| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|
| 1 |
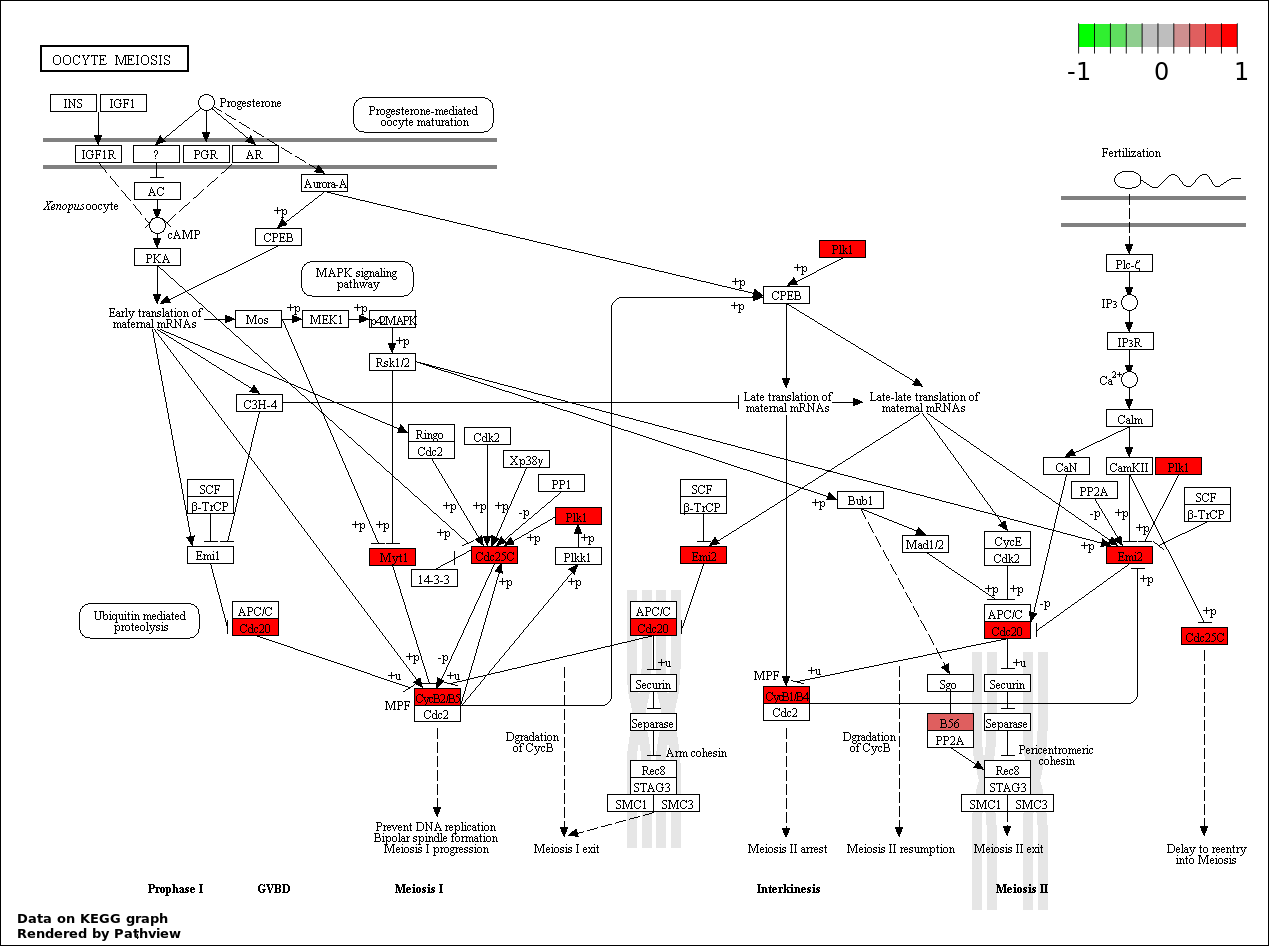
CELL CYCLE |
42 |
1316 |
1.281e-17 |
5.962e-14 |
| 2 |
CELL CYCLE PROCESS |
38 |
1081 |
2.863e-17 |
6.66e-14 |
| 3 |
MITOTIC CELL CYCLE |
29 |
766 |
5.041e-14 |
7.818e-11 |
| 4 |
CELL CYCLE PHASE TRANSITION |
18 |
255 |
1.797e-13 |
2.09e-10 |
| 5 |
NEGATIVE REGULATION OF CELL CYCLE |
22 |
433 |
2.549e-13 |
2.372e-10 |
| 6 |
CELL CYCLE G2 M PHASE TRANSITION |
14 |
138 |
7.215e-13 |
5.595e-10 |
| 7 |
REGULATION OF CYTOSKELETON ORGANIZATION |
22 |
502 |
4.813e-12 |
3.199e-09 |
| 8 |
REGULATION OF CELL CYCLE |
29 |
949 |
1.025e-11 |
5.961e-09 |
| 9 |
REGULATION OF MICROTUBULE BASED PROCESS |
16 |
243 |
1.235e-11 |
6.385e-09 |
| 10 |
REGULATION OF ORGANELLE ORGANIZATION |
32 |
1178 |
1.571e-11 |
7.312e-09 |
| 11 |
NEGATIVE REGULATION OF ORGANELLE ORGANIZATION |
18 |
387 |
1.897e-10 |
8.023e-08 |
| 12 |
REGULATION OF PROTEIN MODIFICATION PROCESS |
36 |
1710 |
8.423e-10 |
3.266e-07 |
| 13 |
REGULATION OF TRANSFERASE ACTIVITY |
26 |
946 |
1.374e-09 |
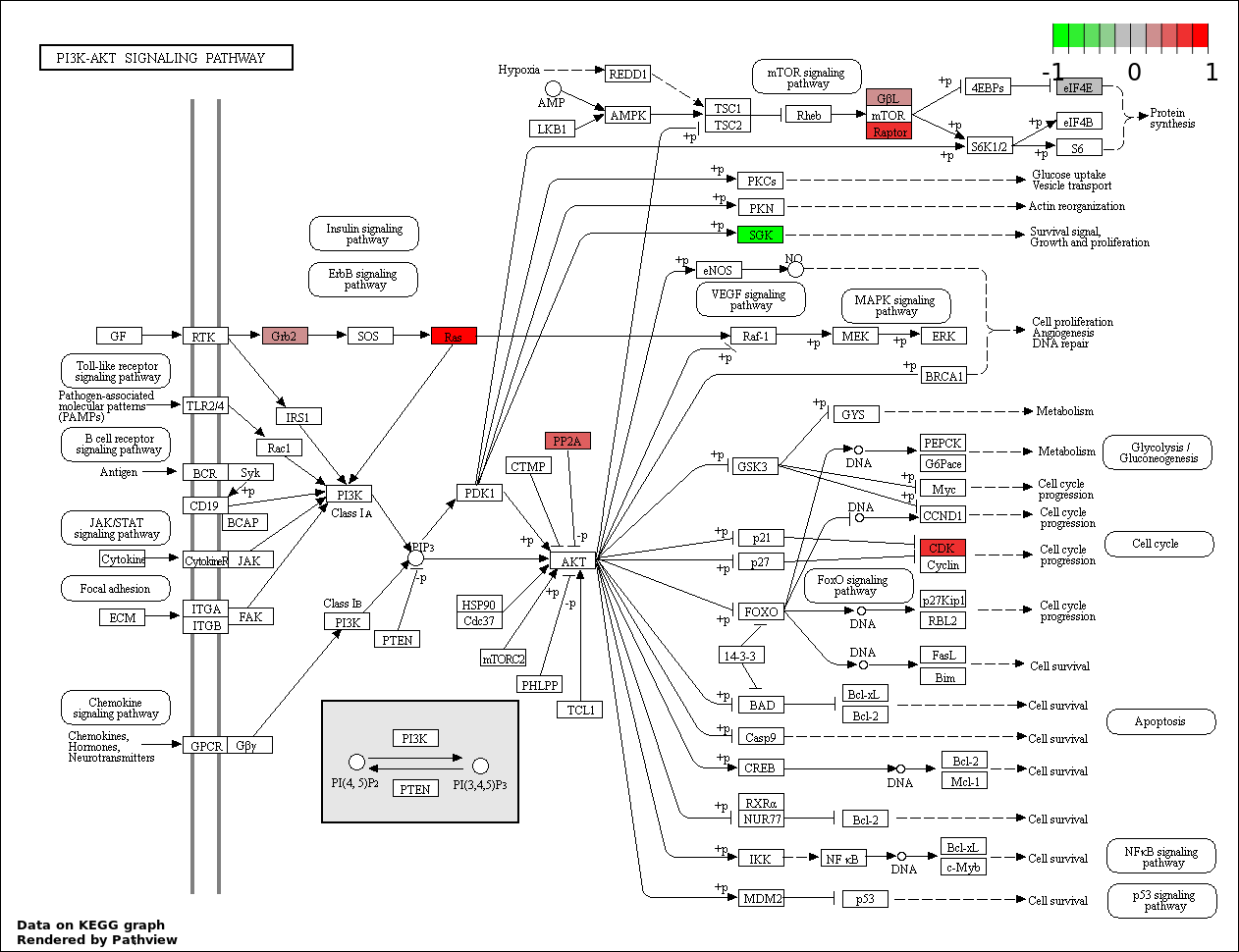
4.919e-07 |
| 14 |
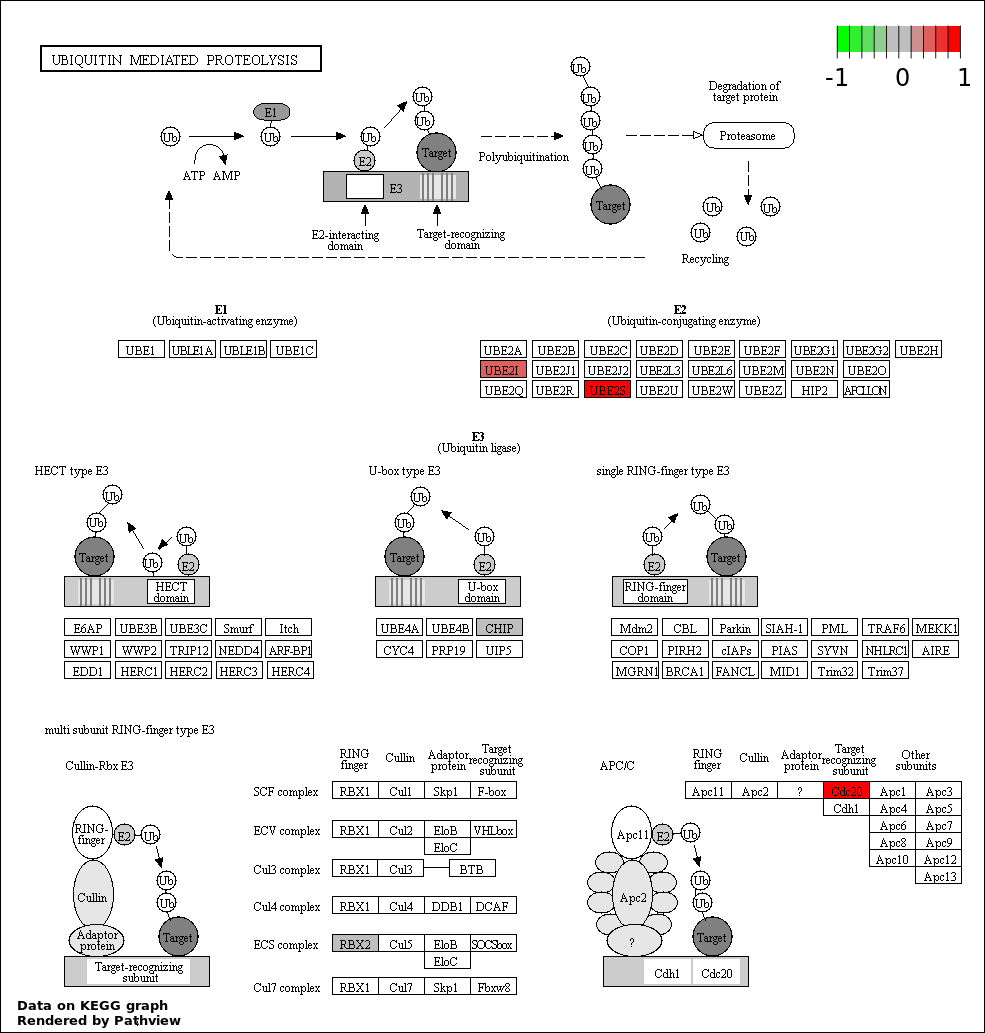
NEGATIVE REGULATION OF CELL CYCLE PROCESS |
13 |
214 |
3.047e-09 |
1.013e-06 |
| 15 |
REGULATION OF MICROTUBULE POLYMERIZATION OR DEPOLYMERIZATION |
12 |
178 |
3.917e-09 |
1.215e-06 |
| 16 |
NEGATIVE REGULATION OF CYTOSKELETON ORGANIZATION |
13 |
221 |
4.499e-09 |
1.308e-06 |
| 17 |
REGULATION OF CELL CYCLE PROCESS |
19 |
558 |
1.011e-08 |
2.533e-06 |
| 18 |
CELL CYCLE CHECKPOINT |
12 |
194 |
1.035e-08 |
2.533e-06 |
| 19 |
NEGATIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION |
21 |
684 |
9.646e-09 |
2.533e-06 |
| 20 |
CELL DIVISION |
17 |
460 |
1.963e-08 |
4.349e-06 |
| 21 |
MICROTUBULE CYTOSKELETON ORGANIZATION |
15 |
348 |
1.891e-08 |
4.349e-06 |
| 22 |
REGULATION OF MITOTIC CELL CYCLE |
17 |
468 |
2.525e-08 |
5.34e-06 |
| 23 |
MITOTIC NUCLEAR DIVISION |
15 |
361 |
3.073e-08 |
6.216e-06 |
| 24 |
ANAPHASE PROMOTING COMPLEX DEPENDENT CATABOLIC PROCESS |
8 |
77 |
5.905e-08 |
1.07e-05 |
| 25 |
ORGANELLE FISSION |
17 |
496 |
5.86e-08 |
1.07e-05 |
| 26 |
REGULATION OF CELL CYCLE ARREST |
9 |
108 |
5.982e-08 |
1.07e-05 |
| 27 |
POSTTRANSCRIPTIONAL REGULATION OF GENE EXPRESSION |
16 |
448 |
8.447e-08 |
1.456e-05 |
| 28 |
NEGATIVE REGULATION OF MITOTIC CELL CYCLE |
11 |
199 |
1.374e-07 |
2.283e-05 |
| 29 |
POSITIVE REGULATION OF CELL CYCLE PROCESS |
12 |
247 |
1.485e-07 |
2.383e-05 |
| 30 |
NEGATIVE REGULATION OF PROTEIN MODIFICATION PROCESS |
18 |
616 |
2.521e-07 |
3.91e-05 |
| 31 |
ERBB2 SIGNALING PATHWAY |
6 |
39 |
2.618e-07 |
3.929e-05 |
| 32 |
REGULATION OF LIGASE ACTIVITY |
9 |
130 |
2.963e-07 |
4.308e-05 |
| 33 |
SIGNAL TRANSDUCTION IN RESPONSE TO DNA DAMAGE |
8 |
96 |
3.334e-07 |
4.701e-05 |
| 34 |
PROTEIN UBIQUITINATION INVOLVED IN UBIQUITIN DEPENDENT PROTEIN CATABOLIC PROCESS |
9 |
134 |
3.835e-07 |
5.249e-05 |
| 35 |
REGULATION OF PHOSPHORUS METABOLIC PROCESS |
30 |
1618 |
4.795e-07 |
6.375e-05 |
| 36 |
MITOTIC CELL CYCLE CHECKPOINT |
9 |
139 |
5.233e-07 |
6.706e-05 |
| 37 |
SPINDLE ASSEMBLY |
7 |
70 |
5.332e-07 |
6.706e-05 |
| 38 |
MICROTUBULE BASED PROCESS |
16 |
522 |
6.586e-07 |
8.064e-05 |
| 39 |
G1 DNA DAMAGE CHECKPOINT |
7 |
73 |
7.124e-07 |
8.499e-05 |
| 40 |
DNA INTEGRITY CHECKPOINT |
9 |
146 |
7.921e-07 |
9.214e-05 |
| 41 |
POSITIVE REGULATION OF LIGASE ACTIVITY |
8 |
110 |
9.5e-07 |
0.0001078 |
| 42 |
NEGATIVE REGULATION OF GENE EXPRESSION |
28 |
1493 |
1.001e-06 |
0.0001109 |
| 43 |
PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS |
33 |
1977 |
1.218e-06 |
0.0001267 |
| 44 |
POSITIVE REGULATION OF TRANSFERASE ACTIVITY |
17 |
616 |
1.225e-06 |
0.0001267 |
| 45 |
REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER |
31 |
1784 |
1.198e-06 |
0.0001267 |
| 46 |
NEGATIVE REGULATION OF MOLECULAR FUNCTION |
23 |
1079 |
1.297e-06 |
0.0001312 |
| 47 |
NEGATIVE REGULATION OF NITROGEN COMPOUND METABOLIC PROCESS |
28 |
1517 |
1.371e-06 |
0.0001358 |
| 48 |
NEGATIVE REGULATION OF PROTEIN METABOLIC PROCESS |
23 |
1087 |
1.471e-06 |
0.0001426 |
| 49 |
POSITIVE REGULATION OF TOR SIGNALING |
5 |
29 |
1.512e-06 |
0.0001435 |
| 50 |
REGULATION OF KINASE ACTIVITY |
19 |
776 |
1.645e-06 |
0.0001531 |
| 51 |
POSITIVE REGULATION OF CELL CYCLE ARREST |
7 |
85 |
2.015e-06 |
0.0001838 |
| 52 |
REGULATION OF PROTEIN COMPLEX DISASSEMBLY |
10 |
217 |
2.713e-06 |
0.0002427 |
| 53 |
NEGATIVE REGULATION OF PROTEIN COMPLEX DISASSEMBLY |
9 |
170 |
2.81e-06 |
0.0002467 |
| 54 |
PROTEASOMAL PROTEIN CATABOLIC PROCESS |
11 |
271 |
2.907e-06 |
0.0002505 |
| 55 |
MACROMOLECULAR COMPLEX ASSEMBLY |
26 |
1398 |
3.059e-06 |
0.0002588 |
| 56 |
POSITIVE REGULATION OF CELL CYCLE |
12 |
332 |
3.371e-06 |
0.0002801 |
| 57 |
REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL |
11 |
280 |
3.977e-06 |
0.0003246 |
| 58 |
NEGATIVE REGULATION OF CELL CYCLE G1 S PHASE TRANSITION |
7 |
98 |
5.242e-06 |
0.0004205 |
| 59 |
NEGATIVE REGULATION OF TRANSFERASE ACTIVITY |
12 |
351 |
5.947e-06 |
0.000465 |
| 60 |
MITOTIC DNA INTEGRITY CHECKPOINT |
7 |
100 |
5.996e-06 |
0.000465 |
| 61 |
POSITIVE REGULATION OF CELLULAR PROTEIN CATABOLIC PROCESS |
9 |
192 |
7.583e-06 |
0.0005697 |
| 62 |
PROTEIN PHOSPHORYLATION |
20 |
944 |
7.592e-06 |
0.0005697 |
| 63 |
NEGATIVE REGULATION OF CELL CYCLE PHASE TRANSITION |
8 |
146 |
7.935e-06 |
0.000586 |
| 64 |
INTRACELLULAR SIGNAL TRANSDUCTION |
27 |
1572 |
8.382e-06 |
0.0006094 |
| 65 |
NEGATIVE REGULATION OF CELL DEATH |
19 |
872 |
8.853e-06 |
0.0006313 |
| 66 |
POSITIVE REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL |
9 |
196 |
8.955e-06 |
0.0006313 |
| 67 |
PHOSPHORYLATION |
23 |
1228 |
1.109e-05 |
0.0007705 |
| 68 |
PROTEIN CATABOLIC PROCESS |
15 |
579 |
1.15e-05 |
0.0007872 |
| 69 |
INTERSPECIES INTERACTION BETWEEN ORGANISMS |
16 |
662 |
1.368e-05 |
0.0009095 |
| 70 |
SYMBIOSIS ENCOMPASSING MUTUALISM THROUGH PARASITISM |
16 |
662 |
1.368e-05 |
0.0009095 |
| 71 |
NEGATIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER |
17 |
740 |
1.395e-05 |
0.0009142 |
| 72 |
REGULATION OF CELL CYCLE PHASE TRANSITION |
11 |
321 |
1.444e-05 |
0.0009331 |
| 73 |
REGULATION OF NUCLEAR DIVISION |
8 |
163 |
1.773e-05 |
0.001119 |
| 74 |
ERBB SIGNALING PATHWAY |
6 |
79 |
1.779e-05 |
0.001119 |
| 75 |
CYTOSKELETON ORGANIZATION |
18 |
838 |
1.887e-05 |
0.00117 |
| 76 |
REGULATION OF CELL DIVISION |
10 |
272 |
1.968e-05 |
0.001205 |
| 77 |
MACROMOLECULE CATABOLIC PROCESS |
19 |
926 |
2.049e-05 |
0.001238 |
| 78 |
REGULATION OF CELLULAR PROTEIN CATABOLIC PROCESS |
10 |
274 |
2.096e-05 |
0.00125 |
| 79 |
REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY |
13 |
470 |
2.315e-05 |
0.001363 |
| 80 |
SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR |
7 |
127 |
2.868e-05 |
0.001668 |
| 81 |
PROTEIN UBIQUITINATION |
15 |
629 |
3.018e-05 |
0.001734 |
| 82 |
PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL |
18 |
873 |
3.237e-05 |
0.001837 |
| 83 |
POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS |
21 |
1135 |
3.36e-05 |
0.001884 |
| 84 |
CELLULAR RESPONSE TO DNA DAMAGE STIMULUS |
16 |
720 |
3.792e-05 |
0.002101 |
| 85 |
POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION |
21 |
1152 |
4.171e-05 |
0.002283 |
| 86 |
REGULATION OF RNA STABILITY |
7 |
139 |
5.114e-05 |
0.002735 |
| 87 |
NEGATIVE REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL |
7 |
139 |
5.114e-05 |
0.002735 |
| 88 |
RIBONUCLEOPROTEIN COMPLEX BIOGENESIS |
12 |
440 |
5.533e-05 |
0.002925 |
| 89 |
RIBOSOME BIOGENESIS |
10 |
308 |
5.642e-05 |
0.00295 |
| 90 |
NEGATIVE REGULATION OF CATALYTIC ACTIVITY |
17 |
829 |
5.834e-05 |
0.003016 |
| 91 |
CELLULAR RESPONSE TO OXYGEN LEVELS |
7 |
143 |
6.123e-05 |
0.003097 |
| 92 |
RESPONSE TO OXYGEN LEVELS |
10 |
311 |
6.118e-05 |
0.003097 |
| 93 |
CHROMOSOME ORGANIZATION |
19 |
1009 |
6.552e-05 |
0.003278 |
| 94 |
REGULATION OF CELL DEATH |
24 |
1472 |
6.631e-05 |
0.003282 |
| 95 |
GLOBAL GENOME NUCLEOTIDE EXCISION REPAIR |
4 |
32 |
6.716e-05 |
0.003289 |
| 96 |
TRANSLATIONAL INITIATION |
7 |
146 |
6.983e-05 |
0.003385 |
| 97 |
REGULATION OF CELL CYCLE G1 S PHASE TRANSITION |
7 |
147 |
7.29e-05 |
0.003497 |
| 98 |
PEPTIDYL SERINE MODIFICATION |
7 |
148 |
7.609e-05 |
0.003613 |
| 99 |
REGULATION OF PROTEIN UBIQUITINATION INVOLVED IN UBIQUITIN DEPENDENT PROTEIN CATABOLIC PROCESS |
6 |
103 |
7.993e-05 |
0.003757 |
| 100 |
POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS |
24 |
1492 |
8.186e-05 |
0.003809 |
| 101 |
REGULATION OF CELLULAR RESPONSE TO STRESS |
15 |
691 |
8.733e-05 |
0.004023 |
| 102 |
POSITIVE REGULATION OF PROTEIN CATABOLIC PROCESS |
9 |
263 |
8.985e-05 |
0.004099 |
| 103 |
POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS |
19 |
1036 |
9.285e-05 |
0.004154 |
| 104 |
POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS |
19 |
1036 |
9.285e-05 |
0.004154 |
| 105 |
CELL CYCLE ARREST |
7 |
154 |
9.766e-05 |
0.004324 |
| 106 |
PROTEIN COMPLEX BIOGENESIS |
20 |
1132 |
9.944e-05 |
0.004324 |
| 107 |
PROTEIN COMPLEX ASSEMBLY |
20 |
1132 |
9.944e-05 |
0.004324 |
| 108 |
REGULATION OF TOR SIGNALING |
5 |
68 |
0.0001069 |
0.004608 |
| 109 |
MITOTIC SPINDLE ORGANIZATION |
5 |
69 |
0.0001147 |
0.004895 |
| 110 |
CHROMOSOME SEGREGATION |
9 |
272 |
0.000116 |
0.004906 |
| 111 |
REGULATION OF CELLULAR PROTEIN LOCALIZATION |
13 |
552 |
0.0001188 |
0.004981 |
| 112 |
REGULATION OF SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR |
7 |
162 |
0.0001339 |
0.005564 |
| 113 |
ACTIVATION OF ANAPHASE PROMOTING COMPLEX ACTIVITY |
3 |
15 |
0.0001376 |
0.005667 |
| 114 |
MEMBRANE ORGANIZATION |
17 |
899 |
0.0001557 |
0.006354 |
| 115 |
ORGANELLE ASSEMBLY |
12 |
495 |
0.0001677 |
0.006785 |
| 116 |
CELLULAR RESPONSE TO STRESS |
24 |
1565 |
0.0001703 |
0.006829 |
| 117 |
MITOTIC SPINDLE ASSEMBLY |
4 |
41 |
0.0001803 |
0.006989 |
| 118 |
MICROTUBULE CYTOSKELETON ORGANIZATION INVOLVED IN MITOSIS |
4 |
41 |
0.0001803 |
0.006989 |
| 119 |
POSITIVE REGULATION OF CANONICAL WNT SIGNALING PATHWAY |
6 |
119 |
0.0001773 |
0.006989 |
| 120 |
REGULATION OF CELLULAR AMIDE METABOLIC PROCESS |
10 |
354 |
0.0001771 |
0.006989 |
| 121 |
CARBOHYDRATE BIOSYNTHETIC PROCESS |
6 |
121 |
0.0001942 |
0.007405 |
| 122 |
PROTEIN DNA COMPLEX SUBUNIT ORGANIZATION |
8 |
229 |
0.0001936 |
0.007405 |
| 123 |
SISTER CHROMATID SEGREGATION |
7 |
176 |
0.0002233 |
0.008446 |
| 124 |
PEPTIDYL AMINO ACID MODIFICATION |
16 |
841 |
0.0002298 |
0.008622 |
| 125 |
NEGATIVE REGULATION OF ERBB SIGNALING PATHWAY |
4 |
44 |
0.0002378 |
0.00878 |
| 126 |
PROTEIN LOCALIZATION |
26 |
1805 |
0.0002387 |
0.00878 |
| 127 |
DNA METABOLIC PROCESS |
15 |
758 |
0.0002396 |
0.00878 |