Browse CHD7 in pancancer
| Summary | |
|---|---|
| Symbol | CHD7 |
| Name | chromodomain helicase DNA binding protein 7 |
| Aliases | KIAA1416; FLJ20357; FLJ20361; CRG; CHARGE association; HH5; IS3; KAL5; ATP-dependent helicase CHD7; chromodo ...... |
| Location | 8q12.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF07533 BRK domain PF00385 Chromo (CHRromatin Organisation MOdifier) domain PF00271 Helicase conserved C-terminal domain PF00176 SNF2 family N-terminal domain |
||||||||||
| Function |
Probable transcription regulator. Maybe involved in the in 45S precursor rRNA production. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0001501 skeletal system development GO:0001654 eye development GO:0001701 in utero embryonic development GO:0001974 blood vessel remodeling GO:0002521 leukocyte differentiation GO:0002790 peptide secretion GO:0002791 regulation of peptide secretion GO:0003007 heart morphogenesis GO:0003013 circulatory system process GO:0003205 cardiac chamber development GO:0003206 cardiac chamber morphogenesis GO:0003208 cardiac ventricle morphogenesis GO:0003215 cardiac right ventricle morphogenesis GO:0003221 right ventricular cardiac muscle tissue morphogenesis GO:0003222 ventricular trabecula myocardium morphogenesis GO:0003223 ventricular compact myocardium morphogenesis GO:0003226 right ventricular compact myocardium morphogenesis GO:0003229 ventricular cardiac muscle tissue development GO:0003231 cardiac ventricle development GO:0003279 cardiac septum development GO:0006338 chromatin remodeling GO:0006364 rRNA processing GO:0006816 calcium ion transport GO:0006874 cellular calcium ion homeostasis GO:0006875 cellular metal ion homeostasis GO:0007159 leukocyte cell-cell adhesion GO:0007204 positive regulation of cytosolic calcium ion concentration GO:0007423 sensory organ development GO:0007507 heart development GO:0007512 adult heart development GO:0007517 muscle organ development GO:0007548 sex differentiation GO:0007605 sensory perception of sound GO:0007626 locomotory behavior GO:0007628 adult walking behavior GO:0007635 chemosensory behavior GO:0008015 blood circulation GO:0008344 adult locomotory behavior GO:0009914 hormone transport GO:0010522 regulation of calcium ion transport into cytosol GO:0010817 regulation of hormone levels GO:0010880 regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum GO:0010959 regulation of metal ion transport GO:0014706 striated muscle tissue development GO:0014808 release of sequestered calcium ion into cytosol by sarcoplasmic reticulum GO:0015833 peptide transport GO:0016072 rRNA metabolic process GO:0021537 telencephalon development GO:0021545 cranial nerve development GO:0021553 olfactory nerve development GO:0021675 nerve development GO:0021772 olfactory bulb development GO:0021988 olfactory lobe development GO:0022613 ribonucleoprotein complex biogenesis GO:0023061 signal release GO:0030072 peptide hormone secretion GO:0030098 lymphocyte differentiation GO:0030217 T cell differentiation GO:0030252 growth hormone secretion GO:0030326 embryonic limb morphogenesis GO:0030534 adult behavior GO:0030540 female genitalia development GO:0030900 forebrain development GO:0032844 regulation of homeostatic process GO:0032845 negative regulation of homeostatic process GO:0034470 ncRNA processing GO:0034762 regulation of transmembrane transport GO:0034765 regulation of ion transmembrane transport GO:0035107 appendage morphogenesis GO:0035108 limb morphogenesis GO:0035113 embryonic appendage morphogenesis GO:0035116 embryonic hindlimb morphogenesis GO:0035137 hindlimb morphogenesis GO:0035239 tube morphogenesis GO:0035264 multicellular organism growth GO:0035904 aorta development GO:0035909 aorta morphogenesis GO:0036302 atrioventricular canal development GO:0040014 regulation of multicellular organism growth GO:0040018 positive regulation of multicellular organism growth GO:0042048 olfactory behavior GO:0042110 T cell activation GO:0042254 ribosome biogenesis GO:0042471 ear morphogenesis GO:0042472 inner ear morphogenesis GO:0042886 amide transport GO:0043010 camera-type eye development GO:0043583 ear development GO:0043584 nose development GO:0044708 single-organism behavior GO:0045927 positive regulation of growth GO:0046660 female sex differentiation GO:0046879 hormone secretion GO:0046883 regulation of hormone secretion GO:0048514 blood vessel morphogenesis GO:0048562 embryonic organ morphogenesis GO:0048568 embryonic organ development GO:0048608 reproductive structure development GO:0048638 regulation of developmental growth GO:0048639 positive regulation of developmental growth GO:0048644 muscle organ morphogenesis GO:0048736 appendage development GO:0048738 cardiac muscle tissue development GO:0048752 semicircular canal morphogenesis GO:0048771 tissue remodeling GO:0048806 genitalia development GO:0048839 inner ear development GO:0048844 artery morphogenesis GO:0050890 cognition GO:0050954 sensory perception of mechanical stimulus GO:0051208 sequestering of calcium ion GO:0051209 release of sequestered calcium ion into cytosol GO:0051235 maintenance of location GO:0051238 sequestering of metal ion GO:0051279 regulation of release of sequestered calcium ion into cytosol GO:0051282 regulation of sequestering of calcium ion GO:0051283 negative regulation of sequestering of calcium ion GO:0051480 regulation of cytosolic calcium ion concentration GO:0051924 regulation of calcium ion transport GO:0055008 cardiac muscle tissue morphogenesis GO:0055010 ventricular cardiac muscle tissue morphogenesis GO:0055074 calcium ion homeostasis GO:0060021 palate development GO:0060041 retina development in camera-type eye GO:0060123 regulation of growth hormone secretion GO:0060173 limb development GO:0060324 face development GO:0060384 innervation GO:0060401 cytosolic calcium ion transport GO:0060402 calcium ion transport into cytosol GO:0060411 cardiac septum morphogenesis GO:0060415 muscle tissue morphogenesis GO:0060537 muscle tissue development GO:0060541 respiratory system development GO:0060840 artery development GO:0060872 semicircular canal development GO:0061383 trabecula morphogenesis GO:0061384 heart trabecula morphogenesis GO:0061458 reproductive system development GO:0070296 sarcoplasmic reticulum calcium ion transport GO:0070486 leukocyte aggregation GO:0070489 T cell aggregation GO:0070509 calcium ion import GO:0070588 calcium ion transmembrane transport GO:0070838 divalent metal ion transport GO:0071593 lymphocyte aggregation GO:0072503 cellular divalent inorganic cation homeostasis GO:0072507 divalent inorganic cation homeostasis GO:0072511 divalent inorganic cation transport GO:0090087 regulation of peptide transport GO:0090276 regulation of peptide hormone secretion GO:0090279 regulation of calcium ion import GO:0090596 sensory organ morphogenesis GO:0090659 walking behavior GO:0097553 calcium ion transmembrane import into cytosol GO:1902656 calcium ion import into cytosol GO:1903169 regulation of calcium ion transmembrane transport GO:1903514 calcium ion transport from endoplasmic reticulum to cytosol GO:1904062 regulation of cation transmembrane transport GO:2000021 regulation of ion homeostasis |
| Molecular Function |
GO:0000978 RNA polymerase II core promoter proximal region sequence-specific DNA binding GO:0000987 core promoter proximal region sequence-specific DNA binding GO:0001159 core promoter proximal region DNA binding GO:0003682 chromatin binding GO:0004386 helicase activity GO:1990841 promoter-specific chromatin binding |
| Cellular Component | - |
| KEGG | - |
| Reactome | - |
| Summary | |
|---|---|
| Symbol | CHD7 |
| Name | chromodomain helicase DNA binding protein 7 |
| Aliases | KIAA1416; FLJ20357; FLJ20361; CRG; CHARGE association; HH5; IS3; KAL5; ATP-dependent helicase CHD7; chromodo ...... |
| Location | 8q12.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
|
| Summary | |
|---|---|
| Symbol | CHD7 |
| Name | chromodomain helicase DNA binding protein 7 |
| Aliases | KIAA1416; FLJ20357; FLJ20361; CRG; CHARGE association; HH5; IS3; KAL5; ATP-dependent helicase CHD7; chromodo ...... |
| Location | 8q12.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | CHD7 |
| Name | chromodomain helicase DNA binding protein 7 |
| Aliases | KIAA1416; FLJ20357; FLJ20361; CRG; CHARGE association; HH5; IS3; KAL5; ATP-dependent helicase CHD7; chromodo ...... |
| Location | 8q12.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |

Differential expression analysis for cancers with more than 10 normal samples
|
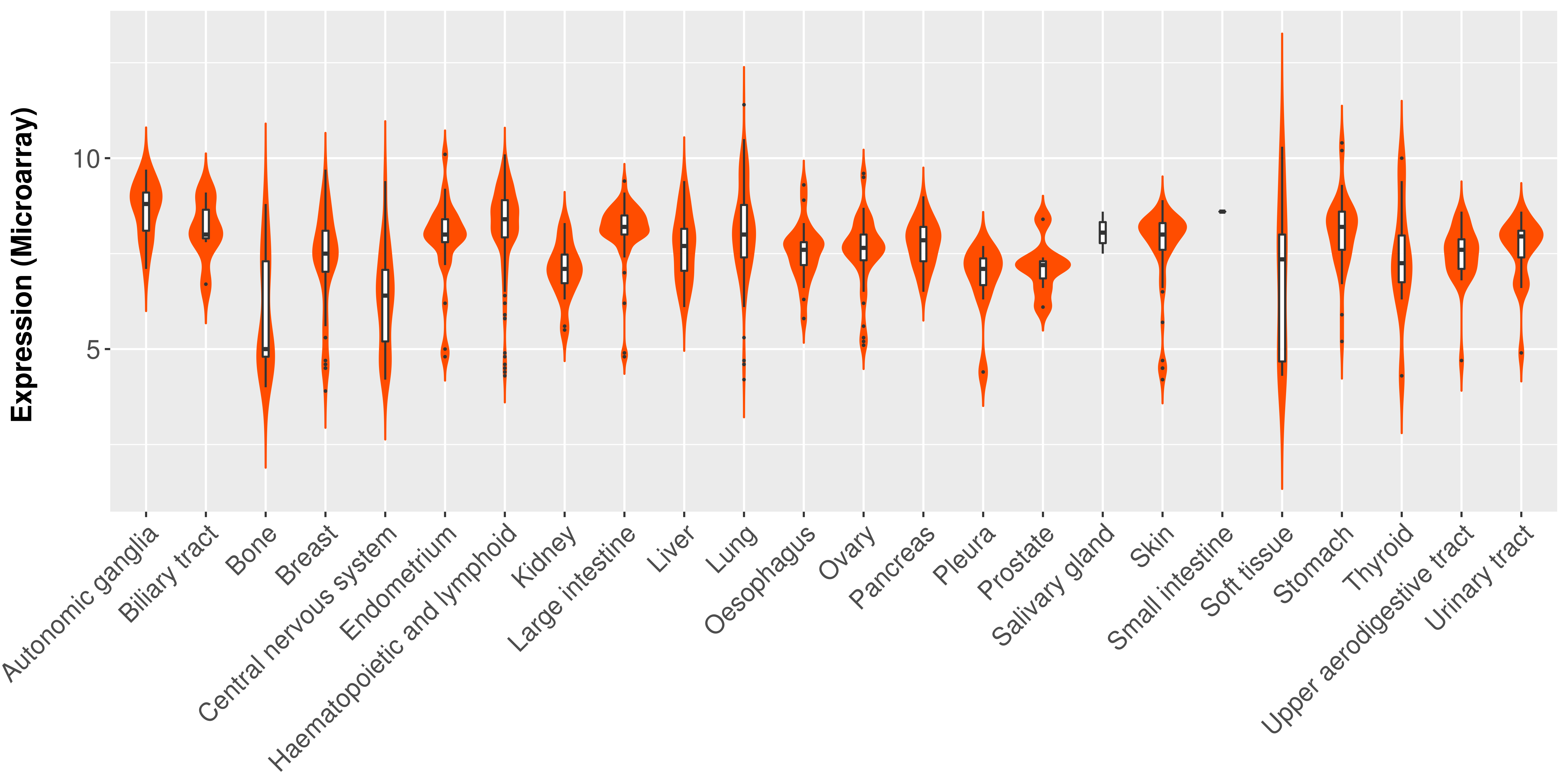
|
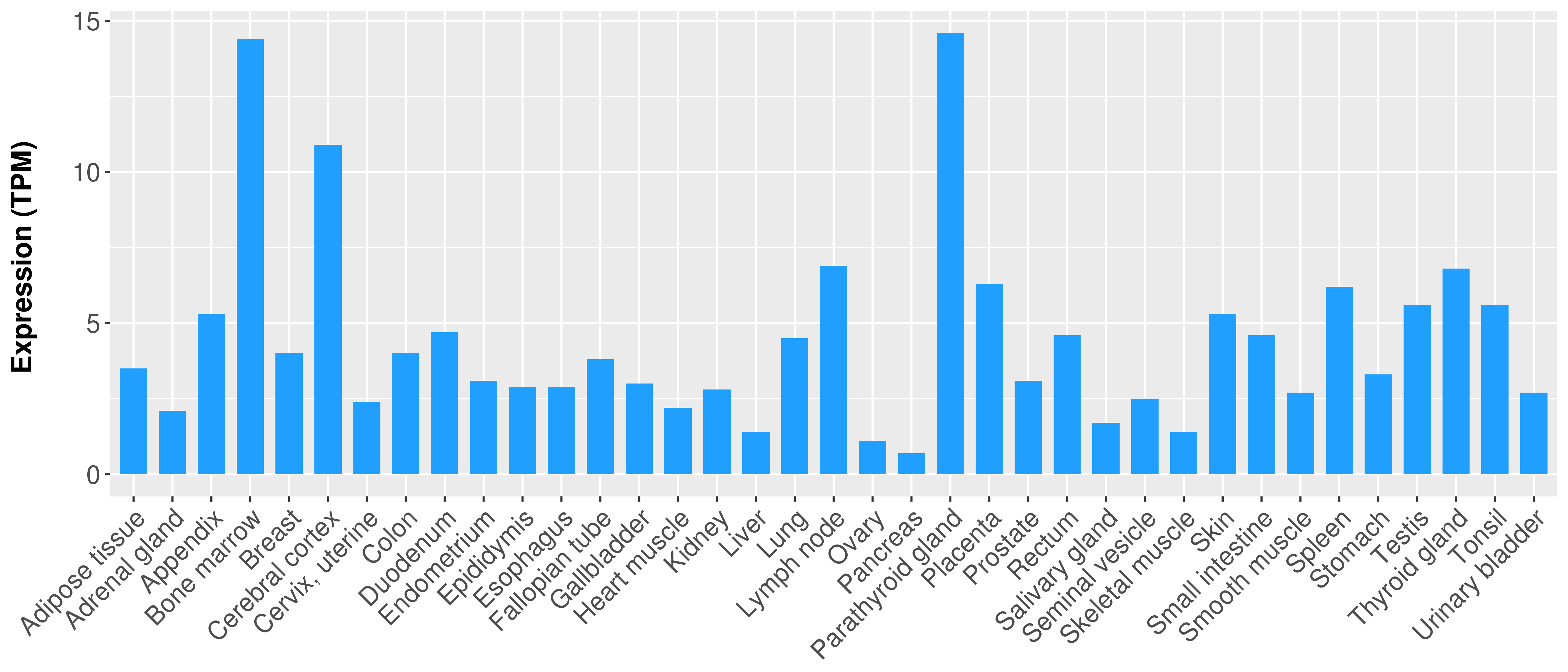
|
|
| Summary | |
|---|---|
| Symbol | CHD7 |
| Name | chromodomain helicase DNA binding protein 7 |
| Aliases | KIAA1416; FLJ20357; FLJ20361; CRG; CHARGE association; HH5; IS3; KAL5; ATP-dependent helicase CHD7; chromodo ...... |
| Location | 8q12.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
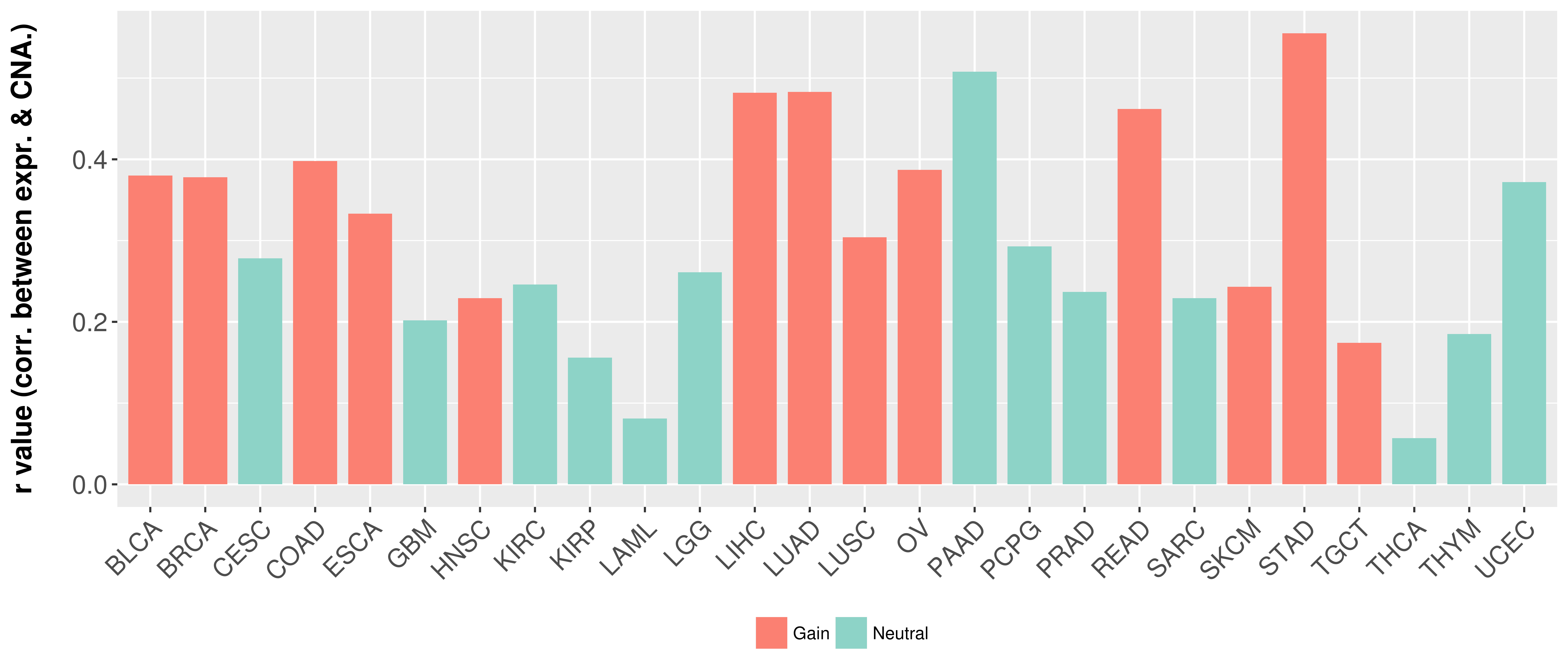
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | CHD7 |
| Name | chromodomain helicase DNA binding protein 7 |
| Aliases | KIAA1416; FLJ20357; FLJ20361; CRG; CHARGE association; HH5; IS3; KAL5; ATP-dependent helicase CHD7; chromodo ...... |
| Location | 8q12.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
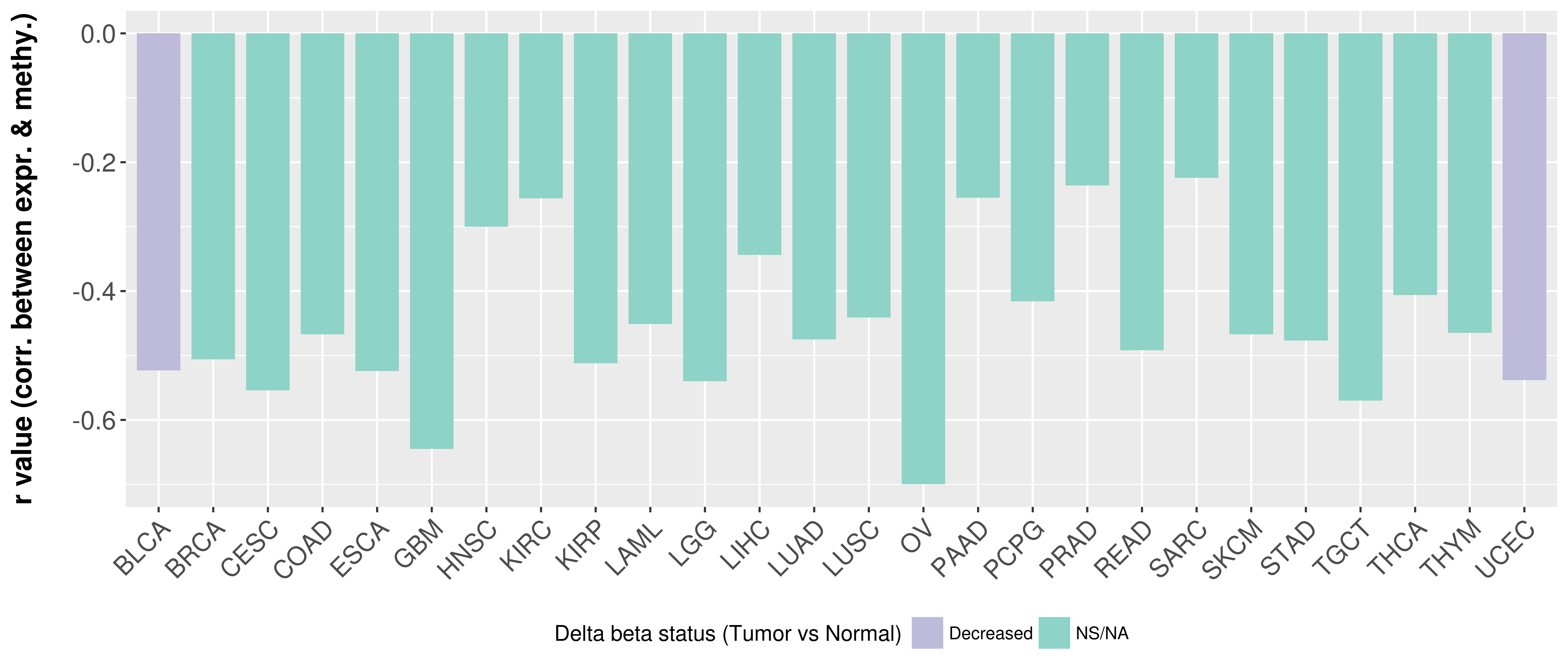
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | CHD7 |
| Name | chromodomain helicase DNA binding protein 7 |
| Aliases | KIAA1416; FLJ20357; FLJ20361; CRG; CHARGE association; HH5; IS3; KAL5; ATP-dependent helicase CHD7; chromodo ...... |
| Location | 8q12.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
| There is no antibody staining data. |
| Summary | |
|---|---|
| Symbol | CHD7 |
| Name | chromodomain helicase DNA binding protein 7 |
| Aliases | KIAA1416; FLJ20357; FLJ20361; CRG; CHARGE association; HH5; IS3; KAL5; ATP-dependent helicase CHD7; chromodo ...... |
| Location | 8q12.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
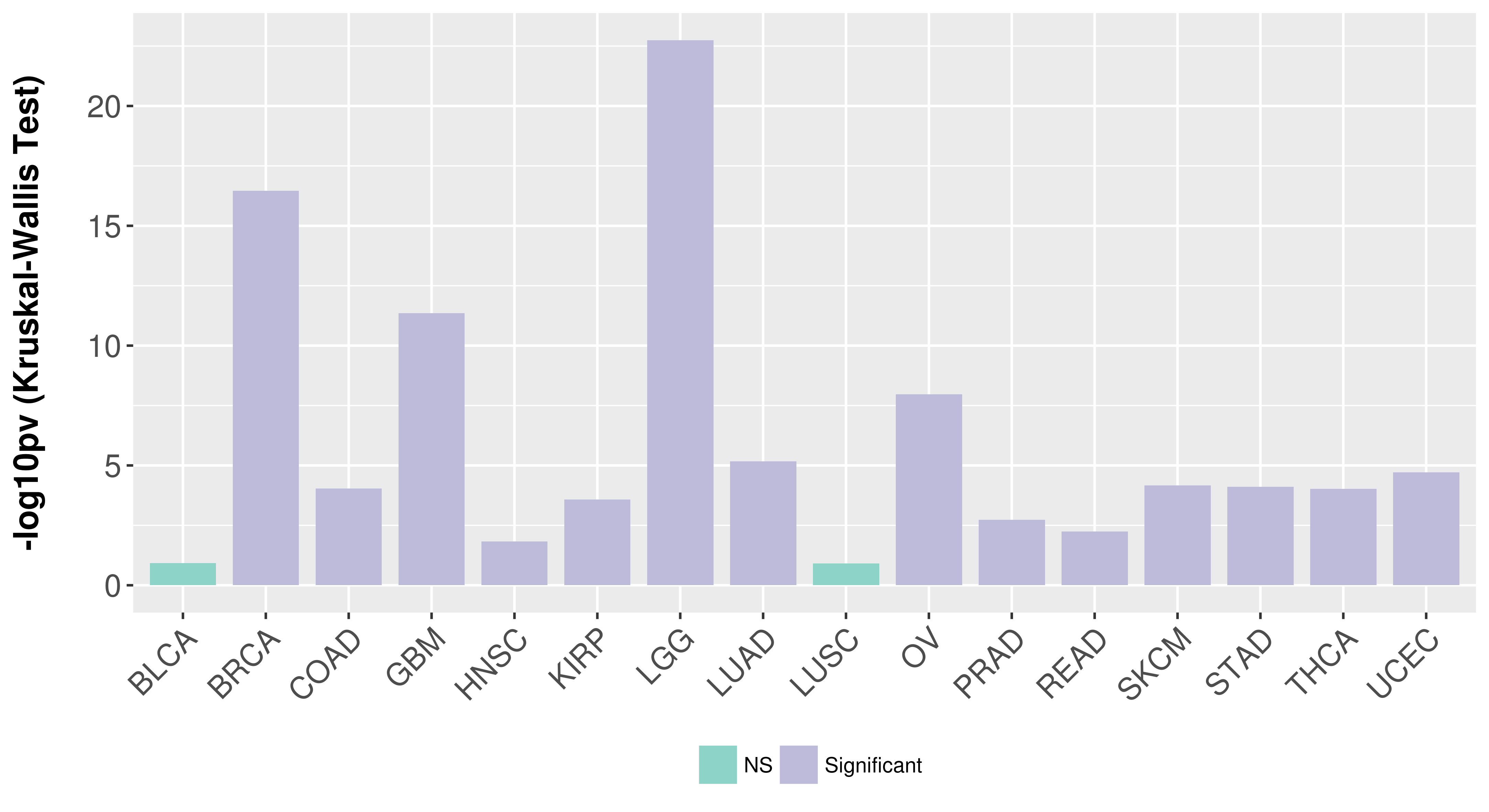
Association between expresson and subtype.
|
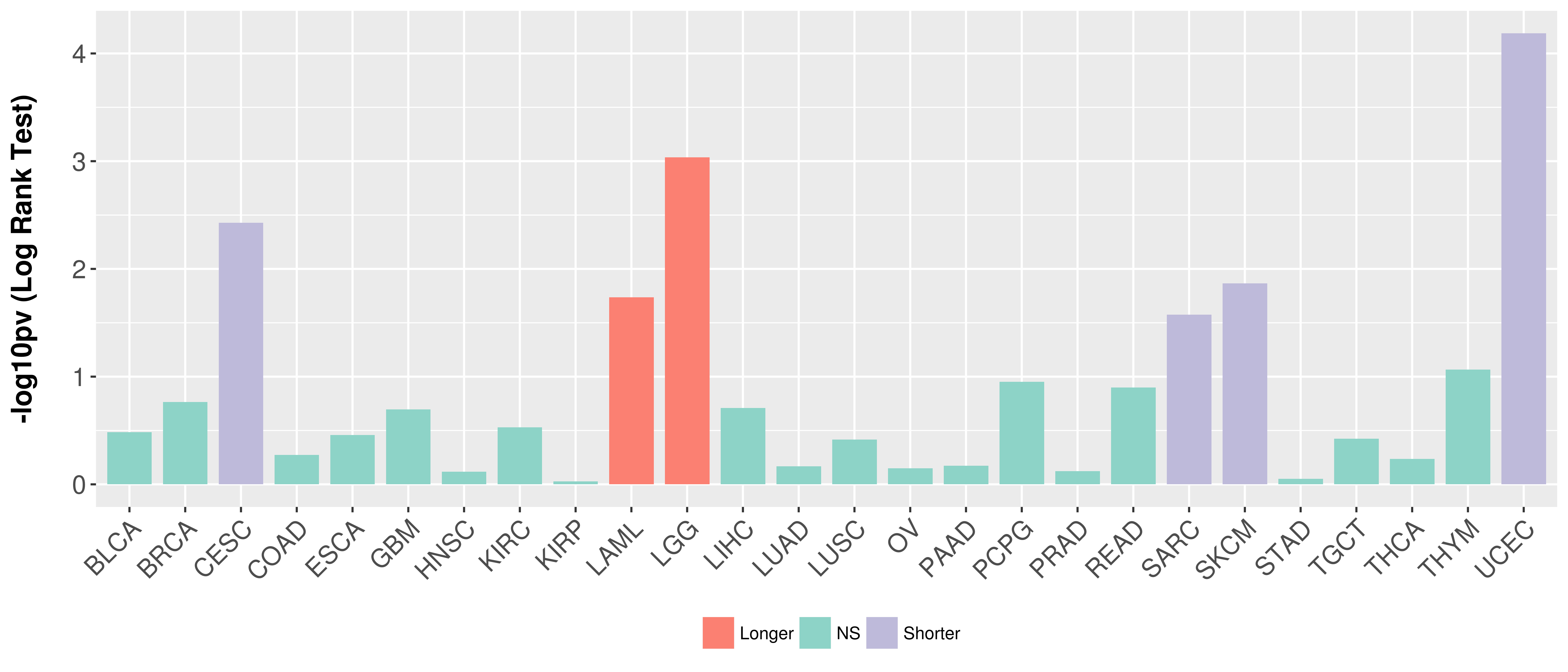
Overall survival analysis based on expression.
|
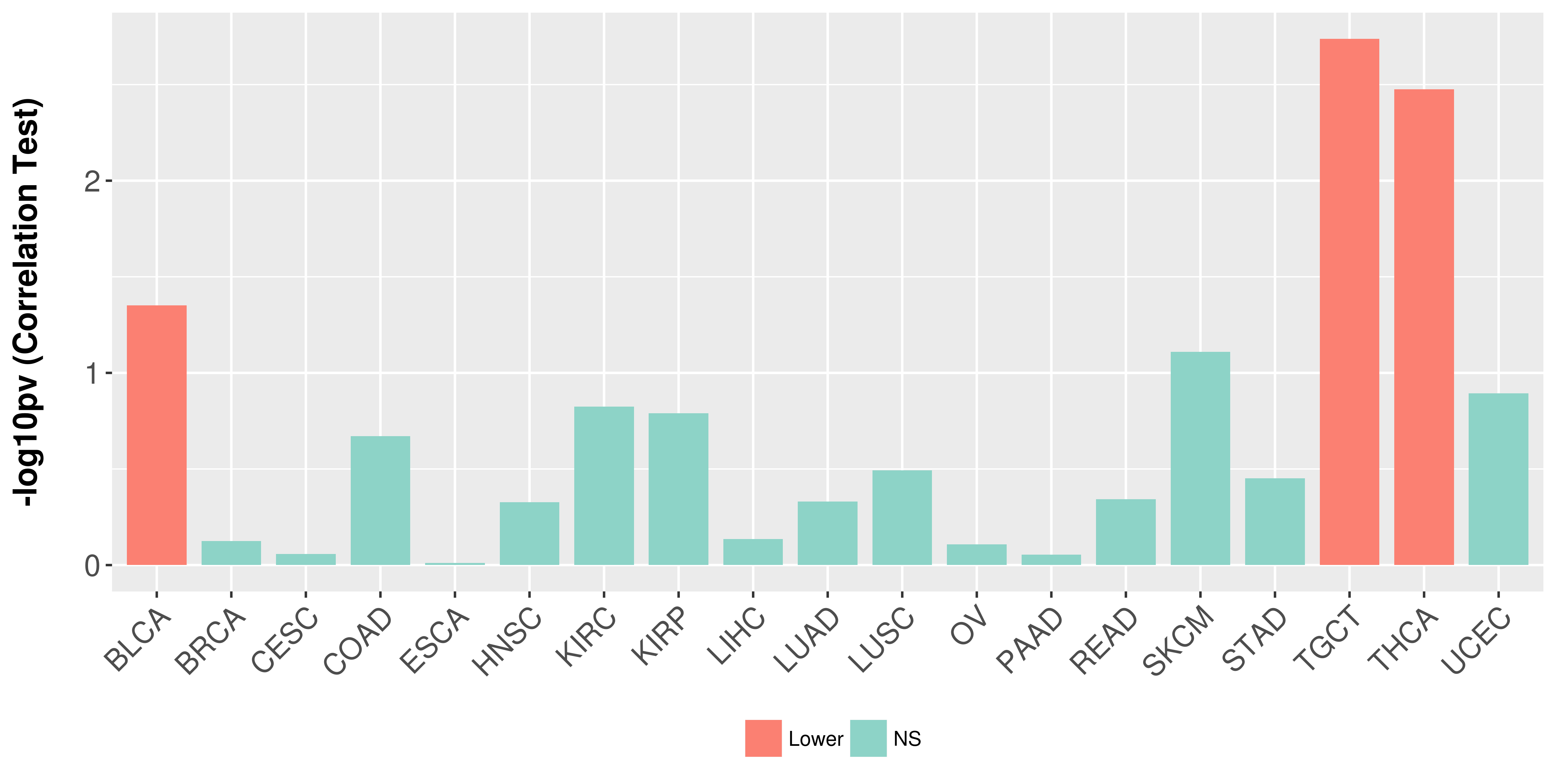
Association between expresson and stage.
|
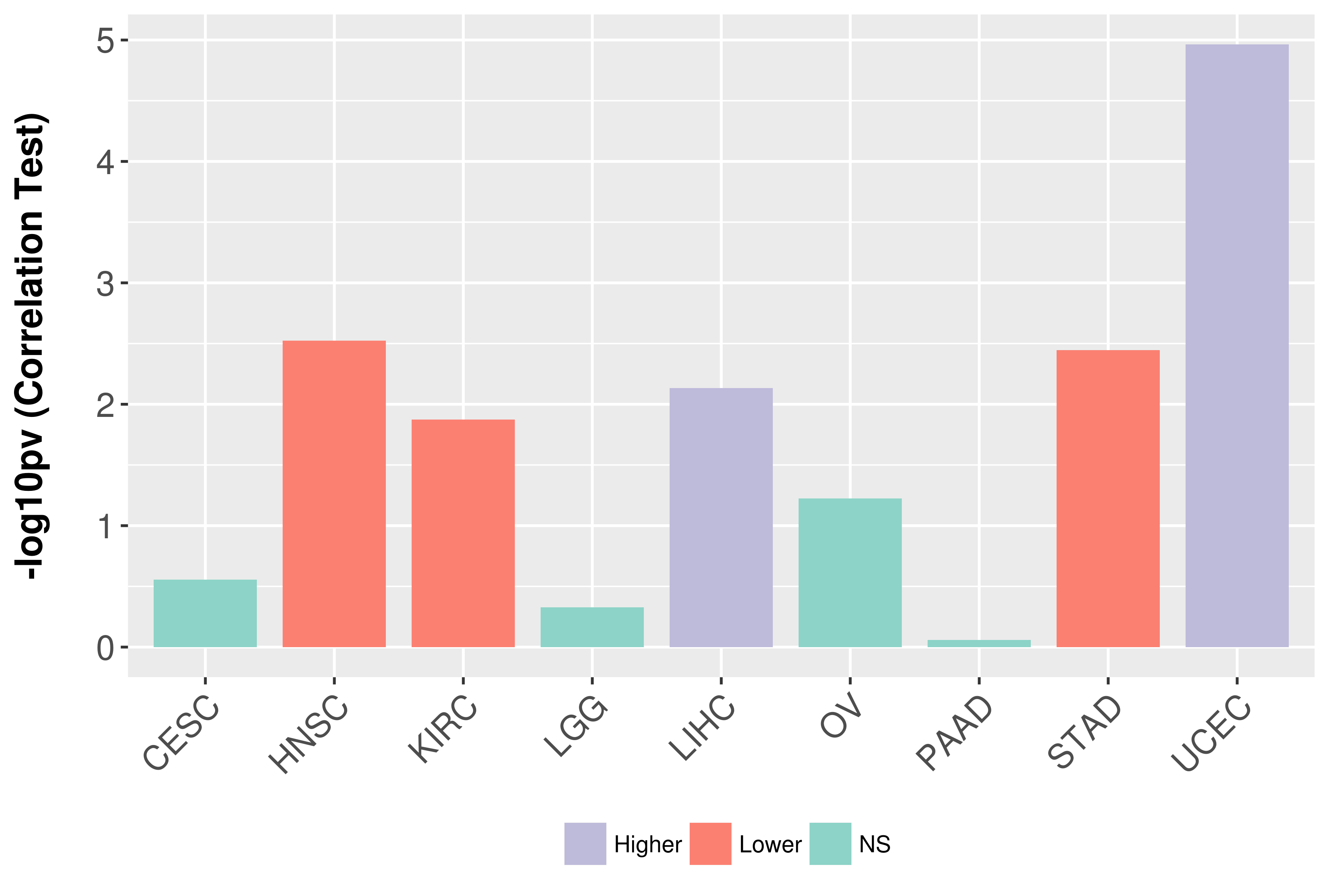
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | CHD7 |
| Name | chromodomain helicase DNA binding protein 7 |
| Aliases | KIAA1416; FLJ20357; FLJ20361; CRG; CHARGE association; HH5; IS3; KAL5; ATP-dependent helicase CHD7; chromodo ...... |
| Location | 8q12.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | CHD7 |
| Name | chromodomain helicase DNA binding protein 7 |
| Aliases | KIAA1416; FLJ20357; FLJ20361; CRG; CHARGE association; HH5; IS3; KAL5; ATP-dependent helicase CHD7; chromodo ...... |
| Location | 8q12.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for CHD7. |
| Summary | |
|---|---|
| Symbol | CHD7 |
| Name | chromodomain helicase DNA binding protein 7 |
| Aliases | KIAA1416; FLJ20357; FLJ20361; CRG; CHARGE association; HH5; IS3; KAL5; ATP-dependent helicase CHD7; chromodo ...... |
| Location | 8q12.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|