Browse CARM1 in pancancer
| Summary | |
|---|---|
| Symbol | CARM1 |
| Name | coactivator associated arginine methyltransferase 1 |
| Aliases | PRMT4; Histone-arginine methyltransferase CARM1 |
| Location | 19p13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF11531 Coactivator-associated arginine methyltransferase 1 N terminal PF05185 PRMT5 arginine-N-methyltransferase |
||||||||||
| Function |
Methylates (mono- and asymmetric dimethylation) the guanidino nitrogens of arginyl residues in several proteins involved in DNA packaging, transcription regulation, pre-mRNA splicing, and mRNA stability. Recruited to promoters upon gene activation together with histone acetyltransferases from EP300/P300 and p160 families, methylates histone H3 at 'Arg-17' (H3R17me), forming mainly asymmetric dimethylarginine (H3R17me2a), leading to activate transcription via chromatin remodeling. During nuclear hormone receptor activation and TCF7L2/TCF4 activation, acts synergically with EP300/P300 and either one of the p160 histone acetyltransferases NCOA1/SRC1, NCOA2/GRIP1 and NCOA3/ACTR or CTNNB1/beta-catenin to activate transcription. During myogenic transcriptional activation, acts together with NCOA3/ACTR as a coactivator for MEF2C. During monocyte inflammatory stimulation, acts together with EP300/P300 as a coactivator for NF-kappa-B. Acts as coactivator for PPARG, promotes adipocyte differentiation and the accumulation of brown fat tissue. Plays a role in the regulation of pre-mRNA alternative splicing by methylation of splicing factors. Also seems to be involved in p53/TP53 transcriptional activation. Methylates EP300/P300, both at 'Arg-2142', which may loosen its interaction with NCOA2/GRIP1, and at 'Arg-580' and 'Arg-604' in the KIX domain, which impairs its interaction with CREB and inhibits CREB-dependent transcriptional activation. Also methylates arginine residues in RNA-binding proteins PABPC1, ELAVL1 and ELAV4, which may affect their mRNA-stabilizing properties and the half-life of their target mRNAs. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000075 cell cycle checkpoint GO:0000077 DNA damage checkpoint GO:0000082 G1/S transition of mitotic cell cycle GO:0001501 skeletal system development GO:0003416 endochondral bone growth GO:0003417 growth plate cartilage development GO:0003419 growth plate cartilage chondrocyte proliferation GO:0003420 regulation of growth plate cartilage chondrocyte proliferation GO:0006479 protein methylation GO:0006977 DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest GO:0007050 cell cycle arrest GO:0007093 mitotic cell cycle checkpoint GO:0007346 regulation of mitotic cell cycle GO:0007568 aging GO:0008213 protein alkylation GO:0009755 hormone-mediated signaling pathway GO:0010721 negative regulation of cell development GO:0010948 negative regulation of cell cycle process GO:0010975 regulation of neuron projection development GO:0010977 negative regulation of neuron projection development GO:0014074 response to purine-containing compound GO:0016358 dendrite development GO:0016570 histone modification GO:0016571 histone methylation GO:0018195 peptidyl-arginine modification GO:0018216 peptidyl-arginine methylation GO:0019919 peptidyl-arginine methylation, to asymmetrical-dimethyl arginine GO:0030330 DNA damage response, signal transduction by p53 class mediator GO:0030518 intracellular steroid hormone receptor signaling pathway GO:0030520 intracellular estrogen receptor signaling pathway GO:0030522 intracellular receptor signaling pathway GO:0031345 negative regulation of cell projection organization GO:0031570 DNA integrity checkpoint GO:0031571 mitotic G1 DNA damage checkpoint GO:0032091 negative regulation of protein binding GO:0032259 methylation GO:0033143 regulation of intracellular steroid hormone receptor signaling pathway GO:0033146 regulation of intracellular estrogen receptor signaling pathway GO:0034969 histone arginine methylation GO:0034970 histone H3-R2 methylation GO:0034971 histone H3-R17 methylation GO:0035246 peptidyl-arginine N-methylation GO:0035247 peptidyl-arginine omega-N-methylation GO:0035265 organ growth GO:0042770 signal transduction in response to DNA damage GO:0043393 regulation of protein binding GO:0043401 steroid hormone mediated signaling pathway GO:0043414 macromolecule methylation GO:0044770 cell cycle phase transition GO:0044772 mitotic cell cycle phase transition GO:0044773 mitotic DNA damage checkpoint GO:0044774 mitotic DNA integrity checkpoint GO:0044783 G1 DNA damage checkpoint GO:0044819 mitotic G1/S transition checkpoint GO:0044843 cell cycle G1/S phase transition GO:0045444 fat cell differentiation GO:0045598 regulation of fat cell differentiation GO:0045600 positive regulation of fat cell differentiation GO:0045665 negative regulation of neuron differentiation GO:0045786 negative regulation of cell cycle GO:0045787 positive regulation of cell cycle GO:0045930 negative regulation of mitotic cell cycle GO:0046620 regulation of organ growth GO:0046683 response to organophosphorus GO:0048545 response to steroid hormone GO:0048638 regulation of developmental growth GO:0048705 skeletal system morphogenesis GO:0050768 negative regulation of neurogenesis GO:0050773 regulation of dendrite development GO:0051098 regulation of binding GO:0051100 negative regulation of binding GO:0051216 cartilage development GO:0051591 response to cAMP GO:0051961 negative regulation of nervous system development GO:0060348 bone development GO:0060349 bone morphogenesis GO:0060350 endochondral bone morphogenesis GO:0060351 cartilage development involved in endochondral bone morphogenesis GO:0061035 regulation of cartilage development GO:0061448 connective tissue development GO:0071156 regulation of cell cycle arrest GO:0071158 positive regulation of cell cycle arrest GO:0071383 cellular response to steroid hormone stimulus GO:0071396 cellular response to lipid GO:0071407 cellular response to organic cyclic compound GO:0072331 signal transduction by p53 class mediator GO:0072395 signal transduction involved in cell cycle checkpoint GO:0072401 signal transduction involved in DNA integrity checkpoint GO:0072413 signal transduction involved in mitotic cell cycle checkpoint GO:0072422 signal transduction involved in DNA damage checkpoint GO:0072431 signal transduction involved in mitotic G1 DNA damage checkpoint GO:0090068 positive regulation of cell cycle process GO:0098868 bone growth GO:1901987 regulation of cell cycle phase transition GO:1901988 negative regulation of cell cycle phase transition GO:1901990 regulation of mitotic cell cycle phase transition GO:1901991 negative regulation of mitotic cell cycle phase transition GO:1902400 intracellular signal transduction involved in G1 DNA damage checkpoint GO:1902402 signal transduction involved in mitotic DNA damage checkpoint GO:1902403 signal transduction involved in mitotic DNA integrity checkpoint GO:1902415 regulation of mRNA binding GO:1902806 regulation of cell cycle G1/S phase transition GO:1902807 negative regulation of cell cycle G1/S phase transition GO:1903010 regulation of bone development GO:1905214 regulation of RNA binding GO:2000027 regulation of organ morphogenesis GO:2000045 regulation of G1/S transition of mitotic cell cycle GO:2000134 negative regulation of G1/S transition of mitotic cell cycle GO:2000171 negative regulation of dendrite development |
| Molecular Function |
GO:0003713 transcription coactivator activity GO:0008013 beta-catenin binding GO:0008168 methyltransferase activity GO:0008170 N-methyltransferase activity GO:0008276 protein methyltransferase activity GO:0008469 histone-arginine N-methyltransferase activity GO:0008757 S-adenosylmethionine-dependent methyltransferase activity GO:0016273 arginine N-methyltransferase activity GO:0016274 protein-arginine N-methyltransferase activity GO:0016741 transferase activity, transferring one-carbon groups GO:0030374 ligand-dependent nuclear receptor transcription coactivator activity GO:0035242 protein-arginine omega-N asymmetric methyltransferase activity GO:0035642 histone methyltransferase activity (H3-R17 specific) GO:0042054 histone methyltransferase activity GO:0042393 histone binding GO:0070577 lysine-acetylated histone binding |
| Cellular Component |
GO:0005667 transcription factor complex GO:0044798 nuclear transcription factor complex GO:0090575 RNA polymerase II transcription factor complex |
| KEGG | - |
| Reactome |
R-HSA-2426168: Activation of gene expression by SREBF (SREBP) R-HSA-1368108: BMAL1 R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-400253: Circadian Clock R-HSA-1266738: Developmental Biology R-HSA-535734: Fatty acid, triacylglycerol, and ketone body metabolism R-HSA-74160: Gene Expression R-HSA-212436: Generic Transcription Pathway R-HSA-1430728: Metabolism R-HSA-556833: Metabolism of lipids and lipoproteins R-HSA-1592230: Mitochondrial biogenesis R-HSA-1852241: Organelle biogenesis and maintenance R-HSA-1989781: PPARA activates gene expression R-HSA-3214858: RMTs methylate histone arginines R-HSA-1368082: RORA activates gene expression R-HSA-1655829: Regulation of cholesterol biosynthesis by SREBP (SREBF) R-HSA-400206: Regulation of lipid metabolism by Peroxisome proliferator-activated receptor alpha (PPARalpha) R-HSA-6791312: TP53 Regulates Transcription of Cell Cycle Genes R-HSA-6804114: TP53 Regulates Transcription of Genes Involved in G2 Cell Cycle Arrest R-HSA-3700989: Transcriptional Regulation by TP53 R-HSA-2151201: Transcriptional activation of mitochondrial biogenesis R-HSA-381340: Transcriptional regulation of white adipocyte differentiation R-HSA-2032785: YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| Summary | |
|---|---|
| Symbol | CARM1 |
| Name | coactivator associated arginine methyltransferase 1 |
| Aliases | PRMT4; Histone-arginine methyltransferase CARM1 |
| Location | 19p13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for CARM1. |
| Summary | |
|---|---|
| Symbol | CARM1 |
| Name | coactivator associated arginine methyltransferase 1 |
| Aliases | PRMT4; Histone-arginine methyltransferase CARM1 |
| Location | 19p13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | CARM1 |
| Name | coactivator associated arginine methyltransferase 1 |
| Aliases | PRMT4; Histone-arginine methyltransferase CARM1 |
| Location | 19p13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
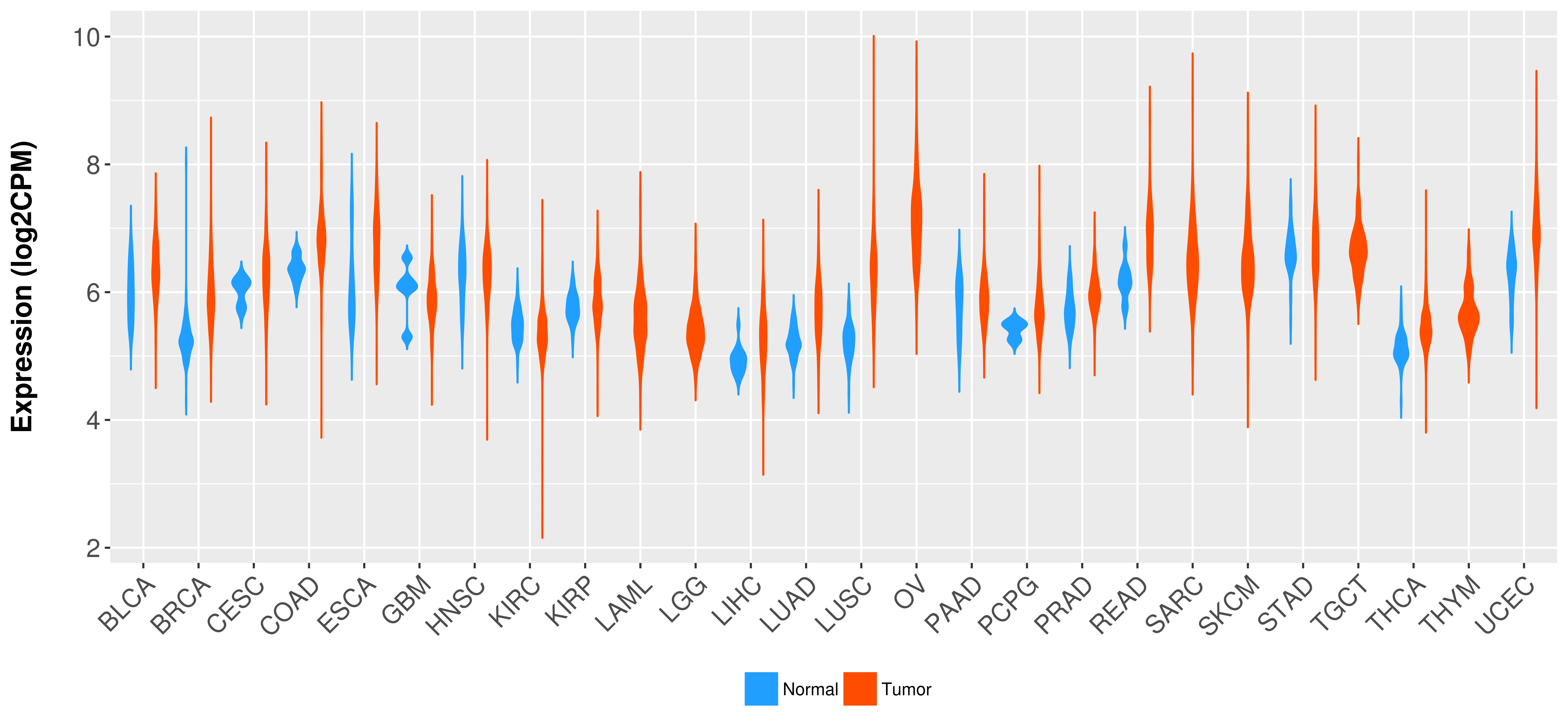
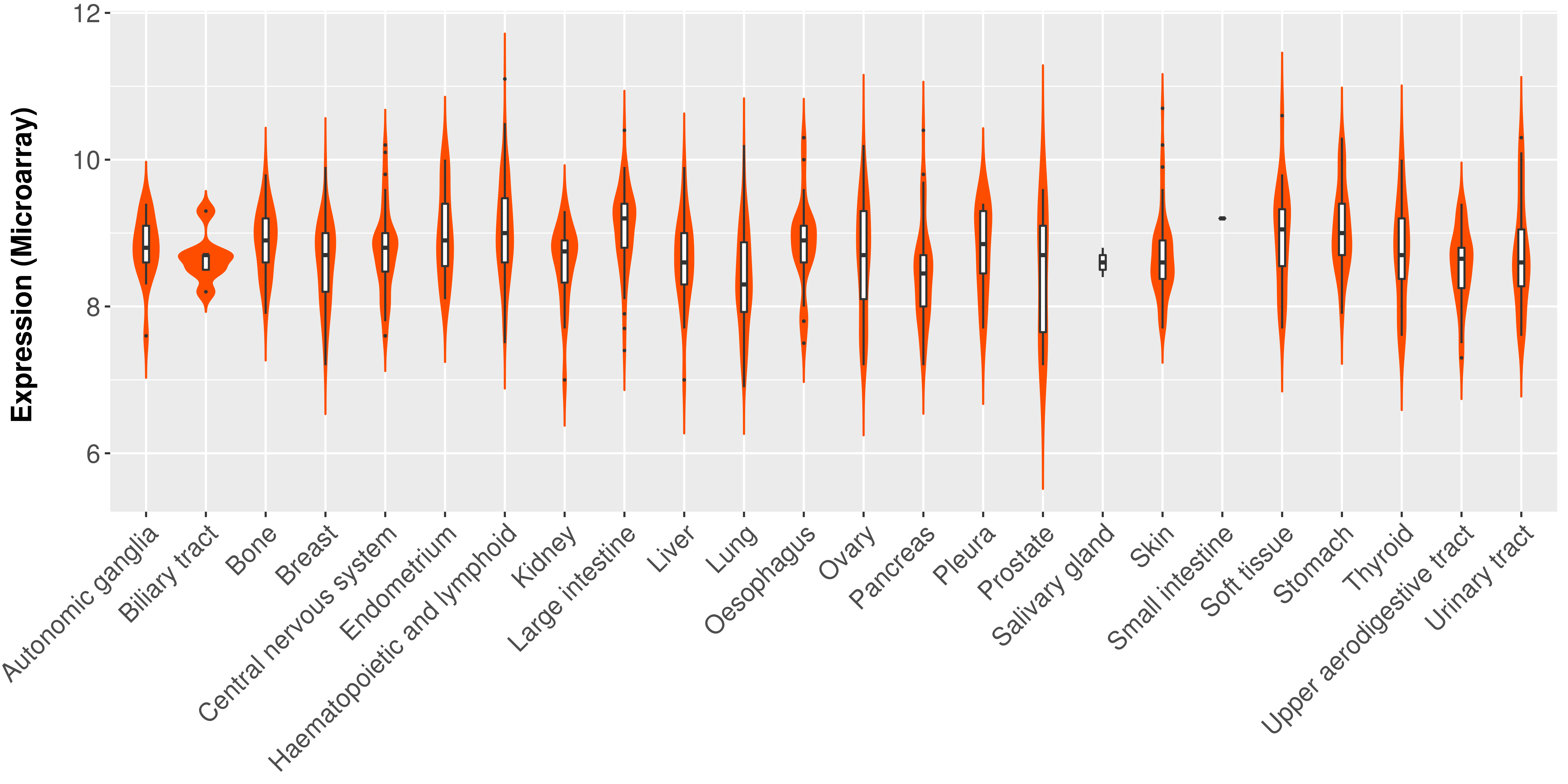
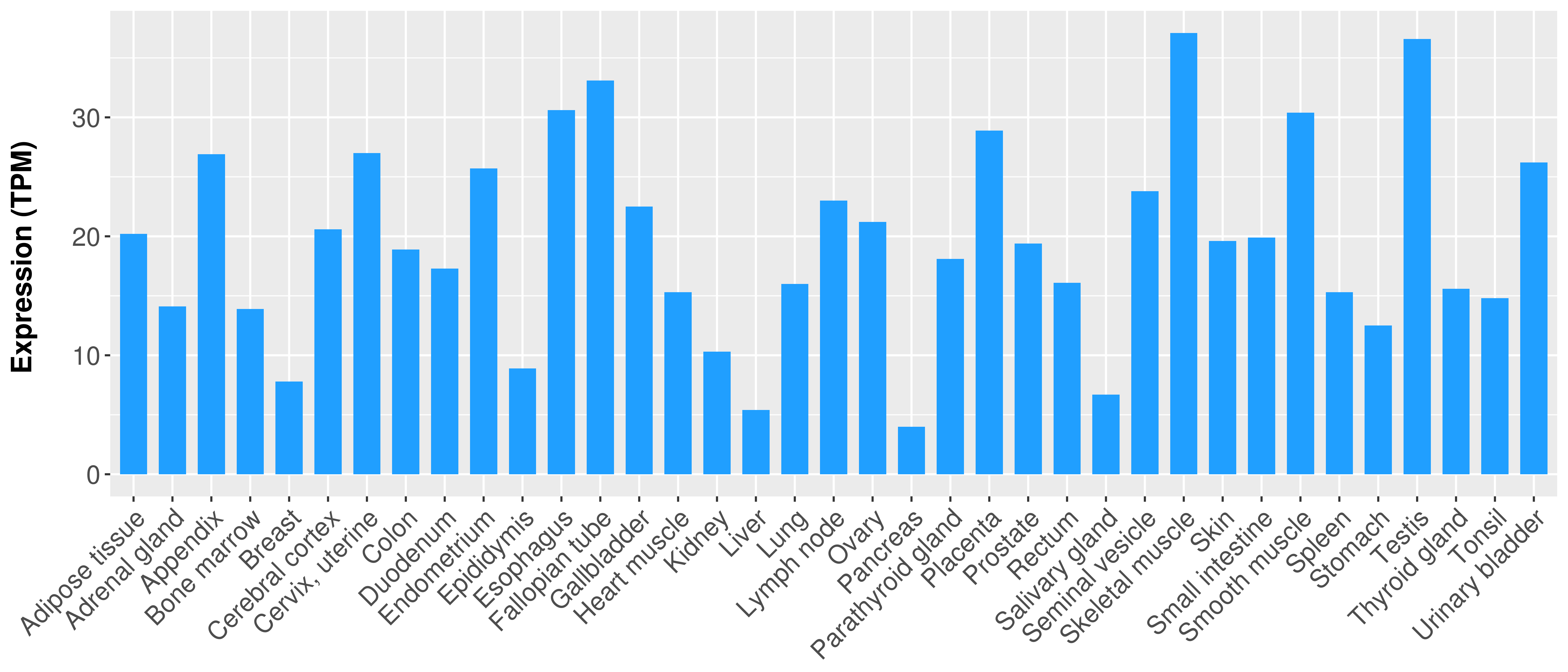
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | CARM1 |
| Name | coactivator associated arginine methyltransferase 1 |
| Aliases | PRMT4; Histone-arginine methyltransferase CARM1 |
| Location | 19p13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
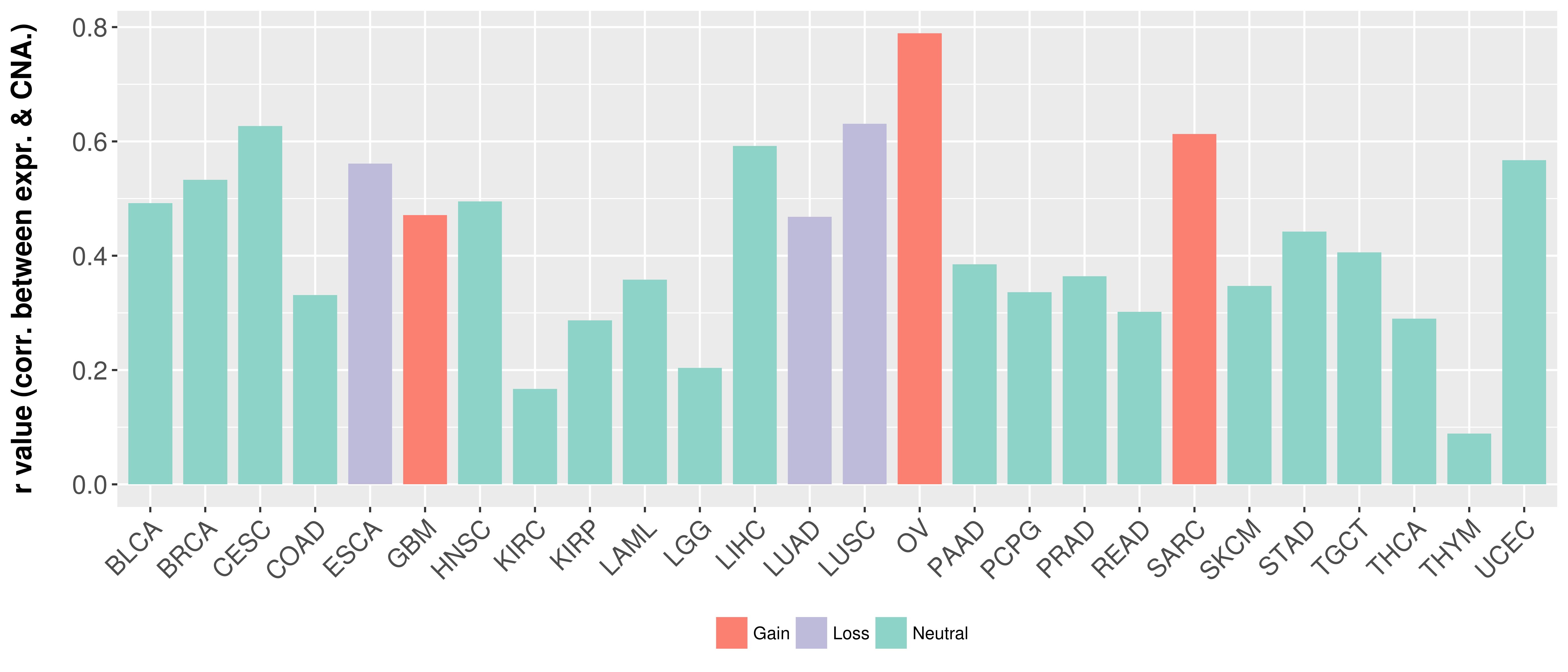
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | CARM1 |
| Name | coactivator associated arginine methyltransferase 1 |
| Aliases | PRMT4; Histone-arginine methyltransferase CARM1 |
| Location | 19p13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
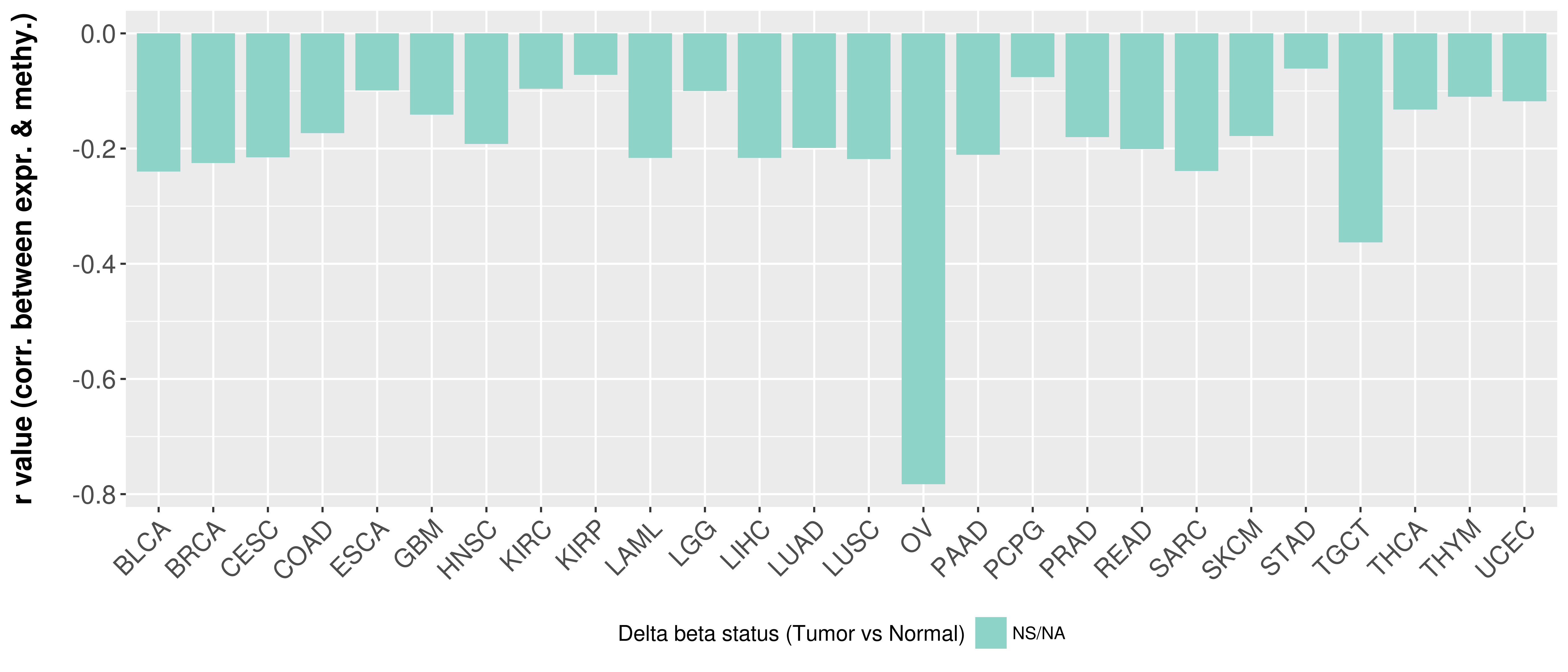
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | CARM1 |
| Name | coactivator associated arginine methyltransferase 1 |
| Aliases | PRMT4; Histone-arginine methyltransferase CARM1 |
| Location | 19p13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | CARM1 |
| Name | coactivator associated arginine methyltransferase 1 |
| Aliases | PRMT4; Histone-arginine methyltransferase CARM1 |
| Location | 19p13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
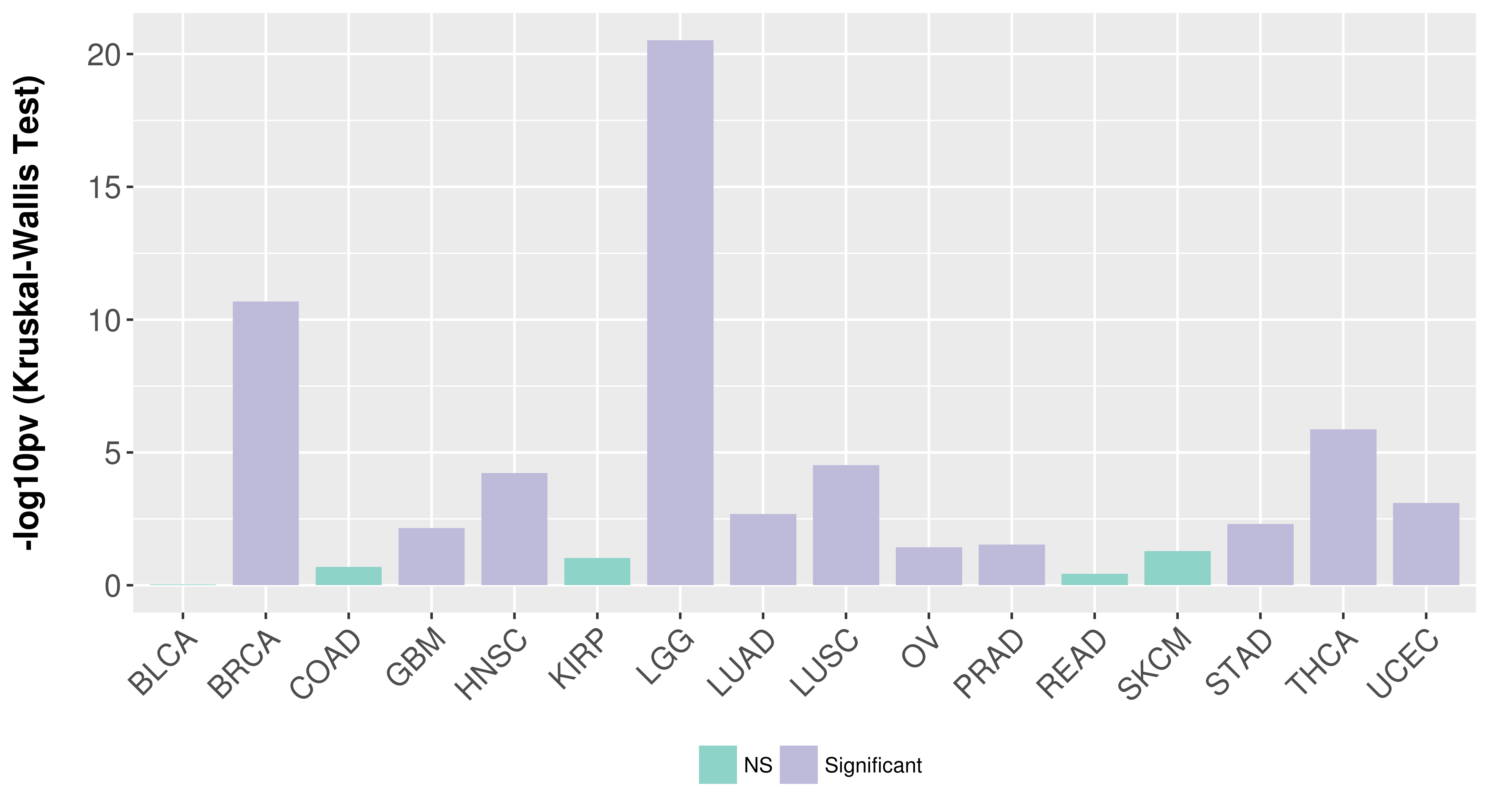
Association between expresson and subtype.
|
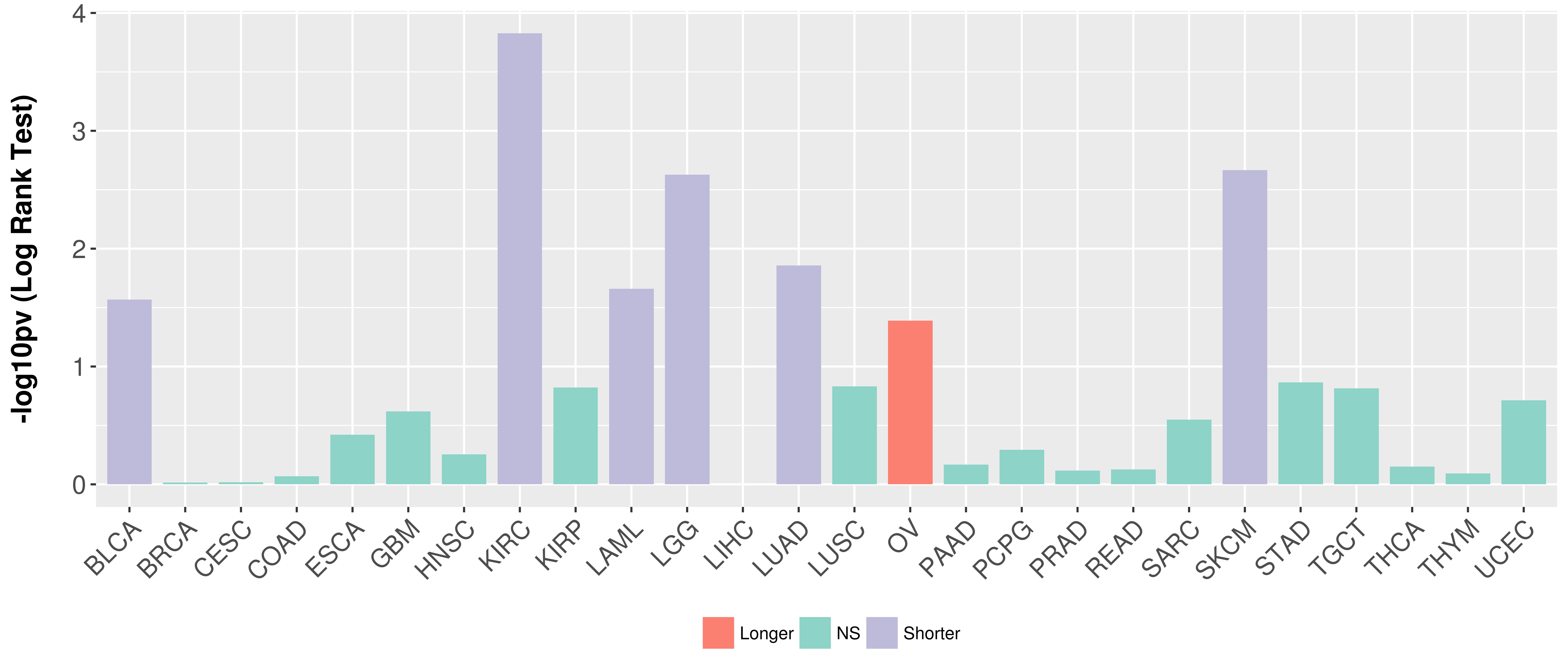
Overall survival analysis based on expression.
|
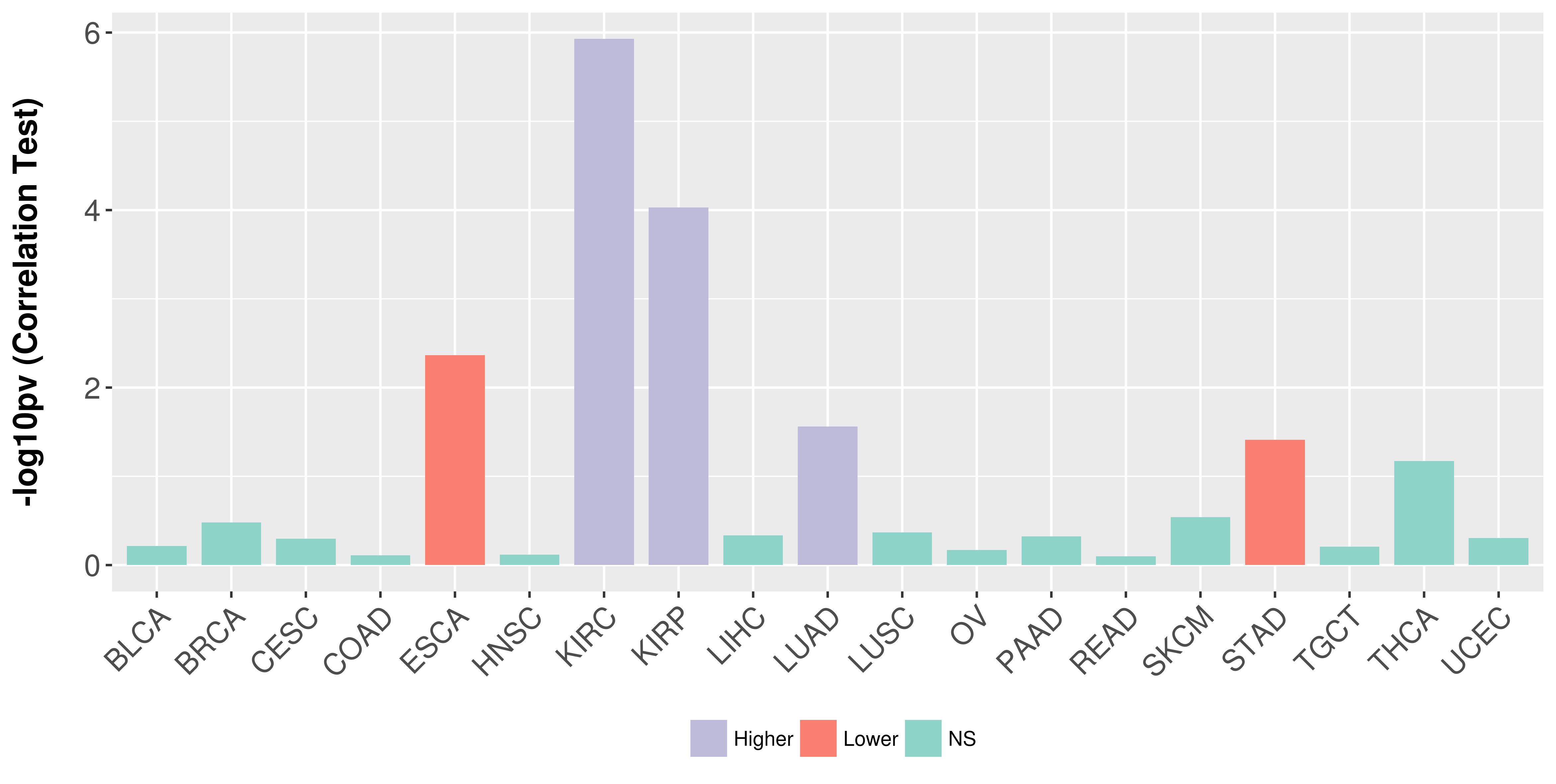
Association between expresson and stage.
|
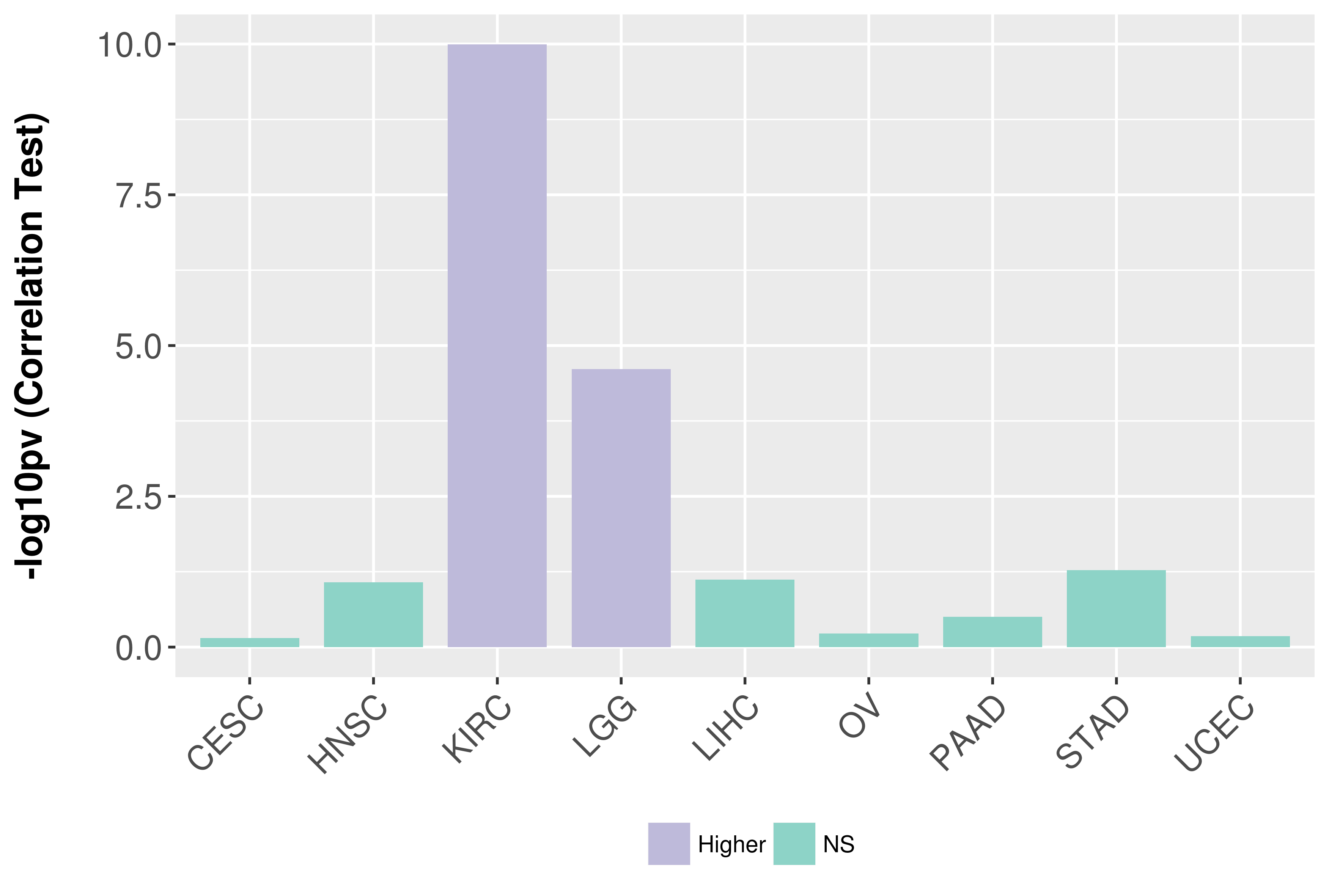
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | CARM1 |
| Name | coactivator associated arginine methyltransferase 1 |
| Aliases | PRMT4; Histone-arginine methyltransferase CARM1 |
| Location | 19p13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | CARM1 |
| Name | coactivator associated arginine methyltransferase 1 |
| Aliases | PRMT4; Histone-arginine methyltransferase CARM1 |
| Location | 19p13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for CARM1. |
| Summary | |
|---|---|
| Symbol | CARM1 |
| Name | coactivator associated arginine methyltransferase 1 |
| Aliases | PRMT4; Histone-arginine methyltransferase CARM1 |
| Location | 19p13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|