Browse BMI1 in pancancer
| Summary | |
|---|---|
| Symbol | BMI1 |
| Name | BMI1 proto-oncogene, polycomb ring finger |
| Aliases | RNF51; PCGF4; polycomb group ring finger 4; B lymphoma Mo-MLV insertion region 1 homolog (mouse); FLVI2/BMI1 ...... |
| Location | 10p12.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF16207 RAWUL domain RING finger- and WD40-associated ubiquitin-like |
||||||||||
| Function |
Component of a Polycomb group (PcG) multiprotein PRC1-like complex, a complex class required to maintain the transcriptionally repressive state of many genes, including Hox genes, throughout development. PcG PRC1 complex acts via chromatin remodeling and modification of histones; it mediates monoubiquitination of histone H2A 'Lys-119', rendering chromatin heritably changed in its expressibility. In the PRC1 complex, it is required to stimulate the E3 ubiquitin-protein ligase activity of RNF2/RING2. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0003002 regionalization GO:0007379 segment specification GO:0007389 pattern specification process GO:0016925 protein sumoylation GO:0018205 peptidyl-lysine modification GO:0031396 regulation of protein ubiquitination GO:0031398 positive regulation of protein ubiquitination GO:0035282 segmentation GO:0040029 regulation of gene expression, epigenetic GO:0045814 negative regulation of gene expression, epigenetic GO:0048144 fibroblast proliferation GO:0048145 regulation of fibroblast proliferation GO:0048146 positive regulation of fibroblast proliferation GO:0051438 regulation of ubiquitin-protein transferase activity GO:0051443 positive regulation of ubiquitin-protein transferase activity GO:1903320 regulation of protein modification by small protein conjugation or removal GO:1903322 positive regulation of protein modification by small protein conjugation or removal |
| Molecular Function |
GO:0003682 chromatin binding GO:0071535 RING-like zinc finger domain binding GO:1990841 promoter-specific chromatin binding |
| Cellular Component |
GO:0000151 ubiquitin ligase complex GO:0000152 nuclear ubiquitin ligase complex GO:0031519 PcG protein complex GO:0035102 PRC1 complex |
| KEGG |
hsa04550 Signaling pathways regulating pluripotency of stem cells |
| Reactome |
R-HSA-2559583: Cellular Senescence R-HSA-2262752: Cellular responses to stress R-HSA-392499: Metabolism of proteins R-HSA-2559580: Oxidative Stress Induced Senescence R-HSA-597592: Post-translational protein modification R-HSA-3108232: SUMO E3 ligases SUMOylate target proteins R-HSA-2990846: SUMOylation R-HSA-3108214: SUMOylation of DNA damage response and repair proteins R-HSA-4570464: SUMOylation of RNA binding proteins |
| Summary | |
|---|---|
| Symbol | BMI1 |
| Name | BMI1 proto-oncogene, polycomb ring finger |
| Aliases | RNF51; PCGF4; polycomb group ring finger 4; B lymphoma Mo-MLV insertion region 1 homolog (mouse); FLVI2/BMI1 ...... |
| Location | 10p12.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
|
|
| Summary | |
|---|---|
| Symbol | BMI1 |
| Name | BMI1 proto-oncogene, polycomb ring finger |
| Aliases | RNF51; PCGF4; polycomb group ring finger 4; B lymphoma Mo-MLV insertion region 1 homolog (mouse); FLVI2/BMI1 ...... |
| Location | 10p12.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
| There is no PTM data |
| Summary | |
|---|---|
| Symbol | BMI1 |
| Name | BMI1 proto-oncogene, polycomb ring finger |
| Aliases | RNF51; PCGF4; polycomb group ring finger 4; B lymphoma Mo-MLV insertion region 1 homolog (mouse); FLVI2/BMI1 ...... |
| Location | 10p12.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
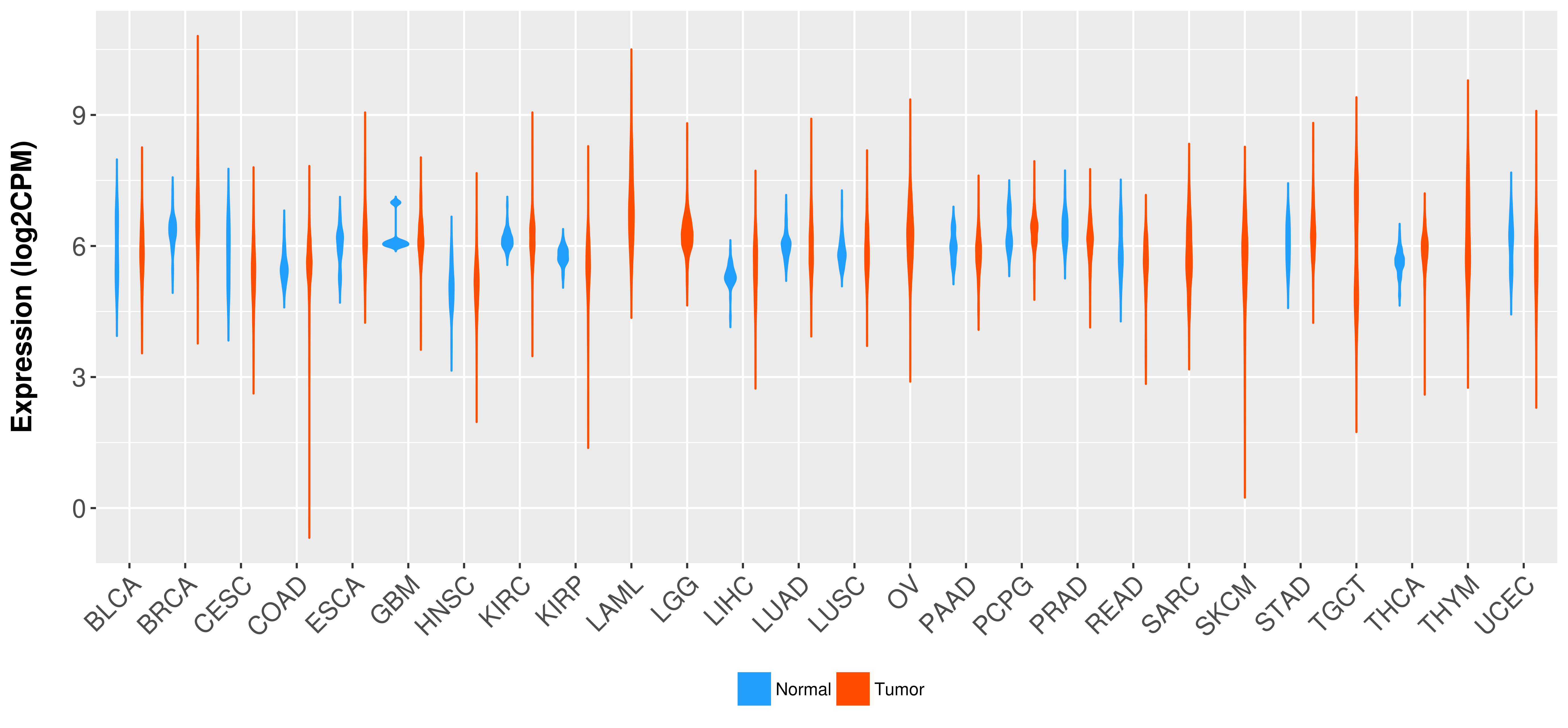
Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
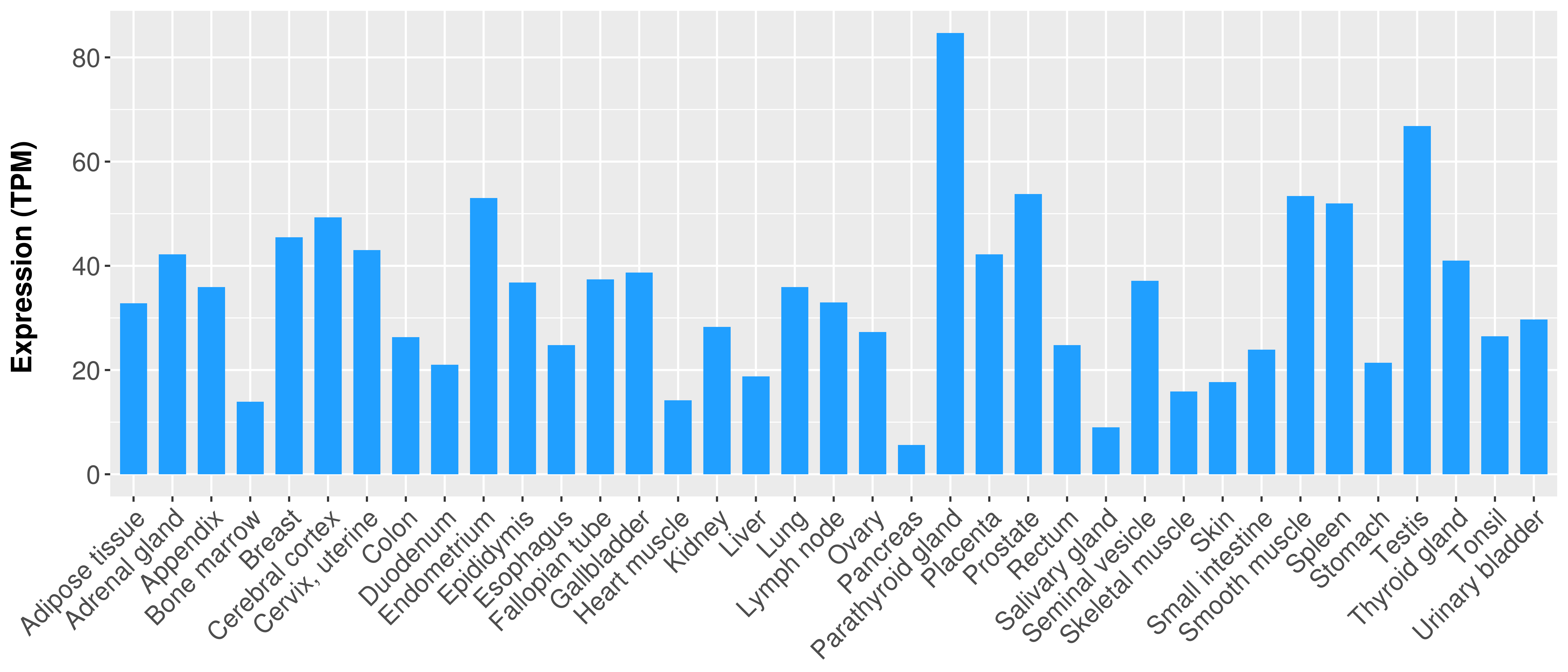
|
|
| Summary | |
|---|---|
| Symbol | BMI1 |
| Name | BMI1 proto-oncogene, polycomb ring finger |
| Aliases | RNF51; PCGF4; polycomb group ring finger 4; B lymphoma Mo-MLV insertion region 1 homolog (mouse); FLVI2/BMI1 ...... |
| Location | 10p12.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
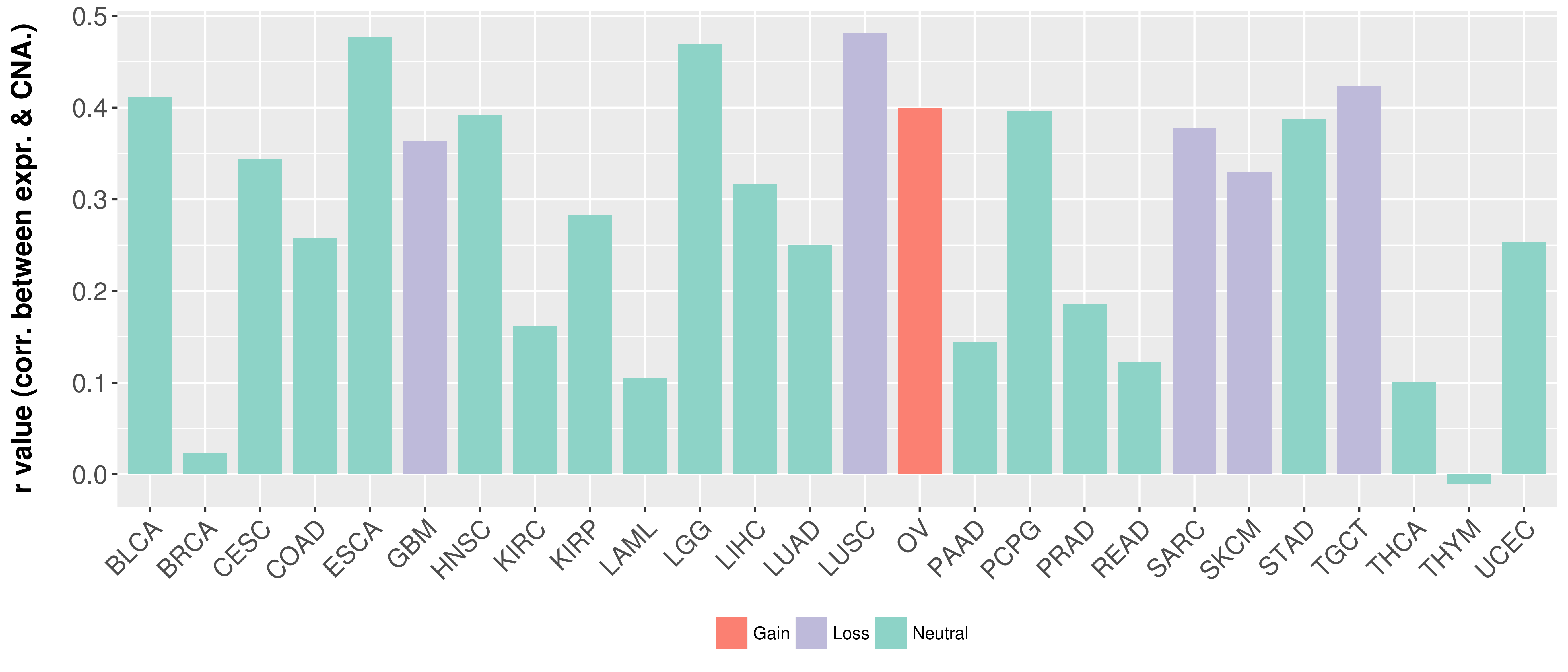
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | BMI1 |
| Name | BMI1 proto-oncogene, polycomb ring finger |
| Aliases | RNF51; PCGF4; polycomb group ring finger 4; B lymphoma Mo-MLV insertion region 1 homolog (mouse); FLVI2/BMI1 ...... |
| Location | 10p12.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
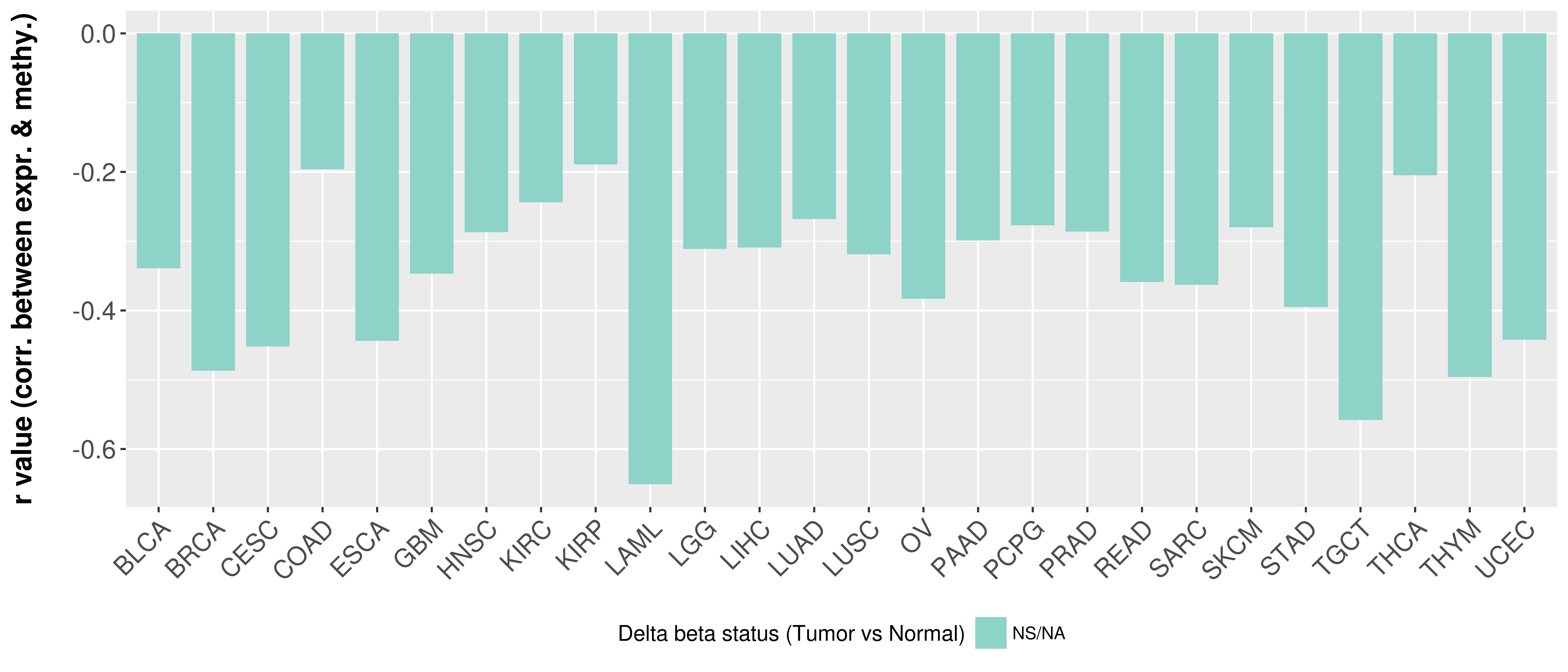
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | BMI1 |
| Name | BMI1 proto-oncogene, polycomb ring finger |
| Aliases | RNF51; PCGF4; polycomb group ring finger 4; B lymphoma Mo-MLV insertion region 1 homolog (mouse); FLVI2/BMI1 ...... |
| Location | 10p12.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
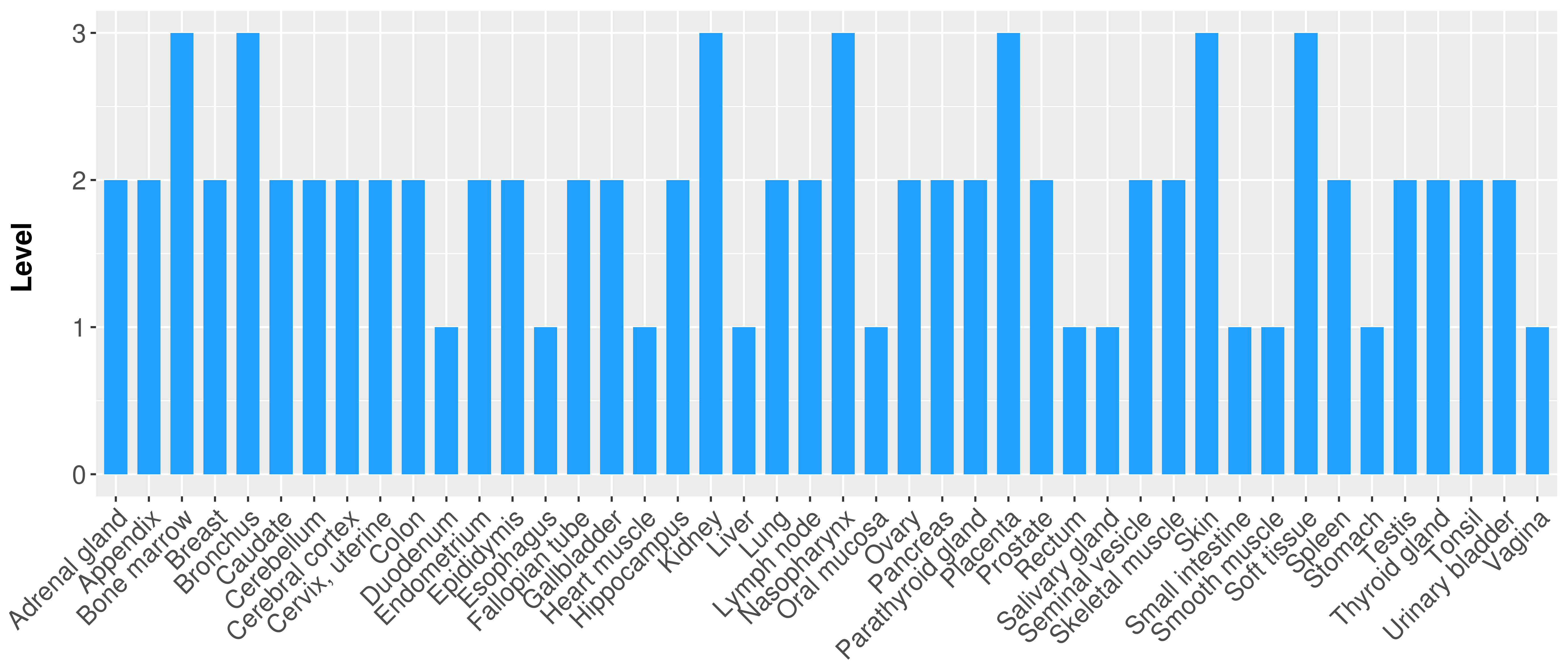
|
| Summary | |
|---|---|
| Symbol | BMI1 |
| Name | BMI1 proto-oncogene, polycomb ring finger |
| Aliases | RNF51; PCGF4; polycomb group ring finger 4; B lymphoma Mo-MLV insertion region 1 homolog (mouse); FLVI2/BMI1 ...... |
| Location | 10p12.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
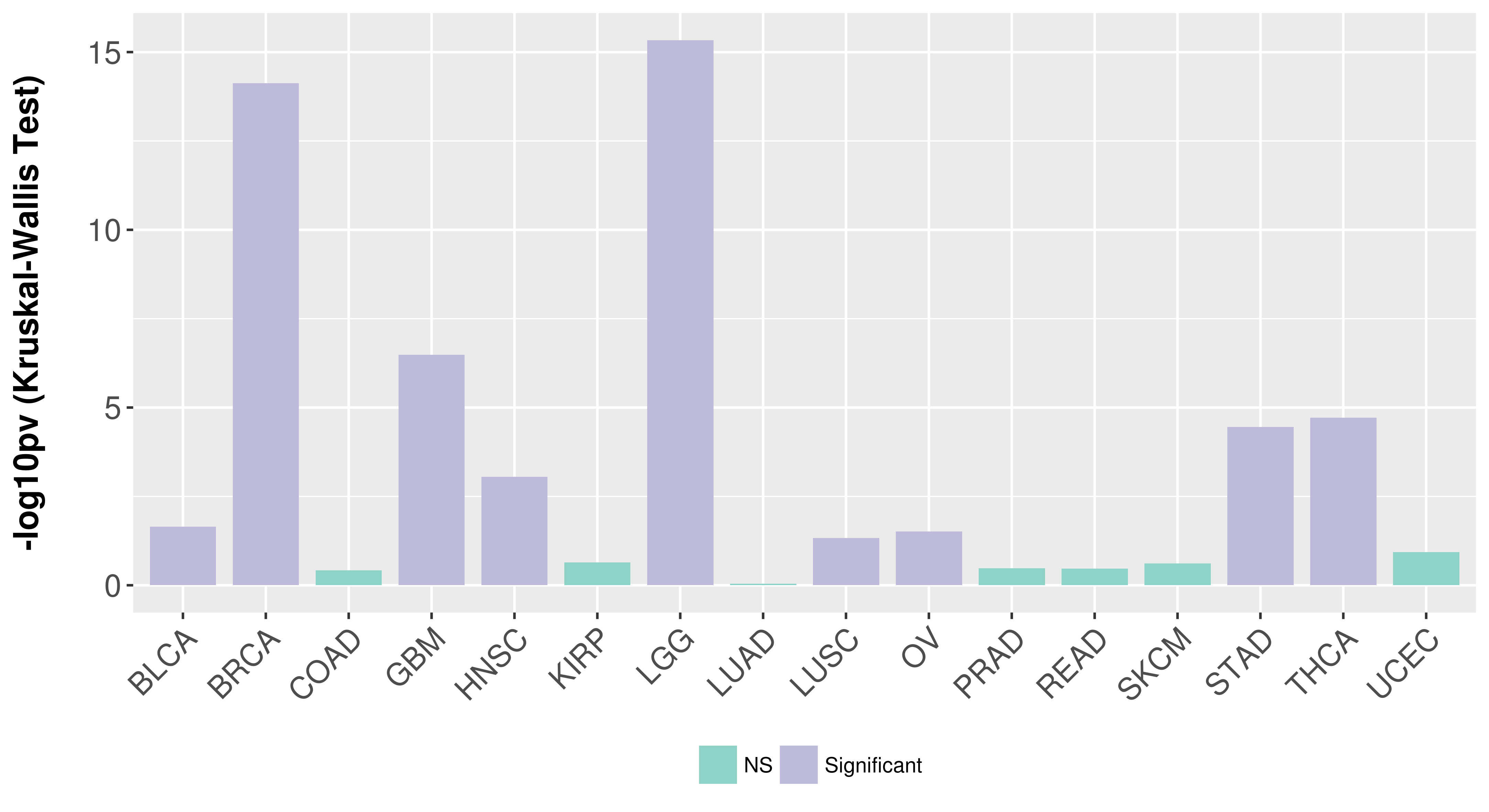
Association between expresson and subtype.
|
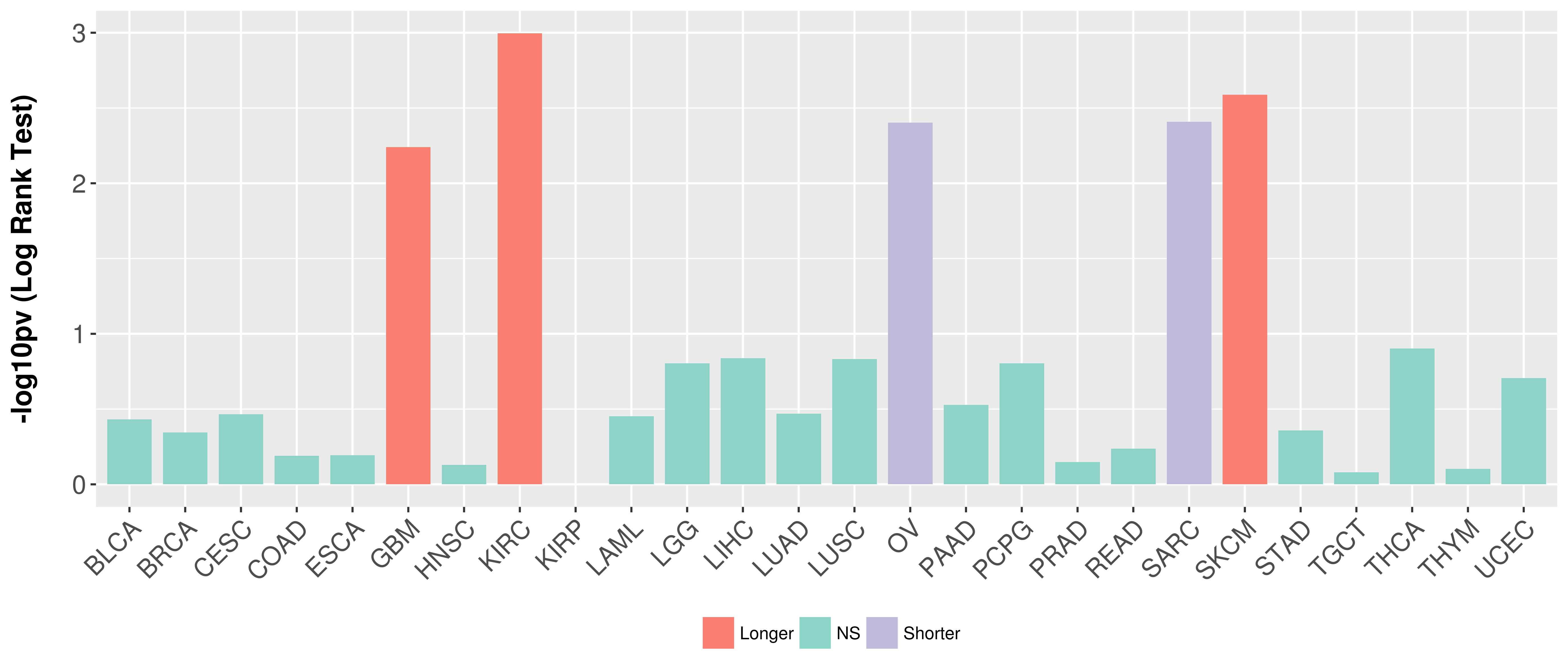
Overall survival analysis based on expression.
|
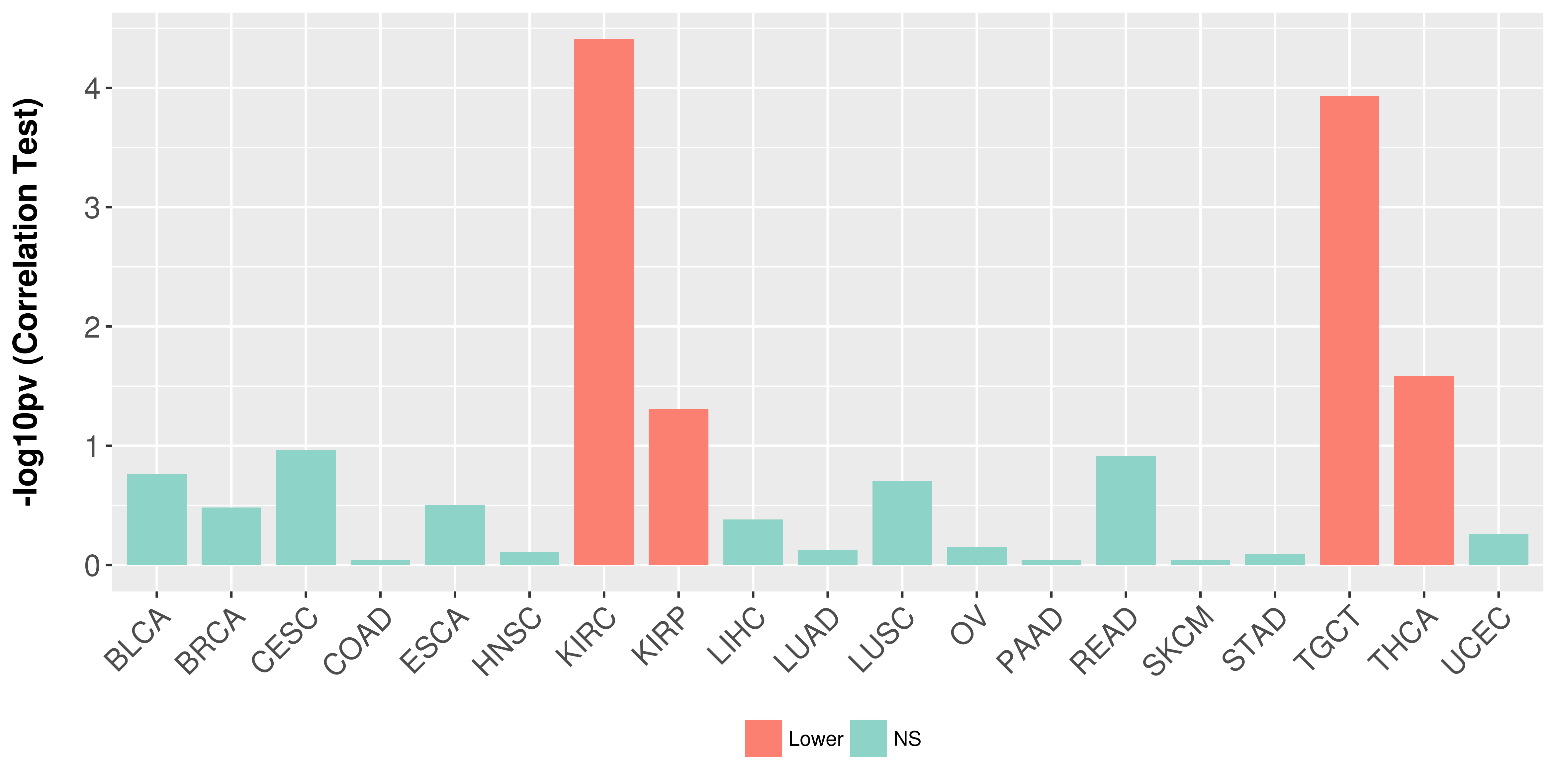
Association between expresson and stage.
|
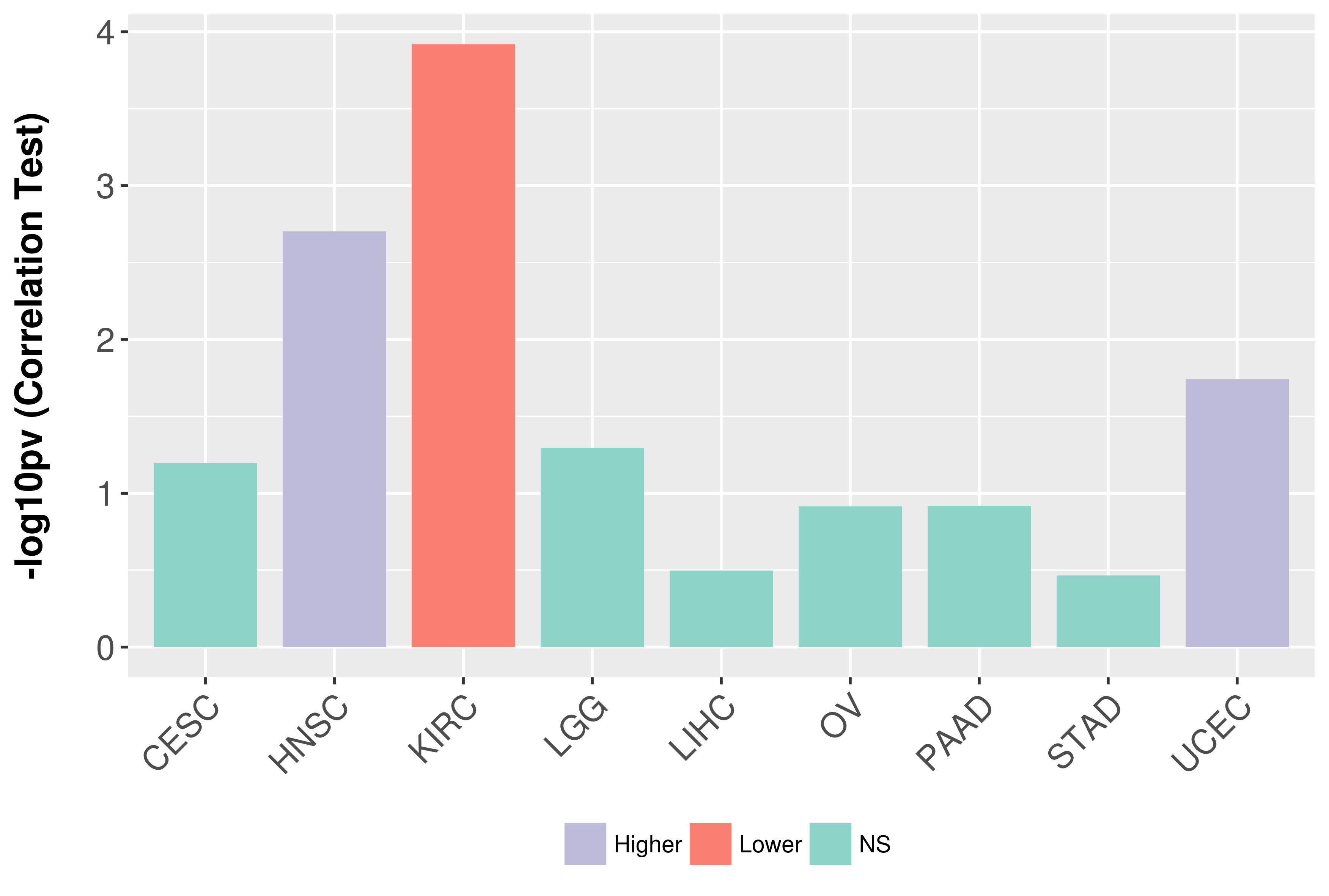
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | BMI1 |
| Name | BMI1 proto-oncogene, polycomb ring finger |
| Aliases | RNF51; PCGF4; polycomb group ring finger 4; B lymphoma Mo-MLV insertion region 1 homolog (mouse); FLVI2/BMI1 ...... |
| Location | 10p12.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | BMI1 |
| Name | BMI1 proto-oncogene, polycomb ring finger |
| Aliases | RNF51; PCGF4; polycomb group ring finger 4; B lymphoma Mo-MLV insertion region 1 homolog (mouse); FLVI2/BMI1 ...... |
| Location | 10p12.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for BMI1. |
| Summary | |
|---|---|
| Symbol | BMI1 |
| Name | BMI1 proto-oncogene, polycomb ring finger |
| Aliases | RNF51; PCGF4; polycomb group ring finger 4; B lymphoma Mo-MLV insertion region 1 homolog (mouse); FLVI2/BMI1 ...... |
| Location | 10p12.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|