Browse CAMKMT in pancancer
| Summary | |
|---|---|
| Symbol | CAMKMT |
| Name | calmodulin-lysine N-methyltransferase |
| Aliases | CLNMT; CaM KMT; C2orf34; chromosome 2 open reading frame 34; KMT |
| Location | 2p21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF10294 Lysine methyltransferase |
||||||||||
| Function |
Catalyzes the trimethylation of 'Lys-116' in calmodulin. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0006479 protein methylation GO:0007602 phototransduction GO:0007603 phototransduction, visible light GO:0008213 protein alkylation GO:0008277 regulation of G-protein coupled receptor protein signaling pathway GO:0009314 response to radiation GO:0009416 response to light stimulus GO:0009581 detection of external stimulus GO:0009582 detection of abiotic stimulus GO:0009583 detection of light stimulus GO:0009584 detection of visible light GO:0016056 rhodopsin mediated signaling pathway GO:0018022 peptidyl-lysine methylation GO:0018205 peptidyl-lysine modification GO:0022400 regulation of rhodopsin mediated signaling pathway GO:0032259 methylation GO:0043414 macromolecule methylation GO:0071214 cellular response to abiotic stimulus GO:0071478 cellular response to radiation GO:0071482 cellular response to light stimulus |
| Molecular Function |
GO:0008168 methyltransferase activity GO:0008170 N-methyltransferase activity GO:0008276 protein methyltransferase activity GO:0008757 S-adenosylmethionine-dependent methyltransferase activity GO:0016278 lysine N-methyltransferase activity GO:0016279 protein-lysine N-methyltransferase activity GO:0016741 transferase activity, transferring one-carbon groups GO:0018025 calmodulin-lysine N-methyltransferase activity |
| Cellular Component | - |
| KEGG |
hsa00310 Lysine degradation |
| Reactome |
R-HSA-2514859: Inactivation, recovery and regulation of the phototransduction cascade R-HSA-392499: Metabolism of proteins R-HSA-597592: Post-translational protein modification R-HSA-8876725: Protein methylation R-HSA-162582: Signal Transduction R-HSA-2514856: The phototransduction cascade R-HSA-2187338: Visual phototransduction |
| Summary | |
|---|---|
| Symbol | CAMKMT |
| Name | calmodulin-lysine N-methyltransferase |
| Aliases | CLNMT; CaM KMT; C2orf34; chromosome 2 open reading frame 34; KMT |
| Location | 2p21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for CAMKMT. |
| Summary | |
|---|---|
| Symbol | CAMKMT |
| Name | calmodulin-lysine N-methyltransferase |
| Aliases | CLNMT; CaM KMT; C2orf34; chromosome 2 open reading frame 34; KMT |
| Location | 2p21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
| There is no PTM data |
| Summary | |
|---|---|
| Symbol | CAMKMT |
| Name | calmodulin-lysine N-methyltransferase |
| Aliases | CLNMT; CaM KMT; C2orf34; chromosome 2 open reading frame 34; KMT |
| Location | 2p21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
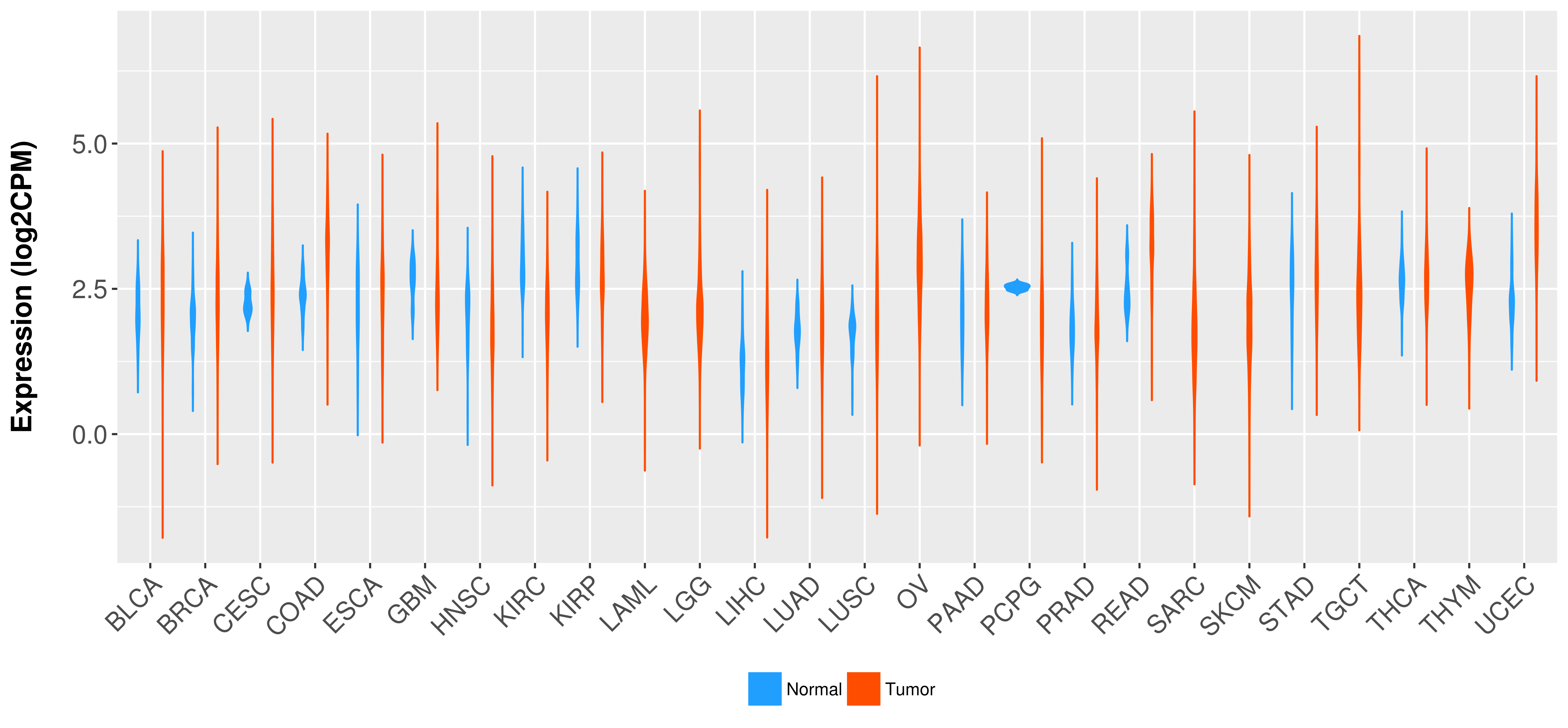
Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
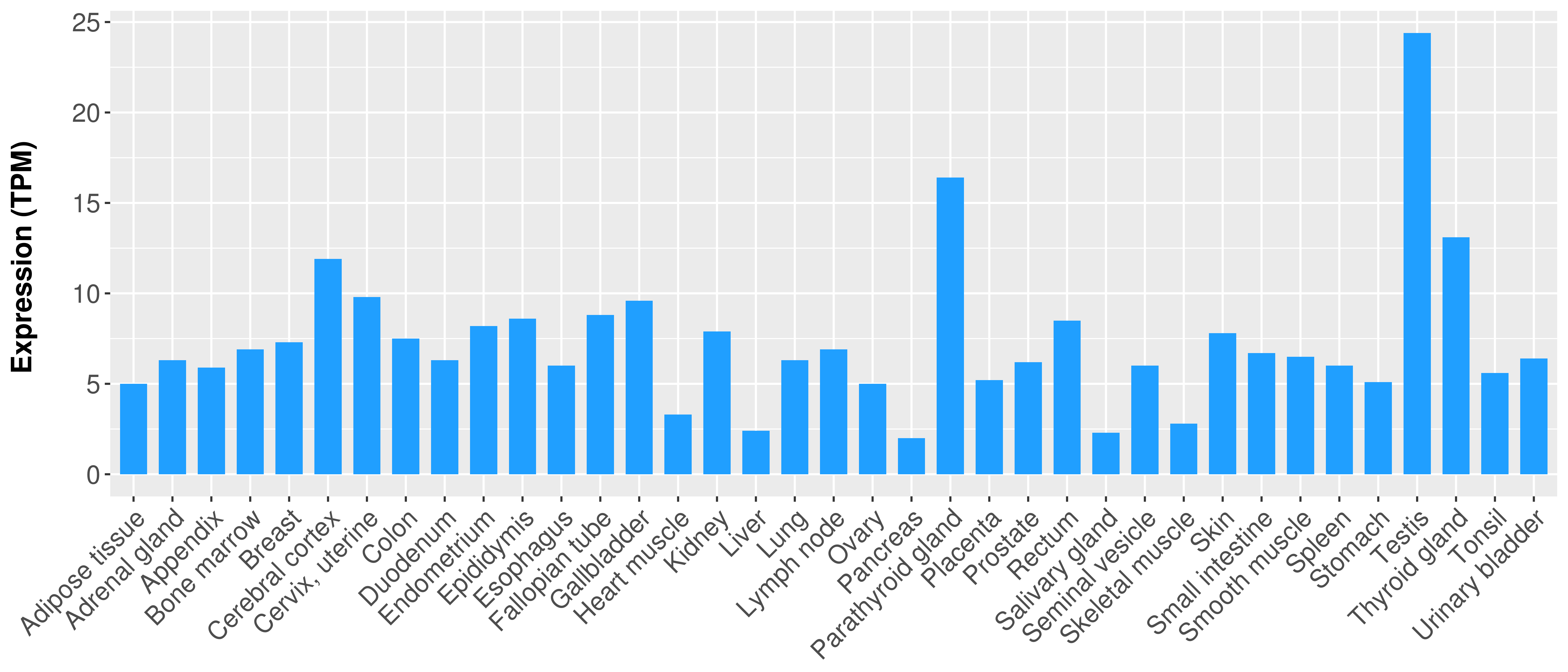
|
| There is no record. |
| Summary | |
|---|---|
| Symbol | CAMKMT |
| Name | calmodulin-lysine N-methyltransferase |
| Aliases | CLNMT; CaM KMT; C2orf34; chromosome 2 open reading frame 34; KMT |
| Location | 2p21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |

Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | CAMKMT |
| Name | calmodulin-lysine N-methyltransferase |
| Aliases | CLNMT; CaM KMT; C2orf34; chromosome 2 open reading frame 34; KMT |
| Location | 2p21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
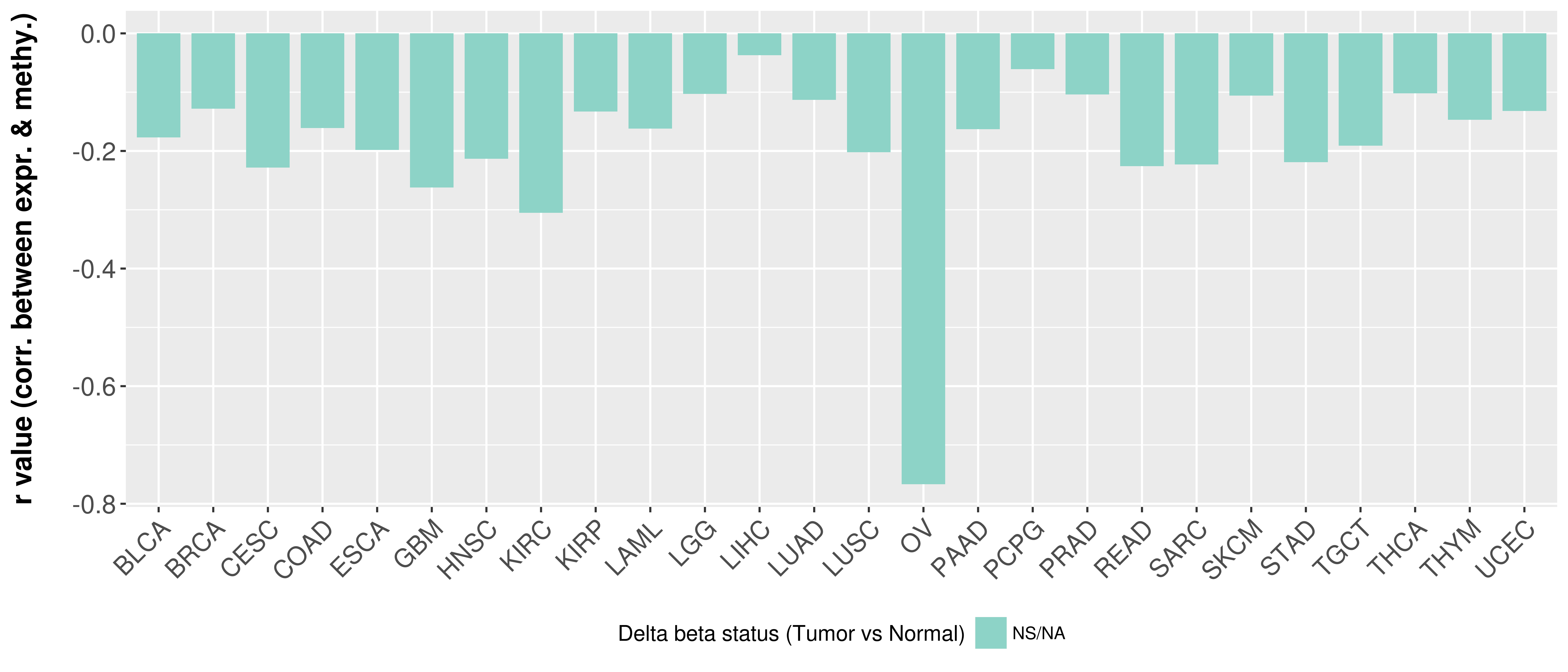
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | CAMKMT |
| Name | calmodulin-lysine N-methyltransferase |
| Aliases | CLNMT; CaM KMT; C2orf34; chromosome 2 open reading frame 34; KMT |
| Location | 2p21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
| There is no antibody staining data. |
| Summary | |
|---|---|
| Symbol | CAMKMT |
| Name | calmodulin-lysine N-methyltransferase |
| Aliases | CLNMT; CaM KMT; C2orf34; chromosome 2 open reading frame 34; KMT |
| Location | 2p21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
Association between expresson and subtype.
|
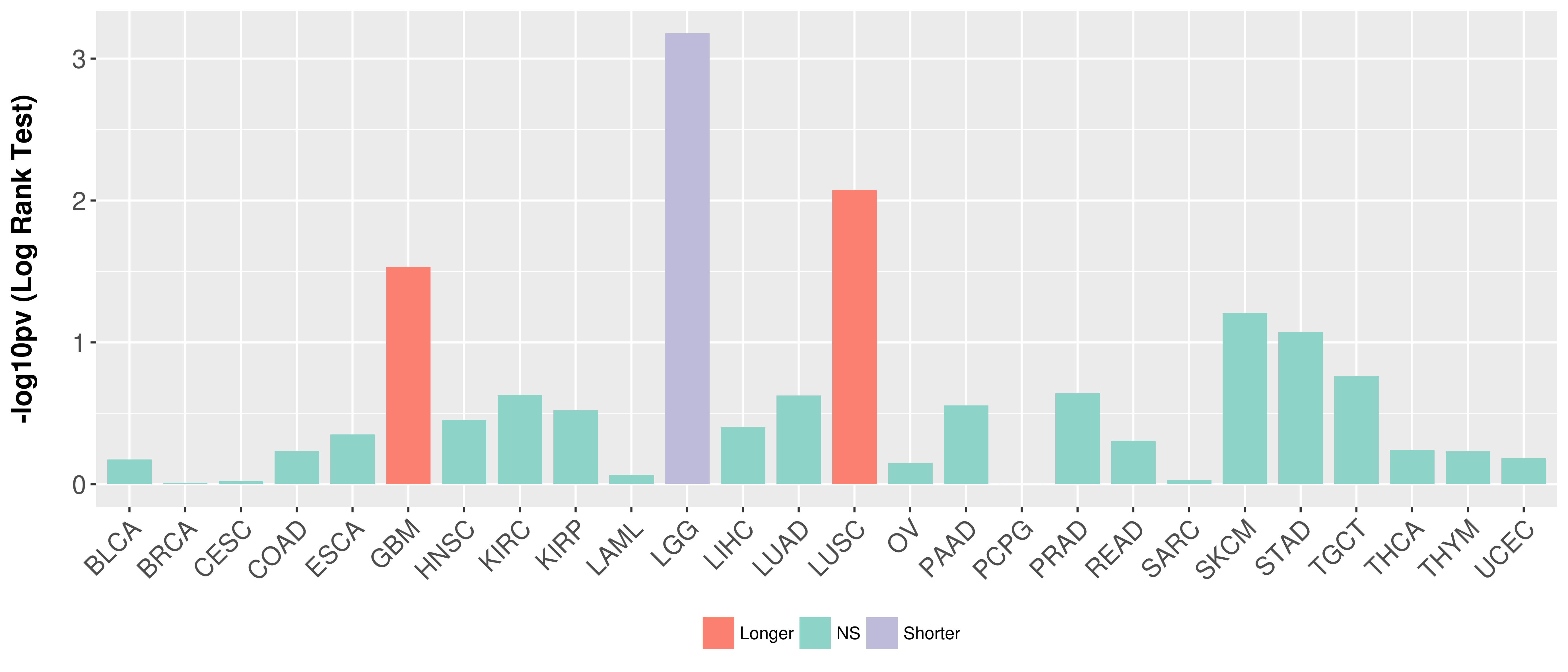
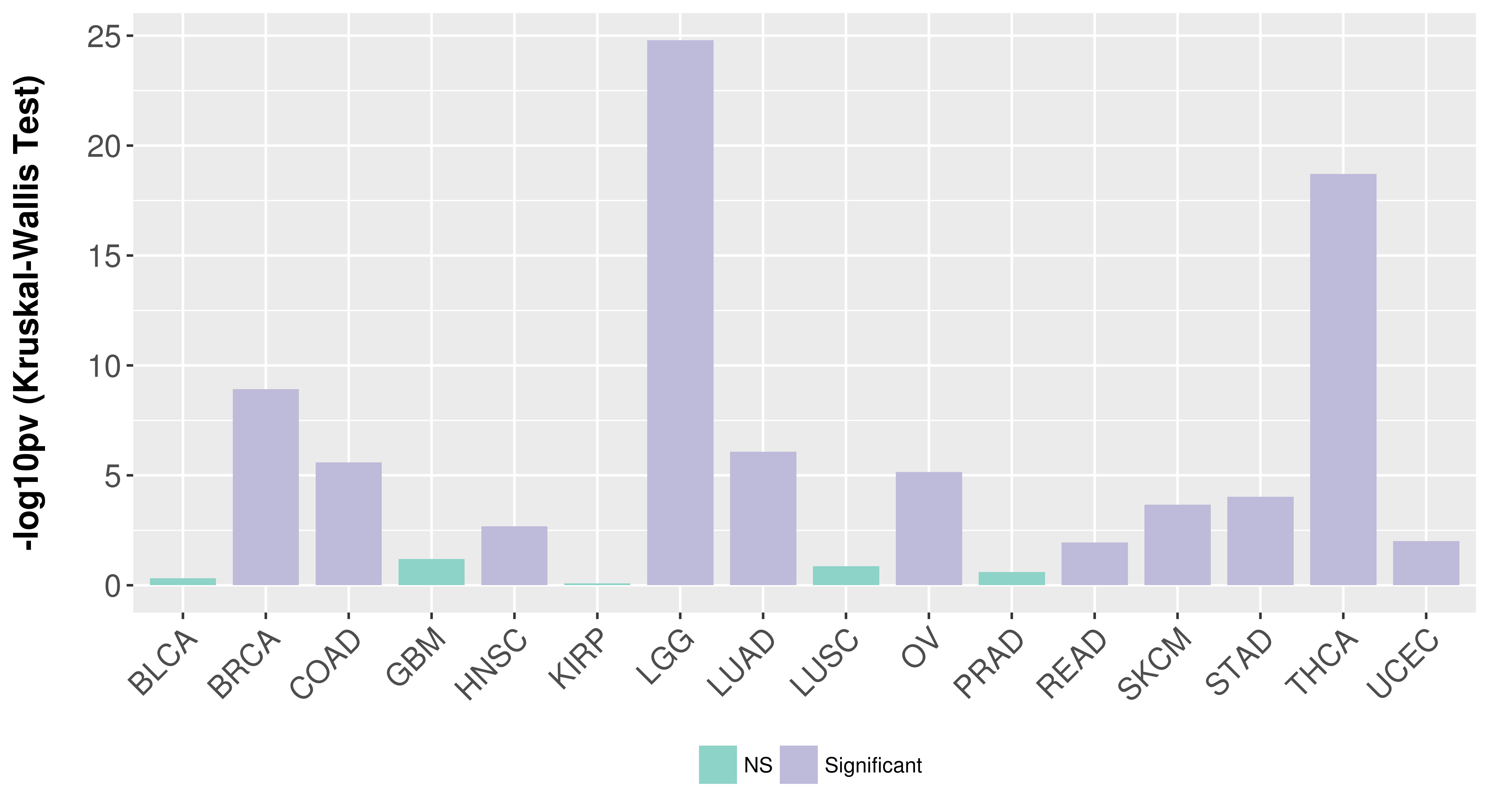
Overall survival analysis based on expression.
|
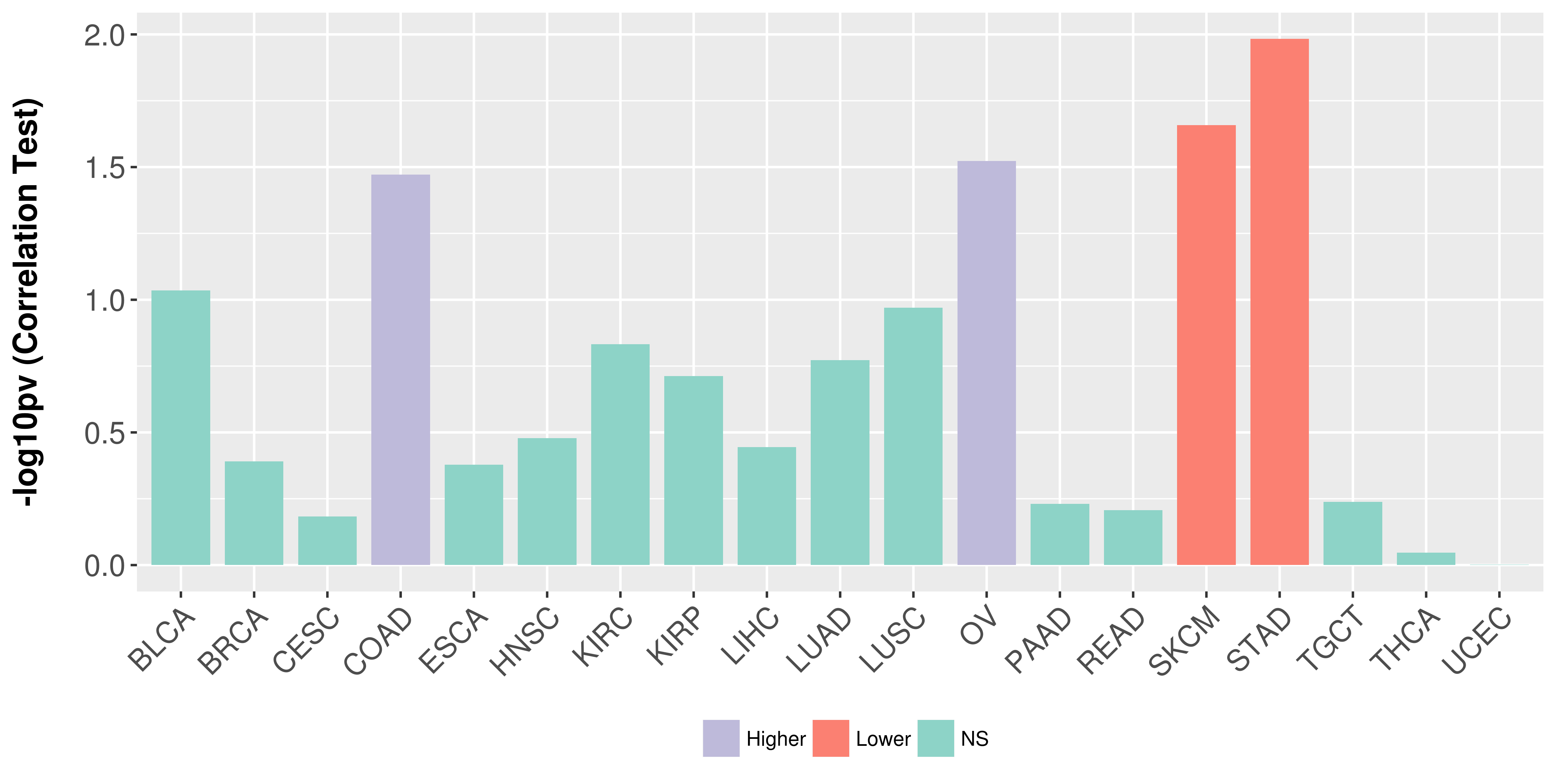
Association between expresson and stage.
|
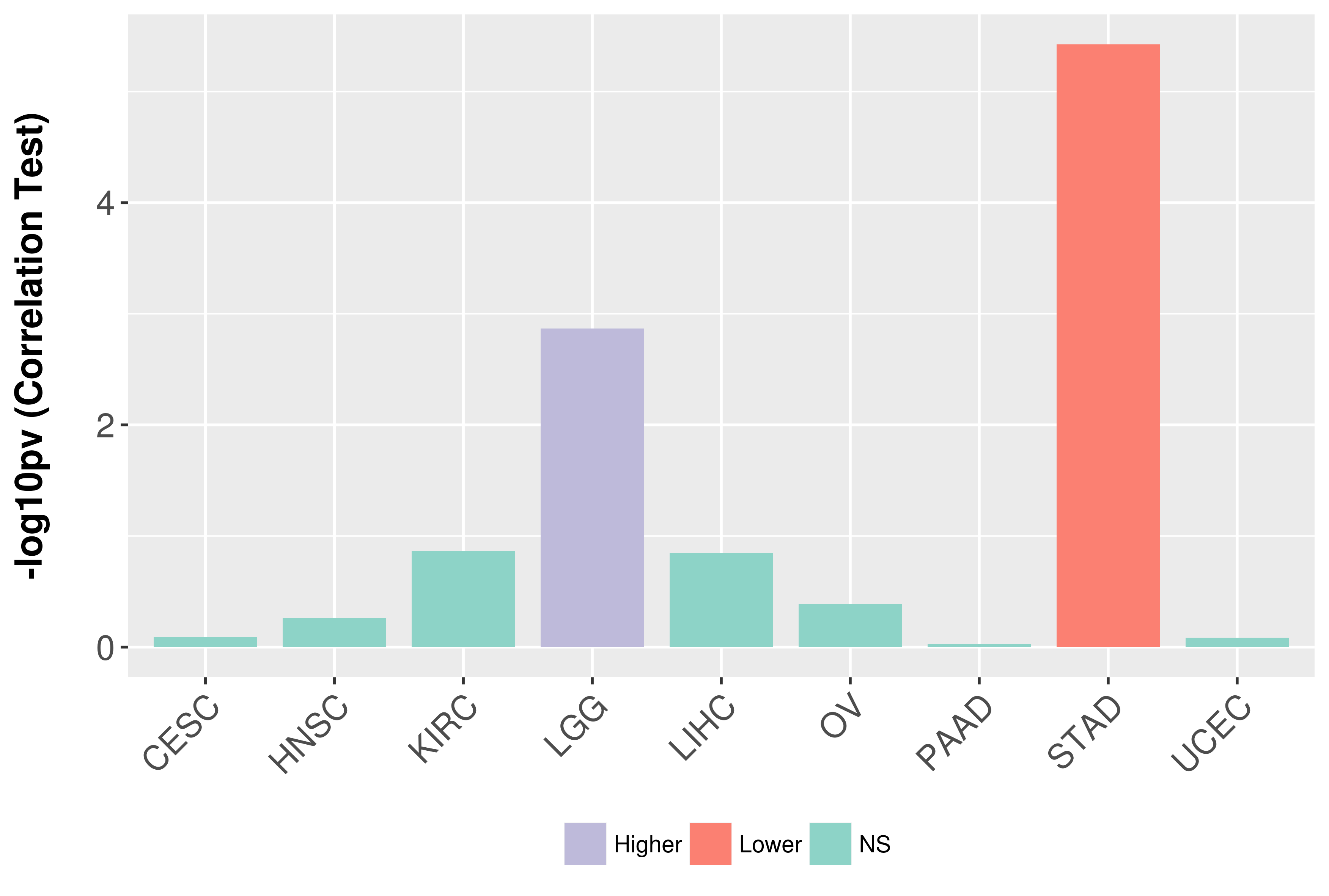
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | CAMKMT |
| Name | calmodulin-lysine N-methyltransferase |
| Aliases | CLNMT; CaM KMT; C2orf34; chromosome 2 open reading frame 34; KMT |
| Location | 2p21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | CAMKMT |
| Name | calmodulin-lysine N-methyltransferase |
| Aliases | CLNMT; CaM KMT; C2orf34; chromosome 2 open reading frame 34; KMT |
| Location | 2p21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for CAMKMT. |
| Summary | |
|---|---|
| Symbol | CAMKMT |
| Name | calmodulin-lysine N-methyltransferase |
| Aliases | CLNMT; CaM KMT; C2orf34; chromosome 2 open reading frame 34; KMT |
| Location | 2p21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for CAMKMT. |
| There is no record for CAMKMT. |