 9 cancers: BRCA; COAD; HNSC; KIRC; KIRP; LIHC; SARC; STAD; UCEC | MirTarget | TCGA BRCA -0.053; TCGA COAD -0.087; TCGA HNSC -0.107; TCGA KIRC -0.147; TCGA KIRP -0.234; TCGA LIHC -0.114; TCGA SARC -0.141; TCGA STAD -0.136; TCGA UCEC -0.109 |
hsa-miR-197-3p | SCAMP1 | 10 cancers: BRCA; KIRC; KIRP; LGG; LUAD; OV; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BRCA -0.113; TCGA KIRC -0.143; TCGA KIRP -0.109; TCGA LGG -0.1; TCGA LUAD -0.126; TCGA OV -0.09; TCGA PRAD -0.105; TCGA SARC -0.12; TCGA STAD -0.174; TCGA UCEC -0.118 |
hsa-miR-197-3p | AFF1 | 10 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.13; TCGA CESC -0.161; TCGA ESCA -0.308; TCGA HNSC -0.159; TCGA KIRC -0.103; TCGA KIRP -0.091; TCGA LUSC -0.101; TCGA SARC -0.252; TCGA STAD -0.183; TCGA UCEC -0.202 |
hsa-miR-197-3p | PTPRT | 9 cancers: BRCA; CESC; COAD; HNSC; KIRC; LIHC; LUSC; PAAD; STAD | mirMAP | TCGA BRCA -1.294; TCGA CESC -0.474; TCGA COAD -0.602; TCGA HNSC -0.443; TCGA KIRC -0.637; TCGA LIHC -0.385; TCGA LUSC -0.519; TCGA PAAD -0.728; TCGA STAD -0.593 |
hsa-miR-197-3p | RALGPS1 | 9 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUSC; OV; SARC | miRNATAP | TCGA BRCA -0.263; TCGA CESC -0.179; TCGA ESCA -0.332; TCGA HNSC -0.258; TCGA KIRC -0.633; TCGA KIRP -0.391; TCGA LUSC -0.159; TCGA OV -0.195; TCGA SARC -0.197 |
hsa-miR-197-3p | UBE3A | 9 cancers: CESC; HNSC; KIRP; LUAD; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA CESC -0.071; TCGA HNSC -0.056; TCGA KIRP -0.098; TCGA LUAD -0.054; TCGA PAAD -0.102; TCGA PRAD -0.051; TCGA SARC -0.098; TCGA STAD -0.07; TCGA UCEC -0.106 |
hsa-miR-197-3p | SGMS1 | 9 cancers: CESC; ESCA; HNSC; KIRC; KIRP; LUSC; PRAD; THCA; UCEC | miRNATAP | TCGA CESC -0.143; TCGA ESCA -0.16; TCGA HNSC -0.126; TCGA KIRC -0.154; TCGA KIRP -0.168; TCGA LUSC -0.144; TCGA PRAD -0.078; TCGA THCA -0.175; TCGA UCEC -0.157 |
hsa-miR-197-3p | AK3 | 10 cancers: ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA ESCA -0.211; TCGA HNSC -0.146; TCGA KIRC -0.282; TCGA KIRP -0.087; TCGA LUAD -0.068; TCGA LUSC -0.15; TCGA PRAD -0.119; TCGA SARC -0.122; TCGA STAD -0.107; TCGA UCEC -0.13 |
Enriched cancer pathways of putative targets