microRNA information: hsa-miR-30a-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-30a-3p | miRbase |
| Accession: | MIMAT0000088 | miRbase |
| Precursor name: | hsa-mir-30a | miRbase |
| Precursor accession: | MI0000088 | miRbase |
| Symbol: | MIR30A | HGNC |
| RefSeq ID: | NR_029504 | GenBank |
| Sequence: | CUUUCAGUCGGAUGUUUGCAGC |
Reported expression in cancers: hsa-miR-30a-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-30a-3p | bladder cancer | downregulation | "We identified a subset of 7 miRNAs miR-145 miR-30a ......" | 19378336 | qPCR |
| hsa-miR-30a-3p | breast cancer | upregulation | "Then five up-regulated miRNAs miR-100 miR-29a miR- ......" | 23994196 | |
| hsa-miR-30a-3p | cervical and endocervical cancer | downregulation | "Has miR 30a regulates autophagic activity in cervi ......" | 26463633 | |
| hsa-miR-30a-3p | colorectal cancer | downregulation | "miRNA 30a miR-30a is a member of the miR-30 family ......" | 25582198 | |
| hsa-miR-30a-3p | colorectal cancer | downregulation | "Heterochromatin protein HP1γ promotes colorectal ......" | 26333808 | |
| hsa-miR-30a-3p | endometrial cancer | deregulation | "A set of EEC-associated miRNAs in tissue and plasm ......" | 24491411 | RNA-Seq |
| hsa-miR-30a-3p | esophageal cancer | downregulation | "A comparison of miRNA signatures from ESCC and our ......" | 21351259 | |
| hsa-miR-30a-3p | gastric cancer | upregulation | "The present study was designed to investigate the ......" | 27729002 | |
| hsa-miR-30a-3p | kidney renal cell cancer | downregulation | "We found that expression of miR-30a was significan ......" | 25712526 | |
| hsa-miR-30a-3p | liver cancer | downregulation | "MicroRNA 30a 3p inhibits tumor proliferation invas ......" | 24290372 | |
| hsa-miR-30a-3p | liver cancer | downregulation | "Effects of microRNA 30a on migration invasion and ......" | 24954667 | |
| hsa-miR-30a-3p | lung cancer | downregulation | "In the consistently reported down-regulated microR ......" | 22672859 | |
| hsa-miR-30a-3p | lung squamous cell cancer | downregulation | "Using a chicken embryonic metastasis assay we foun ......" | 21633953 | |
| hsa-miR-30a-3p | lung squamous cell cancer | downregulation | "Increasing evidences indicate that miR-30 expressi ......" | 25249344 | Reverse transcription PCR; qPCR |
| hsa-miR-30a-3p | lung squamous cell cancer | downregulation | "Expression profile analysis of microRNAs and downr ......" | 26238736 | Microarray |
| hsa-miR-30a-3p | pancreatic cancer | upregulation | "We focused on miR-30a -30b and -30c in the miR-30 ......" | 26965588 | |
| hsa-miR-30a-3p | prostate cancer | downregulation | "MiR-30a and miR-205 are two miRNAs downregulated i ......" | 27160121 |
Reported cancer pathway affected by hsa-miR-30a-3p
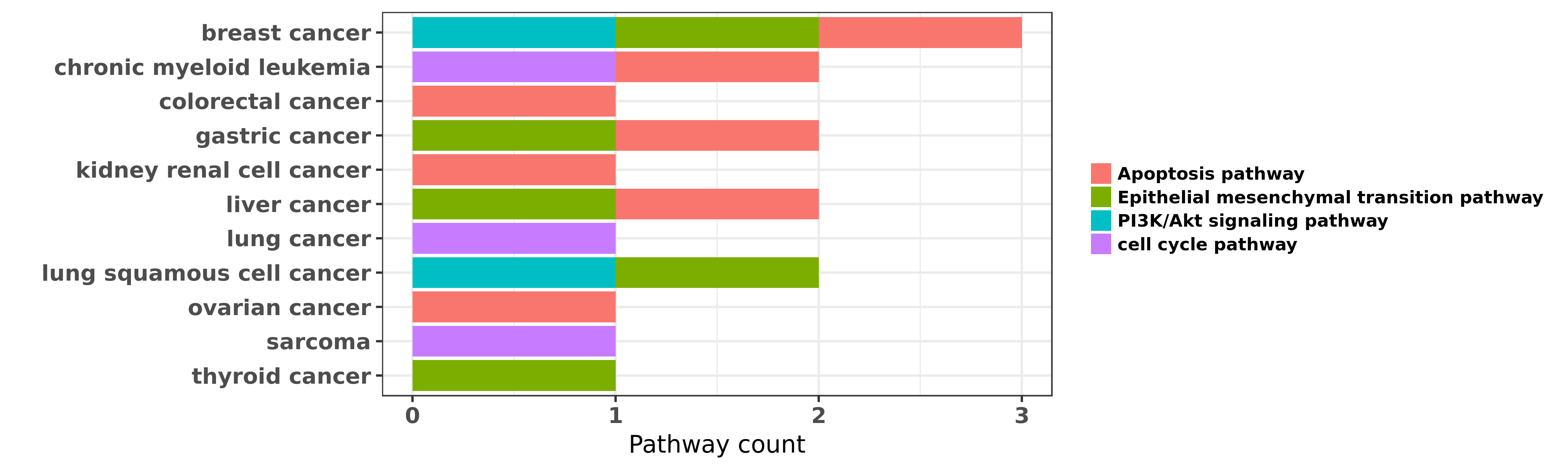
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-30a-3p | breast cancer | Apoptosis pathway | "Mir 30 reduction maintains self renewal and inhibi ......" | 20498642 | |
| hsa-miR-30a-3p | breast cancer | Epithelial mesenchymal transition pathway | "MicroRNA 30a inhibits cell migration and invasion ......" | 22476851 | Western blot; Luciferase |
| hsa-miR-30a-3p | breast cancer | Apoptosis pathway | "MicroRNA miR 30 family regulates non attachment gr ......" | 23445407 | |
| hsa-miR-30a-3p | breast cancer | PI3K/Akt signaling pathway | "Mutation R273H confers p53 a stimulating effect on ......" | 26898459 | |
| hsa-miR-30a-3p | breast cancer | Epithelial mesenchymal transition pathway | "MicroRNA 30a increases tight junction protein expr ......" | 26918943 | |
| hsa-miR-30a-3p | chronic myeloid leukemia | Apoptosis pathway | "Targeting microRNA 30a mediated autophagy enhances ......" | 22395361 | |
| hsa-miR-30a-3p | chronic myeloid leukemia | cell cycle pathway | "Decreased microRNA 30a levels are associated with ......" | 23287430 | |
| hsa-miR-30a-3p | colorectal cancer | Apoptosis pathway | "Association of Beclin 1 and microRNA 30a expressio ......" | 27173217 | |
| hsa-miR-30a-3p | gastric cancer | Epithelial mesenchymal transition pathway | "RUNX3 regulates vimentin expression via miR 30a du ......" | 24447545 | |
| hsa-miR-30a-3p | gastric cancer | Apoptosis pathway | "miR 30 functions as an oncomiR in gastric cancer c ......" | 27729002 | |
| hsa-miR-30a-3p | kidney renal cell cancer | Apoptosis pathway | "MiRNA-30a miR-30a is a potent inhibitor of autopha ......" | 25712526 | |
| hsa-miR-30a-3p | liver cancer | Apoptosis pathway | "MicroRNA 30a 3p inhibits tumor proliferation invas ......" | 24290372 | |
| hsa-miR-30a-3p | liver cancer | Epithelial mesenchymal transition pathway | "Effects of microRNA 30a on migration invasion and ......" | 24954667 | |
| hsa-miR-30a-3p | liver cancer | Apoptosis pathway | "Effect of miR 30a 5p on the proliferation apoptosi ......" | 25654285 | Flow cytometry; Colony formation |
| hsa-miR-30a-3p | liver cancer | Apoptosis pathway | "Overexpression of microRNA 30a 5p inhibits liver c ......" | 26589417 | Flow cytometry; Western blot; Luciferase |
| hsa-miR-30a-3p | lung cancer | cell cycle pathway | "Overexpression of microRNA miR 30a or miR 191 in A ......" | 20169152 | Colony formation |
| hsa-miR-30a-3p | lung cancer | cell cycle pathway | "Overexpression of miR 30a in lung adenocarcinoma A ......" | 26837415 | Flow cytometry; Cell proliferation assay; Cell Proliferation Assay; Wound Healing Assay; Luciferase |
| hsa-miR-30a-3p | lung squamous cell cancer | Epithelial mesenchymal transition pathway | "AEG 1 3' untranslated region functions as a ceRNA ......" | 25484183 | |
| hsa-miR-30a-3p | lung squamous cell cancer | PI3K/Akt signaling pathway | "MiR 30a suppresses non small cell lung cancer prog ......" | 26025408 | Luciferase |
| hsa-miR-30a-3p | ovarian cancer | Apoptosis pathway | "miR 30a inhibits endothelin A receptor and chemore ......" | 26675258 | |
| hsa-miR-30a-3p | sarcoma | cell cycle pathway | "The overexpression of miR 30a affects cell prolife ......" | 26596830 | Colony formation; Flow cytometry; Luciferase |
| hsa-miR-30a-3p | thyroid cancer | Epithelial mesenchymal transition pathway | "We identified two significantly decreased microRNA ......" | 20498632 |
Reported cancer prognosis affected by hsa-miR-30a-3p

| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-30a-3p | breast cancer | poor survival | "To investigate the global expression profile of mi ......" | 18812439 | |
| hsa-miR-30a-3p | breast cancer | staging | "These five miRNAs were measured in an independent ......" | 20490652 | |
| hsa-miR-30a-3p | breast cancer | metastasis; tumorigenesis | "Mir 30 reduction maintains self renewal and inhibi ......" | 20498642 | |
| hsa-miR-30a-3p | breast cancer | drug resistance | "MiRNA microarray analysis identified 299 and 226 m ......" | 21399894 | |
| hsa-miR-30a-3p | breast cancer | cell migration; metastasis; progression; poor survival; staging; recurrence | "MicroRNA 30a inhibits cell migration and invasion ......" | 22476851 | Western blot; Luciferase |
| hsa-miR-30a-3p | breast cancer | progression | "MicroRNA miR 30 family regulates non attachment gr ......" | 23445407 | |
| hsa-miR-30a-3p | breast cancer | metastasis | "MicroRNA 30a suppresses breast tumor growth and me ......" | 23851509 | Luciferase |
| hsa-miR-30a-3p | breast cancer | drug resistance | "In present study miRNA expression profiles of MCF- ......" | 23994196 | |
| hsa-miR-30a-3p | breast cancer | progression | "miR 30a suppresses breast cancer cell proliferatio ......" | 24508260 | |
| hsa-miR-30a-3p | breast cancer | recurrence | "Using microarray-based technology we have performe ......" | 24632820 | |
| hsa-miR-30a-3p | breast cancer | metastasis; malignant trasformation | "MicroRNA 30a increases tight junction protein expr ......" | 26918943 | |
| hsa-miR-30a-3p | chronic myeloid leukemia | progression | "Decreased microRNA 30a levels are associated with ......" | 23287430 | |
| hsa-miR-30a-3p | colorectal cancer | cell migration; metastasis | "miR 30a suppresses cell migration and invasion thr ......" | 23486085 | Luciferase |
| hsa-miR-30a-3p | colorectal cancer | tumorigenesis; staging | "Role of microRNA 30a targeting insulin receptor su ......" | 25582198 | Luciferase |
| hsa-miR-30a-3p | colorectal cancer | malignant trasformation | "MiR-30c belongs to miR-30 family and is involved i ......" | 25799050 | |
| hsa-miR-30a-3p | colorectal cancer | progression | "Heterochromatin protein HP1γ promotes colorectal ......" | 26333808 | |
| hsa-miR-30a-3p | colorectal cancer | drug resistance | "Association of Beclin 1 and microRNA 30a expressio ......" | 27173217 | |
| hsa-miR-30a-3p | colorectal cancer | metastasis | "MiR 30a 5p Suppresses Tumor Metastasis of Human Co ......" | 27576787 | Western blot; Luciferase; Transwell assay |
| hsa-miR-30a-3p | head and neck cancer | progression | "Alcohol dysregulated miR 30a and miR 934 in head a ......" | 26472042 | |
| hsa-miR-30a-3p | liver cancer | metastasis | "MicroRNA 30a 3p inhibits tumor proliferation invas ......" | 24290372 | |
| hsa-miR-30a-3p | liver cancer | worse prognosis; progression; poor survival; cell migration; metastasis | "Effects of microRNA 30a on migration invasion and ......" | 24954667 | |
| hsa-miR-30a-3p | lung cancer | cell migration | "Overexpression of microRNA miR 30a or miR 191 in A ......" | 20169152 | Colony formation |
| hsa-miR-30a-3p | lung cancer | worse prognosis; staging; metastasis; tumor size; poor survival | "Downregulation of MiR 30a is Associated with Poor ......" | 26305739 | |
| hsa-miR-30a-3p | lung cancer | progression; cell migration | "Overexpression of miR 30a in lung adenocarcinoma A ......" | 26837415 | Flow cytometry; Cell proliferation assay; Cell Proliferation Assay; Wound Healing Assay; Luciferase |
| hsa-miR-30a-3p | lung squamous cell cancer | metastasis | "MicroRNA 30a inhibits epithelial to mesenchymal tr ......" | 21633953 | |
| hsa-miR-30a-3p | lung squamous cell cancer | poor survival | "BCL11A overexpression predicts survival and relaps ......" | 23758992 | Western blot; Luciferase |
| hsa-miR-30a-3p | lung squamous cell cancer | tumorigenesis | "Increasing evidences indicate that miR-30 expressi ......" | 25249344 | Luciferase; Western blot; MTT assay |
| hsa-miR-30a-3p | lung squamous cell cancer | metastasis | "AEG 1 3' untranslated region functions as a ceRNA ......" | 25484183 | |
| hsa-miR-30a-3p | lung squamous cell cancer | progression | "MiR 30a suppresses non small cell lung cancer prog ......" | 26025408 | Luciferase |
| hsa-miR-30a-3p | ovarian cancer | poor survival | "The Cox proportional hazards model and the log-ran ......" | 21345725 | |
| hsa-miR-30a-3p | ovarian cancer | staging | "We measured the expression of nine miRNAs miR-181d ......" | 22925189 | |
| hsa-miR-30a-3p | ovarian cancer | drug resistance | "miR 30a inhibits endothelin A receptor and chemore ......" | 26675258 | |
| hsa-miR-30a-3p | sarcoma | worse prognosis | "Association of SOX4 regulated by tumor suppressor ......" | 25572678 | |
| hsa-miR-30a-3p | thyroid cancer | cell migration | "However specific miRNAs are downregulated in ATC s ......" | 25202329 |
Reported gene related to hsa-miR-30a-3p
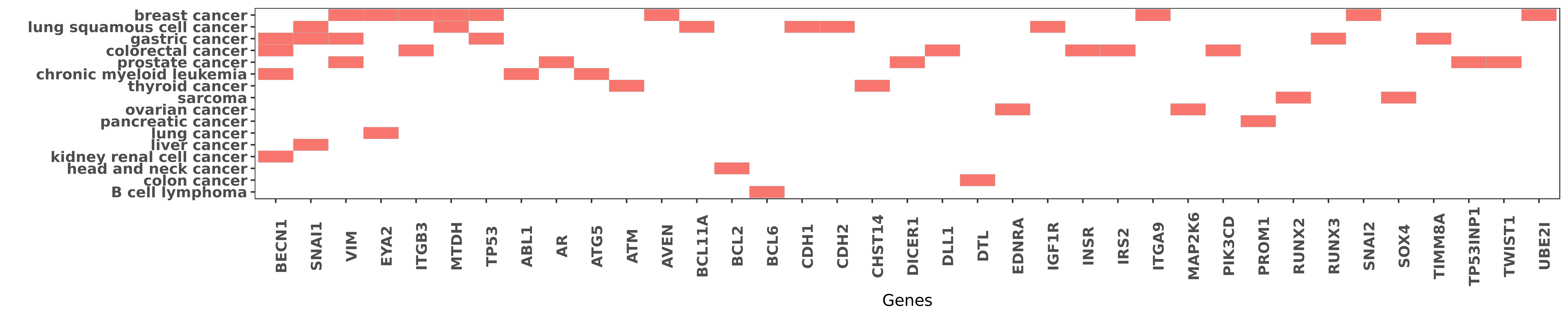
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-30a-3p | chronic myeloid leukemia | BECN1 | "miR-30a is a potent inhibitor of autophagy by down ......" | 22395361 |
| hsa-miR-30a-3p | colorectal cancer | BECN1 | "Association of Beclin 1 and microRNA 30a expressio ......" | 27173217 |
| hsa-miR-30a-3p | gastric cancer | BECN1 | "Fluorouracil induces autophagy related gastric car ......" | 26209295 |
| hsa-miR-30a-3p | kidney renal cell cancer | BECN1 | "MiRNA-30a miR-30a is a potent inhibitor of autopha ......" | 25712526 |
| hsa-miR-30a-3p | breast cancer | VIM | "MicroRNA 30a inhibits cell migration and invasion ......" | 22476851 |
| hsa-miR-30a-3p | gastric cancer | VIM | "RUNX3 regulates vimentin expression via miR 30a du ......" | 24447545 |
| hsa-miR-30a-3p | gastric cancer | VIM | "Knockdown of endogenous miR-30a promoted the elong ......" | 27212164 |
| hsa-miR-30a-3p | prostate cancer | VIM | "Furthermore low-expression levels of miR-30a and h ......" | 25409297 |
| hsa-miR-30a-3p | gastric cancer | SNAI1 | "Knockdown of endogenous miR-30a promoted the elong ......" | 27212164 |
| hsa-miR-30a-3p | liver cancer | SNAI1 | "We identified SNAI1 as a direct target of miR-30a ......" | 24954667 |
| hsa-miR-30a-3p | lung squamous cell cancer | SNAI1 | "MicroRNA 30a inhibits epithelial to mesenchymal tr ......" | 21633953 |
| hsa-miR-30a-3p | breast cancer | EYA2 | "miR 30a suppresses breast cancer cell proliferatio ......" | 24508260 |
| hsa-miR-30a-3p | lung cancer | EYA2 | "Overexpression of miR 30a in lung adenocarcinoma A ......" | 26837415 |
| hsa-miR-30a-3p | breast cancer | ITGB3 | "Enforced constitutive expression of mir-30 in BT-I ......" | 20498642 |
| hsa-miR-30a-3p | colorectal cancer | ITGB3 | "MiR 30a 5p Suppresses Tumor Metastasis of Human Co ......" | 27576787 |
| hsa-miR-30a-3p | breast cancer | MTDH | "MicroRNA 30a suppresses breast tumor growth and me ......" | 23851509 |
| hsa-miR-30a-3p | lung squamous cell cancer | MTDH | "AEG 1 3' untranslated region functions as a ceRNA ......" | 25484183 |
| hsa-miR-30a-3p | breast cancer | TP53 | "Mutation R273H confers p53 a stimulating effect on ......" | 26898459 |
| hsa-miR-30a-3p | gastric cancer | TP53 | "miR 30 functions as an oncomiR in gastric cancer c ......" | 27729002 |
| hsa-miR-30a-3p | chronic myeloid leukemia | ABL1 | "In K562 leukemia cells overexpression of miR-30a r ......" | 23287430 |
| hsa-miR-30a-3p | prostate cancer | AR | "Results uncovered miR-30 family members as direct ......" | 27683042 |
| hsa-miR-30a-3p | chronic myeloid leukemia | ATG5 | "miR-30a is a potent inhibitor of autophagy by down ......" | 22395361 |
| hsa-miR-30a-3p | thyroid cancer | ATM | "However specific miRNAs are downregulated in ATC s ......" | 25202329 |
| hsa-miR-30a-3p | breast cancer | AVEN | "At least one of these targets the anti-apoptotic p ......" | 23445407 |
| hsa-miR-30a-3p | lung squamous cell cancer | BCL11A | "BCL11A overexpression predicts survival and relaps ......" | 23758992 |
| hsa-miR-30a-3p | head and neck cancer | BCL2 | "Overexpression of miR-30a and miR-934 in normal an ......" | 26472042 |
| hsa-miR-30a-3p | B cell lymphoma | BCL6 | "It was also shown that the patients with BCL-6 pro ......" | 23450486 |
| hsa-miR-30a-3p | lung squamous cell cancer | CDH1 | "Consistent with this microRNA-30a expression was f ......" | 21633953 |
| hsa-miR-30a-3p | lung squamous cell cancer | CDH2 | "Consistent with this microRNA-30a expression was f ......" | 21633953 |
| hsa-miR-30a-3p | thyroid cancer | CHST14 | "We identified two significantly decreased microRNA ......" | 20498632 |
| hsa-miR-30a-3p | prostate cancer | DICER1 | "The expression change of miR-30a miR-205 and Dicer ......" | 27160121 |
| hsa-miR-30a-3p | colorectal cancer | DLL1 | "A luciferase reporter assay was performed to deter ......" | 23486085 |
| hsa-miR-30a-3p | colon cancer | DTL | "MiR 30a 5p suppresses tumor growth in colon carcin ......" | 22287560 |
| hsa-miR-30a-3p | ovarian cancer | EDNRA | "miR 30a inhibits endothelin A receptor and chemore ......" | 26675258 |
| hsa-miR-30a-3p | lung squamous cell cancer | IGF1R | "MiR 30a suppresses non small cell lung cancer prog ......" | 26025408 |
| hsa-miR-30a-3p | colorectal cancer | INSR | "Role of microRNA 30a targeting insulin receptor su ......" | 25582198 |
| hsa-miR-30a-3p | colorectal cancer | IRS2 | "Moreover bioinformatic algorithms and luciferase r ......" | 25582198 |
| hsa-miR-30a-3p | breast cancer | ITGA9 | "Downregulation of β3 integrin by miR 30a 5p modul ......" | 26781040 |
| hsa-miR-30a-3p | ovarian cancer | MAP2K6 | "Overexpression of miR-30a decreased Akt and mitoge ......" | 26675258 |
| hsa-miR-30a-3p | colorectal cancer | PIK3CD | "miR 30a suppresses cell migration and invasion thr ......" | 23486085 |
| hsa-miR-30a-3p | pancreatic cancer | PROM1 | "Using miRNA microarray we found that the expressio ......" | 26965588 |
| hsa-miR-30a-3p | sarcoma | RUNX2 | "The overexpression of miR 30a affects cell prolife ......" | 26596830 |
| hsa-miR-30a-3p | gastric cancer | RUNX3 | "RUNX3 regulates vimentin expression via miR 30a du ......" | 24447545 |
| hsa-miR-30a-3p | breast cancer | SNAI2 | "MicroRNA 30a increases tight junction protein expr ......" | 26918943 |
| hsa-miR-30a-3p | sarcoma | SOX4 | "Association of SOX4 regulated by tumor suppressor ......" | 25572678 |
| hsa-miR-30a-3p | gastric cancer | TIMM8A | "In this study we investigated the association betw ......" | 27212164 |
| hsa-miR-30a-3p | prostate cancer | TP53INP1 | "MiR 30a and miR 205 are downregulated in hypoxia a ......" | 27160121 |
| hsa-miR-30a-3p | prostate cancer | TWIST1 | "Furthermore low-expression levels of miR-30a and h ......" | 25409297 |
| hsa-miR-30a-3p | breast cancer | UBE2I | "Enforced constitutive expression of mir-30 in BT-I ......" | 20498642 |
Expression profile in cancer corhorts:
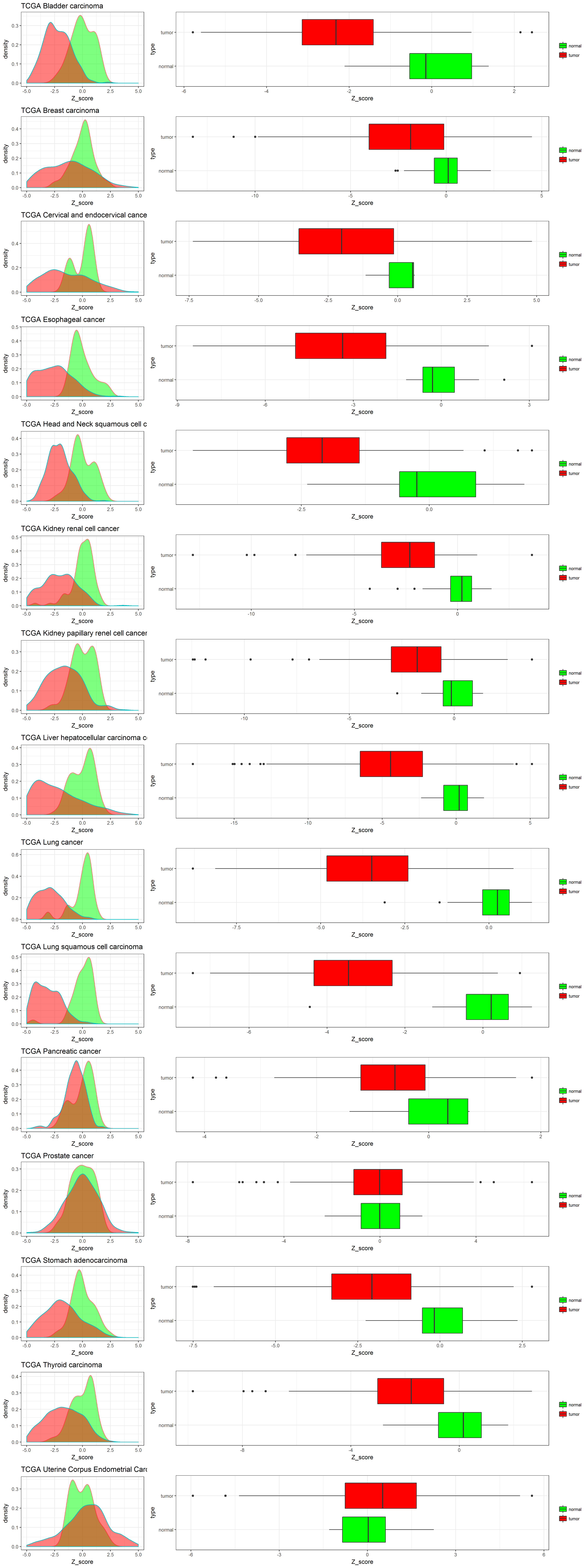
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-30a-3p | EXOSC2 | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.065; TCGA BRCA -0.061; TCGA COAD -0.257; TCGA ESCA -0.117; TCGA HNSC -0.069; TCGA KIRC -0.091; TCGA KIRP -0.058; TCGA LGG -0.182; TCGA LIHC -0.126; TCGA LUAD -0.118; TCGA LUSC -0.21; TCGA SARC -0.078; TCGA STAD -0.155; TCGA UCEC -0.057 |
hsa-miR-30a-3p | LRP8 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.214; TCGA BRCA -0.345; TCGA CESC -0.175; TCGA COAD -0.321; TCGA ESCA -0.37; TCGA HNSC -0.222; TCGA KIRC -0.273; TCGA KIRP -0.392; TCGA LGG -0.109; TCGA LUAD -0.177; TCGA LUSC -0.44; TCGA PRAD -0.103; TCGA SARC -0.191; TCGA STAD -0.538 |
hsa-miR-30a-3p | ANP32E | 9 cancers: BLCA; BRCA; ESCA; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.059; TCGA BRCA -0.096; TCGA ESCA -0.158; TCGA LIHC -0.069; TCGA LUAD -0.229; TCGA LUSC -0.202; TCGA PRAD -0.097; TCGA STAD -0.333; TCGA UCEC -0.17 |
hsa-miR-30a-3p | DVL3 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; PRAD | MirTarget | TCGA BLCA -0.106; TCGA BRCA -0.063; TCGA CESC -0.122; TCGA ESCA -0.112; TCGA HNSC -0.094; TCGA KIRC -0.084; TCGA LGG -0.101; TCGA LIHC -0.083; TCGA LUAD -0.113; TCGA LUSC -0.293; TCGA PRAD -0.133 |
hsa-miR-30a-3p | CACYBP | 14 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.093; TCGA BRCA -0.119; TCGA COAD -0.232; TCGA ESCA -0.16; TCGA KIRP -0.094; TCGA LGG -0.05; TCGA LIHC -0.221; TCGA LUAD -0.161; TCGA LUSC -0.224; TCGA PAAD -0.082; TCGA SARC -0.086; TCGA THCA -0.081; TCGA STAD -0.244; TCGA UCEC -0.189 |
hsa-miR-30a-3p | UNG | 9 cancers: BLCA; COAD; ESCA; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.072; TCGA COAD -0.244; TCGA ESCA -0.086; TCGA LIHC -0.061; TCGA LUAD -0.155; TCGA LUSC -0.295; TCGA PRAD -0.087; TCGA STAD -0.201; TCGA UCEC -0.108 |
hsa-miR-30a-3p | NKRF | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; OV; PAAD; STAD | MirTarget | TCGA BLCA -0.069; TCGA CESC -0.069; TCGA COAD -0.172; TCGA ESCA -0.159; TCGA HNSC -0.08; TCGA LGG -0.137; TCGA LUAD -0.09; TCGA LUSC -0.17; TCGA OV -0.067; TCGA PAAD -0.064; TCGA STAD -0.309 |
hsa-miR-30a-3p | BCL11B | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUSC; STAD | MirTarget; mirMAP | TCGA BLCA -0.173; TCGA BRCA -0.14; TCGA CESC -0.376; TCGA ESCA -0.463; TCGA HNSC -0.279; TCGA KIRC -0.639; TCGA KIRP -0.296; TCGA LGG -0.357; TCGA LUSC -0.299; TCGA STAD -0.266 |
hsa-miR-30a-3p | LUC7L3 | 9 cancers: BLCA; COAD; KIRC; LGG; LIHC; LUAD; LUSC; PRAD; STAD | MirTarget | TCGA BLCA -0.072; TCGA COAD -0.111; TCGA KIRC -0.106; TCGA LGG -0.181; TCGA LIHC -0.076; TCGA LUAD -0.137; TCGA LUSC -0.13; TCGA PRAD -0.073; TCGA STAD -0.187 |
hsa-miR-30a-3p | DONSON | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; SARC; THCA | MirTarget | TCGA BLCA -0.08; TCGA BRCA -0.252; TCGA COAD -0.208; TCGA ESCA -0.115; TCGA HNSC -0.067; TCGA KIRC -0.492; TCGA KIRP -0.189; TCGA LGG -0.07; TCGA LIHC -0.176; TCGA LUAD -0.289; TCGA LUSC -0.33; TCGA PRAD -0.296; TCGA SARC -0.122; TCGA THCA -0.27 |
hsa-miR-30a-3p | TTPAL | 9 cancers: BLCA; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.059; TCGA ESCA -0.114; TCGA HNSC -0.175; TCGA KIRC -0.095; TCGA LUAD -0.094; TCGA LUSC -0.163; TCGA PAAD -0.077; TCGA SARC -0.097; TCGA STAD -0.135 |
hsa-miR-30a-3p | CENPL | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.112; TCGA BRCA -0.147; TCGA CESC -0.083; TCGA COAD -0.157; TCGA ESCA -0.151; TCGA HNSC -0.068; TCGA KIRC -0.234; TCGA KIRP -0.136; TCGA LIHC -0.311; TCGA LUAD -0.22; TCGA LUSC -0.334; TCGA PRAD -0.181; TCGA SARC -0.103; TCGA THCA -0.061; TCGA STAD -0.324; TCGA UCEC -0.071 |
hsa-miR-30a-3p | CNPY2 | 11 cancers: BLCA; BRCA; COAD; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; STAD | MirTarget; miRNATAP | TCGA BLCA -0.083; TCGA BRCA -0.08; TCGA COAD -0.238; TCGA KIRC -0.176; TCGA KIRP -0.16; TCGA LGG -0.11; TCGA LIHC -0.172; TCGA LUAD -0.19; TCGA LUSC -0.107; TCGA PAAD -0.245; TCGA STAD -0.08 |
hsa-miR-30a-3p | TADA1 | 9 cancers: BLCA; CESC; COAD; ESCA; LGG; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.095; TCGA CESC -0.062; TCGA COAD -0.117; TCGA ESCA -0.061; TCGA LGG -0.128; TCGA LUAD -0.078; TCGA LUSC -0.15; TCGA STAD -0.13; TCGA UCEC -0.091 |
hsa-miR-30a-3p | CNIH4 | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; LUSC; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.075; TCGA BRCA -0.09; TCGA ESCA -0.107; TCGA HNSC -0.091; TCGA KIRC -0.096; TCGA LIHC -0.234; TCGA LUSC -0.062; TCGA PRAD -0.062; TCGA THCA -0.163; TCGA STAD -0.06 |
hsa-miR-30a-3p | GORAB | 10 cancers: BLCA; COAD; KIRC; KIRP; LUAD; LUSC; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.074; TCGA COAD -0.084; TCGA KIRC -0.153; TCGA KIRP -0.08; TCGA LUAD -0.064; TCGA LUSC -0.108; TCGA SARC -0.096; TCGA THCA -0.051; TCGA STAD -0.195; TCGA UCEC -0.114 |
hsa-miR-30a-3p | RBM45 | 11 cancers: BLCA; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.072; TCGA COAD -0.065; TCGA ESCA -0.106; TCGA HNSC -0.086; TCGA LGG -0.056; TCGA LIHC -0.063; TCGA LUAD -0.066; TCGA LUSC -0.09; TCGA SARC -0.058; TCGA STAD -0.103; TCGA UCEC -0.071 |
hsa-miR-30a-3p | GLO1 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.059; TCGA BRCA -0.065; TCGA COAD -0.151; TCGA ESCA -0.174; TCGA HNSC -0.13; TCGA LIHC -0.07; TCGA LUSC -0.051; TCGA PRAD -0.148; TCGA STAD -0.124; TCGA UCEC -0.055 |
hsa-miR-30a-3p | CASP2 | 11 cancers: BLCA; CESC; COAD; ESCA; KIRC; LGG; LIHC; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.112; TCGA CESC -0.07; TCGA COAD -0.094; TCGA ESCA -0.081; TCGA KIRC -0.158; TCGA LGG -0.084; TCGA LIHC -0.08; TCGA LUAD -0.079; TCGA LUSC -0.153; TCGA STAD -0.272; TCGA UCEC -0.064 |
hsa-miR-30a-3p | CEP152 | 9 cancers: BLCA; CESC; COAD; ESCA; LIHC; LUAD; LUSC; PRAD; STAD | MirTarget | TCGA BLCA -0.133; TCGA CESC -0.156; TCGA COAD -0.225; TCGA ESCA -0.15; TCGA LIHC -0.135; TCGA LUAD -0.165; TCGA LUSC -0.27; TCGA PRAD -0.196; TCGA STAD -0.335 |
hsa-miR-30a-3p | PPAT | 10 cancers: BLCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.066; TCGA COAD -0.211; TCGA ESCA -0.098; TCGA HNSC -0.074; TCGA LIHC -0.064; TCGA LUAD -0.281; TCGA LUSC -0.357; TCGA SARC -0.113; TCGA STAD -0.383; TCGA UCEC -0.093 |
hsa-miR-30a-3p | LIN9 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; LGG; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.143; TCGA BRCA -0.07; TCGA CESC -0.056; TCGA COAD -0.173; TCGA ESCA -0.144; TCGA LGG -0.243; TCGA LIHC -0.246; TCGA LUAD -0.124; TCGA LUSC -0.263; TCGA PRAD -0.156; TCGA STAD -0.365; TCGA UCEC -0.124 |
hsa-miR-30a-3p | PIN4 | 9 cancers: BLCA; BRCA; COAD; KIRP; LIHC; LUAD; LUSC; PAAD; UCEC | MirTarget | TCGA BLCA -0.091; TCGA BRCA -0.075; TCGA COAD -0.13; TCGA KIRP -0.083; TCGA LIHC -0.082; TCGA LUAD -0.098; TCGA LUSC -0.066; TCGA PAAD -0.103; TCGA UCEC -0.056 |
hsa-miR-30a-3p | TFDP1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; OV; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.091; TCGA BRCA -0.062; TCGA CESC -0.118; TCGA COAD -0.196; TCGA ESCA -0.215; TCGA HNSC -0.079; TCGA LGG -0.091; TCGA LUAD -0.075; TCGA LUSC -0.225; TCGA OV -0.087; TCGA PAAD -0.059; TCGA STAD -0.257; TCGA UCEC -0.083 |
hsa-miR-30a-3p | PDK1 | 13 cancers: BLCA; BRCA; COAD; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.13; TCGA BRCA -0.08; TCGA COAD -0.262; TCGA HNSC -0.12; TCGA KIRC -0.36; TCGA KIRP -0.213; TCGA LUAD -0.211; TCGA LUSC -0.346; TCGA PRAD -0.095; TCGA SARC -0.122; TCGA THCA -0.161; TCGA STAD -0.128; TCGA UCEC -0.17 |
hsa-miR-30a-3p | DARS2 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.106; TCGA BRCA -0.067; TCGA COAD -0.134; TCGA ESCA -0.163; TCGA HNSC -0.07; TCGA LIHC -0.133; TCGA LUAD -0.186; TCGA LUSC -0.265; TCGA PRAD -0.094; TCGA SARC -0.115; TCGA STAD -0.292; TCGA UCEC -0.083 |
hsa-miR-30a-3p | DTL | 17 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.177; TCGA BRCA -0.302; TCGA CESC -0.071; TCGA COAD -0.299; TCGA ESCA -0.259; TCGA HNSC -0.141; TCGA KIRC -0.848; TCGA KIRP -0.542; TCGA LGG -0.445; TCGA LIHC -0.532; TCGA LUAD -0.318; TCGA LUSC -0.521; TCGA PRAD -0.343; TCGA SARC -0.154; TCGA THCA -0.316; TCGA STAD -0.559; TCGA UCEC -0.16 |
hsa-miR-30a-3p | ILF2 | 13 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.075; TCGA BRCA -0.107; TCGA COAD -0.172; TCGA ESCA -0.146; TCGA KIRP -0.102; TCGA LGG -0.178; TCGA LIHC -0.16; TCGA LUAD -0.199; TCGA LUSC -0.196; TCGA PAAD -0.062; TCGA SARC -0.112; TCGA STAD -0.172; TCGA UCEC -0.077 |
hsa-miR-30a-3p | RNF149 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; THCA; STAD | MirTarget | TCGA BLCA -0.09; TCGA BRCA -0.068; TCGA CESC -0.089; TCGA ESCA -0.215; TCGA HNSC -0.199; TCGA KIRC -0.35; TCGA KIRP -0.215; TCGA LIHC -0.089; TCGA THCA -0.185; TCGA STAD -0.15 |
hsa-miR-30a-3p | RCOR2 | 9 cancers: BLCA; BRCA; COAD; ESCA; LGG; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.287; TCGA BRCA -0.333; TCGA COAD -0.456; TCGA ESCA -0.27; TCGA LGG -0.877; TCGA LUAD -0.579; TCGA LUSC -0.615; TCGA STAD -0.254; TCGA UCEC -0.209 |
hsa-miR-30a-3p | FAM104A | 11 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD | MirTarget; miRNATAP | TCGA BLCA -0.054; TCGA COAD -0.079; TCGA ESCA -0.09; TCGA HNSC -0.066; TCGA KIRC -0.203; TCGA KIRP -0.179; TCGA LGG -0.112; TCGA LIHC -0.085; TCGA LUAD -0.111; TCGA LUSC -0.113; TCGA PAAD -0.112 |
hsa-miR-30a-3p | DEPDC1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.224; TCGA BRCA -0.412; TCGA CESC -0.078; TCGA COAD -0.3; TCGA ESCA -0.305; TCGA HNSC -0.14; TCGA KIRC -1.156; TCGA KIRP -0.753; TCGA LIHC -0.629; TCGA LUAD -0.559; TCGA LUSC -0.791; TCGA PRAD -0.265; TCGA SARC -0.174; TCGA THCA -0.357; TCGA STAD -0.629 |
hsa-miR-30a-3p | UBA6 | 10 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; THCA; STAD | mirMAP | TCGA BLCA -0.082; TCGA CESC -0.143; TCGA ESCA -0.148; TCGA HNSC -0.146; TCGA KIRC -0.209; TCGA KIRP -0.122; TCGA LUAD -0.068; TCGA LUSC -0.19; TCGA THCA -0.095; TCGA STAD -0.302 |
hsa-miR-30a-3p | OTUB2 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; SARC | mirMAP | TCGA BLCA -0.135; TCGA BRCA -0.055; TCGA CESC -0.106; TCGA COAD -0.293; TCGA ESCA -0.168; TCGA HNSC -0.211; TCGA LGG -0.201; TCGA LIHC -0.214; TCGA LUAD -0.191; TCGA LUSC -0.187; TCGA SARC -0.138 |
hsa-miR-30a-3p | PTCD1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD | mirMAP | TCGA BLCA -0.085; TCGA BRCA -0.104; TCGA CESC -0.091; TCGA COAD -0.136; TCGA ESCA -0.084; TCGA HNSC -0.056; TCGA KIRC -0.08; TCGA LIHC -0.151; TCGA LUAD -0.169; TCGA LUSC -0.22; TCGA PAAD -0.1 |
hsa-miR-30a-3p | ONECUT2 | 9 cancers: BLCA; BRCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; STAD | mirMAP | TCGA BLCA -0.238; TCGA BRCA -0.663; TCGA HNSC -0.154; TCGA KIRC -0.34; TCGA LGG -0.659; TCGA LIHC -0.148; TCGA LUAD -0.191; TCGA LUSC -0.492; TCGA STAD -0.353 |
hsa-miR-30a-3p | ATAD2B | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.094; TCGA CESC -0.064; TCGA ESCA -0.097; TCGA HNSC -0.05; TCGA KIRC -0.09; TCGA LGG -0.121; TCGA LUAD -0.096; TCGA LUSC -0.138; TCGA PRAD -0.133; TCGA STAD -0.15; TCGA UCEC -0.093 |
hsa-miR-30a-3p | HELLS | 13 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.128; TCGA BRCA -0.198; TCGA COAD -0.325; TCGA ESCA -0.153; TCGA KIRP -0.165; TCGA LGG -0.454; TCGA LIHC -0.536; TCGA LUAD -0.398; TCGA LUSC -0.634; TCGA PRAD -0.309; TCGA SARC -0.172; TCGA THCA -0.325; TCGA STAD -0.574 |
hsa-miR-30a-3p | ODF2L | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LUAD; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.13; TCGA CESC -0.086; TCGA ESCA -0.122; TCGA HNSC -0.14; TCGA KIRC -0.141; TCGA LUAD -0.09; TCGA PRAD -0.156; TCGA THCA -0.13; TCGA STAD -0.186 |
hsa-miR-30a-3p | SOX4 | 10 cancers: BLCA; BRCA; ESCA; LGG; LUAD; LUSC; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.227; TCGA BRCA -0.064; TCGA ESCA -0.128; TCGA LGG -0.682; TCGA LUAD -0.238; TCGA LUSC -0.159; TCGA PRAD -0.116; TCGA THCA -0.63; TCGA STAD -0.348; TCGA UCEC -0.101 |
hsa-miR-30a-3p | SLC38A2 | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUSC; OV | mirMAP | TCGA BLCA -0.07; TCGA CESC -0.124; TCGA ESCA -0.124; TCGA HNSC -0.17; TCGA KIRC -0.208; TCGA KIRP -0.111; TCGA LGG -0.1; TCGA LUSC -0.105; TCGA OV -0.072 |
hsa-miR-30a-3p | FOXK2 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; STAD | mirMAP; miRNATAP | TCGA BLCA -0.054; TCGA BRCA -0.079; TCGA COAD -0.087; TCGA ESCA -0.114; TCGA HNSC -0.07; TCGA KIRP -0.109; TCGA LGG -0.134; TCGA LIHC -0.088; TCGA LUAD -0.139; TCGA LUSC -0.176; TCGA PAAD -0.083; TCGA STAD -0.066 |
hsa-miR-30a-3p | TANC2 | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LGG; LUAD; PRAD; THCA | mirMAP | TCGA BLCA -0.143; TCGA BRCA -0.069; TCGA ESCA -0.13; TCGA HNSC -0.125; TCGA KIRC -0.208; TCGA LGG -0.173; TCGA LUAD -0.079; TCGA PRAD -0.126; TCGA THCA -0.266 |
hsa-miR-30a-3p | LCLAT1 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; STAD | mirMAP | TCGA BLCA -0.097; TCGA BRCA -0.09; TCGA COAD -0.11; TCGA ESCA -0.083; TCGA HNSC -0.096; TCGA LGG -0.09; TCGA LUAD -0.129; TCGA LUSC -0.217; TCGA PAAD -0.11; TCGA STAD -0.288 |
hsa-miR-30a-3p | TBL1XR1 | 10 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; STAD | mirMAP | TCGA BLCA -0.093; TCGA CESC -0.079; TCGA ESCA -0.109; TCGA HNSC -0.093; TCGA KIRC -0.126; TCGA KIRP -0.091; TCGA LGG -0.11; TCGA LUAD -0.076; TCGA LUSC -0.273; TCGA STAD -0.188 |
hsa-miR-30a-3p | BRI3BP | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; STAD | mirMAP | TCGA BLCA -0.143; TCGA BRCA -0.146; TCGA COAD -0.44; TCGA ESCA -0.152; TCGA HNSC -0.092; TCGA LIHC -0.175; TCGA LUAD -0.253; TCGA LUSC -0.363; TCGA PAAD -0.155; TCGA STAD -0.382 |
hsa-miR-30a-3p | FAM111B | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.175; TCGA BRCA -0.317; TCGA CESC -0.107; TCGA COAD -0.195; TCGA ESCA -0.265; TCGA KIRC -0.785; TCGA KIRP -0.433; TCGA LGG -0.389; TCGA LIHC -0.401; TCGA LUAD -0.356; TCGA LUSC -0.505; TCGA PRAD -0.272; TCGA THCA -0.662; TCGA STAD -0.544; TCGA UCEC -0.115 |
hsa-miR-30a-3p | ZNF250 | 10 cancers: BLCA; CESC; COAD; HNSC; LGG; LIHC; LUAD; OV; PRAD; STAD | mirMAP | TCGA BLCA -0.055; TCGA CESC -0.056; TCGA COAD -0.109; TCGA HNSC -0.063; TCGA LGG -0.217; TCGA LIHC -0.106; TCGA LUAD -0.055; TCGA OV -0.1; TCGA PRAD -0.139; TCGA STAD -0.051 |
hsa-miR-30a-3p | HDAC2 | 10 cancers: BLCA; BRCA; ESCA; KIRP; LGG; LIHC; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BLCA -0.071; TCGA BRCA -0.093; TCGA ESCA -0.106; TCGA KIRP -0.056; TCGA LGG -0.238; TCGA LIHC -0.052; TCGA LUAD -0.085; TCGA LUSC -0.182; TCGA STAD -0.174; TCGA UCEC -0.11 |
hsa-miR-30a-3p | TRIM59 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.141; TCGA BRCA -0.163; TCGA CESC -0.125; TCGA COAD -0.139; TCGA ESCA -0.2; TCGA HNSC -0.064; TCGA KIRC -0.313; TCGA KIRP -0.152; TCGA LIHC -0.263; TCGA LUAD -0.235; TCGA LUSC -0.414; TCGA PRAD -0.164; TCGA THCA -0.35; TCGA STAD -0.35 |
hsa-miR-30a-3p | NOL12 | 12 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PAAD | mirMAP | TCGA BLCA -0.078; TCGA BRCA -0.116; TCGA COAD -0.282; TCGA ESCA -0.134; TCGA KIRC -0.231; TCGA KIRP -0.184; TCGA LGG -0.144; TCGA LIHC -0.203; TCGA LUAD -0.091; TCGA LUSC -0.15; TCGA OV -0.059; TCGA PAAD -0.088 |
hsa-miR-30a-3p | LMNB1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.113; TCGA BRCA -0.307; TCGA CESC -0.061; TCGA COAD -0.271; TCGA ESCA -0.214; TCGA KIRC -0.662; TCGA KIRP -0.355; TCGA LGG -0.326; TCGA LIHC -0.286; TCGA LUAD -0.301; TCGA LUSC -0.369; TCGA PRAD -0.398; TCGA THCA -0.348; TCGA STAD -0.463; TCGA UCEC -0.086 |
hsa-miR-30a-3p | VAX2 | 9 cancers: BLCA; BRCA; ESCA; KIRC; LGG; LIHC; LUAD; LUSC; PRAD | miRNATAP | TCGA BLCA -0.232; TCGA BRCA -0.279; TCGA ESCA -0.334; TCGA KIRC -0.427; TCGA LGG -0.648; TCGA LIHC -0.294; TCGA LUAD -0.308; TCGA LUSC -0.379; TCGA PRAD -0.236 |
hsa-miR-30a-3p | DTD1 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LGG; LUSC; PAAD; PRAD; THCA; UCEC | miRNATAP | TCGA BLCA -0.116; TCGA BRCA -0.077; TCGA COAD -0.124; TCGA ESCA -0.194; TCGA HNSC -0.105; TCGA KIRC -0.161; TCGA LGG -0.192; TCGA LUSC -0.096; TCGA PAAD -0.164; TCGA PRAD -0.064; TCGA THCA -0.064; TCGA UCEC -0.065 |
hsa-miR-30a-3p | CLEC2D | 10 cancers: BRCA; KIRC; KIRP; LGG; LIHC; LUSC; OV; PRAD; SARC; THCA | MirTarget | TCGA BRCA -0.062; TCGA KIRC -0.549; TCGA KIRP -0.253; TCGA LGG -0.267; TCGA LIHC -0.209; TCGA LUSC -0.053; TCGA OV -0.07; TCGA PRAD -0.103; TCGA SARC -0.071; TCGA THCA -0.27 |
hsa-miR-30a-3p | COL1A2 | 10 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; THCA; UCEC | MirTarget | TCGA BRCA -0.196; TCGA ESCA -0.262; TCGA HNSC -0.193; TCGA KIRC -0.987; TCGA KIRP -0.308; TCGA LUAD -0.23; TCGA LUSC -0.133; TCGA PRAD -0.266; TCGA THCA -0.834; TCGA UCEC -0.154 |
hsa-miR-30a-3p | SUV39H2 | 12 cancers: BRCA; COAD; ESCA; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; STAD; UCEC | MirTarget | TCGA BRCA -0.137; TCGA COAD -0.284; TCGA ESCA -0.096; TCGA KIRC -0.077; TCGA KIRP -0.133; TCGA LGG -0.297; TCGA LIHC -0.156; TCGA LUAD -0.221; TCGA LUSC -0.288; TCGA SARC -0.089; TCGA STAD -0.315; TCGA UCEC -0.116 |
hsa-miR-30a-3p | ELL3 | 9 cancers: BRCA; COAD; ESCA; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA BRCA -0.134; TCGA COAD -0.371; TCGA ESCA -0.149; TCGA LIHC -0.11; TCGA LUAD -0.096; TCGA LUSC -0.076; TCGA PAAD -0.152; TCGA STAD -0.168; TCGA UCEC -0.064 |
hsa-miR-30a-3p | RALA | 11 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; LIHC; OV; PRAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.058; TCGA ESCA -0.146; TCGA HNSC -0.065; TCGA KIRC -0.183; TCGA KIRP -0.059; TCGA LIHC -0.061; TCGA OV -0.088; TCGA PRAD -0.092; TCGA THCA -0.124; TCGA STAD -0.134; TCGA UCEC -0.089 |
hsa-miR-30a-3p | DUSP9 | 9 cancers: BRCA; CESC; HNSC; LGG; LIHC; LUAD; LUSC; OV; SARC | MirTarget | TCGA BRCA -0.47; TCGA CESC -0.238; TCGA HNSC -0.324; TCGA LGG -0.549; TCGA LIHC -0.937; TCGA LUAD -0.401; TCGA LUSC -0.993; TCGA OV -0.233; TCGA SARC -0.455 |
hsa-miR-30a-3p | SLC25A33 | 9 cancers: BRCA; COAD; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BRCA -0.068; TCGA COAD -0.174; TCGA LIHC -0.171; TCGA LUAD -0.088; TCGA LUSC -0.122; TCGA PAAD -0.183; TCGA PRAD -0.083; TCGA STAD -0.145; TCGA UCEC -0.108 |
hsa-miR-30a-3p | CBFB | 11 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUSC; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.103; TCGA CESC -0.06; TCGA ESCA -0.15; TCGA HNSC -0.063; TCGA KIRC -0.316; TCGA KIRP -0.095; TCGA LGG -0.073; TCGA LUSC -0.126; TCGA THCA -0.223; TCGA STAD -0.332; TCGA UCEC -0.121 |
hsa-miR-30a-3p | ARF6 | 9 cancers: BRCA; CESC; ESCA; HNSC; LUSC; OV; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.051; TCGA CESC -0.064; TCGA ESCA -0.138; TCGA HNSC -0.067; TCGA LUSC -0.087; TCGA OV -0.088; TCGA THCA -0.218; TCGA STAD -0.072; TCGA UCEC -0.071 |
hsa-miR-30a-3p | RELT | 12 cancers: BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PRAD; THCA; STAD | MirTarget | TCGA BRCA -0.279; TCGA COAD -0.165; TCGA ESCA -0.205; TCGA HNSC -0.096; TCGA KIRC -0.897; TCGA KIRP -0.426; TCGA LUAD -0.092; TCGA LUSC -0.062; TCGA OV -0.103; TCGA PRAD -0.224; TCGA THCA -0.571; TCGA STAD -0.183 |
hsa-miR-30a-3p | RAD21 | 12 cancers: BRCA; CESC; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; OV; PRAD; STAD; UCEC | MirTarget | TCGA BRCA -0.088; TCGA CESC -0.075; TCGA ESCA -0.192; TCGA HNSC -0.062; TCGA LGG -0.081; TCGA LIHC -0.1; TCGA LUAD -0.102; TCGA LUSC -0.142; TCGA OV -0.12; TCGA PRAD -0.182; TCGA STAD -0.231; TCGA UCEC -0.089 |
hsa-miR-30a-3p | ANLN | 13 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; THCA; STAD | MirTarget | TCGA BRCA -0.439; TCGA CESC -0.09; TCGA COAD -0.135; TCGA ESCA -0.313; TCGA HNSC -0.173; TCGA KIRC -1.159; TCGA KIRP -0.574; TCGA LIHC -0.564; TCGA LUAD -0.498; TCGA LUSC -0.723; TCGA PRAD -0.527; TCGA THCA -0.626; TCGA STAD -0.533 |
hsa-miR-30a-3p | ADA | 11 cancers: BRCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PRAD; THCA | MirTarget | TCGA BRCA -0.254; TCGA HNSC -0.253; TCGA KIRC -1.184; TCGA KIRP -0.404; TCGA LGG -0.136; TCGA LIHC -0.2; TCGA LUAD -0.175; TCGA LUSC -0.291; TCGA OV -0.117; TCGA PRAD -0.186; TCGA THCA -0.247 |
hsa-miR-30a-3p | SEC61B | 10 cancers: BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD | MirTarget | TCGA BRCA -0.12; TCGA COAD -0.176; TCGA ESCA -0.104; TCGA HNSC -0.053; TCGA KIRC -0.142; TCGA KIRP -0.084; TCGA LIHC -0.104; TCGA LUAD -0.113; TCGA LUSC -0.065; TCGA PAAD -0.128 |
hsa-miR-30a-3p | PAG1 | 9 cancers: BRCA; CESC; HNSC; KIRC; LGG; LIHC; OV; THCA; STAD | MirTarget | TCGA BRCA -0.097; TCGA CESC -0.097; TCGA HNSC -0.079; TCGA KIRC -0.568; TCGA LGG -0.151; TCGA LIHC -0.184; TCGA OV -0.128; TCGA THCA -0.356; TCGA STAD -0.101 |
hsa-miR-30a-3p | PHTF2 | 12 cancers: BRCA; CESC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.095; TCGA CESC -0.073; TCGA KIRC -0.318; TCGA KIRP -0.095; TCGA LGG -0.226; TCGA LIHC -0.057; TCGA LUAD -0.154; TCGA LUSC -0.14; TCGA PRAD -0.155; TCGA THCA -0.096; TCGA STAD -0.128; TCGA UCEC -0.138 |
hsa-miR-30a-3p | COL1A1 | 11 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PRAD; THCA; STAD | mirMAP | TCGA BRCA -0.293; TCGA ESCA -0.435; TCGA HNSC -0.307; TCGA KIRC -1.441; TCGA KIRP -0.649; TCGA LUAD -0.436; TCGA LUSC -0.372; TCGA OV -0.2; TCGA PRAD -0.536; TCGA THCA -1.424; TCGA STAD -0.275 |
hsa-miR-30a-3p | ACTR3 | 10 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; THCA; STAD | mirMAP | TCGA BRCA -0.092; TCGA ESCA -0.147; TCGA HNSC -0.11; TCGA KIRC -0.15; TCGA KIRP -0.118; TCGA LUAD -0.054; TCGA LUSC -0.147; TCGA PRAD -0.068; TCGA THCA -0.105; TCGA STAD -0.085 |
hsa-miR-30a-3p | ENY2 | 12 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PRAD; STAD; UCEC | mirMAP | TCGA BRCA -0.172; TCGA ESCA -0.192; TCGA HNSC -0.096; TCGA KIRC -0.095; TCGA KIRP -0.073; TCGA LIHC -0.159; TCGA LUAD -0.137; TCGA LUSC -0.159; TCGA OV -0.146; TCGA PRAD -0.133; TCGA STAD -0.102; TCGA UCEC -0.058 |
hsa-miR-30a-3p | SOX12 | 11 cancers: BRCA; COAD; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD | mirMAP | TCGA BRCA -0.124; TCGA COAD -0.168; TCGA ESCA -0.126; TCGA HNSC -0.153; TCGA KIRC -0.233; TCGA LGG -0.319; TCGA LIHC -0.279; TCGA LUAD -0.268; TCGA LUSC -0.336; TCGA PAAD -0.194; TCGA PRAD -0.086 |
hsa-miR-30a-3p | COL5A2 | 12 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.196; TCGA ESCA -0.303; TCGA HNSC -0.277; TCGA KIRC -0.908; TCGA KIRP -0.371; TCGA LUAD -0.326; TCGA LUSC -0.214; TCGA OV -0.268; TCGA PRAD -0.414; TCGA THCA -0.57; TCGA STAD -0.218; TCGA UCEC -0.169 |
hsa-miR-30a-3p | TAP2 | 10 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; PRAD; THCA; STAD | mirMAP | TCGA BRCA -0.183; TCGA CESC -0.136; TCGA ESCA -0.191; TCGA HNSC -0.179; TCGA KIRC -0.243; TCGA KIRP -0.112; TCGA LIHC -0.095; TCGA PRAD -0.089; TCGA THCA -0.3; TCGA STAD -0.121 |
hsa-miR-30a-3p | EIF5AL1 | 10 cancers: BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; STAD | mirMAP | TCGA BRCA -0.106; TCGA COAD -0.332; TCGA ESCA -0.162; TCGA HNSC -0.119; TCGA KIRC -0.111; TCGA KIRP -0.07; TCGA LGG -0.07; TCGA LUAD -0.109; TCGA LUSC -0.151; TCGA STAD -0.249 |
hsa-miR-30a-3p | PPP2R5E | 9 cancers: CESC; ESCA; HNSC; LGG; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA CESC -0.084; TCGA ESCA -0.096; TCGA HNSC -0.104; TCGA LGG -0.064; TCGA LUAD -0.058; TCGA LUSC -0.088; TCGA PRAD -0.078; TCGA STAD -0.078; TCGA UCEC -0.065 |
hsa-miR-30a-3p | OTUD6B | 10 cancers: CESC; ESCA; HNSC; LIHC; LUAD; LUSC; OV; PRAD; STAD; UCEC | MirTarget | TCGA CESC -0.064; TCGA ESCA -0.169; TCGA HNSC -0.071; TCGA LIHC -0.105; TCGA LUAD -0.104; TCGA LUSC -0.183; TCGA OV -0.087; TCGA PRAD -0.155; TCGA STAD -0.283; TCGA UCEC -0.101 |
hsa-miR-30a-3p | MAPK6 | 10 cancers: CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA CESC -0.177; TCGA ESCA -0.245; TCGA HNSC -0.236; TCGA KIRC -0.109; TCGA KIRP -0.119; TCGA LGG -0.063; TCGA LUAD -0.088; TCGA LUSC -0.253; TCGA STAD -0.143; TCGA UCEC -0.107 |
hsa-miR-30a-3p | ARMC1 | 11 cancers: CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA CESC -0.078; TCGA COAD -0.094; TCGA ESCA -0.086; TCGA HNSC -0.076; TCGA LGG -0.127; TCGA LIHC -0.068; TCGA LUAD -0.103; TCGA LUSC -0.133; TCGA PRAD -0.076; TCGA STAD -0.097; TCGA UCEC -0.067 |
hsa-miR-30a-3p | PSMD10 | 10 cancers: CESC; ESCA; HNSC; KIRC; LIHC; LUSC; OV; PAAD; STAD; UCEC | MirTarget | TCGA CESC -0.05; TCGA ESCA -0.104; TCGA HNSC -0.077; TCGA KIRC -0.078; TCGA LIHC -0.082; TCGA LUSC -0.12; TCGA OV -0.061; TCGA PAAD -0.108; TCGA STAD -0.057; TCGA UCEC -0.059 |
hsa-miR-30a-3p | SENP1 | 9 cancers: CESC; KIRC; LGG; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA CESC -0.057; TCGA KIRC -0.113; TCGA LGG -0.109; TCGA LIHC -0.053; TCGA LUAD -0.087; TCGA LUSC -0.12; TCGA PRAD -0.106; TCGA STAD -0.133; TCGA UCEC -0.07 |
hsa-miR-30a-3p | POLR3G | 12 cancers: CESC; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA CESC -0.222; TCGA ESCA -0.387; TCGA HNSC -0.216; TCGA KIRC -0.386; TCGA KIRP -0.166; TCGA LGG -0.091; TCGA LIHC -0.093; TCGA LUAD -0.173; TCGA LUSC -0.246; TCGA PRAD -0.213; TCGA STAD -0.479; TCGA UCEC -0.138 |
hsa-miR-30a-3p | PTGFRN | 10 cancers: CESC; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; THCA; UCEC | mirMAP | TCGA CESC -0.106; TCGA ESCA -0.167; TCGA HNSC -0.209; TCGA KIRC -0.41; TCGA KIRP -0.158; TCGA LUAD -0.106; TCGA LUSC -0.349; TCGA PRAD -0.113; TCGA THCA -0.237; TCGA UCEC -0.159 |
hsa-miR-30a-3p | RBAK | 9 cancers: CESC; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA CESC -0.055; TCGA ESCA -0.095; TCGA HNSC -0.078; TCGA KIRC -0.091; TCGA LUAD -0.061; TCGA LUSC -0.163; TCGA PRAD -0.089; TCGA STAD -0.121; TCGA UCEC -0.084 |
hsa-miR-30a-3p | FOXK1 | 9 cancers: CESC; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | mirMAP | TCGA CESC -0.131; TCGA HNSC -0.081; TCGA LIHC -0.178; TCGA LUAD -0.052; TCGA LUSC -0.103; TCGA PAAD -0.203; TCGA PRAD -0.153; TCGA THCA -0.089; TCGA STAD -0.128 |
hsa-miR-30a-3p | TMEM154 | 9 cancers: CESC; ESCA; HNSC; KIRC; KIRP; LUSC; OV; PRAD; THCA | mirMAP | TCGA CESC -0.362; TCGA ESCA -0.341; TCGA HNSC -0.244; TCGA KIRC -0.32; TCGA KIRP -0.367; TCGA LUSC -0.177; TCGA OV -0.169; TCGA PRAD -0.282; TCGA THCA -0.492 |
hsa-miR-30a-3p | NAA25 | 10 cancers: CESC; COAD; ESCA; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; STAD | miRNATAP | TCGA CESC -0.075; TCGA COAD -0.133; TCGA ESCA -0.125; TCGA KIRC -0.198; TCGA KIRP -0.133; TCGA LGG -0.125; TCGA LIHC -0.096; TCGA LUAD -0.182; TCGA LUSC -0.193; TCGA STAD -0.295 |
hsa-miR-30a-3p | C1QBP | 10 cancers: COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | miRNAWalker2 validate | TCGA COAD -0.391; TCGA ESCA -0.158; TCGA HNSC -0.063; TCGA LGG -0.071; TCGA LIHC -0.056; TCGA LUAD -0.084; TCGA LUSC -0.191; TCGA PAAD -0.109; TCGA STAD -0.268; TCGA UCEC -0.108 |
hsa-miR-30a-3p | CCDC117 | 9 cancers: COAD; KIRC; LGG; LIHC; LUSC; OV; SARC; STAD; UCEC | MirTarget | TCGA COAD -0.103; TCGA KIRC -0.062; TCGA LGG -0.129; TCGA LIHC -0.111; TCGA LUSC -0.125; TCGA OV -0.092; TCGA SARC -0.061; TCGA STAD -0.124; TCGA UCEC -0.052 |
hsa-miR-30a-3p | SUB1 | 9 cancers: COAD; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; UCEC | MirTarget | TCGA COAD -0.128; TCGA HNSC -0.101; TCGA LGG -0.121; TCGA LIHC -0.088; TCGA LUAD -0.072; TCGA LUSC -0.097; TCGA PAAD -0.101; TCGA PRAD -0.052; TCGA UCEC -0.071 |
hsa-miR-30a-3p | RPAP3 | 10 cancers: COAD; ESCA; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA COAD -0.078; TCGA ESCA -0.104; TCGA KIRP -0.076; TCGA LIHC -0.072; TCGA LUAD -0.119; TCGA LUSC -0.154; TCGA PAAD -0.061; TCGA PRAD -0.051; TCGA STAD -0.209; TCGA UCEC -0.063 |
hsa-miR-30a-3p | RSL1D1 | 11 cancers: COAD; ESCA; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; STAD; UCEC | MirTarget | TCGA COAD -0.172; TCGA ESCA -0.109; TCGA KIRC -0.155; TCGA KIRP -0.111; TCGA LGG -0.143; TCGA LIHC -0.053; TCGA LUAD -0.051; TCGA LUSC -0.099; TCGA SARC -0.126; TCGA STAD -0.112; TCGA UCEC -0.076 |
hsa-miR-30a-3p | HAUS6 | 10 cancers: COAD; ESCA; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA COAD -0.139; TCGA ESCA -0.124; TCGA KIRC -0.2; TCGA KIRP -0.115; TCGA LIHC -0.073; TCGA LUAD -0.11; TCGA LUSC -0.177; TCGA PRAD -0.111; TCGA STAD -0.348; TCGA UCEC -0.069 |
hsa-miR-30a-3p | TSN | 9 cancers: COAD; ESCA; HNSC; LGG; LUAD; LUSC; THCA; STAD; UCEC | mirMAP; miRNATAP | TCGA COAD -0.108; TCGA ESCA -0.138; TCGA HNSC -0.061; TCGA LGG -0.069; TCGA LUAD -0.058; TCGA LUSC -0.127; TCGA THCA -0.092; TCGA STAD -0.181; TCGA UCEC -0.072 |
hsa-miR-30a-3p | RBMX | 10 cancers: COAD; ESCA; KIRC; LGG; LIHC; LUAD; LUSC; SARC; STAD; UCEC | miRNATAP | TCGA COAD -0.104; TCGA ESCA -0.056; TCGA KIRC -0.083; TCGA LGG -0.18; TCGA LIHC -0.108; TCGA LUAD -0.053; TCGA LUSC -0.07; TCGA SARC -0.066; TCGA STAD -0.13; TCGA UCEC -0.074 |
hsa-miR-30a-3p | NAV1 | 9 cancers: ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; THCA; UCEC | mirMAP | TCGA ESCA -0.248; TCGA HNSC -0.216; TCGA KIRC -0.409; TCGA KIRP -0.201; TCGA LGG -0.229; TCGA LUAD -0.252; TCGA LUSC -0.256; TCGA THCA -0.181; TCGA UCEC -0.147 |
hsa-miR-30a-3p | TWF1 | 9 cancers: ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; THCA; STAD | miRNATAP | TCGA ESCA -0.133; TCGA HNSC -0.143; TCGA KIRP -0.139; TCGA LUAD -0.146; TCGA LUSC -0.221; TCGA PAAD -0.114; TCGA PRAD -0.056; TCGA THCA -0.08; TCGA STAD -0.212 |
hsa-miR-30a-3p | CSNK1E | 12 cancers: ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | miRNATAP | TCGA ESCA -0.108; TCGA HNSC -0.083; TCGA KIRC -0.348; TCGA KIRP -0.133; TCGA LGG -0.305; TCGA LIHC -0.058; TCGA LUAD -0.101; TCGA LUSC -0.096; TCGA PAAD -0.081; TCGA PRAD -0.063; TCGA THCA -0.243; TCGA STAD -0.084 |
hsa-miR-30a-3p | LPGAT1 | 9 cancers: KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA KIRC -0.264; TCGA KIRP -0.174; TCGA LGG -0.063; TCGA LIHC -0.106; TCGA LUAD -0.16; TCGA LUSC -0.107; TCGA PRAD -0.134; TCGA STAD -0.231; TCGA UCEC -0.15 |
Enriched cancer pathways of putative targets