microRNA information: hsa-miR-628-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-628-5p | miRbase |
| Accession: | MIMAT0004809 | miRbase |
| Precursor name: | hsa-mir-628 | miRbase |
| Precursor accession: | MI0003642 | miRbase |
| Symbol: | MIR628 | HGNC |
| RefSeq ID: | NR_030358 | GenBank |
| Sequence: | AUGCUGACAUAUUUACUAGAGG |
Reported expression in cancers: hsa-miR-628-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-628-5p | kidney renal cell cancer | upregulation | "The most relevant differentially expressed microRN ......" | 24475095 | qPCR |
| hsa-miR-628-5p | prostate cancer | downregulation | "Circulatory miR 628 5p is downregulated in prostat ......" | 24477576 | Microarray; qPCR |
Reported cancer pathway affected by hsa-miR-628-5p
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|
Reported cancer prognosis affected by hsa-miR-628-5p

| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-628-5p | kidney renal cell cancer | progression; poor survival | "The most relevant differentially expressed microRN ......" | 24475095 | |
| hsa-miR-628-5p | prostate cancer | worse prognosis | "Circulatory miR 628 5p is downregulated in prostat ......" | 24477576 |
Reported gene related to hsa-miR-628-5p
| miRNA | cancer | gene | reporting | PUBMED |
|---|
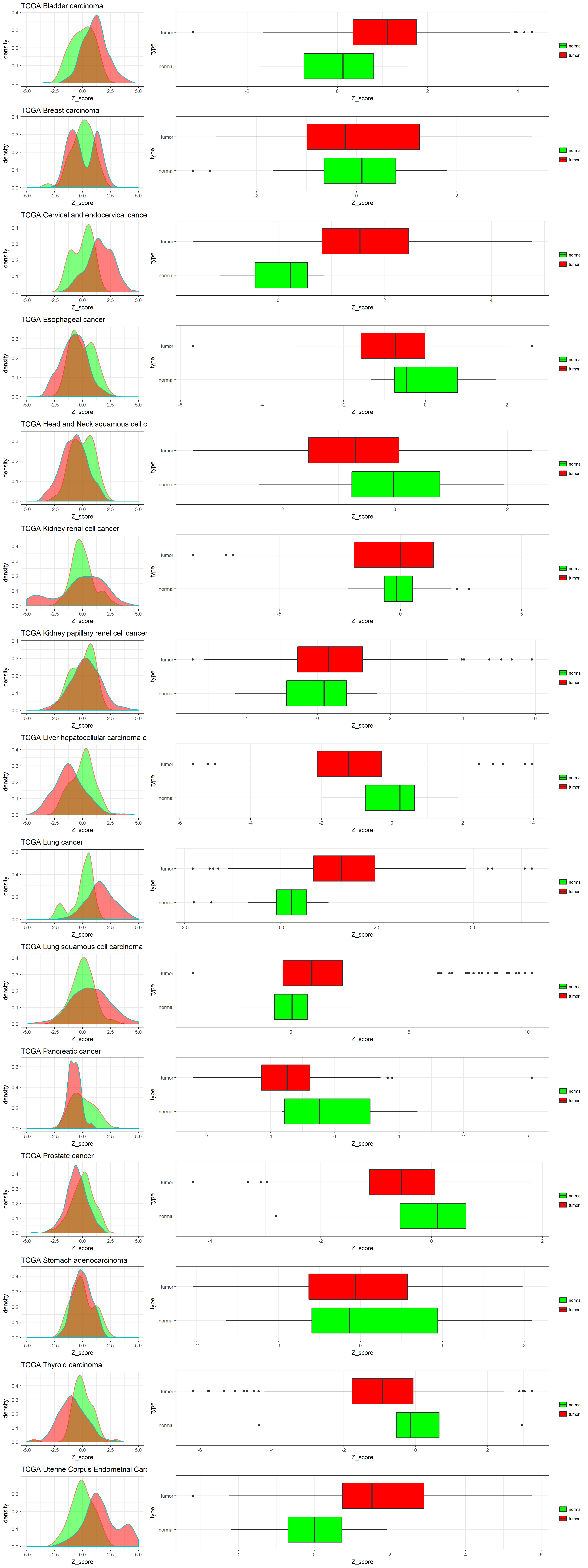
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-628-5p | PLEKHN1 | 11 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUSC; PAAD; PRAD; THCA | MirTarget; miRNATAP | TCGA BLCA -0.37; TCGA BRCA -0.184; TCGA CESC -0.501; TCGA HNSC -0.294; TCGA KIRC -0.277; TCGA KIRP -0.506; TCGA LIHC -0.243; TCGA LUSC -0.223; TCGA PAAD -1.241; TCGA PRAD -0.165; TCGA THCA -1.062 |
hsa-miR-628-5p | ANAPC16 | 9 cancers: BLCA; COAD; KIRP; LGG; LIHC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.12; TCGA COAD -0.14; TCGA KIRP -0.089; TCGA LGG -0.076; TCGA LIHC -0.081; TCGA PRAD -0.058; TCGA SARC -0.084; TCGA STAD -0.155; TCGA UCEC -0.112 |
hsa-miR-628-5p | BCL6 | 10 cancers: BLCA; ESCA; KIRC; KIRP; LGG; LUAD; PAAD; SARC; THCA; STAD | MirTarget; PITA; miRNATAP | TCGA BLCA -0.22; TCGA ESCA -0.266; TCGA KIRC -0.087; TCGA KIRP -0.229; TCGA LGG -0.149; TCGA LUAD -0.122; TCGA PAAD -0.39; TCGA SARC -0.194; TCGA THCA -0.213; TCGA STAD -0.179 |
hsa-miR-628-5p | SH3D19 | 9 cancers: BLCA; CESC; COAD; LGG; LUAD; PAAD; SARC; STAD; UCEC | PITA | TCGA BLCA -0.216; TCGA CESC -0.189; TCGA COAD -0.111; TCGA LGG -0.116; TCGA LUAD -0.137; TCGA PAAD -0.274; TCGA SARC -0.221; TCGA STAD -0.273; TCGA UCEC -0.07 |
hsa-miR-628-5p | PLEKHM1 | 9 cancers: BLCA; BRCA; CESC; HNSC; LGG; LUAD; LUSC; PAAD; THCA | PITA; miRNATAP | TCGA BLCA -0.114; TCGA BRCA -0.056; TCGA CESC -0.13; TCGA HNSC -0.092; TCGA LGG -0.056; TCGA LUAD -0.099; TCGA LUSC -0.089; TCGA PAAD -0.135; TCGA THCA -0.129 |
hsa-miR-628-5p | FLYWCH1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LIHC; PRAD; SARC | mirMAP | TCGA BLCA -0.143; TCGA BRCA -0.181; TCGA CESC -0.173; TCGA COAD -0.122; TCGA ESCA -0.227; TCGA HNSC -0.063; TCGA KIRC -0.092; TCGA LGG -0.051; TCGA LIHC -0.173; TCGA PRAD -0.07; TCGA SARC -0.061 |
hsa-miR-628-5p | RAPGEF3 | 13 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.396; TCGA BRCA -0.259; TCGA CESC -0.212; TCGA HNSC -0.179; TCGA KIRC -0.186; TCGA KIRP -0.247; TCGA LIHC -0.27; TCGA LUAD -0.128; TCGA LUSC -0.167; TCGA PAAD -0.227; TCGA SARC -0.223; TCGA STAD -0.12; TCGA UCEC -0.135 |
hsa-miR-628-5p | HEMK1 | 10 cancers: BLCA; BRCA; KIRC; KIRP; LGG; LIHC; LUSC; PRAD; SARC; UCEC | mirMAP | TCGA BLCA -0.13; TCGA BRCA -0.135; TCGA KIRC -0.133; TCGA KIRP -0.176; TCGA LGG -0.152; TCGA LIHC -0.133; TCGA LUSC -0.106; TCGA PRAD -0.162; TCGA SARC -0.194; TCGA UCEC -0.053 |
hsa-miR-628-5p | LYNX1 | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LUSC; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.401; TCGA BRCA -0.163; TCGA CESC -0.441; TCGA HNSC -0.385; TCGA KIRC -0.132; TCGA KIRP -0.203; TCGA LUSC -0.202; TCGA SARC -0.331; TCGA THCA -0.487; TCGA STAD -0.27 |
hsa-miR-628-5p | MKL1 | 9 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LIHC; LUAD; PAAD; SARC | mirMAP | TCGA BLCA -0.131; TCGA BRCA -0.057; TCGA HNSC -0.053; TCGA KIRC -0.066; TCGA KIRP -0.06; TCGA LIHC -0.055; TCGA LUAD -0.066; TCGA PAAD -0.105; TCGA SARC -0.098 |
hsa-miR-628-5p | FBXO32 | 9 cancers: BLCA; CESC; ESCA; LIHC; PAAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.227; TCGA CESC -0.246; TCGA ESCA -0.353; TCGA LIHC -0.471; TCGA PAAD -0.401; TCGA SARC -0.462; TCGA THCA -0.379; TCGA STAD -0.556; TCGA UCEC -0.249 |
hsa-miR-628-5p | JMJD6 | 10 cancers: BRCA; HNSC; KIRC; KIRP; LGG; LIHC; PRAD; SARC; THCA; UCEC | mirMAP | TCGA BRCA -0.06; TCGA HNSC -0.106; TCGA KIRC -0.07; TCGA KIRP -0.105; TCGA LGG -0.063; TCGA LIHC -0.123; TCGA PRAD -0.178; TCGA SARC -0.112; TCGA THCA -0.058; TCGA UCEC -0.061 |
hsa-miR-628-5p | SEPT9 | 9 cancers: BRCA; COAD; HNSC; KIRC; KIRP; LIHC; PAAD; SARC; THCA | mirMAP | TCGA BRCA -0.067; TCGA COAD -0.077; TCGA HNSC -0.118; TCGA KIRC -0.062; TCGA KIRP -0.096; TCGA LIHC -0.067; TCGA PAAD -0.294; TCGA SARC -0.07; TCGA THCA -0.267 |
hsa-miR-628-5p | GABRE | 11 cancers: BRCA; CESC; COAD; HNSC; KIRC; KIRP; LIHC; LUSC; PAAD; STAD; UCEC | miRNATAP | TCGA BRCA -0.134; TCGA CESC -0.416; TCGA COAD -0.509; TCGA HNSC -0.193; TCGA KIRC -0.381; TCGA KIRP -0.4; TCGA LIHC -0.236; TCGA LUSC -0.402; TCGA PAAD -0.733; TCGA STAD -0.448; TCGA UCEC -0.237 |
hsa-miR-628-5p | CRABP2 | 9 cancers: BRCA; COAD; HNSC; KIRC; KIRP; PAAD; THCA; STAD; UCEC | miRNATAP | TCGA BRCA -0.122; TCGA COAD -0.419; TCGA HNSC -0.392; TCGA KIRC -0.21; TCGA KIRP -0.686; TCGA PAAD -1.243; TCGA THCA -0.718; TCGA STAD -0.417; TCGA UCEC -0.158 |
hsa-miR-628-5p | ARL8B | 9 cancers: CESC; COAD; ESCA; HNSC; LUSC; PAAD; THCA; STAD; UCEC | MirTarget; PITA | TCGA CESC -0.212; TCGA COAD -0.074; TCGA ESCA -0.137; TCGA HNSC -0.098; TCGA LUSC -0.106; TCGA PAAD -0.07; TCGA THCA -0.099; TCGA STAD -0.133; TCGA UCEC -0.129 |
hsa-miR-628-5p | CAV2 | 10 cancers: CESC; ESCA; HNSC; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA CESC -0.358; TCGA ESCA -0.603; TCGA HNSC -0.289; TCGA LUAD -0.227; TCGA LUSC -0.286; TCGA PAAD -0.525; TCGA SARC -0.237; TCGA THCA -0.174; TCGA STAD -0.314; TCGA UCEC -0.185 |
hsa-miR-628-5p | CAV1 | 11 cancers: CESC; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | PITA; miRNATAP | TCGA CESC -0.437; TCGA ESCA -0.686; TCGA HNSC -0.509; TCGA KIRP -0.285; TCGA LUAD -0.41; TCGA LUSC -0.283; TCGA PAAD -0.338; TCGA SARC -0.383; TCGA THCA -0.194; TCGA STAD -0.393; TCGA UCEC -0.239 |
hsa-miR-628-5p | KCTD15 | 9 cancers: CESC; ESCA; HNSC; LGG; LIHC; LUAD; SARC; STAD; UCEC | mirMAP; miRNATAP | TCGA CESC -0.509; TCGA ESCA -0.356; TCGA HNSC -0.23; TCGA LGG -0.086; TCGA LIHC -0.286; TCGA LUAD -0.144; TCGA SARC -0.243; TCGA STAD -0.267; TCGA UCEC -0.223 |
hsa-miR-628-5p | KLF6 | 9 cancers: CESC; HNSC; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA CESC -0.293; TCGA HNSC -0.122; TCGA LUAD -0.164; TCGA LUSC -0.178; TCGA PAAD -0.35; TCGA SARC -0.189; TCGA THCA -0.303; TCGA STAD -0.124; TCGA UCEC -0.237 |
hsa-miR-628-5p | ENAH | 9 cancers: COAD; ESCA; HNSC; LGG; PAAD; SARC; THCA; STAD; UCEC | MirTarget; PITA | TCGA COAD -0.167; TCGA ESCA -0.246; TCGA HNSC -0.233; TCGA LGG -0.087; TCGA PAAD -0.196; TCGA SARC -0.28; TCGA THCA -0.186; TCGA STAD -0.307; TCGA UCEC -0.079 |
hsa-miR-628-5p | CEP164 | 9 cancers: HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; STAD | mirMAP | TCGA HNSC -0.148; TCGA KIRC -0.106; TCGA KIRP -0.159; TCGA LGG -0.162; TCGA LIHC -0.11; TCGA LUAD -0.098; TCGA LUSC -0.061; TCGA SARC -0.076; TCGA STAD -0.125 |
Enriched cancer pathways of putative targets