Browse CBX1 in pancancer
| Summary | |
|---|---|
| Symbol | CBX1 |
| Name | chromobox 1 |
| Aliases | HP1Hs-beta; M31; MOD1; CBX; HP1-BETA; HP1 beta homolog (Drosophila ); chromobox homolog 1 (Drosophila HP1 be ...... |
| Location | 17q21.32 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00385 Chromo (CHRromatin Organisation MOdifier) domain PF01393 Chromo shadow domain |
||||||||||
| Function |
Component of heterochromatin. Recognizes and binds histone H3 tails methylated at 'Lys-9', leading to epigenetic repression. Interaction with lamin B receptor (LBR) can contribute to the association of the heterochromatin with the inner nuclear membrane. |
||||||||||
| Classification |
|
||||||||||
| Biological Process | - |
| Molecular Function |
GO:0003682 chromatin binding GO:1990226 histone methyltransferase binding |
| Cellular Component |
GO:0000775 chromosome, centromeric region GO:0000781 chromosome, telomeric region GO:0000784 nuclear chromosome, telomeric region GO:0000785 chromatin GO:0000790 nuclear chromatin GO:0000792 heterochromatin GO:0001939 female pronucleus GO:0001940 male pronucleus GO:0005720 nuclear heterochromatin GO:0005721 pericentric heterochromatin GO:0005819 spindle GO:0010369 chromocenter GO:0044454 nuclear chromosome part GO:0045120 pronucleus GO:0098687 chromosomal region |
| KEGG | - |
| Reactome | - |
| Summary | |
|---|---|
| Symbol | CBX1 |
| Name | chromobox 1 |
| Aliases | HP1Hs-beta; M31; MOD1; CBX; HP1-BETA; HP1 beta homolog (Drosophila ); chromobox homolog 1 (Drosophila HP1 be ...... |
| Location | 17q21.32 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for CBX1. |
| Summary | |
|---|---|
| Symbol | CBX1 |
| Name | chromobox 1 |
| Aliases | HP1Hs-beta; M31; MOD1; CBX; HP1-BETA; HP1 beta homolog (Drosophila ); chromobox homolog 1 (Drosophila HP1 be ...... |
| Location | 17q21.32 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | CBX1 |
| Name | chromobox 1 |
| Aliases | HP1Hs-beta; M31; MOD1; CBX; HP1-BETA; HP1 beta homolog (Drosophila ); chromobox homolog 1 (Drosophila HP1 be ...... |
| Location | 17q21.32 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
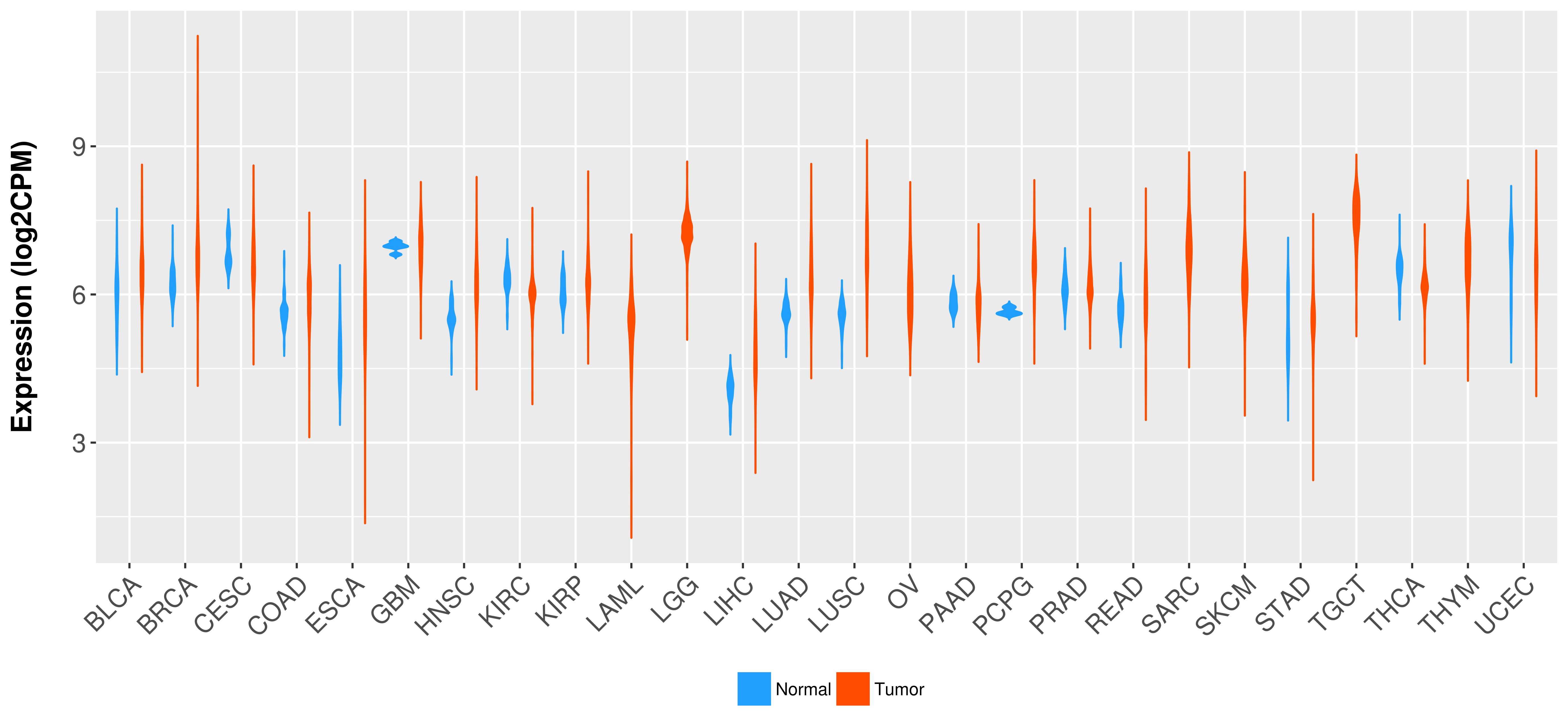
Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |

|
|
| Summary | |
|---|---|
| Symbol | CBX1 |
| Name | chromobox 1 |
| Aliases | HP1Hs-beta; M31; MOD1; CBX; HP1-BETA; HP1 beta homolog (Drosophila ); chromobox homolog 1 (Drosophila HP1 be ...... |
| Location | 17q21.32 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
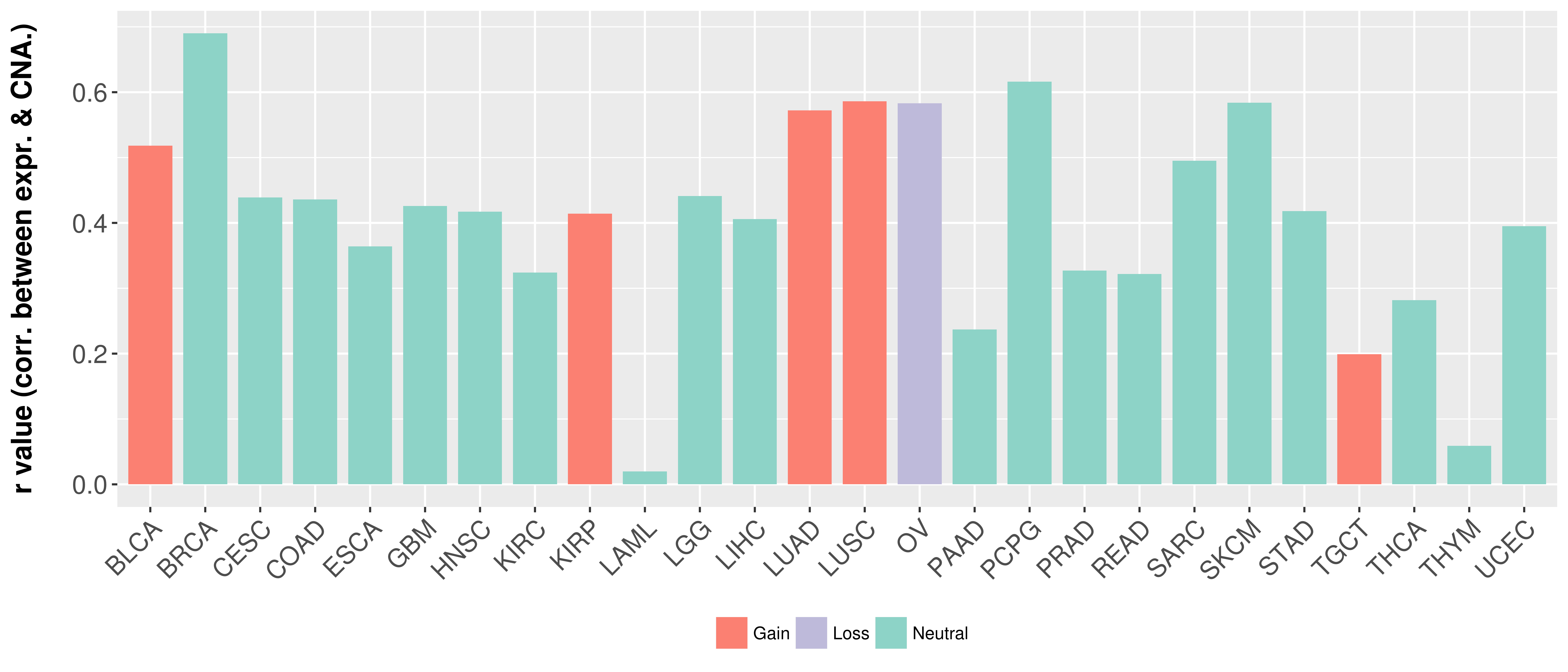
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | CBX1 |
| Name | chromobox 1 |
| Aliases | HP1Hs-beta; M31; MOD1; CBX; HP1-BETA; HP1 beta homolog (Drosophila ); chromobox homolog 1 (Drosophila HP1 be ...... |
| Location | 17q21.32 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
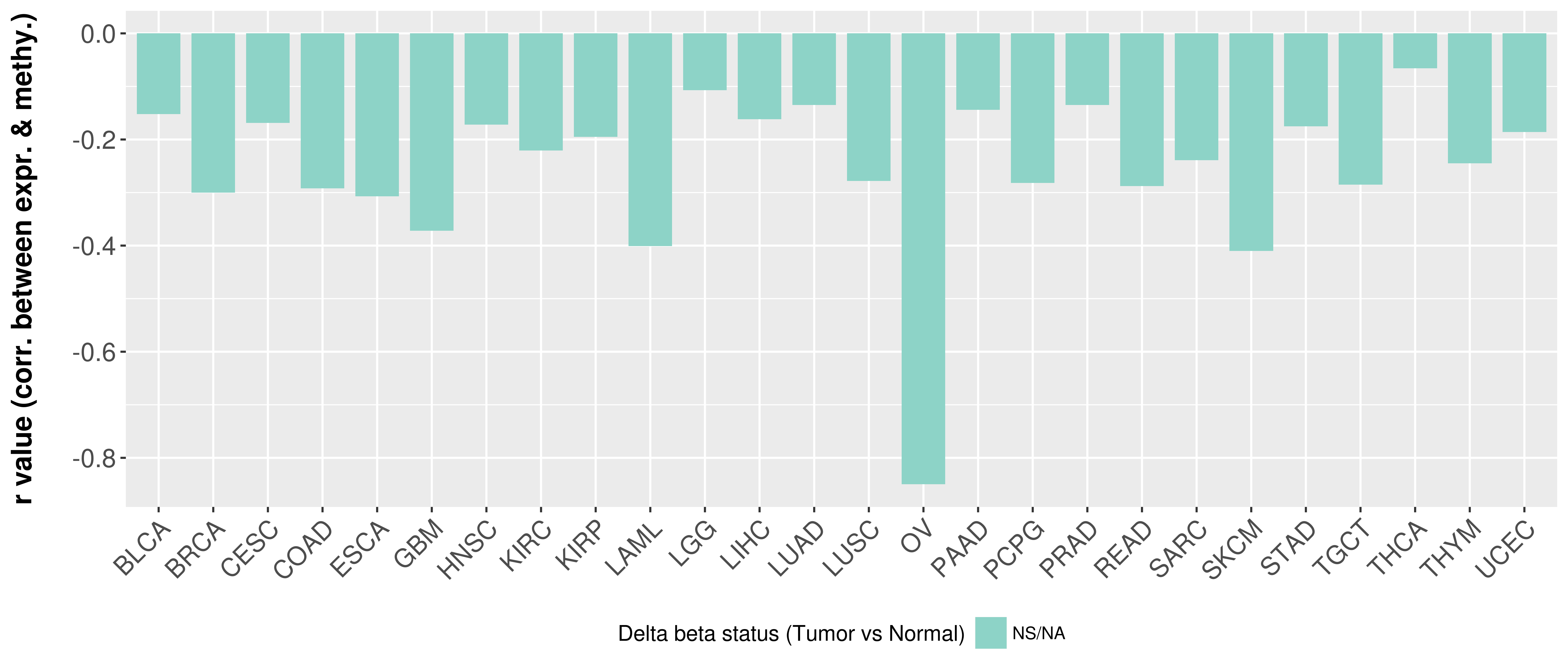
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | CBX1 |
| Name | chromobox 1 |
| Aliases | HP1Hs-beta; M31; MOD1; CBX; HP1-BETA; HP1 beta homolog (Drosophila ); chromobox homolog 1 (Drosophila HP1 be ...... |
| Location | 17q21.32 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
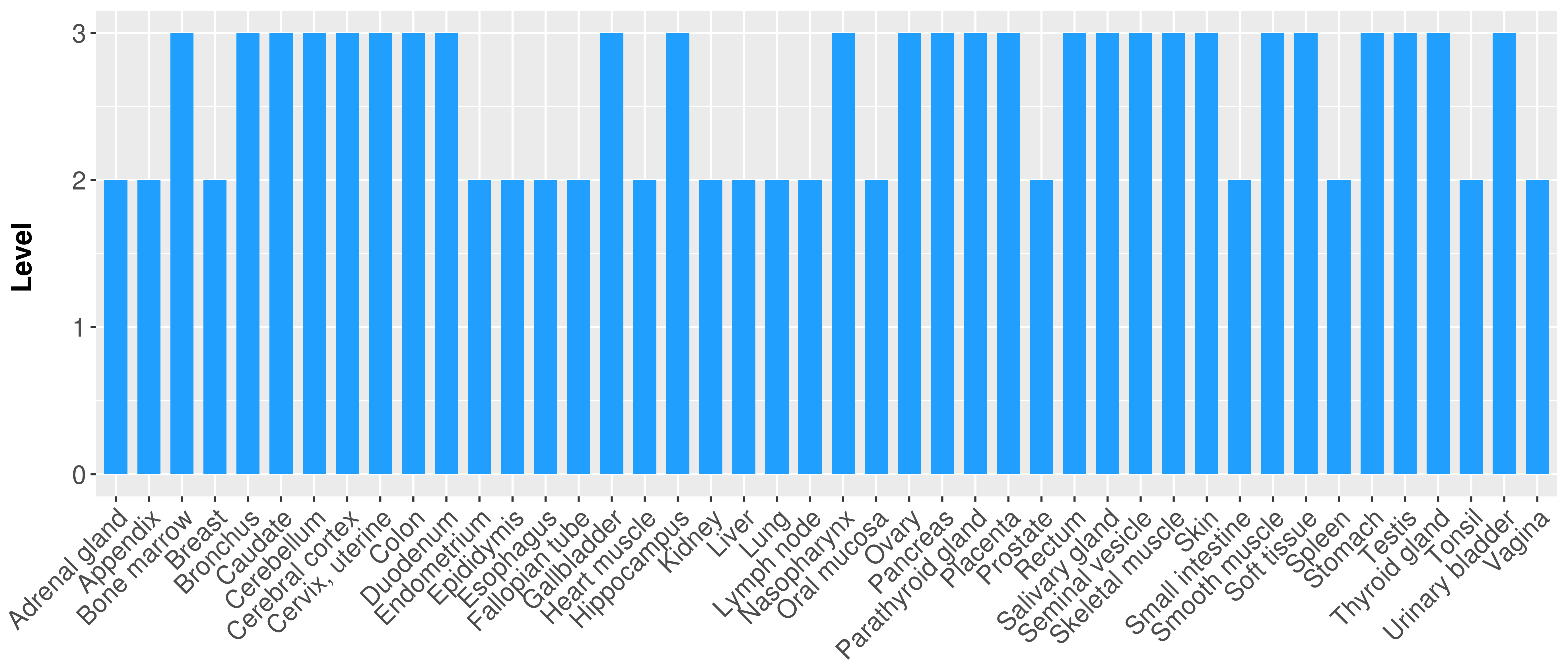
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | CBX1 |
| Name | chromobox 1 |
| Aliases | HP1Hs-beta; M31; MOD1; CBX; HP1-BETA; HP1 beta homolog (Drosophila ); chromobox homolog 1 (Drosophila HP1 be ...... |
| Location | 17q21.32 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |

Association between expresson and subtype.
|
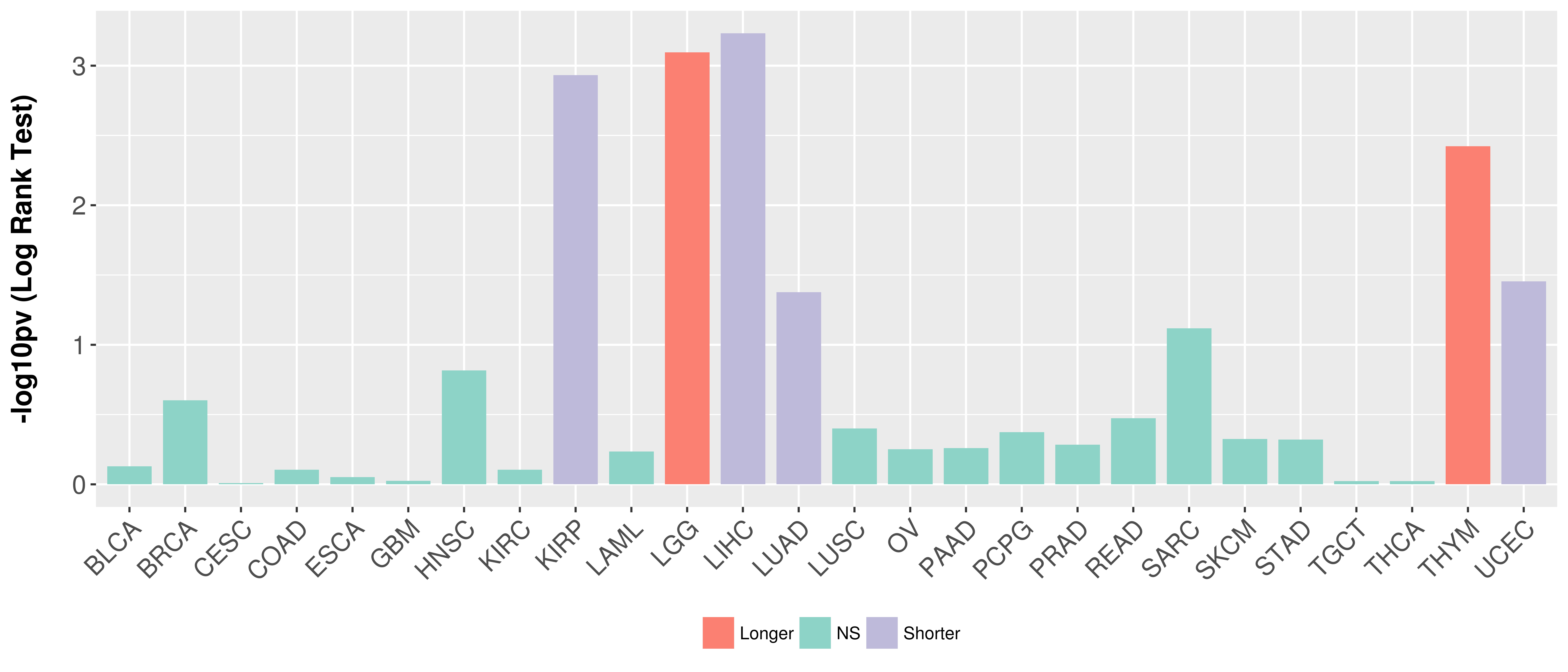
Overall survival analysis based on expression.
|
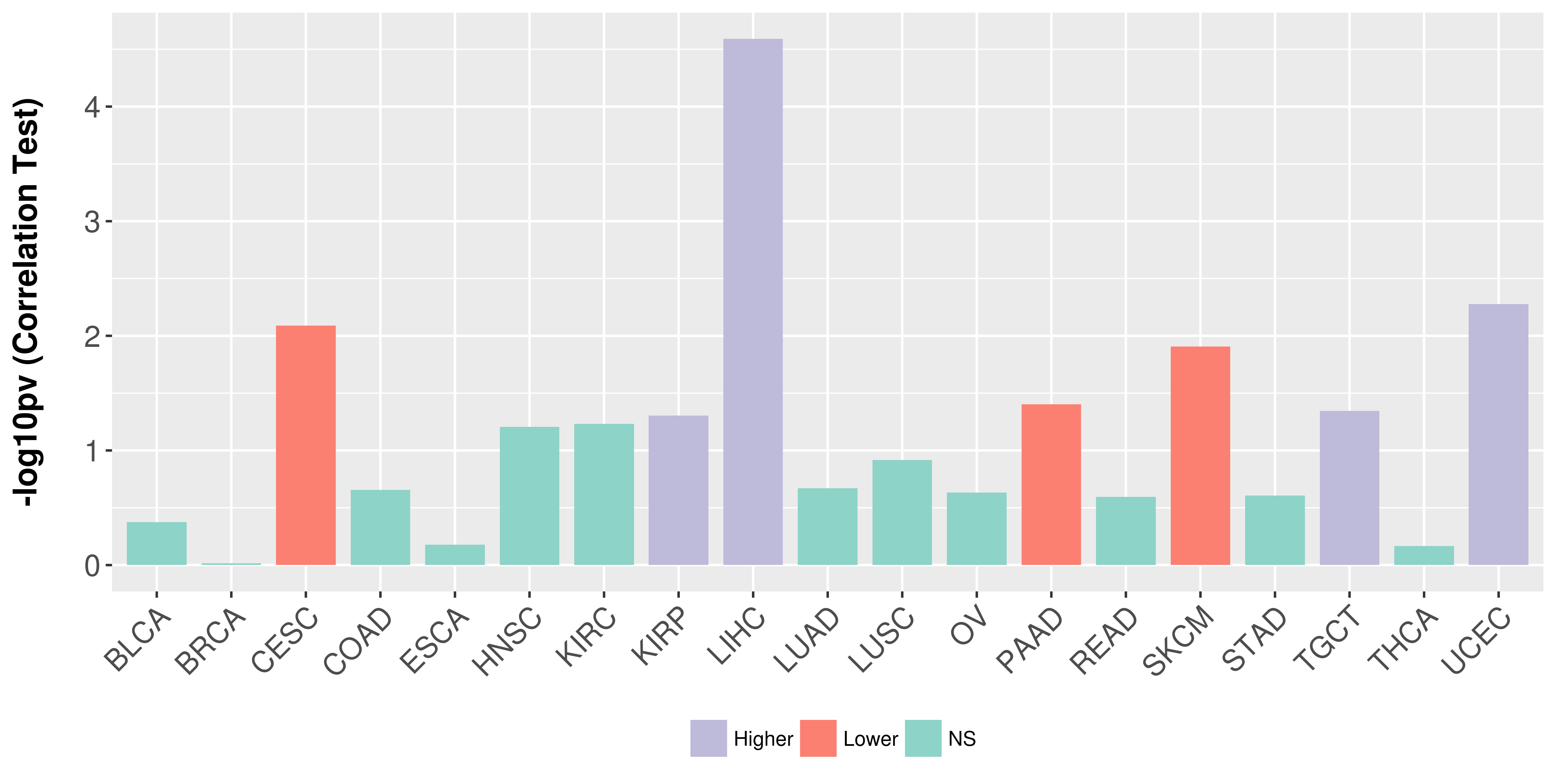
Association between expresson and stage.
|
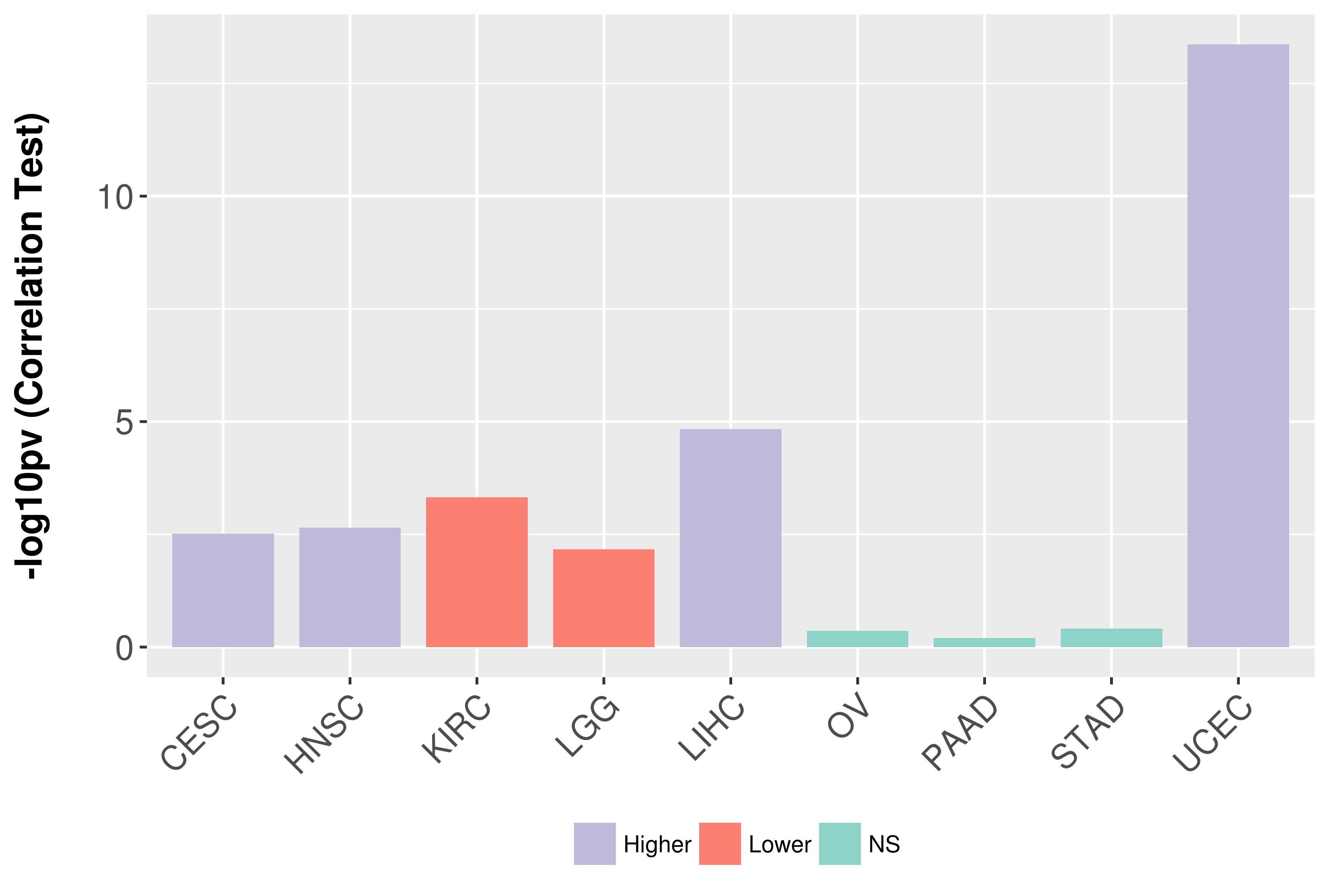
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | CBX1 |
| Name | chromobox 1 |
| Aliases | HP1Hs-beta; M31; MOD1; CBX; HP1-BETA; HP1 beta homolog (Drosophila ); chromobox homolog 1 (Drosophila HP1 be ...... |
| Location | 17q21.32 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | CBX1 |
| Name | chromobox 1 |
| Aliases | HP1Hs-beta; M31; MOD1; CBX; HP1-BETA; HP1 beta homolog (Drosophila ); chromobox homolog 1 (Drosophila HP1 be ...... |
| Location | 17q21.32 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for CBX1. |
| Summary | |
|---|---|
| Symbol | CBX1 |
| Name | chromobox 1 |
| Aliases | HP1Hs-beta; M31; MOD1; CBX; HP1-BETA; HP1 beta homolog (Drosophila ); chromobox homolog 1 (Drosophila HP1 be ...... |
| Location | 17q21.32 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for CBX1. |