microRNA information: hsa-miR-758-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-758-3p | miRbase |
| Accession: | MIMAT0003879 | miRbase |
| Precursor name: | hsa-mir-758 | miRbase |
| Precursor accession: | MI0003757 | miRbase |
| Symbol: | MIR758 | HGNC |
| RefSeq ID: | NR_030406 | GenBank |
| Sequence: | UUUGUGACCUGGUCCACUAACC |
Reported expression in cancers: hsa-miR-758-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|
Reported cancer pathway affected by hsa-miR-758-3p
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|
Reported cancer prognosis affected by hsa-miR-758-3p
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|
Reported gene related to hsa-miR-758-3p
| miRNA | cancer | gene | reporting | PUBMED |
|---|
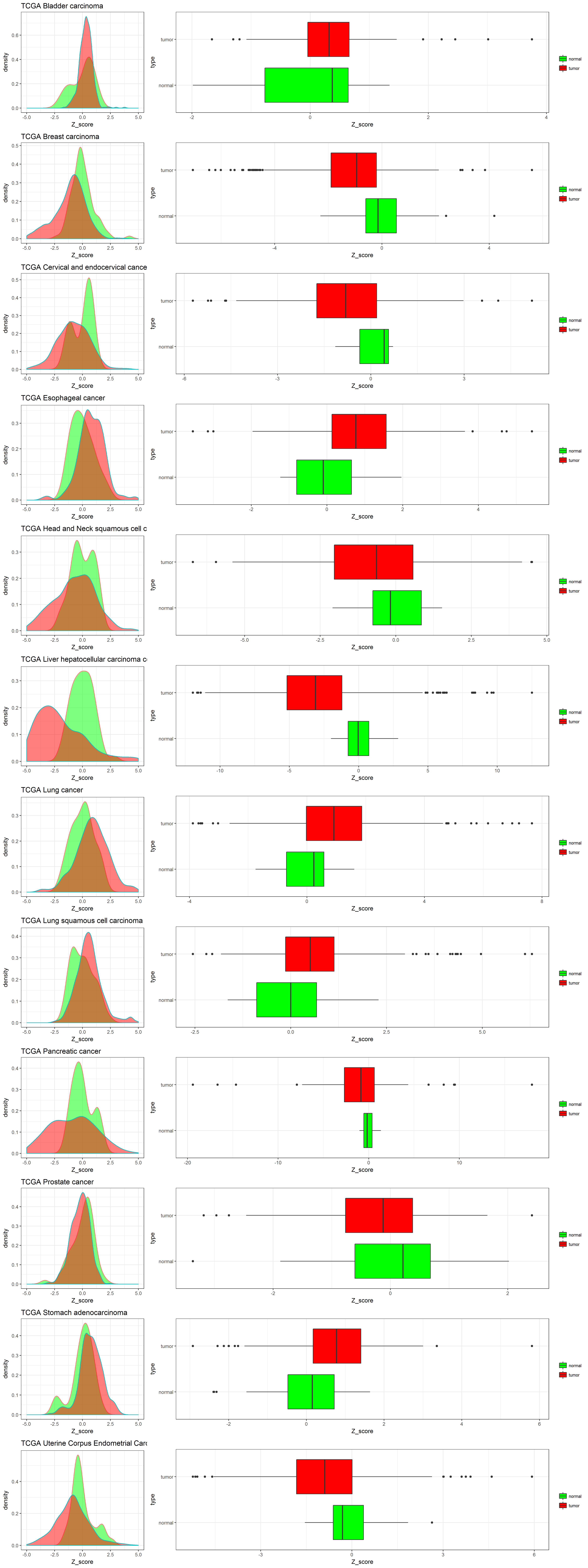
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-758-3p | FAM122C | 9 cancers: BLCA; CESC; ESCA; LGG; LIHC; OV; PAAD; SARC; UCEC | MirTarget | TCGA BLCA -0.052; TCGA CESC -0.069; TCGA ESCA -0.107; TCGA LGG -0.07; TCGA LIHC -0.077; TCGA OV -0.059; TCGA PAAD -0.1; TCGA SARC -0.097; TCGA UCEC -0.101 |
hsa-miR-758-3p | PARP12 | 9 cancers: BRCA; CESC; HNSC; LGG; LUSC; OV; PAAD; PRAD; UCEC | MirTarget | TCGA BRCA -0.195; TCGA CESC -0.121; TCGA HNSC -0.162; TCGA LGG -0.077; TCGA LUSC -0.078; TCGA OV -0.18; TCGA PAAD -0.1; TCGA PRAD -0.067; TCGA UCEC -0.098 |
Enriched cancer pathways of putative targets