microRNA information: hsa-miR-629-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-629-5p | miRbase |
| Accession: | MIMAT0004810 | miRbase |
| Precursor name: | hsa-mir-629 | miRbase |
| Precursor accession: | MI0003643 | miRbase |
| Symbol: | MIR629 | HGNC |
| RefSeq ID: | NR_030714 | GenBank |
| Sequence: | UGGGUUUACGUUGGGAGAACU |
Reported expression in cancers: hsa-miR-629-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-629-5p | gastric cancer | upregulation | "Plasma samples from 3 independent groups comprise ......" | 26607322 | Microarray |
Reported cancer pathway affected by hsa-miR-629-5p
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|
Reported cancer prognosis affected by hsa-miR-629-5p
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|
Reported gene related to hsa-miR-629-5p
| miRNA | cancer | gene | reporting | PUBMED |
|---|
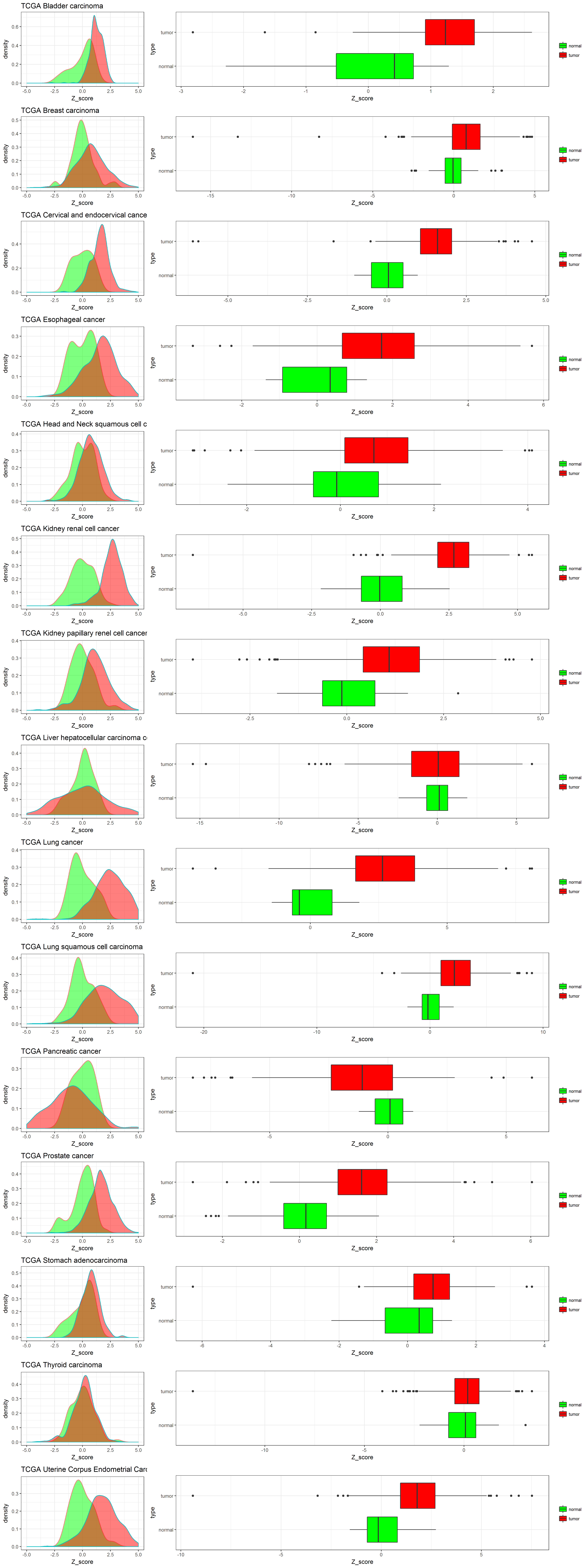
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-629-5p | FGF1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.165; TCGA BRCA -0.283; TCGA CESC -0.203; TCGA COAD -0.497; TCGA ESCA -0.35; TCGA HNSC -0.659; TCGA KIRC -1.385; TCGA LUAD -0.256; TCGA LUSC -0.427; TCGA PAAD -0.327; TCGA PRAD -0.509; TCGA STAD -0.417; TCGA UCEC -0.376 |
hsa-miR-629-5p | TCF4 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LUAD; OV; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.216; TCGA BRCA -0.138; TCGA COAD -0.336; TCGA ESCA -0.213; TCGA HNSC -0.208; TCGA LGG -0.13; TCGA LUAD -0.217; TCGA OV -0.099; TCGA PRAD -0.256; TCGA STAD -0.265; TCGA UCEC -0.213 |
hsa-miR-629-5p | NCAM1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.887; TCGA BRCA -0.308; TCGA CESC -0.196; TCGA COAD -0.571; TCGA ESCA -1.063; TCGA HNSC -0.911; TCGA LGG -0.138; TCGA OV -0.392; TCGA PAAD -0.792; TCGA PRAD -0.929; TCGA SARC -0.638; TCGA THCA -0.494; TCGA STAD -0.819; TCGA UCEC -0.342 |
hsa-miR-629-5p | ZNF135 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUSC; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.605; TCGA CESC -0.272; TCGA COAD -0.417; TCGA ESCA -0.534; TCGA HNSC -0.528; TCGA KIRC -0.256; TCGA LUSC -0.221; TCGA SARC -0.32; TCGA STAD -0.655; TCGA UCEC -0.401 |
hsa-miR-629-5p | GAL3ST1 | 9 cancers: BLCA; BRCA; CESC; KIRP; LGG; LUAD; PAAD; PRAD; UCEC | MirTarget | TCGA BLCA -0.274; TCGA BRCA -0.255; TCGA CESC -0.434; TCGA KIRP -0.36; TCGA LGG -0.403; TCGA LUAD -0.452; TCGA PAAD -0.835; TCGA PRAD -0.192; TCGA UCEC -0.261 |
hsa-miR-629-5p | ANO5 | 16 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.819; TCGA CESC -0.466; TCGA COAD -0.686; TCGA ESCA -0.888; TCGA HNSC -1.276; TCGA KIRC -1.534; TCGA KIRP -0.341; TCGA LGG -0.51; TCGA LUAD -0.591; TCGA LUSC -0.257; TCGA PAAD -0.628; TCGA PRAD -1.032; TCGA SARC -0.953; TCGA THCA -0.312; TCGA STAD -0.983; TCGA UCEC -0.413 |
hsa-miR-629-5p | ZFP3 | 9 cancers: BLCA; COAD; KIRC; KIRP; LUAD; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.221; TCGA COAD -0.214; TCGA KIRC -0.184; TCGA KIRP -0.173; TCGA LUAD -0.157; TCGA PAAD -0.233; TCGA SARC -0.087; TCGA STAD -0.153; TCGA UCEC -0.383 |
hsa-miR-629-5p | KLHL13 | 9 cancers: BLCA; BRCA; COAD; KIRC; KIRP; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.285; TCGA BRCA -0.268; TCGA COAD -0.441; TCGA KIRC -0.738; TCGA KIRP -0.406; TCGA PRAD -0.459; TCGA SARC -0.837; TCGA STAD -0.664; TCGA UCEC -0.25 |
hsa-miR-629-5p | CACNA1D | 10 cancers: BLCA; CESC; KIRC; LGG; LUAD; LUSC; PAAD; SARC; THCA; UCEC | MirTarget | TCGA BLCA -0.359; TCGA CESC -0.523; TCGA KIRC -0.303; TCGA LGG -0.3; TCGA LUAD -0.478; TCGA LUSC -0.38; TCGA PAAD -0.586; TCGA SARC -0.263; TCGA THCA -0.257; TCGA UCEC -0.255 |
hsa-miR-629-5p | RASSF8 | 12 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.14; TCGA COAD -0.309; TCGA ESCA -0.354; TCGA HNSC -0.211; TCGA KIRC -0.441; TCGA KIRP -0.257; TCGA LUAD -0.215; TCGA LUSC -0.145; TCGA PRAD -0.166; TCGA SARC -0.192; TCGA STAD -0.558; TCGA UCEC -0.273 |
hsa-miR-629-5p | TRIL | 11 cancers: BLCA; BRCA; COAD; HNSC; KIRC; KIRP; LIHC; LUAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.281; TCGA BRCA -0.128; TCGA COAD -0.193; TCGA HNSC -0.29; TCGA KIRC -0.148; TCGA KIRP -0.204; TCGA LIHC -0.145; TCGA LUAD -0.347; TCGA PRAD -0.23; TCGA STAD -0.233; TCGA UCEC -0.131 |
hsa-miR-629-5p | TSPAN2 | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.464; TCGA BRCA -0.177; TCGA COAD -0.37; TCGA ESCA -0.703; TCGA HNSC -0.428; TCGA KIRC -0.426; TCGA LUAD -0.223; TCGA LUSC -0.42; TCGA OV -0.231; TCGA PRAD -0.737; TCGA SARC -1.282; TCGA THCA -0.295; TCGA STAD -0.789; TCGA UCEC -0.67 |
hsa-miR-629-5p | KCNK2 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; OV; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.465; TCGA BRCA -0.234; TCGA COAD -0.571; TCGA ESCA -0.83; TCGA HNSC -0.746; TCGA LGG -0.171; TCGA OV -0.299; TCGA PRAD -0.823; TCGA STAD -0.865; TCGA UCEC -0.243 |
hsa-miR-629-5p | ANGPTL7 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; STAD | MirTarget | TCGA BLCA -0.934; TCGA BRCA -0.736; TCGA COAD -0.858; TCGA ESCA -0.694; TCGA HNSC -0.845; TCGA KIRC -1.056; TCGA LUAD -0.743; TCGA LUSC -0.64; TCGA PRAD -0.693; TCGA STAD -1.223 |
hsa-miR-629-5p | PHLDB2 | 9 cancers: BLCA; COAD; ESCA; KIRC; KIRP; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.216; TCGA COAD -0.375; TCGA ESCA -0.278; TCGA KIRC -0.16; TCGA KIRP -0.197; TCGA PRAD -0.557; TCGA SARC -0.509; TCGA STAD -0.556; TCGA UCEC -0.248 |
hsa-miR-629-5p | ATP8B2 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUSC; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.219; TCGA BRCA -0.164; TCGA COAD -0.314; TCGA ESCA -0.43; TCGA HNSC -0.182; TCGA LUSC -0.109; TCGA SARC -0.222; TCGA STAD -0.423; TCGA UCEC -0.254 |
hsa-miR-629-5p | MAGI2 | 18 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.327; TCGA BRCA -0.106; TCGA CESC -0.151; TCGA COAD -0.275; TCGA ESCA -0.394; TCGA HNSC -0.337; TCGA KIRC -0.531; TCGA KIRP -0.314; TCGA LGG -0.132; TCGA LIHC -0.107; TCGA LUAD -0.275; TCGA OV -0.12; TCGA PAAD -0.536; TCGA PRAD -0.152; TCGA SARC -0.258; TCGA THCA -0.088; TCGA STAD -0.666; TCGA UCEC -0.327 |
hsa-miR-629-5p | SARM1 | 11 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUAD; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.286; TCGA CESC -0.155; TCGA ESCA -0.206; TCGA HNSC -0.343; TCGA LGG -0.171; TCGA LUAD -0.242; TCGA PAAD -0.438; TCGA SARC -0.388; TCGA THCA -0.052; TCGA STAD -0.107; TCGA UCEC -0.087 |
hsa-miR-629-5p | IGF1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BLCA -0.694; TCGA BRCA -0.49; TCGA CESC -0.428; TCGA COAD -0.581; TCGA ESCA -0.884; TCGA HNSC -0.473; TCGA KIRC -0.151; TCGA LUAD -0.207; TCGA LUSC -0.276; TCGA STAD -0.559; TCGA UCEC -0.264 |
hsa-miR-629-5p | CELF2 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.477; TCGA BRCA -0.206; TCGA COAD -0.183; TCGA ESCA -0.573; TCGA HNSC -0.218; TCGA LUAD -0.347; TCGA LUSC -0.347; TCGA PRAD -0.408; TCGA SARC -0.13; TCGA STAD -0.549 |
hsa-miR-629-5p | MGLL | 12 cancers: BLCA; BRCA; COAD; ESCA; LGG; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.295; TCGA BRCA -0.162; TCGA COAD -0.158; TCGA ESCA -0.208; TCGA LGG -0.097; TCGA LUAD -0.663; TCGA LUSC -0.268; TCGA PRAD -0.388; TCGA SARC -0.11; TCGA THCA -0.107; TCGA STAD -0.382; TCGA UCEC -0.191 |
hsa-miR-629-5p | JPH4 | 14 cancers: BLCA; BRCA; CESC; COAD; KIRC; LGG; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.362; TCGA BRCA -0.176; TCGA CESC -0.205; TCGA COAD -0.426; TCGA KIRC -0.633; TCGA LGG -0.514; TCGA LUAD -0.602; TCGA LUSC -0.313; TCGA OV -0.253; TCGA PAAD -0.342; TCGA PRAD -0.91; TCGA SARC -0.356; TCGA STAD -0.46; TCGA UCEC -0.727 |
hsa-miR-629-5p | YPEL1 | 9 cancers: BLCA; COAD; HNSC; LGG; LIHC; LUAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.114; TCGA COAD -0.2; TCGA HNSC -0.184; TCGA LGG -0.136; TCGA LIHC -0.215; TCGA LUAD -0.146; TCGA SARC -0.331; TCGA STAD -0.144; TCGA UCEC -0.098 |
hsa-miR-629-5p | CRTC1 | 13 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LUAD; OV; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.152; TCGA BRCA -0.086; TCGA CESC -0.169; TCGA COAD -0.093; TCGA HNSC -0.28; TCGA KIRC -0.323; TCGA KIRP -0.143; TCGA LGG -0.261; TCGA LUAD -0.101; TCGA OV -0.088; TCGA PAAD -0.489; TCGA STAD -0.125; TCGA UCEC -0.118 |
hsa-miR-629-5p | LRP2BP | 12 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; OV; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.138; TCGA CESC -0.162; TCGA ESCA -0.138; TCGA HNSC -0.22; TCGA LGG -0.145; TCGA LUAD -0.521; TCGA LUSC -0.284; TCGA OV -0.225; TCGA PAAD -0.383; TCGA SARC -0.251; TCGA STAD -0.211; TCGA UCEC -0.123 |
hsa-miR-629-5p | TSPAN11 | 9 cancers: BLCA; BRCA; COAD; ESCA; LGG; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.301; TCGA BRCA -0.214; TCGA COAD -0.281; TCGA ESCA -0.353; TCGA LGG -0.235; TCGA LUSC -0.277; TCGA PRAD -0.524; TCGA STAD -0.38; TCGA UCEC -0.14 |
hsa-miR-629-5p | BVES | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRC; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.403; TCGA COAD -0.574; TCGA ESCA -0.568; TCGA HNSC -0.426; TCGA KIRC -0.241; TCGA PRAD -0.659; TCGA SARC -0.406; TCGA THCA -0.253; TCGA STAD -0.9; TCGA UCEC -0.276 |
hsa-miR-629-5p | PDE4D | 12 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUSC; OV; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.34; TCGA COAD -0.144; TCGA ESCA -0.377; TCGA HNSC -0.208; TCGA KIRC -0.134; TCGA KIRP -0.131; TCGA LGG -0.267; TCGA LUSC -0.146; TCGA OV -0.168; TCGA PRAD -0.275; TCGA SARC -0.62; TCGA STAD -0.417 |
hsa-miR-629-5p | MTR | 12 cancers: BLCA; CESC; COAD; ESCA; KIRC; LIHC; LUAD; OV; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.112; TCGA CESC -0.109; TCGA COAD -0.081; TCGA ESCA -0.155; TCGA KIRC -0.082; TCGA LIHC -0.148; TCGA LUAD -0.118; TCGA OV -0.102; TCGA PRAD -0.07; TCGA SARC -0.205; TCGA STAD -0.166; TCGA UCEC -0.094 |
hsa-miR-629-5p | ARHGEF9 | 14 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.279; TCGA COAD -0.081; TCGA ESCA -0.3; TCGA HNSC -0.187; TCGA KIRC -0.195; TCGA KIRP -0.207; TCGA LGG -0.209; TCGA LIHC -0.095; TCGA LUAD -0.23; TCGA PAAD -0.284; TCGA PRAD -0.189; TCGA SARC -0.269; TCGA STAD -0.237; TCGA UCEC -0.09 |
hsa-miR-629-5p | SOX5 | 11 cancers: BLCA; COAD; HNSC; KIRC; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.447; TCGA COAD -0.298; TCGA HNSC -0.348; TCGA KIRC -0.329; TCGA OV -0.283; TCGA PAAD -0.556; TCGA PRAD -0.451; TCGA SARC -0.367; TCGA THCA -0.193; TCGA STAD -0.417; TCGA UCEC -0.168 |
hsa-miR-629-5p | ALPK3 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUSC; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.331; TCGA BRCA -0.116; TCGA COAD -0.332; TCGA ESCA -0.267; TCGA HNSC -0.485; TCGA LUSC -0.196; TCGA PAAD -0.438; TCGA SARC -0.588; TCGA STAD -0.456 |
hsa-miR-629-5p | RALGPS1 | 9 cancers: BLCA; KIRC; KIRP; LGG; LUAD; OV; PAAD; SARC; UCEC | mirMAP | TCGA BLCA -0.236; TCGA KIRC -0.85; TCGA KIRP -0.313; TCGA LGG -0.214; TCGA LUAD -0.258; TCGA OV -0.126; TCGA PAAD -0.612; TCGA SARC -0.241; TCGA UCEC -0.092 |
hsa-miR-629-5p | ENTPD1 | 10 cancers: BLCA; CESC; COAD; ESCA; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.257; TCGA CESC -0.096; TCGA COAD -0.151; TCGA ESCA -0.254; TCGA LUAD -0.075; TCGA LUSC -0.176; TCGA PRAD -0.139; TCGA SARC -0.363; TCGA STAD -0.241; TCGA UCEC -0.111 |
hsa-miR-629-5p | NFIB | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; LIHC; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.219; TCGA BRCA -0.171; TCGA CESC -0.249; TCGA COAD -0.134; TCGA HNSC -0.154; TCGA LIHC -0.128; TCGA PRAD -0.135; TCGA SARC -0.154; TCGA STAD -0.413 |
hsa-miR-629-5p | DST | 9 cancers: BLCA; BRCA; COAD; KIRC; LUAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.151; TCGA BRCA -0.228; TCGA COAD -0.218; TCGA KIRC -0.226; TCGA LUAD -0.327; TCGA PRAD -0.508; TCGA SARC -0.356; TCGA STAD -0.394; TCGA UCEC -0.275 |
hsa-miR-629-5p | JMY | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LGG; LIHC; LUAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.167; TCGA BRCA -0.069; TCGA COAD -0.136; TCGA ESCA -0.263; TCGA KIRC -0.456; TCGA LGG -0.059; TCGA LIHC -0.101; TCGA LUAD -0.078; TCGA SARC -0.321; TCGA STAD -0.249; TCGA UCEC -0.109 |
hsa-miR-629-5p | SHROOM4 | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.246; TCGA BRCA -0.15; TCGA COAD -0.178; TCGA ESCA -0.221; TCGA KIRP -0.114; TCGA LIHC -0.106; TCGA LUAD -0.557; TCGA LUSC -0.325; TCGA PRAD -0.411; TCGA STAD -0.169; TCGA UCEC -0.222 |
hsa-miR-629-5p | EDA | 10 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LGG; LUAD; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.29; TCGA BRCA -0.123; TCGA ESCA -0.298; TCGA KIRC -0.091; TCGA KIRP -0.158; TCGA LGG -0.085; TCGA LUAD -0.521; TCGA PAAD -0.445; TCGA STAD -0.231; TCGA UCEC -0.138 |
hsa-miR-629-5p | NFIA | 10 cancers: BLCA; BRCA; COAD; ESCA; LIHC; LUAD; OV; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.237; TCGA BRCA -0.175; TCGA COAD -0.107; TCGA ESCA -0.164; TCGA LIHC -0.15; TCGA LUAD -0.198; TCGA OV -0.12; TCGA PRAD -0.06; TCGA SARC -0.198; TCGA STAD -0.507 |
hsa-miR-629-5p | ANTXR2 | 10 cancers: BLCA; BRCA; COAD; ESCA; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.449; TCGA BRCA -0.165; TCGA COAD -0.159; TCGA ESCA -0.36; TCGA LUAD -0.167; TCGA LUSC -0.322; TCGA PRAD -0.138; TCGA SARC -0.147; TCGA STAD -0.579; TCGA UCEC -0.316 |
hsa-miR-629-5p | APBB2 | 11 cancers: BLCA; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.134; TCGA ESCA -0.119; TCGA HNSC -0.15; TCGA KIRC -0.086; TCGA LGG -0.108; TCGA LIHC -0.124; TCGA LUAD -0.116; TCGA LUSC -0.211; TCGA THCA -0.082; TCGA STAD -0.124; TCGA UCEC -0.066 |
hsa-miR-629-5p | NLGN2 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.122; TCGA BRCA -0.104; TCGA COAD -0.275; TCGA ESCA -0.188; TCGA HNSC -0.164; TCGA LGG -0.166; TCGA PRAD -0.135; TCGA STAD -0.384; TCGA UCEC -0.168 |
hsa-miR-629-5p | SIK2 | 9 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LIHC; LUSC; SARC; STAD | mirMAP | TCGA BLCA -0.087; TCGA BRCA -0.099; TCGA ESCA -0.184; TCGA KIRC -0.15; TCGA KIRP -0.119; TCGA LIHC -0.173; TCGA LUSC -0.095; TCGA SARC -0.091; TCGA STAD -0.104 |
hsa-miR-629-5p | KCND3 | 10 cancers: BLCA; COAD; ESCA; KIRC; LIHC; LUAD; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.773; TCGA COAD -0.402; TCGA ESCA -0.841; TCGA KIRC -1.056; TCGA LIHC -0.231; TCGA LUAD -0.4; TCGA LUSC -0.215; TCGA SARC -1.27; TCGA STAD -0.719; TCGA UCEC -0.432 |
hsa-miR-629-5p | RGMB | 12 cancers: BLCA; CESC; ESCA; HNSC; LGG; LIHC; LUAD; OV; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.149; TCGA CESC -0.071; TCGA ESCA -0.444; TCGA HNSC -0.176; TCGA LGG -0.336; TCGA LIHC -0.242; TCGA LUAD -0.201; TCGA OV -0.099; TCGA PRAD -0.237; TCGA SARC -0.102; TCGA STAD -0.535; TCGA UCEC -0.148 |
hsa-miR-629-5p | GRIN2A | 9 cancers: BLCA; BRCA; COAD; ESCA; LUAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.509; TCGA BRCA -0.15; TCGA COAD -0.672; TCGA ESCA -0.957; TCGA LUAD -0.911; TCGA PRAD -0.827; TCGA SARC -1.055; TCGA STAD -1.162; TCGA UCEC -0.363 |
hsa-miR-629-5p | SV2B | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LGG; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.329; TCGA BRCA -0.327; TCGA COAD -0.494; TCGA ESCA -0.446; TCGA HNSC -0.332; TCGA KIRC -0.562; TCGA LGG -0.535; TCGA PRAD -0.449; TCGA THCA -0.332; TCGA STAD -0.376; TCGA UCEC -0.424 |
hsa-miR-629-5p | TEAD1 | 11 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LIHC; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.206; TCGA COAD -0.156; TCGA ESCA -0.118; TCGA HNSC -0.113; TCGA KIRC -0.092; TCGA LIHC -0.075; TCGA LUSC -0.099; TCGA PRAD -0.346; TCGA SARC -0.242; TCGA STAD -0.347; TCGA UCEC -0.092 |
hsa-miR-629-5p | ZC3H6 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.165; TCGA BRCA -0.071; TCGA COAD -0.147; TCGA ESCA -0.301; TCGA HNSC -0.122; TCGA LGG -0.064; TCGA LIHC -0.131; TCGA LUAD -0.171; TCGA LUSC -0.128; TCGA SARC -0.135; TCGA STAD -0.315; TCGA UCEC -0.139 |
hsa-miR-629-5p | PTAR1 | 9 cancers: BLCA; CESC; ESCA; KIRC; KIRP; LIHC; LUAD; SARC; UCEC | mirMAP | TCGA BLCA -0.06; TCGA CESC -0.099; TCGA ESCA -0.188; TCGA KIRC -0.085; TCGA KIRP -0.117; TCGA LIHC -0.083; TCGA LUAD -0.109; TCGA SARC -0.116; TCGA UCEC -0.059 |
hsa-miR-629-5p | NUDT16 | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUAD; PAAD; STAD | mirMAP | TCGA BLCA -0.103; TCGA BRCA -0.063; TCGA CESC -0.065; TCGA HNSC -0.18; TCGA KIRC -0.246; TCGA KIRP -0.237; TCGA LIHC -0.08; TCGA LUAD -0.188; TCGA PAAD -0.096; TCGA STAD -0.109 |
hsa-miR-629-5p | BMPR2 | 10 cancers: BLCA; COAD; ESCA; LGG; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.123; TCGA COAD -0.224; TCGA ESCA -0.158; TCGA LGG -0.06; TCGA LUAD -0.134; TCGA LUSC -0.128; TCGA PRAD -0.135; TCGA SARC -0.107; TCGA STAD -0.141; TCGA UCEC -0.084 |
hsa-miR-629-5p | MBD5 | 9 cancers: BLCA; COAD; ESCA; KIRC; LGG; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.206; TCGA COAD -0.138; TCGA ESCA -0.184; TCGA KIRC -0.068; TCGA LGG -0.073; TCGA PRAD -0.162; TCGA SARC -0.15; TCGA STAD -0.238; TCGA UCEC -0.064 |
hsa-miR-629-5p | NEURL1B | 9 cancers: BLCA; COAD; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.159; TCGA COAD -0.153; TCGA LUAD -0.275; TCGA LUSC -0.16; TCGA PRAD -0.385; TCGA SARC -0.592; TCGA THCA -0.119; TCGA STAD -0.31; TCGA UCEC -0.123 |
hsa-miR-629-5p | ALS2CL | 9 cancers: BRCA; KIRC; KIRP; LGG; LUAD; LUSC; PRAD; SARC; UCEC | MirTarget | TCGA BRCA -0.328; TCGA KIRC -0.994; TCGA KIRP -0.199; TCGA LGG -0.131; TCGA LUAD -0.5; TCGA LUSC -0.403; TCGA PRAD -0.501; TCGA SARC -0.319; TCGA UCEC -0.18 |
hsa-miR-629-5p | LETMD1 | 9 cancers: BRCA; CESC; HNSC; KIRP; LGG; LIHC; LUAD; PAAD; THCA | MirTarget | TCGA BRCA -0.089; TCGA CESC -0.066; TCGA HNSC -0.118; TCGA KIRP -0.212; TCGA LGG -0.193; TCGA LIHC -0.058; TCGA LUAD -0.095; TCGA PAAD -0.209; TCGA THCA -0.07 |
hsa-miR-629-5p | CLCN4 | 9 cancers: BRCA; CESC; HNSC; KIRP; LUSC; OV; PRAD; THCA; UCEC | mirMAP | TCGA BRCA -0.237; TCGA CESC -0.238; TCGA HNSC -0.39; TCGA KIRP -0.128; TCGA LUSC -0.189; TCGA OV -0.256; TCGA PRAD -0.216; TCGA THCA -0.173; TCGA UCEC -0.186 |
hsa-miR-629-5p | DZIP1 | 11 cancers: BRCA; CESC; COAD; KIRC; KIRP; LGG; OV; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BRCA -0.157; TCGA CESC -0.207; TCGA COAD -0.452; TCGA KIRC -0.218; TCGA KIRP -0.202; TCGA LGG -0.193; TCGA OV -0.136; TCGA PAAD -0.336; TCGA PRAD -0.524; TCGA STAD -0.55; TCGA UCEC -0.255 |
hsa-miR-629-5p | C16orf52 | 9 cancers: BRCA; ESCA; KIRP; LGG; LIHC; LUSC; OV; STAD; UCEC | mirMAP | TCGA BRCA -0.058; TCGA ESCA -0.155; TCGA KIRP -0.121; TCGA LGG -0.12; TCGA LIHC -0.097; TCGA LUSC -0.06; TCGA OV -0.057; TCGA STAD -0.13; TCGA UCEC -0.111 |
hsa-miR-629-5p | SPIN1 | 9 cancers: COAD; ESCA; KIRC; KIRP; OV; PRAD; SARC; STAD; UCEC | MirTarget | TCGA COAD -0.084; TCGA ESCA -0.094; TCGA KIRC -0.106; TCGA KIRP -0.092; TCGA OV -0.152; TCGA PRAD -0.07; TCGA SARC -0.12; TCGA STAD -0.076; TCGA UCEC -0.102 |
hsa-miR-629-5p | PITPNM3 | 9 cancers: COAD; ESCA; KIRC; KIRP; LGG; LUAD; PRAD; SARC; STAD | mirMAP | TCGA COAD -0.173; TCGA ESCA -0.25; TCGA KIRC -0.709; TCGA KIRP -0.504; TCGA LGG -0.189; TCGA LUAD -0.459; TCGA PRAD -0.39; TCGA SARC -0.572; TCGA STAD -0.684 |
hsa-miR-629-5p | HDHD2 | 9 cancers: KIRC; KIRP; LGG; LIHC; LUAD; PAAD; SARC; THCA; STAD | MirTarget | TCGA KIRC -0.18; TCGA KIRP -0.227; TCGA LGG -0.052; TCGA LIHC -0.064; TCGA LUAD -0.095; TCGA PAAD -0.256; TCGA SARC -0.055; TCGA THCA -0.053; TCGA STAD -0.068 |
Enriched cancer pathways of putative targets