microRNA information: hsa-miR-508-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-508-3p | miRbase |
| Accession: | MIMAT0002880 | miRbase |
| Precursor name: | hsa-mir-508 | miRbase |
| Precursor accession: | MI0003195 | miRbase |
| Symbol: | MIR508 | HGNC |
| RefSeq ID: | NR_030235 | GenBank |
| Sequence: | UGAUUGUAGCCUUUUGGAGUAGA |
Reported expression in cancers: hsa-miR-508-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-508-3p | kidney renal cell cancer | downregulation | "Identification of miR 508 3p and miR 509 3p that a ......" | 22369946 |
Reported cancer pathway affected by hsa-miR-508-3p
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-508-3p | kidney renal cell cancer | Apoptosis pathway | "Identification of miR 508 3p and miR 509 3p that a ......" | 22369946 |
Reported cancer prognosis affected by hsa-miR-508-3p
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-508-3p | esophageal cancer | poor survival | "miR 508 sustains phosphoinositide signalling and p ......" | 25099196 | |
| hsa-miR-508-3p | gastric cancer | drug resistance | "miR 508 5p regulates multidrug resistance of gastr ......" | 23893241 | |
| hsa-miR-508-3p | gastric cancer | drug resistance | "The miR27b CCNG1 P53 miR 508 5p axis regulates mul ......" | 26623719 | |
| hsa-miR-508-3p | kidney renal cell cancer | cell migration | "Identification of miR 508 3p and miR 509 3p that a ......" | 22369946 | |
| hsa-miR-508-3p | melanoma | progression | "MiR 508 5p Inhibits the Progression of Glioma by T ......" | 27003587 | |
| hsa-miR-508-3p | ovarian cancer | staging; progression | "To identify a unique microRNA miRNA pattern associ ......" | 23836287 | |
| hsa-miR-508-3p | pancreatic cancer | poor survival | "Quantitative real-time PCR qRT-PCR was subsequentl ......" | 26819679 |
Reported gene related to hsa-miR-508-3p
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-508-3p | gastric cancer | ABCB1 | "miR 508 5p regulates multidrug resistance of gastr ......" | 23893241 |
| hsa-miR-508-3p | melanoma | TECRL | "MiR 508 5p Inhibits the Progression of Glioma by T ......" | 27003587 |
| hsa-miR-508-3p | gastric cancer | ZNRD1 | "miR 508 5p regulates multidrug resistance of gastr ......" | 23893241 |
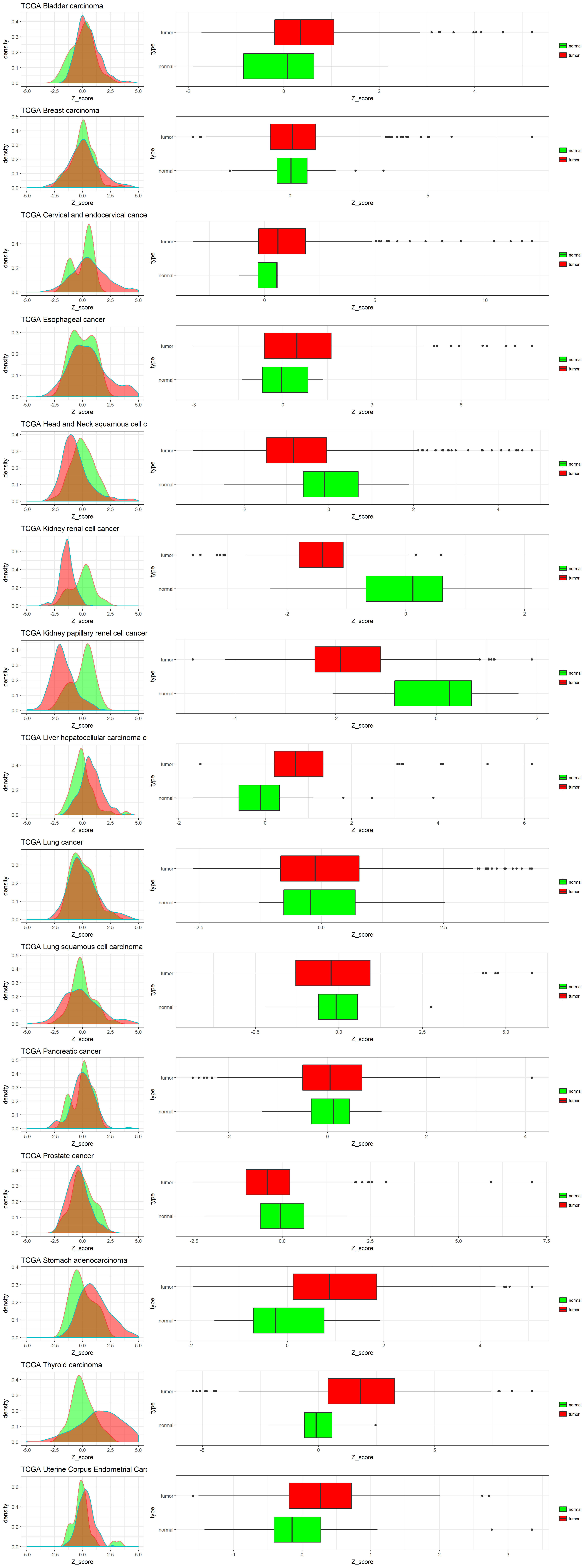
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-508-3p | MITF | 9 cancers: BLCA; BRCA; COAD; KIRP; LGG; LIHC; OV; SARC; STAD | PITA; miRNATAP | TCGA BLCA -0.121; TCGA BRCA -0.09; TCGA COAD -0.155; TCGA KIRP -0.098; TCGA LGG -0.097; TCGA LIHC -0.066; TCGA OV -0.052; TCGA SARC -0.092; TCGA STAD -0.231 |
hsa-miR-508-3p | ELL2 | 9 cancers: BLCA; CESC; COAD; HNSC; OV; PAAD; SARC; THCA; STAD | PITA; miRNATAP | TCGA BLCA -0.079; TCGA CESC -0.112; TCGA COAD -0.056; TCGA HNSC -0.061; TCGA OV -0.103; TCGA PAAD -0.214; TCGA SARC -0.119; TCGA THCA -0.052; TCGA STAD -0.12 |
hsa-miR-508-3p | LOX | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LUSC; OV; PRAD | mirMAP | TCGA BLCA -0.105; TCGA BRCA -0.058; TCGA CESC -0.119; TCGA COAD -0.139; TCGA HNSC -0.087; TCGA KIRC -0.273; TCGA LUSC -0.12; TCGA OV -0.246; TCGA PRAD -0.142 |
hsa-miR-508-3p | MAP7D3 | 9 cancers: BLCA; BRCA; KIRC; KIRP; LGG; PAAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.146; TCGA BRCA -0.093; TCGA KIRC -0.075; TCGA KIRP -0.067; TCGA LGG -0.197; TCGA PAAD -0.106; TCGA SARC -0.069; TCGA THCA -0.119; TCGA STAD -0.109 |
hsa-miR-508-3p | FBXO32 | 11 cancers: BLCA; CESC; COAD; HNSC; LGG; LUAD; LUSC; OV; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.111; TCGA CESC -0.106; TCGA COAD -0.11; TCGA HNSC -0.05; TCGA LGG -0.159; TCGA LUAD -0.073; TCGA LUSC -0.092; TCGA OV -0.06; TCGA SARC -0.133; TCGA STAD -0.22; TCGA UCEC -0.077 |
hsa-miR-508-3p | ATL3 | 9 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUSC; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.07; TCGA CESC -0.062; TCGA ESCA -0.084; TCGA HNSC -0.053; TCGA LGG -0.071; TCGA LUSC -0.094; TCGA PRAD -0.054; TCGA SARC -0.057; TCGA STAD -0.054 |
hsa-miR-508-3p | SAMD4A | 9 cancers: BLCA; BRCA; CESC; COAD; LGG; LUSC; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.147; TCGA BRCA -0.07; TCGA CESC -0.103; TCGA COAD -0.098; TCGA LGG -0.161; TCGA LUSC -0.099; TCGA SARC -0.077; TCGA STAD -0.174; TCGA UCEC -0.118 |
hsa-miR-508-3p | ABLIM3 | 9 cancers: CESC; COAD; ESCA; HNSC; KIRC; LIHC; THCA; STAD; UCEC | MirTarget; PITA; miRNATAP | TCGA CESC -0.156; TCGA COAD -0.202; TCGA ESCA -0.122; TCGA HNSC -0.091; TCGA KIRC -0.167; TCGA LIHC -0.079; TCGA THCA -0.11; TCGA STAD -0.164; TCGA UCEC -0.068 |
hsa-miR-508-3p | GJC1 | 9 cancers: COAD; HNSC; KIRC; LGG; OV; SARC; THCA; STAD; UCEC | mirMAP | TCGA COAD -0.074; TCGA HNSC -0.096; TCGA KIRC -0.338; TCGA LGG -0.092; TCGA OV -0.059; TCGA SARC -0.082; TCGA THCA -0.087; TCGA STAD -0.188; TCGA UCEC -0.066 |
Enriched cancer pathways of putative targets