microRNA information: hsa-miR-584-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-584-5p | miRbase |
| Accession: | MIMAT0003249 | miRbase |
| Precursor name: | hsa-mir-584 | miRbase |
| Precursor accession: | MI0003591 | miRbase |
| Symbol: | MIR584 | HGNC |
| RefSeq ID: | NR_030310 | GenBank |
| Sequence: | UUAUGGUUUGCCUGGGACUGAG |
Reported expression in cancers: hsa-miR-584-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-584-5p | kidney renal cell cancer | downregulation | "Tumour suppressor microRNA 584 directly targets on ......" | 21119662 | Microarray |
| hsa-miR-584-5p | lung cancer | deregulation | "Therefore we conduct to systematically identify mi ......" | 26870998 | Microarray |
Reported cancer pathway affected by hsa-miR-584-5p
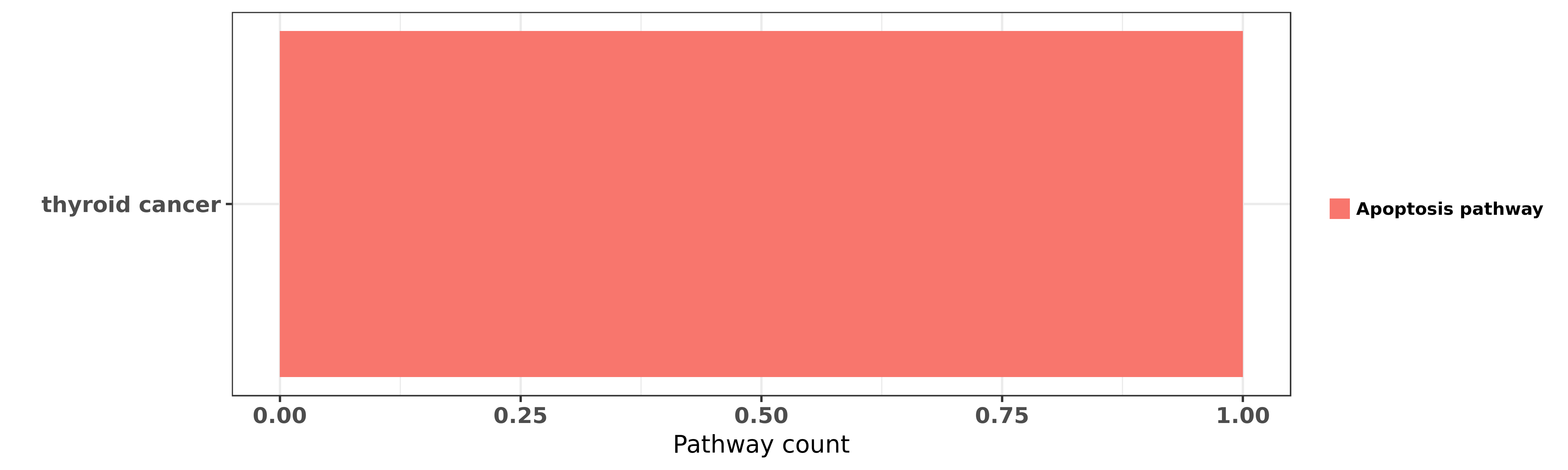
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-584-5p | thyroid cancer | Apoptosis pathway | "Here using miRNome profiling of papillary thyroid ......" | 27661106 | Luciferase |
Reported cancer prognosis affected by hsa-miR-584-5p
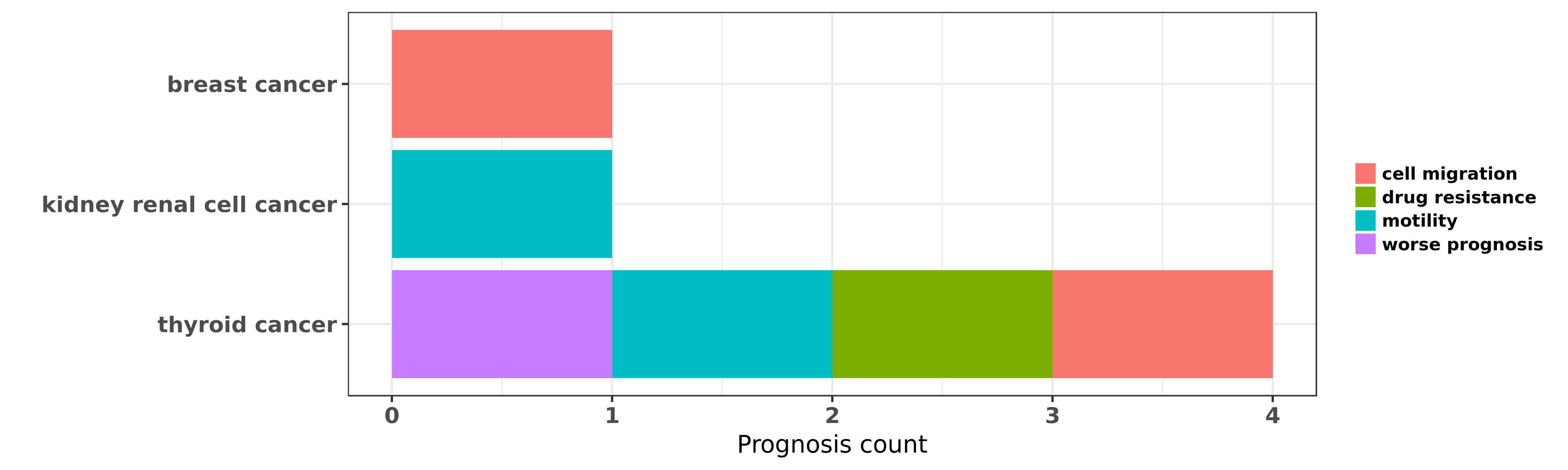
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-584-5p | breast cancer | cell migration | "MicroRNA 584 and the protein phosphatase and actin ......" | 23479725 | |
| hsa-miR-584-5p | kidney renal cell cancer | motility | "Tumour suppressor microRNA 584 directly targets on ......" | 21119662 | Wound Healing Assay; Luciferase |
| hsa-miR-584-5p | thyroid cancer | cell migration; worse prognosis; motility | "miR 584 Suppresses Invasion and Cell Migration of ......" | 26405762 | Cell proliferation assay; Cell Proliferation Assay; Western blot |
| hsa-miR-584-5p | thyroid cancer | drug resistance | "Here using miRNome profiling of papillary thyroid ......" | 27661106 | Luciferase |
Reported gene related to hsa-miR-584-5p
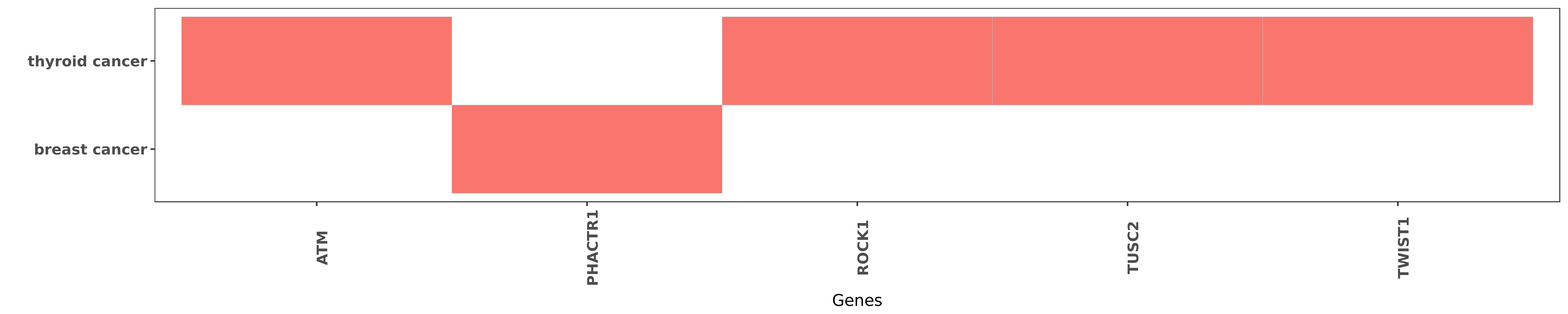
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-584-5p | thyroid cancer | ATM | "Importantly miR-584 was up-regulated in human ATC ......" | 27661106 |
| hsa-miR-584-5p | breast cancer | PHACTR1 | "MicroRNA 584 and the protein phosphatase and actin ......" | 23479725 |
| hsa-miR-584-5p | thyroid cancer | ROCK1 | "miR 584 Suppresses Invasion and Cell Migration of ......" | 26405762 |
| hsa-miR-584-5p | thyroid cancer | TUSC2 | "Using bioinformatics programs we identified TUSC2 ......" | 27661106 |
| hsa-miR-584-5p | thyroid cancer | TWIST1 | "Here using miRNome profiling of papillary thyroid ......" | 27661106 |
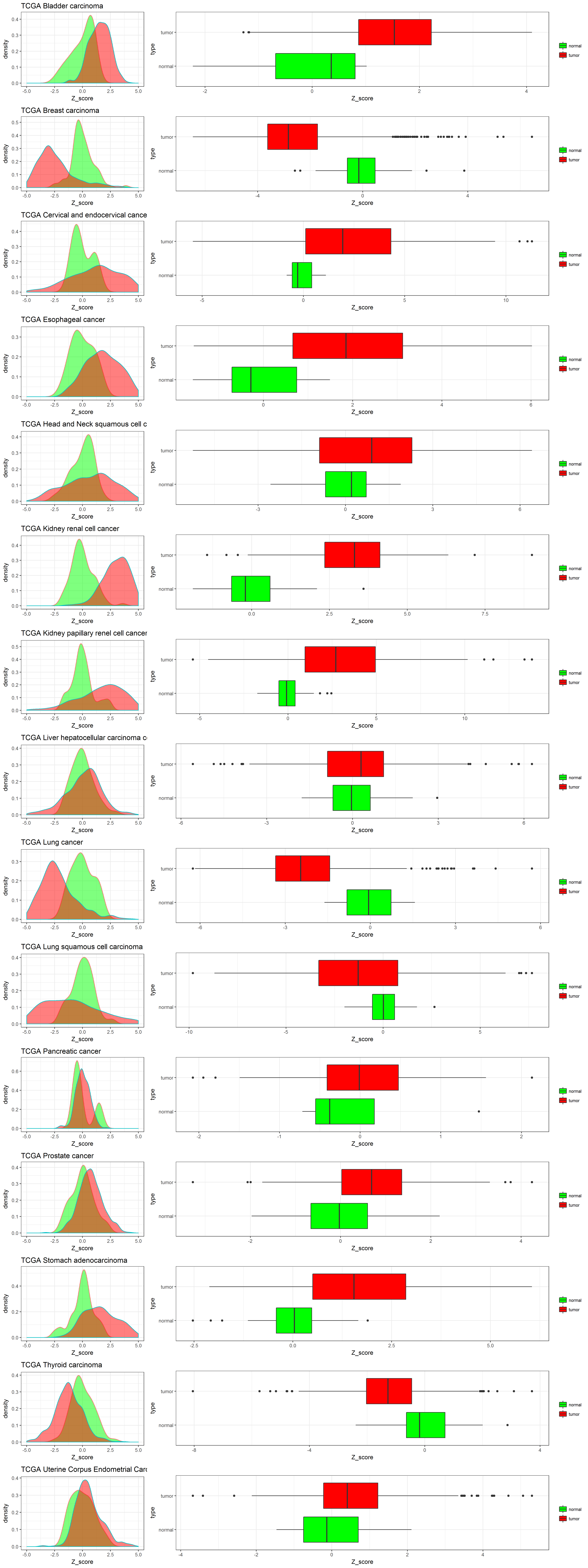
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-584-5p | LMO3 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; PAAD; PRAD; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.52; TCGA CESC -0.506; TCGA COAD -0.702; TCGA ESCA -0.719; TCGA HNSC -0.365; TCGA KIRC -1.141; TCGA LUAD -0.518; TCGA PAAD -0.492; TCGA PRAD -0.179; TCGA SARC -0.621; TCGA THCA -0.381; TCGA STAD -0.734 |
hsa-miR-584-5p | MYRIP | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.282; TCGA BRCA -0.296; TCGA CESC -0.43; TCGA ESCA -0.555; TCGA HNSC -0.396; TCGA PAAD -0.441; TCGA SARC -0.325; TCGA STAD -0.571; TCGA UCEC -0.321 |
hsa-miR-584-5p | NPHP1 | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.118; TCGA BRCA -0.183; TCGA CESC -0.152; TCGA COAD -0.139; TCGA HNSC -0.106; TCGA KIRC -0.07; TCGA KIRP -0.157; TCGA PAAD -0.139; TCGA STAD -0.154; TCGA UCEC -0.11 |
hsa-miR-584-5p | ZNF512 | 14 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; UCEC | MirTarget | TCGA BLCA -0.06; TCGA CESC -0.12; TCGA ESCA -0.123; TCGA HNSC -0.163; TCGA KIRP -0.083; TCGA LGG -0.06; TCGA LIHC -0.085; TCGA LUAD -0.068; TCGA LUSC -0.074; TCGA PAAD -0.115; TCGA PRAD -0.229; TCGA SARC -0.082; TCGA THCA -0.056; TCGA UCEC -0.109 |
hsa-miR-584-5p | ZNF208 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.242; TCGA CESC -0.166; TCGA COAD -0.366; TCGA ESCA -0.653; TCGA HNSC -0.241; TCGA KIRC -0.211; TCGA KIRP -0.3; TCGA LUSC -0.122; TCGA PAAD -0.252; TCGA SARC -0.22; TCGA STAD -0.312; TCGA UCEC -0.202 |
hsa-miR-584-5p | KIAA2022 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.662; TCGA BRCA -0.127; TCGA CESC -0.799; TCGA COAD -0.534; TCGA ESCA -0.928; TCGA HNSC -0.799; TCGA KIRC -1.074; TCGA KIRP -0.202; TCGA LUSC -0.506; TCGA PAAD -0.557; TCGA SARC -0.852; TCGA STAD -0.781; TCGA UCEC -0.249 |
hsa-miR-584-5p | PPP1R12B | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.29; TCGA CESC -0.124; TCGA COAD -0.306; TCGA ESCA -0.402; TCGA HNSC -0.144; TCGA KIRP -0.089; TCGA PRAD -0.186; TCGA SARC -0.843; TCGA STAD -0.495; TCGA UCEC -0.191 |
hsa-miR-584-5p | SORBS1 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.509; TCGA CESC -0.187; TCGA COAD -0.289; TCGA ESCA -0.551; TCGA HNSC -0.163; TCGA KIRP -0.123; TCGA PRAD -0.281; TCGA SARC -0.6; TCGA STAD -0.552 |
hsa-miR-584-5p | TBL1X | 9 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; PAAD; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.124; TCGA BRCA -0.065; TCGA HNSC -0.179; TCGA KIRC -0.068; TCGA KIRP -0.076; TCGA PAAD -0.184; TCGA PRAD -0.073; TCGA SARC -0.209; TCGA STAD -0.116 |
hsa-miR-584-5p | NTRK3 | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.512; TCGA CESC -0.325; TCGA COAD -0.443; TCGA ESCA -0.792; TCGA HNSC -0.508; TCGA KIRC -0.255; TCGA KIRP -0.288; TCGA PRAD -0.311; TCGA SARC -0.305; TCGA THCA -0.668; TCGA STAD -0.629 |
hsa-miR-584-5p | ST6GAL2 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.327; TCGA BRCA -0.316; TCGA CESC -0.222; TCGA COAD -0.485; TCGA ESCA -0.502; TCGA HNSC -0.435; TCGA KIRP -0.284; TCGA LGG -0.081; TCGA LUSC -0.202; TCGA PRAD -0.418; TCGA SARC -0.574; TCGA STAD -0.33; TCGA UCEC -0.166 |
hsa-miR-584-5p | PIK3R1 | 10 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LGG; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.137; TCGA CESC -0.264; TCGA ESCA -0.129; TCGA HNSC -0.159; TCGA KIRP -0.086; TCGA LGG -0.072; TCGA LUSC -0.072; TCGA PRAD -0.15; TCGA STAD -0.09; TCGA UCEC -0.192 |
hsa-miR-584-5p | SSBP2 | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.239; TCGA BRCA -0.102; TCGA CESC -0.3; TCGA ESCA -0.237; TCGA HNSC -0.289; TCGA KIRP -0.137; TCGA LUSC -0.14; TCGA PAAD -0.2; TCGA PRAD -0.198; TCGA SARC -0.259; TCGA THCA -0.206; TCGA STAD -0.423; TCGA UCEC -0.115 |
hsa-miR-584-5p | GABRB2 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUSC; THCA | mirMAP | TCGA BLCA -0.364; TCGA BRCA -0.129; TCGA CESC -0.5; TCGA COAD -0.541; TCGA ESCA -0.412; TCGA HNSC -0.547; TCGA KIRC -0.388; TCGA LUSC -0.199; TCGA THCA -1.624 |
hsa-miR-584-5p | TXLNB | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.432; TCGA BRCA -0.183; TCGA CESC -0.248; TCGA COAD -0.278; TCGA ESCA -0.481; TCGA HNSC -0.262; TCGA LUSC -0.193; TCGA SARC -0.218; TCGA STAD -0.429; TCGA UCEC -0.143 |
hsa-miR-584-5p | KCND3 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.501; TCGA BRCA -0.314; TCGA CESC -0.176; TCGA COAD -0.34; TCGA ESCA -0.346; TCGA KIRC -0.652; TCGA PAAD -0.278; TCGA SARC -0.98; TCGA STAD -0.414 |
hsa-miR-584-5p | THAP2 | 13 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; OV; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.128; TCGA CESC -0.085; TCGA ESCA -0.083; TCGA HNSC -0.093; TCGA KIRC -0.096; TCGA KIRP -0.074; TCGA LGG -0.071; TCGA OV -0.088; TCGA PAAD -0.078; TCGA PRAD -0.097; TCGA THCA -0.053; TCGA STAD -0.064; TCGA UCEC -0.103 |
hsa-miR-584-5p | FAM120C | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; PAAD; THCA; UCEC | mirMAP | TCGA BLCA -0.076; TCGA CESC -0.139; TCGA ESCA -0.096; TCGA HNSC -0.134; TCGA KIRC -0.121; TCGA KIRP -0.056; TCGA PAAD -0.263; TCGA THCA -0.059; TCGA UCEC -0.072 |
hsa-miR-584-5p | PTPRT | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.282; TCGA BRCA -0.865; TCGA CESC -0.46; TCGA COAD -0.444; TCGA ESCA -0.725; TCGA HNSC -0.524; TCGA KIRC -0.761; TCGA PAAD -0.887; TCGA SARC -0.52; TCGA STAD -0.475 |
hsa-miR-584-5p | DOK6 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.401; TCGA BRCA -0.109; TCGA CESC -0.199; TCGA COAD -0.312; TCGA ESCA -0.215; TCGA HNSC -0.188; TCGA KIRP -0.148; TCGA PAAD -0.276; TCGA PRAD -0.165; TCGA STAD -0.241; TCGA UCEC -0.162 |
hsa-miR-584-5p | ZNF627 | 13 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; OV; PAAD; PRAD; SARC; UCEC | MirTarget | TCGA BRCA -0.119; TCGA CESC -0.092; TCGA ESCA -0.082; TCGA HNSC -0.074; TCGA KIRC -0.123; TCGA KIRP -0.063; TCGA LGG -0.093; TCGA LUAD -0.057; TCGA OV -0.17; TCGA PAAD -0.095; TCGA PRAD -0.064; TCGA SARC -0.061; TCGA UCEC -0.111 |
hsa-miR-584-5p | MGAT4A | 9 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; PAAD | mirMAP | TCGA BRCA -0.243; TCGA CESC -0.122; TCGA COAD -0.09; TCGA ESCA -0.166; TCGA HNSC -0.297; TCGA KIRC -0.133; TCGA KIRP -0.128; TCGA LUAD -0.072; TCGA PAAD -0.184 |
hsa-miR-584-5p | ERBB4 | 9 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; OV; PAAD; STAD | mirMAP | TCGA BRCA -1.049; TCGA CESC -0.471; TCGA ESCA -0.664; TCGA HNSC -0.411; TCGA KIRC -1.708; TCGA KIRP -0.562; TCGA OV -0.442; TCGA PAAD -0.472; TCGA STAD -0.363 |
hsa-miR-584-5p | ZNF33A | 9 cancers: CESC; ESCA; HNSC; KIRC; KIRP; LUSC; PAAD; SARC; UCEC | mirMAP | TCGA CESC -0.089; TCGA ESCA -0.131; TCGA HNSC -0.105; TCGA KIRC -0.06; TCGA KIRP -0.055; TCGA LUSC -0.066; TCGA PAAD -0.08; TCGA SARC -0.165; TCGA UCEC -0.11 |
Enriched cancer pathways of putative targets