Browse CTCF in pancancer
| Summary | |
|---|---|
| Symbol | CTCF |
| Name | CCCTC-binding factor |
| Aliases | 11 zinc finger transcriptional repressor; MRD21; 11-zinc finger protein; CTCFL paralog; Transcriptional repr ...... |
| Location | 16q22.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain | - | ||||||||||
| Function |
Chromatin binding factor that binds to DNA sequence specific sites. Involved in transcriptional regulation by binding to chromatin insulators and preventing interaction between promoter and nearby enhancers and silencers. Acts as transcriptional repressor binding to promoters of vertebrate MYC gene and BAG1 gene. Also binds to the PLK and PIM1 promoters. Acts as a transcriptional activator of APP. Regulates APOA1/C3/A4/A5 gene cluster and controls MHC class II gene expression. Plays an essential role in oocyte and preimplantation embryo development by activating or repressing transcription. Seems to act as tumor suppressor. Plays a critical role in the epigenetic regulation. Participates in the allele-specific gene expression at the imprinted IGF2/H19 gene locus. On the maternal allele, binding within the H19 imprinting control region (ICR) mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to IGF2. Plays a critical role in gene silencing over considerable distances in the genome. Preferentially interacts with unmethylated DNA, preventing spreading of CpG methylation and maintaining methylation-free zones. Inversely, binding to target sites is prevented by CpG methylation. Plays a important role in chromatin remodeling. Can dimerize when it is bound to different DNA sequences, mediating long-range chromatin looping. Mediates interchromosomal association between IGF2/H19 and WSB1/NF1 and may direct distant DNA segments to a common transcription factory. Causes local loss of histone acetylation and gain of histone methylation in the beta-globin locus, without affecting transcription. When bound to chromatin, it provides an anchor point for nucleosomes positioning. Seems to be essential for homologous X-chromosome pairing. May participate with Tsix in establishing a regulatable epigenetic switch for X chromosome inactivation. May play a role in preventing the propagation of stable methylation at the escape genes from X- inactivation. Involved in sister chromatid cohesion. Associates with both centromeres and chromosomal arms during metaphase and required for cohesin localization to CTCF sites. Regulates asynchronous replication of IGF2/H19. Plays a role in the recruitment of CENPE to the pericentromeric/centromeric regions of the chromosome during mitosis (PubMed:26321640). |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000819 sister chromatid segregation GO:0006304 DNA modification GO:0006305 DNA alkylation GO:0006306 DNA methylation GO:0006323 DNA packaging GO:0006333 chromatin assembly or disassembly GO:0006349 regulation of gene expression by genetic imprinting GO:0006473 protein acetylation GO:0006475 internal protein amino acid acetylation GO:0006479 protein methylation GO:0007059 chromosome segregation GO:0007062 sister chromatid cohesion GO:0007063 regulation of sister chromatid cohesion GO:0008213 protein alkylation GO:0010216 maintenance of DNA methylation GO:0016570 histone modification GO:0016571 histone methylation GO:0016573 histone acetylation GO:0016584 nucleosome positioning GO:0018205 peptidyl-lysine modification GO:0018393 internal peptidyl-lysine acetylation GO:0018394 peptidyl-lysine acetylation GO:0031056 regulation of histone modification GO:0031060 regulation of histone methylation GO:0031497 chromatin assembly GO:0032259 methylation GO:0033044 regulation of chromosome organization GO:0033045 regulation of sister chromatid segregation GO:0034502 protein localization to chromosome GO:0034728 nucleosome organization GO:0035065 regulation of histone acetylation GO:0040029 regulation of gene expression, epigenetic GO:0040030 regulation of molecular function, epigenetic GO:0043414 macromolecule methylation GO:0043543 protein acylation GO:0044728 DNA methylation or demethylation GO:0051983 regulation of chromosome segregation GO:0070601 centromeric sister chromatid cohesion GO:0070602 regulation of centromeric sister chromatid cohesion GO:0071103 DNA conformation change GO:0071459 protein localization to chromosome, centromeric region GO:0071514 genetic imprinting GO:0071824 protein-DNA complex subunit organization GO:0098813 nuclear chromosome segregation GO:1901983 regulation of protein acetylation GO:1902275 regulation of chromatin organization GO:2000756 regulation of peptidyl-lysine acetylation |
| Molecular Function |
GO:0000978 RNA polymerase II core promoter proximal region sequence-specific DNA binding GO:0000982 transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding GO:0000987 core promoter proximal region sequence-specific DNA binding GO:0001078 transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding GO:0001159 core promoter proximal region DNA binding GO:0001227 transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding GO:0001228 transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding GO:0003682 chromatin binding GO:0003714 transcription corepressor activity GO:0031490 chromatin DNA binding GO:0043035 chromatin insulator sequence binding GO:0043566 structure-specific DNA binding |
| Cellular Component |
GO:0000775 chromosome, centromeric region GO:0000793 condensed chromosome GO:0098687 chromosomal region |
| KEGG | - |
| Reactome |
R-HSA-5619507: Activation of HOX genes during differentiation R-HSA-5617472: Activation of anterior HOX genes in hindbrain development during early embryogenesis R-HSA-1266738: Developmental Biology |
| Summary | |
|---|---|
| Symbol | CTCF |
| Name | CCCTC-binding factor |
| Aliases | 11 zinc finger transcriptional repressor; MRD21; 11-zinc finger protein; CTCFL paralog; Transcriptional repr ...... |
| Location | 16q22.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
|
| Summary | |
|---|---|
| Symbol | CTCF |
| Name | CCCTC-binding factor |
| Aliases | 11 zinc finger transcriptional repressor; MRD21; 11-zinc finger protein; CTCFL paralog; Transcriptional repr ...... |
| Location | 16q22.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | CTCF |
| Name | CCCTC-binding factor |
| Aliases | 11 zinc finger transcriptional repressor; MRD21; 11-zinc finger protein; CTCFL paralog; Transcriptional repr ...... |
| Location | 16q22.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
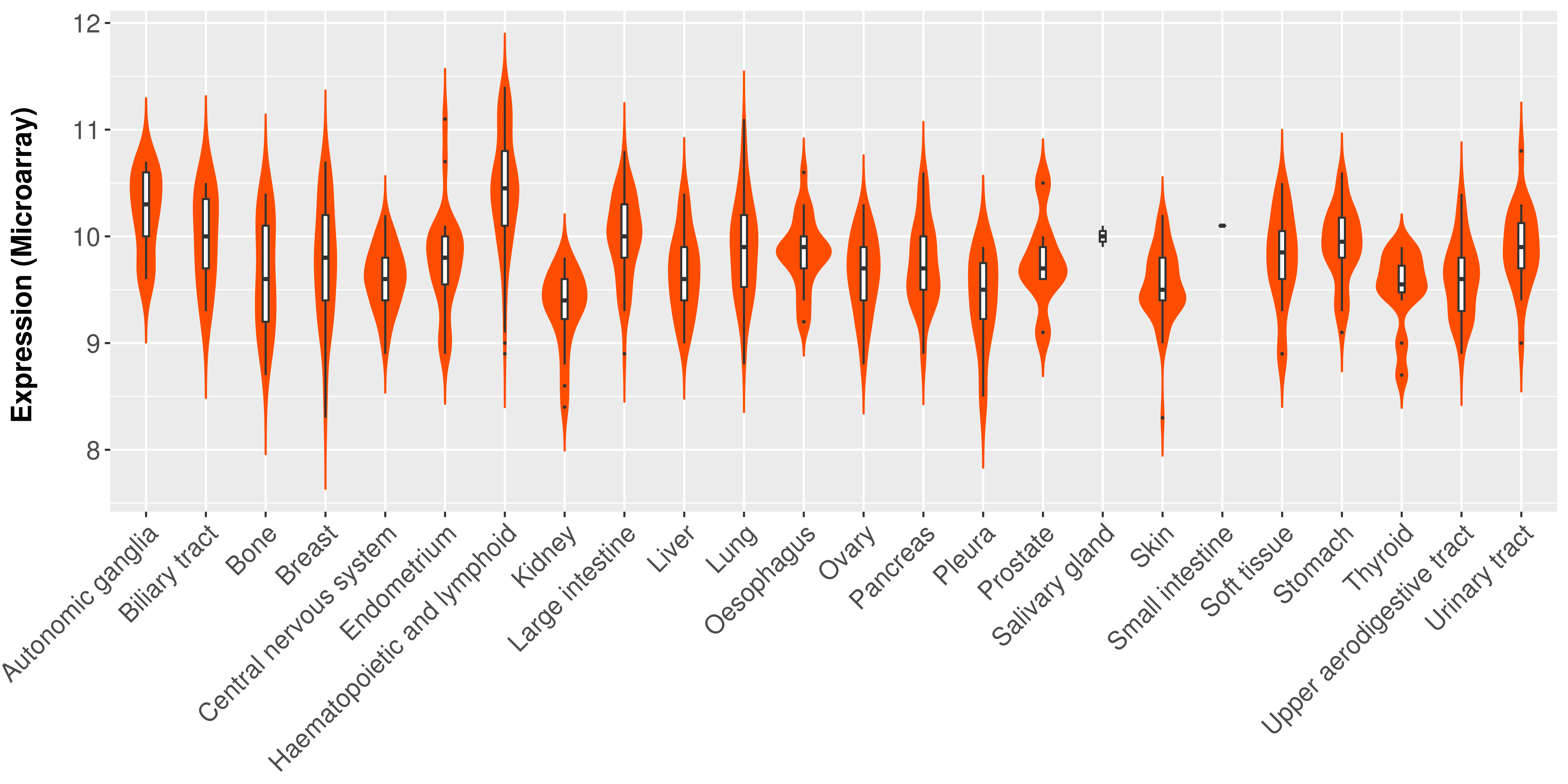
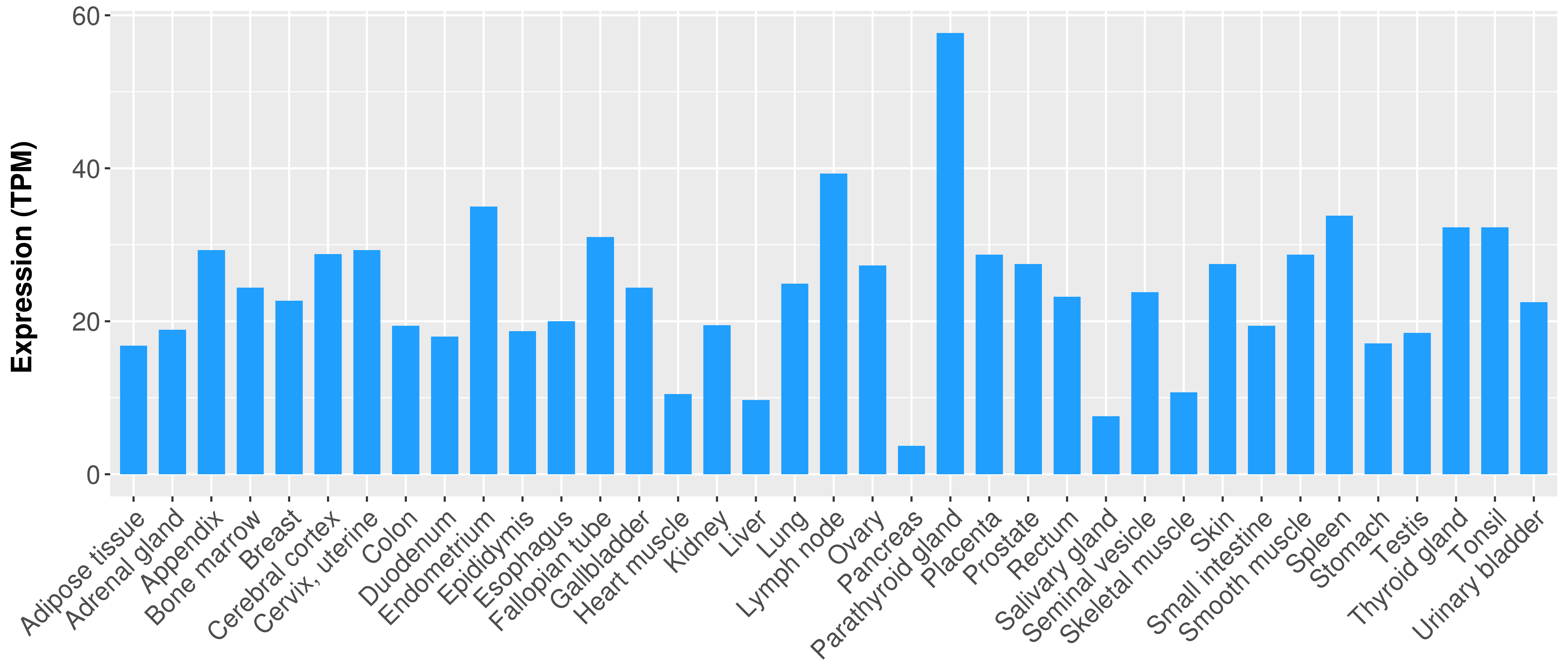
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
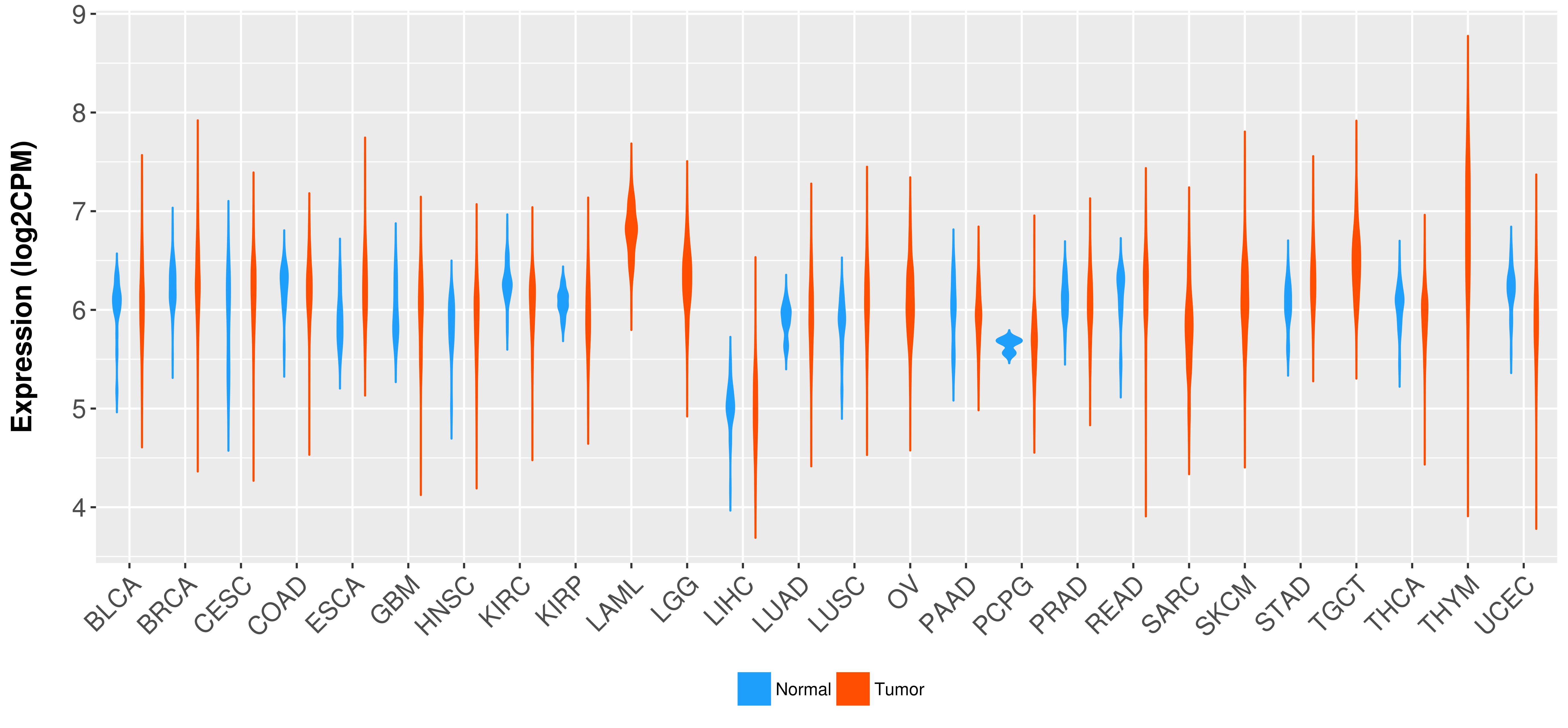
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | CTCF |
| Name | CCCTC-binding factor |
| Aliases | 11 zinc finger transcriptional repressor; MRD21; 11-zinc finger protein; CTCFL paralog; Transcriptional repr ...... |
| Location | 16q22.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
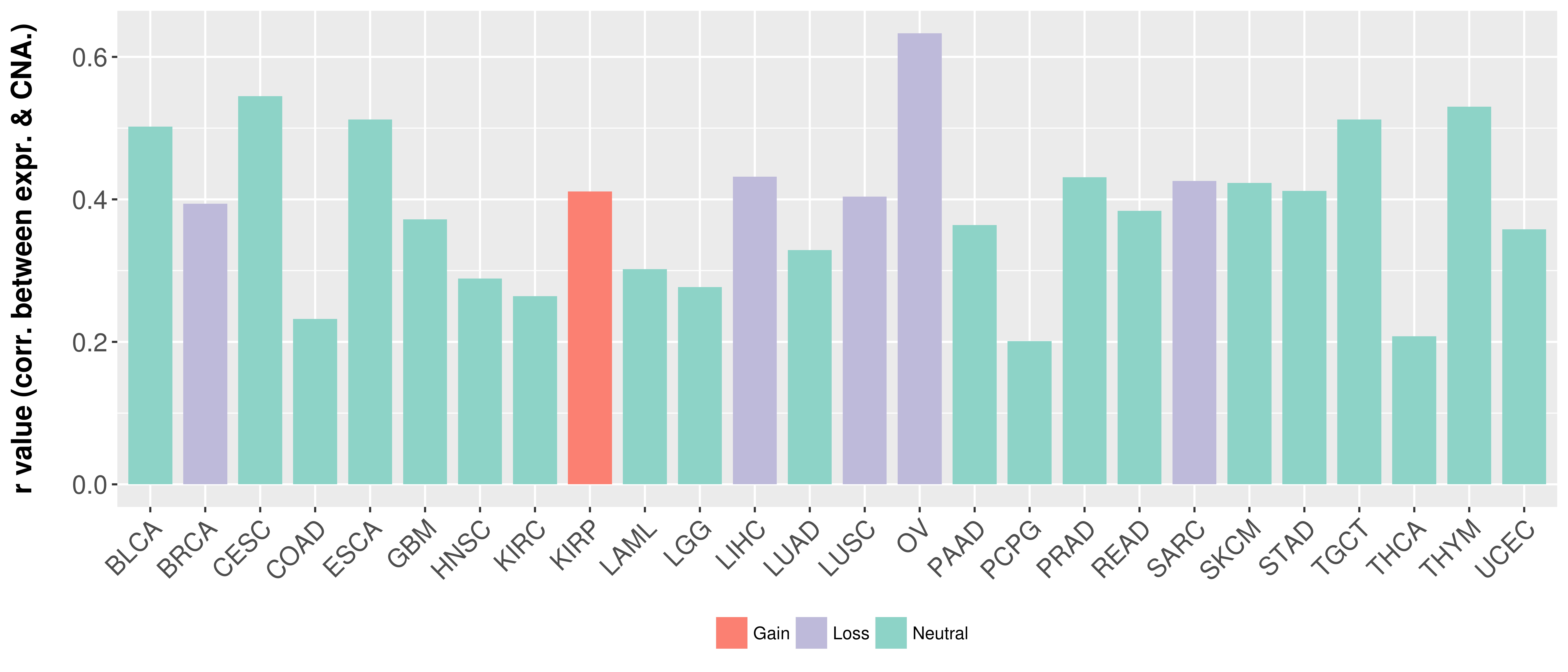
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | CTCF |
| Name | CCCTC-binding factor |
| Aliases | 11 zinc finger transcriptional repressor; MRD21; 11-zinc finger protein; CTCFL paralog; Transcriptional repr ...... |
| Location | 16q22.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
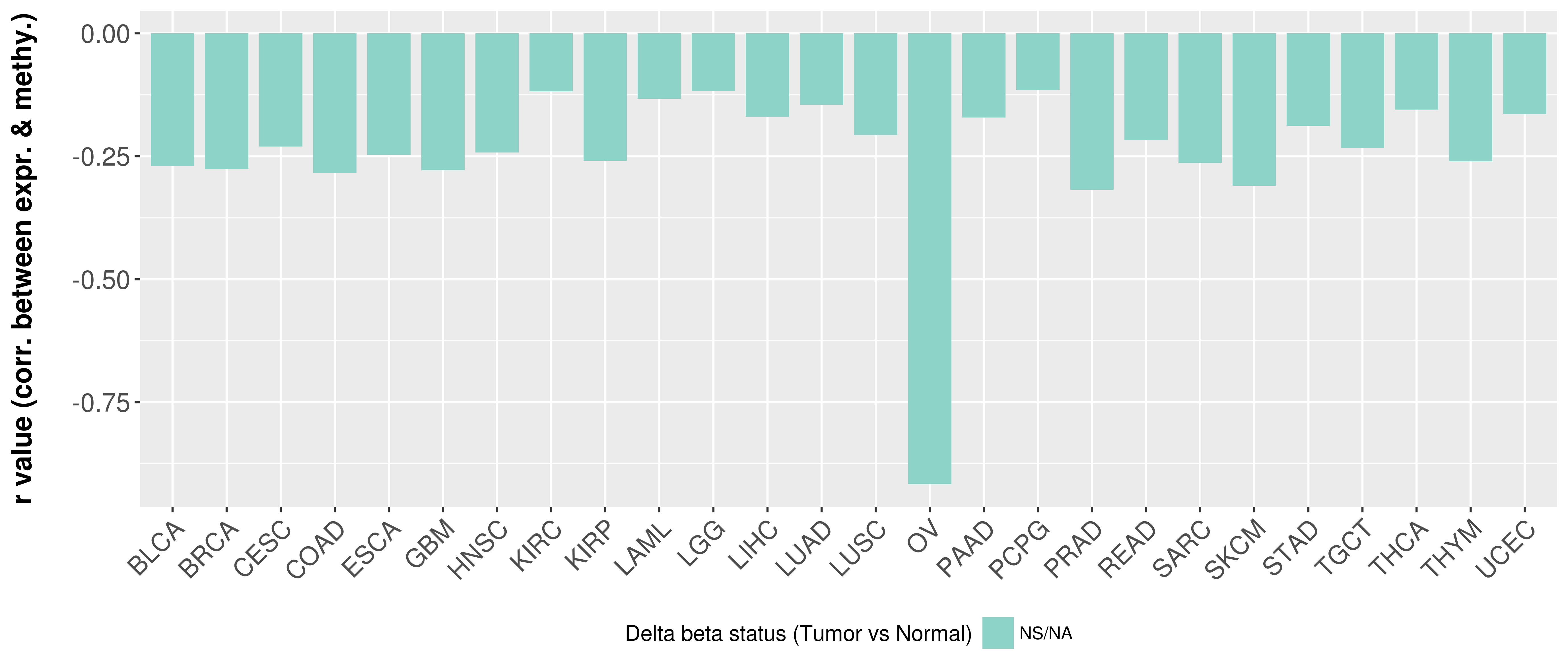
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | CTCF |
| Name | CCCTC-binding factor |
| Aliases | 11 zinc finger transcriptional repressor; MRD21; 11-zinc finger protein; CTCFL paralog; Transcriptional repr ...... |
| Location | 16q22.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | CTCF |
| Name | CCCTC-binding factor |
| Aliases | 11 zinc finger transcriptional repressor; MRD21; 11-zinc finger protein; CTCFL paralog; Transcriptional repr ...... |
| Location | 16q22.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
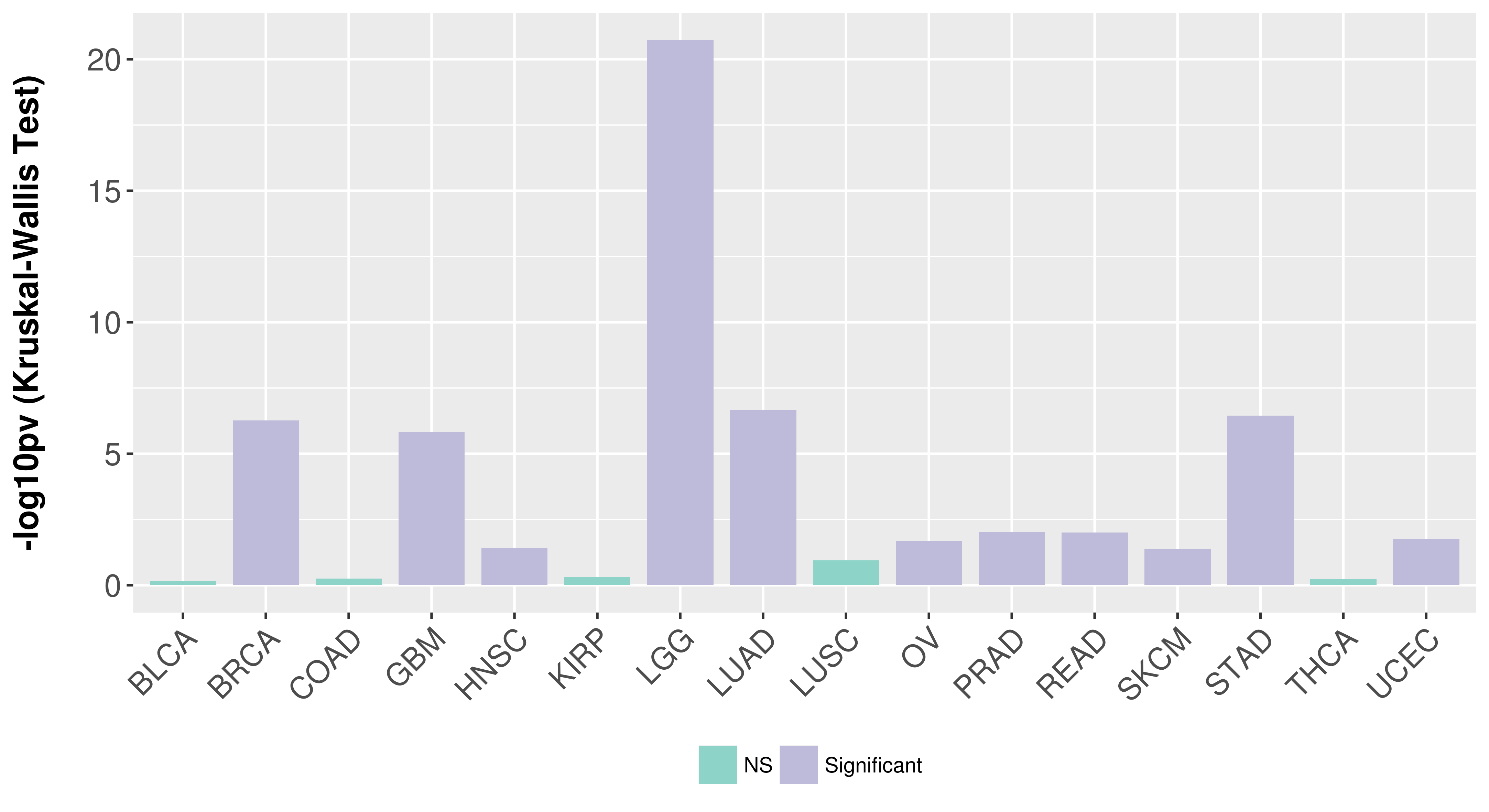
Association between expresson and subtype.
|
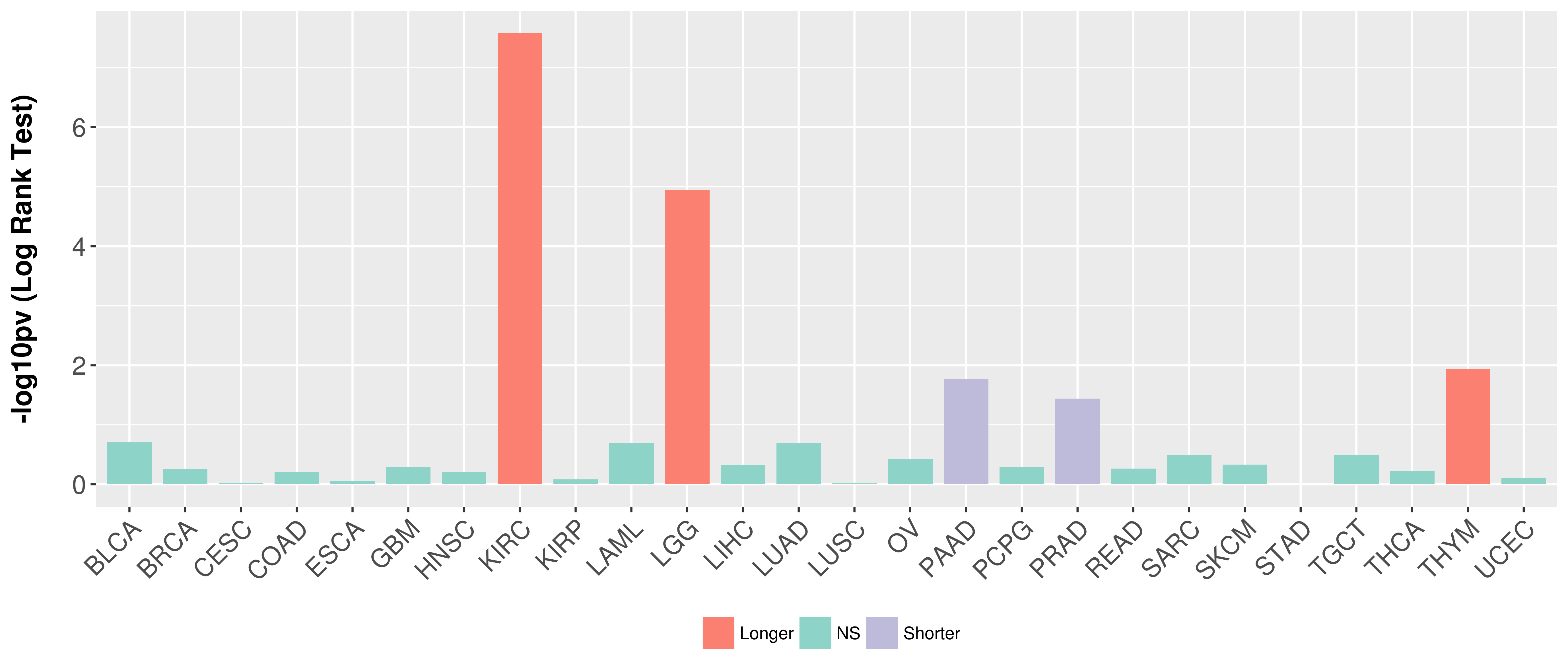
Overall survival analysis based on expression.
|
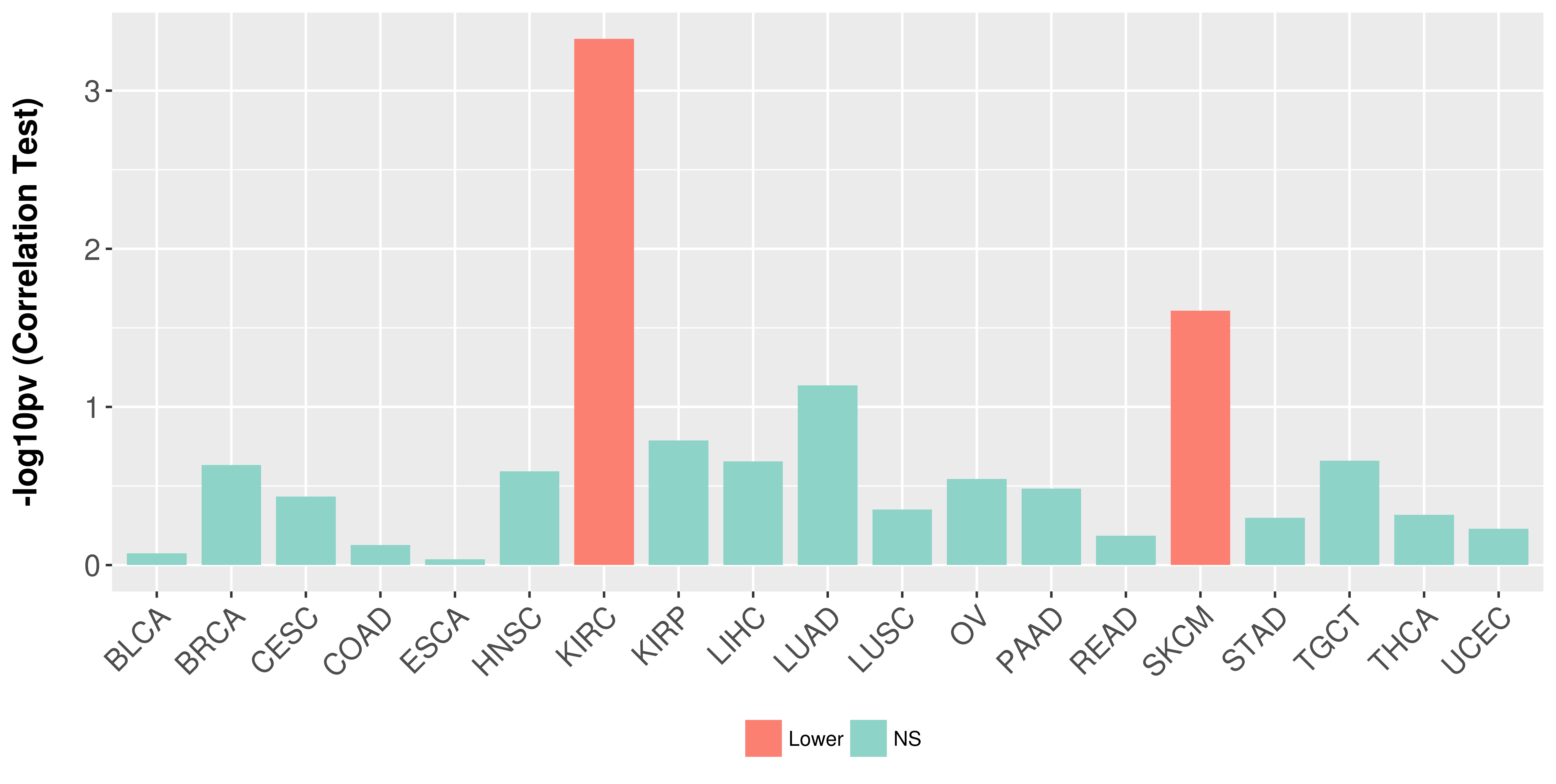
Association between expresson and stage.
|

Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | CTCF |
| Name | CCCTC-binding factor |
| Aliases | 11 zinc finger transcriptional repressor; MRD21; 11-zinc finger protein; CTCFL paralog; Transcriptional repr ...... |
| Location | 16q22.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | CTCF |
| Name | CCCTC-binding factor |
| Aliases | 11 zinc finger transcriptional repressor; MRD21; 11-zinc finger protein; CTCFL paralog; Transcriptional repr ...... |
| Location | 16q22.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for CTCF. |
| Summary | |
|---|---|
| Symbol | CTCF |
| Name | CCCTC-binding factor |
| Aliases | 11 zinc finger transcriptional repressor; MRD21; 11-zinc finger protein; CTCFL paralog; Transcriptional repr ...... |
| Location | 16q22.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|