Browse ATAT1 in pancancer
| Summary | |
|---|---|
| Symbol | ATAT1 |
| Name | alpha tubulin acetyltransferase 1 |
| Aliases | FLJ13158; Em:AB023049.7; MEC17; alpha-tubulin N-acetyltransferase; C6orf134; chromosome 6 open reading frame ...... |
| Location | 6p21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF05301 GNAT acetyltransferase |
||||||||||
| Function |
Specifically acetylates 'Lys-40' in alpha-tubulin on the lumenal side of microtubules. Promotes microtubule destabilization and accelerates microtubule dynamics; this activity may be independent of acetylation activity. Acetylates alpha-tubulin with a slow enzymatic rate, due to a catalytic site that is not optimized for acetyl transfer. Enters the microtubule through each end and diffuses quickly throughout the lumen of microtubules. Acetylates only long/old microtubules because of its slow acetylation rate since it does not have time to act on dynamically unstable microtubules before the enzyme is released. Required for normal sperm flagellar function. Promotes directional cell locomotion and chemotaxis, through AP2A2-dependent acetylation of alpha-tubulin at clathrin-coated pits that are concentrated at the leading edge of migrating cells. May facilitate primary cilium assembly. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000226 microtubule cytoskeleton organization GO:0006473 protein acetylation GO:0006475 internal protein amino acid acetylation GO:0007283 spermatogenesis GO:0018205 peptidyl-lysine modification GO:0018393 internal peptidyl-lysine acetylation GO:0018394 peptidyl-lysine acetylation GO:0021537 telencephalon development GO:0021542 dentate gyrus development GO:0021543 pallium development GO:0021761 limbic system development GO:0021766 hippocampus development GO:0030900 forebrain development GO:0031334 positive regulation of protein complex assembly GO:0032886 regulation of microtubule-based process GO:0043254 regulation of protein complex assembly GO:0043543 protein acylation GO:0044089 positive regulation of cellular component biogenesis GO:0044546 NLRP3 inflammasome complex assembly GO:0045444 fat cell differentiation GO:0045598 regulation of fat cell differentiation GO:0048232 male gamete generation GO:0051493 regulation of cytoskeleton organization GO:0070507 regulation of microtubule cytoskeleton organization GO:0071929 alpha-tubulin acetylation GO:1900225 regulation of NLRP3 inflammasome complex assembly GO:1900227 positive regulation of NLRP3 inflammasome complex assembly |
| Molecular Function |
GO:0004468 lysine N-acetyltransferase activity, acting on acetyl phosphate as donor GO:0008080 N-acetyltransferase activity GO:0016407 acetyltransferase activity GO:0016410 N-acyltransferase activity GO:0016746 transferase activity, transferring acyl groups GO:0016747 transferase activity, transferring acyl groups other than amino-acyl groups GO:0019799 tubulin N-acetyltransferase activity GO:0034212 peptide N-acetyltransferase activity GO:0048037 cofactor binding GO:0050662 coenzyme binding GO:0061733 peptide-lysine-N-acetyltransferase activity |
| Cellular Component |
GO:0005819 spindle GO:0005874 microtubule GO:0005905 clathrin-coated pit GO:0005924 cell-substrate adherens junction GO:0005925 focal adhesion GO:0030055 cell-substrate junction GO:0030424 axon GO:0072686 mitotic spindle GO:0097427 microtubule bundle GO:0098589 membrane region |
| KEGG | - |
| Reactome |
R-HSA-5617833: Cilium Assembly R-HSA-1852241: Organelle biogenesis and maintenance |
| Summary | |
|---|---|
| Symbol | ATAT1 |
| Name | alpha tubulin acetyltransferase 1 |
| Aliases | FLJ13158; Em:AB023049.7; MEC17; alpha-tubulin N-acetyltransferase; C6orf134; chromosome 6 open reading frame ...... |
| Location | 6p21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for ATAT1. |
| Summary | |
|---|---|
| Symbol | ATAT1 |
| Name | alpha tubulin acetyltransferase 1 |
| Aliases | FLJ13158; Em:AB023049.7; MEC17; alpha-tubulin N-acetyltransferase; C6orf134; chromosome 6 open reading frame ...... |
| Location | 6p21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | ATAT1 |
| Name | alpha tubulin acetyltransferase 1 |
| Aliases | FLJ13158; Em:AB023049.7; MEC17; alpha-tubulin N-acetyltransferase; C6orf134; chromosome 6 open reading frame ...... |
| Location | 6p21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
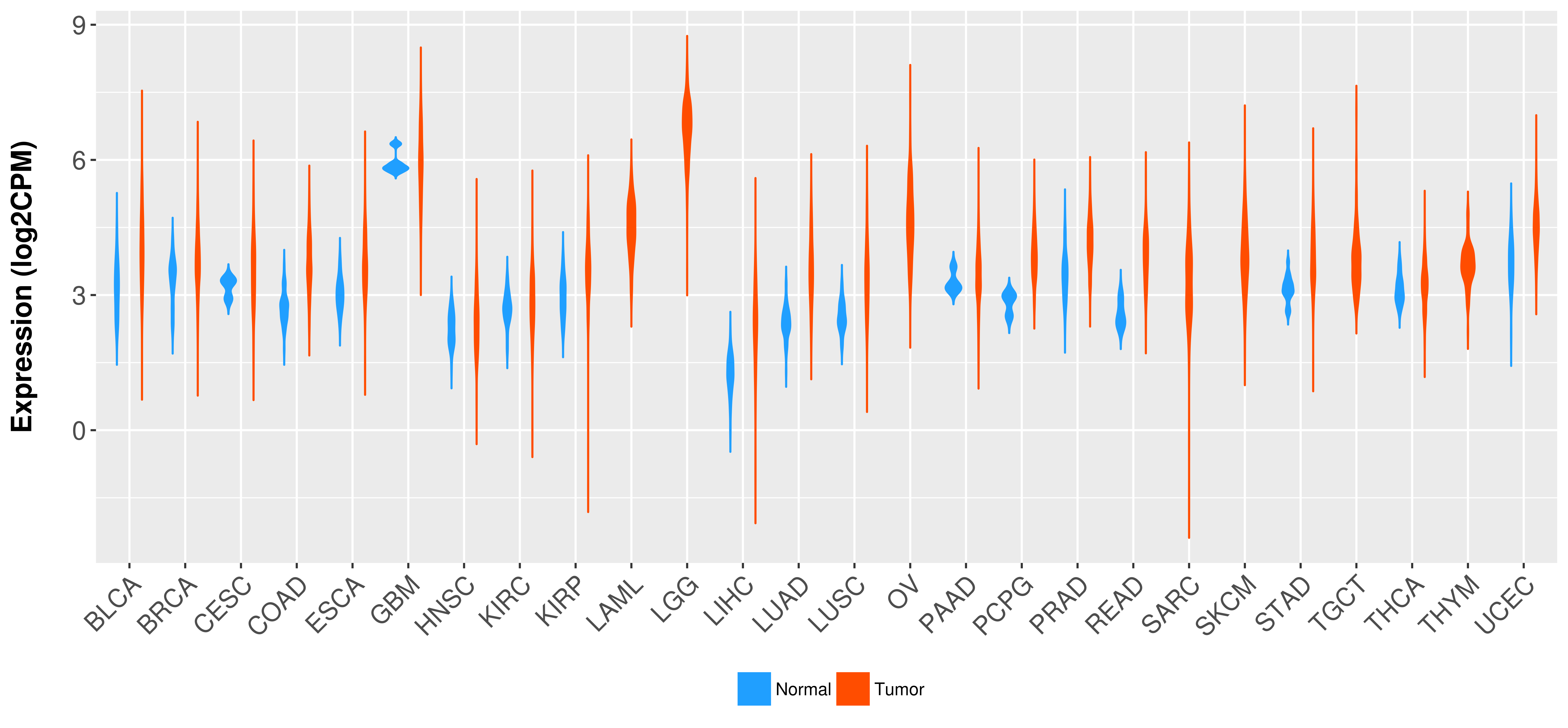
Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
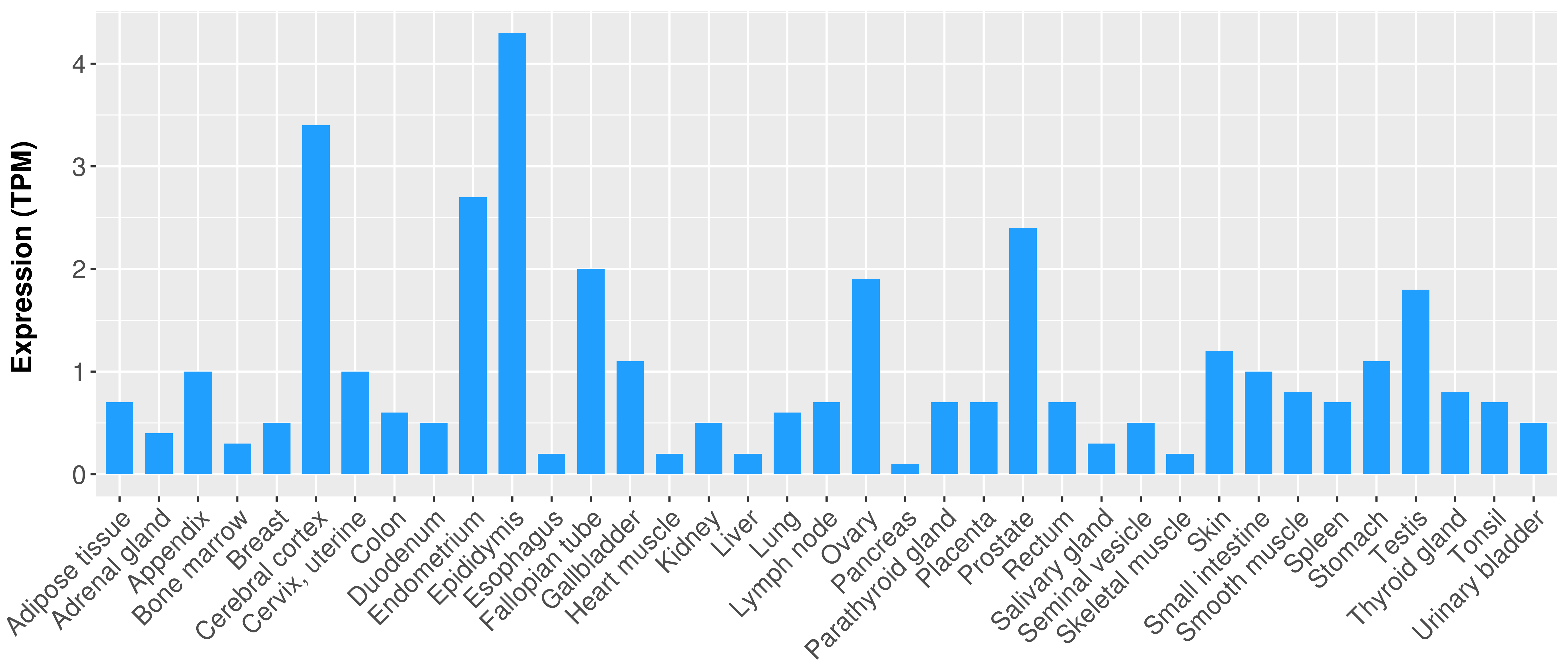
|
|
| Summary | |
|---|---|
| Symbol | ATAT1 |
| Name | alpha tubulin acetyltransferase 1 |
| Aliases | FLJ13158; Em:AB023049.7; MEC17; alpha-tubulin N-acetyltransferase; C6orf134; chromosome 6 open reading frame ...... |
| Location | 6p21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
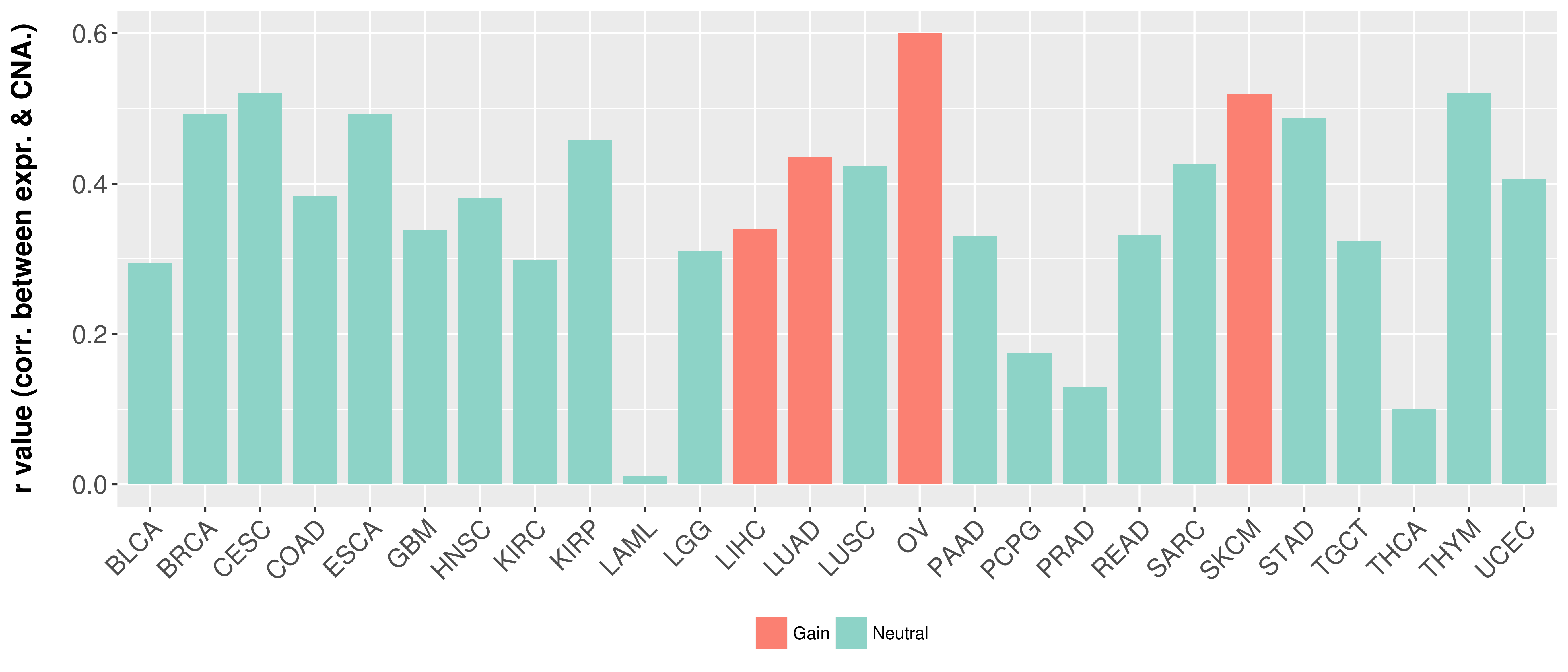
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | ATAT1 |
| Name | alpha tubulin acetyltransferase 1 |
| Aliases | FLJ13158; Em:AB023049.7; MEC17; alpha-tubulin N-acetyltransferase; C6orf134; chromosome 6 open reading frame ...... |
| Location | 6p21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
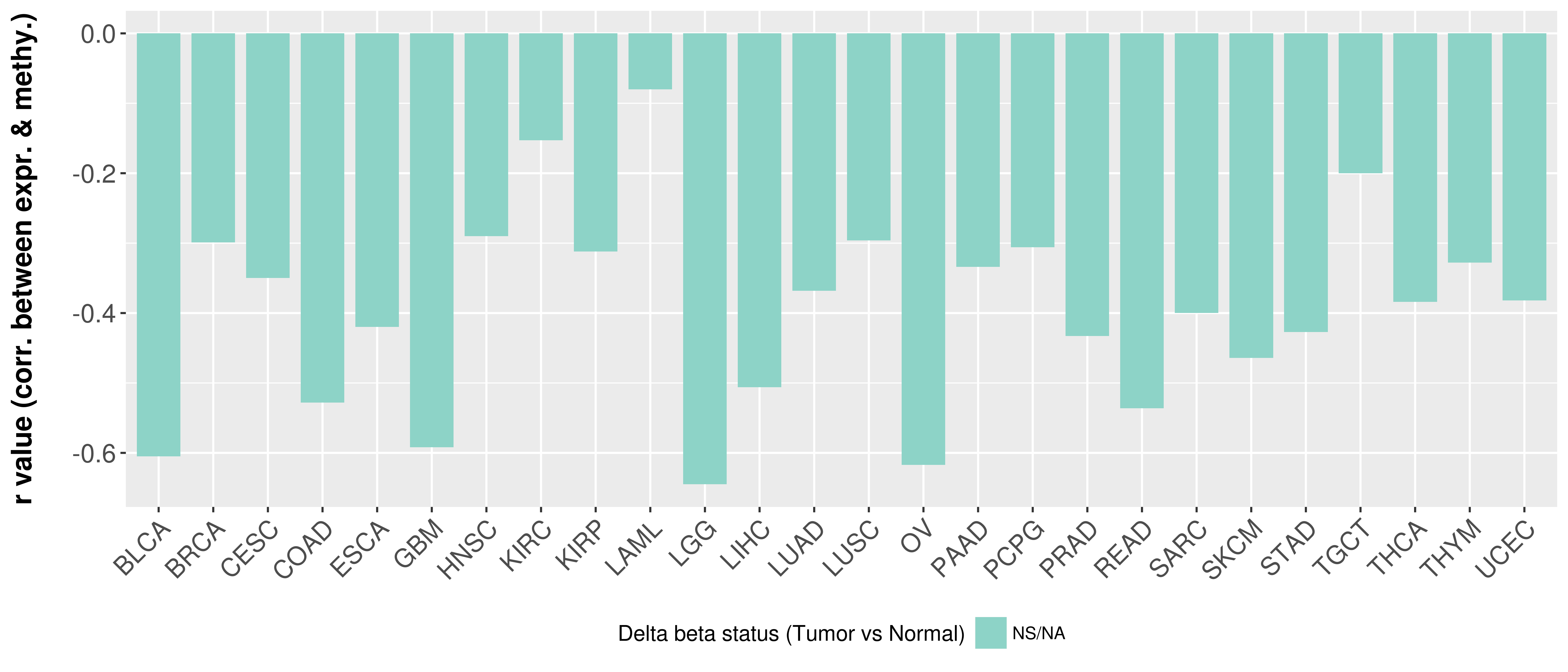
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | ATAT1 |
| Name | alpha tubulin acetyltransferase 1 |
| Aliases | FLJ13158; Em:AB023049.7; MEC17; alpha-tubulin N-acetyltransferase; C6orf134; chromosome 6 open reading frame ...... |
| Location | 6p21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
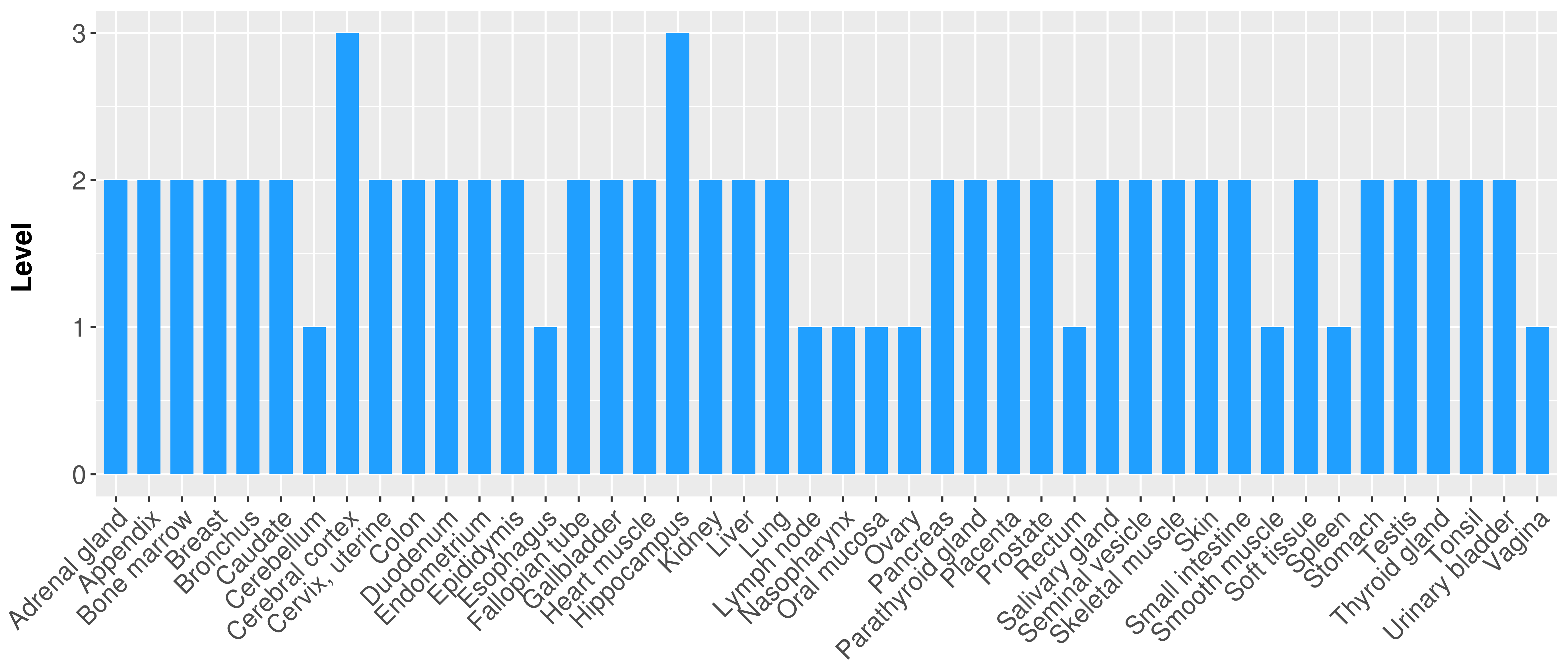
|
| Summary | |
|---|---|
| Symbol | ATAT1 |
| Name | alpha tubulin acetyltransferase 1 |
| Aliases | FLJ13158; Em:AB023049.7; MEC17; alpha-tubulin N-acetyltransferase; C6orf134; chromosome 6 open reading frame ...... |
| Location | 6p21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
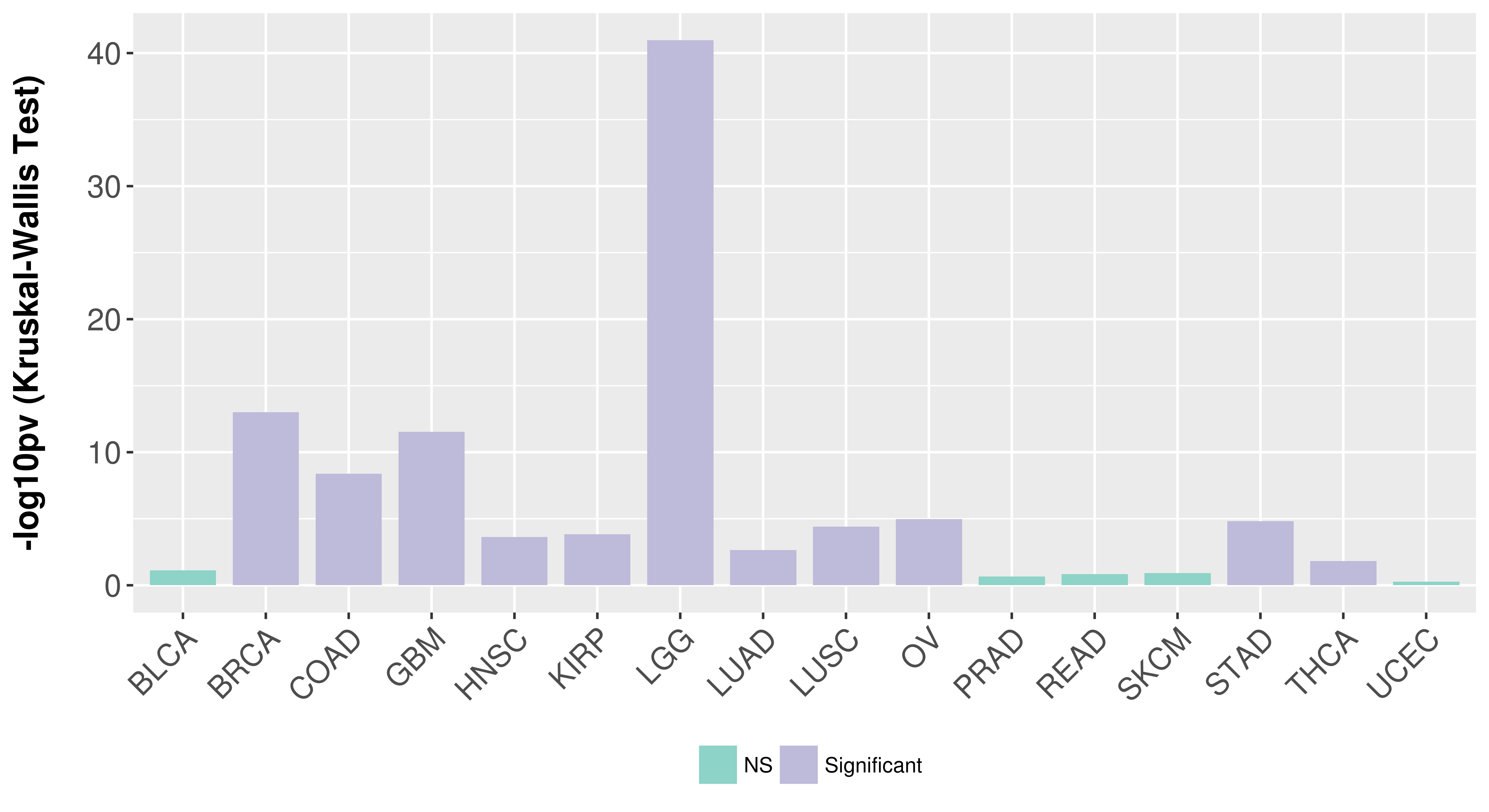
Association between expresson and subtype.
|
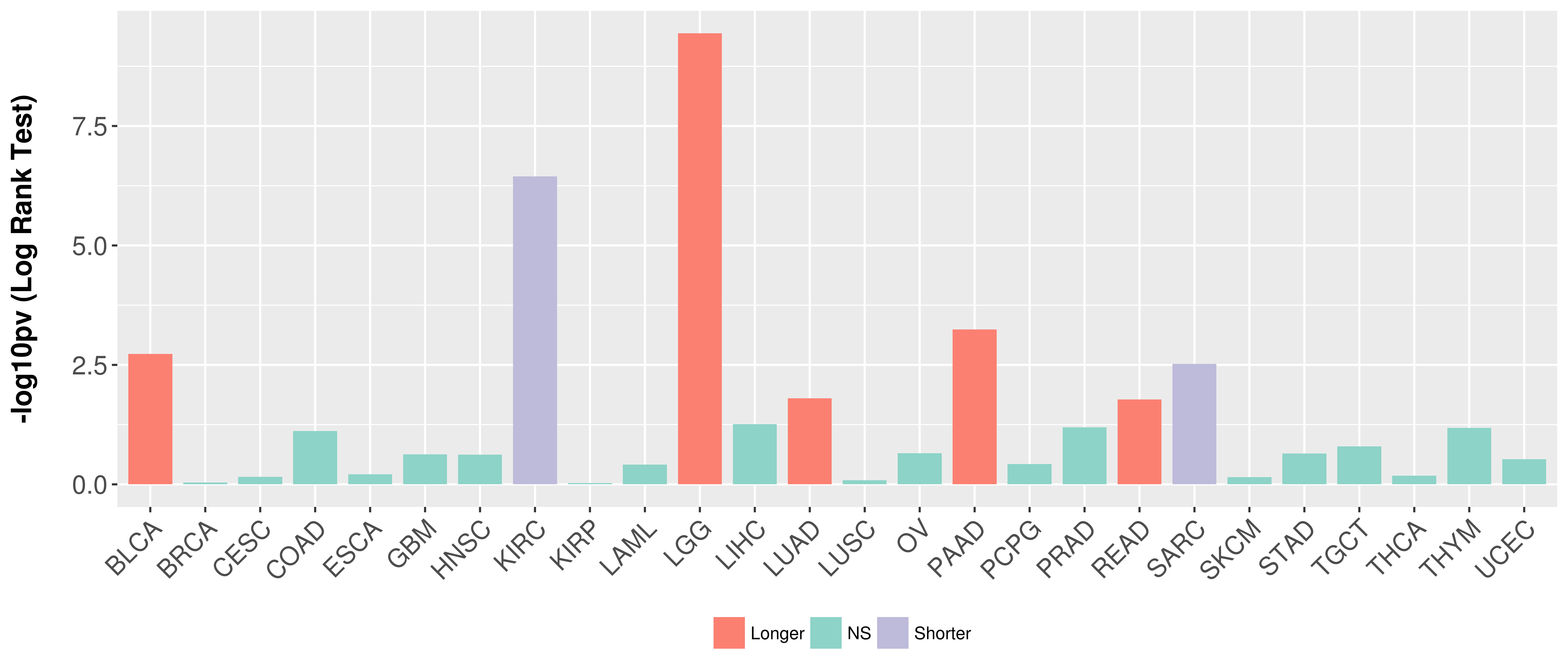
Overall survival analysis based on expression.
|
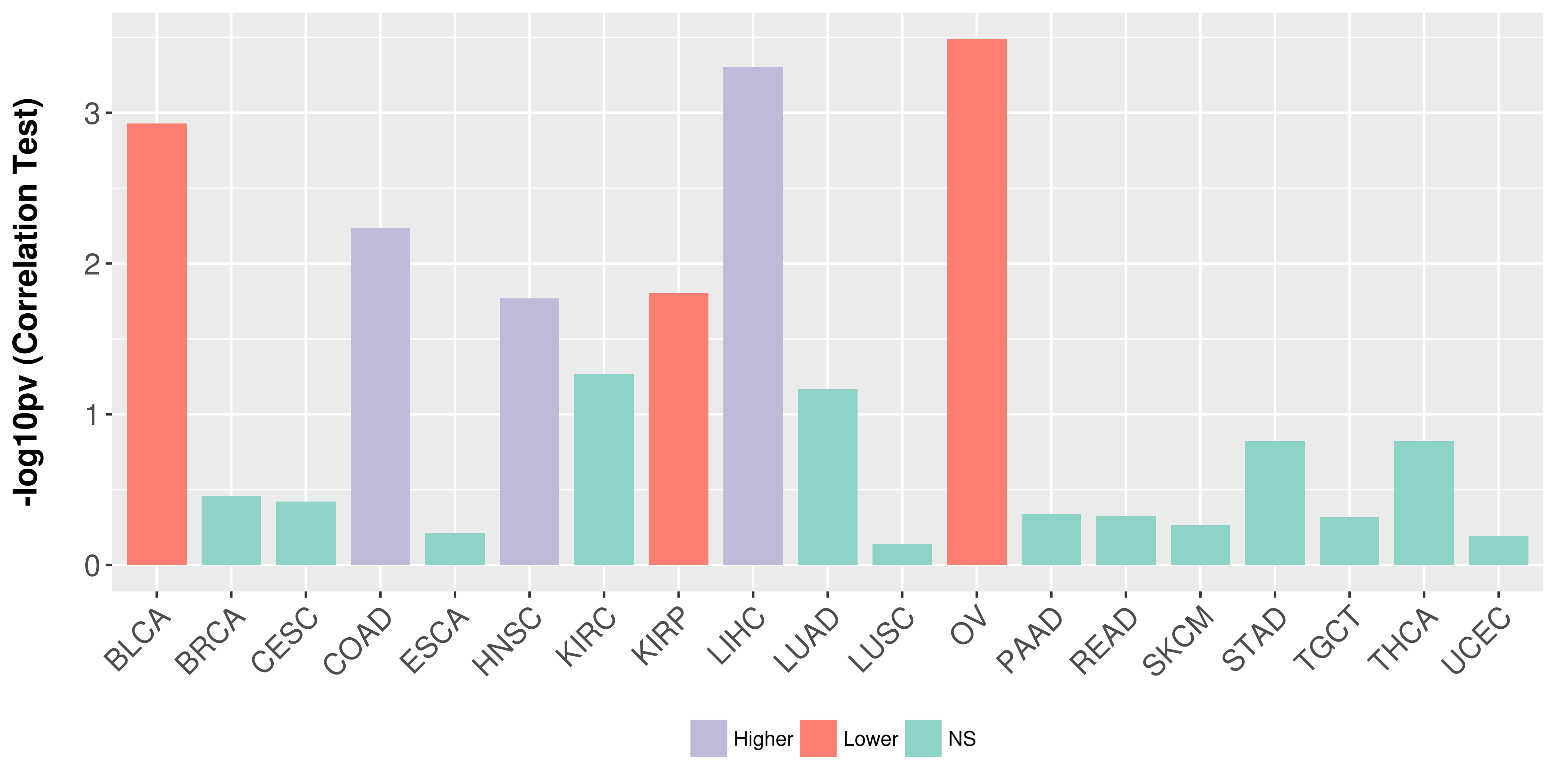
Association between expresson and stage.
|
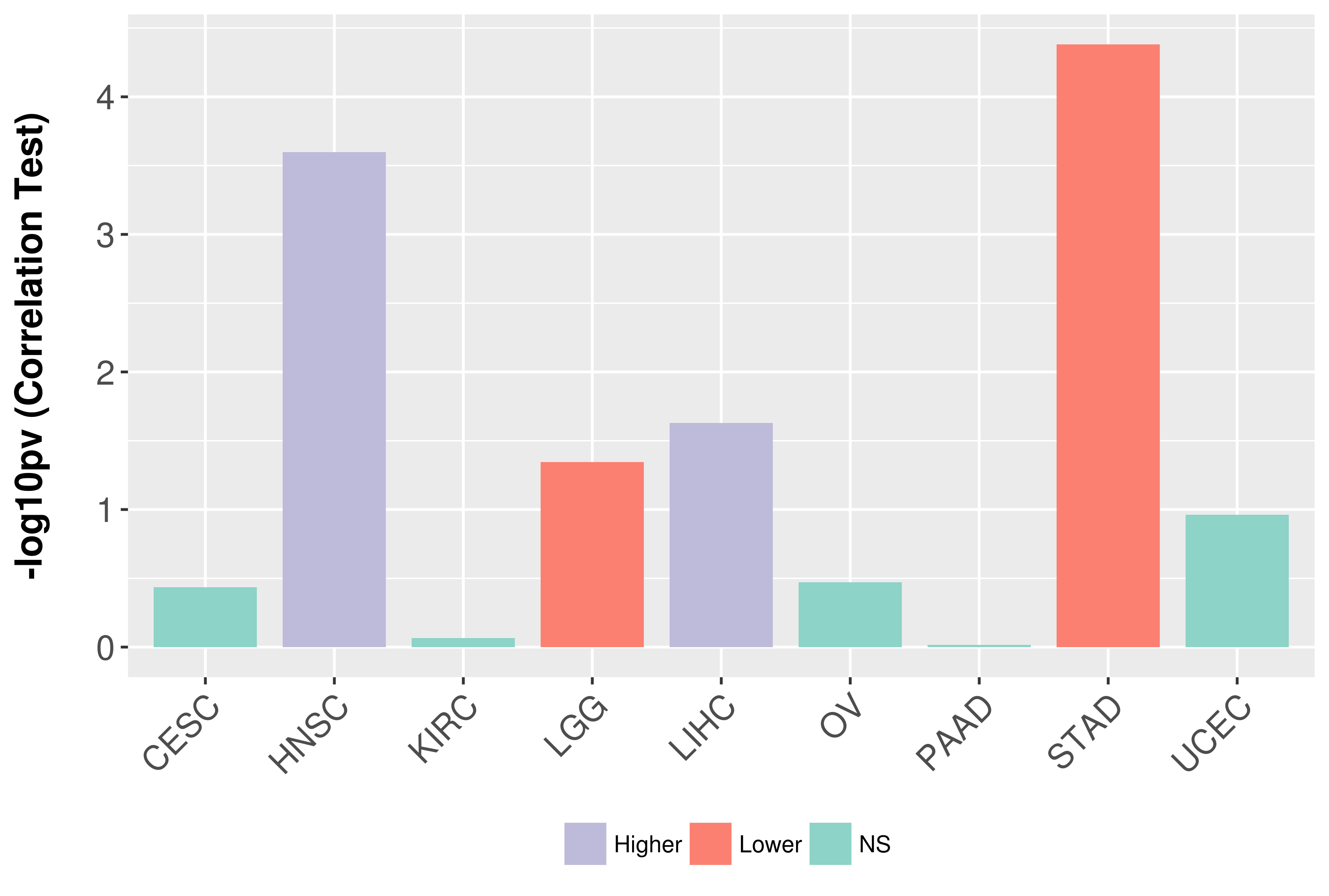
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | ATAT1 |
| Name | alpha tubulin acetyltransferase 1 |
| Aliases | FLJ13158; Em:AB023049.7; MEC17; alpha-tubulin N-acetyltransferase; C6orf134; chromosome 6 open reading frame ...... |
| Location | 6p21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | ATAT1 |
| Name | alpha tubulin acetyltransferase 1 |
| Aliases | FLJ13158; Em:AB023049.7; MEC17; alpha-tubulin N-acetyltransferase; C6orf134; chromosome 6 open reading frame ...... |
| Location | 6p21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for ATAT1. |
| Summary | |
|---|---|
| Symbol | ATAT1 |
| Name | alpha tubulin acetyltransferase 1 |
| Aliases | FLJ13158; Em:AB023049.7; MEC17; alpha-tubulin N-acetyltransferase; C6orf134; chromosome 6 open reading frame ...... |
| Location | 6p21.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|