microRNA information: hsa-miR-744-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-744-5p | miRbase |
| Accession: | MIMAT0004945 | miRbase |
| Precursor name: | hsa-mir-744 | miRbase |
| Precursor accession: | MI0005559 | miRbase |
| Symbol: | MIR744 | HGNC |
| RefSeq ID: | NR_030613 | GenBank |
| Sequence: | UGCGGGGCUAGGGCUAACAGCA |
Reported expression in cancers: hsa-miR-744-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-744-5p | breast cancer | upregulation | "The aim of this study was to investigate the role ......" | 27746365 | qPCR |
| hsa-miR-744-5p | head and neck cancer | deregulation | "A global miRNA profiling was performed on 12 sampl ......" | 21637912 | qPCR; Microarray |
| hsa-miR-744-5p | liver cancer | downregulation | "Increasing evidences indicate that miR-744 deregul ......" | 24991193 | |
| hsa-miR-744-5p | pancreatic cancer | upregulation | "MiR 744 increases tumorigenicity of pancreatic can ......" | 26485754 |
Reported cancer pathway affected by hsa-miR-744-5p

| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-744-5p | cervical and endocervical cancer | cell cycle pathway; Apoptosis pathway | "These include hsa-miR-138 hsa-miR-210 and hsa-miR- ......" | 23012319 | |
| hsa-miR-744-5p | cervical and endocervical cancer | Apoptosis pathway | "MicroRNA 744 inhibited cervical cancer growth and ......" | 27261616 | |
| hsa-miR-744-5p | liver cancer | cell cycle pathway | "Decrease expression of microRNA 744 promotes cell ......" | 24991193 | Luciferase; Western blot |
Reported cancer prognosis affected by hsa-miR-744-5p

| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-744-5p | breast cancer | worse prognosis; drug resistance | "The aim of this study was to investigate the role ......" | 27746365 | |
| hsa-miR-744-5p | cervical and endocervical cancer | progression | "These include hsa-miR-138 hsa-miR-210 and hsa-miR- ......" | 23012319 | |
| hsa-miR-744-5p | cervical and endocervical cancer | progression; metastasis | "MicroRNA 744 inhibited cervical cancer growth and ......" | 27261616 | |
| hsa-miR-744-5p | head and neck cancer | malignant trasformation | "A global miRNA profiling was performed on 12 sampl ......" | 21637912 | |
| hsa-miR-744-5p | liver cancer | poor survival; recurrence; worse prognosis | "miR 744 is a potential prognostic marker in patien ......" | 25543521 | |
| hsa-miR-744-5p | pancreatic cancer | poor survival | "MiR 744 increases tumorigenicity of pancreatic can ......" | 26485754 | |
| hsa-miR-744-5p | pancreatic cancer | metastasis; recurrence; progression; poor survival; drug resistance; worse prognosis | "Plasma microRNA profiles: identification of miR 74 ......" | 26505678 |
Reported gene related to hsa-miR-744-5p
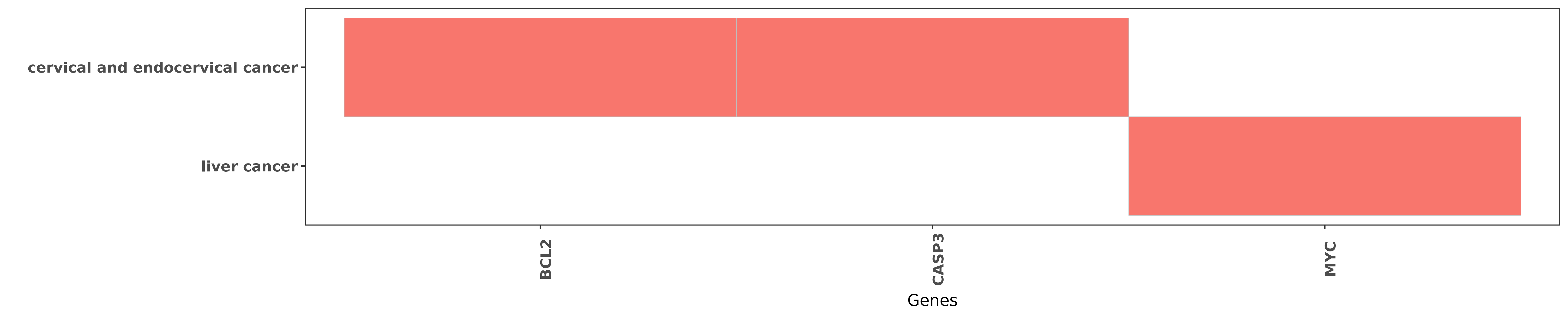
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-744-5p | cervical and endocervical cancer | BCL2 | "MicroRNA 744 inhibited cervical cancer growth and ......" | 27261616 |
| hsa-miR-744-5p | cervical and endocervical cancer | CASP3 | "In addition both up-regulation of miR-744 and down ......" | 27261616 |
| hsa-miR-744-5p | liver cancer | MYC | "Decrease expression of microRNA 744 promotes cell ......" | 24991193 |
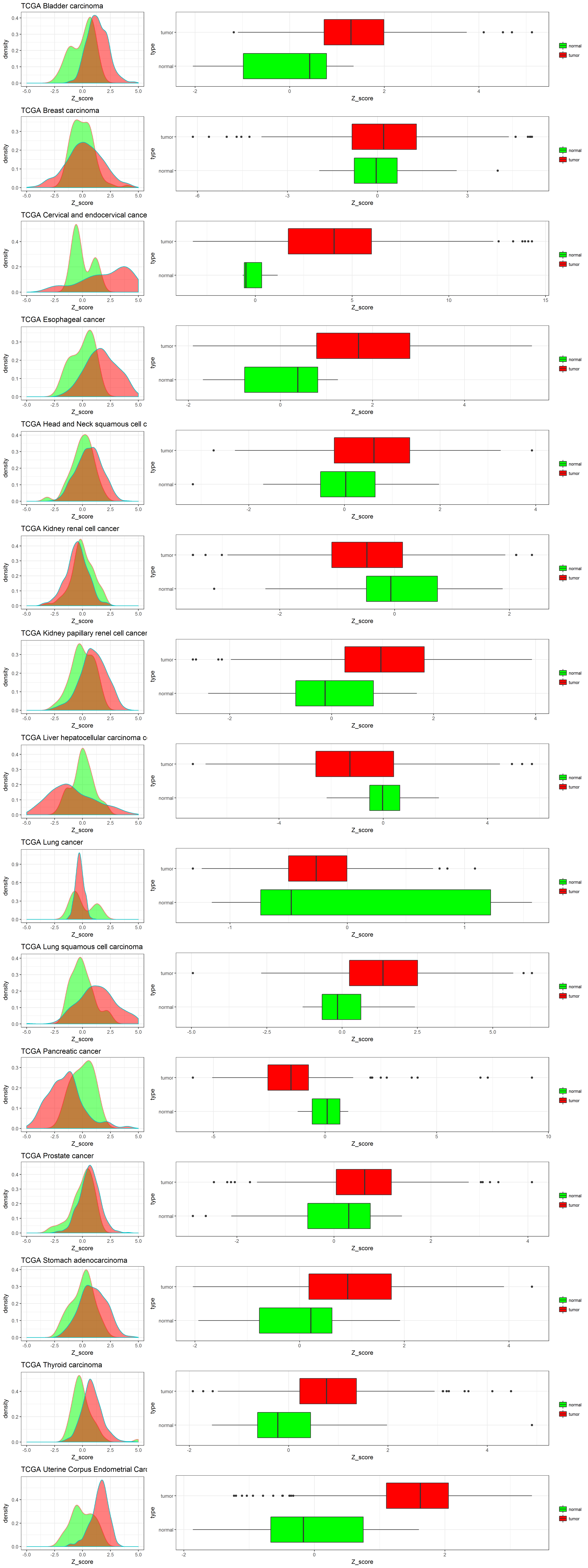
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-744-5p | ABI3 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.395; TCGA BRCA -0.149; TCGA CESC -0.339; TCGA ESCA -0.238; TCGA HNSC -0.285; TCGA LGG -0.181; TCGA LUAD -0.107; TCGA LUSC -0.464; TCGA PRAD -0.204; TCGA THCA -0.205; TCGA STAD -0.13; TCGA UCEC -0.188 |
hsa-miR-744-5p | ADCY9 | 9 cancers: BLCA; BRCA; CESC; LIHC; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.338; TCGA BRCA -0.087; TCGA CESC -0.231; TCGA LIHC -0.179; TCGA LUSC -0.17; TCGA PRAD -0.083; TCGA SARC -0.185; TCGA STAD -0.283; TCGA UCEC -0.43 |
hsa-miR-744-5p | ARRDC2 | 15 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.091; TCGA BRCA -0.133; TCGA CESC -0.314; TCGA ESCA -0.206; TCGA HNSC -0.347; TCGA KIRC -0.326; TCGA KIRP -0.165; TCGA LIHC -0.168; TCGA LUAD -0.117; TCGA LUSC -0.408; TCGA PAAD -0.248; TCGA PRAD -0.249; TCGA SARC -0.183; TCGA STAD -0.092; TCGA UCEC -0.114 |
hsa-miR-744-5p | FRZB | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUSC; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.281; TCGA BRCA -0.275; TCGA COAD -0.43; TCGA ESCA -0.465; TCGA HNSC -0.348; TCGA KIRC -0.459; TCGA KIRP -0.234; TCGA LIHC -0.256; TCGA LUSC -0.336; TCGA PRAD -0.157; TCGA SARC -0.274; TCGA THCA -0.456; TCGA STAD -0.917; TCGA UCEC -0.215 |
hsa-miR-744-5p | IER5 | 9 cancers: BLCA; CESC; HNSC; KIRC; LGG; LUAD; LUSC; PAAD; PRAD | miRNAWalker2 validate | TCGA BLCA -0.104; TCGA CESC -0.143; TCGA HNSC -0.105; TCGA KIRC -0.122; TCGA LGG -0.083; TCGA LUAD -0.087; TCGA LUSC -0.239; TCGA PAAD -0.28; TCGA PRAD -0.284 |
hsa-miR-744-5p | ILK | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.308; TCGA BRCA -0.085; TCGA COAD -0.092; TCGA ESCA -0.095; TCGA HNSC -0.127; TCGA LGG -0.088; TCGA LUSC -0.254; TCGA OV -0.076; TCGA PAAD -0.149; TCGA PRAD -0.14; TCGA SARC -0.23; TCGA STAD -0.426; TCGA UCEC -0.151 |
hsa-miR-744-5p | KIAA1462 | 9 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LUSC; SARC; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.421; TCGA BRCA -0.144; TCGA COAD -0.227; TCGA ESCA -0.408; TCGA KIRP -0.265; TCGA LUSC -0.417; TCGA SARC -0.124; TCGA STAD -0.176; TCGA UCEC -0.551 |
hsa-miR-744-5p | LAIR1 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; THCA | miRNAWalker2 validate | TCGA BLCA -0.635; TCGA BRCA -0.108; TCGA CESC -0.301; TCGA ESCA -0.292; TCGA HNSC -0.275; TCGA KIRC -0.334; TCGA LUAD -0.143; TCGA LUSC -0.612; TCGA PAAD -0.282; TCGA PRAD -0.194; TCGA SARC -0.22; TCGA THCA -0.421 |
hsa-miR-744-5p | LGALS3 | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LUSC; OV; PAAD; PRAD; SARC | miRNAWalker2 validate | TCGA BLCA -0.09; TCGA BRCA -0.22; TCGA ESCA -0.186; TCGA HNSC -0.125; TCGA KIRC -0.445; TCGA LUSC -0.249; TCGA OV -0.151; TCGA PAAD -0.849; TCGA PRAD -0.401; TCGA SARC -0.212 |
hsa-miR-744-5p | LYNX1 | 9 cancers: BLCA; CESC; COAD; KIRP; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.4; TCGA CESC -0.31; TCGA COAD -0.27; TCGA KIRP -0.323; TCGA LUSC -0.233; TCGA PRAD -0.233; TCGA SARC -0.485; TCGA STAD -0.763; TCGA UCEC -0.408 |
hsa-miR-744-5p | PDE4A | 10 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LUSC; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.219; TCGA BRCA -0.129; TCGA ESCA -0.316; TCGA KIRC -0.129; TCGA KIRP -0.304; TCGA LUSC -0.26; TCGA PAAD -0.405; TCGA PRAD -0.384; TCGA STAD -0.123; TCGA UCEC -0.242 |
hsa-miR-744-5p | PPM1M | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.174; TCGA BRCA -0.07; TCGA CESC -0.152; TCGA ESCA -0.109; TCGA HNSC -0.143; TCGA LUSC -0.31; TCGA PAAD -0.111; TCGA PRAD -0.13; TCGA SARC -0.128; TCGA THCA -0.147; TCGA STAD -0.255; TCGA UCEC -0.084 |
hsa-miR-744-5p | RASSF1 | 13 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; OV; PAAD; SARC; THCA | miRNAWalker2 validate | TCGA BLCA -0.103; TCGA BRCA -0.063; TCGA CESC -0.142; TCGA HNSC -0.264; TCGA KIRC -0.131; TCGA KIRP -0.143; TCGA LGG -0.065; TCGA LUAD -0.103; TCGA LUSC -0.318; TCGA OV -0.099; TCGA PAAD -0.099; TCGA SARC -0.072; TCGA THCA -0.196 |
hsa-miR-744-5p | TGFB1 | 10 cancers: BLCA; BRCA; KIRC; KIRP; LGG; LUSC; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.229; TCGA BRCA -0.166; TCGA KIRC -0.4; TCGA KIRP -0.156; TCGA LGG -0.095; TCGA LUSC -0.122; TCGA PAAD -0.229; TCGA PRAD -0.166; TCGA STAD -0.195; TCGA UCEC -0.189 |
hsa-miR-744-5p | TSC22D3 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.446; TCGA BRCA -0.229; TCGA CESC -0.183; TCGA COAD -0.274; TCGA ESCA -0.541; TCGA HNSC -0.195; TCGA KIRC -0.294; TCGA KIRP -0.22; TCGA LIHC -0.4; TCGA LUAD -0.126; TCGA LUSC -0.464; TCGA PRAD -0.088; TCGA THCA -0.247; TCGA STAD -0.595; TCGA UCEC -0.364 |
hsa-miR-744-5p | ZCCHC24 | 10 cancers: BLCA; BRCA; COAD; ESCA; LUSC; PAAD; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.618; TCGA BRCA -0.351; TCGA COAD -0.484; TCGA ESCA -0.511; TCGA LUSC -0.408; TCGA PAAD -0.269; TCGA PRAD -0.124; TCGA THCA -0.195; TCGA STAD -0.843; TCGA UCEC -0.478 |
hsa-miR-744-5p | FBLN2 | 10 cancers: BLCA; BRCA; COAD; ESCA; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.708; TCGA BRCA -0.354; TCGA COAD -0.323; TCGA ESCA -0.472; TCGA LUSC -0.38; TCGA PAAD -0.449; TCGA PRAD -0.367; TCGA THCA -0.68; TCGA STAD -0.522; TCGA UCEC -0.33 |
hsa-miR-744-5p | JUNB | 12 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; THCA; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.226; TCGA BRCA -0.321; TCGA CESC -0.258; TCGA HNSC -0.185; TCGA KIRC -0.185; TCGA KIRP -0.183; TCGA LUAD -0.106; TCGA LUSC -0.271; TCGA PAAD -0.325; TCGA PRAD -0.391; TCGA THCA -0.158; TCGA UCEC -0.241 |
hsa-miR-744-5p | KANK2 | 11 cancers: BLCA; BRCA; COAD; ESCA; LIHC; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.475; TCGA BRCA -0.192; TCGA COAD -0.263; TCGA ESCA -0.507; TCGA LIHC -0.169; TCGA LUSC -0.38; TCGA PAAD -0.212; TCGA PRAD -0.265; TCGA SARC -0.316; TCGA STAD -0.732; TCGA UCEC -0.447 |
hsa-miR-744-5p | PPP2R5B | 9 cancers: BLCA; CESC; ESCA; HNSC; LIHC; LUAD; LUSC; SARC; STAD | MirTarget | TCGA BLCA -0.117; TCGA CESC -0.082; TCGA ESCA -0.111; TCGA HNSC -0.073; TCGA LIHC -0.098; TCGA LUAD -0.078; TCGA LUSC -0.148; TCGA SARC -0.057; TCGA STAD -0.191 |
hsa-miR-744-5p | THY1 | 10 cancers: BLCA; BRCA; COAD; KIRP; LIHC; LUAD; LUSC; PAAD; THCA; UCEC | MirTarget | TCGA BLCA -0.401; TCGA BRCA -0.149; TCGA COAD -0.315; TCGA KIRP -0.482; TCGA LIHC -0.186; TCGA LUAD -0.134; TCGA LUSC -0.327; TCGA PAAD -0.402; TCGA THCA -0.21; TCGA UCEC -0.156 |
hsa-miR-744-5p | NFIX | 11 cancers: BLCA; CESC; COAD; ESCA; KIRC; KIRP; LGG; LIHC; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.433; TCGA CESC -0.242; TCGA COAD -0.137; TCGA ESCA -0.156; TCGA KIRC -0.213; TCGA KIRP -0.176; TCGA LGG -0.06; TCGA LIHC -0.213; TCGA PAAD -0.269; TCGA STAD -0.376; TCGA UCEC -0.285 |
hsa-miR-744-5p | PRDM16 | 10 cancers: BLCA; BRCA; CESC; ESCA; KIRP; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.371; TCGA BRCA -0.372; TCGA CESC -0.458; TCGA ESCA -0.727; TCGA KIRP -1.141; TCGA PAAD -0.345; TCGA PRAD -0.227; TCGA SARC -0.574; TCGA STAD -0.242; TCGA UCEC -0.388 |
hsa-miR-744-5p | PLD4 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.802; TCGA BRCA -0.352; TCGA CESC -0.632; TCGA ESCA -0.571; TCGA HNSC -0.302; TCGA LUAD -0.222; TCGA LUSC -0.535; TCGA PRAD -0.488; TCGA SARC -0.411; TCGA THCA -0.289; TCGA STAD -0.584; TCGA UCEC -0.264 |
hsa-miR-744-5p | SLC12A4 | 11 cancers: BLCA; BRCA; LGG; LIHC; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.216; TCGA BRCA -0.131; TCGA LGG -0.116; TCGA LIHC -0.126; TCGA LUSC -0.247; TCGA PAAD -0.243; TCGA PRAD -0.287; TCGA SARC -0.085; TCGA THCA -0.065; TCGA STAD -0.27; TCGA UCEC -0.116 |
hsa-miR-744-5p | MEF2B | 12 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; THCA; STAD | mirMAP | TCGA BRCA -0.246; TCGA CESC -0.284; TCGA ESCA -0.249; TCGA HNSC -0.162; TCGA KIRC -0.338; TCGA KIRP -0.184; TCGA LGG -0.273; TCGA LUAD -0.216; TCGA LUSC -0.298; TCGA PAAD -0.246; TCGA THCA -0.389; TCGA STAD -0.106 |
Enriched cancer pathways of putative targets