microRNA information: hsa-miR-224-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-224-5p | miRbase |
| Accession: | MIMAT0000281 | miRbase |
| Precursor name: | hsa-mir-224 | miRbase |
| Precursor accession: | MI0000301 | miRbase |
| Symbol: | MIR224 | HGNC |
| RefSeq ID: | NR_029638 | GenBank |
| Sequence: | CAAGUCACUAGUGGUUCCGUU |
Reported expression in cancers: hsa-miR-224-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-224-5p | B cell lymphoma | downregulation | "In this study we analyzed the expression of miR-22 ......" | 25146331 | |
| hsa-miR-224-5p | B cell lymphoma | downregulation | "To investigate the expression and prognostic value ......" | 26301883 | |
| hsa-miR-224-5p | breast cancer | downregulation | "Moreover metastasis as assayed by Transwell migrat ......" | 22809510 | |
| hsa-miR-224-5p | breast cancer | downregulation | "MiRNA expression profiles in 11 PMBCs were analyze ......" | 23691441 | qPCR; Microarray |
| hsa-miR-224-5p | cervical and endocervical cancer | upregulation | "Upregulation of microRNA 224 is associated with ag ......" | 23631806 | Microarray; qPCR |
| hsa-miR-224-5p | cervical and endocervical cancer | upregulation | "Vote-counting analysis showed that up-regulation w ......" | 25920605 | |
| hsa-miR-224-5p | cervical and endocervical cancer | upregulation | "In the current study we demonstrated by real-time ......" | 27626930 | qPCR |
| hsa-miR-224-5p | colorectal cancer | upregulation | "Up regulation of miR 224 promotes cancer cell prol ......" | 24152489 | qPCR |
| hsa-miR-224-5p | colorectal cancer | upregulation | "miR 224 overexpression is a strong and independent ......" | 25420464 | qPCR |
| hsa-miR-224-5p | colorectal cancer | upregulation | "In order to clarify the miRNA profile in CRC tissu ......" | 25421755 | Microarray |
| hsa-miR-224-5p | colorectal cancer | upregulation | "MicroRNA 224 sustains Wnt/β catenin signaling and ......" | 26822534 | qPCR |
| hsa-miR-224-5p | gastric cancer | upregulation | "From the set of the 29 microRNAs of interest we fo ......" | 27081844 | |
| hsa-miR-224-5p | liver cancer | upregulation | "Role of miR 224 in hepatocellular carcinoma: a too ......" | 22122284 | |
| hsa-miR-224-5p | liver cancer | upregulation | "MicroRNA 224 is up regulated in hepatocellular car ......" | 22459148 | |
| hsa-miR-224-5p | liver cancer | upregulation | "MicroRNA 224 upregulation and AKT activation syner ......" | 24923856 | in situ hybridization |
| hsa-miR-224-5p | liver cancer | upregulation | "MicroRNA 224 Induces G1/S Checkpoint Release in Li ......" | 26343737 | Microarray |
| hsa-miR-224-5p | lung cancer | upregulation | "By examining the expression of 22 HCC-related miRN ......" | 22392644 | |
| hsa-miR-224-5p | lung cancer | upregulation | "We recently reported that miR-224 was significantl ......" | 26307684 | |
| hsa-miR-224-5p | lung squamous cell cancer | downregulation | "Decreased microRNA 224 and its clinical significan ......" | 25410592 | qPCR |
| hsa-miR-224-5p | lung squamous cell cancer | downregulation | "Quantitative real-time PCR assay was employed to c ......" | 27073334 | qPCR |
| hsa-miR-224-5p | prostate cancer | downregulation | "Downregulation and prognostic performance of micro ......" | 23136246 | qPCR |
| hsa-miR-224-5p | prostate cancer | downregulation | "Our previous microarray data showed that microRNA- ......" | 24382668 | Microarray |
| hsa-miR-224-5p | prostate cancer | downregulation | "Our recent study of the microRNA expression signat ......" | 24768995 | |
| hsa-miR-224-5p | prostate cancer | downregulation | "Our previous study revealed that microRNA miR-224 ......" | 25532941 | |
| hsa-miR-224-5p | thyroid cancer | upregulation | "We analyzed the expression of nine miRNAs miR-21 m ......" | 22747440 | qPCR |
Reported cancer pathway affected by hsa-miR-224-5p

| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-224-5p | breast cancer | Apoptosis pathway | "Epigenetic changes and nuclear factor κB activati ......" | 26788213 | Western blot; Flow cytometry |
| hsa-miR-224-5p | colorectal cancer | PI3K/Akt signaling pathway | "microRNA 224 promotes cell proliferation and tumor ......" | 23846336 | Luciferase |
| hsa-miR-224-5p | colorectal cancer | Epithelial mesenchymal transition pathway | "MicroRNA 224 is associated with colorectal cancer ......" | 25919696 | |
| hsa-miR-224-5p | esophageal cancer | Apoptosis pathway | "Aberrant expression of miR-224 is associated with ......" | 26245343 | Western blot; Colony formation; Luciferase |
| hsa-miR-224-5p | gastric cancer | Apoptosis pathway | "Effect of antisense miR 224 on gastric cancer cell ......" | 24796455 | Flow cytometry; MTT assay |
| hsa-miR-224-5p | gastric cancer | Apoptosis pathway | "RKIP suppresses gastric cancer cell proliferation ......" | 25017365 | Western blot; MTT assay; Flow cytometry; Cell migration assay; Luciferase |
| hsa-miR-224-5p | gastric cancer | Apoptosis pathway | "MicroRNA 224 aggrevates tumor growth and progressi ......" | 27315344 | Luciferase |
| hsa-miR-224-5p | liver cancer | Apoptosis pathway | "Profiling microRNA expression in hepatocellular ca ......" | 18319255 | |
| hsa-miR-224-5p | liver cancer | Apoptosis pathway | "MicroRNA 224 is up regulated in hepatocellular car ......" | 22459148 | |
| hsa-miR-224-5p | liver cancer | Apoptosis pathway | "Involvement of microRNA 224 in cell proliferation ......" | 22989374 | Western blot |
| hsa-miR-224-5p | liver cancer | PI3K/Akt signaling pathway | "miR 224 functions as an onco miRNA in hepatocellul ......" | 23741247 | Western blot; Luciferase |
| hsa-miR-224-5p | liver cancer | PI3K/Akt signaling pathway | "MicroRNA 224 upregulation and AKT activation syner ......" | 24923856 | |
| hsa-miR-224-5p | liver cancer | cell cycle pathway | "MicroRNA 224 Induces G1/S Checkpoint Release in Li ......" | 26343737 | |
| hsa-miR-224-5p | lung cancer | Apoptosis pathway; cell cycle pathway | "MiR 224 promotes the chemoresistance of human lung ......" | 24921914 | |
| hsa-miR-224-5p | lung cancer | Apoptosis pathway | "MicroRNA 224 is implicated in lung cancer pathogen ......" | 26307684 | |
| hsa-miR-224-5p | lung squamous cell cancer | Apoptosis pathway | "Decreased microRNA 224 and its clinical significan ......" | 25410592 | Cell migration assay |
| hsa-miR-224-5p | lung squamous cell cancer | Apoptosis pathway | "Quantitative real-time PCR assay was employed to c ......" | 27073334 | Flow cytometry; Wound Healing Assay |
| hsa-miR-224-5p | ovarian cancer | Apoptosis pathway | "Upregulated microRNA 224 promotes ovarian cancer c ......" | 27663866 | |
| hsa-miR-224-5p | prostate cancer | Apoptosis pathway | "MicroRNA 224 inhibits progression of human prostat ......" | 24382668 | |
| hsa-miR-224-5p | prostate cancer | cell cycle pathway | "GABRE∼miR-452∼miR-224 promoter methylation was ......" | 24737792 | |
| hsa-miR-224-5p | prostate cancer | cell cycle pathway | "MicroRNA 224 and its target CAMKK2 synergistically ......" | 25394900 |
Reported cancer prognosis affected by hsa-miR-224-5p
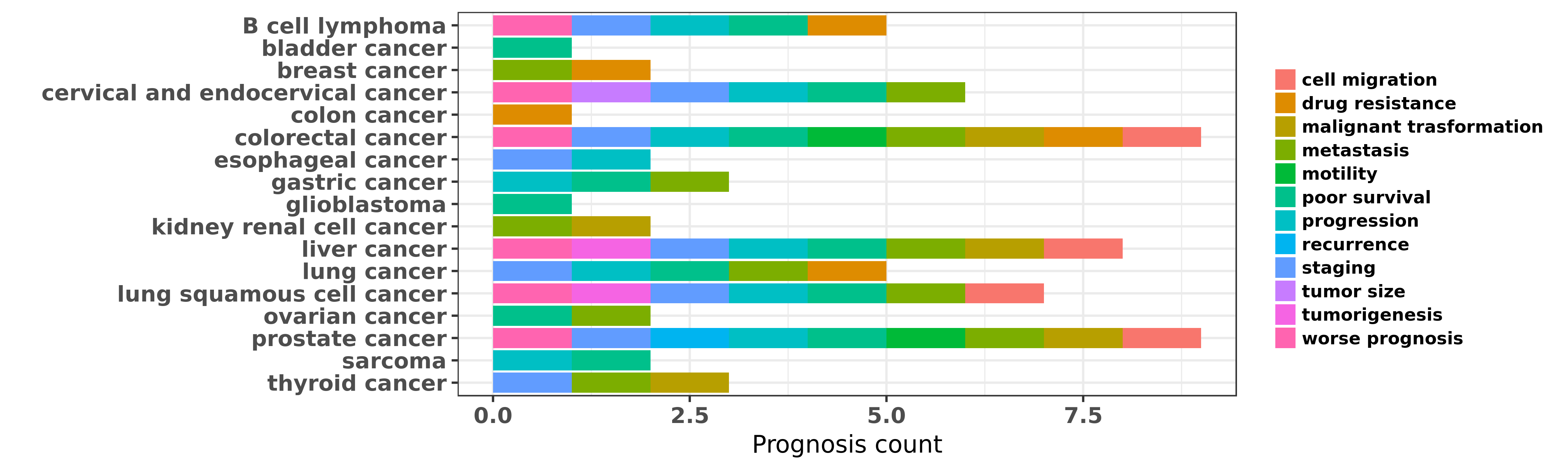
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-224-5p | B cell lymphoma | staging; drug resistance | "In the discovery phase real-time polymerase chain ......" | 24858372 | |
| hsa-miR-224-5p | B cell lymphoma | worse prognosis; staging; progression | "Expression of miR 224 in diffuse large B cell lymp ......" | 25052605 | |
| hsa-miR-224-5p | B cell lymphoma | progression; poor survival; drug resistance | "Deregulated expression of miR 224 and its target g ......" | 25146331 | Luciferase |
| hsa-miR-224-5p | bladder cancer | poor survival | "Uncovering the clinical utility of miR 143 miR 145 ......" | 25804644 | |
| hsa-miR-224-5p | breast cancer | metastasis; drug resistance | "MicroRNA 224 targets RKIP to control cell invasion ......" | 22809510 | |
| hsa-miR-224-5p | breast cancer | drug resistance | "Increased fucosylation has a pivotal role in multi ......" | 26701615 | |
| hsa-miR-224-5p | cervical and endocervical cancer | progression; worse prognosis; metastasis; poor survival | "Upregulation of microRNA 224 is associated with ag ......" | 23631806 | |
| hsa-miR-224-5p | cervical and endocervical cancer | progression; staging; metastasis; tumor size | "Over Expressed miR 224 Promotes the Progression of ......" | 27626930 | Western blot; Luciferase; RNAi |
| hsa-miR-224-5p | colon cancer | drug resistance | "Underexpression of miR 224 in methotrexate resista ......" | 21864507 | |
| hsa-miR-224-5p | colorectal cancer | progression | "Furthermore several of these miRNAs were associate ......" | 19843336 | |
| hsa-miR-224-5p | colorectal cancer | staging | "Genome-wide microarray analysis of miRNA expressio ......" | 23673725 | |
| hsa-miR-224-5p | colorectal cancer | metastasis; staging; poor survival; motility; progression | "Decreased levels of miR 224 and the passenger stra ......" | 23770133 | |
| hsa-miR-224-5p | colorectal cancer | poor survival; worse prognosis | "microRNA 224 promotes cell proliferation and tumor ......" | 23846336 | Luciferase |
| hsa-miR-224-5p | colorectal cancer | poor survival | "Up regulation of miR 224 promotes cancer cell prol ......" | 24152489 | Western blot; Luciferase |
| hsa-miR-224-5p | colorectal cancer | cell migration; progression | "MicroRNA 224 suppresses colorectal cancer cell mig ......" | 24817781 | |
| hsa-miR-224-5p | colorectal cancer | poor survival; malignant trasformation | "miR 224 overexpression is a strong and independent ......" | 25420464 | |
| hsa-miR-224-5p | colorectal cancer | metastasis; poor survival | "The clinical and biological significance of MIR 22 ......" | 25804630 | Western blot; Luciferase |
| hsa-miR-224-5p | colorectal cancer | progression; drug resistance | "MicroRNA 224 is associated with colorectal cancer ......" | 25919696 | |
| hsa-miR-224-5p | colorectal cancer | metastasis | "MicroRNA 224 sustains Wnt/β catenin signaling and ......" | 26822534 | Luciferase; Western blot |
| hsa-miR-224-5p | esophageal cancer | progression; staging | "Aberrant expression of miR-224 is associated with ......" | 26245343 | Western blot; Colony formation; Luciferase |
| hsa-miR-224-5p | gastric cancer | poor survival | "Effect of antisense miR 224 on gastric cancer cell ......" | 24796455 | Flow cytometry; MTT assay |
| hsa-miR-224-5p | gastric cancer | progression; metastasis | "MicroRNA 224 aggrevates tumor growth and progressi ......" | 27315344 | Luciferase |
| hsa-miR-224-5p | glioblastoma | poor survival | "MiR 224 expression increases radiation sensitivity ......" | 24785373 | |
| hsa-miR-224-5p | kidney renal cell cancer | metastasis; malignant trasformation | "miR-122 miR-141 miR-155 miR-184 miR-200c miR-210 m ......" | 23178446 | |
| hsa-miR-224-5p | liver cancer | tumorigenesis | "Profiling microRNA expression in hepatocellular ca ......" | 18319255 | |
| hsa-miR-224-5p | liver cancer | malignant trasformation | "Our study identified and validated miR-224 overexp ......" | 18433021 | |
| hsa-miR-224-5p | liver cancer | tumorigenesis; progression | "Involvement of microRNA 224 in cell proliferation ......" | 22989374 | Western blot |
| hsa-miR-224-5p | liver cancer | tumorigenesis; progression; cell migration | "Autophagy suppresses tumorigenesis of hepatitis B ......" | 23913306 | |
| hsa-miR-224-5p | liver cancer | cell migration | "miR 224 promotion of cell migration and invasion b ......" | 24219032 | Luciferase; Western blot |
| hsa-miR-224-5p | liver cancer | worse prognosis; staging; poor survival; progression | "MicroRNA 224 upregulation and AKT activation syner ......" | 24923856 | |
| hsa-miR-224-5p | liver cancer | poor survival | "Univariate analysis revealed that miR-200a miR-21 ......" | 25275448 | |
| hsa-miR-224-5p | liver cancer | staging; poor survival; metastasis | "Serum miR 224 reflects stage of hepatocellular car ......" | 25688365 | |
| hsa-miR-224-5p | liver cancer | progression | "MicroRNA 224 Induces G1/S Checkpoint Release in Li ......" | 26343737 | |
| hsa-miR-224-5p | liver cancer | malignant trasformation | "Using gene expression microarrays and high-through ......" | 26646011 | |
| hsa-miR-224-5p | lung cancer | staging | "By examining the expression of 22 HCC-related miRN ......" | 22392644 | |
| hsa-miR-224-5p | lung cancer | drug resistance; poor survival | "MiR 224 promotes the chemoresistance of human lung ......" | 24921914 | |
| hsa-miR-224-5p | lung cancer | metastasis; progression | "MicroRNA 224 is implicated in lung cancer pathogen ......" | 26307684 | |
| hsa-miR-224-5p | lung squamous cell cancer | progression; tumorigenesis; worse prognosis; staging; metastasis; poor survival | "Decreased microRNA 224 and its clinical significan ......" | 25410592 | Cell migration assay |
| hsa-miR-224-5p | lung squamous cell cancer | progression; metastasis; cell migration | "MicroRNA 224 promotes tumor progression in nonsmal ......" | 26187928 | |
| hsa-miR-224-5p | ovarian cancer | metastasis; poor survival | "Upregulated microRNA 224 promotes ovarian cancer c ......" | 27663866 | |
| hsa-miR-224-5p | prostate cancer | malignant trasformation; staging; progression; poor survival; worse prognosis | "Downregulation and prognostic performance of micro ......" | 23136246 | |
| hsa-miR-224-5p | prostate cancer | progression; staging; metastasis; poor survival; recurrence; worse prognosis | "MicroRNA 224 inhibits progression of human prostat ......" | 24382668 | |
| hsa-miR-224-5p | prostate cancer | malignant trasformation; motility | "GABRE∼miR-452∼miR-224 promoter methylation was ......" | 24737792 | |
| hsa-miR-224-5p | prostate cancer | cell migration | "Tumour suppressive microRNA 224 inhibits cancer ce ......" | 24768995 | |
| hsa-miR-224-5p | prostate cancer | progression; worse prognosis; staging; malignant trasformation | "MicroRNA 224 and its target CAMKK2 synergistically ......" | 25394900 | |
| hsa-miR-224-5p | prostate cancer | progression; poor survival; recurrence; staging; metastasis | "Our previous study revealed that microRNA miR-224 ......" | 25532941 | |
| hsa-miR-224-5p | sarcoma | progression; poor survival | "MicroRNA 224 promotes the sensitivity of osteosarc ......" | 27222381 | |
| hsa-miR-224-5p | thyroid cancer | staging; metastasis | "We analyzed the expression of nine miRNAs miR-21 m ......" | 22747440 | |
| hsa-miR-224-5p | thyroid cancer | malignant trasformation | "A limited set of miRNA have been assessed as part ......" | 22771635 |
Reported gene related to hsa-miR-224-5p

| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-224-5p | colorectal cancer | SMAD4 | "Increased miR-224 diminished Cdc42 and SMAD4 expre ......" | 24817781 |
| hsa-miR-224-5p | colorectal cancer | SMAD4 | "In addition the regulation of SMAD4 by miR-224 was ......" | 24152489 |
| hsa-miR-224-5p | colorectal cancer | SMAD4 | "We identified SMAD4 as a miR-224 target and observ ......" | 25804630 |
| hsa-miR-224-5p | liver cancer | SMAD4 | "Our in vitro study demonstrated that miR-224 playe ......" | 23913306 |
| hsa-miR-224-5p | lung cancer | SMAD4 | "We further demonstrated that miR-224 functions as ......" | 26307684 |
| hsa-miR-224-5p | lung squamous cell cancer | SMAD4 | "Increased miR-224 expression promotes cell migrati ......" | 26187928 |
| hsa-miR-224-5p | breast cancer | PEBP1 | "MicroRNA 224 targets RKIP to control cell invasion ......" | 22809510 |
| hsa-miR-224-5p | breast cancer | PEBP1 | "Bisulphite modification methylation specific-polym ......" | 26788213 |
| hsa-miR-224-5p | gastric cancer | PEBP1 | "RKIP suppresses gastric cancer cell proliferation ......" | 25017365 |
| hsa-miR-224-5p | liver cancer | AFP | "In addition the combined upregulation of miR-224 a ......" | 24923856 |
| hsa-miR-224-5p | liver cancer | AFP | "Serum levels of miR-224 showed significant relatio ......" | 25688365 |
| hsa-miR-224-5p | glioblastoma | API5 | "MiR-224 expression in glioblastoma cells resulted ......" | 24785373 |
| hsa-miR-224-5p | liver cancer | API5 | "Profiling microRNA expression in hepatocellular ca ......" | 18319255 |
| hsa-miR-224-5p | liver cancer | MMP9 | "We also performed real-time PCR to measure miR-224 ......" | 23988648 |
| hsa-miR-224-5p | liver cancer | MMP9 | "iii miR-224 promoted expression of the tumor invas ......" | 24219032 |
| hsa-miR-224-5p | colorectal cancer | PHLPP1 | "microRNA 224 promotes cell proliferation and tumor ......" | 23846336 |
| hsa-miR-224-5p | esophageal cancer | PHLPP1 | "miR-224 expression was associated with advanced cl ......" | 26245343 |
| hsa-miR-224-5p | colorectal cancer | PHLPP2 | "microRNA 224 promotes cell proliferation and tumor ......" | 23846336 |
| hsa-miR-224-5p | esophageal cancer | PHLPP2 | "miR-224 expression was associated with advanced cl ......" | 26245343 |
| hsa-miR-224-5p | lung cancer | TNFAIP1 | "We further demonstrated that miR-224 functions as ......" | 26307684 |
| hsa-miR-224-5p | lung squamous cell cancer | TNFAIP1 | "Increased miR-224 expression promotes cell migrati ......" | 26187928 |
| hsa-miR-224-5p | prostate cancer | TPD52 | "miR-224 functions as a tumour suppressor by target ......" | 27070713 |
| hsa-miR-224-5p | prostate cancer | TPD52 | "Tumour suppressive microRNA 224 inhibits cancer ce ......" | 24768995 |
| hsa-miR-224-5p | prostate cancer | APLN | "In the current study apelin APLN was identified as ......" | 25532941 |
| hsa-miR-224-5p | gastric cancer | BCL2 | "In addition the expressions of Bcl2 mRNA and prote ......" | 24796455 |
| hsa-miR-224-5p | colorectal cancer | BRAF | "5-FU chemosensitivity was significantly increased ......" | 25919696 |
| hsa-miR-224-5p | prostate cancer | CAMKK2 | "MicroRNA 224 and its target CAMKK2 synergistically ......" | 25394900 |
| hsa-miR-224-5p | lung cancer | CASP3 | "MicroRNA 224 is implicated in lung cancer pathogen ......" | 26307684 |
| hsa-miR-224-5p | lung cancer | CASP7 | "MicroRNA 224 is implicated in lung cancer pathogen ......" | 26307684 |
| hsa-miR-224-5p | liver cancer | CCNE1 | "We next identified p21 p15 and CCNE1 as downstream ......" | 26343737 |
| hsa-miR-224-5p | B cell lymphoma | CD59 | "Deregulated expression of miR 224 and its target g ......" | 25146331 |
| hsa-miR-224-5p | colorectal cancer | CDC42 | "MicroRNA 224 suppresses colorectal cancer cell mig ......" | 24817781 |
| hsa-miR-224-5p | colorectal cancer | CDKN1A | "Interestingly cell-unique downregulation of miR-22 ......" | 26634414 |
| hsa-miR-224-5p | lung cancer | CDKN3 | "p21WAF1/CIP1 a potent cyclin-dependent kinase inhi ......" | 24921914 |
| hsa-miR-224-5p | liver cancer | CHRFAM7A | "miR 224 promotion of cell migration and invasion b ......" | 24219032 |
| hsa-miR-224-5p | B cell lymphoma | DDIT3 | "Deregulated expression of miR 224 and its target g ......" | 25146331 |
| hsa-miR-224-5p | liver cancer | DLL1 | "2-Delta Delta CT method was used for quantitative ......" | 19403413 |
| hsa-miR-224-5p | liver cancer | EP300 | "Notably in HCC tumors that significantly overexpre ......" | 22459148 |
| hsa-miR-224-5p | breast cancer | FUT4 | "Increased fucosylation has a pivotal role in multi ......" | 26701615 |
| hsa-miR-224-5p | breast cancer | FZD1 | "Collectively these data indicated that miR-224 dow ......" | 27323393 |
| hsa-miR-224-5p | breast cancer | FZD5 | "Collectively these data indicated that miR-224 dow ......" | 27323393 |
| hsa-miR-224-5p | liver cancer | HOXD10 | "A luciferase reporter assay was used to confirm th ......" | 24219032 |
| hsa-miR-224-5p | kidney renal cell cancer | KLK1 | "Transfection of miR-224 into HEK-293 cells resulte ......" | 20180642 |
| hsa-miR-224-5p | ovarian cancer | KLK10 | "When we co-transfected cells with pMIR-KLK10 and e ......" | 20354523 |
| hsa-miR-224-5p | ovarian cancer | KLLN | "Upregulated microRNA 224 promotes ovarian cancer c ......" | 27663866 |
| hsa-miR-224-5p | colorectal cancer | KRAS | "MicroRNA 224 is associated with colorectal cancer ......" | 25919696 |
| hsa-miR-224-5p | colorectal cancer | MBD2 | "Decreased levels of miR 224 and the passenger stra ......" | 23770133 |
| hsa-miR-224-5p | liver cancer | MMP2 | "We also performed real-time PCR to measure miR-224 ......" | 23988648 |
| hsa-miR-224-5p | gastric cancer | MTOR | "MicroRNA 224 aggrevates tumor growth and progressi ......" | 27315344 |
| hsa-miR-224-5p | colon cancer | MTX1 | "The underexpression of miR-224 was also observed i ......" | 21864507 |
| hsa-miR-224-5p | lung cancer | PARP1 | "In addition miR-224 attenuated TNF-α induced apop ......" | 26307684 |
| hsa-miR-224-5p | liver cancer | PPP2R1B | "These data suggest that miR-224 plays a significan ......" | 23741247 |
| hsa-miR-224-5p | cervical and endocervical cancer | PTGIS | "Bioinformatic analysis indicated that RASSF8 RAS-a ......" | 27626930 |
| hsa-miR-224-5p | sarcoma | RAC1 | "MicroRNA 224 promotes the sensitivity of osteosarc ......" | 27222381 |
| hsa-miR-224-5p | breast cancer | RAF1 | "Epigenetic changes and nuclear factor κB activati ......" | 26788213 |
| hsa-miR-224-5p | cervical and endocervical cancer | RASSF8 | "Over Expressed miR 224 Promotes the Progression of ......" | 27626930 |
| hsa-miR-224-5p | cervical and endocervical cancer | RB1CC1 | "miR 224 3p inhibits autophagy in cervical cancer c ......" | 27615604 |
| hsa-miR-224-5p | colorectal cancer | SERPINB5 | "Decreased levels of miR 224 and the passenger stra ......" | 23770133 |
| hsa-miR-224-5p | colorectal cancer | SFRP2 | "Bioinformatics analysis combined with in vivo and ......" | 26822534 |
| hsa-miR-224-5p | lung cancer | SSB | "The aim of our study was to investigate the roles ......" | 24921914 |
| hsa-miR-224-5p | lung cancer | TIMM8A | "Here we showed that miR-224 could promote the in v ......" | 24921914 |
| hsa-miR-224-5p | prostate cancer | TRIB1 | "MicroRNA 224 inhibits progression of human prostat ......" | 24382668 |
| hsa-miR-224-5p | kidney renal cell cancer | VHL | "Importantly both VHL and the hypoxia-inducible fac ......" | 22326755 |
Expression profile in cancer corhorts:

Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-224-5p | CXCR4 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; THCA; STAD | miRNAWalker2 validate; miRTarBase | TCGA BLCA -0.132; TCGA CESC -0.192; TCGA COAD -0.411; TCGA ESCA -0.168; TCGA HNSC -0.204; TCGA LUAD -0.055; TCGA LUSC -0.285; TCGA THCA -0.146; TCGA STAD -0.094 |
hsa-miR-224-5p | AFF3 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.326; TCGA BRCA -0.489; TCGA CESC -0.271; TCGA COAD -0.367; TCGA ESCA -0.299; TCGA HNSC -0.434; TCGA LUAD -0.248; TCGA LUSC -0.421; TCGA PAAD -0.365; TCGA PRAD -0.198; TCGA SARC -0.183; TCGA STAD -0.424; TCGA UCEC -0.196 |
hsa-miR-224-5p | GK | 10 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; THCA | MirTarget | TCGA BLCA -0.053; TCGA ESCA -0.084; TCGA HNSC -0.071; TCGA KIRC -0.301; TCGA KIRP -0.14; TCGA LIHC -0.072; TCGA LUAD -0.075; TCGA LUSC -0.103; TCGA PRAD -0.057; TCGA THCA -0.178 |
hsa-miR-224-5p | RALGPS1 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; PAAD; PRAD; THCA | MirTarget | TCGA BLCA -0.091; TCGA BRCA -0.154; TCGA CESC -0.101; TCGA ESCA -0.302; TCGA HNSC -0.232; TCGA KIRC -0.553; TCGA KIRP -0.246; TCGA PAAD -0.166; TCGA PRAD -0.052; TCGA THCA -0.123 |
hsa-miR-224-5p | FAM46C | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.167; TCGA BRCA -0.123; TCGA CESC -0.223; TCGA COAD -0.242; TCGA ESCA -0.398; TCGA HNSC -0.383; TCGA KIRC -0.245; TCGA LIHC -0.148; TCGA LUAD -0.113; TCGA LUSC -0.336; TCGA PAAD -0.327; TCGA SARC -0.16; TCGA THCA -0.265; TCGA STAD -0.187; TCGA UCEC -0.204 |
hsa-miR-224-5p | SH3BP5 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUSC; THCA; STAD | MirTarget | TCGA BLCA -0.103; TCGA CESC -0.223; TCGA COAD -0.155; TCGA ESCA -0.099; TCGA HNSC -0.363; TCGA LIHC -0.085; TCGA LUSC -0.285; TCGA THCA -0.184; TCGA STAD -0.076 |
hsa-miR-224-5p | TXNIP | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.073; TCGA COAD -0.224; TCGA ESCA -0.185; TCGA HNSC -0.225; TCGA KIRC -0.082; TCGA KIRP -0.112; TCGA LIHC -0.078; TCGA LUSC -0.225; TCGA STAD -0.245; TCGA UCEC -0.101 |
hsa-miR-224-5p | KCTD12 | 11 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; STAD | MirTarget | TCGA BLCA -0.106; TCGA COAD -0.377; TCGA ESCA -0.112; TCGA HNSC -0.188; TCGA KIRC -0.098; TCGA KIRP -0.174; TCGA LIHC -0.068; TCGA LUAD -0.057; TCGA LUSC -0.267; TCGA PAAD -0.126; TCGA STAD -0.183 |
hsa-miR-224-5p | SLC4A4 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUSC; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.165; TCGA CESC -0.5; TCGA COAD -0.599; TCGA ESCA -0.849; TCGA HNSC -0.587; TCGA KIRC -0.095; TCGA KIRP -0.117; TCGA LIHC -0.25; TCGA LUSC -0.491; TCGA PRAD -0.233; TCGA THCA -0.912; TCGA STAD -0.184 |
hsa-miR-224-5p | CBX7 | 11 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.134; TCGA COAD -0.113; TCGA ESCA -0.097; TCGA HNSC -0.203; TCGA KIRC -0.118; TCGA KIRP -0.091; TCGA LUSC -0.097; TCGA PAAD -0.272; TCGA SARC -0.174; TCGA STAD -0.236; TCGA UCEC -0.107 |
hsa-miR-224-5p | RNF38 | 9 cancers: BLCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PRAD; SARC | MirTarget | TCGA BLCA -0.08; TCGA HNSC -0.093; TCGA KIRC -0.11; TCGA KIRP -0.063; TCGA LUAD -0.053; TCGA LUSC -0.055; TCGA OV -0.054; TCGA PRAD -0.062; TCGA SARC -0.131 |
hsa-miR-224-5p | PJA2 | 11 cancers: BLCA; CESC; COAD; HNSC; LIHC; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.056; TCGA CESC -0.065; TCGA COAD -0.158; TCGA HNSC -0.087; TCGA LIHC -0.054; TCGA LUSC -0.087; TCGA PAAD -0.089; TCGA PRAD -0.058; TCGA SARC -0.062; TCGA STAD -0.139; TCGA UCEC -0.068 |
hsa-miR-224-5p | ACAT1 | 15 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.067; TCGA CESC -0.088; TCGA ESCA -0.119; TCGA HNSC -0.178; TCGA KIRC -0.385; TCGA KIRP -0.244; TCGA LIHC -0.135; TCGA LUAD -0.083; TCGA LUSC -0.081; TCGA PAAD -0.234; TCGA PRAD -0.054; TCGA SARC -0.104; TCGA THCA -0.191; TCGA STAD -0.06; TCGA UCEC -0.099 |
hsa-miR-224-5p | DENND5B | 12 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; SARC; THCA | MirTarget | TCGA BLCA -0.158; TCGA CESC -0.226; TCGA ESCA -0.404; TCGA HNSC -0.321; TCGA KIRP -0.057; TCGA LIHC -0.072; TCGA LUAD -0.083; TCGA LUSC -0.135; TCGA OV -0.089; TCGA PAAD -0.207; TCGA SARC -0.103; TCGA THCA -0.138 |
hsa-miR-224-5p | RELN | 14 cancers: BLCA; CESC; COAD; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.406; TCGA CESC -0.324; TCGA COAD -0.41; TCGA HNSC -0.406; TCGA KIRC -0.472; TCGA KIRP -0.792; TCGA LIHC -0.271; TCGA LUAD -0.235; TCGA LUSC -0.477; TCGA PAAD -0.52; TCGA PRAD -0.246; TCGA SARC -0.258; TCGA THCA -0.891; TCGA STAD -0.339 |
hsa-miR-224-5p | EFR3B | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; OV; PAAD | MirTarget | TCGA BLCA -0.13; TCGA BRCA -0.166; TCGA CESC -0.365; TCGA ESCA -0.086; TCGA HNSC -0.133; TCGA KIRC -0.165; TCGA LUAD -0.115; TCGA LUSC -0.307; TCGA OV -0.225; TCGA PAAD -0.546 |
hsa-miR-224-5p | SLMAP | 9 cancers: BLCA; COAD; ESCA; KIRC; KIRP; SARC; THCA; STAD; UCEC | MirTarget; mirMAP | TCGA BLCA -0.092; TCGA COAD -0.106; TCGA ESCA -0.095; TCGA KIRC -0.168; TCGA KIRP -0.067; TCGA SARC -0.267; TCGA THCA -0.198; TCGA STAD -0.169; TCGA UCEC -0.069 |
hsa-miR-224-5p | PCDH10 | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LUSC; OV; PAAD; SARC; STAD | MirTarget | TCGA BLCA -0.183; TCGA BRCA -0.143; TCGA CESC -0.204; TCGA HNSC -0.295; TCGA KIRP -0.635; TCGA LUSC -0.28; TCGA OV -0.223; TCGA PAAD -0.416; TCGA SARC -0.352; TCGA STAD -0.6 |
hsa-miR-224-5p | DTNA | 11 cancers: BLCA; CESC; COAD; HNSC; KIRP; LUAD; LUSC; OV; PAAD; SARC; STAD | MirTarget | TCGA BLCA -0.218; TCGA CESC -0.221; TCGA COAD -0.289; TCGA HNSC -0.394; TCGA KIRP -0.307; TCGA LUAD -0.109; TCGA LUSC -0.113; TCGA OV -0.269; TCGA PAAD -0.442; TCGA SARC -0.342; TCGA STAD -0.26 |
hsa-miR-224-5p | PACRG | 10 cancers: BLCA; CESC; KIRC; KIRP; LIHC; LUSC; OV; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.211; TCGA CESC -0.31; TCGA KIRC -0.651; TCGA KIRP -0.364; TCGA LIHC -0.255; TCGA LUSC -0.234; TCGA OV -0.266; TCGA PAAD -0.54; TCGA STAD -0.365; TCGA UCEC -0.156 |
hsa-miR-224-5p | NPR1 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; STAD | MirTarget | TCGA BLCA -0.165; TCGA CESC -0.187; TCGA COAD -0.286; TCGA ESCA -0.233; TCGA HNSC -0.275; TCGA KIRC -0.078; TCGA KIRP -0.265; TCGA LIHC -0.111; TCGA LUAD -0.088; TCGA LUSC -0.455; TCGA PAAD -0.248; TCGA STAD -0.193 |
hsa-miR-224-5p | PDGFRA | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LIHC; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.183; TCGA CESC -0.181; TCGA ESCA -0.202; TCGA HNSC -0.292; TCGA KIRC -0.561; TCGA LIHC -0.144; TCGA LUSC -0.251; TCGA STAD -0.076; TCGA UCEC -0.137 |
hsa-miR-224-5p | ITPR1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.177; TCGA BRCA -0.112; TCGA CESC -0.094; TCGA COAD -0.261; TCGA ESCA -0.141; TCGA HNSC -0.257; TCGA KIRC -0.167; TCGA LUAD -0.092; TCGA LUSC -0.222; TCGA PAAD -0.22; TCGA SARC -0.401; TCGA THCA -0.568; TCGA STAD -0.22; TCGA UCEC -0.154 |
hsa-miR-224-5p | ADAM22 | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRC; LUSC; PAAD; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.149; TCGA BRCA -0.058; TCGA CESC -0.21; TCGA HNSC -0.397; TCGA KIRC -0.102; TCGA LUSC -0.298; TCGA PAAD -0.204; TCGA SARC -0.19; TCGA THCA -0.774; TCGA STAD -0.138 |
hsa-miR-224-5p | CCDC141 | 11 cancers: BLCA; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget; mirMAP | TCGA BLCA -0.16; TCGA ESCA -0.214; TCGA HNSC -0.441; TCGA LIHC -0.104; TCGA LUAD -0.127; TCGA LUSC -0.483; TCGA PAAD -0.246; TCGA PRAD -0.228; TCGA THCA -0.343; TCGA STAD -0.113; TCGA UCEC -0.163 |
hsa-miR-224-5p | SNX29 | 9 cancers: BLCA; COAD; HNSC; KIRC; KIRP; LUSC; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.062; TCGA COAD -0.13; TCGA HNSC -0.089; TCGA KIRC -0.113; TCGA KIRP -0.085; TCGA LUSC -0.077; TCGA PRAD -0.07; TCGA THCA -0.104; TCGA STAD -0.099 |
hsa-miR-224-5p | RUNX1T1 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.316; TCGA CESC -0.264; TCGA COAD -0.242; TCGA ESCA -0.187; TCGA HNSC -0.447; TCGA LUAD -0.112; TCGA LUSC -0.44; TCGA THCA -0.299; TCGA STAD -0.265; TCGA UCEC -0.172 |
hsa-miR-224-5p | SLC1A2 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUSC; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.122; TCGA BRCA -0.254; TCGA CESC -0.328; TCGA COAD -0.193; TCGA ESCA -0.246; TCGA HNSC -0.429; TCGA KIRC -0.081; TCGA LIHC -0.326; TCGA LUSC -0.115; TCGA PAAD -0.405; TCGA THCA -0.518; TCGA STAD -0.189; TCGA UCEC -0.097 |
hsa-miR-224-5p | BVES | 10 cancers: BLCA; CESC; COAD; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; STAD | mirMAP | TCGA BLCA -0.217; TCGA CESC -0.254; TCGA COAD -0.331; TCGA HNSC -0.211; TCGA KIRC -0.087; TCGA LIHC -0.093; TCGA LUAD -0.079; TCGA LUSC -0.183; TCGA PAAD -0.182; TCGA STAD -0.259 |
hsa-miR-224-5p | IGFBP5 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; OV; STAD | mirMAP | TCGA BLCA -0.226; TCGA CESC -0.392; TCGA COAD -0.26; TCGA ESCA -0.095; TCGA HNSC -0.439; TCGA KIRC -0.189; TCGA LIHC -0.144; TCGA LUAD -0.102; TCGA OV -0.213; TCGA STAD -0.209 |
hsa-miR-224-5p | PAIP2B | 10 cancers: BLCA; HNSC; KIRC; KIRP; LIHC; LUAD; OV; PAAD; PRAD; THCA | mirMAP | TCGA BLCA -0.051; TCGA HNSC -0.121; TCGA KIRC -0.354; TCGA KIRP -0.178; TCGA LIHC -0.259; TCGA LUAD -0.132; TCGA OV -0.095; TCGA PAAD -0.231; TCGA PRAD -0.241; TCGA THCA -0.325 |
hsa-miR-224-5p | THRA | 10 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LUSC; OV; PAAD; SARC | mirMAP | TCGA BLCA -0.063; TCGA CESC -0.114; TCGA ESCA -0.218; TCGA HNSC -0.166; TCGA KIRC -0.152; TCGA KIRP -0.138; TCGA LUSC -0.1; TCGA OV -0.074; TCGA PAAD -0.161; TCGA SARC -0.168 |
hsa-miR-224-5p | AIF1L | 10 cancers: BLCA; COAD; HNSC; KIRC; KIRP; LUSC; OV; PAAD; PRAD; THCA | mirMAP | TCGA BLCA -0.128; TCGA COAD -0.45; TCGA HNSC -0.12; TCGA KIRC -0.746; TCGA KIRP -0.293; TCGA LUSC -0.091; TCGA OV -0.063; TCGA PAAD -0.225; TCGA PRAD -0.074; TCGA THCA -0.237 |
hsa-miR-224-5p | PGPEP1 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUSC; OV; PAAD; SARC | mirMAP | TCGA BLCA -0.054; TCGA BRCA -0.123; TCGA CESC -0.172; TCGA ESCA -0.317; TCGA HNSC -0.256; TCGA KIRC -0.114; TCGA KIRP -0.07; TCGA LUSC -0.089; TCGA OV -0.054; TCGA PAAD -0.146; TCGA SARC -0.091 |
hsa-miR-224-5p | KIAA1614 | 11 cancers: BLCA; CESC; COAD; HNSC; KIRC; KIRP; LUSC; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.168; TCGA CESC -0.173; TCGA COAD -0.105; TCGA HNSC -0.14; TCGA KIRC -0.083; TCGA KIRP -0.204; TCGA LUSC -0.062; TCGA PAAD -0.138; TCGA THCA -0.28; TCGA STAD -0.123; TCGA UCEC -0.105 |
hsa-miR-224-5p | ADCYAP1 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.272; TCGA CESC -0.327; TCGA COAD -0.504; TCGA ESCA -0.152; TCGA HNSC -0.183; TCGA KIRC -0.152; TCGA KIRP -0.167; TCGA LIHC -0.109; TCGA LUAD -0.139; TCGA LUSC -0.317; TCGA PAAD -0.573; TCGA STAD -0.413; TCGA UCEC -0.144 |
hsa-miR-224-5p | LRRC27 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LUSC; OV; PAAD; UCEC | mirMAP | TCGA BLCA -0.074; TCGA BRCA -0.118; TCGA CESC -0.136; TCGA ESCA -0.184; TCGA HNSC -0.194; TCGA KIRP -0.082; TCGA LUSC -0.177; TCGA OV -0.07; TCGA PAAD -0.12; TCGA UCEC -0.099 |
hsa-miR-224-5p | LIX1L | 9 cancers: BLCA; CESC; COAD; HNSC; LUSC; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.095; TCGA CESC -0.139; TCGA COAD -0.308; TCGA HNSC -0.153; TCGA LUSC -0.179; TCGA PAAD -0.096; TCGA THCA -0.106; TCGA STAD -0.195; TCGA UCEC -0.096 |
hsa-miR-224-5p | KCNE4 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.225; TCGA BRCA -0.302; TCGA CESC -0.31; TCGA COAD -0.293; TCGA ESCA -0.094; TCGA HNSC -0.119; TCGA LIHC -0.092; TCGA LUAD -0.212; TCGA LUSC -0.161; TCGA SARC -0.247; TCGA STAD -0.222; TCGA UCEC -0.194 |
hsa-miR-224-5p | GNAQ | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUSC; PRAD; STAD | mirMAP | TCGA BLCA -0.087; TCGA CESC -0.068; TCGA COAD -0.11; TCGA ESCA -0.134; TCGA HNSC -0.062; TCGA KIRP -0.082; TCGA LIHC -0.055; TCGA LUSC -0.135; TCGA PRAD -0.089; TCGA STAD -0.085 |
hsa-miR-224-5p | FYCO1 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.08; TCGA CESC -0.086; TCGA COAD -0.119; TCGA ESCA -0.137; TCGA HNSC -0.187; TCGA KIRC -0.147; TCGA KIRP -0.067; TCGA LUSC -0.117; TCGA SARC -0.202; TCGA THCA -0.052; TCGA STAD -0.153; TCGA UCEC -0.051 |
hsa-miR-224-5p | PARM1 | 10 cancers: BLCA; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.144; TCGA ESCA -0.252; TCGA HNSC -0.388; TCGA KIRC -0.232; TCGA LUAD -0.175; TCGA LUSC -0.272; TCGA PAAD -0.19; TCGA SARC -0.281; TCGA THCA -0.221; TCGA UCEC -0.141 |
hsa-miR-224-5p | LONRF2 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.421; TCGA BRCA -0.417; TCGA CESC -0.497; TCGA COAD -0.761; TCGA ESCA -0.253; TCGA HNSC -0.711; TCGA KIRC -0.617; TCGA LIHC -0.139; TCGA PAAD -0.553; TCGA PRAD -0.126; TCGA SARC -0.458; TCGA STAD -0.357; TCGA UCEC -0.123 |
hsa-miR-224-5p | BCL2 | 11 cancers: BLCA; BRCA; COAD; HNSC; KIRP; LUAD; OV; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.12; TCGA BRCA -0.241; TCGA COAD -0.302; TCGA HNSC -0.354; TCGA KIRP -0.206; TCGA LUAD -0.138; TCGA OV -0.071; TCGA PAAD -0.186; TCGA THCA -0.418; TCGA STAD -0.2; TCGA UCEC -0.101 |
hsa-miR-224-5p | GNG7 | 14 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUSC; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.214; TCGA CESC -0.154; TCGA COAD -0.226; TCGA ESCA -0.242; TCGA HNSC -0.415; TCGA KIRC -0.308; TCGA KIRP -0.143; TCGA LIHC -0.087; TCGA LUSC -0.363; TCGA PAAD -0.464; TCGA SARC -0.28; TCGA THCA -0.156; TCGA STAD -0.299; TCGA UCEC -0.155 |
hsa-miR-224-5p | ZNF154 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUSC; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.16; TCGA CESC -0.14; TCGA COAD -0.106; TCGA ESCA -0.17; TCGA HNSC -0.268; TCGA LUSC -0.209; TCGA PAAD -0.187; TCGA SARC -0.158; TCGA STAD -0.145 |
hsa-miR-224-5p | SFMBT2 | 9 cancers: BLCA; COAD; HNSC; LIHC; LUAD; LUSC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.138; TCGA COAD -0.377; TCGA HNSC -0.224; TCGA LIHC -0.107; TCGA LUAD -0.111; TCGA LUSC -0.189; TCGA THCA -0.301; TCGA STAD -0.165; TCGA UCEC -0.163 |
hsa-miR-224-5p | PCM1 | 13 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; OV; PAAD; STAD; UCEC | MirTarget | TCGA BRCA -0.066; TCGA CESC -0.093; TCGA COAD -0.092; TCGA ESCA -0.085; TCGA HNSC -0.171; TCGA KIRC -0.057; TCGA LIHC -0.086; TCGA LUAD -0.102; TCGA LUSC -0.12; TCGA OV -0.061; TCGA PAAD -0.117; TCGA STAD -0.061; TCGA UCEC -0.077 |
hsa-miR-224-5p | AMN1 | 9 cancers: BRCA; CESC; COAD; KIRC; OV; PAAD; PRAD; SARC; STAD | MirTarget | TCGA BRCA -0.111; TCGA CESC -0.076; TCGA COAD -0.109; TCGA KIRC -0.091; TCGA OV -0.074; TCGA PAAD -0.076; TCGA PRAD -0.058; TCGA SARC -0.117; TCGA STAD -0.05 |
hsa-miR-224-5p | DUSP16 | 9 cancers: BRCA; CESC; ESCA; HNSC; KIRC; LIHC; OV; PAAD; PRAD | MirTarget | TCGA BRCA -0.085; TCGA CESC -0.071; TCGA ESCA -0.083; TCGA HNSC -0.057; TCGA KIRC -0.081; TCGA LIHC -0.114; TCGA OV -0.099; TCGA PAAD -0.122; TCGA PRAD -0.11 |
hsa-miR-224-5p | ARMCX2 | 10 cancers: BRCA; CESC; COAD; HNSC; KIRC; LIHC; LUSC; PAAD; SARC; STAD | MirTarget | TCGA BRCA -0.11; TCGA CESC -0.212; TCGA COAD -0.21; TCGA HNSC -0.354; TCGA KIRC -0.122; TCGA LIHC -0.065; TCGA LUSC -0.125; TCGA PAAD -0.229; TCGA SARC -0.134; TCGA STAD -0.191 |
hsa-miR-224-5p | HS6ST3 | 10 cancers: BRCA; ESCA; KIRC; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.331; TCGA ESCA -0.225; TCGA KIRC -0.905; TCGA LUAD -0.122; TCGA PAAD -0.78; TCGA PRAD -0.243; TCGA SARC -0.323; TCGA THCA -0.594; TCGA STAD -0.346; TCGA UCEC -0.223 |
hsa-miR-224-5p | POLI | 12 cancers: BRCA; CESC; COAD; HNSC; KIRP; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.082; TCGA CESC -0.077; TCGA COAD -0.065; TCGA HNSC -0.111; TCGA KIRP -0.144; TCGA LUAD -0.068; TCGA PAAD -0.103; TCGA PRAD -0.06; TCGA SARC -0.126; TCGA THCA -0.149; TCGA STAD -0.07; TCGA UCEC -0.098 |
hsa-miR-224-5p | ZNF528 | 9 cancers: BRCA; CESC; HNSC; KIRP; LUSC; OV; PRAD; STAD; UCEC | MirTarget | TCGA BRCA -0.126; TCGA CESC -0.308; TCGA HNSC -0.218; TCGA KIRP -0.108; TCGA LUSC -0.193; TCGA OV -0.12; TCGA PRAD -0.065; TCGA STAD -0.09; TCGA UCEC -0.086 |
hsa-miR-224-5p | ZNF516 | 9 cancers: BRCA; CESC; COAD; LUAD; OV; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.159; TCGA CESC -0.093; TCGA COAD -0.171; TCGA LUAD -0.062; TCGA OV -0.116; TCGA PAAD -0.226; TCGA SARC -0.209; TCGA STAD -0.121; TCGA UCEC -0.122 |
hsa-miR-224-5p | CPEB4 | 10 cancers: BRCA; COAD; KIRC; LIHC; LUSC; PAAD; PRAD; SARC; THCA; UCEC | mirMAP | TCGA BRCA -0.06; TCGA COAD -0.114; TCGA KIRC -0.192; TCGA LIHC -0.094; TCGA LUSC -0.1; TCGA PAAD -0.107; TCGA PRAD -0.126; TCGA SARC -0.126; TCGA THCA -0.269; TCGA UCEC -0.053 |
hsa-miR-224-5p | ALDH6A1 | 12 cancers: BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.059; TCGA COAD -0.129; TCGA ESCA -0.117; TCGA HNSC -0.2; TCGA KIRC -0.682; TCGA KIRP -0.227; TCGA LIHC -0.242; TCGA PAAD -0.089; TCGA PRAD -0.238; TCGA THCA -0.245; TCGA STAD -0.096; TCGA UCEC -0.062 |
hsa-miR-224-5p | FBXL17 | 9 cancers: BRCA; HNSC; KIRC; KIRP; LUSC; PAAD; PRAD; THCA; STAD | mirMAP | TCGA BRCA -0.064; TCGA HNSC -0.111; TCGA KIRC -0.115; TCGA KIRP -0.058; TCGA LUSC -0.075; TCGA PAAD -0.141; TCGA PRAD -0.052; TCGA THCA -0.096; TCGA STAD -0.088 |
hsa-miR-224-5p | MSI2 | 9 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; PRAD; SARC | mirMAP | TCGA BRCA -0.163; TCGA CESC -0.122; TCGA ESCA -0.081; TCGA HNSC -0.07; TCGA KIRC -0.338; TCGA KIRP -0.118; TCGA LUAD -0.055; TCGA PRAD -0.109; TCGA SARC -0.203 |
hsa-miR-224-5p | ZSWIM5 | 9 cancers: BRCA; ESCA; KIRP; LUSC; OV; PAAD; PRAD; SARC; UCEC | mirMAP | TCGA BRCA -0.232; TCGA ESCA -0.277; TCGA KIRP -0.147; TCGA LUSC -0.131; TCGA OV -0.093; TCGA PAAD -0.259; TCGA PRAD -0.103; TCGA SARC -0.205; TCGA UCEC -0.158 |
hsa-miR-224-5p | TOM1L2 | 9 cancers: BRCA; COAD; KIRC; KIRP; OV; PAAD; SARC; THCA; UCEC | mirMAP | TCGA BRCA -0.058; TCGA COAD -0.093; TCGA KIRC -0.263; TCGA KIRP -0.109; TCGA OV -0.052; TCGA PAAD -0.159; TCGA SARC -0.088; TCGA THCA -0.056; TCGA UCEC -0.085 |
hsa-miR-224-5p | SLC35E2B | 9 cancers: BRCA; CESC; ESCA; HNSC; KIRP; LUSC; OV; PAAD; PRAD | mirMAP | TCGA BRCA -0.082; TCGA CESC -0.076; TCGA ESCA -0.111; TCGA HNSC -0.099; TCGA KIRP -0.085; TCGA LUSC -0.067; TCGA OV -0.069; TCGA PAAD -0.061; TCGA PRAD -0.058 |
hsa-miR-224-5p | KIAA0040 | 9 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUSC; STAD | mirMAP | TCGA BRCA -0.16; TCGA CESC -0.111; TCGA COAD -0.16; TCGA ESCA -0.105; TCGA HNSC -0.08; TCGA KIRC -0.092; TCGA LIHC -0.051; TCGA LUSC -0.229; TCGA STAD -0.054 |
hsa-miR-224-5p | PEBP1 | 10 cancers: CESC; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; THCA | miRNAWalker2 validate; miRTarBase | TCGA CESC -0.056; TCGA HNSC -0.08; TCGA KIRC -0.253; TCGA KIRP -0.093; TCGA LIHC -0.061; TCGA LUAD -0.072; TCGA LUSC -0.066; TCGA PAAD -0.184; TCGA SARC -0.127; TCGA THCA -0.066 |
hsa-miR-224-5p | ANKRD45 | 9 cancers: CESC; COAD; ESCA; KIRC; KIRP; LUSC; OV; PAAD; SARC | MirTarget | TCGA CESC -0.302; TCGA COAD -0.379; TCGA ESCA -0.196; TCGA KIRC -0.279; TCGA KIRP -0.224; TCGA LUSC -0.232; TCGA OV -0.249; TCGA PAAD -0.243; TCGA SARC -0.229 |
hsa-miR-224-5p | PPP2R1B | 9 cancers: CESC; ESCA; KIRC; LIHC; LUAD; PAAD; PRAD; THCA; UCEC | MirTarget | TCGA CESC -0.07; TCGA ESCA -0.07; TCGA KIRC -0.088; TCGA LIHC -0.071; TCGA LUAD -0.076; TCGA PAAD -0.052; TCGA PRAD -0.095; TCGA THCA -0.063; TCGA UCEC -0.072 |
hsa-miR-224-5p | TMEM47 | 11 cancers: CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; THCA; STAD; UCEC | MirTarget | TCGA CESC -0.288; TCGA COAD -0.171; TCGA ESCA -0.125; TCGA HNSC -0.34; TCGA KIRP -0.134; TCGA LIHC -0.155; TCGA LUAD -0.095; TCGA LUSC -0.278; TCGA THCA -0.104; TCGA STAD -0.225; TCGA UCEC -0.19 |
hsa-miR-224-5p | ZNF615 | 9 cancers: CESC; ESCA; HNSC; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA CESC -0.119; TCGA ESCA -0.128; TCGA HNSC -0.153; TCGA LUSC -0.108; TCGA OV -0.078; TCGA PAAD -0.111; TCGA PRAD -0.182; TCGA STAD -0.09; TCGA UCEC -0.071 |
hsa-miR-224-5p | ATP8A1 | 12 cancers: CESC; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA CESC -0.196; TCGA ESCA -0.503; TCGA HNSC -0.38; TCGA KIRC -0.083; TCGA KIRP -0.211; TCGA LUAD -0.14; TCGA LUSC -0.261; TCGA PAAD -0.301; TCGA PRAD -0.2; TCGA THCA -0.082; TCGA STAD -0.122; TCGA UCEC -0.119 |
hsa-miR-224-5p | MSRA | 9 cancers: CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUSC; SARC; UCEC | MirTarget | TCGA CESC -0.085; TCGA COAD -0.085; TCGA ESCA -0.214; TCGA HNSC -0.132; TCGA KIRC -0.269; TCGA LIHC -0.135; TCGA LUSC -0.236; TCGA SARC -0.139; TCGA UCEC -0.056 |
hsa-miR-224-5p | NCOR1 | 12 cancers: CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; OV; PRAD; THCA; UCEC | MirTarget | TCGA CESC -0.078; TCGA COAD -0.12; TCGA ESCA -0.107; TCGA HNSC -0.065; TCGA KIRC -0.127; TCGA KIRP -0.125; TCGA LIHC -0.112; TCGA LUAD -0.075; TCGA OV -0.067; TCGA PRAD -0.128; TCGA THCA -0.083; TCGA UCEC -0.056 |
hsa-miR-224-5p | UBE2J1 | 9 cancers: CESC; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | MirTarget | TCGA CESC -0.111; TCGA ESCA -0.131; TCGA HNSC -0.084; TCGA LUAD -0.054; TCGA LUSC -0.082; TCGA PAAD -0.064; TCGA PRAD -0.146; TCGA THCA -0.084; TCGA STAD -0.058 |
hsa-miR-224-5p | SETDB2 | 9 cancers: CESC; ESCA; HNSC; KIRC; KIRP; LUSC; PAAD; THCA; UCEC | MirTarget | TCGA CESC -0.073; TCGA ESCA -0.116; TCGA HNSC -0.094; TCGA KIRC -0.12; TCGA KIRP -0.115; TCGA LUSC -0.105; TCGA PAAD -0.123; TCGA THCA -0.116; TCGA UCEC -0.08 |
hsa-miR-224-5p | ABI2 | 11 cancers: CESC; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD | mirMAP | TCGA CESC -0.1; TCGA HNSC -0.122; TCGA KIRC -0.097; TCGA KIRP -0.056; TCGA LUAD -0.056; TCGA LUSC -0.087; TCGA OV -0.083; TCGA PAAD -0.109; TCGA PRAD -0.053; TCGA SARC -0.11; TCGA STAD -0.054 |
hsa-miR-224-5p | ZNF585B | 10 cancers: CESC; ESCA; HNSC; KIRC; KIRP; LUSC; OV; PRAD; THCA; STAD | mirMAP | TCGA CESC -0.206; TCGA ESCA -0.216; TCGA HNSC -0.149; TCGA KIRC -0.058; TCGA KIRP -0.126; TCGA LUSC -0.151; TCGA OV -0.111; TCGA PRAD -0.122; TCGA THCA -0.127; TCGA STAD -0.088 |
hsa-miR-224-5p | SMAD4 | 9 cancers: COAD; HNSC; KIRC; KIRP; LUAD; PAAD; PRAD; SARC; STAD | miRNAWalker2 validate; miRTarBase; MirTarget | TCGA COAD -0.162; TCGA HNSC -0.104; TCGA KIRC -0.079; TCGA KIRP -0.08; TCGA LUAD -0.09; TCGA PAAD -0.146; TCGA PRAD -0.051; TCGA SARC -0.055; TCGA STAD -0.084 |
Enriched cancer pathways of putative targets