microRNA information: hsa-miR-331-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-331-3p | miRbase |
| Accession: | MIMAT0000760 | miRbase |
| Precursor name: | hsa-mir-331 | miRbase |
| Precursor accession: | MI0000812 | miRbase |
| Symbol: | MIR331 | HGNC |
| RefSeq ID: | NR_029895 | GenBank |
| Sequence: | GCCCCUGGGCCUAUCCUAGAA |
Reported expression in cancers: hsa-miR-331-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-331-3p | acute myeloid leukemia | deregulation | "Expression of microRNA 331 can be used as a predic ......" | 25620533 | Reverse transcription PCR |
| hsa-miR-331-3p | colorectal cancer | downregulation | "However little research has been conducted on the ......" | 26718987 | |
| hsa-miR-331-3p | glioblastoma | downregulation | "Here we investigate the functional role of miR-331 ......" | 24142150 | |
| hsa-miR-331-3p | liver cancer | upregulation | "We discovered miR-331-3p as one of most significan ......" | 24825302 | |
| hsa-miR-331-3p | liver cancer | upregulation | "Serum miR 182 and miR 331 3p as diagnostic and pro ......" | 25903466 | qPCR |
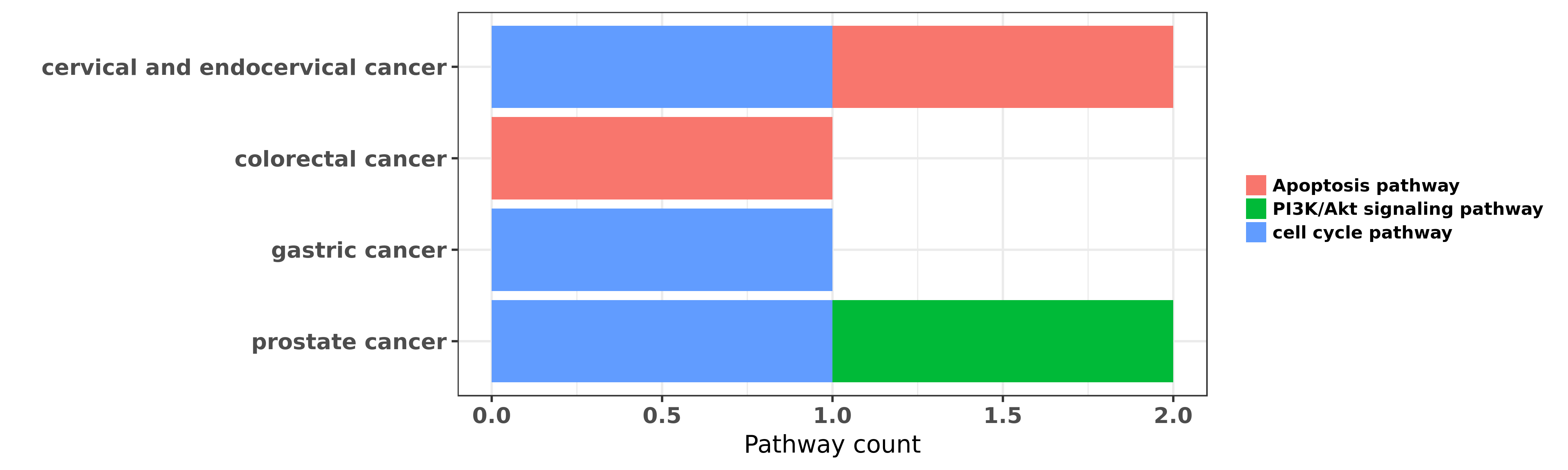
Reported cancer pathway affected by hsa-miR-331-3p
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-331-3p | cervical and endocervical cancer | Apoptosis pathway; cell cycle pathway | "MicroRNA 331 3p Suppresses Cervical Cancer Cell Pr ......" | 27548144 | Cell Proliferation Assay; Luciferase |
| hsa-miR-331-3p | colorectal cancer | Apoptosis pathway | "MiR 331 3p inhibits proliferation and promotes apo ......" | 26718987 | |
| hsa-miR-331-3p | gastric cancer | cell cycle pathway | "Here we report a member of miR-331 family miR-331- ......" | 20510161 | Colony formation |
| hsa-miR-331-3p | prostate cancer | PI3K/Akt signaling pathway | "miR 331 3p regulates ERBB 2 expression and androge ......" | 19584056 | |
| hsa-miR-331-3p | prostate cancer | cell cycle pathway | "Interestingly 5 of the 7 differentially expressed ......" | 19996289 |
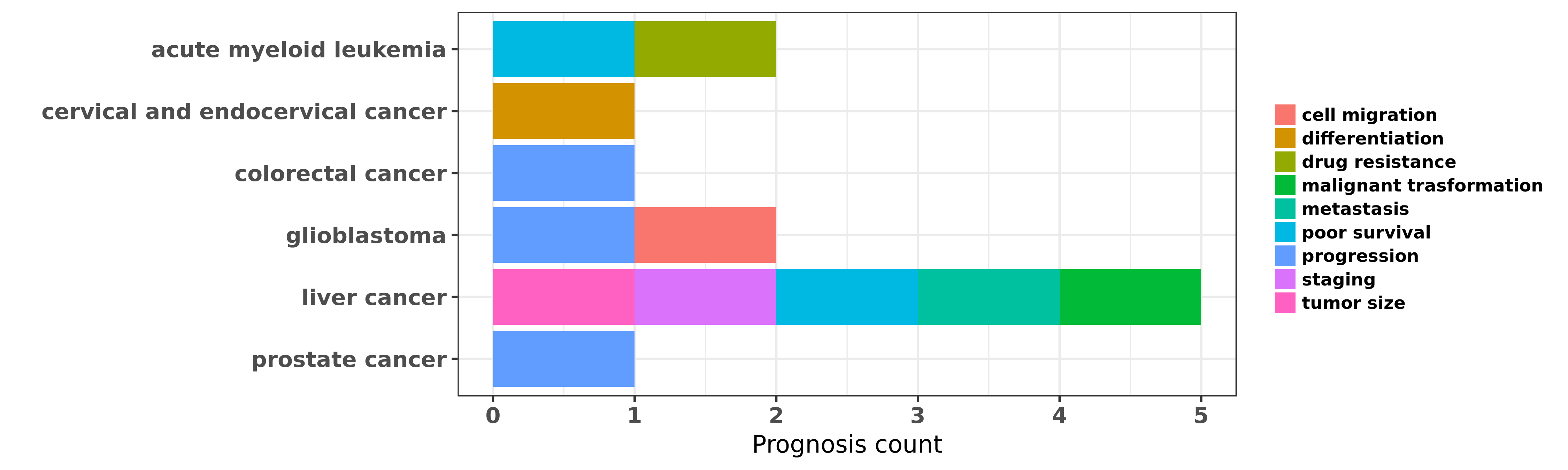
Reported cancer prognosis affected by hsa-miR-331-3p
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-331-3p | acute myeloid leukemia | poor survival; drug resistance | "Expression of microRNA 331 can be used as a predic ......" | 25620533 | |
| hsa-miR-331-3p | cervical and endocervical cancer | differentiation | "MicroRNA 331 3p Suppresses Cervical Cancer Cell Pr ......" | 27548144 | Cell Proliferation Assay; Luciferase |
| hsa-miR-331-3p | colorectal cancer | progression | "MiR 331 3p inhibits proliferation and promotes apo ......" | 26718987 | |
| hsa-miR-331-3p | glioblastoma | cell migration; progression | "miR 331 3p regulates expression of neuropilin 2 in ......" | 24142150 | RNAi |
| hsa-miR-331-3p | liver cancer | metastasis; poor survival | "MicroRNA 331 3p promotes proliferation and metasta ......" | 24825302 | |
| hsa-miR-331-3p | liver cancer | malignant trasformation; staging; tumor size; poor survival | "Serum miR 182 and miR 331 3p as diagnostic and pro ......" | 25903466 | |
| hsa-miR-331-3p | prostate cancer | progression | "miR 331 3p regulates ERBB 2 expression and androge ......" | 19584056 |
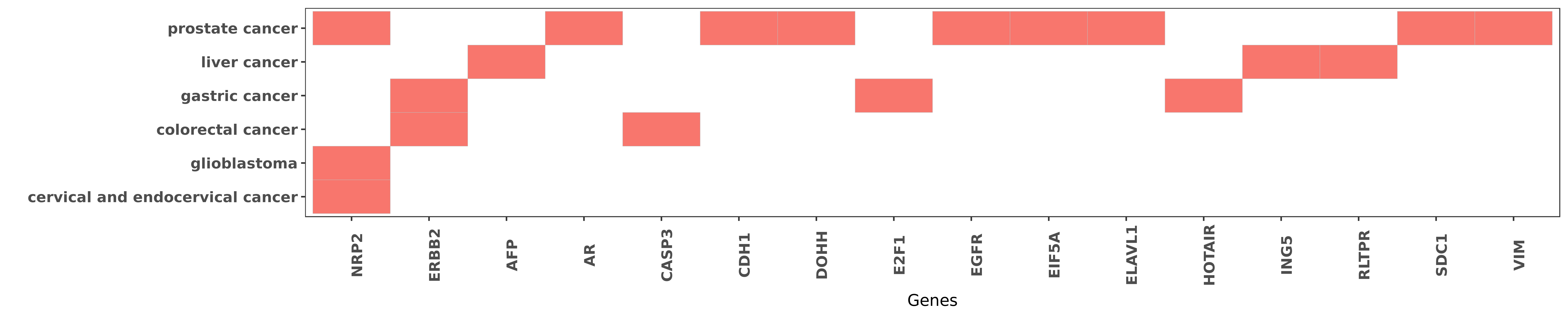
Reported gene related to hsa-miR-331-3p
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-331-3p | cervical and endocervical cancer | NRP2 | "MicroRNA 331 3p Suppresses Cervical Cancer Cell Pr ......" | 27548144 |
| hsa-miR-331-3p | glioblastoma | NRP2 | "miR 331 3p regulates expression of neuropilin 2 in ......" | 24142150 |
| hsa-miR-331-3p | prostate cancer | NRP2 | "We identified Neuropilin 2 and nucleus accumbens-a ......" | 26259043 |
| hsa-miR-331-3p | prostate cancer | EGFR | "The RNA binding protein HuR opposes the repression ......" | 21971048 |
| hsa-miR-331-3p | prostate cancer | EGFR | "miR 331 3p regulates ERBB 2 expression and androge ......" | 19584056 |
| hsa-miR-331-3p | colorectal cancer | ERBB2 | "MiR 331 3p inhibits proliferation and promotes apo ......" | 26718987 |
| hsa-miR-331-3p | gastric cancer | ERBB2 | "Lnc RNA HOTAIR functions as a competing endogenous ......" | 24775712 |
| hsa-miR-331-3p | liver cancer | AFP | "Serum miR-182 was positively correlated with serum ......" | 25903466 |
| hsa-miR-331-3p | prostate cancer | AR | "miR 331 3p regulates ERBB 2 expression and androge ......" | 19584056 |
| hsa-miR-331-3p | colorectal cancer | CASP3 | "Overexpression of miR-331-3p by transfection with ......" | 26718987 |
| hsa-miR-331-3p | prostate cancer | CDH1 | "In situ hybridization and immunohistochemistry of ......" | 26259043 |
| hsa-miR-331-3p | prostate cancer | DOHH | "Using bioinformatic predictions and reporter gene ......" | 22908221 |
| hsa-miR-331-3p | gastric cancer | E2F1 | "In this study we screened the expressing levels of ......" | 20510161 |
| hsa-miR-331-3p | prostate cancer | EIF5A | "Our results suggest a novel role for miR-331-3p an ......" | 22908221 |
| hsa-miR-331-3p | prostate cancer | ELAVL1 | "The RNA binding protein HuR opposes the repression ......" | 21971048 |
| hsa-miR-331-3p | gastric cancer | HOTAIR | "Lnc RNA HOTAIR functions as a competing endogenous ......" | 24775712 |
| hsa-miR-331-3p | liver cancer | ING5 | "Upregulated in Hepatitis B virus associated hepato ......" | 26497554 |
| hsa-miR-331-3p | liver cancer | RLTPR | "MicroRNA 331 3p promotes proliferation and metasta ......" | 24825302 |
| hsa-miR-331-3p | prostate cancer | SDC1 | "Syndecan 1 up regulates microRNA 331 3p and mediat ......" | 26259043 |
| hsa-miR-331-3p | prostate cancer | VIM | "In situ hybridization and immunohistochemistry of ......" | 26259043 |
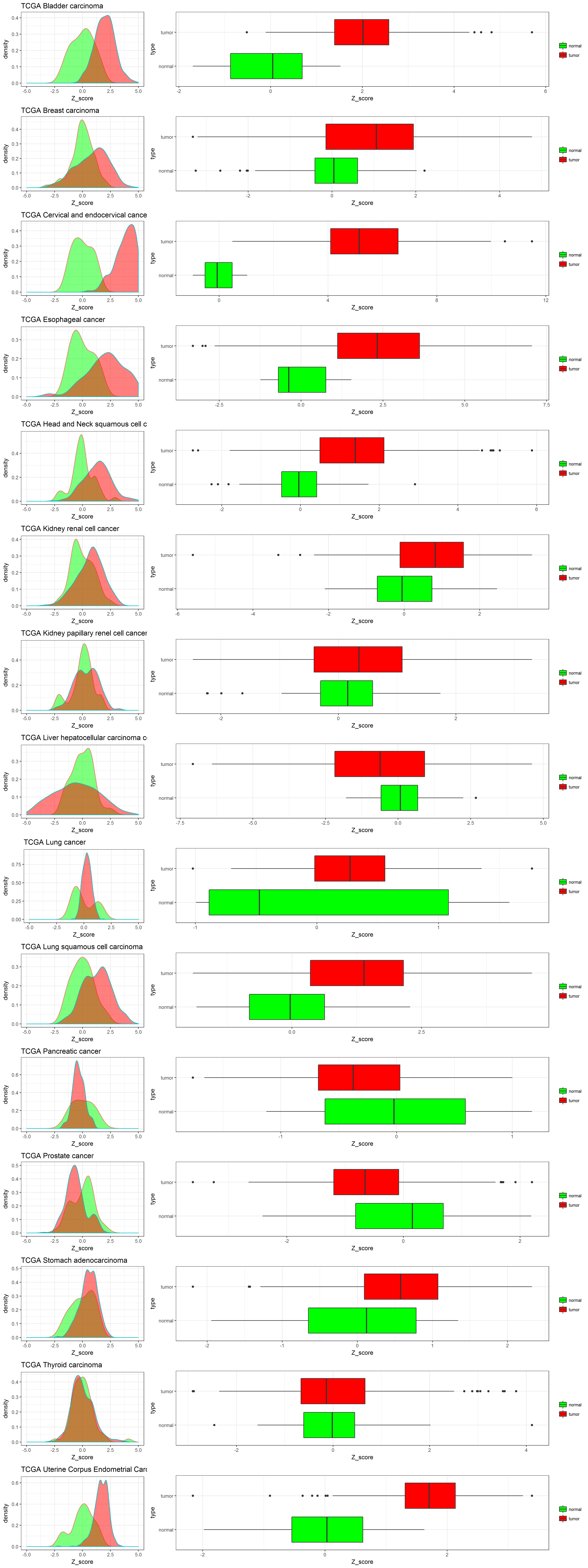
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-331-3p | ADCY9 | 9 cancers: BLCA; CESC; COAD; LIHC; LUSC; OV; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.398; TCGA CESC -0.14; TCGA COAD -0.098; TCGA LIHC -0.103; TCGA LUSC -0.172; TCGA OV -0.118; TCGA SARC -0.275; TCGA STAD -0.163; TCGA UCEC -0.333 |
hsa-miR-331-3p | APBB1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.581; TCGA BRCA -0.198; TCGA CESC -0.29; TCGA COAD -0.455; TCGA ESCA -0.325; TCGA HNSC -0.303; TCGA KIRC -0.125; TCGA KIRP -0.115; TCGA LUSC -0.275; TCGA SARC -0.131; TCGA THCA -0.067; TCGA STAD -0.408; TCGA UCEC -0.364 |
hsa-miR-331-3p | ATF3 | 9 cancers: BLCA; BRCA; KIRC; LIHC; LUSC; PRAD; SARC; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.381; TCGA BRCA -0.278; TCGA KIRC -0.412; TCGA LIHC -0.198; TCGA LUSC -0.203; TCGA PRAD -0.226; TCGA SARC -0.268; TCGA THCA -0.222; TCGA UCEC -0.32 |
hsa-miR-331-3p | ATP8B2 | 9 cancers: BLCA; BRCA; COAD; ESCA; LUSC; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.566; TCGA BRCA -0.114; TCGA COAD -0.429; TCGA ESCA -0.318; TCGA LUSC -0.117; TCGA SARC -0.18; TCGA THCA -0.227; TCGA STAD -0.312; TCGA UCEC -0.269 |
hsa-miR-331-3p | BOK | 9 cancers: BLCA; BRCA; HNSC; LIHC; LUAD; LUSC; PAAD; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.204; TCGA BRCA -0.162; TCGA HNSC -0.139; TCGA LIHC -0.373; TCGA LUAD -0.115; TCGA LUSC -0.226; TCGA PAAD -0.317; TCGA THCA -0.143; TCGA STAD -0.147 |
hsa-miR-331-3p | ENTPD1 | 10 cancers: BLCA; CESC; COAD; ESCA; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.345; TCGA CESC -0.121; TCGA COAD -0.258; TCGA ESCA -0.249; TCGA LUAD -0.058; TCGA LUSC -0.198; TCGA SARC -0.266; TCGA THCA -0.131; TCGA STAD -0.216; TCGA UCEC -0.205 |
hsa-miR-331-3p | FBXL5 | 9 cancers: BLCA; COAD; ESCA; LIHC; LUAD; LUSC; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.071; TCGA COAD -0.138; TCGA ESCA -0.138; TCGA LIHC -0.224; TCGA LUAD -0.104; TCGA LUSC -0.091; TCGA SARC -0.082; TCGA STAD -0.185; TCGA UCEC -0.107 |
hsa-miR-331-3p | FLT4 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; LGG; LIHC; LUSC; UCEC | miRNAWalker2 validate | TCGA BLCA -0.146; TCGA BRCA -0.257; TCGA CESC -0.181; TCGA COAD -0.402; TCGA ESCA -0.186; TCGA LGG -0.108; TCGA LIHC -0.118; TCGA LUSC -0.293; TCGA UCEC -0.375 |
hsa-miR-331-3p | PKIG | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; LUSC; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.464; TCGA BRCA -0.083; TCGA COAD -0.199; TCGA ESCA -0.238; TCGA HNSC -0.277; TCGA LIHC -0.095; TCGA LUSC -0.278; TCGA SARC -0.213; TCGA STAD -0.352; TCGA UCEC -0.191 |
hsa-miR-331-3p | PTPRT | 10 cancers: BLCA; BRCA; CESC; COAD; KIRC; LUAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; mirMAP | TCGA BLCA -0.435; TCGA BRCA -0.267; TCGA CESC -0.5; TCGA COAD -0.638; TCGA KIRC -0.648; TCGA LUAD -0.388; TCGA SARC -0.421; TCGA THCA -0.31; TCGA STAD -0.425; TCGA UCEC -0.519 |
hsa-miR-331-3p | RPS12 | 9 cancers: BLCA; BRCA; HNSC; LGG; LUAD; PAAD; PRAD; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.114; TCGA BRCA -0.076; TCGA HNSC -0.187; TCGA LGG -0.182; TCGA LUAD -0.097; TCGA PAAD -0.251; TCGA PRAD -0.135; TCGA THCA -0.117; TCGA UCEC -0.207 |
hsa-miR-331-3p | ZBTB4 | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; LUAD; LUSC; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.154; TCGA BRCA -0.097; TCGA COAD -0.185; TCGA ESCA -0.157; TCGA KIRC -0.136; TCGA KIRP -0.073; TCGA LUAD -0.066; TCGA LUSC -0.123; TCGA SARC -0.12; TCGA STAD -0.273; TCGA UCEC -0.228 |
hsa-miR-331-3p | CRIM1 | 13 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LIHC; LUAD; LUSC; OV; SARC; THCA; STAD; UCEC | MirTarget; PITA; miRNATAP | TCGA BLCA -0.141; TCGA BRCA -0.085; TCGA COAD -0.138; TCGA ESCA -0.196; TCGA KIRC -0.117; TCGA LIHC -0.285; TCGA LUAD -0.112; TCGA LUSC -0.096; TCGA OV -0.118; TCGA SARC -0.209; TCGA THCA -0.113; TCGA STAD -0.18; TCGA UCEC -0.304 |
hsa-miR-331-3p | CSDC2 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUSC; PAAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -1.335; TCGA BRCA -0.486; TCGA CESC -0.636; TCGA COAD -0.613; TCGA ESCA -0.746; TCGA HNSC -0.372; TCGA KIRC -0.478; TCGA LGG -0.273; TCGA LUSC -0.539; TCGA PAAD -0.549; TCGA THCA -0.865; TCGA STAD -0.383; TCGA UCEC -0.775 |
hsa-miR-331-3p | THSD4 | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; LUSC; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.277; TCGA BRCA -0.117; TCGA COAD -0.18; TCGA ESCA -0.387; TCGA KIRC -0.248; TCGA KIRP -0.175; TCGA LUSC -0.167; TCGA SARC -0.393; TCGA THCA -0.179; TCGA STAD -0.186; TCGA UCEC -0.22 |
hsa-miR-331-3p | UBL3 | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.126; TCGA CESC -0.242; TCGA COAD -0.206; TCGA ESCA -0.504; TCGA HNSC -0.118; TCGA KIRC -0.174; TCGA LUAD -0.104; TCGA LUSC -0.219; TCGA OV -0.09; TCGA STAD -0.171; TCGA UCEC -0.29 |
hsa-miR-331-3p | ARHGEF37 | 10 cancers: BLCA; BRCA; CESC; ESCA; KIRP; LIHC; LUSC; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.397; TCGA BRCA -0.195; TCGA CESC -0.355; TCGA ESCA -0.31; TCGA KIRP -0.123; TCGA LIHC -0.249; TCGA LUSC -0.316; TCGA SARC -0.424; TCGA STAD -0.375; TCGA UCEC -0.131 |
hsa-miR-331-3p | TEF | 11 cancers: BLCA; BRCA; CESC; COAD; KIRC; KIRP; LIHC; LUAD; SARC; STAD; UCEC | MirTarget; mirMAP | TCGA BLCA -0.152; TCGA BRCA -0.15; TCGA CESC -0.166; TCGA COAD -0.139; TCGA KIRC -0.303; TCGA KIRP -0.176; TCGA LIHC -0.128; TCGA LUAD -0.152; TCGA SARC -0.261; TCGA STAD -0.246; TCGA UCEC -0.318 |
hsa-miR-331-3p | MAP3K14 | 9 cancers: BLCA; BRCA; ESCA; LIHC; LUSC; OV; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.178; TCGA BRCA -0.148; TCGA ESCA -0.153; TCGA LIHC -0.106; TCGA LUSC -0.101; TCGA OV -0.071; TCGA THCA -0.281; TCGA STAD -0.107; TCGA UCEC -0.09 |
hsa-miR-331-3p | CAMK2A | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -1.158; TCGA BRCA -0.108; TCGA CESC -0.307; TCGA ESCA -0.395; TCGA HNSC -0.446; TCGA KIRC -0.463; TCGA LUAD -0.156; TCGA LUSC -0.709; TCGA PAAD -0.565; TCGA STAD -0.617; TCGA UCEC -0.756 |
hsa-miR-331-3p | KCNJ5 | 9 cancers: BLCA; CESC; COAD; ESCA; LIHC; LUAD; LUSC; OV; THCA | MirTarget; mirMAP | TCGA BLCA -0.499; TCGA CESC -0.233; TCGA COAD -0.394; TCGA ESCA -0.39; TCGA LIHC -0.331; TCGA LUAD -0.203; TCGA LUSC -0.258; TCGA OV -0.147; TCGA THCA -0.519 |
hsa-miR-331-3p | CADM3 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget; mirMAP | TCGA BLCA -1.535; TCGA BRCA -0.689; TCGA CESC -0.636; TCGA COAD -1.246; TCGA ESCA -1.217; TCGA HNSC -0.351; TCGA LUAD -0.279; TCGA LUSC -0.342; TCGA PAAD -0.787; TCGA PRAD -0.174; TCGA SARC -0.545; TCGA THCA -1.149; TCGA STAD -0.891; TCGA UCEC -0.608 |
hsa-miR-331-3p | CBX7 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.439; TCGA BRCA -0.358; TCGA CESC -0.442; TCGA COAD -0.234; TCGA ESCA -0.478; TCGA HNSC -0.16; TCGA KIRC -0.226; TCGA KIRP -0.121; TCGA LIHC -0.25; TCGA LUAD -0.196; TCGA LUSC -0.27; TCGA OV -0.192; TCGA SARC -0.607; TCGA THCA -0.07; TCGA STAD -0.497; TCGA UCEC -0.592 |
hsa-miR-331-3p | FBLN7 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; LUSC; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.249; TCGA BRCA -0.264; TCGA COAD -0.276; TCGA ESCA -0.347; TCGA HNSC -0.139; TCGA LIHC -0.239; TCGA LUSC -0.253; TCGA SARC -0.39; TCGA STAD -0.123; TCGA UCEC -0.119 |
hsa-miR-331-3p | GIMAP8 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.387; TCGA BRCA -0.178; TCGA CESC -0.221; TCGA COAD -0.41; TCGA ESCA -0.342; TCGA HNSC -0.107; TCGA LIHC -0.143; TCGA LUAD -0.175; TCGA LUSC -0.432; TCGA STAD -0.18; TCGA UCEC -0.478 |
hsa-miR-331-3p | ELN | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUSC; PAAD; THCA; STAD; UCEC | MirTarget; PITA | TCGA BLCA -0.925; TCGA BRCA -0.526; TCGA CESC -0.744; TCGA COAD -0.505; TCGA ESCA -0.848; TCGA HNSC -0.388; TCGA LGG -0.108; TCGA LUSC -0.353; TCGA PAAD -0.496; TCGA THCA -0.311; TCGA STAD -0.36; TCGA UCEC -0.257 |
hsa-miR-331-3p | RASL12 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.761; TCGA BRCA -0.277; TCGA CESC -0.431; TCGA COAD -0.421; TCGA ESCA -0.554; TCGA HNSC -0.286; TCGA LUAD -0.17; TCGA LUSC -0.424; TCGA PAAD -0.319; TCGA SARC -0.721; TCGA STAD -0.327; TCGA UCEC -0.588 |
hsa-miR-331-3p | KLHL30 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; PAAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.55; TCGA BRCA -0.381; TCGA COAD -0.387; TCGA ESCA -0.377; TCGA HNSC -0.367; TCGA KIRC -0.268; TCGA KIRP -0.436; TCGA LUSC -0.3; TCGA PAAD -0.33; TCGA SARC -0.611; TCGA THCA -0.339; TCGA STAD -0.257; TCGA UCEC -0.257 |
hsa-miR-331-3p | LRRC4B | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRP; LUSC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.36; TCGA BRCA -0.612; TCGA CESC -0.448; TCGA COAD -0.276; TCGA ESCA -0.303; TCGA KIRP -0.467; TCGA LUSC -0.283; TCGA STAD -0.204; TCGA UCEC -0.481 |
hsa-miR-331-3p | FAM109B | 9 cancers: BLCA; BRCA; HNSC; KIRC; LUSC; PAAD; SARC; THCA; UCEC | MirTarget | TCGA BLCA -0.202; TCGA BRCA -0.061; TCGA HNSC -0.101; TCGA KIRC -0.122; TCGA LUSC -0.112; TCGA PAAD -0.286; TCGA SARC -0.146; TCGA THCA -0.202; TCGA UCEC -0.107 |
hsa-miR-331-3p | KCNC4 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUSC; SARC; UCEC | PITA | TCGA BLCA -0.349; TCGA BRCA -0.168; TCGA CESC -0.218; TCGA ESCA -0.224; TCGA HNSC -0.164; TCGA KIRC -0.267; TCGA LUSC -0.101; TCGA SARC -0.204; TCGA UCEC -0.204 |
hsa-miR-331-3p | SHANK3 | 9 cancers: BLCA; BRCA; CESC; COAD; LIHC; LUSC; PAAD; STAD; UCEC | PITA | TCGA BLCA -0.168; TCGA BRCA -0.293; TCGA CESC -0.13; TCGA COAD -0.143; TCGA LIHC -0.158; TCGA LUSC -0.172; TCGA PAAD -0.212; TCGA STAD -0.126; TCGA UCEC -0.252 |
hsa-miR-331-3p | MEIS1 | 9 cancers: BLCA; BRCA; CESC; COAD; KIRC; LUAD; PAAD; THCA; STAD | PITA; miRNATAP | TCGA BLCA -0.301; TCGA BRCA -0.157; TCGA CESC -0.326; TCGA COAD -0.486; TCGA KIRC -0.132; TCGA LUAD -0.113; TCGA PAAD -0.253; TCGA THCA -0.126; TCGA STAD -0.592 |
hsa-miR-331-3p | MEIS2 | 9 cancers: BLCA; BRCA; CESC; COAD; KIRC; PRAD; SARC; STAD; UCEC | PITA | TCGA BLCA -0.312; TCGA BRCA -0.26; TCGA CESC -0.231; TCGA COAD -0.46; TCGA KIRC -0.169; TCGA PRAD -0.111; TCGA SARC -0.229; TCGA STAD -0.308; TCGA UCEC -0.339 |
hsa-miR-331-3p | DTX1 | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; PAAD; THCA; STAD | PITA; miRNATAP | TCGA BLCA -0.473; TCGA BRCA -0.32; TCGA ESCA -0.409; TCGA HNSC -0.161; TCGA KIRC -0.411; TCGA KIRP -0.293; TCGA PAAD -0.415; TCGA THCA -0.259; TCGA STAD -0.255 |
hsa-miR-331-3p | EGR3 | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | PITA | TCGA BLCA -0.743; TCGA BRCA -0.318; TCGA COAD -0.262; TCGA ESCA -0.386; TCGA KIRC -0.389; TCGA LUAD -0.148; TCGA LUSC -0.341; TCGA PRAD -0.215; TCGA THCA -0.273; TCGA STAD -0.247; TCGA UCEC -0.139 |
hsa-miR-331-3p | CLDN11 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; THCA; STAD; UCEC | PITA | TCGA BLCA -0.479; TCGA BRCA -0.545; TCGA CESC -0.309; TCGA COAD -0.548; TCGA ESCA -0.691; TCGA HNSC -0.375; TCGA LUAD -0.145; TCGA LUSC -0.256; TCGA THCA -0.456; TCGA STAD -0.375; TCGA UCEC -0.327 |
hsa-miR-331-3p | MYRIP | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; SARC; STAD; UCEC | PITA; miRNATAP | TCGA BLCA -0.318; TCGA BRCA -0.224; TCGA CESC -0.748; TCGA ESCA -0.932; TCGA HNSC -0.35; TCGA KIRC -0.449; TCGA LIHC -0.892; TCGA LUAD -0.26; TCGA LUSC -0.288; TCGA SARC -0.748; TCGA STAD -0.545; TCGA UCEC -0.289 |
hsa-miR-331-3p | PKNOX2 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; STAD; UCEC | PITA; mirMAP | TCGA BLCA -0.439; TCGA BRCA -0.28; TCGA CESC -0.294; TCGA COAD -0.692; TCGA ESCA -0.609; TCGA HNSC -0.382; TCGA KIRC -0.366; TCGA LUAD -0.242; TCGA LUSC -0.42; TCGA PAAD -0.471; TCGA STAD -0.592; TCGA UCEC -0.47 |
hsa-miR-331-3p | YPEL4 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUSC; PAAD; SARC; THCA; STAD; UCEC | PITA; miRNATAP | TCGA BLCA -0.235; TCGA BRCA -0.428; TCGA CESC -0.23; TCGA COAD -0.576; TCGA ESCA -0.252; TCGA HNSC -0.113; TCGA LGG -0.113; TCGA LUSC -0.316; TCGA PAAD -0.234; TCGA SARC -0.165; TCGA THCA -0.525; TCGA STAD -0.258; TCGA UCEC -0.459 |
hsa-miR-331-3p | MDGA1 | 10 cancers: BLCA; BRCA; CESC; COAD; KIRC; LIHC; LUAD; PAAD; STAD; UCEC | PITA; miRNATAP | TCGA BLCA -0.574; TCGA BRCA -0.15; TCGA CESC -0.221; TCGA COAD -0.321; TCGA KIRC -0.177; TCGA LIHC -0.249; TCGA LUAD -0.186; TCGA PAAD -0.329; TCGA STAD -0.22; TCGA UCEC -0.315 |
hsa-miR-331-3p | STAC2 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; OV; PAAD; SARC; UCEC | PITA | TCGA BLCA -0.423; TCGA BRCA -0.895; TCGA CESC -0.379; TCGA ESCA -0.404; TCGA HNSC -0.565; TCGA KIRC -1.533; TCGA KIRP -0.436; TCGA OV -0.368; TCGA PAAD -0.531; TCGA SARC -0.457; TCGA UCEC -0.281 |
hsa-miR-331-3p | NFIX | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; LGG; LIHC; LUAD; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.501; TCGA BRCA -0.112; TCGA CESC -0.27; TCGA COAD -0.123; TCGA ESCA -0.214; TCGA LGG -0.104; TCGA LIHC -0.23; TCGA LUAD -0.174; TCGA PAAD -0.22; TCGA SARC -0.116; TCGA THCA -0.075; TCGA STAD -0.235; TCGA UCEC -0.221 |
hsa-miR-331-3p | EVC | 10 cancers: BLCA; BRCA; COAD; KIRC; LUSC; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.529; TCGA BRCA -0.057; TCGA COAD -0.505; TCGA KIRC -0.26; TCGA LUSC -0.194; TCGA PAAD -0.274; TCGA SARC -0.182; TCGA THCA -0.171; TCGA STAD -0.277; TCGA UCEC -0.236 |
hsa-miR-331-3p | MGLL | 10 cancers: BLCA; BRCA; ESCA; HNSC; LUAD; LUSC; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.432; TCGA BRCA -0.145; TCGA ESCA -0.497; TCGA HNSC -0.185; TCGA LUAD -0.118; TCGA LUSC -0.33; TCGA SARC -0.166; TCGA THCA -0.093; TCGA STAD -0.159; TCGA UCEC -0.123 |
hsa-miR-331-3p | RAPGEF3 | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LIHC; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.228; TCGA BRCA -0.466; TCGA CESC -0.217; TCGA ESCA -0.28; TCGA HNSC -0.133; TCGA KIRC -0.576; TCGA LIHC -0.134; TCGA LUSC -0.39; TCGA PAAD -0.232; TCGA PRAD -0.088; TCGA SARC -0.156; TCGA STAD -0.137; TCGA UCEC -0.191 |
hsa-miR-331-3p | PRX | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.324; TCGA BRCA -0.431; TCGA CESC -0.125; TCGA ESCA -0.255; TCGA HNSC -0.18; TCGA KIRC -0.153; TCGA LUAD -0.132; TCGA LUSC -0.404; TCGA PAAD -0.286; TCGA THCA -0.157; TCGA STAD -0.179; TCGA UCEC -0.443 |
hsa-miR-331-3p | RASA4 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUSC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.307; TCGA BRCA -0.235; TCGA CESC -0.28; TCGA COAD -0.173; TCGA ESCA -0.299; TCGA HNSC -0.22; TCGA LGG -0.167; TCGA LUSC -0.177; TCGA THCA -0.195; TCGA STAD -0.209; TCGA UCEC -0.191 |
hsa-miR-331-3p | AGFG2 | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUSC; THCA; UCEC | mirMAP | TCGA BLCA -0.104; TCGA BRCA -0.112; TCGA CESC -0.175; TCGA HNSC -0.199; TCGA KIRC -0.235; TCGA KIRP -0.214; TCGA LIHC -0.168; TCGA LUSC -0.088; TCGA THCA -0.074; TCGA UCEC -0.14 |
hsa-miR-331-3p | NRXN2 | 9 cancers: BLCA; BRCA; CESC; COAD; LGG; LIHC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.841; TCGA BRCA -0.424; TCGA CESC -0.448; TCGA COAD -0.415; TCGA LGG -0.09; TCGA LIHC -0.397; TCGA PAAD -0.432; TCGA STAD -0.503; TCGA UCEC -0.518 |
hsa-miR-331-3p | TSPAN11 | 9 cancers: BLCA; BRCA; COAD; ESCA; LGG; LUSC; PAAD; THCA; STAD | mirMAP | TCGA BLCA -0.449; TCGA BRCA -0.257; TCGA COAD -0.422; TCGA ESCA -0.432; TCGA LGG -0.177; TCGA LUSC -0.343; TCGA PAAD -0.446; TCGA THCA -0.397; TCGA STAD -0.266 |
hsa-miR-331-3p | TFEB | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LGG; LUSC; PAAD; PRAD; UCEC | mirMAP; miRNATAP | TCGA BLCA -0.322; TCGA BRCA -0.223; TCGA CESC -0.131; TCGA ESCA -0.167; TCGA HNSC -0.171; TCGA KIRP -0.111; TCGA LGG -0.12; TCGA LUSC -0.202; TCGA PAAD -0.166; TCGA PRAD -0.137; TCGA UCEC -0.127 |
hsa-miR-331-3p | KCNIP3 | 9 cancers: BLCA; BRCA; ESCA; HNSC; LGG; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.49; TCGA BRCA -0.198; TCGA ESCA -0.303; TCGA HNSC -0.137; TCGA LGG -0.174; TCGA LUSC -0.264; TCGA PAAD -0.233; TCGA STAD -0.422; TCGA UCEC -0.274 |
hsa-miR-331-3p | CHD5 | 10 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LUSC; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.362; TCGA BRCA -0.116; TCGA ESCA -0.891; TCGA KIRC -1.013; TCGA KIRP -0.658; TCGA LUSC -0.228; TCGA SARC -0.694; TCGA THCA -0.381; TCGA STAD -0.572; TCGA UCEC -0.256 |
hsa-miR-331-3p | MEF2D | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.185; TCGA BRCA -0.079; TCGA CESC -0.085; TCGA COAD -0.111; TCGA ESCA -0.157; TCGA LUSC -0.121; TCGA SARC -0.149; TCGA STAD -0.096; TCGA UCEC -0.2 |
hsa-miR-331-3p | PDLIM2 | 11 cancers: BLCA; BRCA; COAD; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; THCA; UCEC | mirMAP | TCGA BLCA -0.408; TCGA BRCA -0.217; TCGA COAD -0.141; TCGA HNSC -0.205; TCGA LGG -0.09; TCGA LIHC -0.146; TCGA LUAD -0.159; TCGA LUSC -0.287; TCGA PAAD -0.212; TCGA THCA -0.154; TCGA UCEC -0.11 |
hsa-miR-331-3p | PDE1B | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.625; TCGA BRCA -0.209; TCGA CESC -0.41; TCGA COAD -0.473; TCGA ESCA -0.698; TCGA HNSC -0.198; TCGA LUAD -0.198; TCGA LUSC -0.483; TCGA PAAD -0.258; TCGA SARC -0.291; TCGA STAD -0.401; TCGA UCEC -0.478 |
hsa-miR-331-3p | SLC12A4 | 10 cancers: BLCA; BRCA; COAD; LGG; LIHC; LUSC; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.203; TCGA BRCA -0.19; TCGA COAD -0.13; TCGA LGG -0.151; TCGA LIHC -0.13; TCGA LUSC -0.271; TCGA PAAD -0.208; TCGA SARC -0.175; TCGA STAD -0.151; TCGA UCEC -0.145 |
hsa-miR-331-3p | THRA | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LGG; SARC; UCEC | mirMAP | TCGA BLCA -0.336; TCGA BRCA -0.111; TCGA CESC -0.199; TCGA HNSC -0.096; TCGA KIRC -0.241; TCGA KIRP -0.168; TCGA LGG -0.084; TCGA SARC -0.12; TCGA UCEC -0.197 |
hsa-miR-331-3p | WTIP | 12 cancers: BLCA; BRCA; COAD; HNSC; KIRC; KIRP; LUSC; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.166; TCGA BRCA -0.332; TCGA COAD -0.215; TCGA HNSC -0.221; TCGA KIRC -0.285; TCGA KIRP -0.13; TCGA LUSC -0.299; TCGA PAAD -0.504; TCGA PRAD -0.085; TCGA THCA -0.335; TCGA STAD -0.19; TCGA UCEC -0.203 |
hsa-miR-331-3p | ADAMTS10 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUSC; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.627; TCGA BRCA -0.366; TCGA CESC -0.442; TCGA COAD -0.331; TCGA ESCA -0.527; TCGA HNSC -0.212; TCGA LGG -0.177; TCGA LUSC -0.437; TCGA PAAD -0.447; TCGA SARC -0.176; TCGA THCA -0.286; TCGA STAD -0.295; TCGA UCEC -0.253 |
hsa-miR-331-3p | GPSM1 | 9 cancers: BLCA; BRCA; COAD; KIRP; LGG; LUSC; PAAD; THCA; UCEC | mirMAP | TCGA BLCA -0.131; TCGA BRCA -0.166; TCGA COAD -0.243; TCGA KIRP -0.127; TCGA LGG -0.256; TCGA LUSC -0.203; TCGA PAAD -0.328; TCGA THCA -0.075; TCGA UCEC -0.089 |
hsa-miR-331-3p | KLHL21 | 9 cancers: BLCA; BRCA; CESC; KIRC; LIHC; OV; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.174; TCGA BRCA -0.135; TCGA CESC -0.175; TCGA KIRC -0.211; TCGA LIHC -0.247; TCGA OV -0.105; TCGA SARC -0.259; TCGA STAD -0.146; TCGA UCEC -0.156 |
hsa-miR-331-3p | MXRA8 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUSC; PAAD; PRAD; THCA; UCEC | mirMAP | TCGA BLCA -0.694; TCGA BRCA -0.262; TCGA CESC -0.293; TCGA COAD -0.381; TCGA ESCA -0.393; TCGA HNSC -0.296; TCGA LUSC -0.308; TCGA PAAD -0.396; TCGA PRAD -0.071; TCGA THCA -0.66; TCGA UCEC -0.409 |
hsa-miR-331-3p | NFASC | 10 cancers: BLCA; CESC; COAD; ESCA; KIRC; LUAD; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.458; TCGA CESC -0.438; TCGA COAD -0.398; TCGA ESCA -0.492; TCGA KIRC -0.334; TCGA LUAD -0.145; TCGA LUSC -0.375; TCGA SARC -0.826; TCGA STAD -0.702; TCGA UCEC -0.441 |
hsa-miR-331-3p | SYNPO | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUAD; LUSC; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.455; TCGA BRCA -0.2; TCGA CESC -0.16; TCGA COAD -0.275; TCGA ESCA -0.28; TCGA LUAD -0.19; TCGA LUSC -0.425; TCGA SARC -0.364; TCGA THCA -0.33; TCGA STAD -0.264; TCGA UCEC -0.303 |
hsa-miR-331-3p | ZHX3 | 9 cancers: BLCA; CESC; ESCA; KIRC; LIHC; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.178; TCGA CESC -0.106; TCGA ESCA -0.3; TCGA KIRC -0.189; TCGA LIHC -0.125; TCGA LUSC -0.109; TCGA SARC -0.219; TCGA STAD -0.221; TCGA UCEC -0.122 |
hsa-miR-331-3p | SCN4B | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.462; TCGA BRCA -0.454; TCGA CESC -0.364; TCGA COAD -0.596; TCGA ESCA -0.488; TCGA HNSC -0.192; TCGA LIHC -0.271; TCGA LUAD -0.28; TCGA LUSC -0.4; TCGA PAAD -0.32; TCGA STAD -0.49; TCGA UCEC -0.592 |
hsa-miR-331-3p | PTRF | 13 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.693; TCGA BRCA -0.275; TCGA COAD -0.309; TCGA ESCA -0.2; TCGA KIRP -0.173; TCGA LUAD -0.16; TCGA LUSC -0.267; TCGA PAAD -0.277; TCGA PRAD -0.071; TCGA SARC -0.16; TCGA THCA -0.249; TCGA STAD -0.393; TCGA UCEC -0.483 |
hsa-miR-331-3p | LYNX1 | 13 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LUSC; OV; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.46; TCGA BRCA -0.291; TCGA CESC -0.527; TCGA COAD -0.38; TCGA HNSC -0.347; TCGA KIRC -0.307; TCGA KIRP -0.229; TCGA LUSC -0.305; TCGA OV -0.297; TCGA SARC -0.472; TCGA THCA -0.386; TCGA STAD -0.48; TCGA UCEC -0.383 |
hsa-miR-331-3p | CBX6 | 9 cancers: BLCA; BRCA; CESC; COAD; KIRC; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.176; TCGA BRCA -0.109; TCGA CESC -0.201; TCGA COAD -0.285; TCGA KIRC -0.144; TCGA SARC -0.324; TCGA THCA -0.164; TCGA STAD -0.286; TCGA UCEC -0.234 |
hsa-miR-331-3p | SORCS2 | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.91; TCGA BRCA -0.293; TCGA COAD -0.455; TCGA ESCA -0.505; TCGA KIRC -0.392; TCGA LUAD -0.313; TCGA LUSC -0.492; TCGA PAAD -0.384; TCGA THCA -0.587; TCGA STAD -0.262; TCGA UCEC -0.437 |
hsa-miR-331-3p | MAP3K3 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LUSC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.266; TCGA BRCA -0.073; TCGA CESC -0.142; TCGA COAD -0.169; TCGA ESCA -0.172; TCGA KIRC -0.111; TCGA LUSC -0.183; TCGA THCA -0.169; TCGA STAD -0.186; TCGA UCEC -0.224 |
hsa-miR-331-3p | CCL21 | 12 cancers: BLCA; BRCA; CESC; COAD; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -1.721; TCGA BRCA -0.76; TCGA CESC -0.394; TCGA COAD -1.026; TCGA LUAD -0.2; TCGA LUSC -0.283; TCGA PAAD -0.836; TCGA PRAD -0.127; TCGA SARC -0.793; TCGA THCA -0.822; TCGA STAD -0.418; TCGA UCEC -0.833 |
hsa-miR-331-3p | ZCCHC24 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.709; TCGA BRCA -0.309; TCGA CESC -0.293; TCGA COAD -0.331; TCGA ESCA -0.581; TCGA HNSC -0.248; TCGA LGG -0.131; TCGA LIHC -0.144; TCGA LUAD -0.09; TCGA LUSC -0.376; TCGA PAAD -0.364; TCGA THCA -0.169; TCGA STAD -0.474; TCGA UCEC -0.473 |
hsa-miR-331-3p | GRASP | 11 cancers: BLCA; BRCA; CESC; COAD; HNSC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.404; TCGA BRCA -0.484; TCGA CESC -0.189; TCGA COAD -0.21; TCGA HNSC -0.279; TCGA LUAD -0.179; TCGA LUSC -0.215; TCGA PAAD -0.337; TCGA PRAD -0.184; TCGA STAD -0.21; TCGA UCEC -0.275 |
hsa-miR-331-3p | NTN1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LGG; LIHC; PRAD; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.744; TCGA BRCA -0.13; TCGA CESC -0.322; TCGA COAD -0.366; TCGA ESCA -0.47; TCGA KIRC -0.403; TCGA LGG -0.137; TCGA LIHC -0.225; TCGA PRAD -0.185; TCGA SARC -0.355; TCGA THCA -0.2; TCGA STAD -0.595 |
hsa-miR-331-3p | KCTD15 | 9 cancers: BLCA; CESC; COAD; KIRC; LGG; LUAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.296; TCGA CESC -0.2; TCGA COAD -0.317; TCGA KIRC -0.134; TCGA LGG -0.08; TCGA LUAD -0.116; TCGA SARC -0.265; TCGA STAD -0.284; TCGA UCEC -0.311 |
hsa-miR-331-3p | SLC17A7 | 9 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; PAAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.307; TCGA BRCA -0.443; TCGA ESCA -0.353; TCGA KIRC -0.269; TCGA KIRP -0.223; TCGA PAAD -0.535; TCGA THCA -0.257; TCGA STAD -0.341; TCGA UCEC -0.188 |
hsa-miR-331-3p | DNAJB5 | 10 cancers: BLCA; BRCA; COAD; ESCA; LGG; LIHC; LUSC; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.63; TCGA BRCA -0.067; TCGA COAD -0.477; TCGA ESCA -0.243; TCGA LGG -0.112; TCGA LIHC -0.072; TCGA LUSC -0.156; TCGA SARC -0.348; TCGA STAD -0.655; TCGA UCEC -0.207 |
hsa-miR-331-3p | CXXC5 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUSC; SARC; THCA | miRNATAP | TCGA BLCA -0.252; TCGA BRCA -0.099; TCGA CESC -0.347; TCGA ESCA -0.214; TCGA HNSC -0.153; TCGA KIRC -0.107; TCGA LUSC -0.17; TCGA SARC -0.159; TCGA THCA -0.136 |
hsa-miR-331-3p | JPH4 | 12 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.239; TCGA BRCA -0.307; TCGA CESC -0.453; TCGA COAD -0.516; TCGA HNSC -0.17; TCGA KIRC -0.342; TCGA LUAD -0.229; TCGA LUSC -0.344; TCGA PAAD -0.412; TCGA THCA -0.326; TCGA STAD -0.354; TCGA UCEC -0.89 |
hsa-miR-331-3p | LTBP3 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; PAAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.308; TCGA BRCA -0.267; TCGA COAD -0.216; TCGA ESCA -0.249; TCGA HNSC -0.167; TCGA KIRC -0.202; TCGA KIRP -0.255; TCGA LGG -0.203; TCGA PAAD -0.292; TCGA SARC -0.149; TCGA THCA -0.271; TCGA STAD -0.201; TCGA UCEC -0.152 |
hsa-miR-331-3p | ZC3H12B | 10 cancers: BLCA; BRCA; CESC; ESCA; KIRC; KIRP; LUAD; LUSC; STAD; UCEC | miRNATAP | TCGA BLCA -0.14; TCGA BRCA -0.144; TCGA CESC -0.35; TCGA ESCA -0.6; TCGA KIRC -0.266; TCGA KIRP -0.126; TCGA LUAD -0.094; TCGA LUSC -0.328; TCGA STAD -0.381; TCGA UCEC -0.437 |
hsa-miR-331-3p | ADAMTS15 | 9 cancers: BLCA; BRCA; ESCA; LUAD; LUSC; PAAD; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.759; TCGA BRCA -0.257; TCGA ESCA -0.408; TCGA LUAD -0.189; TCGA LUSC -0.453; TCGA PAAD -0.477; TCGA SARC -0.311; TCGA THCA -0.295; TCGA STAD -0.271 |
hsa-miR-331-3p | ARID5A | 10 cancers: BLCA; BRCA; HNSC; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.372; TCGA BRCA -0.302; TCGA HNSC -0.089; TCGA LUSC -0.129; TCGA PAAD -0.372; TCGA PRAD -0.166; TCGA SARC -0.227; TCGA THCA -0.207; TCGA STAD -0.104; TCGA UCEC -0.183 |
hsa-miR-331-3p | SH2D3C | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUAD; LUSC; PAAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.269; TCGA BRCA -0.223; TCGA CESC -0.222; TCGA COAD -0.277; TCGA ESCA -0.176; TCGA LUAD -0.124; TCGA LUSC -0.343; TCGA PAAD -0.23; TCGA STAD -0.109; TCGA UCEC -0.239 |
hsa-miR-331-3p | KCNMA1 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUSC; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.946; TCGA BRCA -0.09; TCGA CESC -0.323; TCGA COAD -0.851; TCGA ESCA -0.783; TCGA LUSC -0.174; TCGA SARC -0.567; TCGA THCA -0.429; TCGA STAD -0.912; TCGA UCEC -0.553 |
hsa-miR-331-3p | MITF | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.664; TCGA CESC -0.28; TCGA COAD -0.496; TCGA ESCA -0.685; TCGA HNSC -0.207; TCGA KIRP -0.135; TCGA LUAD -0.124; TCGA LUSC -0.347; TCGA SARC -0.308; TCGA THCA -0.074; TCGA STAD -0.356; TCGA UCEC -0.389 |
hsa-miR-331-3p | FHIT | 9 cancers: BRCA; ESCA; HNSC; KIRC; LGG; LUSC; OV; PRAD; STAD | miRNAWalker2 validate; miRTarBase | TCGA BRCA -0.121; TCGA ESCA -0.502; TCGA HNSC -0.319; TCGA KIRC -0.128; TCGA LGG -0.1; TCGA LUSC -0.254; TCGA OV -0.116; TCGA PRAD -0.084; TCGA STAD -0.175 |
hsa-miR-331-3p | HEMK1 | 11 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; SARC | MirTarget | TCGA BRCA -0.149; TCGA ESCA -0.174; TCGA HNSC -0.202; TCGA KIRC -0.158; TCGA KIRP -0.111; TCGA LGG -0.11; TCGA LIHC -0.11; TCGA LUAD -0.063; TCGA LUSC -0.207; TCGA OV -0.168; TCGA SARC -0.196 |
hsa-miR-331-3p | FAM78B | 10 cancers: BRCA; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.134; TCGA ESCA -0.335; TCGA HNSC -0.223; TCGA LIHC -0.152; TCGA LUAD -0.173; TCGA LUSC -0.185; TCGA PAAD -0.2; TCGA THCA -0.2; TCGA STAD -0.167; TCGA UCEC -0.285 |
hsa-miR-331-3p | FNDC4 | 9 cancers: BRCA; COAD; ESCA; HNSC; KIRP; PAAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.296; TCGA COAD -0.366; TCGA ESCA -0.226; TCGA HNSC -0.452; TCGA KIRP -0.261; TCGA PAAD -0.263; TCGA THCA -0.31; TCGA STAD -0.146; TCGA UCEC -0.149 |
hsa-miR-331-3p | MAFK | 9 cancers: BRCA; CESC; KIRC; KIRP; LUSC; PRAD; SARC; THCA; UCEC | PITA | TCGA BRCA -0.279; TCGA CESC -0.14; TCGA KIRC -0.199; TCGA KIRP -0.186; TCGA LUSC -0.239; TCGA PRAD -0.071; TCGA SARC -0.181; TCGA THCA -0.076; TCGA UCEC -0.145 |
hsa-miR-331-3p | ATOH8 | 15 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | PITA; miRNATAP | TCGA BRCA -0.719; TCGA CESC -0.377; TCGA ESCA -0.594; TCGA HNSC -0.487; TCGA KIRC -0.309; TCGA KIRP -0.305; TCGA LGG -0.426; TCGA LIHC -0.354; TCGA LUAD -0.246; TCGA LUSC -0.416; TCGA PAAD -0.394; TCGA PRAD -0.172; TCGA THCA -0.181; TCGA STAD -0.487; TCGA UCEC -0.517 |
hsa-miR-331-3p | P2RX6 | 9 cancers: BRCA; HNSC; KIRC; LGG; LUAD; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.44; TCGA HNSC -0.462; TCGA KIRC -0.214; TCGA LGG -0.263; TCGA LUAD -0.244; TCGA LUSC -0.465; TCGA SARC -0.34; TCGA STAD -0.284; TCGA UCEC -0.229 |
hsa-miR-331-3p | CDH23 | 10 cancers: BRCA; COAD; ESCA; HNSC; LIHC; LUSC; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.406; TCGA COAD -0.329; TCGA ESCA -0.574; TCGA HNSC -0.199; TCGA LIHC -0.425; TCGA LUSC -0.359; TCGA PAAD -0.35; TCGA THCA -0.353; TCGA STAD -0.322; TCGA UCEC -0.253 |
hsa-miR-331-3p | CLN8 | 9 cancers: BRCA; CESC; ESCA; HNSC; KIRC; LUSC; PAAD; THCA; STAD | mirMAP | TCGA BRCA -0.105; TCGA CESC -0.175; TCGA ESCA -0.155; TCGA HNSC -0.089; TCGA KIRC -0.135; TCGA LUSC -0.12; TCGA PAAD -0.147; TCGA THCA -0.063; TCGA STAD -0.075 |
hsa-miR-331-3p | GABBR1 | 9 cancers: BRCA; ESCA; KIRC; KIRP; LUSC; PAAD; SARC; THCA; UCEC | mirMAP | TCGA BRCA -0.191; TCGA ESCA -0.263; TCGA KIRC -0.386; TCGA KIRP -0.594; TCGA LUSC -0.26; TCGA PAAD -0.254; TCGA SARC -0.289; TCGA THCA -0.18; TCGA UCEC -0.412 |
hsa-miR-331-3p | KIAA0141 | 10 cancers: BRCA; CESC; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; OV; STAD | miRNATAP | TCGA BRCA -0.075; TCGA CESC -0.093; TCGA ESCA -0.138; TCGA HNSC -0.069; TCGA KIRC -0.062; TCGA LIHC -0.143; TCGA LUAD -0.104; TCGA LUSC -0.203; TCGA OV -0.165; TCGA STAD -0.066 |
hsa-miR-331-3p | PIM1 | 9 cancers: BRCA; CESC; HNSC; LIHC; LUSC; PAAD; PRAD; THCA; UCEC | miRNATAP | TCGA BRCA -0.093; TCGA CESC -0.165; TCGA HNSC -0.188; TCGA LIHC -0.172; TCGA LUSC -0.151; TCGA PAAD -0.188; TCGA PRAD -0.189; TCGA THCA -0.258; TCGA UCEC -0.203 |
hsa-miR-331-3p | UBE2G2 | 9 cancers: HNSC; KIRP; LGG; LUAD; LUSC; PAAD; SARC; STAD; UCEC | MirTarget | TCGA HNSC -0.076; TCGA KIRP -0.116; TCGA LGG -0.133; TCGA LUAD -0.07; TCGA LUSC -0.06; TCGA PAAD -0.076; TCGA SARC -0.063; TCGA STAD -0.074; TCGA UCEC -0.07 |
Enriched cancer pathways of putative targets