microRNA information: hsa-miR-22-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-22-3p | miRbase |
| Accession: | MIMAT0000077 | miRbase |
| Precursor name: | hsa-mir-22 | miRbase |
| Precursor accession: | MI0000078 | miRbase |
| Symbol: | MIR22 | HGNC |
| RefSeq ID: | NR_029494 | GenBank |
| Sequence: | AAGCUGCCAGUUGAAGAACUGU |
Reported expression in cancers: hsa-miR-22-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-22-3p | breast cancer | upregulation | "miR-22 is up-regulated in numerous metastatic canc ......" | 26512777 | |
| hsa-miR-22-3p | breast cancer | upregulation | "Aberrant Expression of Breast Development Related ......" | 27382390 | qPCR |
| hsa-miR-22-3p | colorectal cancer | downregulation | "Clinical significance of miR 22 expression in pati ......" | 22492279 | qPCR |
| hsa-miR-22-3p | gastric cancer | downregulation | "Reduced expression of miR 22 in gastric cancer is ......" | 23786758 | qPCR |
| hsa-miR-22-3p | gastric cancer | downregulation | "microRNA miR-22 has been reported to be downregula ......" | 25323629 | in situ hybridization |
| hsa-miR-22-3p | gastric cancer | downregulation | "MiR-22 was previously reported to act as tumor sup ......" | 26610210 | |
| hsa-miR-22-3p | kidney renal cell cancer | downregulation | "Accumulating evidence demonstrates that microRNA-2 ......" | 26499759 | qPCR |
| hsa-miR-22-3p | kidney renal cell cancer | downregulation | "MicroRNA miR-22 has previously been reported to be ......" | 27082730 | |
| hsa-miR-22-3p | liver cancer | downregulation | "microRNA 22 downregulated in hepatocellular carcin ......" | 20842113 | qPCR |
| hsa-miR-22-3p | liver cancer | downregulation | "MicroRNA 22 is down regulated in hepatitis B virus ......" | 23582783 | |
| hsa-miR-22-3p | lung cancer | upregulation | "Expression level of miR-22 in lung cancer specimen ......" | 22484852 | qPCR |
| hsa-miR-22-3p | lung squamous cell cancer | upregulation | "Circulating miR 22 miR 24 and miR 34a as novel pre ......" | 23794259 | |
| hsa-miR-22-3p | lymphoma | deregulation | "In addition we derived an 11-miRNA signature 4 upr ......" | 23801630 | |
| hsa-miR-22-3p | melanoma | deregulation | "Comparative microarray analysis of microRNA expres ......" | 23111773 | Microarray; RNA-Seq; qPCR |
| hsa-miR-22-3p | ovarian cancer | downregulation | "Down regulated miR 22 as predictive biomarkers for ......" | 25257702 | qPCR |
| hsa-miR-22-3p | retinoblastoma | upregulation | "Transfection of miR-22 was performed using Lipofec ......" | 22510010 |
Reported cancer pathway affected by hsa-miR-22-3p
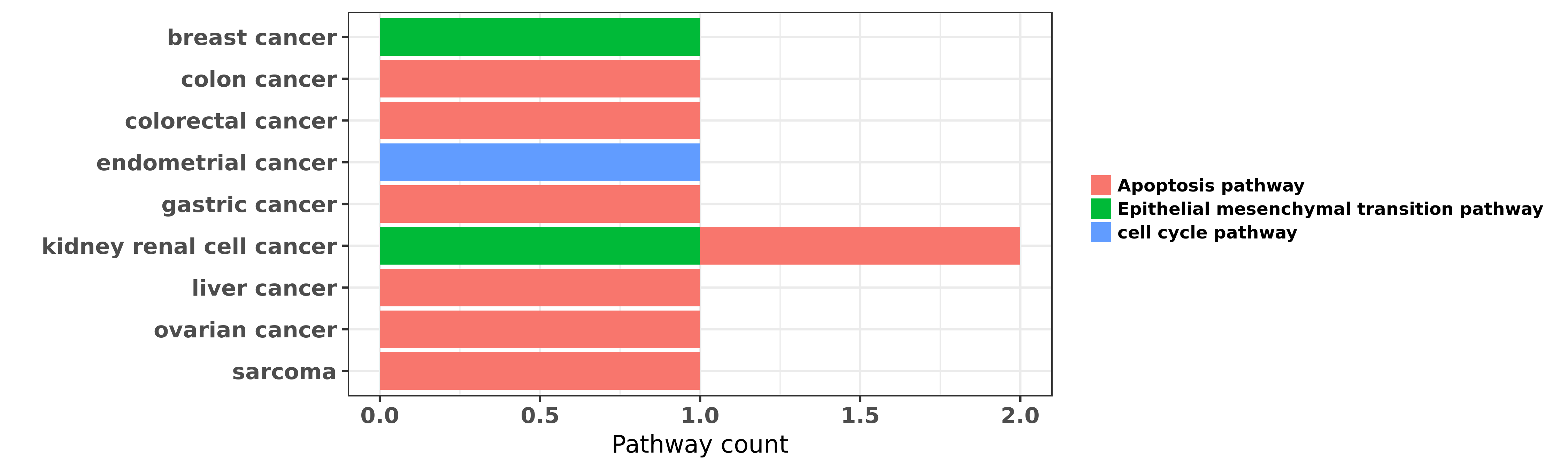
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-22-3p | breast cancer | Epithelial mesenchymal transition pathway | "Here we find miR-22 triggers epithelial-mesenchyma ......" | 23830207 | |
| hsa-miR-22-3p | colon cancer | Apoptosis pathway | "Overexpression of miR 22 reverses paclitaxel induc ......" | 21594648 | |
| hsa-miR-22-3p | colorectal cancer | Apoptosis pathway | "MiR 22 regulates 5 FU sensitivity by inhibiting au ......" | 25449431 | |
| hsa-miR-22-3p | endometrial cancer | cell cycle pathway | "miR 22 inhibits proliferation and invasion in estr ......" | 24715036 | |
| hsa-miR-22-3p | gastric cancer | Apoptosis pathway | "Diallyl disulfide suppresses proliferation and ind ......" | 23851184 | |
| hsa-miR-22-3p | gastric cancer | Apoptosis pathway | "EDD1 predicts prognosis and regulates gastric canc ......" | 27124677 | Colony formation |
| hsa-miR-22-3p | kidney renal cell cancer | Apoptosis pathway; Epithelial mesenchymal transition pathway | "MicroRNA 22 functions as a tumor suppressor by tar ......" | 26499759 | Luciferase |
| hsa-miR-22-3p | liver cancer | Apoptosis pathway | "microRNA 22 downregulation of galectin 9 influence ......" | 26239725 | Western blot; Luciferase |
| hsa-miR-22-3p | ovarian cancer | Apoptosis pathway | "We performed deep sequencing to comprehensively pr ......" | 20081105 | |
| hsa-miR-22-3p | ovarian cancer | Apoptosis pathway | "An inhibitory effect of miR 22 on cell migration a ......" | 20869762 | |
| hsa-miR-22-3p | sarcoma | Apoptosis pathway | "miR 22 inhibits tumor growth and metastasis by tar ......" | 27317765 | |
| hsa-miR-22-3p | sarcoma | Apoptosis pathway | "Deep Sequencing Reveals a Novel miR 22 Regulatory ......" | 27569217 |
Reported cancer prognosis affected by hsa-miR-22-3p

| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-22-3p | breast cancer | metastasis | "Control of EVI 1 oncogene expression in metastatic ......" | 21057539 | |
| hsa-miR-22-3p | breast cancer | metastasis | "Here we find miR-22 triggers epithelial-mesenchyma ......" | 23830207 | |
| hsa-miR-22-3p | breast cancer | metastasis; staging | "A regulatory loop involving miR 22 Sp1 and c Myc m ......" | 24906624 | |
| hsa-miR-22-3p | breast cancer | staging; metastasis; poor survival | "miR 22 as a prognostic factor targets glucose tran ......" | 25304371 | |
| hsa-miR-22-3p | breast cancer | progression; differentiation | "Systematic integration of molecular profiles ident ......" | 26477310 | |
| hsa-miR-22-3p | breast cancer | progression; metastasis; poor survival | "TIP60 miR 22 axis as a prognostic marker of breast ......" | 26512777 | |
| hsa-miR-22-3p | breast cancer | poor survival | "In this multicenter study a 10-miRNA classifier in ......" | 27566954 | |
| hsa-miR-22-3p | colon cancer | drug resistance; poor survival | "Overexpression of miR 22 reverses paclitaxel induc ......" | 21594648 | |
| hsa-miR-22-3p | colon cancer | cell migration | "MicroRNA 22 is induced by vitamin D and contribute ......" | 22328083 | |
| hsa-miR-22-3p | colorectal cancer | tumorigenesis; poor survival; metastasis | "Clinical significance of miR 22 expression in pati ......" | 22492279 | |
| hsa-miR-22-3p | endometrial cancer | progression | "miR 22 inhibits proliferation and invasion in estr ......" | 24715036 | |
| hsa-miR-22-3p | esophageal cancer | cell migration | "miR 22 is down regulated in esophageal squamous ce ......" | 25516722 | Wound Healing Assay |
| hsa-miR-22-3p | gastric cancer | cell migration; malignant trasformation | "miR 22 is down regulated in gastric cancer and its ......" | 23529765 | Luciferase |
| hsa-miR-22-3p | gastric cancer | worse prognosis; tumorigenesis; staging; metastasis; poor survival; malignant trasformation | "Reduced expression of miR 22 in gastric cancer is ......" | 23786758 | |
| hsa-miR-22-3p | gastric cancer | motility | "MiR 22 suppresses the proliferation and invasion o ......" | 24495805 | Colony formation |
| hsa-miR-22-3p | gastric cancer | metastasis; staging | "microRNA 22 acts as a metastasis suppressor by tar ......" | 25323629 | Luciferase |
| hsa-miR-22-3p | gastric cancer | metastasis; poor survival; cell migration | "MicroRNA 22 inhibits tumor growth and metastasis i ......" | 26610210 | |
| hsa-miR-22-3p | gastric cancer | worse prognosis | "EDD1 predicts prognosis and regulates gastric canc ......" | 27124677 | Colony formation |
| hsa-miR-22-3p | kidney renal cell cancer | staging | "This study aims to profile dysregulated microRNA m ......" | 25938468 | |
| hsa-miR-22-3p | kidney renal cell cancer | tumorigenesis; staging; metastasis | "MicroRNA 22 functions as a tumor suppressor by tar ......" | 26499759 | Luciferase |
| hsa-miR-22-3p | kidney renal cell cancer | cell migration | "MicroRNA 22 is downregulated in clear cell renal c ......" | 27082730 | Western blot; Luciferase |
| hsa-miR-22-3p | liver cancer | worse prognosis; poor survival; progression; tumorigenesis | "microRNA 22 downregulated in hepatocellular carcin ......" | 20842113 | |
| hsa-miR-22-3p | liver cancer | metastasis; differentiation; poor survival; worse prognosis | "MicroRNA 22 expression in hepatocellular carcinoma ......" | 23766411 | |
| hsa-miR-22-3p | lung cancer | progression | "Tumor suppressor miR 22 suppresses lung cancer cel ......" | 22484852 | |
| hsa-miR-22-3p | lung cancer | malignant trasformation | "Diagnostic Value of Circulating Extracellular miR ......" | 24851110 | |
| hsa-miR-22-3p | lung squamous cell cancer | staging; progression; drug resistance | "Circulating miR 22 miR 24 and miR 34a as novel pre ......" | 23794259 | |
| hsa-miR-22-3p | lung squamous cell cancer | progression | "We found that miR-34b and miR-520h might play impo ......" | 25702651 | |
| hsa-miR-22-3p | lymphoma | cell migration; metastasis | "In this study the precise role of miRNA-22 miR-22 ......" | 23440286 | Western blot |
| hsa-miR-22-3p | lymphoma | malignant trasformation | "Jak3 STAT3 and STAT5 inhibit expression of miR 22 ......" | 26244872 | |
| hsa-miR-22-3p | melanoma | metastasis; malignant trasformation | "Comparative microarray analysis of microRNA expres ......" | 23111773 | |
| hsa-miR-22-3p | ovarian cancer | cell migration; metastasis | "An inhibitory effect of miR 22 on cell migration a ......" | 20869762 | |
| hsa-miR-22-3p | ovarian cancer | metastasis | "miRNA microarray was applied to compare the miRNA ......" | 21122376 | |
| hsa-miR-22-3p | ovarian cancer | metastasis | "the differential expression of ezrin in two paired ......" | 21176563 | Western blot |
| hsa-miR-22-3p | ovarian cancer | worse prognosis; tumorigenesis; staging; metastasis; progression; poor survival | "Down regulated miR 22 as predictive biomarkers for ......" | 25257702 | |
| hsa-miR-22-3p | ovarian cancer | progression; cell migration | "Gene expression profiling analysis of the role of ......" | 27565323 | |
| hsa-miR-22-3p | pancreatic cancer | staging | "Blood-based circulating microRNAs were profiled by ......" | 24578785 | |
| hsa-miR-22-3p | pancreatic cancer | staging | "Plasma miR 22 3p miR 642b 3p and miR 885 5p as dia ......" | 27631726 | |
| hsa-miR-22-3p | prostate cancer | tumorigenesis; progression | "Previous network analysis and miR arrays suggested ......" | 26544868 | Luciferase |
| hsa-miR-22-3p | sarcoma | drug resistance | "miR 22 targets the 3' UTR of HMGB1 and inhibits th ......" | 24609901 | |
| hsa-miR-22-3p | sarcoma | worse prognosis; poor survival; metastasis; recurrence; drug resistance | "Downregulation of miR 22 acts as an unfavorable pr ......" | 25953260 | |
| hsa-miR-22-3p | sarcoma | metastasis | "miR 22 inhibits tumor growth and metastasis by tar ......" | 27317765 | |
| hsa-miR-22-3p | sarcoma | differentiation; drug resistance | "Deep Sequencing Reveals a Novel miR 22 Regulatory ......" | 27569217 |
Reported gene related to hsa-miR-22-3p
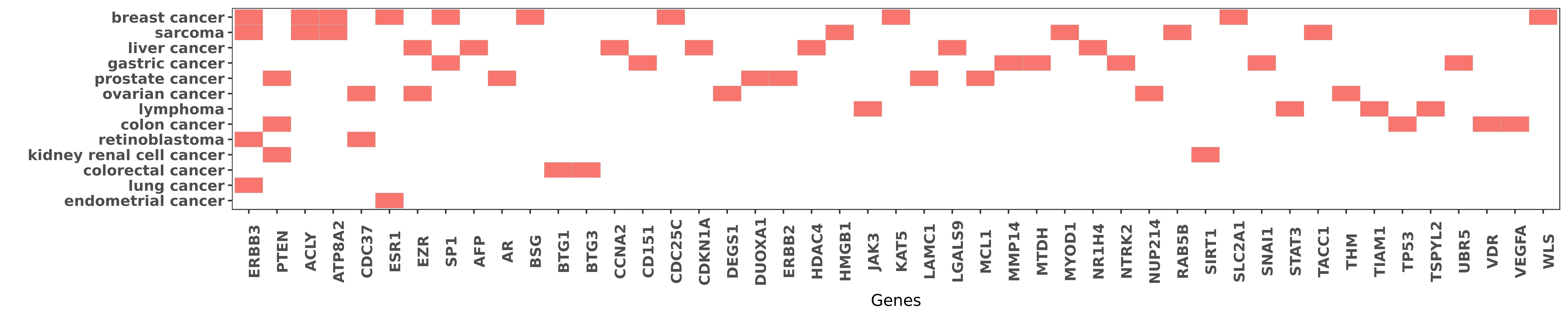
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-22-3p | breast cancer | ERBB3 | "These metastatic cells expressed higher levels of ......" | 21057539 |
| hsa-miR-22-3p | lung cancer | ERBB3 | "Tumor suppressor miR 22 suppresses lung cancer cel ......" | 22484852 |
| hsa-miR-22-3p | retinoblastoma | ERBB3 | "Transfected miR-22 Y79 cells inhibited the cell pr ......" | 22510010 |
| hsa-miR-22-3p | sarcoma | ERBB3 | "Finally we demonstrated the ability of miR-22 to i ......" | 27569217 |
| hsa-miR-22-3p | colon cancer | PTEN | "Overexpression of miR 22 reverses paclitaxel induc ......" | 21594648 |
| hsa-miR-22-3p | kidney renal cell cancer | PTEN | "MicroRNA 22 is downregulated in clear cell renal c ......" | 27082730 |
| hsa-miR-22-3p | prostate cancer | PTEN | "Relevant luciferase+3'-UTR expression studies conf ......" | 26544868 |
| hsa-miR-22-3p | breast cancer | ACLY | "Within this cluster the cancer-associated and card ......" | 26477310 |
| hsa-miR-22-3p | sarcoma | ACLY | "Herein four different types of tumor cells includi ......" | 27317765 |
| hsa-miR-22-3p | breast cancer | ATP8A2 | "Within this cluster the cancer-associated and card ......" | 26477310 |
| hsa-miR-22-3p | sarcoma | ATP8A2 | "miR 22 inhibits tumor growth and metastasis by tar ......" | 27317765 |
| hsa-miR-22-3p | ovarian cancer | CDC37 | "Furthermore overexpression of the putative tumor s ......" | 20081105 |
| hsa-miR-22-3p | retinoblastoma | CDC37 | "Transfected miR-22 Y79 cells inhibited the cell pr ......" | 22510010 |
| hsa-miR-22-3p | breast cancer | ESR1 | "An estrogen receptor alpha suppressor microRNA 22 ......" | 20180843 |
| hsa-miR-22-3p | endometrial cancer | ESR1 | "miR 22 inhibits proliferation and invasion in estr ......" | 24715036 |
| hsa-miR-22-3p | liver cancer | EZR | "MicroRNA 22 expression in hepatocellular carcinoma ......" | 23766411 |
| hsa-miR-22-3p | ovarian cancer | EZR | "2 By a functional screen using miRNA microarray co ......" | 21176563 |
| hsa-miR-22-3p | sarcoma | HMGB1 | "However possibly as a compensatory effect miR-22 w ......" | 24609901 |
| hsa-miR-22-3p | sarcoma | HMGB1 | "In this study we determined the targeting role of ......" | 24752578 |
| hsa-miR-22-3p | breast cancer | SP1 | "We also determined that miR-22 could indirectly pa ......" | 24906624 |
| hsa-miR-22-3p | gastric cancer | SP1 | "miR 22 is down regulated in gastric cancer and its ......" | 23529765 |
| hsa-miR-22-3p | liver cancer | AFP | "However using miRNA panel of miR-22 and miR-199a-3 ......" | 27271989 |
| hsa-miR-22-3p | prostate cancer | AR | "A direct AR regulation of miR-22 miR-29a and miR-1 ......" | 26052614 |
| hsa-miR-22-3p | breast cancer | BSG | "We discovered that miR-22 repressed CD147 expressi ......" | 24906624 |
| hsa-miR-22-3p | colorectal cancer | BTG1 | "B-cell translocation gene 1 BTG1 was identified as ......" | 25449431 |
| hsa-miR-22-3p | colorectal cancer | BTG3 | "B-cell translocation gene 1 BTG1 was identified as ......" | 25449431 |
| hsa-miR-22-3p | liver cancer | CCNA2 | "The sequence of miR-22 which is conserved in mice ......" | 25596928 |
| hsa-miR-22-3p | gastric cancer | CD151 | "MiR 22 suppresses the proliferation and invasion o ......" | 24495805 |
| hsa-miR-22-3p | breast cancer | CDC25C | "These metastatic cells expressed higher levels of ......" | 21057539 |
| hsa-miR-22-3p | liver cancer | CDKN1A | "Overexpression of miR-22 in vitro effectively supp ......" | 23582783 |
| hsa-miR-22-3p | ovarian cancer | DEGS1 | "Nine DEGs as target genes were identified to be re ......" | 27565323 |
| hsa-miR-22-3p | prostate cancer | DUOXA1 | "We previously detected several miRNAs for example ......" | 22815235 |
| hsa-miR-22-3p | prostate cancer | ERBB2 | "Relevant luciferase+3'-UTR expression studies conf ......" | 26544868 |
| hsa-miR-22-3p | liver cancer | HDAC4 | "Furthermore histone deacetylase 4 HDAC4 known to h ......" | 20842113 |
| hsa-miR-22-3p | lymphoma | JAK3 | "Jak3 STAT3 and STAT5 inhibit expression of miR 22 ......" | 26244872 |
| hsa-miR-22-3p | breast cancer | KAT5 | "TIP60 miR 22 axis as a prognostic marker of breast ......" | 26512777 |
| hsa-miR-22-3p | prostate cancer | LAMC1 | "The identification of laminin gamma 1 LAMC1 and my ......" | 26052614 |
| hsa-miR-22-3p | liver cancer | LGALS9 | "microRNA 22 downregulation of galectin 9 influence ......" | 26239725 |
| hsa-miR-22-3p | prostate cancer | MCL1 | "The identification of laminin gamma 1 LAMC1 and my ......" | 26052614 |
| hsa-miR-22-3p | gastric cancer | MMP14 | "MicroRNA 22 inhibits tumor growth and metastasis i ......" | 26610210 |
| hsa-miR-22-3p | gastric cancer | MTDH | "microRNA 22 acts as a metastasis suppressor by tar ......" | 25323629 |
| hsa-miR-22-3p | sarcoma | MYOD1 | "miR-22 was physiologically induced during normal m ......" | 27569217 |
| hsa-miR-22-3p | liver cancer | NR1H4 | "Among the studied primary and secondary bile acids ......" | 25596928 |
| hsa-miR-22-3p | gastric cancer | NTRK2 | "NTRK2 is an oncogene and associated with microRNA ......" | 27662840 |
| hsa-miR-22-3p | ovarian cancer | NUP214 | "In addition to these specific effects reversing th ......" | 20081105 |
| hsa-miR-22-3p | sarcoma | RAB5B | "Moreover restoring miR-22 expression blocked tumor ......" | 27569217 |
| hsa-miR-22-3p | kidney renal cell cancer | SIRT1 | "MicroRNA 22 functions as a tumor suppressor by tar ......" | 26499759 |
| hsa-miR-22-3p | breast cancer | SLC2A1 | "In this study we found that GLUT1 is a direct targ ......" | 25304371 |
| hsa-miR-22-3p | gastric cancer | SNAI1 | "MicroRNA 22 inhibits tumor growth and metastasis i ......" | 26610210 |
| hsa-miR-22-3p | lymphoma | STAT3 | "Jak3 STAT3 and STAT5 inhibit expression of miR 22 ......" | 26244872 |
| hsa-miR-22-3p | sarcoma | TACC1 | "Moreover restoring miR-22 expression blocked tumor ......" | 27569217 |
| hsa-miR-22-3p | ovarian cancer | THM | "Furthermore overexpression of the putative tumor s ......" | 20081105 |
| hsa-miR-22-3p | lymphoma | TIAM1 | "Computational in silico analysis predicted that T- ......" | 23440286 |
| hsa-miR-22-3p | colon cancer | TP53 | "Overexpression of miR 22 reverses paclitaxel induc ......" | 21594648 |
| hsa-miR-22-3p | lymphoma | TSPYL2 | "In conclusion we provide the first evidence that d ......" | 26244872 |
| hsa-miR-22-3p | gastric cancer | UBR5 | "EDD1 predicts prognosis and regulates gastric canc ......" | 27124677 |
| hsa-miR-22-3p | colon cancer | VDR | "In 39 out of 50 78% human colon cancer patients mi ......" | 22328083 |
| hsa-miR-22-3p | colon cancer | VEGFA | "Over-expression of miR-22 inhibits HIF-1α express ......" | 21629773 |
| hsa-miR-22-3p | breast cancer | WLS | "Control of EVI 1 oncogene expression in metastatic ......" | 21057539 |
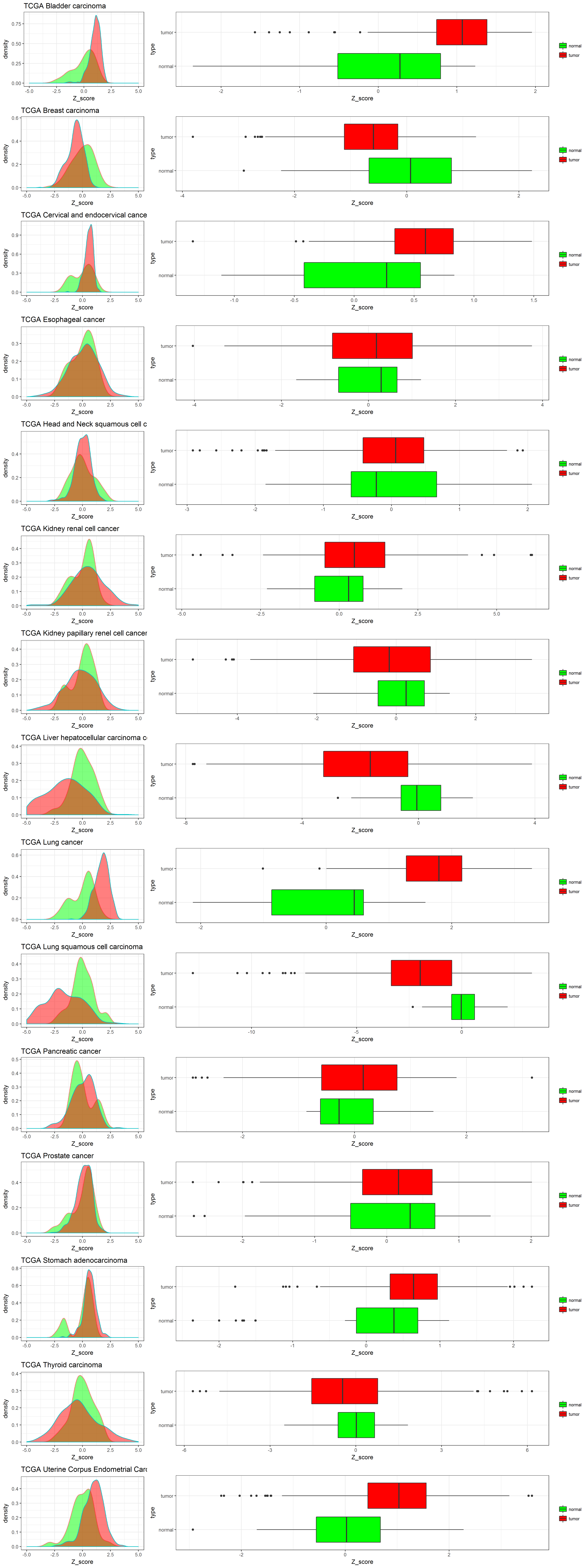
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-22-3p | ALMS1 | 14 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.21; TCGA BRCA -0.104; TCGA CESC -0.172; TCGA COAD -0.118; TCGA HNSC -0.133; TCGA KIRC -0.141; TCGA KIRP -0.138; TCGA LGG -0.142; TCGA LIHC -0.231; TCGA LUAD -0.144; TCGA LUSC -0.342; TCGA SARC -0.159; TCGA THCA -0.258; TCGA UCEC -0.161 |
hsa-miR-22-3p | BTF3 | 10 cancers: BLCA; BRCA; HNSC; LGG; LIHC; LUAD; OV; PRAD; SARC; UCEC | miRNAWalker2 validate | TCGA BLCA -0.098; TCGA BRCA -0.203; TCGA HNSC -0.133; TCGA LGG -0.247; TCGA LIHC -0.107; TCGA LUAD -0.142; TCGA OV -0.221; TCGA PRAD -0.3; TCGA SARC -0.064; TCGA UCEC -0.101 |
hsa-miR-22-3p | CCNT2 | 13 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LGG; LIHC; LUAD; LUSC; OV; PRAD; SARC; THCA | miRNAWalker2 validate | TCGA BLCA -0.157; TCGA BRCA -0.096; TCGA COAD -0.203; TCGA ESCA -0.203; TCGA KIRP -0.286; TCGA LGG -0.112; TCGA LIHC -0.128; TCGA LUAD -0.075; TCGA LUSC -0.245; TCGA OV -0.094; TCGA PRAD -0.103; TCGA SARC -0.204; TCGA THCA -0.116 |
hsa-miR-22-3p | H3F3B | 12 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; THCA; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.096; TCGA BRCA -0.132; TCGA HNSC -0.09; TCGA KIRC -0.383; TCGA KIRP -0.291; TCGA LGG -0.23; TCGA LIHC -0.17; TCGA LUAD -0.126; TCGA LUSC -0.176; TCGA PRAD -0.181; TCGA THCA -0.219; TCGA UCEC -0.159 |
hsa-miR-22-3p | HDAC4 | 9 cancers: BLCA; COAD; KIRC; LGG; LIHC; OV; SARC; STAD; UCEC | miRNAWalker2 validate; miRTarBase; miRNATAP | TCGA BLCA -0.442; TCGA COAD -0.199; TCGA KIRC -0.127; TCGA LGG -0.364; TCGA LIHC -0.39; TCGA OV -0.124; TCGA SARC -0.225; TCGA STAD -0.344; TCGA UCEC -0.292 |
hsa-miR-22-3p | HMGB1 | 14 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; OV; PRAD; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.087; TCGA CESC -0.122; TCGA COAD -0.242; TCGA ESCA -0.203; TCGA HNSC -0.115; TCGA KIRC -0.128; TCGA KIRP -0.219; TCGA LGG -0.15; TCGA LUAD -0.102; TCGA LUSC -0.199; TCGA OV -0.113; TCGA PRAD -0.173; TCGA THCA -0.232; TCGA UCEC -0.103 |
hsa-miR-22-3p | LEMD3 | 11 cancers: BLCA; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; THCA | miRNAWalker2 validate | TCGA BLCA -0.101; TCGA COAD -0.195; TCGA HNSC -0.09; TCGA KIRC -0.116; TCGA KIRP -0.068; TCGA LGG -0.055; TCGA LIHC -0.108; TCGA LUAD -0.111; TCGA LUSC -0.151; TCGA OV -0.105; TCGA THCA -0.081 |
hsa-miR-22-3p | NUP214 | 9 cancers: BLCA; BRCA; COAD; KIRC; KIRP; LGG; LIHC; PRAD; THCA | miRNAWalker2 validate | TCGA BLCA -0.079; TCGA BRCA -0.056; TCGA COAD -0.101; TCGA KIRC -0.224; TCGA KIRP -0.16; TCGA LGG -0.117; TCGA LIHC -0.117; TCGA PRAD -0.102; TCGA THCA -0.11 |
hsa-miR-22-3p | PEX5 | 10 cancers: BLCA; BRCA; CESC; COAD; LGG; LUAD; LUSC; OV; PAAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.079; TCGA BRCA -0.064; TCGA CESC -0.147; TCGA COAD -0.165; TCGA LGG -0.155; TCGA LUAD -0.09; TCGA LUSC -0.164; TCGA OV -0.143; TCGA PAAD -0.158; TCGA UCEC -0.117 |
hsa-miR-22-3p | RPS4X | 14 cancers: BLCA; BRCA; CESC; COAD; HNSC; LGG; LIHC; LUAD; LUSC; OV; PAAD; PRAD; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.102; TCGA BRCA -0.174; TCGA CESC -0.137; TCGA COAD -0.251; TCGA HNSC -0.203; TCGA LGG -0.325; TCGA LIHC -0.252; TCGA LUAD -0.146; TCGA LUSC -0.136; TCGA OV -0.265; TCGA PAAD -0.234; TCGA PRAD -0.263; TCGA THCA -0.459; TCGA UCEC -0.16 |
hsa-miR-22-3p | SP1 | 9 cancers: BLCA; KIRC; KIRP; LGG; LIHC; LUSC; PRAD; SARC; THCA | miRNAWalker2 validate | TCGA BLCA -0.093; TCGA KIRC -0.107; TCGA KIRP -0.292; TCGA LGG -0.063; TCGA LIHC -0.168; TCGA LUSC -0.107; TCGA PRAD -0.087; TCGA SARC -0.062; TCGA THCA -0.154 |
hsa-miR-22-3p | TCEAL1 | 10 cancers: BLCA; BRCA; CESC; COAD; LGG; LUAD; OV; SARC; STAD; UCEC | miRNAWalker2 validate; miRTarBase | TCGA BLCA -0.245; TCGA BRCA -0.198; TCGA CESC -0.185; TCGA COAD -0.407; TCGA LGG -0.06; TCGA LUAD -0.131; TCGA OV -0.14; TCGA SARC -0.633; TCGA STAD -0.48; TCGA UCEC -0.245 |
hsa-miR-22-3p | ATP9A | 10 cancers: BLCA; CESC; COAD; ESCA; KIRP; LIHC; LUAD; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.263; TCGA CESC -0.247; TCGA COAD -0.559; TCGA ESCA -0.477; TCGA KIRP -0.167; TCGA LIHC -0.316; TCGA LUAD -0.233; TCGA SARC -0.528; TCGA THCA -0.122; TCGA STAD -0.19 |
hsa-miR-22-3p | BCL9 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LGG; LIHC; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.169; TCGA BRCA -0.284; TCGA CESC -0.288; TCGA COAD -0.258; TCGA ESCA -0.377; TCGA KIRC -0.287; TCGA KIRP -0.179; TCGA LGG -0.08; TCGA LIHC -0.608; TCGA LUSC -0.435; TCGA OV -0.132; TCGA PRAD -0.137; TCGA SARC -0.333; TCGA THCA -0.595; TCGA STAD -0.176; TCGA UCEC -0.152 |
hsa-miR-22-3p | WDR33 | 17 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PAAD; PRAD; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.068; TCGA BRCA -0.056; TCGA CESC -0.059; TCGA COAD -0.105; TCGA HNSC -0.056; TCGA KIRC -0.077; TCGA KIRP -0.116; TCGA LGG -0.125; TCGA LIHC -0.085; TCGA LUAD -0.07; TCGA LUSC -0.177; TCGA OV -0.068; TCGA PAAD -0.076; TCGA PRAD -0.076; TCGA SARC -0.054; TCGA THCA -0.114; TCGA STAD -0.064 |
hsa-miR-22-3p | MAT2A | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; LIHC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.197; TCGA BRCA -0.153; TCGA CESC -0.133; TCGA COAD -0.126; TCGA ESCA -0.303; TCGA LIHC -0.15; TCGA LUAD -0.106; TCGA LUSC -0.207; TCGA PAAD -0.36; TCGA SARC -0.197; TCGA STAD -0.097; TCGA UCEC -0.092 |
hsa-miR-22-3p | DPF2 | 16 cancers: BLCA; BRCA; CESC; COAD; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.118; TCGA BRCA -0.127; TCGA CESC -0.087; TCGA COAD -0.183; TCGA KIRC -0.137; TCGA KIRP -0.114; TCGA LGG -0.216; TCGA LIHC -0.289; TCGA LUAD -0.071; TCGA LUSC -0.207; TCGA OV -0.106; TCGA PRAD -0.087; TCGA SARC -0.171; TCGA THCA -0.203; TCGA STAD -0.147; TCGA UCEC -0.092 |
hsa-miR-22-3p | TLK2 | 10 cancers: BLCA; BRCA; COAD; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; UCEC | MirTarget | TCGA BLCA -0.059; TCGA BRCA -0.132; TCGA COAD -0.115; TCGA KIRP -0.119; TCGA LGG -0.135; TCGA LIHC -0.234; TCGA LUAD -0.122; TCGA LUSC -0.238; TCGA PRAD -0.06; TCGA UCEC -0.071 |
hsa-miR-22-3p | MSL2 | 14 cancers: BLCA; BRCA; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; SARC; THCA; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.116; TCGA BRCA -0.092; TCGA COAD -0.213; TCGA HNSC -0.148; TCGA KIRC -0.09; TCGA KIRP -0.123; TCGA LGG -0.175; TCGA LIHC -0.084; TCGA LUAD -0.156; TCGA LUSC -0.293; TCGA PRAD -0.151; TCGA SARC -0.118; TCGA THCA -0.131; TCGA UCEC -0.127 |
hsa-miR-22-3p | RBMXL1 | 11 cancers: BLCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PRAD; SARC; THCA; UCEC | MirTarget | TCGA BLCA -0.207; TCGA HNSC -0.19; TCGA KIRC -0.31; TCGA KIRP -0.076; TCGA LUAD -0.148; TCGA LUSC -0.154; TCGA OV -0.141; TCGA PRAD -0.102; TCGA SARC -0.213; TCGA THCA -0.127; TCGA UCEC -0.107 |
hsa-miR-22-3p | CDCA7L | 14 cancers: BLCA; CESC; COAD; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.6; TCGA CESC -0.293; TCGA COAD -0.325; TCGA HNSC -0.244; TCGA KIRC -0.445; TCGA KIRP -0.806; TCGA LIHC -0.658; TCGA LUAD -0.228; TCGA LUSC -0.467; TCGA OV -0.185; TCGA PRAD -0.306; TCGA SARC -0.298; TCGA STAD -0.391; TCGA UCEC -0.229 |
hsa-miR-22-3p | IL17RD | 15 cancers: BLCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.501; TCGA CESC -0.597; TCGA COAD -0.389; TCGA HNSC -0.331; TCGA KIRC -0.514; TCGA KIRP -0.306; TCGA LGG -0.172; TCGA LIHC -0.301; TCGA LUAD -0.189; TCGA LUSC -0.587; TCGA OV -0.3; TCGA SARC -0.468; TCGA THCA -1.443; TCGA STAD -0.439; TCGA UCEC -0.343 |
hsa-miR-22-3p | TMSB15B | 9 cancers: BLCA; CESC; KIRP; LGG; LUSC; OV; SARC; THCA; UCEC | MirTarget | TCGA BLCA -0.244; TCGA CESC -0.509; TCGA KIRP -0.241; TCGA LGG -0.358; TCGA LUSC -0.874; TCGA OV -0.355; TCGA SARC -0.343; TCGA THCA -0.659; TCGA UCEC -0.439 |
hsa-miR-22-3p | DGKE | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUSC; SARC; STAD | MirTarget | TCGA BLCA -0.17; TCGA BRCA -0.269; TCGA CESC -0.278; TCGA ESCA -0.444; TCGA HNSC -0.429; TCGA KIRC -0.391; TCGA LUSC -0.229; TCGA SARC -0.097; TCGA STAD -0.271 |
hsa-miR-22-3p | PHACTR4 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; SARC | MirTarget | TCGA BLCA -0.126; TCGA BRCA -0.136; TCGA CESC -0.135; TCGA ESCA -0.2; TCGA HNSC -0.089; TCGA KIRC -0.16; TCGA KIRP -0.118; TCGA LUAD -0.104; TCGA LUSC -0.172; TCGA PRAD -0.069; TCGA SARC -0.091 |
hsa-miR-22-3p | CREB1 | 11 cancers: BLCA; COAD; KIRC; KIRP; LGG; LIHC; LUAD; OV; PRAD; SARC; UCEC | MirTarget | TCGA BLCA -0.186; TCGA COAD -0.091; TCGA KIRC -0.166; TCGA KIRP -0.158; TCGA LGG -0.072; TCGA LIHC -0.207; TCGA LUAD -0.137; TCGA OV -0.081; TCGA PRAD -0.112; TCGA SARC -0.078; TCGA UCEC -0.09 |
hsa-miR-22-3p | CENPV | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LUSC; OV; PAAD; PRAD; SARC; UCEC | MirTarget | TCGA BLCA -0.313; TCGA CESC -0.777; TCGA ESCA -1.029; TCGA HNSC -0.469; TCGA KIRC -1.046; TCGA LUSC -0.588; TCGA OV -0.263; TCGA PAAD -0.569; TCGA PRAD -0.396; TCGA SARC -0.282; TCGA UCEC -0.487 |
hsa-miR-22-3p | DNAJC27 | 11 cancers: BLCA; BRCA; CESC; COAD; KIRC; LUAD; LUSC; OV; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.292; TCGA BRCA -0.073; TCGA CESC -0.113; TCGA COAD -0.202; TCGA KIRC -0.269; TCGA LUAD -0.188; TCGA LUSC -0.139; TCGA OV -0.112; TCGA SARC -0.109; TCGA STAD -0.253; TCGA UCEC -0.189 |
hsa-miR-22-3p | PIP4K2B | 13 cancers: BLCA; CESC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.174; TCGA CESC -0.152; TCGA KIRC -0.134; TCGA KIRP -0.169; TCGA LGG -0.095; TCGA LIHC -0.28; TCGA LUAD -0.114; TCGA LUSC -0.172; TCGA OV -0.096; TCGA SARC -0.109; TCGA THCA -0.179; TCGA STAD -0.139; TCGA UCEC -0.2 |
hsa-miR-22-3p | ANGEL2 | 13 cancers: BLCA; BRCA; COAD; HNSC; KIRP; LGG; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.114; TCGA BRCA -0.185; TCGA COAD -0.183; TCGA HNSC -0.089; TCGA KIRP -0.204; TCGA LGG -0.116; TCGA LUAD -0.065; TCGA LUSC -0.188; TCGA OV -0.078; TCGA PRAD -0.096; TCGA SARC -0.219; TCGA THCA -0.064; TCGA STAD -0.1 |
hsa-miR-22-3p | H3F3C | 9 cancers: BLCA; BRCA; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; THCA | MirTarget | TCGA BLCA -0.122; TCGA BRCA -0.139; TCGA KIRC -0.438; TCGA KIRP -0.169; TCGA LGG -0.157; TCGA LIHC -0.113; TCGA LUAD -0.101; TCGA LUSC -0.289; TCGA THCA -0.281 |
hsa-miR-22-3p | ZMYM4 | 10 cancers: BLCA; CESC; COAD; ESCA; KIRC; KIRP; LIHC; LUSC; PRAD; STAD | MirTarget | TCGA BLCA -0.161; TCGA CESC -0.106; TCGA COAD -0.104; TCGA ESCA -0.135; TCGA KIRC -0.118; TCGA KIRP -0.175; TCGA LIHC -0.192; TCGA LUSC -0.196; TCGA PRAD -0.123; TCGA STAD -0.101 |
hsa-miR-22-3p | PRPF38A | 17 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.153; TCGA BRCA -0.137; TCGA CESC -0.166; TCGA COAD -0.113; TCGA ESCA -0.273; TCGA HNSC -0.243; TCGA KIRC -0.386; TCGA KIRP -0.325; TCGA LGG -0.127; TCGA LIHC -0.318; TCGA LUAD -0.126; TCGA LUSC -0.299; TCGA OV -0.097; TCGA PRAD -0.177; TCGA SARC -0.09; TCGA THCA -0.213; TCGA STAD -0.123 |
hsa-miR-22-3p | SCYL3 | 9 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LUSC; OV; PRAD; THCA | MirTarget | TCGA BLCA -0.102; TCGA BRCA -0.231; TCGA HNSC -0.152; TCGA KIRC -0.115; TCGA KIRP -0.24; TCGA LUSC -0.299; TCGA OV -0.107; TCGA PRAD -0.16; TCGA THCA -0.071 |
hsa-miR-22-3p | FAM53C | 12 cancers: BLCA; CESC; COAD; HNSC; LIHC; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.152; TCGA CESC -0.097; TCGA COAD -0.086; TCGA HNSC -0.07; TCGA LIHC -0.14; TCGA LUAD -0.134; TCGA LUSC -0.191; TCGA OV -0.052; TCGA PRAD -0.098; TCGA SARC -0.261; TCGA THCA -0.106; TCGA STAD -0.113 |
hsa-miR-22-3p | BRSK2 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LGG; LIHC; LUSC; OV; PAAD; SARC; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.544; TCGA BRCA -0.498; TCGA COAD -0.985; TCGA ESCA -1.37; TCGA HNSC -0.579; TCGA KIRC -0.474; TCGA LGG -0.322; TCGA LIHC -1.065; TCGA LUSC -0.729; TCGA OV -0.304; TCGA PAAD -1.245; TCGA SARC -0.843; TCGA UCEC -0.34 |
hsa-miR-22-3p | SIRT1 | 12 cancers: BLCA; CESC; COAD; ESCA; KIRC; LGG; LUAD; OV; PRAD; SARC; THCA; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.291; TCGA CESC -0.207; TCGA COAD -0.145; TCGA ESCA -0.339; TCGA KIRC -0.233; TCGA LGG -0.248; TCGA LUAD -0.259; TCGA OV -0.095; TCGA PRAD -0.121; TCGA SARC -0.096; TCGA THCA -0.198; TCGA UCEC -0.163 |
hsa-miR-22-3p | PBX1 | 15 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; OV; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.499; TCGA BRCA -0.313; TCGA CESC -0.347; TCGA COAD -0.603; TCGA HNSC -0.3; TCGA KIRC -0.453; TCGA KIRP -0.219; TCGA LGG -0.106; TCGA LUAD -0.209; TCGA LUSC -0.338; TCGA OV -0.194; TCGA SARC -0.679; TCGA THCA -0.346; TCGA STAD -0.624; TCGA UCEC -0.402 |
hsa-miR-22-3p | AGBL5 | 12 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LGG; LIHC; LUSC; OV; PAAD; SARC | MirTarget; miRNATAP | TCGA BLCA -0.133; TCGA BRCA -0.203; TCGA CESC -0.349; TCGA COAD -0.313; TCGA HNSC -0.161; TCGA KIRC -0.119; TCGA LGG -0.11; TCGA LIHC -0.326; TCGA LUSC -0.361; TCGA OV -0.161; TCGA PAAD -0.245; TCGA SARC -0.304 |
hsa-miR-22-3p | RSBN1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; OV; PRAD; SARC; THCA; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.225; TCGA BRCA -0.086; TCGA CESC -0.12; TCGA COAD -0.128; TCGA ESCA -0.19; TCGA HNSC -0.131; TCGA KIRC -0.332; TCGA KIRP -0.155; TCGA LUAD -0.251; TCGA OV -0.167; TCGA PRAD -0.109; TCGA SARC -0.1; TCGA THCA -0.196; TCGA UCEC -0.159 |
hsa-miR-22-3p | USP37 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; SARC; THCA | MirTarget; miRNATAP | TCGA BLCA -0.211; TCGA BRCA -0.173; TCGA CESC -0.145; TCGA COAD -0.314; TCGA ESCA -0.405; TCGA HNSC -0.252; TCGA KIRC -0.284; TCGA KIRP -0.116; TCGA LGG -0.116; TCGA LIHC -0.426; TCGA LUAD -0.157; TCGA LUSC -0.367; TCGA OV -0.055; TCGA SARC -0.253; TCGA THCA -0.241 |
hsa-miR-22-3p | PHC1 | 16 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PRAD; SARC; THCA; UCEC | MirTarget | TCGA BLCA -0.22; TCGA BRCA -0.233; TCGA CESC -0.494; TCGA COAD -0.213; TCGA HNSC -0.147; TCGA KIRC -0.439; TCGA KIRP -0.37; TCGA LGG -0.338; TCGA LIHC -0.186; TCGA LUAD -0.144; TCGA LUSC -0.317; TCGA OV -0.17; TCGA PRAD -0.085; TCGA SARC -0.286; TCGA THCA -0.08; TCGA UCEC -0.334 |
hsa-miR-22-3p | ZBTB39 | 15 cancers: BLCA; BRCA; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PRAD; SARC; THCA; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.209; TCGA BRCA -0.092; TCGA COAD -0.272; TCGA HNSC -0.132; TCGA KIRC -0.206; TCGA KIRP -0.088; TCGA LGG -0.181; TCGA LIHC -0.183; TCGA LUAD -0.15; TCGA LUSC -0.469; TCGA OV -0.137; TCGA PRAD -0.13; TCGA SARC -0.111; TCGA THCA -0.224; TCGA UCEC -0.238 |
hsa-miR-22-3p | PHF8 | 11 cancers: BLCA; BRCA; COAD; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; SARC; THCA | MirTarget; miRNATAP | TCGA BLCA -0.128; TCGA BRCA -0.21; TCGA COAD -0.218; TCGA HNSC -0.223; TCGA KIRC -0.214; TCGA KIRP -0.191; TCGA LGG -0.114; TCGA LUAD -0.154; TCGA LUSC -0.347; TCGA SARC -0.116; TCGA THCA -0.237 |
hsa-miR-22-3p | WDR82 | 10 cancers: BLCA; COAD; ESCA; KIRP; LIHC; LUAD; OV; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.146; TCGA COAD -0.099; TCGA ESCA -0.184; TCGA KIRP -0.077; TCGA LIHC -0.109; TCGA LUAD -0.113; TCGA OV -0.092; TCGA SARC -0.125; TCGA THCA -0.162; TCGA STAD -0.078 |
hsa-miR-22-3p | HUNK | 11 cancers: BLCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; SARC; UCEC | MirTarget | TCGA BLCA -0.833; TCGA CESC -0.625; TCGA COAD -0.773; TCGA HNSC -0.422; TCGA KIRC -1.009; TCGA KIRP -0.459; TCGA LGG -0.215; TCGA LIHC -0.971; TCGA LUAD -0.384; TCGA SARC -0.468; TCGA UCEC -0.237 |
hsa-miR-22-3p | WASF1 | 10 cancers: BLCA; KIRP; LGG; LIHC; LUSC; OV; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.216; TCGA KIRP -0.229; TCGA LGG -0.165; TCGA LIHC -0.49; TCGA LUSC -0.564; TCGA OV -0.223; TCGA SARC -0.239; TCGA THCA -0.508; TCGA STAD -0.361; TCGA UCEC -0.415 |
hsa-miR-22-3p | KDM3A | 11 cancers: BLCA; BRCA; COAD; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; THCA | MirTarget; miRNATAP | TCGA BLCA -0.144; TCGA BRCA -0.077; TCGA COAD -0.141; TCGA KIRC -0.161; TCGA KIRP -0.227; TCGA LGG -0.141; TCGA LIHC -0.295; TCGA LUAD -0.081; TCGA LUSC -0.39; TCGA SARC -0.221; TCGA THCA -0.195 |
hsa-miR-22-3p | UNC119B | 10 cancers: BLCA; HNSC; KIRC; LIHC; LUAD; OV; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.23; TCGA HNSC -0.23; TCGA KIRC -0.195; TCGA LIHC -0.517; TCGA LUAD -0.199; TCGA OV -0.232; TCGA PRAD -0.105; TCGA SARC -0.13; TCGA STAD -0.178; TCGA UCEC -0.223 |
hsa-miR-22-3p | SYT13 | 9 cancers: BLCA; BRCA; ESCA; LIHC; LUSC; OV; PRAD; SARC; UCEC | MirTarget | TCGA BLCA -0.559; TCGA BRCA -0.96; TCGA ESCA -1.474; TCGA LIHC -0.503; TCGA LUSC -0.977; TCGA OV -0.339; TCGA PRAD -0.589; TCGA SARC -0.469; TCGA UCEC -0.544 |
hsa-miR-22-3p | UNK | 17 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUSC; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.121; TCGA BRCA -0.25; TCGA CESC -0.225; TCGA COAD -0.146; TCGA HNSC -0.138; TCGA KIRC -0.125; TCGA KIRP -0.257; TCGA LGG -0.283; TCGA LIHC -0.414; TCGA LUSC -0.299; TCGA OV -0.061; TCGA PAAD -0.284; TCGA PRAD -0.061; TCGA SARC -0.081; TCGA THCA -0.06; TCGA STAD -0.166; TCGA UCEC -0.179 |
hsa-miR-22-3p | MDC1 | 10 cancers: BLCA; BRCA; COAD; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; SARC | MirTarget | TCGA BLCA -0.146; TCGA BRCA -0.11; TCGA COAD -0.151; TCGA KIRC -0.346; TCGA KIRP -0.304; TCGA LGG -0.182; TCGA LIHC -0.422; TCGA LUAD -0.128; TCGA LUSC -0.316; TCGA SARC -0.129 |
hsa-miR-22-3p | STAG2 | 13 cancers: BLCA; BRCA; COAD; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; OV; PRAD; SARC; THCA | MirTarget; miRNATAP | TCGA BLCA -0.2; TCGA BRCA -0.074; TCGA COAD -0.264; TCGA HNSC -0.121; TCGA KIRC -0.338; TCGA KIRP -0.1; TCGA LGG -0.153; TCGA LUAD -0.208; TCGA LUSC -0.165; TCGA OV -0.07; TCGA PRAD -0.088; TCGA SARC -0.198; TCGA THCA -0.114 |
hsa-miR-22-3p | MAGI2 | 9 cancers: BLCA; CESC; KIRC; KIRP; LUAD; OV; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.463; TCGA CESC -0.285; TCGA KIRC -0.23; TCGA KIRP -0.23; TCGA LUAD -0.131; TCGA OV -0.143; TCGA SARC -0.344; TCGA STAD -0.398; TCGA UCEC -0.46 |
hsa-miR-22-3p | CBFA2T2 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; OV; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.194; TCGA BRCA -0.249; TCGA CESC -0.184; TCGA COAD -0.408; TCGA ESCA -0.254; TCGA HNSC -0.196; TCGA KIRP -0.208; TCGA LIHC -0.526; TCGA LUAD -0.126; TCGA LUSC -0.492; TCGA OV -0.064; TCGA PRAD -0.138; TCGA SARC -0.123; TCGA STAD -0.164 |
hsa-miR-22-3p | DIDO1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRP; LGG; LIHC; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.111; TCGA BRCA -0.156; TCGA CESC -0.128; TCGA COAD -0.234; TCGA ESCA -0.196; TCGA KIRP -0.205; TCGA LGG -0.096; TCGA LIHC -0.196; TCGA LUSC -0.164; TCGA OV -0.064; TCGA PAAD -0.169; TCGA PRAD -0.067; TCGA SARC -0.182; TCGA STAD -0.102; TCGA UCEC -0.094 |
hsa-miR-22-3p | CLUAP1 | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; OV; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.299; TCGA BRCA -0.11; TCGA CESC -0.299; TCGA HNSC -0.208; TCGA KIRC -0.157; TCGA KIRP -0.187; TCGA OV -0.223; TCGA SARC -0.152; TCGA STAD -0.152; TCGA UCEC -0.088 |
hsa-miR-22-3p | PRX | 10 cancers: BLCA; CESC; KIRC; LIHC; LUAD; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.193; TCGA CESC -0.168; TCGA KIRC -0.376; TCGA LIHC -0.361; TCGA LUAD -0.273; TCGA PAAD -0.277; TCGA SARC -0.136; TCGA THCA -0.401; TCGA STAD -0.255; TCGA UCEC -0.347 |
hsa-miR-22-3p | PCBD2 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; LGG; LUSC; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.12; TCGA BRCA -0.231; TCGA CESC -0.161; TCGA COAD -0.196; TCGA ESCA -0.188; TCGA LGG -0.098; TCGA LUSC -0.155; TCGA PAAD -0.242; TCGA SARC -0.212; TCGA STAD -0.12; TCGA UCEC -0.067 |
hsa-miR-22-3p | WNK2 | 15 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LIHC; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.534; TCGA BRCA -0.488; TCGA CESC -0.591; TCGA COAD -0.415; TCGA HNSC -0.98; TCGA KIRC -0.712; TCGA KIRP -1.302; TCGA LIHC -1.353; TCGA LUSC -1.245; TCGA OV -0.23; TCGA PAAD -1.234; TCGA PRAD -0.388; TCGA SARC -0.704; TCGA STAD -0.919; TCGA UCEC -0.18 |
hsa-miR-22-3p | ZFP41 | 11 cancers: BLCA; BRCA; COAD; KIRP; LGG; LIHC; LUSC; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.126; TCGA BRCA -0.29; TCGA COAD -0.375; TCGA KIRP -0.165; TCGA LGG -0.202; TCGA LIHC -0.494; TCGA LUSC -0.191; TCGA PAAD -0.44; TCGA SARC -0.26; TCGA STAD -0.135; TCGA UCEC -0.134 |
hsa-miR-22-3p | RBM33 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; SARC; UCEC | mirMAP | TCGA BLCA -0.093; TCGA BRCA -0.113; TCGA COAD -0.16; TCGA ESCA -0.155; TCGA HNSC -0.079; TCGA KIRP -0.367; TCGA LGG -0.113; TCGA LIHC -0.171; TCGA LUAD -0.082; TCGA LUSC -0.223; TCGA PAAD -0.18; TCGA SARC -0.134; TCGA UCEC -0.073 |
hsa-miR-22-3p | PURB | 12 cancers: BLCA; BRCA; COAD; KIRC; KIRP; LGG; LIHC; LUAD; OV; PRAD; SARC; THCA | miRNATAP | TCGA BLCA -0.101; TCGA BRCA -0.09; TCGA COAD -0.195; TCGA KIRC -0.151; TCGA KIRP -0.28; TCGA LGG -0.081; TCGA LIHC -0.201; TCGA LUAD -0.128; TCGA OV -0.082; TCGA PRAD -0.146; TCGA SARC -0.15; TCGA THCA -0.135 |
hsa-miR-22-3p | RNF38 | 11 cancers: BLCA; COAD; KIRC; LGG; LIHC; LUAD; OV; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.233; TCGA COAD -0.225; TCGA KIRC -0.286; TCGA LGG -0.075; TCGA LIHC -0.202; TCGA LUAD -0.266; TCGA OV -0.078; TCGA SARC -0.413; TCGA THCA -0.165; TCGA STAD -0.194; TCGA UCEC -0.18 |
hsa-miR-22-3p | HNRNPH1 | 12 cancers: BLCA; BRCA; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; SARC | miRNATAP | TCGA BLCA -0.107; TCGA BRCA -0.195; TCGA COAD -0.09; TCGA HNSC -0.135; TCGA KIRC -0.158; TCGA KIRP -0.326; TCGA LGG -0.142; TCGA LIHC -0.333; TCGA LUAD -0.087; TCGA LUSC -0.27; TCGA OV -0.05; TCGA SARC -0.11 |
hsa-miR-22-3p | NPNT | 9 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LIHC; LUAD; SARC; UCEC | miRNATAP | TCGA BLCA -0.327; TCGA BRCA -0.502; TCGA ESCA -0.448; TCGA KIRC -0.493; TCGA KIRP -0.642; TCGA LIHC -0.591; TCGA LUAD -0.276; TCGA SARC -0.889; TCGA UCEC -0.412 |
hsa-miR-22-3p | CUL3 | 9 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LUAD; OV; PRAD; THCA | miRNATAP | TCGA BLCA -0.099; TCGA BRCA -0.064; TCGA COAD -0.182; TCGA ESCA -0.201; TCGA KIRC -0.105; TCGA LUAD -0.104; TCGA OV -0.108; TCGA PRAD -0.055; TCGA THCA -0.077 |
hsa-miR-22-3p | NFIB | 10 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; PRAD; THCA | miRNATAP | TCGA BLCA -0.457; TCGA CESC -0.376; TCGA ESCA -0.515; TCGA HNSC -0.189; TCGA KIRC -0.163; TCGA KIRP -0.115; TCGA LGG -0.366; TCGA LUAD -0.299; TCGA PRAD -0.282; TCGA THCA -0.203 |
hsa-miR-22-3p | MECP2 | 12 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; LIHC; LUAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.179; TCGA BRCA -0.063; TCGA COAD -0.123; TCGA ESCA -0.173; TCGA KIRC -0.127; TCGA KIRP -0.085; TCGA LIHC -0.142; TCGA LUAD -0.119; TCGA SARC -0.156; TCGA THCA -0.11; TCGA STAD -0.171; TCGA UCEC -0.11 |
hsa-miR-22-3p | PRRT2 | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LIHC; LUSC; OV; PAAD; SARC; THCA; UCEC | miRNATAP | TCGA BLCA -0.503; TCGA BRCA -0.707; TCGA CESC -0.458; TCGA ESCA -0.596; TCGA HNSC -0.244; TCGA KIRP -0.881; TCGA LIHC -0.593; TCGA LUSC -0.415; TCGA OV -0.237; TCGA PAAD -0.417; TCGA SARC -0.943; TCGA THCA -0.339; TCGA UCEC -0.587 |
hsa-miR-22-3p | FMR1 | 11 cancers: BLCA; BRCA; CESC; COAD; KIRP; LGG; LUAD; LUSC; SARC; THCA; UCEC | RAID | TCGA BLCA -0.087; TCGA BRCA -0.083; TCGA CESC -0.129; TCGA COAD -0.139; TCGA KIRP -0.087; TCGA LGG -0.066; TCGA LUAD -0.133; TCGA LUSC -0.202; TCGA SARC -0.144; TCGA THCA -0.236; TCGA UCEC -0.154 |
hsa-miR-22-3p | FUS | 13 cancers: BLCA; BRCA; COAD; KIRC; KIRP; LGG; LIHC; LUSC; OV; PAAD; PRAD; THCA; UCEC | RAID | TCGA BLCA -0.082; TCGA BRCA -0.282; TCGA COAD -0.132; TCGA KIRC -0.158; TCGA KIRP -0.157; TCGA LGG -0.214; TCGA LIHC -0.296; TCGA LUSC -0.358; TCGA OV -0.138; TCGA PAAD -0.196; TCGA PRAD -0.15; TCGA THCA -0.167; TCGA UCEC -0.152 |
hsa-miR-22-3p | EDC3 | 11 cancers: BRCA; COAD; KIRC; KIRP; LGG; LIHC; LUSC; OV; PRAD; SARC; THCA | miRNAWalker2 validate; MirTarget | TCGA BRCA -0.131; TCGA COAD -0.098; TCGA KIRC -0.069; TCGA KIRP -0.147; TCGA LGG -0.232; TCGA LIHC -0.315; TCGA LUSC -0.23; TCGA OV -0.081; TCGA PRAD -0.108; TCGA SARC -0.128; TCGA THCA -0.188 |
hsa-miR-22-3p | ERBB3 | 11 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUSC; PRAD; THCA | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | TCGA BRCA -0.876; TCGA CESC -0.347; TCGA COAD -0.197; TCGA ESCA -1.184; TCGA HNSC -0.403; TCGA KIRC -0.279; TCGA KIRP -0.366; TCGA LIHC -0.458; TCGA LUSC -0.207; TCGA PRAD -0.147; TCGA THCA -1.735 |
hsa-miR-22-3p | FRAT2 | 12 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUSC; PAAD; PRAD | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BRCA -0.282; TCGA CESC -0.265; TCGA COAD -0.19; TCGA ESCA -0.34; TCGA HNSC -0.416; TCGA KIRC -0.16; TCGA KIRP -0.098; TCGA LGG -0.172; TCGA LIHC -0.233; TCGA LUSC -0.398; TCGA PAAD -0.412; TCGA PRAD -0.171 |
hsa-miR-22-3p | LRRC1 | 10 cancers: BRCA; HNSC; KIRC; KIRP; LGG; LIHC; LUSC; PRAD; SARC; THCA | miRNAWalker2 validate; miRNATAP | TCGA BRCA -0.318; TCGA HNSC -0.25; TCGA KIRC -0.177; TCGA KIRP -0.126; TCGA LGG -0.288; TCGA LIHC -1.338; TCGA LUSC -0.17; TCGA PRAD -0.189; TCGA SARC -0.127; TCGA THCA -0.128 |
hsa-miR-22-3p | MYO6 | 9 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; PRAD; THCA | miRNAWalker2 validate | TCGA BRCA -0.299; TCGA ESCA -0.459; TCGA HNSC -0.183; TCGA KIRC -0.181; TCGA KIRP -0.111; TCGA LIHC -0.167; TCGA LUAD -0.109; TCGA PRAD -0.367; TCGA THCA -0.18 |
hsa-miR-22-3p | RBM39 | 12 cancers: BRCA; CESC; COAD; ESCA; KIRP; LGG; LIHC; LUSC; OV; PAAD; SARC; UCEC | miRNAWalker2 validate | TCGA BRCA -0.221; TCGA CESC -0.108; TCGA COAD -0.25; TCGA ESCA -0.19; TCGA KIRP -0.331; TCGA LGG -0.109; TCGA LIHC -0.175; TCGA LUSC -0.263; TCGA OV -0.07; TCGA PAAD -0.125; TCGA SARC -0.081; TCGA UCEC -0.094 |
hsa-miR-22-3p | RPL24 | 11 cancers: BRCA; COAD; HNSC; LGG; LIHC; LUSC; OV; PAAD; PRAD; THCA; UCEC | miRNAWalker2 validate | TCGA BRCA -0.119; TCGA COAD -0.15; TCGA HNSC -0.136; TCGA LGG -0.286; TCGA LIHC -0.152; TCGA LUSC -0.13; TCGA OV -0.278; TCGA PAAD -0.332; TCGA PRAD -0.27; TCGA THCA -0.146; TCGA UCEC -0.139 |
hsa-miR-22-3p | RPL35A | 11 cancers: BRCA; CESC; COAD; HNSC; LGG; LIHC; LUSC; OV; PRAD; THCA; UCEC | miRNAWalker2 validate | TCGA BRCA -0.114; TCGA CESC -0.174; TCGA COAD -0.261; TCGA HNSC -0.241; TCGA LGG -0.239; TCGA LIHC -0.292; TCGA LUSC -0.368; TCGA OV -0.236; TCGA PRAD -0.292; TCGA THCA -0.143; TCGA UCEC -0.203 |
hsa-miR-22-3p | RPS2 | 10 cancers: BRCA; COAD; HNSC; LGG; LIHC; LUSC; OV; PAAD; PRAD; THCA | miRNAWalker2 validate | TCGA BRCA -0.257; TCGA COAD -0.158; TCGA HNSC -0.167; TCGA LGG -0.316; TCGA LIHC -0.315; TCGA LUSC -0.133; TCGA OV -0.189; TCGA PAAD -0.309; TCGA PRAD -0.408; TCGA THCA -0.297 |
hsa-miR-22-3p | RPSA | 11 cancers: BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; OV; PAAD; PRAD; THCA | miRNAWalker2 validate | TCGA BRCA -0.162; TCGA CESC -0.217; TCGA COAD -0.197; TCGA ESCA -0.223; TCGA HNSC -0.274; TCGA LGG -0.326; TCGA LIHC -0.441; TCGA OV -0.198; TCGA PAAD -0.28; TCGA PRAD -0.279; TCGA THCA -0.255 |
hsa-miR-22-3p | NUSAP1 | 9 cancers: BRCA; COAD; HNSC; KIRP; LGG; LIHC; LUSC; SARC; THCA | MirTarget | TCGA BRCA -0.4; TCGA COAD -0.203; TCGA HNSC -0.33; TCGA KIRP -0.216; TCGA LGG -0.319; TCGA LIHC -1.02; TCGA LUSC -0.966; TCGA SARC -0.161; TCGA THCA -0.255 |
hsa-miR-22-3p | GPBP1 | 14 cancers: BRCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PRAD; THCA; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.112; TCGA CESC -0.115; TCGA COAD -0.071; TCGA HNSC -0.11; TCGA KIRC -0.082; TCGA KIRP -0.199; TCGA LGG -0.073; TCGA LIHC -0.143; TCGA LUAD -0.141; TCGA LUSC -0.159; TCGA OV -0.108; TCGA PRAD -0.099; TCGA THCA -0.088; TCGA UCEC -0.065 |
hsa-miR-22-3p | MTF2 | 12 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PRAD; THCA | MirTarget; miRNATAP | TCGA BRCA -0.19; TCGA ESCA -0.168; TCGA HNSC -0.152; TCGA KIRC -0.087; TCGA KIRP -0.322; TCGA LGG -0.069; TCGA LIHC -0.421; TCGA LUAD -0.09; TCGA LUSC -0.297; TCGA OV -0.058; TCGA PRAD -0.171; TCGA THCA -0.187 |
hsa-miR-22-3p | TRIM46 | 9 cancers: BRCA; KIRP; LIHC; LUSC; OV; PAAD; SARC; THCA; UCEC | MirTarget | TCGA BRCA -0.485; TCGA KIRP -0.517; TCGA LIHC -0.56; TCGA LUSC -0.64; TCGA OV -0.115; TCGA PAAD -0.721; TCGA SARC -1.007; TCGA THCA -1.025; TCGA UCEC -0.297 |
hsa-miR-22-3p | POGK | 13 cancers: BRCA; COAD; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PRAD; SARC; THCA; UCEC | MirTarget | TCGA BRCA -0.18; TCGA COAD -0.141; TCGA KIRC -0.17; TCGA KIRP -0.161; TCGA LGG -0.123; TCGA LIHC -0.447; TCGA LUAD -0.13; TCGA LUSC -0.261; TCGA OV -0.106; TCGA PRAD -0.111; TCGA SARC -0.173; TCGA THCA -0.212; TCGA UCEC -0.083 |
hsa-miR-22-3p | SEPHS1 | 9 cancers: BRCA; CESC; COAD; HNSC; KIRP; LGG; LIHC; LUSC; OV | MirTarget; miRNATAP | TCGA BRCA -0.106; TCGA CESC -0.149; TCGA COAD -0.208; TCGA HNSC -0.137; TCGA KIRP -0.125; TCGA LGG -0.224; TCGA LIHC -0.097; TCGA LUSC -0.349; TCGA OV -0.111 |
hsa-miR-22-3p | LIG3 | 10 cancers: BRCA; CESC; COAD; HNSC; LGG; LIHC; LUAD; LUSC; OV; SARC | MirTarget | TCGA BRCA -0.328; TCGA CESC -0.109; TCGA COAD -0.178; TCGA HNSC -0.103; TCGA LGG -0.142; TCGA LIHC -0.227; TCGA LUAD -0.111; TCGA LUSC -0.485; TCGA OV -0.122; TCGA SARC -0.227 |
hsa-miR-22-3p | RFXANK | 10 cancers: BRCA; CESC; HNSC; KIRP; LGG; LIHC; LUSC; OV; PAAD; PRAD | MirTarget | TCGA BRCA -0.278; TCGA CESC -0.191; TCGA HNSC -0.194; TCGA KIRP -0.143; TCGA LGG -0.092; TCGA LIHC -0.353; TCGA LUSC -0.22; TCGA OV -0.088; TCGA PAAD -0.505; TCGA PRAD -0.111 |
hsa-miR-22-3p | TET2 | 10 cancers: BRCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PRAD; SARC; THCA | MirTarget; miRNATAP | TCGA BRCA -0.078; TCGA HNSC -0.122; TCGA KIRC -0.198; TCGA KIRP -0.194; TCGA LGG -0.226; TCGA LUAD -0.109; TCGA LUSC -0.172; TCGA PRAD -0.104; TCGA SARC -0.166; TCGA THCA -0.185 |
hsa-miR-22-3p | COPS7B | 13 cancers: BRCA; CESC; COAD; ESCA; KIRP; LGG; LIHC; LUSC; OV; PAAD; PRAD; SARC; THCA | MirTarget; miRNATAP | TCGA BRCA -0.15; TCGA CESC -0.114; TCGA COAD -0.119; TCGA ESCA -0.149; TCGA KIRP -0.251; TCGA LGG -0.088; TCGA LIHC -0.344; TCGA LUSC -0.351; TCGA OV -0.073; TCGA PAAD -0.243; TCGA PRAD -0.061; TCGA SARC -0.106; TCGA THCA -0.118 |
hsa-miR-22-3p | WRNIP1 | 9 cancers: BRCA; COAD; KIRC; KIRP; LGG; LIHC; LUSC; OV; PAAD | MirTarget; miRNATAP | TCGA BRCA -0.21; TCGA COAD -0.159; TCGA KIRC -0.18; TCGA KIRP -0.063; TCGA LGG -0.073; TCGA LIHC -0.186; TCGA LUSC -0.172; TCGA OV -0.059; TCGA PAAD -0.193 |
hsa-miR-22-3p | WARS2 | 9 cancers: BRCA; CESC; COAD; HNSC; KIRC; LIHC; LUAD; OV; PRAD | MirTarget | TCGA BRCA -0.075; TCGA CESC -0.204; TCGA COAD -0.135; TCGA HNSC -0.104; TCGA KIRC -0.152; TCGA LIHC -0.152; TCGA LUAD -0.086; TCGA OV -0.139; TCGA PRAD -0.09 |
hsa-miR-22-3p | TJAP1 | 12 cancers: BRCA; CESC; COAD; KIRP; LGG; LIHC; LUSC; OV; PAAD; SARC; THCA; UCEC | MirTarget | TCGA BRCA -0.197; TCGA CESC -0.15; TCGA COAD -0.112; TCGA KIRP -0.168; TCGA LGG -0.107; TCGA LIHC -0.29; TCGA LUSC -0.221; TCGA OV -0.061; TCGA PAAD -0.382; TCGA SARC -0.096; TCGA THCA -0.055; TCGA UCEC -0.132 |
hsa-miR-22-3p | SOX13 | 12 cancers: BRCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; OV; PAAD; PRAD; SARC; UCEC | MirTarget | TCGA BRCA -0.228; TCGA HNSC -0.241; TCGA KIRC -0.529; TCGA KIRP -0.161; TCGA LGG -0.22; TCGA LIHC -0.272; TCGA LUAD -0.234; TCGA OV -0.137; TCGA PAAD -0.409; TCGA PRAD -0.108; TCGA SARC -0.226; TCGA UCEC -0.178 |
hsa-miR-22-3p | NHP2 | 11 cancers: BRCA; CESC; COAD; HNSC; LGG; LIHC; LUSC; OV; PAAD; PRAD; THCA | MirTarget | TCGA BRCA -0.291; TCGA CESC -0.162; TCGA COAD -0.127; TCGA HNSC -0.151; TCGA LGG -0.141; TCGA LIHC -0.119; TCGA LUSC -0.198; TCGA OV -0.158; TCGA PAAD -0.383; TCGA PRAD -0.139; TCGA THCA -0.091 |
hsa-miR-22-3p | UTP18 | 10 cancers: BRCA; CESC; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; PRAD; THCA | MirTarget | TCGA BRCA -0.176; TCGA CESC -0.102; TCGA HNSC -0.197; TCGA KIRC -0.083; TCGA LGG -0.101; TCGA LIHC -0.345; TCGA LUAD -0.102; TCGA LUSC -0.273; TCGA PRAD -0.109; TCGA THCA -0.224 |
hsa-miR-22-3p | BSPRY | 10 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; PAAD; PRAD; SARC | MirTarget | TCGA BRCA -0.69; TCGA CESC -0.444; TCGA ESCA -0.874; TCGA HNSC -0.979; TCGA KIRC -1.118; TCGA KIRP -0.437; TCGA LIHC -0.857; TCGA PAAD -0.842; TCGA PRAD -0.135; TCGA SARC -0.824 |
hsa-miR-22-3p | PLEKHG4B | 10 cancers: BRCA; CESC; KIRC; KIRP; LIHC; LUSC; OV; PRAD; SARC; UCEC | mirMAP | TCGA BRCA -0.284; TCGA CESC -0.533; TCGA KIRC -0.906; TCGA KIRP -1.008; TCGA LIHC -0.393; TCGA LUSC -1.047; TCGA OV -0.297; TCGA PRAD -0.313; TCGA SARC -0.379; TCGA UCEC -0.526 |
hsa-miR-22-3p | ADAMTS13 | 9 cancers: BRCA; CESC; KIRP; LGG; LUSC; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.581; TCGA CESC -0.296; TCGA KIRP -0.725; TCGA LGG -0.268; TCGA LUSC -0.662; TCGA PAAD -1.087; TCGA SARC -0.336; TCGA STAD -0.281; TCGA UCEC -0.254 |
hsa-miR-22-3p | LRRC14 | 10 cancers: BRCA; CESC; COAD; KIRP; LGG; LIHC; LUSC; PAAD; SARC; STAD | mirMAP | TCGA BRCA -0.331; TCGA CESC -0.211; TCGA COAD -0.283; TCGA KIRP -0.257; TCGA LGG -0.176; TCGA LIHC -0.338; TCGA LUSC -0.335; TCGA PAAD -0.609; TCGA SARC -0.205; TCGA STAD -0.165 |
hsa-miR-22-3p | RAB11FIP4 | 12 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; THCA | miRNATAP | TCGA BRCA -0.62; TCGA CESC -0.333; TCGA COAD -0.146; TCGA ESCA -0.755; TCGA HNSC -0.175; TCGA KIRP -0.113; TCGA LIHC -0.613; TCGA LUAD -0.142; TCGA LUSC -0.313; TCGA PAAD -0.448; TCGA PRAD -0.121; TCGA THCA -0.451 |
hsa-miR-22-3p | FBRS | 10 cancers: BRCA; COAD; HNSC; KIRP; LGG; LIHC; LUSC; PAAD; PRAD; THCA | miRNATAP | TCGA BRCA -0.249; TCGA COAD -0.143; TCGA HNSC -0.075; TCGA KIRP -0.166; TCGA LGG -0.06; TCGA LIHC -0.158; TCGA LUSC -0.191; TCGA PAAD -0.419; TCGA PRAD -0.078; TCGA THCA -0.129 |
hsa-miR-22-3p | JARID2 | 9 cancers: BRCA; COAD; KIRC; KIRP; LGG; LIHC; LUSC; OV; PRAD | miRNATAP | TCGA BRCA -0.21; TCGA COAD -0.188; TCGA KIRC -0.099; TCGA KIRP -0.067; TCGA LGG -0.21; TCGA LIHC -0.357; TCGA LUSC -0.197; TCGA OV -0.175; TCGA PRAD -0.11 |
hsa-miR-22-3p | PLAGL2 | 10 cancers: BRCA; COAD; HNSC; LIHC; LUAD; LUSC; OV; PRAD; THCA; UCEC | miRNATAP | TCGA BRCA -0.148; TCGA COAD -0.578; TCGA HNSC -0.207; TCGA LIHC -0.494; TCGA LUAD -0.154; TCGA LUSC -0.232; TCGA OV -0.123; TCGA PRAD -0.088; TCGA THCA -0.325; TCGA UCEC -0.111 |
hsa-miR-22-3p | DGCR8 | 10 cancers: BRCA; CESC; KIRC; KIRP; LGG; LIHC; LUSC; PAAD; SARC; STAD | RAID | TCGA BRCA -0.153; TCGA CESC -0.097; TCGA KIRC -0.091; TCGA KIRP -0.252; TCGA LGG -0.147; TCGA LIHC -0.163; TCGA LUSC -0.339; TCGA PAAD -0.359; TCGA SARC -0.13; TCGA STAD -0.097 |
hsa-miR-22-3p | PTBP1 | 9 cancers: BRCA; COAD; KIRP; LGG; LIHC; LUSC; PAAD; PRAD; THCA | RAID | TCGA BRCA -0.215; TCGA COAD -0.137; TCGA KIRP -0.118; TCGA LGG -0.16; TCGA LIHC -0.222; TCGA LUSC -0.165; TCGA PAAD -0.232; TCGA PRAD -0.077; TCGA THCA -0.064 |
hsa-miR-22-3p | PTP4A3 | 9 cancers: CESC; COAD; LGG; LIHC; PAAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA CESC -0.217; TCGA COAD -0.535; TCGA LGG -0.145; TCGA LIHC -0.733; TCGA PAAD -0.404; TCGA SARC -0.148; TCGA THCA -0.39; TCGA STAD -0.422; TCGA UCEC -0.313 |
hsa-miR-22-3p | TRIM13 | 9 cancers: COAD; ESCA; KIRC; KIRP; LGG; LUAD; OV; PRAD; THCA | MirTarget | TCGA COAD -0.172; TCGA ESCA -0.222; TCGA KIRC -0.135; TCGA KIRP -0.129; TCGA LGG -0.18; TCGA LUAD -0.093; TCGA OV -0.113; TCGA PRAD -0.111; TCGA THCA -0.106 |
hsa-miR-22-3p | ISY1 | 10 cancers: COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUSC; PRAD; THCA; UCEC | mirMAP | TCGA COAD -0.098; TCGA HNSC -0.082; TCGA KIRC -0.073; TCGA KIRP -0.052; TCGA LGG -0.131; TCGA LIHC -0.181; TCGA LUSC -0.273; TCGA PRAD -0.07; TCGA THCA -0.115; TCGA UCEC -0.079 |
hsa-miR-22-3p | SLC39A10 | 9 cancers: COAD; ESCA; KIRC; KIRP; LIHC; LUSC; OV; THCA; UCEC | miRNATAP | TCGA COAD -0.18; TCGA ESCA -0.238; TCGA KIRC -0.238; TCGA KIRP -0.183; TCGA LIHC -0.488; TCGA LUSC -0.235; TCGA OV -0.186; TCGA THCA -0.214; TCGA UCEC -0.184 |
Enriched cancer pathways of putative targets